Abstract
The interaction of exoribonuclease (ExoN) nonstructural protein (NSP14) with NSP10 co-factors is crucial for high-fidelity proofreading activity of coronavirus replication and transcription. Proofreading function is critical for maintaining the large genomes to ensure replication proficiency; therefore, while maintaining the viral replication fitness, quick resistance has been reported to the nucleotide analogue (NA) drugs. Therefore, targeting the NSP14 and NSP10 interacting interface with small molecules or peptides could be a better strategy to obstruct replication processes of coronaviruses (CoVs). A comparative study on the binding mechanism of NSP10 with the NSP14 ExoN domain of SARS-CoV-2, SARS-CoV, MERS-CoV, and four SARS-CoV-2 NSP14mutant complexes has been carried out. Protein–protein interaction (PPI) dynamics, per-residue binding free energy (BFE) analyses, and the identification of interface hotspot residues have been studied using molecular dynamics simulations and various computational tools. The BFE of the SARS-CoV NSP14–NSP10 complex was higher when compared to novel SARS-CoV-2 and MERS. However, SARS-CoV-2 NSP14mutant systems display a higher BFE as compared to the wild type (WT) but lower than SARS-CoV and MERS. Despite the high BFE, the SARS-CoV NSP14–NSP10 complex appears to be structurally more flexible in many regions especially the catalytic site, which is not seen in SARS-CoV-2 and its mutant or MERS complexes. The significantly high residue energy contribution of key interface residues and hotspots reveals that the high binding energy between NSP14 and NSP10 may enhance the functional activity of the proofreading complex, as the NSP10–NSP14 interaction is essential in maintaining the stability of the ExoN domain for the replicative fitness of CoVs. The factors discussed for SARS-CoV-2 complexes may be responsible for NSP14 ExoN having a high replication proficiency, significantly leading to the evolution of new variants of SARS-CoV-2. The NSP14 residues V66, T69, D126, and I201and eight residues of NSP10 (L16, F19, V21, V42, M44, H80, K93, and F96) are identified as common hotspots. Overall, the interface area, hotspot locations, bonded/nonbonded contacts, and energies between NSP14 and NSP10 may pave a way in designing potential inhibitors to disrupt NSP14–NSP10 interactions of CoVs especially SARS-CoV-2.
1. Introduction
The emergence of various coronaviruses (CoVs) has been causing serious epidemic diseases to humans, viz., severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and coronavirus disease-2019 (COVID-19, SARS-CoV-2), posing serious concerns.1 Like the cellular replicative DNA polymerase that has high fidelity, viral RNA-dependent RNA polymerase (RdRp), including the CoVs RdRp, does not have a proofreading exoribonuclease (ExoN) domain for high-fidelity replication and transcription.2−5 RNA virus replication is an error-prone process (or low viral fidelity) resulting in a different population of genomic mutants or ″quasispecies″.6 Low replication fidelity RNA viruses lead to an increased chance of error in the transcription process resulting in the extinction of the virus, which suggests the need for stability between quasispecies type and replication fitness for the virulence and evolution of viruses.7,8 In SARS-CoV-2 and other CoVs, replication and transcription occur by the viral RdRp, NSP12.9 The lack of proofreading activity in RdRp is challenging for the replication of CoVs. The ExoN enhances the fidelity for the synthesis of RNA by correcting errors in nucleotide incorporation made by the RdRp. To diminish the low fidelity of RdRp, all CoVs have nonstructural protein 14 (NSP14) consisting of a 3′-to-5′ N-terminal ExoN domain (res. 1–289)10,11 that forms a complex with NSP10 crucial for ExoN activity and acting as a co-factor. Additionally, NSP14 has a C-terminal guanine N7 methyl transferase (N7-MTase) whose function is different from the proofreading ExoN activity.10,11 Reports have also shown that ExoN inactivation disrupts SARS-CoV-2 and MERS-CoV replication that displays apart from transcription, ExoN is also involved in CoV replication.
It has been observed that mutations in SARS-CoV-2, SARS-CoV, MERS-CoV, and murine hepatitis virus (MHV) NSP14 display a strong relation with increasing mutational load in the viral genome,12−15 and genetically engineered inactivation of ExoN often results in 15–20-fold increased mutation rates, while knockout ExoN and CoVs produce crippled but viable viruses resulting in mutant phenotypes.16−19 The NSP14 ExoN proofreading function is critical for maintaining and extending large genomes of CoVs to ensure replication proficiency.7,20 Because of its role in enhancing the fidelity of replication while maintaining viral replication fitness, quick resistance can be developed to nucleotide analogue drugs, which can promote antiviral drug resistance.20 Therefore, instead of targeting the active site, targeting the NSP14 and NSP10 interacting interface with small molecules or peptides could be a better strategy to disrupt transcription and replication processes of SARS-CoV-2.
Recently, the structure of electron microscopy has revealed the molecular mechanism of how the ExoN NSP14–NSP10 complex interacts with double-stranded RNA consisting of a 5′ overhang and a one-nucleotide mismatch at the 3′ end.21 In the narrow ExoN active site, the mismatched base enters and interacts with catalytic conserved residues via its 3′-hydroxy and 2′-hydroxy groups. The double-stranded RNA portions interact with both NSP10 N-terminal regions, and NSP14-ExoN residues interact outside the catalytic site.21 The strong interaction of the cofactor NSP10 with NSP14 may be traced to the stability of the ExoN domain and enhances NSP14 ExoN activity. This information provides direct structural visualization of recognition of ExoN to its chosen mismatched RNA substrate.21,22
The fast spread of the SARS-CoV-2 and its deadly consequences have emphasized the need for additional viral inhibitors with more specific targeting. The key target is thought to be the NSP12 inhibition through nucleoside analog viral inhibitors,23 but NSP14 is thought to be less important.22 It is also hypothesized that the regulation of CoV genome fidelity may depend completely on NSP10/12/14.24 Inhibition of PPI between NSP14 and NSP10 is essential to abrogate the transcription and replication of viral RNA, thereby controlling the COVID19 disease.24
Unveiling the protein–protein interaction (PPI) at the atomic level is a crucial step for designing potential PPI inhibitors to obstruct protein–protein interactions.25−32 At the PPI interface, amino acids interact with each other, and a couple of them contribute high binding energy toward the stabilization and formation of a PPI complex that offers specificity to the binding sites and those residues considered to be hotspots.33−43 The hotspot residues are those, when mutated to alanine, that significantly increase the BFE (ΔΔG) to the tune of 1.5 to 2.0 kcal/mol.33,39−46 Hotspot residues at the PPI interface are present together as clusters, and they are conserved, which were noticed to be more buried compared to the other interface residues of the PPI interface.33,39−46 Besides the presence of hotspots, other parameters such as interacting interface area, polarity, flatness, and buriedness have been considered to characterize PPI interfaces.47−49 These parameters including hotspot residues at interfaces of the SARS-CoV-2 NSP14–NSP10 proofreading complex may help in understanding NSP14–NSP10 interaction in a better way to modulate the interface area by designing PPI modulators or inhibitors, thereby controlling the replication and transcription of COVID-19.
Extensive MD simulations were performed on the NSP14–NSP10 complex of SARS-CoV-2, SARS-CoV, and MERS-CoV and four NSP14mutant complexes to obtain information on the dynamics of the PPI interface. The interacting interface area including hotspots can be targeted by small molecules or biologics to gain valuable insights on the therapeutic indications.50−55 Further, the total binding free energy (BFE) was computed using the molecular mechanics Poisson–Boltzmann surface area (MM-PBSA)56,57 approach followed by per-residue energy contribution analysis.57 The PDBsum server was used to analyze the PPI profile,58 and hotspot residues at the NSP14–NSP10 interface were identified using different computational approaches implemented in web servers including the KFC2,59 DrugScorePPI,60 and Robetta servers along with per-residue energy contribution analysis.61,62 A single method may not give a significant result; thus, these methods were considered for accuracy improvement for hotspot identification. Various computational approaches have been employed to decipher the active site and hotspot residues of macromolecules such as proteins and DNA for drug targeting.63−65 While targeting the PPI interface remains an issue, in designing and discovery of small molecule inhibitors or modulators because the PPI interface are highly dynamic in nature. Therefore, various attempts have been made by different groups to understand the molecular mechanism of PPI at the atomic level through MD simulation studies to predict the hotspot residues for drug design.26,66−70
In the current study, we noticed a significant difference in the dynamic behavior of the NSP14–NSP10 complex among the selected viruses, wherein, overall, the SARS-CoV NSP14–NSP10 complex was observed to have more structural flexibility especially at the catalytic region as opposed to wild-type (WT) SARS-CoV-2, NSP14mutant complexes, and MERS-CoV. Potential hotspot residues were also identified and contributed more energy toward the formation of the complex. The lesser fluctuation in the SARS-CoV-2/mutant proofreading complex may be necessary for maintaining the structural stability of the ExoN domain of SARS-CoV-2 (WT/mutant). This may be responsible for efficient NSP14 ExoN activity that is crucial in expanding and maintaining the large genome of CoVs for high replication proficiency, which may also significantly lead to stable genome mutation and evolution of a new variant. The experimental Alanine Scanning Mutagenesis (ASM)44 method is very tedious; hence, identification of potential locations of hotspots at the NSP14 and NSP10 interface by in silico approaches may be convenient for the researchers to perform their experimental ASM for only those amino acids that are being predicted as hotspots.
2. Methodology
2.1. Protein Complex Structure Preparation and Sequence Comparison
The SARS-CoV-2 NSP14 consists of an N-terminal (ExoN) domain (residues 1–289) that is involved in proofreading activity and a C-terminal region consisting of N7-MTase domain (290–527) involved in mRNA capping. Three-dimensional (3D) NSP14–NSP10 complexes of SARS-CoV-2 and SARS-CoV were retrieved from PDB (PDB ID: 7MC5 with 1.64 Å resolution and 5C8U with 3.40 Å resolution), and MERS-CoV NSP14 was modeled in the SWISS MODEL server as the crystal structure is not available71 and docked with NSP10 in the PatchDock server.72 The best docked model based on global energy was chosen for further analysis. In addition, based on the literature study, four SARS-CoV-2 NSP14 mutant PPI complexes were prepared using PyMol.73 The mutations are P203L, F233L, L177F, and a combination of these three considered as triple mutations in this study; these mutations display a strong relation with increased mutational load in the SARS-CoV-2 genome.12,13 Then all the NSP14–NSP10 complexes were cleaned in UCSF Chimera74 by removing solvent molecules, ions, and other heteroatoms, and the complexes were subjected to MD simulations. The arrangement of the selected CoV NSP14–NSP10 complexes is presented in Figure 1. In the present study, we mainly focus on the N-terminal ExoN domain (res. 1–289) interaction with NSP10 because the SARS-CoV-2 NSP14 N7-MTase function does not depend on ExoN activity and NSP10.21 The multiple sequence alignment for NSP14 and NSP10 of the selected three viruses was carried out by the Clustal Omega server (https://www.ebi.ac.uk/Tools/msa/clustalo/).
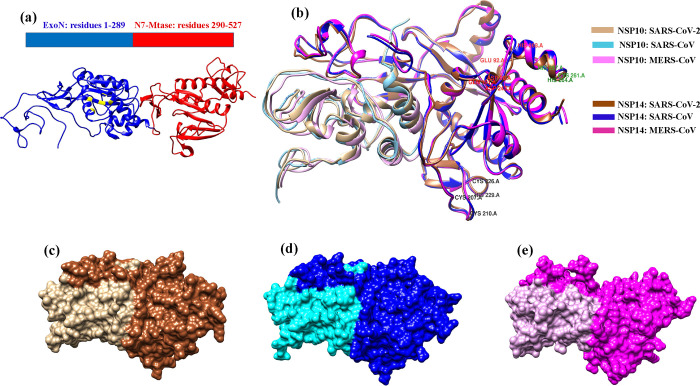
Figure 1.
Cartoon representation of (a) the SARS-CoV-2 NSP14 structure containing an ExoN domain (blue) and MTase domain (red). (b) Superimposed initial complex structure of NSP14–NSP10 SARS-CoV-2 (PDB ID: 7MC5), SARS-CoV (PDB ID: 5C8U), and MERS-CoV (modeled). (c) Surface representation of the NSP14–NSP10 complex of SARS-CoV-2 (light brown: NSP10, dark brown: NSP14). (d) Surface representation of the NSP14–NSP10 complex of SARS-CoV (cyan: NSP10, navy blue: NSP14). (e) Surface representation of the NSP14–NSP10 complex of MERS-CoV (pink: NSP10, magenta: NSP14).
2.2. Molecular Dynamics Simulations
MD simulations in triplicate have been performed on the heterodimer NSP14–NSP10 complex of SARS-CoV-2, SARS-CoV, and MERS-CoV and SARS-CoV-2 NSP14mutant complexes using the Gromacs 5.0.4 package.75 The CHARMM force field (version: charmm36-Jul2020)76 was used for PPI complexes, and the SPC water model was used to solvate the complex systems.77 The periodic boundary conditions (PBCs) were set up with a cubic box by keeping 1.0 nm from the edge for the MD simulations. The box size information for SARS-CoV-2, SARS-CoV, and MERS-CoV NSP14–NSP10 PPI complex is presented in Table S1. The systems were neutralized by incorporating the counterions accordingly into the solvated box followed by energy minimization by the 5000 steepest descent method to remove hindrances and clash in the solvated system.
