Abstract
The nucleotide sequence of the pathogenic spirochete Brachyspira hyodysenteriae bit (for “Brachyspira iron transport”) genomic region has been determined. The bit region is likely to encode an iron ATP-binding cassette transport system with some homology to those encountered in gram-negative bacteria. Six open reading frames oriented in the same direction and physically linked have been identified. This system possesses a protein containing ATP-binding motifs (BitD), two hydrophobic cytoplasmic membrane permeases (BitE and BitF), and at least three lipoproteins (BitA, BitB, and BitC) with homology to iron periplasmic binding proteins. These periplasmic binding proteins exhibit lipoprotein features. They are labeled by [3H]palmitate when tested in recombinant Escherichia coli, and their signal peptides are typical for substrates of the type II secretory peptidase. The FURTA system and Congo red assay indicate that BitB and BitC are involved in iron binding. The Bit system is detected only in B. hyodysenteriae and is absent from B. innocens and B. pilosicoli.
In Gram-negative bacteria, active transport of molecules from the periplasm to the cytosol occurs via well-conserved systems (28). These specialized high-affinity transport systems are members of the ATP-binding cassette (ABC) superfamily of transporters, which are also responsible for the transport of a wide variety of solutes, including sugars, iron, amino acids, ions, proteins, and polysaccharides (19, 28). The common denominator of ABC transporters is the presence of high-homology stretch of about 200 amino acids in one of the polypeptides. This stretch contains a highly conserved ATP-binding motif consisting of two conserved sites (A and B) that form an ATP-binding pocket, also called the Walker motif (14). The classical ABC transporters possess a common structure consisting of four domains that can be on separate polypeptides; two are hydrophobic integral membrane proteins (permeases), and two are hydrophilic ATP-binding proteins, which can act as homodimers and couple ATP hydrolysis to import movement. In the bacterial ABC transporters-importers, the cytoplasmic permease and the nucleotide-binding protein activities exist on separate polypeptides (21, 28). The periplasmic proteins of importer systems are solute specific and determine the substrate specificity of the transport systems. The periplasmic substrate-binding protein interacts with and concentrates the specific incoming substrate and then presents it to the import complex formed by the cytoplasmic membrane permease and the nucleotide-binding protein.
Cytoplasmic permeases are energized by ATP (reviewed by Ames in 1990 [4]). Common amino acid sequences (EAA---G-----------I-LP) were observed in the hydrophilic region of cytoplasmic permeases (33). These sequences, called EAA loops, have been proposed to interact directly with portions of the nucleotide binding site of the ATP-binding protein (33).
Transport of free iron from the periplasmic space into the cytoplasm is proposed to occur by a classic active-transport (import) process involving a periplasmic binding protein, a specific cytoplasmic permease, and an energy-supplying nucleotide binding protein. This active-transport system is widely distributed among gram-negative pathogenic bacteria.
Spirochetes form a distinct bacterial group within the eubacterial kingdom (61, 70). Brachyspira hyodysenteriae (Serpulina hyodysenteriae [48]) is a member of the spirochete family and is the agent of swine dysentery (26). B. hyodysenteriae is characterized by its strong beta-hemolytic activity and its enteropathogenicity in swine (34). Different virulence factors of B. hyodysenteriae have been cloned and identified. Inactivation of flagellar proteins (FlaA1 and FlaB1) resulted in a strong reduction in virulence and colonizing ability, indicating that motility is an essential virulence factor of B. hyodysenteriae (32, 55, 56). Different hemolysins have also been cloned and are considered important for the virulence of B. hyodysenteriae (29, 43, 67, 68). A B. hyodysenteriae mutant with a mutation of one of the hemolysins (tlyA) showed a reduced virulence in mice in a swine dysentery model (29, 68). Furthermore, B. hyodysenteriae possesses a NADH oxidase (Nox) (63) that reduces chemically the oxygen in water. The NADH oxidase may play an essential role in the colonization of the mammalian intestinal tract by diverse bacterial species.
In this study, we describe the cloning and characterization of an ABC importer system of B. hyodysenteriae implicated in iron import (the Bit system). The Bit system possesses all the features characteristic of bacterial periplasmic importers; it bears strong primary sequence and secondary-structure similarities to the importer systems, suggesting that it has a similar mechanism of action. We suggest that in B. hyodysenteriae the periplasm-to-cytosol transport of iron proceeds by an analogous mechanism to those observed in gram-negative bacteria. The Bit system has similarities to the periplasmic binding protein-dependent iron transport systems in Actinobacillus pleuropneumoniae (afuABC) (13), Haemophilus influenzae (hitABC) (58), Neisseria meningitidis (fbpABC) (1), and Serratia marcescens (sfuABC) (6).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
B. hyodysenteriae, B. innocens, and Escherichia coli strains, as well as phages and plasmids used in this study, are listed in Table 1. Bacteria were grown on solid medium by using blood agar base no. 2 (Oxoid Ltd., Basingstoke, England) containing 5% bovine blood. The plates were incubated anaerobically at 37°C for 4 days in jars (Oxoid) under a GasPak Plus generator atmosphere (BBL, Becton Dickinson and Co., Cockeysville, Md.). E. coli LE392, used for phage multiplication, was grown in Luria-Bertani broth (57) supplemented with 0.2% maltose and 10 mM MgSO4 to an optical density at 600 nm of 0.6. E. coli XL1 Blue (Table 1) was used as the host and phagemid pBluescript II KS(+) (Stratagene, La Jolla, Calif.) was used as the vector for subcloning. E. coli strains were grown in Luria-Bertani medium under antibiotic selection with ampicillin (50 μg/ml). Congo red (Sigma, St. Louis, Mo.) was added to 0.01% to Trypticase soy agar plus 10% IsoVitaleX (BBL, Becton Dickinson and Co.) for determination of the Crb (Congo red-binding) phenotype (50).
TABLE 1.
Strains, phages, and plasmids used in this study
| Strain, phage, or plasmid | Genotype | Reference or source |
|---|---|---|
| Strains | ||
| B. hyodysenteriae | ||
| B78T | Type strain (ATCC 31212) | 7 |
| FM88-90 | Reference strain serotype 8 (ATCC 49887) | 38 |
| B. pilosicoli | ||
| P43/6/78T | Type strain | 69 |
| B. innocens | ||
| B256T | Type strain (ATCC 29796) | 64 |
| E. coli K-12 | ||
| LE392 | F− hsdR514 (rK−mK+) supE44 supF58 lacY1 | Promega |
| XL1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIq ZΔM15 Tn10 (Tetr)] | Stratagene |
| Phage | ||
| λGEM-11 | Bacteriophage vector, Spi selection | Promega |
| Plasmids | ||
| pBluescriptII KS(+) | Apr Plasmid vector | Stratagene |
| pDJ1 | pKS(+) bitA′BCDEF | This study |
| pDJ2 | pKS(+) bitA′B | This study |
| pDJ3 | pKS(+) bitDEF | This study |
| pDJ4 | pKS(+) bitF | This study |
| pDJ5 | pKS(+) bitCDEF | This study |
| pDJ6 | pKS(+) bitA′BDEF | This study |
| pDJ7 | pKS(+) bitA′ | This study |
| pDJ8 | pKS(+) bitA′ | This study |
| pDJ9 | pKS(+) bitBC | This study |
Construction and immunological screening of the genomic library.
A DNA library of B. hyodysenteriae serotype 8 strain FM88-90 (ATCC 49887) was constructed in phage LambdaGEM-11 (Promega Biotec, Madison, Wis.) by the method recommended by the manufacturer (53). Briefly, total cellular DNA of B. hyodysenteriae FM88-90 was isolated as previously described (20) and partially digested with Sau3AI. The partially digested genomic DNA and XhoI-digested λGEM-11 arms were halfway filled in to make them compatible and ligated.
