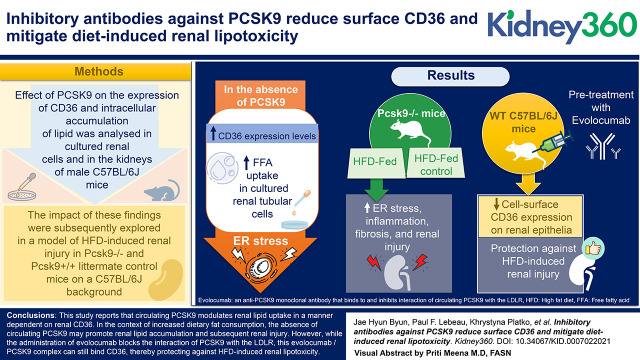
Key Points
PCSK9 protects against diet-induced renal injury in both cultured cells and mice by enhancing the degradation of renal cluster of differentiation 36 (CD36).
PCSK9 mAbs (evolocumab) do not block the ability of PCSK9 to bind to surface CD36, unlike its effects on degrading hepatic LDL receptor.
PCSK9 mAbs (evolocumab) protect against diet-induced renal injury by enhancing the degradation of surface CD36.
Keywords: renal physiology, basic science, cardiovascular disease, CD36, chronic kidney disease, endoplasmic reticulum stress, hypercholesterolemia, lipid accumulation, lipids, monoclonal antibodies, PCSK9, renal injury
Visual Abstract
Abstract
Background
PCSK9 modulates the uptake of circulating lipids through a range of receptors, including the low-density lipoprotein receptor (LDLR) and CD36. In the kidney, CD36 is known to contribute to renal injury through pro-inflammatory and -fibrotic pathways. In this study, we sought to investigate the role of PCSK9 in modulating renal lipid accumulation and injury through CD36 using a high fat diet (HFD)-induced murine model.
Methods
The effect of PCSK9 on the expression of CD36 and intracellular accumulation of lipid was examined in cultured renal cells and in the kidneys of male C57BL/6J mice. The effect of these findings was subsequently explored in a model of HFD-induced renal injury in Pcsk9−/− and Pcsk9+/+ littermate control mice on a C57BL/6J background.
Results
In the absence of PCSK9, we observed heightened CD36 expression levels, which increased free fatty acid (FFA) uptake in cultured renal tubular cells. As a result, PCSK9 deficiency was associated with an increase in long-chain saturated FFA-induced ER stress. Consistent with these observations, Pcsk9−/− mice fed a HFD displayed elevated ER stress, inflammation, fibrosis, and renal injury relative to HFD-fed control mice. In contrast to Pcsk9−/− mice, pretreatment of WT C57BL/6J mice with evolocumab, an anti-PCSK9 monoclonal antibody (mAb) that binds to and inhibits the function of circulating PCSK9, protected against HFD-induced renal injury in association with reducing cell surface CD36 expression on renal epithelia.
Conclusions
We report that circulating PCSK9 modulates renal lipid uptake in a manner dependent on renal CD36. In the context of increased dietary fat consumption, the absence of circulating PCSK9 may promote renal lipid accumulation and subsequent renal injury. However, although the administration of evolocumab blocks the interaction of PCSK9 with the LDLR, this evolocumab/PCSK9 complex can still bind CD36, thereby protecting against HFD-induced renal lipotoxicity.
Introduction
The global prevalence of chronic kidney disease (CKD) is increasing at an alarming rate and is associated with a substantial burden on patients and the health care system (1). The progression of CKD in affected individuals commonly develops as a result of interstitial fibrosis, proteinuria, and tubular atrophy, which compromises the overall filtration capacity of the kidney (2). Dyslipidemia and obesity are considered prominent risk factors in CKD (3–6). Increased free fatty acid (FFA) uptake due to excess consumption of diets rich in fats promotes intrarenal lipid accumulation in several animal models and patients at various stages of CKD (7–10). As a result, excess renal uptake of FFAs damages podocytes, mesangial cells, and proximal tubular epithelial cells through various mechanisms, including increased reactive oxygen species production and lipid peroxidation (11,12). This in turn promotes mitochondrial dysfunction and tissue inflammation, resulting in the formation of glomerular and tubular lesions (13,14).
Among the underlying mechanisms of FFA-induced CKD progression is the induction of endoplasmic reticulum (ER) stress (15). Changes in the lipid composition of the ER membrane antagonizes the sarco/ER Ca2+-ATPase pump, leading to the depletion of ER Ca2+ levels (16). In support of this evidence, intracellular lipid accumulation is associated with ER stress (17–21). Because the chaperones expressed in the ER lumen that function to fold de novo proteins properly are dependent on ER Ca2+ as an essential cofactor, this state of Ca2+ depletion commonly leads to the accumulation and subsequent aggregation of misfolded proteins in the ER.
The ER is responsible for the synthesis and proper folding of secretory, transmembrane, and ER luminal proteins and addresses the accumulation of misfolded protein via a highly conserved signaling cascade known as the unfolded protein response (UPR) (15). Activation of the UPR is dependent on the dissociation of ER chaperones such as the glucose-regulated protein of 78 kDa (GRP78) from three transmembrane sensors in the ER lumen: the activating transcription factor 6 (ATF6), the protein kinase RNA–like ER kinase, and the inositol-requiring enzyme I. Chronic activation of these transmembrane proteins and their downstream signaling pathways result in cellular apoptosis, inflammation, and fibrosis in the context of CKD progression (15,22).
The scavenger receptor known as the cluster of differentiation 36 (CD36) promotes intrarenal lipid accumulation, inflammatory signaling, cellular apoptosis, and profibrotic signaling pathways (23–25). CD36 is a multifunctional receptor that mediates the cellular uptake of long-chain FFAs and oxidized lipoproteins and is abundantly expressed in proximal and distal tubular epithelium, podocytes, mesangial cells, and interstitial macrophages (11–13,26). Previous studies have reported that CD36-dependent pathways can modulate the development of kidney fibrosis. In murine proximal tubular epithelial cells, overexpression of tubular CD36 increases intracellular lipid accumulation and induces the expression of profibrotic genes and markers of UPR activation (24). In podocytes, CD36-dependent uptake of palmitic acid dose dependently increases ER stress, mitochondrial reactive oxygen species production, ATP depletion, and apoptosis (12). Thus, CD36 stands as a key driver of renal damage, and its potential use as a therapeutic target for the management of renal disease has yet to be fully elucidated.
Patients with renal disease also are at a substantially higher risk for atherosclerotic cardiovascular diseases (CVD) (27). In the general population and in CKD patients, circulating LDL cholesterol (LDLc) is a well-known driver of atherosclerotic lesion development and CVD progression. It is well established that circulating PCSK9 binds to and degrades the LDL receptor (LDLR) (28–30), thereby increasing LDLc levels and the risk of CVD. PCSK9 also degrades several other receptors that promote extracellular lipid uptake into tissue such as the LDLR-related protein-1, the very low-density lipoprotein receptor, and CD36 (31,32). Given the abundant expression of CD36 in renal tubular epithelium, deficiency of circulating PCSK9 may affect the expression of this receptor, thereby contributing to renal lipid accumulation and injury via the enhancement of FFA and lipoprotein uptake. In this study, we examined the role of circulating PCSK9 on the expression of renal CD36 and its ability to increase lipid uptake. We also explored the effect of evolocumab—an inhibitory antibody targeting PCSK9—on renal CD36 expression and diet-induced renal lipotoxicity.
Materials and Methods
Cell Culture, Transfections, and FFA Treatments
Human immortalized proximal tubule epithelial (HK-2), hepatocyte (HuH-7), and embryonic kidney (HEK293) cell lines and Sprague–Dawley primary rat mesangial cells were used (passages 6 through 15). All cells were cultured in DMEM (Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS (Sigma–Aldrich, St. Louis, MO), 50 IU/ml of penicillin and 100 μg/ml streptomycin (Sigma–Aldrich), and maintained in 5% CO2 at 37°C. Cells were plated at a confluence of 60% for transfection experiments. The transfection cocktail consisted of the following ratio: 1 µg DNA/3 µl X-tremeGENE HP DNA reagent/100 µl Opti-MEM. The cDNA for human wild-type (WT) PCSK9 and the PCSK9Q152H loss-of-function ER retention variant was cloned into pIRES-EGFP (Clontech Laboratories, Mountain View, CA) that contains a C-terminal V5 epitope, allowing protein detection with a V5 mAb (Invitrogen, Carlsbad, CA), as described (33). Cells were treated with either LDL (Lee Biosolutions, Maryland Heights, OH), oxidized LDL; (oxLDL; Alfa Aesar, Ward Hill, MA), oleate (Alfa Aesar), or palmitate (PA; Sigma–Aldrich) that were conjugated to FA-free BSA. All treatments were carried out overnight for 24 hours unless specified otherwise.
Animal Studies
Six-week-old male Pcsk9−/− mice on a C57BL/6J background and age-matched WT C57BL/6J controls (Jackson Laboratories, Bar Harbor, ME) were placed on either a normal control diet (NCD; n=10) or a HFD (60% fat/Kcal; ENVIGO #TD06414; n=10) for 12 weeks with ad libitum access to food and water. Animals were fasted overnight before euthanasia. Pcsk9−/− mice were a generous gift from Dr. Nabil G. Seidah and were generated as described (34). All animal procedures were approved by the McMaster University Animal Research Ethics Board.
PCSK9 mAbs Administration
Ten-week-old male WT mice on a C57BL/6J background were administered the PCSK9 mAb evolocumab (n=10) or saline (n=10) at 30 mg/kg via orbital injection and euthanized 10 days later. All animal procedures were approved by the McMaster University Animal Research Ethics Board.
Immunoblotting
Total protein lysates were electrophoretically resolved on 7%–10% polyacrylamide gels and transferred onto nitrocellulose membranes. Membranes were then blocked using 5% w/v skimmed milk in Tris-buffered saline containing Tween-20 for 1 hour and incubated with primary antibody overnight for 18 hours at 4°C. To detect bound primary antibodies, horse radish peroxidase–conjugated antibodies were used (goat antirabbit, Bio-Rad Laboratories, Hercules, CA; donkey anti-goat, Santa Cruz Biotechnology, Santa Cruz, CA; goat antimouse, Bio-Rad Laboratories) and developed with a chemiluminescent reagent (FroggaBio, Toronto, Canada). Quantification of immunoblots was assessed after normalization to β-actin (Sigma–Aldrich). See Supplemental Table 1 for a list of the antibodies used for this study.
Real-Time PCR
Total RNA from tissues and cultured cells was isolated using an RNA purification kit according to the manufacturer’s instructions (Thermo Fisher Scientific). A total of 2 µg of RNA was reverse transcribed to cDNA using SuperScript Vilo IV cDNA synthesis kit (Thermo Fisher Scientific). Quantitative RT-PCR (qRT-PCR) assessment of different mRNA species was used in conjunction with FAST SYBR Green (Thermo Fisher Scientific). All primer sequences are listed in Supplemental Table 2.
