Abstract
Articular cartilage has low regenerative capacity despite permanent stress. Irreversible cartilage lesions characterize osteoarthritis (OA); this is not followed by tissue repair. Lin28a, an RNA binding protein, is detected in damaged cartilage in humans and mice. We investigated the role of LIN28a in cartilage physiology and in osteoarthritis. Lin28a-inducible conditional cartilage deletion up-regulated Mmp13 in intact mice and exacerbated the cartilage destruction in OA mice. Lin28a-specific cartilage overexpression protected mice against cartilage breakdown, stimulated chondrocyte proliferation and the expression of Prg4 and Sox9, and down-regulated Mmp13. Lin28a overexpression inhibited Let-7b and Let-7c miRNA levels while RNA-sequencing analysis revealed five genes of transcriptional factors regulated by Let-7. Moreover, Lin28a overexpression up-regulated HMGA2 and activated SOX9 transcription, a factor required for chondrocyte reprogramming. HMGA2 siRNA knockdown inhibited the cartilage protective effect of Lin28a overexpression. This study provides insights into a new pathway including the Lin28a-Let7 axis, thus promoting chondrocyte anabolism in injured cartilage in mice.
Lin28a facilitates HMGA2 and Sox9 transcription, and stimulates chondrocyte reprogramming and cartilage regeneration in osteoarthritis.
INTRODUCTION
Cell reprogramming is a biological process that allows for regenerating tissues with low regenerative capacity. The absence of cartilage repair characterizes joint diseases such as osteoarthritis (OA), in which cartilage breakdown is promoted by mechanical overload, metabolic processes, or aging (1–6). In OA, the catabolic activity of chondrocytes is promoted in part by the loss of hypoxia, which triggers the transcription of matrix metalloproteinase 13 (MMP13) as well as cartilage loss (5). OA chondrocytes are committed to hypertrophic specification and produce metalloproteases such as MMPs, which are catabolic factors that are not compensated by an anabolic process. Hence, OA chondrocytes fail to promote matrix production, as observed in undamaged articular chondrocytes. In such chondrocytes, collagen type 2 (Col2), aggrecan, and lubricin [proteoglycan 4 (PRG4)] are synthesized by the activation of the transcription factor SRY-box transcription factor 9 synthase 9 (SOX9). Furthermore, reactivation of chondrocyte proliferation is limited by the low number of chondrocyte progenitors in OA (7) located at the surface of the articular cartilage and expressing SOX9 and PRG4. These cells have a low proliferation rate (8) and high capacity for colony formation as well as the expression of mesenchymal stem cell markers (9, 10). PRG4-expressing cells could form a fully differentiated articular matrix as observed at the surface of osteophytes, the bone spur that develops with OA (11). However, the progenitors are the first cells targeted in OA matrix and rapidly disappear at the onset of cartilage catabolism.
Other stem cells are present in the articular cavity such as in the synovial membrane (12), the Ranvier groove (13), and the subchondral bone marrow (14). To promote tissue repair, several strategies have been developed with local or heterotopic stem cells, alone or by tissue engineering. Repair results in the replacement of local cartilage defects by fibrocartilage (15), but the new cartilage fails to recapitulate the mechanical properties and articular function of native cartilage and thus might be a source of long-term pain and disability (16). Therefore, repair is limited and strategies are needed to enhance cartilage regeneration by activating stem cells or reprogramming chondrocytes to promote fully functional cartilage.
Here, we investigated the effect of Lin28a on the physiology of articular cartilage homeostasis and in cartilage repair. Lin28a is an RNA-binding protein expressed in early developmental stages that orchestrates cell differentiation or proliferation in various tissues, notably in the embryonic germ bud (17). In human adults, Lin28a is expressed in early stem cells such as germinal cells (18) and dental pulp (19). It is involved in tissue regeneration in zebrafish (20) and is required to generate induced pluripotent stem cells in vitro. Reprogramming requires the activation of octamer-binding transcription factor 4 (OCT4), NANOG, and SOX2 in addition to Lin28a (21). In chondrocytes, Lin28a modulates the differentiation at the growth plate with the ability to inhibit microRNA (miRNA) activity. Specifically, Lin28a inhibits the Let-7 miRNA family, which plays a critical role in chondrocyte differentiation. Let-7 miRNA expression is associated with reduced SOX9 expression (22). Hence, SOX9 is the transcription factor required for the differentiation of mesenchymal stem cells into chondrocytes but is also required for maintaining chondrocyte progenitors and reprogramming mature chondrocytes to chondrocyte progenitors (23).
We aimed to investigate Lin28a as a target for inducing cartilage regeneration in vivo. We show that in mouse or human OA cartilage samples, chondrocytes re-express Lin28a. The inducible-specific deletion of Lin28a in chondrocytes up-regulated Mmp13 expression in sham mice and enhanced cartilage degradation in OA mice; inducible-specific overexpression limited cartilage degradation. The induction of Lin28a sustained matrix integrity and allowed for re-expression of chondrocyte anabolic factors and increased cell proliferation. We also show that Lin28a specifically inhibited Let-7b and Let-7c miRNA levels and the expression of high mobility group A2 (HMGA2), a chromatin remodeling factor that binds to the SOX9 promoter, thereby increasing its expression and promoting chondrocyte reprogramming. Last, HMGA2 knockdown by small interfering RNA (siRNA) aggravated OA in mice. These data show that reactivation of Lin28a in chondrocytes could be a relevant therapeutic target to regenerate damaged tissue such as in OA.
RESULTS
Lin28a is required for cartilage maintenance
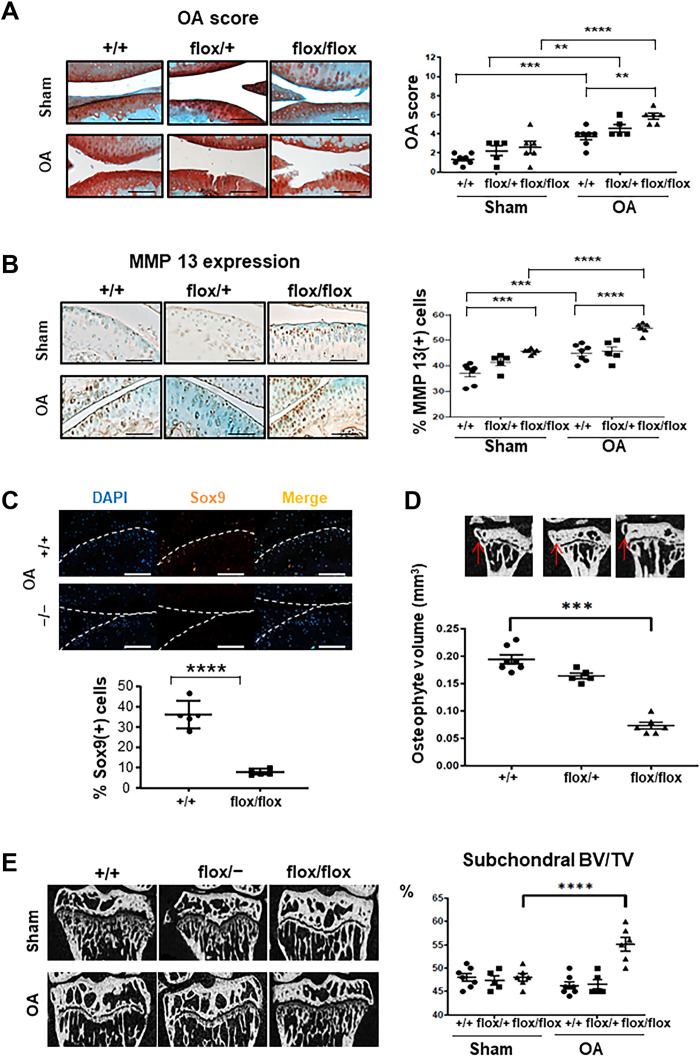
Because Lin28 is involved in many regeneration processes, we asked whether Lin28a could be re-expressed in cartilage injuries and analyzed whether the reactivation occurs in damaged cartilage. Immunohistochemistry revealed the presence of clusters of Lin28a-positive chondrocytes in damaged cartilage as compared with healthy tissue in patients and mice with OA (fig. S1A). To investigate the function of Lin28a in OA, we generated mice with inducible cartilage-specific knockout of Lin28atm1.2Gqda tm1.2Gqda/Col2a1-CreER (hereafter called flox/flox) or haplo-insufficiency in Lin28atm1.2Gqda/+/Col2a1-CreER (hereafter called flox/+) by using recombination induced by tamoxifen administration at age 9 weeks. Col2a1-CreER mice were controls (CTs) (hereafter Lin28a+/+ mice). After confirming LIN28A knockdown in Lin28aflox/flox mice (fig. S1B), OA was induced by destabilization of the medial meniscus (DMM) in 10-week-old mice, which were euthanized 8 weeks later (18 weeks old). Safranin-O staining revealed a higher OA score in Lin28aflox/flox than in Lin28a+/+ mice (Fig. 1A), along with higher MMP13 expression (Fig. 1B). Moreover, the number of MMP13+ cells was lower in Lin28a+/+ than in Lin28aflox/flox in both sham and OA mice (Fig. 1B), and the number of apoptotic chondrocytes was higher (fig. S1C), which suggests a role for Lin28a in cartilage physiology. SOX9 expression was lower in Lin28aflox/flox than in Lin28a+/+ OA mice (Fig. 1C). Osteophyte volume was markedly lower in Lin28aflox/flox than in Lin28a+/+ OA mice (Fig. 1D), but subchondral bone volume (BV/TV) was increased in Lin28aflox/flox OA mice (Fig. 1E). Together, these data demonstrate that Lin28a is required to sustain cartilage integrity in the physiological condition and in OA.
Fig. 1. Lin28a-deficient chondrocytes exacerbate cartilage degradation in mice with OA.
Ablation of Lin28a in chondrocytes was induced in Lin28atm1.2Gqda/Col2a1-CreER knockout homozygous (flox/flox) and heterozygous (flox/+) mice by intraperitoneal tamoxifen injection in 9-week-old mice; Col2a1-CreER (+/+) littermates were controls (CTs). OA was induced at age 10 weeks, and then mice were euthanized 8 weeks after OA induction and analyzed at age 18 weeks. (A) Safranin-O staining of sham and OA joints (scale bars, 100 μm). Graph represents OA score in sham and OA joints. (B) Immunohistochemistry of MMP13 content (scale bars, 100 μm). Graph represents the percentage of MMP13-positive cells in sham and OA mice. (C) Immunofluorescence of SOX9 in OA mice (scale bars, 200 μm). Graph represents the percentage of Sox9-positive cells. (D) Osteophyte volume analyzed by microtomography in OA mice. (E) Subchondral bone volume to total volume (BV/TV) analyzed by microtomography in sham and OA mice and quantification. Data are means ± SEM. **P < 0.01, ***P < 0.005, and ****P < 0.001.