The DNA was then packaged by using the Packagene in vitro packaging system (Promega) and transfected into E. coli LE392. A 100-μl volume of E. coli LE392 culture was infected with 100 μl of phages in phage dilution buffer (20 mM Tris-HCl [pH 7.4], 100 mM NaCl, 10 mM MgSO4) and grown in top agarose overnight (0.7% agarose, 1% Bacto Tryptone, 0.5% NaCl). Phage plaques immunoreactive with a rabbit anti-B. hyodysenteriae sera were removed with Pasteur pipettes, transferred to phage dilution buffer, and stored at 4°C. Plaques were first adsorbed onto nitrocellulose filters (Schleicher & Schuell, Inc., Dassel, Germany) and further incubated for 2 h at room temperature in rabbit polyclonal antiserum prepared by injecting animals with formalin-killed B. hyodysenteriae FM88-90 whole cells (41). The antiserum was preabsorbed overnight at 4°C with B. innocens reference strain B256 and E. coli LE392. Bound antibodies were detected with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Jackson, West Grove, Pa.) and 4-chloro-1-naphthol color reagent (Sigma) as described by Sambrook et al. (57). Selected positive clones were purified by several rounds of single-plaque isolation. These purified phages were then tested for their nonreactivity with an antiserum of B. innocens type strain B256 by Western blotting and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE); the positive ones were discarded. Phage 42 has been selected for further studies.
DNA purification.
Phage DNA was extracted from each of the positive phages that reacted only with the B. hyodysenteriae antiserum, by scraping the top agarose containing nearly confluent plaques in dilution buffer. After 3 h of elution at room temperature, the agarose and bacterial cells were pelleted by centrifugation. The DNA was then extracted from the supernatant by an extraction method involving Phase Lock Gel II (5Prime-3Prime, Boulder, Colo.). Plasmid DNA was isolated from E. coli by alkaline extraction (57).
Southern blotting and hybridization.
DNA was transferred to Zeta-Probe blotting membranes (Bio-Rad Laboratories Ltd, Mississauga, Ontario, Canada). DNA probes were labeled with [α-32P]dCTP (DuPont Canada, Mississauga, Ontario, Canada) by using a random oligolabelling kit (Pharmacia-LKB, Biotechnology Inc., Dorval, Canada) as previously described (25). The blots were hybridized with DNA probes as recommended by the supplier of the Zeta-Probe blotting membranes and exposed for autoradiography.
SDS-PAGE and Western blot (immunoblot) analysis.
Proteins were purified by scraping the top agarose from plates containing nearly confluent plaques into phage dilution buffer. After a 3-h elution period, the agarose was pelleted and proteins in the supernatant were recovered by precipitation with 10% cold trichloroacetic acid as previously described (27), washed twice in phosphate-buffered saline, suspended in Laemmli sample buffer (37), and subjected to SDS-PAGE. Protein profiles of subclones were analyzed for the expression of B. hyodysenteriae proteins by Western immunoblot analysis with whole bacterial cells.
DNA sequencing.
Restriction fragments of insert DNA from recombinant phages were ligated into pBluescriptII KS plasmid. Plasmids carrying the appropriate inserts were subjected to double-stranded DNA sequencing with both strands as template by the dideoxyoligonucleotide method of Sanger et al. (59) with the AutoRead sequencing kit (Pharmacia-LKB). Oligonucleotides were synthesized on a Cyclone nucleic acid synthesizer (Millipore Corp., Bedford, Mass.). DNA sequence analysis was performed with the GeneWorks software package (IntelliGenetics, Inc., Mountain View, Calif.). DNA sequence information was analyzed with the Intelligenetics Suite package and programs from the University of Wisconsin Genetics Computer Group software sequence analysis package (18).
Analysis of hydropathy and sequence homology of proteins.
Average hydropathy profiles of the proteins were obtained by the method of Kyte and Doolittle (36) with the GeneWorks program (IntelliGenetics), using a window length of 9. Amino acid comparisons were effected with the BESTFIT program of the University of Wisconsin Genetics Computer Group software (18) with the National Center for Biotechnology Information databases. Comparisons using BESTFIT were performed with standard default parameters of gap weight 3.0 and length weight 0.1.
[3H]palmitate labeling and immunoprecipitation of BitB and BitC lipoproteins.
The transformants were labeled in vivo with [3H]palmitate as previously described (54). Briefly, transformants XL1 containing pDJ2, pDJ3, and pDJ5 were grown aerobically at 37°C in Luria-Bertani medium to exponential phase (optical density at 600 nm = 0.5). The cells were then labeled with [3H]palmitate (0.05 mCi/ml) (Amersham) plus 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), harvested and immunoprecipitated. The bacterial suspension was pelleted, and the pellets were washed three times in 1× phosphate-buffered saline. After the last wash, the pellet was resuspended in 100 μl of 0.1 M KH2PO4 (pH 6.2)–50 μg of phenylmethylsulfonyl fluoride per ml, 25 μg of leupeptin per ml–0.7 μg of pepstatin per ml and incubated for 30 min at 37°C. After the suspensions were frozen, thawed, and centrifuged at 10,000 rpm in an Eppendorf 1754C centrifuge for 5 min, 40-μl volumes of supernatants were collected and 4 μl of rabbit polyclonal B. hyodysenteriae antiserum was added to each; the mixtures were incubated for 1 h 30 min at 4°C with agitation. Then 20 μl of blocked Pansorbin cells (Calbiochem) was added. The suspension was incubated for 1 h 30 min at 4°C with agitation and centrifuged for 1 min at 13,000 rpm in a Eppendorf 1754 C centrifuge, and the pellets were washed vigorously three times with 1 ml of RIPA buffer (1.0% Triton X-100, 1.0% sodium deoxycholate, 0.1% SDS, 0.001% aprotinin, 0.15 M sodium chloride, 0.01 M Tris-HCl [pH 7.4]) and once with 0.05 M sodium chloride–0.01 M Tris-HCl [pH 7.4]–1 mM EDTA. The pellets were then resuspended in SDS-PAGE sample buffer, heated for 3 min at 95°C, and analyzed by SDS-PAGE.
Fur titration assay (FURTA).
To determine whether putative Fur-box sequences can function as a binding site for the Fur protein, we introduced plasmids pDJ1 to pDJ9 into E. coli H1717. This strain contains an iron-regulated fhuF::lacZ fusion which is very sensitive to small changes in the iron concentration in the medium because the fhuF promoter region has a weak binding affinity for the Fur-Fe2+ repressor (65). The presence of a multicopy plasmid containing a Fur box nucleotide sequence will deplete the pool of the Fur protein, allowing transcription of the fhuF::lacZ. The lacZ expression assay was performed with MacConkey lactose plates (Difco) supplemented with 25 to 50 μM FeCl3 · 6H2O.
Immunoaffinity purification of antibodies.
To detect the presence of the cloned proteins in B. hyodysenteriae, the rabbit antiserum raised against B. hyodysenteriae was enriched for antibodies to the Bit proteins by the method of Oaks et al. (47). This enriched antiserum was used to localize the proteins on the bacteria by immunoelectron microscopy. Rabbit anti-B. hyodysenteriae polyclonal antiserum was incubated with nitrocellulose onto which clone 42 plaques had first been adsorbed and blocked with 2% skim milk–Tris-buffered saline. After a 3-h incubation period, the nitrocellulose was washed first in Tris-buffered saline with 0.05% Tween 20, then in Tris-buffered saline, and then in 0.15 M NaCl. Finally, the bound antibodies were eluted in 0.2 M glycine–0.15 M NaCl (pH 2.8). The antibody solution was then neutralized with 0.75 M Tris (pH 8.8) and used undiluted for electron microscopy.
Electron microscopy and immunogold labeling.
The electron microscopy and immunogold labeling were done as previously described (39). Briefly, a drop of bacterial suspension was placed on a Formvar-coated grid. After blocking for 5 min with 1% egg albumin, the grids were incubated for 30 min with undiluted antibody-enriched solution (described above) and rinsed with distilled water five times. The grids were then incubated for 30 min with protein A-gold particles (10 nm; Sigma), rinsed, and negatively stained with 1% phosphotungstate for 10 s. They were examined under an electron microscope (Philips 201) at an accelerating voltage of 60 kV.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper appears in the GenBank Database Library under accession no. U75349.
RESULTS
Construction and screening of the library.
Chromosomal DNA isolated from strain FM88-90 of B. hyodysenteriae serotype 8 was partially digested with Sau3AI, and the sites were partially filled with dGTP and dATP. A library was then constructed in λGEM-11 phage. We obtained approximately 3 × 103 recombinants per μg of DNA. To obtain clones expressing B. hyodysenteriae specific proteins, the library was screened with anti-B. hyodysenteriae rabbit sera adsorbed against B. innocens B256 reference strain and E. coli LE392. Five recombinant phages positive for B. hyodysenteriae and nonreactive with B. innocens were chosen. We report here the characterization of the recombinant bacteriophage 42.
