Abstract
Cardiac muscle damage-induced loss of cardiomyocytes (CMs) and dysfunction of the remaining ones leads to heart failure, which nowadays is the number one killer worldwide. Therapies fostering effective cardiac regeneration are the holy grail of cardiovascular research to stop the heart failure epidemic. The main goal of most myocardial regeneration protocols is the generation of new functional CMs through the differentiation of endogenous or exogenous cardiomyogenic cells. Understanding the cellular and molecular basis of cardiomyocyte commitment, specification, differentiation and maturation is needed to devise innovative approaches to replace the CMs lost after injury in the adult heart. The transcriptional regulation of CM differentiation is a highly conserved process that require sequential activation and/or repression of different genetic programs. Therefore, CM differentiation and specification have been depicted as a step-wise specific chemical and mechanical stimuli inducing complete myogenic commitment and cell-cycle exit. Yet, the demonstration that some microRNAs are sufficient to direct ESC differentiation into CMs and that four specific miRNAs reprogram fibroblasts into CMs show that CM differentiation must also involve negative regulatory instructions. Here, we review the mechanisms of CM differentiation during development and from regenerative stem cells with a focus on the involvement of microRNAs in the process, putting in perspective their negative gene regulation as a main modifier of effective CM regeneration in the adult heart.
Keywords: cardiac stem cells, microRNA, myogenesis, regeneration
An introduction to cell differentiation: mesoderm specification into cardiac mesoderm
Cell-based regenerative medicine aims at the development of therapeutical approaches with cells capable to efficiently differentiate into specialized cells. Directed differentiation of specific lineages of stem/precursor cells, such as embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs) or multipotent adult stem/progenitor cells, including endogenous/resident cardiac stem/progenitor cells (CSCs), is therefore the first critical step toward the development of cellular therapy. Thus, to define efficient protocols of myocardial regeneration depends on our understanding of the gene regulatory networks responsible for the differentiation of cardiomyocytes (CMs) during embryonic development as well as in adulthood and disease.
Cell differentiation is the process by which dividing cells change their functional or phenotypical type, becoming specialized. All cells presumably derive from stem cells and obtain their functions as they mature. Organismal cellular diversity is often modeled as a hierarchical scheme with totipotent stem cells at the top of the hierarchy. Totipotency means the potential to become any cell type and is a potential observed in the fertilized egg and the cells at the beginning of the differentiation process. This broad potential slowly becomes restricted as the cell potential gains specificity limited to one of a group of related cell types. Stem cell differentiation dramatically alters the cell shape, size, and energy requirements. This process is not irreversible until terminal differentiation occurs. Differentiation requires sequentially selecting a subset of genetic information to be expressed in a spatiotemporal fashion in response to different mechanical and chemical stimuli.
The sequential activation of transcription factors that determines the cell fate within the heart and drives its formation from the mesoderm is likely to be controlled in the same way in stem cells as it is in embryos. This is the reason why many differentiation protocols of stem cells in vitro are often guided by our understanding of cardiac development. To this aim, unravelling the essential signals for cardiac lineage decisions during embryonic development may help to induce stem/progenitor cells differentiation into cardiac muscle fate, the main goal of regenerative cardiology.
The heart is the first mesoderm-derived functional embryonic organ developed after gastrulation. During embryonic cardiogenesis, a hierarchy of growth/transcription factors, sequentially induced in cardiac progenitor cells ultimately lead to the formation of CMs [1], the major cell types of the heart. We can recognize atrial and ventricular CMs, and cardiac conduction cells such as those residing in the sinoatrial/atrioventricular nodes, in the HIS bundle or in the and Purkinje fibers. CMs, together with cardiac fibroblasts, vascular smooth muscle cells and endothelial cells, cooperate to produce a functional heart to pump oxygen- and nutrient-rich blood throughout the body.
Cardiac muscle embryogenesis requires extracellular instructive signals finally orchestrated by molecules that are synergically regulated in time and space. These signals, coupled with intracellular genetic programs, produce their specification/commitment and a progressive cell fate restriction. Thus, heart development is governed by a core set of evolutionarily conserved transcription factors that regulate each other’s expression, serving to stabilize and reinforce the cardiac gene program [2,3].
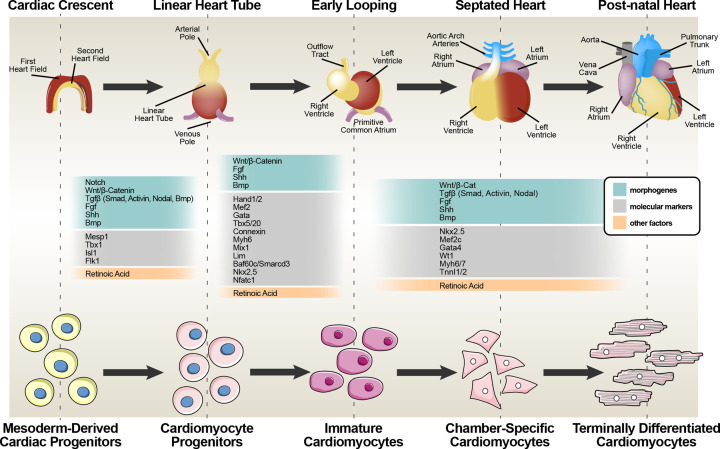
Mesoderm, one of the three germ layers, emerges from the primitive steak during gastrulation (Figure 1). In mouse, around embryonic life 6.5 dpc (day post coitum), prospective myocardial cells migrate cephalad from the primitive steak forming two groups of cells, one on each side of the midline of the epiblast (the cardiogenic areas) which will give rise to cardiac progenitor cells specification [4,5]. These presumptive cardiac cells, which will contribute to the myocardium and endocardium, appear fuse to form the so-called ‘cardiac crescent’, underlying the head folds, at embryonic life 7.5 dpc. Soon after gastrulation, as the embryo grows, the cardiac crescent fuses ventrally at the midline to form the beating primitive cardiac tube at embryonic life 8 dpc which subsequently growths and folds to the right in a S-shaped structure. The primitive cardiac tube is the first identifiable functional organ. Its early function is essential for the circulation of nutrients and waste removal in the embryo grown too large to be fed by diffusion [2,5,6]. The heart tube expands, essentially by two mechanisms: cell proliferation and recruitment of additional cells leading to the formation of recognizable septated cardiac chambers (embryonic life 14.5 dpc) [5]. This primitive structure further expands through significant increase in cell size [7,8]. Growth of the heart depends on the further addition of progenitor cells that mainly occurs in a dorsal/medial position to contribute to the arterial and venous pole of the cardiac tube. The cardiogenic mesoderm harbors two myocardial cell lineages that segregate early taking part in heart formation: the first and second heart fields (FHF and SHF) progenitors [9]. The left ventricle mainly derives from FHF progenitors [2,10–13] that are exclusively committed to a cardiomyogenic cell fate [14,15]. SHF progenitors mainly contribute to the outflow tract (OFT), the right ventricle and a large portion of the inflow region (atria) and provide a platform for subsequent heart growth. The proepicardial organ (PEO), a transitory mesenchymal structure localized at the posterior end of the heart tube, grows over the myocardium of the heart to form the outer layer of the epicardium. Some of these epicardial cells undergo an epithelial–mesenchymal transition (EMT) and then constitute the smooth muscle of the coronary blood vessels and the population of cardiac fibroblasts/interstitial cells [16]. The functional form of the heart also depends on the contribution of neural crest cells contributing to septum and valve formation and septation of the outflow track into the pulmonary trunk and aorta [16] ensuring separation of the venous and arterial circulation.
Figure 1. Overview of five major steps of the heart development.
The schematic representation shows the stage specific main cardiac morphogens and molecular markers composing the regulatory signaling pathways of cardiomyocyte formation (commitment, specification, differentiation) during heart development. From left, cardiac crescent formation through mesoderm-derived cardiac progenitor activation, consisting of the first heart field (FHF) and second heart field (SHF) cardiac progenitors. Following is the linear heart tube formation and cardiomyocyte progenitors appearance and sequentially heart looping with initiation of chamber formation that ultimate with the formation of the septated heart. The last stage of the adult heart coincides with cardiomyocyte terminal differentiation.
The molecular mechanisms involved in mesoderm specification into cardiac mesoderm and cardiac progenitor cells, are controlled by three families of extracellular signaling molecules: Wnt (as above), FGF and many members of the transforming growth factor β (TGFβ) family proteins. FHF formation, requires first the inhibitory Wnt signaling from the ectoderm [17], such as Wnt3a, followed by BMP4, Nodal and activin A, which provide the initial signals for heart creation (Figure 1).
Germline deletion of a particular gene and the consequent disruption of its downstream signaling, reveals the role of that gene in cardiac development [18]. In the case of BMP, its deletion impairs gastrulation and the formation of primitive mesoderm leading to death before embryonic 9.5 dpc [19,20]. Activin and Nodal, like BMPs, are members of the TGFβ family, and may serve as a surrogate for an authentic Nodal signal which is essential for the establishment of anterior-posterior and left-right axes, gastrulation, for the formation of the primitive streak, the creation of mesoderm and for the later cardiac myogenesis in mouse embryos [21,22]. BMP signaling usually promotes heart formation and cardiogenesis in vertebrate embryos, whereas wingless in Drosophila and related Wnt proteins are involved in cardiac specification. Mesodermal cells within the primitive streak express the T-box transcription factor brachyury (Bry, also T), a direct target of the Wnt pathway [23] that comprise a complex set of events: it may inhibit or promote differentiation depending on the spatiotemporal context and on the activation of the canonical Wnt pathway (acting via β-catenin/GSK3 to repress cardiogenesis) or the non-canonical Wnt pathway (acting via protein kinase C/c-Jun N-terminal kinase to promote cardiogenesis). β-Catenin has been shown to be essential for mesoderm formation since no mesodermal or head structures are formed in β-catenin-null mice where TbxT is not expressed [24]. Canonical Wnt signaling and secreted IGF-1 from the epicardium, are involved in controlling the number and size of CMs in the mouse embryo and in the postnatal heart [25–27]. They act via GSK3β to stabilize β-catenin and together with the Hippo signaling pathway they mediate cardiac growth and size [28–30]. FGFs and BMPs, as well as Notch signaling, also have a proliferative effect on CMs during development [31–33]. FGFs appear to provide positional cues to cells for specification and together with BMP2 probably synergize to drive mesodermal cells into myocardial differentiation. Distinct subsets of mesodermal progenitors are characterized by differential expression of Flk1 and PDGFRα and concomitantly, in these cells the expression of Tbx decreases [34,35]. Studies in the chick and mouse have shown that in addition to these pathways, retinoic acid (RA) signaling plays a crucial role in early cardiac development of the atria. Indeed, inhibition of RA signaling resulted in atrial malformation, whereas excessive signaling led to the development of enlarged atria at the expense of ventricular chamber formation (Figure 1) [36,37].
Thus, multiple complex interactions among these highly conserved regulatory networks of transcription factors control CM differentiation, their proliferation, and maturation. These factors have also been used as markers of emerging CMs in differentiating cultures of stem cells.
Cardiomyocyte differentiation in vivo: cardiac progenitors and cell fate determination
Once mesoderm cells have received the appropriate signals, they start to express a highly conserved heart-specific combination of transcription factors that together establish the cardiac transcriptional program. Over the last few years, the transcriptional regulators of the cardiac transcription program directing the morphogenesis of the cardiac progenitors have been uncovered.
The mesoderm-derived of the FHF and SHF are the predominant sources of CMs in the heart. One of the first markers expressed by mesoderm cells within the FHF, is Mesp1 which is required for the formation of a crescent-shaped structure that constitutes the first morphological sign of heart development (Figure 1) [5,38]. The primordial importance of Mesp1 in promoting cardiovascular cell fate in the presence of Dkk1 (Dickkopf, WNT Signaling Pathway Inhibitor 1) has been demonstrated through the injection of Mesp1 RNA into Xenopus embryos where an endodermal induction of cardiogenesis formation of an ectopic heart has been observed [39,40]. Mesp1 has been implicated in the EMT required for the exit of cells from the streak to form the mesoderm layer, and it is thereafter rapidly down-regulated [41]. Experiments conducted using ESCs have demonstrated the importance of this transcription factor acting as a master regulator of cardiovascular cell fates [41], down-regulating pluripotency and early mesodermal genes. Subsequently, a subset of Mesp1 positive cells start to co-express the key cardiac transcription factors Nkx2.5, Tbx5, Mixl, Isl1, and LIM, which are early markers of the cardiac lineage activated during the formation of the two heart fields [41–45]. Cells expressing Mesp1 and Islet1 also contribute to smooth muscle at the arterial pole of the heart and Tbx1 expression regulates this derivative, as well as myocardium [46–49]. Although Tbx5 has been associated with FHF, specific markers are still lacking for this field, whereas Isl1 has been considered a specific marker for SHF [2] and genetic tracing in the mouse embryo suggests that cells that had expressed Islet1, Nkx2.5 and activated the Mef2c expression in the SHF, contribute to endocardium and myocardium (Figure 1). Tbx5, Hand1/2, and Mef2c are implicated in the differentiation of cells in the cardiac crescent and in the heart. These transcriptional regulators activate the expression of atrial natriuretic factor and connexin and, in association with members of the GATA family, activate cardiac structural genes, such as actin, myosin light chain, myosin heavy chain, troponins and desmin [50]. GATA-4 promotes the development of cardiac muscle and regulates the expression of genes such as α-myosin heavy chain, cardiac troponin C and atrial natriuretic peptide [51]. It has been shown that disruption of the GATA-4 gene in mice leads to early embryonic lethality because of specific defects in ventral heart tube formation [51]. In particular, GATA-4 and Tbx5 are master regulators in cardiogenesis: in the presence of the chromatin remodeling component, Baf60c/Smarcd3, these factors induce beating myocardial tissue formation [52]. GATA-4 and Baf60c induce Nkx2.5 expression which acts with GATA-4 to initiate the cardiac program; Tbx5 is instead required for full differentiation [52]. The importance of Nkx-2.5 is demonstrated by the fact that mutations in its sequence are involved in congenital human heart diseases and affect cardiac developmental pathways impairing normal heart architecture in patients with heart diseases [53]. The transcription factors GATA-4 and Nkx-2.5 mediate the expression of BMP-2, a cardiac-specific protein. Indeed, the application of BMP-2 in vivo induces ectopic expression of Nkx-2.5 and GATA as well as cardiac-specific proteins such as ventricular myosin heavy chain [54,55].
BMPs expression in the mouse appears to be more complex: deletion of BMPs seems to have a late effect on cardiogenesis where mutants present cardiac defects and are embryonic lethal despite the fact that cardiac mesoderm specification still occurs [56]. Furthermore, administration of BMP-2 or BMP-4 to explant cultures of stage 5-7 anterior medial mesoderm, induces full cardiac differentiation of this tissue that normally is not cardiogenic [54].
The MEF2 family of transcription factors are expressed in many types of cells, including immune system cells, neurons, and striated muscle. At E7.5 dpc, Mef2b and Mef2c are initially expressed in the cardiac mesoderm, followed a day later by expression of Mef2a and Mef2d. Mef2c transcripts are detected in the somites at E8.5 dpc, concomitant with the onset of myocyte differentiation in the myotome. Disruption of Mef2c produce death of the murine embryo at E9.5 dpc due to arrested cardiac looping and right ventricular formation during embryogenesis [57]. Members of the MEF2 family also play key roles in CM differentiation by regulating cardiac muscle structural genes.
Another level of factor interaction is illustrated by Tbx20 which synergizes with Islet1 and GATA-4 to activate the Mef2c enhancer and an Nkx2.5 cardiac enhancer [58]. Moreover, Flk1 expressing progenitors contribute to both tissues and Nfatc1 provides a marker for endocardium that distinguishes these cells from other endothelial cells [48,59–63].
