Abstract
Conjugative transfer of IncN plasmid pKM101 is mediated by the TraI-TraII region-encoded transfer machinery components. Similar to the case for the related Agrobacterium tumefaciens T-complex transfer apparatus, this machinery is needed for assembly of pili to initiate cell-to-cell contact preceding DNA transfer. Biochemical and cell biological experiments presented here show extracellular localization of TraC, as suggested by extracellular complementation of TraC-deficient bacteria by helper cells expressing a functional plasmid transfer machinery (S. C. Winans, and G. C. Walker, J. Bacteriol. 161:402–410, 1985). Overexpression of TraC and its export in large amounts into the periplasm of Escherichia coli allowed purification by periplasmic extraction, ammonium sulfate precipitation, and column chromatography. Whereas TraC was soluble in overexpressing strains, it partly associated with the membranes in pKM101-carrying cells, possibly due to protein-protein interactions with other components of the TraI-TraII region-encoded transfer machinery. Membrane association of TraC was reduced in strains carrying pKM101 derivatives with transposon insertions in genes coding for other essential components of the transfer machinery, traM, traB, traD, and traE but not eex, coding for an entry exclusion protein not required for DNA transfer. Cross-linking identified protein-protein interactions of TraC in E. coli carrying pKM101 but not derivatives with transposon insertions in essential tra genes. Interactions with membrane-bound Tra proteins may incorporate TraC into a surface structure, suggested by its removal from the cell by shearing as part of a high-molecular-weight complex. Heterologous expression of TraC in A. tumefaciens partly compensated for the pilus assembly defect in strains deficient for its homolog VirB5, which further supported its role in assembly of conjugative pili. In addition to its association with high-molecular-weight structures, TraC was secreted into the extracellular milieu. Conjugation experiments showed that secreted TraC does not compensate transfer deficiency of TraC-deficient cells, suggesting that extracellular complementation may rely on cell-to-cell transfer of TraC only as part of a bona fide transfer apparatus.
Conjugative transfer of genetic information plays a major role in bacterial adaptation to changing environmental conditions, as exemplified by the rapid spread of antibiotic resistance markers and of determinants for detoxification of xenobiotic compounds (10, 44, 45). A better understanding of this natural process is necessary to devise strategies for environmental release of genetically modified microorganisms for sustainable ecosystem management, e.g., for detoxification of xenobiotics, biocontrol of plant pathogens, or enhanced nitrogen fixation in symbiotic bacterium-plant associations (18, 42, 49, 50). Conjugative plasmids are frequently used as tools for such applications, and analysis of their biology may allow construction of improved vectors.
Studies on plasmids from different incompatibility groups (F episome, IncF [13]; pKM101, IncN [32]; pRP1/4, IncP [26]; pR388, IncW [38]; and the Ti plasmid from Agrobacterium tumefaciens [27, 51]) reveal striking similarities in their transfer mechanisms. First, DNA processing involves several enzymes forming a relaxosome at the nicking site, with concomitant covalent attachment of the relaxase to the transferred DNA (25). Second, a family of proteins homologous to TraG from IncP plasmid RP4 may link the relaxosome to the membrane-bound transfer machinery; exchange of such linkage components between broad-host-range plasmids of different incompatibility groups illustrates their evolutionarily conserved function (7). Third, components of the transfer machineries were identified, and sequence analysis suggested export as well as membrane association (21, 26, 32, 39, 48). Sequence comparison revealed significant similarities between components from different plasmid transfer systems, suggesting an evolutionarily conserved mechanism for cell-to-cell trafficking of DNA-protein complexes (9, 27, 51, 55). Fourth, the transfer machineries determine assembly of pili, which presumably mediate cell-to-cell contact preceding DNA transfer and serve as binding sites for pilus-specific bacteriophages (3, 6, 12, 14, 15, 23, 28, 55).
With the exception of the F pilus, the assembly and composition of conjugative pili remained enigmatic until recently. TraA, the major subunit of the F pilus, shows similarity to components of several macromolecular transfer systems, predicted to exert similar roles (13, 16, 37). Minor pilus components, e.g., as tip-localized adhesins in P and CS1 pili (20, 34, 36), play important roles in adhesive pili, but so far, only indirect evidence suggests minor components in conjugative pili (1, 13). Compositional analyses of conjugative pili are needed to understand the molecular basis of cell-to-cell recognition and macromolecular transfer. VirB2 was recently identified as major constituent of the T pilus from A. tumefaciens (23), confirming earlier predictions of VirB2 as a major pilus subunit, based on its sequence similarity to the F pilus major subunit TraA (37).
pKM101 is a 35.4-kb incompatibility group N plasmid resulting from a natural deletion of resistance plasmid R46 originally isolated from Enterobacter cloacae (24). Expression of plasmid-encoded genes mucA and mucB increases sensitivity of plasmid-carrying strains to toxic chemicals, and pKM101 is included in Salmonella typhimurium strains used for the Ames test (30, 31). Conjugative transfer relies on the tra regions of pKM101, and 11 Tra proteins show significant sequence similarity to VirB1 to VirB11, components of the membrane-bound T-complex transfer machinery of the A. tumefaciens Ti plasmid. pKM101 derivatives carrying nonpolar transposon insertions in traC as well as in several other tra genes cannot undergo independent conjugative transfer. However, helper strains expressing a functional transfer machinery can partly compensate for the conjugative defect of insertions in traC but not in any other tra gene (8, 52). This extracellular complementation suggested an extracellular function of TraC, possibly as a pilus component, allowing its intercellular transfer to deficient strains (51).
