Abstract
Microaerophiles like Campylobacter jejuni must resist oxidative stresses during transmission or infection. Growth of C. jejuni 81116 under iron limitation greatly increased the expression of two polypeptides of 26 and 55 kDa. The identification of these proteins by N-terminal amino acid sequencing showed both to be involved in the defense against oxidative stress. The 55-kDa polypeptide was identical to C. jejuni catalase (KatA), whereas the N terminus of the 26-kDa polypeptide was homologous to a 26-kDa Helicobacter pylori protein. The gene encoding the C. jejuni 26-kDa protein was cloned, and the encoded protein showed significant homology to the small subunit of alkyl hydroperoxide reductase (AhpC). The upstream region of ahpC encoded a divergent ferredoxin (fdxA) homolog, whereas downstream sequences contained flhB and motB homologs, which are involved in flagellar motility. There was no evidence for an adjacent homolog of ahpF, encoding the large subunit of alkyl hydroperoxide reductase. Reporter gene studies showed that iron regulation of ahpC and katA is achieved at the transcriptional level. Insertional mutagenesis of the ahpC gene resulted in an increased sensitivity to oxidative stresses caused by cumene hydroperoxide and exposure to atmospheric oxygen, while resistance to hydrogen peroxide was not affected. The C. jejuni AhpC protein is an important determinant of the ability of this microaerophilic pathogen to survive oxidative and aerobic stress.
Campylobacter jejuni is a curved, microaerophilic, fastidious gram-negative pathogen of humans. Infection with C. jejuni is one of the most common bacterial causes of diarrhea in the developed world, as well as a major cause of diarrhea in developing countries, and as such represents a major health and economic burden (26). Serious complications associated with infection include reactive arthritis, Reiter’s syndrome (21), and Guillain-Barré syndrome (13).
The production of reactive oxygen intermediates in any aerobically metabolizing cell has to be dealt with continuously to avoid damage to DNA, RNA, proteins, and lipids. Therefore, bacteria have evolved a variety of responses to oxidative stress (9). Microaerophiles like C. jejuni are particularly vulnerable to the detrimental effects of oxidative stress and must utilize mechanisms that will eliminate toxic oxygen products. Oxidative stress defense genes of C. jejuni that have been characterized include an iron-containing superoxide dismutase (sodB) gene (20, 22) and a catalase gene, katA (10).
In their natural environment, whether in the gastrointestinal tracts of avian or mammalian hosts or in the external environment during transmission, it is likely that C. jejuni cells are faced with growth-limiting or potentially lethal conditions, such as iron limitation and osmotic and oxidative stresses. In almost all organisms, iron is essential for electron transport functions and as a cofactor for enzymes. There is currently very little information indicating how C. jejuni overcomes these limitations and stresses, either during the disease process through expression of virulence factors or by protective responses to stress during transmission from one host to another. In this study, we investigated the response of C. jejuni cells to iron-restricted and iron-replete conditions by comparing protein profiles. One of the proteins found to be expressed at a significantly higher level in cells grown under iron-restricted conditions than in iron sufficiency was identified as the small subunit protein, AhpC, of the alkyl hydroperoxide reductase enzyme (Ahp). Ahp is positively regulated in Enterobacteriaceae (9) and other organisms such as mycobacteria (8) by the oxyR gene product. It has been shown in Salmonella typhimurium and Escherichia coli (24) to confer resistance to alkyl hydroperoxides by reducing these compounds to alcohols (11).
In this study we describe the identification of an iron-repressible AhpC protein of C. jejuni, characterize its regulation, and demonstrate its involvement in the aerotolerance and defense against oxidative stress of this important microaerophilic pathogen.
MATERIALS AND METHODS
Strains, media, and growth conditions.
C. jejuni 81116 (NCTC 11828; National Collection of Type Cultures, Colindale, London, United Kingdom) (17) and 480 (NCTC 12744) (12) were routinely maintained on Mueller-Hinton (MH) media (Oxoid, Basingstoke, United Kingdom) at 37°C in a variable atmosphere incubator (VAIN; Don Whitley Scientific, Shipley, United Kingdom) in 10% CO2, 5% O2, and 85% N2. Iron-restricted and iron-replete conditions were achieved by supplementing MH media with 20 μM desferrioxamine-B (Desferal; Sigma Chemical Co.) and 40 μM Fe(III)SO4, respectively. Minimal essential medium, alpha modification (MEMα; Gibco-BRL), which contains no added iron source, was used as the defined iron-restricted medium. Iron-replete conditions were achieved by supplementing MEMα with Fe(III)SO4 to a final concentration of 40 μM. E. coli strains used were XL1-Blue MRF′ (Stratagene, La Jolla, Calif.) for plating lambda clones, E. coli XLOLR as a host for excised pBK-CMV derived phagemids (Stratagene), and DH5α F− (Gibco-BRL) as a host for general cloning experiments. Antibiotics used were ampicillin (100 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (20 μg/ml).
