Abstract
MicroRNA-124a (miR-124a) is one of the most abundantly expressed microRNAs in the central nervous system and is encoded in mammals by the three genomic loci miR-124a-1/2/3; however, its in vivo roles in neuronal development and function remain ambiguous. In the present study, we investigated the effect of miR-124a loss on neuronal differentiation in mice and in embryonic stem (ES) cells. Since miR-124a-3 exhibits only background expression levels in the brain and we were unable to obtain miR-124a-1/2/3 triple knockout (TKO) mice by mating, we generated and analyzed miR-124a-1/2 double knockout (DKO) mice. We found that these DKO mice exhibit perinatal lethality. RNA-seq analysis demonstrated that the expression levels of proneural and neuronal marker genes were almost unchanged between the control and miR-124a-1/2 DKO brains; however, genes related to neuronal synaptic formation and function were enriched among downregulated genes in the miR-124a-1/2 DKO brain. In addition, we found the transcription regulator Tardbp/TDP-43, loss of which leads to defects in neuronal maturation and function, was inactivated in the miR-124a-1/2 DKO brain. Furthermore, Tardbp knockdown suppressed neurite extension in cultured neuronal cells. We also generated miR-124a-1/2/3 TKO ES cells using CRISPR-Cas9 as an alternative to TKO mice. Phase-contrast microscopic, immunocytochemical, and gene expression analyses showed that miR-124a-1/2/3 TKO ES cell lines were able to differentiate into neurons. Collectively, these results suggest that miR-124a plays a role in neuronal maturation rather than neurogenesis in vivo and advance our understanding of the functional roles of microRNAs in central nervous system development.
Keywords: microRNA(miRNA), brain, mouse, CRISPR/Cas, embryonic stem cell, neuronal development, neuronal differentiation, neuronal maturation, microRNA-124, Tardbp/TDP-43
Abbreviations: CDM, chemically defined medium; CNS, central nervous system; DHet, double heterozygous; DKO, double knockout; ES, embryonic stem; FPKM, fragments per kilobase of exon per million mapped fragment; FTD, frontotemporal dementia; IPA, ingenuity pathway analysis; miR-124a, MicroRNA-124a; pri-miR-124a, primary miR-124a; TBS, tris-buffered saline; THet, triple heterozygous; TKO, triple knockout
miRNAs are small noncoding RNA molecules regulating the gene expression of a variety of biological processes in plants and animals. A large number of diverse miRNAs are expressed in the vertebrate central nervous system (CNS). MicroRNA-124a (miR-124a) is one of the most abundantly expressed miRNAs in mouse and human brains (1). The nucleotide sequence of miR-124a and its nervous system-specific expression pattern are evolutionarily highly conserved from Caenorhabditis elegans and Drosophila melanogaster to all studied vertebrates, including human. Both in human and mouse genomes, miR-124as are encoded by three loci: miR-124a-1, miR-124a-2, and miR-124a-3. Among the three primary miR-124as (pri-miR-124as), pri-miR-124a-1 is predominantly expressed, while pri-miR-124a-3 is barely expressed in the mouse brain (2). In mice, retinal non-coding RNA 3 (RNCR3) functions as a pri-miR-124a-1 precursor. We previously identified RNCR3 in the mouse retina and generated RNCR3-deficient mice (RNCR3−/−) by deleting the entire 4.5 kb region harboring RNCR3 (2). In the RNCR3−/− brain, the level of mature miR-124a was reduced by 60 to 80% compared with that in the WT control brain. RNCR3−/− mice showed abnormalities in the CNS such as overextension of the dentate gyrus granule neuron axons and apoptosis of cone photoreceptor cells in the retina (2). Since in our previous study we confirmed that a loss of miR-124a is responsible for the RNCR3−/− abnormalities by in vivo rescue experiments, we will refer to RNCR3−/− mice as miR-124a-1−/− mice in the current study.
In C. elegans miR-124 mutants, neurons are normally generated and no overt phenotype is observed (3, 4). Drosophila miR-124 mutants exhibit abnormalities in neuroblast proliferation and neuronal maturation (5, 6). In vertebrates, consistent with its expression pattern in the developing CNS, miR-124a has been reported to be essential for neurogenesis (7, 8), maturation (2, 9, 10, 11), and progenitor proliferation (7). However, it should be noted that conflicting results on miR-124a function have been reported. It was reported that neither inhibition nor overexpression of miR-124a affects neuronal differentiation or progenitor proliferation in the chick neural tube (9). Overexpression of transcription factors Neurog1 and Neurog2 and a small molecule-based culture condition can induce neuronal differentiation from human-induced pluripotent stem cells deficient for miR-124a (12). On the other hand, other studies reported that miR-124a promotes both embryonic and adult neurogenesis (7, 13). In the chick spinal cord, miR-124a was implicated in the stimulation of neuronal differentiation by suppressing the antineuronal REST–SCP1 pathway. In addition, miR-124a induces neurogenesis in P19 mouse embryonic cells (7). In the adult mouse subventricular zone, miR-124a was shown to induce adult neurogenesis through the regulation of Sox9 (13). In the current study, we investigated the role of miR-124a in neuronal development by generating and analyzing miR-124a-1/2 double knockout (DKO) mice and miR-124a-1/2/3 triple knockout (TKO) cells.
