Abstract
Herein we describe a designed protein building block whose self-assembly behaviour is dually gated by the redox state of disulphide bonds and the identity of exogenous metal ions. This protein construct is shown–through extensive structural and biophysical characterization–to access five distinct oligomeric states, exemplifying how the complex interplay between hydrophobic, metal-ligand, and reversible covalent interactions could be harnessed to obtain multiple, responsive protein architectures from a single building block.
Graphical Abstract
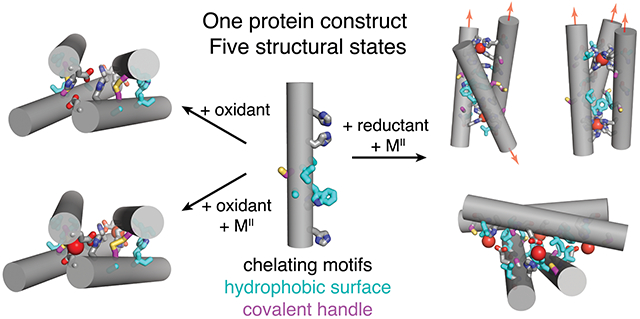
The propensity of a single protein sequence to form multiple conformations or assembly states has been crucial for the generation of structural and functional diversity during evolution.1–3 For instance, protein folds such as the Rossman, four-helix bundle, and βαβββ motifs have been repeatedly used as modular building blocks for larger architectures or quaternary assemblies with a wide variety of functions.4–6 Similarly, obtaining multiple structural outcomes from a single protein sequence also is a prerequisite for building switchable systems that transduce external stimuli into functionally relevant changes to their tertiary folds or quaternary assembly states.7–10 Inspired by such natural examples, there has been great interest in designing proteins that can alter their conformations or alter their assembly states in response to different stimuli, such as ligand binding,11, 12 metal coordination,13, 14 phosphorylation,15, 16 and cysteine oxidation/reduction.17, 18 While there have indeed been several examples of such artificial multi-state systems,3, 11–20 the ability to design proteins that respond to more than one type of stimulus or to obtain more than two structurally distinct states from a single protein sequence/structure has been limited (Figure 1a).21 This is primarily due to the fact that most protein design strategies involve the implementation of extensive noncovalent interactions (in particular, hydrophobic packing) to obtain single, stable structures that correspond to deep free energy minima.21–24 This strategy not only restricts the potential of structural diversification but lowers the potential for the resulting protein architecture to be stimuli-responsive and reconfigurable.
Fig. 1.
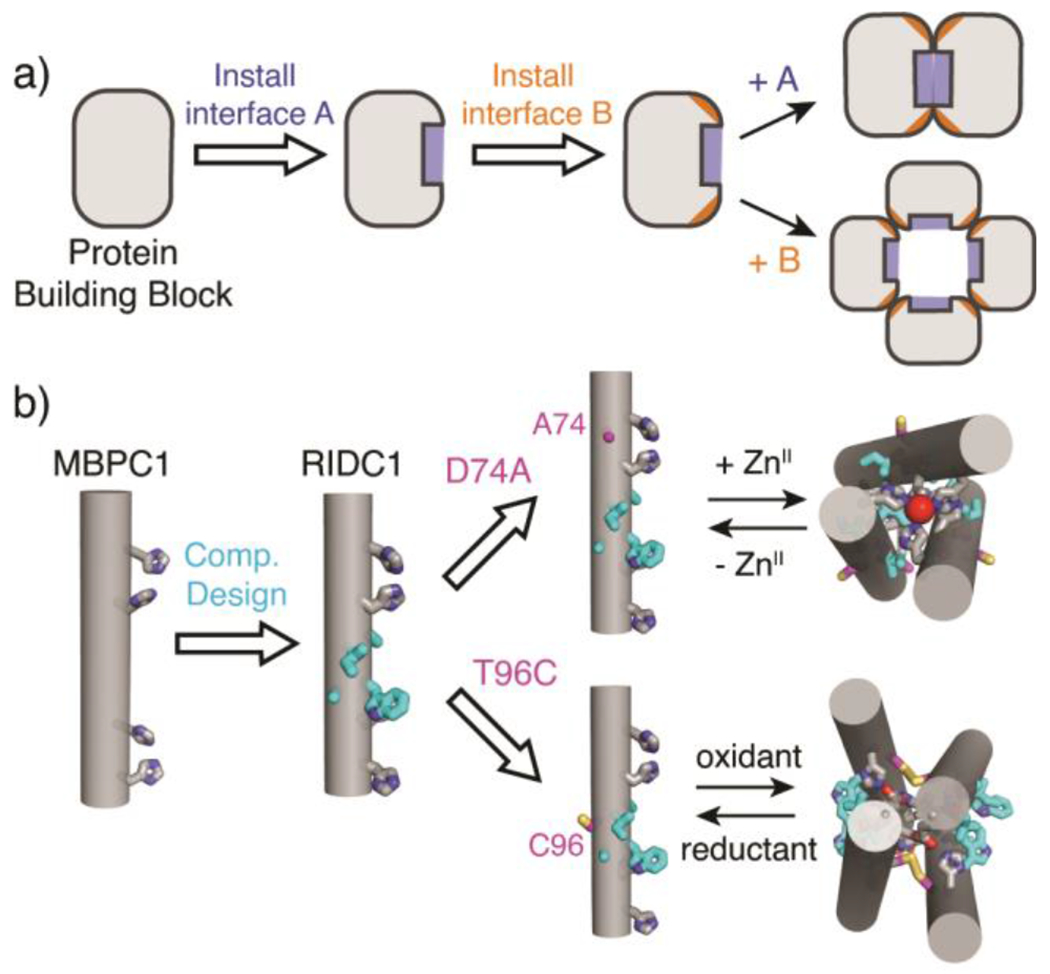
(a) General workflow to design a multi-stimuli responsive protein construct. A and B represent different stimuli. (b) Cartoon schemes of previously designed cytochrome cb562 variants.
