Abstract
Autophagy is a vital cellular process whose role in T immune cells is poorly understood, specifically, in its regulation of allo-immunity. Stimulation of wild type T cells in vitro and in vivo with allo-antigens enhances autophagy. To assess the relevance of autophagy to T cell allo-immunity, we generated T cell specific ATG5 knock-out mice. Deficiency of ATG5 dependent autophagy reduced T cell proliferation, increased apoptosis following in vitro and in vivo allo-stimulation. The absence of ATG5 in allo-stimulated T cells enhanced their ability to release effector cytokines and cytotoxic functions, uncoupling their proliferation and effector functions. Absence of autophagy reduced intracellular degradation of cytotoxic enzymes such as granzyme B, thus enhancing the cytotoxicity of T cells. In several in vivo models of allo-HSCT, ATG5-dependent dissociation of T cell functions contributed to significant reduction in graft-versus-host disease (GVHD) but retained sufficient graft versus tumor (GVT) response. Our findings demonstrate that ATG5 dependent autophagy uncouples T cell proliferation from its effector functions and offers a potential new strategy to enhance outcomes after allo-HSCT.
Keywords: cytotoxicity, graft-versus-host disease, ATG5, graft-versus-leukemia
Introduction
Macroautophagy is an adaptive response that permits cell response to stress and is involved in cellular differentiation, programed cell death. Its dysfunction is associated with a broad range of diseases 1, 2. During autophagy, cells engulf intracellular organelles, cytoplasmic aggregates and proteins in a double-membrane vesicle called an autophagosome, that subsequently fuse with the lysosome and promotes their degradation and recycling 3, 4. ATG5 plays a key role in this process of autophagy and is a critical component of the membrane extension process of the autophagosome. It is conjugated to ATG12 in a complex that catalyzes the conjugation of microtubule-associated protein 1 light chain 3 (LC3–1) to phosphatidylethanolamine (LC3-PE or LC3-II) on the extension membrane 5, 6, 7, 8, 9, 10. Mice deficient in ATG5 have been shown to succumb to neuronal dysfunction 11 in vivo, but its role in immunity and specifically with in vivo T cell responses to clinically relevant antigens remain unknown. Several other components of the autophagic machinery, besides ATG5, have been shown to affect T cell proliferation, cytokine production, and memory cell generation 5. Whether they differentially affect the various responses, and the impact on in vivo clinically relevant non-infectious immunity remains unknown.
Allogeneic hematopoietic cell transplantation (allo-HCT) is a curative therapy against hematological malignancies. The curative potential is realized by the donor T cell mediated graft versus tumor (GVT) responses after allo-HCT. However, the concomitant occurrence of T cell dependent graft-versus-host disease (GVHD), the major complication of allo-HCT, has precluded its efficacy. Strategies that can reduce GVHD while persevering sufficient GVT would be a major advance in the field. Donor T cell responses that cause GVHD and GVT share critical pathways including shared antigens, proliferation and effector functions. However, whether uncoupling these shared responses, such as proliferation versus cytotoxic effector functions, might provide a window of opportunity to meaningfully separate GVHD from GVT remains unclear.
Herein we show that allo-stimulation promotes autophagic response in T cells, in vitro and in vivo. Deficiency of ATG5 in donor T cells diminished induction of autophagy, decreased proliferation, increased effector cytokine and CTL functions after in vitro and in vivo allo-stimulation. Mechanistic studies suggest that the lack of degradation of the CD8+ cytotoxic molecules such as granzyme B in the autophagosome enhanced the cytotoxicity of the ATG5−/− CD8+ T cells. This uncoupling of the proliferation from effector responses reduced GVHD but still retained sufficient GVT responses.
Material and methods
Reagents
Rapamycin (MilliporeSigma, St. Louis, MO) was reconstituted in dimethylsulfoxide (DMSO) at a concentration of 100 mM, further dilutions were made in RPMI media. Chloroquine diphosphate salt (MilliporeSigma, St. Louis, MO) was reconstituted in PBS at a concentration of 100 mM, further dilutions were made in RPMI media. 3-Methyladenine (3-MA) (MilliporeSigma, St. Louis, MO) was reconstituted in DMSO at a concentration of 100 mM with further dilutions made in RPMI media.
Mice
C57BL/6 (B6, H-2b) and BALB/c (H-2d) mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) and Charles River Laboratories (Wilmington, MA, USA). Previously described 29 15 B6-background ATG5flox/flox mice (B6.129S-ATG5<tm1Myok>, #RBRC02975, Riken RBC, Ibaraki, Japan) were bred to Lck-Cre (B6 Cg-Tg(Lck-Cre)1Cwi N9) transgenic mice (Taconic, #004197, Hudson, New York). All animals were cared for according to regulations reviewed and approved by the University Committee on Use and Care of Animals of the University of Michigan, based on University Laboratory Animal Medicine guidelines.
BM transplantation (BMT)
BMTs were performed as previously described 35 36. Briefly, splenic T cells from donors were enriched using the Pan T Cell Isolation Kit II and manual MACS separation with LS columns (Miltenyi Biotec Inc., San Diego, CA). BM was depleted of T cells by manual MACS separation utilizing CD90.2 microbeads (Miltenyi Biotec). We used the well-established MHC-mismatched B6→BALB/c BMT model 37. BALB/c animals were used as recipients and received 8Gy (137Cs source) on day −1 and then 0.5×106 CD90.2+ T cells along with 5×106 T cell-depleted BM (TCD-BM) cells from either syngeneic (BALB/c) or allogeneic (B6 or ATG5flox/flox Lck-Cre+) donors on day 0.
Systemic analysis of GVHD
We monitored survival of transplanted animals daily and assessed the degree of clinical GVHD weekly, as described previously 38. (Supplemental Table 1)
Human T-cell isolation
Human peripheral blood was obtained via routine venipuncture according to IRB protocol UMCC 2001.0234 (HUM00043287), Long-Term Evaluation of the Biology and Outcomes of Hematopoietic Stem Cell Transplantation, which collects clinical data and blood samples serially throughout the transplant course as well as at the time of significant clinical complications, such as GVHD, at our center. Alternatively, heparinized whole blood was purchased from Innovative Research (Novi, MI). Peripheral blood mononuclear cells (PBMC) were purified from heparinized whole blood via density centrifugation with Ficoll-Paque Premium (GE Healthcare) per manufacturer’s recommendations. PBMCs were then treated with red blood cell (RBC) lysis buffer (MilliporeSigma) per manufacturer’s instructions, and cryopreserved in heat-shocked fetal calf serum (Gibco) containing 10% DMSO (MilliporeSigma). PBMCs were thawed in a 37° C water bath and immediately mixed with complete cell media (RPMI 1640, 10% HS-FBS, penicillin, streptomycin, L-glutamine) supplemented with 50 U/mL of Benzonase (EMDMillipore). Cells were rinsed once with media and then T-cells were isolated via negative magnetic selection with pan T-cell kits according to the manufacturer’s protocol (Miltenyi Biotec).
Human In vitro proliferation assays
For mixed lymphocyte reaction (MLR), with allogeneic activation, human T-cells (2×105/200μl/well) were co-cultured with an equal amount of lethally irradiated (30 Gy) allogeneic monocytes (pool of 2–3 unrelated donors) derived from the cell fraction bound to the Pan T cell negative selection column. Human MLRs were incubated for 6–7 days in complete cell media (RPMI 1640, 10% heat-shocked FBS, penicillin, streptomycin, L-glutamine, 2-mercaptoethanol). For nonspecific TCR stimulation, T cells (2×105/200μl/well) were cultured with soluble 2μg/ml anti-CD28 (clone: 37.51) and plate-bound 10μg/ml anti-CD3ε (clone: 145–2C11) purified anti-mouse antibodies (BioLegend) in 96-well flat bottom plate for 48h. To assess T-cell proliferation, incorporation of 3H-thymidine (1 μCi/96 well) during the final 6 hours (CD3/CD28 stimulation) or 15 hours (allogeneic stimulation) of culture was measured by a TopCount (PerkinElmer)
Mouse In vitro proliferation assays
For in vitro experiments comparing B6-WT and B6-ATG5−/− T cells, T cells were first isolated from the spleens using the Pan T Cell Isolation Kit II (Miltenyi Biotec) with manual MACS separation and LS columns. To increase the purity to >95%, T cells were then positively sorted with a CD90.2-PerCP/Cy5.5 and negatively for CD25 antibody (Biolegend, San Diego, CA) with MoFlo Astrios or Synergy Head cell sorters. For nonspecific TCR stimulation, sorted T cells (2×105/200μl/well) were cultured with soluble 2μg/ml anti-CD28 (clone: 37.51) and plate-bound 10μg/ml anti-CD3ε (clone: 145–2C11) purified anti-mouse antibodies (BioLegend) in 96-well flat bottom plate for 48h or 60h. For mixed lymphocyte reaction (MLR), 2×105 ATG5−/− and WT-B6 T cells (2×105/100 μl/well) were co-cultured at a 20:1 ratio with bone marrow-derived dendritic cells (BMDCs) from syngeneic B6 or allogeneic BALB/c mice (1×104/100 μl/well) in 96-well flat bottom plate for 4 or 6 days. To obtain BMDCs, BM cells were cultured with murine recombinant GM-CSF (20ng/ml; PeproTech Inc., Rocky Mill, NJ) for 7 days, then DCs were purified using CD11c microbeads and the autoMACS separator (Miltenyi Biotec). Before being put in culture, BMDCs were irradiated with 20Gy. The incorporation of 3H-thymidine (1μCi/well; PerkinElmer, Waltham, MA) by proliferating T cells during the final 6 hours (CD3/28 stimulation) or 16 hours (MLR) of culture was measured by a β-plate reader TopCount (PerkinElmer).
