Keywords: bladder, fibroblast, interstitial cell, PDGFRA, teolcyte
Abstract
Fibroblasts are crucial to normal and abnormal organ and tissue biology, yet we lack basic insights into the fibroblasts that populate the bladder wall. Candidates may include bladder interstitial cells (also referred to as myofibroblasts, telocytes, and interstitial cells of Cajal-like cells), which express the fibroblast-associated marker PDGFRA along with VIM and CD34 but whose form and function remain enigmatic. By applying the latest insights in fibroblast transcriptomics, coupled with studies of gene expression, ultrastructure, and marker analysis, we observe the following: 1) that mouse bladder PDGFRA+ cells exhibit all of the ultrastructural hallmarks of fibroblasts including spindle shape, lack of basement membrane, abundant endoplasmic reticulum and Golgi, and formation of homotypic cell-cell contacts (but not heterotypic ones); 2) that they express multiple canonical fibroblast markers (including Col1a2, CD34, LY6A, and PDGFRA) along with the universal fibroblast genes Col15a1 and Pi16 but they do not express Kit; and 3) that PDGFRA+ fibroblasts include suburothelial ones (which express ACTA2, CAR3, LY6A, MYH10, TNC, VIM, Col1a2, and Col15a1), outer lamina propria ones (which express CD34, LY6A, PI16, VIM, Col1a2, Col15a1, and Pi16), intermuscular ones (which express CD34, VIM, Col1a2, Col15a1, and Pi16), and serosal ones (which express CD34, PI16, VIM, Col1a2, Col15a1, and Pi16). Collectively, our study revealed that the ultrastructure of PDFRA+ interstitial cells combined with their expression of multiple canonical and universal fibroblast-associated gene products indicates that they are fibroblasts. We further propose that there are four regionally distinct populations of fibroblasts in the bladder wall, which likely contribute to bladder function and dysfunction.
NEW & NOTEWORTHY We currently lack basic insights into the fibroblasts that populate the bladder wall. By exploring the ultrastructure of mouse bladder connective tissue cells, combined with analyses of their gene and protein expression, our study revealed that PDGRA+ interstitial cells (also referred to as myofibroblasts, telocytes, and interstitial cells of Cajal-like cells) are fibroblasts and that the bladder wall contains multiple, regionally distinct populations of these cells.
INTRODUCTION
One of the major, but least understood, components of the bladder wall is its connective tissue, an amalgam of cells and extracellular matrix (ECM) components that include collagens, elastic fibers, and proteoglycans (1–3). Key resident cells of connective tissues are fibroblasts, which synthesize, organize, and remodel the ECM (4). Other fibroblast functions include the release of cytokines, adipokines, and growth factors, which serve as signals to recruit cells or modulate the activity of nearby ones; production of bactericidal peptides; differentiation into specialized cell types such as adipocytes and osteoblasts; promotion of wound healing; and deposition of excess matrix proteins in pathological states such as fibrosis and cancer, diseases that impact the bladder (4–18). Despite the recent creation of genomic atlases that indicate multiple fibroblast clusters may exist in the mouse and human bladder (19–22), the validation of these cell populations remains incomplete, and thus we still have limited insights into the fibroblast constituents of the bladder wall.
Historically, it has been difficult to define fibroblast-specific markers and subtypes in part because of significant intraorgan diversity (multiple fibroblast subtypes are described in the skin, lungs, skeletal muscle, and heart) as well as interorgan variability (<12% of fibroblast-enriched genes are shared between the mouse heart, skeletal muscle, intestine, and bladder) (4, 5, 20, 23). And although no single marker can define a fibroblast, Muhl et al. (20) recently described 45 shared “canonical gene markers” among the fibroblasts of four organs, including the bladder, and which collectively can be used to differentiate fibroblasts from mural cells. Interestingly, S100A4 (common name: fibroblast-specific protein 1) is not one of these. Moreover, a recent analysis of single-cell transcriptomic data from multiple databases, from multiple organs, and from mouse and human samples revealed the existence of two universal fibroblast subtypes: one that expresses the gene encoding Pi16 (common name: proteinase inhibitor 16) and the other that expresses Col15a1 (common name: collagen type XV-α1) (23). These universal fibroblasts also express the gene encoding Dpt (common name: dermatopontin), which, in turn, marks most cells expressing Pdgfra (common name: platelet-derived growth factor-α), a well-known gene (and protein) marker of fibroblasts or cells in the fibroblast lineage (20, 23).
In the case of the bladder, candidate fibroblasts could include so-called interstitial cells (also referred to as myofibroblasts, telocytes, or interstitial cell of Cajal-like cells), which reside in the connective tissue of the bladder wall but whose form and function remains mysterious (24–26). Much of our understanding of these cells is driven by Smet et al.’s early hypothesis that they may function in a manner analogous to the interstitial cells of Cajal (ICC) (27), gut-associated cells that act as pacemakers to regulate peristalsis (28–30). Early evidence in support of this premise included reports that the canonical ICC marker KIT is expressed by bladder interstitial cells (31–37), but these findings are now controversial and disputed by many (24, 38–44). One characteristic of bladder interstitial cells is that they express the canonical fibroblast markers PDGFRA and CD34, often in combination with VIM (common name: vimentin) (20, 39, 41–43, 45–49). Using the latest insights into fibroblast gene expression, combined with electron microscopy (EM) and light microscopy, we sought to test the hypothesis that bladder interstitial cells are fibroblasts. Based on their ultrastructure and their expression of canonical and universal fibroblast gene products, our study revealed that mouse bladder PDGFRA+ interstitial cells are fibroblasts and that multiple regionally distinct populations of these cells are found in the bladder wall. The implications of our findings are discussed.
METHODS
Reagents Including Antibodies
Unless otherwise specified, all chemicals were reagent grade or better and obtained from Sigma-Millipore (St. Louis, MO). The primary antibodies used in this study are shown in Table 1. They were chosen based on their prior validation in numerous publications. Primary antibodies were diluted 1:1 with glycerol and stored at −20°C. The working dilution of these stock antibody solutions is described in Table 1. Secondary antibodies against the chicken, goat, rabbit, rat, and sheep, conjugated to Alexa 488, Dylight 549, Cy3, or Cy5 dyes and with minimal cross reactivity to other host species, were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). After reconstitution, they too were mixed with an equal volume of glycerol and stored at −20°C. These stocks were used at a working dilution of 1:100 (Alexa 488 and Cy5 conjugates) or 1:3,000 (Dylight 549 or Cy3 conjugates). Donkey anti-rabbit 10-nm gold was purchased from Electron Microscopy Science (Hatfield, PA).
Table 1.
Primary antibodies used in the present study
| Antigen | Host Species | Source | Catalog Number | Dilution of stock |
|---|---|---|---|---|
| ACTA2 | Rabbit | Proteintech | 14395-1-AP | 1:100 |
| CAR3 | Rabbit | Proteintech | 15197-1-AP | 1:100 (1:100 for FISHID) |
| CD34 | Sheep | R&D Systems | AF6518 | 1:100 (1:50 for FISHID) |
| CD34 | Rat (clone MEC14.7) |
Invitrogen (ThermoFisher) | MA1-22646 | 1:200 |
| CSPG4 | Rabbit | Millipore Sigma | AB5320 | 1:100 |
| GFP | Rabbit | Abcam | Ab6556 | 1:75 for EM |
| KIT (CD117) | Rat (clone ACK2) |
Stemcell | 60034.1 | 1:100 |
| KRT20 | Rabbit | Abcam | ab53120 | 1:100 |
| KRT5 | Chicken | BioLegend | 905901 | 1:400 |
| LY6A | Rat | BioLegend | 108101 | 1:100 |
| MYH11 | Rabbit | Proteintech | 21404-1-AP | 1:100 (1:100 for FISHID) |
| PDGFRA | Goat | R&D Systems | AF1062 | 1:100 (1:50 for FISHID) Preferred antibody used in most experiments |
| PECAM1 (CD31) | Rat (clone 390) |
Millipore Sigma | CBL1337-I | 1:100 |
| PI16 | Goat | R&D Systems | AF4929-SP | 1:100 |
| PI16 | Rabbit | US Biological | 141796 | 1:100 |
| PTPRC (CD45) | Rat (clone IBL-3/16) |
Bio-Rad | MCA1388 | 1:100 |
| TNC | Rat (clone no. 578) |
R&D Systems | MAB2138 | 1:400 |
| UCHL1 (PGP9.5) | Rabbit | Genetex | GTX109637 | 1:100 |
| VIM | Rat | BioLegend | 699301 | 1:100 |
FISHID, fluorescent in situ hybridization with immunodetection.
Animals
Most experiments used wild-type virgin female C57Bl/6J mice (Jackson Laboratory, Bangor, ME), which were 8–12 wk of age. They were group housed up to five animals per cage in standard caging with an automatic water dispenser and dry mouse chow ad libitum. The bottom of the cage was covered in standard bedding and included a plastic igloo and a small square of paper for shredding. Animals were held under a 12:12-h day-night cycle with 7:00 AM being zeitgeber time = 0. Where appropriate, animals were genotyped using Jackson Laboratory guidelines, and heterozygotes group housed up to five females per cage, whereas male breeders were housed individually. PdgfraH2B-EGFP+/– reporter mice (Cat. No. 007669, Jackson Laboratory) were maintained by breeding PdgfraH2B-EGFP+/– mice with wild-type C57Bl6/J mice. In experiments examining expression of Col1a2, we harem bred female hemizygous Col1a2Cre-ERT+/– mice (Cat. No. 029567, Jackson Laboratory) with male homozygous RosamT/mG+/+ mice (Cat. No. 07676, Jackson Laboratory). At 8 wk of age, female experimental Col1a2Cre-ERT+/–;RosamT/mG+/– or control Col1a2Cre-ERT–/–; RosamT/mG+/– mice were injected once daily for 5 consecutive days with 100 mg/kg tamoxifen injected interperitoneally. Tamoxifen was dissolved in corn oil. Mice were then allowed to recover for 10 days before euthanasia.
Euthanization of mice was accomplished by inhalation of CO2 gas followed by a thoracotamy or cervical dislocation. All animal experiments were performed in accordance with relevant guidelines/regulations of the Public Health Service Policy on Humane Care and Use of Laboratory Animals and the Animal Welfare Act and under the approval of the University of Pittsburgh Institutional Animal Care and Use Committee.
Alkaline Maceration of Bladder Tissue, Freeze Cracking, and Field Emission Scanning Electron Microscopy
Mice were euthanized, and their bladder was exposed via a midline laparotomy that extended toward the pubic bone. The bladder was excised by cutting below its neck region with sharp scissors, and the released bladder was rinsed in Krebs solution [110 mM NaCl, 25 mM NaHCO3, 5.8 mM KCl, 1.2 mM MgSO4, 1.2 mM KH2PO4, 11 mM glucose, and 2 mM CaCl2, pH 7.4 when gassed with 5% (vol/vol) CO2] and then gently cut open along its midline. The tissue was then carefully pinned out on a rubber mat (mucosal surface facing upward) with minimal stretching to maintain the rougal folds. The tissue was then fixed in 2% (vol/vol) glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA) and 2% (wt/vol) EM-grade paraformaldehyde [Polysciences, Warrington, PA, prepared as a 40% (wt/vol) stock] in 100 mM sodium cacodylate buffer (CB; pH 7.4) for 60 min at room temperature. To reveal the collagen matrix, alkaline maceration was performed as previously described (50–52). Briefly, the fixed tissue was washed with water three times for 5 min and then incubated for 6 days in a freshly prepared aqueous solution of 10% (wt/vol) NaOH. The NaOH solution was exchanged on days 1 and 3 with fresh NaOH solution. After maceration, the tissue, including the blood vessels, became transparent and colorless. The tissue was rinsed with H2O and then incubated overnight at room temperature in H2O. Samples were then incubated for 4 h at room temperature with a freshly prepared aqueous solution of 1% (wt/vol) tannic acid (filtered through a 0.2-µm filter before use, Electron Microscopy Sciences), rinsed overnight in H2O, rinsed three times for 5 min with CB, and then incubated for 2 h at room temperature in 1% (wt/vol) OsO4 in CB. After being rinsed with H2O, samples were dehydrated in 70%, 80%, 90%, and 100% (vol/vol) ethanol. Pieces of bladder tissue were then transferred to a rectangular ice bucket in which an aluminum plate (20 × 20 cm, 6 mm thick) was submerged ∼1 cm in liquid nitrogen. The tissue was placed on the aluminum plate and cracked by releasing a precooled metal Leica 12-cm C-type microtome blade onto the tissue, fracturing it in two. The cracked tissue was transferred to dried 100% ethanol, which was prepared by adding 50 g of dried molecular sieve beads (4 Å, 8–12 mesh) per 100-mL ethanol as previously described (53). After an additional exchange with dried 100% ethanol, samples were critical point dried in a Samdri-PVT-3D critical point dryer (Tousimis, Rockville, MD) using bone-dry CO2 fed by a siphon tube. Samples were mounted on aluminum stubs to which double-sided conductive copper foil was attached (Ted Pella, Redding, CA), sputter coated with gold/palladium using a 108Auto sputter coater (Cressington, Watford, UK), and then viewed in a JSM6335F field emission scanning electron microscope equipped with a digital camera (Jeol, Peabody, MA). The contrast of the images was corrected in Photoshop CC2022 (Adobe, San Jose, CA) and composite images were prepared in Adobe Illustrator CC2022.
Transmission Electron Microscopy
Excised bladder tissue was rinsed in Krebs solution and then fixed in 2.5% (vol/vol) glutaraldehyde and 2% (wt/vol) EM-grade paraformaldehyde in CB for 60 min at room temperature. Samples were then rinsed with CB three times and then incubated for 60 min at room temperature with 1% (wt/vol) OsO4, dissolved in CB, and containing 1.5% (wt/vol) K4Fe(CN)6. The tissue was rinsed with water several times and then stained en bloc overnight at 4°C in freshly prepared 0.5% (wt/vol) aqueous uranyl acetate (filtered through a 0.2-µm filter before use, Electron Microscopy Services). The tissue was rinsed three times with water, cut into 1-mm-thick strips, dehydrated through a graded series of ethanol, treated with propylene oxide, and then embedded in EPON substitute LX112 (Ladd Research, Williston, VT). After polymerization at 60°C for 48 h, the blocks were trimmed with a glass knife and then sectioned using a diamond knife (Diatome, Hatfield, PA) mounted on an Ultracut R ultramicrotome (Leica Microsystems, Wood Dale, IL). Sections, pale gold to silver in color, were collected on Butvar-coated nickel slot grids and contrasted using 2.0% (wt/vol) uranyl acetate and then Reynold’s lead citrate. Samples were viewed in a Jeol 1400 transmission electron microscope equipped with an Advanced Microscopy Techniques (Woburn, MA) 4 K digital imaging system. The contrast of the images was corrected in Adobe Photoshop CC2022 using the levels command, or contrast was increased using the clarity function of Adobe Lightroom. Composite images were prepared in Adobe Illustrator CC2022.
Fixation of Tissue, Immunofluorescence, and Confocal Microscopy
Fixed-frozen preparation.
Excised bladders were fixed by injecting ∼50 µL of 4% (wt/vol) EM-grade paraformaldehyde (dissolved in CB) into the bladder lumen and then immersing the bladder in the same fixative for a total of 60 min at room temperature. Injection of fixative into the bladder lumen was performed using the plastic catheter portion of 24-gauge × ¾” Jelco Safety IV catheter (Smiths Medical ASD, Southington, CT) attached to a 1-mL syringe prefilled with fixative. The tissue was rinsed with PBS (2.7 mM KCl, 1.5 mM KH2PO4, 136.9 mM NaCl, and 8.9 mM Na2HPO4), transferred to 30% (wt/vol) sucrose dissolved in PBS at 4°C for 5 min to remove excess PBS, and then transferred to new sucrose/PBS solution, and incubated at 4°C on a rotator until the tissue lost its buoyancy. The sucrose-impregnated tissues were immersed in OCT solution (Tissue-Tek, Sakura Finetek, Torrance, CA), and OCT was injected into the lumen of the fixed bladder using a catheter (see description above) until the OCT back flowed. The bladder was then transferred to OCT-filled 10 × 10 × 5 mm Tissue-Tek Cryomolds (Sakura Finetek), oriented with the bladder neck region facing downward. The molds were placed in a −80°C freezer to freeze the OCT compound, and the blocks were stored at −80°C in sealed plastic bags until they were cryosectioned.
Fresh-frozen preparation.
For experiments exploring KIT expression or for in situ hybridization (see protocol below), excised bladders or the colon were rinsed with 37°C Krebs solution and then placed in a 10-cm square dish filled with the same buffer. OCT was instilled via catheter into the bladder lumen as described above. The bladder was dipped into a beaker containing OCT, transferred to OCT-filled Cryomolds, oriented with neck facing downward, frozen, and stored as described above. Colon tissue was cut into 5-mm cross sections using a sharp scalpel, OCT was introduced into the lumen using the same technique as used with the bladder, and the cross sections were placed in the OCT-filled Cryomolds (cut surface facing upward), frozen, and then stored at −80°C.
