Abstract
Double-stranded RNA adenosine deaminase 1 (ADAR1) is significantly down-regulated in fibroblasts derived from Idiopathic Pulmonary Fibrosis (IPF) patients, and its overexpression restored levels of miRNA-21, PELI1, and SPRY2. There are two ADAR1 isoforms in humans, ADAR1-p110 and ADAR1-p150, generated by an alternative promoter. Let-7d is considered an essential microRNA in Pulmonary Fibrosis (PF). In silico analysis revealed COL3A1 and SMAD2, proteins involved in the development of IPF, as Let-7d targets. We analyzed the role of ADAR1-p110 and ADAR1-p150 isoforms in the regulation of Let-7d maturation and the effect of this regulation on the expression of COL3A1 and SMAD2 in IPF fibroblast. We demonstrated that differential expression and subcellular distribution of ADAR1 isoforms in fibroblasts contribute to the up-regulation of pri-miR-Let-7d and down-regulation of mature Let-7d. Induction of overexpression of ADAR1 reestablishes the expression of pri-miR-Let-7d and Let-7d in lung fibroblasts. The reduction of mature Let-7d upregulates the expression of COL3A1 and SMAD2. Thus, ADAR1 isoforms and Let-7d could have a synergistic role in IPF, which is a promising explanation in the mechanisms of fibrosis development, and the regulation of both molecules could be used as a therapeutic approach in IPF.
Keywords: ADAR, RNA edition, miRNA, IPF
1. Introduction
Idiopathic Pulmonary Fibrosis (IPF) is the most frequent and aggressive interstitial lung disease. It is considered a consequence of several risk factors such as smoking habits, genetic determinants, environmental factors, gastroesophageal reflux, viral infections, and aging; however, its origin and further progression are not fully understood [1,2,3,4,5]. In situ, fibroblasts differentiate into myofibroblasts and form fibroblastic foci, which are responsible for secreting excessive amounts of extracellular matrix (ECM) components such as fibrillar collagens [6,7,8]. The excessive accumulation of these proteins entails the destruction of the pulmonary architecture [1,2,3,4,5].
The molecular mechanisms of IPF have not been fully described. Some evidence suggests the possible participation of some microRNAs [9,10,11,12,13]. Several editing microRNAs by Adenosine Deaminase acting on RNA (ADAR) are affected in their maturation process mediated by Drosha and Dicer, or they are redirecting to new mRNAs target because their seed region nucleotides are modified [14,15,16,17,18,19,20,21,22,23]. This edition represents a specific tissue regulatory mechanism of microRNA expression [22].
ADAR1 has two isoforms generated by the alternative promoter, ADAR1-p110 and ADAR1-p150. ADAR1-p110 is the amino-terminally truncated isoform located in nuclear regions and constitutively expressed. ADAR1-p150 is the full-length isoform, which is interferon-inducible and is preferentially located in cytoplasmic regions [14,21]. ADAR1 isoforms have 2–3 double-stranded RNA binding domains (dsRNA) in the amino-terminal region, followed by a deaminase domain in the carboxyl-terminal region and defined domains of subcellular localization that allow them to shift between the nucleolus, nucleus, and cytoplasm [19]. Sub-cellular location differences suggest that ADAR1 isoforms display differential RNA regulation [3]. ADAR1 is significantly down-regulated in IPF fibroblasts, and the over-expression of ADAR1 and ADAR2 reestablishes the expression levels of miRNA-21, PELI1, and SPRY2 in fibroblasts from IPF patients [14]. Different works showed a Let-7d down-regulation in lung fibroblasts of IPF patients [9,10,12]. SMAD2 and 3, positive regulator of the TGF-β1 pathway, represents one of the main Let-7d targets, while Let-7d low expression promotes an increase in mesenchymal markers such as HMGA2, vimentin, N-cadherin, and ACTA2 [10,13]. In leukemia stem cells, ADAR1 can edit pri-miRNA-Let-7d transcript forming a secondary double-stranded structure that induces a negative regulation of Let-7d and self-renewal [23].
Dysregulation of the microRNA maturation processing caused by ADAR1 editing, and its effects on pulmonary fibroblasts, are not well known. We studied the role of the ADAR1-p110 and ADAR1-p150 isoforms in the regulation of Let-7d maturation and the effect of these modifications on the expression of collagen-3, a putative Let-7d target related to IPF.
