Abstract
Although transcription by RNA polymerase (RNAP) is highly processive, elongation can be transiently halted by RNAP pausing. Pausing provides time for diverse regulatory events to occur such as RNA folding and regulatory factor binding. The transcription elongation factors NusA and NusG dramatically affect the frequency and duration of RNAP pausing, and hence regulation of transcription. NusG is the only transcription factor conserved in all three domains of life; its homolog in archaea and eukaryotes is Spt5. This review focuses on NusG-dependent pausing, which is a common occurrence in Bacillus subtilis. B. NusG induces pausing about once per 3 kb at a consensus TTNTTT motif in the non-template DNA strand within the paused transcription bubble. A conserved region of NusG contacts the TTNTTT motif to stabilize the paused transcription elongation complex (TEC) in multiple catalytically inactive RNAP conformations. The density of NusG-dependent pause sites is 3-fold higher in untranslated regions, suggesting that pausing could regulate the expression of hundreds of genes in B. subtilis. We describe how pausing in 5’ leader regions contributes to regulating the expression of B. subtilis genes by transcription attenuation and translation control mechanisms. As opposed to the broadly accepted view that NusG is an anti-pausing factor, phylogenetic analyses suggest that NusG-dependent pausing is a widespread mechanism in bacteria. This function of NusG is consistent with the well-established role of its eukaryotic homolog Spt5 in promoter-proximal pausing. Since NusG is present in all domains of life, NusG-dependent pausing could be a conserved mechanism in all organisms.
Keywords: NusG, Spt5, RNA polymerase pausing, gene regulation, riboswitch, transcription attenuation, translation control
Introduction
Transcription by RNA polymerase (RNAP) is punctuated by ubiquitous and regulated pausing events. Pausing can regulate co-transcriptional folding of nascent RNA or provide time for the binding of regulatory factors (Pan et al. 1999; Yakhnin and Babitzke 2002; Yakhnin et al. 2020). Pausing is triggered by interactions of RNAP with components of the transcription elongation complex (TEC). Pause signals within the nascent RNA and DNA template cause isomerization of RNAP such that it enters into an elemental pause state in which elongation is reversibly inhibited (reviewed in Zhang and Landick 2016). Elemental pause sites are frequent, have a short duration, and exhibit limited nucleic acid sequence specificity, while other pauses are long-lived and have more extensive sequence requirements and/or RNA secondary structure (Herbert et al. 2006; Zhang and Landick 2016). A limited in vivo consensus sequence for the elemental pause sites was identified for Escherichia coli and Bacillus subtilis, with pausing occurring every 100–200 nt on average (Larson et al. 2014; Vvedenskaya et al. 2014). Although entry into an elemental pause state is a fundamental property of RNAP, the majority of these pause sites are unlikely to play key regulatory roles due to their stochastic nature and short duration (Saba et al. 2019). However, elemental pauses likely serve as precursors for long-lived pauses.
Transcription factors can influence both elemental and long-lived pause events. Interaction of transcription factors with the TEC can dramatically alter the duration of a pause (pause half-life) and/or the frequency that RNAP pauses at a particular site (pause efficiency) (Landick et al. 1996). NusA and NusG are two general transcription elongation factors that stimulate pausing of B. subtilis RNAP in response to specific signals in the DNA template and nascent transcript (Yakhnin and Babitzke 2002; Yakhnin et al. 2006; Yakhnin et al. 2008; Ma et al. 2015; Yakhnin et al. 2016). In E. coli, NusA stimulates pausing whereas NusG suppresses pausing (Artsimovitch and Landick 2000).
NusA consists of two KH domains, an S1 domain, and an RNAP interacting domain. Although intact NusA is only found in bacteria, NusA might be split into two polypeptides in archaeal species, with one polypeptide containing the KH domains and a subunit of RNAP (Rpo7) containing the S1 and RNAP interacting domains (reviewed in Fouqueau et al. 2018). NusA-stimulated pausing was first discovered in E. coli during studies of the trp operon attenuation mechanism (Winkler and Yanofsky 1981; Farnham et al. 1982; Fisher and Yanofsky 1983; Landick and Yanofsky 1984). Hairpin-stimulated pause signals include an RNA hairpin, the RNA sequence between the hairpin and 3′ end of the transcript, and the downstream DNA sequence (Zhang and Landick 2016). E. coli NusA increases the pause half-life by stimulating a basal interaction between the pause hairpin and the β flap domain of RNAP (Artsimovitch and Landick 2000; Toulokhonov et al. 2001). The pause hairpin also blocks backtracking of RNAP, which prevents conversion of the transient pause into a poorly reversible backtracked state (Artsimovitch and Landick 2000). As NusA from E. coli and B. subtilis can substitute for one another to stimulate pausing in vitro (Yakhnin et al. 2008), it is apparent that their mechanism of action is essentially identical and widely conserved in bacteria. In contrast to NusA, NusG from these organisms have opposite functions on pausing. B. subtilis NusG greatly stimulates pausing by making sequence-specific contacts with the non-template DNA (ntDNA) strand within the paused transcription bubble (Yakhnin et al. 2008; Yakhnin and Babitzke 2010; Yakhnin et al. 2016), whereas E. coli NusG increases the transcription elongation rate by suppressing pausing (Herbert et al. 2010; Mooney, Schweimer, et al. 2009; Yakhnin et al. 2008; Sevostyanova et al. 2011). However, our preliminary analysis suggests that, similar to E. coli NusG, B. subtilis NusG may inhibit short elemental pauses at sites lacking the cognate TTNTTT motif in ntDNA.
NusG is the only universally conserved transcription factor and is called Spt5 in archaeal and eukaryotic species (Tomar and Artsimovitch 2013). NusG was originally discovered as an E. coli cofactor of bacteriophage N protein required for lytic infection of bacteriophage lambda. Together with NusA and other Nus factors [NusB, NusE (S10)], NusG is part of an N protein-mediated transcription antitermination complex that transcribes phage lytic genes past multiple terminators (Olson et al. 1982; Mason and Greenblatt 1991; Zhou et al. 2002). A similar antitermination complex containing NusA and NusG plays a critical role during the transcription of rRNA operons (Zellars and Squires 1999; Huang et al. 2020). Bacterial NusG consists of two domains including the N-terminal NGN domain that binds to the clamp helices of the β′ subunit of RNAP, and the KOW domain that is free to interact with various regulatory partners (Figure 1(A)) (Liu and Steitz 2017). Regions of RNAP that interact with NusG and with the RNAP σ factor overlap, thus binding of these two factors are mutually exclusive. Not surprisingly, ChIP experiments identified enrichment of σ at promoter-proximal regions of operons and enrichment of NusG inside protein-coding regions of genes (Mooney, Davis, et al. 2009). The KOW domain of NusG recruits additional factors to the TEC such as the Rho termination factor, leading to increased termination efficiency at a subset of Rho-dependent termination sites in E. coli (reviewed in Peters et al. 2011). Rho protein is an RNA helicase that binds to untranslated nascent RNA and translocates along the RNA toward RNAP to cause termination. In addition, the interaction of E. coli NusG and ribosomal protein S10 (NusE) is involved in the coupling of transcription and translation, which protects RNAP transcribing protein-coding regions from premature termination by Rho (Burmann et al. 2010).
Figure 1.
