Abstract
The trb operon from pTiC58 is one of three loci that are required for conjugal transfer of this Ti plasmid. The operon, which probably codes for the mating bridge responsible for pair formation and DNA transfer, contains 12 genes, 11 of which are related to genes from other members of the type IV secretion system family. The 12th gene, traI, codes for production of Agrobacterium autoinducer (AAI). Insertion mutations were constructed in each of the 12 genes, contained on a full-length clone of the trb region, using antibiotic resistance cassettes or a newly constructed transposon. This transposon, called mini-Tn5Ptrb, was designed to express genes downstream of the insertion site from a promoter regulated by TraR and AAI. Each mutation could trans complement downstream Tn3HoHo1 insertions in the trb operon of full-sized Ti plasmids. When marker-exchanged into the transfer-constitutive Ti plasmid pTiC58ΔaccR mutations in trbB, -C, -D, -E, -L, -F, -G, and -H abolished conjugal transfer from strain UIA5, which lacks the 450-kb catabolic plasmid pAtC58. However, these mutants retained residual conjugal transfer activity when tested in strain NT1, which contains this large plasmid. The trbJ mutant failed to transfer at a detectable frequency from either strain, while the trbI mutant transferred at very low but detectable levels from both donors. Only the trbK mutant was unaffected in conjugal transfer from either donor. Transfer of each of the marker-exchange mutants was restored by a clone expressing only the wild-type allele of the corresponding mutant trb gene. An insertion mutation in traI abolished the production of AAI and also conjugal transfer. This defect was restored by culturing the mutant donor in the presence of AAI. We conclude that all of the trb genes except trbI and trbK are essential for conjugal transfer of pTiC58. We also conclude that mutations in any one of the trb genes except traI and trbJ can be complemented by functions coded for by pAtC58.
The Ti plasmids of Agrobacterium tumefaciens code for two distinct conjugal transfer systems. One, mediated by the Vir system, transfers T-DNA into the plant cells but also can mobilize transfer of a suitable plasmid to recipient bacteria (for a recent review, see reference 17). The second, which constitutes the major pathway for Ti plasmid transfer, operates through a functionally and physically separated system called Tra. Expression of the Tra system on at least two Ti plasmids is tightly regulated at the transcriptional level through a complex signalling circuitry that involves opines produced by the crown gall tumors plus a LuxR-LuxI-type quorum-sensing mechanism (5, 22, 31, 37, 38, 46).
The Tra system of pTiC58 consists of two physically separated gene sets, tra and trb, which contain all of the genes essential for conjugal transfer (15). The tra region encodes the origin of conjugal transfer (oriT) and two sets of genes organized as divergently expressed operons (14, 24). Three of the six genes flanking the oriT region are related to essential tra genes from IncP and IncQ plasmids. The products of some of these genes comprise the DNA transfer and replication (Dtr) function of the Ti plasmid Tra system and most probably form the relaxosome complex at the oriT site. The second region, trb, is located at the 2 o’clock position on the plasmid and is flanked by noc, the locus conferring catabolism of nopaline, and oriV/rep, the locus for vegetative replication.
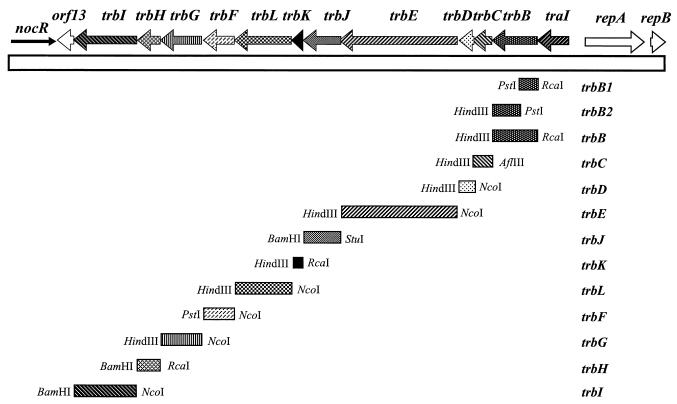
The trb genes are believed to encode the mating pair formation (Mpf) apparatus required for the physical translocation of DNA from donors to recipients. Sequence analysis and genetic studies have shown that this region contains 12 genes, traI and trbB, -C, -D, -E, -J, -K, -L, -F, -G, -H, and -I, organized in a single operon (42). Expression of this operon is controlled by the quorum-sensing activator, TraR, and the acyl-homoserine lactone signal, Agrobacterium autoinducer [AAI; N-(3-oxooctanoyl)-l-homoserine lactone], which is synthesized by the gene product of traI (38, 42, 43). The trb genes of pTiC58 are closely related in sequence and organization to the 11 trb genes from the tra2 core region of the IncP plasmids RP4 and R751. Genes of the trb system also are related to those of several other bacterial conjugation or protein secretion systems (42), including the VirB system of A. tumefaciens (49), the Ptl system of Bordetella pertussis (21, 55), the Tra system of plasmid F (27), the cag system of Helicobacter pylori (10, 36, 50), and the Dot system of Legionella pneumophila (48, 53).
In RP4, all but one of the 11 core trb genes are required for conjugal transfer (34). Similarly, 10 of the 11 virB genes are essential for the transfer of T-DNA to plant cells (8). Although the trb system of the Ti plasmid is related to these two systems, which of the Ti plasmid trb genes are essential for conjugal transfer remains unknown. In this report we describe the construction of a minitransposon carrying the promoter region of the traI-trb operon and the use of this element to generate complementable mutations in the trb genes of pTiC58. Results from matings with these mutants indicate that unlike the case for the RP4 trb system, only 9 of the 11 Ti plasmid trb genes are required for conjugal transfer. Our results also indicate that the large catabolic plasmid pAtC58 harbored by A. tumefaciens C58 not only can mobilize the IncQ plasmid RSF1010 at low frequency (14) but also can complement the mutations in most of the trb genes of the Ti plasmid.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
The strains of A. tumefaciens and Escherichia coli and the plasmids used in this study are listed in Table 1. A. tumefaciens strains were grown at 28°C in L broth (LB [47]) (Gibco-BRL, Gaithersburg, Md.), in AB minimal medium (11), or on nutrient agar plates (Difco Laboratories, Detroit, Mich.). Mannitol or glucose, at a final concentration of 0.2%, was used as the sole carbon source in the minimal medium. E. coli strains were grown at 37°C in LB or on L agar plates. Antibiotics were added at the following concentrations when required: for A. tumefaciens, carbenicillin at 100 or 200 μg/ml, gentamicin at 30 μg/ml, kanamycin at 100 μg/ml, rifampin at 50 μg/ml, streptomycin at 200 μg/ml, erythromycin at 100 μg/ml, chloramphenicol at 30 μg/ml, and tetracycline at 2 μg/ml; for E. coli, ampicillin at 100 μg/ml, kanamycin at 50 μg/ml, rifampin at 50 μg/ml, and tetracycline at 10 μg/ml. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Gibco-BRL) was included in media at 40 μg/ml to assess the production of β-galactosidase.
