Abstract
RNA polymerase (RNAP) purified from Methanobacterium thermoautotrophicum ΔH has been shown to initiate transcription accurately in vitro from the hmtB archaeal histone promoter with either native or recombinant forms of the M. thermoautotrophicum TATA-binding protein and transcription factor TFB. Efforts to obtain transcription initiation from hydrogen-regulated methane gene promoters were, however, unsuccessful. Two previously unrecognized archaeal RNAP subunits have been identified, and complex formation by the M. thermoautotrophicum RNAP and TFB has been demonstrated.
The biochemistry and molecular biology of methanogenesis from CO2 and H2 have been established primarily through studies of Methanobacterium thermoautotrophicum ΔH and Marburg (26). Several steps in this pathway are catalyzed by isoenzymes or pairs of functionally equivalent enzymes, and the availability of H2 has been shown to determine which of these alternative enzymes are synthesized (19). This regulation occurs at the level of methane gene transcription; however, the molecular mechanisms by which H2 availability is titrated and communicated intracellularly into promoter activation or inactivation remain unknown. As the essential next step in furthering this investigation, both to understand the regulation of methanogenesis and to determine how an archaeon senses and signals environmental change and regulates promoter function, we report here the establishment and characterization of in vitro transcription systems, using native and recombinant transcription factors and RNA polymerase (RNAP) from M. thermoautotrophicum ΔH. Two previously unrecognized archaeal RNAP subunits have been identified, and complex formation by M. thermoautotrophicum RNAP and archaeal transcription factor TFB has been demonstrated.
Purification and identification of subunits of M. thermoautotrophicum ΔH RNAP.
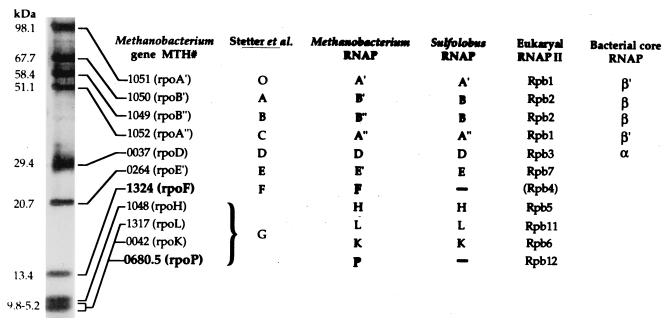
RNAP was purified under anaerobic conditions from M. thermoautotrophicum cells resuspended (0.5 g [wet weight]/ml) in TMK buffer (50 mM Tris HCl, 10 mM MgCl2, 50 mM KCl, 20% [vol/vol] glycerol [pH 8]). The cells were ruptured by passage twice through a French pressure cell at 20,000 lb/in2, and following centrifugation of the resulting lysate at 100,000 × g for 1 h at 4°C, the supernatant obtained was loaded onto a DEAE-cellulose column (Whatman, Fairfield, N.J.). The column was washed with TMK buffer, bound proteins were eluted with a linear gradient of 50 to 525 mM KCl in TMK buffer, and the fractions collected were assayed for nonspecific transcription activity as previously described (7). Fractions that contained this activity were combined, diluted with 50 mM Tris HCl (pH 8) to ∼75 mM KCl, loaded onto a heparin-Sepharose column (Pharmacia, Piscataway, N.J.), and washed with TMK buffer, and the bound proteins were eluted with a 50 mM-to-1 M KCl gradient in TMK buffer. Fractions with RNAP activity were again combined, diluted with 50 mM Tris HCl to reduce the KCl concentration, loaded onto a Mono-Q column (Pharmacia), and washed with TMK buffer, and the bound proteins were eluted with a 50 mM-to-1 M KCl gradient in TMK buffer. Fractions that contained RNAP activity were combined and loaded onto a HiLoad 16/60 Superdex 200 gel filtration column (Pharmacia) equilibrated with TMK buffer containing 300 mM KCl, and the RNAP obtained from this column was used in promoter-specific in vitro transcription reactions. The polypeptides present in such a M. thermoautotrophicum ΔH RNAP preparation, silver stained following separation by tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (tricine-SDS-PAGE), are shown in Fig. 1. Their identities as specific MTH gene products were determined either following trypsin digestion by matrix-assisted laser-desorption/ionization-time of flight (MALDI-ToF) mass spectrometry (3) and/or following transfer to polyvinylidene difluoride membranes (Bio-Rad Laboratories, Hercules, Calif.) by Edman degradation and N-terminal amino acid sequencing at the University of California (Davis, Calif.) protein sequencing facility.