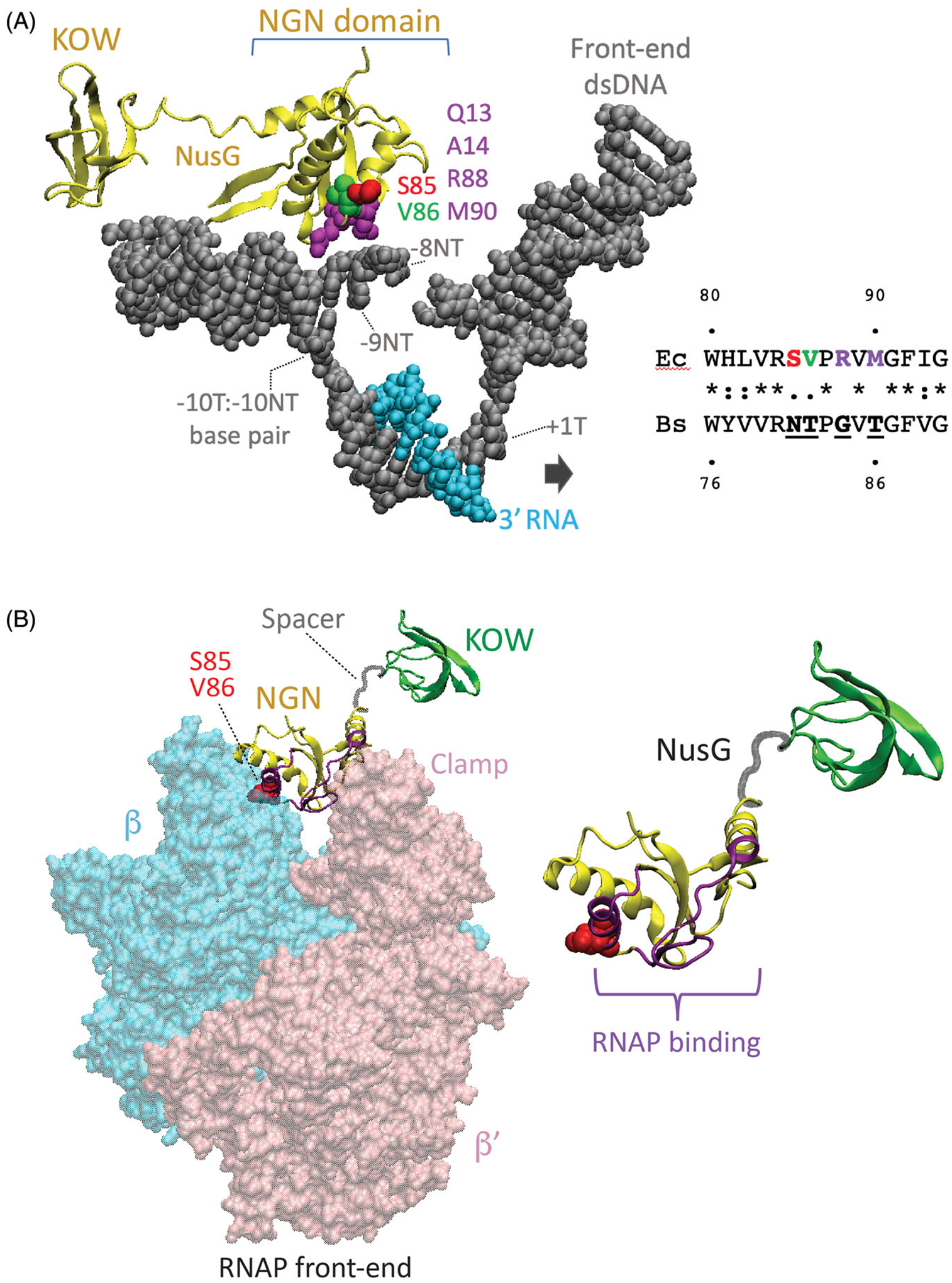
Structural foundation for NusG-dependent pausing. (A) Cryo-EM structure of the E. coli TEC containing Nus factors and N protein of bacteriophage lambda (PDB: 6GOV, Krupp et al. 2019). All Nus factors and RNAP subunits were removed for clarity. E. coli NusG shows the KOW and NGN domains with the NGN domain bound to a nucleic acid scaffold containing DNA (gray) and RNA (cyan). The residues in the template (T) and the non-template (NT) DNA strands in the vicinity of the NGN domain are shown with the numbers indicating their distance from the RNA 3’ end. Surface exposed residues of the NusG NGN domain in close proximity to the ntDNA strand within the transcription bubble are color-coded. Residues of the B. subtilis NGN domain that are vital for pausing (N81 and T82) correspond to S85 and V86 of E. coli NusG, respectively. (B) Binding site for the NGN domain on RNAP (PDB: 6GOV). NGN domain, yellow; KOW domain, green; Spacer, gray. Only β (cyan) and β’ (pink) subunits of RNAP are shown. Surface of the NGN domain and the S85/V86 residues at the interface with RNAP are colored in magenta and red, respectively.
Mechanism of NusG-dependent pausing
Pause-stimulating or anti-pausing activity of NusG varies between organisms and sequence motifs
In E. coli, NusG accelerates the elongation rate and suppresses pausing by stimulating forward translocation and preventing backtracking of RNAP (Sevostyanova et al. 2011; Turtola and Belogurov 2016). In contrast, NusG stimulates pausing in the B. subtilis trpEDCFBA, tlrB (yxjB) and ribDEAHT operons (Yakhnin et al. 2008; Yakhnin et al. 2019; Yakhnin et al. 2020). Perhaps in a similar fashion, NusG from Thermus thermophilus slows down transcript elongation (Sevostyanova and Artsimovitch 2010) and mycobacterial NusG exhibits a termination-stimulating activity (Czyz et al. 2014). E. coli NusG does not affect B. subtilis RNAP pausing, but competitively inhibits B. subtilis NusG-stimulated pausing when both factors are present together in vitro, indicating that they bind to the same surface of RNAP (Yakhnin et al. 2008). Mutational analysis of NusG-dependent pause sites revealed the importance of a hairpin structure in the nascent RNA (pause hairpin) and a stretch of T residues in the ntDNA strand upstream of the pause site. NusG protects these single-stranded T residues from permanganate oxidation when they are present within the transcription bubble in the paused TEC (Yakhnin et al. 2008). In vitro reconstitution of paused TECs using a variety of nucleic acid scaffolds consisting of DNA and RNA oligonucleotides and wild type (WT) and mutant NusG proteins revealed that these single-stranded T residues are recognized by a cluster of surface-exposed amino acid residues of NusG including a critical N81 T82 motif (Figure 1(A)) (Yakhnin et al. 2016). NusG was shown to crosslink to one of these single-stranded T residues, providing direct evidence that it binds to the T-rich tract in the bubble (Yakhnin et al. 2016). Binding of B. subtilis NusG to E. coli RNAP does not stimulate pausing at the B. subtilis trp pause site in vitro (Yakhnin et al. 2008), indicating that there are additional elements within these T-rich pause sites that are recognized by B. subtilis core RNAP. Eukaryotic Spt5 protein stimulates promoter-proximal pausing of RNA polymerase II by interaction with negative elongation factor NELF (Yamaguchi et al. 1999). Since yeast Spt5 also interacts with the ntDNA strand within the transcription bubble (Crickard et al. 2016), NusG/Spt5-dependent pausing might be a universally conserved mechanism.
Genome-wide mapping of nascent RNA 3′ ends using RNET-seq (Imashimizu et al. 2015) identified 1600 NusG-dependent pause sites in the B. subtilis genome (Yakhnin et al. 2020). Bioinformatic analysis of the RNET-seq data revealed a conserved TTNTTT sequence in the transcriptional bubble that is necessary, but not sufficient for NusG-dependent pausing. Each of the characterized NusG-dependent pause sites that regulate expression of the trpEDCFBA, tlrB and ribDEAHT operons possess this short motif (Figure 2(A)). The degree of conservation of each T residue in the TTNTTT motif identified in vivo closely parallels the relative importance of each T residue previously determined for the trp leader pause site in vitro (Yakhnin and Babitzke 2010). NusG-dependent pausing was confirmed in vitro for 10 NusG-dependent pause sites identified in vivo. However, this motif is not sufficient for NusG-dependent pausing. Only a small fraction of the TTNTTT sequences present in the B. subtilis genome induce RNAP pausing in vivo, which was also confirmed in vitro (Yakhnin et al. 2020). Competition between WT and pause-deficient NusG mutants capable of binding to RNAP revealed that NusG binds to TECs with low affinity such that RNAP-bound and free NusG readily exchange with one another. Sequence-specific interaction with the pause motif in the ntDNA strand slows down the exchange, indicating that the affinity of NusG is higher for a TEC carrying the TTNTTT sequence in the transcription bubble (Yakhnin et al. 2016).
Figure 2.
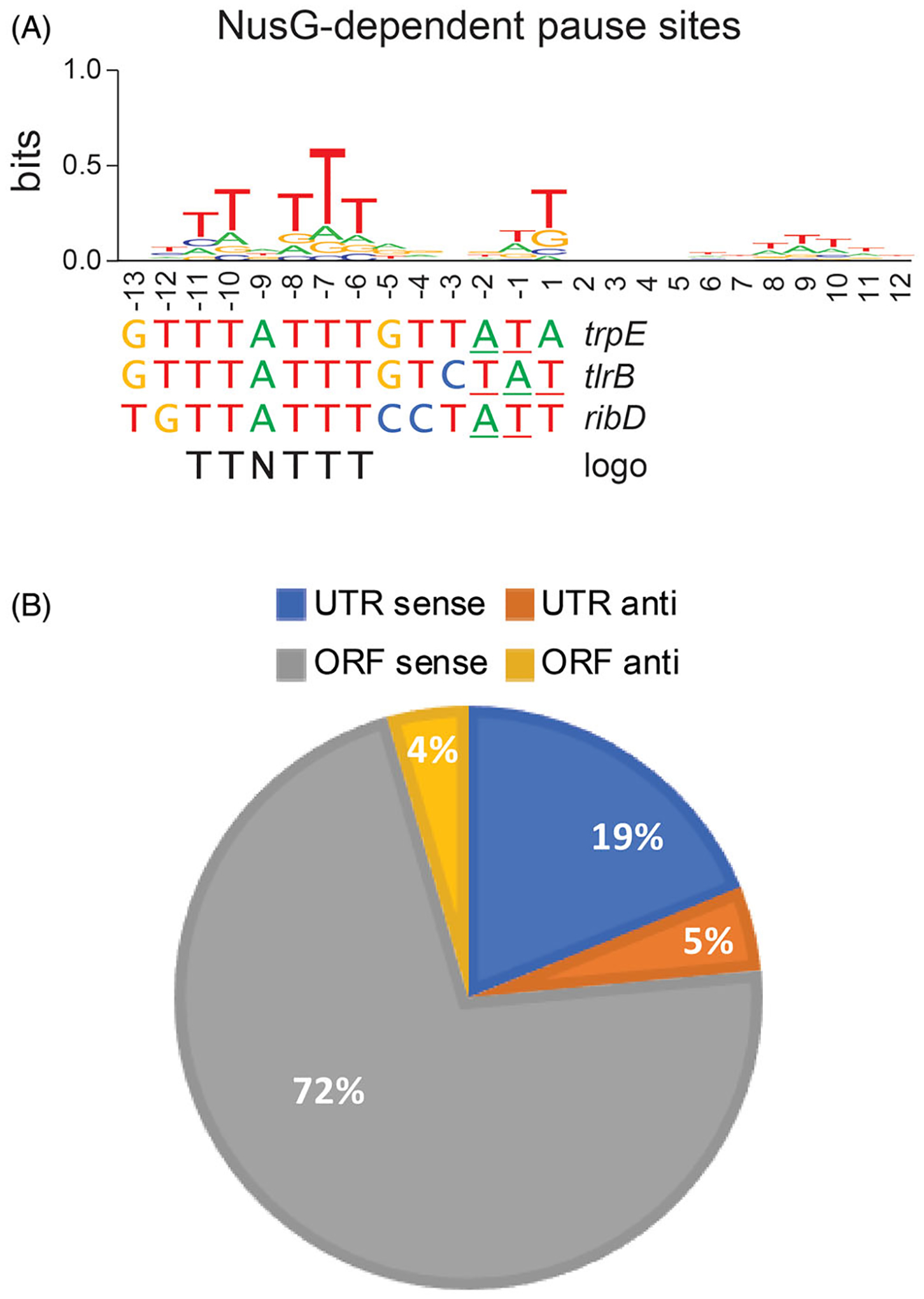
NusG-dependent pausing is sequence-specific and occurs throughout the B. subtilis genome. (A) TTNTTT sequence logo of the 1600 NusG-dependent pause sites identified in vivo by RNET-seq. This logo is shared by the trpE, tlrB and ribD pause sites that have been characterized in vitro. 3’ ends of paused RNA are underlined. (B) Genome-wide distribution of NusG-dependent pause sites in untranslated (UTR) and coding (ORF) regions in the sense (sense) and antisense (anti) directions.