Sectioning.
Sections (8–15 µm) along the axial axis of the bladder were cut using a Leica CM1950 cryostat (chamber temperature of −20°C and a knife temperature of −18°C), collected on Superfrost Plus microscope slides (ThermoFisher Scientific, Waltham, MA), and “dried” by storing them in the cryostat chamber for 30 min. Samples were generally cut just before immunolabeling, although it was possible to store them at −80°C for several weeks.
Immunolabeling of sectioned tissue.
For fresh-frozen samples, the slide-mounted tissue was fixed by the addition of 4% (wt/vol) paraformaldehyde in 100 mM phosphate buffer (pH 7.4) for 10 min at room temperature. For both fresh-frozen or fixed-frozen tissues, the sample was incubated for 10 min at room temperature in quench buffer [75 mM NH4Cl and 20 mM glycine, pH 8.0 dissolved in PBS, containing 0.1% (vol/vol) Triton X-100] and rinsed three times quickly with PBS and then three times for 5 min with PBS, followed by incubation in Block solution [PBS containing 0.6% (wt/vol) fish skin gelatin and 0.05% (wt/vol) saponin] supplemented with 5% (vol/vol) donkey or goat serum for 30–60 min at room temperature. Samples were subsequently incubated with primary antibodies (diluted in Block solution) overnight at 4°C in a humid chamber. Control samples lacked primary antibodies. Samples were washed three times quickly and three times for 5 min with Block solution and then incubated with minimal cross-reactivity, fluorophore-labeled secondary antibodies, diluted in Block solution, for 60 min at room temperature. Nuclei were counterstained with DAPI (1:1,000, ThermoFisher Scientific) The labeled tissues were then rinsed three times quickly and three times for 5 min with Block solution, rinsed with PBS, and then postfixed in 4% (wt/vol) paraformaldehyde dissolved in 0.1 mM phosphate buffer (pH 7.4) for 5–10 min at room temperature. Slides were rinsed with PBS, and samples were mounted under borosilicate coverslips (No. 1.5, 0.17 mm thickness, 24 × 50 mm, ThermoFisher) using SlowFade Diamond Antifade mounting medium (ThermoFisher). The edges of the coverslip were sealed with clear nail polish, and slides were stored at −20°C until image acquisition was performed.
Whole mount tissue preparation and staining.
Excised bladders were rinsed with 37°C Krebs solution, and a peeled bladder preparation was made as previously described (54); however, we retained both the dissected mucosa and dissected muscular tissue/serosa. Both tissue preparations were submerged in Krebs solution, pinned out, and then fixed by immersion in 4% (wt/vol) EM-grade paraformaldehyde (dissolved in CB) at room temperature for a total of 60 min. Whole mounted tissue was stained and mounted as previously described (54).
Image capture.
Images were captured using a Leica HCX PL APO ×20, 0.75 numerical aperture dry objective or a Leica HCX PL APO CS ×40, 1.25 numerical aperture oil objective and the appropriate laser lines of a Leica Microsystems SP8 Stellaris confocal microscope outfitted with a 405-laser diode and a white light laser. The signal from the Power HyD detectors was optimized using the Q-LUT option, and 8-bit images were collected at 600 Hz using 2-line averages combined with 2 frame averages. Cross talk between channels was prevented by use of spectral detection coupled with sequential scanning. Stacks of images (1,024 × 1,024, 8-bit) were collected using system-optimized parameters for the z-axis. Images were processed using the 3 D visualization package of Leica LASX software and exported as TIFF files. If necessary, contrast of the images was corrected in Photoshop CC2022, and composite images were prepared in Adobe Illustrator CC2022.
Control samples without primary antibody lacked specific staining. However, the tunica media of bladder blood vessels is autofluorescent, as are the endolysosomes of umbrella cells, which accumulate lipofuscin pigments (55). Control samples were acquired using identical parameters and processed identically to experimental samples. Representative images from multiple animals and experiments are shown in each figure.
Tissue Cytometry
To perform tissue cytometry, we used version 1.0.3 of the Volumetric Tissue Exploration and Analysis (VTEA) plugin for Fiji/NIH ImageJ (https://github.com/icbm-iupui/volumetric-tissue-exploration-analysis). VTEA is a software package, created at Indiana University by Winfree et al., which was previously used to analyze marker distribution and identify cell populations in kidney samples (56). Random multichannel confocal Z-stacks (1,024 × 1,024 pixels, each pixel = 0.75 µm, ×20 objective, zoom = 0.75) were acquired as described above. The files were opened in Fiji and converted to image stacks. The VTEA plugin was launched, and the image stack was imported using the Load Image function. The image files were preprocessed by applying a background subtraction of 15 to the non-DAPI signals using the Process Image function. In the resulting Segment Objects tab, a filter was created with the following parameters: Connect 2D/3D with KDtree option, the channel containing the nuclei was selected, low threshold of 40, centroid offset of 5, minimum volume 20, and maximum volume 1,250. The Find Objects command was then executed. Visual inspection confirmed that this operation resulted in the selection of >99% of nuclei, which were cleanly segmented from one another. In the resulting scatterplot, the markers of interest were chosen (PDGFRA on the y-axis and the other marker on the x-axis), and the mean intensity for both channels was plotted. The resulting scatterplots were gated into four quadrants, and the resulting numbers of positive nuclei (i.e., cells) were recorded from each gated region: nuc1, nuclei with values of <30 on the x-axis and >30 on the y-axis; nuc2, nuclei with values of >30 on the x-axis and >30 on the y-axis; nuc3, values of <30 on the x-axis and <30 on the y-axis; and nuc4, values of >30 on the x-axis and <30 on the y-axis. A coefficient of colocalization with respect to PDGFRA (CCPDGFRA) was calculated as follows:
A coefficient of colocalization with respect to the other marker (CCmarker) was similarly calculated using the following equation:
A feature of VTEA is the ability to choose nuclei in the gated regions of the scatterplot, which are then immediately highlighted on the original image (see Supplemental Fig. S3 for examples; all Supplemental Material is available at https://doi.org/10.6084/m9.figshare.19719856). A visual inspection of the resulting nuclei within each gated region (which appear red in these images) along with the corresponding markers confirmed that gating was working as intended (see Supplemental Fig. S3).
Detection of Gene Expression Using Fluorescent In Situ Hybridization, Including Fluorescent In Situ Hybridization Coupled With Immunodetection
To analyze gene expression in the bladder wall, we used the RNAscope multiplex fluorescent kit (ACD, Newark, CA) and the following probes: Mm-Col1a2 in channel 3 (Cat. No. 585461-C3), Mm-Col15a1 in channel 1 (Cat. No. 1092391-C1), Mm-cKit in channel 3 (Cat. No. 314151-C3), Mm-Pdgfra in channel 2 (Cat. No. 480661-C2), and Mm-Pi16 in channel 3 (Cat. No. 451311-C3). Probe diluent was used when the channel 1 probe was not included in the staining protocol. The three-plex negative control probe against Bacillus subtilis dapB (Cat. No. 320871) was used to detect nonspecific signal. Procedures were performed using gloves and sterilized equipment to prevent RNAse contamination.
Fluorescent in situ hybridization protocol.
Fresh-frozen bladder tissue was prepared and sectioned at 8–10 µm as described above for performing immunofluorescence. The manufacturer’s protocol for performing RNAscope Multiplex Fluorescent v2 was used with the following changes. Slides containing sectioned tissue were removed from the −80°C freezer and fixed for 15 min at 4°C with neutral buffered formalin [29 mM NaH2PO4, 45.8 mM Na2HPO4, and 4.0% (wt/vol) EM-grade paraformaldehyde]. Tissue was treated with protease IV for 5 min at room temperature. In all experiments, slides incubated with negative control probes were run side by side with those incubated with the fibroblast probes and treated identically. Labeled tissue was mounted using ProLong Gold antifade mountant (ThermoFisher) and cured overnight at room temperature in the dark, and slides were subsequently stored at 4°C.
Fluorescent in situ hybridization coupled with immunodetection protocol.
We followed the manufacturer’s workflow recommendations for combining the RNAscope Multiplex Fluorescent v2 assay with immunofluorescence detection. The additions to the general fluorescent in situ hybridization (FISH) protocol included the following: before protease IV treatment, samples were incubated overnight at 4°C with primary antibodies diluted in codetection antibody diluent (Cat. No. 323160) and then postfixed for 30 min at room temperature with neutral buffered formalin. After the last treatment with horseradish peroxidase blocker, samples were incubated for 30 min at room temperature with fluorophore-conjugated secondary antibodies (in codetection antibody diluent) and DAPI was then added for 30 s and slides were mounted as described above.
Ultrathin Cryo-EM
Ultrathin cryo-EM was performed as follows. Bladder tissue was excised, rinsed with Krebs solution, loosely pinned out on a rubber mat, and then fixed for 60 min at 37°C with 4% (wt/vol) EM-grade paraformaldehyde, 0.05% (vol/vol) glutaraldehyde dissolved in PIPES-HEPES-MgCl2-EGTA buffer (120 mM PIPES, 50 mM HEPES, 4 mM MgCl2, and 20 mM EGTA, titrated to a pH of 6.9 using KOH). The fixed tissue was cut into 2 × 2-mm squares and cryoprotected in polyvinylpyrollidone-sucrose (prepared by mixing 80 mL of 3.2 mM sucrose dissolved in 0.1 M phosphate buffer, pH 7.4 with 20 g of 40 K average molecular weight polyvinyl pyrollidone dissolved in 4 mL of 1.1 M NaH2CO3). After the tissue sank, it was then mounted on stainless steel stubs with a cut surface facing upward. Samples were frozen by plunging in liquid nitrogen and then stored in liquid nitrogen until used. Sections were cut on a Leica Ultracut UCT ultramicrotome with a Leica EM-FCS cryokit attachment outfitted with a diamond cryo-knife (Diatome). Sections were picked up using a 30-gauge nichrome wire loop (internal diameter of 2.5 mm, attached to a wooden dowel) filled with a 1:1 mixture of 3.2 M sucrose (dissolved in 0.1 M phosphate buffer, pH 7.4) and 2% (wt/vol) methylcellulose and transferred to Butvar-coated nickel grids with a 50 lines/in. square mesh or 50 lines/in. parallel bars (EMS).
Immunogold labeling of ultrathin cryosections.
The following steps were performed by placing grids on the top of drops resting on parafilm. Grid transfers were made using loops as described above. Samples were washed with PBS, followed by PBS containing 0.15% (wt/vol) glycine, and then three times for 5 min with EM block solution [0.5% (wt/vol) BSA and 0.15% (wt/vol) glycine dissolved in PBS]. After an additional 30-min incubation in EM block solution, sections were incubated with rabbit anti-green fluorescent protein (GFP) primary antibody (diluted in EM block solution) for 60 min at room temperature. Sections were then washed three times with EM block solution for 5 min and then incubated with donkey anti-rabbit 10-nm gold (diluted 1:25 in EM block solution) for 30 min at room temperature. Sections were washed three times with EM block solution for 5 min, rinsed with PBS three times, and postfixed for 5 min with 1.0% (vol/vol) glutaraldehyde dissolved in 0.1 M phosphate buffer (pH 7.4). Sections were then washed three times with PBS, washed four times with distilled water, and then incubated for 7 min at room temperature in 2.0% (wt/vol) neutral uranyl acetate. Sections were washed with distilled water (2 times) and incubated for 5 min on drops of 2% (wt/vol) methylcellulose containing 4% (wt/vol) aqueous uranyl acetate, followed by a 1-min incubation on a drop of 4% (wt/vol) aqueous uranyl acetate. Excess uranyl acetate was removed by carefully dragging the EM grid across filter paper, and the grid was allowed to dry overnight on its associated loop. Samples were viewed in a transmission electron microscope, photographed, and processed using the methods described above in Transmission Electron Microscopy.
Statistics
Data for tissue cytometry are presented as means ± SD.
RESULTS
Ultrastructural Analysis Confirms the Presence of Fibroblasts in the Mouse Bladder Wall
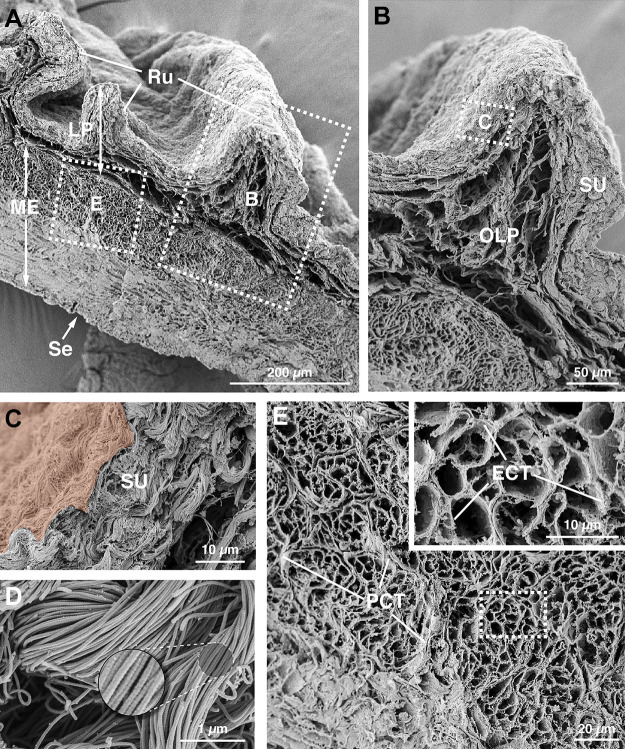
We first sought to examine the fibrillar collagen matrix of the mouse bladder, as these collagens are synthesized by fibroblasts (4) and their presence can serve as a surrogate of fibroblast activity and localization. Using a technique previously applied to human bladders (52), we performed alkaline maceration, which removes all cellular material and extracts all of the ECM components except for fibrillar collagen (50–52). When the resulting tissue was inspected by high-resolution field emission scanning EM, we observed a dense network of fibrillar collagen within the bladder wall (Fig. 1A). In the innermost portions of the lamina propria, which we refer to as the suburothelial region in this report, we observed layers of closely packed and woven bundles of collagen fibrils (Fig. 1, B and C). At high magnifications, the collagen fibrils (in this and in all regions) had a rope-like appearance with a characteristic striated appearance (periodicity of ∼70 nm; Fig. 1D) (57). Below the suburothelial region was the outer lamina propria, which was composed of a looser network of collagen fibrils, particularly in the central portions of the rougae (mucosal folds; Fig. 1B). Collagen was also found in the intermuscular region of the muscularis externa, where it was a major component of the perimysial connective tissue surrounding fascicles of smooth muscle cells and of the endomysial connective tissue that encased individual myocytes (Fig. 1E). In the case of the latter, collagen fibrils were oriented perpendicular to the long axis of the muscle fiber, but because of the lack of fibroblasts in this region [see transmission EM (TEM) experiments below], this collagen is likely secreted by the smooth muscle cells themselves. Collagen fibrils were also found in the serosa-associated connective tissue, a thin layer of tissue sandwiched between the muscularis externa and the mesothelium (visceral peritoneum), which encapsulates the rodent bladder.
Figure 1.
Fibrillar collagen matrix of the mouse bladder wall. A: ultrastructure of the bladder fibrillar collagen matrix was revealed by alkaline maceration and field emission scanning electron microscopy. The dashed boxed regions are magnified in B and E. B: collagen matrix within the region of the lamina propria, which can be subdivided into the suburothelial (SU) region and outer lamina propria (OLP). The dashed boxed region is magnified in C. C: suburothelial collagen matrix. The urothelium, which is extracted during the alkaline maceration, would be found just above the region false colored in orange. D: ultrastructure of the collagen fibrils. E: intermuscular portion of the bladder wall. The dashed boxed region is magnified in the inset. Smooth muscle cells (extracted) would sit within the lacunae created by the endomysial connective tissue (ECT). Fascicles of smooth muscle cells are surrounded by perimysial connective tissue (PCT). Images are representative of bladders from two mice. LP, lamina propria; ME, muscularis externa; Ru, rugae; Se, serosa.
