Abstract
An abundant metal in the human body, iron is essential for key biological pathways including oxygen transport, DNA metabolism, and mitochondrial function. Most iron is bound to heme but it can also be incorporated into iron-sulfur clusters or bind directly to proteins. Iron’s capacity to cycle between Fe2+ and Fe3+ contributes to its biological utility but also renders it toxic in excess. Heme is an iron-containing tetrapyrrole essential for diverse biological functions including gas transport and sensing, oxidative metabolism, and xenobiotic detoxification. Like iron, heme is essential yet toxic in excess. As such, both iron and heme homeostasis are tightly regulated. Here we discuss molecular and physiologic aspects of iron and heme metabolism. We focus on dietary absorption; cellular import; utilization; and export, recycling, and elimination, emphasizing studies published in recent years. We end with a discussion on current challenges and needs in the field of iron and heme biology.
Keywords: heme, tetrapyrrole, porphyrin, iron, trafficking, metabolism
1. INTRODUCTION
An abundant metal in the human body, iron is essential for key biological pathways (18, 21, 54, 79, 94). While most of the body’s iron complement is found in heme, iron can also be incorporated into iron-sulfur clusters or bind directly to proteins. In addition to its role in oxygen transport as a component of hemoglobin, iron is critical for multiple biological pathways including DNA metabolism and mitochondrial function. Iron’s capacity to cycle between ferrous (Fe2+) and ferric (Fe3+) states contributes to its utility in biological systems but also renders it toxic when present in excess. As such, both iron deficiency and excess are detrimental to human health.
Iron deficiency is a common nutritional deficiency worldwide (94). General causes of iron deficiency include inadequate dietary iron levels, insufficient dietary iron absorption, and blood loss. If severe enough, iron deficiency leads to anemia. Iron excess is also a common condition. If not bound to factors such as the serum iron-binding protein transferrin or the iron storage protein ferritin, iron can promote oxidative damage at the macromolecule, organelle, cell, and organ level. In contrast to iron deficiency, iron excess is typically caused by genetic or acquired defects rather than nutritional excess. If untreated, iron excess can lead to dysfunction in multiple organs including the liver, pancreas, heart, and endocrine glands.
Heme (iron-protoporphyrin IX) is an iron-containing tetrapyrrole that is a requisite for diverse biological functions. It plays crucial roles in oxygen binding and transport (globins), oxidative metabolism (cytochrome c oxidase, succinate dehydrogenase), xenobiotic detoxification (cytochrome P450), gas sensing (guanyl cyclases, nitic oxide synthase), and microRNA processing (DGCR8). Heme can also bind to transcription factors and regulate transcription of target genes in varied pathways including circadian rhythm, antioxidant stress response, apoptosis, cell proliferation, ion channel activity, and mitochondrial respiration (13).
While heme plays a pivotal role in biology, it is also cytotoxic and hydrophobic. Free heme has peroxidase activity and can generate reactive oxygen species, thus leading to oxidative stress. A hydrophobic molecule, heme can intercalate into membranes and bind nonspecifically to proteins (17). A tight regulation system is necessary to keep in check the concentration and bioavailability of heme. Heme homeostasis in an organism can be controlled at different levels: import, utilization, synthesis, degradation, and export. While heme synthesis and degradation are extensively characterized, heme trafficking remains poorly understood.
In this review, we focus on molecular and physiologic aspects of iron and heme metabolism, with an emphasis on regulation of organismal heme homeostasis. We pay particular attention to recently published results, most notably those published in the last two to three years. With regard to nonheme iron, we cover dietary absorption; cellular iron uptake; and intracellular iron trafficking, utilization, storage, and recycling. With regard to heme, we cover dietary absorption; heme synthesis and intracellular trafficking; and systemic heme transport, sequestration, degradation, and elimination, then end with a discussion of cellular heme import and export. For details on well-established aspects of iron and heme metabolism, we provide references to reviews on these areas. We do not cover the role of iron and heme metabolism in immunity, infection, cancer, or ferroptosis and defer to other sources for coverage of these topics.
2. NONHEME IRON
2.1. Gastrointestinal Nonheme Iron Absorption
Dietary iron is present in heme and nonheme forms. Nonheme iron is present in the ferric state and is found in cereals, vegetables, legumes, and fruits. Heme iron is more abundant in animal than plant matter and more efficiently absorbed than nonheme iron. A key initial step in nonheme iron absorption is secretion of gastric acid in the form of hydrochloric acid by the stomach (116). Gastric acid secretion is regulated by gastrin and other hormones (116). Gastric acid solubilizes nonheme iron, rendering it available for reduction by ascorbic acid and reductases and subsequent absorption by the small intestine. Gastric acid also maintains ascorbic acid in its more active reduced form. Additionally, ascorbic acid chelates soluble iron, preventing its precipitation. As humans cannot synthesize ascorbic acid, they must acquire it from the diet.
Dietary nonheme iron absorption occurs in the proximal small intestine and involves several steps (54) (Figure 1). First, nonheme iron is reduced to the ferrous form in the gut lumen by gastric acid or ferrireductases such as duodenal cytochrome b reductase 1 (DCYTB). Second, iron is transported across the apical surface of enterocytes by divalent metal transporter 1 (DMT1) (SLC11A2). Ferrous iron bound to nicotianamine, an organic small molecule found in plants, can also be transported by the amino acid transporter SLC36A1, although the overall contribution of SLC36A1 to iron absorption has yet to be established (85). Third, within enterocytes, iron is either utilized for iron-dependent processes, stored in ferritin, or trafficked to the basolateral surface. Fourth, iron is exported from enterocytes into the blood by the basolateral iron transport protein ferroportin (SLC40A1). Iron transport by ferroportin is coupled to antitransport of two H+ ions (92). Fifth, iron is oxidized to the ferric form by the multicopper oxidase hephaestin. Sixth, ferric iron binds to transferrin, a serum iron-binding protein produced largely by the liver (145). [Ferroportin-mediated iron export also plays an essential role in release of iron from red blood cell (RBC)–scavenging macrophages, as discussed below.]
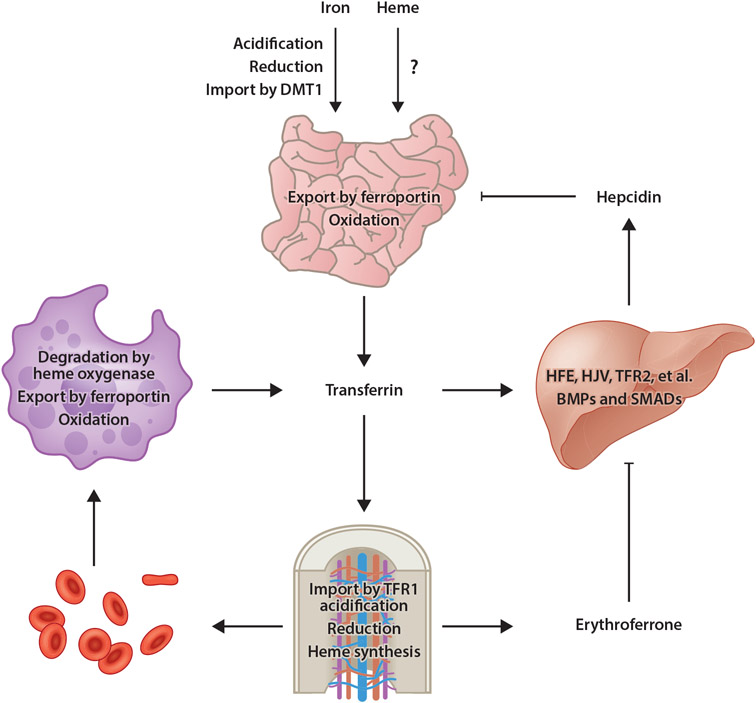
Figure 1.
Model of mammalian iron homeostasis. Both nonheme and heme iron are absorbed in the small intestine. Transporters involved with heme iron absorption have yet to be identified. Nonheme iron is acidified in the stomach, reduced by gastric acid or DCYTB in the small intestine, then imported into enterocytes by DMT1. Iron is then utilized by or stored in enterocytes or exported into circulation by ferroportin. After oxidation by hephaestin, iron binds to transferrin for distribution throughout the body. Most transferrin-bound iron is imported by TFR1-mediated endocytosis into erythroid precursors, a process that requires acidification to liberate iron from transferrin then export into the cytosol followed by reduction by STEAP3. Iron is then utilized for heme synthesis. Senescent or damaged RBCs are degraded by RES macrophages, in which heme oxygenase degrades heme, thereby liberating iron for export by ferroportin into circulation. Under conditions of iron excess, transferrin (and other factors) stimulates BMP expression in sinusoidal endothelial cells. BMPs then stimulate hepcidin expression by hepatocytes in a pathway dependent upon HFE, HJV, TFR2, and other membrane factors, as well as SMAD1/5/8 transcription factors. Hepcidin then posttranslationally inhibits ferroportin activity and expression. Under conditions of anemia, erythroid progenitors produce the hormone erythroferrone, which inhibits BMP-dependent hepcidin expression, thereby ensuring continued dietary iron absorption for erythropoiesis. Abbreviations: BMP, bone morphogenetic protein; DCYTB, duodenal cytochrome b reductase 1; DMT1, divalent metal transporter 1; HFE, homeostatic iron regulator; HJV, hemojuvelin; RBC, red blood cell; RES, reticuloendothelial system; TFR1, transferrin receptor 1; TFR2, transferrin receptor 2. Figure adapted from images created with BioRender.com.
Regulation of ferroportin is an active area of research. Central to this regulation is hepcidin, a peptide hormone synthesized mainly by hepatocytes (18) (Figure 1). Hepcidin regulates iron absorption by binding to ferroportin and inducing its internalization and lysosomal degradation. Hepcidin has a greater affinity for iron-replete than iron-poor ferroportin and binds in a manner that physically occludes iron transport by ferroportin (8). Internalized ferroportin is targeted for proteasomal degradation by the E3 ubiquitin ligase RNF217 in a process that also requires the E1 enzyme UBA6 and adaptor protein NDFIP1 (44, 130). Downregulation of ferroportin by hepcidin also increases enterocyte iron levels, which in turn increases iron-dependent degradation of hypoxia-inducible factor 2, a transcription factor that stimulates expression of genes required for iron absorption including DMT1 and DCYTB (117).
Like ferroportin, hepcidin is highly regulated (18) (Figure 1). Expression increases under conditions of iron excess and inflammation and decreases under hypoxia or conditions of iron demand such as increased erythropoietic activity. Key to this regulation is a bone morphogenetic protein (BMP)-dependent signaling pathway involving BMP2 and BMP6, both of which are produced by liver sinusoidal endothelial cells (139). Multiple proteins on the cell surface of hepatocytes are required to positively and negatively regulate hepcidin expression in response to BMPs as well as diferric transferrin. These factors include homeostatic iron regulator (HFE); hemojuvelin (HJV); transferrin receptor 2 (TFR2); BMP receptor proteins ALK2, ALK3, and BMPR-II; neogenin; and TMPRSS6 (18, 30). Notably, inherited mutations in HFE, HJV, and TFR2 lead to hereditary hemochromatosis, a disease of iron excess resulting in dysfunction of multiple organs including liver, pancreas, heart, and endocrine glands. SMAD1/5/8 transcription factors transduce the BMP signal to the nucleus to stimulate hepcidin gene expression (132). Additionally, the transcription factor nuclear factor E2-related factor-2 (NRF2) stimulates BMP-dependent hepcidin expression under conditions of iron-induced oxidative stress (67). Finally, the regulation of hepcidin expression by erythropoietic activity is mediated by the hormone erythroferrone (128). Erythroferrone is synthesized by bone marrow erythroblasts and suppresses hepcidin expression by inhibiting BMP signaling in hepatocytes. Erythroferrone-dependent hepcidin suppression ensures adequate dietary iron absorption to supply erythropoietic demand for iron. Notably, erythroferrone, hepcidin, and TFR2 also play roles in bone homeostasis. Erythroferrone expression by osteoblasts modulates BMP signaling in osteoblasts and serves an osteoprotective effect to prevent bone loss under conditions of enhanced erythropoiesis (11). Hepcidin deficiency leads to bone loss by perturbing Wnt/β-catenin signaling (61) and suppressing bone formation (60). TFR2 also inhibits bone formation by modulating BMP and Wnt signaling (109).
