Abstract
Meiosis entry during spermatogenesis requires reprogramming from mitotic to meiotic gene expression profiles. Transcriptional regulation has been extensively studied in meiosis entry, but gain of function for master transcription factors is insufficient to down-regulate mitotic genes. RNA helicase YTHDC2 and its partner MEIOC emerge as essential posttranscriptional regulators of meiotic entry. However, it is unclear what governs the RNA binding specificity of YTHDC2/MEIOC. Here, we identified RNA binding protein RBM46 as a component of the YTHDC2/MEIOC complex. Testis-specific Rbm46 knockout in mice causes infertility with defective mitotic-to-meiotic transition, phenocopying global Ythdc2 or Meioc knockout. RBM46 binds to 3′ UTR of mitotic transcripts within 100 nucleotides from YTHDC2 U-rich motifs and targets these transcripts for degradation. Dysregulated RBM46 expression is associated with human male fertility disorders. These findings establish the RBM46/YTHDC2/MEIOC complex as the major posttranscriptional regulator responsible for down-regulating mitotic transcripts during meiosis entry in mammalian spermatogenesis, with implications for understanding meiosis-related fertility disorders.
The RBM46/YTHDC2/MEIOC complex is the major posttranscriptional regulator that regulates the mitosis-to-meiosis transition.
INTRODUCTION
Meiosis is a unique cell division process in sexually reproducing organisms. The meiotic cell cycle begins with one round of DNA replication, followed by two successive rounds of cell divisions, thus creating haploid germ cells from diploid progenitors. After DNA replication, the meiosis-specific synaptonemal complex physically links homologous chromosomes to pair, allowing the exchange of genetic materials and accurate chromosome segregation at the first meiotic division (1, 2). The assembly of the synaptonemal complex and numerous meiosis-specific chromosomal events occur in the meiotic prophase I. Thus, the entry into the meiotic prophase I is a crucial step of spermatogenesis. Meiotic entry is regulated by retinoic acid through the meiotic initiation factor STRA8 (3, 4). However, STRA8 and its protein partners are necessary but not sufficient to initiate meiotic entry, indicating that multiple layers of regulation underlie the commitment to meiotic initiation.
N6-methyladenosine (m6A) is the most abundant internal modification of mRNAs and has emerged as a key posttranscriptional mechanism regulating gene expression (5–8). m6A methylation occurs at the consensus RRACH motif and is controlled by opposing functions of two classes of enzymes: RNA methyltransferase and RNA demethylases. In mammals, METTL3 and METTL4 form a heterodimer complex to catalyze m6A RNA methylation (9–13), while two m6A demethylases, FTO (14) and ALKBH5, remove it (15). m6A modifications are recognized and bound by m6A reader proteins, and their networks are implicated in many steps of RNA metabolism. The YT521-B homology (YTH) domain–containing proteins are a family of m6A reader proteins that use an aromatic cage to accommodate the methyl group on m6A (16–18). Cytoplasmic YTH domain family 2 (YTHDF2) accelerates mRNA degradation by localizing bound transcripts to processing bodies in the cytoplasm (19) and recruiting the CCR4-NOT deadenylation complex (20). The other two cytoplasmic readers, YTHDF1 (21) and YTHDF3, regulate the translation of methylated mRNAs (22, 23), and the nuclear reader YTHDC1 regulates m6A-dependent pre-mRNA splicing (24). YTHDF3 is also shown to promote mRNA degradation in cooperation with YTHDF2, and m6A-modified transcripts are markedly accumulated in cells that were deficient in all three YTHDF proteins (23, 25), suggesting that YTHDF members function redundantly to mediate mRNA degradation.
YTHDC2 is the fifth member of the YTH family that expresses abundantly in male germ cells. YTHDC2 belongs to the DExH family of RNA-dependent helicases. Crystal structure analyses show that the YTH domain from YTHDC2 resembles other YTH domains with a key feature to accommodate m6A-containing substrates (26, 27), and biochemical studies confirmed its direct recognition to m6A-modified transcripts (26–29). Recombinant human YTHDC2 protein has 3′→5′ RNA helicase activity because it is an RNA-dependent adenosine triphosphatase (ATPase) that unwinds the RNA duplex with a 3′ overhang (26, 27). Unexpectedly, recent genetic evidence demonstrated that m6A-binding capacity of YTHDC2 is not required for spermatogenesis (30, 31). The loss of YTHDC2 3′→5′ RNA helicase activity by a point mutation in the ATPase motif causes infertility with defects in meiotic prophase I progression (30, 31). Male germ cells mutant for Ythdc2 initiate meiosis but fail to maintain prolonged meiotic prophase I with a premature exit into aberrant metaphase (26–28, 32). Recently, YTHDC2 was also shown to modulate the prolonged pachytene stage of meiotic prophase I using an inducible genetic strategy, a function beyond its early role at the meiotic entry (33). In the mouse testis, YTHDC2 associates with the 5′→3′ exoribonuclease XRN1 (26, 29, 30) and the meiotic-specific protein MEIOC (26, 34, 35). The interaction of YTHDC2 to XRN1 is direct, and biochemical assays reveal that the helicase activity of YTHDC2 is enhanced by its interaction with XRN1 (30). Ythdc2 and Meioc have similar mutant phenotypes, and YTHDC2 protein directly interacts with MEIOC in the mouse germ line. Meioc-depleted germ cells enter meiosis but fail to maintain prolonged meiotic prophase I, resulting in a premature transition into abnormal metaphase (34, 35). On the basis of the shared meiotic phenotypes and their physical interaction, it is proposed that YTHDC2 and MEIOC function together to regulate the switch from the mitotic proliferation program to the meiotic program.
In this study, we isolated endogenous YTHDC2 complexes in mouse testis and subsequent mass spectrometry analysis identified RBM46 as an essential YTHDC2-interacting partner. These results are in agreement with recently published data showing that RBM46 protein was identified as a new component of the mouse YTHDC2 complex (30). RBM46 is the vertebrate ortholog of the Drosophila Tut. RBM46 was first reported as a factor that is required for spermatogenesis in zebrafish (36). It is also shown to modulate the expression of embryonic differentiation–related genes (37). Here, we propose that the mouse RBM46-MEIOC-YTHDC2 complex, paralleling the Drosophila Tut-Bam-Bgcn complex, forms an ancient posttranscriptional mechanism that ensures a successful transition into the meiotic program at the meiotic entry.
RESULTS
RBM46 is associated with YTHDC2 and MEIOC
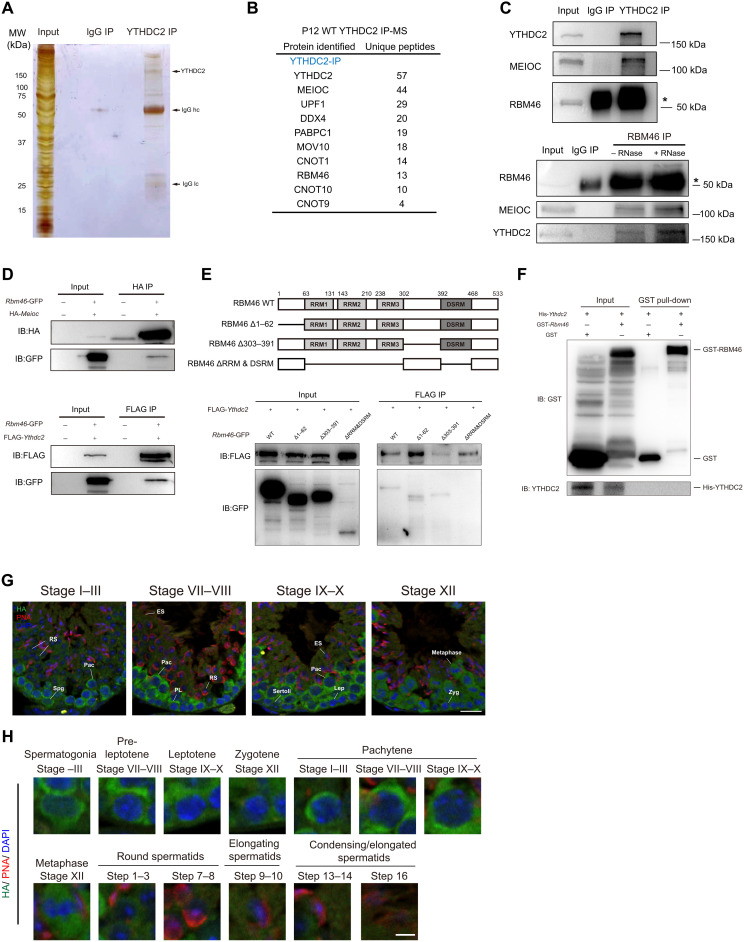
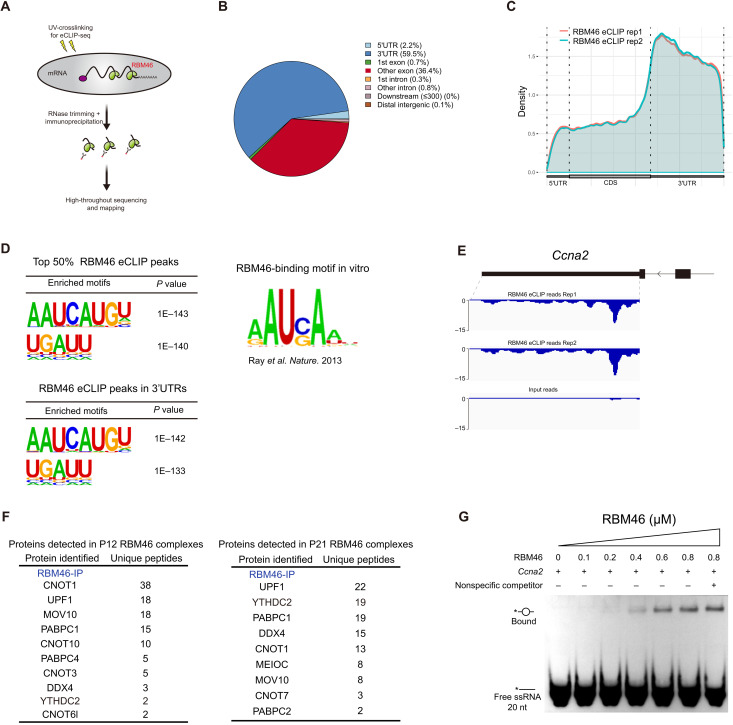
YTHDC2 and MEIOC proteins act together in a core RNA-protein complex for the switch from mitosis to meiosis. To identify other essential factors in this regulatory network, we performed anti-YTHDC2 immunoprecipitations in mouse testes at postnatal day 12 (P12) and P21, and subjected the isolated YTHDC2 complexes to mass spectrometry. As expected, YTHDC2 band migrated near its predicted molecular weight of 160 kDa (Fig. 1A). Gene ontology (GO) analysis based on all detected YTHDC2-associated proteins showed a strong enrichment for categories linked to RNA binding and ribonucleoprotein granules, consistent with the role of YTHDC2 in RNA binding and processing (fig. S1, A and B). MEIOC, a meiosis-specific protein that interacts with YTHDC2, is also present in endogenous mouse YTHDC2 complexes in both P12 and P21 testis (Fig. 1B and fig. S1C). Notably, mass spectrometric analysis further revealed that RBM46, a putative RNA binding protein, is one of the top interaction partners (Fig. 1B and fig. S1C). Rbm46 is an evolutionarily conserved gene, which encodes putative RNA binding domains of unknown function (fig. S2A).
Fig. 1. RBM46 associates with YTHDC2 and MEIOC in mouse testis.
