Abstract
Background
Lung adenocarcinoma (LUAD) is one of the most frequent types of lung cancer, with a high mortality and recurrence rate. This study aimed to design a RiskScore to predict the prognosis and immunotherapy response of LUAD patients due to a lack of metabolic and immune-related prognostic models.
Methods
To identify prognostic genes and generate a RiskScore, we conducted differential gene expression analysis, bulk survival analysis, Lasso regression analysis, and univariate and multivariate Cox regression analysis using TCGA-LUAD as a training subset. GSE31210 and GSE50081 were used as validation subsets to validate the constructed RiskScore. Following that, we explored the connection between RiskScore and clinicopathological characteristics, immune cells infiltration, and immunotherapy. In addition, we investigated into RiskScore's biological roles and constructed a Nomogram model.
Results
A RiskScore was identified consisting of five genes (DKK1, CCL20, NPAS2, GNPNAT1 and MELTF). In the RiskScore-high group, LUAD patients showed decreased overall survival rates and shorter progression-free survival. Multiple clinicopathological characteristics and immune cells infiltration in TME, in particular, have been linked to RiskScore. Of note, RiskScore-related genes have been implicated to substance metabolism, carcinogenesis, and immunological pathways, among other things. Finally, the C-index of the RiskScore-based Nomogram model was 0.804 (95% CI: 0.783–0.825), and time-dependent ROC predicted probabilities of 1-, 3- and 5-year survival for LUAD patients were 0.850, 0.848 and 0.825, respectively.
Conclusion
The RiskScore, which integrated metabolic and immunological features with DKK1, CCL20, NPAS2, GNPNAT1, and MELTF, could reliably predict prognosis and immunotherapy response in LUAD patients. Moreover, the RiskScore-based Nomogram model had a promising clinical application.
Keywords: Lung adenocarcinoma, Prognostic, Tumor immunity, DKK1, CCL20, NPAS2, GNPNAT1, MELTF
Lung adenocarcinoma; Prognostic; Tumor immunity; DKK1; CCL20; NPAS2; GNPNAT1; MELTF
1. Introduction
Despite breakthroughs in early detection, lung cancer remains the most common cause of cancer death globally [1]. LUAD is the most frequent histologic subtype of primary lung cancer, accounting for almost 40% of all cases, and it is also one of the most aggressive and swiftly deadly tumor types, with an overall survival rate of fewer than 5 years [2]. Surgical resection of early primary adenocarcinoma remains the preferred treatment approach, with a minimal risk of recurrence [3]. Unfortunately, because lung adenocarcinoma is generally identified in advanced stages or even in the presence of metastases, only conventional chemotherapy, radiotherapy, or immunotherapy can be used, but drug resistance still leads to a poor prognosis [2]. As a result, identifying novel LUAD biomarkers that can be used to predict patient survival and immunotherapy response is critical. These biomarkers could provide valuable clinical information which could be used to assess a patient's overall health and provide personalized treatment for precision medicine.
In contrast to the once-dominant tumor-centered concept of cancer, the tumor microenvironment (TME) is becoming incredibly prominent [4]. Tumor genesis, growth, invasion, metastasis, and response to therapy can all be reprogrammed by the stromal cells and non-cellular components of the TME [5]. It is, nevertheless, genetically stable, making it a promising therapeutic target for reducing treatment resistance and the probability of tumor recurrence [6]. The detailed investigation of infiltrating immune cells in TME, in particular, contributes to the discovery of cancer immune evasion mechanisms, allowing for the creation of new therapeutic options [7].
With the constant advancement of sequencing technology in recent years, a large amount of expression profile data has amassed, and its re-examination and identification can surely aid in the advancement of medicine. Two main databases were utilized in this study: the Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) databases. First, the TCGA-LUAD cohort was used to screen prognostic genes, and a RiskScore was generated using the Cox regression coefficient. Second, the GSE31210 and GSE50081 cohorts were used to validate the findings. The link between RiskScore and clinicopathological characteristics, immune cells infiltration in TME, and immunotherapy response was then investigated. Finally, a Nomogram model was constructed, and the model's predictive capacity was confirmed using time-dependent receiver operator characteristic curve (ROC), calibration curve, and decision curve analysis (DCA).
2. Materials and methods
2.1. Data source
RNA-seq data and clinical information from 59 normal and 535 lung adenocarcinoma patients were retrieved from the TCGA-LUAD dataset (https://portal.gdc.cancer.gov/) as of March 2022. GSE31210, GSE50081 and GSE135222 datasets obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). GSE31210 and GSE50081 contained RNA-seq data and corresponding clinical information for 226 and 127 LUAD patients, respectively, while GSE135222 comprised 25 patients with non-small cell lung cancer (NSCLC) who were treated with anti-PD-1/PD-L1. Missing values were defined as missing or unknown clinical characteristics. This study analyzed data from publicly available databases and did not necessitate a re-evaluation of medical ethics.
2.2. Data processing
Transcriptome expression data and corresponding clinical information were downloaded from the TCGA database in the data format HTSeq-Counts for differential gene expression analysis between LUAD and normal lung tissue and between RiskScore-high and -low. Then, data formatted as HTSeq FPKM was downloaded and converted to TPM for subsequent analyses. In addition, log 2 conversion was performed on all RNA-seq data prior to all analyses.
2.3. Screening for prognostic genes
Differential gene expression analysis was performed using 535 LUAD tissues and 59 paracancer tissues from the TCGA-LUAD cohort using the R package “DESeq2” and the R package “ggplot2” for volcano plotting with a threshold value of |log 2 FC|≥1 and P < 0.05. For the bulk survival analysis, the R package “survivor” was used. The Lasso regression analysis was performed using the R package “glmnet” and “survival” with seed number 2022, and the method used ten-fold cross-validation, selecting the lambda value corresponding to the smallest mean value of the cross-validation error as the truncation value. Univariate and multivariate Cox regression analyses were performed using the R package “survival” and the R package “ggplot2” was used to visualize the forest plots, and a RiskScore was constructed from the regression coefficients.
2.4. Prognostic value of RiskScore
The 526 LUAD patients in the TCGA-LUAD cohort served as the training subset, 226 LUAD patients from the GSE31210 cohort as the validation subset 1 and 127 LUAD patients from the GSE50081 cohort as the validation subset 2. LUAD patients were divided into RiskScore-high and -low groups according to the median of RiskScore. The scatter plot was visualized by the R package “ggplot2”, Kaplan-Meier survival analysis was performed by the R package “survival”, and the R package “survminer” was used for visualization. Time-dependent ROC curve was analyzed by the R package “timeROC” and visualized by the R package “ggplot2”.
2.5. Biological functions of RiskScore
First, LUAD patients in the TCGA database were divided into high and low groups according to the median of RiskScore, and then differential gene expression analysis was performed with a threshold of |log 2 FC|≥1 and P < 0.05. The volcano plot was visualized using the R package “ggplot2”. Gene ontology (GO) functional analysis was conducted to identify the unique biological properties, including biological processes (BP), cellular components (CC), and molecular functions (MF). All up-regulated genes were extracted for GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, analyzed by R package “clusterProfiler” and visualized by R package “ggplot2”.
Next, correlation analysis of these five genes included in the RiskScore with oncogenes (KRAS, BRAF, EGFR, ERBB2, PIK3CA, FGFR1, DDR2, RET, MYC, RB1, NF1, ROS1) driving lung cancer progression was performed using the R package “ggplot2” for visualization using the TCGA-LUAD cohort. In addition, KRAS and TP53 mutation data were downloaded from TCGA database through the UCSC Xena web tool (https://xenabrowser.net/datapages/).
