Abstract
Increased antimicrobial resistance (AMR) has been reported for pathogenic and commensal Escherichia coli (E. coli), hampering the treatment, and increasing the burden of infectious diarrhoeal diseases in children in developing countries. This study focused on exploring the occurrence, patterns, and possible drivers of AMR E. coli isolated from children under-five years in Zambia. A hospital-based cross-sectional study was conducted in the Lusaka and Ndola districts. Rectal swabs were collected from 565 and 455 diarrhoeic and healthy children, respectively, from which 1020 E. coli were cultured and subjected to antibiotic susceptibility testing. Nearly all E. coli (96.9%) were resistant to at least one antimicrobial agent tested. Further, 700 isolates were Multi-Drug Resistant, 136 were possibly Extensively-Drug Resistant and nine were Pan-Drug-Resistant. Forty percent of the isolates were imipenem-resistant, mostly from healthy children. A questionnaire survey documented a complex pattern of associations between and within the subgroups of the levels of MDR and socio-demographic characteristics, antibiotic stewardship, and guardians’ knowledge of AMR. This study has revealed the severity of AMR in children and the need for a community-specific-risk-based approach to implementing measures to curb the problem.
Keywords: antimicrobial resistance, Escherichia coli, children, risk factors, Zambia
1. Introduction
The emergence of antimicrobial resistance (AMR) is a global public health threat [1]. Increased resistance to commonly used antibiotics has been reported for various pathogenic and commensal bacteria, hampering the treatment, and increasing the burden of infectious diarrhoeal disease in general, especially in children under five years [2,3]. Resistance against all available antimicrobial agents has been reported [4], including the last line of antibiotics reserved for treating infectious diarrhoeal diseases, which is medically alarming [5].
In developing countries, the burden of childhood infectious diseases remains high [6], partly due to inadequate health care systems [7]. Infectious diarrhoeal disease is one of the significant causes of mortality and morbidity in children under-five years in developing countries, including Zambia [8,9]. Pathogenic Escherichia coli is a major cause of bacterial diarrhoeal disease in this age group [10] and AMR increases the burden of such diarrhoeal diseases even further [3].
The rapidly growing trend of AMR is a multifaceted problem driven by several interlinked factors, including inherent microbial characteristics, selective pressures of antimicrobial use, and changes in society and technology that enhance the transmission of drug-resistant organisms [11,12]. A significant driver of AMR development is the over- and misuse of antimicrobials as therapeutics in human and veterinary medicine, agriculture growth promotors, and disinfectants in households [13,14,15,16]. Many of these compounds end up in the environment [17,18,19], hence contributing to the spread of AMR in animals, the environment, and directly or indirectly to humans [20,21,22,23]. Further, other factors such as behavioural, e.g., self-prescription of antibiotics [24]; sanitation and demographic e.g., crowded settings, poor cleanliness [25,26]; socio-economic e.g., poverty [27,28], have been implicated in the spread of AMR in communities.
Increased consumption of antibiotics during the last decades, especially in developing countries [29] has led to an increase in the occurrence of AMR [30,31]. Due to the lack of strict regulations on the use of antibiotics in many developing countries, the population has easy access to these compounds, even without prescription [32,33,34]. In addition, despite the World Health Organization (WHO) recommendation to reserve antibiotics only in cases of bloody diarrhoea [35], antibiotics are readily used to treat any form of diarrhoea in children [36].
Most studies have focused on AMR in pathogenic E. coli collected from diarrhoeic children, as antimicrobial susceptibility patterns govern the treatment and AMR pathogenic bacteria is a direct public health threat. However, samples from healthy children may reveal resistant commensal E. coli that might act as significant reservoirs for resistance genes [37] hence, playing a considerable role in spreading the resistance within a community [38]. Tenover and McGowan (1996) suggested that exposure of commensal bacteria like E. coli to antibiotics increases the carriage levels of resistant organisms and, if plasmid-mediated, resistance might be transmitted to a more virulent acquired organism [39].
Therefore, this study aimed to explore the occurrence and patterns of antimicrobial-resistant E. coli isolated from children under five years old in Zambia and identify possible drivers for AMR in the study population. We used E. coli since it is commonly found in humans and animals, can cause diseases in both host categories, and might serve as markers of antibiotic resistance spread to pathogens and the remaining gut microbiota [40,41].
2. Materials and Methods
2.1. Study Design, Sites, and Population
A hospital-based cross-sectional study was conducted in 12 purposively selected health centres (hospitals) and the children’s hospital in Lusaka and Ndola districts, respectively. The study sites are the provincial headquarters of the most populated provinces of Zambia and host heterogeneous populations from different cultures and social backgrounds [42] (Figure 1).
Figure 1.
Map of Zambia showing the study sites.
2.2. Sample Size and Sampling Strategy
The population of children under five years of age in the Lusaka and Ndola districts in 2019 were projected at 425,000 and 89,000, respectively [43]. If 50% of these children sought health services [44], a fraction of 0.5% would be considered representative, and the chosen sample size would be large enough to detect rare varieties of AMR. The targeted sample size was, therefore, estimated to be 1287 children (diarrhoea and non-diarrhoeic). Sampling was proportionally distributed in both districts using the under-five population as a weighing proxy factor. The standard World Health Organization (WHO) definition of diarrhoea was used [45], while healthy children were those without any symptomatic disease at the time of visiting the clinic. Children undergoing antibiotic treatment at the time of sampling and non-concerting parents were excluded.
2.3. Sample Collection and Epidemiological Survey
A rectal swab specimen was aseptically collected from each study participant and transported chilled (<4 °C) in Cary-Blair enteric transport media (Oxoid, Basingstoke, UK) to the Bacteriology Laboratory at the University Teaching Hospital in Lusaka for analysis. Health status, food habits, socio-demographic characteristics, antibiotic stewardship, and awareness of AMR of guardians were investigated through a pre-tested structured questionnaire administered to the child’s guardian. The final questionnaire used is provided as a Supplementary Material File.
2.4. Laboratory Analysis
2.4.1. Isolation and Identification of E. coli
Rectal swabs were pre-enriched in buffered peptone water (Oxoid, Basingstoke, UK) and aerobically incubated at 37 °C for 24 h. The enriched broth was plated onto MacConkey agar plates (Oxoid, Basingstoke, UK) and incubated aerobically for an additional 24 h at 37 °C. Lactose fermenting colonies were then sub-cultured onto Eosin Methylene Blue (EMB) agar plates (Oxoid, Basingstoke, UK) and incubated aerobically at 37 °C for 24 h. Presumptive E. coli colonies displaying a green metallic sheen were then purified on nutrient agar (Oxoid, Basingstoke, UK). One colony from each plate was further confirmed by phenotypic characterization and standard biochemical tests using triple sugar iron (Oxoid, Basingstoke, UK), Sulphur Indole Motility (Oxoid, Basingstoke, UK) and citrate agar (Oxoid, Basingstoke, UK). For additional taxonomic confirmation, 323 isolates were randomly selected for further identification by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) using the VITEK® MS—SARAMIS® KB V4.16 (bioMérieux, Lyon, France).
2.4.2. Antimicrobial Susceptibility Testing (AST)
The AST was performed by the Kirby-Bauer disc diffusion method using the Clinical Laboratory Standards Institute (CLSI) guidelines on Müeller-Hinton agar plates (Oxoid, Basingstoke, UK) [46]. Suspensions of 0.5 McFarland were prepared from pure colonies of isolated E. coli and inoculated onto Müller-Hinton agar plates (Oxoid, Basingstoke, UK). The susceptibility pattern of the isolates was determined for a panel of ten (10) antibiotics (Table 1). A standard culture of E. coli (ATCC 25922) was used as positive control culture with each batch of antimicrobial susceptibility testing.
Table 1.