Immunofluorescent Staining
Cells were fixed using 4% paraformaldehyde and either nonpermeabilized or permeabilized with 0.025% Triton-X in PBS and blocked with 1% BSA for 30 minutes. Cells were then stained with anti-LDLR (catalog no. AF2148; R&D Systems, Minneapolis, MN), anti-CD36 (catalog no. NB400–144; Novus Biologicals, Centennial, CO), or anti-ATF6 (catalog no. NBP1–40256; Novus Biologicals) for 1 hour. After primary antibody incubation, cells were fluorescently labeled with either Alexa 647 (catalog no. 21245; Thermo Fisher Scientific), Alexa 594 (catalog no. A11058, Thermo Fisher Scientific), or Alexa 488 (catalog no. A10468) and DAPI. Slides were mounted using PermaFluor and visualized using the EVOS FL color imaging system.
Oil Red O Staining
Cells were fixed using 4% paraformaldehyde and stained with Oil Red O (ORO; Sigma–Aldrich) for 5 minutes. After washing, cells were counterstained with Gills hematoxylin (no. 5; Sigma–Aldrich) and mounted on glass slides. For quantification purposes, ORO stain was extracted from cells using isopropanol for 20 minutes on an orbital shaker at 37°C. The optical density of ORO-exposed isopropanol extracts was quantified using a spectrophotometer (Molecular Devices, Sunnyvale, CA) at a wavelength of 520 nm. Sections of murine OCT-embedded kidneys 10 µm thick were washed with propylene glycol and stained with ORO for 10 minutes. After staining, tissues were repeatedly washed with propylene glycol and counterstained using alum hematoxylin.
DiI-LDL/oxLDL Uptake Assay
Approximately 3.0 × 104 cells were seeded per well in black clear-bottom 96-well plates for 24 hours before treatment at the respective time points. During the last 5 hours of treatment, cells were exposed to DiI-LDL/oxLDL (10 μg/ml) and then washed with prewarmed (37°C) HBSS containing 20 mM HEPES before analysis. The fluorescence intensity of DiI was subsequently quantified using the SpectraMax GeminiEM fluorescent spectrophotometer (Molecular Devices; ex 554/em 571).
Thioflavin-T/S Staining
Thioflavin staining is commonly used for the visualization of misfolded protein aggregates as a result of ER stress (35). Cells were fixed using 4% paraformaldehyde and stained with Thioflavin-T (Sigma–Aldrich) for 15 minutes at 37°C. For tissue, sections of murine OCT-embedded kidneys 10 µm thick were stained with Thioflavin-S staining for 15 minutes. All images and quantifications were collected using a fluorescent microscope (EVOS FL; Thermo Fisher Scientific).
Trigylceride Quantification
Renal cortical tissue was assessed for triglyceride and cholesterol content using a triglyceride assay kit (Abcam, Cambridge, United Kingdom) and colorimetric cholesterol assay kit (Wako Diagnostics, Mountain View, CA), respectively, according to the manufacturers’ instructions. All data were normalized to tissue weight.
PCSK9 ELISA
Quantification of PCSK9 in media and throughout the in vivo studies was determined using the R&D human and murine Quantikine PCSK9 ELISA kits, respectively.
Cystatin C ELISA
Quantification of serum murine Cystatin C was determined using the Abcam murine Cystatin C ELISA kit as per the manufacturer’s instructions.
Immunohistochemical Staining
Sections 4-µm-thick were deparaffinized, blocked in 5% v/v serum, and subsequently incubated with primary antibodies for 18 hours at 4°C. Sections were incubated with either biotin-labeled secondary antibodies (Vector Laboratories, Burlingame, CA) or Alexa Fluor secondaries (Invitrogen) for fluorescent imaging. Streptavidin-labeled horse radish peroxidase solution (Vector Laboratories) and NovaRed developing solution (Vector Laboratories) were used to visualize the staining. Slides were examined using a Nikon light microscope (model no. DS-Ri2). Relative staining intensity was quantified using ImageJ software (n=10). For Picrosirius Red staining, kidney sections were exposed to saturated picric acid solution and stained with Sirius red F3B (Color Index 35782). Please refer to Supplemental Table 1 for a list of the antibodies used for this study.
Statistical Analyses
Statistical analysis for two-group comparisons was conducted using unpaired Student’s t test, and for multiple group comparison, one-way ANOVA was used. Statistically significant differences were considered at P<0.05. All error bars are represented as SD of the mean.
Results
Secreted PCSK9 Regulates Surface CD36 and LDLR in HK-2 Cells
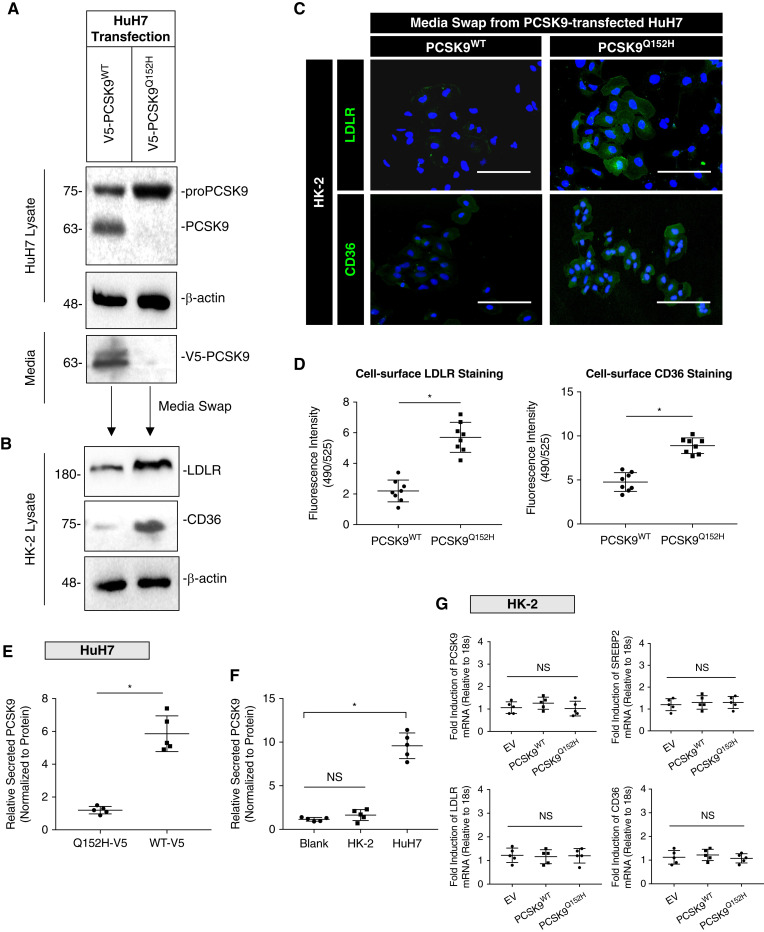
Renal proximal tubular cells express a variety of receptors targeted by PCSK9. Given that PCSK9 is secreted into the blood almost exclusively by the liver (36), we developed an in vitro model to mimic the effect of liver-secreted PCSK9 on HK-2 cells. In these experiments, cultured HuH7 cells were transfected with either WT PCSK9 or the PCSK9Q152H loss-of-function variant that fails to be secreted from cells (37). PCSK9-containing medium from PCSK9WT-transfected cells or control medium from PCSK9Q152H-transfected cells (devoid of secreted PCSK9) was harvested and used to culture HK-2 cells. Western blot and immunofluorescence analysis demonstrated that HK-2 cells exposed to PCSK9-containing medium express reduced levels of LDLR and CD36 (Figure 1, A–C). To confirm these observations, immunofluorescent staining intensity was quantified using ImageJ software (Figure 1D; P<0.05). Secreted PCSK9 was also measured in the medium from HuH7 cells, along with nontransfected HuH7 and HK-2 cells. Using ELISAs, the majority of secreted PCSK9 originated from hepatocytes (Figure 1, E and F; P<0.05). To show that these findings were the direct result of PCSK9, Western blot analysis revealed that the reduction of LDLR and CD36 in HK-2 cells treated with recombinant human PCSK9 (rhPCSK9) were comparable to the media harvested from HuH7 cells overexpressing WT PCSK9 at equal concentrations of PCSK9 (Supplemental Figure 1C; P<0.05). qRT-PCR analysis revealed that relative mRNA transcripts of CD36, sterol regulatory element-binding protein 2 (SREBP2), LDLR, and PCSK9 in HK-2 cells were not significantly affected by the media swap compared with empty vector controls (Figure 1G; NS). Collectively, these data demonstrate that PCSK9 secreted from hepatocytes can downregulate CD36 expression on the surface of renal-derived cells.
Figure 1.
Secreted PCSK9 regulates the LDL receptor (LDLR) and cluster of differentiation 36 (CD36) in cultured renal cells. (A) Whole-cell lysates and media immunoblotted for PCSK9 from HuH7 cells transfected with either PCSK9WT or PCSK9Q152H expression plasmids. (B) Relative protein expression of CD36 and LDLR in whole-cell lysates from HK-2 cells exposed to media harvested from HuH7 cells. (C and D) Immunofluorescence microscopy staining for surface CD36 and LDLR in HK-2 cells post media swap (*P<0.05). (E) Relative secreted PCSK9 measured in HuH7 cells transfected with either PCSK9WT or PCSK9Q152H after 24 hours (*P<0.05). (F) Secreted PCSK9 was also measured in HuH7 and HK-2 cells to confirm that renal cells secrete less PCSK9 relative to hepatocytes in vitro (*P<0.05, NS). (G) mRNA transcript levels of PCSK9, SREBP2, LDLR, and CD36 in HK-2 cells exposed to harvested media from HuH7-transfected cells confirming that modulation of surface receptors on HK-2 cells were through exogenously added PCSK9 from hepatocytes (NS). Data are represented as the mean, and errors are represented as the SD. Differences between groups were determined using t tests or ANOVAs. Scale bars, 200 µm.