2.6. Role of RiskScore in the TME
First, the relative infiltration levels of immune cells in the TME were quantified using the ssGSEA algorithm in the R package “GSVA”, and the specific markers and classifications of the 24 immune cells were identified in a paper by Bindea et al. [8]. Based on the GSVA algorithm, LUAD patients in the TCGA database were divided into high and low score groups according to the median immune cell enrichment scores, and survival analysis was performed using the R package “survival” and the R package “survminer” for visualization. In addition, transcriptome data from the TCGA-LUAD cohort were uploaded to the CIBERSORTx web tool (https://cibersortx.stanford.edu/index.php) to obtain immune cell infiltration scores based on the cibersort algorithm. Data on the immune subtypes of LUAD patients in TCGA were obtained from the paper published by Thorsson V. et al. [9].
2.7. RiskScore response to immunotherapy and tumor relapse
First, the differential expression of four immunosuppressive checkpoints (PD-1, PD-L1, PD-L2 and CTLA4) were compared in the RiskScore-high and -low groups. Then, RNA-seq and corresponding clinical information for NSCLC patients receiving anti-PD-1/PD-L1 therapy in the GSE135222 dataset were downloaded. Patients were divided into two groups for survival analysis based on the median RiskScore. Finally, LUAD patients in the RiskScore-high group were further divided into four groups based on quartiles of RiskScore using the TCGA database, with the top 25% being the extremely high-risk group (n = 67) and the bottom 25% being the high-risk group (n = 66) for relapse-free survival (RFS) analysis.
2.8. Construction and evaluation of nomogram model
Based on the TCGA-LUAD cohort, univariate and multivariate Cox regressions were used to screen independent predictors of prognosis, and then Nomogram model and calibration curve were constructed and evaluated using the R packages “rms” and “survival”. Time-dependent ROC curve was analyzed using the R package “timeROC” and visualized using the R package “ggplot2”. Decision curve analysis was analyzed and visualized using the R package “survival” and the “stdca.R”.
2.9. Statistical analysis
All statistical analyses were processed on R Studio software and P < 0.05 was considered statistically significant. In this study, patients with LUAD were classified into RiskScore-high and –low groups based on the median RiskScore of each separate cohort as the cut-off value. Weltch't test and Wilcoxon rank sum test were used for comparison of two groups. Spearman's test was applied in all correlation analysis. Cox regression and Log-rank P were performed for survival analysis.
3. Results
3.1. Screening for prognostic genes in lung adenocarcinoma
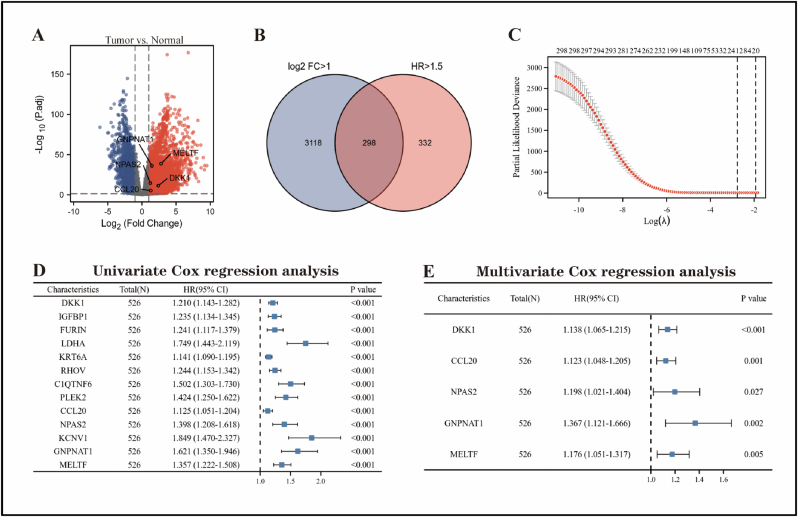
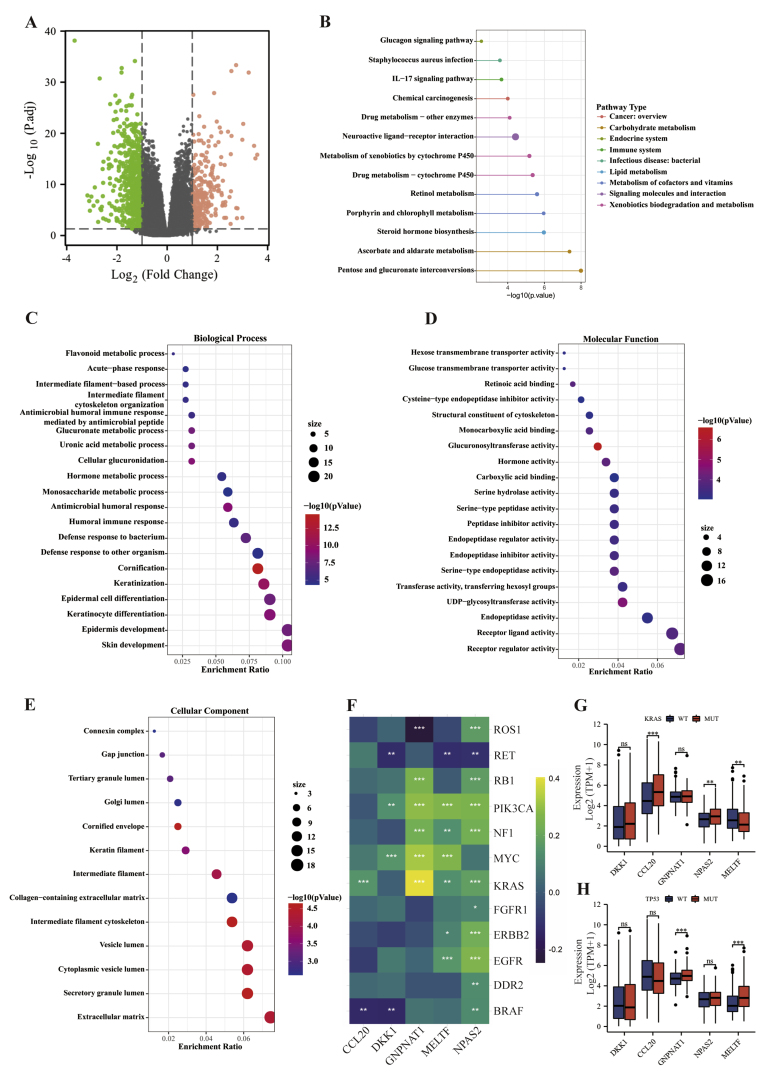
The overview of the process used in our study was shown in Figure 1. In LUAD tissues, differential gene expression analysis revealed that 3416 genes were up-regulated and 1966 genes were down-regulated (Figure 2A). Bulk survival analysis showed that the number of genes meeting HR > 1.5 and P < 0.05 was 630. All up-regulated genes were extracted and intersected with potential prognostic genes, yielding a total of 298 genes up-regulated in LUAD and linked to prognosis (Figure 2B). Then, a total of 13 candidate genes were identified using Lasso regression analysis (Figure 2C). Finally, univariate and multivariate Cox regression analyses identified DKK1, CCL20, NPAS2, GNPNAT1, and MELTF as LUAD-associated prognostic genes (Figure 2D, E). We generated a RiskScore using the TCGA training subset based on the regression coefficients of these five genes. The following formula was used to calculate the RiskScore:
| RiskScore = 0.13 × DKK1 + 0.12 × CCL20 + 0.18 × NPAS2 + 0.31 × GNPNAT1 + 0.16 × MELTF |
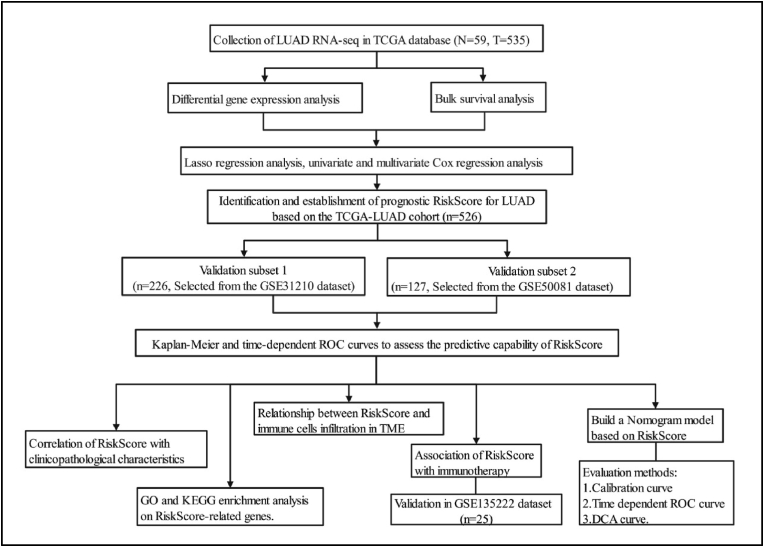
Figure 1.
