Abstract
Ω4499 is the site of a Tn5 lac insertion in the Myxococcus xanthus chromosome that fuses lacZ expression to a developmentally regulated promoter. Cell-cell interactions that occur during development, including C signaling, are required for normal expression of Tn5 lac Ω4499. The DNA upstream of the Ω4499 insertion has been cloned, and the promoter has been localized. Analysis of the DNA sequence downstream of the promoter revealed one complete open reading frame and a second partial open reading frame that is interrupted by Tn5 lac Ω4499. The predicted products of these open reading frames are highly similar to reductase and oxidase components of bacterial cytochrome P-450 systems, which allow catabolism or anabolism of unusual compounds. However, the function of the gene products of the Ω4499 locus remains unclear because M. xanthus containing Tn5 lac Ω4499 exhibits no apparent defect in growth, developmental aggregation, fruiting body formation, or sporulation. Deletion analysis of the Ω4499 regulatory region showed that multiple DNA elements spanning more than 500 bp upstream of the transcriptional start site contribute to developmental promoter activity. At least two DNA elements, one downstream of −49 bp and one between −49 and −218 bp, boosted activity of the promoter in response to intercellular C signaling. Three sequences in the Ω4499 promoter region, centered at −55, −33, and −1 bp, nearly match a 7-bp sequence found in other C signal-dependent promoters. We propose that these sequences, matching the consensus sequence 5′-CAYYCCY-3′, be called C box sequences, and we speculate that these sequences are cis-acting regulatory elements important for the expression of M. xanthus genes that depend upon intercellular C signaling during development.
Myxococcus xanthus is a gram-negative soil bacterium that undergoes multicellular development (11). When starved at a high cell density on a solid surface, cells move into aggregation centers, forming mound-shaped fruiting bodies that each contain approximately 105 cells. Within the fruiting bodies, rod-shaped cells differentiate into dormant, ovoid spores that are resistant to heat and desiccation. This developmental process depends on extracellular signals known as the A, B, C, D, and E signals (9, 18). Mutants defective in the production of any one of these signals are arrested in development at a particular stage, but development is restored by mixing with wild-type cells or cells defective in the production of a different signal.
To study the role of cell-cell interactions in controlling gene expression during M. xanthus development, Tn5 lac, a transposon containing a promoterless lacZ gene near one end, has been used to identify developmentally regulated genes (36). By examining the expression of transcriptional fusions to lacZ created by Tn5 lac and the appearance of other developmental markers in signaling-defective mutants, it has been shown that A and B signaling are required at the onset of development, D and E signaling are required slightly later, at 3 to 5 h into development, and C signaling is required at about 6 h for normal developmental gene expression (9, 10, 23).
Considerable progress has been made toward elucidating the A and C signaling and response mechanisms. The A signal is a mixture of peptides and amino acids apparently generated by extracellular proteases (38, 50) and is used to determine whether cells are at a sufficiently high density to initiate multicellular development (39). When A signal reaches a critical threshold concentration, a two-component signal transduction system composed of the SasS sensor histidine kinase and the SasR response regulator appears to trigger expression of 4521 and presumably other early developmental genes (67, 68). Since SasR is NtrC-like in its amino acid sequence and since the 4521 promoter sequence is ς54-like (25), an attractive model is phosphorylated SasR binding to the 4521 regulatory region, which extends at least 146 bp upstream of the transcriptional start site (17) and activates transcription by ς54 RNA polymerase (26). In the case of C signaling, the cell surface-associated CsgA protein is required (19, 29, 30, 41, 58), as is motility (28, 33), which brings cells into alignment (27). The alignment of cells during the early stages of aggregation permits C signaling, which is necessary for the completion of aggregation (42). Within fruiting bodies, densely packed cells are thought to participate in efficient C signaling, and the higher level of C signaling appears to be necessary for sporulation (31, 42, 53). Different levels of C signaling are also required for expression of different developmental genes (31). Hence, C signaling seems to couple morphogenesis of the fruiting body with expression of genes at the proper times and differentiation of cells into spores. Recently, FruA, a response regulator in the FixJ family, has been shown to be involved in the response to intercellular C signaling (12, 46, 59). Mutational analysis suggests that FruA must be phosphorylated to act (12), but neither an upstream kinase nor an immediate downstream target gene in this signaling pathway has been identified. Two potential targets for direct regulation by phosphorylated FruA are promoters identified by Tn5 lac insertions Ω4403 (13) and Ω4400 (6). These are among the first known promoters to be expressed in response to C signaling. The promoter sequences are not ς54-like, but they do share the sequence 5′-CATCCCT-3′, centered at −49 bp.
Here, we report the identification of the Ω4499 regulatory region. The Ω4499 promoter is also a candidate for direct regulation by phosphorylated FruA because expression begins at about 6 h into development and depends strongly upon C signaling (35). Interestingly, the Ω4499 promoter has three sequences that nearly match the sequence found in the other two C signal-dependent promoters, suggesting that these sequences may be important cis-acting regulatory elements. Our results also show differences between the Ω4499 promoter and the two promoters characterized previously. For full expression, DNA more than 500 bp upstream of the Ω4499 transcriptional start site is necessary, yet a low level of developmental expression remains even when upstream DNA is deleted to −49 bp. Hence, the Ω4499 regulatory region spans more than 500 bp, and expression may involve several transcription factors that respond to different developmental cues by binding to upstream DNA elements and activating transcription.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids that were used in this work are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | φ80 ΔlacZΔM15 ΔlacU169 recA1 endA1 hsdR17 supE44 thi-1 gyrA relA1 | 20 |
| JM83 | ara Δlac-pro strA thi φdlacZ ΔM15 | 44 |
| M. xanthus | ||
| DK1622 | Wild type | 22 |
| DK4499 | Tn5 lac (Kmr) Ω4499 | 36 |
| MMF1727 | attB::pREG1727 | 13 |
| MEB1 | attB::pREG4499 | This work |
| MMF21 | attB::pMF21 | This work |
| MMF51 | attB::pMF51 | This work |
| MDB01 | attB::pDB01 | This work |
| MMF25 | attB::pMF25 | This work |
| MMF521 | attB::pMF521 | This work |
| DK5208 | csgA::Tn5-132 (Tcr)a Ω205 | 57 |
| DK5246 | csgA::Tn5-132 (Tcr) Ω205 Tn5 lac (Kmr) Ω4499 | 35 |
| MDB01C | csgA::Tn5-132 (Tcr) Ω205 attB::pDB01 | This work |
| MMF25C | csgA::Tn5-132 (Tcr) Ω205 attB::pMF25 | This work |
| MMF521C | csgA::Tn5-132 (Tcr) Ω205 attB::pMF521 | This work |
| Plasmids | ||
| pGEM7Zf | Aprlacα | Promega |
| pIP110 | Apr Kmr (pGEM7Zf)b; 20.4-kb HindIII fragment from DK4499 | This work |
| pMF002 | Apr (pGEM7Zf)b; 4.5-kb SmaI-BamHI fragment from pIP110 | This work |
| pUC19 | Aprlacα | 69 |
| pMF0051 | Apr (pUC19)b; 3.2-kb PstI-BamHI fragment from pMF002 | This work |
| pDB001 | Apr (pUC19)b; 2.9-kb NarI-BamHI fragment from pMF0051 | This work |
| pMF2.3 | Apr (pUC19)b; 2.1-kb NcoI-BamHI fragment from pMF0051 | This work |
| pREG1666 | Apr Kmr P1-inc attP ′lacZ | 13 |
| pREG1727 | Apr Kmr P1-inc attP ′lacZ | 13 |
| pREG4499 | Apr Kmr (pREG1666)b; 9.4-kb HindIII-BamHI fragment from pIP110 | This work |
| pMF21 | Apr Kmr (pREG1666)b; 4.5-kb XbaI-BamHI fragment from pMF002 | This work |
| pMF51 | Apr Kmr (pREG1666)b; 3.2-kb HindIII-BamHI fragment from pMF0051 | This work |
| pDB01 | Apr Kmr (pREG1727)b; 2.9-kb HindIII-BamHI fragment from pDB001 | This work |
| pMF25 | Apr Kmr (pREG1727)b; 2.7-kb SfuI-BamHI fragment from pMF0051 | This work |
| pMF521 | Apr Kmr (pREG1727)b; 2.1-kb HindIII-BamHI fragment from pMF2.3 | This work |
Tcr, tetracycline resistant.