2. Results
2.1. Expression of ADAR1 Isoforms in Controls and IPF Fibroblasts
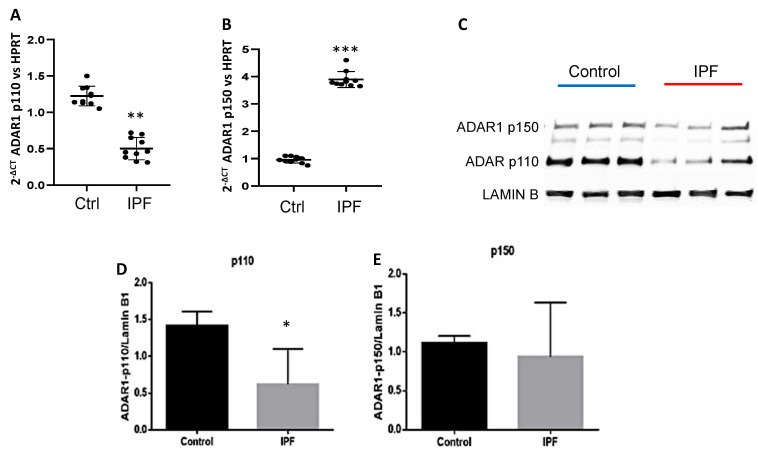
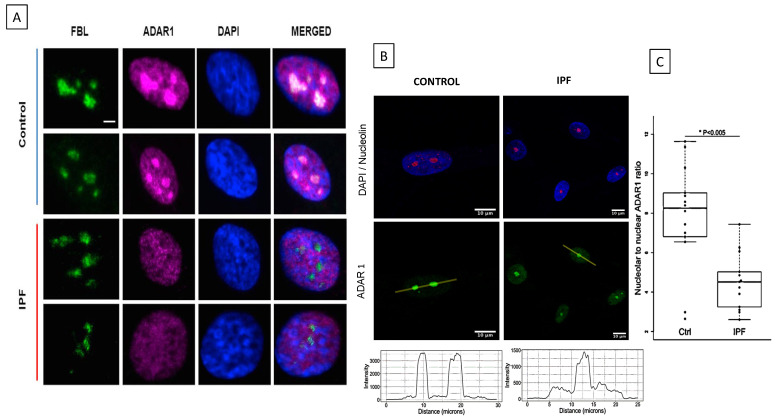
ADAR1-p110 isoform in IPF fibroblasts (n = 10) was downregulated (** p = 0.008, Figure 1A), whereas ADAR1-p150 isoform was overexpressed (*** p = 0.00054; Figure 1B), compared with control fibroblasts (n = 10). Additionally, we verified that there were no differences in the basal levels of ADAR1 expression between both control and fibrotic fibroblasts (Figure S1). We further confirmed these observations at the protein level by Western blot analysis, and we observed that ADAR1-p110 isoform levels were less than 66% (* p = 0.05; Figure 1C,D) in total protein extracts in IPF fibroblasts compared to controls. The ADAR1-p150 isoform did not show changes (Figure 1C,E). Different distribution of fluorescence intensity of ADAR1 was detected in the nuclei of the IPF fibroblasts (n = 6) compared to control cells (n = 6) (Figure 2A). ADAR1 was in the nucleolar region mainly in control fibroblasts, whereas in IPF cells, ADAR1 was exclusively detected in the nucleoplasm Figure 2A,B). These preliminary observations suggested an altered subnuclear location of ADAR1 in IPF. It was confirmed by the analysis of fluorescence intensity. The ratio of ADAR1 nucleolar to nuclear expression is significantly higher in control fibroblasts than in IPF fibroblasts (Figure 2C).
Figure 1.
Expression levels of ADAR1-p110 and ADAR1-p150 in control compared with fibrotic fibroblasts. (A) ADAR1-p110 is significantly down-regulated in IPF fibroblasts (** p < 0.0001). (B) ADAR1-p150 is significantly overexpressed in IPF fibroblasts (*** p < 0.0001). (C) Analysis of the protein levels of the isoforms of ADAR1 in total protein extracts corroborated the decrease in ADAR1-p110 (* p < 0.05), whereas ADAR1-p150 did not show significant differences. (D) Densitometric analysis of the protein levels of ADAR1 (C) corroborates the decrease in ADAR1-p110 in fibrotic cells and no difference in ADAR1-p150 (E).
Figure 2.
Immunolocalization of ADAR1 in the nucleus on control and fibrotic pulmonary fibroblasts. (A) Fibrillarin (FBL, green; lane 1) was used as a specific marker for the nucleolus. ADAR1 was observed in magenta color (lane 2). Nuclei-stained with DAPI (lane 3). In control fibroblasts, ADAR1 is in the nucleolus and nucleoplasm (lane 2), whereas in fibrotic fibroblasts, the distribution of ADAR1 occurs mainly in the nucleoplasm. Merge image for FBL, ADAR1 and DAPI (lane 4). Bar = 5 μm. (B) ADAR1 (shown in green) seems to be predominantly detected in the nucleolus in both control and IPF fibroblasts, as shown by its similar distribution in the nucleolus marker Nucleolin (shown in red). However, ADAR1 expression is reduced in the nucleoli of IPF fibroblasts compared to control fibroblasts; in contrast, its expression is enriched in the nucleoplasm, as shown in the intensity profile (yellow line, intensity values plotted below the micrographs). (C) The ratio of ADAR1 nucleolar to nuclear expression is significantly higher in control fibroblasts than in IPF fibroblasts (mean of ratios in Ctrl group = 7.84 {n = 14}, the mean of ratios in the IPF group = 4.52 {n = 13}, p = 0.0015, Wilcoxon rank-sum test).
2.2. ADAR1 Is Hyperactive in IPF Fibroblasts
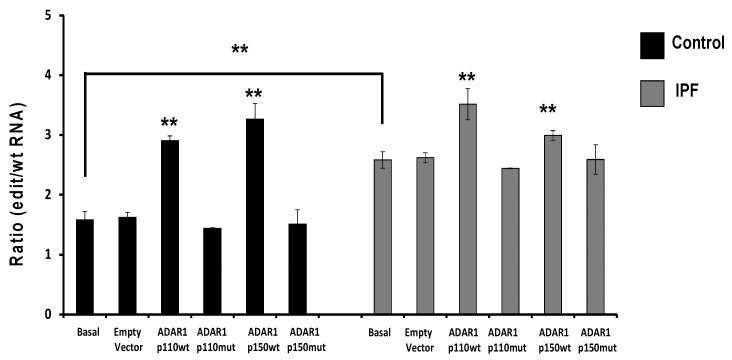
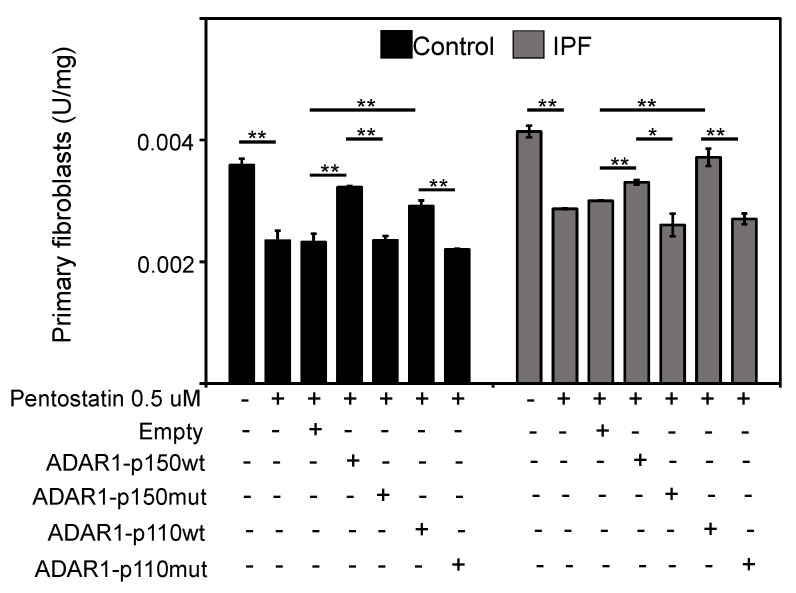
