Summary
Copy-number variations (CNV) are believed to play an important role in a wide range of complex traits, but discovering such associations remains challenging. While whole-genome sequencing (WGS) is the gold-standard approach for CNV detection, there are several orders of magnitude more samples with available genotyping microarray data. Such array data can be exploited for CNV detection using dedicated software (e.g., PennCNV); however, these calls suffer from elevated false-positive and -negative rates. In this study, we developed a CNV quality score that weights PennCNV calls (pCNVs) based on their likelihood of being true positive. First, we established a measure of pCNV reliability by leveraging evidence from multiple omics data (WGS, transcriptomics, and methylomics) obtained from the same samples. Next, we built a predictor of omics-confirmed pCNVs, termed omics-informed quality score (OQS), using only PennCNV software output parameters. Promisingly, OQS assigned to pCNVs detected in close family members was up to 35% higher than the OQS of pCNVs not carried by other relatives (p < 3.0 × 10−90), outperforming other scores. Finally, in an association study of four anthropometric traits in 89,516 Estonian Biobank samples, the use of OQS led to a relative increase in the trait variance explained by CNVs of up to 56% compared with published quality filtering methods or scores. Overall, we put forward a flexible framework to improve any CNV detection method leveraging multi-omics evidence, applied it to improve PennCNV calls, and demonstrated its utility by improving the statistical power for downstream association analyses.
Keywords: anthropometric traits, copy-number variation, gene expression, methylation, multi-omics, PennCNV, structural variation, whole genome sequencing
Lepamets et al. presented a customizable framework to estimate copy-number variation quality based on independent omics data layers, as well as a prebuilt omics-informed quality score (OQS) model for widely used PennCNV software. In association analyses, OQS outperformed the previously published CNV filtering approaches or quality scores.
Introduction
Copy-number variations (CNV) are unbalanced structural variations that alter the dosage of genomic regions via deletion and duplication events. Approximately 9.5% of the human genome is subject to CNVs,1 which vary in length, ranging from a few dozens to several millions of base pairs (bp) in length. CNVs tend to have more severe phenotypic consequences compared with single-nucleotide variations (SNVs) as, due to their larger size, they can encompass entire coding regions.
CNVs have been associated with a number of conditions including autism,2 schizophrenia,3 neurodegenerative disorders,4,5 and cancer.6 A number of large recurrent deletions and duplications have been combined into the DECIPHER CNV syndromes list.7 Importantly, incomplete penetrance of several syndromic CNVs has been established by studying large population biobanks, where CNV load was shown to increase the risk of obesity, physical or cognitive impairment, and congenital malformations while lowering educational attainment and socio-economic status.8, 9, 10, 11 In parallel, CNV genome-wide association studies (GWASs) have been conducted on numerous diagnoses12,13 and medically relevant continuous traits,11,14,15 including a large meta-analysis on anthropometric measurements,16 revealing the important role of CNVs in shaping the human phenome.
Over the years, multiple CNV detection algorithms have been developed for CNV detection from SNV genotyping microarray probe intensities.17 Currently, PennCNV18 is the most widely used software for genotyping array-based calling. For each sample, a hidden Markov model (HMM)-based algorithm uses overall signal intensity and continuous allelic intensity at polymorphic probes to estimate the probability of a hidden copy-number state at this genomic location. Unfortunately, CNV regions found by different array-based detection methods only agree in about 20% of cases,19 indicating the high likelihood of false-positive calls. To counter this, various filtering strategies have been employed, usually by setting cut-off values to combinations of parameters including number of CNVs per sample, minimum CNV length, probe density, and PennCNV confidence score.9,12,18,20,21 Filtering based on arbitrary thresholds is suboptimal and a continuous CNV quality score that predicts the probability that a CNV region is a consensus (≥70% call overlap) between PennCNV, QuantiSNP,22 and CNVpartition (an Illumina developed GenomeStudio software plug-in, web resources) has been proposed.19 We refer to this as consensus-based quality score (cQS).
Still, cQS relies only on a single input dataset (i.e., microarray data). An alternative strategy to improve CNV calling can be to incorporate various types of omics datasets. Previously, software have been developed to infer CNVs from high-density DNA methylation arrays23,24 or RNA sequencing data of highly and stably expressed genes.25 While promising, none of these approaches were developed with the intent of performing scalable and reliable genome-wide CNV detection in large biobanks.
To fill this gap, we propose a method to improve the detection of false-positive CNV calls among PennCNV output by discriminating between high-quality (true) and low-quality (false) CNV regions based on multi-omics data (Figure 1A). Specifically, we checked if PennCNV calls (pCNVs) (1) are detectable by whole-genome sequencing (WGS), (2) alter the expression levels of overlapping genes in the expected direction (i.e., decreased by deletions, increased by duplications), and/or (3) alter the total methylation probe intensity of overlapping CpG sites in the expected direction. We built a predictor of CNV quality inferred from WGS, transcriptomics, and methylomics, solely based on PennCNV software output parameters in these samples assayed by multiple omics technologies. Predicted omics-informed quality scores (OQSs) distinguish high- from low-quality CNVs even in samples for which only SNV genotyping microarray data are available. We show that OQS reduces false discovery rate and improves CNV-trait association discovery compared with both raw pCNVs and cQSs19 in regions with variable CNV quality.
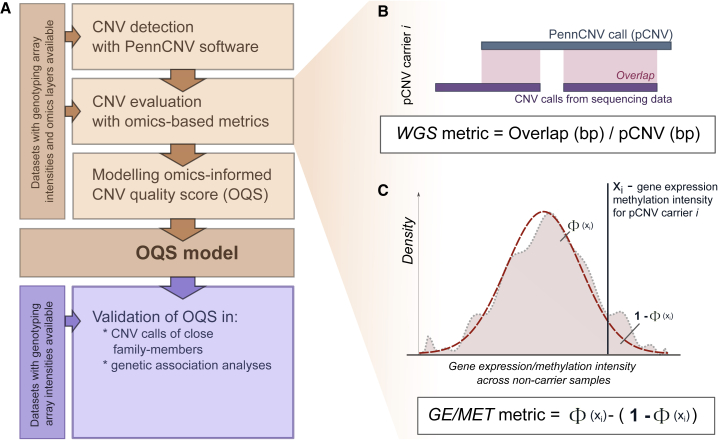
Figure 1.
Workflow overview
(A) Quality estimation and modeling pipeline for PennCNV copy-number variation calls (pCNVs).
(B and C) The pCNV quality metrics are estimated based on (B) whole-genome sequencing (WGS) data and (C) gene expression (GE) and/or overall methylation (MET) intensity of genes/CpG sites overlapping the corresponding CNV calls.
(B) WGS metric is a fraction of pCNV that can be mapped to WGS CNVs of the same individual.
