Keywords: gut microbiome, intestinal inflammation, necrotizing enterocolitis
Abstract
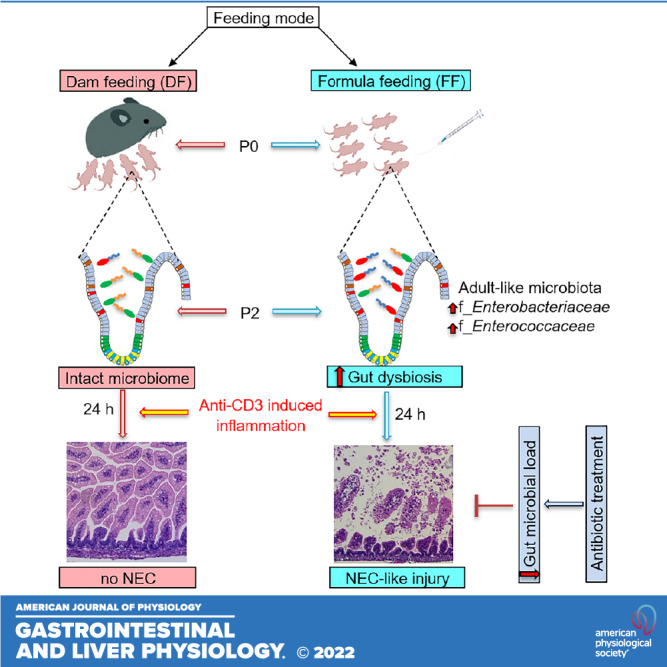
Feeding modes influence the gut microbiome, immune system, and intestinal barrier homeostasis in neonates; how feeding modes impact susceptibility to neonatal gastrointestinal (GI) diseases is still uncertain. Here, we investigated the impact of dam feeding (DF) and formula feeding (FF) on features of the gut microbiome and physiological inflammation during the first 2 days of postnatal development and on the susceptibility to intestinal injury related to the inflammatory state in neonatal mouse pups. 16S rRNA sequencing data revealed microbiome changes, lower α-diversity, and a distinct pattern of β-diversity including expansion of f_Enterobacteriaceae and f_Enterococcaceae in the ileum of FF pups compared with DF pups by postnatal day (P)2. Together with gut dysbiosis, the FF cohort also had greater ileal mucosa physiological inflammatory activity compared with DF pups by P2 but maintained normal histological features. Interestingly, FF but not DF mouse pups developed necrotizing enterocolitis (NEC)-like intestinal injury within 24 h after anti-CD3 mAb treatment, suggesting that FF influences the susceptibility to intestinal injury in neonates. We further found that NEC-like incidence in anti-CD3 mAb-treated FF neonatal pups was attenuated by antibiotic treatment. Collectively, our data suggest that FF predisposes mouse pups to anti-CD3 mAb-induced intestinal injury due to abnormal f_Enterobacteriaceae and f_Enterococcaceae colonization. These findings advance our understanding of FF-associated microbial colonization and intestinal inflammation, which may help inform the development of new therapeutic approaches to GI diseases like NEC in infants.
NEW & NOTEWORTHY This report shows that a feeding mode profoundly affects gut colonization in neonatal mice. Furthermore, our results demonstrate that formula feeding predisposes mouse pups to anti-CD3 mAb-induced necrotizing enterocolitis (NEC)-like intestinal injury upon inadequate microbial colonization. The study suggests the role of the combined presence of formula feeding-associated dysbiosis and mucosal inflammation in the pathogenesis of NEC and provides a new mouse model to study this disease.
INTRODUCTION
Although breastfeeding (BF) and formula feeding (FF) are common feeding modes for infants, the choice of feeding mode is an important modifiable risk factor for various inflammatory diseases in neonates. The most effective way for newborns to get nutrition is through BF, which also has additional health benefits; for example, BF supports health-promoting microbial communities and stimulates the maturation of the neonatal gut microbiome (1). It also facilitates immune system maturation and metabolic activities in neonates and can shorten the hospitalization of premature infants in the neonatal intensive care unit (2). The World Health Organization recommends at least 6 mo of exclusive BF, followed by continued BF for up to 1 yr or longer as complementary foods are initiated (2, 3). FF is an alternative feeding method for moms who cannot breastfeed or decide not to. In developed countries, FF has become more common than BF, which has a negative impact on infant health (4), yet there is mounting evidence that formula-fed infants exhibit an increased risk of gastroenteritis and diarrhea and have a higher risk of neonatal diseases compared with exclusively breastfed infants during the first 6 mo of life (1, 5). Among premature babies, not receiving breast milk is associated with an increased risk of necrotizing enterocolitis (NEC), a life-threatening intestinal disease (6, 7).
After birth, the neonatal gastrointestinal (GI) system undergoes colonization of microbiota, which promotes postnatal development of the immune system and improves intestinal barrier homeostasis. The gut microbiome and its barrier function play an important role in human health and disease (8). It is believed that gut colonization of microbiota usually begins at birth and that feeding mode (e.g., BF vs. FF) has a significant impact on shaping and remodeling the microbiome that colonizes the gut at birth (9–11). Previous studies have demonstrated that BF shapes the microbiome in breastfed infants with health-promoting bacteria linked to improved immune status (12). Human milk has anti-inflammatory and immunosuppressive properties that modulate neonatal immune function (13, 14). Taylor et al. (15) previously reported that the intestinal permeability of preterm infants is significantly decreased in those receiving human milk versus formula in a dose-dependent manner during the first postnatal month. In contrast, studies have shown that formula diets modify anti-inflammatory responses and induce inflammatory responses (4). Formula-fed infants display a distinct microbiome in the gut (16, 17), as well as exhibiting higher GI tract permeability compared with breastfed neonates, and are thus susceptible to infection and inflammation (18). Despite the observed differential effects of breast milk versus formula on GI development and function, how FF modulates susceptibility to GI diseases in neonates remains unknown.
NEC is a devastating GI disease that affects preterm infants. Evidence shows that dysregulated inflammation and abnormal bacterial colonization contribute to the NEC pathogenesis (19, 20). Using mouse models, we and others showed that NEC-like intestinal injury develops through an inflammatory event that involves TLR4 signaling pathways with infiltration and activation of inflammatory cells such as macrophages and lymphocytes (21, 22). Research has shown that anti-CD3 mAb treatment results in T cell-mediated small intestinal inflammation and leads to self-limited “cytokine release syndrome” of fever, enteritis, and diarrhea in adult mice, mimicking features of intestinal inflammatory diseases (23, 24). However, little is known about how neonatal mice respond to anti-CD3 challenge and pathological characteristics of anti-CD3 mAb-treated neonatal mouse small intestine. In the present study, we examined how feeding modes including dam feeding (DF) and FF affect gut microbiome characteristics and physiological inflammation during the first 2 days of postnatal development in neonatal mice. We further examined whether feeding mode affects the susceptibility of neonatal mice to anti-CD3 mAb-induced intestinal injury during the inflammatory state and whether the gut microbiome mediates this effect. Our study provides new insight into the role of feeding mode in characteristics of the neonatal gut microbiome and susceptibility to intestinal inflammation in infants.
MATERIALS AND METHODS
Animals
C57BL/6 wild-type (WT) mice (stock no. 000664) were purchased from Jackson Laboratory (Bar Harbor, ME). All adult mice were housed under a 12:12-h light-dark cycle with unlimited water and standard rodent chow in a specific pathogen-free environment. Mice aged 8–12 wk were used as breeders to produce pups for the FF and DF cohorts. All studies were conducted with the ethical approval of our Institutional Animal Care and Use Committee and following all relevant ethical regulations for animal testing and research.
Experimental Design
Within 4–6 h after birth [postnatal day (P)0], neonatal pups without identification of sex from multiple litters were processed for measuring body weight. P0 pups were excluded if they were born small (<1 g) and sick or had a small litter size (n ≤ 2 pups/litter). Then, they were randomly divided into appropriate experimental groups. We did not confirm the sex characteristics of pups at the end of the experiments. Therefore, sex variations between groups were unknown in our study. Pups assigned to dam feeding (DF) groups continued to be housed with the dam and nursed throughout the study period. Pups assigned to formula feeding (FF) groups were separated from the dams on P0 and cohoused in a 37°C humidified human neonatal incubator (Air-Shield Vickers Medical, Hatboro, PA). These pups were gavage fed with Esbilac formula (PetAg, Inc., Hampshire, IL) every 3 h (Esbilac, 200 mL/kg/day, prepared on a daily basis) for 72 h (25, 26). To account for somatic growth, feeds were started at 0.03 mL every 3 h beginning 4–6 h after birth and gradually increased to 0.06 mL per feeding by the third day of life. Pups were excluded if they experienced discomfort or were injured in the trachea while gavage feeding in this study.
To assess the impact of feeding mode on gut microbiota composition and physiological inflammatory mediators, ileum tissue samples from P0, P1, and P2 neonatal DF and FF pups were collected and subjected to 16S ribosomal RNA (rRNA) sequencing and real-time quantitative PCR (RT-qPCR), respectively.
Evidence shows that T cells play a role in NEC pathogenesis (22). It has been demonstrated that treatment with anti-CD3 mAb induces T-cell activation in mouse intestinal mucosa, which is associated with small intestinal inflammation in adult mice (23, 24). To evaluate the influence of feeding mode on anti-CD3 mAb-induced intestinal injury, the DF and FF neonatal pups on P2 were treated with 10 mg/kg anti-CD3ε mAb (145-2C11; BioLegend) or isotype IgG (controls) subcutaneously. The anti-CD3 mAb was diluted with endotoxin-free saline for preparation of antibody working solution at 1 mg/mL before administration to pups. After treatment, the pups were observed for clinical signs including severe abdominal distension, apnea, and lethargy, and the survival rate of the pups was noted. The pups were humanely euthanized by decapitation after 24 h at P3 or if they developed signs of distress, and the small intestine was collected for analysis. The tissues were not collected if the pups were noticed to have already died. The severity of the anti-CD3 mAb-induced intestinal injury was determined based on intestinal permeability, body weight change, histological features, expression of inflammatory mediators, and infiltration of inflammatory cells. Unlike classic rodent NEC models (21, 25–27), the pups in our model were not subjected to additional stressors such as commensal bacterial inoculation, asphyxia, hypothermia, and lipopolysaccharide administration.
To examine the significance of FF-mediated dysbiosis in anti-CD3 mAb-induced intestinal injury, Esbilac formula was prepared with/without streptomycin (STM, 1 mg/mL; Sigma) and gavage fed to pups every 3 h from P0 to P3 (30–60 µg/pup/feeding; streptomycin), as indicated above. These pups were treated with anti-CD3 mAb treatment on P2. In another experimental approach, DF pups were subjected to streptomycin every 3 h during the postnatal days, followed by anti-CD3 mAb treatment on P2. In these experiments, survival rate, body weight, intestinal permeability, and intestinal histology were all recorded.