RESS-qPCR analysis [24] was performed to determine RNA editing mediated by ADAR1 in lung fibroblasts. We analyzed six controls and six IPF fibroblasts; IPF fibroblasts possess increased editing activity in specific adenosine of APOBEC3D compared with control fibroblasts (* p = 0.0193; Figure 3). The editing activity on APOBEC3D in control and fibrotic fibroblasts transfected with ADAR1 isoforms showed that the catalytic activity is increased after transfection with active plasmids. In contrast, the mutant plasmids do not modify the activity. In control fibroblasts (black bars), ADAR1-p110 wt shows a significant increase in editing activity (** p = 0.003737). A remarkable increase in editing activity is observed with the ADAR1-p150 isoform (** p = 0.000139), while the most significant increase the editing activity in IPF fibroblasts are observed after overexpression of the ADAR1-p110 (** p = 0.007030). An increase in ADAR1-p150 wt activity was also significant (** p = 0.009158). Editing-deficient ADAR-p110 mt and ADAR-p150 mt constructs did not affect the editing activity. ADAR editing activity on healthy control and fibrotic fibroblasts considerably decreases upon Pentostatin treatment, which is a specific inhibitor of the editing activity of Adenosine Deaminases (ADA) (Figure 4). Pentostatin, as a specific ADA inhibitor, decreases the catalytic activity in control fibroblasts (** p = 0.0059) and IPF fibroblasts (** p = 0.0014). Over-expression of ADAR1-p150 wt (** p = 0.0058) and ADAR1-p110 wt (** p = 0.0018) increased the catalytic activity of ADAR in control fibroblasts. On the contrary, the overexpression of ADAR1-p150 (p = 0.8) and ADAR1-p110 mutant (p = 0.72) did not induce significant changes in the catalytic activity in the control fibroblasts. However, ADAR1-p150 wt over-expression in IPF fibroblasts increases its activity (** p = 0.0042), similar to the overexpression of ADAR1-p110 (** p = 0.0067). Over-expression of ADAR1 mutant plasmids does not modify the activity. ADAR1-p150 mutant (p = 0.2) and ADAR1-p110 mutant (p = 0.02).
Figure 3.
The catalytic activity of ADAR1 on APOBEC3D in control fibroblasts compared with fibrotic fibroblasts. ADAR1 catalytic activity in basal conditions in IPF fibroblasts concerning control fibroblasts (** p = 0.0018). Editing activity increase is shown as the ratio of the edition divided by the ratio of wild-type (n = 6). The catalytic activity is increased after transfection with the catalytically active plasmids, whereas the mutant plasmids do not modify the activity. In control fibroblasts (black bars), ADAR1 p110 wt shows a significant increase in editing activity (** p = 0.003737). Remarkable increase in editing activity is observed with the ADAR1-p150 isoform (** p = 0.000139), while the greatest increase the editing activity in IPF fibroblasts are observed after over-expression of the ADAR1-p110 (** p = 0.007030). An increase in ADAR1-p150 wt activity was also significant (** p = 0.009158).
Figure 4.
The specific catalytic activity of ADAR1 is increased in primary fibroblasts of IPF. Pentostatin, as a specific ADA inhibitor, significantly decreased the catalytic activity in control fibroblasts (** p = 0.0059) and IPF fibroblasts (** p = 0.0014). Over-expression of ADAR1-p150 wt (** p = 0.0058) and ADAR1-p110 wt (** p = 0.0018) significantly increased the catalytic activity of ADAR in control fibroblasts. Over-expression of ADAR1-p150 (* p = 0.8) and ADAR1-p110 mutant (* p = 0.72) did not induce significant changes in the catalytic activity in the control fibroblasts. ADAR1-p150 wt over-expression in IPF fibroblasts significantly increases its activity (** p = 0.0042) like the over-expression of ADAR1-p110 (** p = 0.0067). Over-expression of ADAR1 mutant plasmids does not modify the activity. ADAR1-p150 mutant (* p = 0.2) and ADAR1-p110 mutant (* p = 0.02).
2.3. Basal Expression of Let7d and pri-miR-Let7d in Normal and IPF Fibroblasts and Correlation with ADAR1p110 and p150
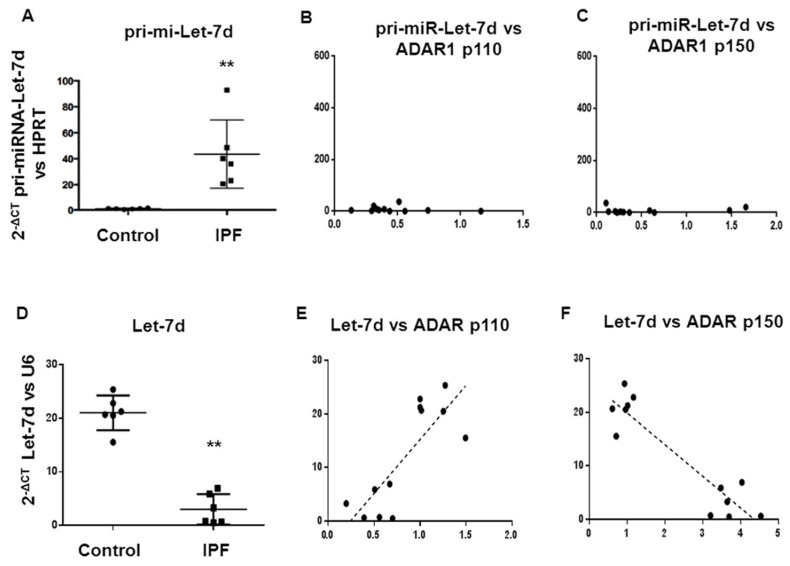
The basal expression of pri-miR-Let7d is higher in IPF fibroblasts than in normal fibroblasts (p = 0.05, Figure 5A). On the other hand, Let7d expression is lower in IPF fibroblasts than in Normal fibroblasts (p = 0.0028, Figure 5D). The correlation of ADAR1 isoforms does not show significant differences between ADAR1-p110 (Figure 5B) and ADAR1-p150 (Figure 5C). Let7d displayed a negative correlation with ADAR1-p150 (p < 0.0001) (Figure 5E) and a positive correlation with ADAR1-p110 (p = 0.0014) (Figure 5F).