NusG shifts RNAP to the post-translocation register
After each NMP incorporation, RNAP alternates between the pre-translocation and the post-translocation register by rapidly translocating back and forth at one base-pair along the DNA template. The transient forward translocation vacates the active site (i + 1) for binding of the next incoming NTP, followed by phosphodiester bond formation and establishment of a new pre/post-translocation equilibrium at the next template position. Because transcription elongation requires that RNAP and the DNA move with respect to one another, simultaneous NusG interaction with RNAP and the DNA template was initially assumed to stimulate pausing by preventing forward translocation of RNAP (reviewed in Yakhnin and Babitzke, 2014). A NusG-dependent pausing model was proposed in which recognition of the TTNTTT motif occurred via T-base flipping and their subsequent capture by the NGN domain of NusG (Yakhnin et al. 2016). This model predicted that RNAP would be in the pre-translocation register at NusG-dependent pause sites, but that model was not supported by in vivo RNET-seq results. RNET-seq is a unique method that provides genome-wide assessment of the translocation state of RNAP at NusG-dependent and NusG-independent pause sites in vivo (Imashimizu et al. 2015). Surprisingly, RNET-seq identified a higher fraction of RNAP in the post-translocation register at NusG-dependent pause sites compared to NusG-independent pauses (Yakhnin et al. 2020), in agreement with the promotion of forward translocation observed for E. coli NusG in vitro (Bar-Nahum et al. 2005; Herbert et al. 2010; Sevostyanova et al. 2011). During normal elongation, RNAP translocation relative to the DNA template and nascent RNA occurs simultaneously. For many NusG-dependent sites identified by RNET-seq, RNAP paused at several consecutive positions separated by 1–2 base pairs of DNA. The predominant post-translocated state of RNAP was preferentially observed at the first pause position, while this state was progressively less populated for the downstream positions, indicating that the paused TEC adopted a series of spatially related conformational states while transcribing through the TTNTTT motif in the presence of NusG (Yakhnin et al. 2020). This pattern indicated that NusG caused multiple catalytically inactive RNAP conformations depending on a spatial location of a TTNTTT motif relative to the 3′ RNA end, rather than serving as a general passive block to translocation. NusG likely inactivates RNAP by inducing partially translocated intermediates where the RNA 3′ end occupies the posttranslocated register while the DNA is pre-translocated, which prevents proper alignment of the incoming NTP with the template DNA base in the active site. A similar conformation was observed in the RNA hairpin- and NusA-stabilized E. coli his pause site (Kang, Mishanina, et al. 2018; Guo et al. 2018). In this model, the simultaneous binding of NusG to the TTNTTT sequence in ntDNA and to RNAP mechanically inhibits forward translocation of DNA, but not the nascent RNA. Interestingly, the amino acid residues of NusG that interact with the TTNTTT motif localize at the interface between the NGN domain of NusG and the clamp domain of RNAP (Krupp et al. 2019). Loosening or swiveling of the clamp domain was implicated in pausing of E. coli RNAP (Hein et al. 2014; Kang, Mishanina, et al. 2018; Saba et al. 2019). This raises an intriguing possibility that binding of B. subtilis NusG to the bubble may directly interfere with the clamp closure leading to the asynchronous translocation (Figure 1(B)).
Distribution of NusG-dependent pause sites in the genome
The identification of 1600 strong NusG-dependent pause sites revealed that NusG is a global pause-stimulating factor in B. subtilis (Yakhnin et al. 2020). Most of these pause sites are located in protein-coding regions distributed relatively evenly along open reading frames (Figure 2(B)). Although not yet tested, it is possible that these pause sites are involved in the coupling of transcription and translation to prevent Rho-dependent termination during transcription in ORFs, or for slowing down translation to stimulate proper folding of the emerging polypeptide chain. While lower in overall frequency (~25%), the density of pause sites is higher in untranslated regions (Figure 2(B)), the majority of which are in 5′ leaders and they could contribute to regulation via transcription attenuation and translation control mechanisms (see below). Interestingly, NusG-dependent pause sites were not identified in rRNA operons, and these operons were significantly depleted of the TTNTTT pause motifs compared to the other B. subtilis genes. This finding suggests that the highly transcriptionally active rRNA operons evolved to be protected from transcriptional inhibition by NusG. Strikingly, no queue of trailing RNAPs behind a leading paused RNAP (so-called traffic jam) was observed even for exceptionally strong NusG-dependent pauses. The lack of a queue suggests that a trailing RNAP is capable of restoring elongation of the leading paused RNAP, or that the paused RNAP resumes transcription prior to the arrival of the trailing RNAP. The frequency of these two possibilities would be dictated by the strength of the pause and the rate of transcription initiation.
The B. subtilis type of NusG is widespread among bacteria
Amino acid residues N81 and T82 in B. subtilis NusG are involved in recognizing the TTNTTT pause motif. In contrast, E. coli NusG has residues S85 and V86 at these positions and does not stimulate pausing at the TTNTTT sequence (Yakhnin et al. 2016). The structure of a transcription antitermination complex confirmed close proximity of V86 in E. coli NusG to the −8 residue in the ntDNA strand; this residue corresponds to the underlined T of the TTNTTT pause motif (Figure 2(A)) (Said et al. 2017; Krupp et al. 2019; Huang et al. 2020). Notably, the T residue in the −7 position of a NusG-dependent pause site crosslinked to NusG (Yakhnin et al. 2016). Phylogenetic analysis of the taxonomic relationship of bacterial NusG-like homologs was performed based on the presence of either B. subtilis-like (NT) or E. coli-like (SV) amino acid residues. This search indicated that the B. subtilis type of NusG is widespread among bacteria, whereas the E. coli version of NusG is restricted primarily to γ-proteobacteria (Yakhnin et al. 2020). The phylogenetic search also revealed occasional horizontal transfer of nusG genes between bacterial and archaeal species.
Comparison of σ (SigA)- and NusG-dependent pausing
Although best known for its role in promoter recognition during transcription initiation, σ factor-mediated promoter-proximal pausing has been identified in bacteria (Ring et al. 1996). Since the binding sites of the σ factor and NusG on core RNAP overlap, the binding of these proteins is mutually exclusive. Notably, both σ and NusG recognize T-rich hexanucleotide sequence motifs (TATAAT and TTNTTT, respectively) in unpaired regions of the ntDNA strand (Ring et al. 1996; Yakhnin et al. 2016; Yakhnin et al. 2020). Moreover, ChIP-seq analysis showed that σ and NusG are associated with promoter-proximal and more distal regions of transcription units, respectively (Mooney, Davis, et al. 2009). As a result, NusG-dependent pausing is more frequent in promoter-distal regions of the transcriptome (Yakhnin et al. 2020), whereas σ-dependent pausing prevails in promoter-proximal regions (Ring et al. 1996). Together, σ and NusG may cause RNAP pausing anywhere inside a transcription unit. In addition, unstable σ and NusG association with the TEC, and its stabilization by recognition of specific sequences, may allow exchange of these auxiliary factors during transcription depending on the frequency of their recognition motifs and/or the presence of additional elongation factors such as NusA; the NusA and NusG binding sites on core RNAP overlap two different portions of the σ binding site.