Microbiome Analysis
Total DNA was isolated from ileum tissue samples of P0, P1, and P2 neonatal DF and FF pups with the ZymoBiomics DNA Microprep Kit (Zymo Research) and was used to amplify the target V3 and V4 hypervariable regions of 16S ribosomal RNA (rRNA) gene with the Quick-16S NGS Library Prep Kit (Zymo Research) according to the manufacturer’s manuals. The DNA fragments were sequenced with a 300-bp paired-end sequencing protocol on an Illumina MiSeq platform (Illumina, San Diego, CA) at the Research Resources Center, University of Illinois in Chicago (UIC). Bioinformatics analysis was performed at the UIC Research Informatics Core. Sequences were merged with PEAR (28) and then trimmed on a quality threshold of P = 0.01 (29). Chimeric sequences were removed with a comparison to Silva v132 reference sequence using the USEARCH algorithm (30, 31). Amplicon sequence variants were identified with DADA2 (32) and then annotated taxonomically using the Naive Bayesian classifier included in DADA2 with the Silva v132 training set. Operational taxonomic units (OTUs) were assigned, and their relative abundance was computed. The 16S rRNA gene sequencing data sets generated during the present study were uploaded to the BioProject database at the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA780308. Bacterial diversity was measured within samples (α-diversity) with Chao-1 and Shannon indexes and between samples (β-diversity) with principal coordinate analysis and permutational multivariate analysis of variance (PERMANOVA) of weighted and unweighted UniFrac distance matrixes. The DESeq2 procedure was used to calculate differentially abundant taxonomic units between DF and FF groups. Taxonomic units tested with a false discovery rate (FDR) < 0.1 were considered significant (33).
Intestinal Permeability
To evaluate gut barrier function, the pups in both DF and FF groups were fasted for 2 h at the end of the experiment as described in our previous publications (21, 25). The pups were administered fluorescein isothiocyanate (FITC)-dextran (400 mg/kg, FD-10S; Sigma) by gavage (19). Four hours later, the FITC fluorescence signal was measured with a Synergy HTX multimode reader (BioTek, Winooski, VT) in serum samples obtained after euthanasia of the pups.
Histological Examination
Small intestines were collected and fixed overnight in 10% buffered formalin and then embedded in paraffin. Tissue sections (4 μm) were stained by hematoxylin-eosin (H&E) for histological examination. H&E-stained slides were examined under a light microscope. Intestinal injury was graded in a blinded fashion on a 5-point scale using a histological scoring system modified from our previous publications (21): grade 0: no injury; grade 1: injury limited to the tip of the villi; grade 2: midvillous disruption and moderate lamina propria (LP) separation; grade 3: complete villous disruption and edema in submucosa; grade 4: transmural injury. For presentation, we acquired images with the Leica Thunder Microscope Imager System equipped with a DMC2900 Color Camera (Wetzlar, Germany) followed by processing and assembling with Adobe Photoshop 2021 (San Jose, CA).
Flow Cytometry
Flow cytometry (FACS) analysis was performed in the ileum tissue samples as described in Managlia et al. (21), with some modifications to characterize the infiltration of immune cells during the development of anti-CD3 mAb-induced intestinal injury. Briefly, the ileum samples were washed with phosphate-buffered saline (PBS) and cut longitudinally under a dissection microscope to carefully remove mesenteric vasculature and pancreatic tissues. The first digestion was performed in the tissue pieces with PBS containing 5 mmol/L EDTA, 15 mmol/L HEPES, 1 mmol/L dithiothreitol, and 10% fetal bovine serum (FBS) for 30 min at 37°C. After the supernatants were discarded, the tissues were washed in PBS and subjected to second digestion with Dulbecco’s modified Eagle’s medium (DMEM) containing 1 mg/mL collagenase VIII (Sigma), 15 mmol/L HEPES, 5 mmol/L CaCl2, and 2% FBS for 15 min at 37°C. After filtration (40 μm) and centrifugation, 1 × 106 cells in suspension were stained with viability dye FVD-eFluor-506 (Thermo Fisher Scientific) in PBS for 15 min on ice, followed by Fc block (Miltenyi Biotec, San Diego, CA) in staining buffer containing PBS and 2% FBS for 15 min on ice. Cells were incubated with the antibodies listed in Table 1 in staining buffer for 20 min on ice, followed by cell fixation using 2% paraformaldehyde. FACS data were acquired on a BD FACSymphony A5 Cell Analyzer (Indianapolis, IN) and analyzed with FlowJo version 10.7.1 (FlowJo LLC, Ashland, OR). Live CD45+ cells were characterized for further immune cell subtypes.
Table 1.
Antibodies used for flow cytometry
| Antigen | Fluorochrome | Concentration | Vendor | Catalog No. |
|---|---|---|---|---|
| CD8 | BB700 | 1:100 | BD | 566409 |
| CD3 | APC/Cy7 | 1:20 | BD | 557596 |
| CD4 | AF700 | 1:200 | BD | 557956 |
| CD64 | A647 | 1:20 | BD | 558539 |
| MHCII | PE/Cy7 | 1:800 | BioLegend/Thermo Fisher | 107630 |
| SigF | PE/CF594 | 1:20 | BD | 562757 |
| Ly6G | BV711 | 1:50 | BD | 563979 |
| CD45 | BV570 | 1:100 | BioLegend/Thermo Fisher | 103136 |
| Fixable viability dye | eFluor 506 | 1:1000 | eBioscience | 65086614 |
| Ly6C | BV421 | 1:200 | BioLegend/Thermo Fisher | 128032 |
| CD11b | BUV737 | 1:400 | BD | 612800 |
| CD19 | BUV563 | 1:50 | BD | 749028 |
| CD11c | BUV395 | 1:20 | BD | 564080 |
Note that the specificity of antibodies was verified based on information from datasheets provided by vendors.
RT-qPCR
Ileum tissue samples were collected, directly snap frozen, and stored at −80°C for RT-qPCR studies. Total RNA was extracted from the ileum samples with TRIzol reagent (Life Technologies) according to the manufacturer’s instructions. The purity and concentration of RNA were measured with a NanoDrop spectrophotometer (Agilent Technologies, Santa Clara, CA). Reverse transcription was performed with the iScript cDNA Synthesis Kit (Bio-Rad). RT-qPCR was performed with PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) according to the manufacturer’s manuals on the QuantStudio 6 real-time PCR system (Thermo Fisher Scientific). Primers were synthesized by Integrated DNA Technologies and are listed in Table 2. The relative quantification of target mRNA expression levels was calculated and normalized to the expression of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (Gapdh). The fold change in target gene expression levels between samples was calculated with the method, where CT is threshold cycle.
Table 2.
Primer sequences used for RT-qPCR
| Target Gene | Forward Sequence (5′–3′) | Reverse Sequence (5′–3′) |
|---|---|---|
| Tnf-α | CCACCACGCTCTTCTGTCTA | AGGGTCTGGGCCATAGAACT |
| Il1β | GAAATGCCACCTTTTGACAGTG | TGGATGCTCTCATCAGGACAG |
| Il2 | AGCAGCTGTTGATGGACCTA | CGCAGAGGTCCAAGTTCAT |
| Il6 | GTTCTCTGGGAAATCGTGGA | TTCTGCAAGTGCATCATCGT |
| Il10 | TGAATTCCCTGGGTGAGAAG | TGGCCTTGTAGACACCTTGG |
| Il12p40 | GGAAGCACGGCAGCAGAATAA | CTTGAGGGAGAAGTAGGAATG |
| Il15 | CATCCATCTCGTGCTACTTGTGTT | CATCTATCCAGTTGGCCTCTGT |
| Il17 | AAGGCAGCAGCGATCATCC | GGAACGGTTGAGGTAGTCTGAG |
| Il18 | GCCTCAAACCTTCCAAATCA | TGGATCCATTTCCTCAAAGG |
| Nos2 | ACATCGACCCGTCCACAGTAT | CAGAGGGGTAGGCTTGTCTC |
| Ifn-γ | TGCTGATGGGAGGAGATGTCT | TTTCTTTCAGGGACAGCCTGTT |
| Ccl2 | AGGTCCCTGTCATGCTTCTG | TCTGGACCCATTCCTTCTTG |
| Ccl3 | TGAAACCAGCAGCCTTTGCTC | AGGCATTCAGTTCCAGGTCAGTG |
| Ccl4 | AAACCTAACCCCGAGCAACA | CCATTGGTGCTGAGAACCCT |
| Ccl5 | ACACCACTCCCTGCTGCTTT | GACTGCAAGATTGGAGCACTTG |
| Ccl7 | AAGATCCCCAAGAGGAATCTCAAG | CAGACTTCCATGCCCTTCTTTG |
| Cxcl1 | GACCATGGCTGGGATTCACC | CCAAGGGAGCTTCAGGGTCA |
| Ccr2 | ACAGCTCAGGATTAACAGGGACTTG | ACCACTTGCATGCACACATGAC |
| Ccr5 | AGGCCATGCAGGCAACAG | TCTCTCCAACAAAGGCATAGATGA |
| Cxcr3 | CAGCCTGAACTTTGACAGAACCT | GCAGCCCCAGCAAGAAGA |
| Gapdh | AACTTTGGCATTGTGGAAGG | ACACATTGGGGGTAGGAACA |
Statistical Analysis
All experiments were performed at least twice with duplicate samples. All data are shown as means ± standard deviation (SD). Statistical analysis was performed with GraphPad Prism 8 (GraphPad Software, San Diego, CA). The independent Student’s t test and one-way or two-way analysis of variance (ANOVA) followed by Tukey posttest were used to analyze the statistical significance of the group differences. χ2 Analysis was used to evaluate differences in the incidence of NEC (grade ≥ 2). The significance level was set at P < 0.05, P < 0.01, P < 0.001, or P < 0.0001.
Statement about How Antibodies Were Validated for Specificity
All antibodies used in this work were purchased from reliable biotech companies as stated above. We chose to use these commercially available antibodies based on previous publications of other investigators. The companies provided datasheets regarding species reactivity of antibodies. However, our laboratory did not perform additional experiments to further verify the specificity of these antibodies.