Lin28a overexpression promoted anabolism and inhibited catabolism in chondrocytes
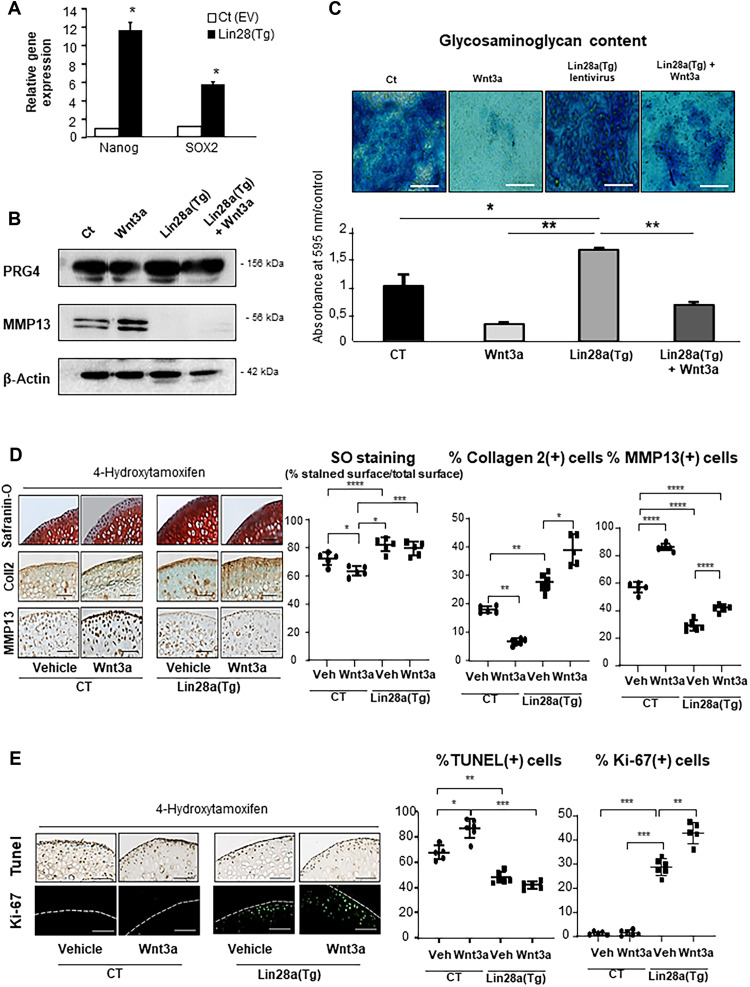
To better decipher the role of Lin28a in chondrocytes, primary murine chondrocytes were transduced with Lin28a by using a lentiviral vector (fig. S2A) and cultured under hypoxic (1% O2) conditions with or without Wnt3a conditioned medium (CM-Wnt3a) to activate chondrocyte catabolism. Lin28a overexpression promoted the expression of NANOG and SOX, markers of stemness in chondrocytes (Fig. 2A). Lin28a overexpression also enhanced the basal level of PRG4 and inhibited the protein expression of MMP13 in primary chondrocytes (Fig. 2B). The expression profile remained the same upon Wnt3a treatment (Fig. 2B), which indicates sustained activation of PRG4 and inhibition of MMP13 even in a pro-catabolic environment.
Fig. 2. Lin28a overexpression increased extracellular matrix production in vitro.
Wild-type mouse primary chondrocytes were transduced with Lin28a lentivirus [Lin28a(TG)] or empty vector [Ct (EV)] and cultured for 48 hours or 1 week at 1% O2 in the presence of Wnt3a conditioned medium (Wnt3a-CM) to trigger chondrocyte catabolism. (A) RT-qPCR analysis of mRNA levels of stemness genes. (B) Western blot analysis of PRG4 and MMP13 protein expression in primary chondrocytes transduced with Ct (EV), Ct (EV), with Wnt3a-CM (wnt3a), Lin28a(TG), and Lin28a lentivirus + Wnt3a-CM [Lin28a(TG) + Wnt]. (C) Alcian Blue staining and spectrophotometry quantification of sulfated glycosaminoglycans in primary chondrocytes after 1 week of culture (scale bars, 200 μm). Femoral explants were harvested from 10-week-old TgLin28aflox/flox/Col2a1-CreER [Lin28a(Tg)] or Col2a1-CreER (CT) mice and treated with Wnt3a-CM (Wnt3a) or not (vehicle). 4-Hydroxitamoxifen was used to induce in vitro recombination. (D) Safranin-O staining and immunohistochemistry (collagen 2, MMP13) of CT and Lin28a(Tg) explants (scale bars, 100 μm). Graphs show the percentage of ratio of Safranin-O–unstained cartilage to Safranin-O–positive cartilage and the percentage of MMP13-positive and collagen 2–positive cells. (E) Apoptosis was assessed by terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) assay and proliferation by Ki-67 immunofluorescence staining. Data are means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001.
We further determined whether the anabolic effect of Lin28a increased the content of glycosaminoglycans (GAGs) and production of matrix. Lin28a overexpression stimulated the production of GAGs as revealed by Alcian blue assay (Fig. 2C) and partially rescued the Wnt3a-induced pro-catabolic effect. We then harvested cartilage explants from Lin28aflox/flox/Col2a1-CreER mice and induced the ex vivo recombination with 4-hydroxytamoxifen. Explants of Lin28a-overexpressing chondrocytes are hereafter called Lin28a (Tg), and explants from Col2a1-CreER mice were the CT. Immunohistochemistry revealed that 1.42% chondrocytes were Lin28a-positive in CT explants, whereas 30.43% chondrocytes are positive in Lin28a(Tg) explants (P < 0.005; fig. S2B). Lin28a(Tg) explants showed higher GAG content as assessed by Safranin-O staining, increased Col2 level, and reduced MMP13 level as compared with CTs at baseline (Fig. 2D). This effect was maintained with Wnt3a stimulation. In addition, Lin28a overexpression reduced the number of apoptotic cells and promoted the proliferation of chondrocytes (Fig. 2E). These data show that Lin28a activated the stemness and maintained chondrocyte anabolism, even under pro-catabolic conditions.
To compare the effect in hypoxic conditions, we evaluated the impact of Lin28a overexpression under 21% of oxygen. Western blot analysis of PRG4 and MMP13 revealed the same ability of Lin28a to sustain anabolic gene expression and to repress catabolic genes even under catabolic stress induced by Wnt3a (fig. S3A). To further understand the protective function of Lin28a overexpression, we asked whether Lin28a interfered directly with Wnt canonical pathway in addition to inhibiting the Wnt3a-induced chondrocyte gene expression. RNA-sequencing (RNA-seq) analysis revealed, at the basal level, that Lin28a overexpression does not interfere with the expression of Wnt/β-catenin target genes such SLC7a5 (solute carrier family 7 member 5), TCF (T cell factor), LEF (lymphoid enhancer factor), and Axin2 (fig. S3B). However, we observed a significant increase of the four genes in the presence of Wnt3a. RNA-seq also showed that Lin28a alone have no effect on gene expression, but Lin28a overexpression had a different effect according to the gene in the presence of Wnt3a. Hence, Lin28a did not change SLC7a5 and axin2 gene expression under Wnt3a stimulation but inhibited TCF and LEF gene expression (fig. S3C). Because of inconsistent results in gene expression, we analyzed the effects of Lin28a overexpression in β-catenin stabilization induced by Wnt3a. Thus, we assessed the quantity of β-catenin by Western blot as a direct marker of the activation of the Wnt canonical pathway. Wnt3a promoted increased β-catenin content, and this expression seems to be not affected by the Lin28a overexpression (fig. S3D). Together, these results demonstrated that Lin28a did not alter the Wnt/β-catenin canonical pathway but was able to modulate the transcriptional activity of β-catenin. Albeit Wnt signaling promotes osteoarthritis, other cytokines activate chondrocyte metabolism. We therefore analyzed the effect of interleukin-1 (IL-1) stimulation in CT or Lin28a-overexpressing chondrocytes. IL-1 induced pro-catabolic gene expression (MMP-13) and reduced the pro-anabolic genes expression (SOX9 and PRG4) (fig. S4). This effect was strongly inhibited with Lin28a overexpression. Moreover, HMGA2 gene expression induced by Lin28a overexpression was not altered by IL-1. Together, these results indicated that Lin28a was able to limit and reverse the transition of control chondrocytes to OA chondrocytes under the stimulation of several OA inducers.
Lin28a overexpression prevented cartilage damage in mice
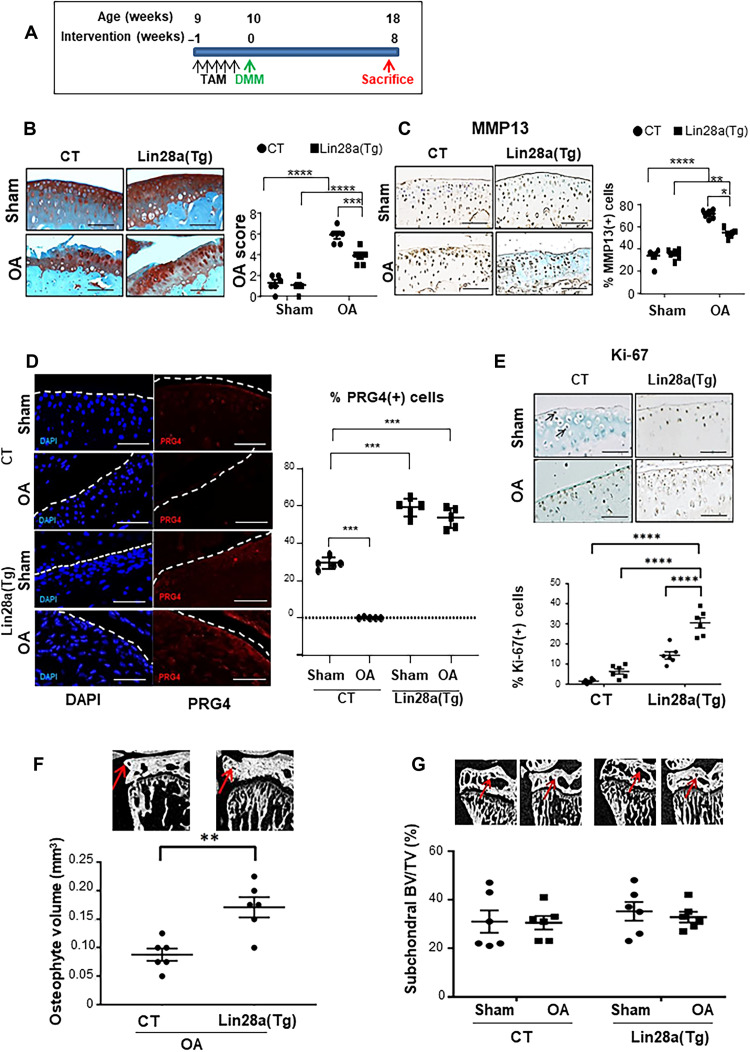
Given that deletion of Lin28a accelerated cartilage degradation and Lin28a overexpression maintained chondrocyte anabolism in cells and explants, we investigated whether the overexpression of Lin28a could prevent cartilage breakdown or regenerate tissue after cartilage damage. To first test whether Lin28a protects against cartilage loss, we used Lin28(Tg) mice with Lin28a expression stimulated at age 9 weeks by tamoxifen administration. OA was then induced in 10-week-old mice, which were euthanized at 18 weeks (Fig. 3A). Col2a1-CreER littermates were OA CT. Overexpression of Lin28a was confirmed in Lin28a(Tg) cartilage, and immunohistochemistry revealed 2.53% of positive cells in Col2a1-CreER (CT) mice and 60.47% in Lin28atm1.2Gqda/tm1.2Gqda/Col2a1-CreER [Lin28a(Tg)] (P < 0.001; fig. S5A). OA score and MMP13 level were lower in Lin28(Tg) OA mice than in OA CTs (Fig. 3, B and C). The anabolic factor PRG4 was slightly detectable in sham-operated CT mice and absent in OA CT mice, but the expression was noted again at the superficial layers of articular cartilage in sham-operated OA mice and further in Lin28(Tg) OA mice (Fig. 3D). Analysis of joints of Lin28(Tg) OA mice revealed increased chondrocyte proliferation (Fig. 3E) in parallel with increased osteophyte volume (Fig. 3F) but no change in subchondral bone volume (Fig. 3G).