PCSK9 Blocks Oleic Acid- and LDL-Induced Lipid Droplet Accumulation in Renal Cells
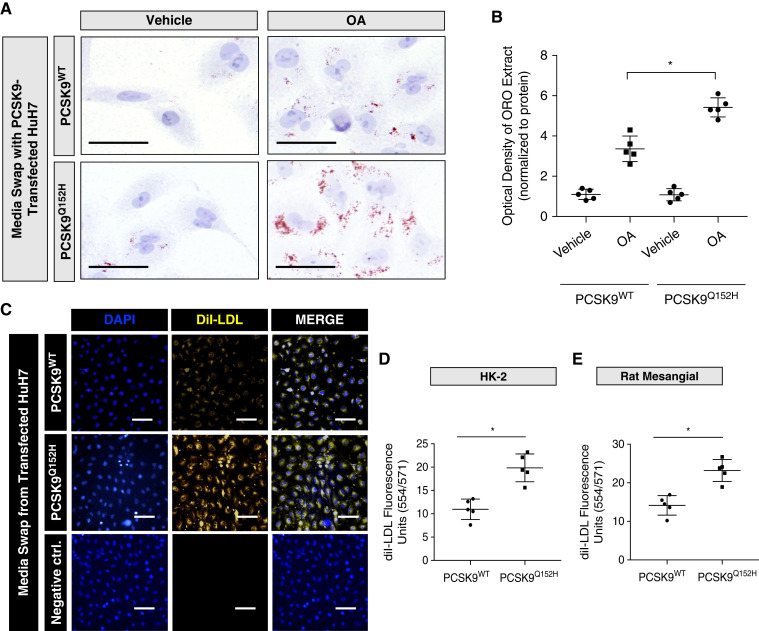
We next examined whether exogenous PCSK9 could regulate lipid uptake in a variety of renal-derived cell lines. HK-2 cells were exposed to medium harvested from PCSK9-transfected HuH7 cells, treated with oleic acid (OA), and stained with the ORO lipid stain. HK-2 cells exposed to medium from WT PCSK9–transfected HuH7 cells exhibited reduced lipid accumulation relative to cells exposed to control medium from the PCSK9Q152H-transfected HuH7 cells (Figure 2A). Consistent with visual observations, densitometric quantification of ORO staining via densitometry of isopropanol extract at 520 nm suggests that PCSK9 blocks OA-induced lipid accumulation in HK-2 cells (Figure 2B; P<0.05). To confirm this observation further, both HK-2 and primary mesangial cells exposed to PCSK9-containing medium were treated with fluorescently labeled DiI-LDL in order to visualize LDL uptake. A reduction in uptake of DiI-labeled LDL was observed in HK-2 cells exposed to PCSK9-containing medium (Figure 2C)—a finding confirmed using quantitative spectrophotometry (Figure 2, D and E; P<0.05). To examine whether the observed difference in lipid accumulation occurred due to exogenous V5-labeled PCSK9, anti-V5 antibodies were added to the media to block the binding of V5-labeled PCSK9 to CD36. Compared with IgG controls, the addition of anti-V5 antibody located on the C-terminus of PCSK9 (33,38) reversed the attenuating effects of lipid accumulation by exogenous V5-PCSK9 in the media (Supplemental Figure 1A; P<0.05), which was also quantified (Supplemental Figure 1B; P<0.05). Altogether, these observations demonstrate that secreted PCSK9 modulates lipid uptake in renal-derived cell lines.
Figure 2.
PCSK9 blocks oleic acid (OA)- and LDL-induced lipid droplet accumulation in renal cells. (A and B) Oil Red O (ORO) staining of HK-2 cells exposed to media harvested from HuH7 cells and treated with OA (200 μM) for 18 hours in the presence or absence of PCSK9 from media (*P<0.05). (C–E) HK-2 and rat mesangial cells were treated with fluorescently labeled DiI-LDL cholesterol and quantified (*P<0.05). Data are represented as the mean, and errors are represented as the SD. Differences between groups were determined using t tests or ANOVAs. Scale bars: (A) 10 µm and (C)100 µm.
PCSK9 Attenuates Lipid Accumulation in Renal-Derived Cells in a Manner Dependent on CD36
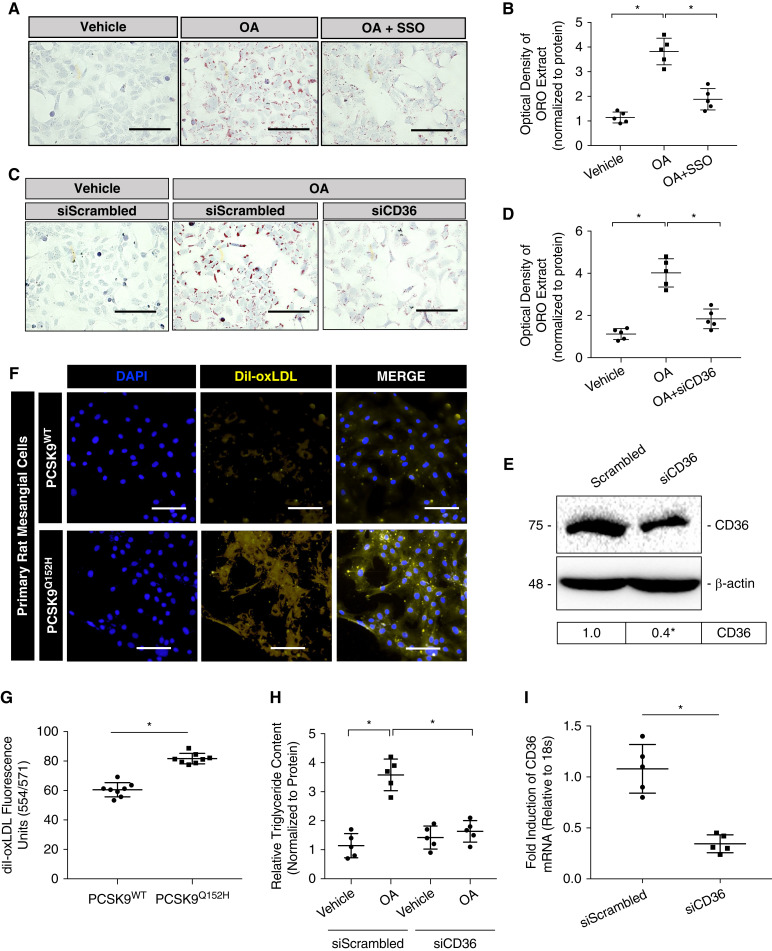
Given that PCSK9 regulates CD36 expression (31) and that CD36 is an established scavenger receptor for FFA in renal cells (11), we examined whether PCSK9 regulates FFA uptake and subsequent accumulation in a manner dependent on CD36. First, ORO staining and quantification was carried out in HK-2 cells treated with sulfo-N-succinimidyl oleate, a well-established blocker of CD36 (Figure 3, A and B; P<0.05) (39). Then, HK-2 cells were treated with OA in the presence of either scrambled small interfering (si)RNA (siScrambled) or siRNA targeted against CD36 (siCD36), pretreated for 18 hours prior. Treatment with siCD36 was able to attenuate OA-induced lipid accumulation (Figure 3, C and D; P<0.05). The effectiveness of siRNA-mediated knockdown of CD36 was assessed via immunoblotting (Figure 3E; P<0.05) and qRT-PCR (Figure 3I; P<0.05). Next, primary rat mesangial cells were treated with DiI-oxLDL for 5 hours. Visualization and fluorescent staining quantification revealed that exogenous PCSK9 was able to attenuate DiI-oxLDL uptake (Figure 3, F and G; P<0.05). Lastly, as ORO staining measures for both esterified cholesterol and triglyceride (40), intracellular triglyceride content was measured in HK-2 cells after CD36 knockdown and OA treatment. Results revealed that knockdown of CD36 expression using siCD36 was able to attenuate relative triglyceride content in HK-2 cells compared with the scrambled siRNA control (Figure 3H; P<0.05). In addition to regulating CD36 expression, these data demonstrate that PCSK9 also regulates the uptake and accumulation of FFA in cultured renal cells.
Figure 3.
PCSK9 regulates lipid accumulation in renal-derived cells in a manner dependent on CD36. (A and B) ORO staining of HK-2 cells treated with OA and or sulfo-N-succinimidyl oleate (10 μM), a well-established blocker of CD36 activity, for 18 hours (*P<0.05). (C and D) The effect of a small interfering RNA (siRNA) targeted against CD36 was also assessed via ORO staining with the treatment of OA in HK-2 cells (*P<0.05). (E) Immunoblot of CD36, confirming knockdown of its expression using siRNA targeted against CD36 (*P<0.05). (F and G) Uptake of fluorescently labeled oxidized LDL (oxLDL) was also measured in HK-2 cells and quantified using a spectrophotometer (*P<0.05). (H) Relative intracellular triglyceride levels measured in HK-2 cells treated with OA with the modulation of CD36 expression (*P<0.05). (I) Knockdown of CD36 via siRNA was also confirmed using qRT-PCR (*P<0.05). Data are represented as the mean, and errors are represented as the SD. Differences between groups were determined using t tests or ANOVAs. Scale bars, 200 µm.
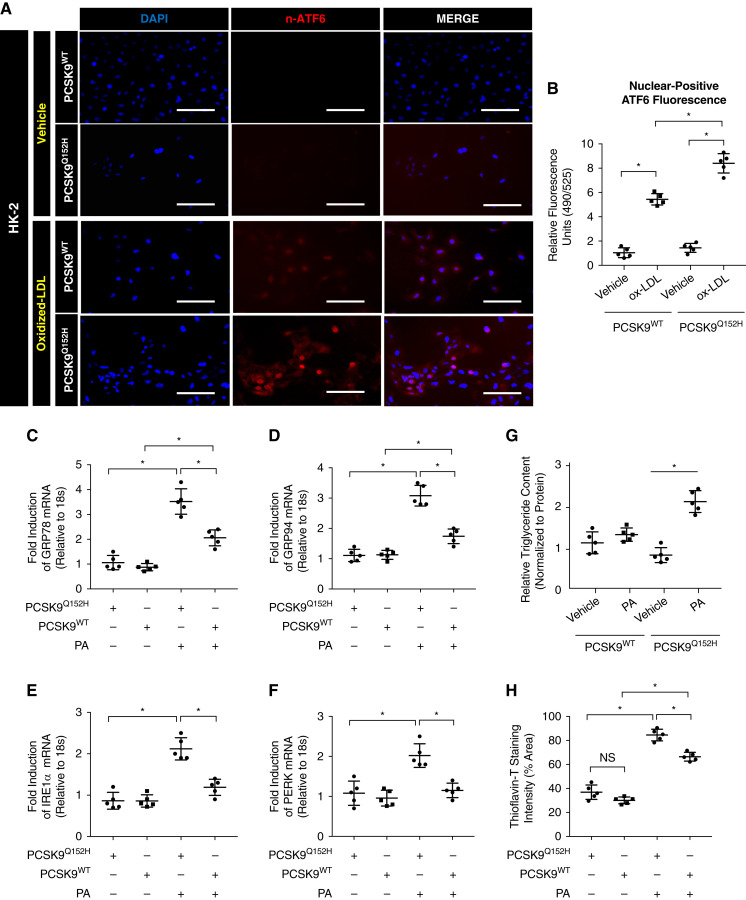
PCSK9 Attenuates PA- and oxLDL-Induced ER Stress In Vitro
CD36 contributes to ER stress in proximal tubular cells in a manner dependent on excess uptake of ER stress-inducing lipoproteins and FFAs, such as oxLDL and PA, respectively (11). Therefore, we next examined whether PCSK9 could protect against ER stress in the presence of oxLDL and PA resulting from the attenuation of CD36 expression. Immunofluorescent staining of the UPR marker nuclear ATF6 (n-ATF6) demonstrated that PCSK9-containing medium reduces oxLDL-induced ER stress (Figure 4A), which was also quantified (Figure 4B; P<0.05). Next, qRT-PCR assessment revealed that PCSK9-containing medium attenuated relative UPR activation induced by PA (Figure 4, C–F; P<0.05). Interestingly, palmitoleic acid (PoA) had no effect on UPR activation in either the presence or absence of HuH7-secreted circulating PCSK9 (Supplemental Figure 2C; NS). To further correlate the lipotoxic effects of PA on the observed UPR activation, relative triglyceride content was measured in HK-2 cells treated with PA in the presence or absence of exogenously added PCSK9. The presence of PCSK9 led to a significant reduction in triglyceride uptake in HK-2 cells with PA (Figure 4G; P<0.05). Lastly, protein aggregate accumulation, a hallmark characteristic of ER stress, was assessed via quantification of live staining in HK-2 cells with Thioflavin-T. Reduced fluorescence in Thioflavin-T staining was observed in HK-2 cells exposed to exogenous PCSK9 treated with PA, indicative of a reduction in misfolded protein aggregates (Figure 4H; P<0.05, NS). Together, these observations suggest that PCSK9 is able to protect against PA and oxLDL-induced ER stress by reducing extracellular FFA uptake.