A number of signaling pathways that impact the formation of the two heart fields have been shown to promote proliferation. This is the case for FGF signaling that seems to increase the extent of the SHF and the number of CMs [64,65]. Wnt/β-Catenin signaling presents a biphasic function in cardiogenesis displaying an inhibitory effect in the FHF cell proliferation while it stimulates cell proliferation in the SHF. Indeed, Wnt signaling in cardiac progenitors induces their proliferation and the maintenance of undifferentiated state prior to entering the heart tube, while Wnt/β-catenin inhibition is required to instruct progenitors to leave the SHF proliferative state and to start differentiating. Hedgehog (Hh) signaling is also often associated with proliferation; particularly in the chick embryo, it is clearly important for maintaining progenitor cell proliferation in the critical time frame which precedes addition of cells to the heart tube [66].
Overall, the heart development insights described above have been used in efforts to direct stem cell differentiation in vitro, using exogenous growth factors, toward the cardiac muscle lineage. Many of the successful protocols developed to induce cardiomyogenesis in stem/progenitor cells are based on activating or inhibiting these known signaling pathways. Although new layers of epigenetic control have come to be understood in cardiac biology, much remains to be learned mechanistically about where and how the instructive cardiogenic pathways intersect with other essential regulators networks and chromatin modification.
Cardiomyocyte differentiation from embryonic and cardiac stem cells
At present, there is a growing consensus about the capacity of the adult mammalian heart to generate new CMs [67–73] although their origin is still debated: some studies propose they originate from the proliferation of mature CMs, whereas others, including us, suggest as the endogenous cardiac stem/progenitor cells (CSCs) as the source of cells differentiating in fully mature CMs [70–74].
Comparative analyses of genes involved in cardiac development and their interconnected regulatory elements have highlighted the conservation of genetic pathways that direct cardiogenesis during the embryo development which requires extracellular instructive molecules known as morphogens or cardiopoietic growth factors. As above, Wnt/β-catenin and TGF-β/SMAD signaling pathways have well-defined and distinct functions in mammalian cardiogenesis. Progenitors with different potential are specified during gastrulation to give rise to different regions of the heart. Therefore, the signaling pathways that regulate these early developmental decisions, including Nodal, Wnt, BMPs, and FGFs, are likely to also play a role in establishing these cardiac fates from embryonic and/or cardiac stem cells.
Activin A, together with BMPs, has been successfully used to induce cardiac myogenesis in several cell lines including mouse ESCs (mESCs), human ESCs (hESCs), human iPSCs, and cardiac progenitor cell [71,75–78]. Homozygous-null ESCs for the Activin A/nodal pathway lack beating cardiomyocytes, cardiac proteins and cardiac mRNAs. A recombinant Cripto rescued the defect in null ESCs, confirming an essential role of this pathway in the myogenic specification [79]. Activin and FGF2 maintain pluripotency in hESCs and the combined use of Activin, FGF2, BMP4, and vascular endothelial growth factor promotes the formation of multipotent progenitor cells, able to generate all mesoderm cell types, including CMs, smooth muscle cells, and endothelial cells [80,81].
Another important driver of the cardiac differentiation process is the so-called ‘canonical Wnt pathway’ that include Wnt1, -2a, -3a, and -8, acting through β-catenin for signal transduction. Positive and negative modulators of the Wnt/β-catenin axis act through respectively stabilizing or degrading β-catenin and its LEF/TCF-dependent transcriptional activity. When Wnt binds the Frizzled receptors, it triggers a variety of intracellular responses leading to the decreased phosphorylation of β-catenin, allowing its stabilization, cytoplasmic accumulation, and its eventual nuclear translocation [82] producing the transcriptional activation of the Wnt target genes. Secreted Frizzled-related proteins (sFRPs), FrzB, Crescent, and the Fz ectodomain are all broad-spectrum inhibitors of Wnt signaling. The extracellular protein Dkk and constitutively active GSK3 are both selective inhibitors for canonical Wnts [83]. Cre-dependent expression of stable β-catenin in Isl1-determined cells produced hyperplasia of the right ventricle. On the other hand, ablating β-catenin in this cell lineage causes failure of right ventricular development [84]. P19CL6 embryonic cancer cells provided the first evidence that the canonical Wnt pathway is required for mammalian cardiomyocyte differentiation from pluripotent cells [85]. The induction of myogenic transcription factors was blocked by the Wnt inhibitor Frz8/Fc or constitutively active GSK3β [85]. Conversely, the expression of Nkx2-5, Gata4, Mef2c, Tbx5, and Myh6 was enhanced by Wnt3A-conditioned medium or by lithium, a GSK3β inhibitor [86]. The chemical molecule SU5402 inhibited FGF signaling and significantly increased BMP2-dependent cardiogenic commitment, as seen by a 3- to 10-fold increase in mesodermal and cardiac markers [87].
hESCs, as well as mESCs, are isolated from blastocysts cultured from in vitro fertilization. They are capable of unlimited self-renewal and can differentiate into derivatives of all three germ layers providing a source of cells for research and clinical application [88]. Initial efforts to differentiate hESCs into CMs employed the embryoid body (EB) technique, monolayer differentiation, and inductive differentiation [89]. Cells in EBs are able to differentiate into derivatives of the three primary germ layers [90]. Usually, EBs were highly variable in composition and structure; a fraction of them exhibited spontaneously contracting regions that contained CMs. This method proved to be inefficient, only yielding a low percentage of CMs (∼1–5%). Differentiation protocols for EBs were developed using a variety of methods including the use of serum-free media and a range of growth factors implicated in normal cardiac development, including BMP4, Activin A, FGF2, Wnt agonists and antagonists, and vascular endothelial growth factor. An alternative method to differentiate ESCs starts with cells grown as monolayers. This method does not require the replating steps typical of EB protocols and the use of tissue culture supplies but needs of cytokines in order to induce cardiac mesoderm, specifically BMP4, Activin A, and WNT3A. This differentiation process, as the EB-based methods, is again critically dependent on the ratio of BMP4 to activin A, the time of addition, and the time of removal of growth factors [76].
The inductive differentiation approach was first described by Christine Mummery and colleagues [91] and consists in derivation of CMs from hPSCs by co-culture with visceral endoderm-like cells. Visceral endoderm-like cells play an important inductive role in the differentiation of cardiogenic precursor cells in developing embryos. This protocol is particularly useful for testing the ability of hPSCs to form cardiac mesoderm cells and functional CMs. Co-culture is convenient because it requires a few number of cells, it is simple and rapid to perform, and it yields CMs in sufficient numbers and quality of differentiated CMs [92].
Further studies revealed that the timed addition of defined extracellular molecules, either inhibiting or activating specific intracellular signaling pathways, improved the differentiation efficiency into CMs [83]. mESCs treated with RA differentiate into neural progenitor cells and then treated with antagonist of the Shh pathway differentiate into motor neurons [93]. Using the PI3-K inhibitor LY294002 it is possible to promote the differentiation of ESCs into insulin-producing β cells, and these cells can improve insulin levels in vivo, prevent weight loss, and control blood glucose concentration [94]. Ascorbic acid and many other small molecules have been found to effectively enhance the differentiation of ESCs into CMs [95,96]. Multistage protocols entailing the sequential use of BMP4, Activin A, and FGF followed by Dkk1 (to terminate the canonical Wnt pathway inhibition), or these factors plus VEGF and FGF2 (as mitogens) [71,97] or the sequential administration of Activin A followed by BMP4 [76,87] have been used. In pioneering studies, low concentrations of BMP4 and high concentrations of Activin A were found to induce α-myosin heavy chain in mouse ESCs [98]. The temporary and dose-controlled addition of activin A and BMP4 efficiently induced cardiac mesoderm formation in mouse and human EBs characterized by co-expression of Flk1 and PDGFRα or by the generation of a cardiovascular progenitor characterized by co-expression of KDR and c-KIT [97,99]. This series of steps efficiently induces ESCs to generate CMs, smooth muscle cells, and endothelial cells both in vitro and in vivo [97]. Implementation of conditioned medium with defined factors, such as FGF2, VEGFA, and DKK1, and inhibition of Nodal or TGFβ receptor 2 signaling, enhanced the efficiency of CM differentiation to greater than 50% [62,76,97,99,100].
As the production of hESCs involves the destruction of human embryos, there are many ethical controversies about their use [101]. Many other cell types have been explored too as source of CMs with the aim to overcome these ethical concerns. Among them, human iPSCs (hiPSCs) and adult multipotent stem cells. iPSCs are reprogrammed somatic cells with the capacity to differentiate into cells of all three embryonic germ layers. To develop iPSCs it’s necessary to overexpress, in a somatic cell, genes that reprogram the cell to its pluripotent ESC-like state [95]. hiPSC differentiation protocols yield CMs that display an immature phenotype representative of the fetal stage of development [102,103]. Several approaches have been explored to overcome this limitation in hPSC-derived CM production, as recapitulating the metabolic changes that occur within the CMs at birth and the use of tissue engineering to expose the developing CMs to electromechanical forces present in the heart. Currently, the purity of hPSC-derived CMs following differentiation can reach over 90% [99,104] and the phenotype of hPSC-derived CMs resembles that of fetal CMs: they are morphologically small, spontaneously beat, lack T-tubules, and have inefficient calcium handling [105]. However, to achieve optimal results, the differentiation process needs to be monitored and optimization of protocols is required for individual cell lines and for the timely addition of factors. In order to generate a relatively pure CM culture (up to 98%), further enrichment steps included cell sorting with an antibody against signal-regulatory protein α (SIRPA) [106], or cell purification based on biochemical differences in the metabolism of glucose and lactate between CMs and non-CMs [107]. Other methods of cell purification are based on the relatively high content of mitochondria in CMs using mitochondria-specific fluorescent dyes [108]. The main goal is to transplant enough hiPSC-derived CMs in patients with a MI in order to muscularized the damaged region of the heart and to replace the fibrotic scar that develops following a MI with new myocardium generated from the transplanted hPSC-derived CMs. The workability of this type of approach was documented by the transplantation of primary neonatal rat CMs in recipient rat hearts where neonatal rat CMs survived and engrafted [109,110]. Since then, several investigators have shown that mESC-derived CMs, as well as hPSC-derived CMs could engraft the hearts of mice or rats following transplantation [76,111,112]. Teratoma formation was not reported in these studies. Therefore, injected CMs contained few, if any, undifferentiated hiPSCs. Although improvements in ventricular function were documented following transplantation, the effects of hPSC-derived CMs, were likely due, at least in part, to paracrine mechanisms and not to the contractile function of the graft because the transplanted cells were not electrically coupled with the host myocardium, probably due to the difference in heart rate in rats, mice, and humans (400, 600, and 60–80 b.p.m., respectively). Transplanted hPSC-derived CMs into guinea pigs that have a slower heart rate between 200 and 250 b.p.m. were able to electrically integrate with the host myocardium, supporting the concept that the engrafted cells can function to provide new contractile force. Nevertheless, transient arrhythmias developed in the larger animal recipients of the hPSC-derived CMs has been documented [113–115]. Another major challenge is that of immune rejection of the graft. The use of patient-specific iPSCs could circumvent this problem but this strategy has been largely abandoned due to the time and cost of producing clinical grade cells specific for each patient [116]. An alternative approach is the use of allogeneic cells that allow for the development of a ‘off-the-shelf’ product for transplantation. To this end, hPSCs have been engineered to remove human leukocyte antigens (HLAs), with the goal of generating a ‘universal donor cell’ [117–119].
To date, numerous different types of stem/progenitor cells expressing c-kit have been reported as a source of new CMs in the adult heart: cardiac stem cells, epicardium-derived cells, endothelial progenitor cells, Isl1 positive cardiac progenitor cell and CardioSphere-derived cells are examples of source of cells evaluated for their capacity to replace lost CMs and recover the myocardial tissue by transplantation into the post-infarcted myocardium [70–73,120–124]. It has been extensively demonstrated that c-Kit is a relatively ubiquitous marker (monocytes within the tissue, endothelial or hematopoietic cells express c-kit), and that its expression alone is insufficient to define a specific cardiac progenitor c-Kit positive cell population [72]. Therefore, better cell surface marker signatures are still needed to resolve c-kit positive cardiac cell heterogeneity [71,72]. Adult endogenous cardiac stem/progenitor cells (CSCs) have been shown to contribute to the cardiomyogenic cell lineage both in vitro and in vivo when delivered after injury; they also can be reactivated in situ to induce cardiac regeneration following cardiac injury [70–72,123]. Through gain- and loss-of-function experiments we have shown the involvement of Wnt/β-catenin and TGF-β/SMAD signaling in growth promotion and cardiomyogenic specification of the CSCs [71,72]. We have also shown that CSCs respond in vitro to these known cardiac morphogens: the inhibition of the Wnt canonical pathway and TGF-β family activation, each independently, promotes cardiomyogenic commitment. Wnts improve CSCs expansion and clonogenicity while canonical Wnt pathway inhibition by Dkk1 or by constitutively active GSK3β signaling induces myogenic commitment of CSCs by promoting differentiation with the inhibition of proliferation and clonogenicity potential [71,125,126]. On the other hand, conditional expression of β-catenin, results in expansion of CSCs preventing their myogenic commitment. In response to this combination of signaling factors and downstream transcriptional events, it is possible to generate functional CMs able to contract synergically in vitro. Indeed, sequential, and step-by-step administration of BMP-4, Activin A, β-FGF, and DKK-1 to clonal CSCs, increase the expression of myogenic genes and the number of troponin or myosin positive cells [71,78,127]. We have recently shown that, when CSCs are induced in vitro to differentiate using well-established protocols, CSCs are robustly cardiomyogenic through the activation of the entire gene network characteristic of the cardiomyocyte phenotype. Nevertheless, like iPSCs-derived CMs, those derived from CSCs, still have an immature cardiac muscle phenotype, closely resembling fetal/neonatal CMs, lacking the levels of expression of the sarcomeric genes typical of the adult terminally differentiated phenotype, even though spontaneously contracting [78]. To that, the whole transcriptome of CSCs, CSC-derived CMs and CMs isolated from neonatal and adult mice hearts, has been analyzed showing changes in mRNA and miRNA underlying the transition from replicating CSCs to terminally differentiated but still immature CMs [78].
microRNAs and cardiomyocyte differentiation in vivo
As summarized above, the transcriptional regulation of CM differentiation is an evolutionary highly conserved process that requires sequential activation and/or repression of different genetic programs [3]. Very broadly, CM differentiation follows progressive maturation steps that require the switch-on of the cardiac muscle gene network, and the repression of the cell cycle gene machinery. Overall, CM differentiation and specification has been depicted for years as a mainly positive process, where stepwise specific gene activation by chemical and mechanical stimuli induce complete myogenic terminal differentiation during development into adulthood. Yet, the demonstration that microRNAs are sufficient to direct ESC differentiation into CMs [89,91] and that four specific miRNAs can reprogram fibroblasts into CMs [128] show that the CM specification and differentiation cascade can also be triggered by suppressing the expression of gene cohorts by the negative instructions of specific miRNAs.
The switch from pluripotent to lineage-specified cells involves the downregulation of pluripotency markers with decrease of the self-renewal capacity of cells together with the activation of lineage-specific genes. All these events are orchestrated by the up and/or down-regulation of several conserved non-coding RNAs [129–131]. Endogenous non-coding RNAs (ncRNAs) including microRNAs (miRNAs, miRs), long non-coding RNAs (lncRNAs), or piwi-interacting protein RNAs (piRNAs) are regulatory elements consisting in RNA molecules without an open reading frame that can control the expression of target genes at the post-transcriptional level [131] and have been reported to engage in cardiac regeneration and repair. miRNAs exert crucial functions in development, differentiation, cell-fate specification, and pathogenesis. Many miRNAs are expressed in a tissue-specific and developmentally specific manner revealing a novel mechanism by which the proteome is regulated during the dynamic events of cell-lineage specification and morphogenesis. Approximately 500 miRNAs are transcribed as longer precursors and subsequently processed by the enzymes Drosha and Dicer to ultimately yield mature miRNAs of ∼20–22 nucleotides [132]. Mature non-coding RNAs typically base pair to specific target messenger RNAs (mRNAs) by partial sequence matching after becoming incorporated into the RNA-induced silencing complex (RISC). The result is the degradation of the mRNA transcript and/or its translational inhibition [131]. Disruption of Dicer in mouse cardiac progenitor cells by knockout of the miRNA-processing proteins Dicer1 or Dgcr8, results in embryonic lethality due to cardiac failure. Moreover, Dicer-deficient ESC, which lack mature miRNAs, are defective in differentiation both in vitro and in vivo, and do not form the three germ layers normally found in embryoid bodies derived from ESC. Thus, non-coding RNAs are essential during early development [129,133–137].
