Abstract
The two transducers in the phototaxis system of the archaeon Halobacterium salinarum, HtrI and HtrII, are methyl-accepting proteins homologous to the chemotaxis transducers in eubacteria. Consensus sequences predict three glutamate pairs containing potential methylation sites in HtrI and one in HtrII. Mutagenic substitution of an alanine pair for one of these, Glu265-Glu266, in HtrI and for the homologous Glu513-Glu514 in HtrII eliminated methylation of these two transducers, as demonstrated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis autofluorography. Photostimulation of the repellent receptor sensory rhodopsin II (SRII) induced reversible demethylation of HtrII, while no detectable change in the extent of methylation of HtrI was observed in response to stimulation of its cognate sensory rhodopsin, the attractant receptor SRI. Cells containing HtrI or HtrII with all consensus sites replaced by alanine still exhibited phototaxis responses and behavioral adaptation, and methanol release assays showed that methyl group turnover was still induced in response to photostimulation of SRI or SRII. By pulse-chase experiments with in vivo l-[methyl-3H]methionine-labeled cells, we found that repetitive photostimulation of SRI complexed with wild-type (or nonmethylatable) HtrI induced methyl group turnover in transducers other than HtrI to the same extent as in wild-type HtrI. Both attractant and repellent stimuli cause a transient increase in the turnover rate of methyl groups in wild-type H. salinarum cells. This result is unlike that obtained with Escherichia coli, in which attractant stimuli decrease and repellent stimuli increase turnover rate, and is similar to that obtained with Bacillus subtilis, which also shows turnover rate increases regardless of the nature of the stimulus. We found that a CheY deletion mutant of H. salinarum exhibited the E. coli-like asymmetric pattern, as has recently also been observed in B. subtilis. Further, we demonstrate that the CheY-dependent feedback effect does not require the stimulated transducer to be methylatable and operates globally on other transducers present in the cell.
In the archaeon Halobacterium salinarum, phototaxis is mediated by two photoreceptive protein complexes, sensory rhodopsin I (SRI)-HtrI and sensory rhodopsin II (SRII)-HtrII. SRI is a photoreceptor for attractant (orange) and repellent (near-UV) stimuli, and SRII is a photoreceptor for repellent (blue-green) stimuli (8). HtrI (35) and HtrII (37) are integral membrane proteins bound to SRI and SRII, respectively. The Htr proteins belong to a family of transducer proteins that are highly homologous in their cytoplasmic domains. The best studied are the methyl-accepting chemotaxis receptor-transducers (MCPs) of Escherichia coli and Salmonella typhimurium (5, 30). As in the enteric eubacteria, the conserved cytoplasmic domains control a two-component regulatory system with the histidine kinase CheA and its protein substrate, the response regulator CheY (20).
In the chemotaxis system of E. coli, behavioral adaptation is correlated with the reversible methylation of the carboxyl side chains of glutamyl residues (carboxylmethylesterification) (10, 11, 19, 29, 32). A positive stimulus (i.e., addition of an attractant or removal of a repellent) causes an increase in the level of methylation, and a negative stimulus causes a reduction in the number of methylated residues on the MCP protein through which the stimulus signal is transmitted. Changes in the steady-state methylation of MCPs upon stimulation have been detected as changes in their migration rate determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (2, 6). The rate of methyl group turnover in vivo has been measured as the release of volatile radioactive methyl groups by cells since a product of carboxylmethyl glutamate hydrolysis is methanol (10). Genes encoding a methyl esterase, CheB (20), and a methyltransferase, CheR (9a), have been identified in H. salinarum.
The methylation system in H. salinarum is similar in some respects to that of enteric bacteria and in others to the gram-positive B. subtilis. As in E. coli, methionine starvation eliminates swimming direction reorientation (tumbling in E. coli and swimming reversals in H. salinarum) and tactic responses (27). A cheB gene deletion mutant in H. salinarum has been shown to exhibit a phenotype (frequent swimming reversals) similar to that of the corresponding mutant in E. coli (21), and loss of methylation attributed to a methyltransferase (CheR) mutation results in a smooth swimming phenotype as in E. coli (24, 31). Unlike E. coli, but as for B. subtilis (12), H. salinarum cells undergo increased turnover of methyl groups after both a positive and a negative stimulus when exposed to both chemical and light stimuli (1, 25). Increases in the extent of methylation on gels following chemostimulation with histidine and leucine (attractants) and decreases after chemostimulation with phenol (repellent) in cells labeled to steady state have been observed in H. salinarum (1). Changes in methylation upon photostimulation were detected in HtrII in the work reported here.
In this study, we have investigated the nature of methylation changes in the response to light stimuli in H. salinarum. We identified the sites of methylation in HtrI and HtrII and found similarities to and differences from the methylation system in E. coli chemotaxis.
MATERIALS AND METHODS
Recipient strain and plasmids.
The recipient strain for plasmid transformation was Pho81Wr− (bacteriorhodopsin− halorhodopsin− SRI− SRII− HtrI− HtrII−; carotenoid deficient and lacking a restriction activity) (36). The plasmid pKJ306 is a shuttle vector able to replicate in both H. salinarum and E. coli (9, 15), and genes encoding SRI apoprotein and HtrI were expressed from their native promoter. A modified form of the same plasmid, the expression vector pPR5, was used to express genes encoding SRII and HtrII under the control of the htrI promoter as described previously (26).
PCR mutagenesis.
