Abstract
Purpose:
FGFR2 amplification is associated with poor prognosis in advanced gastric cancer and its subclonal heterogeneity has been revealed. Here, we examined whether circulating tumor DNA (ctDNA) was useful for detecting FGFR2 amplification and co-occurring resistance mechanisms in advanced gastric cancer.
Experimental Design:
We assessed genomic characteristics of FGFR2-amplified advanced gastric cancer in a nationwide ctDNA screening study. We also analyzed FGFR2 amplification status in paired tissue and plasma samples with advanced gastric cancer. In addition, we examined patients with FGFR2-amplified advanced gastric cancer identified by ctDNA sequencing who received FGFR inhibitors.
Results:
FGFR2 amplification was more frequently detected by ctDNA sequencing in 28 (7.7%) of 365 patients with advanced gastric cancer than by tissue analysis alone (2.6%–4.4%). FGFR2 amplification profiling of paired tissue and plasma revealed that FGFR2 amplification was detectable only by ctDNA sequencing in 6 of 44 patients, which was associated with a worse prognosis. Two patients in whom FGFR2 amplification was detected by ctDNA sequencing after tumor progression following previous standard chemotherapies but not by pretreatment tissue analysis had tumor responses to FGFR inhibitors. A third patient with FGFR2 and MET co-amplification in ctDNA showed a limitation of benefit from FGFR inhibition, accompanied by a marked increase in the MET copy number.
Conclusions:
ctDNA sequencing identifies FGFR2 amplification missed by tissue testing in patients with advanced gastric cancer, and these patients may respond to FGFR inhibition. The utility of ctDNA sequencing warrants further evaluation to develop effective therapeutic strategies for patients with FGFR2-amplified advanced gastric cancer.
Translational Relevance.
The efficacy of FGFR inhibition has not been clear for FGFR2-amplified advanced gastric cancer with significant genomic heterogeneity, because adequate testing to detect FGFR2 amplification for this population has not been established before FGFR treatment. In this study, we demonstrated that the utility of circulating tumor DNA (ctDNA) to evaluate targetable genomic alterations including FGFR2 amplification, and to guide targeted therapy in advanced gastric cancer. In addition, we also suggested that ctDNA sequencing may be useful for assessing other concurrent genomic alterations including resistance alterations for guiding the management of this type of cancer.
Introduction
Gastric cancer remains an important cancer, being the fifth most frequently diagnosed cancer and the third leading cause of cancer death worldwide (1). Despite the advance in systemic therapy, the prognosis of patients with advanced gastric cancer is still poor with median survival time of approximately 1 year. Molecularly targeted therapeutic strategies have been attempted for patients with advanced gastric cancer, but have frequently failed to improve overall survival (OS) due to its nature of molecular heterogeneity (2–10).
FGFRs (FGFR1, FGFR2, FGFR3, and FGFR4) are transmembrane receptor tyrosine kinases, and FGF/FGFR signaling can be aberrantly activated by altered FGFR genes in cancers (11). Approximately 5% of patients with gastric cancer have FGFR2 amplification (12), which is associated with poor prognosis (13–15). The relevance of high-level FGFR2 amplification in gastric cancer to the response to FGFR inhibitors has been suggested in preclinical studies (16–18); however, a randomized phase II trial (SHINE) failed to demonstrate improved progression-free survival (PFS) with the pan-FGFR tyrosine kinase inhibitor (TKI) AZD4547 compared with paclitaxel in the second-line treatment of advanced gastric cancer with FGFR2 amplification confirmed by tissue testing (19).
The analysis of circulating tumor DNA (ctDNA) has been demonstrated to be able to detect genomic alterations in tumor cells throughout the body and has been suggested as a method to assess heterogeneous resistance mechanisms (20, 21). A translational study of patients with FGFR2-amplified advanced gastric cancer treated with AZD4547 indicated that ctDNA sequencing identified high-level FGFR2 amplifications in responders (16). A utility of ctDNA sequencing for identifying heterogeneous FGFR2 amplification within spatially distinct regions of the primary tumor and distant metastases has also been suggested (22). In addition, previous studies suggested the utility of ctDNA sequencing for identifying genomic resistance mechanisms in advanced gastric cancer harboring ERBB2, MET, and EGFR amplifications (23–26). These observations suggest that ctDNA sequencing may be useful in guiding therapy for FGFR2-amplified advanced gastric cancer by detecting FGFR2 amplification, including cases missed by single-lesion tumor biopsies, and by identifying heterogeneous resistance mechanisms. Indeed, recently, a randomized phase II trial reported that the addition of bemarituzumab, a mAb against FGFR2b, to chemotherapy improved survival in patients with FGFR2b-overexpressing or FGFR2-amplified gastric cancer identified by tissue or ctDNA analysis (NCT03343301; ref. 27).
Here, we evaluated the utility of ctDNA sequencing compared with tissue analysis for detecting FGFR2 amplification in advanced gastric cancer as well as other genomic alterations, including resistance alterations, and for guiding the management of this type of cancer. The ctDNA sequencing revealed that some patients with FGFR2-amplified advanced gastric cancer in ctDNA may benefit from FGFR inhibition but cannot be identified by current tissue testing practices and clarified resistance mechanisms.
Materials and Methods
GI-SCREEN and GOZILA study design and patient selection
SCRUM-Japan GI-SCREEN is a nationwide tumor tissue cancer genomic profiling study involving 26 core cancer institutions in Japan (28), which aims to characterize the genomic landscape for all gastrointestinal cancers and accelerate development and improve care in this area by matching patients to suitable clinical trials. The key inclusion criteria included the following: (i) histopathologically confirmed unresectable or metastatic gastrointestinal cancer, (ii) receipt (or planned receipt) of systemic therapy, (iii) age ≥20 years, (iv) an Eastern Cooperative Oncology Group performance status (ECOG PS) of 0–1, (v) adequate organ function, and (vi) available tumor tissue. Eligible patients provided written informed consent. The genotyping of archival or fresh tumor tissue samples from enrolled patients was performed using the Oncomine Comprehensive Assay (OCA; Thermo Fisher Scientific), which is described in more detail below. This study was initiated in February 2015 and completed enrollment in April 2019.