Due to their simultaneous strength and reversibility, metal-ligand and disulphide bonding interactions represent promising conduits for the design of protein constructs that can access multiple structural states in a stimuli-responsive manner..18, 25–28 We have previously exploited metal coordination, disulphide bonding, and hydrophobic packing to construct cytochrome (cyt) cb562-based assemblies with diverse conformations, oligomeric states and metal coordination environments.13, 29–34 MBPC1, an early designed variant of cyt cb562, was shown to form different assemblies with distinct oligomeric states and structures based on the coordination preferences of exogenously added metal ions.13, 29 One of these assemblies, the Zn-directed tetramer Zn4:MBPC14, served as a structural template for the computational design of a hydrophobic interface (highlighted in cyan in Figure 1b) on the surface of MBPC1.31 Owing to the designed interactions between the hydrophobic surface residues, the resulting variant, RIDC1, formed a considerably more stable Zn-directed tetramer (Zn4:RIDC14) with a nearly identical structure to that of Zn4:MBPC14.31 Importantly, Zn4:RIDC14 served as a starting point for designing assemblies with functions that ranged from selective metal binding and metal-based allostery to in vivo enzymatic activity.32, 35, 36
In the course of our previous studies, we observed that single mutations of the RIDC1 construct alter the assembly outcomes.37, 38 One RIDC1 variant, C96RIDC1, formed a redox-dependent but metal-independent tetramer (C96RIDC14) stabilized by both hydrophobic and Cys96-Cys96 disulphide bonding (Figure 1b, S1a).37 A second RIDC1 variant, A74RIDC1 (wherein a metal binding residue Asp74 was mutated to Ala) assembled into a Zn-dependent trimer (Zn2:A74RIDC13) stabilized by hydrophobic packing interactions and tetrahedral, Zn:His4 coordination sites that had not been observed in other RIDC1 variants (Figure 1b, S1b).38 Here, with the aim of developing a protein construct that can respond to redox and metal-based stimuli to access multiple structural states, we combined the A74 and C96 mutations to generate A74/C96RIDC1 (Figure 2). We found that the interplay between hydrophobic, metal-ligand, and covalent interactions enabled this variant to form five discrete structural states in a redox- and metal-responsive fashion (Figure 2).
Fig. 2.