Flow chart of this study.
Figure 2.
Screening for prognostic genes in LUAD using the TCGA-LUAD cohort. (A) Volcano map showing differentially expressed genes in LUAD tissue and normal lung tissue. (B) Venn diagram showing genes that are up-regulated in LUAD tissue and associated with prognosis. Lasso regression (C), univariate (D) and multivariate Cox regression analyses (E) were performed to screen candidate genes.
3.2. Validating the prognostic value of RiskScore
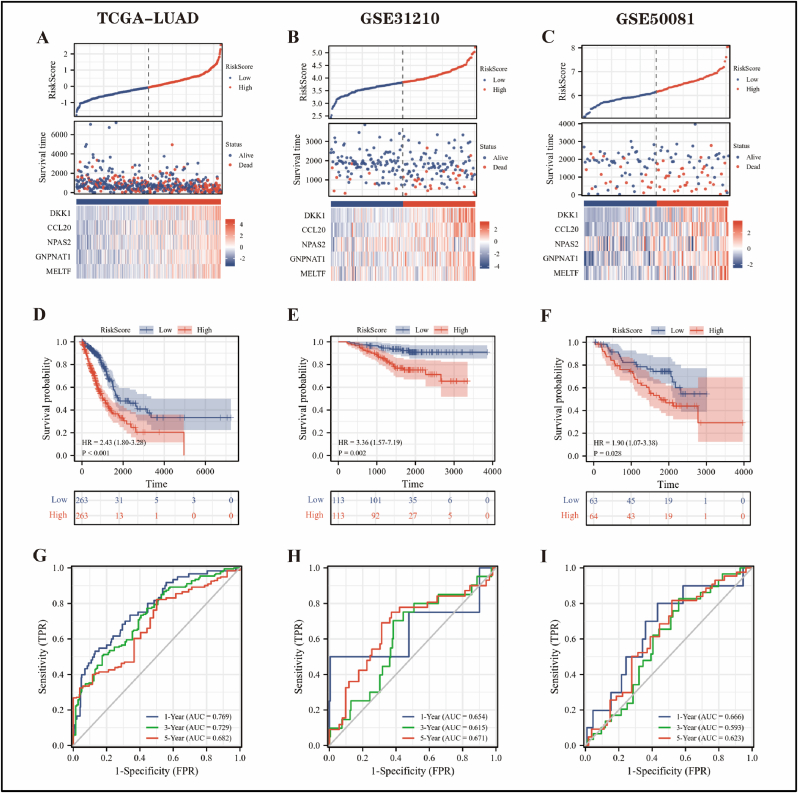
To investigate the prognostic value of RiskScore, the TCGA-LUAD cohort was employed as the training subset, GSE31210 and GSE50081 were applied as the validation subset 1 and validation subset 2, respectively. First, scatter plots were performed to estimate the survival status and expression of five prognostic genes in LUAD patients in the RiskScore-high and -low groups, and the result was that patients in the RiskScore-high group had more deaths and slightly shorter survival (Figure 3A–C). According to Kaplan-Meier curves, LUAD patients in the RiskScore-high group had a worse overall survival rate than those in the -low group (Figure 3D–F). In the TCGA-LUAD cohort, time-dependent ROC curves revealed that the probability of RiskScore predicting 1-, 3-, and 5-year survival in LUAD patients was 0.769, 0.729, and 0.682, respectively (Figure 3G). In addition, the prediction efficiency of RiskScore was almost always above 0.600 in all cases of the GSE31210 and GSE50081 cohorts (Figure 3H, I), indicating that the prediction results were more reliable.
Figure 3.
Prognostic value of RiskScore. The scatterplot of RiskScore, TCGA-LUAD cohort was the training subset (A), GSE31210 (B) and GSE50081 (C) cohorts were the validation subset 1 and validation subset 2, respectively. (D–F) Kaplan–Meier curves demonstrated the overall survival of LUAD patients in the RiskScore-high and -low groups, and statistical tests were performed using Cox regression analysis. (G–I) Time-dependent ROC curves demonstrated the ability of RiskScore to predict 1-, 3-, and 5-year overall survival in LUAD patients.
3.3. Relationship between RiskScore and clinicopathological characteristics
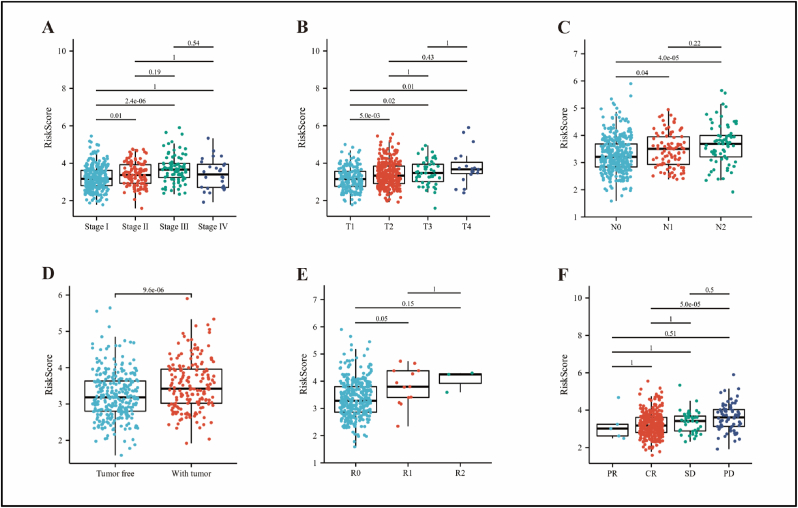
Next, we evaluated the relationship between clinicopathological characteristics and RiskScore using TCGA-LUAD cohort. As shown in Figure 4, high-RiskScore was associated with pathological stage (stage II vs. stage I, P = 0.01; stage III vs. stage I, P < 0.001), T-stage (T2 vs. T1, P = 0.005; T3 vs. T1, P = 0.02; T4 vs. T1, P = 0.01), N-stage (N1 vs. N0, P = 0.04), tumor status (with tumor vs. tumor free, P < 0.001), residual tumor (R1 vs. R0, P = 0.05), and treatment outcome (PD vs. CR, P < 0.001).
Figure 4.
Correlation analysis of RiskScore with clinicopathological characteristics using the TCGA-LUAD cohort. RiskScore was related to pathological stage (A), T-stage (B), N-stage (C), tumor status (D), residual tumor (E), and treatment outcome (F). Statistical tests were performed using the Wilcoxon rank sum test. CR, complete remission/response; PD, progressive disease; PR, partial remission/response; SD, stable disease.
3.4. Identification of the biological functions of RiskScore-related genes
To explore the biological function of RiskScore-related genes, we separated patients in the TCGA-LUAD cohort into two groups based on the median RiskScore. In total, 274 up-regulated genes and 631 down-regulated genes were identified (Figure 5A). Following that, we extracted all up-regulated genes for KEGG pathway enrichment analysis and found RiskScore-related genes were mainly implicated in substance metabolism, including retinol metabolism, ascorbate and aldarate metabolism, porphyrin and chlorophyll metabolism, and drug metabolism. This was followed by oncogenic and immune pathways, such as chemical carcinogenesis and the IL-17 signaling pathway (Figure 5B). Furthermore, GO analysis indicated that RiskScore-related genes mainly participated in keratinization and epidermal cell differentiation, followed by still metabolism-related pathways, and was also associated with humoral immune response (Figure 5C). The majority of molecular functions were found in the activities of several metabolic enzymes and membrane transporters (Figure 5D). Transcription proteins were mostly located in the extracellular matrix and various lumens (Figure 5E).