Role of specialized NusG paralogs in pausing
Bacterial NusG has several paralogs that are not general transcription factors, but instead regulate expression of specific regions of the genome (reviewed in Artsimovitch and Knauer 2019). The best-studied NusG paralog is RfaH, an E. coli protein that activates expression of regions acquired by horizontal gene transfer (Kang, Mooney, et al. 2018). NusG and its paralogs are recruited to the TEC in markedly different ways. NusG binds to the TEC after dissociation of the σ factor either directly or via delivery by the 30S ribosomal subunit during the process of coupled transcription and translation (Saxena et al. 2018). NusG recruitment does not appear to rely on a specific nucleic acid sequence, and NusG remains loosely bound through elongation unless additional factors result in the formation of a stable NusG-containing TEC. In contrast, specialized NusG paralogs such as RfaH are recruited at specific structured sequence elements and remain tightly bound to the TEC throughout elongation (Zuber et al. 2018). Apparently, the short single-stranded stretch of ntDNA is sufficient to provide flexible non-overlapping specificity for distinct NusG paralogs. In E. coli, RfaH is recruited at ops sites that form a short hairpin structure, although this sequence is not similar to the B. subtilis TTNTTT motif (Artsimovitch and Landick 2002; Zuber et al. 2018). However, homologous regions of RhaH and B. subtilis NusG participate in the binding of the corresponding ntDNA sequence elements (Belogurov et al. 2010; Yakhnin et al. 2016). As a part of the TEC, NusG may suppress or stimulate pausing in response to specific signals. In contrast, specialized RfaH stimulates pausing only at the site of recruitment and suppresses downstream pausing and Rho-dependent termination signals as a component of the TEC (Artsimovitch and Landick 2002).
NusG-dependent pausing regulates gene expression
Regulation of tryptophan biosynthesis
NusG-dependent pausing was discovered during our studies of the B. subtilis trpEDCFBA operon. This operon contains six of the seven genes required for tryptophan biosynthesis (reviewed in Babitzke 2004). The 11 subunit trp RNA-binding attenuation protein (TRAP) is responsible for regulating expression of the trpEDCFBA operon in response to tryptophan by transcription attenuation and translation control mechanisms (Babitzke and Yanofsky 1993; Merino et al. 1995; Antson et al. 1995). In the attenuation mechanism, TRAP is responsible for the decision to terminate transcription in the leader region or to allow transcription to proceed into the trp operon structural genes (Figure 3). An antiterminator structure can form just upstream of an intrinsic terminator. Since these two structures overlap by 3 nt their formation is mutually exclusive. When activated by tryptophan, TRAP binds to 11 trinucleotide repeats in the leader region of the nascent trp transcript, which prevents the formation of the antiterminator (Babitzke et al. 1996; Antson et al. 1999). As a consequence, the terminator can form and RNA is released upstream of the trpE coding sequence. In limiting tryptophan growth conditions TRAP is not activated and does not bind to the nascent transcript. Under these conditions, the antiterminator forms and the operon is expressed (Figure 3). A hairpin-dependent pause site was identified at position U107 just upstream of the critical overlap between the antiterminator and terminator structures. Pausing at this position, which is stimulated by both NusA and NusG in vitro, provides additional time for TRAP to bind and promote termination (Figure 3) (Yakhnin and Babitzke 2002; Mondal et al. 2017). However, since we did not identify this pause site in vivo by RNET-seq (Yakhnin et al. 2020), it is not clear whether pausing participates in the trp operon attenuation mechanism in vivo.
Figure 3.
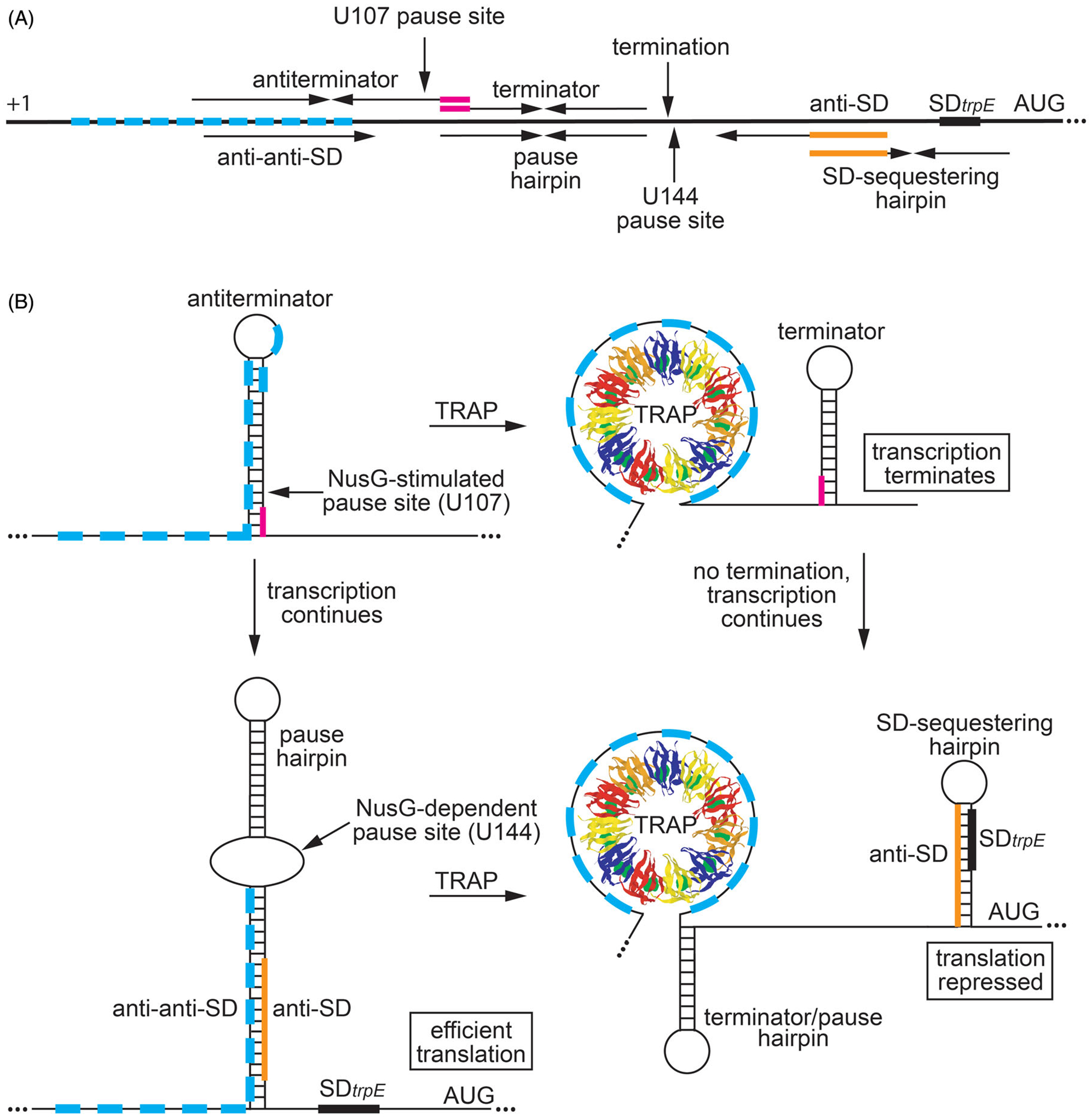
Regulation of trpEDCFBA operon in response to tryptophan availability by transcription attenuation and translational control mechanisms. (A) Schematic representation of the trpEDCFBA operon leader region. The thick black line represents the leader region between the start of transcription (+1) and the AUG start codon for trpE. The 11 trinucleotide repeats within the TRAP binding are shown as cyan boxes. (Top) Components involved in transcription attenuation. Inverted repeats for the overlapping antiterminator and terminator structures (3-nt overlap in magenta), the U107 pause site, and the point of termination (G140) are labeled. (Bottom) Components involved in translation control. The pause hairpin and U144 pause site are labeled. Inverted repeats for the large secondary structure and the overlapping SD-sequestering hairpin are shown (overlap in orange). The anti-anti-SD, anti-SD (orange), and SD sequences are also labeled. (B) Transcription attenuation model (top). During transcription, RNAP pauses at the NusG-dependent pause site at position U107. Under tryptophan-limiting conditions, TRAP does not bind to trp leader RNA, and RNAP eventually overcomes the pause and transcription resumes. In this case, formation of the antiterminator structure prevents the formation of the terminator hairpin, resulting in transcription readthrough into the trp operon structural genes. Under tryptophan-excess conditions, tryptophan-activated TRAP binds to the 11 trinucleotide repeats while RNAP is paused at U107, which prevents the formation of the antiterminator structure. As a consequence, the terminator hairpin forms and transcription terminates at G140. Thus, NusG-stimulated pausing at U107 provides additional time for TRAP to bind and promote termination. trpE translation control model (bottom). During transcription of trp operon readthrough transcripts, RNAP pauses at the NusG-dependent pause site at position U144, which provides a second opportunity for TRAP to bind to the nascent trp transcript. Under tryptophan-limiting conditions, TRAP does not bind to the nascent trp leader transcript, and RNAP eventually overcomes the pause and resumes transcription. In this case, the RNA adopts a structure such that the trpE SD sequence is single-stranded, resulting in efficient translation. Under tryptophan-excess conditions, TRAP can bind to the paused transcript. RNAP eventually overcomes the pause and resumes transcription, which leads to the formation of the trpE SD sequestering hairpin and repression of translation. Note that the same structure functions as the terminator and U144 pause hairpin. Color coding is the same as in (A).
