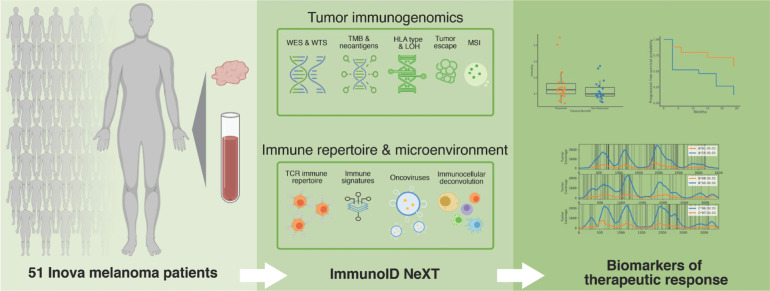
Abstract
Purpose:
While immune checkpoint blockade (ICB) has become a pillar of cancer treatment, biomarkers that consistently predict patient response remain elusive due to the complex mechanisms driving immune response to tumors. We hypothesized that a multi-dimensional approach modeling both tumor and immune-related molecular mechanisms would better predict ICB response than simpler mutation-focused biomarkers, such as tumor mutational burden (TMB).
Experimental Design:
Tumors from a cohort of patients with late-stage melanoma (n = 51) were profiled using an immune-enhanced exome and transcriptome platform. We demonstrate increasing predictive power with deeper modeling of neoantigens and immune-related resistance mechanisms to ICB.
Results:
Our neoantigen burden score, which integrates both exome and transcriptome features, more significantly stratified responders and nonresponders (P = 0.016) than TMB alone (P = 0.049). Extension of this model to include immune-related resistance mechanisms affecting the antigen presentation machinery, such as HLA allele-specific LOH, resulted in a composite neoantigen presentation score (NEOPS) that demonstrated further increased association with therapy response (P = 0.002).
Conclusions:
NEOPS proved the statistically strongest biomarker compared with all single-gene biomarkers, expression signatures, and TMB biomarkers evaluated in this cohort. Subsequent confirmation of these findings in an independent cohort of patients (n = 110) suggests that NEOPS is a robust, novel biomarker of ICB response in melanoma.
Translational Relevance.
This work evaluates the predictive strength of a composite biomarker, neoantigen presentation score (NEOPS), in a cohort of patients with late-stage melanoma. NEOPS incorporates damaging events in the antigen presentation machinery with predicted neoantigens to stratify patient response to immunotherapy. Others have previously reported that tumor mutational burden and RNA-based signatures are associated with therapeutic response, albeit with limited reproducibility. Data presented here demonstrate that our integrative approach outperformed such single-analyte biomarkers, suggesting that more complex models capturing multiple aspects of tumor escape may provide more robust stratification of patient response. We also demonstrate that data intensive biomarkers such as NEOPS are clinically practical, with comprehensive tumor profiling in our clinical cohort achieved using limited tumor tissue. These findings provide a novel composite biomarker of response in patients with late-stage melanoma, as well as evidence supporting the use of whole-exome and whole-transcriptome data in a clinical setting.
Introduction
Checkpoint inhibitor therapy has demonstrated meaningful, if varied, antitumor activity, with patient response influenced by a variety of biological factors, including complex interactions between the tumor, tumor microenvironment (TME), and immune system (1–5). Numerous biomarkers of response to immune checkpoint blockade (ICB) have been proposed, including PD-L1 expression, IFNγ based signatures, tumor mutational burden (TMB), mismatch repair deficiency, genetic alterations including those within the antigen presentation machinery (APM), HLA LOH, and T-cell repertoire diversity (6–12).
Owing to the diversity of biological features that can influence response to ICB therapy, there has been increasing effort toward identifying biomarkers that integrate multiple biological features to better predict response to immunotherapy (13). Work integrating immunogenicity and neoantigen clonal structures predicted response to ICB and prognosis in patients with melanoma, lung cancer, and kidney cancers, suggesting broad applicability of the biomarker (14). While these studies have yielded many positive results, challenges remain; investigation of individual biomarkers can overlook the cumulative impact of rare events, and more extensive integrative studies have been impeded by the difficult task of assembling the broad input data required for analysis.
To address these challenges, we used a validated, enhanced exome- and transcriptome-based tumor profiling platform to generate the broad tumor immunogenomic inputs required for an integrative biomarker approach. We optimized this platform to elucidate tumor mutational information as well as neoantigen prediction, immune repertoire, gene expression, and HLA mutations/type from a single formalin-fixed paraffin-embedded (FFPE) tumor sample and paired normal sample. We then used this platform to profile a cohort of patients with advanced melanoma (n = 51) treated with anti-PD-1 ICB, allowing us to integrate a broad set of biological features. From these data, we developed a novel composite framework for predicting ICB response that models biological mechanisms driving response and resistance to cancer therapy.
Materials and Methods
Experimental design
Paired pretreatment FFPE tumor and normal blood sample was collected and profiled using Personalis' ImmunoID NeXT platform; an augmented exome/transcriptome platform and analysis pipeline, which produces comprehensive tumor mutation information, gene expression quantification, neoantigen characterization, HLA (typing, mutation, and LOH), T-cell receptor (TCR) repertoire profiling, and TME profiling. These data were then analyzed together with clinical outcome, and a composite neoantigen score computed for each patient along with additional biomarkers such as TMB.
Study population
A total of 51 patients with unresectable, stage III/IV melanoma who underwent treatment at the Inova Schar Cancer Institute (Annandale, VA) were enrolled retrospectively without randomization or blinding. Patients were treated with either nivolumab (480 mg i.v. every 4 weeks or 240 mg i.v. every 2 weeks), a combination of nivolumab and ipilimumab (1 mg/kg i.v. and 3 mg/kg i.v., respectively, every 3 weeks), or pembrolizumab (200 mg i.v. every 3 weeks). Solid tumor and blood samples were collected within 3 months prior to treatment start. CT scans were performed 10–12 weeks after treatment start, with follow-up scans every 3 months. Responders were defined as complete response or partial response. Nonresponders were defined as stable disease or progressive disease. This study was conducted in accordance with recognized ethical guidelines including the Belmont Report and U.S. Common Rule, and was approved by the Human Research Protections Program at Inova; protocol number 16-2427. In addition, written informed consent was obtained from each patient.
Whole-exome sequencing
Whole-exome library preparation and sequencing was performed as described previously (15, 16). Whole-exome capture libraries were constructed using DNA from tumor and blood. Target probes were used to enhance coverage of biomedically and clinically relevant genes. Protocols were modified to yield an average library insert length of approximately 250 bp. In addition, we used KAPA HiFi DNA Polymerase (Kapa Biosystems) instead of Herculase II DNA polymerase (Agilent). Paired-end sequencing was performed using either HiSeq 2500 or NovaSeq instrumentation (Illumina).
Alignment
Reads were mapped to the hs37d5 reference genome build. The pipeline performs alignment, duplicate removal, and base quality score recalibration (BQSR) using best practice guidelines recommended by the Broad Institute (17, 18). The pipeline uses the Picard toolkit (RRID:SCR_006525) for duplicate removal and Genome Analysis Toolkit (GATK, RRID:SCR_001876) to improve sequence alignment and to correct base quality scores (BQSR). Aligned sequence data are then returned in BAM format according to the SAM (RRID:SCR_01095) specification. A complete list of all major tools used for processing and analysis can be found in Supplementary Table S1.