Fig. 3. Lin28a overexpression prevented OA development in mice.
(A) Design of the prevention model applied to Col2a1-CreER (CT) and Tg Lin28aflox/flox/Col2a1-CreER [Lin28a(Tg)] mice. OA was induced in mice at age 10 weeks, and then mice were euthanized 8 weeks after OA induction and analyzed at age 18 weeks. (B) Safranin-O staining and quantification of articular cartilage in sham and OA CT and Lin28a(Tg) mice (scale bars, 100 μm). (C) Immunohistochemistry of MMP13 and quantification (scale bars, 100 μm). (D) Immunofluorescence of PRG4 and quantification (scale bars, 100 μm). (E) Immunohistochemistry of Ki-67 expression and quantification (scale bars, 100 μm). (F) Osteophyte volume analyzed by microtomography. (G) Subchondral BV/TV ratio analyzed by microtomography. Data are means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001.
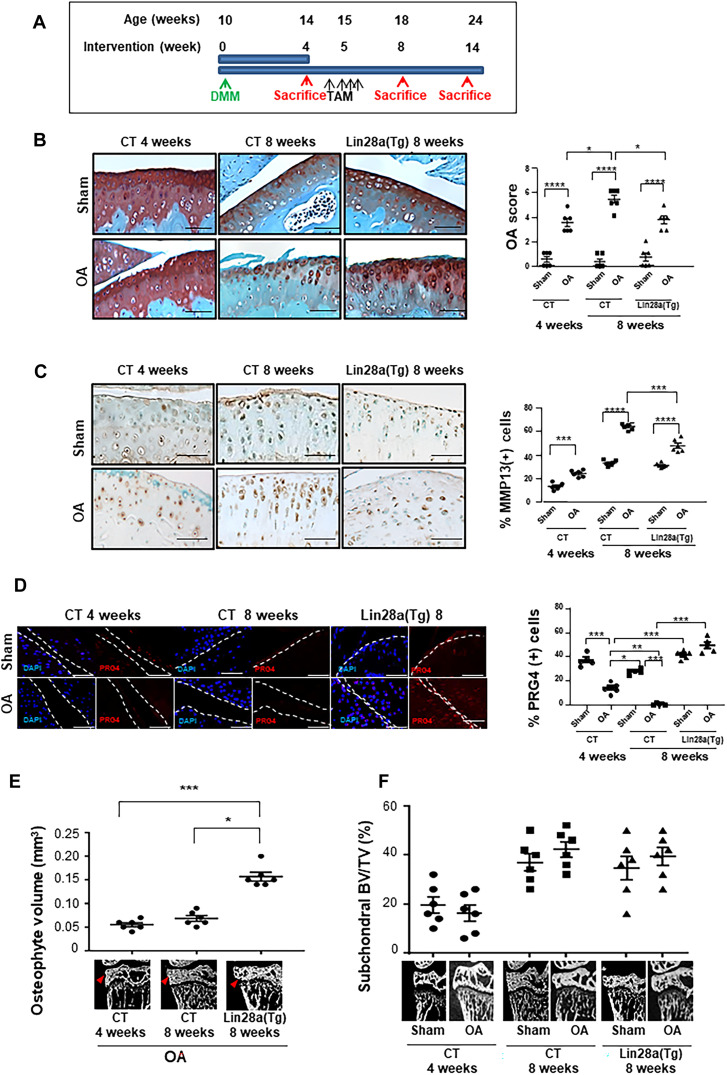
The induction of Lin28a in chondrocytes and explants suggested chondrocyte reprogramming revealed by the expression of PRG4 and SOX9. We then hypothesized that Lin28a may promote cartilage regeneration in damaged joints. To investigate whether Lin28a could reverse OA changes, we used a regenerative model of OA induced in 10-week-old mice at baseline (hereafter called week 0). In this experiment, the expression of Lin28a was induced after the onset of OA (4 week after DMM) to allow cartilage damage to occur. Mice were euthanized 4 weeks later (designated as 8 weeks) (Fig. 4A). In contrast to CT OA mice, which showed significantly increased OA score between 4 and 8 weeks, OA scores for Lin28a(Tg) mice were similar at week 8 to those for CT mice at week 4 (Fig. 4B), which suggests that Lin28a overexpression in cartilage limited the progression of cartilage damage. Furthermore, 14 weeks after DMM induction in CT mice, the OA score peaked, which meant that more than 75% of the superficial cartilage layer was lost. In contrast, Lin28a(Tg) OA mice showed a lower cartilage loss (OA score about 7); thus, the protective effect was sustained long term after Lin28a activation (fig. S4C). Accordingly, MMP13 level increased between 4 and 8 weeks in CT mice but not Lin28a(Tg) OA mice (Fig. 4C).
Fig. 4. Lin28a overexpression abolished cartilage destruction and restored anabolism in OA.
(A) Schematic representation of regenerative protocol applied to Col2a1-CreER (CT) and Lin28aflox/flox/Col2a1-CreER [Lin28a(Tg)] mice. OA was induced in mice at age 10 weeks, then mice were euthanized at 4, 8, and 14 weeks after OA induction. (B) Safranin-O staining and quantification of OA score for articular cartilage in sham and OA CT and Lin28a(Tg) mice (scale bars, 100 μm). (C) Immunohistochemistry of MMP13 and quantification (scale bars, 100 μm). (D) Immunofluorescence of PRG4 and quantification (scale bars, 100 μm). (E) Osteophyte volume analyzed by microtomography. (F) Subchondral BV/TV analyzed by microtomography. Data are means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001.
Moreover, the anabolic factor PRG4 was not expressed at 4 weeks in OA mice but was reactivated at 8 weeks only in Lin28a(Tg) OA mice (Fig. 4D). This was associated with a significantly increased chondrocyte proliferation index in Lin28a(Tg) OA mice (fig. 5B), which suggests that Lin28a allows for reprogramming early precursor chondrocytes committed to an OA program. In this regenerative model, the osteophyte volume was higher in Lin28a(Tg) OA mice than in OA CTs (Fig. 4E), with no change in subchondral bone volume (Fig. 4F). Last, at 14 weeks, osteophyte volume of Lin28a(Tg) OA mice was comparable to that of OA CTs (fig. S5D), with no difference in subchondral bone volume (fig. S5E). These results suggest that Lin28a overexpression allows OA chondrocytes to reverse the anabolism/catabolism balance and protect cartilage against degradation.
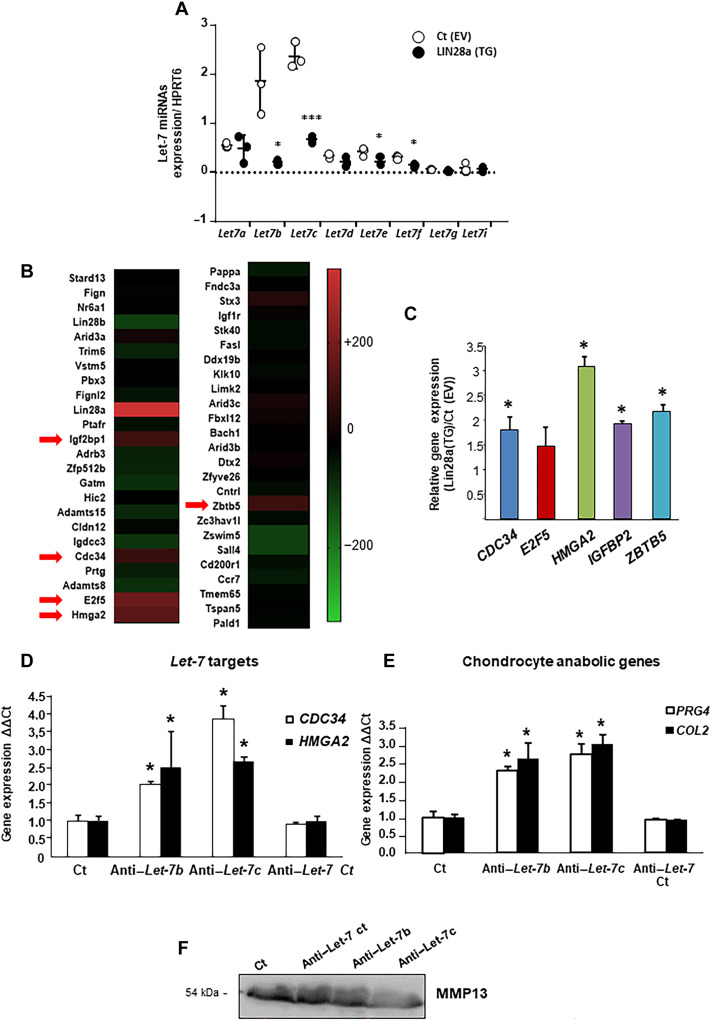
Lin28a-inhibited Let7-b/let-7c–reduced chondrocyte catabolism via HMGA2
Given that Lin28a promoted the emergence of stemness factors in chondrocytes and considering that the Let-7 miRNA family is one of the principal targets of Lin28a, we assessed whether cartilage integrity is driven by regulation of the Let-7 miRNA family. At baseline, control chondrocytes expressed low levels of miRNAs except for Let-7b and Let-7c; conversely, Lin28a-overexpressing chondrocytes showed significantly decreased levels of Let-7b, Let-7c, Let-7e, and Let-7f, but the other Let-7 members were not affected (Fig. 5A). We then performed RNA-seq of Lin28a-overexpressing chondrocytes (transfected with Lin28a vector) and chondrocytes transfected with empty vector as the CT condition. Using the miRDP database, we selected the 50 first mRNAs predicted to interact with Let-7b and Let-7c. Heatmap analysis revealed that the expression of five Let-7b and Let-7c mRNA targets was significantly overrepresented, above 200%, in Lin28a-overexpressing chondrocytes compared to control chondrocytes: CDC34, E2F5, HMGa2, IGFBP2, and ZBTB5 (Fig. 5B). Reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis further confirmed that overexpression of Lin28 activated the expression of these genes, predominantly the expression of HMGA2 (Fig. 5C). Therefore, we inhibited Let-7b and Let-7c miRNA in primary chondrocytes and found that anti–Let-7b and anti–Let-7c promoted the mRNA levels of HMGA2 and CDC34 in transfected but not anti–Let-7 control chondrocytes (Fig. 5D). In addition, inhibition of Let-7b or Let-7c promoted the mRNA expression of COL2 or PRG4 (Fig. 5E) and reduced the secretion of MMP13 (Fig. 5F). Together, the inhibition of Let-7b and Let-7c mediated the effect of Lin28a overexpression, thereby restoring the anabolism/catabolism balance.
