Abstract
The cellulolytic and hemicellulolytic complex of Clostridium thermocellum, termed cellulosome, consists of up to 26 polypeptides, of which at least 17 have been sequenced. They include 12 cellulases, 3 xylanases, 1 lichenase, and CipA, a scaffolding polypeptide. We report here a new cellulase gene, celK, coding for CelK, a 98-kDa major component of the cellulosome. The gene has an open reading frame (ORF) of 2,685 nucleotides coding for a polypeptide of 895 amino acid residues with a calculated mass of 100,552 Da. A signal peptide of 27 amino acid residues is cut off during secretion, resulting in a mature enzyme of 97,572 Da. The nucleotide sequence is highly similar to that of cbhA (V. V. Zverlov et al., J. Bacteriol. 180:3091–3099, 1998), having an ORF of 3,690 bp coding for the 1,230-amino-acid-residue CbhA of the same bacterium. Homologous regions of the two genes are 86.5 and 84.3% identical without deletion or insertion on the nucleotide and amino acid levels, respectively. Both have domain structures consisting of a signal peptide, a family IV cellulose binding domain (CBD), a family 9 glycosyl hydrolase domain, and a dockerin domain. A striking distinction between the two polypeptides is that there is a 330-amino-acid insertion in CbhA between the catalytic domain and the dockerin domain containing a fibronectin type 3-like domain and family III CBD. This insertion, missing in CelK, is responsible for the size difference between CelK and CbhA. Upstream and downstream flanking sequences of the two genes show no homology. The data indicate that celK and cbhA in the genome of C. thermocellum have evolved through gene duplication and recombination of domain coding sequences. celK without a dockerin domain was expressed in Escherichia coli and purified. The enzyme had pH and temperature optima at 6.0 and 65°C, respectively. It hydrolyzed p-nitrophenyl-β-d-cellobioside with a Km and a Vmax of 1.67 μM and 15.1 U/mg, respectively. Cellobiose was a strong inhibitor of CelK activity, with a Ki of 0.29 mM. The enzyme was thermostable, after 200 h of incubation at 60°C, 97% of the original activity remained. Properties of the enzyme indicated that it is a cellobiohydrolase.
Clostridium thermocellum secretes into the cultural medium a multiprotein complex, termed cellulosome, capable of efficient hydrolysis of highly ordered crystalline cellulose (3, 15). It contains 14 to 26 different polypeptides and possesses endo- and exoglucanase, xylanase, mannanase, lichenase, and feruloyl esterase activities (8, 23, 36). All cellulosomal components have modular structures (5, 38). The enzymatically active components are composed of at least a catalytic domain and a highly conservative type I dockerin domain (5). Some of the enzymes are more complex and include cellulose binding domains (CBD), S-layer-homologous domains, and domains of unknown functions (38). The largest cellulosome subunit is a 210-kDa enzymatically inactive scaffolding protein, CipA (17). It is composed of nine highly similar cohesin domains interacting with dockerin domains of catalytic subunits, (42), a family III CBD binding the cellulosome to the cellulose, and a special dockerin type II domain attaching the complex to the cell surface (29). A high degree of homology between CipA cohesin domains (17) together with studies on the interactions between different cohesin domains and some catalytic subunits suggest that binding of the catalytic subunits to CipA occurs on random basis (21, 27, 32). This seems to indicate that the incorporation of a specific catalytic subunit into the cellulosome depends on its relative amount and that predominant enzymes play important roles in the cellulosome.
Many genes encoding cellulosomal components have been cloned, and their products have been characterized (3, 5, 15). Surprisingly, a 98-kDa protein, the presence of which, in relatively large amounts, in the cellulosome was described by Choi and Ljungdahl (10), has been neither sequenced nor characterized.
This report describes in detail the cloning and sequencing of celK. CelK, the product of celK, has a high degree of homology with CbhA (51), a 138-kDa cellobiohydrolase from the same bacterium. CelK expressed in Escherichia coli was purified, and its enzymatic properties indicate strongly that it is a cellobiohydrolase. Thus, the cellulosome of C. thermocellum contains at least three cellobiohydrolases, CelS, CbhA, and CelK. (A preliminary report covering some properties of CelK was given at the MIE BIOFORUM 98 conference on the Genetics, Biochemistry and Ecology of Cellulose Degradation [22].)
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
C. thermocellum JW20, described by Freier et al. (16), was used for isolation of genomic DNA and cellulosomes. Culture conditions were as described by Wiegel (49); 1% (wt/vol) cellobiose and 5% (wt/vol) Avicel PH-101 were used as carbon sources. E. coli INVaF′ (Invitrogen Inc., Carlsbad, Calif.) and JM109 (Stratagene Cloning Systems, La Jolla, Calif.), used as cloning hosts for pCR2.1 (Invitrogen) and pBluescript SK(+) (Stratagene), respectively, were grown in Luria-Bertani medium supplemented with ampicillin (100 μg/ml).
Isolation and internal peptide sequencing of CelK.
Cellulosomes (100 μg) purified from 3-day-old-culture as described earlier (10) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (26). The concentration of acrylamide was 7.5% (wt/vol). After electrophoresis, the proteins were transferred to a polyvinylidene difluoride membrane and stained with Ponceau S dye. The CelK band (∼98 kDa) was identified according to the banding pattern of cellulosomal proteins on gels used for SDS-PAGE (10), excised with a razor blade, rinsed with 0.5 ml of distilled water, and then digested with protease Lys-C as specified by the supplier (Boehringer Mannheim, Indianapolis, Ind.). Residual peptides were separated by means of Hewlett-Packard (Wilmington, Del.) 1100 series high-pressure liquid chromatography (HPLC) control module equipped with a V8 reverse-phase column. Peptide peaks were monitored by UV absorption at 280 nm. N-terminal amino acid sequences of selected peptides were determined by using an Applied Biosystems model 477A gas-phase sequencer with an automatic on-line phenylthiohydantion analyzer.
Isolation of genomic DNA from C. thermocellum.
