Summary
m6A modification plays an important role in regulating mammalian neurogenesis. However, whether and how the major cytoplasmic m6A readers, YTHDF1, YTHDF2, and YTHDF3 mediate this process is still not clear. Here, we demonstrate that Ythdf1 and Ythdf2 double deletion but not individual knockout recapitulates the phenotype of Mettl14 knockout in cortex. In addition, we find that Mettl14 knockout in retina causes protracted proliferation of retinal progenitors, decreased numbers of retinal neurons, and disturbed laminar structure. This phenotype is only reproduced when Ythdf1, Ythdf2, and Ythdf3 are knocked out simultaneously in retina. Analysis of YTHDF target mRNAs in mouse cortex and retina reveals abundant overlapping mRNAs related to neurogenesis that are recognized and regulated by both YTHDF1 and YTHDF2. Together our results demonstrate that the functionally redundant YTHDFs mediate m6A regulation of cortical and retinal neurogenesis.
Subject areas: Molecular biology, Molecular Genetics, Neuroscience, Molecular neuroscience
Graphical abstract
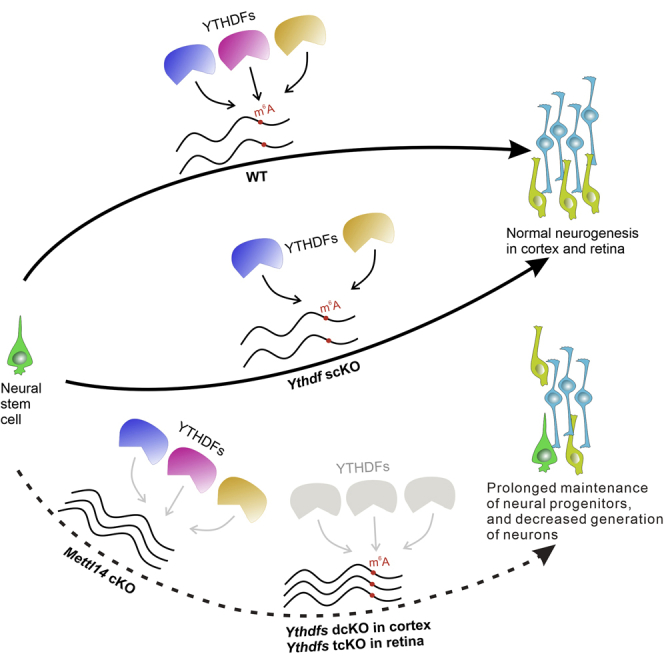
Highlights
-
•
YTHDF1 and YTHDF2 have redundant functions in m6A regulation of cortical neurogenesis
-
•
m6A modification plays a critical role in retinal neurogenesis
-
•
Only Ythdf1, Ythdf2, and Ythdf3 triple deletion disrupts retinal neurogenesis
-
•
YTHDF1 and YTHDF2 share a large pool of target mRNAs related to neurogenesis
Molecular biology; Molecular Genetics; Neuroscience; Molecular neuroscience.
Introduction
N6-methyladenosine (m6A) modification, as the most abundant internal modification on mRNA, is dynamically regulated by its “writers” and “erasers”, and recognized by its “readers” (Meyer and Jaffrey, 2017). These readers include the YTH domain-containing proteins such as YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2, and bind the m6A site directly to exert multiple functions such as mRNA splicing, alternative polyadenylation, nucleus export, degradation, and translation (He and He, 2021; Zaccara et al., 2019). However, the functional relations among different YTHDFs readers remain controversial: initial studies suggested that different YTHDFs have distinct functions and working mechanisms (Shi et al., 2017; Wang et al., 2014, 2015b); later studies argued that different YTHDFs work redundantly (Kontur et al., 2020; Lasman et al., 2020; Zaccara and Jaffrey, 2020); a recent study showed that YTHDF1 and YTHDF2 could work synergistically to regulate the same pathway and process (Yu et al., 2021). Thus, further clarification is needed to elucidate the functions of YTHDFs.
Neurogenesis is the process of neural stem cell proliferation, migration, and differentiation that is precisely regulated by diverse factors and mechanisms. In neocortical neurogenesis, self-renewal radial glial cells in the ventricular zone (VZ) differentiate into the intermediate progenitors in the subventricular zone (SVZ) and then give rise to neurons that migrate into the specified cortical layers in an “inside-out” order (Fishell and Kriegstein, 2003; Woodworth et al., 2012). In retinal neurogenesis, all retinal neurons are derived from the retinal progenitors with a particular order and organized into the proper laminar structure from embryonic to postnatal stages (Cepko, 2014).
Two studies reported that deletion of the m6A writer METTL14 results in severe defects in cortical neurogenesis (Wang et al., 2018; Yoon et al., 2017). However, these two studies using the same Nestin-cre to ablate Mettl14 came up with two contrary mechanisms to explain their results (Wang et al., 2018; Yoon et al., 2017), suggesting that further exploration and clarification is needed to decide the role of m6A modification in cortical neurogenesis. In addition, whether and how the m6A readers mediate m6A modification in cortical neurogenesis remain to be studied. Moreover, whether m6A modification and its readers work similarly or distinctly to regulate neurogenesis in different brain regions remain elusive.
Here, we used Mettl14 conditional knockout (cKO) by Nestin-cre and Six3-cre to examine the functions of m6A modification in cortical and retinal neurogenesis, respectively. By generating single, double, or triple deletion of Ythdf1, Ythdf2 and Ythdf3, and comparing their phenotypes with that of Mettl14 cKO in cortex and retina, we investigated how YTHDFs mediate m6A modification and whether they are redundant in cortical and retinal neurogenesis. Finally, we identified target mRNAs of YTHDFs. Together, our results reveal mechanisms by which the m6A readers YTHDFs mediate m6A modification in neurogenesis of different brain regions.
Results
Mettl14 deletion delays cortical neurogenesis and reduces production of neurons
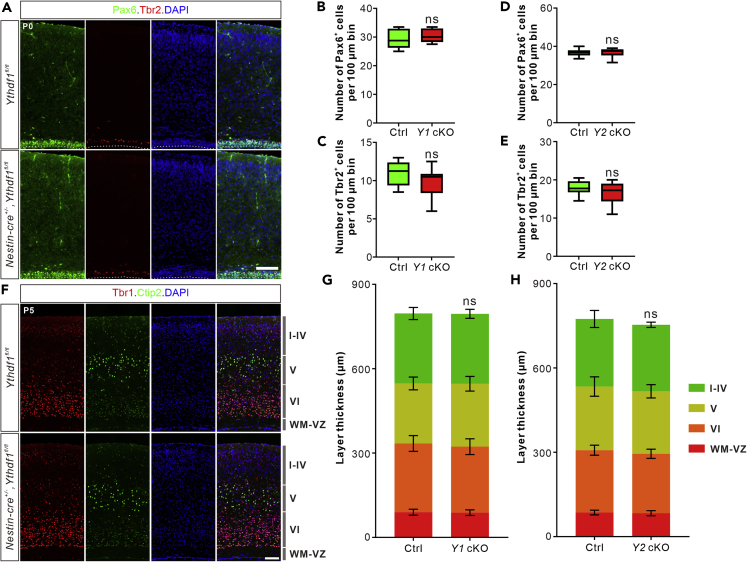
To further explore and clarify the function of m6A modification in cortical neurogenesis, we first checked m6A modification level in the embryonic cortex. Immunostaining with a widely used m6A antibody revealed high m6A level in the developing cortex (Figure S1A). Abundant m6A modification in the radial glia cells and intermediated progenitors supports the function of m6A modification in the cortical neurogenesis. To test this, we generated Mettl14 conditional knockout (cKO) by crossing Mettl14 fl/fl with Nestin-cre+/− to get Nestin-cre+/−, Mettl14 fl/fl. The knockout efficiency of METTL14 protein in the cKO mouse was verified by anti-METTL14 immunostaining, which demonstrated that METTL14 was ablated successfully in the cKO mouse cortex (Figure S1B). The numbers of Pax6+ radial glia cells in the ventricular zone (VZ) and Tbr2+ intermediate progenitors were dramatically larger in the Mettl14 cKO cortex compared with control at P5 (postnatal day 5) (Figures S1C–S1E). We continued to use Tbr1 to label layer VI neurons and Ctip2 to label layer V neurons (Woodworth et al., 2012). The quantification and measurement also revealed significant increase in the thickness of layers WM-SVZ-VZ including white matter (WM), subventricular zone (SVZ) composed by intermediate progenitors, and VZ made up of radial glia cells (Figures S1F and S1G). Larger numbers of radial glia cells and intermediate progenitors and thicker SVZ and VZ indicate that the maintenance of radial glia cells is abnormally extended to postnatal stages in the Mettl14 cKO cortex. In contrast, dramatic decreases in the thickness of layers VI, V, and I–IV were observed in the cKO cortex, indicating the defects of neuron production in the Mettl14 cKO cortex (Figures S1F and S1G). These results demonstrate that knockout of the m6A writer METTL14 could delay cortical neurogenesis to postnatal stage and lead to defects in the production of neurons.