Fig. 5. Inhibition of Let-7b and Let-7c by Lin28a promoted chondrocyte reprogramming.
(A) Let-7 miRNA expression was assessed by RT-qPCR in primary chondrocytes transduced with control empty vector [Ct(EV)] or Lin28a lentivirus [Lin28a(TG)] and cultured at 1% O2. *P < 0.05 and ***P < 0.005 compared with empty vector control. (B) Heatmap representing the fold increase in the 50 most changed Let-7b and Let-7c targets in Lin28a-overexpressing chondrocytes compared to control chondrocytes in RNA-seq analysis. Red arrows indicate the genes with increase in expression ≥200%. (C) Let-7 target gene expression (CDC34, E2F5, HMGA2, IGFBP2, and ZBTB5) measured by RT-qPCR in primary chondrocytes transduced with [Lin28a(TG)]; *P < 0.05 compared with control empty vector [Ct(EV)]. Primary chondrocytes were treated with anti–Let-7b, anti–Let-7c, and anti–Let-7 antibody control siRNA. (D) Let-7 target gene expression (CDC34 and HMGA2) measured by RT-qPCR; *P < 0.05 compared with control. (E) Chondrocyte anabolic gene expression (Col2 and PRG4) measured by real-time qPCR; *P < 0.05 compared with controls. (F) Western blot analysis of MMP13 level in conditioned medium. Data are means ± SEM.
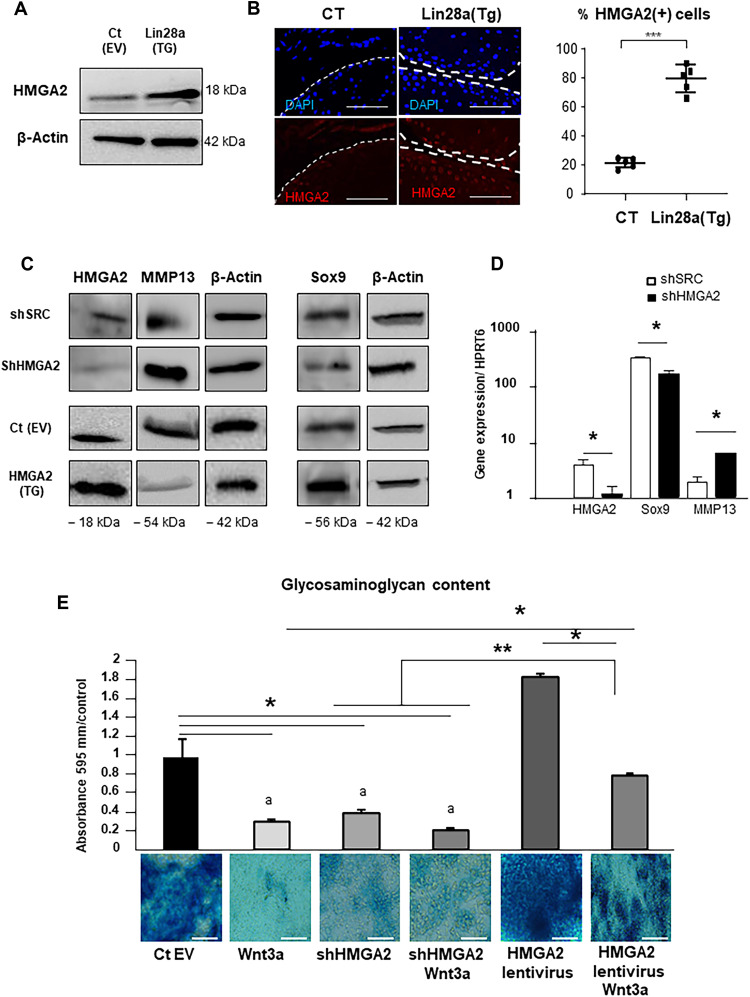
HMGA2 mediated Lin28a anabolic and catabolic effects in chondrocytes
HMGA2 is a Let-7 chondrocyte target and regulates the fate of growth-plate chondrocytes; thus, we hypothesized that chondrocyte reprogramming induced by Lin28a is mediated by HMGA2. First, the transduction of Lin28a stimulated the expression of HMGA2 in primary chondrocytes as compared with the empty vector (Fig. 6A) and in Lin28a(Tg) mice as compared with CT mice (Fig. 6B). To further address whether the activity of Lin28a was mediated by HMGA2, we used a transduction strategy in murine primary chondrocytes. Inhibiting HMGA2 by shRNA increased MMP13 and reduced SOX9 for both protein and mRNA expression. Conversely, the overexpression of HMGA2 inhibited the chondrocyte catabolism, as illustrated by reduced MMP13 protein level, and activated chondrocyte anabolism, as shown by increased SOX9 protein level (Fig. 6, C and D). Last, GAG content was lower in chondrocytes transduced with shHMGA2 but was higher with HMGA2 overexpression as revealed by Alcian blue staining (Fig. 6E). The activation of the anabolic pathway was sustained even under pro-catabolic conditions in the presence of Wnt3a. Together, these findings suggest that HMGA2 mediated the anabolism and catabolism induced by Lin28a in chondrocytes.
Fig. 6. HMGA2 expression increased anabolism and reduced catabolism in chondrocytes.
(A) Western blot analysis of HMGA2 protein expression in primary chondrocytes transduced with control empty vector [Ct(EV)] or Lin28a lentivirus [Lin28a(TG)] at 1% O2. (B) Immunofluorescence staining and quantification of HMGA2 expression in cartilage from mice (scale bars, 100 μm). (C) Western blot analysis of HMGA2, MMP13, and SOX9 protein levels in primary chondrocytes transduced with scramble vector (shSRC), empty vector [Ct(EV)], shRNA against HMGA2 (shHMGA2), or HMGA2 lentivirus [HMGA2(TG)] at 1% O2. (D) RT-qPCR analysis of HMGA2, MMP13, and SOX9 mRNA levels in primary chondrocytes treated with shSRC or shHMGA2 at 1% O2. (E) Alcian blue staining of sulfated glycosaminoglycans (GAGs) in chondrocytes treated or not with shHMGA2 or lentivirus. Wnt3a was used to induce chondrocyte catabolism (scale bars, 200 μm). Data are means ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.005.
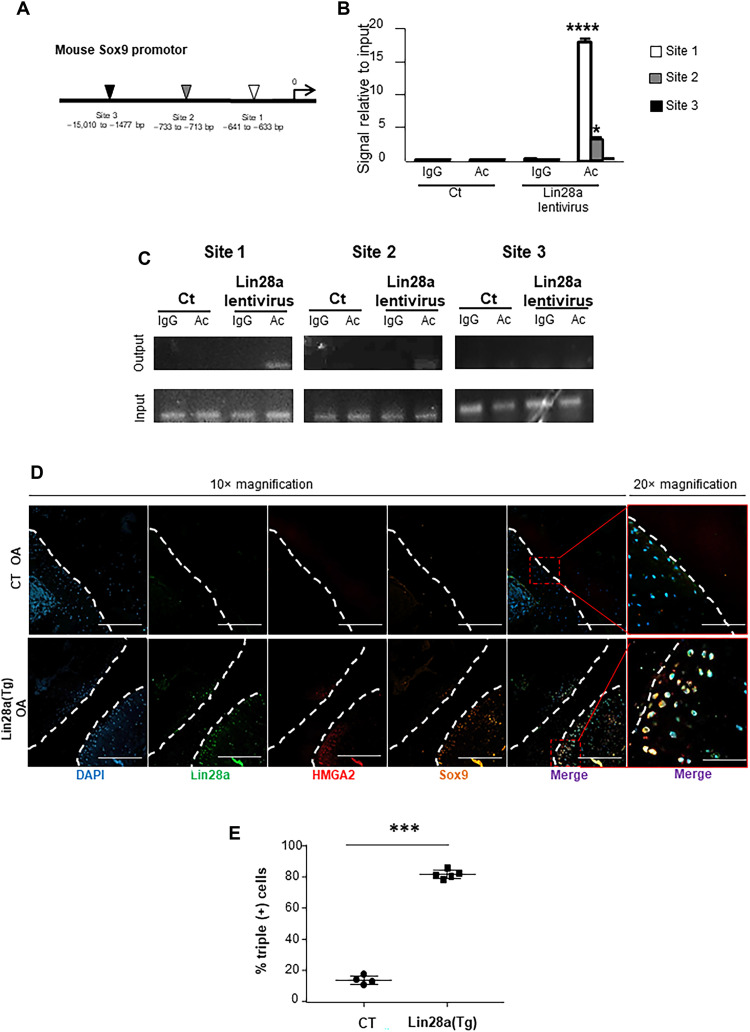
HMGA2 activated chondrocyte reprogramming by binding to SOX9 promoter regulatory regions
We then explored the mechanisms by which HMGA2 mediated chondrocyte function. HMGA2 promotes chromatin rearrangement by binding to DNA-specific sequences and then facilitates transcription by increasing DNA accessibility (24). Computational analyses showed that the SOX9 promoter contains three putative sequences that may interact with HMGA2 and thus may facilitate SOX9 transcription (Fig. 7A). Chromatin immunoprecipitation assay revealed that HMGA2 bound preferentially to the more proximal site, less to the medial site, but not to the distal site (Fig. 7, B and C). To assess whether Lin28a induced SOX9 expression via HMGA2 in mice, we used triple immunostaining of Lin28a, HMGA2, and SOX9 in Lin28a(Tg) OA mice. Concomitant expression of HMGA2 and SOX9 was weak (12%) in CT mice but was ≥80% in Lin28a(Tg) mice (Fig. 7, D and E). Therefore, the concomitant expression of Lin28a, HMGA2, and SOX9 suggested activation of SOX9 in the pro-anabolic effect of HMGA2 induced by Lin28a.