FIG. 1.
Identities of M. thermoautotrophicum ΔH RNAP subunits silver stained after separation by tricine-SDS-PAGE. Stetter et al. (24) identified eight subunits (designated by the letters O and A through G) in RNAP preparations from M. thermoautotrophicum Winter. Based on estimated sizes and very similar patterns of SDS-PAGE resolution, O and A through F appear to have been homologs of the M. thermoautotrophicum ΔH subunits here designated A′, B′, B", A", D, E′, and F, and subunit G was a mixture that contained the four polypeptides designated subunits H, L, K, and P. With the exception of subunits F and P, the M. thermoautotrophicum ΔH subunits are homologs of subunits identified previously for Sulfolobus acidocaldarius RNAP (14, 15). M. thermoautotrophicum ΔH RNAP preparations did not contain a homolog of the S. acidocaldarius subunit G, and the MTH0265 and MTH0040 gene products, predicted to be RNAP subunits E" (6.7 kDa) and N (6.4 kDa) (23), were not detected, although their presence may have been masked within the cluster of similarly sized, small subunits. Eucaryal and bacterial homologs of the M. thermoautotrophicum ΔH subunits are listed based on motif conservation and sequence alignments (5, 16, 21, 22). Members of the MTH1324 family (subunit F) have limited similarity to eucaryal Rpb4, a nonessential subunit of RNAPII that in yeast participates in transcription under stress conditions and at temperature extremes (4, 20). All members of the MTH0680.5 family (subunit P) contain four cysteinyl residues arranged in a manner consistent with a C-4 zinc finger motif and homology to Rpb12 (see Fig. 2C). .
For MALDI-ToF analysis, gel fragments containing the individual polypeptides were excised and cut into ∼1-mm cubes that were washed with water, dehydrated with acetonitrile, rehydrated in 100 mM ammonium bicarbonate containing 10 mM dithiothreitol (DTT), and incubated at 50°C for 30 min. The gel fragments were then again dehydrated with acetonitrile, rehydrated with 100 mM ammonium bicarbonate containing 50 mM iodoacetamide, incubated in the dark for 20 min, washed with ammonium bicarbonate, dehydrated with acetonitrile, blotted dry, and rehydrated in 50 mM ammonium bicarbonate containing 5 mM CaCl2 and 6.25 ng of trypsin/μl (Boehringer Mannheim, Indianapolis, Ind.). Following incubation on ice for 30 min, excess trypsin solution was removed and replaced with 50 mM ammonium bicarbonate containing 5 mM CaCl2, and incubation was continued at 37°C overnight. Tryptic peptides were eluted from the gel fragments by two sequential 30-min incubations in 100 mM ammonium bicarbonate, and the eluates were combined and acetic acid was added to 1%. The peptides were adsorbed onto a C18 reverse-phase resin, washed with 2% acetonitrile–1% acetic acid, eluted with 65% acetonitrile–1% acetic acid, and analyzed by MALDI-ToF mass spectrometry, using 0.5 μl of a matrix solution that contained 20 mg of α-cyano-4-hydroxy-trans-cinnamic acid (Sigma, St. Louis, Mo.) dissolved in 1 ml of 50% acetone–50% isopropanol–1% acetic acid on a Voyager Elite spectrometer (PerSeptive Biosystems, Framingham, Mass.) equipped with delayed extraction, a timed ion selector, and an ion reflector. Data were interpreted with the assistance of the search engine and nonredundant database maintained by Zhang and Chait (28).
