Abstract
A 20S proteasome, composed of α1 and β subunits arranged in a barrel-shaped structure of four stacked rings, was purified from a halophilic archaeon Haloferax volcanii. The predominant peptide-hydrolyzing activity of the 600-kDa α1β-proteasome on synthetic substrates was cleavage carboxyl to hydrophobic residues (chymotrypsin-like [CL] activity) and was optimal at 2 M NaCl, pH 7.7 to 9.5, and 75°C. The α1β-proteasome also hydrolyzed insulin B-chain protein. Removal of NaCl inactivated the CL activity of the α1β-proteasome and dissociated the complex into monomers. Rapid equilibration of the monomers into buffer containing 2 M NaCl facilitated their reassociation into fully active α1β-proteasomes of 600 kDa. However, long-term incubation of the halophilic proteasome in the absence of salt resulted in hydrolysis and irreversible inactivation of the enzyme. Thus, the isolated proteasome has unusual salt requirements which distinguish it from any proteasome which has been described. Comparison of the β-subunit protein sequence with the sequence deduced from the gene revealed that a 49-residue propeptide is removed to expose a highly conserved N-terminal threonine which is proposed to serve as the catalytic nucleophile and primary proton acceptor during peptide bond hydrolysis. Consistent with this mechanism, the known proteasome inhibitors carbobenzoxyl-leucinyl-leucinyl-leucinal-H (MG132) and N-acetyl-leucinyl-leucinyl-norleucinal (calpain inhibitor I) were found to inhibit the CL activity of the H. volcanii proteasome (Ki = 0.2 and 8 μM, respectively). In addition to the genes encoding the α1 and β subunits, a gene encoding a second α-type proteasome protein (α2) was identified. All three genes coding for the proteasome subunits were mapped in the chromosome and found to be unlinked. Modification of the methods used to purify the α1β-proteasome resulted in the copurification of the α2 protein with the α1 and β subunits in nonstoichometric ratios as cylindrical particles of four stacked rings of 600 kDa with CL activity rates similar to the α1β-proteasome, suggesting that at least two separate 20S proteasomes are synthesized. This study is the first description of a prokaryote which produces two separate 20S proteasomes and suggests that there may be distinct physiological roles for the two different α subunits in this halophilic archaeon.
ATP-dependent proteolysis is central to the breakdown of the majority of proteins, including short-lived regulatory and metabolic enzymes in prokaryotic and eukaryotic cells (reviewed in references 21, 34, and 37). A variety of ATP-dependent proteases have been identified, and these include Lon, FtsH (HflB), ClpAP, ClpXP, HslUV (ClpYQ), and the 26S proteasome (reviewed in references 7 and 54). Although the ATP-dependent proteases have limited primary sequence homology, their quaternary structures have converged to a universal self-compartmentalized barrel-like complex with the proteolytic active sites located in a central chamber (reviewed in references 7 and 54). Channels leading to the central chamber form physical barriers which limit substrate access to the active sites and, thus, protect cytoplasmic proteins from nonspecific degradation (53, 81, 82). In addition, these structures prevent the dissociation of partially digested substrate proteins to insure a processive mechanism of proteolysis (1, 78). The high concentration of active sites in the central proteolytic chamber and the narrow portal to the chamber are presumed to play a role in determining the size of peptide products generated (47, 48).
The 20S proteasome, ClpP, and HslV complexes all catalyze ATP-independent proteolysis of unfolded proteins in vitro and have been shown to associate with ATPase subunits to form larger, energy-dependent proteases (reviewed in references 7 and 54). Of these, however, the 20S proteasome has the most elaborate structure, consisting of four stacked seven-membered rings. Although the number of different subunits which form the 20S complex varies among organisms, the primary sequence of all proteasome subunits classify them to α and β superfamilies (20). The α-type subunits form the outer rings and are presumed to limit the access of protein substrates into and out of the central proteolytic chamber formed by the two inner rings of β-type subunits (36, 53). The α subunits can self assemble into heptameric rings and are presumed to provide scaffolding for assembly as well as autocatalytic processing of the β subunits (16, 55, 67, 70). Several of the β-type subunits belong to the N-terminal hydrolase family (9) and have an active site N-terminal threonine residue which is exposed after the autocatalytic removal of a prosequence (29, 36, 53, 55, 69, 70). The 20S proteasome appears to be restricted to the Eucarya, Archaea, and gram-positive actinomycetes of the Bacteria, and, based on genomic sequence data, it is likely that ATPase subunits of the 26S proteasome are also conserved in these organisms (reviewed in reference 7).
Among the halophilic Archaea, numerous extracellular serine proteases have been characterized. These include the 41- and 66-kDa proteases from Halobacterium salinarium (39, 46, 65), the halolysins of the thermitase branch of subtilases isolated from Haloferax mediterranei strains R4 and 1539 (42, 74) as well as the haloalkaliphile Natrialba asiatica (43), and the 60-kDa ESP4 protease of Halobacterium sp. strain TuA4 (68). In addition, extracellular and intracellular protease activities have been reported in the haloarchaea (3, 33, 61). However, halophilic self-compartmentalizing proteases, such as proteasomes, have not been described. In fact, attempts to identify genomic DNA which may encode 20S proteasomes from the halophile Haloferax volcanii by using DNA cross-hybridization techniques with proteasome gene fragments from Thermoplasma acidophilum (63) or Methanosarcina thermophila (54a) as probes were unsuccessful. In addition, preliminary attempts to purify H. volcanii proteasomes suggested that the complex may not be synthesized in this organism (63).
In this study, we report the biochemical and genetic analyses of 20S proteasomes from H. volcanii, a halophile growing optimally in salt concentrations of 2 M NaCl and 1.4 M MgCl2, similar to the mud of the Dead Sea where it was isolated (57). This is the first description of an archaeon which synthesizes two different α subunits, α1 and α2, which copurify with a β subunit as at least two separate 20S proteasomes. Currently, H. volcanii is one of the few halophilic archaea which is known to have a stable genome (52), and the organism is readily transformed with linear and circular DNA (13, 17). In addition, a variety of shuttle and expression vectors which are compatible with Escherichia coli (38, 60), transcription reporter systems (24, 62), and a complete set of overlapping cosmid clones covering the genome are available for genetic studies of this organism (14). This study, therefore, provides a genetic and biochemical characterization of archaeal 20S proteasomes in a microbial system where a variety of genetic tools are available to analyze the role of the proteasome in bacterial cell physiology.
MATERIALS AND METHODS
Materials.
Biochemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.). Other organic and inorganic chemicals were from Fisher Scientific (Atlanta, Ga.) and were of analytical grade. Restriction endonucleases and DNA-modifying enzymes were from New England Biolabs (Beverly, Mass.) or Promega (Madison, Wis.) unless otherwise indicated. Oligonucleotides were from Genemed Synthesis (San Francisco, Calif.). Digoxigenin-11-dUTP (2′-deoxyuridine-5′-triphosphate coupled by an 11-atom spacer to digoxigenin) and alkaline phosphatase conjugate antibody raised against digoxigenin were from Boehringer Mannheim (Indianapolis, Ind.). Polyvinylidene difluoride and positively charged nylon membranes were from MicroSeparations (Westborough, Mass.) and Ambion (Austin, Tex.), respectively.
Purification of halophilic 20S proteasomes.
H. volcanii DS2 was grown aerobically in a complex medium (ATCC 974) at 42°C in a shaker at 200 rpm. Cells from log to stationary phases of growth (A600 ≅ 0.29 to 3.6 U) were harvested by centrifugation at 10,800 × g for 25 min at 4°C. Cell pellets were resuspended in a 2.5-fold-larger volume of 2 M NaCl-Tris buffer (50 mM Tris-HCl buffer containing 1 mM dithiothreitol [DTT] with 2 M NaCl at pH 7.2) and passed through a French pressure cell at 20,000 psi. Extract was clarified by centrifugation for 30 min at 15,600 × g at 4°C. Polyethylene glycol (PEG) 8000 was added to the supernatant at a final concentration of 5%, and the solution was gently stirred for 15 min at 21°C. The pellet was discarded after centrifugation at 10,800 × g for 15 min at 4°C. The concentration of PEG 8000 in the supernatant was increased to 15%, and the proteasome fraction was precipitated. After centrifugation at 10,800 × g for 15 min, the protein pellet was resuspended in 2 M NaCl-Tris buffer to a final concentration of 10 mg of protein ml−1. The sample was heated to 90°C for 5 min, chilled on ice for 15 min, and centrifuged at 4°C for 15 min at 15,600 × g. This heat treatment removed most of the contaminating proteins. The resulting supernatant was slowly diluted threefold with 50 mM Tris-HCl buffer containing 3 M (NH4)2SO4, 1 M NaCl, and 1 mM DTT at pH 7.2. The sample was centrifuged at 4°C for 15 min at 15,600 × g, and the supernatant was filtered through glass wool to remove membrane vesicles and contaminating proteins. The filtrate was applied to a DE52 cellulose column (1.5 by 30 cm) that was equilibrated in 50 mM Tris-HCl buffer containing 1.96 M (NH4)2SO4, 1.74 M NaCl, and 1 mM DTT at pH 7.2. The column was washed with 200 ml of the equilibration buffer, and the sample was eluted with 50 mM Tris-HCl buffer containing 1.84 M (NH4)2SO4, 1.86 M NaCl, and 1 mM DTT at pH 7.2. The fractions catalyzing hydrolysis of succinyl-Leu-Leu-Val-Tyr-4-methyl-coumaryl-7-amide (Suc-LLVY-Amc) were pooled and dialyzed against 2 M NaCl-Tris buffer at 4°C for 16 h at a final concentration of ∼0.01 to 0.03 mg ml−1. The proteasome fractions were concentrated by 15% PEG 8000 precipitation and dialyzed against 2 M NaCl-Tris buffer. Concentrated samples were applied to a Superose 6 HR (Pharmacia) 10/30 column (2.5 by 28.5 cm) equilibrated with 2 M NaCl-Tris buffer, and the high-molecular-mass fractions (600 kDa) were collected. Proteasome fractions were determined to be pure by reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (51).