List of antibiotics used in AST.
| Antibiotics | Class of Antibiotic | Description | Source * |
|---|---|---|---|
| Amoxicillin-clavulanate | Penicillin + beta-lactamase inhibitor | 20 µg | Oxoid |
| Ampicillin | Penicillin (Beta-lactam) | 10 µg | Oxoid |
| Cefotaxime | Third Generation Cephalosporin (Beta-lactam) | 30 µg | Oxoid |
| Chloramphenicol | Phenicols | 30 µg | Oxoid |
| Ciprofloxacin | Fluoroquinolone | 5 µg | Oxoid |
| Gentamicin | Aminoglycosides | 10 µg | Oxoid |
| Nalidixic acid | Quinolones | 30 µg | Oxoid |
| Imipenem | Carbapenems | 10 µg | Oxoid |
| Tetracycline | Tetracycline | 30 µg | Oxoid |
| Trimethoprim-Sulphamethoxazole | Folate Pathway Antagonist | 25 µg | Oxoid |
* Source: Oxoid, Basingstoke, UK.
The plates were incubated for 16–18 h at 37 °C. The zones of inhibition were read using a digital Vernier Calliper and interpreted as Susceptible (S), Intermediate (I), and Resistant (R) based on the CLSI guidelines [46]. Multi-Drug Resistant (MDR), Extensively-Drug Resistant (XDR), and Pan-Drug-Resistant (PDR) isolates were identified, with MDR defined as non-susceptibility to at least one antibiotic in three antimicrobial classes tested; XDR as non-susceptibility to at least one antibiotic in all but two or fewer antimicrobial classes (i.e., E. coli isolates remain susceptible to only one or two classes); PDR as non-susceptibility to all antibiotics in all antimicrobial classes tested [47]. Since only one antimicrobial agent was tested for each antimicrobial class, the concepts possible XDR and possible PDR were used as per the international expert proposal for interim standard definitions for resistance recommendations [47].
2.5. Data Analysis
The measured diameters of the zones of inhibition for AST were analysed using the WHONET 2021® software. The resistance profile for all antibiotics was reported, and tables and graphs were produced in WHONET. Epidemiological data and certain outputs from WHONET® 2021 software were summarized and then entered into a database using Excel 2016®. Further statistical analyses were completed using Stata (StataCorp, College Station, TX) version 16.0 for Windows.
Initially, descriptive statistics focused on describing the categorical variables from the questionnaire focusing on demographic and hygienic factors, as shown in Table 2 and Table 3. The potential associations between the hypothesized categorical risk factors and the dichotomous outcomes (MDR, XDR, and PDR patterns displayed by E. coli) were assessed using Chi-square analyses. A new variable was created as an ordinal outcome with 0 = AMR, 1 = ANY AMR, 2 = MDR, 3 = XDR, and 4 = PDR. Explanatory variables showing a p-value < 0.20 from one of the outcome models were selected as candidate variables and taken into the multivariable logistic and ordinal regression models. The multivariable models were built using a backward selection strategy, using a p-value of <0.05 of the likelihood ratio test as inclusion criteria. The model fit was assessed using the Hosmer Lemeshow test, lroc and lsens procedures in Stata for logistic models, and graphical methods for the ordinal model. Finally, the potential effects of the random effect of Health Centres were assessed for all models using the melogit procedure in Stata.
Table 2.
Socio-demographic characteristics of study participants.
| Variables | Categories | N (1020) | Percent |
|---|---|---|---|
| Health status | Healthy | 455 | 44.61 |
| Diarrhoeic | 565 | 55.39 | |
| Gender | Female | 499 | 48.92 |
| Male | 521 | 51.08 | |
| Age | 0–5 months | 322 | 31.57 |
| 6–11 months | 239 | 23.43 | |
| 12–35 months | 359 | 58.92 | |
| 36–59 months | 100 | 9.80 | |
| Guardian’s level of education | None | 51 | 5.00 |
| Primary | 253 | 24.80 | |
| Secondary | 601 | 30.29 | |
| Tertiary | 115 | 11.27 | |
| Population density in the area of habitation | Low density | 36 | 3.53 |
| Medium density | 190 | 18.63 | |
| High density | 794 | 77.84 | |
| Size of the household | Below 5 people | 422 | 41.37 |
| Equal or above 5 people | 598 | 58.63 | |
| Keeping animals at the household level | No | 864 | 84.71 |
| Yes | 156 | 15.29 | |
| Types of animals kept at household level * (N = 156) | Livestock | 11 | 7.05 |
| Poultry | 84 | 53.85 | |
| Pets | 82 | 52.56 | |
| Other animals | 8 | 5.13 |
* Variable with multiple responses.
Table 3.
Hygienic and child feeding characteristics of study participants.
| Variables | Categories | N (1020) | Percent |
|---|---|---|---|
| Source of water for drinking * | Pipe borne (council water) | 859 | 84.22 |
| Borehole | 147 | 14.41 | |
| River/Pond/Dam | 18 | 1.76 | |
| Sachet/Bottled/Filtered | 11 | 1.08 | |
| Treatment of drinking water | No | 366 | 35.88 |
| Sometimes | 215 | 21.08 | |
| Yes | 439 | 43.04 | |
| Washing hands before cooking and feeding the child | No | 51 | 5.00 |
| Sometimes | 241 | 23.63 | |
| Yes | 728 | 71.37 | |
| Washing hands after disposing of the child’s faeces | No | 55 | 5.39 |
| Sometimes | 182 | 17.84 | |
| Yes | 783 | 76.76 | |
| Types of toilets * | Flush toilet | 495 | 48.53 |
| Pit latrine | 536 | 52.55 | |
| Disposing of solid waste * | Bin | 749 | 73.43 |
| Pit | 238 | 23.33 | |
| Roadside | 36 | 3.53 | |
| Storage of prepared food for the child | No | 284 | 27.84 |
| Yes | 736 | 72.16 | |
| Storage methods of prepared food for the child (N = 736) | At room temperature | 331 | 44.97 |
| In a cold chain | 83 | 11.28 | |
| In a warmer | 322 | 43.75 | |
| Exclusive breastfeeding | No | 92 | 9.02 |
| Partially | 737 | 72.25 | |
| Exclusively | 191 | 18.73 | |
| Child feeding methods * | Spoon | 736 | 72.16 |
| Fingers/hands | 623 | 61.08 | |
| Bottle feeding | 3 | 0.29 |
* Variable with multiple responses.
2.6. Ethical Consideration
The study was approved by the Zambian ERES Converge Institutional Review Board (Ref. No. 2020-Aug-006) and the National Health Research Authority (NHRA00010/3/09/2020). Further, the Provincial and District Health Offices were informed about the study. Informed consent was obtained before the study, and only the under-five-year-old children, as defined above, whose guardians consented to participate in the study were included.
3. Results
3.1. Characteristics of Study Participants
In total, 1020 children were included in this study, of which 565 (55.39%) were diarrhoeic and 455 (44.61%) healthy. Boys were slightly more represented than girls (n = 521, 51.08%). The median age (IQR) of study participants was 10 (4–21) months, with the minimum and maximum age range between one and 59 months, respectively. Most of the study participants lived in high-density population areas (77.84%), and 58.92% of their guardians had attained up to a secondary level of education. Further, more than half, 598 (58.62%), of the households had five or more members (Table 2).
With regards to the variables connected to hygiene and behavioural characteristics, 859 (84.22%) households used council water (pipe-borne), and 439 (43.04%) further treated water intended for drinking. Most guardians, 728 (71.37%) and 783 (76.76%) reported washing their hands before cooking or feeding the child and after disposing of the child’s faeces, respectively. The pit latrine was used slightly more (52.55%) than the flush toilet, while the bin (73.43%) was the commonly used disposal method of solid waste. Only 191 (18.73%) of the participant’s guardians complied with the exclusive breastfeeding of children below six months. Most guardians, 736 (72.16%), stored prepared food for children for further use, mostly at room temperature (44.97%) or in a warmer (43.75%), and the spoon (72.16%) was commonly used to feed children (Table 3).
The knowledge and characteristics of antibiotics, AMR, and diarrhoea are summarized in Table 4. Most guardians, 531 (77.75%), perceived the consumption of contaminated food as the cause of their children’s diarrhoea. Diarrhoea with mucus 301/565 (53.46%) and fever 347/565 (61.63%) were the most frequently reported symptoms in children with diarrhoea. Further, most guardians reported that they lacked knowledge of antibiotics and AMR. Of these, 110/175 and 26/37 correctly understood antibiotics and AMR, respectively.
