Graphical abstract
Keywords: mRNA delivery, Cell reprogramming, Tissue engineering, Regenerative medicine, Clinical translation
Abstract
After thirty years of intensive research shaping and optimizing the technology, the approval of the first mRNA-based formulation by the EMA and FDA in order to stop the COVID-19 pandemic was a breakthrough in mRNA research. The astonishing success of these vaccines have brought the mRNA platform into the spotlight of the scientific community. The remarkable persistence of the groundwork is mainly attributed to the exceptional benefits of mRNA application, including the biological origin, immediate but transitory mechanism of action, non-integrative properties, safe and relatively simple manufacturing as well as the flexibility to produce any desired protein. Based on these advantages, a practical implementation of in vitro transcribed mRNA has been considered in most areas of medicine. In this review, we discuss the key preconditions for the rise of the mRNA in the medical field, including the unique structural and functional features of the mRNA molecule and its vehicles, which are crucial aspects for a production of potent mRNA-based therapeutics. Further, we focus on the utility of mRNA tools particularly in the scope of regenerative medicine, i.e. cell reprogramming approaches or manipulation strategies for targeted tissue restoration. Finally, we highlight the strong clinical potential but also the remaining hurdles to overcome for the mRNA-based regenerative therapy, which is only a few steps away from becoming a reality.
1. Introduction
For more than 5000 years, medicinal drugs were discovered by an empirical approach based on screening of biological extracts usually used in traditional medicine. However, this cost- and time-intensive method of drug development delivered hardly predictable compounds with unknown targets and pharmacological properties, which were often unsuitable for clinical application [1], [2], [3]. Consequentially, in the second half of the 20th century, the scientific community has turned its attention to the target itself and devised a rational methodology for target-based drug design, which is still dominant in the field of drug research. Generally, two categories of novel pharmaceuticals emerged that are widely used in all medical sectors: (1) synthetic drugs manufactured via chemical processes and (2) biopharmaceuticals, also known as biologicals, which are commonly generated from living cells [4]. Yet, over the past decades, a paradigm shift from implementation of chemically derived drugs towards treatment with biological compounds (such as proteins, peptides or nucleic acids; Table 1 ) has occurred leading to a prominent and rapid expansion of the biopharmaceutical market [5], [6]. The use of synthetic drugs is commonly associated with toxicity concerns due to their broad chemical reactivity with organic molecules [7], while biologicals show fewer or no off-target activity due to higher specificity for their targets [8], [9]. Today, the vast majority of commercially approved biologicals includes recombinant proteins, which are prevalently utilized as therapeutic antigens or antibodies for treatment of cancer, inflammatory or infectious diseases [4], [10]. In context of biological drug development, another class of therapeutic molecules has come into focus – nucleic acids, one of the most ancient biopolymers [11]. Represented by DNA and RNA, nucleic acids are responsible for the transfer of biological information within and between living organisms as stated in the central dogma of molecular biology by Francis Crick in 1970 [12].
Table 1.
Benefits and drawbacks of DNA-, RNA- or protein-based biologics.
| Type | Advantages | Disadvantages |
|---|---|---|
| DNA |
|
|
| RNA |
|
|
| Proteins |
|
|
As a carrier for the genetic code, nucleic acids offer the advantage of in situ production of target proteins [13]. Unlike DNA, ribonucleic acids possess a much wider spectrum of functions and include not only protein-coding sequences, but also non-coding types that participate in protein synthesis (e.g. rRNA, tRNA), regulation of gene expression (snoRNA, snRNA, lncRNA) or post-transcriptional gene silencing (miRNA or anti-sense RNA). However, in contrast to DNA-based therapeutics, the importance of protein encoding messenger RNA (mRNA) for drug research was not immediately recognized because of the high instability of the molecule, potential immunogenicity and suboptimal delivery approaches [14]. Due to the great advances in science and technology, it was possible to evade most of these issues. The rapid degradation of mRNA caused by enzymes was minimized by establishing nuclease-free working conditions. The poor translatability of synthetic mRNAs was drastically improved with the discovery of the mRNA cap structure, which can be replaced by various translation-promoting cap modifications [15], [16]. Furthermore, structural mRNA optimization with modified nucleosides additionally hampered recognition by the immune system [17]. In addition, innovations in drug delivery offered novel vehicles based on polymeric, lipid or peptide materials to improve the mRNA transfer [18], [19].
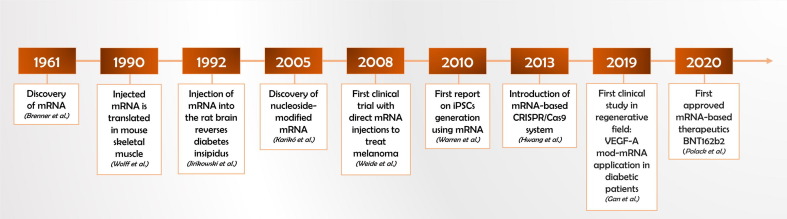
Thus, it was not until three decades after the discovery of mRNA [20] that Wolff et al. (1990) pioneered a direct injection of pure in vitro transcribed, cap-optimized mRNA into the skeletal muscle of mice and evidenced a transient expression pattern of encoded reporter protein (Fig. 1 ) [21]. At this point, the exploration of the therapeutic use of mRNA got a second boost. In 1992, modified mRNA (mod-mRNA) was successfully deployed in vivo as a protein substitution therapy to treat genetic condition of diabetes insipidus in Brattleboro rats lacking vasopressin expression in hypothalamic neurons [22]. In 1993, the first mRNA-based vaccination of mice against influenza virus was demonstrated as an effective immunization strategy preventing the risk of infectious diseases [23]. The next milestone in the history of mRNA-therapeutics was reached in the field of cancer research. In 1996, Boczkowski et al. introduced mRNA encoding tumor-enriched protein ovalbumin to dendritic cells (DCs). Following injection of these mRNA-pulsed DCs into tumor-bearing mice, a significant reduction of the tumor burden was observed indicating an efficient mRNA-mediated sensitization of the immune system [24]. The stunning success of this anti-cancer approach accelerated the immediate clinical translation of mRNAs in the same year, when mRNA-modulated DCs were applied to patients suffering treat prostate cancer [25]. About ten years later, a direct injection of tumor-specific mRNA was administered for the first time as a cell-free vaccine to medicate metastatic melanoma patients [26], [27]. Today, although mRNA biologicals are considered to possess a powerful immunomodulatory potential in anticancer therapy, they still reside in extensive clinical research [28], [29]. Nonetheless, the biopharmaceutical industry recently achieved a spectacular breakthrough in the field of mRNA-based medicine. As a global pandemic caused by the coronavirus disease 2019 (COVID-19) overwhelmed the world [30], BioNTech/Pfizer (BNT162b2) and Moderna (mRNA-1273) have developed the first, highly potent mRNA-based vaccines within an extraordinary short period of time – only a year since COVID-19 outbreak [31], [32]. These mRNA vaccines demonstrated remarkable safety and showed the most potent efficacy (95% and 94.1%, respectively) [33], [34] outranging the traditional viral-vectored vaccines, e.g. Oxford-AstraZeneca chimpanzee-derived adenovirus vaccine (ChAdOx1 or AZD1222; 62% efficacy) [35].
Fig. 1.
Milestones of mRNA application in regenerative medicine. Time history chart showing the key discoveries and breakthrough events that contributed to the rise of mRNA technology in regenerative therapy [17], [20], [21], [22], [26], [33], [134], [222], [263].
The winning profile of mRNA COVID-19 vaccines highlights again the advantages of mRNA over virus-encapsulated or plasmid DNA that largely rely on fundamental laws of protein origin. The route of the eukaryotic protein biosynthesis starts in the nucleus, where DNA is transcribed to mRNA that in turn enters the cytoplasm to be translated into a protein. Thus, while mRNA therapeutics only need to reach the cytoplasm in order to perform its function, DNA-based drugs are compelled to overcome a tight nuclear membrane that is naturally eased during cell division. This additional barrier is a major issue limiting the efficiency of DNA delivery, whereas mRNA transport into the cell is not affected by nuclear boundaries. As for safety concerns, DNA and viral vectors are prone to incorporate into the host genome posing considerable risk of mutagenesis. In contrast, mRNA offers great safety as it does not interact with genetic material, acts as a short-lived gene transcript for cytoplasmic protein production and can be degraded completely via physiological metabolic pathways. Furthermore, mRNA does not contain additional foreign gene information, allows in vitro synthesis under cell-free conditions avoiding bacterial contamination, can encode any kind of protein and, therefore, potentially represent a therapeutic tool for any disease. Taking into account that mRNA manufacturing is a relatively simple and inexpensive process, further rising interest for mRNA technology exceeding the field of vaccine design can be expected in the near future [14], [36], [37], [38].
For instance, numerous preclinical studies have recently demonstrated the promising potential of mRNA to treat a wide range of protein malfunction diseases including hemophilia type A [39] and B [40], Fabry (lysosomal storage) disease [41], glycogen storage disease type Ia [42] and other hepatic abnormalities [43], [44], [45]. In 2017, a novel biological MRT5005 (Translate Bio) has marked an important milestone for mRNA-based protein replacement therapy by entering the phase I/II of clinical trial (NCT03375047). This compound contains mRNA of CFTR (cystic fibrosis transmembrane conductance regulator) – a protein either misfolded or depleted in cystic fibrosis patients due to mutations in CFTR gene [46]. Very recently, two more mRNA therapeutics have attained the clinical sector: mRNA-3704 (Moderna) restoring the function of MUT (mitochondrial enzyme methylmalonic-CoA mutase) in rare cases of methylmalonic acidemia (NCT03810690) and ARCT-810 (Arcturus Therapeutics), targeting patients with ornithine transcarbamylase deficiency (NCT04416126).
However, despite the encouraging results demonstrated in preclinical and clinical studies, mRNA therapeutics are still facing certain downsides that all innovative drugs have in common – an urgent need for optimized pharmacokinetics and pharmacodynamics. To understand the current challenges of clinical mRNA application, it is crucial to take a closer look into the nature of mRNA molecule, its structure and functions. The following chapters will also address the importance of mRNA modifications as well as recent advances in mRNA delivery methods. In this review, we will focus on the relevance of the mRNA platform for cellular reprogramming and its purpose specifically in regenerative medicine.
2. mRNA: molecular composition and function
The cellular synthesis of an mRNA molecule takes place during a process called transcription, when RNA polymerase reads out and duplicates the template (antisense) strand of a protein-coding DNA sequence in 3 → 5‘direction, thereby switching every thymine nucleobase for uracil in the transcript. Thus, the resulting precursor mRNA (pre-mRNA) represents a single-stranded copy of a complementary gene segment with opposite direction (5́ to 3́end) and uracil replacing thymine [11]. However, the premature mRNA needs further processing to become a functional mRNA. This includes essential steps of co-transcriptional 5́end capping, 3́end rearrangement as well as splicing [47], important processes that will be discussed in the subsection below (Section 2.1.). Additionally, we will highlight the current advanced approaches to enhance stability, translatability and overall life-time of in vitro synthesized mRNA drugs (Section 2.2.).
2.1. Native mRNA structure
A mature eukaryotic mRNA is composed of a nucleotide sequence with four distinct regions: cap structures at the 5́end, polyadenylated site at the 3́end and a protein-coding sequence flanked by untranslated regions (UTRs) at both 5́and 3́end [48].
The 5́capping of the nascent pre-mRNA is the initial modification step, executed during transcription as soon as the first 25–30 nucleotides (nt) are synthesized [49]. Chemically, it is defined as a 7-methylguanosine (m7G), coupled with the first transcribed nucleotide via reversed 5́ to 5́ triphosphate bridge. Three enzymes mediate m7G incorporation: (1) a triphosphatase cleaves the terminal 5́phosphate from the nucleotide; (2) a guanylyltransferase establishes a chemical bond between the inverted guanosine monophosphate and diphosphate creating G cap (GpppNp); (3) a methyltransferase conveys a methyl residue to the seventh position of G cap completing the “cap0” as m7GpppNp [50], [51]. This cap0 structure is involved in several biological processes as it mediates pre-mRNA processing such as 3́polyadenylation and splicing, ensures molecule stability, prevents the mRNA from degradation by 5́exonucleases and facilitates mRNA export from nucleus into the cytoplasm [52], [53]. However, the most prominent cap0 feature is the high affinity to the eukaryotic translation initiation factor eIF4e, which promotes ribosome recruitment, thereby elevating transcript turnover [54]. Furthermore, an additional m7G-specific methylation of the 2́-OH group within adjacent second and third ribose molecules forms cap1 (m7GpppNmp) and cap2 (m7GpppNmpNmp), respectively, inherent for higher eukaryotes [55]. Recent evidence demonstrated that foreign mRNA lacking 2́-O methylation activates the host innate immune system via induction of interferon signaling. Subsequently, the exogenous mRNA missing 2́-O methylation is assigned to immediate degradation [56], [57], [58] suggesting cap1 as a crucial “self” recognition mark.
The 3́end of the pre-mRNA is shaped by complex enzymatic events supported by an interplay of more than eighty different proteins [59]. Here, we provide a highly simplified overview of 3́end processing, but refer to recent comprehensive review articles that focus on enzymatic mechanism of action responsible for 3́end formation [60], [61], [62], [63]. The first steps towards maturation of the 3́end are initiated co-transcriptionally, when the 3′ processing machinery scavenges the nascent transcripts for polyadenylation site (PAS) sequence. Once the PAS motif (AAUAAA) is recognized, a protein complex with endonuclease activity catalyzes the pre-mRNA cleavage reaction 10–30 nt downstream of the PAS [64], [65]. Finally, multiple adenosine molecules (poly(A)) are added to the cleaved 3́end by a nuclear poly(A)-polymerase (PAP) that extends the pre-mRNA chain by 50–100 nt, leading to formation of a 3́poly(A)-tail [66]. The importance of accurate 3́end handling becomes particularly apparent when disruption of its elements promotes or results in impaired health conditions such as increased risk of cancer, immunodeficiency, severe hematological diseases and muscle dystrophies [67], [68]. Indeed, the main functions of the poly(A)-tail encompass many vital aspects of the mRNA life cycle: it provokes transcription termination [69], enhances stability [70], assists in nuclear export [71], favors translation [72] and determines degradation of the mRNA molecule [66], [73]. Noteworthy, the poly(A) sequence is recognized by several polyadenosyl binding proteins (PABPs), which in turn interact with the eIF4 complex recruited by the 5́cap. Thus, the poly(A)-tail can synergistically mediate translation initiation by attracting eIF4 via PABPs [54].
Furthermore, two components of the mRNA chain, located upstream and downstream of the coding sequence, fulfil a considerable regulatory function – the 5́untraslated region (UTR) and 3́UTR, respectively. Although both UTRs contain elements responsible for molecule stabilization and translation control, their impact thereon is rather diverse, which is probably attributed to a different composition of these regions. For example, the average human 5́UTR is about five times shorter than 3́UTR (∼210 nt and ∼1027 nt, respectively) and has greater G + C content (∼60% vs. ∼45%) [74], [75]. Most of the elements within the 5́UTR (hairpin, internal ribosome entry sites, binding sites for regulatory proteins, upstream open reading frames (ORFs) and alternative start codons) are crucial for translation initiation [76]. Additionally, Jia et al. (2020) recently demonstrated that the 5́UTR has a notable impact on the stability, translation efficiency and turnover of mRNA and even minor (10-nucleotide) alterations within the 5́UTR sequence have considerable relevance [77]. On the other hand, the regulatory potential of the 3́UTR appears to be even more diverse. Accumulated evidence suggests that 3́UTR mediates nuclear export [78], subcellular mRNA localization [79], poly(A)-status [80], stability and translation dynamics [81]. Overall, the UTRs are indispensable parts of mRNA but their detailed function remains elusive due to the complexity of its molecular interactivity.
Lastly, mRNA maturation is fully accomplished when the transcribed non-coding intron sequences are removed by a spliceosome, a massive (∼4.8 mega Dalton) molecular mRNA splicing machinery [82], thereafter the mRNA chain is primed for subsequent translation.
2.2. Optimization of synthetic mRNA for therapeutic applications
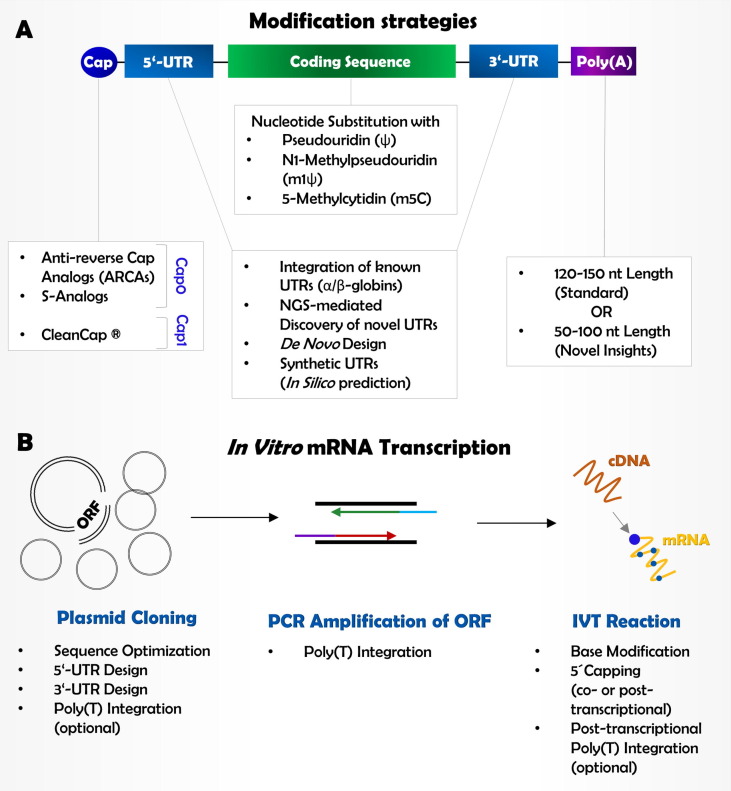
In contrast to native cellular mRNA, synthetic mRNA can be perceived by a host immune system as a foreign hazardous molecule that has to be eliminated, thereby causing preliminary mRNA breakdown. Additional drawbacks are caused by low stability of artificial transcripts, insufficient protein output rates as well as decreased protein half-life. Over the last years, several strategies utilizing chemical mRNA modifications have been developed to circumvent these issues (Fig. 2 A). In this regard, we address the recent advances in the present section.
Fig. 2.
Molecular composition and modification strategies for in vitro mRNA synthesis. (A) Several strategies have been applied to improve mRNA stability and translational efficiency, including cap optimization, nucleotide substitution and UTR-modifications. (B) In vitro synthesis of modified mRNA starts with plasmid cloning, followed by PCR amplification of the respective ORF. Lastly, cDNA is transcribed into mRNA, containing the optimized molecular composition.
Generally, the process of mRNA synthesis consists of following core steps (Fig. 2B): (1) cloning of a plasmid containing the ORF of the target gene; (2) amplification of the ORF inserts by proof-reading polymerase chain reaction (PCR); (3) in vitro transcription (IVT) reaction of PCR product [83].
2.2.1. UTRs design
The first step is a rather simple procedure as a tremendous variety of both premade and customized plasmids, coding a known ORF, are commercially available. Moreover, plasmid amplification is a commonly applied, well-established and inexpensive technique in molecular biology, available in almost any standard laboratories. Although technically uncomplicated, this initial step includes engineering of suitable UTRs, necessary for fine-tuning of the future mRNA molecule. In general, UTR sequences of strongly expressed human genes encoding long-living proteins, such as human serum globins [84] or cytochrome enzymes, have been used for mRNA synthesis [85], [86]. However, new NGS technology offering a tool to comprehensively analyze the whole cellular mRNA pool, now enables detection of novel naturally occurring UTRs that can outperform the latter in terms of molecular stability and protein half-life. For example, a study by Schwanhaeusser et al. (2011) delivered an extensive data set of gene expression in fibroblasts, which was grouped in four classes depending on the stability of mRNA related to the stability of the resulting protein. Based on these data, the identified genes coding for the most abundant long-living mRNAs and corresponding proteins are mainly involved in cellular metabolic processes (glycolysis, gluconeogenesis, tricarboxylic acid cycle etc.), respiration and translation [87]. Consequently, utilizing UTRs of these housekeeping genes could potentially prolong the half-life of synthetic mRNAs as well as their products. Recently, Zeng et al. (2020) described a more sophisticated concept for UTR integration that is based on reconstruction of endogenous UTRs. Here, the sequence length and the nucleotide composition were adjusted, the inhibitory microRNA sites were removed and additional protein binding motifs that promote translation were included. This approach, based on de novo design of endogenous UTRs, enhanced the translation efficiency of the introduced mRNA in vivo by five to ten-fold compared to commonly used and control UTRs [88]. Alternatively, Cao et al. (2020) presented major advances in bioinformatics and particularly in machine learning that allow to completely recreate highly efficient UTRs in silico using predicting algorithms that generate unique synthetic motifs [89]. Taken together, UTR optimization is an essential part of mRNA drug development that has the potential to greatly increase the potency of mRNA-based therapeutics. However, some authors suggested that the impact of UTRs on translatability may differ between tissues and depends on the disease status of the target organ [90]. Unfortunately, this delicate question has not been examined yet, and should be considered in the upcoming research.
