Abstract
Infertility is a complex multifactorial problem that affects about 7% of men and 15% of couples worldwide. Many molecular mechanisms involved in male infertility. Destructive effects of infertility on the next generations are not well understood. Approximately 60-75% of male infertility cases have idiopathic causes, and there is a need for additional investigations other than routine examinations. Molecular factors that surround DNA, which are mitotically stable and independently regulate genome activity of DNA sequences, are known as epigenetics. The known epigenetic mechanisms are DNA methylation, histone modifications and non-coding RNAs. Prevalence of metabolic diseases has been increased dramatically because of changes in lifestyle and the current levels of inactivity. Metabolic disorders, such as obesity and diabetes, are prevalent reasons for male infertility; despite the association between metabolic diseases and male infertility, few studies have been conducted on the effects of epigenetic alterations associated with these diseases and sperm abnormalities. Diabetes can affect the reproductive system and testicular function at multiple levels; however, there are very few molecular and epigenetic studies related to sperm from males with diabetes. On the other hand, obesity has similar conditions, while male obesity is linked to notable alterations in the sperm molecular architecture affecting both function and embryo quality. Therefore, in this review article, we presented new and developed technologies to study different patterns of epigenetic changes, and explained the exact mechanisms of epigenetic changes linked to metabolic diseases and their relationship with male infertility.
Keywords: Diabetes, DNA Methylation, Epigenetics, Male Infertility, Obesity
Introduction
The concept of epigenetics was presented approximately about 30 years ago, after which researchers attributed cell inheritance to gene regulatory feedback loops, chromatin modifications (DNA methylation and histone modifications), and non-coding RNA (ncRNA) molecules, which are collectively referred to as "epigenomes" (1). Epigenetics is broadly defined as "molecular factors and processes that surround DNA, independently regulate genome activity of DNA sequence and they are mitotically stable" (2).
The term of "epigenetics" was first introduced by the English developmental biologist Conrad Hal Waddington in 1942. He published a research paper titled "The Genetic Assimilation of the Bithorax Phenotype" in 1956 and stated that environmental stimuli play important roles in the inheritance of an acquired trait in a population (3, 4). Gametes contain epigenetic information that plays a key role in embryonic development and any perturbation in the epigenome of gametes could alter the phenotype of the next generation offspring through epigenetic inheritance (5). Epigenetic mechanisms may be the main mediators in the occurrence and development of metabolic disorders and subsequent diseases (6). Study of the impact of epigenetic alterations on male infertility and complications in the next generation is an undeniable need and studies on this topic have recently been initiated.
Male infertility, as a complex multifactorial problem that affects about 7% of men and 15% of couples worldwide. It is commonly reported that both genetic and epigenetic factors play a role in infertility. Chronic diseases such as inflammations, obesity and infections as well as lifestyle choices and environmental factors play major roles in incidence and prevalence of infertility in men (7). Abnormal sperm epigenetic profiles are related to semen analysis parameters and reproductive function defined by embryo quality and abortion rate (8), In spermatozoa, DNA methylation and post-translational histone modifications are carriers of epigenetic signals. In addition, sperm-transmitted small RNA (sRNA) may also contribute to epigenetic inheritance (5). Abnormal epigenetic mechanisms play an important role in the pathology of infertility; therefore, new frontiers can be set in the search for infertility causes and their associated clinical manifestations. Here, we intended to review association of epigenetic alterations and metabolic diseases with male infertility.
Sperm genome and epigenome
Sperm nucleus has a very complex architecture. Sperm DNA methylation is substantially reduced in comparison with somatic cells. DNA methylation occurs at the cytosine residue of CpG dinucleotides in gene control regions, which are named CpG islands. These islands are frequently located at gene promoters. DNA methylation is an epigenetic landmark that results in regulation of gene transcription (9).
DNA is associated with histones and shapes the nucleosome structure in somatic cells; however, in mature sperm, only 5-10% of DNA wrap around histone octamers. In the rest, sperm DNA histones are replaced by protamines; thus, 90-95% of sperm DNA is bound to protamine. Only 5%-10% of the male genetic content is packed with retained paternal histones (10) and assist in sperm chromatin compression, which promotes motility, fertilization and sperm DNA protection. The remaining 5-10% of histones are essential and part of the sperm’s epigenetic signatures (11, 12). These histones could be targeted by biochemical modifications. Their alterations have the potential to activate or suppress expressions of genes organized with these histones.
In addition, mature spermatozoa contain small RNAs. These RNAs are implicated in regulation of gene transcription. The exact mechanism by which small RNAs act as another part of the epigenetic profile and lead to the regulation of gene expression is not known (9).
Effects of epigenetics on male infertility
Studies over the past decade have shown that in addition to effective genetic differences, epigenetic alterations such as DNA methylation, histone modification and RNA Significantly contributed to male infertility and health problems of their offspring (13). In the following, we explained each epigenetic changes in greater detail. Table 1 presents an overview of these changes.
Table 1.
