Abstract
Acute myeloid leukaemia (AML) remains difficult to treat and requires new therapeutic approaches. Potent inhibitors of the chromatin-associated protein MENIN have recently entered human clinical trials, opening new therapeutic opportunities for some genetic subtypes of this disease. Using genome-scale functional genetic screens we identified IKZF1/IKAROS as an essential transcription factor in MLL1-rearranged (MLL-r) AML that maintains leukaemogenic gene expression while also repressing pathways for tumour suppression, immune regulation, and cellular differentiation. Furthermore, IKAROS displays an unexpected functional cooperativity and extensive chromatin co-occupancy with MLL1/MENIN and MEIS1 and an extensive hematopoietic transcriptional complex involving HOXA10, MEIS1 and IKAROS. This dependency could be therapeutically exploited by inducing IKAROS protein degradation with immunomodulatory imide drugs (IMiDs). Finally, we demonstrate that combined IKAROS degradation and MENIN inhibition effectively disrupts leukaemogenic transcriptional networks resulting in synergistic killing of leukaemia cells and providing a paradigm for improved drug-targeting of transcription, and an opportunity for rapid clinical translation.
KMT2A/MLL1 gene chromosomal translocations (MLL-r) define a distinct subset of Acute Myeloid Leukaemia (AML)1,2 characterized by well-defined pathologic features and a dismal prognosis. MLL-r AML serves as a paradigm for malignant disease that is driven by disordered gene expression3. Normal haematopoiesis is governed by a hierarchy of transcription factors (TFs) that mediate cell fate decisions while co-operating within densely interconnected co-regulatory circuits4. The MLL-fusion drives leukaemia through deregulation of these critical transcriptional networks, functioning within multi-component protein complexes containing key structural proteins and enzymes, including MENIN5, LEDGF6, DOT1L7,8, and the super-elongation complex (SEC)9; each of which is important for MLL-fusion function. Acting within these complexes, the MLL-fusion drives the abnormal expression of numerous haematopoietic TFs such as MEF2C10,11, CDK612, HOXA913, PBX314, and MEIS115 which, in turn, mediate downstream transcriptional de-regulation. These TFs interact with additional myeloid transcriptional regulatory proteins such as CEBPα16, RUNX117, KMT2C/MLL318, SPI1/PU.119 and JMJD1C20, thereby impacting diverse cellular processes. Of these downstream oncogenic collaborators, HOXA cluster genes and MEIS1, which act together, are considered among the most important due to their ability to recapitulate much of the oncogenic function of the MLL-fusion protein13,21. Considered broadly, the MLL-fusion acts within a complex network of transcriptional regulators that are essential for its leukaemogenicity, and elucidating strategies to shut down these pathways has been a major focus of therapeutic development for this disease. Furthermore, mechanisms that govern MLL-fusion driven gene expression are also critical for leukemia development in other subtypes of AML; of particular importance, high-level HOX/MEIS gene expression is observed in NPM1-mutant (NPM1c) AML22.
Deregulated transcription is an established hallmark of malignant disease23 and a major target for the development of cancer therapeutics. MLL-r leukaemia serves as a model disease for this approach. Highly specific and potent small molecule inhibitors have been developed to target the MENIN-MLL1 protein-protein interaction (VTP-5046924, MI-503/46325 and MI-345426), and also the enzymatic function of the H3K79 methyltransferase, DOT1L (EPZ-567627,28), as a means to target the MLL-fusion-containing complex and downstream leukaemia-promoting transcription. These drugs exhibit potent antileukemic effects in pre-clinical models of both MLL-r and NPM1c leukaemia22,24,26,29. Clinical-grade compounds related to VTP-50469 and MI-3454 have now entered Phase I/II clinical studies (Syndax/SNDX-5613 and Kura/KO-539, respectively). The DOT1L inhibitor, EPZ-5676 (Epizyme/Pinometostat), has completed a Phase I/II clinical study30, demonstrating on-target activity in humans but achieving only modest therapeutic effect, which highlights the need to understand resistance mechanisms and develop drug combinations to improve efficacy. Combination therapies are typically superior to monotherapy because they can abrogate resistance mechanisms, which is of particular importance for therapeutic targeting of transcriptional networks due to the presence of large multi-protein complexes containing cooperating TFs with functional redundancies that may be inefficiently targeted by a single drug alone31. With MENIN and DOT1L inhibitors entering clinical trials as single agent therapies, we sought to investigate how mechanisms of resistance may limit their clinical application and inform the rational development of more effective drug combinations. Remarkably, we identified IKAROS/IKZF1 as an essential transcriptional regulator in MLL-r and NPM1c AML that can be immediately exploited using the immunomodulatory imide drugs (IMiDs); furthermore, we found that combined therapeutic targeting of IKAROS and MENIN leads to synergistic anti-proliferative effects and propose this as a novel therapeutic strategy for AML.
Results
IKZF1/IKAROS modulates dependency on MLL1-MENIN and DOT1L.
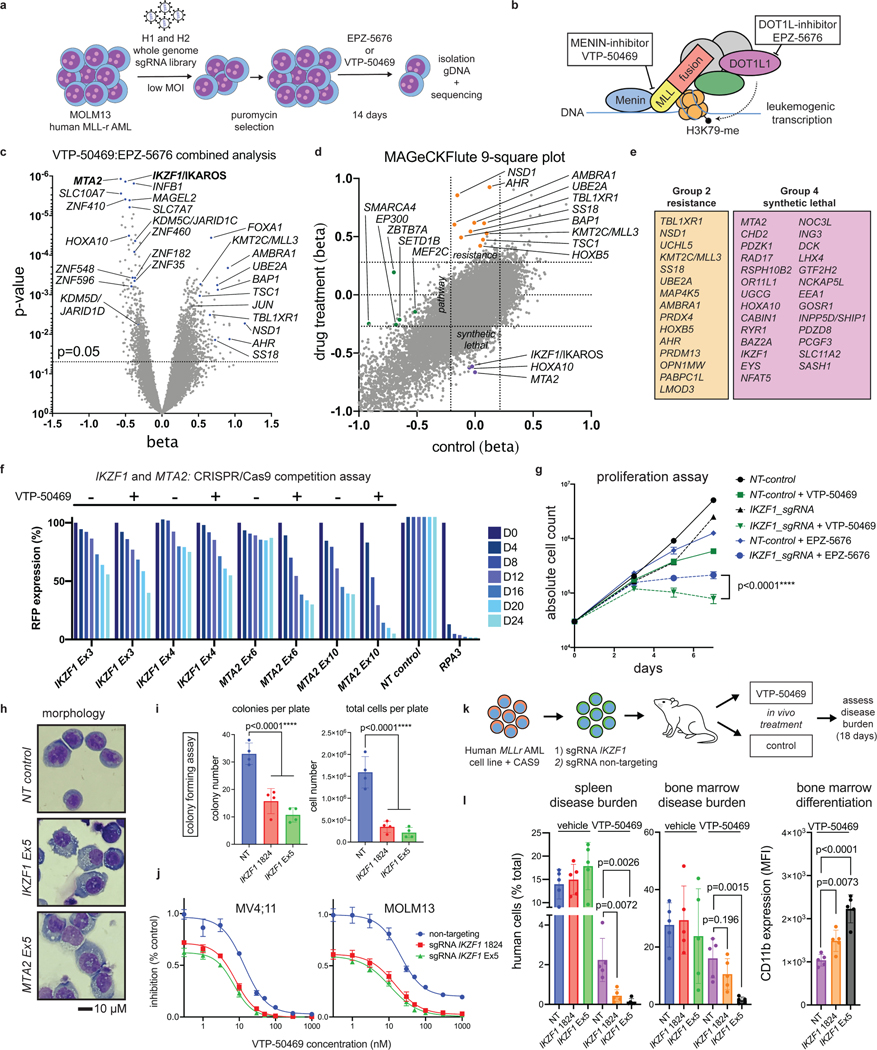
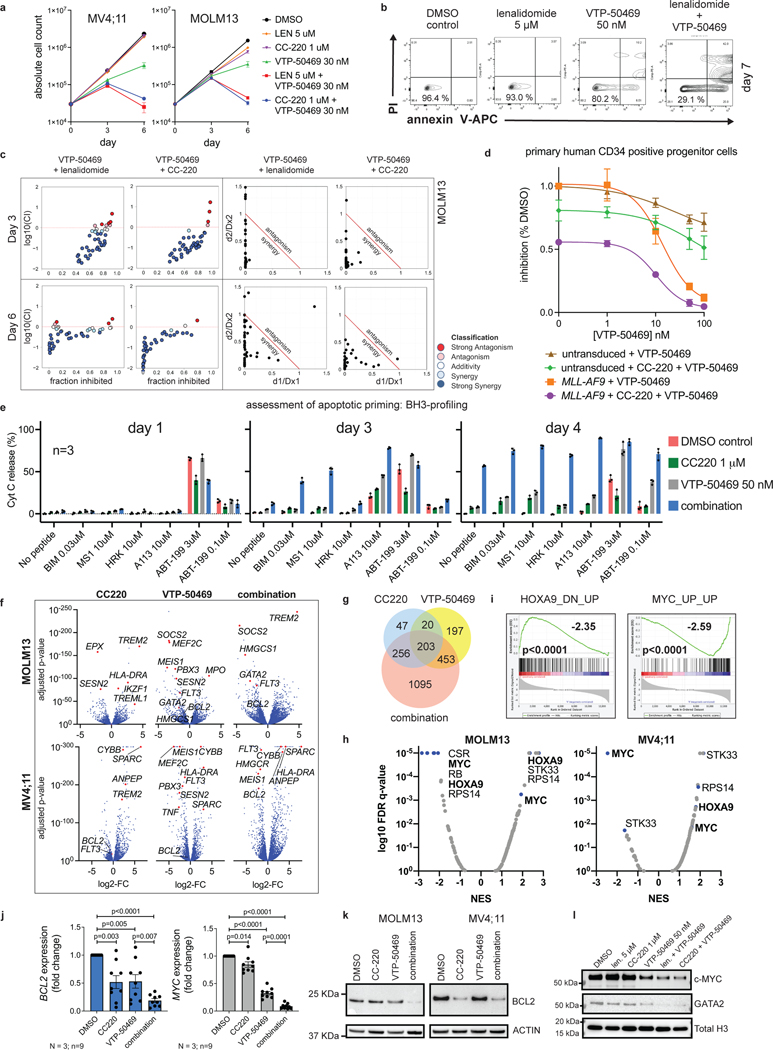
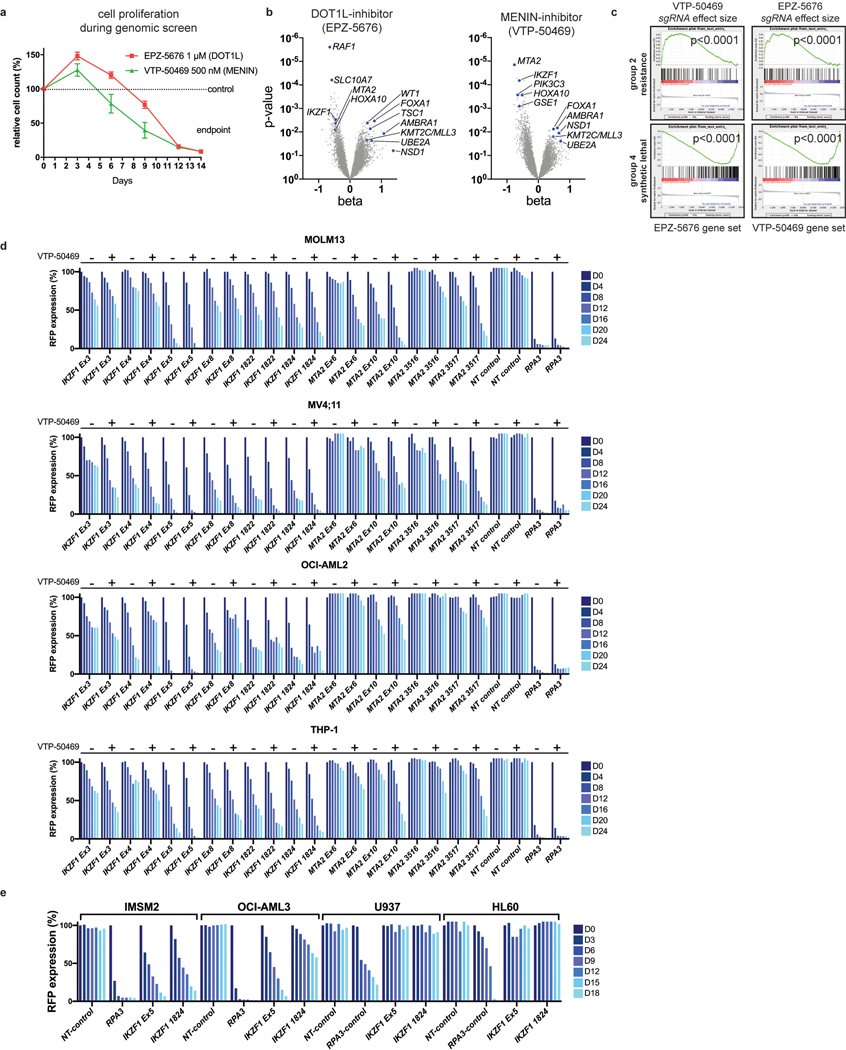
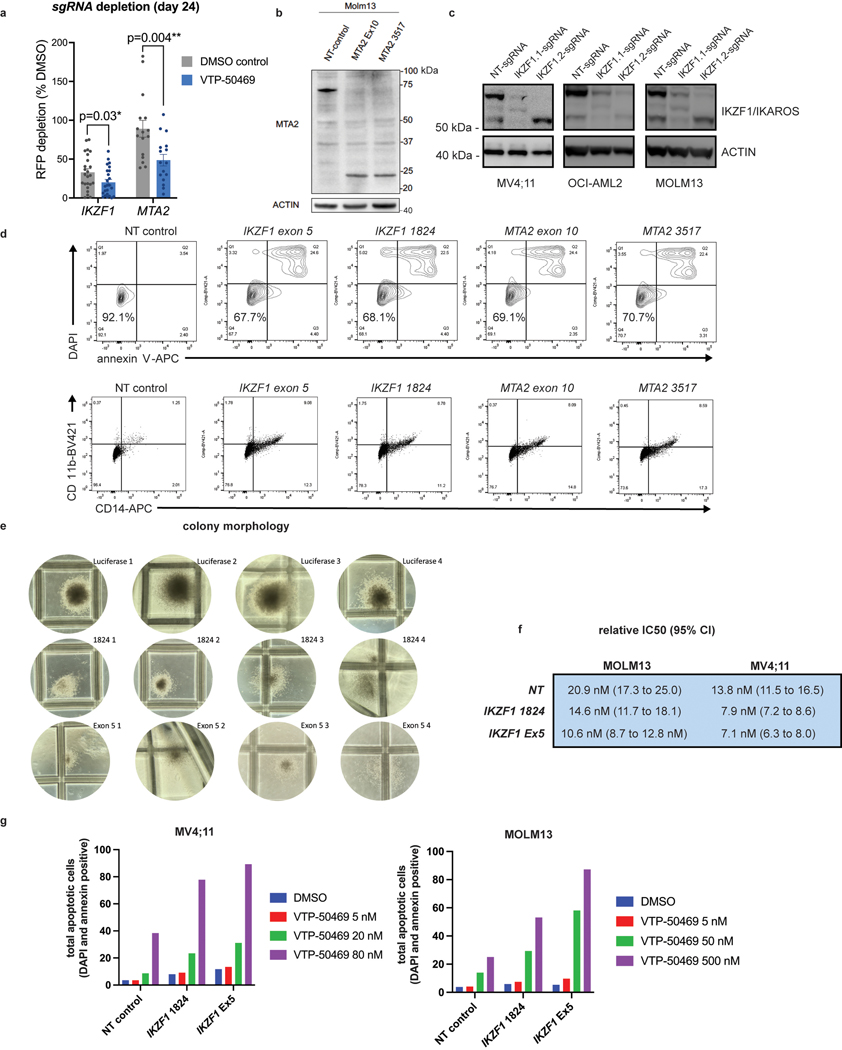
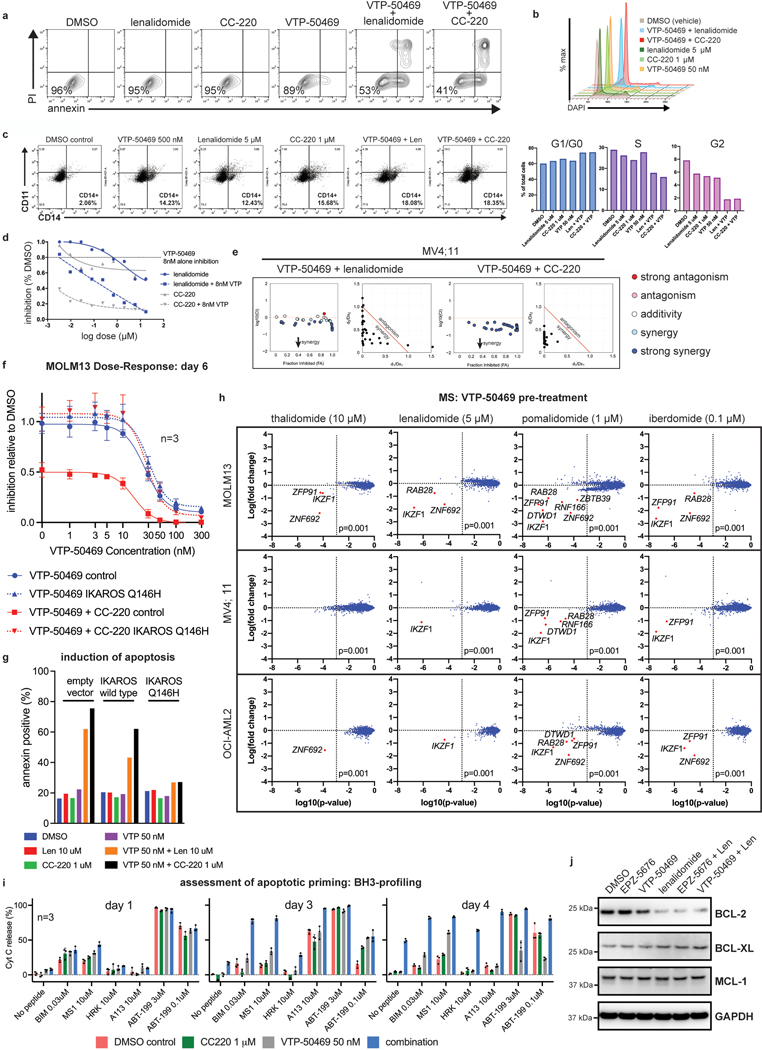
Genome-scale, CRISPR/Cas9-based functional genetic screening was used to characterize resistance mechanisms and synthetic lethal interactions for small molecule inhibitors of DOT1L or MENIN (EPZ-5676 and VTP-50469, respectively) using the human MLL-r AML cell line, MOLM13 (Fig. 1a). These drugs target the MLL-fusion complex3 (Fig. 1b). Cell proliferation was monitored during the screen (Extended Data Fig. 1a), showing strong inhibition. Comparison of the drug-treatment group to the vehicle-control on Day 14 (Supplementary Table 1) revealed selection for sgRNAs targeting transcriptional regulators (Extended Data Fig. 1b), and a strong overlap between the two drugs (Extended Data Fig. 1c). Leading edge analysis, focusing on resistance genes (Group 2) and synthetic lethal genes (Group 4), according to the MAGeCKFlute computational pipeline32, highlighted genes exerting a large effect size in both screens (Fig. 1e) and common mechanisms of resistance. A combined analysis of the two screens highlighted these common pathways (Fig. 1c) (Supplementary Table 1); IKZF1/IKAROS and MTA2 were the two highest-scoring negatively-selected genes, which are notable as constituents of the nucleosome remodelling and histone deacetylase complex (NuRD)33. Other high-ranking genes that were depleted or enriched included members of the SWI/SNF complex (SMARCA4, SS18), mTOR pathway (TSC1, TSC2, PTEN), protein ubiquitination regulators (UBE2A, BAP1, TBL1XR1, AMBRA1, UCHL5, RNF138, NOSIP), regulators of histone methylation (KDM5D/JARID1D, KDM5C/JARID1C, SETD1B, NSD1, KMT2C/MLL3, PRDM13), transcription factors (FOXA1, HOXA10, HOXB5, MEF2C, AHR) and zinc-finger proteins (ZNF410, ZNF460, ZNF182, ZNF548, ZNF596). MAGeCKFlute was utilized to correct cell cycle biases and classify hits into functional categories (Fig. 1d)32. This highlighted IKZF1, MTA2, and HOXA10 as synthetic lethal gene targets, because they were disproportionately depleted in drug-treated cells, and identified these genes as therapeutic targets that may increase sensitivity to inhibition of MENIN and DOT1L. IKAROS is an established therapeutic target in lymphoproliferative disorders, but a role in MLL-r AML has not been previously reported. Notably, IKZF1 exhibits high-level gene expression in human AML34,35 and is also detected as an AML dependency in the Broad DepMap (p = 3.8 × 10−12); across all cancer cell lines in the DepMap, there is a significant correlation of dependency between MEN1 and IKZF1 (Pearson 0.15, p-value 5.3 × 10−7) and between DOT1L and IKZF1 (Pearson 0.19, p-value 5.7 × 10−10)35. We next evaluated the impact of genetic targeting of both IKZF1 and MTA2 using CRISPR/Cas9-based competition assays, using lentiviral vectors co-expressing a sgRNA and red fluorescence protein (RFP), in four different Cas9-expressing human MLL-r AML cell lines (MOLM13, MV4;11, OCI-AML2 and THP-1), revealing strong depletion of sgRNAs targeting IKZF1 and MTA2 (Fig. 1f and Extended Data Fig. 1d). Addition of VTP-50469 enhanced sgRNA depletion (Fig. 1f and Extended Data Fig. 2a), as predicted by the genetic screen. To generalize this finding to other leukaemia subtypes, two NPM1c cell lines (IMSM2 and OCI-AML3) and two non-MLL-rearranged leukaemias (U937 and HL60) were examined (Extended Data Fig. 1e); importantly, U937 and HL60 did not display IKAROS dependency and, notably, are also insensitive to MENIN-inhibition24, highlighting a context-dependent role for IKAROS. In contrast, both NPM1c cell lines exhibited strong IKAROS dependency in accordance with their overlapping gene expression profile with MLL-r AML36 and dependency on DOT1L/MENIN22, suggesting that IKAROS and MENIN may be co-dependencies. CRISPR/Cas9-mediated depletion of IKAROS and MTA2 protein was confirmed by Western blot (Extended Data Fig. 2b,c). Cell proliferation was measured following IKZF1 knock-out, with and without MENIN or DOT1L inhibition, revealing a modest reduction in cell proliferation that was enhanced when combined with either the MENIN or DOT1L inhibitor (Fig. 1g); IKZF1 knockout in combination with the MENIN inhibition resulted in the strongest overall anti-proliferative effect, shifting of the IC50 of MENIN inhibition (Fig. 1j, Extended Data Fig. 2f), and substantially increasing the amount of apoptosis induced with MENIN inhibition (Extended Data Fig. 2g). Remarkably, loss of either IKZF1 or MTA2 led to cellular differentiation, induction of apoptosis (Extended Data Fig. 2d), and morphologic changes consistent with monocytic differentiation (Fig. 1h), indicating that IKZF1 and the NuRD complex have a fundamentally important role in leukaemia maintenance. Additionally, IKZF1 loss impaired colony formation in colony forming assays. (Fig. 1i, Extended Data Fig. 2e) and in a MOLM13 xenograft experiment, IKZF1 knockout enhanced MENIN inhibition in terms of reducing burden of disease and increasing differentiation markers in vivo (Fig. 1k,l), further implicating IKZF1 as essential to MLL-r leukemia maintenance.
