Abstract
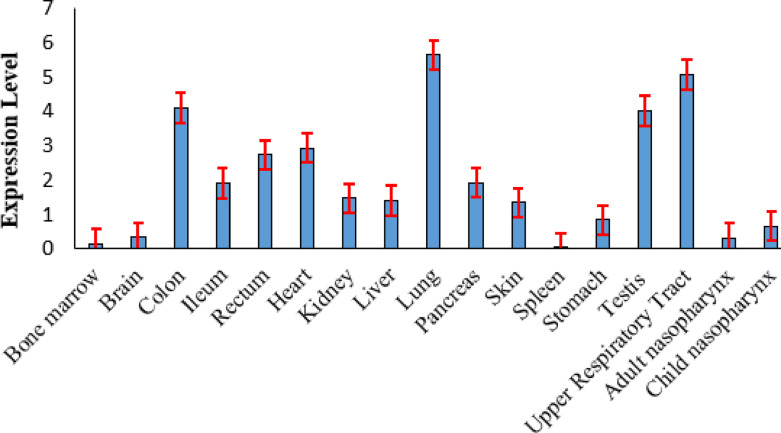
Coronavirus disease (COVID-19) is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and has infected millions worldwide. SARS-CoV-2 spike protein uses Angiotensin-converting enzyme 2 (ACE2) and Transmembrane serine protease 2 (TMPRSS2) for entering and fusing the host cell membrane. However, interaction with spike protein receptors and protease processing are not the only factors determining coronaviruses’ entry. Several proteases mediate the entry of SARS-CoV-2 virus into the host cell. Identifying receptor factors helps understand tropism, transmission, and pathogenesis of COVID-19 infection in humans. The paper aims to identify novel viral receptor or membrane proteins that are transcriptionally and biologically similar to ACE2 and TMPRSS2 through a fuzzy clustering technique that employs the Grey wolf optimizer (GWO) algorithm for finding the optimal cluster center. The exploratory and exploitation capability of GWO algorithm is improved by hybridizing mutation and crossover operators of the evolutionary algorithm. Also, the genetic diversity of the grey wolf population is enhanced by eliminating weak individuals from the population. The proposed clustering algorithm’s effectiveness is shown by detecting novel viral receptors and membrane proteins associated with the pathogenesis of SARS-CoV-2 infection. The expression profiles of ACE2 protein and its co-receptor factor are analyzed and compared with single-cell transcriptomics profiling using the Seurat R toolkit, mass spectrometry (MS), and immunohistochemistry (IHC). Our advanced clustering method infers that cell that expresses high ACE2 level are more affected by SARS-CoV-infection. So, SARS-CoV-2 virus affects lung, intestine, testis, heart, kidney, and liver more severely than brain, bone marrow, skin, spleen, etc.
We have identified 58 novel viral receptors and 816 membrane proteins, and their role in the pathogenicity mechanism of SARS-CoV-2 infection has been studied. Besides, our study confirmed that Neuropilins (NRP1), G protein-coupled receptor 78 (GPR78), C-type lectin domain family 4 member M (CLEC4M), Kringle containing transmembrane protein 1 (KREMEN1), Asialoglycoprotein receptor 1 (ASGR1), A Disintegrin and metalloprotease 17 (ADAM17), Furin, Neuregulin-1,(NRG1), Basigin or CD147 and Poliovirus receptor (PVR) are the potential co-receptors of SARS-CoV-2 virus. A significant finding is that heparin derivative glycosaminoglycans could block the replication of SARS-CoV-2 virus inside the host cytoplasm. The membrane protein N-Deacetylase/N-Sulfotransferase-2 (NDST2), Extostosin protein (EXT1, EXT2, and EXT3), Glucuronic acid epimerase (GLCE), and Xylosyltransferase I, II (XYLT1, XYLT2) could act as the therapeutic target for inhibiting the spread of SARS-CoV-2 infection. Drugs such as carboplatin and gemcitabine are effective in such situations.
Keywords: Single cell RNA sequencing data, Clustering, COVID-19, SARS-CoV-2, ACE2, TMPRSS2, Fuzzy clustering, Grey wolf optimizer, Differential evolution algorithm, Evolution population dynamics
1. Introduction
Coronaviruses (CoVs) are highly diverse groups of single stranded RNA (ssRNA) viruses. Seven variants of human coronaviruses have been reported till now. They are human coronavirus variant such as alpha coronavirus (229E), alpha coronavirus (NL63), beta coronavirus (OC43) and beta coronavirus (HKU1). The other human coronaviruses are Middle east respiratory syndrome (MERS), Severe acute respiratory syndrome (SARS), and the novel coronavirus or COVID-19 (SARS-CoV-2). Human CoVs cause common cold and respiratory illnesses. However, in early 2000, SARS and MERS CoVs were identified. SARS coronavirus was first identified in 2003 and infected 8000 people with a fatality case of 9.6%. More recently, in 2004, MERS CoVs had infected 2519 people, and the fatality rate was 34.3%. People infected with SARS virus and MERS virus usually suffer from fever, chill, headache, muscle ache, and diarrhea. More severe cases cause a severe respiratory syndrome that reduces lung functionality, increases the risk of atrial fibrillation, and even death. A novel strain of coronavirus known as SARS-CoV-2 was identified in Wuhan city, China, in December 2019. SARS-CoV-2 has infected more than 280 million people and has a fatality rate of 5.41%. Although the fatality rate of SARS-CoV-2 is far less than that of MERS and SARS coronaviruses, its transmission and severity are very high, making it difficult to curb the disease.
Structurally, SARS-CoV-2 is composed of spike (S) glycoprotein, envelope (E) glycoprotein, membrane (M), and nucleocapsid (N). The S glycoprotein is present on the outer surface of the viral particle and is composed of an amino terminal S1 subunit and a carboxyl-terminal S2 subunit. The S1 subunit binds the virus into a host cell, and the S2 subunit attaches the virus to the cell membrane. The S1 subunit splits into a receptor-binding domain (RBD) and an N-terminal domain (NTD). The RBD binds to the host receptor protein ACE2 [1] and initiate infection in the host cell. Hoffmann et al. [2] show that SARS-CoV-2 required ACE2 protein for binding to its spike protein, and TMPRSS2 protease cleaves the S2 subunit of the spike protein. Besides this receptor factor, SARS-CoV-2 also depends on some other receptor factor or membrane protein to initiate an infection to the host cell [2]. Therefore, it is necessary to identify the viral receptor protein and membrane protein that allows the binding and entry of SARS-CoV-2 and causes COVID-19 infection in the host cell. It will help better understand COVID-19 disease and the development of novel therapeutics and vaccines instead of experimental therapies and drug repositories.
In early 2020, Gordon et al. identified the human protein interacting with SARS-CoV-2 protein using affinity-purification mass spectrometry (AP-MS) [3]. They study SARS-CoV-2 protein and human protein interaction in the infected human embryonic kidney (HEK) 293 cells. The study identified 332 human proteins associated with protein trafficking, transcription, translation, and ubiquitination. The work provides insight into the pathway causing SARS-CoV-2 infection and predicts the possible drug target. Refs. [4], [5], [6] also applied the MS proteomics approach to study the interaction between SARS-CoV-2 protein and human protein. The virus protein-host protein interaction helps to reveal the pathogenesis pathway of the SARS-CoV-2 viral protein and provides a strategy to search for a novel antiviral treatment by targeting the host protein [3], [4], [5]. However, it is observed that the experimental design for the viral protein-host protein interaction network does not provide the functioning and environment of protein processing, accessory protein, etc. Therefore, enrichment pathway analysis, validation process, and biological functioning of the predicted SARS-CoV-2 viral protein and host protein interaction are needed to confirm the pathogenesis pathway of COVID-19 disease and explore the natural and functional significance [4].
Numerous researchers have investigated the pathogenesis of COVID-19 transmission by surveying the gene expression pattern of host receptors associated with SARS-CoV-2 protein using transcriptomics profiling of single-cell RNA sequencing (scRNA-Seq) technology [1], [7], [8], [9], [10], [11]. The scRNA-Seq technology provides the method to understand the cellular, biological and molecular process of SARS-CoV-2 infection from the expression pattern of genes on various human cells. Some studies pointed out that SARS-CoV-2 requires other receptors to infect specific types of human cells. Like other human coronavirus and SARS coronavirus, SARS-CoV-2 utilize multiple viral receptor factor such as CD209 [12], CLEC4G [13], CLEC4M [14], etc. to enter the host cell. The protease cathepsins (CTSL/M) [15] and FURIN [16] cleave the spike protein of SARS-CoV-2 virus [8]. To the best of our knowledge, no previous study had identified the SARS-CoV-2 viral entry-associated gene and examined the expression pattern of these viral receptor factors or membrane proteins using machine learning techniques. To be noted, Furong et al. demonstrate that ANPEP, ENPEP, and DPP4 exhibit similar expression profiles with ACE2 using hierarchical clustering and correlation coefficient [7]. The research provides a foundation for utilizing other unsupervised clustering approaches to identify the potential co-receptor showing similar expression patterns with ACE2 and TMPRSS2.
The current paper attempts to organize a group of genes (i.e., membrane protein or ssRNA viral protein) with a similar expression pattern to ACE2 and TMPRSS2 protein utilizing the fuzzy clustering technique. At the end of the fuzzy clustering technique, an improved metaheuristic algorithm (GWO) is implemented to find the optimal cluster center in the search space with less computational time [17], [18]. Several classical metaheuristic algorithms have been implemented to solve real-life clustering. But the problem with the classical approaches is that it gets trapped at local minima without giving the best solution. So efforts should always lie in applying a modified version of the classical metaheuristic algorithm when adapting to a real-life domain. This work develops an improved GWO algorithm by hybridizing the mutation and crossover operator of the Differential evolution (DE) algorithm [19]. Later, the worst search agent from the population is removed, and its position is reinitialized around the best search agent. It is observed that the improved GWO algorithm balanced the exploratory and exploitative stage of the classical GWO algorithm and performed a local search around the best solution vector [20].
The advantage of the fuzzy clustering technique is its ability to associate a gene showing more than one type of co-regulation into multiple clusters. It helps monitor the expression level of thousands of genes at a time [21]. In the fuzzy clustering technique, a gene point is associated with every other cluster with a membership function. The membership function measures the degree to which a gene point relates to a cluster group. The higher the membership value, a gene point is associated more strongly with a cluster. It gives the expressive level of a gene point in a cluster [21], [22]. For example, the expression pattern vector of ACE2 in the upper respiratory tract is [0.3796, 0.067, 0.091, 0.489], ACE2 is expressed with 37.96% in the non-ciliated secretory cell, 6.7% in the basal cell, 9.1% in the goblet cell, and 48.9% in the ciliated cell. Thus, ACE2 is highly expressed in the ciliated cell and is the primary site of SARS-CoV-2 infection.
We then identify a gene group that shares similar gene expression patterns with ACE2 and TMPRSS2. The study successfully predicts 58 ssRNA viral and 816 membrane proteins (or genes) significantly co-expressed with SARS-CoV-2 receptor protein (ACE2 and TMPRSS2). Finally, the predicted viral and membrane protein are analyzed by protein–protein interaction (PPI) network, Gene Ontology (GO) terms, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The analysis help understand the biological functioning of these predicted proteins involved in the pathogenesis of SARS-CoV-2 infection.
1.1. Current existing work
In early 2020, many studies identified human proteins and SARS-CoV-2 protein interaction using AP-MS, BioID, and MS approaches, etc.[3], [4], [5], [6]. Such studies have analyzed the interaction between SARS-CoV-2 protein and human protein using PPIs network [1], [3], [23]. In Ref. [23] authors computationally analyze the interaction of SARS-CoV-2 viral protein and human protein in HEK 293 cells to identify the processes affected by the SARS-CoV-2 infection. The authors implement the GoNet algorithm to determine human proteins and SARS-CoV-2 protein interaction. The GONet algorithm detects the GO term of protein clustered in the STRING-extended PPI network. However, the problem with this algorithm is that it can return a significantly large amount of overlapping GO terms that might be difficult to interpret the functionality of the protein sets.
Different machine learning algorithms have been investigated in mid-2020 to study the pathogenesis of COVID-19 disease. Recently there has been a surge in research to identify the viral co-receptors and human protein interacting with SARS-CoV-2 protein using the unsupervised clustering approach. In Ref. [7], Furong et al. analyzed the co-expression pattern of 51 RNA viral receptors and 400 membrane proteins. Through hierarchical clustering, the paper observed that the peptidases ANPEP, DPP4 and ENPEP show similar expression patterns with ACE2 protein. To further analyze the co-expression relationship, Pearson correlation coefficient (PCC) is calculated between ACE2 protein with all the viral receptors and membrane protein. Furong et al. confirmed that ANPEP, DPP4 and ENPEP could act as candidate receptors for causing COVID-19 infection. But apart from the peptidases, glutamine, leucine, asparagine, amino acid, and phenylalanine also facilitate the binding of SARS-CoV-2 spike protein to ACE2 protein receptor and membrane protein mediates the entry of enveloped virus on the host cell [1], [7], [24]. The above observation suggests that other co-receptor factors might also mediate and restrict the entry of SARS-CoV-2 host receptors and must be identified to develop an effective combination of drugs.
Manvendra et al. [8] initially created a list of 28 host receptors associated with coronavirus infection from the scRNA-Seq data of various human tissues. The authors then study the expression level of these host receptors to predict the subset of cells or tissue vulnerable to SARS-CoV-2 infection. The scRNA-Seq gene expression matrix analysis is performed using an unsupervised clustering approach (Seurat package implemented in R environment). Cell type identification is performed using the default “Findclusters” function implemented in the Seurat R package. The study concludes that SARS-CoV-2 infection affects the heart, lung, kidney, central nervous system, liver, gastrointestinal tract, etc. The paper does not clearly state the factors considering the SARS-CoV-2 28 potential receptor factors.
Similarly, Zou et al. [11], [25] used the default “Findclusters” function implemented in the Seurat R package to identify different cell types. The expression distribution of ACE2 across different cell types of human organoids is evaluated, and the organs vulnerable to SARS-CoV-2 infection are placed according to ACE2 expression level. However, there is still a discordant report on considering ACE2 expression range in some tissue to determine the organ at high risk and low risk for SARS-CoV-2 infection. Other studies determine ACE2 expression levels to find the organs or cell types vulnerable to SARS-CoV-2 infection. The higher the ACE2 expression level, the more vulnerable the organ to SARS-CoV-2 infection. However, severe COVID-19 illness in immunocompromised patients might result from increased ACE2 expression levels or the underlying health condition. This fact is still unclear now. Sungnak et al. analyzed the expression level or patterns of ACE2, TMPRSS2, and other associated viral receptor proteins used by coronaviruses and influenza viruses [9]. The standard clustering tool Scanpy (implemented in Python) is used to identify cell types [26].
Current studies commonly use Seurat [27] and Scanpy [26] package to identify cell type from the single-cell transcriptome data. Seurat and Scanpy, by default, implement a graph-based clustering approach with an optimization algorithm to organize transcriptionally similar cells. Marker genes in each cell cluster are determined using logistic regression. The cell cluster is assigned manually based on the knowledge of the previous cell type marker gene. However, the main drawback to this approach is that the obtained cluster number depends on a resolution parameter assigned by the user. A high-resolution value generates more clusters, and a less resolution value produces fewer clusters. Thus, it may not reflect the correct cell type.
1.2. Motivation
The main factors that inspired us to develop an unsupervised fuzzy clustering approach utilizing the GWO algorithm are described as follows:
-
•
The hierarchical clustering algorithm tends to form crisp clusters that are not appropriate for some scRNA-Seq datasets. Integration of the ‘fuzziness’ concept in the clustering algorithm eliminates the challenges often created by extensive dimensional scRNA-seq data. It allows the cluster to grow in its natural structure and form.
-
•
Detection of correct cluster number from single-cell transcriptome data remains challenging. It has motivated us to develop a novel unsupervised fuzzy clustering technique utilizing the GWO algorithm to find the optimal cluster number from scRNA-Seq data.
-
•
Some authors implement the GoNet algorithm to study SARS-CoV-2 protein and human protein interaction. But GoNet algorithm returns a large number of similar clustered GO terms. But in addition to the clustering, we also need to learn the expression level of a gene in each cluster group.
-
•
All the previous methods do not explicitly justify how the expression level of ACE2 protein is determined or calculated. The current work finds the expression level of each gene point using the fuzzy clustering technique.
-
•
Some previous work created an initial gene list of the host receptors from the published articles and used it in their work. The reason for considering the receptor factors in analyzing the co-expression level with ACE2 protein is still unclear. The current paper attempts to identify novel receptor factors required for causing SARS-CoV-2 infection. This receptor factor might not have been reported in the published articles.
1.3. Key contribution
The key contributions of the current paper are summarized as follows:
-
•
The paper proposes an unsupervised fuzzy clustering with an optimization algorithm to analyze single-cell transcriptomes data. The primary purpose of the fuzzy clustering technique is to associate genes to multiple clusters to study the regulatory relationship between the genes. Through this, genes that regulate various signaling pathways in the pathogenicity of SARS-CoV-2 infection can be identified.
-
•
The fuzzy clustering technique determines the expression level of a gene point associated with a multiple cluster group. In the fuzzy clustering algorithm, each gene is related to every cluster by a membership function. The membership function expresses the strength a gene point is associated with a cluster group. This help finds the expression level of ACE2 protein and its co-receptor genes.
-
•
A comparative analysis of ACE2 expression profiles is conducted with the proposed clustering method and MS, single-cell transcriptomics profiling, and Immunohistochemistry (IHC) experiment. A similar kind of inference established by the previous experiment is observed in this work. A cell or tissue with high ACE2 expression is more affected by SARS-CoV-2 infection than a cell with less ACE2 expression. It is observed that the lung, kidney, testis, heart, upper respiratory tract, and gastrointestinal tract are more affected than the brain, bone marrow, spleen, and skin organoids.
-
•
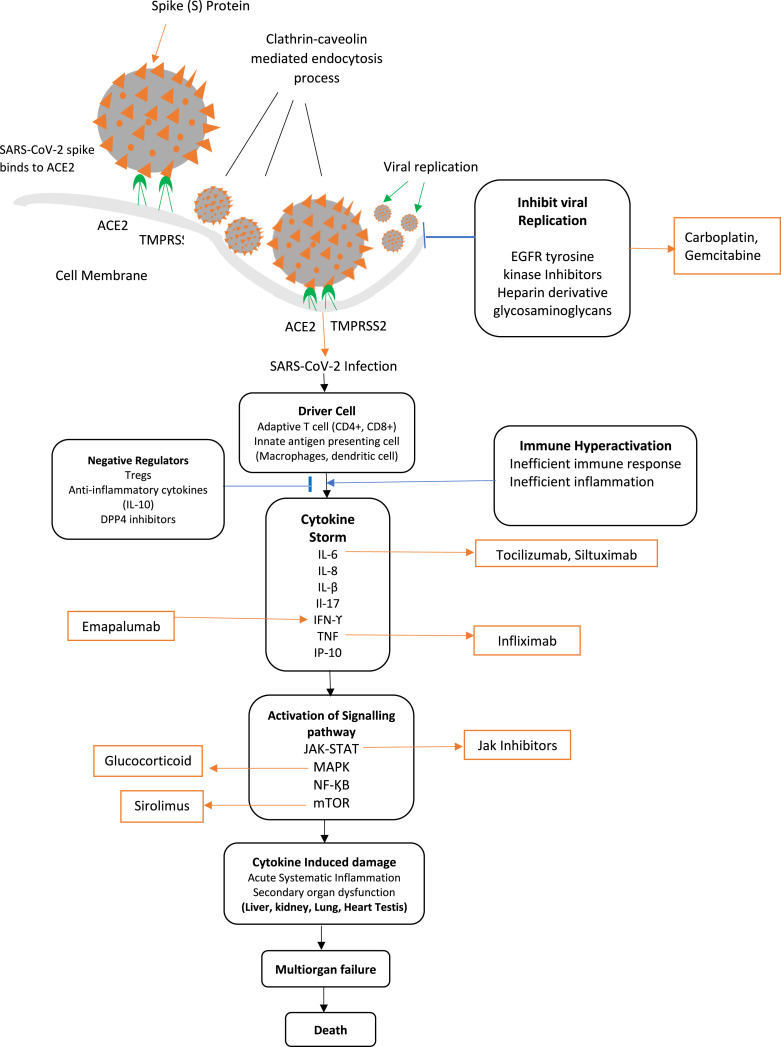
During India’s second wave of COVID-19 infection, children developed a better immune response against SARS-CoV-2 infection than adults. So, children experience much mild symptoms and are less affected by SARS-CoV-2 disease. In children’s nasopharyngeal samples, expression of progenitor Fc Receptor-Like 6 (FCRL6) is detected in the B cell. The B cell differentiates during early fetal development and produces “natural” antibodies to neutralize the invading pathogens in children. SLAMF1 positively regulate the pathway of B-1 cell to make specific antigen against the SARS-CoV-2 virus.