Protein analysis.
Overnight cultures grown in MH broth were harvested, washed in phosphate-buffered saline (PBS), and gently resuspended to an optical density at 600 nm (OD600) of 0.05 in iron-restricted or iron-replete MH or MEMα broth. Cells were shaken at 37°C and at intervals 1-ml samples were taken, washed with phosphate-buffered saline, and resuspended to an OD600 of 2 in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer (23). Twelve-microliter samples were subjected to sodium dodecyl phosphate-polyacrylamide gel electrophoresis on 12% gels (23) and subsequently stained with Coomassie brilliant blue. N-terminal amino acid sequences were determined by Edman degradation (Alta Bioscience, University of Birmingham, Birmingham, United Kingdom) from proteins transferred to polyvinylidene difluoride (PVDF) membrane.
Construction and screening of C. jejuni genomic library.
A C. jejuni genomic phage library was constructed by using a partial Sau3AI digest of C. jejuni 81116 genomic DNA with the Lambda Zap Express system (Stratagene) according to the manufacturer’s instructions. Degenerate oligonucleotide primers, containing a BamHI restriction endonuclease site (underlined), were designed from a reverse translation of the N-terminal sequence of the 26-kDa protein (MIVTKKALDF; 5′-CGGGGATCC AA[A/G] AA[A/G] GC[A/T] [C/T]T[A/T] GA[C/T] TT-3′) and from a reverse-translated, inverse complementary sequence from a conserved motif in the C terminus of bacterial AhpC proteins (GEVCPA; 5′-CGGGGATCC GC[A/T] GG[A/G] CA[A/T] ACT TC[A/T] CC-3′) (see Fig. 2). Homologous sequences in chromosomal C. jejuni DNA were amplified by PCR under the following conditions: 2 min at 95°C, followed by 35 cycles of 30 s at 95°C, 1 min at 50°C, and 1 min at 72°C, followed by 10 min at 72°C. The resulting PCR product (∼500 bp) was cloned into the BamHI site of pBluescript II SK(−) (Stratagene), sequenced, and labelled with digoxigenin-dUTP (Boehringer Mannheim) for use as a C. jejuni ahpC-specific probe. This probe was used to screen the C. jejuni Lambda Zap Express library, and one positive clone, designated pMLB4.2, was used for further analysis.
FIG. 2.
Alignment of C. jejuni AhpC protein (Cj) with AhpC proteins of H. pylori (Hp), Legionella pneumophila (Lp), and E. coli (Ec). Asterisks denote identical residues, and dots indicate conservative substitutions. Regions used for the design of degenerate primers are boxed, and the directions of the resulting primers are indicated by arrows.
Primer extension analysis.
The transcriptional start site of C. jejuni ahpC was determined by using total RNA extracted from a 50-ml culture of C. jejuni grown overnight in MEMα (to induce a high level of ahpC expression) by the hot acid phenol method (1). A 32P-labelled oligonucleotide (5′-TCC CAA TAC TGC TGG AGC AG-3′) was annealed to 5 μg of total RNA in 30 μl of buffer (40 mM piperazine-N,N′-bis[2-ethanesulfonic acid] [PIPES], 1 mM EDTA, 400 mM NaCl, 80% formamide) by heating the mixture at 85°C for 5 min followed by cooling to 45°C overnight. Primer extension was performed at 42°C for 1 h by using 400 U of Superscript reverse transcriptase (Gibco-BRL) in a buffer with 1 mM deoxynucleotide triphosphates, 10 mM dithiothreitol, and 40 U of RNasin (RNase inhibitor; Promega). cDNA was precipitated, heated at 70°C for 5 min, and loaded onto a 6% sequencing gel alongside the products of a sequencing reaction of pMLB4.2 with the primer D15210B. Products were visualized with a PhosphorImager and ImageQuant software (Molecular Dynamics).
Reporter gene assays.
The promoter regions of the ahpC and the katA genes were amplified with the Expand polymerase kit (Boehringer Mannheim) and cloned into the BamHI site of pMW10 (33) upstream of a promoterless lacZ gene. The resulting constructs were transformed into C. jejuni 480 (12) by using electroporation under conditions described previously (30). The expression of β-galactosidase was assayed as described previously (16, 23, 33) from cultures grown to logarithmic phase in MH broth supplemented with Desferal or Fe(III)SO4 to final concentrations of 20 and 40 μM, respectively. The negative control was plasmid pMW10, whereas the positive control consisted of plasmid 23E5, which is a pMW10 derivative with the promoter of the housekeeping gene metK upstream of the promoterless lacZ gene (33); this promoter is not iron regulated.
Construction of a C. jejuni ahpC mutant.