Assays of protein and peptide degradation.
Protein hydrolysis was measured with bovine oxidized insulin B chain or β-casein as a substrate (Sigma). Purified proteasome (0.035 mg) and 145 μM substrate, in a final volume of 1 ml containing 2 M NaCl-Tris buffer and 6% (vol/vol) dimethyl sulfoxide, were incubated at 60°C. The rate of hydrolysis of oxidized insulin B chain was linear within the first 15 min and was dependent on proteasome concentration. Samples of 2 to 20 μl were removed from the reaction mixture at intervals of 0 to 15 min for oxidized insulin B chain and 0 to 135 min for β-casein and were added to a mixture of 100 μl of 0.1 M sodium phosphate buffer (pH 6.8) and 50 μl of fluorescamine (0.3 mg per ml of acetone) according to Akopian et al. (1). After thorough mixing for 1 min (vortex mixing), the volume was increased to 1 ml by addition of dH2O. The α-amino groups generated by peptide bond hydrolysis were measured by fluorescence with an excitation wavelength of 370 nm and an emission of 480 nm with leucine as the standard (79) (Aminco-Bowman series 2 luminescence spectrometer; Spectronic Instruments, Rochester, N.Y.).
Peptide-hydrolyzing activity was assayed by fluorimetric measurement of the release of 7-amino-4-methylcoumarin or the colorimetric measurement of the release of β-naphthylamine (β-Na) by using synthetic peptide substrates as previously described (55). Specific activities were reported as nanomoles of product per min per mg of protein. Protein concentration was determined by the Bradford method (8) using bovine serum albumin as the standard.
Molecular mass and amino acid sequence determination.
Molecular masses of the purified proteasome subunits were determined by reducing and denaturing SDS-PAGE using 12% polyacrylamide gels (51) which were stained with Coomassie blue R-250. The molecular mass standards for SDS-PAGE were phosphorylase b (97.4 kDa), serum albumin (66.2 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), trypsin inhibitor (21.5 kDa), and lysozyme (14.4 kDa).
For determination of the native molecular mass and stability of the proteasomes, samples were dialyzed against 1,000 volumes of 50 mM Tris-HCl buffer containing 1 mM DTT at pH 7.2 (Tris buffer) in the presence and absence of 2 M NaCl at 4°C for 16 h. The dialyzed samples were applied to a Superose 6 column equilibrated in 2 M NaCl Tris buffer and eluted with 2 M NaCl-Tris buffer to allow an appropriate comparison of the H. volcanii proteins. The molecular mass standards were applied to the same column equilibrated in 50 mM Tris-HCl buffer with 150 mM NaCl and 1 mM DTT at pH 7.2 to avoid denaturation of the nonhalophilic proteins. These standards included serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), β-amylase (200 kDa), apoferritin (443 kDa), and thyroglobulin (669 kDa).
The N-terminal sequence of the β subunit was determined from purified protein which had been separated by SDS-PAGE and electroblotted onto a polyvinylidene difluoride (Immobilon-P) membrane. The N-terminally blocked α1 and α2 proteins were separated by SDS-PAGE, eluted into 0.2 M ammonium biocarbonate (pH 8.4), and incubated with trypsin or endoproteinase Lys-C. The protein fragments were separated by C18 reversed-phase high-pressure liquid chromatography analysis, and three internal fragments for each α protein were sequenced. The amino acid sequences of the N terminus of the β subunit and the internal fragments of the α1 and α2 subunits were determined by automated Edman degradation (26) at the protein chemistry core facility of the University of Florida Interdisciplinary Center for Biotechnology Research.
Cloning and sequencing the proteasome-encoding DNA.
Degenerate oligonucleotides based on the partial amino acid sequences determined for the α1 and β subunits were used to generate DNA probes to screen genomic DNA for the genes encoding these proteins. In addition, the codon bias of H. volcanii (80) was used in the oligonucleotide design. The primers 5′-CGS CTS GTS CAG GTS GAR TAC GC-3′ (primer 1) and 5′-CC SSW SGG GTC SGT YTC GTA SAG-3′ (primer 2) were based on the internal α1-subunit protein sequences RLVQVEYAR and GSPDTEYL (where S indicates C plus G, R indicates A plus G, W indicates A plus T, and Y indicates T plus C) which shared identity with other α-type proteasome proteins. The PCR used to generate an internal α1-gene (psmA) fragment contained 10 mM Tris-HCl at pH 8.8, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100, 250 μM deoxynucleoside triphosphates (dNTPs), 1.5 μM each of primer 1 and 2, 500 ng of genomic DNA, and 0.02 U of Vent (exo−) DNA polymerase per μl. Cycling conditions were 45 s at 94°C, 45 s at 37°C, and 45 s at 72°C (for five cycles) followed by 45 s at 94°C, 45 s at 52°C, and 45 s at 72°C (for 30 cycles) by using a Gene Cycler (BioRad, Hercules, Calif.). The resulting 0.4-kb PCR product was purified by 0.8% agarose gel electrophoresis, was phosphorylated using T4 polynucleotide kinase, and was cloned into the HincII site of pUC19. The cloned DNA fragment was sequenced by the dideoxy chain termination method (66) to confirm that the cloned DNA sequence was indeed an α-type proteasome gene fragment (DNA Sequencing Facility, Department of Microbiology and Cell Science, University of Florida). The α-subunit gene fragment was excised from the pUC19-based plasmid with the restriction enzymes EcoRI and HindIII and was used as a template (100 ng) in a linear labeling reaction containing 10 mM Tris-HCl buffer at pH 9.0 with 50 mM KCl, 0.1% Triton X-100, 6 mM MgCl2, 20 μM digoxigenin-11-dUTP, 30 μM each of dATP and dTTP, 45 μM each of dGTP and dCTP, 3 μM primer 2, and 0.05 U of Taq DNA polymerase per μl. Cycling conditions were 45 s at 95°C, 1 min at 60°C, and 2 min at 72°C (for 40 cycles). The linear DNA fragment labeled with digoxigenin was used as an α1-gene (psmA) probe in the hybridization experiments described below. The β-subunit sequence TTTVGIKTEEGVVLATDMRA was used to synthesize a β-gene probe (5′-ACS ACS ACS GTS GGY ATC AAG ACS GAR GAR GGY GTS GTS CTS GCS ACS GAC ATG CGC GC-3′) which was directly 3′-labeled by using terminal transferase with digoxigenin-11-dUTP and dATP as recommended by the supplier (Boehringer Mannheim).
For Southern analysis, genomic DNA was isolated from stationary-phase cultures of H. volcanii according to the method of Jolley et al. (40) with modifications according to Ausubel et al. (5). The purified genomic DNA was cleaved with HincII, EcoRI, or SalI, was separated by 0.8% agarose electrophoresis, and was transferred by downward capillary action to positively charged nylon membranes (73). Membranes were equilibrated at 60°C for 2 h in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 1% blocking reagent (Boehringer Mannheim), 0.1% N-lauroylsarcosine, and 0.02% SDS. The α-specific probe was added to a final concentration of 25 ng/ml and was incubated at 60°C. The β-specific probe was added to a final concentration of 1.25 pmol/ml along with 0.1 mg of Poly(A) per ml, and the membrane was incubated at 68°C. After 14 h, both membranes were washed twice with a solution containing 2× SSC and 0.1% SDS for 5 min per wash. The membrane incubated with the α-specific probe was then washed twice with a solution containing 0.5× SSC and 0.1% SDS for 15 min per wash at 60°C. The membrane incubated with the β probe was washed twice with a solution containing 0.1× SSC and 0.1% SDS for 15 min per wash at 70°C. Signals were visualized colorimetrically according to supplier instructions (Boehringer Mannheim). For isolation of cosmids 5G7, 547, 2D7, and 564 from a set of overlapping genomic clones (14), similar hybridization conditions were used with [α32-P]-labeled probes according to the method of Charlebois et al. (12, 18). The α- and β-proteasome gene fragments isolated from cosmid clones were ligated into pUC19 and were sequenced using the Sanger dideoxy method (66).
Electron microscopy.
The 20S proteasomes purified from H. volcanii were placed on 200-mesh grids coated with Formvar or carbon film, were fixed with 2% cacodylate-buffered glutaraldehyde, were briefly stained with 1% aqueous uranyl acetate, and were treated with 0.01% bacitracin to improve spreading (35). Samples were viewed and photographed on a Zeiss EM-10CA transmission electron microscope operated at 80 kV.
Protein sequence analyses.
GenBank, EMBL, and SwissProt databases were searched at the National Center for Biotechnology Information, Bethesda, Md. with the BLAST network server (2). CLUSTALW version 1.7 (77) was used for alignment of protein sequences accessible through the National Center for Biotechnology Information.
Nucleotide sequence accession numbers.