Table 4.
Children’s guardians’ knowledge of antibiotics and characteristics of diarrhoea.
| Variables | Categories | N | Percent |
|---|---|---|---|
| Knowledge of antibiotics (N = 1020) | No | 845 | 82.84 |
| Yes | 175 | 17.16 | |
| Correct knowledge of antibiotics by examples (N = 175) |
No | 65 | 37.14 |
| Yes | 110 | 62.86 | |
| Awareness of AMR (N = 1020) | No | 983 | 96.37 |
| Yes | 37 | 3.63 | |
| Correct awareness of AMR by the concept definition (N = 37) |
No | 11 | 29.73 |
| Yes | 26 | 70.27 | |
| Use of antibiotics suggested by unauthorized personnel |
No | 740 | 72.55 |
| Sometimes | 135 | 13.24 | |
| Yes | 145 | 14.22 | |
| Knowledge of causes of diarrhoea (N = 1020) | No | 337 | 33.04 |
| Yes | 683 | 66.96 | |
| Perceived causes of diarrhoea * (N = 683) | Poor hygiene | 261 | 38.21 |
| Food likely to be contaminated | 531 | 77.75 | |
| Teething | 134 | 19.62 | |
| Undercooked food | 18 | 2.64 | |
| Complimentary food before six months | 33 | 4.83 | |
| Change of diet | 5 | 0.73 | |
| Germs | 13 | 1.90 | |
| Others | 26 | 3.81 | |
| Symptoms of diarrhoeic children * (N = 565) | Bloody diarrhoea | 33 | 5.86 |
| Diarrhoea with mucus | 301 | 53.46 | |
| Fever | 347 | 61.63 | |
| Vomiting | 269 | 47.78 |
* Variable with multiple responses.
3.2. Antimicrobial Susceptibility Patterns
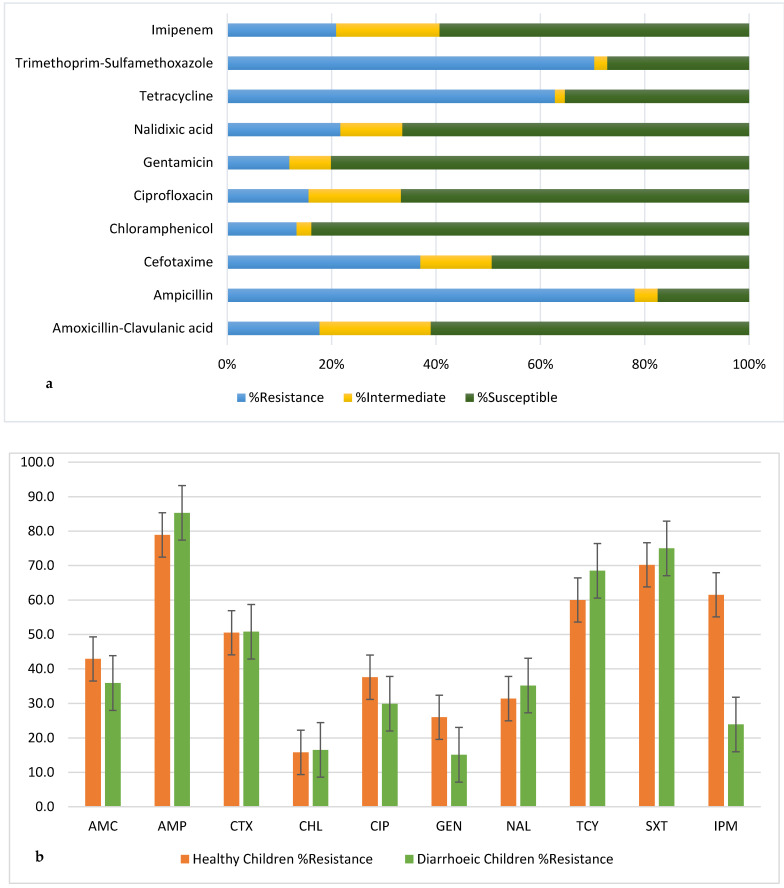
The identification of isolates using standard biochemical tests was reliable since their confirmation using MALDI-TOF MS showed 98.8% accuracy. All E. coli isolates were subjected to AST using the above-mentioned panel of antibiotics (Table 1). Most E. coli were resistant to at least one antibiotic (988/1020), nearly equally distributed between healthy and diarrhoeic children with 95.4% (434/455) and 98.1% (554/565), respectively (S1). The E. coli isolates displayed the highest resistance against ampicillin (78.0%), trimethoprim-sulfamethoxazole (70.4%), and tetracycline (62.8%), while they were more susceptible to chloramphenicol (83.8%) and gentamicin (80.1%) (Figure 2a). This trend was the same in both healthy and diarrhoeic children, although at different percentage levels. However, a significant difference in the susceptibility and resistance profiles of the isolates to imipenem was observed between the healthy and diarrhoeic children. Nearly 62% and 24% of isolates from healthy and diarrhoeic children were resistant to imipenem, respectively (Figure 2b).
Figure 2.
Resistance profile of 1020 E. coli isolates; (a) Overall and (b) healthy versus diarrhoeic children; AMC: Amoxicillin-Clavulanic acid; AMP: Ampicillin; CTX: Cefotaxime; CHL: Chloramphenicol; CIP: Ciprofloxacin; GEN: Gentamicin; NAL: Nalidixic acid; TCY: Tetracycline; SXT: Trimethoprim-Sulfamethoxazole; IPM: Imipenem.
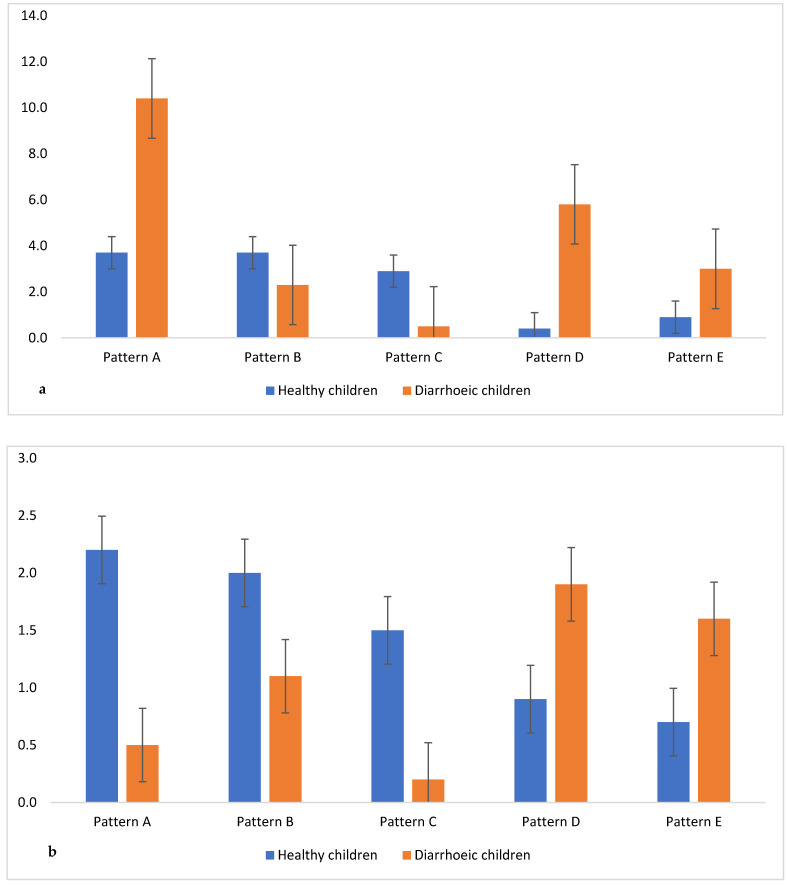
Of the 1020 E. coli isolates, 82.8% (845) were MDR, with 136 and 9 isolates being possible XDR and possible PDR, respectively. The MDR profiles were grouped in 251 different patterns consisting of 208 MDR patterns (resistance to at least one antibiotic in three antimicrobial classes tested), 41 possible XDR patterns (resistance to at least one antibiotic in all, except in one or two, antimicrobial classes tested) and two possible PDR patterns (resistance to all antibiotics in all antimicrobial classes tested) (Tables S1–S3). The five most frequent MDR and possible XDR patterns occurred differently between both groups, with patterns A and D being significant for MDR (Figure 3a). The frequency of the five possible XDR patterns was significantly different between the groups (Figure 3b), while possible PDR E. coli were isolated from healthy children only.