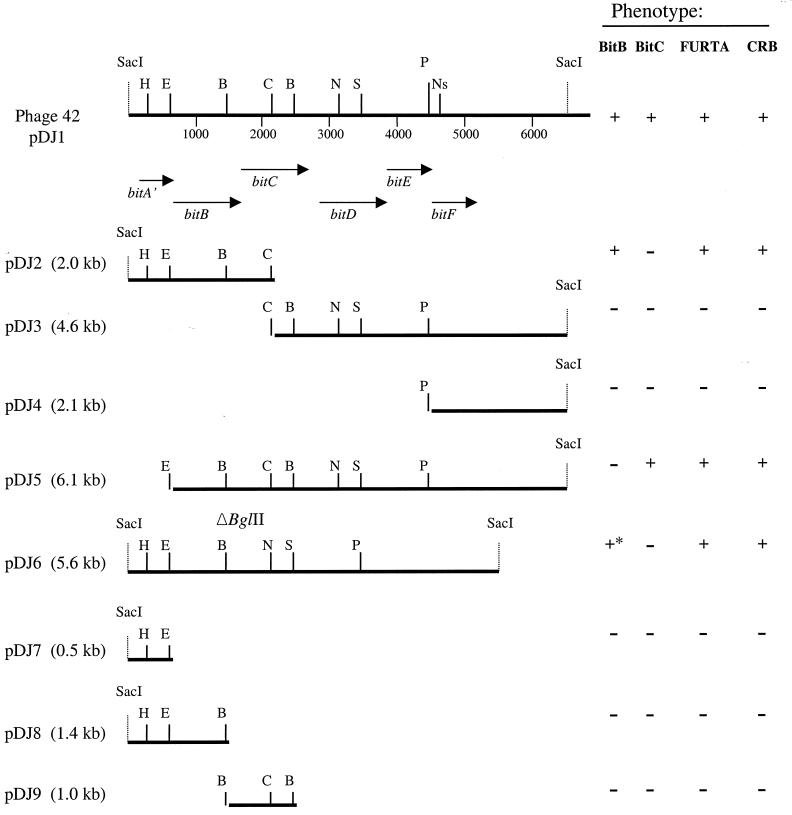
Restriction mapping and expression of recombinant proteins.
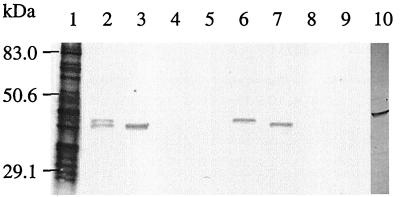
The recombinant bacteriophage 42 consisted of the vector λGEM-11 and a genomic insert of 6.6 kb. The insert was then subcloned in the phagemid pBluescriptII KS as a SacI-SacI fragment (Fig. 1). The plasmid was named pDJ1. Western blot analysis of pDJ1 revealed two proteins with similar molecular masses of 42 and 43 kDa that reacted with the anti-B. hyodysenteriae serum (Fig. 2, lane 2). The recombinant plasmid pDJ2 comprising the 2-kb SacI-ClaI fragment of the insert revealed only a 42-kDa immunoreactive protein (lane 3). Plasmids pDJ3, pDJ4, pDJ7, and pDJ8 did not reveal any immunoreactive proteins. Plasmids pDJ5 and pDJ6, containing an EcoRV and a BglII deletion, respectively, expressed a 43- and a 42-kDa immunoreactive protein, respectively (lanes 6 and 7). In contrast to E. coli carrying pDJ1, where two proteins with apparent molecular masses of 43 and 42 kDa reacted with antiserum from proteins expressed by pDJ1, in B. hyodysenteriae only a band with apparent molecular mass of 45 kDa was detected (Fig. 2).
FIG. 1.
Restriction map of the Bit A′BCDEF system and the different subclones. The pDJ1 insert was subcloned from the SacI sites of phage λGEM-11 clone 42 into the pBluescriptII KS plasmid. The pDJ2 plasmid corresponds to the SacI-ClaI fragment of pDJ1. pDJ3, pDJ4, pDJ5, and pDJ6 correspond, respectively, to the ClaI, PstI, EcoRV, and BglII restriction fragments subcloned from pDJ1. pDJ7 and pDJ8 correspond, respectively, to the SacI-EcoRV and SacI-BglII fragments of pDJ1 ligated in pBluescriptII KS. Finally, pDJ9 corresponds to the BglII-BglII fragment of pDJ1. The parenthetical sizes refer to the length of the cloned DNA fragment and do not include the vector length. The arrows indicate the directions and lengths of the ORFs. The Western blot results with anti-B. hyodysenteriae serum are shown on the right. Restriction sites: B, BglII; C, ClaI; E, EcoRV; H, HindIII; N, NdeI; Ns, NsiI; P, PstI, S, StuI. In FURTA, + indicates that LacZ from fhuF::lacZ is expressed and − indicates that LacZ is not expressed. In Congo red binding (CRB), + indicates that the colonies are red on Trypticase soy agar plates containing Congo red and − indicates that the Congo red is not bound by the strains (white). The asterisk indicates that the immunoreactive protein is composed of the first 296 amino acids of BitB and the last 46 amino acids of BitC.
FIG. 2.
Western blot analysis. Expression of recombinant proteins of pDJ plasmids as determined by Western blot analysis using a rabbit antiserum directed against B. hyodysenteriae serotype 8. Lanes: 1, B. hyodysenteriae serotype 8; 2, pDJ1; 3, pDJ2; 4, pDJ3; 5, pDJ4; 6, pDJ5; 7, pDJ6; 8, pDJ7; 9, pDJ8; 10, detection of Bit-responsive proteins in B. hyodysenteriae with an antiserum against the recombinant BitB-expressing E. coli.
DNA sequences and homologies of the gene products.
The sequence of a 6.6-kb B. hyodysenteriae insert from pDJ1 was determined. Sequence analysis revealed six potential open reading frames (ORFs) reading from left to right. The G+C content was 30.1%, in agreement with the estimated G+C content of the whole genome of B. hyodysenteriae, which is 3.2 × 106 bp and possesses a G+C content of 26 ± 1 mol% (72, 73).
In pDJ1, the bitA gene corresponds to the first bit ORF (bitA′) and is truncated with an ORF encoding a protein of 175 amino acids. The 5′ portion of bitA was sequenced by gene-walking PCR. The procedure is based on single-specific-primer PCR amplification of chromosomal DNA adjacent to the known sequence (46). The sequence bitA (1,008 nucleotides) encodes a putative polypeptide of 336 amino acids with a deduced molecular mass of 37,027 Da. BitA has homology to many proteins involved in iron periplasmic binding, such as AfuA (56% similarity) in A. pleuropneumoniae (12) and a putative periplasmic iron-binding protein (51.6%) of Sinorhizobium meliloti (GenBank accession no. AAC64671). BitA has homology of 49.4% to a thiamin-binding lipoprotein of a member of the Archaea, Pyrococcus horikoshii (31). Downstream of bitA, a 1,026-nucleotides ORF, bitB, encodes a putative polypeptide of 342 amino acids (BitB), with a deduced molecular mass of 37,262 Da. BitB has homology to many proteins involved in iron periplasmic binding, such as AfuA (51.3% similarity) in A. pleuropneumoniae (13) and HitA (45.5% similarity) from the system HitABC in H. influenzae. The third (1,017-nucleotide) ORF, bitC, encodes a putative polypeptide of 339 amino acids (BitC) with a deduced molecular mass of 36,885 Da. The amino acid sequences of BitA, BitB, and BitC showed homology to each other (approximately 90% similarity). The genes encoding BitB and BitC expressed a 42- and a 43-kDa protein, respectively, as revealed by Western blotting with anti-B. hyodysenteriae serum (Fig. 2).
The fourth ORF, bitD (of 1,122 nucleotides) encodes a putative polypeptide of 374 amino acids (BitD) with a deduced molecular mass of 42,589 Da. BitD has homology to many proteins belonging to the superfamily of the ABC transporter proteins and contained an ATP-binding domain consisting of two highly conserved amino acid sequences known as the Walker motif A (amino acids 37 to 53) and Walker motif B (amino acids 134 to 167 and 188 to 194). More specifically, the BitD protein has homology to a hypothetical ABC transporter of H. influenzae, Hi0126 (67.1% similarity), to PotA (63.7 similarity) of H. influenzae, to AttA1 (63.6% similarity) of Agrobacterium tumefaciens, to AfuC (62.8% similarity) of A. pleuropneumoniae, and to HitC (58% similarity) of H. influenzae.