The nucleotide sequences of the H. volcanii proteasome genes have been deposited in the GenBank database under accession no. AF126260 (psmA, encoding the α1 subunit), AF126262 (psmB, encoding the β subunit), and AF126261 (psmC, encoding the α2 subunit).
RESULTS AND DISCUSSION
Purification of a 20S proteasome of α1 and β subunits from H. volcanii.
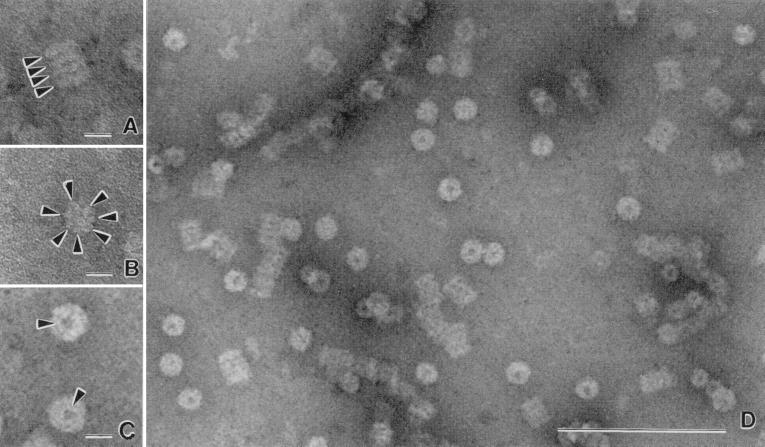
A 600-kDa proteasome was purified from H. volcanii and was found to be composed of α1 and β subunits with relative molecular masses of 37.5 and 30 kDa, respectively (Fig. 1). Electron micrographs of the negatively stained proteasomes reveal barrel-shaped structures which measure 12 nm in diameter by 17 nm in height (Fig. 2A to D). The rectangular side views of the purified protein (Fig. 2A) suggest that the complex is composed of four stacked rings of uniform diameter. In addition, some of the protein particles are positioned in end-on views which reveal discrete protein domains located in the outer ring of the complex (Fig. 2B). The direct end-on views of the purified proteasome (Fig. 2C) reveal an outer pore which, based on analogy to other self-compartmentalized proteases, is presumed to be the passageway for substrate proteins. The distinct structural features and size of the purified α1β-proteasomes suggest that the four-stacked heptameric ring structure which is characteristic of other 20S proteasomes is conserved in H. volcanii (6, 22, 55, 58, 71, 75).
FIG. 1.
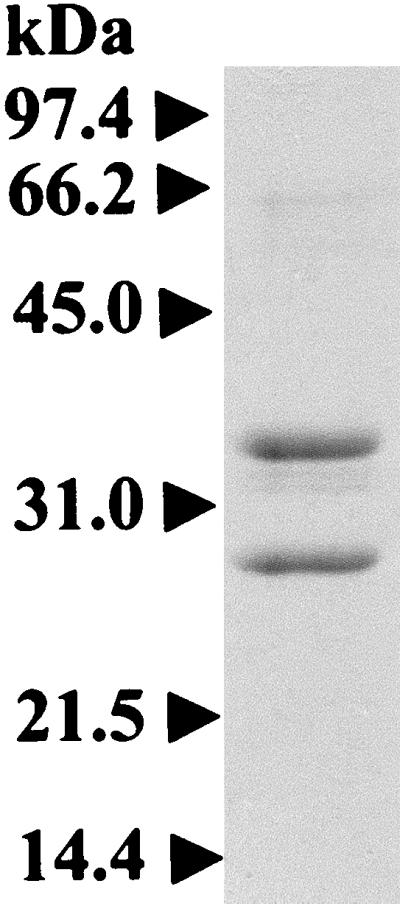
Reducing SDS-PAGE of a 20S proteasome (2.3 μg) purified from H. volcanii.
FIG. 2.
Electron micrograph of negatively stained H. volcanii proteasomes of α1 and β subunits. (A) Side view of proteasome cylinder with arrowheads indicating the four stacked rings. (B) End-on view revealing protein domains within a ring-like structure indicated by arrowheads. (C) Direct end-on view of central channels indicated by arrowheads. (D) Barrel-shaped cylindrical structures of purified proteasomes. Bars, 100 nm.
Catalytic properties.
The purified H. volcanii proteasome comprised of α1 and β subunits catalyzed the hydrolysis of oxidized bovine insulin B-chain protein with a specific activity of 82 nmol of leucine equivalents per h per mg of protein in 2 M NaCl-Tris buffer at 60°C. Hydrolysis of bovine β-casein by the H. volcanii α1β-proteasome was undetectable under similar assay conditions. These results contrast with the 20S proteasomes of T. acidophilum and M. thermophila which hydrolyze bovine β-casein in low-concentration salt buffers at denaturing temperatures, including 60°C (23, 55). This difference in the rate of β-casein hydrolysis between the halophilic and related archaeal 20S proteasomes is probably due to aggregation of the nonhalophilic substrate protein at the high concentrations of salt required to maintain the H. volcanii α1β-proteasome as an active complex (see below). Since 20S proteasomes require the peptide chain of substrate proteins to pass through a narrow pore of ∼13 Å (82), aggregation of β-casein, which is otherwise unfolded at 60°C, would be expected to eliminate hydrolysis by the H. volcanii α1β-proteasome.
The peptide-hydrolyzing activities of the purified proteasome of H. volcanii were examined with a variety of fluorogenic (amido-methyl-coumarin [Amc] linked) and chromogenic (β-Na linked) small-peptide substrates to identify sites of hydrolysis of the peptide chain (Table 1). The highest measured activity of the proteasome was cleavage carboxyl to the hydrophobic residues phenylalanine, tyrosine, and tryptophan (chymotrypsin-like [CL] activity) as determined by using Suc-Ala-Ala-Phe-Amc (Suc-AAF-Amc), Suc-LLVY-Amc, and Suc-Ile-Ile-Trp-Amc. Among the three substrates, when the phenylalanine and tyrosine residues preceded Amc, the activity was the highest. Substantially lower activities were measured with the peptidyl-glutamyl peptidase substrates Cbz-Leu-Leu-Glu-βNa (Cbz-LLE-βNa) and Ala-Glu-Amc, which were at 8 and 3%, respectively, of the highest measured CL activity (Suc-AAF-Amc hydrolysis). Synthetic substrates which are cleaved by trypsin were not hydrolyzed by the H. volcanii proteasome. The substrate preference of the H. volcanii proteasome is similar to the activities of proteasomes from the archaebacteria T. acidophilum (1, 23) and Pyrococcus furiosus (6) as well as the eubacterial actinomycetes Rhodococcus erythropolis (75) and Streptomyces coelicolor (58). However, this substrate preference contrasts with the proteasome from the methanogenic archaeon M. thermophila which hydrolyzes Cbz-LLE-βNa at a significantly higher rate than the CL substrates Suc-LLVY-Amc and Suc-AAF-Amc (55).
TABLE 1.
Peptide-hydrolyzing activities of a 20S proteasome of H. volcanii
| Activitya | Substrateb | Sp act (nmol min−1 mg−1) |
|---|---|---|
| PGPH | Cbz-Leu-Leu-Glu-βNa | 28 |
| Ala-Glu-Amc | 10 | |
| Ac-Tyr-Val-Ala-Asp-Amc | UDc | |
| CL | Suc-Ala-Ala-Phe-Amc | 370 |
| Suc-Leu-Leu-Val-Tyr-Amc | 340 | |
| Suc-Ile-Ile-Trp-Amc | 110 | |
| TL | Boc-Phe-Ser-Arg-Amc | UD |
| Gly-Gly-Arg-Amc | UD |
PGPH, peptidyl-glutamyl peptide hydrolyzing; TL, trypsin-like. Peptide hydrolysis was measured at 60°C by using 100 μM substrate and 0.01 mg of protein in 1 ml of Tris-2 M NaCl buffer with 6% (vol/vol) dimethyl sulfoxide.
Cbz, carbobenzoxy; Ac, acetyl; Suc, succinyl; Boc, tert-butyloxycarbonyl.
UD, undetectable.
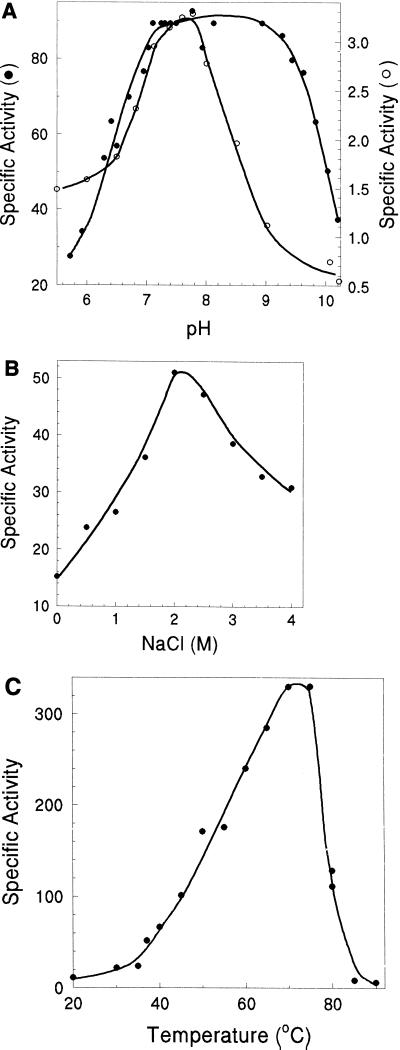
The Suc-LLVY-Amc-hydrolyzing activity of the H. volcanii proteasome had a broad pH optimum, between pH 7.0 and 9.3 (Fig. 3A). This broad pH optimum is comparable to the T. acidophilum 20S proteasome (23) and differs from the narrower optimum of pH 7.2 to 7.8 for the M. thermophila 20S proteasome (55) (Fig. 3A). Although the ability to hydrolyze synthetic substrates is not indicative of the peptide products generated during hydrolysis of large protein substrates by the proteasome, the observed differences in substrate specificities and pH optima suggest that there are distinct differences in the ability of archaeal 20S proteasomes to bind short peptide ligands.