Figure 1. IKZF1/IKAROS and MTA2 exhibit synthetic lethal interaction with pharmacologic inhibition of MENIN and DOT1L.
a, Functional genetic screen schematic: cells treated with EPZ-5676 (1 μM), VTP-50469 (500 nM) or vehicle. b, Drug targeting of MENIN (VTP-50469) and DOT1L (EPZ-5676) disrupts MLL-fusion complex function. c, Volcano plot depicting Wald p-value and beta value calculated using MAGeCK MLE for the VTP-50469 and EPZ-5676 combined analysis comparing vehicle-treated (n=2) and drug-treated states (n=4) on Day 14. d, MAGeCKFlute 9-square correlation plot using the beta value calculated between Day 0 and Day 14 for vehicle-treatment as compared to drug treatment (VTP-50469 and EPZ-5676 screens combined). Regions corresponding to pathway (group1), resistance (group2) and synthetic lethal (group4) genetic hits are indicated. e, Leading edge analysis from GSEA showing overlapping genetic hits for each screen considered separately. f, CRISPR/Cas9-based competition assays targeting either IKZF1 or MTA2 monitoring sgRNA-RFP expression over time in Cas9-expressing MOLM13 AML cell line +/− VTP-50469. g, Proliferation assay on sorted, sgRNA-RFP-expressing cells +/− VTP-50469 or EPZ-5676. Data represent mean+/−SEM (n=3) with p-value calculated using paired, two-tail t-test. h, Cell morphology assessed using light microscopy (100X) 7 days following CRISPR/Cas9-mediated deletion of IKZF1 or MTA2, compared to non-targeting sgRNA control i, Colony forming assay comparing MOLM13 cells with sgRNA targeting Luciferase (Non-Targeting) control versus two different sgRNAs targeting IKZF1 (n =4 per sgRNA). Data represent mean +/− SEM. p-value determined by unpaired, two-tailed t-test. j, Proliferation assay comparing MOLM13 cells with sgRNA targeting Luciferase (NT control) and two sgRNAs targeting IKZF1 (n=3 per sgRNA) upon treatment with VTP-50469 for 5 days. Data represent mean +/− SEM. k, Schematic for in vivo, MOLM13 xenograft experiment: 200,000 MOLM13 cells with sgRNA targeting Luciferase (NT control) versus two different sgRNAs targeting IKZF1 (n = 5 per group) were injected via tail vein at Day 0, treatment began with VTP-50469 chow at Day 9, and mice were sacrificed at Day 18 when they became ill. l, MOLM13 xenograft experiment, burden of disease and differentiation markers in spleen and bone marrow after 9 days of VTP-50469 treatment. Data represent mean +/− SEM. p-value determined by unpaired, two-tailed t-test.
IMiDs efficiently drive IKAROS protein degradation in AML.
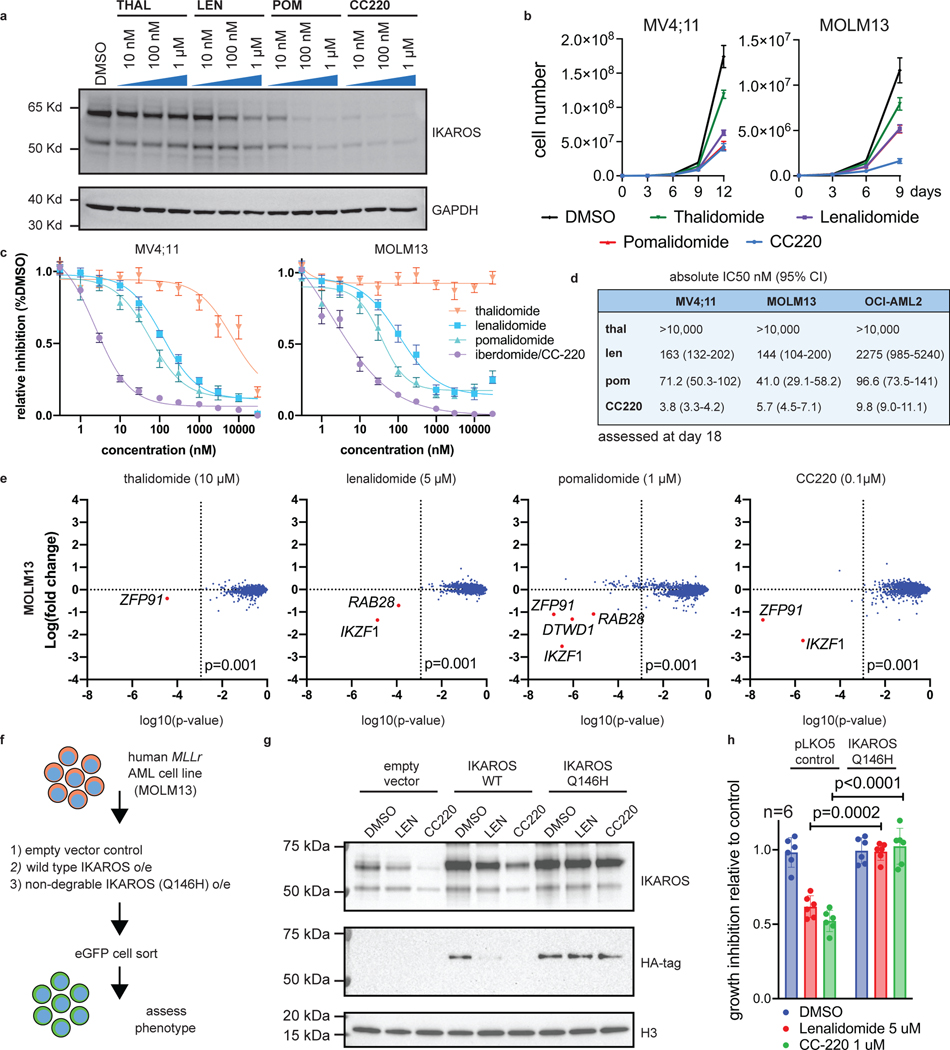
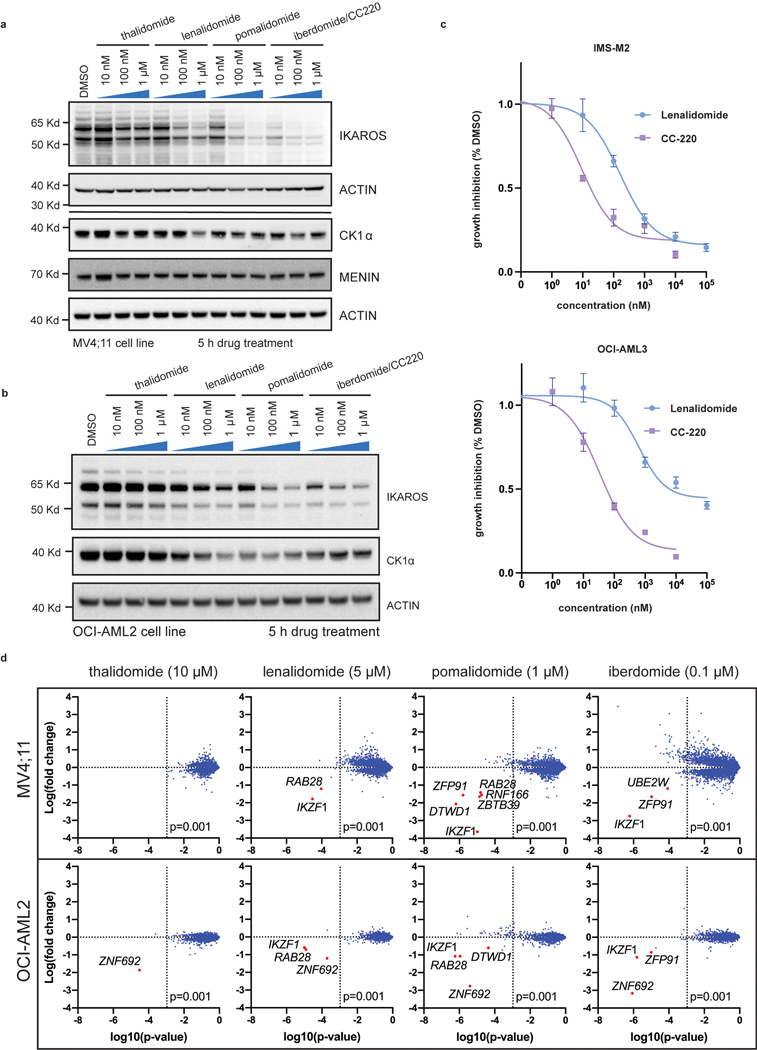
We next sought to leverage this finding by targeting IKAROS protein in AML using IMiDs. Lenalidomide, and related thalidomide-analogues, promote Cereblon (CRBN)-mediated degradation of the neo-substrates IKAROS, AIOLOS37,38, Casein Kinase 1α (CK1α)39 and others. Thalidomide (THAL), and newer generation IMiDs, lenalidomide (LEN), pomalidomide (POM) and iberdomide (CC220), display differing neo-substrate profiles and potency of IKAROS protein degradation40. Detailed evaluation of these drugs in MLL-r AML has not been previously reported. Thus, we evaluated all 4 drugs for potency of IKAROS degradation in three MLL-r human AML cell lines (Fig. 2a and Extended Data Fig. 3a,b); increasing potency of IKAROS degradation was observed for THAL, LEN, POM and CC220, respectively. Each IMiD impaired cell proliferation in a manner correlating with the depth of IKAROS degradation (Fig. 2b) and, notably, a defect in proliferation was not observed until 6–9 days. The therapeutic dose-response relationship was strongly correlated with the degree of IKAROS degradation (CC220>>POM>LEN>>THAL) (Fig. 2c,d). In accordance with genetic deletion, IMiD treatment also impaired proliferation of the NPM1c AML cell lines, IMSM2 and OCI-AML3 (Extended Data Fig. 3c). A detailed neo-substrate profile for each drug in all 3 MLL-r cell lines tested (MV4;11, MOLM13, OCI-AML2) was determined using multiplexed mass spectrometry (MS)-based global proteomics. This confirmed IKAROS as the primary neo-substrate (Fig. 2e and Extended Data Fig. 3d; Supplementary Table 2) without obvious novel neo-substrate degradation detected; notably, ZFP91 and ZNF692 are not detected as AML dependencies in the Depmap (data not shown). CK1α peptides were not efficiently detected by MS but confirmed by Western blot analysis to only be a substrate of LEN (Extended Data Fig. 3a,b); CC220 elicited the most potent inhibitory effect on cell growth while exhibiting profound IKAROS degradation and minimal effect on CK1α, consistent with IKAROS being the primary substrate mediating these effects. Further, overexpression of a non-degradable IKAROS mutant with a single amino acid substitution rendering it not capable of being degraded (Q146H)38, rescued the antiproliferative effects of IMiDs (Fig. 2f,g,h). Altogether, these data support IMiDs as a potential novel therapeutic approach for MLL-r AML through their ability to degrade the IKAROS protein.
Figure 2. IMiDs effectively target IKAROS protein for degradation and show therapeutic efficacy in MLL-r AML.
a, Western blot analysis for IKAROS protein following treatment of the MOLM13 cell line for 5 h with increasing doses of THAL, LEN, POM and CC220, using GAPDH as a loading control. b, Proliferation assay measuring cell number over time for the MV4;11 and MOLM13 cell lines treated with vehicle-control (DMSO), THAL, LEN, POM and CC220. Data represent mean+/−SD (n=3). c, Dose response curves for MV4;11 and MOLM13 cell lines using THAL, LEN, POM and CC220. Data represent mean+/−SD (n=6) with non-linear regression curve fit shown. Percentage indicated as a proportion. d, Calculated absolute IC50 and 95% confidence intervals for MOLM13, OCI-AML2 and MV4;11 AML cell lines, assessed by proliferation assay at Day 18. e, Scatterplot for MS determination of IMiD substrates 5 h after indicated drug treatment in the MOLM13 cell line. Data represent the Log-fold change in abundance and log10(p-value). p-value determined by moderated t-test as implemented by the Bioconductor Limma package. f, Schematic depicting generation of MOLM13 cell lines with IKAROS overexpression constructs treated with DMSO, LEN (5 uM), and CC220 (1 uM) for 5 hours. g, Western Blot showing IKAROS overexpression constructs, h, Proliferation assay showing rescue of antiproliferative effects with overexpressed, non-degradable IKAROS (Q146H). Data represent mean +/− SEM. p-value determined by paired, two-tail t-test.
IKAROS is a core transcriptional regulator in MLL-r AML.
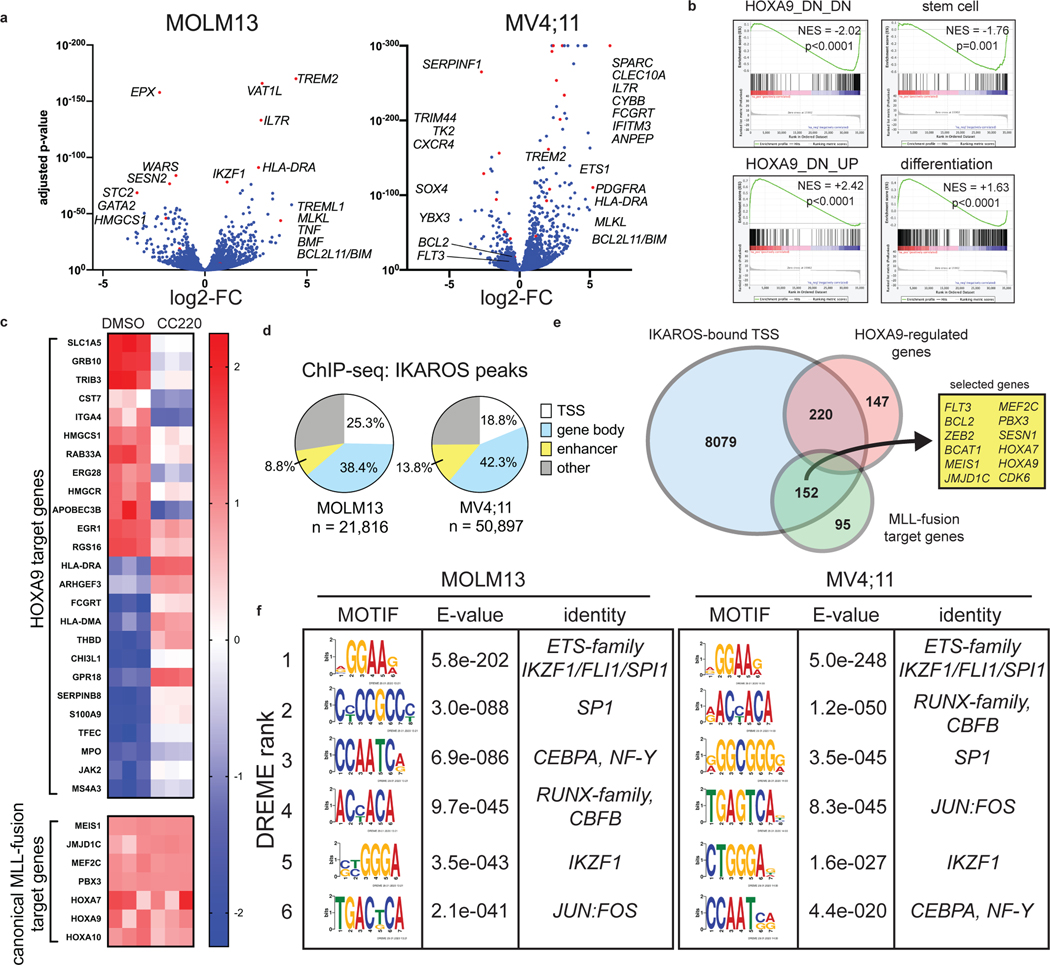
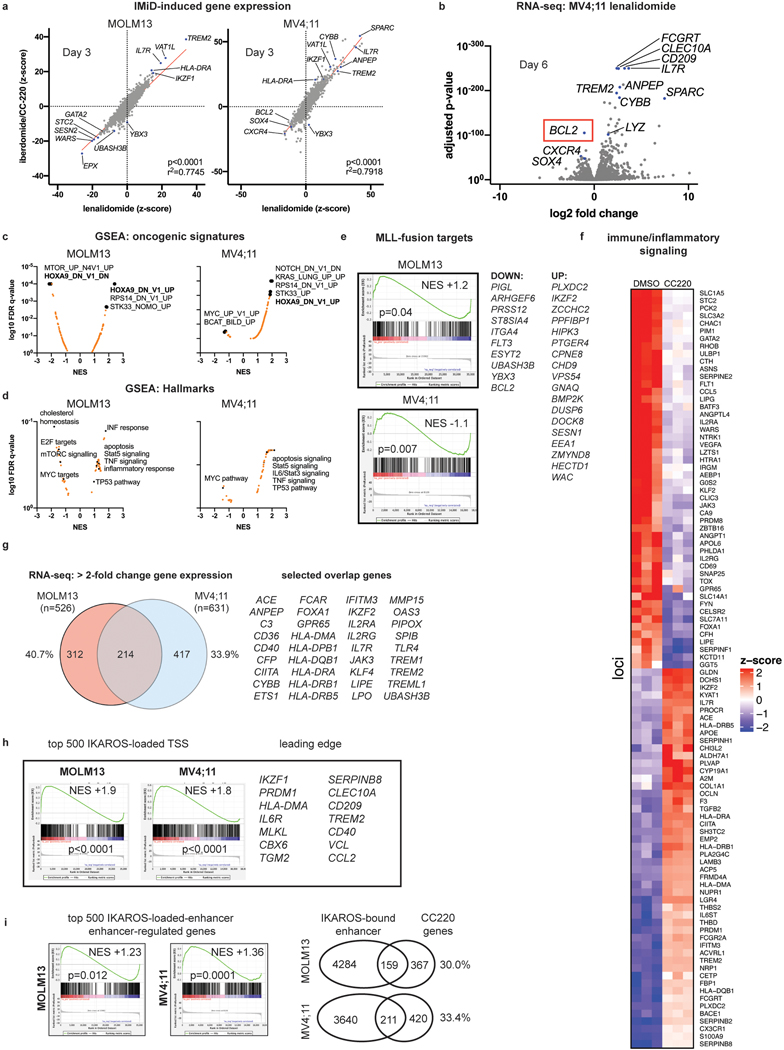
We next determined the transcriptional changes following IKAROS degradation in MLL-r AML using RNA-sequencing (RNA-seq) (Fig. 3a; Supplementary Table 3). LEN and CC220 displayed highly correlated gene expression changes (Extended Data Fig. 4a). Gene set enrichment analysis (GSEA) highlighted gene expression changes related to HOXA9 (Fig. 3b), stem cell biology, myeloid differentiation, MTOR pathway, inflammatory/immune signalling and cell death regulators (Extended Data Fig. 4c,d). Remarkably, after 6 days of LEN treatment, the critical pro-survival gene, BCL2, was the most significantly down-regulated gene in the MV4;11 cell line (Extended Data Fig. 4b; Supplementary Table 3). IKAROS degradation resulted in induction of gene expression associated with myelomonocytic differentiation and loss of stem cell-associated genes (Fig. 3b), demonstrating therapeutic response at the level of transcription and indicating that IKAROS is required to sustain the differentiation block. IMiD treatment perturbed the expression of HOXA9 target genes without impacting the expression of the HOXA9 gene itself, or other canonical MLL-fusion target genes (Fig, 3c), implicating a functional interaction between IKAROS and HOXA/MEIS1. The HOXA/MEIS1 transcriptional program is considered critical for the oncogenicity of the MLL-fusion protein13,21 and is a shared feature with NPM1c AML36. More broadly, IKAROS loss impacted non-canonical MLL-fusion target genes, including FLT3 and BCL2 (Extended Data Fig. 4e)8,41. Prominently, IMiD treatment strongly activated pathways involving the immune response (SOX4, SOCS2, TREM signalling, the TNF pathway, IL6/STAT-signalling, HLA gene expression) (Extended Data Fig. 4f), also suggesting an important role for IKAROS in immune-regulation in AML.