Ythdf1 or Ythdf2 conditional knockout mice show no defect in cortical neurogenesis
To identify which reader(s) directly mediate functions of m6A modification in cortical neurogenesis, we first investigated the expression patterns of YTHDF1 and YTHDF2 in the developing cortex. The co-staining of YTHDF1 or YTHDF2 with Pax6 showed YTHDF1 and YTHDF2 are highly expressed in the radial glia cells in embryonic cortex from E11 (embryonic day 11) to E13.5 (Figures 1A and 1B), suggesting an early role of YTHDFs in cortical neurogenesis. With decreases of radial glia cells, the expression of YTHDFs also show decreases from E15.5 to P0 (Figures 1A and 1B). We first generated conditional knockouts of Ythdf1 and Ythdf2 respectively, by crossing Ythdf1fl/fl and Ythdf2fl/fl with Nestin-cre to get Nesin-cre+/−,Ythdf1fl/fl (Ythdf1 scKO) and Nesin-cre+/−,Ythdf1fl/fl (Ythdf2 scKO) mice. Anti-YTHDF1 and YTHDF2 immunostaining showed that these reader proteins were started to be ablated from E13.5 and were completely knocked out after E15.5 (Figures 1A and 1B).
Figure 1.
YTHDF1 and YTHDF2 are highly expressed and efficiently ablated in the Ythdf1 and Ythdf2 cKO cortices
(A and B) Representative images showing immunostaining of YTHDF1 (A) or YTHDF2 (B) and Pax6 in coronal sections of Ythdf1 (A) or Ythdf2 (B) scKO and littermate controls at E11 or E11.5, E13.5, E15.5, E17.5, and P0. As shown, YTHDF1 and YTHDF2 are highly expressed in the radial glia cells from E11. Partial knockout of YTHDF1 and YTHDF2 could be observed from E13.5, and complete knockout was achieved from E15.5. Scale bars, 50 μm.
We then checked the cortical neurogenesis of Ythdf1 and Ythdf2 scKO mice. The numbers of either Pax6-labeled radial glia cells or Tbr2-marked intermediate progenitors showed no difference between the scKO and its corresponding control at P0 (Figures 2A–2E). The scKO cortex also exhibited no difference in thickness of cortical layers identified by Tbr1/Ctip2 co-staining at P5 (Figures 2F–2H). Together, these results indicate that knockout of Ythdf1 or Ythdf2 individually does not affect cortical neurogenesis, and thus cannot recapitulate the phenotypes of Mettl14 cKO.
Figure 2.
Ythdf1 or Ythdf2 conditional knockout mice show no defect in cortical neurogenesis
(A) Representative images of coronal sections of P0 Ythdf1fl/fl and Nestin-cre+/−,Ythdf1fl/fl cortices immunostained with Pax6 and Tbr2 to label radial glia cells and intermediate progenitors, respectively.
(B–E) Quantification of Pax6+ radial glial cells (B and D) and Tbr2+ intermediated progenitors (C and E). Numbers of corresponding cells per 100 μm bin of cortical coronal sections were quantified and showed no difference between the Ythdf1 scKO (Y1 cKO, n = 8 sections) or Ythdf2 scKO (Y2 cKO, n = 8 sections) and their corresponding littermate controls (Ctrl, n = 8 sections) at P0. Data are represented as box and whisker plots: p = 0.43 for B, p = 0.21 for C; p = 0.96 for D, p = 0.29 for E; ns, not significant; by unpaired Student’s t test.
(F) Representative images of coronal sections of P5 Ythdf1fl/fl and Nestin-cre+/−,Ythdf1fl/fl cortices immunostained with Tbr1 and Ctip2 to distinguish different cortical layers.
(G and H) Measurement and quantification of cortical layer thickness. The thickness of layers WM-VZ, VI, V, I–IV, and whole cortex was measured and showed no difference between the scKO (n = 30 sections for Y1 cKO in G, n = 21 sections for Y2 cKO in (H)) and littermate controls (n = 30 sections for Ctrl in G, n = 21 sections for Ctrl in H) at P5. Data are represented as accumulative histogram: in G, p = 0.56 for layers WM-VZ, p = 0.20 for layer VI, p = 0.13 for layer V, p = 0.96 for layers I–IV, p = 0.94 for whole cortex; in H, p = 0.38 for layers WM-VZ, p = 0.067 for layer VI, p = 0.66 for layer V, p = 0.60 for layers I–IV, p = 0.069 for whole cortex; ns, not signification; by unpaired Student’s t test. Scale bars, 100 μm.
Double knockout of Ythdf1 and Ythdf2 causes cortical neurogenesis defects
Because individual deletion of Ythdf1 or Ythdf2 could not affect cortical neurogenesis as Mettl14 cKO, we further generated the double conditional knockout (dcKO) of Ythdf1 and Ythdf2 (Nesin-cre+/−, Ythdf1fl/fl, Ythdf2fl/fl, Y1Y2 dcKO) to ablate Ythdf1 and Ythdf2 simultaneously and examine the effect of loss-of-function of both readers on cortical neurogenesis. We first verified the efficient knockout of both YTHDF1 and YTHDF2 in the dcKO mice with anti-YTHDF1 and YTHDF2 immunostaining (Figures 3A and 3B).
Figure 3.
Simultaneous knockout of Ythdf1 and Ythdf2 causes cortical neurogenesis defects
(A and B) Representative images of coronal sections of Ythdf1fl/fl,Ythdf2fl/fl (Ctrl) and Nestin-cre+/−,Ythdf1fl/fl,Ythdf2fl/fl (Y1Y2 dcKO) cortices immunostained with YTHDF1 (A) and YTHDF2 (B) at P0. YTHDF1 and YTHDF2 were knocked out efficiently in the dcKO mice.
(C and D) Quantification of Pax6+ radial glial cells (C) and Tbr2+ intermediated progenitors (D). Numbers of corresponding cells per 100 μm bin of cortical coronal sections were quantified and showed increases in the dcKO (n = 10 sections) compared with the littermate control (Ctrl, n = 10 sections) at P0. Data are represented as box and whisker plots: ∗∗∗∗p = 3.27 × 10−5 for C, ∗∗∗p = 0.00018 for D; by unpaired Student’s t test.
(E) Representative images of Ki67 and BrdU co-labeling of coronal cortical sections at E17.5. Pregnant mice were injected with BrdU 24 h before collecting embryos. The arrowheads indicate the Ki67+BrdU+ cells and the arrows point to the Ki67−BrdU+ cells.
(F) Quantification of results in E. The percentage of Ki67−BrdU+/BrdU+, representing the ratio of cells exiting cell cycle, decreased in the dcKO (n = 10 sections) compared with littermate controls (n = 10 sections). Data are represented as box and whisker plots: ∗∗p = 0.0091, by unpaired Student’s t test.
(G) Measurement and quantification of the thickness of cortical layers at P5. The thickness of layers WM-VZ and layer VI showed no difference between the dcKO and control mice. The thickness of layer V, layers I-IV, and whole cortex decreased in the dcKO cortex (n = 27 sections) compared with littermate controls (n = 23 sections). Data are represented as an accumulative histogram: p = 0.10 for layers WM-VZ, p = 0.44 for layer VI, ∗p = 0.025 for layer V, ∗∗∗∗p = 9.10E-12 for layers I–IV, ∗∗∗∗p = 2.98E-05 for whole cortex; ns, not signification; by unpaired Student’s t-test. Scale bars, 100 μm (A,B) and 50 μm (E).
Next, we examined the radial glia cells and the intermediate progenitors in the dcKO cortex. Significant increases in the numbers of Pax6+ radial glia cells and Tbr2+ intermediate progenitors were observed in the dcKO cortex at P0 (Figures 3C and 3D). To determine whether cell cycle exit of radial glia cells was affected in the dcKO cortex, we performed co-staining of BrdU and Ki67 in E17.5 cortical sections collected after 24 h of BrdU injection (Figure 3E). There is a significant decrease in the percentage of Ki67−BrdU+/BrdU+, suggesting a delay in cell cycle exit of radial glial cells in the dcKO cortex (Figure 3F). Then we assessed the cortical layer thickness. Although the thickness of layers WM-VZ and earlier born layer VI did not change, the thickness of layer V and later born layers I-IV significantly decreased (Figure 3G).
Together, our findings demonstrate that the dcKO of Ythdf1 and Ythdf2 causes prolonged cell cycle and maintenance of radial glia cells, and decreased generation of neurons in the cortex, recapitulating the phenotypes of Mettl14 cKO. These results suggest that YTHDF1 and YTHDF2 have redundant functions in mediating m6A regulation of cortical neurogenesis.
We continued to check the cortical neurogenesis of the Ythdf3 cKO mice using Nestin-cre. We found no difference in the numbers of Pax6-labeled radial glia cells or Tbr2-marked intermediate progenitors between Ythdf3 cKO and littermate control at P0 (Figures S2A and S2B). The Ythdf3 cKO cortex also showed no difference in thickness of cortical layers identified by Tbr1/Ctip2 co-staining at P5 (Figure S2C). Together, these results suggest that Ythdf3 cKO does not affect cortical neurogenesis.