Further, the complexes were heated under NVT from absolute zero to room temperature for 100 ps using a modified Berendsen thermostat followed by a 100 ps equilibration run under NPT.78,79
Finally, the complex was simulated with no constraints for a production run of 100 ns. For restraining the bond lengths, the LINCS algorithm80 was employed, and long-range electrostatics were calculated by employing PME81 along with the SETTLE algorithm for the solvent molecules. Using the g_mmpbsa package of GROMACS, the MMPBSA method was utilized for binding free energy calculation between interacting NSPs (NSP14 and NSP10) by extracting the last 20 ns MD trajectory.
By employing the MM-PBSA method, the total BFE was calculated by incorporating the explicit solvation model for the estimation of binding free energy ΔGbinding.
| 1 |
The ensuing equations describe the MM-PBSA protocol:
| 2 |
| 3 |
| 4 |
where ΔEMM = molecular mechanics energy system in a vacuum, Eint = internal energy, EvdW = van der Waals forces, ΔEele = electrostatic energy, ΔGPBSA = sum total of polar solvation free energy of the Poisson–Boltzmann model (ΔGPB) and the nonpolar/surface solvation free energy (ΔGsurf), and TΔS = entropy.
The entropy term is the most difficult to compute; therefore, in the above calculation, entropy (TΔS) was neglected because this study mainly focused on calculating only the total binding energy contribution of each amino acid toward the complexation process.
The final 20 ns trajectory was extracted (i.e., 80–100 ns) for predicting binding energy by employing MMPBSA using the inbuilt MmPbSaStat.py program and for per-residue energy decomposition analysis, MmPbSaDecomp.py was used to extract the residue wise energy contribution during protein−protein binding.56,57
The MMPBSA methods are commonly used to calculate binding affinities of protein–protein and protein–ligand interactions at a reasonable computational cost. Although this method has provided lots of valuable predictive results in different types of studies,82−84 it is less accurate than few of the computationally expensive methods, for example, free energy perturbation and thermodynamic integration methods.
2.3. Principal Component Analysis
Through PCA, the protein motion can be analyzed by considering the combined essential motion of the protein throughout the MD simulation trajectories in the protein. PCA was carried out in two steps: (1) constructing a covariance matrix using protein C-α atoms and (2) diagonalization of the covariance matrix. By utilizing the GROMACS software package, PCA was done following the standard protocol.85 The motion in the PPI complexes was analyzed by projecting the first two eigenvectors.
In the current study, PCA was done to analyze the conformational projection of the NSP14–NSP10 complex of SARS-CoV-2, SARS-CoV, and MERS-CoV and SARS-CoV2 NSP14mutant complexes using the protein C-α atoms. Based on the covariance matrix, PCA is calculated by following equation:86
where xi/xj signifies the Cartesian coordinate of the ith/jth atom and <−> signifies the ensemble average. All the MD simulation results are plotted using the Xmgrace tool.87
2.4. Free Energy Landscape (FEL) Analysis
FEL analysis is beneficial to characterize the mechanism of protein folding.88 For a protein structure, FEL can provide a quantitative description of protein folding dynamics. FELs were prepared and evaluated for all the three viral complexes and NSP14mutant complexes from the 100 ns MD trajectories. The FELs of the complex systems were created by employing the gmx sham utility of GROMACS utilizing the following formula:
where KB indicates the Boltzmann constant; T indicates the temperature, that is, 300 K; and P(X) signifies the probability distribution.
2.5. Detection of Hotspots
The interaction profile of heterodimer NSP14–NSP10 complexes was studied using the PDBsum server. The server provides a pictographic summary of the macromolecular structure and their important information as well as results obtained by PROCHECK and diagrammatic representation molecular contacts of biomolecular complexes.58 Three online servers—KFC2,59 DrugScorePPI,60 and Robetta servers61, 62—were utilized to identify hotspots. The KFC2 server implements machine learning (ML) techniques for in silico ASM by considering atomic contacts, hydrogen bonding, and the size of residue for hotspot detection.59 The DrugscorePPI server also utilizes the computational alanine-scanning technique, which has a knowledge-based scoring function for predicting hotspots across protein–protein interacting interfaces.60 In the Robetta server, for interaction free energy calculation, different parameters such as H-bonds and implicit solvation are utilized, along with other interactions like solvation, packing, and Lennard–Jones. This server (Robetta) can precisely predict the hotspot residues by 79% with a 1.0 kcal/mol cutoff value.61,62
3. Results and Discussion
3.1. Structural and Sequence Comparison
The superimposed NSP14–NSP10 complex of SARS-CoV-2, SARS-CoV, and MERS (modeled docked complex) is depicted in Figure 1. The all-atom root mean square deviation (RMSD) between SARS-CoV-2 and SARS-CoV, and SARS-CoV-2 and MERS-CoV was found to be the same, that is, 0.87 Å. The all-atom RMSD between SARS-CoV and MERS-CoV NSP14–NSP10 complex was observed to be 0.44 Å. Multiple sequence alignment (MSA) analysis of the NSP14 exon domain provides the percentage identity matrix. For SARS-CoV-2 and SARS-CoV, the percentage identity matrix was observed to be 97.12%, while it was 61.67% for SARS-CoV-2 and MERS and 61.59% for SARS-CoV and MERS-CoV.
The percentage identity matrix for NSP10 SARS-CoV-2 and NSP10 SARS-CoV was observed to be 97.12%; for SARS-NSP10 CoV/CoV-2 and MERS-CoV, it was 58.99%. The multiple sequence alignment is depicted in Figures S1 and S2. The hydrophobicity map for all the three viruses has been depicted in Figure S3. Hydrophobicity analyses categorized the surfaces into nonpolar (orange) containing hydrophobic groups, polar (blue), and neutral (white) regions. The majority of the interacting interface of NSP14 SARS-CoV-2, SARS-CoV, and MERS-CoV contains stronger nonpolar patches (orange) and weak polar patches (blue) with few neutral regions (white).
3.2. Molecular Dynamics Simulation and Binding Free Energy Analysis
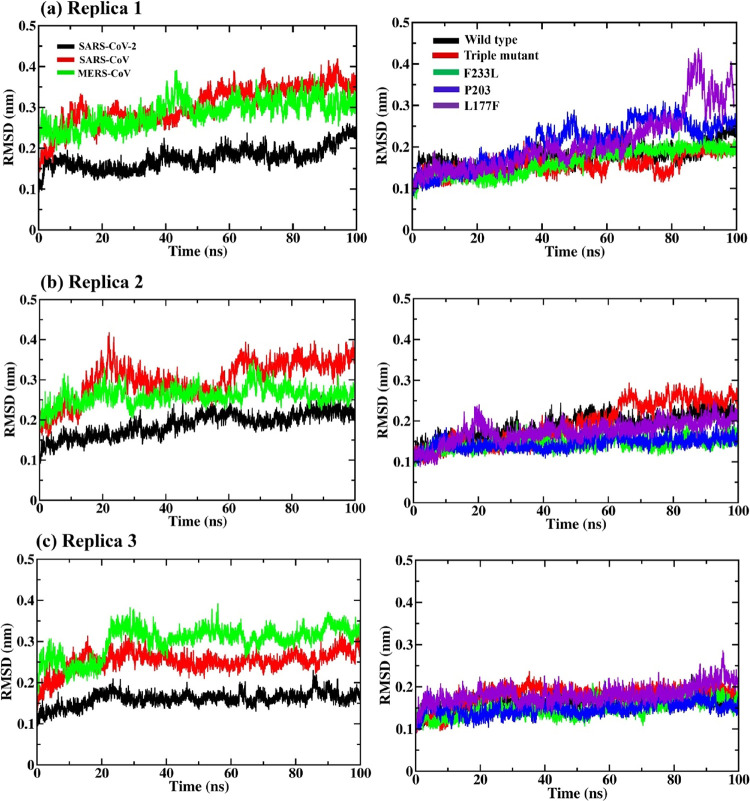
MD simulations were performed for the selected PPI complexes to check the stability of the PPI structures in a dynamic system for which 100 ns MD simulations were carried out in triplicate (three sets). The NSP14–NSP10 SARS-CoV-2 complex in all the three sets of MD simulations attain structural stability with less fluctuation from the initial conformer with an RMSD value of 0.15 nm as shown in Figure 2, and for SARS-CoV and MERS-CoV, the complex converged with a high RMSD value of around 0.3 nm or more in all the three sets of MD simulations. Overall, the NSP14–NSP10 complex of SARS-CoV-2 is rigid and slightly more stable in comparison with that of SARS-CoV and MERS-CoV. The mutant structure of SARS-CoV-2 NSP14L117F in complex with NSP10 shows a slight fluctuation after 30 ns till 100 ns with an RMSD of 0.25 nm in the case of replica 1 (see Figure 2a). However, in replicas 2 and 3, NSP14L117F was found to converge with a 0.2 nm RMSD value (see Figure 2b,c).
Figure 2.
Root mean square deviation (RMSD) graphs for (a–c) SARS-CoV-2, SARS-CoV, and MERS-CoV NSP14–NSP10 complexes (left side) and SARS-CoV-2 NSP14mutant complexes (right side) along the 100 ns MD simulations in triplicate. The triple mutant (in red, right-side plots) is the combination of the three (F233L, P203L, and L177F) mutations.
The mutant complex does not show any huge difference in RMSD and is more like the WT, attaining stability and convergence at around 0.2 nm (see Figure 2). In the case of replica 2, the triple mutation (P203L, L177F, and F233L) has shown convergence after 60 ns with an RMSD of 0.25 nm (see Figure 2).
To analyze the compactness in the PPI complexes throughout the MD simulation, the radius of gyration (Rg) was measured. In the dynamic system, Rg signifies the overall dimension of the protein computing the mass weight of RMSD by collecting all atoms from the center of mass. In the case of replica 1, the Rg value for the SARS-CoV-2 NSP14–NSP10 complex is around 2.35 nm; for SARS-CoV, there is a slight decrease in Rg with fluctuation after 40 mn; for MERS-CoV, the Rg value is 2.35 nm, which is higher than SARS-CoV-2. In the case of the mutant complex, the Rg value is more for L177F and F233L, which signifies a change in protein folding and its compactness, while for P203L and the triple mutant, the Rg value is almost the same as WT, i.e., 2.35 nm (see Figure S4).We could not see any huge difference in the Rg plots in the triplicate; overall, the convergence pattern in Rg for all the three sets of MD simulations is the same.
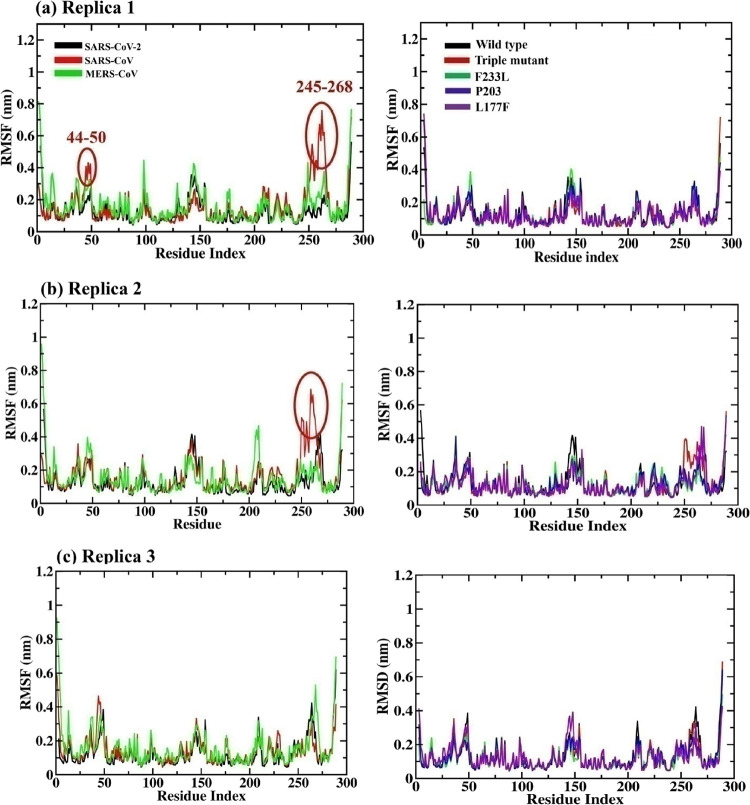
The stability of C-α atoms and residues can be analyzed by the RMSF plot (see Figures 3 and 4). For SARS-CoV NSP14, flexibility is more in comparison to SARS-CoV-2 and MERS-CoV and SARS-CoV-2 NSP14mutant complexes with high peaks in many positions such as for residue ranges 44–50 and 245–268 in the first and second replica, as shown in Figure 4a,b (left side). Meanwhile, in replica 3 at position 245–268, MERS-CoV has shown fluctuation with an RMSF value of 0.5. In the case of the SARS-CoV-2 NSP14 mutant, slight fluctuations can be seen in L177F and P203L in various positions in all the triplicates.