TABLE 1.
Bacterial strains and plasmids used
| Bacterial strain or plasmid | Relevant genotype, phenotype, or characteristic(s)a | Source or reference(s) |
|---|---|---|
| Escherichia coli | ||
| DH5α | supE44 ΔlacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 47 |
| S17-1 | pro Res− Mod+ Mob+ Ampr Cmr Tpr Strr | 15 |
| S17-1λ-pir | pro Res− Mod+ Mob+ Ampr Cmr Tpr Strr λ::pir | 24 |
| 2174(pPH1JI) | met pro Gmr Spr, used to isolate homogenotes | 24 |
| Agrobacterium tumefaciens | ||
| C58 | Wild-type pathogenic strain carrying pTiC58 and pAtC58 | Our collection |
| NT1 | Ti plasmid-cured C58 | 15 |
| NT1(pTiC58ΔaccR) | NT1 harboring a Trac mutant of pTiC58 | 6 |
| UIA5 | Ti plasmid- and cryptic plasmid-cured C58, Rifr Strr | 14 |
| C58C1RS | Ti plasmid-cured C58, Rifr Strr | 15 |
| C58C1EC | Ti plasmid-cured C58, Eryr Cmr | Our collection |
| Plasmids | ||
| pCF1 | 3,680-bp BamHI fragment 13 encoding traI, trbB, and repA from pTiC58 cloned in pTZ18U, Ampr | 38 |
| pDCI41E33 | traG::lacZ traR in pDSK519, Kmr, autoinducer reporter plasmid | 15 |
| pDEK-mb | pTiC58ΔaccR::Tn3HoHo1 marker-exchanged mutants, Cbr | 42 |
| pDSK519 | Broad-host-range IncQ cloning vector, Kmr | 41 |
| pHM1 | Delivery plasmid for mini-Tn5Ptrb, Ampr Kmr | This study |
| pHM25 | HindIII fragment 8 containing Tn3HoHo1 from pPLE2-25 cloned in pRK415, Tcr | 38 |
| pHM25-70 | pHM25::mini-Tn5Ptrb, Tcr Kmr | This study |
| pJB3 | Broad-host-range IncP cloning vector, Ampr | 9 |
| pKK38 | Broad-host-range IncP cloning vector, tac promoter, Tcr | David Nunn |
| pKK38ASH | Derivative of pKK38 with extra cloning sites, Tcr | This study |
| pPLE2-25 | pPLE2 with Tn3HoHo1 insertion in trbE, Cbr | 38, 42 |
| pPLKmPtrb | Km-Ptrb cassette from mini-Tn5Ptrb cloned in pBluescript SK(+), Ampr Kmr | This study |
| pPLK-nc | pTiC58ΔaccR::mini-Tn5Ptrb marker-exchanged mutants, Kmr | This study |
| pPLKΔC | pTiC58ΔaccR trbC::Km-Ptrb marker-exchanged mutant, Kmr | This study |
| pPLKΔK | pTiC58ΔaccR trbK::nptI marker-exchanged mutant, Kmr | This study |
| pPLtra | tra region and traR from pTiC58 cloned in pDSK519, Tcr | This study |
| pPLtrbXd | PCR-generated individual trb gene ORFs cloned in pKK38ASH or pKK38, Tcr | This study |
| pRK415 | Broad-host-range IncP cloning vector, Tcr | 41 |
| pRKtrb | 11,003-bp BglII-XbaI fragment containing the trb region of pTiC58 cloned in pRK415, Tcr | This study |
| pRKtrb-n | pRKtrb::mini-Tn5Ptrb mutants, Tcr Kmr | This study |
| pRKtrbΔC | pRKtrb with a Km-Ptrb cassette in trbC, Tcr Kmr | This study |
| pSB315 | pUC4K derivative containing nptI, Kmr | 32 |
| pSVB33 | 1.8-kb EcoRI fragment encoding traR from pTiC58 cloned in pSa152, Kmr Gmr | 46 |
| pTra17-52 | pTiC58ΔaccR nocR::Tn5 17-52, Noc− Trac Kmr | 6 |
| pUTmini-Tn5Km | Delivery plasmid for mini-Tn5Km, Ampr Kmr | 20 |
Abbreviations: Rifr, rifampin resistance; Strr, streptomycin resistance; Tcr, tetracycline resistance; Cbr, carbenicillin resistance; Kmr, kanamycin resistance; Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Gmr, gentamicin resistance; Emr, erythromycin resistance; Spr, spectinomycin resistance; Tpr, trimethoprim resistance; Trac, transfer constitutive.
m refers to the allele number of the Tn3HoHo1 insertion as shown in Fig. 3C.
n refers to the allele number of the mini-Tn5Ptrb insertion as shown in Fig. 3B.
trbX refers to trb genes. X can be B, C, D, E, J, K, L, F, G, H, or I.
DNA manipulation and plasmid constructions.
Ti plasmids were isolated as described by Hayman and Farrand (35). Other plasmids were isolated by an alkaline lysis method (47). Standard recombinant DNA techniques were used as described by Sambrook et al. (47). Digestions with restriction endonucleases were conducted according to the manufacturers’ instructions, and digestion products were separated by electrophoresis in agarose gels, using Tris-borate-EDTA buffer.
PCR.
The traI-trb promoter region was amplified from pCF1 by using AmpliTaq DNA polymerase (Perkin-Elmer, Foster City, Calif.) and oligomers 5′-GGGCGGCCGCCCGATTCTTCAAATGC-3′ and 5′-GAGCGGCCGCATCGTAATCTCCGC-3′. Pfu DNA polymerase (Stratagene, La Jolla, Calif.) was used to amplify each of the 11 trb genes, and the products were cloned into pKK38ASH or pKK38. In each case, the 5′ primer was designed to generate an NcoI, RcaI, StuI, or AflIII site allowing for in-frame fusion of the second codon of the trb gene to an ATG initiation codon provided by the vector. The sequences of the primers used for these amplifications are available upon request.
Mini-Tn5 mutagenesis and homogenotization.