RESULTS
DF and FF Mouse Pups Differ in Microbial Composition in the Ileum within 48 h after Birth
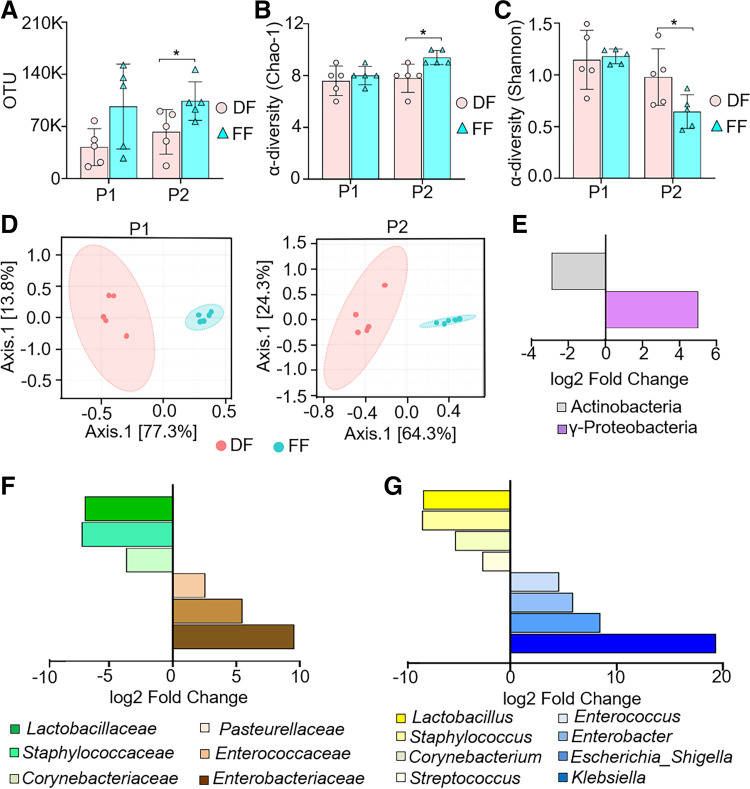
To assess the impact of feeding mode on bacterial communities of the ileum in the early postnatal period, we sequenced V3 and V4 amplicons of 16S rRNA genes in mouse ileum tissue samples at birth [postnatal day (P)0], P1, and P2. After trimming, assembly, and quality filtering, we obtained a total of 4,712,964 reads in all 25 sequenced samples, with an average of 188,518 reads per sample. The total number of operational taxonomic units (OTUs) was 309,537 at >99% similarity level. Rarefaction analysis with Faith’s phylogenetic diversity index was used to validate adequate sequencing capture of microbiota gene diversity for both DF and FF cohorts (Supplemental Fig. S1; see https://doi.org/10.6084/m9.figshare.17077328) First, we found that the total number of OTUs was significantly higher in the ileum of FF pups compared with DF pups on P2 (Fig. 1A). Next, we evaluated α-diversity based on the Chao-1 and Shannon indexes. The Chao-1 index revealed that the number of bacterial species in the ileum of DF pups did not change during the first two postnatal days (Supplemental Fig. S2A; see https://doi.org/10.6084/m9.figshare.19658412). In contrast, the Chao-1 index of FF pups showed a significant increase in the number of bacterial species in the ileum compared with the DF group on P2 (Fig. 1B). We also noted that FF pups on P2 showed an increase in Chao-1 index compared with FF pups on P1 (Supplemental Fig. S2A). The Shannon index (for variety and equitability of OTUs) revealed that pups in both feeding cohorts exhibited a marked decrease in the abundance and evenness of the gut microbiome in a time-dependent manner during the first two postnatal days (Supplemental Fig. S2B). Interestingly, we found a significant decrease in the Shannon index of FF pups compared with DF pups on P2 (Fig. 1C). The unweighted UniFrac β-diversity index showed distinct patterns of bacterial diversity in FF and DF neonatal pups respective to their postnatal ages in P1 and P2 cohorts (Fig. 1D). Together, our data suggest that FF causes adverse feeding effects including higher total number of bacterial species and lower α-diversity with distinct bacterial colonization compared with the DF cohort, which appears to play a role in NEC pathogenesis (34, 35).
Figure 1.
Formula feeding (FF) induces microbiome changes and dysbiosis of f_Enterobacteriaceae and f_Enterococcaceae in the ileum of mouse pups. Neonatal mice [postnatal day (P)0] from multiple litters were subjected to dam feeding (DF) or FF for 2 days. Ileum tissue samples were harvested on P0, P1, and P2 and processed for 16S rRNA gene sequencing followed by bioinformatics analysis. Ileum samples from 2 independent experiments were used for 16S sequencing in each group. A: total number of operational taxonomic units (OTUs) between DF and FF pups on the indicated postnatal days. B and C: Chao-1 index (B) and Shannon index (C) were used to measure the bacterial α-diversity. A–C: statistical significance was calculated by Student’s t test between DF and FF groups on the indicated postnatal days. Data are presented as means ± SD; n = 5 in each group. *P < 0.05 was considered statistically significant. D: β-diversity of the gut microbiota was measured with principal coordinate analysis (PCA) based on unweighted UniFrac distance matrixes between DF (orange) and FF (green) groups on the indicated postnatal days; n = 5 in each group. E–G: fold change (FC log2) in the relative abundance of significantly different (DESeq2) OTUs between groups and their normalized mean counts. A subset of OTUs, grouped by class, family, or genus, with a raw count at least of 100 was kept. The fold changes in microbial abundance at class (E), family (F), and genus (G) levels were calculated between DF and FF pups on P2; n = 5 in each group. Taxonomic units tested with P < 0.05 and FDR < 0.1 were considered significant.
FF Mouse Pups Display Expansion of the Enterobacteriaceae and Enterococcaceae Families in the Ileum
Because both α-diversity and β-diversity of bacteria in the ileum significantly differed between FF and DF pups on P2, we compared the microbiota composition between DF and FF in P2 groups using the DESeq2 method at different taxonomic levels. Analysis at the class level showed an increase in γ-proteobacteria and a decrease in Actinobacteria in FF pups compared with DF pups at P2 (Fig. 1E). At the family level, Enterobacteriaceae, Enterococcaceae, and Pasteurellaceae were significantly increased, whereas Corynebacteriaceae, Staphylococcaceae, and Lactobacillaceae were markedly reduced in FF pups compared with DF pups by P2 (Fig. 1F). At the genus level, Klebsiella, Escherichia/Shigella, Enterobacter, and Enterococcus were significantly increased, whereas Lactobacillus, Staphylococcus, and Corynebacterium were markedly decreased in FF pups compared with DF pups by P2 (Fig. 1G). The relative abundance at class, family, and genus levels of DF and FF pups during the postnatal days (P0–P2) is shown in Supplemental Fig. S2, C–E. Collectively, our data demonstrate that the microbial composition in the ileum of DF and FF mouse pups differs in the first 2 days of life. The increased abundance of genera in Enterobacteriaceae and Enterococcaceae families in the FF neonatal gut represents a dysbiotic change that is often noted in neonatal patients with inflammatory diseases.
FF Mouse Pups Exhibit Profound Low-Grade Postnatal Inflammation in the Ileum without Intestinal Mucosal Injury
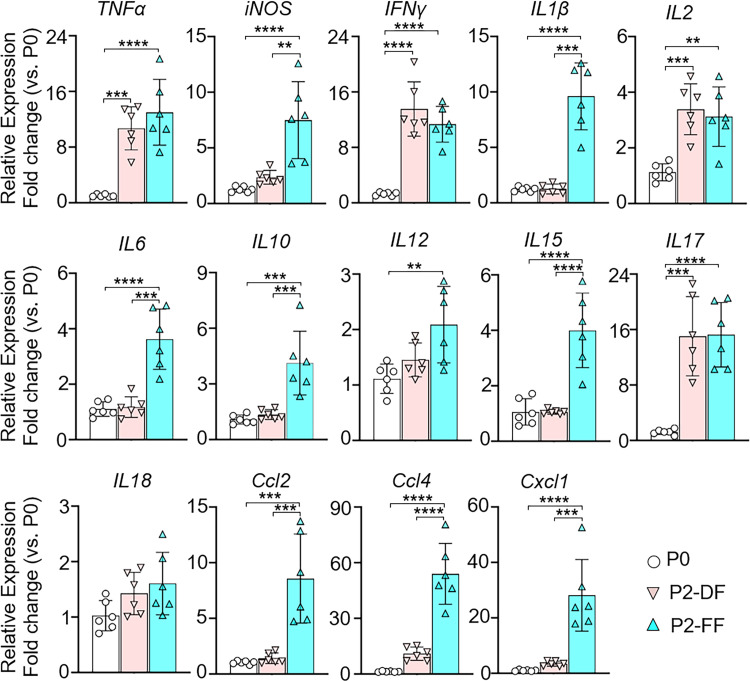
The f_Enterobacteriaceae and f_Enterococcaceae dysbiosis in the ileum of FF pups led us to examine the inflammatory mediator profiles within the ileum in each group of mice on P2. The expression levels of various inflammatory mediators in the ileum of P0 mouse pups and P2 DF and FF pups were measured. We found that P2 pups in both DF and FF cohorts displayed significantly higher expression levels of Tnfα, Ifnγ, Il2, and Il17 in the ileum compared with P0 pups (Fig. 2), suggesting that microbiome colonization and exposure to environmental factors establish a state of physiological inflammation in the ileal mucosa of P2 mice, regardless of feeding mode. We also noted that FF pups exhibited higher expression of several other inflammatory mediators including Nos2, Il1β, Il6, Il10, Il12, Il15, Ccl2, Ccl4, and Cxcl1 in the ileum compared with both P2 DF pups and P0 pups (Fig. 2). Taken together, pups in the FF cohort with f_Enterobacteriaceae and f_Enterococcaceae dysbiosis in the ileum by P2 also had higher levels of physiological inflammation in the ileal mucosa compared with DF pups, suggesting that they experience a low grade of postnatal gut inflammation. However, histological examination on P2 demonstrated that FF-induced postnatal physiological inflammation in the ileal mucosa did not result in intestinal mucosal injury in the ileum. Interestingly, we noted that the ileum of FF pups displayed vacuoles in enterocytes (a feature of fetal-type enterocytes) compared with DF pups (Supplemental Fig. S3; see https://doi.org/10.6084/m9.figshare.17077880).
Figure 2.
Formula feeding (FF) mouse pups have a greater inflammation in the ileum compared with dam feeding (DF) mouse pups. Mouse pups [postnatal day (P)0] from multiple litters were subjected to DF or FF for 2 days. Ileum tissues were processed for RT-qPCR to analyze gene expression levels of the indicated inflammatory cytokines and chemokines. P0 pups were used as the baseline. PCR reactions were run in duplicate for each sample. Data are presented as means ± SD; n = 6 in each group. Data represent 3 independent experiments and were analyzed by 1-way ANOVA with Tukey posttest. **P < 0.01, ***P < 0.001, ****P < 0.0001. Changes are considered statistically significant if P < 0.05.
