Abstract
COVID-19 breakthrough cases among vaccinated individuals demonstrate the value of measuring long-term immunity to SARS-CoV-2 and its variants. We demonstrate that anti-spike T-cell responses and IgG antibody levels are maintained but decrease over time and are lower in BNT162b2- versus mRNA-1273–vaccinated individuals. T-cell responses to the variants are relatively unaffected.
Keywords: SARS-CoV2 mRNA vaccine, T-cell response, T-cell immunity, mRNA-1273, BNT162b2
BNT162b2 (Pfizer-BioNTech) and mRNA-1273 (Moderna) mRNA vaccines are highly protective against severe disease and hospitalization from coronavirus disease 2019 (COVID-19) while also reducing the risk of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection [1]. As time postvaccination lengthens, the vaccines remain protective against severe disease, but breakthrough cases are becoming more frequent—especially in BNT162b2-vaccinated individuals [2]. This could stem from the emergence of SARS-CoV-2 variants of concern (VOCs) that are more infectious and resistant to vaccine-induced immunity and/or general waning of immunity postvaccination. Both vaccines induce strong neutralizing antibody and T-cell responses against SARS-CoV-2 within 2 weeks of full vaccination, which decline over time [3–8]. Vaccine-induced antibodies only partially neutralize certain VOCs (B.1.1.7, B.1.351, P.1, B.1.617.2, and B.1.1.529) compared with the original SARS-CoV-2 virus [5–7, 9–12].
To date, long-term T-cell responses to SARS-CoV-2 and VOCs have mostly been assessed by flow cytometry using previously cryopreserved samples [6, 7, 11, 13] and measuring absolute frequencies at the lowest limits of quantifiable detection. In contrast, ELISpot assays have higher sensitivity and dynamic range at low frequencies and are the clinical standard for quantifying low levels of functional memory T cells (eg, the clinical test for prior exposure to Mycobacterium tuberculosis). Here, we developed an interferon-gamma (IFN-γ) ELISpot assay as a laboratory-developed test and used it to measure T-cell immunity over time following vaccination and in response to SARS-CoV-2 VOCs that were salient at the time.
METHODS
Study Participants
All donors participated voluntarily and were consented according to our approved Partners Institutional Review Board protocol. The vaccination cohort (Supplementary Table 1) included 13 donors (6 male, 7 female) after their first dose (post-V1; 2 BNT162b2, 11 mRNA-1273) with a median age of 35.6 (range: 23.0–61.6) years; 30 donors (15 male, 15 female) following their second dose (post-V2; 14 BNT162b2, 16 mRNA-1273) with a median age of 35.6 (23.0–81.0) years; and 32 donors for long-term follow-up: 15 BNT162b2 (7 male, 8 female) with a median age of 59.0 (21.6–73.3) years and 17 mRNA-1273 (7 male, 10 female) with a median age of 35.3 (23.0–73.0) years. The antibody quantification cohorts were the same, except the post-V2 follow-up containing 30 donors (14 male, 16 female; 13 BNT162b2, 17 mRNA-1273; median 57.5 days after initial dose; range: 38–91 days) with a median age of 35.6 (23.0–81.0) years. No donors in the vaccinated cohorts had confirmed history of COVID-19 or tested positive by polymerase chain reaction (PCR) or rapid antigen test during the study. The healthy donor and convalescent cohorts used to establish the positive cutoff for the ELISpot are described in the Supplementary Methods.