Transcription in vitro by M. thermoautotrophicum ΔH RNAP with native and/or recombinant M. thermoautotrophicum TATA-binding protein (TBP) and TFB.
The template DNA was released from plasmid pRT74 (25) by DdeI digestion. Transcription initiated 24 bp downstream from the TATA box element of the hmtB promoter, resulting in a 193-nucleotide runoff transcript and extension of a 20-mer primer that hybridized to hmtB transcripts confirmed that hmtB transcription initiation occurred at the same site in vivo and in vitro (Fig. 3A). In vitro transcription reaction mixtures contained (in 100 μl) 20 mM Tris HCl (pH 8), 10 mM MgCl2, 120 mM KCl, 30 μM ATP, 30 μM CTP, 30 μM GTP, 2 μM UTP, 2 mM DTT, 2.2 μCi of [α-32P]UTP (3 kCi/mmol), 1 μg of DdeI-digested pRT74 DNA (25), 10 μl of RNAP, and 10 μl of partially purified native TBP (nTBP) and nTFB or 100 ng of purified recombinant TBP (rTBP) and 600 ng of rTFB. Following incubation for 30 min at 58°C, the proteins present were removed by phenol-chloroform extraction, and the RNA products were characterized by electrophoresis and autoradiography, as previously described (7, 9).
FIG. 3.
(A) Template DNA, obtained by DdeI digestion of plasmid pRT74 (25), and origin of the 193-nt runoff transcript. (B) Autoradiogram of in vitro-synthesized transcripts. A plus sign indicates that a reaction mixture contained the protein listed. Lane S, size standards; nt, nucleotides.
To obtain nTBP, fractions from the DEAE-cellulose column used to purify M. thermoautotrophicum RNAP were assayed for activity in an in vitro transcription system derived from Methanococcus thermolithotrophicus that lacked TBP (7). Active fractions were pooled, diluted with 50 mM Tris HCl (pH 8) to ∼115 mM KCl, and loaded onto a Q-Sepharose ion-exchange column. Unbound proteins were eluted with TMK buffer, and bound proteins were then eluted with TMK buffer containing a 50 mM-to-1 M gradient of KCl. The fractions containing TBP activity were pooled and loaded onto a HiLoad 16/60 Superdex 200 gel filtration column, and the partially purified nTBP that eluted from this column in TMK buffer containing 300 mM KCl was used in this study.
To obtain nTFB, M. thermoautotrophicum cells (0.5 g [wet weight]/ml) were suspended in TK buffer (50 mM Tris HCl [pH 8], 50 mM KCl, 20% [vol/vol] glycerol) and ruptured by passage twice through a French pressure cell at 20,000 lb/in2. The supernatant obtained from this lysate by centrifugation at 100,000 × g for 1 h at 4°C was loaded onto a phosphocellulose column (Whatman), unbound proteins were washed from the column with TK buffer, and the bound proteins were eluted with a 50 mM-to-1 M gradient of KCl in TK buffer. The fractions that contained the partially purified nTFB used in this study were identified by their activation of specific transcription when added to reaction mixtures that contained M. thermoautotrophicum RNAP and partially purified nTBP.