A method based on mutagenesis with a mini-Tn5 transposon as described by de Lorenzo and Timmis (20) was used to transpose mini-Tn5Ptrb from pHM1 into the promoterless trb reporter clone pHM25 and into the full-length trb clone pRKtrb. The transposon delivery strain S17-1λ-pir(pHM1) was mated with the target strain, DH5α(pHM25) or DH5α(pRKtrb), on a 0.22-μm-pore-size filter, and the filter was incubated at 37°C for 6 h on the surface of an L agar plate. Following this incubation, the cells on the filter were suspended in 3 ml of LB, serial dilutions were prepared, and 0.1-ml volumes were spread on L agar plates containing kanamycin and tetracycline. The plates were incubated at 37°C, and colonies that appeared were combined. Plasmid DNA was extracted from the pool and used to transform E. coli DH5α with selection for resistance to kanamycin and tetracycline on L agar plates. Independent colonies were isolated and purified, and the locations and orientations of insertions of mini-Tn5Ptrb in the target plasmid were mapped by restriction endonuclease analysis. Insertion mutations of interest in pRKtrb were homogenotized into pTiC58ΔaccR by using pPH1JI as the eviction plasmid as previously described (24). Proper marker exchanges in the Ti plasmids were confirmed by restriction endonuclease analysis. pPH1JI was cured from these strains by continuous growth of the strain in LB without gentamicin, the selection marker of pPH1JI. Alternatively, the marker-exchanged Ti plasmid was isolated and introduced into A. tumefaciens NT1 via electroporation. Transformants resistant to kanamycin but remaining susceptible to gentamicin were retained for further study.
β-Galactosidase assay.
Quantitative assays for β-galactosidase activity were conducted as described previously (37). Each sample was analyzed in triplicate, and activity was expressed as units of β-galactosidase per 109 CFU.
Conjugation assays.
Conjugal transfer of the Ti plasmid and of the oriT-tra plasmids, pFRtra and pPLtra, of the binary transfer system (15) to the A. tumefaciens recipient strains C58C1RS and C58C1EC was assayed by a filter mating method as described previously (14). Samples were plated in triplicate, and the values obtained were used to calculate the average number of transconjugants that arose for each mating. Transfer frequencies were expressed as numbers of transconjugants obtained per input donor cell. Each set of matings was repeated once or twice. Although absolute transfer frequencies usually differed, the patterns of transfer were similar from one experiment to the next. Thus, in each case we present data from a single experiment in which all of the matings shown were conducted in parallel.
Analysis of AAI production.
AAI production was assayed by the semiquantitative plate method using A. tumefaciens NT1(pDCI41E33) as the indicator strain as previously described (15). A diffuse blue zone on the assay plate indicates the production of an active acyl-homoserine lactone by the strain being tested.
RESULTS
Construction and evaluation of mini-Tn5Ptrb.
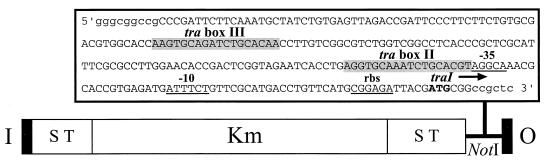
To construct a mini-Tn5 transposon carrying the traI-trb promoter, a 236-bp fragment containing the promoter region of the traI-trb operon and the first two codons of traI was amplified by PCR using primers containing a NotI site as described in Materials and Methods. The amplified fragment was cloned into the unique NotI site in pUTmini-Tn5Km (20), and the fidelity of the sequence and orientation of the insert were confirmed by nucleotide sequencing. The resulting plasmid is designated pHM1, and the minitransposon is designated mini-Tn5Ptrb (Fig. 1).
FIG. 1.
Structure of mini-Tn5Ptrb. The 236-bp traI-trb promoter region was amplified from pCF1 by PCR, and the product was cloned into the NotI site of mini-Tn5Km (19, 20). tra box II and tra box III, the 18-bp almost perfect inverted repeat sequences that are conserved in LuxRI-type quorum sensing regulatory systems (29, 38); −10 and −35, the promoter elements of the traI-trb operon identified in pTiR10 (29); rbs, the putative ribosomal binding site of traI; S, translational stop; T, transcriptional terminator; Km, kanamycin resistance gene; I and O, the 19-bp I and O ends of Tn5.
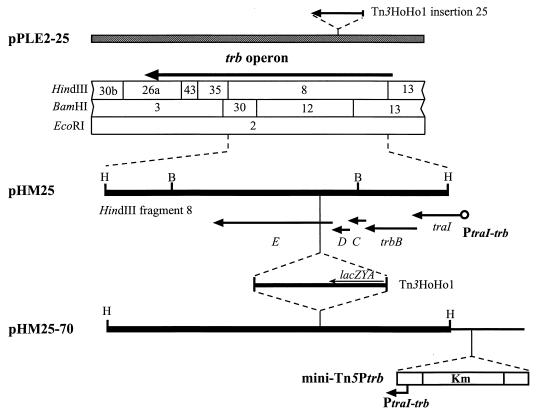
We tested this transposon by mutagenizing a promoterless reporter plasmid, pHM25 (Fig. 2). This plasmid, which is a derivative of pPLE2-25, contains HindIII fragment 8 of pTiC58 with a Tn3HoHo1 insertion in trbE but lacks the 5′ end of traI and the entire upstream traI-trb promoter region (38). Although the lacZ of Tn3HoHo1 is oriented in the proper direction, the construct does not express β-galactosidase activity (38). Following mutagenesis of pHM25 with mini-Tn5Ptrb, we identified an insertion derivative, pHM25-70, in which the transposon is located just upstream of traI and is oriented such that Ptrb (the trb promoter) can drive expression of trb. Strains harboring this plasmid expressed β-galactosidase activity but only when both TraR and AAI were provided (Table 2). Thus, the cloned promoter in this newly constructed minitransposon, when inserted in the proper orientation, can express downstream genes, and this expression is dependent on the quorum-sensing regulators, TraR and AAI.
FIG. 2.
Physico genetic organization of pPLE2-25 and construction of pHM25 and pHM25-70. The restriction map is according to the published sequence (42). pPLE2-25 contains the entire trb region of pTiC58 and a Tn3HoHo1 insertion in trbE. pHM25 is derived from pPLE2-25 by cloning the HindIII fragment 8 including the Tn3HoHo1 insertion into pRK415 (38). The open circle represents the traI-trb promoter region, which is just upstream of the HindIII site. pHM25-70 contains a mini-Tn5Ptrb insertion just upstream of the truncated traI gene. The transposons are not drawn to scale.
TABLE 2.
Mini-Tn5Ptrb restores TraR-AAI-dependent lacZ expression of a promoterless trbE::lacZ reporter fusion
| Test straina | β-Galactosidase activity (U/109 CFU)
|
|||
|---|---|---|---|---|
| −TraR
|
+TraRb
|
|||
| −AAI | +AAIc | −AAI | +AAI | |
| pPLE2-25 | 3 | 4 | 146 | NTd |
| pHM25 | 6 | 7 | 5 | 6 |
| pHM25-70 | 5 | 5 | 4 | 85 |
All plasmids are harbored in NT1.
TraR was supplied by the traR-expressing clone pSVB33.