The microbiome also contributes to regulation of dietary iron absorption. Intestinal microbes produce metabolites that inhibit iron absorption, thereby ensuring an adequate supply for their own metabolism (25). 3-Hydroxypropionaldehyde is produced by Limosilactobacillus reuteri but its role in the gut microbiome is not well understood. 1,3-Diaminopropane is produced by several bacterial species in the gut but can also come from dietary sources. Both metabolites impair expression of DCYTB, DMT1, and ferroportin by inhibiting activity of hypoxia-inducible factor 2 (HIF-2) in enterocytes. This results in decreased iron import into enterocytes and export from enterocytes into blood. These metabolites also stimulate expression of ferritin, which stores iron within enterocytes and limits its availability for export from enterocytes, ultimately returning iron to the gut when the enterocytes are sloughed.
2.2. Cellular Nonheme Iron Uptake
The majority of circulating diferric transferrin is taken up by erythroid precursors for use in erythropoiesis (Figure 1). Under physiologic conditions, cellular iron uptake is mediated by the transferrin cycle, in which diferric transferrin binds to transferrin receptor 1 (TFR1) and is internalized into the cell via endocytosis (79). Endosomal acidification leads to release of ferric iron from transferrin. Iron is then reduced to the ferrous state by the reductase STEAP3 and transported into the cytoplasm by DMT1. TFR1 can also bind the heavy chain of serum ferritin, representing a potential transferrin-independent means of cellular iron uptake (80). Another transferrin-independent mechanism of cellular iron uptake was recently identified in cells undergoing the epithelial-mesenchymal transition, in which the transmembrane glycoprotein CD44 mediates the endocytosis of iron-bound hyaluronates (83).
Transferrin was discovered two decades ago, but our understanding of its role in iron homeostasis continues to evolve. While transferrin has an iron-binding site in each of its two lobes, mice with mutations preventing iron loading in either the N or C lobe of transferrin do not display similar sensitivities to erythropoietin, indicating that these iron-binding sites are not functionally homologous (93). Novel roles for TFR1 also continue to be identified. Tfr1 deficiency in mice protects them from metabolic dysfunction induced by high-fat diets by impairing intestinal lipid absorption, although the underlying mechanism of the adipocyte-intestine link is not fully understood (149). Notably, some novel roles for TFR1 may not involve iron transport. For example, Tfr1 deficiency in adipocytes in mice leads to impaired thermogenesis, increased insulin resistance, inflammation, iron deficiency, and mitochondrial dysfunction (62). However, mice on an iron-deficient diet do not develop all the characteristics observed in adipocyte Tfr1-deficient tissues.
Cellular iron uptake is highly regulated by RNA binding proteins known as iron regulatory proteins (IRPs) (79) (Figure 2). IRPs regulate translation of mRNAs encoding TFR1, DMT1, ferroportin, and ferritin light and heavy chain. IRP binding to iron-responsive elements (IREs) in the 3′ untranslated region (UTR) of mRNAs increases mRNA stability and translation, while IRP binding to IREs in the 5′ UTR inhibits translation. Degradation of mRNAs not stabilized by IRP activity may be mediated by the RNA binding protein roquin, which destabilizes TFR1 mRNA in several cell lines (22). IRP1 is regulated by iron-sulfur cluster binding: Only when devoid of iron-sulfur clusters does IRP1 bind RNA. IRP2 activity is regulated by protein degradation mediated by an iron-stabilized E3 ligase FBXL5. Through these mechanisms, IRP-dependent regulation of gene expression maintains cellular iron at sufficient but nontoxic levels. Notably, some IRP-regulated mRNAs encoding factors such as DMT1 and ferroportin can also be expressed as IRE-free isoforms in some but not all cell types, indicating that the impact of IRPs on gene expression is not identical across all tissues. Additionally, a recent study on the glycogen branching enzyme AGBE in Drosophila indicates that AGBE binds to holo-IRP1 and modulates its translocation to the nucleus, suggesting that holo-IRP1 can also regulate gene expression (41).
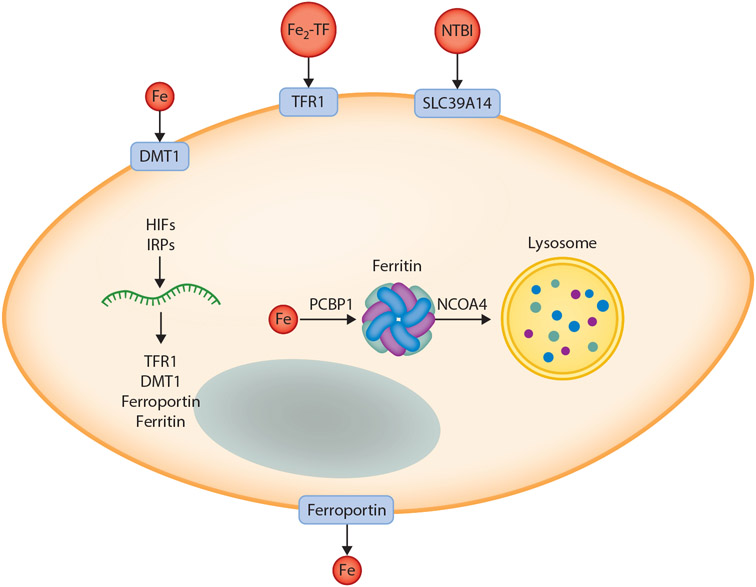
Figure 2.
Model of main pathways of cellular iron import and export and intracellular distribution. Iron can be imported into cells via one of several pathways including import of ferrous iron by DMT1; import of Fe2-TF by TFR1-mediated endocytosis, followed by reduction by STEAP3 and export into the cytoplasm; and import of NTBI by SLC39A14. Iron is exported by ferroportin, the only known mammalian iron export protein, then oxidized by hephaestin or ceruloplasmin and bound to transferrin for distribution to other cellular targets. Within the cell, expression of iron transporters, the iron storage protein ferritin, and other factors is regulated by HIFs and IRPs to ensure sufficient but nontoxic levels of iron for cellular metabolism and adequate storage in settings of iron excess. The iron metallochaperone PCBP1 delivers iron to multiple protein targets within the cell including ferritin. Under conditions of iron limitation, ferritin is mobilized to the lysosome by NCOA4 for degradation and liberation of stored iron. For simplicity, not all pathways of iron import are shown, nor are all known intracellular destinations or targets for iron indicated. Abbreviations: DMT1, divalent metal transporter 1; Fe2-TF, diferric transferrin; HIF, hypoxia-inducible factor; IRP, iron regulatory protein; NCOA4, nuclear receptor coactivator 4; NTBI, nontransferrin-bound iron; PCBP1, poly(rC)-binding protein; TFR1, transferrin receptor 1. Figure adapted from images created with BioRender.com.
The transferrin cycle is not the only means of cellular iron import. Under conditions of iron excess, transferrin becomes saturated and excess iron circulates in the blood as a toxic species known as nontransferrin-bound iron (NTBI). The biochemical nature of NTBI has yet to be firmly established. NTBI import into liver and pancreas is dependent upon SLC39A14 (ZIP14), a protein that transports manganese under physiologic conditions and iron under conditions of iron excess (79).
2.3. Intracellular Iron Trafficking, Utilization, Storage, and Recycling
A significant fraction of iron imported into cells is directed to mitochondria (35). Mitochondrial iron uptake is an active area of research, with several pathways shown to occur in different cell lines including a kiss-and-run model in which the endosome transfers iron to mitochondria via direct contact and uptake of iron from the labile iron pool, which consists of low-molecular-weight species of redox-active iron. Within mitochondria, iron is either stored or used for synthesis of heme and iron-sulfur clusters. (Heme synthesis is discussed below.) Iron-sulfur cluster biogenesis is a multistep process requiring a sulfur donor, iron supply, and a cohort of proteins that enable synthesis, trafficking, and targeting of clusters to proteins residing throughout the cell (9). Biogenesis can occur in mitochondria and cytoplasm. Iron-sulfur clusters are essential for electron transfer in the respiratory chain and serve as cofactors for proteins required for DNA metabolism, oxygen sensing, and other critical cellular activities. Additional factors essential for iron-sulfur cluster biogenesis continue to be identified as relevant to human health and disease. For example, mutations in heat shock cognate B, which mediates transfer of iron-sulfur clusters to target proteins, lead to a form of congenital sideroblastic anemia (23). Additionally, impairments in lysosomal acidification lead to impaired cellular iron uptake, iron-sulfur cluster depletion, mitochondrial dysfunction, and impaired cell viability (135, 141). Finally, some defects in iron-sulfur cluster biosynthesis can be attenuated by hypoxia. Severity of disease in a mouse model of Friedreich’s ataxia, caused by mutation in a protein essential for iron-sulfur cluster biogenesis known as frataxin, is attenuated in mice grown under 11% oxygen (6).
Iron not directed to mitochondria or utilized for biological processes within the cytoplasm or other cellular locales is stored in the ferric state in ferritin, which is composed of 24 subunits of ferritin heavy and light chains (79) (Figure 2). Ferritin heavy chains possess ferroxidase activity. Iron is delivered to ferritin by poly(rC)-binding protein (PCBP1). PCBP1 is a multifunctional protein that possesses both RNA- and iron-binding activity, with these activities physically separable yet both essential (95). PCBP1 binds iron and delivers it to ferritin and other iron-dependent proteins. Mice with liver-specific Pcbp1 deficiency develop oxidative stress, lipid peroxidation, and steatosis, indicating that PCBP1 is essential for limiting the toxicity of cytoplasmic iron (104). Although ferritin was discovered years ago, new roles have been discovered for this intriguing molecule. In Drosophila, oligodendrocytes secrete ferritin heavy chain in extracellular vesicles to attenuate iron-mediated oxidative stress (82). Another recent study demonstrated that exosomal excretion of ferritin-rich vesicles involves IRP-dependent regulation of the extracellular vesicle marker CD63 (142).
Mobilization of iron stored in ferritin is achieved by ferritinophagy, an autophagy process mediated by nuclear receptor coactivator 4 (NCOA4) (Figure 2). Recent studies have elaborated upon the essential role for NCOA4 in systemic and tissue-specific iron homeostasis (63, 113). NCOA4 expression is upregulated under conditions of iron deficiency via HIFs and downregulated under conditions of iron excess by proteasomal degradation. Acute Ncoa4 deficiency in mice leads to tissue ferritin and iron accumulation, serum iron deficiency, and anemia. Erythroid compartment-specific Ncoa4 deficiency also leads to anemia, consistent with a role for ferritin iron mobilization in erythropoiesis. Hepatocyte-specific Ncoa4 deficiency leads to impaired mobilization of hepatocyte ferritin iron after blood loss. In cell culture, NCOA4 deficiency leads to impaired mitochondrial function (33).
2.4. Iron Elimination
Dietary iron absorption is a key determinant of body iron levels. In contrast, iron excretion is attributed to passive pathways not regulated by iron levels or other parameters relevant to iron homeostasis (54). These pathways include sloughing of intestinal epithelial cells, exfoliation of skin cells, and physiologic blood loss due to menstruation or minor trauma to epithelial linings. However, evidence of iron excretion in rodent models has been reported recently. Transferrin treatment of a transferrin-deficient mouse model of iron excess normalizes iron levels by gastrointestinal iron excretion (78). Additionally, hepatocyte uptake of NTBI by SLC39A14 is an essential prerequisite for biliary excretion of iron-loaded ferritin, although the significance of biliary iron excretion to iron homeostasis is not clear (103). Finally, a recent study was conducted in anemic children to evaluate the use of stable iron isotopes in long-term assessment of iron absorption and loss during administration of iron supplements (127). Surprisingly, iron losses increased during supplementation. The authors speculatively attributed iron losses to occult gastrointestinal bleeding, although the study was not designed to assess routes of iron loss.