Mettl14 ablation in retina protracts retinal neurogenesis and leads to reduced generation of late-born neurons
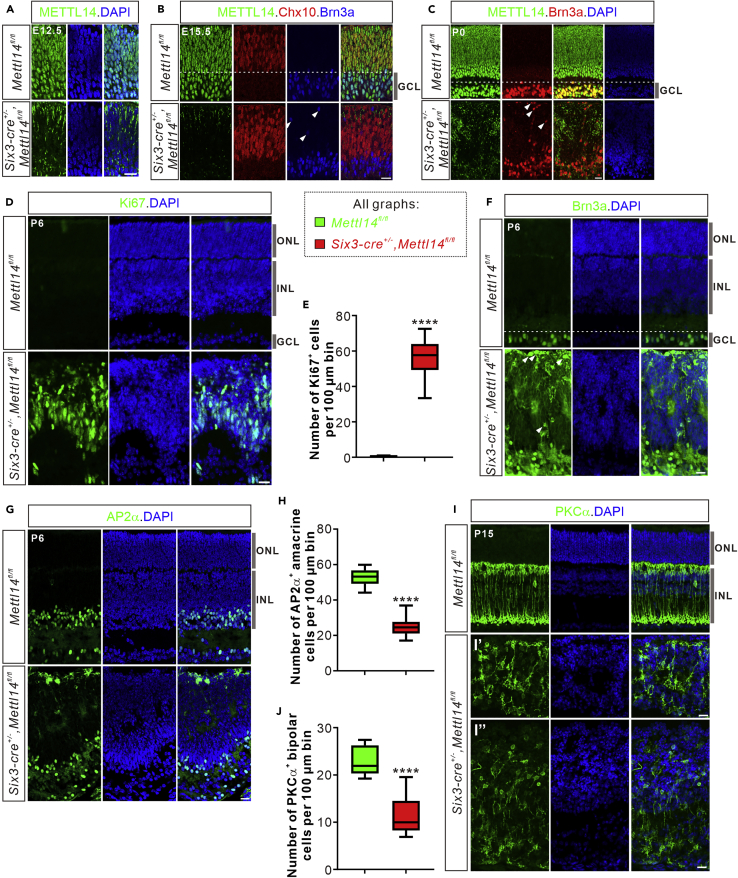
Next we tested whether and how m6A modification regulates neurogenesis in other brain regions. To explore whether m6A modification mediates retinal neurogenesis, we first checked the m6A modification level in the embryonic retina. Abundant m6A modification was detected in the retinal progenitors and retinal ganglion cells (RGCs) at E14.5 (Figure S3A). Then, we generated Mettl14 cKO mouse by crossing Mettl14 fl/fl with Six3-cre mouse which has been widely used in the studies of retinal neurogenesis (Furuta et al., 2000; Krishnaswamy et al., 2015; Luo et al., 2012; Sapkota et al., 2014). METTL14 immunostaining in E12.5, E15.5, and P0 retinal vertical sections revealed that METTL14 expression was high in the developing retinas (control retinas in Figures 4A–4C), and was efficiently ablated in the Mettl14 cKO retina (Figures 4A–4C). Furthermore, Mettl14 cKO retina exhibited significantly lower m6A levels compared with littermate control (Figure S3A), suggesting that m6A modification is largely restricted in retina after Mettl14 deletion.
Figure 4.
Mettl14 ablation in retina protracts retinal neurogenesis and leads to reduced generation of late-born neurons
(A–C) Representative images of vertical sections from Mettl14 cKO (Six3-cre+/−,Mettl14fl/fl) and control (Mettl14fl/fl) retinas immunostained with METTL14 at E12.5 (A), E15.5 (B), and P0 (C). METTL14 expression is high in the control retinas and is efficiently ablated in the Mettl14 cKO retinas. The residual bright signals in the cKO sections are non-specific immunofluorescence signals with this METTL14 Ab. Chx10 and Brn3a are the markers for retinal progenitor cells and retinal ganglion cells, respectively. The arrowheads indicate the misplaced Brn3a+ RGCs (B and C). GCL, ganglion cell layer.
(D) Representative images of P6 Mettl14 cKO and control retinas immunostained with the proliferation cell marker Ki67. ONL, outer nuclear layer; INL, inner nuclear layer.
(E) Quantification of Ki67+ retinal progenitor cell numbers in (D). The cell numbers per 100 μm bin of retinal vertical sections were quantified. The retinal progenitors were abnormally maintained in the Mettl14 cKO retina at P6. Data are represented as box and whisker plots: n = 12 sections for Mettl14 cKO and control, respectively; ∗∗∗∗p = 7.00E-15; by unpaired Student’s t test.
(F) Representative images of P6 Mettl14 cKO and control retinas immunostained with Brn3a. Misplaced Brn3a+ retinal ganglion cells (arrowheads) were found in the Mettl14 cKO retina.
(G) Representative images of P6 Mettl14 cKO and control retinas immunostained with AP2α. AP2α marks the amacrine cells.
(H) Quantification of AP2α+ amacrine cell numbers in G. Numbers of AP2α+ amacrine cells per 100 μm bin of retinal vertical sections were quantified and decreased in the Mettl14 cKO (n = 12 sections) compared with littermate controls (n = 12 sections) at P6. Data are represented as box and whisker plots: ∗∗∗∗p = 1.27E-11; by unpaired Student’s t test.
(I) Representative images of the vertical sections immunostained with the bipolar cell marker PKCα from Mettl14 cKO and control retinas at P15. Please note that the cKO retinas are severely deformed at P15, and their thicknesses are not even with some areas becoming much thinner (I’) and others thicker (I’’) compared with control retinas. The organization of bipolar cells was totally messed up.
(J) Quantification of PKCα+ bipolar cell numbers in I. Numbers of PKCα+ bipolar cells per 100 μm bin of retinal vertical sections were quantified and decreased in the Mettl14 cKO (n = 17 sections) compared with littermate controls (n = 15 sections) at P15. Data are represented as box and whisker plots: ∗∗∗∗p = 6.96E-11; by unpaired Student’s t test. Scale bars, 20 μm (A–C, D, F, G, and I).
To check the retinal neurogenesis in Mettl14 cKO, we examined the major retinal cell and neuron types by immunostaining of vertical retinal sections with different markers: Ki67, a proliferation marker; Brn3a, a marker for retinal ganglion cells (Brodie-Kommit et al., 2021); AP2α, a marker for amacrine cells (Hicks et al., 2018); PKCα, a marker for rod bipolar cells (Lu et al., 2013). During mouse retinal neurogenesis, retinal progenitors are largely depleted by P6 (control retina in Figure 4D). However, a substantial number of Ki67+ cells were still present in the Mettl14 cKO retina (Figures 4D and 4E). Retinal ganglion cells, as the early born neuron type in the retina, are normally located at the basal ganglion cell layer (GCL) after being differentiated from retinal progenitors at E15.5 and P0 (control retina in Figures 4B and 4C) (Cepko, 2014). However, in the Mettl14 cKO retina, some of retinal ganglion cells were dislocated at the apical retina revealed by Brn3a immunostaining (Figures 4B and 4C). In addition, the distribution of Brn3a+ retinal ganglion cells in the basal retina was also disturbed in the Mettl14 cKO retina at P6 (Figure 4F). AP2α+ amacrine cells dramatically decreased in the Mettl14 cKO retina compared with control retina at P6 (Figures 4G and 4H). Generation and patterning of retinal bipolar cells were severely impaired (Figures 4I and 4J). Besides the decreased numbers of retinal neurons, the structure of the Mettl14 cKO retinas was deformed and the retinal layers were disorganized compared with control retinas (Figures 4C, 4D, 4F, 4G, and 4I). All together, these results indicate that Mettl14 cKO in retina leads to extended retinal neurogenesis, decreased generation of late-born neurons, and deformed retinal patterning.
Single or double conditional knockout of Ythdf1 and Ythdf2 in retina does not affect retinal neurogenesis
Next, we wanted to identify which readers mediate m6A functions in retinal neurogenesis. We first focused on YTHDF1 and YTHDF2 which are expressed in the developing retinas (Niu et al., 2022) (control retinas in Figure S4A). We generated Ythdf1 and Ythdf2 single conditional knockouts in retina using Six3-cre: Six3-cre+/−, Ythdf1fl/fl (Ythdf1 scKO) and Six3-cre+/−,Ythdf2fl/fl (Ythdf2 scKO). We have previously shown that although retinal ganglion cell dendrite branching increases, retinal neurogenesis is not affected in the Ythdf2 scKO (Niu et al., 2022).
We continued to check the Ythdf1 scKO. Immunostaining of YTHDF1 confirmed that YTHDF1 was efficiently knocked out in the Ythdf1 scKO retinas (Figure S4A). Immunostaining of Brn3a, AP2α and PKCα on the retinal vertical sections showed that generation of retinal ganglion cells, amacrine cells, or bipolar cells was not affected in the Ythdf1 scKO retinas at P6 and P15 (Figures S4B–S4F). These results and our previous findings (Niu et al., 2022) together suggest that knockout of Ythdf1 or Ythdf2 alone does not affect retinal neurogenesis.
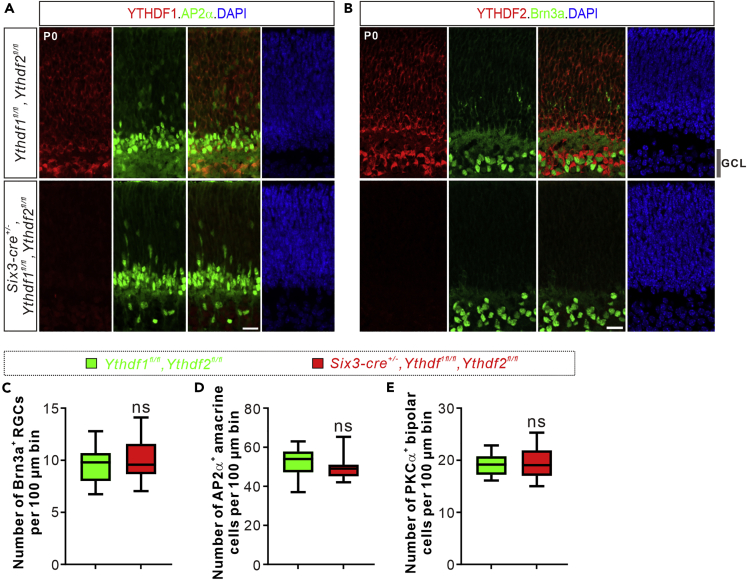
Because YTHDF1 and YTHDF2 function redundantly to mediate m6A modification in regulating cortical neurogenesis (Figures 2 and 3), we wondered if there is a similar mechanism in the retinal neurogenesis. So we generated double conditional knockout of Ythdf1 and Ythdf2 (Y1Y2 dcKO) in retina (Six3-cre+/−, Ythdf1fl/fl, and Ythdf2fl/fl) to ablate Ythdf1 and Ythdf2 in the retina simultaneously (Figures 5A and 5B). However, we surprisingly found that numbers of retinal ganglion cells, amacrine cells, or bipolar cells were not changed in the Y1Y2 dcKO retinas (Figures 5C–5E). Together, these data suggest that single or double conditional knockout of Ythdf1 and Ythdf2 in retina has no effect on retinal neurogenesis.