Figure 3. IKAROS is a core transcriptional regulator in MLL-r AML.
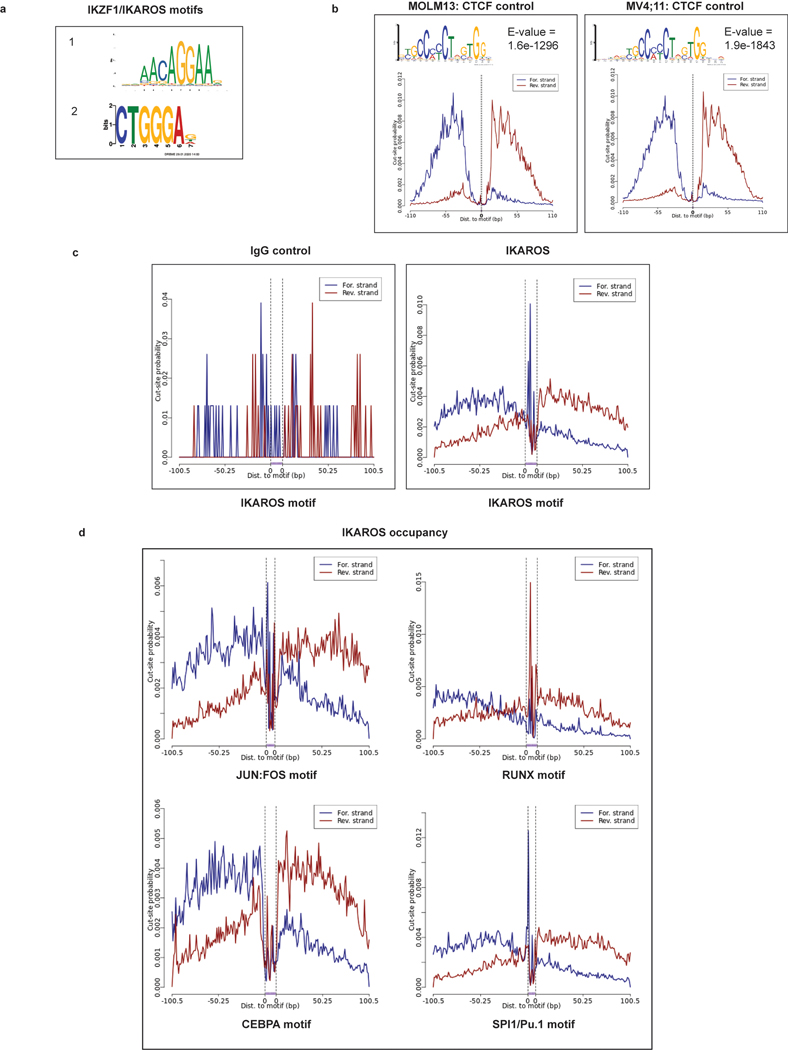
a, Volcano plots displaying RNA-seq data for MOLM13 (n=3) and MV4;11 (n=3) cell lines treated with iberdomide/CC220 1 μM for 3 days. Selected genes are indicated. b, Bar code plots depicting GSEA results for HOXA9-regulated genes, stem cell genes and myeloid differentiation in the MOLM13 cell line. Normalized enrichment score and family-wise error rate p-value determined by GSEA computational method. c, Heatmap depicting gene expression z-scores for canonical MLL1-fusion target genes and selected HOXA9-regulated genes following 3 days treatment with CC220 compared to DMSO-control in MOLM13 cell line. d, Percentage of ChIP-seq IKAROS peaks with 5-fold enrichment over background at regulatory regions, as indicated. e, Venn diagram depicting overlap between all TSS-proximal IKAROS peaks with genes within the MLL-fusion and HOXA/MEIS1 network. f, CUT&RUN de novo DREME motif detection results for the MOLM13 and MV4;11 cell lines. Top 6 motifs detected for IKAROS, in order of statistical significance are shown. Motif, statistical E-value and predicted motif identity are indicated.
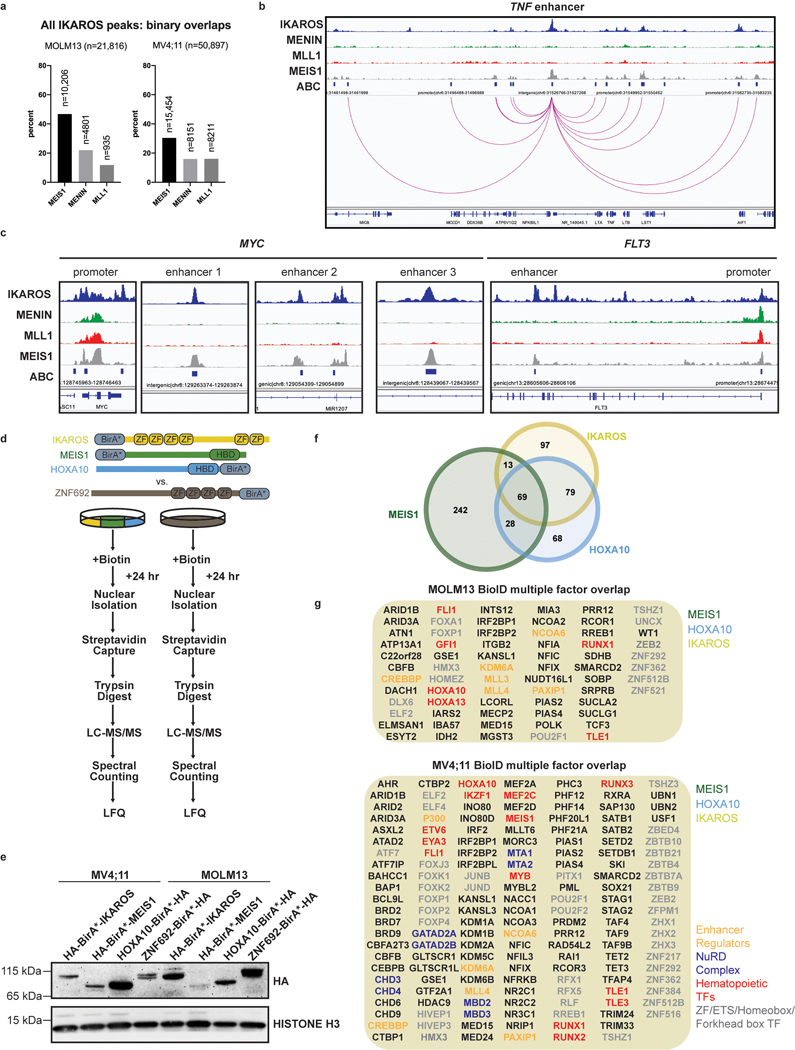
We next defined genome-wide chromatin occupancy for IKAROS, using a combination of chromatin immunoprecipitation followed by next-generation DNA-sequencing (ChIP-seq) and the Cleavage Under Target and Release Using Nuclease (CUT&RUN)42–44. To delineate IKAROS occupancy at functional regulatory regions, we defined a genome-wide map of enhancers and enhancer-promoter connections using the activity-by-contact (ABC) model45. ChIP-seq revealed widespread IKAROS-binding in MOLM13 (21,816 peaks showing >5-fold enrichment (FE) over background) and MV4;11 (50,897 peaks at >5FE) with frequent occupancy in proximity to transcriptional start sites (TSS) (<1kb from the TSS) and at enhancers (Fig. 3d) (also see Fig. 6b,c and Extended Data 8b,c; Supplementary Table 4). In MOLM13, 8079 genes exhibited an IKAROS peak in proximity to the TSS, including many MLL-fusion- and HOXA9-regulated genes (Fig. 3e). A context-dependent role for IKAROS was observed with substantial cell line-specific gene expression changes (Extended Data Fig. 4g). Globally, IKAROS-bound TSSes were strongly predictive of gene expression changes (Extended Data Fig. 4h) with a modest predominance of activated gene expression. Extensive IKAROS occupancy was also noted at enhancers (including those for MYC, BCL2, FLT3, MYB, and TNF) and this also correlated with gene expression changes using enhancer-gene connection predictions by the ABC model. Notably, many genes exhibited IKAROS peaks at their TSS as well as a connecting enhancer (Fig. 6b,c and Extended Data 8b,c) and approximately 30% of all genes deregulated following IMiD treatment were found to have an associated IKAROS-bound enhancer (Extended Data Fig. 4i). We next used CUT&RUN to define the repertoire of TF binding motifs present at IKAROS-bound sites. Discriminative Regular Expression Motif Elicitation (DREME) detected two motifs with homology to reported IKZF1 motifs (Extended Data Fig. 5a) among the highest-ranking motifs in both cell lines (Fig. 3f); CTCF and IgG were used as positive and negative controls, respectively (Extended Data Fig. 5b,c). IKAROS peaks were bound at motifs for critical myeloid TFs (Fig. 3f and Extended Data Fig. 5d) including those for the ETS-family (FLI1/SPI1/Pu.1), RUNX1/2/3/CBFB, SP1, CEBPα/NF-Y, and JUN:FOS. Notably, all of these TFs play an important role in transcriptional deregulation by the MLL-fusion, placing IKAROS within this critical transcriptional network. Overall, these findings position IKAROS as a core transcriptional regulator in MLL-r AML.
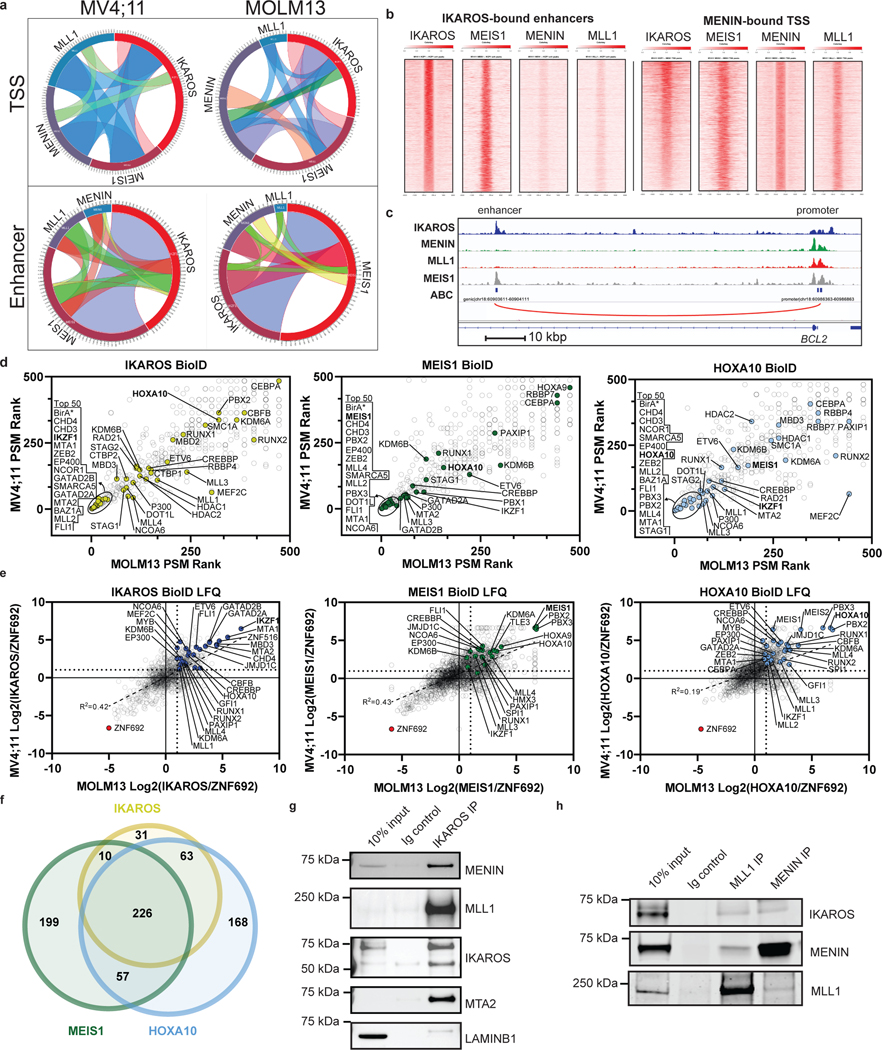
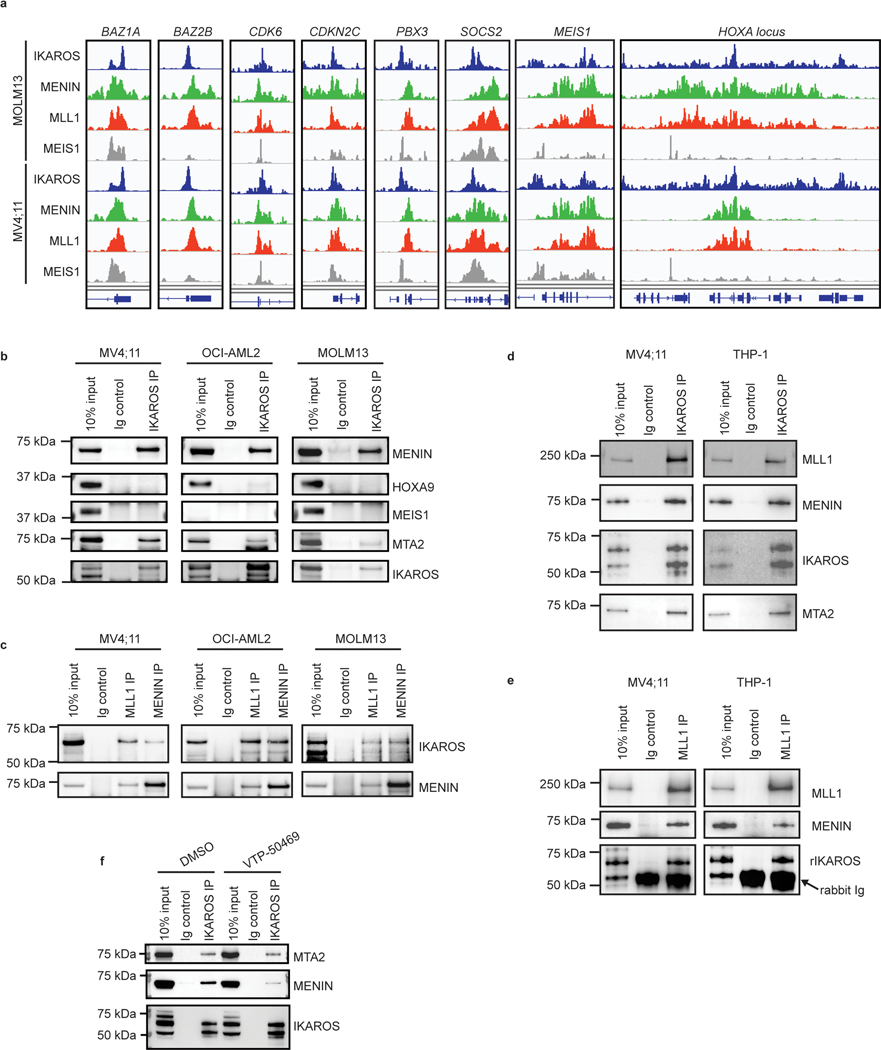
Figure 6. Chromatin co-occupancy and proximal protein-protein interactions of IKAROS and the MLL-fusion network.
a, Circos plots displaying co-occupancy between IKAROS, MEIS1, MENIN and MLL1 determined by ChIP-seq at all TSSes and active enhancers bound by at least one factor, genome wide. b, Tornado plots depicting chromatin occupancy as determined by ChIP-seq from the MOLM13 cell line at the indicated regulatory regions of interest: IKAROS-bound enhancers and MENIN-bound TSSes. c, IGV tracks depicting binding of IKAROS, MENIN, MLL1 and MEIS1 at the BCL2 gene. The promoter and a selected intragenic enhancer are indicated (genomic location indicated in Hg19). The predicted enhancer-promoter connection is indicated (red line). d, Dot plots displaying PSM rank values from spectral counting on proteins identified in IKAROS, MEIS1 and HOXA10 BioID in MV4;11 vs. MOLM13 cell lines by LC-MS/MS. Notable proteins ranked in the top 50 are circled and labelled on the left of each plot in order of lowest rank to highest (n=2). e, Dot plots displaying the Log2 abundance ratios of IKAROS/ZNF692, MEIS1/ZNF692 and HOXA10/ZNF692 BioID in MV4;11 vs. MOLM13 cell lines derived from label free quantitation analysis of LC-MS/MS data (n=2). f, Venn diagram displaying the number of proteins identified in label free quantitation analysis of the MV4;11 IKAROS, MEIS1 and HOXA10 BioID LC-MS/MS data. All proteins included in Venn diagram have a >2-fold enrichment over the ZNF692 control. g, Direct protein Co-IP using IKAROS as the bait and probed for MLL1, MENIN, MEIS1, MTA2 and LAMINB1 for a negative control, in the MV4;11 cell line. h, Direct protein Co-IP using MLL1 and MENIN as the bait and probed for MENIN, MLL1 and IKAROS, in the MV4;11 cell line.
Co-targeting MENIN and IKAROS drives synergistic cell death.
We next evaluated combined IKAROS degradation using IMiDs and MENIN inhibition using VTP-50469. Combined treatment resulted in more potent antiproliferative effects compared to VTP-50469 alone (Fig. 4a). VTP-50469 drives a cellular response typified by cell cycle arrest and differentiation24; however, when used in combination with IMiDs, early loss of cell number was observed due to marked, early induction of apoptosis (Fig. 4b and Extended Data Fig. 6a), representing a potent switch from VTP-50469-induced differentiation to apoptotic cell death. IMiD treatment alone was sufficient to produce immunophenotypic evidence of differentiation and cell cycle arrest, and this was enhanced with combination treatment (Extended Data Fig. 6b,c). The IMiD-VTP-50469 combination displayed synergy, with the response correlative to the potency of IKAROS degradation (CC220>LEN) (Fig. 4c and Extended Data Fig. 6d,e). The IMiD effect was rescued by overexpression of a non-degradable IKAROS mutant (Extended Data Fig. 6f,g). To assess for toxicity to normal hematopoietic cells, we treated non-transformed CD34+ cells and MLL-AF9 transformed CD34+ cells, demonstrating a potential therapeutic window (Fig. 4d). Given the profound apoptotic response, we used MS to exclude the possibility that concomitant VTP-50469 treatment may alter the IMiD neo-substrate profile (Extended Data Fig. 6h; Supplementary Table 2). To characterize the mechanism of enhanced apoptosis, we evaluated apoptotic priming using BH3-profiling46. At baseline, the MV4;11 and MOLM13 cell lines were primarily dependent on BCL-2 for survival, based on mitochondrial sensitivity to BCL-2 selective ABT-199 (Day 1; Fig. 4e and Extended Data Fig. 6i). With combination treatment, the cells became globally primed for apoptosis (increasing BIM sensitivity on Day 3), which progressed to a state of initiated apoptosis (no peptide) by Day 4. On day 3, cells showed a dramatic increase in sensitivity to targeting of MCL-1 and BCL-XL (MS1 for MCL-1; HRK and A113 for BCL-XL), which is in keeping with loss of BCL2-expression following IKAROS degradation (Extended Data Fig. 6j) while MCL1 and BCL-XL protein levels remain unchanged. Remarkably, this combination converts two small molecules that primarily induce differentiation into potent inducers of cell death.
Figure 4. Combined targeting of MENIN and IKAROS results in synergistic induction of apoptosis and cooperative deregulation of gene expression.
a, Proliferation assay measuring cell number over time for the MV4;11 and MOLM13 cell lines following drug treatment. Data represent mean+/−SD (n=3). b, Analysis of annexin V staining (apoptosis) following drug treatment. Viable cell percentage indicated. Representative experiment shown. c, Proliferation assay in MOLM13 cells treated with VTP-50469 combined with either LEN or CC220. Synergy assessment by isobolgrams (left) and Chou-Talalay (right) d, Proliferation assay after 6 day treatment in human CD34 progenitor cells that were non-transformed versus CD34 progenitor cells with MLL-AF9 transformation. Data represent mean+/−SEM (n=3) relative to DMSO control. e, BH3-profiling in MOLM13 cells following drug treatments. Cytochrome C (Cyt C) release was measured. Data represent mean+/−SEM (n=3). f, Volcano plots displaying RNA-seq data for MOLM13 and MV4;11 cells following 3 days of drug treatment (n=3 per condition). g, Venn diagram for overlapping transcriptional changes between CC220, VTP-50469 and the combination (total number of genes indicated with >2-fold change and p-adj<0.05 as determined by DESeq2 package). h, GSEA results querying the Broad “oncogenic signatures” comparing combination treatment with VTP-50469 monotherapy. Results are displayed as dot plot of the log10 false discovery rate (FDR) q-value and normalized enrichment score (NES). i, Bar code plots depicting selected GSEA results in MOLM13 cells from (h). Depicts additive effect of the combination compared to VTP-50469 therapy alone. Normalized enrichment score and family-wise error rate p-value determined by GSEA computational method. j, BCL2 and MYC gene expression assessed by RT-qPCR following treatment with CC220 1 μM, VTP-50469 250 nM and the combination. Data represent mean+/−SEM for combined anlaysis of 3 AML cell lines (MOLM13, MV4;11 and OCI-AML2) with n=3 replicates each (N=3; n=9). p-value determined by paired, two-tailed t-test. k, Western blot for BCL2 protein following treatment of MOLM13 and MV4;11 cells for 72 h with CC220 1 μM, VTP-50469 50 nM and in combination, using ACTIN as a loading control. l, Western blot for MYC and GATA2 protein following drug treatment of MOLM13 cells for 72 hours. Histone H3 as a loading control.
MENIN and IKAROS support leukaemia-promoting transcription.