Figure 4.
PCSK9 protects from palmitate and oxidized LDL-induced endoplasmic reticulum (ER) stress in vitro. (A and B) Immunofluorescence microscopy of HK-2 cells stained for n-ATF6 treated with oxLDL either in the presence or absence of PCSK9 (*P<0.05). (C–F) Quantitative RT-PCR (qRT-PCR) analysis of unfolded protein response activation markers in HK-2 cells (*P<0.05). (G) Relative intracellular triglyceride levels measured in HK-2 cells treated with palmitate (PA) in the presence or absence of PCSK9 (*P<0.05). (H) Quantification of Thioflavin-T fluorescent staining in HK-2 cells treated with PA in media harvested from transfected HuH7 cells (*P<0.05, NS). Data are represented as the mean, and errors are represented as the SD. Differences between groups were determined using t tests or ANOVAs. Scale bars, 200 µm.
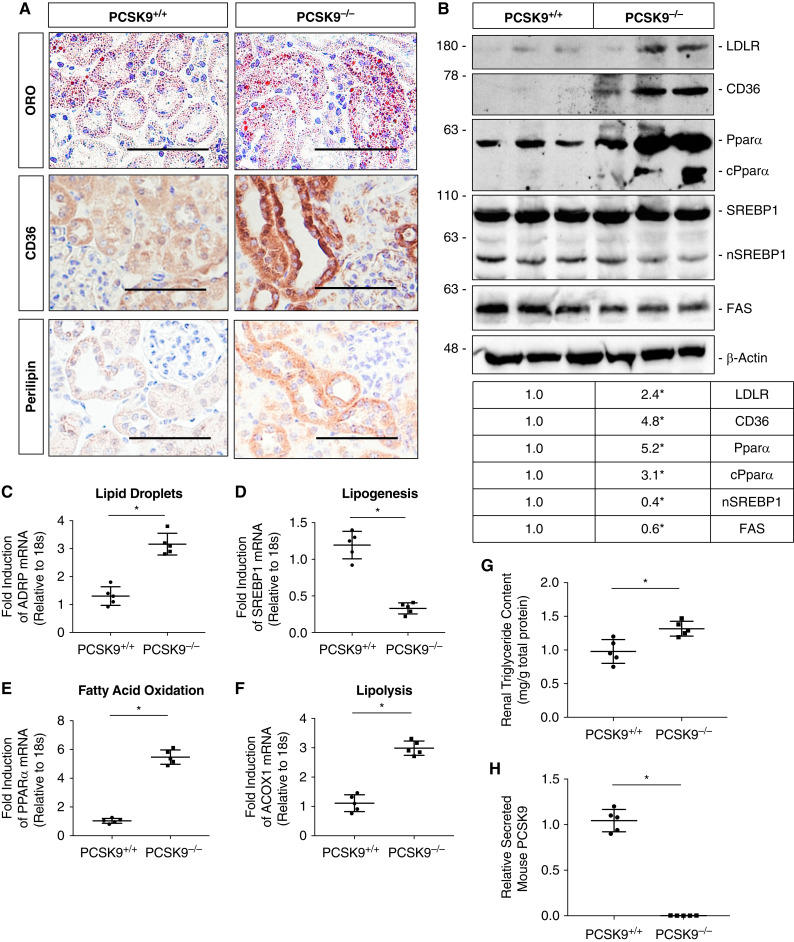
Pcsk9−/− Mice Exhibit Increased Renal Lipid Accumulation
To gain insight into the ability of PCSK9 to modulate renal lipid uptake/accumulation in vivo, male Pcsk9−/− mice and age-matched WT C57BL/6J Pcsk9+/+ controls fed NCD were euthanized at 12 weeks of age. Immunohistochemical (IHC) analysis of the renal cortex revealed that Pcsk9−/− mice exhibited increased tubular lipid accumulation assessed via ORO staining relative to the controls (Figure 5A). This observation was consistent with increased immunostaining for CD36 and the lipid droplet marker, perilipin, staining in Pcsk9−/− mice. Immunoblot analysis also demonstrated that Pcsk9−/− mice had increased protein expression of CD36 and LDLR—receptors known to promote lipid uptake in a variety of tissues, including the kidney (Figure 5B; P<0.05) (23–25). Interestingly, as a compensatory response, Pcsk9−/− mice also exhibited increased β oxidation through increased expression and cleavage of the peroxisome proliferator-activated receptor α. Simultaneously, the nuclear form of sterol regulatory element-binding protein 1 (SREBP1) and FA synthase (FAS) expression were reduced, which are well-established markers of lipogenesis (Figure 5B) (41). Interestingly, there were no significant differences in relative SREBP2 mRNA levels between the two groups (Supplemental Figure 1D; NS). In line with these observations, qRT-PCR analysis also showed a decrease in lipogenesis accompanied by marked increase in FA oxidation and lipolysis (Figure 5, C–F; P<0.05, NS). Renal triglyceride content in the cortex was also significantly higher in Pcsk9−/− mice relative to controls (Figure 5G; P<0.05). As expected, Pcsk9−/− mice exhibited undetectable circulating PCSK9 levels compared with their respective controls (Figure 5H; P<0.05). Similar to our in vitro findings, these data suggest that PCSK9 deficiency increases renal CD36 expression and drives renal lipid levels in vivo.
Figure 5.
Pcsk9−/− mice exhibit increased renal lipid accumulation on a normal control diet (NCD). (A) Sections from the renal cortex in Pcsk9−/− mice were staining for ORO via cryosections and immunostained for CD36 and perilipin via paraffin-embedded sections and used to measure relative lipid accumulation in the renal cortex in vivo. (B) Immunoblot of lysates from the renal cortex in Pcsk9−/− mice and its relative controls for different markers of lipid metabolism (*P<0.05). (C–F) qRT-PCR analysis of different genes involved in lipid droplet formation, lipogenesis, fatty acid oxidation, and lipolysis (*P<0.05, NS). (G) Relative intrarenal triglyceride content assessed in Pcsk9−/− mice relative to controls (*P<0.05). (H) Serum PCSK9 levels were measured via ELISA in Pcsk9−/− mice compared with controls (*P<0.05). Data are represented as the mean, and errors are represented as the SD. Differences between groups were determined using t tests or ANOVAs. Scale bars, 100 µm.
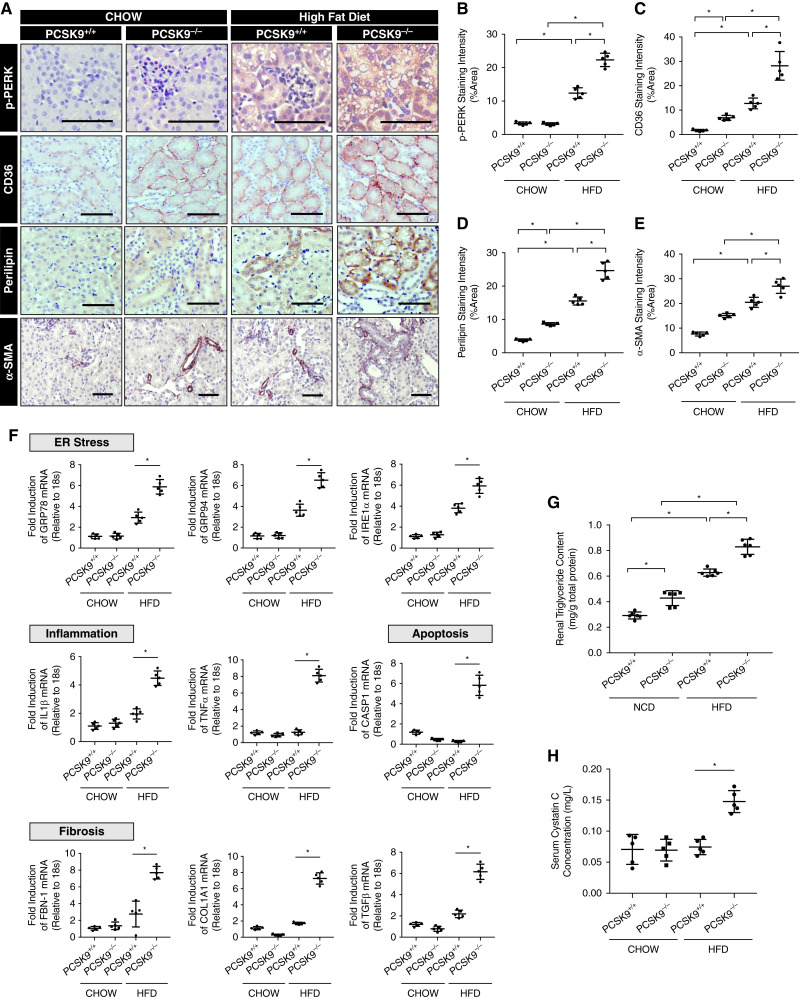
Pcsk9−/− Mice Exhibit Increased Renal ER Stress, Fibrosis, and Inflammation on a HFD
To further investigate the role of PCSK9 on renal lipid accumulation in vivo, 6-week-old male Pcsk9−/− mice and age-matched C57BL/6J controls were fed either a NCD or HFD for 12 weeks before euthanasia. IHC analysis revealed an increase in the expression of the ER stress marker phosphorylated protein kinase RNA–like ER kinase in the kidneys of HFD-fed Pcsk9−/− mice compared with HFD-fed controls (Figure 6, A and B; P<0.05). CD36 and perilipin expression was induced by a HFD and elevated in the Pcsk9−/− mice fed both a NCD and HFD (Figure 6, A, C, and D; P<0.05). In addition, we observed an increase in the expression of the profibrotic marker α-smooth muscle actin (Figure 6, A and E; P<0.05) in Pcsk9−/− mice on a HFD relative to controls. Along with these observations, qRT-PCR also revealed an increase in mRNA abundance of ER stress and inflammatory, apoptotic, and fibrotic markers in Pcsk9−/− mice compared with controls (Figure 6F; P<0.05). In support of these findings, renal triglyceride content was significantly higher in Pcsk9−/− mice on both NCD and HFD (Figure 6G; P<0.05). To assess renal function in these mice, we observed that Pcsk9−/− mice on a HFD exhibited significantly increased serum Cystatin C levels relative to WT controls (Figure 6H; P<0.05). Lastly, Thioflavin-S (35) fluorescence was increased in the renal cortex of HFD-fed Pcsk9−/− mice relative to HFD-fed controls (Supplemental Figure 1E). Altogether, these data suggest that PCSK9 is able to attenuate HFD-induced ER stress by modulating lipid uptake within the cortex of the kidney.
Figure 6.
Pcsk9−/− mice exhibit increased renal ER stress on a high-fat diet (HFD). (A–E) Immunohistochemical (IHC) analysis and quantification of the renal cortex of Pcsk9−/− mice and relevant controls on a HFD for phosphorylated protein kinase RNA–like ER kinase, CD36, perilipin, and α-smooth muscle actin (*P<0.05). (F) qRT-PCR analysis of ER stress, inflammatory, apoptotic, and fibrotic markers in the renal cortex of the mice (*P<0.05). (G) Relative intrarenal triglyceride content assessed in Pcsk9−/− mice fed either a NCD or a HFD relative to controls (*P<0.05). (H) Serum Cystatin C levels were measured and assessed in Pcsk9−/− mice fed either a NCD or a HFD relative to controls (*P<0.05). Data are represented as the mean, and errors are represented as the SD. Differences between groups were determined using t tests or ANOVAs. Scale bars, 100 µm.