The vector is indicated in parentheses.
Growth and development.
Escherichia coli cells were grown at 37°C in Luria-Bertani medium (54) containing 50 μg of ampicillin or 50 μg of kanamycin per ml when necessary. M. xanthus was grown at 32 to 34°C in CTT medium (21) (1% casitone, 10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4-K2HPO4, 8 mM MgSO4 [final pH 7.6]) in liquid cultures or on agar (1.5%) plates. Forty micrograms of kanamycin or 12.5 μg of oxytetracycline per ml was used when required for selective growth. Fruiting body development was performed on TPM (10 mM Tris-HCl [pH 8.0], 1 mM KH2PO4-K2HPO4, 8 mM MgSO4 [final pH 7.6]) agar (1.5%) plates as described previously (36).
Molecular cloning and construction of plasmids.
Recombinant DNA work was performed by using standard techniques (54). Plasmid DNA was prepared from E. coli DH5α or JM83.
To clone the DNA upstream of Tn5 lac Ω4499, chromosomal DNA was prepared (40) from M. xanthus DK4499 and digested with HindIII, the fragments were ligated to HindIII-digested pGEM7Zf, and the mixture was transformed into E. coli DH5α, selecting for both ampicillin resistance (Apr) of the vector and kanamycin resistance (Kmr) of the desired insert. One transformant with a plasmid bearing an insert of the expected size was characterized further. Restriction fragments of M. xanthus DNA from this plasmid, pIP110, were gel purified and ligated into vectors as indicated in Table 1. In these and the subsequent subcloning steps described in Table 1, vectors were digested with the same restriction enzymes used to produce the fragments, except as indicated below.
To construct pDB001, the 2.9-kb NarI-BamHI fragment from pMF0051 was gel purified and ligated to pUC19, which had been digested with AccI and BamHI.
pMF2.3 was constructed by digesting pMF0051 to completion with PstI, followed by partial digestion with NcoI. The 4.8-kb fragment containing the pUC19 vector and 2.1 kb of Ω4499 upstream DNA was gel purified, digested with mung bean nuclease, and circularized by ligation.
To construct pMF25, the insert fragment was generated by digesting pMF0051 with SfuI, filling in the ends with the Klenow fragment of DNA polymerase I, and then digesting it with BamHI. The 2.7-kb fragment was gel purified and directionally cloned into pREG1727, which had been digested with HindIII, subjected to end filling, digested with BamHI, and gel purified.
pREG1666 and pREG1727 were nearly identical, except that the terminator-containing fragment in pREG1666 was slightly larger than expected (13). This difference did not affect developmental lacZ expression in a direct comparison (13).
DNA sequencing.
DNA fragments to be sequenced were cloned into either pUC19 or pGEM7Zf and sequenced on both strands with synthetic oligonucleotide primers made by the Michigan State University Macromolecular Structure Facility. Double-stranded sequencing was performed by the Sanger method (55), using a Sequenase kit (United States Biochemical). To resolve regions of compression, 7-deaza-dGTP reaction mixes (United States Biochemical) were used. DNA and protein sequence analyses were done with the University of Wisconsin Genetics Computer Group software package.
Construction of M. xanthus strains.
Strains containing pREG1666 or pREG1727, or derivatives of these plasmids, integrated at the Mx8 phage attachment site (designated attB in Table 1) were constructed by P1-specialized transduction from the rec+ E. coli strain JM83 (15) or by electroporation (24) of the wild-type M. xanthus strain DK1622 or the csgA mutant strain DK5208. Except where noted otherwise, at least three derivatives, each containing a single copy of integrated plasmid, were identified by Southern blot analysis (data not shown) as described previously (13).
RNA analysis.
RNA was prepared as described previously (4) from M. xanthus DK1622. S1 nuclease protection assays were performed with two probes. Fragments from pDB001 digested with SphI (a site in the vector) and either EagI or NcoI were gel purified, phosphatase treated, and 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. RNA was precipitated with approximately 0.1 μg of labeled probe. The pellet was resuspended in 30 μl of hybridization buffer {80% formamide, 20 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 0.4 mM NaCl} and incubated for 10 min at 85°C followed by 3 h at 52°C. S1 nuclease buffer (0.03 M sodium acetate [pH 4.6], 0.05 NaCl, 1 mM ZnSO4, 5% glycerol) and 500 U of S1 nuclease (Boehringer Mannheim) were added, giving a final volume of 300 μl. After 30 min, the mixtures were extracted with phenol-chloroform (300 μl), precipitated with ethanol, and resuspended in formamide loading buffer (80% formamide, 10 mM EDTA, 0.01% xylene cyanol, 0.01% bromophenol blue). The protected products were resolved on a 5% polyacrylamide–8 M urea gel and visualized by autoradiography.
Primer extension analysis was performed as described previously (5), using the oligonucleotide 5′-GGTTGGAGTCGTACAGCAGC-3′, which corresponds to a sequence located about 100 bp downstream of the putative transcriptional start site mapped by S1 nuclease protection. The reaction products were analyzed on a 5% polyacrylamide-urea sequencing gel next to dideoxy sequencing reactions (55) that utilized the same primer.
Nucleotide sequence accession number.
The DNA sequence of 2,877 bp of M. xanthus DNA immediately upstream of the Ω4499 insertion site has been deposited in the GenBank database under accession no. AF111947.
RESULTS
Cloning DNA upstream of Ω4499 and testing it for promoter activity.
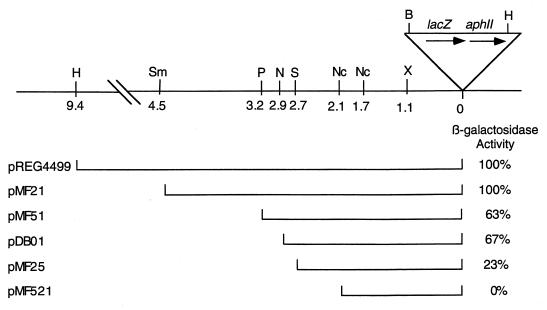
To clone the DNA upstream of the developmentally regulated Tn5 lac insertion Ω4499, we made use of the previous finding that a HindIII restriction site is located approximately 9.4 kb upstream of Ω4499 in the M. xanthus chromosome (36) (Fig. 1). Digestion with HindIII of chromosomal DNA from M. xanthus DK4499 was expected to yield a 20.4-kb fragment because Tn5 lac has a HindIII site located approximately 11 kb from the left end (34) (Fig. 1). This fragment would confer Kmr when cloned in E. coli, due to the presence of the aphII gene encoding aminoglycoside phosphotransferase (Fig. 1). The 20.4-kb fragment was cloned as described in Materials and Methods. The structure of the resulting plasmid, pIP110, was confirmed by restriction mapping, based on the positions of restriction sites in DNA upstream of Ω4499 that had been mapped previously by Southern blotting (36) and on the positions of known restriction sites in Tn5 lac and the vector (data not shown). Figure 1 shows the restriction map of DNA upstream of Ω4499.