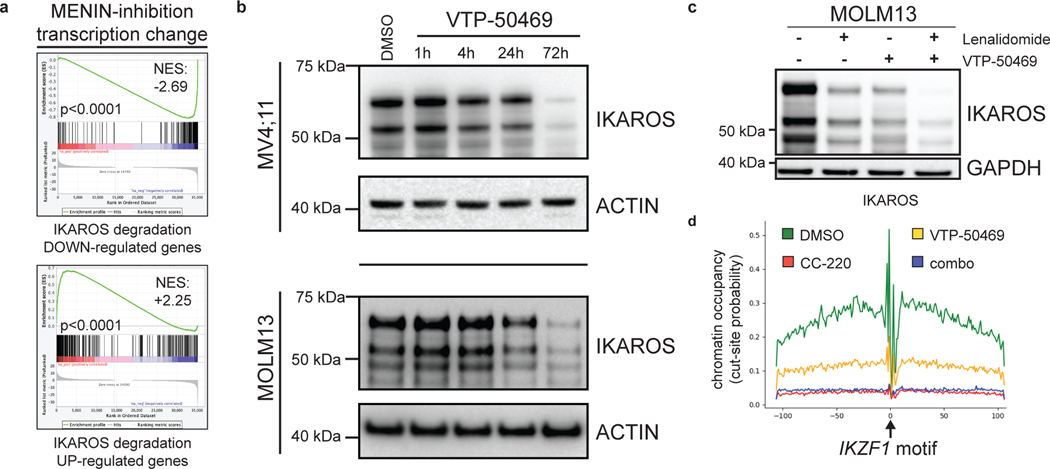
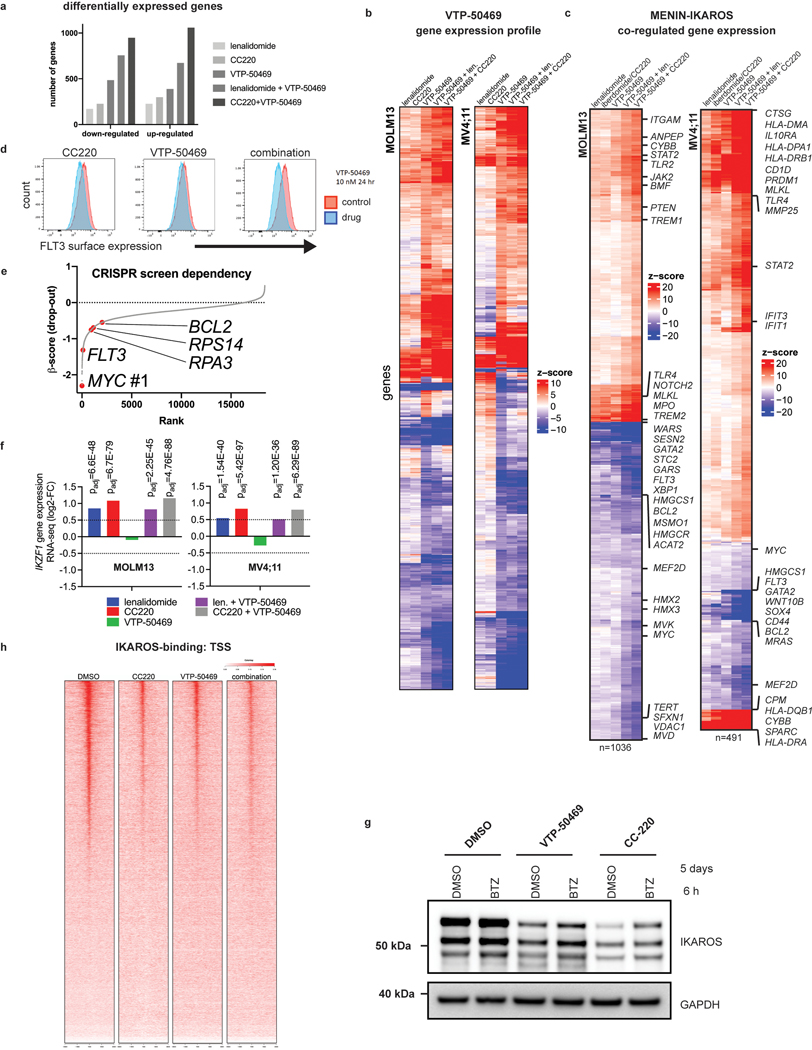
We next evaluated the impact of IKAROS degradation on the transcriptional response to MENIN inhibition (Fig. 4f). Surprisingly, IMiD and VTP-50469 treatments produced strongly overlapping transcriptional changes (Fig. 4f,g) and a large number of genes differentially expressed only with the combination. For MOLM13, 42% of all differentially expressed genes by CC220 (>2 FC) were shared with VTP-50469 and 1095 genes (54.6% of the combination drug response) were unique to the drug combination (Extended Data Fig. 7a). Therefore, numerous genes are commonly regulated by IKAROS and MENIN (Extended Data Fig. 7b,c) and many are only perturbed under combination treatment. GSEA identified HOXA9 and MYC among pathways accentuated by the combination (Fig. 4h,i) as compared to VTP-50469 monotherapy. This finding demonstrates that although VTP-50469 drives rapid downregulation of MEIS1 gene expression, combined IKAROS targeting results in more profound disruption of the downstream HOX/MEIS-regulated network; importantly, combined IKAROS-targeting did not impact the VTP-50469 effect on MEIS1, HOXA9 or HOXA10 gene expression. Combination treatment led to further down-regulation of MYC and BCL2 gene expression (Fig. 4j). Reduction in BCL2, MYC and GATA2 protein levels was confirmed by Western blot (Fig. 4k,l), and there was loss of cell surface expression of FLT3 protein (Extended Data Fig. 7d). Notably, MYC, BCL2 and FLT3 were all identified as strong dependencies in MOLM13 (Extended Data Fig. 7e). MENIN inhibition significantly altered expression of genes also deregulated following IKAROS degradation (Fig. 5a) prompting examination of IKAROS protein levels under VTP-50469 treatment. VTP-50469 caused substantial loss of IKAROS protein over 72 h (Fig. 5b) that was additive with the IMiD-VTP-50469 combination (Fig. 5c) and associated with compensatory increase in IKZF1 gene expression (Extended Data Fig. 7f). VTP-50469-induced loss of IKAROS protein was partially rescued using the proteosome inhibitor, bortezomib (BTZ) (Extended Data Fig. 7g), in keeping with IKAROS loss via proteasomal degradation. Furthermore, MENIN inhibitor treatment led to loss of IKAROS binding proximal to TSSes (Extended Data Fig. 7h) and loss of the IKAROS footprint over the IKZF1 motif (Fig. 5d), directly linking MENIN with the regulation of IKAROS protein stability.
Figure 5. Inhibition of the MENIN-MLL1 protein-protein interaction leads to proteasomal degradation of IKAROS protein.
a, Bar code plots depicting the effect of MENIN-inhibition on the set of genes deregulated following IKAROS degradation by CC220 according to RNA-seq data on Day 3. Normalized enrichment score and family-wise error rate p-value determined by the GSEA computational method. b, Western blot analysis for IKAROS protein following treatment of the MV4;11 and MOLM13 cell lines for 1, 4, 24 and 72 hours, using ACTIN as a loading control. c, Western blot analysis for IKAROS protein following treatment of MOLM13 human MLL-r AML cell line for 72 hours with LEN, VTP-50469 and in combination, using GAPDH as a loading control. d, IKAROS protein chromatin binding at the IKZF1 motif displayed as the cut-site probability +/−100bp around the IKZF1 motif under each drug treatment, as indicated.
IKAROS, MENIN/MLL1 and MEIS1/HOXA chromatin co-occupancy.
We next profiled the genome-wide chromatin binding of IKAROS in relation to the key regulators of these transcriptional programs: MEIS1, MLL1, and MENIN. Significant overlap was observed between IKAROS and each of the other factors (Extended Data Fig. 8a), with a high level of co-occupancy between IKAROS and MENIN proximal to TSSes and co-occupancy of IKAROS and MEIS1 at enhancers (Fig. 6a,b,c). MENIN-bound TSSes showed co-occupancy with IKAROS at 62.0% and 85.2%, for MOLM13 and MV4;11, respectively, while IKAROS-bound enhancers showed co-occupancy with MEIS1 at 81.4% and 62.8%, for MOLM13 and MV4;11, respectively. IKAROS co-occupancy peaks with MEIS1 were detected at enhancers with predicted connections to genes exhibiting IKAROS peaks proximal to the TSS (Fig. 6c and Extended Data Fig. 8b,c). Thus, IKAROS is extensively intertwined on the chromatin with MENIN, MLL1 and MEIS1. To investigate how IKAROS may be engaged in protein complexes at these locations, we examined the IKAROS, MEIS1 and HOXA10 proximal interactome using global proteomic proximity-based in-cell labelling (BioID)47. BioID utilizes an engineered promiscuous biotin ligase, BirA* (~10 nm labelling radius), fused to a protein of interest enabling biotinylation of direct and proximal interacting proteins; IKAROS, MEIS1, HOXA10 and, as a control, ZNF692 were cloned with BirA* to produce fusion proteins that were constitutively expressed in MV4;11 and MOLM13 (Extended Data Fig. 8d,e). ZNF692 is a zinc finger containing transcription factor used as a control because it is not a dependency in MLL-r leukaemia and was considered an inert chromatin binding control. Stable BioID cell lines were grown in the presence of biotin for 24 hours prior to nuclear isolation, lysis, streptavidin capture of biotinylated proteins, on-bead tryptic digestion and LC-MS/MS analysis. Proteines were first ranked by peptide spectral matches (PSM) (Fig. 6d and Supplementary Table 7); IKAROS BioID strongly detected CHD4, CHD3, MTA2, MTA1, GATAD2A (p66α) and GATAD2B (p66β), confirming its association with the NuRD complex. IKAROS BioID analysis also detected interactions with KMT2B/MLL2, DOT1L and KMT2A/MLL1, suggesting proximity between IKAROS and MLL1-containing protein complexes. IKAROS BioID enriched for proteins associated with active enhancers (P300, CREBBP, KMT2C/MLL3, KMT2D/MLL4, NCOA6, and KDM6A/UTX) and myeloid TFs (HOXA10, CEBPA, RUNX1, RUNX2, ETV6 and PBX2). Complementary to these findings, HOXA10 and MEIS1 BioID both enriched for IKAROS and components of the NuRD complex. Protein abundances were also determined using label free quantitation (LFQ) and expressed relative to the control construct, ZNF692 (Fig. 6e, Extended Data Fig. 8d and Supplementary Table 8), with findings consistent with the PSM analysis. Striking overlap among enriched proteins by the IKAROS, MEIS1 and HOXA10 BioID systems was observed (Fig. 6f and Extended Data Fig. 8f,g), which included numerous TFs (IKAROS, MEF2A/C/D, ETV6, RUNX1/2/3, TLE1/3, ZEB2, GFI1, FLI1, MYB, and HOXA10). The BioID data confirms, at the level of protein-protein proximity, that IKAROS is physically intertwined within the core transcriptional circuitry of MLL-r leukaemia. MENIN was poorly detected by LC-MS/MS so we further explored the possibility that IKAROS may be a constituent of an isolatable MENIN-containing chromatin complex using co-immunoprecipitation (Co-IP). Co-IPs using antibodies specific to 1) MENIN or MLL1, and 2) IKAROS, resulted in enrichment of IKAROS and MENIN, respectively (Fig. 6g,h and Extended data Fig. 9b,c,d,e), suggesting the presence of shared protein complex(es); in contrast, co-immunoprecipitation using IKAROS as bait did not enrich for MEIS1 and HOXA9 under these experimental conditions. Finally, we found that inhibition of the MENIN-MLL1 protein-protein interaction by VTP-50469 disrupted MENIN-IKAROS co-immunoprecipitation (Extended Data Fig. 9f). We propose that dual targeting of this putative protein complex containing MENIN and IKAROS contributes to the observed drug synergy.
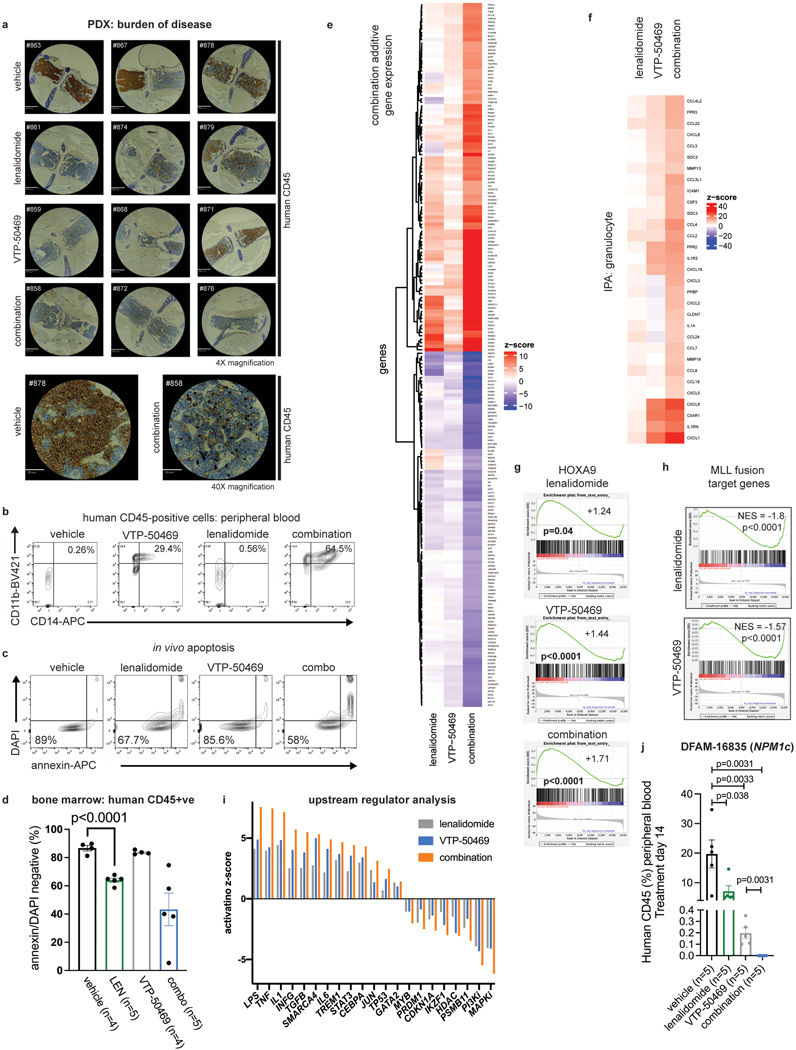
Combined therapeutic effect of VTP-50469 and LEN in vivo.
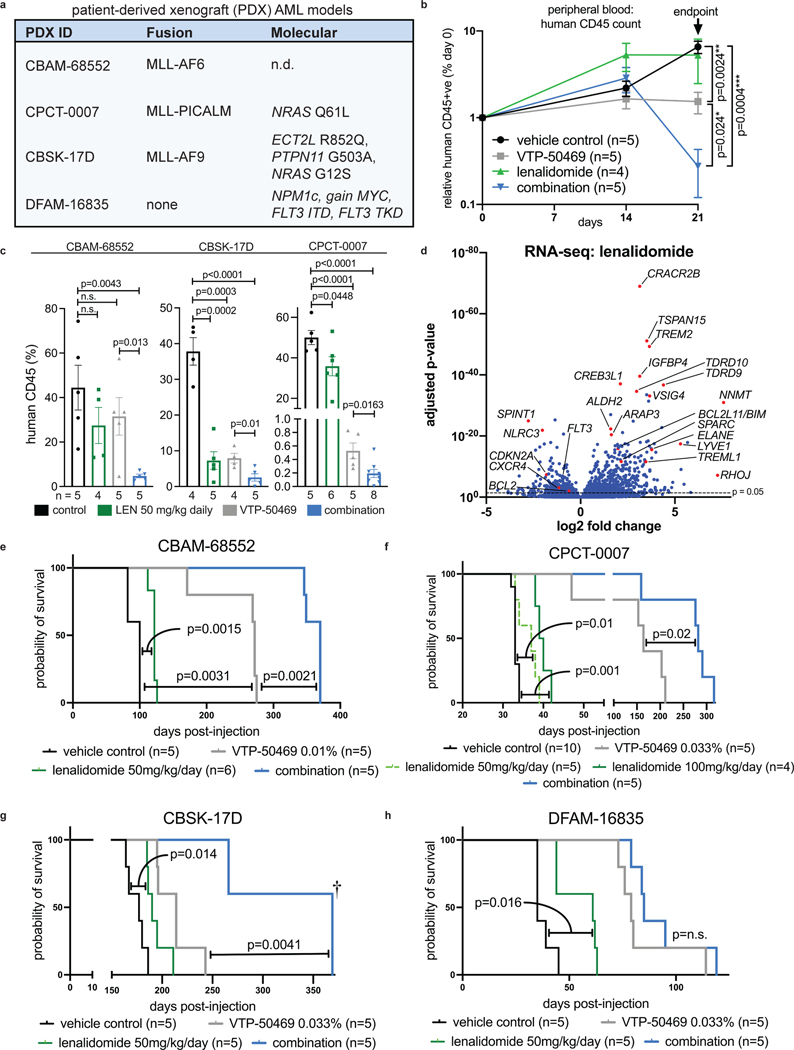
LEN monotherapy and the VTP-50469 combination were then evaluated for in vivo efficacy. We evaluated LEN because it is supported by clinical experience, good safety profile, and multiple FDA approvals48, providing opportunity for rapid clinical translation. LEN exhibits high oral bioavailability and has been used in mouse models49; in contrast, CC220, although a more potent IKAROS degrader, exhibits poor oral bioavailability and has more limited use in vivo. We utilized three MLL-r and one NPM1c AML patient-derived xenograft (PDX) models (Fig. 7a). Human PDX cells were transplanted into non-conditioned immunodeficient recipient mice and, following engraftment, treatment was commenced with vehicle-control, LEN 50 mg/kg daily PO, continuous VTP-50469 in rodent special diet or the drug combination. Combination treatment led to more rapid clearance of leukaemia cells from peripheral blood compared with VTP-50469 alone (p=0.024) (Fig. 7b) and induced strong myelomonocytic differentiation (Fig. Extended Data Fig. 10b). After 7–21 days of treatment, according to VTP-50469 sensitivity of each PDX model, mice were sacrificed for analysis. LEN monotherapy reduced bone marrow disease burden (Fig. 7c), demonstrating in vivo efficacy for IMiD monotherapy in MLL-r AML. Furthermore, mice receiving combination treatment displayed a further reduction in bone marrow infiltration compared to either monotherapy (Fig. 7c and Extended Data Fig. 10a) with morphologic evidence of differentiation. LEN and combination-treated mice exhibited evidence of apoptosis induction in accordance with improved clearance of leukaemia (Extended Data Fig. 10c,d). We used RNA-seq to evaluate gene expression changes for each drug in vivo. This confirmed a strong in vivo effect of LEN in altering transcription (Fig. 7d; Supplementary Table 3). LEN elicited expression of differentiation-associated genes (Fig. 7d) and enhanced the VTP-50469 effect when used in combination (Extended Data Fig. 10e,f). An enhanced effect on the HOXA9 transcriptional program was observed in vivo (Extended data Fig. 10g) and LEN resulted in downregulation of MLL-fusion target genes (Extended data Fig. 10h). Ingenuity upstream regulator analysis using IPA showed upregulation of pathways associated with myeloid differentiation, immune activation/inflammation (LPS, TNF, IL1, INFG) and tumour suppressor pathways (TP53) (Extended Data Fig. 10i). Finally, we evaluated whether LEN monotherapy and the VTP-50469 combination improves survival. LEN monotherapy at 50 mg/kg once daily resulted in a modest survival advantage (Fig. 7e,f,g,h), which could be improved by increasing the dose to 50 mg/kg twice daily (Fig. 7f); combination treatment extended survival of the mice compared to VTP-50469 monotherapy in all three MLL-r models. In the NPM1c PDX, survival was extended by LEN monotherapy, indicating efficacy for the IMiDs in this distinct AML sub-type, but the drug combination did not significantly extend survival compared to VTP-50469 monotherapy although it resulted in more rapid clearance of leukemia cells from the peripheral blood (Extended Data Fig. 10j). These studies demonstrate in vivo efficacy using a series of human PDX AML models, supporting this approach as a novel treatment strategy for patients with MLL-r AML.
Figure 7. Combination therapy with VTP-50469 and LEN results in synergistic anti-leukemic activity in vivo.
a, Four PDX models used indicating PDX identification number (ID), MLL-fusion present and additional molecular features. b, Peripheral blood circulating human leukaemia cells indicated by human CD45 positive cells using flow cytometry during treatment with vehicle, LEN 50 mg/kg daily, VTP-50469 continuously and the drug combination in CBAM-68552. Data represent mean+/−SEM with pair-wise p-value calculated using the two-tail t-test. c, Analysis of bone marrow from transplanted mice following 3 weeks (CBAM-68552), 2 weeks (CBSK-17D) or 1 week (CPCT-0007) of drug treatment for leukaemia burden (human CD45 +ve), as assessed by flow cytometry. Data represent mean+/−SEM with p-value calculated using the two-tail t-test (n=4–5 mice in each group). d, Volcano plot depicting RNA-seq results from human leukaemia cells isolated from CBAM-68552 mouse bone marrow following in vivo treatment with LEN (n=4) compared to vehicle control (n=5). Selected genes are indicated. e, Kaplan-Meier Survival curves for CBAM-68552 following 4 weeks treatment with vehicle, LEN 50 mg/kg/day, VTP-50469 0.1% continuously; p-value by Log-Rank (Mantel-Cox) test. f, Kaplan-Meier Survival curves for CPCT-0007 following 2 weeks treatment with vehicle, LEN 50 mg/kg/day, LEN 100 mg/kg/day, VTP-50469 0.033% continuously, or the combination of LEN 50 mg/kg/day with VTP-50469 0.033%; p-value by Log-Rank (Mantel-Cox) test. g, Kaplan-Meier Survival curves for CBSK-17D following 2 weeks treatment with vehicle, LEN 50 mg/kg/day, VTP-50469 0.033% continuously; p-value by Log-Rank (Mantel-Cox) test. 3 combination-treated mice were sacrificed after 350 days without evidence for leukemia by flow cytometry of bone marrow (†) h, Kaplan-Meier Survival curves for DFAM-16835 following 2 weeks treatment with vehicle, LEN 50 mg/kg/day, VTP-50469 0.033% continuously; p-value by Log-Rank (Mantel-Cox) test.