Figure 3.
The top five patterns in healthy and diarrhoeic children ((a) MDR and (b) XDR). MDR patterns A (AMP TCY SXT); B (AMP TCY SXT IPM); C (AMP SXT IPM); D (AMP CTX TCY SXT); E (AMC AMP CTX TCY SXT); XDR patterns A (AMC AMP CTX CIP GEN NAL TCY SXT IPM); B (AMC AMP CTX CIP NAL TCY SXT IPM); C (AMC AMP CTX CIP TCY SXT IPM); D (AMC AMP CTX CHL CIP GEN NAL TCY SXT); E (AMC AMP CTX CIP GEN NAL TCY SXT).
3.3. Potential Risk Factors Associated with AMR
The dichotomous outcome variables MDR, possible XDR, and possible PDR were first analysed individually and merged into a single ordinal outcome variable called Levels of AMR (LAMR). Each outcome variable was analysed into three categories, diarrhoeic, healthy, and all children. In the univariable analysis, all variables with a p < 0.20 (Table 5) were selected to build the multivariable logistic regression and ordered logistic regression models.
Table 5.
Standard multivariable logistic regression and adjusted for health centres (HC) models for risk factors associated with MDR, possible XDR, and possible PDR in children.
| MDR | OR (95% C.I) | p > |z| | OR Adjusted for HC (95% C.I) | p > |z| |
|---|---|---|---|---|
| All children | ||||
| Household in a high-density area | 0.13 (0.02–0.99) | 0.050 | ||
| Storing prepared food for the child | 0.65 (0.44–0.96) | 0.031 | 0.65 (0.43–0.98) | 0.040 |
| Disposing of solid waste in a pit | 1.60 (1.04–2.45) | 0.033 | ||
| Disposing of solid waste on the roadside | 8.80 (1.19–64.96) | 0.033 | ||
| Diarrhoeic children | ||||
| Disposing of solid waste in a bin | 0.46 (0.27–0.79) | 0.006 | ||
| Knowledge of antibiotics | 1.96 (1.00–3.83) | 0.049 | ||
| Healthy children | ||||
| Gender | 0.51 (0.31–0.86) | 0.011 | 0.57 (0.32–0.98) | 0.041 |
| Storing prepared food for the child | 0.60 (0.37–0.98) | 0.046 | ||
| Knowledge of antibiotics | 0.53 (0.28–1.02) | 0.057 | 0.48 (0.23–0.99) | 0.049 |
| Possible XDR | OR (95% C.I) | p > |z| | OR adjusted for HC (95% C.I) | p > |z| |
| All children | ||||
| Age group 6–11 months | 0.77 (0.44–1.34) | 0.365 | 0.47 (0.28–0.79) | 0.005 |
| Age group 12–35 months | 1.02 (0.63–1.65) | 0.918 | 0.55 (0.35–0.88) | 0.013 |
| Age group 36–59 months | 0.28 (0.11–0.74) | 0.010 | 0.14 (0.05–0.39) | 0.000 |
| Child feeding with a spoon | 0.61 (0.39–0.95) | 0.028 | ||
| Size of the household | 1.57 (1.08–2.29) | 0.019 | ||
| Diarrhoeic children | ||||
| Child feeding with fingers/hands | 2.47 (1.02–5.95) | 0.044 | 2.90 (1.16–7.19) | 0.022 |
| Keeping poultry at the household level | 2.67 (1.18–6.01) | 0.018 | 2.54 (1.09–5.87) | 0.029 |
| Using ATB given by non-professional | 1.36 (0.98–1.88) | 0.058 | ||
| Healthy children | ||||
| Storing prepared food in a warmer | 0.48 (0.23–0.99) | 0.049 | ||
| Keeping pets at household | 0.28 (0.08–0.93) | 0.038 | 0.19 (0.05–0.71) | 0.013 |
| Treatment of drinking water | 1.26 (0.95–1.66) | 0.107 | ||
| Possible PDR | OR (95% C.I) | p > |z| | OR adjusted for HC (95% C.I) | p > |z| |
| Healthy children | ||||
| Awareness of AMR | 10.33 (1.93–55.04) | 0.006 | 9.06 (1.48–55.45) | 0.017 |
Results from the standard multivariable logistic regression analysis are shown in Table 5. Different sets of variables emerged in the different models, and many variables were removed when adjusting for the random effect of health centres. All the logistic regression models adequately fit the data (Hosmer and Lemeshow test), but with limited explanatory power, with ROC areas around 0.60 (a ROC area of 0.50 indicated no explanatory power). Substantial model improvements were observed for the random effect (LR test, p < 0.001).
In the MDR-adjusted models, only storing prepared food for the child was significantly linked to MDR in all children, while gender and guardians’ awareness of antibiotics remained in the healthy children model. For XDR, after adjusting for the effect of health centres, five variables were removed, while age groups 6 to 11 and 12 to 35 months became significantly associated with possible XDR isolates (Table 5). For PDR, the only variable remaining for healthy children was awareness of AMR.
Table 6 shows results for LAMR, the combined ordinal variable. While several factors were identified in the standard multivariable model, adjusting for clustering removed most variables from the model. Only feeding a child with stored prepared food remained.
Table 6.
Ordered logistic regression model for risk factors associated with LAMR in children.
| Variables | OR (95% C.I) | p > |z| | OR Adjusted for HC (95% C.I) | p > |z| |
|---|---|---|---|---|
| All children | ||||
| Age group 36–59 months | 0.58 (0.34–0.98 | 0.041 | ||
| Household in a medium-density area | 0.47 (0.22–0.99) | 0.049 | ||
| Household in a high-density area | 0.48 (0.24–0.98) | 0.045 | ||
| Storing prepared food for the child | 0.61 (0.42–0.87) | 0.007 | 0.65 (0.43–0.98) | 0.040 |
| Disposing solid waste in a bin | 0.66 (0.48–0.89) | 0.006 | ||
| Diarrhoeic children | ||||
| Disposing solid waste in a bin | 0.57 (0.38–0.85) | 0.006 | ||
| Keeping poultry in the household | 2.37 (1.17–4.80) | 0.016 | ||
| Knowledge of antibiotics | 1.70 (1.06–2.72) | 0.026 | ||
| Healthy children | ||||
| Storing prepared food for the child | 0.62 (0.42–0.91) | 0.015 | ||
| SWH after disposing of the child’s faeces | 0.26 (0.08–0.81) | 0.021 |
SHW: Sometimes Hands washing.
4. Discussion
This study investigated the occurrence and patterns of AMR—E. coli in healthy and diarrhoeic children and established risk factors for AMR in children below five years. Nearly all E. coli (96.9%) were resistant to at least one antimicrobial tested, slightly above the 83% reported in Nigeria [48], and resistant E. coli were equally common among healthy and diarrhoeic children. This near absence of susceptible E. coli is an alarming sign, foreshadowing a future public health crisis. Indeed, AMR is frequently observed in many low- and middle-income countries, as reported in Ethiopia [49], Bolivia, Peru [50], and Vietnam [51], including in children’s commensal E. coli in Kenya [52]. Some studies on commensal E. coli isolated from children from Asia found a lower occurrence of resistance [51,53]. However, these studies were conducted in rural areas, which could explain their lower reporting rates of AMR as access to and therefore misuse of antibiotics are more prominent in urban areas [54,55]. Equally, resistant E. coli isolated from diarrhoeic children were common in Burkina Faso [56], South Africa [5], and Ethiopia [57] corroborating our findings, while relatively different trends were observed in Taiwan [58] and Pakistan [59]. The difference in the setting of these two studies could justify this disparity.