(C) To calculate GE/MET metrics, the reference distribution of expression/intensity based on non-carriers (pink area) is approximated to standard normal distribution (red dashed line), and the Z score of the expression/intensity of each pCNV carrier (xi) is compared with it one at a time. The metric is a difference between the fraction of non-carriers with the corresponding value ≤xi and those with the corresponding value >xi and captures how extreme xi is compared with the reference distribution of non-carriers. In case a pCNV overlaps with several genes/CpG sites, the metric values are averaged over them.
Material and methods
Cohorts
Estonian Biobank (EstBB; data freeze January 8, 2021; Note S1; Table 1) is an Estonian population-based cohort that consists of ∼200,000 adults (≥18 years of age at recruitment).26 About 7,750 individuals are genotyped on Illumina Infinium OmniExpress-24 genotyping array (∼730,000 markers). A subset of these samples (referred to as EstBB-MO) has one or more of the following omics datasets available: 30× coverage WGS, RNA sequencing,27 and/or methylation data (Illumina Infinium Human Methylation 450 k Beadchip). Additionally, the full EstBB cohort is genotyped on Illumina Global Screening Array (GSA; ∼760,000 markers). All participants signed a broad informed consent, and analyses were carried out under ethical approval 1.1-12/624 from the Estonian Committee on Bioethics and Human Research and data release N05 from the EstBB.
Table 1.
Overview of datasets and final sample sizes used in the analyses.
| Dataset | n | Sample counts per data type |
Analysis steps |
|||||
|---|---|---|---|---|---|---|---|---|
| WGS | Methyl. | RNA-seq | Omics-based metrics calculation | Model building | Model selection and validation | CNV associations | ||
| Estonian OmniExpress sample set (N = 7,750) | ||||||||
| EstBB-MO | 1,066 | 983 | 295 | 382 | + | + | – | – |
| First-degree relatives | 504a | N/A | N/A | N/A | – | – | + | – |
| Lifelines deep (N = ∼1,500) | ||||||||
| LLDeep | 1,383 | N/A | 768 | 1,098 | + | + | – | – |
| Swiss Kidney Project on Genes in Hypertension (N = 1,128) | ||||||||
| SkiPOGH | 466 | N/A | 148 | 405 | + | – | – | – |
| Parent-child pairs | 319 | N/A | N/A | N/A | – | – | + | – |
| Estonian Biobank GSA sample set (N = ∼200,000) | ||||||||
| EstBB-GSA (unrelated) | 89,516 | N/A | N/A | N/A | – | – | – | + |
| MZ twins | 312 | N/A | N/A | N/A | – | – | + | – |
| First-degree relatives | 79,903 | N/A | N/A | N/A | – | – | + | – |
| UK Biobank (N = ∼500,000) | ||||||||
| UKB (unrelated British) | 331,522 | N/A | N/A | N/A | – | – | – | + |
| MZ twins | 302 | N/A | N/A | N/A | – | – | + | – |
| First-degree relatives | 42,032 | N/A | N/A | N/A | – | – | + | – |
N/A, not applicable.
Estonian OmniExpress first-degree relatives do not overlap with EstBB-MO samples.
LifeLines Deep (LLDeep; Note S2; Table 1) is a deeply phenotyped ∼1,500 individual subset of the Dutch population cohort LifeLines.28 LLDeep samples are genotyped on HumanCytoSNP-12 array (∼300,000 markers), and the majority of them have either RNA sequencing29 or methylation data (Illumina Infinium Human Methylation 450 k Beadchip)30 available. The LLDeep study was approved by the ethics committee of the University Medical Centre Groningen. All participants provided a written informed consent.
Swiss Kidney Project on Genes in Hypertension (SkiPOGH; Note S3; Table 1) is a Swiss family- and population-based cohort of 1,128 individuals from 273 families recruited to study the genetic determinants of blood pressure.31 The samples were genotyped on Illumina 2.5 array (∼2,500,000 markers). RNA sequencing and methylation array (Illumina Infinium Human Methylation 450 k) data were available for a subset of participants.32 The study was approved by the competent institutional ethics committees in Bern, Geneva, and Lausanne. All participants signed a written informed consent.
UK Biobank (UKB; phenotype data freeze March 22, 2018; Note S4; Table 1) is a cohort of ∼500,000 individuals from the UK.33 The majority of samples (∼450,000) are genotyped on Affymetrix UKB Axiom array, while the rest (∼50,000) are genotyped on Affymetrix UK BiLEVE Axiom array (both arrays have ∼820,000 markers). Participants signed a broad informed consent, and the data are accessed through application numbers 17085 and 16389.
Data preparation
Sample sets
We included three independent datasets—LLDeep, SkiPOGH, and a subset of Estonian samples (EstBB-MO)—in CNV quality calculations and modeling. Each of these datasets had additional omics data (WGS, methylation arrays, and/or RNA sequencing) available. The summary of PennCNV output parameters for each quality-controlled cohort are shown in Table S1. For model selection and validation steps, we extracted monozygotic (MZ) twins and first-degree relatives from the EstBB and the UKB and parent-child pairs from the SkiPOGH. Finally, we extracted unrelated quality-controlled EstBB-GSA and UKB samples for CNV association analyses. Datasets and their usage for various analyses are summarized in Table 1. Sample quality control steps are summarized in Notes S1–S4.
CNV detection
We used PennCNV18 as our main CNV detection algorithm due to its popularity (PubMed citations: PennCNV: 885; QuantiSNP:22 279; Birdsuite:34 466; September 8, 2021). We detected putative autosomal CNV regions (pCNV) for EstBB, LLDeep, SkiPOGH, and UKB datasets as previously described19 (Notes S1–S4). For each sample, we obtained the pCNV together with the values of four CNV-specific and nine sample-specific parameters described in Table S2. In all datasets, we filtered out samples with more than 200 pCNVs and with pCNVs larger than 10 Mbp, as these are likely to be either samples with poor genotyping quality or extreme cases that might distort the analysis. Additionally, we detected CNVs from EstBB WGS reads (WGS-CNVs) using the Genome STRiP35 discovery pipeline (v.2.00.1611; Note S5). All genomic coordinates are in GRCh37 build version.
Methylation and RNA sequencing data preprocessing
We obtained methylation intensities (Infinium Human Methylation 450 k Beadchip) and RNA sequencing data for EstBB-MO, LLDeep, and SkiPOGH datasets. The data preparation is described in detail in Notes S6 and S7. Briefly, where applicable, after the quality control step, we corrected for age, sex, batch, blood cell counts, and population stratification based on four principal components (PCs) calculated from pruned SNP genotypes (minor allele frequency >1%). Additionally, we corrected for PCs calculated based on methylation/gene-expression data (Figures S1 and S2). Gene-expression residuals were further corrected for independent expression quantitative trait loci (eQTL) within 500 kbp of the gene.