Electron Microscopy
Mice underwent normal transplant procedures with 1×106 C57BL/6 90.2 donor T cells and 5.0×106 TCD bone morrow into C57BL/6 or Balb/c recipients. 12–14 days’ post-transplant the recipient mice were euthanized and spleens collected. T cells were purified from either C57BL/6 syngeneic or Balb/c allogeneic mice. After initial purification, donor T cells were then isolated with H-2Kb beads (Miltenyi Biotec) from allogeneic recipients. Donor T cell samples were pooled, pelleted and then washed with PBS. Cells were pelleted and fixed with glutaraldehyde and taken to the Biomedical Research Core Facilities Microscopy Core for processing and sectioning onto grids.
Cytotoxic T Lymphocyte (CTL) assay
2×106cells/ml/well purified T cells from B6-WT and B6-ATG5−/− spleens were incubated with 5×106cells/ml/well whole splenocytes previously irradiated with 30Gy in a bulk MLR in 24-well plate to produce activated CD8+ effector T cells. At day 6, cells were harvested, analyzed by flow cytometry for the percentage of CD4 and CD8+ cells and re-suspend at a concentration of 4×106 CD8+ T cells/ml. P815 (H-2d) mastocytoma cell line derived from a DBA/2 mouse was used as target. Briefly, 2×106 tumor cells were re-suspended in 200 μl RPMI with 10% FCS and incubated for 2h with 12μCi 51Cr (PerkinElmer). After being washed 3 times, 4000 tumor cells were incubated in 96-well round-bottom plate with activated CD8+ T cells at effector-to-target ratios from 100:1 to 12.5:1 in quadruplicates. Supernatants were harvested after 4h and 51Cr activity was determined in an autogamma counter (Packard, Meridian, CT, USA). Maximal and background release were determined by the addition of 4% Triton X-100 or media alone to targets, respectively. The percentage of specific lysis was calculated as following: 100x (sample count-background count)/(maximal count-background count).
Carboxyfluorescein diacetate succinimidyl ester (CFSE) and apoptosis analysis
Purified T cells were washed and suspended at a concentration of 1×106 cells/ml in phosphate-buffered saline (PBS) containing 0.5% bovine serum albumin (BSA) and 2mM EDTA. CFSE (Thermo Scientific, Rockford, IL) in dimethylsulfoxide (DMSO) (5mM) was added to the cell suspension at a final concentration of 2μM and incubated at 37°C for 10 minutes. The staining was quenched by the addition of ice-cold Roswell Park Memorial Institute (RPMI) medium containing 10% fetal calf serum (FCS). The cells were then washed according to the manufacturer’s instructions and used for the proliferation studies. At the end of the proliferation, T cells were stained with a PE-conjugated anti-Annexin-V antibody (BD Biosciences) in the dark for 10 minutes at room temperature in labeling buffer and analyzed using Accuri C6 cytometer (Accuri, Ann Arbor, MI, USA).
FACS analysis
For the staining of LC3, purified T cells previously marked with CFSE were harvested after stimulation, washed twice with FACS wash buffer (0.2% BSA in PBS), fixed with 1x BD FACSTM Lysing Solution (BD Biosciences, San Jose, CA) and permeabilized for 10 min at 4°C with 1x Permeabilization Buffer (eBioscience, San Diego, CA) in the presence of 1:400 rat anti-mouse FcR mAb 2.4G2 to block nonspecific FcR binding of labeled antibodies. Primary rabbit-LC3 antibody (Novus Biological) diluted 1:200 in Permeabilization Buffer was incubated for 30 minutes at 4°C, cells were washed once and secondary anti-rabbit Alexa Fluor®633 goat anti-rabbit IgG antibody (Thermo Scientific, Rockford, IL) was also incubated for 30 minutes at 4°C in Permeabilization Buffer at the same concentration. Cells were then re-suspended in FACS wash buffer and analyzed using Accuri C6 cytometer. In assessing autophagy in unstimulated and stimulated human T cells, the CYTO-ID® Autophagy Detection Kit 2.0 (Enzo Life Sciences, Farmingdale, NY) was used for flow analysis using the Attune NXT cytometer (Thermo Scientific) For other staining experiments, see the Supplemental Methods.
Confocal Microscopy Analysis
Acquisition of all samples were stained using identical conditions and mounted in Prolong Gold (RI=1.5). Fluorescence was imaged onto GaAsP detectors using a Nikon A1 confocal scan head and 60× 1.4NA oil objective. Identical image acquisition settings were used for all samples. Image Analysis for ‘Co-localization’ was quantified using Mander’s Overlap Coefficients, k1 and k2 (Manders, J Microscopy, 1993). This approach was chosen because we hypothesized that the amount of overlap in the spatial distribution of protein concentrations would change due to treatment. Calculations were performed on z-stack images using the FIJI JACoP plugin (Bolte, J Microscopy, 2006). Background pixels were excluded from the calculation based on a manual threshold. The signals were quantified using ImageJ software with a JaCoP plugin. The JaCoP plugin takes two signals A (eGFP) and B (RFP) that it labels as ‘Image A’ and ‘Image B’. The interpretation of k1 is then “the amount (measured as intensity) of Signal B that overlaps with (occupies sample pixels) Signal A”. k2 is the amount of Signal A that overlaps with Signal B.
Cytokine enzyme-linked immunosorbent assay (ELISA)
Concentrations of IFNγ, IL-2 and IL-17α were measured in the culture supernatants or in the serum by ELISA with specific anti-mouse MoAbs for capture and detection utilizing OptEIA™ (IFNγ, IL-2; BD Biosciences), ELISA MAX™ (IL-17α; Biolegend). Assays were performed according to the manufacturer’s protocol and read at 450 nm using a microplate reader (Model 3550; Bio-Rad Labs, Hercules, CA).
Western blotting
T cells, unstimulated or stimulated for 24 hours (mouse) or 48 hours (human) with anti-CD3 (4μg/ml) and CD28 (2μg/ml) with or without chloroquine (10μM) or 3-MA (5 mM) were harvested and washed twice with PBS. Then whole cells were lysed in RIPA lysis extraction buffer (Thermo Scientific). Equal amounts of protein lysates quantified with the BCA Protein Assay Kit (Thermo Scientific) were separated with 10% or 12.5% SDS-PAGE. Rabbit-LC3 antibody (Novus Biologicals, Littelton, CO) was used according to manufacturer’s instructions. Secondary antibody conjugated to horseradish peroxidase (Jackson ImmunoResearch, West Grove, PA) was used to detect the primary antibody. Densitometric analysis was performed using ImageJ software.
Statistical analysis
Survival curves were analyzed with the Wilcoxon test. An unpaired T test was performed for the statistical analysis of all other data. A P value <.05 was considered to be statistically significant (*P<.05, **P<.01, ***P<.001, ****P<.0001).
Results
Allo-stimulation induces autophagy in conventional T cells (Tcon)
We first analyzed T cells for induction of autophagy following allo-stimulation. Purified splenic T (Tcon) cells from C57BL/6 (B6-WT) mice were harvested and stimulated for 4 days with irradiated splenic cells from an allogeneic BALB/c mouse. Unstimulated B6-WT Tcon cells, rapamycin (Rapa; induces autophagy), hydroxy-chloroquine (CQ; autophagy inhibitor) and 3-Methyladenine (3-MA) treatments were utilized as controls. The inhibitors, CQ and 3-MA, regulate distinct stages of autophagy (Figure S1a). Specifically, CQ is commonly used as a positive control for LC3-II staining because it blocks formation of the autophosomes when the conversion to LC3-II has already taken place and autophagosomes accumulate in the cell (Figure S1b), while 3-MA blocks autophagy proximally, thus preventing formation of LC3-II and leading to reduction in both LC3-II and LC3-I. This accumulation of autophagosomes can be observed with CQ inhibiting autophagy during B6-WT Tcon cell stimulation with allogeneic BALB/c splenocytes by EM (Figure S1c).