Genomic DNA was purified from a 0.5-liter culture by the method of Marmur (33), with the following modifications. After treatment of the cells with lysozyme (5 mg/ml) for 4 h, SDS (0.1%, wt/vol) and proteinase K (500 μg/ml) were added. The solution was incubated at 37°C and then dialyzed for 24 h against 1 liter of 100 mM Tris-HCl buffer (pH 7.5) containing 10 mM EDTA and 150 mM NaCl. The dialysate was incubated with DNase-free RNase (50 μg/ml) for 30 min at 37°C. Genomic DNA was extracted repeatedly (30 min for each extraction) with phenol-chloroform-isoamyl alcohol (12:12:1) at 37°C. DNA from the upper phase was precipitated with 0.1 volume of 3 M of sodium acetate (pH 5.2) and 2.5 volume of cold ethanol. After incubation at −20°C for 30 min, DNA was collected by centrifugation (7,000 × g, 30 min) and then carefully resuspended in 2.5 ml of distilled water. The suspension was then incubated at 37°C overnight for slow hydration. Purity and size of the genomic DNA were determined on the basis of absorption at 260 and 280 nm and by 1% agarose gel electrophoresis in the presence of ethidium bromide.
Primer design, PCR, and cloning.
Degenerate oligonucleotides were designed according to protein (peptide) sequences (Table 1) and synthesized with an Applied Biosystems DNA synthesizer. Using the oligonucleotides in combination as primers and purified genomic DNA as a template, PCRs were done on a model 480 thermal cycler (Perkin-Elmer, Norwalk, Conn.). All reagents were purchased from Perkin-Elmer and used as instructed. Annealing temperatures were 42 and 54°C with degenerate and specific primers, respectively; extension time was from 1 to 4 min, depending on the length of amplified fragments. PCR products (10 μl) were analyzed on agarose gels in the presence of ethidium bromide. They were either sequenced directly or cloned into the pCR2.1 vector with a TA cloning kit (Invitrogen) and then sequenced (see below).
TABLE 1.
Oligonucleotide primers used for amplification of the celK gene by PCR
| Name | Peptide | Sequencea | Orientation | Positionb |
|---|---|---|---|---|
| CelK1F | KLPDYKND | 5′-AARYTICCIGAYTAYAARAAYGA-3′ | Forward | 1242–1265 |
| CelK1R | IPIEMPYA | 5′-GCRTAIGGCATYTCDATIGGDAT-3′ | Reverse | 2119–2142 |
| CelK2F | 5′-CAGTGCTGATATTTACTCCA-3′ | Forward | 2051–2071 | |
| CelK2R | GDVNDDG | 5′-CCRTCRTCRTTRCARTCICC-3′ | Reverse | 3639–3658 |
| CelK3F | 5′-GCAGGCGGCATTAAAGCATG-3′ | Forward | 2732–2751 | |
| CelK3R | 5′-ATGTGATTTCGCTGTTGTTGAT-3′ | Reverse | 1337–1358 | |
| CelK4F | 5′-AGACTCATGGTCAACCAACGA-3′ | Forward | 3506–3526 | |
| CelK5R | 5′-CATTATATGGCAGTTTTTTAT-3′ | Reverse | 3827–3847 |
D, A, G, or T; R, A or G; Y, C or T.
Numbering corresponds to that of GenBank accession no. AF039030.
DNA sequencing and sequence analysis.
PCR products were purified by using either Microcon tubes (Amicon, Beverly, Mass.) or a Geneclean Spin kit (Bio101) before subjected to sequencing. Plasmids were purified from overnight-grown E. coli cultures by using a QIAprep Spin Plasmid Miniprep kit (Qiagen, Valencia, Calif.). DNA fragments were sequenced, using universal and sequence specific primers by means of an automatic PCR sequencer (Applied Biosystems). The Genetics Computer Group (GCG) program (version 8; GCG, University of Wisconsin Biotechnology Center, Madison) on the VAX/VMS system of the BioScience Computing Resource at the University of Georgia was used to analyze sequence data.
Southern blot analysis and colony hybridization.
Genomic DNA of C. thermocellum was digested with a single restriction enzyme and with enzymes in combinations. The digested fragments were separated by electrophoresis and transferred to a nylon membrane with a Turboblotter (Schleicher & Schuell, Keene, N.H.). Transfer of colonies to nylon membranes was done as described by Sambrook et al. (39). DNA was cross-linked to the membrane with a UV cross-linker (Stratagene). Hybridization probes for DNA were generated by PCR amplification in the presence of digoxigenin-labeled dUTP (Boehringer Mannheim). Hybridization with the labeled probe, stringency washing, and detection of positive bands or colonies were done as instructed by Boehringer Mannheim.
CelK purification.
A 10-liter culture of E. coli BL21(DE3)(pLys) harboring pET-21b(+) containing celK lacking the dockerin domain coding region (nucleotide residues 3594 to 3829) was grown to an optical density at 600 nm of approximately 0.8. After addition of 2 mM isopropyl-β-d-thiogalactopyranoside, the culture was incubated at 37°C for 5 h. The cells were collected, washed with 50 mM sodium phosphate buffer (pH 7.8), and broken in a French press. Cell debris was removed by centrifugation. The clear supernatant was mixed at 4°C with an increasing amount of Ni-nitrilotriacetic acid resin (Xpress protein purification System; Invitrogen) until the activity of CelK in the supernatant was negligible. The suspension was packed into a column. Washing and elution conditions were as recommended by the supplier. Fractions containing p-nitrophenyl-β-d-cellobioside (PNP-cellobioside) activity were combined and dialyzed against 20 mM Tris-HCl buffer, pH 7.5. The dialyzate was applied to a Mono Q HR10/10 column, and proteins were eluted with a gradient from 0 to 0.6 M NaCl. Fractions containing CelK were concentrated and chromatographed on a Superose 12 HR10/30 column in the presence of 0.1 M NaCl.
Enzyme assay and analytical methods.
The activity of CelK was assayed at 65°C in 50 mM sodium citrate buffer, pH 6.0. The concentration of PNP-cellobioside was 5 mM. Hydrolysis of PNP-cellobioside was determined by the release of p-nitrophenol. One enzyme unit was defined as amount of the enzyme releasing 1 μmol of p-nitrophenol per min. To test the dependence of CelK activity on pH, the following buffers (50 mM each) were used: histidine-HCl (pH 5.0 to 6.0), sodium phosphate (pH 6.0 to 7.0), and Tris-HCl (pH 7.0 to 9.0). The Km for PNP-cellobioside and Ki for cellobiose were determined at 65°C in 50 mM sodium citrate buffer (pH 6.0).
Nucleotide sequence accession number.
The nucleotide sequence of celK of C. thermocellum JW20 has been assigned accession no. AF039030 in the GenBank database.
RESULTS
Both celK and cbhA are present in the C. thermocellum genome.