Figure 5.
Double conditional knockout of Ythdf1 and Ythdf2 in retina does not disturb retinal neurogenesis
(A and B) Representative images of the retinal vertical sections co-stained with YTHDF1 and AP2α (A), YTHDF2 and Brn3a (B) from Ythdf1fl/fl,Ythdf2fl/fl and Six3-cre+/−,Ythdf1fl/fl,Ythdf2fl/fl (Y1Y2 dcKO) retinas at P0.
(C–E) Quantification of numbers of Brn3a+ retinal ganglion cells (C), AP2α+ amacrine cells (D), and PKCα+ bipolar cells (E) in the Y1Y2 dcKO and control retinas. Numbers of corresponding cell types per 100 μm bin of retinal vertical sections were quantified and showed no difference between the dcKO and control retinas. All quantification data are represented as box and whisker plots: in (C), n = 16 sections for dcKO, n = 16 sections for control, p = 0.67; in D, n = 16 sections for dcKO, n = 16 sections for control, p = 0.18; in E, n = 22 sections for dcKO, n = 12 sections for control, p = 0.94; ns, not signification; by unpaired Student’s t test. Scale bars, 20 μm (A and B).
Triple knockout of Ythdf1, Ythdf2, and Ythdf3 in retina extends retinal progenitor proliferation and decreases generation of late-born retinal neurons
Because single or double conditional knockout of Ythdf1 and Ythdf2 cannot recapitulate the phenotype of Mettl14 cKO in retina, we continued to check another m6A reader, YTHDF3. We first checked expression of Ythdf3 in retina using in situ hybridization (ISH), which detected signals in the developing retina (control retina in Figure S5A). Then we generated Ythdf3 cKO mice (Figure S5B), and Six3-Cre induced efficient knockout of Ythdf3 in retina (Six3-cre+/−, Ythdf3fl/fl, and Ythdf3 cKO) (Figure S5A). We further checked retinal neurogenesis of Ythdf3 cKO in retina. As shown in Figures S5C–S5E, generation of retinal ganglion cells, amacrine cells, or bipolar cells was not affected in the Ythdf3 cKO retinas.
We continued to generate triple conditional knockout of Ythdf1, Ythdf2, and Ythdf3 (Y1Y2Y3 tcKO) in retina (Six3-cre+/−, Ythdf1fl/fl, Ythdf2fl/fl, and Ythdf3fl/fl) which exhibited efficient knockout of YTHDF1 and YTHDF2 by immunostaining (Figures 6A and 6B) and Ythdf3 by ISH (Figure 6C) at P0. Then we checked retinal neurogenesis in the Y1Y2Y3 tcKO mice. Ki67+ retinal progenitors were abnormally maintained in the tcKO retinas at P6 (Figures 6D and 6E). Retinal ganglion cells showed disordered location in the tcKO retinas at P6 (Figure 6F). Numbers of AP2α+ amacrine cells at P6 and PKCα+ bipolar cells at P15 were decreased in the tcKO retinas (Figures 6G–6J). In addition, retinal layers were also disorganized and neurons were misplaced in the tcKO retinas (Figures 6D, 6F, 6G, and 6I). These results demonstrate that Y1Y2Y3 tcKO leads to prolonged retinal neurogenesis and impaired generation of late-born retinal neurons.
Figure 6.
Triple knockout of Ythdf1, Ythdf2, and Ythdf3 in retina extends retinal progenitor proliferation and decreases generation of late-born retinal neurons
(A and B) Representative images of the vertical sections immunostained with YTHDF1 and YTHDF2 showing efficient knockout of Ythdf1 and Ythdf2 in theY1Y2Y3 triple cKO retinas at P0.
(C) Representative ISH images of the vertical sections showing efficient ablation of Ythdf3 in the Y1Y2Y3 triple cKO retinas at P0.
(D) Representative images of the retinal vertical sections immunostained with the retinal progenitor marker Ki67. While Ki67+ cells were absent from the control (Ythdf1fl/fl, Ythdf2fl/fl, and Ythdf3fl/fl) retinas at P6, there were still a lot of Ki67+ cells in the Y1Y2Y3 tcKO (Six3-cre+/−, Ythdf1fl/fl, Ythdf2fl/fl, and Ythdf3fl/fl) retinas at P6.
(E) Quantification of numbers of Ki67+ retinal progenitors in (A). Numbers of Ki67+ retinal progenitors per 100 μm bin of retinal vertical sections were quantified and abnormally maintained in the Y1Y2Y3 triple cKO (n = 20 sections) compared with littermate controls (n = 13 sections). Data are represented as box and whisker plots: ∗∗∗∗p = 1.67E-09, by unpaired Student’s t test.
(F) Representative images of the retinal vertical sections immunostained with Brn3a, the retinal ganglion cell marker. The location and organization of Brn3a+ retinal ganglion cells were disturbed in the Y1Y2Y3 tcKO retinas at P6. Arrowheads indicate the misplaced Brn3a+ retinal ganglion cells.
(G) Representative images of the vertical sections with amacrine cells marked by AP2α in the Y1Y2Y3 tcKO and control retinas at P6. AP2α+ amacrine cells were decreased and disorganized in the Y1Y2Y3 tcKO retinas. Arrowheads indicate the misplaced AP2α+ amacrine cells.
(H) Quantification of numbers of AP2α+ amacrine cells in (D). Numbers of AP2α+ amacrine cells per 100 μm bin of retinal vertical sections were quantified and showed decreases in the Y1Y2Y3 tcKO (n = 20 sections) compared with littermate controls (n = 14 sections). Data are represented as box and whisker plots: ∗∗∗∗p = 8.01E-09, by unpaired Student’s t test.
(I) Representative images of the vertical sections with bipolar cells labeled by PKCα in the Y1Y2Y3 tcKO and control retinas at P15. The organization of PKCα+ bipolar cells was severely disturbed.
(J) Quantification of numbers of PKCα+ bipolar cells in (F). Numbers of PKCα+ bipolar cells per 100 μm bin of retinal vertical sections were quantified and showed decreases in the Y1Y2Y3 tcKO (n = 20 sections) compared with littermate controls (n = 24 sections). Data are represented as box and whisker plots: ∗p = 0.011, by unpaired Student’s t test. Scale bars, 20 μm (A–C, D, F, G, and I).
All these data suggest that YTHDF1, YTHDF2 and YTHDF3 function redundantly to mediate m6A modification in retinal neurogenesis, and simultaneous knockout of Ythdf1, Ythdf2, and Ythdf3 can recapitulate the retinal neurogenesis defects caused by Mettl14 conditional knockout in retina.
We next continued to test whether other YTHDFs were upregulated to compensate the single Ythdf cKO. By RT-qPCR analysis, the expression levels of Ythdf2 and Ythdf3 mRNAs were not changed after Ythdf1 knockout (Figures S6A and S6B). In the Ythdf2 cKO retina, the expression levels of Ythdf1 and Ythdf3 mRNAs were also not affected (Figures S6D and S6E). Similarly, the expression levels of Ythdf1 and Ythdf2 mRNAs were not changed in the Ythdf3 cKO retina (Figures S6G and S6H). Next, we also checked the protein levels of YTHDF1 and YTHDF2 in the Ythdf scKO retinas by IF. Similarly as its mRNA level, YTHDF2 protein expression also exhibited no difference between Ythdf1 scKO and Ctrl (Figure S6C). Similarly, YTHDF1 protein expression was also not changed in Ythdf2 scKO (Figure S6F). In addition, YTHDF1 and YTHDF2 protein levels were not affected in Ythdf3 scKO (Figures S6I and S6J). These results suggest that single cKO of each Ythdfs does not affect mRNA or protein levels of other YTHDFs.