This study aims to elucidate the function of TraC in conjugative plasmid transfer and the molecular basis of extracellular complementation. Biochemical analyses of E. coli carrying pKM101, and transposon insertions in essential tra genes demonstrated that membrane attachment of TraC is mediated by protein-protein interactions with other components of the plasmid transfer machinery. TraC was partly secreted but also colocalized with high-molecular-weight structures, which could be isolated from transfer-competent cells by shearing and high-speed centrifugation. Mating experiments showed that helper strains expressing an intact DNA transfer machinery partly compensated for the conjugative defect of strains carrying pKM101traC-insertion derivatives. However, external supply of large amounts of TraC did not exert such an effect, suggesting that TraC-mediated extracellular complementation requires its association with an intact plasmid transfer machinery.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used are listed in Table 1. E. coli FM433 and derivatives were grown in Luria-Bertani (LB) media supplemented with streptomycin (100 μg/ml), spectinomycin (100 μg/ml), ampicillin (100 μg/ml), chloramphenicol (20 μg/ml), and kanamycin (50 μg/ml) for plasmid propagation or selection of transconjugants. A. tumefaciens carrying pTrc200 and derivatives was grown on YEB media containing streptomycin (100 μg/ml) and spectinomycin (300 μg/ml) for plasmid propagation (2).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype or description | Reference |
|---|---|---|
| E. coli strains | ||
| JM109 | endA1 gyrA96 thi hsdR17 supE44 recA1 relA1 Δ(lac-proAB) (F′ traD36 proAB+ lacIqlacZΔM15), cloning host | 53 |
| FM433 | SpcraraD139 Δ(argF-lac)U169 ptsF25 deoC1 relA1 flbB5301 rpsE13 Δ(srl-recA)306::Tn10, conjugation donor | 54 |
| WL400 | Cmr StrraraD139 Δ(argF-lac)U169, ptsF25 deoC1 relA1 flbB5301 rpsL150 ΔselD204::cat, conjugation recipient | 25a |
| A. tumefaciens strains | ||
| C58 | Wild type, pTiC58 | 43 |
| CB1005 | pTiC58 carrying in-frame deletion of virB5 | This study |
| Plasmids | ||
| pGW2137 | Cmr, BglII fragment with TraI-TraII region from pKM101 cloned in pBR322 BamHI site | 52 |
| pGVO310 | Ampr, pBR322 derivative carrying the virA-virB region and part of virG from pTiC58 | 11 |
| pTrc200 | Strr, Spcr, pVS1 derivative, LacIq, trc promoter expression vector | This study |
| pTrcTraC | pTrc200, traC PCR fragment cloned downstream of the trc promoter | This study |
| pTrcB5 | pTrc200, virB5 PCR fragment cloned downstream of the trc promoter | This study |
| pBCSK+.Nde | Cmr, cloning vector pBCSK+ (Stratagene) carrying an NdeI restriction site at the first ATG codon of lacZ′ | 4 |
| pB56 | Cmr, pBCSK+.Nde carrying a 2.5-kb BamHI/HindIII fragment from pGVO310 carrying virB5 and virB6 | This study |
| pdelB56 | Cmr, pB56 carrying deletion of virB5 | This study |
| pBB50 | Cmr, pBCSK+.Nde carrying a 3-kb fragment determining resistance to kanamycin (nptII) and sensitivity to sucrose (sacB) cloned into the polylinker BamHI site | 4 |
| pBB50-delB5 | Kmr, pBB50 carrying a 2.5-kb virB5 deletion fragment from pdelB56 | This study |
| pKM101 | Ampr, mucA, mucB, TraI-TraIII region for DNA processing, DNA transfer, and entry exclusion | 24 |
| pKM101traL | Ampr, Kmr, pKM101traL53::Tn5 | 52 |
| pKM101traM | Ampr, Kmr, pKM101traM363::TnphoA | 52 |
| pKM101traB | Ampr, Kmr, pKM101traB1100::Tn5 | 52 |
| pKM101traC | Ampr, Kmr, pKM101traC1134::Tn5 | 52 |
| pKM101eex | Ampr, Kmr, pKM101eex1232::Tn5 | 52 |
| pKM101traD | Ampr, Kmr, pKM101traD1141::Tn5 | 52 |
| pKM101traE | Ampr, Kmr, pKM101traE1228::Tn5 | 52 |
A. tumefaciens vir genes were induced by growth in AB minimal medium (10 g of glucose, 4 g of morpholinoethanesulfonic acid (MES), 2 g of NH4Cl, 0.3 g of MgSO4 · 7H2O, 0.15 g of KCl, 0.01 g of CaCl2, and 0.0025 g of FeSO4 · 7H2O per liter, 1 mM potassium phosphate [pH 5.5]) at 20°C by the addition of acetosyringone at a final concentration of 200 μM. For isolation of pili, cells were induced for 3 or 4 days at 20°C on AB medium solidified with 2% agar and further processed as described elsewhere (23). For induction of the LacI-repressed trc promoter in pTrc200 constructs, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM.
Quantitation of conjugative DNA transfer.
Escherichia coli strains were grown in liquid LB at 37°C in the presence of antibiotics for plasmid propagation to an A600 of 0.8 to 1, sedimented by centrifugation, and resuspended in an appropriate volume of LB medium without antibiotics. Equal amounts of donor, recipient, and helper strain (10 μl of each, corresponding to 107 cells) were mixed on a prewarmed LB agar plate and incubated for 1 h at 37°C; the spot was washed from the plate three times with 300 μl of LB medium. To quantitate conjugative transfer, dilutions were plated on LB media containing appropriate antibiotics for selection of plasmid-containing recipients.
DNA manipulations.
DNA preparation, modification, and cloning were performed by standard procedures (29) using enzymes purchased from MBI Fermentas and New England Biolabs. The DNA sequence of PCR-amplified genes was confirmed by sequencing on an ABI Prism 377 sequencer.
pTrc200 was designed as a tool for IPTG-inducible expression of genes in a wide variety of gram-negative bacteria. The broad-host-range plasmid backbone of pPZP200 was combined with a region coding for the LacIq repressor and the trc promoter (fusion of trp and lac promoter) followed by a polylinker sequence and strong transcriptional terminators from the 5S rRNA operon of E. coli. pPZP200 derivative pBP2N was cleaved with Ecl136II and ScaI and ligated to a 2.6-kb ScaI/NdeI fragment from pTrc99A (Pharmacia), which had been treated with Klenow enzyme to generate blunt ends. Genes cloned into the NcoI site show strong IPTG-inducible expression from the trc promoter followed by an efficient Shine-Dalgarno sequence.
The TraC coding region was PCR amplified with Goldstar DNA polymerase (Eurogentec) from 1 ng of pGW2137 template, using oligonucleotides C5 (5′-GGGGCCATGGCAAAATCACTTACGGCAGT-3′) and C3 (5′-GAAAGTACTCAGTTAATTGAAGGTGA-3′) and the following cycle conditions: denaturation (one cycle) at 95°C for 2 min; 30 cycles at 44°C for 1 min, 72°C for 2 min, and 95°C for 30 s; strand completion (one cycle) at 44°C for 1 min and 72°C for 5 min; and termination at 4°C. The resulting 0.7-kb fragment was cleaved with NcoI and ScaI (underlined in sequences above) and ligated with NcoI/SmaI-cleaved pTrc200, resulting in plasmid pTrcTraC.
Similar conditions were used for PCR amplification of virB5 from plasmid target pGVO310; the fragment was then cleaved with AflIII and ScaI (underlined in sequence below) and ligated with NcoI/SmaI-cleaved pTrc200, resulting in plasmid pTrcB5. Oligonucleotides used for amplification were B55 (5′-CCACATGTCGATCATGCAACTTGTTGC-3′) and B53 (5′-GAAAGTACTCAGGGGACGGCCC-3′).
Construction of virB5 deletion strain CB1005.
Strain CB1005 carrying an in-frame deletion in the virB5 gene on the Ti plasmid of strain C58 was constructed as described by Berger and Christie (4) as follows. Briefly, a QuickChange site-directed mutagenesis kit (Stratagene) was used with oligonucleotides dB5-1 (5′-GATCAAAGGTGGGGAACTATGAATTTCACGATCCCGGCGC-3′) and dB5-2 (5′-GCGCCGGGATCGTGAAATTCATAGTTCCCCACCTTTGATC-3′) for deletion of virB5 in plasmid pB56. A fragment carrying the deletion was then excised from pdelB56 (BamHI/HindIII); overhanging ends were filled in with Klenow enzyme and then ligated with ScaI-cleaved pBB50, resulting in suicide vector pBB50delB5. The deletions were then introduced in the Ti plasmid. First, recombinant pBB50delB5 was transformed by electroporation into strain C58. The replication origin of pBB50 derivatives is nonfunctional in A. tumefaciens; selection of transformants for resistance to kanamycin (100 μg/ml on LB plates) therefore identifies strains carrying cointegrates formed via virB-homologous regions on their Ti plasmid. Second, several independent strains were grown in LB medium without antibiotics and then plated on LB agar containing 5% sucrose to select for loss of pBB50 carrying the sacB gene (expression of levan sucrase SacB is lethal on sucrose-containing medium) via a second recombination event. Western and Southern blotting identified those strains which had lost virB5 due to successive crossovers on either side of the deletion in pBB50delB5.