Results
Generation of multiple miR-124a KO mice
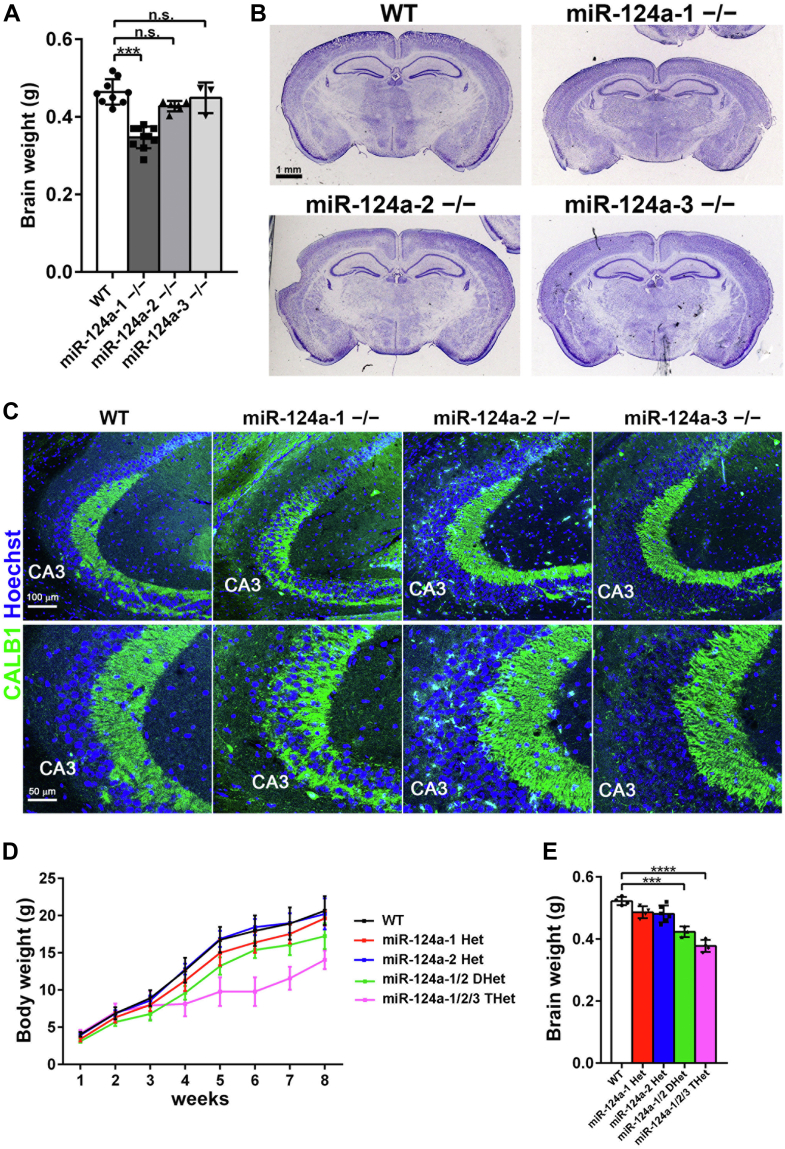
To investigate the in vivo function of miR-124a paralogs, we first generated miR-124a-2−/− mice, which harbor a deletion that includes the entire pre-miR-124a-2 sequence on mouse chromosome 3 (Fig. S1A). We also generated miR-124a-3−/− mice by deleting a fragment containing the entire pre-miR-124a-3 sequence on chromosome 2 (Fig. S1B). We confirmed deletions by Southern blot analysis using genomic DNA from these mice (Fig. S1, C–F). Both miR-124a-2−/− and miR-124a-3−/− mice were viable and fertile. In contrast to the previously described decreased brain weight of miR-124a-1−/− mice (2), miR-124a-2−/− and miR-124a-3−/− mice exhibited no brain weight change at 2 months of age (2M) (Fig. 1A). While coronal sections of miR-124a-2−/− and miR-124a-3−/− brains showed no obvious morphological changes compared with the WT control brain, thickness of the cerebral cortex decreased in the miR-124a-1−/− brain (Figs. 1B, S2, and S3). Aberrant outgrowth of mossy fibers stained with Calbindin into the hippocampal CA3 region was observed in miR-124a-1−/− mice as previously described (Fig. 1C) (2). In contrast, the morphology of mossy fibers in the miR-124a-2−/− and miR-124a-3−/− hippocampi were not substantially different from that in the WT control hippocampus (Fig. 1C). These results suggest that miR-124a-2 and miR-124a-3 have a minor role in brain development compared with miR-124a-1.
Figure 1.
Anatomical analysis of the miR-124a mutant mouse brain.A, brain weight of WT control, miR-124a-1−/−, miR-124a-2−/−, or miR-124a-3−/− mice at 2M. Data are presented as the mean ± SD. ∗∗∗p < 0.001 (one-way ANOVA followed by Tukey-Kramer test), n = 3 to 10 per genotype. B, Nissl-stained coronal sections of the brain from WT control, miR-124a-1−/−, miR-124a-2−/−, or miR-124a-3−/− mice at 2M. Thinning of the cerebral cortex was observed in the miR-124a-1−/− brain. C, immunofluorescent staining of hippocampi from WT control, miR-124a-1−/−, miR-124a-2−/−, and miR-124a-3−/− mice at 2M. Mossy fibers were immunostained with an anti-CALB1 antibody (green). D, body weight of WT control, miR-124a-1+/−, miR-124a-2+/−, miR-124a-1/2 DHet, and miR-124a-1/2/3 THet mice was measured weekly for 8 weeks after birth. Data are presented as the mean ± SD. n = 4 to 11 per genotype. E, brain weight of WT control, miR-124a-1+/−, miR-124a-2+/−, miR-124a-1/2 DHet, and miR-124a-1/2/3 THet mice was measured at 2M. Data are presented as the mean ± SD. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 (one-way ANOVA followed by Tukey-Kramer test), n = 3 to 6 per genotype. DHet, double heterozygous; miR-124a, MicroRNA-124a; n.s., not significant; THet, triple heterozygous.
Next, to investigate the effect of the complete loss of miR-124a in vivo by analyzing miR-124a-1/2/3 TKO mice, we generated miR-124a-1/2 double heterozygous (DHet) and then miR-124a-1/2/3 triple heterozygous (THet) mice by mating. We examined whether miR-124a paralog multiple heterozygosity affects postnatal growth. We measured body weights of miR-124a-1/2 DHet and miR-124a-1/2/3 THet mice and found that miR-124a-1/2 DHet and miR-124a-1/2/3 THet mice both exhibit growth retardation (Fig. 1D). At 8 weeks (wks), miR-124a-1/2 DHet and miR-124a-1/2/3 THet mice were 83% and 68% smaller than WT control mice, respectively. Brain weights of miR-124a-1/2 DHet and miR-124a-1/2/3 THet mice at 2M were significantly smaller than those of WT control mice (81% and 72%, respectively) (Fig. 1E). Although we first attempted to generate miR-124a-1/2/3 TKO mice, we could not obtain progeny by mating miR-124a-1/2/3 THet mice. Since pri-miR-124a-2 is the second major source of miR-124a after pri-miR-124a-1, and pri-miR-124a-3 is barely expressed in the brain (2), we next tried to generate miR-124a-1/2 DKO mice by mating miR-124a-1/2 DHet mice. Both male and female miR-124a-1/2 DHet mice were fertile. We found that miR-124a-1/2 DKO mice show perinatal lethality, suggesting severe abnormalities in the CNS of miR-124a-1/2 DKO mice.
Embryonic brain development is perturbed in miR-124a-1/2 DKO mice
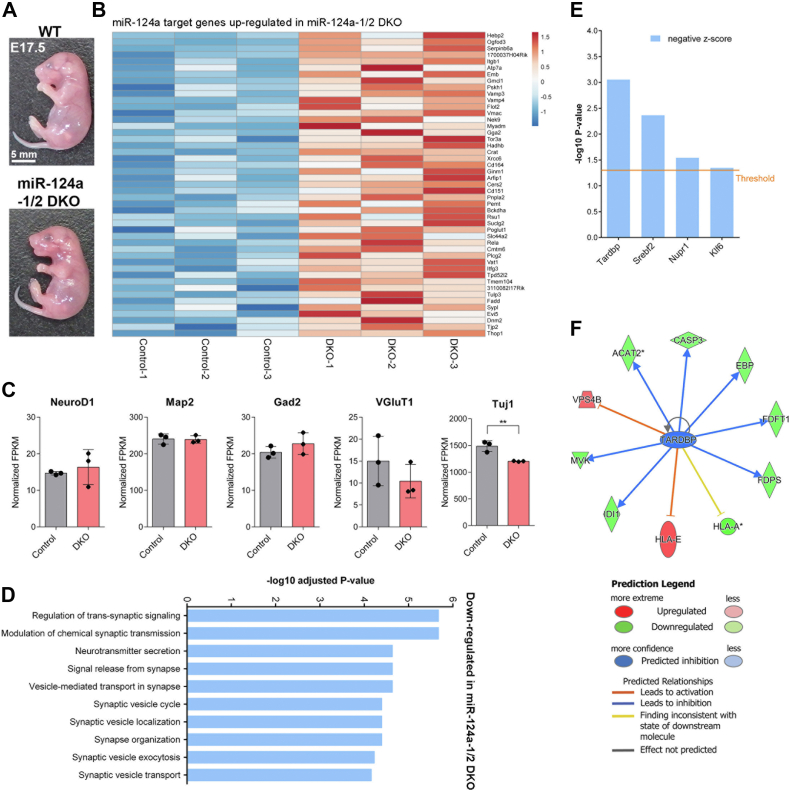
To assess the consequence of miR-124a-1/2 deficiency on CNS development, we performed RNA-seq analysis using total RNAs purified from three control (miR-124a-1+/−) and three miR-124a-1/2 DKO mouse brains at embryonic day 17.5 (E17.5) (Fig. 2). There were no obvious differences between WT control and miR-124a-1/2 DKO embryos at E17.5 (Fig. 2A). We confirmed that miR-124a target genes are enriched in the upregulated genes in the miR-124a-1/2 DKO brain (adjusted p = 1.8 × 10−7) (Fig. 2B). We first examined the gene expression of NeuroD1 (a proneural marker), Map2 (a mature neuronal marker), Gad2 (a GABAergic neuronal marker), VGluT1 (a glutamatergic neuronal marker), and Tuj1 (an early neuronal marker) in the control and miR-124a-1/2 DKO brains. Although the expression level of Tuj1 was significantly but slightly decreased in the miR-124a-1/2 DKO brain, that of other examined markers was not significantly changed, suggesting that neurogenesis occurs in the miR-124a-1/2 DKO brain (Fig. 2C). We also examined the gene expression of Gfap, Vim, S100b, and Aldh1l1 (radial glia and/or astrocyte markers). We found that there are no significant differences between the control and miR-124a-1/2 DKO brains (Fig. S4).