One of the most abundant subunits of the C. thermocellum cellulosome (here designated CelK) has a mass of 98 kDa with an N-terminal amino acid sequence of LEDKSSKLPDYKNDLLYE (10). Database search revealed that of the 19 amino acid residues, there were three mismatches (in boldface) with that of the N-terminal region of CbhA (formerly Cbh3; accession no. X80993), which contains 1,230 amino acid residues with molecular mass of 138,007 Da. To clarify the relationship between the 98-kDa subunit and CbhA amino acid sequences, purified CelK was digested with Lys-C protease. Two internal peptides, EYYFK and IPIEMPYAGGEQ, were obtained after separation by HPLC. The first short sequence was not found in the deduced protein sequence of CbhA, although two YYF sites (residues 284 to 286 and 1044 to 1046) were identified. The second internal sequence matched perfectly with amino acid residues of 326 to 337 of CbhA. The differences with respect to size and partial amino acid sequence indicated that the 98-kDa component and CbhA may be encoded by two distinct genes of C. thermocellum.
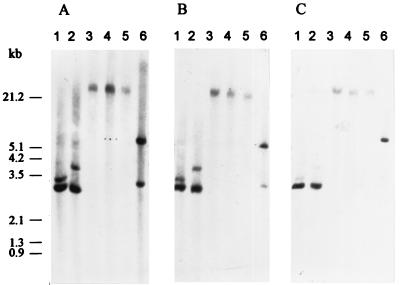
Degenerate oligonucleotides CelK1F and CelK1R (Table 1), corresponding to the less identical sites of the 98-kDa N-terminal sequence and the longer internal peptide, were used to amplify the putative gene coding for the 98-kDa protein. A 0.9-kb DNA fragment was obtained (data not shown) after amplification. This size is in agreement with the corresponding region of CbhA. Southern blotting using the amplified 0.9-kb fragment as a hybridization probe should detect at least two signals, assuming that the two genes celK and cbhA have high level of homology (Fig. 1). Indeed, two signals were obtained with genomic DNA digested with EcoRI (lane 1), DraI (lane 2), or BamHI (lane 6) under low-stringency washing (A). As the stringency washing temperature was increased from 42°C (A) to 55°C (B) and 65°C (C), one of the two signals became less intense or disappeared (e.g., 3.0-, 3.9-, and 3.0-kb bands with EcoRI, DraI, and BamHI, respectively [Fig. 1B and C]). The sizes of these bands matched those found in the sequenced cbhA. The results strongly suggest the presence of two separate genes encoding CelK and CbhA in the genome of C. thermocellum. Only one signal was found with genomic DNA digested with ApaI (lane 3), NotI (lane 4), and AscI (lane 5) at the three washing temperatures, and all of these bands were larger than 21.2 kb. This means that there are no restriction sites involving these enzymes in the DNA regions flanking the celK and cbhA genes or, alternatively, that these enzymes failed to digest the DNA sample efficiently.
FIG. 1.
Southern analysis of C. thermocellum genomic DNA. Genomic DNA was digested with EcoRI (lane 1), DraI (lane 2), ApaI (lane 3), NotI (lane 4), AscI (lane 5), and BamHI (lane 6) and fractionated on an agarose gel (1%). DNA fragments in the gel were transferred to a nylon membrane. The hybridization probe was the 0.9-kb PCR fragment amplified by using the CelK1F and CelK1R (Table 1). Digoxigenin was incorporated into the fragment during PCR amplification in the presence of digoxigenin-conjugated dUTP (Boehringer Mannheim). Stringency washing was done twice for 15 min at 42°C (A), 55°C (B), and 65°C (C). DNA standards labeled with digoxigenin were run under identical conditions, and their positions of migration are shown on the left.
The 0.9-kb PCR product was cloned into pCR2.1, and its nucleotide sequence was determined. Sequence analysis demonstrated that this fragment was different from but highly similar to the 5′ half of the cbhA open reading frame (ORF). Identity on the nucleotide level between the two ORFs was 74.2%. The deduced amino acid sequence of CelK revealed 74.1% identity with that of CbhA without any deletion or insertion. Furthermore, the amino acid sequences of the N terminus and the two internal peptides of the 98-kDa protein all matched those of the deduced amino acid sequence of the PCR product except for one residue: EYYFK was obtained by protein sequencing, whereas GYYFK was obtained by deduction. This mismatch may be attributable to experimental error during the protein sequencing or PCR cloning. Combined, the data clearly show that two highly similar genes are present in the genome of C. thermocellum. We propose the name celK for the gene coding for the 98-kDa cellulosomal subunit, and CelK for its encoded polypeptide, consistent with the nomenclature for the recently published cellulase gene, celJ (1).
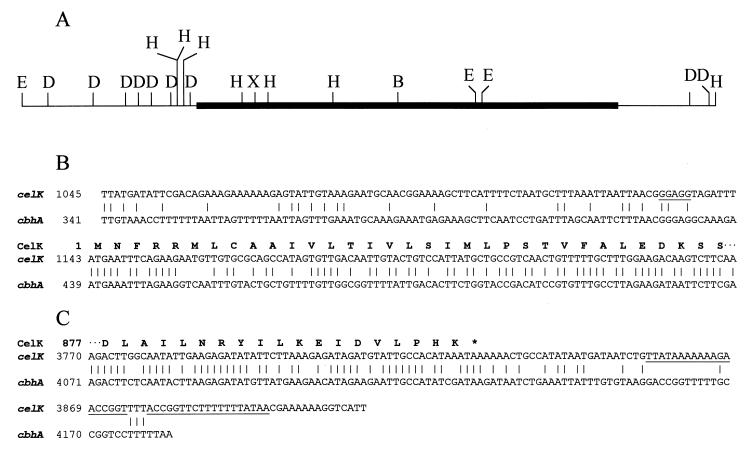
Several steps were taken to obtain the complete nucleotide sequence of celK and its upstream and downstream regions. First, we took advantage of the fact that CelK is a subunit of the cellulosome and therefore its dockerin domain, particularly the first peptide of that domain, should be highly similar to that of many other subunits (10). A 1.7-kb DNA fragment was amplified by PCR using a pair of primers, CelK2F (with low homology to the corresponding site of cbhA) and CelK2R (a degenerate primer corresponding to the first conserved region of the dockerin domain) (Table 1). The nucleotide sequence of the PCR fragment allowed us to generate two hybridization probes corresponding to the 5′ and 3′ halves of celK and then obtain two separate plasmid clones spanning the complete ORF plus 1.1-kb 5′ and 0.6-kb 3′ ends. Therefore, the nucleotide sequence determined included a total of 4.187 kb of the celK locus. A restriction map of celK is shown in Fig. 2A.