Discussion
Genome-scale functional genetic screening in MLL-r AML identified mechanisms of drug resistance to clinically relevant DOT1L (EPZ-5676) and MENIN (VTP-50469) inhibitors, and guided the design of a highly effective combination drug treatment using IMiDs. EPZ-5676 and VTP-50469 therapeutically target MLL-r-driven transcriptional dependencies. We identified pathways toward drug resistance involving transcriptional regulators and chromatin remodelling complexes (e.g., NuRD and SWI/SNF), suggesting that cells can undergo transcriptional alterations to escape the effect of these drugs. Complementary to this finding, we found that concomitant targeting of transcriptional regulators that support leukaemogenic transcription, such as IKZF1/IKAROS, enhance the therapeutic effect of these drugs.
We found a diverse role for IKAROS in supporting leukaemia-promoting gene expression, including HOXA/MEIS1-driven gene expression, and repressing pathways for tumour suppression, immune signalling and differentiation. IKAROS function during haematopoiesis has been extensively studied in the lymphoid lineage due to its role as a tumour suppressor in acute lymphoblastic leukaemia and therapeutic target in multiple myeloma; however, less is known about its role in myeloid neoplasia. The IKZF1 gene is expressed and tightly regulated throughout myeloid maturation, and loss of IKZF1/IKAROS during normal haematopoiesis impairs myeloid maturation50. Patients with germline dominant negative IKZF1 gene mutations exhibit multi-lineage haematopoietic defects including abnormal myeloid maturation51. Furthermore, IKAROS is a critical regulator of haematopoietic stem/progenitor cell function, implying a potential role AML stem cell function52. Loss of IKAROS in a cell line model of del(5q) myelodysplastic syndrome results in megakaryocytic differentiation53, reflecting features that we observed in MLL-r AML. In MLL-r AML, we found that IKAROS regulates diverse, essential transcriptional processes while displaying extensive chromatin co-occupancy with key regulators of the MLL-r transcriptional program: MLL1, MENIN and MEIS1. This is in keeping with known functions of IKAROS: IKAROS can mediate transcriptional repression and activation via recruitment of the NuRD complex54, it can support active transcription through recruitment of the SWI/SNF complex54 and SEC55, and can also act as a pioneer TF with enhancer remodelling activity56. Thus, we identify IKAROS is a central component of a TF-driven circuitry in MLL-r AML, and likely other AML subtypes that are dependent on HOX/MEIS1 driven gene expression like NPM1c AML.
IKAROS displays extensive chromatin co-occupancy with MENIN, MLL1, and MEIS1, with a pattern of overlap suggesting a role for co-occupancy of IKAROS and MEIS1 at enhancers and a distinct co-occupancy involving MENIN, MLL1 and IKAROS proximal to the TSS. Of particular note, proximity labelling experiments using BioID revealed that IKAROS, MEIS1 and HOXA10 share a similar proximal protein environment populated by important transcriptional regulatory protein complexes such as NuRD and MLL3/4 COMPASS as well as transcription factors well known for their role in hematopoietic development and leukemia. This nominates IKAROS as a critical regulator of myeloid leukemia associated gene expression. Furthermore, these data define the proximal protein network that comprises the HOXA10/MEIS1 regulatory complex, which is central to hematopoietic development and multiple AML subtypes. Notably, MEIS1/HOXA9 has been reported to play a key role in enhancer regulation18 and MENIN is thought to primarily act proximal to the TSS57, whereas IKAROS has diverse described functions at both the TSS and enhancer54 supporting a role for IKAROS at both of these locations in MLL-r AML. Importantly, chromatin co-occupancy at the TSS by IKAROS and MENIN raises the possibility that IKAROS may participate in the MLL-fusion protein complex. In support of this possibility, IKAROS and other components of the NuRD complex have been previously reported to co-immunoprecipitate with seven different MLL-fusion proteins58. Interestingly, as a sequence-specific DNA-binding protein, IKAROS could potentially play a role in directing the genomic localization of the MLL-fusion and other components of its transcriptional network. It is also important to note that outside of its well-defined protein-protein interaction with KMT2A/MLL159 and the MLL1-fusion protein5, MENIN is reported to interact with a number of TFs including c-MYC60 and JUND61,62, which may be akin to our observations for IKAROS. We also found post-translational loss of IKAROS protein following treatment with the MENIN inhibitor, similar to what has been described for MENIN24,63 and partially attributed to its eviction from stabilizing chromatin-bound complexes; we hypothesize that IKAROS may be similarly stabilized within such chromatin-bound complexes involving MENIN/MLL1 that then become destabilized following MENIN inhibitor treatment. Indeed, we find that IKAROS exhibits a physical interaction with both MENIN and MLL1 (albeit this may be direct or through interaction with a larger multi-protein complex) using a series of targeted Co-IPs.
From the perspective of clinical translation, these findings support a novel therapeutic approach for the treatment of MLL-r AML, using drugs that target IKAROS for degradation in combination with small molecule inhibitors of MENIN and DOT1L. IMiDs have already been evaluated in numerous clinical trials for AML64 with encouraging but variable evidence for efficacy, however these trials have not evaluated specific leukaemia sub-types. The rationale for using these drugs in the setting of AML has been attributed to targeting CK1α65, immune modulation66 and activation of calpain-dependent67 apoptosis. In addition to these important features of the IMiD mode of action, we present data that therapeutic targeting of IKAROS is a critical determinant of the drug’s efficacy in AML, supporting the optimization of IMiD-induced IKAROS degradation as a goal for improved AML therapeutics. Having defined overlapping functions of MENIN and IKAROS in MLL-r AML, we show that combined therapeutic targeting of these two AML dependencies leads to profound anti-leukemic effects both in vitro and in vivo, supporting this as a novel therapeutic approach and highlighting an essential functional interaction between MENIN and IKAROS in leukemic transcriptional regulation. This serves as an example of dual therapeutic targeting of a robust transcriptional network, and putative chromatin-bound protein complex, that more effectively shuts down its oncogenic function compared to either drug alone. Furthermore, combining MENIN inhibition and IKAROS degradation converts two small molecules that primarily induce AML differentiation into a combination that potently induces apoptosis, an effect that is predicted to provide therapeutic benefit.
Online Methods
Animals and patient-derived cells
Animal experiments were approved by Dana-Farber Cancer Institute’s (DFCI) Institutional Animal Care and Use Committee (IACUC) (DFCI protocol number: 16–021; Principal Investigator: Scott Armstrong). Patient-derived xenograft (PDX) models were provided by the Center for Pediatric Cancer Therapeutics (CPCT) at DFCI. CPCT-0007 is a newly generated model: primary human leukemic cells obtained from the Pediatric LEAP Consortium were injected intravenously (IV) to sub-lethally irradiated (200Gy, 4 hrs prior to injection) 6–8 week old NSG-S mice (genotype: NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ) (Jackson Laboratory; Stock #013062). When animals developed signs of leukaemia, bone marrow and spleen were harvested, and cells were frozen for banking purposes. After secondary transplantation, samples were analysed using a targeted next-generation sequencing panel (Rapid Heme Panel, Brigham and Women’s Hospital)68. The source patient (0.75 y male) had refractory MLL-r AML. CBAM-6855224,69 and DFAM-16835 were previously described29, and CBSK-17D was a kind gift from Dr. Ross Levine, Memorial Sloan Kettering Cancer Center. Laboratory mice are housed in solid-bottom, polysulfone 75 sq. in. microisolator cages. The cages are used in conjunction with the Optimice® rack systems with integrated automatic watering. Temperature and humidity in the rodent facilities are controlled at 72 +/− 2°F and a target range of 35–55% relative humidity. A standard photoperiod of 12 hours light/12 hours dark is controlled by an automated system.
Cell lines
Cell lines were acquired from American Type Culture Collection (ATCC) or the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), as indicated: HEK293T cells (ATCC #CRL-3216), MOLM13 (DSMZ Cat#ACC554), MV4;11 (ATCC Cat#CRL-9591), OCI-AML2 (DSMZ Cat#ACC-99), THP-1 (ATCC Cat#TIB-202), OCI-AML3 (DSMZ Cat#ACC-582), U937 (ATCC Cat#CRL-1593.2), and HL60 (ATCC Cat#CCL-240). The IMS-M2 cell line (NPM1c) was a kind gift from Dr. Daniel Tenen, Harvard Medical School, with original source as previously described70. Identification cell lines was independently confirmed by cytogenetics profiling. Routine mycoplasma testing was negative. Cells were cultured in RPMI 1640 (Gibco Cat #11875–093) supplemented with 10% foetal bovine serum, 100U/mL penicillin (Gibco), 100 μg/mL streptomycin (Gibco), and L-glutamine 2 mM at 37°C and 5% CO2.
Genome-scale functional genetic screen
Functional genetic screening was carried out as previously described71 using paired whole genome libraries (denoted H1 and H2) (Addgene Cat#1000000132). Each library contains approximately 90,000 sgRNAs with 5 sgRNAs per gene and a panel of AAVS1 negative-control sgRNAs. The H1 and H2 libraries, combined, target over 18,000 genes with a total of 185,634 sgRNAs. The sgRNA vector combines both Cas9 and sgRNA expression along with puromycin resistance cassette in a single construct. The H1 and H2 libraries were prepared by electroporation (10 μF, 600 Ohms, 1800V) of E. cloni high efficiency bacteria (Lucigen) and culture on agar plates overnight prior to DNA purification using maxi-prep DNA purification columns (Invitrogen Cat #K210017). Prepared libraries were sequenced prior to performing the screen, showing a Gini index <0.1, zero sgRNA count <0.5% and sgRNA representation of the parental library >99.5%. Virus production was carried out on 10 cm plates using HEK293T cells with 10 μg library plasmid per 10 cm plate along with packaging plasmids psPAX2 (Addgene#12260) (7.5 μg) and pMD2.G (Addgene#12259) (3 μg). X-tremeGENE 9 DNA transfection reagent (Sigma-Aldrich) was used to deliver DNA in HEK293T cells, in OPTI-MEM media (Gibco#31985062). Viral supernatant was collected at 24 and 48 h, and pooled. Viral supernatant was titrated to produce a multiplicity of infection (MOI) of approximately 0.2. For each experimental replicate, 50×106 cells were infected with an MOI of 0.2 resulting in ~10×106 transduced cells, providing for 100x coverage of the sgRNA library. Infections were carried out as a 2 h co-incubation with viral supernatant followed by 2 h centrifugation at 800g, 37°C. After 48 h recovery time from infection, cells were placed in puromycin (1 μg/mL) for 72 h to enrich for sgRNA-transduced cells. Cells were then collected and placed in fresh medium to recover for a further 48 hr prior to commencing the screen. Library coverage of 300X (30×106 cells) was maintained throughout the experiment. Cell pellets were obtained at Day 0 and Day 14 containing 30×106 cells. Cells were washed in phosphate-buffered saline (PBS) and lysed in 8 mL lysis buffer (NaCl 300 mM, SDS 0.2%, EDTA 1mM, Tris-HCL pH 8.0 10 mM). Samples were then incubated with RNAase A (100 μg/mL) for 1 h at 65°C and then overnight with proteinase K (100 μg/mL) at 55°C on constant rotation. Genomic DNA was then purified by phenol:chloroform extraction. Amplicon sequencing libraries were then produced using 200 μg of genomic DNA from each experimental replicate, as follows: a first-round PCR reaction, in 32 separate 100 μL reactions, using Q5 High-Fidelity DNA Polymerase (NEB) to amplify the region of the sgRNA between U6 and EF-1α using primers: forward: AATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG and reverse: TCTACTATTCTTTCCCCTGCACTGTACCTGTGGGCGATGTGCGCTCTG; the product was pooled from the primary PCR reaction. a second PCR was carried out to incorporate Illumina adaptors and a 6bp barcode for identification of samples (Supplementary Table 5). A third PCR reaction was carried out to enrich for full-length amplicon using primers: forward AATGATACGGCGACCACCGAGATC and reverse CAAGCAGAAGACGGCATACGAGAT. Final amplicon libraries were purified by gel electrophoresis and extraction of the library followed by gel purification using the QiaQuick Gel Extraction Kit (Qiagen Cat#28704). Sequencing carried out by Novogene Incorporated, Inc., and analysed using the MAGeCK and MAGeCKFlute computational pipeline32,72. FASTQ files were converted to read count tables using the MAGeCK count command. Each genetic screen was carried out in n=2 replicates for each of the H1 and H2 libraries (total n=4 for each drug and control condition). H1 and H2 libraries were combined into a single analysis using the MAGECK maximum likelihood estimation (MLE) function using read-normalized count tables and AAVS1 normalization. MLE calculations were then analysed using the MAGeCKFlute package in R Studio to apply corrections for cell cycle bias and categorize genetic hits as pathway (group 1), resistance (group 2) and synthetic lethal (group 4). For the combined EPZ-5676/VTP-50469 analysis, each set of screens were treated as experimental replicates and analysed using the MAGeCK MLE function followed by MAGeCKFLUTE.
Drug treatment studies: in vivo
For PDX experiments, 1×106 human cells were injected into the tail vein of non-irradiated NOG-F mice (NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac) (Taconic BioSciences, USA). For the cell line xenograft experiment, MOLM13 Cas9-expressing cell lines with sgRNA against Luciferase (non-targeting, NT) and two sgRNAs against IKZF1 were used. All transplanted mice were female, aged between 8–10 weeks on delivery to the facility, and injected 1–2 weeks thereafter. Drug treatment began after engraftment was confirmed by flow cytometric detection of human CD45 expressing cells in the peripheral blood. VTP-50469-containing chow (0.1% and 0.03%, as indicated) was provided by Syndax pharmaceuticals. Lenalidomide (MedChem Express) was reconstituted in 0.5% carboxymethylcellulose (Sigma C4888), 0.25% Tween 80 (Sigma P1754), in water, and sonicated until forming a homogenous suspension immediately prior to dosing by oral gavage. Peripheral blood leukaemia burden was monitored by blood sampling. Blood samples were red cell lysed using Pharm Lyse (BD Cat#555899) prior to staining with phycoerythrin (PE)-conjugated anti-human CD45 antibody (BioLegend Cat#304058) 5 uL antibody per 100 uL solution, BV421-conjugated anti-CD11b (Biolegend) ) 5 uL antibody per 100 uL solution and APC-conjugated anti-CD14 (BioLegend) ) 5 uL antibody per 100 uL solution, as indicated. Samples were then analysed on an LSR Fortessa flow cytometer (Becton Dickinson). For survival experiments, mice were sacrificed when showing signs of illness. We adhered to DFCI standard operating procedure for humane experimental endpoints, which includes tumours measuring 2 cm, ulceration or infection of the tumor site, tumours which compromise mobility, tumours that have become necrotic, weight loss exceeding 15% of the animal’s body weight, interference with the animal’s ability to eat, drink, urinate or defecate, and anorexia. Leukaemia burden was assessed by measuring human CD45 positive cells by flow cytometry or by immunohistochemistry (ServiceBio, Inc., Boston, MA).
Drug treatment studies: in vitro
All drugs were diluted from stock solutions into full tissue culture medium (RPMI+10% FBS). Bortezomib (Thermo Fisher) was reconstituted in DMSO at 10 mM stock solution and used at a final working concentration of 50 nM. QVD-OPh (Abcam) 10 mM stock solution was diluted to a final working concentration of 10 μM. All IMiD drugs were obtained from MedChemExpress: Thalidomide (MedChem Cat#HY-14658), Pomalidomide (MedChem Cat#HY-10984), Lenalidomide (MedChem Cat#HY-A0003), and Iberdomide/CC220 (MedChem Cat#HY-101291) were made into single-use aliquots at 10 mM in DMSO, stored at −80°C, and diluted. An equivalent proportion of Dimethyl Sulfoxide (DMSO) was added to all control samples.
Cell viability, differentiation, apoptosis, cell cycle and morphology analysis
Cell proliferation assays: MOLM13, MV4;11, OCI-AML-2 and IMSM2 cells were plated in 96-well plates. IMiD drug dilutions were made on a half-log scale and plated in triplicate. Cells were split 1:10 on Days 3, 6, 9, 12, and 15 into fresh drug-containing media. Viable cells were measured using an LSR Fortessa (Becton Dickinson) every 3 days with DAPI used as viability stain. Cell counts were normalized to the DMSO control. Apoptosis was measured using the Annexin V Apoptosis Detection Kit (eBioScience Cat#88–8007074). For cell cycle analysis, cells were collected after indicated drug treatment, washed in PBS and fixed using the Cytofix/Cytoperm kit (BD Bioscience Cat#554714) prior to DAPI staining and analysis by flow cytometry. For assessment of differentiation, after indicated drug treatment or CRISPR/Cas9 modification, cells were stained with BV421-conjugated anti-CD11b (BioLegend Cat#301324) and APC-conjugated anti-CD14 (BioLegend Cat#367118), each at 5 uL antibody per 100 uL solution. Samples were then analysed on an LSR Fortessa flow cytometer. For morphology assessment, cells were collected after CRISPR-modification, centrifuged onto slides using the CytoCentrifuge (Thermo Scientific) and stained for microscopy using the JorVet DipQuick Stain (Jorgensen Laboratories, Inc. Cat#JO322). FLT3 cell surface expression assessed by flow cytometry using anti-CD135/FLT3/Flk-2 antibody (BioLegend Cat#313306) using 5 uL antibody per 100 uL solution.
Lentivirus production and CRISPR/Cas9-mediated cell modification
Sequences for selected small guide RNAs (sgRNAs) targeting human IKZF1 and MTA2 were extracted from the H1/H2 sgRNA libraries and additional sgRNAs were designed using the BROAD Genetic Perturbation Platform (GPP) web tool73. Sequences used are listed in Supplementary Table 5. Oligonucleotides for sgRNAs were synthesized by Integrated DNA Technologies (IDT) and cloned into the lentiviral vector, improved-scaffold-pU6-sgRNA-EF1alpha_PURO-T2A-RFP (ipUSEPR) (Gift from Dr Yadira Soto-Feliciano and Dr. David Allis, Rockefeller University). Constitutive Cas9 expression was achieved using pLentiCas9-Blast (Addgene #52962). Lentivirus was produced in 293T cells following transient co-transfection with ipUSEPR or pLentiCas9 (10 μg), pMD2.G (3 μg) and psPAX2 (5 μg) per 10 cm tissue culture plate. Viral supernatant was collected at 24 and 48 h post transfection and passed through a 0.45 μm filter. Viral supernatant produced for pLentiCas9-Blast was concentrated approximately 30-fold by ultracentrifugation in an SW-28 rotor at 112,700g for 90 min. Viral supernatant for ipUSEPR was used neat. All human AML cell lines were transduced by co-incubation of 105 cells with viral supernatant supplemented with polybrene 0.8μg/mL followed by centrifugation at 800g, 37°C for 2h. Cells infected with pLentiCas9-Blast were selected in Blasticidin 10 μg/mL for 6 days. For competition assays, Cas9-expressing human AML cell lines were infected with the respective sgRNA construct, were not sorted (yielding a mixed population of RFP+ and RFP- cells), and RFP expression was measured over time by flow cytometry. For assessment of proliferation, apoptosis and morphology, sgRNA-transduced cell populations were sorted using a FACSMelody Cell Sorter (BD) to produce a uniform population of high-RFP-expressing cells prior to commencing the assay; experiments were performed on bulk cell populations. For all experiments, a control non-targeting sgRNA construct was utilized for comparison (Supplementary Table 5).