2.5. Maternal-Fetal Iron Homeostasis
Maternal-fetal iron homeostasis is a special area within the broader field of iron homeostasis (59). Over the course of a pregnancy, maternal and fetal needs for iron must both be met to ensure the well-being of the mother and embryo/fetus/baby during gestation and the postnatal period. Sufficient iron must be available to accommodate increased maternal red cell mass, fetal growth, and placental metabolism and to compensate for maternal iron loss during delivery. Recent studies in animal models have expanded our understanding of the mechanisms of maternal-fetal iron homeostasis.
Maternal iron deficiency is a common condition and can have adverse effects on the mother and baby. In mice, maternal iron deficiency induced through dietary means results in aberrant cardiovascular development and embryonic lethality (48). Cardiovascular defects may reflect impaired differentiation and increased retinoic acid signaling in cardiac progenitor cells, as they are attenuated by maternal administration of iron early in gestation or retinoic acid signaling inhibitors in mid-gestation. Maternal iron deficiency can also exacerbate the severity of placental insufficiency in rats administered alcohol, while maternal iron administration attenuates such severity (58). Finally, maternal iron deficiency impacts gene expression in the placenta, an organ essential for gas and nutrient exchange between mother and fetus. Maternal iron deficiency increases TFR1 expression at the maternal face of the placenta and decreases ferroportin expression at the fetal face of the placenta in an IRP1-dependent manner (112).
Hepcidin plays a critical role in maternal-fetal iron homeostasis. Maternal hepcidin expression is suppressed during pregnancy, which ensures increased dietary iron absorption and release of iron stores to ensure adequate iron delivery to the fetus. Maternal hepcidin is a key determinant of iron homeostasis in mouse embryos and placentas (111). In contrast, fetal hepcidin impacts fetal but not placental iron homeostasis. Mouse fetuses expressing a hepcidin-resistant mutant form of ferroportin globally or only within the liver have decreased liver iron and ferroportin levels, although ferroportin levels are unchanged in the placenta (49).
3. HEME
3.1. Dietary Heme Absorption
Heme is a readily bioavailable from of iron in our diet. Meat, poultry, and seafood are rich in heme primarily derived from hemoglobin and myoglobin. The alkaline pH in the lumen of the small intestine makes heme iron more bioavailable by preventing its polymerization. While the mechanism of nonheme iron uptake has been well characterized, the mechanisms governing heme iron absorption from the intestine remain poorly understood (Figure 1). Studies suggest two potential paradigms, receptor-mediated endocytosis and membrane transport (136). Early studies showed the presence of a high-affinity heme-binding protein on the microvillus membrane of the upper small intestine in pigs and humans that was pH-dependent, saturable, and susceptible to trypsin digestion (37). Another study demonstrated that iron deficiency increased intestinal heme uptake as well as binding in rat duodenal enterocytes (55). Localization assays revealed heme accumulation at the surface of microvilli as well as endosomal compartments in the apical cytoplasm (138). These studies did not rule out the possibility of heme transport across the apical plasma membrane via a membrane transporter.
Heme carrier protein 1 (HCP1) (SLC46A1) was initially identified as an intestinal heme importer but later shown to serve as a high-affinity, pH-dependent folate transporter and renamed as proton-coupled folate transporter (105, 121). HCP1 has a Km of 0.8 μM for 5-methyltetrahydrate folate and 125 μM for hemin. Loss-of-function splicing mutations in SLC46A1 cause hereditary folate malabsorption in humans. Folate supplementation resolves symptoms in affected children, who showed no defects in iron metabolism. Hence, the mechanism of intestinal heme transport remains unknown, a question compounded by the difficulty of a suitable genetic model organism as mice are poor absorbers of heme (31). A compelling candidate for intestinal heme absorption is heme responsive gene (HRG) HRG1, a four-transmembrane-domain heme transporter present on erythrophagosomal membranes that imports heme into the cytosol of macrophages during iron recycling from RBCs (137). HRG1 is expressed in the human small intestine, where it could function as a heme importer via endocytic compartments (107).
As mentioned above, the intestinal microbiome contributes to regulation of dietary nonheme absorption. Dietary heme can also impact the microbiome by increasing the population of gram-negative bacteria (42). Whether the microbiome influences dietary heme absorption remains to be determined.
3.2. Heme Synthesis
Heme biosynthesis involves a series of eight highly conserved enzymatic reactions that occur in the mitochondria and cytosol and are shared across all metazoans. Animals, fungi, and α-protobacteria synthesize heme via the Shemin/C4 pathway (119). The first committed step is condensation of succinyl-coenzyme A (succinyl-CoA) and glycine to form 5-aminolevulinate (ALA) by the enzyme ALA synthase (ALAS). This rate-limiting step takes place in the mitochondrial matrix. ALAS has two isoforms, the ubiquitous ALAS1 and the erythroid-specific ALAS2. ALAS1 is negatively regulated by heme at transcriptional, translational, and posttranslational levels, whereas ALAS2 is not affected by heme (150). Heme affects ALAS1 mRNA stability, leading to RNA degradation, and impedes the processing of ALAS1 from its pro to mature form. Heme also inhibits the mitochondrial import of ALAS1 (84). (The regulation of ALAS2 is described in Section 3.4.) Once ALA is made, it crosses the inner and outer mitochondrial membranes into the cytosol by a yet-to-be identified transporter. SLC25A38 and SLC25A39 have been implicated but they are also involved in glycine transport and iron-sulfur cluster biogenesis (38, 69, 87). In the cytosol, two molecules of ALA combine to from porphobilinogen, catalyzed by the zinc-dependent enzyme porphobilinogen synthase or ALA dehydratase. ALA dehydratase was recently shown to contain a Fe4S4 cluster instead of zinc (68). Four prophobilinogen molecules are then assembled into a chemically reactive, linear tetrapyrrole, hydroxymethylbilane, by hydroxymethylbilane synthase or porphobilinogen deaminase. Hydroxymethylbilane is then cyclized to form uroporphyrinogen III by uroporphyrinogen synthase. Uroporphyrinogen decarboxylase then catalyzes decarboxylation of uroporphyrinogen III to form coproporphyrinogen III, which is transported back into the mitochondria. Coproporphyrinogen III is then converted to protoporphyrinogen IX by coproporphyrinogen oxidase. Protoporphyrinogen IX undergoes oxidation by protoporphyrinogen oxidase to form protoporphyrin IX (PPIX). The final step involves insertion of Fe2+ into the protoporphyrin ring to form heme. This reaction is catalyzed by ferrochelatase (FECH) on the inner membrane side of the matrix (119). Iron delivery to FECH occurs via mitoferrin 1 and 2. Proteomic analysis has revealed that FECH is involved in protein-protein interactions with different enzymes of the heme synthesis pathway (ALAS and protoporphyrinogen oxidase), as well as α-ketoglutarate dehydrogenase, succinyl-CoA-synthase, and the porphyrinogen transporter TMEM14C (28, 39). FECH also forms complexes with mitoferrin, ATP-binding cassette (ABC) transporters ABCB7 and ABCB10, and heme chaperones progesterone receptor membrane component 1 and 2 (PGRMC1 and PGRMC2) (71, 99). (ABCB proteins are discussed in Section 3.8.) These findings support the notion that a metabolon for heme synthesis machinery exists to ensure efficient substrate channeling without exposing toxic intermediates to the cellular milieu.
The control of heme homeostasis in an organism begins with regulation of heme synthesis at multiple levels. Defects in heme synthesis can lead to toxic accumulation of intermediates, ultimately culminating in porphyrias (119). Hence it is necessary to ensure efficient shuttling and partitioning of these intermediates between the mitochondria and cytosol. Additionally, heme synthesis is tightly coupled to other metabolic pathways in the mitochondria including oxidative phosphorylation, the tricarboxylic acid cycle, and iron-sulfur cluster biogenesis (32). A recent investigation indicated that flux to the tricarboxylic acid cycle and oxidative phosphorylation is modulated by heme synthesis via ALAS1 and heme export by feline leukemia virus subgroup C receptor-related (FLVCR) protein FLVCR1a, thus impacting ATP distribution between the mitochondria and cytosol. (FLVCR is discussed in Sections 3.7 and 3.8.) Increased heme synthesis dampens the activity of the electron transport chain, which ensures an adequate heme supply. Another functional study showed that ALA dehydratase coordinates with an Fe4S4 cluster essential for enzymatic activity of ALA dehydratase (68). The crosstalk between heme biosynthesis and other metabolic pathways emphasizes the necessity for tight regulation of the former, since any errors in the heme synthesis pathway can have other negative downstream consequences.
3.3. Intracellular Heme Trafficking
After synthesis in mitochondria, heme must cross the inner and outer mitochondrial membranes for incorporation into various hemoproteins residing in cellular compartments such as the cytosol, nucleus, endoplasmic reticulum (ER), Golgi complex, peroxisomes, and plasma membrane (Figure 3). The mechanism of intracellular heme trafficking is poorly understood. Here we discuss this issue, first in the context of known heme-binding proteins.
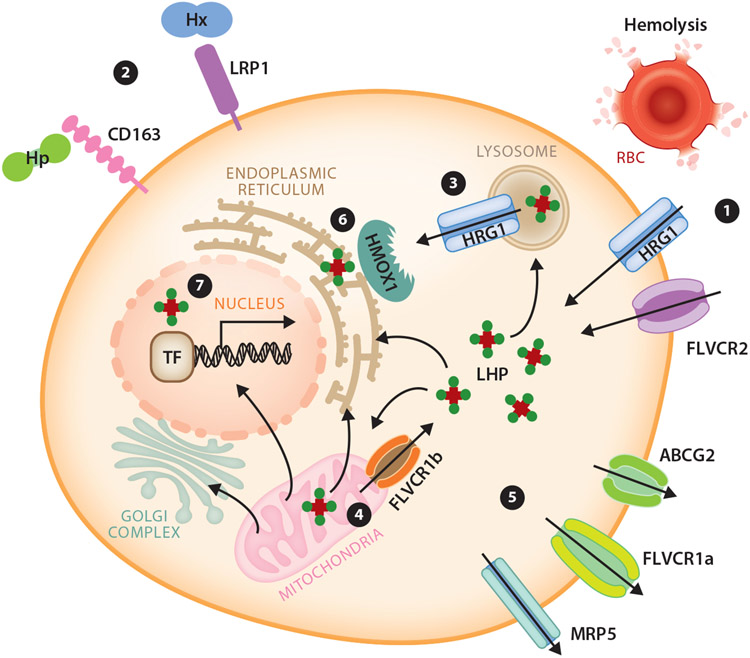
Figure 3.
Model of known metazoan heme transport mediators and pathways for intracellular heme trafficking. A cell can autonomously meet its heme requirements by de novo synthesis in mitochondria. (①) It can also uptake heme through the plasma membrane heme importer HRG1, FLVCR2, or via endocytosis of senescent RBCs. Additionally, (②) Hp and Hx are scavenged by membrane-bound CD163 and LRP1 receptor-mediated endocytosis, respectively. Intracellular heme can be imported into the cytosol via HRG1 on endolysosomal membranes (③) and FLVCR1b on mitochondrial membranes (④). (⑤) Export of heme can be mediated by plasma membrane exporters FLVCR1a, ABCG2, and MRP5. To prevent free heme cytotoxicity, heme is degraded by HMOX1 (⑥. Cellular LHP may facilitate intracellular heme trafficking. (⑦) In the nucleus, TFs bind heme to regulate transcription of downstream target genes. Abbreviations: ABCG2, ATP-binding cassette subfamily G member 2; FLVCR, feline leukemia virus subgroup C receptor; HMOX1, heme oxygenase 1; Hp, haptoglobin; HRG, heme responsive gene; Hx, hemopexin; LHP, labile heme pool; LRP1, LDL receptor related protein 1; MRP, multidrug resistance protein; RBC, red blood cell; TF, transcription factor. Figure adapted from images created using smart.servier.com and motifolio.com.