(A) Identification of YTHDC2-associated partners by immunoprecipitation (IP) experiments with either YTHDC2 or normal immunoglobulin antibodies from P12 mouse testes. Immunoprecipitated YTHDC2 protein complexes were separated by SDS-PAGE, and the gel was stained with silver before mass spectrometry (MS). Testis lysates were prepared from mouse at P12 (n = 8). (B) Selective YTHDC2-associated protein candidates identified by MS analysis from P12 mouse testes. (C) Immunoprecipitation of YTHDC2 from mouse testis lysates at P21 and Western blot with anti-YTHDC2, anti-MEIOC, and anti-RBM46 antibodies (top). RBM46 protein was immunoprecipitated with anti-RBM46 antibodies with or without RNases, followed by Western blot analysis with indicated antibodies (bottom). (D) Full-length GFP-Rbm46 and FLAG-Ythdc2 or HA-Meioc expression constructs were cotransfected in HEK293T cells. Cell lysates were immunoprecipitated with anti-FLAG or anti-HA antibodies and Western blot analysis with indicated antibodies. IB, immunoblotting. (E) GFP-tagged RBM46 mutants with deleted fragments were coexpressed with FLAG-YTHDC2 in HEK293T cells. Cell lysates were immunoprecipitated with anti-FLAG antibodies before Western blot analysis with GFP antibodies. (F) GST pull-down assay showing that in vitro translated YTHDC2 does not bind to GST-RBM46. GST-RBM46 was expressed in E. coli and purified with glutathione beads. YTHDC2 with N-terminal His tag was in vitro translated. (G and H) Immunofluorescence analysis of Rbm46-HA knock-in mice with anti-HA antibodies and the acrosome marker PNA that defines different stages of spermatogenesis. DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bars, 20 μm. Lower panels show magnification image of single cell in the upper panels. Spg, spermatogonia; PL, pre-leptotene; Lep, leptotene; Zyg, zygotene; Pac, pachytene; RS, round spermatids; ES, elongating spermatids. Scale bars, 5 μm.
To verify the association of YTHDC2 with RBM46 and MEIOC, we performed immunoprecipitation combined with Western blot analysis and confirmed that MEIOC was present in the protein complex immunoprecipitated with the anti-YTHDC2 antibody in testicular extracts (Fig. 1C). The abundant form of RBM46 protein is at its predicted size of ~50 kDa, close to the heavy chain; coimmunoprecipitation analysis with the anti-RBM46 antibody confirmed that endogenous YTHDC2 was present in RBM46 protein complexes in mouse testis (Fig. 1C). Moreover, the interaction between RBM46 and MEIOC was observed in mouse testis (Fig. 1C). We further treated the immunoprecipitates with ribonucleases (RNases) and found that both YTHDC2 and MEIOC proteins were present in RBM46 immunoprecipitates, indicating that their interactions are not dependent on the presence of RNA (Fig. 1C). We next overexpressed green fluorescent protein (GFP)–tagged RBM46 with either FLAG-tagged YTHDC2 or hemagglutinin (HA)–tagged MEIOC in human embryonic kidney (HEK) 293T cells. Coimmunoprecipitation analysis demonstrated that YTHDC2 and MEIOC proteins pulled down exogenously expressed RBM46 (Fig. 1D). We then mapped the interaction sites of RBM46 with YTHDC2 by a series of coimmunoprecipitations using tagged truncation constructs (Fig. 1E). Deletions of both RNA recognition motifs (RRM1, RRM2, and RRM3) and double-stranded RNA binding domain (DSRM) abolished its association with YTHDC2 (Fig. 1E), whereas the RBM46-YTHDC2 interaction was not affected upon the deletion of either the N terminus (amino acids 1 to 62) or amino acids 303 to 391 of RBM46 (Fig. 1E). However, RBM46 protein containing the deletion of both RRMs and DSRM was expressed at a much lower level. We cannot rule out the possibility that this abolished interaction results from such a low expression level. To further determine whether the interaction between YTHDC2 and RBM46 is direct, a full-length RBM46 protein fused with glutathione S-transferase (GST) tag was expressed in Escherichia coli and purified, and full-length His-YTHDC2 was in vitro translated. Unexpectedly, our GST pull-down assay showed that in vitro translated YTHDC2 did not bind to GST-RBM46 (Fig. 1F), suggesting that RBM46 does not interact directly with YTHDC2. Together, RBM46 associates with MEIOC and YTHDC2 in mouse testes in an RNA-independent manner, but the interaction between RBM46 and YTHDC2 is indirect.
Up-regulation of RBM46 protein at the onset of meiosis
Rbm46 is an evolutionarily conserved gene, which encodes putative RNA binding domains of unknown function (fig. S2A). To understand the biological function of RBM46 protein, we examined its expression pattern in mice. We found that Rbm46 is highly expressed in testis at both the mRNA and protein levels (fig. S2, B and C). Next, we collected mouse testes at different time points corresponding to defined developmental stages of spermatogenesis and monitored Rbm46 transcripts. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis showed that Rbm46 mRNA was detectable at P7 and up-regulated substantially in testis at P10 (fig. S2D), when germ cells initiate meiosis to become early meiotic spermatocytes at the leptotene stage. The second wave of Rbm46 transcript up-regulation occurred during meiotic prophase I and reached its highest level in testis at P21 (fig. S2D), when late stages of spermatocytes are enriched in the entire germ cell population. Western blot analyses showed that RBM46 protein increased in testis at P10 and expressed highly in P21 testis (fig. S2E), confirming that this protein first accumulates when meiosis initiates and abundantly expresses during late stages of meiotic prophase I. Compared with RBM46 protein dynamics during spermatogenesis, the levels of YTHDC2 and MEIOC proteins were lower in spermatogonia (fig. S2E). Both proteins were expressed at a much higher level in spermatocytes than in spermatogonia (fig. S2E).
We next examined the cell type–specific expression of RBM46, C-terminal Rbm46-HA knock-in mice were generated, and mouse testis was immunostained with HA antibodies together with peanut agglutinin (PNA), an acrosome marker that defines different spermatogenic stages of seminiferous tubules. Immunostaining showed that RBM46 localized predominantly to the cytoplasm of spermatocytes in the testis (Fig. 1G). Detailed analysis indicated that RBM46 protein was detectable in spermatogonia and the preleptotene spermatocytes in stage VII and VIII seminiferous tubules (Fig. 1, G and H) but was not observed in Sertoli cells (Fig. 1G). During meiosis, RBM46 was present in the cytoplasm of early spermatocytes from the leptotene stage and reached its highest level in late stages of meiotic prophase I, from the pachytene to diplotene stage (Fig. 1H), as well as in metaphase I and secondary spermatocytes (Fig. 1H). RBM46 protein starts to decline in postmeiotic spermatids (Fig. 1H) and disappeared in round spermatids from step 4 and elongated spermatids (Fig. 1H).
To further confirm the localization pattern of RBM46 in mouse testis, mouse testes were immunostained with specific antibodies against RBM46 and PNA. Immunofluorescence analysis revealed that RBM46 protein was present in spermatogonia and preleptotene spermatocytes, abundant in late spermatocytes, and absent from round spermatids at step 4 (fig. S2, F and G), a pattern similarly observed with the HA antibodies in C-terminal–tagged Rbm46-HA mice. Hence, RBM46 starts to up-regulate at the onset of meiosis and accumulates highly throughout meiotic prophase I.
Rbm46-deficient mice are infertile with an early meiotic arrest
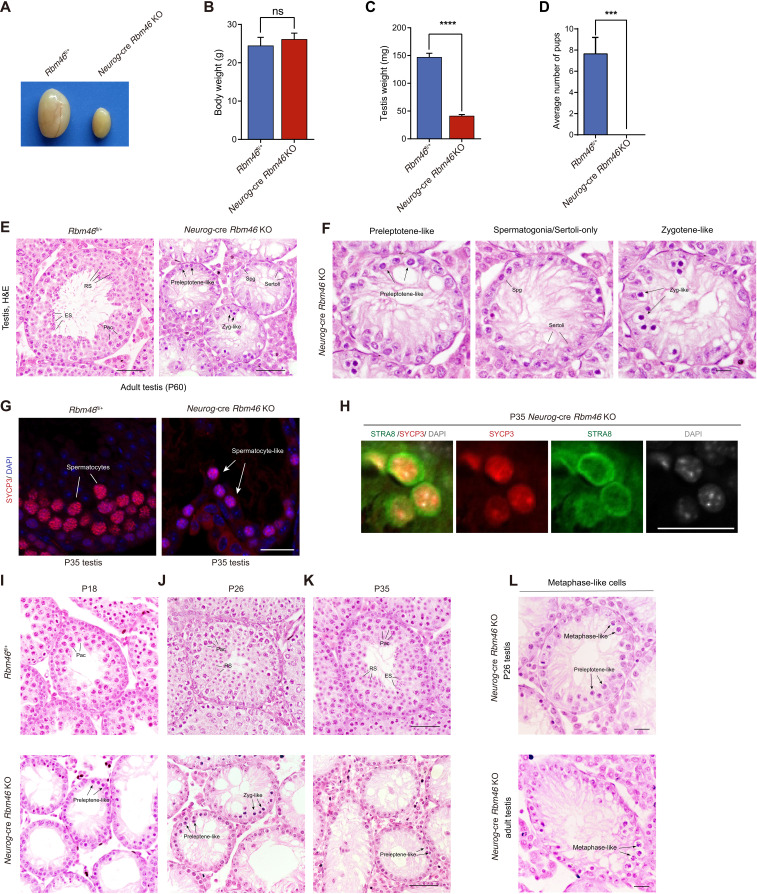
To better understand the physiological function of mammalian RBM46 in spermatogenesis, we generated mice with a floxed Rbm46 allele (Rbm46fl) through homologous recombination in embryonic stem cells (ESCs). Sequence analysis showed that the mouse Rbm46 gene contains five exons and spans a genomic region of 40 kb on chromosome 3. In the target allele, LoxP sites were inserted into the introns flanking exons 3 and 4, which contain the putative RRMs and RNA binding domains (fig. S3A). To generate testis-specific Rbm46 knockouts (KOs), we bred mice with a floxed Rbm46 allele with Neurog-cre mice, in which Cre recombinase starts to express in differentiation spermatogonia and mediates complete recombination in early meiotic spermatocytes (38, 39). The deletion of exons 3 and 4 [468 and 783 base pairs (bp), respectively] would be expected to disrupt 78% of the coding sequence for RBM46. Western blot analysis revealed the absence of RBM46 protein in testes of Neurog-cre–mediated Rbm46 KOs (referred to as Neurog-cre Rbm46 KO hereafter) at P21 (fig. S3B), when late stages of spermatocytes are enriched in the testis. Neurog-cre Rbm46 KO mice grow normally into adulthood but exhibit highly atrophied testes (Fig. 2A). The testes of Neurog-cre Rbm46 KO at 7 to 8 weeks old weighed 72% less than those of littermate control (Fig. 2, B and C), and Neurog-cre Rbm46 KO males were not able to father litters when bred with wild-type (WT) females (Fig. 2D). Therefore, Neurog-cre Rbm46 KO mice are infertile, suggesting that RBM46 is required for spermatogenesis.
Fig. 2. Testis-specific Rbm46 knockouts are infertile with defects in meiotic progression.
(A to C) Testis size, body weight, and testis weight of WT and Neurog-cre Rbm46 knockout (KO) mice at 8 weeks old. Data are represented as means ± SD (n = 3). ns, not significant. (D) Number of litters when WT or Neurog-cre Rbm46 KO males were mated with WT females. n = 3 for each genotype. (E) Histological images of 8-week-old WT and Neurog-cre Rbm46 KO. Scale bar, 50 μm. H&E, hematoxylin and eosin. (F) Enlarged images of the Neurog-cre Rbm46 KO tubule. Scale bar, 20 μm. (G) Immunostaining was performed using SYCP3 antibody on P35 testis sections from WT and Neurog-cre Rbm46 KO mice. Scale bar, 20 μm. (H) Frozen sections prepared from P35 Neurog-cre Rbm46 KO testes were immunolabeled with STRA8 and SYCP3 antibodies. Scale bar, 20 μm. (I to K) Histological analysis of WT and Neurog-cre Rbm46 KO testes at P18 (I), P26 (J), and P35 (K). Arrows indicate atypical zygotene spermatocytes with condensed nuclei and preleptotene-like cells. Scale bar, 50 μm. (L) Abnormal metaphase-like cells with condensed chromosomes were observed in P26 and adult Neurog-cre Rbm46 KO testes. Scale bar, 20 μm.