CNV quality metrics based on multi-omics data
WGS quality metric
WGS data were available for a subset of EstBB-MO samples (n = 979). For each pCNV in these individuals, we defined a WGS metric as the fraction of the pCNV (in bps) overlapping with WGS-CNVs in the same sample (Figure 1B). Metric calculation was restricted to pCNV deletions and duplications longer than 1 and 2 kb, respectively, as we did not detect shorter WGS-CNVs (Note S5). Additionally, these samples were used to estimate the fraction and distribution of false negative (FN) and false positive (FP) pCNVs. High-confidence FN calls were defined as WGS-CNVs (copy numbers between 0 and 4) with less than 10% bp overlap with pCNVs in the same sample. pCNVs with WGS metric <0.1 were defined as high-confidence FPs.
MET quality metric
Analogously to SNP arrays, higher or lower total signal intensity captured by methylation array at a CpG site indicates excess or lack of DNA material, respectively, regardless of the methylation status of the region. Exploiting this phenomenon, CpG site intensity data can be used to validate duplications (i.e., excess genetic material leading to increased total intensity) and deletions (i.e., reduced genetic material leading to decreased total intensity). This approach was used to assess CNV quality in EstBB-MO, LLDeep, and SkiPOGH datasets. For each methylation probe passing the preprocessing steps (Note S6), we used the samples with no pCNVs overlapping the corresponding CpG site (i.e., non-carriers) to construct the approximately Gaussian reference distribution of site overall intensity (sum of the methylated and un-methylated intensities). For each carrier, we then transformed its CpG site overall intensity into a Z score by using the mean and standard deviation of the constructed reference distribution. We denoted the quality metric based on the methylation data for the -th pCNV () across all its overlapping CpG sites as and calculated its value as
where is the total number of CpG sites overlapping , Φ is the cumulative distribution function of the standard normal distribution, and is the Z score calculated for the -th CpG site overlapping (Figure 1C). The proposed measure captures how extreme an observed methylation intensity is compared with that of the bulk of the samples (assumed to be copy neutral), equivalent to a signed two-sided tail probability. We expected for deletions and for duplications. If this was not the case, was set to zero. Finally, was converted to its absolute value such that .
Gene expression (GE) quality metric
GE levels from RNA sequencing data were used to assess CNV quality in the EstBB-MO, LLDeep, and SkiPOGH datasets. We extracted all the genic regions from Ensemble database (GRCh37) using biomaRt.36 To avoid penalizing genes whose transcript levels are not affected by CNVs, we only retain genes for which expression is positively correlated (Pearson R > 0.1) to the copy number of the gene in an independent dataset.25 After preprocessing steps (Note S7), we retained 10,786 genes. Additionally, over 80% of the genic region was required to overlap the pCNV for the gene to be included in the quality calculations of that pCNV. Requiring higher gene overlap did not show considerable improvement (Figure S3). Expression values of genes with overlap below 80% were marked as missing. Analogously to , we constructed the expression reference distribution based on non-carriers and used its mean and standard deviation to calculate an expression Z score for each carrier. We calculated , the quality metric based on GE across all its overlapping genes (analogously to the metric for methylations), as
where is the number of genes overlapping at least 80% of the and is the Z score calculated for the -th gene overlapping (Figure 1C). We set zero values for deletions with and duplications with and converted all scores to their absolute values such that . Although we hypothesized that duplications could alter GE in either direction through either triplication or disruption of gene sequence,37 this was not observed in our results (Figure S3).
Combined metric
Let us define the collection of quality metrics Further metrics can be defined by their mean, maximum, and the measure furthest away from 0.5 (i.e., most extreme; denoted as ). We chose as our final combined metric, the motivation being that if one metric clearly indicated the truth status of a pCNV, then we would use that metric (even if other metrics are unsure) (Figure S4). Note that EXTR values are only calculated for pCNVs that have at least two out of three metrics available.
CNV quality prediction models
In order to assess CNV quality in samples with no complementary omics data, we fitted prediction models for the values of the previously defined four omics metrics, summarized by .
The set of possible explanatory variables included CNV- and sample-specific parameters from PennCNV output (CNV length, number of overlapping probes, PennCNV-specific CNV confidence score, number of pCNVs per sample [and its derivations, see Figure S5], mean and standard deviation of the allelic intensity ratios and B allele frequencies of a sample, signal waviness factor; Table S2) and their interaction terms. CNV coordinates were not included as explanatory variables. We fitted a generalized linear regression model separately on each column of :
where is the -th column of (representing one of the four quality metrics), is the intercept term, and is the effect estimate of explanatory variable . We used a quasi-binomial link function since instead of being strictly binary, our response was a bimodal continuous variable ranging from zero to one.
In order to choose the best subset of , we implemented a forward stepwise model selection (starting with an empty parameter set) using custom R scripts. Briefly, in each round, a parameter was added if the resulting model minimized the 10-fold cross-validation mean square error (MSE). If adding any of the remaining parameters did not improve the average MSE, the algorithm stopped and returned the existing model. We tested model building with eight different sets of conditions/parameters to choose from and repeated the procedure separately for deletions and duplications. The modeling process and the eight parameter sets are characterized in detail in Note S8 and Table S3.
The model coefficients can be used to predict omics-informed CNV quality scores (OQSs) as
For not included in the final model, is set to be equal to zero.
Selection of the best OQS and comparison with other CNV calls
CNV quality models were fitted as described above for deletions and duplication from each multi-omics dataset separately (Table 1). To determine the best models, we incorporated family information. We reasoned that the set of familial pCNVs present in at least two close family members (Jaccard index based on overlapping bp count of at least 0.9) contains a higher fraction of true-positive CNV calls than the non-familial set (no overlap in a relative). Partially overlapping pCNVs with a Jaccard index lower than 0.9 were discarded from the calculations. We predicted and compared the OQS values of all familial and non-familial pCNVs from MZ twins from the UKB and EstBB-GSA and parent-child pairs from the SkiPOGH. To select the best models, we maximized the difference of mean OQS values between the two groups, averaged over the three datasets. We validated our best models on first-degree relative pCNVs from Estonian OmniExpress samples, EstBB-GSA and UKB. To further reduce the likelihood of two relatives carrying overlapping FP calls by chance in our validation sets, we restricted the analysis to rare (frequency <0.1%) familial pCNVs, with frequency being calculated as the fraction of samples in the full cohort with a pCNV overlapping the region in question. As a comparison, we estimated the quality of non-familial and familial pCNVs using the previously published cQSs.19
CNV association analyses
We compared OQS with raw pCNV, four previously published PennCNV output filtering approaches18,20,38,39 (Table S4), and cQS in an association analysis setting by incorporating them into the association models analogously to SNV dosages.16 We used 89,516 and 331,522 quality-controlled unrelated European individuals from the EstBB-GSA and UKB data, respectively (Notes S1 and S4). We considered 21 CNV-trait pairs (Table S5) involving four continuous anthropometric traits—body mass index (BMI), height, weight, and waist-to-hip ratio (WHR)—and 13 CNV regions that had previously16 shown association p <1 × 10−4. Importantly, these associations were obtained using cQSs in the first wave of UKB genotype data samples (n = 119,873). All phenotypes were inverse normal transformed and corrected for batch, sex, age, age,2 and PCs prior further analysis (Notes S1 and S4).