Figure 5.
Explore the biological functions of RiskScore using the TCGA-LUAD cohort. (A) Volcano plot showing differentially expressed genes (DEGs) in the RiskScore-high and -low groups. (B) KEGG analysis of DEGs in the RiskScore-high group. GO analysis of DEGs in the RiskScore-high group, BP (C), MF (D), CC (E). (F) Correlation analysis of DKK1, CCL20, NPAS2, GNPNAT1 and MELTF with critical oncogenes in LUAD respectively, statistical tests were performed by Spearman's test. (G, H) DKK1, CCL20, NPAS2, GNPNAT1 and MELTF were differentially expressed in KRAS and TP53 mutant and wild-type groups, and statistical tests were performed using the Wilcoxon rank sum test.
Next, to investigate whether these five genes show co-expression with critical oncogenes, we selected a subset of key genes driving lung cancer progression and performed a correlation analysis. The results were that NPAS2 was positively associated with almost all oncogenes, followed by MELTF and GNPNAT1, but DKK1 and CCL20 were not related to the vast majority of oncogenes. Interestingly, the oncogenes KRAS and PIK3CA were closely linked to these five genes (Figure 5F). Among them, KRAS is a common mutation site in LUAD, and almost 30% of LUAD are driven by activated KRAS mutations [10]. Here, we found that the expression of CCL20 and NPAS2 were significantly higher in the KRAS mutant group than in the wild-type group, while the opposite was true for MELTF (Figure 5G). Furthermore, tumor suppressors undergoing genomic alterations, such as TP53, have also emerged as central determinants of oncogene-driven molecular and clinical heterogeneity in subgroups of lung cancer [11]. Strikingly, the expression of GNPNAT1 and MELTF was significantly elevated when TP53 was mutated (Figure 5H).
3.5. Relationship between RiskScore and immune cells infiltration in TME
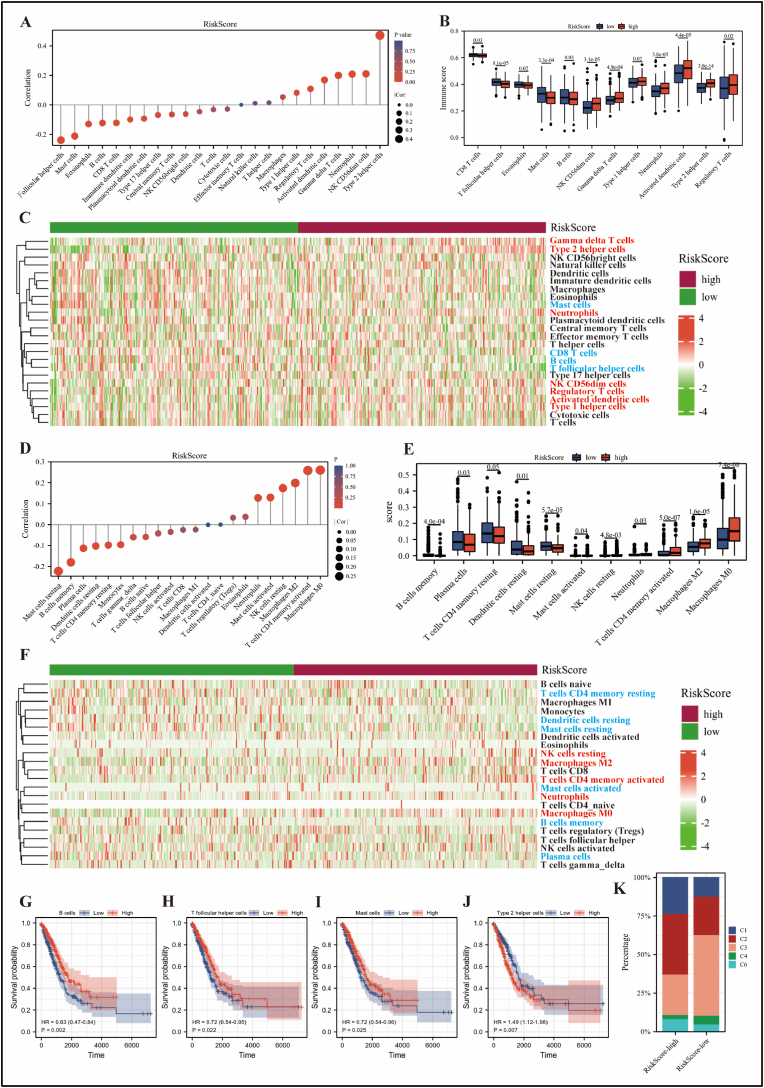
Given that RiskScore-related genes were linked to immunological pathways, such as the IL-17 signaling pathway and humoral immune response. Therefore, we proceeded to explore the relationship between RiskScore and immune cells in TME. Firstly, based on the GSVA algorithm, RiskScore was mainly positively correlated with Th2 cells and negatively correlated with T follicular helper cells and mast cells as shown in Figure 6A and Table 1. In addition, neutrophils, aDC, NK CD56dim, Tgd, Th1, Th2 and Treg cells were significantly enriched in the RiskScore-high group, while B cells, CD8+ T cells, eosinophils, mast cells and T follicular helper cells were more abundant in the -low group (Figure 6B, C). Second, based on the cibersort algorithm, RiskScore was mainly positively correlated with macrophage M0 and T cells CD4 memory activated, and negatively correlated with mast cells resting and B cells memory as shown in Figure 6D and Table 1. Moreover, macrophages M0, macrophages M2, T cells CD4 memory activated, neutrophils, and NK cell resting were more infiltrated in the RiskScore-high group, while mast cells resting, B cells memory, plasma cells, T cells CD4 memory resting, dendritic cells resting, and mast cells activated were more abundant in the -low group (Figure 6D, E). Overall, the results based on these two algorithms were mostly consistent, but there were still differences. Thus, a beneficial combination of the two may contribute to a clearer understanding of the role of RiskScore in TME.
Figure 6.
Role of RiskScore in the TME was analyzed using the TCGA-LUAD cohort. Based on GSVA algorithm: (A) Relationship between RiskScore and immune cells infiltration in TME, and statistical tests were performed by Spearman’ test. (B) The infiltration abundance of immune cells was differentially distributed between RiskScore-high and -low groups, and statistical tests were performed using the Wilcoxon rank sum test. (C) Heat map showing the enrichment score of immune cells in the RiskScore-high and -low groups. Red font indicates that immune cells are infiltrated in higher abundance in the RiskScore-high group than in the low group. The opposite is true for blue font. Based on cibersort algorithm: (D) Relationship between RiskScore and immune cells infiltration in TME, and statistical tests were performed by Spearman's test. (E) The infiltration abundance of immune cells was differentially distributed between RiskScore-high and -low groups, and statistical tests were performed using the Wilcoxon rank sum test. (F) Heat map showing the enrichment score of immune cells in the RiskScore-high and -low groups. B cells (G), T follicular helper cells (H), mast cells (I) and Th2 cells (J) were associated with overall survival in LUAD patients according to the GSVA algorithm, and statistical tests were performed using Cox regression analysis. (K) Differences in immune subtypes between the RiskScore-high and -low groups. C1: wound healing, C2: IFN-gamma dominant, C3: inflammatory, C4: lymphocyte depleted, C6: TGF-β dominant.
Table 1.