SNV and indel calling
GATK's HaplotypeCaller was used to generate the core set of single-nucleotide variant (SNV) calls and their accompanying quality metrics. The pipeline then uses GATK's variant quality score recalibration module, which stratifies SNVs by their likelihood of representing false positive calls, and in-house SNV accuracy software, which incorporates both genomic context and sequence alignment information into a model that corrects miscalled variants. All calls were made on BAM files recalibrated by GATK's BAM processing tools. MuTect (RRID:SCR_000559) was used to call somatic SNVs and indels, with Vardict used for calling small somatic insertions or deletions (<50 bp). Somatic SNV and indel calls were then combined and analyzed through a comprehensively tested set of filters based on (i) alignment metrics, such as sequence coverage and read quality, (ii) positional features, such as proximity to a gap region, and (iii) likelihood of presence in normal tissue.
Transcriptomic analysis
Whole-transcriptome sequencing was aligned using STAR (RRID:SCR_015899; ref. 19) and normalized expression values in transcripts per million (TPM) calculated by an in-house tool, Expressionist. For RNA sequencing and alignment quality control, we evaluated the following metrics: average read length, percentage of uniquely mapped reads, average mapped read pair length, number of splice sites, mismatch rate per base, deletion/insertion rate per base, mean deletion/insertion length, and anomalous read pair alignments including inter-chromosomal and orphaned reads. Three samples were withheld from sequencing and analysis due to poor-quality RNA.
Immune infiltrate signatures
Immune infiltration scores were calculated using transcriptome data from ImmunoID NeXT. Semiquantitative scores representing the enrichment of gene sets in single samples (19, 20) were calculated using single-sample gene set enrichment analysis (21). We utilized a publicly available set of reference gene expression signatures representing 17 cell types (22). With this approach, enrichment scores for the same cell type (gene set) can be compared across samples, profiling immune infiltration across the cohort.
TCRβ clonality
We profiled TCRβ clones using the ImmunoID NeXT transcriptome, which provides augmented (approximately a 100× increase over a standard transcriptome) coverage of TCRβ. Only patients with sufficient remaining evaluable material were included in this analysis (n = 28). We identified clones using MiXCR (RRID:SCR_018725; ref. 23). Nonproductive clones which have a frameshift or premature stop codon in the CDR3 sequence were filtered out, as well as low-confidence clones which have an alignment score below threshold for the V or J hit. Clonality was then calculated as 1-Pielou's evenness (24).
Differential expression analyses
Counts per million (CPM) was calculated by normalizing read counts by the total number of reads per sample. Only genes with CPM > 0 in 25% or more of the samples were included for analysis. Remaining data were then rlog transformed and differential gene expression analyzed using the DESeq2 package (RRID:SCR_000154; ref. 25). Genes with an Padjusted < 0.05, and a minimum log2 fold change of ←0.5 or >1 were considered differentially expressed. Biological significance of differentially expressed genes (DEG) was explored at the pathway level using MSigDB (Molecular Signatures Database, RRID:SCR_016863) hallmark gene sets and Kyoto Encyclopedia of Genes and Genomes (RRID:SCR_012773) gene sets (26, 27).
Allele-specific HLA LOH
HLA LOH was detected using a machine learning–based algorithm called DASH (Deletion of Allele-Specific HLAs; ref. 28). A set of 720 heterozygous genes from 279 patients were used to train DASH. The features used for training the algorithm were gathered with the following steps:
1. All tumor and normal reads were mapped to the patient's allele-specific HLA references sequences. No mismatches were allowed except for alleles with an identified somatic mutation. Reads with >20% of their total length soft clipped were removed.
2. Homologous alleles were aligned to find all patient-specific mismatch positions.
3. Coverage values were calculated by Samtools at each mismatch position (SNP and indel) for each homologous allele
- 4. Features were collected for each heterozygous gene pair. The features included:
- (i) Adjusted b-allele frequency: For each position of difference (SNP or indel) between the homologous alleles, a b-allele frequency (BAF) was calculated for both the normal and tumor reads. To adjust for differences in probe capture of the two alleles, the tumor BAF was divided by the normal BAF. To attain a single feature, the median adjusted BAF was taken within each 150 bp bin. Then, the median was taken across all bins.
- (ii) Allele-specific coverage ratios: For each position of difference between the homologous alleles, the ratio between the tumor and normal coverage for each specific allele was calculated. The values were normalized by the genome wide ratio. The minimum value from the two alleles was taken at each position. The median within each bin was calculated and then the median across the bins was used as the feature.
- (iii) Consistency of coverage: For each position of difference between the homologous alleles, a 0 or 1 was assigned for each allele depending on if it has higher coverage than the other allele. The median value within all bins is calculated and then the median value across all bins. The higher value of the two alleles is taken for the feature value.
- (iv) Tumor purity: The tumor purity value was called by Sequenza.
- (v) Tumor ploidy: The tumor ploidy value was called by Sequenza.
- (vi) Deletion of flanking regions: This is a binary feature—1 if Sequenza calls a deletion in the 10,000 bp flanking the gene and a 0 otherwise.
HLA deletion labels were manually curated through visualization for all heterozygous genes. An XGBoost model was trained using these features and labels to learn to predict occurrences of HLA LOH. To increase specificity at low tumor purities with poor training data, we implemented a secondary check that requires the allele-specific coverage ratio to be <0.98 and the adjusted BAF to be > 0.02 for HLA LOH to be detected. If HLA LOH was detected by DASH, the allele with the lower coverage was labeled as deleted. DASH was validated using cell line dilutions and allele-specific digital PCR and demonstrated higher sensitivity and specificity than LOH HLA. For every patient in this study, DASH was used to detect HLA LOH for all heterozygous alleles. Additional details describing this methodology can be found in Supplementary Materials and Methods S1.
Neoantigen prediction and composite NEOPS
Putative neoantigens were generated using tumor-specific genomic events (SNVs, indels, and fusions) that were verified using transcriptomic data. For each tumor-specific genomic event, all peptides containing the event were generated. All candidate peptides were scored using SHERPA, a machine learning tool for predicting MHC class I presentation (29).
SHERPA is a composite model that is trained on large-scale monoallelic immunopeptidomics data of >70 alleles (HLA-A/HLA-B/HLA-C). Monoallelic data were generated through stable transfections to the K562 HLA-null cell line. All immunopeptidomics data were processed through PEAKs. Negative examples were generated by randomly selecting peptides from the human proteome. Twenty times as many negatives as positives were used to train the model. SHERPA is a pan-allelic, pan-length model that incorporates the following features:
1. Peptide sequence: The peptide sequence (8- to 11-mers). All peptides were adjusted to a length of 11 amino acids by adding padding to the middle of the sequence. The peptide was encoded as using a BLOSUM62 matrix, resulting in an 11 × 10 matrix.
2. Peptide length: A number denoting the number of amino acids in the peptide.
3. Allele binding pocket sequence: The binding pocket is represented by the 34-mer amino acid sequence that this within 4 Å from the peptide. The sequence is encoded using a BLOSUM62 substitution matrix.