FIG. 3.
Effect of pH (A), salt (B), and temperature (C) on the CL activity of H. volcanii proteasome of α1 and β subunits (●). (A) Purified enzyme at 0.0232 mg per ml of 2 M NaCl-Tris buffer was diluted 60-fold in 50 mM 2-[N-morpholino]-ethanesulfonic acid (pH 5.7–6.7), 50 mM n-Tris[hydroxymethyl]methyl-2- aminoethanesulfonic acid (pH 6.8 to 8.2), and 50 mM 3-(cytohexylamino)-2-hydroxy-1-propanesulfonic acid (pH 8.9 to 10.2) buffers with 2 M NaCl. The sample was equilibrated for 15 min at 21°C and then incubated at 37°C with 20 μM Suc-LLVY-Amc and 0.8% dimethylsulfoxide. The pH optimum for the Suc-LLVY-Amc hydrolyzing activity of a M. thermophila 20S proteasome (○) which was previously reported (55) is included for comparison. (B) Purified enzyme at 0.0232 mg per ml of 2 M NaCl-Tris buffer was diluted 60-fold to the final concentrations of NaCl indicated and was incubated for 15 min at 21°C before assaying for activity at 37°C with 20 μM Suc-LLVY-Amc and 6% (vol/vol) dimethylsulfoxide. (C) Peptide hydrolysis was assayed at the temperatures indicated by using 20 μM Suc-LLVY-Amc and 0.4 μg of protein per ml of 2 M NaCl-Tris buffer with 6% (vol/vol) dimethylsulfoxide.
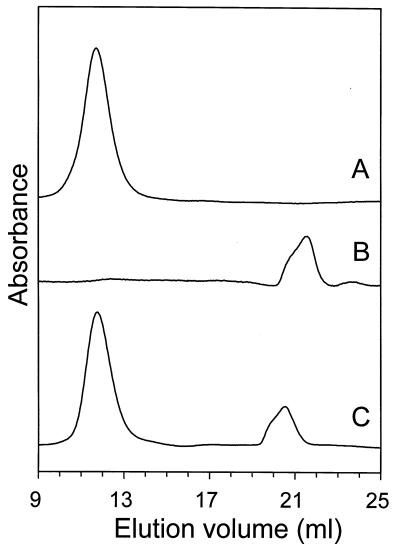
The CL activity of the H. volcanii proteasome was optimal at 2 M NaCl (Fig. 3B). Incubation of the enzyme in buffer without NaCl for less than 30 min resulted in greater than 70% loss of Suc-LLVY-Amc-hydrolyzing activity (Fig. 3B). Removal of salt by dialysis resulted in the complete loss of CL activity and the dissociation of the 20S proteasome complex into monomers, as determined by gel filtration chromatography (Fig. 4A and B). Interestingly, reequilibration of the inactivated proteasome monomers by rapid dilution in 2 M NaCl-Tris buffer resulted in the formation of a 600-kDa proteasome complex (Fig. 4C) of α1 and β subunits with measured specific activity of Suc-LLVY-Amc hydrolysis similar to the α1β-proteasome. Although dissociated monomers are still evident in the reequilibrated mixture, approximately 70% of fully active α1β-proteasome is recovered. Storage of the H. volcanii α1β-proteasome without salt for about 1 week at 4°C in 50 mM Tris-Cl buffer (pH 7.2) containing 1 mM DTT, however, results in autohydrolysis and irreversible inactivation of the proteasome subunits, which contrasts with the high stability of the enzyme observed when stored in the presence of salt (data not shown). These results suggest that the H. volcanii proteasome requires ∼2 M salt concentrations, similar to the extracellular environment and cytoplasm of this organism, for complex stability and optimal activity. This is similar to many other intracellular and extracellular proteins of the haloarchaea, which also require these unusually high concentrations of salt for activity and stability (reviewed in reference 27). Furthermore, these results suggest that assembly of the halophilic α1β-proteasome from dissociated monomers does not require the 49-amino-acid β-subunit propeptide. Similar propeptide-independent assembly has been observed for other archaeal proteasomes which have shorter β-propeptides, of eight to nine amino acids, as well as the β1 and β2 subunits of the yeast proteasome with 19 and 29 residue propeptides, respectively (4, 55, 70). However, studies demonstrate that the 59- to 75-residue β-propeptides of the eucaryotic β5 and β5i subunits, as well as the Rhodococcus β subunits, are critical for initial folding and/or final maturation of the 20S proteasome (11, 16, 83). It remains to be determined whether the β-propeptide of H. volcanii is required for initial folding and/or facilitates assembly of the proteasome in the cell, since the folding state of the dissociated monomers is not known and 30% of the proteasome proteins remain disassociated throughout the in vitro assembly reaction (Fig. 4C).
FIG. 4.
Salt-dependent dissociation and reassociation of the α1β-proteasome. Purified α1β-proteasome (250 μg) in 2 M NaCl-Tris buffer was directly applied to a Superose 6 column equilibrated in 2 M NaCl-Tris buffer (line A), was dialyzed against 1,000 volumes of Tris buffer without salt for 16 h and then applied to a Superose 6 column equilibrated in 2 M NaCl-Tris buffer (line B), and was dialyzed as described above, rapidly diluted 30-fold in 2 M NaCl-Tris buffer, incubated for 2 h at 21°C, concentrated by dialysis against PEG 8000, and applied to a Superose 6 column equilibrated in 2 M NaCl-Tris buffer (line C).
The temperature optimum for the CL activity of the H. volcanii proteasome was 75°C (Fig. 3C), which is 30°C higher than the optimum growth temperature. In fact, at the optimum temperature for the growth of the organism (45°C) (57), the proteasome activity was less than 20% of the maximum observed activity. This is not surprising, however, since thermal stability is a commonly observed trait for proteins from the halophilic Archaea and probably reflects the same structural forces that stabilize these proteins at high concentrations of salt (45). The presence of 2 M NaCl also could lead to the stabilization of the protein at the higher temperature. Many of the bacterial proteasomes which have been characterized are also thermostable, including a 20S proteasome from the mesophilic, nonhalophilic S. coelicolor which hydrolyzes Suc-LLVY-Amc at an optimum temperature which is 25°C higher than the growth temperature optimum for the organism (58). When the H. volcanii proteasome was frozen at liquid nitrogen temperatures and then assayed at 60°C, the CL activity was only 60 to 75% of the original activity, suggesting that the halophilic proteasome is not stable at temperatures below 4°C (data not shown).
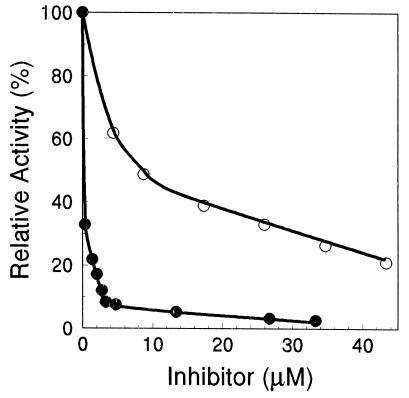
Carbobenzoxyl-leucinyl-leucinyl-leucinal-H (MG132) was found to be a potent inhibitor of the CL activity of the H. volcanii proteasome (>90% inhibition by 3 μM of inhibitor; Ki = 0.2 μM) (Fig. 5). Significant inhibition of CL activity was also observed for the known proteasome inhibitor N-acetyl-leucinyl-leucinyl-norleucinal-H (calpain inhibitor I) (Ki = 8 μM). Similar inhibition patterns have been observed for 20S proteasomes isolated from the domains Archaea, Eubacteria, and Eucarya (36, 53, 55, 58, and other references), and this inhibition is consistent with the novel proteolytic mechanism proposed for these enzymes (29, 53, 69).
FIG. 5.
Inhibition of the H. volcanii proteasome of α1 and β subunits. Purified proteasome (0.9 μg of protein per ml) was preincubated at 21°C for 60 min with N-acetyl-leucinyl-leucinyl-norleucinal-H (○) or carbobenzoxyl-leucinyl-leucinyl-leucinal-H (●) in 2 M NaCl-Tris buffer with 0.1% (vol/vol) ethanol or 5% (vol/vol) dimethylsulfoxide, respectively. Peptide hydrolysis was then assayed at 60°C with 20 μM Suc-LLVY-Amc at 2.5% (vol/vol) dimethylsulfoxide.
Cloning the genes encoding the 20S proteasome subunits.