The fifth ORF (bitE) (of 831 nucleotides) encodes a polypeptide of 277 amino acids (BitE) with a deduced molecular mass of 30,853 Da. The sixth ORF (bitF) (of 771 nucleotides) encodes a polypeptide of 257 amino acids (BitF) with a deduced molecular mass of 28,343 Da. Both BitE and BitF proteins have homology to hydrophobic permeases and show homology to each other (48.7% similarity). BitE has homology to the first half of AfuB (54.4% similarity) (A. pleuropneumoniae) and to the first half of HitB (58.4% similarity) (H. influenzae). BitF has homology to the second half of AfuB (57.8% similarity) and to the second half of HitB (48.6% similarity). Both AfuB and HitB are permeases involved in iron transport. BitE and BitF proteins also have homology to a putative sulfate permease, NifC, of H. influenzae, also named Hi0129 (22). BitE has homology (55.0% similarity) to the first half of NifC, and BitF has homology to the second half (54.6% similarity). BitE and BitF are very hydrophobic proteins, their mean hydrophobicity (36) being 0.89 and 0.91, respectively. Their hydropathy profiles indicate that BitE and BitF contain six putative transmembrane-spanning segments linked by hydrophilic segments of variable length (not shown). A sequence matching the consensus permease EAA motif (EAA---G-----------I-LP) (33, 60) was found in both BitE and BitF (amino acids 177 to 198 for BitE and 157 to 178 for BitF).
A homology search using the default parameters of Blastp at the National Center for Biotechnology Information and restricted to the amino acid sequences of the spirochete protein has been applied to BitABCDEF. BitA showed homology to a thiamine ABC transporter of Treponema pallidum (accession no. AAC65133), 47% similarity on a stretch of 78 amino acids. BitB showed homology to the surface protein VspA of Borrelia turicatae (12), 37% similarity on a stretch of 216 amino acids. BitC, like BitA, showed homology to a thiamine ABC transporter of T. pallidum (AAC65133), 43% similarity on a stretch of 138 amino acids. BitD showed homology to an ATP-binding protein (PotA) of the spermidine/putrescine ABC transporter of T. pallidum (AAC65627), 63% homology on a stretch of 296 amino acids. BitE showed homology to a permease protein (PotB) of the spermidine/putrescine ABC transporter of Borrelia burgdorferi (AAB91526), 45% homology on a stretch of 269 amino acids. Finally, BitF showed homology to another permease protein (PotC) of the spermidine/putrescine ABC transporter of B. burgdorferi (AAB91527), 48% homology on a stretch of 263 amino acids.
Preceding the putative ATG start codon of all ORFs, a ribosome-binding site characteristic of gram-negative bacteria was found. The first 20 amino acids of BitB and the first 21 amino acids of BitC, the putative proteins homologous to the periplasmic protein-binding substrate, have putative lipoprotein cleavage signal sequences (GCG software). No typical signal peptide sequence was found for BitD, the putative ATP-binding protein, or for BitE or BitF, which are homologous to hydrophobic permeases. Moreover, −35 and −10 consensus sequences (24) were found upstream of the sequence of bitB and bitC. A potential rho-independent termination site has been found just downstream the last ORF of the Bit system, bitF. Furthermore, the 84-bp intragenic region between the bitC stop codon and the first ATG codon of bitD includes a strong stem-loop structure with the potential to form 26 hydrogen bonds with a ΔG of −14.6 kcal/mol as determined with Oligo version 5.0 distributed by National Biosciences (Hamel, Minn.).
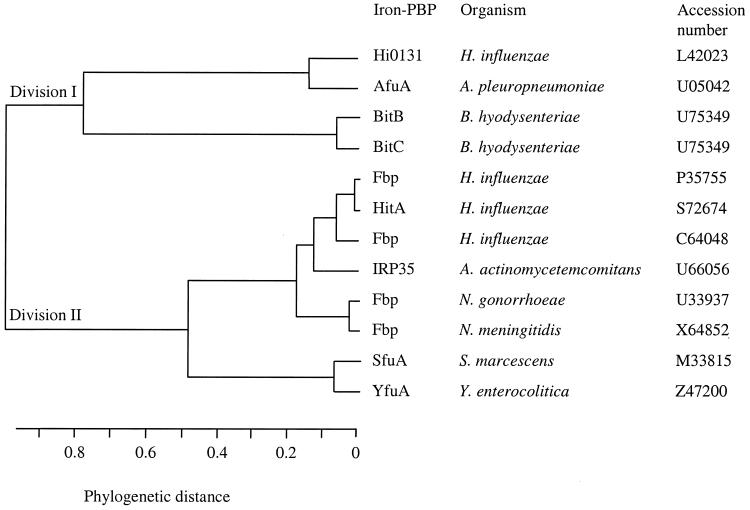
Thus, sequence analysis indicates that the Bit polypeptides belong to an ABC transporter system where BitD has the characteristic feature of an ABC transporter and BitE and BitF have the characteristics of permeases and homologies to permeases. Moreover, the homologies of Bit periplasmic binding proteins consistent with many periplasmic binding proteins involved in iron periplasmic binding and the overall homology of BitD, BitE, and BitF to the ABC transporter and permeases of an iron importer system suggest that the Bit system could be involved in iron import. A dendrogram was constructed by using amino acids sequences of periplasmic iron binding (PIB) proteins present in different gram-negative bacteria (Fig. 3). Two clusters were obtained. The PIB proteins sequences of Bit system were grouped with those of some members of the Pasteurellaceae, H. influenzae and A. pleuropneumoniae, in division I. The majority of the sequences of the PIB proteins of gram-negative bacteria were clustered in division II. Division II was formed by the sequences of the PIB proteins belonging to FbpABC, HitABC, and YfuABC systems.
FIG. 3.
Possible evolutionary tree for the periplasmic binding proteins (PBP). Homologous sequences were aligned with Clustal, and the relationship between sequences were calculated by the unweighted pair group method with arithmetic mean with the GeneWorks program (IntelliGenetics, Inc.).
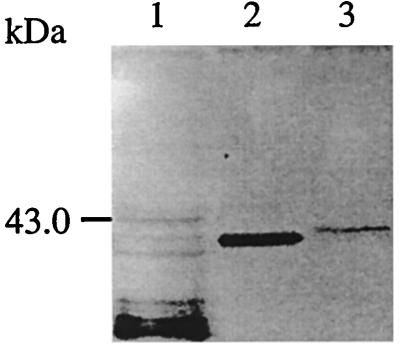
BitB and BitC are lipoproteins.
The signal sequences of the BitA, BitB, and BitC proteins are typical type II signal sequences that could be cleaved by the type II signal peptidase. Transformant cells harboring pBluescript II KS(+) (negative control), pDJ2 (BitB), and pDJ5 (BitC) were labeled in vitro with [3H]palmitate (Fig. 4). The size of the labeled lipoproteins expressed by pDJ2 and pDJ5 corresponded to the size of BitB and BitC proteins when compared in Western blots where these recombinant proteins were revealed by antiserum against B. hyodysenteriae. These lipoproteins were absent from the cells containing the pBluescript II KS without insert. Thus, BitB and BitC are periplasmic binding lipoproteins. When the prebleed antiserum was used, no palmitate-labeled lipoproteins were immunoprecipitated from the different transformants (data not shown). Furthermore, when B. hyodysenteriae antiserum was used, no palmitate-labeled lipoproteins were immunoprecipitated from transformant cells harboring pBluescript II KS without insert (data not shown).
FIG. 4.
[3H]palmitate labeling of BitB and BitC. Lanes: 1, whole-protein content of recombinant E. coli XL1 Blue containing the pBluescriptII KS plasmid; 2 and 3, pDJ5 and pDJ2 transformants, respectively, immunoprecipitated as described in Materials and Methods. Between 5 and 10 μg of proteins was added to each well.
Characterization of the BitA′BC system by FURTA and Congo red binding.