We calculated association Z statistics (estimated effect size over its standard error) and p values in a probe-by-probe manner across all probes that overlapped with >5 pCNVs inside the 13 regions of interest. The analysis was conducted separately for deletions, duplications, and mirroring effects. To model the mirroring effect (i.e., both deletions and duplications have similar effects but in opposite directions), the OQS values for deletions were negated. Associations were run using linear regression (lm function) in custom R scripts. We used a Bonferroni-corrected p value threshold of 0.05/21 = 2.38 × 10−3 to determine significance. All regions containing significantly associated probes—except for the 18q21.32 region, which in the EstBB-GSA did not contain the previously reported CNV (Figure S6)—were included in the final association comparison step.
Finally, for both datasets (i.e., UKB and EstBB-GSA) and all four phenotypes, we estimated the change in explained variance when applying the OQS model, as compared with raw PennCNV values, four filtering approaches, or cQS model. We started by clumping probes originating from significant CNV regions with using snp_clumping from the bigsnpr R package.40 Clumping usually prioritizes probes based on association summary statistics or allele frequencies, which in our case are heavily dependent on the applied CNV quality measure. To avoid any bias, we generated a random probe priority order instead. If after clumping we retained probes, we calculated
where is an array of Z statistics (of clumped probes) from association analysis using OQS, is the corresponding array from the comparison analysis (either raw PennCNV, filtering approaches, or cQS), and is a probe correlation matrix calculated based on raw pCNV. Under the null hypothesis, follows an F distribution with both degrees of freedom equal to . Since depends on the probes retained after the randomized clumping process, we repeated the random clumping 20 times and used the average F value.
Results
Omics-based metrics for CNV quality
We estimated pCNV quality based on methylation (MET), GE, and WGS (only available in EstBB-MO dataset) data, which resulted in up to three independent omics-based CNV quality metrics in three independent datasets (EstBB-MO, LLDeep, and SkiPOGH; Tables 1 and S6). Within all datasets, the metrics were positively correlated with each other (Figures 2A, 2B, S7, and S8). Both MET and GE metrics had high correlations (Pearson R ≥ 0.7) with the WGS metric. The correlations between MET and GE metrics ranged between 0.59 and 0.80 for deletions and 0.33 and 0.57 for duplications. The correlations between all three metrics and previously published cQSs19 ranged between 0.17 and 0.55 for deletions and 0.21 and 0.63 for duplications, depending on the dataset.
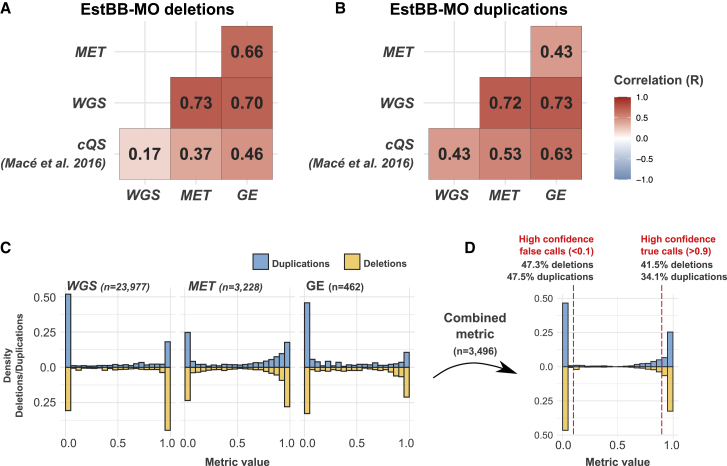
Figure 2.
Overview of CNV quality metrics in EstBB-MO
(A and B) Omics-based metrics— WGS, MET, and GE—and cQS19 Pearson correlations for EstBB-MO deletions (A) and duplication (B). Note that the number of pCNVs used in correlation calculations is not identical in each group of metric pairs (Figure S9).
(C and D) Bimodal distribution of WGS, MET, and GE metrics (C), as well as their combined metric (see material and methods) (D) for duplications (blue) and deletions (yellow). The combined metric is calculated for pCNVs that have at least two omics-based metrics available (n = 3,496) and the fractions of high-confidence false (combined metric <0.1) and true (combined metric >0.9) calls are reported.
All three metrics had bimodal distributions with modes near 0 and 1, which indicates clear differentiation between true and false calls for the majority of pCNVs (Figures 2C, S7, and S8). To retain just one quality metric per pCNV (i.e., combined metric; Figure 2D), we retained the metric that was furthest from 0.5 (denoted as EXTR in material and methods). Detailed composition of this metric is characterized in Figure S10.
We estimated the precision of pCNV based on the WGS and the combined metric. In the EstBB-MO dataset, out of 3,496 pCNVs evaluated with the combined metric (1,750 deletions, 1,746 duplications), 47.3% of deletions and 47.5% of duplications had values inferior to 0.1, most likely reflecting FP calls. In LLDeep and SkiPOGH, the percentages corresponding to high-confidence FP calls were 50.5%/28.3% and 70.9%/59.4% for deletions/duplications, respectively (Table S7). When considering a larger EstBB-MO set of 15,063 deletions and 8,914 duplications with the WGS metrics available, 31.6% (n = 4,762) of deletions and 53.2% (n = 4,745) of duplications could be labeled as FPs. These results illustrate the need for CNV quality filtering prior further analyses.
Using this EstBB-MO set, we studied the distribution of FP calls across the genome (Figure S11). Overall, 120,395 probes (17% of all Illumina OmniExpress autosomal probes, 77.5% of probes overlapping pCNV) had an FP rate (FPR) >0. There was a modest negative correlation between FPR and pCNV frequency (Pearson R = −0.12) indicating that if a pCNV is detected in multiple samples, it is more likely to be true. Still, for 939 probes, FP pCNVs were discovered in ≥10 samples (FP frequency >1% in 979 samples) with 816 (86.9%) of them having a FPR >0.9. These FP “hot spots” contributed to 5.1% (488/9,507) of all FP calls.
Additionally, we estimated the fraction of FN pCNV calls based on the overlap with WGS-CNVs to be 97.3%. When only considering CNVs that overlapped ≥3 genotyping array probes (n = 69,889), thus meeting the minimum requirement of the PennCNV discovery algorithm, this number dropped to 75.7% (66.6% for deletions only, and 84.1% for duplications only). These percentages further decreased with increasing number of overlapping probes, remaining considerably higher for duplications, compared with deletions (Figure S12). We further studied the genome-wide distribution of FNs on a restricted set on WGS-CNVs with a ≥3 probe overlap (Figure S13). We observed that despite a large fraction of FNs, only 12,065 probes (1.7% of all probes, 40.0% of probes overlapping WGS-CNVs) had an FN rate (FNR) >0. Unlike the FPR, the FNR was positively correlated to WGS-CNV frequency (Pearson R = 0.19), and CNVs with >50% frequency contributed to 31.8% (22,196/69,889) of all FN CNVs.