In contrast to WT tubules full of germ cells representative of different stages, including mitotic spermatogonia, meiotic spermatocytes, postmeiotic round spermatids, elongating spermatids, and spermatozoa, histological analysis showed severely impaired spermatogenesis with a complete absence of postmeiotic germ cells and a marked decrease of spermatocytes in meiotic prophase I in adult (P60) Neurog-cre Rbm46 KO testes (Fig. 2E). Most tubules in the Neurog-cre Rbm46 KO were arrested at the preleptotene-like stages or contained a single layer of spermatogonia/Sertoli cells (Fig. 2F). Some Neurog-cre–mediated KO germ cells progressed to zygotene-like spermatocytes (Fig. 2F). Consistent with the histological observation, the staining for SYCP3, a component of the synaptonemal complex during meiotic prophase I, was only detected in fewer cells in the Neurog-cre Rbm46 KO testes (Fig. 2G).
To further investigate the timing of the primary defects in meiosis, juvenile WT and Neurog-cre Rbm46 KO testes were collected for histological analysis. Meiotic spermatocytes at the pachytene stage were observed in WT at P18 (Fig. 2I). However, most germ cells in the Neurog-cre Rbm46 KO testes were arrested at the preleptotene-like stages (Fig. 2I). In WT testes, germ cells in some seminiferous tubules progressed into elongating spermatids at P26 (Fig. 2J) and differentiated into mature sperm at P35 (Fig. 2K). In contrast, postmeiotic spermatids and late stages of spermatocytes were completely absent in the seminiferous tubules of Neurog-cre Rbm46 KO mice at P26 and P35, with germ cells arrested mostly at the preleptotene-like stages (Fig. 2, J and K). Similar to the phenotype in adult Neurog-cre Rbm46 KO, some germ cells in young Neurog-cre Rbm46 KO mice at P26 and P35 also advanced to zygotene-like spermatocytes (Fig. 2, J and K). We noticed that a large population of preleptotene-like cells in P35 Neurog-cre Rbm46 KO tubules expressed STRA8 (Fig. 2H), a key factor that promotes meiotic initiation, but exhibited dense aggregates of SYCP3 signal (Fig. 2H). Notably, most of the Rbm46 KO preleptotene-like cells contained patchy aggregates of DNA (Fig. 2H), with increased chromatin density, suggesting that these cells underwent chromatin condensation.
A subpopulation of spermatocyte-like cells in P26 Neurog-cre Rbm46 KO appeared to be metaphase-like cells with condensed chromosomes (Fig. 2L). Approximately 10% of P26 Neurog-cre Rbm46 KO tubules contained these aberrant metaphase-like cells. Consistently, similar phenomena were observed in P12, P18, and adult (P60) Neurog-cre Rbm46 KO (Fig. 2L and fig. S3C), albeit at a lower frequency compared to Neurog-cre Rbm46 KO at P26. Notably, these advanced metaphase–like cells appeared frequently lying adjacent to preleptotene-like cells (Fig. 2L), suggesting that these cells that underwent precocious chromosome condensation were early meiotic spermatocytes at the preleptotene stage before the leptotene and zygotene stages. Such abnormal metaphase-like cells were similarly observed in mutant germ cells from Meioc KOs (34, 35) or Ythdc2 null testes (26–28, 32). Collectively, these data indicate that germ cells in Neurog-cre Rbm46 KO mice enter meiosis but fail to proceed through meiosis, with a predominant arrest at the preleptotene-like stage. These preleptotene-like cells underwent precocious chromatin condensation, resulting in some germs rapidly and prematurely exiting meiotic prophase I and entering a metaphase-like state.
Rbm46 KO spermatocytes exhibit defective synapsis and recombination
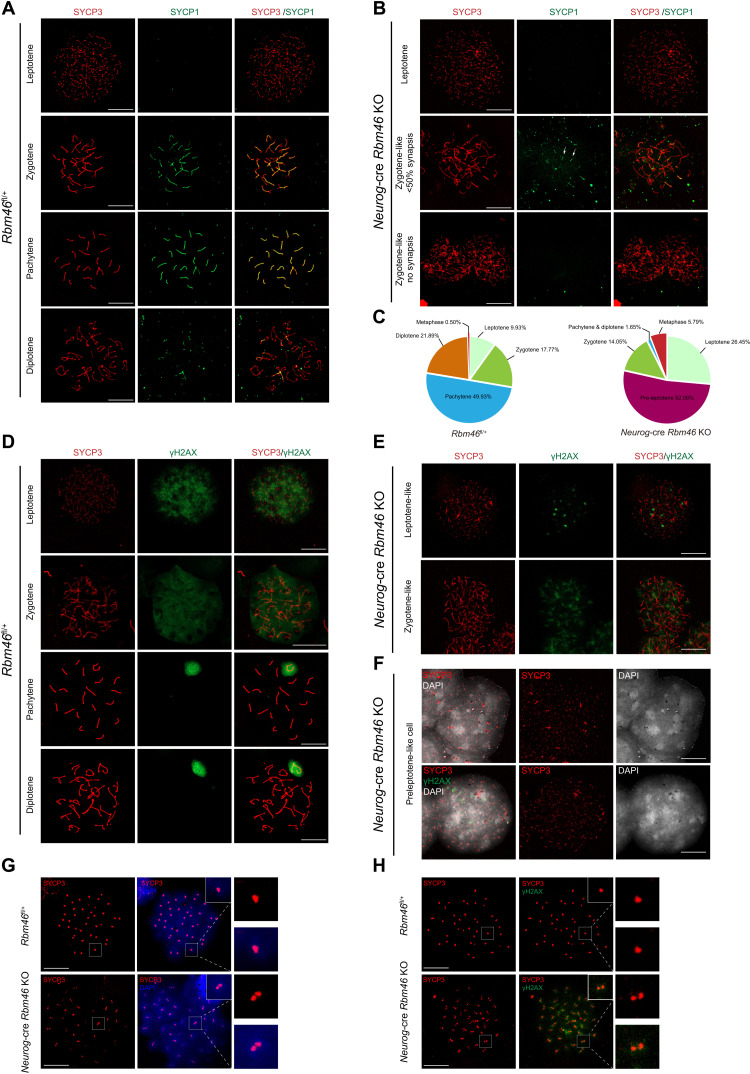
Since some germ cells in Neurog-cre Rbm46 KO advanced to the zygotene stage of meiosis, we analyzed the process of chromosomal synapsis by immunostaining axial (SYCP3) (40) and central elements (SYCP1) of synaptonemal complexes (41). In the leptotene and zygotene stages, when homologous chromosomes start to synapse following meiotic DNA double-strand breaks (DSBs), SYCP3 was normally assembled into lateral elements of synaptonemal complexes in Rbm46-deficient spermatocytes (Fig. 3B), as in the WT (Fig. 3A). However, the localization of SYCP1, a central element of the synaptonemal complex that localized to synapsed regions of chromosomes at the zygotene and pachytene stages in normal meiosis (Fig. 3A), was abolished in most Rbm46-deficient spermatocytes that advanced to the zygotene stage (Fig. 3B). The remaining zygotene-like spermatocytes contained SYCP1 signal but showed extremely short stretches of SYCP1 staining on a few condensing chromosomes (Fig. 3B). These results demonstrate that these zygotene-like spermatocytes in Neurog-cre Rbm46 KO lacked synapsis. Spermatocyte progression to later stages than to the zygotene-like stage failed, with only a few pachytene/diplotene spermatocytes that were occasionally observed.
Fig. 3. Neurog-cre Rbm46 KO mice exhibit early meiotic defects.
(A) Spread nuclei were prepared from P35 WT testes followed by immunostaining with antibodies against synaptonemal complex proteins SYCP3 (red) and SYCP1 (green). Meiotic prophase I stages (leptotene, zygotene, pachytene, and diplotene) were identified by the localization pattern of SYCP1 and SYCP3. Scale bar, 20 μm. (B) Immunostaining of spread nuclei from P35 Neurog-cre Rbm46 KO with anti-SYCP3 (red) and anti-SYCP1 (green). Representative images of zygotene-like spermatocytes in Neurog-cre Rbm46 KO were shown. SYCP3 formed long fibers in zygotene spermatocytes but lacked SYCP1 signal or only with very short stretches of SYCP1 (arrows). Scale bar, 20 μm. (C) Quantitative analysis of meiotic spermatocytes at the indicated stages in WT and Neurog-cre Rbm46 KO. Rbm46fl/+, n = 2015; Neurog-cre Rbm46 KO, n = 121. (D and E) Immunostaining of spread nuclei from P35 WT (D) and Neurog-cre Rbm46 KO (E) with γH2AX. γH2AX signal condensates in regional chromatin. Scale bar, 20 μm. (F) Spread nuclei of preleptotene-like cells from Neurog-cre Rbm46 KO were immunostained with the indicated antibodies. Preleptotene-like cells contained prominent condensed DNA. SYCP3 (red) aggregates with the presence of γH2AX signal (green). (G and H) Spread nuclei of metaphase-like cells from Neurog-cre Rbm46 KO were immunostained with the indicated antibodies. Scale bar, 20 μm.
Meiotic recombination is tightly associated with the progression of homologous chromosome synapsis. We next examined the process of meiotic recombination by immunostaining the spread nuclei with γH2AX, a phosphorylated form of the histone variant H2AX that is generated in response to meiotic DNA DSBs (42). During normal meiosis, the first wave of γH2AX accumulation occurs extensively on the entire chromatin at the leptotene and zygotene stages. Still, it is confined to the unsynapsed X and Y chromosomes as the sex body when homologous synapsis is completed at the pachytene stage of meiosis (42). Analysis of chromosome spreads from Neurog-cre Rbm46 KO spermatocytes revealed that γH2AX signal appeared in Rbm46 KO spermatocytes at the leptotene and zygotene stages but in an altered pattern (Fig. 3E). In contrast to the distribution of γH2AX across bulk chromatin in the WT (Fig. 3D), γH2AX signals were not observed throughout the entire chromatin; rather, it existed in distinct regions (Fig. 3E). Moreover, the sex body marked by γH2AX, correlating with the pachytene stage, was found in the WT (Fig. 3D), but only one or two spermatocytes containing the sex body were observed in Neurog-cre Rbm46 KO (Fig. 3C). These data were in agreement with synapsis analysis as revealed by SYCP1 and SYCP3, showing that Neurog-cre Rbm46 KO spermatocytes rarely progressed to the pachytene stage.
Analysis of chromosome spreads revealed that two aberrant populations of spermatocytes, preleptotene- and metaphase-like cells, were frequently observed in Neurog-cre Rbm46 KO. Quantification of all the meiotic spermatocytes observed showed that preleptotene- and metaphase-like cells comprised 52.06 and 5.79% of the spermatocyte pool (Fig. 3C), respectively, whereas the percentages of zygotene-like and pachytene spermatocytes were markedly reduced (Fig. 3C). This is unusual and prompts us to take a closer look at these two aberrant cell populations. In the WT, preleptotene spermatocytes can only be observed in stage VII and VIII tubules with a small number. Analysis of chromosome spreads revealed that most of the Rbm46 KO preleptotene-like cells showed prominent condensed chromatin, based on the presence of patchy aggregates of DNA (Fig. 3F). These spermatocyte-like cells exhibited punctate aggregates of SYCP3 signal, and γH2AX signal precociously appeared (Fig. 3F). Thus, meiotic DSBs were partially generated in the preleptotene-like stage, whereas the first wave of meiotic DSBs normally occurs until spermatocytes reach the leptotene stage. Assessment of these metaphase-like cells from Neurog-cre Rbm46 KO testes showed normal disassembly of synaptonemal complexes, as SYCP3 disappeared from the chromosome arms and localized to foci at ends of chromosomes (Fig. 3G), presumably centromeric regions. However, these metaphase-like cells did not form condensed metaphase chromosomes with 40 SYCP3 foci; instead, condensed chromosomes with 80 SYCP3 foci per cell were observed (Fig. 3G). The presence of abnormal doublets of SYCP3 foci is consistent among all the metaphase-like cells observed, indicating that sister chromatids prematurely separated at metaphase I likely resulted from defective sister chromatid cohesion. Although this population morphologically resembles metaphase I spermatocytes, γH2AX signal persisted in chromatin (Fig. 3H). The strong γH2AX staining is consistent with the observation that these cells have initiated meiotic recombination but rarely proceed to late stages of meiotic prophase I. These data further support the notion that this metaphase transition likely occurs rapidly, probably in a time window before recombination and synapsis of chromosomes.
