Abstract
The morphological plasticity of Candida albicans is an important determinant of pathogenicity, and nonfilamentous mutants are avirulent. HWP1, a hypha-specific gene, was identified in a genetic screen for developmentally regulated genes and encodes a cell surface protein of unknown function. Heterozygous and homozygous deletions of HWP1 resulted in a medium-conditional defect in hyphal development. HWP1 expression was blocked in a Δefg1 mutant, reduced in an Δrbf1 mutant, and derepressed in a Δtup1 mutant. Therefore, HWP1 functions downstream of the developmental regulators EFG1, TUP1, and RBF1. Mutation of CPH1 had no effect on HWP1 expression, suggesting that the positive regulators of hyphal development, CPH1 and EFG1, are components of separate pathways with different target genes. The expression of a second developmentally regulated gene, ECE1, was similarly regulated by EFG1. Since ECE1 is not required for hyphal development, the regulatory role of EFG1 apparently extends beyond the control of cell shape determinants. However, expression of ECE1 was not influenced by TUP1, suggesting that there may be some specificity in the regulation of morphogenic elements during hyphal development.
Candida albicans is a significant opportunistic fungal pathogen and causes superficial mucosal disease as well as life-threatening systemic infections in immunocompromised patients. C. albicans is described as a dimorphic fungus that can grow as a budding yeast or a true hyphal form but can also adopt a range of intermediate pseudohyphal morphologies. C. albicans strains defective in the ability to form hyphae are avirulent in a mouse model of systemic infection (22). Thus, hyphal development is essential to pathogenicity, but its precise role in this process is as yet undefined.
Hyphal development is under both positive and negative control. TUP1 is a general transcriptional repressor in Saccharomyces cerevisiae (40). Deletion of the C. albicans homolog of TUP1 results in a constitutive pseudohyphal phenotype under all growth conditions, suggesting that TUP1 is responsible for maintenance of the yeast morphology through repression of genes required for filamentous growth (6). RBF1 (for “RPG binding factor 1”) encodes a putative transcription factor of C. albicans that binds to the same consensus sequence as the S. cerevisiae transcription factor encoded by RAP1 (14). Mutation of RBF1 results in a stimulation of filamentous growth, suggesting that RBF1 also plays a negative regulatory role in the yeast-to-hyphal-form transition (15).
Positive control of hyphal development is effected in part by EFG1, which encodes a basic helix-loop-helix-type transcription factor (36). Reduced expression or deletion of EFG1 results in rod-like, elongated cells under the conditions tested and in the inability to form true hyphae (22, 36). Overexpression results in enhanced pseudohyphal growth (36). Additional positive control is signaled via a mitogen-activated protein (MAP) kinase cascade analogous to that which controls pseudohyphal development in S. cerevisiae (21, 28). Mutation in the kinase components encoded by HST7, CST20, or CEK1, or the terminal transcription factor encoded by CPH1, results in a medium-conditional defect in hyphal development (8, 17, 18, 20). Conversely, mutation of the MAP kinase phosphatase CPP1 results in a hyperhyphal phenotype (7). This kinase cascade cooperatively controls hyphal development in conjunction with EFG1. Mutation of either EFG1 or CPH1 alone partially compromises filamentation, while a cph1/cph1 efg1/efg1 double mutant is restricted entirely to the yeast growth form under standard growth conditions (22).
The molecular interactions between these various regulators leads to the coordinate control of hyphal development, but these interactions remain incompletely defined since no downstream target genes have been identified. Three developmentally expressed genes, ECE1, HYR1, and HWP1, have been reported, but neither ECE1 nor HYR1 is required for hyphal morphogenesis (2, 5, 35). Here we report the morphogenic role of HWP1. Expression of HWP1 is dependent upon EFG1 but does not require CPH1. TUP1 represses its expression, while RBF1 appears to act as an inducer. Examination of ECE1 expression demonstrated that this nonmorphogenic function is similarly regulated by EFG1 but is not affected by TUP1. These results suggest that EFG1 and CPH1 function within largely independent control pathways and regulate distinct sets of morphology-related functions. In addition, the regulatory function of EFG1 extends beyond the control of cell shape.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this study are listed in Table 1. They were routinely cultured on YPD medium (33) or YNB medium (2% glucose, 0.17% Difco yeast nitrogen base without amino acids or ammonium sulfate, 0.5% ammonium sulfate) at 30°C. Medium 199 (Gibco BRL, Gaithersburg, Md.) containing Earle’s salts and glutamine but lacking sodium bicarbonate was buffered with 150 mM Tris (pH 7). Spider medium was prepared as described previously (20). The medium of Lee et al. was prepared as described previously (19). N-Acetylglucosamine induction medium was prepared as described by Shepherd et al. (32). Media were solidified with 1.5% agar and supplemented with 25 μg of uridine per ml for growth of Urd3− strains. Germ tube induction was assessed at 37°C following inoculation of stationary-phase cells into prewarmed medium at a density of 5 × 106 cells/ml. Negative controls were incubated at 25°C. Filamentation on agar-solidified media was assessed by diluting stationary-phase cells to 2 × 108 cells/ml in water, spotting 1 × 106 cells (5 μl) onto the plate, and incubating them at 37°C. Invasive hyphal growth was assessed after washing the agar plates with sterile water to remove surface growth (28).