FF but Not DF Mouse Pups Are Susceptible to Anti-CD3 mAb-Induced Intestinal Injury
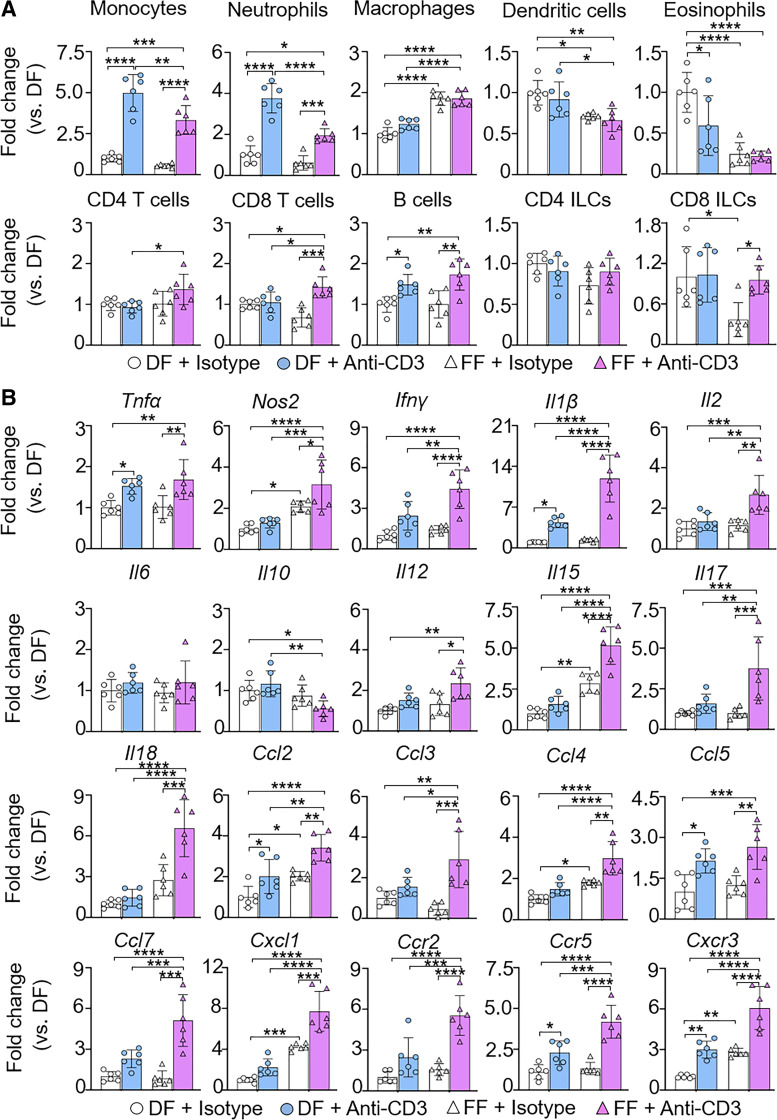
Treatment with monoclonal antibody against CD3 induces injury in the small intestine of adult mice (23, 24). Interestingly, when DF mouse pups (P2) were treated with anti-CD3 mAb at the dose range of 0.1–25 mg/kg sc, no histological intestinal injury was observed as opposed to adult mice (7 wk old, male) treated with anti-CD3 mAb (5 mg/kg sc) (Supplemental Fig. S4; see https://doi.org/10.6084/m9.figshare.17077883). We then tested the hypothesis that FF neonatal mice had increased susceptibility to anti-CD3 mAb-induced intestinal injury. P0 mice were subjected to DF or FF for 3 days. On the postnatal day 2, pups were treated with anti-CD3 mAb (10 mg/kg sc) or isotype IgG (as a negative control) and monitored for an additional 24 h. First, we measured the infiltration of inflammatory cells in our model (Fig. 3A). We found that the FF-alone cohort exhibited significantly higher infiltration of macrophages and lower infiltration of dendritic cells, eosinophils, and CD8+ innate lymphocyte cells (ILCs) in the ileum compared with the DF-alone group. Within the same feeding mode cohort, treatment with anti-CD3 mAb profoundly induced infiltration of monocytes, neutrophils, and B cells in the ileum of both DF and FF pups compared with isotype-treated pups; furthermore, the ileum of FF mouse pups but not DF pups also showed a significant increase in cytotoxic T cells and CD8+ ILCs. Compared with anti-CD3 mAb-treated DF pups, FF pups showed markedly less increase in monocyte, neutrophil, and dendritic cell infiltration after anti-CD3 mAb treatment (Fig. 3A). Interestingly, we found that the ileum of FF mouse pups showed a significant higher increase in macrophages after anti-CD3 mAb treatment compared with anti-CD3 mAb-treated DF pups (Fig. 3A). In addition, the flow cytometric analysis shows that anti-CD3 mAb treatment induced intestinal infiltration of CD4+ T cells and cytotoxic T cells in FF but not DF pups (Fig. 3A).
Figure 3.
Formula feeding (FF) mouse pups exhibit higher infiltration of inflammatory cells and expression of inflammatory mediators than dam feeding (DF) pups after anti-CD3 mAb treatment. Mouse pups [postnatal day (P)0] from multiple litters were subjected to DF and FF for 3 days. On P2, all mice were treated with anti-CD3 mAb (10 mg/kg sc) or isotype IgG (control). Pups were euthanized at P3. A: ileum tissue was processed for multiple-color FACS to profile infiltrating inflammatory cells. Live CD45+ cells were characterized for immune cell phenotypes. The FACS gating strategy is shown in Supplemental Fig. S8 (see https://doi.org/10.6084/m9.figshare.19658460). Data are presented as means ± SD; n = 6 in each group. Data represent 2 independent experiments and were analyzed by 2-way ANOVA with Tukey posttest. ILCs, innate lymphocyte cells. B: expression of indicated cytokines and chemokines in the ileum by RT-qPCR. PCR reactions were run in duplicate for each sample. Data are presented as means ± SD; n = 6 in each group. Data represent 3 independent experiments and were analyzed by 2-way ANOVA with Tukey posttest. *P < 0.05 was considered statistically significant. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Second, we focused on the expression of inflammatory mediators (Fig. 3B). Compared with the DF-alone cohort, the FF-alone group showed a significant increase in the expression levels of Nos2, Il15, Ccl2, Ccl4, Cxcl1, and Cxcr3 inflammatory mediators in the ileum. Both DF and FF mouse pups exhibited significantly elevated expression of inflammatory mediators including Tnfα, Il1β, Ccl2, Ccl5, Ccr5, and Cxcr3 in the ileum at 24 h after anti-CD3 mAb treatment compared with their isotype IgG-treated respective feeding controls; furthermore, the ileum of FF pups but not DF pups also showed a profound increase in various inflammatory mediators including Nos2, Ifnγ, Il2, Il12, Il15, Il17, Il18, Ccl3, Ccl4, Ccl7, Cxc11, and Ccr2 at 24 h after anti-CD3 treatment. The expression levels of these inflammatory mediators were also significantly higher in FF mouse pups compared with the DF cohort at 24 h after anti-CD3 treatment, whereas Il10 was found significantly lower in FF pups after anti-CD3 treatment (Fig. 3B).
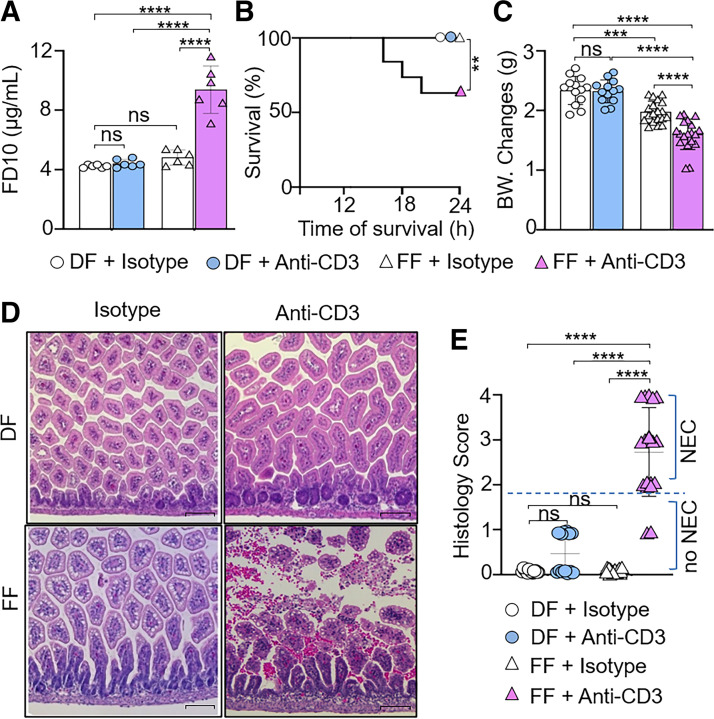
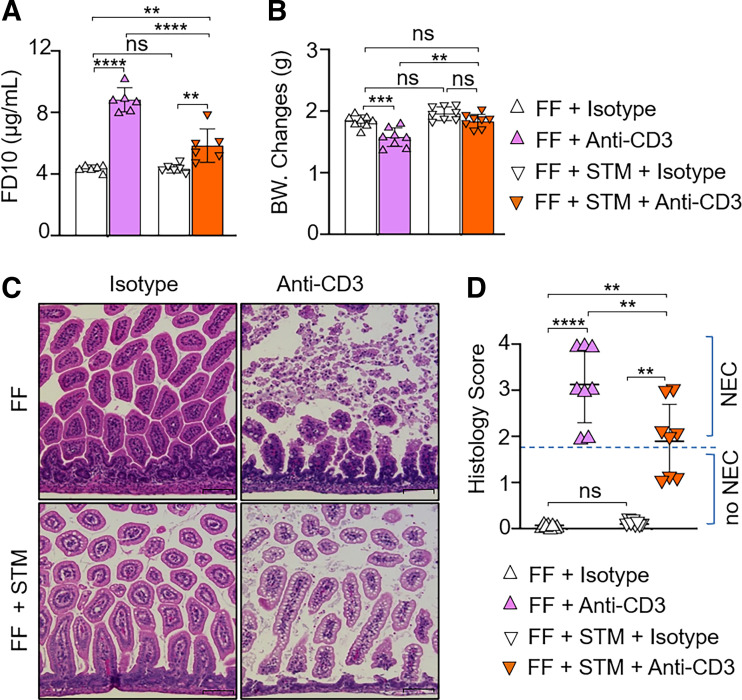
Finally, we examined changes in intestinal permeability and histology following anti-CD3 mAb treatment. Through analysis of initial body weight, we noted that there were no significant differences in weight at the start of the experiment between the groups (Supplemental Fig. S5A; see https://doi.org/10.6084/m9.figshare.19918387). At 24 h of sham treatment (i.e., control IgG), FF pups exhibited a decrease in weight without any changes in intestinal permeability and survival rate compared with the DF + control IgG cohort (Fig. 4, A–C). Anti-CD3 mAb-treated FF but not DF pups had significantly higher intestinal permeability (Fig. 4A), decrease in 24-h survival rate (Fig. 4B), and lower body weight (Fig. 4C) compared with their isotype IgG-treated respective feeding controls as well as anti-CD3 mAb-treated DF pups. Histological examination revealed that FF pups developed NEC-like intestinal injury in the small intestine upon anti-CD3 mAb treatment (Fig. 4, D and E; Supplemental Fig. S6, see https://doi.org/10.6084/m9.figshare.19658445). Collectively, our data suggest that FF but not DF mouse pups were predisposed to anti-CD3 mAb-induced NEC-like intestinal injury.
