Abstract
The Roundabout (Robo) receptors, located on growth cones of neurons, induce axon repulsion in response to the extracellular ligand Slit. The Robo family of proteins controls midline crossing of commissural neurons during development in flies. Mono- and bi-allelic variants in human ROBO1 (HGNC: 10249) have been associated with incomplete penetrance and variable expressivity for a breath of phenotypes, including neurodevelopmental defects such as strabismus, pituitary defects, intellectual impairment, as well as defects in heart and kidney. Here, we report two novel ROBO1 variants associated with very distinct phenotypes. A homozygous missense p.S1522L variant in three affected siblings with nystagmus; and a monoallelic de novo p.D422G variant in a proband who presented with early-onset epileptic encephalopathy. We modeled these variants in Drosophila and first generated a null allele by inserting a CRIMIC T2A-GAL4 in an intron. Flies that lack robo1 exhibit reduced viability but have very severe midline crossing defects in the central nervous system. The fly wild-type cDNA driven by T2A-Gal4 partially rescues both defects. Overexpression of the human reference ROBO1 with T2A-GAL4 is toxic and reduces viability, whereas the recessive p.S1522L variant is less toxic, suggesting that it is a partial loss-of-function allele. In contrast, the dominant variant in fly robo1 (p.D413G) affects protein localization, impairs axonal guidance activity and induces mild phototransduction defects, suggesting that it is a neomorphic allele. In summary, our studies expand the phenotypic spectrum associated with ROBO1 variant alleles.
Introduction
Roundabouts (Robos) are single-pass transmembrane proteins that belong to the immunoglobulin superfamily of cell adhesion molecules. Robo receptors are highly conserved from invertebrates to mammals. The Drosophila robo1 gene was first identified in a mutant screen for genes that control axonal guidance of the midline in the embryonic central nervous system (CNS) (1,2). Robos act as axon guidance receptors, upon interaction with the soluble secreted extracellular ligand Slit proteins. They regulate proper formation of neuronal connectivity and play roles in variety of neuronal developmental processes. Indeed, they are also involved in angiogenesis and organogenesis of muscle, kidney, lungs, heart (3,4) and limbs (5).
Here, we report probands from two families who carry unreported pathogenic mutations in ROBO1 (MIM: 602430). A homozygous p.S1522L variant was identified from three affected siblings who present with nystagmous; in contrast, a de novo heterozygous p.D422G variant was identified in a patient with an early-onset epileptic encephalopathy (EOEE). Previously, biallelic variants in ROBO1 were reported in patients with congenital anomalies of the kidney and urinary tract (CAKUT) (6). Both biallelic and monoallelic variants in ROBO1 were reported in patients with congenital heart disease (6,7), as well as neurodevelopmental disorders including strabismus, pituitary stalk interruption syndrome and intellectual impairment (6,8–11). The molecular underpinnings associated with ROBO1 variants remains elusive partly due to the incomplete penetrance as well as the variable expressivity of diverse clinical features associated with different ROBO1 variants. This study aims to define the function of two novel ROBO1 variants with irrelevant clinical presentations by employing Drosophila as in vivo model organism.
There are four Robo paralogs (1–4) in mammals and three Robos (1–3) in Drosophila due to gene duplication in evolution. Drosophila robo1 is the closest homolog of human ROBO1, ROBO2 and ROBO3 with DIOPT scores (12) of 9/16, 11/16 and 8/16, respectively. It is well established that Drosophila robos differentially control axonal guidance: robo1 loss-of-function (LoF) causes axonal roundabout phenotype of the midline of the ventral nerve cord (VNC). In contrast, robo1 gain-of-function (GoF) causes axonal repulsion from the midline of the VNC (13). robo1 was also shown to play roles in development of dendrites (14), heart tube (15) and trachea (16).
Here, we show that robo1 is not essential for survival, but either LoF or GoF significantly reduce fly viability. robo1 is expressed in neurons but not in glia of CNS. In the visual system, robo1 exhibits broad expression in adult optic neurons and our data show that it plays a role in modulating adult phototransduction. We characterized the nature of the ROBO1 variants identified in probands with a de novo dominant and with a biallelic variant. The recessive variant is a partial LoF allele whereas the dominant variant is a neomorphic allele that leads to protein mislocalization, loss of the midline guidance activity and defects in phototransduction. Our data show that these variants are associated with a phenotypic expansion and affect the function of the protein in a very different manner.
Results
Clinical profiles of probands
We identified two novel ROBO1 variants associated with distinct phenotypes. In family #1, there are three affected males who present with isolated nystagmus (Table 1 and Supplementary Material, Fig. S1A). The parents are first degree cousin. They had one female sibling who died when she was 18 years. She had severe hypoxic ischemic encephalopathy resulting in cerebral palsy and profound developmental delay. All affected siblings (individual 1.1, 1.2, 1.3) were born at term via normal delivery after an uncomplicated pregnancy. They developed normally, graduated from colleges, currently working full time and maintaining a normal life. On clinical evaluation, they had normal anthropometric measurements. Physical examination revealed bilateral horizontal nystagmus but no other neurological symptoms were observed. They had no additional neurological or other system anomalies including normal finger to nose and heel shin test for cerebellar examination. Diagnostic work up including blood count, comprehensive metabolic panel, urine organic acid and plasma amino acid were normal. Brain magnetic resonance imaging (MRI) of two subjects (individual 1.1 and 1.3) did not reveal any abnormality. Pentad exome of the three individuals and parents revealed biallelic missense variant (NM_002941: c.4565C > T, p.S1522L) in ROBO1 that segregated with the nystagmus phenotype (Supplementary Material, Fig. S1A). The variant was surrounded by an absence of heterozygosity (AOH) block ranging from 23.6 Mb to 57.3 Mb (Supplementary Material, Fig. S1B). The recessive p.S1522L variant is present in the population database gnomAD, reported as heterozygous in 0.058% of individuals and homozygous in one individual of African origin. These data are compatible with a rare hypomorphic allele with a CADD score of 22.9.
Table 1.
Clinical and genetic features of affected individuals with ROBO1 variants
| Family | Family 1 | Family 2 | ||
|---|---|---|---|---|
| Ethnicity | Turkish | Chinese | ||
| Proband | 1.1 (BAB7196) | 1.2 (BAB7197) | 1.3 (BAB7198) | 2 |
| Gender | Male | Male | Male | Male |
| Age | 42 years | 37 years | 29 years | 3 months |
| ROBO1 variants | 3:78656062, G > A c.4565C > T p.S1522L | 3:78656062, G > A c.4565C > T p.S1522L | 3:78656062, G > A c.4565C > T p.S1522L | 3: 78734973, T > C c.1265A > G p.D413G |
| Zygosity | homozygous | homozygous | homozygous | heterozygous |
| Inheritance | AR, inherited | AR, inherited | AR, inherited | AD, de novo |
| Allele frequency (gnomAD) | 0.058% | Not listed | ||
| CADD score | 22.9 | 28 | ||
| Clinical phenotypes | ||||
| Seizures | No | No | No | EOEE |
| Ophthalmologic defects | Nystagmus | Nystagmus | Nystagmus | No |
| Developmental delay | No | No | No | Yes |
| Dysmorphism | No | No | No | No |
| Brian MRI | Normal | NA | Normal | Normal |
AD: autosomal dominant; AR: autosomal recessive; NA: not available.
In family #2, a proband presented with a severe EOEE (Table 1). The proband was born full-term to non-consanguineous parents. Typical infantile spasms with ‘nodding and holding ball’ movements accompanied with loss of consciousness were noticed at 3 months of age. An electroencephalogram (EEG) showed a large number of high-amplitude sharp waves, spikes, irregular slow waves firing in bilateral central, parietal and mid-posterior temporal regions during both awake and asleep states. These epileptic discharges were more obvious in the left hemisphere and were able to spread to all channels. Multiple isolated as well as clustered seizures during wakefulness were observed. Anti-epileptic treatments including valproate, topiramate, adrenocorticotropic hormone (ACTH) as well as a ketogenic diet unable to control the seizures. A brain MRI did not reveal abnormalities and no obvious dysmorphic features were observed at the time.
The individual is delayed in developmental milestones. He could not lift his head at 3 months of age, and has no ability to stand without support or follow objects with his eyes at the age of 4. His height is 102 cm (27th percentile), his weight is 15.4 kg (22.8th percentile) and his head circumference 50 cm (39.7th percentile). Trio-exome sequencing identified a monoallelic de novo variant (NM_002941: c.1265A > G, p.D422G) that was confirmed with Sanger sequencing (Supplementary Material, Fig. S1C). The dominant p.D422G variant is absent from the population databases (ExAC, gnomAD and 1000genomes) and a patient database (ClinVar). It is predicted to be conserved and pathogenic by multiple algorithms (see Material and Methods) with a CADD score of 28.
