Abstract
How novel gene functions evolve is a fundamental question in biology. Mucin proteins, a functionally but not evolutionarily defined group of proteins, allow the study of convergent evolution of gene function. By analyzing the genomic variation of mucins across a wide range of mammalian genomes, we propose that exonic repeats and their copy number variation contribute substantially to the de novo evolution of new gene functions. By integrating bioinformatic, phylogenetic, proteomic, and immunohistochemical approaches, we identified 15 undescribed instances of evolutionary convergence, where novel mucins originated by gaining densely O-glycosylated exonic repeat domains. Our results suggest that secreted proteins rich in proline are natural precursors for acquiring mucin function. Our findings have broad implications for understanding the role of exonic repeats in the parallel evolution of new gene functions, especially those involving protein glycosylation.
Mucin proteins provide mechanistic insights into how genes can evolve to gain novel functions.
INTRODUCTION
Parallel independent evolution resulting in similar genetic variants has been discussed as a common driver of convergent response to adaptive pressures (1). This line of inquiry is exciting because instances of parallel evolution provide a natural framework to study the relative contributions of selection and mutational constraints to genomic variation. Recent studies provided evidence that parallel evolution is widespread in all branches of life (2). A considerable number of reported cases of parallel evolution involve recurrent structural variants, originating through convergent expansions of gene families as a response to similar adaptive pressures. Examples include the recurrent duplications of amylase genes among animals consuming starch-rich diets (3), recurrent mutations in innate immune system proteins (4), species-specific gene duplications involved in caffeine synthesis in coffee and tea plants (5), and venom evolution through gene duplications in reptiles (6) and mammals (7).
Recent studies have implied that mucin genes, which are grouped on the basis of their function rather than evolutionary commonality, may have been particularly prone to convergent evolution (8, 9). Mucins are a group of functionally characterized glycoproteins, defined by the presence of repeated proline (P)-, threonine (T)-, and serine (S)-rich O-linked glycosylation sites (10) known as PTS repeats. Functionally, mucins play crucial roles in mediating signaling between epithelial cells, in forming mucous layers to lubricate various organs, and in providing a protective barrier against environmental insult (11). In addition, mucins form an interface with commensal and pathogenic microbes, thus contributing to both colonization by a physiological microflora and host defense against pathogens (12). In a disease-related context, mucins have been shown to play roles in the pathology of cystic fibrosis (13) and other lung diseases (14) as well as in various malignancies (15). Despite the widespread and growing interest in the functional and biomedical aspects of mucin proteins (16), the evolution of mucin genes is not well understood.
Most genes with similar functions originate from duplication of a shared ancestral gene (17). They are identical by descent. However, mucin genes in the human genome do not all share common ancestry. Instead, most genes with well-described mucin function in humans belong to two gene families: secreted gel-forming mucins and membrane-bound mucins that likely evolved independently (8). Other mucins (MUC7, MUC22, and MUC16), not belonging to these two major families, were named “orphans” by Dekker and coworkers (8) because they represent no apparent orthology to other genes, including other mucins. The presence of two evolutionarily distinct mucin gene families, as well as the existence of scattered orphan mucins in the human genome, suggests that recurrent, lineage-specific evolution of mucin function may be a widespread evolutionary phenomenon in this functionally homologous, but genetically heterogeneous, group of genes. Thus, mucins provide an excellent model to study the independent evolution of specific gene functions for shedding light on the functional potential of nonconserved sequences. By studying the evolution of mucin genes in mammals, this study puts forward an evolutionary model for generation of new gene functions, especially pertaining to glycosylation.
RESULTS AND DISCUSSION
Multiple instances of de novo mucin evolution in the SCPP locus
To build a foundation for studying mucin evolution, we constructed a simple but conservative bioinformatic approach to identify potential mucin genes in a given genome by searching available gene annotations, and confirming mucin function by verifying the existence of exonic repeats that are rich in proline (P), threonine (T), and serine (S) amino acids. Using this approach, we searched for mucin genes in the genomes of human, mouse, cow, and ferret. These genomes are available as chromosome-level assemblies and can serve as representatives of primates, rodents, ungulates, and carnivores. We found that most mucins are ancestrally shared among these mammalian genomes (Fig. 1). However, we also detected at least one lineage-specific mucin gene in each species (table S1). For example, we found MUC22 only in the human genome with no orthologs in mouse, cow, or ferret reference genomes. More notably, the ferret genome harbors six unique mucin genes that are not present in the other genomes. Evolution of de novo gene functions that does not involve neofunctionalization is rare (18). Thus, the fact that our stringent search identified multiple lineage-specific mucins that are not results of whole-gene duplications was unexpected.
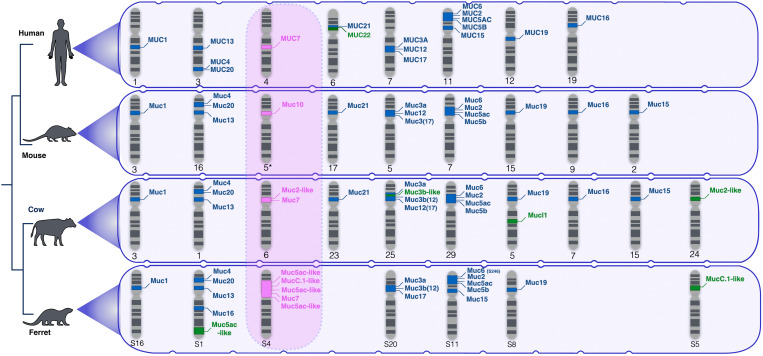
Fig. 1. Novel and previously known mucin genes in select mammalian species.
Phylogeny on the left represents the relationship between the species analyzed here [human (hg38), mouse (mm10), cow (bosTau9), and ferret (musFur1)]. Schematic karyotypes show the chromosomes in each species that harbor mucin genes. Mucin gene locations are indicated on each chromosome. Ancestral mucin genes that are orthologous in the four genomes are indicated in blue fonts. Lineage-specific mucins are indicated in green fonts. Mucin genes found within the SCPP gene family, all of which, except for MUC7, are lineage-specific are indicated in pink fonts. Note: Some of the orthologous genes carry different names in different species. For example, rodent Muc3 is orthologous to human MUC17. For those genes, we indicated in parentheses following the official gene annotation the name of the likely human ortholog based on sequence similarity and synteny. In ferrets, the “S” proceeding the putative chromosome number indicates on which Hi-C scaffold the mucin genes were found.
We found that four of the ferret-specific mucins are localized within the secretory calcium-binding phosphoprotein (SCPP) locus (bordered in humans by CSN1s1 on the 5′ end and ENAM on the 3′ end). This locus also harbors another lineage-specific mucin that we identified in the cow reference genome. Last but not least, the salivary MUC7 gene and its functional counterpart, Muc10, in mice are both located within the SCPP locus as well. The multiple occurrences of lineage-specific mucins among the SCPP genes prompted us to extend our investigation by focusing on the SCPP locus and including additional mammalian species.
Orphan mucin genes within the SCPP locus have evolved independently
The evolution of genes within the SCPP locus has been discussed within the context of calcium-binding proteins important for bone and tooth mineralization as well as major protein components in milk and saliva (19). Furthermore, this locus was highlighted as a major example for “twilight zone of sequence conservation” (20) where lineage-specific adaptive evolution leads to nonconserved sequence variation while retaining important functions. Most relevant to this study, this locus harbors multiple lineage-specific orphan mucins. As mentioned before, orphan mucins are those that do not belong to known mucin gene families that are identical by descent, while lineage-specific mucins are those that have evolved only in a given branch of the mammalian phylogeny. One mechanism for a lineage-specific mucin to evolve is through whole-gene duplication of another mucin. In this case, we expect the ancestral and duplicated mucin genes to share sequence similarity and form a gene family. Given that orphan mucins do not show such sequence similarity to other mucins, we hypothesize that lineage-specific orphan mucins evolve through a mechanism other than whole-gene duplication. Therefore, we investigated the presence of mucin functional domains within the SCPP locus in 49 mammalian reference genomes (fig. S1; see Materials and Methods for details). Next, we searched for orthologs of these genes using a combination of BLAST-based sequence similarity and manual verification of gene synteny across mammals (see Materials and Methods). Using this approach, we identified 28 putative mucin genes within the SCPP locus that only appear in certain mammalian lineages but not in others (table S1). Furthermore, we identified 15 independent, lineage-specific events explaining the origin of all 28 mucins found within the SCPP locus (fig. S2). All of these putative lineage-specific mucin genes were found in a confined region flanked by the CSN3 and AMTN genes. These two genes are conserved across all mammals and provided robust locational anchors of synteny for our study, marking a relatively short segment, ranging from ~250 to 300 kb, depending on the species.
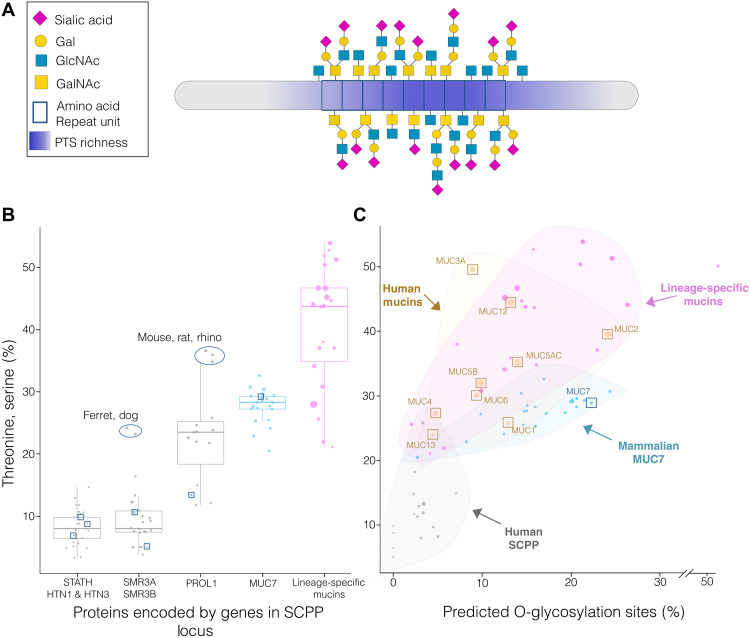
Next, we asked whether the putative mucin genes that we identified encode for proteins with functional mucin properties (Fig. 2A). To investigate this question, we first analyzed the percentage of threonines (T) and serines (S) within the protein products of these genes (Fig. 2B). These amino acids are of particular importance because they act as anchoring sites for O-glycans, which are hallmarks of mucin function (21). Our analysis showed that most proteins encoded by SCPP genes have approximately 10% T and S content, independent of species of origin. In comparison, MUC7, which is a well-described mucin in humans (22), has at least 20% T and S content in all the species where it is present. The lineage-specific putative mucins that we found in the SCPP locus harbor a significantly higher percentage of T and S amino acids than proteins from nonmucin genes in this locus (Wilcoxon test; P < 6.811 × 10−10). Furthermore, we found that the TS richness (T and S percentage of the total number of amino acids for a given protein) correlated with the number of predicted O-glycosylation sites (Fig. 2C). In summary, the analyses support that the identified genes encode for proteins with mucin characteristics (fig. S3).