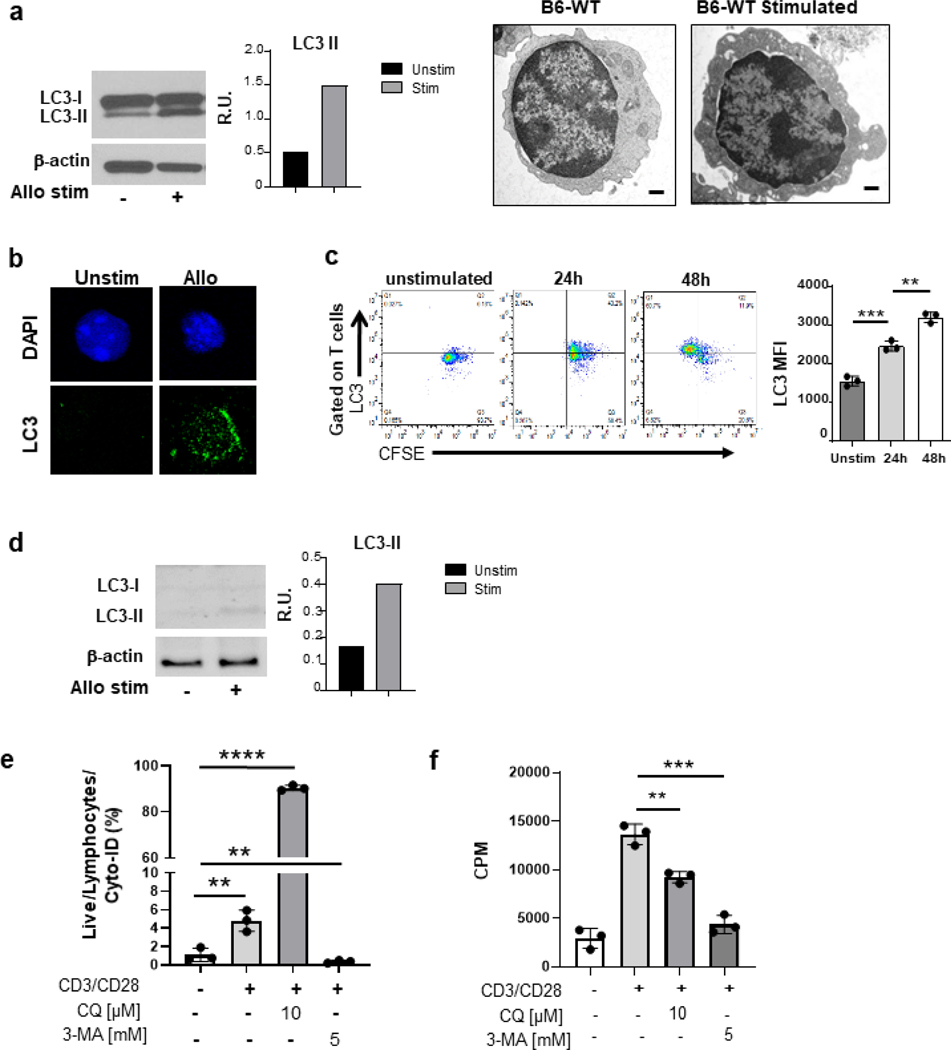
B6-WT Tcon cells activated by allo-BALB/c cells demonstrated greater conversion to the autophagosome-bound LC3-II form when compared with controls (right panel) and seen visually by EM of increased cytoplasmic activity of activated cells (left panel) (Figure 1a). This increase in LC3-II observed in mouse allogeneic stimulated Tcon cells was further confirmed by confocal microscopy (Figure 1b). The increase in autophagy in Tcon cells increased in a time dependent manner (Figure 1c). A similar trend was observed when B6-WT Tcon cells were stimulated with anti-CD3/CD28 antibodies, consistent with previous reports12 (Figure S1d). We next determined whether inhibition of autophagy in Tcon cells alters their morphological and functional response to allo-stimulation. As shown in Figure S1e, blocking autophagy with increased doses of CQ reduced Tcon cell proliferation in a dose and time dependent manner while showing a reciprocal increase in apoptosis. (Figure S1f).
Fig. 1. Autophagy is induced following allogeneic TCR stimulation in vitro.
Whole-cell lysates were prepared from purified T cells after stimulation for 4–6 days in a mixed lymphocytic reaction (MLR) with allogeneic irradiated whole mouse splenocytes or human T cell depleted PBMC’s (a) After 4 days of allogenic stimulation, Tcon and activated T cells were processed for electron microscopy preparation.(right) as well as for western blot analysis for the presence of LC3-I and LC3-II bands (left). Purified Tcon cells were used as controls. Graph of the relative intensities of LC3 II bands in relative units (R.U.) after their quantification and normalization to β-actin. (b) After 4 days of allogeneic stimulation, C57BL/6 purified splenic T cells were stained with LC3-FITC antibody and prepared for confocal microscopy. Purified Tcon cells were used as controls. (c) Gated with CD90.2 for T cells, representative FACS plots (left) of CFSEhigh/low and LC3high/low. Graph of the LC3 mean fluorescence intensity (right) of T cells (MFI ± standard error of the mean (SEM). (d) After 6 days of allogeneic stimulation, from a pool of 3 allogenic donors, purified human T cells were prepared for western blot analysis with similar analysis for the presence of LC3-I and LC3-II bands as in (a). (e) Purified human T cells were stimulated with plate-bound anti-CD3 and anti-CD28 antibodies, then analyzed for T cell proliferation based on 3H-thymidine incorporation at 48 hours. The data in counts per minute (CPM) are the mean ± STD of triplicates and are representative of 3 independent donors. (f) After 48 hr stimulation with CD3/CD28, cells were gated on live lymphocytes positive for Cyto-ID staining to analyze for autophagy induction. Cells were treated with 10 uM CQ and 5 mM 3-MA for positive and negative controls. (c) analyzed with two-tailed, unpaired t-test. (a) This graph is representative of 3 separate experiments. (c) Data are a summary of triplicates representative of 2 different experiments. Data in *P< 0.05; **P< 0.01; ***P< 0.001; ****P< 0.0001.
We next analyzed the induction of autophagy on activation of human T cells and further determined the impact of inhibiting autophagy on their responses in MLR. Human T cells were isolated from whole blood from 3 individual healthy volunteers and stimulated in an allogeneic MLR as in Methods. Western blot (Figure 1d) and flow cytometry analysis (Figure 1e) 6 days after stimulation of human T cells in an MLR caused induction of autophagy in T cells, similar to murine T cells. We next assessed the functional relevance of autophagy induction in human T cells. We utilized 3-MA and CQ to inhibit distinct stages of autophagy (Figure S1a) to determine whether inhibition of autophagy regulates the proliferative responses of human T cells. As shown in Figure 1f, inhibition of autophagy with both 3-MA and CQ significantly reduced the proliferation of human T cells. Collectively these data demonstrate that Tcon cells upon in vitro stimulation by allo-APC’s undergo autophagy, which when inhibited, mitigated their proliferative responses but increased their apoptosis.
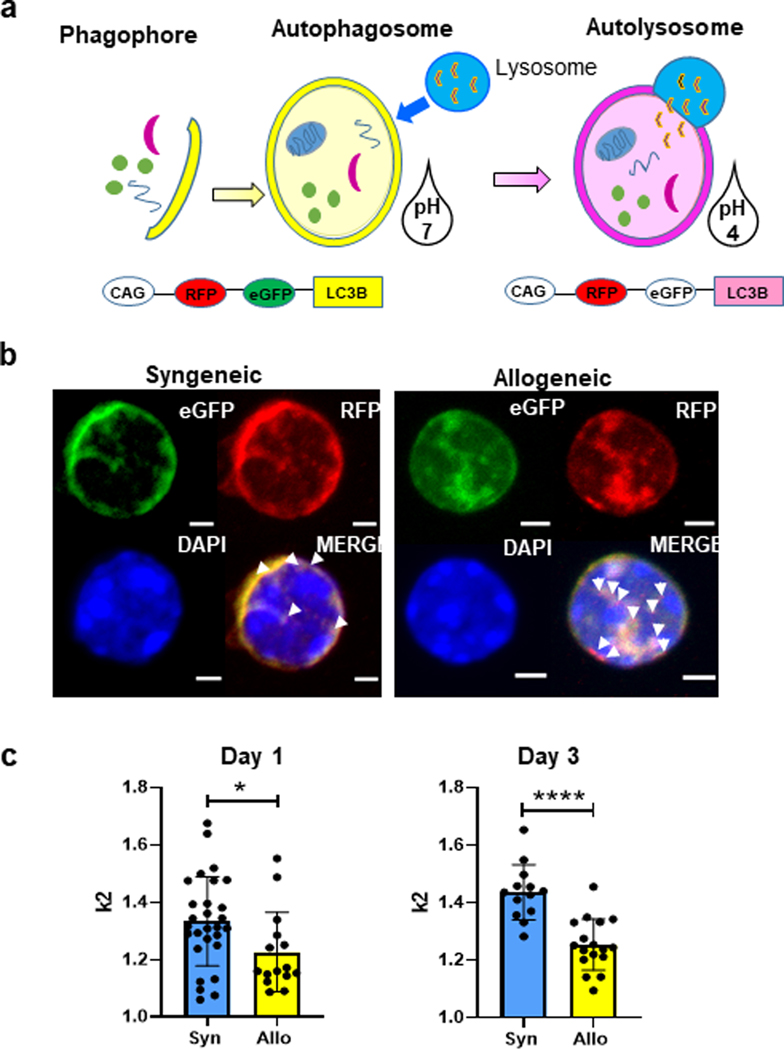
We next explored whether autophagic induction following in vitro allo-stimulation was observed in vivo. To this end, we utilized donor Tcon cells from the autophagy reporter CAG-RFP-eGFP-LC3 mice which express a tandem RFP-eGFP LC3 fusion protein 13. Autophagosomes found in the cytoplasm would express both the RFP (red) and eGFP (green) simultaneously thus producing a yellow color. Once the autophagosomes fuse with the lysosomes, indicative of autophagy, then the eGFP emission is lost due to the acidic pH of the autophagolysosome thereby leaving only the red color (RFP) to be visible (Figure 2a). Splenic Tcon cells from these mice were mixed with T cell depleted bone marrow (TCD-BM) from WT-B6 and transferred to BALB/c recipients following myeloablative conditioning as described in Methods. Congenic C57BL/6 recipients served as syngeneic controls. On days 1 and 3, recipient mice from each group were sacrificed and their spleens were harvested and analyzed for donor T cells. After magnetic bead purification and cytospin preparations, cells were imaged with a confocal microscope (Figure 2b) in order to quantitate the amount of combined fluorescence signaling of eGFP plus RFP verses RFP signaling alone. The imaging and their analysis were performed in an operator-blinded manner. As shown in Figure 2b, when compared to syngeneic controls, donor T cells harvested from allo-recipients showed significantly greater loss of eGFP, i.e. increased autophagy on both days, 1 and 3 (Figure 2c). These data collectively demonstrate that autophagy is induced in Tcon cells following stimulation by allo-antigens in vitro and in vivo.
Fig. 2. Verification of autophagy induction with allogeneic stimulation in vivo.