Mutations introduced into the sequences of HtrI and HtrII were created by PCR site-directed mutagenesis (3). A 759-bp SacI/XhoI fragment from the native htrI gene containing the sites to be mutated was cloned in pBluescript KS− (Stratagene, La Jolla, Calif.), and the resulting plasmid was used as a template for PCR. Similarly, a 659-bp NotI/SmaI fragment from the native htrII gene cloned in pBluescript KS− was used to make the mutations in this gene. Mutagenized fragments were reintroduced in the respective original expression plasmids. The T3 and T7 promoters and synthetic oligonucleotides (Bioserve, Laurel, Md.) were used to introduce the following pairs of mutations: Q258A-Q259A, E265A-E266A, E315A-Q316A, E336A-Q337A, E485A-E486A, E265A, and E266A in HtrI and E513A-E514A in HtrII. The triple mutant in HtrI contains the mutation pairs Q258A-Q259A, E265A-E266A, and E485A-E486A. It was made by mutating each pair in three different PCR mutagenesis cycles. Pfu DNA polymerase (Stratagene) was used for PCR amplification. The mutations introduced in HtrI and HtrII were confirmed by DNA sequencing.
Motion analysis.
A computerized cell-tracking system (Motion Analysis, Santa Rosa, Calif.) was used to monitor the swimming behavior of wild-type and mutant H. salinarum strains (28). Cells were grown to early stationary phase, diluted 1:10 in fresh complete medium (CM), and incubated for 1 h at 37°C with shaking. Cells containing SRI-HtrI were stimulated with 600-nm attractant light in an infrared background and white light for repellent stimulation. Swimming reversals in response to a 4-s step down in this light were recorded and analyzed on an SPARC-IPC workstation (Sun Microsystems, Mountain View, Calif.). The SRII-HtrII photostimulus consisted of 9 s of illumination with a 500-nm light. Photostimuli were delivered from a Nikon 100-W Hg-Xe lamp beam with appropriate 40-nm band pass interference filters (Corion, Holliston, Mass.).
Autofluorography and immunoblotting.
Cells were labeled in vivo with l-[methyl-3H]methionine in the presence of puromycin as described previously (24). The absolute amount of l-[methyl-3H]methionine uptake varied in different cultures, but the relative amounts of label of specific bands corresponding to specific methyl-accepting proteins was reproducible. Cells were precipitated in acetone by the protocol described by Spudich and Spudich (23) except that the drying of the samples was done with a Speed-Vac (Savant Instruments, Inc., Farmingdale, N.Y.). Acetone-precipitated cells or membrane proteins were separated by SDS-PAGE (16) and analyzed by autofluorography of dried gels. Immunoblot analysis was performed by using a polyclonal antibody to the signaling domain of halobacterial transducers (HC23 antibody) as described previously (38).
Assay for volatile [3H]methyl group production.
Cells radiolabeled as described above and immobilized on a 0.45-μm-pore-size nitrocellulose filter (Nalgene, Rochester, N.Y.) were used in accordance with the same protocol as described previously (25), except a flow rate of 1 ml/min was used and the void volume of the filter and outlet tubing was 0.4 ml.
Pulse-chase methyl turnover experiments.
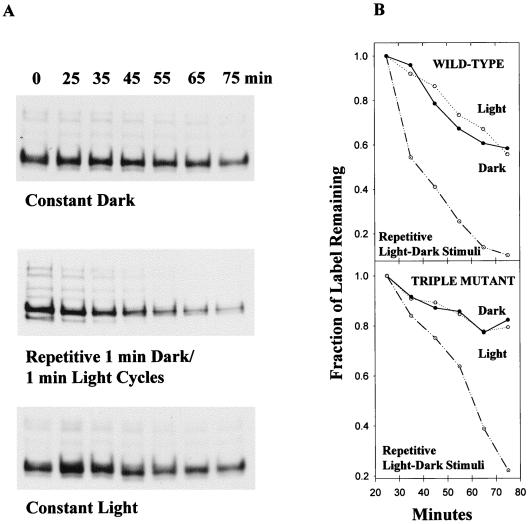
Cells were radiolabeled in vivo as described above. The radioactivity was chased with a 10× excess of nonradioactive l-methionine (130 μM). Two-milliliter volumes of the cell suspensions were then transferred to a disposable cuvette (1 cm by 1 cm by 4.5 cm) held in a removable sample chamber taken from an SLM Aminco DW-2000 spectrophotometer (SLM Instruments, Urbana, Ill.) and maintained at a constant temperature of 37°C with stirring throughout the experiment. Three different conditions were used in three separate experiments of 75 min each: constant dark, repetitive cycles of 1 min of light and 1 min of dark, and constant light. Cells were illuminated with light from a 100-W tungsten-halogen lamp passed through 4-mm-thick heat-absorbing glass (Edmund Scientific, Barrington, N.J.) and a 600-nm interference filter. Two-hundred-microliter samples were withdrawn at each time point, precipitated in cold acetone, and processed for SDS-PAGE. Quantitation of the remaining label on autofluorographs of SDS-PAGE was done with SigmaScan (Jandel Scientific, San Rafael, Calif.).
RESULTS
Light induces large methylation changes in HtrII but not in HtrI transducers.