Fig. 7. SOX9 promoter is a target for HMGa2 DNA regulation.
(A) Representation of HMGA2 binding sites on SOX9 promoter. (B) qPCR analysis of HMGA2 binding to SOX9 regulatory regions; significance is compared with control antibody. (C) ChIP analysis of HMGA2 binding to the three SOX9 regulatory regions. IgG, mouse IgG (negative control). (D) Triple staining of Lin28a, HMGA2, and SOX9 by confocal immunofluorescence in CT and Lin28a(Tg) OA mice. Images show 10× magnification (scale bars, 500 μm) and 20× magnification (scale bars, 200 μm). (E) Quantification of immunofluorescence percentage of cells triple positive for Lin28a, HMGA2, and SOX9 staining. Data are means ± SEM. *P < 0.05, ***P < 0.005, and ****P < 0.001 compared with CT.
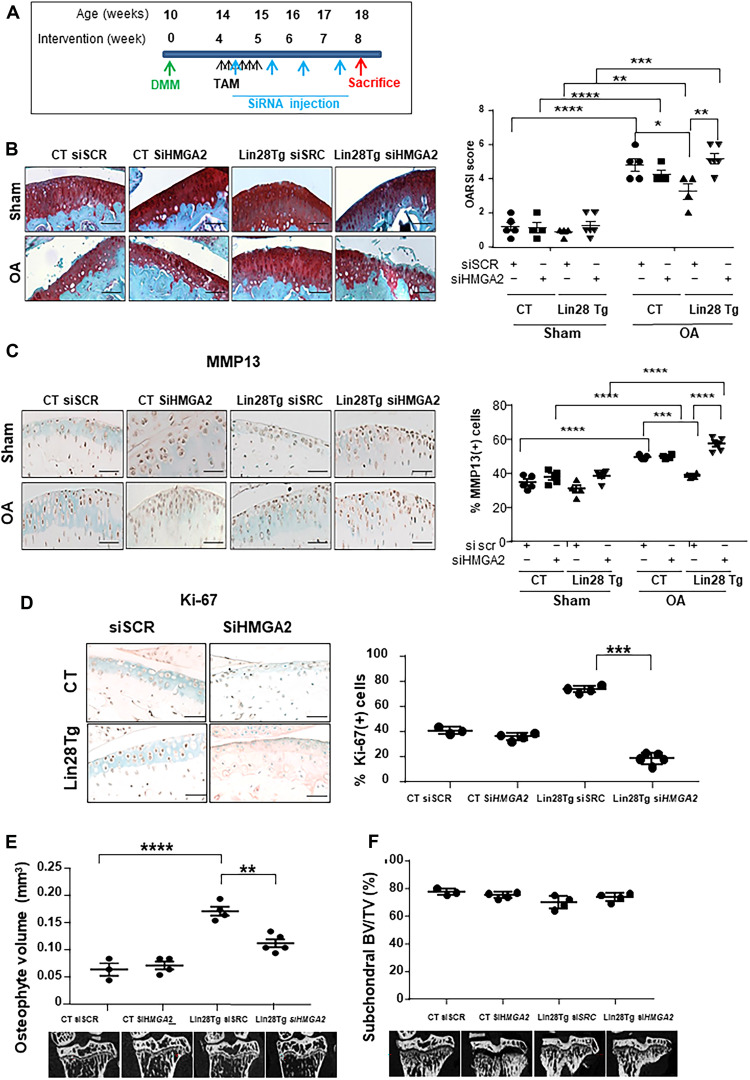
Lin28a-induced reprogramming is mediated by HMGA2 in mice
To assess whether HMGA2 mediated the anabolic effect of Lin28a in OA, we inhibited HMGA2 expression by siRNA in the regenerative model. Four weeks after OA induction, Lin28a(Tg) and CT mice received weekly intra-articular injections of siRNA against HMGA2 and were euthanized at 8 weeks (Fig. 8A and fig. S6). As compared with CT mice, Lin28a(Tg) mice receiving siRNA showed a lower OA score, but the score was higher after siHMGA2 administration (Fig. 8B). A similar profile was found for MMP13 level (Fig. 8C). Moreover, the chondrocyte proliferation in OA induced by Lin28a was totally abolished with HMGA2 knockdown, as illustrated by Ki-67 assay (Fig. 8D). These findings suggest that the effect of Lin28a depended on HMGA2 in cartilage. Last, HMGA2 knockdown reduced the osteophyte volume in Lin28a(Tg) OA mice, unlike the siRNA control (Fig. 8E), with no change in subchondral bone (Fig. 8F). These data confirm the major role of Lin28a in regulating Let-7, mediated by HMGA2 in chondrocyte reprogramming and cartilage repair.
Fig. 8. Protective effects of Lin28a depend on HMGa2.
(A) Schematic representation of the regenerative protocol applied to Col2a1-CreER (CT) and Lin28aflox/flox/Col2a1-CreER [Lin28a(Tg)] mice. OA was induced in mice at age 10 weeks, and then mice were euthanized at 4 and 8 weeks after. All mice received weekly intra-articular injection of scramble siRNA (siSCR) or siRNA against HMGA2 (siHMGA2). (B) Safranin-O staining and quantification of OA score of articular cartilage (scale bars, 100 μm). (C) Immunohistochemistry of MMP13 level and quantification of MMP13-positive cells (scale bars, 100 μm). (D) Immunohistochemistry of Ki-67 expression and quantification in mouse joints with OA (scale bars, 100 μm). (E) Osteophyte volume analyzed by microtomography in mice with OA. (F) Subchondral BV/TV analyzed by microtomography in mouse joints with OA. Data are means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001.
DISCUSSION
OA remains a chronic disease with high prevalence and economic burden but with no effective therapy (25–27). The current challenge is the development of anabolic molecules to limit cartilage loss and promote cartilage regeneration. Here, we took advantage of the regenerative properties of Lin28a to demonstrate the role of the Lin28a–Let-7 axis in cartilage physiology and the ability of Lin28a to protect and regenerate cartilage even in damaged joints. We showed that Lin28a overexpression reactivated the expression of early anabolic factors expressed by chondrocyte precursors, which are lost in the early stages of OA.
Lin28a could reactivate PRG4, which synthesizes lubricin (28), thus indicating the capacity to produce cartilage matrix. Moreover, the anabolic effect of Lin28a in chondrocytes is associated with increased proliferation and acquisition of stemness markers such as NANOG and SOX2, in line with the described effect of Lin28a generating induced pluripotent stem cells (21) and driving stemness properties in several tissues (29, 30). Our results are consistent with the ability of Lin28a to induce reprogramming of differentiated chondrocytes into immature proliferative chondrocytes in OA cartilage. We also showed that Lin28a up-regulated HMGA2, a nuclear factor that activates the transcription of SOX9, another crucial transcription factor induced in chondrocyte reprogramming. Thus, the recovery of stemness features may also explain the ability of Lin28a to trigger chondrocyte proliferation in Lin28a(Tg) mice, an essential function that is lacking in mature articular chondrocytes.
One of the main actions of Lin28a is to control Let-7 miRNA biogenesis in different tissues by inhibiting their maturation (31–33). Previous works showed that inhibiting Let-7c allows for restoring cell functions and recovery after injury (34). Here, Lin28a was also able to down-regulate Let-7b and Let-7c, the inhibition of which promoted chondrocyte anabolism. This effect was observed in mice, an in vivo situation in which cartilage is under low oxygen concentrations, and in cells and explants cultured under hypoxia, mimicking the hypoxic environment of undamaged cartilage. Therefore, the activation of Lin28a might be required in situations of low oxygen tension, a condition for chondrocyte survival and reprogramming. Hence, we might speculate that with the loss of hypoxia in OA cartilage (5), the inhibition of Let-7 is reduced, thereby affecting the induction of the reprogramming effect of the endogenous Lin28a and the poor regeneration of cartilage in OA. Even if Lin28a reactivation is not sufficient for complete restitution of cartilage, Lin28a helped protect against cartilage degeneration, as shown by the exacerbation of cartilage catabolism activity and the lesions observed in mice with Lin28a deleted in chondrocytes.
We highlight the mediation of the Let-7 effect by HMGA2, whose activation resulted in chondrocyte anabolism and catabolism, thereby reversing the OA features. By binding to DNA sequences and modifying chromatin condensation (35), HMGA2 modulates gene transcription. Previous reports showed that HMGA2 directly induces SOX9 transcription to maintain the pool of stem cells in intestinal niches (36). Accordingly, our results show that HMGA2 activated Sox9 transcription by binding to the regulatory sequences of its promoter in chondrocytes. In addition, SOX9 activates the expression of Col2 and aggrecan (37, 38), thus promoting chondrocyte anabolism along with HMGA2 activation. We also found reduced chondrocyte catabolism with Lin28a overexpression. Because HMGA2 is able to modulate Wnt-β–catenin signaling (39, 40) and, here, proteoglycan production, the reduction in chondrocyte catabolism might be Wnt dependent and explain the down-regulation of MMP13 observed under hypoxia (7). Further, these results have been confirmed in IL-1–priming chondrocytes, which indicate that Lin28a blocked not only Wnt effect but also other pathways involved in OA.
We observed a strong increase in osteophyte volume in Lin28a(Tg) mice along with less cartilage degradation. This is an intriguing observation because osteophyte development is usually associated with more severe cartilage loss (41, 42). This finding may indicate an uncoupling effect of Lin28a on bone and articular cartilage, within the joint and under environmental conditions. Our approach in mice and under ex vivo hypoxic conditions would facilitate the effect of Lin28a, favoring chondrocyte reprogramming and the emergence of progenitors able to form osteophytes at the bone-cartilage junction. This finding further demonstrated that the effect of Lin28a could depend on oxygen tension.
In conclusion, our results provide evidence that the Lin28a–Let-7 axis contributes to chondrocyte reprogramming via HMGA2 and prevents the development of OA in mice. More specifically in chondrocytes, Lin28a regulates HMGA2 expression, thus driving the reprogramming of OA chondrocytes by chromatin structural changes and SOX9 transcription. Lin28a may represent a key pathophysiological factor in OA chondrocytes and pave the way for future therapeutics targeting OA.