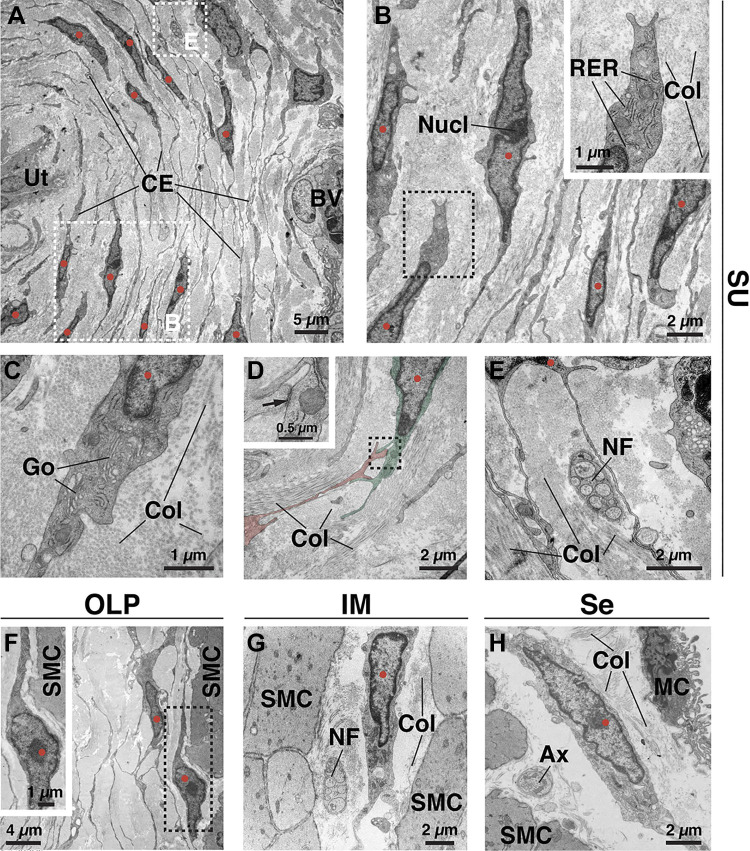
Although previous studies have described the ultrastructure of “interstitial cells” in the human bladder, particularly those in the lamina propria (31, 42, 43, 58–61), we explored the ultrastructure of the connective tissue cells in the entirety of the mouse bladder wall using TEM (Fig. 2). In the suburothelial region, we observed a large population of cells that were stacked in sheet-like layers (typically 3–5 layers thick) with a distance between layers on the order of 2–4 µm. Consistent with the dense suburothelial collagen network shown in Fig. 1, these suburothelial fibroblasts were surrounded by intervening bundles of collagen (Fig. 2, A–E). The ultrastructure of these cells bore all of the well-established hallmarks of fibroblasts examined in cross section (62). These include the lack of a basement membrane and spindle-shaped cell bodies containing oblong, irregularly shaped nuclei (with peripheral heterochromatin) sometimes with a prominent nucleolus (Fig. 2B). Other fibroblast features included the presence of a rough endoplasmic reticulum (Fig. 2, B and C) and Golgi stacks (Fig. 2C). Although mitochondria were present, they were not abundant, and networks of cytoskeletal filaments were not apparent. Caveolae and coated pits were occasionally observed at their surfaces, but we found no evidence of fibronexus formation (a fibronectin-dependent cell-ECM junction and defining feature of myofibroblasts) (62). In cross-section, long thin processes (150–400 nm wide and up to 100 µm in length) were extended from both poles of these spindle-shaped cells (Fig. 2, A and B). These extensions were generally oriented parallel to the bladder lumen, were oftentimes branched, and were observed to form homotypic adherence junctions with one another (Fig. 2, A and D). Although the extensions of the suburothelial fibroblasts came into close proximity to the basal cells of the urothelium, blood vessels, and nerve termini/fascicles, they were separated by collagen fibrils or by the epithelium-, endothelium-, or Schwann cell-associated basement membrane and did not form obvious cell-cell junctions with these tissues (Fig. 2, A and E, and Supplemental Fig. S1, A and B).
Figure 2.
Ultrastructure of fibroblasts in the bladder wall as revealed by transmission electron microscopy. A: cross section through the suburothelial (SU) region showing the layered nature of fibroblasts under the urothelium (Ut) and their numerous cell extensions (CE) and protrusions. A portion of a blood vessel (BV) is found at the right. Surrounding the cells and their extensions is a collagen-rich matrix (light gray material). The dashed boxed regions are magnified in B and E. In all images, the nuclei of presumptive fibroblasts are marked with red circles. B: suburothelial fibroblast with prominent rough endoplasmic reticulum (RER) in its cytoplasm. The nucleolus (Nucl) of a fibroblast nucleus is marked. Bundles of collagen (Col) are found in the matrix. The dashed boxed region is magnified in the inset. C: suburothelial fibroblast with readily discernable Golgi (Go). The cell is surrounded by collagen fibrils. D: cell-cell adhesion complex between two fibroblasts (false colored red or green). The dashed boxed region is magnified in the inset. The site of cell-cell adhesion, including cytoplasmic densities on either side, is indicated by the arrow. Collagen surrounds the cells. E: close proximity of fibroblast extensions to a nerve fascicle (NF) composed of nonmyelinated axons and surrounded by collagen. F: fibroblast in the outer lamina propria (OLP) near the border of smooth muscle cells (SMC). The dashed boxed region is magnified in the inset. G: fibroblast in the intermuscular region (IM) between adjacent smooth muscle cells, surrounded by collagen, and near an unmyelinated nerve fascicle (NF). H: serosal (Se) fibroblast within the thin collagen matrix between smooth muscle cells and mesothelial cells (MC), which form the mesothelium. A free axon (Ax) is present. Images are representative of experiments performed in three mice.
The fibroblasts in the other regions of the bladder wall were not as closely apposed or as obviously organized as those in the suburothelial region; however, they had a similar ultrastructure including a close association with collagen fibrils (Fig. 2, G and H, and Supplemental Fig. S1, C and D) and elongate shaped cell bodies with irregularly shaped heterochromatic nuclei (some with a nucleolus). The thin band of cytoplasm surrounding the nucleus contained the rough endoplasmic reticulum and occasional mitochondria. In cross section, the cells extended long processes from either pole of their spindle-shaped bodies. Like suburothelial ones, the cell bodies and associated processes did not form interactions with closely apposed nerve bundles/axons (Fig. 2, G and H) or smooth muscle cells (Fig. 2, G and H, and Supplemental Fig. S1, C and D), although they did approach closely in some cases (Fig. 2F and Supplemental Fig. S1, C and D). Although fibroblasts were the chief component of the bladder wall connective tissue, we also observed blood vessels and associated cells (endothelial cells, pericytes, and smooth muscle cells), Schwann cells with associated axons, and immune cells.
Taken together, our ultrastructural analysis confirmed that a large population of cells in the mouse bladder connective tissue shared a similar morphology, one most consistent with these cells being classified as fibroblasts. Furthermore, although fibroblasts formed homomeric interactions with each other, we did not observe that they formed direct heterotypic contacts with closely apposed nerve fibers, smooth muscles cells, epithelial cells, or mural cells.
PDGFRA+ Cells of the Mouse Bladder Wall Are Fibroblasts
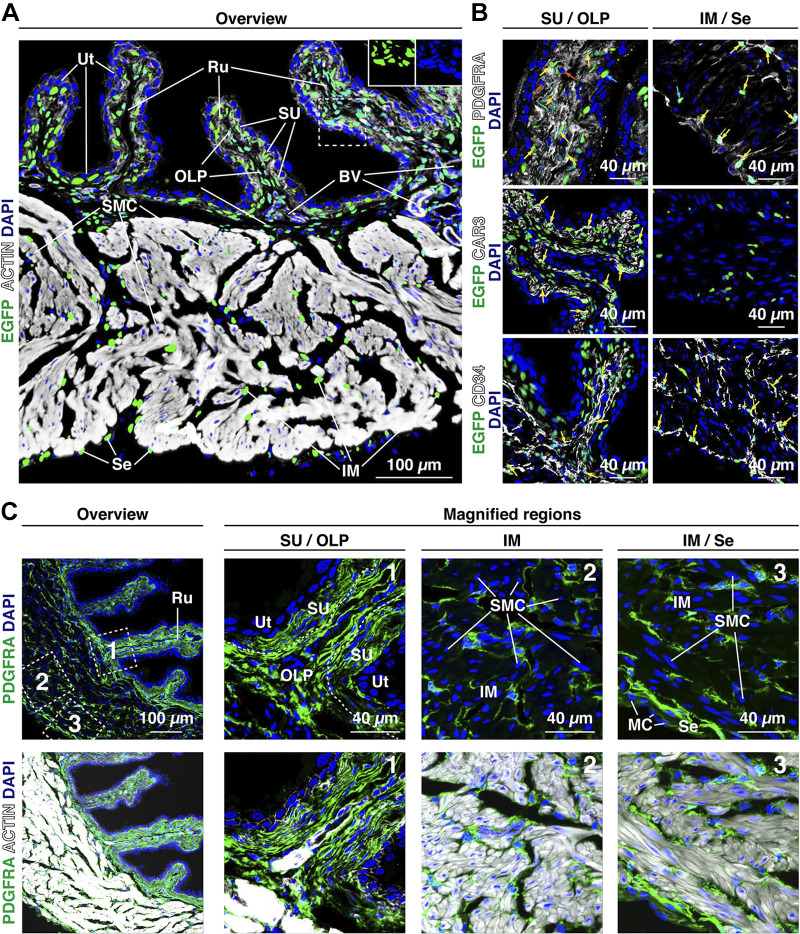
PDGFRA, a canonical fibroblast marker, has been previously reported to be expressed by mouse, guinea pig, and human bladder interstitial cells (20, 39, 43, 45, 48, 49). To gain insights into the cells that express the Pdgfra gene in the mouse bladder, we used PdgfraH2B-EGFP reporter mice. In these mice, one copy of the Pdgfra gene is ablated and instead expresses nuclear-localized H2B-enhanced GFP (EGFP) under the control of the native Pdgfra promoter (63). We observed relatively large numbers of closely apposed H2B-EGFP+ cell nuclei in the suburothelial region and outer lamina propria, whereas in the intermuscular region and near the serosal surface H2B-EGFP+ cell nuclei appeared more scattered (Fig. 3A). When bladder tissue from PdgfraH2B-EGFP reporter mice was colabeled with antibodies to PDGFRA, we observed the expected overlap between H2B-EGFP+ cell nuclei and the PDGFRA signal (yellow arrows in Fig. 3B, top). However, we also observed small numbers of scattered cells that were PDGFRA+ but did not express H2B-EGFP (see orange arrows in Fig. 3B), possibly indicating incomplete penetrance or low expression of the H2B-EGFP transgene. In addition, a small number of H2B-EGFP+;PDGFRA− cells were also noted (see cyan arrows in Fig. 3B).
Figure 3.
Distribution of mouse bladder Pdgfra/PDGFRA+ fibroblasts. A: expression of Pdgfra in the bladder wall of PdgfraH2B-EGFP reporter mice as revealed by confocal microscopy. Tissue was counterstained with phalloidin to reveal the actin cytoskeleton (white) and DAPI (blue) to reveal cell nuclei. Actin was associated with the cortical cytoskeleton of most cells but strongly labeled the actin filaments that abound in the cytoplasm of smooth muscle cells. As expected for a nucleus-localized reporter, the H2B-enhanced green fluorescent protein (EGFP) signal overlapped completely with the DAPI signal (see insets for example of the DAPI signal alone and EGFP signal alone). Representative confocal micrographs from two mice are shown. B: bladder tissue from PdgfraH2B-EGFP reporter mice was counterstained with antibodies to PDGFRA, CAR3, or CD34. Representative confocal micrographs from two mice are shown. Examples of H2B-EGFP+ nuclei that were also positive for PDGFRA, CAR3, or CD34 are marked with yellow arrows. In the top images, a small number of cells were found that were PDGFRA+ but H2B-EGFP– (nuclei marked with orange arrows) or were H2B-EGFP+ but PDGFRA– (nuclei labeled with cyan arrows). C: PDGFRA+ cells (green) were found in the following regions: the suburothelial region (SU); in the subjacent outer lamina propria (OLP), which forms the core of the rougae (Ru) and extends below suburothelial cells in the unfolded regions; within the perimysial connective tissue in the intermuscular region (IM); and in the serosa (Se), where a single layer of cells was sandwiched between the smooth muscle and mesothelial cell (MC) layer. The tissue was counterstained with DAPI (blue) and phalloidin (white; bottom images). The boxed regions in the top left images are magnified in the adjacent images. Representative confocal micrographs from four experiments (each with 2–3 mice) are shown.
As PDGFRA is a surface-expressed protein, it allowed us to better visualize the overall cellular architecture of Pdgfra+ cells. In general, PDGFRA staining closely mimicked the features and distribution of the fibroblasts described in our TEM studies (e.g., compare Fig. 3C with Fig. 2 and Supplemental Fig. S1). In the suburothelial region, we observed elongate PDGFRA+ cells, typically three to five cell layers thick (in cross section), that ran parallel to the lumen, closely following the shape of the rougae (Fig. 3, B and C). Their spindle-shaped morphology combined with their layered arrangement indicated these cells correspond to the suburothelial fibroblasts shown in Fig. 2, A–E. To confirm this possibility, we immunogold labeled ultrathin cryosections of bladder tissue obtained from PdgfraH2B-EGFP reporter mice with anti-GFP antibodies (Fig. 4A). These experiments revealed that cells with GFP+ nuclei (see Fig. 4A, examples 1 and 2) had a fibroblast-like ultrastructure identical to that observed in standard transmission electron micrographs. However, as in Fig. 3B, we occasionally observed fibroblast-like cells that were unlabeled (see Fig. 4A, example 3). In these EM experiments, the nuclei (and cell bodies) of epithelial cells, endothelial cells, and smooth muscle cells lacked nuclear staining, and no staining was observed in samples incubated in the absence of primary antibody.
Figure 4.
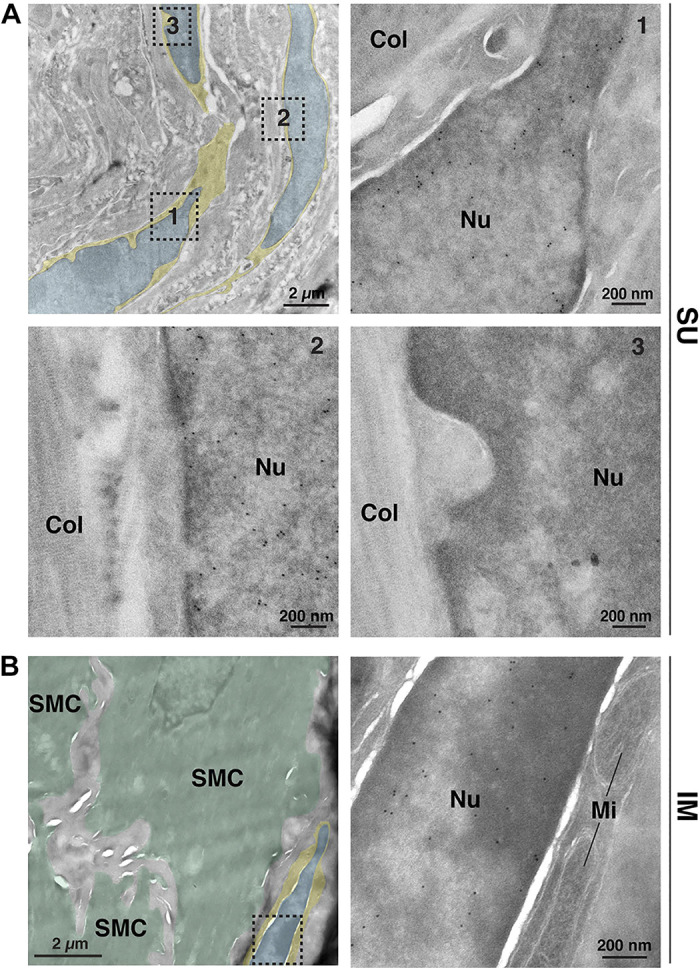
Bladder fibroblasts express Pdgfra. A and B: ultrathin cryosections obtained from PdgfraH2B-EGFP mouse bladders were immunogold labeled to reveal Pdgfra+ cells expressing the nucleus-localized H2B-enhanced green fluorescent protein (EGFP) marker. As cryo-electron microscopy (EM) generates relatively low-contrast images, we color coded the cytoplasm of the fibroblasts yellow and their nuclei blue. A: EM images of suburothelial fibroblasts. The dashed boxed regions in the top left image are magnified in their indicated images. Positive nuclei (Nu) contained gold particles. Collagen fibers (Col) were found in the matrix surrounding the cells. B: EM image of an intermuscular fibroblast closely apposed to smooth muscle cells (SMC), which are false colored green in the left micrograph. The dashed boxed region in the left image is magnified in the right image. Mitochondria (Mi) are marked. Data are representative of images acquired from two mice. Nuclei of nonfibroblast cells were negative, and control experiments performed in the absence of primary antibody also exhibited no staining.
To better understand the morphology of PDGFRA+ suburothelial cells, we performed confocal microscopy on whole mounted tissue viewed en face (Supplemental Fig. S2A). This analysis revealed that suburothelial fibroblasts (in this case labeled with CAR3) were stellate shaped, extending numerous projections from their outer margins. Although it was difficult to assess whether extensions from adjacent cells were forming direct contacts using this technique, our EM analysis indicated that this was the case (Fig. 2D). We further observed that suburothelial fibroblasts were layered in the z-axis (Supplemental Fig. S2A), confirming the stacked nature of the fibroblasts we observed in our immunofluorescence experiments and electron microscopic analysis.