TABLE 1.
C. albicans strains used in this study
| Strain | Parent | Genotype | Source or reference |
|---|---|---|---|
| SGY243 | Δura3::ADE2/Δura3::ADE2 | 16 | |
| SC5314 | Clinical isolate | 12 | |
| CAI4 | CAF2-1 | Δura3::imm434/Δura3::imm434 | 10 |
| CAL1 | CAI4 | Δhwp1::hisG-URA3-hisG/HWP1 Δura3::imm434/Δura3::imm434 | This work |
| CAL2 | CAL1 | Δhwp1::hisG/HWP1 Δura3::imm434/Δura3::imm434 | This work |
| CAL3 | CAL2 | Δhwp1::hisG/Δhwp1::hisG-URA3-hisG Δura3::imm434/Δura3::imm434 | This work |
| CAL4 | CAL3 | Δhwp1::hisG/Δhwp1::hisG Δura3::imm434/Δura3::imm434 | This work |
| CAL5 | CAL4 | Δhwp1::hisG/Δhwp1::hisG HWP1::URA3 Δura3::imm434/Δura3::imm434 | This work |
| CAL6 | CAL4 | Δhwp1::hisG/Δhwp1::hisG EF1α::HWP1::URA3 Δura3::imm434/Δura3::imm434 | This work |
| BCa2-10 | Δtup1::hisG/Δtup1::hisG-URA3-hisG | 6 | |
| JKC19 | Δcph1::hisG/Δcph1::hisG Δura3::imm434/Δura3::imm434 | 22 | |
| HLC52 | Δefg1::hisG/Δefg1::hisG Δura3::imm434/Δura3::imm434 | 22 | |
| HLC54 | Δcph1::hisG/Δcph1::hisG Δefg1::hisG/Δefg1::hisG Δura3::imm434/Δura3::imm434 | 22 | |
| 1161KR | LYS/lys1 ARG4/arg4 ura3/ura3 ser57/ser57 gal1/gal1 MPAr | 14 | |
| 1161K1R13 | lys1/lys1 arg4/arg4 ura3/ura3 ser57/ser57 gal1/gal1 rbf1::LYS1/rbf1::ARG4 MPAr | 14 |
Gene isolation.
cDNA clones of hypha-expressed genes and their corresponding genomic clones were isolated as previously described (5). A 4.3-kb EcoRI genomic fragment corresponding to cDNA 8 was subcloned from plasmid pSMS22 into pUC18. The gene was tentatively called ECE2.
DNA sequence analysis.
Nucleotide sequences were determined by the dideoxy chain termination method with Sequenase 2.0 T7 DNA polymerase (United States Biochemical, Cleveland, Ohio). Reactions were primed with the T3 and T7 primers and custom-designed oligonucleotide primers. Sequence analysis was performed with DNA Strider (23). Homology searches were conducted with the BLAST algorithm (1) and SCAN (26). Sequence alignments were performed with MACAW (31) and ClustalW 1.6 (13).
Construction of mutant strains.
The insert from plasmid pSMS22 was subcloned into the EcoRI site of a pBSK+ (Stratagene, La Jolla, Calif.) derivative in which the BamHI site had been destroyed by filling in with Klenow DNA polymerase. The resulting plasmid, pELS-1, contained a unique BamHI site within HWP1. A 434-bp BglII-BamHI fragment in the 5′ coding region was deleted and replaced with the 3.8-kb HindIII-BglII hisG-URA3-hisG fragment from plasmid pMB7 (10), generating plasmid pELS-2. The BamHI and HindIII ends were blunt-end ligated following a filling-in reaction. pELS-2 was digested with SmaI and HindIII, releasing a 5.4-kb fragment containing the hisG-URA3-hisG fragment flanked by HWP1 sequences. This DNA was used for the sequential disruption of both HWP1 alleles in strain CAI4 by previously published methods (10).