The effect of some epigenetic changes on male fertility
|
| |||
|---|---|---|---|
| Epigenetic changes | Alterations in infertile individuals | Impact on male fertility | Reference |
|
| |||
| DNA methylation | Hypermethylation of MTHFR promoter | Idiopathic infertility | (14) |
| In repetitive sequences LINE-1, Alu Yb8, NBL2, D4Z4 | Control of the functional capacity of germ cells | (15) | |
| Hypermethylation of the RPS6KA2, APCS, JAM3/NCAPD3 and ANK2 genes | Oligospermia, abnormalities in some of the sperm chromosomes; reduced fertility | (16) | |
| Decreased methylation in H19 gene and increased methylation in the MEST and SNRPN genes | Associated with male infertility | (17) | |
| Methylation level of MEST, GNAS, LINE-1 | FSH and LH level | (18) | |
| Abnormal methylation of IGF-2, KCNQ-1 | Sperm DNA damage and impaired fertility | (19) | |
| Hypermethylation of SPATA4, SPATA5, SPATA6 | Oligozoospermia and infertility | (13) | |
| Hypomethylation of H19 | Multiple sperm defects, Infertility biomarker | (20) | |
| Alterations in methylation of the DEFB126, TPI1P3, PLCH2 and DLGAP2 genes | Abnormal embryo growth | (21) | |
| Histone modification | H3K4me2 activation; H3K27me3 suppressive changes | Fetal growth and formation | (22) |
| H2A ubiquitination (ubH2A) and histone 3 K18 acetylation (H3AcK18); acetylation of four histones: H4 K5, K8, K12 and K16 (H4tetraAck) | Disruption of protamine 1 (Prm1) deposition in the testes | (23) | |
| testis specific histone H2B variant (TH2B) | Abnormal nucleus regeneration during spermiogenesis | (24) | |
| Non coding RNAs | Reduction of HOTTIP expression | Promotes proliferation of testicular embryonal carcinoma cells | (25) |
| high expression of lnc32058, lnc09522 and lnc98497 | Immotile sperm | (26) | |
|
| |||
FSH; Follicle - stimulating hormone and LH; Luteinizing hormone.
DNA methylation
DNA methylation can regulate gene expression using different mechanisms. DNA methylation plays a key role in gene expression; therefore, coordination of the increase, maintenance and elimination of methylation between tissues must be carefully controlled. This regulation is mediated by function of the methylation enzymes, such as DNA methyltransferases (DNMT1, DNMT3A, DNMT3B and DNMT3L) and TET protein family (TET1, TET2 and TET3). TET protein family was recently discovered to mediate active demethylation processes, particularly after fertilization (27).
DNA methylation may potentially influence postfertilization processes. Most signs of paternal methylation are actively corrected during epigenetic reprogramming after fertilization. However, some areas affected by incorrect methylation escape reprogramming and may pass this methylation status to the developing embryo (9, 28). Hypermethylation can suppress gene expression as methyl groups prevent recruitment of transcription factors and DNA polymerases. On the other hand, hypomethylation up-regulates gene expression (20). The results of a study showed that most epigenetic changes occurred at the pachytene stage of spermatocytes, which might lead to a large change in DNA methylation at this growth stage. Observations suggested that primary germ cells, prospermatogonia and spermatogonial stem cells had DNA methylation profiles correlating with the epigenetic programming cascade (29). Accordingly, inappropriate hypomethylation and hypermethylation can affect proper sperm function.
Genomic imprinting is an epigenetic modification process that allows gene to be expressed in a certain way by parents and it plays an essential role in normal growth and development. Expressions of imprinted genes are determined by the parents (30). Studies on DNA methylation in infertility have mostly concentrated on imprinted genes like H19, IGF2, MEST, PEG3, LIT1, SNRPN and KCNQ1 in addition to some non-imprinted genes, like MTHFR and DAZL. These studies showed that alterations in DNA methylation of these genes are associated with abnormal semen parameters and spermatogenesis. Recently, upon analyzing methylation across the genome, a study found varying levels of methylation in a number of genes, including some of the SPATA family members (SPATA4, SPATA5 and SPATA6). These researchers reported that hypermethylation of these genes is associated with infertility of oligozoospermia men (13, 17). In this context, the results of several studies have shown a close relationship between changes of cytosine methylation and H19 gene expression in males with infertility. Therefore, H19 hypomethylation is a proposed epigenomic infertility biomarker that could assess oligospermia in men with various sperm defects (20). Moreover, methylation of the MEST, GNAS and LINE1 genes is significantly associated with sperm concentration, blood follicle stimulating hormone (FSH) and luteinizing hormone (LH) levels (18). Thus, degree of methylation of the MEST and GNAS genes is significantly associated with increased levels of LH. Likewise, LINE1 methylation is remarkably correlated by increased FSH levels (18). Abnormal methylation of the IGF2 and KCNQ1 genes is associated with DNA damage in sperm and consequent impaired fertility (19). Taken together, these studies suggested that abnormal methylation patterns in imprinted and non-imprinted genes or epimutation may play role in male infertility by causing abnormal spermatogenesis.
Histone modifications
Despite their relatively short history, histone modifications and histone methylation are one of the most important parts of epigenetic research, due to their important role in regulating transcription of gene expression in various organisms. During spermatogenesis, structure of chromatin changes due to histone modifications (31). Histones are sensitive to post-translational modification (PTM). These modifications include acetylation, methylation, phosphorylation, ubiquitination, sumoylation, and glycosylation among the others. Histone PTMs collectively act as "epigenetic codes" to activate transcription, suppression and coordination of chromatin structure in a more orderly fashion (10).
Environmental factors appear to alter histone modifications to some degree by directly regulating level and/or activity of histone-modifying enzymes. For example, hypoxia and nickel exposure increase H3K9me2 levels by inhibiting the histone demethylase JMJD1A (32).
Some cellular metabolites directly regulate expression of metabolic genes through histone modification. Lysinespecific demethylase-1-dependent flavin adenosine dinucleotide (LSD1-dependent FAD) is an enzyme which can degrade histones and regulate cellular energy levels by suppressing genes involved in mitochondrial respiration and energy consumption (33). In human spermatozoa, about 5-10% of histones are conserved and precise substitution of histone by protamine is critical for normal sperm production. The relationship between intracellular metabolites, histone signatures and their effect on gene transcription may play a role in disease progression. Therefore, some infertility and sterility problems may be attributed to abnormal histone modifications in spermatozoa.