2.2.2. Length of the poly(A)-tail
The length of the poly(A)-motif is another parameter that determines the fate of the future mRNA as it hampers transcript degradation and affects translation initiation. While some research groups tend to believe that a longer poly(A)-sequence (150–200 nt) is more efficient for enhanced translation [44], [91], [92], [93], [94], the newest reports strongly indicate that the most abundant proteins may be actually derived from much shorter-tailed (50–100 nt) transcripts [95]. In fact, recent studies arguing for shorter motifs heavily rely on data of different species by technically more advanced genome-wide RNA sequencing platforms, such as TAIL-seq [66], [96], FLAM-seq [97] and PAL-seq [98]. Moreover, the global poly(A)-tail profiling revealed a weak or even negative correlation between the tail length, transcript half-life and ribosome binding with an exception of zygotic mRNA [98].
To clarify the inconsistency, several studies brought forward the arguments that explain the possible disadvantage of long poly(A)-sequences [99], [100], [101]. Here, the reasoning is based on a concept of cytoplasmic poly(A)-driven mRNA disintegration. According to the authors, the protruding poly(A)-fragments that are not covered with PABPs (typical for tails longer than 150 nt) trigger the cytoplasmic deadenylation complex, predominantly CCR4-NOT (CNOT). The CAF1 catalytic subunit of CNOT trims exposed poly(A)-sites (in units of 27 nt) until its passage is blocked by the proximally seated PABP. However, the presence of PABP activates the CCR4 subunit of CNOT, which is able to extrude the bound PABPs, thereby gaining access to underlying poly(A)-segments. As CCR4 proceeds with the deadenylation process, the PABP complex completely dissociates, leaving the remaining poly(A)-unit (∼25 nt) uncovered. Subsequently, these short motifs are recognized by mRNA uridylation enzymes TUT4 and TUT7 that mark the transcript for decay [102]. Although the depicted process supports the estimation that longer poly(A)-motifs might reduce the transcript turnover, it still remains elusive whether shorter motifs are indeed a wiser choice. Certainly, the poly(A)-tail is a dynamic indispensable structure controlled by a variety of protein complexes, which nature is not yet fully understood and needs more in-depth research to solve the mentioned disagreements.
For mRNA synthesis, the poly(A)-elements can be included in various ways: (1) integration of the poly(T)-sequence into the DNA template; (2) extension during the second (template amplification) step of IVT by using a reverse primer with attached poly(T)-tail; (3) extension on already synthesized IVT mRNA by recombinant poly(A)-polymerase (PAP). In practice, it was shown that PAP does not generate equally long poly(A)-tails, i.e. the resulting length varies with each reaction [103], [104]. Therefore, the poly(A)-parts directly derived from the fixed poly(T)-sequence in the DNA template are believed to have more consistent lengths [105], although PCR-driven, primer-coupled poly(T)-extension of the template likely gives similar results [83].
2.2.3. Base editing
Besides optimization of UTRs and poly(A)-tail, the most unique and pivotal feature of IVT mRNA setting the term “modified” (mod-mRNA) relies on replacement of natural prime nucleosides (A, T, G, C, U) with modified base derivatives that are partly presented in multiple types of RNA (mRNA, rRNA, tRNA and viral RNA). The discovery of mod-mRNA goes back to 2005, as Karikó et al. linked the immune-stimulatory potential of native mRNA with activation of human Toll-like receptors (TLRs), namely TLR3, TLR7 and TLR8 [17]. In contrast, IVT mRNA containing substituted nucleosides, such as 5-methylcytidine (m5C), N6-methyladenosine (m6A), 5-methyluridine (m5U), 2-thiouridine (s2U) or pseudouridine (Ψ), did not provoke TLR recognition. Few years later, the same group evidenced an outstanding Ψ-mediated enhancement of translation capacity (9-fold) followed by m5C (4-fold) in vitro, whereas translation of mRNAs incorporating m6A, m5U or s2U was comparable to the unmodified control transcripts. These observations were confirmed in vivo in the same study [106]. Further research has introduced the N1-methylpseudouridine (m1Ψ) as an even more potent, less cytotoxic and less immunogenic nucleoside exchange than Ψ, which was demonstrated in vitro and in vivo [107]. A newly published study by Leppek et al. (2021), who aimed to secure the stability of mRNA beyond the cell borders, e.g. in hydrophilic solution, confirmed that IVT mRNA stabilization can be vastly improved if Ψ or m1Ψ were integrated into the sequence [108]. Thus, the most favorable nucleoside modifications today are m1Ψ, Ψ, m5C or combinations thereof (m1Ψ/m5C or Ψ/m5C) [48], [107], [108], [109], [110].
2.2.4. 5́Cap optimization
Another component of mRNA able to impede the immune response is the 5́cap. Uncapped RNA is characteristic for some viruses and can be recognized by human RIG-I (retinoic acid–inducible protein I) that stimulates human antiviral defense systems [111]. The 5́capping of IVT mRNA is therefore an essential procedure that can be performed co-transcriptionally by RNA polymerase. Yet, the conventional cap0 (m7GpppGp) is prone to incorrect attachment to the mRNA body. According to Pasquinelli et al. (1995), nearly one-third of produced IVT mRNA is capped in reverse direction (Gpppm7G) when using cap0. This inversion prevents proper binding of eIF4 and thereupon the overall translational activity of such mRNA products is diminished [112]. However, the emergence of anti-reverse cap analogs (ARCAs) managed to overcome the problem of inaccurate cap integration. Additionally to m7G, ARCAs are methylated either at the 2́- or 3́-position (m2 7,2́-OGpppG or m2 7,3́-OGpppG, respectively) [113]. The finding that artificial cap modifications can radically affect the translational outcome led to further chemical improvements of ARCAs. For example, phosphorothioate substitutions in β-position of 5́-5́ triphosphate bridge named as β-S-ARCA or S-analogs (m2 7,2′- OGppSpG) [114] ensured additional molecule stabilization, higher affinity to eIF4 and resistance to the decapping pyrophosphatase Dcp2/Dcp1 [115], [116]. In the follow-up study, the authors introduced 1,2-dithiodiphosphate moiety at α,β- or β,γ-position of a triphosphate bridge, thereby defining a new class of synthetic caps as 2S analogs that is proclaimed to be even more superior than the previous modifications [117]. Alternatively, BH3 cap analog bearing a β-boranophosphate within the phosphate bridge (m2 7,2′- OGppBH3pG) has been described as a compound that has comparable properties to S-analogs [118]. Lastly, it is conspicuous that ARCAs represent a modified cap0 but do not include the additional 2́-O methylation at the second nucleotide that is typical for eukaryotic cap1. However, cap1 is of utter relevance for mRNA drug engineering as it is capable to restrain the immune response to external mRNA. Today, some companies already offer premade cap1 reagents (e.g. CleanCap by TriLink) for co-transcriptional capping. Otherwise, cap0 can be easily upgraded to cap1 post-transcriptionally using 2́O-methyltransferase [119].
Furthermore, 5́capping can also be performed post-transcriptionally using the corresponding enzymatic machinery (RNA triphosphatase, guanylyltransferase, guanine N7-methyltransferase). However, the co-transcriptional method is eminently more advantageous as it allows for more experimental freedom regarding the chemical composition of the cap, thereby enhancing the affinity to translation initiation factors and simultaneously hiding the cap from the scavenging decapping complexes without disturbing the translational activity [120].
2.2.5. Purification of IVT mRNA
Finally, to guarantee high-purity of produced IVT mRNA, it is necessary to dispose all possible contaminants that might be potentially immunogenic. These include DNA template remnants, double-stranded or fragmented mRNA as well as residual nucleotides, which are commonly removed by means of high performance liquid chromatography (HPLC) [121]. Indeed, purified mRNA was associated with reduced inflammatory responses and increased protein yields [122].
3. Systems for mRNA drug delivery in therapeutic applications
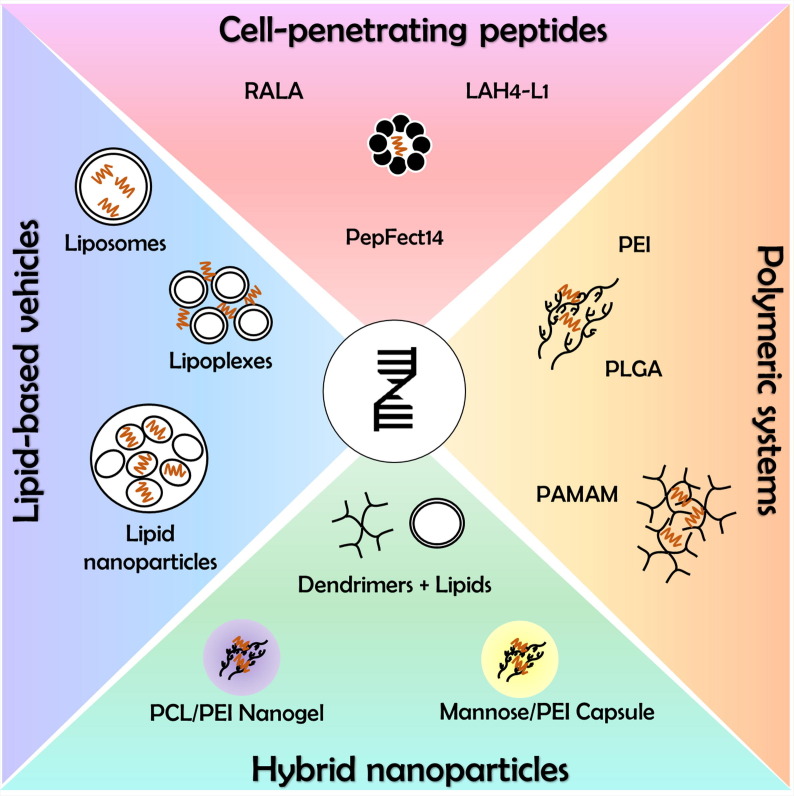
To unlock the full therapeutic potential, an IVT mRNA needs to be delivered to its final destination – the cytosol, overcoming many obstacles along the way. Despite numerous precautions, IVT mod-mRNA remains a relatively unstable molecule that is unprotected against enzymatic degradation by serum endonucleases or elimination by the host immune system. Moreover, being a large, hydrophilic and negatively charged compound, it is quite challenging for mRNA to enter the cell [123]. With an exception of skin-residual dendritic cells that are capable of effective mRNA uptake via micropinocytosis [124], the “naked” mRNA is believed to be poorly engulfed by most cell types and even if internalized, it might be trapped within endosomes with slender chances to escape lysosomal digestion [125], [126]. Thus, diverse carrier systems have been developed to secure the mRNA in blood stream, permit extravasation, prevent renal clearance, ensure specific delivery to target cells, facilitate cellular uptake and promote lysosomal release [127]. Although viral vectors could be also used as a natural transport system, their application might be contentious due to the risks of genome integration or cytotoxicity [128]. In this chapter, we will preferentially recapitulate non-viral delivery methods that mainly employ formulations based on lipid, polymer, peptide or hybrid vectors (Fig. 3 ; Table 2 ). Additionally, we will discuss possible routes of mRNA administration in accordance with a carrier system.
Fig. 3.
Non-viral carrier system for delivery of modified mRNA. Since mRNAs are large, hydrophilic, negatively charged molecules, entering the cell is challenging. Therefore, various carrier systems have been developed to improve cellular uptake and protecting the mRNA from degradation. Lipid-based delivery represents the most commonly applied technique to facilitate cellular entry. In addition, positively charged peptides can be used for complexation of mRNA, enabling access to the cell via an endocytotic mechanism. Similarly, polymers like polyethylenimine, polylactide-co-glycolide, polyamidoamine have been utilized to encapsulate mRNA molecules before cellular transfection. Novel delivery concepts aim to exploit the benefits of existing carrier systems by a combination of nanoparticles, lipids and proteins leading to hybrid nanoparticles.
Table 2.
Advantages and disadvantages of selected carrier systems for mRNA delivery.
| Carrier System | Advantages | Disadvantages |
|---|---|---|
| Lipid –based vehicles |
|
|
| Peptid-based vehicles |
|
|
| Polymer-based vehicles |
|
|
3.1. Naked mod-mRNA
A carrier-free mod-mRNA can be advantageous for locally applied treatments. For example, mod-mRNA targeting antigen presenting cells (APCs) can improve the outcome of a tumor case. Here, intradermal or intranodal delivery is highly recommended to prime T-cells since these tissues are rich in APCs (macrophages, dendritic cells or B cells) [129]. The proposed mechanism of action relies on active uptake of exogenous mRNA by dendritic cells via micropinocytosis [124], which in turn interact and sensitize both CD4+ and CD8+ T-cells against tumor-associated antigens (TAAs) [130]. In 2011, a proof-of-concept (phase I/II) clinical study conducted on patients suffering from stage IV renal cell cancer demonstrated overall safety of such mRNA-based vaccination as well as a moderate clinical response [131]. In 2012, Van Lint et al. developed a mixed formulation of mod-mRNAs called TriMix that encodes for CD40 Ligand, constitutively active Toll-like receptor 4 and CD70. The TriMix was then combined with a TAA mod-mRNA and injected into lymph nodes of mice. Captured and processed by DCs, this mod-mRNA combination created a niche that attracts T-cells and, thus, facilitates recognition of a TAA [132]. Subsequently, the same group aimed for treatment of cervical cancer caused by human papilloma virus (HPV) infection and applied the TriMix technology in conjunction with mod-mRNA of HPV16-E7 oncoprotein. Although E7-TriMix vaccine was able to specify the T-cells against tumor tissue, the therapeutic efficacy remained insufficient due to the protective microenvironment within the tumor. The most effective way to weaken the tumor microenvironment utilizes chemotherapeutics, e.g. cisplatin, which promoted the immunomodulatory activity of E7-TriMix vaccine. This kind of synergistic relationship has led to a tumor rejection in more than 85% of examined mice, thereby providing a groundwork for further combinatory implementation of mRNA-based drugs in anti-cancer research [133].
In the field of regenerative medicine, a recent investigation by Gan et al. (2019) evaluated safety and efficacy of carrier-free vascular endothelial growth factor A (VEGF-A) mod-mRNA, administrated intradermally in male patients with type 2 diabetes mellitus (NCT02935712). The results indicated well tolerability and local enhancement of blood flow with signs of induced neovascularization, which might be beneficial for care of diabetes-related wounds in the future [134]. Apart from this, the VEGF-A mod-mRNA (invented and registered as AZD8601 by AstraZeneca) may also improve the condition of ischemic cardiovascular disease. In this respect, the safety evaluation of AZD8601 injected into epicardium of patients with heart failure has already been initiated and currently remains in phase II clinical trial (NCT03370887).
Over the last few years, an alternative route of mRNA administration has gained increased attention – a non-invasive aerosol-based delivery to the mucosal tissues. In order to hamper respiratory viral infections, Tiwari et al. (2018) designed naked mod-mRNA, encoding for membrane-anchored neutralizing antibodies, that was solved in nuclease free water or saline, nebulized and dispersed in the trachea of mice. Evidently, this formulation successfully prevented viral uptake in transfected cells of respiratory epithelium, which was evidenced by reduced in vitro titer and in vivo virus copies by 99.7% and 89.6%, respectively [135]. Additionally, the authors demonstrated that nearly half of the mod-mRNA, delivered via airways, has escaped the endosomal compartment in target cells. This observation was contrary to previous assumptions, suggesting a poor cytosolic availability of vehicle-free mRNA [125], [126]. In fact, it is reasonable to assume that the success of cytosolic mRNA entry might depend on the method of pharmacological distribution (fluid vs. gasiform; injected vs. dispersed). Although the delivery of aerosolized mRNA was not tested in humans yet, the newly published studies performed on large animals (horse or sheep) ensured overall safety and success of this transfection technique, which might serve as a prophylactic treatment of pathogen-exposed mucosal areas (airway, genital or rectal tract) in the future [136], [137].
3.2. Lipid-based vehicles
Lipid-based formulations represent the most popular and developed technology for systemic (intravenous) delivery of mRNA. Among them, prevalently lipoplexes, liposomes and lipid nanoparticles (LNPs) possess the utmost potency to form a stable unit that shields the mRNA from enzymatic degradation, facilitates cellular entry and subsequent endosomal release [138].
Lipoplexes were the earliest lipid systems utilized for delivery of nucleic acids [139]. These particles consist of cationic lipids whose positively charged heads electrostatically attract the negatively charged backbone of the mRNA. In contrast to other vectors, lipoplexes do not enclose the mRNA within their inner core. Instead, the mRNA is captured between the surfaces of multiple lipoplexes [140]. Therefore, this system is in great demand especially for in vitro transfection as it allows an effortless and fast procedure of mRNA encapsulation. For instance, the most known and efficient, commercially available lipoplex reagent is Lipofectamine (Invitrogen) that is considered to be the “gold standard” for in vitro experiments due to its low cytotoxicity and optimal intracellular trafficking [141], [142]. However, as reported by Sultana et al. (2017), in vivo mod-mRNA delivery into mouse heart assisted by Lipofectamine or its in vivo analog Invivofectamine (Invitrogen) turned out to be significantly less effective compared to injection of naked mod-mRNA solved in sucrose-citrate buffer [143]. On the other hand, the fact that the exact composition of commercialized transfection systems is confidential prevents the possibility to optimize the transfection procedure, which might depend on the target cell type. Regarding this issue, a self-made lipoplex formulation could be more beneficial. In general, a lipoplex particle contains a cationic lipid, e.g. dioctadecenyl-trimethylammonium-propane (DOTMA) or dioleoyl-trimethylammonium-propane (DOTAP), which is usually combined with a “helper” lipid dioleoyl-phosphoethanolamine (DOPE) to neutralize the excessive cationic charge. Further, cholesterol is added to increase stability of the complex [144]. Such kind of lipids were used in a study by Kranz et al. (2016), who investigated the impact of net lipoplex charge on efficiency of RNA delivery to lymphoid-resident DCs in vivo by changing the ratio of lipoplexes to the RNA. Surprisingly, strongly positive charge (5:1 ratio) facilitated the uptake of lipoplexed RNA (RNA-LPX) in lungs, whereas negative charge (1.3:2) promoted the internalization in a spleen [145]. Thus, to achieve a therapeutic effect in the target organ or tissue, it might be helpful to carefully define the optimal chemical composition of an RNA-LPX formulation. Noteworthy, such RNA-LPX compound denoted as FixVac (BNT111) is currently under evaluation in a Phase I clinical trial (Lipo-MERIT; NCT02410733) conducted on patients suffering from advanced melanoma. Although this study is still ongoing, the first reports have been recently presented by Loquai et al. (2020) and Sahin et al. (2020) [146], [147]. Briefly, FixVac was indicated as a potent and well-tolerated anticancer vaccine that holds a promising potential for future immunotherapy of tumor-associated indications.
Alternatively, liposome structures are broadly utilized as well. Unlike lipoplexes, cationic liposomes enclose the mRNA within an aquatic core, surrounded by a uni- or multilamellar bilayer of amphiphilic lipids. For a liposome formation, the most frequently applied lipid materials include DOPE in conjunction with cholesterol [148], [149], [150]. For example, Michel et al. (2017) assessed the cytotoxicity, immunogenicity, stability and transfection efficacy of the liposome-loaded mRNA in vitro. This study demonstrated maintained viability of transfected cells, low immune response (measured by interferon expression) and efficient translation of introduced alpha-1-antitrypsin mRNA. Moreover, protein yield in cells transfected with liposome-mRNA was 3 times greater (72 h after transfection) compared to delivery with a standard Lipofectamine agent. The loaded liposomes were storable for up to 80 days at 4 °C, without impairing transfection efficiency, and were proved to be haemocompatible, which is a necessary criterion of International Organization for Standardization (ISO) 10993-4 for future clinical use [151]. A similar formulation comprising dipalmitoyl-glycero-phosphocholine (DPPC), DOPE and cholesterol was recently developed by Dhaliwal et al. (2020), who aimed to deliver liposome-loaded mRNA into a mouse brain via intranasal route. A distinct expression of reporter protein was detected in the brain tissue, the intensity of which was significantly higher than in vehicle-free (naked) transfected group. Furthermore, the biodistribution of in vivo administered mRNA was restricted to the cerebral area pointing out the brain-targeted delivery. Although slight accumulation of reporter protein was detected in liver and lungs, this extent of systemic exposure was considered to be harmless [152]. Hence, these results might place the liposome-mediated complexation of mRNA into a closer perspective for a clinical translation to treat brain-related and other diseases.