Figure 3.
Root mean square fluctuation (RMSF) graphs in triplicate for (a–c) NSP14 SARS-CoV-2, SARS-CoV, and MERS-CoV (left side) and SARS-CoV-2 mutants (NSP14) along the 100 ns MD simulation. The triple mutant (in red, right-side plots) is the combination of the three (F233L, P203L, and L177F) mutants.
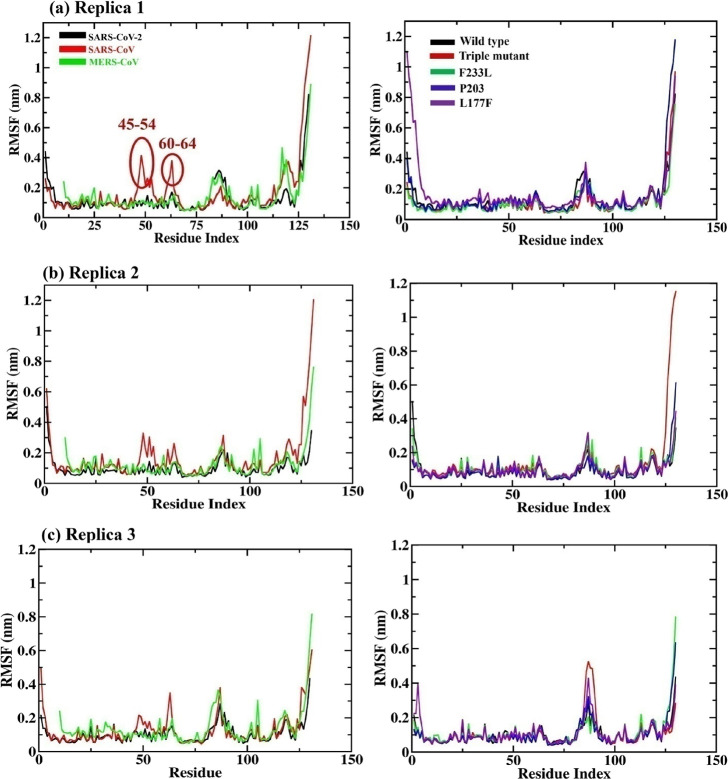
Figure 4.
Root mean square fluctuation (RMSF) graphs in triplicate for (a–c) NSP10 SARS-CoV-2, SARS-CoV, and MERS-CoV (left side) and SARS-CoV-2 mutants (in complex with NSP14) along the 100 ns MD simulation. The triple mutant (in red) is the combination of the three (F233L, P203L, and L177F) mutants.
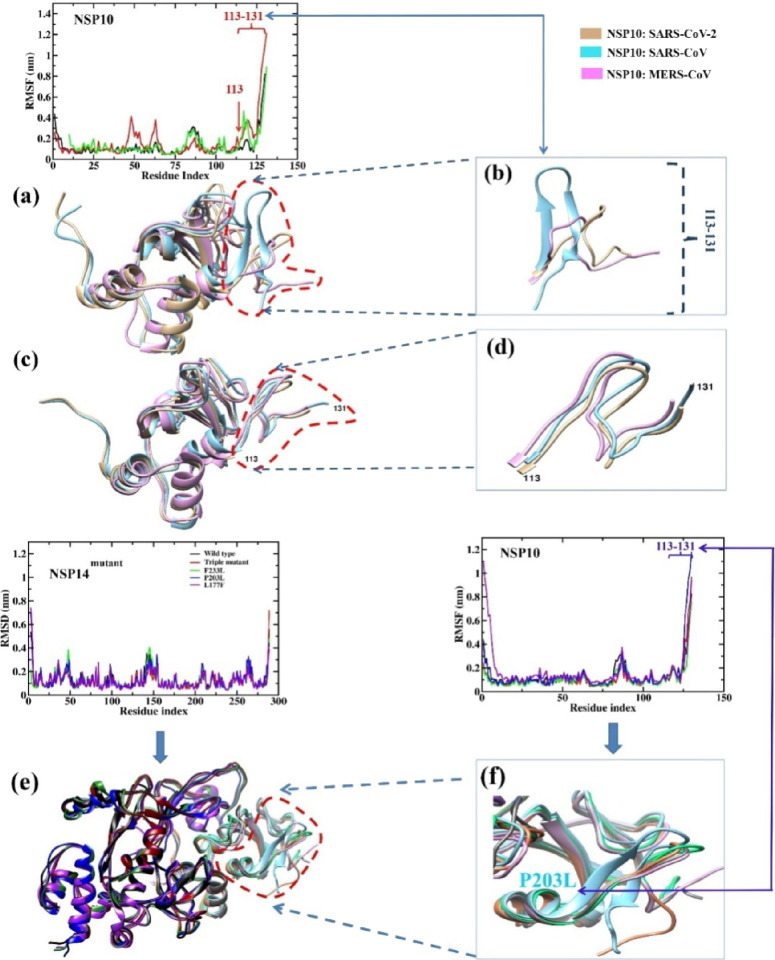
In the case of NSP10, SARS-CoV residue fluctuation can be seen at positions 45–54 and 60–64 in all the three sets of MD simulations. Additionally, the SARS-CoV NSP10 C-terminal region (113–131) fluctuates more in replicas 1 and 2. Overall, SARS-CoV and MERS-CoV NSP10 show slightly more residue fluctuation than SARS-Cov-2.
The conformational change in the average PPI complexes of the three viruses can be observed in Figure 5. The all-atom RMSD between SARS-CoV-2 and SARS-CoV is observed to be 3.13 Å, while the all-atom RMSD value is 2.18 Å in SARS-CoV-2 and MERS-CoV and 3.57 Å in SARS-CoV and MERS-CoV (see Figure 5).
Figure 5.
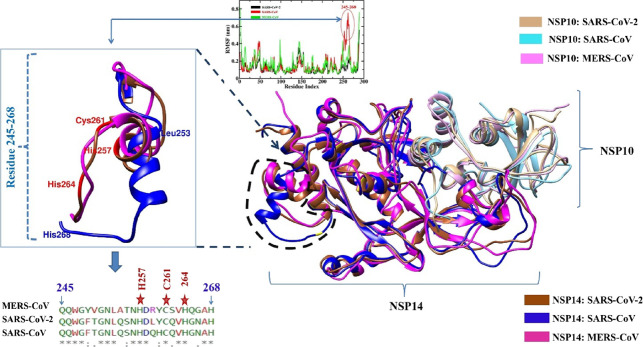
Superimposed average structures (collected from the trajectories of last 20 ns) of the NSP14–NSP10 complex of SARS-CoV-2 with SARS-CoV and MERS-CoV. The square box (on the left side) depicts the C-terminal fluctuating residues ranging from 245 to 268. NSP14 SARS-CoV is depicted in blue color, SARS-CoV-2 is in brown, and MERS-CoV is in magenta.
In Figure 5, fluctuations in the residue range 245–268 of SARS-CoV NSP14 ExoN were observed in replicas 1 and 2 (see Figure S5). Within this region, His257, Cys261, and His264 are the conserved residues of the second zinc finger region, while H268 is one of the conserved catalytic residues among the three selected viruses. It has been suggested that the mutation of the second zinc finger residue Cys261 to Ala or His264 to Arg disrupts the enzymatic activity, suggesting its major role in catalysis. The mutation of His268 to Ala shows a lack of RNA degradation ability, hence suggesting the importance of these residues in the nucleotide excision process.89,90 The residue range 245–268 of SARS-CoV NSP14 has shifted more from its original position in comparison to SARS-CoV-2, MERS-CoV, and SARS-CoV-2 NSP14 mutant complexes. It has been recently reported that in the narrow ExoN active site, a mismatched base enters and interacts with catalytic conserved residues.21 The fluctuation in this important region that includes the key catalytic residues may have a significant impact on the transcription and replication process of SARS-CoV. Thus, the SARS-CoV NSP14–NSP10 complex displays a pronounced structural flexibility compared to SARS-CoV-2, MERS-CoV, and SARS-CoV-2 mutant complexes. Recently, Gribble et al., suggested that RNA proofreading ExoN is accountable for the generation of recombination frequency and also the change in the recombination products in vitro; they demonstrated that a recombination event is crucial for generating CoVs’ diversity.90 A stable complex is required for the recombination, which may be essential for the high fidelity and replicative fitness of SARS-CoV-2 and perhaps for the evolution of new stable variants of SARS-CoV-2.
Additionally, in all the three sets of MD simulations, SARS-CoV NSP10 residues 113–131 at the C-terminal display greater fluctuation compared to the other two CoVs, as shown in Figure 6 and Figure S6. There is a conformational change at position 113–131 during the MD simulations in all the three sets of SARS-CoV NSP10 from coil to beta strand, as shown in Figure 6b and Figure S6. In all the mutant complexes, slight residual flexibility has been observed in several positions of NSP10 with the mutation L177F and P203L in replica 1, and in the P203L mutant complex, a slight fluctuation at the C-terminal region of NSP10 has been noticed (res. 113–131) where conformational change was observed from coil to beta strand as that of SARS-CoV NSP10 (Figure 6e,f). Similarly, in replica 2, conformational change has been observed in the same C-terminal region of NSP10 in case of the NSP14triple mutant complex (Figure S7b), whereas in replica 3, none of the NSP10 mutant complexes displayed any conformational change as that of replica 1 (in P203L) and replica 2 (in triple mutant), as shown in Figure S7a,b.
Figure 6.
(a) Superimposed average structures (from last 20 ns) of NSP10 SARS-CoV-2 (light brown), SARS-CoV (cyan), and MERS-CoV (pink). (b) The C-terminal residues of NSP10 SARS-CoV (cyan) are highlighted, showing more residue fluctuation and change in the initial conformation (coil) to beta strand. (c) Superimposed initial structures (before MD) of NSP10 SARS-CoV-2, SARS-CoV, and MERS-CoV. (d) The C-terminal region of the three viruses is highlighted. (e) Superimposed mutant NSP14–NSP10 complexes (all). (f) Superimposed structure of NSP10 of mutant complexes. In one of the mutant complexes (P203L), NSP10 shows a conformational change from coil to beta strand.
To evaluate the folding dynamics during simulations, SASA has been a useful analysis. Based on the SASA plot, SARS-CoV-2 NSP14–NSP10 showed SASA values of 190–220 nm2 in all the three sets of MD simulations; however, SARS-CoV and MERS-CoV SASA values were found to be higher and fluctuate within 200–220 nm2 as shown in Figure S8. The SARS-CoV-2 NSP14 mutants do not display a huge difference with the wild type and maintain consistency throughout the three, but the values were lower than those of SARS-CoV and MERS-CoV in the three sets of MD simulation, showing that the change in surface residues may be accessible to the solvent.
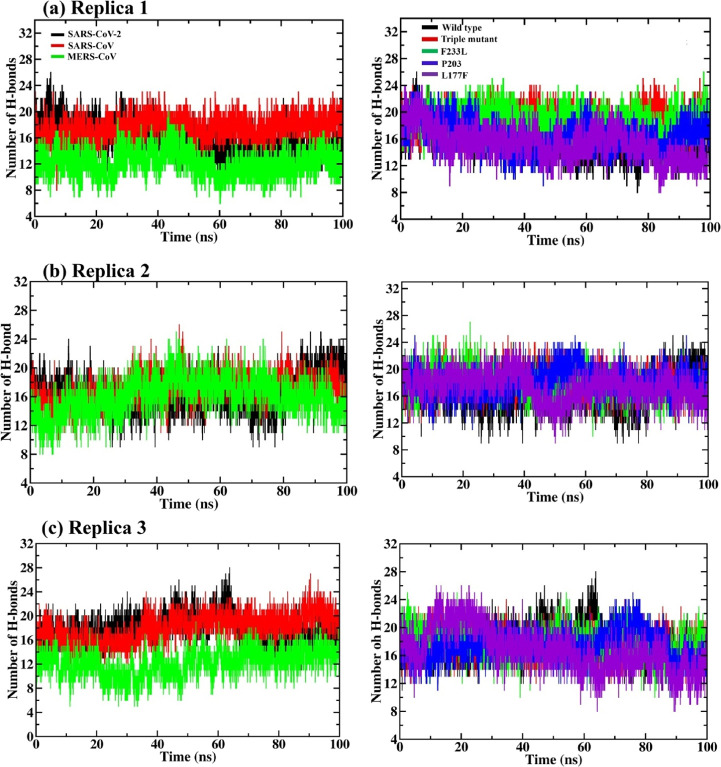
Hydrogen bonds are considered to play a crucial role in the stabilization of PPI conformation in the dynamic system. The formation of intermolecular H-bonds between NSP14 and NSP10 in 100 ns MD trajectory for all the three sets of simulation can be seen in Figure 7. A perusal of the trajectories of MD results reveals that around 18 H-bonds formed at the NSP14 and NSP10 interface of SARS-CoV. In the case of SARS-CoV-2 and MERS-CoV, 12–16 H-bonds formed after 60 to 100 ns. Almost a similar pattern of H-bonds can be seen during the three sets of MD simulations.