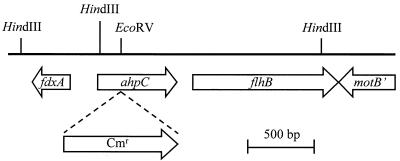
A C. jejuni ahpC mutant was constructed by insertional mutagenesis with a chloramphenicol resistance gene. The ahpC sequence present in pMLB4.2 was disrupted by the insertion of a chloramphenicol resistance gene (34) from pAV35 (31) inserted into the unique EcoRV site of pMLB4.2. This construct, called pAV103, was then transformed by electroporation into C. jejuni 81116, and the resulting C. jejuni ahpC mutant was designated AV32.
Oxidative stress and aerotolerance assays.
Oxidative stress resistance was determined by a disk inhibition assay. C. jejuni bacteria were grown overnight microaerophilically in MH broth at 37°C with chloramphenicol supplementation as necessary. Culture samples were adjusted to an OD600 of 0.4, and 200-μl samples were then added to 4 ml of soft MH agar, which was subsequently poured onto MH plates. Samples (3 μl) of 3% hydrogen peroxide in water or 3% cumene hydroperoxide (CHP) in dimethyl sulfoxide were applied to 6-mm-diameter 3M Whatman paper disks placed on the soft agar surface. The zone of killing (no growth visible) was measured after incubating the disks for 24 h at 37°C. The aerotolerance of C. jejuni was assayed by growing 10-ml MH broth cultures of C. jejuni 81116 and ahpC mutant AV32 overnight at 37°C with shaking to an OD600 of approximately 0.4. Cultures were then shaken at 200 rpm in 25-ml flasks with cotton wool plugs at 37°C in either atmospheric aerobic conditions or in microaerobic conditions in the VAIN. Samples were removed from the cultures and diluted, and 5 μl per dilution was spotted onto agar plates to assess viability.
Nucleotide sequence accession number.
The sequence of the genomic region containing the C. jejuni fdxA, ahpC, flhB, and motB genes has been deposited in the GenBank database with accession no. AF044271.
RESULTS
Identification and characterization of iron-repressible polypeptides of C. jejuni.
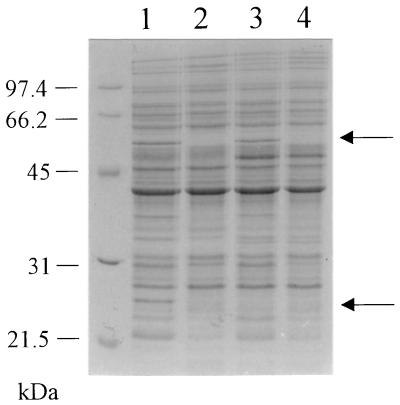
The response of C. jejuni 81116 to various iron levels in the culture medium was studied by growing cells grown in an iron-depleted tissue culture medium (MEMα). A comparison of total protein profiles of cells grown in MEMα and in complex medium (MH broth) revealed two polypeptides of approximately 26 and 55 kDa, which were expressed at higher levels in the MEMα-grown cells (Fig. 1). In the absence of any added iron source in MEMα, iron deprivation appeared a possible reason for this phenotype, and this was confirmed by supplementing MEMα with iron, which suppressed the expression of both proteins (Fig. 1, lanes 1 and 2). Similar results were obtained in iron-restricted MH media (data not shown). The iron-repressed 26- and 55-kDa polypeptides were identified by N-terminal amino acid sequencing. The N-terminal amino acid sequence of the 55-kDa polypeptide (MKKLTNDFGN) was identical to that of the C. jejuni catalase (KatA) protein (10). The N-terminal amino acid sequence of the 26-kDa protein (MIVTKKALDF) showed significant homology (seven amino acids identical, one conserved) with the N-terminal amino acid sequence (MLVTKLAPDF) of a 26-kDa protein of Helicobacter pylori (19), which is homologous to the alkyl hydroperoxide reductase small subunit AhpC protein found in several bacteria, including E. coli (24).
FIG. 1.
Effects of iron restriction on total protein profiles of C. jejuni 81116 and AV32 (ahpC mutant) strains. Lane 1, C. jejuni 81116 grown in MEMα (iron restricted); lane 2, C. jejuni 81116 grown in MEMα supplemented with 40 μM Fe(III)SO4 (iron replete); lane 3, C. jejuni AV32 grown in MEMα (iron limited); lane 4, C. jejuni AV32 grown in MEMα supplemented with 40 μM Fe(III)SO4 (iron replete). Molecular mass markers (in kilodaltons) are shown on the left. Arrows on the right indicate the 55-kDa KatA and 26-kDa AhpC polypeptides.
Characterization of the C. jejuni ahpC gene.