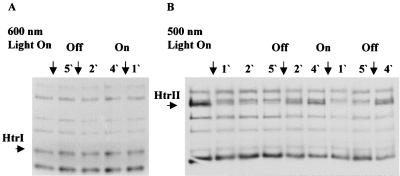
Cells containing either SRI-HtrI or SRII-HtrII complexes were labeled to a steady-state level in vivo with l-[methyl-3H]methionine. Fluorography of SDS-PAGE was used to detect changes in methylation of both HtrI and HtrII upon specific light stimulation of the respective coupled receptors. SRI stimulation with 600-nm light did not produce detectable changes in the level of methylation of HtrI protein or other methylated proteins in the gel (Fig. 1A). SRII stimulation with 500-nm light caused a reduction of label in the band corresponding to the HtrII protein evident after 1 min of illumination and further decreased after 5 min. Remethylation of HtrII protein was observed after 2 min in the dark, and it was almost completed after 4 min. Exposing the cells to a second cycle of illumination reproduced the demethylation response observed in the first cycle (Fig. 1B).
FIG. 1.
Autofluorograms of SDS–8% polyacrylamide gels of protein from l-[methyl-3H]methionine-labeled H. salinarum cells. (A) HtrI-containing cells; (B) HtrII-containing cells. Cells were exposed to dark and light cycles as shown at the top of the fluorograms. Light at 600 and 500 nm was used as stimuli for HtrI and HtrII, respectively. Acetone-precipitated cells were processed for SDS-polyacrylamide gels as described previously (23). Arrows on the left of the gels point to HtrI and HtrII migration positions.
HtrI and HtrII contain putative methylation sites.
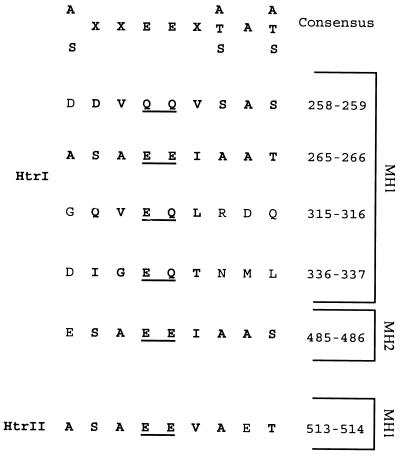
Sequence comparison of HtrI, HtrII, and other Htr proteins with the eubacterial chemoreceptors identifies glutamates or glutamines at positions Q258-Q259, E265-E266, E315-Q316, E336-Q337 in MH1 (methylation helix 1) (30) and E485-E486 in MH2 of HtrI. HtrII shows in its MH1 glutamate residues E513-E514 that correspond to E265-E266 of HtrI. In HtrI, Q258-Q259 and E265-E266 are homologous to Q297-Q298 and E303-E304 of Tsr, respectively. Residues E315-Q316 and E336-Q337 are conserved among all the H. salinarum Htr proteins whose sequences are available to date. A recognition sequence (consensus in Fig. 2) for the methyltransferase enzyme, CheR, as reported for the eubacterial chemotransducers (33), is present in the region flanking residues Q258-Q259, E265-E266, and E485-E486 of HtrI and E513-E514 of HtrII. Both HtrI and HtrII are missing at their C terminus the pentapeptide sequence (NWET/SF), which is conserved among the major chemotransducers Tsr, Tar, and Tcp and found to be the site of binding of CheR (34).
FIG. 2.
Predicted methylation sites. At the top of the figure in bold letters is the consensus sequence for the methyltransferase CheR reported for enteric bacteria (33). In HtrI and HtrII, the potential methylation sites are underlined and the residues that are common to the consensus are in bold. Residues E265 and E266 in HtrI and their flanking sequences perfectly match the consensus sequence. Sequences flanking residue pairs Q258-Q259 and E485-E486 in HtrI and E513-E514 in HtrII differ from the consensus sequence by only one residue. MH1 and MH2 are the first and second methylation helices from the N terminus.
Methylation of HtrI is lost when residues at positions 265 and 266 are mutated to alanines.
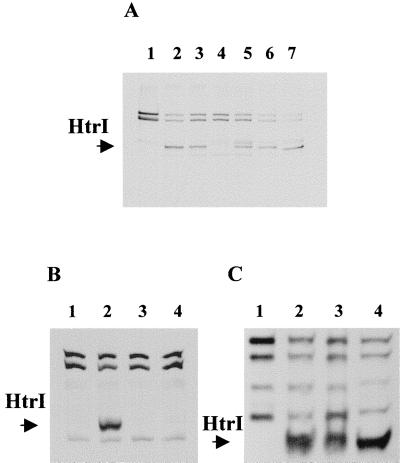
Single-residue pairs on HtrI were mutated to alanines by PCR site-directed mutagenesis. Mutant cells were labeled in vivo with l-[methyl-3H]methionine, membrane proteins were isolated, and the pattern of methylated bands was analyzed by SDS-PAGE fluorography (Fig. 3A). Pho81Wr−, an HtrI− SRI− strain, did not show any methylated band corresponding to the HtrI protein (Fig. 3A, lane 1). A distinct methyl-labeled band corresponding to HtrI was observed in the wild-type strain (Fig. 3A, lane 2). Alanine substitutions of glutamates 265 and 266 caused loss of this methylated band (Fig. 3A, lane 4). The other mutants did not show detectable band differences from that of the wild-type (Fig. 3A, lane 3, 5, 6, and 7). Mutations at the three sites containing the methyltransferase recognition sequence were combined in a single construct (the triple mutant). In Fig. 3B, the triple mutant in lane 4, like the one with the mutations E265A and E266A in lane 3, did not show any methylation corresponding to the HtrI band. The amounts of HtrI proteins in these mutants detected by immunoblot analysis were similar to that of the wild-type strain (Fig. 3C).