Several studies have reported high resistance to commonly used antibiotics, e.g., ampicillin, trimethoprim-sulfamethoxazole, and tetracycline, against enteric bacterial infections [49,58,59,60]. The easy accessibility and affordability of these drugs have led to their overuse by the population [29,33] but also in the animal production chain, as evidenced by recent studies completed in Zambia [22,23,61]. Further, in countries with a high prevalence of HIV infection, like Zambia [62], trimethoprim-sulfamethoxazole has been heavily used for the past decades as prophylaxis against opportunistic infections in HIV-infected and/or exposed individuals [63,64]. The above factors could explain the maintenance of this resistance through selection pressure.
We found that most E. coli were susceptible to chloramphenicol and gentamicin. An earlier study from Zambia [65] also found a low prevalence of chloramphenicol and gentamicin-resistant E. coli isolates. This observation implies that the strains have not developed more resistance against these two drugs since the last study in Zambia by Chiyangi [65]. However, increased resistance to antibiotics recommended in the Zambia standard treatment guidelines for infectious diseases in children, including diarrhoeal diseases [66], limits treatment options.
There were few observable differences in resistance patterns between E. coli in healthy and diarrhoeic children. Surprisingly, E. coli from healthy children were more frequently resistant to commonly used antimicrobial like amoxicillin-clavulanic acid, ciprofloxacin, and gentamicin compared to diarrhoeic children. This points to the community-level circulation of resistance as opposed to hospital-acquired resistance or resistance associated with disease management. Of special worry was the high percentage of imipenem-resistant isolates observed in healthy children; imipenem-resistant E. coli were almost three times more commonly isolated from healthy compared to diarrhoeic children. The high prevalence of resistant strains against imipenem is not unique to this study [5]. It is especially worrisome as the drug is a last resort classified as critically important and a high-priority antimicrobial agent to treat severe bacterial infections in humans [67]. This scenario supports previous postulates that healthy children carrying commensal E. coli could serve as reservoirs of resistance genes with a possible transmission, if plasmid-mediated, to more virulent bacteria [37,39]. Therefore, future infections in these children could be more challenging to treat if the AMR in commensal E. coli is transferred to pathogenic bacteria.
Many of the E. coli were MDR, including possible XDR and PDR. This contrast earlier studies from Taiwan [58], China [68], Nigeria [69], and India [53]. The robustness of our study in terms of its considerable sample size and heterogeneous population could have allowed the capture of rare patterns of MDR as compared to the above-mentioned studies. The indicative significant difference in the patterns of MDR and possible XDR between the isolates from diarrhoeic and healthy children is of great public health concern considering the recent report implicating commensal E. coli in the maintenance and transfer of XDR plasmid to Shigella sonnei which could increase disease morbidity [41].
Importantly, the presence of all possible PDR E. coli in healthy children signals a public health threat since the commensal flora is a highly populated ecosystem which may be harbouring bacterial resistant genes and transfer these to other members of the microbiota, including pathogens [70]. This may result in an increased probability of acquiring clinical infections with AMR pathogens.
Performing risk factor analysis for AMR has been challenging because its occurrence is a complex phenomenon linked to many factors [26,71,72,73]. Some authors have considered analysing only one antimicrobial agent at a time in univariable and rarely on multivariable models [51,74]. The interpretation of such models might not reflect the true situation in which an isolate might have multiple resistance and would not potentially capture the multiple effects of epidemiological factors on the occurrence of AMR. Analysing the association between potential risk factors for AMR must be completed carefully, considering a broad view of the combination of different patterns observed. A multiple resistance patterns approach in a multivariable model is therefore preferable. One of the drawbacks observed in the literature is that many authors discuss results from univariable statistical analyses. In principle, these estimates of Odds Ratio (OR) are unreliable, and more elaborate models need to be developed.
In the present study, we found a strong communal effect shown through the improved explanatory power of the random-effect models used. The multivariable risk factors analysis revealed an inconsistent association between the multiple resistance patterns outcomes (MDR, possible XDR, possible PDR, and LAMR) and the independent variables. Further, within the subgroups (all children, diarrhoeic and healthy children) of each outcome, no single variable could consistently predict AMR. This variability shows the complexity and heterogeneity of risk factors linked to AMR in the community [11,75,76]. It further implies that the significant variables retained in the final multivariable logistic and ordinal regression models could be a result of random chance and would vary in other communities based on their socio-demographic, economic, behavioural, and environmental characteristics. For instance, four variables in this study, namely residence in a high-density area, feeding the child with stored prepared food, disposing of the solid waste in a pit and by the roadside, predicted the occurrence of MDR in all children in the multivariable logistic model. However, when split between diarrhoeic and healthy children’s subgroups, two different variables (disposing of solid waste in a bin and knowledge of antibiotics) and none of the four from the main model predicted the occurrence of MDR in diarrhoeic children. Equally, one other variable (gender of the child) and only one of the four from the main model predicted the outcome in healthy children.
Further, after adjusting for the effect of the health centres, which are proxy measures of the residential areas of the study participants, only feeding the child with stored prepared food (adjusted OR: 0.65; p = 0.040) remained significantly associated with MDR occurrence in all children. Furthermore, all variables in the diarrhoeic children were insignificant, while gender (adjusted OR: 0.57; p = 0.041) and the guardians’ knowledge of antibiotics (adjusted OR: 0.48; p = 0.049) remained in the healthy children model. Interestingly, in the ordinal logistic regression model, the level of AMR after adjusting for the effect of the health centres was only significantly influenced by feeding the child with stored prepared food for all children, while in the subgroups, all variables became none-significant. A multivariable analysis and interpretation as shown in this study give a holistic understanding of the interdependence and multiple effects of predictors of the AMR in the community.
Although the large sample size and many antimicrobial agents were used in this study to assess different patterns of AMR, we only used one isolate and one colony from each child’s sample. There is a possibility that some resistant or susceptible colonies were missed, under- or overestimating the true prevalence of resistant E. coli and/or certain resistance patterns. Further, linking the potential epidemiological drivers and patterns of AMR to the geographical location of each community should be explored to enable effective surveillance.
5. Conclusions
This study has revealed a high occurrence of MDR in healthy and diarrhoeic children with several distinct patterns involving classified antibiotics that are critically important and high-priority antimicrobials for treating serious bacterial infections in humans. It further identified possible PDR—E. coli carriage in healthy children, highlighting the role that this group could play in harbouring and transferring resistant genes to other pathogens.
The limited explanatory power of all logistic models (ROC around 0.60), the different factors popping up in different models, and the strong random effect visualised in the final model point toward a situation where the spread of AMR is linked to the widespread use of antibiotics in the communities involved and cannot be attributed to special exposures. This is also a warning for society, as AMR profiles strongly linked to specific risk factors would be possible to control by focusing on specific hygienic measures within the family unit. If AMR is a community-based problem, interventions also need to be communal—a more fundamental challenge. The considerable sample size of the study should indicate that the patterns observed are representative of the study provinces of Zambia.
Acknowledgments
We would like to thank Eden University for its support. Further gratitude also goes to Patrick Katemangwe, Fred Tembo, and Velu Milomba Rachel for the help rendered during sample processing, data collection, and data management, respectively. We equally thank Mainda Geoffrey and Mercy Mukuma for their valuable contributions to this work.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10081684/s1, File S1: Questionnaire for guardians; Table S1: Overall AMR E. coli patterns; Table S2: Healthy children AMR E. coli patterns; Table S3: Diarrhoeic children AMR E. coli patterns.