An alignment of bacterial AhpC sequences (Fig. 2) was used to design degenerate oligonucleotide primers required for the cloning and further characterization of C. jejuni ahpC. These degenerate primers produced a fragment of ∼500 bp when used in a PCR with C. jejuni genomic DNA. This PCR product was cloned and sequenced, and it contained one continuous open reading frame (ORF) encoding a protein homologous to bacterial AhpC proteins. The PCR product was used to screen a C. jejuni 81116 Lambda Zap Express library, and one of three positive clones (pMLB4.2) was selected for further characterization of the genomic region containing the C. jejuni ahpC homolog. The sequence upstream of ahpC contained a divergent ORF encoding a ferredoxin homolog designated fdxA (28) (Fig. 3). Sequences downstream of ahpC contained a full-length ORF encoding an flhB homolog and a truncated ORF without a start codon encoding a motB homolog (Fig. 3). The flhB homolog is independently transcribed (15). The FlhB protein is involved in both the biogenesis and regulation of biogenesis of flagella (14).
FIG. 3.
Restriction and gene map of the C. jejuni genomic region containing the ahpC gene. Only a part of the motB gene is shown. The place and direction of insertion of the chloramphenicol resistance gene (Cmr) used to create the C. jejuni ahpC mutant are also indicated.
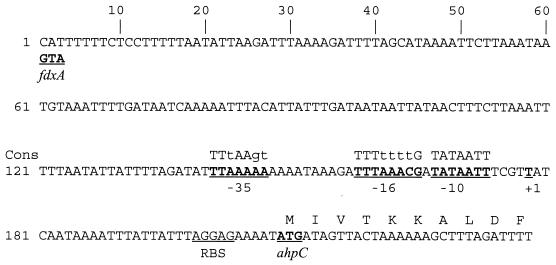
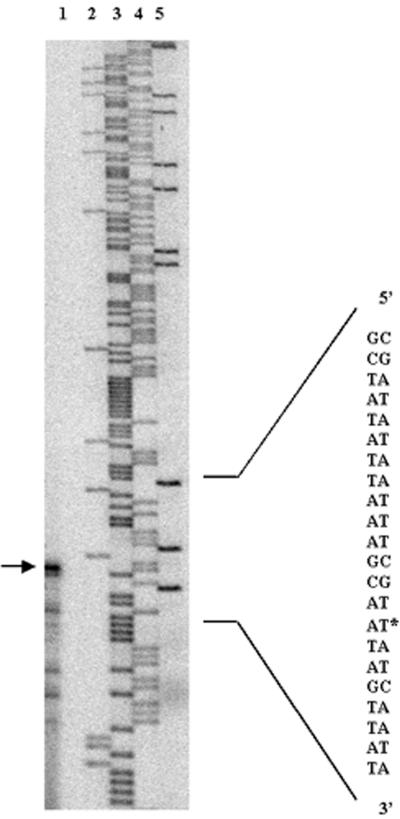
The transcriptional start point of ahpC was determined by primer extension. Figure 4, left panel, shows the most prominent band representing a reverse transcript which ran level with a band in the adenine track corresponding to a T residue in the template sequence, 31 base pairs upstream of the start codon. Sequences resembling the C. jejuni ς70 consensus sequence (33) were present at the correct distances from the transcriptional start site and are indicated in the upper panel of Fig. 4. There was a perfect match with the C. jejuni −10 sequence (7 of 7) and good matches with the −16 and −35 sequences (4 of 8 and 4 of 7, respectively) (33).
FIG. 4.
Analysis of the promoter region of the C. jejuni ahpC gene. The mapping of the transcriptional start site of ahpC is shown (left panel). Lane 1, primer extension of C. jejuni mRNA; lanes 2 to 5, sequencing reaction of pMLB4.2 (in the order C, T, A, and G). The right-hand column shows the complementary template sequence, and the transcriptional start site is indicated by an asterisk. The sequence of the ahpC promoter region is shown in the upper panel. The start codons of ahpC and the divergent fdxA gene are indicated, as well as the transcriptional start site (+1) of ahpC. Sequences closely resembling the C. jejuni ς70 consensus sequence (Cons) are indicated.