Overexpression, purification of TraC, and generation of antisera.
For overexpression of TraC, pTrcTraC-carrying strain JM109 was grown in 1 liter of liquid LB medium with streptomycin (50 μg/ml) and spectinomycin (50 μg/ml) at 37°C to late log phase; expression of the trc promoter was induced by addition of 0.2 mM IPTG followed by growth for 2.5 h under the same conditions. Cells were sedimented by centrifugation, washed in phosphate-buffered saline (8 g NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 per liter [pH 7.2]), and frozen at −20°C.
For periplasmic extraction, the cell sediment was suspended in TEX buffer (50 mM Tris-HCl [pH 8.0], 3 mM EDTA, 0.1% Triton X-100), incubated on ice for 30 min, and then subjected to centrifugation (41). The supernatant was subjected to differential ammonium sulfate precipitation; concentrations between 40 and 50% saturation precipitated the highest amounts of TraC. The pellet was resuspended in buffer A (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 0.5 mM dithiothreitol) containing 0.1% Triton X-100, dialyzed in 5 liters of buffer A, and applied to a Mono Q HR5/5 column (Pharmacia) in a Pharmacia FPLC system. Whereas most proteins bound to the column under these conditions, TraC was strongly enriched in the flowthrough containing only minor impurities. Gel filtration chromatography on a Superdex 75 column (Pharmacia) in buffer A was applied as final purification followed by dialysis in buffer A containing 50% glycerol and storage at −20°C. Proteins for column calibration were ferritin (450 kDa), aldolase (158 kDa), bovine serum albumin (68 kDa), chicken albumin (45 kDa), chymotrypsinogen (25 kDa), and cytochrome c (12.5 kDa) (Roche).
TraC-specific antisera were generated by injection of 500 μg of purified protein in rabbits following standard protocols of Eurogentec (Seraing, Belgium). Unspecific cross-reactions of the antisera were reduced by incubation with polyvinylidene difluoride membrane-fixed antigen and elution of specific antibodies in 15 mM NaOH according to standard procedures (19).
VirB2-specific antiserum was generated by injection of 500 μg of purified inclusion bodies of phage T7 gene 10 protein fused to amino acids 9 to 121 of VirB2 in New Zealand White rabbits for immunization as described previously (40).
SDS-PAGE and protein analysis.
Proteins in cell lysates were detected after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 10% acrylamide-containing gels (22) followed by Western blotting, incubation with TraC-specific polyclonal antisera, and detection with anti-rabbit horseradish peroxidase-conjugated secondary antibody (Bio-Rad), using a chemiluminescence-based detection system (NEN).
Subcellular fractionation and preparation of macromolecular surface structures.
E. coli strains carrying pKM101 and derivatives were grown in liquid culture in LB at 37°C to late log phase (A600 = 0.6 to 0.8); 1 ml of culture was plated on LB agar plates (diameter, 15 cm) and incubated at 28°C for 18 to 24 h. Subcellular fractions (total cell lysates, soluble proteins, and membrane fraction) were prepared in 50 mM potassium phosphate buffer, pH 7 (buffer N), followed by separation of inner and outer membrane by centrifugation through isopycnic sucrose gradients essentially as described previously (2). Extracellular macromolecular structures were removed from cells grown on LB agar plates by shearing in buffer N and were sedimented by high-speed centrifugation as described for the T pilus from A. tumefaciens (23). To assess the molecular weight of TraC-containing macromolecules, pellets obtained by high-speed centrifugation were suspended in 200 μl of buffer N and applied to a Superose 6 column (Pharmacia) for chromatography at a flow rate of 0.25 ml/min. Reference proteins for calibration of the column are indicated in the legend to Fig. 6.
FIG. 6.
Analysis of TraC-containing high-molecular-weight structures by gel filtration chromatography. Surface structures isolated from FM433/pKM101 (A) and FM433 (B) were subjected to gel filtration on a Superose 6 column. Shown is analysis of column fractions by SDS-PAGE followed by Western blotting with specific antiserum; the arrow indicates TraC in a high-molecular-weight complex. Molecular masses of reference proteins for calibration of the gel filtration column: F, ferritin (440 kDa); B, bovine serum albumin (68 kDa); C, cytochrome c (12 kDa).
Cross-linking.
To monitor protein-protein interactions in E. coli, cells were washed twice with buffer N and suspended in 500 μl of the same buffer followed by addition of bis(sulfosuccinimidyl) suberate (BS3; Pierce) to a final concentration of 1 mM and incubation for 1 h at 28°C. A. tumefaciens cells were treated similarly in 50 mM potassium phosphate buffer (pH 5.5), followed by addition of formaldehyde to a final concentration of 1% and incubation for 1 h at 20°C. Addition of 100 μl 1 M Tris-Cl (pH 6.8) to stop the reaction was followed by centrifugation, washing, and freezing at −20°C.
Image processing.
Gels and chemoluminographs were digitalized with a UMAX UC840 MaxVision scanner. Images were further processed on a Power Macintosh computer using Adobe Photoshop 3 software and printed on an Epson Stylus Photo printer.
RESULTS
Expression and purification of soluble TraC from the periplasm of E. coli.
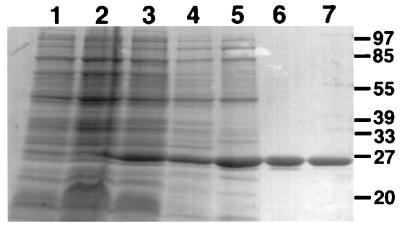
The traC coding sequence was PCR amplified and cloned behind the Shine-Dalgarno sequence of pTrc200, resulting in strong IPTG-inducible expression from the trc promoter in strains transformed with the resulting vector pTrcTraC (Fig. 1). Soluble TraC was released from cells by periplasmic extraction using different protocols as described by Thorstenson et al. (41), confirming its export into the periplasm predicted by its protein sequence. Triton X-100-containing buffer (TEX) was chosen as the most efficient method of extraction from the periplasm, and TraC was further purified by differential ammonium sulfate precipitation, anion-exchange chromatography, and Superdex 75 gel filtration chromatography (Fig. 1). Elution from the gel filtration column was compared to that of reference proteins, showing a molecular mass of 43 kDa for purified TraC. Since protein sequence analysis predicted a molecular mass of 26 kDa, confirmed by its mobility in SDS-PAGE, this may indicate an abnormal shape or purification of TraC as a dimer under nondenaturing conditions. Purified TraC was used for immunization of rabbits to generate antisera for further biochemical analyses of its function in plasmid transfer.
FIG. 1.
Purification of TraC. Coomassie-stained SDS-polyacrylamide gel showing steps leading to purification of TraC from an overexpressing strain. Lanes: 1 to 3, pTrcTraC-carrying strains JM109 before (lane 1) and after induction with IPTG for 45 (lane 2) and 90 min (lane 3); 4, supernatant resulting from extraction of the periplasm with Triton X-100-containing buffer; 5, 40 to 50% ammonium sulfate precipitation; 6, Mono Q anion-exchange chromatography; 7, Superdex 75 gel filtration chromatography. In all figures, positions of size standards are indicated in kilodaltons.