In addition to the attenuation mechanism, TRAP regulates translation of trpE by binding to the same trinucleotide repeat region of trp operon readthrough transcripts that fail to terminate in the leader region. In this case, TRAP binding promotes the formation of an RNA structure that sequesters the trpE Shine-Dalgarno (SD) sequence, resulting in repression of trpE translation (Merino et al. 1995; Du and Babitzke 1998). In the absence of bound TRAP, a large RNA secondary structure forms such that the trpE SD sequence is single-stranded and available for ribosome binding (Figure 3). A hairpin-stimulated NusG-dependent pause site was identified at position U144 that is also stimulated by NusA in vitro (Yakhnin and Babitzke 2002; Yakhnin et al. 2008). Pausing at this position participates in the trpE translation control mechanism (Yakhnin et al. 2006). Importantly, our RNET-seq studies identified a strong NusG-dependent pause site at this position in vivo (Yakhnin et al. 2020). Thus, NusG-dependent pausing participates in the trpE translation control mechanism by providing a second opportunity for TRAP to bind to the nascent trp transcript, which in this case results in the formation of the trpE SD-sequestering hairpin.
Regulation of tylosin resistance
The strongest NusG-dependent pause site in the B. subtilis genome was identified in the leader region of tlrB (yxjB). A pause hairpin was also identified for this pause site (Yakhnin et al. 2019). tlrB encodes RlmAII, which is an enzyme that methylates the N1 position of G748 in 23S rRNA (Liu and Douthwaite 2002; Yakhnin et al. 2019). Methylation of this residue confers resistance to tylosin, a macrolide antibiotic produced by Streptomyces fradiae. Tylosin binds in the ribosome exit tunnel and inhibits peptidyl transferase activity, while methylation of G748 inhibits tylosin interaction (Liu and Douthwaite 2002; Vázquez-Laslop and Mankin 2018; Yakhnin et al. 2019). Bound tylosin causes the translating ribosome to stall when an appropriately positioned macrolide arrest motif, such as R/K-X-R/K, is encountered in the nascent polypeptide (Davis et al. 2014). In the absence of tylosin, tlrB expression is repressed by transcription attenuation and translation repression mechanisms (Figure 4). However, growth in the presence of sub-inhibitory concentrations of tylosin induces tlrB expression by alleviating these two repression mechanisms, resulting in increased rRNA methylation and increased resistance (Yakhnin et al. 2019). Induction requires tylosin-dependent ribosome stalling at an RYR arrest motif at the carboxy-terminus of a leader peptide encoded upstream of the tlrB coding sequence. This mechanism of translation arrest is an example of translation attenuation mediated by macrolide antibiotics (Vázquez-Laslop and Mankin 2018). The induction mechanism requires NusG-dependent RNAP pausing upstream of the NusA-dependent termination site in the tlrB leader region (Mondal et al. 2016; Yakhnin et al. 2019). Pausing provides sufficient time for a ribosome to initiate translation of the leader peptide. In the absence of tylosin, as the ribosome approaches the stop codon it results in the release of paused RNAP such that transcription resumes, resulting in transcription termination about 50% of the time (transcription attenuation). Transcripts that fail to terminate result in the formation of a tlrB SD-sequestering hairpin (translation repression) (Figure 4). However, in the presence of tylosin the translating ribosome stalls at the RYR arrest motif. The position of the stalled ribosome simultaneously prevents completion of the terminator hairpin and formation of the tlrB SD-sequestering hairpin, resulting in increased transcription and translation of tlrB, increased 23S rRNA methylation, and hence tylosin resistance (Yakhnin et al. 2019) (Figure 4). This complex mechanism ensures that methylation of G748 in 23S rRNA only occurs when tylsoin is present. This tight control over tlrB expression presumably minimizes a fitness cost associated with methylation of 23S rRNA when tylosin is not present. Since both B. subtilis and S. fradiae are soil microorganisms, the ability of B. subtilis to protect itself from the harmful effects of tylosin produced by S. fradiae provides a distinct growth advantage relative to other soil microbes that are unable to induce resistance to this antibiotic (Yakhnin et al. 2019).
Figure 4.
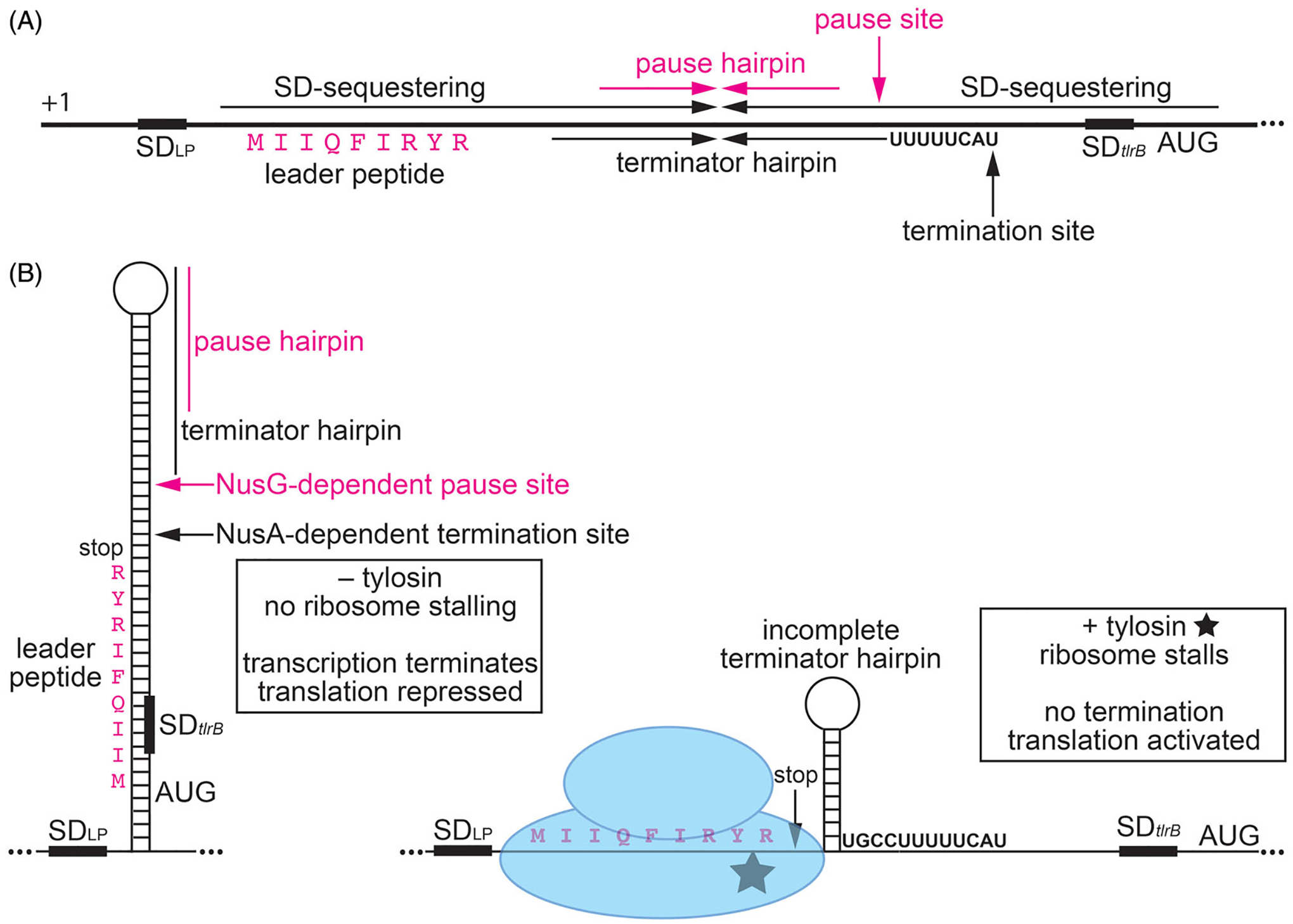
NusG-dependent pausing participates in tylosin-dependent induction of tlrB expression, resulting in antibiotic resistance. The ribosomes translating the leader peptide is simultaneously a sensor of the antibiotic and an effector that effects the structure of the nascent RNA in response to antibiotic challenge. (A) Schematic representation of the tlrB leader region. The thick black line represents the leader region between the start of transcription (+1) and the translation initiation region of tlrB. Inverted repeats for the long SD-sequestering hairpin, terminator hairpin, and pause hairpin (magenta) are labeled. Positions of the NusG-dependent pause site, NusA-dependent termination site, SD sequence (SDLP) for the leader peptide, the leader peptide (magenta), and the SD sequence and AUG start codon for tlrB are also labeled. (B) Model of tylosin-dependent induction of tlrB expression. NusG-dependent RNAP pausing provides time for translation initiation of the leader peptide. In the absence of tylosin (left), the ribosome releases at the stop codon. As a consequence, the terminator hairpin can form and transcription terminates about 50% of the time. For transcripts that fail to terminate, the long tlrB SD-sequestering hairpin forms and represses both tlrB translation and further rounds of leader peptide translation. In the presence of tylosin (right), the ribosome stalls at the C-terminal RYR motif of the leader peptide such that the ribosome remains bound to the nascent transcript. Once RNAP resumes transcription the position of the stalled ribosome prevents completion of the terminator hairpin such that transcription continues into the tlrB coding sequence. The stalled ribosome also prevents the formation of the tlrB SD-sequestering hairpin. Thus, the tlrB SD sequence is single-stranded and translation is activated. TlrB methylates 23S rRNA, leading to tylosin resistance. Color coding is the same as in (A).