C. jejuni ahpC and katA expression is iron regulated at the transcriptional level.
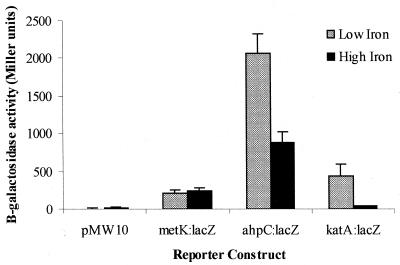
A reporter gene system based on a promoterless lacZ gene on the C. jejuni shuttle vector pMW10 (33) was used to demonstrate that ahpC and katA expression are iron regulated at the transcriptional level. The 204-bp intergenic region between ahpC and fdxA and the region 444 bp upstream of the katA gene were amplified from C. jejuni 81116 and cloned into the BamHI site of pMW10, upstream of its promoterless lacZ gene with the ahpC and katA start codons in the same orientation as lacZ. The constructs were then introduced into C. jejuni 480, and the expression of lacZ was measured under iron-restricted and iron-replete conditions. C. jejuni 480 was used, as this strain accepts shuttle vectors isolated from E. coli, whereas C. jejuni 81116 does not (30, 32). The iron regulation of AhpC and KatA is identical in C. jejuni 480 and 81116 (data not shown). The expression of β-galactosidase in the ahpC::lacZ fusion was two- to threefold higher under iron-restricted conditions than that under iron-replete conditions (Fig. 5), whereas the expression of the katA::lacZ fusion was more than 10-fold higher under iron-restricted conditions (Fig. 5). The expression level of the positive control, which consisted of the promoter of the metK housekeeping gene (33), was not significantly altered by the different iron concentrations.
FIG. 5.
β-Galactosidase assays of C. jejuni strain transformants grown under iron-restricted and iron-replete conditions, confirming that iron regulation of ahpC and katA expression is achieved at the transcriptional level. Error bars represent standard deviations.
Mutational analysis of C. jejuni ahpC.
As a first step towards defining a role for AhpC in C. jejuni, an ahpC mutant was made by the insertion of a chloramphenicol resistance cassette into the unique EcoRV site of pMLB4.2 to generate pAV103 (Fig. 3). This construct was used to transform C. jejuni 81116, and an ahpC-deficient mutant was generated via allelic exchange. The chloramphenicol-resistant C. jejuni ahpC mutant was named AV32. The correct replacement of the wild-type ahpC gene with the mutated copy was confirmed by Southern hybridization (data not shown). Further evidence that the 26-kDa protein was the product of the ahpC gene was obtained by comparing the total protein profiles of the C. jejuni wild type and ahpC mutant grown in MEMα. The iron-regulated 26-kDa protein was not detected in the ahpC mutant, while the cells still showed an upregulation of catalase under iron-restricted conditions (Fig. 1, lanes 3 and 4).
The C. jejuni ahpC mutant has decreased aerotolerance and resistance to oxidative stress.
The role of the C. jejuni AhpC protein was further investigated by the comparison of oxidative stress resistance and aerotolerance of the wild type and ahpC mutant strains. Resistance to the oxidative stress inducers CHP and hydrogen peroxide (H2O2) was measured with a disk inhibition assay (24), in which the zone of killing is measured. The C. jejuni ahpC mutant was found to be hypersensitive to CHP, with a significantly greater zone of killing (50.2 ± 8.5 mm in diameter compared to a 22.6 ± 1.8-mm zone for the wild type) (P < 0.005; paired t test [n = 5]). The ahpC mutant strain was also found to be more susceptible to killing (shown by plate count) by CHP than the parental strain, when various concentrations of this inhibitory agent were added to liquid cultures (data not shown). Sensitivity to H2O2, however, was not affected by the ahpC mutation, as no significant difference was apparent between the C. jejuni wild type and ahpC mutant with zones of killing of 20 ± 0.8 and 21 ± 2.8 mm, respectively (nonsignificant by paired t test).
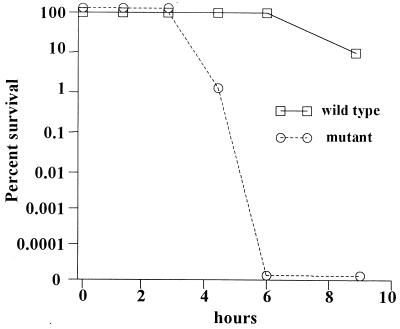
We also compared the effect of the ahpC mutation on the aerotolerance of C. jejuni, an important phenotype for this microaerophile. Aerotolerance was measured by exposing stationary-phase cells to atmospheric levels of oxygen and measuring the viability of the cells. The ahpC mutant had a significantly decreased ability to survive in air compared to the wild type. A representative experiment is illustrated in Fig. 6. In six different experiments, there was in all cases at least a 103-fold difference in survival for at least one time point between 3 and 6 h. In all aerotolerance experiments, the ahpC mutant lost viability substantially earlier than the wild type. Variation in time to onset of loss of viability in repeat experiments precluded analysis of means and variances at fixed time points. In control cultures incubated in parallel in microaerobic conditions, neither mutant nor wild type showed any decrease in viability during the entire incubation period. To minimize any possible effects due to nutrient starvation, stationary-phase cells were also resuspended in fresh media before exposure to air. Under such conditions both strains were able to survive longer in air (the onset of death was increased by approximately 10 h), but the ahpC mutant still lost its viability more rapidly than the wild type (data not shown). The addition of desferrioxamine to the cultures before their exposure to air did not enhance the survival of the wild type, despite the increase in AhpC expression (data not shown).