Anyone miRNA may have numerous targets exerting robust control of complex cellular processes through modulation of multiple interrelated targets. On the other hand, a single mRNA can be commonly targeted by multiple miRNAs, so the combinatorial miRNA:mRNA interactions can produce very complex and alternative regulatory networks in response to different endogenous and exogenous inputs [138,139].
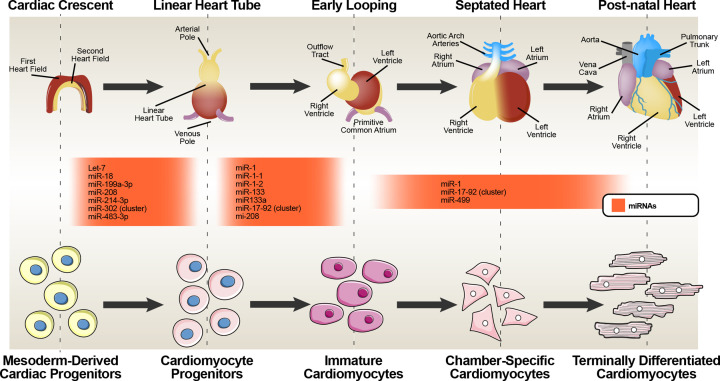
During heart development miRNAs are also involved in regulation of CM proliferation (Figure 2). Several miRNAs endogenously expressed in CMs, have been shown to suppress their proliferation. Among miRNAs involved in suppressing CM proliferation let-7, miR-1, miR-15 family, miR-29a/b, miR-34a, miR-128, miR-128 and miR-133 are the most abundant and better studied in the heart [140]. Let-7 was one of the first isolated and characterized miRNAs. Its overexpression is sufficient to inhibit CM proliferation, whereas its suppression induces CM proliferation improving cardiac function in response to myocardial infarction [141]. miR-1 and miR-133 are highly conserved and are transcriptionally regulated by MyoD, Mef2, and SRF [142,143]. Overexpression of miR-1 in the developing heart leads to decreased CM proliferation where miR-1 deletion increases CM proliferation in the adult [142]. Similar to miR-1, overexpression of miR-133a restricts CM proliferation in zebrafish [144], while miR-133a inhibition increases CM proliferation in adult hearts [145].
Figure 2. Summary of miRNAs differentially expressed in five major steps of heart development.
The schematic representation shows the most characterized miRNAs regulating cardiac development and cardiomyocyte formation.
miR-1 is the most abundantly expressed miRNA in the heart, and in the mouse heart it accounts for almost 40% of all miRs. It is the first miRNA documented as essential for cardiac development with a pattern of expression specific to cardiac and skeletal muscle [146]. During early stages of embryonic development, miR-1, together with miR-133, promote mesoderm induction and suppress its differentiation into the ectodermal or endodermal lineages [147]. These two miRs cooperate to promote mesoderm formation from ESCs and to suppress non-muscle gene expression [148]. The miR-1/miR-133 clusters are regulated through feedback loops where repression of HDAC4 by miR-1 activates MEF2 which in turn activates miR-1expression. SRF regulates the expression of miR-133, which in turn represses SRF expression [145,148]. Furthermore, while miR-1-1 is initially expressed in the inner curvature of the heart loop and in atria, two segments of the developing heart less proliferative, miR-1-2 is expressed mainly in the ventricles [149]. Gain- and loss-of function experiments revealed that miR-1/miR-133 play crucial roles in mouse hearts. miR-1 has been shown to be involved in controlling cycling myocardial cells. Indeed, its overexpression under the control of the βMHC cardiac-specific promoter in transgenic mice results in embryonic lethality at embryonic life 13.5dpc with a phenotype characterized by thin-walled ventricles due to decreased ventricular proliferation and expansion caused by the CMs premature differentiation and early withdrawal from the cell cycle [142]. In contrast, miR-1-2 null-mice result in 50% embryonic lethality accompanied by thickened chamber walls from prolonged hyperplasia and ventricle septal defects and increase CM proliferation resulting in severe heart defects [142]. Surviving miR-1-2 null mice often die suddenly due to conduction system defects and the expression of Hand2, one of the validated targets of miR-1 in the heart. Hand2, involved in the expansion of ventricular CMs, is elevated in miR-1-2 null mice [142,146]. This miRNA binds to the promoters of several essential cardiac transcription factors such as Nkx2.5, GATA4, and Mef2 as well as genes involved in cardiac development such as Hand2 and TMSB4X, GJA1, and KCNJ2 [146,150].Thus, miR-1 is essential in the balance between CM proliferation and differentiation, repressing mouse cardiac progenitor cell proliferation [142]. Deletion of miR-133a-1 or miR-133a-2 alone does not produce cardiac defect whereas deletion of both may cause embryonic lethality due to ventricular septal defects (VSDs). However, surviving miR-133a null mice display dilated cardiomyopathy and heart failure with excessive CM proliferation due to unregulated expression of cyclin D2, a direct target of miR-133a [145]. Consequently, overexpression of miR-133a driven by the βMHC promoter inhibits CM proliferation and causes embryonic lethality characterized by a thinner ventricular wall and VSDs [145]. Nonetheless, mice with simultaneous targeted inactivation of individual miR-1/133a clusters are viable and fertile and show no gross morphological aberrations [151]. In addition to being expressed in cardiac and skeletal muscle during development, miR-1 and miR-133 are expressed in CM derived from ESCs [148]. In vitro overexpression of miR-1 and miR-133 in ESCs results in the suppression of their differentiation into endodermal or ectodermal lineages and promotes CM differentiation [148]. These effects demonstrate the critical importance of correct timing and dosage of miRNAs during heart development.
Like miR-1 and miR-133, the miR-15 family (including miR-15a and b, miR-16-1/2, miR-195, and miR-497), modulates neonatal heart growth and regeneration through inhibition of postnatal CM proliferation by repressing the cell cycle genes, resulting in postnatal loss of cardiac regenerative capacity. Thus, the inhibition of miR-15 family from the early postnatal period until adulthood promotes CM proliferation and enhances cardiac function in response to MI [152,153]. Overexpression of miR-195 in the embryonic heart suppresses CM proliferation and impairs the regeneration capability of neonatal heart after MI through the repression of cell cycle genes [153,154]. Inhibition of miR-26a increased proliferation of mouse neonatal CM in vitro and in vivo via regulation of cell cycle inhibitors, while overexpression of miR-29a suppresses CM proliferation and its inhibition enhances cell division [155,156]. Like miR-29a, inhibition of miR-29b promotes CM proliferation by inactivation of notch receptor in vitro and in vivo [157]. miR-34a, a regulator of age-associated physiology, also suppresses CM proliferation and its inhibition leads to enhanced CM proliferation and improves cardiac function in response to a MI by targeting specific factors such as Sirt1, Bcl2, and Cyclin D1 [158]. Inhibition of miR-128 promotes CM proliferation by activating positive cell cycle regulators and improves cardiac function in response to a MI [159].
Regulation of embryonic germ layer specification during gastrulation, involved other miRNAs clusters. The miR-371-373 cluster it’s involved in the maintenance of pluripotency and in the ectoderm development, probably by modulation of the Nodal/TGF-β signaling. Similar functions have been demonstrated for the miR-302-367 cluster [130,160,161]. Daichi Ishikawa and colleagues [162] revealed different roles of miRNAs during mesoderm formation where miR-199a-3p, miR-214-3p, and miR-483-3p influence mesoderm development and the mesodermal progenitor pool (Figure 2). These miRNAs are potentially regulated by Wnt/β-catenin signaling [163,164]. The induction of mesodermal genes requires BMP4 signaling or Nodal/TGF-β antagonism, while the up-regulation of endodermal genes depends on Nodal/TGF-β signaling [165]. This pathway seems to be regulated by several targets of miR-199a-3p and miR-302 cluster, as these miRNAs lower Nodal/TGF-β signaling to permit mesoderm differentiation [130,161,162]. miR-199a-3p, miR-214-3p, and miR-483-3p have been described as strongly induced exclusively in mesodermal cells, indicating that they are responsible for the mesoderm differentiation of human ESCs [162] These data demonstrate that non-coding RNAs affect development by interference with the pluripotent transcriptional network. Using a lineage-tracking system to isolate Mesp1 positive cardiac progenitor cells, Islas JF's lab and others identified several miRNAs indispensable in the early regulators of cardiac fate. These miRNAs specify cardiac mesoderm and cardiac development (let-7, miR-18, and miR-302; miR-17-miR-92 superfamily) cardiac mesoderm formation and cardiac program initiation [166,167], and display a preeminent role during embryogenesis by regulating cardiac muscle and CM differentiation. They probably also contribute to gender-specific differences in cardiac development and diseases [168].
miRNAs signaling is also involved in controlling muscle gene expression and performance of contractile proteins such as myosin genes. Myosin heavy chain (MHC) is the major contractile protein of cardiac and skeletal muscle cells and the primary regulator of muscle strength and contractility. Cardiac contractility depends on the expression of two MHC genes, α- and β-MHC (also known as Myh6 and Myh7, respectively). The ratio of α- and β-MHC expression is species specific and varies in response to developmental, physiological, and pathological signaling. During development in rodents, β-MHC, a slow ATPase, is highly expressed in the embryonic heart, whereas α-MHC, a fast ATPase, is expressed at relatively low levels in the prenatal heart and then up-regulated during the early postnatal period [169]. In this regard, miR-208, is only expressed in the heart and it is required for the up-regulation of slow β-MHC in the adult heart in response to stress and hypothyroidism [170]. miR-208a, -208b, and -499 are encoded by the Myh6, Myh7, and Myh7b genes, respectively. Because their expression follows the expression of their respective host myosin genes during development, they were named myo-miRs [171]. Indeed, the expression levels of miR-208 b/Myh7 are relatively high in embryonic heart and decline rapidly after birth, whereas miR-208a/Myh6 are highly expressed in adult heart and miR-499/Myh7b are constitutively expressed at a high level in the heart [172]. CM proliferation in mammalian heart is robust during the fetal period but is switched off early after birth [173], and several miRNAs have been identified as responsible of this process by inducing or inhibiting it in vitro and in vivo. High-throughput screening approach to generate a whole-genome miRNA library identified miR-199a, miR-302b, miR-518, miR-590, and miR-1825 as inducer of cell proliferation by EdU-incorporation, Ki-67, and phospho-histone H3 (pHH3) expression in mouse and rat CMs [174]. miR-199a and miR-590 were linked to cell cycle activation and progression enhancing CM proliferation in vitro and in vivo leading to improved cardiac function and decrease cardiac fibrosis in response to a myocardial infarction [174].
The miR-322/-503 cluster, have a strong inhibitory effect on neural differentiation. miR-322/-503 (miR-424 is the human ortholog of miR-322) is implicated in angiogenesis [175,176], myotube formation [177], and regulating cardiac muscle differentiation [168]. Null mutants of miR-322/-503 were not embryonically defective but had adult-onset anomalies (white fat accumulation) [178]. Thus, the miR-322/-503 cluster specifically regulates the CM program through the regulation of the most important targets of these miRNAs.
miR-322/-503, an X-chromosome miR cluster, was identified as the most enriched miRs in the earliest Msep1 positive cardiac progenitor cells [168]. Its ectopic expression specifically drives precocious CM formation suggesting that miR-322/-503 acts as a potent inducer of CM specification [168].
Thus, miRNAs, together with their mRNA gene regulatory networks, regulate CM differentiation and maintain the balance between CM proliferation and their differentiation. For this reason, they are the main RNA candidates tools for producing mature fully functional CMs in vitro as well as in vivo to replace lost and damaged CMs.
microRNAs and stem cell cardiomyocyte differentiation
The data summarized above support the concept that heart development is a complex process requiring many temporally and spatially controlled gene regulatory networks with many intersecting points among them. In vitro differentiation of stem cells such as human/mouse ESCs, iPSCs and adult cardiac progenitor cells into functional CMs can recapitulate different stages of heart development and may serve as an in vitro model for the study of cardiogenesis. Dissecting the molecular players involved during embryonic CM differentiation and maturation might be the key for better understanding the molecular mechanisms involved in cardiac development and disease. The identification of miRNAs expressed in specific cardiac cell types has led to the discovery of their regulatory roles during CM differentiation, cell cycle and hypertrophy in the adult heart, indicating that miRNAs are important in controlling cardiac gene expression. Cardiac signaling and transcriptional pathways involved in cardiac development, maturation, function, and disease have been shown to be intimately regulated by miRNAs.
From embryonic to adult life, mammalian CMs derived from cardiac mesoderm stem/progenitors, undergo a replication, differentiation and maturation process characterized by their permanent exit from the cell cycle, binucleation, increased metabolic demand, development of sarcomeric structures, changes in Ca2+ release and sequestration. Several groups, including ours, have developed different protocols for differentiating stem cells into mature CMs. However, until now all the employed protocols have generated differentiated cells with an immature morphological and functional phenotype. This is not surprising, given that in vivo neonatal human CMs require years of maturation (till 10 years) before reaching the typical adult phenotype, whereas CMs derived from different stem cell sources are differentiated for only few weeks. Indeed, a mature adult CM is significantly larger, rod-shaped, with highly organized sarcomeres, polarized connexin-43 and a more negative resting potential (around −90 mV). Moreover, adult CMs are also multinucleated [179] with regularly spaced T tubules, which are essential for proper excitation–contraction coupling and action potential generation [180]. In contrast, immature PSC-derived CMs exhibits a round cell shape, a disorganized contractile apparatus, a single nucleus, limited calcium-handling ability, and a resting membrane potential of around −60 mV.
As mentioned above, all the main molecular pathways involved in CM differentiation are transcriptionally regulated by specific myogenic mRNAs and have been used in vitro to differentiate ESCs, iPSCs as well as multipotent adult stem cells such as CSCs into CMs [72,181,182].
Several miRs have been discovered and used for stem cell-derived CM differentiation and maturation, particularly those miRs implicated in the maturation of the adult from fetal development. miR-125 b-5p, -199a-5p, -221, and -222 release into human ESC-derived CMs, as prototypical example, resulted in enhanced sarcomere alignment and Ca2+-handling, as well as higher expression of markers widely expressed in mature and terminally differentiated adult CMs [183,184]. Human ESC-derived CMs, as well as human fetal and adult CMs, were found to have significant levels of Let-7 expression [185]. Although Let-7 is not necessary for early CM commitment from human ESCs, it is essential to improve a variety of functional features associated with CM maturity, such as the switch from glucose to fatty acid metabolism [185]. However, despite these findings, in vitro-generated CMs from ESC- or iPSC still reveal immature properties and do not fully recapitulate the adult phenotype [186]. Using an integrative computational approach to assess the transcriptomic profiles of miRs and their regulatory networks, Poon and colleagues have recently identified a miRNA involved in cardiomyogenesis of hESCs [187]. They tested the hypothesis that miRNAs expressed in hESCs, and down-regulated during the differentiation process, could act through inhibition of pro-cardiomyogenic factors. They identify miR-200c, its target genes and five transcriptional factors (GATA4, SRF, TBX5, HAND1-, and NKX2.5) as molecules progressively down-regulated both during hESC differentiation into CMs and in heart development. They postulated that miR-200c and its targets are implicated in the control of Ca2+ handling, contraction, and heart development as repressor of cardiomyogenesis targeting in particular CACNA1C, a subunit of the L-type Ca2+ channel. In addition, overexpression of miR-200c increased beating frequency and abnormal Ca2+-handling, suggesting that negative regulation of genes encoding ion channels and Ca2+-handling proteins by miR-200c may directly contribute to the immature Ca2+-handling and electrophysiological properties in ESC-derived CMs. Thus, miR-200c is a strong repressor of human ESC differentiation into CMs and their maturation.