Figure 7.
(a–c) Hydrogen bonds observed at the PPI interface across NSP14 and NSP10 of SARS-CoV-2, SARS-CoV, MERS-CoV (left side), and SARS-CoV-2 NSP14mutants (right side) complexes along the three sets of 100 ns MD simulation. The triple mutant (in red, in left-side plots) is the combination of the three (F233L, P203L, and L177F) mutants.
By using the PDBsum server, the PPI profile of the last 20 ns average structure was analyzed wherein, in the first set of MD simulation, the average structure showed 12 H-bonds and the initial PPI complex had 22 H-bonds. In the case of SARS-CoV and MERS-CoV, the H-bonds in the initial structure were 14 and 8, respectively; however, after MD simulation, the average structure showed 17 and 10 H-bonds, respectively (see Tables S2–S4). For replicas 2 and 3, the PPI profiles of all the complex systems are presented in Tables S3 and S4. In the case of the SARS-CoV-2 NSP14mutant, the L177F mutation shows a reduction in the H-bond formation after 40 ns, but in other mutants (F233L, P203L, and the triple mutant), there is an increase in the H-bond formation in the mutant structures as compared to WT (see Figure 7 and Tables S2–S4). There are two common interfaces of H-bond between the three viruses; that is, NSP14 Lys61 retains a H-bond with NSP10 Ser15 throughout the MD simulation, and NSP14 Ile201 retains two H-bonds with NSP10 Phe19 and Val21 in all the three viruses as well as in mutant PPI complexes throughout the three sets of MD simulation. Further, H-bond interactions are tabulated in Tables S5–S28 and salt bridges in Tables S29 and S30.
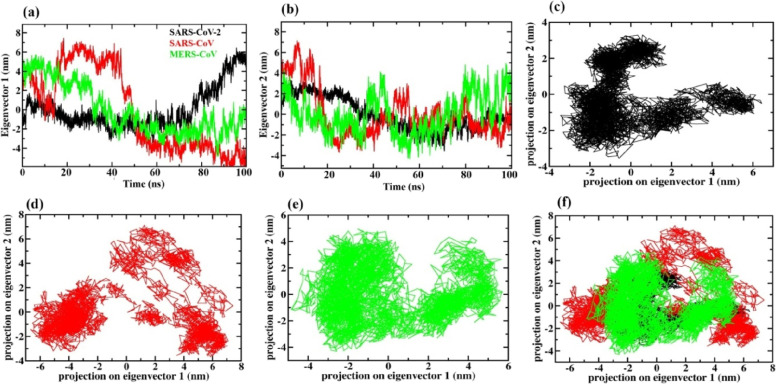
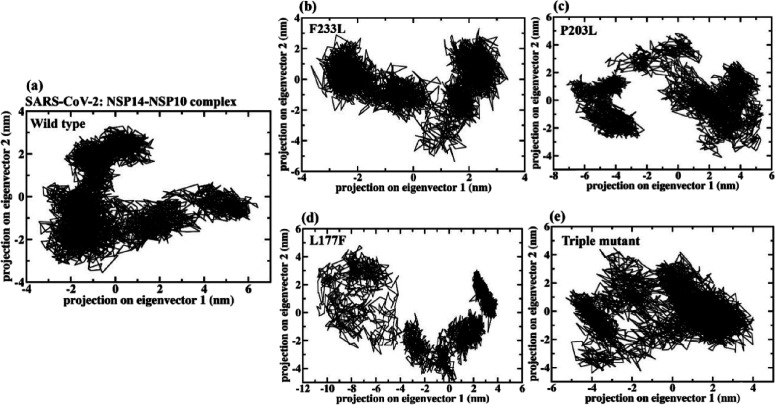
Taking the first two eigenvectors (EVs), PCA was performed to investigate the PPI collective motions (see Figure 8). The scatter plot generated for PPI complexes is shown in Figures 8 and 9. A significant difference has been observed in the conformational projections between all the three PPI complexes of the selected viruses. It has been noticed that the projection of the SARS-CoV-2 and MERS-CoV NSP14–NSP10 PPI complex contracted on both the EVs during the MD simulation (see Figure 8) as compared to SARS-CoV and the three SARS-CoV-2 NSP14 mutants (triple, P203L, and L177F) shown in Figure 9. The NSP14–NSP10 PPI complex of the SARS-CoV-2 and MERS-CoV system explored less phase space compared to the WT NSP14–NSP10 SARS-CoV-2 complex and the three mutant complexes (see Figure 9).
Figure 8.
Time evaluation of conformational projections on (a) eigenvector 1 and (b) eigenvector 2. (c–e) 2D projection plot showing the conformation sampling of the NSP14–NSP10 complex of SARS-CoV-2, SARS-CoV, and MERS-CoV on eigenvector 1 and eigenvector 2. (f) Superimposed 2D projection plots showing the difference in the conformation sampling of PPI complexes.
Figure 9.
2D projection plots showing the conformation sampling of NSP14–NSP10 complexes of (a) wild-type NSP14 in complex with NSP10 of SARS-CoV-2, (b) NSP14F233L mutant, (c) NSP14F203L mutant, (d) NSP14L177F mutant, and (e) NSP14 triple mutant in complex with NSP10. The triple mutant is the combination of the three (F233L, P203L, and L177F) mutants.
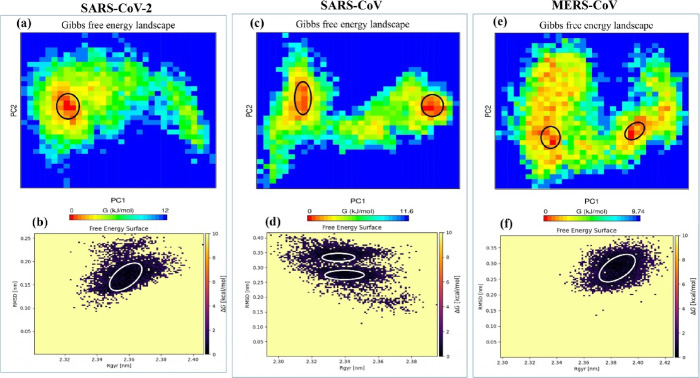
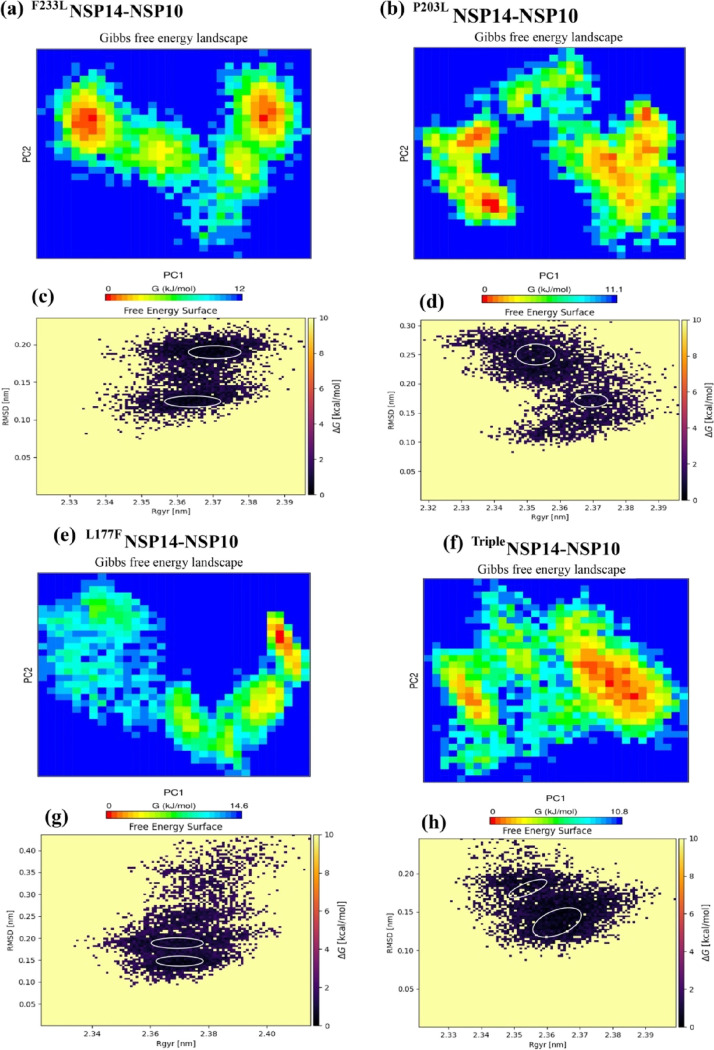
The free energy landscape of (FEL) provided the global minima of backbone atoms of PPI complexes with respect to RMSD and Rg presented in Figures 10 and 11. Distinguishable local basins from free energy surface have been shown in red color (see Figures 10 and 11). The NSP14–NSP10 PPI complex of SARS-CoV-2, SARS-CoV, and MERS-CoV achieved the global minima (lowest free energy state) between RMSD values of 0.15 and 0.20 nm with Rg 2.35–2.37 nm (see Figure 10). For SARS-CoV, there are two positions of global minima conformations at RMSD of 0.35 with Rg 2.33–3.36 and in between RMSD 0.23 and 0.30 nm with Rg at around 2.33–2.357 (see Figure 10). In the case of MERS-CoV, the global minima structure was obtained around RMSD 0.24–0.32 nm with Rg between 2.378 and 3.396 nm (Figure 10). It has been observed that the PC1 and PC2 motion of the mutated systems of SARS-CoV-2 spanned larger ranges than those of the WT system, signifying the rearrangements in the conformation caused by the mutations (see Figure 11).
Figure 10.
Free energy landscape of PC1 and PC2 displaying the achievement of distinguishable local minima basins (in ΔG, kJ/mol) of NSP14–NSP10 complexes of (a) SARS-CoV-2, (c) SARS-CoV, and (e) MERS-CoV and corresponding free energy surface plots of (b) SARS-CoV-2, (d) SARS-CoV, and (f) MERS-CoV complexes with respect to their RMSD (nm) and Rg (nm).
Figure 11.
Free energy landscape of PC1 and PC2 (a, b, e, f) NSP14–NSP10 mutant PPI complexes displaying the achievement of distinguishable local minima basins (in ΔG, kJ/mol) and their corresponding free energy surface plots (c, d, g, h) with respect to their RMSD (nm) and Rg (nm). The triple mutant (in red) is the combination of the three (F233L, P203L, and L177F) mutants.
Simultaneously, for protein surfaces, electrostatic potential was observed (Figures S9 and S10). The electrostatic potential categorized the surface into positively charged patches in blue, negatively charged patches in red, and neutral patches in white. Figure S9 shows an electrostatic potential surface comparison of the three viruses’ initial NSP14–NSP10 complexes with the average structure derived during the last 20 ns, and Figure S10 shows a comparison of WT and mutant NSP14–NSP10 complexes. The interacting interface region mostly consists of neutral amino acids that are of two categories: (a) Nonpolar amino acids containing hydrophobic groups attain the interior of the protein or PPI interface (e.g., Ile, Val, Ala Trp, Leu, Gly, Met, Pro, and Phe). Most of our predicted hotspot residues belong to this category. (b) Polar uncharged amino acids with side chain functional groups contain N, S, and O involved in the formation of H-bonds with water or other molecules. The e.g. of polar uncharged amino acids are Thr, Cys, Tyr, Glu, Ser, and Asn. This category of amino acids (Thr, Asp, Tyr, Ser, and Phe) was found to form a H-bond at the PPI interface in our study (see Tables S5–S28).
For all the triplicates, by employing MM-PBSA, the BFEs were predicted for NSP14–NSP10 complexes, and results are presented in Table 1. The BFE of SARS-CoV NSP14–NSP10 (−463.06, −446.15, and −453.29 kJ/mol) is higher in all the three sets of MD as opposed to SARS-CoV-2 (−227.99, −392.89, and −365.76 kJ/mol); however, in the case of MERS-CoV in the first set of MD simulations, the binding energy is less (−311.00 kJ/mol) as compared to SARS-CoV but higher in replicas 1 and 2 (−726.54 and −695.57 kJ/mol). In the case of SARS-CoV-2 NSP14mutant complexes, most of the mutant complexes show a high binding affinity (BFE) between NSP14 and NSP10 as compared to WT, with the exception of L177F (see Table 1).