RBM46 is required for a proper exit from the mitotic cell cycle program
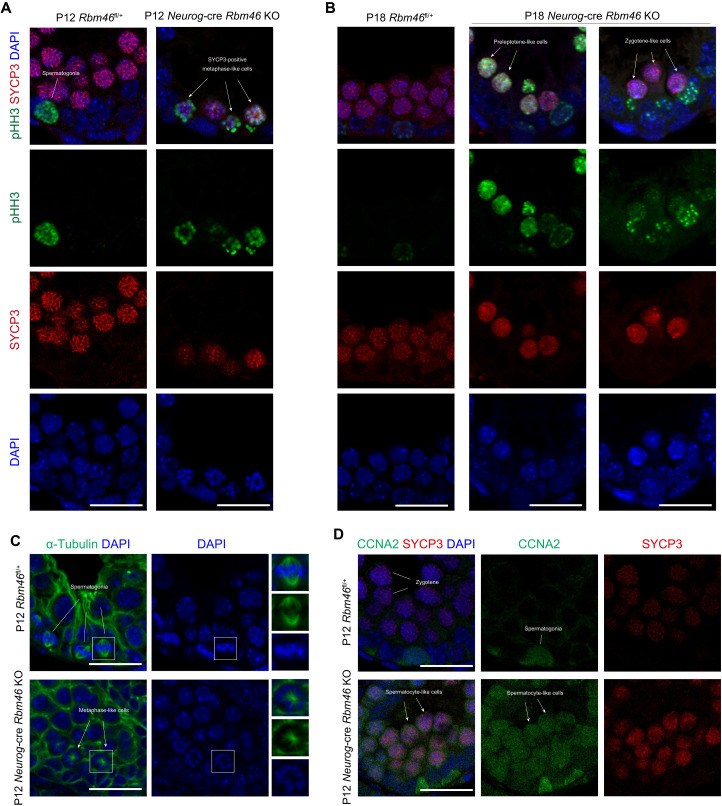
The aforementioned experiments demonstrated that male germ cells from Neurog-cre Rbm46 KO mice fail to properly execute meiotic prophase, with defects in chromosomal synapsis and recombination. Nevertheless, Rbm46 KO germ cells express hallmark meiotic proteins and initiate recombination but with premature entry into aberrant metaphase. We next addressed the mechanism underlying this premature metaphase transition. We first assessed the molecular characteristics of aberrant metaphase-like cells with condensed chromosomes. During the meiotic prophase I to metaphase I transition, histone H3 is phosphorylated at Ser10 (pHH3) across the condensed chromatin, and the chromosomes are aligned at the metaphase plate by a bipolar spindle. In the WT at P12, H3Ser10 phosphorylation was exclusively observed in the germ cells along the periphery of tubules (Fig. 4A), largely corresponding to the mitotic spermatogonia at metaphase. The pHH3 signal was not detected in SYCP3-positive spermatocytes as expected (Fig. 4A), because WT germ cells do not reach metaphase I at this age. In Neurog-cre Rbm46 KO mice at P12, the pHH3 signal was observed in SYCP3-positive metaphase-like cells with condensed chromosomes (Fig. 4A). Notably, pHH3 signals frequently appeared in SYCP3-positive spermatocytes at leptotene- or zygotene-like stages in Rbm46 KO testes at P18 (Fig. 4B), with the earliest signal detected in preleptotene-like cells (Fig. 4B), suggesting that Rbm46 KO germ cells underwent precocious chromatin condensation as early as at the preleptotene-like stage. A similar pattern of H3Ser10 phosphorylation was observed in P35 Neurog-cre Rbm46 KO testes (fig. S4A), whereas pHH3 signals were restricted to mitotic spermatogonia and metaphase I spermatocytes in stage XII WT tubules (fig. S4A). Most Neurog-cre Rbm46 KO metaphase-like cells assembled a monopolar spindle based on tubulin-positive microtubule filaments emanating from a single spindle pole body (Fig. 4C). These apparently monopolar spindles contrasted the typical bipolar spindles observed in the metaphase I spermatocytes from the WT (Fig. 4C).
Fig. 4. RBM46-deficient spermatocytes fail to repress mitotic program.
(A) Testis sections from P12 WT and Neurog-cre Rbm46 KO were immunolabeled with pHH3 (phosphorylated histone H3 at Ser10) and SYCP3, a component of the synaptonemal complex. Aberrant metaphase-like cells with pHH3 signal are indicated by white arrows. Scale bar, 20 μm. (B) Immunofluorescence analysis with pHH3 and SYCP3 antibodies were performed in Neurog-cre Rbm46 KO at P18. pHH3-positive signals were observed in preleptotene- and zygotene-like cells in Neurog-cre Rbm46 KO. (C) Frozen sections from P12 WT and Neurog-cre Rbm46 KO testes were immunostained with α-tubulin to visualize the spindle (green). Monopolar spindles were indicated by arrows. Scale bar, 20 μm. (D) Immunofluorescence of SYCP3 and CCNA2 on testis sections prepared from P12 WT and Neurog-cre Rbm46 KO testis. SYCP3-positive zygotene-like cells that express CCNA2 are indicated by arrows. Scale bar, 20 μm.
To identify the trigger for the premature metaphase entry of Rbm46 KO spermatocytes, we examined the activity of maturation-promoting factor (MPF) complex, which consists of cyclin-dependent kinase I (CDK1) and cyclin B1 (CCNB1). During the transition to metaphase of both meiosis and mitosis, the CDK1 kinase is activated by its binding to CCNB1, phosphorylation at Thr161 (pT161-CDK1), and removal of inhibitory phosphorylations at Thr14 (pT14-CDK1) and Tyr15 (pY15-CDK1). In Neurog-cre Rbm46 KO testes, the amount of phosphorylation of CDK1 at Thr16 remained at a similar level (fig. S4B), and phosphorylation of CDK1 at Tyr15 was not significantly affected (fig. S4B). Mitogen-activated protein kinases extracellular signal–regulated kinase 1 (ERK1) and ERK2, which phosphorylate protein kinases and phosphatases, were slightly diminished in Neurog-cre Rbm46 KO testes (fig. S4B). These data indicate that MPF activity was largely preserved in Neurog-cre Rbm46 KO testes.
We next determined the expression of cyclins that closely correlates with cell cycle status. The level of CCNB1, a cyclin subunit that binds and activates CDK1, was not different between WT and Neurog-cre Rbm46 KO testes (fig. S4B). In WT testes, mitotic cyclin A2 (CCNA2) is expressed in mitotic spermatogonia (Fig. 4D), is down-regulated as cells enter meiosis, and becomes undetectable in leptotene and zygotene spermatocytes. In Neurog-cre Rbm46 KO testes, CCNA2 was aberrantly expressed in SYCP3-positive spermatocyte-like cells in Neurog-cre Rbm46 KO (Fig. 4D). The retention of CCNA2 was also present in SYCP3-positive metaphase-like cells (fig. S4C), indicating that RBM46-deficient spermatocytes lack a proper exit from the mitotic cell cycle program.
RBM46 predominantly targets 3′UTRs
To identify direct RNA targets that underlie RBM46 function, we carried out RBM46 eCLIP-seq (enhanced cross-linking immunoprecipitation coupled with sequencing) (43) in WT testes at P12 to P14 (Fig. 5A), a time point that enables us to investigate its early role at meiotic entry. We prepared biological replicate eCLIP-seq libraries of RBM46 and observed a good correlation between replicates (Pearson correlation coefficient r = 0.89) (fig. S5A). Using eCLIP-seq cluster-finding algorithm CLIPper, we identified 28,292 peaks in 4887 genes and 31,571 peaks in 4997 genes (with P ≤ 0.001, fold change ≥ 8) in two replicates, respectively, with 24,010 binding sites and 4413 protein-coding genes bound in both replicates (fig. S5B).
Fig. 5. RBM46 selectively binds to 3’UTRs of mRNA targets.
(A) Schematic diagram of eCLIP-seq method to identify RNA targets for RNA binding proteins. (B) Pie chart depicting genomic distribution of RBM46-binding sites identified by eCLIP-seq. (C) Distribution of replicate RBM46 eCLIP peaks along the length of mRNA transcripts. CDS, coding sequence. (D) RBM46-binding motifs identified by HOMER from the top 50% of RBM46 eCLIP peaks in different genic regions. This motif is similar to the consensus sequence identified in an in vitro RNA binding assay for recombinant human RBM46 protein. (E) Genome browser tracks showing RBM46 eCLIP-seq reads enriched on the 3′UTRs of the mitotic Rad21 transcripts. (F) RBM46-associated protein candidates identified by MS analysis from P12 and P21 mouse testes. (G) In vitro gel-shift assay of RBM46-RNA interactions using single-stranded RNA (ssRNA) corresponding to Ccna2 3′UTR enriched with eCLIP-seq reads, and it contains a consensus sequence AAUCAU. This 5′ end-labeled ssRNA is indicated by asterisk, and the RBM46-RNA complex formation was not inhibited by the addition of RNA oligonucleotides without the RBM46-binding motif.
Genomic annotation of eCLIP reads showed that RBM46 is predominantly bound to the 3′ untranslated regions (3′UTRs, 60%) and coding sequence (37%) (Fig. 5B). We next analyzed read density across the mRNA transcript length and found that RBM46 eCLIP reads were increased on the last exon, with a marked enrichment over the following 3′UTRs (Fig. 5C). This genomic distribution with a preferential binding over 3′UTRs is similarly observed for m6A peaks on mRNA transcripts (5, 26, 44).
Motif analysis identified the top enriched motif within the top 50% of the RBM46-binding sites containing the core consensus sequence “AAUCAUGU” (Fig. 5D), and “UGAUU” is the second top motif. The same top two motifs were repeatedly identified within RBM46-binding sites located in 3′UTRs (Fig. 5D). Notably, these top two motifs containing consensus sequence “UG(C)AU” were preferentially recognized by the recombinant human RBM46 protein in an in vitro biochemical assay (Fig. 5D), in which RNA binding proteins were incubated with a complex pool of RNA probes comprising all possible nine-base nucleotide sequences (45). Within the top 50% of reproducible RBM46 peaks, target GO analysis revealed biological process categories linked to RNA processing and chromosome events (fig. S5C). Among the most significantly enriched terms were chromosome segregation and organization, RNA catabolic process, nuclear division, and sister chromatid segregation (fig. S5C).
RBM46 binds mitotic transcripts and interacts with crucial P-body proteins
Our immunofluorescence analysis reveals that the mitotic cell cycle factor CCNA2 is aberrantly expressed in spermatocyte-like cells from either MEIOC KO, YTHDC2 KO, or Neurog3-cre RBM46 KO (Fig. 4D). RBM46 is preferentially located at the 3′UTRs of Ccna2 gene with multiple RBM46-binding motifs embedded (Fig. 5E), indicating that Ccna2 is a direct target of RBM46. We also observed significant enrichment of RBM46 eCLIP reads on the mitotic/meiotic cohesion–related transcript Rad21 (fig. S5D), which has been identified as one of the YTHDC2 targets by RNA immunoprecipitation sequencing (RIP-seq) (32).