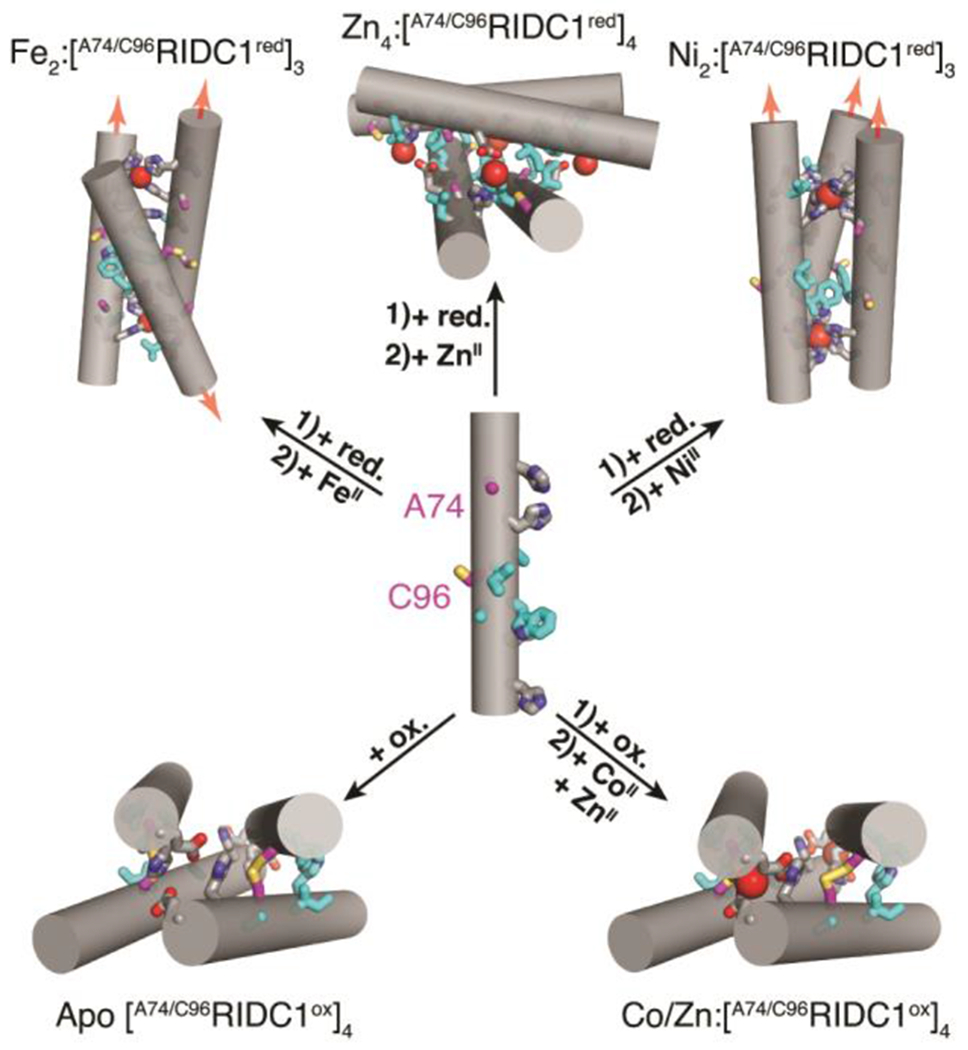
Structural states of A74/C96RIDC1 obtained through the addition of redox and/or metal-based stimuli. Hydrophobic mutations are highlighted in cyan. Ox. = oxidant, red. = reductant.
We surmised that in the oxidized state of A74/C96RIDC1 (A74/C96RIDC1ox), the Cys96-Cys96 disulphide bonds would enforce tetramerization, as observed in the case of C96RIDC1.37 Indeed, both in the absence and presence of metal ions (CoII, NiII, CuII, ZnII), A74/C96RIDC1ox formed a tetrameric species in solution in near quantitative yields as determined by sedimentation velocity-analytical ultracentrifugation (SV-AUC) measurements (Figure S2). The crystal structures of CoII-, and ZnII-bound [A74/C96RIDC1ox]4 are nearly identical to one another, with a root-mean-square deviation (RMSD) of 1.24 Å between all α-C’s (Figure S3–S5). On average, the buried surface area (BSA) of the metal-bound tetramers is about 40% smaller (1018 Å2) than that of the apo structure (1388 Å2) (Table S1). This indicates that the tetrameric assembly undergoes a significant structural change upon metal binding, with an average RMSD of 2.55 Å between apo and metal-bound structures (Figure S3). Both Co and Zn-bound [A74/C96RIDC1ox]4 tetramers contain two C2-symmetry-related coordination sites, with E81 and H77 residues from two different monomers serving as ligands. (Figure S4–7). The [A74/C96RIDC1ox]4 structures illustrate that simultaneously exploiting the flexibility of and structural constraints imposed by disulphide bonds and hydrophobic packing interactions can engender a flexible protein assembly with well-defined metal coordination sites (Figure 2, S4–7).