To gather information about the gene, we queried the Model organism Aggregated Resources or Rare Variant ExpLoration (MARRVEL) (17). ROBO1 is not haploinsufficient with a pLI score of 0 with o/e (observed/expected) ratio of 0.46 (18), and many LoF variants are present in population databases including gnomAD, ExAC and chromosomal deletions database (DGV) of reference individuals (19). ROBO1 is not constrained to missense variation with a Z score of 1.04 based on gnomAD and o/e ratio of 0.90 (18). These data indicate that ROBO1 is not haploinsufficient. Hence, the variant in patient #2 may correspond to a GoF or a dominant negative variant allele.
Developmental loss of robo1 is not lethal in Drosophila
To investigate the function of the ROBO1 variants, we modeled the variants in Drosophila melanogaster. The Slit-Robo1 signaling pathways control the crossing of the midline of some neurons during embryonic CNS development in Drosophila (1,2,13). The closest homolog of human ROBO1 in the fly is robo1. The protein sequences of human ROBO1 and fly Robo1 share 48% similarity and 33% identity, and the overall protein structures are very similar (1,20).
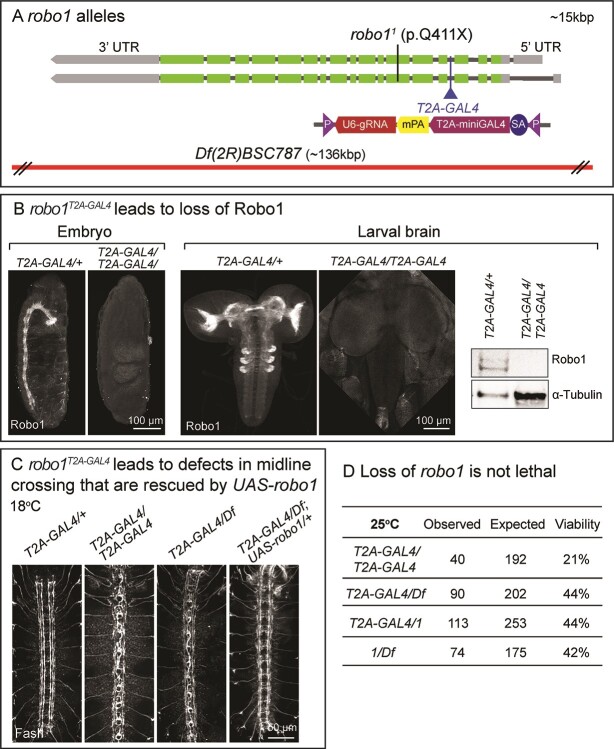
We generated a robo1T2A-GAL4 allele by CRISPR-Mediated Integration Cassette (CRIMIC) that truncates the Robo1 protein and expresses T2A-miniGAL4 in a similar pattern as endogenous robo1 (Fig. 1A) (21,22). This allele leads to a complete loss of Robo1 staining in embryos and larval brain based on whole-mount immunostaining as well as immunoblotting (Fig. 1B). Flies that carry homozygous robo1T2A-GAL4/robo1T2A-GAL4 as well as robo1T2A-GAL4 over a deficiency allele Df (2R)BSC787 (Df for) show the typical midline crossing defects of the axons when labeled by anti-Fasciclin II (FasII, labels three longitudinal tracts on each side of the midline). This phenotype is fully penetrant in the embryonic VNC, and is rescued by expression of the fly robo1 cDNA (UAS-robo1) at 18°C (Fig. 1C). Note that the temperature strongly affects the expression level of the UAS-cDNA as there is very low expression at 18°C and very high levels of expression at 28°C (23). These data indicate that robo1T2A-GAL4 is a null allele and that T2A-GAL4 drives UAS-robo1 expression. Interestingly, homozygous robo1T2A-GAL4/robo1T2A-GAL4 mutants are not lethal as ~ 20% of the flies survive to adulthood at 25°C (Fig. 1D). Previously, robo11/robo11 (p.Q411Term) and robo11/robo18 mutants were reported to be embryonic lethal (24). In contrast, robo11/robo12 and robo12/robo18 mutants escape as adults with severe midline crossing defects in first instar (25). However, the molecular nature of the lesions in robo12 and robo18 have not been established. To determine whether robo1 is essential for viability, we performed complementation tests of different null alleles. The Df allele combined with either robo1T2A-GAL4 or robo11 leads to ~ 40% transheterozygous viable flies and a similar survival rate was also observed for robo1T2A-GAL4/robo11 mutants (Fig. 1D). Together, these data indicate that loss of robo1 does not necessarily causes lethality even when severe axonal guidance defects are observed in embryonic development.
Figure 1.
Loss of robo1 reduces survival rate. (A) Structure of the fly robo1 gene and alleles. The CRIMIC T2A-miniGAL4 sequence is inserted into a shared intron of all robo1 transcripts, truncating the transcript and protein while expressing T2A-miniGAL4 (21). The nonsense robo11 (p.Q411Term) allele (2) as well as the chromosomal deficiency Df(2R)BSC787 allele are indicated. (B) Confocal images of Robo1 immunostaining in stage 16–17 embryos as well as 3rd instar larval (L3) brains of robo1T2A-GAL4/+ and robo1T2A-GAL4/robo1T2A (left). Immunoblot of Robo1 extracted from L3 larval brains of robo1T2A-GAL4/+ and robo1T2A-GAL4/robo1T2A. α-Tubulin served as a loading control (right). (C) Confocal images of FasII immunostaining in stage 16 embryo VNC. (D) Viability rates of adult robo1 mutants with different null alleles.
Drosophila robo1 is expressed in optic neurons and modulates phototransduction
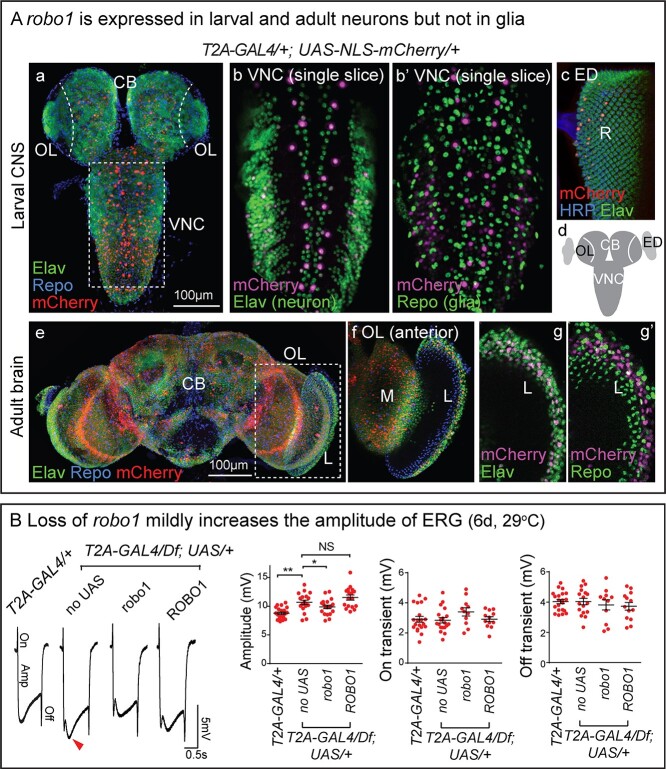
The fly Robo1 protein is widely expressed but it is enriched in the CNS neuropil in embryos and third instar larvae (Fig. 1B). It is also expressed in many neurons in adult neuropils (26). To identify the cells that express robo1, we used the T2A-GAL4 to drive UAS-NLS-mCherry (see Fig. 1A) and compared its expression to the pan-neuronal marker Elav as well as the glial marker Repo. In the third instar larval CNS, mCherry (robo1) is expressed in a very defined subset of neurons but not in glia (Fig. 2Aa and b’). It is expressed sparsely in the optic lobe and eye imaginal disc (Fig. 2Aa), in which most of the Elav-positive cells are immature optic neurons, including medulla and lamina neurons in the optic lobe, retinal cells in eye disc (27). In adults, mCherry (robo1) is expressed in numerous neurons of the central brain, optic lobe and peripheral lamina, but rarely in glia (Fig. 3Ae–g’). Specifically, robo1 shows much broader expression in adult optic neurons than in the larval neuropil. This is consistent with single cell RNA-Seq (Supplementary Material, Fig. S2, Fly Cell Atlas) (28,29).