Heme insertion into the cytosolic hemoproteins nitric oxide synthase and soluble guanylyl cyclase require the glycolytic enzyme glyceraldehyde phosphate dehydrogenase (GAPDH) (24). While the kinetics and key residues involved in heme binding to GAPDH have been elucidated, its mechanism of interaction with target hemoproteins and transfer of heme remains unknown. Glutathione-S-transferases, which conjugate glutathione to electrophilic compounds, also interact with heme and porphyrins (97). p22HBP is a cytosolic heme-binding protein that binds to PPIX while HBP23/PRX1 is a thiol peroxidase that upon binding to heme loses its activity (134). Fatty acid binding protein is also known to bind heme (131). The progesterone receptor membrane component (PGRMC) proteins, PGRMC1, PGRMC2, neudesin, and neuferricin, all contain a heme-binding domain with cyt-b5-like heme/steroid-binding domains. PGRMC1 and PGRMC2 form complexes with FECH, the terminal heme biosynthetic enzyme (77). In vitro heme transfer experiments provide evidence for heme donation from PGRMC1 to apo-cytochrome b5 and further suggest that FECH activity is regulated by PGRMC1 by controlling heme release (100). The ER-based PGRMC2 is predicted to function in heme mobilization to nuclear transcription factors during thermogenesis (34). Cytochromes P450 are tethered to the ER membrane, but their heme-containing globular domain faces the cytosol, from where they probably acquire heme. For most of these cytosolic heme-binding proteins, the cellular functions of their heme-dependent activities and their precise role in heme trafficking and homeostasis are unclear, although broadly their primary function is to prevent the toxic effects of heme by sequestering or transferring heme. Finally, the cytosol constitutes the largest labile heme pool ranging from 20 to 340 nM, while the nucleus and mitochondria have less than 1 nM (40, 147). A recent study illustrated that glutathione binds free heme in the cytosol in a 1:1 ratio and the heme is converted to hematin (89). In this manner, glutathione prevents heme-induced oxidative damage and cytotoxicity.
Several transcription factors are known to bind heme. The nuclear heme-binding proteins include transcription factors HAP1p, BACH1, and REV-ERB-α and the microRNA processing protein DGCR8 (28). When bound to heme, REV-ERB-α functions as a transcriptional repressor impacting its stability and gas-sensing activity. REV-ERB-α contains Fe3+-heme (115). Tight binding of the gases carbon monoxide or nitric oxide is coupled to heme reduction to form Fe2+-heme, thus providing evidence for ligand-driven redox coupling that can modulate the heme- and gas-regulated nuclear receptor to control a multitude of biological processes.
Heme is also required in the secretory pathway. Hemoproteins synthesized, processed, and folded in the ER and Golgi complex need to be targeted to the plasma membrane, lysosomes, or peroxisomes. ER-based hemoproteins include prostaglandin synthases COX1 and COX2, myeloperoxidase in lysosome-like azurophil granules, catalase in peroxisomes, eosinophil peroxidase in eosinophil granules, and thyroperoxidase and ferric reductase on the plasma membrane. Secreted hemoproteins include lactoperoxidase, peroxidasin, fungal ligninase and chloroperoxidase, and Caenorhabditis elegans HRG-3 (14, 55). The mechanism underlying heme trafficking through the secretory pathway and other organelles is unresolved. Inter-organelle membrane contact sites (MCSs) are physical contacts formed between two membrane-bound organelles held together by tethering forces, characterized by a well-defined lipidome and proteome while serving a specific biological function such as molecular transport or signaling (118). The majority of MCSs mediate exchange between the ER and other structures including mitochondria, plasma membrane, Golgi complex, endosomes, peroxisomes, and lipid droplets. Mitochondria can also form MCSs with the ER, lysosomes, lipid droplets, and peroxisomes. Since heme synthesis occurs in the mitochondria, the membranes of which are tethered to the ER, we have proposed that heme is transferred via the ER-mitochondrial network within the cell (119). Studies of ER-mitochondrial contact sites [also known as ER-mitochondria encounter structures (ERMESs)] in Saccharomyces cerevisiae using genetically encoded fluorescent heme sensors revealed that heme is trafficked to the nucleus via the ER-MCS and also identified GTPases that regulate nuclear heme trafficking (73). Heme mobilization from mitochondria to various organelles occurs in parallel, not sequentially. Another recent study described the role of the mitochondrial contact site and cristae organization system (MICOS) in yeast (27). Yeast ferrochelatase forms an oligomeric complex with a MICOS component, Mic60. Ferrochelatase activity is impaired in the absence of Mic60 and causes toxic accumulation of tetrapyrrole precursors, leading to oxidative stress. This stress is mitigated in Mic60-deficient cells by synthetic reconstruction of intermembrane connectivity. These studies establish the potential role of yeast ERMESs in heme synthesis and mobilization to different cellular compartments. In mammals, the ER-mitochondrial membrane sites are less defined and are called mitochondria-associated membranes (MAMs).
Originally identified in C. elegans, the intestinal heme exporter MRP-5 modulates heme levels in the secretory pathway (96). Worms harboring deletions in mrp-5 show a heme-dependent lethality since extraintestinal tissues are heme deprived. In contrast, mice lacking Mrp5 do not show any overt heme-related phenotype, although mice lacking both Mrp5 and a related homolog Mrp9 show male reproductive defects, mitochondrial dysfunction, and aberrant levels of heme precursor succinyl-CoA (12). Notably, both Mrp5 and Mrp9 localize to MAMs in testis. The potential role of MRP5 and MRP9 in heme transport in MAMs has yet to be elucidated.
Mitochondria-derived vesicles (MDVs) are another potential mechanism of heme trafficking from mitochondria to other organelles (13, 55). MDVs are single- or double-membrane-bound vesicles 70–100 nm in diameter derived from mitochondria that contain mitochondria-anchored protein ligase. MDVs traffic in a cargo-specific manner and are targeted to lysosomes and peroxisomes (86, 126). They have been implicated in peroxisomal biogenesis (52). Packaging of heme within MDVs would enable intracellular heme trafficking while protecting the cell from toxicity of free heme.
3.4. Heme in Red Blood Cell Biology
Erythropoiesis generates 2.5 billion erythrocytes per second in humans (81). Hematopoietic stem cells in bone marrow or fetal liver form burst-forming unit erythroid progenitors, colony-forming unit erythroid progenitors, and proerythroblasts, then basophilic, polychromatic, and orthochromatic erythroblasts. The latter cell type then undergoes enucleation and loss of other organelles to form reticulocytes, which enter the circulation to become mature RBCs.
The proerythroblast-erythroblast transition occurs in a specialized extravascular niche called the erythroblastic island, which consists of a central nurse macrophage surrounded by a ring of RBC precursors. This phase is characterized by increased iron requirements for heme and hemoglobin synthesis for the developing RBCs. However, the molecular mechanisms by which iron and heme regulate erythropoiesis is unclear. The erythroid lineage is characterized by abundant expression of a heme-sensitive protein termed heme-regulated inhibitor (HRI)/heme-regulated eIF2α kinase (15). HRI balances globin mRNA translation with heme availability for hemoglobin production. Heme deficiency activates HRI, which phosphorylates eIF2α, thereby inhibiting translation of globin mRNAs. This hinders proteotoxicity by preventing precipitation of globins.
Enucleation and mitochondria loss are executed by the same transcription factor FOXO (65). During murine erythroblast maturation, mitochondria aggregate near the nucleus, which is crucial for enucleation to occur. Pyruvate is the primary mitochondrial metabolite driving this enucleation process. RBCs retain mitochondria even after enucleation so that heme synthesis and other mitochondrial metabolic pathways continue until terminal stages of erythroblast maturation. Recent studies have also implicated the selenoprotein glutathione peroxidase 4 (GPX4) in enucleation and red cell maturation. Independent of its role in ferroptosis, GPX4 mediates lipid raft organization during enucleation (90). Mice with hematopoetic Gpx4 deficiency exhibit perturbed reticulocyte maturation (2).
Given the pivotal role of heme in hemoglobin production, RBCs possess unique strategies to control heme synthesis. ALAS2 is regulated at translational and posttranslational levels to modulate heme synthesis. In the absence of iron, apo-IRP1 inhibits ALAS2 translation by binding to an IRE in the 5′ UTR of ALAS2 mRNA. However, when iron levels are adequate, holo-IRP1, bound to an iron-sulfur cluster, functions as a cytoplasmic aconitase that catalyzes the interconversion of citrate to isocitrate (46). Posttranslationally, ALAS2 turnover is regulated by the mitochondrial protease CLPXP, which is composed of an unfoldase CLPX and a protease CLPP. In erythroid cells, CLPX controls ALAS2 turnover; CLPX also regulates levels of terminal heme synthesis enzymes protoporphyrinogen oxidase and FECH as well as mitochondrial iron metabolism (110).
Erythroid cells also possess a PPIX importer in the inner mitochondrial membrane, called TMEM14C, which mediates PPIX entry into the mitochondrial matrix for terminal heme synthesis (144). TMEM14C plays a crucial role in primitive and definitive erythropoiesis. Functional genomic screens identified the vacuolar-type H+ ATPase CCDC115 as a regulator of heme and iron trafficking in erythroid cells (124). K562 cells deficient in CCDC115 show impaired transferrin-bound-iron uptake and heme trafficking, although the mechanisms for how CCDC115 impacts erythroid differentiation are unclear.
3.5. Systemic Heme Recycling, Transport, Sequestration, Degradation, and Elimination
The majority of the iron requirement for erythropoiesis is fulfilled by iron recycling of senescent RBCs by splenic red pulp macrophages and liver Kupffer cells (Figure 1). This process is termed erythrophagocytosis, in which senescent RBCs are engulfed by macrophages and billions of molecules of heme and hemoglobin are degraded (102). During erythrophagocytosis, the heme importer HRG1 on phagolysosomal membranes mediates heme import into the cytosol (137). Heme is then catabolized by heme oxygenase (HO) tethered to the ER membrane with the active site facing the cytosol. [Heme oxygenases 1 (HO-1) and 2 (HO-2), respectively encoded by HMOX1 and HMOX2, are discussed in more detail below.] The iron liberated from degraded heme is either incorporated into ferritin or exported out of the cell via ferroportin for de novo erythropoiesis. HRG1, heme oxygenase, and ferroportin are all transcriptionally upregulated during erythrophagocytosis (51). The mechanism for heme import via HRG1 for degradation by heme oxygenase is still unknown. Hmox1-deficient mice are embryonic lethal whereas Hrg1-deficient mice survive but accumulate large amounts of heme (96). Mice deficient in Hmox1 and Hrg1 are not viable, but heterozygosity for an Hrg1 null allele in Hmox1-deficient mice results in 40% lethality. This indicates that Hmox1 and Hrg1 interact genetically, but further investigation needs to be done to examine if they interact physically.
The mechanosensitive cation channel PIEZO1 was recently implicated in macrophage iron biology (70). Mice expressing a gain-of-function Piezo1 mutation associated with the blood disorder hereditary xerocytosis develop late-onset iron overload. Mice carrying this mutation only in macrophages developed a similar phenotype. Mutant mice exhibited increased RBC turnover, enhanced erythropoiesis, increased erythroferrone expression, and decreased hepcidin levels. Intriguingly, a mild gain-of-function PIEZO1 allele known as E756del is present in one-third of individuals of African descent and is associated with increased plasma iron (70). An additional study demonstrated that expression of gain-of-function PIEZO1 mutants in hepatocyte cell lines led to increased calcium influx and ERK pathway signal transduction activity leading to inhibition of BMP-SMAD signaling and hepcidin expression (3).