Fig. 2. Verifying mucin function of putative lineage-specific mucins based on threonine (T) and serine (S) richness and O-glycosylation potential.
(A) Simplified model of a mammalian mucin protein. (B) Box plot representing the percentage of T and S amino acids in the overall amino acid sequence of the proteins encoded by different categories of SCPP genes. Proteins are categorized into histatins (HTN1 and HTN3), statherin (STATH), submaxillary gland androgen-regulated proteins (SMR3A and SMR3B), proline-rich, lacrimal 1 (PROL1), mucin 7 (MUC7), and lineage-specific mucins. The y axis shows T and S amino acids as a percentage of all amino acids composing the protein. Individual proteins of each species are indicated by dots, with their diameters corresponding to protein length. The squares indicate human proteins. The two ellipses highlight cases of species that show unusually high levels of T and S percentage for SMR3A, SMR3B, and PROL1 proteins. Further analysis revealed that these proteins gained lineage-specific mucin-like repeat domains as discussed later in the manuscript. (C) Scatterplot comparing lineage-specific mucin proteins to representative human mucins highlighted in (8) (brown), mammalian MUC7 proteins (blue), and human SCPP locus proteins (gray) as a comparison. The y axis represents TS richness, and the x axis is the percentage of predicted O-glycosylation sites (as predicted by SPRINT-Gly) within the full-length protein sequence. Lineage-specific mucins are represented as pink dots. The broader areas in the plot containing the majority of each protein and mucin category are shaded in corresponding colors and labeled with arrows. The sizes of the dots indicate the length of the proteins, and boxed dots correspond to human proteins.
Muc10 as a case example for de novo mucin evolution
The identification of multiple novel mucin genes within the SCPP locus provides a unique opportunity to address the question whether these genes have evolved through neofunctionalization after gene duplication (17), as de novo genes from noncoding sequences (23–25), or through some other mechanism (Fig. 3A). The evolutionary histories of two salivary mucins, MUC7 in humans and Muc10 in mouse and rat, may allow deeper insight into these questions. MUC7 is expressed abundantly in submandibular and sublingual salivary glands in humans (26), as well as in the saliva of nonhuman primates (27), and is shared by most placental mammals (28). However, the MUC7 gene is absent in the rat and mouse genomes (28). Despite the absence of MUC7, mouse saliva contains an abundant amount of MUC10, which is a similarly small-sized, but distinct mucin protein (29). The evolutionary history of MUC10 is unknown.
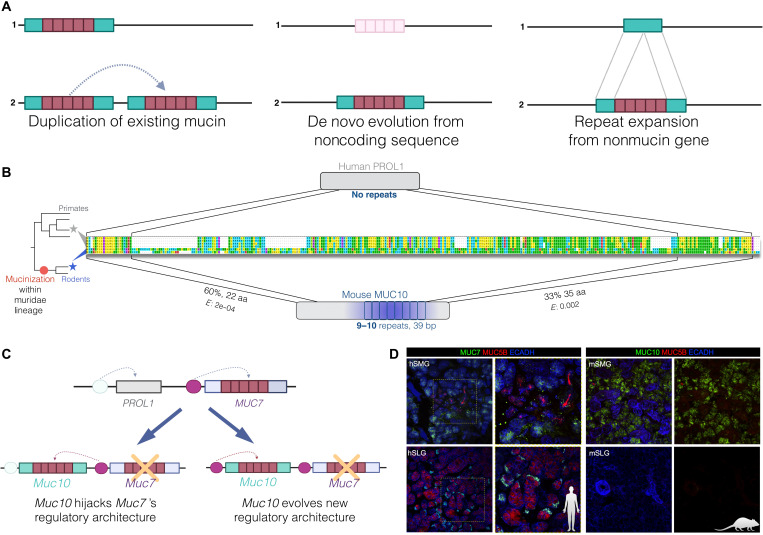
Fig. 3. A protein rich in proline has evolved into a salivary mucin.
(A) Plausible evolutionary mechanisms that were considered during our interrogation: gene duplication, evolution of coding sequence from already repeated noncoding regions of the genome, and gain of repeats from existing proteins. (B) PROL1 (top) has gained exonic PTS-rich repeats and, thus, potential mucin function (mucinization) in mice and rats (bottom). The phylogeny on the left represents the species investigated to construct the alignment shown in this figure (primates: human, chimpanzee, rhesus, and green monkey; rodents: rat and mouse). Repeats in the Muc10 gene are designated by blue boxes below the sequence alignment, and PTS richness is indicated by blue shading. The red dot in the phylogenetic tree indicates the lineage location where mucin function likely evolved. (C) Two scenarios through which Muc10 could have gained salivary expression in mice. (D) Immunofluorescent localization of MUC7 in human (left) and MUC10 in mouse (right) sublingual and submandibular salivary glands. Left-side images of each panel show MUC7 or MUC10 (green), MUC5B (red), and E-cadherin (blue) immunostaining. Right side of the human image shows a magnified view of the demarcated areas (dotted square) in the same glandular region. Right-side image for the mouse gland shows the same image without cadherin (blue) immunostaining for clarity.
Following the potential models of mucin evolution that we summarized (Fig. 3A), we first asked if Muc10 is a product of a recent duplication event involving Muc7. Muc7 and Muc10 are synthetic in the sense that they are located in the SCPP locus flanked by the Amtn gene on their 3′ side. If Muc10 has evolved through duplication of Muc7, we expect to find significant sequence homology between these genes. We found no such homology, thus rejecting neofunctionalization from a Muc7 duplicate as a mechanism for the evolution of Muc10. Instead, we found that the 5′ and 3′ sections of mouse and rat Muc10 show homology to the primate PROL1 sequences (Fig. 3B). In humans and other primates, the PROL1 gene flanks MUC7 on its 5′ side, but the protein lacks the characteristic PTS repeats of a mucin, and is expressed primarily in lacrimal glands (30) and only spuriously in saliva (26). Thus, the most plausible scenario is that the ancestral mammalian Prol1 has seeded the new mucin gene Muc10 in the rat and mouse lineages by gaining PTS repeats (Fig. 3A, “mucinization”) and becoming abundantly expressed in the salivary glands of these rodent species.
We tested this hypothesis first by manual alignment of human PROL1 with mouse and rat MUC10 (Prol1 gene) peptide sequences, which showed that these proteins exhibit ~60 and 33% homology at their 5′ and 3′ ends, respectively, the former corresponding to the signal peptide (Fig. 3B). Homology does not extend to the middle region of the MUC10 protein, which has at least nine repeats of 39 base pairs (bp) (13 amino acids) in length that are approximately 85% identical to each other. These exonic repeats do not exist in any of the primate PROL1 proteins (Fig. 3B). Further investigation showed that these repeats are rich in T and S amino acids (Fig. 2B), thus elevating the overall PTS richness of mouse and rat PROL1. To ensure the validity of our observations, we amplified and sequenced the repeated section of Prol1 from a mouse sample (C57BL/6J strain). This repeat sequence aligns perfectly with the mouse reference genome sequence (sequence file S1) but has no homologs in nonrodent genomes, further supporting the notion that these repeats were gained in the ancestor of mouse and rat.
In parallel to gaining mucin function, tissue expression patterns have also significantly shifted for the orthologous PROL1 and Muc10 genes. Specifically, PROL1 is expressed primarily in lacrimal glands in humans with little or no expression in other tissues. In contrast, MUC10, in mouse and rat, is expressed abundantly in saliva (31) and has little expression in lacrimal glands (32). It appears that the sequences that regulate Muc10 have evolved to gain strong salivary gland–specific expression in the mouse and rat ancestor.
To explain the expression trends of Muc10 in mice, we considered two scenarios (Fig. 3C). First, it is plausible that Muc10, which evolved from the PROL1 precursor, may have adopted the regulatory machinery of MUC7 after it was lost in mouse and rat lineages. Under this scenario, we expect the tissue and cellular expression of MUC7 in humans and Muc10 in mice to be similar. Second, it is possible that the salivary expression of Muc10 has evolved independently in the mouse and rat lineage, leading to expression trends that are distinct from those of human MUC7. To discriminate between these two scenarios, we conducted immunohistochemical staining for MUC10 and MUC7 in mouse and human salivary gland tissues, respectively (Fig. 3D). Consistent with previous studies (31), we found that MUC10 in the mouse is expressed only in the submandibular gland, while MUC7 in humans is expressed in both submandibular and sublingual glands. Furthermore, while MUC10 is expressed in all cell types in the mouse submandibular gland, MUC7 is expressed by specific cell populations within glands. Overall, at both tissue and cellular levels, the expression patterns of MUC10 and MUC7 are different, suggesting that the Muc10 regulatory machinery likely evolved independently in the mouse lineage.