YTHDF1 and YTHDF2 share a large pool of target mRNAs related to neurogenesis in mouse cortex and retina
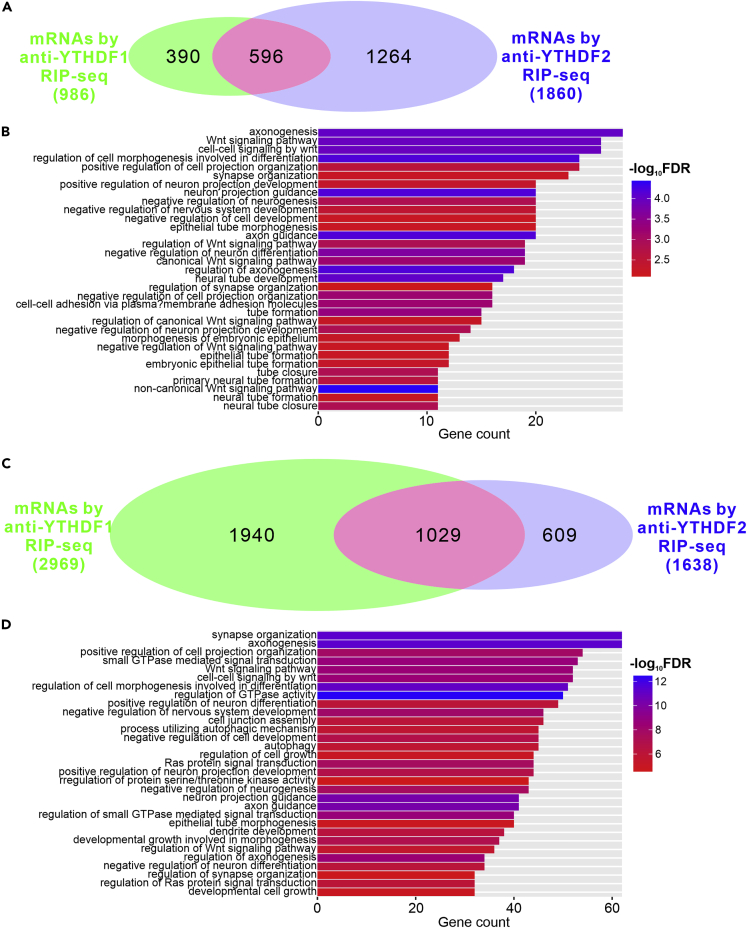
To further explore the mechanisms by which the m6A readers mediate m6A modification in regulating neurogenesis, we continued to identify the target mRNAs of YTHDFs. We performed anti-YTHDF1 and anti-YTHDF2 RNA immunoprecipitation followed by RNA sequencing of the elutes (RIP-seq) in the developing cortex and retina. From the embryonic cortex, we identified 986 and 1860 mRNAs by anti-YTHDF1 and anti-YTHDF2 RIP-seq, respectively (Figure 7A and Table S1). 596 mRNAs were identified to be common targets of YTHDF1 and YTHDF2 (Figure 7A and Table S1). Gene Ontology (GO) analysis of those target mRNAs revealed enrichment of genes related to regulation of nervous system development, regulation of neuron differentiation, regulation of neurogenesis et al. (Figures 7B, S7A, and S7B), which is consistent with the redundant functions of YTHDF1 and YTHDF2 in mediating m6A regulation of cortical neurogenesis.
Figure 7.
YTHDF1 and YTHDF2 share a large pool of target mRNAs related to neurogenesis in mouse cortex and retina
(A) Venn diagram showing numbers of mRNAs identified by anti-YTHDF1 and anti-YTHDF2 RNA immunoprecipitation followed by RNA sequencing (RIP-seq) in wildtype E12.5 cortex.
(B) Gene ontology (GO) analysis of overlapping target mRNAs of YTHDF1 and YTHDF2 in cortex. The GO terms in Biological Process are shown.
(C) Venn diagram showing numbers of mRNAs identified by anti-YTHDF1 and anti-YTHDF2 RIP-seq in wildtype P0 retina.
(D) GO analysis of overlapping target mRNAs of YTHDF1 and YTHDF2 in retina. The GO terms in Biological Process are shown.
We have previously shown that anti-YTHDF2 RIP-seq in mouse retina identified 1638 mRNAs (Niu et al., 2022). We also carried out anti-YTHDF1 RIP-seq in mouse retina and identified 2969 mRNAs (Figure 7C and Table S2). By comparing these two groups of mRNAs, we found that 1029 mRNAs are common targets of YTHDF1 and YTHDF2 in retina (Figure 7C and Table S2). Similarly as cortex, GO analysis of those target mRNAs revealed enrichment of genes related to neuron differentiation, nervous system development, neurogenesis et al. (Figures 7D and S7C).
Together, those results demonstrated that YTHDF1 and YTHDF2 share a large pool of target mRNAs related to neurogenesis in mouse cortex and retina, supporting their redundant functions in mediating m6A regulation of cortical and retinal neurogenesis.
Next we wanted to narrow down and identify the driver targets. By overlapping the common cortical targets of YTHDF1 and YTHDF2 with the m6A-enriched cortical transcripts from Yoon et al. (2017), 146 mRNAs were identified as m6A-enriched common targets of YTHDF1 and YTHDF2 (Table S3).
We further overlapped the common cortical target mRNAs of YTHDF1/2 with the differentially expressed genes in cortical neural progenitor cells (NPCs) of Mettl14 cKO mouse from Yoon et al. (2017). 107 mRNAs were identified, and 95 of them were upregulated after Mettl14 deletion (Table S4). We further focused on Arid1b, Atoh8, and Sema4c. ARID1B, a subunit of the BAF chromatin remodeling complex, was linked to autism spectrum disorder (ASD) and intellectual disability (Moffat et al., 2019). Recently, ARID1B was reported to regulate the proliferation and differentiation of NPCs (Moffat et al., 2021; Yang et al., 2021). Atoh8, also known as Math6, was accumulated in the developing brain and modulated neuron differentiation (Li et al., 2019; Wang et al., 2015a). Sema4c is a transmembrane protein which could interacts with PSD95, was expressed abundantly in NPCs and regulated adult neurogenesis induced by cerebral ischemia (Wu et al., 2009). We next performed RT-qPCR to check the expression levels of these targets in E14 Y1Y2 dcKO mouse. Arid1b, Atoh8, and Sema4c mRNA levels were dramatically increased in the Y1Y2 dcKO cortex compared with controls (Figures S7D–S7F), suggesting that they might be the driver targets of YTHDF1 and YTHDF2 to mediate m6A regulation of cortical neurogenesis.
Discussion
In this study, we revealed the redundant functions of the m6A readers YTHDFs to mediate m6A modification in regulating cortical and retinal neurogenesis.
m6A modification has been found to control mammalian cortical neurogenesis (Wang et al., 2018; Yoon et al., 2017). One study found that conditional knockout of the writer METTL14 caused prolonged cell cycle and remnant of radial glia cells (Yoon et al., 2017). Another study reported that Mettl14 conditional knockout led to decreased proliferation and premature differentiation (Wang et al., 2018). This obvious inconsistency calls for further exploration and clarification of the functions of m6A modification in cortical neurogenesis. Here we found that Mettl14 deletion causes abnormal maintenance of radial glia cells in the postnatal stages, delays cortical neurogenesis, and reduces production of neurons, supporting the findings by Yoon et al. (2017).
We further showed that m6A regulation of neurogenesis is working similarly in other brain regions. Deletion of Mettl14 in retina extends maintenance of proliferating retinal progenitors, decreases production of late-born neurons, and impairs organization of retinal layers. These findings support a general role of m6A modification in regulating neurogenesis.
The YTHDF family proteins, YTHDF1, YTHDF2, and YTHDF3 are the major cytosolic m6A reader proteins to mediate m6A functions by regulating degradation and translation of target mRNAs (Kontur et al., 2020; Lasman et al., 2020; Shi et al., 2017; Wang et al., 2014, 2015b; Zaccara and Jaffrey, 2020). However, which m6A readers and how they mediate m6A modification in neurogenesis are still unknown. Here we found that single conditional knockout of Ythdf1, Ythdf2 or Ythdf3 has no effect on cortical neurogenesis. Double deletion of Ythdf1 and Ythdf2 leads to protracted cortical neurogenesis, delayed cell-cycle exit, and reduced late-born neurons, which is consistent with the neurogenesis phenotypes of Mettl14 conditional knockout in cortex. In retina, single or double knockout of Ythdf1, Ythdf2 does not disturb retinal neurogenesis. Triple conditional knockout of Ythdf1, Ythdf2, and Ythdf3 exhibits extended neurogenesis, disordered lamination, and decreased late-born neurons, resembling the neurogenesis phenotypes of Mettl14 conditional knockout in retina. Identification of common target mRNAs recognized by both YTHDF1 and YTHDF2 further supports the redundant functions of YTHDFs in mediating cortical and retinal neurogenesis.
This functional redundancy of YTHDFs seems to have tissue-specificity because double conditional knockout of Ythdf1 and Ythdf2 can recapitulate the phenotypes of Mettl14 conditional knockout in cortex, whereas only triple conditional knockout of Ythdf1, Ythdf2 and Ythdf3 can reproduce those of Mettl14 conditional knockout in retina. Further studies might be needed to explain this tissue-specificity.
Although YTHDFs are redundant in mediating m6A modification in neurogenesis, they have unique and specific roles in postmitotic neurons. Conditional knockout of Ythdf1 or Ythdf2 in cerebellar granule cells (GC) promoted GC axon growth because YTHDF1 and YTHDF2 can synergistically regulate Wnt5a/PCP pathway through controlling local translation of Dvl1 and Wnt5a, respectively (Yu et al., 2021). Conditional knockout of Ythdf1 in dorsal spinal commissural neurons impaired translation of Robo3.1 mRNA, and disturbed commissural axon guidance (Zhuang et al., 2019). Conditional knockout of Ythdf2 in retinal ganglion cells (RGCs) increased RGC dendrite branching and improved visual acuity (Niu et al., 2022). Thus, the m6A readers YTHDFs have sequential functions in neuronal development, initially mediating m6A modification in neurogenesis redundantly, and then exerting unique and specific roles to regulate axon and dendrite development of postmitotic neurons.