GOZILA is a nationwide plasma genomic profiling study in Japan based on the SCRUM-Japan GI-SCREEN platform, which, like GI-SCREEN, aims to effectively identify patients with gastrointestinal cancers who might benefit from targeted therapy, with 31 institutions including the above 26 sites (28). The key inclusion criteria were similar to those of GI-SCREEN: (i) histopathologically confirmed unresectable or metastatic gastrointestinal cancer, (ii) age ≥20 years, and (iii) a life expectancy of at least 12 weeks. To avoid the suppression of ctDNA shedding due to chemotherapy, patients were included only if they showed disease progression during systemic chemotherapy and had not started the subsequent therapy at the time of blood sampling. Eligible patients provided written informed consent, and ctDNA genotyping was performed using Guardant360 (Guardant Health, Inc.), which is described in additional detail below. This study was launched in January 2018.
Both studies were conducted in accordance with the Declaration of Helsinki and the Japanese Ethical Guidelines for Medical and Health Research Involving Human Subjects. Each study protocol was approved by the institutional review board of each participating institution and registered in the University Hospital Medical Information Network (UMIN) Clinical Trials Registry (protocol IDs UMIN000016344 for GI-SCREEN and UMIN000029315 for GOZILA).
FGFR2 profiling concordance study design and patient selection
A retrospective study was performed to evaluate the concordance of FGFR2 amplification between tissue and plasma in patients with advanced gastric cancer between January 2015 and December 2018 at the National Cancer Center Hospital East. On the basis of the recommended formalin-fixed, paraffin-embedded (FFPE) sample storage period from our previous study (29), the tissue samples collected within 4 years were used. Patients who met the following criteria were included: (i) presence of histologically confirmed gastric adenocarcinoma, (ii) receipt of systemic treatment for advanced disease, and (iii) an available plasma sample collected near the time of tumor biopsy and before the initiation of systemic treatment. Patients with tumors previously known to harbor FGFR2 amplification in GI-SCREEN or GOZILA who had available matched plasma and tissue samples were preferentially included. Tumor responses were assessed according to RECIST v1.1 (30).
This study was conducted in accordance with the Declaration of Helsinki and the Japanese Ethical Guidelines for Medical and Health Research Involving Human Subjects. The study protocol was approved by the Institutional Review Board at the National Cancer Center (UMIN000041008). Written informed consent was obtained from patients who were alive at the time of the study. For deceased patients and their relatives, we disclosed the study design on the website of the National Cancer Center and gave the families a chance to express the will of the decedents.
Tissue-based next-generation sequencing (NGS) analysis
For patients enrolled in GI-SCREEN and the retrospective concordance study, the NGS analysis of tumor tissue was performed using OCA v1 and OCA v3 at the Life Technologies Clinical Services Lab, a Clinical Laboratory Improvement Amendments (CLIA)-certified, College of American Pathologists (CAP)-accredited laboratory, as described previously (31). These assays examined 143 (OCA v1) and 161 (OCA v3) cancer-related genes and detected relevant single-nucleotide variants (SNV), copy-number variations, gene fusions, and indels in one streamlined workflow. Briefly, tumor DNA and RNA were isolated from FFPE sections, and DNA/RNA libraries were prepared. Purified libraries were sequenced using Ion Torrent PGM (Thermo Fisher Scientific). Sequence reads were aligned to the hg19 assembly and were called using Ion Reporter Software version 4.4 (for OCA v1) and v5.0 (for OCA v3) to detect alterations.
ctDNA-based NGS analysis by Guardant360
For patients enrolled in GOZILA and the retrospective concordance study, the NGS analysis of ctDNA was performed using Guardant360 at Guardant Health, a CLIA-certified, CAP-accredited, New York State Department of Health–approved laboratory, as described previously (32). Guardant360 detects SNVs, indels, fusions, and copy-number alterations in 74 genes with a reportable range of ≥0.04%, ≥0.02%, ≥0.04%, and ≥2.12 copies, respectively. For GOZILA patients, 2 × 10-mL whole-blood samples were collected from enrolled patients in Streck Cell-Free DNA blood collection tubes (BCT; Streck, Inc) and sent to Guardant Health. For the other patients, 3 mL of frozen plasma prepared from whole blood collected in EDTA tubes was sent for analysis. Five to 30 ng of cell-free DNA (cfDNA) isolated from plasma was labeled with nonredundant oligonucleotides (“molecular barcoding”), enriched using targeted hybridization capture, and sequenced on an Illumina NextSeq 550 platform (Illumina, Inc.). Base call files generated by Illumina's RTA software version 2.12 were demultiplexed using bcl2fastq version 2.19 and processed as described previously (32). Somatic cfDNA alterations were identified using a proprietary bioinformatics pipeline.
IHC
FGFR2 IHC was performed using a rabbit anti-FGFR2 polyclonal antibody (18601; IBL) at Geneticlab, and MET IHC was performed using a rabbit anti-c-MET mAb (790–4430; Ventana) at National Cancer Center (Kashiwa, Japan). FGFR2 IHC results were scored according to the intensity and percentage of positively stained carcinoma cells, as follows: 0, no positive cells; 1, weak staining and ≥10%; 2, strong staining and <10%; 3, strong staining and 10% to 49%; and 4, strong staining and ≥50%.
FISH
The assessment of FGFR2 amplification by FISH was conducted using an FGFR2/CEP10 probe (Geneticlab; Supplementary Table S1) for 20 tumor nuclei per sample at Geneticlab. FGFR2 amplification was defined as an FGFR2 copy number ≥4.0 signals per cell and FGFR2/CEP10 ratio ≥2.0. If the FGFR2 copy number was ≥4.0 signals per cell and the FGFR2/CEP10 ratio was <2.0, the case was defined as polysomy.
Statistical analysis
Associations of the FGFR2 status with clinicopathological factors and the variant allelic frequency (VAF) were analyzed using Fisher exact test or Mann–Whitney U test. OS was defined as the interval from the first day of the first-line chemotherapy to the day of death or the most recent follow-up visit. Kaplan–Meier curves were constructed, and statistical significance was determined using the log-rank test. A P value of <0.05 was considered significant. JMP software (ver. 14.0; SAS Institute Inc.) was used to perform statistical analyses.