Vascular hemolysis releases free heme and hemoglobin into the circulation that are cleared by acute phase sequestration proteins hemopexin and haptoglobin, respectively (123) (Figure 3). The hemopexin-heme complex is endocytosed via membrane-bound CD91/LRP1 receptors expressed in hepatocytes, macrophages, and neurons. The haptoglobin-hemoglobin complex is scavenged by CD163 receptor-mediated endocytosis in macrophages. While hemopexin is recycled after heme delivery, the haptoglobin-hemoglobin complex is degraded (17). Heme can also bind to albumin, the most abundant serum protein, and the heme-albumin complex can be taken up via transferrin receptor-mediated endocytosis (43). All these heme-scavenging proteins alleviate heme-induced toxicity and lipid oxidation. Heme import into cells also activates heme oxygenase 1, which further combats cytotoxicity and oxidative damage. Hemopexin has been widely studied in the pathophysiology of hemolytic disorders such as sickle cell anemia (5). Both hemopexin and haptoglobin have neuroprotective functions (131) that help counter ischemia and intracerebral hemorrhage (148).
Low-density lipoproteins (LDL) and high-density lipoproteins (HDL), lipocalins, alpha-1-microglobulin, and alpha-1-antitryptin are additional heme-binding proteins (13, 55). Immediately after hemolysis, when heme appears in the plasma, more than 80% of heme binds to LDL and HDL, while the remaining 20% is postulated to bind albumin and hemopexin (17). Albumin and hemopexin gradually clear the heme from LDL and HDL. Albumin and hemopexin bind heme with Kd values of 10−8 and 10−9 M, respectively. LDL and HDL bind heme with Kd values of 10−11 and 10−10 M (4). The role of transient heme binding by LDL and HDL is not known, but initial and rapid binding of heme to LDL and HDL may represent a buffering period so that hemopexin and albumin can steadily bind heme (1, 26).
Once imported into liver, heme undergoes degradation by heme oxygenases HO-1 and HO-2 (Figure 3). HO-1 is an inducible isozyme induced by cellular stress, while HO-2 is constitutively expressed (19). Heme oxygenase cleaves Fe3+-PPIX to form biliverdin, water, carbon monoxide, and iron. Iron is stored within ferritin. Biliverdin is converted to bilirubin by biliverdin reductase using NADPH as a cofactor. Carbon monoxide modulates signaling pathways and binds iron within heme pockets of hemoproteins. Heme oxygenase is expressed primarily in the macrophages of the reticuloendothelial system (RES) (spleen, liver, and bone marrow), but is induced under conditions of stress in many if not all cells (75). The HMOX1 promoter has enhancer sequences that can bind many transcription factors responsible for tissue redox homeostasis including activator protein-1, nuclear factor kappa B, HIFs, and NRF2 (76). HMOX1 transcription is negatively regulated by cellular heme levels by BACH1, a heme-binding protein. Although the structure, gene regulation, and importance of HOs have been extensively studied, both in normal and diseased conditions, there are gaps in knowledge about how heme oxygenases get their heme and which proteins donate heme to heme oxygenases for degradation.
Heme oxygenases have been implicated to play a protective role in a broad range of human diseases including cardiovascular, neurodegenerative, neoplastic, metabolic, and inflammatory diseases (140). Diseases involving severe hemolysis such as sickle cell disease release large amounts of heme and have low levels of circulating hemopexin (5). Alpha-1-microglobulin acts as a secondary scavenger by directing heme to the kidney, which is the main site for hemoglobin and myoglobin clearance and degradation during such conditions (7). Hemoglobin binds to renal brush border epithelial membranes, specifically to the endocytic receptors megalin and cubulin, residing in the renal proximal tubule. Studies using knockout mice indicate that megalin mediates reabsorption of hemoglobin under physiological conditions, whereas cubulin is involved during hemoglobinuria (36). Hemoglobin is nephrotoxic due to its direct toxicity to proximal tubular epithelium, hemoglobin precipitation impairing distal tubule function, and vasoconstriction caused by nitric oxide scavenging. Furthermore, proximal tubules are rich in mitochondria that upon injury release their cytochrome heme content, thereby inducing more damage. Hemolytic stress and rhabdomyolysis cause acute kidney injury, which accounts for up to 15% of mortality associated with sickle cell disease. Together, these findings emphasize the importance of kidney in heme metabolism, but the detailed mechanism and its exact role in mammalian heme homeostasis are still unclear.
3.6. Heme Tolerance and Toxicity
In the absence of a heme degradation system, hemoparasites such as Plasmodium sequester heme in the form of a chemically stable crystal called hemozoin. Hemozoin is formed with the help of histidine-rich proteins in an acidic environment (20). Lipids can also contribute to hemozoin formation by forming hydrogen bonds between propionate side chains of heme. A parasite-derived lipocalin-like protein, PV5, has been implicated in hemozoin formation (74). While the structure of hemozoin is known, the molecular mechanisms for hemozoin formation in vivo are still elusive. Mice lacking the heme importer Hrg1 accumulate hemozoin in enlarged lysosomes within RES macrophages, thus conferring heme tolerance (96). It is not clear if hemozoin heme is a bioavailable form of heme iron that can be used during iron deficiency. If so, pharmacologic targeting of hemozoin formation may represent a novel treatment for hemolytic anemias. Furthermore, Hrg1-deficient mice have enlarged lysosomes reminiscent of lysosomal storage disorders, raising the possibility that lysosomal storage disorders may instigate hemozoin formation.
3.7. Heme Import
The concept of heme importers and exporters is controversial for metazoans (101). All cells can synthesize their own heme and have a heme degradation system to maintain intracellular heme homeostasis. Then why do cells need to import or export heme if they can achieve this autonomously? Even though RBCs are the major iron consumer for hemoglobin synthesis, they still export iron (148). It is reasonable to assume that cells may import or export heme, in spite of their ability to synthesize heme, for regulation of heme homeostasis. Since heme synthesis and heme transport are tightly coupled, it has been challenging to track heme trafficking and transport, without confounding factors from heme synthesis.
Hydrophobic and cytotoxic, free heme cannot freely diffuse within cells and across cell membranes (17). C. elegans is an excellent animal model to identify heme importers as it is a heme auxotroph and completely reliant on environmental heme for survival (122). Genome-wide screens in C. elegans have identified more than 200 HRGs (120). Intestinal heme uptake in worms is mediated by HRG-1 and its paralogs HRG-4, −5, and −6 (Figure 3). HRG-4 is present in the apical membrane and is involved in direct heme uptake from the intestinal lumen. HRG-1 resides in endolysosomal compartments where it imports heme into the cytosol (146). Interestingly, mammals only have HRG1 as a heme importer, thus indicating a redundancy in the heme uptake system in C. elegans as they cannot synthesize their own heme. HRG1 interacts with the c subunit of vacuolar proton ATPase pump and binds to heme in a pH-dependent manner (88, 107). Genetic, biochemical, and functional assays in yeast, worms, Xenopus oocytes, zebrafish, and murine erythroleukemia cells have established the essential role of HRG1 in iron and heme homeostasis at the organismal level. HRG1 has been identified as a target of the transcriptional repressor BACH1, a basic leucine zipper transcription factor that can bind heme and derepress transcription (91, 133). Mechanistic studies have also characterized the key amino acid residues positioned in the exoplasmic, cytoplasmic, and transmembrane regions of HRG1 that facilitate heme translocation across the membrane (146). HRG1 is expressed in RES macrophages and is upregulated in the presence of heme and during erythrophagocytosis. A P36L variant in HRG1 found in four human subjects shows inability to transport heme (137). HRG1 aids in the process of iron recycling by importing heme into the cytosol for subsequent degradation by heme oxygenase. The mechanism by which heme binds to HRG1 is not known. Hrg1-deficient mice showed the first evidence of mammalian heme tolerance, since they accumulated hemozoin crystals resulting in enlarged lysosomes (96). All the RES organs in these mice were dark in color due to hyperaccumulation of heme. Additionally, HRG1 is highly expressed in non-RES tissues including brain, intestine, lung, kidney, and smooth muscle, but its function in these organs is yet to be identified (13).
The FLVCR protein FLVCR2 (MFSD7C) was postulated as a cell surface heme importer (29) (Figure 3). This protein has been associated with Fowler syndrome in humans, characterized by proliferative vasculopathy in the brain (47, 114). FLVCR2 binds to hemin-conjugated agarose, and FLVCR2 silencing using small-interfering RNA inhibits heme import. Furthermore, cells expressing FLVCR2 show increased heme uptake and are more sensitive to heme toxicity. However, recent studies localized FLVCR2 to mitochondria, implying a role for this protein in thermogenesis in response to heme (64). Importantly, FLVCR2 expression in yeast does not restore growth of heme-deficient yeast strains (146). Thus, further investigation needs to be done to ascertain FLVCR2 localization and heme transport function.
3.8. Heme Export
Like heme import, heme export is a controversial topic. But with the discovery of heme exporters there is growing awareness that a network of heme transporters could enable tissues and organs to share heme to maintain systemic heme homeostasis.
FLVCR1 was identified as a plasma membrane heme exporter (Figure 3). Cats develop pure red cell aplasia due to arrested erythropoiesis when infected with feline leukemia virus subgroup C (50). Heme transport assays using a fluorescent heme analog and 55Fe-hemin validated the heme export function of FLVCR1 in a rat renal epithelial cell line and K562 cells (106). Flvcr1-deficient mice have defective erythropoiesis and die at midgestation. FLVCR1 mediates heme export from macrophages that engulf senescent RBCs and hence aid in heme-iron recycling. Mutant mice also show craniofacial and limb deformities, which were later attributed to an essential role for Flvcr in sensory neuron maintenance and T cell development (98). An isoform of FLVCR1, termed FLVCR1b, localizes to mitochondria and effluxes mitochondrial heme. Loss of FLVCR1b results in heme excess in mitochondria and termination of erythroid differentiation, while overexpression increases cytosolic heme levels. FLVCR1b is postulated to regulate erythropoiesis by mediating mitochondrial heme export, while FLVCR1a prevents hemorrhages and edema (16). FLVCR also interacts directly with hemopexin, which facilitates heme export (143). However, there is a significant gap in our knowledge about the mechanism of heme export by FLVCR1a and FLVCR1b. For example, it is unclear whether FLVCR1b is located on the inner or outer mitochondrial membrane (129).
MRP-5/ABCC5 was identified as a heme exporter in C. elegans (Figure 3). MRP-5 localizes to the basolateral membranes of worm intestine and its deficiency leads to embryonic lethality. MRP5 belongs to a family of multidrug resistant proteins within the ABC transporter superfamily that have been extensively implicated in their role in cancer drug resistance. Knockdown of mrp5 in zebrafish causes severe anemia. MRP5 is present on the plasma membrane, Golgi complex, and recycling endosomes, thus supporting its role in heme export and for heme delivery to hemoproteins in the secretory pathway (56). MRP9, a close homolog of MRP5 in mammals, has also been implicated to play a compensatory role in heme homeostasis (12).