Table 1. MM-PBSA Binding Free Energy Calculations of NSP14–NSP10 SARS-CoV-2, SARS-CoV, MERS-CoV, and SARS-CoV-2 NSP14mutated Complexesa.
| human CoV NSP14–NSP10 complex | van der Waals energy | electrostatic energy | polar solvation energy | SASA energy | binding energy |
|---|---|---|---|---|---|
| Replica 1 | |||||
| SARS-CoV-2 | –704.73 ± 346.62 | –628.03 ± 315.97 | 1184.08 ± 581.53 | –79.31 ± 39.11 | –227.99 ± 137.38 |
| SARS-CoV | –934.45 ± 33.92 | –1034.14 ± 63.24 | 1608.86 ± 93.14 | –103.33 ± 3.04 | –463.06 ± 89.90 |
| MERS-CoV | –392.14 ± 371.75 | –543.71 ± 518.92 | 668.52 ± 613.89 | –43.66 ± 41.23 | –311.00 ± 337.02 |
| SARS-CoV-2F233L | –959.63 ± 33.32 | –774.45 ± 86.45 | 1516.18 ± 154.28 | 104.24 ± 3.98 | –322.14 ± 90.90 |
| SARS-CoV-2P203L | –912.90 ± 34.15 | –699.34 ± 68.38 | –1417.66 ± 107.47 | –99.08 ± 3.98 | –293.65 ± 74.37 |
| SARS-CoV-2L177F | –773.79 ± 32.71 | –646.11 ± 103.47 | 1198.85 ± 143.88 | –84.39 ± 3.62 | –305.44 ± 108.38 |
| SARS-CoV-2Triple | –946.44 ± 32.82 | –699.77 ± 66.16 | 1419.22 ± 80.84 | –103.72 ± 2.92 | –330.72 ± 72.02 |
| Replica 2 | |||||
| SARS-CoV-2 | –913.42 ± 32.20 | –717 ± 74.22 | 1337.18 ± 104.99 | –98.73 ± 3.52 | –392.89 ± 75.26 |
| SARS-CoV | –901.61 ± 29.66 | –935.91 ± 96.43 | 1490.98 ± 108.62 | –99.60 ± 3.21 | –446.15 ± 97.82 |
| MERS-CoV | –768.63 ± 30.67 | –1209.81± | 1338.12 ± 109.71 | –86.22 ± 3.57 | –726.54 ± 86.03 |
| SARS-CoV-2F233L | –928.56 ± 36.73 | –633.29 ± 98.69 | 1249.84 ± 102.65 | –99.04 ± 3.80 | –411.06 ± 55.40 |
| SARS-CoV-2P203L | –961.15 ± 31.53 | –732.10 ± 64.25 | 1469.84 ± 92.22 | –105.03 ± 3.15 | –328.45 ± 75.23 |
| SARS-CoV-2L177F | –966.01 ± 28.27 | –643.65 ± 77.75 | 1473.20 ± 99.73 | –104.52 ± 2.88 | –240.99 ± 82.14 |
| SARS-CoV-2Triple | –954.59 ± 29.38 | –780.03 ± 63.07 | 1519.38 ± 101.84 | –105.19 ± 3.29 | –320.43 ± 75.81 |
| Replica 3 | |||||
| SARS-CoV-2 | –912.92 ± 32.83 | –559.06 ± 71.93 | 1205.36 ± 105.18 | –99.14 ± 3.76 | –365.76 ± 65.81 |
| SARS-CoV | –933.03 ± 34.90 | –1012.12 ± 63.11 | 1594.67 ± 96.58 | –102.82 ± 3.83 | –453.29 ± 79.11 |
| MERS-CoV | –768.70 ± 33.16 | –1151.44 ± 85.77 | 1309.04 ± 103.27 | –84.47 ± 2.90 | –695.57 ± 84.87 |
| SARS-CoV-2F233L | –962.58 ± 29.78 | –681.60 ± 60.08 | 1415.92 ± 89.49 | –101.45 ± 2.82 | –329.72 ± 70.07 |
| SARS-CoV-2P203L | –909.33 ± 35.79 | –632.04 ± 53.22 | 1449.00 ± 93.58 | –103.93 ± 3.29 | –196.31 ± 84.50 |
| SARS-CoV-2L177F | –873.85 ± 40.85 | –764.27 ± 90.15 | 1458.46 ± 128.21 | –97.92 ± 3.90 | –277.58 ± 82.14 |
| SARS-CoV-2Triple | –933.09 ± 28.34 | –650.72 ± 68.34 | 1399.72 ± 107.29 | –102.87 ± 3.11 | –286.96 ± 106.96 |
The energy terms (in kJ/mol) were calculated from the data obtained from the last 20 ns trajectory from the three sets of 100 ns MD simulation.
The nonpolar interaction energies including (ΔEvdW) + (SASA) were found to have a higher contribution to ΔGbind, which mean that hydrophobic interaction played a major role in the formation of protein–protein complexes. ΔGbind components signify that ΔEvdW and ΔEelec contributions are greater in the NSP14–NSP10 interaction.
Takada et al. found that viruses containing NSP14L177F or NSP14P203L mutation showed higher nucleotide substitution rates in the spike, membrane, and envelop genes per year than the viruses with the WT NSP14,13 and Eskieret al. reported that the NSP14F233L mutation showed a high prediction capacity for membrane glycoprotein and envelope glycoprotein genes and an increased mutation density.12
In the current study, these NSP14 mutant complexes have been observed to display higher binding affinity (BFE) between NSP14 and NSP10 as compared to WT SARS-CoV-2 NSP14–NSP10 complexes. The BFE of NSP14L177F, NSP14P203L, NSP14F233L, and NSP14triple mutant are −305.44, −293.65, −322.14, and – 330.72 kJ/mol, respectively, much higher than the WT (−227.99 kJ/mol). While minor fluctuations have been noticed between the various replicas, the overall structural pattern is virtually similar in all the replicas. In general, replica 1 BFE was observed to be slightly higher in mutants as compared to WT, while in replica 3, the wild-type SARS-CoV-2 complex (see Table 1) BFE is higher than mutants.
Eckerle et al. (2007 and 2010) also suggested that the mutation of NSP14 ExoN in MHV and SARS-CoV is responsible for 20-fold more mutations in the whole genome than their WT.14,17 The function of NSP14 ExoN proofreading activity is considered to be an important feature in expanding and maintaining the large genomes of CoVs to ensure replication fitness and proficiency in SARS-CoV.3,7
The residue around position 203 has been analyzed, and it was observed that the residue range of NSP14 ExoN 199 to 203 (VKIGP) is a part of interacting interface. Lys200, Ile201, and Gly202 are conserved among all the three selected CoVs except 203. The comprehensive analysis of the PPI complex of three viruses showed that the residues NSP14 Lys200 and Ile201 alone have direct interaction with the residues of NSP10 F19 and V21, which are also well conserved among the three selected viruses, as shown in Tables S5–S13.The comparison of per-residue energy contribution of these residues of all the seven complexes in triplicate MD simulation is presented in Table 2. Most of the mutated SARS-CoV-2 NSP14–NSP10 complexes display higher per-residue contributions in all the three sets of MD simulations as compared to WT, suggesting the significant strong interactions leading to mutant complex stability. Although the total BFE of the mutants in replica 3 is slightly lower than the WT SARS-CoV-2, the per-residue energy contributions of the key interface residue are higher for SARS-CoV-2 NSP14 Lys200 and Ile201 and NSP10 F19 and V21. The strong interaction of NSP10 with NSP14 reflects the important role of NSP10 in maintaining the stability of the ExoN domain structure to fully support the NSP14 ExoN activity.22 Throughout the three sets of 100 ns MD simulation, Lys200 of NSP14 SARS-CoV-2 maintained nonbonded interaction with NSP10 Phe19 and Val21, and NSP14 and Ile201 maintained two H-bonds, one with NSP10 Phe19 and another with NSP10Val21(see Tables S5–S7). The same kinds of interactions have been observed for SARS-CoV as well (see Tables S8–S10) In the case of MERS-CoV, Lys200 was involved in nonbonded interaction with NSP10 Phe19 and NSP10 Asn18, while NSP14 Ile201 formed one H-bond with NSP10 Phe19 and nonbonded interaction with NSP10 Val21 till the end of the simulations in the first replica (see Table S11). However, for replicas 2 and 3, Ile201 formed two H-bonds, one with Phe19 and another with V21 (see Tables S12 and S13).
Table 2. The Per-Residue Energy Decomposition Results of Key Interacting Residues of SARS-CoV-2, SARS-CoV, MERS-CoV, and SARS-CoV-2 Mutant NSP14–NSP10 Complexesa.
| PPI complex systems | per-residue energy
contribution (kJ/mol) |
||||
|---|---|---|---|---|---|
| NSP14-K200 | NSP14-I201 | NSP10-F19 | NSP10-V21 | ||
| replica 1 | wild-type SARS-CoV-2 | –21.50 ± 0.91 | –6.71 ± 0.27 | –25.03 ± 0.89 | –13.16 ± 0.48 |
| SARS-CoV-2/NSP14P203L | –26.81 ± 0.56 | –8.17 ± 0.18 | –31.01 ± 0.20 | –16.48 ± 0.15 | |
| SARS-CoV-2/NSP14F233L | –17.98 ± 1.14 | –7.26 ± 0.18 | –30.34 ± 0.21 | –15.82 ± 0.15 | |
| SARS-CoV-2/NSP14L177F | –23.50 ± 0.70 | –7.71 ± 0.19 | –29.16 ± 0.23 | –15.40 ± 0.19 | |
| SARS-CoV-2/NSP14triple | –28.49 ± 0.59 | –7.76 ± 0.19 | –31.15 ± 0.19 | –15.54 ± 0.15 | |
| SARS-CoV | –44.59 ± 0.67 | –5.61 ± 0.21 | –27.66 ± 0.20 | –17.08 ± 0.17 | |
| MERS-CoV | –6.64 ± 0.68 | –3.82 ± 0.33 | –14.02 ± 0.95 | –9.46 ± 0.66 | |
| replica 2 | wild-type SARS-CoV-2 | –22.95 ± 0.51 | –9.05 ± 0.17 | –29.76 ± 0.20 | –16.09 ± 0.16 |
| SARS-CoV-2/NSP14P203L | –26.61 ± 0.49 | –5.29 ± 0.16 | –30.63 ± 0.20 | –15.22 ± 0.17 | |
| SARS-CoV-2/NSP14F233L | –23.03 ± 0.44 | –5.38 ± 0.17 | –32.51 ± 0.17 | –16.24 ± 0.16 | |
| SARS-CoV-2/NSP14L177F | –13.61 ± 0.55 | –8.03 ± 0.19 | –30.50 ± 0.22 | –15.27 ± 0.15 | |
| SARS-CoV-2/NSP14triple | –29.50 ± 0.67 | –5.49 ± 0.21 | –31.08 ± 0.19 | –15.56 ± 0.14 | |
| SARS-CoV | –40.68 ± 0.83 | –5.53 ± 0.16 | –29.92 ± 0.23 | –16.69 ± 0.16 | |
| MERS-CoV | –24.20 ± 0.62 | –2.51 ± 0.21 | –25.83 ± 0.21 | –16.64 ± 0.16 | |
| replica 3 | wild-type SARS-CoV-2 | –22.28 ± 0.67 | –5.10 ± 0.16 | –30.63 ± 0.17 | –13.41 ± 0.17 |
| SARS-CoV-2/NSP14P203L | –10.85 ± 0.61 | –9.09 ± 0.16 | –29.09 ± 0.20 | –14.52 ± 0.13 | |
| SARS-CoV-2/NSP14F233L | –26.70 ± 0.73 | –7.68 ± 0.17 | –28.75 ± 0.19 | –15.34 ± 0.16 | |
| SARS-CoV-2/NSP14L177F | –21.46 ± 46 | –8.65 ± 0.16 | –30.81 ± 0.22 | –15.11 ± 0.18 | |
| SARS-CoV-2/NSP14triple | –21.39 ± 0.71 | –7.85 ± 0.16 | –30.48 ± 0.21 | –15.62 ± 0.13 | |
| SARS-CoV | –22.30 ± 0.80 | –8.85 ± 0.16 | –30.35 ± 0.24 | –16.41 ± 0.15 | |
| MERS-CoV | –25.53 ± 0.76 | –5.23 ± 0.76 | –31. 90 ± 0.21 | –17.74 ± 0.16 | |
The per-residue energy decomposition analysis was carried out using the last 20 ns MD trajectory of the three sets of MD simulation.
3.3. Profiling the Interaction Complex
The interface statics or profiles of the PPI complex of the initial conformer (before MD) of heterodimer NSP14–NSP10 complex of SARS-CoV-2, SARS-CoV, and MERS-CoV and all the NSP14mutants complexes were analyzed and compared with the average complex structure obtained from the last 20 ns (80–100 ns) MD trajectory. The change in the PPI interface from the initial to average PPI structures obtained from MD simulations can be observed from the results obtained. The PPI profiles such as interface area, nonbonded or noncovalent interactions, H-bond, and salt bridge information of all the complex systems are summarized in Tables S2–S4. In the initial NSP14–NSP10 SARS-CoV-2 complex, 50 total interface residues were observed at NSP14 and 45 at the NSP10 interface, and the interface area was 2135 Å2 (in NSP14) and 2320 Å2 (in NSP10). For replica 1, at the end of MD simulation, the average structure was found to have less interacting residues, that is, 36 at NSP14 and 35 at NSP10 interface; the interface area was 1847 Å2 (in NSP14) and 1994 Å2 (in NSP10). Interestingly, for SARS-CoV-2 NSP14mutant complexes F233L and triple mutant NSP10, the interface area increased to 2042 and 2099 Å2, respectively (see Table S2). In the average PPI structure, there is a reduction in the number of H-bond formation and nonbonded contact, but the salt bridge remains the same till the end of the MD simulations of replica 1, while in replicas 2 and 3, there is a loss of salt bridges in the case of SARS-CoV-2 (see Tables S3 and S4). The details of H-bond and salt bridge information of all the PPI complex systems along with SARS-CoV-2 mutants for the three sets of MD simulation are summarized in Tables S5–S30.