FURTA was used to determine if the bit genes contain fur boxes but also can reveal the presence of an iron-binding system. E. coli H1717 contains an iron-regulated fhuF::lacZ fusion which is very sensitive to small changes in the iron concentration in the medium because the fhuF promoter region has a weak binding affinity for the Fur-Fe2+ repressor. The repressor Fur in presence of iron binds to the fhuF promoter region, and therefore the lacZ reporter gene is not expressed and colonies are white on MacConkey lactose plates. However, Fur will not bind to the fhuF promoter region in the absence of iron or in the presence of Fur binding sequences carried on a multicopy plasmid that will also titrate out the Fur protein from binding sites within fhuF. In both of these latter cases, the lacZ reporter gene will be expressed and the colonies will be red on MacConkey lactose plates. Three putative Fur boxes were identified by homology to the putative Fur-box of E. coli (44). Two of them are localized between or near the −10 and −35 sites of bitB. One sequence which shares 12 of 19 positions with the consensus Fur-binding site overlaps the promoter of bitB (nucleotides 1048 to 1066), which is 100% identical to the promoter of fepA of E. coli. Similarly the fepA −35 promoter sequence is also localized into a putative Fur box. The FepA protein is part of the ferric enterobactin transport system in E. coli (49). The second Fur box (nucleotides 1159 to 1177) is localized just downstream of the ribosome-binding site of bitB (13 nucleotides identical to the 19 nucleotides of the Fur box). The third Fur box (nucleotides 3302 to 3320) has been found between BitC and BitD (14 nucleotides identical to the 19 nucleotides of the E. coli Fur box).
To determine whether these Fur boxes were binding the Fur protein, we introduced constructs pDJ1, pDJ2, pDJ4, pDJ5, pDJ6, pDJ7, pDJ8, pDJ9, pBluescript KS, and a positive control pBCFUR into E. coli H1717. For these tests, two different concentrations of iron (FeCl3) were tested; 25 and 50 μM. No LacZ expression was observed when strain H1717 was transformed with plasmids carrying a putative Fur box as deduced by sequence homology (pDJ8, pDJ9, and pDJ3) (Fig. 1). This indicates that the sequence carried by these recombinant plasmids did not titrate out the Fur repressor from binding to fhu-binding sites. However, colonies of strain H1717 transformed with plasmids expressing BitB and/or BitC (pDJ2, pDJ5, and pDJ6) were red on MacConkey plates. This observation suggests that in the presence of the periplasmic binding proteins, E. coli Fur repressor is not binding the fhuF promoter region, because BitB and BitC are also competing for iron. Thus, BitB and BitC are periplasmic iron-binding proteins and the Bit system is involved in iron transport. Furthermore when the constructs pDJ1, pDJ2, pDJ4, pDJ5, pDJ6, pDJ7, pDJ8, pDJ9, and pBluescript KS were transformed into E. coli XL1 Blue, only colonies transformed with plasmids expressing BitB and/or BitC (pDJ2, pDJ5, and pDJ6) were red on Congo red-containing plates (Fig. 1). This indicates that these proteins have the capacity to bind Congo red.
Localization of the Bit proteins in B. hyodysenteriae.
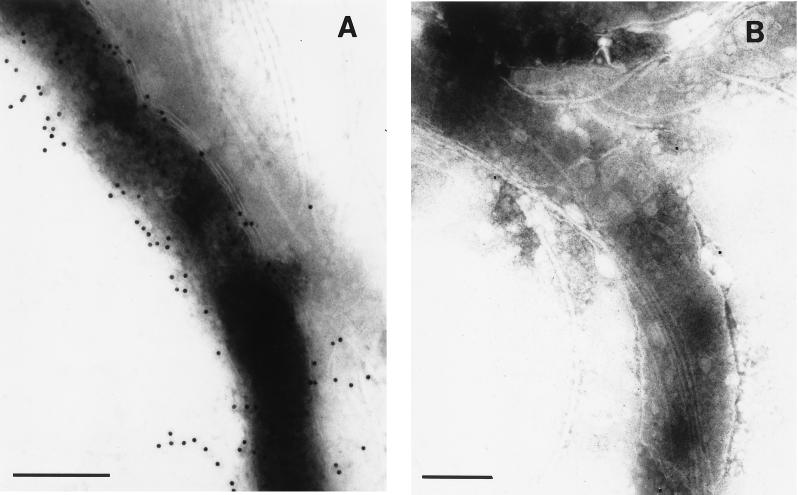
To locate the proteins expressed from phage 42, an immunogold-labeling experiment was performed on B. hyodysenteriae cells grown on blood agar plates. The antiserum against B. hyodysenteriae was affinity purified from phage 42 expressing the Bit proteins by using the method of Oaks et al. 1987 (47). Rabbit anti-B. hyodysenteriae polyclonal antiserum was incubated with nitrocellulose adsorbed with phage 42. The antiserum did not react with E. coli cells used for the phage infection. These antibodies specific to the proteins expressed by phage 42 were used to localize the Bit proteins on the bacteria by immunogold electron microscopy. The affinity-purified antiserum was tested in electron microscopy on whole cells of B. hyodysenteriae and B. innocens. The antigens were localized external to the cytoplasmic membrane of B. hyodysenteriae cells and could be detected when the outer sheet was disrupted (Fig. 5A). Intact B. hyodysenteriae cells were poorly labeled with the anti-Bit protein antibodies (data not shown).
FIG. 5.
Electron micrograph of negatively stained B. hyodysenteriae (A) and B. innocens (B) cells incubated with anti-Bit antibodies and protein A-colloidal gold.
Genetic characterization of the Bit system.
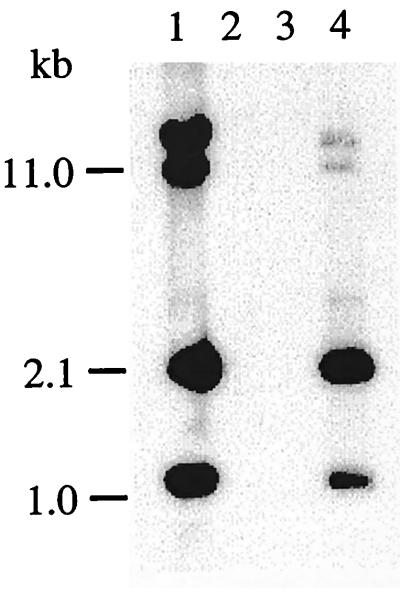
The Bit system of B. hyodysenteriae was characterized genetically by Southern hybridization analysis of genomic DNAs of B. hyodysenteriae and B. innocens. The probe bit consisted of a SacI-ClaI fragment including bitA, bitB, and bitC genes. The probe hybridized with BglII-digested DNAs from the B. hyodysenteriae parental strain and from the type strain of B. hyodysenteriae (Fig. 6). The bit probe detected 4 BglII fragments of 1.0, 1.9, 10, and >12 kb in each strain of B. hyodysenteriae tested. The 1.0-kb fragment corresponded to the BglII internal fragment of the bit DNA (Fig. 1). Since two BglII sites are present in the bit DNA sequenced, three fragments should hybridize with the bit probe. The additional fragment hybridizing with the bit probe indicates that there is an additional sequence to homologous bit periplasmic genes in the B. hyodysenteriae genome. Furthermore, the bit probe did not hybridize with B. innocens, a nonpathogenic spirochete, or with E. coli HB101. B. hyodysenteriae serotype 1 to 9 reference strains, as well as B. hyodysenteriae B78T, B. innocens B256T, and B. pilosicoli P43/6/78T, were tested by Southern blotting with the bit probe (results not shown). Only B. hyodysenteriae strains hybridized with the bit probe. Furthermore, by immunodot blotting with anti-Bit serum purified by an immunoaffinity assay as described in Materials and Methods, only B. hyodysenteriae strains reacted with anti-Bit serum (results not shown). Thus, these results suggest that the Bit system is specific to B. hyodysenteriae.
FIG. 6.
Hybridization of BglII fragments of genomic DNAs by using probe 42 derived from the ClaI-SacI fragment of pDJ2. Lanes: 1, B. hyodysenteriae serotype 8 ATCC 49887; 2, E. coli HB101; 3, B. innocens; 4, B. hyodysenteriae B78.
DISCUSSION
A unique feature of the bacterial ABC importer systems is that they all have a periplasmic substrate-binding protein that interacts with the incoming substrate, binds to it, and presents it to the import complex formed by the permease and the ATP-binding protein in the inner membrane (28). This import complex has the ATP-binding domain and the membrane-spanning domains present on separate polypeptide (21, 28). The Bit system presents characteristics of a periplasmic iron transport system. It includes periplasmic proteins encoded by at least three genes, bitA, bitB, and bitC, that are proposed to be the iron-binding proteins. These genes belong to a gene cluster purported to encode a classic import system that includes BitA, BitB, and BitC; a nucleotide-binding protein, BitD; and two cytoplasmic membrane permeases, BitE and BitF. The characteristics and comparison of the predicted protein products of bitDEF with the products derived from the afuBC, fbpBC, hitBC, sfuBC and yfuBC genes illustrate conserved similarity of the Bit system with respect to the general class of ABC iron periplasmic importers.