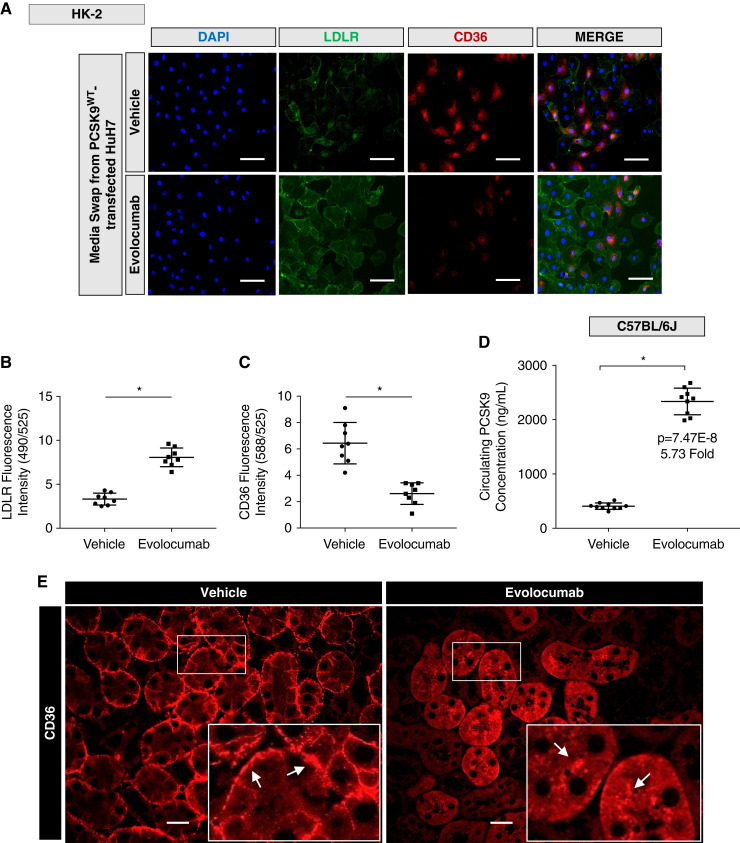
Evolocumab Increases Surface LDLR but Reduces Surface CD36 in Both Cultured Cells and Mice
Evolocumab is a commercially available fully human mAb against PCSK9 that prevents the interaction between circulating PCSK9 and the LDLR (42). Unlike genetic ablation of PCSK9, these anti-PCSK9 antibodies do not affect tissue expression or secretion of PCSK9. Importantly, it is not known whether binding of these anti-PCSK9 antibodies would also block PCSK9 from binding to CD36 because the epitope in PCSK9 that interacts with CD36 is not fully elucidated. To explore the outcome of this important question, cultured HK-2 cells were treated with evolocumab and media swapped with Huh7-incubated media. Immunofluorescence microscopy and subsequent fluorescence quantification revealed that evolocumab was able to reduce surface CD36 fluorescence significantly while upregulating surface LDLR levels (Figure 7, A–C; P<0.05). Consistent with previous reports, ELISA data demonstrate that evolocumab significantly increased circulating PCSK9 levels in mice because circulating PCSK9 is no longer cleared by the LDLR (Figure 7D; P<0.05). Consequently, immunuofluorescent microscopy of evolocumab-treated mice displayed reduced basolateral surface CD36 with a concurrent increased fluorescence localized in the tubules compared with vehicle-treated mice (Figure 7E), suggesting internalization of CD36. In contrast, there was no discernable change in the fluorescence staining of total surface receptor expression of ATP1A1 (Supplemental Figure 2D). Additionally, the administration of evolocumab was confirmed and detected in the renal cortex using a fluorescently labeled anti-human antibody (Supplemental Figure 2E). Lastly, to confirm the established effects of evolocumab administration, we observed that evolocumab (30 mg/kg) upregulated surface hepatic LDLR (Supplemental Figure 2, A and B; P<0.05). Altogether, these findings suggest that the circulating evolocumab/PCSK9 complex increases circulating levels of PCSK9 (likely bound to the antibody) and does not alter the ability of PCSK9 to bind to cell surface CD36, unlike its neutralizing effect against the LDLR.
Figure 7.
Evolocumab increases surface LDLR but reduces surface CD36 in both cultured cells and mice. (A-C) Immunofluorescence microscopy and quantifications of HK-2 cells co-stained for LDLR and CD36 treated with evolocumab exposed to media harvested from HuH7 cells (*P<0.05). (D) Relative secreted PCSK9 measured in wild-type mice treated with either vehicle or evolocumab (*P<0.05). (E) Immunofluorescence microscopy of mice treated with evolocumab stained for CD36 compared with controls. Differences between groups were determined using t tests or ANOVAs. Scale bars: 100 µm (A) and 20 µm (E).
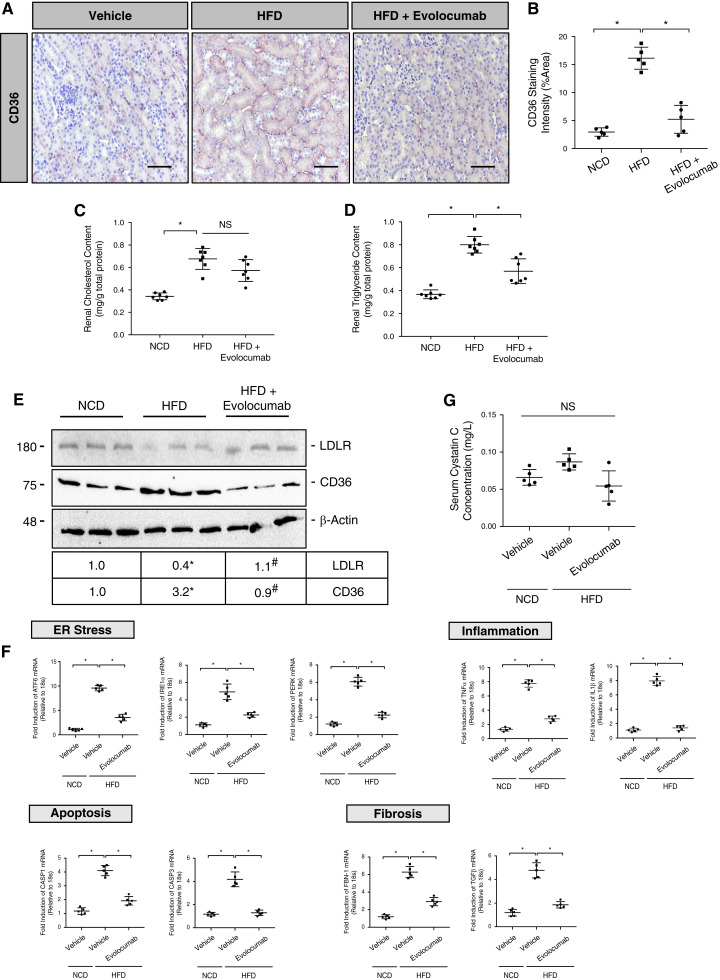
Evolocumab Protects against HFD-Induced Renal Injury by Modulating Surface Expression of CD36 on Renal Epithelia
To assess the effects of evolocumab on diet-induced renal injury via CD36, male C57BL/6J mice (n=10) were placed on a HFD and treated with evolocumab (30 mg/kg) once weekly. IHC analysis demonstrated a significant reduction in HFD-induced surface and total staining of CD36 on renal epithelia (Figure 8, A and B; P<0.05). Interestingly, administration of evolocumab reduced renal triglyceride levels but not cholesterol (Figure 8, C and D; P<0.05, NS). To confirm these findings, evolocumab treatment also reduced HFD-induced CD36 protein levels while upregulating surface LDLR levels (Figure 8E; P<0.05). At the mRNA level, qRT-PCR demonstrated that markers of ER stress, inflammation, apoptosis, and fibrosis were significantly reduced after the administration of evolocumab compared with nontreated controls placed on a HFD (Figure 8F; P<0.05). Consistent with these observations, qRT-PCR analysis demonstrated that evolocumab failed to reduce similar markers of ER stress, inflammation, and fibrosis induced by PA in HK-2 cells deficient in CD36 via siRNA (Supplemental Figure 3, A–C; P<0.05, NS). However, the administration of evolocumab failed to reduce serum Cystatin C levels in mice on a HFD (Figure 8G; NS). Lastly, a visual summary has been supplemented, which compares the difference in mechanism of HFD-induced renal lipotoxicity in both Pcsk9−/− mice and WT mice treated with evolocumab (Supplemental Figure 4, A–C). Altogether, unlike the HFD studies using Pcsk9−/− mice, our findings suggest that evolocumab protects against HFD-induced renal injury by enhancing the degradation of cell surface CD36 while increasing circulating PCSK9 levels independent of its binding to PCSK9.
Figure 8.
Evolocumab protects against HFD-induced renal stress by modulating surface expression of CD36 on renal epithelia. (A and B) IHC analysis and quantification of CD36 in the renal cortex of HFD mice treated with evolocumab compared with relevant controls (*P<0.05). (C and D) Total renal cholesterol and triglyceride content was measured in the renal cortex of these mice (*P<0.05, NS). (E) Immunoblot of CD36 and LDLR protein expression in the renal cortex with quantifications (*P<0.05). (F) qRT-PCR analysis of ER stress, fibrotic, inflammatory, and apoptotic markers in the renal cortex (*P<0.05). (G) Serum Cystatin C levels measured and assessed in evolocumab-treated mice on a HFD relative to NCD controls (NS). Data are represented as the mean, and errors are represented as the SD. Differences between groups were determined using t tests or ANOVAs. Scale bars, 100 µm.
Discussion
The lipid nephrotoxicity hypothesis, first postulated by Moorhead and colleagues in 1982 (43), suggests that dyslipidemia promotes CKD progression by inducing oxidative, inflammatory, and ER stress (7). Since this report, several studies have bridged the gap between renal lipid accumulation and kidney disease using a variety of in vitro and in vivo models and clinical observations (5–8,44). Herein, we report that Pcsk9−/− mice exhibit increased expression of renal tubular CD36 and LDLR, which was associated with increased renal lipid accumulation. Consistent with our observations, PCSK9 is known to interact with surface receptors expressed on renal epithelia, although not to the same extent as the liver (45). Previous studies have highlighted the pathologic role of CD36 in promoting intrarenal oxidative and ER stress by driving the uptake of modified lipoproteins and FFAs. Given that HFD is reported to induce renal ER stress, inflammation, and fibrosis (43, 46), we sought to investigate the effect of a HFD challenge in the context of elevated renal CD36 expression occurring as a result of PCSK9 deficiency. Throughout this study, we have observed that PCSK9 may have a protective effect against HFD-induced renal stress, inflammation, fibrosis, and apoptosis by modulating the levels of receptors known to promote lipid uptake from the circulation.