Figure 2.
robo1 is expressed in some neurons and increases phototransduction. (A) Confocal images of CNS from robo1T2A-GAL4/+; UAS-UAS-NLS-mCherry/+. The animals were raised at 25°C. (a) Projection image of L3 larval CNS co-stained with neuronal marker anti-Elav and glial marker anti-Repo. mCherry fluorescent signal was amplified by anti-mCherry. (b, b’) Single slice images of ventral nerve cord (VNC) co-stained with anti-Elav and anti-Repo. (c) Projection image of L3 Imaginal eye disc (ED) labeled with anti-Elav and anti-HRP, a few retinal cells (R) are mCherry-positive. (d) Schematic of L3 CNS indicating the ventral nerve cord (VNC), central brain (CB), optic lobe (OL) and imaginal eye disc (ED). (e) Projection image of adult brain co-stained with anti-Elav and anti-Repo. (f) The anterior side of the optic lobe (OL) which includes the medulla (M) and lamina (L) is shown. (g, g’) Images of higher magnification show lamina (L) co-stained with anti-Elav (g) and anti-Repo (g’). (B) Electroretinograms (ERGs) of flies at 6 days post eclosion (dpe). Amplitudes, On and Off transients were quantified. Error bar: s.e.m. NS, P > 0.05; *P < 0.05, **P < 0.01 by one-way ANOVA with Turkey’s multiple comparison test between each indicated genotype.
Figure 3.
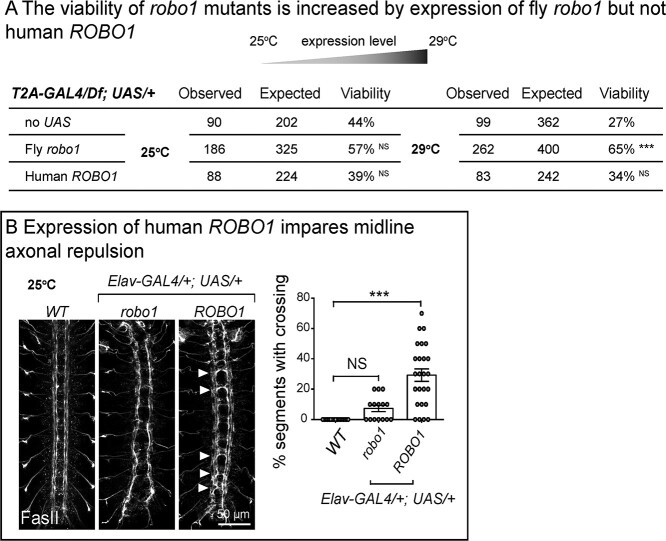
Fly robo1 partially recues but human ROBO1 fails to rescue the loss of fly robo1. (A) Viability analysis of adult flies with indicated genotypes at different temperatures. The viability of robo1 mutants is increased by expression of fly robo1 but not human ROBO1. NS, P > 0.05; ***P < 0.001 by chi-square test between each genotype to corresponding control. (B) Confocal images of FasII immunostaining in stage 16 embryonic VNC. Expression of fly robo1 leads to midline axonal repulsion and widening of the FasII labeled tracts whereas human ROBO1 impairs repulsion and leads to midline crossing. The percentages of segments with midline crossing of each embryo were quantified. Error bar: s.e.m. NS, P > 0.05; ***P < 0.001 by one-way ANOVA with Turkey’s multiple comparison test between each indicated genotype.
The Slit and Robo proteins have been documented to be involved in the development of Drosophila visual system (30), as well as synaptogenesis in the CNS of adult mice (31). To examine whether robo1 is required for proper phototransduction in adult flies, we performed electroretinograms (ERGs) on robo1 mutants. The amplitudes of the ERG traces represent the depolarization of photoreceptors in the retina upon light exposure, while the ON/OFF transients provide a measure of synaptic transmission between photoreceptors and the post-synaptic neurons in the lamina (32). The amplitudes, but not the ON/OFF transients, are mildly but significantly increased in robo1T2A-GAL4/Df mutants when compared to the robo1T2A-GAL4/+ heterozygous controls. Expression of UAS-robo1 diminishes the increase (Fig. 2B), suggesting that robo1 plays a role in modulating retinal activity, without significantly affecting the postsynaptic response of neurons in the lamina. robo1 is expressed in all photoreceptors as revealed by single cell RNA-Seq (Supplementary Material, Fig. S2), suggesting a possible cell-autonomous regulation of robo1 in retinal neurons. To assess the function of human ROBO1, we generated transgenic flies that carry UAS-ROBO1 cDNA. Expression of human ROBO1 in the robo1T2A-GAL4/Df mutants did not restore the amplitude increase (Fig. 2B).
Human ROBO1 does not rescue the loss of fly robo1
To further assess the function of human ROBO1, we tested whether the expression of UAS-ROBO1 under con-trol of robo1T2A-GAL4 alters the viability of robo1T2A-GAL4/Df mutants. Expression of fly UAS-robo1 at 25°C causes a non-significant increase in viability of the robo1T2A-GAL4/Df mutants, whereas human UAS-ROBO1 does not increase the viability (Fig. 3A). When the temperature is increased to 29°C, the fly UAS-robo1 significantly increased the viability rate from 27 to 65% but human UAS-ROBO1 did not (Fig. 3A). Fly robo1 GoF by pan-neuronal GAL4s leads to a failure in midline crossing and a repulsion of the midline axons in embryonic VNC (33) and we observe a similar phenotype (Fig. 3B). In contrast, pan-neuronal overexpression of human UAS-ROBO1 causes frequent ectopic midline crossings as well as mild midline repulsion (Fig. 3B), consistent with a previous finding (34). In summary, the human ROBO1 reference cDNA does not rescue the loss of fly robo1 and may interfere with the normal function of fly Robo1 causing a dominant negative effect.
ROBO1 p.S1522L variant is less toxic than the human reference gene when expressed in Drosophila
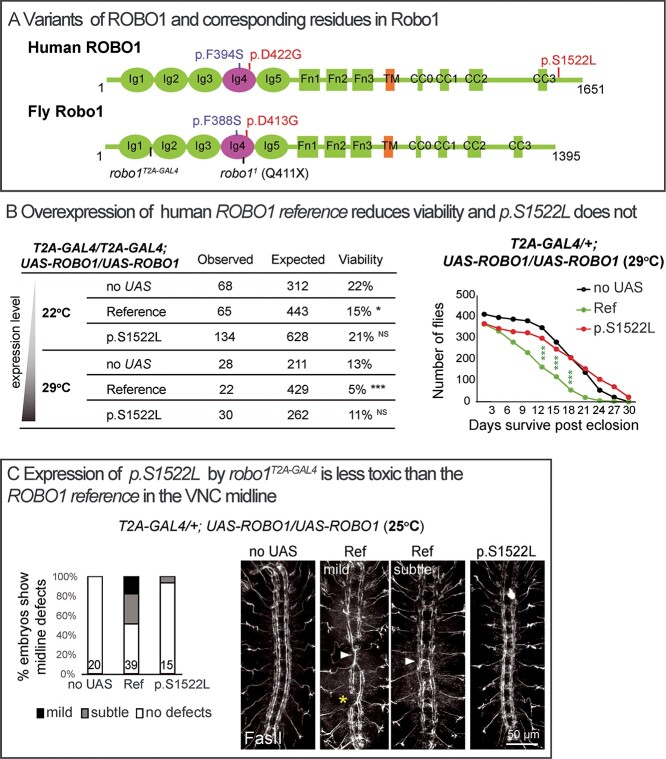
We next aimed to determine whether the proband-associated variants in ROBO1 alter the function of the encoded protein in vivo. Given that the p.S1522L affects a residue that is not conserved in robo1 (Supplementary Material, Fig. S3), this variant was modeled in human UAS-ROBO1 (Fig. 4A), and we assessed its function using GoF assays. The robo1T2A-GAL4/robo1T2A-GAL4 flies show a viability rate of 22% at 22°C and 13% at 29°C (Fig. 1D), and expression of the UAS-ROBO1 reference in robo1T2A-GAL4/robo1T2A-GAL4 flies further decreased the viability rates significantly to 15 and 5%, respectively, whereas p.S1522L did not significantly alter the rates (Fig. 4B). Similarly, expression of the UAS-ROBO1 reference driven by one copy of robo1T2A-GAL4/+ significantly reduced the life span of adult flies, but p.S1522L did not (Fig. 4B). Moreover, expression of the UAS-ROBO1 reference in robo1T2A-GAL4/+ flies leads to midline defects of longitudinal axons and ectopic crossing in some segments, whereas these phenotypes are rarely observed in p.S1522L-expressing embryos (Fig. 4C). These three assays confirm that overexpression of human reference ROBO1 has a toxic effect in Drosophila and p.S1522L is less toxic, suggesting that the p.S1522L variant is a LoF allele.