Figure 2.
RNA-seq analysis of the miR-124a-1/2 DKO mouse brain.A, images of WT control and miR-124a-1/2 DKO mouse embryos at E17.5. B, heatmaps of the miR-124a target genes upregulated in miR-124a-1/2 DKO mouse brains at E17.5 compared with those in the control (miR-124a-1+/−) mouse brains (fold change > 1.2; p < 0.05, unpaired t test). Gene expression values are visualized using a color scale from blue to red. Fragments per kilobase of exon per million–mapped fragment values from the RNA-seq dataset were used for the heatmap visualization. C, gene expression levels of NeuroD1, Map2, Gad2, VGluT1, and Tuj1 (neuronal markers) in the control and miR-124a-1/2 DKO brains at E17.5. Data are presented as the mean ± SD. ∗∗p < 0.01 (unpaired t test), n = 3 per genotype. D, the top ten most significantly enriched biological processes determined by gene ontology enrichment analysis for downregulated genes in the miR-124a-1/2 DKO brain compared with those in the control brain using iDEP. X-axis indicates –log10 adjusted p-value, and Y-axis indicates biological processes. E, IPA to predict the upstream transcription regulators affecting gene expression changes (p < 0.05, unpaired t test) in the miR-124a-1/2 DKO brain. The transcription regulators with activation Z scores <−2 are shown. F, IPA networks showing the transcription factor Tardbp as an upstream regulator. Tardbp is predicted to be inactivated in the miR-124a-1/2 DKO brain. DKO, double knockout; IPA, ingenuity pathway analysis; miR-124a, MicroRNA-124a.
Classification of the downregulated genes in the miR-124a-1/2 DKO brain into functional categories according to the gene ontology term enrichment for the Biological Process showed that these genes are associated with processes related to neuronal synaptic formation and function (Fig. 2D). These results imply that miR-124a plays a role in neuronal maturation and function rather than neurogenesis.
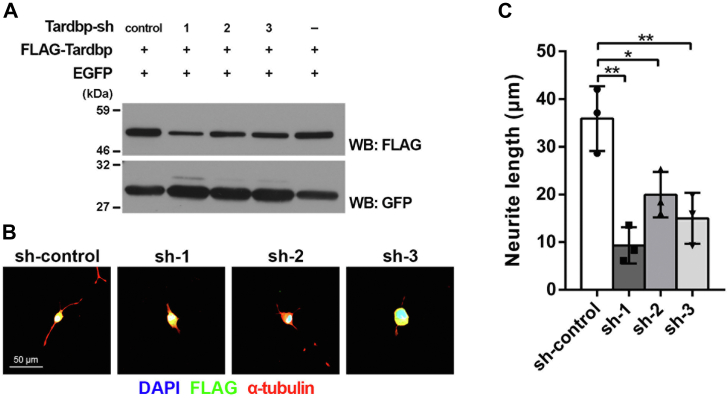
We next searched for the upstream transcription regulators that affect gene expression changes observed in the miR-124a-1/2 DKO brain. Ingenuity pathway analysis (IPA) revealed that the transcription regulators Tardbp/TDP-43, Srebf2, Nupr1, and Klf6 are inactivated by miR-124a-1/2 deficiency (Z scores < −2) (Fig. 2, E and F). We examined the gene expression of Tardbp, Srebf2, Nupr1, and Klf6 by qRT-PCR and found that there are no significant differences between the control and miR-124a-1/2 DKO brains (Fig. S5), suggesting that the transcription regulators Tardbp, Srebf2, Nupr1, and Klf6 are downregulated at the protein level. Among the genes encoding these transcription regulators, loss of function of Tardbp is known to lead to neuronal axon defects, impaired synaptic transmission, neuronal death, and motor deficit in vivo (14, 15). Furthermore, Tardbp KO mice exhibit embryonic lethality (16, 17, 18). To examine the roles of Tardbp in neuronal maturation, we performed knockdown experiments using Neuro2a cells. We first constructed shRNAs to knockdown Tardbp and confirmed that the Tardbp expression levels decrease in the cells expressing Tardbp-shRNA1, Tardbp-shRNA2, and Tardbp-shRNA3 (Fig. 3A). We transfected these constructs into Neuro2a cells and observed the neurites by immunostaining using anti-α-tubulin and anti-FLAG antibodies. We found that Tardbp-shRNAs significantly suppress neurite extension (Fig. 3, B and C). Inactivation of Tardbp in the brain may be at least partly a cause of perinatal lethality of miR-124a-1/2 DKO mice.
Figure 3.
Roles of Tardbp on neurite extension in cultured neuronal cells.A, inhibition efficacy of shRNA expression constructs for Tardbp knockdown. ShRNA-control, Tardbp-shRNA1, Tardbp-shRNA2, or Tardbp-shRNA3 expression plasmids were cotransfected with plasmids expressing a FLAG-tagged Tardbp and a GFP into HEK293T cells. Western blot analysis was performed using anti-FLAG and anti-GFP antibodies. GFP was used as an internal transfection control. Tardbp-shRNA1, Tardbp-shRNA2, and Tardbp-shRNA3 suppressed Tardbp expression. B and C, ShRNA-control, Tardbp-shRNA1, Tardbp-shRNA2, or Tardbp-shRNA3 expression plasmids were cotransfected with a plasmid expressing FLAG-tagged EGFP into Neuro2a cells. Cells were immunostained with anti-FLAG and anti-α-tubulin antibodies. Nuclei were stained with DAPI (B). The length of the longest neurite of the transfected cells was measured (C). Data are presented as the mean ± SD. ∗p < 0.05, ∗∗p < 0.01 (one-way ANOVA followed by Tukey-Kramer test), n = 3 experiments. A total of 63, 25, 59, and 55 cells were measured in shRNA-control, Tardbp-shRNA1, Tardbp-shRNA2, and Tardbp-shRNA3, respectively.