Components of the pKM101 transfer machinery confer membrane localization of TraC.
Sequence analysis predicted soluble as well as membrane-associated Tra components, which may associate via protein-protein interactions to form the plasmid transfer apparatus (32). For example, sequence analysis predicted that TraM may be a pilus component like VirB2 from A. tumefaciens. TraB may be an ATPase like VirB4 supplying energy for plasmid transfer or assembly of the transfer machinery. TraD, a hydrophobic protein containing several membrane-spanning domains like VirB6, may form the transfer pore (9). E. coli FM433 carrying pKM101 and derivatives with transposon insertions in traM, traB, traC, eex, traD, or traE were grown on LB agar plates and lysed in a French pressure cell, and subcellular fractions were analyzed for localization of TraC. FM433/pKM101eex, defective in entry exclusion but not in plasmid transfer (33), was included as control for a nonpolar Tn5 insertion.
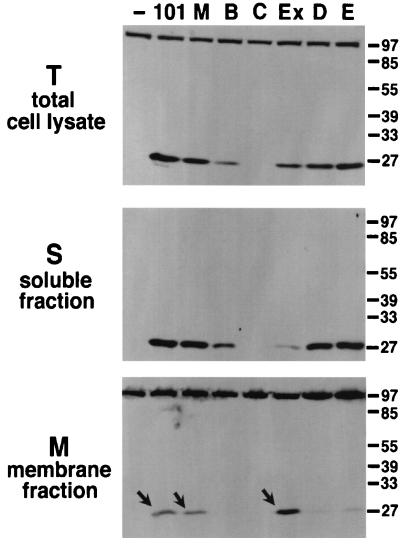
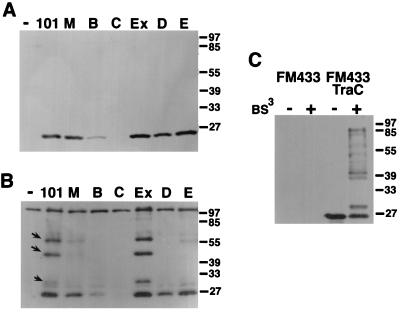
TraC was not detected in total cell lysates of FM433/pKM101traC, and its level was strongly reduced in FM433/pKM101traB, whereas all other strains contained TraC in amounts similar to those in the wild type (Fig. 2). High-speed centrifugation separated soluble and membrane fractions; membrane association of TraC was detected in FM433 carrying pKM101, pKM101eex, and pKM101traM, whereas it remained mostly soluble when other tra genes were disrupted (Fig. 2). Thus, the pKM101-encoded plasmid transfer machinery confers membrane association of TraC. Next, centrifugation through an isopycnic sucrose gradient was used to separate inner and outer membranes of pKM101-carrying cells. To assess the quality of the separation, measurement of NADH oxidase activity served to identify inner membrane fractions (not shown) and Coomassie dye stained porins characteristic for the outer membrane (Fig. 3A). Western blot analysis with TraC-specific antiserum showed preferential association of TraC with the inner membrane in pKM101-carrying cells (Fig. 3B).
FIG. 2.
TraC associates with the membranes in strains carrying a functional pKM101 transfer machinery. Lanes represent Western blot analysis with TraC-specific antiserum after SDS-PAGE of subcellular fractions from strain FM433 without plasmid (−) or carrying pKM101 (101) or derivatives with transposon insertions in genes encoding TraM (M), TraB (B), TraC (C), Eex (Ex), TraD (D), or TraE (E). Arrows indicate membrane-associated TraC.
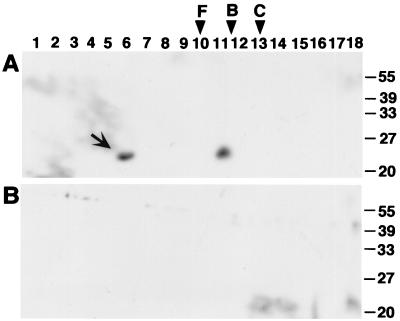
FIG. 3.
Sucrose gradient centrifugation shows preferential association of TraC with the inner membrane. Membranes from strains FM433/pKM101 and FM433 were subjected to centrifugation through isopycnic sucrose gradients, and fractions (1 through 16) were collected from the top of the gradient. Inner membrane-containing fractions were identified by NADH oxidase activity detected in fractions 1 through 5. (A) SDS-PAGE and Coomassie staining identified porins in the outer membrane-containing fractions. (B and C) Western blot analysis with TraC-specific antiserum after SDS-PAGE of fractions from FM433/pKM101 (B) and FM433 (C). The arrowhead indicates porins of the outer membrane, and the arrow indicates TraC.
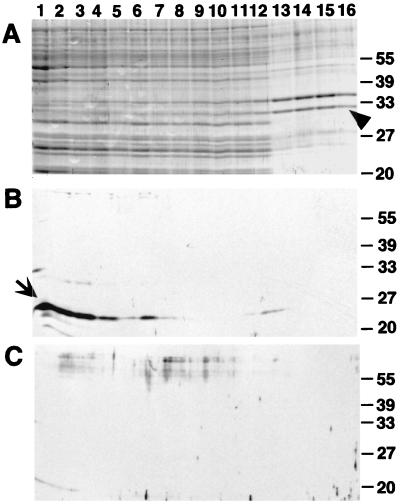
To detect protein-protein interactions of TraC, cells carrying pKM101 or transposon-inserted derivatives were incubated in the presence of the cross-linking agent BS3. Exposure to the cross-linking agent resulted in covalent linkage of TraC into complexes of higher molecular weight in cells carrying transfer-proficient plasmids pKM101 and pKM101eex. Formation of these complexes is strongly reduced or absent in transfer-deficient derivatives inserted in tra genes (Fig. 4B). Since steady-state levels of TraC were not affected by tra mutations except traB (Fig. 4A), cross-linking probably monitors specific interactions in a functional plasmid transfer complex. In contrast, in an overexpressing strain (FM433/pTrcTraC), cross-linking results in multiple TraC-containing complexes differing in molecular weight from the wild type, probably reflecting nonspecific associations in the periplasm (Fig. 4C).
FIG. 4.
Cross-linking identifies interactions of TraC with the pKM101 transfer machinery. Shown are Western blot analyses with TraC-specific antiserum after SDS-PAGE of total cell lysates of FM433 (lane −) and FM433 carrying pKM101 (lane 101) or derivatives with transposon insertions in traM, traB, traC, eex, traD, or traE (lanes B, C, Ex, D, and E, respectively) (A), total cell lysates of the same strains after cross-linking with BS3 (B), and cell lysates and cross-linked samples of FM433 and FM433/pTrcTraC (C). Arrows indicate higher-molecular-weight complexes formed after cross-linking of TraC.
Association of TraC with an extracellular macromolecular structure depends on a functional plasmid transfer machinery.