FIG. 1.
Physical map of the Ω4499 insertion region and summary of deletions tested for promoter activity. The upper part of the diagram depicts the restriction sites in Tn5 lac and upstream M. xanthus chromosomal DNA that were used in this study. Distances of restriction sites from the Tn5 lac Ω4499 insertion are given in kilobases. Site abbreviations: B, BamHI; H, HindIII; N, NarI; Nc, NcoI; P, PstI; S, SfuI; Sm, SmaI; X, XhoI. The lower part of the diagram depicts the segments of Ω4499 upstream DNA fused to lacZ to test for promoter activity. Plasmid designations are indicated on the left (Table 1). Derivatives of wild-type M. xanthus DK1622 containing a single copy of each plasmid integrated at Mx8 attB were constructed as described in Materials and Methods. β-Galactosidase specific activities during 48-h developmental time courses were measured as described previously (36). As controls, activities of DK4499 containing Tn5 lac Ω4499 and MMF1727 containing pREG1727 (with no insert of Ω4499 upstream DNA) integrated at Mx8 attB were also determined. The maximum β-galactosidase specific activity of MMF1727 (10 U) was subtracted from the maximum activities observed for the other strains, and the remaining activity for derivatives with different Ω4499 upstream segments was expressed as a percentage of that remaining for DK4499. For each plasmid, the number is the average for three derivatives, except in the case of pMF521, for which only one derivative was tested because all derivatives failed to show developmental β-galactosidase activity in a qualitative test (36).
To test the cloned 9.4 kb of Ω4499 upstream DNA for promoter activity, the HindIII-BamHI restriction fragment from pIP110, which includes 9.4 kb of M. xanthus DNA and approximately 60 bp of the left end of the Tn5 lac (Fig. 1), was subcloned into HindIII-BamHI-digested pREG1666 to construct pREG4499. Since the BamHI site of pREG1666 is located immediately upstream of the same lacZ-containing segment as that found in Tn5 lac (13), pREG4499 contains Ω4499 upstream DNA that is fused to a promoterless lacZ gene in exactly the same way as it is in the chromosome of Tn5 lac Ω4499-containing M. xanthus DK4499. pREG4499 was transduced from E. coli JM83 into the wild-type M. xanthus strain DK1622 by using bacteriophage P1-specialized transduction (15). Due to the presence of the attP segment from myxophage Mx8, pREG4499 integrated efficiently into the M. xanthus chromosome at attB (60, 61). Transductants containing a single copy of pREG4499 integrated at Mx8 attB were identified by Southern blot hybridization (data not shown). Several of these transductants were assayed for β-galactosidase activity during development and showed similar expression to that of M. xanthus DK4499 containing Tn5 lac Ω4499 (Fig. 1), indicating that the 9.4-kb Ω4499 upstream DNA segment contains a promoter that is able to drive development-specific expression of a lacZ reporter gene.
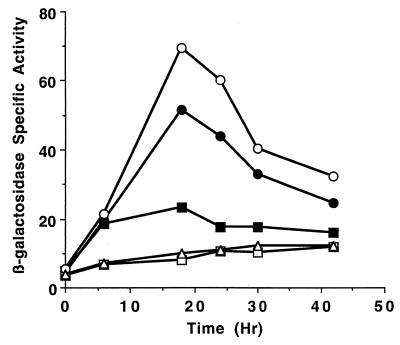
To determine the approximate location of the promoter in the 9.4-kb Ω4499 upstream DNA, smaller fragments were tested for promoter activity after fusion to lacZ and integration at Mx8 attB as described above. The 4.5-kb Ω4499 upstream DNA segment also directed lacZ expression similar to that of Tn5 lac Ω4499-containing M. xanthus DK4499 (Fig. 1). The 3.2-kb (Fig. 1) or 2.9-kb (Fig. 1 and 2) Ω4499 upstream DNA segments directed lacZ expression with a timing similar to that of the strain containing Tn5 lac Ω4499, but the expression reached only about 65% of the maximum level. Derivatives containing the 2.7-kb Ω4499 upstream segment produced even less β-galactosidase activity, reaching only 23% of the maximum level observed in DK4499 (Fig. 1 and 2). A strain containing the 2.1-kb Ω4499 upstream segment showed a level of lacZ expression that was similar to those of transductants containing the vector with no insert of M. xanthus DNA (Fig. 1 and 2). These results indicate that an element essential for developmental promoter activity lies between 2.7 and 2.1 kb upstream of Ω4499 and that DNA beyond 2.7 kb upstream of Ω4499 contributes significantly to promoter activity.
FIG. 2.
Developmental expression of lacZ under the control of the Ω4499 promoter. The β-galactosidase specific activity during development on TPM agar was measured as described previously (36) and is expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. Shown are the activity of DK4499 (○), which contains Tn5 lac Ω4499, and the averages of three independently isolated derivatives of DK1622, each containing a single copy of the 2.9-kb (●) or 2.7-kb (■) Ω4499 upstream DNA fused to lacZ and integrated at Mx8 attB. The activity of DK1622 derivatives containing a single copy of the 2.1-kb (□) Ω4499 upstream DNA or a control with no insert of Ω4499 upstream DNA (▵) is also shown.
Localization of an mRNA 5′ end upstream of Ω4499.
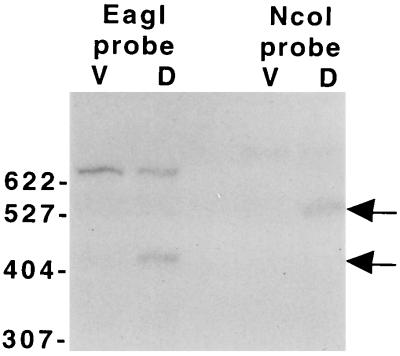
To test whether an mRNA 5′ end maps between 2.7 and 2.1 kb upstream of Ω4499, S1 nuclease protection assays were performed with two probes, one labeled at the NcoI site 2.1 kb upstream of Ω4499 (Fig. 1) and the other labeled at an EagI site approximately 2.2 kb upstream of Ω4499. The probes were hybridized to RNA from M. xanthus DK1622 and subjected to S1 nuclease protection analysis (Fig. 3). RNA from 24-h developing cells, but not from growing cells, protected fragments about 530 and 420 bases in length with the NcoI and EagI probes, respectively. These results indicate that a development-specific mRNA is transcribed from the region upstream of the Ω4499 Tn5 lac insertion and that its 5′ end is approximately 2.6 kb upstream of the insertion site. Together with the deletion analysis (Fig. 1), the results suggest that a promoter lies 2.6 kb upstream of Ω4499 and that the mRNA 5′ end reflects the transcriptional start site.
FIG. 3.
S1 nuclease protection analysis of Ω4499 mRNA. RNA (50 μg) from DK1622 cells grown vegetatively (lanes V) or cells that had undergone 24 h of development (lanes D) was hybridized to DNA probes that were radioactively labeled at either the EagI site (660-bp probe) or the NcoI site (770-bp probe) upstream of Ω4499 and digested with S1 nuclease as described in Materials and Methods. Protected fragments (denoted by arrows) were electrophoresed alongside end-labeled, MspI-digested pBR322 marker fragments (length in bases and positions indicated on the left).