Figure 5.
pri-miRNA-Let-7d is over-expressed in IPF fibroblasts, and Let-7d is downregulated in IPF fibroblasts and fibrotic mice lugs. The expression of pri-miRNA-Let-7d is higher in IPF fibroblasts than in control fibroblasts (p = 0.05) (A). The correlation of ADAR1 isoforms does not show significant differences between ADAR1-p110 (B) and ADAR1-p150 (C). The expression of Let-7d is lower in IPF fibroblasts compared to the expression levels of control fibroblasts (** p = 0.0028) (D). Let-7d displayed a negative correlation with ADAR1-p150 (p < 0.0001) (E) and a positive correlation with ADAR1-p110 (p = 0.0014) (F).
2.4. The Effect of ADAR1 Isoforms on Let-7d
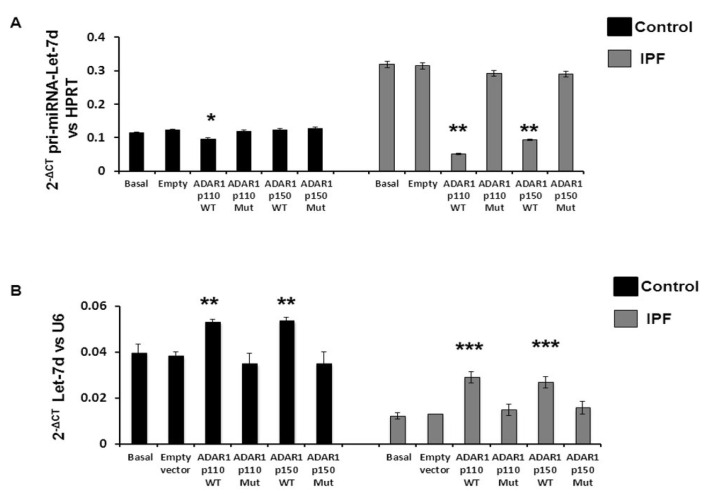
To determine the functional role of ADAR1 isoforms in fibroblasts, we transfected control and IPF fibroblasts with ADAR1-p110 wt, ADAR1-p150 wt, and their respective mutated vectors to analyze the levels of primary and mature Let-7d. ADAR1-p110 wt transfection in control fibroblasts favored the decrease in pri-miR-Let-7d (p = 0.054479, Figure 6A), whereas in IPF fibroblasts, both isoforms promoted a significant decrease in pri-miR-Let-7d, (ADAR1-p110 wt, ** p = 0.0002108, and ADAR1-p150 wt, ** p = 0.00038554, Figure 6A). Isoforms over-expression favored the increase in Let-7d in control (ADAR1-p110 wt, ** p = 0.001477 and ADAR1-p150 wt, ** p = 0.001946, Figure 6B) and in fibrotic fibroblasts (ADAR1-p110-wt, *** p = 0.000151 and ADAR1-p150-wt, *** p = 0.000855, Figure 6B), in contrast with the mutant vectors it remained unaltered (Figure 6). Correlation analysis between the expression levels of ADAR1, pri-miR-Let-7d, and Let-7d after ADAR1 overexpression did not show significant differences (data not shown). Moreover, IPF fibroblasts were treated with Pentostatin, an inhibitor of ADAR, and downregulation of pri-miRLet-7d (p < 0.0001) expression was observed with an upregulation of Let-7d (p < 0.0001) (Figure S2).
Figure 6.
The expression of pri-miR-Let-7d is modified by the over-expression of ADAR1-p110 wt and ADAR1-p150 wt. In (A), the over-expression of ADAR1-p110 wt in control fibroblasts significantly decreases the expression of pri-miR-Let-7d (* p = 0.054479, black bars). In IPF fibroblasts, both isoforms promote the decrease in pri-miR-Let-7d. ADAR1-p110 wt (** p = 0.0002108, gray bars) and ADAR1-p150 wt (** p = 0.00038554) in a higher proportion compared to the control fibroblasts. The expression of Let-7d in (B) was increased in the fibroblasts transfected with ADAR1. In control fibroblasts transfected with ADAR1, the expression of Let-7d is increased significantly ADAR1-p110 wt (** p = 0.001477, black bars) and ADAR1-p150 wt (** p = 0.001946, black bars). In IPF fibroblasts, the over-expression of catalytically active isoforms promotes a significant increase in Let-7d. ADAR1-p110 wt (*** p = 0.000151, gray bars) and ADAR1-p150 wt (*** p = 0.000855, gray bars).