-
•
The article establishes that SARS-CoV-2 virus requires a clathrin-dependent endocytosis process to insert the viral particle into the host cell membrane. SARS-CoV-2 virus penetrates through the endocytic membrane of the host cell to establish an infection. It is observed that the membrane proteins AP2A2, APLP1, DNM2, EPS15, EPN1, EPN2, LDLR, LY75, MRC2, SNX5 mediate the clathrin-dependent endocytosis process in SARS-CoV-2 infection. These membrane proteins formed clathrin-coated pits in the host cell’s cytoplasmic membrane. The roles of these membrane proteins in SARS-CoV-2 pathogenesis can be investigated in the future to find antiviral treatment.
2. Dataset
2.1. Data sources
The publicly available scRNA-Seq data of various human tissue are downloaded from Gene expression omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). The gene expression matrices of human tissue such as brain, adult and child nasopharyngeal count matrices, lung, upper respiratory tract, liver, heart, kidney, stomach, ileum, rectum, colon, human pancreas, adult testis, skin, spleen, and bone marrow are collected. The GEO accession No. and source of each scRNA-Seq data are provided in Table 1.
Adult and children’s upper airway transcriptional profiles are acquired as GEO accession no. GSE179277 [28]. The children’s nasopharyngeal gene expression data contains 38 samples with SARS-CoV-2, 11 samples are from other respiratory viruses, and 34 have no virus. The adult nasopharyngeal gene expression data contains 45 samples with SARS-CoV-2, 28 samples of other respiratory viruses, and 81 samples containing no virus. The upper respiratory tract is obtained as GEO accession no. GSE154564 [29]. The expression matrices of lung (GSE130148) tissue are obtained from four patients who died of lung parenchymal tumor [30]. Five samples of hepatic (or liver) donors are acquired under GSE115469 [31]. Healthy samples of the human heart are obtained as GSE106118 [32]. Samples of human pancreatic islets are collected from four human donors and acquired as GSE84133 [33]. The adult testicular samples are collected from three healthy men of reproductive age and acquired under GSE112013 [34]. The gastritis samples are collected from GSE134520 [35]. Epithelial cells of human ileum, colon and rectum are collected under GSE125970 [32]. Three samples of kidneys are obtained from healthy donors under GSE131685 [36]. Fresh skin samples are acquired from GSE119562 [37]. Six samples of spleen cells are acquired under GSE119562 [38]. Samples of human bone marrow donor cells are acquired from GSE119562 [39]. The expression matrix of human brain are acquired under GSE67835 [40].
Table 1.
Summary of single-cell RNA sequencing data used in this study.
2.2. Dataset preprocessing and analysis
The gene expression matrices of various human tissue generated from scRNA-Seq technology are considered for the experiment. The expression count matrix is of size, , where and . Each row represents a gene point, and the column corresponds to a cell or sample. The expression count matrix stores the number of RNA molecules detected within a sample in each gene. It records the feature count of every gene in each sample of the expression count matrix.
The following steps are executed to process the scRNA-Seq data and are summarized as follows:
-
•
Cell filtering and quality control: Low-quality cells, doublets, or multiple cells are filtered out. Firstly, all rows and columns with zero feature count are removed. The cell with a unique feature count of over 2500 or less than 200 is removed.
-
•
Normalization: The normalization process normalizes the feature expression count of each cell by the total expression count. It then multiplies the normalized value by a scale factor (10,000 default), and each feature count is transformed by applying log10.
-
•Feature selection: The following steps are executed to select variables subset of feature and summarized as follows:
-
–Initially, the mean and variance are calculated for each gene from the normalized count matrix.
-
–A curve is then fitted to predict the variance of each gene (i.e., independent variable) as a function of its mean (dependent variable). The fit gives the regularized estimate of variance given the mean of its feature.
-
–Given the expected variances, the following transformation is applied to standardize each feature count. The transformation is described as:
(1) Where is the standardized value of a feature k in a cell , is the raw value of a feature in a cell j, is the mean of feature , and is the standard deviation (or variance) of feature obtained from the mean–variance function. -
–The standardized variance is computed for all genes (i.e., row) across each cell (i.e., column). The feature is then ranked according to standardized variance value. A high standardized variance value shows that the feature is highly variable, and the top features are selected.
-
–
-
•
Scaling: Scaling is a linear transformation technique applied before reducing the data dimension. It shifts and scales the expression count of each gene such that the mean expression and variance across each cell is 0 and 1, respectively.
-
•
Dimension reduction: The principal component analysis (PCA) algorithm is executed on the normalized expression count matrix to reduce data dimension. The PCA algorithm transforms the large set of variables into a small group of variables, preserving the original information of the dataset.
Finally, the above preprocessing process transforms the high dimension scRNA-Seq data into a lower form suitable for applying the proposed clustering algorithm, as explained in the following section.
3. Proposed methodology
In this work, a variable-length solution vector acting as a search agent in a grey wolf population is implemented to detect the optimal clustering solution automatically. The main objective of the proposed clustering approach is: (1) To optimize multiple cluster validity indexes, (2) to detect the optimal cluster number and cluster center (3) Study the expression distribution pattern of each gene point to every other cluster at a time point.
In Fig. 1(A), the detailed step of preprocessing the expression count matrix of scRNA-Seq data is outlined. Fig. 1(B) explains the stages of the proposed fuzzy-based improved GWO clustering algorithm graphically. The downstream cluster analysis process and prediction of host receptor for causing SARS-CoV-2 infection are shown in Fig. 1(C).
Fig. 1.
A data flow diagram of the proposed Fuzzy based improved GWO clustering algorithm for predicting the potential host receptor for causing SARS-CoV-2 infection in human.
3.1. About grey wolf optimizer algorithm
GWO is a population-based meta-heuristic optimization algorithm that simulates the leadership and hunting mechanism of grey wolves. The social hierarchy of the grey wolf population is classified into four groups. The dominant wolf in the pack is the alpha (), followed by beta (), delta (), and omega () wolves. The main phase of the grey wolf hunting process is enumerated as follows:
-
•Encircling: During this process, the grey wolves surround the prey once its location is determined. The encircling operation of grey wolves is represented as:
(2) (3) Where and refer to the position of prey and the wolves at current iteration , gives the approximate distance between the target prey and grey wolves. refers to the probable position of a grey wolf at the next iteration. The coefficient vector and is defined as:(4) The component vector decreases linearly from [2,0] and . The component vector contains a random value from the range [0,2]. and are the vector generated randomly from the range [0, 1].
-
•Hunting: In the hunting process of the GWO algorithm, it is assumed that (best solution vector), , and have better knowledge of the location of the target prey. Therefore, the position of the best three search agent is saved and oblige the other search agents (including ) to update their position randomly around , , and in each iteration. Mathematically, the hunting operation of grey wolves is achieved using the following Eqs. (5)–(10).
(5) (6) (7) -
•
Attacking: The grey wolves finish the hunting process by attacking the prey. To mathematically simulate the exploitation phase of the grey wolf, the component vector is decreased from 2 to 0. When , grey wolves attack the prey (exploitation). But when, , grey wolves diverge from each other to find a better target (exploration). The component also favors the exploration process. When , the grey wolf repeatedly attacks the prey, and stops the attacking.
3.2. Proposed clustering algorithm
In the proposed clustering scheme, the GWO algorithm is assimilated with the evolutionary operators to balance the exploratory and exploitation phases of the classical GWO algorithm. The evolutionary search pattern of mutation and crossover operators of the DE algorithm is integrated into the classical GWO algorithm to avoid stagnation at the local optima during the optimization process. The evolutionary population dynamics (EPD) operation is executed on the grey wolf population to eliminate the worst search agent for the next generation. The proposed clustering approach is referred to as fuzzy-based improved GWO clustering algorithm in the paper.
The step adopted for fuzzy-based improved GWO clustering algorithm is explained below.
3.2.1. Population initialization and solution vector representation
Initially, the grey wolves population is composed of search agent. Each search agent represent a solution vector in the search space. Each solution vector in initialized with a set of distinct real numbers chosen randomly from the given dataset, where . The solution vector encodes number of possible cluster center. The minimum and maximum value of is 2 and , where and n is the total gene point in a given dataset. Now the possible number of is obtained as: where the gives a random integer value. Therefore, the possible number of cluster center that can be encoded in a solution vector is between 2 to .
Let a solution vector encodes cluster center in ‘’ dimensional search space, then the length of each solution vector is . The first d position represent the dimension of the first cluster center, the next d position represents the dimension of the second cluster center and so on. For example, in 4-d, a solution vector encodes the cluster center , and respectively.
After initializing each solution vector with a random cluster center, steps of the Fuzzy c-means (FCM) clustering algorithm [22], [21] are executed so that the centers get separated at the initial stage.
3.2.2. Performing steps of FCM algorithm
The FCM algorithm aims to partition the dataset into C fuzzy clusters. The fuzzy clustering algorithm optimizes the objective criteria and simultaneously updates the membership value and cluster centers of a gene point associated with a cluster [22], [21].
Suppose there are n finite gene point in a dataset X, and be the gene point in d-dimension, where . represents the th feature value of th gene point, where . Let denotes the set of C fuzzy clusters and gives the set of C cluster centers in d-dimension i.e, . Suppose gives the membership value of a gene point in cluster j and gives the Euclidean distance between the th gene point and th cluster center.
The FCM algorithm is composed of the following steps and is described as follows:
-
1.
The initial fuzzy membership matrix, is initialized according to the degree of membership constraint
-
2.At the current step , cluster center is calculated, with the membership matrix according to the Eq. (12):
(12) -
3.The fuzzy membership matrix is then updated using Eq. (13):
(13) -
4.
If then FCM algorithm terminate successfully otherwise it return from step (2). Here, the terminating criteria .
3.2.3. Computation of objective function
The fuzzy-based improved GWO clustering algorithm simultaneously minimizes the cluster validity indexes, namely the J measure and Xie-Beni (XB) index. The objective functions are described as follows:
-
•
J measure:
measure [21] gives the variance within the cluster. It is defined as:(14) gives the Euclidean distance of th data point from th cluster center. A low value of measure results in a compact cluster. Thus fuzzy-based improved GWO clustering algorithm aims to minimize the measure.
-
•
XB index:
XB index [41] is the ratio of fuzzy compaction to its cluster separation.(15) The goal of the XB objective function is to minimize the numerator (i.e., compactness of fuzzy partition) and maximize the separation between the clusters. Thus, the fuzzy-based improved GWO clustering algorithm tries to minimize the XB index.
3.2.4. GWO algorithm with evolutionary operators
This subsection discusses how the GWO algorithm is incorporated with mutation and crossover operators of the DE algorithm. The main reason for choosing the DE algorithm is because it is easy to transform the continuous problem structure of the GWO algorithm into a combinatorial problem. Incorporating mutation and crossover operators in the GWO algorithm balances the exploratory and exploitative search mechanism. In turn, the hybridization process will produce a more stable recombinant offspring between different hierarchies or levels of the grey wolves. A detailed description of the process of hybridizing the mutation and crossover operators of the DE algorithm with the GWO algorithm is explained below:
-
•Mutation operation: In the DE process, a mutant vector is created for every solution vector in the population. The mutant vector is obtained by taking the difference between any two parent or solution vectors and multiplying it with a scaling factor F. The resulting term is then added to another third solution vector. The mutant vector of DE is generated using the following equation:
(16) Where , , express the index number in the scope from the current solution vector index and . F [0, 1] control the scaling of the two differential vector and is the mutant vector produced at next iteration.
In the mutation process of the fuzzy-based improved GWO algorithm, and wolves are chosen as the two target parents and combined with wolves to introduce variation in the population. So the mutation process is achieved using the following equation:(17) The variation factor F balances the exploration and exploitation process of the improved GWO algorithm. The variation factor is defined as:(18) Where and represent the minimum and maximum value of the scaling factor, . and denotes the maximum iteration and current iteration of the fuzzy-based improved GWO algorithm.
From Eq. (18), it is observed that is large in the beginning stage of the improved GWO algorithm. It enhances the exploration capability of the GWO algorithm, thereby preventing it from falling into the local optima. As the improved algorithm continues to iterate, the variation factor F decreases to improve the exploitation ability and prevent premature convergence.
-
•
Crossover operation: The crossover operation aims to introduce diversity in the population. The crossover operation is performed with the mutant offspring and current search agent to generate a recombinant search agent . For the crossover operation to achieve, the crossover probability factor (CR) determines the index whose value should be copied from the mutant offspring or current search agent.
The crossover operation is achieved using the following equation:(19) Where and . The function generates a uniform random number from the range [0, 1], CR gives the crossover or recombination probability from the range of and returns any index from the range . gives the real value present at the th index of current search agent.
From Eq. (19), it is observed that if value is large, the mutant offspring contribute more to the generation of recombinant search agent. When but if , current search agent contribute more to the generation of recombinant search agent.
-
•
Selection operation: The mutation and crossover operations are performed for all the search agent to generate a recombinant search agent for the new population. The objective function value ( measure and XB index) are calculated for all the recombinant search agent in the new population. All the search agent in the old and new populations are combined to perform the selection operation. The best search agent are selected from the combined population, while the rest search agent are discarded in the next iteration. The selection operation is performed using the non-dominated sorting and crowding distance operator of the Non-dominated sorting genetic algorithm (NSGA-II) [42].
The non-dominated sorting approach [42] divides all the search agents in the population into different non-domination levels. It distributes the search agent into R-different Pareto front such that
contains a higher-ranked search agent with assigned rank 1, and contains a lower-ranked search agent. The top-ranked search agent is selected to fill the position of the new population for the next iteration. This process is continued until search agents up to rank r are copied into the new population. Then the remaining number of positions the search agent can fill in the new population is determined as:
Where denotes the set of search agents at rank p, and P is the total number of search agents in the population. If , it means that all the search agent at rank cannot be added to the new population. Now to select only the exact number of search agents from rank that can be filled up in the remaining position, the crowding distance (CD) operator [42] is executed on the set of search agents at rank . The CD operator first sorts the search agent of rank according to each m objective value (m = 2). It then computes the crowding distance value of each search agent. Thus the remaining search agent is chosen based on lower rank and least CD value. In this manner, P best search agents are selected for the next iteration.(20)
3.2.5. Eliminating worst search agent from the population
The concept of EPD is to eliminate the weak search agent from the population. EPD promotes exploration of the GWO algorithm in a good search direction and resolves the problem of getting trapped at the local optima. EPD eliminate half of the worst solution vector and repositions them randomly around the best solution obtained so far [20].
To re-position the weak search agent around the location of , the following equation is used:
| (21) |
To re-position the weak search agent around the location of , the following equation is used:
| (22) |
To re-position the weak solution vector around the location of , the following equation is used:
| (23) |
To re-position the weak solution vector in a random position around the search space, the following equation is used:
| (24) |
Where ub and lb indicate the upper and lower bound of the search space, respectively, is a random number generated from the range of [0, 1].
The process of mutation, crossover, selection, and elimination of weak individuals from the grey wolves population continues for many iteration. At the end of the iteration, a set of search agent are generated on the final Pareto front. The final position of wolf give the optimal cluster number and cluster center for the clustering purpose.
3.3. Cluster analysis process
The downstream clustering analysis process begins by finding a set of differentially expressed genes (DEGs) for each scRNA-Seq dataset. The DEGs help annotate each cluster to a cell type from the published work. Finally, the DEGs are utilized further to study the biological process and their role in the pathogenesis of SARS-CoV-2 illness.
3.3.1. Annotation of cell clusters
The developed clustering method can determine the correct class label of a gene point and assign it to a proper cluster group. The t-test statistic is used to compare the mean of a group (cluster) but at different times. The t-test statistic is calculated as:
| (25) |
Where is the mean of a cluster group, give the mean hypothesis, gives the standard deviation and is the total gene point in a cluster,
A -value gives the probability value to indicate that the result from the experiment (or a sample group or cluster) occurred by chance. A low -value suggests that the data do not happen by chance and is valid data. The function
is used to compute the -value of each corresponding t-score for each group of cluster. The function computes the right-tailed student’s t distribution taking two arguments, i.e., t-score and degree of freedom (). The -value is then adjusted using the Benjamini Hochberg method to decrease the false discovery rate (FDR) [43].
The DEGs or gene markers are identified for each group cluster. The marker genes are determined using the criteria: A gene is said to be up-regulated if its -value is less than 0.05 and fold change greater than 2, whereas a gene is down-regulated if its -value is less than 0.05 and fold change greater than 0.5. Each cluster is then annotated to a known cell type based on the identified marker genes or DEGS.
3.4. Entire process
The following steps are adopted to predict the potential co-receptors of ACE2 protein that facilitate SARS-CoV-2 infection:
-
1.
An initial population of P search agents is created and represented as the solution vector.
-
2.
Step of FCM algorithm is executed on each solution vector to partition the given data into ‘C’ fuzzy cluster centers [22].
-
3.
For each solution vector in the population, the objective values [22] and XB [44] are determined.
-
4.
The non-dominated sorting and crowding distance operators of the NSGA-II algorithm [42] are executed to rank each solution vector in different non-domination levels.
-
5.
Save the best solution vector, second best solution vector and third best solution vector as position of alpha, beta and delta search agent. = position of , = position of , = position of
-
6.
Initialize control parameters: , and . Current iteration
-
7.
Update the position of the rest search agent according to Eqs. (5)–(11).
-
8.
Use , and position vector to perform mutation and crossover operation of DE algorithm according to Eqs. (16)–(19) [20].
-
9.
Update the position of , , and rest search agent.
-
10.
Eliminate half of the worst search agent and reposition them randomly around the position of , and [20].
-
11.
Update the control parameters and current iteration
-
12.
Repeat step (3–10) until the terminating criteria is satisfied.
-
13.
A set of solution vectors representing the position of search agents are generated in the final Pareto front.
-
14.
Return the position of i.e, as the optimal cluster center for clustering any scRNA-Seq data.
-
15.
Identify the gene marker or DEGs in each cluster group and annotate the cluster to cell types
-
16.
The gene sets (membrane protein or ssRNA viral receptor) that are transcriptionally similar to ACE2 protein are identified
-
17.
The expression distribution pattern of ACE2 and its co-receptor is studied.
-
18.
The target proteins that plays a crucial role in the pathogenesis mechanism of SARS-CoV-2 infection is reviewed through GO and KEGG pathway enrichment analysis
-
19.
The target protein is queried through the drug-gene interaction database, and its drug combination is identified.
4. Experiment
4.1. Experimental parameters
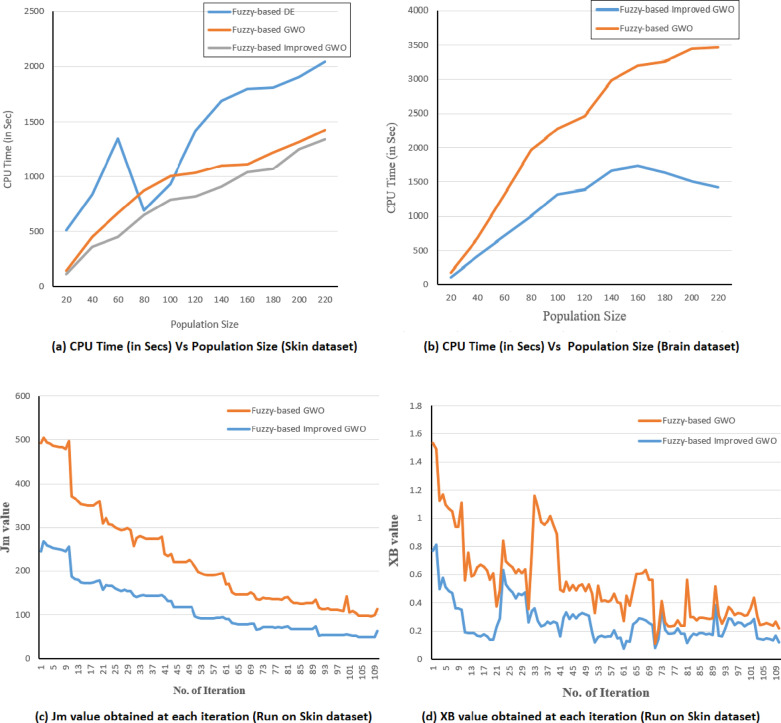
The proposed clustering algorithm (Fuzzy-based improved GWO) is implemented using Python 3.3 and runs on Spyder integrated development environment (IDE). All the experiments are conducted on an Intel Core i7 processor operating at 2.90 GHz and having 8.00 GB RAM under the Windows 10 platform. Other clustering techniques utilizing different optimization methods are also developed and tested on various scRNA-Seq data of human tissue.