Figure 4.
Formula feeding (FF) enhances the susceptibility of mouse pups to anti-CD3 mAb-induced intestinal injury. Neonatal mice [postnatal day (P)0] from multiple litters were subjected to dam feeding (DF) and FF. On P2, both DF and FF pups were treated with anti-CD3 mAb (10 mg/kg) or isotype IgG subcutaneously. Pups were monitored for 24 h after treatment (P3). A: relative levels of FD-10S in serum as a measure of intestinal permeability. The fluorescent marker was administered orally 4 h before the end of the experiment. Intestinal permeability assay was run in duplicate for each sample. Data are presented as means ± SD; n = 6 in each group. Data represent 2 independent experiments and were analyzed by 2-way ANOVA with Tukey posttest. B: analysis of the 24-h survival rate after indicated treatment. Log-rank test was used for survival curve. C: analysis of body weight (BW) changes after indicated treatment. D: representative microscopic images of hematoxylin-eosin (H&E) staining of ileum tissue samples of DF and FF pups with or without anti-CD3 mAb treatment. Scale bars, 100 μm. E: histological scores using a necrotizing enterocolitis (NEC) scoring system as indicated in materials and methods. NEC-like injury was defined as histological grade ≥ 2. Histological score was analyzed by χ2 test. C and E: data are presented as means ± SD; n = 13 DF + isotype group, n = 13 DF + anti-CD3 group, n = 19 FF + isotype group, n = 19 FF + anti-CD3 group. Data represent 3 independent experiments and were analyzed by 2-way ANOVA with Tukey posttest. ns, No significance. **P < 0.01, ****P < 0.0001. Changes are considered statistically significant if P < 0.05.
Disturbing Gut Bacterial Colonization with Antibiotic Treatment Attenuates Anti-CD3 mAb-Induced Intestinal Injury in FF Neonatal Mice
Antibiotic treatment leads to reduction of the gut bacterial load and has been shown to prevent the severity of NEC-like intestinal injury (36). To determine whether changes in the microbiome underlie the effect of formula feeding on predisposition of mouse pups to anti-CD3 mAb-induced intestinal injury, DF and FF pups were subjected to treatment with or without streptomycin (STM, a broad-spectrum antibiotic) to inhibit postnatal colonization of both gram-positive and gram-negative bacteria before anti-CD3 mAb challenge. In the antibiotic cohort, pups were gavaged with formula and streptomycin (P0–P3; 30–60 µg/pup/feeding) in 3-h intervals for 3 days. Littermate control pups were subjected to gavage with formula plus saline. On P2, pups were treated with anti-CD3 mAb and monitored for NEC severity for 24 h. There were no significant differences in initial body weight between the groups at the start of the experiment (Supplemental Fig. S5B). At the end of the experiment, we found that all FF pups that were pretreated with streptomycin survived after anti-CD3 mAb treatment. In the saline cohort, FF pups treated with anti-CD3 mAb developed NEC (Fig. 5). FF pups pretreated with streptomycin displayed less increase in intestinal permeability and less reduction of body weight upon anti-CD3 mAb challenge compared with the FF pups without pretreatment with streptomycin group upon anti-CD3 mAb challenge (Fig. 5, A and B). Pretreatment with streptomycin decreased severity of NEC-like injury induced by anti-CD3 mAb in FF pups (Fig. 5, C and D). In contrast to the FF cohort, pups nursed by dam feeding were resistant to anti-CD3 mAb-induced intestinal injury. Pretreatment with streptomycin did not affect their susceptibility to anti-CD3 mAb challenge (Supplemental Fig. S7; see https://doi.org/10.6084/m9.figshare.19658457). Collectively, the data suggest that inadequate postnatal bacterial colonization may mediate the development of NEC-like injury in FF pups upon anti-CD3 mAb treatment.
Figure 5.
Anti-CD3 mAb-induced injury is reduced in streptomycin-treated formula feeding (FF) pups. Neonatal mice from multiple litters were gavaged with formula milk with/without streptomycin [STM; postnatal day (P)0–P3: 30–60 µg/pup/feeding] every 3 h for 72 h. On P2, the pups were treated with anti-CD3 mAb (10 mg/kg) or isotype IgG subcutaneously. Pups were monitored for 24 h after treatment (P3). A: intestinal permeability expressed as fluorescence readings in serum 4 h after gavage with FITC-dextran. Intestinal permeability assay was run in duplicate for each sample. Data are presented as means ± SD; n = 6 in each group. B: analysis of body weight (BW) changes after indicated treatment. Data are presented as means ± SD; n = 8 in each group. C: representative microscopic images of hematoxylin-eosin (H&E) staining of ileum tissue samples from pups with or without antibiotics treated with anti-CD3 mAb. Scale bars, 100 μm. D: histological scores using a necrotizing enterocolitis (NEC) scoring system, with NEC-like injury defined as histological grade ≥ 2. Histological score was analyzed by χ2 test, and data are presented as means ± SD; n = 8 in each group. A–D: data represent 2 independent experiments and were analyzed by 2-way ANOVA with Tukey posttest. ns, No significance. **P < 0.01, ***P < 0.001, ****P < 0.0001. Changes are considered statistically significant if P < 0.05.
DISCUSSION
Our study shows that microbiota in the ileum during the first 2 days of life differ at a taxonomic level in DF and FF mouse pups. We noted that FF pups have a distinct gut microbiome characterized by increased f_Enterococcaceae and f_Enterobacteriaceae in the ileum. Together with this microbiota change, FF pups exhibited profound inflammatory responses in the ileum. We provide the first evidence that anti-CD3 mAb treatment induces NEC-like injury in FF but not DF neonatal mouse pups. The disruption of gut bacterial colonization in FF pups with antibiotic pretreatment partially attenuated the severity of the anti-CD3 mAb-induced intestinal injury in FF pups, suggesting that FF-induced dysbiosis affects the susceptibility of intestine injury during the inflammatory state in neonates.
The intestines of newborns mount an effective immune response and tolerance against pathogens and food antigens when exposed to milk and microbes shortly after birth (37). The development of the gut microbiome is affected by the type of infant feeding (BF or FF). Several studies have described distinct microbiomes in breastfed infants compared with formula-fed infants (4, 38–40). In the present study, we found that DF pups displayed expansion of f_Lactobacillaceae, f_Staphylococcaceae, and f_Corynebacteriaceae abundance on P2, whereas FF pups exhibited an increase in total number of bacterial species, low species diversity, and the presence of potential pathogens that are associated with NEC development in neonates (34, 35). Evidence suggests that formula-fed infants exhibit proinflammatory bacterial class γ-Proteobacteria, whereas breastfed infants have protective bacterial class Actinobacteria (38, 39, 41). Increase in postnatal colonization of γ-Proteobacteria likely precedes NEC development (42, 43). BF maintains a lower abundance of facultative anerobic microorganisms, such as f_Enterobacteriaceae and f_Enterococcaceae, in the gut (16, 17, 44, 45). It has been reported that preterm infants are frequently colonized with Klebsiella sp., Enterobacter, and Enterococcus, whereas Lactobacillus are less prevalent compared with in-term breastfed infants (34, 46). We also found a rapid shift in the bacterial profile with the increased abundance of genera in f_Enterobacteriaceae and f_Enterococcaceae in the ileum of FF pups compared with DF pups at P2. Premature infants had higher levels of potentially pathogenic bacteria such as Enterococcus sp. and Klebsiella sp. (47). Furthermore, evidence shows the onset of Enterobacteriaceae dysbiosis and expansion of Klebsiella and Enterobacter before NEC (48–50). Zhou et al. (51) reported that Escherichia/Shigella was significantly higher in late-onset NEC cases than control subjects 6 days before NEC onset. Importantly, our study noted that FF mouse pups exhibited a dysbiotic pattern that mimics the proinflammatory microbial composition in the gut of preterm infants (52, 53).
Next, we investigated whether FF influences the mucosal physiological inflammatory profile, as the gut microbiota plays a significant role in regulating the intestinal immune system (54). We found increased expression of inflammatory factors in the ileum of P2 DF cohorts compared with P0, indicating the presence of a controlled inflammatory response against colonizing bacteria (55). Previous studies have demonstrated that FF animals exhibit a significant increase in inflammatory markers and a decrease in anti-inflammatory mediators in several animal models (4, 56, 57). We also found exaggerated inflammatory responses in the ileal mucosa of FF pups compared with DF pups at P2. FF-induced inflammatory mediators in the ileal mucosa did not impact histological features of the small intestine in our study; however, we note that abnormal colonization of f_Enterobacteriaceae and f_Enterococcaceae has been found to contribute to neonatal intestinal inflammation such as NEC (54). Thus, it seems that FF induces dysbiosis associated with an increase in genera of f_Enterobacteriaceae and f_Enterococcaceae in the gut that may predispose infants to intestinal inflammation and gut pathology such as NEC.