miR-124a-1/2/3 TKO ES cells differentiate into neurons
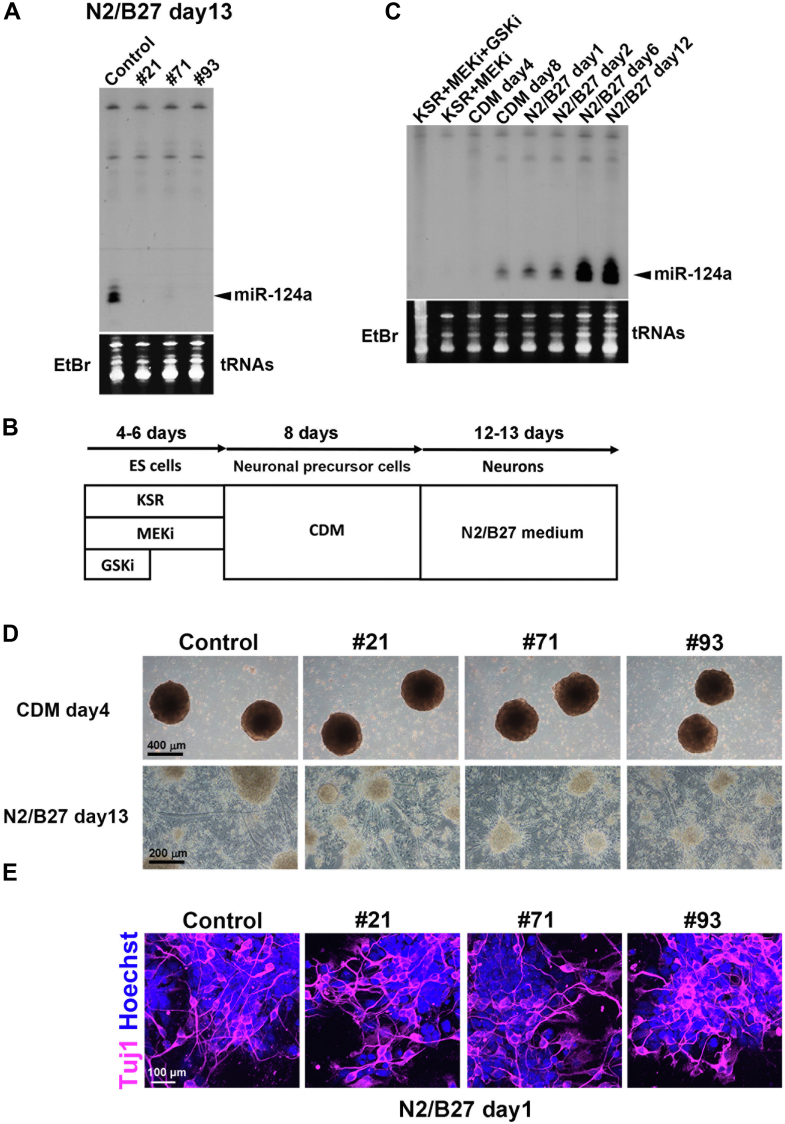
To investigate the effect of complete loss of miR-124a in neuronal development, we generated miR-124a-1/2/3 TKO embryonic stem (ES) cells and tried to induce their differentiation into neurons. To create multiplex deletions of miR-124a-1/2/3 loci in mouse ES cells, we used CRISPR/Cas9-mediated genome editing. Three clones (#21, #71, and #93) were identified as having deletions or insertions in both alleles of these three loci (Fig. S6). We tested whether these miR-124a TKO ES cells would differentiate into neurons by a previously established, chemically defined medium (CDM) culture system (19, 20). To confirm that miR-124a-1/2/3 TKO cells do not express miR-124a, we performed Northern blot analysis and found that the expression of miR-124a is abolished in the miR-124a-1/2/3 TKO cells (Fig. 4, A and B). We also performed Northern blot analysis using a miR-124a probe and found that miR-124a expression increases along with neuronal development in the control ES cells (Fig. 4C). The phase-contrast view shows that all three miR-124a-1/2/3 TKO ES cell lines differentiated into aggregates of neural precursor cells in CDM and into neurons in N2/B27 medium similarly to the control ES cells (Figs. 4D and S7). We also observed that the control and miR-124a-1/2/3 TKO cells at N2/B27 day 1 contain Tuj1-positive cells (Fig. 4E). These results suggest that miR-124a is not essential for neurogenesis.
Figure 4.
Neuronal differentiation of miR-124a-1/2/3 TKO ES cells.A, Northern blot analysis of miR-124a in the control and miR-124a-1/2/3 TKO cells. B, scheme of neuronal differentiation induction from mouse ES cells. C, Northern blot analysis of miR-124a in the control cells at the indicated time points. D, phase-contrast view of the control and miR-124a-1/2/3 TKO cells at the indicated time points. E, immunostaining of the control and miR-124a-1/2/3 TKO cells at N2/B27 day 1 using an anti-Tuj1 antibody (a marker for neurons). ES, embryonic stem; GSKi, GSK3 inhibitor; MEKi, MEK inhibitor; miR-124a, MicroRNA-124a; TKO, triple knockout.
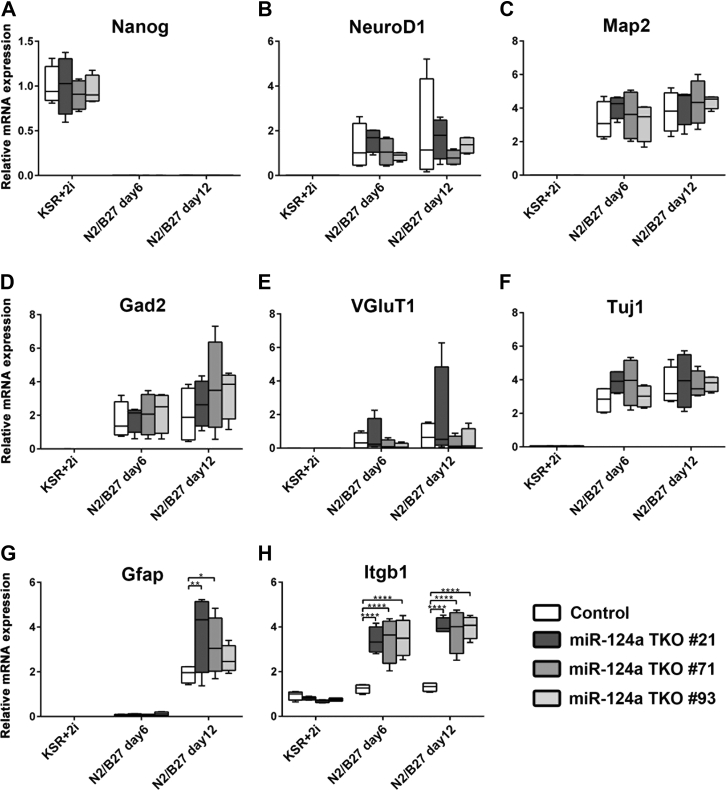
To further investigate the nature of miR-124a-1/2/3 TKO cells, we examined the expression profiles of developmental markers in ES cells and cells at day 6 or 12 of culturing in N2/B27 medium. We observed that the expression of a stem cell marker, Nanog, is lost both in the control and miR-124a-1/2/3 TKO cells at days 6 and 12 of culture in N2/B27 medium (Fig. 5A). In contrast, the expression of NeuroD1, Map2, Gad2, VGluT1, and Tuj1 increased to similar levels both in the control and miR-124a-1/2/3 TKO cells at days 6 and 12 of culture in N2/B27 medium (Fig. 5, B–F). The expression levels of S100b and Aldh1l1 increased to similar levels both in the control and miR-124a-1/2/3 TKO cells at days 6 and/or 12 of culture in N2/B27 medium (Fig. S8). The expression of Gfap was increased in the control and miR-124a-1/2/3 TKO cells at day 12 in N2/B27 medium, although its expression levels in two of three miR-124a-1/2/3 TKO cell lines were significantly higher than those in the control cell line (Fig. 5G). The expression levels of Vim increased in the control and miR-124a-1/2/3 TKO cells at days 6 and 12 in N2/B27 medium, although its expression levels in one or two of three miR-124a-1/2/3 TKO cell lines were significantly higher than those in the control cell line (Fig. S8). We also observed that the expression levels of one of the miR-124a targets, Itgb1, were significantly increased in miR-124a TKO cells compared with those in the control cells at days 6 and 12 of culture in N2/B27 medium (Fig. 5H), supporting miR-124a dysfunction in miR-124a TKO cells. Itgb1 is a major Integrin β subunit and functions in cerebral cortex development by forming heterodimers with various α subunits (21, 22). For example, the interaction between α3β1 integrin receptor and its ligand, Reelin, regulates cortical neuronal migration (23). Taken together, these results suggest that miR-124a is dispensable for neurogenesis.