The fuzzy-based DE clustering approach utilizes the DE optimization algorithm [45] and the fuzzy-based GWO clustering approach uses the classical GWO algorithm [18]. All the methods are based on the fuzzy clustering technique and simultaneously optimize measure and XB index. The required parameter values for executing fuzzy-based DE, fuzzy-based GWO, and the proposed fuzzy-based improved GWO clustering approaches are provided in Table 2.
Table 2.
The setting of control parameters used in developing the clustering approach.
| Clustering method | Control parameters |
|---|---|
| Fuzzy-based DE | F = 0.8, CR = 0.5, |
| Fuzzy-based GWO | , , |
| Fuzzy-based improved GWO | CR = 0.5, , , |
| , , | |
The entire code of the fuzzy clustering approach utilizing the classical GWO algorithm and proposed fuzzy-based improved GWO clustering approaches are uploaded to the github url as given: https://github.com/achomamika01/Metaheuristics_Fuzzy_based_Clustering-Algorithm
4.2. Performance metrics
The following cluster evaluation metrics are chosen to measure the goodness of the obtained gene cluster. It is described as follows:
-
•Silhouette Coefficient (SC): The SC [45] measures the closeness of each gene point in a cluster to the other gene point in the neighboring clusters. SC measures the average distance between each gene point within a cluster (a) and the average distance between all clusters (b). The silhouette value is calculated based on ‘a’ and ‘b’ parameters and is defined as:
(26) Now, the SC is calculated as the average silhouette values over all the gene points. The SC value varies from −1 to +1, and an SC closer to +1 signifies a better clustering result.
The SC can describe the performance of an entire population with a single value. So, we use the SC values to find the optimal cluster number in scRNA-Seq data.
-
•Calinski–Harabasz Index (CHI): CHI [45] measures the within-cluster dispersion (i.e., cohesion) and the dispersion between-cluster (i.e., separation). The cohesion is calculated based on the distances of the gene point in a cluster to its cluster center, and separation is estimated based on the distance of the cluster center from the global cluster center. Thus CHI is defined as:
(27) Here, and are the number of gene point and cluster center of th cluster respectively. is the best or global cluster center, n is the total gene point in a dataset, and C is the cluster number. A higher value of CHI means the clusters are well separated and dense.
-
•
Davies–Bouldin Index (DBI): DBI [45] is defined as the ratio of within-cluster distances to between-cluster distances. DBI maximizes the inter-cluster distance and minimizes the intra-cluster distance. DBI is based on the calculation of cohesion and separation values. Cohesion measures the closeness of a gene point in a cluster to the cluster center, while separation measures the distance between the cluster center (or centroid).
The cohesion value is calculated using the Sum of Square Within Cluster (SSW) equation, and the separation value is calculated using the Sum of Square Between Cluster (SSB) equation. SSW is defined as:
where = Number of gene point in cluster and = Distance between a gene point and cluster center .(28) SSB is defined as:
where = Distance between the cluster center and .(29) The ratio measures the similarity of cluster and cluster and is defined as:
Now, DBI is the average of the similarity measures of each cluster with a cluster most similar to it:(30) (31) It is observed from Eq. (31), lower the average similarity values, the gene clusters are well separated. Thus, a minimum value of DBI gives a good clustering solution.
4.3. Find ACE2 expression distribution pattern and identify its co-receptor
To identify the potential host receptor i.e, viral receptor/membrane protein of ACE2 that facilitate SARS-CoV-2 infection into human cells, membrane proteomes are extracted from the Membranome database of single-helix transmembrane proteins.1 Also, the viral receptor genes are downloaded from the Viral Receptor database.2 The viral receptor database comprises 332 interactions of mammalian virus–host receptors, including 142 unique viral species and 150 receptors. We only extract ssRNA viral receptor genes because coronaviruses are highly diverse positive sense ssRNA viruses.
The proposed fuzzy-based improved GWO clustering technique identifies gene sets (membrane protein/ssRNA viral receptors) showing similar expression patterns with ACE2 protein in all 16 human tissue. We then analyzed the tissue-specific expression pattern of ACE2 in 16 different human tissues. In single-cell gene expression data, each gene point is an element, and the vector of each gene corresponds to its expression pattern. The FCM algorithm aims to organize a group of genes having similar expression patterns in a cluster [21] . This means that genes in the same cluster are co-regulated and involved in the same biological function. The FCM algorithm can arrange genes showing more than one type of co-regulation nature into multiple clusters. So many authors have utilized the FCM algorithm to analyze the expression levels of thousands of genes at a time [46], [47]. In the FCM algorithm, each gene is associated with every cluster by a membership function. The membership function expresses the gene’s strength or degree of association with a particular cluster. Therefore, we have computed the membership degree of each gene point in a cluster using the above Eq. (13) of the FCM algorithm.
For example, ACE2 has a membership or expression value of 0.0136, 0.0010, 0.00028, 0.98209, 0.002927 when associated with the goblet, basal/suprabasal cells, tuft cell, ciliated cell, and neuroendocrine cell in the adult nasopharyngeal data. A particular gene is said to be potentially associated with a biological process or a cluster if its membership value, in Ref. [48] experiment. It can infer that ACE2 is associated more strongly with the adult nasopharynx’s ciliated cell (expression value 0.98).
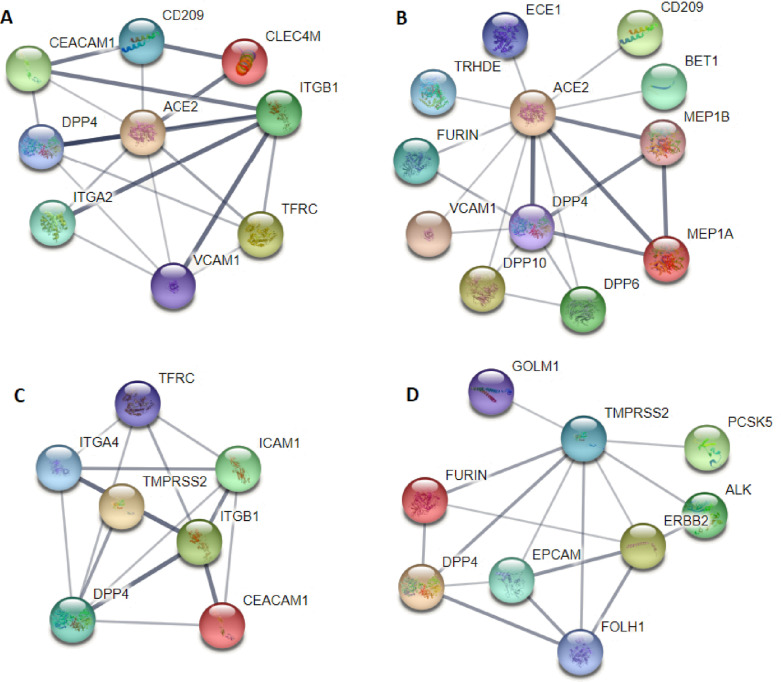
Further to validate the result of obtained clustering results, PCC is calculated between ACE2 protein and membrane protein or ssRNA viral receptor. It is observed that ACE2 is strongly correlated with viral receptors such as CD209, GPR78, ADAM17, NRP1, ICAM1, AXL, LDLR, EGFR, CLEC4M, and FCGRT, etc. (). While ADAM7, ADAM9, NRP1, NRG1, FCRL6, LRP1, FURIN, FGFR1, EFNB1, and CLDN1 are the top membrane protein strongly correlated with ACE2 protein. These may act as the potential co-receptor of ACE2 that facilitates the entry and binding mechanism of SARS-CoV-2 infection.
5. Result and discussion
5.1. Cell type identification and analysis of ACE2 expression pattern and role of host receptor in various human tissue
-
•
Adult nasopharyngeal dataset: Cells in the adult nasopharyngeal gene expression dataset are organized into five major clusters. Cluster 0 is annotated as goblet cell with the marker genes SYT8, GP2 and ANPEP. Cluster 1 is annotated as the basal or supra-basal cell with the marker genes NRP1, LDLR and ITGB1. Cluster 2 is annotated as the tuft cell using the canonical marker genes PTPRC and FXYD6. Cluster 3 is annotated as ciliated with the marker genes CDHR3 and SYT5. Cluster 4 is annotated as neuroendocrine cell with the marker genes SYT1 (as shown in Supplementary (Suppl) Fig. 1(A)).
ACE2 has an expression value 0.0136, 0.0010, 0.00028, 0.98209, 0.002927 when associated with the goblet, basal or suprabasal cells, tuft cell, ciliated cell, and neuroendocrine cell, respectively. Thus, ACE2 is associated more strongly with the adult nasopharynx’s ciliated cell (expression value 0.98).
SIGLEC1, CD209 and CLEC4M are the viral genes that are significantly correlated with ACE2 (PCC > 0.65) in adult nasopharynx. SIGLEC1 plays an important role in the antiviral and antibacterial host response to SARS-CoV-2 infection and HIV infection. Type I interferon is the key antiviral mediator of SARS-CoV-2 infection. Activation of type I interferon signaling increases the expression of SIGLEC1 on the circulating cell of monocyte and macrophages. CD209 and CLEC4M are also responsible for the autoimmune response in SARS-CoV-2 infection. NRCAM, MLN-4760, ADAM7, ASGR1 and TFR2 are the top membrane protein that are significantly correlated with ACE2 protein (). NRCAM protein is reported to induce an inflammatory response to SARS-CoV-2 infection. MLN-4760 protein binds to the enzymatic active site of ACE2 protein with high affinity and can alter the conformation of ACE2 protein. The increased activity of ADAM7 cleaves ACE2 ectodomain and other pro-inflammatory molecules, thereby reinforcing the inflammatory process during SARS-CoV-2 infection. ASGR1 neutralizes the antibodies targetting the S protein of ACE2. TFR acts as another receptor for SARS-CoV-2 infection entry and exerts significant antiviral effects. The expression profile of ACE2 and its co-receptor is shown in Suppl Fig. 1(B).
-
•
Child nasopharyngeal dataset: Cells are organized into five main clusters in child nasopharyngeal gene expression data. Cluster 0 is annotated as the neuroendocrine cell using the marker gene SYT1, and cluster 1 is annotated as the ciliated cell with the marker gene FMO3, SYT5. Cluster 2 is annotated as a goblet cell with the marker gene SYT8, GP2. Cluster 3 is annotated as a basal/suprabasal cell with the canonical marker gene PLS3, MET, OSMR, CLIC4. Cluster 4 is annotated as tuft cell with the marker gene PTPRC, SDC4 (as shown in Suppl Fig. 1(C)).
ACE2 protein has an expression value of 0.056, 0.188, 0.6860, 0.0530, and 0.0162 when associated with the neuroendocrine, ciliated, goblet, basal/suprabasal, and tuft cells. ACE2 exhibits both the nature of goblet and ciliated cells because the membership values in both the cell type are 0.188 and 0.6860. The expression profiles of ACE2 and its co-receptor are shown in Suppl Fig. 1(D).
CD209, DPP4 and GPR78 are the viral receptors strongly correlated with ACE2 protein. In Ref. [49], it is established that CD209 mediates entry of the SARS-CoV-2 infection through the heterodimerization process, and our work also confirms the same observation. DPP4 interacts with the S1 domain of the viral spike glycoprotein. DPP4 is suggested to be the alternate receptor of SARS-CoV-2 infection [2], [50]. GPR78 protein is mainly expressed in the upper airway cell and overexpressed in SARS-CoV-2 infection. During the infection, the SARS-CoV-2 virus migrates to the GPR78 cell surface and promotes viral entry. The virus-induced endoplasmic reticulum (ER) stress increased the surface expression of GPR78 to enhance the viral entry. In turn, the viral infection sets up a positive feedback cycle and hijacks the chaperone molecule for signaling multiple molecules acting as the co-receptor for viral entry.
GDF15, CD8B2, FCRL6 , CD244 and SLAMF1 are the membrane protein strongly correlate with ACE2 protein (). GDF15 modulates immunity in COVID-19 infection via its iron metabolism [51]. Expression profiles of host receptor protein are shown in (as shown in Suppl Fig. 1(D)). SLAMF1 and FCRL6 protein expression is detected in children’s nasopharyngeal swab samples. It is observed that children experience a milder clinical symptoms of COVID-19 disease than adults. This is because children can neutralize the antibody after the onset of infection [52]. CD244 and SLAMF1 are present in activated B and cell and responsible for signal transduction and viral entry. FCRL6 acts as the major histocompatibility complex II receptor (MHC II) for mediating viral entry. SLAMF1 positively regulates the production antigen (Ag) specific immune response in the B cell of children. The above observance proves that children with neutralizing antibodies have a lower viral load count and faster virus clearance.
-
•
Upper respiratory tract: Four major cell types are detected in the upper respiratory tract (nose and oropharynx): ciliated cells, non-ciliated secretory cells, basal or suprabasal cells, and goblet cells. Cluster 0 is annotated as non-ciliated secretory cell with the marker genes SRPRB and CNMD. Cluster 1 is annotated as basal cell with the marker genes MEGF9, APMAP, KRT5 and PCDH7. Cluster 2 is annotated as goblet cell using the marker genes CXCL10, IDO1, SLC26A4 and ANPEP. Cluster 3 is annotated as multi-ciliated cell with the marker genes CXCL13, CCDC78, SCGB3A1. The cell cluster are shown in Suppl Fig. 1(E).
From the result, it is observed that ACE2 has expressive values 0.3796, 0.067, 0.091, 0.489 associated with non-ciliated cell secretory, basal cell, goblet, and ciliated cell. This means that ACE2 protein is found to express in all the cell types in some extent because . But it is observed that ACE2 is highly expressed in the ciliated cell compared to secretory and goblet cells. AXL, EGFR, FCGRT, LDLR, KREMEN1 and ASGR1 are the viral receptors coexpressed with ACE2 protein in the ciliated cell of upper airway. The protein receptor ASGR1 and KREMEN1 are found to co-expressed with ACE2 protein in the non-ciliated cell (secretory cell). This means the relative expression of ACE2, ASGR1 and KREMEN1 is much higher in SARS-CoV-2 infected cell than the uninfected cell. ASGR1 and KREMEN1 specific antibodies can block the binding and entry mechanism of SARS-CoV-2 S protein into the cell and reduce the spread of infection in lung organoids [53]. From our result, the AXL protein significantly correlates with ACE2 protein () and promotes viral infection and reproduction in the upper respiratory system [54]. Through experiment model and cell culture, it is observed that SARS-CoV-2 infection-induced pulmonary infection in older patients due to hyperactive response to lung injury mediated by epidermal growth factor receptor (EGFR) signaling. The reason for the activation of EGFR signaling is the release of ligands such as epigen (EGN), heparin-binding EGF-like growth factor (HB-EGF), amphi-regulin (AREG), epiregulin (EREG), etc from the damaged cell to bind EGFR and activate the would healing response in COVID-19 patients [55].
-
•
Lung dataset: A total of 13 primary cluster cells are detected in human lungs. The cell clusters are annotated as: Cluster 0 is annotated as basal cell using the marker genes ISLR, SNCA and PCDH7. Cluster 1 is annotated as endothelial cell using the marker genes ANXA3, CALCRL and FOXF1. Cluster 2 is annotated as alveolar type 1 (AT1) cell using the canonical marker genes AGER, MYRF, PDPN. Cluster 3 is annotated as B cell using the marker genes CD14 and CD4. Cluster 4 is annotated as alveolar type 2 (AT2) cell using the marker genes TCF7L2, CYP4B1, LRP5. Cluster 5 is annotated as the smooth muscle cell using the marker genes ACTA2. Cluster 6 is annotated as ciliated cell using the canonical marker genes SERPINB4, PDZK1IP1 and KRT4. Cluster 7 is annotated as mesothelial cell using the marker genes PLXNA1 and PLXNA2. Cluster 8 is annotated as dendritic cell using the canonical marker genes VEGFA, EREG, IGSF21 and APOE. Cluster 9 is annotated as NK and cell using the marker genes CD11B, CD56 and CD45RO. Cluster 10 is annotated as pericytes using the marker genes ACTA2, TAGLN and COL1A2. Cluster 11 is annotated as macrophages from the marker genes FABP4 and MCEMP1. Cluster 12 is annotated as fibroblast or stromal cell using the marker genes PGS5, TAGLN and MYH11.
ACE2 protein has an expressive value of 0.0203, 0.0387, 0.2171, 0.0947, 0.1906, 0.0053, 0.1065, 0.0377, 0.1767, 0.0717, 0.0120, 0.0077, 0.0203 associated to basal cell, endothelial cell, AT1 cell, B cell, AT2, smooth muscle cell, ciliated cell, mesothelial cell, myeloid and dendritic cell, NK and cell, pericytes, macro-phages, and fibroblast cell respectively. ANPEP, DPP4, CD209, EGFR, MMP14 are the viral receptors co-expressed with ACE2 protein in AT2 cell of human lung. The expression profile of ACE2 and its co-receptor protein is shown in Suppl Fig. 1(F).
The coronaviruses use peptidases such as ANPEP and DPP4 to enter host cells [2], [56]. ACE2 interacts with DPP4 and ANPEP peptidases in AT2 cell (). CD209L is seen to co-expressed with ACE2 protein in AT2 cells. CD209L and CD209 interact with the receptor-binding domain (RBD) of ACE2 protein and mediates viral entry into human cells. EGFR receptor enhances the spread of the SARS-CoV-2 infection by stimulating cell motility. SARS-CoV-2 activates the epidermal growth factor receptor (EGFR), leading to the suppression of interferon regulating factor 1 (IFR1) dependent interferon () and decreased antiviral defense in the upper airway epithelial cell. Matrix metalloproteinases (MMPs) play a key role in lung immunity against SARS-CoV-2 infection by facilitating inflammatory cell influx and modulating the chemokines and cytokines signaling pathway.
-
•
Heart dataset: A total of 10 cell clusters are obtained in the human heart. The cell clusters are annotated as follows: cluster 0 is annotated as endothelial cell using the marker genes PDPN, cluster 1 is annotated as atrial cardiomyocytes using the marker gene ALDH1A2, cluster 2 is annotated as ventricular cardiomyocytes using the marker gene MYH2 and MYH7, cluster 3 is annotated as macrophages using the marker genes MRC1, cluster 4 is annotated as the pericytes using the marker genes ABCC9, cluster 5 is annotated as the adipocytes using the marker genes LAMA2, cluster 6 is annotated as fibroblast following the marker gene CD63, cluster 7 is annotated as mesothelial cell using the marker gene VT1, cluster 8 is annotated as immune cell using the prominent marker genes ICAM1, cluster 9 is annotated as neuronal cell using the marker genes NRXN1 and PLP1 (as shown in Suppl Fig. 2(A)).
ACE2 has an expression value 0.0014, 0.0228, 0.8634, 0.0154, 0.02162, 0.0138, 0.0039, 0.0079, 0.0069, 0.0423 when associated to endothelial cell, atrial cardiomyocytes, ventricular cardiomyocytes, macrophages, pericytes, adipocytes, fibroblast, mesothelial cell, immune cell and neuronal cell (shown in Suppl Fig. 1(B)).
The membrane protein ADAM9, VCAM1, ICAM1, ERBB2, NRG1 and ERAP1 coexpressed with ACE2 protein in cardiomyocytes cell. ADAM9 mediates the entry of the encephalomyocarditis (EMCV) virus. EMCV virus is associated with myocarditis and encephalitis. EMCV infection causes acute myocarditis due to a direct infection in cardiomyocytes cell by the SARS-CoV-2 virus. The comorbidities caused an imbalance in the renin-angiotensin system (RAS) mediated by the interaction between ACE2 protein and ADAM, along with some factors associated with TMPRSS2 expression. ERAP1 and ERAP2 are the key regulator of RAS and a key component of the MHC class I antigen processing system. Because of their involvement in RAS, the dysfunction of the ERAP1 enzyme exacerbate the effect of SARS-CoV-2 infection.
-
•
Testis dataset: A total of seven cell clusters are detected in adult male testis. Cluster 0 is annotated as a myoid cell using the marker gene ACTA2, VIM. Cluster 1 is annotated as Sertoli cell using the marker gene RHOX8, APOA1. Cluster 2 is annotated as spermatid cell using the marker gene SPAG6, ZPBP. Cluster 3 is annotated as germ cell using the marker genes ID4. Cluster 4 is annotated as Leydig cell using the marker genes CYP11A1, VIM. Cluster 5 is annotated as spermatogonial stem cell (SSC) using the marker gene NEUROG3, ID4. Cluster 6 is annotated as spermatogonia (SPG) cell cluster using the marker genes MAGEA4, KIT. The cell clusters are shown in Suppl Fig. 2(C).