FIG. 3.
Autofluorograms of l-[methyl-3H]methionine-labeled membrane proteins and immunoblot. (A) Methylation pattern of membrane proteins from the following strains: Pho81Wr− (lane 1), Pho81Wr−/pKJ306WT (lane 2), Pho81Wr−/pKJ306Q258A-Q259A (lane 3), Pho81Wr−/pKJ306E265A-E266A (lane 4), Pho81Wr−/pKJ306E315A-Q316A (lane 5), Pho81Wr−/pKJ306E336A-Q337A (lane 6), Pho81Wr−/pKJ306E485A-E486A (lane 7). (B) Methylation pattern of membrane proteins. Lanes 1, 2, and 3 contain the same strains as lanes 1, 2, and 4 in panel A, respectively. Lane 4 contains the Pho81Wr−/pKJ306 triple mutant (see Materials and Methods). (C) Immunoblot with HC23 antibody of membrane proteins. Lanes 1, 2, 3, and 4 contain the same strains as those lanes in panel B.
Behavioral analysis of methylation mutants of HtrI.
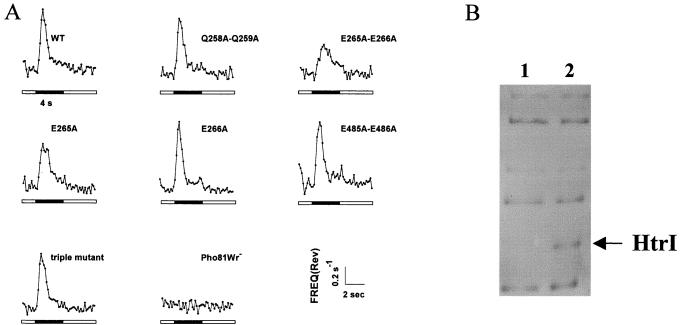
Light stimuli control the motility behavior of the cells by modulating their frequency of reversal of swimming direction. An increase in the intensity of orange (attractant) light suppresses reversal probability, inducing the cells to swim smoothly towards higher intensities of the light. A step down in intensity of the same light presents a negative stimulus to which the cells react by increasing their reversal frequency and is commonly used as a measure of function of the attractant signaling system. Wild-type cells respond to an orange step down stimulus by increasing their reversal frequency (excitation). Within the time the light remains off (4 s), their reversal frequency returns to near the prestimulus level (adaptation) (Fig. 4). All the mutants analyzed, as well as the wild type, adapted to a step down in orange light. The mutation pair E265A-E266A, which caused loss of methylation of HtrI on SDS gels, produced a reduced response compared to that of the wild type. Single alanine substitution at positions E265 and E266 established that glutamate 265 is responsible for the behavioral phenotype seen in the double mutant (Fig. 4A). In vivo labeling of the same mutants showed complete loss of methylation by the mutation E265A but not by E266A (Fig. 4B). The triple mutant in which all the methylation sites with the methyl transferase recognition sequence were changed to alanines exhibited the same behavior as did the wild type (Fig. 4). Analysis of responses of pairs of methylation site mutants showed that specifically E485A-E486A restores the E265A-E266A reduced response (data not shown). The reversal frequencies of each of these mutants, in the dark and after 2 to 5 min of light, was indistinguishable from that of the wild-type strain (1.3 min−1).
FIG. 4.
(A) Reversal frequency responses to photostimuli. Phototaxis responses of wild-type cells and HtrI mutants in response to a 4-s step down (dark bar under each graph) in orange (600-nm) light. Stimuli were delivered at 26-s intervals and three or more sets of 16 stimuli each were averaged to produce the final data. (B) Autofluorograms of l-[methyl-3H]methionine labeled membrane proteins. Lane 1, Pho81Wr−/pKJ306E265A; lane 2, Pho81Wr−/pKJ306E266A.
Stimuli through nonmethylatable HtrI induce volatile methyl group release from cells.
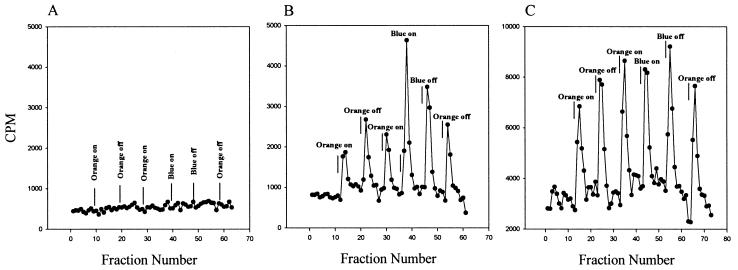
A vapor phase transfer assay was used to measure stimulus-induced SRI-HtrI-dependent release of methyl groups. The light-induced peaks have been shown to be due to [3H]methanol (18) consistent with carboxylmethyl ester hydrolysis. Transient increases in the rate of release of volatile [3H]methyl groups were detected after both attractant and repellent photostimuli in both wild-type and triple-mutant cells (Fig. 5B and C). Light-induced methyl release occurs from the triple mutant even though no steady-state labeling of HtrI was observed on gels. Therefore, photoactivated SRI-induced turnover of methyl groups in the cell does not depend on the methylated sites on the HtrI transducer.