We next turned to the reduced form of our construct, A74/C96RIDC1red, with the hypothesis that the lack of disulphide-mediated interfacial constraints could allow it to access different oligomeric states upon metal coordination. A74/C96RIDC1red was obtained by adding 5-fold excess of the reductant tris(3-hydroxylpropyl) phosphine (THPP). At a protein concentration of ≥200 μM and up to 5-fold excess of metal ions, FeII, NiII and CuII addition to A74/C96RIDC1red led primarily to trimeric species in solution, whereas ZnII and CoII addition yielded tetrameric or higher-order assemblies (Figure S8, Table S2). All metal-directed A74/C96RIDC1red oligomers could be completely disassembled by the addition of a mixture of ethylenediaminetetraacetic acid (EDTA) and dipicolinic acid (DPA) (Figure S8).
The crystal structures of FeII-, NiII-, CuII-, and ZnII-directed assemblies of A74/C96RIDC1red were determined at resolutions of 1.6 Å to 2.7 Å (Table S3). These structures revealed a correspondence between the oligomerization states observed in solution and crystals for the FeII (n=3), NiII (n=3) and ZnII (n=4) complexes (Figure S5b, S9–10). By contrast, there was a deviation in the case of the CuII-directed A74/C96RIDC1red assembly (n=4 in crystals vs. n=3 in solution) (Figure S11). A closer look at the latter structure showed that all four Cu centres in the tetrameric assembly adopted a tetrahedral coordination geometry, strongly suggesting that they were in the +1 oxidation state and thus reduced by the excess THPP present in the crystallization solution (Figure S7a, S11, Table S4). In light of the complex redox equilibrium that exists between Cu ions, Cys-disulphide bonds, and THPP, we decided to focus our further analyses on FeII, NiII and ZnII complexes of A74/C96RIDC1red.