To reintroduce a functional copy of HWP1, plasmid pELS-1 was digested with PshAI and XhoI, filled in, and ligated to create plasmid pELS-5. This removed a 2.0-kb fragment of 3′-flanking sequence containing two HindIII restriction sites. A 1.4-kb XbaI-PstI fragment containing URA3 (30) was added to create pELS-6. pELS-6 was linearized at the unique HindIII site located 3′ of the BamHI site within HWP1 to target integration to the HWP1 locus.
Constitutive expression of HWP1 was effected with the TEF2 promoter (37). A ClaI site was introduced 5 bp upstream of the HWP1 open reading frame in plasmid pELS-5 by QuikChange mutagenesis (Stratagene). The HWP1 promoter was removed by EcoRI-ClaI digestion and replaced with the 0.75-kb EcoRI-ClaI fragment of the TEF2 promoter from plasmid pEF1-Fow (30). The XbaI-PstI fragment of URA3 was added, resulting in plasmid pELS-10. Plasmid DNA was digested with BspEI prior to transformation to target integration to the HWP1 locus. The integration events in all the transformations were verified by Southern blot analysis.
Southern and Northern blot analyses.
Southern and Northern blotting were conducted as described previously (25). Blots were hybridized with the HWP1 4.3-kb EcoRI insert from plasmid pSMS22, the 1.6-kb BamHI-EcoRV ECE1 fragment from plasmid pCAN4, or ACT1 DNA as a control. Northern blots were quantitated with a Molecular Dynamics 445SI PhosphorImager and associated software.
Nucleotide sequence accession number.
The sequence of ECE2 was entered into GenBank under accession no. AF001978.
RESULTS
Isolation and identification of HWP1.
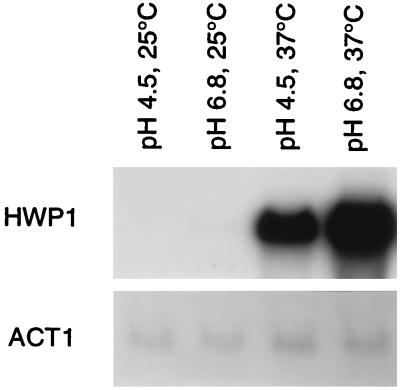
Hyphal development is presumably effected by differential expression of morphogenic functions in response to external signals. Previously, we isolated several cDNA clones of genes differentially expressed during the yeast-to-hyphal-form transition (5). One of these clones, cDNA 8, hybridized to a 2.4-kb mRNA expressed only in pseudohyphae and hyphae (Fig. 1). The expression pattern was similar to that previously described for ECE1 (5); thus, the gene was tentatively designated ECE2. Expression was independent of the medium used to induce filamentation, and the expression pattern in the medium-conditional strain SGY243 (16) demonstrated that expression was morphology specific rather than being a response to the inducing environment.
FIG. 1.
Differential expression of cDNA 8. RNA was isolated from strain SC5314 grown in medium 199 at the indicated pH and temperature. A Northern blot of the sample was hybridized with cDNA 8 (HWP1) or ACT1.
Sequence analysis of the genomic clone identified an open reading frame of 1,902 bp encoding a 634-residue protein with four distinct repeat domains (Fig. 2). The amino terminus contained a potential 27-residue secretory leader sequence (27), and a second hydrophobic region was located at the carboxy terminus. The latter region was preceded by residues G613, A614, and G615, which forms a potential glycosylphosphatidylinositol (GPI) attachment site according to the ω, ω+1, and ω+2 rule (39). The protein also contained three consensus sites for N-linked glycosylation (3).
FIG. 2.
Predicted amino acid sequence encoded by HWP1. Identical residues are boxed. Dashes indicate gaps introduced to maximize the alignment. The line above the sequence indicates the predicted leader. The arrow indicates the potential cleavage site. The broken line indicates the C-terminal hydrophobic region. Potential GPI anchor sites are marked with stars.
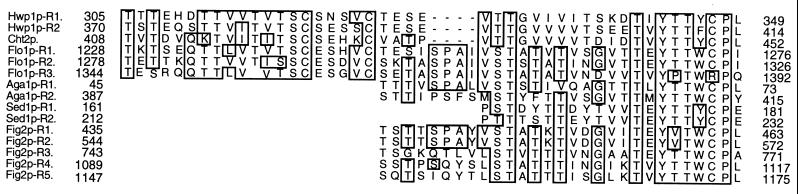
A BLAST (1) comparison revealed that ECE2 was >99% identical to the independently isolated and characterized HWP1 (for “hyphal wall protein”) (35). No obvious homologs were identified. However, a region of the third repeat domain of Hwp1p was conserved in CHT2, encoding a C. albicans chitinase (24), the S. cerevisiae flocculation protein Flo1p (4), the α-agglutinin subunit encoded by AGA1 (29), the pheromone-regulated protein Fig2p (9), and the SED1-encoded cell wall protein (11) (Fig. 3). All of these proteins are either known or predicted to be GPI-anchored cell surface proteins. This conserved domain was characterized by the consensus motif YTTWCPL, present in one to five copies.