Five let-7 family miRNAs (let−7f−5p, miR−98−5p, let−7b−5p, let−7d−5p, and let−7i−5p) are up-regulated during the maturation of hESC-derived CMs [188,189]. miR-1 promotes CM differentiation from ESCs by the down-regulation of Notch ligand Delta-like 1, and miR-133 inhibits differentiation of ESCs into CMs [148]. miR-1 and miR-133 are differentially expressed in ESC-derived CMs compared with ESCs. Transduction of ESC-derived CMs with miR-1 facilitates electrophysiological and Ca2+-handling maturation of CMs [183]. Overexpression of miR-145 inhibits hESC self-renewal and induces differentiation with an increased expression of mesodermal and ectodermal genes [190]. Transduction of ESC-CMs with miR-499 significantly increase the yield of the ventricular lineage CMs by upregulation of Myh6, Myh7, Mlc2, and TNNT2, suggesting its role in ventricular specification [183]. Given these data, it is not surprising that a combination of miR-1, miR-133, miR-208, and miR-499 is able to directly induce the cellular reprogramming of fibroblasts into CMs in vitro [191].
Several reports have shown that miR−455−5p under hypoxic conditions in vitro increased 4-fold suggesting a role in CM differentiation [189,192,193]. The heart exclusively expresses miR−208b-3p, which regulates the myosin heavy chain switch. This miRNA is up-regulated in hiPSC-generated CMs [189]. Taken together, all these data indicate that in vitro CM specification of ESCs involves the same regulatory mechanism that govern in vivo embryo development and lineage specification.
iPSCs are somatic cells reprogrammed to become pluripotent, self-renewal and totipotent that can differentiate into all somatic cell types [95]. Several studies have shown that human ESCs and iPSCs are different because they display distinct gene expression profiles and regulatory mechanism [194,195]. miRNAs can directly reprogram somatic cells into iPSCs. For example, miR-302 promotes mesoendoderm at the expense of neuroectoderm in human ESCs and the miR-302/-367 cluster, highly enriched in iPSCs, can reprogram human skin cancer cells into a pluripotent state [196,197]. Also, direct transfection of the mature double-stranded miR-200c, miR-302, and miR-369 family can reprogram mouse and human somatic cells into pluripotent states [198]. The miR-200 family (miR-200b and miR-200c), the miR-106a-363, miR-302-367 cluster, and miR-93/106b (activated by Oct4, Sox2, Klf4 or Oct4, Sox2, Klf4, c-myc) can promote the epithelial–mesenchymal transition process during iPSC initiation and could be used in the activation and maintenance of pluripotency. Also, down-regulated miR-30/let-7 family, up-regulated miR-17, miR-19, miR-290 and miR-8 family miRNAs play important roles in maintaining the pluripotent state [199,200]. Furthermore, miRNAs participate in the reprogramming process by regulating cell cycle factors. For instance, the miR-25 and miR-130/301/721 families target p21, a cell cycle inhibitor, to promote reprogramming efficiency [201,202]. Depletion of miR-34a significantly promotes the somatic cell reprogramming process, while miR-34a and p21 together regulate reprogramming efficiency by targeting p53 [203].
Using a cell lineage-tracking approach, Xiaopeng Shen and colleagues [168] isolated miRNAs from Mesp1 positive CPCs. Most of the identified miRNAs do not have established roles in cardiac development but those identified in the early stage of cardiac fate specification were involved with cardiac mesoderm formation (let-7, miR-18, and miR-302) and cardiac program initiation (the miR-17-miR-92 superfamily). This superfamily is essential for cardiac development whereby its loss produces severe cardiac defects and embryonic death [204]. On the other hand, none of the heart- and skeletal muscle-specific miRNAs such as miR-1, -133, -206, -208, and -499 were not expressed in Mesp1 CPCs and were shown not to be a resource for dissecting early cell fate and CM differentiation (Figure 2) [168]. Thus, it is still unknown which miRNAs act at the precursor steps and which are directly involved in cardiac fate establishment.
The majority of predicted targets of the Mesp1 miRNAs are down-regulated in Mesp1 CPCs supporting the hypothesis that Mesp1 miRNAs main function is to inhibit specification of other cell lineages in order to foster cardiac-lineage specification.
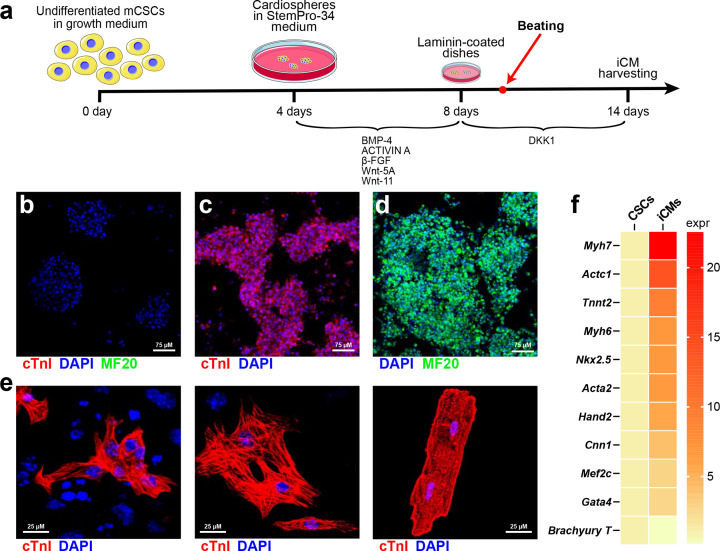
We have intensively investigated the biology of multipotent CSC and their regenerative potential in vitro as well as in animal models of myocardial injury [71,72,205–210]. CSCs robustly differentiate into functional beating CMs with a transcriptome/miRnome which closely resembles that of adult CMs further supporting the true myogenic stem/precursor nature of these cells [78]. In particular, we have demonstrated that a population of clonogenic cardiac stem cells that is c-kit positive and negative for surface markers typically expressed by already committed cells [71], when exogenously transplanted in an injured heart, efficiently differentiate [71,72] and progressively mature over time acquiring a fully mature CM architecture and function [70]. We demonstrated that a protocol of sequential and step-by-step administration of BMP-4, Activin A, β-FGF, and DKK-1 to clonal CSCs increase the expression of myogenic genes and the number of troponin and myosin positive cells, generating spontaneously beating cardiomyocytes in vitro (Figure 3) [78]. We used a genome-wide small RNA sequencing approach to identify mRNA and miRNA changes in CSC differentiation in vitro to identify specific miRNAs/mRNA network involved with CSC differentiation into CMs. Comparison of the miRNome profiles of CSCs with those of CSC-derived CMs and CMs isolated from adult hearts identified miRNAs that potentially play a role in the development of the CM phenotype. Interestingly, CSC-derived CMs express a miRNA profile related to cell cycle progression, cardiac function and maturation which is similar to the miRs set typical of adult CMs. Thus, whole gene expression profiling by mRNASeq analysis of CSCs and CSC-derived CMs revealed that the CSC-derived CMs closely resemble adult terminally differentiated CMs. Also, comparison of the miRNome profile of CSCs with those of CSC-derived CMs and CMs isolated from adult hearts, identified miRNAs that potentially play a role in the development of the adult CM phenotype. CSCs differentiation into CMs is characterized by miRNA/mRNA networks operative in the adult CMs and two miRs from these miRNA/mRNA networks, miR-1 and miR-499, individually and together enhance myogenic commitment, increasing the expression of sarcomere genes, inducing terminal differentiation of CSC-derived CMs as previously reported for differentiation of ESCs and adult cardiac progenitors into CMs [211]. miR-1 and miR-499, independently and together, reduced the expression of positive cell cycle regulatory genes, such as CyclinA2 and Cyclin D, while increasing the expression of negative cell cycle regulatory genes, such as Rb1, crucial for CM terminal differentiation [212,213]. Thus, the up-regulation of miR-1 and miR-499 can be used to enhance CM generation in vivo and in vitro.
Figure 3. Cardiac stem/progenitor cell derived cardiomyocyte formation*.
(A) Schematic representation shows the differentiation protocol to derive beating cardiomyocytes from cloned CSCs in vitro. (B) Representative confocal image of mouse cloned CSCs-derived cardiospheres (CS) before myogenic differentiation induction (left), showing no cTnI or MF20 expressing cells and of functional cardiomyocytes derived from CS differentiation, showing a homogenous expression of cTnI (red) and MF20 (green). (C) Confocal microscopy examples of CSC-derived iCMs, neoCMs and adult CMs labeled with cTnI (red). (D) Heatmap showing qPCR analysis of main contractile genes in cardiosphere-derived CSCs (Actc1, Tnnt2, Myh7, Actc1, Myh6, Acta2, Cnn1) and cardiac transcription factors (Mef2c, Gata4, Nkx2.5, Hand 2 and Brachyury T) after myogenic differentiation. *Adapted from reference [78].
Despite all the advances listed above, it remains to be demonstrated whether the differentiation of CSCs into CMs undergoes the transcriptional switch typical of cell cycle exit during cardiac myocyte differentiation which up-regulates the known myo-miRs and more generally the cohort of miRs which are more abundant in the adult CMs.
microRNAs for myocardial regeneration
For a long time, the mammalian heart has been considered a post-mitotic organ for the lack of an efficient or at least well-defined cardiac muscle regeneration potential in health and in response to injury [214,215]. It has been repeatedly reported that the total number of CMs is permanently established shortly after birth and that the lifespan of a CM is equal to the lifespan of the body because no CM are formed, the heart lacking an endogenous regenerative potential [216,217].
Notwithstanding the latter, during the first postnatal week, the mouse heart still possesses the robust regenerative capacity characteristic of the late fetal life [154,216,218]. It has been indeed shown that after partial cardiac excision, the hearts of 1-day-old neonatal mice can regenerate the lost CMs, but this potential is gone after 7 days from birth [219]. Similarly, neonatal human hearts have the ability to repair myocardial damage and fully recover cardiac function following a MI due to coronary anomaly [219].
During cardiogenesis, after the appearance of a beating cardiac tube at 9.5 dpc, these newly generated CMs, following cardiomyocyte lineage specification, undergo several rounds of proliferation before their post-natal terminal differentiation and exit from the cell cycle in order to produce sufficient numbers of mature CMs sufficient to maintain the cardiac output required for adult circulatory homeostasis. Paracrine signals from the epicardium and endocardium play a crucial role in orchestrating this growth process. This fetal growth and regenerative capacity which continues during the first postnatal week is rapidly lost and the CMs undergo terminal differentiation and permanently exit the cell cycle [218,220,221]. Changes in hormone concentrations and availability of metabolites promote a switch from glycolysis to fatty acid oxidation to meet the increased energy demands of the neonatal heart [222,223]. As consequence, CMs become acytokinetic a process that coincides with their polyploidization and multinucleation [68,110,224–226]. However, an ever-increasing body of evidence suggests that continuous turnover of CMs occurs in the adult mammalian heart. Bergmann et al. [67,227] revealed that the yearly turnover of terminally differentiated CMs progressively diminishes from a 1% at 20 years old, to 0.3% at 75 years old, while other cardiac cells, such as endothelial and mesenchymal cells, keep up with an yearly turnover pace of roughly 20%. If this was the case, it would suggest an exceptionally restricted proliferative and regenerative limit of the CM pool in the adult heart that would be totally useless to replace lost CMs in order to counteract not even partially for cardiac dysfunction after myocardial injury [217].
Modulating the normal miRNA content, by either overexpression of synthetic miRNA, through the delivery of miRNA mimics or viral vector-mediated miRNAs, or knocking them down with specific antagomirs to suppress their function, could yield additional insight into the role miRNA plays in development and disease pathogenesis [228]. The regulatory effects of miRNAs in myocardium development and homeostasis are involved in all aspects of cardiac biology: myocyte proliferation, differentiation, myocyte survival, and their reprogramming. Among all the described above, miR-222, miR-17-92, miR-590, miR-199a, and miR-302-367 have been reported to promote CM proliferation while miR-1 and miR-133 suppress it [142,167]. A high-throughput screen of a human whole-genome miRNA library identified miR-590 and miR-199a, as inducers of CM proliferation in the neonatal mouse and rat [174] whereby, after myocardial infarction in mice, these two miRNAs stimulated robust cardiac regeneration, decreased cardiac fibrosis and substantially improved cardiac function. These two miRNAs promoted CM proliferation both in vitro and in vivo thought the activation of the cell cycle [174]. The miR-17-92 cluster is also a critical regulator of cardiac proliferation. Indeed, its overexpression induces CM proliferation in embryonic, postnatal, and even adult hearts and reverses cardiac dysfunction and cardiac fibrosis after myocardial infarction [167]. The miR302-367 cluster also promotes CM proliferation during developmental and adult stages and its transient overexpression leads to increased CM proliferation, decreased cardiac fibrosis and improved cardiac function after injury by targeting the Hippo signaling pathway [229]. miR-222, up-regulated in several models of exercise, is necessary for exercise-induced cardiac proliferation and overexpression of miR-222 in the heart confers resistance to cardiac remodeling and dysfunction after ischemia/reperfusion injury. The inhibition of miR-222 blocks cardiac growth in response to exercise [230]. There are also miRNAs that suppress CM proliferation. In neonatal mice, the miR-15 family, together with the transcription factor homeobox Meis 1 (MEIS1), suppresses numerous cell cycle genes and plays a role in the postnatal loss of CM proliferation and cell cycle arrest [154]. MEIS1 was identified as a transcriptional activator of the cyclin-dependent kinase (CDK) inhibitors p15, p16 and p21 that synergistically inhibit cell cycle progression [153,220]. Thus, the miR-15 family by repressing postnatal CM proliferation leads to the postnatal loss of cardiac regenerative capacity. Conversely, its inhibition from the early postnatal to adult stages increases CM proliferation and improves left ventricular function after myocardial infarction [153].
miR-499 and miR-1 are involved in promoting cardiac stem cell differentiation into CMs [183] while miR-133 inhibits it [148]. miR-1 inhibits CM proliferation when overexpressed during mouse development [142,146]. On the other hand, deletion of miR-133a results in aberrant CM proliferation and its overexpression produces a diminishes CM proliferation, which is consistent with the conclusion that miR-133a represses CM proliferation [145]. Recently, the involvement on an additional miRNA in cardiac repair/remodeling has been shown. Expression of human microRNA-199a, delivered through an adeno-associated viral vector, stimulates cardiac repair one month after myocardial infarction with an improvement in global and regional contractility, increased muscle mass and reduced scar size [231].
Despite all the interesting results outlined here, reactivation of the CM cell cycle in the injured adult mammalian heart through the use of miRNAs independently or together with manipulation of pathways involved in cell proliferation has been insufficient and non-curative practice. The better explored option to replace the CMs lost by injury has been the use of cells from different sources which are able to home to the injured myocardium and differentiate into CMs or that through a paracrine effect foster the activation of resident cardiac stem cells.
Transplantation of mouse ESCs overexpressing miR-1 into the border zone of myocardial infarction protects against ischemia-induced apoptosis and promotes reprogramming and CM differentiation of cardiac cells. miR-133 appears to inhibit cardiac cell differentiation [145,232], and its overexpression suppresses cardiac markers in mouse and human ESCs. miR-499 is also strongly associated with cardiac differentiation. Similar to miR-1, overexpression of miR-499 reduces cardiac stem/progenitor cell proliferation, promotes the formation of beating embryoid bodies of mouse ESCs and improves in vitro CSCs differentiation [78,233]. Concomitantly, cardiomyocyte-specific genes such as cardiac troponin T, Actinin and Mef2 are also up-regulated [78].