Figure 5.
Gene expression analysis of neuronal differentiation–induced miR-124a-1/2/3 TKO cells. Expression levels of genes for a pluripotent cell marker (Nanog) (A), neuronal markers (NeuroD1 (B), Map2 (C), Gad2 (D), VGluT1 (E), Tuj1 (F)), a glial marker (Gfap) (G), and the miR-124a target gene Itgb1 (H) in miR-124a-1/2/3 TKO cells at the indicated time points were analyzed. RNA was purified from cells cultured in ES medium (KSR+2i) and neuronal differentiation medium (N2/B27) for 6 or 12 days. Box–whisker plots present the median (center line), ±1.5 interquartile range (box), and minimal and maximal values (whiskers). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001 (two-way ANOVA followed by Bonferroni test), n = 4 per cell line. 2i, MEKi and GSKi. ES, embryonic stem; KSR, knockout serum replacement; miR-124a, MicroRNA-124a; TKO, triple knockout.
Discussion
In the current study, we first generated miR-124a-2−/− and miR-124a-3−/− mice. Although miR-124a-1−/− mice exhibited reduced brain weight and aberrant outgrowth of mossy fibers in the hippocampus as previously described (2), miR-124a-2−/− and miR-124a-3−/− mice did not show these abnormalities, suggesting that miR-124a-2 and miR-124a-3 have minor roles in the CNS development compared with miR-124a-1. We also found that thickness of the cerebral cortex decreased in the miR-124a-1−/− mice, which is probably due to increased apoptosis in the miR-124a-1−/− cerebral cortex (2). Since pri-miR-124a-1 is highly expressed in the CNS compared with pri-miR-124a-2 and pri-miR-124a-3 (2), abnormalities observed in the miR-124a-1−/−, but not miR-124a-2−/− or miR-124a-3−/− brain, may reflect their expression levels. We next attempted to investigate the consequence of the complete loss of miR-124a in vivo. While we could generate miR-124a-1/2 DKO mice, in which almost all sources of miR-124a are depleted, we could not obtain miR-124a-1/2/3 TKO mice by mating miR-124a-1/2/3 THet mice. We thus generated miR-124a-1/2/3 TKO ES cells using the CRISPR-Cas9 system. We observed that the expression levels of the proneural and neuronal marker genes were almost unchanged in the miR-124a-1/2 DKO mouse brain and miR-124a-1/2/3 TKO cells. Phase-contrast microscopic and immunocytochemical analyses showed that neurons can differentiate from the miR-124a-1/2/3 TKO ES cells. These results suggest that miR-124a is dispensable for neurogenesis, which is consistent with previous studies (2, 3, 4, 6, 9, 12). On the other hand, we observed that the expression levels of Gfap and Vim increase in one or two of three miR-124a-1/2/3 TKO cell lines compared with those in the control cell line at days 6 and/or 12 of culture in N2/B27 medium, suggesting that miR-124a is required for suppressing astrogenesis. miR-124a-1/2 DKO mice and miR-124a-1/2/3 TKO ES cells can be a useful resource for a range of researchers interested in neuronal development and miRNA biology.
Our RNA-seq analysis demonstrated that genes associated with neuronal synaptic formation and function are enriched in downregulated genes in the miR-124a-1/2 DKO brain, implying that miR-124a is involved in neuronal maturation and function rather than neurogenesis. Notably, it was previously reported that miR-124a is involved in neuronal synaptic plasticity in Aplysia and rodents (24, 25, 26). In addition, the transcription factor Tardbp, which is required for neuronal maturation and function in vivo (14, 15, 27), was inactivated in the miR-124a-1/2 DKO brain. We observed that Tardbp knockdown suppresses neurite extension. Notably, loss of TARDBP function is linked to the neurodegenerative disease frontotemporal dementia (FTD) (28, 29). Since the expression level of miR-124a decreases in brain tissues from subjects with FTD (30), the TARDBP inactivation caused by downregulation of miR-124a may contribute to the pathogenesis of FTD. Together, previous studies and our results suggest that miR-124a plays a role in neuronal maturation. Although we cannot rule out other explanations, upregulated expression of a variety of miR-124a target genes may abrogate neuronal synaptic formation and function, inhibit Tardbp activity, and then lead to perinatal lethality in miR-124a-1/2 DKO mice. Future analyses, such as electrophysiological and histological evaluation, are needed to uncover how deficiency of both miR-124a-1 and -2 influences CNS development.
In contrast to our results, it was previously reported that miR-124a is required for neurogenesis (5, 7, 8, 13). This discrepancy might be due to the differences in experimental systems. However, we cannot exclude the possibility that miR-124a is involved in the differentiation of specific neuronal subtypes. Furthermore, despite the low expression level, miR-124a-3 may be able to instruct neurogenesis in miR-124a-1/2 DKO mice. It would be helpful to evaluate in detail the effects of miR-124a-1/2/3 loss on neuronal differentiation in mice using conditional approaches such as the Cre/loxP system.
Experimental procedures
Animal care
All procedures were approved by the Institutional Safety Committee on Recombinant DNA Experiments (approval ID 04220-4) and Animal Experimental Committees of the Institute for Protein Research at Osaka University (approval ID 29-01-4). These procedures were performed in compliance with the institutional guidelines. Mice were housed in a temperature-controlled room at 22 °C with a 12 h light/dark cycle. Fresh water and rodent diet were available at all times.
Generation of miR-124a-2 and miR-124a-3 mutant mice
We obtained miR-124a-2 and miR-124a-3 genomic clones from a screening of the 129S6/SvEvTac mouse genomic DNA library. We obtained ∼6.1-kb and ∼5.1-kb fragments for miR-124a-2 and ∼8.2-kb and ∼5.7-kb fragments for miR-124a-3 from the genomic clones. We subcloned them into a modified pPNT vector (31). We transfected the linearized targeting construct into a 129S6/SvEvTac-derived TC1 ES cell line (31). The culture, electroporation, and selection of ES cells were performed as previously described (32). ES cells that were heterozygous for the targeted gene disruption were microinjected into C57BL/6 blastocysts to obtain chimeric mice. We mated this mouse line with a CAG-Cre transgenic 129S6/SvEvTac mouse line (33), which expresses Cre recombinase under the control of the CAG promoter, to obtain null alleles of miR-124a-2 and miR-124a-3. We used miR-124a-1 (2), miR-124a-2, and miR-124a-3 129S6/SvEvTac mice to generate multiple miR-124a mutant mice.