MATERIALS AND METHODS
Mice
Lin28atm1.2Gqda/Col2a1-CreER (KO) mice were purchased from the Jackson Laboratory (43), and Col2a1-CreER and TgLin28aflox/flox/Col2a1-CreER [Lin28a(Tg)] mice were a gift from T. Kobayashi (Harvard Medical School, Boston, MA, USA) (22). We generated mice with inducible cartilage-specific knockout of Lin28atm1.2Gqda tm1.2Gqda/Col2a1-CreER (hereafter called flox/flox) or haplo-insufficiency in Lin28atm1.2Gqda/+/Col2a1-CreER (hereafter called flox /+) by using recombination induced by tamoxifen administration at age 9 weeks. Col2a1-CreER mice were CTs (thereafter Lin28a+/+ mice). All animal experiments were performed according to procedures approved by the local animal ethics committee and were approved by the French Ministry of Higher Education and Research (APAFIS#9563 and APAFIS#3817).
Experimental OA animal models
OA was induced in mice by DMM of the right knee as previously described (44); a sham operation was performed on the contralateral knee by incision of the cutaneous and muscular planes at baseline. Mice underwent two types of induction of DNA recombination. In the first experiment, the preventive experiment, 9-week-old transgenic Lin28a(Tg) mice received a 75-mg/kg daily injection of tamoxifen (Sigma-Aldrich, Darmstadt, Germany) for 5 days. Then, DMM was performed at 10 weeks and mice were euthanized at 8 weeks after the surgery. The second protocol was designed for the regenerative aspects and used knockout and transgenic mice. Ten-week-old mice underwent DMM, received tamoxifen at 15 weeks, and were euthanized at 4, 8, and 14 weeks after surgery. Dissected joints were processed for microtomography and histopathological or molecular analysis.
DMM was performed in CT, Lin28a-transgenic mice [Lin28a(Tg)], and KO (−/−) mice at 10 weeks. Recombination was induced 1 week before (prevention model) or 4 weeks after DMM (regenerative model) by daily injection of tamoxifen (75 mg/kg) for 5 days. Then, 10 nmol/3 μl siRNA for HMGA or siCtrl (Dharmacon, Cambridge, UK) dissolved in phosphate-buffered saline (PBS) was administered weekly in the left and right knees of mice for 4 weeks.
Isolation of human cartilage
Human cartilage samples were harvested from patients who were undergoing total knee replacement surgery (OA) or amputation (control). Samples were obtained in accordance with the guidelines and regulations of the French National Authority Legislation for the collection of human tissues. Collections were approved by the ethics committee of the institution; informed consent was obtained from patients. Cartilage samples were collected from the femoral condyle at the posterior surface of the knee. Samples were collected from a zone that appeared undamaged, as defined by white and shiny cartilage without lesions, and in a zone that appeared damaged, as defined by a discoloration and a degraded surface. All samples were fixed in 4% paraformaldehyde (PFA) and embedded in paraffin until immunohistochemistry for Lin28a.
Isolation and culture of primary chondrocytes
Chondrocytes from 6-day-old mice were isolated from cartilage fragments dissected from knee joint and femoral heads. The cartilage fragments were minced and digested with Liberase (0.52 U/ml; Sigma Darmstadt, Germany) at 37°C for 16 hours. Cells were suspended in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, CA, USA) containing 10% fetal bovine serum (FBS; HyClone, Villebon-sur-Yvette, France). Only first-passage cells were used for experiments. Primary chondrocytes were used for Let-7 siRNA treatment. Cells at 50 to 60% confluency were treated with siRNA against miRNA–Let-7b or Let-7c or siRNA control (ON-TARGETplus SMARTpool, Dharmacon, Cambridge, UK) for 48 hours. During culture, cells were maintained in a humidified incubator containing 5% CO2 at 37°C, in 21 and 1% O2.
Infection and transduction of primary chondrocytes
Human embryonic kidney 293T cells were cultured in DMEM with 10% FBS at 37°C and 5% CO2. Ready-to-use packaging plasmid mix (Cellectar, Florham Park, NJ, USA) and ExGen 500 (Euromedex, Souffelweyersheim, France) transfection reagents were used for plasmid transfection. Plasmids (2 μg) for Lin-28a (pMSCV-mLin28A, Addgene plasmid no. 26357) and HMGA2 (pBabe zeo HMGA2, Addgene plasmid no. 17411; pLKO.shHmga2, Addgene plasmid no. 32399) or empty vector (CMV500 empty vector, Addgene plasmid no. 33348) were transfected into cells. Viral particles were obtained and stored at −80°C. Chondrocytes at 50 to 60% confluency were infected with viral particles (1 ml) and Polybrene (Sigma-Aldrich, Darmstadt, Germany) at 4 μg/ml for 48 hours. During culture, cells were maintained in a humidified incubator containing 5% CO2 at 37°C and 3% O2.
Alcian blue staining
After lentivirus infection, plates were washed twice with PBS (Gibco, CA, USA) and fixed in 4% PFA for 2 hours at room temperature and washed twice with PBS. Plates were treated with Alcian blue stain (1% Alcian blue; Sigma-Aldrich, Darmstadt, Germany) in 3% glacial acetic acid solution for 15 min and washed twice with PBS. Alcian blue was extracted and quantified with overnight incubation in 6 M guanidine-HCl. Absorbance was measured at 630 nm with a Bio-Rad Model 680 microplate reader and images were obtained by using an EVOSxl microscope.
Culture of mice femoral explants
Femoral heads were harvested from 10-week-old WT and transgenic mice. Explants were deposited in flat-bottom culture cell plates with 0% FBS DMEM and then, 24 hours later, were treated with (Z)-4-hydroxytamoxifen (Sigma-Aldrich, Darmstadt, Germany) at 0.02 mg/ml for 24 hours to induce recombination. CTs received cell culture media. After 24 hours, explants were treated with Wnt3a diluted at 50% in cell culture media obtained from the L cell line (5). During culture, explants were maintained in a humidified incubator containing 5% CO2 at 37°C and 1% O2. Explants were then washed with PBS, fixed with 4% PFA. Decalcification involved using EDTA. Mice explants were included in OCT. Frozen sections 5 μm thick were stored at −80°C. Samples were stained with Safranin-O, and immunohistochemistry involved using antibodies for Lin28a (Abcam, Cambridge, UK; 1:500), type II collagen (Abcam, 1:200, France), and MMP13 (Abcam, 1:150), and immunofluorescence involved using an antibody for Ki-67 (Abcam, 1:150). Safranin-O staining was evaluated semi-automatically by using Archimed image analyzer software from Microvison Company (France).
RNA extraction and RT-PCR analysis
Total RNA was isolated by using TRIzol reagent (Invitrogen, Villebon-sur-Yvette, France). Purified RNA (1 μg) was reverse-transcribed by using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Villebon-sur-Yvette, France). Real-time RT-PCR involved using the SensiFAST Sybr No-Rox Mix (Bioline, Memphis, TN, USA). Relative gene levels are expressed as fold change, calculated by the 2−ΔΔCt formula. Values were normalized to the expression of hypoxanthine phosphoribosyl transferase 6 (HPRT6).
The following primer sequences were used: Lin28a, 5′CTTTTGCCAAAGCATCAACC3′ (forward) and 5′GGGCTGTGGATCTCTTCCTC3′ (reverse); HMGA2, 5′ATCCAACTTTCTCCCCGTTC3′ (forward) and 5′AGGTATTGCCACAAGCAAGC3′ (reverse); CDC34, 5′TGGCACCCAAACATCTATGA34 (forward) and 5′AAGGTCTTGGGCTCATTCAG (reverse); HPRT6, 5′GGTGGATATGCCCTTGACTATAATGA3′ (forward) and 5′CAACATCAACAGGACTCCTCGTATT3′ (reverse); COL2A2, 5′CCGTCATCGAGTACCGATCA3′ (forward) and 5′CAGGTCAGGTCAGCCATTCA3′ (reverse); PRG 4, 5′ATGGTAAGCCAGTGGATGGA3′ (forward) and 5′GGTAATTCTGCGTGGTGGAG3′ (reverse); AXIN 2, 5′TGACTCTCCTTCCAGATCCCA3′ (forward) and 5′TGCCCACACTAGGCTGACA3′ (reverse); SLc7a5, 5′CAGCTCCCTGAGTATGAAAGC3′ (forward) and 5′CCATTCCAGTAGACACCCCTTC3′ (reverse); LEF, 5′AGAAATGAGAGCGAATGTCGTAG3′ (forward) and 5′ CTTTGCACGTTGGGAAGGA 3′ (reverse); and TCF, 5′ACAGTGCTCTAGGCTGTCC3′ (forward) and 5′CGACCTGAGAATGTTGGTGCT3′ (reverse).
RNA extraction and RT-PCR analysis for miRNA Let-7 expression
Total RNA was isolated by using TRIzol reagent (Invitrogen, Villebon-sur-Yvette, France). cDNA was created by using the miScript II RT kit (QIAGEN, Les Ulis, France). Specifically, the pre-miRNA sequences were quantified by creating cDNA with the miScript II RT Kit with miScript HiFlex Buffer, and then amplified by qPCR with primers that flank the mature miRNA sequence within the pre-miRNA molecule. The mature miRNA sequences were quantified by creating cDNA from mature miRNA with the miScript II RT kit with miScript HiSpec buffer, and then amplified by qPCR with a primer that targets the miRNA sequence. The miScript II kit was used to selectively amplify the short miRNA and not the longer product that may be created by ligation of the 3′ adaptor to pre-miRNA.
The following primer sequences were used: Let-7a, 5′TGAGGTAGTAGGTTGTATAGTT3′ (forward); Let-7b, 5′TGAGGTAGTAGGTTGTGTGGTT3′ (forward); Let-7c, 5′TGAGGTAGTAGGTTGTATGGTT3′ (forward); Let-7e, 5′TGAGGTAGGAGGTTGTATAGTT3′ (forward); Let-7g, 5′TGAGGTAGTAGTTTGTACAGTT3′ (forward); and Let-7i, 5′TGAGGTAGTAGTTTGTGCTGTT3′. Reverse primer was provided in the kit and corresponded to the adaptor.
RNA-seq
RNA-seq was performed in duplicate for the following conditions: primary murine chondrocytes were transduced for control conditions with empty vector (CMV500 empty vector; Addgene plasmid no. 33348) and for Lin28 overexpression with pMSCV-mLin28A (Addgene plasmid no. 26357). Total RNA was extracted and transferred for sequencing to IntegraGen SA Co. (Evry, France). Libraries were prepared with the NEBNext Ultra II directional RNA library prep kit for the Illumina protocol according to the supplier’s recommendations. Briefly, the successive stages of this protocol are the purification of PolyA containing mRNA molecules using poly-T oligo attached magnetic beads from 1 μg of total RNA (with the magnetic mRNA isolation kit from New England Biolabs), fragmentation using divalent cations under elevated temperature to obtain approximately 300–base pair (bp) pieces, double-strand cDNA synthesis, and, last, Illumina adapter ligation and cDNA library amplification by PCR for sequencing. Sequencing then involves paired-end 100-bp reads with Illumina NovaSeq. Image analysis and base calling involved using Illumina Real-Time Analysis (3.4.4) with default parameters.