We next assessed the influence of RBM46 binding on target transcripts, and RNA sequencing (RNA-seq) was performed to assess gene expression alterations resulting from Rbm46 deletion. Because the primary germ cell arrest occurs at the preleptotene-like stage in Neurog-cre Rbm46 KO mice, young WT and RBM46-deficient testes at P10 (when germ cells initiate meiosis) were collected to characterize the earliest transcriptional changes without major alterations in cellular composition in the testis. However, RNA-seq analysis revealed only a few genes to be differentially expressed in Neurog-cre Rbm46 KO compared to the WT (fig. S6A). The Ccna2 transcript was significantly up-regulated in the P14 Neurog-cre Rbm46 KO testes (fig. S6B), but we cannot rule out the possibility that this expression alteration could be largely indirect from the developmental arrest at the preleptotene-like stage in mutant testes. A mild change on the entire transcriptome is similarly observed in Ythdc2 KOs from P8 (26) and P12 testis (32); at both stages, germ cell types remain similar. The cytological observation showed that Rbm46-deficient germ cells initiate meiosis but lack a proper exit from the mitotic cell cycle program. Because the number of mutant spermatocytes with a mixed mitotic and meiotic identity in P10 Rbm46 KO testes is limited at the meiotic entry, transcript changes in stage-specific germ cell populations may not be captured by the entire testis expression analysis.
To gain a mechanistic understanding of RBM46-mRNA interactions, we set out to identify proteins that associate with RBM46 in mouse testis. Endogenous RBM46 protein complexes in both P12 and P21 testis were prepared from mouse testes, followed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining (fig. S7A). Quantitative mass spectrometry analysis further revealed the presence of MEIOC and YTHDC2 (Fig. 5F). In addition to YTHDC2 and MEIOC, mass spectrometry analysis of all the RBM46-associated proteins showed the presence of several key components in cytoplasmic processing bodies (P bodies), an RNA processing center in germ cells. For example, our analysis revealed interactions between RBM46 and CCR4-NOT complex subunits (CNOT1, CNOT6l, and CNOT7), polyadenylate [poly(A)]–binding complex members (PABPC1 and PABPC4), MOV10, DDX4, and UPF1 (Fig. 5F), all of which have been implicated in RNA binding and processing related with P-body biology. CNOT1 is a major cytoplasmic mRNA deadenylase and UPF1 is the key component of the nonsense-mediated mRNA decay pathway, and both are known as RNA clearance factors to mediate mRNA degradation and mRNA decay–coupled translational regulation. Subsequent immunoprecipitation combined with Western blotting confirmed that RBM46 interacts with several key P-body components in mouse testis, e.g., MOV10, CNOT6l, DDX4, PABPC1, and UPF1 (fig. S7B). Human YTHDC2 protein not only distributes throughout the cytoplasm but also concentrates in P bodies, and it colocalizes with the decapping enzyme DCP1A in spermatocytes (32). Endogenous YTHDC2 associates with two RNA granule components MSY2 and MIWI (32). Using the observation that RBM46 and YTHDC2 interact with P-body components, including deadenylation and decapping complexes, we propose that RBM46 and YTHDC2 may function collectively in a specialized RNA germ granule for processing and/or translation of target RNAs.
RBM46 interacts with RNA close to YTHDC2-binding sites
YTHDC2 has been reported to function as an RNA-induced ATPase that exhibits 3′→5′ RNA helicase activity (26, 27). However, in vitro biochemical experiments show that recombinant YTHDC2 protein binds and unwinds RNA targets without sequence preference (26). To address whether RBM46 contributes more to the RNA binding specificity, we next tested whether recombinant RBM46 protein has the ability to recognize and directly bind sequence-specific RNA using the electrophoretic mobility shift assay (EMSA). Analysis of top 50% RBM46 eCLIP peaks by HOMER identified a consensus sequence, AAUCAUGU, as the most enriched motif (Fig. 5D). Incubation of RBM46 protein with a consensus RNA sequence from Ccna2 3′UTRs identified in eCLIP analyses resulted in a specific RNA-protein complex (Fig. 5G). This RBM46-RNA interaction was absent when the RBM46 protein was incubated with an RNA sequence without the RBM46-binding motif (fig. S5E). Moreover, the RBM46-RNA complex was not observed when incubated with RNA oligonucleotides containing this U-rich motif only (fig. S5F), indicating that RBM46 protein binds specific RNA sequences.
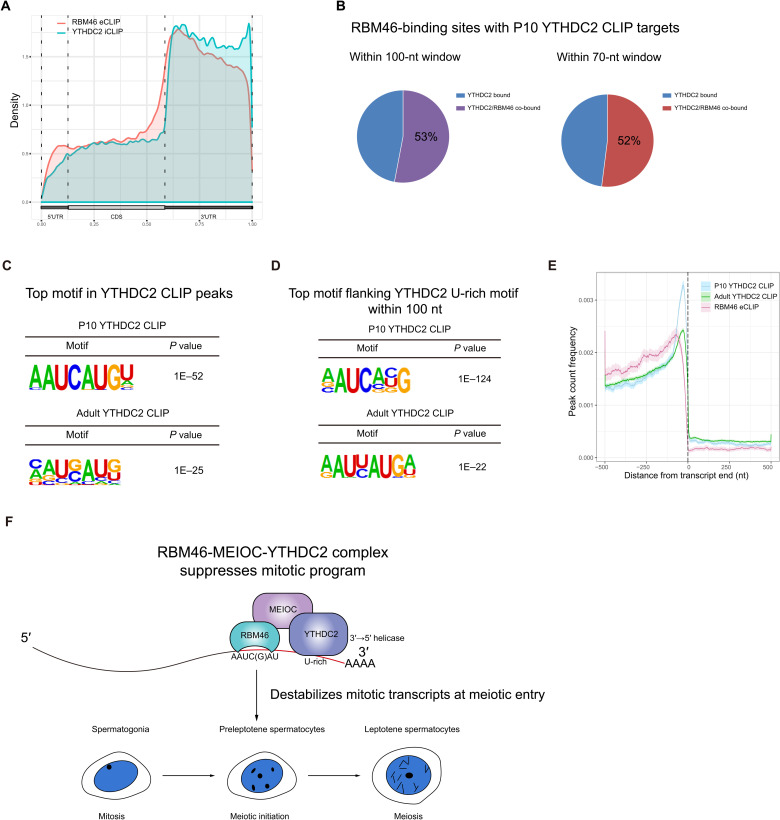
Next, we compared RBM46 eCLIP peaks with the published YTHDC2 and MEIOC RNA targets identified by RIP-seq. Within 523 YTHDC2 target mRNA transcripts (28), 284 transcripts (54.3%) were bound by RBM46 (fig. S8A). When comparing the RBM46-bound mRNAs with MEIOC target transcripts (34), we observed a similar overlap (53.8%; fig. S8B). To precisely evaluate a possible coordinated role on their target mRNAs, we compared RBM46-binding sites by eCLIP-seq with YTHDC2 targets by individual-nucleotide resolution CLIP-seq (iCLIP-seq) from FLAG-tagged Ythdc2 knock-in mice at P14 (30) and a recently published YTHDC2 CLIP-seq in WT using YTHDC2 antibodies (31). YTHDC2 predominantly targets the 3′UTRs (Fig. 6A) (30, 31), coinciding with the distribution pattern of RBM46 along the length of mRNA. Analysis of YTHDC2 CLIP-seq identified 3764 and 21,313 YTHDC2-binding sites in P10 and adult mouse testis (31), respectively. Among 511 and 398 YTHDC2 top targets in P10 and adult mouse testis listed in these published YTHDC2 CLIP-seq data [with filter peak height (PH) ≥ 10 and PH ≥ 40 for P10 and adults, respectively, according to sequencing depth of CLIP libraries], more than half (272, 53%) of P10 THDC2-binding sites were bound by RBM46 within a 100-nucleotide (nt) window (Fig. 6B), with most binding sites (267, 52%) predominantly located within a 70-nt window (Fig. 6B). A similar overlap in their binding sites was observed with adult YTHDC2 CLIP targets in the testis (fig. S8C). GO analysis of these shared targets showed a strong enrichment for mRNA binding and metabolic process, including mRNA 3′UTR binding, poly(A) binding, and mRNA catabolic process (fig. S8D).
Fig. 6. RBM46 interacts with RNA close to YTHDC2-binding sites.
(A) RBM46 and YTHDC2 exhibit a similar distribution pattern along the length of mRNA. (B) RBM46 eCLIP mRNA targets substantially overlapped with YTHDC2 mRNA targets by CLIP-seq in P10 testis. In total, 53 and 52% of P10 YTHDC2–binding sites were co-occupied by RBM46 within a 100- and 70-nt window, respectively. (C) Top motif at best-scored YTHDC-binding sites from P10 and adult YTHDC2 CLIP-seq using HOMER. Sequences within ±100 nt relative to YTHDC2-binding sites were used for de novo motif analysis. (D) RBM46-binding motif AAUCAUGU is the top motif within a 100-nt window centered on YTHDC2 U-rich motifs in both P10 and adult testis. Motif analysis by HOMER was performed with best-scored P10 and adult YTHDC2 CLIP targets. (E) Position and distance between RBM46 and YTHDC2 binding on their shared targets. (F) Schematic illustrations for the function of RBM46-MEIOC-YTHDC2 complex during spermatogenesis. RNA binding protein RBM46 forms an ancient posttranscriptional network with MEIOC and YTHDC2 to recognize and destabilize mitotic transcripts for a successful meiotic entry.
Analysis of YTHDC2-binding sites in YTHDC2 iCLIP-seq by HOMER showed the U-rich motifs (fig. S8E) (30), and this nucleotide feature is recaptured in YTHDC2 CLIP-seq (31). Notably, the top enriched motif found at best-scored YTHDC2-binding sites from P10 YTHDC2 CLIP-seq is the consensus sequence AAUCAUGU (Fig. 6C), which is the RBM46-binding motif (Fig. 5D). The same motif was enriched as the top at YTHDC2-binding sites from adult YTHDC2 top targets (Fig. 6C). To determine the positional relationship of RBM46- and YTHDC2-binding sites, we first scanned for the occurrence of additional motifs within an adjacent window (±100 nt) flanking the YTHDC2 U-rich motifs within 3′UTRs of their shared mRNA targets. Motif analysis revealed the presence of the RBM46 consensus sequence AAUCAUGU, which is identified as the top enriched motif (Fig. 6D). This RBM46-binding motif also came up on top within a 100-nt window centered on YTHDC2 U-rich motifs in adult testis (Fig. 6D). YTHDC2 CLIP tags are significantly enriched near the ends of 3′UTRs (31), and we observed increased RBM46 binding to a region located 5′ upstream of YTHDC2 sites (Fig. 6E). Vice versa, the U-rich motifs were present within a 100-nt window flanking the RBM46 motif (fig. S8F). RIP-qPCR analysis further revealed that YTHDC2 binding at the previously determined sites was significantly lower in testes from Neurog-cre Rbm46 KO mice than from WT mice (fig. S8G). This observation suggests that the RNA binding sites where RBM46 and YTHDC2 are bound are in close proximity to each other, and they may function together within the same protein complex to target their shared targets.