FIG. 5.
Volatile [3H]methyl group release upon SRI-HtrI photostimulation. (A) Pho81Wr− (SRI− HtrI−); (B) Pho81Wr−/pKJ306WT; (C) Pho81Wr−/pKJ306 triple mutant. Each point corresponds to volatile counts per minute contained in a 0.5-ml fraction collected in 30 s. The variation in the ordinate scale is due to the variation in extent of l-[methyl-3H]methionine uptake in different strains and on different days.
SRI-HtrI photostimulation causes global methylation turnover.
The methanol release assay showed that there must be another source of methyl groups different from the stimulated transducer HtrI. To identify this source, wild-type and triple-mutant cells were radiolabeled with l-[methyl-3H]methionine, excess nonradioactive methionine was added to the cells, and stimulus-induced chase of the methylation on gels was monitored. Cells were exposed to three different conditions: constant dark, cycles of 1 min of dark and 1 min of light, and constant light in three separate assays. The rate of turnover of methyl groups on HtrI and on other transducer proteins was accelerated by cycles of stimulation in comparison to those of the constant-dark or constant-light conditions, showing that the stimulus and not the light per se accelerates the turnover (Fig. 6). Quantitation of the HtrI band in the three different experimental conditions showed that labeled methyl groups in the protein in the dark and in the light are chased to 60 to 80% of the starting level of radioactivity in 75 min. Repetitive light and dark stimuli induced a higher level of turnover that resulted in an almost complete chase of radioactivity to 10 to 20% in the same time period (Fig. 6B).
FIG. 6.
Pulse-chase experiment with l-[methyl3H]methionine-labeled Pho81Wr−/pKJ306WT and triple mutant cells. (A) Autofluorogram. Radioactivity was chased by adding a 10× excess of nonradioactive l-methionine to cells labeled in vivo as described in Materials and Methods. Wild-type cells were exposed to the dark, to the light, and to alternating light and dark as described in Materials and Methods, and acetone-precipitated samples from each time point were processed for SDS-PAGE. (B) Amount of labeling corresponding to the HtrI band was quantified by using SigmaScan for each condition used. For the triple mutant, the amount of labeling corresponding to the second band from the top (Fig. 3) was plotted. The values at 25 min were maximal and were therefore defined as 100% labeling.
The residue pair E513-E514 of HtrII is responsible for most of the methylation seen in this protein.
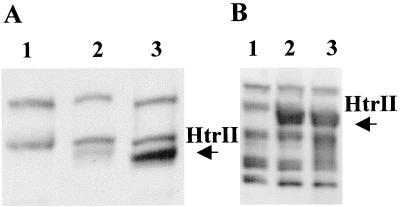
In HtrII, the residue pair E513-E514 is homologous to E265-E266 in HtrI and contains the only putative methylation site in HtrII with the transferase recognition consensus sequence (Fig. 2). When these residues in HtrII were mutated to a pair of alanines, nearly all methylation was lost (Fig. 7A, lane 2), even though the protein was expressed at the same level as the wild type (Fig. 7B, lanes 2 and 3, respectively). However, motility analysis showed that alanine substitutions at these sites did not impair the ability of the cells to respond to a repellent stimulus (a step-up in 500-nm light for 9 s) and to adapt to it. Furthermore, the reversal frequencies of the mutant in the dark and after 2 to 5 min in the light were indistinguishable from those of the wild type (3 min−1 in this particular experiment). As in SRI-HtrI, release of methyl groups upon SRII-HtrII-specific stimulation is not affected by the mutations at these sites (Fig. 8B).
FIG. 7.
Autofluorograms of l-[methyl-3H]methionine-labeled membrane proteins and corresponding immunoblot. (A) Methylation patterns of membrane proteins from the following strains: Pho81Wr− (lane 1), Pho81Wr−/pPR5E513AE514A (lane 2), Pho81Wr−/pPR5WT (lane 3). (B) Immunoblot. Samples were loaded in the same order as in panel A, and the blot was labeled with HC23 antibody.
FIG. 8.
Volatile [3H]methyl group release upon SRII-HtrII photostimulation. (A) Pho81Wr/pPR5WT; (B) Pho81Wr−/pPR5E513AE514A.
Methyl group turnover is regulated by a CheY-dependent pathway.