Certainly, among all different kinds of lipid-based vectors, the so-called lipid nanoparticles (LNPs) are the most advanced and popular tool for mRNA transport. In fact, the LNP system was implemented in the world’s first approved mRNA-based therapeutics developed by BioNTech/Pfizer (BNT162b2) and Moderna (mRNA-1273) in 2020. Following intramuscular injection, these mRNA vaccines remarkably prevented the SARS-CoV-2 infection with ∼95% efficacy during the COVID-19 pandemic [33], [34]. The basic structure of such LNP includes several lipid components located in the outer shell: (1) Cationic ionizable lipids, e.g. DOTMA, containing an amino head group that provides neutral charge at physiological pH (7.4) and protonation at acidic pH, thereby facilitating initial enclosure of anionic mRNA, cellular uptake and subsequent endosomal escape [153]. (2) Helper lipids, e.g. DOPE, cholesterol or distearoyl-phosphocholine (DSPC), which provide additional stabilization of the LNPs [154]. (3) Polyethylene glycol (PEG)-conjugated lipids, e.g. dimyristoyl-methoxypolyethylene glycol (DMG-PEG), to prolong circulation time and control the nanoparticle size [123], [155]. However, the LNP composition is constantly undergoing further enhancements. For example, a rather “classical” DOTMA has been replaced with novel ionizable lipid materials, such as ALC-0315 lipid (used in BNT162b2), SM-102 (in mRNA-1273) or DLin-MC3-DMA [156], [157], [158]. Besides, the helper lipid cholesterol might experience certain improvements as well in the future. As reported by Herrera et al. (2021), LNPs containing β-sitosterol component (plant cholesterol analog) intensified the endosomal escape of an mod-mRNA at 10-fold rate compared to the standard cholesterol formulation [159], thereby increasing the bioavailability of an mRNA drug. Furthermore, the LNP structure gives a unique opportunity to promote tissue-selective delivery. Firstly, an adjustment of particle size through variation of PEG-lipids content supports targeting the tissue of interest. For example, LNPs with low PEG percentage (0.5%; ∼150 nm in size) achieved highest transfection efficiencies in the eye [155], whereas liver cells preferably internalized smaller particles (1.5% PEG; ∼68 nm) [160]. Secondly, it is possible to incorporate cell-specific antibodies or receptor ligands into the exterior layer of an LNP. Thus, Li et al. (2020) succeed in targeting murine caveolae (part of endothelial membrane in lung capillaries) using antibody-conjugated LNPs. Although these antibody-modified LNPs were applied systemically, a tremendous increase of protein expression (40-fold rate related to unconjugated control) was detected specifically in lungs [161]. Another example of a successful selective delivery was provided by Kim et al. (2021), who accomplished specific transfection of liver sinusoidal endothelial cells by interlinking mannose molecules with PEG lipids [160].
The peculiar advantage of an LNP system lays in almost unlimited latitude of modification possibilities. At the same time, the complexity of these particles makes it difficult to predict the actual molecular organization of its ingredients. Whereas some groups believe that LNPs build a lipid bilayer [162], thereby PEG-lipids reside at the outside and DSPCs are turned towards the inside of the particle [163], other scientists tend to consider a monolayer arrangement of outward lipids [158], [164]. The latter point of view assumes that the DSPC layer diverges upon attraction to the mRNA molecules, which leads to a formation of multiple aqueous “bleds” containing mRNA within the core of an LNP [165]. Yet, the precise location of these mRNA vesicles remains elusive [166]. Therefore, despite the already proven efficacy and safety of LNP technology at least in the field of vaccine development, it might be beneficial to further elucidate the nature of LNP-mRNA arrangement. Such insights into the chemical organization of LNP-mRNA therapeutics could possibly promote the required optimization of the technology, which would allow its application in other branches of medicine.
3.3. Peptide-based delivery
Positively charged peptides are eminently suitable for complexation of anionic mRNA. The most established peptide-based vectors are the cell-penetrating peptides (CPP), which gain access to the cell via endocytosis-mediated translocation. In 2017, Udhayakumar et al. utilized synthetic peptides RALA (rich on arginine) to transfect DCs with eGFP mod-mRNA, thereby ∼35% of transfected cells were measurably eGFP-positive [167]. In the following animal experiments, mice were immunized (i.d.) with ovalbumin mod-mRNA/RALA formulation. A significant expansion of CD8+ T-cells with strong ovalbumin-specific cytolytic activity was observed indicating a successful RALA-mediated vaccination, which even outperformed a conventional DOPE/DOTAP lypoplex vector.
As an alternative to arginine-rich motifs, histidine-rich CPPs for mRNA transport were described in the literature as well [168]. Indeed, Coolen et al. (2019) identified LAH4-L1 peptide as a superior CPP to RALA in terms of mRNA vaccine delivery to DCs in vitro [169]. According to the authors, the LAH4-L1 vector was more potent to stimulate innate immune responses than RALA by acting through an activation of pattern recognition receptors (PRRs). Moreover, humoral and adaptive immune responses were intensified as well.
Furthermore, a PepFect 14 (PF14) peptide was analyzed with regard to mRNA delivery by Van den Brand et al. (2019), who explored the potency of mRNA/PF14 complex to target a xenograft of an ovarian cancer in mice [170]. Following an intraperitoneal administration, a translation of introduced mRNA coding for reporter protein was found specifically in the tumor microenvironment including tumor cells, fibroblasts and immune cells. No expression was detected outside the abdominal area. Noteworthy, application of naked mRNA or in complex with Lipofectamine MessengerMax did not lead to observable protein translation. Potentially, PF14-based carrier could be suitable for clinical therapy of ovarian cancer. However, a proof-of-concept study employing antigen-coding mRNA instead of reporter protein might be required to ensure the feasibility of PF14 system.
Although CPPs represent an interesting and effective delivery approach, their clinical application is hindered by a lack of toxicity studies (e.g. side effects in liver or kidney) and high production costs, which might be associated with vast expenses in future therapies. However, the therapy costs could be justified in case of short-termed treatments, thereby anticancer vaccination would be the main application area for a CPP-mediated mRNA delivery [171]. Lastly, as the reader might appreciate, we refer to several up-to-date comprehensive reviews on modern peptide-based delivery tools for further reading [172], [173], [174].
3.4. Polymer-based systems
Apart from lipid nanomaterials, the delivery of mRNA can be guaranteed by implementation of polymeric substances, thereof most systems are based on either linear, branched or dendrimeric polyethylenimine (PEI). The multiple amine groups in PEI units confer a positive charge to the molecule, which makes them suitable for complexation of nucleic acids. Several studies successfully applied PEI to encapsulate and deliver DNA [175], [176], [177], siRNA [178] and self-replicating RNA in vitro [179]. However, nanoparticles based solely on PEI were associated with considerable cytotoxicity values [180]. To provide greater safety, Debus et al. (2010) engineered PEI/PEG co-polymer that not only was able to eliminate the toxicity issues, but also led to a significant improvement of transfection efficiency in comparison to single-component particles [180]. Hence, a combinatory approach for polymer-based nanocarrier design has been preferably used in the following studies.
For example, Sharifnia et al. (2019) decided to use a polylactide-co-glycolide (PLGA) to deliver GFP mod-mRNA into human monocyte-derived DCs in vitro. PLGA is an FDA approved, non-toxic and biodegradable polymer that is metabolized via Krebs cycle, but its interaction with anionic sites is limited. To overcome this drawback, PEI was chosen to drive the net surface charge into a positive range [181]. The resulting PLGA/PEI system was sufficient to transfect about 70% of analyzed cells, thereby keeping low cytotoxicity profile. As announced by the authors, a subsequent in vivo study should further evaluate the safety of PLGA/PEI vehicle.
Furthermore, the encapsulation of mRNA can be performed using dendrimers – a highly branched macromolecular polymeric system that usually employs cationic polyamidoamine (PAMAM) [182]. In 2016, Chahal et al. suggested modified PAMAM nanoparticles as a vehicle for an mRNA-based immunization. In this work, a complete protection against several lethal infections (Ebola virus, H1N1 influenza and Toxoplasma gondii) was achieved in vivo following single-dose intramuscular immunization of mice [183]. Although dendrimers appear as a valuable approach, their translation into the clinical sector is still limited. Pursuant to the current state of the art, ensuring the sterility of dendrimeric formulations is rather complicated, which causes serious safety concerns for patients [184]. Thus, in order to promote the clinical entry, further research should focus on additional improvement of this technology.
Yet, mRNA delivery based on mere polymeric systems is not sufficient to leave the area of preclinical research. On this account, the prevailing projects have been focused on development of hybrid technology that implements the use of polymers with non-polymeric compounds. We provide an up-to-date insight to these novel concepts in the next section.
3.5. Hybrid nanoparticles
New findings and progressive exploration in the field of nanomaterials constantly inspire scientist with new ideas for carrier engineering. To give an example, Son et al. (2020) pursued a vaccination strategy using sugar-nanocapsules built up of mannan polysaccharides that mimic the microbial cell wall [185]. In this nanostructure, the mRNA resides underneath the mannan shell and is captured by PEI-coated silica core. Additionally, pathogen associated molecular patterns (PAMPs) were anchored on the surface, thereby providing the capsule with typical antigens for PRRs located within the membrane of DCs. A subcutaneous application of the nanocapsules loaded with ovalbumin mRNA led to strong antitumor immune responses in mice suffering from melanoma. Hence, the coworkers emphasized the great potential of this carrier system for immunotherapeutic applications.
Another fascinating in vitro project was presented by Huang et al. (2020), who designed a cross-linked nanogel with imbedded mRNA [186]. Here, the polycaprolactone was grafted with poly(T)20-segments (T20-g-PCL) to capture the mRNA poly(A)-tail. Once the corresponding site is hybridized, DNA linkers are added to the mRNA/ PCL complexes leading to the formation of a nanogel. Lastly, the mRNA-nanogel was coated with PEI to switch the zeta potential from negative (−3.0 mV) to positive values (+3.2 mV). The demonstrated transfection results were comparable to the performance of the commercial Lipofectamine. Although the fraction of transfected cells remained lower (37% by nanogel vs. 44% by Lipo), further improvement of this novel technology is likely to occur soon. As proposed by the authors, the nanogel system might be particularly useful for CRISPR/Cas9-mediated genome editing (see Section 4.) to prevent abnormal disease-related protein expression.
Furthermore, the latest invention of Xiong et al. (2020) employs nanoparticles consisting of both dendrimers and lipids (DLNPs) [187]. In this complex, lipid components involve DOPE, cholesterol and newly developed PEGylated BODIPY dyes (PBD) lipids. In contrast to DMG-PEG lipids (commonly used for LNP formulations), the PBD lipids allow non-invasive near-infrared imaging in vitro and in vivo due to their photoactive core. Thus, the DLNPs provide not only a vehicle for mRNA delivery, but also an opportunity to track the accumulation of the particles in cells or tissues. Moreover, the animal experiments showed significantly stronger protein expression (5- to 35-fold) when the DLNP carrier included PBD lipids instead of DMG-PEG lipids. In addition, the mRNA/DLNP complex injected intravenously into mice with subcutaneous breast cancer xenografts demonstrated high uptake in tumor tissue and liver. Therefore, this technology could be possibly beneficial for therapeutic and diagnostic applications to treat particularly hepatic pathologies.
Recently, an investigation by Liu et al. (2021) implemented a highly exciting and innovative delivery tool based on graphene quantum dots (GQD) for mRNA transport [188]. The major advantage of GQDs is their exceptional stability, tunable surface structure and responsiveness to physical stimuli (magnetic fields, ultrasound, light). Perspectively, these features might enable a precise tissue targeting, which poses the GQDs as a highly advanced system for therapeutic intentions. In this study, the GQDs were additionally functionalized with PEI (FGQDs) in order to enhance the mRNA binding. Unfortunately, despite this necessary modification, the FGQDs still lack general in vitro optimization owing to the general novelty of this platform. Here, the most eminent downsides that require special attention are a moderate cytotoxicity and low transfection efficiency (∼25%).
Taking together, many interesting preparation methods have been developed in order to improve the previously described carrier systems. This led to a rise of various hybrid solutions for a vehicle design that all hold a great promise for diverse branches of medicine. Yet, most of these novel nanotools are still under development and require extensive research to be allowed for clinical use.
3.6. Exosomes
Beside in vitro produced lipid-based vesicles, mRNA can also be found in exosomes produced by eukaryotic cells, thereby playing a physiological role in cell–cell communication and protein synthesis [189], [190]. Considering that exosomes represent a physiological carrier system for mRNA transfer between cells and tissue, exosomes have emerged a promising delivery tool for therapeutic mRNAs [191], [192]. Moreover, exosomes have been found to possess optimal permeation to physiological barriers, low immunogenicity and favorable pharmacokinetic properties [193], [194].
To generate exosomes for a desired therapeutic application, the specific mRNA needs to be incorporated into the vesicle. This requires transfection of exosome producer cells, followed by purification and concentration of released exosomes [194]. However, large scale production, which is crucial for clinical trials, is associated with high costs and low efficiency [191]. Recently, Kojima and colleagues presented an approach for customized production of mRNA-containing exosomes using genetically modified producer cells that demonstrated improved exosome production and mRNA packaging as well as enhanced delivery to the cytosol [192]. Likewise, an electrical nanoporation technique was applied to increase the yield for producing large quantities of exosomes containing selected mRNA [195].
While a high production efficiency is crucial for the clinical translation, adequate targeting remains another challenge for exosome-based mRNA therapies. The target distribution of exosomes is closely related to the surface-derived molecules, provided by the donor cell [196]. Hence, guided distribution can be achieved by tailored exosomes containing specific surface molecules that mediate anchoring to the target cell. For example, incorporation of a rabies virus glycoprotein into exosomes allowed for successful targeting of mice brain to deliver therapeutic molecules for the treatment of cerebral ischemia or Parkinson disease [192], [197].
Despite these recent advances for exosomal mRNA delivery, some obstacles remain to be solved on the way to clinical translation, including optimized drug loading capacity, sufficient yield and purification efficiency as well as the development of appropriate targeting strategies.
4. mRNA-assisted CRISPR/Cas9 gene editing
The CRISPR (clustered, regularly interspaced, short palindromic repeats)/CRISPR-associated protein 9 (Cas9) system is a highly precise, easy to design, third generation gene-editing platform that was awarded with a Nobel Prize in Chemistry in 2020. The excellent functionality of CRISPR/Cas9 technology is determined by two components – the endonuclease enzyme Cas9 representing molecular DNA scissors and the single guide RNA (sgRNA) that defines the target gene sequence [198]. Today, besides the basic cutting function, the Cas9 protein can also act as a gene repressor (CRISPRi), also known as a catalytically deactivated Cas9 (dCas9). On the other hand, dCas9 can adopt the role of a gene activator (CRISPRa), if its acting sites are fused with transcription factor-derived activating domains [199]. These meaningful refinements extended the original range of CRISPR/Cas9 applicability, thereby allowing for a precise gene regulation apart from classical gene knock-out. However, the clinical use of CRISPR/Cas9 in patients is limited by the drawbacks of its delivery medium. The various aspects of existing viral and non-viral transport vehicles were systematically reviewed elsewhere [200]. Here, we would like to exclusively point out the novel virus-free Cas9 delivery method using IVT mRNA, which has considerable advantages (e.g. low risks of mutagenesis, low immunoreactivity and transient activity) over competitor virus- or protein-based formulations and, therefore, great chances for a clinical translation [201].
The first successful attempt to co-deliver Cas9-coding mRNA along with corresponding sgRNA was reported by Miller et al. in 2017 [202], who developed a lipid-based vehicle consisting of zwitterionic amino lipids with a great loading capacity (Cas9 mRNA is approx. 4500 nt long). The proof of concept was demonstrated by an induced expression of tdTomato in the organs (liver, kidneys, lungs) of transgenic mouse resulting from a Cas9/sgLoxP-mediated removal of the loxP-flanked stop cassette. Later, Cheng et al. (2020) presented a valuable tissue-specific Cas9/sgRNA delivery concept termed Selective ORgan Targeting (SORT) [203]. In this study, the authors developed variously composed LNPs containing a SORT molecule – an essential lipid component that determines the targeted uptake in lungs, liver or spleen. To indicate the high targeted therapeutic potency of this system, the researches performed a CRISPR/Cas9-induced knock-out of a PCSK9 (proprotein convertase subtilisin/kexin type 9) gene, whose abnormally high expression is associated with increased risks of familial hypercholesterolemia and atherosclerotic cardiovascular disease [204]. Following a triple intravenous administration of liver SORT LNPs carrying both Cas9 mRNA and sgPCSK9 into mice, a 60% Indel at PCSK9 locus was detected by TIDE analysis (tracking of indels by decomposition) accompanied by a complete abolishment of PCSK9 protein in liver and serum. A similar approach to treat hypercholesterolemia with mRNA-based CRISPR/Cas9 machinery delivered by LNPs was introduced by Qiu et al. (2021), who targeted the Angptl3 (angiopoietin-like 3) gene with Cas9 mRNA/sgAngptl3 in order to reduce the plasma lipoprotein levels [205]. A single injection of this formulation led to 38.5% genomic editing efficiency of Angptl3 locus and a decreased serum ANGPTL3 levels by 65.2%. The observed therapeutic effect persisted for at least 100 days, thereby neither off-target occasions nor organ toxicity could be identified.
Apparently, CRISPR/Cas9 can be of great use to treat inherited genetic disorders especially in cases of a high disease severity with extremely low therapeutic options. Concerning this matter, Hsu et al. (2019) elaborated a substantial report on the utility of CRISPR/Cas9 tool explicitly in the area of regenerative medicine, thereby exemplifying its potential for function restoration in a morbid muscle, liver, eye, brain, bone and cartilage tissue using either a stem cell-based or a cell-free approach [206]. Yet, to our knowledge, no reports on utilization of mRNA-based CRISPR/Cas9 particularly for regenerative directions are existing except for a recently published pilot study by Abbasi et al. (2021) [207]. Here, the Cas9 mRNA co-encapsulated with sgRNA in polyplex micelles showed a great ability to target parenchymal cells (neurons, microglia, astrocytes) in the brain of Ai9 (Rosa26-floxed stop tdTomato) mice. The authors strongly underlined the potential of their newly established mRNA-based CRISPR/Cas9 system to treat genetically conditioned neurodegenerative diseases (Alzheimer, Huntington’s disease or fragile X syndrome) in the future. Therefore, it is justified to assume a greater role of mRNA for CRISPR/Cas9 gene editing in regenerative field in the near future.
5. mRNA in cellular (re-)programming – state of the art
5.1. Generation of induced pluripotent stem cells
In the last decades, stem cell-based approaches have emerged as a novel concept to replace or repair dysfunctional organs, tissues or cells. A broad spectrum of human pathologies could be treated by means of stem cells: neurodegenerative and ocular diseases, implications of diabetes mellitus, dental tissue degradation, cardiovascular or severe skin disorders [208], [209], [210]. For this wide field of applications, the central role is held by pluripotent stem cells, which source comprised exclusively embryonic stem cells (ESCs) until 2006. This year was designated by a groundbreaking scientific event – the discovery of a cocktail of transcription factors that enable the reprogramming of adult somatic cells into a pluripotent state, the so-called Yamanaka factors (ct3/4, Sox2, Klf4 and c-Myc) [211], [212]. The resulting induced pluripotent stem cells (iPSCs) can give rise to any cell type while offering the opportunity of patient-specific derivation. By this means, manipulated iPSC derivatives could serve as personalized therapeutic cells [213]. However, iPSC generation usually involves viral vectors as vehicle for reprogramming factors. Unfortunately, viral components raise risk of tumorigenicity, thereby restricting clinical approval of these cells [214].
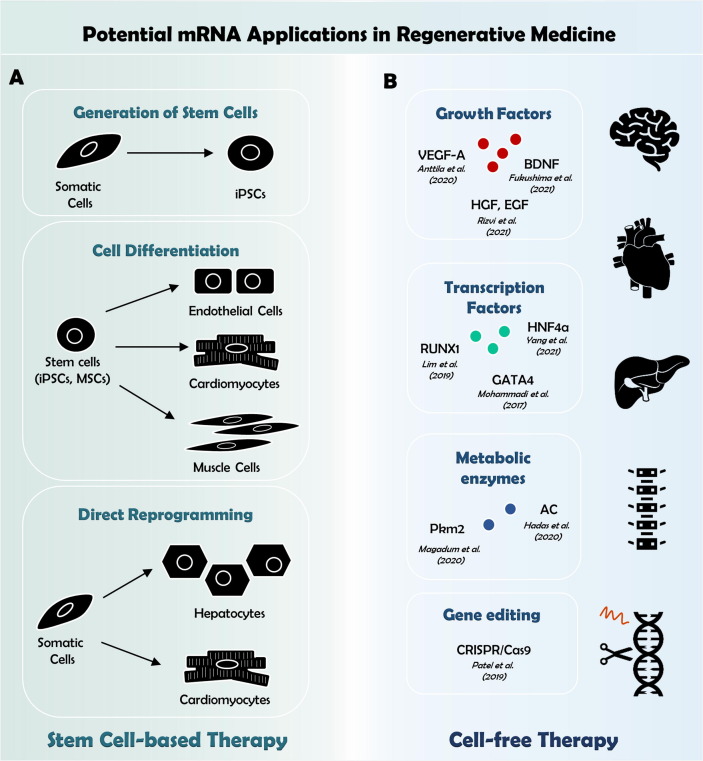
Although emerging integration-free approaches, such as Sendai virus [215], episomal [216] or plasmid DNA [217] and cell permeant proteins [218] minimized risks of genome insertion, cell reprogramming using these tools appeared insufficient [219]. Here, mRNA offers an exceptional benefit of transient and efficient protein expression with tight control over dosing, stoichiometry and time course while posing no risks of genome intrusion. Hence, mRNA-based approaches are becoming progressively more prevalent to achieve “footprint-free” reprogramming in the field of developmental and cell-based regenerative biology (Fig. 4 A).