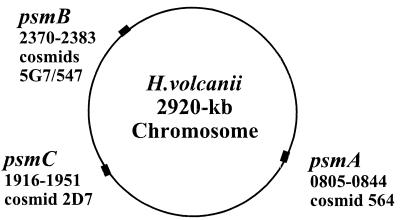
The availability of the pure α1β-proteasome allowed the construction of DNA-hybridization probes for the isolation of corresponding genes. To accomplish this objective, the N-terminal amino acid sequence of the β subunit and two internal peptide sequences of the α1 subunit of the purified proteasome were used to generate DNA probes with the codon bias of H. volcanii for hybridization to the genomic DNA of this organism. This approach enabled the isolation of the psmA and -B genes which encode the α1 and β subunits of the purified 20S proteasome, respectively, from a set of overlapping cosmid clones which cover the entire genome of H. volcanii (14). The cosmid clones also allowed mapping of the two genes in the genome. The psmA gene is located between nucleotide positions 805 and 844 while the psmB gene is located between positions 2370 and 2383 of the 2,920-kb chromosome (Fig. 6). This unlinked distribution of genes coding for subunits of the proteasome is apparently common in archaea, including T. acidophilium (85), M. thermophila (56), Methanococcus jannaschii (10), Methanobacterium thermoautotrophicum ΔH (72), Archaeoglobus fulgidus (49), and Pyrococcus horikoshii (44).
FIG. 6.
Location of the proteasome genes on the chromosome of H. volcanii. psmA, psmB, and psmC encode the α1, β, and α2 subunits of 20S proteasomes of H. volcanii. Cosmid numbers from a set of overlapping clones covering the genome of H. volcanii DS2 (14) and chromosome positions are indicated.
When the α-subunit DNA probe was used for hybridization to the H. volcanii genome, a second DNA fragment which hybridized to the probe was also identified and designated psmC. This gene was located at a third position in the chromosome, between positions 1916 and 1951 (Fig. 6).
Deduced sequence of the β subunit.
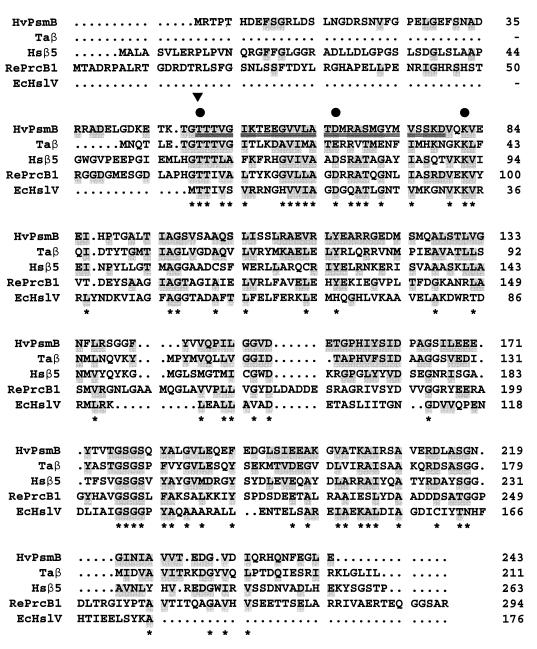
Based on the DNA sequence, the psmB gene is predicted to encode a putative protein of 243 amino acids (PsmB) with a calculated pI of 4.33 and an anhydrous molecular mass of 25,934 Da (Fig. 7, HvPsmB). The N-terminal threonine of the purified β subunit was located at amino acid position 50 in the DNA-deduced amino acid sequence (Fig. 7). The 29 amino acids following the threonine of the pure β subunit, as determined by sequencing the protein, were identical to amino acids 51 to 78, confirming that the 243-amino-acid precursor of the β subunit was processed to yield a mature protein of 194 amino acids. The 49-residue propeptide of the immature PsmB protein is significantly larger than any of the known archaeal propeptides, which range from only 6 to 11 residues. However, large propeptides are not uncommon among the β precursors of the eukaryotes and actinomycetes (15, 75, and other references). Although the calculated anhydrous molecular mass of the mature β subunit was 20,585 Da, the purified protein migrated as a 28,000-Da protein during SDS-PAGE (Fig. 1). This discrepancy is apparently related to the acidic nature of the protein (15% glutamate and aspartate compared to 7.2% lysine and arginine). The predominance of acidic residues in most halophilic proteins results in their abnormal behavior during SDS-PAGE (39).
FIG. 7.
Multiple-amino-acid alignment of the PsmB protein (β subunit) of H. volcanii with the β-type proteasome and HslV proteins. Double-underlined amino acid residues are identical to the N-terminal protein sequence of the purified β subunit of a 20S proteasome from H. volcanii. The cleavage site used during maturation of the β-type protein is indicated by an arrowhead above the protein sequence alignment. Conserved residues proposed to be involved in peptide bond hydrolysis are indicated by a filled-in circle above the protein sequence alignment. Abbreviations: Hv, H. volcanii; Ta, T. acidophilum; Hs, Homo sapiens; Re, R. erythropolis; Ec, E. coli. Taβ, Hsβ5 (ɛ, X, MB1), RePrcB1, and EcHslV(ClpQ) sequences were accessed through GenBank protein loci numbers 130867, 4506201, 847769, and 417158, respectively. Residues identical or functionally similar to HvPsmB are shaded. Highly conserved residues are indicated with an asterisk below the protein sequence alignment.
Multiple-amino-acid sequence alignment of the deduced PsmB protein sequence with other known sequences reveals high identity to β-type proteasome subunits and significant similarity to the HslV proteins of eubacteria (Fig. 7). The Thr1, Asp17, and Lys33 residues of the mature β subunit of H. volcanii are highly conserved with other β-type proteasomes and HslV proteins which catalyze peptide bond hydrolysis. Thus, by analogy, the Thr1 hydroxyl group of the mature β subunit of H. volcanii is proposed to be the nucleophile, and the Thr1 amino group is thought to be the primary proton acceptor in peptide bond hydrolysis (4, 29, 36, 53, 55, 69). In addition, the Lys33 and Asp17 residues of the β subunit of the H. volcanii proteasome may form a salt bridge which accepts the side chain proton of Thr1 through a charge relay system, similar to the T. acidophilum proteasome (53, 69).
Deduced sequences of the α1 and α2 subunits.
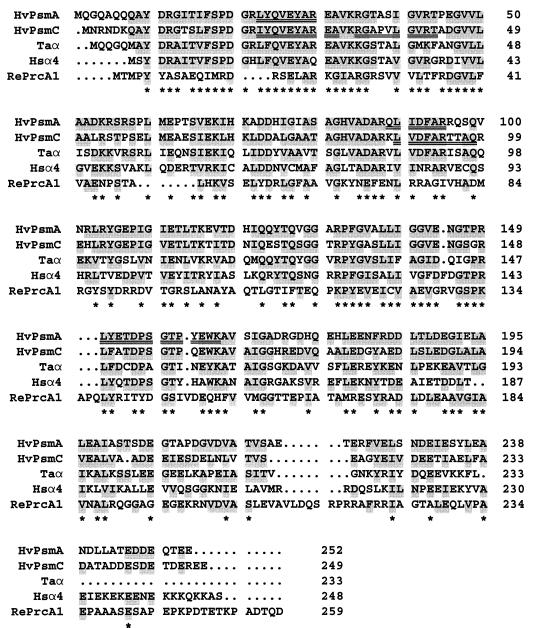
The psmA and psmC genes encode putative proteins (PsmA and PsmC) of 252 and 249 amino acids, respectively, with calculated relative pIs of 4.25 and 4.07 and anhydrous molecular masses of 27,613 and 26,727 Da (Fig. 8). The internal protein sequences 23-LYQVEYAR-30, 89-QLIDFAR-95, and 150-LYETDPSGTPYEWK-163 of the purified α1 protein are identical to the PsmA protein sequence which was deduced from the DNA sequence and are composed of residues distinctly characteristic of the PsmA protein and not of PsmC (underlined amino acids are found only in PsmA). This confirms that PsmA is indeed the α1 subunit of the 20S proteasome described above. Although the α1 subunit migrated at ∼10 kDa larger than predicted during SDS-PAGE (Fig. 1), the discrepancy is probably related to the acidic nature of the α1 protein (19.4% glutamate and aspartate compared to 9.1% lysine and arginine) and is similar to the discrepancy observed in the β subunit.
FIG. 8.
Multiple-amino-acid alignment of the PsmA (α1 subunit) and PsmC (α2 subunit) proteins of H. volcanii with α-type proteasome subunits. Double-underlined amino acid residues are identical to the internal protein sequences determined by Edman degradation. Abbreviations are the same as those used in Fig. 7. Taα, Hsα4 (XAPC-7), and RePrcA1 sequences were accessed through GenBank protein loci numbers 585729, 2555136, and 847770. Residues identical or functionally similar to HvPsmA and HvPsmC are shaded. Highly conserved residues are indicated with an asterisk below the protein sequence alignment.
The paralogous pair of α-type proteasome genes from H. volcanii (psmA and psmC) are 69.4% identical, which contrasts with the two paralogous superoxide dismutase (sod) genes of 99.5% identity from this halophile (41). Although this is the first example of paralogous α-type proteasome genes found in Archaea, genomic sequencing of the hyperthermophilic archaeon P. horikoshii (44) has revealed two paralogous β-type proteasome genes of 55% identity. The other archaeal genomes which have been sequenced apparently encode only one α-type and one β-type subunit (10, 49, 72). Thus, the number of genes encoding 20S proteasome subunits is not uniformly distributed in Archaea, similar to what has been observed in Eubacteria. The gram-positive actinomycete R. erythropolis has two paralogous operons of α- and β-type genes with 74% identity (75), whereas Mycobacterium spp. and S. coelicolor apparently have only single α- and β-type genes (19, 50, 58).