Synthetic AAI was added to the culture at a concentration of 25 nM.
NT, not tested. The strain itself produces AAI.
Phenotype of pRKtrb::mini-Tn5Ptrb mutants.
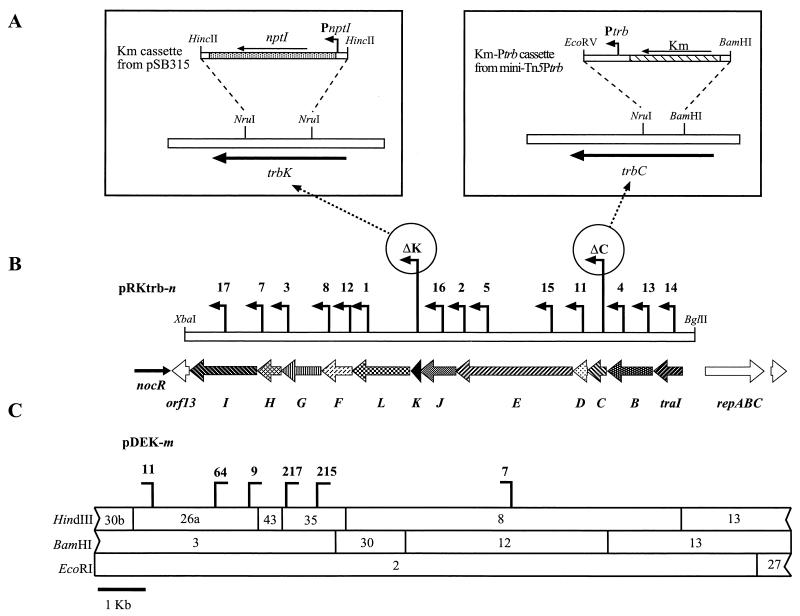
pRKtrb, which contains the entire trb region, was mutagenized with mini-Tn5Ptrb as described in Materials and Methods. Using restriction endonuclease analysis, we identified 14 independent insertions, all oriented in the correct direction and representing at least one insertion in 9 of the 11 trb genes (Fig. 3B). To generate nonpolar mutations in the two remaining genes, trbC and trbK, the following cloning strategies were used (Fig. 3A). For trbC, a cassette containing Ptrb and the nptII gene was constructed from mini-Tn5Ptrb. This cassette, called Km-Ptrb, retains the kanamycin resistance gene and the traI promoter region but lacks some restriction sites and the insertion sequence elements of the transposon. The cassette was cloned between the internal BamHI and NruI sites in trbC, thus replacing 112 bp of the gene with the Km-Ptrb cassette. Subsequent cloning resulted in the replacement of wild-type trbC by the trbC deletion-insertion allele within the full-length trb clone, pRKtrb, to generate pRKtrbΔC. For trbK, an nptI cassette coding for resistance to kanamycin was excised from pSB315 and cloned between the internal NruI sites within the gene. This cassette lacks a transcriptional terminator, and the promoter of the nptI gene is known to express genes downstream of the insertion (32). This resulted in an allele of trbK deleted for 133 internal residues and containing the nptI cassette oriented such that the trb genes downstream from trbK will be expressed from the promoter of nptI.
FIG. 3.
Mutational analysis of the trb genes of pTiC58. (A) Cloning strategies for constructing nonpolar insertions in trbC and trbK. (B) Locations of mini-Tn5Ptrb insertions in pRKtrb. Each vertical bar represents an independent insertion, and the horizontal arrow indicates the orientation of the traI-trb promoter of the transposon. (C) Restriction map of the trb region and locations of the Tn3HoHo1 insertions in pTiC58ΔaccR::Tn3HoHo1 which were used in complementation assays to test polarity of the mini-Tn5Ptrb insertion mutants of pRKtrb.
Each mutation was assessed for any strong polar effects by testing its ability to complement a Ti plasmid derivative with a Tn3HoHo1 insertion in a downstream trb gene (Fig. 3C) (42). These Tn3HoHo1 insertion derivatives do not transfer at detectable frequencies, but transfer can be restored to wild-type levels (∼10−2 transconjugant per input donor) by introducing a full-length trb clone such as pPLE2 (42) or pRKtrb (data not shown). Among the mini-Tn5Ptrb derivatives of pRKtrb tested, 13, including at least one in each trb gene, complemented the Tn3HoHo1 insertion mutations located downstream in the tester Ti plasmids (Table 3). pRKtrb-2 with a mini-Tn5Ptrb insertion in trbE failed to complement the test plasmid, suggesting that the insertion in this mutant exerts a strongly polar effect on expression of downstream trb genes. On the other hand, another trbE mutant, pRKtrb-5 restored transfer of the test plasmid to a reasonable level. These results indicate that in most cases the mini-Tn5Ptrb insertion and the nptI cassette mutants express trb genes located downstream of the insertion sites at levels allowing formation of a functional trb transporter.
TABLE 3.
Complementation analysis of mini-Tn5Ptrb mutants of pRKtrb
| pRKtrb::mini-Tn5Ptrb mutant | Gene mutated | pTiC58ΔaccR:: Tn3HoHo1 mutanta | Transfer frequency of pTiC58ΔaccR:: Tn3HoHo1 derivative |
|---|---|---|---|
| None | None | pDEK-11b | 9.0 × 10−2 |
| pRKtrb-14 | traI | pDEK-7 | 3.1 × 10−2 |
| pRKtrb-4 | trbB | pDEK-7 | 2.3 × 10−1 |
| pRKtrb-13 | trbB | pDEK-7 | 2.6 × 10−1 |
| pRKtrbΔCc | trbC | pDEK-7 | 2.2 × 10−3 |
| pRKtrb-11 | trbD | pDEK-7 | 8.8 × 10−2 |
| pRKtrb-2 | trbE | pDEK-215 | <10−7 |
| pRKtrb-5 | trbE | pDEK-215 | 4.1 × 10−3 |
| pRKtrb-15 | trbE | pDEK-215 | 1.6 × 10−4 |
| pRKtrb-16 | trbJ | pDEK-215 | 9.4 × 10−5 |
| pRKtrb-1 | trbL | pDEK-217 | 1.4 × 10−3 |
| pRKtrb-8 | trbF | pDEK-217 | 4.1 × 10−4 |
| pRKtrb-12 | trbF | pDEK-217 | 5.1 × 10−4 |
| pRKtrb-3 | trbG | pDEK-9 | 3.9 × 10−3 |
| pRKtrb-7 | trbH | pDEK-9 | 4.4 × 10−3 |
All derivatives of pTiC58ΔaccR::Tn3HoHo1 except pDEK-11 failed to transfer at a detectable frequency in the absence of the complementing plasmids.
pDEK-11 is a derivative of pTiC58ΔaccR with a Tn3HoHo1 insertion in nocR (42).