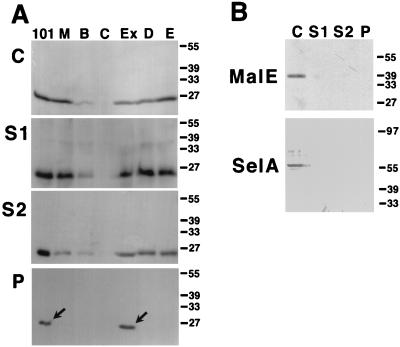
Extracellular complementation suggested that TraC may be a component of the pKM101-determined conjugative pilus (51, 52). To test this hypothesis, cells carrying pKM101 and mutant derivatives were grown on LB agar plates, and macromolecular surface structures were stripped from the cells by shearing through a needle (23, 35). The supernatant was subjected to high-speed centrifugation to sediment high-molecular-weight structures in the pellet, and TraC content of the different fractions was analyzed by SDS-PAGE and Western blotting. TraC was detected in total cell lysates and supernatants (after shearing and high-speed centrifugation) from all strains except negative control FM433/pKM101traC::Tn5 (Fig. 5A). High-speed centrifugation, however, sedimented TraC-containing macromolecules only from FM433 carrying pKM101 and pKM101eex::Tn5 (Fig. 5A). Analyses with periplasmic protein MalE-specific and cytoplasmic protein SelA-specific antisera showed that the above procedure does not release significant amounts of periplasmic or cytoplasmic proteins from E. coli (Fig. 5B). Thus, in strains carrying transfer-proficient pKM101 derivatives, TraC assembles into a high-molecular-weight structure.
FIG. 5.
TraC associates with an extracellular macromolecular structure in FM433/pKM101. Macromolecular surface structures were isolated from FM433 carrying pKM101 and mutant derivatives. Cells were grown on LB agar plates and subjected to shearing, and the resulting samples were analyzed by SDS-PAGE and Western blotting with TraC-specific antiserum. (A) C, Total cell lysates; S1, supernatants after shearing; S2, supernatants after high-speed centrifugation; P, pellets after high-speed centrifugation. TraC detected in the pellet fractions is indicated by arrows. Lanes are labeled as in Fig. 4. (B) Analysis for content of periplasmic and cytoplasmic proteins with MalE- and SelA-specific antisera.
The molecular weight of TraC-containing structures was next characterized by gel filtration chromatography. High-molecular-weight structures were isolated from strains FM433 and FM433/pKM101 as described above; pellets obtained after ultracentrifugation were suspended in buffer N and subjected to gel filtration on a Superose 6 column followed by SDS-PAGE. Western blot analysis revealed that TraC elutes in two fractions, indicating its association in complexes of different molecular weights (Fig. 6). Comparison with the elution volume of reference proteins demonstrates a molecular mass larger than 440 kDa (ferritin) for the TraC-containing complex detected in fraction 6. TraC detected in fraction 11, however, elutes from the Superose 6 column like TraC purified by TEX extraction from the periplasm (not shown).
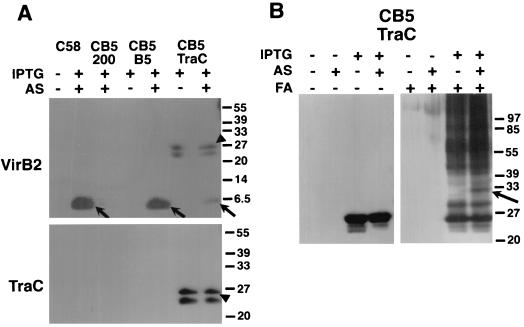
To further substantiate its role in pilus assembly, TraC was expressed in A. tumefaciens strain CB1005, which carries a deletion of the gene coding for its homolog VirB5 on the Ti plasmid and does not assemble VirB2-containing T pili on its surface (23). Strain CB1005 carrying VirB5- or TraC-expressing plasmids was grown on agar medium either in the presence of IPTG to induce plasmid-coded genes or in the presence of IPTG and acetosyringone to induce expression of plasmid-coded genes and vir genes, respectively. Surface structures were isolated as described previously (23) and monitored by SDS-PAGE followed by Western blotting with VirB2- and TraC-specific antisera. Extracellular pilus assembly of VirB2 was observed in vir-induced strain CB1005 expressing VirB5 or TraC, albeit at strongly reduced levels in the latter case (Fig. 7A), but the strain did not elicit tumors after wounding and infection of Kalanchoë diagremontiana (not shown). However, this finding raised the possibility of a specific interaction of TraC with VirB components leading to partial restoration of pilus formation. To test this possibility, formaldehyde was added to strain CB1005 expressing TraC alone or in the presence of Vir proteins to analyze its interactions with VirB components. Exposure to chemical cross-linking agent resulted in covalent association of TraC with higher-molecular-weight complexes, but a complex of 32 kDa is observed only in vir-induced cells, suggesting that TraC may interact with possibly one (or few) components of the VirB transmembrane machinery (Fig. 7B), thereby mediating assembly of VirB2 into the T pilus.
FIG. 7.
Extracellular pilus assembly of VirB2 in A. tumefaciens and cross-linking of cell-associated proteins suggest interaction of TraC with VirB components. (A) Pili were isolated from wild-type C58, virB5 deletion mutant CB1005 (CB5) carrying cloning vector pTrc200 (200), and CB1005 expressing VirB5 (B5) and TraC alone (+IPTG) or in the presence of Vir proteins (+AS [acetosyringone], +IPTG), and analyzed with VirB2- and TraC-specific antisera. Arrows indicate VirB2 content of extracellular high-molecular-weight structures. TraC is secreted in large amounts of pTrcTraC-carrying cells, partly associates with the pellets obtained after high-speed centrifugation of surface structures, and is also detected with VirB2-specific antiserum (arrowheads). (B) Cells of strain CB1005 carrying pTrcTraC were grown on AB agar plates without inducer, in the presence of IPTG or IPTG and acetosyringone (AS), followed by cross-linking with 1% formaldehyde (FA) and analysis of TraC content with specific antiserum.
TraC is a secreted protein in E. coli and A. tumefaciens.
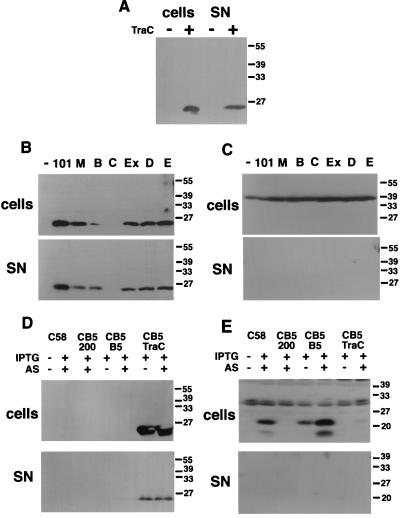
TraC-overexpressing liquid cultures accumulate TraC in the supernatant (Fig. 8A), but this may be caused by periplasmic leakage in strains expressing TraC at nonphysiological levels. We next analyzed TraC secretion in liquid-grown FM433 carrying pKM101 or its mutant derivatives. TraC was detected in concentrated culture supernatants of all strains, and the ratio of cell-bound to secreted protein was approximately equal (Fig. 8B). As a control for periplasmic leakage, we monitored localization of MalE, which was detected exclusively in cell lysates (Fig. 8C). Thus, TraC is partly secreted from E. coli independent of a functional plasmid transfer machinery. Gel filtration chromatography determined a molecular mass of 43 kDa for periplasmic and secreted TraC (not shown), implying that secretion of TraC in liquid-grown cells does not lead to its incorporation into a high-molecular-weight structure.