DNA sequence of the Ω4499 upstream region.
We sequenced 2,877 bp of M. xanthus DNA immediately upstream of the Ω4499 insertion site and deposited the sequence in GenBank. The first base pair of the NarI site 2.9 kb upstream of Ω4499 (Fig. 1) was designated position 1 in the sequence. One complete open reading frame (ORF) and one partial ORF interrupted by Ω4499 were inferred from the sequence. The two ORFs exhibit a codon preference typical of M. xanthus genes, including a strong bias toward the use of guanine or cytosine at the third codon position (56). The first ORF, beginning with ATG at position 243 and ending with a TGA stop codon at position 1878, is predicted to encode a 545-amino-acid polypeptide. The second ORF, beginning 59 bp downstream of the first one, continues uninterrupted for at least 313 amino acids, extending to the Ω4499 insertion site at the end of the sequenced region. Upstream of each ORF is a potential ribosome binding site (AGGAGG, 10 bp upstream of the first ORF, and AAGGAGA, 7 bp upstream of the second ORF) (48).
The deduced amino acid sequences encoded by the ORFs upstream of Ω4499 exhibit high similarity to proteins that are components of bacterial cytochrome P-450 systems. A BLAST search (1) with the first ORF revealed significant (P < 10−30) similarity to several reductases that transfer electrons to cytochrome P-450 oxidases (data not shown). Among these, the best known is putidaredoxin reductase (49), to which the product of the first ORF exhibits 32% amino acid identity and 40% amino acid similarity over a 398-amino-acid stretch. Putidaredoxin reductase is involved in camphor catabolism in Pseudomonas putida (49). It transfers electrons from NADH to putidaredoxin, an iron-sulfur protein that in turn transfers the electrons to cytochrome P-450cam, which hydroxylates camphor. All three proteins are encoded in an operon, as is often the case for bacterial cytochrome P-450 systems (45).
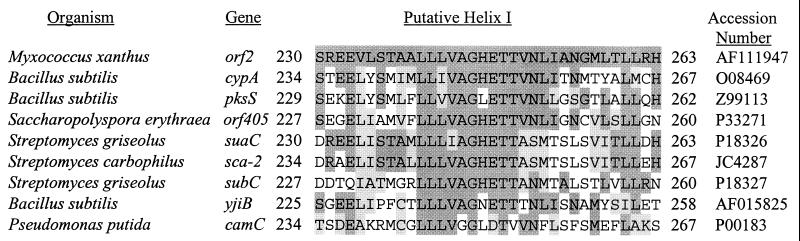
A BLAST search with the second (partial) ORF revealed significant (P < 10−25) similarity to several cytochromes P-450, including PksS, which is thought to be a hydroxylase involved in polyketide synthesis in Bacillus subtilis (37), and SuaC (P-450 SU1) and SubC (P-450 SU2), which enable Streptomyces griseolus to metabolize sulfonylurea herbicides (47). The partial ORF showed significant but lower similarity to many other bacterial cytochromes P-450, including P-450cam of P. putida, which was the first P-450 to have its structure determined (51). An α-helix called helix I of P-450cam contains residues that contact heme and substrates. Helix I is a strongly conserved structural element in other cytochromes P-450 (45). Figure 4 shows the putative helix I of the second ORF in the Ω4499 upstream region aligned with the corresponding regions of the seven sequences showing the highest similarity in a BLAST search with the 313-amino-acid sequence of the partial ORF and aligned with helix I of P-450cam (encoded in the camC gene). The high degree of conservation with respect to both sequence and position in the protein strongly supports the idea that the second ORF in the Ω4499 upstream region encodes a cytochrome P-450 oxidase.
FIG. 4.
Alignment of putative helixes I of cytochromes P-450. Numbers to the left and right of each sequence indicate the position of the sequence in the protein. Dark shading indicates amino acids identical to the second ORF upstream of Ω4499 (orf2), and light shading indicates similar amino acids according to the following groupings: GP, ACILMV, FWY, NQST, RHK, and DE.
Based on the DNA sequence analysis, we propose that Tn5 lac insertion Ω4499 disrupts the second gene in a developmentally regulated operon encoding reductase and oxidase components of a cytochrome P-450 system. The function of this putative cytochrome P-450 system is unknown. M. xanthus cells bearing Tn5 lac Ω4499 show no discernible growth or developmental defect (36).
Precise mapping of the Ω4499 mRNA 5′ end.
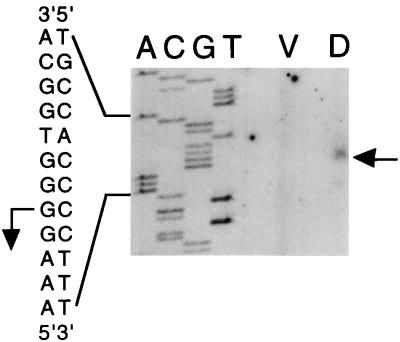
The DNA sequence of the Ω4499 upstream region and the results of the S1 nuclease protection assays (Fig. 3) allowed us to design a primer for precise mapping of the Ω4499 mRNA 5′ end by primer extension analysis. The primer spanned positions 335 to 354 in the sequence. Figure 5 shows that the 5′ end of Ω4499 mRNA from 24-h developing cells mapped to a cytosine base at position 221 in the sequence. This result is in good agreement with the S1 nuclease protection results (Fig. 3), since it predicts protected fragments of 537 and 431 bases with probes labeled at the NcoI and EagI sites, respectively. No primer extension product was generated with mRNA prepared from growing M. xanthus cells (Fig. 5).
FIG. 5.
Primer extension analysis of Ω4499 mRNA. RNA (10 μg) from DK1622 cells grown vegetatively (lane V) or cells that had undergone 24 h of development (lane D) was subjected to primer extension analysis as described in Materials and Methods. The straight arrow at the right indicates the position of the primer extension product in lane D. The same primer was used to sequence Ω4499 upstream DNA, and a portion of the sequence is indicated at the left. The bent arrow indicates the position of the mRNA 5′ end in the sequence.
Comparison of the Ω4499 promoter with known M. xanthus promoters.
Inspection of the DNA sequence upstream of the apparent Ω4499 transcriptional start site revealed sequences similar to those found in two other M. xanthus promoters. Figure 6 shows the positions of three sequences in the Ω4499 promoter region that are similar to the sequence 5′-CATCCCT-3′, which is found in the Ω4403 and Ω4400 promoters, centered at −49 bp (6, 13). The similar sequences in the Ω4499 promoter region are centered at −55, −33, and −1 bp. The position of the SfuI site used in our deletion analysis (Fig. 1 and 2) is also shown in Fig. 6. The fusion with its upstream end at the SfuI site (−49 bp) does not contain the sequence centered at −55 bp. This fusion produced considerably less β-galactosidase than the fusion with its upstream end at the NarI site (−218 bp). Whether this difference depends on the sequence centered at −55 bp or on other sequences between −49 and −218 bp remains to be tested. Two inverted repeats are present upstream of −60 bp (Fig. 6), and these could be sites recognized by dimeric DNA-binding proteins. Also shown in Fig. 6 is a 9-bp sequence centered at −65 bp that matches a sequence in the Ω4400 promoter in eight of nine positions. The sequence in the Ω4400 promoter is in the opposite orientation and is centered at −80 bp, within a region (between −101 and −76 bp) that was shown previously to be essential for activity of this promoter (6).