Fig. 4.
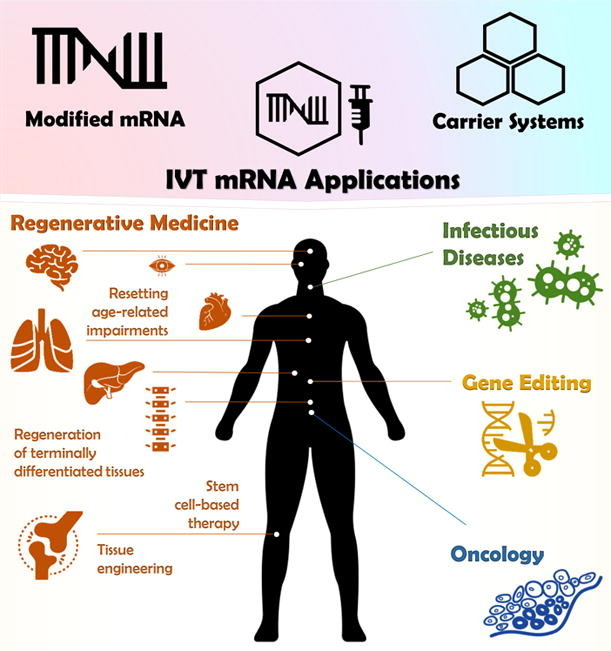
Potential mRNA applications in regenerative medicine. mRNAs can be applied to generate cells in vitro that are supposed to be transplanted for tissue repair following injury. In this regard, mRNA-based techniques are (A) either utilized to reprogram somatic cells into iPSCs, which can be further differentiated into any desired cell type. On the other hand, mRNA, transferred into stem cells or somatic cells, enable guided differentiation to obtain cells suitable for cell replacement therapy. For cell free strategies (B), mRNA technology enables to deliver signaling molecules into the tissue of interest. mRNAs encoding growth or transcription factors have been successfully applied in vivo to promote tissue regeneration [162], [246], [248], [251], [252], [255], [256], [264].
In theory, every desired mRNA can be produced in vitro for the induction of target protein biosynthesis in each cell type. Practically, the scientific community encountered major hurdles early on as synthetic mRNAs provoked antiviral defense pathways, which in turn hampered efficient protein translation. A cascade of cytotoxic and cytostatic responses interfered with reprogramming attempts. Although double-stranded RNA was thought to be the major agonist, single-stranded RNA transcripts triggered the innate immune system as well and type I interferons even sensitized cells for upcoming mRNA transfections [220].
Yakubov et al. (2010) were the first to generate iPSCs using in vitro generated mRNAs for the four Yamanaka factors [221]. However, despite successful transfection rates of at least 70% of human fibroblasts 24 h after the first transfection, conversion efficiency into iPSCs was low due to the aforementioned limitations. In the same year, Warren et al. synthesized mRNAs of the transcription factors Oct4, Sox2, Klf4, cMyc and Lin28 containing the modified nucleosides 5-methylcytidine and pseudouridine (Ψ) for transfection of fibroblasts [222]. Moreover, they employed the interferon inhibitor B18R as media additive to further bypass the innate anti-viral immune response. By these means, conversion efficiency peaked at 4.4%, thus being substantially higher than for virus-based approaches. Going even further, they afterwards differentiated murine C3H10T1/2 cells into the myogenic lineage using MyoD mRNA with modified ribonucleotides for increased stability.
Later this approach was adjusted to convert blood-derived endothelial progenitor cells into iPSCs. Refraining from mod-mRNAs, Poleganov et al. (2015) employed mRNAs for reprogramming (Oct4, Sox2, Klf4, cMyc, Nanog, and Lin28) in combination with mRNAs that were supposed to facilitate immune evasion (E3, K3, and B18R) [223]. When adding mature miRNAs302a–d and 367 to enhance the reprogramming capability, they reported successful reprogramming within ten days, thereby demonstrating that RNA cocktails containing non-modified mRNAs could outperform mod-mRNA given the right circumstances.
Although much emphasis is put on the reduction of immunogenicity for mRNA therapeutics, it soon became apparent that some inflammatory signaling is required for nuclear reprogramming by exogenous mRNA [224]. Activation of the innate immunity alters the expression of epigenetic modifiers such as histone acetyltransferases and increases the chromatin accessibility thereby facilitating the access to consensus sequences for transcriptional activators. A further contribution in this process is attributed to reactive oxygen and nitrogen species [225].
5.2. Reprogramming for regenerative medicine and cell therapies
After a rapid progression of protocols, iPSCs generated by mRNA based reprogramming could give rise to therapeutically relevant cell types. For example, cardiomyocytes derived from human iPSCs demonstrated typical molecular, morphological and functional features, thereby implicating the option to generate cells from patient-specific iPSCs in large scale that are only of limited regenerative capacity in vivo. However, optimal cell delivery methods are still to be established and improving the retention of injected cells at sites of injury will be crucial for effective cardiac regenerative medicine [226]. Other cells with severely limited regenerative capacity are neurons. Employing IVT mRNAs coding Atoh1 and Ngn2, iPSCs could be differentiated into highly pure midbrain dopaminergic neurons within only five days [227]. This method might be implemented in patient specific disease modeling or could constitute the basis for cell replacement strategies for neuronal regeneration. Due to their limited regenerative capacity, both cardiac and neuronal tissues are particularly susceptible for the degenerative effects of aging. Intriguingly, rather than replacing cells recent approaches for regenerative medicine and cell therapies were expanded more and more towards rejuvenating them.
Apart from the generation of iPSCs, reprogramming using Yamanaka factors was demonstrated to ameliorate structural and functional hallmarks of aging. By resetting DNA methylation patterns and transcriptomes to youthful states without resetting cellular identity, partial reprogramming was show to increase lifespan in a mouse model of premature aging [228] while it promoted axon regeneration and reversed vision loss in another aged mice model [229]. Employing IVT mRNAs for a transient expression of nuclear reprogramming factors, similar age ameliorating effects could be transferred to human cells. Transfection with mRNAs expressing OCT4, SOX2, KLF4, c-MYC, LIN28 and NANOG for four consecutive days was reported to reset the epigenetic clock, reduce the inflammatory status in chondrocytes, and restore regenerative potential in aged, human muscle stem cells [230]. Another approach to rejuvenate cellular phenotypes was based on human telomerase (hTERT), an enzyme counteracting telomere attrition underlying cellular senescence. In fibroblasts derived from Hutchinson-Gilford progeria syndrome patients, a disorder characterized by severely accelerated aging, treatment with IVT hTERT mRNA increased telomere length, proliferative capacity and cellular lifespan [231]. Both approaches are sufficient to reset certain age-related impairments and thus hold promise as therapeutic for age-related diseases.
A number of studies highlighted the cell reprogramming potential of mRNA including not only the direct conversion of fibroblasts to pluripotent stem cells [221], [222] but also the differentiation into other cell types such as endothelial cells [232], myoblasts [233] and cardiomyocytes [234]. Moreover, direct transdifferentiation of human pancreatic duct-derived cells into insulin producing β-cells could be accomplished using a single synthetic mod-mRNA encoding for the pancreatic transcription factor MafA [235]. Transfected cells were able to secrete insulin and reduce the blood glucose level in a diabetic mice model one week after transplantation. As the transfected cells replace lost β-cells, this approach describes not only a protein replacement therapy but can be considered a tissue regeneration therapy.
Alternatively, cellular programming could be supported using mRNA-based CRISPR/Cas9 system. In 2020, Chen et al. transduced differentiating human mesenchymal stem cells (hMSCs) with doxycycline-inducible CRISPRi or CRISPRa lentivirus repressing or activating the target alkaline phosphatase (ALP) gene, respectively. The researchers demonstrated a great efficacy of their approach to regulate gene transcription (60–90% ALP expression inhibition in hMSC-CRISPRi; 20-fold expression increment in hMSC-CRISPRa) with no off-target occasions as the differentiation outcome was not affected by a transduction in the manipulated cell line vs. control group [236]. Thus, the described technique could be a valuable tool to direct the differentiation process towards the desired cell type. In case when the target gene requires more stable intervention, CRISPR system might even be a wiser choice in comparison to transiently active transcription factors encoded directly by IVT mRNA. On the other hand, the delivery of Cas9 machinery should be further refined with viral-free vehicles (e.g. LNPs) to ensure firstly the safety and secondly the conformity with IVT mRNA transport medium. Although such comparative analysis has not been carried out yet, it would be intriguing to examine these questions in the future.
One focus of heart regeneration therapy lies in the replacement of contractile cells. The myogenic differentiation factor MyoD1 acts as a master regulator for muscle cell fate. Myoblasts can differentiate into cardiomycytes and, hence, are highly relevant for cardiac regeneration. Hausburg et al. (2015) could achieve direct cell conversion of murine C3H10T1/2 into myoblast-like cells in three daily transfections with modified MyoD mRNA in B18R supplemented medium [233]. Adapting the protocol from Hausburg et al., Preskey et al. (2016) demonstrated the successful transdifferentiation of human foreskin fibroblasts into myoblast‐like cells [237]. The generation of myoblast‐like cells was accomplished in seven days with only four daily transfections with IVT MyoD1 mRNA. Conversion efficiency increased with higher amounts of mRNA but did not exceed 0.4%. Similarly, our group applied mRNAs coding for the transcription factors Tbx3, Shox2 and Tbx18 to induce reprogramming of iPSC-derived cardiomyocytes towards a pacemaker-like phenotype. Using microarray data, we detected changes in gene expression in transfected cells, suggesting the possibility to use mRNA for the subtype specific generation of cardiac cells (unpublished data). While levels of success are currently limited, further optimization of IVT mRNAs and carrier systems will unequivocally improve translational efficiency and thus conversion efficiency in the future.
In an attempt to directly generate cardiomyocytes, Lee et al. (2015) used a construct of heart-targeting peptide (CRPPR-R9) and lipofectamine to repeatedly transfect murine cardiac fibroblasts with mRNAs encoding Gata4, Mef2C and Tbx5 [234]. High expression levels of α-actin and cardiomyocyte specific marker genes such as Actc1, Actn2, Gja1, Hand2 and Tnnt2 after two weeks of transfection indicated a successful transdifferentiation. Interestingly, they reported an influence of reprogramming factor stoichiometry on the marker gene expression such as higher expression of Gja1 and Hand2 upon excess Tbx5 (1:1:3) and higher expression of Nppa and Tnnt2 upon excess Gata4 (3:1:1).
Another attempt in 2019 found that MSCs derived from adipose tissue can partly be directed towards the cardiac lineage using mod-mRNAs for Gata4, Mef2C, Tbx5 and Mesp1, while cells isolated from bone marrow, and dental follicle demonstrated only weak reprogramming efficiency [238]. Although full maturity of cardiomyocytes could not be reached in the study, the data suggest that adult MSCs can acquire a cardiac-like phenotype and might serve as source for cell replacement strategies in heart regeneration.
MSCs are also a main target in mRNA-based bone regeneration therapies, which often aim to recruit these cells to the sites of bone defect by transplantation of scaffolds loaded with osteogenic mRNAs. Scaffolds releasing mRNA encoding human BMP-2 have been demonstrated to stimulate bone regeneration in femur [239] and calvarial bone [240] defect models. A comparative study found that complexes of PEI and BMP-9 mRNA performed even better in some aspects of bone regeneration such as enhanced osteogenic differentiation of BMSCs and increased density of the regenerated bone [241]. Still, due to its robust performance BMP-2 is the most widely used therapeutic in mRNA-based bone regeneration therapy and recent research concentrates more on the delivery systems than on the exploration of other osteogenic compounds [242], [243].
IVT mRNAs also conquered many other fields of application. For example, they were early recognized as tool for immunotherapy of cancer. In 2009, RNAs encoding T cell receptors either specific against ErbB2 or the carcinoembryonic antigen (CEA) were utilized to render T cells transiently cytotoxic for cancer cells expressing the respective antigens. CD4+ as well as CD8+ T cells electroporated with the immunoreceptor-encoding mRNAs secreted cytokines upon antigen contact and in case of CD8+ T cells specifically lysed tumor cells. This activity was maximal at day 2 and persisted no longer than nine days, thereby avoiding the persistence of unintended autoaggression [244]. Recently, another group reprogrammed tumor-associated macrophages that typically express an M2 phenotype and exert immunosuppressive effects toward an M1 phenotype using a nanocarrier for IVT mRNA delivery [245]. Nanoparticles formulated with mRNAs encoding interferon regulatory factor 5 in combination with its activating kinase IKKβ were used in models of ovarian cancer, melanoma, and glioblastoma to transiently trigger anti-tumor immunity in macrophages without eliciting a systemic inflammation.
6. Direct cell-free regenerative approaches using mRNA
Another possibility of tissue restoration is given by a precise regulation of signaling cascades, gene transcription or even metabolic pathways (Fig. 4B). Regarding this, the most favorable protein candidates are the descendants of a growth factor family. In 2021, Rizvi et al. took the advantage of hepatocyte-growth-factor (HGF) and epidermal-growth-factor (EGF) to repair acute liver damage after induced injury in vivo [246]. The corresponding mod-mRNAs were loaded in LNP and intravenously administered to liver-injured mice. A single injection of HGF/EGF mod-mRNA was sufficient to repeal the functional and morphological morbid condition two days post injection, thereby the ALT (alanine aminotransferase; a biomarker of hepatic inflammation) levels dropped back to baseline and incipient steatosis was revoked. Recently, Yang et al. (2021) aimed to attenuate the fibrotic spread in a diseased mice liver caused by cholestasis and an exposure to hepatotoxins. In this study, LNP-encapsulated IVT mRNA encoding the transcription factor HNF4a (hepatocyte nuclear factor alpha) has demonstrated very promising therapeutic properties and significantly diminished the fibrogenesis [247]. Taking together, these studies obtained the first solid, preclinical evidence on considerable therapeutic effects of IVT mRNA to treat diverse liver injuries.
In contrast to hepatic applications, mod-mRNA coding for VEGF-A is already undergoing early phase clinical trials in: (1) type 2 diabetic patients receiving intradermal VEGF-A mod-mRNA injections to induce local angiogenesis (NCT02935712) [134]; (2) patients with impaired systolic function that are epicardially administered with VEGF-A mod-mRNA during coronary artery bypass grafting surgery to restore cardiac activity (NCT03370887) [248]. While the latter study is still under investigation, the results from the first trial have already been published and demonstrated great tolerability and safety of VEGF-A mod-mRNA at both local and systemic level [134]. However, further clinical examination must ensure the efficiency of VEGF-A mod-mRNA treatment, which might be challenging. The previous intents to improve ischemic cardiac outcome with VEGF-A have failed to gain a therapeutic significance [249]. On the other hand, the introduction of VEGF-A in these first-generation trials has been mediated trough a DNA platform. Concerning this matter, Zangi et al. (2013) assessed the impact of intramyocardial VEGF-A administration encoded either by DNA or mod-mRNA in a mouse myocardial infarction (MI) model [250]. When comparing the outcomes, the group evidenced that VEGF-A DNA-treated hearts displayed obvious edema and increased mortality (even in comparison to vehicle controls), although both treatments reduced infarct size and apoptotic cell frequency while increasing capillary density. Hence, this clear benefit of mod-mRNA might be the determining factor for VEGF-A to revolutionize the cardiac domain in the regenerative medicine. Noteworthy, an alternative novel concept for mRNA-based MI treatment has been presented by Magadum et al. in 2020 [251], who explored the role of glycolytic pyruvate kinase muscle isozyme 2 (Pkm2) in the mouse heart. As proposed by the authors, Pkm2 positively affects the proliferation of cardiomyocytes and counteracts oxidative stress damage supposedly by regulation of β-catenin (non-enzymatic) and enzymatic anabolic pathway (via glucose-6-phosphat-dehydrogenase) post ischemic injury, respectively. In addition, Magadum et al. provided the evidence that Pkm2 is a convenient contender with a strong therapeutic potential for MI treatment, which bears an attractive clinical prospect. However, Pkm2 is not the only metabolite capable of cardio-protection post MI. Hadas et al. (2020) hypothesized that a precise shift in sphingolipid metabolism could improve the MI-induced cardiac complications. Here, the group took the advantage of an acid ceramidase (AC) – a hydrolytic enzyme that produces sphingosines, which in turn are phosphorylated to the sphingosine-1-phosphate (S1P) [252]. S1P is believed to possess anti-apoptotic, cyto-protective properties that might be highly beneficial for a prevention of MI-conditioned heart failure [253], [254]. Hence, the naked mod-mRNA (in citrate buffer) encoding AC was injected directly into infarcted mice hearts. Following the treatment, overall cell death post infarction has decreased, thereby leading to improved cardiac function as well as extended long-term survival in mice.
There are plenty of other fascinating examples for utility of the mRNA platform in the regenerative field: Lin et al. (2019) aimed to treat the degenerative occurrence in the intervertebral disk with direct disk injections of mod-mRNA coding for a transcription factor RUNX1 (runt-related transcription factor-1) [255]; Fukushima et al. (2021) sought to preserve the brain tissue from neuronal death caused by ischemic attack through an intraventricular administration of brain-derived neurotrophic factor (BDNF) mod-mRNA [256]; Patel et al. (2019) presented their first successful attempt to deliver a mod-mRNA coding for a reporter protein to the retinal pigmented epithelium, thereby highlighting a prospect for treatment of retinal degenerative diseases [257]. Yet, an evident limitation that these studies have in common is the extremely elaborate technique required for drug administration. Moreover, local injections into the brain, vertebral disk or eye are highly invasive, which further impedes the potential clinical translation. Lastly, the gene regulation (by transcription factors) or gene editing (e.g. with Cas9 mRNA [205]) is still associated with certain safety concerns due to not fully predictable outcomes of genetic intervention. Hence, future research should focus on finding an appropriate method to solve the depicted problems.
7. Towards clinical approval in regenerative medicine: challenges and perspectives
The fundamental principles underlying regenerative medicine rely heavily on tissue repair and engineering, thereby the most advanced approaches include targeted cell programming. Here, stem cell-based tissue replacement can imply the mRNA to direct the cell differentiation towards the desired cell type (see section 4), which would subsequently undergo either allogeneic or autologous transplantation to rehabilitate damaged regions in a patient. However, stem cells and their derivatives still face difficult hurdles for their clinical approval. At this point, we would like to emphasize the recent thorough articles that carefully review the reasoning behind the problematic clinical translation of manipulated stem cells for regenerative purposes [208], [258], [259], [260]. In case of iPSCs, potential complications are attributed to their pluripotent properties that could lead to tumor formation. Additionally, highly promising results of preclinical studies could not be comparatively reproduced in patients, which attracted even more adverse criticism. Lastly, meeting all requirements of GMP (good manufacturing practice), strict controlling during the production process as well as product up-scaling for clinical use can be extremely difficult to manage. Notwithstanding these discouraging challenges, advancing innovations in technology and research constantly present novel elegant solutions to ensure highly standardized manufacturing, warranting safety and efficacy of the stem cell-based therapies. Therefore, we expect that this approach could enter the clinical sector in the foreseeable future.
Concerning the cell-free regenerative approaches, the mRNA-based therapy holds a great potential particularly in the area of degenerative hepatic pathologies as it was demonstrated that the LNP-loaded mod-mRNA is predominantly metabolized in the liver when applied intravenously [261], [262]. Thus, targeted liver repair might apparently profit from such an active LNP uptake by hepatocytes, especially because additional elaborate LNP design for tissue-specific delivery can be omitted. Considering the current success of already approved COVID-19 mRNA vaccines formulated in LNPs, we could soon expect the mRNA to obtain a clinical permission for safety and efficacy evaluation in patients with hepatic deficiency. Unfortunately, although the regenerative properties of the IVT mRNA are also highly potent for organs such as bone, eye or brain, application routes for these tissues represent the major limitation. It is therefore crucial to develop tissue-specific delivery systems, which could transport the therapeutic mRNA directly to the organ of interest following an intravenous administration. Until then, the IVT mRNA might prevail in hepatic or cardiac regenerative applications.