Regulation of riboflavin biosynthesis
A strong NusG-dependent pause site was identified by RNET-seq in the leader region of the B. subtilis ribDEAHT operon involved in flavin biosynthesis. A pause hairpin was also identified for this pause site (Yakhnin et al. 2020). This pause site is within the ribD riboswitch that binds the flavin nucleotides FMN and FAD (Mironov et al. 2002; Winkler et al. 2002). Pausing at this position was previously identified by in vitro transcription using E. coli RNAP (Wickiser et al. 2005). The ribD pause site lies between the FMN-binding RNA structure (aptamer) and an intrinsic terminator (Figure 5). Bound FMN stabilizes an anti-antiterminator structure. An alternative antiterminator structure can form by pairing of a region within the anti-antiterminator and the terminator hairpin. Thus, bound FMN favors transcription termination by preventing the formation of the antiterminator. NusG-dependent pausing increases the FMN-dependent termination efficiency, resulting in reduced ribD expression (Yakhnin et al. 2020). Thus, pausing provides additional time for co-transcriptional binding of FMN to the nascent RNA, resulting in increased termination in the ribD leader region, and hence reduced expression of the ribDEAHT operon and reduced flavin biosynthesis. Pausing in 5′UTRs appears to be essential for precise sensing of growth conditions by B. subtilis cells by providing additional time for binding of ligands such as FMN to their targets in nascent RNA. This mechanism may underlie proper transcription readout of changing environmental conditions.
Figure 5.
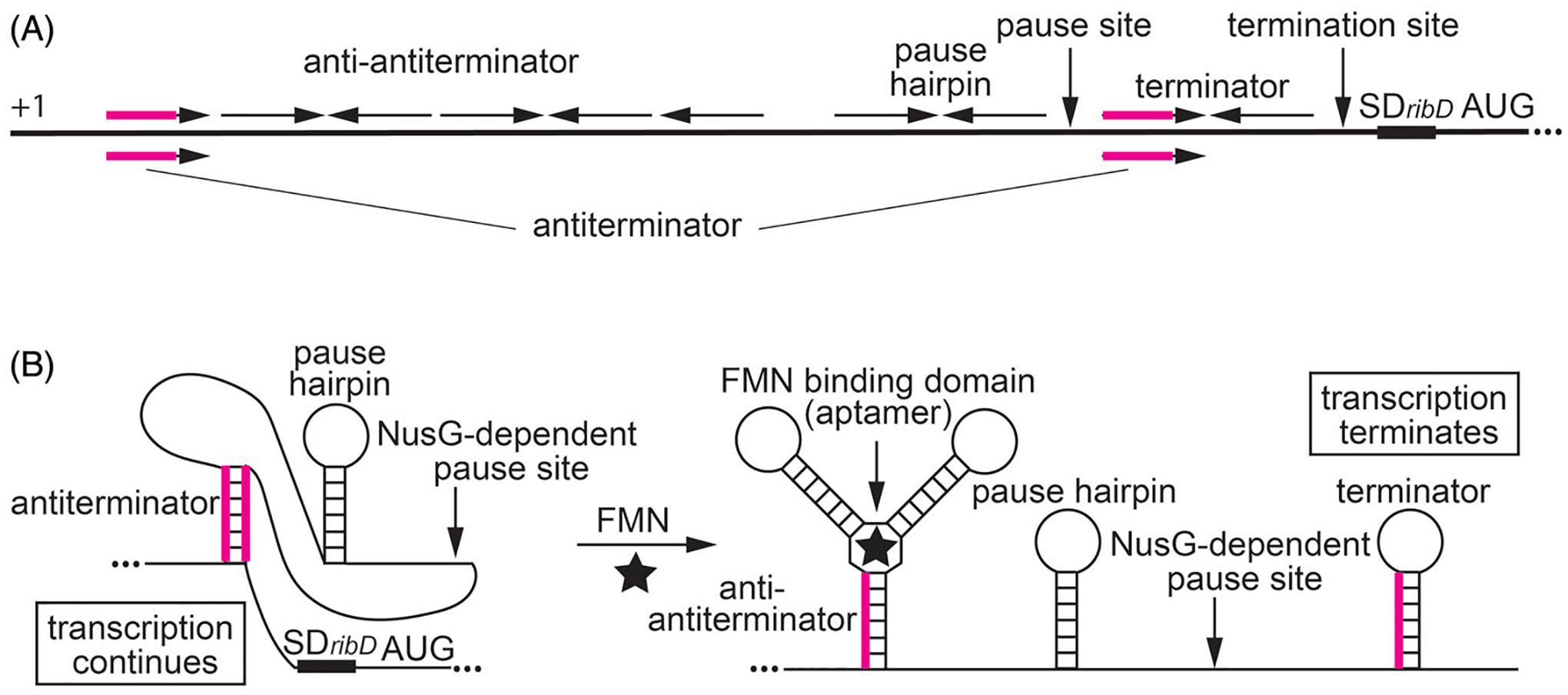
NusG-dependent pausing participates in the regulation of the ribDEAHT operon by an FMN-sensing riboswitch. (A) Schematic representation of the ribDEAHT leader region. The thick black line represents the leader region between the start of transcription (+1) and the AUG start codon for ribD. Inverted repeats for the anti-antiterminator, pause hairpin, and the terminator hairpin are shown above the line. The antiterminator that overlaps the anti-antiterminator and terminator structures is shown below the line (overlap in magenta). Positions of the NusG-dependent pause site, termination site, and the ribD SD sequence and AUG start codon are also labeled. (B) Model of FMN-dependent transcription attenuation. During transcription, RNAP pauses at a NusG-dependent pause site. Under FMN-limiting conditions (left), FMN does not bind to the nascent transcript. RNAP eventually overcomes the pause and resumes transcription. In this case, formation of the antiterminator structure prevents the formation of the terminator hairpin, resulting in transcription readthrough. Under FMN-excess conditions (right), FMN binds to the aptamer and stabilizes the anti-antiterminator structure, which prevents the formation of the antiterminator structure. As a consequence, formation of the terminator hairpin causes transcription to terminate. NusG-dependent pausing provides additional time for FMN binding and reduces the concentration of FMN required for termination. Color coding is the same as in (A).
Proposed integrative role of pausing that links NTP pools and specific metabolites in regulating gene expression
NusG-dependent pausing only affects the expression of the rib operon by two-fold (Yakhnin et al. 2020). This mild effect may be connected to the nucleotide pools in the cell. Pausing in the ribD riboswitch provides additional time for co-transcriptional binding of FMN to the nascent RNA to promote termination. Growing cells must balance requirements for a particular metabolic process with resources available to perform that process. The most common resource consumed by transcription and all other metabolic pathways is energy in the form of NTPs. The influence of intracellular NTP concentrations on gene expression has been demonstrated both in E. coli (Schneider et al. 2002) and B. subtilis (Kriel et al. 2014). The duration of a transcriptional pause is dramatically influenced by the concentration of NTP(s) that need to be incorporated for pause escape (Landick et al. 1996). Therefore, pause sites are natural sensors perfectly suited to measure intracellular NTP concentrations and to carry out the appropriate regulatory responses. According to this model, cellular NTP concentrations can vary over a larger range than certain other metabolites, and pause sites integrate inputs from a pathway-specific metabolite with the pathway-independent availability of NTPs (Figure 6). In vivo expression of reporter fusions is routinely performed in exponentially growing cells that are not limited in NTPs. As a result, expression of a reporter depends almost exclusively on the availability of a pathway-specific metabolite, while pausing modulates the range of regulatory concentrations of that metabolite. In other growth conditions, stressed cells could have low intracellular NTP pools such that gene expression may depend primarily on NTP concentrations, while the pathway-specific metabolite modulates the range of regulatory concentrations of NTPs. For example, low levels of ATP and UTP in stressed cells likely make pausing in the ribD leader region long enough that low FMN concentrations become sufficient for cotranscriptional binding to the riboswitch in nascent RNA. Similarly, prolonged pausing in the trpE leader region when the ATP and UTP pools are low may provide time for TRAP binding to nascent RNA at low cellular concentrations of tryptophan. However, this model would not be applicable to all regulatory switches. For example, despite making pausing extremely long, low intracellular GTP concentrations may inhibit translation of the tlrB leader peptide to such an extent that it would prevent tlrB induction even in tylosin-challenged cells (Yakhnin et al. 2019). Note that the level of either metabolites or NTPs never drops below some minimal level in living cells, thus setting quantitative limits in concentration-dependent regulation. Pause sites may regulate expression in response to the level of one or several NTPs depending on the sequence context of the pause and the number of pause positions (Figure 6).
Figure 6.