ACE2 is associated with the myoid cell, Sertoli cell, spermatids, germ cell, Leydig cell, SSC, and SPG cell with an expression value of 0.0556, 0.2940, 0.0341, 0.1653, 0.0351, 0.2858, 0.1297 respectively. ACE2 is highly expressed in spermatid, Leydig cells, SPG, and SSC. GGT5, GT7, JAG2, JAM2, PLD6, SPAG4, SPEM1, SGPL1, AXL, BAX, KIT, MERTK, ROS1, SUN5, TYRO3, CADM1, GGT1 are the potential co-receptor of ACE2 in testis. The expression profiles of ACE2 and its co-receptor are shown in Suppl Fig. 2(D).
Testicular damage is one of the clinical damage caused by SARS-CoV-2 infection. The main reason for the testicular damage is the direct invasion of ACE2 receptors into the testicular tissue. This is due to a persistent rise in temperature, other secondary inflammation such as autoimmune response, and unexpected side effects such as steroid and oxidative stress from COVID-19 medications. Infertility in males may be the possible long-term effect of COVID-19 infection.
-
•
Liver dataset:
Six cell clusters are detected in the human liver. Cluster 0 is annotated as hepatocyte cell with the corresponding marker genes CYP1A2, JUN. Cluster 1 is annotated as cholangiocyte cell with the marker genes EPCAM, ONECUT1. Cluster 2 is annotated as an endothelial cell using the marker genes CLEC14A and SPARCL1. Cluster 3 is annotated as hepatic stellate cell with the marker genes BAMBI, CSF1, HEXIM1. Cluster 4 is annotated as macro-phage from the marker genes HMOX1, MERTK, and MS4A7. Cluster 5 is annotated as lymphoid cell following the marker genes CD8A, IL7R. The cell cluster are shown in Suppl Fig. 2(E).
ACE2 has an expression value 0.4636, 0.1032, 0.0289, 0.3899, 0.0037, and 0.0105 when associated with hepatocyte cell, cholangiocyte cell, endothelial cell, hepatic stellate cell, macrophage, and lymphoid cell. A high expression value of ACE2 in hepatocyte cells (0.466) indicates that it is the leading site of infection of SAR-CoV-2 infection. CEACAM1, IGF1R, BAX, LEPR, INSR, XBP1, LRP5, HFE, MET, COX7B, COX8C, COX8A, CADM1, CLDN1 are the possible co-receptor of ACE2 and TMPRSS2 membrane protein. The expression profiles of ACE2 and its co-receptor protein are shown in Suppl Fig. 2(F).
There is a close association between SARS-CoV-2 infection and liver disease. Liver injury, chronic liver disease (CLD), liver cirrhosis, inflammation, and viral hepatitis are the possible outcome of COVID-19 illness. An elevated level of alanine aminotransferase (ALT), gamma-glutamyltransferase (GGT), and aspartate aminotransferase (AST) as an impact of cytokine storm could damage the liver and produce more inflammation. Elevation of AST and GGT cause ischemia and liver cirrhosis and has been associated with cytokine-mediated injury. Tocilizumab is now approved to treat severe lung injury in COVID-19 disease.
-
•
Kidney dataset: In the kidney organoid, six main clusters are identified. Cluster 0 is annotated as distal tubule cell using the marker genes GATA3 and EGF, cluster 1 is annotated as glomerular parietal epithelial cell using the marker gene PECAM1 and PDGFRB, cluster 2 is annotated as immune cell using the marker gene IL1RL1, cluster 3 is annotated as collecting duct principal cell using the marker gene KCNE1, cluster 4 is annotated as proximal tubule cell, using the marker gene SLC22A8, CUBN. Cluster 5 is annotated as collecting duct intercalated cell using the marker gene SLC26A7 and FOXL1. The cell cluster are shown in Suppl Fig. 4(A)).
ACE2 has an expression value of 0.00344, 0.03650, 0.00828, 0.01373, 0.9081, and 0.02984 associated with distal tubule cell, glomerular parietal epithelial cell, immune cell, collecting principal duct cell, proximal tubule cell and collecting duct intercalated cell. ACE2 expression is primarily expressive in proximal tubule cell (shown in Suppl Fig. 4(B)).
ICAM1, CX3CR1 and CD81 are the viral receptors correlated with ACE2 protein. Acute renal ischemic injury is one of the common features observed in comorbidities of COVID-19 patients. Ischemic injury upregulates proinflammatory mediators such as cytokines and arachidonic acid metabolism. This increases the expression of CD11/CD81 on leukocytes and ICAM1 on endothelial cells. A monoclonal antibody directed against ICAM1 prevents functional impairment of renal failure. Acute renal ischemic is characterized by loss of renal function and accumulation of end product of nitrogen metabolism. Several inflammatory responses, such as chemokines, promote the recruitment of immune cells to the injured kidney. The chemokines receptor CX3CR1 recruits monocyte or macrophage, induces chemotaxis towards kidney tissue damage, and initiates the repair process. The exosomes rich in tetraspanins (CD9, CD63, and CD81) and heat shock and Rab proteins act as a shuttle to transfer biologically active proteins, lipids, and RNAs. The plasma from COVID-19 recovered exosomes reproduce molecular patterns to develop immune responses and activate coagulation and complement pathways in the damaged tissue.
LRP1, NRP1, JAG1 and NOTCH2 are the membrane protein strongly correlated with ACE2 in renal proximal tubular cells. SARS-CoV-2 infection initiates cytokine storms in renal proximal tubule cells and activates multiple genetic programs leading to kidney dysfunction. Acute kidney injury has been the main cause of cytokine storms. It is reported in article [57] that type I interferon lead to renal damage after acute kidney injury. The type I interferons upregulates interleukins (IL), toll-like receptors (TLR2, TLR4), interferon regulatory factors (IRF1, IRF7, IRF9), interferon-induced proteins (IFIT1, IFIT2, IFIT3, IFI44), and chemoattractants (CXCL10, CXCL11) enhancing ACE2 protein expression. To counteract the effect, Interferon (IFN) simulate immune response through JAK/STAT pathway in COVID-19 patient. LRP1 or CD91 membrane protein is responsible for initiating cell migration, proliferation and differentiation process. It also regulates multiple immune signaling pathways such as JAK/STAT and ERK1/ERK2 in renal COVID-19 patients. NRP1 is highly expressed in diabetic kidney patients with podocytes cells. A strong correlation of ACE2 protein with NRP1 suggests the increased risk of COVID-19 and the development of diabetic nephropathy disease condition. The research article [58] suggests that notch signaling in renal tubular epithelial cells (RTECS) induces the development of fibrosis in the kidney. JAG1 and NOTCH2 are significantly correlated with ACE2 protein in renal tubule cell. JAG1 along with NOTCH2 reprogrammed the metabolic activity of RTEC via mitochondrial transcription factor A (TFAM). It results in cell proliferation, differentiation, and ultimately developed fibrosis in RTECS [58]. Ischemic acute renal failure is one of the common effects of SARS-CoV-2 infection. Expression profiles of some ACE2 co-receptor proteins are shown in Fig. 8(B).
-
•
Pancreas dataset: A total of eight-cell clusters are detected in the pancreas. The cell clusters are alpha, beta, delta, epsilon, pancreatic polypeptide, acinar, ductal, and endothelial cells. The cell cluster are shown in Fig. 3(C).
The studies in Ref. [59] show a close association between SARS-CoV-2 infection and the development of diabetes. SARS-CoV-2 infection induces pancreatic cell death through several mechanisms such as programmed cell death, inflammation, autoimmunity against cell, direct cell lysis, etc. The receptor proteins DPP4, NRP1 and HMGB1 along with ACE2 protein facilitates SARS-CoV-2 viral entry in cell. Type 2 diabetes mellitus (T2DM) develops due to cell dysfunction in the presence of insulin resistance. DPP4 plays a significant role in glucose metabolism, neuropeptide, and cytokine activity. DPP4 inhibitors could reduce the severity of COVID-19 disease and prevent lung inflammation and injury.
NRP1 acts as the co-receptor that enhance SARS-CoV-2 virus infectivity when co-expressed with ACE2 protein. SARS-CoV-2 uses spike protein (S) to facilitate cell entry, and its cleavage allows attachment to NRP1 membrane protein. Therefore, tissue with increased NRP1 expression levels may raise infection risk. NRP1 exists in two isoforms: one is secreted form of NRP1 (sNRP1), and the other is the transmembrane form that interacts with SARS-CoV-2 infection. sNRP1 inhibits the interaction of vascular endothelial growth factor A (VEGF-A) or other growth factors with some specific receptor and membrane protein NRP1. NRP1 interacts with RAS to protect from hypertension-induced angiotensin II. T2DM is a feature associated with severe SARS-Cov-2 infection and acute respiratory distress syndrome (ARDS). Diabetic patients have overactive RAS due to increased ACE2 expression in the kidney. Thus, activating RAS increases the levels of sNRP1 and its associated ligands (VEGF-A) in hypoglycemia T2DM patients hospitalized with COVID-19. The expression profiles and ACE2 and its co-receptor are shown in Fig. 3(D).
-
•
Gastrointestinal tract: Stomach, Ileum, colon and rectum
The SARS-CoV-2 virus replicates inside the gastrointestinal tract cell inferring the intestine as the main site of SARS-CoV-2 infection. It is observed that CD147 or basigin correlates strongly with ACE2 protein in the intestinal epithelial cell. An increased expression of ACE2 and CD147 damage the vascular endothelium and cause thrombosis in COVID-19 patients. The SARS-CoV-2 infection elevates vascular endothelial growth factor (VEGF) and its receptor VEGFR-1 and VEGFR-2 in COVID-19 patients. VEGF supplies adequate oxygen and nutrient to the gastrointestinal tissue and removes its metabolic toxins. Elevated serum VEGF level is seen in COVID-19 patients with intestinal edema. SAR-CoV-2 spike protein promotes VEGF production by activating mitogen-activated protein kinases (MAPK) or extracellular signal-regulated kinase 1/2 (ERK) signaling in enterocytes cell and induces permeability and inflammation. The ERK/VEGF pathway blockage reduces intestinal inflammation and allows vascular permeability.
Besides ACE2 and CD147, NRP1 also promotes the entry of SARS-CoV-2 infection into the gastric cell. NRP1 is critical in tumor progression, cell invasion, migration, and angiogenesis. NRP1 promotes tumor angiogenesis of gastric cancer by interacting with VEGF and its receptor. In the tumor microenvironment (TME), tumor cells interact with immune cells, stromal cells, and fibroblast cells, providing an environment of tumor immune escape, resulting in malignancies. In TME of gastric cancer, macrophages produce a variety of cytokines, proteases, and growth factors to regulate tumor immunity. In addition to macrophages, the regulatory (Treg) cell acts on the innate immune cell to suppress immune responses by secreting cytokines and TGF-. Thus, NRP1 could work as a prognostic marker in gastric cancer by predicting the infiltration of Treg cells and macrophages. The cell cluster and expression profile of ACE2 and its co-receptor in stomach are shown in Suppl Fig. 3(A) and (B).
-
•
Skin dataset: A total of seven cell clusters are detected in skin tissue, and the obtained cluster are shown in Suppl Fig. 3(E).
A few cases of cutaneous manifestations have been reported as the outcome of COVID-19 disease. Few cutaneous symptoms arising from COVID-19 disease are atopic dermatitis, urticarial eruptions, acral ischaemia, retiform purpura, papular dermatoses, etc [60].
ADAM17, GPR78, CD147, CD209, DPP4 are the receptors that manifest skin infection as a consequence of COVID-19 disease. ADAM17 is important in skin protection and acts as an intestinal barrier during adulthood. ADAM17 cleaves the ectodomain of transmembrane protein such as heparin-binding epidermal growth factor (HB-EGF). Because of that, it activates EGFR and promotes cell proliferation. ADAM17 encourages the shedding of ACE2 receptors from the membrane to cytosol forming soluble ACE2 (sACE2). sACE2 potentially blocks the spike protein and protects from cell infection [60]. Recently, a patient with a homozygous loss of function mutation of the ADAM17 gene presented with repeated skin infections. The gene ADAM17 depends on rhomboid-related protein 1 (RHBDL1/RHBDL2) for maturation and functioning. A slight mutation in RHBDL2 causes tylosis, a rare hereditary disorder characterized by hyperkeratosis of the palms and soles. Keratinocytes sample from this patient is characterized by EGFR signaling, which is not detected in a normal person [61].
During the second wave of the COVID-19 pandemic, many incidences of mucormycosis as a result of post-COVID-19 symptoms rises in India. Mucormycosis is a life-threatening fungal infection caused by Rhizopus oryzae. The factor that caused COVID-19-associated mucormycosis is the injudicious use of steroids in hyperglycemia patients with a history of glucocorticoid therapy. Mucorales use GPR78 as a host receptor to enter the endothelial cell, and tissues [62], [63]. Studies conducted in Ref. [63] show an interaction between the receptor binding domain (RBD) of SARS-CoV-2 spike protein and GPR78. SARS-CoV-2 viral infection induces endoplasmic reticulum (ER) stress by accumulating excess unfolded protein in the ER lumen and activating unfolded protein response (UPR) signaling pathway. The UPR pathway upregulates the GPR78 synthesis process to overcome the unfolded protein. In this situation, GPR78 is exported out oF ER lumen and expressed on the cell surface. The increased GPR78 expression enhances the viral entry by positive feedback cycle [62], [63].
The expression profile of ACE2 and its co-receptor are shown in Suppl Fig. 3(F).
-
•
Lymphatic tissue: Bone marrow
Eight cell populations are detected in the human bone marrow. The cell clusters are B cell, NK/NKT cell, erythrocytes, hematopoietic stem cells (HSCs), endothelial progenitor cells (EPCs), monocytes, dendritic cells, and myeloid cells. It is shown in Suppl Fig. 4(E).
ACE2 is expressed in bone marrow-derived in HSCs and EPCs. This shows that SARS-Cov-2 infection infects and damages the stem cell. It is observed from the result that NRP1 is strongly correlated with ACE2 protein in an immune cell derived from macrophages. NRP1 mediates SARS-CoV-2 infection in bone marrow-derived macrophages (BMMs). The entry of SARS-CoV-2 infection on BMMs depends on the expression of NRP1 rather than ACE2 expression. SARS-CoV-2 infection hinders the differentiation process of BMM to osteoclast. COVID-19 disease is associated with a disorder in calcium metabolism and osteoporosis. Severe COVID-19 patients have lower blood calcium and phosphorous levels than moderate COVID-19 patients. Approaches such as the knockdown of NRP1 expression or blockage of NRP1 expression can inhibit SARS-CoV-2 infection in BMMs [64]. A recent study in Ref. [65] observed that SARS-CoV-2 envelope protein activates NLRP3 inflammasome, thereby inducing interleukin-1 (IL-1) secretion. IL-1 induces an inflammatory response by activating nuclear factor-B (NF-B) and the c-Jun N-terminal kinase signaling pathway. As a result, many cytokines are released in acute inflammatory disease and are associated with more severity in COVID-19 patients. The expression profile of some ACE2 co-receptors is plotted in the form of a dot matrix and is shown in Suppl Fig. 4(F).
-
•
Brain dataset: A total of seven cell clusters are detected in the human brain. Cluster 0 is annotated as the astrocyte cell with the prominent marker genes FGFR3. Cluster 1 is annotated as the microglial cell with the known marker genes CSF1R and CD83. Cluster 2 is annotated as neurons using the marker genes SLC10A4, C14ORF37. Cluster 3 is annotated as the oligodendrocyte precursor cell (OPC) using the prominent marker genes MEGF11. Cluster 4 is annotated as the vascular cell using the marker genes GRM8, TRPM3. Cluster 5 is annotated as oligodendrocytes with the canonical marker genes MAG. Cluster 6 is annotated as endothelial cell using the marker genes TM4SF1, ICAM1, VCAM1. The cell clusters are shown in Suppl Fig. 4(C).
ACE2 protein is detected mainly in the astrocytes and microglial cells to a small extent. This means astrocytes and microglial cells both express ACE2 protein. The receptor responsible for causing SARS-CoV-2 infection are co-expressed in the astrocyte and microglial cells [66]. ACE2 is associated with astrocyte cell, microglial cell, neuron cell, oligodendrocytes precursor cell, vascular pericytes cell, oligodendrocytes and endothelial cell with an expression value 0.9912, 0.0039, 0.0007, 0.0008, 0.00034, 0.0019, 0.0009 respectively. A high ACE2 expression value shows that the SARS-CoV-2 virus initially infects the astrocytes cell after crossing the blood–brain barrier and impairs neuronal viability. A similar kind of observation has also been reported in Ref. [67]. Besides, ICAM1, VCAM1, DAG1, LDLR and MXRA8 are the ssRNA receptors correlated significantly with ACE2 protein in human brain cell.
Endothelial cells (EC) are the primary site of leukocyte trafficking from the circulating blood into the areas of infection and inflammation. During SARS-CoV-2 infection, early cytokines response interleukin 1 receptor type 1 (IL1R1) and tumor necrosis factor-alpha (TNF-) initiates various kinase cascades and activates transcription molecules such as ICAM1, E-selectin, P-selectin and VCAM1. VCAM1 mediates the recruitment of monocytes to infection and injury sites. ICAM1 mediates the transmigration of monocytes and lymphocytes to active infection sites.
The membrane protein ADAM9, FGFR1, EFNB1, NRP1, FURIN and CD147 co-expressed with ACE2 protein in astrocytes cell of the brain. ADAM9 and FGFR1 facilitate the binding and genome translation of encephalomyocarditis virus (EMCV) to the cell surface. It is involved in inflammation and tumorigenesis and causes meningitis or encephalitis. EFNB1 initiates cell exhaustion during SARS-CoV-2 viral infection. SARS-CoV-2 infection impacts cells, and lymphopenia is its common cause. A reduction in the number of cells causes severe diseases. NRP1 mediates the entry of the SARS-CoV-2 virus into the brain through the olfactory epithelium. The highest expression of NRP1 is found in the astrocytes cell. NRP1 induces multiple effects such as cell proliferation, angiogenesis, and axon control. NRP1 is involved in various neurological symptoms such as encephalomyelitis and stroke in COVID-19 patients. Both CD147 and NRP1 mediate the entry of SARS-CoV-2 infection into the human brain cell. The expression profiles of ACE2 co-receptor are displayed in the form of a dot plot in Suppl Fig. 4(D).
Fig. 3.
(A) A PPI network of ACE2 and predicted viral receptor protein (B) A PPI network of ACE2 and predicted membrane protein (C) A PPI network of TMPRSS2 and predicted viral receptor protein (D) A PPI network of TMPRSS2 and predicted membrane protein.
5.2. Comparison of ACE2 expression profiles with other methods
The expression level of ACE2 at each specific cell type is analyzed for all human tissue as described in the previous subsection 4.3. Suppose the expression pattern vector of ACE2 in the upper respiratory tract is 0.3796, 0.067, 0.091, 0.489 when associated with the goblet, basal cell, non-ciliated secretory cell, and ciliated cell, respectively. ACE2 has the highest expression level in ciliated and goblet cells and is 2%–3% and 4%–5%. A similar result has also been observed in recent studies [68], [69], [11] where ACE2 expresses primarily in ciliated and goblet cells (2%–3% and 4%–5%). In adult testis, ACE2 expression pattern vector is 0.055, 0.294, 0.0341, 0.165, 0.035, 0.285, and 0.130 when associated with the myoid cell, sertoli cell, spermatids, germ, Leydig cell, SSC and SPG cell. ACE2 is highly expressed in Sertoli and Leydig cells (> 2.9%). Similar observation has also been reported in Ref. [68]. ACE2 protein showed a high expression value of > 3% in Leydig or Sertoli cells.
A comparative study is conducted to analyze the expression profiles of ACE2 protein in various human tissues. We have compared expression profiles of ACE2 based on IHC [68], scRNA-seq transcriptomics profiling using Seurat tool (Seurat tool) [68], MS [68] and proposed clustering technique. Table 3 presents a comparative account of ACE2 expression profiles in different human tissue using IHC, MS, and Seurat packages. In IHC based expression profile, the highest level of ACE2 expression is detected in the small intestine (93.724), testis (26.895), kidney (30.81), and heart (12.309). A medium value of ACE2 expression is detected in the colon (4.695), liver (1.294), and stomach (1.177), while very low ACE2 expression level is detected in bone marrow (0.049), brain (0.045), and spleen (0.007) (as refer from Table 3).
Table 3.