FIG. 6.
Percentage survival of C. jejuni 81116 wild-type and ahpC mutant stationary-phase cells kept under normal atmospheric oxygen conditions at 37°C. The graph shows results of a single representative experiment typical of the six that were carried out.
DISCUSSION
When bacteria are subjected to iron limitation they respond in a variety of ways. We have shown that when C. jejuni cells are in an iron-restricted environment, the production of at least two polypeptides is induced to a high level—becoming among the most abundant proteins and likely therefore to be highly significant to the organism. These two proteins were identified as the C. jejuni KatA and AhpC homologs. The biochemical characterization of S. typhimurium alkyl hydroperoxide reductase showed that the enzyme consists of two subunits (11), a small (26-kDa) subunit, AhpC, and a large (56-kDa) subunit, AhpF. The reduced form of the AhpC subunit reduces alkyl hydroperoxides to alcohols and in the process is oxidized, while the AhpF subunit transfers electrons from NAD(P)H to oxidized AhpC and thus rereduces it.
AhpC homologs have been shown to be present in organisms from all kingdoms. The alignment of the amino acid sequences reveals two highly conserved cysteine residues toward the N and C termini (5), which in the C. jejuni AhpC protein are found at positions 49 and 169. The sequence surrounding the conserved cysteine residue at position 49 is FVCP, which is also the case for the other two-cysteine AhpC family members. The N-terminal cysteine is conserved in all members of this family of antioxidant enzymes, and it has been shown, by the genetic and biochemical alteration of this residue, to be important in the antioxidant activity of both S. typhimurium alkyl hydroperoxide reductase and yeast thiol-specific antioxidant (5, 11).
Although ahpF, the gene that encodes the large subunit of the alkyl hydroperoxide reductase enzyme, has been shown to be part of an ahp operon with ahpC in S. typhimurium, E. coli, and Bacillus subtilis (3, 24), no ahpF homolog was found in the vicinity of the ahpC gene. Analysis of the recently completed genome sequence of C. jejuni NCTC 11168 (4a) did not reveal the presence of an ahpF homolog. Therefore, it is likely that C. jejuni utilizes an alternative system for the reduction of oxidized AhpC. As ferredoxins are powerful reducing agents, one possibility is that the AhpC subunit receives electrons from the reduced form of the ferredoxin protein that is encoded adjacent to ahpC. An ahpF gene homolog is also absent from the genome sequence of the closely related microaerophilic pathogen H. pylori (2, 27), indicating that Campylobacter and Helicobacter species may use similar mechanisms to recycle oxidized AhpC.
Reactive oxygen intermediates such as superoxide, hydroxide radicals, and peroxides are formed within cells because of aerobic metabolism (9). The presence of such reactive oxygen species causes damage to nucleic acids, proteins, and lipids, in some cases by peroxidation to hydroperoxide derivatives. Furthermore, alkyl hydroperoxides are able to initiate and propagate free-radical chain reactions that result in DNA and membrane damage and therefore may be particularly deleterious. Alkyl hydroperoxide reductase can both destroy toxic hydroperoxide intermediates and repair the damaged molecules where they have been peroxidized and may thus play an important role in detoxification following oxidative stress.
The increase in production of AhpC in C. jejuni in response to iron limitation is not unique to this organism. The AhpC proteins of Corynebacterium diphtheriae (25) and B. subtilis (3, 6) are also iron regulated. Whereas the expression of ahpC is usually mediated through the OxyR protein in response to oxidative stress (7–9), it has been shown that in B. subtilis the expression is regulated through a Fur homolog, named PerR (4). In C. jejuni we have previously demonstrated that the iron-responsive expression of AhpC and KatA is not Fur dependent (31). The availability of the genome sequence has allowed the identification of a second fur homolog, designated PerR by analogy to B. subtilis PerR, which has been shown to regulate ahpC and katA expression in C. jejuni (29). There are indications that the expression of C. jejuni ahpC is not solely regulated by PerR, as has been shown for KatA (29). The promoter regions of both ahpC and katA contain one or more sequences closely resembling the B. subtilis PerR binding sequences (Per boxes) as well as a sequence resembling the E. coli Fur binding sequence (Furbox). Whether these sequences indeed interact with either regulator should be further investigated using gel shift assays and DNase footprinting.