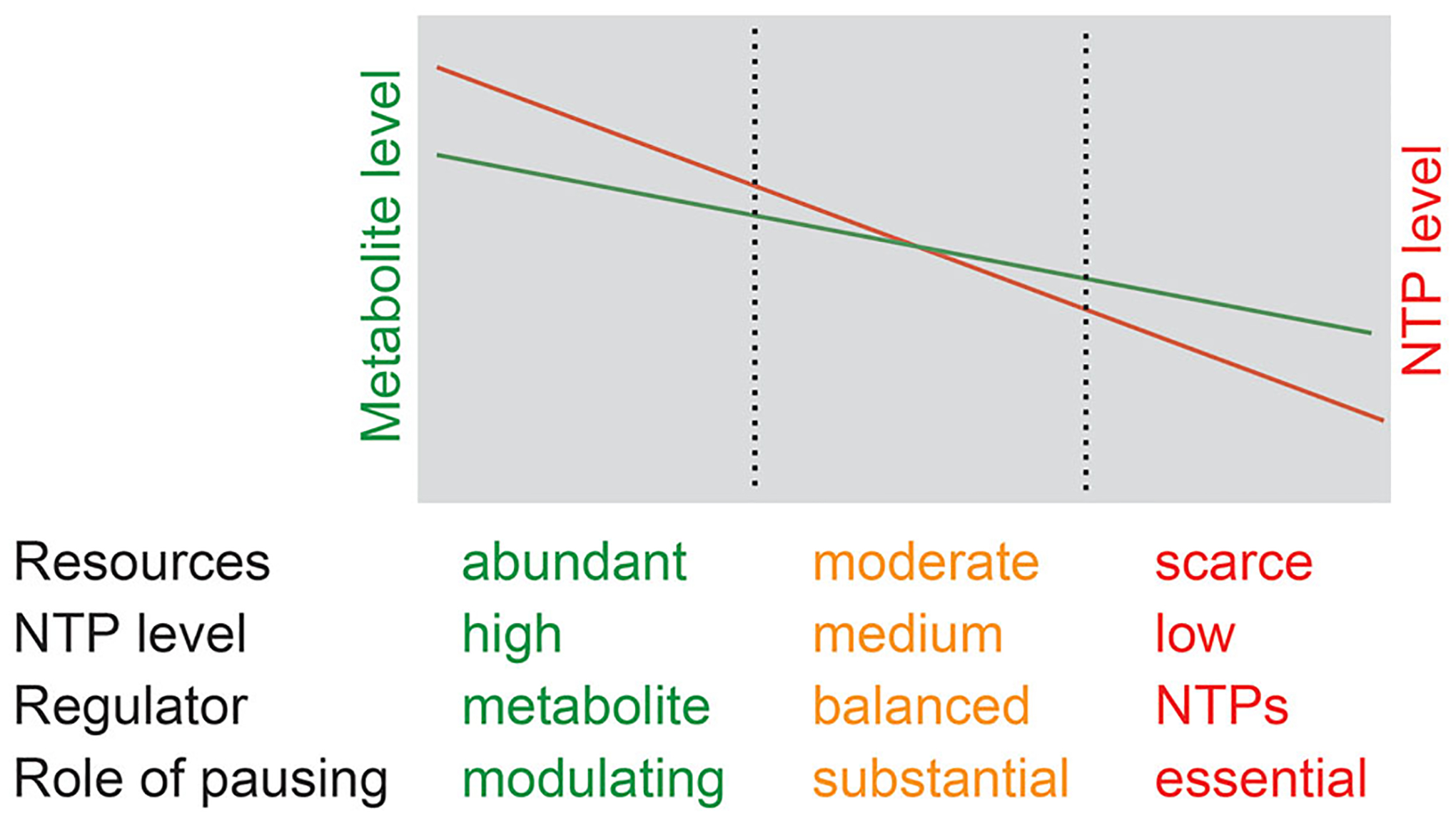
Model of pause sites as NTP sensors that integrate inputs from a pathway-specific metabolite and pathway-independent availability of energy. Exponentially growing cells have short pause duration because of high NTP levels, and therefore regulate gene expression in response to the availability of specific metabolites (shown in green). Resource-limited cells have intermediate NTP levels and pause duration regulates gene expression by balancing the need for metabolites with the availability of energy required for metabolite production (shown in yellow). Stressed cells have low NTP levels, long pause duration, and regulate gene expression primarily via their response to the availability of energy (shown in red). Cells may not be able to grow under extreme energy stress conditions.
Conclusions and perspectives
Recent studies firmly established the role of RNAP pausing in transcription attenuation that was proposed nearly 40 years ago (Winkler and Yanofsky 1981). Furthermore, modulation of pausing by general transcription elongation factors was instrumental in demonstrating the importance of synchronizing the position of RNAP with the timing of regulatory factor binding in controlling gene expression. However, many questions remain regarding the stimulation of pausing by NusG. What determines the sequence-specific interaction of NusG with ntDNA? Does the NusG-recognition motif form a short secondary structure as for RfaH–ntDNA interaction (Zuber et al. 2018; Kang, Mooney, et al. 2018)? Is flipping of T-bases toward NusG involved? Do NusG proteins from other species recognize the same, similar, or unrelated nucleic acid sequence motifs? High-resolution structures of RNAP paused at NusG-dependent sites may answer such questions. Distribution of pause sites also remains unexplained. Although many of the NusG-dependent pause sites in 5′ leader regions are probably involved in controlling expression of the downstream genes, what is the function of the pause sites identified in ORFs? Perhaps these pause sites are involved in the coupling of transcription and translation to prevent Rho-dependent termination, or for slowing down translation to stimulate proper folding of the emerging polypeptide chain. Also, it will be important to determine the features that convert some TTNTTT sequence motifs into NusG-dependent pause sites but not others. Moreover, a TTNTTT motif can simultaneously function as a NusG-dependent pause site and as part of the U-tract of an intrinsic terminator. Indeed, NusG increases termination by mycobacterial RNAP at suboptimal intrinsic terminators in vitro (Czyz et al. 2014). Interestingly, our Term-seq results in B. subtilis indicate that NusG functions as a general intrinsic termination factor in vivo (Mandell et al. 2020).
Funding
This work was supported by The National Institutes of Health under Grant [GM098399] to Paul Babitzke, and the Intramural Research Program of the National Institutes of Health/National Cancer Institute to Mikhail Kashlev.
Footnotes
Disclosure statement
The authors declare that this review was written in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Antson AA, Dodson EJ, Dodson G, Greaves RB, Chen X, Gollnick P. 1999. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 401(6750): 235–242. [DOI] [PubMed] [Google Scholar]
- Antson AA, Otridge J, Brzozowski AM, Dodson EJ, Dodson GG, Wilson KS, Smith TM, Yang M, Kurecki T, Gollnick P. 1995. The structure of trp RNA-binding attenuation protein. Nature. 374(6524):693–700. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, Knauer SH. 2019. Ancient transcription factors in the news. mBio. 10(1):e01547–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. 2000. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci USA. 97(13):7090–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Landick R. 2002. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 109(2):193–203. [DOI] [PubMed] [Google Scholar]
- Babitzke P 2004. Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: the Bacillus subtilis TRAP protein. Curr Opin Microbiol. 7(2):132–139. [DOI] [PubMed] [Google Scholar]
- Babitzke P, Yanofsky C. 1993. Reconstitution of Bacillus subtilis trp attenuation in vitro with TRAP, the trp RNA-binding attenuation protein. Proc Natl Acad Sci USA. 90(1): 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P, Yealy J, Campanelli D. 1996. Interaction of the trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis with RNA: effects of the number of GAG repeats, the nucleotides separating adjacent repeats, and RNA secondary structure. J Bacteriol. 178(17):5159–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. 2005. A ratchet mechanism of transcription elongation and its control. Cell. 120(2):183–193. [DOI] [PubMed] [Google Scholar]
- Belogurov GA, Sevostyanova A, Svetlov V, Artsimovitch I. 2010. Functional regions of the N-terminal domain of the antiterminator RfaH. Mol Microbiol. 76(2):286–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, Rösch P. 2010. A NusE:NusG complex links transcription and translation. Science. 328(5977):501–504. [DOI] [PubMed] [Google Scholar]
- Crickard JB, Fu J, Reese JC. 2016. Biochemical analysis of yeast suppressor of Ty 4/5 (Spt4/5) reveals the importance of nucleic acid interactions in the prevention of RNA polymerase II arrest. J Biol Chem. 291(19):9853–9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyz A, Mooney RA, Iaconi A, Landick R. 2014. Mycobacterial RNA polymerase requires a U-tract at intrinsic terminators and is aided by NusG at suboptimal terminators. mBio. 5(2):e00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AR, Gohara DW, Yap MN. 2014. Sequence selectivity of macrolide-induced translational attenuation. Proc Natl Acad Sci USA. 111(43):15379–15384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Babitzke P. 1998. trp RNA-binding attenuation protein-mediated long distance RNA refolding regulates translation of trpE in Bacillus subtilis. J Biol Chem. 273(32): 20494–20503. [DOI] [PubMed] [Google Scholar]
- Farnham PJ, Greenblatt J, Platt T. 1982. Effects of NusA protein on transcription termination in the tryptophan operon of Escherichia coli. Cell. 29(3):945–951. [DOI] [PubMed] [Google Scholar]
- Fisher R, Yanofsky C. 1983. A complementary DNA oligomer releases a transcription pause complex. J Biol Chem. 258(15):9208–9212. [PubMed] [Google Scholar]
- Fouqueau T, Blombach F, Cackett G, Carty AE, Matelska DM, Ofer S, Pilotto S, Phung DK, Werner F. 2018. The cutting edge of archaeal transcription. Emerg Top Life Sci. 2(4): 517–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Myasnikov AG, Chen J, Crucifix C, Papai G, Takacs M, Schultz P, Weixlbaumer A. 2018. Structural basis for NusA stabilized transcriptional pausing. Mol Cell. 69(5):816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein PP, Kolb KE, Windgassen T, Bellecourt MJ, Darst SA, Mooney RA, Landick R. 2014. RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat Struct Mol Biol. 21(9): 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. 2006. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 125(6): 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KM, Zhou J, Mooney RA, Porta AL, Landick R, Block SM. 2010. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol. 399(1):17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Hilal T, Loll B, Bürger J, Mielke T, Böttcher C, Said N, Wahl MC. 2020. Structure-based mechanisms of a molecular RNA polymerase/chaperone machine required for ribosome biosynthesis. Mol Cell. 79(6):1024–1036.e5. [DOI] [PubMed] [Google Scholar]
- Imashimizu M, Takahashi H, Oshima T, McIntosh C, Bubunenko M, Court DL, Kashlev M. 2015. Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome Biol. 16(1):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Mishanina TV, Bellecourt MJ, Mooney RA, Darst SA, Landick R. 2018. RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol Cell. 69(5):802–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Mooney RA, Nedialkov Y, Saba J, Mishanina TV, Artsimovitch I, Landick R, Darst SA. 2018. Structural basis for transcript elongation control by NusG family universal regulators. Cell. 173(7):1650–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriel A, Brinsmade SR, Tse JL, Tehranchi AK, Bittner AN, Sonenshein AL, Wang JD. 2014. GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes. J Bacteriol. 196(1):189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp F, Said N, Huang YH, Loll B, Bürger J, Mielke T, Spahn CMT, Wahl MC. 2019. Structural basis for the action of an all-purpose transcription anti-termination factor. Mol Cell. 74(1):143–157.e5. [DOI] [PubMed] [Google Scholar]