The development of strategies that boost cell survival is required to improve the survival of the transplanted cells into injured myocardium.
miR-21 and miR-199a improve cardiac survival while miR-34a and miR-1 exhibit opposite effects. In vitro gain- and loss-of-function studies of miR-21 confirm that it protects against ischemia-induced CM apoptosis and its overexpression in rats decreases cardiac fibrosis by 29% at 24 h, reduces ventricular dimension at 2 weeks and decreases cell apoptosis in the infarct or border area after MI [234]. miR-24 shares similar functions with miR-21, reducing cardiac apoptosis and increasing cell number and its overexpression in MI models is protective against cardiac apoptosis [235]. Moreover, cardiac stem/progenitor cells transfected with a cocktail of miR-21, miR-24 and miR-221 exhibit higher viability in serum-free medium compared with untreated cells and in vivo the same cells transfected with the same cocktail decreases cardiac dysfunction by up-regulating Bim [236]. Thus, this cocktail of microRNAs may also improve the survival of transplanted cardiac stem/progenitor cells, improving cardiac function.
As mentioned above, the regulatory effects of ncRNAs in the myocardium are also involved in cardiac reprogramming. Indeed, miR-1, miR-133, miR-208, and miR-499 are able of reprogramming fibroblasts to cardiomyocyte-like cells and miR-284, miR-302, miR-93, and miR-106b are able to enhance cardiac reprogramming [128,191]. The miR-290 family is highly expressed in ESCs and significantly increases the efficiency of reprogramming of mouse embryonic fibroblasts (MEFs) [237,238]. Conversely, the inhibition of miR-290 preclude cell reprogramming. miR-17-92, miR-106b-25, and the miR-106a-363 clusters are highly expressed when reprogramming fibroblasts into CMs [201].
Several studies have observed the appearance of ventricular arrhythmia following transplantation of stem cell-derive CMs, as in the case of iPSC-derived CMs. This untoward event represents a significant obstacle in the application of a cell source to translational medicine [113,115]. This complication is often due to the immaturity of the pluripotent stem cell derived CMs used. Several approaches have been used to enhance the maturation toward the adult phenotype of these in vitro-generated CMs. It has been demonstrated that miR-1 can enhance the electrophysiological maturation of CMs while let-7 enhances their metabolic maturation with an increase in fatty acid metabolism, size and contraction force, while decreasing PI3/AKT/insulin [183,185].
Overall, these studies have convincingly showed that miRNA manipulation might improve the outcome of cell-based therapies either directly or through the enhancement of the endogenous cardiac repair processes. Therefore, exploring non-coding RNA-based methods to enhance cardiac regeneration remains instrumental for devising new effective therapies against cardiovascular diseases.
Conclusions
microRNAs are post-transcriptional regulators of gene expression emerging as central players in the establishment of cellular programs which modulate cell-type commitment, proliferation and differentiation during development as well as in adult tissues. Because of their mode of action, degradation of their target mRNAs, they directly suppress the expression of certain genes while enhance the expression of others by degrading their RNA inhibitors.
Heart muscle develops from two cohort of mesodermal cells organized into two heart fields which give raise to the two poles of the heart. These progenitors proliferate and differentiate to establish a complex and interconnected 3D structure, through a specific gene expression program highly influenced by the interplay of transcription factors and microRNAs. Although the mammalian heart has conventionally been viewed as a post-mitotic organ, it has been shown recently that the adult myocardium has some regenerative potential based on the existence of a small population of multipotent cardiac stem cells capable to differentiate into CMs, endothelial and smooth vascular cells and connective tissue cells. Although proliferation of adult cardiac stem cells and the ability of mature cardiomyocytes to re-enter the cell cycle have been proposed by some to be involved in the regenerative process, there is no available conclusive experimental evidence. The role of microRNAs in the control of the cardiac regeneration processes, and their potential applications for the treatment of cardiac injury could become an additional tool to ameliorate the functional cardiomyocyte deficit at the root of most heart diseases. Future analyses of gene expression profiles from different cell types, developmental, physiological, and pathological states should further expand the list of ncRNAs and their role (see Table 1 for an updated list) in the development of the myocardium, its homeostasis, and mechanisms to improve its repair from wear and tear and after injury.
Table 1. List of the miRNAs (involved with myogenesis) that are differently or are commonly expressed in cardiac development and in adult myocardial regeneration.
| miRNA | Cardiac linked biological process |
|---|---|
| Let-7 | Cardiac development |
| miR-18 | Cardiac development |
| miR-199a-3p | Cardiac development and cardiac reprogramming/regeneration |
| miR-208 | Cardiac development and cardiac reprogramming |
| miR-214-3p | Cardiac development |
| miR-302 (cluster) | Cardiac development and cardiac reprogramming |
| miR-483-3p | Cardiac development |
| miR-1 | Cardiac development and cardiac regeneration |
| miR-1-1 | Cardiac development |
| miR-1-2 | Cardiac development |
| miR-133 | Cardiac development and cardiac regeneration |
| miR133a | Cardiac development and cardiac regeneration |
| miR-17-92 (cluster) | Cardiac development and cardiac reprogramming/regeneration |
| miR-208 | Cardiac development and cardiac reprogramming/regeneration |
| miR-499 | Cardiac development and cardiac regeneration |
| miR-15 | Cardiac development and cardiac regeneration |
| miR-29a/b | Cardiac development |
| miR-34a | Cardiac development |
| miR-128 | Cardiac development |
Perspective
Importance of the field: The discovery of microRNAs as main modulators of cardiac development and cardiomyocyte differentiation has revolutionized developmental and adult cardiac biology
Summary of the current thinking: CM differentiation and specification has been depicted for years as a mainly positive process, where stepwise specific gene activation by chemical and mechanical stimuli induce complete myogenic terminal differentiation during development into adulthood. Yet, the demonstration that microRNAs are sufficient to direct stem cell differentiation into CMs and that specific miRNAs can reprogram fibroblasts into CMs shows that the CM specification and differentiation cascade can also be triggered by solely suppressing the expression of gene cohorts by the negative instructions of specific miRNAs.
Future directions: Unraveling the precise role of the different miRNAs in the regulation of cardiomyocyte specification, differentiation, maturation, and proliferation is set to provide a key approach for the field of regenerative medicine in the treatment of cardiovascular disease. microRNA-induced CM differentiation and maturation from different regenerative cells such as ESCs, iPSCs or multipotent adult stem cells as CSCs represents the way forwards to obtain efficient cardiac remuscularization in the clinical scenario.
Abbreviations
- CM
cardiomyocyte
- CSC
cardiac stem/progenitor cell
- HLA
human leukocyte antigen
- MEF
mouse embryonic fibroblast
- MHC
myosin heavy chain
- RA
retinoic acid
- TGFβ
transforming growth factor β
- VSD
ventricular septal defect
Contributor Information
Eleonora Cianflone, Email: cianflone@unicz.it.
Daniele Torella, Email: dtorella@unicz.it.
Data Availability
Data sharing is not applicable to the paper
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This research was funded by Grants from the Ministry of University and Research [grant numbers PRIN2017NKB2N4_005, PRIN20203YAY9B_005, and PRIN2020L45ZW4_005].
CRediT Author Contribution
Eleonora Cianflone: Writing—original draft, Writing—review & editing. Mariangela Scalise: Writing—original draft. Fabiola Marino: Writing—original draft. Luca Salerno: Visualization. Nadia Salerno: Writing—review & editing. Konrad Urbanek: Writing—review & editing. Daniele Torella: Conceptualization, Writing—original draft, Writing—review & editing.
References
- 1.Tzahor E. and Lassar A.B. (2001) Wnt signals from the neural tube block ectopic cardiogenesis. Genes Dev. 15, 255–260 10.1101/gad.871501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buckingham M., Meilhac S. and Zaffran S. (2005) Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 6, 826–835 10.1038/nrg1710 [DOI] [PubMed] [Google Scholar]
- 3.Srivastava D. and Olson E.N. (2000) A genetic blueprint for cardiac development. Nature 407, 221–226 10.1038/35025190 [DOI] [PubMed] [Google Scholar]
- 4.Tam P.P. and Behringer R.R. (1997) Mouse gastrulation: the formation of a mammalian body plan. Mech. Dev. 68, 3–25 10.1016/S0925-4773(97)00123-8 [DOI] [PubMed] [Google Scholar]
- 5.Vincent S.D. and Buckingham M.E. (2010) How to make a heart: the origin and regulation of cardiac progenitor cells. Curr. Top. Dev. Biol. 90, 1–41 10.1016/S0070-2153(10)90001-X [DOI] [PubMed] [Google Scholar]
- 6.Brand T. (2003) Heart development: molecular insights into cardiac specification and early morphogenesis. Dev. Biol. 258, 1–19 10.1016/S0012-1606(03)00112-X [DOI] [PubMed] [Google Scholar]
- 7.Soufan A.T., van den Berg G., Ruijter J.M., de Boer P.A., van den Hoff M.J. and Moorman A.F. (2006) Regionalized sequence of myocardial cell growth and proliferation characterizes early chamber formation. Circ. Res. 99, 545–552 10.1161/01.RES.0000239407.45137.97 [DOI] [PubMed] [Google Scholar]
- 8.van den Berg G., Abu-Issa R., de Boer B.A., Hutson M.R., de Boer P.A., Soufan A.T.et al. (2009) A caudal proliferating growth center contributes to both poles of the forming heart tube. Circ. Res. 104, 179–188 10.1161/CIRCRESAHA.108.185843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meilhac S.M., Esner M., Kelly R.G., Nicolas J.F. and Buckingham M.E. (2004) The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev. Cell. 6, 685–698 10.1016/S1534-5807(04)00133-9 [DOI] [PubMed] [Google Scholar]
- 10.Mjaatvedt C.H., Nakaoka T., Moreno-Rodriguez R., Norris R.A., Kern M.J., Eisenberg C.A.et al. (2001) The outflow tract of the heart is recruited from a novel heart-forming field. Dev. Biol. 238, 97–109 10.1006/dbio.2001.0409 [DOI] [PubMed] [Google Scholar]
- 11.Waldo K.L., Kumiski D.H., Wallis K.T., Stadt H.A., Hutson M.R., Platt D.H.et al. (2001) Conotruncal myocardium arises from a secondary heart field. Development 128, 3179–3188 10.1242/dev.128.16.3179 [DOI] [PubMed] [Google Scholar]
- 12.Kelly R.G. (2012) The second heart field. Curr. Top. Dev. Biol. 100, 33–65 10.1016/B978-0-12-387786-4.00002-6 [DOI] [PubMed] [Google Scholar]
- 13.Kelly R.G., Buckingham M.E. and Moorman A.F. (2014) Heart fields and cardiac morphogenesis. Cold Spring Harb. Perspect. Med. 4, a015750, 10.1101/cshperspect.a015750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Später D., Abramczuk M.K., Buac K., Zangi L., Stachel M.W., Clarke J.et al. (2013) A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat. Cell Biol. 15, 1098–1106 10.1038/ncb2824 [DOI] [PubMed] [Google Scholar]
- 15.Liang X., Wang G., Lin L., Lowe J., Zhang Q., Bu L.et al. (2013) HCN4 dynamically marks the first heart field and conduction system precursors. Circ. Res. 113, 399–407 10.1161/CIRCRESAHA.113.301588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krainock M., Toubat O., Danopoulos S., Beckham A., Warburton D. and Kim R. (2016) Epicardial epithelial-to-mesenchymal transition in heart development and disease. J. Clin. Med. 5, 27 10.3390/jcm5020027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lickert H., Kutsch S., Kanzler B., Tamai Y., Taketo M.M. and Kemler R. (2002) Formation of multiple hearts in mice following deletion of beta-catenin in the embryonic endoderm. Dev. Cell. 3, 171–181 10.1016/S1534-5807(02)00206-X [DOI] [PubMed] [Google Scholar]
- 18.Olson E.N. and Schneider M.D. (2003) Sizing up the heart: development redux in disease. Genes Dev. 17, 1937–1956 10.1101/gad.1110103 [DOI] [PubMed] [Google Scholar]
- 19.Winnier G., Blessing M., Labosky P.A. and Hogan B.L. (1995) Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 9, 2105–2116 10.1101/gad.9.17.2105 [DOI] [PubMed] [Google Scholar]
- 20.Mishina Y., Suzuki A., Ueno N. and Behringer R.R. (1995) Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 9, 3027–3037 10.1101/gad.9.24.3027 [DOI] [PubMed] [Google Scholar]
- 21.Conlon F.L., Lyons K.M., Takaesu N., Barth K.S., Kispert A., Herrmann B.et al. (1994) A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development 120, 1919–1928 10.1242/dev.120.7.1919 [DOI] [PubMed] [Google Scholar]
- 22.Schier A.F. (2003) Nodal signaling in vertebrate development. Annu. Rev. Cell Dev. Biol. 19, 589–621 10.1146/annurev.cellbio.19.041603.094522 [DOI] [PubMed] [Google Scholar]
- 23.Showell C., Binder O. and Conlon F.L. (2004) T-box genes in early embryogenesis. Dev. Dyn. 229, 201–218 10.1002/dvdy.10480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huelsken J., Vogel R., Brinkmann V., Erdmann B., Birchmeier C. and Birchmeier W. (2000) Requirement for beta-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 148, 567–578 10.1083/jcb.148.3.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buikema J.W., Mady A.S., Mittal N.V., Atmanli A., Caron L., Doevendans P.A.et al. (2013) Wnt/β-catenin signaling directs the regional expansion of first and second heart field-derived ventricular cardiomyocytes. Development 140, 4165–4176 10.1242/dev.099325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerkela R., Kockeritz L., Macaulay K., Zhou J., Doble B.W., Beahm C.et al. (2008) Deletion of GSK-3beta in mice leads to hypertrophic cardiomyopathy secondary to cardiomyoblast hyperproliferation. J. Clin. Invest. 118, 3609–3618 10.1172/JCI36245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li P., Cavallero S., Gu Y., Chen T.H., Hughes J., Hassan A.B.et al. (2011) IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development 138, 1795–1805 10.1242/dev.054338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heallen T., Morikawa Y., Leach J., Tao G., Willerson J.T., Johnson R.L.et al. (2013) Hippo signaling impedes adult heart regeneration. Development 140, 4683–4690 10.1242/dev.102798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Gise A., Lin Z., Schlegelmilch K., Honor L.B., Pan G.M., Buck J.N.et al. (2012) YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc. Natl. Acad. Sci. U.S.A. 109, 2394–2399 10.1073/pnas.1116136109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xin M., Kim Y., Sutherland L.B., Qi X., McAnally J., Schwartz R.J.et al. (2011) Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci. Signal. 4, ra70 10.1126/scisignal.2002278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collesi C., Zentilin L., Sinagra G. and Giacca M. (2008) Notch1 signaling stimulates proliferation of immature cardiomyocytes. J. Cell Biol. 183, 117–128 10.1083/jcb.200806091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hotta Y., Sasaki S., Konishi M., Kinoshita H., Kuwahara K., Nakao K.et al. (2008) Fgf16 is required for cardiomyocyte proliferation in the mouse embryonic heart. Dev. Dyn. 237, 2947–2954 10.1002/dvdy.21726 [DOI] [PubMed] [Google Scholar]
- 33.Qi X., Yang G., Yang L., Lan Y., Weng T., Wang J.et al. (2007) Essential role of Smad4 in maintaining cardiomyocyte proliferation during murine embryonic heart development. Dev. Biol. 311, 136–146 10.1016/j.ydbio.2007.08.022 [DOI] [PubMed] [Google Scholar]