FF is an alternative for preterm or low-birth-weight infants to obtain fat, energy, protein content, and nutrients when sufficient maternal breast milk is not available. On the other hand, FF is a well-recognized risk factor for NEC in premature babies (6, 7). However, it is uncertain why FF compared with breast milk feeding shows a higher risk of developing NEC. Recently, it has been found that human NEC is rich in intestinal CD4+ Th17 lymphocytes and reduction of Treg, suggesting an association between T-cell activation and NEC (22). To examine whether FF impacts the susceptibility of neonatal intestines to inflammatory stress derived from T-cell activation, we applied a well-characterized adult mouse model of anti-CD3 mAb-induced intestinal injury to neonatal pups in this study. Research has shown that anti-CD3 mAb induces a cytokine storm and local inflammation primarily in the small intestine of adult mice (23, 58–60). TNF-α, IFN-γ, Fas/Fas ligand interactions, and perforin are involved in mediating mucosal damage in anti-CD3 mAb-induced T-cell activation-associated intestinal damage in adult mice (58, 61, 62). In addition, other investigators reported that treatment of adult mice with anti-CD3 mAb induces prominent villus shortening, crypt degeneration, mucosal flattening, and epithelial cell apoptosis in the small intestine (24, 63). In the present study, we confirmed previous observations that adult mice are susceptible to anti-CD3 mAb-induced small bowel injury (Supplemental Fig. S4). Surprisingly, we found that DF neonatal mouse pups are highly resistant to anti-CD3 mAb-induced intestinal injury, although the treatment results in elevated inflammatory markers and infiltration of inflammatory cells in the ileum. In contrast, FF mouse pups were vulnerable to anti-CD3 mAb-induced intestinal injury, suggesting that the feeding mode influences the susceptibility of neonatal mouse intestine to inflammatory injury upon T-cell activation. Currently, it is unclear why treatment with anti-CD3 mAb is unable to induce intestinal injury in DF mouse pups. Evidence shows the protective role of breast milk and its nutritional and immune factors such as maternal IgA and human milk oligosaccharides and beneficial microbiome (64–66). Breastfeeding mediates mother-to-infant transfer of bacterial strains including genera Lactobacillus and Staphylococcus (67). Breastfed infants are highly characterized with predominant organisms such as Lactobacilli (68). We found that DF pups displayed expansion of Lactobacillus and Staphylococcus abundance and reduction of anti-CD3 mAb-induced mucosal damage, suggesting the beneficial effect of these microbes as well as prebiotics within breast milk (67). Thus, it is likely that DF promotes a healthy GI system that prevents anti-CD3 mAb-induced inflammatory injury through complex mechanisms that remain to be investigated.
Evidence suggests that formula-fed infants are deprived of both the nutritional benefits and the immune benefits of breast milk, which places them at higher risk of developing neonatal GI diseases (5). Interestingly, we found that FF pups were highly vulnerable to anti-CD3 mAb stimulation. We report for the first time that FF influences the susceptibility of anti-CD3 mAb-induced increase in intestinal permeability and intestinal injury in neonatal pups. We noted that FF pups developed human NEC-like intestinal injury upon anti-CD3 mAb treatment, which includes mucosal edema, inflammatory cell infiltration, and hemorrhagic/coagulative necrosis (69). Anti-CD3 mAb treatment also resulted in higher mortality rate in FF pups in addition to inducing NEC-like intestinal injury. NEC is frequently complicated by perforation, peritonitis, septicemia, and multiple organ failure (70, 71). We expect that these complications may contribute to death occurring in anti-CD3-treated FF pups in our study.
Emerging evidence shows that formula-fed infants resemble the gut microbiome in adults (45). Formula-fed infants were highly colonized with adult-like microbiota that displayed f_Enterobacteriaceae and f_Enterococcaceae compared with breastfed infants (40, 45, 72). The predominant organisms such as Lactobacillus in breastfed infants are less dominant in formula-fed and premature infants who are at the greatest risk of NEC (68). The microbiota pattern in FF mice in our study mimics the prominent gut dysbiosis seen in preterm infants with inflammatory diseases. NEC develops in premature infants upon gut dysbiosis with the expansion of f_Enterobacteriaceae after birth (48, 73–75). The gut of neonates with NEC is colonized mainly with f_Enterobacteriaceae, as well as f_Enterococcaceae (53, 73, 76). We found that FF pups but not DF pups are vulnerable to anti-CD3 mAb-induced NEC-like intestinal injury. This discovery suggests that FF promotes dysbiosis of genera in f_Enterobacteriaceae and f_Enterococcaceae with adult-like microbiota and increases the susceptibility to anti-CD3 mAb-mediated inflammatory injury. It has been reported that, in several animal models of NEC, intestinal injury does not occur in germ-free or broad-spectrum antibiotic-treated animals (35, 36, 77, 78). A recent study showed the significance of microbial sensing by Nod2 to control mucosal damage after anti-CD3 mAb injection in NOD2−/− adult mice (62). In the present study, we observed that streptomycin treatment attenuated NEC severity in anti-CD3 mAb-treated FF pups. Our finding advances the potential link between NEC-like intestinal injury and FF-mediated abnormal bacterial colonization in neonates. However, we did not examine whether and how streptomycin treatment resulted in changes in gut bacterial profiles in our experiments. This limitation opens an opportunity for future studies.
Previously, Egan et al. (22) reported that T cell-mediated IL-17RA/IL-17 signaling is an important mechanism by which NEC-associated postnatal stresses cause intestinal injury in neonatal mice. In our study, we observed the infiltration of cytotoxic CD8 T lymphocytes in the ileum of anti-CD3-treated FF pups but not in DF pups, supporting the notion that lymphocytes play a crucial role in the pathogenesis of NEC. Furthermore, our novel finding that DF pups lack infiltration of cytotoxic T cells in the intestinal mucosa after anti-CD3 mAb treatment suggests the role of breast milk in regulation of T cell-associated mucosal immune response. Breast milk-derived components maintain TH1/TH2 balance with activation of T reg cells and decrease exaggerated inflammatory responses in the intestinal mucosa (2, 55, 79). In addition to lymphocytes, we observed that the number of myeloid cells in ileum of DF-alone pups differs from that in the FF-alone cohort. Compared with the DF plus anti-CD3 group, pups in the FF plus anti-CD3 group also exhibited changes in myeloid cells in ileum. Together, our findings suggest that the postnatal homeostasis of gut mucosal inflammatory cells is affected by feeding mode in neonates. However, how a feeding mode influences these inflammatory cells and how FF-mediated dysbiosis enhances the susceptibility of T cell-mediated intestinal injury remains an open question. The mechanisms by which breastfeeding attenuates the effect of T-cell activation-induced intestinal injury and NEC development in neonates need to be further investigated.
In summary, we found that FF promotes adult-like microbiota with an abundance of genera in f_Enterobacteriaceae and f_Enterococcaceae that increases the susceptibility to inflammatory injury in mouse pups, mimicking the pathophysiological events observed in premature infants in humans. We showed for the first time that anti-CD3 mAb-induced intestinal injury in FF mouse pups and FF-associated dysbiosis contribute to increased susceptibility to intestinal NEC-like injury related to anti-CD3 mAb-induced inflammation. Future studies are needed to better understand the role of feeding-associated changes in bacterial colonization and intestinal inflammation and injury, so therapeutic approaches to prevent NEC in neonates can be developed.
SUPPLEMENTAL DATA
Supplemental Fig. S1: https://doi.org/10.6084/m9.figshare.17077328.
Supplemental Fig. S2: https://doi.org/10.6084/m9.figshare.19658412.
Supplemental Fig. S3: https://doi.org/10.6084/m9.figshare.17077880.
Supplemental Fig. S4: https://doi.org/10.6084/m9.figshare.17077883.
Supplemental Fig. S5: https://doi.org/10.6084/m9.figshare.19918387.
Supplemental Fig. S6: https://doi.org/10.6084/m9.figshare.19658445.
Supplemental Fig. S7: https://doi.org/10.6084/m9.figshare.19658457.
Supplemental Fig. S8: https://doi.org/10.6084/m9.figshare.19658460.
GRANTS
X.-D.T. is funded by National Institutes of Health (NIH) Grants (GM117628, GM122406, DK064240, DK123826, DK129960) and a Department of Veterans Affairs Merit Review Award (I01BX001690). I.G.D.P. is funded by NIH Grant R01DK116568. Additional support was provided by the Dorothy M. and Edward E. Burwell Professorship (to X.-D.T.).
DISCLAIMERS
The funders had no role in study design, data collection and analysis, interpretation of data, decision to publish, or preparation of the manuscript.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
S. Subramanian and X.-D.T. conceived and designed research; S. Subramanian, H.G., C.D., and H.-F.B. performed experiments; S. Subramanian, H.G., C.D., P.M.C., S.C.T., and S. Swaminathan analyzed data; S. Subramanian, H.G., C.D., P.M.C., S. Swaminathan, J.M.R., and X.-D.T. interpreted results of experiments; S. Subramanian, H.G., and X.-D.T. prepared figures; S. Subramanian and X.-D.T. drafted manuscript; S. Subramanian, H.G., C.D., P.M.C., H.-F.B., X.W., S. Swaminathan, J.M.R., I.G.D.P, and X.-D.T. edited and revised manuscript; S. Subramanian, H.G., C.D., P.M.C., H.-F.B., X.W., S. Swaminathan, S.C.T., J.M.R., I.G.D.P., and X.-D.T. approved final version of manuscript.
ACKNOWLEDGMENTS
Histology services were provided by the Microscopy and Histology Group at Stanley Manne Children’s Research Institute, Chicago, IL. Bioinformatics analysis was performed by the UIC Research Informatics Core, supported in part by the National Center for Advancing Translational Science (NCATS) through Grant UL1TR002003.