Correlation analysis of RiskScore and immune cells infiltration in TME using the TCGA database.
| Immune cells | Cor/P-value | Immune cells | Cor/P-value | Immune cells | Cor/P-value |
|---|---|---|---|---|---|
| GSVA algorithm | |||||
| T follicular helper cells | −0.239/∗∗∗ | Central memory T cells | −0.065/ns | Macrophages | 0.053/ns |
| Mast cells | −0.211/∗∗∗ | NK CD56 bright cells | −0.062/ns | Type 1 helper cells | 0.082/ns |
| Eosinophils | −0.130/∗∗ | Dendritic cells | −0.046/ns | Regulatory T cells | 0.109/∗ |
| B cells | −0.123/∗∗ | T cells | −0.032/ns | Activated dendritic cells | 0.169/∗∗∗ |
| CD8 T cells | −0.122/∗∗ | Cytotoxic cells | −0.029/ns | Gamma delta T cells | 0.200/∗∗∗ |
| Immature dendritic cells | −0.099/∗ | Effector memory T cells | 0/ns | Neutrophils | 0.209/∗∗∗ |
| Plasmacytoid dendritic cells | −0.093/∗ | Natural killer cells | 0.011/ns | NK CD56dim cells | 0.211/∗∗∗ |
| Type 17 helper cells | −0.069/ns | T helper cells | 0.015/ns | Type 2 helper cells | 0.471/∗∗∗ |
| Cibersort algorithm | |||||
| Mast cells resting | −0.222/∗∗∗ | T cells follicular helper | −0.042/ns | Neutrophils | 0.128/∗∗ |
| B cells memory | −0.180/∗∗∗ | NK cells activated | −0.035/ns | Mast cells activated | 0.130/∗∗ |
| Plasma cells | −0.113/∗∗ | T cells CD8 | −0.025/ns | NK cells resting | 0.174/∗∗∗ |
| Dendritic cells resting | −0.102/∗ | Macrophages M1 | −0.024/ns | Macrophages M2 | 0.199/∗∗∗ |
| T cells CD4 memory resting | −0.098/∗ | Dendritic cells activated | −0.001/ns | T cells CD4 memory activated | 0.258/∗∗∗ |
| Monocytes | −0.096/∗ | T cells CD4_naive | 0/ns | Macrophages M0 | 0.260/∗∗∗ |
| T cells gamma_delta | −0.060/ns | T cells regulatory | 0.033/ns | ||
| B cells naïve | −0.059/ns | Eosinophils | 0.037/ns | ||
∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001; ns, no statistical significance.
According to the GSVA algorithm, infiltration of B cells, T follicular helper cells and mast cells favored prolonged overall survival in LUAD patients (Figure 6G–I), whereas Th2 cells were detrimental (Figure 6J). Next, we explored the correlation between the RiskScore-high and -low groups and the six previously reported pan-cancer immune subtypes (C1–C6), in which LUAD was mainly concentrated in C1, C2, C3, C4 and C6 [9]. As shown in Figure 6K, the proportion of immune subtypes C1 and C2 was higher and the proportion of C3 was lower in the RiskScore-high group compared to the -low group. Of these, C3 was associated with a better prognosis, while C1 and C2 indicated a poorer prognosis. These results correspond to a longer survival of LUAD patients in the RiskScore-low than in the -high group. Interestingly, immune cells in C1 and C2 subtypes were predominantly Th2 cells and macrophages, respectively, which further validated the conclusion based on GSVA and cibersort algorithms that the abundance of Th2 cells and macrophages was higher in the RiskScore-high group.
3.6. RiskScore prediction of response to immunotherapy and tumor relapse
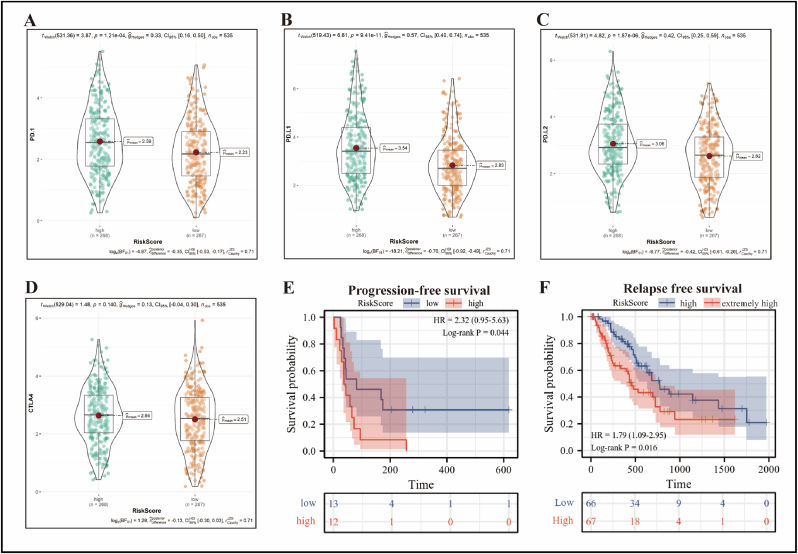
To explore the response of RiskScore to immunotherapy, we first evaluated its relationship with clinically significant immunosuppressive checkpoints. Of note, the expression of PD-1, PD-L1 and PD-L2 was significantly higher in the RiskScore-high group than in the -low group (P < 0.001) (Figure 7A–C), while CTLA4 expression was not statistically different between the two groups (Figure 7D). Subsequently, we included 25 patients with NSCLC receiving anti-PD-1/PD-L1 immunotherapy in the GSE135222 dataset. Strikingly, Kaplan-Meier curve demonstrated that patients with a low RiskScore had a greater progression-free survival rate than those with a high RiskScore (Figure 7E).
Figure 7.
Relationship of RiskScore to immunosuppressive checkpoints and response to immunotherapy and tumor relapse. PD-1 (A), PD-L1 (B), PD-L2 (C), and CTLA4 (D) were differentially expressed in the RiskScore-high and -low groups using the TCGA-LUAD cohort, and statistical tests were performed using Weltch't test. (E) Kaplan-Meier curve showed the difference in progression-free survival between 25 patients receiving anti-PD-1/PD-L1 therapy in the RiskScore-high and -low groups using the GSE135222 cohort, and statistical test was performed using Log-rank P test. (F) LUAD patients in the RiskScore-high group were further divided into four groups based on quartiles of RiskScore using the TCGA database, with the top 25% being the extremely high-risk group (n = 67) and the bottom 25% being the high-risk group (n = 66) for RFS analysis, and statistical test was performed using Log-rank P test.
Next, to investigate the effect of RiskScore on relapse-free survival in LUAD patients, we further divided LUAD patients in the RiskScore-high group into four groups according to the quartiles of RiskScore by using the TCGA database, with the top 25% being the extremely high-risk group (n = 67) and the bottom 25% being the high-risk group (n = 66) for RFS analysis. The result was that patients in the extremely high-risk group had a significantly shorter relapse-free survival than those in the high-risk group (Figure 7F).