Contains a replacement mutation in trbC as described in the text.
The mini-Tn5Ptrb, Km-Ptrb, or nptI insertion allele of each trb gene was marker exchanged into pTiC58ΔaccR, and each Ti plasmid was tested for its conjugal properties. With two exceptions, all such donors exhibited reduced but detectable transfer frequencies compared to that of the transfer-constitutive (Trac) Ti plasmid (Table 4). The mini-Tn5Ptrb insertion in trbJ completely abolished conjugal transfer, while the nptI cassette in trbK had virtually no effect on transfer frequencies. We considered the possibility that pPH1JI, the R751 derivative used as the eviction plasmid in the marker exchange, was complementing the trb mutations in the Ti plasmids. However, when tested in a donor lacking pPH1JI, each mutant Ti plasmid except the trbJ mutant continued to transfer at a low but detectable frequency (Table 4). Again, transfer of the trbK mutant occurred at near-wild-type frequencies. Strain NT1 harbors a 450-kb catabolic plasmid called pAtC58 (23). To determine whether this plasmid was contributing transfer functions, we introduced each of the mutant Ti plasmids into UIA5, a derivative of NT1 cured of pAtC58. When these strains were used as donors, all except those with a mutation in trbK or trbI failed to transfer at a detectable frequency (Table 4). The trbI mutant showed an approximately 3- to 4-orders-of-magnitude decrease in transfer frequency compared to the parent Ti plasmid, whereas the mutation in trbK had no effect on the transfer frequency. To confirm that the trbI mutation does not abolish conjugal transfer, we tested pDEK-9 and pDEK-64, two derivatives of pTiC58ΔaccR with independent Tn3HoHo1 insertions in trbI (Fig. 3C). We previously reported that these Ti plasmids failed to transfer (42). However, when we increased the sensitivity of the assay by plating less diluted samples of the mating mix, we observed transfer of pDEK-9 and pDEK-64 at frequencies of 9.4 × 10−8 and 1.5 × 10−7 respectively.
TABLE 4.
Conjugal transfer frequency of pTiC58ΔaccR::mini-Tn5Ptrb mutants and complementation of these mutants with trb gene clones
| Mutant Ti plasmid | trb gene mutated | Transfer frequency when mated from:
|
|||
|---|---|---|---|---|---|
| NT1 (Ti plasmid only) | UIA5
|
||||
| Ti plasmid only | +pKK38ASH | +trb ORF clone | |||
| pTra17-52 | None | 1.6 × 10−1 | 2.4 × 10−2 | NTa | NT |
| pPLK-4 | trbB | 7.6 × 10−4 | <10−8 | <10−8 | 8.3 × 10−5 |
| pPLKΔC | trbC | 6.4 × 10−4 | <10−8 | <10−8 | 4.4 × 10−2 |
| pPLK-11 | trbD | 1.2 × 10−5 | <10−8 | <10−8 | 5.6 × 10−3 |
| pPLK-5 | trbE | 2.1 × 10−6 | <10−8 | <10−8 | 2.0 × 10−3 |
| pPLK-16 | trbJ | <10−7 | <10−8 | <10−8 | 3.8 × 10−6 |
| pPLKΔK | trbK | 5.5 × 10−2 | 2.9 × 10−3 | 1.2 × 10−4 | 1.2 × 10−3 |
| pPLK-1 | trbL | 3.3 × 10−6 | <10−8 | <10−8 | 2.0 × 10−3 |
| pPLK-12 | trbF | 6.6 × 10−4 | <10−8 | <10−8 | 1.8 × 10−2 |
| pPLK-3 | trbG | 4.7 × 10−6 | <10−8 | <10−8 | 1.1 × 10−4 |
| pPLK-7 | trbH | 3.7 × 10−4 | <10−8 | <10−8 | 2.0 × 10−4 |
| pPLK-17 | trbI | 8.7 × 10−5 | 5.0 × 10−6 | 2.8 × 10−6 | 4.1 × 10−4 |
NT, not tested.
Complementation analysis.
Each of the trb genes was amplified by PCR using primers containing NcoI, RcaI, StuI, or AflIII sites, depending on the 5′-end sequence of the gene, and either HindIII, PstI, or BamHI sites immediately following the stop codon of the gene. These PCR products were cloned into pKK38ASH, which is a derivative of pKK38 into which we inserted extra cloning sites for HindIII, PstI, and BamHI (Fig. 4). Each of the trb open reading frame (ORF) clones was introduced in trans into UIA5 harboring a derivative of pTiC58ΔaccR with a mini-Tn5Ptrb insertion in the corresponding trb gene. When mated with C58C1EC, all complemented donor strains transferred the mutant Ti plasmids, although the efficiency of complementation varied for different mutations (Table 4). Complemented donors with mutations in trbC, trbD, trbE, trbL, and trbF transferred the test plasmid at near-wild-type levels. On the other hand, the trbB and trbJ mutants were only poorly complemented, while mutants with insertions in trbG, trbH, and trbI transferred their Ti plasmids at intermediate frequencies (Table 4).
FIG. 4.
Construction of individual trb gene ORF clones. PCR products corresponding to each of the trb genes were cloned into the expression vector as described in Materials and Methods. Due to the lack of suitable restriction sites, the trbB clone was obtained by first amplifying the 5′ and 3′ halves of the gene separately to generate trbB1 and trbB2 and then cloning trbB2 into trbB1. All PCR products were cloned in pKK38ASH except for trbK, which was cloned in pKK38.
Conjugal transfer of a nonpolar traI mutant can be restored by adding AAI.
We also identified a mini-Tn5Ptrb insertion in traI, the first gene of the trb operon (Fig. 3B). An Agrobacterium strain harboring this mutant plasmid, pRKtrb-14, does not produce AAI at detectable levels even in the presence of TraR (Table 5). Complementation assays using pTiC58ΔaccR::Tn3HoHo1 indicated that this insertion mutation is not strongly polar (Table 3). Attempts to marker exchange this mutation in pTiC58ΔaccR were not successful. Thus, we assessed the effect of the disruption of traI on conjugal transfer by testing the ability of pRKtrb-14 to mobilize pPLtra, which is a pDSK519 derivative containing the tra operons, the Ti plasmid oriT, and traR of the Ti plasmid, in a binary transfer system. NT1 harboring both pPLtra and pRKtrb-14 mobilizes the tra plasmid from the Ti plasmid oriT only at very low frequency (Table 5). This transfer rate is similar to that observed from strain NT1(pPLtra), which lacks the trb component, and is about 3 orders of magnitude lower than that observed from NT1(pPLtra, pRKtrb), which contains the wild-type Trb system of the Ti plasmid (Table 5). This basal level of mobilization of pPLtra is commonly observed when an RSF1010 derivative is harbored in strain NT1 (14). However, the mobilization frequency of the tra plasmid in NT1(pPLtra, pRKtrb-14) was restored to that of NT1(pPLtra, pRKtrb) by adding exogenous AAI (Table 5). Mobilization of the oriT plasmid from NT1(pPLtra) was not stimulated by addition of AAI.