Lineage-specific mucins evolved from precursors rich in proline
On the basis of the insights from the Prol1 to Muc10 transition in the rodent lineage, we hypothesized that the other novel mucins may have also evolved from proteins that are rich in proline. Specifically, we were interested in three genes, i.e., PROL1 (recently called OPRPN), SMR3A (previously PROL5), and SMR3B (previously PROL3), that are situated adjacent to one another in the SCPP locus and are likely identical by descent. To test whether these genes constitute precursors to the novel mucins, we searched for sequence homology between these three proteins and the newly identified 28 lineage-specific mucins. We found at least five instances among closely related species where the lineage-specific mucins show significant sequence similarity with nonmucin proteins that are rich in proline (Fig. 4, fig. S4, and table S1). We also found that they retain the signal peptide from their precursors (60 to 84% amino acid homology) but evolved TS-rich repeats in a lineage-specific manner (Fig. 4). For example, similar to the situation in mouse and rat, PROL1 in the rhinoceros harbored significantly higher T and S amino acid content than in other species (Wilcoxon test; P < 0.002198) (Fig. 2B). However, PROL1 in rhinoceros and MUC10 in mouse and rat lineages shared little sequence homology, suggesting that the T and S richness in these proteins is unlikely to be identical by descent. The emergence of a novel gene function is generally considered a rare phenomenon. Thus, it is remarkable that in two distant mammalian lineages, rhinoceros and mouse, evolution generated a novel mucin gene from the same ancestral gene, Prol1. These observations are concordant with the evolutionary scenario where the ancestral secreted protein rich in proline, PROL1, has independently gained mucin function in two different lineages through de novo gain of TS repeats rather than through neofunctionalization after whole-gene duplication or de novo gene evolution from noncoding sequences.
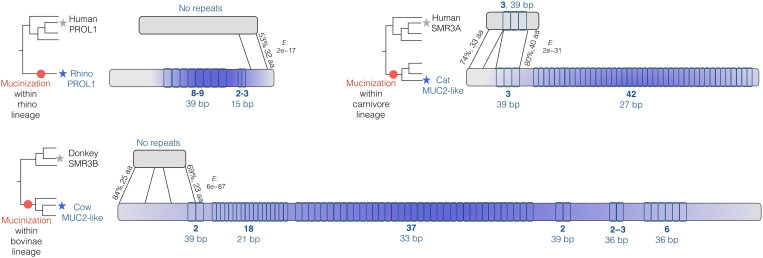
Fig. 4. Evolution of lineage-specific mucins from proteins that are rich in proline (mucinization).
Three examples of lineage-specific mucinization events are shown. The branches in the phylogeny where mucinization likely occurred are indicated by red dots. The homologous regions are shown with lines, and BLAST e values are provided. The proposed mechanism of how a nonmucin precursor protein (top) gives rise to its orthologous mucin protein (bottom) is schematically shown for the three examples, i.e., rhinoceros (rhino), cat, and cow (indicated by stars on the phylogeny). Exonic repeats are indicated by small boxes. Number of repeats and number of nucleotides per repeat are indicated below the designated repeat sections in bold. Blue color intensity indicates approximate PTS richness.
Our observations provide several venues for future research. For example, we found two novel mucins in the pangolin genome, which show homology to the human PROL1 and SMR3A/B genes, respectively, but gained exonic T- and S-rich repeats in the pangolin (fig. S4). This is an interesting observation because these lineage-specific mucins may contribute to the unusual sticky property of pangolin saliva, a trait likely selected to accommodate the animal’s insectivore feeding habits (33). Our findings thus suggest that the evolution of mucin genes repeatedly uses the mechanism that we outlined for the evolution of MUC10 in the mouse and rat lineages, where T- and S-rich exonic repeats are gained by a protein that is secreted and already rich in proline (Fig. 3A). Collectively, we argue that the evolution of mucins is facilitated by the existence of secreted proteins rich in proline in the SCPP locus.
Rapid evolution of mucin exonic repeats
In our prior analysis of Muc7 in mammals, we found that its exonic repeats retain their T and S content but vary widely in copy number within and between species (28). Our results for Muc7 stay in contrast to other exonic repeats in the genome, which occur in more than 10% of all protein-coding genes and are often highly conserved both at the nucleotide and copy number levels (34, 35). On the basis of these results, we hypothesized that exonic repeats in mucins vary in copy number as a response to adjust the overall glycosylation of the mucin protein to variable selective pressures including dietary and pathogenic changes. If this hypothesis is true, we expect that we will observe considerable levels of copy number variation for mucin repeats among species and that T and S content of the individual repeats will be conserved over evolutionary time.
We first investigated the copy number variation of mucin repeats among mammalian species (Fig. 4 and table S1). We found that the numbers of mucin repeats range substantially, from 3 in seal Muc19-like to 42 in carnivore Muc2-like/Smr3a independent of the length of the repeat (fig. S5) or the mechanism of copy number change (fig. S6). Furthermore, we have several instances where copy number variation of certain repeats evolved in a species-specific manner. For example, we found that a maximum-likelihood tree of the individual repeats from mouse and rat Muc10 can separate the repeats from each species into distinct clusters with high confidence (fig. S6). This finding suggests independent expansions of exonic repeat copy number in both mouse and rat lineages. We previously reported on lineage-specific copy number gains and losses in MUC7 in primates (28). Collectively, the considerable copy number variation of the exonic mucin repeats we observed is concordant with the adaptive hypothesis that we described above.
Next, we investigated our second expectation, namely, that the T and S content of the mucin exonic repeats is retained over evolutionary time. We focused on Muc10 in rodents and Muc2-like in felines where reasonable alignments of the individual repeat units to each other are possible. Measuring the number of synonymous and nonsynonymous nucleotide differences between repeat units, we observed that nonsynonymous changes pertaining to T and S amino acids occur less often than expected based on the number of synonymous changes (R2 < 0.15; fig. S7). This finding suggests that the T and S content of the repeats remains at similar levels and does not follow neutral expectations. For amino acids other than T and S, we observed the expected neutral rate of nonsynonymous differences (R2 > 0.65; fig. S7). Overall, as exemplified by Muc7 (28), Muc10, and Muc2-like exonic repeats, mucin repeats have adaptively retained their T and S amino acid content, suggesting that lineage-specific mucins evolve under selective constraint to retain O-glycosylation.
Lineage-specific mucins contribute to variation in the mammalian salivary glycoproteome
Previous work on mucins, mostly in humans, categorized mucins as either membrane bound or secreted (36, 37). Given that the SCPP gene family primarily comprises genes that encode secreted proteins, we hypothesized that lineage-specific mucins that evolve in this locus will also have secretory properties. We bioinformatically tested this hypothesis and found that all novel lineage-specific mucins are predicted to be secreted (see Materials and Methods; table S1). Furthermore, we found no transmembrane domains in any of the lineage-specific mucins, supporting that they may be secreted proteins.
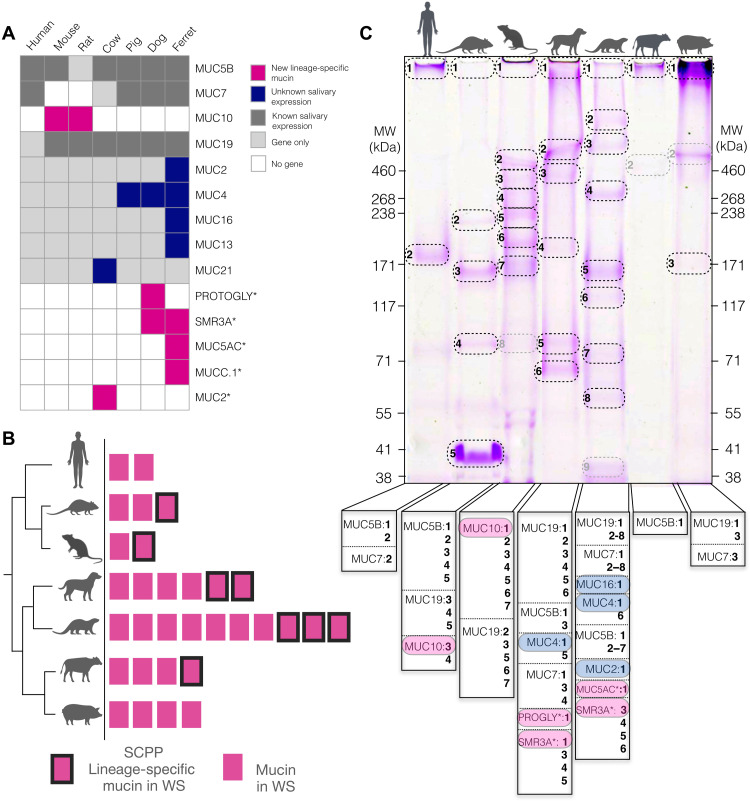
We verified previous work (26) showing that the SCPP mucins, MUC7 and Muc10, are expressed abundantly and specifically in the salivary glands of humans and mice, respectively (Fig. 3D). Thus, we investigated whether other lineage-specific mucins are also expressed in salivary glands. Other than MUC7 in humans and MUC10 in mice, it was difficult to conduct immunohistochemistry or Western blot analysis of lineage-specific mucins due to a lack of commercially available, validated antibodies. Nevertheless, although cross-species expression data from salivary glands are limited, we were able to detect salivary gland expression of some of the lineage-specific mucins, including bat MucC.1-like, cow Muc2-like, and pangolin new gene_9802, using available RNA sequencing (RNA-seq) data (figs. S4 and S8). To further investigate the salivary expression of mucin genes, we conducted liquid chromatography–mass spectrometry (LC-MS) analysis on the whole saliva from humans, mice, rats, pigs, cows, dogs, and ferrets (see Materials and Methods; Fig. 5A). Besides mucins already known to be expressed in saliva, such as MUC5b, MUC7, MUC19, and MUC10, we found a number of additional previously known mucins for which salivary expression had not been demonstrated, such as MUC4, MUC21, MUC13, MUC2, and MUC16 (Fig. 5A). In addition, we found that 8 of 11 lineage-specific mucins are secreted in saliva of dog, ferret, and cow (Fig. 5, A and B, and table S2).
Fig. 5. Comparison of mucins in saliva of diverse mammalian species.