- 34.Takakura N., Yoshida H., Ogura Y., Kataoka H., Nishikawa S. and Nishikawa S. (1997) PDGFR alpha expression during mouse embryogenesis: immunolocalization analyzed by whole-mount immunohistostaining using the monoclonal anti-mouse PDGFR alpha antibody APA5. J. Histochem. Cytochem. 45, 883–893 10.1177/002215549704500613 [DOI] [PubMed] [Google Scholar]
- 35.Fehling H.J., Lacaud G., Kubo A., Kennedy M., Robertson S., Keller G.et al. (2003) Tracking mesoderm induction and its specification to the hemangioblast during embryonic stem cell differentiation. Development 130, 4217–4227 10.1242/dev.00589 [DOI] [PubMed] [Google Scholar]
- 36.Heine U.I., Roberts A.B., Munoz E.F., Roche N.S. and Sporn M.B. (1985) Effects of retinoid deficiency on the development of the heart and vascular system of the quail embryo. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 50, 135–152 10.1007/BF02889897 [DOI] [PubMed] [Google Scholar]
- 37.Yutzey K.E., Rhee J.T. and Bader D. (1994) Expression of the atrial-specific myosin heavy chain AMHC1 and the establishment of anteroposterior polarity in the developing chicken heart. Development 120, 871–883 10.1242/dev.120.4.871 [DOI] [PubMed] [Google Scholar]
- 38.Brade T., Pane L.S., Moretti A., Chien K.R. and Laugwitz K.L. (2013) Embryonic heart progenitors and cardiogenesis. Cold Spring Harb. Perspect. Med. 3, a013847 10.1101/cshperspect.a013847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.David R., Brenner C., Stieber J., Schwarz F., Brunner S., Vollmer M.et al. (2008) MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat. Cell Biol. 10, 338–345 10.1038/ncb1696 [DOI] [PubMed] [Google Scholar]
- 40.Lindsley R.C., Gill J.G., Murphy T.L., Langer E.M., Cai M., Mashayekhi M.et al. (2008) Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 3, 55–68 10.1016/j.stem.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bondue A., Lapouge G., Paulissen C., Semeraro C., Iacovino M., Kyba M.et al. (2008) Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 3, 69–84 10.1016/j.stem.2008.06.009 [DOI] [PubMed] [Google Scholar]
- 42.Wu S.M., Chien K.R. and Mummery C. (2008) Origins and fates of cardiovascular progenitor cells. Cell 132, 537–543 10.1016/j.cell.2008.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lints T.J., Parsons L.M., Hartley L., Lyons I. and Harvey R.P. (1993) Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119, 419–431 10.1242/dev.119.2.419 [DOI] [PubMed] [Google Scholar]
- 44.Bruneau B.G., Logan M., Davis N., Levi T., Tabin C.J., Seidman J.G.et al. (1999) Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev. Biol. 211, 100–108 10.1006/dbio.1999.9298 [DOI] [PubMed] [Google Scholar]
- 45.Horb M.E. and Thomsen G.H. (1999) Tbx5 is essential for heart development. Development 126, 1739–1751 10.1242/dev.126.8.1739 [DOI] [PubMed] [Google Scholar]
- 46.Chen L., Fulcoli F.G., Tang S. and Baldini A. (2009) Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circ. Res. 105, 842–851 10.1161/CIRCRESAHA.109.200295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saga Y., Kitajima S. and Miyagawa-Tomita S. (2000) Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc. Med. 10, 345–352 10.1016/S1050-1738(01)00069-X [DOI] [PubMed] [Google Scholar]
- 48.Moretti A., Caron L., Nakano A., Lam J.T., Bernshausen A., Chen Y.et al. (2006) Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127, 1151–1165 10.1016/j.cell.2006.10.029 [DOI] [PubMed] [Google Scholar]
- 49.Sun Y., Liang X., Najafi N., Cass M., Lin L., Cai C.L.et al. (2007) Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev. Biol. 304, 286–296 10.1016/j.ydbio.2006.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuo C.T., Morrisey E.E., Anandappa R., Sigrist K., Lu M.M., Parmacek M.S.et al. (1997) GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 11, 1048–1060 10.1101/gad.11.8.1048 [DOI] [PubMed] [Google Scholar]
- 51.Svensson E.C., Tufts R.L., Polk C.E. and Leiden J.M. (1999) Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc. Natl. Acad. Sci. U.S.A. 96, 956–961 10.1073/pnas.96.3.956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takeuchi J.K. and Bruneau B.G. (2009) Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459, 708–711 10.1038/nature08039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benson D.W., Silberbach G.M., Kavanaugh-McHugh A., Cottrill C., Zhang Y., Riggs S.et al. (1999) Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J. Clin. Invest. 104, 1567–1573 10.1172/JCI8154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schultheiss T.M., Burch J.B. and Lassar A.B. (1997) A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 11, 451–462 10.1101/gad.11.4.451 [DOI] [PubMed] [Google Scholar]
- 55.Simon M.C. (1995) Gotta have GATA. Nat. Genet. 11, 9–11 10.1038/ng0995-9 [DOI] [PubMed] [Google Scholar]
- 56.Zhang H. and Bradley A. (1996) Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development 122, 2977–2986 10.1242/dev.122.10.2977 [DOI] [PubMed] [Google Scholar]
- 57.Lin Q., Schwarz J., Bucana C. and Olson E.N. (1997) Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276, 1404–1407 10.1126/science.276.5317.1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takeuchi J.K., Mileikovskaia M., Koshiba-Takeuchi K., Heidt A.B., Mori A.D., Arruda E.P.et al. (2005) Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development 132, 2463–2474 10.1242/dev.01827 [DOI] [PubMed] [Google Scholar]
- 59.Verzi M.P., McCulley D.J., De Val S., Dodou E. and Black B.L. (2005) The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev. Biol. 287, 134–145 10.1016/j.ydbio.2005.08.041 [DOI] [PubMed] [Google Scholar]
- 60.Motoike T., Markham D.W., Rossant J. and Sato T.N. (2003) Evidence for novel fate of Flk1+ progenitor: contribution to muscle lineage. Genesis 35, 153–159 10.1002/gene.10175 [DOI] [PubMed] [Google Scholar]
- 61.Misfeldt A.M., Boyle S.C., Tompkins K.L., Bautch V.L., Labosky P.A. and Baldwin H.S. (2009) Endocardial cells are a distinct endothelial lineage derived from Flk1+ multipotent cardiovascular progenitors. Dev. Biol. 333, 78–89 10.1016/j.ydbio.2009.06.033 [DOI] [PubMed] [Google Scholar]
- 62.Cai C.L., Liang X., Shi Y., Chu P.H., Pfaff S.L., Chen J.et al. (2003) Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell. 5, 877–889 10.1016/S1534-5807(03)00363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stanley E.G., Biben C., Elefanty A., Barnett L., Koentgen F., Robb L.et al. (2002) Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3'UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int. J. Dev. Biol. 46, 431–439 [PubMed] [Google Scholar]
- 64.Watanabe Y., Miyagawa-Tomita S., Vincent S.D., Kelly R.G., Moon A.M. and Buckingham M.E. (2010) Role of mesodermal FGF8 and FGF10 overlaps in the development of the arterial pole of the heart and pharyngeal arch arteries. Circ. Res. 106, 495–503 10.1161/CIRCRESAHA.109.201665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keren-Politansky A., Keren A. and Bengal E. (2009) Neural ectoderm-secreted FGF initiates the expression of Nkx2.5 in cardiac progenitors via a p38 MAPK/CREB pathway. Dev. Biol. 335, 374–384 10.1016/j.ydbio.2009.09.012 [DOI] [PubMed] [Google Scholar]
- 66.Dyer L.A. and Kirby M.L. (2009) The role of secondary heart field in cardiac development. Dev. Biol. 336, 137–144 10.1016/j.ydbio.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bergmann O., Bhardwaj R.D., Bernard S., Zdunek S., Barnabé-Heider F., Walsh S.et al. (2009) Evidence for cardiomyocyte renewal in humans. Science 324, 98–102 10.1126/science.1164680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bergmann O., Zdunek S., Frisén J., Bernard S., Druid H. and Jovinge S. (2012) Cardiomyocyte renewal in humans. Circ. Res. 110, e17–e18, author reply e19-21 10.1161/CIRCRESAHA.111.259598 [DOI] [PubMed] [Google Scholar]
- 69.Senyo S.E., Steinhauser M.L., Pizzimenti C.L., Yang V.K., Cai L., Wang M.et al. (2013) Mammalian heart renewal by pre-existing cardiomyocytes. Nature 493, 433–436 10.1038/nature11682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ellison G.M., Vicinanza C., Smith A.J., Aquila I., Leone A., Waring C.D.et al. (2013) Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell 154, 827–842 10.1016/j.cell.2013.07.039 [DOI] [PubMed] [Google Scholar]
- 71.Vicinanza C., Aquila I., Scalise M., Cristiano F., Marino F., Cianflone E.et al. (2017) Adult cardiac stem cells are multipotent and robustly myogenic: c-kit expression is necessary but not sufficient for their identification. Cell Death Differ. 24, 2101–2116 10.1038/cdd.2017.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vicinanza C., Aquila I., Cianflone E., Scalise M., Marino F., Mancuso T.et al. (2018) Kit(cre) knock-in mice fail to fate-map cardiac stem cells. Nature 555, E1–E5 10.1038/nature25771 [DOI] [PubMed] [Google Scholar]
- 73.Cianflone E., Cappetta D., Mancuso T., Sabatino J., Marino F., Scalise M.et al. (2020) Statins stimulate new myocyte formation after myocardial infarction by activating growth and differentiation of the endogenous cardiac stem cells. Int. J. Mol. Sci. 21, 7927 10.3390/ijms21217927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scalise M., Marino F., Cianflone E., Mancuso T., Marotta P., Aquila I.et al. (2019) Heterogeneity of adult cardiac stem cells. Adv. Exp. Med. Biol. 1169, 141–178 10.1007/978-3-030-24108-7_8 [DOI] [PubMed] [Google Scholar]
- 75.Flaim C.J., Teng D., Chien S. and Bhatia S.N. (2008) Combinatorial signaling microenvironments for studying stem cell fate. Stem Cells Dev. 17, 29–39 10.1089/scd.2007.0085 [DOI] [PubMed] [Google Scholar]
- 76.Laflamme M.A., Chen K.Y., Naumova A.V., Muskheli V., Fugate J.A., Dupras S.K.et al. (2007) Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 25, 1015–1024 10.1038/nbt1327 [DOI] [PubMed] [Google Scholar]
- 77.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K.et al. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 78.Scalise M., Marino F., Salerno L., Mancuso T., Cappetta D., Barone A.et al. (2021) In vitro CSC-derived cardiomyocytes exhibit the typical microRNA-mRNA blueprint of endogenous cardiomyocytes. Commun. Biol. 4, 1146 10.1038/s42003-021-02677-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parisi S., D'Andrea D., Lago C.T., Adamson E.D., Persico M.G. and Minchiotti G. (2003) Nodal-dependent Cripto signaling promotes cardiomyogenesis and redirects the neural fate of embryonic stem cells. J. Cell Biol. 163, 303–314 10.1083/jcb.200303010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vallier L., Alexander M. and Pedersen R.A. (2005) Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 118, 4495–4509 10.1242/jcs.02553 [DOI] [PubMed] [Google Scholar]
- 81.Evseenko D., Zhu Y., Schenke-Layland K., Kuo J., Latour B., Ge S.et al. (2010) Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 107, 13742–13747 10.1073/pnas.1002077107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.MacDonald B.T., Tamai K. and He X. (2009) Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 17, 9–26 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Noseda M., Peterkin T., Simões F.C., Patient R. and Schneider M.D. (2011) Cardiopoietic factors: extracellular signals for cardiac lineage commitment. Circ. Res. 108, 129–152 10.1161/CIRCRESAHA.110.223792 [DOI] [PubMed] [Google Scholar]
- 84.Qyang Y., Martin-Puig S., Chiravuri M., Chen S., Xu H., Bu L.et al. (2007) The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 1, 165–179 10.1016/j.stem.2007.05.018 [DOI] [PubMed] [Google Scholar]
- 85.Nakamura T., Sano M., Songyang Z. and Schneider M.D. (2003) A Wnt- and beta -catenin-dependent pathway for mammalian cardiac myogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 5834–5839 10.1073/pnas.0935626100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Deb A., Davis B.H., Guo J., Ni A., Huang J., Zhang Z.et al. (2008) SFRP2 regulates cardiomyogenic differentiation by inhibiting a positive transcriptional autofeedback loop of Wnt3a. Stem Cells 26, 35–44 10.1634/stemcells.2007-0475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tomescot A., Leschik J., Bellamy V., Dubois G., Messas E., Bruneval P.et al. (2007) Differentiation in vivo of cardiac committed human embryonic stem cells in postmyocardial infarcted rats. Stem Cells 25, 2200–2205 10.1634/stemcells.2007-0133 [DOI] [PubMed] [Google Scholar]
- 88.Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S.et al. (1998) Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- 89.Mummery C.L., Zhang J., Ng E.S., Elliott D.A., Elefanty A.G. and Kamp T.J. (2012) Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circ. Res. 111, 344–358 10.1161/CIRCRESAHA.110.227512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Doetschman T.C., Eistetter H., Katz M., Schmidt W. and Kemler R. (1985) The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J. Embryol. Exp. Morphol. 87, 27–45 10.1242/dev.87.1.27 [DOI] [PubMed] [Google Scholar]
- 91.Mummery C., Ward-van Oostwaard D., Doevendans P., Spijker R., van den Brink S., Hassink R.et al. (2003) Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation 107, 2733–2740 10.1161/01.CIR.0000068356.38592.68 [DOI] [PubMed] [Google Scholar]
- 92.Mummery C.L., Ward D. and Passier R. (2007) Differentiation of human embryonic stem cells to cardiomyocytes by coculture with endoderm in serum-free medium. Curr. Protoc. Stem Cell Biol. Chapter 1, Unit 1F.2 10.1002/9780470151808.sc01f02s2 [DOI] [PubMed] [Google Scholar]
- 93.Calder E.L., Tchieu J., Steinbeck J.A., Tu E., Keros S., Ying S.W.et al. (2015) Retinoic acid-mediated regulation of gli3 enables efficient motoneuron derivation from human ESCs in the absence of extrinsic SHH activation. J. Neurosci. 35, 11462–11481 10.1523/JNEUROSCI.3046-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hori Y., Rulifson I.C., Tsai B.C., Heit J.J., Cahoy J.D. and Kim S.K. (2002) Growth inhibitors promote differentiation of insulin-producing tissue from embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 99, 16105–16110 10.1073/pnas.252618999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takahashi K. and Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 96.Wu X., Ding S., Ding Q., Gray N.S. and Schultz P.G. (2004) Small molecules that induce cardiomyogenesis in embryonic stem cells. J. Am. Chem. Soc. 126, 1590–1591 10.1021/ja038950i [DOI] [PubMed] [Google Scholar]
- 97.Yang L., Soonpaa M.H., Adler E.D., Roepke T.K., Kattman S.J., Kennedy M.et al. (2008) Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 453, 524–528 10.1038/nature06894 [DOI] [PubMed] [Google Scholar]
- 98.Johansson B.M. and Wiles M.V. (1995) Evidence for involvement of activin A and bone morphogenetic protein 4 in mammalian mesoderm and hematopoietic development. Mol. Cell. Biol. 15, 141–151 10.1128/MCB.15.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kattman S.J., Witty A.D., Gagliardi M., Dubois N.C., Niapour M., Hotta A.et al. (2011) Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 8, 228–240 10.1016/j.stem.2010.12.008 [DOI] [PubMed] [Google Scholar]