In the Bit periplasmic transport system, the BitA, BitB, and BitC proteins correspond to the substrate-binding lipoproteins homologous to periplasmic iron-binding protein AfuA of the A. pleuropneumoniae afuABC operon (13), similar to HI0131, which is suggested to be an iron-binding periplasmic protein of H. influenzae (66), and to HitA of the H. influenzae HitABC periplasmic iron import operon (58). The dendrogram obtained from the homologies of the substrate-binding proteins BitB and BitC showed that the periplasmic iron-binding proteins of the Bit system were distant but clustered with the other periplasmic iron-binding proteins. We chose to derive the dendrogram from the homologies and the differences between the iron periplasmic protein of the different iron transport systems since these proteins have a strong amino acid sequence heterogeneity, varying from species to species. Interestingly, the BitB and BitC proteins from the Bit system are grouped with those of another swine pathogen, A. pleuropneumoniae, the causative agent of porcine pleuropneumonia. Spirochetes form a distinct group within the eubacterial kingdom (61). The homologies and differences observed across the Bit system argue that the afu, fbp, hit, sfu, and yfu operons and the bit system have evolved separately but seem to function similarly in the periplasm-to-cytosol transport of iron. The homologies between the BitA, BitB, and BitC amino acid sequences suggest a duplication phenomenon. The sequencing and hybridization results suggested that there are possibly more than three duplications of the periplasmic binding substrate gene in B. hyodysenteriae. This feature has not been observed in homologous iron transport systems of gram-negative bacteria. In H. influenzae at least two related periplasmic iron-binding proteins physically distant on the genome have been identified (58, 66). In B. hyodysenteriae, a family of genes with 45 to 95% homology have been found in a locus called VspA-H (variable surface protein) (23). Furthermore, in spirochetes, gene duplication has been observed, particularly in Borrelia hermsii (35). The consequence of gene duplication in B. hermsii is to evade the host immune response through multiphasic antigenic variation by sequential expression of genes of variable major proteins (52). For the Bit system, the reason for this gene duplication is unclear and the minor amino acid sequence differences observed between the Bit periplasmic binding proteins might reflect a difference in the affinity for iron binding or for different iron sources.
The only proteins identified by Western blotting with B. hyodysenteriae antiserum were the two hydrophilic periplasmic binding lipoproteins BitB and BitC exposed to the periplasm. BitD, BitE, and BitF are more hydrophobic proteins and are hidden into the cytoplasmic membrane and probably not accessible to the anti-whole-cell B. hyodysenteriae antiserum. Interestingly, a strong stem-loop structure was found between the bitC and bitD. This structure was also found in the fbp, hit, and sfu operons, between the gene coding for the periplasmic substrate-binding protein and the gene coding for the ATP-binding protein (1). It has been proposed (1) that since stem-loop structures could function in mRNA stability (51), the presence of this structure might allow bitB and bitC expression at higher levels than that of bitD, bitE, and bitF. Therefore, the products of bitDEF could be less strongly expressed and not detectable. The BitABC transport system includes genes that are physically linked. The different plasmid constructions might indicate that two promoters are likely to exist in the bit gene cluster, since deletion of the 5′ region upstream of bitC, including bitB, did not affect its expression. However, vector promoters can be responsible for the expression observed.
BitD is the ABC transporter polypeptide with two highly conserved sequences, called Walker motifs A and B, which are believed to form the ATP-binding pocket involved in active transport (28).
BitE and BitF are the inner membrane permeases with membrane-spanning segments (six for each protein) that form the pathway through which substrate crosses the membrane. They bear strong homology to each other, indicating that they originated by a gene duplication, as suggested for other cytoplasmic membrane permeases, HisQ and HisM (3). BitE and BitF are supposed to form a pseudodimer that is closely associated with BitD. Similar to the Bit periplasmic iron transport system, the Yfe periplasmic iron transport system of Yersinia pestis possessed two cytoplasmic membrane permeases, YfeC and YfeD (8). The conserved EAA loop motifs (33) observed in BitE and BitF probably interact directly with the BitD protein.
Iron can exist in two redox states which are dependent upon pH and oxygen availability. Naturally abundant iron is not readily available to microbes in aerobic aqueous environments at neutral pH, where it exists as insoluble ferric complexes. Some gram-negative bacteria such as E. coli solubilize and scavenge ferric iron [Fe(III)] by means of excreted siderophores molecules with high affinities for Fe(III). In anaerobic environment, the iron is in the ferrous [Fe(II)] form (45). An Fe(II) uptake system has been characterized in E. coli (30). Although B. hyodysenteriae has been shown to express at least three iron-regulated proteins when grown under iron-restricted conditions (40), it was shown to be unable to use siderophores such as catechol and hydroxamate. Like other anaerobes, this anaerobic spirochete may not secrete siderophore. Uptake of the metal can occur via a ferrous transport system, from host iron-binding proteins such as degraded products of hemoglobin, or from ferrated transferrin or ferrated lactoferrin. It could be envisioned that degraded products of hemoglobin such as hemin or heme are readily available because the hemolytic activity of B. hyodysenteriae could serve as a source of iron. It was observed for a nontypeable H. influenzae strain that mutation of hitC from the hitABC operon prevents this strain to utilize these sources of iron, demonstrating that this system is implicated in the uptake of iron from transferrin or iron chelates (58). It is conceivable that the Bit system described in this report is similarly implicated in iron transport from these host iron sources. The presence of different periplasmic iron-binding proteins in the Bit system tends to support this hypothesis.
We propose a model for the BitABC system. Periplasmic iron-binding proteins (BitA, BitB, or BitC) bind iron from different sources, Fe(III) or hemin, and change conformation. Then the protein-iron complex interacts with the cytoplasmic membrane complex formed by the ABC transporter protein (BitD) and the permeases (BitE and BitF). The importation of iron into the cytoplasm of the bacterial cell is then coupled with ATP hydrolysis.
The intracellular level of iron is carefully controlled in the bacterial cell. Storage of iron will reduce the growth of bacteria, whereas a high concentration of the metal can be toxic. Therefore, the expression of iron acquisition systems is regulated in response to iron, being increased under iron limitation. Iron-dependent regulation of genes involved in iron acquisition is under the transcriptional control of the holorepressor composed of the Fur protein and the cofactor Fe(II) in a large number of gram-negative bacteria (16, 62). The operator consensus palindromic sequence (iron box or Fur box) has been identified on promoter regions of a number of iron-controlled genes including iron transport-related genes (17). Analysis of the results indicated that the presence or absence of a putative Fur box in different recombinant clones did not affect the level of β-galactosidase expression of the fhuF::lacZ fusion. Although the presence of the bit sequences did not titrate out the E. coli Fur protein from the FURTA system, we cannot conclude that there are no Fur-binding sequences in their insert. We can only conclude that if these sequences are present, the E. coli Fur protein does not recognize them. Fur regulation might not act on the regulation of bit gene expression as demonstrated for the N. gonorrhoeae iron periplasmic import system, fbpABC (17). In spirochetes, no Fur-regulated genes have been identified. Interestingly, a DtxR homologue has been cloned in Treponema pallidum, indicating that regulation of iron in spirochetes might function similarly to some gram-positive bacteria. The first example of a gram-positive iron-regulated promoter-operator was that of the diphtheria toxin gene, toxA, by an Fe(II)-dependent DNA-binding repressor, DtxR (11). It is now clear that DtxR functions as a global iron-sensitive regulatory element in the control of gene expression in some gram-negative bacteria (15). It would be interesting to investigate if the DtxR homologue is present in B. hyodysenteriae.
A correlation has been established between the synthesis of an iron periplasmic transport system and virulence of bacterial pathogens such as N. gonorrhoeae (1, 10), H. influenzae (2, 58), Serratia marcescens (5, 71), A. pleuropneumoniae (13), Yersinia enterocolitica (56a), and Y. pestis (8). Furthermore, the ability to compete for iron in the host environment correlates with pathogenicity of Neisseria species. Indeed, the iron periplasmic binding protein Nfbp is expressed at antigenically detectable levels only in the microbial pathogens N. gonorrhoeae and N. meningitidis but not in commensal Neisseria species such as N. sicca or N. perflava when propagated under iron stress (9, 42). The Bit system is detected only in B. hyodysenteriae and is absent in B. innocens and B. pilosicoli. The Bit system might correlate with the pathogenicity of B. hyodysenteriae. Inactivating the Bit system in B. hyodysenteriae will enable us to better understand its role in the pathogenicity of this spirochete.