- Landick R, Wang D, Chan CL. 1996. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: his leader pause site as paradigm. Meth Enzymol. 274: 334–353. [DOI] [PubMed] [Google Scholar]
- Landick R, Yanofsky C. 1984. Stability of an RNA secondary structure affects in vitro transcription pausing in the trp operon leader region. J Biol Chem. 259(18):11550–11555. [PubMed] [Google Scholar]
- Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. 2014. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 344(6187): 1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Steitz TA. 2017. Structural insights into NusG regulating transcription elongation. Nucleic Acids Res. 45(2): 968–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Douthwaite S. 2002. Resistance to the macrolide antibiotic tylosin is conferred by single methylations at 23S rRNA nucleotides G748 and A2058 acting in synergy. Proc Natl Acad Sci USA. 99(23):14658–14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Mobli M, Yang X, Keller AN, King GF, Lewis PJ. 2015. RNA polymerase-induced remodelling of NusA produces a pause enhancement complex. Nucleic Acids Res. 43(5): 2829–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell ZF, Oshiro RT, Yakhnin AV, Kashlev M, Kearns DB, Babitzke P. 2020. NusG is an intrinsic transcription termination factor that stimulates motility and coordinates global gene expression with NusA. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason SW, Greenblatt J. 1991. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 5(8):1504–1512. [DOI] [PubMed] [Google Scholar]
- Merino E, Babitzke P, Yanofsky C. 1995. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J Bacteriol. 177(22): 6362–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. 2002. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 111(5):747–756. [DOI] [PubMed] [Google Scholar]
- Mondal S, Yakhnin AV, Babitzke P. 2017. Modular organization of the NusA- and NusG-stimulated RNA polymerase pause signal that participates in the Bacillus subtilis trp operon attenuation mechanism. J Bacteriol. 199(14): e00223–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S, Yakhnin AV, Sebastian A, Albert I, Babitzke P. 2016. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat Microbiol. 1: 15007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. 2009. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 33(1):97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Schweimer K, Rösch P, Gottesman M, Landick R. 2009. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol. 391(2): 341–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson ER, Flamm EL, Friedman DI. 1982. Analysis of nutR: a region of phage lambda required for antitermination of transcription. Cell. 31(1):61–70. [DOI] [PubMed] [Google Scholar]
- Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. 1999. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc Natl Acad Sci USA. 96(17):9545–9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Vangeloff AD, Landick R. 2011. Bacterial transcription terminators: the RNA 3’-end chronicles. J Mol Biol. 412(5):793–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring BZ, Yarnell WS, Roberts JW. 1996. Function of E. coli RNA polymerase σ factor σ70 in promoter-proximal pausing. Cell. 86(3):485–493. [DOI] [PubMed] [Google Scholar]
- Saba J, Chua XY, Mishanina TV, Nayak D, Windgassen TA, Mooney RA, Landick R. 2019. The elemental mechanism of transcriptional pausing. Elife. 8:e40981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said N, Krupp F, Anedchenko E, Santos KF, Dybkov O, Huang YH, Lee CT, Loll B, Behrmann E, Bürger J, et al. 2017. Structural basis for λN-dependent processive transcription antitermination. Nat Microbiol. 2:17062 [DOI] [PubMed] [Google Scholar]
- Saxena S, Myka KK, Washburn R, Costantino N, Court DL, Gottesman ME. 2018. Escherichia coli transcription factor NusG binds to 70S ribosomes. Mol Microbiol. 108(5): 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, Gaal T, Gourse RL. 2002. NTP-sensing by rRNA promoters in Escherichia coli is direct. Proc Natl Acad Sci USA. 99(13):8602–8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A, Artsimovitch I. 2010. Functional analysis of Thermus thermophilus transcription factor NusG. Nucleic Acids Res. 38(21):7432–7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevostyanova A, Belogurov GA, Mooney RA, Landick R, Artsimovitch I. 2011. The β subunit gate loop is required for RNA polymerase modification by RfaH and NusG. Mol Cell. 43(2):253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar SK, Artsimovitch I. 2013. NusG-Spt5 proteins-Universal tools for transcription modification and communication. Chem Rev. 113(11):8604–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulokhonov I, Artsimovitch I, Landick R. 2001. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 292(5517):730–733. [DOI] [PubMed] [Google Scholar]
- Turtola M, Belogurov GA. 2016. NusG inhibits RNA polymerase backtracking by stabilizing the minimal transcription bubble. Elife. 5:e18096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Laslop N, Mankin AS. 2018. How macrolide antibiotics work. Trends Biochem Sci. 43(9):668–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, Ebright RH, Nickels BE. 2014. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science. 344(6189): 1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. 2005. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 18(1):49–60. [DOI] [PubMed] [Google Scholar]
- Winkler ME, Yanofsky C. 1981. Pausing of RNA polymerase during in vitro transcription of the tryptophan operon leader region. Biochemistry. 20(13):3738–3744. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Cohen-Chalamish S, Breaker RR. 2002. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA. 99(25):15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Babitzke P. 2002. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism invitro. Proc Natl Acad Sci USA. 99(17):11067–11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Babitzke P. 2010. Mechanism of NusG-stimulated pausing, hairpin-dependent pause site selection and intrinsic termination at overlapping pause and termination sites in the Bacillus subtilis trp leader. Mol Microbiol. 76(3): 690–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Babitzke P. 2014. NusG/Spt5: are there common functions of this ubiquitous transcription elongation factor? Curr Opin Microbiol. 18:68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, FitzGerald PC, McIntosh C, Yakhnin H, Kireeva M, Turek-Herman J, Mandell ZF, Kashlev M, Babitzke P. 2020. NusG controls transcription pausing and RNA polymerase translocation throughout the Bacillus subtilis genome. Proc Natl Acad Sci USA. 117(35):21628–21636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Murakami KS, Babitzke P. 2016. NusG is a sequence-specific RNA polymerase pause factor that binds to the non-template DNA within the paused transcription bubble. J Biol Chem. 291(10):5299–5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Yakhnin H, Babitzke P. 2006. RNA polymerase pausing regulates translation initiation by providing additional time for TRAP-RNA interaction. Mol Cell. 24(4): 547–557. [DOI] [PubMed] [Google Scholar]
- Yakhnin AV, Yakhnin H, Babitzke P. 2008. Function of the Bacillus subtilis transcription elongation factor NusG in hairpin-dependent RNA polymerase pausing in the trp leader. Proc Natl Acad Sci USA. 105(42):16131–16136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin H, Yakhnin AV, Mouery BL, Mandell ZF, Karbasiafshar C, Kashlev M, Babitzke P. 2019. NusG-dependent RNA polymerase pausing and tylosin-dependent ribosome stalling are required for tylosin resistance by inducing 23S rRNA methylation in Bacillus subtilis. mBio. 10(6): e02665–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. 1999. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 274(12): 8085–8092. [DOI] [PubMed] [Google Scholar]
- Zellars M, Squires CL. 1999. Antiterminator-dependent modulation of transcription elongation rates by NusB and NusG. Mol Microbiol. 32(6):1296–1304. [DOI] [PubMed] [Google Scholar]
- Zhang J, Landick R. 2016. A two-way street: regulatory interplay between RNA polymerase and nascent RNA structure. Trends Biochem Sci. 41(4):293–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Filter JJ, Court DL, Gottesman ME, Friedman DI. 2002. Requirement for NusG for transcription antitermination in vivo by the lambda N protein. J Bacteriol. 184(12): 3416–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber PK, Artsimovitch I, NandyMazumdar M, Liu Z, Nedialkov Y, Schweimer K, Rösch P, Knauer SH. 2018. The universally-conserved transcription factor RfaH is recruited to a hairpin structure of the non-template DNA strand. Elife. 7:e36349. [DOI] [PMC free article] [PubMed] [Google Scholar]