FIG. 6.
DNA sequence of the Ω4499 promoter region. Sequences similar to the sequence 5′-CATCCCT-3′ are underlined. The SfuI site used to make the deletion to −49 bp is indicated. Arrows indicate inverted repeats, and boldface type highlights a 9-bp sequence similar to a sequence in the Ω4400 promoter region.
Dependence of expression on C signaling.
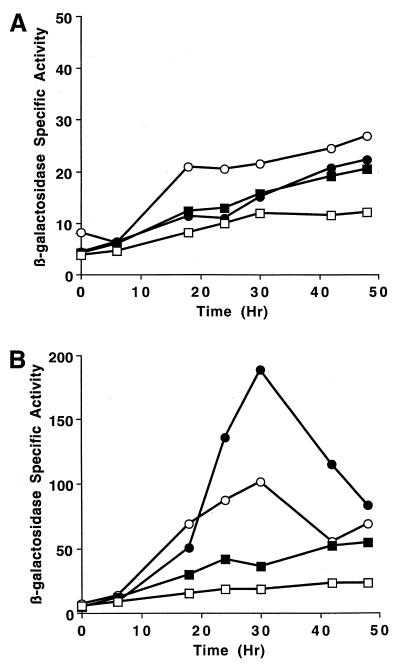
Intercellular C signaling is mediated by the product of csgA (19, 29, 30, 41, 58) and is required for the expression of nearly all M. xanthus genes that begin to be expressed after 6 h into development (35). Introduction of a csgA mutation into cells containing Tn5 lac Ω4499 severely reduced developmental lacZ expression (35). To determine whether the Ω4499 promoter region defined by our deletion analysis also exhibits dependence on C signaling, we measured expression of fusions integrated at Mx8 attB in csgA mutant cells. Figure 7A shows that DNA extending to −49 bp (the SfuI restriction site [Fig. 1]) or −218 bp (the NarI restriction site [Fig. 1]) directed a low level of developmental lacZ expression, eventually reaching about the same level as that observed for DK5246 containing Tn5 lac Ω4499 in a csgA mutant background. Similar results (data not shown) were observed for fusions with Ω4499 upstream DNA extending to the PstI or SmaI restriction site (Fig. 1). These results clearly show that the developmental promoter downstream of −218 bp is dependent upon csgA for full activity, because the fusion with its upstream end at the NarI site was expressed at a considerably higher level in wild-type cells (Fig. 2) than in csgA mutant cells (Fig. 7A). It is not clear from these results whether DNA downstream of −49 bp exhibits csgA dependence, because the fusion with its upstream end at the SfuI site was expressed at a slightly higher level in wild-type cells (Fig. 2) than in the csgA mutant (Fig. 7A) at 6 to 18 h of development but not later in development. A fusion with 2.1 kb of Ω4499 upstream DNA, which does not include the Ω4499 promoter (Fig. 1 and 2), failed to express lacZ above a low background level in csgA mutant cells (Fig. 7A), just as in wild-type cells (Fig. 2).
FIG. 7.
Developmental expression of lacZ under the control of the Ω4499 promoter in a csgA background in the absence (A) or presence (B) of extracellular complementation. (A) Developmental β-galactosidase specific activities were measured as described previously (36) for DK5246 (○), which contains Tn5 lac Ω4499 in a csgA background, and for three independently isolated derivatives (the average is plotted), each containing a single copy of the 2.9-kb (●; extending to −218 bp), 2.7-kb (■; extending to −49 bp), or 2.1-kb (□) Ω4499 upstream DNA fused to lacZ and integrated at Mx8 attB in the same csgA background. (B) The strains used for panel A were codeveloped with an equal number of wild-type DK1622 cells, which express csgA but not lacZ, and the β-galactosidase specific activity was determined as described previously for extracellular complementation (35). β-Galactosidase specific activity is expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein.
To determine whether extracellular C signaling could restore Ω4499 promoter activity of the various fusions in csgA mutant cells, we used wild-type cells as C signal donors. csgA cells with a fusion integrated at Mx8 attB were mixed with an equal number of wild-type DK1622 cells and allowed to codevelop. The specific activity of β-galactosidase in the mixture was determined at different times of development. Figure 7B shows that C signaling restored Ω4499 promoter activity in csgA mutant cells. Expression from Tn5 lac Ω4499 was restored to a slightly higher level in csgA mutant cells than in wild-type cells (Fig. 2). Expression from the Ω4499 promoter region extending to −218 bp unexpectedly reached a higher level in the mixtures than did expression from Tn5 lac Ω4499 (Fig. 7B). Although we do not understand the reason for the higher expression, clearly DNA downstream of −218 bp is sufficient for the Ω4499 promoter to be stimulated by extracellular C signaling. Similarly, DNA downstream of −49 bp was responsive to C signaling because expression from csgA mutant cells containing this fusion was higher during codevelopment with wild-type cells (Fig. 7B) than when the csgA cells were not mixed with wild-type cells (Fig. 7A). In contrast, expression from csgA mutant cells containing the fusion without the Ω4499 promoter remained at a low level during codevelopment with wild-type cells (Fig. 7B). Because C signaling rescued expression of the fusions with upstream endpoints at −49 and −218 bp and because expression was higher from the −218 bp fusion than from the −49 bp fusion, we conclude that C signal responsive elements lie both upstream and downstream of −49 bp in the Ω4499 promoter region.
DISCUSSION
We have cloned the DNA upstream of Tn5 lac insertion Ω4499 and identified a promoter that depends upon intercellular C signaling for expression during M. xanthus development. Promoter activity was lost in a graded fashion in a 5′ deletion series, suggesting that multiple upstream elements contribute to activity, but a deletion to −49 bp still permitted developmental regulation and was responsive to C signaling. A deletion to −218 bp responded more strongly to C signaling than the deletion to −49 bp, indicating that promoter elements responsive to C signaling lie both upstream and downstream of −49 bp. Interestingly, sequences similar to a sequence found in two other C signal-dependent promoters were present both upstream and downstream of −49 bp in the Ω4499 promoter region. These sequences may define a cis-acting regulatory element important for the expression of C signal-dependent genes.
Downstream of the Ω4499 promoter are one complete ORF and one partial ORF (interrupted by Tn5 lac Ω4499) that very likely encode reductase and oxidase components, respectively, of a cytochrome P-450 system. In bacteria, cytochrome P-450 systems are most often involved in the catabolism of lipophilic substrates (45). In many cases, these systems allow the utilization of unusual organic compounds as nutrient sources. Other bacterial cytochromes P-450 participate in the biosynthesis of compounds with antibiotic properties. Both catabolic and anabolic enzymes are represented among the P-450 oxidases most similar in sequence to the second (partial) ORF downstream of the Ω4499 promoter (Fig. 4). Possible functions of the P-450 system encoded at the Ω4499 locus include metabolism of compounds produced by developing cells or potentially present in the environment or biosynthesis of a signal molecule, which could be an autocide or an antibiotic targeted toward competitors. Function of this P-450 system is apparently not essential for growth, developmental aggregation, or sporulation, because M. xanthus cells containing Tn5 lac Ω4499 appear to grow and develop normally (36), and it is very likely that the insertion disrupts gene function. The P-450 oxidases to which the second (partial) ORF is similar have a conserved cysteine residue in the C-terminal region which in P-450cam serves as a heme ligand (45, 51). The 313-amino-acid partial product of the second ORF would be missing the critical C-terminal region. Moreover, since most bacterial P-450 systems include an iron-sulfur protein that shuttles electrons between the reductase and the oxidase and this protein is typically encoded in the same operon (45), it is possible that a third ORF is normally expressed from the Ω4499 promoter but fails to be expressed in cells containing Tn5 lac Ω4499. The lack of an apparent developmental defect of cells containing the Ω4499 insertion could be explained if the products of the Ω4499 locus are functionally redundant with other M. xanthus proteins (36). However, no genes were observed to cross-hybridize with DNA probes from the Ω4499 locus in high-stringency Southern blots of M. xanthus chromosomal DNA (data not shown). We cannot rule out the possibility that cells containing Tn5 lac Ω4499 have a subtle defect in developmental aggregation and/or sporulation that escaped detection.