(A) Mucin proteins in whole saliva of different mammalian species identified by LC-MS analysis. Mucins previously not known to be expressed in saliva are colored in dark blue boxes. Lineage-specific mucins identified in this study are boxes in magenta. Gray boxes indicate mucins with previously known salivary expression, while light gray indicates that the gene is present in the species’ genome with no expression detected in saliva. Empty boxes indicate that the species does not have the corresponding gene. Gene annotations were used as provided by the respective assemblies. Longer gene names indicated by an asterisk were shortened (PROGLY: PROTEOGLYCAN-LIKE; MUC2: MUC2-LIKE; MUC5AC: MUC5AC-LIKE; MUCC.1: MUCC.1-LIKE). (B) Graphical representation of the data in (A) to indicate the total number of mucin proteins expressed in whole saliva (WS) of human, mouse, rat, dog, ferret, cow, and pig (magenta rectangles). Lineage-specific mucins found within the SCPP locus are indicated by a black border. (C) Whole saliva of the above mammalian species separated by SDS-PAGE and stained with periodic acid–Schiff to reveal glycosylated proteins. Gel bands that were analyzed by LC-MS are circled. Gray circles indicate bands where a mucin could not be identified. Vertical banners below the gel lanes show the identified mucins with numbers corresponding to bands in the gel. Magenta highlights indicate lineage-specific orphan mucins, while blue highlights indicate known mucin proteins that were not previously identified in saliva. MW, molecular weight.
To experimentally verify whether the retention of T and S amino acids in lineage-specific mucins observed at the sequence level translates into protein glycosylation, we conducted tris-acetate–based SDS–polyacrylamide gel electrophoresis (PAGE) separation of salivary proteins followed by periodic acid–Schiff (PAS) staining, which reveals glycosylated proteins (see Materials and Methods; Fig. 5C) (27, 29). Comparing the electrophoretic banding pattern among pig, cow, ferret, dog, rat, mouse, and human salivary proteins, we detected a high level of diversity for glycosylated protein bands among the species tested. To confirm at the amino acid sequence level that the strongly stained bands represent mucins, we excised PAS-stained bands individually and conducted mass spectrometric analysis (see Materials and Methods; Fig. 5C). We were able to confirm substantial salivary expression of most mucins that we identified by LC-MS (Fig. 5C and table S2). Of the lineage-specific mucins, besides MUC7 and MUC10, we could identify SMR3A in dog and ferret saliva, proteoglycan-like in dog saliva, and MUC5AC-LIKE in ferret saliva, within strongly PAS-stained bands, supporting our bioinformatic predictions that these proteins are likely glycosylated.
One of the unexpected but interesting results from the SDS-PAGE analysis is the high level of variation of glycosylated protein content among mammalian saliva samples. Our current methods are limited in distinguishing between mucins and other glycoproteins. Thus, linking the glycoprotein variation among mammals to mucins remains an assumption and needs to be investigated further, perhaps using recently available mucin purification methods (38). Having said that, previous work showed that the primary glycosylated proteins most intensely stained by PAS in human saliva within the size range of our SDS-PAGE are MUC5B and MUC7 (27, 39, 40). Thus, our results provide correlative evidence that at least some of these observed differences are driven by mucins. Ferret saliva, for example, yields at least four times as many glycosylated bands as human saliva (Fig. 5B). This is concordant with our finding that ferrets harbor the highest number of lineage-specific mucins among the species we investigated (Fig. 1). In addition to lineage-specific mucins, we found that multiple mucin genes with orthologs in virtually all mammals are expressed in a species-specific manner in ferret saliva. These observations in ferrets contribute an additional piece of evidence to suggest that the high level of diversity in salivary mucin proteins among mammals has evolved by both gaining novel mucin genes and repurposing existing mucins to be expressed and secreted in saliva (Fig. 5B).
Toward a model of mucin evolution
We documented multiple instances of independent evolution of mucin function in different mammals and showed that most of these newly found mucins are located within the SCPP locus. Such recurrent evolution of gene function in a specific locus that does not occur through duplication of a whole gene is unusual. Thus, we constructed a model of mucin evolution (Fig. 6) where nonmucin genes that code for secreted proteins rich in proline serve as building blocks for novel mucins. The hypothesis makes biological sense because proteins rich in proline are similar to mucin proteins structurally (rigidity due to proline richness) and functionally (secreted proteins). They are distinct from mucin proteins only because they lack T- and S-rich exonic repeats—prime targets for O-glycosylation. Thus, these genes carry the potential to rapidly gain mucin function through repeated addition of exonic repeats. Our study provides an initial and conservative map of such occurrences with a focus on the SCPP locus. We conducted a parallel analysis of a recently available, biochemically guided “mucinome” database, reaching similar conclusions but identifying additional candidates for lineage-specific mucin formation (fig. S9). Thus, a more exhaustive effort will be needed to extend this analysis to other species and loci.
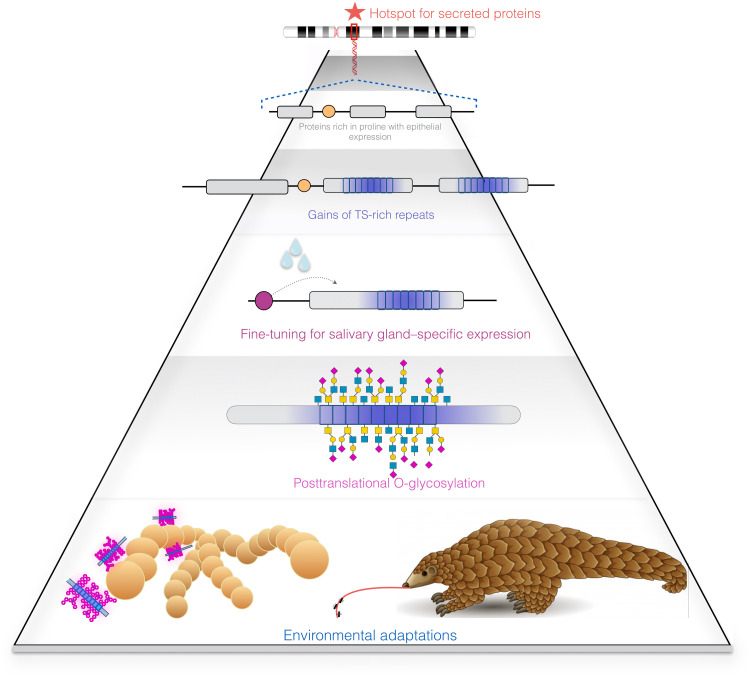
Fig. 6. Evolutionary assembly line of mucinization.
The chromosome at the top shows a hypothetical secretory protein locus where the overall regulatory architecture leads to expression in glandular and secretory tissues. In the case of the SCPP locus, in addition to being expressed in glandular tissues, these genes code for secreted proteins rich in proline. The following step is the gain of repeats that code for peptides rich in serine and threonine (gray and blue boxes). Next, existing posttranslational modification machinery attaches O-glycans to the newly formed TS-rich repeats. Last, the novel gene functions are maintained in the population provided that they lead to environmental adaptations such as pathogen clearance, or in the unique case of the pangolin to increased stickiness of saliva as an adaptation to its specific dietary niche, namely, trapping ants with its long sticky tongue.
The model of mucin evolution that we propose has three broader implications. First, it places exonic repeats as major drivers for rapid evolution and functional diversity (41). Second, it reveals proteins rich in proline as precursors for mucin generation. Third, it identifies glycosylation as a likely force in adaptive mammalian evolution (42). Our model is concordant with the growing appreciation of recurrence, convergence, and reversal as common themes in molecular evolution (43).
Beyond mechanistic insights, our results also beg the question: What are the adaptive forces that led to the retention of novel mucin genes? One clue comes from the salivary expression of these mucins. In humans, mucin function in saliva has been linked to pathogen binding, mucus layer formation, facilitating digestion, and providing viscosity and lubricity to the salivary fluid. Thus, it is safe to argue that novel mucins may have beneficial roles pertaining to immunity, diet, and mechanical properties of saliva. Previous work, including ours, has shown that O-glycans on mucin proteins interact with pathogens (39). Secreted mucins have been discussed as decoys (21) that saturate pathogen receptors in secreted fluids, thereby preventing their binding to tissue surfaces. They can also “tame” the pathogenic behavior and promote more commensal interactions between the microbes and the host organism (44, 45). The overall density, size, structure, and sterical presentation of mucin O-glycans shape the range of interaction with pathogens (39, 46) such that individual mucins have likely evolved to target specific microbes (47). For example, sialic acid residues as terminal components of mucin O-glycans provide molecular motifs for recognition by specific pathogens (48, 49), and these motifs often change in an evolutionary arms race (49, 50). Thus, it is plausible that lineage-specific mucins may bind to, or are bound by, particular pathogens in a lineage-specific manner, and that copy number variation of their exonic repeats fine-tunes the glycosylation that may help to keep pace with ever-evolving pathogenic pressures.
Mucin evolution could also be related to digestion and perception of varied diets in different species. The mucin content of saliva can directly interact with food and alter perception (51, 52). Furthermore, mucins can interact and potentially alter the microbiome composition of the gastrointestinal tract (53), thereby affecting digestion (54). It has been argued that the oral and intestinal microbes are in competition in their interaction with mucins in the gastrointestinal tract (55). Therefore, some of the mucins may have been adaptively maintained in specific lineages due to selective pressures shaped by diet in concert with the gastrointestinal microbiome. Mucins also play critical roles in determining the physical properties of bodily fluids and their function in forming tissue barriers. Thus, one exciting future venue of research will be to study the salivary activity of novel mucins in conjunction with the physical properties of saliva, such as viscosity, lubricity, and Spinnbarkeit (56).
In sum, our study establishes mechanisms how common functional and structural properties of a gene cluster can promote recurrent birth of mucin function among otherwise evolutionarily unrelated genes. Our results provide mechanistic insights into de novo formation of mucins and how it generates diversity in the mucinome. We also open up several avenues for future work to delineate the function, mechanism of formation, and adaptive impact of mucin proteins and, at a broader level, the evolution of novel gene function.