A comparative account of ACE2 expression profiles based on IHC, MS, and Seurat tool with Fuzzy based Improved GWO clustering method.
| Dataset | Immunohistochemistry (IHC) | Mass spectrometry (MS) | Single-cell transcriptomics profiling (Seurat) | Fuzzy-based Improved GWO Clustering method |
|---|---|---|---|---|
| Bonemarrow | 0.049 | 0.65 | 0.16 | 0.14910 |
| Brain | 0.045 | 0.48 | 0.31 | 0.33808 |
| Colon | 4.695 | 4.5 | 1.53 | 4.09998 |
| Heart | 12.309 | 3.4 | 2.01 | 2.94387 |
| Kidney | 30.81 | 4.8 | 2.14 | 1.48309 |
| Liver | 1.294 | 1.45 | 0.23 | 1.40624 |
| Lung | 0.345 | 2.5 | 1.61 | 5.65387 |
| Pancreas | 0.199 | 4.3 | 0.35 | 1.92722 |
| Skin | 0.089 | 0.65 | 0.28 | 1.34841 |
| Small Intestine | 93.724 | 4.6 | 1.53 | 2.74684 |
| Spleen | 0.007 | 0.21 | 0.26 | 0.01932 |
| Stomach | 1.177 | 2.9 | 0.51 | 0.84124 |
| Testis | 26.895 | 4.75 | 1.72 | 4.02471 |
Based on the MS study, a high ACE2 expression level is observed in the kidney (4.8), testis (4.75), small intestine (4.6), and pancreas (4.3). Low ACE2 expression is detected in bone marrow (0.16), brain (0.31) and spleen (0.26) (Referring Table 3). Single-cell transcriptomics profiling using the Seurat tool detects high ACE2 expression levels in the heart (2.01), kidney (2.14), small intestine (1.53), testis (1.72), lung (1.61), and colon (1.53). Low ACE2 expression level is detected in bone marrow (0.16), brain (0.31), pancreas (0.35), skin (0.28), spleen (0.26) and stomach (0.51).
Fuzzy-based improved GWO clustering method detects a high ACE2 expression level in the lung (5.653), upper respiratory tract (5.073), testis (4.024), colon (4.0998), kidney (1.483). Low ACE2 expression level is detected in brain (0.338), spleen (0.019) and bone marrow (0.149) (as Refer from Table 3 and Fig. 2).
Fig. 2.
ACE2 expression level of proposed clustering in all human tissue.
In all the previous studies and proposed fuzzy-based improved GWO clustering technique, it is observed that intestinal cells, heart, kidney, testis, and lung show elevated ACE2 expression. In contrast, low ACE2 expression is detected in the stomach, lymphatic tissue, skin, and brain.
5.3. Interaction of SARS-CoV-2 protein with other membrane protein and ssRNA viral receptor
To identify the gene set (ssRNA viral receptor or membrane) that interact with SARS-CoV-2 protein, PPI network is constructed using string database https://string-db.org/, with a confidence score of . The list of predicted ssRNA viral receptors and membrane protein are then queried from STRING database with ACE2 and TMPRSS2 protein as the hub genes. The PPI network is then visualized using the open cytoscape software available at: https://cytoscape.org/.
CD209, CEACAM1, CLEC4M, DPP4, ITGA2, ITGB1, TFRC, VCAM1 are the top ssRNA viral receptor protein interacts strongly with ACE2 protein. PCC is calculated between ACE2 protein and CD209, CEACAM1, CLEC4M, DPP4, ITGA2, ITGB1, TFRC, VCAM1. Also, ACE2 interact with the membrane protein BET1, DPP10, DPP4, DPP6, ECE1, FURIN, MEP1A, MEP1B, TRHDE and VCAM1. Table 4 gives the confidence score and PCC obtained between ACE2 and ssRNA viral receptor. Table 5 gives the confidence score and PCC between ACE2 and membrane protein.
Table 4.
Confidence score and PCC of ssRNA protein with ACE2.
| SARS-CoV-2 protein | ssRNA viral receptor | Confidence score | PCC |
|---|---|---|---|
| ACE2 | CD209 | 0.455 | 0.8642 |
| ACE2 | CEACAM1 | 0.420 | 0.8660 |
| ACE2 | CLEC4M | 0.420 | 0.7421 |
| ACE2 | DPP4 | 0.898 | 0.715 |
| ACE2 | ITGA2 | 0.530 | 0.728 |
| ACE2 | ITGB1 | 0.591 | 0.7914 |
| ACE2 | TFRC | 0.618 | 0.9743 |
| ACE2 | VCAM1 | 0.410 | 0.8156 |
Table 5.
Confidence score and PCC of membrane protein with ACE2.
| SARS-CoV-2 protein | Membrane protein | Confidence score | PCC |
|---|---|---|---|
| ACE2 | BET1 | 0.422 | 0.8660 |
| ACE2 | CD209 | 0.455 | 0.8642 |
| ACE2 | DPP10 | 0.420 | 0.7437 |
| ACE2 | DPP4 | 0.980 | 0.8010 |
| ACE2 | DPP6 | 0.420 | 0.7157 |
| ACE2 | ECE1 | 0.445 | 0.8620 |
| ACE2 | FURIN | 0.525 | 0.8750 |
| ACE2 | MEP1A | 0.925 | 0.5814 |
| ACE2 | MEP1B | 0.878 | 0.9569 |
| ACE2 | TRHDE | 0.408 | 0.9088 |
| ACE2 | VCAM1 | 0.410 | 0.8156 |
DPP4, TFRC, CEACAM1, ICAM1, ITGA4 and ITGB1 are the viral receptor protein correlate strongly with TMPRSS2 protein. The membrane protein that interacts with TMPRSS2 are ALK, DPP4, EPCAM, ERBB2, FOLH1, FURIN, GOLM1 and PCSK5. Table 6 gives the confidence score and PCC between TMPRSS2 and ssRNA viral receptor. Table 7 gives the confidence score and PCC between TMPRSS2 and membrane protein. The PPI network of the predicted receptor protein and SARS-CoV-2 protein are given in Fig. 3(A)–(D).
Table 6.
Confidence score and PCC of ssRNA protein with TMPRSS2.
| SARS-CoV-2 protein | ssRNA viral receptor | Confidence score | PCC |
|---|---|---|---|
| TMPRSS2 | DPP4 | 0.685 | 0.8229 |
| TMPRSS2 | TFRC | 0.500 | 0.974 |
| TMPRSS2 | CEACAM1 | 0.526 | 0.7451 |
| TMPRSS2 | ICAM1 | 0.474 | 0.9608 |
| TMPRSS2 | ITGA4 | 0.488 | 0.6721 |
| TMPRSS2 | ITGB1 | 0.975 | 0.7914 |
Table 7.
Confidence score and PCC of membrane protein with TMPRSS2.
| SARS-CoV-2 protein | Membrane protein | Confidence score | PCC |
|---|---|---|---|
| TMPRSS2 | ALK | 0.510 | 0.9091 |
| TMPRSS2 | DPP4 | 0.685 | 0.8229 |
| TMPRSS2 | EPCAM | 0.451 | 0.983 |
| TMPRSS2 | ERBB2 | 0.441 | 0.8581 |
| TMPRSS2 | FOLH1 | 0.568 | 0.883 |
| TMPRSS2 | FURIN | 0.663 | 0.866 |
| TMPRSS2 | GOLM1 | 0.422 | 0.673 |
| TMPRSS2 | PCSK5 | 0.449 | 0.885 |
5.4. Experimental comparisons
-
•
Computational complexity: With the simulation in Fig. 4(a) and (b), the population size is varied between 20 to 220, and the CPU execution time is noted for all the repeated experiments. Table 8 shows the maximum execution time for all the experimental runs. It is observed from Fig. 4. that fuzzy-based improved GWO consume less CPU time to execute all the operation when compared to fuzzy-based DE and fuzzy-based GWO algorithm.
-
•
Convergence analysis: With the simulation result in Fig. 4(c) and (d), the objective value is noted at each iteration of fuzzy-based GWO and fuzzy-based improved GWO algorithm. It is seen that the fuzzy-based improved GWO algorithm (blue curve line) minimizes the Jm and XB objective function effectively at each iteration compared to the fuzzy-based GWO algorithm (orange curve line).
In Table 8, we have reported the performance metrics scores achieved by fuzzy-based DE, fuzzy-based GWO, and fuzzy-based Improved GWO algorithm when executed on different datasets. It is observed that our proposed fuzzy-based improved GWO clustering algorithm gives a good SC on brain, heart, kidney, lung and testis dataset. Least DBI value is achieved in brain, small intestine, heart, kidney, pancreas, skin, stomach and testis data. A high CHI value is also achieved in brain, small intestine, heart, kidney, pancreas, lung, skin and stomach dataset.
Fig. 4.
Performance comparisons of different clustering algorithms.
Table 8.
Comparing the result of clustering method applied to the scRNA-Seq data.
| Dataset | Clustering method | Optimal cluster No. | SC | CHI | DBI | CPU execution time (in s) |
|---|---|---|---|---|---|---|
| Brain | Fuzzy-based DE | 6 | 0.40111 | 129.2800 | 1.0981 | 4324.1590 |
| Fuzzy-based GWO | 7 | 0.51035 | 199.4293 | 0.8838 | 3468.0252 | |
| Fuzzy-based Improved GWO | 7 | 0.518334 | 203.3781 | 0.9540981 | 1421.2521 | |
| Small Intestine | Fuzzy-based DE | 7 | 0.28990 | 47.1200 | 0.9250 | 3170.5995 |
| Fuzzy-based GWO | 7 | 0.32706 | 166.5812 | 1.1416 | 2404.980 | |
| Fuzzy-based Improved GWO | 7 | 0.32172 | 160.070 | 1.0754 | 1885.6229 | |
| Heart | Fuzzy-based DE | 9 | 0.16777 | 106.9702 | 1.2495 | 2624.3186 |
| Fuzzy-based GWO | 9 | 0.2503 | 105.3301 | 1.38 | 2735.812 | |
| Fuzzy-based Improved GWO | 10 | 0.26988 | 121.28160 | 1.30915 | 2636.09339 | |
| Kidney | Fuzzy-based DE | 9 | 0.18920 | 174.2728 | 1.30155 | 5443.17944 |
| Fuzzy-based GWO | 8 | 0.2378 | 187.7814 | 1.2034 | 5064.9636 | |
| Fuzzy-based Improved GWO | 6 | 0.24466 | 122.8541 | 0.96017 | ||
| Pancreas | Fuzzy-based DE | 7 | 0.1578 | 52.59440 | 1.375780 | 1881.48332 |
| Fuzzy-based GWO | 9 | 0.14269 | 50.4459 | 1.3097 | 1770.2553 | |
| Fuzzy-based Improved GWO | 9 | 0.17971 | 50.78220 | 1.25620 | 1301.22989 | |
| Lung | Fuzzy-based DE | 11 | 0.1660 | 25.55930 | 1.25809 | 281.25560 |
| Fuzzy-based GWO | 10 | 0.230 | 32.394 | 1.072 | 290.8825 | |
| Fuzzy-based Improved GWO | 13 | 0.29951 | 38.41294660 | 1.051319 | 215.390135 | |
| Skin | Fuzzy-based DE | 6 | 0.296880 | 133.97489 | 1.04126 | 2042.38863 |
| Fuzzy-based GWO | 7 | 0.22799 | 60.4798 | 1.20528 | 1422.42468 | |
| Fuzzy-based Improved GWO | 7 | 0.21182 | 100.504201 | 1.12764 | 1342.23049 | |
| Stomach | Fuzzy-based DE | 7 | 0.363190 | 181.50296 | 1.01722 | 2157.03084 |
| Fuzzy-based GWO | 7 | 0.31017 | 59.4220 | 1.17966 | 1365.365395 | |
| Fuzzy-based Improved GWO | 11 | 0.201755 | 105.90519 | 1.16535 | 1522.731694 | |
| Testis | Fuzzy-based DE | 7 | 0.31205 | 180.6621 | 1.2418 | 2997.6370 |
| Fuzzy-based GWO | 9 | 0.33008 | 203.9592 | 1.1515 | 2914.64364 | |
| Fuzzy-based Improved GWO | 9 | 0.334947 | 160.96580 | 0.88774 | 3152.23961 | |
6. Pathway enrichment analysis
In this work, we have predicted 58 potential viral receptors that mediate SARS-CoV-2 infection in different human organoids. These are: AXL, CD55, CD151, CD209, CD46, CD74, CD80, CD86, CEACAM1, CLDN1, CLEC4G, CXADR, CX3CR1, CACNA1C, CD300LD, CR1, CR2, DAG1, DPP4, EPHA2, EFNB2, EFNB3, FCGRT, F11R, GPR78, GPC5, GRM2, MRC1, MERTK, MXRA8, MOG, ICAM1, ITGB1, ITGB3, ITGB6, ITGB8, ITGA2, KREMEN1, PHB, NGFR, NCAM1, NECTIN4, PVR, RPSA, SLC10A1, OCLN, SCARB1, SCA-RB2, SLAMF1, LAMP1, LDLR, HAVCR1, HLA-DRA, TFRC, TYRO3, VCAM1, CLEC4M and CLEC5A.
816 membrane proteins that are co-expressed with SARS-CoV-2 receptor protein (ACE2 and TMPRSS2) are also predicted in this work. The molecular mechanism of the identified host receptors is studied to investigate their role in the pathogenesis of SARS-CoV-2 infection using GO term and KEGG pathway enrichment analysis.
6.1. Gene ontology term enrichment analysis
The GO term enrichment analysis examines the functional characteristics of the predicted 816 membrane protein and 58 ssRNA viral receptors. The GO annotation term of the predicted genes is collected from DAVID bioinformatics resources [70]. The GO term enrichment analysis is performed to know the biological function (BP) of the predicted membrane protein at the cellular (CC) and molecular level (MF). Those genes involved in similar biological processes or molecular functions are expected to interrelate.
Some of the enriched CC terms of GO analysis process are membrane, plasma membrane, lysosome membrane, endosome membrane, nuclear membrane, endoplasmic reticulum membrane, mitochondrial membrane, cell surface, cytosol, cytoplasm, nucleoplasm and extracellular exosome, etc. The GO-CC term refers to the locations relative to the cellular structure where a gene performs a function, either cellular compartment (e.g., mitochondria) or a part of stable macromolecular complexes (e.g., ribosomes), etc. The SARS-CoV-2 virus invades the host nucleus or cytoplasmic cell and causes severe respiratory complications such as pneumonitis leading to upper acute respiratory distress syndrome (ARDS). Therefore, the membrane proteins involved in the CC terms mediate the entry of coronaviruses into the host cell membrane.
Some of the enriched BP terms of the GO process are regulation of membrane protein, ectodomain proteolysis, cell–cell adhesion mediated by integrin, adaptive and innate immune response, positive regulation of host by the replication of the viral genome, fusion of membrane, regulation of I-kappaB kinase/NF-kappaB signaling pathway, processing of antigen and representation of peptide antigen through major histocompatibility complex (MHC) I, regulate the inflammatory response to the stimulus of antigens, regulation of host morphology or physiology through the virus, mediate receptor endocytosis, activate mitogen-activated protein kinase (MAPK) activity, toll-like receptor signaling pathway, involved in protein ubiquitination catabolic process, activate the protein tyrosine kinase transmembrane receptor protein, etc
A few of the top MF terms of the GO process are binding of receptor, protein, complex molecular, and cell adhesion, endopeptidase activity, binding of adenosine triphosphate (ATP), guanosine triphosphate (GTP) and antigen, binding of lipid antigen, endogenous lipid antigen, and exogenous lipid antigen, amide binding, peptide binding, lipopeptide binding, virion binding, etc.
6.2. KEGG pathway enrichment analysis
The KEGG enrichment pathway analysis of the predicted viral receptor and membrane protein helps identify the target protein that mediates or restricts SARS-CoV-2 infection. It helps understand the pathogenesis mechanism of SARS-CoV-2 infection and identify the target protein for developing effective drugs and therapeutics for COVID-19 disease. We have determined 816 membrane proteins involved in the SARS-CoV-2 condition. The pathogenicity mechanism or pathway of SARS-Cov-2 infection identified through GO and KEGG enrichment analysis is shown in Fig. 5.
Fig. 5.
Schematic flow of SARS-CoV-2 pathogenesis pathway analysis.
Initially, the SARS-CoV-2 virus uses its spike glycoprotein to interact with the host cell surface. The glycosylated protein attaches to ACE2 receptor via glycosaminoglycans (GAGs) and induces a conformational change on the host cell surface. The protein CHST1, CHST2, CHST3, CHST4, CHST17, CHPF2, GLCE, EXT1, EXTL1, EXTL2, NDST2, NDST3, NDST4, XYLT1 and XYLT2 synthesize GAGs. SARS-CoV-2 then penetrates the endocytic membrane of the host cell to create an infection. Research conducted in Ref. [71] shows that the SARS-CoV-2 S1 receptor can bind to heparin derivative GAGs. Because the SARS-CoV-2 protein envelope contains positively charged amino acid and are prone to interact with the negatively charged heparin sulfate proteoglycans group. The membrane proteins NDST1, NDST2, NDST3, NDST4, EXT1, EXTL1, EXTL2, GLCE, XYLT1 and XYLT2 synthesized glycosaminoglycan heparin sulfate or heparin that inhibit SARS-associated coronavirus cell invasion. Ref. [72] also demonstrate that GAGs heparin derivative could prevent the spread of SARS-CoV-2 infection and hence can use as an anticoagulant drug against any other members of coronaviridae. The antiviral drugs carboplatin and gemcitabine could act as therapeutic agents to prevent SARS-CoV-2 infection.
After interacting the viral spike protein with the host cell membrane, the SARS-CoV-2 virus uses clathrin and caveolin-dependent endocytosis to insert the viral particle into the host cell membrane. The SARS-CoV-2 virus then penetrates through the endocytic membrane of the host cell to establish an infection. It then transfers its viral RNA particles from the lumen to the cell cytosol of the endosomal system. [73] demonstrates that the knockdown of clathrin-dependent heavy chain process could reduce virus infectivity. The viral plasma protein low-density lipoprotein receptor (LDLR) is required to regulate plasma lipoprotein levels. LDLR internalizes lipoprotein cargo through the clathrin or caveolin-mediated endocytosis process. LDLR protein level is regulated by an inducible degrader of the LDLR (IDOL) and proprotein convertase subtilisin/kexin type 9 (PCSK9). IDOL, an E3-ubiquitin ligase, promotes the degradation of LDLR through the ubiquitination process. PCSK9 induces LDLR internalization by forming clathrin-coated pits similar to the binding of lipoprotein ligands. LDLR is important lipid metabolism and risk associated with cardiovascular disease. A better investigation into the pathway of degrading LDLR levels could provide a new therapeutic target. It is found that other membrane proteins AP2A2, APLP1, DNM2, EPS15, EPN1, EPN2, LY75, MRC2, SNX5 mediate the clathrin-dependent endocytosis process in SARS-CoV-2 infection. These membrane proteins are responsible for forming clathrin-coated pits in the host cell’s cytoplasmic membrane. The protein involved in the clathrin-dependent endocytosis pathway can be extensively studied in the future to find antiviral therapy’s target.
The ubiquitin–proteasome interaction is also essential for the various stages of the coronavirus infection cycle [74]. The membrane protein FBXW11 mediates the ubiquitination process and degrade the target protein in SARS-CoV-2 infection. The protein ligase namely: HERC2, HERC4, HERC1, WWP1, MGRN1, NEDD4, NEDD4L, UBE2D2, UBE2E1, UBE2G1, UBE2K and UBA2 accept ubiquitin from a conjugating ubiquitin enzyme and transfers the ubiquitin directly to the target substrate. Epithelial growth factor receptor (EGFR) plays a vital role in the internalization process of coronaviruses. SARS-CoV-2 infection can over-activate the EGFR signaling pathway and consequently produce inflammation in the lung. Ref. [71] shows the possible way of preventing SARS-CoV-2 disease is by downregulating the signaling pathway that promotes the endocytosis process. EGFR tyrosine kinase inhibitors (TKIs) inhibit the endocytosis of the SARS-CoV-2 virus through EGFR. Imatinib, an inhibitor, can inhibit the replication process of SARS-CoV-2 infection before their reproduction [71].
When the SARS-CoV-2 virus invades the human cell, viral proteins trigger an immune response to counteract the virus. These viral antigens are recognized by the B cell and presented by MHC to the cell for developing innate immunity. It results in natural antibody production and enhances cytokine secretion and cytolytic activity in the initial phase of infection. During the innate immune response, pattern-recognition receptors (PRRs) are activated to recognize the molecular structure of the invading pathogens [75]. Once PPRs identify the pathogen molecular pattern, several signaling pathways and transcription factors are activated via Janus kinase-signal transducer and activator of transcription (JAK-STAT) pathway [76]. The transcription factors induce gene expression that encodes pro-inflammatory cytokines, chemokines, and several adhesion molecules. Interleukin-1 (IL-1), interleukin-6 (IL-6), type I interferon (IFN-I), and TNF- are the necessary pro-inflammatory cytokines response. The mast cell, macrophages, endothelial and epithelial cells generate pro-inflammatory cytokines during the immune response. The sudden increase in the circulating level of pro-inflammatory cytokines results in the cytokine storm [77]. The cytokines storm caused the influx of various immune cells such as cells, neutrophils, and macrophages from the blood capillary to the infection site, which inflames the injury and promotes ARDS [77], [76].