Alignment of the deduced PsmA and PsmC protein sequences with other known sequences revealed high identity to α-type proteasome proteins, including the characteristic highly conserved N-terminal extension (Fig. 8) (residues 9 to 36 of PsmA and 8 to 35 of PsmC) which is not present in the evolutionarily related β-type proteasome subunits. The predicted α-helices of the N-terminal region spanning residues 24 to 35 of PsmA and 23 to 34 of PsmC are similar to the α helix of the α subunit of the Thermoplasma proteasome, which is required for assembly of the protein into heptameric rings (86). In addition, based on the 3.4-Å structure of the T. acidophilum proteasome, the conserved N-terminal extension of α-type proteasome subunits is located at ends of the 20S core close to the antechamber entrance and may be important for substrate translocation and/or interaction with regulatory complexes (reviewed in reference 7). The PsmA sequence, and not the PsmC sequence, contains a consensus nuclear localization signal (NLS) sequence of K(K/R)-X-(K/R) (residues 54 to 57), acidic residues complementary to the NLS sequence (cNLS) (EDDEXXEE; residues 245 to 252), and a potential tyrosine (Tyr125) autophosphorylation site. All of these sequence motifs are relatively conserved among the α-type subunits, including the α subunit of the T. acidophilum proteasome (87). Why some archaeal proteasomes would retain NLS-cNLS sequence motifs is unclear; however, studies suggest that the NLS sequence of the T. acidophilum α subunit is sufficient to localize proteins to the nucleus of eukaryotic cells (59). In addition, many eukaryotic α-type proteasome subunits have been shown to be phosphorylated at tyrosine or serine residues (reviewed in reference 64). Although phosphorylation of archaeal α-type subunits has not been studied, the α subunit of the Thermoplasma proteasome is actually composed of two isoforms with different pIs, suggesting that the α subunit from this organism is modified posttranscriptionally (85).
To further understand the unusual salt requirements of the halophilic proteasome proteins, the quaternary structures of two hypothetical 20S proteasomes containing the α1 and β as well as the α2 and β subunits were modeled on available X-ray crystallographic structure coordinates for the yeast and Thermoplasma proteasomes with assistance from M. C. Bewley and J. M. Flanagan at Brookhaven National Laboratory (data not shown). The results suggest that the acidic residues of the halophilic proteasome subunits are located primarily on the surface of the 20S cylinder(s), whereas the central proteolytic chamber, which is predicted to be self-compartmentalized from the cytosol, is relatively neutral. This is consistent with current insights into protein adaptation to environments with high concentrations of salt (25, 27, 28, 30) and suggests that acidic domains on the surfaces of the halophilic 20S proteasomes provide extra carboxylates for solvation of the enzymes and prevent the proteolytic complex from nonspecific aggregation.
Purification of 20S proteasomes of α1, α2, and β subunits from H. volcanii.
Although DNA encoding a second α-type proteasome protein was identified in the H. volcanii genome, the 20S proteasome described above contained only the α1 and β subunits as determined by SDS-PAGE and protein sequencing (Fig. 1). A second α subunit (α2) encoded by the psmC gene did not appear to copurify at significant levels with the α1β-proteasome. In order to evaluate the possibility that the α2 subunit is expressed, the initial purification procedure was modified to increase the yield of purified 20S proteasomes from about 100 to 500 μg of protein per 15 g of cellular wet weight. Instead of precipitating the proteasomes from a dilute protein sample, fractions with CL activity were concentrated by dialysis against PEG 8000. The 600-kDa fraction of proteasomes purified from H. volcanii cells grown to mid-log or stationary phase using this modified procedure catalyzed the hydrolysis of Suc-LLVY-Amc at rates similar to that obtained with the α1β-proteasome. However, SDS-PAGE analysis of the newly purified proteasomes revealed the presence of a third, 34.5-kDa protein which was present in a nonstoichometric ratio with the α1 and β proteasome subunits (Fig. 9). Since the 34.5-kDa protein was N-terminally blocked, three internal amino acid sequences of the protein were determined from endoproteinase Lys-C fragments. The internal protein sequences 22-IYQVEYAREA-31, 34-RGAPVLGVRT-43, and 89-LVDFARTTAQ-98 of the purified 34.5-kDa protein are identical to the sequence of the deduced PsmC protein and are composed of residues which are distinctly characteristic of PsmC and not of the PsmA protein (Fig. 8) (underlined amino acids are found only in PsmC). These results confirm that the α2 protein copurifies with the α1 and β subunits as a 600-kDa fraction with CL activity rates similar to the proteasomes composed of only α1 and β subunits.
FIG. 9.
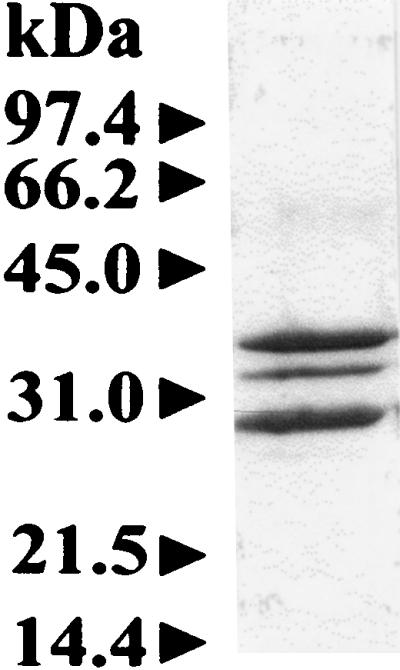
Reducing SDS-PAGE of a 600-kDa fraction (3 μg) of α1-, α2-, and β-proteasome subunits purified from H. volcanii. The additional band at 34.5 kDa represents the α2 subunit which is not present in the α1β-proteasome of Fig. 1.
Electron microscopic analysis of several hundred particles of the 600-kDa fraction composed of α1, α2, and β proteins revealed cylindrical barrel-shaped protein complexes of uniform size (12 by 17 nm) and shape to those observed for the proteasomes of α1 and β subunits described in Fig. 2 (data not shown). These results suggest that the α2 protein is assembling into a core 20S proteasome of four stacked rings and that the 600-kDa fraction which eluted from the Superose 6 gel filtration column as a uniform peak is not a nonspecific aggregation of α1, α2, and β proteins. In addition, it does not appear that the α2 protein was a substrate for proteases in the initial purification method, since incubation of the purified 600-kDa fraction (2.5 μg) in the presence of freshly prepared H. volcanii cell lysate (2 to 10 μg) for 24 h at 25 to 37°C did not result in any detectable reduction in the levels of the α1, α2, or β subunits. Studies are currently in progress to determine whether the α1 and α2 proteins are in the same complex, two separate 20S proteasome complexes, or both; however, these results suggest that at least two types of 20S proteasomes coexist in H. volcanii: an α1β proteasome and either a proteasome(s) containing all three subunits (α1, α2, and β) or a separate α2β-proteasome. It is also possible that a particle composed entirely of α2 subunits copurifies with the α1β-proteasome and cannot be distinguished from 20S proteasome-like structures by electron microscopy. However, the largest reported particles composed entirely of α subunits appear to be restricted to double hexameric rings, and these structures have only been observed when α subunits of proteasomes are heterologously produced in E. coli (31, 32, 55, 86).
Interestingly, two α-type and two β-type subunits copurify as 20S proteasomes with equimolar subunit stoichiometry from the eubacterium R. erythropolis (75). Zühl et al. (84) further investigated the structure of these proteasomes by using a heterologous expression system which enabled the production of four different 20S complexes composed of α1β2, α2β2, α1β1, and α2β1 subunits. The distinct differences in the molecular masses of these recombinant complexes compared to the proteasome fraction isolated from R. erythropolis suggest that a single population of proteasomes built equally from all four subunits exists in this eubacterium (84). Similarly, the eucaryotic 20S proteasome is built equally from 14 different subunits; the outer seven subunits classify to the α superfamily, and the inner seven classify to the β superfamily (reviewed in reference 7). Of the seven β subunits, three are processed to expose an active site N-terminal Thr1, and these subunits include β1, β2, and β5, designated according to the current nomenclature (7). The only confirmed example of multiple 20S proteasomes is in higher vertebrates, where gamma interferon induces the synthesis of three β-type subunits (β1i, β2i, and β5i) which replace their constitutively synthesized counterparts (β1, β2, and β5) to generate the immunoproteasome, a 20S complex which is more efficient at generating dominant T-cell epitopes (reviewed in reference 76). Whether the two α subunits of H. volcanii are differentially regulated remains to be determined. However, the synthesis of at least two 20S proteasomes with different α-subunit compositions in H. volcanii allows the investigation of the physiological role of the α-type proteasome subunits in a microbial system where the powerful tools of molecular biology and genetics can be brought to bear on this important question.
Conclusion.
The presence of 20S proteasomes in H. volcanii confirms that this cytoplasmic protease is widespread among the Archaea and can be observed in organisms which survive in extreme salt environments, such as the Dead Sea. The 20S proteasomes described in this study are the first intracellular proteases purified from the haloarchaea and have unusual salt requirements which distinguish them from any proteasome which has been previously characterized. Our results support the presence of at least two 20S proteasomes with different α-subunit compositions in H. volcanii, a feature which is not common in the prokaryotes.
ACKNOWLEDGMENTS
We thank Mark S. Ou for technical assistance, Donna Williams for help with transmission electron micrographs of the 20S proteasomes, Robert Charlebois and Andrew St. Jean for identifying cosmid clones containing the proteasome genes from an H. volcanii DS2 genomic library, Francis Davis and Jack Shelton for DNA sequencing, John Flanagan and Maria Bewley for assistance in modeling the quaternary structure of 20S proteasomes, and Richard Shand for generously supplying H. volcanii DS2.