Breast cancer resistance protein (ABCG2) has been proposed as a cell surface heme exporter (45) (Figure 3). Mice lacking Abcg2 show extreme photosensitivity attributed to accumulation of pheophorbide, which is structurally similar to PPIX (119). While ABCG2 has broad substrate specificity including various drugs and xenobiotics, its true substrate is unknown. Structural studies indicate that it can efflux heme, PPIX, and other porphyrins (13). Cryoelectron microscopy of human ABCG2 has delineated the structural dynamics and conformational changes involved in the transport cycle (72). In-depth proteomic analysis has revealed eight ABC transporters on the RBC membrane, of which several are implicated in porphyrin efflux (10). ABCB6, −7, −8, and −10 are all located in mitochondria (53). ABCB6 has been associated with the outer mitochondrial membrane and was initially shown to facilitate transport of coproporphyrinogen III (57). This has been challenged since ABCB6 localizes to the plasma membrane of mature erythrocytes, endolysosomal compartments, and exosomes secreted from reticulocytes. Cryoelectron microscopy of human ABCB6 has recently delineated the structural mechanism of ABCB6 for porphyrin transport, although the specific substrate for this transporter is debatable since it has been implicated in cadmium detoxification (108, 125). Human ABCB7, a homology of yeast Atm1p, can restore the phenotypic defects of an Atm1-deficient yeast mutant, including restoration of normal cytochrome and iron levels (66). Defects in ABCB7 are associated with hereditary X-linked sideroblastic anemia and cerebral ataxia due to mitochondrial iron accumulation. ABCB7 has been suggested to positively regulate heme biosynthesis via its interaction with FECH. ABCB10 has also been demonstrated to interact with FECH as well as the mitochondrial iron importer mitoferrin (71, 99, 129).
4. CONCLUDING REMARKS
Both iron and heme are essential for health but potentially toxic when present in excess. As such, body iron and heme levels are tightly regulated to maintain these factors at sufficient but nontoxic levels. Our understanding of the molecular basis of mammalian iron homeostasis exceeds that of heme homeostasis. However, recent years have seen advances in our understanding of molecular determinants of heme transport. Nevertheless, significant challenges and questions still remain. For example, why do RBCs require such an efficient porphyrin clearance system? Do they function in different stages of red cell differentiation to ensure a precise control of porphyrin/heme efflux? What is the mechanism of dietary heme absorption? How is heme delivered to heme oxygenases for degradation? How is hemozoin formed in vivo? Given the progress made in our understanding of both iron and heme metabolism, we are confident that these and other questions will be answered in due time. In turn, this will expand our understanding of iron and heme homeostasis and their role in human health and disease.
DCYTB:
duodenal cytochrome reductase 1
DMT1:
divalent metal transporter 1
BMP:
bone morphogenetic protein
TFR1:
transferrin receptor 1
IRP:
iron regulatory protein
NTBI:
nontransferrin-bound iron
HRG:
heme-responsive gene
ALA:
5-aminolevulinate
ALAS:
ALA synthase
PPIX:
Protoporphyrin IX
FECH:
ferrochelatase
ABC:
ATP-binding cassette
PGRMC:
progesterone receptor membrane component
FLVCR:
feline leukemia virus subgroup C receptor-related
MCS:
membrane contact site
ERMES:
ER-mitochondria encounter structure
MAM:
Mitochondria-associated membrane
MDV:
mitochondria-derived vesicle
HO:
heme oxygenase
RES:
reticuloendothelial system
ACKNOWLEDGMENTS
This work was supported in part by funding from the National Institutes of Health (DK85035, DK125740, and DK074797 to I.H.; DK110049 to T.B.B.).
Footnotes
DISCLOSURE STATEMENT
I.H. is the President and Founder of Rakta Therapeutics Inc. (College Park, Maryland), a company involved in the development of heme transporter-related diagnostics. He declares no other competing financial interests. S.D. and T.B.B. declare no other competing interests.
LITERATURE CITED
- 1.Adams PA, Berman MC. 1980. Kinetics and mechanism of the interaction between human serum albumin and monomeric haemin. Biochem. J 191(1):95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altamura S, Vegi NM, Hoppe PS, Schroeder T, Aichler M, et al. 2020. Glutathione peroxidase 4 and vitamin E control reticulocyte maturation, stress erythropoiesis and iron homeostasis. Haematologica 105(4):937–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andolfo I, Rosato BE, Manna F, Rosa GD, Marra R, et al. 2020. Gain-of-function mutations in PIEZO1 directly impair hepatic iron metabolism via the inhibition of the BMP/SMADs pathway. Am. J. Hematol 95(2):188–97 [DOI] [PubMed] [Google Scholar]
- 4.Ascenzi P, Bocedi A, Visca P, Altruda F, Tolosano E, et al. 2005. Hemoglobin and heme scavenging. IUBMB Life 57(11):749–59 [DOI] [PubMed] [Google Scholar]
- 5.Ashouri R, Fangman M, Burris A, Ezenwa MO, Wilkie DJ, Doré S. 2021. Critical role of hemopexin mediated cytoprotection in the pathophysiology of sickle cell disease. Int. J. Mol. Sci 22(12):6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ast T, Meisel JD, Patra S, Wang H, Grange RMH, et al. 2019. Hypoxia rescues frataxin loss by restoring iron sulfur cluster biogenesis. Cell 177(6):1507–21.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balla J, Zarjou A. 2021. Heme burden and ensuing mechanisms that protect the kidney: insights from bench and bedside. Int. J. Mol. Sci 22(15):8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billesbølle CB, Azumaya CM, Kretsch RC, Powers AS, Gonen S, et al. 2020. Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms. Nature 586(7831):807–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braymer JJ, Freibert SA, Rakwalska-Bange M, Lill R. 2021. Mechanistic concepts of iron-sulfur protein biogenesis in Biology. Biochim. Biophys. Acta Mol. Cell Res 1868(1):118863. [DOI] [PubMed] [Google Scholar]
- 10.Bryk AH, Wiśniewski JR. 2017. Quantitative analysis of human red blood cell proteome. J. Proteome Res 16(8):2752–61 [DOI] [PubMed] [Google Scholar]
- 11.Castro-Mollo M, Gera S, Ruiz-Martinez M, Feola M, Gumerova A, et al. 2021. The hepcidin regulator erythroferrone is a new member of the erythropoiesis-iron-bone circuitry. eLife 10:e68217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers I, Kumar P, Lichtenberg J, Wang P, Yu J, et al. 2021. MRP5 and MRP9 play a concerted role in male reproduction and mitochondrial function. PNAS 119(6):e2111617119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers IG, Willoughby MM, Hamza I, Reddi AR. 2021. One ring to bring them all and in the darkness bind them: the trafficking of heme without deliverers. Biochim. Biophys. Acta Mol. Cell Res 1868(1):118881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C, Samuel TK, Sinclair J, Dailey HA, Hamza I. 2011. An intercellular heme-trafficking protein delivers maternal heme to the embryo during development in C. elegans. Cell 145(5):720–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J-J, Zhang S 2019. Heme-regulated eIF2α kinase in erythropoiesis and hemoglobinopathies. Blood 134(20):1697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiabrando D, Marro S, Mercurio S, Giorgi C, Petrillo S, et al. 2012. The mitochondrial heme exporter FLVCR1b mediates erythroid differentiation. J. Clin. Investig 122(12):4569–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiabrando D, Vinchi F, Fiorito V, Mercurio S, Tolosano E. 2014. Heme in pathophysiology: a matter of scavenging, metabolism and trafficking across cell membranes. Front. Pharmacol 5:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colucci S, Marques O, Altamura S. 2021. 20 years of Hepcidin: How far we have come. Semin. Hematol 58(3):132–44 [DOI] [PubMed] [Google Scholar]
- 19.Consoli V, Sorrenti V, Grosso S, Vanella L. 2021. Heme oxygenase-1 signaling and redox homeostasis in physiopathological conditions. Biomolecules 11(4):589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coronado LM, Nadovich CT, Spadafora C. 2014. Malarial hemozoin: from target to tool. Biochim. Biophys. Acta Gen. Subj 1840(6):2032–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corradini E, Buzzetti E, Pietrangelo A. 2020. Genetic iron overload disorders. Mol. Aspects Med 75:100896. [DOI] [PubMed] [Google Scholar]
- 22.Corral VM, Schultz ER, Eisenstein RS, Connell GJ. 2021. Roquin is a major mediator of iron-regulated changes to transferrin receptor-1 mRNA stability. iScience 24(4):102360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crispin A, Guo C, Chen C, Campagna DR, Schmidt PJ, et al. 2020. Mutations in the iron-sulfur cluster biogenesis protein HSCB cause congenital sideroblastic anemia. J. Clin. Investig 130(10):5245–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai Y, Sweeny EA, Schlanger S, Ghosh A, Stuehr DJ. 2020. GAPDH delivers heme to soluble guanylyl cyclase. J. Biol. Chem 295(24):8145–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das NK, Schwartz AJ, Barthel G, Inohara N, Liu Q, et al. 2020. Microbial metabolite signaling is required for systemic iron homeostasis. Cell Metab. 31(1):115–30.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delanghe JR, Langlois MR. 2001. Hemopexin: a review of biological aspects and the role in laboratory medicine. Clin. Chim. Acta 312(1–2):13–23 [DOI] [PubMed] [Google Scholar]
- 27.Dietz JV, Willoughby MM, Piel RB, Ross TA, Bohovych I, et al. 2021. Mitochondrial contact site and cristae organizing system (MICOS) machinery supports heme biosynthesis by enabling optimal performance of ferrochelatase. Redox Biol. 46:102125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donegan RK, Moore CM, Hanna DA, Reddi AR. 2019. Handling heme: the mechanisms underlying the movement of heme within and between cells. Free Radic. Biol. Med 133:88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duffy SP, Shing J, Saraon P, Berger LC, Eiden MV, et al. 2010. The Fowler syndrome-associated protein FLVCR2 is an importer of heme. Mol. Cell. Biol 30(22):5318–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enns CA, Jue S, Zhang A-S. 2021. Hepatocyte neogenin is required for hemojuvelin-mediated hepcidin expression and iron homeostasis in mice. Blood 138(6):486–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fillebeen C, Gkouvatsos K, Fragoso G, Calvé A, Garcia-Santos D, et al. 2015. Mice are poor heme absorbers and do not require intestinal Hmox1 for dietary heme iron assimilation. Haematologica 100(9):e334–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiorito V, Allocco AL, Petrillo S, Gazzano E, Torretta S, et al. 2021. The heme synthesis-export system regulates the tricarboxylic acid cycle flux and oxidative phosphorylation. Cell Rep. 35(11):109252. [DOI] [PubMed] [Google Scholar]
- 33.Fujimaki M, Furuya N, Saild S, Amo T, Imamichi Y, Hattori N. 2019. Iron supply via NCOA4-mediated ferritin degradation maintains mitochondrial functions. Mol. Cell. Biol 39(14):e00010–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galmozzi A, Kok BP, Kim AS, Montenegro-Burke JR, Lee JY, et al. 2019. PGRMC2 is an intracellular haem chaperone critical for adipocyte function. Nature 576(7785):138–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao J, Zhou Q, Wu D, Chen L. 2021. Mitochondrial iron metabolism and its role in diseases. Clin. Chim. Acta 513:6–12 [DOI] [PubMed] [Google Scholar]
- 36.Gburek J, Verroust PJ, Willnow TE, Fyfe JC, Nowacki W, et al. 2002. Megalin and cubilin are endocytic receptors involved in renal clearance of hemoglobin. J. Am. Soc. Nephrol 13(2):423–30 [DOI] [PubMed] [Google Scholar]
- 37.Gräsbeck R, Kouvonen I, Lundberg M, Tenhunen R. 1979. An intestinal receptor for heme. Scand. J. Haematol 23(1):5–9 [DOI] [PubMed] [Google Scholar]
- 38.Guernsey DL, Jiang H, Campagna DR, Evans SC, Ferguson M, et al. 2009. Mutations in mitochondrial carrier family gene SLC25A38 cause nonsyndromic autosomal recessive congenital sideroblastic anemia. Nat. Genet 41(6):651–53 [DOI] [PubMed] [Google Scholar]