An additional population of PDGFRA+ cells was found in the outer lamina propria (Fig. 3C), just below the suburothelial region. Within rougae, these outer lamina propria cells were also arranged in linear arrays, typically two to four cell layers thick, that were deep to the suburothelial ones (Fig. 3C); however, at the base of the rougae, where larger blood vessels were often found, outer lamina propria-associated PDGFRA+ cells were less obviously organized. In those relatively flat regions between rougae, outer lamina propria cells also appeared stacked (1−3 cell layers thick) and were sandwiched between the suburothelial fibroblasts and underlying fascicles of smooth muscle cells. Also consistent with our EM analysis, we observed a third population of PDGFRA+ cells that were intramuscular and were found in the perimysium surrounding the fascicles of smooth muscle cells (Fig. 3, B and C). In whole mounts, PDGFRA+ cells appeared to be elongate, had irregular borders, and were intercalated between adjacent smooth muscle cells (Supplemental Fig. S2B). These cells sent long processes, which appeared to interconnect adjacent cells. Immunogold labeling confirmed that the nuclei of intermuscular fibroblasts were also H2B-EGFP+ (Fig. 4B). Finally, a fourth, but small, population of PDGFRA+ cells, typically one cell layer thick in cross section, was found in the thin serosal connective tissue layer between the outermost smooth muscle bundles and the mesothelium (Fig. 3, B and C). In whole mounted tissue, these cells were observed to be irregularly shaped, their cell extensions were organized in a reticulum, and the cells abutted the ends of smooth muscle bundles (Supplemental Fig. S2C).
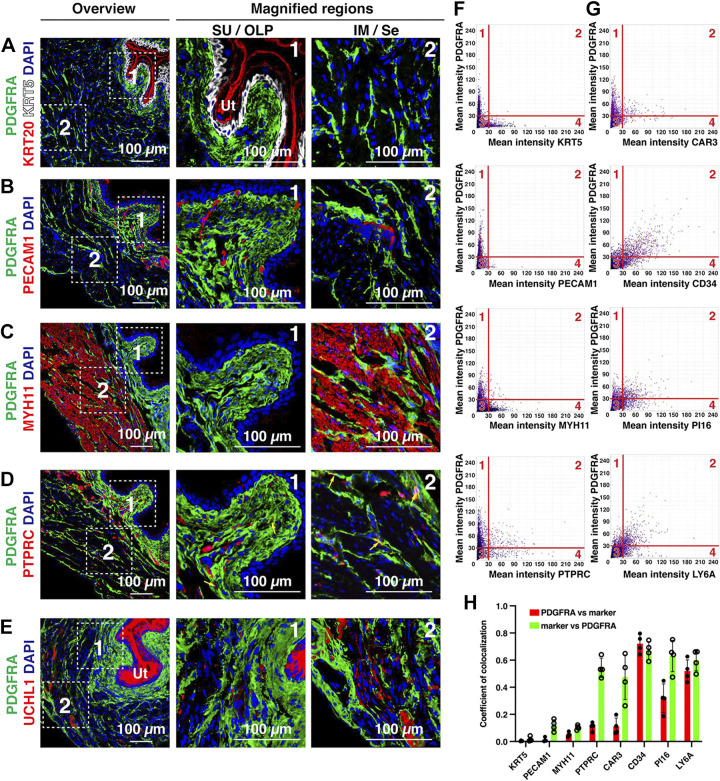
To determine whether PDGFRA+ cells were distinct from other cell types in the bladder wall, we colocalized PDGFRA with the following: KRT5 or KRT20, markers of basal/intermediate cells or umbrella cells of the urothelium, respectively (Fig. 5A); PECAM1, a marker of endothelial cells and associated blood vessels, which are prevalent in the lamina propria (Fig. 5B); MYH11, which labels smooth muscle cells (Fig. 5C); PTPRC (common name: CD45), a protein expressed by all immune cells, including lymphocytes, granulocytes, and mast cells (Fig. 5D); and UCHL1 (common name: PGP9.5), a general marker of nerve fibers (Fig. 5E), although Uchl1 is reportedly expressed in umbrella cells (19). We found limited evidence of colocalization between PDGFRA+ cells and those positive for these other markers by visual inspection, although some PTPRC+ cells were also PDGFRA+ (see yellow arrows in Fig. 5D). We used tissue cytometry to further assess the nature of PDGFRA+ cells including measurements of marker expression. The resulting data are presented in a scatterplot similar to that used for flow cytometry (Fig. 5F and Supplemental Fig. S3) (56). We observed few cells that were positive for PDGFRA and also positive for KRT5, PECAM, or MYH11 (see quadrant 2 of Fig. 5F and quantitation in Fig. 5H), and although the vast majority of PDGFRA+ cells were PTPRC–, ∼45% of PTPRC+ cells were also PDGFRA+ (Fig. 5H). The nature of these latter cells was not explored further, although fibrocytes are CD45+;PDGFRA+. As there are no neuronal cell bodies in the mouse bladder wall, we were unable to perform this colocalization analysis in samples labeled with UCHL1.
Figure 5.
Distribution of PDGFRA+ fibroblasts relative to other tissue types, cells, and markers in the bladder wall. A–E: representative confocal micrographs showing the distribution of PDGFRA+ fibroblasts with respect to KRT5- and KRT20-labeled urothelium (A), PECAM1-labeled endothelial cells (B), MYH11-labeled smooth muscle cells (C), PTPRC-labeled immune cells (D), or UCHL1-labeled nerve fibers (E). Note that the urothelium (Ut; umbrella cells in particular) was also UCHL1+. The dashed boxed regions in the left images are magnified in the middle and right images. Representative confocal micrographs from 4−6 mice are shown. F and G: nuclei in confocal Z-stacks were detected, and the mean fluorescent intensity of the indicated markers associated with each nucleus was measured and plotted using Volumetric Tissue Exploration and Analysis tissue cytometry software. Each dot represents a single nucleus and its associated mean intensity for the indicated markers. The data were gated into quadrants with quadrant 1 representing cells that were just PDGFRA+, quadrant 2 representing cells that were gated as positive for both markers, quadrant 3 representing cells that were “negative” for both markers, and quadrant 4 representing cells positive for just the indicated marker. H: the colocalization coefficient for cells positive for both PDGFRA and the indicated marker, or vice versa, was calculated and plotted. Analysis was performed on random sections taken from the bladders of 4 animals. Data are means ± SD.
Next, we determined if PDGFRA+ cells expressed gene products associated with fibroblasts including VIM, a protein sometimes referred to as the “fibroblast intermediate filament,” although VIM can be expressed by other cell types (e.g., endothelial cells and mesothelial cells). Bladder “interstitial cells” are reportedly VIM+ (27, 36, 39, 41–43, 45–49, 60, 64, 65). On visual inspection, we observed considerable overlap between VIM and PDGFRA in all cell populations, although there were PDGFRA+;VIM– cells, particularly in the intermuscular region (Supplemental Fig. S4).
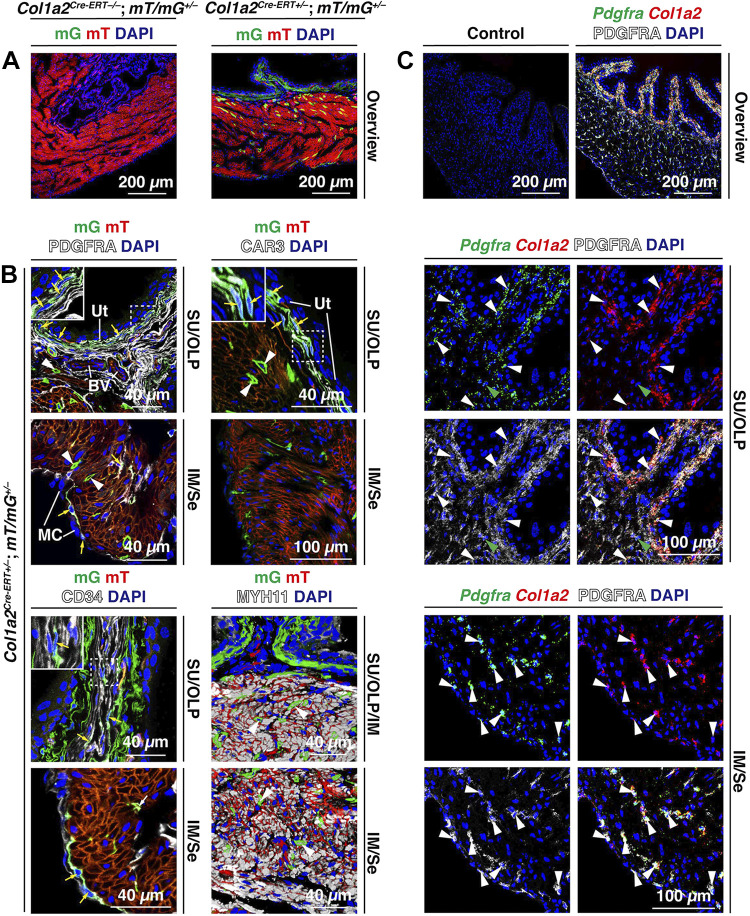
In transcriptomic experiments, the canonical fibroblast marker and collagen gene Col1a2 is reported to be expressed by putative bladder fibroblasts (4, 19, 20, 66). Although bladder smooth muscle cells can also express collagens (19, 67), they are PDGFRA– (Fig. 5C). To examine Col1a2 expression in young adult mice, we treated 8-wk-old Col1a2Cre-ERT–/–;RosamT/mG+/– control mice or Col1a2Cre-ERT+/–;RosamT/mG+/– experimental mice with tamoxifen. RosamT/mG+/– mice constitutively express the gene encoding membrane-bound tandem dimer-Tomato (mT) in all cells, but in cells expressing Col1a2, Cre-mediated recombination removes the mT cassette and associated stop codon allowing expression of the downstream membrane-bound EGFP (mG) cassette. In control mice, Col1a2-driven mG expression was not observed in the urothelium or lamina propria (Fig. 6A); however, smooth muscle cells of these mice expressed low levels of mG (not shown). In experimental mice, suburothelial cells were strongly mG+ and colabeling experiments confirmed that these cells were PDGFRA+ (Fig. 6B). Scattered PDGFRA+;mG+ cells were also found in the outer lamina propria, intermuscular region, and serosa (Fig. 6, A and B); however, the presence of PDGFRA+;mG– cells in these regions indicated that not all PDGFRA+ cells in adult bladders are expressing Col1a2 at the time of tamoxifen induction or that Col1a2Cre-ERT-mediated recombination is inefficient. We also observed a small population of MYH11+ smooth muscle cells that exhibited high amounts of Col1a2-Cre-driven mG expression (white arrowheads in Fig. 6B). The nature of these high-expressing smooth muscle cells was not studied further.
Figure 6.
PDGFRA+ cells express Col1a2. A: expression of Col1a2-driven mG (membrane-bound enhanced green fluorescent protein) in tamoxifen-treated experimental Col1a2Cre-ERT+/–;RosamT/mG+/– or control Col1a2Cre-ERT–/–;RosamT/mG+/– mice. B: coexpression of mG in the PDGFRA+, CAR3+, or CD34+ cells of the indicated regions of the bladder wall [suburothelial (SU), outer lamina propria (OLP), intermuscular (IM), and serosal (Se)]. Examples of cells with visible nuclei coexpressing these markers are marked with yellow arrows. Coexpression of MYH11 and mG was also assessed in the bottom right images. Examples of smooth muscle cells expressing mG are indicated by white arrowheads. C: use of fluorescent in situ hybridization with immunodetection (FISHID) to assess coexpression of PDGFRA, Pdgfra, and Col1a2 in the indicated regions of the bladder wall. Examples of cells (i.e., nuclei) that expressed all three markers are labeled with white arrowheads; cells that were PDGFRA+;Pdgfra+ are marked with green arrowheads. The image marked “Control” was performed in the absence of anti-PDGFRA antibody and using the three-plex negative control probe against Bacillus subtilis dapB. All images are confocal micrographs and representative of images taken from two separate experiments each using two mice. BV, blood vessels; MC, mesothelial cells; Ut, urothelium.
To better understand Col1a2 expression in the bladder wall at steady state, we used a high-sensitivity approach that coupled FISH with immunodetection (FISHID). Consistent with the cell tracing experiments described above, the most extensive Col1a2 signal was observed in the suburothelial region (Fig. 6C). However, we readily detected Pdgfra+;Col1a2+;PDGFRA+ cells in the other three regions and more consistently than with the cell tracing approach. Control incubations lacked signal (Fig. 6C).
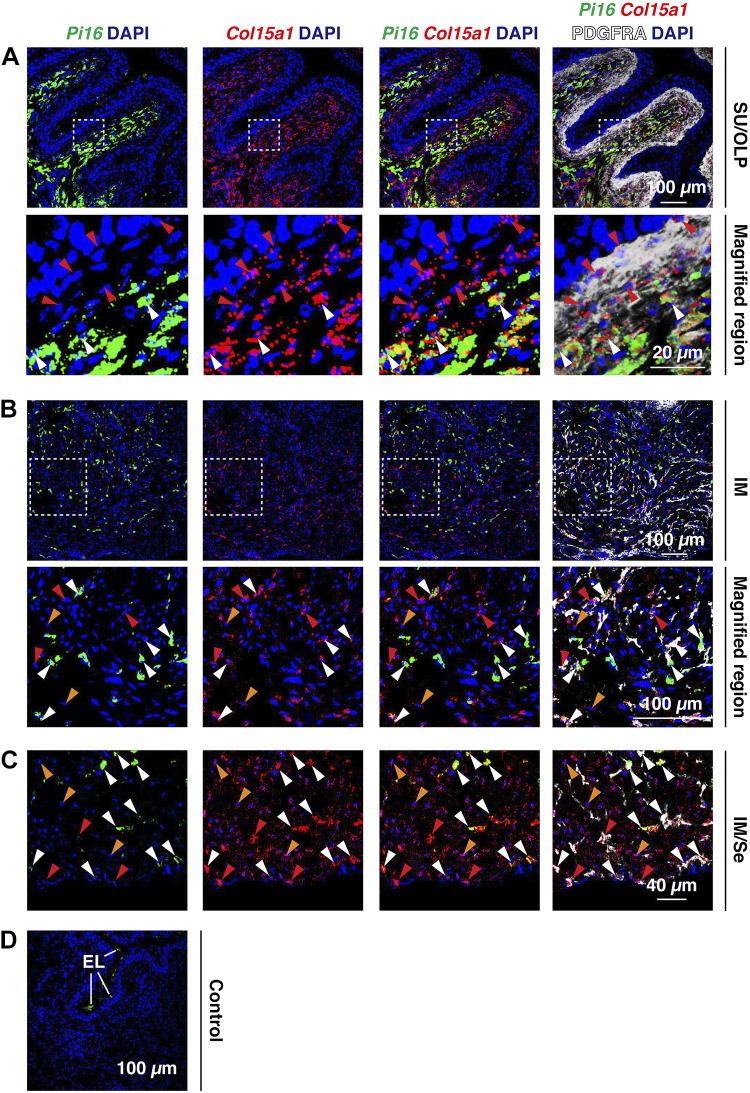
We subsequently used FISHID to determine whether PDGFRA+ cells expressed the universal fibroblast genes Pi16 or Col15a1 at steady state (Fig. 7). Interestingly, the suburothelial population of PDGFRA+ cells was Col15a1+ but Pi16– (red arrowheads in Fig. 7A). In contrast, in the outer lamina propria, most of the PDGFRA+ cells were Col15a1+;Pi16+ (white arrowheads in Fig. 7A). In the intermuscular region (Fig. 7B), we observed many PDGFRA+ cells that were also Col15a1+;Pi16+ (white arrowheads); however, we also observed scattered PDGFRA+ cells that were Col15a1+;Pi16– (red arrowheads) or Col15a1–;Pi16+ (not shown). We also observed that PDGFRA– cells, which because of their localization in the muscularis externa were likely smooth muscle cells, expressed Col15a1 (orange arrowheads in Fig. 7, B and C). Finally, serosal PDGFRA+ cells were either Col15a1+;Pi16+ or Col15a1+;Pi16– (white or red arrowheads, respectively; Fig. 7C). Control incubations lacked specific signal (Fig. 7D).
Figure 7.
Bladder PDGFRA+ fibroblasts express the universal fibroblasts genes Pi16 and Col15a1. A–D: fluorescent in situ hybridization with immunodetection was used to assess coexpression of Col15a1 and Pi16 in PDGFRA+ fibroblasts of the indicated regions of the bladder wall [suburothelial (SU), outer lamina propria (OLP), intermuscular (IM), and serosal (Se)]. Cell nuclei that were PDGFRA+;Col15a1+ are indicated by red arrowheads; cell nuclei that were PDGFRA+;Col15a1+;Pi16+ are indicated by white arrowheads. Cell nuclei that were PDGFRA–;Col15a1+, likely smooth muscle cells, are indicated by orange arrowheads. D: a control incubation was performed in the absence of anti-PDGFRA antibody and using the three-plex negative control probe against Bacillus subtilis dapB. Otherwise, the sample was treated identically to experimental samples. Endolysosomes (EL) in the umbrella cell layer of the urothelium exhibited autofluorescence. All images are confocal micrographs and representative of images taken from two mice.