Results
ctDNA sequencing detects FGFR2 amplification and a unique genomic profile in patients with FGFR2-amplified advanced gastric cancer
To evaluate the utility of ctDNA compared with tissue samples for detecting FGFR2 amplification in advanced gastric cancer, we reviewed ctDNA sequencing results of advanced gastric cancer from the GOZILA and GI-SCREEN studies as well as from publicly available tissue-based databases [The Cancer Genome Atlas (TCGA) and Memorial Sloan Kettering Cancer Center (MSKCC) databases]. FGFR2 amplification was detected in 28 (7.7%) of 365 patients with advanced gastric cancer enrolled in the GOZILA study between January 2018 and January 2020 (Fig. 1A). This prevalence was significantly higher than that detected by tissue sequencing in GI-SCREEN and publicly available tissue-based databases (GI-SCREEN: 3.4%, P = 0.00080; TCGA: 4.4%, P = 0.049; and MSKCC: 2.6%, P = 0.0027; Fig. 1A), which is consistent with a previous study reporting higher incidences of amplification of receptor tyrosine kinase genes in ctDNA than tissue (33). Of 365 patients from GOZILA study, no ctDNA alterations were detected in 54 (14.8%) patients. Very weak correlation between the FGFR2 plasma copy number (pCN) and ctDNA maximum VAF (max VAF) was observed (r2 = 0.15; P = 0.041; Fig. 1B). These findings indicated that ctDNA sequencing may identify FGFR2 amplification that cannot be detected by conventional tissue analysis.
Figure 1.
Genomic characteristics of advanced gastric cancer with FGFR2 amplification based on ctDNA analysis. A, Prevalence of FGFR2 amplification in advanced gastric cancer in GOZILA, GI-SCREEN, and the TCGA and MSKCC databases. B, Correlations [coefficient of determination (r2)] between FGFR2 plasma copy number (pCN) and max VAF in FGFR2-amplified samples from GOZILA (n = 28). C, Prevalence of co-alterations in FGFR2-amplified (n = 28) versus nonamplified patients (n = 337) in GOZILA. Green and blue bars indicate prevalence of mutations and copy-number variations co-altered with FGFR2 amplification, respectively. Prevalence in patients without FGFR2 amplification is highlighted in light colors. cnv, copy-number variation; max VAF, maximum variant allele frequency; mt, mutation.
Next, we evaluated if ctDNA could be used to identify other genomic features in FGFR2-amplified advanced gastric cancer. To this end, we compared concurrent genomic alterations between patients with FGFR2-amplified and nonamplified advanced gastric cancer. In FGFR2-amplified advanced gastric cancer, co-occurring amplifications of PIK3CA, MYC, CDK6, CCND1, BRAF, and CDK4 and mutations in ARID1A, BRCA2, and RHOA were detected at a significantly higher frequency than in gastric cancer without FGFR2 amplification (Fig. 1C). The co-occurring amplification of ERBB2, MET, and EGFR was detected in 1 (3.6%), 3 (10.7%), and 6 (21.4%) of 28 patients with FGFR2 amplification, respectively. These suggest that ctDNA FGFR2-amplified advanced gastric cancer has a distinct profile of concurrent genomic alterations.
ctDNA can detect FGFR2 amplification missed by tissue analyses
The increased prevalence of FGFR2 amplification in ctDNA suggests that ctDNA sequencing may detect heterogeneous FGFR2 amplification that tissue biopsy fails to identify. To assess whether ctDNA analysis can identify FGFR2 amplification missed by tissue analysis, we determined FGFR2 amplification in pretreatment tissue biopsy samples by IHC and FISH and in paired plasma samples obtained near the time of tissue biopsy (median 2 days, interquartile range 1–4 days) by ctDNA sequencing in 44 patients with advanced gastric cancer (Fig. 2A). No ctDNA genomic alteration was identified in 4 (9.1%) patients. FGFR2 amplification was detected by both tissue and ctDNA analysis in 6 patients and ctDNA analysis detected FGFR2 amplification in 6 additional patients, whereas no FGFR2 amplification was detected by tissue analysis only (Fig. 2B). The pCN was not significantly different between tissue+ctDNA+ versus tissue−ctDNA+ (P = 0.18; Fig. 2B). No correlation of the CN detected in tissue and ctDNA analysis was observed in tissue+ctDNA+ and tissue−ctDNA+ patients (r2 = 0.0025; P = 0.88; Fig. 2C). Patients with FGFR2 amplification in ctDNA (tissue+ctDNA+ or tissue−ctDNA+) had a significantly shorter OS than those without FGFR2 amplification [tissue−ctDNA−; median, 13.7 months vs. 27.8 months; HR = 2.2; 95% confidence interval (CI), 1.0–4.9; P = 0.047; Supplementary Fig. S1]. The OS of tissue−ctDNA+ patients was significantly shorter than that of tissue+ctDNA+ patients (median, 12.0 months vs. 14.6 months; HR = 10.1; 95% CI, 1.1–90.8; P = 0.014; Fig. 2D). All of these patients received standard systemic chemotherapy, but not FGFR-targeted therapy. No statistically significant differences were observed in the clinicopathological characteristics among tissue+ctDNA+ and tissue−ctDNA+ patients (Supplementary Table S2). In addition, the median max VAF was 12.8 in the tissue+ctDNA+ and 10.6 in the tissue−ctDNA+ groups with no significant difference (P = 0.52; Supplementary Fig. S2). These findings suggest that ctDNA can identify FGFR2 amplification missed by tissue analyses, which is associated with a poorer prognosis.
Figure 2.
Comparative analysis of paired tissue and plasma samples in patients with advanced gastric cancer. A, Schematic depicting analyses of paired synchronous primary tissue and plasma samples in 44 patients with advanced gastric cancer. B, FGFR2 amplification status based on IHC (score), FISH (FGFR2/CEP10 ratio), and Guardant360 (pCN). Yellow boxes indicate high FGFR2 expression for tissue IHC score and FGFR2 amplification for tissue FISH or ctDNA sequencing. Low FGFR2 expression and no FGFR2 amplification are indicated by blue boxes. C, Correlations [coefficient of determination (r2)] between FGFR2 pCN and tissue CN for patients with FGFR2 amplification detected in ctDNA. D, OS based on the Kaplan–Meier method for patients with FGFR2 amplification detected in tissue+ctDNA+ versus in tissue−ctDNA+. max VAF, maximum variant allele frequency.
Patients with FGFR2 amplification identified by ctDNA analysis can benefit from FGFR inhibition therapy
Previous studies had shown contradictory results regarding the efficacy of FGFR inhibitor therapy in patients with FGFR2-amplified advanced gastric cancer; because our results had shown that FGFR2 amplification might be missed by analysis of single-lesion tumor biopsies probably due to tumor heterogeneity, we next examined whether patients with FGFR2 amplification detected in ctDNA could achieve clinical benefit from an FGFR inhibitor. To this end, we highlight 2 patients who had FGFR2 amplification not detected by tissue analysis but identified by ctDNA sequencing and received an FGFR inhibitor.