FIG. 2.
(A) Restriction map of celK. D, E, H, and X, restriction sites for DraI, EcoRI, HindIII, and XhoI, respectively. (B and C) Comparisons between partial sequences of celK and cbhA. Sequences compared include 5′ (B) and 3′ (C) ends of the two ORFs, together with some untranslated regions. Numbers on the left correspond to those in the databases. The deduced amino acid sequence for celK is shown as boldface; putative ribosome binding and transcription terminator sites are underlined.
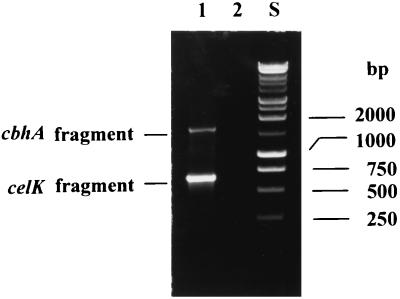
To verify the presence of the two highly similar genes encoding CelK and CbhA in the strain of C. thermocellum JW20 used for genomic DNA extraction, we amplified the regions of the 3′-terminal halves of the two genes where cbhA has about a 900-bp addition in comparison with celK (see below). Two bands with sizes of about 650 and 1,600 bp (Fig. 3) were detected after PCR amplification. These two bands matched the sizes of the corresponding regions of celK and cbhA, respectively, supporting the existence of both genes in the genome of the C. thermocellum JW20. The band of celK was more intensive than the one corresponding to cbhA, probably because the reverse primer was less homologous to the sequence of cbhA and/or the PCR conditions favored the amplifications of shorter products (celK). The presence of two bands, 150 and 98 kDa, recognized with anti-CbhA antibodies in the cellulosome indicates that C. thermocellum F7, a Russian isolate, also contains the two highly homologous genes (51).
FIG. 3.
Analysis of PCR products amplified from C. thermocellum genomic DNA, using CelK4F and Celk2R (Table 1) as primers. The gel was loaded with PCR products amplified in the presence (lane 1) and absence (lane 2) of template. Lane 3 was loaded with 1-kb DNA standards (Promega).
Characterization of celK and CelK.
Sequence analysis revealed that celK had an ORF of 2,685 nucleotides. The G+C content of the ORF was 43.3%, characteristic of C. thermocellum genes sequenced to date (5, 27). The start codon was determined based on the fact that there is a stop codon preceding the ATG codon and that the signal peptide had all of the properties found for signal peptides of bacterial extracellular enzymes (48). A putative ribosome binding site (GGAGG) was found 8 bp before the ATG codon. Upstream of the coding region are possible promoter sequences, TTGATG for the −35 region and TAATTT for the −10 region. A region of 20-bp palindrome sequences downstream of the TAA stop codon might serve as a transcription terminator. Analysis of 1,142-bp upstream and 356-bp downstream sequences failed to detect any ORF. These data suggest that celK, like most genes coding for hydrolytic enzymes of C. thermocellum, is monocistronic (4).
The deduced CelK contained 895 amino acid residues with a molecular mass of 100,713 Da. Residues 28 to 45 matched perfectly the N-terminal amino acid sequence determined for the 98-kDa subunit of the cellulosomes (10). Thus, residues 1 to 27 of CelK served as the signal peptide, and the calculated mass 97,572 Da of the mature CelK was consistent with that determined by SDS-PAGE (10), indicating that glycosylation of the 98-kDa polypeptide was, if present, negligible. The low degree of glycosylation of the CelK could be explained by the absence of long P/T-rich linkers characteristic for CipA and shown to be highly glycosylated (18, 27). The agreement of amino acid sequences determined by protein sequencing and by deduction from nucleotide sequence for CelK confirmed the differences between CelK and CbhA on the nucleotide level and therefore excluded the possibility that the 98-kDa subunit revealed by SDS-PAGE was a proteolytic product of CbhA.
Homology studies.
The complete amino acid sequences of CelK and CbhA were compared by using the GCG program BESTFIT (Fig. 4). Very high sequence identity (83.4%) and similarity (90.5%) between their homologous regions were observed with the addition of only one amino acid residue (position 687) in CbhA. Identity on the nucleotide level between the homologous regions was 86.5%. The 5′ and 3′ untranslated sequences of the genes revealed substantially lower sequence identity (Fig. 2B and C, respectively). A 328-amino-acid-residue region corresponding to residues 815 to 1143 of CbhA was absent in CelK. This region in CbhA explains the size difference (98 versus 130 kDa) between the two proteins.
FIG. 4.
Comparison between the deduced amino acid sequences of celK and cbhA. The sequence in CbhA containing a family III CBD is underlined. The signal peptide in CelK is in boldface; amino acids in CelK determined by protein sequencing are italicized.
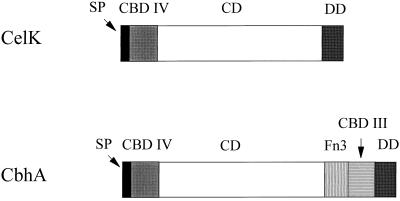
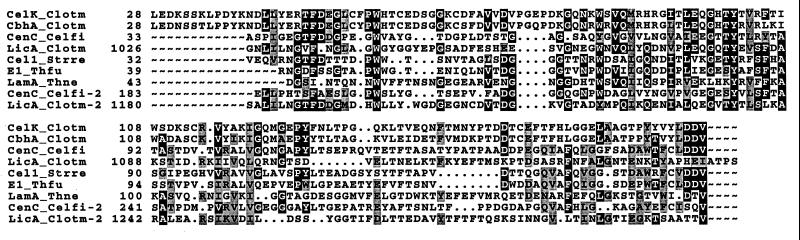
In a recent publication (51), the multidomain structure of CbhA was reported. Analysis of the amino acid sequence of CelK deduced from its nucleotide sequence revealed a domain structure of the polypeptide similar to that of CbhA (Fig. 5). An N-terminal signal peptide was followed by a fragment of 160 amino acids (positions 40 to 200) that showed some homology to family IV bacterial CBDs (Fig. 6). It displayed sequence identities of 77% to the CBD of C. thermocellum CbhA (51), 30% to the CBD N1 of Cellulomonas fimi CenC (11), 28% to the CBD I of C. thermocellum LicA (direct submission; accession no. X89732), 27% to the CBD of Streptomyces reticuli Cel1 (40), 22% to the CBD of Thermomonospora fusca E1 (28), 25% to the CBD of Thermotoga neapolitana LamA (12), 24% to the CBD N2 of C. fimi CenC (11), and 24% to the CBD II of C. thermocellum LicA.