Limitations of the study
Our current study demonstrates that the functionally redundant YTHDFs mediate m6A regulation of cortical and retinal neurogenesis. We found that Ythdf1 and Ythdf2 double deletion, or only Ythdf1, Ythdf2, and Ythdf3 triple deletion recapitulates the phenotype of Mettl14 knockout in cortical or retinal neurogenesis, respectively. In addition, we identified a few target mRNAs which might be the driver targets mediating the redundant functions of YTHDFs in neurogenesis. However, further mechanistic studies are needed to elucidate the tissue-specificity of the YTHDFs redundancy, and the functions of the YTHDFs targets in neurogenesis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-YTHDF1 | Proteintech | Cat#: 17479-1-AP; RRID: AB_2217473 |
| Rabbit polyclonal anti-YTHDF2 | Proteintech | Cat#: 24744-1-AP; RRID: AB_2687435 |
| Rabbit polyclonal anti-Mettl14 | Sigma-Aldrich | Cat#: HPA038002; RRID: AB_10672401 |
| Rabbit polyclonal anti m6A | Synaptic Systems | Cat# 202003; RRID: AB_2279214 |
| Mouse monoclonal anti-Pax6 | BD Biosciences | Cat#: 561462; RRID: AB_10715442 |
| Rat monoclonal anti-BrdU | Abcam | Cat#: ab6326; RRID: AB_305426 |
| Rabbit monoclonal anti-Ki67 | Cell Signaling | Cat#: 12202S; RRID: AB_2620142 |
| Rabbit polyclonal anti-Tbr1 | Abcam | Cat#: ab31940; RRID: AB_2200219 |
| Rat monoclonal anti-Ctip2 | Abcam | Cat#: ab18465; RRID: AB_2064130 |
| Rabbit monoclonal anti-Tbr2 | Abcam | Cat#: ab183991; RRID: AB_2721040 |
| Mouse monoclonal anti-AP2α | DSHB | Cat#: 3B5; RRID: AB_2313947 |
| Mouse monoclonal anti-Brn3a | Millipore | Cat#: MAB1585; RRID: AB_94166 |
| Rabbit polyclonal anti-PKCα | Cell Signaling | Cat#: CST-2056 |
| Guinea pig polyclonal anti-Isl1/2 | (Ji et al., 2009) | N/A |
| Sheep polyclonal anti-Chx10 | Exalpha | Cat#: X1179P; RRID: AB_2889828 |
| Alexa 488 donkey anti-g. pig IgG | Jackson Immunoresearch | Cat#: 706-545-148; RRID: AB_2340472 |
| Alexa 488 donkey anti-mouse IgG | Thermo Fisher Scientific | Cat#: A-21202; RRID: AB_141607 |
| Alexa 488 donkey anti-rabbit IgG | Thermo Fisher Scientific | Cat#: A-21206; RRID: AB_141708 |
| Alexa 488 donkey anti-rat IgG | Thermo Fisher Scientific | Cat#: A-21208; RRID: AB_2535794 |
| Alexa 555 donkey anti-mouse IgG | Thermo Fisher Scientific | Cat#: A-31570; RRID: AB_2536180 |
| Alexa 555 donkey anti-rabbit IgG | Thermo Fisher Scientific | Cat#: A-31572; RRID: AB_162543 |
| Alexa 555 donkey anti-sheep IgG | Thermo Fisher Scientific | Cat# A-21436; RRID: AB_2535857 |
| Alexa 647 donkey anti-mouse IgG | Thermo Fisher Scientific | Cat#: A-31571; RRID: AB_162542 |
| Critical commercial assays | ||
| EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit | Millipore | Cat#: 17-701 |
| Experimental models: Organisms/strains | ||
| Mouse: Mettl14fl/fl | (Huang et al., 2022) | N/A |
| Mouse: Ythdf1fl/fl | (Zhuang et al., 2019) | N/A |
| Mouse: Ythdf2fl/fl | (Yu et al., 2021) | N/A |
| Mouse: Ythdf3fl/fl | This paper | N/A |
| Mouse: Tg(Nes-cre)1Kln/J | Jackson Laboratory | Cat#: JAX_003771 |
| Mouse: Tg(Six3-cre)69Frty/GcoJ | Jackson Laboratory | Cat#: JAX_019755 |
| Oligonucleotides | ||
| ISH Probe primers of mouse Ythdf3: Fwd: TGCTCCGTCCATTGGATT Rev: CGAGGAGCTACCCAACGA | This paper | N/A |
| qPCR primers of mouse Gapdh Fwd: TTGTCAGCAATGCATCCTGCACCACC Rev: CTGAGTGGCAGTGATGGCATGGAC | (Niu et al., 2022) | N/A |
| qPCR primers of mouse Ythdf1 Fwd: GGACAGTCCAATCCGAGTAACA Rev: CCTCGCTGAGGGAGTAAGGA | This paper | N/A |
| qPCR primers of mouse Ythdf2: Fwd: GAGCAGAGACCAAAAGGTCAAG Rev: CTGTGGGCTCAAGTAAGGTTC | (Niu et al., 2022) | N/A |
| qPCR primers of mouse Ythdf3: Fwd: GCCATGCGAAGGGAGAGAAA Rev: AGGCATTTCCAGAGTCTACATCAAG | This paper | N/A |
| qPCR primers of mouse Arid1b: Fwd: CCGATGGACCCAATGGTGATG Rev: TGGCCTTGCTGGCAGTAGCC | (Yang et al., 2021) | N/A |
| qPCR primers of mouse Atoh8: Fwd: AGCCAAGAAACGGAAGGAGTGAC Rev: TGGCAGCATCTTGAGGAAGAC | This paper | N/A |
| qPCR primers of mouse Sema4c: Fwd: ACCCCAGCCTAGGCCCC Rev: CAGGGGATGCCCACAAATGA | This paper | N/A |
| Mouse genotyping primers for Mettl14loxP site 1: CTGCCTGAACCTCTTGAGAACTGA and GCAGACAAGTGAGGAAATAAGCAAG | This paper | N/A |
| Mouse genotyping primers for Mettl14loxP site 2: GGAGGTGAACCTGATGAGCATT and CAACTGTAAGTGGCCGCTGGA | This paper | N/A |
| Mouse genotyping primers for Ythdf1loxP site 1: TAGTGCATTGTTAAGGCTGTCCTCGT and CTGCTGTCTCAAAGCACAAAGCCT | (Zhuang et al., 2019) | N/A |
| Mouse genotyping primers for Ythdf1loxP site 2: CTTAGAAATCAGTGTTTGTGGCCCA and CCTGCCTCAACACACCATTCTCTTT | (Zhuang et al., 2019) | N/A |
| Mouse genotyping primers for Ythdf2loxP site 1: GCTTGTAGTTATGTTGTGTACCAC and GCAGCTCTGACTATTCTAAAACCTCC | (Niu et al., 2022) | N/A |
| Mouse genotyping primers for Ythdf2loxP site 2: CTCATAACATCCATAGCCACAGG and CCAAGAGATAGCTTTCCTAATG | (Niu et al., 2022) | N/A |
| Mouse genotyping primers for Ythdf3loxP site 1: CAAACCAGAACAACGTAAGGCC and CTGGGTATGAAGCCCCACTGAT | This paper | N/A |
| Mouse genotyping primers for Ythdf3loxP site 2: ATTCCTTGCATGCTGTTCTAAAGT and AGTTCATATATAGAGCTGGAGAGC | This paper | N/A |
| Mouse genotyping primers for Nestin-cre: TTGCTAAAGCGCTACATAGGA (WT F) and CCTTCCTGAAGCAGTAGAGCA (Mut F) and GCCTTATTGTGGAAGGACTG (Common R) | This paper | N/A |
| Mouse genotyping primers for Six3-cre: CCTTCCTCCCTCTCTATGTG and GAACGAACCTGGTCGAAATC | (Niu et al., 2022) | N/A |
| Software and algorithms | ||
| GraphPad Prism 9.0 | GraphPad | https://www.graphpad.com; RRID: SCR_002798 |
| ImageJ (Fiji) | (Schindelin et al., 2012) | http://fiji.sc; RRID:SCR_002285 |
| STAR v2.5 | (Dobin et al., 2013) | https://github.com/alexdobin/STAR/ |
| HTSeq | (Anders et al., 2015) | https://pypi.org/project/HTSeq/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sheng-Jian Ji (jisj@sustech.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals
Mettl14fl/fl, Ythdf1fl/fl and Ythdf2fl/fl mice were reported previously (Huang et al., 2022; Yu et al., 2021; Zhuang et al., 2019). For generation of Ythdf3 conditional knockout mice, exon 3 of mouse Ythdf3 was targeted and loxP sites were inserted flanking Ythdf3 exon 3. Nestin-cre (The Jackson Laboratory, # 003,771) and Six3-cre (Furuta et al., 2000) (The Jackson Laboratory, # 019,755) were originally from The Jackson Laboratory. For timed pregnancy, embryos were identified as E0.5 when a copulatory plug was observed. Genotyping primers are as follows: the first Mettl14-loxp site: 5′-CTGCCTGAACCTCTTGAGAACTGA-3′ and 5′-GCAGACAAGTGAGGAAATAAGCAAG-3′; the second Mettl14-loxp site: 5′-GGAGGTGAACCTGATGAGCATT-3′ and 5′-CAACTGTAAGTGGCCGCTGGA-3′. The first Ythdf1-loxp site: 5′-TAGTGCATTGTTAAGGCTGTCCTCGT-3′ and 5′-CTGCTGTCTCAAAGCACAAAGCCT-3′; the second Ythdf1-loxp site: 5′-CTTAGAAATCAGTGTTTGTGGCCCA-3′ and 5′-CCTGCCTCAACACACCATTCTCTTT-3′. The first Ythdf2-loxp site: 5′-GCTTGTAGTTATGTTGTGTACCAC-3′ and 5′-GCAGCTCTGACTATTCTAAAACCTCC-3′; the second Ythdf2-loxp site: 5′-CTCATAACATCCATAGCCACAGG-3′ and 5′-CCAAGAGATAGCTTTCCTAATG-3′. The first Ythdf3-loxp site: 5′-CAAACCAGAACAACGTAAGGCC-3′ and 5′-CTGGGTATGAAGCCCCACTGAT-3′; the second Ythdf3-loxp site: 5′-ATTCCTTGCATGCTGTTCTAAAGT-3′ and 5′-AGTTCATATATAGAGCTGGAGAGC-3′. Nestin-cre site: 5′-CCTTCCTGAAGCAGTAGAGCA-3′ (Mut F) and 5′-GCCTTATTGTGGAAGGACTG-3′ (Common R) and 5′-TTGCTAAAGCGCTACATAGGA-3′ (WT F). Six3-cre site: 5′-CCTTCCTCCCTCTCTATGTG-3′ and 5′-GAACGAACCTGGTCGAAATC-3′. All experiments using mice were carried out following the animal protocols approved by the Laboratory Animal Welfare and Ethics Committee of Southern University of Science and Technology.