Our main interest in the Ω4499 locus is its mechanism of regulation by intercellular C signaling. The Ω4499 promoter is likely to be developmentally regulated at the level of transcription initiation, because β-galactosidase activity from a lacZ fusion to the promoter increases markedly during development from a low background level present in growing cells (Fig. 2) and because Ω4499 mRNA was readily detected in developing cells (Fig. 3 and 5) but was undetectable in growing cells. We cannot rule out the possibility of differential Ω4499 mRNA stability in growing and developing cells, but this would require that Ω4499-lacZ fusion mRNA also exhibit differential stability (to explain increased β-galactosidase activity in developing cells with lacZ fused to the Ω4499 promoter), since β-galactosidase itself does not appear to be more stable in developing cells than in growing cells (36). A simple model to explain regulation of the Ω4499 promoter is that one or more essential transcription factors are made or become active during development.
Our deletion analysis of the Ω4499 promoter region indicates that multiple DNA elements spanning more than 500 bp upstream of the transcriptional start site contribute to developmental promoter activity (Fig. 1 and 2). The deletion of DNA between SmaI and PstI sites located at approximately 1,800 and 500 bp, respectively, upstream of the start site reduced developmental lacZ expression by about 35% (Fig. 1). Therefore, DNA more than 500 bp upstream of the start site is necessary for full Ω4499 promoter activity. Likewise, deletion of DNA between NarI and SfuI sites located at −218 and −49 bp, respectively, reduced developmental lacZ expression an additional 40% (Fig. 1 and 2), indicating the importance of DNA in this region. The activity of certain other developmentally regulated M. xanthus promoters has also been shown to be influenced by multiple upstream DNA elements (8). For example, expression of the csgA gene is controlled by an upstream region spanning more than 700 bp (42). Expression of csgA was lost in a graded fashion as 5′ deletion endpoints approached the transcriptional start site. It was proposed that multiple transcription factors bind to the upstream region and regulate csgA expression in response to nutrient levels, peptidoglycan components, and B signaling. Similarly, expression of the Ω4499 promoter may involve several transcription factors that respond to different developmental cues by binding to upstream DNA elements and activating transcription. At least two DNA elements, one upstream and one downstream of −49 bp, respond to intercellular C signaling, based on the results obtained from the use of wild-type cells as C signal donors to csgA mutant cells containing different Ω4499 upstream segments fused to lacZ (Fig. 7B).
The Ω4499 promoter is the first example of a C signal-dependent promoter that retains some developmental activity despite the loss of DNA upstream of −49 bp. Two other M. xanthus promoters that depend upon intercellular C signaling have been examined in detail. DNA between −80 and −72 bp in the Ω4403 promoter region (13) and between −101 and −76 bp in the Ω4400 promoter region (6) is essential for developmental expression. Since RNA polymerase typically protects the −50 to 20 bp region of promoters, these results suggested that upstream activator proteins are required for transcription of the Ω4403 and Ω4400 genes. In the case of the Ω4499 promoter, perhaps the synthesis or activation of a new ς factor is sufficient for a low level of developmental transcription, and one or more activator proteins bind to DNA upstream of −49 bp and boost expression. Alternatively, it is possible that a transcriptional activator binds downstream of −49 bp, within the region normally bound by RNA polymerase.
It is unknown what form of RNA polymerase transcribes C signal-dependent M. xanthus genes. It was noted previously that the Ω4403 and Ω4400 promoters exhibit a match for five of six positions in the −10 regions (6); however, the Ω4499 promoter does not match this sequence very well, the best match being three of six positions for the Ω4400 promoter. All three promoters have the sequence 5′-CACCC-3′, centered at −8, −6, and −2 in the Ω4403, Ω4400, and Ω4499 promoters, respectively, but its variable position makes it unlikely to be important for RNA polymerase binding. The three promoters do not share a similar sequence in the −35 regions. Since the Ω4499 promoter does not match the M. xanthus vegA promoter in the −10 and −35 regions (32), it was not surprising that ςA RNA polymerase partially purified from growing M. xanthus cells (5) failed to transcribe from the Ω4499 promoter in vitro (data not shown). The Ω4499 promoter also does not resemble the mbhA (52), 4521 (25), pilA (66), or sdeK (14) promoters, which are likely to be transcribed by ς54 RNA polymerase (26), or the carQRS promoter (43), which appears to be transcribed by ςCarQ RNA polymerase (7, 16). Five other putative M. xanthus ς factors have been described, SigB (2), SigC (3), SigD (63), SigE (62), and RpoE1 (65), but promoters recognized by RNA polymerase containing these ς factors have not been identified. We showed previously that a null mutation in sigB or sigC does not affect expression of Tn5 lac Ω4499 (6). The Ω4499 promoter might be transcribed by RNA polymerase containing SigD, SigE, RpoE1, or a ς factor yet to be discovered.
Examination of the DNA sequence in the Ω4499 promoter region revealed three sequences that nearly match a 7-bp sequence found in both the Ω4403 (13) and Ω4400 (6) promoters, centered at −49 bp (Fig. 6). An alignment of these sequences is shown in Table 2. All five sequences match the degenerate consensus 5′-CAYYCCY-3′ (Y means pyrimidine). Below the consensus in Table 2 are four additional sequences found in promoters that have been reported to be C signal dependent. Two of these sequences occur in the Ω4400 promoter region, centered at −5 and −80 bp (6). The other two sequences are centered at −51 and −63 bp in the fruA (46) and csgA (42) promoters, respectively. To our knowledge, only one of the sequences shown in Table 2 has been mutated and the effect on developmental promoter activity been examined. Changing 5′-CATCCCT-3′ centered at −49 bp in the Ω4400 promoter to 5′-ACGAAAG-3′ abolished the activity of the promoter (64). Based on this result and the finding of sequences matching the 5′-CAYYCCY-3′ consensus in the C signal-dependent promoters reported so far, we propose that the consensus be called the C box, and we speculate that C box sequences are cis-acting regulatory elements important for the expression of M. xanthus genes that depend upon intercellular C signaling during development.
TABLE 2.
Alignment of sequences found in C signal-dependent promoters
| Promoter or consensus | Positiona | Sequenceb |
|---|---|---|
| Ω4403 | −49 | CATCCCT |
| Ω4400 | −49 | CATCCCT |
| Ω4499 | −55 | CATTCCC |
| Ω4499 | −33 | CATTCCT |
| Ω4499 | −1 | CACCCCT |
| C box consensus | CAYYCCYc | |
| Ω4400 | −5 | CACCCCC |
| Ω4400 | −80 | CACCCCC |
| fruA | −51 | CACTCCC |
| csgA | −63 | CACTCCC |
The position of the center of the sequence relative to the transcriptional start site. All sequences are on the nontranscribed DNA strand, except the sequence centered at −80 bp in the Ω4400 promoter.