FIG. 3.
Alignment of sequences with the conserved WCPL motif. Numbers to the left and right indicate the location within the amino acid sequence. R1, R2, etc., indicate consecutive repeats within the same protein. Residues conserved in at least 60% of the sequences within the region of overlap are boxed. The proteins and gene accession numbers are as follows: Cht2p, chitinase 2, U15800; Flo1p, flocculation protein, X78160; Aga1p, a-agglutinin core subunit, M60590; Sed1p, suppressor of erd2, X66838; Fig2p, factor induced gene, YCR089W.
Construction of deletion mutants.
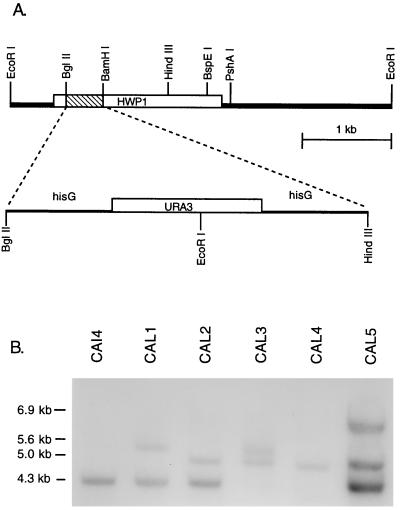
Expression of HWP1 correlated with cell elongation and filamentation, and this suggested a potential role in morphogenesis. Deletion mutants were constructed to test this possibility. A 434-bp segment of the HWP1 open reading frame was replaced with the hisG-URA3-hisG cassette (10) in the Urd− strain CAI4, as illustrated in Fig. 4A. Integration at the HWP1 locus occurred in 22 of 23 Urd+ transformants. The wild-type allele yields a 4.3-kb EcoRI band in Southern blots (Fig. 4B, lane CAI4). The disrupted allele contains an EcoRI restriction site located in URA3 and yields a hybridization band of 5.6 kb, as seen in the representative heterozygous mutant CAL1 (Fig. 4B). Loss of the URA3 gene and one of the hisG repeats in the Urd− segregant strain CAL2 generated a 5.0-kb band (Fig. 4B). Transformation of CAL2 resulted in replacement of the remaining wild-type allele in 8 of 36 transformants to generate strain CAL3. In these transformants, the 4.3-kb band of the wild-type allele was replaced by a 5.6-kb band representing the newly disrupted allele (Fig. 4B). Northern analysis confirmed the absence of HWP1 mRNA in the null mutant.
FIG. 4.
Construction of HWP1 null mutants. (A) Schematic representation of the HWP1 disruption scheme. (B) Southern blot analysis of representative HWP1 disrupted strains. Genomic DNA was digested with EcoRI and hybridized with labeled HWP1 DNA.
Phenotype of the mutants.
HWP1 was nonessential as evidenced by the viability of the null mutant. No differences in the rate or frequency of germ tube formation were observed when the heterozygous or homozygous mutants were inoculated into Spider medium, the medium of Lee et al., 10% serum, or 25 mM N-acetylglucosamine. In medium 199, the mutant exhibited normal germ tube induction, but in approximately half the trials the germ tubes failed to extend into true hyphae and instead developed a pseudohyphal morphology. We were unable to identify the source of this variability. No differences were noted in the heterozygote.
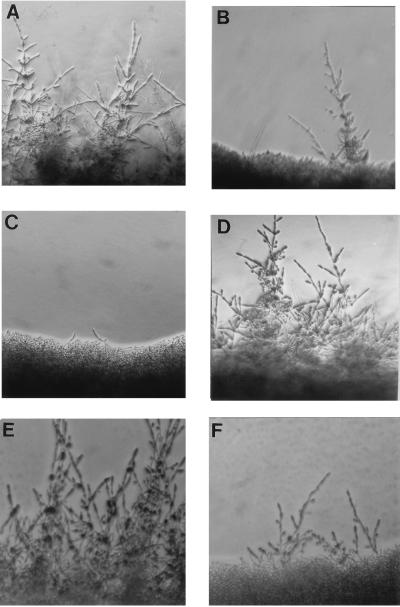
The mutations had a more significant consequence for hyphal development on solid media. Strain SC5314 showed prolific hyphal growth, with lateral extension of the hyphae beyond the border of the colony (Fig. 5A). The heterozygous mutant, CAL1, exhibited greatly reduced hyphal growth, (Fig. 5B), and the null mutant, CAL3, exhibited a nearly complete loss of hyphal growth (Fig. 5C). The same results were seen on solid Spider medium and the medium of Lee et al. for seven independent heterozygotes and eight null mutants. Although the lateral extension of hyphae was compromised, all mutants maintained the ability to invade the agar directly beneath the colony. Microscopic examination revealed that these invasive filaments had a hyphal morphology.