As the H-bond plays a crucial role in stabilizing the PPI complex, we analyzed the common H-bond forming residue among the three selected viruses within the NSP14 and NSP10 interface, presented in Table 3. It may be noticed that there is a reduction in the number of H-bond and nonbonded contacts in the average complex along the MD trajectory, while the number of interactions in the SARS-CoV average PPI structure is more than that of SARS-CoV-2. The SARS-CoV PPI complex contains 42 and 39 interface residues, respectively, in NSP14 and NSP10 in the initial complex, but at the end of the first set of 100 ns simulation, the number of interface residues dropped to 36 in the latter with no change in the number of the former. Similar analysis on the MERS-CoV-2 PPI complex reveals that the initial structure contains 42 and 39 interacting interface residues for NSP14 and NSP10, same as SARS-CoV. However, the end of simulation trajectory analysis reveals that there is an increase in the average H-bonds from 8 to 10, loss in one salt bridge, and a slight reduction in the nonbonded contacts (see Tables S2–S4b). The common H-bond forming residues among all the three viruses at the NSP14 interface are Lys61 and Ile210; at the NSP10 interface, the residues are Ser15, Phe19, Val21, Asp29, Leu45, Cys93, and Gly94. For SARS-CoV and SARS-CoV, the common H-bond forming residues at the NSP14 interface are Thr21, Asp41,Lys61, Asn67, Tyr69, and Ile201; at the NSP10 interface, the residues are Ser15, Phe19, Val21, Asp29, Ser33, Leu45, Cys93, Gly94, and Tyr96. The common H-bond forming residues among SARS-CoV-2 and MERS-CoV at the NSP14 interface are Thr127, Ile201 and Lys61; at the NSP10 interface, the residues are Ser15, Phe19, Val21, Asp29, Ser33, Leu45, Cys93, and Gly94. The common H-bond forming residues among SARS-CoV and MERS-CoV at the NSP14 interface are Asp126, Ile201, and Lys61; at the NSP10 interface, the residues are Ser15, Phe19, Val21, Asp29, Ser33, Asn40, Lys43, Leu45, Cys90, Cys93, and Cys96 (see Table 3).
Table 3. Common H-Bond Forming Residues across NSP14 and NSP10 among SARS-CoV-2, SARS-CoV, and MERS-CoVa.
| NSP14 | ||
|---|---|---|
| SARS-CoV-2 | SARS-CoV | MERS-CoV |
| Thr21 | Thr21 | Ala21 |
| Asp41 | Asp41 | |
| Lys61* | Lys61* | Lys61* |
| Asn67 | Asn67 | Pro69 (x) |
| Tyr69 | Tyr69 | Tyr69 (x) |
| Asp126 (x) | Asp126 | Asp126 |
| Thr127 | Thr127 (x) | Thr127 |
| Thr131 | Thr131 (x) | Thr131 |
| Ile201* | Ile201* | Ile201* |
| NSP10 | ||
| SARS-CoV-2 | SARS-CoV | MERS-CoV |
| Ser15* | Ser15* | Ser15* |
| Phe19* | Phe19* | Phe19* |
| Val 21* | Val 21* | Val 21* |
| Asp29* | Asp29* | Asp29* |
| Ser33* | Ser33* | Ser33* |
| Asn40 | Asn40 | |
| Lys43 (x) | Lys43 | Lys43 |
| Leu45* | Leu45* | Leu45* |
| Thr58 | Thr58 | |
| Cys90 (x) | Cys90 | Cys90 |
| Cys93* | Cys93* | Cys93* |
| Gly94* | Gly94* | Gly94* |
| Tyr96 | Tyr96 | Tyr96 (x) |
The average PPI complex extracted from the last 20 ns MD trajectory was subjected to the PDBsum server to get the interaction profile including H-bonds. The boldface residues are involved in H-bond formation. Unbold residues with an ″x″ mark do not form a H-bond. Residues with an asterisk (*) are common H-bond forming residues among all the viruses.
3.4. Hotspot Residue Prediction
The summary of the results obtained from the four methods (KFC2 server, DrugscorePPI, and Robetta servers and residue wise energy contribution) is presented in Tables S31–S33. The result obtained from the comparison of four methods suggests that among the key interface residues, a couple of them are predicted to be hotspots upon alanine mutation in the Robetta server and DrugScorePPI with ΔΔG values >1 kcal/mol or nearly 1 kcal/mol. The hotspot residues and their energy contributions across PPI interface are tabulated in Tables 4–6.
Table 4. List of Interacting Residues at the Protein–Protein Interacting Interface of SARS-CoV-2 NSP14 (Chain A) and NSP10 (Chain B)a.
| residues | KFC | Robetta ΔΔG (kcal/mol) | DrugScore PPI ΔΔG (kcal/mol) | per-residue energy contribution (kJ/mol) |
|---|---|---|---|---|
| LEU-7A | HS | 0.71 | 0.65 | –6.5 |
| PHE-8A | HS | 2.88 | 1.08 | –15.76 |
| THR-21A | HS | 1.16 | 0.54 | –9.06 |
| ASP-41A | HS | 3.81 | 3.49 | 14.47 |
| PHE-60A | HS | 1.53 | 0.56 | –9.73 |
| MET-62A | HS | 1.11 | 0.46 | –10.24 |
| TYR-64A | 1.14 | 1.27 | –7.46 | |
| VAL-66A | HS | 1.58 | 1.89 | –11.93 |
| TYR-69A | 2.69 | 2.33 | –8.95 | |
| THR-127A | –3.3 | |||
| ASN-130A | HS | 0.36 | 1.03 | 1.58 |
| ILE-201A | HS | 1.61 | 2.03 | –6.71 |
| THR-5B | HS | 1.38 | 0.44 | –5.48 |
| PHE-16B | HS | 3.23 | 1.07 | –16.49 |
| PHE-19B | HS | 4.55 | 1.34 | –4.03 |
| VAL-21B | 1.19 | 1.21 | –13.16 | |
| ASP-29B | 1.13 | –49.77 | ||
| VAL-42B | HS | 1.48 | 1.12 | –11.8 |
| MET-44B | 1.25 | 0.55 | –14.07 | |
| LEU-45B | –7.81 | |||
| THR-58B | –2.85 | |||
| HIS-80B | HS | 1.47 | 0.17 | –1.83 |
| LYS-93B | HS | 3.49 | 1.55 | 88.01 |
| TYR-96B | HS | 5.34 | 3.07 | –5.54 |
Interface hotspot residues are predicted using three computational methods implemented in the KFC, DrugScorePPI, and Robetta web servers. The per-residue energy decomposition analysis was carried out using the last 20 ns MD trajectory. HS: hotspot. H-bond forming hotspot residues are depicted in boldface.
Table 6. List of Interacting Residues at the Protein–Protein Interacting Interface of MERS-CoV NSP14 (Chain A) and NSP10 (Chain Ba.
| residues | KFC | Robetta ΔΔG (kcal/mol) | DrugScore PPI ΔΔG (kcal/mol) | per-residue energy contribution (kJ/mol) |
|---|---|---|---|---|
| TYR-22A | 1.07 | 1.91 | –6.21 | |
| LEU-39A | 1.37 | 1.11 | –7.69 | |
| LEU-62A | HS | 1.43 | 0.89 | –5.71 |
| VAL-66A | HS | 1.23 | 0.93 | –5.68 |
| TYR-69A | 1.92 | 1.00 | –1.77 | |
| ASP-126A | HS | 0.77 | 1.22 | –5.98 |
| ASN-131A | HS | 1.88 | 0.65 | –1.17 |
| LYS-200A | 1.34 | 0.84 | –6.64 | |
| ILE-201A | HS | 0.85 | 1.47 | –3.82 |
| SER-15B | HS | 0.81 | 0.59 | –0.43 |
| LEU-16B | HS | 1.01 | 0.33 | –6.99 |
| PHE-19B | HS | 3.85 | 1.20 | –14.02 |
| THR-20B | HS | 0.82 | 0.38 | –1.39 |
| VAL-21B | HS | 1.49 | 1.67 | –9.46 |
| ASN-40B | –3.43 | |||
| VAL-42B | HS | 1.84 | 1.37 | –9.37 |
| LYS-43B | –6.92 | |||
| MET-44B | 1.29 | 0.50 | –8.23 | |
| HIS-80B | HS | 2.47 | 0.29 | –3.07 |
| LYS-93B | HS | 1.43 | 1.30 | 40.99 |
| PHE-96B | 1.11 | 0.54 | –4.03 |
Interface hotspot residues are predicted using three computational methods implemented in the KFC, DrugScorePPI, and Robetta web servers. The per-residue energy decomposition analysis was carried out using the last 20 ns MD trajectory. HS: hotspot.
At the PPI interface, the energy of the residues is not uniformly distributed, which is one of the key features of the PPI interface. Some of them are highly responsible for binding energy toward the formation of the PPI complex and considered to be hotspot residues.33,38−43 Studies have suggested that hotspots have a tendency to form clusters at the center of the interface.33,38−43 Most of our predicted potential hotspots are observed in clusters and mostly at the center of the PPI interface. At the NSP14–NSP10 interface of SARS-CoV-2 and SARS-CoV, the common predicted hotspot residues at the NSP14 (chain A) interface are Thr21, Asp41, Met62, Thr64, Asn67, Ile201, Val66, Tyr69, and Asp126, and at the NSP10 (chain B) interface, Phe16, Phe19, Val21, Val42, Met44, His80, Lys93, and Tyr96 are predicted as common hotspot residues among SARS-CoV-2 and SARS-CoV (Figure 12 and Tables 4–6). The MERS-CoV NSP14 and NSP10 sequence has 61 and 58% identity with SARS-CoV-2 and SARS-CoV; therefore, many interacting interface residues of MERS NSP14 and NSP10 are different. However, few residues are conserved among all the three viruses, and among them, four residues (Val66, Thr69, Asp126, and Ile201) of MERS NSP14 and nine residues of MERS-CoV NSP10 (Leu16, Phe19, Val21, Val42, Met44, His80, Lys93, and Phe96) are identified as common hotspot residues among all the three viruses, which can be considered to be the important residues toward the formation of the NSP14–NSP10 complex in all the three viruses. Among all the interacting interface residues, the hotspot residues at the NSP14 and NSP10 interface are in boldface in Tables S31–S33, encircled in Figure 12, and depicted as spheres in 3D form in Figure S11.
Figure 12.
Representation of interacting interface residues at NSP14 (chain A) and NSP10 (chain B) of (a) SARS-CoV-2, (b) SARS-CoV, and (c) MERS-CoV obtained from the PDBsum server. The average conformer extracted from the last 20 ns MD trajectory was subjected to the PDBsum server. Interface hotspot residues were predicted using three computational methods implemented in the KFC2, DrugScorePPI, and Robetta web servers along with per-residue energy decomposition analysis. The encircled residues are identified hotspot residues.
Table 5. List of Interacting Residues at the Protein–Protein Interacting Interface of SARS-CoV NSP14 (Chain A) and NSP10 (Chain B)a.
| residues | KFC | Robetta ΔΔG (kcal/mol) | DrugScore PPI ΔΔG (kcal/mol) | per-residue energy contribution (kJ/mol) |
|---|---|---|---|---|
| VAL-4A | 1.05 | 1.28 | –11.36 | |
| THR-5A | HS | 1.04 | 0.41 | –9.63 |
| LEU-7A | HS | 2.07 | 1.31 | –16.56 |
| PHE-8A | HS | 2.10 | 0.62 | –13.63 |
| THR-21A | HS | 0.74 | 0.45 | –10.65 |
| THR-25A | HS | 2.49 | 0.85 | –14.09 |
| SER-28A | HS | 0.93 | 0.24 | –2.87 |
| LEU-38A | 1.29 | 1.13 | –14.12 | |
| ASP-41A | 1.25 | 3.52 | 23.56 | |
| ILE-55A | HS | 0.64 | 0.93 | –4.54 |
| PHE-60A | HS | 0.82 | 0.29 | –2.3 |
| MET-62A | HS | 1.14 | 0.53 | –11.56 |
| TYR-64A | 0.92 | 0.91 | –9.21 | |
| VAL-66A | HS | 1.58 | 1.94 | –15.78 |
| ASN-67A | –1.25 | |||
| TYR-69A | HS | 3.35 | 2.54 | –10.15 |
| ASP-126A | HS | 0.72 | 0.61 | 12.15 |
| ILE-201A | HS | 0.70 | 0.94 | –5.61 |
| PHE-217A | –3.79 | |||
| GLU-6B | HS | 7.92 | –0.61 | –18.3 |
| PHE-16B | HS | 3.16 | 1.03 | –20.09 |
| PHE-19B | HS | 3.40 | 1.02 | –27.66 |
| VAL-21B | HS | 1.46 | 2.08 | –17.08 |
| ASP-29B | 1.37 | 1.51 | –28.04 | |
| VAL-42B | HS | 1.87 | 1.98 | –18.43 |
| MET-44B | 1.21 | 0.51 | –18.26 | |
| ARG-78B | HS | 2.54 | 0.91 | 10.65 |
| HIS-80B | HS | 1.28 | 0.08 | –0.36 |
| LYS-93B | HS | 2.28 | 2.04 | 61.15 |
| TYR-96B | HS | 2.38 | 2.84 | –5.52 |
Interface hotspot residues are predicted using three computational methods implemented in the KFC, DrugScorePPI, and Robetta web servers. The per-residue energy decomposition analysis was carried out using the last 20 ns MD trajectory. HS: hotspot. H-bond forming hotspot residues are depicted in boldface.