Figure 4.
ROBO1 p.S1522L is a partial LoF allele. (A) Variants of ROBO1 (NP_002932.1) and corresponding residues in fly Robo1 (NP_476899.1). Robo receptors contain five immunoglobulin (Ig) domains, three Fibronectin (Fn) type III domains, a transmembrane (TM) domain and a large unstructured intracellular region typically containing four conserved cytoplasmic (CC) motifs. (B) Viability of adult flies as well as life span when human ROBO1 cDNAs are overexpressed. UAS-ROBO1 reference but not p.S1522L reduces viability. NS, P > 0.05; *P < 0.05, ***P < 0.001 by chi-square test between each genotype to corresponding controls (no UAS). (C) Confocal images of FasII immunostaining in the VNC of stage 16 embryos. Expression of p.S1522L is less toxic than the ROBO1 reference in the VNC midline. Segments showing ectopic crossing are indicated by arrowheads, longitudinal axon disruption is indicated by a star. The severity of the VNC defects were quantified, subtle defects correspond to single aberrant segments in the VNC, mild defects correspond to two or three defective segments in a VNC. The number of embryos is indicated at the bottom of the column.
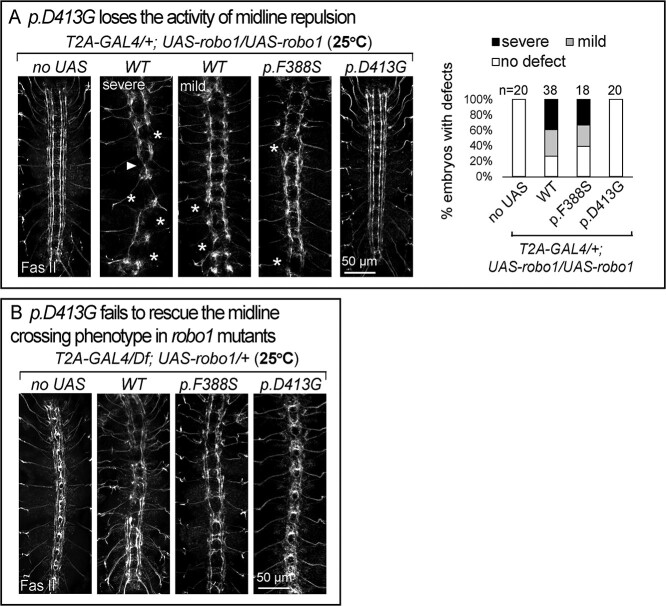
p.D413G affects midline crossing
ROBO1 p.D422G is a dominant mutation (de novo) that affects an amino acid in the Ig4 domain, a highly conserved domain of the Robo family of proteins (Fig. 4A and Supplementary Material, Fig. S3). Loss of this Ig4 domain leads to a loss of Robo1 function phenotype in Caenorhabditis elegans mechanosensory AVM neuron (35). However, loss of the Ig4 domain in flies does not alter Robo1 activity in the VNC of embryos (36), but other phenotypes associated with loss of this domain were not assessed in flies. The Ig4 domain has been shown to be required for the homo-dimerization between Robo receptors in vitro (35,37). Moreover, mutating a conserved phenylalanine in the domain that mediates the dimerization in C. elegans Robo/Sax-3 (p.F360R) leads to loss of Robo activity (35). Hence, it is not obvious how the dominant p.D422G variant may cause a phenotype, nor which processes dependent on Robo1 in flies may be affected by Ig4. We therefore decided to not only model p.D422G but also variants associated in human databases that affect the phenylalanine that is required for Sax3 function in C. elegans (p.F360R) (Supplementary Material, Fig. S3). Heterozygous variants p.F394S have been documented in three individuals in GnomAD and Geno2MP. To assess and compare the function of these two amino acids in Ig4, we modeled p.D422G and p.F394S in fly robo1, p.D413G and p.F388S, respectively (Fig. 4A).
First we examined midline axonal guidance in embryos by overexpression of the fly UAS-robo1. Overexpression of wild-type robo1 (UAS-robo1-WT) driven by robo1T2A-GAL4/+ at 25°C leads to an array of defects in 80% of the embryos, ranging from severe defects, including midline repulsion of longitudinal axons as well as axonal loss, to somewhat milder defects, including repulsion and collapse of the three longitudinal tracts (Fig. 5A). These phenotypes mimic the GoF phenotype of fly robo1. Embryos expressing UAS-robo1-p.F388S display very similar phenotypes as UAS-robo1-WT suggesting no or a very mild LoF. In contrast, UAS-robo1-p.D413G did not cause midline phenotype (Fig. 5A). This suggests that p.D413G is a LoF allele. Next we performed rescue assay by expressing the UAS-cDNAs in the absence of robo1 (robo1T2A-GAL4/Df) at 25°C. UAS-robo1-WT and UAS-robo1-p.F388S rescue the axonal crossing phenotype but both are associated with some mild defects similar to a mild GoF phenotype of robo1. However, UAS-robo1-p.D413G did not rescue the roundabout phenotype in robo1T2A-GAL4/Df mutants (Fig. 5B), again suggesting the p.D413G mutant has no obvious activity in this assay.
Figure 5.
p.D413G affects midline guidance. Confocal images of FasII immunostaining in the VNC of stage 16 embryos. (A) GoF assays by expression of UAS-robo1 cDNAs in robo1T2A-GAL4/+. UAS-robo1-WT or p.F388S causes midline repulsion phenotypes in the VNC, but p.D413G does not cause this GoF phenotype. Segments showing abnormal crossing are indicated by arrowheads, disruption of axonal fascicles are indicated by stars. The severity of the VNC defects was quantified: mild defects correspond to two or three defective segments, severe defects correspond to more than three defective segment of the VNC. The number of quantified embryos is indicated at the top of the columns. (B) Rescue assays with UAS-robo1 cDNAs in robo1T2A-GAL4/Df mutants. p.D413G fails to rescue the roundabout phenotypes in the VNC.
p.D413G affects Robo1 protein localization
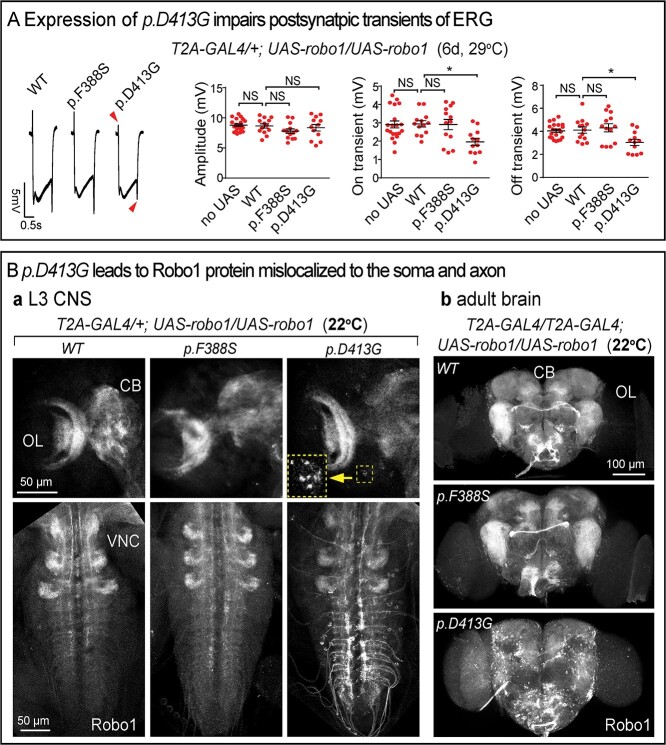
As previously shown, robo1 is expressed in photoreceptors as well as the postsynaptic lamina neurons (L1, 2, 3, 4 and 5; Supplementary Material, Fig. S2). We therefore tested the effect of expression of UAS-robo1-WT, p.F388S and p.D413G driven by robo1T2A-GAL4/+ at 29°C on ERG amplitudes and On/Off transients. None of the proteins affected the ERG amplitudes but expression of p.D413G significantly decreased the amplitude of the On/Off transients (Fig. 6A). This suggests that expression of p.D413G is toxic in this assay.
Figure 6.
p.D413G affects Robo1 protein localization. (A) ERGs of flies expressing UAS-robo1 cDNAs in robo1T2A-GAL4/+ at 6 dpe. UAS-robo1-p.D413G impairs postsynaptic On/Off transients. Amplitudes, On and Off transients were quantified. Error bar: s.e.m. NS, P > 0.05; *P < 0.05, **P < 0.01 by one-way ANOVA with Turkey’s multiple comparison test between each indicated genotype. (B) Confocal images of Robo1 immunostaining in Drosophila CNS. (a) L3 larval brains and VNCs expressing UAS-robo1-cDNAs by robo1T2A-GAL4/+. Arrowhead indicates the soma of neurons that have accumulated Robo. The inset shows the enlarged image. (b) Adult brains (anterior view) expressing UAS-robo1-cDNAs in robo1T2A-GAL4/robo1T2A. CB: central brain; OL: optic lobe. UAS-robo1-p.D413G leads to highly aberrant protein accumulations in soma and axon. This is not observed when UAS-robo1-WT or p.F388S is expressed.