Quantification of gene expression
STAR was used to obtain the number of reads associated with each gene in the Gencode vM24 annotation (restricted to protein-coding genes, antisense, and lincRNAs). Raw counts for each sample were imported into R statistical software. The extracted count matrix was normalized for library size and coding length of genes to compute fragments per kilobase of transcript per million mapped reads (FPKM) expression levels. For the analysis, the Bioconductor edgeR package was used to import raw counts into R statistical software and compute normalized log2 counts per million mapped reads by using weighted trimmed mean of M values as a normalization procedure. The normalized expression matrix from the 1000 most variant genes (based on SD) was used to classify the samples according to their gene expression patterns by using principal components analysis (PCA), hierarchical clustering, and consensus clustering. PCA involved the FactoMineR::PCA function with “ncp = 10, scale.unit = FALSE” parameters. Hierarchical clustering involved the stats::hclust function (with Euclidean distance and ward.D method). Consensus clustering involved the Consensus ClusterPlus::ConsensusClusterPlus function to examine the stability of the clusters. We established consensus partitions of the dataset in K clusters (for K = 2, 3 to 8) on the basis of 1000 resampling iterations (80% of genes and 80% of sample) of hierarchical clustering, with Euclidean distance and ward.D method. Then, the cumulative distribution functions (CDFs) of the consensus matrices were used to determine the optimal number of clusters (e.g., K = 3), considering both the shape of the functions and the area under the CDF curves. tSNE analysis involved using the Bioconductor Rtsne package applied with the PCA object (theta = 0.0, perplexity = 30, and max_iter = 1000).
Differential expression analysis
The Bioconductor edgeR package was used to import raw counts into R statistical software. Differential expression analysis involved using the Bioconductor limma package and the voom transformation. To improve the statistical power of the analysis, only genes expressed in at least one sample (FPKM ≥ 1) were considered. A qval threshold ≤0.05 and a minimum fold change 1.2 were used to define differentially expressed genes. Using the miRDP database (http://mirdb.org/mirdb/index.html), we selected the 50 first mRNA predicted to interact with let-7b and let-7c (http://mirdb.org/cgi-bin/search.cgi?searchType=miRNA&searchBox=mmu-let-7b-5p&full=1) by using the Galileo dynamic gene expression analysis software (Integragen, Paris, France).
Micro–computed tomography and histomorphometry
Mice were euthanized by cervical dislocation at age 18 weeks. The whole knee joints of mice were excised and fixed in 4% PFA for 2 days. Samples were analyzed by microtomography (SkyScan1272). Micro–computed tomography imaging and analyses were performed with a SkyScan 1172 system (Bruker microCT, Belgium), with a voxel size of 6 μm, a voltage of 100 kV, an intensity of 100 μA, and an exposure of 1250 ms. NRecon reconstruction and DataViewer software were used for three-dimensional reconstruction and analyses. Analysis of structural parameters involved using CTAn and CTVox (Bruker microCT, Belgium). The specimens were then decalcified for 10 days and dehydrated in a graded ethanol series, embedded in paraffin, cut into 5-μm-thick sections, and stained with Safranin-O. The Osteoarthritis Research Society International grading system was used to score histopathologic changes in OA cartilage. For immunofluorescence analysis, sections were dewaxed and treated with HIER citrate buffer, pH 6.0 (Zymoted) for 4 hours at 70°C. Permeabilization was performed with PBS–Triton X-100 0.1% for 10 min at room temperature, and blocking epitopes was achieved in PBS–1% bovine serum albumin (BSA)–1% glycine with the first and secondary antibodies. Antibodies against Lin28a (Abcam, 1:500), HMGA2 (Abcam, 1:400), SOX9 (1:200), and PRG4 (Abcam, 1:100) were used. 4′,6-Diamidino-2-phenylindole (DAPI) staining was used to identify nuclei. Fluorescent secondary antibodies were Alexa Fluor 488 (Invitrogen, 1:100) for Lin28a, DyLight 550 (Invitrogen, 1:100) for SOX9, and DyLight 650 (Invitrogen, 1:100) for PRG4. Fluorescent signals were preserved by using DAKO mounting fluorescence medium. Image acquisitions involved using the ApoTome optical sectioning system (Zeiss) with an inverted microscope (Zeiss Axio Observer ZI) and an LSM 880 confocal laser scanning microscope (Zeiss). For immunohistochemistry, sections were dewaxed and treated with HIER citrate buffer, pH 6.0, for 4 hours at 70°C. Permeabilization was performed with chondroitinase 10% for 15 min at room temperature, and blocking epitopes was achieved in PBS–horse serum–5% BSA. Antibodies against Lin28a (Abcam, 1:500), Ki-67 (Abcam, 1:150), and MMP13 (Abcam, 1:150) were used. The secondary antibody was ImmPRESS (Abcam) HRP Reagent kit anti-rabbit antibody (Vector, France). Results were revealed with the 3,3′-diaminobenzidine (DAB) peroxidase substrate kit (Vector, Burlingame, CA, USA). Methyl green was used to identify nuclei. The DAB signal was preserved with Entellan mounting medium (Sigma Darmstadt, Germany) and image acquisitions involved using the Microvision instrument system.
Western blots
Cells were lysed with (RIPA) lysis buffer, separated by SDS–polyacrylamide gel electrophoresis, and transferred to polyvinylidene fluoride membranes that were incubated with primary antibodies overnight at 4°C and secondary antibody at room temperature for 1 hour and visualized by using Clarity Western ECL Substrate (Bio-Rad, Hercules, CA, USA). Proteins were analyzed with antibodies recognizing HMGA2 (Abcam, 1:1000), PRG4 (Abcam, 1:500), SOX9 (Abcam, 1:400), MMP13 (Abcam 1:3500), and β-actin (Abcam, 1:3500). Chemiluminescent signals were detected with the Azure Imaging System.
Chromatin immunoprecipitation assay
Primary chondrocytes were cultured until confluence in 12-well plates (106 cells per well). During the cultures, cells were maintained in a humidified incubator containing 5% CO2 at 37°C and 1% of oxygen. Fixation of cultured cells and their processing for chromatin immunoprecipitation (ChIP) followed the manufacturer’s instructions (Abcam). Immunoprecipitation with Pierce protein A/G magnetic beads followed the manufacturer’s instructions (Thermo Fisher Scientific, Villebon-sur-Yvette, France). Immunoprecipitation and detection involved a rabbit primary antibody (Abcam, France) and rabbit immunoglobulin (IgG) antibody (Diagomic, Blagnac, France). Results are average Ct values reported to input Ct values. The following primer sequences were used: site 1, 5′ACACCAGCTTCGTTGAACCAGAG3′ (forward) and 5′GGAAGCAAATGTTTGGGTGACTCA3′ (reverse); site 2, 5′ACTTGTCAGTTCAAGGTCGGCGTC3′ (forward) and 5′TGTGGTGACTGGAGCTTCTGCTG3′ (reverse); and site 3, 5′CGAGCTTTTCAAAAGCATCCCAAAGA3′ (forward) and 5′TGATAAAGCGAATCGGCCTGTATC3′ (reverse).
Statistical analysis
Data are presented as means ± SEM. Every experiment was performed at least three times. Statistical analysis involved the Mann-Whitney test for comparing two independent samples and two-way analysis of variance (ANOVA) for multiple comparisons controlling for false discovery rate. P < 0.05 was considered statistically significant. Statistical analyses involved using GraphPad Prism 7.00 (GraphPad Software, La Jolla, CA, USA).
Acknowledgments
We thank T. Kobayashi for providing the Lin28aflox/flox mice. The Lin28aflox/flox mice can be provided by T. Kobayashi pending scientific review and a completed material transfer agreement. Requests for the Lin28aflox/flox mice should be submitted to T. Kobayashi. We thank the T. Jacks laboratory for providing pBabe zeo HMGA2 (Addgene, plasmid no. 17411) and pLKO.s hHmga2 (Addgene, plasmid no. 32399). The pBabe zeo HMGA2 and pLKO.s hHmga2 can be provided by Addgene pending scientific review and a completed material transfer agreement. Requests for pBabe zeo HMGA2 and pLKO.s hHmga2 should be submitted to Addgene. We also thank G. Delay for providing pMSCV-mLin28A (Addgene, plasmid no. 26357). pMSCV-mLin28A can be provided by Addgene pending scientific review and a completed material transfer agreement. Requests for pMSCV-mLin28A should be submitted to Addgene.
Funding: This work was funded by the ROAD network and Inserm.