IFN-γ ELISpot Assay
Freshly collected peripheral blood mononuclear cells (PBMCs) were isolated from whole blood (Supplementary Methods), resuspended in serum-free T-cell assay media (ImmunoSpot) at 2.5 × 106/mL, and 100 μL/well was added to a human IFN-γ single-color ELISpot plate (ImmunoSpot). Cells were incubated with 1:1 dimethyl sulfoxide:phosphate-buffered saline (DMSO:PBS; negative control); 2 μg/mL of the spike A, spike B, or nucleocapsid peptide pools (Supplementary Methods); or 5 μg/mL phytohemagglutinin (PHA; positive control; Sigma-Aldrich). Plates were incubated at 37oC for 16–20 hours, developed according to the manufacturer’s instructions, and air dried before counting on an ImmunoSpot CoreS6 ELISpot counter (ImmunoSpot). A laboratory member not involved in the assay set-up performed quality control of the automated count data. The background spot-forming unit (SFU) count was subtracted from the SFU of antigen wells. The average SFU for spike pool A, spike pool B, and the nucleocapsid pool are reported. In some instances, the responses to spike pool A and B were added together to better reflect responses to the entire spike protein. In cases where there were no spot responses to antigen, the PHA well was always strongly positive. The median background in the negative control was 0 SFU/2.5 × 105 cells (range: 0–6). A positive threshold of 6 SFU per 2.5 × 105 PBMCs was determined via receiver operator characteristic (ROC) curve analysis (Supplementary Methods and Supplementary Figure 1E).
Statistical Analysis
Statistical analysis was performed using Prism version 8.0 (GraphPad Software, Inc). Normality of data was evaluated using the Shapiro-Wilk test. The differences between groups were compared using the Mann-Whitney U test.
RESULTS
To establish a positivity threshold for SARS-CoV-2 spike-reactive T cells, we compared responses in prevaccinated healthy donors with no known SARS-CoV-2 exposure (Supplementary Table 2) with COVID-19 convalescent donors (Supplementary Table 3). The PBMCs were stimulated with overlapping SARS-CoV-2 wild-type (WT) spike peptide pools (Supplementary Figure 1A–D) within 8 hours of collection, without freezing the cells, and reactivity was quantified by IFN-γ ELISpot. An average SFU of 6 provided the optimal balance between sensitivity (92.0%) and specificity (90.0%) (Supplementary Figure 1E).
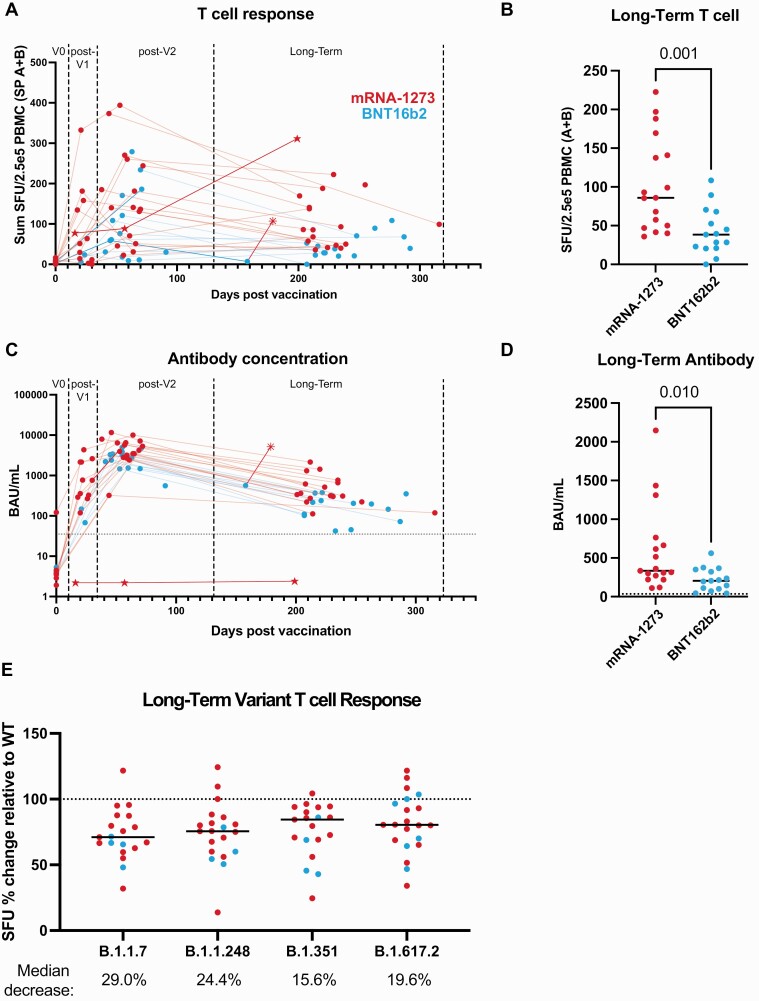
We then measured functional T-cell responses to spike protein in BNT162b2 and mRNA-1273 vaccine recipients (Supplementary Table 1) before vaccination (V0), between the first and second dose (post-V1; median of 22.0 days; mRNA-1273: 21.5 days; BNT162b2: 22.5 days), following the second dose (post-V2; median of 58 days after the first dose; mRNA-1273: 59 days; BNT162b2: 55 days), and at long-term follow-up (median of 223 days after the first dose; mRNA-1273: 220 days; BNT162b2: 230 days). Following vaccination, spike-reactive T cells increased after the first dose and peaked following the second dose (Figure 1A). Fully vaccinated individuals had similar T-cell responses compared with convalescent individuals who had mild symptoms and were at a similar long-term time point after COVID-19 diagnosis (median of 223 days) (Supplementary Figure 1F). The median decrease in T-cell response between post-V2 and long-term follow-up was 35.1% (range: −216.7% to 100% change, with 8 of 24 individuals having higher or the same T-cell response). At long-term follow-up, T-cell responses in BNT162b2-vaccinated individuals were lower than in mRNA-1273-vaccinated individuals (median SFU: 38.5 vs 86.0, respectively) (Figure 1B).