ACKNOWLEDGMENTS
This project was supported by grants from Conseil de Recherche en Pêches et Agro-alimentaire du Québec (JH2983) and from the Ministère de l’Industrie, des Sciences et Technologies, Canada.
We thank J. Coulton (McGill University) for the kind gift of E. coli SG303 and MC4100 and I. Stojijlkovic and K. Hantke for strain H1717. We also thank Julie Couture, Céline Forget, Bernadette Foiry, and Nathalie Boudreault for excellent technical assistance.
REFERENCES
- 1.Adhikari P, Berish S A, Nowalk A J, Veraldi K L, Morse S A, Mietzner T A. The fbpABC locus of Neisseria gonorrhoeae functions in the periplasm-to-cytosol transport of iron. J Bacteriol. 1996;178:2145–2149. doi: 10.1128/jb.178.7.2145-2149.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adhikari P, Kirby S D, Nowalk A J, Veraldi K L, Schyvers A B, Mietzner T A. Biochemical characterization of a Haemophilus influenzae periplasmic iron transport operon. J Biol Chem. 1995;270:25142–25149. doi: 10.1074/jbc.270.42.25142. [DOI] [PubMed] [Google Scholar]
- 3.Ames G F. Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu Rev Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- 4.Ames G F-L. Energetics of periplasmic transport systems. In: Krulwich T A, editor. Bacterial energetics. New York, N.Y: Academic Press; 1990. pp. 225–245. [Google Scholar]
- 5.Angerer A, Klupp B, Braun V. Iron transport systems of Serratia marcescens. J Bacteriol. 1992;174:1378–1387. doi: 10.1128/jb.174.4.1378-1387.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angerer A, Klupp B, Braun V. Nucleotide sequences of the sfuA, sfuB, and sfuC genes of Serratia marcescens suggests a periplasmic-binding-protein-dependent iron transport mechanism. J Bacteriol. 1990;172:572–578. doi: 10.1128/jb.172.2.572-578.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baum D H, Joens L A. Serotypes of beta-hemolytic Treponema hyodysenteriae. Infect Immun. 1979;25:792–796. doi: 10.1128/iai.25.3.792-796.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bearden S W, Staggs T M, Perry R D. An ABC transporter system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J Bacteriol. 1998;180:1135–1147. doi: 10.1128/jb.180.5.1135-1147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berish S A, Chen C-Y, Mietzner T A, Morse S A. Expression of a functional nesserial fbp gene in Escherichia coli. Mol Microbiol. 1992;6:2607–2615. doi: 10.1111/j.1365-2958.1992.tb01438.x. [DOI] [PubMed] [Google Scholar]
- 10.Berish S A, Mietzner T A, Mayer L W, Genco C A, Holloway B P, Morse S A. Molecular cloning and characterization of the structural gene for the major iron-regulated protein expressed by Neisseria gonorrhoeae. J Exp Med. 1992;171:1535–1546. doi: 10.1084/jem.171.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyd J, Oza M N, Murphy J R. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc Natl Acad Sci USA. 1990;87:5968–5972. doi: 10.1073/pnas.87.15.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cadavid D, Pennington P M, Kerentseva T A, Bergstrom S, Barbour A G. Immunologic and genetic analyses of VmpA of a neurotropic strain of Borrelia turicatae. Infect Immun. 1997;65:3352–3360. doi: 10.1128/iai.65.8.3352-3360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin N, Frey J, Chang C-F, Chang Y-F. Identification of a locus involved in the utilization of iron by Actinobacillus pleuropneumoniae. FEMS Microbiol Lett. 1996;143:1–6. doi: 10.1111/j.1574-6968.1996.tb08452.x. [DOI] [PubMed] [Google Scholar]
- 14.Covitz K M, Panagiotidis C H, Hor L I, Reyes M, Treptow N A, Shuman H A. Mutations that alter the transmembrane signalling pathway in an ATP binding cassette (ABC) transporter. EMBO J. 1994;13:1752–1759. doi: 10.1002/j.1460-2075.1994.tb06439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desai P J, Angerer A, Attardo Genco C. Analysis of Fur binding to operator sequences within the Neisseria gonorrhoeae fbpA promoter. J Bacteriol. 1996;178:5020–5023. doi: 10.1128/jb.178.16.5020-5023.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doige C A, Ames G F. ATP-dependent transport systems in bacteria and humans: relevance to cystic fibrosis and multidrug resistance. Annu Rev Microbiol. 1993;47:291–319. doi: 10.1146/annurev.mi.47.100193.001451. [DOI] [PubMed] [Google Scholar]
- 20.Dugourd D, Jacques M, Bigras-Poulin M, Harel J. Characterization of Serpulina hyodysenteriae isolates of serotypes 8 and 9 by random amplification of polymorphic DNA analysis. Vet Microbiol. 1996;48:305–314. doi: 10.1016/0378-1135(95)00149-2. [DOI] [PubMed] [Google Scholar]
- 21.Fath J M, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, R. K A, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 23.Gabe J D, Dragon E, Chang R J, McCaman M T. Identification of a linked set of genes in Serpulina hyodysenteriae (B204) predicted to encode closely related 39-kilodalton extracytoplasmic proteins. J Bacteriol. 1998;180:444–448. doi: 10.1128/jb.180.2.444-448.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gribskov M, Devereux J. Sequence analysis primer. W. H. Madison, Wis: Freeman & Co.; 1992. [Google Scholar]
- 25.Harel J, Daigle F, Maiti S, Desautels C, Labigne A, Fairbrother J M. Occurrence of pap-, sfa-, and afa-related sequences among F165-positive Escherichia coli from diseased animals. FEMS Microbiol Lett. 1991;66:177–182. doi: 10.1016/0378-1097(91)90329-9. [DOI] [PubMed] [Google Scholar]
- 26.Harris D J, Glock R D. Swine dysentery and spirochaetal disease. In: Leman A D, Straw B, Glock R D, Mengeling W L, Penny R H C, Scholl E, editors. Diseases of swine. 6th ed. Ames: Iowa State University Press; 1986. pp. 494–507. [Google Scholar]
- 27.Hendrix L R, Mallavia L P, Samual J E. Cloning and sequencing of Coxiella burnetti outer membrane protein gene com1. Infect Immun. 1993;61:470–477. doi: 10.1128/iai.61.2.470-477.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 29.Hyatt D R, ter Huurne A A, van der Zeijst B A, Joens L A. Reduced virulence of Serpulina hyodysenteriae hemolysin-negative mutants in pigs and their potential to protect pigs against challenge with a virulent strain. Infect Immun. 1994;62:2244–2248. doi: 10.1128/iai.62.6.2244-2248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kammler M, Schön C, Hantke K. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol. 1993;175:6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 32.Kennedy M J, Rosey E L, Yancey R J., Jr Characterization of flaA− and flaB− mutants of Serpulina hyodysenteriae: both flagellin subunits, FlaA and FlaB, are necessary for full motility and intestinal colonization. FEMS Microbiol Lett. 1997;153:119–128. doi: 10.1111/j.1574-6968.1997.tb10472.x. [DOI] [PubMed] [Google Scholar]
- 33.Kerpola R E, Ames G F-L. Topology of the hydrophobic membrane-bound components of the histidine periplasmic permease. J Biol Chem. 1992;267:2329–2336. [PubMed] [Google Scholar]
