Abstract
X-ray fluorescence (XRF) imaging is a highly sensitive non-invasive imaging method for detection of small element quantities in objects, from human-sized scales down to single-cell organelles, using various X-ray beam sizes. Our aim was to investigate the cellular uptake and distribution of Q10, a highly conserved coenzyme with antioxidant and bioenergetic properties. Q10 was labeled with iodine (I2-Q10) and individual primary human skin cells were scanned with nano-focused beams. Distribution of I2-Q10 molecules taken up inside the screened individual skin cells was measured, with a clear correlation between individual Q10 uptake and cell size. Experiments revealed that labeling Q10 with iodine causes no artificial side effects as a result of the labeling procedure itself, and thus is a perfect means of investigating bioavailability and distribution of Q10 in cells. In summary, individual cellular Q10 uptake was demonstrated by XRF, opening the path towards Q10 multi-scale tracking for biodistribution studies.
Keywords: X-ray fluorescence (XRF) imaging, Q10, uptake, single skin cells
1. Introduction
A detailed understanding of the biodistribution of entities such as medical drug compounds, immune cells or antibodies is essential in life sciences. Molecular imaging modalities contribute to a profound understanding of biological processes at the full body, cellular, and even subcellular level. At the same time, imaging technologies can reveal information on the spatial and temporal biodistribution of molecules, and thus, an idea of the biological response to be expected [1]. In order to visualize and quantitatively determine uptake and localization of structural or chemical components into specific organs or cells, a non-invasive method offering both high resolution and sensitivity is advantageous. X-ray fluorescence (XRF) imaging meets all the major requirements for highly scientific evaluations and insights into specific modes of action: high spatial resolution (down to the nanometer scale), high sensitivity (detection of elements down to fg/cell-levels), as well as temporal resolution. The latter is of the highest relevance, because the markers applied do not decay over time, such as in single-photon emission computed tomography (SPECT) imaging or positron emission tomography (PET) [2,3,4,5,6,7,8,9,10,11].
PET/SPECT are inappropriate for single-cell assessments with sub-cellular spatial resolution, and in contrast with the commonly used method of inductively coupled plasma-mass spectrometry (ICP-MS) [12], XRF is non-destructive and thus well suited even for in vivo longitudinal studies [2,3,4,5,6,7,8,9]. Finally, dedicated spatial filtering enables the possibility of overcoming the major challenge of intrinsic multiple Compton scattering when translating XRF from in vitro to large study objects, such as human organs or humans [4,8].
In order to make dedicated entities of interest, e.g., medical drug compounds or biomarkers, detectable via XRF, they need to be labeled with either molecular contrast agents or nanoparticles. The choice of the marker requires profound consideration, and depends on several factors, such as the synthetic processes required for the labeling, object size and incident X-ray energy, which has to be higher than the absorption edge of the used element. Furthermore, one has to ensure that naturally occurring elements in the scanned sample do not show an overlap of signals and, of course, marker elements have to be non-toxic and not interfere with or influence biological processes.
Coenzyme Q10 (Q10), also referred to as ubiquinone, is an essential and highly conserved cellular component mainly present in cellular and mitochondrial membranes, where it acts as a cofactor of mitochondrial respiratory chain complexes supporting cellular bioenergetics in the process of oxidative phosphorylation [13]. In its reduced form (known as ubiquinol), it exerts antioxidant activities. The skin, which is exposed to the external environment like no other organ, is confronted by damaging pro-oxidant stimuli, ultraviolet radiation, and pollution, which are all triggers of cellular oxidative stress and senescence processes. Consequences are skin ageing features such as loss of skin’s firmness and elasticity, wrinkle formation, and susceptibility to inflammatory processes which accelerate the ageing phenotype outcome [14].
With increasing age, Q10 content decreases [15] resulting in significant inhibition of the mitochondrial respiration and leading to increased oxidative stress [16,17,18]. For instance, exogenous Q10 supplementation significantly increased respiration parameters in ex vivo human epidermis and antioxidant capacity in vitro and in vivo, and also led to improved features of aged skin in healthy human subjects [15,17,19,20]. Skin Q10 supplementation strategies have been accepted as anti-ageing approaches [20].
As Q10 is an essential cellular component and is highly conserved across tissues and species, also other organs suffer from an age-, but also disease-dependent decrease in Q10 content. In fact, for conditions associated both with mitochondrial dysfunction and oxidative stress, such as degenerative pathologies and ageing-relevant physiological conditions, a decrease in Q10 content was detected with organ-specific differences [21] both in animal models [22] and in humans [23,24]. Notably, a beneficial impact for Q10 supplementation was shown in evidence-based studies for several of these conditions, such as Parkinson’s disease, Alzheimer’s disease, and heart conditions such as heart failure [25,26,27,28], demonstrating the impact of Q10 in proper function of organisms.
Biodistribution studies using imaging mass spectrometry [29] and PET [30] have been used to reveal the localization of Q10 in the mouse brain as well as the kinetics of commercially available Q10 supplements. In order to further understand Q10 uptake on a cellular level, XRF represents an ideal modality due to its high spatial resolution and the quantitative nature of the modality.
In this study, we reveal two findings:
-
(a)
The feasibility of labeling a relatively small biomolecule, i.e., Q10.
-
(b)
Cellular uptake and distribution of Q10 by means of XRF imaging.
To the best of our knowledge, no detailed quantitative uptake studies exist in the scientific literature. Our new findings obviously can pave the way towards a more detailed understanding of Q10 supplementation in human skin cells and, in addition, provide insights into the mode of action underlying its beneficial effects following topical treatment.
2. Materials and Methods
2.1. Analytical Pre-Assessment of I2-Q10
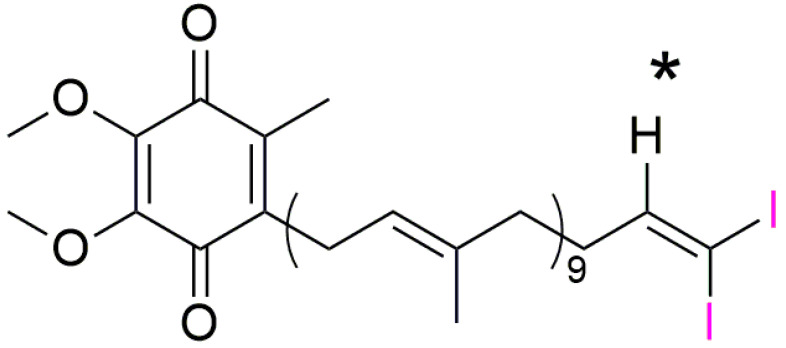
As iodine-labeled Q10 (I2-Q10) has not been described in the literature yet, it had to be newly synthesized in a multistep synthesis starting from standard Q10 (Kaneka Pharma). The identity of the product was confirmed by 1H, 13C and 2D NMR-analysis, as well as mass spectrometry. The chemical shift of the prominent methine group (marked with an asterisk in the molecular structure shown in Figure 1) next to the introduced iodine moiety was found to be 6.90 ppm (1H-NMR) and 153.34 ppm (13C-NMR). This is in very good accordance with similar substructures described in the literature [31]. The position of all other signals, e.g., from the benzoquinone and the isoprene units, remained unchanged. Quantitative NMR assigned the purity to 83% (Ref: Tecnazene, (1,2,4,5 Tetrachlor-3-nitrobenzene)).