Prediction models for omics-informed CNV quality scores (OQSs)
We built logistic regression models to predict the previously calculated CNV quality metrics based solely on PennCNV output parameters to enable pCNV evaluation in samples lacking multi-omics measurements. Due to a smaller fraction of true-positive calls when compared with other datasets, we omitted SkiPOGH from the model-building step but retained it for model validations. Models were evaluated based on their ability to discriminate between pCNVs that were shared (familial) or not (non-familial) between MZ twins in the UKB and EstBB-GSA and parent-child pairs in the SkiPOGH (Tables S8 and S9), as pCNVs detected across multiple family members are less likely to be FPs and, thus, should act as a set of likely true-positive calls suitable for model selection and validation. For both deletions and duplications, the best model was built based on the LLDeep dataset using the combined metric. Models are characterized in Tables S10 and S11. We refer to the CNV quality measure (ranging from 0 to 1) predicted by the best models as the omics-informed CNV quality score (OQS).
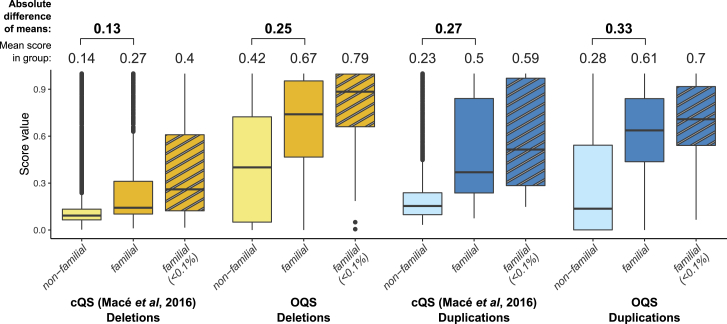
To validate the OQS, we performed a familial versus non-familial pCNV comparison on first-degree relatives from the Estonian OmniExpress, EstBB-GSA, and UKB that did not overlap with individuals used for the CNV quality estimation and model-building steps (i.e., samples with other omics data; Figures 3 and S14). The average OQS for familial calls ranged between 0.67 and 0.82 for deletions and between 0.48 and 0.70 for duplications, which was significantly higher (paired Wilcoxon test p < 1.4 × 10−21) than for cQS (0.27–0.32 in deletions and 0.42–0.53 in duplications). As some genomic regions are more prone to FP pCNVs, resulting in shared false calls between close relatives by chance, we executed a similar analysis using only rare (frequency <0.1%) familial pCNVs. This further increased the average OQS values, which ranged between 0.76 and 0.83 for deletions and between 0.56 and 0.76 for duplications. In all cases except EstBB-GSA duplications, we observed significantly higher score values for rare pCNVs with OQS compared with cQS (0.33–0.40 for deletions, 0.56–0.66 for duplications; Wilcoxon p < 0.046). Furthermore, OQS distinguished well between familial and non-familial pCNVs. The difference in OQSs between two groups were between 0.22 and 0.35 depending on the dataset (0.16–0.25 for deletions and 0.12–0.48 for duplications; Wilcoxon p < 3.0 × 10−90). Only in the case of EstBB-GSA duplications was the average difference larger with cQS.
Figure 3.
Comparison of quality scores on pCNVs of closely related Estonian samples
Consensus-based (cQS)19 and omics-informed (OQS) CNV quality scores of non-familial and familial (found in two or more family members) deletions (yellow) and duplications (blue) calculated on a subset of Estonian OmniExpress samples (n = 504; do not overlap with EstBB-MO). Familial pCNVs are likely true positives, while non-familial group contains both true and false positives. We included rare (frequency <0.1%, striped background) familiar pCNVs as a subset of CNVs less likely to validate in a relative by chance. The mean score of each pCNV group and their pairwise difference are shown on top of the figure. Compared with cQS, the OQS shows higher values for familial pCNVs and larger differences between non-familial and familial pCNV quality. All differences for both scores are significant with p < 1 × 10−16 (Wilcoxon test).
As the best models were built on the LLDeep dataset, we could use EstBB-MO for out-of-sample validations (Figure S15). We found that Pearson correlation coefficients between the combined metric and predicted OQSs were 0.70 and 0.57 for deletions and duplication, respectively. The area under the receiver operating characteristic curve (AUC) values were 0.91 for deletions and 0.87 for duplications (Figure S16).
Associations between CNV and anthropometric traits
We compared the association results obtained using raw pCNV, four quality filtering parameter sets,18,20,38,39 cQS,19 and OQS. Of 21 previously established associations between CNVs and four anthropometric traits (BMI, height, weight, and WHR),16 we replicated (p < 2.38 × 10−3) 10 in the EstBB-GSA and 18 in the UKB cohort (Tables S5, S12, and S13). For both datasets, we calculated the change in variance explained per phenotype when using OQS compared with the other six approaches.
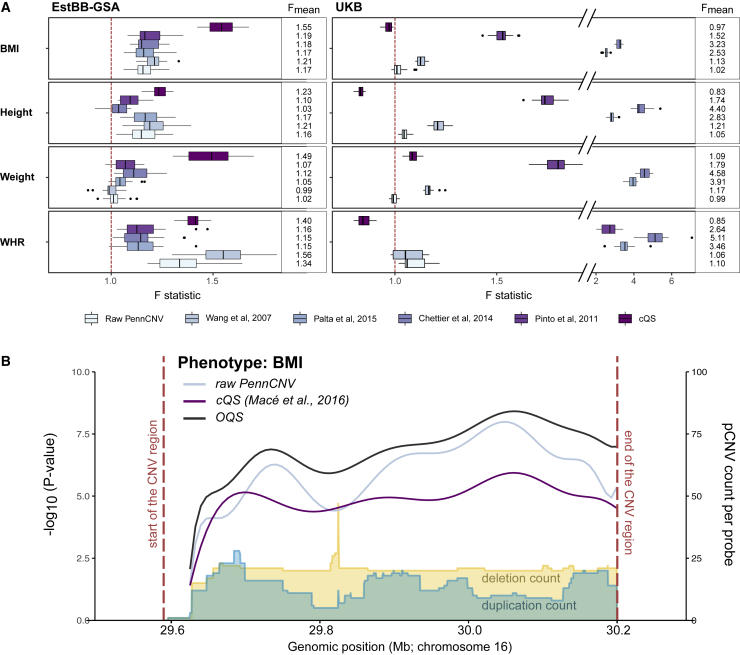
First, we tested mirror-type associations where deletions and duplications have similar effects but opposite effect directions. We found that in the EstBB-GSA, OQS led to a relative increase of 2%–34% and 23%–55% in the explained variance compared with raw PennCNV and cQS, respectively, depending on the phenotype (Figure 4A; Table S14). A good example is an association between the 16p11.2 BP4-BP5 CNV status and BMI (Figure 4B), for which alone the relative variance explained increased by 26% and 40% compared with raw PennCNV and cQS, respectively. Compared with published quality filtering approaches, the relative increase of variance explained was between 3% and 56%, depending on the approach and the phenotype (except for Wang et al.18 and weight, for which the explained variance decreased). For deletion-only associations, the relative increase was equally good, up to 33%, 42%, and 46% compared with raw PennCNV, filtering approaches, and cQS, respectively (Figure S17). For duplication-only analysis, only one BMI-associated region was included, and it showed an up to 71% relative increase in explained variance compared with the other approaches. In the UKB, OQS showed improvement compared with raw PennCNV in three out of four phenotypes and compared with conventional filtering approaches in all four phenotypes. The greatest improvements were over Palta et al.38 with >100% gain in explained variance and Chettier et al. 20 with, in some cases, even >400% gain in explained variance. However, compared with the cQS, the explained variance was decreased in most cases. This was to be expected, as the associations incorporated in this study were originally detected using the cQS in a dataset where over 60% of samples were from UKB.16 None of the changes were statistically significant, as the number of independent CNV regions per phenotype was very low, ranging from one to seven.