The co-receptor protein EPHB2, KIT, MCL1, CRLF2, EGF, ERBB4, EPOR, GHR, IL21R, IL23R, IL5RA, IL9R and LEPR of ACE2 mediate cytokine–cytokine interaction via proximal JAK-STAT signaling pathway. Suppression of interferon pathway is a common approach used by the virus to degrade innate antiviral immunity. Targeting the viral mediators of immune evasion may help block virus replication in patients with COVID-19. The deficiency of human interleukin 21 receptors (IL21R) cause chronic cholangitis and liver disease in severe SARS-CoV-2 infected patients. Thus, IL21 is a new therapeutic target for maintaining immune homeostasis [78].
Other possible immunopathological manifestations of SARS-CoV-2 infection are neutrophilia, dysregulation of monocytes and macrophages, delayed IFN-I response, lymphopenia, etc. Severe COVID-19 patients usually have an increased neutrophil count than a mild case and average person. Neutrophils protect against infection by producing neutrophil extracellular traps (NETs). However, excessive activation of neutrophils in SARS-CoV-2 patients can damage the surrounding cell and dissolve connective tissues [75]. Treatment using the NETs approach decreased the pulmonary hyperinflammation caused due to severe COVID-19 infection.
Macrophages and monocytes are the primary immune cell involved in infection and inflammation. At the initial stages of acute lung injury, an immune response is triggered by macrophages and dendritic cells by activating antigen-presenting cells, which produce pro-inflammatory cytokines, prostaglandins, and histamine. The increased permeability of microcirculatory into the infection site obstructs the blood-air barrier and promotes pulmonary hemorrhages, and ARDS [77], [76], [75]. Dysfunction of endothelial cells and pulmonary tissue oxygenation may promote bacterial pneumonia and sepsis. Ref. [65] article suggests DPP4 inhibitors could suppress the production of interleukin and interferon, reducing cell proliferation. DPP8/9 inhibitor reduced cell activation by reducing the secretion of TNF- and IL-6 from macrophages. Thus, the food and drug administration (FDA) approve using DPP4 inhibitors for managing chronic inflammatory diseases such as atherosclerosis and type II diabetes.
Interferons (IFNs) activate immune cells. IFNs regulate the infiltration of monocyte-derived macrophages in the lung. Delayed IFNs induction in SARS-CoV-2 infection accumulates excessive active macrophages in the lung and causes immunopathology. Impaired production of IFNs during SARS-CoV-2 infection creates an imbalance in upper airway macrophages’ pro-inflammatory and repair function. A delayed IFNs production inhibits cell progression from lymphoid tissue and can cause cell death of cells. Acute lung injury in SARS-CoV-2 patients is due to the failure of cells to activate immunosuppressive mechanism timely [76], [75]. The toll-like receptor membrane protein TLR3 and TLR7 induce macrophage cells to generate innate lung immunity. TLR signaling activates pro-inflammatory cytokine factors such as IL-1, IL-1, IL-4, IL-6, and interferon. Thus, TLRs could act as a potential target for controlling SARS-CoV-2 infection in an early stage of the disease [79], [75].
SARS-CoV-2 infection induce lymphopenia by activating systemic inflammation and directly neutralizing or destroying human lymphoid organs. SARS-CoV-2 infected patients have low lymphocyte count, smaller lymphoid follicles, enhancement of immunoblastic cells, and low zone proliferation. This shows that SARS-CoV-2 infection can cause more severe damage to human lymphoid organs and spleen than hepatitis B virus (HBV) and Epstein–Barr virus (EBV) infection [80]. Lymphatic endothelial cells require Ephrin B4 and Ephrin B2 membrane protein to maintain the integrity of lymph vessels. Ephrin B4 and Ephrin B2 signaling pathways provide a potential therapeutic target to modulate the permeability of lymphatic vessels. The loss of Ephrin B4 and Ephrin B2 signaling increases the vessel leakage in response to bacterial and viral infection [81].
Ref. [82] reported that coronavirus replication induces excessive stress in the endoplasmic reticulum (ER). The excessive synthesis of protein and folding of a viral protein is the main reason for causing ER stress in coronavirus infection. The excessive protein accumulation disrupts the protein synthesis and ER folding capacity balance. This led to the accumulation of excessive unfolded proteins in ER. The membrane proteins EDEM1, XBP1, ATF6B, DERL1, DERL2, HSPA5, NFE2L2, and VCP initiate the unfolding protein response in the ER. The SARS-CoV-2 protein CANX, CALR, HSP90B1, HSPA5, PDIA3 fold a transmembrane protein into ER. We find drugs brefeldin, indapamide, ezogabine, dolasetron, and repaglinide prevent protein assembly, disrupt coatomer protein I (COP–I) transport, and partially block viral RNA synthesis.
The proteins CD28, CD55, CXADR, CYCS, ICAM1, DAG1, HLA-F, SGCA, SGCB, SGCD, and SGCG, are responsible for causing myocardial damage in SARS-CoV-2 infection. Stingi and Cirillo reported that oncogenic might be the long-term secondary effect of SARS-CoV-2 infection [83]. SARS-CoV-2 infection may develop cancer by inhibiting tumor suppressor genes, ST14. Also, SARS-CoV-2 infection induces carcinogenesis via tyrosine kinase receptors [71] by penetrating the blood–brain barrier (BBB). Another potential cause of cancer development and progression in SARS-CoV-2 infection is the activation of IL-6 and JAK/STAT3 signaling pathways in the tumor microenvironment of bladder cancer patients. A high pro-inflammatory cytokine triggers cancer development and progression through tyrosine kinase receptors. The membrane proteins BAK1, BAX, KRAS, CCND1, CDK2, CDKN1A, CDKN1B, and NFKB2 promote mutagenesis in SARS-CoV-2 infection. The identified KEGG signaling pathway of the target membrane protein is shown in Table 9.
Table 9.
The KEGG pathway enrichment analysis of some significant membrane protein.
| KEGG pathway | P value | Predicted protein of SARS-CoV-2 |
|---|---|---|
| Proteosome/Protolysis | 6.9E−2 | VTI1A, ZFPL1, MYRF, MGAT2, CLCA4, PRSS8, |
| ST14, TRHDE, MEP1A, MMP24, PSMC5, PSMD14, | ||
| PSME1, PSMB1, PSMA2, OLR1, POMP | ||
| Ubiquitin mediated proteolysis | 6.9E−2 | FBXW11, HERC2, HERC4, HERC1, WWP1, MGRN1, |
| NEDD4, UBE3A, UBA2, UBE2K, UBE2G1, UBE2E1, | ||
| UBE2D2, NEDD4L | ||
| Clathrin dependent endocytosis | 2.2E−6 | AP2A2, DNM2, APLP1, EPS15, EPN1, EPN2, |
| LDLR, LY75, MRC2, SNX5 | ||
| Endocytosis | 1.2E−8 | FGFR4, IGF1R, IL2RA, PSD2, TPCN2, ARF1, |
| ARAP2, CLTA, CYTH1, RAB11, FIP1, RAB11A, | ||
| SNF8, VPS29, VPS37B, WWP1, ARPC1B, | ||
| GITI, ARPC2, CAPZB, CHMP5, HLA-F, NEDD4, | ||
| PSD2, PDCD6IP, CHMP2A | ||
| Glycosaminoglycan biosynthesis | 2.5E−3 | NDST1, NDST2, NDST3, NDST4, CHST1, CHST2, |
| CHST3, CHST4, CHST17, CHPF2 GLCE, EXT1, | ||
| EXTL1, EXTL2, GLCE, XYLT2, XYLT1 | ||
| Cytokine mediated signaling pathway | 2.6E−5 | BCL2, CD226, CD27, CD276, CD28, FCER2, |
| KIT, LRP8, MCL1, NFAM1, TRIL, VTCN1, | ||
| EDA, EREG, ICAM1, PTPRN, STX1A, STX3 | ||
| Cytokine–cytokine receptor interaction | 7.0E−2 | CD70, ACVR1, AMHR2, BMPR2, CSF1, CRLF2, |
| EDAR, EPOR, GHR, L1R2, IL2RA, IL21R, | ||
| IL23R, IL5RA, IL9R, LEPR, OSMR, PRLR | ||
| T cell receptor signaling pathway | 8.0E−1 | CD8A, CD226, CD276, CD28, CD3D, CD3G, |
| CD8A, CD8B, CTLA4, ICOS, LAT, | ||
| Leukocyte transendothelial migration | 3.9E−3 | THY1, ESAM, ICAM1, JAM2, JAM3, VCAM1 |
| MAPK signaling pathway | 8.9E−4 | FLT3, EPHA8, KIT, MET, AREG, CSF1, |
| EREG, ERBB2, ERBB3, ERBB4, FGFR4, FLT3, | ||
| IGF1R, INSR, NTRK1, NTRK2, PTPRR, TGFA, EPHA2 | ||
| JAK-STAT signaling pathway | 2.9E−2 | KIT, MCL1, CRLF2, EGF, EPOR, GHR |
| , IL2RA, IL21R, IL5RA, IL9R, LEPR, OSMR, | ||
| PRLR, BCL2, | ||
| EGFR tyrosine kinase inhibitor | 8.4E−3 | IGF1R, EGF, ERBB2, ERBB3, IGF1R, NRG1, |
| NRG2, TGFA, BCL2, MET, BAX | ||
| Unfolded protein response | 1.4E−2 | VAPB, XBP1, ATF6B, CREB3, EDEM1 |
| Protein processing in | 1.0E−2 | EDEM1, XBP1, ATF6B, VCP, HSPA5, PDIA6, |
| LMAN1, PREB, RRBP1, | ||
| Endoplasmic reticulum | DNAJB1, SIL1, SEC61G, XBP1, DERL2, PRKCSH, | |
| CAPN1, CALR, CKAP4, DERL1, EIF2AK1, HSP90AB1, | ||
| NFE2L2, PREB, PDIA3, CANX | ||
| Viral carcinogenesis | 6.0E−2 | BAX, BAK1, DCC, REB3, BAD, CREBBP, J |
| UN, KRAS, RASA2, SP100, TRADD, CCND1, | ||
| CCND3, CDK2, CDK6, CDKN1B, GTF2A2, MAPK3 | ||
| SYK, NFKB2, CDKN1A | ||
| Viral myocarditis | 1.4E−2 | DAG1, ICAM1, SGCA, SGCB, SGCD, ICAM1, |
| HLA-F, SGCC, CD28, CD55, CXADR, CYCS | ||
| Antigen processing and presentation | 1.1E−4 | CD8A, CD8B, CD74, HLA-DRA, CD1B, LGMN, |
| HLA-F, | ||
| Carbon metabolism | 9.4E−2 | KIT, MET, FLT3, NTRK1, NTRK3, RET, |
| RAF1, ERBB2 | ||
| Biosynthesis of antibiotics | 3.2E−2 | ACLY, SQLE, NME2, ACAT1, CDC42, FDFT1, |
| FNTA, FMO1, FMO3, FMO4, FMO5, UGT1A1, | ||
| GMPS, MGST1, PGP |
We have identified some of the target membrane proteins related to the pathogenesis of SARS-CoV-2 infection. The membrane protein list is then queried from the drug-gene interaction database3 https://www.dgidb.org/ and its drug combination is found. Table 10, Table 11 list the druggable membrane protein and its composition to treat COVID-19 disease. A list of gene names and their abbreviation used in this article is provided in Tables 12, 13, 14, 15.
Table 10.
List of approved drugs associated with the SARS-CoV-2 membrane protein for the treatment of COVID-19.
| Membrane protein | Drugs composition |
|---|---|
| TLR3 | Rintatolimod, Hiltonol, Hydroxychloroquine, Aspirin |
| Azd-8848 | Imiquimod, Resiquimod, Isatoribine, Loxoribine, Hydroxychloroquine |
| TLR7 | Telratolimod, Vesatolimod, Hydroxychloroquine sulfate, Gsk-2245035 |
| TLR9 | Hydroxychloroquine sulfate, Agatolimod sodium, Tilsotolimod, Emd-1201081 |
| MICB | Ribavirin |
| UGT1A1 | Bilirubin, Indinavir, Tranilast, Nilotinib, 7-Ethyl-10-hydroxycamptothecin |
| Dolutegravir, Letermovir, Raltegravir, Raloxifene, Etoposide | |
| PSMC5 | Carfilzomib, Ixazomib, Bortezomib, Ixazomib citrate, Oprozomib |
| PSMD11 | Carfilzomib, Bortezomib, Ixazomib, Oprozomib |
| PSMD14 | Carfilzomib, Bortezomib, Ixazomib citrate, Oprozomib Sulfuretin |
| PSME1 | Carfilzomib,Bortezomib |
| PSMA2 | Carfilzomib, Bortezomib,Ixazomib citrate, Oprozomib, Marizomib |
| PSMB2 | Carfilzomib, Bortezomib,Ixazomib citrate, Oprozomib, Marizomib, KZR-616 |
| MGAT4A | Bevacizumab, Capecitabine, Oxaliplatin, Cetuximab |
| ALDH2 | Prunetin, Acetaldehyde, Diacetylmorphine, Disulfiram |
| CHST3 | Docetaxel, Thalidomide |
| CHST1 | Imatinib |
| KIT | Imatinib, Quizartinib, Nilotinib, Sunitinib, Amuvatinib |
| GGT1 | Cannabinol, Ditiocarb, Mannitol, Aminoglutethimide, Mestranol |
| VCAM1 | Tamoxifen, Piroxicam, Dexamethasone, Liothyronine Sodium |
| Mercaptopurine, Dexamethasone, Troglitazone, Cyclosporine | |
| CTLA4 | Tremelimumab, Ipilimumab, Zalifrelimab, Abatacept, Atezolizumab, |
| Sirolimus, Wortmannin, Dexamethasone, Methimazole, Antibiotic | |
| LDLR | Cholestyramine, Evolocumab, Mipomersen, Tributyrin, Alirocumab |
| Acetylcysteine, Gemfibrozil, Corticotropin, Retinol | |
| SDC1 | Indatuximab Ravtansine, Heparin |
| BCL2 | Docetaxel, Paclitaxel, Hypoxanthine, Navitoclax |
| Oblimersen, Venetoclax, Obatoclax, Beauvericin, Isosorbide, Protuboxepin A | |
| FOLH1 | Capromab, Technetium TC-99 m Trofolastat Chloride, Mipsagargin, MDX-070 |
| MLN-2704, MLN-591RL,Androstanolone, Methotrexate, Docetaxel, Mercaptopurine | |
| FMO3 | Itopride, Tamoxifen, Nicotine, Rosuvastatin, Tacrolimus |
| FMO1 | Tamoxifen, Nicotine, Olanzapine |
| COL18A1 | Glutamine, Tamoxifen, Aspirin, Collagenase clostridium histolyticum |
| Thrombin, Celecoxib, Ocriplasmin | |
| ERBB3 | Afatinib, Seribantumab, Patritumab,Cetuximab, Lapatinib |
| Pertuzumab, Panitumumab, Erlotinib, Aspirin, Alteplase | |
| ERBB2 | Lapatinib, Afatinib, Trastuzumab, Pertuzumab, Dacomitinib |
| AC-480, Margetuximab, Tucatinib, MM-111, Sapitinib | |
| NRG1 | Afatinib, Seribantumab, Patritumab, Cetuximab, Lapatinib |
| Pertuzumab, Panitumumab, Erlotinib, Aspirin, Alteplase | |
| CD38 | Daratumumab, Isatuximab, Thrombin |
| LEPR | Metreleptin, Atorvastatin, Simvastatin |
Table 11.
List of drugs in clinical trials associated with SARS-CoV-2 membrane protein for the treatment of COVID-19.
| Membrane protein | Drugs composition |
|---|---|
| IL2RA | Basiliximab, Daclizumab, Aldesleukin, Inolimomab, Lmb-2, Lentinan |
| Dinitrochlorobenzene, Denileukin diftitox, Thyroxine, Methimazole | |
| KCNQ1 | Indapamide, Bepridil, Tacrolimus, Celecoxib, Ezogabine, Dolasetron |
| Repaglinide, Insulin, Indomethacin | |
| COL18A1 | Glutamine, Tamoxifen, Aspirin, Collagenase clostridium histolyticum |
| Thrombin, Celecoxib, Ocriplasmin | |
| CDKN1B | Raltitrexed, Epoetin Beta, Celecoxib, Methotrexate, Lapatinib |
| Epoetin Alfa, Tretinoin, Progesterone, Streptozocin | |
| IL23R | Celecoxib |
| CRLF2 | Ruxolitinib |
| PTPRB | Razuprotafib, Sunitinib |
| CR1 | Eculizumab, CDX-1135 |
| CTLA4 | Tremelimumab, Ipilimumab, Zalifrelimab, Abatacept, Atezolizumab |
| Sirolimus, Wortmannin, Dexamethasone, Methimazole, Antibiotic | |
| HLA-DRA | Floxacillin, Amoxicillin, Clavulanic acid, Pembrolizumab, Atezolizumab, Nivolumab |
| NME2 | Zidovudine, Tenofovir, Lamivudine, Progesterone |
| CYYR1 | Cixutumumab, Teprotumumab, Trandolapril, Verapamil, Pioglitazone |
| CD34 | Fludeoxyglucose-F18, Puromycin, Prednisolone, Quercetin |
| TPCN2 | Verapamil |
| FMO3 | Itopride, Tamoxifen, Nicotine, Rosuvastatin, Tacrolimus |
| HSPA8 | Bupivacaine, Denosine Diphosphate, Tretinoin |
| CHST3 | Docetaxel, Thalidomide |
| DPP4 | Sitagliptin, Saxagliptin, Gosogliptin, Vildagliptin, Begelomab |
| Alogliptin, Linagliptin, Bisegliptin, Valacyclovir, Anagliptin | |
| GHR | Pegvisomant, Somatropin, Somatrem, Somatrogon, ACP-001 |
| PRLR | Endostatin, Somatropin, Androstanolone |
Table 12.