In summary, most PDGFRA+ cells are not urothelial, endothelial, muscle, nerve, or immune cells in nature. Instead, their combined expression of VIM, Col1a2, Col15a1, Pdgfra, and Pi16, coupled with their ultrastructure, indicates that they are fibroblasts. Furthermore, their distinct locations within the bladder wall, coupled with different patterns of expression of Col15a1 and Pi16, indicates that regionally distinct populations of fibroblasts likely exist in the bladder wall.
The ICC Marker KIT/Kit Is Not Expressed in PDGFRA+ Fibroblasts of the Bladder Wall
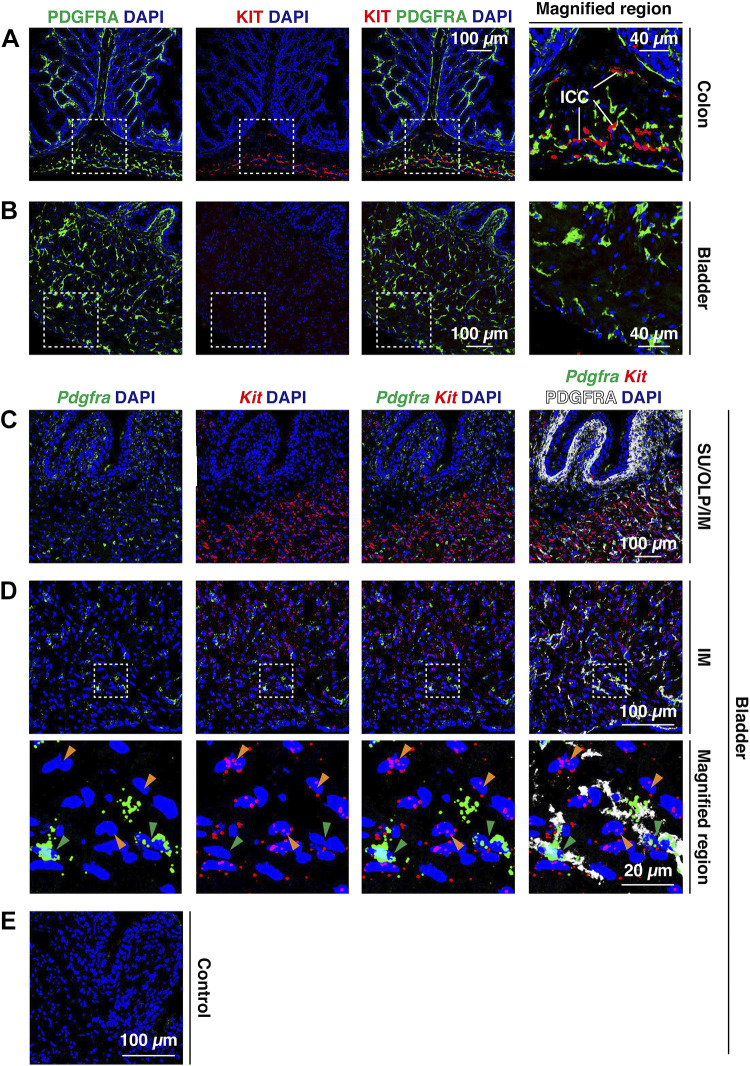
The expression of KIT in bladder interstitial cells is controversial (24, 31–44). To determine if we could detect KIT expression in mouse PDGFRA+ fibroblasts, we used the well-validated function-blocking ACK-2 monoclonal antibody that recognizes ICC in the mouse gut (39, 68, 69). Like Koh et al. (39), we detected KIT+ ICC in colon tissue (Fig. 8A) but detected almost no KIT protein signal in similarly prepared and treated bladder tissue from the same animals (Fig. 8B). In the colon, we observed that KIT+ cells were often in close proximity to those that were PDGFRA+ (see magnified region in Fig. 8A), which is consistent with reports demonstrating that although closely associated, PDGFRA+ cells in the gut are distinct from those that are KIT+ (29, 70–73). We also explored Kit gene expression in these tissues using FISH or FISHID. Consistent with a previous report, Kit expression was observed in the mucosa of the colon by what are likely to be crypt-associated goblet cells (Supplemental Fig. S5A) (74). We also observed presumptive Kit+ ICC within the muscular regions of the colon wall, which had a similar distribution to KIT+ ICC (compare Supplemental Fig. S5A with Fig. 8A). In the mouse bladder, limited Kit expression was found in the urothelium or lamina propria; however, we observed Kit signal in the muscularis externa (Fig. 8, C and D, and Supplemental Fig. S5B). These Kit+ cells (and associated nuclei) were distinct from Pdgfra+/PDGFRA+ ones (compare orange versus green arrowheads in Fig. 8, C and D, and Supplemental Fig. S5B) and FISHID confirmed that much of the Kit signal in the intermuscular region of the mouse bladder was associated with MYH11+ smooth muscle cells and not Pdgfra+/PDGFRA+ fibroblasts (Supplemental Fig. S5C). Low levels of Kit gene expression in bladder smooth muscle cells have been previously described (19). The lack of corresponding KIT protein expression indicates that Kit mRNA is apparently not being translated, KIT protein expression is below the level of detection using our methods, or that KIT protein is rapidly turned over. Taken together, our study indicates that PDGFRA+ fibroblasts of the mouse bladder do not express significant amounts of KIT protein or Kit message.
Figure 8.
Expression of Kit/KIT in the mouse bladder and colon. A and B: localization of PDGFRA and KIT in the colon (A) and bladder wall (B). Presumptive colon-associated interstitial cells of Cajal (ICC) are marked in A. The dashed boxed regions are magnified in the right images. C and D: use of fluorescent in situ hybridization with immunodetection to assess Pdgfra and Kit expression in PDGFRA+ fibroblasts. The dashed boxed regions in D are magnified in the bottom images. Note that most Kit expression was associated with presumptive smooth muscle cell nuclei (orange arrowheads), whereas most Pdgfra expression was associated with PDGFRA+ fibroblast nuclei (green arrowheads). E: a control incubation was performed in the absence of anti-PDGFRA antibody and using the three-plex negative control probe against Bacillus subtilis dapB. All images are confocal micrographs and representative of images taken from two separate experiments each with two animals.
The Mouse Bladder Wall Has Multiple Fibroblast Subtypes
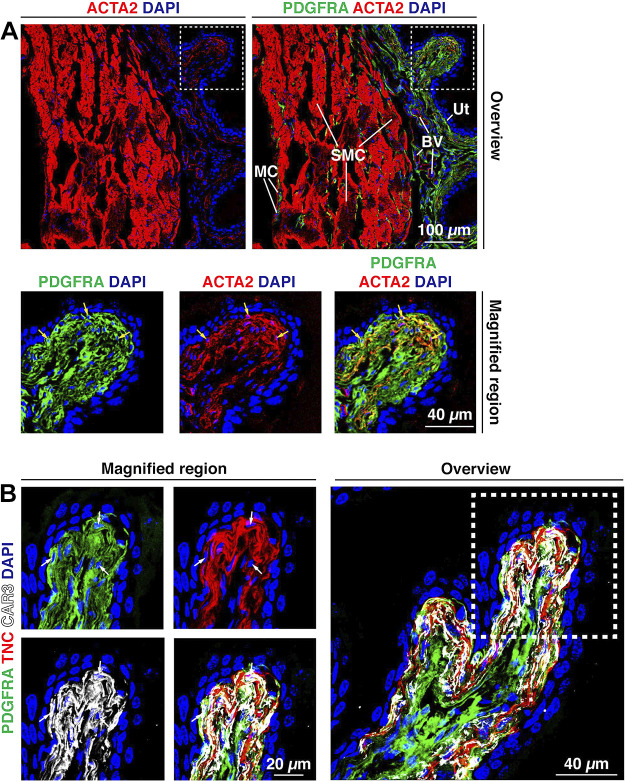
To further characterize PDGFRA+ fibroblasts, we sought to identify markers that could be used to classify fibroblasts in the four regions of the bladder wall. In the case of suburothelial fibroblasts, we looked for expression of MYH10, which we have previously reported is expressed in suburothelial PDGFRA+ cells (75) (and which we confirmed in Supplemental Fig. S6). Mouse and human bladder fibroblasts/interstitial cells also reportedly express Acta2 and Tnc/TNC (19, 20, 43, 49, 58–60). We corroborated these findings by showing that suburothelial PDGFRA+ fibroblasts expressed both TNC and ACTA2 (Fig. 9, A and B). However, we sought to identify other markers, as ACTA2 is a well-known marker of smooth muscle cells (and myofibroblasts) and TNC is a secreted protein that accumulates in the ECM. One candidate was the cytosolic enzyme CAR3, whose gene is reportedly expressed in Acta2+/TNC+ fibroblasts (20, 22), which we validated using an antibody to this protein (Fig. 9B). Although strong expression was observed in suburothelial fibroblasts, weaker expression was also noted in smooth muscle cells. Furthermore, we observed that CAR3+ fibroblasts express Col1a2 (Fig. 6B), and, using FISHID, we confirmed that CAR3+ suburothelial fibroblasts expressed Col15a1 but not Pi16 (Supplemental Fig. S7A). Consistent with its limited distribution, tissue cytometry revealed ∼10% of PDGFRA+ cells were also CAR3+, whereas the majority of CAR3+ cells were also PDGFRA+ (Fig. 5, G and H).
Figure 9.
PDGFRA+ suburothelial fibroblasts express ACTA2, CAR3, and TNC. A: ACTA2 was expressed by smooth muscle cells (SMC) in the muscularis externa and blood vessels (BV) within the lamina propria. A weaker, but reproducible, ACTA2 signal was also observed in suburothelial fibroblasts, subjacent to the urothelium (Ut). The dashed boxed region in the top right image is magnified in the bottom images. Nuclei of PDGFRA+;ACTA2+ cells are indicated by yellow arrows. B: expression of TNC and CAR3 by PDGFRA+ suburothelial fibroblasts. The dashed boxed region in the right image is reproduced in the left images. Cells with visible nuclei expressing all three markers are indicated by white arrows. Note that PDGFRA is a membrane protein, CAR3 is a cytoplasmic protein, and TNC is a secreted protein that is deposited in the space between adjacent fibroblasts. All images are confocal micrographs and representative of images taken from three-five separate experiments each with one-two mice.
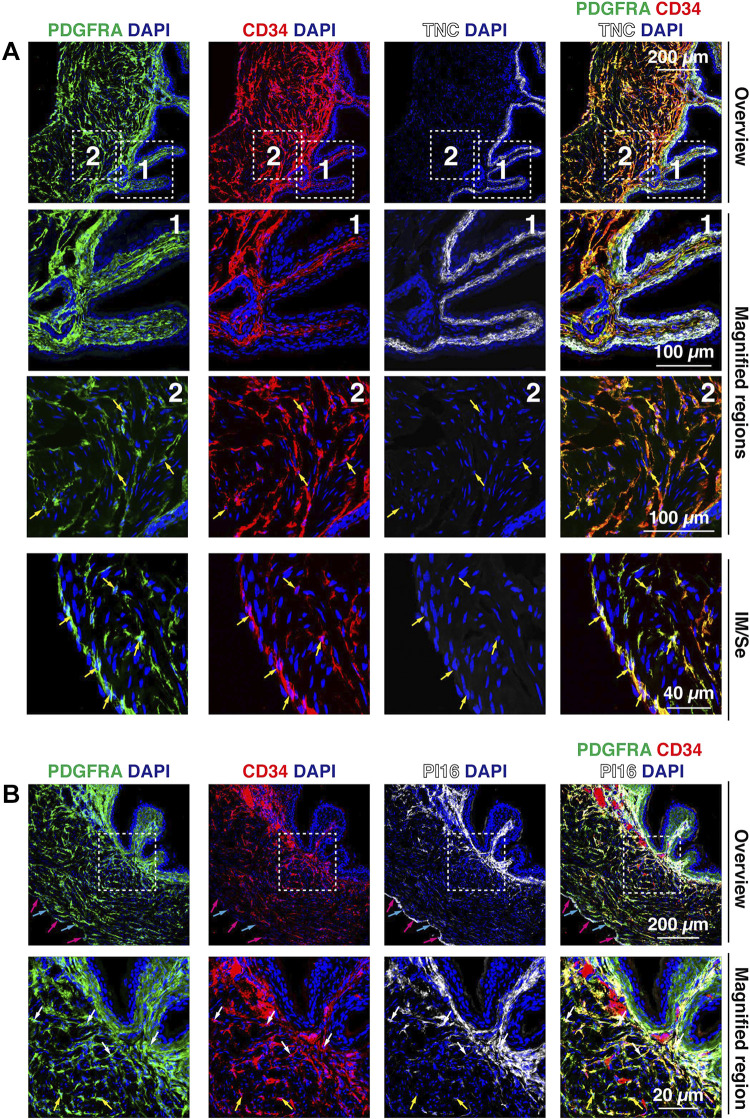
CD34, an additional canonical fibroblast marker, is also reportedly expressed by a subset of fibroblasts/interstitial cells in the bladder wall (19, 20, 43, 46, 48). Although CD34 is also expressed by endothelial and mesothelial cells, these cells are PDGFRA–. In our experiments, the majority of PDGFRA+ cells in the outer lamina propria, intermuscular region, and serosal region were CD34+ (Fig. 10A); however, suburothelial fibroblasts, in this case marked with TNC, were CD34– (Fig. 10A). These observations are in agreement with tissue cytometry, which showed the highest degrees of colocalization between PDGFRA and CD34 (Fig. 5, G and H). Furthermore, and consistent with universal fibroblast gene expression in PDGFRA+ cells (Fig. 7), CD34+ fibroblasts in the outer lamina propria were Pi16+ and Col15a1+ (Supplemental Fig. S7B), whereas in the intermuscular region, fibroblasts were CD34+;Col15a1+, CD34+;Pi16+, or CD34+;Pi16+;Col15a1+ (red, green, or white arrowheads, respectively, in Supplemental Fig. S7B). We also explored PI16 protein expression and distribution (Fig. 10B). PI16 was enriched in those CD34+ fibroblasts in the outer lamina propria, whereas in the intermuscular regions, PI16 reactivity was limited to those CD34+ fibroblasts that were closer to the lamina propria. A population of serosal PDGFRA+ fibroblasts were Pi16+ (Fig. 7C) or PI16+ (see cyan arrows in Fig. 10B), and mesothelial cells were PI16+ (magenta arrows in Fig. 10B). Again, these observations mimic the tissue cytometry analysis: a subset of PDGFRA+ cells (∼30%) were PI16+, whereas the majority of PI16+ cells were PDGFRA+. Unfortunately, we were unable to identify antibodies to COL15A1 that worked reliably in mouse tissues.
Figure 10.
Expression of CD34, PI16, and TNC in bladder fibroblasts. A: coexpression of PDGFRA, CD34, and TNC in the suburothelial region of the mouse bladder. The dashed boxed regions in the top images are magnified in the bottom images. Examples of cells (with visible nuclei) that coexpressed CD34 and PDGFRA are indicated by yellow arrows. B: expression of PDGFRA, CD34, and PI16 in the mouse bladder wall. PI16 was expressed by fibroblasts in the lamina propria, a fraction of fibroblasts in the innermost portions of the intermuscular region (i.e., the region closest to the urothelium), and in serosal fibroblasts. In the top images, nuclei of PDGFRA+; PI16+ serosal cells are indicated by cyan arrows and nuclei of PI16+; PDGFRA– mesothelial cells are marked with magenta arrows. In the bottom images, nuclei of cells coexpressing PDGFRA, CD34, and PI16 are indicated by white arrows and nuclei of PDGFRA+, CD34+, PI16– cells are marked with yellow arrows. All images are confocal micrographs and representative of images taken from three separate experiments each with two mice.