4. Gene expression: The TPM of the gene from which the neoantigen is derived.
5. Peptide flanking region sequences: The five amino acids upstream and downstream of the peptide. The sequences are encoded using a BLOSUM62 matrix.
6. Propensity of a gene to engender presented peptides: Publicly available multi-allelic immunopeptidomics data were systematically reprocessed in a uniform pipeline. The number of peptides mapping to each transcript-associated protein were calculated. An expectation was determined using the TPM, the length of the transcript, and the total number of peptides observed. The feature was calculated as the observed count over the expected value for each protein.
7. Propensity of a within-gene region to engender presented peptides: Peptides were mapped on to all transcript-associated proteins. To assign this feature, the peptide of interest was mapped to its respective protein and the coverage of all amino acids for that peptide were averaged.
SHERPA uses an XGBoost machine learning model. Two models were trained—a binding model, consisting of only the peptide and binding pocket features, and a presentation model, consisting of all features. Both SHERPA models return a rank score, similar to NetMHCpan, that can be interpreted as the percentile of binding or presentation probability compared with other random peptides with the same allele. SHERPA was benchmarked against NetMHCpan 4.0 on an independent set of immunopeptidomics data from tumor samples and attained higher overall sensitivity and specificity. Neoantigen burden score (NBS) was calculated by filtering out peptides with a presentation rank >0.5 (equivalent to NetMHCpan rank value), and dividing by footprint size in Mb. Additional details surrounding this methodology can be found in Supplementary Materials and Methods S1.
To calculate the composite NEOPS, we developed an approach which adjusts NBS to account for patient-specific tumor alterations which may impair neoantigen presentation, including alterations to the MHC and antigen presentation machinery, and HLA LOH. Predicted neoantigens are filtered using the following criteria: (i) Tumors harboring high impact (mutations called by MuTect, and impact defined by SnpEff) mutations in beta-2-microglobulin (B2M) are assigned a score of 0. (ii) High impact somatic variants (called by Polysolver) in HLA-A, HLA-B, and HLA-C are identified, and all neoantigens predicted to be presented by the damaged HLA are removed from consideration. (iii) Allele-specific HLA LOH is determined using DASH, and neoantigens predicted to be presented by the lost HLA allele(s) are removed from consideration. All remaining neoantigens are then summed, and divided by exome footprint in Mb to yield the final NEOPS value.
Validation cohorts
Replication of our findings was conducted using publicly available next-generation sequencing (NGS) data collected from patients with advanced melanoma who underwent ICB therapy (30). Whole-exome and RNA sequencing (RNA-seq) data from this study were obtained from dbGaP (NCBI database of Genotypes and Phenotypes, RRID:SCR_002709; study accession: phs000452.v3.p1). Patients with mixed responses to therapy (n = 2) and low purity tumors (n = 7) were excluded from the analysis, leaving (n = 110) evaluable patients for validation. Clinical characteristics for the validation cohort are provided in the original study.
Statistical analysis
The Kaplan–Meier method was used to estimate progression-free survival (PFS) and overall survival (OS). Univariate CoxPH models were fit to compare the relative effects of increasing NBS and NEOPS on survival using the rms package for R. Objective response rate was reported as proportion along with Clopper–Pearson exact confidence intervals (CI). Fisher exact test and χ2 test were used to test for associations between groups, and categorical variables. When considering the variance between more than two groups, the Kruskal–Wallis H test was used. The Wilcoxon Mann–Whitney rank-sum test (MWW) was used for numeric pairwise comparisons. Benjamini–Hochberg correction was used to adjust P values as listed. The Kolmogorov–Smirnov statistic was used for RNA pathway analyses. Correlations between continuous variables were determined using Kendall tau. Predictive models were generated using logistic regression, and area under the receiver operating characteristic (AUROC) used to determine ability to differentiate between response and nonresponse using published methods and code (30). All tests were two sided; FDR values of <0.1 for pathway analyses, and P values of <0.05 for all other tests were considered statistically significant.
Data availability
Raw sequencing and cohort-level data are available in dbGaP (accession number: phs2388.v1.p1). Mutation, RNA count data, and patient-level data are provided as Supplementary Tables. Code used for analysis and plotting is available at https://codeocean.com/capsule/1428816/tree/v1.
Results
Cohort clinical and genomic characteristics
We identified 51 patients with unresectable melanoma treated with ICB at the Inova Schar Cancer Institute (Annandale, VA) for inclusion in this study (Fig. 1). Median follow-up for the cohort was 24 months, with 33 of 51 patients (50%, 95% Clopper–Pearson CI of 50%–78%) presenting an objective response at first evaluation by RECIST 1.1. The observed response rate is significantly higher than that found in other studies, possibly due to sample size. Within the cohort, tumors originated in the head and neck region (31%), trunk (31%), extremities (25%), acral areas (6%), mucosa (4%), and 2% from occult regions. In addition to these data, sex, age and other subject demographics information is presented in Supplementary Table S2. There were no statistically significant differences in objective response rate between sites of disease origin. A total of 11 patients (22%) had progressed following prior treatment with a checkpoint inhibitor, whereas 40 (78%) were naive to ICB. Patients were administered either pembrolizumab (n = 29, 57%), nivolumab (n = 15, 29%), or a combination of nivolumab and ipilimumab (n = 7, 14%).
Figure 1.
Study schema. Pretreatment blood normal and tumor samples were collected from 51 patients with unresectable, stage III/IV melanoma who underwent anti–PD-1 therapy. Samples were profiled using Personalis' ImmunoID NeXT platform, an enhanced exome/transcriptome platform and analysis pipeline, which produces comprehensive tumor mutation information, gene expression quantification, neoantigen characterization, HLA (typing, mutation, and LOH), TCR repertoire profiling, MSI detection, oncovirus identification, and TME profiling. These data were then analyzed together with clinical outcome, and a composite neoantigen score computed for each patient along with additional biomarkers, such as TMB.
Mutations associated with responding and nonresponding tumors were investigated, revealing no significant single-gene predictors of response following multiple hypothesis correction (patient-level mutation data; Supplementary Table S3). Next, we investigated genetically disrupted pathways (31, 32). The most frequently disrupted pathways include RTK-RAS and WNT pathways (disrupted in 73% and 51% of our cohort, respectively; Supplementary Fig. S1A). Mutations were detected throughout the RTK-RAS pathway: numerous RTKs were mutated, including ROS1 and ERBB4, RAS family genes including NRAS, BRAF, and MAPK1 and MAPK2 (Supplementary Fig. S1B).