DISCUSSION
Meiotic entry decision is a major germ cell fate transition. Upon sensing the transition signaling molecule, how germ cells quickly exit from the preexisting mitotic program and coordinate the cell cycle with meiotic prophase-specific chromosome events remains unclear. Dynamic associations of RNA binding protein networks with their RNA targets constitute an essential posttranscriptional mechanism that influences the fate of associated RNAs. Here, we used a proteomics strategy to identify YTHDC2-associated proteins and found that RNA binding protein RBM46 is part of YTHDC2-containing complex. RBM46 has an essential role in promoting a switch from the mitotic to the meiotic program during mammalian spermatogenesis. Mutant germ cells from Neurog-cre Rbm46 KO mice initiate meiosis based on the observation that part of the meiotic program expresses and several meiotic proteins are assembled into the synaptonemal complex, and chromosomes start recombination. Nevertheless, developmental stages later than zygotene are not observed in Neurog-cre Rbm46 KO testis, and most of the germ cells prematurely exit meiosis into aberrant metaphase. This rapid progression to a metaphase-like state occurs as early as at the preleptotene stage, and the chromatin condensation–associated factor pHH3 is frequently present in early meiotic Rbm46-deficient spermatocytes. High-throughput eCLIP analysis revealed that the most significant biological processes of RBM46 RNA targets in mouse testis are chromosome segregation and the mitotic cell cycle transition, in line with the precocious metaphase entry, thus indicating that RBM46 binding and its coordinated role with RNA processing cofactors like YTHDC2/MEIOC is essential for a proper exit from the mitotic program at the meiotic entry (Fig. 6F). Analysis of human Rbm46 by single-cell RNA-seq of germ cells from non-obstructive azoospermia (NOA) patients and fertile adults indicated that Rbm46 levels decreased significantly in mitotic spermatogonia derived from idiopathic NOA patients or NOA patients with the genetic cause of azoospermia, such as Y chromosome azoospermia factor (AZF) region deletion (fig. S8H) (46), although this Rbm46 down-regulation could be a secondary consequence of unknown primary mutations in azoospermic patients.
Our data show that most RBM46 reads are enriched within 3′UTRs, exhibiting a marked similarity with the binding pattern of YTHDC2 on the germline transcripts. Furthermore, the consensus motifs of RBM46 and YTHDC2 coexist on the same transcripts within 100 nt, suggesting that primary contact sites of RBM46 are in close proximity to those of YTHDC2. RBM46 protein contains the putative RNA recognition and binding domains, and our in vitro analyses show that RBM46 directly binds RNA substrates with sequence preference. YTHDC2 is already reported to physically interact with MEIOC (34, 35), a highly conserved meiosis-specific protein, with a coiled-coil domain. Both YTHDC2 and MEIOC are identified in our mass spectrometry analysis of RBM46 immunoprecipitates, and immunoprecipitation analysis confirmed that RBM46 is an essential component within this protein complex. YTHDC2 contains several RNA binding domains, including the conserved helicase domain and a YTH domain that binds m6A-containing RNAs (26–29). RNA helicases function as adenosine triphosphate (ATP)–dependent enzymes that remodel RNA-protein complexes and/or resolve secondary RNA structures, allowing processing cofactors to act further on their target transcripts. Biochemical assays demonstrate that human YTHDC2 has an ATP-dependent 3′→5′ RNA unwinding activity (26, 27), and this RNA helicase activity is required for YTHDC2 biological function in vivo (30, 31). Ribosome profiling assays further indicate that Ythdc2 KO does not substantially alter translation of its binding mRNA targets (31). Our data show that RBM46 binding was more often located 5′ upstream of YTHDC2 sites on the 3′UTRs of mRNA targets. We propose that RBM46 mainly contributes to specification and recognition of RNA targets, and YTHDC2 unwinds secondary RNA structures and/or removes proteins for subsequent loading of processing factors, i.e., UPF1 before mRNA degradation.
RBM46 is the vertebrate ortholog of Drosophila Tut. In the Drosophila adult stem cell lineage, stem cell daughters undergo several transit-amplifying divisions before terminal differentiation. The Bam gene was first identified for its specific role in the regulation of germline stem cell lineage differentiation. Male germ cells mutant for Bam undergo several extra rounds of mitotic amplification and fail to switch to the meiotic program (47–49). In the Drosophila, Bgcn is a Bam-interacting protein (50–52) and acts as an essential DExH-box RNA helicase in this complex. Similar to Bam mutants, Bgcn mutant male germ stem cells fail to differentiate into meiosis, causing an accumulation of germline stem cell–like cells (49, 51). Tut is a putative RNA binding protein and is identified as a component of the Drosophila Bam-Bgcn protein complex (53). In vitro immunoprecipitation assay showed that Bam protein bridges Tut and Bgcn together to form a complex via its N terminus and C terminus (53), respectively. Similar with the role of Bgcn and Bam on target transcripts (54), Tut also preferentially binds 3′UTRs of target mRNAs (55) and inhibits the expression of Mei-P26, which is a TRIM-NHL (tripartite motif and Ncl-1, HT2a, and Lin-41 domain) tumor suppressor homolog required for germline stem cell differentiation (53). Likewise, Tut deficiency in Drosophila causes spermatogonia to undergo extra mitotic amplification with a failure to exit from mitotic divisions for differentiation (53), suggesting that Tut functions in the same pathway as that of the Bam-Bgcn complex. Thus, in Drosophila germ line, Bam and Bgcn form a protein complex with the RNA binding protein Tut and function together as a core machinery to facilitate the transition from mitosis to meiosis at the posttranscriptional level.
In mammals, YTHDC2 is the mouse homolog of Drosophila Bgcn, and it has a conserved role as a critical regulator of the transition from mitosis to meiosis in mouse germ line. MEIOC protein physically interacts with YTHDC2, and the C-terminal coiled-coil domain of MEIOC mainly mediates the direct interaction with YTHDC2 (34, 35). Although MEIOC is not orthologous to Drosophila Bam, mouse Meioc and Ythdc2 mutants have a shared phenotype, and both are required for germ cells to switch into meiosis and progress through the meiotic prophase I (26–28, 32, 34, 35). The present evidence supports the notion that MEIOC is the functional counterpart of Drosophila Bam in the mouse. Consistent with the hypothesis that Tut acts in concert with Drosophila Bgcn-Bam complex, we found that its mouse ortholog RBM46 interacts with YTHDC2-MEIOC complex. It is possible that MEIOC brings RBM46 and YTHDC2 together into the same protein complex, as the N terminus of Drosophila Bam does in the assembly of Bgcn-Tut complex in Drosophila germ line (53). Consistent with our finding, RBM46 is identified as an interacting partner of YTHDC2 in mouse testis (30). Notably, the N terminus of MEIOC has a large intrinsically disordered region that lacks a well-defined structure. The intrinsically disordered regions have been shown to promote lipid-liquid phase separation and drive the formation of intracellular membrane-less organelles. Endogenous YTHDC2 protein is concentrated in perinuclear puncta (32), and both YTHDC2 and RBM46 interact with several known components of nuage granules. Thus, it is possible that MEIOC recruits YTHDC2 and RBM46 into a dynamic posttranscriptional regulatory complex for highly efficient and robust RNA processing. Future studies using protein biochemistry and structural biology should examine the properties of the mammalian YTHDC2-MEIOC-RBM46 complex.
MATERIALS AND METHODS
Immunoprecipitation and mass spectrometry
Tissue mass (80 mg) was dounced with 30 strokes, lysed in 500 μl of lysis buffer [20 mM tris-Cl (pH 7.4), 150 mM NaCl, 0.5% Triton X-100, 0.5% sodium deoxycholate, 1 mM dithiothreitol (DTT), and 1× protease inhibitor], and diluted with 500 μl of dilution buffer [20 mM tris (pH 7.4), 150 mM NaCl, 0.5% Triton X-100, 1 mM DTT, and protease inhibitor]. After centrifugation, the supernatant was precleared with protein A beads for 2 hours before incubation with indicated primary antibodies overnight. Protein A beads were added and rotated for 3 hours. The immunoprecipitated protein complexes were eluted off the beads into sample loading buffer, run on a 4 to 12% gradient 1× SDS-PAGE gel, and then stained with silver before mass spectrometry analysis. The following primary antibodies were used for immunoprecipitation: anti-YTHDC2 (ab176846, Abcam), anti-RBM46 (A16605, ABclonal), anti-HA (M180-3, MBL), and anti-FLAG (F1804, Sigma-Aldrich).
Targeted disruption of the Rbm46 gene
This floxed Rbm46 allele (Rbm46fl) was generated using homologous recombination in ESCs. Homologous arms and middle fragments were amplified by high-fidelity PCR using an Rbm46-containing bacterial artificial chromosome clone (RP23-324F23). In the targeted allele, one loxP site was inserted in intron 2, and the floxed HyTK (hygromycin and thymidine kinase) double selection cassette was inserted in intron 4. The V6.5 ESCs were electroporated with the linearized construct, and five Rbm463lox clones were identified after screening 196 ESC clones. The Cre-expressing plasmid pOG231 was used to remove the HyTK cassette. Two Rbm46fl ESC clones were injected into C57BL/6 blastcysts, and the Rbm46fl allele was transmitted through the germ line in chimeric mice. Rbm46fl mice were crossed with Neurog-cre mice to specifically delete Rbm46-floxed genomic region in the mouse germ cells. All offsprings were genotyped by PCR with primers TGAGACCAGAGCAGGAGGAC and TTGGTGAGCATAAAGGTATGTC for Rbm46fl allele (556 bp) and WT allele (362 bp). The KO allele was confirmed by PCR using the primers AGCACCCCAACCAGTTTCAG and TTGGTGAGCATAAAGGTATGTC (699 bp). All animals were maintained and used according to the guidelines of the Institutional Animal Care and Use Committee of Nanjing Medical University.
Rbm46-HA knock-in mice
The C-terminal HA-tagged Rbm46 (Rbm46-HA) mice were generated by the Genome Tagging Project Center of Chinese Academy of Sciences in Shanghai. Cas9 mRNA, donor single-stranded oligodeoxyribonucleotide with 1× HA tag-coding sequence at the C terminus of Rbm46, and single-guide RNAs (sgRNAs) were introduced into haploid ESCs, which were derived from haploid blastocysts with the paternal genome only. These gene-modified haploid ESCs were further injected into oocytes for the generation of semi-cloned Rbm46-HA–tagged mice. These pups were used to establish a stable Rbm46-HA knock-in mouse line by breeding with WT C57BL/6J mice. All animals were transferred and maintained according to the guidelines of the Institutional Animal Care and Use Committee of Nanjing Medical University.
Histological and chromosome spread analyses
Fresh samples were fixed in Bouin’s solution (SLBJ3855V, Sigma-Aldrich) overnight, dehydrated in graded ethanol (50, 70, 95, and 100%), embedded in paraffin, and sectioned using a microtome. Sections were stained with hematoxylin and eosin. For nuclear spread analysis, germ cells from P35 testes were collected using a method previously described (56). Antibodies used for spread analysis were as follows: anti-SYCP1 (1:100; ab15090, Abcam), anti-SYCP3 (1:100; ab97672, Abcam), and γH2AX (1:800; 16-202A, Millipore).
Immunoblotting and immunofluorescence assay
Testes or 293T cells were homogenized in radioimmunoprecipitation assay buffer supplemented with protease inhibitor cocktail tablets. The lysates were rotated at 4°C for 1 hour and centrifuged at 12,800 rpm for 30 min at 4°C, and the supernatant was collected. For Western blot analysis, 20 μg of proteins was separated by SDS-PAGE. The following antibodies were used for Western blot: anti-UPF1 (A5071, ABclonal), anti-PABPC1 (A14872, ABclonal), anti-MEIOC (HPA027266, Atlas), anti-MOV10 (ab80613, Abcam), anti–phospho-CDK1-T161 (AP0324, ABclonal), and anti–phospho-CDK1-Y15 (AP0016, ABclonal). For immunofluorescence analysis, tissues were fixed in 4% paraformaldehyde at 4°C overnight. Paraffin-embedded sections were prepared, incubated with the blocking buffer [10% fetal bovine serum (FBS), 1% bovine serum albumin, 1% Triton X-100, and 0.05% Tween-20 in phosphate-buffered saline (PBS)] for 1 hour, and then incubated with primary antibodies at 4°C overnight. The following primary antibodies were used for immunofluorescence: anti-RBM46 (1:200; A16605, ABclonal), anti-HA [1:100; C29F4, Cell Signaling Technology (CST)], anti-SYCP3 (1:200; ab97672, Abcam), anti-STRA8 (1:200; 49602, Abcam), anti-pHH3 (1:200; 9701s, CST), anti-CCNA2 (1:200; ab32386, Abcam), and PNA (fluorescein isothiocyanate–conjugated PNA, CL-1073-1, Vector Labs).