Figure 1.
Molecular structure of I2-Q10 showing the chemical shift of the prominent methine group marked with an asterisk next to the introduced iodine-moiety (marked in pink).
The chemical structure and purity of the I2-Q10 was assessed by Nuclear Magnetic Resonance (NMR) measurements, revealing the following analytical data:
1H-NMR (CDCl3, 400 MHz): δ [ppm] = 6.90 (t, 3J = 6.4 Hz, 1H, CH = CI2), 5.23–4.87 (m, 9H, CH2-CH = C), 3.99 (s, 3H, OMe), 3.98 (s, 3H, OMe), 3.18 (d, 2H, 3J = 7.0 Hz, Ar-CH2), 2.52–1.83 (m, 39H, C-CH2-CH2-CH & Ar-CH3), 1.78–1.17 (m, 27H, C = C (CH3)-CH2). 13C-NMR (CDCl3, 100 MHz): δ [ppm] = 153.34 (assigned by 2D) CH = I2, 142.49* CH2-CH = C, 118.98 CH2-CH = C, 61.28 OMe, 39.56*, 26.59*, 25.30 Ar-CH2, 15.90* C = C(CH3)-CH2.
*Broad signal
HRMS (ESI): m/z calc. [C57H84I2O4 + H]+ = 1087.4537, found: 1087.4509 [M + H]+, 1109.4335 [M + Na]+, 1125.4142 [M + K]+.
Instruments: NMR: Bruker Avance I 400 MHz (Bruker, Billerica, MA, USA), HRMS: Agilent 6224 ESI-TOF, coupled Agilent HPLC 1200 Series (Agilent, Santa Clara, CA, USA).
2.2. Cell Culture
Primary human keratinocytes were purchased from Alphenyx (Marseille, France) and processed as described in [32]. Cells cultured at passage 2-5 were used for the experiments and cultured in EpiLife (ThermoFisher Scientific, Waltham, MA, USA) supplemented with human keratinocytes growth supplements (HKGS; ThermoFisher Scientific, Waltham, MA, USA), 1% penicillin streptomycin, and maintained in standard culture conditions at 37 °C and 5% CO2. For cell culture maintenance, the medium was refreshed every 2–3 days and cells were passaged or used for experiments at 80% confluence.
Q10 and I2-Q10 were solubilized in water using a mixture of glycerol and the emulsifying agent PEG-hydrogenated castor oil (EUMULGIN CO 40; BASF, Ludwigshafen, Germany) (Q10/I2-Q10:glycerol:HCO = 0.4:0.6:1) [33].
2.3. Cell Vitality
Briefly, cells were treated with 10, 25 or 50 µM I2-Q10 for 24 h before 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma Cat# M2003) or fluorescein diacetate (FDA; Sigma Cat#F7378) assays were conducted. The MTT assay was carried out according to the manufacturer’s instructions, except the plate was left on a shaker for 5 min instead of incubating overnight. Additionally, the FDA assay was conducted according to the manufacturer’s instructions. Blank values were subtracted and data normalized to untreated medium control.
2.4. Bioavailability
Cellular (I2-) Q10 content was quantified as previously described [34]. Briefly, cell suspensions from keratinocytes treated with 50 µM (I2-) Q10 for 24 h were extracted with isopropanol. Centrifuged isopropanol extracts were analyzed by means of LC-MS/MS (Agilent 1200 HPLC coupled to an Agilent 6490 triple quadrupole mass spectrometer; separation column: YMC-Pack Pro C8 (YMC-Group)) using adjusted multi-reaction transitions and external standard calibration. Due to the low quantity of I2-Q10, its content was measured for six technical replicates of one donor; for native unlabeled standard Q10 n = 5 donors, each n = 1 technical replicate was analyzed. The (I2-) Q10 content was normalized to total protein content; its content mean value is expressed as ng Q10/mg total protein content. For native unlabeled standard Q10, endogenous Q10 content (solvent control) was subtracted.
2.5. Cell Preparation for XRF Measurements
For cell pellet measurements, cells were treated with 50 µM I2-Q10 for 24 h. After treatment, cells were washed with 1× PBS twice, trypsinized and 1× PBS washed again before 400,000–750,000 cells were transferred to glass capillaries (1.0 × 80 × 0.01 mm; WJM-Glas/Müller GmbH, Berlin, Germany). Cells were centrifuged (12,000 rpm, 5 min) and kept on ice until measurement.
For single-cell measurements, silicon nitride membrane windows (Silson Ltd., Warwickshire, UK) were transferred to 6-well plates with their flat (non-etched) side up. Membranes were treated with UV-light for 1 h, before 10 µL collagen (90 µg/mL; collagen, type I solution from rat tail; Sigma-Aldrich, Saint Louis, MO, USA) was added onto the membranes and left in the incubator to dry for 1 h. Residual coating was removed by a pipette before 3,500 cells were pipetted onto each membrane. Cells were left in the incubator for adherence for 3 h before medium was added to the wells. On the next day, treatment was started by adding 50 µM I2-Q10 to the cells. After 24 h treatment incubation, cells were chemically fixated. For this, membranes were washed with 1x D-PBS before 4% paraformaldehyde was added. After 20 min incubation at room temperature, membranes were washed with 1x D-PBS and rinsed in Tris-glucose buffer (261 mM glucose, 9 mM acetic acid, 10 mM Tris buffer, pH 7.4) twice. Membranes were air dried and kept in a desiccator until measurement.
2.6. XRF Cell Uptake and Single Cell Measurements
Experimental Setup
The experiments were performed at the beamlines P21.1 and P06 at the PETRA III synchrotron at DESY, Hamburg, Germany. At the P21.1 beamline, an incident photon energy of 53.1 keV was used in a 1 mm × 1 mm beam of about 1011 photons/s in order to excite iodine K X-ray fluorescence lines which do not suffer from line overlap with other elements. The flux at this beamline is measured with silicon PIN diodes before and after the target area, which allows continuous monitoring.
Cell pellets after different treatments and substances were filled into glass capillaries of 1 mm diameter and 0.01 mm wall thickness and mounted on a custom-built holder to allow scanning of the samples through the X-ray beam at the P21.1 beamline. Silicon drift detectors (X-123FASTSDD and XR100FASTSDD, Amptek Inc., Bedford, MA, USA) of 17/50 mm2 collimated area and 0.5/1.0 mm sensor thickness were used to record the emitted characteristic X-ray fluorescence photons. The choice of 53.1 keV incident energy has the advantage that only a small number of background photons arising from Compton scattering can reach the iodine Kα line energy at around 28.6 keV. In addition, the used 0.5/1.0 mm thick SDD does only absorb a small fraction of Compton scattered photons due to the low efficiency at 45 keV, which in turn minimizes scan times by reducing the total count rate.