MATERIALS AND METHODS
Initial identification of candidate mucins in select species
Gene and protein annotations were downloaded from the National Center for Biotechnology Information (NCBI) Index of Genomes database available at ftp://ftp.ncbi.nih.gov/genomes/. Putative mucins were extracted from this dataset, by searching for the keywords “muc,” “mucin,” “mucin-like,” and “mucin-domain containing” (accessed 26 May 2021). Each species queried (human, mouse, cow, and ferret) contained several putative mucin genes that were not annotated by the mucin database available at www.medkem.gu.se/mucinbiology/databases/ (accessed 26 May 2021).
BLAST search for homologous sequences
Once we had obtained the list of candidate mucin genes by the keyword search described above, NCBI BLAST was used to determine the presence or absence of the candidate mucins in each human, mouse, cow, and ferret reference genome. This step allowed us to verify the annotations as well as to distinguish between lineage-specific and orthologous genes. Briefly, protein sequences were downloaded from UniProt and NCBI. These sequences were searched in each individual species using BLASTp (nonredundant protein sequences). Scoring parameters for the BLASTp (57) algorithm were as follows: matrix, BLOSUM62; gap costs, existence 11 extension 1; compositional adjustments, composition score matrix adjustment as described elsewhere (58). BLAST hits were assessed on the basis of maximum score, total score, query cover (>30%), e value (<0.01), and identity percentage (>20%). Next, we identified gene annotations in respective reference genomes that correspond to the genomic regions with the highest homology to the candidate protein sequences. Furthermore, using NCBI and UCSC Genome Browsers, we compared the genomic locations of these putative genes relative to each other and other known mucin genes to establish syntenic positions. We report our combined efforts for these four species in Fig. 1. It is important to note that our pipeline is conservative and relies on the accuracy of gene annotations and assembly qualities. We believe that, although our main observations remain the same, further verification is needed to construct a final map of mucin content in mammals. Tandem repeats, for example, are particularly difficult to assemble and thus may be missing in some reference genomes. The recently released human T2T Consortium assembly (59), which is arguably the most accurate mammalian reference genome, identifies two new mucins in the human genome, MUC3B and MUC22-like. These are not included in our dataset. Thus, it is clear that future long-read sequence–based assemblies in other mammals will remedy these shortcomings and expand our understanding of mucins.
Investigating mucin properties
We organized a two-pronged pipeline to confirm mucin properties among these putative mucin candidates. One defining characteristic of mucins is their repetitive open reading frame sequences confined into domains (8). In our pipeline, we searched for repeats on our candidate mucins in all four of our mammalian query species using the Tandem Repeats Finder (60). This algorithm identifies repeating motifs in a given sequence. One issue is that motifs are difficult to define (e.g., we may have multiple repeating motifs within a tandem repeat array) (e.g., fig. S6). For consistency, we report all of the motifs (repeat tandems ≥ 3) in table S1 and use the longest motif unit in our analyses.
Next, we located domains rich in proline, threonine, and serine, a defining feature of a mucin protein. We used a Perl script algorithm called PTSpred (61). PTSpred uses a sliding window (50 to 200 amino acids) along a given protein sequence to count the percentage of proline, threonine, and serine amino acids within that window. We used the recommended thresholds to identify PTS domains. Novel (lineage-specific) mucin characteristics were determined by requiring all of the following features: presence of a >4% predicted O-glycosylation sites per length of the peptide, presence of >20% of TS richness within peptide sequences, presence of repeats contained within a domain of the gene, and, finally, presence of proline-, threonine-, and serine-rich amino acid sequences clustering within exonic repeats.
Determining the secretory potential of proteins
To establish signal peptides on protein sequences, we used SignalP 5.0 (62), available at www.cbs.dtu.dk/services/SignalP/, using the standard parameters for prediction. In addition, we searched for known mucin domains [such as von Willebrand factor–like, epidermal growth factor–like, sperm protein enterokinase, and agrin domains (8)] using Pfam 32.0 (https://pfam.xfam.org/) (63). This algorithm uses multiple sequence alignments and hidden Markov models to predict such domains. In parallel, we searched for the presence of transmembrane helices in novel mucins using TMHMM (www.cbs.dtu.dk/services/TMHMM/) (64). In addition, to determine the likelihood of novel mucins being secreted, we used SRTpred server (65) available at https://webs.iiitd.edu.in/raghava/srtpred/home.html. Briefly, this database uses a machine learning algorithm to measure the secretory potential of proteins where positive values indicate secretion. In parallel, we also verified these results in the OutCyte database (available at www.outcyte.com/) (66), which also involves machine learning to estimate secretion potential. Specifically, a score of 0.5 or higher indicates likely secretion. Both SRTpred and OutCyte results are reported in table S1.
Determining O-glycosylation potential of proteins
O-Glycosylation sites were predicted using SPRINT-Gly (available at https://doi.org/10.1093/bioinformatics/btz215) (67). This deep neural network approach predicts the likelihood of a T or S peptide being O-glycosylated based on the surrounding repertoire of amino acids in each given window. Briefly, the algorithm scans each protein sequence for T and S amino acids and produces a window comprising four amino acids upstream and four amino acids downstream around the identified T or S amino acid. Then, it assigns a probability of O-glycosylation based on this window and previously verified O-glycosylated peptides in humans and mice. To further support potential O-glycan sites predicted by SPRINT-Gly, we used Net-O-Glyc 4.0 (available at www.cbs.dtu.dk/services/NetOGlyc/) (68), which can estimate potential O-glycosylation across mammalian species trained on experimental prediction of O-glycosylation in human cell lines. The results of both algorithms coincide. However, we found that using the SPRINT-Gly provided a more stringent prediction for O-glycosylation, and thus, we chose to use the results from this more conservative algorithm in our figures.
Identification of additional lineage-specific mucins and their likely orthologs
As described in the main text, we identified the 250- to 300-kb region (depending on the species) between CSN3 and AMTN genes within the SCPP locus as a hotspot for lineage-specific mucins. Then, we expanded our search for lineage-specific genes within this locus in additional mammals (49 mammals total). Specifically, we identified gene annotations within this hotspot region and downloaded protein sequences. We then used these protein sequences to categorize genes using our mucin determining pipeline, involving the determination of exonic repeats and the O-glycosylation potential of these repeats, as described above. Next, we used BLAST search, with the same parameters used for our initial screen described above, to search for orthologs of each candidate mucin in other mammalian species. This process allowed us to identify 28 lineage-specific mucins described in table S1.
Identifying precursors for lineage-specific mucins
We wanted to test the hypothesis that at least some lineage-specific mucins have evolved from existing genes, which do not have TS-rich repeats, as is exemplified by the evolution of MUC10 from an ancestral proline-rich protein precursor (Fig. 3). For this purpose, we conducted a thorough search for protein sequences of each of the 28 lineage-specific mucins among mammals using a combination of gene annotations, BLAST searches, and RNA-seq maps. It is noteworthy that every single precursor we identified was a proline-rich protein. Because of the repetitive nature of the lineage-specific proteins, our search was not straightforward. First, the repeat content adds uncertainty to the BLAST similarity search and hence lowers the statistical power. Second, because of PTS-rich repeats, there is a possibility of false-positive BLAST hits. Thus, to avoid including repeat sections for our initial BLAST searches, we used the first 30 amino acids, which roughly coincide with the signal peptides in secreted proteins. Next, we manually aligned lineage-specific mucins with putatively ancestral homologs to identify specific regions of sequence similarity as reported in Fig. 4. We describe the specifics of our search for each lineage-specific protein for which we identified a proline-rich precursor in detail below. Overall, our pipeline is conservative and other lineage-specific mucins may also have proline-rich precursors that we did not detect in this study.
Carnivore MUC2-like
To identify the ancestral origins of the carnivore lineage-specific mucin (called MUC2-like in cats, but SMR3A in ferrets and dogs; fig. S2, seventh row), we BLASTed the first 30 amino acids of MUC2-like protein sequence from cats (Felis catus, felCat9) to humans (taxid: 9606, hg38). We started with a BLAST to the human genome because the gene annotations and the protein sequence accuracy are optimal for humans and can have unknown biases in other species. We found significant hits to SMR3A and SMR3B genes (e = 6 × 10−8). We then manually aligned the human SMR3A and SMR3B to cat MUC2-like protein sequences and found that SMR3A has two regions of high sequence similarity and SMR3B has only one region. We then use BLAST again to verify these individual trimmed regions (see sequence file S1 for the alignments and Fig. 4 for the e values, e < 10−30). Incidentally, the new assembly that was updated during the time of the revision now annotates this gene in cats as SMR3A.
Ungulate MUC2-like
We were able to trace one of the lineage-specific mucins found in even-toed ungulates (cows, sheep, camels, alpacas, and antelope; fig. S2, first row) to the ancestral proline-rich SMR3B protein. Similar to the above pipeline, we first BLASTed the first 30 amino acids of this lineage-specific mucin protein to humans and found a significant hit to SMR3B gene (e = 0.001). Then, we narrowed our search to the outgroup lineage, odd-toed ungulates (taxid: 9787). The most significant hit was SMR3B in the donkey (e = 3 × 10−12). We verified that the donkey SMR3B does not have repeats. Next, we aligned cow MUC2-like and donkey SMR3B sequences manually and retrieved BLAST e values for the nonrepetitive sections as reported in Fig. 4 and sequence file S1.
Rodent MUC10
We found that the first 30 amino acids of this protein BLAST to human PROL1 (e = 0.046). As per previous examples, we aligned mouse and human amino acid sequences and identified the similarities and assessed the uniqueness using BLAST search. We found that doing this same process with the rat produced lower e values. These are now reported in Fig. 3B and sequence file S1. It is of note that gene annotations contribute to the confusion of the evolutionary origins of these genes. For example, concordant with our results, the latest gene annotation of MUC10 in the mouse reference genome refers to this gene as PROL1. However, the latest human gene annotation update now refers to PROL1 in humans as OPRPN.
Rhinoceros PROL1
We could not find any significant hit when we BLASTed the first 30 amino acids of Rhino PROL1 to humans. Instead, trusting the gene annotation in the reference genome, we aligned Rhino PROL1 with human PROL1 (now OPRPN). We found multiple well-aligned sections, which we interrogated in detail using BLAST, and found significant hits for some of these sections (e < 10−6). We report these in Fig. 4 and sequence file S1.