3.7. Construction and evaluation of nomogram model based on RiskScore
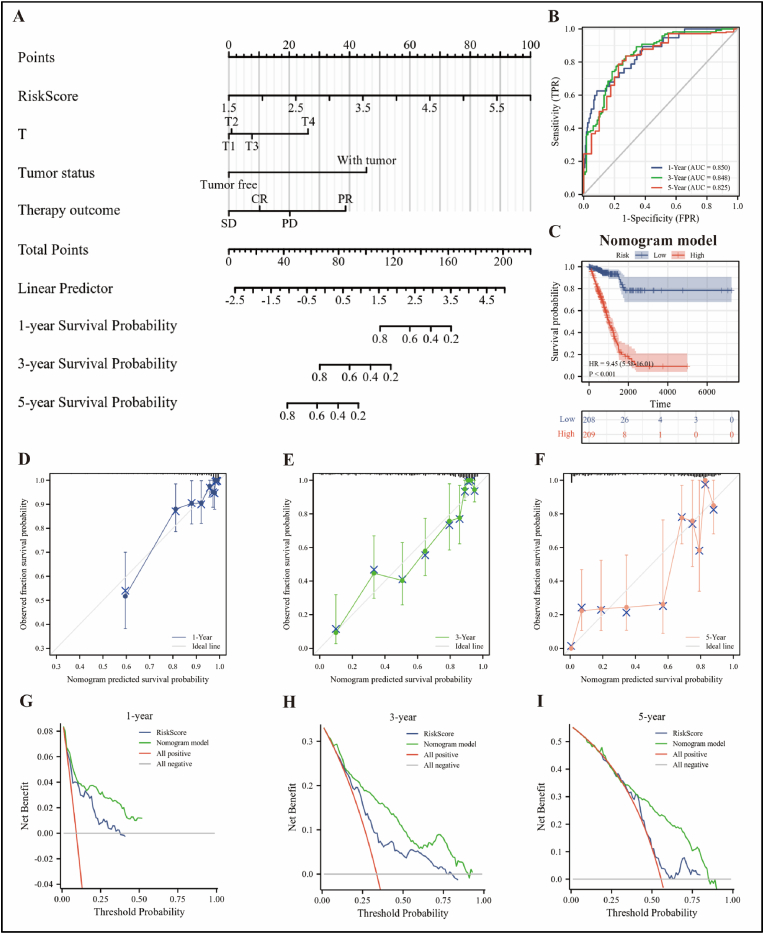
To find independent predictors of prognosis, we performed univariate and multivariate Cox regression analyses using the RiskScore and clinical data of LUAD patients based on TCGA-LUAD cohort. As shown in Table 2, RiskScore, T-stage, tumor status, and treatment outcome were key independent predictors of LUAD prognosis and used to construct a Nomogram model (Figure 8A), which allows for a more accurate and personalized assessment of the probability of survival in LUAD patients.
Table 2.
| Univariate and multivariate Cox regression analysis of clinical data of LUAD patients using the TCGA database (OS).
| Characteristics | Total (N) | Univariate Cox regression analysis |
Multivariate Cox regression analysis |
||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| RiskScore | 526 | 2.718 (2.197–3.363) | <0.001 | 1.993 (1.395–2.848) | <0.001 |
| Pathologic stage | 518 | ||||
| Stage I | 290 | Reference | |||
| Stage II | 121 | 2.418 (1.691–3.457) | <0.001 | 0.388 (0.140–1.071) | 0.068 |
| Stage III | 81 | 3.544 (2.437–5.154) | <0.001 | 0.488 (0.079–3.035) | 0.442 |
| Stage IV | 26 | 3.790 (2.193–6.548) | <0.001 | 0.644 (0.199–2.077) | 0.461 |
| T stage | 523 | ||||
| T1 | 175 | Reference | |||
| T2 | 282 | 1.521 (1.068–2.166) | 0.020 | 1.011 (0.528–1.935) | 0.974 |
| T3 | 47 | 2.937 (1.746–4.941) | <0.001 | 2.863 (0.938–8.734) | 0.065 |
| T4 | 19 | 3.326 (1.751–6.316) | <0.001 | 8.445 (1.673–42.637) | 0.010 |
| N stage | 510 | ||||
| N0 | 343 | Reference | |||
| N1 | 94 | 2.381 (1.695–3.346) | <0.001 | 2.534 (0.955–6.727) | 0.062 |
| N2 | 71 | 3.108 (2.136–4.521) | <0.001 | 3.332 (0.651–17.061) | 0.149 |
| N3 | 2 | 0.000 (0.000–) | 0.994 | 0.000 (0.000–) | 0.995 |
| Residual tumor | 363 | ||||
| R0 | 347 | Reference | |||
| R1 | 13 | 3.255 (1.694–6.251) | <0.001 | 1.333 (0.441–4.026) | 0.610 |
| R2 | 3 | 11.085 (3.443–35.689) | <0.001 | ||
| Therapy outcome | 439 | ||||
| CR | 326 | Reference | |||
| SD | 37 | 1.126 (0.566–2.240) | 0.736 | 0.575 (0.198–1.670) | 0.309 |
| PD | 71 | 3.710 (2.584–5.326) | <0.001 | 3.016 (1.597–5.696) | <0.001 |
| PR | 5 | 2.606 (0.639–10.637) | 0.182 | 14.925 (3.175–70.149) | <0.001 |
| M stage (M1 vs. M0) | 377 | 2.136 (1.248–3.653) | 0.006 | ||
| Tumor status (with tumor vs. tumor free) | 472 | 6.430 (4.418–9.359) | <0.001 | 7.572 (4.080–14.053) | <0.001 |
| Gender (female vs. male) | 526 | 0.934 (0.701–1.245) | 0.642 | ||
| Age (≥65 vs. <65) | 516 | 1.143 (0.854–1.530) | 0.369 | ||
Bold text indicates statistical significance.
Figure 8.
Diagnostic efficiency of the Nomogram model. (A) A nomogram model constructed based on the TCGA-LUAD cohort was used to assess the 1-, 3-, and 5-year overall survival of LUAD patients. (B) Time-dependent ROC curve demonstrated the ability of the model to predict overall survival at 1-, 3- and 5-year in LUAD patients. (C) Kaplan-Meier curve showing overall survival of LUAD patients in high and low risk groups in the Nomogram model, and statistical tests were performed using Cox regression analysis. (D–F) The calibration curves of the model were shown for 1-, 3- and 5-year, respectively. (G–I) The DCA curves for Nomogram model and RiskScore were shown for 1-, 3- and 5-year, respectively.
To assess the reliability of the model, we first employed time-dependent ROC curves to show that the model predicted 1-, 3-, and 5-year survival rates for LUAD patients with probabilities of 0.850, 0.848, and 0.825, respectively (Figure 8B). Kaplan-Meier curve showed that LUAD patients in the high-risk group had significantly lower survival rates than those in the low-risk group (Figure 8C). Then, we calculated the C-index of the model as 0.804 (95% CI: 0.783–0.825) and plotted the calibration curves for 1-, 3-, and 5-year (Figure 8D–F). In addition, ROC curves can only assess the goodness of a model by sensitivity and specificity, whereas decision analysis curves consider the clinical utility or patient benefit aspects of the model. Thus, we further evaluated the reliability of the model using DCA curves and found that the curves of the Nomogram model were found to be higher than all positive and negative control lines within a certain range, and all were consistently higher than the curves of the RiskScore (Figure 8G–I), further suggesting that the Nomogram model has better clinical application in predicting the overall survival of patients than using RiskScore alone.
4. Discussion
The most frequent subtype of lung cancer is lung adenocarcinoma, which has a dismal 5-year survival rate. Evidence currently demonstrated that biomarkers development and implementation can provide potential prognostic value in order to guide sensible clinical treatment [12]. Consequently, understanding the evolution of lung adenocarcinoma necessitates the screening and identification of biomarkers linked to etiology and prognosis. RiskScore, consisting of five genes, DKK1, CCL20, NPAS2, GNPNAT1 and MELTF, was discovered to be an ideal predictive predictor for LUAD in this study. The TNM stage of the tumor was positively connected with RiskScore, implying that RiskScore may have a pro-tumorigenic effect, hastening the progression of LUAD patients to advanced stages of the disease. What's more, LUAD patients with a high RiskScore had a worse survival rate, and the Nomogram model based on RiskScore possessed a high prediction accuracy as measured by numerous methodologies, suggesting that it could be a valuable tool for clinical diagnosis.
Recent studies revealed that DKK1 promoted the migration and invasion of non-small cell lung and ovarian cancers through the β-catenin and P-JNK1 signaling pathways, respectively [13, 14], and was associated with poor prognosis in pancreatic ductal adenocarcinoma, bladder cancer, and hepatocellular carcinoma [15, 16, 17]. In addition, CCL20 exerted tumor-promoting effects in the tumor microenvironment. High expression of CCL20 in liver cancer tissues facilitated angiogenesis and leaded to enhance tumor recurrence and decrease patient survival [18, 19], and CCL20 potentiated the invasion of breast cancer cells and resistance to the chemotherapeutic agent taxane [20, 21], and was also implicated in poor prognosis in colorectal, prostate, and lung cancers [22, 23, 24]. NPAS2 was demonstrated to be a potential prognostic biomarker in colorectal and breast cancers [25, 26], and facilitated cell survival in hepatocellular carcinoma through trans-activation of CDC25A [27]. Several studies had collectively indicated that GNPNAT1 was closely related to poor prognosis in lung adenocarcinoma [28, 29], and MELTF can be considered as a prognostic marker in lung adenocarcinoma and gastric cancer [30, 31]. In summary, many studies have shown that five genes, DKK1, CCL20, NPAS2, GNPNAT1 and MELTF, are engaged in the pathogenesis and progression of multiple tumors.