Transcriptomic features are associated with response to ICB
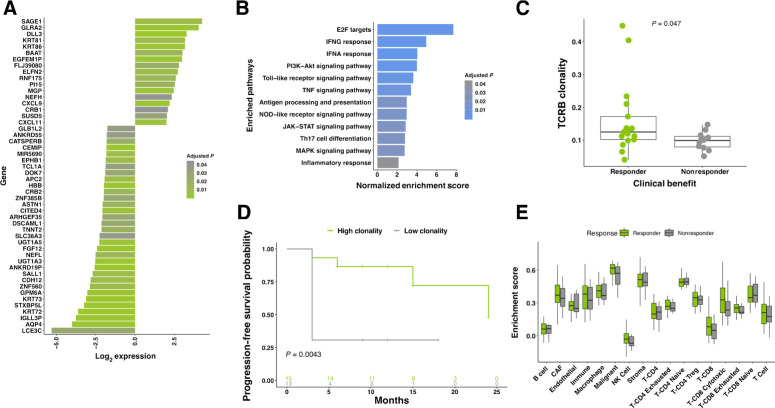
We next investigated transcriptomic profiles within the cohort (raw count data; Supplementary Table S4). From the RNA-seq data, we identified 121 DEGs in responding patients (n = 48 evaluable patients; Supplementary Table S5; Padjusted ≤ 0.05, log fold change > 2 or ←0.5). Enrichment was observed in 29 of these genes, while reduced expression was observed in 92 (top 50 largest fold change genes, Fig. 2A; patient-level heatmap, Supplementary Fig. S1C). Among the most strongly upregulated genes (log2 fold change = 3.28; FDR-adjusted P = 0.0005) that we detected was delta-like ligand 3 (DLL3), an inhibitory Notch ligand that exhibits high expression in small cell lung cancer (SCLC) and other tumors tissues (33). Because of its low cytoplasmic expression in normal tissue, compared with elevated, homogeneous cell surface expression in tumors, it is currently under investigation as a possible therapeutic target (34). Validation of DESeq2 results for DLL3 using MWW confirmed significance (MWW P = 0.02). Though not significantly enriched at a cohort level, IDO1 expression was detected at very high levels in three individuals (median IDO1 TPM = 10.36; outlier IDO1 TPM = 1,955, 661, and 451; Supplementary Fig. S1D). Two of the patients overexpressing IDO1 failed to achieve complete response to therapy, possibly due to an IDO1-driven immunosuppressive environment (35).
Figure 2.
Transcriptomic features associated with response. A, Top 50 DEG. Fold change shown comparing responding patients to nonresponding patients. Benjamini–Hochberg corrected P values below 0.05 are shown, n = 48. B, GSEA identified significant enrichment of pathways related to immune function among genes upregulated in responding patients. Benjamini–Hochberg corrected P values below 0.05 are shown. C, TCRβ clonality is elevated in responding patients, compared with nonresponders (n = 28; MWW; P = 0.047). D, Significantly longer PFS was observed in high-clonality patients when compared with those with low clonality (n = 28; two-sided KM log-rank test; P = 0.0043). High/low stratification was calculated independently for old/young populations (median cohort age used as cut-off point). E, Characterization of tumor-infiltrating lymphocytes. TREG, regulatory T cell; NK cell, natural killer cell; CAF, cancer-associated fibroblast. All boxplots in C and E cover the IQR from 25th percentile at their lower bound to the 75th percentile at their upper bound, with median indicated by a horizontal line. The upper whisker includes the largest value within 1.5× IQR above the 75th percentile. The lower whisker includes the smallest value within 1.5× IQR below the 25th percentile.
Next, we performed gene set enrichment analysis (GSEA) to identify differentially regulated pathways (26, 27). Inflammatory signaling cascades were among the most highly enriched of those profiled (Fig. 2B; significance set as FDR < 0.1; Supplementary Table S3). Activation of these pathways likely results from other enriched pathways; cellular differentiation of Th17 is driven by the cytokine TGFβ, which induces RORγt in Th17 cells, and along with IL6, induces the Th17 lineage (36, 37). The observed enrichment of Th17 may also be positively regulated by the observed increase in STAT3 signaling, which serves to promote Th17 differentiation.
Immune repertoire and immune cell associations with therapeutic response
The adaptive immune system is able to respond to a broad array of antigens due to its large repertoire of unique TCRs. To characterize the pretreatment tumor-immune landscape, we profiled and analyzed TCRβ repertoire diversity using ImmunoID NeXT in a subset of patients (n = 28 patients). Clonality was determined for the clonal abundance of all productive TCRβ sequences using 1-Pielou's evenness. As intratumoral heterogeneity is thought to be a determinant of immune response (38), we compared mutant-allele tumor heterogeneity scores (39), which estimate tumor heterogeneity, and TCRβ clonality. Here, we identified a significant association (MWW, P = 0.014) between high tumor heterogeneity and clonal diversity of the TCRβ repertoire (Supplementary Fig. S1E). TCRβ clonality was found to be significantly associated with therapy outcome (MWW, P = 0.047; Fig. 2C), and PFS (two-sided Kaplan–Meier (KM) log-rank test, P = 0.0043; Fig. 2D), but not treatment history (Supplementary Table S6).
Characterization of immune and stromal cell populations within the TME in our cohort was carried out using publicly available gene sets (22), which were used to produce semiquantitative immune infiltration scores. Using this approach, we found that responding and nonresponding patients largely shared similar distributions of immune cells (Fig. 2E).
TMB as a biomarker of response to ICB
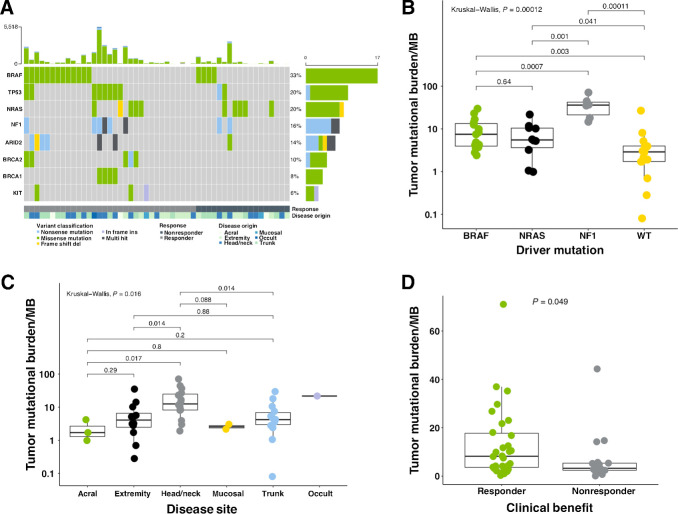
In the discovery cohort, median nonsynonymous TMB was 4.07 mutations/Mb [interquartile range (IQR), 0.95–12.455] (Fig. 3A), consistent with values observed in prior literature and The Cancer Genome Atlas datasets (Supplementary Fig. S2A). C>T transitions make up the bulk of identified SNVs (76%; Supplementary Fig. S2B), and mutational signatures (40) found in the cohort most strongly associated with UV-induced DNA damage (Supplementary Fig. S2C and S2D). The most commonly identified driver mutation occurred in BRAF, in 33% of patients, followed by 20% NRAS and 16% NF1 in the study population. Response rate for the different genomic subtypes did not significantly vary from the expected response rate (Supplementary Fig. S2E). The elevated number of triple wild-type (WT) patients likely arises from the reduced frequency of BRAF, which are typically observed at higher rates (41).
Figure 3.