To compare the localization of the WT and mutant proteins, we examined their distribution by driving their expression with robo1T2A-GAL4/+. Interestingly, in third instar larvae, the p.D413G protein is obviously mislocalized to the soma and axon of VNC neurons. This aberrant localization is not observed in UAS-robo1-WT or p.F388S expressing larvae (Fig. 6Ba). Finally, we performed similar experiments in adult brains by driving UAS-cDNAs with robo1T2A-GAL4/robo1T2A-GAL4. UAS-robo1-WT or p.F388S are broadly distributed in the adult brain. However, the p.D413G protein is mislocalized and accumulates in soma and axons of numerous neurons (Fig. 6Bb). Hence, the p.D413G mutant clearly affects Robo1 protein localization which may underlie the toxic effect discussed above.
Discussion
ROBO1 variants have been associated with very diverse clinical features including neuronal, cardiac and renal developmental defects with incomplete penetrance and phenotypic heterogeneity (6,8–11). Here, we describe two probands with previously undocumented variants and phenotypes that are due to a recessive as well as a de novo variant allele. Functional studies in Drosophila indicate that these two novel missense variants cause very different neurodevelopmental phenotypes via distinct mechanisms.
We created a null allele of robo1 by inserting the T2A-GAL4 in an early coding intron. This allowed us to express the human ROBO1 gene in the proper spatial and temporal expression pattern. The fly robo1 gene fully rescues the phenotypes observed in the midline of the VNC in embryos caused by loss of robo1 (Fig. 1C). It also partially rescues the decreased viability in robo1 LoF flies, in contrast, the reference human ROBO1 does not. Moreover, the expression of reference human ROBO1 is toxic as it affects viability as well as axonal guidance in the embryonic VNC (Fig. 3). The VNC defects with ectopic midline crossing caused by expression of the reference human ROBO1 suggest a dominant negative effect (Fig. 3B) (34). The simplest interpretation is that human ROBO1 poisons the function of the fly robo1 gene/protein or its signaling. This may be due to various causes which include titrating away Slit ligand, affecting the downstream effectors which participate in cytoskeleton modulation such as the Scar/WAVE complex (4,38) or forming non-productive dimers with Robo1. The latter is less likely as our data and those of others (36) indicate that the dimerization domain, which has been mapped to Ig4, is not required for the VNC axonal guidance. These observations do not allow us to determine the function of the newly discovered variants in a LoF context (robo1 null mutants). However, the observed toxicity can be used to determine if specific human variants affect the toxic/dominant negative function of ROBO1 when expressed in flies using the T2A-GAL4.
The p.S1522L variant identified in family #1 maps to the C-terminal cytodomain which is critical to transduce the signal of intracellular effectors (4). This domain is less conserved in the Robo family when compared to its ectodomain and the p.S1522 is not conserved in flies. This variant is inherited in a recessive manner. A comparison of the toxicity induced by p.S1522L with reference ROBO1 with respect to viability and midline axonal guidance, clearly indicate that the p.S1522L variant has reduced toxicity in the both assays (Fig. 4), suggesting that it is a partial LoF variant. We do not know whether the apparent LoF caused by p.S1522L is due to lowered protein activity or protein level. The mechanisms by which p.S1522L affects ROBO1 activity remains to be investigated.
The ROBO family of proteins play important roles in regulating eye movement in mammals. Mouse Robo1 and Robo2 are expressed in oculomotor neuron and regulate their migration in embryos (39). ROBO3 (HGNC:13433) is associated with a diagnosis of recessive horizontal gaze palsy with progressive scoliosis-1 (HGPPS, MIM: 607313) (40,41) and many of the patients have nystagmus (42). Strabismus is one of the most common phenotype in the spectrum of ROBO1-associated disease (6,8,10,11), but nystagmus has not yet been associated with ROBO1 variants. Control of eye movements, including horizontal and vertical movements, as well as vergence, require a complex circuit that involves the brainstem, cerebellum and forebrain. Horizontal eye movements are generated by the lateral and medial rectus muscles which are controlled by the abducens and oculomotor nuclei, respectively. The medial longitudinal fasciculus interconnects the right and left oculomotor, trochlear, as well as the abducens and vestibular nuclei. Any abnormality that affects the connection of these nerves can result in horizontal nystagmus.
Our data indicate that the homozygous missense p.S1522L variant, which is a partial LoF based on our fly studies can cause nystagmus. The variant is within ~24 Mb AOH block which was shared by all three affected siblings (Supplementary Material, Fig. S1), driven by identity-by-descent due to consanguinity between the parents. This variant was reported in one individual in gnomAD in a homozygous state. Given the relatively mild phenotype (isolated nystagmus) without any other associated symptoms, the individual might have been overlooked in gnomAD or the variant was not penetrant. Our study provides a resource for modeling ROBO1 variants by evaluating variants in Drosophila.
ROBO1 is not haploinsufficient as its pLI score is 0 (gnomAD); however, monoallelic LoF variants in ROBO1 have been associated with neurodevelopmental and cardiac phenotypes (7,11), suggesting a low penetrance of dominant variants. Incomplete penetrance was also observed in other genes implicated in the Slit-Robo pathway, including SLIT2 (HGNC:11086), ROBO2 (HGNC:10250) and the effector SRGAP1 (HGNC:17382). Renal defects associated with variants in the three genes are dominant, but the identified variants also presented in healthy carriers (43–47). It is possible that an individual gene in the Slit-Robo signaling is one of the many permissive factors that are required for specific developmental processes. A dosage reduction in individual genes may not reach a phenotypic threshold but sensitize the process, and variants in other components that involved the same process may strengthen the phenotypic outcome (48,49). However, the dominant p.D422G in individual #2 associated with EOEE is likely to act via a different mechanism. None of the reported patients with ROBO1 variants has the phenotypes displayed by individual #2. Our data show that the fly robo1 p.D413G variant causes a very aberrant mislocalization of Robo1 in soma and axons (Fig. 6B), implicating defective trafficking of Robo1. The abnormal protein distribution may cause a LoF and/or affect other interacting proteins. Although the variant maps to the conserved Ig4 domain, this domain is not required for the midline guidance of Drosophila VNC (36). The p.D413G mutant affects axonal guidance activity (Fig. 5A) which is unlikely due to the LoF of Ig4, but is likely due to the aberrant Robo1 localization. The expression of the p.D413G mutant also creates defects in phototransduction that are not observed in robo1 LoF or GoF (WT) flies (Fig. 6A). Hence, fly p.D413G behaves as a neomorphic allele and the human p.D422G variant is highly likely to be pathogenic. We therefore propose that the EOEE phenotype associated with this allele is due to the toxic effects of the mislocalized protein. It remains to be established whether the p.D422G leads to mislocalization of ROBO1 in human neurons and the mechanisms by which p.D422G affects ROBO1 protein localization remains to be further investigated.
Material and Methods
Diagnosis and human genetics
Three brothers of consanguineous parents (Family 1) presented to the genetics clinic in Balikesir (Turkey) due to abnormal eye movements. The three siblings and parents were enrolled into the Baylor-Hopkins Center for Mendelian Genomics research initiative (IRB number: H-29697). Pentad exome sequencing and analysis were performed according to previously described methods (50). Orthogonal Sanger dideoxy sequencing was performed for variant confirmation and segregation purposes. To identify absence of heterozygosity (AOH) genomic regions, we used BafCalculator to calculate the B-allele frequency (ratio of variant reads to total reads) from exome data (51).
The genetic and clinical data of family 2 (the proband and parents) were collected in the Maternal and Child Health Hospital of Hunan Province (China). The diagnosis of EOEE was made according to widely accepted criteria (52). Genomic DNA from peripheral blood leukocytes of the family trio was captured using the IDT xGen Exome Research Panel (Integrated DNA Technologies, San Diego, CA, USA) and was sequenced on the Novaseq 6000 platform (Illumina, San Diego, CA, USA). Bioinformatic analyses were performed according to the standard protocol (53). Human population databases including gnomAD (54), ExAC (18). 1000genomes (55) were used for variant parsing and filtration. GERP++, phyloP, phastCons and SiPhy were used for variant conservation prediction. In silico prediction algorithms including CADD (56), SIFT (57), Polyphen-2 (58), PROVEAN (59), M-CAP (60) and MutationTaster (61), were used to assess variant pathogenicity. Sanger sequencing was performed for variant validation. All participants signed informed consent forms and the study was approved by the Ethics Committee of the Maternal and Child Health Hospital of Hunan Province (2020-S003).