FIG. 5.
Domain organizations of C. thermocellum CelK and CbhA. Assignments of domains are based on sequence similarities to domains of known functions. Symbols: SP, signal peptide; CBD IV, CBD of family IV; CD, catalytic domain; DD, dockerin domain; CBD III, CBD of family III; Fn3, fibronectin type 3-like domain.
FIG. 6.
Alignment of the CBD of CelK with CBDs of family IV found in microbial cellulases. Abbreviations: Celk_Clotm, C. thermocellum CelK; Cbha_Clotm, C. thermocellum CbhA; Cenc_Celfi, Cellulomonas fimi CenC; Lica_Clotm, C. thermocellum LicA; Cel1_Strre, S. reticuli Cel1; E1_Thfu, T. fusca E1; Lama_Thne, Thermotoga neapolitana LamA.
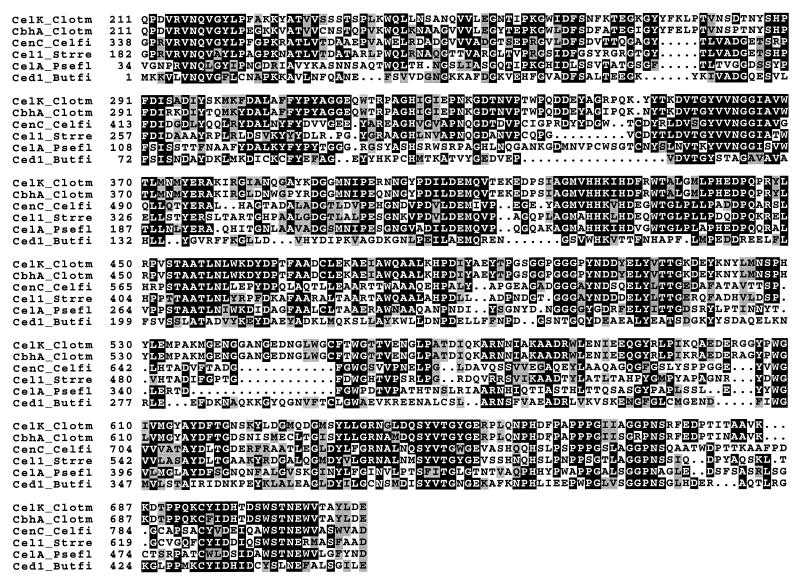
The catalytic domain of CelK was next to the CBD of family IV. The amino acid sequence up to about residue 600 was homologous to a number of cellulases (Fig. 7) belonging to glycosyl hydrolase family 9 (38). This region of CelK displayed sequence identities of 90% to C. thermocellum CbhA (51), 47% to Cellulomonas fimi CenC (11), 46.5% to S. reticuli CelA (40), 44.5% to Pseudomonas fluorescens CelA (19), and 31.1% to Butyrivibrio fibrosolvens Ced1 (6). All those except CelK and CbhA listed in Fig. 7 have been demonstrated to be endoglucanases with inverting mechanisms (38), whereas CbhA expressed in E. coli has been shown to be a cellobiohydrolase (41). It is not clear why CbhA and CelK (see below) from C. thermocellum cellulosome differ in enzymatic properties from the other homologous cellulolytic enzymes. Clearly, the sequence differences are sufficient to distinguish their catalytic modes. This is the case between fungal type II cellobiohydrolases and bacterial endoglucanases all belonging to glycosyl hydrolase family 6 (38). This hypothesis may be confirmed by performing deletions from the ends of CbhA or CelK when expressed in E. coli and/or by comparison of three-dimensional structures of the two types of enzymes. CelK as well as CbhA displayed only weak homology to CelS, a well-established cellobiohydrolase in the C. thermocellum cellulosome (24, 25, 47), suggesting that CelK and CbhA represent a second type of cellobiohydrolase in the cellulosome.
FIG. 7.
Alignment of the catalytic domain of CelK with those of other cellulases. Amino acid sequences that showed over 30% identity to the entire catalytic domain of CelK include CbhA of the same organism (CbhA_Clotm), CenC of Cellulomonas fimi (CenC_Celfi), CelA of S. reticuli (Cela_Strre), and CelA of B. fibrosolvens (Cela_Butfi).
Located on the C terminus of CelK is the dockerin domain found in a number of enzymes of the C. thermocellum cellulosome. This domain allows the catalytic subunits to interact with the cohesins of CipA, the scaffolding protein of the complex (42). Thus, all available evidence indicates that CelK is a component of the cellulosomal complex.
A summary of the putative domain composition of CelK and CbhA presented in Fig. 5 demonstrates that the 328-amino-acid fragment of CbhA located between its catalytic and dockerin domains, missing in CelK, is composed of a Fn3 domain and a CBD of family III.
Properties of CelK.
CelK devoid of the dockerin domain expressed in E. coli has a molecular mass of 94 kDa (Fig. 8), which is in a good agreement with deduced molecular mass of the enzyme. This truncated CelK had a temperature optimum of 65°C and an optimum pH of 6.0. The enzyme was thermostable; after 200 h of incubation at 60°C, 97% of the original activity remained. CelK did not hydrolyze xylan, glucomannan, or cellobiose. It was active toward PNPC, with a Km and Vmax of 1.67 μM and 15.1 U/mg, respectively. Cellobiose was an efficient inhibitor of CelK, with a Ki of 0.29 mM. We have demonstrated that CelK did not decrease the viscosity of carboxymethylcellulose and that it hydrolyzed carboxymethylcellulose to cellobiose, cellotriose to glucose and cellobiose, cellotetraose to cellobiose, and cellopentaose to glucose and cellobiose (22). All of these results strongly indicate that CelK is a cellobiohydrolase.
FIG. 8.
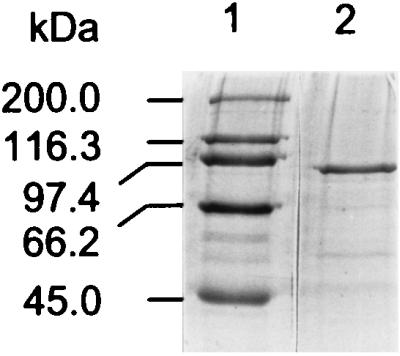
SDS-PAGE of purified CelK. Lane 1, molecular mass standards; lane 2, CelK.