rTBP and rTFB preparations were generated and purified to establish a defined in vitro transcription system and to confirm that the partially purified nTBP and nTFB preparations contained these activities. MTH1627 was amplified by PCR from M. thermoautotrophicum ΔH genomic DNA by using primers with the sequences 5′-CATTGTCAAGATCGAAAACTGCAGGTT and 5′-GGGAGGTCTCGAGTTGACAG. The 604-bp product was digested with XhoI and PstI and ligated with XhoI-plus-PstI-digested pTrcHisA, resulting in plasmid pTD105, which was transformed into Escherichia coli Top 10 (Invitrogen, San Diego, Calif.). Isopropyl-β-d-thiogalactoside (IPTG; final concentration, 1 mM) was added to exponentially growing cultures of E. coli Top 10 (pTD105), and after 5 h of incubation at 37°C, the E. coli cells were concentrated by centrifugation, resuspended in a solution containing 50 mM sodium phosphate and 300 mM NaCl (pH 8), and lysed by passage at 20,000 lb/in2 through a French pressure cell. His tag-labeled rTBP was purified from this lysate by Ni-nitrilotriacetic acid (NTA) Superflow Ni2+ affinity chromatography (Qiagen, Chatsworth, Calif.) by following the manufacturer’s protocol. The same procedure was used to obtain His-tagged rTFB, except that MTH0885 was PCR amplified from genomic DNA using primers with the sequences 5′-GTTCTCTAACCTGCAGAAATTA and 5′-TGTGGATCCATGGGGGCGAAG, and the resulting 998-bp product was digested with PstI and BamHI and cloned into PstI-plus-BamHI-digested pTrcHisA, resulting in plasmid pTD103. Samples of the purified recombinant transcription factors were supplied to ICN Biochemicals (Cleveland, Ohio) to obtain rabbit anti-TBP and anti-TFB antibodies.
Sucrose gradient cosedimentation of M. thermoautotrophicum ΔH RNAP and TFB and immunoprecipitation of RNAP by anti-TFB antibodies.
Under anaerobic conditions, M. thermoautotrophicum cells resuspended (1 g [wet weight]/ml) in 50 mM Tris HCl (pH 8)–10 mM MgCl2–130 mM KCl were lysed by passage at 10,000 lb/in2 through a French pressure cell, and the resulting lysate was centrifuged at 14,000 rpm for 4 min in an Eppendorf microcentrifuge. An aliquot (500 μl) of the cleared supernatant was loaded on top of an anoxic 10-ml sucrose gradient (15 to 40% sucrose dissolved in 50 mM Tris HCl [pH 8]–10 mM MgCl2–130 mM KCl) and centrifuged at 25,000 rpm for 16.5 h at 4°C in a Beckman SW41 rotor. Fractions (500 μl) were collected and assayed for RNAP activity by measuring poly(dA-dT) directed [32P]UTP incorporation into trichloroacetic acid (TCA)-precipitable material and for the presence of TFB and TBP by Western blotting by using antisera raised against purified rTFB and rTBP.
Immunoprecipitation experiments.
Protein A-Sepharose preparations lacking antibodies or coupled to anti-TBP or anti-TFB immunoglobulin G antibodies were mixed and incubated for 3 h at 4°C with aliquots of cleared lysates of M. thermoautotrophicum cells. The matrices were collected by centrifugation, washed four times with TMK buffer containing 0.1% Tween 20 and 2 mM DTT, and then washed with TMK buffer containing 1 M KCl to disrupt protein-protein complexes. RNAP activity in the 1 M KCl eluates was measured by assaying poly(dA-dT)-directed incorporation of [32P]UTP into TCA-precipitable material.