Gene name and its abbreviation used in the article.
| Gene name | Abbreviation | Gene name | Abbreviation |
|---|---|---|---|
| DPP10 | Dipeptidyl peptidase 10 | EPN2 | Epsin 2 |
| ADAM9 | Disintegrin and metalloproteinase domain 9 | MYH7 | Myosin 7 |
| ADAM17 | Disintegrin and metalloproteinase domain 17 | LAMA2 | Laminin subunit alpha 2 |
| CLDN1 | Claudin | MRC1 | Macrophage mannose receptor 1 |
| PLXNA2 | Plexin A2 | PRSS8 | Prostasin preproprotein |
| CD63/CD151 | Tetraspanin | SCARB2 | Scavenger receptor class B member 2 |
| EPAP2 | Endoplasmic reticulum aminopeptidase 2 | GRM2 | Metabotropic glutamate receptor 2 |
| CYP1A2 | Cytochrome P450 family 1 family A member 2 | GPC5 | Glypican 5 |
| CLEC14A | C-type lectin domain family 14 member A | EFNB2 | Ephrin-B2 |
| HLA-DRA | Histocompatibility antigen, DR alpha chain | EFNB3 | Ephrin-B3 |
| NECTIN4 | Nectin cell adhesion molecule 4 | CR1/CR2 | Complement receptor 1/2 |
| ITGB3 | Integrin beta-3 | ST14 | Suppression of tumorigenicity 14 |
| ITGB6 | Integrin beta-6 | CXCL11 | Chemokine (C-X-C motif) ligand 11 |
| ITGB8 | Integrin beta-8 | SLC22A8 | Solute carrier family 22 member 8 |
| CACNA1C | Voltage dependent L-type calcium subunit alpha | SLC26A7 | Solute carrier family 26, member 7 |
| CD81/CD9 | Tetraspanin | COX8C | Cytochrome c oxidase subunit 8C |
| ERK1/ERK2 | Mitogen-activated protein kinase | COX8A | Cytochrome c oxidase subunit 8A |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A | CDKN1B | Cyclin-dependent kinase inhibitor 1B |
| HSP90B1 | Heat shock protein 90 beta family member 1 | DERL2 | Derlin-2 |
| IL23R | Interleukin 23 receptor | IL5RA | Interleukin-5 receptor subunit alpha |
| UBE2D2 | Ubiquitin conjugating enzyme E2 D2 | UBE2E1 | Ubiquitin conjugating enzyme E2 E1 |
| UBE2G1 | Ubiquitin conjugating enzyme E2 G1 | UBE2K | Ubiquitin-conjugating enzyme E2 |
| HERC1 | HECT, RLD domain E3 ubiquitin ligase 1 | EXTL1 | Exostosin like glycosyltransferase 1 |
| HERC4 | HECT, RLD domain E3 ubiquitin ligase 4 | EXTL2 | Exostosin like glycosyltransferase 2 |
| CHST2 | Carbohydrate sulfotransferase 2 | CHST3 | Carbohydrate sulfotransferase 3 |
| CHST4 | Carbohydrate sulfotransferase4 | MEP1B | Meprin B subunit |
| ABCC9 | ATP binding cassette family C member 9 | MMP24 | Matrix metallopeptidase 24 |
| VTI1A | Vesicle transport interaction with t-SNARE 1A | ZFPL1 | Zinc finger protein-like 1 |
| MGAT2 | Mannoside acetylglucosaminyltransferase 2 | PSMC5 | Proteasome 26S subunit, ATPase 5 |
| CLCA4 | Calcium activated chloride channel regulator 4 | PSMD14 | Proteasome 26S subunit, nonATPase 14 |
| PSME1 | Proteasome activator subunit 1 | PSMB1 | Proteasome subunit beta 1 |
| OLR1 | Oxidized low-density lipoprotein receptor 1 | POMP | Proteasome maturation protein |
| FBXW11 | F-box and WD repeat domain containing 11 | FGFR4 | Fibroblast growth factor receptor |
| IL2RA | Interleukin-2 receptor subunit alpha | TPCN2 | Two pore segment channel 2 |
| PSD2 | Phosphatidylserine decarboxylase proenzyme 2 | ARF1 | ADP-ribosylation factor |
| CYTH1 | Cytohesin-1 | CLTA | Clathrin light chain |
| RAB11A | Ras-related protein Rab-11A isoform 1 | SNF8 | Vacuolar-sorting protein SNF8 |
| VPS37B | Vacuolar protein sorting 37B | ARPC2 | Arp2/3 complex 34 kDa subunit |
| CAPZB | F-actin-capping protein subunit beta | CHMP2A | Charged multivesicular body protein 2A |
| BCL2 | B-cell CLL/lymphoma 2 | FCER2 | Fc fragment of IgE receptor II |
| LRP8 | Low density lipoprotein receptor protein 8 | NFAM1 | NFAT activation molecule 1 |
| VTCN1 | V-set domain cell activation inhibitor 1 | PTPRB | Protein tyrosine phosphatase receptor B |
| STX1A | Syntaxin 1A | HSPA8 | Heat shock protein 8 |
| KCNQ1 | Potassium voltage-gated channel family Q1 | CYYR1 | Cysteine and tyrosine-rich protein 1 |
| COL18A1 | Collagen type XVIII alpha 1 | ALDH2 | Aldehyde dehydrogenase 2 family member |
| MICB | MHC class I polypeptide-related sequence B | PGP | Phosphoglycolate phosphatase |
| TLR3/7/9 | Toll like receptor-3/7/9 | MGST1 | Microsomal glutathione S-transferase 1 |
| UGT1A1 | UDP-glucuronosyltransferase | FDFT1 | Farnesyl-diphosphate farnesyltransferase 1 |
| ACAT1 | Acetyl-CoA acetyltransferase 1 | NME2 | Nucleoside diphosphate kinase |
| SQLE | Squalene epoxidase | ACLY | ATP citrate lyase |
| NTRK3 | Neurotrophic tyrosine kinase receptor, type 3a | MAPK3 | Mitogen-activated protein kinase |
| TRADD | Tumor receptor-associated DEATH domain | CAPN12 | Calpain 12 |
| EIF2AK1 | Eukaryotic initiation factor 2-alpha kinase 1 | SEC61G | SEC61 translocon subunit gamma |
| CREB3 | cAMP responsive element binding protein 3 | PRLR | Prolactin receptor |
Table 13.
Gene name and its abbreviation used in the article.
| Gene name | Abbreviation | Gene name | Abbreviation |
|---|---|---|---|
| STX3 | Syntaxin 3 | CD70 | CD70 molecule |
| ACVR1 | Receptor protein serine/threonine kinase | AMHR2 | Anti-Muellerian hormone type-2 receptor |
| BMPR2 | Receptor protein serine/threonine kinase | EDAR | Ectodysplasin A receptor |
| LIR2 | Leukocyte immunoglobuli-like receptor 2 | PRPL | Plastid ribosomal protein L24 |
| CD3D | T-cell surface glycoprotein CD3 delta chain | CTLA4 | Cytotoxic T-lymphocyte protein 4 |
| CD3G | T-cell surface glycoprotein CD3 gamma chain | ICOS | Inducible cell costimulator |
| LAT | Linker for activation of cells | THY1 | Thymus cell antigen 1, theta |
| ESAM | Endothelial cell-selective adhesion molecule | JAM3 | Junctional adhesion molecule 3 |
| FLT3 | Fms related receptor tyrosine kinase 3 | EPHA8 | Ephrin type-A receptor 8 |
| AREG | Amphiregulin | ERBB3 | Receptor protein-tyrosine kinase |
| NTRK1 | Neurotrophic tyrosine kinase, receptor 1 | NTRK2 | neurotrophic tyrosine kinase, receptor 2 |
| PTPRR | Receptor-type tyrosine-protein phosphatase R | TGFA | Transforming growth factor alpha |
| EPHA2 | EPH receptor A2 | NRG2 | Neuregulin 2 |
| VAPB | Vesicle-associated membrane protein B | PDIA6 | Protein disulfide-isomerase A6 |
| LMAN1 | Lectin, mannose binding 1 | PREB | Prolactin regulatory element binding |
| RRBP1 | Ribosome binding protein 1 | SIL1 | Nucleotide exchange factor SIL1 |
| PRKCSH | Protein kinase C substrate 80K-H | CKAP4 | Cytoskeleton associated protein 4 |
| HSP90AB1 | Heat shock protein HSP 90-beta isoform A | DCC | Development and carotenogenesis control-1 |
| BAD | Betaine aldehyde dehydrogenase 2 | CREBBP | Histone acetyltransferase |
| RASA2 | Ras GTPase-activating protein 2 | SP100 | Nuclear autoantigen Sp-100 |
| CCN1 | Cellular communication network factor 1 | CND3 | Condensin complex non-SMC subunit |
| CDK6 | Cyclin dependent kinase 6 | CDKN1B | Cyclin-dependent kinase inhibitor 1B |
| SYK | Spleen associated tyrosine kinase | CD74 | Thyroglobulin type-1 domain protein |
| CD1B | T-cell surface glycoprotein CD1b | RET | Ret proto-oncogene |
| RAF1 | RuBisCO accumulation factor 1 | FNTA | Farnesyltransferase, CAAX box, alpha |
| GMPS | Guanine monophosphate synthase | FMO4 | Flavin containing monooxygenase 4 |
| FMO5 | Flavin containing monooxygenase 5 | PSMD11 | Proteasome 26S subunit, non-ATPase 11 |
| CD34 | Hematopoietic progenitor cell antigen CD34 | CD38 | ADP-ribosyl cyclase 1 |
| CD47 | Leukocyte surface antigen CD47 | CD74 | Histocompatibility antigen gamma chain |
| CD80/86 | Cluster of differentiation 80/86 | CD300LD | CD300 molecule like family member D |
| NDST2 | N-deacetylase and N-sulfotransferase 2 | NDST3 | N-deacetylase and N-sulfotransferase 3 |
| NDST4 | N-deacetylase and N-sulfotransferase 4 | XYLT2 | Xylosyltransferase 2 |
| EXT2 | Exostosin glycosyltransferase 2 | EXT3 | Exostosin glycosyltransferase 3 |
| IL2RA | Interleukin-2 receptor subunit alpha | IL23R | Interleukin 23 receptor |
| IL5RA | Interleukin 5 receptor subunit alpha | IL9R | Interleukin-9 receptor |
Table 14.
Gene name and its abbreviation used in the article.
| Gene name | Abbreviation | Gene name | Abbreviation |
|---|---|---|---|
| CD209 | C-type lectin domain containing protein | CLEC4G | C-type lectin domain family 4 member G |
| CLEC4M | C-type lectin domain family 4 member M | CLEC5A | C-type lectin domain family 5 member A |
| FURIN | Furin, paired basic amino acid cleaving enzyme | DNM1 | Dynamin 1 |
| ANPEP | Aminopeptidase | ENPEP | Glutamyl aminopeptidase |
| DPP4 | Dipeptidyl peptidase 4 | DPP6 | Dipeptidyl peptidase 6 |
| SLAMF1 | Signaling lymphocytic activation molecule | APLP1 | Amyloid beta precursor like protein 1 |
| AP2A2 | Adaptor protein complex 2 subunit alpha 2 | EPS15 | Epidermal pathway substrate 15 |
| EPN1 | Epsin 1 | LDLR | Low density lipoprotein receptor |
| LY75 | Lymphocyte antigen 75 | MRC2 | Mannose receptor, C type 2 |
| ADAM7 | Disintegrin and metalloproteinase protein 7 | SNX5 | Sorting nexin-5 |
| NRP1 | Neuropilin | ICAM1 | Intercellular adhesion molecule 1 |
| EGFR | Epidermal growth factor receptor | AXL | Alpha-xylosidase |
| FCGRT | Fc gamma receptor and transporter | NRG1 | Neuregulin 1 |
| FCRL6 | Fc receptor like 6 | LRP1 | lipoprotein receptor-related protein 1 |
| FGFR1 | Fibroblast growth factor receptor | EFNB1 | Ephrin B1 |
| KREMEN1 | Kringle containing transmembrane protein 1 | ASGR1 | Asialoglycoprotein receptor 1 |
| ISLR | Immunoglobulin superfamily leucine-rich | SNCA | Alpha-synuclein |
| PCDH7 | Protocadherin 7 | ANXA3 | Annexin |
| CD14 | Monocyte differentiation antigen 14 | PDPN | Podoplanin |
| CALRL | Calreticulin | FOXF1 | Forkhead box F1 |
| AGER | Advanced glycosylation end-product receptor | MYRF | Myelin regulatory factor |
| TCF7L2 | Transcription factor 7 like 2 | LRP5 | Low density lipoprotein receptor protein |
| CYP4B1 | Cytochrome P450 family 4 subfamily B member 1 | ACTA2 | Actin alpha 2 |
| SERPINB4 | Serpin B4 | KRT4 | Keratin 4 |
| PLXNA1 | Plexin A1 | VEGFA | Vascular endothelial growth factor |
| EREG | Epiregulin | IGSF21 | Immunoglobin superfamily member 21 |
| APOE | Apolipoprotein E | TAGLN | Transgelin |
| COL1A2 | Collagen type I alpha 2 chain | FABP4 | Fatty acid-binding protein 4 |
| MCEMP1 | Mast cell expressed membrane protein 1 | MYH11 | Myosin heavy chain 11 |
| MMP14 | Matrix metallopeptidase 14 | ALDH1A2 | Aldehyde dehydrogenase 1 family, A2 |
| NRXN1 | Neurexin-1 | MYH2 | Myosin 2 |
| PLP1 | Proteolipid protein 1 | VCAM1 | Vascular cell adhesion protein 1 |
| ERBB2 | Receptor protein-tyrosine kinase | ERAP1 | Endoplasmic reticulum aminopeptidase 1 |
| JUN | Jun proto-oncogene | EPCAM | Epithelial cell adhesion molecule |
| ONECUT1 | One cut domain family member | SPARCL1 | SPARC-like protein 1 |
| BAMBI | BMP and activin membrane-bound inhibitor | CSF1 | Colony stimulating factor 1 |
| HEXIM1 | Hexamethylene bisacetamide inducible 1 | HMOX1 | Heme oxygenase 1 |
| MERTK | MER proto-oncogene, tyrosine kinase | MS4A7 | Membrane spanning 4-domains A7 |
| CD8A | T-cell surface glycoprotein CD8 alpha | IL7R | Interleukin 7 receptor |
| FGFR3 | Fibroblast growth factor receptor | CSF1R | Colony stimulating factor 1 receptor |
| SLC10A4 | Solute carrier family 10 member 4 | MEGF11 | Multiple epidermal growth factor 11 |
| GRM8 | Glutamate metabotropic receptor 8 | TRPM3 | Transient receptor potential member 3 |
| TM4SF1 | Transmembrane 4 L six family member 1 | DAG1 | Dystroglycan 1 |
| MXRA8 | Matrix remodeling associated 8 | CD147 | basigin or BSG |
| ITGA2 | Integrin subunit alpha 2 | ITGB1 | Integrin beta |
| TFRC | Transferrin receptor | BET1 | BET1 isoform 4 |
| ECE1 | Endothelin converting enzyme 1 | MEP1A | Meprin A subunit |
| TRHDE | Thyrotropin releasing hormone degrading enzyme | ITGA4 | Integrin subunit alpha 4 |
| ALK | Aurora-like kinase | FOLH1 | Folate hydrolase 1 |
| GOLM1 | Golgi membrane protein 1 | CHST1 | Carbohydrate sulfotransferase 1 |
| PCSK5 | Proprotein convertase subtilisin/kexin type 5 | CHPF2 | Chondroitin polymerizing factor 2 |
| GLCE | Glucuronic acid epimerase | EXT1 | Exostosin glycosyltransferase |
| EXTL | Exostosin like glycosyltransferase | NDST1 | N-deacetylase and N-sulfotransferase 1 |
| XYLT1 | Xylosyltransferase 1 | DNM2 | Dynamin 2 |
Table 15.
Gene name and its abbreviation used in the article.
| Gene name | Abbreviation | Gene name | Abbreviation |
|---|---|---|---|
| HERC2 | HECT, RLD domain E3 ubiquitin ligase 2 | WWP1 | WW domain E3 ubiquitin protein ligase 1 |
| MGRN1 | Mahogunin, ring finger 1 | NEDD4 | Neural precursor cell, downregulated 4 |
| UBE2 | Ubiquitin conjugating enzyme E2 | UBA2 | Ubiquitin-activating enzyme E1 2 |
| EPHB2 | Ephrin type-B receptor 2 | MCL1 | Myeloid leukemia cell differentiation |
| CRLF2 | Cytokine receptor-like factor 2 | ERBB4 | Receptor protein-tyrosine kinase |
| EPOR | Erythropoietin receptor | IL21R | Interleukin 21 receptor |
| LEPR | Leptin receptor | XBP1 | X-box binding protein 1 |
| EDEM1 | ER degradation enhancer, mannosidase alpha 1 | ATF6B | Activating transcription factor 6 beta |
| DERL1/2 | Derlin-1/2 | VCP | Valosin containing protein |
| HSPA5 | Heat shock protein 90 beta family member 1 | CANX | Calnexin |
| NFE2L2 | Nuclear factor erythroid 2 factor 2 isoform 1 | CALR | Putative calreticulin |
| PDIA3 | Protein disulfide-isomerase A3 | CD55 | Complement decay-accelerating factor |
| CXADR | Coxsackievirus and adenovirus receptor | SGC | Sarcoglycan |
| CYCS | Cytochrome c, somatic | BAK1 | BCL2 antagonist/killer 1 |
| HLA-f | HLA class I histocompatibility antigen, F | BAX | BCL domain-containing protein |
| KRAS | GTPase KRas isoform X1 | SYT | Synaptotagmin |
| CCND1 | Cyclin N-terminal domain-containing protein | SIGLEC1 | Sialic acid binding Ig like lectin 1 |
| CDKN | Cyclin-dependent kinase inhibitor | MLN-4760 | Promotilin-4760 |
| FMO3 | Flavin dimethylaniline monoxygenase 3 | NRCAM | Neuronal cell adhesion molecule |
| NFKB2 | Nuclear factor kappa B subunit 2 | TFR2 | Transferrin receptor 2 |
| CD46 | Membrane cofactor protein | GP2 | Glycoprotein 2 |
| PTPRC | Protein tyrosine phosphatase receptor type C | MET | Methyltransferase |
| FXYD6 | FXYD domain-ion transport regulator | OSMR | Oncostatin M receptor |
| CDHR3 | Cadherin related family member 3 | CLIC4 | Chloride intracellular channel protein |
| SRPRB | Signal recognition particle receptor subunit beta | SDC1/SDC4 | Syndecan -1/Syndecan-4 |
| PLS3 | Plastin 3 | GDF15 | Growth differentiation factor 15 |
| CD8B2 | T-cell surface glycoprotein CD8 beta-2 | CNMD | Chondromodulin-I |
| CD244 | Ig-like domain-containing protein | CXCL10 | C-X-C motif chemokine ligand 10 |
| MEGF9 | Multiple epidermal growth factor 9 | KRT5 | Keratin 5 |
| APMAP | Adipocyte plasma membrane-associated protein | IDO1 | Indoleamine 2,3-dioxygenase 1 |
| CXCL13 | Chemokine (C-X-C motif) ligand 13 | CCDC78 | Coiled-coil domain protein 78 |
| SCGB3A1 | Secretoglobin family 3A member 1 | IGF1R | Insulin-like growth factor 1 receptor |
| CEACAM1 | Carcinoembryonic antigen cell adhesion molecule 1 | INSR | Insulin receptor activity |
| HFE | Homeostatic iron regulator | COX7B | Cytochrome c oxidase subunit 7B |
| CADM1 | Cell adhesion molecule 1 | VIM | Vimentin |
| RHOX8 | Reproductive homeobox 8 | APOA1 | Apolipoprotein A-I |
| SPAG6 | Sperm-associated antigen 6 | ZPBP | Zona pellucida binding protein |
| ID4 | Inhibitor of DNA binding 4 | NEUROG3 | Neurogenin-3 |
| CYP11A1 | Cholesterol side-chain cleavage enzyme | MAGEA4 | Melanoma-associated antigen 4 |
| GGT5 | Gamma-glutamyltransferase 5 | GT7 | Putative glycosyltransferase 7 |
| JAM2 | Junctional adhesion molecule 2 | PLD6 | Phospholipase D family, member 6 |
| SPEM1 | Spermatid maturation protein 1 | SGPL1 | Sphingosine-1-phosphate lyase 1 |
| ROS1 | Tyrosine-protein kinase receptor | TYRO3 | Receptor protein-tyrosine kinase |
| GGT1 | Gamma-glutamyltransferase 1 | EGF | Epidermal growth factor |
| PECAM1 | Platelet endothelial cell adhesion molecule 1 | IL1RL1 | Interleukin 1 receptor like 1 |
| PDGFRB | Platelet-derived growth factor receptor beta | CUBN | Cubilin |
| KCNE1 | Potassium voltage-gated channel family E member 1 | FOXL1 | Forkhead box L1 |
| JAG | Jagged canonical Notch ligand | CX3CR1 | Chemokine (C-X3-C motif) receptor 1 |
| NOTCH2 | Neurogenic locus notch homolog protein 2 | HMGB1 | High mobility group box 1 |
| HAVCR1 | Hepatitis A virus cellular receptor 1 | OCLN | Occludin |
| LAMP1 | Lysosomal associated membrane protein 1 | PVR | Poliovirus receptor |
| SCARB1 | Scavenger receptor class B member 1 | RPSA | 30S ribosomal protein S1 |
| SLC10A1 | Solute carrier family 10 member 1 | PHB | Prohibitin |
| NCAM1 | Neural cell adhesion molecule 1 | NGFR | Nerve growth factor receptor |
7. Conclusion
For the first time, the paper discusses a metaheuristic fuzzy-based clustering approach for predicting the potential host receptor that either mediates or restricts SARS-CoV-2 infection in humans. The main reason for identifying the host receptor of SARS-CoV-2 infection from single-cell gene expression data is to study the role of the receptor in the pathogenesis of COVID-19 disease. It helps investigate the effect of these target receptors in the search for treatment against COVID-19 illness.
The proposed fuzzy-based clustering approach utilizes the GWO algorithm concept to find the optimal cluster number and centroid from the scRNA-Seq data. The exploratory and exploitatory search mechanism of the classical GWO algorithm is improved by hybridizing a set of mutation, crossover, and selection operators of the evolutionary algorithm. Towards the end of the optimization algorithm, the weak search agents are removed from the population and reinitialized around the position of the best search agent randomly to evolve through a better individual in the next generation. The fuzzy-based improved GWO clustering algorithm is then executed on various scRNA-Seq data of human tissue to identify a set of transcriptionally and biologically similar genes (membrane protein or ssRNA viral receptor protein) with ACE2. Also, PCC is calculated between ACE2 protein and membrane protein or viral receptor protein to validate the co-expressed genes. The interaction of the predicted receptor protein with the SARS-CoV-2 protein (ACE2 or TMPRSS2) is also analyzed through the PPI network. Previous work using hierarchy clustering had confirmed that the peptidases: DPP4, ANPEP and ENPEP are the co-receptor of ACE2 protein [7]. But our study successfully identified 816 membrane proteins and 58 viral receptors that play a vital role in the pathogenesis of SARS-CoV-2 infection.