Interestingly, the FeII- and NiII-directed assemblies, while both trimeric, adopt different structural conformations and metal coordination environments (Figure 3). The Fe2:[A74/C96RIDC1red]3 complex features two protein monomers with their C-termini projecting downward and one monomer with its C-terminus projecting upward, resulting in an antiparallel, “up-up-down” arrangement similar to that observed for Zn2:A74RIDC13 (Figure 2, 3a, S9, S12).38 The two FeII centres, termed Fe1 and Fe2, are distinct from one another, with Fe1 in a square pyramidal geometry formed by five His residues and Fe2 in a similar geometry but with three His and two aqua ligands (Figure 3a, S6a, S9).
Fig. 3.
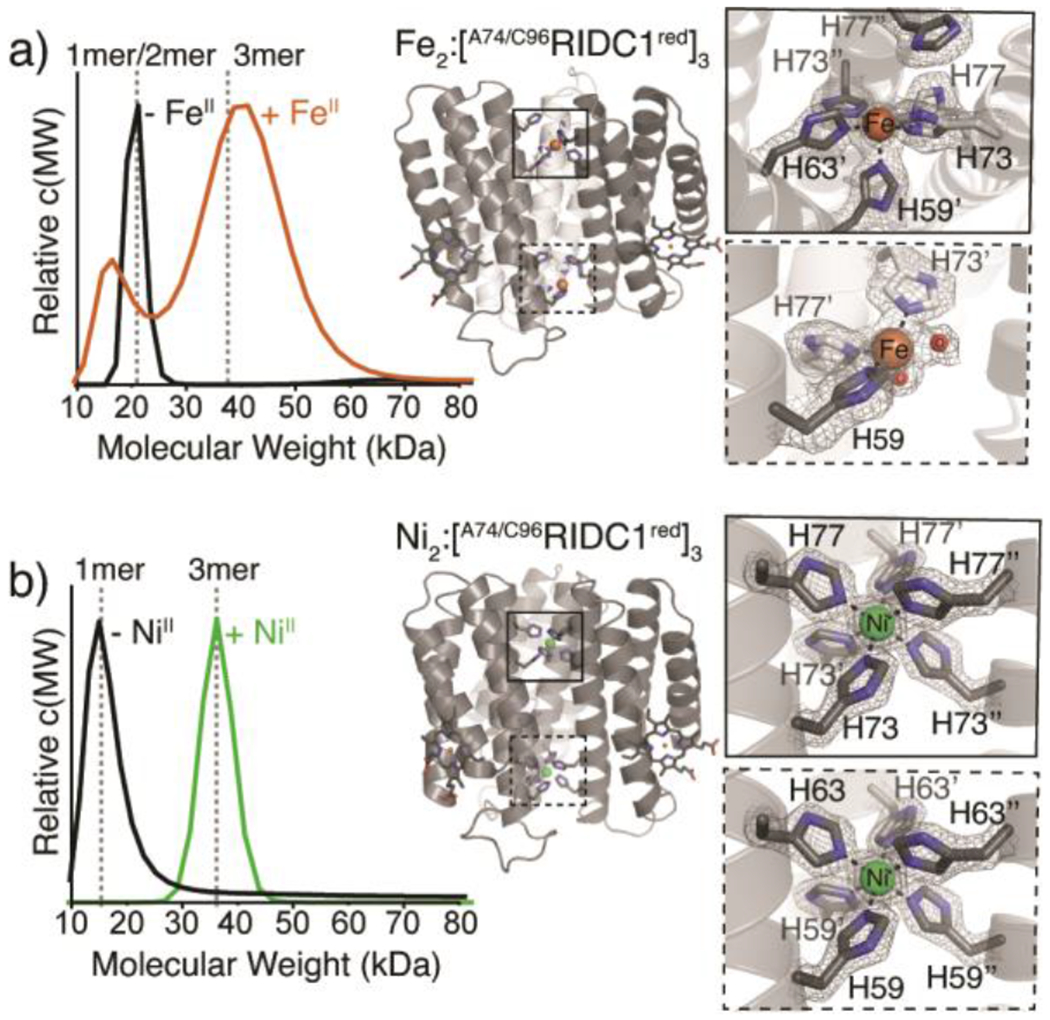
Assembly properties of Fe- and Ni-directed A74/C96RIDC1 trimers. (a) SV-AUC profile of 200 μM A74/C96RIDC1 following the addition/removal of 5 equiv. FeII/monomer (left). Crystal structure of Fe2:[A74/C96RIDC1red]3 (PDB ID: 7RWY), highlighting His3 and His5 coordination sites (right). (b) SV-AUC profile of 200 μM A74/C96RIDC1 following the addition/removal of 1 equiv. NiII/monomer (left). Crystal structure of Ni2:[A74/C96RIDC1red]3 (PDB ID: 7RWU), highlighting the His6 coordination sites (right).
By contrast, the Ni2:[A74/C96RIDC1red]3 assembly has C3 symmetry with an all-parallel, “up-up-up” arrangement of protein monomers and two octahedral, hexa-His-coordinated NiII centres (Figure 2, 3b, S6c, and S10). The structure of Ni2:[A74/C96RIDC1red]3 is nearly identical to that of the previously characterized Ni2:MBPC13 assembly (RMSD = 0.66 Å, Figure S12).13 While buried surface area and Rosetta interface calculations predict that the “up-up-up” trimer is less stable compared to the “up-up-down” configuration based purely on interfacial hydrophobic interactions (Table S1, S5), we propose based on DFT calculations of the metal coordination sites that the stability of the two octahedral NiII:His6 coordination motifs is sufficiently high to favour the assembly of the “up-up-up” trimer (Figure S13–14, Table S6; also see Supplementary Discussion).