Figure 4.
Impact of OQS on CNV-trait associations
(A) Change of variance explained in mirror-type model when using OQS over raw PennCNV, four published quality filtering approaches,18,20,38,39 or cQS19 in the EstBB-GSA and UKB, depicted as distribution of F statistics calculated by randomizing the probe pruning priority order 20 times (see material and methods). Explained variance is increased when F >1 and decreased when F <1. Larger F values indicate greater improvement in statistical power when using OQS over the given reference approach.
(B) Locus plot of a CNV region in 16p11.2 BP4-BP5 (red dashed lines: chr16:29,590,000–30,200,000 in GRCh37) associated with BMI in EstBB-GSA dataset. The lines indicate the –log10 association p values using mirror model with raw PennCNV calls (light blue), cQS (purple), and OQS (black). The yellow and blue areas illustrate the frequency of PennCNV deletion and duplication counts, respectively, across the region.
Discussion
Genotyping microarray data are frequently used for CNV calling and analyses, but up to 48% of the calls from commonly used software, such as PennCNV, are not supported by other omics measures and are, therefore, likely FPs. To counter this, a quality score based on results overlap between three detection software tools has been developed (cQS).19 We aimed at improving the discriminatory capacity of this score by devising an omics-informed CNV quality score—OQS—that incorporates independent omics-based sources of evidence to identify high-quality PennCNV CNV calls (pCNVs).
Datasets included in the development of our OQS include GE levels from RNA sequencing (GE metric) and summed methylated and unmethylated intensities at CpG sites (MET metric), as well as CNVs detected from WGS reads (WGS metric). Each of these three approaches yielded a quality metric between 0 and 1 for every pCNV, all of which showed high concordance. We found that the correlation between WGS and the other two metrics was ≥0.7, suggesting that the use of MET and GE data for CNV quality assessment is a suitable alternative if WGS data are not available.
Still, out of three omics layers used in our study, GE is the noisiest, as the expression changes can have various biological and technical causes. We assumed deletions to always decrease and duplication to increase the expression levels. Duplications, however, can also disrupt the gene sequence, resulting in decreased expression levels instead.37 To minimize the contribution of this scenario to our analysis set, we required the gene (1) to be almost fully (>80%) covered by a CNV region and (2) to show positive correlation (Pearson R > 0.1) to its copy number in an independent study.25 Note that no significant improvement in results was observed when requesting higher gene-duplication overlap or allowing duplications to alter expression in both directions.
Interestingly, the correlation between WGS and previously published cQSs is quite low for comparison (0.17 for deletions, 0.43 for duplications), illustrating the potential benefit of incorporating omics data in CNV quality assessment compared with simple overlap between several detection software (which are all prone to the same weaknesses, such as prioritization of longer and discarding shorter CNV regions).
Using GE, MET, and WGS metrics, we built predictive models relying only on output parameters of PennCNV that allow estimating CNV quality in datasets where no additional omics data are available. Although larger omics data sizes can lead to better CNV quality models, we believe that even modest sample sizes can be used in case the assessed set of CNVs are a good representative of the final CNV set in the analysis. In our study, the best-quality models were built only on 441 pCNVs from an LLDeep dataset having both GE and MET metrics calculated.
In validation sets of close relatives, OQS clearly discriminated between familial(true positives) and non-familial pCNVs, the former being attributed to a higher OQS compared with cQS. This effect was consistent over all independent tested datasets. Based on out-of-sample AUC and correlations, predicting quality of deletions was easier than predicting that of duplications. Possible explanations include better detection of deletions by PennCNV due to larger relative difference in allelic intensity ratio between one and two copies compared with two and three copies or stronger effect of deletions on GE and MET. In a second step, we compared OQS with raw pCNV, four previously published sets of CNV quality filtering thresholds, and cQS through an association analysis exercise aiming at replicating previously established CNV-trait associations. We found that OQS systematically increases (up to 34% in the EstBB-GSA and 10% in the UKB) the amount of explained variance when compared with raw PennCNV. The increase was even higher (up to 56% in the EstBB-GSA and >400% in the UKB) when compared with sets of filtering thresholds. This indicates that, especially in the UKB, conventional filtering methods are too strict and result in a major loss of power. Compared with cQS, we observed a strong improvement in explained variance in the EstBB-GSA (up to 55%) but not in the UKB (down by 18%). As the associations we aimed at replicating were originally detected in the UKB using cQS approach, cQS-based associations suffer from winner’s curse, which distorts the effect magnitudes in favor of the cQS. Alternatively, different quality scores might perform better in different datasets, and combining the two might be a good option (e.g., by incorporating the maximum of the two scores in the analyses).
It is to be expected that the improvements offered by the OQS is small when studying strong associations in a well-known and -detectable genomic region, as we have done. We expect to see greater improvement in intermediate-quality CNV regions for which previous studies have lacked statistical power for CNV-trait association detection. As observed, when excluding a small number of FP hot-spot regions, false pCNVs are distributed randomly and uniformly across most of the genome and, thus, only introduce a modest amount of noise per probe/region. However, given the difficulty to detect CNVs, which themselves tend to be rare, even slight improvement in statistical power to detect CNV associations can be beneficial.
Although we strived to optimize our models for different genotyping array types and densities, our results may still be specific to the arrays we explored. Furthermore, while the OQS helps to reduce FP calls, it does not improve the FNR, which remains high, especially for shorter CNVs and CNVs in regions with low array probe density. Still, unlike FP, FN load mainly originates from a few specific high-frequency CNV regions.
Using only PennCNV output parameters as predictors is a limiting factor in itself, as their prediction ability can vary from dataset to dataset. Furthermore, the PennCNV detection algorithm considers each sample separately and does not exploit between-sample similarities, which was shown to improve the detection of short (and frequent) CNVs considerably.15 Still, our omics-informed CNV quality assessment approach is not limited to PennCNV but can be used with any CNV detection method that produces multiple output parameters.