Western blot analysis and protein immunoprecipitation
For Western blots, cells were collected, washed once in PBS and the protein was collected by lysis in 1X Laemmli buffer containing 10 mM DTT, sonicated and denatured for 5 min at 95°C. For MLL1/KMT2A, samples were loaded onto a 3–8% Tris-Acetate polyacrylamide gel (ThermoFisher) and separated by electrophoresis in 1X Tris-acetate running buffer (Novex#LA0041). Protein was then transferred to PVDF membrane using wet transfer in 1X Transfer Buffer (Novex#NP0006–1) containing 10% methanol for 2 h at 32V and 4°C. Western blotting for all other proteins was carried out using 10% Bis-Tris polyacrylamide gels and then transferred onto nitrocellulose membrane using the iBlot 2 Gel Transfer Device (ThermoFisher). The membranes were blocked in 5% milk in TBS-Tween buffer (TBST) for 1 hr and incubated overnight, as indicated, rabbit anti-IKAROS (Cell Signaling Cat#14859S) 1:1000 dilution, mouse anti-IKAROS (Invitrogen Cat#MA5–28613) 1:2000 dilution, anti-KMT2A/MLL1 (Bethyl Cat#A300–086A) 1:5000 dilution, anti-MENIN (Bethyl Cat#A300–105A) 1:5000 dilution, anti-HOXA9 (Abcam cat#ab140631) 1:5000 dilution, anti-MEIS1 (Abcam Cat#ab19867) 1:1000 dilution, anti-MTA2 (Abcam Cat#ab8106) 1:1000 dilution, anti-cMYC (Abcam Cat#ab32072) 1:1000 dilution, anti-BCL2 (Cell Signaling Cat#15071) 1:1000 dilution, anti-BCL-XL (Cell Signaling Cat#2764S) 1:1000 dilution, anti-MCL1 (Cell Signaling Cat#94296S) 1:1000 dilution, anti-Casein Kinase 1-alpha (Abcam Cat#ab108296) 1:1000 dilution, anti-Histone H3 (Abcam Cat#ab1791) 1:2500 dilution, anti-beta-ACTIN (Cell Signaling Cat#12620S) 1:1000 dilution, anti-LaminB1 (Abcam Cat#ab16048) 1:1000 dilution, or anti-GAPDH (Cell Signaling Cat#97166S) 1:1000 dilution, were used to immunostain the membrane. Membranes were washed in TBST >6 times and developed using a secondary HRP-linked anti-rabbit IgG, (Thermo Fisher Cat#45000682) 1:10000 or secondary HRP-linked anti-mouse IgG (Cell Signaling Cat#7076S) 1:2000 and chemiluminescence kit (Pierce). Precision Plus Dual Color Protein Standard (BioRad) was used to determine molecular weight. Blots were imaged using the ChemiDoc MP Imaging System (BioRad). Protein co-immunoprecipitation (Co-IP) was carried out using 20 million cells per Co-IP according to two different approaches, 1) High salt nuclear extraction with NP40 wash74 (with modification): cells washed in PBS, then washed in hypotonic buffer (HB) (10 mM Tris pH 7.5, 10 mM KCl, 1.5 mM MgCl2), then incubated for 10 min on ice in HB prior to dounce homogenization and centrifugation at 2600g 10 min, then resuspended in low salt buffer (LSB) (20 mM Tris pH 7.5, 12.5% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA pH 8.0), then an equal volume of high salt buffer (20 mM Tris pH 7.5, 12.5% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA pH 8.0, 1.2M KCl) is added dropwise while continuously mixing, then agitated at 4°C for 25min prior to centrifugation at 17,000g for 25min; the supernatant containing extracted protein is then diluted 4-fold in LSB prior to proceeding with immunoprecipitation. The resulting protein extract was then incubated with antibody-bound beads at 4°C (as described below). After overnight incubation, protein-bound beads were then washed three times in wash buffer (20 mM Tris pH 7.5, 150 mM NaCl, 0.25% NP40, 0.2 mM PMSF). 2) High glycerol wash75: cells were collected and washed in PBS, then incubated in nuclear lysis buffer (50 mM KCl, 10 mM MgSO4.7H2O, 5mM HEPES, 0.05% NP40, 1mM PMSF, 3mM DTT, supplemented with Complete protease inhibitor), followed by wash of nuclei in nuclei wash buffer (NaCl 300mM, Tris pH7.5 50mM, MgCl2 1mM) and subsequent disruption using dounce homogenization in buffer EB300 (Tris pH 7.5 50 mM, EDTA 1 mM, NP-40 1%, NaCl 300mM) and centrifugation at 17,000g for 15min. The resulting nuclear extract was supplemented with 250U benzonase (Sigma) and then incubated with antibody-bound beads overnight at 4°C. Protein-bound beads were washed in buffer BC200 (20 mM Tris (pH 7.9), 0.2 mM EDTA, 1 mM DTT, 0.2 mM PMSF, 20% glycerol, and 200 mM of KCl) three times. In both protocols, protein was eluted from beads by incubating in 1X Laemmli buffer at 95°C for 10 min prior to Western blot analysis. Immunoprecipitation of each protein of interest carried out using 5 μg antibody with Protein A Dynabeads (for rabbit antibody) or Protein G Dynabeads (for mouse antibody). Beads washed three times (0.5% Bovine serum albumin, Triton X-100 0.05% in PBS) and incubated with antibody overnight with constant turning; 5μg of antibody was used for each IP. Beads washed again three times prior to addition of protein preparation. Anti-bodies used were: MENIN antibody (Bethyl), IKAROS antibody (Invitrogen-mouse antibody), KMT2A/MLL1 antibody (Bethyl), normal rabbit IgG control (Bethyl), or normal mouse IgG control (Cell Signaling). All buffers were supplemented with 1X Complete protease inhibitor tablets (Roche#11836145001).
RNA isolation, quantitative real-time PCR and RNA-seq
For all cell line experiments, RNA was isolated from cells following the indicated drug treatments using the RNeasy Mini Kit (Qiagen). RNA samples from in vivo experiments were obtained by isolating human cells from mouse marrow using the EasySep mouse-human chimera kit (Stem Cell Technologies) followed by RNA extraction using the PicoPure RNA isolation kit (Thermo Fisher). For RT-qPCR, cDNA synthesis was carried using the Azuraflex cDNA Synthesis Kit (Azura Genomics). Taqman gene expression assay for BCL2 (ThermoFisher Cat #4331182) and MYC (ThermoFisher Cat #4331182) was used for RT-qPCR reaction on the ViiA 7 Real-Time PCR system (Applied Biosystems). GAPDH was used a housekeeping control. For next-generation RNA-sequencing, RNA quality was assessed using the Agilent Tapestation 4200 (Agilent Technologies). Library preparation was performed using the NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs). Sequencing was performed on an Illumina Nextseq 550 (Illumina) acquiring 37 bp, paired-end reads.
Chromatin immunoprecipitation and next-generation sequencing (ChIP-seq)
ChIP-seq was performed in MV4;11 and MOLM13: cells were crosslinked in 1% methanol-free formaldehyde (ThermoFisher) for 7 min at room temperature followed by quenching of the formaldehyde using 100 mM Tris pH 8.0 and 250 mM Glycine. Cells were then lysed using 50 mM Tris-HCl pH 8.0, 100 mM NaCl, 5 mM EDTA, 1% SDS for 10 min at ambient temperature and chromatin was collected by centrifugation at 10,000 3xg for 10 min. Chromatin was then resuspended in 66mM Tris-HCl pH 8.0, 100mM NaCl, 5mM EDTA, 1.7% Triton X-100, 0.5% SDS and sheared using an E100S sonicator (Covaris) to chromatin fragments of 200–400 base-pair DNA size. Sheared chromatin from the 20 million cells was used in each immunoprecipitation using the respective antibodies: anti-MENIN (Bethyl Cat#A300–105A), KMT2A/MLL1 (Bethyl Cat#A300–086A), MEIS1 (Abcam Cat#ab19867) and IKAROS (Cell Signaling Cat#14859S) antibodies and protein-A magnetic beads (Dynal). Immunoprecipitated DNA fragments were eluted and de-crosslinked in 100 mM NaHCO3, 100 mM NaCl, 1% SDS, and quantified by TapeStation 4200 (Agilent) and Qubit (ThermoFisher). 1–10 ng of DNA was used in preparation of Illumina compatible libraries using SMARTer ThruPLEX DNA-Seq Kit (Takara) followed by sequencing using NextSeq550 (Illumina) to obtain paired-end reads (R1: 37bp, R2: 37bp).
CUT&RUN sequencing
The protein-A-MNase fusion protein (pA-MNase) was produced by transforming the pK19pA-MN plasmid (Addgene #86973) into Rosetta (DE3) competent cells (Novagen). Expression of the pA-MNase was induced in liquid LB culture at an optical density of 0.7 with IPTG treatment for 2 h. Following cell lysis, pA-MNase was purified using IgG Sepharose 6 Fast Flow beads (GE Healthcare Cat#17–0969-01) followed by concentration in a 20,000 MW Slide-A-Lyzer cassette (Thermo Fisher #66012). The pA-MNase product was assessed by BCA quantification and Western blot analysis. CUT&RUN was performed as previously described42. In short, 5×105 cells were washed twice in PBS, then once in a wash buffer (20mM HEPES pH 8.0, 150mM NaCl, 0.5mM spermidine) followed by immobilization on Concanavalin A magnetic beads activated in binding buffer (20mM HEPES pH 8.0, 10mM KCl, 1mM CaCl2, 1mM MnCl2) (Polyscience). The immobilized cells were permeabilized by washing in wash buffer supplemented with 0.1% Triton-X100 and incubated with anti-IKAROS antibody (Cell Signaling Cat#14859S), rabbit IgG control (Bethyl Cat#P120–201), anti-CTCF (Cell Signaling Cat#3418S) or anti-H3K27Ac (Diagenode Cat#15410196) at 4°C with addition of EDTA to 2mM. After 2-hour incubation the cells were washed twice and incubated with Protein-A-MNase conjugate at 4°C. After 4 h, the cells were washed twice and MNase was activated by addition of CaCl2 to 20mM. The DNA digestion was stopped by addition of EDTA and EGTA, both to 10mM. DNA fragments were collected in supernatant and purified using 1.8x AMPure XP beads (Beckman Coulter) which were then converted to Illumina compatible libraries using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England BioLabs) followed by sequencing using NextSeq550 (Illumina). For the drug treatment experiment, carry-over E.coli DNA from the pA-MNase preparation was used for sample normalization during bioinformatic analysis.
ATAC-sequencing
Five million cells from MV4;11 and MOLM13 were washed in PBS and resuspend in a hypotonic buffer (50 mM KCl, 10 mM MgSO4, 5 mM HEPES, 0.05% NP-40, 1 mM PMSF, 3 mM DTT) to generate intact nuclei and washed three times in a nuclear wash buffer (10 mM Tris HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2). 50,000 nuclei per cell line were used for the transposition reaction. Nuclei were incubated in 1X TD Buffer (Illumina) and 2.5 μM Transposase enzyme (Illumina) for 30 minutes at 37 °C. Transposed DNA was then extracted from nuclei using the MiniElute PCR purification Kit (Qiagen). Transposed DNA fragments were amplified using NEBNext® High-Fidelity PCR kit (NEB) with a universal Nextera primer and a unique indexing primer and sequenced using NextSeq550 (Illumina) obtaining paired end reads (R1: 75bp; R2: 75bp) with 57 million and 48 million reads obtained for the MOLM13 and MV4;11 cell lines, respectively.
Sample preparation for quantitative mass spectrometry based global protein measurements
Ten million cells from each cell line (MV4;11, MOLM13 and OCI-AML2) were treated with DMSO, thalidomide 10 μM, lenalidomide 5 μM, pomalidomide 1 μM or iberdomide/CC220 0.1 μM for 5 h and cells were harvested by centrifugation and washed three times in PBS. For combination drug treatment, cells were first treated in DMSO or VTP-50469 50 nM for 5 days prior to IMiD treatment. Lysis buffer (8 M Urea, 50 mM NaCl, 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (EPPS) pH 8.5, Protease and Phosphatase inhibitors) was added to the cell pellets and manually homogenized by 20 passes through a 21-gauge (1.25 in. long) needle and resulting cell lysate was clarified by centrifugation. Bradford assay (Bio-Rad) was used to determine protein concentration. 200 μg of protein for each sample was reduced, alkylated, LysC/Trypsin digested and TMT labelled as previously described40. Each of the sample channels were combined in a 1:1 ratio, desalted using C18 solid phase extraction cartridges (Waters) and analysed by LC-MS/MS for quality control analysis and channel ratio comparison. Equally combined samples were then offline fractionated into 96 fractions by high pH reverse-phase HPLC (Agilent LC1260) through an aeris peptide xb-c18 column (phenomenex) with mobile phase A containing 5% acetonitrile and 10 mM NH4HCO3 in LC-MS grade H2O, and mobile phase B containing 90% acetonitrile and 5 mM NH4HCO3 in LC-MS grade H2O (both pH 8.0) and resulting fractions were then pooled into 24 fractions, desalted, and submitted for mass spectrometry analysis.
Sample preparation for BioID interaction analysis
To generate BioID systems, coding sequences for IKAROS, MEIS1, HOXA10, and ZNF692 were synthesized and cloned by Twist Biosciences to produce N- and C-terminally BirA* tagged pTwist-Lenti-SFFV-Puro-WPRE constructs. Stably expressing MV4;11 and MOLM13 cell lines were then generated and the near-equal biotin labelling was determined by Western blotting with streptavidin-HRP (data not shown) and N-terminally BirA* tagged IKAROS, C-terminally BirA* tagged MEIS1, C-terminally BirA* tagged HOXA10 and C-terminally BirA* tagged ZNF692. MV4;11 and MOLM13 cell lines were chosen for subsequent experiments. Cell lines were expanded to large volumes and 50 μM Biotin was added to culture medium 24 h prior to harvest. Cells were harvested and washed 3 times with PBS and then resuspended in a hypotonic buffer (50 mM KCl, 10 mM MgSO4, 5 mM HEPES, 0.05% NP-40, 1 mM PMSF) followed by 3 washes in a nuclear wash buffer (10 mM Tris HCl pH 7.5, 10 mM NaCl, 3 mM MgCl2) to generate intact nuclei. Nuclei were lysed in BioID Lysis buffer (50 mM Tris, pH 7.5, 500 mM NaCl, 0.4% SDS, 2% Triton X-100 supplemented with Roche protease inhibitor tablets), followed by probe tip sonication (3 rounds, duty cycle 65%, 30 s pulses). Samples were clarified by centrifugation and lysates were quantified by bicinchoninic acid (BCA) assay. Ten mg of protein per replicate, two replicates per cell line, were incubated with 80 μL of streptavidin sepharose high performance beads (Millipore Sigma) overnight at 4 °C. After incubation beads were washed with 2 mL of 2% SDS, 2 mL of BioID lysis buffer three times, 10 mL of 50 mM Tris six times, and 5 mL of 50 mM triethylammonium bicarbonate (TEABC) once. Streptavidin bound proteins then underwent on-bead tryptic digestion by adding 1 μg of sequencing grade trypsin (Promega) and incubating beads overnight at 37 °C. After digestion, the supernatant was separated from the streptavidin beads and acidified to 1% Formic acid. Acidified peptides were then de-salted and purified using SOLAμ™ Solid phase extraction plates (Thermo Fisher Scientific).
Mass spectrometry data collection
All mass spectrometry data were collected using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) coupled with a Proxeon EASY-nLC 1200 LC pump (Thermo Fisher Scientific). Peptides were separated on an EasySpray ES803a 75 μm inner diameter microcapillary column (Thermo Fisher Scientific). For global quantitative proteomics and BioID interaction proteomics, peptides were separated using a 190 min/140 min gradient of 6 – 27%/5 – 34% buffer B (95% acetonitrile) in 1.0% formic acid with a flow rate of 300 nL/min, respecitvely. For global quantitative proteomics, each analysis used a MS3-based TMT method as described previously76. For global quantitative proteomics and BioID interaction proteomics the data were acquired using a mass range of m/z 340 – 1350/375 – 1500, resolution 120,000, AGC target 5 × 105, maximum injection time 100/50 ms, dynamic exclusion of 120 seconds for the peptide measurements in the Orbitrap, respectively. For global quantitative proteomics and BioID interaction proteomics, data dependent MS2 spectra were acquired in the ion trap with a normalized collision energy (NCE) set at 35%, AGC target set to 1.8 × 104/4 × 105 and a maximum injection time of 120/50 ms, respectively. MS3 scans were acquired in the Orbitrap with HCD collision energy set to 55%, AGC target set to 2 × 105, maximum injection time of 150 ms, resolution at 50,000 and with a maximum synchronous precursor selection (SPS) precursors set to 10. For BioID interaction proteomics, data dependent MS/MS spectra were acquired in the ion trap in centroid mode with the collision energy set at 30%, AGC target set to 1 × 104 and a maximum injection time of 30 ms.
Mass spectrometry data analysis
Proteome Discoverer 2.2 (Thermo Fisher Scientific) was used for .RAW file processing and controlling peptide and protein level false discovery rates, assembling proteins from peptides, and protein quantification from peptides. MS/MS spectra were searched against a Swissprot human database (December 2016) with both the forward and reverse sequences. For BioID data searches a FASTA entry was included for the protein sequence of BirA*. Database search criteria was conducted as previously described.75 Reporter ion intensities were normalized and scaled using in-house scripts in the R framework (R Development Core Team, 2014). Statistical analysis was carried out using the limma package within the R framework (Ritchie et al., 2015). For BioID data spectral counting analysis, individual Proteome Discoverer searches were conducted on IKAROS, MEIS1 and HOXA10 BioID .RAW files, combining both replicates. The protein identities and their corresponding PSM values were then merged between MV4;11 and MOLM13 cell lines to create a unified dataset with proteins uniquely detected in MV4;11 or MOLM13 cells excluded. The protein identities were then referenced against the CRAPome repository77 to remove contaminants common to affinity purification MS experiments. The dataset was further filtered by removal of proteins that met either of the following two criteria; if the number of “experiments detected” in the CRAPome repository was greater than the average of the number of “experiments detected” values across our BioID datasets or if the “average spectral count” reported in the CRAPome repository was greater than the average of the “average spectral count” values detected in our BioID datasets. NuRD complex member proteins are highly prevalent in the CRAPome and because of the biological relevancy of this study, we made a special exemption for removal. After CRAPome proteins were removed, the resulting proteins were ranked by descending number of PSMs and plotted in rank order. For Label Free Quantitation (LFQ), analysis was carried out using Proteome Discoverer 2.2. Separate analyses were made for MV4;11 and MOLM13 cells using corresponding IKAROS, MEIS1, HOXA10 and ZNF692 BioID .RAW files that were searched together and the Minora Feature Detector was used to generate abundance values for proteins across the different samples. Normalization was applied in the Precursor Ions Quantifier tool using the Total Peptide Amount feature. Retention time alignment was set at 15 minutes. Log2 abundance ratios were generated for IKAROS, MEIS1 and HOXA10 over ZNF692 and a threshold was set to define enrichment as being greater than 2-fold (Log2 ratio=1). Based on Western blot analysis and the median value of Log2 ratios called in the MOLM13 experiment (IKAROS/ZNF692=−1.64, MEIS1/ZNF692=−0.73, and IKAROS/ZNF692=−1.95) we concluded that ZNF692-BirA* induced significantly higher labelling than IKAROS/MEIS1/HOXA10. Thus, MOLM13 BioID data was further normalized by subtracting the median value of all Log2 ratios in each comparison from each individual value. No extra normalization was applied to the MV4;11 LFQ data. PSM values from individual searches described above were added, and any protein with less than 2 PSMs was excluded from the LFQ analysis. Of those proteins that had 1 or more isoform detected, the isoform with the most PSMs was retained and the others were removed.
BH3 profiling
BH3 profiling was done as described previously46. Cells were permeabilized with 0.001% digitonin suspended in MEB2 buffer (150 mM mannitol, 10 mM HEPES-KOH pH 7.5, 150 mM KCI, 1 mM EGTA, 1 mM EDTA, 0.1% BSA, 5 mM succinate) and treated with different BH3- only peptides in 384-well plate. After 60mins incubation at 25°C, cells were fixed with 4% formaldehyde for 10 mins at RT followed by neutralization by adding N2 buffer (1.7 M Tris base, 1.25 M glycine, pH 9.1). Cells were stained overnight at 4°C with Hoechst 33342 (H3570, Invitrogen) and anti–cytochrome° c–Alexa Fluor 488 (6H2.B4/612308). The rate of loss of cytochrome c was analysed using Intellicyt iQue flow cytometer in response to each BH3 peptide. Assay were conducted in triplicates. DMSO and alamethicin were used as negative and positive control for cytochrome c release.
IKAROS overexpression cell lines
IKAROS overexpression vectors were a kind gift from Dr. William G. Kaelin and Dr. Vidyasagar Koduri, Dana-Farber Cancer Institute: 1. Wild type IKAROS overexpression vector (CAG-IKZF1-WT-FLAG-IRES-GFP, https://www.addgene.org/107387/), 2. Non-degradable mutant IKAROS overexpression vector (CAG-IKZF1-V1-Q146H-FLAG-IRES-GFP; https://www.addgene.org/69048/), and 3. Empty vector control (pLKO5-GFP, https://www.addgene.org/57822). Lentivirus produced as described above. The MOLM13 cell line was transduced as described above and sorted using a FACSMelody Cell Sorter (BD) to produce a uniform population of GFP-expressing cells.
Human CD34+ cell isolation and MLL-AF9 transduction
Production of MLL-AF9 transduced human CD34+ cells has been described.78 Human CD34+ cells were isolated from human cord blood (New York Blood Center) using the EasySep™ Human Cord Blood CD34 Positive Selection Kit II (Stemcell Technologies). For pre-enrichment, Lymphoprep (Stemcell Technologies) and SepMate™ columns (Stemcell Technologies) were used. Viral supernatants were generated by co-transfection of HEK293-T cells with retroviral (pMSCV-MLL-AF9-IRES-GFP)78 expression vector with packaging and envelope vectors (Human Retro: pUMVC and VSV-G) and X-tremeGene transfection reagent (Millipore). The viral supernatant was filtered through 0.45μm and was concentrated using Amicon Ultra centrifugal filters (Millipore). Human CD34+ cells were plated on retronectin-coated plates (Takara) and were spin infected at a 1:1 dilution of virus:media at 800g at 37°C for 1.5h. The cells were dissociated from the plates using enzyme-free dissociation buffer (Gibco) and were plated in fresh media. Cells were maintained in IMDM with 20% FBS, 1x penicillin/streptomycin (Gibco), 1xß-mercaptoethanol (Gibco), 6 μg/mL hIL3, 10 μg/mL hIL6, 10 μg/mL hSCF, 10 μg/mL TPO, and 10 μg/mL FLT3 (Stemcell Technologies) at 37°C and 5% CO2. Two days after transduction, GFP positive cells were sorted using FACSAria cell sorter (BD Bioscience).
Colony-formation assay
Colony-forming unit (CFU) assays were performed by plating 200 MOLM-13 CRISPR-modified cells into 3 mL Methocult GF M3434 (STEMCELL Technologies). For each sgRNA, four separate methylcellulose plates were used. Colonies were quantified 9 and 12 days after plating by manual counting. On the 13th day of the experiment, each plate was washed with PBS, cells were spun down and resuspended in quadruplicate to run on LSR Fortessa to quantify cells per plate.