DISCUSSION
Two genes coding for two cellulosomal subunits, CelK and CbhA, with highly similar catalytic domains are present in the genome of C. thermocellum. The regions of the catalytic domains have identities of about 90% on both nucleotide and amino acid levels, a level of homology that has not been found between any two genes of C. thermocellum sequenced to date (5). Identities between cellulases or between xylanases of C. thermocellum have never exceeded 50%. We propose that catalytic sites of celK and cbhA arose from a common ancestral gene by duplication. Duplication of genes coding for cellulases and other hydrolases is more common for anaerobic fungi (31). Thus, highly similar mannanases (14, 35) and cellulases (9, 30) are present in the same species of the anaerobic fungi; examples are the several multiple cellulases of the anaerobic fungus Orpinomyces strain PC-2, which contain highly similar catalytic domains. Gene duplication has been found to be common in organisms from bacteria to mammals and believed to be critical for evolution but appears unusual for genes of C. thermocellum. The fact that the catalytic domains of CelK and CbhA were duplicated and then favorably selected suggests that these catalytic domains may be essential for the bacterium to engage in cellulose degradation. In addition to the catalytic domains, CelK and CbhA have very similar dockerin domains. The striking distinction between the enzymes is the 328-amino-acid region located close to the C terminus. This region, which contains a combination of fibronectin type 3-like domain and a CBD of family III (51), is absent in CelK. The domain organization shown in Fig. 4 implies that sequences coding for different domains of the cellulosomal subunits evolved independently and then combined to code for the complete polypeptides. If this is true, then possibly celK was formed by combining regions of the family IV CBD and catalytic domain from one side with the dockerin domain from another and then was inserted by the sequences coding for the fibronectin-like domain and the family III CBD to yield cbhA. Alternatively, cbhA was first present and then lost the fragment described to become celK. Nevertheless, duplication and rearrangement involving sequences encoding separate domains were needed to yield the two complete genes.
CBDs are believed to be important in increasing local concentration of the catalytic domains to the substrate and/or in disrupting the hydrogen bonds between cellulose chains. These domains play important roles in free-acting enzymes (20, 43, 46). The role of CBDs found in some cellulosomal catalytic components is not yet clear. The cellulosomal enzymes are attached to the cellulose surface by means of CipA containing a CBD of family III and in fact do not need their own CBDs. However, CBDs are found in several cellulosomal cellulolytic subunits, including CelF (containing a CBD of family III) (44), CelE (CBD of family VII) (38), CbhA (two CBDs of families III and IV) (51), and CelK (CBD of family IV). Another subunit of the cellulosome, XynZ, contains a CBD of family VI (44). It has been demonstrated that the family III CBD of CipA binds to both amorphous and crystalline cellulose, but its binding capacity with amorphous cellulose is 20 times higher (37). CelK CBD binds efficiently to acid-swollen cellulose and weakly to Avicel (22). Properties of CBDs of the catalytic cellulosome components have not been studied. These domains belong to at least four different families, and a particular type of CBD is usually associated with a particular type of catalytic domain (38, 44): CBDs of family III of CelF and CbhA as well as CBDs of family IV of CelK and CbhA with family 9 catalytic domains; CBD of family VII of CelE with a family 5 catalytic domain; and CBD of family VI of XynZ with a family 10 catalytic domain (38). The presence of different CBDs in several cellulosomal enzymes suggests that these domains play significant and specific roles in cellulose degradation. The chemical simplicity of cellulose belies its structural complexity. The diversity of CBDs found in cellulosomal subunits may be necessary for binding of the complex to various regions of cellulose regardless of the degree of its crystallinity and other peculiarities of its structure. Perhaps CBDs are in some way involved in the hydrolysis process (13).
For a long time it has been believed that the cellulosome contains mostly endoglucanases (34). The discoveries of CelS (24), CbhA (51), and, as reported here, CelK indicate that cellobiohydrolases play an important role in cellulose degradation by the cellulosome of C. thermocellum. This idea is further supported by the fact that CelS and CelK are the most abundant components of the cellulosome. The synergism between cellulases in terms of cellulose hydrolysis is not limited to between cellobiohydrolases and endoglucanases but also occurs between two classes of cellobiohydrolases, one cleaving cellobiose from the reducing ends and the other doing so from the nonreducing ends of cellulose chains (2). High homology observed between CelK and CbhA together with lack of significant homology between these enzymes and CelS may reflect the presence of two different types of cellobiohydrolases in the cellulosome.
ACKNOWLEDGMENTS
This work was funded by grant DE-FG02-93ER20127 from the Department of Energy. Support by a Georgia Power Distinguished Professorship in Biotechnology (to L.G.L.) is also gratefully acknowledged.
REFERENCES
- 1.Ahsan M M, Kimura T, Karita S, Sakka K, Ohmiya K. Cloning, DNA sequencing and expression of the gene encoding Clostridium thermocellum cellulase CelJ, the largest catalytic component of the cellulosome. J Bacteriol. 1996;178:5732–5740. doi: 10.1128/jb.178.19.5732-5740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr B K, Hsieh Y-L, Ganem B, Wilson D B. Identification of two different classes of exocellulases. Biochemistry. 1996;35:586–592. doi: 10.1021/bi9520388. [DOI] [PubMed] [Google Scholar]
- 3.Bayer E A, Morag E, Shoham Y, Tormo J, Lamed R. The cellulosome: a cell surface organelle for the adhesion to and degradation of cellulose. In: Fletcher M, editor. Bacterial adhesion: molecular and ecological diversity. New York, N.Y: Wiley-Liss Inc.; 1996. pp. 155–182. [Google Scholar]