FIG. 5.
Effect of HWP1 mutation on hyphal formation. Strains were cultured for 48 h on medium 199 (pH 7) at 37°C (A to D) or for 24 h on 10% serum (E and F). The strains used are SC5314 (A and E), CAL1 (B), CAL3 (C and F), and CAL5 (D). Magnification, ×10.
When plated on 10% serum, a strong inducer of hyphal formation, all strains produced peripheral hyphae within 24 h. However, the hyphae extending from the null mutant colony were significantly reduced in both number and length compared to those in the wild type (Fig. 5E and F). By 48 h, the null mutant exhibited extensive filamentation around the colony, although the hyphae extended only half the distance from the colony as did those of the wild type. This is in contrast to incubation on medium 199, where the mutant failed to develop hyphae even with extended incubation.
To confirm that the mutant phenotype was directly associated with the loss of HWP1, the wild-type allele was introduced into the Urd− homozygous null mutant CAL4 (Fig. 4B) to produce CAL5. Southern blot analyses confirmed proper integration of the functional allele in 8 of 15 transformants (Fig. 4B). Introduction of HWP1 restored hyphal development in all eight of the revertants (Fig. 5D), verifying the correspondence between phenotype and genotype. However, the revertants, which contained only one functional allele, did not exhibit the partial defects evident in the heterozygous parent of the null mutant. Instead, they resembled the homozygous wild type. This phenotype was evident in several other independently constructed revertants and may indicate that the fragment of the promoter driving expression of the reintroduced allele is inadequate for complete regulation.
To test whether HWP1 was simply necessary or both necessary and sufficient for hypha production, the HWP1 promoter was replaced with the constitutive promoter from TEF2 (37) and integrated into the null mutant strain CAL4. Constitutive expression of HWP1 was confirmed by Northern analysis. Constitutive expression restored the ability of the mutant to form hyphae on medium 199 plates. However, when cells were incubated under conditions that repress filamentation (medium 199 plates at pH 4.5 and 25°C), no hyphae were formed. Thus, HWP1 is insufficient to initiate or promote hyphal development.
Regulation of HWP1 and ECE1.
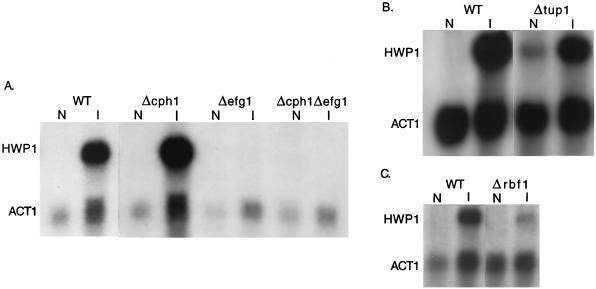
Because of the developmental expression pattern of HWP1 and its involvement in filamentation, it was potentially under the control of one or more of the known regulators of this process. Their influence on the expression of HWP1 was assessed by examining HWP1 induction in the corresponding null mutants. Induction of HWP1 was completely blocked by deletion of EFG1, either alone or in combination with CPH1 (Fig. 6A). Deletion of CPH1 alone had no effect. Thus, HWP1 expression is under the positive regulatory control of EFG1 but is independent of the MAP kinase cascade.
FIG. 6.
Effect of regulatory mutations on HWP1 expression. RNA was prepared from cells grown under noninducing conditions (N) or hypha-inducing conditions (I), and the blots were probed with HWP1 and ACT1. The relevant genotypes are indicated above the lanes. (A and B) SC5314 was the wild-type strain (WT). (C) The parental strain of the Δrbf1 mutant, 1161KR, was used as the control.
The EFG1 dependence of HWP1 expression suggested the possibility that the defective hyphal development of Δefg1 mutants was due to the absence of HWP1p. To determine if forced expression of HWP1 could suppress the Δefg1 mutant phenotype, HWP1, under the control of the constitutive promoter from the TEF2 gene, was transformed into the Δefg1 background. No changes in the phenotype were observed.
The repressors encoded by both TUP1 (6) and RBF1 (15) modulated HWP1 expression. In a TUP1 null mutant, HWP1 was expressed even under noninducing conditions (Fig. 6B). However, expression was lower than the induced level. Alternatively, deletion of RBF1 resulted in a 70% reduction of HWP1 expression under inducing conditions (Fig. 6C).