Sequence amplification and validation
Mouse Prol1/Muc10 genomic sequence was polymerase chain reaction (PCR)–amplified and Sanger-sequenced using standard methods. Primer sequences and sequencing results are found in sequence file S1. We found no differences between our sequenced region and the mouse (mm10) reference genome for repeat number and for nucleotides.
Phylogenetic and synonymous versus nonsynonymous site analysis
The lineage-specific mucin sequences found in rodents (Muc10) and felines (Muc2-like) were downloaded from NCBI. Repeats contained within the repeat domain were manually compiled in TextWrangler and aligned using CLUSTALW (69) in MEGA (70). A maximum-likelihood phylogenetic tree was constructed using 100 bootstrap replicates. Repeat sequences were then analyzed in MEGA’s pairwise distance computer for synonymous versus nonsynonymous site changes within and between species for rodents and felines independently.
RNA-seq data mining
RNA-seq data used to construct fig. S8 were taken from the expression exonic coverage track on NCBI Genome Data Viewer (www.ncbi.nlm.nih.gov/genome/gdv/). This database houses comprehensive RNA-seq data from multiple tissues and species. To determine whether a gene had observable tissue expression, we used a “housekeeping” RNA expression gene, PSMB2, which is known to be expressed in all tissues of all placental mammals (71). If a gene is expressed at the same order of magnitude as PSMB2, we deemed this gene to be “expressed” in that tissue.
Saliva collection
Saliva samples from human, mouse, rat, pig, cow, dog, and ferret individuals were collected and stored at −80°C. Human subjects: Saliva from humans was collected by passive drooling following the protocol approved by the University at Buffalo Human Subjects Institutional Review Board (IRB) board (study #030-505616). Informed consent was obtained from all human participants. The samples from other mammals were collected in collaboration with colleagues and other research institutions. A more detailed description of the collection methods used for the different mammalian species is provided in (3).
SDS-PAGE separation of salivary proteins and PAS staining of glycosylated components
Samples were denatured under reducing conditions by adding 4× tris-acetate sample buffer (NuPAGE, Invitrogen, Carlsbad, CA), 2.5% β-mercaptoethanol by sample volume, and boiling in water for 10 min. Equal amounts of total protein (15 μg per lane) were subjected to separation by SDS-PAGE using 3 to 8% gradient tris-acetate mini gels (NuPAGE, Invitrogen, Carlsbad, CA). Glycosylated protein bands were revealed using PAS stain as previously described (40). Stained gels were imaged using a flat-bed scanner in the transparent mode (ImageScanner III, GE Healthcare).
Saliva sample preparation for mass spectrometry
Saliva samples were prepared using a surfactant-aided precipitation/on-pellet digestion protocol (71). Briefly, 50 μg of protein was aliquoted from each saliva sample and spiked with SDS to a final concentration of 0.5%. Samples were sequentially reduced by 10 mM dithiothreitol (DTT) at 56°C for 30 min and alkylated by 25 mM iodoacetamide (IAM) at 37°C for 30 min, both of which were performed with constant shaking in a covered thermomixer (Eppendorf). A total of six volumes of chilled acetone were then added to the samples under vigorous vortexing, and the mixture was incubated at −20°C for 3 hours. After centrifugation at 18,000g, 4°C for 30 min, samples were decanted and the pelleted protein was gently washed with 500 μl of methanol. After air-drying for 1 min, a volume of 40 μl of 50 mM (pH 8.4) tris–formic acid (FA) was added to the pellet, and a total volume of 10 μl of trypsin [0.25 μg/μl, dissolved in 50 mM (pH 8.4) tris-FA] was added for 6-hour tryptic digestion at 37°C with constant shaking. Digestion was terminated by the addition of 0.5 μl of FA, and protein digest was centrifuged at 18,000g, 4°C for 30 min. The supernatant was carefully transferred to LC vials for analysis.
Excision of bands from protein gels and preparation for mass spectrometry
Excised gel band samples were prepared using an in-gel digestion protocol. Gel bands were first cut into smaller cubes (1 to 2 mm in each dimension) using a clean scalpel and transferred to new LoBind tubes (Eppendorf). Gel cubes were dehydrated by incubating in 500 μl of acetonitrile (ACN) for 5 min with constant vortexing, and liquid was discarded (all dehydration steps below followed the same procedure, if not specified otherwise). After incubation in 500 μl of 50% ACN in 50 mM tris-FA (pH 8.4) overnight, gel cubes were then dehydrated three times and kept at 37°C in a thermomixer for 5 min to completely evaporate the remaining ACN. Samples were sequentially reduced in 100 μl of 10 mM DTT at 56°C for 30 min and alkylated in 100 μl of 25 mM IAM at 37°C for 30 min, both of which were performed with constant shaking in a covered thermomixer. Gel cubes were then dehydrated three times and incubated in 200 μl of trypsin (0.0125 μg/μl) (in tris-FA) on ice for 30 min. Excess trypsin was then removed and replaced by 200 μl of tris-FA, and samples were incubated at 37°C overnight with constant shaking. Digestion was terminated by addition of 20 μl of 5% FA and incubation for 15 min with constant vortexing, and liquid was transferred to new LoBind tubes. Gel bands were dehydrated by sequential incubation with 500 μl of 50% ACN in 50 mM tris-FA and 500 μl of ACN, each for 15 min with constant vortexing, and liquid from all three steps was combined. The protein digest was dried in a SpeedVac and reconstituted in 50 μl of 1% ACN and 0.05% trifluoroacetic acid in ddH2O with 10-min gentle vortexing. Samples were centrifuged at 18,000g, 4°C for 30 min, and the supernatant was carefully transferred to LC vials for analysis.
LC-MS analysis
The LC-MS system consisted of a Dionex UltiMate 3000 nano LC system, a Dionex UltiMate 3000 micro LC system with a WPS-3000 autosampler, and an Orbitrap Fusion Lumos mass spectrometer. A large–inner diameter (i.d.) trapping column (300-μm i.d. × 5 mm) was implemented before the nano LC column (75-μm i.d. × 65 cm, packed with 2.5-μm Xselect CSH C18 material) for high-capacity sample loading, cleanup, and delivery. For each sample, 4 μl of derived peptides was injected for LC-MS analysis. Mobile phase A and B were 0.1% FA in 2% ACN and 0.1% FA in 88% ACN. The 180-min LC gradient profile was 4% for 3 min, 4 to 11% for 5 min, 11 to 32% B for 117 min, 32 to 50% B for 10 min, 50 to 97% B for 1 min, and 97% B for 17 min and then equilibrated to 4% for 27 min. The mass spectrometer was operated under data-dependent acquisition mode with a maximal duty cycle of 3 s. MS1 spectra were acquired by Orbitrap under 120k resolution for ions within the mass/charge ratio (m/z) range of 400 to 1500. Automatic gain control and maximal injection time were set at 175% and 50 ms, and dynamic exclusion was set at 60 s, ±10 parts per million (ppm). Precursor ions were isolated by quadrupole using an m/z window of 1.2 Th and were fragmented by high-energy collision dissociation at 30% energy. MS2 spectra were acquired by ion trap under rapid scan rate with a maximal injection time of 35 ms. Detailed LC-MS settings and relevant information are described in a previous publication by Shen et al. (72).
LC-MS files were searched against UniProt protein sequence databases plus putative mucin sequences that were predicted in this study (sequence file S1) for corresponding species (Swiss-Prot: Homo sapiens, Mus musculus; Swiss-Prot + TrEMBL: Rattus norvegicus, Bos taurus, Canis lupus familiaris, Mustela putorius furo, and Sus scrofa) using Sequest HT embedded in Proteome Discoverer 1.4 (Thermo Fisher Scientific). Target-decoy searching approach using a concatenated forward and reverse protein sequence database was applied for false discovery rate (FDR) estimation and control purposes. Searching parameters include (i) precursor ion mass tolerance: 20 ppm; (ii) product ion mass tolerance: 0.8 Da; (iii) maximal missed cleavages per peptide: 2; (iv) fixed modifications: carbamidomethylation of cysteine; and (v) dynamic modifications: oxidation of methionine, acetylation of peptide N-terminals. Peptide filtering, protein inference and grouping, and FDR control were accomplished by Scaffold v5.0.0 (Proteome Software Inc.). Criteria for protein identification included 1% protein/peptide FDR and ≥2 peptides per protein. Protein lists containing relative protein abundance (spectral counts) and sequence coverage were exported from Scaffold and manually curated by R using a customized script. The parameters described here, including the 0.8-Da mass tolerance for their MS2s, have been routinely used in the field [see, e.g., (73)]. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (74) partner repository with the dataset identifier PXD033197.
Parallel examination of lineage-specific mucin evolution using the mucinome database
Our pipeline uses a general definition of mucins, i.e., proteins that harbor highly O-glycosylated T- and S-rich repeats, as a bioinformatic starting point. However, recently, biochemically guided classification of mucins (38) has been published and thus provides an alternative starting database for human mucins. We conducted a parallel analysis of the genes that are scored in the top 50 for “mucin” properties in this database. Specifically, among these 50 genes, we identified 28 genes that fit our definition of mucins in humans (i.e., harboring T- and S-rich tandem repeats). All of these genes were previously identified to have very high levels of O-glycosylation, and thus, we have not conducted additional analysis on that. Of the 28 putative mucin genes, 15 of them were already in our previous analysis and include well-described canonical human mucin genes, such as MUC5B and MUC2. In addition, we identified 13 genes that are not previously annotated as mucin genes but exhibit all characteristics of mucin genes based on both our definition and the biochemical characterizations of the mucinome database. Furthermore, we found that, of these 13 genes, 6 have conserved mucin repeat domains across mammals that we investigated, while 7 may have evolved the mucin repeat domains in a lineage-specific manner (fig. S9). These results provide additional candidates for exciting future studies to verify the functional and evolutionary relevance of these putative mucin genes.
Statistical information
Wilcoxon test was used to determine P values in Fig. 2 and fig. S9. All other statistics performed are mentioned in the above appropriate methods sections.