Construction of expression plasmids and cell culture
Full-length Rbm46, truncated Rbm46 expression constructs with the deletion of both RRMs and DSRM, individual deletions of N terminus (amino acids 1 to 62) or amino acids 303 to 391, full-length Ythdc2, and Meioc were generated by PCR amplification using mouse testis cDNA, followed by purification with 1% agarose gel electrophoresis. The PCR products were subcloned into the Eco RI and Bam HI sites of the pEGFP-N1 plasmids to yield expression constructs before transfection into 293T cells. For the generation of Ythdc2-FLAG and Meioc-HA, corresponding PCR products were cloned into the FLAG and HA plasmids, respectively. The 293T cells were maintained in Dulbecco’s modified Eagle’s medium/high-glucose medium with 10% FBS (10270106, Gibco) and penicillin-streptomycin and seeded into 10-cm dishes. Expression plasmids (10 μg) were transfected in 293T cells using a standard calcium phosphate method. Cells were collected 48 hours after transfection, and coimmunoprecipitation assay was performed with anti-FLAG and anti-HA antibodies.
qRT-PCR assay and RNA-seq
Testes samples were collected from WT and Neurog-cre Rbm46 KO at P10, and RNA was extracted using TRIzol reagent (Invitrogen). cDNA was prepared from 1 μg of total RNA through reverse transcription using a PrimeScriptRT Master Mix (TaKaRa). Diluted cDNA (1 μl) was used for each reaction using SYBR Green Premix Ex Taq II (RR820A, TaKaRa). A standard 20-μl reaction volume contained forward and reverse primers (200 nM), 1 μl of cDNA, and 10 μl of SYBR Green Mix. For RNA-seq, strand-specific libraries were prepared using the TruSeq Stranded Total RNA Sample Preparation Kit (Illumina) according to the manufacturer’s instructions before submitting to the Illumina NovaSeq 6000 system. The adaptor sequence and sequences with low quality were subsequently removed, and clean reads were then mapped to the mouse genome (mm10) with STAR (v 2.7.6A) with a GTF file downloaded from GENCODE database (57). Gene expression was evaluated using HTSeq, followed by DESeq2 normalization to evaluate gene expression. To define differentially expressed genes, the parameters (P < 0.05, fold change > 1.5) were used. RNA-seq data have been deposited in the Gene Expression Omnibus database (GSE197282).
eCLIP-seq and data analysis
eCLIP-seq was performed in mouse testis as previously described (43). Briefly, 150 mg of mouse testes was collected from WT mice at P12 to P14 and triturated by a loose glass pestle. The tissue was transferred to a cell culture dish and covered with a minimum amount of ice-cold PBS, and then the suspension was cross-linked by ultraviolet irradiation three times at 254 nm (400 mJ/cm2) and lysed in eCLIP lysis buffer with RNase I (LifeTech) and Turbo deoxyribonuclease (LifeTech). RBM46-RNA complexes were immunoprecipitated using 10 μg of anti-RBM46 antibodies and incubated at 25°C for 3 hours. Besides, a parallel size-matched input library was generated. RBM46-RNA complexes were dephosphorylated by FastAP (LifeTech), followed by 3′ RNA adapter ligation using T4 RNA Ligase (NEB). RBM46-RNA complexes were run on SDS-PAGE gel and transferred to a nitrocellulose membrane at 15 V for 70 min in 1× transfer buffer (LifeTech) with 10% methanol. The membrane between 50 and 125 kDa was collected and subsequently treated with urea/proteinase K (NEB). RNAs were extracted with acid phenol/chloroform/isoamyl alcohol, followed by purification with RNA Clean & Concentrator (Zymo Research). After purification, RNAs were reverse-transcribed into cDNA with AffinityScript (Agilent) and cleaned with ExoSAP-IT reagent, and free RNAs were further removed by NaOH/HCl treatment. The adapter was ligated to the 3′ end of cDNA with T4 RNA Ligase (NEB). Libraries were then PCR-amplified using Q5 Polymerase. PCR products were run on 3% low–melting temperature agarose gel (Seakem GTG LMP) electrophoresis, and size-selected libraries were purified with a MinElute gel extraction kit (Qiagen). eCLIP data were analyzed following ENCODE standard eCLIP pipeline (ENCODE Project Consortium, 2012). Upon trimming adapters and 3′ adapter dimers using cutadapt (V1.14) and removing PCR duplicates using a custom python script, we first filtered out reads aligned to the repeat elements from UCSC with STAR (V2.7.6a) and then mapped the rest to mm10 reference genome (GENCODE vM23). CLIPPER was used to call peak regions, whose fold enrichment was estimated as previously described (43). In the end, we identified significantly enriched peaks as those with irreproducible discovery rate cutoff of 0.01 as well as P ≤ 0.001 and fold enrichment ≥ 8. Binding motif among enriched peaks was predicted by HOMER (58). eCLIP sequencing data were deposited in the Gene Expression Omnibus database (GSE197282).
RIP-qPCR assay
RIP-qPCR analysis with YTHDC2 antibodies was carried out on P10 to P12 testes from WT and Neurog-cre Rbm46 KO. Testis (100 mg) was lysed in 1 ml of lysis buffer [50 mM tris-Cl (pH 8.0), 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 0.5% sodium deoxycholate, 1 mM DTT, 1× protease inhibitor, tRNA (50 μg/ml), and 2 mM Vanadyl RC]. After centrifugation, the supernatants were precleared for 1 hour at 4°C with protein A beads. Protein A beads were incubated with 10 μg of anti-YTHDC2 antibody (Abcam, ab220160) at 4°C overnight. For immunoprecipitation, 1 ml of lysates was incubated with 100 μl of antibody-bound beads with rotation for 4 hours at 4°C. The bead complexes were then washed five times with washing buffer [10 mM tris-HCl (pH 8.0), 150 mM NaCl, 0.01% NP-40, 1 mM MgCl2, and 5% glycerol]. The immunoprecipitates and input samples were treated with proteinase K before RNA extraction. For qPCR analysis, immunoprecipitated RNA was reverse-transcribed using PrimeScriptRT Master Mix (TaKaRa) and analyzed using SYBR Green Premix Ex Taq II (RR820A, TaKaRa). A standard 20-μl reaction volume contained forward and reverse primers (200 nM), 2 μl of cDNA, and 10 μl of SYBR Green Mix.
Protein purification
Rbm46 was amplified by PCR from mouse testis cDNA and cloned into the pQE60 expression vector. The base vector was engineered to include a 3′ 6× histidine tag. The RBM46-pQE60 expression construct was confirmed by sequencing and transformed into E. coli BL21 (DE3) cells. RBM46 expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 19°C overnight. Harvested cells were resuspended in 10 ml of lysis buffer [50 mM tris-HCl (pH 7.0), 500 mM NaCl, 2 mM MgCl2, 10% glycerol, 0.1% Triton X-100, lysozyme (1 mg/ml), and 1 mM phenylmethylsulfonyl fluoride (PMSF)] supplemented with 10 mM imidazole and sonicated (output power 20%, 30 cycles of 10 s on, 15 s off). The lysates were centrifuged at 4000g for 30 min at 4°C, and the supernatant was incubated with 500 μl of Ni–nitrilotriacetic acid (NTA) agarose beads (Qiagen, 30210) that had been preequilibrated with five volumes of the binding buffer [50 mM tris-HCl (pH 7.0), 500 mM NaCl, 2 mM MgCl2, 10% glycerol, 0.1% Triton X-100, and 10 mM imidazole]. After 3 hours of rotation, Ni-NTA bead–protein complexes were extensively washed with the washing buffer containing different concentrations of imidazole (40 and 60 mM). Proteins were eluted with washing buffer containing 250 mM imidazole and concentrated by centrifugal filter units (molecular weight cutoff, 10,000; Thermo Fisher Scientific). The elution was exchanged with the sample buffer [20 mM tris-HCl (pH 7.0) and 10% glycerol] at 4°C.
GST pull-down assay
GST-RBM46 was expressed in E. coli BL21, and YTHDC2 protein with N-terminal His tag was produced by in vitro translation using the TNT quick coupled transcription/translation systems (Promega). GST-RBM46 fusion protein or GST alone in the lysates was immobilized on precleared MagneGST particles (Promega) and then incubated with in vitro translated YTHDC2 protein in binding (Promega). After incubation with rotation for 1 hour at room temperature, the MagneGST particle–protein complexes were washed extensively with washing buffer before addition of SDS-PAGE loading buffer. The following antibodies were used for Western blot: anti-GST (ABclonal, AE001) and anti-YTHDC2 (Abcam, ab220160).
Electrophoretic mobility shift assay
Single-stranded RNA probes were designed on the basis of the sequences with enriched peaks in RBM46 eCLIP-seq. Purified RBM46 protein at different concentrations (1, 5, and 10 nM) was incubated with 5 nM biotin-labeled RNA substrates with or without the RBM46-binding motif sequence in binding buffer [400 mM tris-HCl (pH 7.5), 5 mM MgCl2, 150 mM NaCl, 0.1% NP-40, 1 mM DTT, and 1 U of RNase inhibitor] at 23°C for 30 min, and this reaction was quenched with the stopping buffer [80 mM EDTA (pH 8.0), 50% glycerol, and diethyl pyrocarbonate (DEPC) H2O]. RBM46-RNA complexes and RNA substrates were separated on 12% Native TBE (tris-borate EDTA) PAGE gel [DEPC H2O, 5 × TBE, 50% glycerol, 30% acrylamide, 10% ammonium persulphate, and tetramethylethylenediamine (TEMED)] at 160 V for 70 min at room temperature and then transferred to nylon membrane through electrophoresis at 90 mA for 50 min. Signals from the nylon membrane were analyzed using Tanon4500 SF. The RNA probe from Ccna2 transcript was (motif sequence underlined) 5′-UUGUGAAACUAAAACCCGAC-3′, and the RNA probe with RBM46-binding motif absence was 5′-UGUUUGAAACAGGAGUCGCU-3′.
Quantification and statistical analysis
All data are reported as means ± SD unless otherwise noted in the figure legends. Significance was tested by using the two-tailed unpaired Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001) using Prism 7.0 (GraphPad Software, La Jolla, CA, USA).
Acknowledgments
We thank all the members in Ye laboratories.
Funding: This project was funded by the National Natural Science Foundation of China (31871503 and 32070843) and the Excellent Youth Foundation of Jiangshu Scientific Committee (BK20211532) to L.Y.; research grants from the National Natural Science Foundation of China (81773001) and the Top Talent Support Program for young and middle-aged people of Wuxi Health Committee (BJ2020086) to J.Zho.
Author contributions: B.Q. carried out most of the experiments and data analysis. S.H. and C.X. organized most of the figures. B.Q. and Z.B. performed genotyping experiments, immunoprecipitation, and Western blot analysis. J.Zho. and B.Z. designed and generated the Rbm46fl/fl mice. B.Q. and C.X. performed immunofluorescence and nuclear spread analysis. Y.L., B.Q., and S.H. performed eCLIP experiments. R.Y., R.W., J.Zha., and M.L. analyzed RNA-seq and eCLIP-seq data. J.G. cultured 293T cells and performed immunoprecipitation assay. B.Q. and S.H. purified RBM46 protein and designed RNA probes. Z.Y. and S.R. performed EMSA experiments. Q.H. drafted the introduction. L.Y., B.Q., and J.Zho. conceived the hypothesis and analyzed the data. L.Y. and Z.S. prepared and wrote the manuscript with input from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq and eCLIP sequencing data were deposited in the Gene Expression Omnibus database (GSE197282).