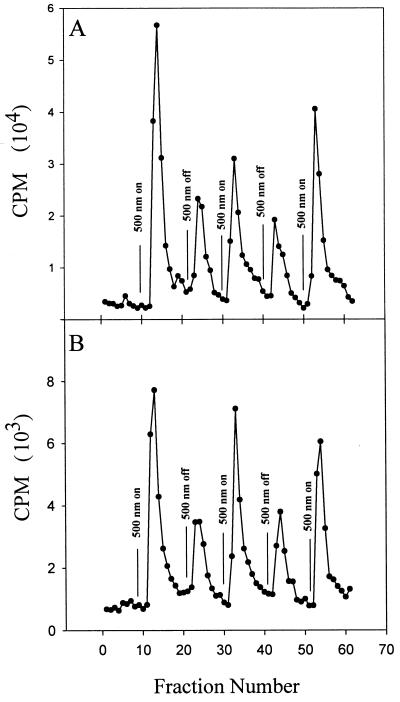
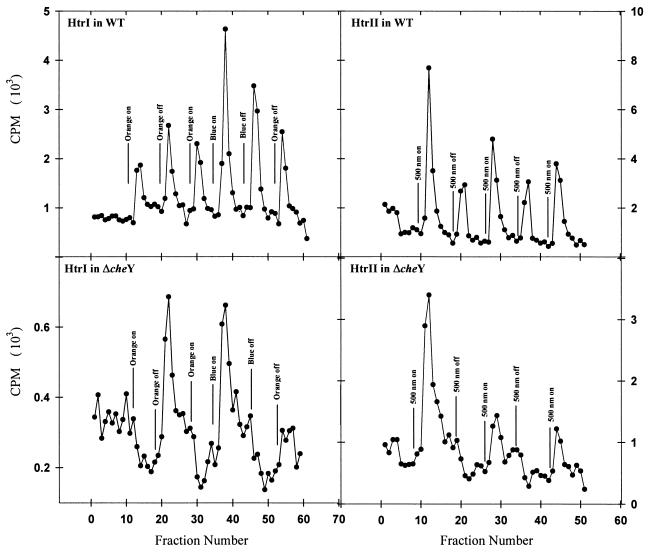
The response regulator gene cheY was deleted in the strain Pho81Wr− by using a gene replacement technique (14). The deletion mutant did not form a swarm ring in soft agar plates, as was observed previously for a similar cheY deletion mutant (21). The phenotype of the ΔcheY strain was smooth swimming both in the dark and in the light, and the reversal frequency of this strain was restored to wild-type levels by plasmid expression of CheY. Expression of the SRI-HtrI or SRII-HtrII complexes in the cheY deletion mutant, as expected, did not restore spontaneous reversals but did restore light-stimulated changes in methyl turnover rates. Volatile methanol release assay of the ΔcheY strains expressing the SRI-HtrI and the SRII-HtrII complexes showed in both cases reduction of turnover of methyl groups upon positive stimuli (Fig. 9). Thus, the deletion of the cheY gene converted the pattern of stimulus-induced methanol release from symmetric to asymmetric (Fig. 9).
FIG. 9.
Volatile [3H]methyl release group of SRI-HtrI and SRII-HtrII-containing cells in Pho81Wr− (top panels) and Pho81Wr−ΔcheY background (bottom panels).
DISCUSSION
In the generally accepted model of bacterial chemotaxis, adaptation to chemostimuli is brought about by methylation and demethylation of glutamate residues on the receptor-transducer (MCP) protein (4, 5, 30). A change in the level of methylation on the stimulated MCP cancels the effect of ligand occupancy and therefore resets the flagellar motor switch to its prestimulus bias. We found that H. salinarum cells carrying transducers mutated at their methylation sites, which consequently cannot be methylated, are still able to adapt to light stimuli. Our results prove that adaptation to phototaxis stimuli in H. salinarum, defined as a return to the prestimulus flagellar bias within the time the stimulus persists, does not require changes in methylation of the transducer through which the stimulus is sent. This finding may seem initially to be fundamentally different from the E. coli chemotaxis paradigm, but upon deeper analysis the difference may be a kinetic rather than a mechanistic one. For large and abrupt temporal gradient stimuli with chemotaxis effectors, the time required to change the level of methylation of the MCPs is, under many experimental conditions, the rate-limiting step for adaptation to occur (30). Evidently, this is not the case for the analogous temporal gradient photostimuli used here since the elimination of Htr methylation sites does not alter the kinetics of recovery to near the prestimulus reversal frequency. In their analysis, Stock and Surette (30) point out that in natural spatial gradients much faster responses and faster adaptation mechanisms are necessary than would be possible from the relatively slow methylation system in E. coli. Therefore, methylation is likely to be one of several processes resetting flagellar motor switch bias in the fluctuating natural environment of the cells. The other processes may dominate the behavior of H. salinarum in the short time window.
Methylation changes on the stimulated transducer are clearly not required in H. salinarum in the short time in which most of the recovery from a step up or step down in light intensity occurs. However, methylation extent does bias the flagellar motor switch in a similar manner as in E. coli. Demethylation by methionine starvation or cheR mutation causes smooth swimming, and overmethylation by cheB deletion causes a high swimming reversal frequency. Furthermore, the transducer HtrII undergoes demethylation in response to a repellent stimulus as do MCPs in E. coli. We did not observe a methylation increase in response to attractant stimulation of HtrI, but small changes may have been below our level of detection. Given that methylation regulates the flagellar motor switch bias, it is likely to play a role in the adaptation of the cells over longer time scales, but transducer-specific methylation is not required.
We observed photostimulus-induced turnover of methyl groups monitored by methanol evolution, and pulse-chase measurements demonstrated that transducers other than that stimulated undergo methylation-demethylation reactions. Similarly, it has been shown in E. coli that attractant stimulation of one MCP transiently increases the methylation level of another and that the inhibition of the methyl esterase is responsible for this effect (22). A global methylation process was observed in our studies in all cases, including stimulation of a nonmethylatable transducer, and may be needed for altering the conformation of a cluster of transducers to contribute to resetting the kinase activity of the CheA protein. Such “adaptational cross-talk” has been suggested for E. coli, in which chemotaxis mediated by the ribose and galactose transducer Trg lacking its methylation sites was restored by expressing the mutated transducer in the presence of other chemoreceptors (7). Also, transducer protein clustering, which would provide a mechanism for conformational coupling of stimulated and unstimulated transducers, has been reported (17).