Results. By using assays of poly(dA-dT) transcription resulting in [14C]ATP or [32P]UTP incorporation into TCA-precipitable material, RNAP preparations were isolated from several strains of M. thermoautotrophicum during the 1980s (1, 13, 24, 27). Consistent with the RNAPs isolated in parallel from other archaea (8, 27), these M. thermoautotrophicum enzymes were reported to contain 8 to 10 subunits and to more closely resemble eucaryal than bacterial RNAPs, but they could not be shown to initiate transcription accurately in vitro from methanogen promoters. The basis for this deficiency was apparently revealed when subsequent studies with other archaeal in vitro transcription systems demonstrated that archaeal homologs of both the eucaryal TBP and general transcription factor IIB, designated TFB in Archaea, were needed in addition to archaeal RNAP for accurate promoter-dependent transcription initiation (7, 9, 14, 18). In the M. thermoautotrophicum ΔH genome report (23), MTH1627 and MTH0885 were annotated as encoding TBP and TFB, respectively, and MTH genes encoding RNAP subunits A′, A", B′, B", D, E′, E", H, K, L, and N were so designated based on sequence similarities to RNAP subunits characterized previously from other archaea, primarily from S. acidocaldarius (14, 15). None of the MTH genes, however, encoded amino acid sequences related to those reported for S. acidocaldarius RNAP subunits F and G (15). As illustrated in Fig. 1, RNAP preparations purified from M. thermoautotrophicum ΔH contained seven polypeptides that were readily resolved by tricine-SDS-PAGE and four smaller polypeptides that migrated with similar electrophoretic mobilities. Each of these polypeptides was identified as the product of a specific MTH gene by MALDI-ToF analysis of tryptic peptides (3) and/or by Edman degradation and N-terminal amino acid sequencing. The six largest were confirmed as RNAP subunits A′, B′, B", A", D, and E′, encoded as predicted by MTH1051, MTH1050, MTH1049, MTH1052, MTH0037, and MTH0264, respectively (23), but the seventh largest was encoded by MTH1324, a gene not previously recognized as encoding an RNAP subunit. Similarly, three of the smaller polypeptides were confirmed as RNAP subunits H, L, and K, encoded as predicted by MTH1048, MTH1317 and MTH0042, respectively, but the fourth was encoded by a previously unannotated open reading frame located between MTH0680 and MTH0681, here designated MTH0680.5 (Fig. 2). Based on their consistent presence in near-stoichiometric amounts and their limited but detectable sequence similarities to eucaryal RNAP subunits Rpb4 and Rpb12 (16), the MTH1324 and MTH0680.5 gene products have been designated as RNAP polymerase subunits F and P, respectively (Fig. 1). Additional circumstantial support for this functional designation is provided by the genomic locations of MTH1324 and MTH0680.5 (both are directly downstream and potentially cotranscribed with ribosomal protein-encoding genes) and by the conservation of MTH1324 and MTH0680.5 homologs in all completed archaeal genome sequences (Fig. 2) (2, 6, 11, 12). The genomic organization of MTH1324 and MTH0680.5 homologs, directly downstream of rpL21- and rpL37a-encoding genes, respectively, is also conserved in Methanococcus jannaschii, Archaeoglobus fulgidus, and Pyrococcus horikoshii (2, 11, 12). The MTH1324 homolog and rpL21-encoding gene are also adjacent in the Pyrobaculum aerophilum genome but are transcribed divergently, and although an MTH0680.5 homolog and an rpL37a-encoding gene are present, they are not adjacent in this crenarchaeal genome (Fig. 2) (6).
FIG. 2.
Organization and conservation of the rpoF (A) and rpoP (B) regions in the genomes of M. thermoautotrophicum (MT), A. fulgidus (AF), M. jannaschii (MJ), P. horikoshii (PH), and P. aerophilum (PA) (2, 6, 11, 12, 23). Arrows indicate directions of transcription, shading patterns show homology between genes, and broken lines indicate nonadjacent locations. MTH1323 and MTH0681 and their homologs are predicted to encode rpL21 and rpL37a, respectively. MTH1325 and MTH0680 gene products and their homologs are unknown. Amino acid sequences (C) of MTH0680.5 (MT) and MJ0593.5 (MJ) aligned with their archaeal homologs and the sequences of Rpb12 from Saccharomyces cerevisiae (SC), Schizosaccharomyces pombe (SP), and Homo sapiens (HS) (21, 22). Identical and similar (indicated by asterisks) amino acid residues are identified in the conserved sequence. Four cysteinyl residues predicted to form a C-4 type of zinc finger are boxed. The numbers of amino acid residues present, but not shown, at the N termini of the eucaryal proteins are indicated by the numbers in parentheses. The PH homolog may have longer N-terminal sequences initiated 28 codons upstream at an in-frame ATG (11).