Genomic features and the tumor mutational landscape in melanoma patients. A, Mutation in driver genes of patients receiving anti–PD-1 therapy. Top bar plot represents mutational load. Tiled plot shows mutated genes (rows) by sample (columns), with tile color indicating mutation type. The bar plot to the right represents the number of patients with mutations in the specified gene, colored to indicate mutation type. Under the tiled plot, the first line represents therapeutic response, as either response (partial or complete response; dark green; n = 33), or nonresponse (black; n = 18). B, Comparison of TMB in tumors harboring different driver mutations revealed significant variation (KW; P = 0.00012). Values are plotted on log10 scale. C, Comparison of TMB in different melanoma types, and sites of origin revealed significant global variation (KW; P = 0.016), with significant variation found in comparison with melanomas originating in the head and neck. Values are plotted on log10 scale. D, Comparison of TMB in responding versus nonresponding patients revealed significant associations (MMW; P = 0.049). All boxplots in B–D cover the IQR from 25th percentile at their lower bound to the 75th percentile at their upper bound, with median indicated by a horizontal line. The upper whisker includes the largest value within 1.5× IQR above the 75th percentile. The lower whisker includes the smallest value within 1.5× IQR below the 25th percentile.
TMB varied significantly between tumors harboring different driver mutations (Kruskal–Wallis, P = 0.00012; Fig. 3B), different sites of disease origin (Kruskal–Wallis, P = 0.016; Fig. 3C), as well as between responding and nonresponding patients (P = 0.049; Fig. 3D). The relatively small variance between TMB in responding and nonresponding patients in this cohort (Fig. 3D) could be due to the confounding effects of melanoma subtype, and varying tumor purity, as these measures have recently been shown to limit TMB's effectiveness as a predictive biomarker (30, 42).
A neoantigen-based biomarker approach achieves stronger correlation with response to ICB
We developed two different neoantigen models, one score based on neoantigen burden (NBS), and another that extended this model to account for impairment to neoantigen presentation and other established resistance markers, creating a composite NEOPS.
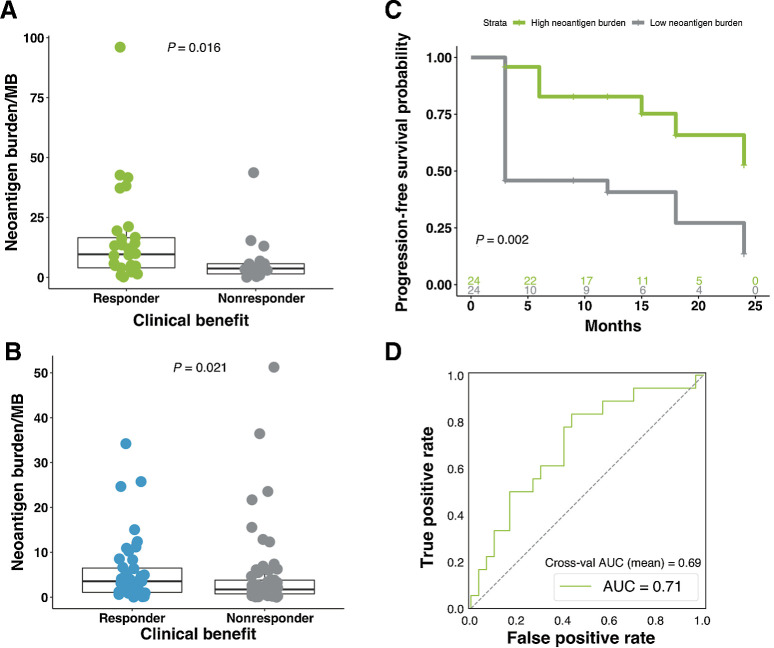
To calculate NBS, we developed an integrative computational pipeline that utilized broad exome- and transcriptome-derived features. Putative neoepitopes were predicted from SNVs, indels, and fusions detected from both exome and transcriptome sequencing. To improve MHC class I neoantigen prediction, we generated mass spectrometry–based peptide binding data from monoallelic HLA-transfected cell lines. These data were used to train an improved machine learning algorithm which integrates HLA binding, proteasomal cleavage, and gene expression information to improve neoantigen prediction. Using this approach, we surveyed the neoantigen landscape across different driver mutations in this cohort, revealing significant variation among subtypes (Kruskal–Wallis, P = 1e-04; Supplementary Fig. S2F). Counter to what was observed with TMB, we did not detect a significant association across disease sites of origin (Kruskal–Wallis, P = 0.08; Supplementary Fig. S2G), suggesting that neoantigen burden may be robust to these influences, despite being strongly correlated with TMB (Kendall tau = 0.8625637, P < 2.2e-16; Supplementary Table S6). In addition, no significant variance was observed between treatment-naive and treatment-experienced patients (Supplementary Table S6). We found that neoantigen burden was significantly elevated in patients that responded to therapy in the current cohort (MWW, P = 0.016; Fig. 4A), and confirmed these findings in an independent validation cohort of 110 patients with advanced melanoma (30) who received ICB (MWW, P = 0.021; Fig. 4B). Patients with above median (high) predicted neoantigens had longer PFS than those with low neoantigen load (two-sided KM log-rank test, P = 0.002; Fig. 4C). While PFS of high neoantigen burden patients was not significantly longer than those with low neoantigen burden in the validation cohort, marked improvements to OS were observed (two-sided KM log-rank test, P = 0.085, Supplementary Fig. S2H and P = 0.005, Supplementary Fig. S2I). AUROC for the NBS model was 0.71 and the cross-validation AUC mean was 0.69 (log-likelihood ratio P = 0.0329; Fig. 4D).
Figure 4.
Neoantigen burden is associated with response to therapy. A, Neoantigen burden is significantly higher in responding patients compared with nonresponding patients (n = 48; MWW; P = 0.016). Boxplot covers the IQR from the 25th percentile at its lower bound to the 75th percentile at its upper bound, with median indicated by a horizontal line. The upper whisker includes the largest value within 1.5× IQR above the 75th percentile. The lower whisker includes the smallest value within 1.5× IQR below the 25th percentile. B, Similar findings were observed in the validation cohort, with patients who responded to therapy presenting significantly higher neoantigen burden (MWW; P = 0.021). C, Significantly longer PFS was observed in patients with high neoantigen burden when compared with those with low neoantigen burden (two-sided KM log-rank test; P = 0.002). D, AUROC for the neoantigen model was 0.71, and the cross-validation AUROC mean was 0.69 (log-likelihood ratio P = 0.0329).