REFERENCES
- 1. Radlowski EC, Wang M, Monaco MH, Comstock SS, Donovan SM. Combination-feeding causes differences in aspects of systemic and mucosal immune cell phenotypes and functions compared with exclusive sow-rearing or formula-feeding in piglets. Nutrients 13: 1097, 2021. doi: 10.3390/nu13041097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Selma-Royo M, Calvo Lerma J, Cortés-Macías E, Collado MC. Human milk microbiome: from actual knowledge to future perspective. Semin Perinatol 45: 151450, 2021. doi: 10.1016/j.semperi.2021.151450. [DOI] [PubMed] [Google Scholar]
- 3. Fan HS, Fong DY, Lok KY, Tarrant M. Association between expressed breast milk feeding and breastfeeding duration in Hong Kong mothers. Women Birth 35: e286–e293, 2022. doi: 10.1016/j.wombi.2021.06.007. [DOI] [PubMed] [Google Scholar]
- 4. Yeruva L, Spencer NE, Saraf MK, Hennings L, Bowlin AK, Cleves MA, Mercer K, Chintapalli SV, Shankar K, Rank RG, Badger TM, Ronis MJ. Formula diet alters small intestine morphology, microbial abundance and reduces VE-cadherin and IL-10 expression in neonatal porcine model. BMC Gastroenterol 16: 40, 2016. doi: 10.1186/s12876-016-0456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taye AA, Asegidew W, Taderegew MM, Bizuwork YG, Zegeye B. Formula feeding practice and associated factors among mothers with infants 0-6 months of age in Addis Ababa, Ethiopia: a community-based cross-sectional study. Ital J Pediatr 47: 55, 2021. doi: 10.1186/s13052-021-01010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lucas A, Cole TJ. Breast milk and neonatal necrotising enterocolitis. Lancet 336: 1519–1523, 1990. doi: 10.1016/0140-6736(90)93304-8. [DOI] [PubMed] [Google Scholar]
- 7. Schanler RJ, Shulman RJ, Lau C. Feeding strategies for premature infants: beneficial outcomes of feeding fortified human milk versus preterm formula. Pediatrics 103: 1150–1157, 1999. doi: 10.1542/peds.103.6.1150. [DOI] [PubMed] [Google Scholar]
- 8. Wang Z, Neupane A, Vo R, White J, Wang X, Marzano SL. Comparing gut microbiome in mothers’ own breast milk- and formula-fed moderate-late preterm infants. Front Microbiol 11: 891, 2020. doi: 10.3389/fmicb.2020.00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jones RB, Berger PK, Plows JF, Alderete TL, Millstein J, Fogel J, Iablokov SN, Rodionov DA, Osterman AL, Bode L, Goran MI. Lactose-reduced infant formula with added corn syrup solids is associated with a distinct gut microbiota in Hispanic infants. Gut Microbes 12: 1813534, 2020. doi: 10.1080/19490976.2020.1813534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yasmin F, Tun HM, Konya TB, Guttman DS, Chari RS, Field CJ, Becker AB, Mandhane PJ, Turvey SE, Subbarao P, Sears MR; CHILD Study Investigators, Scott JA, Dinu I, Kozyrskyj AL. Cesarean section, formula feeding, and infant antibiotic exposure: separate and combined impacts on gut microbial changes in later infancy. Front Pediatr 5: 200, 2017. doi: 10.3389/fped.2017.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guaraldi F, Salvatori G. Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol 2: 94, 2012. doi: 10.3389/fcimb.2012.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jost T, Lacroix C, Braegger CP, Chassard C. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS One 7: e44595, 2012. doi: 10.1371/journal.pone.0044595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Castellote C, Casillas R, Ramírez-Santana C, Pérez-Cano FJ, Castell M, Moretones MG, López-Sabater MC, Franch A. Premature delivery influences the immunological composition of colostrum and transitional and mature human milk. J Nutr 141: 1181–1187, 2011. doi: 10.3945/jn.110.133652. [DOI] [PubMed] [Google Scholar]
- 14. Moore RE, Townsend SD. Temporal development of the infant gut microbiome. Open Biol 9: 190128, 2019. doi: 10.1098/rsob.190128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Taylor SN, Basile LA, Ebeling M, Wagner CL. Intestinal permeability in preterm infants by feeding type: mother’s milk versus formula. Breastfeed Med 4: 11–15, 2009. doi: 10.1089/bfm.2008.0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Baumann-Dudenhoeffer AM, D’Souza AW, Tarr PI, Warner BB, Dantas G. Infant diet and maternal gestational weight gain predict early metabolic maturation of gut microbiomes. Nat Med 24: 1822–1829, 2018. doi: 10.1038/s41591-018-0216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma J, Li Z, Zhang W, Zhang C, Zhang Y, Mei H, Zhuo N, Wang H, Wang L, Wu D. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: a study of 91 term infants. Sci Rep 10: 15792, 2020. doi: 10.1038/s41598-020-72635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boudry G, Morise A, Seve B, Huërou-Luron IL. Effect of milk formula protein content on intestinal barrier function in a porcine model of LBW neonates. Pediatr Res 69: 4–9, 2011. doi: 10.1203/PDR.0b013e3181fc9d13. [DOI] [PubMed] [Google Scholar]
- 19. MohanKumar K, Namachivayam K, Song T, Jake Cha B, Slate A, Hendrickson JE, Pan H, Wickline SA, Oh JY, Patel RP, He L, Torres BA, Maheshwari A. A murine neonatal model of necrotizing enterocolitis caused by anemia and red blood cell transfusions. Nat Commun 10: 3494, 2019. doi: 10.1038/s41467-019-11199-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Call L, Stoll B, Oosterloo B, Ajami N, Sheikh F, Wittke A, Waworuntu R, Berg B, Petrosino J, Olutoye O, Burrin D. Metabolomic signatures distinguish the impact of formula carbohydrates on disease outcome in a preterm piglet model of NEC. Microbiome 6: 111, 2018. doi: 10.1186/s40168-018-0498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Managlia E, Liu SX, Yan X, Tan XD, Chou PM, Barrett TA, De Plaen IG. Blocking NF-κB activation in Ly6c+ monocytes attenuates necrotizing enterocolitis. Am J Pathol 189: 604–618, 2019. doi: 10.1016/j.ajpath.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Egan CE, Sodhi CP, Good M, Lin J, Jia H, Yamaguchi Y, Lu P, Ma C, Branca MF, Weyandt S, Fulton WB, Niño DF, Prindle T Jr, Ozolek JA, Hackam DJ. Toll-like receptor 4-mediated lymphocyte influx induces neonatal necrotizing enterocolitis. J Clin Invest 126: 495–508, 2016. doi: 10.1172/JCI83356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ferran C, Sheehan K, Dy M, Schreiber R, Merite S, Landais P, Noel LH, Grau G, Bluestone J, Bach JF. Cytokine-related syndrome following injection of anti-CD3 monoclonal antibody: further evidence for transient in vivo T cell activation. Eur J Immunol 20: 509–515, 1990. doi: 10.1002/eji.1830200308. [DOI] [PubMed] [Google Scholar]
- 24. Radojevic N, McKay DM, Merger M, Vallance BA, Collins SM, Croitoru K. Characterization of enteric functional changes evoked by in vivo anti-CD3 T cell activation. Am J Physiol Regul Integr Comp Physiol 276: R715–R723, 1999. doi: 10.1152/ajpregu.1999.276.3.R715. [DOI] [PubMed] [Google Scholar]
- 25. Yan X, Managlia E, Liu SX, Tan XD, Wang X, Marek C, De Plaen IG. Lack of VEGFR2 signaling causes maldevelopment of the intestinal microvasculature and facilitates necrotizing enterocolitis in neonatal mice. Am J Physiol Gastrointest Liver Physiol 310: G716–G725, 2016. doi: 10.1152/ajpgi.00273.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yan X, Managlia E, Zhao YY, Tan XD, De Plaen IG. Macrophage-derived IGF-1 protects the neonatal intestine against necrotizing enterocolitis by promoting microvascular development. Commun Biol 5: 320, 2022. doi: 10.1038/s42003-022-03252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan X, Managlia E, Tan XD, De Plaen IG. Prenatal inflammation impairs intestinal microvascular development through a TNF-dependent mechanism and predisposes newborn mice to necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol 317: G57–G66, 2019. doi: 10.1152/ajpgi.00332.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30: 614–620, 2014. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12, 2011. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 30. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461, 2010. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 31. Glöckner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, Bruns G, Yarza P, Peplies J, Westram R, Ludwig W. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol 261: 169–176, 2017. doi: 10.1016/j.jbiotec.2017.06.1198. [DOI] [PubMed] [Google Scholar]
- 32. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13: 581–583, 2016. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gonzalez E, Brereton NJ, Li C, Lopez Leyva L, Solomons NW, Agellon LB, Scott ME, Koski KG. Distinct changes occur in the human breast milk microbiome between early and established lactation in breastfeeding Guatemalan mothers. Front Microbiol 12: 557180, 2021. doi: 10.3389/fmicb.2021.557180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cilieborg MS, Boye M, Sangild PT. Bacterial colonization and gut development in preterm neonates. Early Hum Dev 88: S41–S49, 2012. doi: 10.1016/j.earlhumdev.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 35. Sangild PT, Siggers RH, Schmidt M, Elnif J, Bjornvad CR, Thymann T, Grondahl ML, Hansen AK, Jensen SK, Boye M, Moelbak L, Buddington RK, Weström BR, Holst JJ, Burrin DG. Diet- and colonization-dependent intestinal dysfunction predisposes to necrotizing enterocolitis in preterm pigs. Gastroenterology 130: 1776–1792, 2006. doi: 10.1053/j.gastro.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 36. Jensen ML, Thymann T, Cilieborg MS, Lykke M, Mølbak L, Jensen BB, Schmidt M, Kelly D, Mulder I, Burrin DG, Sangild PT. Antibiotics modulate intestinal immunity and prevent necrotizing enterocolitis in preterm neonatal piglets. Am J Physiol Gastrointest Liver Physiol 306: G59–G71, 2014. doi: 10.1152/ajpgi.00213.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pan X, Zhang D, Nguyen DN, Wei W, Yu X, Gao F, Sangild PT. Postnatal gut immunity and microbiota development is minimally affected by prenatal inflammation in preterm pigs. Front Immunol 11: 420, 2020. doi: 10.3389/fimmu.2020.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe 17: 478–482, 2011. doi: 10.1016/j.anaerobe.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 39. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118: 511–521, 2006. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 40. Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr 30: 61–67, 2000. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 41. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17: 852, 2015. doi: 10.1016/j.chom.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 42. Morrow AL, Lagomarcino AJ, Schibler KR, Taft DH, Yu Z, Wang B, Altaye M, Wagner M, Gevers D, Ward DV, Kennedy MA, Huttenhower C, Newburg DS. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome 1: 13, 2013. doi: 10.1186/2049-2618-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brower-Sinning R, Zhong D, Good M, Firek B, Baker R, Sodhi CP, Hackam DJ, Morowitz MJ. Mucosa-associated bacterial diversity in necrotizing enterocolitis. PLoS One 9: e105046, 2014. doi: 10.1371/journal.pone.0105046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, Adisetiyo H, Zabih S, Lincez PJ, Bittinger K, Bailey A, Bushman FD, Sleasman JW, Aldrovandi GM. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr 171: 647–654, 2017. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roger LC, McCartney AL. Longitudinal investigation of the faecal microbiota of healthy full-term infants using fluorescence in situ hybridization and denaturing gradient gel electrophoresis. Microbiology (Reading) 156: 3317–3328, 2010. doi: 10.1099/mic.0.041913-0. [DOI] [PubMed] [Google Scholar]