- 100.Willems E., Cabral-Teixeira J., Schade D., Cai W., Reeves P., Bushway P.J.et al. (2012) Small molecule-mediated TGF-β type II receptor degradation promotes cardiomyogenesis in embryonic stem cells. Cell Stem Cell. 11, 242–252 10.1016/j.stem.2012.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lo B. and Parham L. (2009) Ethical issues in stem cell research. Endocr. Rev. 30, 204–213 10.1210/er.2008-0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiang Y., Park P., Hong S.M. and Ban K. (2018) Maturation of cardiomyocytes derived from human pluripotent stem cells: current strategies and limitations. Mol. Cells 41, 613–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scuderi G.J. and Butcher J. (2017) Naturally engineered maturation of cardiomyocytes. Front. Cell Dev. Biol. 5, 50 10.3389/fcell.2017.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang J., Klos M., Wilson G.F., Herman A.M., Lian X., Raval K.K.et al. (2012) Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ. Res. 111, 1125–1136 10.1161/CIRCRESAHA.112.273144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oikonomopoulos A., Kitani T. and Wu J.C. (2018) Pluripotent stem cell-derived cardiomyocytes as a platform for cell therapy applications: progress and hurdles for clinical translation. Mol. Ther. 26, 1624–1634 10.1016/j.ymthe.2018.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dubois N.C., Craft A.M., Sharma P., Elliott D.A., Stanley E.G., Elefanty A.G.et al. (2011) SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat. Biotechnol. 29, 1011–1018 10.1038/nbt.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tohyama S., Hattori F., Sano M., Hishiki T., Nagahata Y., Matsuura T.et al. (2013) Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 12, 127–137 10.1016/j.stem.2012.09.013 [DOI] [PubMed] [Google Scholar]
- 108.Hattori F., Chen H., Yamashita H., Tohyama S., Satoh Y.S., Yuasa S.et al. (2010) Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat. Methods 7, 61–66 10.1038/nmeth.1403 [DOI] [PubMed] [Google Scholar]
- 109.Soonpaa M.H., Koh G.Y., Klug M.G. and Field L.J. (1994) Formation of nascent intercalated disks between grafted fetal cardiomyocytes and host myocardium. Science 264, 98–101 10.1126/science.8140423 [DOI] [PubMed] [Google Scholar]
- 110.Li R.K., Mickle D.A., Weisel R.D., Zhang J. and Mohabeer M.K. (1996) In vivo survival and function of transplanted rat cardiomyocytes. Circ. Res. 78, 283–288 10.1161/01.RES.78.2.283 [DOI] [PubMed] [Google Scholar]
- 111.Caspi O., Huber I., Kehat I., Habib M., Arbel G., Gepstein A.et al. (2007) Transplantation of human embryonic stem cell-derived cardiomyocytes improves myocardial performance in infarcted rat hearts. J. Am. Coll. Cardiol. 50, 1884–1893 10.1016/j.jacc.2007.07.054 [DOI] [PubMed] [Google Scholar]
- 112.van Laake L.W., Passier R., Monshouwer-Kloots J., Verkleij A.J., Lips D.J.et al. (2007) Human embryonic stem cell-derived cardiomyocytes survive and mature in the mouse heart and transiently improve function after myocardial infarction. Stem Cell Res. 1, 9–24 10.1016/j.scr.2007.06.001 [DOI] [PubMed] [Google Scholar]
- 113.Chong J.J., Yang X., Don C.W., Minami E., Liu Y.W., Weyers J.J.et al. (2014) Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 510, 273–277 10.1038/nature13233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu Y.W., Chen B., Yang X., Fugate J.A., Kalucki F.A., Futakuchi-Tsuchida A.et al. (2018) Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat. Biotechnol. 36, 597–605 10.1038/nbt.4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shiba Y., Gomibuchi T., Seto T., Wada Y., Ichimura H., Tanaka Y.et al. (2016) Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 538, 388–391 10.1038/nature19815 [DOI] [PubMed] [Google Scholar]
- 116.Chakradhar S. (2016) An eye to the future: researchers debate best path for stem cell-derived therapies. Nat. Med. 22, 116–119 10.1038/nm0216-116 [DOI] [PubMed] [Google Scholar]
- 117.Deuse T., Hu X., Gravina A., Wang D., Tediashvili G., De C.et al. (2019) Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat. Biotechnol. 37, 252–258 10.1038/s41587-019-0016-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gornalusse G.G., Hirata R.K., Funk S.E., Riolobos L., Lopes V.S., Manske G.et al. (2017) HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat. Biotechnol. 35, 765–772 10.1038/nbt.3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Riolobos L., Hirata R.K., Turtle C.J., Wang P.R., Gornalusse G.G., Zavajlevski M.et al. (2013) HLA engineering of human pluripotent stem cells. Mol. Ther. 21, 1232–1241 10.1038/mt.2013.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bu L., Jiang X., Martin-Puig S., Caron L., Zhu S., Shao Y.et al. (2009) Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature 460, 113–117 10.1038/nature08191 [DOI] [PubMed] [Google Scholar]
- 121.Chong J.J., Chandrakanthan V., Xaymardan M., Asli N.S., Li J., Ahmed I.et al. (2011) Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell. 9, 527–540 10.1016/j.stem.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Smith R.R., Barile L., Cho H.C., Leppo M.K., Hare J.M., Messina E.et al. (2007) Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation 115, 896–908 10.1161/CIRCULATIONAHA.106.655209 [DOI] [PubMed] [Google Scholar]
- 123.Aquila I., Cianflone E., Scalise M., Marino F., Mancuso T., Filardo A.et al. (2019) c-kit Haploinsufficiency impairs adult cardiac stem cell growth, myogenicity and myocardial regeneration. Cell Death Dis. 10, 436 10.1038/s41419-019-1655-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Beltrami A.P., Barlucchi L., Torella D., Baker M., Limana F., Chimenti S.et al. (2003) Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 114, 763–776 10.1016/S0092-8674(03)00687-1 [DOI] [PubMed] [Google Scholar]
- 125.Torella D., Ellison G.M., Karakikes I. and Nadal-Ginard B. (2007) Growth-factor-mediated cardiac stem cell activation in myocardial regeneration. Nat. Clin. Pract. Cardiovasc. Med. 4, S46–S51 10.1038/ncpcardio0772 [DOI] [PubMed] [Google Scholar]
- 126.Mii Y. and Taira M. (2009) Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Development 136, 4083–4088 10.1242/dev.032524 [DOI] [PubMed] [Google Scholar]
- 127.Cianflone E., Aquila I., Scalise M., Marotta P., Torella M., Nadal-Ginard B.et al. (2018) Molecular basis of functional myogenic specification of Bona Fide multipotent adult cardiac stem cells. Cell Cycle 17, 927–946 10.1080/15384101.2018.1464852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Paoletti C., Divieto C., Tarricone G., Di Meglio F., Nurzynska D. and Chiono V. (2020) MicroRNA-mediated direct reprogramming of human adult fibroblasts toward cardiac phenotype. Front. Bioeng. Biotechnol. 8, 529 10.3389/fbioe.2020.00529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T.et al. (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19, 489–501 10.1101/gad.1248505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rosa A. and Brivanlou A.H. (2009) MicroRNAs in early vertebrate development. Cell Cycle 8, 3513–3520 10.4161/cc.8.21.9847 [DOI] [PubMed] [Google Scholar]
- 131.Bartel D.P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Berezikov E., Cuppen E. and Plasterk R.H. (2006) Approaches to microRNA discovery. Nat. Genet. 38, S2–S7 10.1038/ng1794 [DOI] [PubMed] [Google Scholar]
- 133.Eliasson L. and Esguerra J.L. (2014) Role of non-coding RNAs in pancreatic beta-cell development and physiology. Acta. Physiol. (Oxf.) 211, 273–284 10.1111/apha.12285 [DOI] [PubMed] [Google Scholar]
- 134.Sayed D. and Abdellatif M. (2011) MicroRNAs in development and disease. Physiol. Rev. 91, 827–887 10.1152/physrev.00006.2010 [DOI] [PubMed] [Google Scholar]
- 135.Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z.et al. (2003) Dicer is essential for mouse development. Nat. Genet. 35, 215–217 10.1038/ng1253 [DOI] [PubMed] [Google Scholar]
- 136.Wang J., Chen J. and Sen S. (2016) MicroRNA as biomarkers and diagnostics. J. Cell. Physiol. 231, 25–30 10.1002/jcp.25056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fiedler J., Baker A.H., Dimmeler S., Heymans S., Mayr M. and Thum T. (2018) Non-coding RNAs in vascular disease - from basic science to clinical applications: scientific update from the working group of myocardial function of the european society of cardiology. Cardiovasc. Res. 114, 1281–1286 10.1093/cvr/cvy121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mourelatos Z., Dostie J., Paushkin S., Sharma A., Charroux B., Abel L.et al. (2002) miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16, 720–728 10.1101/gad.974702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stefani G. and Slack F.J. (2008) Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9, 219–230 10.1038/nrm2347 [DOI] [PubMed] [Google Scholar]
- 140.Braga L., Ali H., Secco I. and Giacca M. (2021) Non-coding RNA therapeutics for cardiac regeneration. Cardiovasc. Res. 117, 674–693 10.1093/cvr/cvaa071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hu Y., Jin G., Li B., Chen Y., Zhong L., Chen G.et al. (2019) Suppression of miRNA let-7i-5p promotes cardiomyocyte proliferation and repairs heart function post injury by targetting CCND2 and E2F2. Clin. Sci. (Lond.) 133, 425–441 10.1042/CS20181002 [DOI] [PubMed] [Google Scholar]
- 142.Zhao Y., Ransom J.F., Li A., Vedantham V., von Drehle M., Muth A.N.et al. (2007) Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129, 303–317 10.1016/j.cell.2007.03.030 [DOI] [PubMed] [Google Scholar]
- 143.Srivastava D. (2006) Making or breaking the heart: from lineage determination to morphogenesis. Cell 126, 1037–1048 10.1016/j.cell.2006.09.003 [DOI] [PubMed] [Google Scholar]
- 144.Yin V.P., Lepilina A., Smith A. and Poss K.D. (2012) Regulation of zebrafish heart regeneration by miR-133. Dev. Biol. 365, 319–327 10.1016/j.ydbio.2012.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu N., Bezprozvannaya S., Williams A.H., Qi X., Richardson J.A., Bassel-Duby R.et al. (2008) microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 22, 3242–3254 10.1101/gad.1738708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao Y., Samal E. and Srivastava D. (2005) Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436, 214–220 10.1038/nature03817 [DOI] [PubMed] [Google Scholar]
- 147.Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M.et al. (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 10.1038/ng1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ivey K.N., Muth A., Arnold J., King F.W., Yeh R.F., Fish J.E.et al. (2008) MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2, 219–229 10.1016/j.stem.2008.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Catalucci D., Latronico M.V., Ellingsen O. and Condorelli G. (2008) Physiological myocardial hypertrophy: how and why? Front. Biosci. 13, 312–324 10.2741/2681 [DOI] [PubMed] [Google Scholar]
- 150.Yang B., Lin H., Xiao J., Lu Y., Luo X., Li B.et al. (2007) The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 13, 486–491 10.1038/nm1569 [DOI] [PubMed] [Google Scholar]
- 151.Wystub K., Besser J., Bachmann A., Boettger T. and Braun T. (2013) miR-1/133a clusters cooperatively specify the cardiomyogenic lineage by adjustment of myocardin levels during embryonic heart development. PLoS Genet. 9, e1003793 10.1371/journal.pgen.1003793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hullinger T.G., Montgomery R.L., Seto A.G., Dickinson B.A., Semus H.M., Lynch J.M.et al. (2012) Inhibition of miR-15 protects against cardiac ischemic injury. Circ. Res. 110, 71–81 10.1161/CIRCRESAHA.111.244442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Porrello E.R., Mahmoud A.I., Simpson E., Johnson B.A., Grinsfelder D., Canseco D.et al. (2013) Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl. Acad. Sci. U.S.A. 110, 187–192 10.1073/pnas.1208863110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Porrello E.R., Johnson B.A., Aurora A.B., Simpson E., Nam Y.J., Matkovich S.J.et al. (2011) MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 109, 670–679 10.1161/CIRCRESAHA.111.248880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Crippa S., Nemir M., Ounzain S., Ibberson M., Berthonneche C., Sarre A.et al. (2016) Comparative transcriptome profiling of the injured zebrafish and mouse hearts identifies miRNA-dependent repair pathways. Cardiovasc. Res. 110, 73–84 10.1093/cvr/cvw031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cao X., Wang J., Wang Z., Du J., Yuan X., Huang W.et al. (2013) MicroRNA profiling during rat ventricular maturation: A role for miR-29a in regulating cardiomyocyte cell cycle re-entry. FEBS Lett. 587, 1548–1555 10.1016/j.febslet.2013.01.075 [DOI] [PubMed] [Google Scholar]
- 157.Yang Q., Wu F., Mi Y., Wang F., Cai K., Yang X.et al. (2020) Aberrant expression of miR-29b-3p influences heart development and cardiomyocyte proliferation by targeting NOTCH2. Cell Prolif. 53, e12764 10.1111/cpr.12764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yang Y., Cheng H.W., Qiu Y., Dupee D., Noonan M., Lin Y.D.et al. (2015) MicroRNA-34a Plays a Key Role in Cardiac Repair and Regeneration Following Myocardial Infarction. Circ. Res. 117, 450–459 10.1161/CIRCRESAHA.117.305962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Huang W., Feng Y., Liang J., Yu H., Wang C., Wang B.et al. (2018) Loss of microRNA-128 promotes cardiomyocyte proliferation and heart regeneration. Nat Commun. 9, 700 10.1038/s41467-018-03019-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rosa A., Papaioannou M.D., Krzyspiak J.E. and Brivanlou A.H. (2014) miR-373 is regulated by TGFβ signaling and promotes mesendoderm differentiation in human Embryonic Stem Cells. Dev. Biol. 391, 81–88 10.1016/j.ydbio.2014.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Barroso-delJesus A., Lucena-Aguilar G., Sanchez L., Ligero G., Gutierrez-Aranda I. and Menendez P. (2011) The Nodal inhibitor Lefty is negatively modulated by the microRNA miR-302 in human embryonic stem cells. FASEB J. 25, 1497–1508 10.1096/fj.10-172221 [DOI] [PubMed] [Google Scholar]
- 162.Ishikawa D., Diekmann U., Fiedler J., Just A., Thum T., Lenzen S.et al. (2017) miRNome profiling of purified endoderm and mesoderm differentiated from hESCs reveals functions of miR-483-3p and miR-1263 for cell-fate decisions. Stem Cell Rep. 9, 1588–1603 10.1016/j.stemcr.2017.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mendjan S., Mascetti V.L., Ortmann D., Ortiz M., Karjosukarso D.W., Ng Y.et al. (2014) NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell. 15, 310–325 10.1016/j.stem.2014.06.006 [DOI] [PubMed] [Google Scholar]
- 164.Lian X., Zhang J., Azarin S.M., Zhu K., Hazeltine L.B., Bao X.et al. (2013) Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 8, 162–175 10.1038/nprot.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Faial T., Bernardo A.S., Mendjan S., Diamanti E., Ortmann D., Gentsch G.E.et al. (2015) Brachyury and SMAD signalling collaboratively orchestrate distinct mesoderm and endoderm gene regulatory networks in differentiating human embryonic stem cells. Development 142, 2121–2135 10.1242/dev.117838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Islas J.F., Liu Y., Weng K.C., Robertson M.J., Zhang S., Prejusa A.et al. (2012) Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc. Natl. Acad. Sci. U.S A. 109, 13016–13021 10.1073/pnas.1120299109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chen J., Huang Z.P., Seok H.Y., Ding J., Kataoka M., Zhang Z.et al. (2013) mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ. Res. 112, 1557–1566 10.1161/CIRCRESAHA.112.300658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shen X., Soibam B., Benham A., Xu X., Chopra M., Peng X.et al. (2016) miR-322/-503 cluster is expressed in the earliest cardiac progenitor cells and drives cardiomyocyte specification. Proc. Natl. Acad. Sci. U.S.A. 113, 9551–9556 10.1073/pnas.1608256113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Morkin E. (2000) Control of cardiac myosin heavy chain gene expression. Microsc. Res. Tech. 50, 522–531 [DOI] [PubMed] [Google Scholar]
- 170.van Rooij E., Sutherland L.B., Qi X., Richardson J.A., Hill J. and Olson E.N. (2007) Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579 10.1126/science.1139089 [DOI] [PubMed] [Google Scholar]