Integrating antigen presentation into a composite neoantigen score strengthens association with ICB response
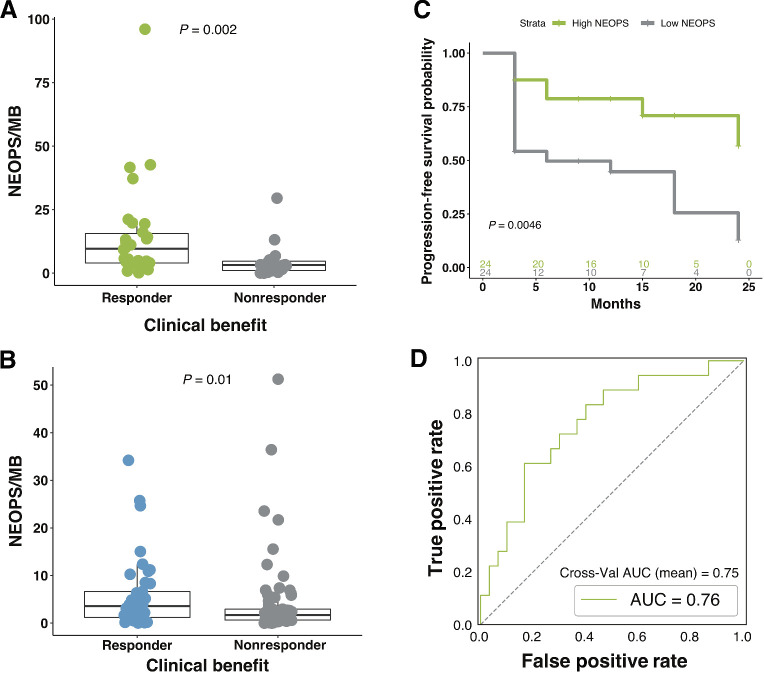
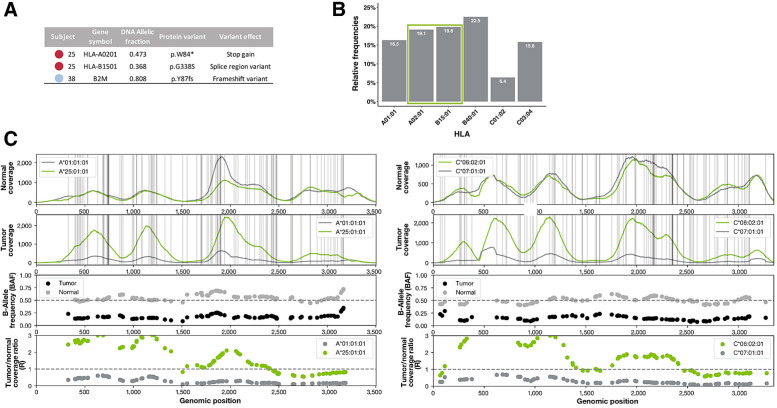
We hypothesized that accounting for alterations in the APM that could interfere with neoantigen presentation could improve the performance of the model as these events have been noted individually to impact patient response to ICB. To explore this, we created NEOPS, a computational model that adjusts the NBS to account for patient specific tumor alterations that could interfere with neoantigen presentation, including HLA mutations, HLA LOH, and B2M mutations. Contribution of each NEOPS feature to model performance is presented in Supplementary Table S7. Analysis of patients in this cohort using NEOPS resulted in improved prediction of therapy outcome, when compared with neoantigen burden and TMB alone (MWW, P = 0.002; Fig. 5A), a finding which was replicated in our validation cohort (MWW, P = 0.01; Fig. 5B). In addition, NEOPS was significantly associated with longer PFS (two-sided KM log-rank test, P = 0.0046; Fig. 5C). Model performance is improved with the inclusion of the additional features: AUROC for NEOPS increased to 0.76 and the cross-validation AUC mean was 0.75 (log-likelihood ratio P = 0.0057; Fig. 5D). In contrast to what was found for neoantigen burden in the validation cohort, PFS of patients with high NEOPS was significantly longer than those with low NEOPS (two-sided KM log-rank test, P = 0.05; Supplementary Fig. S2J). Greater significance was also achieved when analyzing OS, which was significantly longer in patients with high NEOPS (two-sided KM log-rank test, P = 0.002; Supplementary Fig. S2K). Comparison of log relative hazard predicted for NBS and NEOPS demonstrates greater proportional reduction in relative hazard with increasing NEOPS (HR = 0.399, P = 0.026) compared with NBS (HR = 0.545, P = 0.046; Supplementary Fig. S2L). The improvement with NEOPS can be understood biologically with the finding that 23.5% of patients in the discovery cohort, and 17.27% of patients in the validation cohort had at least one mechanism potentially affecting antigen presentation, suggesting these features may frequently influence therapy response. A review of damaging HLA mutations across the cohort revealed deleterious variants in many patients (patients 25 and 38 highlighted in Fig. 6A). We identified two distinct somatic HLA mutations in patient 25; a stop gain mutation in HLA-A02:01, and a splice region variant in HLA-B15:01 (allele fraction = 0.473 and 0.368, respectively), that can lead to the loss of surface expression of HLA-A02:01 and possible misfolding of HLA-B15:01. 38.9% of neoantigens in this patient were predicted to bind to the damaged alleles (Fig. 6B), suggesting potentially severe impairment of neoantigen presentation. Of note, this patient was an outlier in the nonresponding cohort, with much higher neoantigen burden, suggesting impaired neoantigen presentation beyond that which is captured in NEOPS may be a contributing factor to ICB resistance. In another outlier patient (high neoantigen burden, nonresponder), a damaging frameshift variant was detected in B2M at a high allelic fraction (Fig. 6A), also potentially impacting antigen presentation. HLA LOH was also examined in this cohort, as it can potentially impact neoantigen presentation. HLA LOH is an acquired resistance mechanism that facilitates immune escape by reducing capacity for presentation of tumor neoantigens to the immune system (15). As the process of HLA loss is governed by selective pressures within the TME, particularly at later stages of tumor evolution, we hypothesized that within our cohort of patients with late-stage melanoma allele-specific HLA LOH could contribute to reduced therapeutic response despite apparent elevated neoantigen burden. We found that HLA LOH was the most prevalent form of HLA disruption, occurring in 19.6% of evaluable patients (10/51), with three individuals presenting LOH across all non-homozygous HLAs (these data and other patient-level values are presented in Supplementary Table S8). Highlighting one such case, we see that matched normal tissue from the patient generally presents even allele-specific coverage across HLAs A and C (Fig. 6C, HLA-A at left, HLA-C at right). In contrast, tumor tissue from this patient exhibits broad imbalances in allele-specific coverage spanning large portions of each HLA, with low levels of coverage in HLA-A01:01 and HLA-C07:01. BAF shows absolute difference from the normal. Consistently, lower ratio of coverage is observed in the lost alleles (Fig. 6C, bottom plots), which are predicted to present approximately 54% of this patient's neoantigens, likely reducing capacity for presentation to the immune system.
Figure 5.
Composite neoantigen presentation score is more strongly associated with response to therapy than neoantigen burden alone. A, Composite NEOPS is significantly higher in responding patients compared with nonresponding patients (n = 48; MWW; P = 0.002). B, Similar findings were observed in the validation cohort, with high responding patients presenting significantly higher NEOPS (n = 110; MWW; P = 0.010). C, Significantly longer PFS was observed in patients with high NEOPS when compared with those with low NEOPS (two-sided KM log-rank test; P = 0.0046). Boxplots in A and B cover the IQR from the 25th percentile at its lower bound to the 75th percentile at its upper bound, with median indicated by a horizontal line. The upper whisker includes the largest value within 1.5× IQR above the 75th percentile. The lower whisker includes the smallest value within 1.5× IQR below the 25th percentile. D, AUROC for the NEOPS model was 0.76, and the cross-validation AUROC mean was 0.75 (log-likelihood ratio P = 0.0057).
Figure 6.