M. thermoautotrophicum RNAP initiated transcription accurately in vitro, using templates that carried the promoter for the M. thermoautotrophicum archaeal histone-encoding gene hmtB (25) when supplied with either partially purified preparations of nTBP and nTFB and/or recombinant His-tagged versions of these archaeal transcription factors purified from E. coli (Fig. 3). Primer extension experiments confirmed that hmtB transcription initiated at the same site, 24 nucleotides downstream from the TATA box element of the hmtB promoter, both in vivo and in vitro, and changing the TATA box sequence from 5′-TTTATATA to 5′-TTTGGATA eliminated transcription initiation in vitro. Accurate transcription initiation also occurred in vitro in reaction mixtures supplied with the heterologous templates that carried the tRNAVal and archaeal histone hmfB promoters from Methanococcus vannielii and Methanothermus fervidus, respectively, used previously in archaeal in vitro transcription systems (7, 9). Transcription initiation was not, however, detected in reaction mixtures supplied with templates that carried the M. thermoautotrophicum H2-regulated mcr, mrt, and ftr methane gene promoters (19) or with templates that contained the upstream intergenic and coding regions of the M. thermoautotrophicum TBP and TFB genes (results not shown). Based on these observations, it seemed likely that one or more additional factors were required to activate transcription from these promoters, but assays of many fractions obtained from M. thermoautotrophicum cell lysates by several different chromatographic procedures failed to detect such an activating factor. MTH1314 is predicted to encode a polypeptide related to eucaryal RNAPII subunit Rpb9 (also designated transcription elongation factor TFIIS [16, 23]), and this polypeptide was not present in the M. thermoautotrophicum RNAP preparations. MTH1314 was therefore PCR amplified, cloned, and expressed in E. coli; however, addition of the recombinant MTH1314 gene product, purified by His tag affinity chromatography from E. coli, also did not activate transcription from the H2-regulated mcr methane gene promoter in vitro.
Because very large, multicomponent RNAPII holoenzyme complexes are required for eucaryal transcription activation in vitro (10, 16, 17), it seemed possible that transcription initiation from the methane gene promoters might be detected if less-purified RNAP preparations, in which the RNAP activity remained part of a larger complex, were used. Such complexes exhibiting RNAP activity were therefore isolated by phosphocellulose chromatography, sucrose gradient sedimentation, and immunoprecipitation, but they did not exhibit detectable transcription initiation in vitro from the methane gene promoters. Considerable effort was made to minimize the exposure of cell extracts, complexes, and fractions to air, but the addition of 1 mM DTT was still needed to obtain transcription in vitro in reaction mixtures provided with the hmtB promoter-containing templates. In this regard, it is noteworthy that the C-terminal region of the subunit D (MTH0037) of M. thermoautotrophicum RNAP contains eight cysteinyl residues, consistent with the presence of a ferredoxin-like [4Fe-4S] center, and it remains possible that transient exposure to oxidizing conditions irreversibly inactivated the factor(s) needed for transcription in vitro from other M. thermoautotrophicum promoters. The ferredoxin-like motif is also present in subunit D of A. fulgidus and S. acidocaldarius RNAPs (19a).
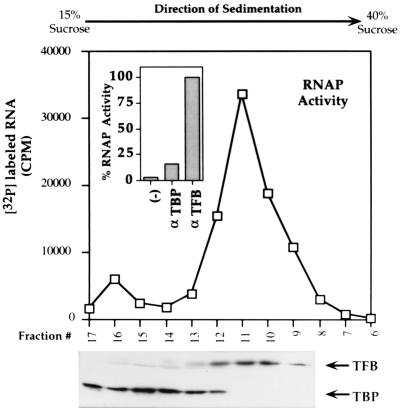
Further characterization of the large RNAP-containing complexes led to the discovery that M. thermoautotrophicum TFB and RNAP associate independently of TBP. They cosedimented through sucrose gradients, apparently within a complex that does not contain TBP (Fig. 4), and RNAP activity was immunoprecipitated from M. thermoautotrophicum cell lysates by protein A-Sepharose carrying anti-TFB antibodies, whereas very little RNAP activity bound to protein A-Sepharose carrying anti-TBP antibodies (Fig. 4). RNAP activity was also removed from M. thermoautotrophicum cell lysates by affinity to His-tagged TFB immobilized on Ni-NTA Superflow, whereas only background levels of RNAP activity bound to His-tagged TBP immobilized on Ni-NTA Superflow (results not shown).