In conclusion, we developed a modular and customizable omics-based quality score framework that can be used for both genome-wide and smaller-scale CNV analyses. The OQS developed in the current study is independent of CNV coordinates or genome build and can be applied directly to filter out high-confidence FP pCNVs using a hard cutoff (i.e., OQS <0.5) or plugged into dosage-based association models, eliminating the need for an arbitrary CNV quality threshold. In turn, lower FNRs increase statistical power to detect associations between CNVs and complex traits. Alternatively, with at least one suitable multi-omics measurement available for a subset of the samples, researchers can use our framework to build their own custom models, which could be applied to any CNV detection software, leading to further improved results for follow-up analyses.
Data and code availability
Access to the UKB Resource is available by application (http://www.ukbiobank.ac.uk/). LLDeep RNA sequencing and MET data can be accessed via European Genome-Phenome Archive (accession code EGAS00001001077). EstBB, SkiPOGH, and LLDeep individual-level data are available upon request. Custom R code and Genome STRiP pipeline commands are available on GitHub: https://github.com/maarjl/CNV_OQS.
Consortia
The members of Estonian Biobank Research Team are Andres Metspalu, Tõnu Esko, Mari Nelis, and Lili Milani.
Acknowledgments
We thank participants of EstBB, UKB, SkiPOGH, and LLDeep for their data provision. The scholarships for PhD student mobility of M.L. and K.L. to work with SkiPOGH (University of Lausanne, Switzerland) and LLDeep (University of Groningen, the Netherlands) data and funding for EstBB data acquisition were provided by the European Regional Development Fund (project nos. 2014-2020.4.01.16-0125 and 2014-2020.4.01.15-0012 and EXCITE). This project has benefited from the funding of European Union’s Horizon 2020 Research and Innovation Programme under grant agreement nos. 101016775 and 633666 and of the Estonian Research Council grant PUT (PRG687, PRG555, PRG1095, and PUT JD817). Z.K. was supported by the Department of Computational Biology at the University of Lausanne. M.N. was supported by Jacobs Foundation Research Fellowship (grant 2016 1217 09, to Dr. Katrin Männik, Health 2030 Genome Center, Geneva, Switzerland). The SKIPOGH study was supported by grants from the Swiss National Science Foundation (33CM30-124087 and 33CM30-140331). EstBB and part of the UKB computations were carried out on the High-Performance Computing (HPC) Center, University of Tartu. UKB association study was carried out on the JURA server, University of Lausanne. SkiPOGH computations were carried out on the HPC1 computational server of the University Hospital of Lausanne. We thank UMCG Genomics Coordination Center, the UG Center for Information Technology, and their sponsors BBMRI-NL and TarGet for storage and compute infrastructure for LLDeep data. Finally, we thank Natàlia Pujol Gualdo and Triin Laisk for critical reading of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2022.100133.
Contributor Information
Maarja Lepamets, Email: maarja.lepamets@ut.ee.
Zoltán Kutalik, Email: zoltan.kutalik@unil.ch.
Web Resources
CNVPartition, https://support.illumina.com/downloads/genomestudio-2-0-plug-ins.html.
Genome STRiP, https://software.broadinstitute.org/software/genomestrip/.
QTLtools v.1.1, https://qtltools.github.io/qtltools/.
Supplemental information
References
- 1.Zarrei M., MacDonald J.R., Merico D., Scherer S.W. A copy number variation map of the human genome. Nat. Rev. Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 2.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S., et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall C.R., Howrigan D.P., Merico D., Thiruvahindrapuram B., Wu W., Greer D.S., Antaki D., Shetty A., Holmans P.A., Pinto D., et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41, 321 subjects. Nat. Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman J., Rees E., Harold D., Ivanov D., Gerrish A., Sims R., Hollingworth P., Stretton A., GERAD1 Consortium. Holmans P., et al. A genome-wide study shows a limited contribution of rare copy number variants to Alzheimer’s disease risk. Hum. Mol. Genet. 2013;22:816–824. doi: 10.1093/hmg/dds476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentile G., La Cognata V., Cavallaro S. The contribution of CNVs to the most common aging-related neurodegenerative diseases. Aging Clin. Exp. Res. 2021;33:1187–1195. doi: 10.1007/s40520-020-01485-4. [DOI] [PubMed] [Google Scholar]
- 6.Shlien A., Malkin D. Copy number variations and cancer. Genome Med. 2009;1:62. doi: 10.1186/gm62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Männik K., Mägi R., Macé A., Cole B., Guyatt A.L., Shihab H.A., Maillard A.M., Alavere H., Kolk A., Reigo A., et al. Copy number variations and cognitive phenotypes in unselected populations. JAMA. 2015;313:2044–2054. doi: 10.1001/jama.2015.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huguet G., Schramm C., Douard E., Jiang L., Labbe A., Tihy F., Mathonnet G., Nizard S., Lemyre E., Mathieu A., et al. Measuring and estimating the effect sizes of copy number variants on general intelligence in community-based samples. JAMA Psychiatr. 2018;75:447–457. doi: 10.1001/jamapsychiatry.2018.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford K., Bracher-Smith M., Owen D., Kendall K.M., Rees E., Pardiñas A.F., Einon M., Escott-Price V., Walters J.T.R., O’Donovan M.C., et al. Medical consequences of pathogenic CNVs in adults: analysis of the UK Biobank. J. Med. Genet. 2019;56:131–138. doi: 10.1136/jmedgenet-2018-105477. [DOI] [PubMed] [Google Scholar]
- 11.Auwerx C., Lepamets M., Sadler M.C., Patxot M., Stojanov M., Baud D., Mägi R., Porcu E., Reymond A., Kutalik Z. The individual and global impact of copy number variants on complex human traits. medRxiv. 2021 doi: 10.1101/2021.08.10.21261839. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguirre M., Rivas M.A., Priest J. Phenome-wide burden of copy-number variation in the UK biobank. Am. J. Hum. Genet. 2019;105:373–383. doi: 10.1016/j.ajhg.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y.R., Glessner J.T., Coe B.P., Li J., Mohebnasab M., Chang X., Connolly J., Kao C., Wei Z., Bradfield J., et al. Rare copy number variants in over 100, 000 European ancestry subjects reveal multiple disease associations. Nat. Commun. 2020;11:255. doi: 10.1038/s41467-019-13624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinnott-Armstrong N., Tanigawa Y., Amar D., Mars N., Benner C., Aguirre M., Venkataraman G.R., Wainberg M., Ollila H.M., Kiiskinen T., et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 2021;53:185–194. doi: 10.1038/s41588-020-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hujoel M.L.A., Sherman M.A., Barton A.R., Mukamel R.E. Influences of rare copy number variation on human complex traits. bioRxiv. 