Non-coding RNAs (ncRNAs)
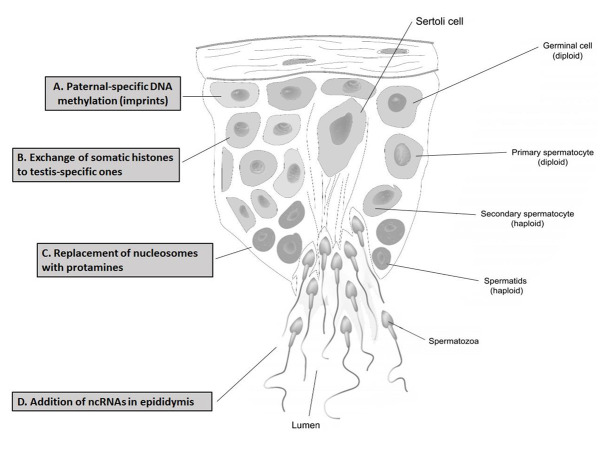
Mammalian sperm RNAs is a source of paternal hereditary information beyond DNA. Environmental factors that include imbalanced diet, mental stress and exposure to toxins can alter sperm RNAs and cause phenotypes related to paternal environmental stress in offspring (34). Among the different types of RNA, messenger RNA (mRNA), transporter RNA (tRNA) and ribosomal RNA (rRNA) are the best-known RNAs in all organisms. In addition to these, RNAs can be broadly divided into coding RNA (cRNA) and ncRNA (35). Mature spermatozoa have several types of small ncRNAs that include silencing RNAs (siRNAs) and microRNAs (miRNAs), which vary in length and generally lack an open reading frame. miRNAs are associated with molecular mechanisms regulating spermatogenesis, particularly endogenous genes in germline cells which regulate their complex process of renewal and/or differentiation. Recent studies identified another new class of siRNAs, PIWI-interacting RNAs (piRNAs), which are expressed in testes during spermatogenesis (36). Hypomethylation of repetitive elements in the male germline was associated with an increase in miR-29, proposed to reduce DNMT3a, a protein required for genomic methylation (37). In a study of male infertility, researchers identified 9879 long noncoding RNAs (lncRNAs) with differential expressions; only three (lnc32058, lnc09522 and lnc98497) showed high expressions in immotile spermatozoa compared to normal motile spermatozoa. Several lncRNAs (Mrhl, Drm, Spga-lncRNAs, NLC1-C, HongrES2, Tsx, lncRNA-tcam1, Tug1, Tesra, AK015322, Gm2044 and lncRNA033862) were confirmed to have functionally distinct roles in spermatogenesis (26). Recent evidence suggested that small RNAs (tsRNAs) derived from sperm tRNA, as a carrier of paternal epigenetic information, may mediate intergenerational inheritance (38). In the paternal high fat diet (HFD) mouse model, a subset of tsRNAs, with size range of 30-34 nucleotides, led to changes in the RNA expression profile and modifications. Injection of sperm tsRNA from HFD male mice into normal zygotes also caused metabolic disorders in F1 offspring and impaired the expressions of metabolic pathway genes in early embryos. Thus, sperm tsRNAs represent a paternal epigenetic factor that might cause intergenerational inheritance of dietary metabolic disorders (39). Based on these findings, it can be concluded that many lncRNAs have potential impacts on male spermatogenesis and infertility, but very few have thus far been identified and confirmed. Comparing the results of these studies may lead to recognition of important lncRNAs for spermatogenesis. This can also be used as markers for infertility in men with metabolic diseases. Figure 1 provides a summary of the epigenetics events in the testis.
Fig 1.
The seminiferous tubule regions of testis and possible epigenetics events in male fertility. Each process of epigenetics occurs in specific cells and regions of the testis: A. Paternal-specific DNA methylation in spermatogonia, B. Exchange of somatic histones to testis-specific ones in primary spermatocyte, C. Replacement of nucleosomes with protamines in spermatid, and D. Addition of non-coding RNAs in the epididymis.
Metabolic syndrome
Metabolic syndrome (MetS) is a complex pathophysiological state caused by an imbalance between calorie intake and consumption. Urbanization and lack of physical activity as well as an excessive and poor quality diet can lead to MetS. This metabolic syndrome is defined as a cluster of at least three out of five medical indicators: abdominal obesity, high blood pressure, high blood sugar, increased triglyceride levels and reduced high-density lipoprotein (HDL) level. MetS increases risk of stroke, cardiovascular disease and type 2 diabetes mellitus (40).
MetS is associated with disruption of hypothalamicpituitary-testes (HPT) axis, as well as reduction of gonadotropins and steroidogenesis, specifically testosterone levels in men. In MetS, systemic chronic inflammation mediates hypogonadism, erectile dysfunction and poor spermatogenesis. Actually, oxidative stress cause inflammation in MetS. It finally leads to loss of DNA integrity, improper packaging and aberrant modification of sperm DNA. In other words, there is a change in MetS and sperm epigenetics of obese individuals. This change can be passed to the next generation. Thus, offspring will be susceptible to metabolic and reproductive problems (40, 41). The idea of paternal effects on offspring health is an exciting area for research and clinical studies.
Metabolic disorders and male infertility: epigenetics approaches
Study of the association of MetS with epigenetic aspects of male infertility is a spectacular, emerging topic for clinical and basic research. The main problem is multifactorial nature of these disorders. Metabolic disorders may be only one of the common causes of male infertility; however, infertile men suffering metabolic disorders during middle age. On the other hand, father lifestyle plays a key role in influencing spermatogenesis for future of child, as it has been proven that obesity, unhealthy diet, inactivity and pollution are factors that can alter sperm counts and sperm quality through epigenetics (42).
Previous studies supported the hypothesis that metabolic changes related to environmental factors can be passed from father to the offspring. These metabolic disorders acquired in offspring may be partly explained by potential genetic information carriers like epigenetic agents and their processes (38).
Diabetes
Association of type 2 diabetes with reproductive dysfunction is a serious challenge, due to the high prevalence in young people. Diabetes can affect reproductive function in the testes at multiple levels, including changes in sperm quality and spermatogenesis, and it may lower testosterone levels. Epigenetic alterations are important risk factors for diabetic complications (43, 44).