FIG. 4.
Sucrose gradient cosedimentation of M. thermoautotrophicum ΔH RNAP and TFB and immunoprecipitation of RNAP by anti-TFB antibodies. Fractions containing TFB and TBP were identified by Western blotting. The inset shows the results of immunoprecipitation experiments with protein A-Sepharose preparations lacking antibodies (−) or coupled to anti-TBP (α-TBP) or anti-TFB (α-TFB). The average values for four separate experiments are shown, with the RNAP activity eluted from each matrix calculated as a percentage of the RNAP activity eluted from the matrix carrying anti-TFB antibodies.
Acknowledgments
This research was supported by grants from the U.S. Department of Energy (DE-FGO2-87ER13731), the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, the Medical Research Council (MRC) of Canada, and NATO (collaborative research grant 950733). T.J.D. was supported by a U.S. Air Force Predoctoral Fellowship, and A.M.E. was supported as an MRC scientist.
We thank J. Kane for providing the tRNA-expression plasmid pRI952 and S. Fitz-Gibbon and J. H. Miller for providing Pyrobaculum aerophilum genome data prior to publication.
REFERENCES
- 1.Brown J W, Reeve J N. Transcription initiation and a RNA polymerase binding site upstream of the purE gene of the archaebacterium Methanobacterium thermoautotrophicum strain ΔH. FEMS Microbiol Lett. 1989;60:131–136. doi: 10.1016/0378-1097(89)90495-3. [DOI] [PubMed] [Google Scholar]
- 2.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 3.Courchesne P L, Luethy R, Patterson S D. Comparison of in-gel and on-membrane digestion methods at low to sub-pmol level for subsequent peptide and fragment-ion mass analysis using matrix-assisted laser-desorption/ionization mass spectrometry. Electrophoresis. 1997;18:369–381. doi: 10.1002/elps.1150180311. [DOI] [PubMed] [Google Scholar]
- 4.Edwards A M, Kane C M, Young R A, Kornberg R D. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J Biol Chem. 1991;266:71–75. [PubMed] [Google Scholar]
- 5.Eloranta J J, Kato A, Teng M S, Weinzierl R O J. In vitro assembly of an archaeal D-L-N RNA polymerase subunit complex reveals a eukaryote-like structural arrangement. Nucleic Acids Res. 1998;26:5562–5567. doi: 10.1093/nar/26.24.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitz-Gibbon, S., and J. H. Miller. 1999. Personal communication.