Author Contributions
Conceptualization: F.N.B. and J.B.M.; methodology: F.N.B., A.-K.L., E.S. and J.B.M.; software: F.N.B. and E.S.; validation: F.N.B., A.-K.L., E.S., P.B.M. and J.B.M.; formal analysis: F.N.B. and E.S.; investigation: F.N.B.; resources: F.N.B., E.S., B.M.H. and J.B.M.; data curation: F.N.B.; writing—original draft preparation: F.N.B.; writing—review and editing: F.N.B., A.-K.L., E.S., P.M., S.M., B.M.H., P.B.M. and J.B.M.; visualization: F.N.B., A.-K.L., E.S., P.B.M. and J.B.M.; supervision: A.-K.L., E.S., B.M.H., P.B.M. and J.B.M.; project administration: F.N.B.; funding acquisition: F.N.B., E.S., B.M.H. and J.B.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The essential data supporting the reported results are contained in this study. All other supplementary data can be made available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This study was funded by the WHO-Advisory Group for Integrated Surveillance of Antimicrobial Resistance (AGISAR) Project (grant number 204954). The work was also supported by the Africa Centre of Excellence for Infectious Disease of Humans and Animals (ACEIDHA) project (grant number P151847) funded by the World Bank, the project NORPART-2018/10213 funded by the Norwegian Agency for International Cooperation and Quality Enhancement in Higher Education.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO . Fact Sheets. World Health Organization; Geneva, Switzerland: 2021. [(accessed on 15 April 2022)]. Antimicrobial resistance. Available online: https://www.who.int/en/news-room/fact-sheets/detail/antimicrobial-resistance. [Google Scholar]
- 2.Ingle D.J., Levine M.M., Kotloff K.L., Holt K.E., Robins-Browne R.M. Dynamics of antimicrobial resistance in intestinal Escherichia coli from children in community settings in South Asia and sub-Saharan Africa. Nat. Microbiol. 2018;3:1063–1073. doi: 10.1038/s41564-018-0217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medernach R.L., Logan L.K. The growing threat of antibiotic resistance in children. Infect. Dis. Clin. 2018;32:1–17. doi: 10.1016/j.idc.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russo T.A., Johnson J.R. Medical and economic impact of extraintestinal infections due to Escherichia coli: Focus on an increasingly important endemic problem. Microbes Infect. 2003;5:449–456. doi: 10.1016/S1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 5.Omolajaiye S., Afolabi K., Iweriebor B. Pathotyping and antibiotic resistance profiling of Escherichia coli isolates from children with acute diarrhea in Amatole district municipality of Eastern Cape, South Africa. Biomed Res. Int. 2020;2020:4250165. doi: 10.1155/2020/4250165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roser M., Ritchie H. Our World in Data. Global Change Data Lab; Oxford, UK: 2021. [(accessed on 10 April 2022)]. Burden of disease. Available online: https://ourworldindata.org/burden-of-disease?fbclid=IwAR0I88KzppGueXUvnzb84O86C86NsCzk81r89Ng79SpYq-TbtfH_x84G86Jm83_k_c80#citation. [Google Scholar]
- 7.Iskandar K.M.L., Hallit S., Sartelli M., Hardcastle T.C., Haque M., Lugova H., Dhingra S., Sharma P., Islam S. Surveillance of antimicrobial resistance in low-and middle-income countries: A scattered picture. Antimicrob. Resist. Infect. Control. 2021;10:63. doi: 10.1186/s13756-021-00931-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ugboko H.U., Nwinyi O.C., Oranusi S.U., Oyewale J.O. Childhood diarrhoeal diseases in developing countries. Heliyon. 2020;6:e03690. doi: 10.1016/j.heliyon.2020.e03690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubbard S.C., Meltzer M.I., Kim S., Malambo W., Thornton A.T., Shankar M.B., Adhikari B.B., Jeon S., Bampoe V.D., Cunningham L.C. Household illness and associated water and sanitation factors in peri-urban Lusaka, Zambia, 2016–2017. NPJ Clean Water. 2020;3:26. doi: 10.1038/s41545-020-0076-4. [DOI] [Google Scholar]
- 10.Mogasale V.V., Saldanha P., Pai V., Rekha P., Mogasale V. A descriptive analysis of antimicrobial resistance patterns of WHO priority pathogens isolated in children from a tertiary care hospital in India. Sci. Rep. 2021;11:5116. doi: 10.1038/s41598-021-84293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramaniam G., Girish M. Antibiotic resistance—A cause for reemergence of infections. Indian J. Pediatr. 2020;87:937–944. doi: 10.1007/s12098-019-03180-3. [DOI] [PubMed] [Google Scholar]
- 12.Kostyanev T., Can F. Antimicrobial Stewardship. Elsevier; Amsterdam, The Netherlands: 2017. The global crisis of antimicrobial resistance; pp. 3–12. [DOI] [Google Scholar]
- 13.Bloomfield S. Significance of biocide usage and antimicrobial resistance in domiciliary environments. J. Appl. Microbiol. 2002;92:144S–157S. doi: 10.1046/j.1365-2672.92.5s1.15.x. [DOI] [PubMed] [Google Scholar]
- 14.McEwen S.A., Fedorka-Cray P.J. Antimicrobial use and resistance in animals. Clin. Infect. Dis. 2002;34((Suppl. 3)):S93–S106. doi: 10.1086/340246. [DOI] [PubMed] [Google Scholar]
- 15.Vidaver A.K. Uses of antimicrobials in plant agriculture. Clin. Infect. Dis. 2002;34((Suppl. 3)):S107–S110. doi: 10.1086/340247. [DOI] [PubMed] [Google Scholar]
- 16.Wise R., Soulsby E. Antibiotic resistance—An evolving problem. Vet. Rec. 2002;151:371–372. [PubMed] [Google Scholar]
- 17.Ngigi A.N., Magu M.M., Muendo B.M. Occurrence of antibiotics residues in hospital wastewater, wastewater treatment plant, and in surface water in Nairobi County, Kenya. Environ. Monit. Assess. 2020;192:18. doi: 10.1007/s10661-019-7952-8. [DOI] [PubMed] [Google Scholar]
- 18.Bilal M., Mehmood S., Rasheed T., Iqbal H.M. Antibiotics traces in the aquatic environment: Persistence and adverse environmental impact. Curr. Opin. Environ. Sci. Health. 2020;13:68–74. doi: 10.1016/j.coesh.2019.11.005. [DOI] [Google Scholar]
- 19.Polianciuc S.I., Gurzău A.E., Kiss B., Ştefan M.G., Loghin F. Antibiotics in the environment: Causes and consequences. Med. Pharm. Rep. 2020;93:231. doi: 10.15386/mpr-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimera Z.I., Mshana S.E., Rweyemamu M.M., Mboera L.E., Matee M.I. Antimicrobial use and resistance in food-producing animals and the environment: An African perspective. Antimicrob. Resist. Infect. Control. 2020;9:37. doi: 10.1186/s13756-020-0697-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanna N., Sun P., Sun Q., Li X., Yang X., Ji X., Zou H., Ottoson J., Nilsson L.E., Berglund B. Presence of antibiotic residues in various environmental compartments of Shandong province in eastern China: Its potential for resistance development and ecological and human risk. Environ. Int. 2018;114:131–142. doi: 10.1016/j.envint.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Muonga E., Mainda G., Mukuma M., Kwenda G., Hang’ombe B., Bumbangi F., Phiri N., Mwansa M., Munyeme M., Muma J. Antimicrobial Resistance of Escherichia coli and Salmonella isolated from Raw Retail Broiler Chicken Carcasses in Zambia. J. Epidemiol. Res. 2020;6:35–43. doi: 10.5430/jer.v6n1p35. [DOI] [Google Scholar]
- 23.Phiri N., Mainda G., Mukuma M., Sinyangwe N., Banda L., Kwenda G., Muligisa-Muonga E., Flavien B., Mwansa M., Yamba K. Antibiotic-resistant Salmonella species and Escherichia coli in broiler chickens from farms, abattoirs and open markets in selected districts of Zambia. J. Epidemiol. Res. 2020;6:13–21. doi: 10.5430/jer.v6n1p13. [DOI] [Google Scholar]
- 24.Larson E. Community factors in the development of antibiotic resistance. Annu. Rev. Public Health. 2007;28:435–447. doi: 10.1146/annurev.publhealth.28.021406.144020. [DOI] [PubMed] [Google Scholar]