Glucose metabolism plays a vital role in spermatogenesis. Numerous studies in humans and animals have confirmed the effects of diabetes on sexual function, semen parameters, nuclear DNA and chromatin quality (45, 46). But there are few studies on the pattern of DNA methylation in spermatozoa from type 2 diabetic patients at the genome level (47). In 2019, Chen et al. (48) investigated the whole-genome DNA methylation profile of human spermatozoa by comparing eight individuals with type 2 diabetes and nine healthy individuals using the whole-genome bisulfite sequencing method. The results showed that ratio of methylated cytosine in the whole genome of type 2 diabetics was lower than the control group. They also identified differential methylation of 10 genes: IRS1, PRKCE, FTO, PPARGC1A, KCNQ1, ATP10A, GHR, CREB1, PRKAR1A and HNF1B in men with type 2 diabetes.
The results of previous studies have shown that abnormal levels of some lncRNAs can accelerate progression of diabetic retinopathy (49). Therefore, expression profiles of lncRNAs in the sperm of diabetic mice were studied to determine genes that might be related to reproduction and their association with the onset of diabetes. lncRNAs functions were assessed by subgroup analysis and their physical or functional relationships with the corresponding mRNAs. Expression profiles from six microarray evaluations of diabetic mice spermatozoa showed differential expressions of lncRNAs and mRNAs (4134 up-regulation and 3407 down-regulation of lncRNAs, 2590 up-regulation and 3507 down-regulation of coding-mRNAs, in sperm samples from mice diabetic mice group and control group, respectively). Genetics and pathway analysis revealed that function of mRNAs with differential expression was closely related to many processes associated with development of diabetes. In addition, study of lncRNAs and mRNAs identified potential nuclear genes that might play a substantial role in the pathogenesis of diabetes-related infertility (50).
Molecular studies that involved sperm of diabetic men are generally rare. Majority of men are unaware of their current status and develop diabetes later in life (47). Therefore, it is postulated that future of research on epigenetics and male infertility should focus on identifying the putative molecular mechanisms of effects of diabetes on epigenetic abnormalities during spermatogenesis.
Obesity
The interaction between obesity and diabetes is currently established; but more research has been conducted in the field of infertile obese men epigenetics rather than investigations on epigenetics of infertile diabetics. Male obesity is associated with notable alterations in sperm molecular composition, not only impairing the sperm function, but also posing risks to embryo (37). Many studies have shown abnormalities in sperm quality, particularly decreased sperm concentrations in over weight patients with high body mass index (BMI) of 25- 29 Kg/m2 . Two recent meta-analyses, including 14 and 21 studies, reported an increased risk of azoospermia or oligozoospermia in obese men. In this regard, results of one study showed association of oxidative stress and sperm DNA fragmentation with sperm DNA methylation in men (51). In conclusion, it has been shown that spermatozoa epigenetic pattern changes in men with high BMI resulted in alteration of sperm DNA methylation. Thus, this abnormality is directly related to male infertility, low embryo quality and a decrease in fertility (52).
Obesity changes methylation status of DNA in other tissues. Soubry et al. compared sperm DNA methylation in 12 differentially methylated regions (DMRs) of 23 obese compared to 44 normal weight men. Percentage of DNA methylation was decreased in MEG3, SNRPN and SGCE/PEG10, while it was increased in MEG3-intergenic differentially methylated region (MEG3-IG DMR) and H19 DMR in the sperm of the overweight/obese men (37, 53). Donkin et al. (54) focused on the regeneration of sperm methylation after weight loss by gastric bypass (GBP) in obese patients (mean BMI=31.8 Kg/m2). The results showed that methylation status of 1509 unique genes was changed one week after surgery. In addition, 3910 unique genes had a different methylation status one year after GBP rather than before GBP. Additionally, sperm histone status was not changed in the lean and obese men, and sperm from the obese men altered small non-coding RNAs (sncRNAs) expression. These findings suggested that weight loss caused a gradual change in the sperm epigenome. Future efforts to determine epigenetic profile of human sperm exposed to other environmental factors, such as exercise or smoking, might reveal specific signatures which could influence metabolic health of future generations in positive or detrimental ways.
A recent study examined epigenetic changes of sperm, in relation to BMI. In this study, 144 samples (48.97%) were "normal weight" (BMI: 19.00-24.90) and 149 samples (50.68%) were "pre-obese/obese" (BMI: 25.00- 40.30). Methylation levels of paternally imprinted genes (H19-IG DMR, IGF2-DMR0 and MEG3-IG DMR) and non-imprinted gene regions associating with obesity were reported. Regression analysis showed positive correlation between BMI and MEG3-IG DMR methylation in sperm DNA (55). These results suggested that obesity is related to sperm DNA methylation programming in humans and it may influence epigenome of the next generation.
Changes in histone of sperm DNA is another parameter that is affected by obesity. Expression of related genes to sperm motility, histone 3 and 4 modifications and posttranslation global modification process in the testes of obese mice were reduced compared to the control group. In addition, the results of quantitative Western blot analysis showed a decrease in H3K23pr, H4K8cr, H3K122ac and H4K8ac in the testes of mice that consumed a HFD. These findings suggested that altered gene expression and PTM are associated with impaired reproductive function in obese males (56).
In addition, abnormal lncRNA expression from HFD also causes epigenetic changes. There was a significant difference between lncRNAs and mRNA expressions in the sperm of obese mice fed HFD compared to mice that received normal diets. Neat1 and Malat1 expression levels were lower in obese mice compared to normal weight mice. NEAT1 is expressed in human embryonic stem cells and plays role in spermatogenesis regulation. Decreased expression of Neat1 is associated with reduced sperm quality and fertility, and its expression is negatively regulated by Malat1 (57). Malat1, a long non-coding RNA, is present in active loci and it utilizes splicing factors. An increase in Malat1 is recognized in all sperm samples and extra-nuclear areas. Enrichment of Malat1 in sperm is due to the triple helix structure at its 3ʹ end. This RNA is associated with histone bound DNA of sperm and it plays role in chromatin remodeling (58).