Statistical Analysis and Reproducibility
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications24. Single experiments shown are representative of at least 3 independent experimental replicates. Data collection and analysis were not performed blind to the conditions of the experiments. For PDX studies, animals were randomized according to weight and leukaemia burden prior to treatment using Excel (Microsoft). Animals injured by gavage and required to be sacrificed were excluded from the analysis. For proliferation assays, dose-response curves were calculated in a variable slope model as a four-parameter dose-response curve (GraphPad Software). Absolute IC50s were calculated by setting the maximal inhibition baseline parameter to 0% and constraining the minimal inhibition top parameter to 100%. The calculation was done via non-linear regression of the log of the dose (GraphPad Software). Kaplan-Meier survival curves and paired T-test were preformed using the statistical software, GraphPad PRISM. Gene set enrichment analysis (GSEA v3.0 software/Broad Institute)79,80 was used to identify functional associations from the described RNA-seq profiles. Global analysis was performed on raw RNA-seq counts; specific query of gene sets of interest was performed in “pre-ranked” mode using gene lists ranked according to DESeq statistical z-score. Significance cut-offs were assessed on the basis of the GSEA standard recommendations: absolute NES ≥ 1, P ≤ 0.05, Benjamini–Hochberg FDR ≤ 0.25. Gene sets used in presented data are listed in Supplementary Table 6. Bliss independence and Loewe additivity models were used to assess synergy, as described previously81. The Bliss independence model calculates a quantitative measure, called the excess over Bliss, which represents the difference between the observed combined effect and the expected combined effect of the two compounds. The Chou-Talalay combination index for the Loewe additivity model is a dose-effect approach to estimate the effect of combining two compounds based upon the dose of each individual compound that produces the same quantitative effect. All data from flow cytometry was analysed using Flojo software, Becton Dickinson (v10.6.2).
Bioinformatics Analysis
Raw Illumina sequencer output was converted to FASTQ format using bcl2fastq (v2.20.0.422) (Illumina, Inc.). Reads (paired-end 37-mers) were aligned to the human genome (hg19) using STAR (v2.7.5a)82, sorted and duplicates marked/removed with picard pipeline tools (v2.9.4). Final “deduped” .BAM files were indexed using SAMtools (v1.95)83. Files for data visualization were produced using IGVtools (TDF signal pileups; v2.3.75)84 and ngs.plot (pileup heatmaps; ChIP-seq only). ChIPseq signal peaks were called using MACS2 (v2.1.4). For RNA-seq, raw counts were calculated with HTSeq (htseq-count, v0.11.2)85. Raw counts were used to calculate differential expression values with the R Bioconductor DESeq2 package (v1.24.0)86. Results were filtered to exclude non-protein-coding genes (based on Ensembl gene biotype annotations) and low-expressed genes (those with average values < 1 in rlog scale for both treatments in comparison). Expression differentials with adjusted pValue < 0.05 were considered significant with the additional requirement of a greater than two-fold change, where indicated. RNA-seq data were further analysed through the use of Ingenuity Pathway Analysis (IPA) (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis). For CUT&RUN-seq analysis, FASTQ files were processed using a version of the published CUT&RUNTools suite44 (release date 19th August, 2019), modified to run on a local computer cluster. Briefly, reads were trimmed using trimmomatic and aligned to the human genome (hg19) with Bowtie2 (v2.3.4.3)87. Resulting BAM files were sorted, duplicates marked and filtered for fragments <120bp (all with SAMtools, v1.95), and peaks called (with MACS2, v2.1.4). ATAC-seq data was processed as described above for ChIPseq. Enhancer prediction using the activity-by-contact model (ABC) was carried out as previously described45. Heatmap displays for RNA-seq data were generated using the Bioconductor package ComplexHeatmap (v3.14)88 using RStudio (v1.4.1106) with R (v3.6.0). GNU parallel (v20161222)89 was used to facilitate simultaneous processing and analysis of samples.
Publicly Available Data Usage
The Cancer Dependency Map (DepMap) from the Broad Institute of MIT and Harvard, Boston, MA, USA,35 was used to examine cancer dependencies on IKZF1, MEN1, DOT1L and ZFP91 (Broad(2021): DepMap21Q4Public. figshare.Dataset doi:10.6084/m9.figshare.12280541.v3). The cBio Cancer Genomics Portal (cBIOPortal)34,90 was used to examine IKZF1 gene expression across a range cancer subtypes utilizing data from The Cancer Genome Atlas (TCGA) (https://www.cancer.gov/tcga). For ABC enhancer predictions, average Hi-C matrix data was utilized, as recommended45, and obtained from the Broad Institute of MIT and Harvard (https://github.com/broadinstitute/ABC-Enhancer-Gene-Prediction) as well as an additional publicly available HiC dataset for MV4;1191.
Data Availability
Raw and analysed data for ChIP-seq, CUT&RUN-seq, RNA-seq, ATAC-seq and ABC model predictions have been deposited at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under the accession number GSE168463; deposited data correspond to Figures 3, 4, 5, 6, 7 and associated extended data figures. All mass spectrometry data and search results have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD007862) via the PRIDE partner repository with the data set identifiers PXD025227, PXD025228, PXD025229, PXD025230, PXD025231, PXD025232, PXD025271 and PXD025272. Additional data that support the findings of this study are available from the corresponding author upon reasonable request.
Extended Data
Extended Data Fig. 1. Chromatin remodeling complexes modulate the cellular response to therapeutic targeting of MLL-r driven gene expression.
a, Proliferation assay conducted in real-time during the functional genomic screen showing cell number over time. Data represent mean+/−SEM (n=4). b, Volcano plots depicting Wald p-value and beta value calculated using MAGeCK MLE for VTP-50469 and EPZ-5676, comparing the vehicle-treated and drug-treated state on Day 14. c, Gene set testing comparing the behaviour of group 2 (resistance) and group 4 (synthetic lethal) genetic hits between each screen. Family-wise error rate p-value determined by GSEA computational method. d, Expanded panels of CRISPR/Cas9-based competition assays targeting either IKZF1 or MTA2 monitoring sgRNA-RFP expression over time in Cas9-expressing in 4 MLL-r human AML cell lines (MOLM13, MV4;11, OCI-AML2 and THP-1), with and without concurrent treatment with the MENIN inhibitor, VTP-50469. e, CRISPR/Cas9-based competition assays targeting IKZF1 monitoring sgRNA-RFP expression over time in 4 non-MLL-fusion human AML cell lines (IMSM2, OCI-AML3, U937 and HL60) expressing Cas9. IMSM2 and OCI-AML3 carry the NPM1c mutation.
Extended Data Fig. 2. Using CRISPR/Cas9 to genetically target IKZF1 and MTA2 and validate genomic screen findings.
a, Combined analysis of sgRNA depletion across samples, depicted in Extended Data Fig. 1d, showing enhanced sgRNA depletion in the presence of the MENIN-inhibitor, VTP-50469, despite the disadvantage of drug-induced cell cycle arrest. Data represent mean+/−SEM with p-value by two-tail t-test evaluated for n=4 cell lines and n=6 sgRNAs targeting IKZF1 or n=4 sgRNAs targeting MTA2. b, Western blot analysis for MTA2 protein in sgRNA-RFP sorted cells comparing MTA2-targeted bulk cell populations with non-targeting control and ACTIN used as a loading control. c, Western blot analysis for IKAROS protein in sgRNA-RFP sorted cells comparing IKZF1-targeted bulk cell populations with non-targeting control and ACTIN used as loading control. d, Analysis of annexin V staining (apoptosis) and CD11b/CD14 (monocytic differentiation) using flow cytometry 7 days following CRISPR/Cas9-mediated deletion of either IKZF1 or MTA2. Representative experiment is shown. e, Colony forming assay comparing MOLM13 cells with sgRNA targeting Luciferase (Non-Targeting) control versus two different sgRNAs targeting IKZF1 (n =4 per sgRNA), representative colony morphology is shown. f, Relative IC50 for proliferation assays, pertaining to Fig. 1j, comparing MOLM13 cells with sgRNA targeting Luciferase (Non-Targeting control) and two sgRNAs targeting IKZF1 (n=3 per sgRNA) upon treatment with MENIN inhibition. g, Apoptosis assay in MOLM13 and MV4;11 cell lines testing the impact of sgRNAs targeting Luciferase (NT control) versus two sgRNAs targeting IKZF1 on the response to MENIN inhibition with VTP-50469, using doses indicated. Apoptosis was assessed by DAPI exclusion (viability) and annexin V staining with 50,000 cells analysed per sample. Representative example of multiple independent experiments.
Extended Data Fig. 3. IMiDs effectively target IKAROS protein for degradation in human MLL-r AML.
a, Western blot analysis for IKAROS, CK1α and MENIN protein following treatment of MV4;11 human MLL-r AML cell line for 5 hours with increasing doses of THAL, LEN, POM and CC220, using ACTIN as a loading control. b, Western blot analysis for IKAROS and CK1α following treatment of OCI-AML2 human MLL-r AML cell line for 5 hours with increasing doses of THAL, LEN, POM and CC220, using ACTIN as a loading control. c, LEN and CC220 dose-response curves on: Day 9 of treatment for the NPM1-mutant (NPM1c) IMSM2 human AML cell line cell (Absolute IC50 267.0 nM [95% CI 189.6 to 381.0] for LEN, 17.3 nM [95% CI 9.3 to 37.8] for CC220) and Day 18 of treatment for the NPM1-mutant (NPM1c) OCI-AML3 human AML cell line (Absolute IC50 4626 nM [95% CI could not be calculated] for LEN, 57.7 nM [95% CI 33.1 to 101.5] for CC220). Data represent mean+/−SEM with absolute IC50 indicated (n=3). d, Scatterplot for MS determination of IMiD substrates 5 hours after drug treatment in the MV4;11 and OCI-AML2 cell lines. Data represent the log-fold change in abundance and log10(p-value). p-value determined by moderated t-test as implemented by the Bioconductor Limma package.
Extended Data Fig. 4. IKAROS degradation perturbs diverse cellular pathways.
a, RNA-seq dot-plot with linear regression showing z-score for differential gene expression correlating LEN (5 μM) and CC220 (1 μM) treatment for 3 days. b, RNA-seq volcano plot for MV4;11 treated with LEN for 6 days. c, GSEA results for “oncogenic signatures” for RNA-seq data on Day 3. Dot plot is shown of the log10 false discovery rate (FDR) q-value and normalized enrichment score (NES) as determined by GSEA computational method. d, GSEA results for “Hallmarks” signatures. Dot plot is shown of the log10 false discovery rate (FDR) q-value and normalized enrichment score (NES) as determined by the GSEA computational method. e, Bar code plots created using the GSEA tool for putative MLL-fusion target genes with selected genes from leading edge analysis indicated with adj-p-value<0.05 in MOLM13 or MV4;11. Normalized enrichment score and family-wise error rate p-value as determined by GSEA computational method. f, Gene expression z-score heatmap for selected genes within immune regulatory and inflammatory pathways under treatment with CC220 (n=3) compared to DMSO-treated control (n=3). g, Venn diagram for genes displaying > 2-fold change in expression following CC-220 treatment comparing cell lines. Gene number and percentage overlap is indicated. Selected genes deregulated in both cell lines are indicated. h, Correlation of IKAROS-bound gene TSSes, determined by ChIP-seq, with RNA-seq changes. Bar code plot showing gene expression changes following treatment with CC220 (3 days) for the top 500 genes bound by IKAROS (by peak enrichment over background). Selected genes from leading edge analysis indicated. Normalized enrichment score and family-wise error rate p-value determined by GSEA computational method. i, Correlation of IKAROS-bound enhancers, determined by ChIP-seq and the ABC prediction tool, with RNA-seq gene expression changes. Bar code plots display gene expression changes following treatment with CC-220 (3 days) for the top 500 genes predicted to be regulated by IKAROS-bound enhancers (by peak enrichment over background at enhancers). Normalized enrichment score and family-wise error rate p-value determined by GSEA computational method. Venn diagram for overlap between gene expression change and genes predicted to be regulated by the top 1000 IKAROS-bound enhancers.
Extended Data Fig. 5. IKAROS CUT&RUN motif foot printing.
a, IKZF1 DNA-binding motifs. b, Foot printing for CTCF protein over the CTCF motif for each cell line; used as a positive control for motif detection. Result for CUT&RUNTools de novo DREME motif detection is shown with highly significant discovery of the known CTCF motif. MEME and E-value is indicated. c, CUT&RUNTools foot printing for IKAROS and IgG control over the IKZF1 motif in the MOLM13 cell line. Cut site probability distribution as determined by CUT&RUNTools. d, CUT&RUNTools foot printing for IKAROS over the JUN:FOS motif (centrally bound), CEBPα motif (centrally bound), SPI/Pu.1 motif (centrally bound) and the RUNX motif (non-centrally bound), as indicated, in the MOLM13 cell line. Cut site probability distribution as determined by CUT&RUNTools.
Extended Data Fig. 6. Combined targeting of MENIN and IKAROS.
a, Annexin V staining (apoptosis) by flow cytometry following 6 days treatment with VTP-50469, LEN, CC220 and combination (MOLM13). b, Cell cycle analysis: histogram for DAPI stain and bar graphs displaying cell cycle stage (%) are shown for each condition. Representative experiment shown. (MOLM13) c, CD11b/CD14 expression measured by flow cytometry 6 days following treatment with VTP-50469, LEN, CC220 and combination (MOLM13). Representative experiment shown. d, Dose-response curves for LEN and CC220, with and without VTP-50469 (8 nM), compared to VTP-50469 alone (dotted line) measured by Cell-Titer Glo in MV4;11 for synergy studies. e, Chou-Talalay synergy analysis for VTP-50469 in combination with LEN or CC-220 (MV4;11). Chou-Talalay Combination Index (CI) plots (left panel) and normalized isobolograms (right panel) are shown for each IMiD-VTP-50469 combination (day 6). Line of additivity is shown (red). f, CC-220 and MENIN inhibitor dose response curves (day 6) with and without overexpression of non-degradable IKAROS (Q146H) in MOLM13. Data represent mean+/−SD with non-linear regression curve fit shown. g, IMiD and MENIN inhibitor apoptosis after 6 days treatment, with and without overexpression of a wild-type IKAROS or non-degradable IKAROS mutant (Q146H) in MOLM13. h, Scatterplots for mass spectrometry (MS) results on MOLM13, MV4;11 and OCI-AML2 cell lines after 5 days pre-treatment with VTP-50469 and then treatment with THAL, LEN, POM and CC-220 (iberdomide), for 5 h. Data represent the log-fold change in abundance and log10(p-value). p-value determined by moderated t-test as implemented by the Bioconductor Limma package. i, BH3-profiling in MV4;11 under drug treatments indicated. Cytochrome C (Cyt C) release was measured. Data represent mean+/−SEM (n=3). j, Western blot analysis for BCL2, MCL1 and BCL-XL protein following treatment of OCI-AML2 human MLL-r AML cell line for 72 hours with LEN, EPZ-5676, VTP-50469 and each combination, using GAPDH as a loading control.
Extended Data Fig. 7. MENIN and IKAROS display overlapping functions in regulation of gene expression.
a, Number of differentially expressed genes (greater than 2-fold change and adjusted p-value <0.05) from RNA-seq data for MOLM13 and MV4;11 cell lines treated with CC220 1 μM (n=3), VTP-50469 50 nM (n=3) and the combination (n=3) for 3 days. b, Heatmap showing DESeq2 statistical z-score for all genes deregulated under treatment with VTP-50469 alone across all other treatment groups. c, Heatmap of DESeq2 statistical z-score for MENIN and IKAROS co-regulated gene expression. Showing genes with shared or additive regulation. Selected genes are indicated. d, Cell surface expression of FLT3 as measured by flow cytometry in the MOLM13 cell line. Cells were pre-treated for 2 days with CC220 or DMSO control and then VTP-50469 was added for 24 hours. e, CRISPR screen beta scores from MAGeCK MLE comparing sgRNA representation at day 0 compared to day 14 in the DMSO-treated control samples for the MOLM13 cell line. Negative beta scores for MYC, FLT3, RPA3 (common essential gene), RPS14, and BCL2 are indicated. f, Gene expression for IKZF1 as determined by RNA-seq in MOLM13 and MV4;11, indicating log-2-fold change value and adjusted p-value determined using DESeq2. g, Western blot analysis for IKAROS protein in the MOLM13 cell line following 5 days treatment with either VTP-50469 or iberdomide/CC220, followed by rescue treatment with bortezomib (BTZ) for 6 hours, using GAPDH as a loading control. Performed in the presence of the broad-spectrum caspase inhibitor, QV-D-OPH, to prevent cell death. h, Tornado plots depicting global IKAROS chromatin binding at TSSes, as determined by CUT&RUN in the MOLM13 cell line.
Extended Data Fig. 8. IKAROS protein interactome in MLL-r AML.
a, Binary overlap percentage between all IKAROS peaks enriched 5-fold over background, genome-wide, with each of the other 3 factors, MEIS1 (at 15FE), MENIN (at 5FE) and MLL1 (at 5FE) according ChIP-seq data. b, IGV tracks depicting binding of IKAROS, MENIN, MLL1 and MEIS1, as determined by ChIP-seq, in the region of the TNF gene. Promoters and enhancers, predicted from the ABC tool, are indicated with genomic location indicated (Hg19). c, IGV tracks depicting binding of IKAROS, MENIN, MLL1 and MEIS1 at the MYC and FLT3 genes as determined by ChIP-seq. Promoters and selected enhancers, predicted from the ABC tool, are indicated with genomic location indicated (Hg19). d, Schematic diagrams of protein domains and experimental workflow of BioID system. IKAROS, MEIS1, HOXA10 and ZNF692 proteins were tagged with the biotin ligase BirA*, the left side of the protein schematic diagram denotes N-terminal tagging and the right-side, C-terminal. MV4;11 and MOLM13 cells expressing BioID constructs, 8 total cell lines, were cultured separately. ZF=Zinc finger domain, HBD=homeobox domain, LFQ=label free quantitation, LC-MS/MS=liquid chromatography coupled tandem mass spectrometry. e, Western Blot analysis of the expression of BirA* and HA tagged fusion proteins in MV4;11 and MOLM13 cells, using total Histone H3 as a loading control. f, Venn diagram displaying the number of proteins identified in label free quantitation analysis of the MOLM13 IKAROS, MEIS1 and HOXA10 BioID LC-MS/MS data. All proteins included in Venn diagram have a >2-fold enrichment over the ZNF692 control. g, List of proteins common to IKAROS, MEIS1, HOXA10 BioID identified in LFQ analysis having >2-fold enrichment over the ZNF692 control in the MV4;11 and MOLM13 cell lines.
Extended Data Fig. 9. IKAROS and MENIN co-reside within an isolatable protein complex.
a, IGV ChIP-seq tracks for IKAROS, MENIN, MEIS1 and MLL1 at selected gene TSSes. Tracks are derived from the same ChIP-seq experiments depicted in Fig. 6 and Extended Data Fig. 8. b, Direct protein Co-IP using IKAROS as the bait and probed for MENIN, HOXA9, MEIS1, MTA2 and IKAROS in the MV4;11, OCI-AML2 and MOLM13 cell lines using high salt nuclear extraction and detergent wash. c, Direct protein Co-IP using MLL1 and MENIN as the bait and probed for MENIN and IKAROS, in the MV4;11, OCI-AML2 and MOLM13 cell lines using high salt nuclear extraction and detergent wash. d, Direct protein Co-IP using IKAROS as the bait and probed for MENIN, MLL1, IKAROS and MTA2 in the MV4;11 and THP-1 cell lines using benzonase-treated nuclear extract and high glycerol wash. e, Direct protein Co-IP using MLL1 (rabbit antibody) and MENIN (rabbit antibody) as the bait and probed for MLL1, MENIN and rabbit IKAROS antibody (rIKAROS), in the MV4;11 and THP-1 cell lines using benzonase-treated nuclear extract and high glycerol wash. Rabbit immunoglobulin is seen overlying the IKAROS Western blot bands (indicated in the figure) due to the use of an anti-rabbit secondary antibody. f, Direct protein Co-IP using IKAROS as the bait, with and without prior VTP-50469 treatment (250 nM for 48 hr), and probed for MENIN, MTA2, and IKAROS, in the THP-1 cell line.
Extended Data Fig. 10. Combination therapy with VTP-50469 and LEN results in additive anti-leukemic activity in vivo.
a, Immunohistochemistry for human CD45 antigen on bone marrow sections (sternum) on PDX/CBAM-68552 following 3 weeks treatment with vehicle, LEN 50 mg/kg daily, VTP-50469 0.1% rodent diet and the drug combination. b, Analysis of differentiation in circulating leukaemia cells using CD11b and CD14 expression assessed by flow cytometry in CBAM-68552 after 3 weeks drug treatment. Percentage of double-positive cells is indicated. Representative examples from individual mice are shown. c, Assessment of apoptosis in human CD45-positive cells from bone marrow of drug-treated mice using the CBSK-17D model following 2 weeks drug treatment. The percentage of total viable, human cells are indicated. Representative examples from individual mice are shown. d, Quantitation of apoptotic cells from bone marrow of mice transplanted with the CBSK-17D PDX model following 2 weeks drug treatment. Data represent mean+/−SEM with p-value by unpaired two-tail t-test. e, RNA-seq heatmap from PDX/CBAM-68552 following 3 weeks treatment with vehicle, LEN 50 mg/kg daily, VTP-50469 0.1% rodent diet and the drug combination. Heatmap displaying RNA-Seq DESeq2 statistical z-score for the co-regulated gene network between VTP-50469 and LEN f, RNA-seq heatmap displaying DESeq2 statistical z-score for genes detected in the “granulocyte” pathway as additively deregulated using QIAGEN Ingenuity Pathway Analysis (IPA) analysis. g, Bar code plots using the gene set for HOXA9 regulated genes under each drug treatment from RNA-seq in vivo using the PDX/CBAM-68552 model. Normalized enrichment score and family-wise error rate p-value determined by the GSEA computational method. h, Gene set testing for reported MLL-fusion gene targets for LEN and VTP-50469. Family-wise p-value and normalized enrichment score (NES) determined by the GSEA computational method. i, IPA upstream regulator analysis. Graph displays selected activation z-scores for detected pathways with p-value <0.05. j, Measurement of peripheral blood circulating human CD45 positive cells from PDX mice transplanted with the DFAM-16835 PDX model (NPM1c) after two weeks of drug treatment in vivo. Data represent mean+/−SEM with p-value determined using unpaired, two-tail t-test.