FIG. 8.
Secretion of TraC in E. coli and A. tumefaciens. Cells were grown in liquid medium to late logarithmic growth phase and sedimented by centrifugation followed by trichloroacetic acid precipitation of secreted proteins. Cell-bound and secreted proteins (fivefold concentrated) were subjected to SDS-PAGE and Western blotting with TraC-specific (A, B, and D), MalE-specific (C), and VirB5-specific (E) antisera. (A) E. coli FM433 (lanes −) and FM433/pTrcTraC (lanes +); (B and C) FM433 (lanes −) and FM433 carrying pKM101 (lanes 101) and derivatives pKM101traM, pKM101traB, pKM101traC, pKM101eex, pKM101traD, and pKM101traE (remaining lanes, left to right); (D and E) A. tumefaciens wild-type C58 and virB5 deletion strain CB1005 (CB5) carrying pTrc200, pTrcB5, and pTrcTraC grown in the presence of IPTG in vir gene-inducing (+AS [acetosyringone]) or noninducing (−AS) conditions. SN, supernatant.
Secretion of TraC was analyzed in A. tumefaciens and compared to that of its homolog VirB5. Wild-type strain C58, virB5 deletion strain CB1005, and CB1005 transformed with TraC- and VirB5-expressing plasmids pTrcTraC and pTrcB5 were grown in liquid AB medium in virulence gene-inducing or noninducing conditions. Cells were sedimented followed by analysis of cell-bound proteins and supernatants for VirB5 and TraC. As in E. coli, TraC was detected in cells and supernatants of TraC-expressing agrobacteria in the presence and in the absence of virulence gene induction (Fig. 8D). In contrast, VirB5 was detected exclusively in cells (Fig. 8E).
A functional plasmid transfer machinery is necessary for extracellular complementation of TraC defects.
TraC undergoes partial secretion, suggesting that extracellular complementation may rely on external supply of soluble TraC from the helper strain, allowing assembly of the conjugative pilus of a TraC-deficient recipient. Mating experiments were performed to directly assess this possibility. Donor and recipient (and sometimes a third helper strain) from liquid-grown cultures were mixed and incubated on LB agar without antibiotics for 1 h and washed from the plates, and different dilutions were plated on LB agar containing antibiotics for selection of transconjugants (Table 2). Conjugative transfer of pKM101 from donor FM433 to recipient WL400 was 109-fold more efficient than that of pKM101traC. Extracellular complementation by helper cells (pGW2137) carrying the TraI-TraII region encoding plasmid transfer but not DNA processing functions from pKM101 inserted in pACYC184 (52), resulted in 400-fold more efficient conjugative transfer of pKM101traC. In contrast, cells carrying pKM101traM or pKM101traD could not exert helper function, showing that an intact plasmid transfer machinery is required for extracellular complementation.
TABLE 2.
Compensation of TraC deficiency by extracellular complementation
| Donor, FM433 carrying: | Helper, FM433 carrying: | Recipient | Transconjugants/ donor · ha |
|---|---|---|---|
| pKM101 | WL400 | 1.1 × 10−1 | |
| pKM101traC::Tn5 | WL400 | ≤10−10 | |
| pKM101traC::Tn5 | pGW2137 | WL400 | 3.7 × 10−8 |
| pKM101traC::Tn5 | pACYC184 | WL400 | ≤10−10 |
| pKM101traC::Tn5 | pKM101traM::TnphoA | WL400 | ≤10−10 |
| pKM101traC::Tn5 | pKM101traD::Tn5 | WL400 | ≤10−10 |
| pKM101traC::Tn5 | pTrc200 | WL400 | ≤10−10 |
| pKM101traC::Tn5 | pTrcTraC | WL400 | ≤10−10 |
| pKM101traC::Tn5 | WL400 pTrc200 | ≤10−10 | |
| pKM101traC::Tn5 | WL400 pTrcTraC | ≤10−10 |
Results from two to five independent experiments.
These results argue against a role of secreted TraC in extracellular complementation, as insertions in traM and traD affect plasmid transfer and presumably pilus assembly but not steady-state levels and secretion of TraC (see above). Further experiments were performed to supply large amounts of external TraC to stimulate pKM101traC transfer (Table 2). First, when donor FM433/pKM101traC and recipient WL400 were mixed with TraC-overproducing (and secreting) helper strain FM433/ pTrcTraC on a plate, there was no effect on conjugative transfer of pKM101traC. Second, pTrcTraC was introduced into WL400 to analyze whether overexpression of TraC in the recipient promotes conjugative transfer. TraC overexpression failed to increase conjugative transfer in this experiment as well. Third, purified TraC (0.1, 1, and 10 ng [equivalent to 500 to 50,000 molecules per donor cell]) or TraC-containing high-molecular-weight structures isolated by shearing and ultracentrifugation were added to FM433 pKM101traC and recipient WL400 on a plate, but changes in the efficiency of conjugative transfer were not observed (not shown).
DISCUSSION
Cells expressing the pKM101 transfer machinery partly compensate for conjugative defects of cells carrying transposon insertions in the traC gene, a phenomenon termed extracellular complementation (52). It was suggested that TraC may localize at the cell exterior, e.g., as a pilus component, allowing transfer to the deficient strain and incorporation into its plasmid transfer machinery (51). Here, the basis of this complementation was analyzed in detail.
By analogy to the T-complex transfer machinery from A. tumefaciens, the TraI-TraII region-coded products from pKM101 were predicted to form a membrane-associated plasmid trafficking complex. Indeed, here we show that these predictions hold true. Whereas expression of TraC in the absence of other TraI-TraII region gene products resulted in a soluble periplasmic protein, it associated with the membranes in pKM101- and pKM101eex-carrying bacteria, suggesting protein-protein interactions with membrane-bound components of functional transfer machineries. Membrane association was also detected in strains carrying transfer-deficient pKM101traM. TraM may not be required for membrane attachment of TraC but exert other functions in plasmid transfer, e.g., as a pilus component (32). Membrane association of TraC was not observed in cells carrying pKM101 with transposon insertions located in traB, traD, and traE, implying involvement of their gene products in assembly or stabilization of the plasmid transfer complex. Interestingly, steady-state levels of TraC were strongly reduced in pKM101traB-carrying cells, similar to effects of deletions in some virB genes on the stability of the T-complex transfer machinery (4), but transcriptional polarity due to transposon insertion in the upstream gene may also account for this phenomenon.
The different Tra proteins probably exert specialized functions in assembly and/or stabilization of the membrane-bound plasmid transfer complex and conjugative pilus. Cross-linking directed TraC to high-molecular-weight complexes in E. coli carrying transfer-proficient but not tra-defective plasmids. The lack of cross-linking in strains carrying transposon-inserted pKM101 derivatives, probably reflecting misassembly or destabilization of the plasmid transfer machinery, correlates well with their deficiency in conjugative transfer. Cross-linking of TraC may therefore constitute a biochemical assay for assembly of a functional plasmid transfer complex, which will be useful for further analyses of Tra protein function(s).