PCSK9 is primarily expressed in the liver, the small intestine, and the kidney (33). Despite its presence in the kidney, little is currently known about its potential role in kidney function and renal disease. Preliminary studies demonstrate that circulating PCSK9 is significantly elevated in multiple in vivo models of renal pathology (47,48). Recent studies have also shown a connection between circulating PCSK9 and its role in nephrotic syndrome (49). The primary pathology associated with nephrotic syndrome is damage to the glomeruli and podocytes. Patients with this disease are also at an increased risk of atherosclerosis and thromboembolism due to a well-established dyslipidemia observed in most patients with CKD (50). Haas et al. demonstrated that podocyte damage-induced nephrotic syndrome increases plasma PCSK9 levels and leads to dyslipidemia that was markedly attenuated in PCSK9 knockout mice (49). The upregulation of circulating PCSK9 in multiple pathophysiologic conditions is also consistent with our previous findings that diet-induced hepatic steatosis increases plasma PCSK9 concentrations (48). In other studies, Zhang et al. further strengthened the connection between PCSK9 and lipid-induced renal injury, reporting that inflammation-induced lipid deposition in the kidney was induced by primarily downregulating renal PCSK9 levels (51). However, as PCSK9 binds to circulating LDLc, the causal association of increased PCSK9 and dyslipidemia in renal disease remains unclear.
Accumulating evidence suggest that inhibition of PCSK9 has a protective effect against dyslipidemia in patients with renal disease (52–54). As circulating PCSK9 concentrations positively correlate with plasma cholesterol levels, these important findings highlight the role of PCSK9 as a driver of dyslipidemia in patients with CKD (55). In our studies, we demonstrate for the first time that the genetic ablation of PCSK9 in mice promotes renal lipid deposition, likely occurring as a result of increased expression in receptors known to endocytose lipids from the circulation, such as CD36 and the LDLR. Accordingly, PCSK9 deficiency further exacerbated HFD-induced renal ER stress, inflammation, and fibrosis. In addition, we have previously reported that these Pcsk9−/− mice also display increased insulin resistance and circulating glucose levels in HFD conditions (45). These observations therefore suggest that PCSK9 may play an important role in regulating cholesterol and triglyceride levels in circulation and in target tissues via CD36. As such, it is crucial that future studies further investigate the influence of circulating PCSK9 on diet-induced renal injury by placing CD36−/− mice on a HFD and assessing their relative renal function. Additionally, it is worth considering in later studies whether the paracrine or autocrine effects of secreted PCSK9 by renal cells may contribute to this homeostatic effect. Although our findings are consistent with several others regarding the pathogenic role of renal CD36 (11–13,24–26), Garbacz et al. reported that hepatic overexpression of CD36 was surprisingly able to attenuate HFD-induced hepatic steatosis and insulin resistance (56). These findings warrant further investigation because the expression of major genes involved in lipogenesis and FA oxidation in their study was not significantly different between HFD-fed transgenic CD36 mice and WT controls.
Clinically, mAbs against PCSK9 approved by the Food and Drug Administration, such as alirocumab and evolocumab, bind to the EGFa-binding domain of PCSK9 in order to prevent the interaction of PCSK9 with the LDLR (36). Consequently, these mAbs increase circulating PCSK9 levels seven-fold as a result of the reduced rate of LDLR-mediated clearance of PCSK9 from the circulation (57). Additionally, familial hypercholesterolemic patients with loss-of-function mutations in the LDLR also exhibit elevated levels of circulating PCSK9 (58). As such, our findings suggest that using Pcsk9−/− mice, which are devoid of circulating PCSK9 levels, lower LDLc via a fundamentally different mechanism than evolocumab. As a result, we investigated whether the compensatory increase in circulating PCSK9 by treating mice with evolocumab could enhance the degradation of renal CD36 and protect against lipid-induced renal injury. Additionally, the epitope binding domain in which PCSK9 interacts with CD36 currently remains unclear and may potentially differ from the LDLR. As such, we now report for the first time that evolocumab is able to reduce surface expression of CD36 on the renal epithelia. These data strongly suggest that (1) despite the ability of this circulating evolocumab/PCSK9 complex to mitigate the interaction of PCSK9 with the LDLR, it does not affect its binding to cell surface CD36, and (2) the binding domains in which PCSK9 uses to interact and degrade both the LDLR and CD36 are distinct. Thus, a reduction in surface CD36 expression on the basolateral epithelia (59), as a result of exposure to the circulating evolocumab-PCSK9 complex, protects against diet-induced renal ER stress, inflammation, fibrosis, and apoptosis. Hence, the pathophysiologic changes that we observed in Pcsk9−/− mice challenged on a HFD do not accurately represent the additional clinical benefit when compared with PCSK9 mAb therapy. We have supplemented a visual diagram comparing both HFD models to clarify our findings reported from this study.
Since the successful introduction of PCSK9 mAb therapy to patients, there has been a rapid development focused on generating new approaches at reducing circulating PCSK9 levels. One such approach is inclisiran, an siRNA targeted against PCSK9, which has recently undergone phase 3 clinical trials (60). Although the siRNA approach has demonstrated great efficacy in reducing circulating LDLc, its potential effects on increasing surface CD36 levels on hepatocytes has not yet been investigated. This is akin to our findings when comparing the Pcsk9−/− mice and evolocumab-treated mice on a HFD; although both approaches may significantly lower LDLc levels, the specific mechanism in which it lowers LDLc may display significantly different off-target effects due to the ability of PCSK9 to modulate a wide range of receptors that play a role beyond LDLc regulation. Herein, our data provide a novel yet crucial insight, suggesting that the administration of PCSK9 mAbs may have additional clinical benefits in alleviating diet-induced renal stress and injury.
Overall, we report for the first time that PCSK9 has a direct effect on renal function beyond hepatic lipid homeostasis by modulating the LDLR and CD36 on renal epithelia. Given this effect, we observed that in turn PCSK9 is able to attenuate CD36-driven ER stress, inflammation, and fibrosis in the context of diet-induced renal disease. Data demonstrating the extent to which the observed renal injury in Pcsk9−/− mice occurs as a result of excess diet-induced lipid uptake represent a limitation of this study. Recently, we have identified a mechanism in which the loss-of-function PCSK9Q152H variant acts as a co-chaperone within liver hepatocytes (61) and in kidney tubules (Byun and Austin, unpublished data), by stabilizing the protein levels of several critical ER chaperones, including GRP78 and GRP94. This raises the question as to whether the observed increased ER stress in HFD-fed Pcsk9−/− may have occurred as a result of (1) increased lipid uptake and (2) increased sensitivity to ER stress due to a lack of ER stress-response chaperones expressed in the kidney. Furthermore, our HFD murine model may portray significantly different lipid metabolic processes in comparison with other models such as in CD36−/−, Lpl−/−, and MTTP−/− mice (62). To clarify these findings further, future studies will focus on determining whether intracellular or extracellular expression of PCSK9 acts to mitigate ER stress in CKD, and to what extent the clinically approved mAbs against PCSK9 protect against renal stress through cell surface CD36 degradation.
Disclosures
R.C. Austin reports being an editorial board member for the Journal of Biological Chemistry. All remaining authors have nothing to disclose.
Funding
This work was supported in part by research grants to R.C. Austin from the Canadian Institutes of Health Research (FRN173520) and to N.G. Seidah from the CIHR Foundation (grant 148363) and Canada Research Chair (216684 and 231335). Financial support from the Research Institute of St. Joe’s Hamilton and Amgen Canada is acknowledged. R.C. Austin is a Career Investigator of the Heart and Stroke Foundation of Ontario and holds the Amgen Canada Research Chair in the Division of Nephrology at St. Joseph’s Healthcare and McMaster University.
Footnotes
See related editorial, “The Many Lives of PCSK9: Therapeutic Implications,” on pages 1296–1298.
Author Contributions
R.C. Austin was responsible for resources; R.C. Austin and J.H. Byun wrote the original draft of the manuscript; R.C. Austin, J.H. Byun, R.E. Carlisle, J. Chen, J.G. Dickhout, M. Faiyaz, S.A. Igdoura, J.C. Krepinsky, P.F. Lebeau, E.G. Lynn, M.E. MacDonald, Y. Makda, K. Platko, N.G. Seidah, and T. Yousof reviewed and edited the manuscript; R.C. Austin, J.H. Byun, R.E. Carlisle, M. Faiyaz, S.A. Igdoura, J.C. Krepinsky, P.F. Lebeau, E.G. Lynn, M.E. MacDonald, Y. Makda, K. Platko, N.G. Seidah, and F. Weaver were responsible for the formal analysis; R.C. Austin, J.H. Byun, J.G. Dickhout, S.A. Igdoura, J.C. Krepinsky, P.F. Lebeau, K. Platko, and N.G. Seidah were responsible for the investigation; R.C. Austin, J.H. Byun, J.G. Dickhout, J.C. Krepinsky, E.G. Lynn, and N.G. Seidah were responsible for supervision; R.C. Austin, J.H. Byun, J.G. Dickhout, P.F. Lebeau, K. Platko, and N.G. Seidah were responsible for conceptualization; R.C. Austin, J.H. Byun, P.F. Lebeau, K. Platko, and N.G. Seidah were responsible for project administration; R.C. Austin, J.G. Dickhout, and N.G. Seidah were responsible for funding acquisition; J.H. Byun, R.E. Carlisle, J. Chen, M. Faiyaz, S.A. Igdoura, P.F. Lebeau, M.E. MacDonald, K. Platko, N.G. Seidah, and F. Weaver were responsible for data curation; J.H. Byun and P.F. Lebeau were responsible for the methodology; and J.H. Byun and T. Yousof were responsible for validation.
Supplemental Material
This article contains supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0007022021/-/DCSupplemental.
Antibodies used for immunoblotting and immunohistochemical analysis. Download Supplemental Table 1, PDF file, 8.8 MB (8.8MB, pdf) .
Primers used for quantitative RT-PCR. Download Supplemental Table 2, PDF file, 8.8 MB (8.8MB, pdf) .
Staining and quantification of HK-2 cells, Immunoblot of LDLR and CD36, Transcript levels of SREBP2, and Thioflavin-S staining in the renal cortex. Download Supplemental Figure 1, PDF file, 8.8 MB (8.8MB, pdf) .
Immunohistochemical analysis and quantification of LDLR, qRT-PCR analysis of ER stress markers, Immunofluorescence microscopy of ATP1A1, and Immunofluorescence microscopy co-stained for CD36 and antihuman antibody. Download Supplemental Figure 2, PDF file, 8.8 MB (8.8MB, pdf) .
qRT-PCR analysis of ER stress, inflammatory, and fibrotic markers in HK-2 cells. . Download Supplemental Figure 3, PDF file, 8.8 MB (8.8MB, pdf) .
Visual summary outlining the underlying mechanisms of HFD-induced renal lipotoxicity reported within this study. Download Supplemental Figure 4, PDF file, 8.8 MB (8.8MB, pdf) .