A CAG-RFP-eGFP-LC3 mouse strain was purchased from Jackson Laboratories and purified splenic T cells from these mice were used for a MHC major mismatch allogeneic bone marrow transplant allo-HCT transplants into BALB/c recipients with C57BL/6 donor TCD bone marrow. (a) Schematic of CAG-RFP-eGFP-LC3 mice with dual fluorescent expression capabilities which allows for autophagosomes to be distinguished from autolysosomes, depending on acidity of the environment. RFP is stable in acidic pH (pKa 4.5) while EGFP (pKa 5.9) is quenched. (b) Syngeneic and allogeneic transplanted animals with CAG-RFP-eGFP-LC3 T cells were harvested 4 days’ post-transplant. Co-expression of eGFP (green) and RFP (red) indicated by yellow punctate when LC3 in cytoplasm and autophagosome. Increase in red punctate alone, indicated by arrows indicates LC3 in autophagolysosome. (Scale bar, 2mm) (c) Quantification of the ratio of yellow fluorescent vs red fluorescence between syngeneic and allogeneic was used for indication of autophagy flux.
Generation of T cell specific ATG5 deficient mice
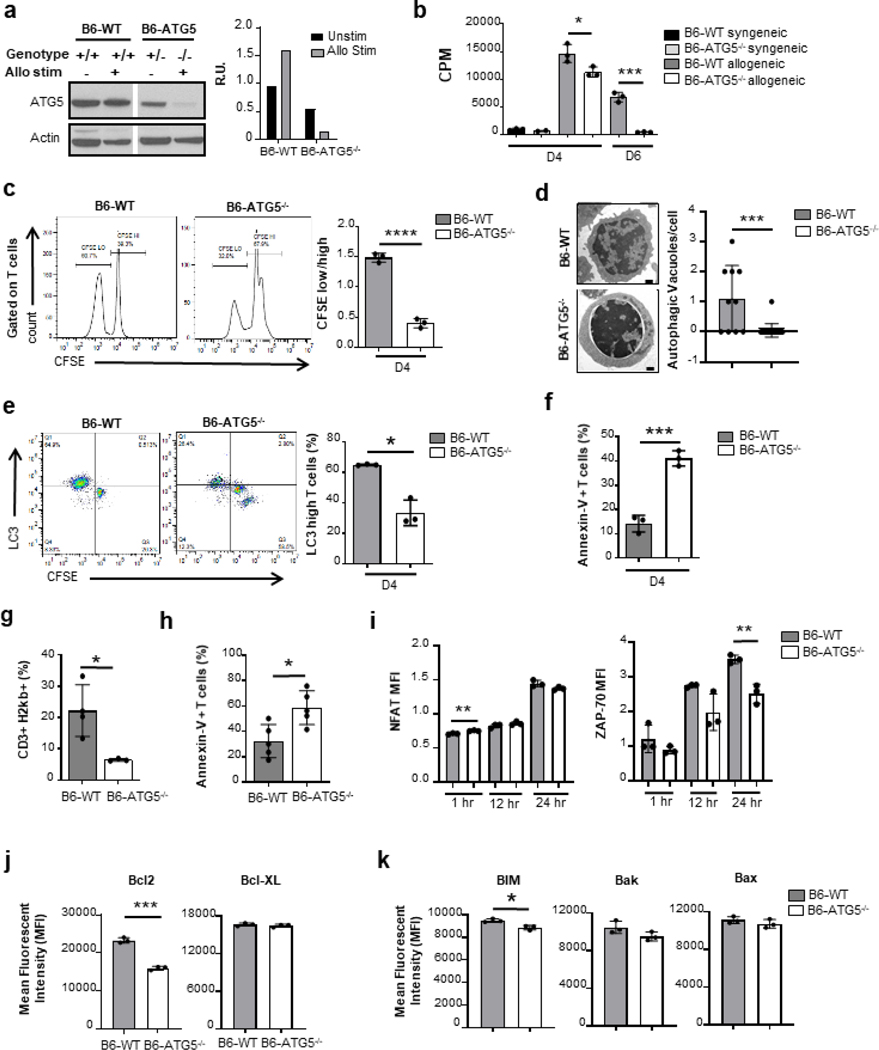
In order to study the impact of the loss of autophagy, specifically in Tcon cells, we developed an ATG5 conditional knockout mouse (B6-ATG5−/−) (Figure S2a). We confirmed deficiency of ATG5 in T cells and also concurrently assessed the impact of loss of ATG5 on induction of autophagy in T cells following allo-stimulation. Tcon cells from B6-WT, and B6-ATG5−/− animals were harvested and stimulated with splenic cells from BALB/c mice for 4 days and analyzed by immunoblot for ATG5 expression. Un-stimulated T cells were used as controls. As shown in Figure 3a, T cells from the ATG5 knockout mice demonstrated minimal ATG5. This loss therefore correlates with a reduction in autophagy when compared with B6-WT T cells.
Fig. 3. Autophagy induction is required for T cell proliferation from ATG5−/− mice.
CD90.2+ purified splenic T cells from C57BL/6 and ATG5−/− mice underwent allogenic stimulation for 4 or 6 days in an MLR and then harvested. (a) Confirmation of loss of ATG5 in the KO animals by WB (left) compared to heterozygote littermate and wildtype C57BL/6 animals. Quantification of WB bands (right). (b) Purified T cell proliferation were quantitated based on 3H-thymidine incorporation at day 4 and day 6. The data in CPM are the mean ± SEM of triplicates and are representative of 2 independent experiments. (c) T cells were labeled with CFSE prior to stimulation and analyzed for CFSE dilution at day 4. Representative histograms of CFSE dilution (left). The mean of the ratios CFSElow/ CFSEhigh ± SEM of triplicates. Data is representative of 2 independent experiments. (d) After 4 days of allogenic stimulation, activated B6-WT and B6-ATG5−/− T cells were processed for electron microscopy preparation (scale bars, 500 nm) individual cells were quantitated for autophagic vacuoles (right panel). (e) Gated with CD90.2 for T cells, (left) graph of the LC3 mean fluorescence intensity of T cells (MFI ± standard error of the mean (SEM) Data are a summary of triplicates representative of 2 different experiments. (f) After 4 days of allogenic stimulation, activated B6-WT and B6-ATG5−/− T cells were stained with Annexin-V and analyzed by flow cytometry. BALB/c recipient mice underwent MHC major mismatch allogeneic bone marrow transplant with C57BL/6 donor TCD bone marrow with purified splenic T cells from B6-WT or B6-ATG5−/− donors. Isolated the splenocytes of recipients at day 7 post-BMT and analyzed them by flow cytometry. We observed a decrease in the expansion of B6-ATG5−/− T cells (g) as well as an increased apoptosis (h) when compared to WT-B6. Data is representative of 3 experiments Whole splenocytes from C57BL/6 and ATG5−/− mice were cultured with anti-CD3/CD28 antibodies and harvested for analysis by flow cytometry at 1h, 12h and 24h. Unstimulated splenocytes were used as controls. (i) Gated on CD90.2-positive T cells, they were analyzed for the MFI of intracellular NFAT (left panel) and histogram overlay on left for unstimulated and 24 h time point (right panel) (j) Intracellular Zap-70. Data are the mean MFI± SEM of triplicates representative of 3 independent experiments. Whole splenocytes from C57BL/6 and ATG5−/− mice were cultured with anti-CD3/CD28 for 48h with additional 5h with Cell Stimulation Cocktail. After stimulation, they were stained and gated on CD90.2-positive T cells, then analyzed by flow cytometry. (k) Left panel: representative histograms of Bcl-2 in CD90.2-positive T cells. Right panel: mean of Bcl-XL MFI ± SEM of triplicates representative of 3 independent experiments. (l) These same CD90.2-positive T cells, then analyzed by flow cytometry for the presence of intracellular staining BIM, Bak, and Bax. Graphs are the mean of MFI ± SEM of triplicates representative of 3 independent experiments. *P< 0.05; **P< 0.01; ***P< 0.001; ****P< 0.0001.
We next determined the impact of ATG5 deficiency on T cell development. T cells from B6-WT and B6-ATG5−/− animals were harvested from thymus and spleen 8 weeks after birth. Deficiency of ATG5 did not affect thymopoiesis as determined by similar numbers and percentages of CD4−/CD8−, CD4+, CD8+ and CD4+/CD8+ subsets in the thymus (Figure S2b). T cells harvested from B6-ATG5−/− spleens had similar splenic cellularity (Figure S2c), as well as CD4+ and CD4+Foxp3 T cells (Figure S2d). However, a reduction in absolute numbers of CD8+ T cells (Figure S2e) was observed, demonstrating that loss of autophagy led to loss of CD8+ T cell in periphery without a significant change in CD4+ subsets and in lymphocytes.
ATG5 regulates Tcon cell proliferation and enhanced apoptosis following activation
We next analyzed the impact of ATG5-dependent autophagy on T cell functional responses to allo-stimulation in vitro and in vivo. B6-ATG5−/− Tcon cells stimulated with allogeneic BALB/c showed a significantly reduced proliferation when compared with WT-B6 controls on day 4 and 6 (Figure 3b). The impact on proliferation was also confirmed with CFSE (Figure 3c). In Figure 3a, B6-WT Tcon cells demonstrated an increase in ATG5 expression which correlates with an observed increase in autophagy via increased autophagosomes observed via electron microscopy. As expected, B6-ATG5−/− Tcon cells demonstrated decreased autophagy (Figure 3d). In order to specifically confirm ATG5 deletion was associated with the autophagy machinery, we stained CFSE pre-marked T cells for intracellular LC3 after stimulation. B6-ATG5−/− Tcon cells showed a lower proportion of LC3high T cells compared to B6-WT Tcon (30% versus 65%) in MLR at day 4 (Figure 3e). Stimulation with anti-CD3/CD28 similarly demonstrated reduction in proliferation of B6-ATG5−/− Tcon cells as determined by H3-thymidine incorporation, CFSE dilution assays and LC3 quantification (Figure S2f–2h).