2.5. Expression of COL3A1 and SMAD2 Targets of Let-7d in IPF
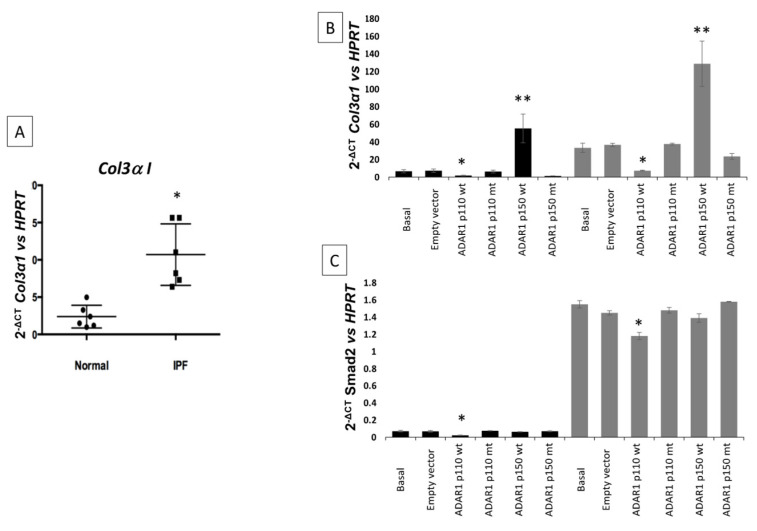
The expression of COL3A1 and SMAD2 mRNA targets of Let-7d potentially involved with the progression of IPF was analyzed in lung fibroblasts of control and IPF samples. The Let-7d target, COL3A1 obtained by in silico analysis, was upregulated (** p = 0.006, Figure 7A) in IPF when Let-7d expression was decreased. Over-expression of ADAR1p110 wt isoform showed a negative effect on the expression levels of COL3A1 (p < 0.05) and SMAD2 (p < 0.05) (Figure 7B,C) in both control and IPF fibroblasts, however, ADARp150 wt showed an increased effect in the expression of COL3A1 in normal and IPF fibroblasts (p < 0.0001) (Figure 7B). However, expression levels of SMAD2 did not show changes by ADARp150 (Figure 7C).
Figure 7.
Over-expression of COL3A1 in IPF fibroblasts (black square) vs Normal fibroblasts (black circle) (* p < 0.05) (A), Over-expression of ADAR1p110 wt isoform showed a negative effect on the expression levels of COL3A1 (* < 0.05) and SMAD2 (* p < 0.05) (B,C) in both control and IPF fibroblasts; however, ADARp150 wt showed an increased effect in the expression of COL3A1 in normal and IPF fibroblasts (** p < 0.0001) (B). The expression of SMAD2 did not show changes by ADARp150 (C).
3. Discussion
The ADAR1-p110 isoform is down-regulated both transcript and protein level, while the ADAR1-p150 isoform is overexpressed in IPF fibroblasts, even though at protein ADAR1-p150 level, there are no differences. This discrepancy may be due in part to the fact that the messenger RNA encoding ADAR1p150 has IRES-type elements in its sequence, which in pathological or stress conditions could respond to the presence of IRES transactivating factors, such as the polypyrimidine track binding protein (PTBP1). This results in the use of different translation starts, producing different isoforms of ADAR1 including ADARp110 [25]. Additionally, the distribution of these proteins is different in control fibroblasts; ADAR1-p110 is found mainly in the nucleolus, while in fibrotic cells, it is found in the nucleoplasm (Figure 2A–C). Previous reports showed that nucleolar ADAR2 location decreases its editing activity, and we hypothesize that the same regulation would be present in ADAR1 since both ADARs have dsRNA-binding motifs (DRBMs) and binding to rRNA [26]. Then, we propose that the ADAR1-p110 isoform is not correctly performing the editing in controls. In the case of fibrotic fibroblasts, this isoform is in the nucleoplasm, a region in which it has been demonstrated that the catalytic activity of this protein is executed [20], and even though the fibrotic fibroblasts have a diminished expression of the ADAR1-p110 isoform, their editing activity is increased. A report has mentioned that the ADAR editing activity is independent of their protein levels and expression levels [21], which supports our hypothesis that the ADAR1-editing activity depends mainly on its subcellular location. The expression levels of the ADAR1 isoforms and their location indicate that the regulatory mechanisms of each isoform are specific and complex. Differential locations for each ADAR1 isoform make us think that ADAR1-p110 is responsible for editing pri-miR-Let-7d and that cytoplasmic ADAR1-p150 for editing Let-7d transcript.
When the basal expression of an ADAR1 isoform is low, as in the case of ADAR1-p110, and is exogenously overexpressed, its edit activity levels do increase. Hence, we could suggest the existence of a negative feedback mechanism, which is essential to maintain specific expression levels in ADAR1 proteins to achieve an optimal function of the cells. However, we observed that the edition activity significantly increased in both control and fibrotic primary cultures when transfected with the wild-type isoforms, suggesting the existence of other regulatory mechanisms involved in the catalytic activity of ADAR in IPF fibroblasts. Analysis in silico showed that pri-miR-Let-7d has more than 70% adenosines edited by ADAR1; it is considered hyper-edition. Hyper-edited primary transcript is accumulated and rapidly degraded by Tudor-SN and is a less efficient substrate for Drosha and Dicer [26,27,28]. Thus, ADAR1 may inhibit the processing of the pri-miR-Let-7d through the loss of its double-stranded secondary structure. Consequently, Dicer and Drosha do not recognize it, redirecting it to another target or its degradation [29,30,31,32]. This degradation or deficiency in the processing of the pri-miR-Let-7d induced by the hyper-edition of the same transcript could explain the decrease in the Let-7d mature transcript in IPF since a down-regulation was observed along with ADAR1 expression.
The COL3A1 and SMAD2 mRNAs, putative targets of Let-7d, were quantified in IPF and controls. High levels of COL3A1 and SMAD2 were corroborated in IPF. A decrease in Let-7d expression induces an increase in COL3A1 and SMAD2 expression, which are involved in the development of pulmonary fibrosis.
When ADAR1 isoforms are over-expressed, the expression levels of COL3A1 and SMAD2 mRNA are modified, ADARp-110 induces downregulation of COL3A1 and SMAD2, and interestingly ADARp-150 induced an increase in COL3A1 on both normal and IPF fibroblasts. In this sense, we propose that the ADAR1-p150 isoform is editing pri-miR-Let-7d, avoiding its maturation to Let-7d. However, this has no effect on SMAD2.