8. Conclusions
The very first use of mRNA to accomplish a protein production in vivo has been recorded nearly 30 years ago [21]. However, it took 18 years to translate the mRNA-based medicine from preclinical stage to the first clinical trials [26] and a world-wide pandemic to obtain a global permit for an mRNA-based therapeutic (an antiviral vaccine) for the first time in history [33], [34]. This long-term development of mRNA platform is mainly attributed to the complex nature of this molecule. Fortunately, extensive explorations of native mRNA structure as well as enormous technical advances have dramatically enhanced the mRNA utility. First, a rational mRNA design now secures a low immunogenicity, sufficient protein translation rate, extended turnover and great molecular stability. Second, numerous delivery approaches allow for a protected mRNA transport in vivo, tissue-specific targeting, facilitated cellular uptake and endosomal escape. These significant improvements led to the emergence of various mRNA-based applications nearly in all branches of medicine including cancer research, infectious diseases, protein dysfunctions and gene editing. Although the most prominent achievements so far have been reached in the development of vaccines, the mRNA platform is also strongly attractive for regenerative approaches that rely on targeted cell or gene manipulation. For instance, a great promise for mRNA technology is given by treatment of degenerative hepatic diseases and myocardial infarction. Here, the therapeutic effect rests upon cell-operated secretion of signaling molecules, e.g. growth factors, which active translation depends on the introduced mRNA. A tissue restoration could be also assisted with mRNA-manipulated stem cells, e.g. MSCs, iPSCs or their derivatives. However, while cell-free regenerative therapies already undergo clinical examination, cell-based approaches have to overcome several concerns regarding their safety, efficacy and manufacturing. Yet, in view of the groundbreaking approval of mRNA-based COVID-19 vaccines in 2020, we expect a massive rise of interest among both commercial and academic facilities in the limitless opportunities that the mRNA platform has to offer. This will lead to further rapid optimization of the mRNA technology and new discoveries that will promote clinical translation. Without much doubt, the mRNA revolution will soon move beyond the scope of infectiology and usher a new chapter across the spectrum of medicine.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The authors are grateful for support by the EU Structural Fund [ESF/14-BM-A55-0024/18], the DFG [DA1296/6-1], the German Heart Foundation, the FORUN Program of Rostock Medical University, the Josef and Käthe Klinz Foundation, the BMBF [VIP+00240] and the European Regional Development Fund [GHS-19-0016].
R.D. and O.C. conceived and directed the presented review. O.C. took the lead in writing the manuscript, with input by A.M.G. and H.L.. O.C. prepared the artwork with help of H.L. and A.M.G.. All authors provided a critical feedback and contributed to the final version of the manuscript.
The following icons were provided by the Noun Project (thenounproject.com) to create the figures in accordance with the credit requirements of the Noun Project.
«Brain» icon by Cédric Villain.
«Bone» icon by WEBTECHOPS LLP.
«Cancer» icon by Lucas Helle.
«DNA» icon by Léa Lortal.
«Eye» icon by Rank Sol.
«Gene Editing» icon by dDara.
«Heart» icon by Clockwise and Philip Hogeboom.
«Human Body» icon by visual world.
«Injection» icon by Debi Alpa Nugraha.
«Liver» icon by Clockwise.
«Lungs» icon by Andi Nur Abdillah.
«Microbe» icon by Mask Icon.
«Plasmid» icon by Cornelia Scheitz and Léa Lortal.
«RNA» icon by Georgiana Ionescu.
«Spinal Column» icon by Smalllike.
«Virus» icon by Teewara soontorn.
References
- 1.Müller K. Drugs: from discovery to approval, second ed., By Rick Ng. ChemMedChem. 2009;4:1546–1547. doi: 10.1002/cmdc.200900232. [DOI] [Google Scholar]
- 2.F. Chast, A history of drug discovery, in: The Practice of Medicinal Chemistry, Elsevier, 2008, pp. 1–62.
- 3.Lounnas V., Ritschel T., Kelder J., McGuire R., Bywater R.P., Foloppe N. Current progress in Structure-Based Rational Drug Design marks a new mindset in drug discovery. Comput. Struct. Biotechnol. J. 2013;5:e201302011. doi: 10.5936/csbj.201302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kesik-Brodacka M. Progress in biopharmaceutical development. Biotechnol. Appl. Biochem. 2018;65:306–322. doi: 10.1002/bab.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deshaies R.J. Multispecific drugs herald a new era of biopharmaceutical innovation. Nature. 2020;580:329–338. doi: 10.1038/s41586-020-2168-1. [DOI] [PubMed] [Google Scholar]
- 6.Moorkens E., Meuwissen N., Huys I., Declerck P., Vulto A.G., Simoens S. The market of biopharmaceutical medicines: a snapshot of a diverse industrial landscape. Front. Pharmacol. 2017;8:314. doi: 10.3389/fphar.2017.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller P.Y., Milton M.N. The determination and interpretation of the therapeutic index in drug development. Nat. Rev. Drug Discov. 2012;11:751–761. doi: 10.1038/nrd3801. [DOI] [PubMed] [Google Scholar]
- 8.Kintzing J.R., Filsinger Interrante M.V., Cochran J.R. Emerging strategies for developing next-generation protein therapeutics for cancer treatment. Trends Pharmacol. Sci. 2016;37:993–1008. doi: 10.1016/j.tips.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubo T. In: Toxins and Drug Discovery. Cruz L.J., Luo S., editors. Springer; Netherlands, Dordrecht: 2017. Random peptide library for ligand and drug discovery; pp. 207–230. [Google Scholar]
- 10.Dimitrov D.S. Therapeutic proteins. Methods Mol. Biol. 2012;899:1–26. doi: 10.1007/978-1-61779-921-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. fourth ed. Garland Science Taylor & Francis Group; New York, NY: 2002. Molecular biology of the cell. [Google Scholar]
- 12.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 13.van Hoecke L., Roose K. How mRNA therapeutics are entering the monoclonal antibody field. J. Transl. Med. 2019;17:54. doi: 10.1186/s12967-019-1804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weng Y., Li C., Yang T., Hu B., Zhang M., Guo S., Xiao H., Liang X.-J., Huang Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020;40:107534. doi: 10.1016/j.biotechadv.2020.107534. [DOI] [PubMed] [Google Scholar]
- 15.Muthukrishnan S., Both G.W., Furuichi Y., Shatkin A.J. 5'-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–37. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- 16.Furuichi Y., Miura K. A blocked structure at the 5' terminus of mRNA from cytoplasmic polyhedrosis virus. Nature. 1975;253:374–375. doi: 10.1038/253374a0. [DOI] [PubMed] [Google Scholar]
- 17.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Papahadjopoulos D., Vail W.J., Jacobson K., Poste G. Cochleate lipid cylinders: formation by fusion of unilamellar lipid vesicles. Biochimica et Biophysica Acta (BBA) – Biomembranes. 1975;394:483–491. doi: 10.1016/0005-2736(75)90299-0. [DOI] [PubMed] [Google Scholar]
- 19.Dimitriadis G.J. Translation of rabbit globin mRNA introduced by liposomes into mouse lymphocytes. Nature. 1978;274:923–924. doi: 10.1038/274923a0. [DOI] [PubMed] [Google Scholar]
- 20.Brenner S., Jacob F., Meselson M. An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature. 1961;190:576–581. doi: 10.1038/190576a0. [DOI] [PubMed] [Google Scholar]
- 21.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 22.Jirikowski G.F., Sanna P.P., Maciejewski-Lenoir D., Bloom F.E. Reversal of diabetes insipidus in Brattleboro rats: intrahypothalamic injection of vasopressin mRNA. Science. 1992;255:996–998. doi: 10.1126/science.1546298. [DOI] [PubMed] [Google Scholar]
- 23.Martinon F., Krishnan S., Lenzen G., Magné R., Gomard E., Guillet J.G., Lévy J.P., Meulien P. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 1993;23:1719–1722. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 24.Boczkowski D., Nair S.K., Snyder D., Gilboa E. Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J. Exp. Med. 1996;184:465–472. doi: 10.1084/jem.184.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy G., Tjoa B., Ragde H., Kenny G., Boynton A. Phase I clinical trial: T-cell therapy for prostate cancer using autologous dendritic cells pulsed with HLA-A0201-specific peptides from prostate-specific membrane antigen. Prostate. 1996;29:371–380. doi: 10.1002/(SICI)1097-0045(199612)29:6<371:AID-PROS5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 26.Weide B., Carralot J.-P., Reese A., Scheel B., Eigentler T.K., Hoerr I., Rammensee H.-G., Garbe C., Pascolo S. Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J. Immunother. 2008;31:180–188. doi: 10.1097/CJI.0b013e31815ce501. [DOI] [PubMed] [Google Scholar]
- 27.Weide B., Pascolo S., Scheel B., Derhovanessian E., Pflugfelder A., Eigentler T.K., Pawelec G., Hoerr I., Rammensee H.-G., Garbe C. Direct injection of protamine-protected mRNA: results of a phase 1/2 vaccination trial in metastatic melanoma patients. J. Immunother. 2009;32:498–507. doi: 10.1097/CJI.0b013e3181a00068. [DOI] [PubMed] [Google Scholar]
- 28.Liang X., Li D., Leng S., Zhu X. RNA-based pharmacotherapy for tumors: From bench to clinic and back. Biomed. Pharmacother. 2020;125:109997. doi: 10.1016/j.biopha.2020.109997. [DOI] [PubMed] [Google Scholar]
- 29.Miao L., Zhang Y., Huang L. mRNA vaccine for cancer immunotherapy. Mol. Cancer. 2021;20:41. doi: 10.1186/s12943-021-01335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K.R., Walsh E.E., Frenck R., Falsey A.R., Dormitzer P.R., Gruber W.C., Şahin U., Jansen K.U. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 32.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O'Dell S., Schmidt S.D., Swanson P.A., Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2 – preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Bibi S., Briner C., Cicconi P., Collins A.M., Colin-Jones R., Cutland C.L., Darton T.C., Dheda K., Duncan C.J.A., Emary K.R.W., Ewer K.J., Fairlie L., Faust S.N., Feng S., Ferreira D.M., Finn A., Goodman A.L., Green C.M., Green C.A., Heath P.T., Hill C., Hill H., Hirsch I., Hodgson S.H.C., Izu A., Jackson S., Jenkin D., Joe C.C.D., Kerridge S., Koen A., Kwatra G., Lazarus R., Lawrie A.M., Lelliott A., Libri V., Lillie P.J., Mallory R., Mendes A.V.A., Milan E.P., Minassian A.M., McGregor A., Morrison H., Mujadidi Y.F., Nana A., O’Reilly P.J., Padayachee S.D., Pittella A., Plested E., Pollock K.M., Ramasamy M.N., Rhead S., Schwarzbold A.V., Singh N., Smith A., Song R., Snape M.D., Sprinz E., Sutherland R.K., Tarrant R., Thomson E.C., Török M.E., Toshner M., Turner D.P.J., Vekemans J., Villafana T.L., Watson M.E.E., Williams C.J., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Pollard A.J., Aban M., Abayomi F., Abeyskera K., Aboagye J., Adam M., Adams K., Adamson J., Adelaja Y.A., Adewetan G., Adlou S., Ahmed K., Akhalwaya Y., Akhalwaya S., Alcock A., Ali A., Allen E.R., Allen L., Almeida T.C.D.S.C., Alves M.P., Amorim F., Andritsou F., Anslow R., Appleby M., Arbe-Barnes E.H., Ariaans M.P., Arns B., Arruda L., Azi P., Azi L., Babbage G., Bailey C., Baker K.F., Baker M., Baker N., Baker P., Baldwin L., Baleanu I., Bandeira D., Bara A., Barbosa M.A., Barker D., Barlow G.D., Barnes E., Barr A.S., Barrett J.R., Barrett J., Bates L., Batten A., Beadon K., Beales E., Beckley R., Belij-Rammerstorfer S., Bell J., Bellamy D., Bellei N., Belton S., Berg A., Bermejo L., Berrie E., Berry L., Berzenyi D., Beveridge A., Bewley K.R., Bexhell H., Bhikha S., Bhorat A.E., Bhorat Z.E., Bijker E., Birch G., Birch S., Bird A., Bird O., Bisnauthsing K., Bittaye M., Blackstone K., Blackwell L., Bletchly H., Blundell C.L., Blundell S.R., Bodalia P., Boettger B.C., Bolam E., Boland E., Bormans D., Borthwick N., Bowring F., Boyd A., Bradley P., Brenner T., Brown P., Brown C., Brown-O'Sullivan C., Bruce S., Brunt E., Buchan R., Budd W., Bulbulia Y.A., Bull M., Burbage J., Burhan H., Burn A., Buttigieg K.R., Byard N., Cabera Puig I., Calderon G., Calvert A., Camara S., Cao M., Cappuccini F., Cardoso J.R., Carr M., Carroll M.W., Carson-Stevens A., Carvalho Y.d.M., Carvalho J.A., Casey H.R., Cashen P., Castro T., Castro L.C., Cathie K., Cavey A., Cerbino-Neto J., Chadwick J., Chapman D., Charlton S., Chelysheva I., Chester O., Chita S., Cho J.-S., Cifuentes L., Clark E., Clark M., Clarke A., Clutterbuck E.A., Collins S.L., Conlon C.P., Connarty S., Coombes N., Cooper C., Cooper R., Cornelissen L., Corrah T., Cosgrove C., Cox T., Crocker W.E., Crosbie S., Cullen L., Cullen D., Cunha D.R., Cunningham C., Cuthbertson F.C., Da Guarda S.N.F., Da Silva L.P., Damratoski B.E., Danos Z., Dantas M.T., Darroch P., Datoo M.S., Datta C., Davids M., Davies S.L., Davies H., Davis E., Davis J., Davis J., de Nobrega M.M., de Oliveira Kalid L.M., Dearlove D., Demissie T., Desai A., Di Marco S., Di Maso C., Dinelli M.I., Dinesh T., Docksey C., Dold C., Dong T., Donnellan F.R., Dos Santos T., dos Santos T.G., Dos Santos E.P., Douglas N., Downing C., Drake J., Drake-Brockman R., Driver K., Drury R., Dunachie S.J., Durham B.S., Dutra L., Easom N.J., van Eck S., Edwards M., Edwards N.J., El Muhanna O.M., Elias S.C., Elmore M., English M., Esmail A., Essack Y.M., Farmer E., Farooq M., Farrar M., Farrugia L., Faulkner B., Fedosyuk S., Felle S., Da Ferreira Silva C., Field S., Fisher R., Flaxman A., Fletcher J., Fofie H., Fok H., Ford K.J., Fowler J., Fraiman P.H., Francis E., Franco M.M., Frater J., Freire M.S., Fry S.H., Fudge S., Furze J., Fuskova M., Galian-Rubio P., Galiza E., Garlant H., Gavrila M., Geddes A., Gibbons K.A., Gilbride C., Gill H., Glynn S., Godwin K., Gokani K., Goldoni U.C., Goncalves M., Gonzalez I.G., Goodwin J., Goondiwala A., Gordon-Quayle K., Gorini G., Grab J., Gracie L., Greenland M., Greenwood N., Greffrath J., Groenewald M.M., Grossi L., Gupta G., Hackett M., Hallis B., Hamaluba M., Hamilton E., Hamlyn J., Hammersley D., Hanrath A.T., Hanumunthadu B., Harris S.A., Harris C., Harris T., Harrison T.D., Harrison D., Hart T.C., Hartnell B., Hassan S., Haughney J., Hawkins S., Hay J., Head I., Henry J., Hermosin Herrera M., Hettle D.B., Hill J., Hodges G., Horne E., Hou M.M., Houlihan C., Howe E., Howell N., Humphreys J., Humphries H.E., Hurley K., Huson C., Hyder-Wright A., Hyams C., Ikram S., Ishwarbhai A., Ivan M., Iveson P., Iyer V., Jackson F., de Jager J., Jaumdally S., Jeffers H., Jesudason N., Jones B., Jones K., Jones E., Jones C., Jorge M.R., Jose A., Joshi A., Júnior E.A., Kadziola J., Kailath R., Kana F., Karampatsas K., Kasanyinga M., Keen J., Kelly E.J., Kelly D.M., Kelly D., Kelly S., Kerr D., Kfouri R.d.Á., Khan L., Khozoee B., Kidd S., Killen A., Kinch J., Kinch P., King L.D., King T.B., Kingham L., Klenerman P., Knapper F., Knight J.C., Knott D., Koleva S., Lang M., Lang G., Larkworthy C.W., Larwood J.P., Law R., Lazarus E.M., Leach A., Lees E.A., Lemm N.-M., Lessa A., Leung S., Li Y., Lias A.M., Liatsikos K., Linder A., Lipworth S., Liu S., Liu X., Lloyd A., Lloyd S., Loew L., Lopez Ramon R., Lora L., Lowthorpe V., Luz K., MacDonald J.C., MacGregor G., Madhavan M., Mainwaring D.O., Makambwa E., Makinson R., Malahleha M., Malamatsho R., Mallett G., Mansatta K., Maoko T., Mapetla K., Marchevsky N.G., Marinou S., Marlow E., Marques G.N., Marriott P., Marshall R.P., Marshall J.L., Martins F.J., Masenya M., Masilela M., Masters S.K., Mathew M., Matlebjane H., Matshidiso K., Mazur O., Mazzella A., McCaughan H., McEwan J., McGlashan J., McInroy L., McIntyre Z., McLenaghan D., McRobert N., McSwiggan S., Megson C., Mehdipour S., Meijs W., Mendonça R.N., Mentzer A.J., Mirtorabi N., Mitton C., Mnyakeni S., Moghaddas F., Molapo K., Moloi M., Moore M., Moraes-Pinto M.I., Moran M., Morey E., Morgans R., Morris S., Morris S., Morris H.C., Morselli F., Morshead G., Morter R., Mottal L., Moultrie A., Moya N., Mpelembue M., Msomi S., Mugodi Y., Mukhopadhyay E., Muller J., Munro A., Munro C., Murphy S., Mweu P., Myasaki C.H., Naik G., Naker K., Nastouli E., Nazir A., Ndlovu B., Neffa F., Njenga C., Noal H., Noé A., Novaes G., Nugent F.L., Nunes G., O'Brien K., O'Connor D., Odam M., Oelofse S., Oguti B., Olchawski V., Oldfield N.J., Oliveira M.G., Oliveira C., Oosthuizen A., O'Reilly P., Osborne P., Owen D.R., Owen L., Owens D., Owino N., Pacurar M., Paiva B.V., Palhares E.M., Palmer S., Parkinson S., Parracho H.M., Parsons K., Patel D., Patel B., Patel F., Patel K., Patrick-Smith M., Payne R.O., Peng Y., Penn E.J., Pennington A., Peralta Alvarez M.P., Perring J., Perry N., Perumal R., Petkar S., Philip T., Phillips D.J., Phillips J., Phohu M.K., Pickup L., Pieterse S., Piper J., Pipini D., Plank M., Du Plessis J., Pollard S., Pooley J., Pooran A., Poulton I., Powers C., Presa F.B., Price D.A., Price V., Primeira M., Proud P.C., Provstgaard-Morys S., Pueschel S., Pulido D., Quaid S., Rabara R., Radford A., Radia K., Rajapaska D., Rajeswaran T., Ramos A.S.F., Ramos Lopez F., Rampling T., Rand J., Ratcliffe H., Rawlinson T., Rea D., Rees B., Reiné J., Resuello-Dauti M., Reyes Pabon E., Ribiero C.M., Ricamara M., Richter A., Ritchie N., Ritchie A.J., Robbins A.J., Roberts H., Robinson R.E., Robinson H., Rocchetti T.T., Rocha B.P., Roche S., Rollier C., Rose L., Ross Russell A.L., Rossouw L., Royal S., Rudiansyah I., Ruiz S., Saich S., Sala C., Sale J., Salman A.M., Salvador N., Salvador S., Sampaio M., Samson A.D., Sanchez-Gonzalez A., Sanders H., Sanders K., Santos E., Santos Guerra M.F., Satti I., Saunders J.E., Saunders C., Sayed A., van der Schim Loeff I., Schmid A.B., Schofield E., Screaton G., Seddiqi S., Segireddy R.R., Senger R., Serrano S., Shah R., Shaik I., Sharpe H.E., Sharrocks K., Shaw R., Shea A., Shepherd A., Shepherd J.G., Shiham F., Sidhom E., Silk S.E., Da Silva Moraes A.C., Silva-Junior G., Silva-Reyes L., Silveira A.D., Silveira M.B., Sinha J., Skelly D.T., Smith D.C., Smith N., Smith H.E., Smith D.J., Smith C.C., Soares A., Soares T., Solórzano C., Sorio G.L., Sorley K., Sosa-Rodriguez T., Souza C.M., Souza B.S., Souza A.R., Spencer A.J., Spina F., Spoors L., Stafford L., Stamford I., Starinskij I., Stein R., Steven J., Stockdale L., Stockwell L.V., Strickland L.H., Stuart A.C., Sturdy A., Sutton N., Szigeti A., Tahiri-Alaoui A., Tanner R., Taoushanis C., Tarr A.W., Taylor K., Taylor U., Taylor I.J., Taylor J., te Water Naude R., Themistocleous Y., Themistocleous A., Thomas M., Thomas K., Thomas T.M., Thombrayil A., Thompson F., Thompson A., Thompson K., Thompson A., Thomson J., Thornton-Jones V., Tighe P.J., Tinoco L.A., Tiongson G., Tladinyane B., Tomasicchio M., Tomic A., Tonks S., Towner J., Tran N., Tree J., Trillana G., Trinham C., Trivett R., Truby A., Tsheko B.L., Turabi A., Turner R., Turner C., Ulaszewska M., Underwood B.R., Varughese R., Verbart D., Verheul M., Vichos I., Vieira T., Waddington C.S., Walker L., Wallis E., Wand M., Warbick D., Wardell T., Warimwe G., Warren S.C., Watkins B., Watson E., Webb S., Webb-Bridges A., Webster A., Welch J., Wells J., West A., White C., White R., Williams P., Williams R.L., Winslow R., Woodyer M., Worth A.T., Wright D., Wroblewska M., Yao A., Zimmer R., Zizi D., Zuidewind P. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. The Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavernier G., Andries O., Demeester J., Sanders N.N., de Smedt S.C., Rejman J. mRNA as gene therapeutic: how to control protein expression. J. Control. Release. 2011;150:238–247. doi: 10.1016/j.jconrel.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 37.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics–developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 38.Zhong Z., Mc Cafferty S., Combes F., Huysmans H., de Temmerman J., Gitsels A., Vanrompay D., Portela Catani J., Sanders N.N. mRNA therapeutics deliver a hopeful message. Nano Today. 2018;23:16–39. doi: 10.1016/j.nantod.2018.10.005. [DOI] [Google Scholar]