Generation of miR-124a TKO ES cell
To target all three miR-124a precursors simultaneously, pX330 plasmids (34) expressing both the mammalian-codon–optimized Cas9 and guide RNAs, which target miR-124a-1/2/3 loci, were transfected into mouse ES cells by electroporation using GENE Pulser II (Bio-Rad). A plasmid containing PGK-neo cassette was cotransfected into cells. Neomycin-resistant cells were selected by culturing in G418-containing medium. To identify clones with biallelic mutations in all three genes, the PCR products of miR-124a-1/2/3 precursor regions from the genomic DNA of these ES lines were sequenced. The oligonucleotide sequences for the guide RNA expression plasmid are shown in Table S1.
Plasmid construction
Plasmids expressing EGFP and FLAG-tagged EGFP were previously constructed (35, 36). Full-length cDNA fragment of mouse Tardbp was amplified by PCR using mouse retinal cDNA as a template and subcloned into the pCAGGSII-3xFLAG vector (37). For Tardbp knockdown, the Tardbp-shRNA and shRNA-control cassette was subcloned into pBAsi-mU6 vector (Takara). The target sequences were as follows: Tardbp-shRNA1, 5′-GGTGTTGCTGTCCACAGTTAC-3′; Tardbp-shRNA2, 5′-GCGATGGTGTGACTGTA AACT-3′; Tardbp-shRNA3, 5′-GCATGCAGAGGGAACCAAATC-3′; and shRNA-control, 5′-GACGTCTAACGGATTCGAGCT-3′ (38). Primer sequences used for amplification are shown in Table S1.
PAGE Northern blot analysis
The PAGE Northern blot analysis was performed as described previously (2). Briefly, total RNAs from mouse tissues were isolated by Trizol (Invitrogen), and 20 μg of the total RNAs were denatured in 5 mM EDTA containing formamide at 80 °C for 5 min. RNAs were separated on 15% denaturing (7 M urea) polyacrylamide gels. RNAs were transferred to a nylon membrane (Pall Corporation Biodyne). LNA-modified anti–miR-124a (Exiqon, 20 pmol) was end-labeled with γ-32P-ATP (Muromachi Yakuhin) using T4 polynucleotide kinase (Takara). The nylon filters were hybridized with the labeled probe in salmon sperm–containing hybridization solution (120 mM sodium phosphate (pH 7.2), 250 mM sodium chloride, 7% SDS, and 50% formamide) at 43 °C overnight. The filters were exposed to X-ray film.
Cell culture and transfection
HEK293T and Neuro2a cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Sigma) containing 10% fetal bovine serum, supplemented with penicillin (100 μg/ml) and streptomycin (100 μg/ml) at 37 °C with 5% CO₂. Transfection was performed with the calcium phosphate method for HEK293T cells or Lipofectamine 3000 (Thermo Fisher Scientific) for Neuro2a cells.
Western blot analysis
Western blot analysis was performed as previously described (39). Briefly, HEK293T cells were washed with Tris-buffered saline (TBS) twice and lysed in an SDS-sample buffer. The samples were resolved by SDS-PAGE and transferred to PVDF membrane using the iBlot system (Invitrogen). The membranes were blocked with blocking buffer (3% skim milk and 0.05% Tween 20 in TBS) and incubated with primary antibodies overnight at 4 °C. The membranes were washed with 0.05% Tween 20 in TBS three times for 10 min each and then incubated with secondary antibodies for 2 h at room temperature. Signals were detected using Chemi-Lumi One L (Nakalai Tesque). We used the following primary antibodies: mouse anti-FLAG M2 (1:10,000, Sigma, F1804) and rabbit anti-GFP (1:2500, MBL, 598). The following secondary antibodies were used: horseradish peroxidase–conjugated anti-mouse IgG (1:10,000, Zymed) and anti-rabbit IgG (1:10,000, Jackson Laboratory).
Cell differentiation induction
Differentiation of Neuro2a cells was performed as previously described (40). After 24 h of transfection, the medium was changed to DMEM with 2% fetal bovine serum containing 20 μM retinoic acid and cultured for 48 h.
Immunofluorescent staining of brain sections and cells
Immunohistochemistry and immunocytochemistry were performed as previously described (41) with certain modifications. For immunohistochemistry, 30-μm brain sections were washed twice in phosphate-buffered saline, permeabilized with 0.1% Triton X-100 in PBS, and then incubated with PBS containing 4% donkey serum for 1 h for blocking. The samples were incubated with a primary antibody at 4 °C overnight. After PBS-washing, these samples were incubated with fluorescent-labeled secondary antibodies at room temperature for 2 h. For immunocytochemistry, cells were fixed with 4% paraformaldehyde in PBS for 5 or 15 min, washed once or twice with PBS, and then incubated in 4% normal donkey serum and 0.05% Triton X-100 in PBS or 5% normal donkey serum and 0.1% Triton X-100 in PBS for blocking. The samples were incubated with a primary antibody at 4 °C overnight. After PBS-washing, these samples were incubated with fluorescent-labeled secondary antibodies at room temperature for 2 h. The specimens were observed under a laser confocal microscope (LSM700, Carl Zeiss). Hoechst (Sigma) or DAPI (Nacalai Tesque) was used for nuclear staining. The primary antibodies were as follows: rabbit anti-CALB1 (1:1000, Sigma, PC253L), mouse anti-Tuj1 (1:500, Covance, MMS-435P), rabbit anti-FLAG (1:1000, Sigma, F7425), and mouse anti-α-tubulin (1:1000, Sigma, DM1A, T9026) antibodies. We used Cy3-conjugated (1:500, Jackson ImmunoResearch Laboratories) and Alexa Fluor 488–conjugated (1:500, Sigma) secondary antibodies.
Nissl staining
Nissl staining was performed as previously described (35). Coronal sections (30 μm thick) from frozen mouse brains at 2M were stained with 0.1% cresyl violet for 5 min, washed in 100% ethanol, and incubated in xylene. Slides were coverslipped with Permount (Fisher Scientific).
Induction of neuronal differentiation from ES cells
ES cells were maintained using mouse primary embryonic fibroblasts (19). We used ES cell culture medium containing Knockout DMEM/F12 (Gibco) supplemented with 20% knockout serum replacement (Gibco), penicillin (100 μg/ml), streptomycin (100 μg/ml), 0.1 mM 2-mercaptoethanol, 1× L-glutamine, 1× nonessential amino acids (Sigma), 10 μM MEK inhibitor (PD 0325901, Cayman Chemical Company), 10 μM GSK3 inhibitor (CHIR99021, Cayman Chemical Company), and 10 ng/ml leukemia inhibitory factor. The procedure of differentiation of ES cells into neurons was described previously (19, 20). In brief, ES cells were suspended in CDM medium containing Iscove's modified Dulbecco's medium/Hams F12 1:1 (Gibco), 1× lipid concentrate (Gibco), penicillin (100 μg/ml), streptomycin (100 μg/ml), transferrin (150 μg/ml final, Sigma), insulin (7 μg/ml, Sigma), 450 μM monothioglycerol (Sigma), and plated onto a 10-cm Lipidure-coated dish (Sumitomo Bakelite). After 8 days, aggregated cells were dissociated using 25% Accumax (Innovate Cell Technologies) in PBS, plated onto a 10-cm dish coated with laminin and poly-L-lysine using N2/B27 medium containing 0.5% N-2 Supplement (Gibco) and 1% B-27 Supplement (Gibco) in DMEM/F12, and cultured for 12 to 13 days.