References
- 1.Hill NR, Fatoba ST, Oke JL, Hirst JA, O’Callaghan CA, Lasserson DS, Hobbs FDR: Global prevalence of chronic kidney disease—A systematic review and meta-analysis. PLoS One 11: e0158765, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Metcalfe W: How does early chronic kidney disease progress? A background paper prepared for the UK Consensus Conference on early chronic kidney disease. Nephrol Dial Transplant 22: ix26–ix30, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Bigot A, Gusto G, Copin N, Sautenet B, Lantieri O, Halimi J-M: Body mass index, fatty liver index and other metabolic disturbances differentially affect albuminuria and glomerular filtration rate in the general population. J Diabetes Metab 05: 1000387, 2014 [Google Scholar]
- 4.Singh GM, Danaei G, Farzadfar F, Stevens GA, Woodward M, Wormser D, Kaptoge S, Whitlock G, Qiao Q, Lewington S, Di Angelantonio E, Vander Hoorn S, Lawes CM, Ali MK, Mozaffarian D, Ezzati M; Global Burden of Metabolic Risk Factors of Chronic Diseases Collaborating Group; Asia-Pacific Cohort Studies Collaboration (APCSC); Diabetes Epidemiology: Collaborative analysis of Diagnostic criteria in Europe (DECODE); Emerging Risk Factor Collaboration (ERFC); Prospective Studies Collaboration (PSC) : The age-specific quantitative effects of metabolic risk factors on cardiovascular diseases and diabetes: A pooled analysis. PLoS One 8: e65174, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Afshin A, Forouzanfar MH, Reitsma MB, Sur P, Estep K, Lee A, Marczak L, Mokdad AH, Moradi-Lakeh M, Naghavi M, Salama JS, Vos T, Abate KH, Abbafati C, Ahmed MB, Al-Aly Z, Alkerwi A, Al-Raddadi R, Amare AT, Amberbir A, Amegah AK, Amini E, Amrock SM, Anjana RM, Ärnlöv J, Asayesh H, Banerjee A, Barac A, Baye E, Bennett DA, Beyene AS, Biadgilign S, Biryukov S, Bjertness E, Boneya DJ, Campos-Nonato I, Carrero JJ, Cecilio P, Cercy K, Ciobanu LG, Cornaby L, Damtew SA, Dandona L, Dandona R, Dharmaratne SD, Duncan BB, Eshrati B, Esteghamati A, Feigin VL, Fernandes JC, Fürst T, Gebrehiwot TT, Gold A, Gona PN, Goto A, Habtewold TD, Hadush KT, Hafezi-Nejad N, Hay SI, Horino M, Islami F, Kamal R, Kasaeian A, Katikireddi SV, Kengne AP, Kesavachandran CN, Khader YS, Khang YH, Khubchandani J, Kim D, Kim YJ, Kinfu Y, Kosen S, Ku T, Defo BK, Kumar GA, Larson HJ, Leinsalu M, Liang X, Lim SS, Liu P, Lopez AD, Lozano R, Majeed A, Malekzadeh R, Malta DC, Mazidi M, McAlinden C, McGarvey ST, Mengistu DT, Mensah GA, Mensink GBM, Mezgebe HB, Mirrakhimov EM, Mueller UO, Noubiap JJ, Obermeyer CM, Ogbo FA, Owolabi MO, Patton GC, Pourmalek F, Qorbani M, Rafay A, Rai RK, Ranabhat CL, Reinig N, Safiri S, Salomon JA, Sanabria JR, Santos IS, Sartorius B, Sawhney M, Schmidhuber J, Schutte AE, Schmidt MI, Sepanlou SG, Shamsizadeh M, Sheikhbahaei S, Shin MJ, Shiri R, Shiue I, Roba HS, Silva DAS, Silverberg JI, Singh JA, Stranges S, Swaminathan S, Tabarés-Seisdedos R, Tadese F, Tedla BA, Tegegne BS, Terkawi AS, Thakur JS, Tonelli M, Topor-Madry R, Tyrovolas S, Ukwaja KN, Uthman OA, Vaezghasemi M, Vasankari T, Vlassov VV, Vollset SE, Weiderpass E, Werdecker A, Wesana J, Westerman R, Yano Y, Yonemoto N, Yonga G, Zaidi Z, Zenebe ZM, Zipkin B, Murray CJL; GBD 2015 Obesity Collaborators : Health effects of overweight and obesity in 195 countries over 25 years. N Engl J Med 377: 13–27, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SM, Park M, Yoon H-J: Association of body mass index with estimated glomerular filtration rate and incident proteinuria. In: Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies, pp 587–590, 2018
- 7.Kuwahara S, Hosojima M, Kaneko R, Aoki H, Nakano D, Sasagawa T, Kabasawa H, Kaseda R, Yasukawa R, Ishikawa T, Suzuki A, Sato H, Kageyama S, Tanaka T, Kitamura N, Narita I, Komatsu M, Nishiyama A, Saito A: Megalin-mediated tubuloglomerular alterations in high-fat diet–induced kidney disease. JAm Soc Nephrol 27: 1996–2008, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aukema HM: Lipids in chronic kidney disease: Alterations and interventions. Lipid Technol 25: 207–209, 2013 [Google Scholar]
- 9.Martino-Costa AL, Malhão F, Lopes C, Dias-Pereira P: Renal interstitial lipid accumulation in cats with chronic kidney disease. J Comp Pathol 157: 75–79, 2017 [DOI] [PubMed] [Google Scholar]
- 10.Kume S, Uzu T, Araki S, Sugimoto T, Isshiki K, Chin-Kanasaki M, Sakaguchi M, Kubota N, Terauchi Y, Kadowaki T, Haneda M, Kashiwagi A, Koya D: Role of altered renal lipid metabolism in the development of renal injury induced by a high-fat diet. J Am Soc Nephrol 18: 2715–2723, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Hua W, Huang HZ, Tan LT, Wan JM, Gui HB, Zhao L, Ruan XZ, Chen XM, Du XG: CD36 mediated fatty acid-induced podocyte apoptosis via oxidative stress. PLoS One 10: e0127507, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruan XZ, Varghese Z, Powis SH, Moorhead JF: Human mesangial cells express inducible macrophage scavenger receptor. Kidney Int 56: 440–451, 1999 [DOI] [PubMed] [Google Scholar]
- 13.Yang P, Xiao Y, Luo X, Zhao Y, Zhao L, Wang Y, Wu T, Wei L, Chen Y: Inflammatory stress promotes the development of obesity-related chronic kidney disease via CD36 in mice. J Lipid Res 58: 1417–1427, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries APJ, Ruggenenti P, Ruan XZ, Praga M, Cruzado JM, Bajema IM, D’Agati VD, Lamb HJ, Pongrac Barlovic D, Hojs R, Abbate M, Rodriquez R, Mogensen CE, Porrini E; ERA-EDTA Working Group Diabesity : Fatty kidney: Emerging role of ectopic lipid in obesity-related renal disease. Lancet Diabetes Endocrinol 2: 417–426, 2014 [DOI] [PubMed] [Google Scholar]
- 15.Inagi R, Ishimoto Y, Nangaku M: Proteostasis in endoplasmic reticulum—New mechanisms in kidney disease. Nat Rev Nephrol 10: 369–378, 2014 [DOI] [PubMed] [Google Scholar]
- 16.Fu S, Yang L, Li P, Hofmann O, Dicker L, Hide W, Lin X, Watkins SM, Ivanov AR, Hotamisligil GS: Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature 473: 528–531, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, Guan Y, Loor JJ, Sha X, Coleman DN, Zhang C, Du X, Shi Z, Li X, Wang Z, Liu G, Li X: Fatty acid-induced endoplasmic reticulum stress promoted lipid accumulation in calf hepatocytes, and endoplasmic reticulum stress existed in the liver of severe fatty liver cows. J Dairy Sci 102: 7359–7370, 2019 [DOI] [PubMed] [Google Scholar]
- 18.Zhang K, Wang S, Malhotra J, Hassler JR, Back SH, Wang G, Chang L, Xu W, Miao H, Leonardi R, Chen YE, Jackowski S, Kaufman RJ: The unfolded protein response transducer IRE1α prevents ER stress-induced hepatic steatosis. EMBO J 30: 1357–1375, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Kaufman RJ: The role of ER stress in lipid metabolism and lipotoxicity. J Lipid Res 57: 1329–1338, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Meng Q, Xiao F, Chen S, Du Y, Yu J, Wang C, Guo F: ATF4 deficiency protects mice from high-carbohydrate-diet-induced liver steatosis. Biochem J 438: 283–289, 2011 [DOI] [PubMed] [Google Scholar]
- 21.Kharroubi I, Ladrière L, Cardozo AK, Dogusan Z, Cnop M, Eizirik DL: Free fatty acids and cytokines induce pancreatic β-cell apoptosis by different mechanisms: Role of nuclear factor-kappaB and endoplasmic reticulum stress. Endocrinology 145: 5087–5096, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Dickhout JG, Carlisle RE, Austin RC: Interrelationship between cardiac hypertrophy, heart failure, and chronic kidney disease: Endoplasmic reticulum stress as a mediator of pathogenesis. Circ Res 108: 629–642, 2011 [DOI] [PubMed] [Google Scholar]
- 23.Kennedy DJ, Chen Y, Huang W, Viterna J, Liu J, Westfall K, Tian J, Bartlett DJ, Tang WHW, Xie Z, Shapiro JI, Silverstein RL: CD36 and Na/K-ATPase-α1 form a proinflammatory signaling loop in kidney. Hypertension 61: 216–224, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Susztak K, Ciccone E, McCue P, Sharma K, Böttinger EP: Multiple metabolic hits converge on CD36 as novel mediator of tubular epithelial apoptosis in diabetic nephropathy. PLoS Med 2: e45, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamura DM, López-Guisa JM, Koelsch K, Collins S, Eddy AA: Atherogenic scavenger receptor modulation in the tubulointerstitium in response to chronic renal injury. Am J Physiol Renal Physiol 293: F575–F585, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Zoccali C: Traditional and emerging cardiovascular and renal risk factors: An epidemiologic perspective. Kidney Int 70: 26–33, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Goldsmith DJ: Cardiovascular disease and chronic kidney disease. In: Oxford Textbook of Clinical Nephrology, 4th Ed., edited by Turner N, Lameire N, Goldsmith DJ, Winearls CG, Himmelfarb J, Remuzzi G, Edinburgh, UK, Oxford University Press, 2015, pp 1–19 [Google Scholar]
- 28.Benjannet S, Rhainds D, Essalmani R, Mayne J, Wickham L, Jin W, Asselin M-C, Hamelin J, Varret M, Allard D, Trillard M, Abifadel M, Tebon A, Attie AD, Rader DJ, Boileau C, Brissette L, Chrétien M, Prat A, Seidah NG: NARC-1/PCSK9 and its natural mutants: Zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J Biol Chem 279: 48865–48875, 2004 [DOI] [PubMed] [Google Scholar]
- 29.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH: Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet 37: 161–165, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Timms KM, Wagner S, Samuels ME, Forbey K, Goldfine H, Jammulapati S, Skolnick MH, Hopkins PN, Hunt SC, Shattuck DM: A mutation in PCSK9 causing autosomal-dominant hypercholesterolemia in a Utah pedigree. Hum Genet 114: 349–353, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Poirier S, Mayer G, Benjannet S, Bergeron E, Marcinkiewicz J, Nassoury N, Mayer H, Nimpf J, Prat A, Seidah NG: The proprotein convertase PCSK9 induces the degradation of low density lipoprotein receptor (LDLR) and its closest family members VLDLR and ApoER2. J Biol Chem 283: 2363–2372, 2008 [DOI] [PubMed] [Google Scholar]