- 7.Gohl H P, Gröndahl B, Thomm M. Promoter recognition in archaea is mediated by transcription factors: identification of transcription factor aTFB from Methanococcus thermolithotrophicus as archaeal TATA-binding protein. Nucleic Acids Res. 1995;23:3837–3841. doi: 10.1093/nar/23.19.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gropp F, Reiter W D, Sentenac A, Zillig W, Schnabel R, Thomm M, Stetter K O. Homologies of components of DNA-dependent RNA polymerases of archaebacteria, eukaryotes and eubacteria. Syst Appl Microbiol. 1986;7:95–101. [Google Scholar]
- 9.Hausner W, Wettach J, Hethke C, Thomm M. Two transcription factors related with the eucaryal transcription factors TATA-binding protein and transcription factor IIB direct promoter recognition by an archaeal RNA polymerase. J Biol Chem. 1996;271:30144–30148. doi: 10.1074/jbc.271.47.30144. [DOI] [PubMed] [Google Scholar]
- 10.Holstege F C P, Young R A. Transcriptional regulation: contending with complexity. Proc Natl Acad Sci USA. 1999;96:2–4. doi: 10.1073/pnas.96.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Aohi K, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:147–155. doi: 10.1093/dnares/5.2.147. [DOI] [PubMed] [Google Scholar]
- 12.Klenk H-P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Krypides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1998;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 13.Knaub S, Klein A. Specific transcription of cloned Methanobacterium thermoautotrophicum transcription units by homologous RNA polymerase in vitro. Nucleic Acids Res. 1990;18:1441–1446. doi: 10.1093/nar/18.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langer D, Hain J, Thuriaux P, Zillig W. Transcription in archaea: similarity to that in Eukarya. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanzendörfer M, Langer D, Hain J, Klenk H-P, Holz I, Arnold-Ammer I, Zillig W. Structure and function of the DNA-dependent RNA polymerase of Sulfolobus. Syst Appl Microbiol. 1994;16:156–164. [Google Scholar]
- 16.Myer V E, Young R A. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 17.Nikolov D B, Burley S K. RNA polymerase II transcription initiation: a structural view. Proc Natl Acad Sci USA. 1997;94:15–22. doi: 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qureshi S A, Bell S D, Jackson S P. Factor requirements for transcription in the archaeon Sulfolobus shibatae. EMBO J. 1997;16:2927–2936. doi: 10.1093/emboj/16.10.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reeve J N, Nölling J, Morgan R M, Smith D R. Methanogenesis: genes, genomes, and who’s on first? J Bacteriol. 1997;179:5975–5986. doi: 10.1128/jb.179.19.5975-5986.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Rodriguez-Monge L, Ouzounis C A. A ferrodoxin-like domain in RNA polymerase 30/40-kDa subunits. Trends Biochem Sci. 1998;23:169–170. doi: 10.1016/s0968-0004(98)01203-1. [DOI] [PubMed] [Google Scholar]
- 20.Rosenheck S, Choder M. Rpb4, a subunit of RNA polymerase II, enables the enzyme to transcribe at temperature extremes in vitro. J Bacteriol. 1998;180:6187–6192. doi: 10.1128/jb.180.23.6187-6192.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakurai H, Ishihama A. Gene organization and protein sequence of the small subunits of Schizosaccharomyces pombe RNA polymerase II. Gene. 1997;196:165–174. doi: 10.1016/s0378-1119(97)00222-9. [DOI] [PubMed] [Google Scholar]
- 22.Shpakovski G V, Acker J, Wintzerith M, Lacroix J-F, Thuriaux P, Vigneron M. Four subunits that are shared by the three classes of RNA polymerase are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4702–4710. doi: 10.1128/mcb.15.9.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum strain ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stetter K O, Winter J, Hartlieb R. DNA-dependent RNA polymerase of the archaebacterium Methanobacterium thermoautotrophicum. Zentbl Bakteriol Hyg I Abt Orig Teil C. 1980;1:201–214. [Google Scholar]
- 25.Tabassum R, Sandman K M, Reeve J N. HMt, a histone-related protein from Methanobacterium thermoautotrophicum ΔH. J Bacteriol. 1992;174:7890–7895. doi: 10.1128/jb.174.24.7890-7895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thauer R K, Hedderich R, Fischer R. Reactions and enzymes involved in methanogenesis from CO2 and H2. In: Ferry J M, editor. Methanogenesis, ecology, physiology, biochemistry and genetics. New York, N.Y: Chapman and Hall; 1993. pp. 209–252. [Google Scholar]
- 27.Thomm M, Madon J, Stetter K O. DNA-dependent RNA polymerases of the three orders of methanogens. Biol Chem Hoppe-Seyler. 1986;367:473–481. doi: 10.1515/bchm3.1986.367.1.473. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, W., and B. T. Chait. Search engine and database. [Online.] http://prowl.rochefeller.edu. [4 May 1999, last date accessed.]