Author contributions: Conceptualization: Y.J., E.H., and M.C.-S. Methodology: Y.J., E.H., H.-K.E., and M.C.-S. Investigation: Y.J., Z.B., B.B.-T., J.S., C.A., A.L., P.R., E.H., and M.C.-S. Funding acquisition: E.H. and M.C.S. Project administration: E.H. and M.C.-S. Supervision: E.H. and M.C.-S. Writing—original draft: E.H., A.L., P.R., and M.C.-S. Writing—review and editing: E.H., A.L., H.-K.E., P.R., and M.C.-S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The datasets generated during and/or analyzed in the current study are available at the following links: https://doi.org/10.5281/zenodo.6553754 and https://doi.org/10.5061/dryad.g1jwstqt6. Lin28a(Tg) mice were provided by T. Kobayashi (with complete material transfer agreement). pLKO.shHmga2 (45) and pBabe zeo HMGA2 (46) are from Addgene deposited by T. Jacks and pMSCV-mLin28A (47) is from Addgene deposited by G. Q. Daley.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
REFERENCES AND NOTES
- 1.Pitsillides A. A., Beier F., Cartilage biology in osteoarthritis—Lessons from developmental biology. Nat. Rev. Rheumatol. 7, 654–663 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Latourte A., Cherifi C., Maillet J., Ea H.-K., Bouaziz W., Funck-Brentano T., Cohen-Solal M., Hay E., Richette P., Systemic inhibition of IL-6/Stat3 signalling protects against experimental osteoarthritis. Ann. Rheum. Dis. 76, 748–755 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Lin H., Hay E., Latourte A., Funck-Brentano T., Bouaziz W., Ea H.-K., Khatib A.-M., Richette P., Cohen-Solal M., Proprotein convertase furin inhibits matrix metalloproteinase 13 in a TGFβ-dependent manner and limits osteoarthritis in mice. Sci. Rep. 8, 10488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funck-Brentano T., Bouaziz W., Marty C., Geoffroy V., Hay E., Cohen-Solal M., Dkk-1-mediated inhibition of Wnt signaling in bone ameliorates osteoarthritis in mice. Arthritis Rheumatol. 66, 3028–3039 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Bouaziz W., Sigaux J., Modrowski D., Devignes C.-S., Funck-Brentano T., Richette P., Ea H.-K., Provot S., Cohen-Solal M., Haÿ E., Interaction of HIF1α and β-catenin inhibits matrix metalloproteinase 13 expression and prevents cartilage damage in mice. Proc. Natl. Acad. Sci. 113, 5453–5458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George B. M., Cabahug-Zuckerman P., Hunter D., Liu C., Singh G., Flamini V., Carrera R., Liu B., Helms J. A., Castillo A. B., Kenny L., Leucht P., Mann K. A., Effects of mechanical loading on cortical defect repair using a novel mechanobiological model of bone healing. Bone 108, 145–155 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Y., Tuan R. S., Origin and function of cartilage stem/progenitor cells in osteoarthritis. Nat. Rev. Rheumatol. 11, 206–212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunziker E. B., Kapfinger E., Geiss J., The structural architecture of adult mammalian articular cartilage evolves by a synchronized process of tissue resorption and neoformation during postnatal development. Osteoarthr. Cartil. 15, 403–413 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Dowthwaite G. P., Bishop J. C., Redman S. N., Khan I. M., Rooney P., Evans D. J. R., Haughton L., Bayram Z., Boyer S., Thomson B., Wolfe M. S., Archer C. W., The surface of articular cartilage contains a progenitor cell population. J. Cell Sci. 117, 889–897 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Hattori S., Oxford C., Reddi A. H., Identification of superficial zone articular chondrocyte stem/progenitor cells. Biochem. Biophys. Res. Commun. 358, 99–103 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roelofs A. J., Kania K., Rafipay A. J., Sambale M., Kuwahara S. T., Collins F. L., Smeeton J., Serowoky M. A., Rowley L., Wang H., Gronewold R., Kapeni C., Méndez-Ferrer S., Little C. B., Bateman J. F., Pap T., Mariani F. V., Sherwood J., Crump J. G., De Bari C., Identification of the skeletal progenitor cells forming osteophytes in osteoarthritis. Ann. Rheum. Dis. 79, 1625–1634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Bari C., Dell’Accio F., Tylzanowski P., Luyten F. P., Multipotent mesenchymal stem cells from adult human synovial membrane. Arthritis Rheum. 44, 1928–1942 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Karlsson C., Thornemo M., Henriksson H. B., Lindahl A., Identification of a stem cell niche in the zone of Ranvier within the knee joint. J. Anat. 215, 355–363 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson K., Zhu S., Tremblay M. S., Payette J. N., Wang J., Bouchez L. C., Meeusen S., Althage A., Cho C. Y., Wu X., Schultz P. G., A stem cell-based approach to cartilage repair. Science 336, 717–721 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Murphy M. P., Koepke L. S., Lopez M. T., Tong X., Ambrosi T. H., Gulati G. S., Marecic O., Wang Y., Ransom R. C., Hoover M. Y., Steininger H., Zhao L., Walkiewicz M. P., Quarto N., Levi B., Wan D. C., Weissman I. L., Goodman S. B., Yang F., Longaker M. T., Chan C. K. F., Articular cartilage regeneration by activated skeletal stem cells. Nat. Med. 26, 1583–1592 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwamoto M., Ohta Y., Larmour C., Enomoto-Iwamoto M., Toward regeneration of articular cartilage. Birth Defects Res. C Embryo Today 99, 192–202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moss E. G., Tang L., Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 258, 432–442 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Lee W.-Y., Park H.-J., Lee R., Lee J.-H., Jhun H., Hur T.-Y., Song H., Analysis of putative biomarkers of undifferentiated spermatogonia in dog testis. Anim. Reprod. Sci. 185, 174–180 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., Dong N., Miao J., Li C., Wang X., Ruan J., Lin28 promotes dental pulp cell proliferation via upregulation of cyclin-dependent proteins and interaction with let-7a/IGF2BP2 pathways. Biomed. Pharmacother. 113, 108742 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Elsaeidi F., Macpherson P., Mills E. A., Jui J., Flannery J. G., Goldman D., Notch suppression collaborates with Ascl1 and Lin28 to unleash a regenerative response in fish retina, but not in mice. J. Neurosci. 38, 2246–2261 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A., Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Papaioannou G., Inloes J. B., Nakamura Y., Paltrinieri E., Kobayashi T., let-7 and miR-140 microRNAs coordinately regulate skeletal development. Proc. Natl. Acad. Sci. U.S.A. 110, E3291–E3300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Driessen B. J. H., Logie C., Vonk L. A., Cellular reprogramming for clinical cartilage repair. Cell Biol. Toxicol. 33, 329–349 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fusco A., Fedele M., Roles of HMGA proteins in cancer. Nat. Rev. Cancer 7, 899–910 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Chen D., Shen J., Zhao W., Wang T., Han L., Hamilton J. L., Im H. J., Osteoarthritis: Toward a comprehensive understanding of pathological mechanism. Bone Res. 5, 16044 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cross M., Smith E., Hoy D., Nolte S., Ackerman I., Fransen M., Bridgett L., Williams S., Guillemin F., Hill C. L., Laslett L. L., Jones G., Cicuttini F., Osborne R., Vos T., Buchbinder R., Woolf A., March L., The global burden of hip and knee osteoarthritis: Estimates from the Global Burden of Disease 2010 study. Ann. Rheum. Dis. 73, 1323–1330 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Karsdal M. A., Michaelis M., Ladel C., Siebuhr A. S., Bihlet A. R., Andersen J. R., Guehring H., Christiansen C., Bay-Jensen A. C., Kraus V. B., Disease-modifying treatments for osteoarthritis (DMOADs) of the knee and hip: Lessons learned from failures and opportunities for the future. Osteoarthr. Cartil. 24, 2013–2021 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Kozhemyakina E., Zhang M., Ionescu A., Ayturk U. M., Ono N., Kobayashi A., Kronenberg H., Warman M. L., Lassar A. B., Identification of a Prg4-expressing articular cartilage progenitor cell population in mice. Arthritis Rheumatol. 67, 1261–1273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan G., Thomson J. A., Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 17, 42–49 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Zhang S., Cui W., Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J. Stem Cells. 6, 305–311 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinton D. A., Chal J., Lummertz da Rocha E., Han A., Yermalovich A. V., Oginuma M., Schlaeger T. M., Sousa P., Rodriguez A., Urbach A., Pourquié O., Daley G. Q., The Lin28/let-7 pathway regulates the mammalian caudal body axis elongation program. Dev. Cell. 48, 396–405.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Tsialikas J., Romer-Seibert J., LIN28: Roles and regulation in development and beyond. Development 142, 2397–2404 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Chen Y., Xie C., Zheng X., Nie X., Wang Z., Liu H., Zhao Y., LIN28/let-7/PD-L1 pathway as a target for cancer immunotherapy. Cancer Immunol. Res. 7, 487–497 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Tolonen A. M., Magga J., Szabó Z., Viitala P., Gao E., Moilanen A. M., Ohukainen P., Vainio L., Koch W. J., Kerkelä R., Ruskoaho H., Serpi R., Inhibition of Let-7 microRNA attenuates myocardial remodeling and improves cardiac function postinfarction in mice. Pharmacol. Res. Perspect. 2, e00056 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozturk N., Singh I., Mehta A., Braun T., Barreto G., HMGA proteins as modulators of chromatin structure during transcriptional activation. Front. Cell Dev. Biol. 2, 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuang W., Ewald A. J., Resar L. M. S., Senger S., Xian L., Huso T., Georgess D., Belton A., Zhang X., Fasano A., Huso D. L., Gu Q., Chang Y.-T., Cope L., HMGA1 amplifies Wnt signalling and expands the intestinal stem cell compartment and Paneth cell niche. Nat. Commun. 8, 15008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sekiya I., Tsuji K., Koopman P., Watanabe H., Yamada Y., Shinomiya K., Nifuji A., Noda M., SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. J. Biol. Chem. 275, 10738–10744 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Bell D. M., Leung K. K. H., Wheatley S. C., Ng L. J., Zhou S., Ling K. W., Sham M. H., Koopman P., Tam P. P. L., Cheah K. S. E., SOX9 directly regulates the type-II collagen gene. Nat. Genet. 16, 174–178 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Xu X., Wang Y., Deng H., Liu C., Wu J., Lai M., HMGA2 enhances 5-fluorouracil chemoresistance in colorectal cancer via the Dvl2/Wnt pathway. Oncotarget 9, 9963–9974 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang S., Gu Y., Wang G., Hu Q., Chen S., Wang Y., Zhao M., HMGA2 regulates acute myeloid leukemia progression and sensitivity to daunorubicin via Wnt/β-catenin signaling. Int. J. Mol. Med. 44, 427–436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Kraan P. M., The interaction between joint inflammation and cartilage repair. Tissue Eng. Regen. Med. 16, 327–334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menkes C. J., Lane N. E., Are osteophytes good or bad? Osteoarthr. Cartil. 12, S53-4 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Zhu H., Ng S. C., Segr A. V., Shinoda G., Shah S. P., Einhorn W. S., Takeuchi A., Engreitz J. M., Hagan J. P., Kharas M. G., Urbach A., Thornton J. E., Triboulet R., Gregory R. I., Altshuler D., Daley G. Q., The Lin28/let-7 axis regulates glucose metabolism. Cell 147, 81–94 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maumus M., Noël D., Ea H. K., Moulin D., Ruiz M., Hay E., Houard X., Cleret D., Cohen-Solal M., Jacques C., Jouzeau J. Y., Lafage-Proust M. H., Reboul P., Sellam J., Vinatier C., Rannou F., Jorgensen C., Guicheux J., Berenbaum F., Identification of TGFβ signatures in six murine models mimicking different osteoarthritis clinical phenotypes. Osteoarthr. Cartil. 28, 1373–1384 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Winslow M. M., Dayton T. L., Verhaak R. G. W., Kim-Kiselak C., Snyder E. L., Feldser D. M., Hubbard D. D., Dupage M. J., Whittaker C. A., Hoersch S., Yoon S., Crowley D., Bronson R. T., Chiang D. Y., Meyerson M., Jacks T., Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar M. S., Erkeland S. J., Pester R. E., Chen C. Y., Ebert M. S., Sharp P. A., Jacks T., Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. U.S.A. 105, 3903–3908 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viswanathan S. R., Powers J. T., Einhorn W., Hoshida Y., Ng T. L., Toffanin S., O’Sullivan M., Lu J., Phillips L. A., Lockhart V. L., Shah S. P., Tanwar P. S., Mermel C. H., Beroukhim R., Azam M., Teixeira J., Meyerson M., Hughes T. P., Llovet J. M., Radich J., Mullighan C. G., Golub T. R., Sorensen P. H., Daley G. Q., Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 41, 843–848 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6