For single-cell micro-XRF element mapping, thin silicon nitride membrane windows (Silson Ltd., Warwickshire, England) with I2-Q10-treated cells were mounted on a custom-made holder and positioned normal to the incident beam at the P06 beamline at the PETRA III synchrotron at DESY, Hamburg, Germany [35]. In total, 3 different incident energies were used to determine the optimum parameters in terms of fluorescence cross-section and background contributions. Experiments at 35 and 40 keV incident energy with CRL lenses and CRL pre-focusing were less successful due to their comparatively low flux and high Compton scatter background. Excitation at 12 keV showed much higher sensitivity and higher spatial resolution, since KB mirrors with and without CRL pre-focusing could be used. The smallest beam size of 330 × 240 nm2 with 1.6 × 1010 photons/s was achieved at 12 keV by using KB mirrors without CRL pre-focusing. With pre-focusing, the flux could be increased up to 6 × 1011 photons/s on the expense of a beam size increase to 2.4 × 2.4 μm2. The different beam sizes allow for coarse and fine scans that are needed to determine the cell-to-cell variation in terms of uptake, but also the intracellular distribution. The iodine concentration was high enough to separate iodine L lines from overlapping calcium K lines for a dwell time per scan pixel of 1 s.
A Vortex-EM silicon drift X-ray detector of 50 mm2 active area and 2 mm chip thickness with an Al collimator was positioned at 10 mm distance and 80° angle to the membrane normal. The scanning positions were determined by an online microscope with 2 μm resolution installed at the beamline.
2.7. Data Acquisition and Analysis
2.7.1. Fit Routine and Calibration in Cell Pellet Measurements
XFI is based on the excitation of tracers inside objects by high energy X-rays. The tracers thereby emit element-characteristic X-rays into the full solid angle , which are detected by energy-discriminating detectors. The incident photon beam with area A and intensity gets attenuated by the target material before interaction with the tracer. The probability of an X-ray fluorescence photon being produced is described by the fluorescence cross-section . Emitted fluorescence photons again need to traverse parts of the surrounding target material and a certain distance through air before hitting the detector. Along this path, they are attenuated due to scattering or absorption processes; hence, only a fraction of the photons with energy reach the detector chip. Therefore, the detected number of fluorescence photons S needs to be corrected for those effects.
Even if multiple detectors are used, only a fraction of the full solid angle ,is covered, which needs to be addressed by the correction factor , as the fluorescence photons are emitted isotropically. The detector chip itself has limited thickness and therefore lacks efficiency at high X-ray energies. Consequently, the number of detected photons further needs to be corrected by . In addition, potential detector deadtime needs to be addressed in case the detector is exposed to a high photon flux. In summary, the tracer mass in the irradiated area can be calculated with:
| (1) |
The fluorescence photon number S is determined by fitting with the curve_fit method from
SciPy in python. The total fit consists of a signal function, a sum of all Gaussian peaks of any elements assumed to be present in the probe, and a background function. The data are fitted in a range of interest around the peaks. Element-specific fluorescence data such as relative line intensity and mean line energy is obtained from the xraylib database [36].
Single line intensities are corrected for energy-dependent detector efficiency and transmission through air or other absorbing material, such that one global free parameter per element is to be fitted: the corrected element amplitude. The mostly flat background is modeled with a linear function and an additional exponential term.
A single-sided hypothesis test is performed to assure that the assumed signal is a valid signal and not only a background fluctuation; hence, the significance is calculated. If the significance exceeds 3 standard deviations, the peak is recognized as a signal. This analysis is performed in an interval of ±3 around the mean energy of the peaks. The signal counts S are then calculated by integration of the fitted Gaussian from -infinity to infinity.
2.7.2. Fit Routine and Calibration in Single-Cell Measurements
For the quantification of elements, X-ray spectra were peak fitted using a program package based on PyMCA, in order to extract the net intensities of fluorescence lines. The net peak intensities were normalized to the intensity of the incoming monochromatic beam. A multi-element standard reference material (RF-200-0205-C10-X, AXO Dresden GmbH, Dresden, Germany) was employed for external standardization. Element areal densities were calculated from the normalized net peak intensities via comparison with the reference.
Quantitative analysis to determine the cell size and mean intensity values in the elemental maps was carried out with ImageJ [37]. Cell-to-cell uptake variation was determined by fitting a linear function to the reconstructed mass per cell vs. projected cell area and calculating the rms value of the fit-subtracted masses.
3. Results
3.1. Biological Pre-Assessment of I2-Q10
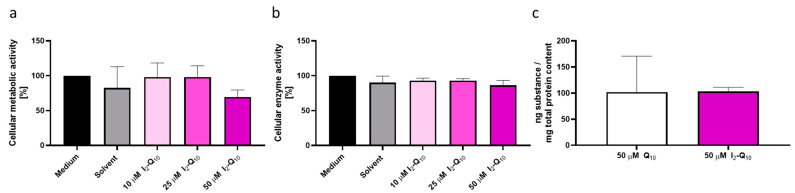
Synthesized and labeled I2-Q10 was qualified and compared to native, non-labeled Q10 by testing both human skin cell viability and bioavailability. For this purpose, primary human keratinocytes obtained from three different donors were treated with 10–50 µM I2-Q10 for 24 h, before cellular metabolic activity (Figure 2a) and cellular enzyme activity were measured (Figure 2b) to ensure non-toxicity of the cells. As standard cell culture Q10 concentrations of 50 µM [38] show no negative side effects on cells, a similar concentration can also be used for I2-Q10. Therefore, this concentration was chosen for further analyses, including bioavailability assessment via liquid chromatography—tandem mass spectrometry (LC-MS/MS). Our results revealed a cellular uptake for I2-Q10 (Figure 2c) which was comparable to native unlabeled standard Q10 (103 ng I2-Q10/mg total protein content vs. 101 ng Q10/mg total protein content).
Figure 2.
I2-Q10 pre-assessment. Both (a) cellular metabolic activity (MTT assay) and (b) cellular enzyme activity (FDA assay) were measured to assess cell vitality at different I2-Q10 concentrations (n = 3, mean ± SD). Bioavailability comparison for (c) Q10 (n = 5 donors, each n = 1 technical replicate) and) I2-Q10 (n = 1 donor, n = 6 technical replicates); 50 µM treatment, LC-MS/MS, mean ± SD.
3.2. Quantitative I2-Q10 Uptake Measurements of Human Skin Cell Pellets
On basis of these positive analytical and biological pre-assessment results, I2-Q10 uptake was further evaluated by X-ray fluorescence imaging. Cell pellets from keratinocytes obtained from three donors, treated with I2-Q10 and centrifuged, were filled into glass capillaries and scanned at the P21.1 beamline at the PETRA III synchrotron at DESY, Hamburg, Germany, with an incident energy of 53.1 keV. The goal of these first measurements was the determination of cellular uptake variations between each of the single treated donors, as well as a comparison with untreated skin cells. A summary of the main scan and sample parameters, as well as the resulting iodine mass per cell, is shown in Table 1.