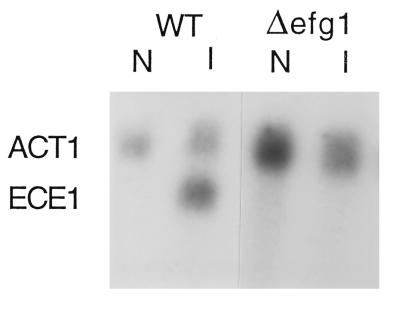
ECE1 exhibits a developmental expression pattern similar to HWP1, but Δece1 mutants are not affected in filamentation ability. However, as with HWP1, expression of ECE1 was abrogated in an EFG1 null mutant (Fig. 7) but occurred normally in a Δcph1 background. TUP1 also had no effect. In contrast, ECE1 was not expressed in the 1161KR strain background in the presence or absence of the RBF1 null mutation.
FIG. 7.
Effect of regulatory mutations on ECE1 expression. RNA was prepared from cells grown under noninducing conditions (N) or hypha-inducing conditions (I), and the blots were probed with ECE1 and ACT1. The relevant genotypes are indicated above the lanes. SC5314 was the wild-type strain (WT).
DISCUSSION
HWP1 was identified in a genetic screen for developmentally regulated genes. This same gene was previously identified in an immunological screen for hypha-specific proteins (35). Deletion analysis revealed that HWP1 is conditionally required for hyphal formation. The ability to form hyphae on solid media was severely reduced in the HWP1 heterozygous mutant and essentially eliminated in the null mutant. In the presence of serum, colonies of the null mutant were able to produce peripheral hyphae, but at reduced levels compared to the wild type. Reintroduction of a functional allele of HWP1 resulted in a return to the wild-type phenotype, confirming that the loss of HWP1 expression was responsible for the defects in hyphal development in the mutant strains. All mutants maintained the ability to invade the agar directly beneath the colony and to form germ tubes in liquid suspension cultures.
Presently, no specific role in hyphal development can be assigned to HWP1. No homologs or functional motifs were identified. HWP1 encodes a putative GPI-linked surface protein and has been localized to the cell surface by immunofluorescence labeling (35). Staab et al. reported that Hwp1p can serve as a substrate for mammalian transglutaminase and can mediate the stable attachment of C. albicans hyphae to human buccal epithelial cells (34). They also reported that HWP1 is required for virulence in a mouse model of systemic infection, a result corroborated by Tsuchimori et al. (34, 38).
Hwp1p may belong to a unique subset of GPI-anchored proteins characterized by the presence of a conserved structural motif, YTTWCPL. The diverse functions of the proteins that contain this motif suggest that it imparts a general property, e.g., interaction with specific surface proteins or wall polysaccharides. The surface localization of Hwp1p is compatible with diverse functions, from cell wall assembly to cell signaling. However, Hwp1p is unlikely to participate in the reception of the initial developmental signal since it is not expressed in the yeast form and is downstream of the developmental regulator EFG1. It is also unlikely to function directly in the formation of the filamentous wall structure. The heterozygous null mutant exhibited a reduction in the frequency of hyphal elements but no alteration in their morphology. Similarly, hyphae formed by the null mutant in the presence of serum, while shorter, appeared otherwise normal. Thus, Hwp1p may be required for hyphal development to proceed or be sustained once the signal has been received. Notably, there are no apparent homologs in the S. cerevisiae genome, and so HWP1 may be one of the developmental components that allows true filamentation in C. albicans versus the pseudohyphal morphology seen in baker’s yeast. Whatever its function, the conditional requirement for HWP1 on solid medium indicates that the function is either unnecessary or redundant in liquid culture.
HWP1 is the first morphogenic target identified that is downstream of the signaling components and regulators of hyphal development. Examining the influence of these regulators on HWP1 expression has elucidated several facets of hyphal development. Previous studies had demonstrated that mutation of either CPH1 or EFG1 alone resulted in partial suppression of development but that deletion of both genes caused complete suppression (22). The simplest interpretation of these results is that CPH1 and EFG1 provide additive inputs in regulating a set of genes required for filamentation. However, expression of HWP1 was entirely EFG1 dependent and was not influenced by CPH1. This is consistent with the phenotype of the HWP1 null mutant, which is compromised in hyphal development on several media on which the Δcph1 mutant is not compromised. These results indicate that CPH1 and EFG1 are components of separate pathways with different target genes. The Δhwp1 and Δefg1 mutants differ in that the Δhwp1 mutant is able to form true hyphae in the presence of serum while the Δefg1 mutant is not. This difference and the inability of constitutive expression of HWP1 to suppress the Δefg1 mutation indicate that EFG1 regulates additional genes required for hyphal development.