RNA-seq and data analysis
RNA-seq analysis was performed as previously described (42), with certain modifications. Total RNAs from the control and miR-124a-1/2 DKO mouse brains at E17.5 were isolated using TRIzol RNA extraction reagent (Invitrogen). Sequencing was performed on an Illumina NovaSeq 6000 platform in the 101-base single-end mode. The raw reads were mapped to the mouse reference genome sequences (mm10) using the software TopHat ver. 2.0.13, in combination with Bowtie2 ver. 2.3.5.1 and SAMtools ver. 1.11. The numbers of reads were 10,275,949 for control-1, 11,664,879 for control-2, 11,416,851 for control-3, 10,954,843 for DKO-1, 13,526,133 for DKO-2, and 12,471,336 for DKO-3. The number of fragments per kilobase of exon per million mapped fragments (FPKMs) was calculated using Cufflinks ver. 2.2.1. Using the cut-off (fold change > 1.2, <−1.2; p < 0.05, unpaired t test), we obtained 223 downregulated and 395 upregulated genes in the miR-124a-1/2 DKO mouse brain. Heatmap visualization was conducted using the web tool ClustVis (43) under default parameters with FPKM values. Pathway analyses were performed using integrated Differential Expression and Pathway analysis v. 0.93 (44) under default parameters with the raw counts, and miRTarBase and gene ontology Biological Process were selected as gene sets. Upstream regulator analysis was carried out using IPA (Qiagen) with FPKM values. IPA was applied to predict the activated or inhibited transcription factors based on the observed differential gene expression profiles. Statistical thresholds were determined through the calculation of the activation z score of gene sets composed of randomly chosen perturbed genes with random sign of fold change that do not lead to significant results on average (ingenuity downstream effects analysis, whitepaper). Adjusted p values (adj. p val) and activated z-scores were used to identify significant pathways and upstream regulators. The adj. p val indicates significance, while z-scores were used to define activation (z-score ≥ 2.0) or inhibition (z-score ≤ −2.0). The strongest predicted activation corresponds to z scores ≥2, and the strongest predicted inhibition corresponds to z scores ≤−2.
qRT-PCR
qRT-PCR was performed as described previously (45). Total RNA was extracted using Trizol reagent (Invitrogen) and reverse transcribed into cDNA using SuperScript II reverse transcriptase (Invitrogen) with random hexamers and Oligo dT (Invitrogen). Quantitative PCR was performed using a SYBR GreenER qPCR SuperMix Universal (Invitrogen) and Thermal Cycler Dice Real Time System Single MRQ TP870 (Takara) in accordance with the manufacturer’s instructions. Quantification was carried out by Thermal Cycler Dice Real Time System software version 2.0 (Takara). Nucleotide sequences of primers are shown in Table S1 (46, 47).
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 6.04 (GraphPad Software). Single comparisons were performed with an unpaired t test, while multiple comparisons were performed using one-way ANOVA with post hoc Tukey–Kramer test. The statistical significance of experiments involving three or more groups and two or more treatments was assessed by two-way ANOVA with post hoc Bonferroni test. Data are reported as the median (center line), ±1.5 interquartile range (box), minimal and maximal values (whiskers), or as the mean ± SD. The analyzed number of samples is indicated in the figure legends. Asterisks indicate significance values as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Data availability
All sequencing data is available on GEO (GSE196356).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank M. Shimada, T. Kozuka, M. Kadowaki, A. Tani, A. Ishimaru, T. Nakayama, S. Gion, M. Wakabayashi, H. Abe, M. Nakamura, and K. Yoshida for technical assistance. We acknowledge the NGS core facility of the Genome Information Research Center at the Research Institute for Microbial Diseases of Osaka University for their support with RNA sequencing and data analysis.
Author contributions
T. F. conceptualization; T. C., Y. M., D. O., S. W., D. M., H. Y., H. K., and T. F. methodology; T. C., Y. M., R. S., D. O., S. W., L. R. V., D. M., H. Y., and T. F. validation; T. C., Y. M., D. O., S. W., L. R. V., D. M., and H. Y. formal analysis; T. C., Y. M., R. S., D. O., S. W., L. R. V., D. M., and H. Y. investigation; T. C., Y. M., R. S., D. O., S. W., L. R. V., D. M., D. G., H. Y., H. K., and T. F. resources; T. C., Y. M., R. S., D. O., S. W., L. R. V., D. M., and H. Y. data curation; T. C. and Y. M., writing–original draft; T. F. writing–review & editing; T. C., Y. M., D. O., S. W., and L. R. V. visualization; T. F. supervision; T. F. project administration; T. C. and T. F. funding acquisition.
Funding and additional information
This work was supported by the Grant-in-Aid for Scientific Research (21H02657, 20K07326) from the Japan Society for the Promotion of Science, AMED-CREST (21gm1510006) from the Japan Agency for Medical Research and Development, Japan Science and Technology Agency Moonshot R&D (JPMJMS2024), The Takeda Science Foundation, and The Uehara Memorial Foundation.