- 39.Hamza I, Dailey HA. 2012. One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim. Biophys. Acta Mol. Cell Res 1823(9):1617–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanna DA, Martinez-Guzman O, Reddi AR. 2017. Heme gazing: illuminating eukaryotic heme trafficking, dynamics, and signaling with fluorescent heme sensors. Biochemistry 56(13):1815–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huynh N, Ou Q, Cox P, Lill R, King-Jones K. 2019. Glycogen branching enzyme controls cellular iron homeostasis via Iron Regulatory Protein 1 and mitoNEET. Nat. Commun 10:5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ijssennagger N, Belzer C, Hooiveld GJ, Dekker J, van Mil SWC, et al. 2015. Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus barrier in colon. PNAS 112(32):10038–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jennifer B, Berg V, Modak M, Puck A, Seyerl-Jiresch M, et al. 2020. Transferrin receptor 1 is a cellular receptor for human heme-albumin. Commun. Biol 3(1):621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang L, Wang J, Wang K, Wang H, Wu Q, et al. 2021. RNF217 regulates iron homeostasis through its E3 ubiquitin ligase activity by modulating ferroportin degradation. Blood 138(8):689–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jonker JW, Buitelaar M, Wagenaar E, van der Valk MA, Scheffer GL, et al. 2002. The breast cancer resistance protein protects against a major chlorophyll-derived dietary phototoxin and protoporphyria. PNAS 99(24):15649–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kafina MD, Paw BH. 2017. Intracellular iron and heme trafficking and metabolism in developing erythroblasts. Metallomics 9(9):1193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalailingam P, Wang KQ, Toh XR, Nguyen TQ, Chandrakanthan M, et al. 2020. Deficiency of MFSD7c results in microcephaly-associated vasculopathy in Fowler syndrome. J. Clin. Investig 130(8):4081–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalisch-Smith JI, Ved N, Szumska D, Munro J, Troup M, et al. 2021. Maternal iron deficiency perturbs embryonic cardiovascular development in mice. Nat. Commun 12(1):3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kämmerer L, Mohammad G, Wolna M, Robbins PA, Lakhal-Litdeton S. 2020. Fetal liver hepeidin secures iron stores in utero. Blood 136(13): 1549–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan AA, Quigley JG. 2013. Heme and FLVCR-related transporter families SLC48 and SLC49. Mol. Aspects Med 34(2–3):669–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kikuchi G, Yoshida T, Noguchi M. 2005. Heme oxygenase and heme degradation. Biochem. Biophys. Res. Commun 338(1):558–67 [DOI] [PubMed] [Google Scholar]
- 52.Kim P 2017. Peroxisome biogenesis: a union between two organelles. Curr. Biol 27(7):R271–74 [DOI] [PubMed] [Google Scholar]
- 53.Kiss K, Brozik A, Kucsma N, Toth A, Gera M, et al. 2012. Shifting the paradigm: The putative mitochondrial protein ABCB6 resides in the lysosomes of cells and in the plasma membrane of erythrocytes. PLOS ONE 7(5):e37378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koleini N, Shapiro JS, Geier J, Ardehali H. 2021. Ironing out mechanisms of iron homeostasis and disorders of iron deficiency. J. Clin. Investig 131(11):e148671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korolnek T, Hamza I. 2014. Like iron in the blood of the people: the requirement for heme trafficking in iron metabolism. Front. Pharmacol 5:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korolnek T, Zhang J, Beardsley S, Scheffer GL, Hamza I. 2014. Control of metazoan heme homeostasis by a conserved multidrug resistance protein. Cell Metab. 19(6): 1008–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krishnamurthy PC, Du G, Fukuda Y, Sun D, Sampath J, et al. 2006. Identification of a mammalian mitochondrial porphyrin transporter. Nature 443(7111):586–89 [DOI] [PubMed] [Google Scholar]
- 58.Kwan STC, Kezer CA, Helfrich KK, Saini N, Huebner SM, et al. 2020. Maternal iron nutriture modulates placental development in a rat model of fetal alcohol spectrum disorder. Alcohol 84:57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lakhal-Littleton S 2021. Advances in understanding the crosstalk between mother and fetus on iron utilization. Semin. Hematol 58(3):153–60 [DOI] [PubMed] [Google Scholar]
- 60.Ledesma-Colunga MG, Baschant U, Fiedler IAK, Busse B, Hofbauer LC, et al. 2020. Disruption of the hepcidin/ferroportin regulatory circuitry causes low axial bone mass in mice. Bone 137:115400. [DOI] [PubMed] [Google Scholar]
- 61.Li G, Zhang H, Wu J, Wang A, Yang F, et al. 2020. Hepcidin deficiency causes bone loss through interfering with the canonical Wnt/β-catenin pathway via Forkhead box O3a. J. Orthop. Translat 23:67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J, Pan X, Pan G, Song Z, He Y, et al. 2020. Transferrin receptor 1 regulates thermogenic capacity and cell fate in brown/beige adipocytes. Adv. Sci. (Weinh.) 7(12):1903366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li X, Lozovatsky L, Sukumaran A, Gonzalez L, Jain A, et al. 2020. NCOA4 is regulated by HIF and mediates mobilization of murine hepatic iron stores after blood loss. Blood 136(23):2691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, Ivica NA, Dong T, Papageorgiou DP, He Y, et al. 2020. MFSD7C switches mitochondrial ATP synthesis to thermogenesis in response to heme. Nat. Commun 11(1):4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang R, Menon V, Qiu J, Arif T, Renuse S, et al. 2021. Mitochondrial localization and moderated activity are key to murine erythroid enucleation. Blood Adv. 5(10):2490–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lill R, Kispal G. 2001. Mitochondrial ABC transporters. Res. Microbiol 152(3–4):331–40 [DOI] [PubMed] [Google Scholar]
- 67.Lim PJ, Duarte TL, Arezes J, Garcia-Santos D, Hamdi A, et al. 2019. Nrf2 controls iron homeostasis in haemochromatosis and thalassaemia via Bmp6 and hepcidin. Nat. Metab 1(5):519–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu G, Sil D, Maio N, Tong W-H, Bollinger JM, et al. 2020. Heme biosynthesis depends on previously unrecognized acquisition of iron-sulfur cofactors in human amino-levulinic acid dehydratase. Nat. Commun 11(1):6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lunetti P, Damiano F, De Benedetto G, Siculella L, Pennetta A, et al. 2016. Characterization of human and yeast mitochondrial glycine carriers with implications for heme biosynthesis and anemia. J. Biol. Chem 291(38)19746–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma S, Dubin AE, Zhang Y, Mousavi SAR, Wang Y, et al. 2021. A role of PIEZO1 in iron metabolism in mice and humans. Cell 184(4):969–82.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maio N, Kim KS, Holmes-Hampton G, Singh A, Rouault TA. 2019. Dimeric ferrochelatase bridges ABCB7 and ABCB10 homodimers in an architecturally defined molecular complex required for heme biosynthesis. Haematologica 104(9):1756–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Manolaridis I, Jackson SM,Taylor NMI, Kowal J, Stahlberg H, Locher KP. 2018. Cryo-EM structures of a human ABCG2 mutant trapped in ATP-bound and substrate-bound states. Nature 563(7731):426–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martinez-Guzman O, Willoughby MM, Saini A, Dietz JV, Bohovyeh I, et al. 2020. Mitochondrial-nuclear heme trafficking in budding yeast is regulated by GTPases that control mitochondrial dynamics and ER contact sites. J. Cell Sci 133(10):jcs237917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matz JM, Drepper B, Blum TB, van Genderen E, Burrell A, et al. 2020. A lipocalin mediates unidirectional heme biomineralization in malaria parasites. PNAS 117(28):16546–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McMahon M, Ding S, Acosta-Jimenez LP, Frangova TG, Henderson CJ, Wolf CR. 2018. Measuring in vivo responses to endogenous and exogenous oxidative stress using a novel haem oxygenase 1 reporter mouse. J. Physiol 596(1):105–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Medina MV, Sapochnik D, Garcia Solá M, Coso O. 2020. Regulation of the expression of heme oxygenase-1: signal transduction, gene promoter activation, and beyond. Antioxid Redox Signal. 32(14):1033–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Medlock AE, Shiferaw MT, Marcero JR, Vashisht AA, Wohlschlegel JA, et al. 2015. Identification of the mitochondrial heme metabolism complex. PLOS ONE 10(8):e0135896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mercadante CJ, Prajapati M, Parmar JH, Conboy HL, Dash ME, et al. 2019. Gastrointestinal iron excretion and reversal of iron excess in a mouse model of inherited iron excess. Haematologica 104(4):678–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mleczko-Sanecka K, Silvestri L. 2021. Cell-type-specific insights into iron regulatory processes. Am. J. Hematol 96(1):110–27 [DOI] [PubMed] [Google Scholar]
- 80.Montemiglio LC, Testi C, Ceci P, Falvo E, Pitea M, et al. 2019. Cryo-EM structure of the human ferritin-transferrin receptor 1 complex. Nat. Commun 10(1):1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Muckenthaler MU, Rivella S, Hentze MW, Galy B. 2017. A red carpet for iron metabolism. Cell 168(3):344–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mukherjee C, Kling T, Russo B, Miebach K, Kess E, et al. 2020. Oligodendrocytes provide antioxidant defense function for neurons by secreting ferritin heavy chain. Cell Metab. 32(2):259–72.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Müller S, Sindikubwabo F, Cañeque T, Lafon A, Versini A, et al. 2020. CD44 regulates epigenetic plasticity by mediating iron endocytosis. Nat. Chem 12(10):929–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Munakata H, Sun J-Y, Yoshida K, Nakatani T, Honda E, et al. 2004. Role of the heme regulatory motif in the heme-mediated inhibition of mitochondrial import of 5-aminolevulinate synthase. J. Biochem 136(2):233–38 [DOI] [PubMed] [Google Scholar]
- 85.Murata Y, Yoshida M, Sakamoto N, Morimoto S, Watanabe T, Namba K. 2021. Iron uptake mediated by the plant-derived chelator nicotianamine in the small intestine. J. Biol. Chem 296:100195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Neuspiel M, Schauss AC, Braschi E, Zunino R, Rippstein P, et al. 2008. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr. Biol 18(2):102–8 [DOI] [PubMed] [Google Scholar]
- 87.Nilsson R, Schultz IJ, Pierce EL, Soltis KA, Naranuntarat A, et al. 2009. Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis. Cell Metab. 10(2):119–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O’Callaghan KM, Ayllon V, O’Keeffe J, Wang Y, Cox OT, et al. 2010. Heme-binding protein HRG-1 is induced by insulin-like growth factor I and associates with the vacuolar H+-ATPase to control endosomal pH and receptor trafficking. J. Biol. Chem 285(1):381–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O’Keeffe R, Latunde-Dada GO, Chen Y-L, Kong XL, Cilibrizzi A, Hider RC. 2021. Glutathione and the intracellular labile heme pool. Biometals 34(2):221–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ouled-Haddou H, Messaoudi K, Demont Y, Lopes Dos Santos R, Carola C, et al. 2020. A new role of glutathione peroxidase 4 during human erythroblast enucleation. Blood Adv. 4(22):5666–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, et al. 1996. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol 16(11):6083–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pan Y, Ren Z, Gao S, Shen J, Wang L, et al. 2020. Structural basis of ion transport and inhibition in ferroportin. Nat. Commun 11:5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parrow NL, Li Y, Feola M, Guerra A, Casu C, et al. 2019. Lobe specificity of iron binding to transferrin modulates murine erythropoiesis and iron homeostasis. Blood 134(17):1373–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pasricha S-R, Tye-Din J, Muckenthaler MU, Swinkels DW. 2021. Iron deficiency. Lancet 397(10270):233–48 [DOI] [PubMed] [Google Scholar]