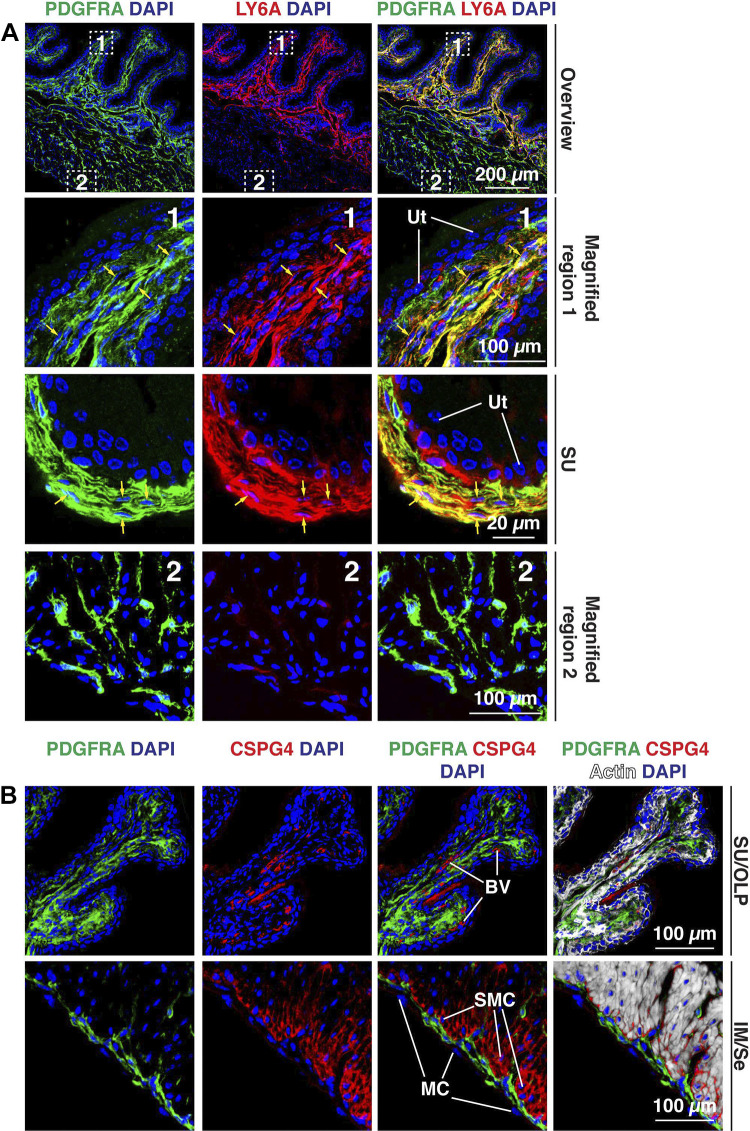
LY6A (common name: stem cell antigen 1) has previously been reported to label “mesenchymal stem cells” in the lamina propria region of the mouse bladder wall (9). However, more recent analysis indicates that Ly6a is a canonical fibroblast marker (20), and Ly6a is often coexpressed with the universal gene markers Col15a1 and Pi16 (23). We observed significant overlap between PDGFRA and LY6A in those fibroblasts located in the lamina propria, including the suburothelial fibroblasts and outer lamina propria-associated ones (Fig. 11A), and LY6A antibodies also labeled those intermuscular fibroblasts that were near the lamina propria. However, most intermuscular and serosal fibroblasts expressed low levels of or no LY6A. Consistent with a recent report (22), we observed that basal urothelial cells, which are PDGFRA−, were also variably LY6A+ (see third row of Fig. 11A). Tissue cytometry analysis revealed that ∼50% of PDGFRA+ cells were LY6A+ (Fig. 5, G and H). Data from Muhl et al. (20) indicated the possible existence of a small population of Cspg4+ (common names include chondroitin sulfate proteoglycan 4 and neural/glial antigen 2) fibroblasts in the bladder wall. CSPG4 is a transmembrane proteoglycan and marker of endothelial cells and pericytes. We confirmed that CSPG4 was associated with blood vessels in the bladder wall (Fig. 11B). We also observed CSPG4 expression in smooth muscle cells, particularly those in the outer longitudinal layer of the muscularis externa (closest to the serosal surface; Fig. 11B). However, there was little overlap between CSPG4+ cells and PDGFRA+ fibroblasts of the suburothelial, outer lamina propria, or intermuscular regions of the bladder wall (Fig. 11B).
Figure 11.
Expression of LY6A and CSPG4 in the mouse bladder wall. A: the canonical fibroblast marker LY6A was expressed by PDGFRA+ fibroblasts in the suburothelial and outer lamina propria regions. The dashed boxed regions in the top images are magnified in the bottom images. A region with more pronounced labeling of PDGFRA+, LY6A+ suburothelial (SU) fibroblasts is shown in the third row. Examples of colocalization are indicated by yellow arrows. B: CSPG4, a transmembrane proteoglycan, was associated with blood vessels (BV) and smooth muscle cells (SMCs), particularly those in the outer longitudinal layer of the muscularis externa. In the muscle tissue, CSPG4 is likely to contribute to the endomysial connective tissue between smooth muscle cells. Mesothelial cells (MCs) are indicated. There was little colocalization of PDGFRA and CSPG4 in the bladder wall. All images are confocal micrographs and representative of images taken from four separate experiments each with two mice. Ut, urothelium.
In summary, we found that the mouse bladder wall includes multiple populations of fibroblasts with overlapping but distinct patterns of marker expression. A synopsis of these fibroblasts, their location, and their markers is shown in Fig. 12.
Figure 12.
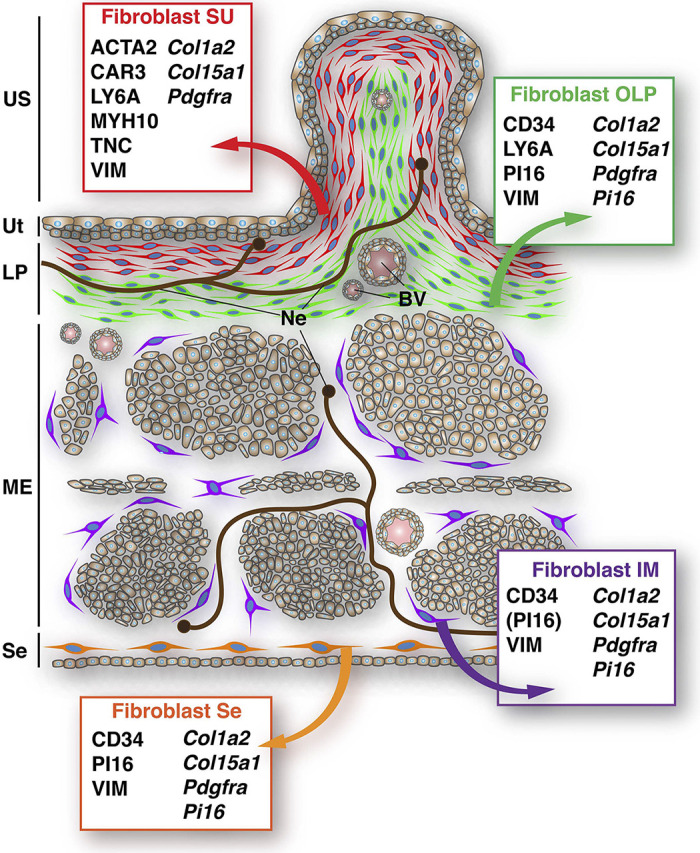
Summary of fibroblast populations and their marker expression in the mouse bladder wall. BV, blood vessel; IM, intermuscular; LP, lamina propria; ME, muscularis externa; Ne, neuron; OLP, outer lamina propria; Se, serosa; SU, suburothelial; US, urinary space; Ut, urothelium.
DISCUSSION
Current studies of bladder connective tissue and fibroblast biology are mired in a more than two-decade-long debate concerning the nature of bladder interstitial cells and whether they function in a manner similar to ICC, pacemaker cells in the gut (24). Based on early reports that bladder interstitial cells generate cGMP in response to nitric oxide donors as well as early evidence that they express markers of ICC (including the canonical ICC marker KIT), some have proposed the existence of bladder ICC-like cells that integrate communication between the urothelium, nervous system, and musculature (27, 31–37, 76). However, expression of KIT in the bladder wall is controversial and the hypothesis of ICC-like cells in the bladder does not comport with the following: reports (including this one) showing limited or no KIT protein expression in the bladder (24, 38–44), our observation that at steady state, Kit gene expression is limited to smooth muscle cells, findings that the majority of KIT+ cells in the human bladder wall are mast cells (31, 38, 41), and evidence that rodents with reduced Kit expression have limited bladder symptoms (34, 77). Additional evidence against a pacemaker role for bladder interstitial cells is the observation that few of these cells generate spontaneous Ca2+ transients ex vivo, and the majority of these Ca2+ transients do not coincide with those of smooth muscle cells (35).
Furthermore, our EM analysis revealed that mouse bladder PDGFRA+ cells, like their human interstitial cell counterparts (31, 42, 43, 58–61), lack the ultrastructural characteristics of ICC (which include a complete or partial basement membrane, abundant mitochondria, smooth endoplasmic reticulum, visible cytoskeletal elements, presence of surface caveolae, close proximity to nerves, and formation of gap junctions with smooth muscle cells) (28, 69, 78). In the case of the bladder, we found no evidence of direct cell-cell contacts (i.e., gap junctions or adherence junctions) between PDGFRA+ cells and other cell types in the bladder wall, something also true of human bladder interstitial cells (42, 61), making it further unlikely that PDGFRA+ cells are ICC-like. This lack of direct heterotypic cell-cell contacts also does not comport with the hypothesis put forward by Koh and colleagues that in response to mechanochemical signals, bladder PDGFRA+ interstitial cells modulate the membrane potential of smooth muscle cells by way of gap junction-mediated coupling (24, 79–81). However, as discussed below, this does not rule out other forms of intercellular communication between these cell types.
If bladder PDGFRA+ interstitial cells are not ICC equivalents, then what are they? Their location in connective tissue along with their archetypal fibroblast-like ultrastructure would be sufficient to characterize these cells as fibroblasts. When coupled with data that they express the mesenchymal marker VIM, that they express several canonical fibroblast markers (Col1a2, CD34, LY6A, and PDGFRA), and that they express the universal fibroblast genes Col15a1 and Pi16, then it is highly probable that PDGFRA+ interstitial cells are fibroblasts. Our study further reveals that multiple populations of fibroblasts likely exist, each with a distinct cellular location and marker expression profile (Fig. 12), and presumably with specialized functions. Lying just below the urothelium is a suburothelial population of fibroblasts that are likely equivalent to the Acta2+ “myofibroblast” cluster proposed by Muhl et al. (20) and Yu et al. (19), although we believe this to be an unfortunate moniker given that myofibroblasts are generally associated with wound healing or disease processes such as fibrosis and cancer (82). Furthermore, mouse suburothelial fibroblasts lack a defining characteristic of myofibroblasts: the fibronexus (83). Although controversial, a small population of myofibroblasts may be found in the lamina propria of human bladder (43). In the case of the mouse bladder, it may be best to describe these cells as suburothelial, CAR3+, or TNC+ fibroblasts. Muhl et al. (20) have identified several other genes expressed by this specialized population of mouse fibroblasts, and exploration of these genes and their products may provide new insights into suburothelial fibroblast form and function. Interestingly, a human bladder population of “upper lamina propria” interstitial cells has been described, which share several features with mouse suburothelial fibroblasts including the layered organization, their ultrastructure, and markers that include ACTA2, PDGFRA, VIM, and CAV1, but not CD34 (42, 43, 49, 59–61). Thus, suburothelial fibroblasts, with common marker profiles and morphologies, likely exist across species.
Additional fibroblast populations can be found in the mouse bladder wall, including those in the outer lamina propria. Based on their marker profile, which includes expression of CD34 (Fig. 12), we believe outer lamina propria fibroblasts to be a subset of the CD34+ fibroblasts that Muhl et al. (20) have proposed. But, how the marker profile of outer lamina propria fibroblasts relates to proposed fibroblast clusters 1–3 in the Yu et al. (19) study is difficult to assess as all three of their fibroblast populations express these gene products, albeit to different degrees. Human bladders also have an outer lamina propria population of fibroblasts, which are CD34+ and VIM+ (but negative for ACTA2) (41, 42, 49). Although the expression of PDGFRA in these fibroblasts is controversial (43, 45), the general marker profile is on its face like that of mouse. The final clusters of PDGFRA+ fibroblasts we identified are found in the intermuscular region and serosa (Fig. 12). Although most intermuscular fibroblasts expressed CD34 and PDGFRA, there was variation in the expression of the universal markers Col15a1 and Pi16, with some cells expressing one or the other gene and some cells expressing both. This may indicate further microdiversity in this population of fibroblasts. Serosal fibroblasts exhibit a similar expression profile to intermuscular ones, although serosal ones are also PI16+ (Fig. 12). These serosal fibroblasts also exhibit marker variability, with some expressing one or both universal gene markers. Other previously described interstitial cell protein markers include the connexin GJA1, caveolin-associated protein CAV1, cadherin CDH2, purinergic receptors, ecto-nucleotidase ENTPD2 (which is also a canonical fibroblast marker), muscarinic receptor CHRM3, ion channels [including transient receptor potential (TRP)V4, TRPA, and KCNN3], and receptors for prostaglandins (47, 49, 65, 79–81, 84–90). Further work will be necessary to define the relationship of these previously described markers and the framework we present in this report.
By recognizing that most or all bladder interstitial cells are fibroblasts, we are poised to apply the known biology of fibroblasts to the bladder. One well-documented function of fibroblasts is the release of bioactive substances such as cytokines and growth factors (4). Thus, bladder fibroblasts could alter the function of the urothelium or adjacent muscle cells, immune cells, or nerve fibers by way of paracrine signaling pathways. Our observation that fibroblasts and their cell extensions are in close proximity to nerve bundles and smooth muscle cells would support such a possibility. This putative communication network could work along the lines of the hypothesis put forward by Fry et al. (26) that interstitial cells function as variable gain amplifiers that can promote or integrate signaling responses from multiple cellular sources in the bladder wall. Unfortunately, these pathways are poorly understood at this moment (1). Interestingly, during chemical injury or bacterial cystitis, urothelium-released SHH (common name: sonic hedgehog) binds to its receptor PTCH1 (common name: Patched1) on so-called stromal cells, possibly fibroblasts, leading to the expression of Gli1 and Ptch1 (91). Inhibition of GLI1 function or Gli1 expression blocks proliferation of both stromal cells and the overlying urothelium and thus prevents reestablishment of the urothelial barrier (91). Release of fibroblast growth factors and bone morphogenetic proteins by suburothelial fibroblasts may also regulate urothelial regeneration (22, 92). Thus, suburothelial fibroblasts may have critical roles in responses to urothelial injury, lending further credence to their hypothesized role in intertissue communication.
An additional function of fibroblasts is to synthesize, degrade, and organize the ECM, which in turn impacts the mechanical properties of the bladder wall. For example, changes in collagen turnover or changes in the collagen type I-to-collagen type III ratio can impact wall stiffness (93, 94), whereas altered synthesis of proteoglycans such as decorin can impact elasticity and collagen assembly (95). Collagen is also likely important for responses of the bladder to strain, particularly during the filling phase of the bladder cycle (57, 96). Suburothelial fibroblasts express ACTA2, an isoform of actin often implicated in contractile responses (97). The function of this expression is unknown, but one hypothesis is that it promotes the refolding of the mucosal surface into rugae after voiding. Suburothelial fibroblasts also produce TNC (20), a multimodal protein that modulates cell signaling, gene expression, adhesion, migration, and proliferation, which may contribute to the specialized functions of the cells (98). The role(s) of the other populations of fibroblasts in the mouse bladder wall remains to be determined, but intramuscular connective tissues (i.e., epimysium, perimysium, and endomysium) are important in the development and function of skeletal muscles (99). By extension, the matrix produced by mouse bladder intermuscular and serosal fibroblasts may have similar roles in governing smooth muscle function. Interestingly, fibroblasts are themselves mechanosensitive (100–102), which comports with previous findings that interstitial cells may be stretch sensitive (103, 104), and our findings (and those of others) that native bladder PDGFRA+ fibroblasts express the stretch-sensitive channel PIEZO1 and also express Piezo2 (54, 104, 105). Thus, fibroblasts are likely able to sense changes in wall stiffness and bladder filling, and the activation of downstream mechanotransduction cascades could contribute to bladder function.
An important pathological role for fibroblasts occurs in the setting of fibrosis, which happens when the functional tissue of an organ is replaced by an excessive and stiff ECM (4, 106, 107). Despite evidence that bladder fibrosis is associated with ketamine abuse (8), outflow obstruction (9–11, 108), radiation-induced cystitis (12–15), spinal cord injury (16), or congenital anomalies (17), there remains limited understanding of how these pathologies impact the fibroblast composition of the affected bladder or how fibroblasts contribute to these conditions. Likewise, tumor-associated fibroblasts are critical components of tumor aggressiveness and disease (109, 110). Thus, a potential benefit of our study is the future identification of fibroblast subtypes and corresponding cell type-specific markers that can be used to define stem cells or progenitor cells that give rise to fibrosis-inducing myofibroblasts or tumor-associated fibroblasts. In turn, this may lead to new ways to target disease processes that impact the urinary bladder.
Finally, in the past decade, the concept of fibroblast-like interstitial cells and telocytes has increasingly pervaded studies of the genitourinary system, gut, skin, and peripheral nervous system (30, 111–115). In light of our findings, we believe a careful analysis of these connective tissue cells, many of which express CD34, may reveal that they too are closely related in form and function to fibroblasts.
SUPPLEMENTAL DATA
Supplemental Figs. S1–S7: https://doi.org/10.6084/m9.figshare.19719856.