Edited by Ronald Wek
Supporting information
References
- 1.Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 2.Sanuki R., Onishi A., Koike C., Muramatsu R., Watanabe S., Muranishi Y., et al. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat. Neurosci. 2011;14:1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- 3.Miska E.A., Alvarez-Saavedra E., Abbott A.L., Lau N.C., Hellman A.B., McGonagle S.M., et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3 doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark A.M., Goldstein L.D., Tevlin M., Tavare S., Shaham S., Miska E.A. The microRNA miR-124 controls gene expression in the sensory nervous system of Caenorhabditis elegans. Nucleic Acids Res. 2010;38:3780–3793. doi: 10.1093/nar/gkq083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weng R., Cohen S.M. Drosophila miR-124 regulates neuroblast proliferation through its target anachronism. Development. 2012;139:1427–1434. doi: 10.1242/dev.075143. [DOI] [PubMed] [Google Scholar]
- 6.Sun K., Westholm J.O., Tsurudome K., Hagen J.W., Lu Y., Kohwi M., et al. Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visvanathan J., Lee S., Lee B., Lee J.W., Lee S.K. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makeyev E.V., Zhang J., Carrasco M.A., Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao X., Pfaff S.L., Gage F.H. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu J.Y., Chung K.H., Deo M., Thompson R.C., Turner D.L. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp. Cell Res. 2008;314:2618–2633. doi: 10.1016/j.yexcr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franke K., Otto W., Johannes S., Baumgart J., Nitsch R., Schumacher S. miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J. 2012;31:2908–2921. doi: 10.1038/emboj.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kutsche L.K., Gysi D.M., Fallmann J., Lenk K., Petri R., Swiersy A., et al. Combined experimental and system-level analyses reveal the complex regulatory network of miR-124 during human neurogenesis. Cell Syst. 2018;7:438–452.e8. doi: 10.1016/j.cels.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng L.C., Pastrana E., Tavazoie M., Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabashi E., Lin L., Tradewell M.L., Dion P.A., Bercier V., Bourgouin P., et al. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum. Mol. Genet. 2010;19:671–683. doi: 10.1093/hmg/ddp534. [DOI] [PubMed] [Google Scholar]
- 15.Diaper D.C., Adachi Y., Sutcliffe B., Humphrey D.M., Elliott C.J., Stepto A., et al. Loss and gain of Drosophila TDP-43 impair synaptic efficacy and motor control leading to age-related neurodegeneration by loss-of-function phenotypes. Hum. Mol. Genet. 2013;22:1539–1557. doi: 10.1093/hmg/ddt005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraemer B.C., Schuck T., Wheeler J.M., Robinson L.C., Trojanowski J.Q., Lee V.M., et al. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 2010;119:409–419. doi: 10.1007/s00401-010-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sephton C.F., Good S.K., Atkin S., Dewey C.M., Mayer P., 3rd, Herz J., et al. TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu L.S., Cheng W.C., Hou S.C., Yan Y.T., Jiang S.T., Shen C.K. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- 19.Bouhon I.A., Kato H., Chandran S., Allen N.D. Neural differentiation of mouse embryonic stem cells in chemically defined medium. Brain Res. Bull. 2005;68:62–75. doi: 10.1016/j.brainresbull.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 20.Kohama C., Kato H., Numata K., Hirose M., Takemasa T., Ogura A., et al. ES cell differentiation system recapitulates the establishment of imprinted gene expression in a cell-type-specific manner. Hum. Mol. Genet. 2012;21:1391–1401. doi: 10.1093/hmg/ddr577. [DOI] [PubMed] [Google Scholar]
- 21.Graus-Porta D., Blaess S., Senften M., Littlewood-Evans A., Damsky C., Huang Z., et al. Beta1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron. 2001;31:367–379. doi: 10.1016/s0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 22.Schmid R.S., Anton E.S. Role of integrins in the development of the cerebral cortex. Cereb. Cortex. 2003;13:219–224. doi: 10.1093/cercor/13.3.219. [DOI] [PubMed] [Google Scholar]
- 23.Dulabon L., Olson E.C., Taglienti M.G., Eisenhuth S., McGrath B., Walsh C.A., et al. Reelin binds alpha3beta1 integrin and inhibits neuronal migration. Neuron. 2000;27:33–44. doi: 10.1016/s0896-6273(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 24.Rajasethupathy P., Fiumara F., Sheridan R., Betel D., Puthanveettil S.V., Russo J.J., et al. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y., Shu X., Liu D., Shang Y., Wu Y., Pei L., et al. EPAC null mutation impairs learning and social interactions via aberrant regulation of miR-124 and Zif268 translation. Neuron. 2012;73:774–788. doi: 10.1016/j.neuron.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou Q., Ruan H., Gilbert J., Wang G., Ma Q., Yao W.D., et al. MicroRNA miR124 is required for the expression of homeostatic synaptic plasticity. Nat. Commun. 2015;6 doi: 10.1038/ncomms10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iguchi Y., Katsuno M., Niwa J., Takagi S., Ishigaki S., Ikenaka K., et al. Loss of TDP-43 causes age-dependent progressive motor neuron degeneration. Brain. 2013;136:1371–1382. doi: 10.1093/brain/awt029. [DOI] [PubMed] [Google Scholar]
- 28.Ma X.R., Prudencio M., Koike Y., Vatsavayai S.C., Kim G., Harbinski F., et al. TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A. Nature. 2022;603:124–130. doi: 10.1038/s41586-022-04424-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown A.L., Wilkins O.G., Keuss M.J., Hill S.E., Zanovello M., Lee W.C., et al. TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature. 2022;603:131–137. doi: 10.1038/s41586-022-04436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gascon E., Lynch K., Ruan H., Almeida S., Verheyden J.M., Seeley W.W., et al. Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat. Med. 2014;20:1444–1451. doi: 10.1038/nm.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng C., Wynshaw-Boris A., Zhou F., Kuo A., Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 32.Muranishi Y., Terada K., Inoue T., Katoh K., Tsujii T., Sanuki R., et al. An essential role for RAX homeoprotein and NOTCH-HES signaling in Otx2 expression in embryonic retinal photoreceptor cell fate determination. J. Neurosci. 2011;31:16792–16807. doi: 10.1523/JNEUROSCI.3109-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakai K., Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem. Biophys. Res. Commun. 1997;237:318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- 34.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaya T., Omori Y., Kuwahara R., Furukawa T. ICK is essential for cell type-specific ciliogenesis and the regulation of ciliary transport. EMBO J. 2014;33:1227–1242. doi: 10.1002/embj.201488175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Omori Y., Chaya T., Katoh K., Kajimura N., Sato S., Muraoka K., et al. Negative regulation of ciliary length by ciliary male germ cell-associated kinase (Mak) is required for retinal photoreceptor survival. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22671–22676. doi: 10.1073/pnas.1009437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irie S., Sanuki R., Muranishi Y., Kato K., Chaya T., Furukawa T. Rax homeoprotein regulates photoreceptor cell maturation and survival in association with Crx in the postnatal mouse retina. Mol. Cell. Biol. 2015;35:2583–2596. doi: 10.1128/MCB.00048-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Itoh Y., Moriyama Y., Hasegawa T., Endo T.A., Toyoda T., Gotoh Y. Scratch regulates neuronal migration onset via an epithelial-mesenchymal transition-like mechanism. Nat. Neurosci. 2013;16:416–425. doi: 10.1038/nn.3336. [DOI] [PubMed] [Google Scholar]
- 39.Tsutsumi R., Chaya T., Tsujii T., Furukawa T. The carboxyl-terminal region of SDCCAG8 comprises a functional module essential for cilia formation as well as organ development and homeostasis. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.101686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugiyama T., Yamamoto H., Kon T., Chaya T., Omori Y., Suzuki Y., et al. The potential role of Arhgef33 RhoGEF in foveal development in the zebra finch retina. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-78452-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Omori Y., Chaya T., Yoshida S., Irie S., Tsujii T., Furukawa T. Identification of G protein-coupled receptors (GPCRs) in primary cilia and their possible involvement in body weight control. PLoS One. 2015;10 doi: 10.1371/journal.pone.0128422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaya T., Ishikane H., Varner L.R., Sugita Y., Maeda Y., Tsutsumi R., et al. Deficiency of the neurodevelopmental disorder-associated gene Cyfip2 alters the retinal ganglion cell properties and visual acuity. Hum. Mol. Genet. 2022;31:535–547. doi: 10.1093/hmg/ddab268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metsalu T., Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge S.X., Son E.W., Yao R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinformatics. 2018;19:534. doi: 10.1186/s12859-018-2486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaya T., Tsutsumi R., Varner L.R., Maeda Y., Yoshida S., Furukawa T. Cul3-Klhl18 ubiquitin ligase modulates rod transducin translocation during light-dark adaptation. EMBO J. 2019;38 doi: 10.15252/embj.2018101409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J., Song X., Kuang F., Zhang Q., Xie Y., Kang R., et al. NUPR1 is a critical repressor of ferroptosis. Nat. Commun. 2021;12:647. doi: 10.1038/s41467-021-20904-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsumoto N., Kubo A., Liu H., Akita K., Laub F., Ramirez F., et al. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood. 2006;107:1357–1365. doi: 10.1182/blood-2005-05-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data is available on GEO (GSE196356).