This work was supported by the University of Florida Institute of Food and Agricultural Sciences’ Center for Biomass Programs and the Florida Agricultural Experiment Station (Journal Series R07043).
REFERENCES
- 1.Akopian T N, Kisselev A F, Goldberg A L. Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J Biol Chem. 1997;272:1791–1798. doi: 10.1074/jbc.272.3.1791. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Anderson H. The reddening of salted hides and fish. Appl Microbiol. 1954;2:64–69. doi: 10.1128/am.2.2.64-69.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arendt C S, Hochstrasser M. Eukaryotic 20S proteasome catalytic subunit propeptides prevent active site inactivation by N-terminal acetylation and promote particle assembly. EMBO J. 1999;18:3575–3585. doi: 10.1093/emboj/18.13.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Sideman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: Green Publishing Associates and Wiley-Interscience; 1987. Preparation and analysis of DNA; pp. 2.0.1–2.9.10. [Google Scholar]
- 6.Bauer M W, Halio S B, Kelly R M. Purification and characterization of a proteasome from the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol. 1997;63:1160–1164. doi: 10.1128/aem.63.3.1160-1164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Brannigan J A, Dodson G, Duggleby H J, Moody P C E, Smith J L, Tomchick D R, Murzin A G. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 10.Bult C J, White O, Olsen G J, Zhou L, Fleischmann D, Sutton G G, Blake J A, FitzGerald M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 11.Cerundolo V, Kelly A, Elliott T, Trowsdale J, Townsend A. Genes encoded in the major histocompatibility complex affecting the generation of peptides for TAP transport. Eur J Immunol. 1995;25:554–562. doi: 10.1002/eji.1830250238. [DOI] [PubMed] [Google Scholar]
- 12.Charlebois R L, Hofman J D, Schalkwyk L C, Lam W L, Doolittle W F. Genome mapping in halobacteria. Can J Microbiol. 1989;35:21–29. doi: 10.1139/m89-004. [DOI] [PubMed] [Google Scholar]
- 13.Charlebois R L, Lam W L, Cline S W, Doolittle W F. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc Natl Acad Sci USA. 1987;84:8530–8534. doi: 10.1073/pnas.84.23.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlebois R L, Schalkwyk L C, Hofman J D, Doolittle W F. Detailed physical map and set of overlapping clones covering the genome of the archaebacterium Haloferax volcanii DS2. J Mol Biol. 1991;222:509–524. doi: 10.1016/0022-2836(91)90493-p. [DOI] [PubMed] [Google Scholar]
- 15.Chen P, Hochstrasser M. Biogenesis, structure, and function of the yeast 20S proteasome. EMBO J. 1995;14:2620–2630. doi: 10.1002/j.1460-2075.1995.tb07260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen P, Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- 17.Cline S W, Schalkwyk L C, Doolittle W F. Transformation of the archaebacterium Halobacterium volcanii with genomic DNA. J Bacteriol. 1989;171:4987–4991. doi: 10.1128/jb.171.9.4987-4991.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen A, Lam W L, Charlebois R L, Doolittle W F, Schalkwyk L C. Localizing genes on the map of the genome of Haloferax volcanii, one of the Archaea. Proc Natl Acad Sci USA. 1992;89:1602–1606. doi: 10.1073/pnas.89.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 20.Coux O, Nothwang H G, Pereira I S, Targa F R, Bey F, Scherrer K. Phylogenic relationships of the amino acid sequences of prosome (proteasome, MCP) subunits. Mol Gen Genet. 1994;245:769–780. doi: 10.1007/BF00297284. [DOI] [PubMed] [Google Scholar]
- 21.Coux O, Tanaka K, Goldberg A L. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 22.Dahlmann B, Kopp F, Kuehn L, Niedel B, Pfeifer G, Hegerl R, Baumeister W. The multicatalytic proteinase (prosome) is ubiquitous from eukaryotes to archaebacteria. FEBS Lett. 1989;251:125–131. doi: 10.1016/0014-5793(89)81441-3. [DOI] [PubMed] [Google Scholar]
- 23.Dahlmann B, Kuehn L, Grizwa A, Zwickl P, Baumeister W. Biochemical properties of the proteasome from Thermoplasma acidophilum. Eur J Biochem. 1992;208:789–797. doi: 10.1111/j.1432-1033.1992.tb17249.x. [DOI] [PubMed] [Google Scholar]
- 24.Danner S, Soppa J. Characterization of the distal promoter element of halobacteria in vivo using saturation mutagenesis and selection. Mol Microbiol. 1996;19:1265–1276. doi: 10.1111/j.1365-2958.1996.tb02471.x. [DOI] [PubMed] [Google Scholar]
- 25.Dym O, Mevarech M, Sussman J L. Structural features that stabilized halophilic malate dehydrogenase from an archaebacterium. Science. 1995;267:1344–1346. doi: 10.1126/science.267.5202.1344. [DOI] [PubMed] [Google Scholar]
- 26.Edman P. Sequence determination. Mol Biol Biochem Biophys. 1970;8:211–255. doi: 10.1007/978-3-662-12834-3_8. [DOI] [PubMed] [Google Scholar]
- 27.Eisenberg H. Life in unusual environments: progress in understanding the structure and function of enzymes from extreme halophilic bacteria. Arch Biochem Biophys. 1995;318:1–5. doi: 10.1006/abbi.1995.1196. [DOI] [PubMed] [Google Scholar]
- 28.Elcock A H, McCammon J A. Electrostatic contributions to the stability of halophilic proteins. J Mol Biol. 1998;280:731–748. doi: 10.1006/jmbi.1998.1904. [DOI] [PubMed] [Google Scholar]
- 29.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 30.Frolow F, Harel M, Sussman J L, Mevarech M, Shoham M. Insights into protein adaptation to a saturated salt environment from the crystal structure of a halophilic 2Fe-2S ferredoxin. Nat Struct Biol. 1996;3:452–458. doi: 10.1038/nsb0596-452. [DOI] [PubMed] [Google Scholar]
- 31.Gerards W L, de Jong W W, Bloemendal H, Boelens W. The human proteasomal subunit HsC8 induces ring formation of other α-type subunits. J Mol Biol. 1998;275:113–121. doi: 10.1006/jmbi.1997.1429. [DOI] [PubMed] [Google Scholar]
- 32.Gerards W L, Enzlin J, Haner M, Hendriks I L, Aebi U, Bloemendal H, Boelens W. The human α-type proteasomal subunit HsC8 forms a double ringlike structure, but does not assemble into proteasome-like particles with the β-type subunits HsDelta or HsBPROS26. J Biol Chem. 1997;272:10080–10086. doi: 10.1074/jbc.272.15.10080. [DOI] [PubMed] [Google Scholar]
- 33.Gibbons N E. The effect of salt concentration on the biochemical reactions of some halophilic bacteria. Can J Microbiol. 1957;3:249–255. doi: 10.1139/m57-029. [DOI] [PubMed] [Google Scholar]
- 34.Gottesman S. Regulation by proteolysis: developmental switches. Curr Opin Microbiol. 1999;2:142–147. doi: 10.1016/S1369-5274(99)80025-3. [DOI] [PubMed] [Google Scholar]
- 35.Gregory D W, Pirie B J. Wetting agents for biological electron microscopy. I. General considerations and negative staining. J Microsc. 1973;99:251–255. doi: 10.1111/j.1365-2818.1973.tb04625.x. [DOI] [PubMed] [Google Scholar]
- 36.Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik H D, Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 37.Hilt W, Wolf D H. Proteasomes: destruction as a programme. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- 38.Holmes M, Pfeifer F, Dyall-Smith M. Improved shuttle vectors for Haloferax volcanii including a dual-resistance plasmid. Gene. 1994;146:117–121. doi: 10.1016/0378-1119(94)90844-3. [DOI] [PubMed] [Google Scholar]
- 39.Izotova L S, Strongin A Y, Chekulaeva L N, Sterkin V E, Ostoslavskaya V I, Lyublinskaya L A, Timokhina E A, Stepanov V M. Purification and properties of serine protease from Halobacterium halobium. J Bacteriol. 1983;155:826–830. doi: 10.1128/jb.155.2.826-830.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jolley K A, Rapaport E, Hough D W, Danson M J, Woods W G, Dyall-Smith M L. Dihydrolipoamide dehydrogenase from the halophilic archaeon Haloferax volcanii: homologous overexpression of the cloned gene. J Bacteriol. 1996;178:3044–3048. doi: 10.1128/jb.178.11.3044-3048.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joshi P, Dennis P P. Characterization of paralogous and orthologous members of the superoxide dismutase gene family from genera of the halophilic archaebacteria. J Bacteriol. 1993;175:1561–1571. doi: 10.1128/jb.175.6.1561-1571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamekura M, Seno Y, Dyall-Smith M. Halolysin R4, a serine proteinase from the halophilic archaeon Haloferax mediterranei: gene cloning, expression and structural studies. Biochim Biophys Acta. 1996;1294:159–167. doi: 10.1016/0167-4838(96)00016-7. [DOI] [PubMed] [Google Scholar]
- 43.Kamekura M, Seno Y, Holmes M L, Dyall-Smith M L. Molecular cloning and sequencing of the gene for a halophilic alkaline serine protease (halolysin) from an unidentified halophilic archaea strain (172P1) and expression of the gene in Haloferax volcanii. J Bacteriol. 1992;174:736–742. doi: 10.1128/jb.174.3.736-742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawarabayasi Y, Sawada M, Horikawa H, Haidawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahasi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Yoshizawa T, Nakamura Y, Robb F T, Horikoshi K, Masuchi Y, Shizuya H, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:147–155. doi: 10.1093/dnares/5.2.147. [DOI] [PubMed] [Google Scholar]
- 45.Keradjopoulos D, Holldorf A W. Thermophilic character of enzymes from extreme halophilic bacteria. FEMS Microbiol Lett. 1977;1:179. [Google Scholar]
- 46.Kim J, Dordick J S. Unusual salt and solvent dependence of a protease from an extreme halophile. Biotech Bioeng. 1997;55:471–479. doi: 10.1002/(SICI)1097-0290(19970805)55:3<471::AID-BIT2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 47.Kisselev A F, Akopian T N, Goldberg A L. Range of sizes of peptide products generated during degradation of different proteins by archaeal proteasomes. J Biol Chem. 1998;273:1982–1989. doi: 10.1074/jbc.273.4.1982. [DOI] [PubMed] [Google Scholar]
- 48.Kisselev A F, Akopian T N, Woo K M, Goldberg A L. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J Biol Chem. 1999;274:3363–3371. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
- 49.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 50.Knipfer N, Shrader T E. Inactivation of the 20S proteasome in Mycobacterium smegmatis. Mol Microbiol. 1997;25:375–383. doi: 10.1046/j.1365-2958.1997.4721837.x. [DOI] [PubMed] [Google Scholar]
- 51.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 52.Lopez-Garcia P, St. Jean A, Amils R, Charlebois R L. Genomic stability in the archaeae Haloferax volcanii and Haloferax mediterranei. J Bacteriol. 1995;177:1405–1408. doi: 10.1128/jb.177.5.1405-1408.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 54.Lupas A, Flanagan J M, Tamura T, Baumeister W. Self-compartmentalizing proteases. Trends Biochem Sci. 1997;22:399–404. doi: 10.1016/s0968-0004(97)01117-1. [DOI] [PubMed] [Google Scholar]
- 54a.Maupin-Furlow, J. A. Unpublished results.