To demonstrate the direct relationship between ADAR1 activity and the expression of Let-7d, we performed the experiment treating fibrotic fibroblasts with pentostatin, an inhibitor of ADAR activity (Figure S2). We were able to observe that Let-7d levels increased and pri-miR-Let-7d levels decreased, which demonstrates that ADAR1 activity directly influences the inhibition of maturation of pri-miR-Let-7d. These observations correlate with what is shown in Figure 4, where we observe a decrease in ADAR activity, which is reestablished with the transfection of the ADAR1 isoforms.
ADAR1-p110 and ADAR1-p150 regulate the processing of the pri-miR-Let-7d and mature Let-7d in such a way that the sub-expression of the latter deregulated, in turn, the expression of COL3A1 and SMAD2, proteins involved in the development of idiopathic pulmonary fibrosis. In summary, the regulation of biosynthesis and the processing of Let-7d using the ADAR1 isoforms should be the subject of new studies and, in the future, could be considered important therapeutic targets for idiopathic pulmonary fibrosis.
Our study has several limitations. At first, we did know how many ADAR1 editing sites have pri-miR-Let-7d and if these sites could be promoting its maturation or its degradation, which has been reported for pri-miR-142, whose degradation is ADAR1-mediated-hyper-edition [31]. On the other hand, we did not explore the role of Let-7d in the conformation of the nucleolus in normal fibroblasts compared with fibrotic fibroblasts because it has been reported that Let-7d takes part in the nucleolar organization in mouse lung fibroblasts (MLg and MFML4), mouse lung epithelial cells (MLE-12), mouse mammary gland epithelial cells (NMuMG), and human lung epithelial carcinoma cells (A549); participate in the epigenetic regulation of several genes [32]. Future work will be necessary to investigate the role of ADAR-2 as a possible regulator of Let-7d and determine the role of ADAR-3, an isoform without catalytic activity but with a role of inhibitor of ADAR1 and 2 because it can form inactive dimers in combination with both proteins.
4. Materials and Methods
4.1. Cell Culture
The study population consisted of primary fibroblasts isolated from control (n = 10) or IPF (n = 10) lung tissue biopsies collected in the frame of the Mexican and the European IPF. All cells were cultured with Ham’s/F-12 medium (Thermo-Fischer, San Francisco, CA, USA) supplemented with 10% Fetal Bovine Serum (Thermo-Fischer, San Francisco, CA, USA). Patient characteristics are shown in Table S1.
4.2. RNA Isolation, Reverse Transcription, Quantitative PCR
The total RNA of fibroblast and mice lung tissue was extracted with Trizol (Thermo-Fischer, San Francisco, CA, USA) and quantified using the Nanodrop system (Thermo-Scientific, San Francisco, CA, USA). The cDNA synthesis was performed using total RNA and the High-Capacity cDNA Reverse Transcriptase kit (Applied Biosystems, San Francisco, CA, USA). Quantitative real-time PCR reactions were performed using SYBR® Green (Applied Biosystems, San Francisco, CA, USA) and TaqMan gene expression Real-Time PCR system (Applied Biosystems, San Francisco, CA, USA) on the Step-One-plus Real-time PCR system (Applied Biosystems, San Francisco, CA, USA) according to the manufacturer’s instructions as previously described [32]. The housekeeping gene HPRT was used to normalize gene expression. All primer pairs and TaqMan assay used are described in Table S2.
4.3. Western Blot
Western blots of total protein were performed as previously described. Proteins were isolated using RIPA lysis buffer (Santa-Cruz Biotechnology, Dallas, TX, USA). Proteins were quantified using the Bradford method (Pierce, CA, USA) in a Beckman DU640 spectrophotometer [32].
All samples were incubated at 95 °C for 5 m with Laemmli buffer, followed by SDS-PAGE. Samples were transferred to nitrocellulose membranes, which were blocked for 1 h at room temperature with 5% fat-free milk in PBS-T 0.05%. Membranes were incubated overnight with a primary antibody at 4 °C (ADAR1, sc-271854; Lamin B, sc-17810, Santa-Cruz Biotechnology), washed with 0.05% PBS-T, and incubated for 1 h at room temperature with an HRP-linked secondary antibody, which was detected with the Super Signal West Dura Extended Duration Substrate detection solutions (Thermo-Scientific, San Francisco, CA, USA). The signal was detected and analyzed with the Chemi-Doc technology system (Bio-Rad, Hercules, CA, USA).
4.4. Immunofluorescence
Immunofluorescence was performed as described previously [12]. Briefly, control and IPF fibroblasts were seeded on glass slides, incubated for 24 h in RPMI at 37 °C, 5% CO2, then fixed with cold methanol, blocked for 20 m with 1× universal blocker (Biogenex, Fremont, CA, USA), and incubated with primary antibodies against ADAR1 (sc-271854) and fibrillarin (sc-374022) (Santa-Cruz Biotechnology, Dallas, TX, USA). Slides were incubated with secondary antibodies coupled to a fluorophore (Jackson-ImmunoResearch, Philadelphia, PA, USA) and analyzed using an FV-1000 confocal microscope (Olympus, Japan).
4.5. Generation of ADAR1-p110 Deletion Mutant
ADAR1-p150 variant was PCR cloned with Platinum-Pfx Polymerase (ThermoFisher, San Francisco, CA, USA) following standard protocols and using ADAR1-p150 wt [18,22] as a template. PCR-Products were gel purified and digested with BamHI (#R0136S NEB, Ipswich, MA, USA) and XbaI (#R0145S, NEB) and ligated overnight at 16 °C into linearized pcDNA3.1-Flag-HA (RRID:Addgene_52335, Cambridge, MA, USA). ADAR1-p110 mutant (catalytically inactive) was generated using ADAR1E912A [22,30] as a template and PCR cloned. All constructs were sequence verified.