Nucleotides identical in all the sequences are in boldface type.
Y means pyrimidine.
If all of the C box sequences noted in Table 2 are important for transcription of the corresponding genes, then the variable positions of these sequences in different promoters suggest that the C box might be recognized by a transcriptional activator protein rather than by RNA polymerase. A candidate for a protein that might recognize the C box is FruA, an apparent response regulator protein that mediates the response to C signaling (12, 46, 59).
In summary, we have identified the regulatory region of an operon encoding a cytochrome P-450 system that is induced during M. xanthus development. Although the function of this P-450 system remains unknown, the regulatory region has three copies of a sequence found in other C signal-dependent promoters. Alignment of these sequences produced a consensus that we call the C box. Future studies will include further mutational analysis of C box sequences and attempts to identify a putative protein that binds to the C box and activates transcription in response to intercellular C signaling.
ACKNOWLEDGMENTS
We thank Y. Cheng, D. Kaiser, and R. Gill for providing bacterial strains and plasmids used in this study. We thank I.-S. Park for constructing pIP110 and E. Borer Meyer for constructing pREG4499 and MEB1.
This research was supported by NIH grant GM47293 and by the Michigan Agricultural Experiment Station.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Apelian D, Inouye S. Development-specific ς-factor essential for late-stage differentiation of Myxococcus xanthus. Genes Dev. 1990;4:1396–1403. doi: 10.1101/gad.4.8.1396. [DOI] [PubMed] [Google Scholar]
- 3.Apelian D, Inouye S. A new putative sigma factor of Myxococcus xanthus. J Bacteriol. 1993;175:3335–3342. doi: 10.1128/jb.175.11.3335-3342.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biran D, Brot N, Weissbach H, Ron E. Heat shock-dependent transcriptional activation of the metA gene of Escherichia coli. J Bacteriol. 1995;177:1374–1379. doi: 10.1128/jb.177.5.1374-1379.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biran D, Kroos L. In vitro transcription of Myxococcus xanthus genes with RNA polymerase containing ςA, the major sigma factor in growing cells. Mol Microbiol. 1997;25:463–472. doi: 10.1046/j.1365-2958.1997.4751843.x. [DOI] [PubMed] [Google Scholar]
- 6.Brandner J P, Kroos L. Identification of the Ω4400 regulatory region, a developmental promoter of Myxococcus xanthus. J Bacteriol. 1998;180:1995–2004. doi: 10.1128/jb.180.8.1995-2004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browning D, Khoshkhoo N, Hodgson D. Abstracts of the 24th Annual Meeting on the Biology of the Myxobacteria. 1997. CarQ is a sigma factor, CarR is an inner membrane protein of Myxococcus xanthus and CarS appears not to be a DNA binding protein; p. 2. [Google Scholar]
- 8.Downard J, Kroos L. Transcriptional regulation of developmental gene expression in Myxococcus xanthus. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C: American Society for Microbiology; 1993. pp. 183–199. [Google Scholar]
- 9.Downard J, Ramaswamy S V, Kil K. Identification of esg, a genetic locus involved in cell-cell signaling during Myxococcus xanthus development. J Bacteriol. 1993;175:7762–7770. doi: 10.1128/jb.175.24.7762-7770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Downard J, Toal D. Branched-chain fatty acids: the case for a novel form of cell-cell signalling during Myxococcus xanthus development. Mol Microbiol. 1995;16:171–175. doi: 10.1111/j.1365-2958.1995.tb02290.x. [DOI] [PubMed] [Google Scholar]
- 11.Dworkin M. Recent advances in the social and developmental biology of the myxobacteria. Microbiol Rev. 1996;60:70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellehauge E, Norregaard-Madsen M, Sogaard-Andersen L. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in Myxococcus xanthus development. Mol Microbiol. 1998;30:807–817. doi: 10.1046/j.1365-2958.1998.01113.x. [DOI] [PubMed] [Google Scholar]
- 13.Fisseha M, Gloudemans M, Gill R, Kroos L. Characterization of the regulatory region of a cell interaction-dependent gene in Myxococcus xanthus. J Bacteriol. 1996;178:2539–2550. doi: 10.1128/jb.178.9.2539-2550.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garza A G, Pollack J S, Harris B Z, Lee A, Keseler I M, Licking E F, Singer M. SdeK is required for early fruiting body development in Myxococcus xanthus. J Bacteriol. 1998;180:4628–4637. doi: 10.1128/jb.180.17.4628-4637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gill R E, Cull M G, Fly S. Genetic identification and cloning of a gene required for developmental cell interactions in Myxococcus xanthus. J Bacteriol. 1988;170:5279–5288. doi: 10.1128/jb.170.11.5279-5288.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorham H, McGowan S, Robson P, Hodgson D. Light-induced carotenogenesis in Myxococcus xanthus: light-dependent membrane sequestration of ECF sigma factor CarQ by anti-sigma factor CarR. Mol Microbiol. 1996;19:171–186. doi: 10.1046/j.1365-2958.1996.360888.x. [DOI] [PubMed] [Google Scholar]
- 17.Gulati P, Xu D, Kaplan H. Identification of the minimum regulatory region of a Myxococcus xanthus A-signal-dependent developmental gene. J Bacteriol. 1995;177:4645–4651. doi: 10.1128/jb.177.16.4645-4651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagen D C, Bretscher A P, Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978;64:284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- 19.Hagen T J, Shimkets L J. Nucleotide sequence and transcriptional products of the csg locus of Myxococcus xanthus. J Bacteriol. 1990;172:15–23. doi: 10.1128/jb.172.1.15-23.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 21.Hodgkin J, Kaiser D. Cell-to-cell stimulation of motility in nonmotile mutants of Myxococcus. Proc Natl Acad Sci USA. 1977;74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaiser D, Kroos L. Intercellular signaling. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C: American Society for Microbiology; 1993. pp. 257–284. [Google Scholar]
- 24.Kalman L V, Cheng Y, Kaiser D. The Myxococcus xanthus dsg gene product performs functions of translation initiation factor IF3 in vivo. J Bacteriol. 1994;176:1434–1442. doi: 10.1128/jb.176.5.1434-1442.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keseler I, Kaiser D. An early A-signal-dependent gene in Myxococcus xanthus has a ς54-like promoter. J Bacteriol. 1995;177:4638–4644. doi: 10.1128/jb.177.16.4638-4644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keseler I, Kaiser D. ς54, a vital protein for Myxococcus xanthus. Proc Natl Acad Sci USA. 1997;94:1979–1984. doi: 10.1073/pnas.94.5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S K, Kaiser D. Cell alignment required in differentiation of Myxococcus xanthus. Science. 1990;249:926–928. doi: 10.1126/science.2118274. [DOI] [PubMed] [Google Scholar]
- 28.Kim S K, Kaiser D. Cell motility is required for the transmission of C-factor, an intercellular signal that coordinates fruiting body morphogenesis of Myxococcus xanthus. Genes Dev. 1990;4:896–905. doi: 10.1101/gad.4.6.896. [DOI] [PubMed] [Google Scholar]