Table 1.
Summary of the main scan and sample parameters for glass capillary measurements containing keratinocyte cell pellets after different treatment at the P21.1 beamline.
| Cell Pellet | Cell Number | Scan Duration [s] | Photons per Pixel | Resolution [mm] | Iodine/Cell [fg] |
|---|---|---|---|---|---|
| Donor 1, 50 μM I2-Q10 | 600,000 | 10 | 1.7 × 1012 | 0.5 | 44.5 |
| Donor 2, 50 μM I2-Q10 | 600,000 | 10 | 1.7 × 1012 | 0.5 | 48 |
| Donor 3, 50 μM I2-Q10 | 400,000 | 10 | 1.7 × 1012 | 0.5 | 68.5 |
| Donor 1, no treatment | 750,000 | 10 | 1.7 × 1012 | 0.5 | 0 |
| Donor 1, 50 μM Iodine | 1,000,000 | 10 | 7.5 × 1011 | 1 | 9.1 |
| Donor 2, 50 μM Iodine | 1,000,000 | 10 | 7.5 × 1011 | 1 | 9.6 |
| Donor 2, 8.5 μM Iodine | 1,000,000 | 10 | 7.5 × 1011 | 1 | 0 |
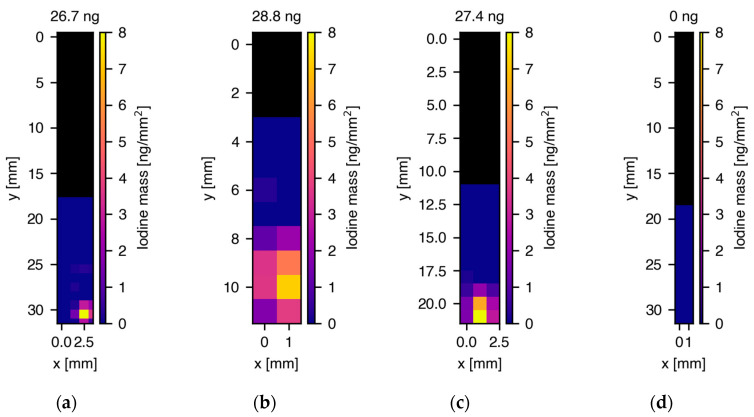
For each capillary, the total iodine mass as well as the iodine distribution in the capillary was reconstructed, as described by Schmutzler and colleagues [2] and in the Methods section. Pixel-by-pixel full spectral analysis consisting of fits to the characteristic iodine K shell fluorescence lines, as well as X-ray fluorescence lines from the detector (Sn, Ba) and background contributions were applied to generate elemental concentration maps. Figure 3a–c shows the two-dimensional iodine distribution maps for the three different donors and d) depicts the map for the untreated skin cell sample from donor 1. The total reconstructed iodine masses in the capillaries were 26.7 ± 4.0 ng, 28.8 ± 4.3 ng and 27.4 ± 4.1 ng for donor 1 (600,000 cells), 2 (600,000 cells) and 3 (400,000 cells), respectively, whereas no iodine could be detected in the untreated cell sample (750,000 cells). In the capillaries from all three donors, iodine was strongly localized at the bottom part of the capillary where the pellet with a height of about 1.5 mm was located. The approx. 9 mm high PBS fluid column above the pellet did not show iodine signals, which was a strong indicator that the cells were still intact during the measurements, without any sign of iodine leakage. Dividing the reconstructed iodine masses by the number of cells per pellet resulted in 44.5, 48 and 68.5 fg iodine/cell for donors 1-3, respectively, in ascending order. In XRF, only control groups containing iodine will yield signals. Since the natural iodine concentration in skin cells was below the detection limit of our XRF experiment, as we could not measure any signals in the untreated skin sample from donor 1 and measurements of the used solutions did not show any iodine impurities, no further control groups were needed.
Figure 3.
Two-dimensional maps of the spatial distribution and reconstructed masses of iodine in glass capillaries containing cell pellets treated with 50 M I2-Q10. (a) 26.7 ng total iodine mass in cell pellet from donor 1, (b) 28.8 ng total iodine mass in cell pellet from donor 2, (c) 27.4 ng total iodine mass in cell pellet from donor 3, (d) no iodine signal in untreated cell sample. The maps in (a–c) show a strong localization of iodine in the bottom part of the capillaries where the cell pellet was located in a PBS fluid column. Above the fluid column, the capillaries were empty, indicated by the black color in the XRF maps. Note the different y-axis scales arising from the varying length and thickness of the hand-blown capillaries.
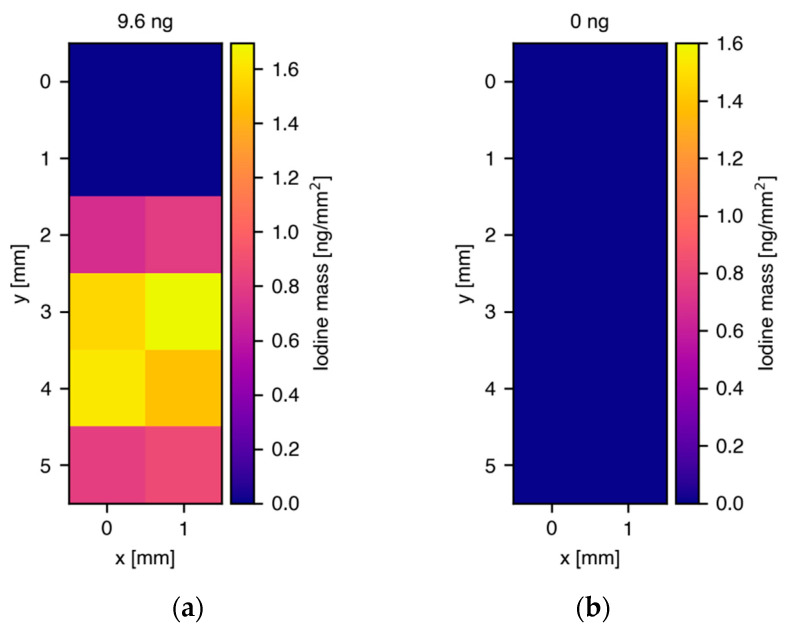
In order to address the question of iodine leakage during the incubation process, skin cells obtained from the same donors were treated with iodine only, without being labeled to Q10. Two different concentrations were chosen for the treatment, 50 M and 8.5 M, of which the first one equals the concentration that was used for the I2-Q10 treatment and the second one takes the I2-Q10 NMR purity of 83% into account, meaning, in the worst case, a maximum of 17% pure iodine residue (17% of 50 M = 8.5 M). Figure 4 shows the reconstructed iodine masses for one cell sample treated with 50 M (a) and one treated with 8.5 M (b), whereby 1 million cells were contained within each pellet. The treatment with 50 M iodine resulted in a total reconstructed mass of 9.6 1.4 ng which equals 9.6 fg/cell, roughly a factor of five lower than for I2-Q10 treatment, while in the case of 8.5 M, no iodine signals were found. This finding is a strong indicator that the uptake is mainly driven by Q10-specific and Q10-related processes of the cells, and that iodine can be regarded as a well-suited labeling element.