The identified variants have been submitted to Clinvar, accession number: SCV002099445, SCV002102599
Drosophila strains
The available stocks were obtained from the Bloomington Drosophila Stock Center (BDSC, Supplementary Material, Table S1). Transgenic stocks were generated as previously described (62). Briefly, a human ROBO1 cDNA (GenBank: BC171855.1; clone: MHS6278–213246291, clone ID 9054509) was purchased from Horizon. Fly robo1 cDNA was produced by RT-PCR using SuperScript IV First-Strand Synthesis System (Invitrogen, CA, USA) from RNA extracted from adult fly heads (yw). RNA isolation was previously described (28). The cDNA was cloned into the entry vector pDONR223 and expression plasmid pGW-attB-HA (63) using Gateway cloning. Variants were generated in the entry plasmid using site-directed mutagenesis followed by Sanger sequencing. The primers used are listed in Supplementary Material, Table S2. The expression constructs were inserted into the VK33 docking site by φ-C31-mediated transgenesis (64).
The robo1 CRIMIC T2A-miniGAL4 allele was generated as previously described (21). The sgRNA to target the robo1 locus (TTATAATCGGAGACAAAGCTGGG) was cloned in pCFD3 vector as previously described (65). The sequence of homology donor construct is in Supplemental information. It contains 100 nts of homology on either side of the cut site and was commercially synthesized in pUC57-Kan vector by Genewiz (South Plainfield, NJ). The homology donor construct was injected together with pCFD3 vector expressing the sgRNA targeting the locus in embryos expressing Cas9 and transgenic lines (22).
Immunochemistry and image collection
For immunostaining of embryos, eggs were collected, dechorionated in 50% bleach for 3 minutes and fixed in 4% paraformaldehyde. For larval or adult brains, we fixed the tissues in 4% paraformaldehyde for 1 hour and washed them in 0.2% Triton X-100 in PBS. Samples were incubated with antibodies as follows: anti-Robo1 (DSHB#13C9; 1:200), anti-FasII (DSHB#7G10; 1:100; Univercity of Iowa, IA, USA), anti-Elav (DSHB#7E8A10; 1:500), anti-Repo (DSHB#8D12; 1:100), anti-mCherry (Genetex#GTX59788; 1:200; CA, USA), anti-HRP (Jackson ImmunoResearch#2314647; 1:200; PA, USA), anti-GFP (Thermo Fisher#A-21311; 1:200; MA, USA). Fluorescent secondary antibodies were used at 1:500 (Jackson ImmunoResearch). Confocal images were collected with a Leica confocal microscope SP8 and LAS X software. Images were processed by Fiji imageJ (66) and brightness, contrast and color were adjusted by Photoshop CC 2019 (Adobe).
For immunoblots, proteins were extracted by lysis buffer with protease inhibitors (ThermoFisher#88266) from brains of third instar larvae and subjected to SDS-PAGE and immunoblotting. Mouse anti-Robo1 (DSHB #13C9, 1:1000) and mouse anti-α-tubulin (Millipore-Sigma#T6074, 1:20 000; MA, USA) were used in these assays.
Drosophila ERG recording
ERGs (electroretinograms) were performed as described (67). In brief, flies were fixed to a slide with Glue. A recording electrode filled with 150 mM NaCl was placed on the eye, and a ground electrode was placed on the upper torso. A one second pulse of light stimulation was given during the recording, and the ERG traces were recorded and analyzed with LabChart 8 software.
Supplementary Material
Acknowledgements
We thank families for their participation into this study. We thank Hongling Pan for injections to create transgenic flies. We thank Dr Thomas A. Ravenscroft for isolation of RNA from fly heads. We thank the Bloomington Drosophila Stock Center for providing stock and the Developmental Studies Hybridoma Bank for antibodies. We thank Hyung-lok Chuang and Shinya Yamamoto for suggestions and discussions of this project. This work was supported in part by the Baylor College of Medicine IDDRC P50HD103555 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development for use of the Microscopy Core facilities. We acknowledge support from HHMI, the Jan and Dan Duncan Neurological Research Institute and the Huffington Foundation to H.J.B. T.M. is supported by the Uehara Memorial Foundation.
Conflict of Interest statement. J.R.L. has stock ownership in 23andMe, is a paid consultant for the Regeneron Genetics Center and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, genomic disorders and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at the Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics (BG) Laboratories. J.R.L. serves on the Scientific Advisory Board of BG. Other authors have no potential conflicts to report.
Contributor Information
Yan Huang, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA; Jan and Dan Duncan Neurological Research Institute, Texas Children’s Hospital, Baylor College of Medicine, Houston, TX 77030, USA.
Mengqi Ma, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA; Jan and Dan Duncan Neurological Research Institute, Texas Children’s Hospital, Baylor College of Medicine, Houston, TX 77030, USA.
Xiao Mao, National Health Commission Key Laboratory for Birth Defect Research and Prevention, Hunan Provincial Maternal and Child Health Care Hospital, Changsha, Hunan 410008, China; Department of Medical Genetics, Maternal and Child Health Hospital of Hunan Province, Changsha, Hunan 410008, China.
Davut Pehlivan, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA; Division of Neurology and Developmental Neuroscience, Department of Pediatrics, Baylor College of Medicine, Houston, TX 77030, USA; Texas Children’s Hospital, Houston, TX 77030, USA.
Oguz Kanca, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA; Jan and Dan Duncan Neurological Research Institute, Texas Children’s Hospital, Baylor College of Medicine, Houston, TX 77030, USA.
Feride Un-Candan, Department of Neuroloy, Balikesir Ataturk Public Hospital, Balikesir 10100, Turkey.
Li Shu, National Health Commission Key Laboratory for Birth Defect Research and Prevention, Hunan Provincial Maternal and Child Health Care Hospital, Changsha, Hunan 410008, China; Department of Medical Genetics, Maternal and Child Health Hospital of Hunan Province, Changsha, Hunan 410008, China.
Gulsen Akay, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
Tadahiro Mitani, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
Shenzhao Lu, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA; Jan and Dan Duncan Neurological Research Institute, Texas Children’s Hospital, Baylor College of Medicine, Houston, TX 77030, USA.
Sukru Candan, Department of Medical Genetics, Balikesir Ataturk Public Hospital, Balikesir 10100, Turkey.
Hua Wang, National Health Commission Key Laboratory for Birth Defect Research and Prevention, Hunan Provincial Maternal and Child Health Care Hospital, Changsha, Hunan 410008, China; Department of Medical Genetics, Maternal and Child Health Hospital of Hunan Province, Changsha, Hunan 410008, China.
Bo Xiao, Neurology Department, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
James R Lupski, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA; Texas Children’s Hospital, Houston, TX 77030, USA.
Hugo J Bellen, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA; Jan and Dan Duncan Neurological Research Institute, Texas Children’s Hospital, Baylor College of Medicine, Houston, TX 77030, USA.
Funding
The Office of Research Infrastructure Programs of the NIH under the award numbers [R24 OD022005 and R24 OD031447 to H.J.B.]; U.S. National Human Genome Research Institute (NHGRI) and National Heart Lung and Blood Institute (NHBLI) to the Baylor-Hopkins Center for Mendelian Genomics (BHCMG, UM1 HG006542 to J.R.L.); U.S. National Institute of Neurological Disorders and Stroke (NINDS, R35NS105078 to J.R.L.); International Rett Syndrome Foundation (IRSF grant #3701-1 to D.P.); National Natural Science Foundation of China (81801136 to X.M.); Major Scientific and Technological Projects for Collaborative Prevention and Control of Birth Defects in Hunan Province (2019SK1010 and 2019SK1014 to H.W. and B.X.); National Key R&D Program of China (2019YFC1005100 to H.W. and B.X.); Baylor College of Medicine IDDRC P50HD103555 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development for use of the Microscopy Core facilities; HHMI; Jan and Dan Duncan Neurological Research Institute; Huffington Foundation to H.J.B.; Uehara Memorial Foundation to T.M.