Method details
Immunofluorescence and immunostaining
For brain sections, mouse brain was dissected and fixed with 4% paraformaldehyde (Sigma) in 0.1 M Phosphate Buffer (PB) overnight at 4°C. After PBS washing, mouse brain was successively dehydrated with 15 and 30% sucrose in 0.1MPB for one day at 4°C, and then embedded with O.C.T. (SAKURA) and cryosectioned at 12 μm with Leica CM1950 Cryostat coronally. Coronal sections of brain were treated with sodium citrate buffer, pH 6.0 (Sigma) at 90°C for 40 min and then equilibrated at room temperature (RT). After PBS washing, brain sections were permeabilized and blocked with 10% donkey serum, 5% BSA (Sigma) and 0.25% Triton X-100 (Sigma) in PBS for 1 h at RT, and then incubated with 5% donkey serum, 5% BSA and 0.25% Triton X-100 in PBS overnight at 4°C with primary antibodies. After equilibrated at RT and three times of PBS washing, sections were incubated with 5% donkey serum, 5% BSA and 0.25% Triton X-100 in PBS for 2 h at RT with secondary antibodies. After two times of PBS washing, sections were mounted with the VECTASHIELD Antifade Mounting Medium with DAPI (Vector Laboratory).
For retinal vertical sections, E12.5 mouse embryos were fixed with 4% paraformaldehyde in 0.1MPB overnight at 4°C; embryonic eyes of later stages were fixed for 20–45 min at RT; eyes of mouse pups were pre-fixed briefly and then eyecups were dissected and fixed for 30–45 min at RT. After PBS washing, tissues were dehydrated with 30% sucrose in 0.1MPB overnight at 4°C, and then the E12.5 embryonic heads or the eyes of other stages were embedded with O.C.T. (SAKURA) and cryosectioned at 12 μm with Leica CM1950 Cryostat. Tissue sections were permeabilized and blocked with 1% BSA (Sigma) and 0.5% Triton X-100 (Sigma) in PBS (PBST) for 1 h at RT and incubated in PBST overnight at 4°C with primary antibodies. After three times of PBS washing, sections were incubated in PBST for 1 h at RT with secondary antibodies and then mounted with the VECTASHIELD Antifade Mounting Medium with DAPI (Vector Laboratory). METTL14 immunostaining experiments were carried out after antigen retrieval by sodium citrate buffer treatment.
Antibodies were used as follows: rabbit anti-YTHDF1 (1:1000, Proteintech 17479-1-AP), rabbit anti-YTHDF2 (1:1000, Proteintech 24744-1-AP), rabbit anti-METTL14 (1:1000, Sigma HPA038002), rabbit anti-m6A (1:200, Synaptic Systems, 202,003), mouse anti-Pax6 (1:500, BD Biosciences 561,462), rat anti-BrdU (1:400, Abcam ab6326), rabbit anti-Ki67 (1:400, Cell Signaling 12202S), rabbit anti-Tbr1 (1:500, Abcam ab31940), rabbit anti-Tbr2 (1:500, Abcam ab183991), rat anti-Ctip2 (1:500, Abcam ab18465), mouse anti-AP2α (1:1000, DSHB 3B5), mouse anti-Brn3a (1:300, Millipore MAB1585), rabbit anti-PKCα (1:1000, Cell Signaling CST-2056), guinea pig anti-Isl1/2 (1:10,000) (Ji et al., 2009), sheep anti-Chx10 (1:1000, Exalpha X1179P); Alexa 488 donkey anti-guinea pig (1:500, Jackson 706-545-148), Alexa 488 donkey anti-mouse (1:500, Thermo A21202), Alexa 488 donkey anti-rabbit (1:500, Thermo A21206), Alexa 488 donkey anti-rat (1:500, Thermo A21208), Alexa 555 donkey anti-mouse (1:1000, Thermo A31570), Alexa 555 donkey anti-rabbit (1:1000, Thermo A31572), Alexa 555 donkey anti-sheep (1:1000, Thermo A21436), Alexa 647 donkey anti-mouse (1:200, Thermo A31571). All antibodies have been previously validated.
All images were captured on Nikon A1R confocal microscope, Zeiss LSM 800 confocal microscope or TissueGnostics TissueFAXS PLUS Upright Fluorescence and Brightfield System with identical settings for each group in the same experiment. The number of neurons in a specific area and the thickness of cortical layers were quantified blindly and manually.
In situ hybridization (ISH)
The PCR primers used for cloning mouse Ythdf3 ISH probe are 5′-TGCTCCGTCCATTGGATT-3′ and 5′-CGAGGAGCTACCCAACGA-3′. The tissue sections for ISH were prepared as the immunofluorescence procedures with RNase-free reagents. Briefly, the tissue sections were fixed with 4% PFA in PBS and treated with Proteinase K. After re-fixation with PFA, the tissue sections were acetylated by acetic anhydride diluted in triethanolamine buffer. After acetylation, the prehybridization of sections were performed with the hybridization buffer for 2 h at 65°C and then the sections were hybridized with probes in fresh hybridization buffer overnight at 65°C. The sections were washed with preheated and RT 2× SSC sequentially, and then treated with 10 μg/mL RNase A in 2× SSC at 37°C for 8 min. After washing with 2× SSC, 0.2× SSC and PBT sequentially, the sections were blocked with 10% heat-inactivated sheep serum (SS) in PBT for 1 h at RT. Anti digoxygenin (Roche) was diluted in PBT with 10% SS, and used to incubate the sections overnight at 4°C. After washing with PBT and alkaline phosphatase buffer, NBT/BCIP (Roche) diluted in alkaline phosphatase buffer was used for color development reactions in darkness. After stopping color development reactions with PBS and briefly rinsing with ddH2O, the sections were then mounted with Permount™ mounting medium. All images were captured on TissueGnostics TissueFAXS PLUS Upright Fluorescence and Brightfield System with identical settings for each group in the same experiment.
BrdU assay
BrdU dissolved in normal saline was injected in the E16.5 pregnant mice with 0.1 mg/g by intraperitoneal injection. The embryonic brains were collected 24 h after BrdU injection and treated as above. After antigen retrieval with sodium citrate buffer, the sections were treated with 2 M HCl at RT for 30 min and then 0.1 M borate buffer at RT for 10 min. After PBS washing, the process of blocking and antibody incubation was performed as immunofluorescence and immunostaining.
RNA immunoprecipitation and sequencing (RIP-seq)
We used EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore) following the manual with minor modifications. 1 × 107 × 1012.5 cortical or P0 retinal neurons were lysed and incubated with YTHDF1 (Proteintech 17479-1-AP) or YTHDF2 antibody (Proteintech 24744-1-AP) overnight at 4°C. After quality control monitoring with Agilent 2100, 100 ng RNA of input and elutes after RIP were used to construct the libraries with TruSeq Stranded RNA Sample Preparation Kit (Illumina) and sequenced on the Illumina HiSeq 3000 platform (Jingneng, Shanghai, China). The filtered reads were then mapped to the mouse reference genome (GRCm38) using STAR v2.5 (Dobin et al., 2013) with default parameters. The resulting bam files were fed to HTSeq tool (Anders et al., 2015) to count the number of RNA-seq reads, which was further normalized to calculate FPKM. Any fold change greater than 2 of the FPKM from RIP elute to input was considered enriched. All enriched genes were used to do Gene Ontology (GO) analyses by the GOseq R package, in which gene length bias was corrected. GO terms with corrected p value less than 0.05 were considered significantly enriched.
RT-qPCR assay
RT-qPCR process was described previously (Niu et al., 2022). Total RNAs of Ythdfs cKO and control cortices or retinas were extracted by TRIzol Reagent (Life) and used for RT-qPCR. Primers used for qPCR are as following: mouse Gapdh: 5′-TTGTCAGCAATGCATCCTGCACCACC-3′ and 5′-CTGAGTGGCAGTGATGGCATGGAC-3′ (Niu et al., 2022); mouse Ythdf1: 5′-GGACAGTCCAATCCGAGTAACA-3′ and 5′-CCTCGCTGAGGGAGTAAGGA-3′; mouse Ythdf2: 5′-GAGCAGAGACCAAAAGGTCAAG-3′ and 5′-CTGTGGGCTCAAGTAAGGTTC-3′ (Niu et al., 2022); mouse Ythdf3: 5′-GCCATGCGAAGGGAGAGAAA-3′ and 5′-AGGCATTTCCAGAGTCTACATCA-3′; mouse Arid1b: 5′-CCGATGGACCCAATGGTGATG-3′ and 5′-TGGCCTTGCTGGCAGTAGCC-3′(Yang et al., 2021); mouse Atoh8: 5′-AGCCAAGAAACGGAAGGAGTGAC-3′ and 5′-TGGCAGCATCTTGAGGAAGAC-3′; mouse Sema4c: 5′-ACCCCAGCCTAGGCCCC-3′ and 5′-CAGGGGATGCCCACAAATGA-3′.