Figure 1.
BNT162b2 (Pfizer-BioNTech) has significantly lower T-cell and antibody responses compared with mRNA-1273 (Moderna) in a long-term follow-up postvaccination. A, T-cell responses from spike peptide pools (sum of the average SFU in response to pool A plus pool B) versus sample collection date in relation to first vaccine dose as evaluated by IFN-γ ELISpot. B, Comparison of long-term follow-up T-cell responses after vaccination with mRNA-1273 and BNT162b2. C, Quantified anti-spike IgG values versus sample collection date in relation to first vaccine dose, determined by ELISA. The dotted line represents the positivity cutoff of 35.2 BAU/mL. D, Comparison of mRNA-1273 and BNT162b2 anti-spike antibody values at long-term follow-up after vaccination. E, ELISpot response to variant peptide pools relative to the individual subject’s WT response at long-term follow-up after vaccination. The dotted line represents response to WT. Statistical comparisons were made using unpaired, 2-tailed Mann-Whitney test. Medians are shown as black lines: mRNA-1273 in red, BNT162b2 in blue; ☆ = rituximab-treated patient; * = received a third vaccination dose. Abbreviations: BAU, binding antibody units; ELISA, enzyme-linked immunosorbent assay; IFN-γ, interferon-gamma; IgG, immunoglobulin G; PBMC, peripheral blood mononuclear cells; SFU, spot-forming units; SP, spike pool; WT, wild-type.
Levels of anti-spike immunoglobulin G (IgG) antibodies in serum were found to be increased post-V2 but decreased by a median of 90.5% (range: 63.0–97.3%) at long-term follow-up (Figure 1C). BNT162b2-vaccinated individuals had lower serum antibody levels over the long term compared with mRNA-1273–vaccinated individuals (Figure 1D). Although there was not a significant difference in age between the vaccine groups (P = .0623), the median age of the BNT162b2 group was higher than that in the mRNA-1273 group; yet, there was no correlation between age and T-cell response or serum antibody levels (Supplementary Figure 2). This may be a feature of our relatively small cohort, since other studies observed decreased antibody titers and/or T-cell responses with increasing age following vaccination [4, 7, 8]. However, another study similarly saw no correlation [5]. One BNT162b2-vaccinated individual had very few SFU at long-term follow-up and received an mRNA-1273 booster, which increased their T-cell response and antibody levels to higher than post-V2. Another individual was receiving rituximab (a B-cell–depleting antibody) treatment at the time of vaccination and failed to develop an antibody response but had a positive T-cell response, which peaked at long-term follow-up (Figure 1A, 1C).