- 4.Béguin P. Molecular biology of cellulose degradation. Annu Rev Microbiol. 1990;44:219–248. doi: 10.1146/annurev.mi.44.100190.001251. [DOI] [PubMed] [Google Scholar]
- 5.Béguin P, Lemaire M. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit Rev Biochem Mol Biol. 1996;31:201–236. doi: 10.3109/10409239609106584. [DOI] [PubMed] [Google Scholar]
- 6.Berger E, Jones W A, Jones D T, Woods D R. Sequence and expression of a cellodextrinase (ced1) gene from Butyrivibrio fibrisolvens H17 cloned in Escherichia coli. Mol Gen Genet. 1990;223:310–318. doi: 10.1007/BF00265068. [DOI] [PubMed] [Google Scholar]
- 7.Bhat S, Goodenough P W, Bhat M K. Isolation of four major subunits from Clostridium thermocellum cellulosome and their synergism in the hydrolysis of crystalline cellulose. Int J Macromol. 1994;16:355–343. doi: 10.1016/0141-8130(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 8.Blum D L, Kataeva I, Li X-L, Ljungdahl L G. Phenolic acid esterase activity of Clostridium thermocellum cellulosome is attributed to previously unknown domains of XynY and XynZ. In: Ohmiya K, Sakka K, Karita S, Hayashi K, Kobayashi Y, Kimura T, editors. Genetics, biochemistry and ecology of cellulose degradation. 1998. p. 478. UniPublishers Co., Tokyo, Japan. [Google Scholar]
- 9.Chen H, Li X-L, Blum D L, Ljungdahl L G. Two genes of the anaerobic fungus Orpinomyces sp. strain PC-2 encoding cellulases with endoglucanase activities may have arisen by gene duplication. FEMS Lett. 1998;159:63–68. doi: 10.1111/j.1574-6968.1998.tb12842.x. [DOI] [PubMed] [Google Scholar]
- 10.Choi S K, Ljungdahl L G. Dissociation of the cellulosome of Clostridium thermocellum in the presence of ethylenediaminetetraacetic acid occurs with the formation of truncated polypeptides. Biochemistry. 1996;35:4897–4905. doi: 10.1021/bi9524629. [DOI] [PubMed] [Google Scholar]
- 11.Coutinho J B, Moser B, Kilburn D G, Warren R A J, Miller R C. Nucleotide sequence of the endoglucanase C gene (cenC) of Cellulomonas fimi, its high-level expression in Escherichia coli, and characterization of its products. Mol Microbiol. 1991;5:1221–1233. doi: 10.1111/j.1365-2958.1991.tb01896.x. [DOI] [PubMed] [Google Scholar]
- 12.Dakhova O N, Kurepina N E, Zverlov V V, Svetlichnyi V A, Velikodvorskaya G A. Cloning and expression in Escherichia coli of Thermonotoga neapolitana genes coding for enzymes of carbohydrate substrate degradation. Biochem Biophys Res Commun. 1993;194:1359–1364. doi: 10.1006/bbrc.1993.1974. [DOI] [PubMed] [Google Scholar]
- 13.Din N, Gilkes N R, Tekant B, Miller R C, Jr, Warren R A J, Kilburn D G. Non-hydrolytic disruption of cellulose fibres by the binding domain of a bacterial cellulase. Bio/Technology. 1991;9:1096–1099. [Google Scholar]
- 14.Fanutti C, Ponyi T, Black G W, Hazlewood G P, Gilbert H J. The conserved noncatalytic 40-residue sequence in cellulases and hemicellulases from anaerobic fungi functions as a protein docking domain. J Biol Chem. 1995;270:29314–29322. doi: 10.1074/jbc.270.49.29314. [DOI] [PubMed] [Google Scholar]
- 15.Felix C, Ljungdahl L G. The cellulosome: the exocellular organelle of Clostridium thermocellum. Annu Rev Microbiol. 1993;47:791–819. doi: 10.1146/annurev.mi.47.100193.004043. [DOI] [PubMed] [Google Scholar]
- 16.Freier D, Mothershed C P, Wiegel J. Characterization of Clostridium thermocellum JW20. Appl Environ Microbiol. 1988;54:204–211. doi: 10.1128/aem.54.1.204-211.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerngross U T, Romaniec M P M, Kobayashi T, Huskisson N S, Demain A L. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal S1-protein reveals an unusual degree of internal homology. Mol Microbiol. 1993;8:325–334. doi: 10.1111/j.1365-2958.1993.tb01576.x. [DOI] [PubMed] [Google Scholar]
- 18.Gerwig G J, Kamerling J P, Vliegenthart J F G, Morag E, Lamed R, Bayer E A. The nature of the carbohydrate-peptide linkage region in glycoproteins from the cellulosomes of Clostridium thermocellum and Bacteroides cellulosolvens. J Biol Chem. 1993;268:26956–26960. [PubMed] [Google Scholar]
- 19.Hall J, Gilbert J. The nucleotide sequence of a carboxymethylcellulase gene from Pseudomonas fluorescens subsp. cellulosa. Mol Gen Genet. 1988;213:112–117. doi: 10.1007/BF00333406. [DOI] [PubMed] [Google Scholar]
- 20.Hefford M A, Laderoute K, Willick G E, Yaguchi M, Seligy V. Bipartite organization of the Bacillus subtilis endo-β-1,4-glucanase revealed by C-terminal mutations. Protein Eng. 1992;5:433–439. doi: 10.1093/protein/5.5.433. [DOI] [PubMed] [Google Scholar]
- 21.Kataeva I, Guglielmi G, Béguin P. Interaction between Clostridium thermocellum endoglucanase CelD and polypeptides derived from the cellulosome-integrating protein CipA: stoichiometry and cellulolytic activity of the complexes. Biochem J. 1997;326:617–624. doi: 10.1042/bj3260617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kataeva I A, Li X-L, Chen H, Ljungdahl L G. CelK—a new cellobiohydrolase from Clostridium thermocellum cellulosome: role of N-terminal cellulose-binding domain. In: Ohmiya K, Sakka K, Karita S, Hayashi K, Kobayashi Y, Kimura T, editors. Genetics, biochemistry and ecology of cellulose degradation. 1998. pp. 454–460. UniPublishers Co., Tokyo, Japan. [Google Scholar]
- 23.Kohring S, Wiegel J, Mayer F. Subunit composition and glycosidic activities of the cellulase complex from Clostridium thermocellum JW20. Appl Environ Microbiol. 1990;56:3798–3804. doi: 10.1128/aem.56.12.3798-3804.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruus K, Lua A C, Demain A L, Wu J H D. The anchorage function of CipA (CelL), a scaffolding protein of the Clostridium thermocellum cellulosome. Proc Natl Acad Sci USA. 1995;92:9254–9258. doi: 10.1073/pnas.92.20.9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruus K, Wang W K, Ching J, Wu J H D. Exoglucanase activities of the recombinant Clostridium thermocellum CelS, a major cellulosome component. J Bacteriol. 1995;117:1641–1644. doi: 10.1128/jb.177.6.1641-1644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Lamed R, Bayer E A. The cellulosome concept—a decade later. In: Shimada K, Hoshino S, Ohmiya K, Sakka K, Kobayashi Y, Karita S, editors. Genetics, biochemistry and ecology of lignocellulose degradation. 1993. pp. 1–12. UniPublishers Co., Tokyo, Japan. [Google Scholar]