TABLE 5.
Conjugal mobilization of pPLtra by the traI::mini-Tn5Ptrb mutant of pRKtrb can be restored by addition of AAI
| Test strain | AAI production | Conjugal transfer frequency (transconjugants/input donor)
|
|
|---|---|---|---|
| −AAI | +AAIa | ||
| NT1(pPLtra) | − | 3.9 × 10−6 | 4.0 × 10−7 |
| NT1(pPLtra, pRKtrb) | +++ | 4.7 × 10−3 | NTb |
| NT1(pPLtra, pRKtrb-14) | − | 3.2 × 10−6 | 4.8 × 10−3 |
Synthetic AAI was added at a concentration of 40 nM, and the culture was grown for an additional 6 h.
NT, not tested.
DISCUSSION
traI and 9 of the 11 trb genes of pTiC58 are essential for conjugation.
Mutations in all but two trb genes resulted in complete loss of conjugal activity of the Ti plasmid (Table 4). The first gene of the trb operon is traI, the only known function of which is the synthesis of AAI, the essential signal for the quorum-sensing regulation of Ti plasmid conjugal transfer (30, 38, 43). Consistent with this, a mini-Tn5Ptrb mutation in traI abolished normal conjugation in a binary transfer assay but the wild-type phenotype could be restored by supplying exogenous AAI (Table 5). Thus, we conclude that the first gene in the trb operon, traI, is essential for Ti plasmid conjugal transfer but only because it is required for synthesis of the quorum-sensing signal. A nonpolar mutation in trbK has virtually no effect on conjugal transfer, suggesting that the product of the gene is not essential for the trb-encoded Mpf apparatus. However, trbK does play a role in conjugation; when present in a recipient, this gene confers entry exclusion against closely related Ti plasmids (44). This is consistent with studies of RP4 in which trbK is not required for conjugal transfer but is responsible for entry exclusion (33, 34). Hence the conservation between these two systems extends, at least in one case, to the function of the individual genes, even though the TrbK proteins from the two plasmids show considerable sequence divergence (42).
Although members of the type IV secretion family share many characteristics, not all systems contain the same sets of genes. For example, the virB system of Ti plasmids and the trb system of RP4 have only six genes in common. Moreover, only trbI/virB10 is present in every known type IV secretion system characterized to date, including the very distantly related systems such as cag of H. pylori, which contains only four trb homologs (10, 36, 50), and dot of L. pneumophila, which contains only two virB homologs (48, 53). trbI of RP4 has been reported to be essential for conjugal transfer (34). Similarly, virB10, the trbI homolog of the Ti plasmid Vir system, apparently is required for T-strand transfer to plants (8, 54) as well as for mobilization of RSF1010 to bacteria (28). Yet our results indicated that disruption of trbI of pTiC58, while severely reducing the frequency, did not abolish conjugal transfer. That transfer could be restored to near-normal levels when we supplied a copy of trbI in trans to the mutant Ti plasmid indicates that only the mutation in trbI is responsible for the decreased conjugal transfer activity. It is conceivable that the insertion of mini-Tn5Ptrb in trbI did not completely destroy the TrbI protein and that the remaining N- or C-terminal portion, or both parts of the protein, still retains partial function. However, our derivatives of pTiC58ΔaccR with Tn3HoHo1 insertions in trbI also exhibit very low but detectable levels of transfer. Furthermore, our results are consistent with the observation that pTiA6NC, an octopine-type Ti plasmid containing a deletion that removes 90% of trbI, conjugally transfers at a very low but detectable frequency (2). Thus, we conclude that trbI of the Ti plasmid is not essential for conjugation but is required for transfer at wild-type efficiencies.
Although the function of TrbI is unknown, VirB10, the TrbI homolog of the Ti plasmid virB system, is believed to play a crucial role in assembling the mating pore complex. Several groups have proposed that VirB10 functions as an anchor by interacting with other VirB proteins, including VirB7 and VirB9, to form a high-molecular-weight complex (3, 4, 8, 13, 25, 26). However, the trb systems of the Ti plasmid and RP4 contain neither a VirB7 nor a VirB9 homolog. Thus, TrbI and VirB10 may play different roles in their two respective Mpf systems.
Mini-Tn5Ptrb as a tool to generate complementable mutations in the trb operon.
Using transposable elements as mobile promoters to study polycistronic transcriptional units has proved to be useful (for a review, see reference 7). Transposons such as Tn5virB (16) and mini-Tn5-lacIq/Ptrc (18) have been successfully applied in the genetic analyses of complex operons in A. tumefaciens and Pseudomonas spp. Our analyses indicate that when inserted in the proper location and correct orientation, mini-Tn5Ptrb can provide a promoter capable of expressing downstream genes, and that expression from this promoter is regulated by TraR and AAI. However, the transcriptional activity from the traI-trb promoter of mini-Tn5Ptrb in pHM25-70 was only about 60% of that observed when the trbE::lacZ fusion was expressed from the native traI-trb promoter in the original clone pPLE2-25 (Table 2). This difference in expression probably is due to the location of the insertion and the distance between the insertion and the immediate downstream gene. It also is possible that early termination of transcription occurs due to translational stops in the other reading frames. Therefore, for any given insertion some degree of polarity may be expected, and examination of more than one insertion in each gene may be necessary to obtain a suitable nonpolar mutation. Such factors may account for the difference in the ability of the two trbE mutants to trans complement a downstream mutation in the trb operon. Polar effects also may arise from the disruption of the preceding gene in a translationally coupled gene cluster as observed in other studies (8, 34). However, the promoter in mini-Tn5Ptrb contains a ribosomal binding site which may allow translational reinitiation of downstream, translationally coupled genes. With respect to our analysis, each of the Ti plasmid trb genes is preceded by a sequence that could serve as a ribosomal binding site (reference 42 and data not shown). Even so, that some of our trb mutants could not be complemented to wild-type levels of transfer suggests that mini-Tn5Ptrb insertions can induce some degree of polarity on the expression of downstream genes. Such effects may account for the relatively weak complementation of the trbB and trbJ mutations by the complementing cloned genes.