- 29.Kim S K, Kaiser D. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell. 1990;61:19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- 30.Kim S K, Kaiser D. Purification and properties of Myxococcus xanthus C-factor, an intercellular signaling protein. Proc Natl Acad Sci USA. 1990;87:3635–3639. doi: 10.1073/pnas.87.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S K, Kaiser D. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J Bacteriol. 1991;173:1722–1728. doi: 10.1128/jb.173.5.1722-1728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komano T, Franceschini T, Inouye S. Identification of a vegetative promoter in Myxococcus xanthus: a protein that has homology to histones. J Mol Biol. 1987;196:517–524. doi: 10.1016/0022-2836(87)90029-5. [DOI] [PubMed] [Google Scholar]
- 33.Kroos L, Hartzell P, Stephens K, Kaiser D. A link between cell movement and gene expression argues that motility is required for cell-cell signaling during fruiting body development. Genes Dev. 1988;2:1677–1685. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- 34.Kroos L, Kaiser D. Construction of Tn5 lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc Natl Acad Sci USA. 1984;81:5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kroos L, Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- 36.Kroos L, Kuspa A, Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986;117:252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- 37.Kunst F, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 38.Kuspa A, Plamann L, Kaiser D. Identification of heat-stable A-factor from Myxococcus xanthus. J Bacteriol. 1992;174:3319–3326. doi: 10.1128/jb.174.10.3319-3326.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuspa A, Plamann L, Kaiser D. A-signalling and the cell density requirement for Myxococcus xanthus development. J Bacteriol. 1992;174:7360–7369. doi: 10.1128/jb.174.22.7360-7369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laue B E, Gill R. Use of a phase variation-specific promoter of Myxococcus xanthus in a strategy for isolating a phase-locked mutant. J Bacteriol. 1994;176:5341–5349. doi: 10.1128/jb.176.17.5341-5349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee B-U, Lee K, Mendez J, Shimkets L. A tactile sensory system of Myxococcus xanthus involves an extracellular NAD(P)+-containing protein. Genes Dev. 1995;9:2964–2973. doi: 10.1101/gad.9.23.2964. [DOI] [PubMed] [Google Scholar]
- 42.Li S-F, Lee B, Shimkets L J. csgA expression entrains Myxococcus xanthus development. Genes Dev. 1992;6:401–410. doi: 10.1101/gad.6.3.401. [DOI] [PubMed] [Google Scholar]
- 43.McGowan S, Gorham H, Hodgson D. Light-induced carotenogenesis in Myxococcus xanthus: DNA sequence analysis of the carR region. Mol Microbiol. 1993;10:713–735. doi: 10.1111/j.1365-2958.1993.tb00943.x. [DOI] [PubMed] [Google Scholar]
- 44.Messing J. A multipurpose cloning system based on the single-stranded DNA bacteriophage M13. Recombinant DNA bulletin, publication no. 71–99. Bethesda, Md: National Institutes of Health; 1979. pp. 43–48. [Google Scholar]
- 45.Munro A W, Lindsay J G. Bacterial cytochromes P-450. Mol Microbiol. 1996;20:1115–1125. doi: 10.1111/j.1365-2958.1996.tb02632.x. [DOI] [PubMed] [Google Scholar]
- 46.Ogawa M, Fujitani S, Mao X, Inouye S, Komano T. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol Microbiol. 1996;22:757–767. doi: 10.1046/j.1365-2958.1996.d01-1725.x. [DOI] [PubMed] [Google Scholar]
- 47.Omer C A, Lenstra R, Litle P J, Dean C, Tepperman J M, Leto K J, Romesser J A, O’Keef D P. Genes for two herbicide-inducible cytochromes P-450 from Streptomyces griseolus. J Bacteriol. 1990;172:3335–3345. doi: 10.1128/jb.172.6.3335-3345.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oyaizu H, Woese C. Phylogenetic relationships among the sulfate respiring bacteria, myxobacteria and purple bacteria. Syst Appl Microbiol. 1985;6:257–263. [Google Scholar]
- 49.Peterson J A, Lorence M C, Amarneh B. Putidaredoxin reductase and putidaredoxin. Cloning, sequence determination, and heterologous expression of the proteins. J Biol Chem. 1990;265:6066–6073. [PubMed] [Google Scholar]
- 50.Plamann L, Kuspa A, Kaiser D. Proteins that rescue A-signal-defective mutants of Myxococcus xanthus. J Bacteriol. 1992;174:3311–3318. doi: 10.1128/jb.174.10.3311-3318.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poulos T L, Finzel B C, Howard A J. High-resolution crystal structure of cytochrome P450cam. J Mol Biol. 1987;159:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 52.Romeo J M, Zusman D R. Transcription of the myxobacterial hemagglutinin gene is mediated by a ς54-like promoter and a cis-acting upstream regulatory region of DNA. J Bacteriol. 1991;173:2969–2976. doi: 10.1128/jb.173.9.2969-2976.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sager B, Kaiser D. Spatial restriction of cellular differentiation. Genes Dev. 1993;7:1645–1653. doi: 10.1101/gad.7.9.1645. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimkets L. The myxobacterial genome. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C: American Society for Microbiology; 1993. pp. 85–107. [Google Scholar]
- 57.Shimkets L J, Asher S J. Use of recombination techniques to examine the structure of the csg locus of Myxococcus xanthus. Mol Gen Genet. 1988;211:63–71. doi: 10.1007/BF00338394. [DOI] [PubMed] [Google Scholar]
- 58.Shimkets L J, Rafiee H. CsgA, an extracellular protein essential for Myxococcus xanthus development. J Bacteriol. 1990;172:5299–5306. doi: 10.1128/jb.172.9.5299-5306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sogaard-Andersen L, Slack F, Kimsey H, Kaiser D. Intercellular C-signaling in Myxococcus xanthus involves a branched signal transduction pathway. Genes Dev. 1996;10:740–754. doi: 10.1101/gad.10.6.740. [DOI] [PubMed] [Google Scholar]
- 60.Stellwag E, Fink J M, Zissler J. Physical characterization of the genome of the Myxococcus xanthus bacteriophage MX-8. Mol Gen Genet. 1985;199:123–132. doi: 10.1007/BF00327521. [DOI] [PubMed] [Google Scholar]
- 61.Stephens K, Kaiser D. Genetics of gliding motility in Myxococcus xanthus: molecular cloning of the mgl locus. Mol Gen Genet. 1987;207:256–266. [Google Scholar]
- 62.Ueki T, Inouye S. Abstracts of the 23rd Annual Meeting on the Biology of the Myxobacteria. 1996. New sigma factors, SigD and SigE, from Myxococcus xanthus; p. 37. [Google Scholar]
- 63.Ueki T, Inouye S. A new sigma factor, SigD, essential for stationary phase is also required for multicellular differentiation in Myxococcus xanthus. Genes Cells. 1998;3:371–385. doi: 10.1046/j.1365-2443.1998.00197.x. [DOI] [PubMed] [Google Scholar]
- 64.Velicer, G., and L. Kroos. Unpublished data.
- 65.Ward M, Lew H, Treuner-Lange A, Zusman D. Regulation of motility behavior in Myxococcus xanthus may require an extracytoplasmic-function sigma factor. J Bacteriol. 1998;180:5668–5675. doi: 10.1128/jb.180.21.5668-5675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu S S, Kaiser D. Regulation of expression of the pilA gene in Myxococcus xanthus. J Bacteriol. 1997;179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang, C., and H. Kaplan. Unpublished data.
- 68.Yang C, Kaplan H B. Myxococcus xanthus sasS encodes a sensor histidine kinase required for early developmental gene expression. J Bacteriol. 1997;179:7759–7767. doi: 10.1128/jb.179.24.7759-7767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yanisch-Perron C, Vieria J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp 18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]