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
REFERENCES AND NOTES
- 1.Handel M. A., Schimenti J. C., Genetics of mammalian meiosis: Regulation, dynamics and impact on fertility. Nat. Rev. Genet. 11, 124–136 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Hunter N., Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koubova J., Menke D. B., Zhou Q., Capel B., Griswold M. D., Page D. C., Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc. Natl. Acad. Sci. U.S.A. 103, 2474–2479 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowles J., Knight D., Smith C., Wilhelm D., Richman J., Mamiya S., Yashiro K., Chawengsaksophak K., Wilson M. J., Rossant J., Hamada H., Koopman P., Retinoid signaling determines germ cell fate in mice. Science 312, 596–600 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., Sorek R., Rechavi G., Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Desrosiers R., Friderici K., Rottman F., Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U.S.A. 71, 3971–3975 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz S., Agarwala S. D., Mumbach M. R., Jovanovic M., Mertins P., Shishkin A., Tabach Y., Mikkelsen T. S., Satija R., Ruvkun G., Carr S. A., Lander E. S., Fink G. R., Regev A., High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155, 1409–1421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu Y., Dominissini D., Rechavi G., He C., Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 15, 293–306 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Liu J. Z., Yue Y. N., Han D. L., Wang X., Fu Y., Zhang L., Jia G. F., Yu M., Lu Z. K., Deng X., Dai Q., Chen W. Z., He C., A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y., Li Y., Toth J. I., Petroski M. D., Zhang Z. L., Zhao J. C., N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., Gong Z., Wang Q., Huang J., Tang C., Zou T., Yin P., Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature 534, 575–578 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Wang P., Doxtader K. A., Nam Y., Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell 63, 306–317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Śledź P., Jinek M., Structural insights into the molecular mechanism of the m6A writer complex. eLife 5, e18434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia G. F., Fu Y., Zhao X., Dai Q., Zheng G. Q., Yang Y., Yi C. Q., Lindahl T., Pan T., Yang Y. G., He C., N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C.-M., Li C. J., Vågbø C. B., Shi Y., Wang W.-L., Song S.-H., Lu Z., Bosmans R. P. G., Dai Q., Hao Y.-J., Yang X., Zhao W.-M., Tong W.-M., Wang X.-J., Bogdan F., Furu K., Fu Y., Jia G. F., Zhao X., Liu J., Krokan H. E., Klungland A., Yang Y.-G., He C., ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu C., Wang X., Liu K., Roundtree I. A., Tempel W., Li Y., Lu Z., He C., Min J., Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Xu C., Liu K., Ahmed H., Loppnau P., Schapira M., Min J., Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 290, 24902–24913 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z., Theler D., Kaminska K. H., Hiller M., de la Grange P., Pudimat R., Rafalska I., Heinrich B., Bujnicki J. M., Allain F. H.-T., Stamm S., The YTH domain is a novel RNA binding domain. J. Biol. Chem. 285, 14701–14710 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Lu Z., Gomez A., Hon G. C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G., Ren B., Pan T., He C., N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., Ma J., Wu L., YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7, 12626 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Zhao B. S., Roundtree I. A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C., N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li A., Chen Y. S., Ping X. L., Yang X., Xiao W., Yang Y., Sun H. Y., Zhu Q., Baidya P., Wang X., Bhattarai D. P., Zhao Y. L., Sun B. F., Yang Y. G., Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 27, 444–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi H., Wang X., Lu Z., Zhao B. S., Ma H., Hsu P. J., Liu C., He C., YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao W., Adhikari S., Dahal U., Chen Y. S., Hao Y. J., Sun B. F., Sun H. Y., Li A., Ping X. L., Lai W. Y., Wang X., Ma H. L., Huang C. M., Yang Y., Huang N., Jiang G. B., Wang H. L., Zhou Q., Wang X. J., Zhao Y. L., Yang Y. G., Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Zaccara S., Jaffrey S. R., A unified model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 181, 1582–1595.e18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wojtas M. N., Pandey R. R., Mendel M., Homolka D., Sachidanandam R., Pillai R. S., Regulation of m6A transcripts by the 3′→5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Molecular Cell 68, 374–387.e12 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Jain D., Puno M. R., Meydan C., Lailler N., Mason C. E., Lima C. D., Anderson K. V., Keeney S., ketu mutant mice uncover an essential meiotic function for the ancient RNA helicase YTHDC2. eLife 7, e30919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu P. J., Zhu Y., Ma H., Guo Y., Shi X., Liu Y., Qi M., Lu Z., Shi H., Wang J., Cheng Y., Luo G., Dai Q., Liu M., Guo X., Sha J., Shen B., He C., Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kretschmer J., Rao H., Hackert P., Sloan K. E., Hobartner C., Bohnsack M. T., The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′-3′ exoribonuclease XRN1. RNA 24, 1339–1350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L., Krasnykov K., Homolka D., Gos P., Mendel M., Fish R. J., Pandey R. R., Pillai R. S., The XRN1-regulated RNA helicase activity of YTHDC2 ensures mouse fertility independently of m6A recognition. Mol. Cell 82, 1678–1690.e12 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Saito Y., Hawley B. R., Puno M. R., Sarathy S. N., Lima C. D., Jaffrey S. R., Darnell R. B., Keeney S., Jain D., YTHDC2 control of gametogenesis requires helicase activity but not m6A binding. Genes Dev. 36, 180–194 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey A. S., Batista P. J., Gold R. S., Chen Y. G., de Rooij D. G., Chang H. Y., Fuller M. T., The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife 6, e26116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu R., Kasowitz S. D., Homolka D., Leu N. A., Shaked J. T., Ruthel G., Jain D., Lin H. J., Keeney S., Luo M. C., Pillai R. S., Wang P. J., YTHDC2 is essential for pachytene progression and prevents aberrant microtubule-driven telomere clustering in male meiosis. Cell Rep. 37, 110110 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soh Y. Q. S., Mikedis M. M., Kojima M., Godfrey A. K., de Rooij D. G., Page D. C., Meioc maintains an extended meiotic prophase I in mice. PLOS Genet. 13, e1006704 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abby E., Tourpin S., Ribeiro J., Daniel K., Messiaen S., Moison D., Guerquin J., Gaillard J. C., Armengaud J., Langa F., Toth A., Martini E., Livera G., Implementation of meiosis prophase I programme requires a conserved retinoid-independent stabilizer of meiotic transcripts. Nat. Commun. 7, 10324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai X., Cheng X., Huang J., Gao Y., Wang D., Feng Z., Zhai G., Lou Q., He J., Wang Z., Yin Z., Rbm46, a novel germ cell-specific factor, modulates meiotic progression and spermatogenesis. Biol. Reprod. 104, 1139–1153 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Wang C., Chen Y., Deng H., Gao S., Li L., Rbm46 regulates trophectoderm differentiation by stabilizing Cdx2 mRNA in early mouse embryos. Stem Cells Dev. 24, 904–915 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Zheng K., Wang P. J., Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLOS Genet. 8, e1003038 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korhonen H. M., Meikar O., Yadav R. P., Papaioannou M. D., Romero Y., Da Ros M., Herrera P. L., Toppari J., Nef S., Kotaja N., Dicer is required for haploid male germ cell differentiation in mice. PLOS ONE 6, e24821 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan L., Liu J. G., Zhao J., Brundell E., Daneholt B., Hoog C., The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell 5, 73–83 (2000). [DOI] [PubMed] [Google Scholar]
- 41.de Vries F. A. T., de Boer E., van den Bosch M., Baarends W. M., Ooms M., Yuan L., Liu J.-G., van Zeeland A. A., Heyting C., Pastink A., Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19, 1376–1389 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahadevaiah S. K., Turner J. M. A., Baudat F., Rogakou E. P., de Boer P., Blanco-Rodriguez J., Jasin M., Keeney S., Bonner W. M., Burgoyne P. S., Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27, 271–276 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Van Nostrand E. L., Pratt G. A., Shishkin A. A., Gelboin-Burkhart C., Fang M. Y., Sundararaman B., Blue S. M., Nguyen T. B., Surka C., Elkins K., Stanton R., Rigo F., Guttman M., Yeo G. W., Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer K. D., Saletore Y., Zumbo P., Elemento O., Mason C. E., Jaffrey S. R., Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray D., Kazan H., Cook K. B., Weirauch M. T., Najafabadi H. S., Li X., Gueroussov S., Albu M., Zheng H., Yang A., Na H., Irimia M., Matzat L. H., Dale R. K., Smith S. A., Yarosh C. A., Kelly S. M., Nabet B., Mecenas D., Li W. M., Laishram R. S., Qiao M., Lipshitz H. D., Piano F., Corbett A. H., Carstens R. P., Frey B. J., Anderson R. A., Lynch K. W., Penalva L. O. F., Lei E. P., Fraser A. G., Blencowe B. J., Morris Q. D., Hughes T. R., A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao L., Yao C., Xing X., Jing T., Li P., Zhu Z., Yang C., Zhai J., Tian R., Chen H., Luo J., Liu N., Deng Z., Lin X., Li N., Fang J., Sun J., Wang C., Zhou Z., Li Z., Single-cell analysis of developing and azoospermia human testicles reveals central role of Sertoli cells. Nat. Commun. 11, 5683 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKearin D. M., Spradling A. C., bag-of-marbles: A Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 4, 2242–2251 (1990). [DOI] [PubMed] [Google Scholar]
- 48.Insco M. L., Leon A., Tam C. H., McKearin D. M., Fuller M. T., Accumulation of a differentiation regulator specifies transit amplifying division number in an adult stem cell lineage. Proc. Natl. Acad. Sci. U.S.A. 106, 22311–22316 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonczy P., Matunis E., DiNardo S., bag-of-marbles and benign gonial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Development 124, 4361–4371 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Ohlstein B., Lavoie C. A., Vef O., Gateff E., McKearin D. M., The Drosophila cystoblast differentiation factor, benign gonial cell neoplasm, is related to DExH-box proteins and interacts genetically with bag-of-marbles. Genetics 155, 1809–1819 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y., Minor N. T., Park J. K., McKearin D. M., Maines J. Z., Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc. Natl. Acad. Sci. U.S.A. 106, 9304–9309 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen R., Weng C. J., Yu J. J., Xie T., eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc. Natl. Acad. Sci. U.S.A. 106, 11623–11628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen D., Wu C., Zhao S. W., Geng Q., Gao Y., Li X., Zhang Y., Wang Z. H., Three RNA binding proteins form a complex to promote differentiation of germline stem cell lineage in Drosophila. PLOS Genet. 10, e1004797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Insco M. L., Bailey A. S., Kim J., Olivares G. H., Wapinski O. L., Tam C. H., Fuller M. T., A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell Stem Cell 11, 689–700 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shan L., Wu C., Chen D., Hou L., Li X., Wang L., Chu X., Hou Y., Wang Z., Regulators of alternative polyadenylation operate at the transition from mitosis to meiosis. J. Genet. Genomics 44, 95–106 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Peters A. H., Plug A. W., van Vugt M. J., de Boer P., A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 5, 66–68 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Frankish A., Diekhans M., Ferreira A. M., Johnson R., Jungreis I., Loveland J., Mudge J. M., Sisu C., Wright J., Armstrong J., Barnes I., Berry A., Bignell A., Sala S. C., Chrast J., Cunningham F., Di Domenico T., Donaldson S., Fiddes I. T., Garcia Giron C., Gonzalez J. M., Grego T., Hardy M., Hourlier T., Hunt T., Izuogu O. G., Lagarde J., Martin F. J., Martinez L., Mohanan S., Muir P., Navarro F. C. P., Parker A., Pei B., Pozo F., Ruffier M., Schmitt B. M., Stapleton E., Suner M. M., Sycheva I., Uszczynska-Ratajczak B., Xu J., Yates A., Zerbino D., Zhang Y., Aken B., Choudhary J. S., Gerstein M., Guigó R., Hubbard T. J. P., Kellis M., Paten B., Reymond A., Tress M. L., Flicek P., GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8