Mini-Tn5Ptrb provides several advantages for studying the trb genes. First, compared to MURFI linker insertion, which involves cloning and subcloning steps (34, 45), or DNA polymerase-directed site-specific deletion, which usually requires extensive in vivo and in vitro DNA manipulations, transposon mutagenesis is a relatively quick and easy way to generate acceptably nonpolar mutations in a large gene cluster. Second, using a cognate promoter such as Ptrb ensures that the same regulatory mechanism controls expression of both the downstream genes, which are transcribed from the transposon, and the upstream genes, which are transcribed from the native promoter of the operon. However, in common with all transposon mutagenesis schemes, insertions of mini-Tn5Ptrb in small genes can be difficult to obtain. Such was the case of trbC and trbK in this study even though the minitransposon insertions appeared to be evenly distributed throughout the trb region.
The role of the catabolic plasmid pAtC58 in conjugal transfer.
In addition to the well-studied Ti plasmids, most isolates of Agrobacterium spp. harbor other extrachromosomal elements. These replicons usually are very large, but little is known concerning traits they confer on their bacterial hosts. A. tumefaciens C58 harbors at least one such plasmid, pAtC58, with a size variously estimated at 450 to 550 kb (1, 12, 39, 40). This plasmid, which codes for catabolism of a set of Amadori compounds produced by rotting vegetation and also by some crown gall tumors (52), is self-conjugal (51) and can mobilize an RSF1010 derivative at low but detectable frequency (14). Our results suggest that components of the transfer system of pAtC58 can substitute for certain of the trb functions of pTiC58. Thus, NT1, which harbors pAtC58, transfers most of our trb mutants at low frequency, while these same mutant plasmids fail to transfer from UIA5, a strain that lacks pAtC58 but otherwise is nearly isogenic to NT1 (Table 4). However, our oriT-tra plasmids such as pFRtra and pDCtra-5 are not mobilized from NT1 in the absence of a functional Ti plasmid trb system (15). Moreover, Ti plasmid trb mutants derived by insertion of Tn3HoHo1, which can exert strong polarity, fail to transfer from an NT1 donor (42). These observations suggest that the Mpf of pAtC58 is not itself able to substitute for that of the Ti plasmid. Rather, we propose that Mpf components of pAtC58 can replace some but not all of those coded for by the trb operon of pTiC58 to form a functional chimeric conjugal transporter. In this regard, only the trbJ mutant of pTiC58 failed to transfer at detectable frequencies from a donor harboring pAtC58 (Table 4). This observation suggests that the function coded for by this gene is an essential component of, and highly specific to, the Ti plasmid transporter and cannot be replaced by the corresponding Mpf component of pAtC58. Such a dependence on the Ti plasmid trbJ product for transfer from the Ti plasmid relaxosome might explain why the Mpf of pAtC58 cannot substitute for that of pTiC58. Interestingly, pPH1JI, which codes for a trb system closely related to that of pTiC58 (42), apparently does not complement any mutations in the Ti plasmid trb operon. Our nonpolar trb mutants do not transfer from a donor harboring pPH1JI at frequencies any higher than from donors lacking this IncP1β plasmid (data not shown). This is reminiscent of our observation that although the Dtr components of pTiC58 and RSF1010 are related, the Ti plasmid will not mobilize the IncQ plasmid (14). Determining the functional and phylogenetic interrelationships of these type IV systems and the points at which specificity is conferred should aid us in understanding how these transporters recognize and translocate their substrates.
ACKNOWLEDGMENTS
This work was supported by grants R01GM52465 from the NIH and AG92-3312-8231 from the USDA to S.K.F. P.-L.L. was supported in part from Hatch project 15-0326 to S.K.F. H.M. was supported by a predoctoral fellowship from the Howard Hughes Medical Institute.
REFERENCES
- 1.Allardet-Servent A, Michaux-Charachon S, Jumas-Bilak E, Karayan L, Ramuz M. Presence of one linear and one circular chromosome in the Agrobacterium tumefaciens C58 genome. J Bacteriol. 1993;175:7869–7874. doi: 10.1128/jb.175.24.7869-7874.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alt-Mörbe J, Stryker J L, Fuqua C, Li P-L, Farrand S K, Winans S C. The conjugal transfer system of Agrobacterium tumefaciens octopine-type Ti plasmids is closely related to the transfer system of an IncP plasmid and distantly related to Ti plasmid vir genes. J Bacteriol. 1996;178:4248–4257. doi: 10.1128/jb.178.14.4248-4257.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banta L M, Bohne J, Lovejoy S D, Dostal K. Stability of the Agrobacterium tumefaciens VirB10 protein is modulated by growth temperature and periplasmic osmoadaption. J Bacteriol. 1998;180:6597–6606. doi: 10.1128/jb.180.24.6597-6606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaupré C E, Bohne J, Dale E M, Binns A N. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J Bacteriol. 1997;179:78–89. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck von Bodman S, Hayman G T, Farrand S K. Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc Natl Acad Sci USA. 1992;89:643–647. doi: 10.1073/pnas.89.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck von Bodman S, McCutchan J E, Farrand S K. Characterization of conjugal transfer functions of Agrobacterium tumefaciens Ti plasmid pTiC58. J Bacteriol. 1989;171:5281–5289. doi: 10.1128/jb.171.10.5281-5289.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg C M, Berg D E. Transposable element tools for microbial genetics. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2588–2612. [Google Scholar]
- 8.Berger B R, Christie P J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blatny J M, Brautaset T, Winther-Larsen H C, Haugan K, Valla S. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl Environ Microbiol. 1997;63:370–379. doi: 10.1128/aem.63.2.370-379.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chilton M D, Currier T C, Farrand S K, Bendich A J, Gordon M P, Nester E W. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci USA. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho K, Fuqua C, Winans S C. Transcriptional regulation and locations of Agrobacterium tumefaciens genes required for complete catabolism of octopine. J Bacteriol. 1997;179:1–8. doi: 10.1128/jb.179.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie P J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook D M, Farrand S K. The oriT region of the Agrobacterium tumefaciens Ti plasmid pTiC58 shares DNA sequence identity with the transfer origins of RSF1010 and RK2/RP4 and with T-region borders. J Bacteriol. 1992;174:6238–6246. doi: 10.1128/jb.174.19.6238-6246.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook D M, Li P-L, Ruchaud F, Padden S, Farrand S K. Ti plasmid conjugation is independent of vir: reconstitution of the tra functions from pTiC58 as a binary system. J Bacteriol. 1997;179:1291–1297. doi: 10.1128/jb.179.4.1291-1297.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dale E M, Binns A N, Ward J E., Jr Construction and characterization of Tn5virB, a transposon that generates nonpolar mutants, and its use to define virB8 as an essential virulence gene in Agrobacterium tumefaciens. J Bacteriol. 1993;175:887–891. doi: 10.1128/jb.175.3.887-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de la Cruz F, Lanka E. Function of the Ti-plasmid Vir proteins: T-complex formation and transfer to the plant cell. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishing; 1998. pp. 281–301. [Google Scholar]