- 55.Maupin-Furlow J A, Aldrich H C, Ferry J G. Biochemical characterization of the 20S proteasome from the methanoarchaeon Methanosarcina thermophila. J Bacteriol. 1998;180:1480–1487. doi: 10.1128/jb.180.6.1480-1487.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maupin-Furlow J A, Ferry J G. A proteasome from the methanogenic archaeon Methanosarcina thermophila. J Biol Chem. 1995;270:28617–28622. doi: 10.1074/jbc.270.48.28617. [DOI] [PubMed] [Google Scholar]
- 57.Mullakhanbhai M S, Larsen H. Halobacterium volcanii spec. nov., a Dead Sea Halobacterium with a moderate salt requirement. Arch Microbiol. 1975;104:207–214. doi: 10.1007/BF00447326. [DOI] [PubMed] [Google Scholar]
- 58.Nagy I, Tamura T, Vanderleyden J, Baumeister W, De Mot R. The 20S proteasome of Streptomyces coelicolor. J Bacteriol. 1998;180:5448–5453. doi: 10.1128/jb.180.20.5448-5453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nederlof P M, Wang H-R, Baumeister W. Nuclear localization signals of human and Thermoplasma proteasomal α subunits are functional in vitro. Proc Natl Acad Sci USA. 1995;92:12060–12064. doi: 10.1073/pnas.92.26.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nieuwlandt D T, Daniels C J. An expression vector for the archaebacterium Haloferax volcanii. J Bacteriol. 1990;172:7104–7110. doi: 10.1128/jb.172.12.7104-7110.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Norberg P, Hofsten B V. Proteolytic enzymes from extremely halophilic bacteria. J Gen Microbiol. 1969;55:251–256. doi: 10.1099/00221287-55-2-251. [DOI] [PubMed] [Google Scholar]
- 62.Palmer J R, Daniels C J. A transcriptional reporter for in vivo promoter analysis in the archaeon Haloferax volcanii. Appl Environ Microbiol. 1994;60:3867–3869. doi: 10.1128/aem.60.10.3867-3869.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pühler G, Pitzer F, Zwickl P, Baumeister W. Proteasomes: multisubunit proteinases common to Thermoplasma and eukaryotes. Syst Appl Microbiol. 1994;16:734–741. [Google Scholar]
- 64.Rivett A J. Intracellular distribution of proteasomes. Curr Opin Immunol. 1998;10:110–114. doi: 10.1016/s0952-7915(98)80040-x. [DOI] [PubMed] [Google Scholar]
- 65.Ryu K, Dordick J S. Catalytic properties and potential of an extracellular protease from an extreme halophile. Enzyme Microb Technol. 1994;16:266–275. doi: 10.1016/0141-0229(94)90165-1. [DOI] [PubMed] [Google Scholar]
- 66.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmidtke G, Kraft R, Kostka S, Henklein P, Frommel C, Löwe J, Huber R, Kloetzel P M, Schmidt M. Analysis of mammalian 20S proteasome biogenesis: the maturation of β-subunits is an ordered two-step mechanism involving autocatalysis. EMBO J. 1996;15:6887–6898. [PMC free article] [PubMed] [Google Scholar]
- 68.Schmitt W, Rdest U, Goebel W. Efficient high-performance liquid chromatographic system for the purification of halobacterial serine protease. J Chromatogr. 1990;521:211–220. [Google Scholar]
- 69.Seemüller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 70.Seemüller E, Lupas A, Baumeister W. Autocatalytic processing of the 20S proteasome. Nature. 1996;382:468–470. doi: 10.1038/382468a0. [DOI] [PubMed] [Google Scholar]
- 71.Shelton E, Kuff E L, Maxwell E S, Harrington J T. Cytoplasmic particles and aminoacyl transferase I activity. J Cell Biol. 1970;45:1–8. doi: 10.1083/jcb.45.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 74.Stepanov V M, Rudenskaya G N, Revina L P, Gryaznova Y B, Lysogorskaya E N, Filippova I Y, Ivanova I I. A serine proteinase of an archaebacterium, Halobacterium mediterranei. A homologue of eubacterial subtilisins. Biochem J. 1992;285:281–286. doi: 10.1042/bj2850281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tamura T, Nagy I, Lupas A, Lottspeich F, Cejka Z, Schoofs G, Tanaka K, DeMot R, Baumeister W. The first characterization of a eubacterial proteasome: the 20S complex of Rhodococcus. Curr Biol. 1995;5:766–774. doi: 10.1016/s0960-9822(95)00153-9. [DOI] [PubMed] [Google Scholar]
- 76.Tanaka K, Kasahara M. The MHC class I ligand-generating system: roles of immunoproteasomes and the interferon-gamma-inducible proteasome activator PA28. Immunol Rev. 1998;163:161–176. doi: 10.1111/j.1600-065x.1998.tb01195.x. [DOI] [PubMed] [Google Scholar]
- 77.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thompson M W, Singh S K, Maurizi M R. Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli: requirement for the multiple array of active sites in ClpB but not ATP hydrolysis. J Biol Chem. 1994;269:18209–18215. [PubMed] [Google Scholar]
- 79.Udenfriend S, Stein S, Bohlen P, Dairman W, Leimgruber W, Weigele M. Fluorescamine: a reagent for assay of amino acids, peptides, proteins, and primary amines in the picomole range. Science. 1972;178:871–872. doi: 10.1126/science.178.4063.871. [DOI] [PubMed] [Google Scholar]
- 80.Wada K, Wada Y, Ishibashi F, Gojobori T, Ikelmura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1992;20:2111–2118. doi: 10.1093/nar/20.suppl.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang J, Hartling J A, Flanagan J M. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 82.Wenzel T, Baumeister W. Conformational constraints in protein degradation by the 20S proteasome. Nat Struct Biol. 1995;2:199–204. doi: 10.1038/nsb0395-199. [DOI] [PubMed] [Google Scholar]
- 83.Zühl F, Seemüller E, Golbik R, Baumeister W. Dissecting the assembly pathway of the 20S proteasome. FEBS Lett. 1997;418:189–194. doi: 10.1016/s0014-5793(97)01370-7. [DOI] [PubMed] [Google Scholar]
- 84.Zühl F, Tamura T, Dolenc I, Cejka Z, Nagy I, DeMot R, Baumeister W, De Mot R. Subunit topology of the Rhodococcus proteasome. FEBS Lett. 1997;400:83–90. doi: 10.1016/s0014-5793(96)01403-2. [DOI] [PubMed] [Google Scholar]
- 85.Zwickl P, Grizwa A, Puhler G, Dahlmann B, Lottspeich F, Baumeister W. Primary structure of the Thermoplasma proteasome and its implications for the structure, function, and evolution of the multicatalytic proteinase. Biochemistry. 1992;31:964–972. doi: 10.1021/bi00119a004. [DOI] [PubMed] [Google Scholar]
- 86.Zwickl P, Kleinz J, Baumeister W. Critical elements in proteasome assembly. Nat Struct Biol. 1994;1:765–770. doi: 10.1038/nsb1194-765. [DOI] [PubMed] [Google Scholar]
- 87.Zwickl P, Lottspeich F, Dahlmann B, Baumeister W. Cloning and sequencing of the gene encoding the large (α-) subunit of the proteasome from Thermoplasma acidophilum. FEBS Lett. 1991;278:217–221. doi: 10.1016/0014-5793(91)80120-r. [DOI] [PubMed] [Google Scholar]