We next analyzed whether ATG5-dependent autophagy in Tcon cells altered activation induced apoptosis. In B6-ATG5−/− Tcon cells following in vitro activation, these cells demonstrated significantly enhanced apoptosis when compared with WT-B6 Tcon cells (Figure 3f, Figure S2i). In vivo, the impact on T cell autophagy could be confounded by T cell extrinsic factors. Therefore, we next validated the reduction in proliferation in the absence of ATG5 in B6-WT and B6-ATG5−/− Tcon cells labeled with CFSE and transplanted with TCD-BM from WT-B6 into either syngeneic B6 or allogeneic BALB/c mice after conditioning as in methods. T cells were harvested from allo-recipient’s spleen on day 7 and assessed using CFSE. Consistent with the in vitro data, allo-stimulation of B6-ATG5−/− Tcon cell donor cells (CD3+H2Kb+) in vivo resulted in a significant reduction in proliferation when compared with donor WT-B6 Tcon cell population (Figure 3g). We further confirmed the enhanced apoptosis as shown in Figure 3h.
We then determined whether the increase in apoptosis and the decrease in proliferation was secondary to impairment of Tcon cell activation in the absence of ATG5. Tcon cells from B6-WT and B6-ATG5−/− were stimulated with CD3/CD28 antibodies and harvested at 1h, 12h and 24h, and stained for NFATc1 and ZAP-70. Unstimulated splenocytes were used as controls. WT-B6 and B6-ATG5−/− Tcon cells showed a similar activation pattern with a progressive increase of their NFATc1 level at 12h and 24h (Figure 3i, Figure S2j, left panel) and of ZAP-70 at 24h (Figure 3i, Figure S2j, right panel). Furthermore, consistent with previous reports 14 15, no differences were observed in expression levels of CD69 upon activation (Figure S2k). These data demonstrate that Tcon cell activation cascade downstream of the TCR was not affected by the absence of ATG5 in T cells.
We next analyzed whether deficiency in autophagy altered the balance between pro- and anti-apoptotic factors. Pro-apoptotic proteins BIM, Bak and Bax, and anti-apoptotic proteins Bcl-2 and Bcl-XL levels were determined 48hr after activation. B6-WT Tcon cells demonstrated greater increase in Bcl-2 anti-apoptotic protein upon stimulation when compared with B6-ATG5−/− Tcon cells while no difference in expression of Bcl-XL was observed (Figure 3j). In contrast, the expression of all three pro-apoptotic proteins BIM, Bak and Bax were also similar with a modest decrease in BIM in WT-B6 and B6-ATG5−/− Tcon cells after stimulation (Figure 3k). Thus, absence of ATG5 in T cell altered the levels of apoptotic proteins.
Deficiency of ATG5 enhances Th1 and Th17 differentiation in vitro and in vivo
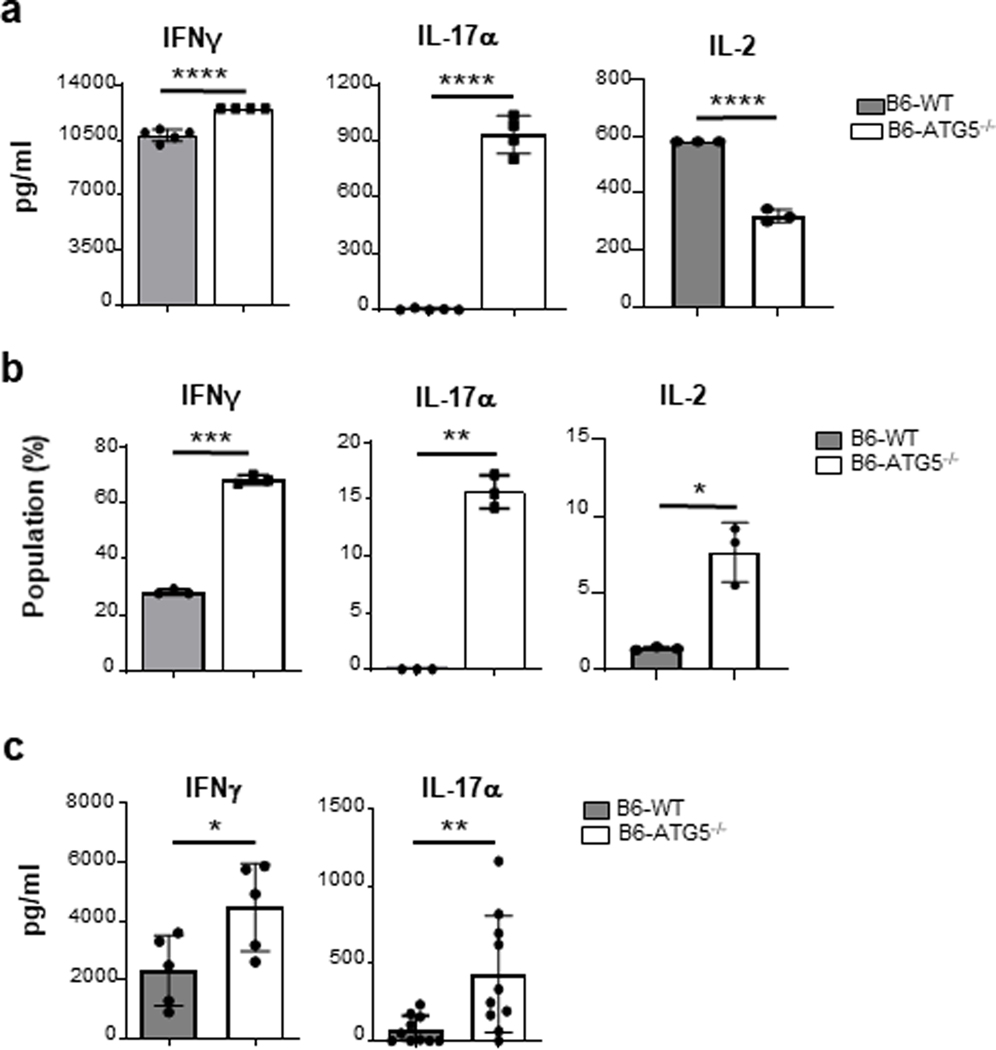
Following allo-stimulation, T cells proliferate and undergo Th differentiation. We therefore examined the impact of T cell ATG5 deficiency on T cell differentiation. Supernatants from allo-stimulated Tcon cells cultured for 4 days were collected and analyzed for the presence of Th1 (IFNγ and IL-2) and Th17 (IL-17α) signature cytokines. We made the surprising observation that production of IL-17α by B6-ATG5−/− Tcon cells was significantly increased as well as IFNγ levels in the supernatants despite the change in proliferation (Figure 4a). Similar results were found in the B6-ATG5−/− Tcon cells with an increase in intracellular IL-17α and IFNγ after in vitro allo-stimulation (Figure 4b). Similar effects were also observed following non-specific activation of cells with CD3/CD28 (Figure S3a–b) demonstrating that ATG5 dependent autophagy increased Th17 differentiation despite reducing proliferation. We next determined whether the increased Th1 and Th17 levels were observed in vivo following B6 to BALB/c allo-HCT. Consistent with in vitro observation, serum levels of IL-17α and IFNγ were significantly greater in allo-recipients of B6-ATG5−/− than in recipients of B6-WT (Figure 4c). These data collectively demonstrate that despite reduction in proliferation and numbers, B6-ATG5−/− Tcon cells demonstrated greater Th1/Th17 differentiation, thus uncoupling T cell proliferation from differentiation following in vivo allo-stimulation.
Fig. 4. ATG5-deficient T cells show an unexpected cytokine profile.
Sorted CD90.2+ B6-WT and B6-ATG5−/− T cells were stimulated for 4 days in an MLR with BMDCs from C57B6 (syngeneic) or BALB/c (allogeneic) mice. (a) Supernatants from the MLR were analyzed with ELISA for IFNγ, IL-17α and IL-2. (b) Cells were harvested and stained for intracellular cytokine and analyzed by flow cytometry. The graphs show the mean of triplicates SD and are representative of 2 independent experiments. (c) Serum levels of Day 7 post-transplant animals for cytokine levels. Data analysis done by ELISA assay. Graph representative of 3 experiments. *P< 0.05; **P< 0.01; ***P< 0.001; ****P< 0.0001.