- 25.Collignon P., Beggs J.J., Walsh T.R., Gandra S., Laxminarayan R. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: A univariate and multivariable analysis. Lancet Planet. Health. 2018;2:e398–e405. doi: 10.1016/S2542-5196(18)30186-4. [DOI] [PubMed] [Google Scholar]
- 26.Aiello A.E., Larson E. Antibacterial cleaning and hygiene products as an emerging risk factor for antibiotic resistance in the community. Lancet Infect. Dis. 2003;3:501–506. doi: 10.1016/S1473-3099(03)00723-0. [DOI] [PubMed] [Google Scholar]
- 27.Alividza V., Mariano V., Ahmad R., Charani E., Rawson T.M., Holmes A.H., Castro-Sanchez E. Investigating the impact of poverty on colonization and infection with drug-resistant organisms in humans: A systematic review. Infect. Dis. Poverty. 2018;7:76. doi: 10.1186/s40249-018-0459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alvarez-Uria G., Gandra S., Laxminarayan R. Poverty and prevalence of antimicrobial resistance in invasive isolates. Int. J. Infect. Dis. 2016;52:59–61. doi: 10.1016/j.ijid.2016.09.026. [DOI] [PubMed] [Google Scholar]
- 29.Klein E.Y., Van Boeckel T.P., Martinez E.M., Pant S., Gandra S., Levin S.A., Goossens H., Laxminarayan R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA. 2018;115:E3463–E3470. doi: 10.1073/pnas.1717295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bebell L.M., Muiru A.N. Antibiotic use and emerging resistance: How can resource-limited countries turn the tide? Glob. Heart. 2014;9:347–358. doi: 10.1016/j.gheart.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun D.S., Kissler S.M., Kanjilal S., Olesen S.W., Lipsitch M., Grad Y.H. Analysis of multiple bacterial species and antibiotic classes reveals large variation in the association between seasonal antibiotic use and resistance. PLoS Biol. 2022;20:e3001579. doi: 10.1371/journal.pbio.3001579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van T.T.H., Yidana Z., Smooker P.M., Coloe P.J. Antibiotic use in food animals worldwide, with a focus on Africa: Pluses and minuses. J. Glob. Antimicrob. Resist. 2020;20:170–177. doi: 10.1016/j.jgar.2019.07.031. [DOI] [PubMed] [Google Scholar]
- 33.Chokshi A., Sifri Z., Cennimo D., Horng H. Global contributors to antibiotic resistance. J. Glob. Infect. Dis. 2019;11:36. doi: 10.4103/jgid.jgid_4110_4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mudenda S., Malama S., Munyeme M., Hang’ombe B.M., Mainda G., Kapona O., Mukosha M., Yamba K., Bumbangi F.N., Mfune R.L. Awareness of Antimicrobial Resistance and Associated Factors among Layer Poultry Farmers in Zambia: Implications for Surveillance and Antimicrobial Stewardship Programs. Antibiotics. 2022;11:383. doi: 10.3390/antibiotics11030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.WHO . Recommendations for Management of Common Childhood Conditions: Evidence for Technical Update of Pocket Book Recommendations: Newborn Conditions, Dysentery, Pneumonia, Oxygen Use and Delivery, Common Causes of Fever, Severe Acute Malnutrition and Supportive Care. World Health Organization; Geneva, Switzerland: 2012. [(accessed on 15 November 2020)]. Available online: https://apps.who.int/iris/handle/10665/44774. [PubMed] [Google Scholar]
- 36.Rhee C., Aol G., Ouma A., Audi A., Muema S., Auko J., Omore R., Odongo G., Wiegand R.E., Montgomery J.M. Inappropriate use of antibiotics for childhood diarrhea case management—Kenya, 2009–2016. BMC Public Health. 2019;19:468. doi: 10.1186/s12889-019-6771-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chopra I., Roberts M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Austin D., Kakehashi M., Anderson R. The transmission dynamics of antibiotic–resistant bacteria: The relationship between resistance in commensal organisms and antibiotic consumption. Proc. R. Soc. London. Ser. B Biol. Sci. 1997;264:1629–1638. doi: 10.1098/rspb.1997.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenover F.C., McGowan J.E., Jr. Reasons for the emergence of antibiotic resistance. Am. J. Med. Sci. 1996;311:9–16. doi: 10.1016/S0002-9629(15)41625-8. [DOI] [PubMed] [Google Scholar]
- 40.Lambrecht E., Van Coillie E., Van Meervenne E., Boon N., Heyndrickx M., Van de Wiele T. Commensal E. coli rapidly transfer antibiotic resistance genes to human intestinal microbiota in the Mucosal Simulator of the Human Intestinal Microbial Ecosystem (M-SHIME) Int. J. Food Microbiol. 2019;311:108357. doi: 10.1016/j.ijfoodmicro.2019.108357. [DOI] [PubMed] [Google Scholar]
- 41.Thanh Duy P., Thi Nguyen T.N., Vu Thuy D., Chung The H., Alcock F., Boinett C., Dan Thanh H.N., Thanh Tuyen H., Thwaites G.E., Rabaa M.A. Commensal Escherichia coli are a reservoir for the transfer of XDR plasmids into epidemic fluoroquinolone-resistant Shigella sonnei. Nat. Microbiol. 2020;5:256–264. doi: 10.1038/s41564-019-0645-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De la Fuente A., Murr A., Rascón E. Mapping Subnational Poverty in Zambia. World Bank Group; Washington, DC, USA: 2015. [(accessed on 10 June 2019)]. Available online: http://hdl.handle.net/10986/21783. [Google Scholar]
- 43.CSO . Zambia Population and Demographic Projections, 2011–2035. Central Statistical Office; Lusaka, Zambia: 2013. [(accessed on 10 June 2019)]. Available online: https://zambia.opendataforafrica.org/ZMPHC2015/population-and-demographic-projections-2011–2035. [Google Scholar]
- 44.Bright T., Felix L., Kuper H., Polack S. A systematic review of strategies to increase access to health services among children in low- and middle-income countries. BMC Health Serv. Res. 2017;17:252. doi: 10.1186/s12913-017-2180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.WHO . Fact Sheets. World Health Organization; Geneva, Switzerland: 2017. [(accessed on 10 January 2019)]. Diarrhoeal disease. Available online: https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease. [Google Scholar]
- 46.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. CLSI supplement M100; Clinical and Laboratory Standard Institute; Berwyn, PA, USA: 2021. [(accessed on 12 December 2021)]. Available online: https://clsi.org/about/press-releases/clsi-publishes-m100-performance-standards-for-antimicrobial-susceptibility-testing-131st-edition/ [Google Scholar]
- 47.Magiorakos A.-P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M., Giske C., Harbarth S., Hindler J., Kahlmeter G., Olsson-Liljequist B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria. An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 48.Dabo N.T., Muhammad B., Saka H.K., Kalgo Z.M., Raheem R.A. Antibiotic resistance pattern of Escherichia coli isolated from diarrhoeic and non-diarrhoeic under-five children in Kano, Nigeria. J. Microbiol. Biotechnol. 2019;4:94–102. [Google Scholar]