Overall, male obesity may affect all aspects of sperm epigenetics including patterns of DNA methylation, histone modifications and ncRNA expression in spermatozoa. It ultimately leads to male infertility. Although the amount of RNA in spermatozoa is not significant, it has potential role in the clinical investigation and diagnosis of male infertility. It is also one of the factors specifically expressed in germ cells and present in mature spermatozoa. Therefore, RNAs are suitable molecular markers for detection of cell lineages in the spermatogenesis pathway. These markers can provide an overview of spermatogenesis status in infertile cases instead of invasive testicular biopsy.
Unexplained infertility
The molecular mechanisms involved in male infertility are not well understood yet and diagnosis of unexplained or idiopathic male infertility is made in cases where standard tests are unable to determine the cause. Such cases account for about 60-75% of male infertility. Uniquely, epigenetic changes such as DNA methylation and histone modifications have significant impact on this problem (14, 59).
Although different DNA methylation patterns of germ cells are associated with changes in sperm quality, few studies focused on the epigenetic investigation of infertile men with normal sperm parameters. For the first time in 2015, a genome-wide study examined sperm DNA methylation in patients with idiopathic infertility compared to fertile men. In this study, approximately 3000 CpG were detected which indicated defective methylation. These results suggested that these changes are precisely related to specific regions of sperm methylation, and it can be concluded that DNA methylation plays a role in controlling germ cell function (15). Results obtained from one study indicated that hypermethylation of MTHFR gene promoter is strongly associated with idiopathic infertility in males (14). Therefore, analysis of promoter methylation in specific genes may provide valuable biomarkers to identify men who are at high risk for infertility. Recently, a molecular experiment was conducted to identify male idiopathic infertility by using DNA methylation variations in sperm. The results of this study showed regions of DMRs in males with idiopathic infertility. In this study, researchers identified an epigenetic DMR signature of male infertility that can be used both as a diagnostic tool and to detect FSH response in the studied patient population (60). It is predicted that further development of this technology will improve diagnosis and management of infertile male patients, overall treatment options and development of therapies.
To date, much attention has been paid to the cellular and molecular mechanisms, in terms of infertility in men with asthenospermia (61, 62). Idiopathic asthenospermia (IAS) falls into this group, but its cause is unknown. IAS is one of the major causes of male infertility diagnosed by decreased sperm motility for which there is no effective treatment (63). Quantification of methylation specific polymerase chain reaction (MS-PCR) data has confirmed the significantly lower methylation level of DAZ3 promoter in IAS patients compared to normozoospermic men (64). Moreover, by studying promoter area methylation changes of the other genes in men with idiopathic infertility, it was found that promoter methylation profiles of the MLH1 and MSH2 genes might be involved in sperm DNA packaging and sperm parameters, respectively (65). However, hypermethylation of MTHFR gene promoter in spermatozoa also appears to be associated with idiopathic male infertility (66). Although idiopathic infertility is associated with an increased risk of developing MetS, obesity, and an increased risk of subsequent cardiovascular disease, idiopathic infertility is controversial because the underlying mechanisms remain unknown (67).
Future perspective
Sperm epigenome is remarkably specific. It offers interesting opportunities for more research and can be assessed in clinical trials. Many attempts have been made to understand nature of this epigenetic perspective, in addition to role of sperm epigenetic patterns in normal sperm development and function, as well as in male fertility. Investigating these factors, along with epigenetic changes in sperm DNA, remains an intriguing target for dissemination of biomarkers that may be able to more accurately identify cause of male infertility.
The finding that "newly fertilized gametes and embryos are sensitive stages for epigenetic changes in the environment" has important implications as changes in lifestyle and reproductive system environment may have long-term consequences for child health, while they have not been fully explored yet. Understanding how and when metabolic diseases -such as obesity and diabetesaffect male fertility and finding ways to modify abnormal epigenetic signatures, in addition to determining the best time frame for reversing abnormal epigenetic symptoms may improve fertility in infertile men.
Currently, most environmental research on the sperm epigenome focuses on DNA methylation. Recently, Abbasi et al. (46) opened a new horizon in terms of the stable, repeatable platform to measure changes in sperm DNA methylation. Uniquely, they suggested that Illumina Infinium platform is highly suitable for diagnostic use in a clinical setting. Progress in identifying other environmentally sensitive epigenetic mechanisms, including histone modifications, three-dimensional conformations and chromatin structure, as well as exosome loading and expression of ncRNA in seminal plasma could be important.
Conclusion
It is well-known that sperm is more than a cell that carries half of the genome and it has a unique epigenome. Any alteration in this epigenome can lead to infertility, abnormal fetal development, and increased risk of certain diseases in the next generation. In vitro fertilization (IVF) treatment cycles were repeatedly failed in some patients carrying obesity and diabetes while they may have normal sperm parameters. To date, new steps have been taken to understand methylation status in patients who refer to infertility centers. Future of this science likely depends on finding epigenetic codes along with epimutations in these individuals. It should be noted that finding epimutations alone is not sufficient and targeted treatment strategies should be sought. Perhaps these efforts can address large gap that exists in the treatment of infertile patients and prevent recurrent miscarriages.
Acknowledgements
This paper was related to a Ph.D. dissertation (research code: 97000124). It was supported by Royan Institute (Tehran, Iran) and the Faculty of Basic Sciences and Advanced Medical Technologies, Royan Institute, ACECR, Tehran, Iran. This study was conducted without any financial support and conflict of interest.
Authors’ Contributions
M.J.; Designed the manuscript as a Ph.D. student, the main contributor and contributed to writing the manuscript's draft. A.R.A.; Designed the manuscript, contributed to the revision process, and performed manuscript editing. M.A.S.G.; Edited and approved the final manuscript. P.E.-Y.; Contributed to prepare the final manuscript. M.Sh.; Contributed to study conception and manuscript editing. A.Sh.; Supervised this project and contributed to approval of the final manuscript. All authors read and approved the final manuscript.