The T-cell response to B.1.1.7 (Alpha), B.1.1.248 (P.1, Gamma), B.1.351 (Beta), and B.1.617.2 (Delta) SARS-CoV-2 VOCs was also measured post-V2 and at long-term follow-up (Supplementary Table 4). The responses were reduced by 29.0%, 24.4%, 15.6%, and 19.6%, respectively at long-term follow-up (Figure 1E). Whether these decreases are enough to have a clinical impact on vaccine effectiveness remains to be determined, but these data demonstrate that T-cell responses to SARS-CoV-2 VOCs are relatively conserved compared with the large decreases in neutralizing antibodies observed against the same variants [5, 6]. The conservation of T-cell response could explain the continued protection against severe COVID-19 in vaccinated individuals.
DISCUSSION
At the time of writing, 99% of COVID-19 cases in the United States were caused by the B.1.617.2 variant. However, the dominant variant has since shifted rapidly with the emergence B.1.1.529 (Omicron). One study indicates that T-cell responses are preserved to the B.1.1.529 variant in most individuals, although a proportion demonstrate a significantly reduced response [14]. Breakthrough cases in vaccinated individuals occur due to a combination of the earlier mentioned factors (increased variant infection rate, waning vaccine immunity, and decreased immune response to variants vs ancestral SARS-CoV-2). Here we demonstrate that humoral and cellular immunity to SARS-CoV-2 are decreased long-term postvaccination with either mRNA vaccine, and appear lower in BNT162b2-vaccinated compared with mRNA-1273–vaccinated individuals, with a greater magnitude of difference in the T-cell response between vaccines compared with antibody levels. Given the relatively small size of our study, it would be valuable to confirm our findings in larger and more age-diverse populations. One group found similar differences at earlier time points postvaccination, with mRNA-1273–vaccinated individuals having higher T-cell responses and anti-spike IgG compared with BNT162b2 [8], although we did not observe a difference between vaccines at early time points. The differences we observed are in contrast to a previous report that used flow cytometry to show similar T-cell responses between the BNT162b2 and mRNA-1273 vaccines 6 and 8 months following vaccination [6]. Our findings could help explain the increased breakthrough cases with BNT162b2 vaccination [2] and highlight the importance of analyzing both the cellular and humoral response to SARS-CoV-2 in populations and in individuals.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. This work was conducted with biostatistical support from Harvard Catalyst. The authors thank Arthur Kim for reviewing the manuscript and offering advice; Zachary J. Manickas Hill for evaluating convalescent patient medical records; Tamina Kienka, Andrea Schmidts, Jane O, and Madeline Polak for phlebotomy; Korneel Grauwet for assistance with SFU counting; Emily Silva and Max Jan for transporting samples, and the Massachusetts General Hospital (MGH) COVID-19 Collection and Processing Team (listed below) for providing convalescent patient samples. The MGH/MassCPR COVID biorepository was supported by a gift from Ms. Enid Schwartz, by the Mark and Lisa Schwartz Foundation, the Massachusetts Consortium for Pathogen Readiness, and the Ragon Institute of MGH, Massachusetts Institute of Technology (MIT), and Harvard. They also thank all the participants for volunteering in the study.
Financial support. This work was supported by the National Cancer Institute at the National Institutes of Health (grant numbers R01CA238268, R01CA252940, R01CA249062 to M. V. M.).