Figures and analyses
All statistical analyses were conducted using R. All data and figures were created using R in RStudio, Keynote, and BioRender.
Ethics
Human subjects: Saliva from humans was collected by passive drooling following the protocol approved by the University at Buffalo Human Subjects IRB board (study #030-505616). Informed consent was obtained from all human participants. Animal experimentation: The samples from other animals were collected in collaboration with colleagues and other research institutions. The samples from all the live animals specifically for this study were collected using minimally invasive methods such as saliva collection kits or from the passive drool. Descriptions of sample sources and collection methods can be found in (3) and in the Acknowledgments section.
Acknowledgments
We thank all individuals and institutions who provided us with saliva samples: J. F. Engelhardt, X. Lui, and X. Sun of University of Iowa, Iowa (ferret saliva); B. McCabe of University at Buffalo, New York (dog saliva); K. Depner and A. Globig at the Friedrich-Loeffler-Institut, Greifswald, Germany (pig saliva); A.-M. Torregrossa of Department of Psychology, University at Buffalo (rat saliva); and J. Kramer of Department of Oral Biology, University at Buffalo, New York (mouse saliva). We are grateful to M. Edgerton, M. Saitou, V. Lynch, and D. Taylor for proofreading the manuscript and discussions of the data. We thank L. Neznanova for technical help.
Funding: This study was funded by NSF grant no. 2049947 (to O.G.), National Institute of Dental and Craniofacial Research (NIDCR) grants R01 DE019807 and R21 DE025826 (to S.R.), National Cancer Institute (NCI) grant U01 CA221244 (to S.R.).
Author contributions: P.P., S.R., and O.G. conceived and designed the study and wrote the paper. P.P. performed all computational analyses and conducted gel electrophoresis experiments. S.S. and J.Q. performed peptide extraction and mass spectrometry analysis. A.J.M. and S.K. conducted immunohistochemistry experiments. All authors edited and approved the final version of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All source data generated and downloaded sequences can be found in the Supplementary Materials. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (75) partner repository with the dataset identifier PXD033197.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
Sequence file S1
REFERENCES AND NOTES
- 1.Stern D. L., The genetic causes of convergent evolution. Nat. Rev. Genet. 14, 751–764 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Sackton T. B., Clark N., Convergent evolution in the genomics era: New insights and directions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20190102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pajic P., Pavlidis P., Dean K., Neznanova L., Romano R.-A., Garneau D., Daugherity E., Globig A., Ruhl S., Gokcumen O., Independent amylase gene copy number bursts correlate with dietary preferences in mammals. eLife 8, e44628 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel M. R., Loo Y.-M., Horner S. M., Gale M. Jr., Malik H. S., Convergent evolution of escape from hepaciviral antagonism in primates. PLOS Biol. 10, e1001282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denoeud F., Carretero-Paulet L., Dereeper A., Droc G., Guyot R., Pietrella M., Zheng C., Alberti A., Anthony F., Aprea G., Aury J.-M., Bento P., Bernard M., Bocs S., Campa C., Cenci A., Combes M.-C., Crouzillat D., Silva C. D., Daddiego L., De Bellis F., Dussert S., Garsmeur O., Gayraud T., Guignon V., Jahn K., Jamilloux V., Joët T., Labadie K., Lan T., Leclercq J., Lepelley M., Leroy T., Li L.-T., Librado P., Lopez L., Muñoz A., Noel B., Pallavicini A., Perrotta G., Poncet V., Pot D., Priyono, Rigoreau M., Rouard M., Rozas J., Tranchant-Dubreuil C., Van Buren R., Zhang Q., Andrade A. C., Argout X., Bertrand B., de Kochko A., Graziosi G., Henry R. J., Jayarama, Ming R., Nagai C., Rounsley S., Sankoff D., Giuliano G., Albert V. A., Wincker P., Lashermes P., The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345, 1181–1184 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Kazandjian T. D., Petras D., Robinson S. D., van Thiel J., Greene H. W., Arbuckle K., Barlow A., Carter D. A., Wouters R. M., Whiteley G., Wagstaff S. C., Arias A. S., Albulescu L.-O., Laing A. P., Hall C., Heap A., Penrhyn-Lowe S., McCabe C. V., Ainsworth S., da Silva R. R., Dorrestein P. C., Richardson M. K., Gutiérrez J. M., Calvete J. J., Harrison R. A., Vetter I., Undheim E. A. B., Wüster W., Casewell N. R., Convergent evolution of pain-inducing defensive venom components in spitting cobras. Science 371, 386–390 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casewell N. R., Petras D., Card D. C., Suranse V., Mychajliw A. M., Richards D., Koludarov I., Albulescu L.-O., Slagboom J., Hempel B.-F., Ngum N. M., Kennerley R. J., Brocca J. L., Whiteley G., Harrison R. A., Bolton F. M. S., Debono J., Vonk F. J., Alföldi J., Johnson J., Karlsson E. K., Lindblad-Toh K., Mellor I. R., Süssmuth R. D., Fry B. G., Kuruppu S., Hodgson W. C., Kool J., Castoe T. A., Barnes I., Sunagar K., Undheim E. A. B., Turvey S. T., Solenodon genome reveals convergent evolution of venom in eulipotyphlan mammals. Proc. Natl. Acad. Sci. U.S.A. 116, 25745–25755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dekker J., Rossen J. W. A., Büller H. A., Einerhand A. W. C., The MUC family: An obituary. Trends Biochem. Sci. 27, 126–131 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Vahdati A. R., Wagner A., Parallel or convergent evolution in human population genomic data revealed by genotype networks. BMC Evol. Biol. 16, 154 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McShane A., Bath J., Jaramillo A. M., Ridley C., Walsh A. A., Evans C. M., Thornton D. J., Ribbeck K., Mucus. Curr. Biol. 31, R938–R945 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linden S. K., Sutton P., Karlsson N. G., Korolik V., McGuckin M. A., Mucins in the mucosal barrier to infection. Mucosal Immunol. 1, 183–197 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McAuley J. L., Linden S. K., Png C. W., King R. M., Pennington H. L., Gendler S. J., Florin T. H., Hill G. R., Korolik V., McGuckin M. A., MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J. Clin. Invest. 117, 2313–2324 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J. D., Dohrman A. F., Gallup M., Miyata S., Gum J. R., Kim Y. S., Nadel J. A., Prince A., Basbaum C. B., Transcriptional activation of mucin by Pseudomonas aeruginosa lipopolysaccharide in the pathogenesis of cystic fibrosis lung disease. Proc. Natl. Acad. Sci. U.S.A. 94, 967–972 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ning Y., Zheng H., Zhan Y., Liu S., Yang Y., Zang H., Luo J., Wen Q., Fan S., Comprehensive analysis of the mechanism and treatment significance of Mucins in lung cancer. J. Exp. Clin. Cancer Res. 39, 162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kufe D. W., Mucins in cancer: Function, prognosis and therapy. Nat. Rev. Cancer 9, 874–885 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malaker S. A., Pedram K., Ferracane M. J., Bensing B. A., Krishnan V., Pett C., Yu J., Woods E. C., Kramer J. R., Westerlind U., Dorigo O., Bertozzi C. R., The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins. Proc. Natl. Acad. Sci. U.S.A. 116, 7278–7287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.S. Ohno, Evolution by Gene Duplication (Springer-Verlag, 1970). [Google Scholar]
- 18.Tautz D., Domazet-Lošo T., The evolutionary origin of orphan genes. Nat. Rev. Genet. 12, 692–702 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Kawasaki K., Weiss K. M., SCPP gene evolution and the dental mineralization continuum. J. Dent. Res. 87, 520–531 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Ponting C. P., Biological function in the twilight zone of sequence conservation. BMC Biol. 15, 71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.A. Varki, R. Cummings, J. Esko, H. Freeze, G. Hart, J. Marth, O-Glycans (Cold Spring Harbor Laboratory Press, 1999). [Google Scholar]
- 22.Bobek L. A., Tsai H., Biesbrock A. R., Levine M. J., Molecular cloning, sequence, and specificity of expression of the gene encoding the low molecular weight human salivary mucin (MUC7). J. Biol. Chem. 268, 20563–20569 (1993). [PubMed] [Google Scholar]
- 23.Van Oss S. B., Carvunis A.-R., De novo gene birth. PLOS Genet. 15, e1008160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLysaght A., Guerzoni D., New genes from non-coding sequence: The role of de novo protein-coding genes in eukaryotic evolutionary innovation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20140332 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weisman C. M., The origins and functions of de novo genes: Against all odds? J. Mol. Evol. 90, 244–257 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitou M., Gaylord E. A., Xu E., May A. J., Neznanova L., Nathan S., Grawe A., Chang J., Ryan W., Ruhl S., Knox S. M., Gokcumen O., Functional specialization of human salivary glands and origins of proteins intrinsic to human saliva. Cell Rep. 33, 108402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thamadilok S., Choi K.-S., Ruhl L., Schulte F., Kazim A. L., Hardt M., Gokcumen O., Ruhl S., Human and nonhuman primate lineage-specific footprints in the salivary proteome. Mol. Biol. Evol. 37, 395–405 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu D., Pavlidis P., Thamadilok S., Redwood E., Fox S., Blekhman R., Ruhl S., Gokcumen O., Recent evolution of the salivary mucin MUC7. Sci. Rep. 6, 31791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denny P. C., Mirels L., Denny P. A., Mouse submandibular gland salivary apomucin contains repeated N-glycosylation sites. Glycobiology 6, 43–50 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Dickinson D. P., Thiesse M., cDNA cloning of an abundant human lacrimal gland mRNA encoding a novel tear protein. Curr. Eye Res. 15, 377–386 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Gao X., Oei M. S., Ovitt C. E., Sincan M., Melvin J. E., Transcriptional profiling reveals gland-specific differential expression in the three major salivary glands of the adult mouse. Physiol. Genomics 50, 263–271 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozyildirim A. M., Wistow G. J., Gao J., Wang J., Dickinson D. P., Frierson H. F. Jr., Laurie G. W., The lacrimal gland transcriptome is an unusually rich source of rare and poorly characterized gene transcripts. Invest. Ophthalmol. Vis. Sci. 46, 1572–1580 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Encyclopedia of Life Sciences (John Wiley & Sons Ltd., 2001), vol. 26, p. 323. [Google Scholar]