Quantification and statistical analysis
Statistical analysis
All experiments were conducted at a minimum of three independent biological replicates in the lab, and for cKO analysis, a minimum of three embryos/pups/mice were analyzed for each genotype. Statistical analysis was performed using GraphPad Prism 9.0. When comparing the means of two groups, an unpaired t-test was performed on the basis of experimental design. The settings for all box and whisker plots are: 25th-75th percentiles (boxes), minimum and maximum (whiskers), and medians (horizontal lines). The settings for all accumulative histograms are: mean (boxes), s.d. (stacked bars). A p value less than 0.05 was considered as statistically significant: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Acknowledgments
We thank Yuxi Sun and Jianhui Liu for technical assistance, and other members of Ji and Ou laboratories for help, technical support, and comments on the manuscript.
This work was supported by National Natural Science Foundation of China (32170955 and 31871038 to S.-J.J.), Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions (2022SHIBS0002), High-Level University Construction Fund for Department of Biology (internal grant no. G02226301), Science and Technology Innovation Commission of Shenzhen Municipal Government (ZDSYS20200811144002008).
Author contributions
Conceptualization: S.J.J., F.N., and P.C.; Methodology: F.N., P.C., Z.Y., and X.O.; Validation: F.N., P.C., Z.Y., and S.J.J.; Formal analysis: F.N., P.C., Z.Y., and L.Y.; Investigation: F.N., P.C., Z.Y., M.Z., J.Z., L.Y., X.O., and S.J.J.; Data curation: F.N. and P.C.; Writing - review & editing: F.N. and S.J.J.; Visualization: F.N. and P.C.; Supervision: S.J.J. and X.O.; Project administration: S.J.J.; Funding acquisition: S.J.J.
Declaration of interests
The authors declare no competing interests.
Published: September 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104908.
Contributor Information
Xijun Ou, Email: ouxj@sustech.edu.cn.
Sheng-Jian Ji, Email: jisj@sustech.edu.cn.
Supplemental information
Data and code availability
The RIPseq data generated in this study has been deposited in GEO under accession number GSE199962. Other data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie-Kommit J., Clark B.S., Shi Q., Shiau F., Kim D.W., Langel J., Sheely C., Ruzycki P.A., Fries M., Javed A., et al. Atoh7-independent specification of retinal ganglion cell identity. Sci. Adv. 2021;7:eabe4983. doi: 10.1126/sciadv.abe4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepko C. Intrinsically different retinal progenitor cells produce specific types of progeny. Nat. Rev. Neurosci. 2014;15:615–627. doi: 10.1038/nrn3767. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G., Kriegstein A.R. Neurons from radial glia: the consequences of asymmetric inheritance. Curr. Opin. Neurobiol. 2003;13:34–41. doi: 10.1016/s0959-4388(03)00013-8. [DOI] [PubMed] [Google Scholar]
- Furuta Y., Lagutin O., Hogan B.L., Oliver G.C. Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis. 2000;26:130–132. [PubMed] [Google Scholar]
- He P.C., He C. m(6) A RNA methylation: from mechanisms to therapeutic potential. EMBO J. 2021;40:e105977. doi: 10.15252/embj.2020105977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks E.A., Zaveri M., Deschamps P.A., Noseworthy M.D., Ball A., Williams T., West-Mays J.A. Conditional deletion of AP-2α and AP-2β in the developing murine retina leads to altered amacrine cell mosaics and disrupted visual function. Invest. Ophthalmol. Vis. Sci. 2018;59:2229–2239. doi: 10.1167/iovs.17-23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Zhang G., Ruan G.X., Li Y., Chen W., Zou J., Zhang R., Wang J., Ji S.J., Xu S., Ou X. Mettl14-Mediated m6A modification is essential for germinal center B cell response. J. Immunol. 2022;208:1924–1936. doi: 10.4049/jimmunol.2101071. [DOI] [PubMed] [Google Scholar]
- Ji S.J., Periz G., Sockanathan S. Nolz1 is induced by retinoid signals and controls motoneuron subtype identity through distinct repressor activities. Development. 2009;136:231–240. doi: 10.1242/dev.028043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontur C., Jeong M., Cifuentes D., Giraldez A.J. Ythdf m(6)A readers function redundantly during zebrafish development. Cell Rep. 2020;33:108598. doi: 10.1016/j.celrep.2020.108598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy A., Yamagata M., Duan X., Hong Y.K., Sanes J.R. Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature. 2015;524:466–470. doi: 10.1038/nature14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasman L., Krupalnik V., Viukov S., Mor N., Aguilera-Castrejon A., Schneir D., Bayerl J., Mizrahi O., Peles S., Tawil S., et al. Context-dependent functional compensation between Ythdf m(6)A reader proteins. Genes Dev. 2020;34:1373–1391. doi: 10.1101/gad.340695.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li H., Zhang L., Xiong S., Wen S., Xia X., Zhou X. Growth/differentiation 5 promotes the differentiation of retinal stem cells into neurons via Atoh8. J. Cell. Physiol. 2019;234:21307–21315. doi: 10.1002/jcp.28735. [DOI] [PubMed] [Google Scholar]
- Lu Q., Ivanova E., Ganjawala T.H., Pan Z.H. Cre-mediated recombination efficiency and transgene expression patterns of three retinal bipolar cell-expressing Cre transgenic mouse lines. Mol. Vis. 2013;19:1310–1320. [PMC free article] [PubMed] [Google Scholar]
- Luo H., Jin K., Xie Z., Qiu F., Li S., Zou M., Cai L., Hozumi K., Shima D.T., Xiang M. Forkhead box N4 (Foxn4) activates Dll4-Notch signaling to suppress photoreceptor cell fates of early retinal progenitors. Proc. Natl. Acad. Sci. USA. 2012;109:E553–E562. doi: 10.1073/pnas.1115767109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Jaffrey S.R. Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017;33:319–342. doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J.J., Jung E.M., Ka M., Smith A.L., Jeon B.T., Santen G.W.E., Kim W.Y. The role of ARID1B, a BAF chromatin remodeling complex subunit, in neural development and behavior. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2019;89:30–38. doi: 10.1016/j.pnpbp.2018.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J.J., Jung E.M., Ka M., Jeon B.T., Lee H., Kim W.Y. Differential roles of ARID1B in excitatory and inhibitory neural progenitors in the developing cortex. Sci. Rep. 2021;11:3856. doi: 10.1038/s41598-021-82974-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu F., Han P., Zhang J., She Y., Yang L., Yu J., Zhuang M., Tang K., Shi Y., Yang B., et al. The m(6)A reader YTHDF2 is a negative regulator for dendrite development and maintenance of retinal ganglion cells. Elife. 2022;11:e75827. doi: 10.7554/eLife.75827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota D., Chintala H., Wu F., Fliesler S.J., Hu Z., Mu X. Onecut1 and Onecut2 redundantly regulate early retinal cell fates during development. Proc. Natl. Acad. Sci. USA. 2014;111:E4086–E4095. doi: 10.1073/pnas.1405354111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Balakrishnan-Renuka A., Napirei M., Theiss C., Brand-Saberi B. Spatiotemporal expression of Math6 during mouse embryonic development. Histochem. Cell Biol. 2015;143:575–582. doi: 10.1007/s00418-014-1305-z. [DOI] [PubMed] [Google Scholar]
- Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li Y., Yue M., Wang J., Kumar S., Wechsler-Reya R.J., Zhang Z., Ogawa Y., Kellis M., Duester G., Zhao J.C. N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 2018;21:195–206. doi: 10.1038/s41593-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth M.B., Greig L.C., Kriegstein A.R., Macklis J.D. SnapShot: cortical development. Cell. 2012;151:918–918.e1. doi: 10.1016/j.cell.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Fan J., Zhu L., Liu S., Wu Y., Zhao T., Wu Y., Ding X., Fan W., Fan M. Sema4C expression in neural stem/progenitor cells and in adult neurogenesis induced by cerebral ischemia. J. Mol. Neurosci. 2009;39:27–39. doi: 10.1007/s12031-009-9177-8. [DOI] [PubMed] [Google Scholar]
- Yang Q.Q., Zhai Y.Q., Wang H.F., Cai Y.C., Ma X.Y., Yin Y.Q., Li Y.D., Zhou G.M., Zhang X., Hu G., Zhou J.W. Nuclear isoform of FGF13 regulates post-natal neurogenesis in the hippocampus through an epigenomic mechanism. Cell Rep. 2021;35:109127. doi: 10.1016/j.celrep.2021.109127. [DOI] [PubMed] [Google Scholar]
- Yoon K.J., Ringeling F.R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., Su Y., Kim N.S., Zhu Y., Zheng L., et al. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell. 2017;171:877–889.e17. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., She Y., Yang L., Zhuang M., Han P., Liu J., Lin X., Wang N., Chen M., Jiang C., et al. The m(6) A readers YTHDF1 and YTHDF2 synergistically control cerebellar parallel fiber growth by regulating local translation of the key Wnt5a signaling components in axons. Adv. Sci. 2021;8:e2101329. doi: 10.1002/advs.202101329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S., Jaffrey S.R. A unified model for the function of YTHDF proteins in regulating m(6)A-modified mRNA. Cell. 2020;181:1582–1595.e18. doi: 10.1016/j.cell.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- Zhuang M., Li X., Zhu J., Zhang J., Niu F., Liang F., Chen M., Li D., Han P., Ji S.J. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 2019;47:4765–4777. doi: 10.1093/nar/gkz157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RIPseq data generated in this study has been deposited in GEO under accession number GSE199962. Other data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.