There are a number of critical residues of the NSP10 interacting interface that have been identified through experimental ASM studies. However, for NSP14, mostly ASM studies have been performed for catalytic residues (Asp90, Glu92, Asp243, Glu191, His268, Asp273), two zinc finger residues (Cys207, Cys210, Cys226, His229), and a second zinc finger (His257, Cys261, His264) in all the three viruses.22,88,89 Only a limited number of ASM studies have been done for the NSP14 interacting interface21 and MERS-CoV NSP14–NSP1089,90 as the complex is not yet explored because there is no crystal structure available.
Recently, Moeller et al. verified the SARS-CoV-2 ExoN activities with single interacting interface amino acid substitutions with Ala for Lys9A, Lys61A, and K139A. In this experiment, all the three Lys-to-Ala mutants exhibited lesser activity than the WT NSP14 ExoN. Particularly, the substitution of K9A and K61A caused more severe defects than K139A.21 NSP14 Lys61 formed one H-bond and few nonbonded interactions with NSP10 Ser15 in all the three viruses, suggesting that this common residue is one of the key residues in the interface.
In addition to Lys61, Met62 was also identified as a hotspot by all the three servers with high per-residue energy contributions, that is, −10.78 kJ/mol in SARS-CoV2 and −11.56 kJ/mol in SARS-CoV NSP14, which are higher than Lys61 (−1.11 kJ/mol in SARS-CoV-2 and −6.75 kJ/mol in SARS-CoV, respectively). Overall, from the triplicate MD analysis, we conclude that, among the three CoVs, the SARS-CoV and MERS-CoV NSP14–NSP10 PPI complex is more flexible with higher binding free energy between NSP14 and NSP10 as compared to SARS-CoV-2 and all the SARS-CoV-2 mutant complexes for all the three sets of MD simulation. SARS-CoV-2 WT and mutant PPI complexes are also observed to be structurally more stable and rigid than the SARS-CoV and MERS-CoV. This structural stability of SARS-CoV-2 NSP14–NSP10 WT and mutants may induce stable mutations throughout the genome by a high-fidelity proofreading mechanism, thereby releasing new possible variants.
4. Conclusions
The present study makes an attempt to delineate the RNA synthesis proofreading mechanism in selected coronaviruses by rigorously analyzing the protein–protein interactions of NSP14–NSP10 in CoVs. A comparative analysis has been carried out by taking SARS-CoV-2, SARS-CoV, and MERS-CoV, as well as the four mutants of SARS-CoV-2 NSP14P203L,L177F,F233L,triple-mutant complexes, through molecular dynamics simulation studies in triplicate. It has been observed that the SARS-CoV NSP14–NSP10 PPI complex had a higher binding affinity in all the three sets of MD simulation as compared to SARS-CoV-2. It is interesting to note that few of theSARS-CoV-2 mutants result in an increase in the BFE in all the triplicates, revealing that the mutations may enhance functionality. The SARS-CoV-2 NSP14–NSP10 complex showed convergence with less RMSD value and less structural flexibility in comparison to SARS-CoV and MERS-CoV according to MD simulations. In the case of the mutant complex, P203L and L177F had significantly higher fluctuations during the simulations than the WT complex, while triple and F233L mutants do not show great differences in RMSD, Rg, and RMSF. All mutant complexes show slightly higher flexibility as compared to WT but are found to be more rigid than the SARS-CoV and MERS-CoV NSP14–NSP10 complex. In this work, more attention is given to the residue range 245–268 of NSP14 ExoN, which shows more structural fluctuations in the case of SARS-CoV NSP14, where some of the catalytic site residues (His257, Cys261, and His264) fall within this region. In addition to this, the SARS-CoV NSP10 C-terminal undergoes conformational change (res. 113–131) from the initial coil to beta strand during the simulation. However, these changes are not observed in NSP14 and NSP10 of SARS-CoV-2, MERS-CoV, and SARS-CoV-2 NSP14mutants. Structural stability is required for efficient proofreading activity by the virus for maintaining the viral replication fitness and proficiency. The study hypothesized that the overall structural stability and rigidity in important regions of SARS-CoV-2 NSP14 and NSP10 and mutant complexes may perhaps be the reason leading to the evolution of new effective variants of SAR-CoV-2. Some of the common NSP14 (Val66, Thr69, Asp126, and Ile201) residues and eight NSP10 residues (Leu16, Phe19, Val21, Val42, Met44, His80, Lys93, and Phe96) are identified as hotspots among all the three viruses. A few of the hotspot residues identified by the series of experiments conducted in this study are not investigated yet by experimental ASM. Moreover, to the best of our knowledge, this is the first attempt where a comparative study has been done on all the three important CoVs. This in silico study may lead the way to predict probable unknown hotspot locations at the interacting protein interface of the SARS-CoV-2 NSP14–NSP10 complex. Experimental ASM is a tedious task as it takes time and is highly expensive. Researchers will be able to minimize the negative results of random ASM experiments by selecting those potential hotspot residue locations identified by in silico approaches. In addition, based on the information on the nature of the interacting interface area and hotspot location, peptide/peptidomimetic, or small molecule can be designed to disturb the PPI between NSP14 and NSP10. This study may also serve as the basis for various other protein–protein interaction studies to identify the interacting protein partners in other disease pathways.
Acknowledgments
We thank the DBT center of excellence project (BT/PR40188/BTIS/137/27/2021) for the financial assistance. We thank the CSIR-Fourth Paradigm Institute for the use of the computational facilities. We thank CSIR project HCP41 for supporting the research.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c03007.
Detailed interacting interface profile of the PPI complexes; Rg and SASA plots of the three sets of MD simulations (in triplicate); and 3D representation of the conformational changes in PPI complexes during the three sets of 100 ns MD simulations (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Sharma A.; Tiwari S.; Deb M. K.; Marty J. L. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): a global pandemic and treatment strategies. Int. J. Antimicrob. Agents 2020, 56, 106054 10.1016/j.ijantimicag.2020.106054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J. W.; Holland J. J. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. 1999, 96, 13910–13913. 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G. M.; Rambaut A.; Pybus O. G.; Holmes E. C. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 2002, 54, 156–165. 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- Denison M. R.; Graham R. L.; Donaldson E. F.; Eckerle L. D.; Baric R. S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011, 8, 270–279. 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A. E.; Enjuanes L.; Ziebuhr J.; Snijder E. J. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006, 117, 17–37. 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R.; Nebot M. R.; Chirico N.; Mansky L. M.; Belshaw R. Viral mutation rates. J. Virol. 2010, 84, 9733–9748. 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. C.; Denison M. R. Implications of altered replication fidelity on the evolution and pathogenesis of coronaviruses. Curr. Opin. Virol. 2012, 2, 519–524. 10.1016/j.coviro.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’kovski P.; Kratzel A.; Steiner S.; Stalder H.; Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E.; Hertzig T.; Gorbalenya A. E.; Campanacci V.; Cambillau C.; Canard B.; Ziebuhr J. Discovery of an RNA virus 3ʹ⃗ 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. 2006, 103, 5108–5113. 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F.; Khan K. S.; Le T. K.; Paris C.; Demirbag S.; Barfuss P.; Rocchi P.; Ng W.-L. Coronavirus RNA proofreading: molecular basis and therapeutic targeting. Mol. Cell 2020, 79, 710–727. 10.1016/j.molcel.2020.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. C.; Denison M. R. Coronaviruses as DNA wannabes: a new model for the regulation of RNA virus replication fidelity. PLoS Pathog. 2013, 9, e1003760 10.1371/journal.ppat.1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskier D.; Suner A.; Oktay Y.; Karakülah G. Mutations of SARS-CoV-2 nsp14 exhibit strong association with increased genome-wide mutation load. Peer J. 2020, 8, e10181 10.7717/peerj.10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K.; Ueda M. T.; Watanabe T.; Nakagawa S.. Genomic diversity of SARS-CoV-2 can be accelerated by a mutation in the nsp14 gene. bioRxiv 2020, 10.1101/2020.12.23.424231. [DOI] [PMC free article] [PubMed]
- Eckerle L. D.; Becker M. M.; Halpin R. A.; Li K.; Venter E.; Lu X.; Scherbakova S.; Graham R. L.; Baric R. S.; Stockwell T. B.; Spiro D. J.; Denison M. R. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010, 6, e1000896 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando N. S.; Zevenhoven-Dobbe J. C.; van der Meer Y.; Bredenbeek P. J.; Posthuma C. C.; Snijder E. J. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. Virol. J. 2020, 94, e01246–e01220. 10.1128/JVI.01246-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. C.; Blanc H.; Vignuzzi M.; Denison M. R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013, 9, e1003565 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle L. D.; Lu X.; Sperry S. M.; Choi L.; Denison M. R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. Virol. J. 2007, 81, 12135–12144. 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F.; Subissi L.; Silveira De Morais A. T.; Le N. T. T.; Sevajol M.; Gluais L.; Decroly E.; Vonrhein C.; Bricogne G.; Canard B.; Imbert I. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. 2018, 115, E162–E171. 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales C.; Domingo E. Antiviral strategies based on lethal mutagenesis and error threshold. Quasispecies: From Theory to Exp. Syst. 2015, 323–339. 10.1007/82_2015_459. [DOI] [PubMed] [Google Scholar]
- Liu C.; Shi W.; Becker S. T.; Schatz D. G.; Liu B.; Yang Y. Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. Science 2021, 373, 1142–1146. 10.1126/science.abi9310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller N. H.; Shi K.; Demir Ö.; Belica C.; Banerjee S.; Yin L.; Durfee C.; Amaro R. E.; Aihara H. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. Proc. Natl. Acad. Sci. 2022, 119, e2106379119 10.1073/pnas.2106379119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.; Wu L.; Shaw N.; Gao Y.; Wang J.; Sun Y.; Lou Z.; Yan L.; Zhang R.; Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc. Natl. Acad. Sci. 2015, 112, 9436–9441. 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien M.; Anderson T. K.; Jockusch S.; Tao C.; Li X.; Kumar S.; Russo J. J.; Kirchdoerfer R. N.; Ju J. Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase, a key drug target for COVID-19. J. Proteome Res. 2020, 19, 4690–4697. 10.1021/acs.jproteome.0c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch S.; Tao C.; Li X.; Anderson T. K.; Chien M.; Kumar S.; Russo J. J.; Kirchdoerfer R. N.; Ju J. A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19. Antiviral Res. 2020, 180, 104857 10.1016/j.antiviral.2020.104857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazer L. L.; Neubig R. R. Small molecule protein–protein interaction inhibitors as CNS therapeutic agents: current progress and future hurdles. Neuropsychopharmacology 2009, 34, 126–141. 10.1038/npp.2008.151. [DOI] [PubMed] [Google Scholar]
- Gurung A. B.; Bhattacharjee A.; Ali M. A.; Al-Hemaid F.; Lee J. Binding of small molecules at interface of protein–protein complex–A newer approach to rational drug design. Saudi J. Biol. Sci. 2017, 24, 379–388. 10.1016/j.sjbs.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenemann M. A.; Sperandio O.; Labbé C. M.; Lagorce D.; Miteva M. A.; Villoutreix B. O. In silico design of low molecular weight protein–protein interaction inhibitors: Overall concept and recent advances. Prog. Biophys. Mol. Biol. 2015, 119, 20–32. 10.1016/j.pbiomolbio.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Panwar D.; Rawal L.; Ali S. Molecular docking uncovers TSPY binds more efficiently with eEF1A2 compared to eEF1A1. J. Biomol. Struct. Dyn. 2015, 33, 1412–1423. 10.1080/07391102.2014.952664. [DOI] [PubMed] [Google Scholar]
- Rognan D. Rational design of protein–protein interaction inhibitors. MedChemComm. 2015, 6, 51–60. 10.1039/C4MD00328D. [DOI] [Google Scholar]