Potential conflicts of interest. M. B. L. privately purchased stock in Moderna and Pfizer (BNT162b2). R. C. L. reports grant support from the National Institutes of Health (NIH; T32 grants) outside the scope of this work and patents related to adoptive cellular therapy with Massachusetts General Hospital. M. V. M. reports grants or contracts from CRISPR Therapeutics, KITE Pharma, Servier, and Novartis; royalties or licenses from Novartis; consulting fees from Adaptimmune, Agenus, Allogene, Arcellx, Astellas, AstraZeneca, Atara, Bayer, BMS, CRISPR therapeutics, EMD Serono, Genocea, Intellia, GSK, Kite Pharma, Micromedicine/BendBio, Neximmune, Novartis, Oncternal, Sanofi, Synthekine, Tmunity, and Werewolf; patents related to adoptive cell therapies, held by Massachusetts General Hospital and University of Pennsylvania (some licensed to Novartis); participation on Scientific Advisory Boards for Cabaletta Bio, Cellectis, In8bio, TCR2, and WindMIL; a leadership or fiduciary role with 2Seventy Bio; and stock or stock options with Century Therapeutics, Genocea, Oncternal, TCR2, and 2Seventy Bio. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
MGH COVID-19 Collection and Processing Team. Collection Team: Kendall Lavin-Parsons,1 Blair Parry,1 Brendan Lilley,1 Carl Lodenstein,1 Brenna McKaig,1 Nicole Charland,1 Hargun Khanna,1 Justin Margolin.1 Processing Team: Anna Gonye,2 Irena Gushterova,2 Tom Lasalle,2 Nihaarika Sharma,2 Brian C. Russo,3 Maricarmen Rojas-Lopez,3 Moshe Sade-Feldman,4 Kasidet Manakongtreecheep,4 Jessica Tantivit,4 Molly Fisher Thomas.4
Massachusetts Consortium on Pathogen Readiness. Betelihem A. Abayneh,5 Patrick Allen,5 Diane Antille,5 Katrina Armstrong,5 Siobhan Boyce,5 Joan Braley,5 Karen Branch,5 Katherine Broderick,5 Julia Carney,5 Andrew Chan,5 Susan Davidson,5 Michael Dougan,5 David Drew,5 Ashley Elliman,5 Keith Flaherty,5 Jeanne Flannery,5 Pamela Forde,5 Elise Gettings,5 Amanda Griffin,5 Sheila Grimmel,5 Kathleen Grinke,5 Kathryn Hall,5 Meg Healy,5 Deborah Henault,5 Grace Holland,5 Chantal Kayitesi,5 Vlasta LaValle,5 Yuting Lu,5 Sarah Luthern,5 Jordan Marchewka (Schneider),5 Brittani Martino,5 Roseann McNamara,5 Christian Nambu,5 Susan Nelson,5 Marjorie Noone,5 Christine Ommerborn,5 Lois Chris Pacheco,5 Nicole Phan,5 Falisha A. Porto,5 Edward Ryan,5 Kathleen Selleck,5 Sue Slaughenhaupt,5 Kimberly Smith Sheppard,5 Elizabeth Suschana,5 Vivine Wilson,5 Galit Alter,6 Alejandro Balazs,6 Julia Bals,6 Max Barbash,6 Yannic Bartsch,6 Julie Boucau,6 Josh Chevalier,6 Fatema Chowdhury,6 Kevin Einkauf,6 Jon Fallon,6 Liz Fedirko,6 Kelsey Finn,6 Pilar Garcia-Broncano,6 Ciputra Hartana,6 Chenyang Jiang,6 Paulina Kaplonek,6 Marshall Karpell,6 Evan C. Lam,6 Kristina Lefteri,6 Xiaodong Lian,6 Mathias Lichterfeld,6 Daniel Lingwood,6 Hang Liu,6 Jinqing Liu,6 Natasha Ly,6 Ashlin Michell,6 Ilan Millstrom,6 Noah Miranda,6 Claire O’Callaghan,6 Matthew Osborn,6 Shiv Pillai,6 Yelizaveta Rassadkina,6 Alexandra Reissis,6 Francis Ruzicka,6 Kyra Seiger,6 Libera Sessa,6 Christianne Sharr,6 Sally Shin,6 Nishant Singh,6 Weiwei Sun,6 Xiaoming Sun,6 Hannah Ticheli,6 Alicja Trocha-Piechocka,6 Daniel Worrall,6 Alex Zhu,6 George Daley,7 David Golan,7 Howard Heller,7 Arlene Sharpe,7 Nikolaus Jilg,8 Alex Rosenthal,8 Colline Wong.8
1Department of Emergency Medicine, Massachusetts General Hospital, Boston, Massachusetts, USA; 2Massachusetts General Hospital Cancer Center, Boston, Massachusetts, USA; 3Division of Infectious Diseases, Department of Medicine, Massachusetts General Hospital, Boston, Massachusetts, USA; 4Massachusetts General Hospital Center for Immunology and Inflammatory Diseases, Boston, Massachusetts, USA; 5Massachusetts General Hospital, Boston, Massachusetts, USA; 6Ragon Institute of Massachusetts General Hospital, MIT, and Harvard, Cambridge, Massachusetts, USA; 7Harvard Medical School, Boston, Massachusetts, USA; and 8Brigham and Women’s Hospital, Boston, Massachusetts, USA.