In contrast to the FeII and NiII-directed assemblies, the crystal structure of the ZnII-directed assembly revealed a tetrameric, D4-symmetric architecture (Zn4:[A74/C96RIDC1red]4) in which the free Cys96 residues coordinate the metal ion (Figure 4a, S5b). The assembly features four identical, tetrahedral His2GluCys coordination sites (Figure 4a, S7b). Each antiparallel dimer is oriented about 75° with respect to the other (Figure 4b). This canted arrangement contrasts with the nearly collinear arrangement (θ = 21°) of antiparallel dimers in Zn4: [A74/C96RIDC1ox]4 (Figure 4b). Taken together, the Zn4:[A74/C96RIDC1red]4 and Zn-bound [A74/C96RIDC1ox]4 structures illustrate the dual functional role of cysteine as a metal coordinating ligand and covalent handle, which is critical to redox signalling in biological systems.10, 39, 40 Our results demonstrate that A74/C96RIDC1red can adopt three unique architectures in the presence of different metal ions (Figure 2–4), exemplifying how metal coordination preferences and hydrophobic packing can collectively influence assembly outcomes.
Fig. 4.
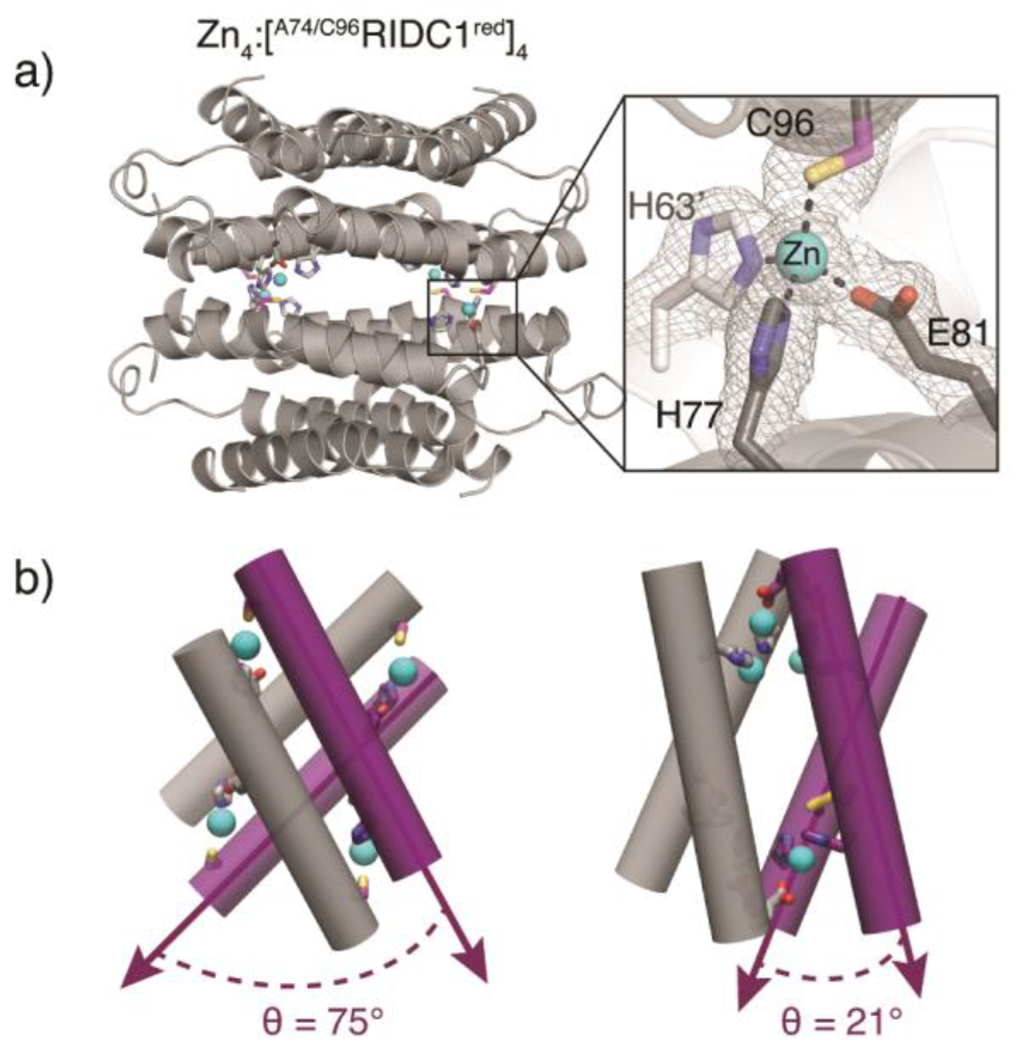
(a) Crystal structure of Zn4:[A74/C96RIDC1red]4 (PDB ID: 7RWX), highlighting one of four identical tetrahedral Zn coordination sites featuring Cys96 in the primary sphere. (b) Cartoon depictions of Zn4:[A74/C96RIDC1red]4 (left) and Zn4:[A74/C96RIDC1ox]4 (right) illustrating significant conformational differences between the two assemblies.