HWP1 expression is also influenced by TUP1 and RBF1, both negative regulators of hyphal development (6, 15). HWP1 was partially derepressed under noninducing conditions in a TUP1 null mutant. The lack of complete derepression indicates that expression of HWP1 is under multiple controls and probably requires an additional positive signal(s). This may be supplied via the EFG1-dependent pathway. The data do not allow us to distinguish whether TUP1 acts through EFG1 or independently. However, it is clear from the results that TUP1 acts independently of CPH1. Previous work had shown that TUP1 mutations are epistatic to a Δcph1 mutation. From this, it could be inferred that TUP1 functions downstream of CPH1, assuming that they act within the same pathway. Since TUP1 influences HWP1 expression but CPH1 does not, CPH1 cannot be acting upstream of TUP1. Unexpectedly, deletion of RBF1 resulted in reduced expression of HWP1 under inducing conditions, indicating that RBF1 is a positive regulator of HWP1.
HWP1 is distinguished from other developmentally regulated genes of C. albicans by its role in filamentation. ECE1 and HYR1 exhibit a similar patterns of expression, but null mutations in these genes do not affect morphogenesis (2, 5). This distinction made it possible to determine whether the similar developmental expression patterns of morphogenic and nonmorphogenic genes reflect a common regulation. This question is relevant to the role of hyphal development in pathogenesis. As demonstrated by Lo et al. (22), nonfilamentous mutants are avirulent. However, it is not known if this reflects the inability of the organism to adopt the filamentous morphology and/or the effect of the mutations on coregulated virulence determinants. Comparison of HWP1 and ECE1 expression demonstrated common and unique regulatory interactions. Expression of both genes was similarly affected by mutation of EFG1 and unaffected by CPH1. However, ECE1 was not affected by mutation of TUP1 and was not induced in strain 1161KR. The lack of ECE1 expression in 1161KR is probably a consequence of the highly mutagenized genetic background of this strain (15). Nonetheless, it demonstrates that there are distinct regulatory features in the developmental expression of HWP1 and ECE1. Although it is not known if ECE1 encodes a virulence attribute, the data demonstrate that the regulatory role of EFG1 extends beyond the control of cell shape determinants. Furthermore, the observed differences in HWP1 and ECE1 regulation offers the possibility that expression of morphogenic and nonmorphogenic functions can be dissociated and their relative contributions to the virulence process can be independently assessed.
ACKNOWLEDGMENTS
We thank Yuhko Aoki, Burk Braun, and Gerald Fink for generously providing strains. L.S. thanks Fritz Muhlschlegal and Shelley Brunt for helpful discussions and Yonghong Zhang for valuable technical assistance.
This work was supported by Public Health Service grants AI-37194 and GM47727 and by the Burroughs Wellcome Fund Molecular Pathogenic Mycology Scholar Award to W.A.F.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bailey D A, Feldmann P J F, Bovey M, Gow N A R, Brown A J P. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol. 1996;178:5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1992;20:2013–2022. doi: 10.1093/nar/20.suppl.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bidard F, Bony M, Blondin B, Dequin S, Barre P. The Saccharomyces cerevisiae FLO1 flocculation gene encodes for a cell surface protein. Yeast. 1995;11:809–822. doi: 10.1002/yea.320110903. [DOI] [PubMed] [Google Scholar]
- 5.Birse C E, Irwin M Y, Fonzi W A, Sypherd P S. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect Immun. 1993;61:3648–3655. doi: 10.1128/iai.61.9.3648-3655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun B R, Johnson A D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- 7.Csank C, Makris C, Meloche S, Schroppel K, Rollinghoff M, Dignard D, Thomas D Y, Whiteway M. Derepressed hyphal growth and reduced virulence in a VH-1 family-related protein phosphatase mutant of the human pathogen Candida albicans. Mol Biol Cell. 1997;8:2539–2551. doi: 10.1091/mbc.8.12.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Csank C, Schroppel K, Leberer E, Harcus D, Mohamed O, Meloche S, Thomas D Y, Whiteway M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erdman S, Lin L, Malczynski M, Snyder M. Pheromone-regulated genes required for yeast mating differentiation. J Cell Biol. 1998;140:461–483. doi: 10.1083/jcb.140.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fonzi W A, Irwin M Y. Isogenic Strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardwick K G, Boothroyd J C, Rudner A D, Pelham H R B. Genes that allow yeast cells to grow in the absence of the HDEL receptor. EMBO J. 1992;11:4187–4195. doi: 10.1002/j.1460-2075.1992.tb05512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilenski L L, Naider F, Becker J M. Polyoxin D inhibits colloidal gold-wheat germ agglutinin labelling of chitin in dimorphic forms of Candida albicans. J Gen Microbiol. 1986;132:1441–1451. doi: 10.1099/00221287-132-6-1441. [DOI] [PubMed] [Google Scholar]