- 28.Lao G, Ghangas G S, Jung E D, Wilson D B. DNA sequences of three beta-1,4-endoglucanase genes from Thermomonospora fusca. J Bacteriol. 1991;173:3397–3407. doi: 10.1128/jb.173.11.3397-3407.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leibovitz E, Béguin P. A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J Bacteriol. 1996;178:3077–3084. doi: 10.1128/jb.178.11.3077-3084.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X-L, Chen H, Ljungdahl L G. Two cellulases, CelA and CelC, from the polycentric anaerobic fungus Orpinomyces strain PC-2 contain N-terminal docking domains for cellulase/hemicellulase complex. Appl Environ Microbiol. 1997;63:4721–4728. doi: 10.1128/aem.63.12.4721-4728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ljungdahl L G, Li X-L, Chen H. Evidence in anaerobic fungi of transfer of genes between them from aerobic fungi, bacteria and animal hosts. In: Wiegel J, Adams W W, editors. Thermophiles: the keys to molecular evolution and the origin of life? Philadelphia, Pa: Taylor and Francis Inc.; 1998. pp. 187–197. [Google Scholar]
- 32.Lytle B, Myers C, Kruus K, Wu J H D. Interactions of the CelS binding ligand with various receptor domains of the Clostridium thermocellum cellulosomal scaffolding protein, CipA. J Bacteriol. 1996;178:1200–1203. doi: 10.1128/jb.178.4.1200-1203.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marmur J. A procedure for the isolation of the deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 34.Mayer F, Coughlan M P, Mori Y, Ljungdahl L. Macromolecular organization of the cellulolytic complex of Clostridium thermocellum as revealed by electron microscopy. Appl Environ Microbiol. 1987;53:2785–2792. doi: 10.1128/aem.53.12.2785-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Millward-Sadler S J, Hall J, Black G W, Hazlewood G P, Gilbert H J. Evidence that the Piromyces gene family encoding endo-1,4-mannanases arose through gene duplication. FEMS Microbiol Lett. 1996;141:183–188. doi: 10.1111/j.1574-6968.1996.tb08382.x. [DOI] [PubMed] [Google Scholar]
- 36.Morag E, Bayer E A, Lamed R. Relationship of cellulosomal and noncellulosomal xylanases of Clostridium thermocellum to cellulose-degrading enzymes. J Bacteriol. 1990;172:6098–6105. doi: 10.1128/jb.172.10.6098-6105.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morag E, Lapidot A, Govorko D, Lamed R, Wilchek M, Bayer E A, Shoham Y. Expression, purification, and characterization of the cellulose-binding domain of the scaffold in subunit from the cellulosome of Clostridium thermocellum. Appl Environ Microbiol. 1995;61:1980–1986. doi: 10.1128/aem.61.5.1980-1986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohmiya K, Sakka K, Karita S, Kimura T. Structure of cellulases and their applications. Biotechnol Genet Eng Rev. 1997;14:365–414. doi: 10.1080/02648725.1997.10647949. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Schlochtermeier A, Walter S, Schroder J, Moorman M, Schrempf H. The gene encoding the cellulase (Avicelase) Cell from Streptomyces reticuli and analysis of protein domains. Mol Microbiol. 1992;6:3611–3621. doi: 10.1111/j.1365-2958.1992.tb01797.x. [DOI] [PubMed] [Google Scholar]
- 41.Singh R N, Akimenko V K. Isolation of a cellobiohydrolase of Clostridium thermocellum capable of degrading of natural crystalline substrates. Biochem Biophys Res Commun. 1993;192:1123–1130. doi: 10.1006/bbrc.1993.1533. [DOI] [PubMed] [Google Scholar]
- 42.Tokatlidis K, Dhurjati P, Béguin P. Properties conferred on Clostridium thermocellum endoglucanase CelC by grafting the duplicated segment of endoglucanase CelD. Protein Eng. 1993;6:947–952. doi: 10.1093/protein/6.8.947. [DOI] [PubMed] [Google Scholar]
- 43.Tomme P, Van Tilbeurgh H, Petterson G, Van Damme J, Vandekerckhove J, Knowles J, Teeri T, Claeyssens M. Studies of the cellulolytic system of Trichoderma reesei QM 9414. Eur J Biochem. 1988;170:575–581. doi: 10.1111/j.1432-1033.1988.tb13736.x. [DOI] [PubMed] [Google Scholar]
- 44.Tomme P, Warren R A J, Miller R C, Jr, Kilburn D G, Gilkes N R. Cellulose-binding domains: classification and properties. In: Saddler J N, Penner M H, editors. Enzymatic degradation of insoluble carbohydrates. Washington, D.C: American Chemical Society; 1995. pp. 142–163. [Google Scholar]
- 45.Tomme P, Creagh A L, Kilburn D G, Haynes C A. Interaction of polysaccharides with the N-terminal cellulose-binding domain of Cellulomonas fimi CenC.1. Binding specificity and calorimetric analysis. Biochemistry. 1996;35:13885–13894. doi: 10.1021/bi961185i. [DOI] [PubMed] [Google Scholar]
- 46.Van Tilbeurgh H, Tomme P, Claeyssens M, Bhikhabhai R, Petterson G. Limited proteolysis of the cellobiohydrolase I from Trichoderma reesei. FEBS Lett. 1986;204:223–227. [Google Scholar]
- 47.Wang W K, Kruus K, Wu H J D. Cloning and DNA sequence of the gene coding for Clostridium thermocellum Ss (CelS), a major cellulosome component. J Bacteriol. 1993;175:1293–1302. doi: 10.1128/jb.175.5.1293-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watson M E E. Compilation of published signal sequences. Nucleic Acids Res. 1984;12:5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiegel J, Dykstra M. Clostridium thermocellum: adhesion and sporulation while adhered to cellulose and hemicellulose. Appl Microbiol Biotechnol. 1984;20:59–65. [Google Scholar]
- 50.Yaron S, Morag E, Bayer E A, Lamed R, Shoham Y. Expression, purification and subunit-binding properties of cohesins 2 and 3 of the Clostridium thermocellum cellulosome. FEBS Lett. 1995;360:121–124. doi: 10.1016/0014-5793(95)00074-j. [DOI] [PubMed] [Google Scholar]
- 51.Zverlov V V, Velokodvorskaya G V, Schwarz W H, Bronnenmeier K, Kellermann J, Staudenbauer W L. Multidomain structure and cellulosomal localization of the Clostridium thermocellum cellobiohydrolase CbhA. J Bacteriol. 1998;180:3091–3099. doi: 10.1128/jb.180.12.3091-3099.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]