Figure 4.
Two-dimensional maps of the spatial distribution and reconstructed masses of iodine in glass capillaries containing cell pellets treated with iodine of different concentrations. (a) 9.6 ng total iodine mass in cell pellet from donor 2 after treatment with 50 M iodine, (b) no iodine signal in cell pellet from donor 2 after treatment with 8.5 M iodine. Compared with the results for I2-Q10 treatment, the localization was not as strong, indicating a rather low free iodine uptake and/or background signal from iodine attached to cell surfaces that distributes in the cell-surrounding PBS. Note that in this measurement, a smaller region around the cell pellet was scanned, which was different to results presented in Figure 3, where the scan areas were larger.
3.3. Single Cell Measurements to Determine I2-Q10 Uptake and Distribution
Having demonstrated the I2-Q10 accumulation within cell pellets using glass capillaries, a follow-up set of measurements was performed on the single-cell level. As the spatial resolution in XRF is determined solely by the diameter of the X-ray beam applied, a different beamline was chosen for those studies: the Hard X-ray Micro/Nano-Probe Beamline P06 at PETRA III, DESY, Hamburg, Germany. This beamline offers beam diameters in the sub-m range, a high photon flux, as well as different incident energies up to 40 keV. A summary of the main parameters used for the measurements is given in Table 2.
Table 2.
Summary of the main beam parameters used for the measurements at the P06 beamline at PETRA III, DESY, Hamburg. The number sets refer to two different X-ray beam conditions (with and without pre-focusing compound refracting lenses) for the measurements shown in Figure 5 and Figure 6.
| P06, DESY, Hamburg | ||
|---|---|---|
| Incident energy [keV] | 12 | |
| High-resolution condition | High-flux condition | |
| Photon flux [photons/sec] | 1.6 × 1010 | 6 × 1011 |
| Beam size [μm2] | 0.33 × 0.24 | 2.40 × 2.40 |
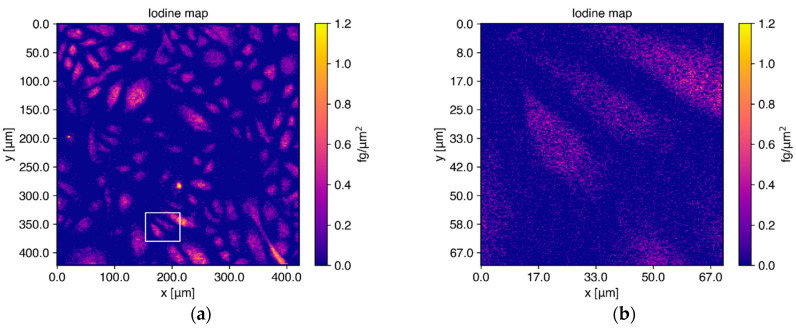
Human keratinocytes from the same donors as those in the previous experiments were chemically fixated on silicon nitride membrane windows (Silson Ltd., Warwickshire, England) after a 24 h incubation with 50 M I2-Q10. Interesting scan areas were determined via high-resolution microscope images of the membranes mounted on a custom-made holder. At the P06 beamline, a membrane with keratinocytes from donor 1 was scanned using a beam with an incident energy of 12 keV, which was chosen due to the optimized background in the iodine L-shell fluorescence region and the high flux achieved in the small beam spot. Figure 5a shows the reconstructed elemental image for iodine resulting from full spectral analysis of all individual pixels in a scan area of 420 × 420 m2 scanned with a resolution of 2 × 2 m2. Panel B) shows the elemental map for three selected cells from the full area shown in A) in an area of 70 × 70 m2, scanned in a subsequent run with higher resolution of 300 × 300 nm2.
Figure 5.
Elemental maps for iodine reconstructed from single-cell measurements after I2-Q10 treatment. (a) Cells from donor 1 on a silicon nitride membrane were scanned with a 12 keV X-ray beam over an area of 420 × 420 m2. Iodine (as labeled to Q10) is distributed homogeneously inside the cells. (b) Elemental maps of 3 cells from the same membrane scanned again with higher resolution, showing the distributions found in (a) in more detail. The scan area is indicated with a white rectangle in the elemental map shown in (a).
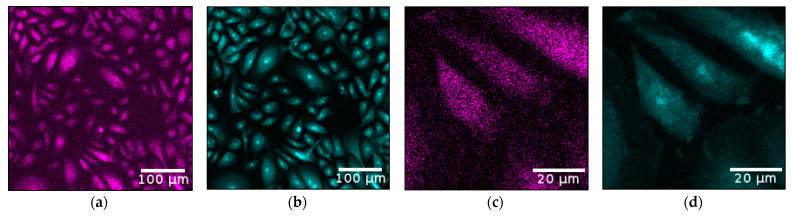
Iodine is distributed homogenously inside the cells, where Q10 is mainly expected, since highest Q10 levels were described for the cell organelles mitochondria, lysosomes and Golgi vesicles [39]. Quantitative analysis of the absolute elemental iodine mass per cell for cells with a size between 100 and 350 m2 yielded a mean value of 46.5 ± 10.70 fg/cell, which is in good agreement with the results from the cell pellet measurements shown in Figure 3. Over the full range of all 100 cells, a mean value of 77.03 ± 21.48 fg iodine/cell (≙ 2.6 × 10−4 ± 0.73 ng Q10/cell) was calculated, with a median value of 49.13 fg iodine/cell (≙ 1.66 × 10−4 ng Q10/cell). In order to emphasize the homogeneous distribution of iodine and hence Q10, the elemental maps of iodine and zinc are compared in Figure 6. The zinc distribution shown in cyan depicted a strong localization in the cell nucleus, different to the homogeneous iodine map colored in magenta. It has to be noted here that the feature of cell nucleus localization is automatically provided in XRF imaging as long as the incident energy is above the K-absorption edge of those elements which are also present in the cell nucleus. Thus, no additional staining is needed, which often alters the kinetics of uptake and distribution and subsequently influences the applicability of final probes [40]. The comparison of the elemental images of zinc and iodine (representing Q10) for the fine scan shown in Figure 6c,d once more demonstrates the homogeneous distribution of iodine among the whole cell area, where Q10 is primarily expected. The concentration map of zinc can be used to define the outline of cell nuclei and to determine the influence of sample preparation on elemental content in mammalian cells, as discussed in [41].
Figure 6.