- 171.Olson E.N. (2006) Gene regulatory networks in the evolution and development of the heart. Science 313, 1922–1927 10.1126/science.1132292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu N. and Olson E.N. (2010) MicroRNA regulatory networks in cardiovascular development. Dev. Cell. 18, 510–525 10.1016/j.devcel.2010.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Günthel M., Barnett P. and Christoffels V.M. (2018) Development, proliferation, and growth of the mammalian heart. Mol. Ther. 26, 1599–1609 10.1016/j.ymthe.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Eulalio A., Mano M., Dal Ferro M., Zentilin L., Sinagra G., Zacchigna S.et al. (2012) Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492, 376–381 10.1038/nature11739 [DOI] [PubMed] [Google Scholar]
- 175.Kim J., Krichevsky A., Grad Y., Hayes G.D., Kosik K.S., Church G.M.et al. (2004) Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc. Natl. Acad. Sci. U.S.A. 101, 360–365 10.1073/pnas.2333854100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ghosh G., Subramanian I.V., Adhikari N., Zhang X., Joshi H.P., Basi D.et al. (2010) Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-α isoforms and promotes angiogenesis. J. Clin. Invest. 120, 4141–4154 10.1172/JCI42980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sarkar S., Dey B.K. and Dutta A. (2010) MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol. Biol. Cell. 21, 2138–2149 10.1091/mbc.e10-01-0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Llobet-Navas D., Rodríguez-Barrueco R., Castro V., Ugalde A.P., Sumazin P., Jacob-Sendler D.et al. (2014) The miR-424(322)/503 cluster orchestrates remodeling of the epithelium in the involuting mammary gland. Genes Dev. 28, 765–782 10.1101/gad.237404.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yang X., Pabon L. and Murry C.E. (2014) Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 114, 511–523 10.1161/CIRCRESAHA.114.300558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ziman A.P., Gómez-Viquez N.L., Bloch R.J. and Lederer W.J. (2010) Excitation-contraction coupling changes during postnatal cardiac development. J. Mol. Cell Cardiol. 48, 379–386 10.1016/j.yjmcc.2009.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.van Vliet P., Roccio M., Smits A.M., van Oorschot A.A., Metz C.H.et al. (2008) Progenitor cells isolated from the human heart: a potential cell source for regenerative therapy. Neth. Heart J. 16, 163–169 10.1007/BF03086138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang Z., Gerstein M. and Snyder M. (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fu J.D., Rushing S.N., Lieu D.K., Chan C.W., Kong C.W., Geng L.et al. (2011) Distinct roles of microRNA-1 and -499 in ventricular specification and functional maturation of human embryonic stem cell-derived cardiomyocytes. PLoS ONE 6, e27417 10.1371/journal.pone.0027417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Alfar E.A., El-Armouche A. and Guan K. (2018) MicroRNAs in cardiomyocyte differentiation and maturation. Cardiovasc. Res. 114, 779–781 10.1093/cvr/cvy065 [DOI] [PubMed] [Google Scholar]
- 185.Kuppusamy K.T., Jones D.C., Sperber H., Madan A., Fischer K.A., Rodriguez M.L.et al. (2015) Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc. Natl. Acad. Sci. U.S.A. 112, E2785–E2794 10.1073/pnas.1424042112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lee D.S., Chen J.H., Lundy D.J., Liu C.H., Hwang S.M., Pabon L.et al. (2015) Defined microRNAs induce aspects of maturation in mouse and human embryonic-stem-cell-derived cardiomyocytes. Cell Rep. 12, 1960–1967 10.1016/j.celrep.2015.08.042 [DOI] [PubMed] [Google Scholar]
- 187.Poon E.N., Hao B., Guan D., Jun Li M., Lu J., Yang Y.et al. (2018) Integrated transcriptomic and regulatory network analyses identify microRNA-200c as a novel repressor of human pluripotent stem cell-derived cardiomyocyte differentiation and maturation. Cardiovasc. Res. 114, 894–906 10.1093/cvr/cvy019 [DOI] [PubMed] [Google Scholar]
- 188.Kimbrel E.A. and Lanza R. (2015) Current status of pluripotent stem cells: moving the first therapies to the clinic. Nat. Rev. Drug Discov. 14, 681–692 10.1038/nrd4738 [DOI] [PubMed] [Google Scholar]
- 189.Kumar N., Dougherty J.A., Manring H.R., Elmadbouh I., Mergaye M., Czirok A.et al. (2019) Assessment of temporal functional changes and miRNA profiling of human iPSC-derived cardiomyocytes. Sci. Rep. 9, 13188 10.1038/s41598-019-49653-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Xu N., Papagiannakopoulos T., Pan G., Thomson J.A. and Kosik K.S. (2009) MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137, 647–658 10.1016/j.cell.2009.02.038 [DOI] [PubMed] [Google Scholar]
- 191.Jayawardena T.M., Egemnazarov B., Finch E.A., Zhang L., Payne J.A., Pandya K.et al. (2012) MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 110, 1465–1473 10.1161/CIRCRESAHA.112.269035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lalevée S., Lapaire O. and Bühler M. (2014) miR455 is linked to hypoxia signaling and is deregulated in preeclampsia. Cell Death Dis. 5, e1408 10.1038/cddis.2014.368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wu C., So J., Davis-Dusenbery B.N., Qi H.H., Bloch D.B., Shi Y.et al. (2011) Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol. Cell. Biol. 31, 4760–4774 10.1128/MCB.05776-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chin M.H., Mason M.J., Xie W., Volinia S., Singer M., Peterson C.et al. (2009) Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 5, 111–123 10.1016/j.stem.2009.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Narsinh K.H., Plews J. and Wu J.C. (2011) Comparison of human induced pluripotent and embryonic stem cells: fraternal or identical twins? Mol. Ther. 19, 635–638 10.1038/mt.2011.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lin S.L., Chang D.C., Chang-Lin S., Lin C.H., Wu D.T., Chen D.T.et al. (2008) Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 14, 2115–2124 10.1261/rna.1162708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Diez-Cuñado M., Wei K., Bushway P.J., Maurya M.R., Perera R., Subramaniam S.et al. (2018) miRNAs that induce human cardiomyocyte proliferation converge on the Hippo pathway. Cell Rep. 23, 2168–2174 10.1016/j.celrep.2018.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Miyoshi N., Ishii H., Nagano H., Haraguchi N., Dewi D.L., Kano Y.et al. (2011) Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 8, 633–638 10.1016/j.stem.2011.05.001 [DOI] [PubMed] [Google Scholar]
- 199.Samavarchi-Tehrani P., Golipour A., David L., Sung H.K., Beyer T.A., Datti A.et al. (2010) Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 7, 64–77 10.1016/j.stem.2010.04.015 [DOI] [PubMed] [Google Scholar]
- 200.Liao B., Bao X., Liu L., Feng S., Zovoilis A., Liu W.et al. (2011) MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J. Biol. Chem. 286, 17359–17364 10.1074/jbc.C111.235960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pfaff N., Fiedler J., Holzmann A., Schambach A., Moritz T., Cantz T.et al. (2011) miRNA screening reveals a new miRNA family stimulating iPS cell generation via regulation of Meox2. EMBO Rep. 12, 1153–1159 10.1038/embor.2011.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Li Z., Yang C.S., Nakashima K. and Rana T.M. (2011) Small RNA-mediated regulation of iPS cell generation. EMBO J. 30, 823–834 10.1038/emboj.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Choi Y.J., Lin C.P., Ho J.J., He X., Okada N., Bu P.et al. (2011) miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat. Cell Biol. 13, 1353–1360 10.1038/ncb2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ventura A., Young A.G., Winslow M.M., Lintault L., Meissner A., Erkeland S.J.et al. (2008) Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875–886 10.1016/j.cell.2008.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cianflone E., Torella M., Biamonte F., De Angelis A., Urbanek K., Costanzo F.S.et al. (2020) Targeting cardiac stem cell senescence to treat cardiac aging and disease. Cells 9, 10.3390/cells9061558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Marino F., Scalise M., Cianflone E., Salerno L., Cappetta D., Salerno N.et al. (2021) Physical exercise and cardiac repair: the potential role of nitric oxide in boosting stem cell regenerative biology. Antioxidants (Basel) 10, 1002 10.3390/antiox10071002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Marino F., Scalise M., Salerno N., Salerno L., Molinaro C., Cappetta D.et al. (2022) Diabetes-induced cellular senescence and senescence-associated secretory phenotype impair cardiac regeneration and function independently of age. Diabetes 10.2337/db21-0536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Scalise M., Marino F., Salerno L., Cianflone E., Molinaro C., Salerno N.et al. (2021) From spheroids to organoids: the next generation of model systems of human cardiac regeneration in a dish. Int. J. Mol. Sci. 22, 13180 10.3390/ijms222413180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Nadal-Ginard B., Cianflone E. and Torella D. (2021) The baby and the bath water: adult cardiac stem cells revisited. Eur. Heart J. 42, 3814–3816 10.1093/eurheartj/ehab335 [DOI] [PubMed] [Google Scholar]
- 210.Di Siena S., Gimmelli R., Nori S.L., Barbagallo F., Campolo F., Dolci S.et al. (2016) Activated c-Kit receptor in the heart promotes cardiac repair and regeneration after injury. Cell Death Dis. 7, e2317 10.1038/cddis.2016.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sluijter J.P., van Mil A., van Vliet P., Metz C.H., Liu J., Doevendans P.A.et al. (2010) MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler. Thromb. Vasc. Biol. 30, 859–868 10.1161/ATVBAHA.109.197434 [DOI] [PubMed] [Google Scholar]
- 212.Bishop S.P., Zhou Y., Nakada Y. and Zhang J. (2021) Changes in cardiomyocyte cell cycle and hypertrophic growth during fetal to adult in mammals. J. Am. Heart Assoc. 10, e017839 10.1161/JAHA.120.017839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.MacLellan W.R., Garcia A., Oh H., Frenkel P., Jordan M.C., Roos K.P.et al. (2005) Overlapping roles of pocket proteins in the myocardium are unmasked by germ line deletion of p130 plus heart-specific deletion of Rb. Mol. Cell. Biol. 25, 2486–2497 10.1128/MCB.25.6.2486-2497.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Doppler S.A., Deutsch M.A., Serpooshan V., Li G., Dzilic E., Lange R.et al. (2017) Mammalian heart regeneration: the race to the finish line. Circ. Res. 120, 630–632 10.1161/CIRCRESAHA.116.310051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lázár E., Sadek H.A. and Bergmann O. (2017) Cardiomyocyte renewal in the human heart: insights from the fall-out. Eur. Heart J. 38, 2333–2342 10.1093/eurheartj/ehx343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Naqvi N., Li M., Calvert J.W., Tejada T., Lambert J.P., Wu J.et al. (2014) A proliferative burst during preadolescence establishes the final cardiomyocyte number. Cell 157, 795–807 10.1016/j.cell.2014.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Yuan T. and Krishnan J. (2021) Non-coding RNAs in cardiac regeneration. Front. Physiol. 12, 650566 10.3389/fphys.2021.650566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Strungs E.G., Ongstad E.L., O'Quinn M.P., Palatinus J.A., Jourdan L.J. and Gourdie R.G. (2013) Cryoinjury models of the adult and neonatal mouse heart for studies of scarring and regeneration. Methods Mol. Biol. 1037, 343–353 10.1007/978-1-62703-505-7_20 [DOI] [PubMed] [Google Scholar]
- 219.Haubner B.J., Schneider J., Schweigmann U., Schuetz T., Dichtl W., Velik-Salchner C.et al. (2016) Functional recovery of a human neonatal heart after severe myocardial infarction. Circ. Res. 118, 216–221 10.1161/CIRCRESAHA.115.307017 [DOI] [PubMed] [Google Scholar]
- 220.Mahmoud A.I., Kocabas F., Muralidhar S.A., Kimura W., Koura A.S., Thet S.et al. (2013) Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature 497, 249–253 10.1038/nature12054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Porrello E.R., Mahmoud A.I., Simpson E., Hill J.A., Richardson J.A., Olson E.N.et al. (2011) Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lopaschuk G.D. and Jaswal J.S. (2010) Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J. Cardiovasc. Pharmacol. 56, 130–140 10.1097/FJC.0b013e3181e74a14 [DOI] [PubMed] [Google Scholar]
- 223.Piquereau J. and Ventura-Clapier R. (2018) Maturation of cardiac energy metabolism during perinatal development. Front. Physiol. 9, 959 10.3389/fphys.2018.00959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Brodsky M., Doria R., Allen B., Sato D., Thomas G. and Sada M. (1992) New-onset ventricular tachycardia during pregnancy. Am. Heart J. 123, 933–941 10.1016/0002-8703(92)90699-V [DOI] [PubMed] [Google Scholar]
- 225.Olivetti G., Cigola E., Maestri R., Corradi D., Lagrasta C., Gambert S.R.et al. (1996) Aging, cardiac hypertrophy and ischemic cardiomyopathy do not affect the proportion of mononucleated and multinucleated myocytes in the human heart. J. Mol. Cell Cardiol. 28, 1463–1477 10.1006/jmcc.1996.0137 [DOI] [PubMed] [Google Scholar]
- 226.Schmid G. and Pfitzer P. (1985) Mitoses and binucleated cells in perinatal human hearts. Virchows Arch. B Cell Pathol. Incl. Mol. Pathol. 48, 59–67 10.1007/BF02890115 [DOI] [PubMed] [Google Scholar]
- 227.Bergmann O., Zdunek S., Felker A., Salehpour M., Alkass K., Bernard S.et al. (2015) Dynamics of cell generation and turnover in the human heart. Cell 161, 1566–1575 10.1016/j.cell.2015.05.026 [DOI] [PubMed] [Google Scholar]
- 228.Ooi J.Y., Bernardo B.C. and McMullen J.R. (2014) The therapeutic potential of miRNAs regulated in settings of physiological cardiac hypertrophy. Fut. Med. Chem. 6, 205–222 10.4155/fmc.13.196 [DOI] [PubMed] [Google Scholar]
- 229.Tian Y., Liu Y., Wang T., Zhou N., Kong J., Chen L.et al. (2015) A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci. Transl. Med. 7, 279ra238 10.1126/scitranslmed.3010841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Liu X., Xiao J., Zhu H., Wei X., Platt C., Damilano F.et al. (2015) miR-222 is necessary for exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell Metab. 21, 584–595 10.1016/j.cmet.2015.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gabisonia K., Prosdocimo G., Aquaro G.D., Carlucci L., Zentilin L., Secco I.et al. (2019) MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature 569, 418–422 10.1038/s41586-019-1191-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Glass C. and Singla D.K. (2011) MicroRNA-1 transfected embryonic stem cells enhance cardiac myocyte differentiation and inhibit apoptosis by modulating the PTEN/Akt pathway in the infarcted heart. Am. J. Physiol. Heart Circ. Physiol. 301, H2038–H2049 10.1152/ajpheart.00271.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wilson K.D., Hu S., Venkatasubrahmanyam S., Fu J.D., Sun N., Abilez O.J.et al. (2010) Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: role for miR-499. Circ. Cardiovasc. Genet. 3, 426–435 10.1161/CIRCGENETICS.109.934281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Dong S., Cheng Y., Yang J., Li J., Liu X., Wang X.et al. (2009) MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J. Biol. Chem. 284, 29514–29525 10.1074/jbc.M109.027896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Qian L., Van Laake L.W., Huang Y., Liu S., Wendland M.F.et al. (2011) miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J. Exp. Med. 208, 549–560 10.1084/jem.20101547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Hu S., Huang M., Nguyen P.K., Gong Y., Li Z., Jia F.et al. (2011) Novel microRNA prosurvival cocktail for improving engraftment and function of cardiac progenitor cell transplantation. Circulation 124, S27–S34 10.1161/CIRCULATIONAHA.111.017954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yang C.S., Li Z. and Rana T.M. (2011) microRNAs modulate iPS cell generation. RNA 17, 1451–1460 10.1261/rna.2664111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yuan K., Ai W.B., Wan L.Y., Tan X. and Wu J.F. (2017) The miR-290-295 cluster as multi-faceted players in mouse embryonic stem cells. Cell Biosci. 7, 38 10.1186/s13578-017-0166-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to the paper