- 39.Chen C.-Y., Tran D.M., Cavedon A., Cai X., Rajendran R., Lyle M.J., Martini P.G.V., Miao C.H. Treatment of hemophilia a using factor VIII messenger RNA lipid nanoparticles. Mol. Ther. Nucleic Acids. 2020;20:534–544. doi: 10.1016/j.omtn.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu X., Yin L., Theisen M., Zhuo J., Siddiqui S., Levy B., Presnyak V., Frassetto A., Milton J., Salerno T., Benenato K.E., Milano J., Lynn A., Sabnis S., Burke K., Besin G., Lukacs C.M., Guey L.T., Finn P.F., Martini P.G.V. Systemic mRNA therapy for the treatment of fabry disease: preclinical studies in wild-type mice, fabry mouse model, and wild-type non-human primates. Am. J. Hum. Genet. 2019;104:625–637. doi: 10.1016/j.ajhg.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roseman D.S., Khan T., Rajas F., Jun L.S., Asrani K.H., Isaacs C., Farelli J.D., Subramanian R.R. G6PC mRNA therapy positively regulates fasting blood glucose and decreases liver abnormalities in a mouse model of glycogen storage disease 1a. Mol. Ther. 2018;26:814–821. doi: 10.1016/j.ymthe.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao J., An D., Galduroz M., Zhuo J., Liang S., Eybye M., Frassetto A., Kuroda E., Funahashi A., Santana J., Mihai C., Benenato K.E., Kumarasinghe E.S., Sabnis S., Salerno T., Coughlan K., Miracco E.J., Levy B., Besin G., Schultz J., Lukacs C., Guey L., Finn P., Furukawa T., Giangrande P.H., Saheki T., Martini P.G.V. mRNA therapy improves metabolic and behavioral abnormalities in a murine model of citrin deficiency. Mol. Ther. 2019;27:1242–1251. doi: 10.1016/j.ymthe.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trepotec Z., Lichtenegger E., Plank C., Aneja M.K., Rudolph C. Delivery of mRNA therapeutics for the treatment of hepatic diseases. Mol. Ther. 2019;27:794–802. doi: 10.1016/j.ymthe.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Truong B., Allegri G., Liu X.-B., Burke K.E., Zhu X., Cederbaum S.D., Häberle J., Martini P.G.V., Lipshutz G.S. Lipid nanoparticle-targeted mRNA therapy as a treatment for the inherited metabolic liver disorder arginase deficiency. Proc. Natl. Acad. Sci. U. S. A. 2019;116:21150–21159. doi: 10.1073/pnas.1906182116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gadsby D.C., Vergani P., Csanády L. The ABC protein turned chloride channel whose failure causes cystic fibrosis. Nature. 2006;440:477–483. doi: 10.1038/nature04712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bentley D.L. Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2 doi: 10.1038/natrevmats.2017.56. [DOI] [Google Scholar]
- 49.Shatkin A.J., Manley J.L. The ends of the affair: capping and polyadenylation. Nat. Struct. Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 50.Moteki S., Price D. Functional coupling of capping and transcription of mRNA. Mol. Cell. 2002;10:599–609. doi: 10.1016/S1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]
- 51.Dunn S., Cowling V.H. Myc and mRNA capping. Biochim. Biophys. Acta, Mol. Cell. Res. 1849;2015:501–505. doi: 10.1016/j.bbagrm.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramanathan A., Robb G.B., Chan S.-H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galloway A., Cowling V.H. mRNA cap regulation in mammalian cell function and fate. Biochim. Biophys. Acta, Gene Regul. Mech. 1862;2019:270–279. doi: 10.1016/j.bbagrm.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mishra R.K., Datey A., Hussain T. mRNA recruiting eIF4 factors involved in protein synthesis and its regulation. Biochemistry. 2020;59:34–46. doi: 10.1021/acs.biochem.9b00788. [DOI] [PubMed] [Google Scholar]
- 55.Furuichi Y., Shatkin A.J. Elsevier; 2000. Viral and Cellular mRNA Capping: Past and Prospects; pp. 135–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.-Y., Schneller S., Zust R., Dong H., Thiel V., Sen G.C., Fensterl V., Klimstra W.B., Pierson T.C., Buller R.M., Gale M., Shi P.-Y., Diamond M.S. 2'-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schlee M. Master sensors of pathogenic RNA – RIG-I like receptors. Immunobiology. 2013;218:1322–1335. doi: 10.1016/j.imbio.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hyde J.L., Diamond M.S. Innate immune restriction and antagonism of viral RNA lacking 2׳-O methylation. Virology. 2015;479–480:66–74. doi: 10.1016/j.virol.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi Y., Di Giammartino D.C., Taylor D., Sarkeshik A., Rice W.J., Yates J.R., Frank J., Manley J.L. Molecular architecture of the human pre-mRNA 3' processing complex. Mol. Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sweet T.J., Licatalosi D.D. 3' end formation and regulation of eukaryotic mRNAs. Methods Mol. Biol. 2014;1125:3–12. doi: 10.1007/978-1-62703-971-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar A., Clerici M., Muckenfuss L.M., Passmore L.A., Jinek M. Mechanistic insights into mRNA 3'-end processing. Curr. Opin. Struct. Biol. 2019;59:143–150. doi: 10.1016/j.sbi.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nourse J., Spada S., Danckwardt S. Emerging roles of RNA 3'-end cleavage and polyadenylation in pathogenesis, diagnosis and therapy of human disorders. Biomolecules. 2020;10 doi: 10.3390/biom10060915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Casañal A., Kumar A., Hill C.H., Easter A.D., Emsley P., Degliesposti G., Gordiyenko Y., Santhanam B., Wolf J., Wiederhold K., Dornan G.L., Skehel M., Robinson C.V., Passmore L.A. Architecture of eukaryotic mRNA 3'-end processing machinery. Science. 2017;358:1056–1059. doi: 10.1126/science.aao6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan S.L., Huppertz I., Yao C., Weng L., Moresco J.J., Yates J.R., Ule J., Manley J.L., Shi Y. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3' processing. Genes Dev. 2014;28:2370–2380. doi: 10.1101/gad.250993.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schönemann L., Kühn U., Martin G., Schäfer P., Gruber A.R., Keller W., Zavolan M., Wahle E. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev. 2014;28:2381–2393. doi: 10.1101/gad.250985.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang H., Lim J., Ha M., Kim V.N. TAIL-seq: genome-wide determination of poly(A) tail length and 3' end modifications. Mol. Cell. 2014;53:1044–1052. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 67.Danckwardt S., Hentze M.W., Kulozik A.E. 3' end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ogorodnikov A., Kargapolova Y., Danckwardt S. Processing and transcriptome expansion at the mRNA 3' end in health and disease: finding the right end. Pflugers Arch. 2016;468:993–1012. doi: 10.1007/s00424-016-1828-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Proudfoot N.J., Furger A., Dye M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 70.Dreyfus M., Régnier P. The Poly(A) tail of mRNAs. Cell. 2002;111:611–613. doi: 10.1016/S0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- 71.Fuke H., Ohno M. Role of poly (A) tail as an identity element for mRNA nuclear export. Nucleic Acids Res. 2008;36:1037–1049. doi: 10.1093/nar/gkm1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sachs A.B., Sarnow P., Hentze M.W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell 89. 1997;831(838) doi: 10.1016/S0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 73.Kühn U., Wahle E. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta, Mol. Cell. Res. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 74.Mignone F., Gissi C., Liuni S., Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pesole G., Liuni S., Grillo G., Saccone C. Structural and compositional features of untranslated regions of eukaryotic mRNAs. Gene. 1997;205:95–102. doi: 10.1016/s0378-1119(97)00407-1. [DOI] [PubMed] [Google Scholar]
- 76.Leppek K., Das R., Barna M. Functional 5' UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018;19:158–174. doi: 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jia L., Mao Y., Ji Q., Dersh D., Yewdell J.W., Qian S.-B. Decoding mRNA translatability and stability from the 5' UTR. Nat. Struct. Mol. Biol. 2020;27:814–821. doi: 10.1038/s41594-020-0465-x. [DOI] [PubMed] [Google Scholar]
- 78.Fan J., Wang K., Du X., Wang J., Chen S., Wang Y., Shi M., Zhang L., Wu X., Zheng D., Wang C., Wang L., Tian B., Li G., Zhou Y., Cheng H. ALYREF links 3'-end processing to nuclear export of non-polyadenylated mRNAs. EMBO J. 2019;38 doi: 10.15252/embj.201899910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.K.M. Yergert, R. O’Rouke, J.H. Hines, B. Appel, Identification of 3’ UTR motifs required for mRNA localization to myelin sheaths in vivo, 2019. [DOI] [PMC free article] [PubMed]
- 80.Conne B., Stutz A., Vassalli J.D. The 3' untranslated region of messenger RNA: a molecular 'hotspot' for pathology? Nat. Med. 2000;6:637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 81.Koh W.S., Porter J.R., Batchelor E. Tuning of mRNA stability through altering 3'-UTR sequences generates distinct output expression in a synthetic circuit driven by p53 oscillations. Sci. Rep. 2019;9:5976. doi: 10.1038/s41598-019-42509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee Y., Rio D.C. Mechanisms and regulation of alternative Pre-mRNA splicing. Annu. Rev. Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Avci-Adali M., Behring A., Steinle H., Keller T., Krajeweski S., Schlensak C., Wendel H.P. In vitro synthesis of modified mRNA for induction of protein expression in human cells. J. Vis. Exp. 2014:e51943. doi: 10.3791/51943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Babendure J.R., Babendure J.L., Ding J.-H., Tsien R.Y. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ferizi M., Aneja M.K., Balmayor E.R., Badieyan Z.S., Mykhaylyk O., Rudolph C., Plank C. Human cellular CYBA UTR sequences increase mRNA translation without affecting the half-life of recombinant RNA transcripts. Sci. Rep. 2016;6:39149. doi: 10.1038/srep39149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Asrani K.H., Farelli J.D., Stahley M.R., Miller R.L., Cheng C.J., Subramanian R.R., Brown J.M. Optimization of mRNA untranslated regions for improved expression of therapeutic mRNA. RNA Biol. 2018;15:756–762. doi: 10.1080/15476286.2018.1450054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 88.Zeng C., Hou X., Yan J., Zhang C., Li W., Zhao W., Du S., Dong Y. Leveraging mRNAs sequences to express SARS-CoV-2 antigens in vivo. bioRxiv. 2020 doi: 10.1101/2020.04.01.019877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.J. Cao, E.M. Novoa, Z. Zhang, W.C. Chen, D. Liu, G.C.G. Choi, A.S.L. Wong, C. Wehrspaun, M. Kellis, T.K. Lu, High-Throughput 5’ UTR Engineering for Enhanced Protein Production in Non-Viral Gene Therapies, 2020. [DOI] [PMC free article] [PubMed]
- 90.Sultana N., Hadas Y., Sharkar M.T.K., Kaur K., Magadum A., Kurian A.A., Hossain N., Alburquerque B., Ahmed S., Chepurko E., Zangi L. Optimization of 5' untranslated region of modified mRNA for use in cardiac or hepatic ischemic injury. Mol. Ther. Methods Clin. Dev. 2020;17:622–633. doi: 10.1016/j.omtm.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kormann M.S.D., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A., Griese M., Bittmann I., Handgretinger R., Hartl D., Rosenecker J., Rudolph C. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 92.Kwon H., Kim M., Seo Y., Moon Y.S., Lee H.J., Lee K., Lee H. Emergence of synthetic mRNA: In vitro synthesis of mRNA and its applications in regenerative medicine. Biomaterials. 2018;156:172–193. doi: 10.1016/j.biomaterials.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 93.Vallazza B., Petri S., Poleganov M.A., Eberle F., Kuhn A.N., Sahin U. Recombinant messenger RNA technology and its application in cancer immunotherapy, transcript replacement therapies, pluripotent stem cell induction, and beyond, Wiley Interdiscip. Rev. RNA. 2015;6:471–499. doi: 10.1002/wrna.1288. [DOI] [PubMed] [Google Scholar]
- 94.Darnell J.E., Philipson L., Wall R., Adesnik M. Polyadenylic acid sequences: role in conversion of nuclear RNA into messenger RNA. Science. 1971;174:507–510. doi: 10.1126/science.174.4008.507. [DOI] [PubMed] [Google Scholar]
- 95.Lima S.A., Chipman L.B., Nicholson A.L., Chen Y.-H., Yee B.A., Yeo G.W., Coller J., Pasquinelli A.E. Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct. Mol. Biol. 2017;24:1057–1063. doi: 10.1038/nsmb.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lim J., Lee M., Son A., Chang H., Kim V.N. mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes Dev. 2016;30:1671–1682. doi: 10.1101/gad.284802.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Legnini I., Alles J., Karaiskos N., Ayoub S., Rajewsky N. FLAM-seq: full-length mRNA sequencing reveals principles of poly(A) tail length control. Nat. Methods. 2019;16:879–886. doi: 10.1038/s41592-019-0503-y. [DOI] [PubMed] [Google Scholar]
- 98.Subtelny A.O., Eichhorn S.W., Chen G.R., Sive H., Bartel D.P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yi H., Park J., Ha M., Lim J., Chang H., Kim V.N. PABP cooperates with the CCR4-NOT complex to promote mRNA deadenylation and block precocious decay. Mol. Cell. 2018;70:1081–1088.e5. doi: 10.1016/j.molcel.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 100.Nicholson A.L., Pasquinelli A.E. Tales of detailed poly(A) tails. Trends Cell Biol. 2019;29:191–200. doi: 10.1016/j.tcb.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Webster M.W., Chen Y.-H., Stowell J.A.W., Alhusaini N., Sweet T., Graveley B.R., Coller J., Passmore L.A. mRNA deadenylation is coupled to translation rates by the differential activities of Ccr4-Not nucleases. Mol. Cell. 2018;70:1089–1100.e8. doi: 10.1016/j.molcel.2018.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lim J., Ha M., Chang H., Kwon S.C., Simanshu D.K., Patel D.J., Kim V.N. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell. 2014;159:1365–1376. doi: 10.1016/j.cell.2014.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Grier A.E., Burleigh S., Sahni J., Clough C.A., Cardot V., Choe D.C., Krutein M.C., Rawlings D.J., Jensen M.C., Scharenberg A.M., Jacoby K. pEVL: a linear plasmid for generating mRNA IVT templates with extended encoded poly(A) sequences. Mol. Ther. Nucleic Acids. 2016;5:e306. doi: 10.1038/mtna.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Holtkamp S., Kreiter S., Selmi A., Simon P., Koslowski M., Huber C., Türeci O., Sahin U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- 105.Arbuthnot P., Ely A., Bloom K. A convenient method to generate and maintain poly(A)-encoding DNA sequences required for in vitro transcription of mRNA. Biotechniques. 2019;66:37–39. doi: 10.2144/btn-2018-0120. [DOI] [PubMed] [Google Scholar]
- 106.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Andries O., Mc Cafferty S., de Smedt S.C., Weiss R., Sanders N.N., Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 217. 2015:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 108.Leppek K., Byeon G.W., Kladwang W., Wayment-Steele H.K., Kerr C.H., Xu A.F., Kim D.S., Topkar V.V., Choe C., Rothschild D., Tiu G.C., Wellington-Oguri R., Fujii K., Sharma E., Watkins A.M., Nicol J.J., Romano J., Tunguz B., Participants E., Barna M., Das R. Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics. bioRxiv. 2021 doi: 10.1101/2021.03.29.437587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Karikó K., Muramatsu H., Keller J.M., Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 2012;20:948–953. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.-K., Schlee M., Endres S., Hartmann G. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 112.Pasquinelli A.E., Dahlberg J.E., Lund E. Reverse 5' caps in RNAs made in vitro by phage RNA polymerases. RNA. 1995;1:957–967. [PMC free article] [PubMed] [Google Scholar]
- 113.Jemielity J., Fowler T., Zuberek J., Stepinski J., Lewdorowicz M., Niedzwiecka A., Stolarski R., Darzynkiewicz E., Rhoads R.E. Novel “anti-reverse” cap analogs with superior translational properties. RNA. 2003;9:1108–1122. doi: 10.1261/rna.5430403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kuhn A.N., Diken M., Kreiter S., Selmi A., Kowalska J., Jemielity J., Darzynkiewicz E., Huber C., Türeci O., Sahin U. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 2010;17:961–971. doi: 10.1038/gt.2010.52. [DOI] [PubMed] [Google Scholar]
- 115.Kowalska J., Lewdorowicz M., Zuberek J., Grudzien-Nogalska E., Bojarska E., Stepinski J., Rhoads R.E., Darzynkiewicz E., Davis R.E., Jemielity J. Synthesis and characterization of mRNA cap analogs containing phosphorothioate substitutions that bind tightly to eIF4E and are resistant to the decapping pyrophosphatase DcpS. RNA. 2008;14:1119–1131. doi: 10.1261/rna.990208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grudzien-Nogalska E., Jemielity J., Kowalska J., Darzynkiewicz E., Rhoads R.E. Phosphorothioate cap analogs stabilize mRNA and increase translational efficiency in mammalian cells. RNA. 2007;13:1745–1755. doi: 10.1261/rna.701307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Strenkowska M., Grzela R., Majewski M., Wnek K., Kowalska J., Lukaszewicz M., Zuberek J., Darzynkiewicz E., Kuhn A.N., Sahin U., Jemielity J. Cap analogs modified with 1,2-dithiodiphosphate moiety protect mRNA from decapping and enhance its translational potential. Nucleic Acids Res. 2016;44:9578–9590. doi: 10.1093/nar/gkw896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kowalska J., Del Wypijewska Nogal A., Darzynkiewicz Z.M., Buck J., Nicola C., Kuhn A.N., Lukaszewicz M., Zuberek J., Strenkowska M., Ziemniak M., Maciejczyk M., Bojarska E., Rhoads R.E., Darzynkiewicz E., Sahin U., Jemielity J. Synthesis, properties, and biological activity of boranophosphate analogs of the mRNA cap: versatile tools for manipulation of therapeutically relevant cap-dependent processes. Nucleic Acids Res. 2014;42:10245–10264. doi: 10.1093/nar/gku757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Muttach F., Muthmann N., Rentmeister A. Synthetic mRNA capping. Beilstein J. Org. Chem. 2017;13:2819–2832. doi: 10.3762/bjoc.13.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jemielity J., Kowalska J., Rydzik A.M., Darzynkiewicz E. Synthetic mRNA cap analogs with a modified triphosphate bridge – synthesis, applications and prospects. New J. Chem. 2010;34:829. doi: 10.1039/C0NJ00041H. [DOI] [Google Scholar]