The ability of a single protein construct to assemble into and interconvert between multiple structural states is crucial for generating functional diversity and the generation of switchable systems.1, 2, 8, 9, 41 Herein, we have demonstrated that a designed protein (A74/C96RIDC1) can be subjected to redox- and metal-based stimuli to obtain five structurally distinct assemblies (Figure 2). The large structural diversity of A74/C96RIDC1 assemblies can be attributed to an intricate interplay between metal-ligand, disulphide bonding, and hydrophobic interactions. While potentially serving as starting points for engineering downstream functions, the dynamic A74/C96RIDC1 assemblies also pave the path to the generation of multistate protein switches.
Supplementary Material
Acknowledgments
This work was funded by NIH (R01-GM138884 and T32-GM112584) and NASA (80NSSC18M0093; ENIGMA: Evolution of Nanomachines in Geospheres and Microbial Ancestors (NASA Astrobiology Institute Cycle 8)). E.G. acknowledges funding by EMBO (ALTF 1336-2015). Portions of this research were carried out at the Stanford Linear Accelerator Center (supported by the DOE, Office of Basic Energy Sciences contract DE-AC02-76SF00515 and NIH P30-GM133894) and the Advanced Light Source at the Lawrence Berkeley National Laboratory (supported by the DOE, Office of Basic Energy Sciences contract DE-AC02-05CH11231and NIH P30-GM124169-01). Coordinate and structure factor files for the crystal structures have been deposited into the Protein Data Bank (www.rcsb.org) with the following accession codes: 7RWV (Apo [A74/C96RIDC1ox]4), 7SU2 (Co2:[A74/C96RIDC1ox]4), 7RWW (Zn4:[A74/C96RIDC1ox]4), 7RWY (Fe2:[A74/C96RIDC1red]3), 7RWU (Fe2:[A74/C96RIDC1red]3), 7TEP (Cu4:[A74/C96RIDC1red]4), and 7RWX (Cu4:[A74/C96RIDC1red]4).
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.James LC and Tawfik DS, Trends Biochem. Sci, 2003, 28, 361–368. [DOI] [PubMed] [Google Scholar]
- 2.Tokuriki N and Tawfik DS, Science, 2009, 324, 203–207. [DOI] [PubMed] [Google Scholar]
- 3.Zhu J, Avakyan N, Kakkis A, Hoffnagle AM, Han K, Li Y, Zhang Z, Choi TS, Na Y, Yu CJ and Tezcan FA, Chem. Rev, 2021, 121, 13701–13796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders BD, Jackson B and Marmorstein R, Biochim. Biophys. Acta, 2010, 1804, 1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chasteen ND and Harrison PM, J Struct Biol, 1999, 126, 182–194. [DOI] [PubMed] [Google Scholar]
- 6.Armstrong RN, Biochemistry, 2000, 39, 13625–13632. [DOI] [PubMed] [Google Scholar]
- 7.Eicken C, Pennella MA, Chen X, Koshlap KM, VanZile ML, Sacchettini JC and Giedroc DP, J. Mol. Biol, 2003, 333, 683–695. [DOI] [PubMed] [Google Scholar]
- 8.Groitl B and Jakob U, Biochim. Biophys. Acta, 2014, 1844, 1335–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenbaum DM, Rasmussen SG and Kobilka BK, Nature, 2009, 459, 356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fillat MF, Arch. Biochem. Biophys, 2014, 546, 41–52. [DOI] [PubMed] [Google Scholar]
- 11.Ghanbarpour A, Pinger C, Esmatpour Salmani R, Assar Z, Santos EM, Nosrati M, Pawlowski K, Spence D, Vasileiou C, Jin X, Borhan B and Geiger JH, J. Am. Chem. Soc, 2019, 141, 17125–17132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karchin JM, Ha JH, Namitz KE, Cosgrove MS and Loh SN, Sci. Rep, 2017, 7, 44388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salgado EN, Lewis RA, Mossin S, Rheingold AL and Tezcan FA, Inorg. Chem, 2009, 48, 2726–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai Y, Luo Q, Zhang W, Miao L, Xu J, Li H and Liu J, J. Am. Chem. Soc, 2013, 135, 10966–10969. [DOI] [PubMed] [Google Scholar]
- 15.Signarvic RS and DeGrado WF, J. Mol. Biol, 2003, 334, 1–12. [DOI] [PubMed] [Google Scholar]