- 32.Demers A, Samami S, Lauzier B, Des Rosiers C, Ngo Sock ET, Ong H, Mayer G: PCSK9 induces CD36 degradation and affects long-chain fatty acid uptake and triglyceride metabolism in adipocytes and in mouse liver. Arterioscler Thromb Vasc Biol 35: 2517–2525, 2015 [DOI] [PubMed] [Google Scholar]
- 33.Seidah NG, Benjannet S, Wickham L, Marcinkiewicz J, Jasmin SB, Stifani S, Basak A, Prat A, Chretien M: The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): Liver regeneration and neuronal differentiation. Proc Natl Acad Sci U S A 100: 928–933, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaid A, Roubtsova A, Essalmani R, Marcinkiewicz J, Chamberland A, Hamelin J, Tremblay M, Jacques H, Jin W, Davignon J, Seidah NG, Prat A: Proprotein convertase subtilisin/kexin type 9 (PCSK9): Hepatocyte-specific low-density lipoprotein receptor degradation and critical role in mouse liver regeneration. Hepatology 48: 646–654, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Bussière T, Bard F, Barbour R, Grajeda H, Guido T, Khan K, Schenk D, Games D, Seubert P, Buttini M: Morphological characterization of Thioflavin-S-positive amyloid plaques in transgenic Alzheimer mice and effect of passive Abeta immunotherapy on their clearance. Am J Pathol 165: 987–995, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roubtsova A, Munkonda MN, Awan Z, Marcinkiewicz J, Chamberland A, Lazure C, Cianflone K, Seidah NG, Prat A: Circulating proprotein convertase subtilisin/kexin 9 (PCSK9) regulates VLDLR protein and triglyceride accumulation in visceral adipose tissue. Arterioscler Thromb Vasc Biol 31: 785–791, 2011 [DOI] [PubMed] [Google Scholar]
- 37.Mayne J, Dewpura T, Raymond A, Bernier L, Cousins M, Ooi TC, Davignon J, Seidah NG, Mbikay M, Chrétien M: Novel loss-of-function PCSK9 variant is associated with low plasma LDL cholesterol in a French-Canadian family and with impaired processing and secretion in cell culture. Clin Chem 57: 1415–1423, 2011 [DOI] [PubMed] [Google Scholar]
- 38.Tveten K, Holla OL, Cameron J, Strøm TB, Berge KE, Laerdahl JK, Leren TP: Interaction between the ligand-binding domain of the LDL receptor and the C-terminal domain of PCSK9 is required for PCSK9 to remain bound to the LDL receptor during endosomal acidification. Hum Mol Genet 21: 1402–1409, 2012 [DOI] [PubMed] [Google Scholar]
- 39.Seidah NG: The PCSK9 revolution and the potential of PCSK9-based therapies to reduce LDL-cholesterol. Glob Cardiol Sci Pract 2017: e201702, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kruth HS: Localization of unesterified cholesterol in human atherosclerotic lesions. Demonstration of filipin-positive, oil-red-O-negative particles. Am J Pathol 114: 201–208, 1984 [PMC free article] [PubMed] [Google Scholar]
- 41.Lebeau PF, Byun JH, Platko K, MacDonald ME, Poon SV, Faiyaz M, Seidah NG, Austin RC: Diet-induced hepatic steatosis abrogates cell-surface LDLR by inducing de novo PCSK9 expression in mice. J Biol Chem 294: 9037–9047, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lebeau PF, Platko K, Byun JH, Makda Y, Austin RC: The emerging roles of intracellular PCSK9 and their implications in endoplasmic reticulum stress and metabolic diseases. Metabolites 12: 215, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moorhead JF, Chan MK, El-Nahas M, Varghese Z: Lipid nephrotoxicity in chronic progressive glomerular and tubulo-interstitial disease. Lancet 2: 1309–1311, 1982 [DOI] [PubMed] [Google Scholar]
- 44.Schmidt RJ, Beyer TP, Bensch WR, Qian Y-W, Lin A, Kowala M, Alborn WE, Konrad RJ, Cao G: Secreted proprotein convertase subtilisin/kexin type 9 reduces both hepatic and extrahepatic low-density lipoprotein receptors in vivo. Biochem Biophys Res Commun 370: 634–640, 2008 [DOI] [PubMed] [Google Scholar]
- 45.Lebeau PF, Byun JH, Platko K, Al-Hashimi AA, Lhoták Š, MacDonald ME, Mejia-Benitez A, Prat A, Igdoura SA, Trigatti B, Maclean KN, Seidah NG, Austin RC: PCSK9 knockout exacerbates diet-induced non-alcoholic steatohepatitis, fibrosis and liver injury in mice. JHEP Rep 1: 418–429, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sekiya M, Yahagi N, Matsuzaka T, Takeuchi Y, Nakagawa Y, Takahashi H, Okazaki H, Iizuka Y, Ohashi K, Gotoda T, Ishibashi S, Nagai R, Yamazaki T, Kadowaki T, Yamada N, Osuga J, Shimano H: SREBP-1-independent regulation of lipogenic gene expression in adipocytes. J Lipid Res 48: 1581–1591, 2007 [DOI] [PubMed] [Google Scholar]
- 47.Sleeman P, Patel NN, Lin H, Walkden GJ, Ray P, Welsh GI, Satchell SC, Murphy GJ: High fat feeding promotes obesity and renal inflammation and protects against post cardiopulmonary bypass acute kidney injury in swine. Crit Care 17: R262, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noeman SA, Hamooda HE, Baalash AA: Biochemical study of oxidative stress markers in the liver, kidney and heart of high fat diet induced obesity in rats. Diabetol Metab Syndr 3: 17, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haas ME, Levenson AE, Sun X, Liao W-H, Rutkowski JM, de Ferranti SD, Schumacher VA, Scherer PE, Salant DJ, Biddinger SB: The role of proprotein convertase subtilisin/kexin type 9 in nephrotic syndrome-associated hypercholesterolemia. Circulation 134: 61–72, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shapiro MD, Miles J, Tavori H, Fazio S: Diagnosing resistance to a proprotein convertase subtilisin/kexin type 9 inhibitor. Ann Intern Med 168: 376–379, 2018 [DOI] [PubMed] [Google Scholar]
- 51.Zhang G, Li Q: Inflammation induces lipid deposition in kidneys by downregulating renal PCSK9 in mice with adriamycin-induced nephropathy. Med Sci Monit 25: 5327–5335, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Charytan DM, Sabatine MS, Pedersen TR, Im K, Park J-G, Pineda AL, Wasserman SM, Deedwania P, Olsson AG, Sever PS, Keech AC, Giugliano RP; FOURIER Steering Committee and Investigators : Efficacy and safety of evolocumab in chronic kidney disease in the FOURIER trial. J Am Coll Cardiol 73: 2961–2970, 2019 [DOI] [PubMed] [Google Scholar]
- 53.Schmit D, Fliser D, Speer T: Proprotein convertase subtilisin/kexin type 9 in kidney disease. Nephrol Dial Transplant 34: 1266–1271, 2019 [DOI] [PubMed] [Google Scholar]
- 54.Agrawal S, Zaritsky JJ, Fornoni A, Smoyer WE: Dyslipidaemia in nephrotic syndrome: mechanisms and treatment. Nat Rev Nephrol 14: 57–70, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sucajtys-Szulc E, Szolkiewicz M, Swierczynski J, Rutkowski B: Up-regulation of liver Pcsk9 gene expression as a possible cause of hypercholesterolemia in experimental chronic renal failure. Mol Cell Biochem 411: 281–287, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garbacz WG, Lu P, Miller TM, Poloyac SM, Eyre NS, Mayrhofer G, Xu M, Ren S, Xie W: Hepatic overexpression of CD36 improves glycogen homeostasis and attenuates high-fat diet-induced hepatic steatosis and insulin resistance. Mol Cell Biol 36: 2715–2727, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Konarzewski M, Szolkiewicz M, Sucajtys-Szulc E, Blaszak J, Lizakowski S, Swierczynski J, Rutkowski B: Elevated circulating PCSK-9 concentration in renal failure patients is corrected by renal replacement therapy. Am J Nephrol 40: 157–163, 2014 [DOI] [PubMed] [Google Scholar]
- 58.Raal F, Panz V, Immelman A, Pilcher G: Elevated PCSK9 levels in untreated patients with heterozygous or homozygous familial hypercholesterolemia and the response to high-dose statin therapy. J Am Heart Assoc 2: e000028, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okamura DM, López-Guisa JM, Koelsch K, Collins S, Eddy AA: Atherogenic scavenger receptor modulation in the tubulointerstitium in response to chronic renal injury. Am J Physiol Renal Physiol 293: F575–F585, 2007 [DOI] [PubMed] [Google Scholar]
- 60.Ray KK, Wright RS, Kallend D, Koenig W, Leiter LA, Raal FJ, Bisch JA, Richardson T, Jaros M, Wijngaard PLJ, Kastelein JJP; ORION-10 and ORION-11 Investigators : Two phase 3 trials of Inclisiran in patients with elevated LDL cholesterol. N Engl J Med 382: 1507–1519, 2020 [DOI] [PubMed] [Google Scholar]
- 61.Lebeau PF, Wassef H, Byun JH, Platko K, Ason B, Jackson S, Dobroff J, Shetterly S, Richards WG, Al-Hashimi AA, Won KD, Mbikay M, Prat A, Tang A, Paré G, Pasqualini R, Seidah NG, Arap W, Chrétien M, Austin RC: The loss-of-function PCSK9Q152H variant increases ER chaperones GRP78 and GRP94 and protects against liver injury. J Clin Invest 131: e128650, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scerbo D, Son N-H, Sirwi A, Zeng L, Sas KM, Cifarelli V, Schoiswohl G, Huggins L-A, Gumaste N, Hu Y, Pennathur S, Abumrad NA, Kershaw EE, Hussain MM, Susztak K, Goldberg IJ: Kidney triglyceride accumulation in the fasted mouse is dependent upon serum free fatty acids. J Lipid Res 58: 1132–1142, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Antibodies used for immunoblotting and immunohistochemical analysis. Download Supplemental Table 1, PDF file, 8.8 MB (8.8MB, pdf) .
Primers used for quantitative RT-PCR. Download Supplemental Table 2, PDF file, 8.8 MB (8.8MB, pdf) .
Staining and quantification of HK-2 cells, Immunoblot of LDLR and CD36, Transcript levels of SREBP2, and Thioflavin-S staining in the renal cortex. Download Supplemental Figure 1, PDF file, 8.8 MB (8.8MB, pdf) .
Immunohistochemical analysis and quantification of LDLR, qRT-PCR analysis of ER stress markers, Immunofluorescence microscopy of ATP1A1, and Immunofluorescence microscopy co-stained for CD36 and antihuman antibody. Download Supplemental Figure 2, PDF file, 8.8 MB (8.8MB, pdf) .
qRT-PCR analysis of ER stress, inflammatory, and fibrotic markers in HK-2 cells. . Download Supplemental Figure 3, PDF file, 8.8 MB (8.8MB, pdf) .
Visual summary outlining the underlying mechanisms of HFD-induced renal lipotoxicity reported within this study. Download Supplemental Figure 4, PDF file, 8.8 MB (8.8MB, pdf) .