T cell cytotoxic functions are enhanced in the absence of ATG5 dependent autophagy
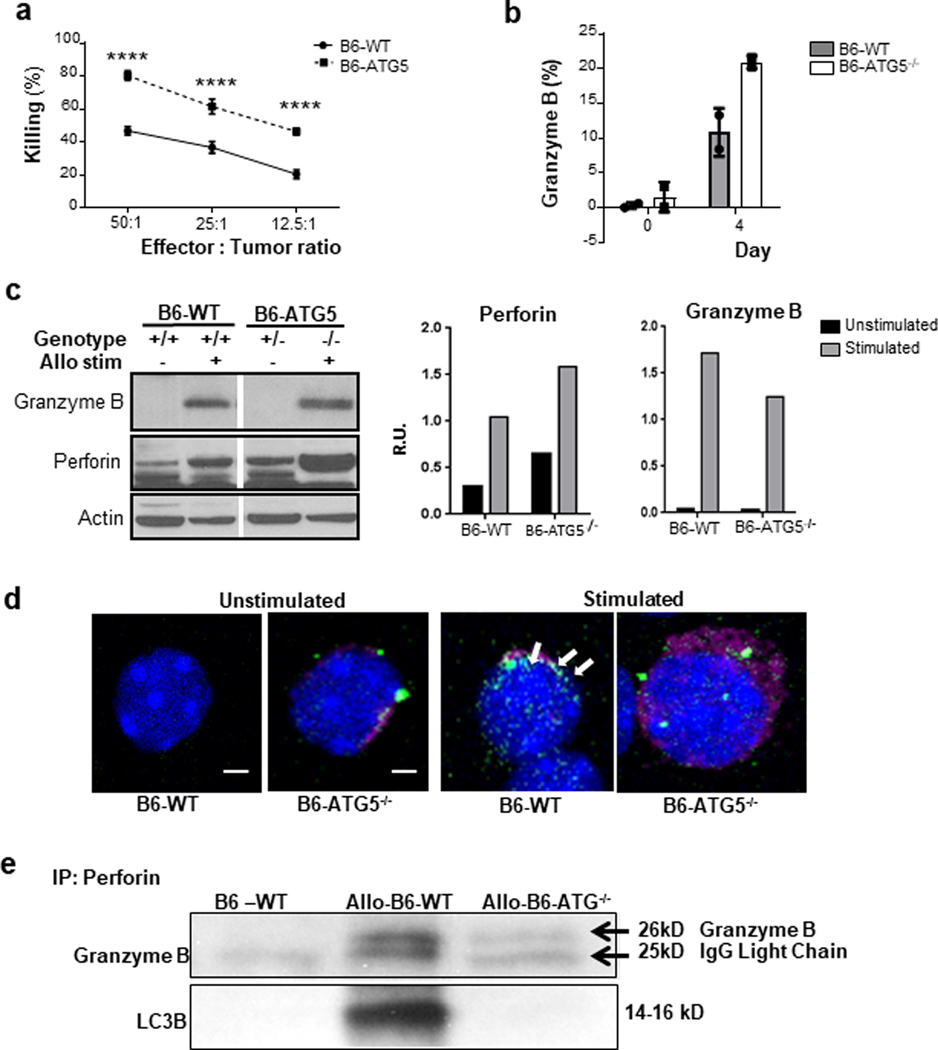
The impact of autophagy on T cell CTL is unknown. In light of normal T cell activation and enhanced differentiation into helper cells, we hypothesized that the cytolytic function of B6-ATG5−/− T cells may be enhanced on a per cell basis despite the reduction in their proliferative capacity. B6-ATG5−/− and B6-WT Tcon were activated in a bulk allogeneic MLR with BALB/c-derived irradiated splenocytes. On day 6, the T cells were harvested and their counts adjusted to the CD8+ fraction as effectors against allogeneic P815 target cells at various effectors to target ratios. Consistent with the hypothesis, the cytotoxic capacity of ATG5−/− CD8+ T cells was significantly greater than that of WT-B6 T cells at every one of the tested ratios (Figure 5a).
Fig. 5. ATG5−/− activated T cells have an increased killing effect.
A cytolytic T lymphocyte assay (CTL) was performed using purified T cells from B6-WT and B6-ATG5−/− mice. Cells were stimulated for 6 days in a bulk MLR with allogeneic irradiated (30Gy) whole splenocytes from BALB/c mice in order to generate activated CD8-positive T cells. (a) The CD8-positive T cells were used as effectors against P815 targets in a CTL assay. The graph shows the percentage of specific killing at various effector-to-target ratios. Data are the mean± SEM of quadruplicates and are representative of 3 independent experiments. (b) CD8-positive T cells were harvested at day 4 from the bulk MLR and stained for intracellular granzyme B (p=0.065). (c) An MLR was performed using purified T cells from B6-WT and B6-ATG5−/− mice. Cells were stimulated for 4 days in a bulk MLR with allogeneic irradiated (30Gy) whole splenocytes from BALB/c mice in order to generate activated CD8-positive T cells. Cells were harvested and pelleted for protein isolation. Control T cells (left panel) were harvest from spleens of naïve mice and processed for protein isolation. Quantitation of protein expression (right panel) was done using ImageJ software. Representative grafts are from duplicate experiments. (d) CD8-positive T cells were analyzed with confocal microscopy in order to identify co-localization of perforin (pink) and LC3 (green) indicated by white punctate (arrows). (e) B6-WT and B6-ATG5−/− splenic T cells were stimulated for 4 days in an MLR reaction, harvested and prepared for protein lysates. Lysates were then immune-precipitated with perforin antibody and then WB for granzyme B and LC3B. *P< 0.05; **P< 0.01; ***P< 0.001; ****P< 0.0001.
Since P815 tumor cells express only MHC class I and are therefore targeted by CD8+ T cells, we reasoned that B6-ATG5−/− CD8+ T cells would demonstrate increased killing through the perforin/granzyme B pathway 16. This result would be due to an increase in perforin and granzyme B content because of a reduction of their clearance by autophagy. To this end, we harvested some B6-WT and B6-ATG5−/− cells following allo-stimulation in an MLR and analyzed CD8+ T cells for their intracellular levels of granzyme B. B6-ATG5−/− CD8+ T cells demonstrated a trend to higher content of granzyme B than B6-WT controls at day 4 (Figure 5b). Western blot analysis from these T cells confirm greater granzyme B and also greater intracellular perforin levels (Figure 5c). We next determined whether the increased perforin and granzyme B following activation in B6-ATG5−/− T cells is directly because of reduction in autophagy. We therefore analyzed for perforin/granzyme B levels following allogeneic stimulation in the presence of CQ, as well as with rapamycin and compared them with diluent treated controls. As shown in Supplemental Figure 4, inhibition of autophagy caused an increase of perforin, granzyme B and LC3 II.
We next further explored the mechanism for the increase in perforin/granzyme B in the absence of ATG5 dependent autophagy. We hypothesized that autophagic vacuoles were critical for degrading and reducing intracellular levels of perforin/granzyme B in T cells. We therefore next determined whether perforin/granzyme B were taken into autophagosomes after T cell activation by analyzing for co-localization of LC3 and perforin/granzyme B. B6-WT and B6-ATG5−/− Tcon cells were stimulated in an allo-MLR and assessed with confocal microscopy for overlapping signaling from LC3 (green) and perforin (pink). While LC3 and perforin overlapped (white arrows, Figure 5d) no such overlap was observed in the negative control stimulated B6-ATG5−/− T cells. To further confirm, we next used perforin antibody for immune-precipitation in allo-stimulated B6-WT cells and then immunoblotted for granzyme B and LC3B antibodies. As shown in Figure 5e, both perforin and granzyme B co-precipitated with LC3B. Collectively, these findings demonstrate that perforin and granzyme B are taken up in autophagosomes and degraded. By contrast, an absence of autophagy in B6-ATG5−/− CD8+ T cells leads to greater accumulation of perforin/granzyme B thus converting them into more efficient cytotoxic cells.
Autophagy-deficient Tcon cells cause less severe GVHD
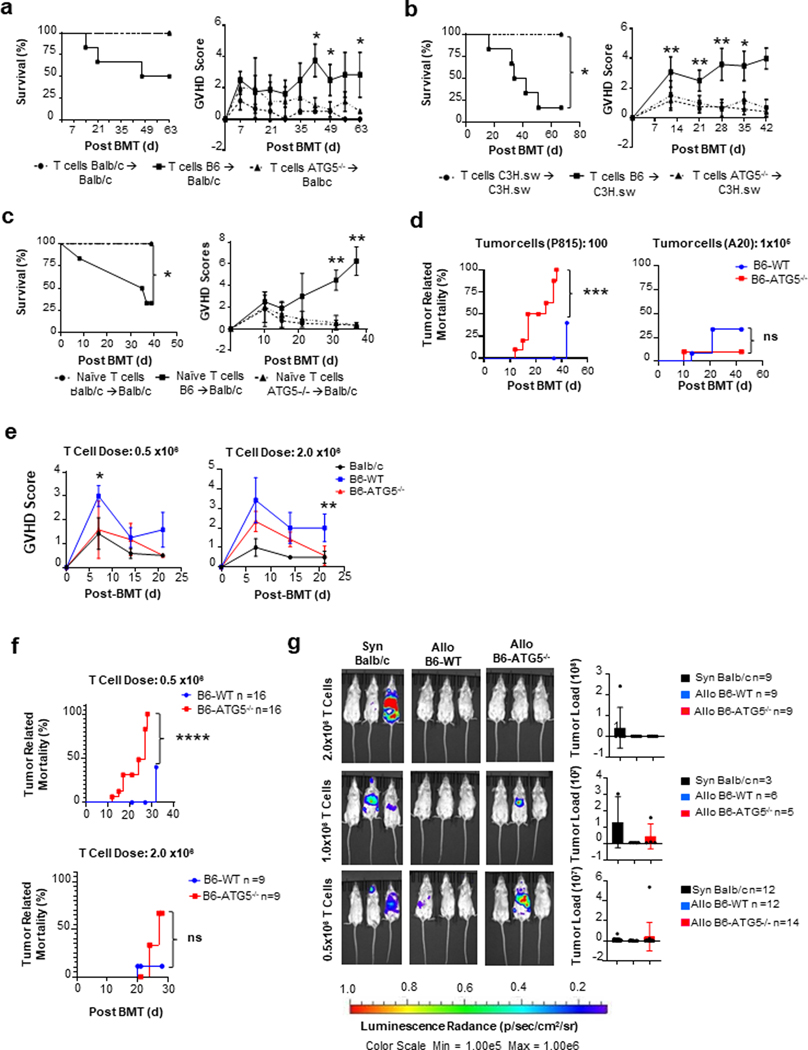
Allo-reactive T cell numbers, Th differentiation and CTL are critical for causing GVHD 17. Allogeneic T cells demonstrated increase in autophagy flux while ATG5 dependent autophagy causes uncoupling of proliferation from Th differentiation and CTL function. We next determined its relevance in well-established in vivo mouse models of experimental GVHD. We once again utilized the clinically relevant B6→BALB/c MHC-major mismatched BMT model as described in methods. Allogeneic recipients receiving total T cells from B6-ATG5−/− donors (ATG5−/− recipients) demonstrated a significantly greater survival and reduced clinical GVHD severity than those receiving WT-B6 T cells (WT-B6 recipients) (Figure 6a).