Images of the zinc (cyan) and iodine (magenta) elemental maps for the full scan (a,b) and the fine scan of 3 selected cells (c,d), indicated by the white rectangle in Figure 5a. As zinc is mainly located in the cell nucleus, its elemental map is ideally suited to indicate the locations of the nuclei. The direct comparison of iodine and zinc once again emphasizes the homogeneous distribution of iodine among the cytoplasm and agrees with the expected Q10 localization.
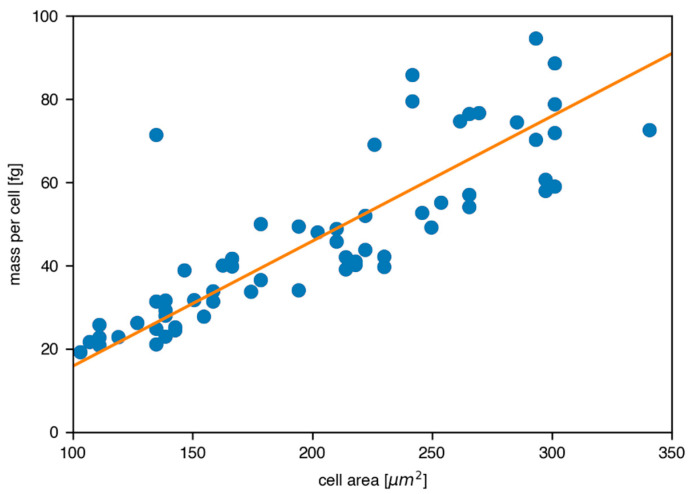
In order to determine the cell-to-cell uptake variation, the total mass of XRF-marker per cell was reconstructed for all the 100 measured cells and plotted against the projected cell area in the range of 100 to 350 m2, containing the majority of data points. The linear fit function in Figure 7 represents a clear correlation of increasing mass per cell, with the projected cell area determined by ImageJ [37].
Figure 7.
Correlation between reconstructed iodine mass per cell and projected cell area. The orange fit function shows a clear trend of uptake increase with area, and the cell-to-cell uptake variation is 10.70 fg iodine/cell, while the mean value was found to read 46.5 fg iodine/cell.
4. Discussion
The results presented in our work clearly demonstrate the feasibility of XRF imaging to quantitatively determine the uptake and subcellular distribution of iodine-labeled Q10. One basic prerequisite of our imaging modality is the development of a labeling strategy, resulting in cellular stability of an element for visualization by XRF imaging. Iodine was found to fulfill the necessary requirements for several reasons: iodine can be bound to Q10 without any observed modification of the coenzyme, and obviously there is no detectable natural iodine background in skin cells (Figure 3); thus, there are no overlapping signals with I2-Q10 treatment. Our reconstructed iodine mass per cell is in good agreement with the expectation value from standard Q10 uptake measurements (Figure 2). Control measurements using free iodine, experiments of I2-Q10 bioavailability via LC-MS/MS as well as the scans around the cell pellets clearly indicate that the measured iodine masses can only be related to the I2-Q10. In addition, there is neither an indication that the iodine marker was disassembled from the Q10 molecule during incubation, nor any noticeable leakage out of the cells.
Our data prove, for the first time, that exogenously applied Q10 is taken up by primary human skin cells. In addition, we were able to quantify the Q10 uptake and visualize spatial distribution per individual skin cell. The large accumulation of Q10, as measured by iodine-labeled Q10 signals, within skin cells is in good agreement with previous work, demonstrating that Q10 supplementation leads to intracellular effects. In this regard, Prahl et al. [20], have shown ex vivo data in mitochondrial membrane potential stabilization (as a parameter of energetic capacity) in isolated keratinocytes following topical Q10 application. Additionally, Schniertshauer et al. [17] demonstrated an increase in mitochondrial respiration, and thus ATP production, after ex vivo treatment of human epidermis samples. Furthermore, inhibition of endogenous Q10 biosynthesis in skin cells resulted in a shift towards a senescent phenotype in vitro, demonstrating that Q10 depletion is not simply a consequence of skin ageing, but also the main cause of it [42]. Experimentally induced senescence phenotypes in these approaches were reverted by Q10 supplementation, replenishing cellular and mitochondrial Q10 content. In addition, molecular markers of senescence (β-galactosidase activity, p21 mRNA and protein expression) were rescued, and a significant upregulation of the gene expression of the structural components of the extracellular matrix, namely elastin and collagen type I, alpha I [33] was observed. Application of Q10 to aged skin in vivo has been shown to improve the phenotypic signs of ageing by acting both as a restorer of mitochondrial function and as an antioxidant [15,17,19,20,43].
XRF provides new insights into the mode of action of Q10 and the beneficial effects of Q10 supplementation reported in the scientific literature. According to our findings reported here, the underlying mechanism of the above-mentioned beneficial effects for human skin and skin cells are mainly facilitated by uptake and insertion of exogenous Q10 to mitochondrial and cellular membranes, confirming the pivotal role of Q10 in regulation and restoration of cutaneous metabolism.
It is noteworthy that iodine can also be used as a labeling element for studies using bigger cellular compositions in three-dimensional contexts, such as skin models. K-shell XRF photons have sufficient energy to penetrate through tissues; thus, using higher-incident X-ray energies with the same marker as demonstrated in this work might be applicable for skin penetration studies as well.
Additionally, the cutting-edge technology of XRF may offer great potential for medical applications as well, reaching from new scientific findings to reducing drug development process efforts, risks and costs. The overall efficiency of the process could be improved by providing a selection of promising candidates and offering insights into the mode of action early on [44]. Thus, exploiting functional and molecular imaging modalities may serve to address and determine pharmacokinetics, drug metabolism, safety aspects and prediction of in vivo efficacy already in the early stages of (pre)clinical studies.
Putative translation from the single-cell level to bigger cellular arrangements such as organ cultures and/or even humans in the future poses challenges for an advanced in vivo XRF imaging approach, as discussed in our previous works [8,9].
Acknowledgments
The authors wish to express their thanks for the highly professional and valuable support of axiom insights GmbH, which enabled this research project. We further acknowledge DESY (Hamburg, Germany), a member of the Helmholtz Association HGF, for the provision of experimental facilities. Parts of this research were carried out at PETRA III, and we would like to thank the beamline teams for assistance in using the P21.1 and P06 beamline.