4.6. ADAR Catalytic Activity Assay
Editing of RNA in control fibroblasts compared with IPF fibroblasts was measured using the RESS-qPCR [30] as follows. As an edition template, APOBEC3D, the ADAR1-mediated editing target, was selected. cDNA of 6 control lines and 6 fibrotic lines (2 μL cDNA per sample) was amplified by qPCR (Table S1), by serial dilutions, and the efficiency of the primers for APOBEC3D wt and APOBEC3D edit was tested, and in a final volume of 10 μL (5 μL #1176202K SYBR GreenER Super Mix Life Technologies, San Francisco, CA, USA. 0.2 μL of primers, calibrated to 10 μL with nuclease-free water). Edition ratios were calculated using the values of the efficiency of the primers in the following way:
| (1) |
Control and IPF primary cultures were transfected with overexpression constructs for ADAR1-p110 wt, ADAR1-p110 mutant, ADAR1-p150 wt, and ADAR1-p150 mutant, as well as the empty vector. Cells were treated with 0.5 μM of pentostatin (#sc-204177 Santa-Cruz Biotechnology, Santa Cruz, CA, USA) 24 h post-transfection to inhibit the catalytic activity of Adenosine Deaminase (ADA) in order to increase the specificity of this assay. Cells were collected 48 h post-transfection and used for the catalytic activity test.
The formation of inosine (from the hydrolysis of adenosine) was measured in aliquots of the proteins extracted from the cells transfected with specific plasmids that were used for each isoform wild-type and mutant of ADAR1, the colorimetric assay Adenosine Deaminase Activity Assay Kit (#K321-100 Biosvision, Milpitas, CA, USA) was used. The protein extract of the cells was prepared by adding 150μL of cold ADA 1× assay buffer with protease inhibitor (#14865751 Thermo-Scientific, San Francisco, CA, USA). Dilutions of inosine standard were used as a standard curve. As a negative control, 50 μL of 1× ADA Assay Buffer was used. The reading plate was incubated at 37 °C for 5 m, after which it was read at a wavelength of 293 nm using the Infinite 200 plate reader (Tecan, Männedorf, Switzerland) at time points 0, 10, 20, and 40 m. All data were analyzed using the following formula:
| (2) |
Inosine standard curves were set at 4 time points for the analysis.
4.7. Selection of Target mRNA
In silico analysis was performed to investigate Let-7d targets, using miRWalk, miRanda, and TargetScan algorithms. Obtained readouts were compared to select the top 250 mRNA with differential expression in IPF, and Collagen III alpha 1 (COL3A1) and SMAD2 were selected.
4.8. Pentostatin Treatment in IPF Fibroblasts
IPF fibroblasts were treated with 0.5 μM of pentostatin (#sc-204177 Santa-Cruz Bio technology, Dallas, TX, USA). At 24 h after, Let-7d and pri-mir-Let-7d expressions were determined by real-time PCR as described before.
4.9. Statistical Methods
All data follow a normal distribution, the Student´s test was used to compare RNA expression, and values are expressed as mean ± SD. All statistical analyses were performed using GraphPad Prism (San Diego, CA, USA).
Acknowledgments
The authors thank Remedios Ramirez Rangel and Jorge A. García-Álvarez, UNAM Facultad de Ciencias, for technical assistance.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23169028/s1.
Author Contributions
V.R., R.M.O.-R. and G.B. designed research; G.D.-P., K.R., E.M., C.B., A.C.-R., A.A.-G. and A.S. conducted research. V.R., R.M.O.-R., G.B, K.R. and H.C.-F. analyzed data. V.R., R.M.O.-R., G.B. and G.D.-P. wrote the paper; all authors read and approved the final manuscript. V.R. and R.M.O.-R. had primary responsibility for the final content. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Mexican samples were obtained under approval by the Scientific and Ethics Committee of Health Research from the Instituto Nacional de Enfermedades Respiratorias (INER), and the European samples were registry and provided by the UGMLC Giessen Biobank (member of the DZL Platform Biobanking) and the protocols approved by the institutional review board and ethics committee of the Faculty of Medicine at the Justus Liebig University in Giessen, Germany (AZ.111/08-eurIPFreg). The study was conducted according to the principles set out in the WMA Declaration of Helsinki.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
Guillermo Barreto was funded by the “Centre National de la Recherche Scientifique” (CNRS, Paris, France), “Délégation Centre-Est” (CNRS-DR6), the “Lorraine Université” (LU, Metz, France) through the initiative “Lorraine Université d’Excellence” (LUE) and the dispositive “Future Leader” and the “Deutsche Forschungsgemeinschaft” (DFG, Bonn, Germany) (BA 4036/4-1). Karla Rubio was funded by the “Consejo de Ciencia y Tecnología del Estado de Puebla” (CONCYTEP, Puebla, Mexico) through the initiative International Laboratory EPIGEN.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fernandez I.E., Eickelberg O. New cellular and molecular mechanisms of lung injury and fibrosis in idiopathic pulmonary fibrosis. Lancet. 2012;380:680–688. doi: 10.1016/S0140-6736(12)61144-1. [DOI] [PubMed] [Google Scholar]
- 2.Martinez F.J., Collard H.R., Pardo A., Raghu G., Richeldi L., Selman M., Swigris J.J., Taniguchi H., Wells A.U. Idiopathic Pulmonary Fibrosis. Nat. Rev. 2017;3:17074. doi: 10.1038/nrdp.2017.74. [DOI] [PubMed] [Google Scholar]
- 3.Selman M., Pardo A., Kaminski N. Idiopathic Pulmonary Fibrosis: Aberrant Recapitulation of Developmental Programs? PLoS Med. 2008;5:373–380. doi: 10.1371/journal.pmed.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee J.S., Collard H.R., Raghu G., Sweet M.P., Hays S.R., Campos G.M., Golden J.A., King T.E., Jr. Does Chronic Microaspiration Cause Idiopathic Pulmonary Fibrosis? Am. J. Med. 2010;123:304–311. doi: 10.1016/j.amjmed.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King T.E., Jr., Pardo A., Selman M. Idiopathic pulmonary fibrosis. Lancet. 2011;378:1949–1961. doi: 10.1016/S0140-6736(11)60052-4. [DOI] [PubMed] [Google Scholar]