Supplementary Material
Supplementary Table 1. MAGeCK MLE and MAGeCKFLUTE computational pipeline output results. Results are listed for MLE calculations for EPZ-5676, VTP-50469 and a combined analysis of EPZ-5676 and VTP-50469. p-value determined using the MAGeCK MLE algorithm (see Methods for details).
Supplementary Table 2. Global mass spectrometry (MS) proteomics analysis for IMiD neo-substrate profiles in the MOLM13, MV4;11 and OCI-AML2 cell lines, with and without VTP-50469 pre-treatment. p-value determined by moderated t-test as implemented by the Bioconductor Limma package (see Methods for details).
Supplementary Table 3. Next-generation RNA-sequencing in MV;411 and MOLM13 cell lines following treatment of MV;411 and MOLM13 cell lines with lenalidomide 5 μM, CC220 1 μM, VTP-50469 50 nM, and the two combinations for 72 hr. Data is also listed for the MV4;11 cell line treated for 6 days with lenalidomide 5 μM. Data is also listed for the CBAM-68552 patient-derived xenograft model treated in vivo for 3 weeks with lenalidomide 50 mg/kg daily, VTP-50469 0.1% in rodent diet and the combination. Results from the DESEQ2 computational pipeline are listed. P-values for the RNA-seq analysis come were derived using the Bioconductor package DESeq2 (see Methods for details).
Supplementary Table 4. ChIP-seq analysis peak-calling and factor co-occupancy for IKAROS, MENIN, MEIS1 and MLL1 in the MV4;11 and MOLM13 cell lines. The presence of a peak at the indicated genomic locus is indicated by a “Y”. Peaks falling within +/−1kb of a TSS are indicated in the “TSS” column. Peaks overlapping an enhancer, predicted using the ABC model, are indicated in the “enhancer” column.
Supplementary Table 5. Oligonucleotide sequences utilized in the methodology are listed.
Supplementary Table 6. Gene lists used in the presented data analysis are listed.
Supplementary Table 7. Spectral Counting for proteins detected by LC-MS/MS for IKAROS, MEIS1 and HOXA10 BioID systems in MV;411 and MOLM13 cell lines.
Supplementary Table 8. Label free quantitation for proteins detected by LC-MS/MS for IKAROS, MEIS1, HOXA10 and ZNF692 BioID systems in MV;411 and MOLM13 cell lines.
Acknowledgements
S.A.A. is supported by NIH grants CA176745, CA206963, CA204639 and CA066996 and by the Leukaemia and Lymphoma Society, USA (LLS). B.J.A. receives a Career Development Fellowship from the LLS. J.A.C. is supported by Ruth L. Kirschstein Postdoctoral Individual National Research Service Award (NIH F32CA250240-02). W.B. is supported by an NIH T32 training grant (5T32HL007574-38) and the Wong Family Award in Translational Oncology. S.G. is supported by the Sara Elizabeth O’Brien Trust Fellowship. X.S.L. is supported by the Breast Cancer Research Foundation (BCRF-20-100). E.S.F. is supported by NIH grants CA214608, CA231637 and CA066996 and by a Damon Runyon-Rachleff Innovator Award DRR-50-18. S.N.O. is supported by the Damon Runyon Cancer Research Foundation (DRSG: 26-18). F.P. is supported by a grant from the German Research Foundation (PE 3217/1-1). A.C. received funding by German Cancer Aid (Deutsche Krebshilfe). We are grateful for support from the St. Baldrick’s Foundation to the Pediatric LEAP Consortium. We thank all members of the Armstrong lab for invaluable discussions. We thank Jennifer Perry, Yana Pikman, and Andrea Pinilla along with the CPCT for the development and use of human PDX models. We are extremely grateful to Andrea Pinilla, Amy Conway, Amanda Robichaud, Prafulla Gokhale and the Experimental Therapeutics Core Facility at Dana-Farber Cancer Institute for assistance with mouse studies. We thank James Healey, Jayant Gadrey and Sayuri Kitajima for technical support. We thank Elizabeth Frank and Kenneth Ross for statistical analyses. We thank Emma Fink, Andrew Guirguis, Quinlan Sievers, and Vidyasagar Koduri for advice on IKAROS studies and in vivo studies. We thank Martin Filipovski for invaluable assistance with CUT&RUN experiments. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health.
Declaration of Competing Interests
S.A.A. has been a consultant and/or shareholder for Neomorph Inc, Imago Biosciences, Vitae/Allergan Pharma, Cyteir Therapeutics, C4 Therapeutics, OxStem Oncology, Accent Therapeutics, and Mana Therapeutics. S.A.A. has received research support from Janssen, Novartis, Syndax, and AstraZeneca. X.S.L. is a cofounder, board member, SAB, and consultant of GV20 Oncotherapy and its subsidiaries, SAB of 3DMedCare, consultant for Genentech, and stockholder of ABT, AMGN, JNJ, MRK, PFE, and receives sponsored research funding from Takeda and Sanofi. G.M.McG. is a shareholder of Syndax Pharmaceuticals. B.J.A. is a former employee of the Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia, and receives proceeds from Royalties and Milestone payments related to the BCL2-inhibitor, ABT-199/venetoclax. E.S.F. is an equity holder and scientific advisor for Civetta therapeutics, Jengu therapeutics (board of directors), and Neomorph Inc, a shareholder in C4 Therapeutics and a consultant to EcoR1 Capital, Sanofi, Astellas, Deerfield and RA Capital. The Fischer lab receives or has received research funding from Novartis, Astellas, Deerfield, and Ajax. All other authors declare no potential conflicts of interest.
References
- 1.Valk PJ et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 350, 1617–1628, doi: 10.1056/NEJMoa040465 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Bullinger L. et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med 350, 1605–1616, doi: 10.1056/NEJMoa031046 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Krivtsov AV, Hoshii T. & Armstrong SA Mixed-Lineage Leukemia Fusions and Chromatin in Leukemia. Cold Spring Harb Perspect Med 7, doi: 10.1101/cshperspect.a026658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novershtern N. et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144, 296–309, doi: 10.1016/j.cell.2011.01.004 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yokoyama A. et al. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell 123, 207–218, doi: 10.1016/j.cell.2005.09.025 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Yokoyama A. & Cleary ML Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell 14, 36–46, doi: 10.1016/j.ccr.2008.05.003 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okada Y. et al. hDOT1L links histone methylation to leukemogenesis. Cell 121, 167–178, doi: 10.1016/j.cell.2005.02.020 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Bernt KM et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20, 66–78, doi: 10.1016/j.ccr.2011.06.010 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith E, Lin C. & Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev 25, 661–672, doi: 10.1101/gad.2015411 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown FC et al. MEF2C Phosphorylation Is Required for Chemotherapy Resistance in Acute Myeloid Leukemia. Cancer Discov 8, 478–497, doi: 10.1158/2159-8290.CD-17-1271 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krivtsov AV et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442, 818–822, doi: 10.1038/nature04980 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Placke T. et al. Requirement for CDK6 in MLL-rearranged acute myeloid leukemia. Blood 124, 13–23, doi: 10.1182/blood-2014-02-558114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faber J. et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood 113, 2375–2385, doi: 10.1182/blood-2007-09-113597 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo H. et al. PBX3 is essential for leukemia stem cell maintenance in MLL-rearranged leukemia. Int J Cancer 141, 324–335, doi: 10.1002/ijc.30739 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Kumar AR et al. A role for MEIS1 in MLL-fusion gene leukemia. Blood 113, 1756–1758, doi: 10.1182/blood-2008-06-163287 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins C. et al. C/EBPalpha is an essential collaborator in Hoxa9/Meis1-mediated leukemogenesis. Proc Natl Acad Sci U S A 111, 9899–9904, doi: 10.1073/pnas.1402238111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson AC et al. RUNX1 is a key target in t(4;11) leukemias that contributes to gene activation through an AF4-MLL complex interaction. Cell Rep 3, 116–127, doi: 10.1016/j.celrep.2012.12.016 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y. et al. HOXA9 Reprograms the Enhancer Landscape to Promote Leukemogenesis. Cancer Cell 34, 643–658 e645, doi: 10.1016/j.ccell.2018.08.018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cusan M. et al. LSD1 inhibition exerts its antileukemic effect by recommissioning PU.1- and C/EBPalpha-dependent enhancers in AML. Blood 131, 1730–1742, doi: 10.1182/blood-2017-09-807024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu N. et al. MLL-AF9- and HOXA9-mediated acute myeloid leukemia stem cell self-renewal requires JMJD1C. J Clin Invest 126, 997–1011, doi: 10.1172/JCI82978 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeisig BB et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol 24, 617–628, doi: 10.1128/mcb.24.2.617-628.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn MW et al. Targeting Chromatin Regulators Inhibits Leukemogenic Gene Expression in NPM1 Mutant Leukemia. Cancer Discov 6, 1166–1181, doi: 10.1158/2159-8290.CD-16-0237 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bradner JE, Hnisz D. & Young RA Transcriptional Addiction in Cancer. Cell 168, 629–643, doi: 10.1016/j.cell.2016.12.013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krivtsov AV et al. A Menin-MLL Inhibitor Induces Specific Chromatin Changes and Eradicates Disease in Models of MLL-Rearranged Leukemia. Cancer Cell 36, 660–673 e611, doi: 10.1016/j.ccell.2019.11.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borkin D. et al. Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell 27, 589–602, doi: 10.1016/j.ccell.2015.02.016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klossowski S. et al. Menin inhibitor MI-3454 induces remission in MLL1-rearranged and NPM1-mutated models of leukemia. J Clin Invest 130, 981–997, doi: 10.1172/JCI129126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daigle SR et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood 122, 1017–1025, doi: 10.1182/blood-2013-04-497644 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perner F. et al. Novel inhibitors of the histone methyltransferase DOT1L show potent antileukemic activity in patient-derived xenografts. Blood 136, 1983–1988, doi: 10.1182/blood.2020006113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uckelmann HJ et al. Therapeutic targeting of preleukemia cells in a mouse model of NPM1 mutant acute myeloid leukemia. Science 367, 586–590, doi: 10.1126/science.aax5863 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stein EM et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 131, 2661–2669, doi: 10.1182/blood-2017-12-818948 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perner F. & Armstrong SA Targeting Chromatin Complexes in Myeloid Malignancies and Beyond: From Basic Mechanisms to Clinical Innovation. Cells 9, doi: 10.3390/cells9122721 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang B. et al. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat Protoc 14, 756–780, doi: 10.1038/s41596-018-0113-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai AY & Wade PA Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer 11, 588–596, doi: 10.1038/nrc3091 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerami E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404, doi: 10.1158/2159-8290.CD-12-0095 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyers RM et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 49, 1779–1784, doi: 10.1038/ng.3984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunetti L. et al. Mutant NPM1 Maintains the Leukemic State through HOX Expression. Cancer Cell 34, 499–512 e499, doi: 10.1016/j.ccell.2018.08.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kronke J. et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305, doi: 10.1126/science.1244851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu G. et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309, doi: 10.1126/science.1244917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kronke J. et al. Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature 523, 183–188, doi: 10.1038/nature14610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donovan KA et al. Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray syndrome. Elife 7, doi: 10.7554/eLife.38430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guenther MG et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev 22, 3403–3408, doi: 10.1101/gad.1741408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skene PJ & Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, doi: 10.7554/eLife.21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meers MP, Bryson TD, Henikoff JG & Henikoff S. Improved CUT&RUN chromatin profiling tools. Elife 8, doi: 10.7554/eLife.46314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Q, Liu N, Orkin SH & Yuan GC CUT&RUNTools: a flexible pipeline for CUT&RUN processing and footprint analysis. Genome Biol 20, 192, doi: 10.1186/s13059-019-1802-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fulco CP et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat Genet 51, 1664–1669, doi: 10.1038/s41588-019-0538-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryan JA, Brunelle JK & Letai A. Heightened mitochondrial priming is the basis for apoptotic hypersensitivity of CD4+ CD8+ thymocytes. Proc Natl Acad Sci U S A 107, 12895–12900, doi: 10.1073/pnas.0914878107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samavarchi-Tehrani P, Samson R. & Gingras AC Proximity Dependent Biotinylation: Key Enzymes and Adaptation to Proteomics Approaches. Mol Cell Proteomics 19, 757–773, doi: 10.1074/mcp.R120.001941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guirguis AA & Ebert BL Lenalidomide: deciphering mechanisms of action in myeloma, myelodysplastic syndrome and beyond. Curr Opin Cell Biol 37, 61–67, doi: 10.1016/j.ceb.2015.10.004 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Fink EC et al. Crbn (I391V) is sufficient to confer in vivo sensitivity to thalidomide and its derivatives in mice. Blood 132, 1535–1544, doi: 10.1182/blood-2018-05-852798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Francis OL, Payne JL, Su RJ & Payne KJ Regulator of myeloid differentiation and function: The secret life of Ikaros. World J Biol Chem 2, 119–125, doi: 10.4331/wjbc.v2.i6.119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boutboul D. et al. Dominant-negative IKZF1 mutations cause a T, B, and myeloid cell combined immunodeficiency. J Clin Invest 128, 3071–3087, doi: 10.1172/JCI98164 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshida T, Ng SY, Zuniga-Pflucker JC & Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol 7, 382–391, doi: 10.1038/ni1314 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez-Hoyer S. et al. Loss of lenalidomide-induced megakaryocytic differentiation leads to therapy resistance in del(5q) myelodysplastic syndrome. Nat Cell Biol 22, 526–533, doi: 10.1038/s41556-020-0497-9 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol 2, 162–174, doi: 10.1038/nri747 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Bottardi S, Mavoungou L. & Milot E. IKAROS: a multifunctional regulator of the polymerase II transcription cycle. Trends Genet 31, 500–508, doi: 10.1016/j.tig.2015.05.003 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Ding Y. et al. Ikaros tumor suppressor function includes induction of active enhancers and super-enhancers along with pioneering activity. Leukemia 33, 2720–2731, doi: 10.1038/s41375-019-0474-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scacheri PC et al. Genome-wide analysis of menin binding provides insights into MEN1 tumorigenesis. PLoS Genet 2, e51, doi: 10.1371/journal.pgen.0020051 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skucha A. et al. MLL-fusion-driven leukemia requires SETD2 to safeguard genomic integrity. Nat Commun 9, 1983, doi: 10.1038/s41467-018-04329-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hughes CM et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell 13, 587–597, doi: 10.1016/s1097-2765(04)00081-4 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Bres V, Yoshida T, Pickle L. & Jones KA SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation. Mol Cell 36, 75–87, doi: 10.1016/j.molcel.2009.08.015 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Agarwal SK et al. Menin interacts with the AP1 transcription factor JunD and represses JunD-activated transcription. Cell 96, 143–152, doi: 10.1016/s0092-8674(00)80967-8 (1999). [DOI] [PubMed] [Google Scholar]
- 62.Huang J. et al. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature 482, 542–546, doi: 10.1038/nature10806 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu Y. et al. Disruption of the menin-MLL interaction triggers menin protein degradation via ubiquitin-proteasome pathway. Am J Cancer Res 9, 1682–1694 (2019). [PMC free article] [PubMed] [Google Scholar]
- 64.Xie CH et al. Efficacy and safety of lenalidomide for the treatment of acute myeloid leukemia: a systematic review and meta-analysis. Cancer Manag Res 10, 3637–3648, doi: 10.2147/CMAR.S168610 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ebert BL & Kronke J. Inhibition of Casein Kinase 1 Alpha in Acute Myeloid Leukemia. N Engl J Med 379, 1873–1874, doi: 10.1056/NEJMcibr1811318 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Le Roy A. et al. Immunomodulatory Drugs Exert Anti-Leukemia Effects in Acute Myeloid Leukemia by Direct and Immunostimulatory Activities. Front Immunol 9, 977, doi: 10.3389/fimmu.2018.00977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fang J. et al. A calcium- and calpain-dependent pathway determines the response to lenalidomide in myelodysplastic syndromes. Nat Med 22, 727–734, doi: 10.1038/nm.4127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Online-only References
- 68.Kluk MJ et al. Validation and Implementation of a Custom Next-Generation Sequencing Clinical Assay for Hematologic Malignancies. J Mol Diagn 18, 507–515, doi: 10.1016/j.jmoldx.2016.02.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Townsend EC et al. The Public Repository of Xenografts Enables Discovery and Randomized Phase II-like Trials in Mice. Cancer Cell 29, 574–586, doi: 10.1016/j.ccell.2016.03.008 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chi HT et al. Detection of exon 12 type A mutation of NPM1 gene in IMS-M2 cell line. Leuk Res 34, 261–262, doi: 10.1016/j.leukres.2009.09.019 (2010). [DOI] [PubMed] [Google Scholar]
- 71.Fei T. et al. Deciphering essential cistromes using genome-wide CRISPR screens. Proc Natl Acad Sci U S A 116, 25186–25195, doi: 10.1073/pnas.1908155116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15, 554, doi: 10.1186/s13059-014-0554-4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doench JG et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32, 1262–1267, doi: 10.1038/nbt.3026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dignam JD, Lebovitz RM & Roeder RG Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11, 1475–1489, doi: 10.1093/nar/11.5.1475 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu X. et al. MTA2/NuRD Regulates B Cell Development and Cooperates with OCA-B in Controlling the Pre-B to Immature B Cell Transition. Cell Rep 28, 472–485 e475, doi: 10.1016/j.celrep.2019.06.029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McAlister GC et al. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Analytical chemistry 86, 7150–7158, doi: 10.1021/ac502040v (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mellacheruvu D. et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 10, 730–736, doi: 10.1038/nmeth.2557 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Olsen SN et al. MLL::AF9 degradation induces rapid changes in transcriptional elongation and subsequent loss of an active chromatin landscape. Mol Cell, doi: 10.1016/j.molcel.2022.02.013 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mootha VK et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273, doi: 10.1038/ng1180 (2003). [DOI] [PubMed] [Google Scholar]
- 81.Iniguez AB et al. Resistance to Epigenetic-Targeted Therapy Engenders Tumor Cell Vulnerabilities Associated with Enhancer Remodeling. Cancer Cell 34, 922–938 e927, doi: 10.1016/j.ccell.2018.11.005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079, doi: 10.1093/bioinformatics/btp352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinson JT et al. Integrative genomics viewer. Nat Biotechnol 29, 24–26, doi: 10.1038/nbt.1754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anders S, Pyl PT & Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, doi: 10.1093/bioinformatics/btu638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Love MI, Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langmead B. & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359, doi: 10.1038/nmeth.1923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gu Z, Eils R. & Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849, doi: 10.1093/bioinformatics/btw313 (2016). [DOI] [PubMed] [Google Scholar]
- 89.Tange O. Gnu parallel-the command-line power tool. [Google Scholar]
- 90.Gao J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1, doi: 10.1126/scisignal.2004088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.David Wang XQ et al. Three-dimensional regulation of HOXA cluster genes by a cis-element in hematopoietic stem cell and leukemia. bioRxiv, 2020.2004.2016.017533, doi: 10.1101/2020.04.16.017533 (2020). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. MAGeCK MLE and MAGeCKFLUTE computational pipeline output results. Results are listed for MLE calculations for EPZ-5676, VTP-50469 and a combined analysis of EPZ-5676 and VTP-50469. p-value determined using the MAGeCK MLE algorithm (see Methods for details).
Supplementary Table 2. Global mass spectrometry (MS) proteomics analysis for IMiD neo-substrate profiles in the MOLM13, MV4;11 and OCI-AML2 cell lines, with and without VTP-50469 pre-treatment. p-value determined by moderated t-test as implemented by the Bioconductor Limma package (see Methods for details).
Supplementary Table 3. Next-generation RNA-sequencing in MV;411 and MOLM13 cell lines following treatment of MV;411 and MOLM13 cell lines with lenalidomide 5 μM, CC220 1 μM, VTP-50469 50 nM, and the two combinations for 72 hr. Data is also listed for the MV4;11 cell line treated for 6 days with lenalidomide 5 μM. Data is also listed for the CBAM-68552 patient-derived xenograft model treated in vivo for 3 weeks with lenalidomide 50 mg/kg daily, VTP-50469 0.1% in rodent diet and the combination. Results from the DESEQ2 computational pipeline are listed. P-values for the RNA-seq analysis come were derived using the Bioconductor package DESeq2 (see Methods for details).
Supplementary Table 4. ChIP-seq analysis peak-calling and factor co-occupancy for IKAROS, MENIN, MEIS1 and MLL1 in the MV4;11 and MOLM13 cell lines. The presence of a peak at the indicated genomic locus is indicated by a “Y”. Peaks falling within +/−1kb of a TSS are indicated in the “TSS” column. Peaks overlapping an enhancer, predicted using the ABC model, are indicated in the “enhancer” column.
Supplementary Table 5. Oligonucleotide sequences utilized in the methodology are listed.
Supplementary Table 6. Gene lists used in the presented data analysis are listed.
Supplementary Table 7. Spectral Counting for proteins detected by LC-MS/MS for IKAROS, MEIS1 and HOXA10 BioID systems in MV;411 and MOLM13 cell lines.
Supplementary Table 8. Label free quantitation for proteins detected by LC-MS/MS for IKAROS, MEIS1, HOXA10 and ZNF692 BioID systems in MV;411 and MOLM13 cell lines.
Data Availability Statement
Raw and analysed data for ChIP-seq, CUT&RUN-seq, RNA-seq, ATAC-seq and ABC model predictions have been deposited at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under the accession number GSE168463; deposited data correspond to Figures 3, 4, 5, 6, 7 and associated extended data figures. All mass spectrometry data and search results have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD007862) via the PRIDE partner repository with the data set identifiers PXD025227, PXD025228, PXD025229, PXD025230, PXD025231, PXD025232, PXD025271 and PXD025272. Additional data that support the findings of this study are available from the corresponding author upon reasonable request.