Changes to APM that may contribute to immune evasion. A, Somatic HLA mutations detected in patient 25 may lead to the loss of surface expression of HLA-A02:01 and possible misfolding of HLA-B15:01. A damaging frameshift variant was detected in B2M in patient 38, possibly impairing all MHC class I presentation in that patient. B, 38.9% of neoantigens predicted to be presented by patient 25 are predicted to bind to the damaged alleles described in A. C, Four panels showing the NGS sequence-based evidence for HLA LOH in HLA-A and HLA-C of patient 54. HLA-B is not shown. The first row shows the raw read coverage of both homologous alleles in the normal sample. The second row shows the raw read coverage of both homologous alleles in the tumor sample. Both plots have vertical gray lines representing the positions of difference between the two alleles. Because of strict mapping parameters requiring all reads to map without mismatch, differences in coverage at the gray lines represent true differences in coverage between the alleles. The third panel shows the BAF from the normal sample (gray) and the tumor sample (black). The BAF in the tumor sample must be considered in light of the BAF in the normal sample because of primer hybridization differences between the alleles. The fourth panel shows the ratio in coverage between the tumor and normal samples for each allele. These values have been normalized by the tumor and normal read depth across the whole exome. The expected value with no copy-number change is 1, shown with a dashed gray line. Both the third and fourth panel show data only for the mismatch positions between the two alleles.
Validation of model performance for both NBS and NEOPS was attempted in the same cohort, though we found reduced performance for both models (AUC 0.63 and 0.65, respectively; Supplementary Table S7). One factor likely contributing to the reduced predictive strength is the relatively poor coverage of HLA A, B, and C genes in the validation cohort (median coverage of 66.95 in the validation cohort vs. 250.01 in the discovery cohort). Despite this, comparison of AUC values between NEOPS and previously described models and transcriptomic signatures highlights the strength of this approach (Supplementary Table S7).
Discussion
In our study, we show that a composite approach like NEOPS, which models both biological mechanisms, and impairment of neoantigen presentation can serve as a strong predictor for ICB therapy response. Here, NEOPS achieved greater separation of ICB therapy responders and nonresponders than TMB and other single analyte/gene, and expression signatures examined in the discovery cohort. We then further demonstrated the value of NEOPS by confirming these findings in a large independent validation cohort. Broadly, we show that integrative, composite biomarker approaches like ours combining both DNA and RNA features, tumor and immune features, and response and resistance biomarkers can better model the complex biological mechanisms underlying ICB therapy response, and potentially improve patient stratification.
These results are consistent with an increasing body of literature across multiple cancer types that have demonstrated that neoantigens can guide immune response, promoting clinical response to immunotherapy (43, 44). While we observed only weak association between response and TMB, stronger association between neoantigen burden and patient response was apparent. Recent work (30) suggests that this finding may be attributed to confounding effects of the distribution of melanoma subtypes within this cohort, which negatively impact the predictive power of TMB, but not neoantigen burden. Future studies leveraging large cohorts such as that found in work by Conway and colleagues (45) will likely reveal further subtleties surrounding this relationship. It is possible that the increased robustness of neoantigen burden as a biomarker is achieved through the inclusion of additional data from subsequent processing steps, as well as RNA expression levels, as this measure has been found to correlate with protein representation in the MHC-bound peptide repertoire (46).
While elevated measures of neoantigen burden predict in part which patients will benefit from immunotherapy, additional resistance mechanisms arising from genetic variation in the antigen presentation machinery, both at a germline as well as somatic level, may further modulate immune response by diminishing capacity for neoantigen presentation (10). We detected a collection of damaging APM alterations that have been previously associated with reduced response to immunotherapy, including HLA class I and B2M mutations, and LOH in HLA class I genes (8, 15, 47, 48). By accounting for these escape mechanisms, and combining them into a composite neoantigen score, we captured a fuller representation of tumor antigen presentation to the immune system compared to simpler models such as TMB, increasing the predictive strength of this biomarker. This approach will likely yield exciting results when applied to non–small cell lung carcinoma and squamous cell carcinoma of the head and neck patient cohorts, as HLA LOH is a prevalent escape mechanism in these, and other tumor types (49). Indeed, work by Filip and colleagues (49) which leverages data from >3,500 tumors found allele-specific expression loss at frequencies above 45% in head and neck, lung adenocarcinoma, pancreatic, and prostate cancers. This, combined with the well-documented prevalence of somatic mutations in class I HLA genes suggests a broad pervasiveness of damaging APM events captured by NEOPS.
In this study, we identified additional factors influencing patient response outside of neoantigen burden. The outlier, non-responding patient in the validation cohort with high NEOPS presents with metastatic desmoplastic melanoma, which is associated with high levels of mutational burden and distinct clinicopathologic and genetic features compared with typical cutaneous melanomas, likely explaining the comparatively high NEOPS (50). While this may explain lack of concordance with predicted outcome in this case, it also highlights some of the limitations of our approach. These limitations are read out as reduced NBS and NEOPS model performance in the validation cohort, where we also observe small relative improvements in PFS and OS. One possible factor impacting performance is reduced coverage of the HLA region in the validation cohort, which contributes critical information for calculating NBS and NEOPS. Discovery cohort samples have significantly increased coverage in the region (median 66.95× coverage in the validation cohort vs. 250.01× in the discovery cohort), and therefore increased confidence in HLA calls. This has wide-ranging effects, from predicting neoantigen presentation to calling HLA LOH. While this makes validation challenging, we believe it also demonstrates the strength of our augmented capture approach for library preparation and sequencing. These observations also suggest that there are opportunities to further expand our composite biomarker approach to model other mechanisms of therapy resistance or response that extend beyond neoantigen presentation.
Given the complex nature of resistance to immunotherapy, as well as potential toxicities associated with treatment, there is a need for biomarkers that can more accurately predict therapeutic response. Here we demonstrate that a composite biomarker approach can significantly improve stratification of patient response. We also demonstrate that data intensive biomarkers like NEOPS can also be clinically practical, with comprehensive tumor profiling in our clinical cohort achieved using very limited tumor tissue. Composite biomarker approaches like this may increasingly serve as important tools for precision immunotherapy as comprehensive exome and transcriptome-scale tumor immunogenomic profiling tests gain clinical adoption.
Authors' Disclosures
C.W. Abbott reports a patent for PCT/US2021/029684 pending. S.M. Boyle reports a patent for PCT/US2021/029684 pending to Personalis. R.M. Pyke reports personal fees from Personalis during the conduct of the study; in addition, R.M. Pyke has a patent for PCT/US2021/029684 pending. E. Levy reports a patent for PCT/US2021/029684 pending. F.C.P. Navarro reports a patent for PCT/US2021/029684 pending. D. Mellacheruvu reports personal fees from Personalis Inc during the conduct of the study; in addition, D. Mellacheruvu has a patent for PCT/US2021/029684 pending to Personalis Inc. S.V. Zhang reports a patent for PCT/US2021/029684 pending. M. Tan reports a patent for PCT/US2021/029684 pending. P. Milani reports a patent for PCT/US2021/029684 pending. G. Bartha reports a patent for PCT/US2021/029684 issued. J. Harris reports a patent for PCT/US2021/029684 pending. R. McClory reports a patent for PCT/US2021/029684 pending. M.P. Snyder reports personal fees from Personalis during the conduct of the study. S. Jang reports grants from Inova Health Foundation during the conduct of the study, as well as personal fees from Bristol-Myers Squibb, Genentech, Sun Biopharma, Novartis, EMD Serono, and Sanofi outside the submitted work. R. Chen reports a patent for PCT/US2021/029684 pending. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
Work by S. Jang was funded by the Inova Health Foundation.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
C.W. Abbott: Data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. S.M. Boyle: Conceptualization, data curation, software, formal analysis, supervision, investigation, methodology, writing–original draft, project administration, writing–review and editing. R.M. Pyke: Formal analysis, visualization, methodology. L.D. McDaniel: Software, Formal analysis. E. Levy: Software, formal analysis. F.C.P. Navarro: Software, formal analysis, investigation. D. Mellacheruvu: Software, formal analysis. S.V. Zhang: Software. M. Tan: Data curation, software. R. Santiago: Data curation, investigation. Z.M. Rusan: Data curation, software, investigation. P. Milani: Software. G. Bartha: Software, methodology. J. Harris: Conceptualization, software, supervision, methodology. R. McClory: Conceptualization, supervision, writing–review and editing. M.P. Snyder: Conceptualization, resources, supervision, investigation, methodology, writing–review and editing. S. Jang: Conceptualization, resources, software, supervision, funding acquisition, investigation, methodology, writing–original draft, writing–review and editing. R. Chen: Conceptualization, resources, software, formal analysis, supervision, funding acquisition, investigation, writing–original draft, writing–review and editing.