- 6.Hinz B., Phan S.H., Thannickal V.J., Galli A., Bochaton-Piallat M.L., Gabbiani G. The myofibroblast: One function, multiple origins. Am. J. Pathol. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selman M., Pardo A. Idiopathic pulmonary fibrosis: An epithelial/fibroblastic cross-talk disorder. Respir Res. 2002;3:3. doi: 10.1186/rr175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kis K., Liu X., Hagood J.S. Myofibroblast differentiation and survival in fibrotic disease. Expert Rev. Mol. Med. 2011;13:e27. doi: 10.1017/S1462399411001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandit K., Milosevic J., Kaminski N. MiRNAs in Idiopathic pulmonary fibrosis. Trans. Res. 2011;157:191–199. doi: 10.1016/j.trsl.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Rubio K., Castillo-Negrete R., Barreto G. Non-coding RNAs and nuclear architecture during epithelial-mesenchymal transition in lung cancer and idiopathic pulmonary fibrosis. Cell Signal. 2020;70:109593. doi: 10.1016/j.cellsig.2020.109593. [DOI] [PubMed] [Google Scholar]
- 11.Yang S., Banerjee S., de Freitas A., Sanders Y.Y., Ding Q., Matalon S., Thannickal V.J., Abraham E., Liu G. Participation of miR-200 in Pulmonary Fibrosis. Am. J. Phatol. 2012;180:484–493. doi: 10.1016/j.ajpath.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huleihel L., Ben-Yehudah A., Milosevic J., Yu G., Pandit K., Sakamoto K., Yousef H., LeJeune M., Coon T.A., Redinger C.J., et al. Let-7d microRNA affects mesenchymal phenotypic properties of lung fibroblasts. Am. J. Physiol. Lung Cell Mol. Physiol. 2014;306:L534–L542. doi: 10.1152/ajplung.00149.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubio K., Singh I., Dobersch S., Sarvari P., Günther S., Cordero J., Mehta A., Wujak L., Cabrera-Fuentes H., Chao C.-M., et al. Inactivation of nuclear histone deacetylases by EP300 disrupts the MiCEE complex in Idiopathic Pulmonary Fibrosis. Nat. Commun. 2019;10:2229. doi: 10.1038/s41467-019-10066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Díaz-Piña G., Ordoñez-Razo R.M., Montes E., Páramo I., Becerril C., Salgado A., Santibañez-Salgado J.A., Maldonado M., Ruiz V. The Role of ADAR1 and ADAR2 in the Regulation of miRNA-21 in Idiopathic Pulmonary Fibrosis. Lung. 2018;196:393–400. doi: 10.1007/s00408-018-0115-9. [DOI] [PubMed] [Google Scholar]
- 15.Narry Kim V. MicroRNA Biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 16.Faller M., Guo F. MicroRNA biogenesis: There’s more than one way to skin a cat. Biochim. Biophys. Acta. 2008;1779:663–667. doi: 10.1016/j.bbagrm.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heale B.S., Keegan L.P., McGurk L., Michlewski G., Brindle J., Stanton C.M., Caceres J.F., O’Connell M.A. Editing independent effects of ADARs on the miRNA/siRNAs pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luciano D.J., Mirsky H., Vendetti N.J., Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson J.B., Samuel C.E. Expression and regulation by interferon of a double stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol. Cell Biol. 1995;15:5376–5388. doi: 10.1128/MCB.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barraud P., Allain F.H. ADAR proteins: Double-stranded RNA and Z-DNA binding domains. Curr. Top Microbiol. Immunol. 2012;353:35–60. doi: 10.1007/82_2011_145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orlandi C., Barbon A., Barlati S. Activity Regulation of Adenosine Deaminases Acting on RNA (ADARs) Mol. Neurobiol. 2012;45:61–75. doi: 10.1007/s12035-011-8220-2. [DOI] [PubMed] [Google Scholar]
- 22.Paul M., Bass B. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zipeto M.A., Court A.C., Sadarangani A., Santos N.P.D., Balaian L., Chun H.J., Pineda G., Morris S.R., Mason C.N., Geron I., et al. ADAR1 Activation Drives Leukaemia Stem Cell Self-Renewal by Impairing Let-7 Biogenesis. Cell Stem Cell. 2016;19:177–191. doi: 10.1016/j.stem.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crews L.A., Jiang Q., Zipeto M.A., Lazzari E., Ali S., Barrett C.L., Barrett C.L., Frazer K.A., Jamieson C.H.M. An RNA editing fingerprint of cancer stem cell reprogramming. J. Transl. Med. 2015;13:52. doi: 10.1186/s12967-014-0370-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang B., Hu P., Lin X., Han W., Zhu L., Tan X., Ye F., Wang G., Wu F., Yin B., et al. PTBP1 induces ADAR1 p110 isoform expression through IRES-like dependent translation control and influences cell proliferation in gliomas. Cell. Mol. Life Sci. 2015;72:4383–4397. doi: 10.1007/s00018-015-1938-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desterro J.M., Keegan L.P., Lafarga M., Berciano M.T., O’Connell M., Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- 27.Brümmer A., Yang Y., Chan T.W., Xiao X. Structure-mediated modulation of mRNA abundance by A-to-I editing. Nat. Commun. 2017;8:1255. doi: 10.1038/s41467-017-01459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmittgen T.D. Regulation of microRNA processing in development, differentiation and cancer. J. Cell Mol. Med. 2008;12:1811–1819. doi: 10.1111/j.1582-4934.2008.00483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sansam C.L., Wells K.S., Emeson R.B. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Nat. Acad. Sci. USA. 2003;100:14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishikura K. Functions and Regulation of RNA Editing by ADAR Deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Connell M.A., Keegan L.P. Drosha versus ADAR: Wrangling over pri-miRNA. Nat. Struct. Mol. Biol. 2006;13:3–4. doi: 10.1038/nsmb0106-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.