The main advantage of the proposed fuzzy-based improved GWO clustering approach is its ability to study the expression level of a gene in every other cluster at one time. Previous work, such as IHC and MS studies, required more detailed pathology information to determine the biomarker of tissue at the microscopic level. As a result, it becomes difficult to study the expression level of a protein at the molecular level. It also requires a lot of effort and time for the specimen collection and laboratory setup. Also, single-cell transcriptomics analysis using the Seurat tool does not give a clear account of the biological functionality of the receptor protein at the molecular level. We have predicted the co-receptor protein of SARS-CoV-2 infection using the unsupervised fuzzy clustering technique with the GWO algorithm and analyzed the biological and cellular functioning of the receptor protein using PPI network, GO term, and KEGG pathway enrichment analysis.
We have identified the set of proteins that either mediates or restricts a biological pathway in the mechanism of SARS-CoV-2 infection. The work has also successfully identified the membrane protein that could inhibit the spread of SARS-CoV-2 infection. Antiviral drugs such as carboplatin and gemcitabine could prevent SARS-CoV-2 disease. Besides, one of the most significant findings is that one of the preventing SARS-CoV-2 infection in the initial stage is by downregulating the signaling pathway that promotes clathrin or caveolin mediated endocytosis process. Drug, imatinib, has been shown to inhibit the replication process of SARS-CoV-2 infection. In future, clathrin or caveolin mediated pathway can be studied to find the root cause of SARS-CoV-2 disease.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The work presented here falls under Research Project Grant No. EEQ/2020/000104 and supported by the Department of Science and Technology (DST) and Science and Engineering Research Board (SERB), Govt. of India .
Footnotes
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.106050.
Appendix A. Supplementary data
The following is the Supplementary material related to this article.
.
References
- 1.Dey L., Chakraborty S., Mukhopadhyay A. Machine learning techniques for sequence-based prediction of viral–host interactions between SARS-CoV-2 and human proteins. Biomed. J. 2020;43(5):438–450. doi: 10.1016/j.bj.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M., Kleine-Weber H., Krüger N., Müller M., Drosten C., Pöhlmann S. 2020. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv. [Google Scholar]
- 3.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J., Guo M., Tian X., Wang X., Yang X., Wu P., Liu C., Xiao Z., Qu Y., Yin Y., et al. Virus-host interactome and proteomic survey reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med. 2021;2(1):99–112. doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.St-Germain J.R., Astori A., Samavarchi-Tehrani P., Abdouni H., Macwan V., Kim D.-K., Knapp J.J., Roth F.P., Gingras A.-C., Raught B. 2020. A SARS-CoV-2 BioID-based virus-host membrane protein interactome and virus peptide compendium: New proteomics resources for COVID-19 research. BioRxiv. [Google Scholar]
- 6.Terracciano R., Preianò M., Fregola A., Pelaia C., Montalcini T., Savino R. Mapping the SARS-CoV-2–host protein–protein interactome by affinity purification mass spectrometry and proximity-dependent biotin labeling: A rational and straightforward route to discover host-directed anti-SARS-CoV-2 therapeutics. Int. J. Mol. Sci. 2021;22(2):532. doi: 10.3390/ijms22020532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh M., Bansal V., Feschotte C. 2020. A single-cell RNA expression map of human coronavirus entry factors. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nature Med. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H., Kang Z., Gong H., Xu D., Wang J., Li Z., Li Z., Cui X., Xiao J., Zhan J., et al. Digestive system is a potential route of COVID-19: An analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut. 2020;69(6):1010–1018. [Google Scholar]
- 11.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020:1–8. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z.-Y., Huang Y., Ganesh L., Leung K., Kong W.-P., Schwartz O., Subbarao K., Nabel G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004;78(11):5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marzi A., Gramberg T., Simmons G., Möller P., Rennekamp A.J., Krumbiegel M., Geier M., Eisemann J., Turza N., Saunier B., et al. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78(21):12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gramberg T., Hofmann H., Möller P., Lalor P.F., Marzi A., Geier M., Krumbiegel M., Winkler T., Kirchhoff F., Adams D.H., et al. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology. 2005;340(2):224–236. doi: 10.1016/j.virol.2005.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simmons G., Zmora P., Gierer S., Heurich A., Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: Cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100(3):605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirjalili S., Mirjalili S.M., Lewis A. Grey wolf optimizer. Adv. Eng. Softw. 2014;69:46–61. [Google Scholar]
- 18.Mirjalili S., Saremi S., Mirjalili S.M., Coelho L.d.S. Multi-objective grey wolf optimizer: A novel algorithm for multi-criterion optimization. Expert Syst. Appl. 2016;47:106–119. [Google Scholar]
- 19.Wang J.-S., Li S.-X. An improved grey wolf optimizer based on differential evolution and elimination mechanism. Sci. Rep. 2019;9(1):1–21. doi: 10.1038/s41598-019-43546-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saremi S., Mirjalili S.Z., Mirjalili S.M. Evolutionary population dynamics and grey wolf optimizer. Neural Comput. Appl. 2015;26(5):1257–1263. [Google Scholar]
- 21.Bezdek J.C. Springer Science & Business Media; 2013. Pattern Recognition with Fuzzy Objective Function Algorithms. [Google Scholar]
- 22.Bezdek J.C., Ehrlich R., Full W. FCM: The fuzzy c-means clustering algorithm. Comput. Geosci. 1984;10(2–3):191–203. [Google Scholar]
- 23.Nadeau R., Shahryari Fard S., Scheer A., Hashimoto-Roth E., Nygard D., Abramchuk I., Chung Y.-E., Bennett S.A., Lavallée-Adam M. Computational identification of human biological processes and protein sequence motifs putatively targeted by SARS-CoV-2 proteins using protein–protein interaction networks. J. Proteome Res. 2020;19(11):4553–4566. doi: 10.1021/acs.jproteome.0c00422. [DOI] [PubMed] [Google Scholar]
- 24.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z., Xu X. Scrna-seq profiling of human testes reveals the presence of the ACE2 receptor, a target for SARS-CoV-2 infection in spermatogonia, leydig and sertoli cells. Cells. 2020;9(4):920. doi: 10.3390/cells9040920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf F.A., Angerer P., Theis F.J. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):1–5. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nature Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mick E., Tsitsiklis A., Spottiswoode N., Caldera S., Serpa P.H., Detweiler A.M., Neff N., Pisco A.O., Li L.M., Retallack H., et al. 2021. Upper airway gene expression reveals a more robust innate and adaptive immune response to SARS-CoV-2 in children compared with older adults. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hui K.P., Cheung M.-C., Perera R.A., Ng K.-C., Bui C.H., Ho J.C., Ng M.M., Kuok D.I., Shih K.C., Tsao S.-W., et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: An analysis in ex-vivo and in-vitro cultures. Lancet Respir. Med. 2020;8(7):687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vieira Braga F.A., Kar G., Berg M., Carpaij O.A., Polanski K., Simon L.M., Brouwer S., Gomes T., Hesse L., Jiang J., et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nature Med. 2019;25(7):1153–1163. doi: 10.1038/s41591-019-0468-5. [DOI] [PubMed] [Google Scholar]
- 31.MacParland S.A., Liu J.C., Ma X.-Z., Innes B.T., Bartczak A.M., Gage B.K., Manuel J., Khuu N., Echeverri J., Linares I., et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nature Commun. 2018;9(1):1–21. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Song W., Wang J., Wang T., Xiong X., Qi Z., Fu W., Yang X., Chen Y.-G. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J. Exp. Med. 2020;217(2) doi: 10.1084/jem.20191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baron M., Veres A., Wolock S.L., Faust A.L., Gaujoux R., Vetere A., Ryu J.H., Wagner B.K., Shen-Orr S.S., Klein A.M., et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter-and intra-cell population structure. Cell Syst. 2016;3(4):346–360. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo J., Grow E.J., Mlcochova H., Maher G.J., Lindskog C., Nie X., Guo Y., Takei Y., Yun J., Cai L., et al. The adult human testis transcriptional cell atlas. Cell Res. 2018;28(12):1141–1157. doi: 10.1038/s41422-018-0099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang P., Yang M., Zhang Y., Xiao S., Lai X., Tan A., Du S., Li S. Dissecting the single-cell transcriptome network underlying gastric premalignant lesions and early gastric cancer. Cell Rep. 2020;30(12):4317. doi: 10.1016/j.celrep.2020.03.020. [DOI] [PubMed] [Google Scholar]
- 36.Liao J., Yu Z., Chen Y., Bao M., Zou C., Zhang H., Liu D., Li T., Zhang Q., Li J., et al. Single-cell RNA sequencing of human kidney. Sci. Data. 2020;7(1):1–9. doi: 10.1038/s41597-019-0351-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim D., Kobayashi T., Voisin B., Jo J.-H., Sakamoto K., Jin S.-P., Kelly M., Pasieka H.B., Naff J.L., Meyerle J.H., et al. Targeted therapy guided by single-cell transcriptomic analysis in drug-induced hypersensitivity syndrome: A case report. Nature Med. 2020;26(2):236–243. doi: 10.1038/s41591-019-0733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madissoon E., Wilbrey-Clark A., Miragaia R., Saeb-Parsy K., Mahbubani K., Georgakopoulos N., Harding P., Polanski K., Huang N., Nowicki-Osuch K., et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol. 2020;21(1):1–16. doi: 10.1186/s13059-019-1906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oetjen K.A., Lindblad K.E., Goswami M., Gui G., Dagur P.K., Lai C., Dillon L.W., McCoy J.P., Hourigan C.S. Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight. 2018;3(23) doi: 10.1172/jci.insight.124928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darmanis S., Sloan S.A., Zhang Y., Enge M., Caneda C., Shuer L.M., Gephart M.G.H., Barres B.A., Quake S.R. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. 2015;112(23):7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saha I., Maulik U., Plewczynski D. A new multi-objective technique for differential fuzzy clustering. Appl. Soft Comput. 2011;11(2):2765–2776. [Google Scholar]
- 42.Deb K., Pratap A., Agarwal S., Meyarivan T. A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans. Evol. Comput. 2002;6(2):182–197. [Google Scholar]
- 43.Ferreira J.A., Zwinderman A.H. On the Benjamini–Hochberg method. Ann. Statist. 2006;34(4):1827–1849. [Google Scholar]
- 44.Xie X.L., Beni G. A validity measure for fuzzy clustering. IEEE Trans. Pattern Anal. Mach. Intell. 1991;(8):841–847. [Google Scholar]
- 45.Maulik U., Bandyopadhyay S., Mukhopadhyay A. Springer Science & Business Media; 2011. Multiobjective Genetic Algorithms for Clustering: aPplications in Data Mining and Bioinformatics. [Google Scholar]
- 46.Dougherty E.R., Barrera J., Brun M., Kim S., Cesar R.M., Chen Y., Bittner M., Trent J.M. Inference from clustering with application to gene-expression microarrays. J. Comput. Biol. 2002;9(1):105–126. doi: 10.1089/10665270252833217. [DOI] [PubMed] [Google Scholar]
- 47.Dembele D., Kastner P. Fuzzy C-means method for clustering microarray data. Bioinformatics. 2003;19(8):973–980. doi: 10.1093/bioinformatics/btg119. [DOI] [PubMed] [Google Scholar]
- 48.Tari L., Baral C., Kim S. Fuzzy c-means clustering with prior biological knowledge. J. Biomed. Inform. 2009;42(1):74–81. doi: 10.1016/j.jbi.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amraei R., Yin W., Napoleon M.A., Suder E.L., Berrigan J., Zhao Q., Olejnik J., Chandler K.B., Xia C., Feldman J., et al. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2. ACS Central Sci. 2021;7(7):1156–1165. doi: 10.1021/acscentsci.0c01537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strollo R., Pozzilli P. DPP4 inhibition: Preventing SARS-CoV-2 infection and/or progression of COVID-19? Diabetes/Metaboli. Res. Rev. 2020;36(8) doi: 10.1002/dmrr.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rochette L., Zeller M., Cottin Y., Vergely C. GDF15: An emerging modulator of immunity and a strategy in COVID-19 in association with iron metabolism. Trends Endocrinol. Metabol. 2021;32(11):875–889. doi: 10.1016/j.tem.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cotugno N., Ruggiero A., Pascucci G.R., Bonfante F., Petrara M.R., Pighi C., Cifaldi L., Zangari P., Bernardi S., Cursi L., et al. Virological and immunological features of SARS-COV-2 infected children with distinct symptomatology. Pediatr. Allergy Immunol. 2021;32(8):1833–1842. doi: 10.1111/pai.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoffmann M., Pöhlmann S. Novel SARS-CoV-2 receptors: ASGR1 and KREMEN1. Cell Res. 2022;32(1):1–2. doi: 10.1038/s41422-021-00603-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S., Qiu Z., Hou Y., Deng X., Xu W., Zheng T., Wu P., Xie S., Bian W., Zhang C., et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31(2):126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Venkataraman T., Coleman C.M., Frieman M.B. Overactive epidermal growth factor receptor signaling leads to increased fibrosis after severe acute respiratory syndrome coronavirus infection. J. Virol. 2017;91(12) doi: 10.1128/JVI.00182-17. e00182–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valencia I., Peiró C., Lorenzo Ó., Sánchez-Ferrer C.F., Eckel J., Romacho T. DPP4 and ACE2 in diabetes and COVID-19: Therapeutic targets for cardiovascular complications? Front. Pharmacol. 2020:1161. doi: 10.3389/fphar.2020.01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jankowski J., Lee H.K., Wilflingseder J., Hennighausen L. JAK inhibitors dampen activation of interferon-activated transcriptomes and the SARS-CoV-2 receptor ACE2 in human renal proximal tubules. Iscience. 2021;24(8) doi: 10.1016/j.isci.2021.102928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang S., Park J., Qiu C., Chung K.W., Li S.-y., Sirin Y., Han S.H., Taylor V., Zimber-Strobl U., Susztak K. Jagged1/Notch2 controls kidney fibrosis via Tfam-mediated metabolic reprogramming. PLoS Biol. 2018;16(9) doi: 10.1371/journal.pbio.2005233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diabetes T.L., et al. COVID-19 and diabetes: A co-conspiracy? Lancet Diabetes Endocrinol. 2020;8(10):801. doi: 10.1016/S2213-8587(20)30315-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ricardo Criado P., Pincelli T.P.H., Criado R.F.J., Abdalla B.M.Z., Belda Junior W. Potential interactions of SARS-CoV-2 with human cell receptors in the skin: Understanding the enigma for a lower frequency of skin lesions compared to other tissues. Exp. Dermatol. 2020;29(10):936–944. doi: 10.1111/exd.14186. [DOI] [PubMed] [Google Scholar]
- 61.Bandsma R.H., van Goor H., Yourshaw M., Horlings R.K., Jonkman M.F., Schölvinck E.H., Karrenbeld A., Scheenstra R., Kömhoff M., Rump P., et al. Loss of ADAM17 is associated with severe multiorgan dysfunction. Hum. Pathol. 2015;46(6):923–928. doi: 10.1016/j.humpath.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gumashta J., Gumashta R. COVID19 associated mucormycosis: Is GRP78 a possible link? J. Infect. Public Health. 2021;14(10):1351–1357. doi: 10.1016/j.jiph.2021.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sabirli R., Koseler A., Goren T., Turkcuer I., Kurt O. High GRP78 levels in Covid-19 infection: A case-control study. Life Sci. 2021;265 doi: 10.1016/j.lfs.2020.118781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao Z.-M., Zhao J. An improved grey wolf optimization algorithm with variable weights. Comput. Intell. Neurosci. 2019;2019 doi: 10.1155/2019/2981282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yalcinkaya M., Liu W., Islam M.N., Kotini A.G., Gusarova G.A., Fidler T.P., Papapetrou E.P., Bhattacharya J., Wang N., Tall A.R. Modulation of the NLRP3 inflammasome by Sars-CoV-2 envelope protein. Sci. Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-04133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Torices S., Cabrera R., Stangis M., Naranjo O., Fattakhov N., Teglas T., Adesse D., Toborek M. Expression of SARS-CoV-2-related receptors in cells of the neurovascular unit: Implications for HIV-1 infection. J. Neuroinflammation. 2021;18(1):1–16. doi: 10.1186/s12974-021-02210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tavčar P., Potokar M., Kolenc M., Korva M., Avšič-Županc T., Zorec R., Jorgačevski J. Neurotropic viruses, astrocytes, and COVID-19. Front. Cell. Neurosci. 2021;15:123. doi: 10.3389/fncel.2021.662578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hikmet F., Méar L., Edvinsson Å., Micke P., Uhlén M., Lindskog C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020;16(7) doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020;202(5):756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(9):1–11. [PubMed] [Google Scholar]
- 71.Purcaru O.-S., Artene S.-A., Barcan E., Silosi C.A., Stanciu I., Danoiu S., Tudorache S., Tataranu L.G., Dricu A. The interference between SARS-CoV-2 and tyrosine kinase receptor signaling in cancer. Int. J. Mol. Sci. 2021;22(9):4830. doi: 10.3390/ijms22094830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mycroft-West C.J., Su D., Li Y., Guimond S.E., Rudd T.R., Elli S., Miller G., Nunes Q.M., Procter P., Bisio A., et al. 2020. Glycosaminoglycans induce conformational change in the SARS-CoV-2 spike S1 receptor binding domain. BioRxiv. [Google Scholar]
- 73.Glebov O.O. Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. FEBS J. 2020;287(17):3664–3671. doi: 10.1111/febs.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raaben M., Posthuma C.C., Verheije M.H., Te Lintelo E.G., Kikkert M., Drijfhout J.W., Snijder E.J., Rottier P.J., De Haan C.A. The ubiquitin-proteasome system plays an important role during various stages of the coronavirus infection cycle. J. Virol. 2010;84(15):7869–7879. doi: 10.1128/JVI.00485-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Borges L., Pithon-Curi T.C., Curi R., Hatanaka E. COVID-19 and neutrophils: The relationship between hyperinflammation and neutrophil extracellular traps. Mediators Inflamm. 2020;2020 doi: 10.1155/2020/8829674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olbei M., Hautefort I., Modos D., Treveil A., Poletti M., Gul L., Shannon-Lowe C.D., Korcsmaros T. SARS-CoV-2 causes a different cytokine response compared to other cytokine storm-causing respiratory viruses in severely ill patients. Front. Immunol. 2021;12:381. doi: 10.3389/fimmu.2021.629193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Diamond M.S., Kanneganti T.-D. Innate immunity: The first line of defense against SARS-CoV-2. Nature Immunol. 2022:1–12. doi: 10.1038/s41590-021-01091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kotlarz D., Zietara N., Milner J.D., Klein C. Human IL-21 and IL-21R deficiencies: Two novel entities of primary immunodeficiency. Curr. Opin. Pediatr. 2014;26(6):704–712. doi: 10.1097/MOP.0000000000000160. [DOI] [PubMed] [Google Scholar]
- 79.Bortolotti D., Gentili V., Rizzo S., Schiuma G., Beltrami S., Strazzabosco G., Fernandez M., Caccuri F., Caruso A., Rizzo R. TLR3 and TLR7 RNA sensor activation during SARS-CoV-2 infection. Microorganisms. 2021;9(9):1820. doi: 10.3390/microorganisms9091820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiang Q., Feng Z., Diao B., Tu C., Qiao Q., Yang H., Zhang Y., Wang G., Wang H., Wang C., et al. SARS-CoV-2 induces lymphocytopenia by promoting inflammation and decimates secondary lymphoid organs. Front. Immunol. 2021;12:1292. doi: 10.3389/fimmu.2021.661052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frye M., Stritt S., Ortsäter H., Vasquez M.H., Kaakinen M., Vicente A., Wiseman J., Eklund L., Martínez-Torrecuadrada J.L., Vestweber D., et al. EphrinB2-Ephb4 signalling provides rho-mediated homeostatic control of lymphatic endothelial cell junction integrity. Elife. 2020;9 doi: 10.7554/eLife.57732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sureda A., Alizadeh J., Nabavi S.F., Berindan-Neagoe I., Cismaru C.A., Jeandet P., Łos M.J., Clementi E., Nabavi S.M., Ghavami S. Endoplasmic reticulum as a potential therapeutic target for covid-19 infection management? Eur. J. Pharmacol. 2020;882 doi: 10.1016/j.ejphar.2020.173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stingi A., Cirillo L. SARS-CoV-2 infection and cancer: Evidence for and against a role of SARS-CoV-2 in cancer onset. BioEssays. 2021 doi: 10.1002/bies.202000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
.