- 18.de Lorenzo V, Eltis L, Kessler B, Timmis K N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 19.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5 and Tn10-derived mini-transposon. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 21.Farizo K M, Cafarella T G, Burns D L. Evidence for a ninth gene, ptlI, in the locus encoding the pertussis toxin secretion system of Bordetella pertussis and formation of a PtlI-PtlF complex. J Biol Chem. 1996;271:31643–31649. doi: 10.1074/jbc.271.49.31643. [DOI] [PubMed] [Google Scholar]
- 22.Farrand S K. Conjugation of Agrobacterium plasmids. In: Clewell D, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 255–291. [Google Scholar]
- 23.Farrand S K. Conjugal plasmids and their transfer. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae. Dordrecht, The Netherlands: Kluwer Academic Publishing; 1998. pp. 199–233. [Google Scholar]
- 24.Farrand S K, Hwang I, Cook D M. The tra region of the nopaline-type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J Bacteriol. 1996;178:4233–4247. doi: 10.1128/jb.178.14.4233-4247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez D, Spudich G M, Zhou X R, Christie P J. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J Bacteriol. 1996;178:3168–3176. doi: 10.1128/jb.178.11.3168-3176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finberg K E, Muth T R, Young S P, Maken J B, Heitritter S M, Binns A N, Banta L M. Interactions of VirB9, −10, and −11 with the membrane fraction of Agrobacterium tumefaciens: solubility studies provide evidence for tight associations. J Bacteriol. 1995;177:4881–4889. doi: 10.1128/jb.177.17.4881-4889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frost L S, Ippen-Ihler K, Skurray R A. An analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev. 1994;58:162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fullner K J. Role of Agrobacterium virB genes in transfer of T complexes and RSF1010. J Bacteriol. 1998;180:430–434. doi: 10.1128/jb.180.2.430-434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuqua C, Winans S C. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J Bacteriol. 1996;178:435–440. doi: 10.1128/jb.178.2.435-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuqua W C, Winans S C. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galán J E, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haase J, Kalkum M, Lanka E. TrbK, a small cytoplasmic membrane lipoprotein, functions in entry exclusion of the IncPα plasmid RP4. J Bacteriol. 1996;178:6720–6729. doi: 10.1128/jb.178.23.6720-6729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haase J, Lurz R, Grahn A M, Bamford D H, Lanka E. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J Bacteriol. 1995;177:4779–4791. doi: 10.1128/jb.177.16.4779-4791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayman G T, Farrand S K. Characterization and mapping of the agrocinopine-agrocin 84 locus on the nopaline Ti plasmid pTiC58. J Bacteriol. 1988;170:1759–1767. doi: 10.1128/jb.170.4.1759-1767.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofreuter D, Odenbreit S, Henke G, Haas R. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol Microbiol. 1998;285:1027–1038. doi: 10.1046/j.1365-2958.1998.00879.x. [DOI] [PubMed] [Google Scholar]
- 37.Hwang I, Cook D M, Farrand S K. A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J Bacteriol. 1995;177:449–458. doi: 10.1128/jb.177.2.449-458.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang I, Li P-L, Zhang L, Piper K R, Cook D M, Tate M E, Farrand S K. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc Natl Acad Sci USA. 1994;91:4639–4643. doi: 10.1073/pnas.91.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jumas-Bilak E, Maugard C, Michaux-Charachon S, Allardet-Servent A, Perrin A, O’Callaghan D, Ramuz M. Study of the organization of the genomes of Escherichia coli, Brucella melitensis and Agrobacterium tumefaciens by insertion of a unique restriction site. Microbiology. 1995;141:2425–2432. doi: 10.1099/13500872-141-10-2425. [DOI] [PubMed] [Google Scholar]
- 40.Jumas-Bilak E, Michaux-Charachon S, Bourg G, Ramuz M, Allardet-Servent A. Unconventional genomic organization in the alpha subgroup of the Proteobacteria. J Bacteriol. 1998;180:2749–2755. doi: 10.1128/jb.180.10.2749-2755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 42.Li P-L, Everhart D M, Farrand S K. Genetic and sequence analysis of the trb locus on pTiC58, a mating-pair formation system related to members of the type IV secretion family. J Bacteriol. 1998;180:6164–6172. doi: 10.1128/jb.180.23.6164-6172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moré M I, Finger L D, Stryker J L, Fuqua C, Eberhard A, Winans S C. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 44.Nelson, C. D., C. E. Bratis, and S. K. Farrand. Unpublished data.
- 45.Perlman D, Halvorson H O. The MURFI linker for multiple reading frame insertion of a sense or nonsense codon into DNA. Nucleic Acids Res. 1986;14:2139–2155. doi: 10.1093/nar/14.5.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piper K R, Beck von Bodman S, Farrand S K. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature (London) 1993;362:448–450. doi: 10.1038/362448a0. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 48.Segal G, Purcell M, Shuman H A. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci USA. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shirasu K, Morel P, Kado C I. Characterization of the virB operon of an Agrobacterium tumefaciens Ti plasmid: nucleotide sequence and protein analysis. Mol Microbiol. 1990;4:1153–1163. doi: 10.1111/j.1365-2958.1990.tb00690.x. [DOI] [PubMed] [Google Scholar]
- 50.Tummuru M K R, Sharma S A, Blaser M J. Helicobacter pylori picB, a homologue of the Bordetella pertussis toxin secretion protein, is required for induction of IL-8 in gastric epithelial cells. Mol Microbiol. 1995;18:867–876. doi: 10.1111/j.1365-2958.1995.18050867.x. [DOI] [PubMed] [Google Scholar]
- 51.van Montagu M, Schell J. The plasmids of Agrobacterium tumefaciens. In: Timmis K N, Pühler A, editors. Plasmids of medical, environmental and commercial importance. Amsterdam, The Netherlands: Elsevier/North Holland Biomedical Press; 1979. pp. 71–95. [Google Scholar]
- 52.Vaudequin-Dransart V, Petit A, Chilton W S, Dessaux Y. The cryptic plasmid of Agrobacterium tumefaciens cointegrates with the Ti plasmid and cooperates for opine degradation. Mol Plant-Microbe Interact. 1998;11:583–591. [Google Scholar]
- 53.Vogel J P, Andrews H L, Wong S K, Isberg R R. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 54.Ward J E, Jr, Dale E M, Christie P J, Nester E W, Binns A N. Complementation analysis of Agrobacterium tumefaciens Ti plasmid virB genes by use of a vir promoter expression vector: virB9, virB10, and virB11 are essential virulence genes. J Bacteriol. 1990;172:5187–5199. doi: 10.1128/jb.172.9.5187-5199.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss A A, Johnson F D, Burns D L. Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci USA. 1993;90:2970–2974. doi: 10.1073/pnas.90.7.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]