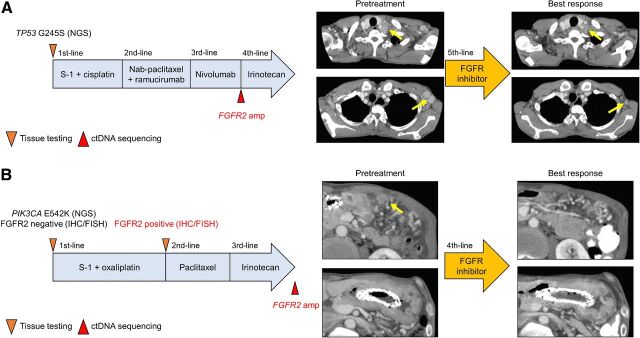
Patient 1 was a 54-year-old man who had recurrent advanced gastric cancer with multiple lymph node metastases. NGS analysis of a tissue sample collected before primary tumor resection identified only a mutation in the TP53 gene. This patient was treated with tegafur/gimeracil/oteracil (S-1) plus cisplatin as first-line, nanoparticle albumin-bound (nab)-paclitaxel plus ramucirumab as second-line, nivolumab as third-line, and irinotecan as fourth-line treatment. At the time of progression on nivolumab, ctDNA analysis using Guardant360 in GOZILA identified FGFR2 amplification with a pCN of 24.2 as well as lower-level amplifications of ERBB2, CDK4, CCND1, and CCNE1, a subclonal FGFR2-TACC2 fusion, and mutations in NF1 and TP53 (Supplementary Table S3). Accordingly, following progression on irinotecan, the patient received an FGFR TKI based on the results of the ctDNA analysis. The patient achieved a −73.6% response with shrinkage of the metastatic cervical and axillary lymph nodes as target lesions (Fig. 3A). The treatment was continued for 3 months, at which point disease progression occurred.
Figure 3.
Treatment history and tumor evaluation by CT before treatment and best response in patients with FGFR2 amplification detected only by ctDNA who had tumor responses to FGFR inhibition with the shrinkage of target lesions (yellow arrows). A, Patient 1. B, Patient 2.
Patient 2 was a 57-year-old man who had unresectable gastric cancer with lymph node metastases and peritoneal dissemination. A pretreatment biopsy of the primary tumor showed only a PIK3CA mutation. This patient was treated with S-1 plus oxaliplatin as first-line, paclitaxel as second-line, and irinotecan as third-line treatment. The analysis of ctDNA using Guardant360 at the time of progression on irinotecan detected FGFR2 amplification with a pCN of 6.0 and mutations in APC and ARID1A (Supplementary Table S3). After progression on irinotecan, he received an FGFR TKI, which lead to the shrinkage of the thickened gastric wall and peritoneal dissemination with a decrease in carbohydrate antigen 19–9 (CA 19–9; Fig. 3B). To investigate changes in FGFR2 amplification patterns during chemotherapy, we retrospectively performed IHC and FISH analysis of tissue samples taken before treatment and after progression on S-1 plus oxaliplatin. Although FGFR2 amplification was not detected in the pretreatment biopsy, the IHC and FISH analysis of the tissue sample obtained after S-1 plus oxaliplatin showed the emergence of high FGFR2 expression (score 4) and FGFR2 amplification with a CN of 32.9 (Supplementary Fig. S3), suggesting that FGFR2 amplification emerged during the treatment or was missed by the initial single-site tissue biopsy in this case and was successfully identified by ctDNA-based sequencing.
These clinical responses to FGFR inhibitors suggest that patients with FGFR2 amplification identified by ctDNA sequencing but not detected by tissue analysis due to heterogeneity potentially benefit from treatment with FGFR inhibitors.
Concurrent MET amplification limited the clinical efficacy of FGFR inhibition therapy
Concurrent genomic alterations in FGFR2-amplified advanced gastric cancer shown in our study may be associated with the resistance to FGFR inhibition. We next report 1 patient with FGFR2 amplification and concurrent MET amplification in ctDNA who was treated with an FGFR inhibitor.
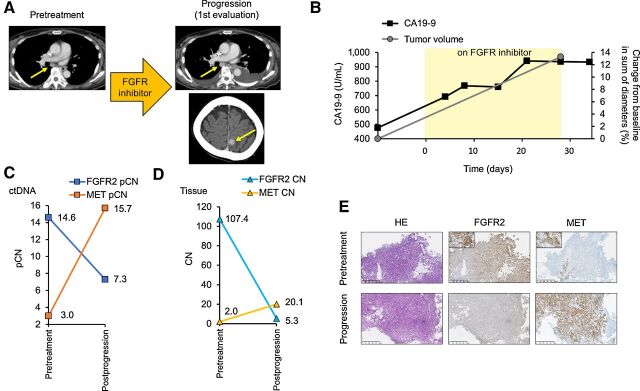
Patient 3 was a 64-year-old woman with unresectable gastric cancer with lymph node metastases and pleural and peritoneal dissemination. This patient was treated with 5-fluorouracil/leucovorin plus oxaliplatin as first-line and nab-paclitaxel plus ramucirumab as second-line treatment. ctDNA sequencing using Guardant360 in GOZILA at the time of progression on nab-paclitaxel plus ramucirumab detected FGFR2 amplification with a pCN of 14.6 with concurrent MET amplification and mutations in TP53, CTNNB1, and ARID1A (Supplementary Table S3). On the basis of ctDNA analysis, she received an FGFR TKI. However, a CT scan at the first evaluation revealed not only progressive lymph node enlargement (+13.3%) but also the emergence of pleural effusion and a new brain metastasis within 30 days after the initiation of the investigational drug (Fig. 4A and B). The tumor marker CA 19–9 increased from 476 to 937 U/mL during this period (Fig. 4B). To identify potential genomic mechanisms of resistance, pre- and postprogression tissue and postprogression plasma were analyzed using OCA and Guardant360, respectively. Compared with the pretreatment pCNs, the pCN of FGFR2 amplification decreased after FGFR inhibitor therapy, falling from 14.6 to 7.3, whereas the pCN of MET amplification markedly increased from 3.0 to 15.7 (Fig. 4C; Supplementary Table S3). Paired tissue NGS analysis also showed similar decreases in the FGFR2 CN and the emergence of MET amplification (Fig. 4D). These dynamic changes in FGFR2 and MET amplification were confirmed by the IHC analysis of protein expression in the paired tissue samples (Fig. 4E). Thus, in the patients with concurrent MET amplification, the clinical benefit of FGFR inhibitors may be limited by the outgrowth of MET-amplified clones that are not sensitive to FGFR inhibition.
Figure 4.