E. coli, B. subtilis, and H. salinarum all exhibit an enhanced rate of methanol evolution caused by stimuli that activate the histidine kinase activity of CheA (repellent stimuli in H. salinarum) (20). This is attributable to the activation of the methyl esterase activity of CheB by phosphorylation in each case. E. coli shows a reduced rate of methanol evolution from stimuli that inhibit CheA activity, as would be expected from the dephosphorylation of CheB. On the other hand, CheA kinase-inhibiting stimuli in B. subtilis and H. salinarum cause an increase in methanol evolution rate even though the activity of CheB is predicted to be decreased (30). A clue to the basis of this anomalous effect has been discovered by Kirby et al. (13), who recently reported that deletion of CheY eliminates the unexpected enhancement of turnover rate in response to removal of the chemoeffector asparagine in B. subtilis. Their explanation is that addition and removal of asparagine each causes transient demethylation of its MCP and that the inability of the CheY null mutant to remethylate the ligand-bound MCP accounts for the lack of methanol production in response to asparagine removal. The authors suggest a mechanism for the CheY-P feedback effect, namely, that the CheY-P interaction with the ligand-bound MCP affects the topology of the C terminus of the MCP so that the esterase has increased access to an otherwise-less-accessible methylated residue (13).
Our results show that as in B. subtilis, a CheY-dependent process is responsible for the increased methanol evolution rate following CheA-inhibiting stimuli in H. salinarum. In a CheY deletion mutant of H. salinarum, repellent and attractant stimuli result in increases and decreases, respectively, in the methanol evolution (methyl group turnover) rate, as expected from the respective phospho-activation and -deactivation of CheB, and as is observed in wild-type E. coli. However, the explanation offered for B. subtilis does not seem to apply to H. salinarum. First, we do not observe remethylation of HtrII in the continuous presence of light after the protein is demethylated in response to blue light. The methylation changes of HtrII in response to addition and removal of repellent blue light are not transient, but methylation levels are shifted in a manner closely similar to those of E. coli MCPs in response to addition and removal of repellent substances. Second, we found with the triple mutant that activation of a nonmethylatable HtrI still produces a symmetric pattern in release of radiolabeled methanol. Therefore, the feedback effect of CheY is not localized to an effect on the stimulated transducer but modulates methyl group turnover of other transducers in the cell. One possibility is that CheY increases the rate of methylation, either by making methylation sites more accessible or by activation of CheR activity, thereby increasing the turnover rate.
ACKNOWLEDGMENTS
We thank Elena Spudich and Kwang-Hwan Jung for critically reading the manuscript and John R. Kirby for helpful discussions.
This work was supported by National Institutes of Health grant R01-GM27750 (to J.L.S.).
REFERENCES
- 1.Alam M, Lebert M, Oesterhelt D, Hazelbauer G L. Methyl-accepting taxis proteins in Halobacterium halobium. EMBO J. 1989;8:631–639. doi: 10.1002/j.1460-2075.1989.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd A, Simon M I. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J Bacteriol. 1980;143:809–815. doi: 10.1128/jb.143.2.809-815.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen B, Przybyla A E. An efficient site-directed mutagenesis method based on PCR. BioTechniques. 1994;17:161–162. [PubMed] [Google Scholar]
- 4.Eisenbach M. Control of bacterial chemotaxis. Mol Microbiol. 1996;20:903–910. doi: 10.1111/j.1365-2958.1996.tb02531.x. [DOI] [PubMed] [Google Scholar]
- 5.Falke J J, Bass R B, Butler S L, Chervitz S A, Danielson M A. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazelbauer G L, Engstrom P. Multiple forms of methyl-accepting chemotaxis proteins distinguished by a factor in addition to multiple methylation. J Bacteriol. 1981;145:35–42. doi: 10.1128/jb.145.1.35-42.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hazelbauer G L, Park C, Nowlin D M. Adaptational “crosstalk” and the crucial role of methylation in chemotactic migration by Escherichia coli. Proc Natl Acad Sci USA. 1989;86:1448–1452. doi: 10.1073/pnas.86.5.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoff W D, Jung K-H, Spudich J L. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu Rev Biophys Biomol Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 9.Jung K-H, Spudich J L. Suppressor mutation analysis of the sensory rhodopsin I-transducer complex: insights into the color-sensing mechanism. J Bacteriol. 1998;180:2033–2042. doi: 10.1128/jb.180.8.2033-2042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Jung, K.-H., and J. L. Spudich. Unpublished data.