Altered metabolism was a hallmark of cancer, and reprogramming of energy metabolism had historically been considered a common phenomenon in tumors [32], as well as affecting tumor proliferation and migration [33]. In this study, KEGG enrichment analysis revealed genes in the RiskScore-high group involved in energy metabolism, such as glucagon signaling pathway and pentose and glucuronate interconversions, implying that RiskScore may modify the metabolic state of malignancies. Additionally, it was relevant to the drug metabolism.
Chemotherapy and tyrosine kinase inhibitors were the mainstays of lung cancer treatment in the past. Because the mechanism of tumor cell immune escape has been explained in recent years, immune checkpoint inhibitor therapy has emerged as a novel hope for cancer patients who have failed numerous lines of treatment [34]. We speculated that RiskScore may contribute to immune escape of tumors, which leads to immunotherapy failure, because LUAD patients in the RiskScore-high group had significantly higher PD-1 and PD-L1 expression and lower progression-free survival in receiving immunotherapy in the current study. Interestingly, Ming Yi et al. [35] constructed a riskscore consisting of 17 genes that also predicted response to immune checkpoint inhibitors in LUAD patients based on an IPS scoring scheme and concluded that the relative probability of response to anti-PD-1/PD-L1 and anti-CTLA-4 therapy was higher in the low risk score group. In contrast, RiskScore, which was applied to predict immunotherapy response in our model, contained fewer genes and facilitated clinical application. Second, the GSE135222 dataset we utilized contained patient survival information, and the effect of predicting immunotherapy was more intuitively demonstrated by Kaplan-Meier curve. However, there were still some limitations, such as the small number of patients, which could easily introduce bias. The second was that this dataset also has patients with lung squamous carcinoma, which may differ somewhat from the lung adenocarcinoma we studied.
In the present study, we utilized GSVA and cibersort algorithms to evaluate the relationship between immune cells in TME and RiskScore, respectively. The two algorithms were consistent in that RiskScore was positively correlated with neutrophils and NK cells, and negatively associated with mast cells, B cells and dendritic cells. The difference was that RiskScore was significantly positively relevant to Th2 cells based on the GSVA algorithm, while RiskScore was positively linked to macrophages M0 and M2 based on the cibersort algorithm. Overall, the beneficial combination of both algorithms helps to understand more clearly the role of RiskScore in the tumor microenvironment. According to the GSVA algorithm, infiltration of B cells, T follicular helper cells, and mast cells favored survival of LUAD patients, whereas Th2 cells were detrimental. Strikingly, RiskScore was positively correlated with Th2 cells, and negatively correlated with B cells, T follicular helper cells, and mast cells. Several studies have shown that increased abundance of Th2 cells promotes tumor progression, such as in cervical cancer [36] and ovarian cancer [37]. What's more, M2 polarization of macrophages was associated with immunosuppression and tumorigenesis and metastasis [38]. These data suggested that RiskScore may be engaged in the regulation of tumor immunity, resulting in an immunosuppressive microenvironment that favored tumor cell survival and proliferation.
To prevent low sample size and weaken individual differences, this work primarily employed the TCGA and GEO databases for screening and validation of prognostic biomarkers, which were based on big samples and vast data. The combination of numerous genes to determine prognosis is more accurate and superior to standard individual indicators. Not only is prognosis useful, but so is immune efficacy prediction. The use of time-dependent ROC curves, calibration curves, and the DCA curves can considerably improve the accuracy of the results when judging the Nomogram model's prediction abilities. However, limitations remain. First, the vast majority of LUAD patients in the TCGA database are white or African American, and other races using the model may experience racial variances. Second, transcriptome data based on different sequencing platforms will vary somewhat, so basic experiments are needed to verify the expression of these five genes at the protein level. In addition, the molecular pathways involved in RiskScore in this study also require further functional experiments to clarify the underlying mechanisms of the genes. Finally, the relatively small number of patients included in the RiskScore prediction of immunotherapy response may reduce the credibility and generalizability of the results. Despite the fact that this is a retrospective study, it does uncover novel prognostic indicators and treatment options for lung cancer.
In conclusion, we have successfully established a novel metabolic-immune related model to predict the prognosis and response to immunotherapy in LUAD patients.
Declarations
Author contribution statement
Xiaolong Tang: Performed the experiments; Analyzed and interpreted the data; Write the paper.
Chumei Qi and Honghong Zhou: Contributed the analysis tools and data.
Yongshuo Liu: Conceived and designed the experiments.
Funding statement
Assistant professor Yongshuo Liu was supported by National Natural Science Foundation of China [81802400] and China Postdoctoral Science Foundation [2020M670053]. Associate professor Honghong Zhou was supported by National Natural Science Foundation of China [81902519].
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Hutchinson B.D., Shroff G.S., Truong M.T., Ko J.P. Spectrum of lung adenocarcinoma. Semin. Ultrasound CT MR. 2019;40:255–264. doi: 10.1053/j.sult.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Denisenko T.V., Budkevich I.N., Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018;9:117. doi: 10.1038/s41419-017-0063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inamura K. Clinicopathological characteristics and mutations driving development of early lung adenocarcinoma: tumor initiation and progression. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19041259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laplane L., Duluc D., Bikfalvi A., Larmonier N., Pradeu T. Beyond the tumour microenvironment. Int. J. Cancer. 2019;145:2611–2618. doi: 10.1002/ijc.32343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin M.Z., Jin W.L. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct. Target. Ther. 2020;5:166. doi: 10.1038/s41392-020-00280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020;17:807–821. doi: 10.1038/s41423-020-0488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C., Angell H., Fredriksen T., Lafontaine L., Berger A., Bruneval P., Fridman W.H., Becker C., Pages F., Speicher M.R., Trajanoski Z., Galon J. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Y.T., Porta-Pardo E., Gao G.F., Plaisier C.L., Eddy J.A., Ziv E., Culhane A.C., Paull E.O., Sivakumar I., Gentles A.J., Malhotra R., Farshidfar F., Colaprico A., Parker J.S., Mose L.E., Vo N.S., Liu J., Liu Y., Rader J., Dhankani V., Reynolds S.M., Bowlby R., Califano A., Cherniack A.D., Anastassiou D., Bedognetti D., Mokrab Y., Newman A.M., Rao A., Chen K., Krasnitz A., Hu H., Malta T.M., Noushmehr H., Pedamallu C.S., Bullman S., Ojesina A.I., Lamb A., Zhou W., Shen H., Choueiri T.K., Weinstein J.N., Guinney J., Saltz J., Holt R.A., Rabkin C.S., Lazar A.J., Serody J.S., Demicco E.G., Disis M.L., Vincent B.G., Shmulevich I. The immune landscape of cancer. Immunity. 2018;48:812–830. doi: 10.1016/j.immuni.2018.03.023. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uras I.Z., Moll H.P., Casanova E. Targeting KRAS mutant non-small-cell lung cancer: past, present and future. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21124325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skoulidis F., Heymach J.V. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat. Rev. Cancer. 2019;19:495–509. doi: 10.1038/s41568-019-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu F., Huang X., Li Y., Chen Y., Lin L. m(6)A-related lncRNAs are potential biomarkers for predicting prognoses and immune responses in patients with LUAD. Mol. Ther. Nucleic Acids. 2021;24:780–791. doi: 10.1016/j.omtn.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J., Zhang X., Zhao X., Jiang M., Gu M., Wang Z., Yue W. DKK1 promotes migration and invasion of non-small cell lung cancer via beta-catenin signaling pathway. Tumour Biol. 2017;39 doi: 10.1177/1010428317703820. [DOI] [PubMed] [Google Scholar]