From the reporter gene assays conducted on the ahpC fusion, it was clear that under various iron conditions the regulation of expression of ahpC occurred at the transcriptional level. However, it should be noted that pMW10 appears to be a moderately high-copy-number plasmid in Campylobacter (approximately 20 copies per cell [33]); hence, any promoter carried might be incompletely repressed under iron-replete conditions and therefore may not exactly reflect the regulation of a single chromosomal copy. Although expression of ahpC and katA is upregulated under iron-restricted conditions, there is a basal level of expression of ahpC under iron-replete conditions as verified by two-dimensional protein analysis (data not shown). It may be the case that a background level of expression of oxidative stress genes is required under iron-replete growth conditions, a concept compatible with enhanced survival of the wild type over the mutant in iron-replete media as shown in this study. Currently it is not known whether C. jejuni ahpC is upregulated under conditions of oxidative stress, although analysis of expression under such conditions would help delineate the cellular response of C. jejuni to oxidative stress. Such a regulatory pattern has been noted in S. typhimurium, in which the OxyR regulator influences ahpC expression (7).
An increase in production of AhpC under conditions of iron restriction could be advantageous to C. jejuni. Iron is known to participate in the production of reactive oxygen intermediates via the Fenton reaction. When cells are in an iron-restricted environment, often the response is to activate iron acquisition systems such as the production of siderophores and their receptors. With such systems induced, the intracellular iron concentration may transiently increase despite the low extracellular concentration, leading to a raised level of reactive oxygen intermediates. Thus, if the production of alkyl hydroperoxide reductase is coupled to the activation of iron acquisition mechanisms, a coordinated survival strategy is established. Such a mechanism of regulation has been demonstrated previously in E. coli for the control of superoxide dismutase by the iron-responsive gene regulator Fur (18).
We have shown that AhpC plays an important role in protecting C. jejuni against alkyl hydroperoxides, as a mutation of the ahpC gene results in decreased resistance to killing by CHP. This phenotype is not due to polar effects on flhB, as flhB mutants do not exhibit the same phenotype as ahpC mutants (data not shown). The ahpC mutant is also far less able to survive periods of exposure to atmospheric oxygen levels than the wild type, especially during the stationary phase, rather than when there is potential for growth. This suggests that the AhpC protein may play an important role in protecting C. jejuni from oxidative stress damage during its transmission from host to host in foods and water, when it is likely to be in stationary phase. Environments such as milk, possibly fresh meat, eggs, and clean water are relatively depleted of available iron and therefore the upregulation of AhpC under such conditions may contribute significantly to the ability of the organism to survive in these environments during transmission.
Of interest also is the possibility that the organism may be exposed to extracellular oxygen metabolites of stimulated polymorphonuclear phagocytes in the inflamed mucosa of the infected gut. The limited availability of iron in mucosal secretions, in tissues, and within phagocytes, due to the presence of iron-binding lactoferrin and transferrin, is well known. Hence, the iron-regulated expression of AhpC might also be significant in survival of the organism in the infected intestinal epithelium.
ACKNOWLEDGMENTS
This study was supported by a studentship from the Ministry of Agriculture Fisheries and Food to M.-L. A. Baillon and by the Wellcome Trust and a Royal Society University Research Fellowship to J. M. Ketley.
We thank Marc Wösten for donating plasmids pMW10 and 23E5.
REFERENCES
- 1.Aiba H, Adhya S, Decrombrugghe B. Evidence for 2 functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:1905–1910. [PubMed] [Google Scholar]
- 2.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 3.Bsat N, Chen L, Helmann J D. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J Bacteriol. 1996;178:6579–6586. doi: 10.1128/jb.178.22.6579-6586.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bsat N, Herbig A, Casillas-Martinez L, Setlow P, Helmann J D. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol Microbiol. 1998;29:189–198. doi: 10.1046/j.1365-2958.1998.00921.x. [DOI] [PubMed] [Google Scholar]
- 4a.Campylobacter jejuni NCTC 11168 Genome Sequence. 26 January 1999, posting date. Sequence. [Online.] Sanger Centre, Hinxton, Cambridge, United Kingdom. http://www.sanger.ac.uk./projects/C_jejuni. [29 June 1999, last date accessed.]