References
- 1.Horsthemke B. A critical view on transgenerational epigenetic inheritance in humans. Nat Commun. 2018;9(1):1–4. doi: 10.1038/s41467-018-05445-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shnorhavorian M, Schwartz SM, Stansfeld B, Sadler-Riggleman I, Beck D, Skinner MK. Differential DNA methylation regions in adult human sperm following adolescent chemotherapy: potential for epigenetic inheritance. PLoS One. 2017;12(2):e0170085–e0170085. doi: 10.1371/journal.pone.0170085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holliday R. Epigenetics: a historical overview. Epigenetics. 2006;1(2):76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- 4.Noble D. Conrad Waddington and the origin of epigenetics. J Exp Biol. 2015;218(6):816–818. doi: 10.1242/jeb.120071. [DOI] [PubMed] [Google Scholar]
- 5.Ingerslev LR, Donkin I, Fabre O, Versteyhe S, Mechta M, Pattamaprapanont P, et al. Endurance training remodels sperm-borne small RNA expression and methylation at neurological gene hotspots. Clin Epigenet. 2018;10(1):1–11. doi: 10.1186/s13148-018-0446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Nigris F, Cacciatore F, Mancini FP, Vitale DF, Mansueto G, D’Armiento FP, et al. Epigenetic hallmarks of fetal early atherosclerotic lesions in humans. JAMA Cardiol. 2018;3(12):1184–1191. doi: 10.1001/jamacardio.2018.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adiga D, Eswaran S, Sriharikrishnaa S, Khan GN, Kabekkodu SP. Role of epigenetic changes in reproductive inflammation and male infertility. Chem Biol Lett. 2020;7(2):140–155. [Google Scholar]
- 8.Jenkins TG, Turek PJ. In: Male infertility.Springer, Cham. Parekattil S, Esteves S, Agarwal A, editors. Springer, Cham; 2020. Epigenetics and male infertility; pp. 139–146. [Google Scholar]
- 9.James E, Jenkins TG. Epigenetics, infertility, and cancer: future directions. Fertil Steril. 2018;109(1):27–32. doi: 10.1016/j.fertnstert.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Rashki Ghaleno L, Alizadeh A, Drevet JR, Shahverdi A, Valojerdi MR. Oxidation of sperm dna and male infertility. Antioxidants. 2021;10(1):97–97. doi: 10.3390/antiox10010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibrahim Y, Hotaling J. Sperm epigenetics and its impact on male fertility, pregnancy loss, and somatic health of future offsprings. Semin Reprod Med. 2018;36(3-04):233–239. doi: 10.1055/s-0038-1677047. [DOI] [PubMed] [Google Scholar]
- 12.Schon SB, Luense LJ, Wang X, Bartolomei MS, Coutifaris C, Garcia BA, et al. Histone modification signatures in human sperm distinguish clinical abnormalities. Assist Reprod Genet. 2019;36(2):267–275. doi: 10.1007/s10815-018-1354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sujit KM, Singh V, Trivedi S, Singh K, Gupta G, Rajender S. Increased DNA methylation in the spermatogenesis-associated (SPATA) genes correlates with infertility. Andrology. 2020;8(3):602–609. doi: 10.1111/andr.12742. [DOI] [PubMed] [Google Scholar]
- 14.Wu W, Shen O, Qin Y, Niu X, Lu C, Xia Y, et al. Idiopathic male infertility is strongly associated with aberrant promoter methylation of methylenetetrahydrofolate reductase (MTHFR) PLoS one. 2010;5(11):e13884–e13884. doi: 10.1371/journal.pone.0013884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urdinguio RG, Bayón GF, Dmitrijeva M, Toraño EG, Bravo C, Fraga MF, et al. Aberrant DNA methylation patterns of spermatozoa in men with unexplained infertility. Hum Reprod. 2015;30(5):1014–1028. doi: 10.1093/humrep/dev053. [DOI] [PubMed] [Google Scholar]
- 16.Camprubí C, Salas-Huetos A, Aiese-Cigliano R, Godo A, Pons M-C, Castellano G, et al. Spermatozoa from infertile patients exhibit differences of DNA methylation associated with spermatogenesis- related processes: an array-based analysis. Reprod Biomed Online. 2016;33(6):709–719. doi: 10.1016/j.rbmo.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Santi D, De Vincentis S, Magnani E, Spaggiari G. Impairment of sperm DNA methylation in male infertility: a metaanalytic study. Andrology. 2017;5(4):695–703. doi: 10.1111/andr.12379. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W, Li M, Sun F, Xu X, Zhang Z, Liu J, et al. Association of sperm methylation at LINE-1, four candidate genes, and nicotine/ alcohol exposure with the risk of infertility. Front Genet. 2019;10:1001–1001. doi: 10.3389/fgene.2019.01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni W, Pan C, Pan Q, Fei Q, Huang X, Zhang C. Methylation levels of IGF2 and KCNQ1 in spermatozoa from infertile men are associated with sperm DNA damage. Andrologia. 2019;51(5):e13239–e13239. doi: 10.1111/and.13239. [DOI] [PubMed] [Google Scholar]
- 20.Al-Qazzaz HK, Al-Awadi SJ. Epigenetic Alteration in DNA methylation pattern and gene expression level using H19 on oligospermia patients in Iraqi Men. Gene Reports. 2020;20:100780–100780. [Google Scholar]
- 21.Oluwayiose OA, Wu H, Saddiki H, Whitcomb BW, Balzer LB, Brandon N, et al. Sperm DNA methylation mediates the association of male age on reproductive outcomes among couples undergoing infertility treatment. Sci Rep. 2021;11(1):1–14. doi: 10.1038/s41598-020-80857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliva R, Ballescà JL. Altered histone retention and epigenetic modifications in the sperm of infertile men. Asian J Androl. 2012;14(2):239–240. doi: 10.1038/aja.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu L, Wei Y, Li H, Li W, Gu C, Sun J, et al. The ubiquitination and acetylation of histones are associated with male reproductive disorders induced by chronic exposure to arsenite. Toxicol Appl Pharmacol. 2020;408:115253–115253. doi: 10.1016/j.taap.2020.115253. [DOI] [PubMed] [Google Scholar]