- 95.Patel SJ, Protchenko O, Shakoury-Elizeh M, Baratz E, Jadhav S, Philpott CC. 2021. The iron chaperone and nucleic acid-binding activities of poly(rC)-binding protein 1 are separable and independently essential. PNAS 118(25):e2104666118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pek RH, Yuan X, Rietzschel N, Zhang J, Jackson L, et al. 2019. Hemozoin produced by mammals confers heme tolerance. eLife 8:e49503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Perally S, Lacourse EJ, Campbell AM, Brophy PM. 2008. Heme transport and detoxification in nematodes: subproteomics evidence of differential role of glutathione transferases. J. Proteome Res 7(10):4557–65 [DOI] [PubMed] [Google Scholar]
- 98.Philip M, Funkhouser SA, Chiu EY, Phelps SR, Delrow JJ, et al. 2015. Heme exporter FLVCRis required for T cell development and peripheral survival. J. Immunol 194(4):1677–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Piel RB, Dailey HA, Medlock AE. 2019. The mitochondrial heme metabolon: insights into the complexity) of heme synthesis and distribution. Mol. Genet. Metab 128(3):198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Piel RB, Shiferaw MT, Vashisht AA, Marcero JR, Praissman JL,et al. 2016. A novel role for progesterone receptor membrane component 1 (PGRMC1): a partner and regulator of ferrochelatase. Biochemistry 55(37):5204–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ponka P, Sheftel AD, English AM, Scott Bohle D, Garcia-Santos D. 2017. Do mammalian cells really need to export and import heme? Trends Biochem. Sci 42(5):395–406 [DOI] [PubMed] [Google Scholar]
- 102.Pradhan P, Vijayan V, Gueler F, Immenschuh S. 2020. Interplay of heme with macrophages in homeostasis and inflammation. Int. J. Mol. Sci 21(3):E740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Prajapati M, Conboy HL, Hojyo S, Fukada T, Budnik B, Bartnikas TB. 2021. Biliary excretion of excess iron in mice requires hepatocyte iron import by Slc39a14. J. Biol. Chem 297(1):100835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Protchenko O, Baratz E, Jadhav S, Li F, Shakoury-Elizeh M, et al. 2021. Iron chaperone poly rC binding protein 1 protects mouse liver from lipid peroxidation and steatosis. Hepatology 73(3):1176–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Qiu A, Jansen M, Sakaris A, Min SH, Chattopadhyay S, et al. 2006. Identification of an intestinal folate transporter and the molecular basis for hereditary folate malabsorption. Cell 127(5):917–28 [DOI] [PubMed] [Google Scholar]
- 106.Quigley JG, Yang Z, Worthington MT, Phillips JD, Sabo KM, et al. 2004. Identification of a human heme exporter that is essential for erythropoiesis. Cell 118(6):757–66 [DOI] [PubMed] [Google Scholar]
- 107.Rajagopal A, Rao AU, Amigo J, Tian M, Upadhyay SK, et al. 2008. Haem homeostasis is regulated by the conserved and concerted functions of HRG-1 proteins. Nature 453(7198):1127–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rakvács Z, Kucsma N, Gera M, Igriczi B, Kiss K, et al. 2019. The human ABCB6 protein is the functional homologue of HMT-1 proteins mediating cadmium detoxification. Cell. Mol. Life Sci 76(20):4131–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rauner M, Baschant U, Roetto A, Pellegrino RM, Rother S, et al. 2019. Transferrin receptor 2 controls bone mass and pathological bone formation via BMP and Wnt signaling. Nat. Metab 1(1):111–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rondelli CM, Perfetto M, Danoff A, Bergonia H, Gillis S, et al. 2021. The ubiquitous mitochondrial protein unfoldase CLPX regulates erythroid heme synthesis by control of iron utilization and heme synthesis enzyme activation and turnover. J. Biol. Chem 297(2):100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sangkhae V, Fisher AL, Chua KJ, Ruchala P, Ganz T, Nemeth E. 2020. Maternal hepcidin determines embryo iron homeostasis in mice. Blood 136(19):2206–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sangkhae V, Fisher AL, Wong S, Koenig MD, Tussing-Humphreys L, et al. 2020. Effects of maternal iron status on placental and fetal iron homeostasis. J. Clin. Investig 130(2):625–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Santana-Codina N, Gableske S, Quiles del Rey M, Małachowska B, Jedrychowski MP, et al. 2019. NCOA4 maintains murine erythropoiesis via cell autonomous and non-autonomous mechanisms. Haematologica 104(7):1342–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Santander N, Lizama CO, Meky E, McKinsey GL, Jung B, et al. 2020. Lack of Flvcr2 impairs brain angiogenesis without affecting the blood-brain barrier. J. Clin. Investig 130(8):4055–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sarkar A, Carter EL, Harland JB, Speelman AL, Lehnert N, Ragsdale SW. 2021. Ferric heme as a CO/NO sensor in the nuclear receptor Rev-Erbβ by coupling gas binding to electron transfer. PNAS 118(3):e2016717118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schubert ML. 2017. Physiologic, pathophysiologic, and pharmacologic regulation of gastric acid secretion. Curr Opin. Gastroenterol 33 (6):430–38 [DOI] [PubMed] [Google Scholar]
- 117.Schwartz AJ, Das NK, Ramakrishnan SK, Jain C, Jurkovic MT, et al. 2019. Hepatic hepcidin/intestinal HIF-2α axis maintains iron absorption during iron deficiency and overload. J. Clin. Investig 129(1):336–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Scorrano L, De Matteis MA, Emr S, Giordano F, Hajnóczky G, et al. 2019. Coming together to define membrane contact sites. Nat. Commun 10(1):1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Severance S, Hamza I. 2009. Trafficking of heme and porphyrins in metazoa. Chem. Rev 109(10):4596–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Severance S, Rajagopal A, Rao AU, Cerqueira GC, Mitreva M, et al. 2010. Genome-wide analysis reveals novel genes essential for heme homeostasis in Caenorhabditis elegans. PLOS Genet. 6(7):e1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shayeghi M, Latunde-Dada GO, Oakhill JS, Laftah AH, Takeuchi K, et al. 2005. Identification of an intestinal heme transporter. Cell 122(5):789–801 [DOI] [PubMed] [Google Scholar]
- 122.Sinclair J, Hamza I. 2015. Lessons from bloodless worms: heme homeostasis in C. elegans. Biometals 28(3):481–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Smith A,McCulloh RJ. 2015. Hemopexin and haptoglobin: allies against heme toxicity from hemoglobin not contenders. Front. Physiol 6:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sobh A, Loguinov A, Zhou J, Jenkitkasemwong S, Zeidan R, et al. 2020. Genetic screens reveal CCDC115 as a modulator of erythroid iron and heme trafficking. Am. J. Hematol 95(9):1085–98 [DOI] [PubMed] [Google Scholar]
- 125.Song G, Zhang S, Tian M, Zhang L, Guo R, et al. 2021. Molecular insights into the human ABCB6 transporter. Cell Discov. 7(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Soubannier V, McLelland G-L, Zunino R, Braschi E, Rippstein P, et al. 2012. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol 22(2):135–41 [DOI] [PubMed] [Google Scholar]
- 127.Speich C, Wegmüller R, Brittenham GM, Zeder C, Cercamondi CI, et al. 2021. Measurement of long-term iron absorption and loss during iron supplementation using a stable isotope of iron (57Fe). Br. J. Haematol 192(1):179–89 [DOI] [PubMed] [Google Scholar]
- 128.Srole DN, Ganz T. 2021. Erythroferrone structure, function, and physiology: iron homeostasis and beyond. J. Cell. Physiol 236(7):4888–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Swenson SA, Moore CM, Marcero JR, Medlock AE, Reddi AR, Khalimonchuk O. 2020. From synthesis to utilization: the ins and outs of mitochondrial heme. Cells 9(3):E579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Traeger L, Wiegand SB, Sauer AJ, Corman BHP, Peneyra KM, et al. 2021. UBA6 and NDFIP1 regulate the degradation of ferroportin. Haematologica 107(2):478–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vincent SH, Muller-Eberhard U. 1985. A protein of the Z class of liver cytosolic proteins in the rat that preferentially binds heme. J. Biol. Chem 260(27):14521–28 [PubMed] [Google Scholar]
- 132.Wang C-Y, Xiao X, Bayer A, Xu Y, Dev S, et al. 2019. Ablation of hepatocyte Smad1, Smad5, and Smad8 causes severe tissue iron loading and liver fibrosis in mice. Hepatology 70(6):1986–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Warnatz H-J, Schmidt D, Manke T, Piccini I, Sultan M, et al. 2011. The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. J. Biol. Chem 286(26):23521–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Watanabe Y, Ishimori K, Uchida T. 2017. Dual role of the active-center cysteine in human peroxiredoxin 1: peroxidase activity and heme binding. Biochem. Biophys. Res. Commun 483(3):930–35 [DOI] [PubMed] [Google Scholar]
- 135.Weber RA, Yen FS, Nicholson SPV, Alwaseem H, Bayraktar EC, et al. 2020. Maintaining iron homeostasis is the key role of lysosomal acidity for cell proliferation. Mol. Cell 77(3):645–55.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.West A-R, Oates P-S. 2008. Mechanisms of heme iron absorption: current questions and controversies. World J. Gastroenterol 14(26):4101–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.White C, Yuan X, Schmidt PJ, Bresciani E, Samuel TK, et al. 2013. HRG1 is essential for heme transport from the phagolysosome of macrophages during erythrophagocytosis. Cell Metab. 17(2):261–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wyllie JC, Kaufman N. 1982. An electron microscopic study of heme uptake by rat duodenum. Lab. Investig 47(5):471–76 [PubMed] [Google Scholar]
- 139.Xiao X, Dev S, Canali S, Bayer A, Xu Y, et al. 2020. Endothelial bone morphogenetic protein 2 (Bmp2) knockout exacerbates hemochromatosis in homeostatic iron regulator (Hfe) knockout mice but not Bmp6 knockout mice. Hepatology 72(2):642–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yachie A 2021. Heme oxygenase-1 deficiency and oxidative stress: a review of 9 independent human cases and animal models. Int. J. Mol. Sci 22(4):1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yambire KF, Rostosky C, Watanabe T, Pacheu-Grau D, Torres-Odio S, et al. 2019. Impaired lysosomal acidification triggers iron deficiency and inflammation in vivo. eLife 8:e51031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yanatori I, Richardson DR, Dhekne HS, Toyokuni S, Kishi F. 2021. CD63 is regulated by iron via the IRE-IRP system and is important for ferritin secretion by extracellular vesicles. Blood 138(16):1490–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang Z, Philips JD, Doty RT, Giraudi P, Ostrow JD, et al. 2010. Kinetics and specificity of feline leukemia virus subgroup C receptor (FLVCR) export function and its dependence on hemopexin. J. Biol. Chem 285(37):28874–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yien YY, Robledo RF, Schultz IJ, Takahashi-Makise N, Gwynn B, et al. 2014. TMEM14C is required for erythroid mitochondrial heme metabolism. J. Clin. Investig. 124(10):4294–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yu Y, Jiang L, Wang H, Shen Z, Cheng Q, et al. 2020. Hepatic transferrin plays a role in systemic iron homeostasis and liver ferroptosis. Blood 136(6):726–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yuan X, Protchenko O, Philpott CC, Hamza I. 2012. Topologically conserved residues direct heme transport in HRG-1-related proteins. J. Biol. Chem 287(7):4914–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yuan X, Rietzschel N, Kwon H, Walter Nuno AB, Hanna DA, et al. 2016. Regulation of intracellular heme trafficking revealed by subcellular reporters. PNAS 113(35):E5144–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang D-L, Wu J, Shah BN, Greutélaers KC, Ghosh MC, et al. 2018. Erythrocytic ferroportin reduces intracellular iron accumulation, hemolysis, and malaria risk. Science 359(6383):1520–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang Z, Funcke J-B, Zi Z, Zhao S, Straub LG, et al. 2021. Adipocyte iron levels impinge on a fat-gut crosstalk to regulate intestinal lipid absorption and mediate protection from obesity. Cell Metab. 33(8):1624–39.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zheng J, Shan Y, Lambrecht RW, Donohue SE, Bonkovsky HL. 2008. Differential regulation of human ALAS1 mRNA and protein levels by heme and cobalt protoporphyrin. Mol. Cell. Biochem 319(1–2):153–61 [DOI] [PubMed] [Google Scholar]