Fig. 6. ATG5 regulates T cell-dependent GVHD and increases T cell cytotoxic function in a murine model.
BALB/c recipient mice underwent MHC major mismatch allogeneic bone marrow transplant with C57BL/6 donor TCD bone marrow with purified splenic T cells from B6-WT or B6-ATG5−/− donors. Mice were monitored weekly for survival and GVHD score. (a) Left Panel: Survival curve of animal’s observations weekly for survival. Right Panel: GVHD scoring of animals weekly for progression of disease. (b) C3H.sw recipient mice underwent MHC minor mismatch allogeneic bone marrow transplant with C57BL/6 donor TCD bone marrow (5×106) with purified splenic T cells (3×106) from WT-C3H.sw (syngeneic), B6-WT or B6-ATG5−/− donors (allogeneic). Mice were monitored weekly. Left Panel: Survival curve of animal’s observations weekly for survival. Right Panel: GVHD scoring of animals weekly for progression of disease. (c) Animal model of GVHD using only naïve T cells for transplant. Naïve T cells from B6-WT or B6-ATG5−/− mice were transplanted into BALB/c recipients. Left Panel: Survival curve of animal’s observations weekly for survival. Right Panel: GVHD scoring of animals weekly for progression of disease. (d) BALB/c recipient’s data for a Graph versus Tumor (GVL) assay against P815 leukemia tumor line. (e) BALB/c recipient’s data for a Graph versus Tumor (GVL) assay against A20 leukemia tumor line. (f) BALB/c mice transplanted with 0.5 or 2.0 × 106 T cells with 100 P815 tumor cells from BALB/c, B6-WT or B6-ATG5−/− donor animals were analyzed weekly for GVHD score (g) and tumor related mortality. (h) BALB/c mice transplanted with 0.5, 1.0, and 2.0 × 106 T cells from either BALB/c, B6-WT or B6-ATG5−/− donor animals were analyzed at day 14. P815 tumor load was measured using the CCD camera (IVIS, Caliper Life Sciences) imaging machine to quantitate to amount of tumor load per mouse. Images are a representative group of animals. *P< 0.05; **P< 0.01; ***P< 0.001; ****P< 0.0001.
To exclude possible strain related artifacts, we also tested in a second clinically relevant, MHC matched but multiple minor antigen-mismatched B6C3H.sw model of GVHD. Similar protection from GVHD was observed in allo-recipient of B6-ATG5−/− T cell when compared to B6-WT cells (Figure 6b).
Because Tcon showed (a) in vitro and in vivo defects in absence of autophagy, (b) phenotypic analyses of splenic T cells from a B6-ATG5−/− donor demonstrated reduced numbers of CD62L+; CD25− naïve T cells compared to B6-WT T cells (Figure S5) and (c) because naïve T cells cause GVHD, we explored whether GVHD improvement was caused by a reduction of naïve Tcon cells in the transplant inoculum. We transplanted lethally irradiated (800cGy) BALB/c recipient with 5×106 TCD-BM from B6 donors along with 0.5×106 purified allogeneic naïve (CD62L+; CD25−) T cells from either B6-ATG5−/− or B6-WT spleen. Again, B6-ATG5−/− allo-donor T cell recipients demonstrated significantly greater survival and reduced severity GVHD (Figure 6c). These data demonstrated that inhibition of autophagy in donor Tcon cells ameliorates GVHD.
ATG5 deficient donor T cells show sufficient GVT responses
GVHD is tightly linked to the beneficial graft versus tumor (GVT) response. GVT is dependent on IFNγ CTL functions of donor T cells18. Because autophagy deficient T cells showed greater CTL response we hypothesized that despite reduction in T cell proliferation causing less GVHD, the uncoupling of proliferation responses from enhanced CTL function will lead to sufficient preservation of GVT. Utilizing the same mouse model as above for GVHD, we incorporated a mouse DBA/2 mastocytoma cell line, P185, to the BMT cell inoculum. P185 cells were transduced with lentivirus that contained a GFP and luciferase (luc+) construct to facilitate in vivo monitoring of the tumor burden by bioluminescence imaging (BLI) after allo-HCT. Mice were imaged weekly for quantitation of tumor load in each mouse. We found that at a dose of 100 P185 tumor cells mixed with the 0.5 × 106 B6-ATG5−/− T cells, all of the allo-B6-ATG5−/− T cell mice succumbed to tumor (Figure 6d). Suggesting that at this ratio of tumor to effector cells, GVT was not sufficiently preserved by allo-BMT. Next we utilized a less aggressive lymphoma cell line (A20) at a dose of 1 × 105 tumor cells with 0.5 × 106 donor T cells. B6-ATG5−/− T cells (Figure 6e) demonstrated greater reduction in tumor related mortality.
In vivo GVT responses are complex. They are, amongst other factors, likely a function of qualitative effects of T cells (such as individual T cell cytotoxic functions) and also quantitative effects (dependent on the numbers of T cells, clinically demonstrated by DLI studies). Because B6-ATG5−/− T cells have a proliferative/survival deficiency, we hypothesized that enhancing T cell numbers will increase GVT without causing a similar magnitude of increase in GVHD as WT-B6 T cells were able to reduce the P185 aggressive tumor cells. Therefore, to assess the impact on magnitude of GVHD and GVT responses, Balb/c recipients were transplanted with either 0.5 × 106 or 2.0 × 106 doses of T cells from B6-WT or B6-ATG5−/− (allogeneic) T cells while keeping the tumor targets, P815 tumor cells at the same dose per mouse. GVHD severity was determined by clinical score. While increasing the T cell dose increased GVHD in both WT and KO T cell recipients, the ATG5−/− T cells caused significantly reduced GVHD scores when compared to B6-WT animals (Figure 6f) at similar T cell numbers. When these animals for analyzed for GVT responses by quantifying tumor induced mortality and with BLI, the animals that received reduced numbers (0.5×106) of ATG5 and WT T cells showed reduced GVT (greater tumor mortality and BLI) than those that were transplanted with higher numbers (2×106) of T cells (Figures 6g). However, at lower T cell dose the KO T cells demonstrated a reduced magnitude of GVT when compared to WT T cells (6g-h), but this reduction was not apparent at higher T cells dose (6g-h). Thus, ATG5 dependent autophagy uncoupled allo-reactive Tcon cell proliferation responses from their Th differentiation and cytotoxic functions, allowing for in vivo reduction in GVHD without causing a similar magnitude of loss of GVT responses.
Discussion
T cells are essential for the beneficial GVT and the undesirable GVHD effects of allo-HCT. Separation of GVHD from GVT remains an intractable clinical problem. We demonstrate that absence of autophagy uncouples T cell functions. Specifically, it separated T cell proliferation from its effector function such as cytokine release and CTL function. This uncoupling effect caused a reduction in GVHD but retained sufficient GVT effects after allo-HCT. Previous studies have demonstrated that autophagy deficient T cells show defects in proliferation and changes in release of cytokines 1, 6, 7, 10, 19, 20, 21. We confirm similar effect on proliferation and apoptosis in Tcon cells and further extend the role of autophagy in T cell function by demonstrating that ATG5-dependent autophagy causes an alteration in Th differentiation and causes a concomitant increase in cytotoxic (CTL) functions. To our knowledge, this is the first demonstration of uncoupling of critical T cell responses after allo-stimulation, namely proliferation/apoptosis from effector function via release of cytokines and cytotoxic functions. T cell subsets have varying effects on GVHD and GVT. Tmem cells have been shown not to induce GVHD but retain sufficient GVT effects in humans and mice22. The induction of GVHD has been demonstrated to be caused primarily by naïve T cells (Tn) 17. The allo-recipients of purified Tn cells from the ATG5 deficient donors showed significant reduction in GVHD severity when compared with recipients that were transplanted with WT Tn cells. Thus, the changes in the composition of Tn and Tmem cells in the absence of ATG5 when compared with WT donor was not the cause for decrease in GVHD.
GVHD and GVT are caused by T cells but its severity is modulated by many other immune cells. Host dendritic APC’s play an important role in GVHD. Reduction of autophagy by a hypomorph of ATG16L1 in host DC’s has been shown to increase GVHD23. This is in contrast to our observation of the reduction in GVHD when ATG5 dependent autophagy is absent in donor T cells. Similarly, another study demonstrates that autophagy in Treg regulated GVHD24. Thus the net effect on GVHD severity by mitigating autophagy non-specifically in various immune cells could depend on which cellular subset is most significantly affected.