- 24.Patankar A, Gajbhiye R, Surve S, Parte P. Epigenetic landscape of testis specific histone H2B variant and its influence on sperm function. Clin Epigenet. 2021;13(1):1–18. doi: 10.1186/s13148-021-01088-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su Y, Zhou LL, Zhang YQ, Ni LY. Long noncoding RNA HOTTIP is associated with male infertility and promotes testicular embryonal carcinoma cell proliferation. Mol Genet Genomic Med. 2019;7(9):e870–e870. doi: 10.1002/mgg3.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi M, Rajender S. Long non-coding RNAs (lncRNAs) in spermatogenesis and male infertility. Reprod Biol Endocrinol. 2020;18(1):1–18. doi: 10.1186/s12958-020-00660-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcho C, Oluwayiose OA, Pilsner JR. The preconception environment and sperm epigenetics. Andrology. 2020;8(4):924–942. doi: 10.1111/andr.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tunc O, Tremellen K. Oxidative DNA damage impairs global sperm DNA methylation in infertile men. J Assist Reprod Genet. 2009;26(9):537–544. doi: 10.1007/s10815-009-9346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skinner MK, Nilsson E, Sadler-Riggleman I, Beck D, Ben Maamar M, McCarrey JR. Transgenerational sperm DNA methylation epimutation developmental origins following ancestral vinclozolin exposure. Epigenetics. 2019;14(7):721–739. doi: 10.1080/15592294.2019.1614417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishida M, Moore GE. The role of imprinted genes in humans. Mol Aspects Med. 2013;34(4):826–840. doi: 10.1016/j.mam.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 31.Okada Y, Tateishi K, Zhang Y. Histone demethylase JHDM2A is involved in male infertility and obesity. J Androl. 2010;31(1):75–78. doi: 10.2164/jandrol.109.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang L, Wuptra K, Chen D, Li H, Huang S-K, Jin C, et al. Environmental- stress-induced chromatin regulation and its heritability. J Carcinog Mutagen. 2014;5(1):22058–22058. doi: 10.4172/2157-2518.1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hino S, Sakamoto A, Nagaoka K, Anan K, Wang Y, Mimasu S, et al. FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat Commun. 2012;3(1):1–12. doi: 10.1038/ncomms1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Shi J, Rassoulzadegan M, Tuorto F, Chen Q. Sperm RNA code programmes the metabolic health of offspring. Nat Rev Endocrinol. 2019;15(8):489–498. doi: 10.1038/s41574-019-0226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abraham M, Dror O, Nussinov R, Wolfson HJ. Analysis and classification of RNA tertiary structures. RNA. 2008;14(11):2274–2289. doi: 10.1261/rna.853208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shukla K, Chambial S, Dwivedi S, Misra S, Sharma P. Recent scenario of obesity and male fertility. Andrology. 2014;2(6):809–818. doi: 10.1111/andr.270. [DOI] [PubMed] [Google Scholar]
- 37.Palmer NO, Bakos HW, Fullston T, Lane M. Impact of obesity on male fertility, sperm function and molecular composition. Spermatogenesis. 2012;2(4):253–263. doi: 10.4161/spmg.21362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan M, Zhai Q. Sperm tsRNAs and acquired metabolic disorders. J Endocrinol. 2016;230(3):F13–F18. doi: 10.1530/JOE-16-0185. [DOI] [PubMed] [Google Scholar]
- 39.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 40.Leisegang K, Sengupta P, Agarwal A, Henkel R. Obesity and male infertility: Mechanisms and management. Andrologia. 2021;53(1):e13617–e13617. doi: 10.1111/and.13617. [DOI] [PubMed] [Google Scholar]
- 41.Leisegang K, Henkel R, Agarwal A. Obesity and metabolic syndrome associated with systemic inflammation and the impact on the male reproductive system. Am J Reprod Immunol. 2019;82(5):e13178–e13178. doi: 10.1111/aji.13178. [DOI] [PubMed] [Google Scholar]
- 42.Hayden RP, Flannigan R, Schlegel PN. The role of lifestyle in male infertility: diet, physical activity, and body habitus. Curr Urol Rep. 2018;19(7):1–10. doi: 10.1007/s11934-018-0805-0. [DOI] [PubMed] [Google Scholar]
- 43.Abbasihormozi S, Babapour V. Stress hormone and oxidative stress biomarkers link obesity and diabetes with reduced fertility potential. Cell J. 2019;21(3):307–307. doi: 10.22074/cellj.2019.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding GL, Liu Y, Liu ME, Pan JX, Guo MX, Sheng JZ, et al. The effects of diabetes on male fertility and epigenetic regulation during spermatogenesis. Asian J Androl. 2015;17(6):948–953. doi: 10.4103/1008-682X.150844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aeeni M, Razi M, Alizadeh A, Alizadeh A. The molecular mechanism behind insulin protective effects on testicular tissue of hyperglycemic rats. Life Sci. 2021;277:119394–119394. doi: 10.1016/j.lfs.2021.119394. [DOI] [PubMed] [Google Scholar]
- 46.Abbasi M, Smith AD, Swaminathan H, Sangngern P, Douglas A, Horsager A, et al. Establishing a stable, repeatable platform for measuring changes in sperm DNA methylation. Clin Epigenetics. 2018;10(1):1–12. doi: 10.1186/s13148-018-0551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laleethambika N, Anila V, Manojkumar C, Muruganandam I, Giridharan B, Ravimanickam T, et al. Diabetes and sperm DNA damage: Efficacy of antioxidants. SN Compr Clin Med. 2019;1(1):49–59. [Google Scholar]