- 121.Kanavarioti A. HPLC methods for purity evaluation of man-made single-stranded RNAs. Sci. Rep. 2019;9:1019. doi: 10.1038/s41598-018-37642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Selmi A., Vascotto F., Kautz-Neu K., Türeci Ö., Sahin U., von Stebut E., Diken M., Kreiter S. Uptake of synthetic naked RNA by skin-resident dendritic cells via macropinocytosis allows antigen expression and induction of T-cell responses in mice. Cancer Immunol. Immunother. 2016;65:1075–1083. doi: 10.1007/s00262-016-1869-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lorenz C., Fotin-Mleczek M., Roth G., Becker C., Dam T.C., Verdurmen W.P.R., Brock R., Probst J., Schlake T. Protein expression from exogenous mRNA: uptake by receptor-mediated endocytosis and trafficking via the lysosomal pathway. RNA Biol. 2011;8:627–636. doi: 10.4161/rna.8.4.15394. [DOI] [PubMed] [Google Scholar]
- 126.Ginn S.L., Alexander I.E., Edelstein M.L., Abedi M.R., Wixon J. Gene therapy clinical trials worldwide to 2012 – an update. J. Gene Med. 2013;15:65–77. doi: 10.1002/jgm.2698. [DOI] [PubMed] [Google Scholar]
- 127.Meng Z., O'Keeffe-Ahern J., Lyu J., Pierucci L., Zhou D., Wang W. A new developing class of gene delivery: messenger RNA-based therapeutics. Biomater. Sci. 2017;5:2381–2392. doi: 10.1039/C7BM00712D. [DOI] [PubMed] [Google Scholar]
- 128.Bessis N., GarciaCozar F.J., Boissier M.-C. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 2004;11(Suppl 1):S10–S17. doi: 10.1038/sj.gt.3302364. [DOI] [PubMed] [Google Scholar]
- 129.Kashem S.W., Haniffa M., Kaplan D.H. Antigen-presenting cells in the skin. Annu. Rev. Immunol. 2017;35:469–499. doi: 10.1146/annurev-immunol-051116-052215. [DOI] [PubMed] [Google Scholar]
- 130.Erdag G., Schaefer J.T., Smolkin M.E., Deacon D.H., Shea S.M., Dengel L.T., Patterson J.W., Slingluff C.L. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res. 2012;72:1070–1080. doi: 10.1158/0008-5472.CAN-11-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rittig S.M., Haentschel M., Weimer K.J., Heine A., Muller M.R., Brugger W., Horger M.S., Maksimovic O., Stenzl A., Hoerr I., Rammensee H.-G., Holderried T.A.W., Kanz L., Pascolo S., Brossart P. Intradermal vaccinations with RNA coding for TAA generate CD8+ and CD4+ immune responses and induce clinical benefit in vaccinated patients. Mol. Ther. 2011;19:990–999. doi: 10.1038/mt.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.van Lint S., Goyvaerts C., Maenhout S., Goethals L., Disy A., Benteyn D., Pen J., Bonehill A., Heirman C., Breckpot K., Thielemans K. Preclinical evaluation of TriMix and antigen mRNA-based antitumor therapy. Cancer Res. 2012;72:1661–1671. doi: 10.1158/0008-5472.CAN-11-2957. [DOI] [PubMed] [Google Scholar]
- 133.Bialkowski L., van Weijnen A., van der Jeught K., Renmans D., Daszkiewicz L., Heirman C., Stangé G., Breckpot K., Aerts J.L., Thielemans K. Intralymphatic mRNA vaccine induces CD8 T-cell responses that inhibit the growth of mucosally located tumours. Sci. Rep. 2016;6:22509. doi: 10.1038/srep22509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gan L.-M., Lagerström-Fermér M., Carlsson L.G., Arfvidsson C., Egnell A.-C., Rudvik A., Kjaer M., Collén A., Thompson J.D., Joyal J., Chialda L., Koernicke T., Fuhr R., Chien K.R., Fritsche-Danielson R. Intradermal delivery of modified mRNA encoding VEGF-A in patients with type 2 diabetes. Nat. Commun. 2019;10:871. doi: 10.1038/s41467-019-08852-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tiwari P.M., Vanover D., Lindsay K.E., Bawage S.S., Kirschman J.L., Bhosle S., Lifland A.W., Zurla C., Santangelo P.J. Engineered mRNA-expressed antibodies prevent respiratory syncytial virus infection. Nat. Commun. 2018;9:3999. doi: 10.1038/s41467-018-06508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lindsay K.E., Vanover D., Thoresen M., King H., Xiao P., Badial P., Araínga M., Park S.B., Tiwari P.M., Peck H.E., Blanchard E.L., Feugang J.M., Olivier A.K., Zurla C., Villinger F., Woolums A.R., Santangelo P.J. Aerosol delivery of synthetic mRNA to vaginal mucosa leads to durable expression of broadly neutralizing antibodies against HIV. Mol. Ther. 2020;28:805–819. doi: 10.1016/j.ymthe.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Legere R.M., Cohen N.D., Poveda C., Bray J.M., Barhoumi R., Szule J.A., de La Concha-Bermejillo A., Bordin A.I., Pollet J. Safe and effective aerosolization of in vitro transcribed mRNA to the respiratory tract epithelium of horses without a transfection agent. Sci. Rep. 2021;11:371. doi: 10.1038/s41598-020-79855-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wadhwa A., Aljabbari A., Lokras A., Foged C., Thakur A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics. 2020;12 doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Felgner P.L., Ringold G.M. Cationic liposome-mediated transfection. Nature. 1989;337:387–388. doi: 10.1038/337387a0. [DOI] [PubMed] [Google Scholar]
- 140.Aldosari B.N., Alfagih I.M., Almurshedi A.S. Lipid nanoparticles as delivery systems for RNA-based vaccines. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cardarelli F., Digiacomo L., Marchini C., Amici A., Salomone F., Fiume G., Rossetta A., Gratton E., Pozzi D., Caracciolo G. The intracellular trafficking mechanism of Lipofectamine-based transfection reagents and its implication for gene delivery. Sci. Rep. 2016;6:25879. doi: 10.1038/srep25879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dalby B., Cates S., Harris A., Ohki E.C., Tilkins M.L., Price P.J., Ciccarone V.C. Advanced transfection with Lipofectamine, reagent: primary neurons, siRNA, and high-throughput applications. Methods. 2000;33(2004):95–103. doi: 10.1016/j.ymeth.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 143.Sultana N., Magadum A., Hadas Y., Kondrat J., Singh N., Youssef E., Calderon D., Chepurko E., Dubois N., Hajjar R.J., Zangi L. Optimizing cardiac delivery of modified mRNA. Mol. Ther. 2017;25:1306–1315. doi: 10.1016/j.ymthe.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hirsch-Lerner D., Zhang M., Eliyahu H., Ferrari M.E., Wheeler C.J., Barenholz Y. Effect of “helper lipid” on lipoplex electrostatics. Biochim. Biophys. Acta, Mol. Cell. Res. 2005;1714:71–84. doi: 10.1016/j.bbamem.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 145.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H., Grunwitz C., Vormehr M., Hüsemann Y., Selmi A., Kuhn A.N., Buck J., Derhovanessian E., Rae R., Attig S., Diekmann J., Jabulowsky R.A., Heesch S., Hassel J., Langguth P., Grabbe S., Huber C., Türeci Ö., Sahin U. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 146.Loquai C., Hassel J.C., Oehm P., Derhovanessian E., Jabulowsky R.A., Gold M., Schwarck-Kokarakis D., Attig S., Cuk K., Vogler I., Sikorski J., Leierer M., Mitzel-Rink H., Miederer M., Grabbe S., Utikal J., Pinter A., Kaufmann R., Sahin U., Tureci O. A shared tumor-antigen RNA-lipoplex vaccine with/without anti-PD1 in patients with checkpoint-inhibition experienced melanoma. JCO. 2020;38:3136. doi: 10.1200/JCO.2020.38.15_suppl.3136. [DOI] [Google Scholar]
- 147.Sahin U., Oehm P., Derhovanessian E., Jabulowsky R.A., Vormehr M., Gold M., Maurus D., Schwarck-Kokarakis D., Kuhn A.N., Omokoko T., Kranz L.M., Diken M., Kreiter S., Haas H., Attig S., Rae R., Cuk K., Kemmer-Brück A., Breitkreuz A., Tolliver C., Caspar J., Quinkhardt J., Hebich L., Stein M., Hohberger A., Vogler I., Liebig I., Renken S., Sikorski J., Leierer M., Müller V., Mitzel-Rink H., Miederer M., Huber C., Grabbe S., Utikal J., Pinter A., Kaufmann R., Hassel J.C., Loquai C., Türeci Ö. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature. 2020;585:107–112. doi: 10.1038/s41586-020-2537-9. [DOI] [PubMed] [Google Scholar]
- 148.Abraham M.-K., Peter K., Michel T., Wendel H.P., Krajewski S., Wang X. Nanoliposomes for safe and efficient therapeutic mRNA delivery: a step toward nanotheranostics in inflammatory and cardiovascular diseases as well as cancer. Nanotheranostics. 2017;1:154–165. doi: 10.7150/ntno.19449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yang S., Chen J., Di Zhao D., Han X. Chen. Comparative study on preparative methods of DC-Chol/DOPE liposomes and formulation optimization by determining encapsulation efficiency. Int. J. Pharmac. 2012;434:155–160. doi: 10.1016/j.ijpharm.2012.05.041. [DOI] [PubMed] [Google Scholar]
- 150.Michel T., Link A., Abraham M.-K., Schlensak C., Peter K., Wendel H.-P., Wang X., Krajewski S. Generation of cationic nanoliposomes for the efficient delivery of in vitro transcribed messenger RNA. J. Vis. Exp. 2019 doi: 10.3791/58444. [DOI] [PubMed] [Google Scholar]
- 151.Michel T., Luft D., Abraham M.-K., Reinhardt S., Salinas Medina M.L., Kurz J., Schaller M., Avci-Adali M., Schlensak C., Peter K., Wendel H.P., Wang X., Krajewski S. Cationic nanoliposomes meet mRNA: efficient delivery of modified mRNA using hemocompatible and stable vectors for therapeutic applications. Mol. Ther. Nucleic Acids 8. 2017:459–468. doi: 10.1016/j.omtn.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dhaliwal H.K., Fan Y., Kim J., Amiji M.M. Intranasal delivery and transfection of mRNA therapeutics in the brain using cationic liposomes. Mol. Pharm. 2020;17:1996–2005. doi: 10.1021/acs.molpharmaceut.0c00170. [DOI] [PubMed] [Google Scholar]
- 153.Hafez I.M., Maurer N., Cullis P.R. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- 154.Cheng X., Lee R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016;99:129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 155.Ryals R.C., Patel S., Acosta C., McKinney M., Pennesi M.E., Sahay G. The effects of PEGylation on LNP based mRNA delivery to the eye. PLoS ONE. 2020;15:e0241006. doi: 10.1371/journal.pone.0241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021;601:120586. doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tam Y.Y.C., Chen S., Cullis P.R. Advances in lipid nanoparticles for siRNA delivery. Pharmaceutics. 2013;5:498–507. doi: 10.3390/pharmaceutics5030498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yanez Arteta M., Kjellman T., Bartesaghi S., Wallin S., Wu X., Kvist A.J., Dabkowska A., Székely N., Radulescu A., Bergenholtz J., Lindfors L. Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2018;115:3351–3360. doi: 10.1073/pnas.1720542115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Herrera M., Kim J., Eygeris Y., Jozic A., Sahay G. Illuminating endosomal escape of polymorphic lipid nanoparticles that boost mRNA delivery. Biomater. Sci. 2021 doi: 10.1039/D0BM01947J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kim M., Jeong M., Hur S., Cho Y., Park J., Jung H., Seo Y., Woo H.A., Nam K.T., Lee K., Lee H. Engineered ionizable lipid nanoparticles for targeted delivery of RNA therapeutics into different types of cells in the liver. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abf4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Li Q., Chan C., Peterson N., Hanna R.N., Alfaro A., Allen K.L., Wu H., Dall'Acqua W.F., Borrok M.J., Santos J.L. Engineering caveolae-targeted lipid nanoparticles to deliver mRNA to the lungs. ACS Chem. Biol. 2020;15:830–836. doi: 10.1021/acschembio.0c00003. [DOI] [PubMed] [Google Scholar]
- 162.Patel S., Ashwanikumar N., Robinson E., Xia Y., Mihai C., Griffith J.P., Hou S., Esposito A.A., Ketova T., Welsher K., Joyal J.L., Almarsson Ö., Sahay G. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 2020;11:983. doi: 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kulkarni J.A., Witzigmann D., Leung J., van der Meel R., Zaifman J., Darjuan M.M., Grisch-Chan H.M., Thöny B., Tam Y.Y.C., Cullis P.R. Fusion-dependent formation of lipid nanoparticles containing macromolecular payloads. Nanoscale. 2019;11:9023–9031. doi: 10.1039/C9NR02004G. [DOI] [PubMed] [Google Scholar]
- 164.Viger-Gravel J., Schantz A., Pinon A.C., Rossini A.J., Schantz S., Emsley L. Structure of lipid nanoparticles containing siRNA or mRNA by dynamic nuclear polarization-enhanced NMR spectroscopy. J. Phys. Chem. B. 2018;122:2073–2081. doi: 10.1021/acs.jpcb.7b10795. [DOI] [PubMed] [Google Scholar]
- 165.Leung A.K.K., Tam Y.Y.C., Chen S., Hafez I.M., Cullis P.R. Microfluidic mixing: a general method for encapsulating macromolecules in lipid nanoparticle systems. J. Phys. Chem. B. 2015;119:8698–8706. doi: 10.1021/acs.jpcb.5b02891. [DOI] [PubMed] [Google Scholar]
- 166.Brader M.L., Williams S.J., Banks J.M., Hui W.H., Zhou Z.H., Jin L. Encapsulation state of messenger RNA inside lipid nanoparticles. Biophys. J. 2021 doi: 10.1016/j.bpj.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Udhayakumar V.K., de Beuckelaer A., McCaffrey J., McCrudden C.M., Kirschman J.L., Vanover D., van Hoecke L., Roose K., Deswarte K., de Geest B.G., Lienenklaus S., Santangelo P.J., Grooten J., McCarthy H.O., de Koker S. Arginine-rich peptide-based mRNA nanocomplexes efficiently instigate cytotoxic T cell immunity dependent on the amphipathic organization of the peptide. Adv. Healthc. Mater. 2017;6 doi: 10.1002/adhm.201601412. [DOI] [PubMed] [Google Scholar]
- 168.He J., Xu S., Leng Q., Mixson A.J. Location of a single histidine within peptide carriers increases mRNA delivery. J. Gene Med. 2021;23:e3295. doi: 10.1002/jgm.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Coolen A.-L., Lacroix C., Mercier-Gouy P., Delaune E., Monge C., Exposito J.-Y., Verrier B. Poly(lactic acid) nanoparticles and cell-penetrating peptide potentiate mRNA-based vaccine expression in dendritic cells triggering their activation. Biomaterials. 2019;195:23–37. doi: 10.1016/j.biomaterials.2018.12.019. [DOI] [PubMed] [Google Scholar]
- 170.van den Brand D., Gorris M.A.J., van Asbeck A.H., Palmen E., Ebisch I., Dolstra H., Hällbrink M., Massuger L.F.A.G., Brock R. Peptide-mediated delivery of therapeutic mRNA in ovarian cancer. Eur. J. Pharm. Biopharm. 2019;141:180–190. doi: 10.1016/j.ejpb.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 171.Taylor R.E., Zahid M. Cell penetrating peptides, novel vectors for gene therapy. Pharmaceutics. 2020;12 doi: 10.3390/pharmaceutics12030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Boisguérin P., Konate K., Josse E., Vivès E., Deshayes S. Peptide-based nanoparticles for therapeutic nucleic acid delivery. Biomedicines. 2021;9 doi: 10.3390/biomedicines9050583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kang Z., Meng Q., Liu K. Peptide-based gene delivery vectors. J. Mater. Chem. B. 2019;7:1824–1841. doi: 10.1039/c8tb03124j. [DOI] [PubMed] [Google Scholar]
- 174.Ruseska I., Zimmer A. Internalization mechanisms of cell-penetrating peptides. Beilstein J. Nanotechnol. 2020;11:101–123. doi: 10.3762/bjnano.11.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Akinc A., Thomas M., Klibanov A.M., Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 176.Schade A., Müller P., Delyagina E., Voronina N., Skorska A., Lux C., Steinhoff G., David R. Magnetic nanoparticle based nonviral MicroRNA delivery into freshly isolated CD105(+) hMSCs. Stem Cells Int. 2014;2014:197154. doi: 10.1155/2014/197154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Voronina N., Lemcke H., Wiekhorst F., Kühn J.-P., Rimmbach C., Steinhoff G., David R. Non-viral magnetic engineering of endothelial cells with microRNA and plasmid-DNA-An optimized targeting approach. Nanomedicine. 2016;12:2353–2364. doi: 10.1016/j.nano.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 178.Helmschrodt C., Höbel S., Schöniger S., Bauer A., Bonicelli J., Gringmuth M., Fietz S.A., Aigner A., Richter A., Richter F. Polyethylenimine nanoparticle-mediated siRNA delivery to reduce α-synuclein expression in a model of Parkinson's disease. Mol. Ther. Nucleic Acids. 2017;9:57–68. doi: 10.1016/j.omtn.2017.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Démoulins T., Milona P., Englezou P.C., Ebensen T., Schulze K., Suter R., Pichon C., Midoux P., Guzmán C.A., Ruggli N., McCullough K.C. Polyethylenimine-based polyplex delivery of self-replicating RNA vaccines. Nanomedicine. 2016;12:711–722. doi: 10.1016/j.nano.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 180.Debus H., Baumhof P., Probst J., Kissel T. Delivery of messenger RNA using poly(ethylene imine)-poly(ethylene glycol)-copolymer blends for polyplex formation: biophysical characterization and in vitro transfection properties. J. Control. Release. 2010;148:334–343. doi: 10.1016/j.jconrel.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 181.Sharifnia Z., Bandehpour M., Hamishehkar H., Mosaffa N., Kazemi B., Zarghami N. In-vitro transcribed mRNA delivery using PLGA/PEI nanoparticles into human monocyte-derived dendritic cells. Iran. J. Pharm. Res. 2019;18:1659–1675. doi: 10.22037/ijpr.2019.1100872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Palmerston Mendes L., Pan J., Torchilin V.P. Dendrimers as nanocarriers for nucleic acid and drug delivery in cancer therapy. Molecules. 2017;22 doi: 10.3390/molecules22091401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chahal J.S., Khan O.F., Cooper C.L., McPartlan J.S., Tsosie J.K., Tilley L.D., Sidik S.M., Lourido S., Langer R., Bavari S., Ploegh H.L., Anderson D.G. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E4133–E4142. doi: 10.1073/pnas.1600299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Tarach P., Janaszewska A. Recent advances in preclinical research using PAMAM dendrimers for cancer gene therapy. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22062912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Son S., Nam J., Zenkov I., Ochyl L.J., Xu Y., Scheetz L., Shi J., Farokhzad O.C., Moon J.J. Sugar-nanocapsules imprinted with microbial molecular patterns for mRNA vaccination. Nano Lett. 2020;20:1499–1509. doi: 10.1021/acs.nanolett.9b03483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Huang X., Zheng R., Ding F., Yang J., Xie M., Liu X., Li J., Feng J., Zhu X., Zhang C. Efficient delivery of mRNA using crosslinked nucleic acid nanogel as a carrier. ACS Materials Lett. 2020;2:1509–1515. doi: 10.1021/acsmaterialslett.0c00375. [DOI] [Google Scholar]
- 187.Xiong H., Liu S., Wei T., Cheng Q., Siegwart D.J. Theranostic dendrimer-based lipid nanoparticles containing PEGylated BODIPY dyes for tumor imaging and systemic mRNA delivery in vivo. J. Control. Release. 2020;325:198–205. doi: 10.1016/j.jconrel.2020.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Liu Y., Zhao C., Sabirsh A., Ye L., Wu X., Lu H., Liu J. A novel graphene quantum dot-based mRNA delivery platform. ChemistryOpen. 2021 doi: 10.1002/open.202000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Prieto-Vila M., Yoshioka Y., Ochiya T. Biological functions driven by mRNAs carried by extracellular vesicles in cancer. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.620498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kim K.M., Abdelmohsen K., Mustapic M., Kapogiannis D., Gorospe M. RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA. 2017;8 doi: 10.1002/wrna.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Aslan C., Kiaie S.H., Zolbanin N.M., Lotfinejad P., Ramezani R., Kashanchi F., Jafari R. Exosomes for mRNA delivery: a novel biotherapeutic strategy with hurdles and hope. BMC Biotech. 2021;21:20. doi: 10.1186/s12896-021-00683-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kojima R., Bojar D., Rizzi G., Hamri G.C.-E., El-Baba M.D., Saxena P., Ausländer S., Tan K.R., Fussenegger M. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson's disease treatment. Nat. Commun. 2018;9:1305. doi: 10.1038/s41467-018-03733-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Elliott R.O., He M. Unlocking the power of exosomes for crossing biological barriers in drug delivery. Pharmaceutics. 2021;13 doi: 10.3390/pharmaceutics13010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mueller S.K., Nocera A.L., Bleier B.S. Exosome function in aerodigestive mucosa. Nanomedicine. 2018;14:269–277. doi: 10.1016/j.nano.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 195.Yang Z., Shi J., Xie J., Wang Y., Sun J., Liu T., Zhao Y., Zhao X., Wang X., Ma Y., Malkoc V., Chiang C., Deng W., Chen Y., Fu Y., Kwak K.J., Fan Y., Kang C., Yin C., Rhee J., Bertani P., Otero J., Lu W., Yun K., Lee A.S., Jiang W., Teng L., Kim B.Y.S., Lee L.J. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat. Biomed. Eng. 2020;4:69–83. doi: 10.1038/s41551-019-0485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fu S., Wang Y., Xia X., Zheng J.C. Exosome engineering: Current progress in cargo loading and targeted delivery. NanoImpact. 2020;20:100261. doi: 10.1016/j.impact.2020.100261. [DOI] [Google Scholar]