2021 doi: 10.1101/2021.10.21.465308. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macé A., Tuke M.A., Deelen P., Kristiansson K., Mattsson H., Nõukas M., Sapkota Y., Schick U., Porcu E., Rüeger S., et al. CNV-association meta-analysis in 191, 161 European adults reveals new loci associated with anthropometric traits. Nat. Commun. 2017;8:744. doi: 10.1038/s41467-017-00556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dellinger A.E., Saw S.-M., Goh L.K., Seielstad M., Young T.L., Li Y.-J. Comparative analyses of seven algorithms for copy number variant identification from single nucleotide polymorphism arrays. Nucleic Acids Res. 2010;38:e105. doi: 10.1093/nar/gkq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F.A., Hakonarson H., Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macé A., Tuke M.A., Beckmann J.S., Lin L., Jacquemont S., Weedon M.N., Reymond A., Kutalik Z. New quality measure for SNP array based CNV detection. Bioinformatics. 2016;32:3298–3305. doi: 10.1093/bioinformatics/btw477. [DOI] [PubMed] [Google Scholar]
- 20.Chettier R., Ward K., Albertsen H.M. Endometriosis is associated with rare copy number variants. PLoS One. 2014;9:e103968. doi: 10.1371/journal.pone.0103968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kendall K.M., Rees E., Escott-Price V., Einon M., Thomas R., Hewitt J., O’Donovan M.C., Owen M.J., Walters J.T.R., Kirov G. Cognitive performance among carriers of pathogenic copy number variants: analysis of 152, 000 UK biobank subjects. Biol. Psychiatr. 2017;82:103–110. doi: 10.1016/j.biopsych.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Colella S., Yau C., Taylor J.M., Mirza G., Butler H., Clouston P., Bassett A.S., Seller A., Holmes C.C., Ragoussis J. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feber A., Guilhamon P., Lechner M., Fenton T., Wilson G.A., Thirlwell C., Morris T.J., Flanagan A.M., Teschendorff A.E., Kelly J.D., Beck S. Using high-density DNA methylation arrays to profile copy number alterations. Genome Biol. 2014;15:R30. doi: 10.1186/gb-2014-15-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marzouka N.-A.-D., Nordlund J., Bäcklin C.L., Lönnerholm G., Syvänen A.C., Carlsson Almlöf J. CopyNumber450kCancer: baseline correction for accurate copy number calling from the 450k methylation array. Bioinformatics. 2016;32:1080–1082. doi: 10.1093/bioinformatics/btv652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talevich E., Hunter Shain A. 2018. CNVkit-RNA: Copy number inference from RNA-Sequencing data. [Google Scholar]
- 26.Leitsalu L., Haller T., Esko T., Tammesoo M.-L., Alavere H., Snieder H., Perola M., Ng P.C., Mägi R., Milani L., et al. Cohort profile: Estonian biobank of the Estonian genome center, university of Tartu. Int. J. Epidemiol. 2015;44:1137–1147. doi: 10.1093/ije/dyt268. [DOI] [PubMed] [Google Scholar]
- 27.Lepik K., Annilo T., Kukuškina V., eQTLGen Consortium. Kisand K., Kutalik Z., Peterson P., Peterson H. C-reactive protein upregulates the whole blood expression of CD59 - an integrative analysis. PLoS Comput. Biol. 2017;13:e1005766. doi: 10.1371/journal.pcbi.1005766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tigchelaar E.F., Zhernakova A., Dekens J.A.M., Hermes G., Baranska A., Mujagic Z., Swertz M.A., Muñoz A.M., Deelen P., Cénit M.C., et al. Cohort profile: LifeLines DEEP, a prospective, general population cohort study in the northern Netherlands: study design and baseline characteristics. BMJ Open. 2015;5:e006772. doi: 10.1136/bmjopen-2014-006772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhernakova D.V., Deelen P., Vermaat M., van Iterson M., van Galen M., Arindrarto W., van ’t Hof P., Mei H., van Dijk F., Westra H.-J., et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 2017;49:139–145. doi: 10.1038/ng.3737. [DOI] [PubMed] [Google Scholar]
- 30.Bonder M.J., Luijk R., Zhernakova D.V., Moed M., Deelen P., Vermaat M., van Iterson M., van Dijk F., van Galen M., Bot J., et al. Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 2017;49:131–138. doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 31.Alwan H., Pruijm M., Ponte B., Ackermann D., Guessous I., Ehret G., Staessen J.A., Asayama K., Vuistiner P., Younes S.E., et al. Epidemiology of masked and white-coat hypertension: the family-based SKIPOGH study. PLoS One. 2014;9:e92522. doi: 10.1371/journal.pone.0092522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmeli C., Kutalik Z., Mishra P.P., Porcu E., Delpierre C., Delaneau O., Kelly-Irving M., Bochud M., Dhayat N.A., Ponte B., et al. Gene regulation contributes to explain the impact of early life socioeconomic disadvantage on adult inflammatory levels in two cohort studies. Sci. Rep. 2021;11:3100. doi: 10.1038/s41598-021-82714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korn J.M., Kuruvilla F.G., McCarroll S.A., Wysoker A., Nemesh J., Cawley S., Hubbell E., Veitch J., Collins P.J., Darvishi K., et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Handsaker R.E., Van Doren V., Berman J.R., Genovese G., Kashin S., Boettger L.M., McCarroll S.A. Large multiallelic copy number variations in humans. Nat. Genet. 2015;47:296–303. doi: 10.1038/ng.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 37.Newman S., Hermetz K.E., Weckselblatt B., Rudd M.K. Next-generation sequencing of duplication CNVs reveals that most are tandem and some create fusion genes at breakpoints. Am. J. Hum. Genet. 2015;96:208–220. doi: 10.1016/j.ajhg.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palta P., Kaplinski L., Nagirnaja L., Veidenberg A., Möls M., Nelis M., Esko T., Metspalu A., Laan M., Remm M. Haplotype phasing and inheritance of copy number variants in nuclear families. PLoS One. 2015;10:e0122713. doi: 10.1371/journal.pone.0122713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinto D., Darvishi K., Shi X., Rajan D., Rigler D., Fitzgerald T., Lionel A.C., Thiruvahindrapuram B., Macdonald J.R., Mills R., et al. Comprehensive assessment of array-based platforms and calling algorithms for detection of copy number variants. Nat. Biotechnol. 2011;29:512–520. doi: 10.1038/nbt.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Privé F., Aschard H., Ziyatdinov A., Blum M.G.B. Efficient analysis of large-scale genome-wide data with two R packages: bigstatsr and bigsnpr. Bioinformatics. 2018;34:2781–2787. doi: 10.1093/bioinformatics/bty185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Access to the UKB Resource is available by application (http://www.ukbiobank.ac.uk/). LLDeep RNA sequencing and MET data can be accessed via European Genome-Phenome Archive (accession code EGAS00001001077). EstBB, SkiPOGH, and LLDeep individual-level data are available upon request. Custom R code and Genome STRiP pipeline commands are available on GitHub: https://github.com/maarjl/CNV_OQS.