References
- 1. Kidd, T., Brose, K., Mitchell, K.J., Fetter, R.D., Tessier-Lavigne, M., Goodman, C.S. and Tear, G. (1998) Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell, 92, 205–215. [DOI] [PubMed] [Google Scholar]
- 2. Seeger, M., Tear, G., Ferres-Marco, D. and Goodman, C.S. (1993) Mutations affecting growth cone guidance in Drosophila: genes necessary for guidance toward or away from the midline. Neuron, 10, 409–426. [DOI] [PubMed] [Google Scholar]
- 3. Bisiak, F. and McCarthy, A.A. (2019) Structure and function of roundabout receptors. Subcell. Biochem., 93, 291–319. [DOI] [PubMed] [Google Scholar]
- 4. Blockus, H. and Chedotal, A. (2016) Slit-Robo signaling. Development, 143, 3037–3044. [DOI] [PubMed] [Google Scholar]
- 5. Rafipay, A., Dun, X.P., Parkinson, D.B., Erskine, L. and Vargesson, N. (2021) Knockdown of slit signaling during limb development leads to a reduction in humerus length. Dev. Dyn., 250, 1340–1357. [DOI] [PubMed] [Google Scholar]
- 6. Münch, J., Engesser, M., Schönauer, R., Hamm, J.A., Hantmann, E., Akay, G., Pehlivan, D.M.T., Akdemir, Z., Tüysüz, B., Shirakawa, T.et al. (2022) Biallelic pathogenic variants in roundabout guidance receptor 1 associate with syndromic congenital anomalies of the kidney and urinary tract. Kidney Int., in press. https://doi.org/10.1016/ j.kint.2022.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kruszka, P., Tanpaiboon, P., Neas, K., Crosby, K., Berger, S.I., Martinez, A.F., Addissie, Y.A., Pongprot, Y., Sittiwangkul, R., Silvilairat, S.et al. (2017) Loss of function in ROBO1 is associated with tetralogy of Fallot and septal defects. J. Med. Genet., 54, 825–829. [DOI] [PubMed] [Google Scholar]
- 8. Dateki, S., Watanabe, S., Mishima, H., Shirakawa, T., Morikawa, M., Kinoshita, E., Yoshiura, K.I. and Moriuchi, H. (2019) A homozygous splice site ROBO1 mutation in a patient with a novel syndrome with combined pituitary hormone deficiency. J. Hum. Genet., 64, 341–346. [DOI] [PubMed] [Google Scholar]
- 9. Calloni, S.F., Cohen, J.S., Meoded, A., Juusola, J., Triulzi, F.M., Huisman, T., Poretti, A. and Fatemi, A. (2017) Compound heterozygous variants in ROBO1 cause a neurodevelopmental disorder with absence of transverse pontine fibers and thinning of the anterior commissure and corpus callosum. Pediatr. Neurol., 70, 70–74. [DOI] [PubMed] [Google Scholar]
- 10. Liu, Z. and Chen, X. (2020) A novel missense mutation in human Receptor Roundabout-1 (ROBO1) gene associated with pituitary stalk interruption syndrome. J. Clin. Res. Pediatr. Endocrinol., 12, 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bashamboo, A., Bignon-Topalovic, J., Moussi, N., McElreavey, K. and Brauner, R. (2017) Mutations in the human ROBO1 gene in pituitary stalk interruption syndrome. J. Clin. Endocrinol. Metab., 102, 2401–2406. [DOI] [PubMed] [Google Scholar]
- 12. Hu, Y., Flockhart, I., Vinayagam, A., Bergwitz, C., Berger, B., Perrimon, N. and Mohr, S.E. (2011) An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinf., 12, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kidd, T., Bland, K.S. and Goodman, C.S. (1999) Slit is the midline repellent for the robo receptor in Drosophila. Cell, 96, 785–794. [DOI] [PubMed] [Google Scholar]
- 14. Furrer, M.P., Kim, S., Wolf, B. and Chiba, A. (2003) Robo and Frazzled/DCC mediate dendritic guidance at the CNS midline. Nat. Neurosci., 6, 223–230. [DOI] [PubMed] [Google Scholar]
- 15. Qian, L., Liu, J. and Bodmer, R. (2005) Slit and Robo control cardiac cell polarity and morphogenesis. Current Biol., 15, 2271–2278. [DOI] [PubMed] [Google Scholar]
- 16. Englund, C., Steneberg, P., Falileeva, L., Xylourgidis, N. and Samakovlis, C. (2002) Attractive and repulsive functions of Slit are mediated by different receptors in the Drosophila trachea. Development, 129, 4941–4951. [DOI] [PubMed] [Google Scholar]
- 17. Wang, J., Al-Ouran, R., Hu, Y., Kim, S.Y., Wan, Y.W., Wangler, M.F., Yamamoto, S., Chao, H.T., Comjean, A., Mohr, S.E.et al. (2017) MARRVEL: integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am. J. Hum. Genet., 100, 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lek, M., Karczewski, K.J., Minikel, E.V., Samocha, K.E., Banks, E., Fennell, T., O'Donnell-Luria, A.H., Ware, J.S., Hill, A.J., Cummings, B.B.et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature, 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. MacDonald, J.R., Ziman, R., Yuen, R.K., Feuk, L. and Scherer, S.W. (2014) The database of genomic variants: a curated collection of structural variation in the human genome. Nucleic Acids Res., 42, D986–D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beaubien, F., Prince, J.E.A. and Cloutier, J.-F. (2013) Axon guidance Slit–Robo signaling. In: Rubenstein, J.L.R and Rakic, R. (eds.), Cellular Migration and Formation of Neuronal Connections: Comprehensive Developmental Neuroscience. Elsevier Inc. USA, Vol. 2, pp. 105–125. [Google Scholar]
- 21. Kanca, O., Zirin, J., Garcia-Marques, J., Knight, S.M., Yang-Zhou, D., Amador, G., Chung, H., Zuo, Z., Ma, L., He, Y.et al. (2019) An efficient CRISPR-based strategy to insert small and large fragments of DNA using short homology arms. Elife, 8, e51539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanca, O., Zirin, J., Hu, Y., Tepe, B., Dutta, D., Lin, W.W., Ma, L., Ge, M., Zuo, Z., Liu, L.P.et al. (2021) An expanded toolkit for Drosophila gene tagging using synthesized homology donor constructs for CRISPR mediated homologous recombination. BioRxiv. 10.1101/2021.12.24.474112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nagarkar-Jaiswal, S., Lee, P.T., Campbell, M.E., Chen, K., Anguiano-Zarate, S., Gutierrez, M.C., Busby, T., Lin, W.W., He, Y., Schulze, K.L.et al. (2015) A library of MiMICs allows tagging of genes and reversible, spatial and temporal knockdown of proteins in Drosophila. Elife, 4, e05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Spitzweck, B., Brankatschk, M. and Dickson, B.J. (2010) Distinct protein domains and expression patterns confer divergent axon guidance functions for Drosophila Robo receptors. Cell, 140, 409–420. [DOI] [PubMed] [Google Scholar]
- 25. Berni, J. (2015) Genetic dissection of a regionally differentiated network for exploratory behavior in Drosophila larvae. Curr. Biol., 25, 1319–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berni, J., Beckwith, E.J., Fernandez, M.P. and Ceriani, M.F. (2008) The axon-guidance roundabout gene alters the pace of the Drosophila circadian clock. Eur. J. Neurosci., 27, 396–407. [DOI] [PubMed] [Google Scholar]
- 27. Apitz, H. and Salecker, I. (2014) A challenge of numbers and diversity: Neurogenesis in the Drosophila optic lobe. J. Neurogenet., 28, 233–249. [DOI] [PubMed] [Google Scholar]
- 28. Ravenscroft, T.A., Janssens, J., Lee, P.T., Tepe, B., Marcogliese, P.C., Makhzami, S., Holmes, T.C., Aerts, S. and Bellen, H.J. (2020) Drosophila voltage-gated sodium channels are only expressed in active neurons and are localized to distal axonal initial segment-like domains. J. Neurosci., 40, 7999–8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li, H., Janssens, J., De Waegeneer, M., Kolluru, S.S., Davie, K., Gardeux, V., Saelens, W., David, F.P.A., Brbic, M., Spanier, K.et al. (2022) Fly cell atlas: a single-nucleus transcriptomic atlas of the adult fruit fly. Science, 375, eabk2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tayler, T.D., Robichaux, M.B. and Garrity, P.A. (2004) Compartmentalization of visual centers in the Drosophila brain requires Slit and Robo proteins. Development, 131, 5935–5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blockus, H., Rolotti, S.V., Szoboszlay, M., Peze-Heidsieck, E., Ming, T., Schroeder, A., Apostolo, N., Vennekens, K.M., Katsamba, P.S., Bahna, F.et al. (2021) Synaptogenic activity of the axon guidance molecule Robo2 underlies hippocampal circuit function. Cell Rep., 37, 109828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dolph, P., Nair, A. and Raghu, P. (2011) Electroretinogram recordings of Drosophila. Cold Spring Harb. Protoc., 2011, pdb prot5549. [DOI] [PubMed] [Google Scholar]