Compositional analysis of the virulence pilus from A. tumefaciens recently identified VirB2 as its major constituent (23). A similar approach was pursued here to isolate components of extracellular macromolecular structures in E. coli carrying pKM101 or its transfer-deficient derivatives. TraC proved to be a component of a high-molecular-weight structure, which could be isolated from the cells by shearing, and transposon insertion in any of the tra genes abolished its assembly. Gel filtration chromatography confirmed the solubility of a high-molecular-weight TraC-containing complex whose molecular weight was larger than that of the reference protein ferritin (440 kDa). In addition, TraC eluted from the gel filtration column at a position corresponding to that of TraC purified from the periplasm. This may be due to disassembly of the high-molecular-weight complex or contamination of the pellet fraction applied to the column with soluble TraC from the supernatant. To further assess a function of TraC in pilus biogenesis, expression was performed in an A. tumefaciens strain defective for its homolog VirB5, which does not form pili (23). Heterologous expression of TraC partly restored external assembly of VirB2 into the T pilus, and cross-linking suggested that TraC may interact with Vir proteins, thereby substituting VirB5 in T pilus assembly. Thus, TraC is partly functional in a well-defined heterologous system, indicating a role in pilus assembly. Further analyses of the composition of TraC-containing high-molecular-weight structures are necessary to assess whether TraC is a component of the pKM101-determined pilus. Alternatively, TraC could be part of a surface-exposed pilus assembly complex which mediates extracellular polymerization of VirB2-homologous protein TraM to form the conjugative pilus.
In spite of the obvious similarities between the A. tumefaciens- and pKM101-coded transfer systems, the mechanisms of pilus assembly may differ. Whereas its homolog VirB5 from A. tumefaciens is a cell-bound protein, TraC was partly secreted independently of the presence of other components of the pKM101 transfer machinery. Possibly, assembly of the conjugative pilus involves a secreted intermediate of TraC. The assembly mechanism may therefore resemble that of adhesive curli involving secretion of CsgA (curlin) subunits and their extracellular assembly into a pilus mediated by outer membrane-localized nucleator protein CsgB (5). Curli assembly in csgA-mutant strains can be complemented intercellularly by CsgA-secreting helper strains (17), and a similar mechanism may explain extracellular complementation. We directly addressed the possibility of pilus assembly mediated by an external pool of TraC in conjugation experiments with TraC-secreting and overproducing helper and recipient strains or by external addition of large amounts of purified TraC. However, conjugative transfer of pKM101traC was never rescued, indicating that the above model for TraC-mediated pilus assembly is probably not correct. Only helpers expressing an intact plasmid transfer machinery can serve as donors in extracellular complementation. Extracellular complementation may therefore bear similarity to transfer of pilus phenotype in Myxococcus xanthus where social gliding defects of tgl mutants are compensated, presumably by cell-to-cell transfer of type IV pilus components or a pilus assembly protein (46, 47). Similarly, cell-to-cell contact may allow transfer of fragments of the pKM101 pilus from helper to pKM101traC-carrying cells, thereby partly restoring their ability for plasmid transfer. Future studies will address the role of TraC and other Tra proteins either as structural components of the pKM101-coded pilus or as pilus assembly factors to unravel the mechanism of cell-cell recognition during bacterial conjugation.
ACKNOWLEDGMENTS
We thank Peter Christie and Stephen C. Winans for gifts of strains, phages, and plasmids, Michael Ehrmann for donation of MalE-specific antiserum, and August Böck for support, discussions, and donation of SelA-specific antiserum. We are indebted to Bernhard Neuhierl for advice during protein purification and P. C. Zambryski for helpful comments on the manuscript.
This study was supported by grant BA 1416/2-1 from the Deutsche Forschungsgemeinschaft to C.B.
REFERENCES
- 1.Anthony K G, Sherbourne C, Sherburne R, Frost L S. The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F-like plasmids. Mol Microbiol. 1994;13:939–953. doi: 10.1111/j.1365-2958.1994.tb00486.x. [DOI] [PubMed] [Google Scholar]
- 2.Baron C, Llosa M, Zhou S, Zambryski P C. C-terminal processing and cellular localization of VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens. J Bacteriol. 1997;179:1203–1210. doi: 10.1128/jb.179.4.1203-1210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron C, Zambryski P C. Plant transformation: a pilus in Agrobacterium T-DNA transfer. Curr Biol. 1996;6:1567–1569. doi: 10.1016/s0960-9822(02)70773-2. [DOI] [PubMed] [Google Scholar]
- 4.Berger B R, Christie P J. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bian Z, Normark S. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 1997;16:5827–5836. doi: 10.1093/emboj/16.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley D E. Morphology of pili determined by the N incompatibility group plasmid N3 and interaction with bacteriophages PR4 and IKe. Plasmid. 1979;2:632–636. doi: 10.1016/0147-619x(79)90061-1. [DOI] [PubMed] [Google Scholar]
- 7.Cabezon E, Sastre J I, de la Cruz F. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet. 1997;254:400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- 8.Cellini C, Kalogeraki V S, Winans S C. The hydrophobic TraM protein of pKM101 is required for conjugal transfer and sensitivity to donor-specific bacteriophage. Plasmid. 1997;37:181–188. doi: 10.1006/plas.1997.1291. [DOI] [PubMed] [Google Scholar]
- 9.Christie P J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies J. Bacteria on the rampage. Nature. 1996;383:219–220. doi: 10.1038/383219a0. [DOI] [PubMed] [Google Scholar]
- 11.Depicker A, DeWilde M, De Vos G, De Vos R, Van Montagu M, Schell J. Molecular cloning of overlapping segments of the nopaline Ti plasmid of pTiC58 as a means to restriction endonuclease mapping. Plasmid. 1980;31:193–211. doi: 10.1016/0147-619x(80)90109-2. [DOI] [PubMed] [Google Scholar]
- 12.Frost L S. Conjugative pili and pilus-specific phages. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 189–221. [Google Scholar]
- 13.Frost L S, Ippen-Ihler K, Skurray R A. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev. 1994;58:162–210. doi: 10.1128/mr.58.2.162-210.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fullner K J, Lara J L, Nester E W. Pilus assembly by Agrobacterium T-DNA transfer genes. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 15.Grahn A M, Haase J, Lanka E, Bamford D H. Assembly of a functional phage PRD1 receptor depends on 11 genes of the IncP plasmid mating pair formation complex. J Bacteriol. 1997;179:4733–4740. doi: 10.1128/jb.179.15.4733-4740.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haase J, Lanka E. A specific protease encoded by the conjugative DNA transfer systems of IncP and Ti plasmids is essential for pilus synthesis. J Bacteriol. 1997;179:5728–5735. doi: 10.1128/jb.179.18.5728-5735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handelsmann J, Stabb E V. Biocontrol of soilborne plant pathogens. Plant Cell. 1996;8:1855–1869. doi: 10.1105/tpc.8.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harlow E, Lane D, editors. Antibodies: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 20.Hultgren S J, Abraham S, Caparon M, Falk P, St. Geme III J W, Normark S. Pilus and nonpilus bacterial adhesins: assembly and function in cell recognition. Cell. 1993;73:887–901. doi: 10.1016/0092-8674(93)90269-v. [DOI] [PubMed] [Google Scholar]
- 21.Kuldau G A, de Vos G, Owen J, McCaffrey G, Zambryski P C. The virB operon of Agrobacterium tumefaciens pTiC58 encodes 11 open reading frames. Mol Gen Genet. 1990;221:256–266. doi: 10.1007/BF00261729. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lai E-M, Kado C I. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J Bacteriol. 1998;180:2711–2717. doi: 10.1128/jb.180.10.2711-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langer P J, Walker G C. Restriction endonuclease cleavage map of pKM101: relationship to parental plasmid R46. Mol Gen Genet. 1981;182:268–272. doi: 10.1007/BF00269669. [DOI] [PubMed] [Google Scholar]
- 25.Lanka E, Wilkins B M. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–69. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 25a.Leinfelder, W. Unpublished data.