- 13.Hogging D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignments on a microcomputer. Gene. 1988;173:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 14.Ishii N, Yamamoto M, Lahm H-W, Lizumi S, Yoshihara F, Nakayama H, Arisawa M, Aoki Y. A DNA-binding protein from Candida albicans that binds to the RPG box of Saccharomyces cerevisiae and the telomeric repeat sequence of C. albicans. Microbiology. 1997;143:417–427. doi: 10.1099/00221287-143-2-417. [DOI] [PubMed] [Google Scholar]
- 15.Ishii N, Yamamoto M, Yoshihara F, Arisawa M, Aoki Y. Biochemical and genetic characterization of Rbf1p, a putative transcription factor of Candida albicans. Microbiology. 1997;143:429–435. doi: 10.1099/00221287-143-2-429. [DOI] [PubMed] [Google Scholar]
- 16.Kelly R, Miller S M, Kurtz M B, Kirsch D R. Directed mutagenesis in Candida albicans: one-step gene disruption to isolate ura3 mutants. Mol Cell Biol. 1987;7:199–207. doi: 10.1128/mcb.7.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohler J R, Fink G R. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leberer E, Harcus D, Broadbent I D, Clark K L, Dignard D, Ziegelbauer K, Schmidt A, Gow N A R, Brown A J P, Thomas D. Signal transduction through homologs of the STE20p and STE7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee K L, Buckley H R, Campbell C C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Kohler J, Fink G. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 21.Liu H, Styles C A, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 22.Lo H, Kohler J R, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 23.Marck C. “DNA Strider”: a C program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCreath K J, Specht C A, Robbins P W. Molecular cloning and characterization of chitinase genes from Candida albicans. Proc Natl Acad Sci USA. 1995;92:2544–2548. doi: 10.1073/pnas.92.7.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muhlschlegel F A, Fonzi W A. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol Cell Biol. 1997;17:5960–5967. doi: 10.1128/mcb.17.10.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nevill-Manning C G, Wu T D, Brutlag D L. Highly specific protein sequence motifs for genome analysis. Proc Natl Acad Sci USA. 1998;95:5865–5871. doi: 10.1073/pnas.95.11.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Roberts R L, Fink G R. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genet Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 29.Roy A, Lu C F, Marykwas D L, Lipke P N, Kurjan J. The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein α-agglutinin. Mol Cell Biol. 1991;11:4196–4206. doi: 10.1128/mcb.11.8.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saporito-Irwin S M, Birse C E, Sypherd P S, Fonzi W A. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol. 1995;15:601–613. doi: 10.1128/mcb.15.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins Struct Funct Genet. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 32.Shepherd M G, Yin C Y, Ram S P, Sullivan P A. Germ tube induction in Candida albicans. Can J Microbiol. 1980;26:21–26. doi: 10.1139/m80-004. [DOI] [PubMed] [Google Scholar]
- 33.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratories; 1986. [Google Scholar]
- 34.Staab J F, Bradway S D, Fidel P L, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- 35.Staab J F, Ferrer C A, Sundstrom P. Developmental expression of a tandemly repeated, proline- and glutamine-rich amino acid motif on hyphal surfaces of Candida albicans. J Biol Chem. 1996;271:6298–6305. doi: 10.1074/jbc.271.11.6298. [DOI] [PubMed] [Google Scholar]
- 36.Stoldt V R, Sonneborn A, Leuker C E, Ernst J F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sundstrom P, Smith D, Sypherd P S. Sequence analysis and expression of the two genes for elongation factor 1 alpha from the dimorphic yeast Candida albicans. J Bacteriol. 1990;172:2036–2045. doi: 10.1128/jb.172.4.2036-2045.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuchimori, N., L. L. Sharkey, W. A. Fonzi, S. W. French, J. E. Edwards, and S. G. Filler. Interaction of HWP-1 deficient mutants of Candida albicans with host cells in vivo and in vitro. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 39.Udenfriend S, Kodukula K. Prediction of w site in nascent precursor of glycosylphosphatidylinositol protein. Methods Enzymol. 1995;250:571–582. doi: 10.1016/0076-6879(95)50098-7. [DOI] [PubMed] [Google Scholar]
- 40.Williams F E, Trumbly R J. Characterization of TUP1, a mediator of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6500–6511. doi: 10.1128/mcb.10.12.6500. [DOI] [PMC free article] [PubMed] [Google Scholar]