Contributor Information
Kathleen M E Gallagher, Immune Monitoring Laboratory, Cancer Center, Massachusetts General Hospital, Boston, Massachusetts, USA; Cellular Immunotherapy Program, Cancer Center, Massachusetts General Hospital, Boston, Massachusetts, USA; Department of Pathology, Harvard Medical School, Boston, Massachusetts, USAand.
Mark B Leick, Cellular Immunotherapy Program, Cancer Center, Massachusetts General Hospital, Boston, Massachusetts, USA; Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA.
Rebecca C Larson, Cellular Immunotherapy Program, Cancer Center, Massachusetts General Hospital, Boston, Massachusetts, USA; Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA.
Trisha R Berger, Cellular Immunotherapy Program, Cancer Center, Massachusetts General Hospital, Boston, Massachusetts, USA.
Katelin Katsis, Immune Monitoring Laboratory, Cancer Center, Massachusetts General Hospital, Boston, Massachusetts, USA; Cellular Immunotherapy Program, Cancer Center, Massachusetts General Hospital, Boston, Massachusetts, USA.
Jennifer Y Yam, Immune Monitoring Laboratory, Cancer Center, Massachusetts General Hospital, Boston, Massachusetts, USA; Cellular Immunotherapy Program, Cancer Center, Massachusetts General Hospital, Boston, Massachusetts, USA.
Marcela V Maus, Cellular Immunotherapy Program, Cancer Center, Massachusetts General Hospital, Boston, Massachusetts, USA; Department of Medicine, Harvard Medical School, Boston, Massachusetts, USA.
References
- 1. Thompson MG, Burgess JL, Naleway AL, et al. Prevention and attenuation of Covid-19 with the BNT162b2 and mRNA-1273 vaccines. N Engl J Med 2021; 385:320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Center for Disease Control and Prevention (CDC). Rates of COVID-19 cases and deaths by vaccination status. Available at: https://covid.cdc.gov/covid-data-tracker/#rates-by-vaccine-status. Accessed 8 November 2021.
- 3. Woldemeskel BA, Garliss CC, Blankson JN.. mRNA vaccine-elicited SARS-CoV-2-specific T cells persist at 6 months and recognize the delta variant. Clin Infect Dis 2021: doi: 10.1093/cid/ciab915. [DOI] [PubMed] [Google Scholar]
- 4. Guerrera G, Picozza M, D’Orso S, et al. BNT162b2 vaccination induces durable SARS-CoV-2 specific T cells with a stem cell memory phenotype. Sci Immunol 2021: eabl5344. [DOI] [PubMed] [Google Scholar]
- 5. Garcia-Beltran WF, Lam EC, St Denis K, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell 2021; 184: 2372–83, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Collier AY, Yu J, McMahan K, et al. Differential kinetics of immune responses elicited by Covid-19 vaccines. N Engl J Med 2021; 385:2010–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goel RR, Painter MM, Apostolidis SA, et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science 2021; 374:abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Markewitz R, Pauli D, Dargvainiene J, et al. The temporal course of T- and B-cell responses to vaccination with BNT162b2 and mRNA-1273. Clin Microbiol Infect 2021; doi: 10.1016/j.cmi.2021.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi A, Koch M, Wu K, et al. Serum neutralizing activity of mRNA-1273 against SARS-CoV-2 variants. J Virol 2021; 95: e0131321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoffmann M, Arora P, Gross R, et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021; 184:2384–93 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Geers D, Shamier MC, Bogers S, et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci Immunol 2021; 6:eabj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garcia-Beltran WF, St Denis KJ, Hoelzemer A, et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 2022; 185:457–466.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tarke A, Sidney J, Methot N, et al. Impact of SARS-CoV-2 variants on the total CD4(+) and CD8(+) T cell reactivity in infected or vaccinated individuals. Cell Rep Med 2021; 2:100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Naranbhai V, Nathan A, Kaseke C, et al. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all prior infected and vaccinated individuals. medRxiv; doi: 10.1101/2022.01.04.2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.