- Xu J.; Xu J.; Chen H. Interpreting the structural mechanism of action for MT7 and human muscarinic acetylcholine receptor 1 complex by modeling protein–protein interaction. J. Biomol. Struct. Dyn. 2012, 30, 30–44. 10.1080/07391102.2012.674188. [DOI] [PubMed] [Google Scholar]
- Jin L.; Wang W.; Fang G. Targeting protein-protein interaction by small molecules. Annu. Rev. Pharmacol. Toxicol. 2014, 54, 435–456. 10.1146/annurev-pharmtox-011613-140028. [DOI] [PubMed] [Google Scholar]
- Cesa L. C.; Mapp A. K.; Gestwicki J. E. Direct and propagated effects of small molecules on protein–protein interaction networks. Front. Bioeng. Biotechnol. 2015, 3, 119. 10.3389/fbioe.2015.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin O.; Ma B.; Nussinov R. Hot regions in protein–protein interactions: the organization and contribution of structurally conserved hot spot residues. J. Mol. Biol. 2005, 345, 1281–1294. 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- González-Ruiz D.; Gohlke H. Targeting protein-protein interactions with small molecules: challenges and perspectives for omputational binding epitope detection and ligand finding. Curr. Med. Chem. 2006, 13, 2607–2625. 10.2174/092986706778201530. [DOI] [PubMed] [Google Scholar]
- Bogan A. A.; Thorn K. S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998, 280, 1–9. 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Cheung L. S.-L.; Kanwar M.; Ostermeier M.; Konstantopoulos K. A hot-spot motif characterizes the interface between a designed ankyrin-repeat protein and its target ligand. Biophys. J. 2012, 102, 407–416. 10.1016/j.bpj.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clackson T.; Wells J. A. A hot spot of binding energy in a hormone-receptor interface. Science 1995, 267, 383–386. 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- Thorn K. S.; Bogan A. A. ASEdb: a database of alanine mutations and their effects on the free energy of binding in protein interactions. Bioinformatics 2001, 17, 284–285. 10.1093/bioinformatics/17.3.284. [DOI] [PubMed] [Google Scholar]
- Moreira I. S.; Fernandes P. A.; Ramos M. J. Hot spots—A review of the protein–protein interface determinant amino-acid residues. Proteins: Struct., Funct., Bioinf. 2007, 68, 803–812. 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- Caffrey D. R.; Somaroo S.; Hughes J. D.; Mintseris J.; Huang E. S. Are protein–protein interfaces more conserved in sequence than the rest of the protein surface?. Protein Sci. 2004, 13, 190–202. 10.1110/ps.03323604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockless S. W.; Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 1999, 286, 295–299. 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- Schreiber G.; Fersht A. R. Energetics of protein-protein interactions: Analysis of the Barnase-Barstar interface by single mutations and double mutant cycles. J. Mol. Biol. 1995, 248, 478–486. 10.1016/S0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- Thornton J. M. The Hans Neurath Award lecture of The Protein Society: proteins—a testament to physics, chemistry, and evolution. Protein Sci.: Publ. Protein Soc. 2001, 10, 3. 10.1110/ps.10.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham B. C.; Wells J. A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 1989, 244, 1081–1085. 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- Chothia C.; Janin J. Principles of protein–protein recognition. Nature 1975, 256, 705–708. 10.1038/256705a0. [DOI] [PubMed] [Google Scholar]
- Janin J. Principles of protein-protein recognition from structure to thermodynamics. Biochimie 1995, 77, 497–505. 10.1016/0300-9084(96)88166-1. [DOI] [PubMed] [Google Scholar]
- Miller S.; Lesk A. M.; Janin J.; Chothia C. The accessible surface area and stability of oligomeric proteins. Nature 1987, 328, 834–836. 10.1038/328834a0. [DOI] [PubMed] [Google Scholar]
- Argos P. An investigation of protein subunit and domain interfaces. Protein Eng., Des. Sel. 1988, 2, 101–113. 10.1093/protein/2.2.101. [DOI] [PubMed] [Google Scholar]
- Janin J.; Miller S.; Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J. Mol. Biol. 1988, 204, 155–164. 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- Jones S.; Thornton J. M. Protein-protein interactions: a review of protein dimer structures. Prog. Biophys. Mol. Biol. 1995, 63, 31–65. 10.1016/0079-6107(94)00008-W. [DOI] [PubMed] [Google Scholar]
- Arkin M. R.; Randal M.; DeLano W. L.; Hyde J.; Luong T. N.; Oslob J. D.; Raphael D. R.; Taylor L.; Wang J.; McDowell R. S.; Wells J. A.; Braisted A. C. Binding of small molecules to an adaptive protein–protein interface. Proc. Natl. Acad. Sci. 2003, 100, 1603–1608. 10.1073/pnas.252756299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R.; Sagurthi S. R.; Sastry G. N. Elucidating the preference of dimeric over monomeric form for thermal stability of Thermus thermophilus isopropylmalate dehydrogenase: A molecular dynamics perspective. J. Mol. Graphics Modell. 2020, 96, 107530 10.1016/j.jmgm.2020.107530. [DOI] [PubMed] [Google Scholar]
- Sharma R.; Sastry G. N. Deciphering the dynamics of non-covalent interactions affecting thermal stability of a protein: Molecular dynamics study on point mutant of Thermus thermophilus isopropylmalate dehydrogenase. PLoS One 2015, 10, e0144294 10.1371/journal.pone.0144294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyrisch S.; Helms V. What induces pocket openings on protein surface patches involved in protein–protein interactions?. J. Comput.-Aided Mol. Des. 2009, 23, 73–86. 10.1007/s10822-008-9239-y. [DOI] [PubMed] [Google Scholar]
- Eyrisch S.; Medina-Franco J. L.; Helms V. Transient pockets on XIAP-BIR2: toward the characterization of putative binding sites of small-molecule XIAP inhibitors. J. Mol. Model. 2012, 18, 2031–2042. 10.1007/s00894-011-1217-y. [DOI] [PubMed] [Google Scholar]
- Kumari R.; Kumar R.; Lynn A. g_mmpbsa— A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- Srivastava H. K.; Sastry G. N. Efficient estimation of MMGBSA-based BEs for DNA and aromatic furan amidino derivatives. J. Biomol. Struct. Dyn. 2013, 31, 522–537. 10.1080/07391102.2012.703071. [DOI] [PubMed] [Google Scholar]
- Laskowski R. A.; Jabłońska J.; Pravda L.; Vařeková R. S.; Thornton J. M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell S. J.; LeGault L.; Mitchell J. C. KFC Server: interactive forecasting of protein interaction hot spots. Nucleic Acids Res. 2008, 36, W265–W269. 10.1093/nar/gkn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger D. M.; Gohlke H. DrugScorePPI webserver: fast and accurate in silico alanine scanning for scoring protein–protein interactions. Nucleic Acids Res. 2010, 38, W480–W486. 10.1093/nar/gkq471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. E.; Chivian D.; Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, W526–W531. 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortemme T.; Kim D. E.; Baker D. Computational alanine scanning of protein-protein interfaces. Science’s STKE 2004, 2004, pl2–pl2. 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- Kumar N.; Sarma H.; Sastry G. N. Repurposing of approved drug molecules for viral infectious diseases: a molecular modelling approach. J. Biomol. Struct. Dyn. 2021, 1-17, 1. 10.1080/07391102.2021.1905558. [DOI] [PubMed] [Google Scholar]
- Badrinarayan P.; Sastry G. N. Sequence, structure, and active site analyses of p38 MAP kinase: exploiting DFG-out conformation as a strategy to design new type II leads. J. Chem. Inf. Model. 2011, 51, 115–129. 10.1021/ci100340w. [DOI] [PubMed] [Google Scholar]
- Srivastava H. K.; Chourasia M.; Kumar D.; Sastry G. N. Comparison of computational methods to model DNA minor groove binders. J. Chem. Inf. Model. 2011, 51, 558–571. 10.1021/ci100474n. [DOI] [PubMed] [Google Scholar]
- Sarma H.; Jamir E.; Sastry G. N. Protein-protein interaction of RdRp with its co-factor NSP8 and NSP7 to decipher the interface hotspot residues for drug targeting: A comparison between SARS-CoV-2 and SARS-CoV. J. Mol. Struct. 2022, 1257, 132602 10.1016/j.molstruc.2022.132602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarvagalla S.; Cheung C. H. A.; Tsai J.-Y.; Hsieh H. P.; Coumar M. S. Disruption of protein–protein interactions: hot spot detection, structure-based virtual screening and in vitro testing for the anti-cancer drug target–survivin. RSC Adv. 2016, 6, 31947–31959. 10.1039/C5RA22927H. [DOI] [Google Scholar]
- Sarvagalla S.; Lin T.-Y.; Kondapuram S. K.; Cheung C. H. A.; Coumar M. S. Survivin-caspase protein-protein interaction: Experimental evidence and computational investigations to decipher the hotspot residues for drug targeting. J. Mol. Struct. 2021, 1229, 129619 10.1016/j.molstruc.2020.129619. [DOI] [Google Scholar]
- Jha V.; Rameshwaram N. R.; Janardhan S.; Raman R.; Sastry G. N.; Sharma V.; Rao J. S.; Kumar D.; Mukhopadhyay S. Uncovering structural and molecular dynamics of ESAT-6: β2M interaction: asp53 of human β2-microglobulin is critical for the ESAT-6: β2M complexation. J. Immunol. 2019, 203, 1918–1929. 10.4049/jimmunol.1700525. [DOI] [PubMed] [Google Scholar]
- Badrinarayan P.; Sastry G. N. Specificity rendering ‘hot-spots’ for aurora kinase inhibitor design: the role of non-covalent interactions and conformational transitions. PLoS One 2014, 9, e113773 10.1371/journal.pone.0113773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.; Bertoni M.; Bienert S.; Studer G.; Tauriello G.; Gumienny R.; Heer F. T.; de Beer T. A. P.; Rempfer C.; Bordoli L.; Lepore R.; Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D.; Inbar Y.; Nussinov R.; Wolfson H. J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S.; Chan H. C. S.; Hu Z. Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2017, 7, e1298 10.1002/wcms.1298. [DOI] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1-2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Best R. B.; Zhu X.; Shim J.; Lopes P. E. M.; Mittal J.; Feig M.; MacKerell A. D. Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmans M.; Sanders B. P.; Marrink S. J.; de Vries A. H. Effects of bundling on the properties of the SPC water model. Theor. Chem. Acc. 2010, 125, 335–344. 10.1007/s00214-009-0590-4. [DOI] [Google Scholar]
- Berendsen H. J. C.; Postma J. P. M.; Van Gunsteren W. F.; DiNola A. R.; Haak J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. 10.1063/1.448118. [DOI] [Google Scholar]
- Berendsen H. J. C.; van der Spoel D.; van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995, 91, 43–56. 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- Hess B.; Bekker H.; Berendsen H. J. C.; Fraaije J. G. E. M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. . [DOI] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle mesh Ewald: An N· log (N) method for Ewald sums in large systems. J. Chem. Phys 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Xu W.; Amire-Brahimi B.; Xie X. J.; Huang L.; Ji J. Y. All-atomic molecular dynamic studies of human CDK8: insight into the A-loop, point mutations and binding with its partner CycC. Comput. Biol. Chem. 2014, 51, 1–11. 10.1016/j.compbiolchem.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Bao Q.-C.; Xu X.-L.; Jiang F.; Gu K.; Jiang Z.-Y.; Zhang X.-J.; Guo X.-K.; You Q.-D.; Sun H. P. Discovery and identification of Cdc37-derived peptides targeting the Hsp90–Cdc37 protein–protein interaction. RSC Adv. 2015, 5, 96138–96145. 10.1039/C5RA20408A. [DOI] [Google Scholar]
- Shi S.; Zhang S.; Zhang Q. Probing difference in binding modes of inhibitors to MDMX by molecular dynamics simulations and different free energy methods. PLoS One 2015, 10, e0141409 10.1371/journal.pone.0141409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S.; Kollman P. A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. 10.1002/jcc.540130805. [DOI] [Google Scholar]
- Amadei A.; Linssen A. B. M.; Berendsen H. J. C. Essential dynamics of proteins. Proteins: Struct., Funct., Bioinf. 1993, 17, 412–425. 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- Turner P.J. XMGRACE, Version 5.1. 19. Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology, Beaverton; 2005, OR, 2.
- Altis A.; Otten M.; Nguyen P. H.; Hegger R.; Stock G. Construction of the free energy landscape of biomolecules via dihedral angle principal component analysis. J. Chem. Phys. 2008, 128, 06B620 10.1063/1.2945165. [DOI] [PubMed] [Google Scholar]
- Bouvet M.; Imbert I.; Subissi L.; Gluais L.; Canard B.; Decroly E. RNA 3′-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. 2012, 109, 9372–9377. 10.1073/pnas.1201130109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble J.; Stevens L. J.; Agostini M. L.; Anderson-Daniels J.; Chappell J. D.; Lu X.; Pruijssers A. J.; Routh A. L.; Denison M. R. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 2021, 17, e1009226 10.1371/journal.ppat.1009226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.