- 33. Kidd, T., Russell, C., Goodman, C.S. and Tear, G. (1998) Dosage-sensitive and complementary functions of roundabout and commissureless control axon crossing of the CNS midline. Neuron, 20, 25–33. [DOI] [PubMed] [Google Scholar]
- 34. Justice, E.D., Barnum, S.J. and Kidd, T. (2017) The WAGR syndrome gene PRRG4 is a functional homologue of the commissureless axon guidance gene. PLoS Genet., 13, e1006865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barak, R., Yom-Tov, G., Guez-Haddad, J., Gasri-Plotnitsky, L., Maimon, R., Cohen-Berkman, M., McCarthy, A.A., Perlson, E., Henis-Korenblit, S., Isupov, M.N.et al. (2019) Structural principles in Robo activation and auto-inhibition. Cell, 177, 272, e216–285. [DOI] [PubMed] [Google Scholar]
- 36. Reichert, M.C., Brown, H.E. and Evans, T.A. (2016) In vivo functional analysis of Drosophila Robo1 immunoglobulin-like domains. Neural Dev., 11, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yom-Tov, G., Barak, R., Matalon, O., Barda-Saad, M., Guez-Haddad, J. and Opatowsky, Y. (2017) Robo Ig4 Is a dimerization domain. J. Mol. Biol., 429, 3606–3616. [DOI] [PubMed] [Google Scholar]
- 38. Chaudhari, K., Gorla, M., Chang, C., Kania, A. and Bashaw, G.J. (2021) Robo recruitment of the Wave Regulatory Complex plays an essential and conserved role in midline repulsion. Elife, 10, e64474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bjorke, B., Shoja-Taheri, F., Kim, M., Robinson, G.E., Fontelonga, T., Kim, K.T., Song, M.R. and Mastick, G.S. (2016) Contralateral migration of oculomotor neurons is regulated by Slit/Robo signaling. Neural Dev., 11, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jen, J.C., Chan, W.M., Bosley, T.M., Wan, J., Carr, J.R., Rub, U., Shattuck, D., Salamon, G., Kudo, L.C., Ou, J.et al. (2004) Mutations in a human ROBO gene disrupt hindbrain axon pathway crossing and morphogenesis. Science, 304, 1509–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chan, W.M., Traboulsi, E.I., Arthur, B., Friedman, N., Andrews, C. and Engle, E.C. (2006) Horizontal gaze palsy with progressive scoliosis can result from compound heterozygous mutations in ROBO3. J. Med. Genet., 43, e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bosley, T.M., Salih, M.A., Jen, J.C., Lin, D.D., Oystreck, D., Abu-Amero, K.K., MacDonald, D.B., Zayed, Z., alDhalaan, H., Kansu, T.et al. (2005) Neurologic features of horizontal gaze palsy and progressive scoliosis with mutations in ROBO3. Neurology, 64, 1196–1203. [DOI] [PubMed] [Google Scholar]
- 43. Hwang, D.Y., Kohl, S., Fan, X., Vivante, A., Chan, S., Dworschak, G.C., Schulz, J., vanEerde, A.M., Hilger, A.C., Gee, H.Y.et al. (2015) Mutations of the SLIT2-ROBO2 pathway genes SLIT2 and SRGAP1 confer risk for congenital anomalies of the kidney and urinary tract. Hum. Genet., 134, 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zu, S., Bartik, Z., Zhao, S., Sillen, U. and Nordenskjold, A. (2009) Mutations in the ROBO2 and SLIT2 genes are rare causes of familial vesico-ureteral reflux. Pediatr. Nephrol., 24, 1501–1508. [DOI] [PubMed] [Google Scholar]
- 45. Lu, W., vanEerde, A.M., Fan, X., Quintero-Rivera, F., Kulkarni, S., Ferguson, H., Kim, H.G., Fan, Y., Xi, Q., Li, Q.G.et al. (2007) Disruption of ROBO2 is associated with urinary tract anomalies and confers risk of vesicoureteral reflux. Am. J. Hum. Genet., 80, 616–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dobson, M.G., Darlow, J.M., Hunziker, M., Green, A.J., Barton, D.E. and Puri, P. (2013) Heterozygous non-synonymous ROBO2 variants are unlikely to be sufficient to cause familial vesicoureteric reflux. Kidney Int., 84, 327–337. [DOI] [PubMed] [Google Scholar]
- 47. Bertoli-Avella, A.M., Conte, M.L., Punzo, F., deGraaf, B.M., Lama, G., La Manna, A., Polito, C., Grassia, C., Nobili, B., Rambaldi, P.F.et al. (2008) ROBO2 gene variants are associated with familial vesicoureteral reflux. J. Am. Soc. Nephrol., 19, 825–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hiesinger, P.R. (2021) Brain wiring with composite instructions. BioEssays, 43, e2000166. [DOI] [PubMed] [Google Scholar]
- 49. Lupski, J.R. (2021) Clan genomics: From OMIM phenotypic traits to genes and biology. Am. J. Med. Genet. A, Genet. A, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Karaca, E., Harel, T., Pehlivan, D., Jhangiani, S.N., Gambin, T., Coban Akdemir, Z., Gonzaga-Jauregui, C., Erdin, S., Bayram, Y., Campbell, I.M.et al. (2015) Genes that affect brain structure and function identified by rare variant analyses of mendelian neurologic disease. Neuron, 88, 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Karaca, E., Posey, J.E., Coban Akdemir, Z., Pehlivan, D., Harel, T., Jhangiani, S.N., Bayram, Y., Song, X., Bahrambeigi, V., Yuregir, O.O.et al. (2018) Phenotypic expansion illuminates multilocus pathogenic variation. Genet. Med., 20, 1528–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jain, P., Sharma, S. and Tripathi, M. (2013) Diagnosis and management of epileptic encephalopathies in children. Epilepsy Res. Treat., 2013, 501981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ulintz, P.J., Wu, W. and Gates, C.M. (2019) Bioinformatics analysis of whole exome sequencing data. Methods Mol. Biol., 1881, 277–318. [DOI] [PubMed] [Google Scholar]
- 54. Karczewski, K.J., Francioli, L.C., Tiao, G., Cummings, B.B., Alfoldi, J., Wang, Q., Collins, R.L., Laricchia, K.M., Ganna, A., Birnbaum, D.P.et al. (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature, 581, 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Genomes Project, C, Auton, A., Brooks, L.D., Durbin, R.M., Garrison, E.P., Kang, H.M., Korbel, J.O., Marchini, J.L., McCarthy, S., McVean, G.A.et al. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kircher, M., Witten, D.M., Jain, P., O'Roak, B.J., Cooper, G.M. and Shendure, J. (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet., 46, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kumar, P., Henikoff, S. and Ng, P.C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc., 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 58. Adzhubei, I.A., Schmidt, S., Peshkin, L., Ramensky, V.E., Gerasimova, A., Bork, P., Kondrashov, A.S. and Sunyaev, S.R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Choi, Y., Sims, G.E., Murphy, S., Miller, J.R. and Chan, A.P. (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS One, 7, e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jagadeesh, K.A., Wenger, A.M., Berger, M.J., Guturu, H., Stenson, P.D., Cooper, D.N., Bernstein, J.A. and Bejerano, G. (2016) M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet., 48, 1581–1586. [DOI] [PubMed] [Google Scholar]
- 61. Schwarz, J.M., Rodelsperger, C., Schuelke, M. and Seelow, D. (2010) MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods, 7, 575–576. [DOI] [PubMed] [Google Scholar]
- 62. Huang, Y., Mao, X., vanJaarsveld, R.H., Shu, L., Terhal, P.A., Jia, Z., Xi, H., Peng, Y., Yan, H., Yuan, S.et al. (2020) Variants in CAPZA2, a member of an F-actin capping complex, cause intellectual disability and developmental delay. Hum. Mol. Genet., 29, 1537–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bischof, J., Bjorklund, M., Furger, E., Schertel, C., Taipale, J. and Basler, K. (2013) A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development, 140, 2434–2442. [DOI] [PubMed] [Google Scholar]
- 64. Venken, K.J., He, Y., Hoskins, R.A. and Bellen, H.J. (2006) P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science, 314, 1747–1751. [DOI] [PubMed] [Google Scholar]
- 65. Port, F., Chen, H.M., Lee, T. and Bullock, S.L. (2014) Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U. S. A., 111, E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B.et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Verstreken, P., Koh, T.W., Schulze, K.L., Zhai, R.G., Hiesinger, P.R., Zhou, Y., Mehta, S.Q., Cao, Y., Roos, J. and Bellen, H.J. (2003) Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron, 40, 733–748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.