- 16.Harrington L, Fletcher JM, Heermann T, Woolfson DN and Schwille P, Nat. Commun, 2021, 12, 1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballister ER, Lai AH, Zuckermann RN, Cheng Y and Mougous JD, Proc. Natl. Acad. Sci. USA, 2008, 105, 3733–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki Y, Cardone G, Restrepo D, Zavattieri PD, Baker TS and Tezcan FA, Nature, 2016, 533, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei KY, Moschidi D, Bick MJ, Nerli S, McShan AC, Carter LP, Huang PS, Fletcher DA, Sgourakis NG, Boyken SE and Baker D, Proc. Natl. Acad. Sci. USA, 2020, 117, 7208–7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campos LA, Sharma R, Alvira S, Ruiz FM, Ibarra-Molero B, Sadqi M, Alfonso C, Rivas G, Sanchez-Ruiz JM, Romero Garrido A, Valpuesta JM and Munoz V, Nat. Commun, 2019, 10, 5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alberstein RG, Guo AB and Kortemme T, Curr. Opin. Struct. Biol, 2021, 72, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhlman B, O’Neill JW, Kim DE, Zhang KY and Baker D, Proc. Natl. Acad. Sci. USA, 2001, 98, 10687–10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA and Baker D, J. Mol. Biol, 2003, 331, 281–299. [DOI] [PubMed] [Google Scholar]
- 24.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D and Bradley P, Meth. Enzymol, 2011, 487, 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brodin JD, Ambroggio XI, Tang C, Parent KN, Baker TS and Tezcan FA, Nat. Chem, 2012, 4, 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brodin JD, Smith SJ, Carr JR and Tezcan FA, J. Am. Chem. Soc, 2015, 137, 10468–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang M and Song WJ, Nat. Commun, 2019, 10, 5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao SR, Schettler SL and Horne WS, Chempluschem, 2021, 86, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salgado EN, Faraone-Mennella J and Tezcan FA, J. Am. Chem. Soc, 2007, 129, 13374–13375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radford RJ, Nguyen PC, Ditri TB, Figueroa JS and Tezcan FA, Inorg. Chem, 2010, 49, 4362–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salgado EN, Ambroggio XI, Brodin JD, Lewis RA, Kuhlman B and Tezcan FA, Proc. Natl. Acad. Sci. USA, 2010, 107, 1827–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medina-Morales A, Perez A, Brodin JD and Tezcan FA, J. Am. Chem. Soc, 2013, 135, 12013–12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rittle J, Field MJ, Green MT and Tezcan FA, Nat. Chem, 2019, 11, 434–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kakkis A, Gagnon D, Esselborn J, Britt RD and Tezcan FA, Angew. Chem. Int. Ed, 2020, 59, 21940–21944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Churchfield LA, Medina-Morales A, Brodin JD, Perez A and Tezcan FA, J. Am. Chem. Soc, 2016, 138, 13163–13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song WJ and Tezcan FA, Science, 2014, 346, 1525–1528. [DOI] [PubMed] [Google Scholar]
- 37.Brodin JD, Medina-Morales A, Ni T, Salgado EN, Ambroggio XI and Tezcan FA, J. Am. Chem. Soc, 2010, 132, 8610–8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ni TW and Tezcan FA, Angew. Chem. Int. Ed, 2010, 49, 7014–7018. [DOI] [PubMed] [Google Scholar]
- 39.Maret W, Biometals, 2009, 22, 149–157. [DOI] [PubMed] [Google Scholar]
- 40.D’Autreaux B, Pecqueur L, Gonzalez de Peredo A, Diederix RE, Caux-Thang C, Tabet L, Bersch B, Forest E and Michaud-Soret I, Biochemistry, 2007, 46, 1329–1342. [DOI] [PubMed] [Google Scholar]
- 41.Takken FL, Albrecht M and Tameling WI, Curr. Opin. Plant Biol, 2006, 9, 383–390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