- 10.Kehry M R, Dahlquist F W. The methyl-accepting chemotaxis proteins of Escherichia coli. Identification of the multiple methylation sites on methyl-accepting chemotaxis protein I. J Biol Chem. 1982;257:10378–10386. [PubMed] [Google Scholar]
- 11.Kleene S J, Toews M L, Adler J. Isolation of glutamic acid methyl ester from an Escherichia coli membrane protein involved in chemotaxis. J Biol Chem. 1977;252:3214–3218. [PubMed] [Google Scholar]
- 12.Kirby J R, Kristich C J, Feinberg S L, Ordal G W. Methanol production during chemotaxis to amino acids in Bacillus subtilis. Mol Microbiol. 1997;24:869–878. doi: 10.1046/j.1365-2958.1997.3941759.x. [DOI] [PubMed] [Google Scholar]
- 13.Kirby J R, Saulmon M M, Kristich C J, Ordal G W. CheY-dependent methylation of the asparagine receptor, McpB, during chemotaxis in Bacillus subtilis. J Biol Chem. 1999;274:11092–11100. doi: 10.1074/jbc.274.16.11092. [DOI] [PubMed] [Google Scholar]
- 14.Krebs M P, Mollaaghababa R, Khorana H G. Gene replacement in Halobacterium halobium and expression of bacteriorhodopsin mutants. Proc Natl Acad Sci USA. 1993;90:1987–1991. doi: 10.1073/pnas.90.5.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krebs M P, Spudich E N, Khorana H G, Spudich J L. Synthesis of a gene for sensory rhodopsin I and its functional expression in Halobacterium halobium. Proc Natl Acad Sci USA. 1993;90:3486–3490. doi: 10.1073/pnas.90.8.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Maddock J R, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 18.Nordmann B, Lebert M R, Alam M, Nitz S, Kollmannsberger H, Oesterhelt D, Hazelbauer G L. Identification of volatile forms of methyl groups released by Halobacterium salinarum. J Biol Chem. 1994;269:16449–16454. [PubMed] [Google Scholar]
- 19.Nowlin D M, Bollinger J, Hazelbauer G L. Sites of covalent modification in Trg, a sensory transducer of Escherichia coli. J Biol Chem. 1987;262:6039–6045. [PubMed] [Google Scholar]
- 20.Rudolph J, Tolliday N, Schmitt C, Schuster S C, Oesterhelt D. Phosphorylation in halobacterial signal transduction. EMBO J. 1995;14:4249–4257. doi: 10.1002/j.1460-2075.1995.tb00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudolph J, Oesterhelt D. Deletion analysis of the che operon in the archaeon Halobacterium salinarium. J Mol Biol. 1996;258:548–554. doi: 10.1006/jmbi.1996.0267. [DOI] [PubMed] [Google Scholar]
- 22.Sanders D A, Koshland D E., Jr Receptor interactions through phosphorylation and methylation pathways in bacterial chemotaxis. Proc Natl Acad Sci USA. 1988;85:8425–8429. doi: 10.1073/pnas.85.22.8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spudich E N, Spudich J L. Measurement of light regulated phosphoproteins of Halobacterium halobium. Methods Enzymol. 1982;26:213–216. [Google Scholar]
- 24.Spudich E N, Hasselbacher C A, Spudich J L. Methyl-accepting protein associated with bacterial sensory rhodopsin I. J Bacteriol. 1988;170:4280–4285. doi: 10.1128/jb.170.9.4280-4285.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spudich E N, Takahashi T, Spudich J L. Sensory rhodopsins I and II modulate a methylation/demethylation system in Halobacterium halobium phototaxis. Proc Natl Acad Sci USA. 1989;86:7746–7750. doi: 10.1073/pnas.86.20.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spudich E N, Zhang W, Alam M, Spudich J L. Constitutive signaling by the phototaxis receptor sensory rhodopsin II from disruption of its protonated Schiff base-Asp-73 interhelical salt bridge. Proc Natl Acad Sci USA. 1997;94:4960–4965. doi: 10.1073/pnas.94.10.4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spudich J L, Bogomolni R A. Sensory rhodopsins of halobacteria. Annu Rev Biophys Biophys Chem. 1988;17:193–215. doi: 10.1146/annurev.bb.17.060188.001205. [DOI] [PubMed] [Google Scholar]
- 28.Spudich J L, Spudich E N. Selection and screening methods for halophilic archael rhodopsin mutants. In: Robb F T, et al., editors. Archaea: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 23–28. [Google Scholar]
- 29.Stock J B, Koshland D E., Jr A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci USA. 1978;75:3659–3663. doi: 10.1073/pnas.75.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stock J B, Surette M. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1996. pp. 123–145. [Google Scholar]
- 31.Sundberg S A, Bogomolni R A, Spudich J L. Selection and properties of phototaxis-deficient mutants of Halobacterium halobium. J Bacteriol. 1985;164:282–287. doi: 10.1128/jb.164.1.282-287.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terwilliger T C, Koshland D E., Jr Sites of methyl esterification and deamination on the aspartate receptor involved in chemotaxis. J Biol Chem. 1984;259:7719–7725. [PubMed] [Google Scholar]
- 33.Terwilliger T C, Wang J Y, Koshland D E., Jr Surface structure recognized for covalent modification of the aspartate receptor in chemotaxis. Proc Natl Acad Sci USA. 1986;83:6707–6710. doi: 10.1073/pnas.83.18.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu J, Li J, Li G, Long D G, Weis R M. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 35.Yao V J, Spudich J L. Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc Natl Acad Sci USA. 1992;89:11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao V J, Spudich E N, Spudich J L. Identification of distinct domains for signaling and receptor interaction of the sensory rhodopsin I transducer, HtrI. J Bacteriol. 1994;176:6931–6935. doi: 10.1128/jb.176.22.6931-6935.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W, Brooun A, Mueller M M, Alam M. The primary structures of the Archaeon Halobacterium salinarium blue light receptor sensory rhodopsin II and its transducer, a methyl-accepting protein. Proc Natl Acad Sci USA. 1996;93:8230–8235. doi: 10.1073/pnas.93.16.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Brooun A, McCandless J, Banda P, Alam M. Signal transduction in the archaeon Halobacterium salinarium is processed through three subfamilies of 13 soluble and membrane-bound transducer proteins. Proc Natl Acad Sci USA. 1996;93:4649–4654. doi: 10.1073/pnas.93.10.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]