Author Contributions
Conceptualization, J.M.W., T.B. (Thomas Blatt), F.G. and G.F., formal analysis, T.S., O.S., O.D., G.F., D.B., M.L.S., T.B. (Thorsten Burkhardt), J.-P.V., A.V. and V.W.; investigation, T.S., M.L.S., O.S., C.K., V.W., A.V., T.B. (Thorsten Burkhardt), J.-P.V., O.D., G.F. and D.B.; writing—original draft preparation, T.S. and M.L.S.; resources, C.S.-C.; writing—review and editing, J.M.W., T.B., G.F. and F.G.; supervision, J.M.W., T.B. and F.G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by MCIN/AEI/10.13039/501100011033 from grant PID2020-118176RJ-I100. This work was performed under the Maria de Maeztu and Severo Ochoa Centres of Excellence Program run by the Spanish State Research Agency, Grant No. CEX2018-000867-S.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Willmann J.K., van Bruggen N., Dinkelborg L.M., Gambhir S.S. Molecular imaging in drug development. Nat. Rev. Drug Discov. 2008;7:591–607. doi: 10.1038/nrd2290. [DOI] [PubMed] [Google Scholar]
- 2.Schmutzler O., Graf S., Behm N., Mansour W.Y., Blumendorf F., Staufer T., Körnig C., Salah D., Kang Y., Peters J.N., et al. X-ray Fluorescence Uptake Measurement of Functionalized Gold Nanoparticles in Tumor Cell Microsamples. Int. J. Mol. Sci. 2021;22:3691. doi: 10.3390/ijms22073691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahl H., Staufer T., Körnig C., Schmutzler O., Rothkamm K., Grüner F. Feasibility of Monitoring Tumor Response by Tracking Nanoparticle-Labelled T Cells Using X-ray Fluorescence Imaging—A Numerical Study. Int. J. Mol. Sci. 2021;22:8736. doi: 10.3390/ijms22168736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ungerer A., Staufer T., Schmutzler O., Körnig C., Rothkamm K., Grüner F. X-ray Fluorescence Imaging for In Vivo Detection of Gold-Nanoparticle-Labeled Immune Cells: A GEANT4 Based Feasiblity Study. Cancers. 2021;13:5759. doi: 10.3390/cancers13225759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osterode W., Falkenberg G., Wrba F. Copper and Trace Elements in Gallbladder from Patients with Wilson’s Disease Imaged and Determined by Synchrotron X-ray Fluorescence. J. Imaging. 2021;7:261. doi: 10.3390/jimaging7120261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pushie M.J., Pickering I.J., Korbas M., Hackett M.J., George G.N. Elemental and Chemically Specific X-ray Fluorescence Imaging of Biological Systems. Chem. Rev. 2014;114:8499–8541. doi: 10.1021/cr4007297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez-Cano C., Alvarez-Puebla R.A., Abendroth J.M., Beck T., Blick R., Cao Y., Caruso F., Chakraborty I., Chapman H.N., Chen C., et al. X-ray Based Techniques to Study the Nano-Bio Interface. ACS Nano. 2021;15:3754–3807. doi: 10.1021/acsnano.0c09563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grüner F., Blumendorf F., Schmutzler O., Staufer T., Bradbury M., Wiesner U., Rosentreter T., Loers G., Lutz D., Richter B., et al. Localising functionalised gold-nanoparticles in murine spinal cords by X-ray fluorescence imaging and background-reduction through spatial filtering for human-sized objects. Sci. Rep. 2018;8:16561. doi: 10.1038/s41598-018-34925-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Körnig C., Staufer T., Schmutzler O., Bedke T., Machicote A., Liu B., Liu Y., Gargioni E., Feliu N., Parak W.J., et al. In-situ X-Ray Fluorescence Imaging of the Endogenous Iodine Distribution in Murine Thyroids. Sci. Rep. 2022;12:2903. doi: 10.1038/s41598-022-06786-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naganawa M., Gallezot J.-D., Rossano S., Carson R.E. Quantitative PET Imaging in Drug Development: Estimation of Target Occupancy. Bull. Math. Biol. 2019;81:3508–3541. doi: 10.1007/s11538-017-0374-2. [DOI] [PubMed] [Google Scholar]
- 11.Gomes C.M., Abrunhosa A.J., Ramos P., Pauwels E.K.J. Molecular imaging with SPECT as a tool for drug development. Adv. Drug Deliv. Rev. 2011;63:547–554. doi: 10.1016/j.addr.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Wilfschefski S.C., Baxter M.R. Inductively Coupled Plasma Mass Spectrometry: Introduction to Analytical Aspects. Clin. Biochem. Rev. 2019;40:115–133. doi: 10.33176/AACB-19-00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Littarru G.P., Tiano L. Clinical aspects of coenzyme Q10: An update. Curr. Opin. Clin. Nutr. Metab. Care. 2005;8:641–646. doi: 10.1097/01.mco.0000171123.60665.16. [DOI] [PubMed] [Google Scholar]
- 14.Rittié L., Fisher G.J. UV-light-induced signal cascades and skin aging. Ageing Res. Rev. 2002;1:705–720. doi: 10.1016/S1568-1637(02)00024-7. [DOI] [PubMed] [Google Scholar]
- 15.Knott A., Achterberg V., Smuda C., Mielke H., Sperling G., Dunckelmann K., Vogelsang A., Krüger A., Schwengler H., Behtash M., et al. Topical treatment with coenzyme Q10-containing formulas improves skin’s Q10 level and provides antioxidative effects. Biofactors. 2015;41:383–390. doi: 10.1002/biof.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montero R., Sánchez-Alcázar J.A., Briones P., Rodríguez Hernández A., Cordero M.D., Trevisson E., Salviati L., Pineda M., García-Cazorla A., Navas P., et al. Analysis of coenzyme Q10 in muscle and fibroblasts for the diagnosis of CoQ10 deficiency syndromes. Clin. Biochem. 2008;41:697–700. doi: 10.1016/j.clinbiochem.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Schniertshauer D., Gebhard D., Bergemann J. Age-Dependent Loss of Mitochondrial Function in Epithelial Tissue Can Be Reversed by Coenzyme Q10. J. Aging Res. 2018;2018:6354680. doi: 10.1155/2018/6354680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quinzii C.M., Hirano M. Coenzyme Q and Mitochondrial Disease. Dev. Disabil. Res. Rev. 2010;16:183–188. doi: 10.1002/ddrr.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sreedhar A., Aguilera-Aguirre L., Singh K.K. Mitochondria in skin health, aging, and disease. Cell Death Dis. 2020;11:444. doi: 10.1038/s41419-020-2649-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prahl S., Kueper T., Biernoth T., Wöhrmann Y., Münster A., Fürstenau M., Schmidt M., Schulze C., Wittern K.-P., Wenck H., et al. Aging skin is functionally anaerobic: Importance of coenzyme Q10 for anti aging skin care. Biofactors. 2008;32:245–255. doi: 10.1002/biof.5520320129. [DOI] [PubMed] [Google Scholar]