- 48.Chen X, Lin Q, Wen J, Lin W, Liang J, Huang H, et al. Whole genome bisulfite sequencing of human spermatozoa reveals differentially methylated patterns from type 2 diabetic patients. J Diabetes Investig. 2020;11(4):856–864. doi: 10.1111/jdi.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Q, Li J. Research progress of lncRNAs in diabetic retinopathy. Eur J Ophthalmol. 2020;31(4):1606–1617. doi: 10.1177/1120672120970401. [DOI] [PubMed] [Google Scholar]
- 50.Jiang GJ, Zhang T, An T, Zhao DD, Yang XY, Zhang DW, et al. Differential expression of long noncoding RNAs between sperm samples from diabetic and non-diabetic mice. PLoS One. 2016;11(4):e0154028–e0154028. doi: 10.1371/journal.pone.0154028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dupont C, Faure C, Sermondade N, Boubaya M, Eustache F, Clément P, et al. Obesity leads to higher risk of sperm DNA damage in infertile patients. Asian J Androl. 2013;15(5):622–625. doi: 10.1038/aja.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Craig JR, Jenkins TG, Carrell DT, Hotaling JM. Obesity, male infertility, and the sperm epigenome. Fertil Steril. 2017;107(4):848–859. doi: 10.1016/j.fertnstert.2017.02.115. [DOI] [PubMed] [Google Scholar]
- 53.Soubry A, Guo L, Huang Z, Hoyo C, Romanus S, Price T, et al. Obesity- related DNA methylation at imprinted genes in human sperm: results from the TIEGER study. Clin Epigenet. 2016;8(1):1–11. doi: 10.1186/s13148-016-0217-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Donkin I, Versteyhe S, Ingerslev LR, Qian K, Mechta M, Nordkap L, et al. Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metab. 2016;23(2):369–378. doi: 10.1016/j.cmet.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 55.Potabattula R, Dittrich M, Schorsch M, Hahn T, Haaf T, El Hajj N. Male obesity effects on sperm and next-generation cord blood DNA methylation. PLoS One. 2019;14(6):e0218615–e0218615. doi: 10.1371/journal.pone.0218615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang F, Chen H, Chen Y, Cheng Y, Li J, Zheng L, et al. Diet-induced obesity is associated with altered expression of sperm motility- related genes and testicular post-translational modifications in a mouse model. Theriogenology. 2020;158:233–238. doi: 10.1016/j.theriogenology.2020.09.023. [DOI] [PubMed] [Google Scholar]
- 57.An T, Zhang T, Teng F, Zuo JC, Pan YY, Liu YF, et al. Long noncoding RNAs could act as vectors for paternal heredity of high fat diet-induced obesity. Oncotarget. 2017;8(29):47876–47876. doi: 10.18632/oncotarget.18138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson GD, Mackie P, Jodar M, Moskovtsev S, Krawetz SA. Chromatin and extracellular vesicle associated sperm RNAs. Nucleic Acids Res. 2015;43(14):6847–6859. doi: 10.1093/nar/gkv591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang Q, Pan F, Yang J, Fu Z, Lu Y, Wu X, et al. Idiopathic male infertility is strongly associated with aberrant DNA methylation of imprinted loci in sperm: a case-control study. Clin Epigenet. 2018;10(1):1–10. doi: 10.1186/s13148-018-0568-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luján S, Caroppo E, Niederberger C, Arce JC, Sadler-Riggleman I, Beck D, et al. Sperm DNA methylation epimutation biomarkers for male infertility and FSH therapeutic responsiveness. Sci Rep. 2019;9(1):1–12. doi: 10.1038/s41598-019-52903-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mousavi MS, Shahverdi A, Drevet J, Akbarinejad V, Esmaeili V, Sayahpour FA, et al. Peroxisome proliferator-activated receptors (PPARs) levels in spermatozoa of normozoospermic and asthenozoospermic men. Syst Biol Reprod Med. 2019;65(6):409–419. doi: 10.1080/19396368.2019.1677801. [DOI] [PubMed] [Google Scholar]
- 62.Rahimizadeh P, Topraggaleh TR, Nasr-Esfahani MH, Ziarati N, Mirshahvaladi S, Esmaeili V, et al. The alteration of PLCζ protein expression in unexplained infertile and asthenoteratozoospermic patients: A potential effect on sperm fertilization ability. Mol Reprod Dev. 2020;87(1):115–123. doi: 10.1002/mrd.23293. [DOI] [PubMed] [Google Scholar]
- 63.Heidary Z, Saliminejad K, Zaki-Dizaji M, Khorram Khorshid HR. Genetic aspects of idiopathic asthenozoospermia as a cause of male infertility. Hum Fertil. 2020;23(2):83–92. doi: 10.1080/14647273.2018.1504325. [DOI] [PubMed] [Google Scholar]
- 64.Zhang S, Xu L, Yu M, Zhang J. Hypomethylation of the DAZ3 promoter in idiopathic asthenospermia: a screening tool for liquid biopsy. Sci Rep. 2020;10(1):1–7. doi: 10.1038/s41598-020-75110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hekim N, Gunes S, Asci R, Henkel R, Abur U. Semiquantitative promoter methylation of MLH1 and MSH2 genes and their impact on sperm DNA fragmentation and chromatin condensation in infertile men.Andrologia. Andrologia; 2020. pp. e13827–e13827. [DOI] [PubMed] [Google Scholar]
- 66.Saki J, Sabaghan M, Arjmand R, Teimoori A, Rashno M, Saki G, et al. Curcumin as an indirect methylation inhibitor modulates the effects of Toxoplasma gondii on genes involved in male fertility. EXCLI J. 2020;19:1196–1207. doi: 10.17179/excli2020-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saleh AAEW, Amin EM, Elfallah AA, Hamed AM. Insulin resistance and idiopathic infertility: a potential possible link. Andrologia. 2020;52(11):e13773–e13773. doi: 10.1111/and.13773. [DOI] [PubMed] [Google Scholar]