- 197.Yang J., Wu S., Hou L., Zhu D., Yin S., Yang G., Wang Y. Therapeutic effects of simultaneous delivery of nerve growth factor mRNA and protein via exosomes on cerebral ischemia. Mol. Ther. Nucleic Acids. 2020;21:512–522. doi: 10.1016/j.omtn.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Gaj T., Gersbach C.A., Barbas C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kampmann M. CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine. ACS Chem. Biol. 2017;13:406–416. doi: 10.1021/acschembio.7b00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.He Z.-Y., Men K., Qin Z., Yang Y., Xu T., Wei Y.-Q. Non-viral and viral delivery systems for CRISPR-Cas9 technology in the biomedical field. Sci. China Life Sci. 2017;60:458–467. doi: 10.1007/s11427-017-9033-0. [DOI] [PubMed] [Google Scholar]
- 201.Luther D.C., Lee Y.W., Nagaraj H., Scaletti F., Rotello V.M. Delivery approaches for CRISPR/Cas9 therapeutics in vivo: advances and challenges. Expert Opin. Drug Deliv. 2018;15:905–913. doi: 10.1080/17425247.2018.1517746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Miller J.B., Zhang S., Kos P., Xiong H., Zhou K., Perelman S.S., Zhu H., Siegwart D.J. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. Engl. 2016;56:1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kazi D.S., Moran A.E., Coxson P.G., Penko J., Ollendorf D.A., Pearson S.D., Tice J.A., Guzman D., Bibbins-Domingo K. Cost-effectiveness of PCSK9 Inhibitor therapy in patients with heterozygous familial hypercholesterolemia or atherosclerotic cardiovascular disease. JAMA. 2016;316:743–753. doi: 10.1001/jama.2016.11004. [DOI] [PubMed] [Google Scholar]
- 205.Qiu M., Glass Z., Chen J., Haas M., Jin X., Zhao X., Rui X., Ye Z., Li Y., Zhang F., Xu Q. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2020401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hsu M.-N., Chang Y.-H., Truong V.A., Lai P.-L., Nguyen T.K.N., Hu Y.-C. CRISPR technologies for stem cell engineering and regenerative medicine. Biotechnol. Adv. 2019;37:107447. doi: 10.1016/j.biotechadv.2019.107447. [DOI] [PubMed] [Google Scholar]
- 207.Abbasi S., Uchida S., Toh K., Tockary T.A., Dirisala A., Hayashi K., Fukushima S., Kataoka K. Co-encapsulation of Cas9 mRNA and guide RNA in polyplex micelles enables genome editing in mouse brain. J. Control. Release. 2021;332:260–268. doi: 10.1016/j.jconrel.2021.02.026. [DOI] [PubMed] [Google Scholar]
- 208.Aly R.M. Current state of stem cell-based therapies: an overview. Stem Cell Investig. 2020;7:8. doi: 10.21037/sci-2020-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hirsch T., Rothoeft T., Teig N., Bauer J.W., Pellegrini G., de Rosa L., Scaglione D., Reichelt J., Klausegger A., Kneisz D., Romano O., Secone Seconetti A., Contin R., Enzo E., Jurman I., Carulli S., Jacobsen F., Luecke T., Lehnhardt M., Fischer M., Kueckelhaus M., Quaglino D., Morgante M., Bicciato S., Bondanza S., de Luca M. Regeneration of the entire human epidermis using transgenic stem cells. Nature. 2017;551:327–332. doi: 10.1038/nature24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Terashvili M., Bosnjak Z.J. Stem cell therapies in cardiovascular disease. J. Cardiothorac. Vasc. Anesth. 2019;33:209–222. doi: 10.1053/j.jvca.2018.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 212.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 213.Shi Y., Inoue H., Wu J.C., Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lee A.S., Tang C., Rao M.S., Weissman I.L., Wu J.C. Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat. Med. 2013;19:998–1004. doi: 10.1038/nm.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Fusaki N., Ban H., Nishiyama A., Saeki K., Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Yu J., Chau K.F., Vodyanik M.A., Jiang J., Jiang Y. Efficient feeder-free episomal reprogramming with small molecules. PLoS ONE. 2011;6:e17557. doi: 10.1371/journal.pone.0017557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Jia F., Wilson K.D., Sun N., Gupta D.M., Huang M., Li Z., Panetta N.J., Chen Z.Y., Robbins R.C., Kay M.A., Longaker M.T., Wu J.C. A nonviral minicircle vector for deriving human iPS cells. Nat. Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kim D., Kim C.-H., Moon J.-I., Chung Y.-G., Chang M.-Y., Han B.-S., Ko S., Yang E., Cha K.Y., Lanza R., Kim K.-S. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chanda P.K., Sukhovershin R., Cooke J.P. mRNA-enhanced cell therapy and cardiovascular regeneration. Cells. 2021;10 doi: 10.3390/cells10010187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Angel M., Yanik M.F. Innate immune suppression enables frequent transfection with RNA encoding reprogramming proteins. PLoS ONE. 2010;5:e11756. doi: 10.1371/journal.pone.0011756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yakubov E., Rechavi G., Rozenblatt S., Givol D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem. Biophys. Res. Commun. 2010;394:189–193. doi: 10.1016/j.bbrc.2010.02.150. [DOI] [PubMed] [Google Scholar]
- 222.Warren L., Manos P.D., Ahfeldt T., Loh Y.-H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A., Daley G.Q., Brack A.S., Collins J.J., Cowan C., Schlaeger T.M., Rossi D.J. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Poleganov M.A., Eminli S., Beissert T., Herz S., Moon J.-I., Goldmann J., Beyer A., Heck R., Burkhart I., Barea Roldan D., Türeci Ö., Yi K., Hamilton B., Sahin U. Efficient reprogramming of human fibroblasts and blood-derived endothelial progenitor cells using nonmodified RNA for reprogramming and immune evasion. Hum. Gene Ther. 2015;26:751–766. doi: 10.1089/hum.2015.045. [DOI] [PubMed] [Google Scholar]
- 224.Lee J., Sayed N., Hunter A., Au K.F., Wong W.H., Mocarski E.S., Pera R.R., Yakubov E., Cooke J.P. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Meng S., Chanda P., Thandavarayan R.A., Cooke J.P. Transflammation: how innate immune activation and free radicals drive nuclear reprogramming. Antioxid. Redox Signal. 2018;29:205–218. doi: 10.1089/ars.2017.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Mehta A., Verma V., Nandihalli M., Ramachandra C.J.A., Sequiera G.L., Sudibyo Y., Chung Y., Sun W., Shim W. A systemic evaluation of cardiac differentiation from mRNA reprogrammed human induced pluripotent stem cells. PLoS ONE. 2014;9:e103485. doi: 10.1371/journal.pone.0103485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Xue Y., Zhan X., Sun S., Karuppagounder S.S., Xia S., Dawson V.L., Dawson T.M., Laterra J., Zhang J., Ying M. Synthetic mRNAs drive highly efficient iPS cell differentiation to dopaminergic neurons. Stem Cells Transl. Med. 2018;8:112–123. doi: 10.1002/sctm.18-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ocampo A., Reddy P., Martinez-Redondo P., Platero-Luengo A., Hatanaka F., Hishida T., Li M., Lam D., Kurita M., Beyret E., Araoka T., Vazquez-Ferrer E., Donoso D., Roman J.L., Xu J., Esteban C.R., Nuñez G., Delicado E.N., Campistol J.M., Guillen I., Guillen P., Belmonte J.C.I. In Vivo amelioration of age-associated hallmarks by partial reprogramming. Cell. 2016;167:1719–1733.e12. doi: 10.1016/j.cell.2016.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lu Y., Brommer B., Tian X., Krishnan A., Meer M., Wang C., Vera D.L., Zeng Q., Yu D., Bonkowski M.S., Yang J.-H., Zhou S., Hoffmann E.M., Karg M.M., Schultz M.B., Kane A.E., Davidsohn N., Korobkina E., Chwalek K., Rajman L.A., Church G.M., Hochedlinger K., Gladyshev V.N., Horvath S., Levine M.E., Gregory-Ksander M.S., Ksander B.R., He Z., Sinclair D.A. Reprogramming to recover youthful epigenetic information and restore vision. Nature. 2020;588:124–129. doi: 10.1038/s41586-020-2975-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sarkar T.J., Quarta M., Mukherjee S., Colville A., Paine P., Doan L., Tran C.M., Chu C.R., Horvath S., Qi L.S., Bhutani N., Rando T.A., Sebastiano V. Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nat. Commun. 2020;11:1545. doi: 10.1038/s41467-020-15174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Li Y., Zhou G., Bruno I.G., Zhang N., Sho S., Tedone E., Lai T.-P., Cooke J.P., Shay J.W. Transient introduction of human telomerase mRNA improves hallmarks of progeria cells. Aging Cell. 2019;18 doi: 10.1111/acel.12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wang K., Lin R.-Z., Hong X., Ng A.H., Lee C.N., Neumeyer J., Wang G., Wang X., Ma M., Pu W.T., Church G.M., Melero-Martin J.M. Robust differentiation of human pluripotent stem cells into endothelial cells via temporal modulation of ETV2 with modified mRNA. Sci. Adv. 2020;6:eaba7606. doi: 10.1126/sciadv.aba7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hausburg F., Na S., Voronina N., Skorska A., Müller P., Steinhoff G., David R. Defining optimized properties of modified mRNA to enhance virus- and DNA- independent protein expression in adult stem cells and fibroblasts. Cell. Physiol. Biochem. 2015;35:1360–1371. doi: 10.1159/000373957. [DOI] [PubMed] [Google Scholar]
- 234.Lee K., Yu P., Lingampalli N., Kim H.J., Tang R., Murthy N. Peptide-enhanced mRNA transfection in cultured mouse cardiac fibroblasts and direct reprogramming towards cardiomyocyte-like cells. Int. J. Nanomedicine. 2015;10:1841–1854. doi: 10.2147/IJN.S75124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Corritore E., Lee Y.-S., Pasquale V., Liberati D., Hsu M.-J., Lombard C.A., van der Smissen P., Vetere A., Bonner-Weir S., Piemonti L., Sokal E., Lysy P.A. V-Maf musculoaponeurotic fibrosarcoma oncogene homolog a synthetic modified mRNA drives reprogramming of human pancreatic duct-derived cells into insulin-secreting cells. Stem Cells Transl. Med. 2016;5:1525–1537. doi: 10.5966/sctm.2015-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Chen L., Shi K., Qiu W., Aagaard L., Kassem M. Generation of inducible CRISPRi and CRISPRa human stromal/stem cell lines for controlled target gene transcription during lineage differentiation. Stem Cells Int. 2020;2020:8857344. doi: 10.1155/2020/8857344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Preskey D., Allison T.F., Jones M., Mamchaoui K., Unger C. Synthetically modified mRNA for efficient and fast human iPS cell generation and direct transdifferentiation to myoblasts. Biochem. Biophys. Res. Commun. 2016;473:743–751. doi: 10.1016/j.bbrc.2015.09.102. [DOI] [PubMed] [Google Scholar]
- 238.Mueller P., Wolfien M., Ekat K., Lang C.I., Koczan D., Wolkenhauer O., Hahn O., Peters K., Lang H., David R., Lemcke H. RNA-based strategies for cardiac reprogramming of human mesenchymal stromal cells. Cells. 2020;9 doi: 10.3390/cells9020504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Balmayor E.R., Geiger J.P., Aneja M.K., Berezhanskyy T., Utzinger M., Mykhaylyk O., Rudolph C., Plank C. Chemically modified RNA induces osteogenesis of stem cells and human tissue explants as well as accelerates bone healing in rats. Biomaterials. 2016;87:131–146. doi: 10.1016/j.biomaterials.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 240.Elangovan S., Khorsand B., Do A.-V., Hong L., Dewerth A., Kormann M., Ross R.D., Sumner D.R., Allamargot C., Salem A.K. Chemically modified RNA activated matrices enhance bone regeneration. J. Control. Release. 2015;218:22–28. doi: 10.1016/j.jconrel.2015.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Khorsand B., Elangovan S., Hong L., Dewerth A., Kormann M.S.D., Salem A.K. A comparative study of the bone regenerative effect of chemically modified RNA encoding BMP-2 or BMP-9. AAPS J. 2017;19:438–446. doi: 10.1208/s12248-016-0034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Badieyan Z.S., Berezhanskyy T., Utzinger M., Aneja M.K., Emrich D., Erben R., Schüler C., Altpeter P., Ferizi M., Hasenpusch G., Rudolph C., Plank C. Transcript-activated collagen matrix as sustained mRNA delivery system for bone regeneration. J. Control. Release. 2016;239:137–148. doi: 10.1016/j.jconrel.2016.08.037. [DOI] [PubMed] [Google Scholar]
- 243.Leng Q., Chen L., Lv Y. RNA-based scaffolds for bone regeneration: application and mechanisms of mRNA, miRNA and siRNA. Theranostics. 2020;10:3190–3205. doi: 10.7150/thno.42640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Birkholz K., Hombach A., Krug C., Reuter S., Kershaw M., Kämpgen E., Schuler G., Abken H., Schaft N., Dörrie J. Transfer of mRNA encoding recombinant immunoreceptors reprograms CD4+ and CD8+ T cells for use in the adoptive immunotherapy of cancer. Gene Ther. 2009;16:596–604. doi: 10.1038/gt.2008.189. [DOI] [PubMed] [Google Scholar]
- 245.Zhang F., Parayath N.N., Ene C.I., Stephan S.B., Koehne A.L., Coon M.E., Holland E.C., Stephan M.T. Genetic programming of macrophages to perform anti-tumor functions using targeted mRNA nanocarriers. Nat. Commun. 2019;10:3974. doi: 10.1038/s41467-019-11911-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Rizvi F., Everton E., Smith A.R., Liu H., Osota E., Beattie M., Tam Y., Pardi N., Weissman D., Gouon-Evans V. Murine liver repair via transient activation of regenerative pathways in hepatocytes using lipid nanoparticle-complexed nucleoside-modified mRNA. Nat. Commun. 2021;12:613. doi: 10.1038/s41467-021-20903-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Yang T., Poenisch M., Khanal R., Hu Q., Dai Z., Li R., Song G., Yuan Q., Yao Q., Shen X., Taubert R., Engel B., Jaeckel E., Vogel A., Falk C.S., Schambach A., Gerovska D., Araúzo-Bravo M.J., Vondran F.W.R., Cantz T., Horscroft N., Balakrishnan A., Chevessier F., Ott M., Sharma A.D. Therapeutic HNF4A mRNA attenuates liver fibrosis in a preclinical model. J. Hepatol. 2021 doi: 10.1016/j.jhep.2021.08.011. [DOI] [PubMed] [Google Scholar]
- 248.Anttila V., Saraste A., Knuuti J., Jaakkola P., Hedman M., Svedlund S., Lagerström-Fermér M., Kjaer M., Jeppsson A., Gan L.-M. Synthetic mRNA encoding VEGF-A in patients undergoing coronary artery bypass grafting: design of a phase 2a clinical trial. Mol. Ther. Methods Clin. Dev. 2020;18:464–472. doi: 10.1016/j.omtm.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Ylä-Herttuala S. Cardiovascular gene therapy with vascular endothelial growth factors. Gene. 2013;525:217–219. doi: 10.1016/j.gene.2013.03.051. [DOI] [PubMed] [Google Scholar]
- 250.Zangi L., Lui K.O., von Gise A., Ma Q., Ebina W., Ptaszek L.M., Später D., Xu H., Tabebordbar M., Gorbatov R., Sena B., Nahrendorf M., Briscoe D.M., Li R.A., Wagers A.J., Rossi D.J., Pu W.T., Chien K.R. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Magadum A., Singh N., Kurian A.A., Munir I., Mehmood T., Brown K., Sharkar M.T.K., Chepurko E., Sassi Y., Oh J.G., Lee P., Santos C.X.C., Gaziel-Sovran A., Zhang G., Cai C.-L., Kho C., Mayr M., Shah A.M., Hajjar R.J., Zangi L. Pkm2 regulates cardiomyocyte cell cycle and promotes cardiac regeneration. Circulation. 2020;141:1249–1265. doi: 10.1161/CIRCULATIONAHA.119.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Hadas Y., Vincek A.S., Youssef E., Żak M.M., Chepurko E., Sultana N., Sharkar M.T.K., Guo N., Komargodski R., Kurian A.A., Kaur K., Magadum A., Fargnoli A., Katz M.G., Hossain N., Kenigsberg E., Dubois N.C., Schadt E., Hajjar R., Eliyahu E., Zangi L. Altering sphingolipid metabolism attenuates cell death and inflammatory response after myocardial infarction. Circulation. 2020;141:916–930. doi: 10.1161/CIRCULATIONAHA.119.041882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Mann D.L. Sphingosine 1-phosphate as a therapeutic target in heart failure: more questions than answers. Circulation. 2012;125:2692–2694. doi: 10.1161/CIRCULATIONAHA.112.107797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Waeber C., Walther T. Sphingosine-1-phosphate as a potential target for the treatment of myocardial infarction. Circ. J. 2014;78:795–802. doi: 10.1253/circj.cj-14-0178. [DOI] [PubMed] [Google Scholar]
- 255.Lin C.-Y., Crowley S.T., Uchida S., Komaki Y., Kataoka K., Itaka K. Treatment of intervertebral disk disease by the administration of mRNA encoding a cartilage-anabolic transcription factor. Mol. Ther. Nucleic Acids. 2019;16:162–171. doi: 10.1016/j.omtn.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Fukushima Y., Uchida S., Imai H., Nakatomi H., Kataoka K., Saito N., Itaka K. Treatment of ischemic neuronal death by introducing brain-derived neurotrophic factor mRNA using polyplex nanomicelle. Biomaterials. 2021;270:120681. doi: 10.1016/j.biomaterials.2021.120681. [DOI] [PubMed] [Google Scholar]
- 257.Patel S., Ryals R.C., Weller K.K., Pennesi M.E., Sahay G. Lipid nanoparticles for delivery of messenger RNA to the back of the eye. J. Control. Release. 2019;303:91–100. doi: 10.1016/j.jconrel.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Heathman T.R.J., Nienow A.W., McCall M.J., Coopman K., Kara B., Hewitt C.J. The translation of cell-based therapies: clinical landscape and manufacturing challenges. Regen. Med. 2015;10:49–64. doi: 10.2217/rme.14.73. [DOI] [PubMed] [Google Scholar]
- 259.He J.Q., Sussman E.S., Steinberg G.K. Revisiting stem cell-based clinical trials for ischemic stroke. Front. Aging Neurosci. 2020;12:575990. doi: 10.3389/fnagi.2020.575990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Deinsberger J., Reisinger D., Weber B. Global trends in clinical trials involving pluripotent stem cells: a systematic multi-database analysis. NPJ Regen. Med. 2020;5:15. doi: 10.1038/s41536-020-00100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Conway A., Mendel M., Kim K., McGovern K., Boyko A., Zhang L., Miller J.C., DeKelver R.C., Paschon D.E., Mui B.L., Lin P.J.C., Tam Y.K., Barbosa C., Redelmeier T., Holmes M.C., Lee G. Non-viral delivery of zinc finger nuclease mRNA enables highly efficient in vivo genome editing of multiple therapeutic gene targets. Mol. Ther. 2019;27:866–877. doi: 10.1016/j.ymthe.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.-R.J., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Malek Mohammadi M., Kattih B., Grund A., Froese N., Korf-Klingebiel M., Gigina A., Schrameck U., Rudat C., Liang Q., Kispert A., Wollert K.C., Bauersachs J., Heineke J. The transcription factor GATA4 promotes myocardial regeneration in neonatal mice. EMBO Mol. Med. 2017;9:265–279. doi: 10.15252/emmm.201606602. [DOI] [PMC free article] [PubMed] [Google Scholar]