- 49.Singh A.K., Das S., Singh S., Gajamer V.R., Pradhan N., Lepcha Y.D., Tiwari H.K. Prevalence of antibiotic resistance in commensal Escherichia coli among the children in rural hill communities of Northeast India. PLoS ONE. 2018;13:e0199179. doi: 10.1371/journal.pone.0199179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartoloni A., Pallecchi L., Benedetti M., Fernandez C., Vallejos Y., Guzman E., Villagran A.L., Mantella A., Lucchetti C., Bartalesi F. Multidrug-resistant commensal Escherichia coli in children, Peru and Bolivia. Emerg. Infect. Dis. 2006;12:907. doi: 10.3201/eid1206.051258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dyar O.J., Hoa N.Q., Trung N.V., Phuc H.D., Larsson M., Chuc N.T., Lundborg C.S. High prevalence of antibiotic resistance in commensal Escherichia coli among children in rural Vietnam. BMC Infect. Dis. 2012;12:92. doi: 10.1186/1471-2334-12-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Omulo S., Lofgren E.T., Lockwood S., Thumbi S.M., Bigogo G., Ouma A., Verani J.R., Juma B., Njenga M.K., Kariuki S. Carriage of antimicrobial-resistant bacteria in a high-density informal settlement in Kenya is associated with environmental risk-factors. Antimicrob. Resist. Infect. Control. 2021;10:18. doi: 10.1186/s13756-021-00886-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purohit M.R., Lindahl L.F., Diwan V., Marrone G., Lundborg C.S. High levels of drug resistance in commensal E. coli in a cohort of children from rural central India. Sci. Rep. 2019;9:6682. doi: 10.1038/s41598-41019-43227-41591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalyani V., Bisht M., Thapliyal S., Rohilla K.K. Comparison of practice and attitude of self-treatment in rural and urban population in Uttarakhand, India: A comparative study. Natl. J. Physiol. Pharm. Pharmacol. 2020;10:1052–1059. doi: 10.5455/njppp.2020.10.07207202031072020. [DOI] [Google Scholar]
- 55.Nga D.T.T., Chuc N.T.K., Hoa N.P., Hoa N.Q., Nguyen N.T.T., Loan H.T., Toan T.K., Phuc H.D., Horby P., Van Yen N. Antibiotic sales in rural and urban pharmacies in northern Vietnam: An observational study. BMC Pharmacol. Toxicol. 2014;15:6. doi: 10.1186/2050-6511-15-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Konaté A., Dembélé R., Zongo C., Kaboré W.A., Bonkoungou I., Traoré A., Barro N. Occurrence of Multiple Antibiotic Resistances of Escherichia coli Isolated from Diarrheal Children Less Than Five Years in Burkina. Eur. J. Pharm. Med. Res. 2017;4:165–171. [Google Scholar]
- 57.Tola M.A., Abera N.A., Gebeyehu Y.M., Dinku S.F., Tullu K.D. High prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae fecal carriage among children under five years in Addis Ababa, Ethiopia. PLoS ONE. 2021;16:e0258117. doi: 10.1371/journal.pone.0258117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang I.-F., Lee W.-Y., Wang J.-L., Hung C.-H., Hu H.-H., Hung W.-Y., Hung Y.-J., Chen W.-C., Shen Y.-T., Cheng M.-F. Fecal carriage of multidrug-resistant Escherichia coli by community children in southern Taiwan. BMC Gastroenterol. 2018;18:86. doi: 10.1186/s12876-018-0807-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masood F., Abdullah R.M., Anam S., Arshad M.I., Anjum F.R. Occurrence and antibiogram of enteric bacterial isolates from stool samples of gastroenteritis children under 5 years of age in district Faisalabad, Pakistan. PAB. 2019;8:2087–2094. doi: 10.19045/bspab.2019.80153. [DOI] [Google Scholar]
- 60.Eltai N.O., Al Thani A.A., Al Hadidi S.H., Al Ansari K., Yassine H.M. Antibiotic resistance and virulence patterns of pathogenic Escherichia coli strains associated with acute gastroenteritis among children in Qatar. BMC Microbiol. 2020;20:54. doi: 10.1186/s12866-020-01732-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mainda G., Bessell P.R., Muma J.B., McAteer S.P., Chase-Topping M.E., Gibbons J., Stevens M.P., Gally D.L. Prevalence and patterns of antimicrobial resistance among Escherichia coli isolated from Zambian dairy cattle across different production systems. Sci. Rep. 2015;5:12439. doi: 10.1038/srep12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mweemba C., Hangoma P., Fwemba I., Mutale W., Masiye F. Estimating district HIV prevalence in Zambia using small-area estimation methods (SAE) Popul. Health Metr. 2022;20:8. doi: 10.1186/s12963-022-00286-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hutchinson E., Parkhurst J., Phiri S., Gibb D.M., Chishinga N., Droti B., Hoskins S. National policy development for cotrimoxazole prophylaxis in Malawi, Uganda and Zambia: The relationship between context, evidence and links. Health Res. Policy Syst. 2011;9:S6. doi: 10.1186/1478-4505-9-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chintu C., Bhat G., Walker A., Mulenga V., Sinyinza F., Lishimpi K., Farrelly L., Kaganson N., Zumla A., Gillespie S. Co-trimoxazole as prophylaxis against opportunistic infections in HIV-infected Zambian children (CHAP): A double-blind randomised placebo-controlled trial. Lancet. 2004;364:1865–1871. doi: 10.1016/S0140-6736(04)17442-4. [DOI] [PubMed] [Google Scholar]
- 65.Chiyangi H., Muma J.B., Malama S., Manyahi J., Abade A., Kwenda G., Matee M.I. Identification and antimicrobial resistance patterns of bacterial enteropathogens from children aged 0–59 months at the University Teaching Hospital, Lusaka, Zambia: A prospective cross-sectional study. BMC Infect. Dis. 2017;17:117. doi: 10.1186/s12879-017-2232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ministry of Health, Zambia National Formulary Committee . Standard Treatment Guidelines, Essential Medicines List, Essential Laboratory Supplies for Zambia. 5th ed. Ministry of Health, Zambia National Formulary Committee; Lusaka, Zambia: 2020. [(accessed on 20 July 2022)]. Available online: https://www.moh.gov.zm/?wpfb_dl=32. [Google Scholar]
- 67.WHO . WHO List of Critically Important Antimicrobials for Human Medicine (WHO CIA list) World Health Organization; Geneva, Switzerland: 2019. [(accessed on 16 April 2022)]. Available online: https://apps.who.int/iris/handle/10665/325036. [Google Scholar]
- 68.Zhou Y., Zhu X., Hou H., Lu Y., Yu J., Mao L., Mao L., Sun Z. Characteristics of diarrheagenic Escherichia coli among children under 5 years of age with acute diarrhea: A hospital-based study. BMC Infect. Dis. 2018;18:63. doi: 10.1186/s12879-017-2936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saka H.K., Dabo N.T., Muhammad B., García-Soto S., Ugarte-Ruiz M., Alvarez J. Diarrheagenic Escherichia coli pathotypes from children younger than 5 years in Kano State, Nigeria. Front. Public Health. 2019;7:348. doi: 10.3389/fpubh.2019.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Macpherson A.J., Harris N.L. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 71.Byarugaba D. Antimicrobial resistance in developing countries and responsible risk factors. Int. J. Antimicrob. Agents. 2004;24:105–110. doi: 10.1016/j.ijantimicag.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 72.Prestinaci F., Pezzotti P., Pantosti A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health. 2015;109:309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gopal Rao G. Risk factors for the spread of antibiotic-resistant bacteria. Drugs. 1998;55:323–330. doi: 10.2165/00003495-199855030-00001. [DOI] [PubMed] [Google Scholar]
- 74.Shakya P., Barrett P., Diwan V., Marothi Y., Shah H., Chhari N., Tamhankar A.J., Pathak A., Lundborg C.S. Antibiotic resistance among Escherichia coli isolates from stool samples of children aged 3 to 14 years from Ujjain, India. BMC Infect. Dis. 2013;13:477. doi: 10.1186/1471-2334-13-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Graham D.W., Bergeron G., Bourassa M.W., Dickson J., Gomes F., Howe A., Kahn L.H., Morley P.S., Scott H.M., Simjee S. Complexities in understanding antimicrobial resistance across domesticated animal, human, and environmental systems. Ann. N. Y. Acad. Sci. 2019;1441:17–30. doi: 10.1111/nyas.14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rousham E.K., Unicomb L., Islam M.A. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: Integrating behavioural, epidemiological and One Health approaches. Proc. R. Soc. B Biol. Sci. 2018;285:20180332. doi: 10.1098/rspb.2018.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The essential data supporting the reported results are contained in this study. All other supplementary data can be made available on request from the corresponding author.