- 34.Gemayel R., Vinces M. D., Legendre M., Verstrepen K. J., Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 44, 445–477 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Schaper E., Gascuel O., Anisimova M., Deep conservation of human protein tandem repeats within the eukaryotes. Mol. Biol. Evol. 31, 1132–1148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jonckheere N., Skrypek N., Frénois F., Van Seuningen I., Membrane-bound mucin modular domains: From structure to function. Biochimie 95, 1077–1086 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Perez-Vilar J., Hill R. L., The structure and assembly of secreted mucins. J. Biol. Chem. 274, 31751–31754 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Malaker S. A., Riley N. M., Shon D. J., Pedram K., Krishnan V., Dorigo O., Bertozzi C. R., Revealing the human mucinome. Nat. Commun. 13, 3542 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cross B. W., Ruhl S., Glycan recognition at the saliva–oral microbiome interface. Cell. Immunol. 333, 19–33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisenberg E., Levanon E. Y., Human housekeeping genes, revisited. Trends Genet. 29, 569–574 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Hannan A. J., Tandem repeats mediating genetic plasticity in health and disease. Nat. Rev. Genet. 19, 286–298 (2018). [DOI] [PubMed] [Google Scholar]
- 42.P. Gagneux, M. Aebi, A. Varki, in Essentials of Glycobiology, A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, A. G. Darvill, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar, P. H. Seeberger, Eds. (Cold Spring Harbor Laboratory Press, 2017). [PubMed] [Google Scholar]
- 43.Church S. H., Extavour C. G., Null hypotheses for developmental evolution. Development 147, dev178004 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Wheeler K. M., Cárcamo-Oyarce G., Turner B. S., Dellos-Nolan S., Co J. Y., Lehoux S., Cummings R. D., Wozniak D. J., Ribbeck K., Mucin glycans attenuate the virulence of Pseudomonas aeruginosa in infection. Nat. Microbiol. 4, 2146–2154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinzón Martín S., Seeberger P. H., Silva D. V., Mucins and pathogenic mucin-like molecules are immunomodulators during infection and targets for diagnostics and vaccines. Front. Chem. 7, 710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen M., Varki A., Modulation of glycan recognition by clustered saccharide patches. Int. Rev. Cell Mol. Biol. 308, 75–125 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Thamadilok S., Roche-Håkansson H., Håkansson A. P., Ruhl S., Absence of capsule reveals glycan-mediated binding and recognition of salivary mucin MUC7 by Streptococcus pneumoniae. Mol. Oral Microbiol. 31, 175–188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barnard K. N., Alford-Lawrence B. K., Buchholz D. W., Wasik B. R., LaClair J. R., Yu H., Honce R., Ruhl S., Pajic P., Daugherity E. K., Chen X., Schultz-Cherry S. L., Aguilar H. C., Varki A., Parrish C. R., Modified sialic acids on mucus and erythrocytes inhibit influenza a virus hemagglutinin and neuraminidase functions. J. Virol. 94, e01567-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng L., Bensing B. A., Thamadilok S., Yu H., Lau K., Chen X., Ruhl S., Sullam P. M., Varki A., Oral streptococci utilize a Siglec-like domain of serine-rich repeat adhesins to preferentially target platelet sialoglycans in human blood. PLOS Pathog. 10, e1004540 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gagneux P., Varki A., Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology 9, 747–755 (1999). [DOI] [PubMed] [Google Scholar]
- 51.Matsuo R., Role of saliva in the maintenance of taste sensitivity. Crit. Rev. Oral Biol. Med. 11, 216–229 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Çelebioğlu H. Y., Lee S., Chronakis I. S., Interactions of salivary mucins and saliva with food proteins: A review. Crit. Rev. Food Sci. Nutr. 60, 64–83 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Tailford L. E., Crost E. H., Kavanaugh D., Juge N., Mucin glycan foraging in the human gut microbiome. Front. Genet. 6, 81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oliphant K., Allen-Vercoe E., Macronutrient metabolism by the human gut microbiome: Major fermentation by-products and their impact on host health. Microbiome 7, 91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Derrien M., van Passel M. W., van de Bovenkamp J. H., Schipper R. G., de Vos W. M., Dekker J., Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut. Microbes 1, 254–268 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inoue H., Ono K., Masuda W., Inagaki T., Yokota M., Inenaga K., Rheological properties of human saliva and salivary mucins. J. Oral Biosci. 50, 134–141 (2008). [Google Scholar]
- 57.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J., Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altschul S. F., Wootton J. C., Gertz E. M., Agarwala R., Morgulis A., Schäffer A. A., Yu Y.-K., Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272, 5101–5109 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nurk S., Koren S., Rhie A., Rautiainen M., Bzikadze A. V., Mikheenko A., Vollger M. R., Altemose N., Uralsky L., Gershman A., Aganezov S., Hoyt S. J., Diekhans M., Logsdon G. A., Alonge M., Antonarakis S. E., Borchers M., Bouffard G. G., Brooks S. Y., Caldas G. V., Chen N.-C., Cheng H., Chin C.-S., Chow W., de Lima L. G., Dishuck P. C., Durbin R., Dvorkina T., Fiddes I. T., Formenti G., Fulton R. S., Fungtammasan A., Garrison E., Grady P. G. S., Graves-Lindsay T. A., Hall I. M., Hansen N. F., Hartley G. A., Haukness M., Howe K., Hunkapiller M. W., Jain C., Jain M., Jarvis E. D., Kerpedjiev P., Kirsche M., Kolmogorov M., Korlach J., Kremitzki M., Li H., Maduro V. V., Marschall T., McCartney A. M., McDaniel J., Miller D. E., Mullikin J. C., Myers E. W., Olson N. D., Paten B., Peluso P., Pevzner P. A., Porubsky D., Potapova T., Rogaev E. I., Rosenfeld J. A., Salzberg S. L., Schneider V. A., Sedlazeck F. J., Shafin K., Shew C. J., Shumate A., Sims Y., Smit A. F. A., Soto D. C., Sović I., Storer J. M., Streets A., Sullivan B. A., Thibaud-Nissen F., Torrance J., Wagner J., Walenz B. P., Wenger A., Wood J. M. D., Xiao C., Yan S. M., Young A. C., Zarate S., Surti U., McCoy R. C., Dennis M. Y., Alexandrov I. A., Gerton J. L., O’Neill R. J., Timp W., Zook J. M., Schatz M. C., Eichler E. E., Miga K. H., Phillippy A. M., The complete sequence of a human genome. Science 376, 44–53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benson G., Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lang T., Alexandersson M., Hansson G. C., Samuelsson T., Bioinformatic identification of polymerizing and transmembrane mucins in the puffer fish Fugu rubripes. Glycobiology 14, 521–527 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Armenteros J. J. A., Tsirigos K. D., Sønderby C. K., Petersen T. N., Winther O., Brunak S., von Heijne G., Nielsen H., SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423 (2019). [DOI] [PubMed] [Google Scholar]
- 63.El-Gebali S., Mistry J., Bateman A., Eddy S. R., Luciani A., Potter S. C., Qureshi M., Richardson L. J., Salazar G. A., Smart A., Sonnhammer E. L. L., Hirsh L., Paladin L., Piovesan D., Tosatto S. C. E., Finn R. D., The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L., Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- 65.Garg A., Raghava G. P. S., A machine learning based method for the prediction of secretory proteins using amino acid composition, their order and similarity-search. In Silico Biol. 8, 129–140 (2008). [PubMed] [Google Scholar]
- 66.Zhao L., Poschmann G., Waldera-Lupa D., Rafiee N., Kollmann M., Stühler K., OutCyte: A novel tool for predicting unconventional protein secretion. Sci. Rep. 9, 19448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taherzadeh G., Dehzangi A., Golchin M., Zhou Y., Campbell M. P., SPRINT-Gly: Predicting N- and O-linked glycosylation sites of human and mouse proteins by using sequence and predicted structural properties. Bioinformatics 35, 4140–4146 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Steentoft C., Vakhrushev S. Y., Joshi H. J., Kong Y., Vester-Christensen M. B., Schjoldager K. T.-B. G., Lavrsen K., Dabelsteen S., Pedersen N. B., Marcos-Silva L., Gupta R., Bennett E. P., Mandel U., Brunak S., Wandall H. H., Levery S. B., Clausen H., Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 32, 1478–1488 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thompson J. D., Higgins D. G., Gibson T. J., CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stecher G., Tamura K., Kumar S., Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 37, 1237–1239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen S., An B., Wang X., Hilchey S. P., Li J., Cao J., Tian Y., Hu C., Jin L., Ng A., Tu C., Qu M., Zand M. S., Qu J., Surfactant cocktail-aided extraction/precipitation/on-pellet digestion strategy enables efficient and reproducible sample preparation for large-scale quantitative proteomics. Anal. Chem. 90, 10350–10359 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Shen X., Shen S., Li J., Hu Q., Nie L., Tu C., Wang X., Orsburn B., Wang J., Qu J., An ionstar experimental strategy for MS1 ion current-based quantification using ultrahigh-field orbitrap: Reproducible, in-depth, and accurate protein measurement in large cohorts. J. Proteome Res. 16, 2445–2456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McAlister G. C., Nusinow D. P., Jedrychowski M. P., Wühr M., Huttlin E. L., Erickson B. K., Rad R., Haas W., Gygi S. P., MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., Kundu D. J., Prakash A., Frericks-Zipper A., Eisenacher M., Walzer M., Wang S., Brazma A., Vizcaíno J. A., The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma J.-E., Li L.-M., Jiang H.-Y., Zhang X.-J., Li J., Li G.-Y., Yuan L.-H., Wu J., Chen J.-P., Transcriptomic analysis identifies genes and pathways related to myrmecophagy in the Malayan pangolin (Manis javanica). PeerJ 5, e4140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
References
Tables S1 and S2
Sequence file S1