Clinical presentation. A, Tumor evaluation by CT at pretreatment and progression on an FGFR inhibitor with progressive lymph node enlargement and the emergence of a new brain metastasis (yellow arrows) in patient 3. B, Changes in CA 19-9 and sum of diameters of target lesions by CT following treatment with an FGFR inhibitor. C, Change in the ctDNA pCN of FGFR2 and MET amplification before treatment and at progression on an FGFR inhibitor. D, Change in the tissue CN of FGFR2 and MET amplification before treatment and at progression on FGFR inhibitor. E, Hematoxylin and eosin–stained and IHC-stained images with anti-FGFR2 and MET antibodies of biopsy specimens of the primary gastric cancer before treatment and at progression on an FGFR inhibitor.
Discussion
The efficacy of FGFR inhibition has not been clear for FGFR2-amplified advanced gastric cancer with significant genomic heterogeneity, because more effective testing to detect FGFR2 amplification for this population has not been established before FGFR treatment. This study reveals that ctDNA sequencing can more frequently detect FGFR2 amplification than tissue analysis by identifying FGFR2 amplifications that may be missed by conventional tissue analysis. In addition, some patients with FGFR2 amplification identified only by ctDNA sequencing responded to treatment with an FGFR inhibitor. To our knowledge, this is the first report to show the efficacy of FGFR inhibition for advanced gastric cancer with amplified FGFR2 detected only by ctDNA sequencing.
Patients with FGFR2 amplification that is detected in ctDNA but undetectable by tissue analysis in our study might have FGFR2-amplified tumor cells in distant metastatic organs or primary tumors missed by single-lesion biopsy. These patients with tissue−ctDNA+ for FGFR2 amplification tended to have poorer prognosis than those with FGFR2 amplification detectable in tissue, despite no statistically significant differences in the clinicopathologic characteristics and max VAF among these groups. This is consistent with previous studies showing an association between genomic heterogeneity in gastric cancer and poor prognosis (34), although it remains possible that ctDNA shedding due to the tumor burden contributed to this effect. These findings support that the utility of ctDNA sequencing in advanced gastric cancer for identifying genomic heterogeneity reported previously (22) can be applied for FGFR2-amplified disease.
In our case series, ctDNA detected FGFR2 amplification that was not detected with tissue testing, which may have occurred due to the acquisition of FGFR2 amplification in the interim between tissue and ctDNA testing and/or intratumoral heterogeneity that can be missed by single-lesion biopsies. Interestingly, the retrospective testing of a previously untested postprogression tissue sample in patient 2 confirmed the ctDNA finding, suggesting that FGFR2 amplification arose in this patient after initial chemotherapy or was missed by the initial single-lesion biopsy and successfully identified by ctDNA analysis, which integrates tumor cells throughout the body. This finding underscores the importance of genomic profiling of patients with advanced gastric cancer at each instance of disease progression, which can be achieved by ctDNA sequencing with minimal invasiveness in clinical practice.
In patient 3 in our study, ctDNA detected the concurrent amplification of FGFR2 and, to a lesser degree, MET. At primary progression on an FGFR inhibitor, the MET pCN was markedly elevated from baseline, while FGFR2 pCN was reduced, suggesting that two coexisting tumor cell subpopulations driven by different oncogenes responded to FGFR inhibition in opposing manners. Tumor tissue analysis and IHC confirmed that the dominant FGFR2-expressing tumor cell population was replaced by MET-expressing clones upon progression. These findings strongly indicate the relevance of MET amplification in limiting the clinical efficacy of FGFR inhibition. This observation has particular relevance in light of our ctDNA genomic profiling, which revealed the frequent co-occurrence of FGFR2 amplification with other alterations, including ERBB2, EGFR, and MET amplifications. Others have similarly suggested that spatial intratumoral heterogeneity and concurrent genomic alterations in downstream molecules or other signaling pathways could act as resistance mechanisms to targeted therapies in advanced gastric cancer (23–26), leading to the frequent failure of targeted therapies. As such, ctDNA sequencing may also be useful for assessing concurrent genomic alterations to guide treatments.
An important caveat is that our findings were restricted to a Japanese population, although the frequency of tissue FGFR2 amplification in GI-SCREEN was consistent with that in the TCGA or MSKCC data sets. The analysis of paired tissue and plasma was conducted with a limited sample size due to the availability of plasma samples from the same timepoint as tissue collection. Furthermore, the frequency of FGFR2 amplification of this analysis was much higher than previously reported because this population included the patients previously known to have FGFR2 amplification. In addition, ctDNA genotyping potentially underestimated the frequency of FGFR2 amplification because some patients with gastric cancer have insufficient ctDNA shedding to detect genomic alterations in ctDNA, although no patients in our study had FGFR2 amplification only in tissue, suggesting the prevalence of FGFR2 amplification missed by ctDNA analysis due to the low amount of ctDNA may be limited. The efficacy of an FGFR inhibitor for FGFR2 amplification in ctDNA was also shown only in 2 patients. The utility of ctDNA sequencing in detecting FGFR2 amplification in advanced gastric cancer needs to be investigated in prospective studies.
In conclusion, we report the utility of ctDNA sequencing for the detection of FGFR2 amplification that is missed by tissue analysis. Patients with such FGFR2 amplifications have a poor prognosis when treated with standard nontargeted therapies but may benefit from FGFR inhibitor treatment. We also found that concurrent MET amplification detected by ctDNA sequencing was associated with limited clinical efficacy of FGFR inhibition, suggesting that combined FGFR and MET inhibition may be indicated in such cases. Taking advantage of the ability of ctDNA sequencing to detect FGFR alterations, we are currently conducting a phase II basket trial of futibatinib, an irreversible FGFR TKI, for patients with solid tumors harboring FGFR alterations confirmed by Guardant360 (JapicCTI-194624; ref. 35). This trial will provide more validated evidence regarding the utility of ctDNA sequencing for identifying FGFR alterations for FGFR-targeted therapy.