- 5.Chae H Z, Uhm T B, Rhee S G. Dimerization of thiol-specific antioxidant and the essential role of cysteine 47. Proc Natl Acad Sci USA. 1994;91:7022–7026. doi: 10.1073/pnas.91.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Keramati L, Helmann J D. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc Natl Acad Sci USA. 1995;92:8190–8194. doi: 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christman M F, Morgan R W, Jacobson F S, Ames B N. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell. 1985;41:753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 8.Dhandayuthapani S, Mudd M, Deretic V. Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis. J Bacteriol. 1997;179:2401–2409. doi: 10.1128/jb.179.7.2401-2409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant K A, Park S F. Molecular characterization of katA from Campylobacter jejuni and generation of a catalase-deficient mutant of Campylobacter coli by interspecific allelic exchange. Microbiology. 1995;141:1369–1376. doi: 10.1099/13500872-141-6-1369. [DOI] [PubMed] [Google Scholar]
- 11.Jacobson F S, Morgan R W, Christman M F, Ames B N. An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. Purification and properties. J Biol Chem. 1989;264:1488–1496. [PubMed] [Google Scholar]
- 12.King V, Wassenaar T M, van der Zeijst B A M, Newell D G. Variations in Campylobacter jejuni flagellin, and flagellin genes, during in vivo and in vitro passage. Microb Ecol Health Dis. 1991;4:135–140. [Google Scholar]
- 13.Kuroki S, Saida T, Nukina M, Haruta T, Yoshioka M, Kobayashi Y, Nakanishi H. Campylobacter jejuni strains from patients with Guillain-Barré syndrome belong mostly to Penner serogroup 19 and contain beta-N-acetylglucosamine residues. Ann Neurol. 1993;33:243–247. doi: 10.1002/ana.410330304. [DOI] [PubMed] [Google Scholar]
- 14.Kutsukake K. Hook-length control of the export-switching machinery involves a double-locked gate in Salmonella typhimurium flagellar morphogenesis. J Bacteriol. 1997;179:1268–1273. doi: 10.1128/jb.179.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matz, C. M., A. H. M. van Vliet, J. M. Ketley, M. L. A. Baillon, C. Constantinidou, and C. W. Penn. Unpublished data.
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 17.Newell D G, McBride H, Pearson A D. The identification of outer membrane proteins and flagella of Campylobacter jejuni. J Gen Microbiol. 1984;130:1201–1208. doi: 10.1099/00221287-130-5-1201. [DOI] [PubMed] [Google Scholar]
- 18.Niederhoffer E C, Naranjo C M, Bradley K L, Fee J A. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J Bacteriol. 1990;172:1930–1938. doi: 10.1128/jb.172.4.1930-1938.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Toole P W, Logan S M, Kostrzynska M, Wadstrom T, Trust T J. Isolation and biochemical and molecular analyses of a species-specific protein antigen from the gastric pathogen Helicobacter pylori. J Bacteriol. 1991;173:505–513. doi: 10.1128/jb.173.2.505-513.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pesci E C, Cottle D L, Pickett C L. Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect Immun. 1994;62:2687–2694. doi: 10.1128/iai.62.7.2687-2694.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson M C. Rheumatic manifestations of Campylobacter jejuni and C. fetus infections in adults. Scand J Rheumatol. 1994;23:167–170. doi: 10.3109/03009749409103055. [DOI] [PubMed] [Google Scholar]
- 22.Purdy D, Park S F. Cloning, nucleotide sequence and characterization of a gene encoding superoxide dismutase from Campylobacter jejuni and Campylobacter coli. Microbiology. 1994;140:1203–1208. doi: 10.1099/13500872-140-5-1203. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Storz G, Jacobson F S, Tartaglia L A, Morgan R W, Silveira L A, Ames B N. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: genetic characterization and cloning of ahp. J Bacteriol. 1989;171:2049–2055. doi: 10.1128/jb.171.4.2049-2055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tai S S, Zhu Y Y. Cloning of a Corynebacterium diphtheriae iron-repressible gene that shares sequence homology with the AhpC subunit of alkyl hydroperoxide reductase of Salmonella typhimurium. J Bacteriol. 1995;177:3512–3517. doi: 10.1128/jb.177.12.3512-3517.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tauxe R V. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C: American Society for Microbiology; 1992. pp. 9–19. [Google Scholar]
- 27.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 28.van Vliet, A. H. M., M.-L. A. Baillon, C. W. Penn, and J. M. Ketley. 1999. Unpublished data.
- 29.van Vliet, A. H. M., M.-L. A. Baillon, C. W. Penn, and J. M. Ketley.Campylobacter jejuni contains two Fur homologs: characterisation of iron-responsive regulation of oxidative stress genes by the PerR repressor. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 30.van Vliet A H M, Wood A C, Henderson J, Wooldridge K G, Ketley J M. Genetic manipulation of enteric Campylobacter species. In: Williams P, Ketley J, Salmond G, editors. Methods in microbiology. Vol. 27. London, England: Academic Press; 1998. pp. 407–419. [Google Scholar]
- 31.van Vliet A H M, Wooldridge K G, Ketley J M. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J Bacteriol. 1998;180:5291–5298. doi: 10.1128/jb.180.20.5291-5298.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wassenaar T M, Fry B N, van der Zeijst B A M. Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene. 1993;132:131–135. doi: 10.1016/0378-1119(93)90525-8. [DOI] [PubMed] [Google Scholar]
- 33.Wösten M M S M, Boeve M, Koot M G A, van Nuenen A C, van der Zeijst B A M. Identification of Campylobacter jejuni promoter sequences. J Bacteriol. 1998;180:594–599. doi: 10.1128/jb.180.3.594-599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao R, Alm R A, Trust T J, Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 1993;130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]