References
- 1. Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Larkin J, Hodi FS, Wolchok JD. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med 2015;373:1270–1. [DOI] [PubMed] [Google Scholar]
- 3. Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ribas A, Hamid O, Daud A, Hodi FS, Wolchok JD, Kefford R, et al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA 2016;315:1600–9. [DOI] [PubMed] [Google Scholar]
- 5. Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob J-J, Cowey CL, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 2017;377:1345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Herbst RS, Soria J-C, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 2016;167:397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med 2016;375:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roh W, Chen P-L, Reuben A, Spencer CN, Prieto PA, Miller JP, et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Science Transl Med 2017;9:eaah3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sade-Feldman M, Jiao YJ, Chen JH, Rooney MS, Barzily-Rokni M, Eliane J-P, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun 2017;8:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chowell D, Krishna C, Pierini F, Makarov V, Rizvi NA, Kuo F, et al. Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nat Med 2019;25:1715–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 2017;18:248–62. [DOI] [PubMed] [Google Scholar]
- 14. Lu T, Wang S, Xu L, Zhou Q, Singla N, Gao J, et al. Tumor neoantigenicity assessment with CSiN score incorporates clonality and immunogenicity to predict immunotherapy outcomes. Sci Immunol 2020;5:eaaz3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 2017;171:1259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Patwardhan A, Harris J, Leng N, Bartha G, Church DM, Luo S, et al. Achieving high-sensitivity for clinical applications using augmented exome sequencing. Genome Med 2015;7:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Finotello F, Trajanoski Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol Immunother 2018;67:1031–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su M-J, Melms JC, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 2018;175:984–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods 2015;12:380–1. [DOI] [PubMed] [Google Scholar]
- 24. Kirsch I, Vignali M, Robins H. T-cell receptor profiling in cancer. Mol Oncol 2015;9:2063–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanehisa M. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marty Pyke R, Mellacheruvu D, Dea S, Abbott C, Zhang SV, Levy E, et al. Validated machine learning algorithm with sub-clonal sensitivity reveals widespread pan-cancer human leukocyte antigen loss of heterozygosity. bioRxiv [Preprint] May 22, 2021. Available from: https://doi.org/10.1101/2021.05.20.445052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marty Pyke R, Mellacheruvu D, Dea S, Abbott C, Zhang SV, Phillips NA, et al. Precision neoantigen discovery using large-scale immunopeptidomes and composite modeling of MHC peptide presentation. bioRxiv [Preprint]. June 7, 2021. Available from: https://doi.org/10.1101/2021.04.30.442203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Liu D, Schilling B, Liu D, Sucker A, Livingstone E, Jerby-Arnon L, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med 2019;25:1916–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lui VWY, Peyser ND, Ng PK, Hritz J, Zeng Y, Lu Y, et al. Frequent mutation of receptor protein tyrosine phosphatases provides a mechanism for STAT3 hyperactivation in head and neck cancer. Proc Natl Acad Sci U S A 2014;111:1114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 2018;173:321–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sabari JK, Lok BH, Laird JH, Poirier JT, Rudin CM. Unravelling the biology of SCLC: implications for therapy. Nat Rev Clin Oncol 2017;14:549–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Owen DH, Giffin MJ, Bailis JM, Smit M-AD, Carbone DP, He K. DLL3: an emerging target in small cell lung cancer. J Hematol Oncol 2019;12:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jung KH, LoRusso P, Burris H, Gordon M, Bang Y-J, Hellmann MD, et al. Phase I study of the indoleamine 2,3-dioxygenase 1 (IDO1) inhibitor navoximod (GDC-0919) administered with PD-L1 inhibitor (Atezolizumab) in advanced solid tumors. Clin Cancer Res 2019;25:3220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 2006;441:235–8. [DOI] [PubMed] [Google Scholar]
- 37. Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, et al. IL-6 programs TH-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol 2007;8:967–74. [DOI] [PubMed] [Google Scholar]
- 38. Wolf Y, Bartok O, Patkar S, Eli GB, Cohen S, Litchfield K, et al. UVB-induced tumor heterogeneity diminishes immune response in melanoma. Cell 2019;179:219–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mroz EA, Tward AD, Hammon RJ, Ren Y, Rocco JW. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med 2015;12:e1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 2015;161:1681–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Anagnostou V, Niknafs N, Marrone K, Bruhm DC, White JR, Naidoo J, et al. Multimodal genomic features predict outcome of immune checkpoint blockade in non-small-cell lung cancer. Nat Cancer 2020;1:99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Verdegaal EME, de Miranda NFCC, Visser M, Harryvan T, van Buuren MM, Andersen RS, et al. Neoantigen landscape dynamics during human melanoma-T cell interactions. Nature 2016;536:91–5. [DOI] [PubMed] [Google Scholar]
- 44. Łuksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, Solovyov A, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 2017;551:517–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Conway JR, Dietlein F, Taylor-Weiner A, AlDubayan S, Vokes N, Keenan T, et al. Integrated molecular drivers coordinate biological and clinical states in melanoma. Nat Genet 2020;52:1373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Granados DP, Yahyaoui W, Laumont CM, Daouda T, Muratore-Schroeder TL, Côté C, et al. MHC I-associated peptides preferentially derive from transcripts bearing miRNA response elements. Blood 2012;119:e181–91. [DOI] [PubMed] [Google Scholar]
- 47. Gettinger S, Choi J, Hastings K, Truini A, Datar I, Sowell R, et al. Impaired HLA class I antigen processing and presentation as a mechanism of acquired resistance to immune checkpoint inhibitors in lung cancer. Cancer Discov 2017;7:1420–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018;359:582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Filip I, Orenbuch R, Zhao J, Manji G, López de Maturana E, et al. HLA allele- specific expression loss in tumors can shorten survival and hinder immunotherapy. medRxiv [Preprint] October 4, 2020. Available from: https://doi.org/10.1101/2020.09.30.20204875. [Google Scholar]
- 50. Ochoa CE, Joseph RW. Desmoplastic melanoma: a brief review and the efficacy of immunotherapy. Expert Rev Anticancer Ther 2019;19:205–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing and cohort-level data are available in dbGaP (accession number: phs2388.v1.p1). Mutation, RNA count data, and patient-level data are provided as Supplementary Tables. Code used for analysis and plotting is available at https://codeocean.com/capsule/1428816/tree/v1.