Authors' Disclosures
Y. Nakamura reports grants from Taiho Pharmaceutical Co., Ltd., Chugai Pharmaceutical Co., Ltd., Daiichi Sankyo Co., Ltd., Guardant Health, Inc., Genomedia, Inc., and Seagen, Inc. outside the submitted work. K. Shitara reports grants and personal fees from Astellas Pharma, Eli Lilly and Company, Ono Pharmaceutical, Daiichi Sankyo, Taiho Pharmaceutical, and Merck Pharmaceutical; personal fees from Bristol-Myers Squibb, Takeda Pharmaceuticals, Pfizer Inc., Novartis, AbbVie Inc., Yakult, GlaxoSmithKline, Amgen, and Boehringer Ingelheim; and grants from Dainippon Sumitomo Pharma, Chugai Pharma, Medi Science, and Eisai outside the submitted work. H. Bando reports personal fees from Eli Lilly Japan and Taiho Pharmaceutical outside the submitted work. H. Yasui reports grants from MSD, Ono Pharmaceutical; grants and personal fees from Daiichi Sankyo; and personal fees from Taiho Pharmaceutical, Chugai Pharma, Bristol-Myers Squibb Japan, TERUMO, Eli Lilly Japan, Merck Biopharma, and Yakult Honsha outside the submitted work. T. Esaki reports grants and personal fees from MSD, Ono, Daiichi Sankyo, and Merck Serono; grants from Novartis, Dainippon Sumitomo, Astellas, AstellasAmgenBiopharma, BeiGene, Pierre Fabre Medicament, Ignyta, and Array BioPharma; and personal fees from Bayer, Eli Lilly, Taiho, Chugai, Sanofi, and Takeda outside the submitted work. T. Satoh reports grants and personal fees from Ono Pharmaceutical, Chugai Pharmaceutical, Yakult Honsha, Eli-Lilly, MSD, Daiichi-Sankyo, and Taiho Pharmaceutical, as well as grants from Gilead, BeiGene, and Astellas outside the submitted work. T. Nishina reports grants and personal fees from Taiho Pharmaceutical Co., Chugai Pharmaceutical Co., Ono Pharmaceutical Co., Bristol-Myers Squibb, and Lilly Pharma, as well as grants from MSD, Sumitomo Dainippon Pharma Co., and AstraZeneca outside the submitted work. Y. Sunakawa reports grants and personal fees from Takeda, Sanofi, Chugai Pharmaceutical, and Taiho Pharmaceutical; personal fees from Bristol-Myers Squib, Merck Biopharma, and Lilly Japan; and grants from Otsuka Pharmaceutical outside the submitted work. H. Hara reports grants from Astellas, AstraZeneca, Eisai, Elevar, GSK, Incyte, Pfizer, and BeiGene; grants and personal fees from Bayer, Boehringer Ingelheim, Chugai, Daiichi Sankyo, Dainippon Sumitomo, Merck, MSD, Ono, and Taiho; and personal fees from BMS, Kyowa Hakko Kirin, Sanofi, Takeda, and Yakult outside the submitted work. E. Oki reports other support from Taiho Pharm during the conduct of the study, as well as other support from Eli Lilly, Bayer, Chugai Pharm, and Ono Pharm outside the submitted work. T. Ohta reports personal fees from Bristol-Myers Squibb Co., Ltd., Eli Lilly Japan K.K., Novartis Pharma K.K., Chugai Pharmaceutical Co., Ltd., Teijin Pharma Ltd., Takeda Pharmaceutical Co. Ltd., Taiho Pharmaceutical Co. Ltd, and Eisai Co., Ltd. outside the submitted work. T. Kawakami reports personal fees from Bayer, Taiho Pharmaceutical, Takeda, Ono Pharmaceutical, and Bristol-Myers Squibb outside the submitted work. N. Okano reports personal fees from Taiho Pharmaceutical, Eli Lilly Japan, Kyowa Hakko Kirin, Eisai, Bayer Yakuhin, Chugai, J-Pharma, Ono Pharmaceutical, Takeda, and GSK outside the submitted work. T. Yamada reports personal fees from Johnson and Johnson, Taiho, Ono, and Nippon Kayaku outside the submitted work. A. Tsuji reports personal fees from Takeda Pharmaceutical Company Limited, Sanofi Corporation, and Pfizer Japan Inc.; grants and personal fees from Merck Serono Co., Ltd., Chugai Pharmaceutical Co., Ltd., Taiho Pharmaceutical Co., Ltd., Eli Lilly Japan Co., Ltd., Ono Pharmaceutical Co., Ltd., Kyowa Hakko Kirin Co., Ltd., MSD Corporation, Toray Medical Co., Ltd., Daiichi Sankyo Co., Ltd., and Bristol-Myers Squibb Corporation; and grants from Eisai Co., Ltd. outside the submitted work. J.I. Odegaard reports employment with Guardant Health. T. Doi reports grants from Lilly, Merck Serono, Pfizer, BMS, IQVIA, and Eisai; grants and personal fees from MSD, Daiichi Sankyo, Sumitomo Dainippon, Taiho, Novartis, Janssen Pharma, Boehringer Ingelheim, and AbbVie; and personal fees from Takeda, Chugai Pharma, Bayer, Rakuten Medical, Otsuka Pharma, BMS, Ono Pharma, Oncolys Bio Pharma, Amgen, and Astellas Pharma outside the submitted work. T. Yoshino reports grants from Taiho Pharmaceutical, Sumitomo Dainippon Pharma, Ono Pharmaceutical, Chugai Pharmaceutical, Amgen, PAREXEL International, MSD, Daiichi Sankyo, and Sanofi outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
BAC clones used for FISH analysis
Association between the FGFR2 amplification status and clinicopathological features
Alterations detected by ctDNA sequencing
OS based on the Kaplan-Meier method for patients with FGFR2 amplification detected in tissue+ctDNA+ or tissue-ctDNA+ versus in tissue-ctDNA-
Box plots show max VAF in the tissue+ctDNA+ and tissue-ctDNA+ groups
FGFR Immunohistochemical staining and FISH images in biopsy specimens from the primary tumor of Patient 2
Supplementary Figure legend
Acknowledgments
We would like to thank the Translational Research Support Section of the National Cancer Center Hospital East for study management and data center support; Geneticlab Co., Ltd. and Thermo Fisher Scientific, Inc. for data analysis; and Mari Takahashi and Yuka Nakamura for their excellent technical assistance. This work was supported by grants from the Japan Agency for Medical Research and Development (20ck0106447h0002 to T. Yoshino).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