- 26.Lessl M, Balzer D, Pansegrau W, Lanka E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J Biol Chem. 1992;267:20471–20480. [PubMed] [Google Scholar]
- 27.Lessl M, Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994;77:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 28.Llosa M, Zambryski P C. On the origin and function of pili. Trends Microbiol. 1998;6:98–99. doi: 10.1016/s0966-842x(98)01222-0. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis T A, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 30.McCann J, Springar N E, Kobori J, Ames B N. Detection of carcinogens as mutagens: bacterial tester strains with R factor plasmids. Proc Natl Acad Sci USA. 1975;79:979–983. doi: 10.1073/pnas.72.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortelmanns K E, Stocker B A D. Ultraviolet light protection, enhancement of ultraviolet light mutagenesis, and mutator effect of plasmid R46 in Salmonella typhimurium. J Bacteriol. 1976;128:271–282. doi: 10.1128/jb.128.1.271-282.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pohlman R F, Genetti H D, Winans S C. Common ancestry between IncN conjugal transfer genes and macromolecular export systems of plant and animal pathogens. Mol Microbiol. 1994;14:655–668. doi: 10.1111/j.1365-2958.1994.tb01304.x. [DOI] [PubMed] [Google Scholar]
- 33.Pohlman R F, Genetti H D, Winans S C. Entry exclusion of the IncN plasmid pKM101 is mediated by a single hydrophilic protein containing a lipid attachment motif. Plasmid. 1994;31:158–165. doi: 10.1006/plas.1994.1017. [DOI] [PubMed] [Google Scholar]
- 34.Riegman N, Hoschützky H, van Die I, Hoekstra W, Jann K, Bergmanns H. Immunocytochemical analysis of P-fimbral structure: localization of minor subunits and the influence of the minor subunit FsoE on the biogenesis of the adhesin. Mol Microbiol. 1990;4:1193–1198. doi: 10.1111/j.1365-2958.1990.tb00694.x. [DOI] [PubMed] [Google Scholar]
- 35.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E L, Kalkkinen N, Romantschuk M, He S Y. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakellaris H, Balding D P, Scott J R. Assembly proteins of CS1 pili of enterotoxigenic Escherichia coli. Mol Microbiol. 1996;21:529–541. doi: 10.1111/j.1365-2958.1996.tb02562.x. [DOI] [PubMed] [Google Scholar]
- 37.Shirasu K, Kado C I. Membrane localization of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like structure in Agrobacterium tumefaciens. FEMS Microbiol Lett. 1993;111:287–294. doi: 10.1111/j.1574-6968.1993.tb06400.x. [DOI] [PubMed] [Google Scholar]
- 38.Shirasu K, Kado C I. The VirB operon of the Agrobacterium tumefaciens virulence regulon has sequence similarities to B, C and D open reading frames downstream of the pertussis toxin-operon and to the DNA transfer-operons of broad-host-range conjugative plasmids. Nucleic Acids Res. 1993;21:353–354. doi: 10.1093/nar/21.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shirasu K, Morel P, Kado C I. Characterization of the virB operon of an Agrobacterium tumefaciens Ti plasmid: nucleotide sequence and protein analysis. Mol Microbiol. 1990;4:1153–1163. doi: 10.1111/j.1365-2958.1990.tb00690.x. [DOI] [PubMed] [Google Scholar]
- 40.Thorstenson Y R, Kuldau G A, Zambryski P C. Subcellular localization of seven VirB proteins of Agrobacterium tumefaciens: implications for the formation of a T-DNA transport structure. J Bacteriol. 1993;175:5233–5241. doi: 10.1128/jb.175.16.5233-5241.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorstenson Y R, Zhang Y, Olson P S, Mascarenhas D. Leaderless polypeptides efficiently extracted from whole cells by osmotic shock. J Bacteriol. 1997;179:5333–5339. doi: 10.1128/jb.179.17.5333-5339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timmis K N, Steffan R J, Unterman R. Designing microorganisms for the treatment of toxic wastes. Annu Rev Microbiol. 1994;48:525–557. doi: 10.1146/annurev.mi.48.100194.002521. [DOI] [PubMed] [Google Scholar]
- 43.van Larebeke N, Engler G, Holsters M, van den Elsacker S, Zaenen I, Schilperoort R A, Schell J. Large plasmids in Agrobacterium tumefaciens essential for crown gall-inducing ability. Nature. 1974;252:169–170. doi: 10.1038/252169a0. [DOI] [PubMed] [Google Scholar]
- 44.Van Overbeck L, Van Elsas J D. Adaptation of bacteria to soil conditions: applications of molecular physiology in soil microbiology. In: van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker; 1997. pp. 441–477. [Google Scholar]
- 45.van Veen J A, van Overbeek L S, van Elsas J D. Fate and activity of microorganisms introduced into soil. Microbiol Mol Biol Rev. 1997;61:121–135. doi: 10.1128/mmbr.61.2.121-135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wall D, Kaiser D. Alignment enhances the cell-to-cell transfer of pilus phenotype. Proc Natl Acad Sci USA. 1998;95:3054–3058. doi: 10.1073/pnas.95.6.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wall D, Wu S S, Kaiser D. Contact stimulation of Tgl and type IV pili in Myxococcus xanthus. J Bacteriol. 1998;180:759–61. doi: 10.1128/jb.180.3.759-761.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward J E, Akiyoshi D E, Regier D, Datta A, Gordon M P, Nester E W. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid. J Biol Chem. 1990;265:4768. [PubMed] [Google Scholar]
- 49.Whipps J M. Ecological considerations involved in commercial development of biological control agents for soil-borne diseases. In: van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker; 1997. pp. 525–546. [Google Scholar]
- 50.Wilson M, Lindow S E. Release of recombinant microorganisms. Annu Rev Microbiol. 1993;47:913–44. doi: 10.1146/annurev.mi.47.100193.004405. [DOI] [PubMed] [Google Scholar]
- 51.Winans S C, Burns D L, Christie P J. Adaption of a conjugal system for the export of pathogenic macromolecules. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winans S C, Walker G C. Conjugal transfer system of the N incompatibility plasmid pKM101. J Bacteriol. 1985;161:402–410. doi: 10.1128/jb.161.1.402-410.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanisch-Perron C, Viera J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC18 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 54.Zinoni F, Heider J, Böck A. Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc Natl Acad Sci USA. 1990;87:4660–4664. doi: 10.1073/pnas.87.12.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zupan J R, Ward D, Zambryski P C. Assembly of the VirB transport complex for DNA transfer from Agrobacterium tumefaciens to plant cells. Curr Opin Microbiol. 1998;1:649–655. doi: 10.1016/s1369-5274(98)80110-0. [DOI] [PubMed] [Google Scholar]