GRANTS
This work was supported by National Institutes of Health (NIH) pilot project award through Grant P30DK079307 (to M.G.D.), NIH Grant R01DK119183 (to G.A. and M.D.C.), an American Urological Association Career Development award and a Winters Foundation grant (to N.M.), by the Cell Physiology and Model Organisms Kidney Imaging Cores of the Pittsburgh Center for Kidney Research (NIH Grant P30DK079307), and by NIH Grant S10OD028596 (to G.A.), which funded the purchase of the Leica Stellaris confocal system.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
D.R.C., W.G.R., M.G.D., N.M., M.D.C., and G.A. conceived and designed research; D.R.C., W.G.R., M.G.D., N.M., and M.D.C. performed experiments; D.R.C., W.G.R., M.G.D., N.M., M.D.C., and G.A. analyzed data; D.R.C., W.G.R., M.G.D., N.M., M.D.C., and G.A. interpreted results of experiments; D.R.C., W.G.R., M.G.D., and G.A. prepared figures; D.R.C., M.G.D., and G.A. drafted manuscript; D.R.C., M.G.D., N.M., M.D.C., and G.A. edited and revised manuscript; D.R.C., W.G.R., M.G.D., N.M., M.D.C., and G.A. approved final version of manuscript.
REFERENCES
- 1. Dalghi MG, Montalbetti N, Carattino MD, Apodaca G. The urothelium: life in a liquid environment. Physiol Rev 100: 1621–1705, 2020. doi: 10.1152/physrev.00041.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gabella G. Lamina propria: the connective tissue of rat urinary bladder mucosa. Neurourol Urodyn 38: 2093–2103, 2019. doi: 10.1002/nau.24085. [DOI] [PubMed] [Google Scholar]
- 3. Fry CH, Kitney DG, Paniker J, Drake MJ, Kanai A, Andersson KE. Fibrosis and the bladder, implications for function ICI-RS 2017. Neurourol Urodyn 37: S7–S12, 2018. doi: 10.1002/nau.23725. [DOI] [PubMed] [Google Scholar]
- 4. Plikus MV, Wang X, Sinha S, Forte E, Thompson SM, Herzog EL, Driskell RR, Rosenthal N, Biernaskie J, Horsley V. Fibroblasts: origins, definitions, and functions in health and disease. Cell 184: 3852–3872, 2021. doi: 10.1016/j.cell.2021.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. LeBleu VS, Neilson EG. Origin and functional heterogeneity of fibroblasts. FASEB J 34: 3519–3536, 2020. doi: 10.1096/fj.201903188R. [DOI] [PubMed] [Google Scholar]
- 6. Louault K, Li RR, DeClerck YA. Cancer-associated fibroblasts: understanding their heterogeneity. Cancers (Basel) 12: 3108, 2020. doi: 10.3390/cancers12113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Desbois M, Wang Y. Cancer-associated fibroblasts: key players in shaping the tumor immune microenvironment. Immunol Rev 302: 241–258, 2021. [DOI] [PubMed] [Google Scholar]
- 8. Xie X, Liang J, Huang R, Luo C, Yang J, Xing H, Zhou L, Qiao H, Ergu E, Chen H. Molecular pathways underlying tissue injuries in the bladder with ketamine cystitis. FASEB J 35: e21703, 2021. [DOI] [PubMed] [Google Scholar]
- 9. Lilly MA, Kulkulka NA, Firmiss PR, Ross MJ, Flum AS, Santos GB, Bowen DK, Dettman RW, Gong EM. The murine bladder supports a population of stromal Sca-1+/CD34+/lin- mesenchymal stem cells. PLoS One 10: e0141437, 2015. doi: 10.1371/journal.pone.0141437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tassone NM, Li B, Patel MS, Devine MY, Firmiss PR, Gould AD, Kochan KS, Stubbee RA, Bowen DK, Dettman RW, Gong EM. Stem cell antigen/Ly6a protects against bladder fibrosis in mice. Am J Physiol Renal Physiol 317: F1503–F1512, 2019. doi: 10.1152/ajprenal.00160.2019. [DOI] [PubMed] [Google Scholar]
- 11. Tanaka ST, Martinez-Ferrer M, Makari JH, Wills ML, Thomas JC, Adams MC, Brock JW, 3rd, Pope JCt, Bhowmick NA. Recruitment of bone marrow derived cells to the bladder after bladder outlet obstruction. J Urol 182: 1769–1774, 2009. doi: 10.1016/j.juro.2009.02.081. [DOI] [PubMed] [Google Scholar]
- 12. Ikeda Y, Zabbarova IV, Birder LA, Wipf P, Getchell SE, Tyagi P, Fry CH, Drake MJ, Kanai AJ. Relaxin-2 therapy reverses radiation-induced fibrosis and restores bladder function in mice. Neurourol Urodyn 37: 2441–2451, 2018. doi: 10.1002/nau.23721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zwaans BMM, Wegner KA, Bartolone SN, Vezina CM, Chancellor MB, Lamb LE. Radiation cystitis modeling: a comparative study of bladder fibrosis radio-sensitivity in C57BL/6, C3H, and BALB/c mice. Physiol Rep 8: e14377, 2020. doi: 10.14814/phy2.14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zwaans BM, Chancellor MB, Lamb LE. Modeling and treatment of radiation cystitis. Urology 88: 14–21, 2016. doi: 10.1016/j.urology.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 15. Zwaans BMM, Grobbel M, Carabulea AL, Lamb LE, Roccabianca S. Increased extracellular matrix stiffness accompanies compromised bladder function in a murine model of radiation cystitis. Acta Biomater 144: 221–229, 2022. doi: 10.1016/j.actbio.2022.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nagatomi J, Gloeckner DC, Chancellor MB, DeGroat WC, Sacks MS. Changes in the biaxial viscoelastic response of the urinary bladder following spinal cord injury. Ann Biomed Eng 32: 1409–1419, 2004. doi: 10.1114/B:ABME.0000042228.89106.48. [DOI] [PubMed] [Google Scholar]
- 17. Kim KM, Kogan BA, Massad CA, Huang YC. Collagen and elastin in the obstructed fetal bladder. J Urol 146: 528–531, 1991. doi: 10.1016/s0022-5347(17)37844-8. [DOI] [PubMed] [Google Scholar]
- 18. Zhang Z, Liang Z, Li D, Wang L, Chen Y, Liang Y, Jiao W, Niu H. Development of a CAFs-related gene signature to predict survival and drug response in bladder cancer. Hum Cell 35: 649–664, 2022. doi: 10.1007/s13577-022-00673-w. [DOI] [PubMed] [Google Scholar]
- 19. Yu Z, Liao J, Chen Y, Zou C, Zhang H, Cheng J, Liu D, Li T, Zhang Q, Li J, Yang X, Ye Y, Huang Z, Long X, Yang R, Mo Z. Single-cell transcriptomic map of the human and mouse bladders. J Am Soc Nephrol 30: 2159–2176, 2019. doi: 10.1681/ASN.2019040335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muhl L, Genove G, Leptidis S, Liu J, He L, Mocci G, Sun Y, Gustafsson S, Buyandelger B, Chivukula IV, Segerstolpe A, Raschperger E, Hansson EM, Bjorkegren JLM, Peng XR, Vanlandewijck M, Lendahl U, Betsholtz C. Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat Commun 11: 3953, 2020. [Erratum in Nat Commun 11: 4493, 2020]. doi: 10.1038/s41467-020-17740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabula Muris Consortium; Overall coordination; Logistical coordination; Organ collection and processing; Library preparation and sequencing; Computational data analysis; Cell type annotation; Writing group; Supplemental text writing group; Principal investigators. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562: 367–372, 2018. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheng X, Lai H, Luo W, Zhang M, Miao J, Song W, Xing S, Wang J, Gao WQ. Single-cell analysis reveals urothelial cell heterogeneity and regenerative cues following cyclophosphamide-induced bladder injury. Cell Death Dis 12: 446, 2021. doi: 10.1038/s41419-021-03740-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buechler MB, Pradhan RN, Krishnamurty AT, Cox C, Calviello AK, Wang AW, Yang YA, Tam L, Caothien R, Roose-Girma M, Modrusan Z, Arron JR, Bourgon R, Muller S, Turley SJ. Cross-tissue organization of the fibroblast lineage. Nature 593: 575–579, 2021. doi: 10.1038/s41586-021-03549-5. [DOI] [PubMed] [Google Scholar]
- 24. Koh SD, Lee H, Ward SM, Sanders KM. The mystery of the interstitial cells in the urinary bladder. Annu Rev Pharmacol Toxicol 58: 603–623, 2018. doi: 10.1146/annurev-pharmtox-010617-052615. [DOI] [PubMed] [Google Scholar]
- 25. McCloskey KD. Bladder interstitial cells: an updated review of current knowledge. Acta Physiol (Oxf) 207: 7–15, 2013. doi: 10.1111/apha.12009. [DOI] [PubMed] [Google Scholar]
- 26. Fry CH, Sui GP, Kanai AJ, Wu C. The function of suburothelial myofibroblasts in the bladder. Neurourol Urodyn 26: 914–919, 2007. doi: 10.1002/nau.20483. [DOI] [PubMed] [Google Scholar]
- 27. Smet PJ, Jonavicius J, Marshall VR, de Vente J. Distribution of nitric oxide synthase-immunoreactive nerves and identification of the cellular targets of nitric oxide in guinea-pig and human urinary bladder by cGMP immunohistochemistry. Neuroscience 71: 337–348, 1996. doi: 10.1016/0306-4522(95)00453-x. [DOI] [PubMed] [Google Scholar]
- 28. Rumessen JJ, Vanderwinden JM. Interstitial cells in the musculature of the gastrointestinal tract: Cajal and beyond. Int Rev Cytol 229: 115–208, 2003. doi: 10.1016/s0074-7696(03)29004-5. [DOI] [PubMed] [Google Scholar]
- 29. Blair PJ, Rhee PL, Sanders KM, Ward SM. The significance of interstitial cells in neurogastroenterology. J Neurogastroenterol Motil 20: 294–317, 2014. doi: 10.5056/jnm14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Foong D, Zhou J, Zarrouk A, Ho V, O'Connor MD. Understanding the biology of human interstitial cells of Cajal in gastrointestinal motility. Int J Mol Sci 21: 4540, 2020. doi: 10.3390/ijms21124540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnston L, Woolsey S, Cunningham RM, O'Kane H, Duggan B, Keane P, McCloskey KD. Morphological expression of KIT positive interstitial cells of Cajal in human bladder. J Urol 184: 370–377, 2010. doi: 10.1016/j.juro.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCloskey KD, Gurney AM. Kit positive cells in the guinea pig bladder. J Urol 168: 832–836, 2002. [PubMed] [Google Scholar]
- 33. Shafik A, El-Sibai O, Shafik AA, Shafik I. Identification of interstitial cells of Cajal in human urinary bladder: concept of vesical pacemaker. Urology 64: 809–813, 2004. doi: 10.1016/j.urology.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 34. McCloskey KD, Anderson UA, Davidson RA, Bayguinov YR, Sanders KM, Ward SM. Comparison of mechanical and electrical activity and interstitial cells of Cajal in urinary bladders from wild-type and W/Wv mice. Br J Pharmacol 156: 273–283, 2009. doi: 10.1111/j.1476-5381.2008.00006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hashitani H, Yanai Y, Suzuki H. Role of interstitial cells and gap junctions in the transmission of spontaneous Ca2+ signals in detrusor smooth muscles of the guinea-pig urinary bladder. J Physiol 559: 567–581, 2004. doi: 10.1113/jphysiol.2004.065136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Davidson RA, McCloskey KD. Morphology and localization of interstitial cells in the guinea pig bladder: structural relationships with smooth muscle and neurons. J Urol 173: 1385–1390, 2005. doi: 10.1097/01.ju.0000146272.80848.37. [DOI] [PubMed] [Google Scholar]
- 37. Piaseczna Piotrowska A, Rolle U, Solari V, Puri P. Interstitial cells of Cajal in the human normal urinary bladder and in the bladder of patients with megacystis-microcolon intestinal hypoperistalsis syndrome. BJU Int 94: 143–146, 2004. doi: 10.1111/j.1464-410X.2004.04914.x. [DOI] [PubMed] [Google Scholar]
- 38. Gevaert T, Ridder D, Vanstreels E, Daelemans D, Everaerts W, Aa FV, Pintelon I, Timmermans JP, Roskams T, Steiner C, Neuhaus J. The stem cell growth factor receptor KIT is not expressed on interstitial cells in bladder. J Cell Mol Med 21: 1206–1216, 2017. doi: 10.1111/jcmm.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koh BH, Roy R, Hollywood MA, Thornbury KD, McHale NG, Sergeant GP, Hatton WJ, Ward SM, Sanders KM, Koh SD. Platelet-derived growth factor receptor-alpha cells in mouse urinary bladder: a new class of interstitial cells. J Cell Mol Med 16: 691–700, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lagou M, De Vente J, Kirkwood TB, Hedlund P, Andersson KE, Gillespie JI, Drake MJ. Location of interstitial cells and neurotransmitters in the mouse bladder. BJU Int 97: 1332–1337, 2006. doi: 10.1111/j.1464-410X.2006.06203.x. [DOI] [PubMed] [Google Scholar]
- 41. Rasmussen H, Hansen A, Smedts F, Rumessen JJ, Horn T. CD34-positive interstitial cells of the human detrusor. APMIS 115: 1260–1266, 2007. doi: 10.1111/j.1600-0643.2007.00759.x. [DOI] [PubMed] [Google Scholar]
- 42. Rasmussen H, Rumessen JJ, Hansen A, Smedts F, Horn T. Ultrastructure of Cajal-like interstitial cells in the human detrusor. Cell Tissue Res 335: 517–527, 2009. doi: 10.1007/s00441-008-0736-z. [DOI] [PubMed] [Google Scholar]
- 43. Gevaert T, Vanstreels E, Daelemans D, Franken J, Van Der Aa F, Roskams T, De Ridder D. Identification of different phenotypes of interstitial cells in the upper and deep lamina propria of the human bladder dome. J Urol 192: 1555–1563, 2014. doi: 10.1016/j.juro.2014.05.096. [DOI] [PubMed] [Google Scholar]
- 44. Pezzone MA, Watkins SC, Alber SM, King WE, de Groat WC, Chancellor MB, Fraser MO. Identification of c-kit-positive cells in the mouse ureter: the interstitial cells of Cajal of the urinary tract. Am J Physiol Renal Physiol 284: F925–F929, 2003. doi: 10.1152/ajprenal.00138.2002. [DOI] [PubMed] [Google Scholar]
- 45. Monaghan KP, Johnston L, McCloskey KD. Identification of PDGFRα positive populations of interstitial cells in human and guinea pig bladders. J Urol 188: 639–647, 2012. doi: 10.1016/j.juro.2012.03.117. [DOI] [PubMed] [Google Scholar]
- 46. Yu W, Zeidel ML, Hill WG. Cellular expression profile for interstitial cells of Cajal in bladder—a cell often misidentified as myocyte or myofibroblast. PLoS One 7: e48897, 2012. doi: 10.1371/journal.pone.0048897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kuijpers KA, Heesakkers JP, Hafmans TG, Schalken JA. An update of the interstitial cell compartment in the normal human bladder. BioMed Res Int 2014: 464217, 2014. doi: 10.1155/2014/464217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sancho M, Triguero D, Lafuente-Sanchis A, Garcia-Pascual A. Proliferation of interstitial cells in the cyclophosphamide-induced cystitis and the preventive effect of imatinib. BioMed Res Int 2017: 3457093, 2017. doi: 10.1155/2017/3457093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Steiner C, Gevaert T, Ganzer R, De Ridder D, Neuhaus J. Comparative immunohistochemical characterization of interstitial cells in the urinary bladder of human, guinea pig and pig. Histochem Cell Biol 149: 491–501, 2018. doi: 10.1007/s00418-018-1655-z. [DOI] [PubMed] [Google Scholar]
- 50. Ohtani O. The maceration technique in scanning electron microscopy of collagen fiber frameworks: its application in the study of human livers. Arch Histol Cytol 55: 225–232, 1992. [DOI] [PubMed] [Google Scholar]
- 51. Stephenson MK, Lenihan S, Covarrubias R, Huttinger RM, Gumina RJ, Sawyer DB, Galindo CL. Scanning electron microscopy of macerated tissue to visualize the extracellular matrix. J Vis Exp 54005, 2016. doi: 10.3791/54005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Murakumo M, Ushiki T, Abe K, Matsumura K, Shinno Y, Koyanagi T. Three-dimensional arrangement of collagen and elastin fibers in the human urinary bladder: a scanning electron microscopic study. J Urol 154: 251–256, 1995. [PubMed] [Google Scholar]