T. Jogo: Conceptualization, data curation, formal analysis, investigation, visualization, writing–original draft, project administration. Y. Nakamura: Conceptualization, resources, formal analysis, visualization, writing–original draft, project administration. K. Shitara: Resources, writing–review and editing. H. Bando: Resources, writing–review and editing. H. Yasui: Resources, writing–review and editing. T. Esaki: Resources, writing–review and editing. T. Terazawa: Resources, writing–review and editing. T. Satoh: Resources, writing–review and editing. E. Shinozaki: Resources, writing–review and editing. T. Nishina: Resources, writing–review and editing. Y. Sunakawa: Resources, writing–review and editing. Y. Komatsu: Resources, writing–review and editing. H. Hara: Resources, writing–review and editing. E. Oki: Resources, writing–review and editing. N. Matsuhashi: Resources, writing–review and editing. T. Ohta: Resources, writing–review and editing. T. Kato: Resources, writing–review and editing. K. Ohtsubo: Resources, writing–review and editing. T. Kawakami: Resources, writing–review and editing. N. Okano: Resources, writing–review and editing. Y. Yamamoto: Resources, writing–review and editing. T. Yamada: Resources, writing–review and editing. A. Tsuji: Resources, writing–review and editing. J.I. Odegaard: Resources, investigation, writing–review and editing. H. Taniguchi: Conceptualization, writing–review and editing. T. Doi: Writing–review and editing. S. Fujii: Resources, investigation, writing–review and editing. T. Yoshino: Conceptualization, supervision, funding acquisition, project administration, writing–review and editing.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Hecht JR, Bang YJ, Qin SK, Chung HC, Xu JM, Park JO, et al. Lapatinib in combination with capecitabine plus oxaliplatin in human epidermal growth factor receptor 2-positive advanced or metastatic gastric, esophageal, or gastroesophageal adenocarcinoma: TRIO-013/LOGiC–A randomized phase III trial. J Clin Oncol 2016;34:443–51. [DOI] [PubMed] [Google Scholar]
- 3. Tabernero J, Hoff PM, Shen L, Ohtsu A, Shah MA, Cheng K, et al. Pertuzumab plus trastuzumab and chemotherapy for HER2-positive metastatic gastric or gastro-oesophageal junction cancer (JACOB): final analysis of a double-blind, randomised, placebo-controlled phase 3 study. Lancet Oncol 2018;19:1372–84. [DOI] [PubMed] [Google Scholar]
- 4. Satoh T, Xu RH, Chung HC, Sun GP, Doi T, Xu JM, et al. Lapatinib plus paclitaxel versus paclitaxel alone in the second-line treatment of HER2-amplified advanced gastric cancer in Asian populations: TyTAN–a randomized, phase III study. J Clin Oncol 2014;32:2039–49. [DOI] [PubMed] [Google Scholar]
- 5. Thuss-Patience PC, Shah MA, Ohtsu A, Van Cutsem E, Ajani JA, Castro H, et al. Trastuzumab emtansine versus taxane use for previously treated HER2-positive locally advanced or metastatic gastric or gastro-oesophageal junction adenocarcinoma (GATSBY): an international randomised, open-label, adaptive, phase 2/3 study. Lancet Oncol 2017;18:640–53. [DOI] [PubMed] [Google Scholar]
- 6. Waddell T, Chau I, Cunningham D, Gonzalez D, Okines AF, Okines C, et al. Epirubicin, oxaliplatin, and capecitabine with or without panitumumab for patients with previously untreated advanced oesophagogastric cancer (REAL3): a randomised, open-label phase 3 trial. Lancet Oncol 2013;14:481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lordick F, Kang YK, Chung HC, Salman P, Oh SC, Bodoky G, et al. Capecitabine and cisplatin with or without cetuximab for patients with previously untreated advanced gastric cancer (EXPAND): a randomised, open-label phase 3 trial. Lancet Oncol 2013;14:490–9. [DOI] [PubMed] [Google Scholar]
- 8. Dutton SJ, Ferry DR, Blazeby JM, Abbas H, Dahle-Smith A, Mansoor W, et al. Gefitinib for oesophageal cancer progressing after chemotherapy (COG): a phase 3, multicentre, double-blind, placebo-controlled randomised trial. Lancet Oncol 2014;15:894–904. [DOI] [PubMed] [Google Scholar]
- 9. Catenacci DVT, Tebbutt NC, Davidenko I, Murad AM, Al-Batran SE, Ilson DH, et al. Rilotumumab plus epirubicin, cisplatin, and capecitabine as first-line therapy in advanced MET-positive gastric or gastro-oesophageal junction cancer (RILOMET-1): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2017;18:1467–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shah MA, Bang YJ, Lordick F, Alsina M, Chen M, Hack SP, et al. Effect of fluorouracil, leucovorin, and oxaliplatin with or without onartuzumab in HER2-negative, MET-positive gastroesophageal adenocarcinoma: the METGastric randomized clinical trial. JAMA Oncol 2017;3:620–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Katoh M. Fibroblast growth factor receptors as treatment targets in clinical oncology. Nat Rev Clin Oncol 2019;16:105–22. [DOI] [PubMed] [Google Scholar]
- 12. Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin Cancer Res 2016;22:259–67. [DOI] [PubMed] [Google Scholar]
- 13. Jung EJ, Jung EJ, Min SY, Kim MA, Kim WH. Fibroblast growth factor receptor 2 gene amplification status and its clinicopathologic significance in gastric carcinoma. Hum Pathol 2012;43:1559–66. [DOI] [PubMed] [Google Scholar]
- 14. Su X, Zhan P, Gavine PR, Morgan S, Womack C, Ni X, et al. FGFR2 amplification has prognostic significance in gastric cancer: results from a large international multicentre study. Br J Cancer 2014;110:967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuboki Y, Schatz CA, Koechert K, Schubert S, Feng J, Wittemer-Rump S, et al. In situ analysis of FGFR2 mRNA and comparison with FGFR2 gene copy number by dual-color in situ hybridization in a large cohort of gastric cancer patients. Gastric Cancer 2018;21:401–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pearson A, Smyth E, Babina IS, Herrera-Abreu MT, Tarazona N, Peckitt C, et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov 2016;6:838–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jang J, Kim HK, Bang H, Kim ST, Kim SY, Park SH, et al. Antitumor effect of AZD4547 in a fibroblast growth factor receptor 2-amplified gastric cancer patient-derived cell model. Transl Oncol 2017;10:469–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cha Y, Kim HP, Lim Y, Han SW, Song SH, Kim TY. FGFR2 amplification is predictive of sensitivity to regorafenib in gastric and colorectal cancers in vitro. Mol Oncol 2018;12:993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Van Cutsem E, Bang YJ, Mansoor W, Petty RD, Chao Y, Cunningham D, et al. A randomized, open-label study of the efficacy and safety of AZD4547 monotherapy versus paclitaxel for the treatment of advanced gastric adenocarcinoma with FGFR2 polysomy or gene amplification. Ann Oncol 2017;28:1316–24. [DOI] [PubMed] [Google Scholar]