- 21.Kalén A., Appelkvist E.L., Dallner G. Age-related changes in the lipid compositions of rat and human tissues. Lipids. 1989;24:579–584. doi: 10.1007/BF02535072. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi K., Ohsawa I., Shirasawa T., Takahashi M. Early-onset motor impairment and increased accumulation of phosphorylated α-synuclein in the motor cortex of normal aging mice are ameliorated by coenzyme Q. Exp. Gerontol. 2016;81:65–75. doi: 10.1016/j.exger.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 23.Nagase M., Yamamoto Y., Matsumoto N., Arai Y., Hirose N. Increased oxidative stress and coenzyme Q10 deficiency in centenarians. J. Clin. Biochem. Nutr. 2018;63:129–136. doi: 10.3164/jcbn.17.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niklowitz P., Onur S., Fischer A., Laudes M., Palussen M., Menke T., Döring F. Coenzyme Q10 serum concentration and redox status in European adults: Influence of age, sex, and lipoprotein concentration. J. Clin. Biochem. Nutr. 2016;58:240–245. doi: 10.3164/jcbn.15-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaki N.M. Strategies for oral delivery and mitochondrial targeting of CoQ10. Drug Deliv. 2016;23:1868–1881. doi: 10.3109/10717544.2014.993747. [DOI] [PubMed] [Google Scholar]
- 26.Ebadi M., Govitrapong P., Sharma S., Muralikrishnan D., Shavali S., Pellett L., Schafer R., Albano C., Eken J. Ubiquinone (coenzyme q10) and mitochondria in oxidative stress of Parkinson’s disease. Biol. Signals Recept. 2001;10:224–253. doi: 10.1159/000046889. [DOI] [PubMed] [Google Scholar]
- 27.Wadsworth T.L. Evaluation of coenzyme Q as an antioxidant strategy for Alzheimer’s disease. J. Alzheimer’s Dis. 2008;14:225–234. doi: 10.3233/JAD-2008-14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Lorenzo A., Iannuzzo G., Parlato A., Cuomo G., Testa C., Coppola M., D‘Ambrosio G., Alessandro Oliviero D., Sarullo S., Vitale G., et al. Clinical Evidence for Q10 Coenzyme Supplementation in Heart Failure: From Energetics to Functional Improvemen. J. Clin. Med. 2020;9:1266. doi: 10.3390/jcm9051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatsuta Y., Kasai K., Maruyama C., Hamano Y., Matsuo K., Katano H., Taira S. Imaging mass spectrometry analysis of ubiquinol localization in the mouse brain following short-term administration. Sci. Rep. 2017;7:12990. doi: 10.1038/s41598-017-13257-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe K., Nozaki S., Goto M., Kaneko K.-I., Hayashinaka E., Irie S., Nishiyama A., Kasai K., Fujii K., Wada Y., et al. PET imaging of 11C-labeled coenzyme Q10: Comparison of biodistribution between [11C]ubiquinol-10 and [11C]ubiquinone-10. Biochem. Biophys. Res. Commun. 2019;7:611–615. doi: 10.1016/j.bbrc.2019.03.073. [DOI] [PubMed] [Google Scholar]
- 31.Dabdoub M.J., Dabdoub V.B., Baroni A.C.M. Hydrozirconation of Stannylacetylenes: A Novel and Highly Efficient Synthesis of 1,1-Diiodo-, 1,1-Dibromo-, and Mixed (Z)- or (E)-1-Iodo-1-bromo-1-alkenes Using 1,1-Hetero-Bimetallic Reagents. J. Am. Chem. Soc. 2001;123:9694–9695. doi: 10.1021/ja0114660. [DOI] [PubMed] [Google Scholar]
- 32.Roggenkamp D., Falkner S., Stäb F., Petersen M., Schmelz M., Neufang G. Atopic keratinocytes induce increased neurite outgrowth in a coculture model of porcine dorsal root ganglia neurons and human skin cells. J. Invest. Dermatol. 2012;132:1892–1900. doi: 10.1038/jid.2012.44. [DOI] [PubMed] [Google Scholar]
- 33.Marcheggiani F., Kordes S., Cirilli I., Orlando P., Silvestri S., Vogelsang A., Möller N., Blatt T., Weise J.M., Damiani E., et al. Anti-aging effects of ubiquinone and ubiquinol in a senescence model of human dermal fibroblasts. Free Radic. Biol. Med. 2021;165:282–288. doi: 10.1016/j.freeradbiomed.2021.01.032. [DOI] [PubMed] [Google Scholar]
- 34.Silvestri S., Orlando P., Armeni T., Padella L., Brugé F., Seddaiu G., Littaru G.P., Tiano L. Coenzyme Q10 and α-lipoic acid: Antioxidant and pro-oxidant effects in plasma and peripheral blood lymphocytes of supplemented subjects. J. Clin. Biochem. Nutr. 2015;57:21–26. doi: 10.3164/jcbn.14-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falkenberg G., Fleissner G., Fleissner G., Alraun P., Boesenberg U., Spiers K. Large scale high-resolution micro-XRF analysis of histological structures in the skin of the pigeon beak. X-Ray Spectrom. 2017;46:467–473. doi: 10.1002/xrs.2769. [DOI] [Google Scholar]
- 36.Schoonjans T., Brunetti A., Golosio B., Sanchez del Rio M., Armando Sole V., Ferrero C., Vincze L. The xraylib library for X-ray-matter interactions. Recent developments. Spectrochim. Acta Part B At. Spectrosc. 2012;66:776–784. doi: 10.1016/j.sab.2011.09.011. [DOI] [Google Scholar]
- 37.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoppe U., Bergemann J., Diembeck W., Ennen J., Gohla S., Harris I., Jacob J., Kielholz J., Mei W., Pollet D., et al. Coenzyme Q10, a cutaneous antioxidant and energizer. Biofactors. 1999;9:371–378. doi: 10.1002/biof.5520090238. [DOI] [PubMed] [Google Scholar]
- 39.Turunen M., Olsson J., Dallner G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta. 2004;1660:171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 40.Králova J., Jurášek M., Mikšátková L., Marešová A., Fähnrich J., Cihlárová P., Drašar P., Bartunek P., Král V. Influence of fluorophore and linker length on the localization and trafficking of fluorescent sterol probes. Sci. Rep. 2020;10:22053. doi: 10.1038/s41598-020-78085-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin Q., Paunesku T., Lai B., Gleber S.-C., Chen S., Finney L., Vine D., Vogt S., Woloschak G., Jacobsen C. Preserving elemental content in adherent mammalian cells for analysis by synchrotron-based x-ray fluorescence microscopy. J. Microsc. 2017;265:81–93. doi: 10.1111/jmi.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcheggiani F., Cirilli I., Orlando P., Silvestri S., Vogelsang A., Knott A., Blatt T., Weise J.M., Tiano L. Modulation of Coenzyme Q10 content and oxidative status in human dermal fibroblasts using HMG-CoA reductase inhibitor over a broad range of concentrations. From mitohormesis to mitochondrial dysfunction and accelerated aging. Aging. 2019;11:2565–2582. doi: 10.18632/aging.101926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blatt T., Littarru G.P. Biochemical rationale and experimental data on the antiaging properties of CoQ(10) at skin level. Biofactors. 2011;37:381–385. doi: 10.1002/biof.169. [DOI] [PubMed] [Google Scholar]
- 44.Mohs R.C., Greig N.H. Drug discovery and development: Role of basic biological research. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2017;3:651–657. doi: 10.1016/j.trci.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is contained within the article.