- 53. Svitkina T. Imaging cytoskeleton components by electron microscopy. Methods Mol Biol 2364: 25–52, 2022. doi: 10.1007/978-1-0716-1661-1_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dalghi MG, Ruiz WG, Clayton DR, Montalbetti N, Daugherty SL, Beckel JM, Carattino MD, Apodaca G. Functional roles for PIEZO1 and PIEZO2 in urothelial mechanotransduction and lower urinary tract interoception. JCI Insight 6, 2021. doi: 10.1172/jci.insight.152984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Truschel ST, Clayton DR, Beckel JM, Yabes JG, Yao Y, Wolf-Johnston A, Birder LA, Apodaca G. Age-related endolysosome dysfunction in the rat urothelium. PLoS One 13: e0198817, 2018. doi: 10.1371/journal.pone.0198817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Winfree S, Khan S, Micanovic R, Eadon MT, Kelly KJ, Sutton TA, Phillips CL, Dunn KW, El-Achkar TM. Quantitative three-dimensional tissue cytometry to study kidney tissue and resident immune cells. J Am Soc Nephrol 28: 2108–2118, 2017. doi: 10.1681/ASN.2016091027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mienaltowski MJ, Gonzales NL, Beall JM, Pechanec MY. Basic structure, physiology, and biochemistry of connective tissues and extracellular matrix collagens. Adv Exp Med Biol 1348: 5–43, 2021. doi: 10.1007/978-3-030-80614-9_2. [DOI] [PubMed] [Google Scholar]
- 58. Rusu MC, Folescu R, Mănoiu VS, Didilescu AC. Suburothelial interstitial cells. Cells Tissues Organs 199: 59–72, 2014. doi: 10.1159/000360816. [DOI] [PubMed] [Google Scholar]
- 59. Neuhaus J, Schroppel B, Dass M, Zimmermann H, Wolburg H, Fallier-Becker P, Gevaert T, Burkhardt CJ, Do HM, Stolzenburg JU. 3D-electron microscopic characterization of interstitial cells in the human bladder upper lamina propria. Neurourol Urodyn 37: 89–98, 2018. doi: 10.1002/nau.23270. [DOI] [PubMed] [Google Scholar]
- 60. Gevaert T, De Vos R, Van Der Aa F, Joniau S, van den Oord J, Roskams T, De Ridder D. Identification of telocytes in the upper lamina propria of the human urinary tract. J Cell Mol Med 16: 2085–2093, 2012. doi: 10.1111/j.1582-4934.2011.01504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Drake MJ, Hedlund P, Andersson KE, Brading AF, Hussain I, Fowler C, Landon DN. Morphology, phenotype and ultrastructure of fibroblastic cells from normal and neuropathic human detrusor: absence of myofibroblast characteristics. J Urol 169: 1573–1576, 2003. doi: 10.1097/01.ju.0000054928.34777.37. [DOI] [PubMed] [Google Scholar]
- 62. Eyden B. The myofibroblast: a study of normal, reactive and neoplastic tissues, with an emphasis on ultrastructure. Part 1—normal and reactive cells. J Submicrosc Cytol Pathol 37: 109–204, 2005. [PubMed] [Google Scholar]
- 63. Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol 23: 4013–4025, 2003. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sui GP, Rothery S, Dupont E, Fry CH, Severs NJ. Gap junctions and connexin expression in human suburothelial interstitial cells. BJU Int 90: 118–129, 2002. doi: 10.1046/j.1464-410x.2002.02834.x. [DOI] [PubMed] [Google Scholar]
- 65. Grol S, Essers PB, van Koeveringe GA, Martinez-Martinez P, de Vente J, Gillespie JI. M(3) muscarinic receptor expression on suburothelial interstitial cells. BJU Int 104: 398–405, 2009. doi: 10.1111/j.1464-410X.2009.08423.x. [DOI] [PubMed] [Google Scholar]
- 66. Florin L, Alter H, Grone HJ, Szabowski A, Schutz G, Angel P. Cre recombinase-mediated gene targeting of mesenchymal cells. Genesis 38: 139–144, 2004. doi: 10.1002/gene.20004. [DOI] [PubMed] [Google Scholar]
- 67. Upadhyay J, Aitken KJ, Damdar C, Bolduc S, Bagli DJ. Integrins expressed with bladder extracellular matrix after stretch injury in vivo mediate bladder smooth muscle cell growth in vitro. J Urol 169: 750–755, 2003. doi: 10.1097/01.ju.0000051682.61041.a5. [DOI] [PubMed] [Google Scholar]
- 68. Torihashi S, Ward SM, Nishikawa S, Nishi K, Kobayashi S, Sanders KM. c-kit-dependent development of interstitial cells and electrical activity in the murine gastrointestinal tract. Cell Tissue Res 280: 97–111, 1995. doi: 10.1007/BF00304515. [DOI] [PubMed] [Google Scholar]
- 69. Faussone-Pellegrini MS, Thuneberg L. Guide to the identification of interstitial cells of Cajal. Microsc Res Tech 47: 248–266, 1999. doi:. [DOI] [PubMed] [Google Scholar]
- 70. Baker SA, Hennig GW, Salter AK, Kurahashi M, Ward SM, Sanders KM. Distribution and Ca(2+) signalling of fibroblast-like (PDGFR(+)) cells in the murine gastric fundus. J Physiol 591: 6193–6208, 2013. doi: 10.1113/jphysiol.2013.264747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Iino S, Horiguchi K, Horiguchi S, Nojyo Y. c-Kit-negative fibroblast-like cells express platelet-derived growth factor receptor alpha in the murine gastrointestinal musculature. Histochem Cell Biol 131: 691–702, 2009. doi: 10.1007/s00418-009-0580-6. [DOI] [PubMed] [Google Scholar]
- 72. Iino S, Nojyo Y. Immunohistochemical demonstration of c-Kit-negative fibroblast-like cells in murine gastrointestinal musculature. Arch Histol Cytol 72: 107–115, 2009. doi: 10.1679/aohc.72.107. [DOI] [PubMed] [Google Scholar]
- 73. Kurahashi M, Zheng H, Dwyer L, Ward SM, Koh SD, Sanders KM. A functional role for the 'fibroblast-like cells' in gastrointestinal smooth muscles. J Physiol 589: 697–710, 2011. doi: 10.1113/jphysiol.2010.201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rothenberg ME, Nusse Y, Kalisky T, Lee JJ, Dalerba P, Scheeren F, Lobo N, Kulkarni S, Sim S, Qian D, Beachy PA, Pasricha PJ, Quake SR, Clarke MF. Identification of a cKit(+) colonic crypt base secretory cell that supports Lgr5(+) stem cells in mice. Gastroenterology 142: 1195–1205.e1196, 2012. doi: 10.1053/j.gastro.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Eaton AF, Clayton DR, Ruiz WG, Griffiths SE, Rubio ME, Apodaca G. Expansion and contraction of the umbrella cell apical junctional ring in response to bladder filling and voiding. Mol Biol Cell 30: 2037–2052, 2019. doi: 10.1091/mbc.E19-02-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gillespie JI, Markerink-van Ittersum M, de Vente J. cGMP-generating cells in the bladder wall: identification of distinct networks of interstitial cells. BJU Int 94: 1114–1124, 2004. doi: 10.1111/j.1464-410X.2004.05186.x. [DOI] [PubMed] [Google Scholar]
- 77. Okada S, Kojima Y, Kubota Y, Mizuno K, Sasaki S, Kohri K. Attenuation of bladder overactivity in KIT mutant rats. BJU Int 108: E97–F103, 2011. doi: 10.1111/j.1464-410X.2010.09870.x. [DOI] [PubMed] [Google Scholar]
- 78. Komuro T. Comparative morphology of interstitial cells of Cajal: ultrastructural characterization. Microsc Res Tech 47: 267–285, 1999. doi:. [DOI] [PubMed] [Google Scholar]
- 79. Lee H, Koh BH, Peri LE, Corrigan RD, Lee HT, George NE, Bhetwal BP, Xie Y, Perrino BA, Chai TC, Sanders KM, Koh SD. Premature contractions of the bladder are suppressed by interactions between TRPV4 and SK3 channels in murine detrusor PDGFRα(+). Sci Rep 7: 12245, 2017. doi: 10.1038/s41598-017-12561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lee H, Koh BH, Peri LE, Sanders KM, Koh SD. Functional expression of SK channels in murine detrusor PDGFR+ cells. J Physiol 591: 503–513, 2013. doi: 10.1113/jphysiol.2012.241505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lee H, Koh BH, Peri LE, Sanders KM, Koh SD. Purinergic inhibitory regulation of murine detrusor muscles mediated by PDGFRα+ interstitial cells. J Physiol 592: 1283–1293, 2014. doi: 10.1113/jphysiol.2013.267989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Eyden B. The myofibroblast: a study of normal, reactive and neoplastic tissues, with an emphasis on ultrastructure. part 2—tumours and tumour-like lesions. J Submicrosc Cytol Pathol 37: 231–296, 2005. [PubMed] [Google Scholar]
- 83. Drake MJ, Fry CH, Eyden B. Structural characterization of myofibroblasts in the bladder. BJU Int 97: 29–32, 2006. doi: 10.1111/j.1464-410X.2006.05818.x. [DOI] [PubMed] [Google Scholar]
- 84. Ikeda Y, Fry C, Hayashi F, Stolz D, Griffiths D, Kanai A. Role of gap junctions in spontaneous activity of the rat bladder. Am J Physiol Renal Physiol 293: F1018–F1025, 2007. doi: 10.1152/ajprenal.00183.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yu W, Robson SC, Hill WG. Expression and distribution of ectonucleotidases in mouse urinary bladder. PLoS One 6: e18704, 2011. doi: 10.1371/journal.pone.0018704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lee H, Koh BH, Yamasaki E, George NE, Sanders KM, Koh SD. UTP activates small-conductance Ca2+-activated K+ channels in murine detrusor PDGFRα+ cells. Am J Physiol Renal Physiol 309: F569–F574, 2015. doi: 10.1152/ajprenal.00156.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sui GP, Wu C, Roosen A, Ikeda Y, Kanai AJ, Fry CH. Modulation of bladder myofibroblast activity: implications for bladder function. Am J Physiol Renal Physiol 295: F688–F697, 2008. doi: 10.1152/ajprenal.00133.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wu C, Sui GP, Fry CH. Purinergic regulation of guinea pig suburothelial myofibroblasts. J Physiol 559: 231–243, 2004. doi: 10.1113/jphysiol.2004.067934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rahnama'i MS, Biallosterski BT, de Wachter SG, Van Kerrebroeck PE, van Koeveringe GA. The distribution of the prostaglandin E receptor type 2 (EP2) in the detrusor of the guinea pig. Prostaglandins Other Lipid Mediat 99: 107–115, 2012. doi: 10.1016/j.prostaglandins.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 90. Rahnama'i MS, de Wachter SG, van Koeveringe GA, van Kerrebroeck PE, de Vente J, Gillespie JI. The relationship between prostaglandin E receptor 1 and cyclooxygenase I expression in guinea pig bladder interstitial cells: proposition of a signal propagation system. J Urol 185: 315–322, 2011. [DOI] [PubMed] [Google Scholar]
- 91. Shin K, Lee J, Guo N, Kim J, Lim A, Qu L, Mysorekar IU, Beachy PA. Hedgehog/Wnt feedback supports regenerative proliferation of epithelial stem cells in bladder. Nature 472: 110–114, 2011. doi: 10.1038/nature09851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mysorekar IU, Isaacson-Schmid M, Walker JN, Mills JC, Hultgren SJ. Bone morphogenetic protein 4 signaling regulates epithelial renewal in the urinary tract in response to uropathogenic infection. Cell Host Microbe 5: 463–475, 2009. doi: 10.1016/j.chom.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Asgari M, Latifi N, Heris HK, Vali H, Mongeau L. In vitro fibrillogenesis of tropocollagen type III in collagen type I affects its relative fibrillar topology and mechanics. Sci Rep 7: 1392, 2017. doi: 10.1038/s41598-017-01476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ricard-Blum S, Baffet G, Theret N. Molecular and tissue alterations of collagens in fibrosis. Matrix Biol 68-69: 122–149, 2018. doi: 10.1016/j.matbio.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 95. Danielson KG, Baribault H, Holmes DF, Graham H, Kadler KE, Iozzo RV. Targeted disruption of decorin leads to abnormal collagen fibril morphology and skin fragility. J Cell Biol 136: 729–743, 1997. doi: 10.1083/jcb.136.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hornsby J, Daly DM, Grundy D, Cheng F, Robertson AM, Watton PN, Thompson MS. Quantitative multiphoton microscopy of murine urinary bladder morphology during in situ uniaxial loading. Acta Biomater 64: 59–66, 2017. doi: 10.1016/j.actbio.2017.09.029. [DOI] [PubMed] [Google Scholar]
- 97. Wang J, Zohar R, McCulloch CA. Multiple roles of alpha-smooth muscle actin in mechanotransduction. Exp Cell Res 312: 205–214, 2006. doi: 10.1016/j.yexcr.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 98. Midwood KS, Chiquet M, Tucker RP, Orend G. Tenascin-C at a glance. J Cell Sci 129: 4321–4327, 2016. doi: 10.1242/jcs.190546. [DOI] [PubMed] [Google Scholar]
- 99. Purslow PP. The structure and role of intramuscular connective tissue in muscle function. Front Physiol 11: 495, 2020. doi: 10.3389/fphys.2020.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tschumperlin DJ. Matrix, mesenchyme, and mechanotransduction. Ann Am Thorac Soc 12 Suppl 1: S24–S29, 2015. doi: 10.1513/AnnalsATS.201407-320MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Stewart L, Turner NA. Channelling the force to reprogram the matrix: mechanosensitive ion channels in cardiac fibroblasts. Cells 10: 990, 2021. doi: 10.3390/cells10050990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hinz B. Matrix mechanics and regulation of the fibroblast phenotype. Periodontol 2000 63: 14–28, 2013. doi: 10.1111/prd.12030. [DOI] [PubMed] [Google Scholar]
- 103. Wang Y, Fang Q, Lu Y, Song B, Li W, Li L. Effects of mechanical stretch on interstitial cells of Cajal in guinea pig bladder. J Surg Res 164: e213–219, 2010. doi: 10.1016/j.jss.2010.04.040. [DOI] [PubMed] [Google Scholar]
- 104. Liu Q, Sun B, Zhao J, Wang Q, An F, Hu X, Yang Z, Xu J, Tan M, Li L. Increased Piezo1 channel activity in interstitial Cajal-like cells induces bladder hyperactivity by functionally interacting with NCX1 in rats with cyclophosphamide-induced cystitis. Exp Mol Med 50: 1–16, 2018. doi: 10.1038/s12276-018-0088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Dalghi MG, Clayton DR, Ruiz WG, Al-Bataineh MM, Satlin LM, Kleyman TR, Ricke WA, Carattino MD, Apodaca G. Expression and distribution of PIEZO1 in the mouse urinary tract. Am J Physiol Renal Physiol 317: F303–F321, 2019. doi: 10.1152/ajprenal.00214.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Pakshir P, Hinz B. The big five in fibrosis: macrophages, myofibroblasts, matrix, mechanics, and miscommunication. Matrix Biol 68-69: 81–93, 2018. doi: 10.1016/j.matbio.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 107. Shaw TJ, Rognoni E. Dissecting fibroblast heterogeneity in health and fibrotic disease. Curr Rheumatol Rep 22: 33, 2020. doi: 10.1007/s11926-020-00903-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kim SJ, Kim J, Na YG, Kim KH. Irreversible bladder remodeling induced by fibrosis. Int Neurourol J 25: S3–S7, 2021. doi: 10.5213/inj.2142174.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Czekay RP, Cheon DJ, Samarakoon R, Kutz SM, Higgins PJ. Cancer-associated fibroblasts: mechanisms of tumor progression and novel therapeutic targets. Cancers (Basel) 14: 1231, 2022. doi: 10.3390/cancers14051231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Asif PJ, Longobardi C, Hahne M, Medema JP. The role of cancer-associated fibroblasts in cancer invasion and metastasis. Cancers (Basel) 13: 4720, 2021. doi: 10.3390/cancers13184720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sanches BDA, Tamarindo GH, Maldarine JDS, Da Silva ADT, Dos Santos VA, Goes RM, Taboga SR, Carvalho HF. Telocytes of the male urogenital system: interrelationships, possible functions, and pathological implications. Cell Biol Int 45: 1613–1623, 2021. doi: 10.1002/cbin.11612. [DOI] [PubMed] [Google Scholar]
- 112. Diaz-Flores L, Gutierrez R, Garcia MP, Gonzalez-Gomez M, Rodriguez-Rodriguez R, Hernandez-Leon N, Diaz-Flores L Jr, Carrasco JL. Cd34+ stromal cells/telocytes in normal and pathological skin. Int J Mol Sci 22: 7342, 2021. doi: 10.3390/ijms22147342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Diaz-Flores L, Gutierrez R, Garcia MP, Gayoso S, Gutierrez E, Diaz-Flores L Jr, Carrasco JL. Telocytes in the normal and pathological peripheral nervous system. Int J Mol Sci 21: 4320, 2020. doi: 10.3390/ijms21124320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Marini M, Rosa I, Ibba-Manneschi L, Manetti M. Telocytes in skeletal, cardiac and smooth muscle interstitium: morphological and functional aspects. Histol Histopathol 33: 1151–1165, 2018. doi: 10.14670/HH-11-994. [DOI] [PubMed] [Google Scholar]
- 115. Vannucchi MG. The telocytes: ten years after their introduction in the scientific literature. An update on their morphology, distribution, and potential roles in the gut. Int J Mol Sci 21: 4478, 2020. doi: 10.3390/ijms21124478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figs. S1–S7: https://doi.org/10.6084/m9.figshare.19719856.