- 14.Wang S., Zhang S. Dickkopf-1 is frequently overexpressed in ovarian serous carcinoma and involved in tumor invasion. Clin. Exp. Metastasis. 2011;28:581–591. doi: 10.1007/s10585-011-9393-9. [DOI] [PubMed] [Google Scholar]
- 15.Tang Y., Zhang Z., Tang Y., Chen X., Zhou J. Identification of potential target genes in pancreatic ductal adenocarcinoma by bioinformatics analysis. Oncol. Lett. 2018;16:2453–2461. doi: 10.3892/ol.2018.8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun D.K., Wang L., Wang J.M., Zhang P. Serum Dickkopf-1 levels as a clinical and prognostic factor in patients with bladder cancer. Genet. Mol. Res. 2015;14:18181–18187. doi: 10.4238/2015.December.23.5. [DOI] [PubMed] [Google Scholar]
- 17.Desert R., Mebarki S., Desille M., Sicard M., Lavergne E., Renaud S., Bergeat D., Sulpice L., Perret C., Turlin B., Clement B., Musso O. “Fibrous nests” in human hepatocellular carcinoma express a Wnt-induced gene signature associated with poor clinical outcome. Int. J. Biochem. Cell Biol. 2016;81:195–207. doi: 10.1016/j.biocel.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 18.Ding X., Wang K., Wang H., Zhang G., Liu Y., Yang Q., Chen W., Hu S. High expression of CCL20 is associated with poor prognosis in patients with hepatocellular carcinoma after curative resection. J. Gastrointest. Surg. 2012;16:828–836. doi: 10.1007/s11605-011-1775-4. [DOI] [PubMed] [Google Scholar]
- 19.He H., Wu J., Zang M., Wang W., Chang X., Chen X., Wang R., Wu Z., Wang L., Wang D., Lu F., Sun Z., Qu C. CCR6(+) B lymphocytes responding to tumor cell-derived CCL20 support hepatocellular carcinoma progression via enhancing angiogenesis. Am. J. Cancer Res. 2017;7:1151–1163. [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S.K., Park K.K., Kim H.J., Park J., Son S.H., Kim K.R., Chung W.Y. Human antigen R-regulated CCL20 contributes to osteolytic breast cancer bone metastasis. Sci. Rep. 2017;7:9610. doi: 10.1038/s41598-017-09040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W., Qin Y., Wang D., Zhou L., Liu Y., Chen S., Yin L., Xiao Y., Yao X.H., Yang X., Ma W., Chen W., He X., Zhang L., Yang Q., Bian X., Shao Z.M., Liu S. CCL20 triggered by chemotherapy hinders the therapeutic efficacy of breast cancer. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2005869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D., Yuan W., Wang Y., Wu Q., Yang L., Li F., Chen X., Zhang Z., Yu W., Maimela N.R., Cao L., Wang D., Wang J., Sun Z., Liu J., Zhang Y. Serum CCL20 combined with IL-17A as early diagnostic and prognostic biomarkers for human colorectal cancer. J. Transl. Med. 2019;17:253. doi: 10.1186/s12967-019-2008-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghadjar P., Loddenkemper C., Coupland S.E., Stroux A., Noutsias M., Thiel E., Christoph F., Miller K., Scheibenbogen C., Keilholz U. Chemokine receptor CCR6 expression level and aggressiveness of prostate cancer. J. Cancer Res. Clin. Oncol. 2008;134:1181–1189. doi: 10.1007/s00432-008-0403-5. [DOI] [PubMed] [Google Scholar]
- 24.Wang G.Z., Cheng X., Li X.C., Liu Y.Q., Wang X.Q., Shi X., Wang Z.Y., Guo Y.Q., Wen Z.S., Huang Y.C., Zhou G.B. Tobacco smoke induces production of chemokine CCL20 to promote lung cancer. Cancer Lett. 2015;363:60–70. doi: 10.1016/j.canlet.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Yi C., Mu L., de la Longrais I.A., Sochirca O., Arisio R., Yu H., Hoffman A.E., Zhu Y., Katsaro D. The circadian gene NPAS2 is a novel prognostic biomarker for breast cancer. Breast Cancer Res. Treat. 2010;120:663–669. doi: 10.1007/s10549-009-0484-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang S.F., Xu M., Yang H.Y., Li P.Q., Chi X.F. [Expression of circadian gene NPAS2 in colorectal cancer and its prognostic significance] Nan Fang Yi Ke Da Xue Xue Bao. 2016;36:714–718. [PubMed] [Google Scholar]
- 27.Yuan P., Li J., Zhou F., Huang Q., Zhang J., Guo X., Lyu Z., Zhang H., Xing J. NPAS2 promotes cell survival of hepatocellular carcinoma by transactivating CDC25A. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S., Zhang H., Li H., Guo J., Wang J., Zhang L. Potential role of glucosamine-phosphate N-acetyltransferase 1 in the development of lung adenocarcinoma. Aging (Albany NY) 2021;13:7430–7453. doi: 10.18632/aging.202604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W., Jiang K., Wang J., Mei T., Zhao M., Huang D. Upregulation of GNPNAT1 predicts poor prognosis and correlates with immune infiltration in lung adenocarcinoma. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.605754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C., Long Q., Zhang D., Li J., Zhang X. Identification of a four-gene panel predicting overall survival for lung adenocarcinoma. BMC Cancer. 2020;20:1198. doi: 10.1186/s12885-020-07657-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sawaki K., Kanda M., Umeda S., Miwa T., Tanaka C., Kobayashi D., Hayashi M., Yamada S., Nakayama G., Omae K., Koike M., Kodera Y. Level of melanotransferrin in tissue and sera serves as a prognostic marker of gastric cancer. Anticancer Res. 2019;39:6125–6133. doi: 10.21873/anticanres.13820. [DOI] [PubMed] [Google Scholar]
- 32.Tan Y.T., Lin J.F., Li T., Li J.J., Xu R.H., Ju H.Q. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun. 2021;41:109–120. doi: 10.1002/cac2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zanotelli M.R., Zhang J., Reinhart-King C.A. Mechanoresponsive metabolism in cancer cell migration and metastasis. Cell Metab. 2021;33:1307–1321. doi: 10.1016/j.cmet.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qu J., Jiang M., Wang L., Zhao D., Qin K., Wang Y., Tao J., Zhang X. Mechanism and potential predictive biomarkers of immune checkpoint inhibitors in NSCLC. Biomed. Pharmacother. 2020;127 doi: 10.1016/j.biopha.2020.109996. [DOI] [PubMed] [Google Scholar]
- 35.Yi M., Li A., Zhou L., Chu Q., Luo S., Wu K. Immune signature-based risk stratification and prediction of immune checkpoint inhibitor's efficacy for lung adenocarcinoma. Cancer Immunol. Immunother. 2021;70:1705–1719. doi: 10.1007/s00262-020-02817-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin W., Niu Z., Zhang H., Kong Y., Wang Z., Yang X., Yuan F. Imbalance of Th1/Th2 and Th17/Treg during the development of uterine cervical cancer. Int. J. Clin. Exp. Pathol. 2019;12:3604–3612. [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L.H., Wang L.L., Zhang J., Zhang P., Li S.Z. [Th1/Th2 and Treg/Th17 cell balance in peripheral blood of patients with ovarian cancer] Nan Fang Yi Ke Da Xue Xue Bao. 2017;37:1066–1070. doi: 10.3969/j.issn.1673-4254.2017.08.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Essandoh K., Li Y., Huo J., Fan G.C. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock. 2016;46:122–131. doi: 10.1097/SHK.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supp. material/referenced in article.