- 20. Nakamura Y, Yoshino T. Clinical utility of analyzing circulating tumor DNA in patients with metastatic colorectal cancer. Oncologist 2018;23:1310–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nakamura Y, Shitara K. Development of circulating tumour DNA analysis for gastrointestinal cancers. ESMO Open 2020;5:e000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pectasides E, Stachler MD, Derks S, Liu Y, Maron S, Islam M, et al. Genomic heterogeneity as a barrier to precision medicine in gastroesophageal adenocarcinoma. Cancer Discov 2018;8:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kwak EL, Ahronian LG, Siravegna G, Mussolin B, Borger DR, Godfrey JT, et al. Molecular heterogeneity and receptor coamplification drive resistance to targeted therapy in MET-amplified esophagogastric cancer. Cancer Discov 2015;5:1271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sanchez-Vega F, Hechtman JF, Castel P, Ku GY, Tuvy Y, Won H, et al. EGFR and MET amplifications determine response to HER2 inhibition in ERBB2-amplified esophagogastric cancer. Cancer Discov 2019;9:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maron SB, Alpert L, Kwak HA, Lomnicki S, Chase L, Xu D, et al. Targeted therapies for targeted populations: anti-EGFR treatment for EGFR-amplified gastroesophageal adenocarcinoma. Cancer Discov 2018;8:696–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakamura Y, Sasaki A, Yukami H, Jogo T, Kawazoe A, Kuboki Y, et al. Emergence of concurrent multiple EGFR mutations and MET amplification in a patient with EGFR-amplified advanced gastric cancer treated with cetuximab. JCO Precis Oncol 2020;4:PO.20.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wainberg ZA. Randomized double-blind placebo-controlled phase 2 study of bemarituzumab combined with modified FOLFOX6 (mFOLFOX6) in first-line (1L) treatment of advanced gastric/gastroesophageal junction adenocarcinoma (FIGHT). In: Zev A. Wainberg PCEY-KKKYSQK-WLSCOJLHMTACTGGC, xF, o SMYYHCDVTC, UCLA Medical Center - Cancer Care - Santa Monica LACA, Dana-Farber Cancer Institute BMA, Asan Medical Center SSK, et al., editors. 2021; Gastrointestinal Cancers Symposium. American Society of Clinical Oncology. [Google Scholar]
- 28. Nakamura Y, Taniguchi H, Ikeda M, Bando H, Kato K, Morizane C, et al. Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med 2020;26:1859–64. [DOI] [PubMed] [Google Scholar]
- 29. Kuwata T, Wakabayashi M, Hatanaka Y, Morii E, Oda Y, Taguchi K, et al. Impact of DNA integrity on the success rate of tissue-based next-generation sequencing: Lessons from nationwide cancer genome screening project SCRUM-Japan GI-SCREEN. Pathol Int 2020;70:932–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. [DOI] [PubMed] [Google Scholar]
- 31. Lih CJ, Harrington RD, Sims DJ, Harper KN, Bouk CH, Datta V, et al. Analytical validation of the next-generation sequencing assay for a nationwide signal-finding clinical trial: molecular analysis for therapy choice clinical trial. J Mol Diagn 2017;19:313–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Odegaard JI, Vincent JJ, Mortimer S, Vowles JV, Ulrich BC, Banks KC, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res 2018;24:3539–49. [DOI] [PubMed] [Google Scholar]
- 33. Maron SB, Chase LM, Lomnicki S, Kochanny S, Moore KL, Joshi SS, et al. Circulating tumor DNA sequencing analysis of gastroesophageal adenocarcinoma. Clin Cancer Res 2019;25:7098–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oh BY, Shin HT, Yun JW, Kim KT, Kim J, Bae JS, et al. Intratumor heterogeneity inferred from targeted deep sequencing as a prognostic indicator. Sci Rep 2019;9:4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jogo T, Nakamura Y, Komatsu Y, Kato K, Shinozaki E, Bando H, et al. TiFFANY study: a multicenter phase II basket-type clinical trial to evaluate efficacy and safety of pan-FGFR inhibitor TAS-120 for advanced solid malignancies with FGFR alterations identified by circulating tumor DNA. J Clin Oncol 2019;37:TPS3156–TPS. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BAC clones used for FISH analysis
Association between the FGFR2 amplification status and clinicopathological features
Alterations detected by ctDNA sequencing
OS based on the Kaplan-Meier method for patients with FGFR2 amplification detected in tissue+ctDNA+ or tissue-ctDNA+ versus in tissue-ctDNA-
Box plots show max VAF in the tissue+ctDNA+ and tissue-ctDNA+ groups
FGFR Immunohistochemical staining and FISH images in biopsy specimens from the primary tumor of Patient 2
Supplementary Figure legend



![Figure 1. Genomic characteristics of advanced gastric cancer with FGFR2 amplification based on ctDNA analysis. A, Prevalence of FGFR2 amplification in advanced gastric cancer in GOZILA, GI-SCREEN, and the TCGA and MSKCC databases. B, Correlations [coefficient of determination (r2)] between FGFR2 plasma copy number (pCN) and max VAF in FGFR2-amplified samples from GOZILA (n = 28). C, Prevalence of co-alterations in FGFR2-amplified (n = 28) versus nonamplified patients (n = 337) in GOZILA. Green and blue bars indicate prevalence of mutations and copy-number variations co-altered with FGFR2 amplification, respectively. Prevalence in patients without FGFR2 amplification is highlighted in light colors. cnv, copy-number variation; max VAF, maximum variant allele frequency; mt, mutation.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/9d69/9401460/906299765fcd/5619fig1.jpg)
![Figure 2. Comparative analysis of paired tissue and plasma samples in patients with advanced gastric cancer. A, Schematic depicting analyses of paired synchronous primary tissue and plasma samples in 44 patients with advanced gastric cancer. B, FGFR2 amplification status based on IHC (score), FISH (FGFR2/CEP10 ratio), and Guardant360 (pCN). Yellow boxes indicate high FGFR2 expression for tissue IHC score and FGFR2 amplification for tissue FISH or ctDNA sequencing. Low FGFR2 expression and no FGFR2 amplification are indicated by blue boxes. C, Correlations [coefficient of determination (r2)] between FGFR2 pCN and tissue CN for patients with FGFR2 amplification detected in ctDNA. D, OS based on the Kaplan–Meier method for patients with FGFR2 amplification detected in tissue+ctDNA+ versus in tissue−ctDNA+. max VAF, maximum variant allele frequency.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/9d69/9401460/f1b67b6f7254/5619fig2.jpg)