- 46. Hunter CJ, Upperman JS, Ford HR, Camerini V. Understanding the susceptibility of the premature infant to necrotizing enterocolitis (NEC). Pediatr Res 63: 117–123, 2008. doi: 10.1203/PDR.0b013e31815ed64c. [DOI] [PubMed] [Google Scholar]
- 47. Schwiertz A, Gruhl B, Löbnitz M, Michel P, Radke M, Blaut M. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr Res 54: 393–399, 2003. doi: 10.1203/01.PDR.0000078274.74607.7A. [DOI] [PubMed] [Google Scholar]
- 48. Olm MR, Bhattacharya N, Crits-Christoph A, Firek BA, Baker R, Song YS, Morowitz MJ, Banfield JF. Necrotizing enterocolitis is preceded by increased gut bacterial replication, Klebsiella, and fimbriae-encoding bacteria. Sci Adv 5: eaax5727, 2019. doi: 10.1126/sciadv.aax5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Claud EC, Keegan KP, Brulc JM, Lu L, Bartels D, Glass E, Chang EB, Meyer F, Antonopoulos DA. Bacterial community structure and functional contributions to emergence of health or necrotizing enterocolitis in preterm infants. Microbiome 1: 20, 2013. doi: 10.1186/2049-2618-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Millar MR, MacKay P, Levene M, Langdale V, Martin C. Enterobacteriaceae and neonatal necrotising enterocolitis. Arch Dis Child 67: 53–56, 1992. doi: 10.1136/adc.67.1_Spec_No.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou Y, Shan G, Sodergren E, Weinstock G, Walker WA, Gregory KE. Longitudinal analysis of the premature infant intestinal microbiome prior to necrotizing enterocolitis: a case-control study. PLoS One 10: e0118632, 2015. doi: 10.1371/journal.pone.0118632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Arboleya S, Ang L, Margolles A, Yiyuan L, Dongya Z, Liang X, Solís G, Fernández N, de Los Reyes-Gavilán CG, Gueimonde M. Deep 16S rRNA metagenomics and quantitative PCR analyses of the premature infant fecal microbiota. Anaerobe 18: 378–380, 2012. doi: 10.1016/j.anaerobe.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 53. Arboleya S, Binetti A, Salazar N, Fernández N, Solís G, Hernández-Barranco A, Margolles A, de Los Reyes-Gavilán CG, Gueimonde M. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol Ecol 79: 763–772, 2012. doi: 10.1111/j.1574-6941.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 54. Liu Y, Fatheree NY, Mangalat N, Rhoads JM. Lactobacillus reuteri strains reduce incidence and severity of experimental necrotizing enterocolitis via modulation of TLR4 and NF-κB signaling in the intestine. Am J Physiol Gastrointest Liver Physiol 302: G608–G617, 2012. doi: 10.1152/ajpgi.00266.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ward TL, Hosid S, Ioshikhes I, Altosaar I. Human milk metagenome: a functional capacity analysis. BMC Microbiol 13: 116, 2013. doi: 10.1186/1471-2180-13-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. O’Sullivan A, He X, McNiven EM, Haggarty NW, Lönnerdal B, Slupsky CM. Early diet impacts infant rhesus gut microbiome, immunity, and metabolism. J Proteome Res 12: 2833–2845, 2013. doi: 10.1021/pr4001702. [DOI] [PubMed] [Google Scholar]
- 57. Bergholz R, Zschiegner M, Eschenburg G, Wenke K, Tiemann B, Roth B, Appl B, Reinshagen K, Sommerfeldt D, Ridderbusch I. Mucosal loss with increased expression of IL-6, IL-8, and COX-2 in a formula-feeding only neonatal rat model of necrotizing enterocolitis. J Pediatr Surg 48: 2301–2307, 2013. doi: 10.1016/j.jpedsurg.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 58. Merger M, Viney JL, Borojevic R, Steele-Norwood D, Zhou P, Clark DA, Riddell R, Maric R, Podack ER, Croitoru K. Defining the roles of perforin, Fas/FasL, and tumour necrosis factor alpha in T cell induced mucosal damage in the mouse intestine. Gut 51: 155–163, 2002. doi: 10.1136/gut.51.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ferran C, Dy M, Sheehan K, Schreiber R, Grau G, Bluestone J, Bach JF, Chatenoud L. Cascade modulation by anti-tumor necrosis factor monoclonal antibody of interferon-gamma, interleukin 3 and interleukin 6 release after triggering of the CD3/T cell receptor activation pathway. Eur J Immunol 21: 2349–2353, 1991. doi: 10.1002/eji.1830211009. [DOI] [PubMed] [Google Scholar]
- 60. Musch MW, Clarke LL, Mamah D, Gawenis LR, Zhang Z, Ellsworth W, Shalowitz D, Mittal N, Efthimiou P, Alnadjim Z, Hurst SD, Chang EB, Barrett TA. T cell activation causes diarrhea by increasing intestinal permeability and inhibiting epithelial Na+/K+-ATPase. J Clin Invest 110: 1739–1747, 2002. doi: 10.1172/JCI0215695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Miura N, Yamamoto M, Fukutake M, Ohtake N, Iizuka S, Ishige A, Sasaki H, Fukuda K, Yamamoto T, Hayakawa S. Anti-CD3 induces bi-phasic apoptosis in murine intestinal epithelial cells: possible involvement of the Fas/Fas ligand system in different T cell compartments. Int Immunol 17: 513–522, 2005. doi: 10.1093/intimm/dxh231. [DOI] [PubMed] [Google Scholar]
- 62. Zanello G, Goethel A, Rouquier S, Prescott D, Robertson SJ, Maisonneuve C, Streutker C, Philpott DJ, Croitoru K. The cytosolic microbial receptor Nod2 regulates small intestinal crypt damage and epithelial regeneration following T cell-induced enteropathy. J Immunol 197: 345–355, 2016. doi: 10.4049/jimmunol.1600185. [DOI] [PubMed] [Google Scholar]
- 63. Richmond CA, Rickner H, Shah MS, Ediger T, Deary L, Zhou F, Tovaglieri A, Carlone DL, Breault DT. JAK/STAT-1 signaling is required for reserve intestinal stem cell activation during intestinal regeneration following acute inflammation. Stem Cell Reports 10: 17–26, 2018. doi: 10.1016/j.stemcr.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dvorak B, Halpern MD, Holubec H, Dvorakova K, Dominguez JA, Williams CS, Meza YG, Kozakova H, McCuskey RS. Maternal milk reduces severity of necrotizing enterocolitis and increases intestinal IL-10 in a neonatal rat model. Pediatr Res 53: 426–433, 2003. doi: 10.1203/01.PDR.0000050657.56817.E0. [DOI] [PubMed] [Google Scholar]
- 65. Bode L. Human milk oligosaccharides in the prevention of necrotizing enterocolitis: a journey from in vitro and in vivo models to mother-infant cohort studies. Front Pediatr 6: 385, 2018. doi: 10.3389/fped.2018.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gopalakrishna KP, Macadangdang BR, Rogers MB, Tometich JT, Firek BA, Baker R, Ji J, Burr AH, Ma C, Good M, Morowitz MJ, Hand TW. Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat Med 25: 1110–1115, 2019. doi: 10.1038/s41591-019-0480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Soto A, Martín V, Jiménez E, Mader I, Rodríguez JM, Fernández L. Lactobacilli and bifidobacteria in human breast milk: influence of antibiotherapy and other host and clinical factors. J Pediatr Gastroenterol Nutr 59: 78–88, 2014. doi: 10.1097/MPG.0000000000000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Caplan MS, Miller-Catchpole R, Kaup S, Russell T, Lickerman M, Amer M, Xiao Y, Thomson R Jr.. Bifidobacterial supplementation reduces the incidence of necrotizing enterocolitis in a neonatal rat model. Gastroenterology 117: 577–583, 1999. doi: 10.1016/S0016-5085(99)70450-6. [DOI] [PubMed] [Google Scholar]
- 69. Ballance WA, Dahms BB, Shenker N, Kliegman RM. Pathology of neonatal necrotizing enterocolitis: a ten-year experience. J Pediatr 117: S6–S13, 1990. doi: 10.1016/S0022-3476(05)81124-2. [DOI] [PubMed] [Google Scholar]
- 70. Neu J, Walker WA. Necrotizing enterocolitis. N Engl J Med 364: 255–264, 2011. doi: 10.1056/NEJMra1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sylvester KG, Liu GY, Albanese CT. Necrotizing enterocolitis. Pediatr Surg 2012: 1187–1207, 2012. doi: 10.1016/B978-0-323-07255-7.00094-5. [DOI] [Google Scholar]
- 72. Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol 5: e177, 2007. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Heida FH, van Zoonen AG, Hulscher JB, Te Kiefte BJ, Wessels R, Kooi EM, Bos AF, Harmsen HJ, de Goffau MC. A necrotizing enterocolitis-associated gut microbiota is present in the meconium: results of a prospective study. Clin Infect Dis 62: 863–870, 2016. doi: 10.1093/cid/ciw016. [DOI] [PubMed] [Google Scholar]
- 74. Chen Y, Brook TC, Soe CZ, O’Neill I, Alcon-Giner C, Leelastwattanagul O, Phillips S, Caim S, Clarke P, Hall LJ, Hoyles L. Preterm infants harbour diverse Klebsiella populations, including atypical species that encode and produce an array of antimicrobial resistance- and virulence-associated factors. Microb Genom 6: e000377, 2020. doi: 10.1099/mgen.0.000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Paveglio S, Ledala N, Rezaul K, Lin Q, Zhou Y, Provatas AA, Bennett E, Lindberg T, Caimano M, Matson AP. Cytotoxin-producing Klebsiella oxytoca in the preterm gut and its association with necrotizing enterocolitis. Emerg Microbes Infect 9: 1321–1329, 2020. doi: 10.1080/22221751.2020.1773743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. López Sastre JB, Coto Cotallo D, Fernández Colomer B; Grupo de Hospitales Castrillo. Neonatal sepsis of nosocomial origin: an epidemiological study from the “Grupo de Hospitales Castrillo”. J Perinat Med 30: 149–157, 2002. doi: 10.1515/JPM.2002.019. [DOI] [PubMed] [Google Scholar]
- 77. Musemeche CA, Kosloske AM, Bartow SA, Umland ET. Comparative effects of ischemia, bacteria, and substrate on the pathogenesis of intestinal necrosis. J Pediatr Surg 21: 536–538, 1986. doi: 10.1016/S0022-3468(86)80228-7. [DOI] [PubMed] [Google Scholar]
- 78. Waligora-Dupriet AJ, Dugay A, Auzeil N, Huerre M, Butel MJ. Evidence for clostridial implication in necrotizing enterocolitis through bacterial fermentation in a gnotobiotic quail model. Pediatr Res 58: 629–635, 2005. doi: 10.1203/01.PDR.0000180538.13142.84. [DOI] [PubMed] [Google Scholar]
- 79. Walker WA, Iyengar RS. Breast milk, microbiota, and intestinal immune homeostasis. Pediatr Res 77: 220–228, 2015. doi: 10.1038/pr.2014.160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. S1: https://doi.org/10.6084/m9.figshare.17077328.
Supplemental Fig. S2: https://doi.org/10.6084/m9.figshare.19658412.
Supplemental Fig. S3: https://doi.org/10.6084/m9.figshare.17077880.
Supplemental Fig. S4: https://doi.org/10.6084/m9.figshare.17077883.
Supplemental Fig. S5: https://doi.org/10.6084/m9.figshare.19918387.
Supplemental Fig. S6: https://doi.org/10.6084/m9.figshare.19658445.
Supplemental Fig. S7: https://doi.org/10.6084/m9.figshare.19658457.
Supplemental Fig. S8: https://doi.org/10.6084/m9.figshare.19658460.