Our data on Tcon cells are consistent with previous reports demonstrating decreased proliferation of total T cells that lack ATG5-dependent autophagy. However, we also noticed an increase in apoptosis of these cells only after activation in proliferating cells. The increase in apoptosis was observed more in CD8+ than CD4+ T cells. The tight interplay between autophagy and apoptosis might explain the increase in apoptosis 5 25. Notably, dysregulation of organelle homeostasis and the accumulation of mitochondria and endoplasmic reticulum have been linked to cell death, due to the release of reactive oxygen species (ROS) 14. It has also been shown that autophagy can regulate apoptosis through the sequestration of the large caspase-8 subunit in autophagosomes and its subsequent lysosomal degradation 26. Similar results were obtained in T cells deficient in autophagy protein Beclin-1, where levels of pro-caspases, Bim and Bcl-2 were increased27, demonstrating that the balance between pro- and anti-apoptotic proteins can be regulated by autophagy. We similarly observed change in the pro vs. anti-apoptotic levels in ATG5-deficient Tcon cells that may also have contributed to increased apoptosis. In agreement with previous reports, we demonstrated that autophagy is induced upon T cell receptor (TCR) engagement as demonstrated by upregulation of LC312, 28, 29, 30, 31. We also additionally observed that ATG5-deficient Tcon cells displayed the similar activation profile of Zap-70 and NFAT as found in WT controls demonstrating no significant difference in TCR activation. Furthermore, the relevance of autophagy deficiency might be distinct for the survival of various T cell subsets. However, the role of autophagy in induction of a Th response under in vivo physiological conditions is variable and not well characterized32. We observed a remarkable enhancement of the T-helper signature cytokines (INFγ, IL-17α) in ATG5-deficient Tcon cells, thus demonstrating uncoupling of Tcon proliferation from Th differentiation following allo-stimulation. The increase in pathogenic Th1/Th17 differentiation did not cause an increase in GVHD in vivo, likely from the reduction in their total effector pool by the reduced proliferation and increase in apoptosis. Depending on the type of MHC disparity and the context of minor antigen presentation GVHD can be caused by either CD4 or CD8 or both subsets. While the in vitro allo-stimulation proliferation studies are largely reflective of CD4 responses, stimulation with anti-CD3+/28+ assays reflect both CD4 and CD8 effects. Thus the impact of autophagy inhibition regulates both CD4 and CD8 responses.
T cell expansion regulates both GVHD and GVT. This GVT response relies on the cytotoxic T CD8+ T cell response and the Tm cells following allo-HCT33. We made the surprising observation that the absence of ATG5-deficient CD8+ T cells, in the periphery, demonstrated significantly greater killing or cytotoxicity in vitro against allogeneic MHC 1-restricted P815 tumor cells than the B6-WT controls. The increase in CTL was associated with increased granzyme B in CD8+ T cells from B6-ATG5−/− compared to those from B6-WT. Mechanistic studies demonstrate that both granzyme B and perforin co-localize with LC3 demonstrating that they are selected targets for the autophagosomes. This data is consistent with a recent paper reporting that breast cancer cells can degrade granzyme B released from NK cells through the activation of autophagy in a hypoxic environment34. Thus granzyme B degradation is likely regulated by autophagic machinery, which in the absence of ATG5-dependent autophagy, gets accumulated over time in B6-ATG5−/− CD8+ T cells, facilitating their ability for cytolytic activity. Furthermore, it may also explain the greater loss of B6-ATG5−/− CD8+ cells from fratricidal cell death. Consistent with our in vitro data, ATG5-deficient T cells demonstrated similar uncoupling of proliferation from cytotoxic response in vivo after allo-BMT. Specifically, we saw decreased proliferation, increased apoptosis, increased Th cytokines and increased cytotoxicity. The net reduction in allo-reactive T cell numbers from decreased proliferation, increased apoptosis caused less GVHD. These observations were true after both MHC-major mismatch and MHC-matched multiple minor antigen-mismatch transplants, demonstrating that they were neither dependent on the mice strains nor on the degree of inflammation or MHC disparity. Despite decreased GVHD, a substantial preservation in GVT was observed, especially at higher T cell doses. The likely reason for the sufficient preservation of GVT responses, especially when higher T cell doses were utilized is likely from increased CTL/effector cytokine release despite a reduction in T cell numbers. Furthermore, autophagy has been shown to modulate Th9 (IL-9) release and affect antitumor responses39 (Vargas et al, Nat Comm ref). The relative contribution of IL-9, cytotoxity and T cell expansion kinetics will need to be assessed in future studies. However, our results should be interpreted with caveats that are inherent to these model systems. It could be a reflection of the tumor and T cell expansion kinetics, because when tumor to effector cell ratios were changed, the enhanced GVT effect was lost. These data suggest the critical nature of the cellular kinetics in addition to the critical role of CTL/cytotoxicity pathways in controlling tumors.
In summary, our study thus demonstrates that autophagy plays a differential role in T cell functions, separating the proliferative responses from effector Th and cytotoxic responses. Thus T cell specific inhibition of autophagy may be a therapeutic strategy to improve allo-BMT outcomes. Importantly, global inhibition of autophagy in all immune cell subsets, the host tumors and target tissues may result in variable impact. Future studies will need to systematically dissect the relevance of autophagy in various immune cells subsets and non-immune cell targets after allo-HCT.
Supplementary Material
Statement of Significance.
Findings demonstrate that induction of autophagy in donor T cell promotes GVHD, whilst inhibition of T cell autophagy mitigates GVHD without substantial loss of GVL responses.
Acknowledgements
This work was supported by the National Institute of Health grants CA-203542, HL-152605 and HL-149633 awarded to P. Reddy. This research was also supported by the National Cancer Institute of the NIH with award number CA-217156. This research was supported with the help of image acquisition and analysis by the University of Michigan’s Biomedical Research Core Facilities Microscopy Core.
Footnotes
Conflict of Interest: We have no conflicts of interest to disclose.
References
- 1.Mizushima N, Levine B, Cuervo AM & Klionsky DJ Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shintani T. & Klionsky DJ Autophagy in health and disease: a double-edged sword. Science 306, 990–995 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeSelm CJ et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell 21, 966–974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel KK et al. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J 32, 3130–3144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y. & Klionsky DJ The regulation of autophagy - unanswered questions. J Cell Sci 124, 161–170 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deretic V, Saitoh T. & Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol 13, 722–737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine B, Mizushima N. & Virgin HW Autophagy in immunity and inflammation. Nature 469, 323–335 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma Y, Galluzzi L, Zitvogel L. & Kroemer G. Autophagy and cellular immune responses. Immunity 39, 211–227 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Puleston DJ & Simon AK Autophagy in the immune system. Immunology 141, 1–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shibutani ST, Saitoh T, Nowag H, Munz C. & Yoshimori T. Autophagy and autophagy-related proteins in the immune system. Nat Immunol 16, 1014–1024 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Kuma A. et al. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Hubbard VM et al. Macroautophagy regulates energy metabolism during effector T cell activation. J Immunol 185, 7349–7357 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Wang ZV, Hill JA & Lin F. New autophagy reporter mice reveal dynamics of proximal tubular autophagy. J Am Soc Nephrol 25, 305–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sena LA et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephenson LM et al. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy 5, 625–635 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teshima T. et al. IL-11 separates graft-versus-leukemia effects from graft-versus-host disease after bone marrow transplantation. J Clin Invest 104, 317–325 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paczesny S, Hanauer D, Sun Y. & Reddy P. New perspectives on the biology of acute GVHD. Bone Marrow Transplant 45, 1–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matte-Martone C. et al. Differential requirements for myeloid leukemia IFN-gamma conditioning determine graft-versus-leukemia resistance and sensitivity. J Clin Invest 127, 2765–2776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharya A. & Eissa NT Autophagy and autoimmunity crosstalks. Front Immunol 4, 88 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deretic V. Autophagy: an emerging immunological paradigm. J Immunol 189, 15–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cadwell K. Crosstalk between autophagy and inflammatory signalling pathways: balancing defence and homeostasis. Nat Rev Immunol 16, 661–675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W. & Chao NJ Memory T cells: A helpful guard for allogeneic hematopoietic stem cell transplantation without causing graft-versus-host disease. Hematol Oncol Stem Cell Ther 10, 211–219 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Hubbard-Lucey VM et al. Autophagy gene Atg16L1 prevents lethal T cell alloreactivity mediated by dendritic cells. Immunity 41, 579–591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Texier L. et al. Autophagy-dependent regulatory T cells are critical for the control of graft-versus-host disease. JCI Insight 1, e86850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merkley SD, Chock CJ, Yang XO, Harris J. & Castillo EF Modulating T Cell Responses via Autophagy: The Intrinsic Influence Controlling the Function of Both Antigen-Presenting Cells and T Cells. Front Immunol 9, 2914 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou W, Han J, Lu C, Goldstein LA & Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy 6, 891–900 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovacs JR et al. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell Death Differ 19, 144–152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu X. et al. Autophagy is essential for effector CD8 (+) T cell survival and memory formation. Nat Immunol 15, 1152–1161 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pua HH, Dzhagalov I, Chuck M, Mizushima N. & He YW A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med 204, 25–31 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C. et al. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol 177, 5163–5168 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Botbol Y, Patel B. & Macian F. Common gamma-chain cytokine signaling is required for macroautophagy induction during CD4+ T-cell activation. Autophagy 11, 1864–1877 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bronietzki AW, Schuster M. & Schmitz I. Autophagy in T-cell development, activation and differentiation. Immunol Cell Biol 93, 25–34 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Zheng H, Matte-Martone C, Jain D, McNiff J. & Shlomchik WD Central memory CD8+ T cells induce graft-versus-host disease and mediate graft-versus-leukemia. J Immunol 182, 5938–5948 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baginska J. et al. Granzyme B degradation by autophagy decreases tumor cell susceptibility to natural killer-mediated lysis under hypoxia. Proc Natl Acad Sci U S A 110, 17450–17455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy P. et al. Histone deacetylase inhibition modulates indoleamine 2,3-dioxygenase-dependent DC functions and regulates experimental graft-versus-host disease in mice. J Clin Invest 118, 2562–2573 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddy P. et al. A crucial role for antigen-presenting cells and alloantigen expression in graft-versus-leukemia responses. Nat Med 11, 1244–1249 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Amaravadi RK et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest 117, 326–336 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooke KR et al. Tumor necrosis factor- alpha production to lipopolysaccharide stimulation by donor cells predicts the severity of experimental acute graft-versus-host disease. J Clin Invest 102, 1882–1891 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivera Vargas T, Cai Z, Shen Y. et al. Selective degradation of PU.1 during autophagy represses the differentiation and antitumour activity of TH9 cells. Nat Commun 8, 559 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.