- 34.Kinyon J M, Harris D L, Glock R D. Enteropathogenicity of various isolates of Treponema hyodysenteriae. Infect Immun. 1977;15:638–646. doi: 10.1128/iai.15.2.638-646.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitten T, Barrera A V, Barbour A G. Intragenic recombination and a chimeric outer membrane protein in the relapsing-fever agent Borrelia hermsii. J Bacteriol. 1993;175:2516–2522. doi: 10.1128/jb.175.9.2516-2522.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kyte J, Doolittle R F. A simple method for displaying the hydrophobic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli U K. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Belanger M, Jacques M. Serotyping of Canadian isolates of Treponema hyodysenteriae and description of two new serotypes. J Clin Microbiol. 1991;29:2794–2797. doi: 10.1128/jcm.29.12.2794-2797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Z, Dumas F, Dubreuil D, Jacques M. A species-specific periplasmic flagellar protein of Serpulina (Treponema) hyodysenteriae. J Bacteriol. 1993;175:8000–8007. doi: 10.1128/jb.175.24.8000-8007.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z, Foiry B, Jacques M. Growth of Serpulina (Treponema) hyodysenteriae under iron-restricted conditions. Can J Vet Res. 1995;59:149–153. [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, Jensen N S, Belanger M, L’Esperance M C, Jacques M. Molecular characterization of Serpulina (Treponema) hyodysenteriae isolates representing serotypes 8 and 9. J Clin Microbiol. 1992;30:2941–2947. doi: 10.1128/jcm.30.11.2941-2947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mietzner T A, Luginbuhl G H, Sandström E, Morse S A. Identification of an iron-regulated 37,000-dalton protein in the cell envelope of Neisseria gonorrhoeae. Infect Immun. 1984;45:410–416. doi: 10.1128/iai.45.2.410-416.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muir S, Koopman M B, Libby S J, Joens L A, Heffron F, Kusters J G. Cloning and expression of a Serpula (Treponema) hyodysenteriae hemolysin gene. Infect Immun. 1992;60:529–535. doi: 10.1128/iai.60.2.529-535.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neilands J B. Effects of iron deprivation on outer membrane protein expression. Methods Enzymol. 1994;235:344–350. doi: 10.1016/0076-6879(94)35152-x. [DOI] [PubMed] [Google Scholar]
- 45.Neilands J B. Microbial iron compounds. Annu Rev Biochem. 1981;50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 46.Novak J, Novak L, Shah G R, Woodruff W A, Caufield P W. Transposon mutagenesis: cloning of chromosomal DNA from the site of Tn916 insertion using polymerase chain reaction. Biotechnol Tech. 1997;11:51–54. [Google Scholar]
- 47.Oaks E V, Stover C K, Rice R. Molecular cloning and expression of Rickettsia tsutsugamushi genes for two major protein antigens in Escherichia coli. Infect Immun. 1987;55:1156–1162. doi: 10.1128/iai.55.5.1156-1162.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ochiai S, Adachi Y, Mori K. Unification of the genera Serpulina and Brachyspira, and proposals of Brachyspira hyodysenteriae comb. nov., Brachyspira innocens comb. nov., and Brachyspira pilosicoli comb. nov. Microbiol Immunol. 1997;41:445–452. doi: 10.1111/j.1348-0421.1997.tb01877.x. [DOI] [PubMed] [Google Scholar]
- 49.Ozenberger B A, Nahlik M S, McIntosh M A. Genetic organization of multiple fep genes encoding ferric enterobactin transport functions in Escherichia coli. J Bacteriol. 1987;169:3638–3646. doi: 10.1128/jb.169.8.3638-3646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Payne S M, Finkelstein R A. Detection and differentiation of iron-responsive avirulent mutants on Congo red agar. Infect Immun. 1977;18:94–98. doi: 10.1128/iai.18.1.94-98.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petersen C. Multiple determinants of functional mRNA stability: sequence alterations at either end of the lacZ gene affect the rate of mRNA inactivation. J Bacteriol. 1991;173:2167–2172. doi: 10.1128/jb.173.7.2167-2172.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plasterk R H, Simon M I, Barbour A G. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature. 1985;318:257–263. doi: 10.1038/318257a0. [DOI] [PubMed] [Google Scholar]
- 53.Promega. Promega protocols and application guide. 2nd ed. Madison, Wis: Promega Corp.; 1991. pp. 175–198. [Google Scholar]
- 54.Rioux C R, Bergeron H, Lin L, Grothe S, O’Connor-McCourt M, Lau P C. A fusion plasmid for the synthesis of lipopeptide-antigen chimeras in Escherichia coli. Gene. 1992;116:13–20. doi: 10.1016/0378-1119(92)90623-w. [DOI] [PubMed] [Google Scholar]
- 55.Rosey E L, Kennedy M J, Petrella D K, Ulrich R G, Yancey R J. Inactivation of Serpulina hyodysenteriae flaA1 and flaB1 periplasmic flagellar genes by electroporation-mediated allelic exchange. J Bacteriol. 1995;177:5959–5970. doi: 10.1128/jb.177.20.5959-5970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosey E L, Kennedy M J, Yancey R J., Jr Dual flaA1 flaB1 mutant of Serpulina hyodysenteriae expressing periplasmic flagella is severely attenuated in a murine model of swine dysentery. Infect Immun. 1996;64:4154–4162. doi: 10.1128/iai.64.10.4154-4162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56a.Saken E M. GenBank accession no. U50903. 1995. [Google Scholar]
- 57.Sambrook J, Fritsch E J, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 58.Sanders J D, Cope L D, Hansen E J. Identification of a locus involved in the utilization of iron by Haemophilus influenzae. Infect Immun. 1994;62:4515–4525. doi: 10.1128/iai.62.10.4515-4525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saurin W, Köster W, Dassa E. Bacterial binding protein-dependent permeases: characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol Microbiol. 1994;12:993–1004. doi: 10.1111/j.1365-2958.1994.tb01087.x. [DOI] [PubMed] [Google Scholar]
- 61.Schwan T G, Burgdorfer W, Rosa P A. Borrelia. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. pp. 626–635. [Google Scholar]
- 62.Silver S, Walderhaug M. Gene regulation of plasmid- and chromosome-determined inorganic ion transport in bacteria. Microbiol Rev. 1992;56:195–228. doi: 10.1128/mr.56.1.195-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stanton T B, Hanzelka B L, Jensen N S. Survey of intestinal spirochaetes for nadh oxidase by gene probe and by enzyme assay. Microb Ecol Health Dis. 1995;8:93–100. [Google Scholar]
- 64.Stanton T B, Jensen N S, Casey T A, Tordoff L A, Dewhirst F E, Paster B J. Reclassification of Treponema hyodysenteriae and Treponema innocens in a new genus, Serpula gen. nov., as Serpula hyodysenteriae comb. nov. and Serpula innocens comb. nov. Int J Syst Bacteriol. 1991;41:50–58. doi: 10.1099/00207713-41-1-50. [DOI] [PubMed] [Google Scholar]
- 65.Stojiljkovic I, Baumler A J, Hantke K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J Mol Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 66.Tatusov R L, Mushegian A R, Bork P, Brown N P, Hayes W S, Borodovsky M, Rudd K E, Koonin E V. Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli. Curr Biol. 1996;6:279–291. doi: 10.1016/s0960-9822(02)00478-5. [DOI] [PubMed] [Google Scholar]
- 67.Ter Huurne A A H M, Muir S, van Houten M, van der Zeijst B A, Gaastra W, Kusters J G. Characterization of three putative Serpulina hyodysenteriae hemolysins. Microb Pathog. 1994;16:269–282. doi: 10.1006/mpat.1994.1028. [DOI] [PubMed] [Google Scholar]
- 68.Ter Huurne A A H M, Muir S, van Houten M, Koopman M B, Kusters J G, van der Zeijst B A, Gaastra W. The role of hemolysin(s) in the pathogenesis of Serpulina hyodysenteriae. Int J Med Microbiol Virol Parasitol Infect Dis. 1993;278:316–325. doi: 10.1016/s0934-8840(11)80848-0. [DOI] [PubMed] [Google Scholar]
- 69.Trott D J, Stanton T B, Jensen N S, Duhamel G E, Johnson J L, Hampson D J. Serpulina pilosicoli sp. nov., the agent of porcine intestinal spirochetosis. Int J Syst Bacteriol. 1996;46:206–215. doi: 10.1099/00207713-46-1-206. [DOI] [PubMed] [Google Scholar]
- 70.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zimmermann L, Angerer A, Braun V. Mechanistically novel iron(III) transport system in Serratia marcescens. J Bacteriol. 1989;171:238–243. doi: 10.1128/jb.171.1.238-243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zuerner R L. Genetic organization in spirochaetes. In: Hampson D J, Stanton T B, editors. Intestinal spirochaetes in domestic animals and humans. Ames, Iowa: CAB International; 1997. pp. 63–89. [Google Scholar]
- 73.Zuerner R L, Stanton T B. Physical and genetic map of the Serpulina hyodysenteriae B78T chromosome. J Bacteriol. 1994;176:1087–1092. doi: 10.1128/jb.176.4.1087-1092.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]