Abstract
Recent developments in applied developmental physiology have provided well-defined methodologies for producing human stem cell derived cardiomyocytes. The cardiomyocytes produced have become commonplace as cardiac physiology research models. Accessibility has also allowed for the development of tissue engineered human heart constructs for drug screening, surgical intervention, and investigating cardiac pathogenesis. However, cardiac tissue engineering is an interdisciplinary field that involves complex engineering and physiological concepts, which limits its accessibility. Our review provides a readable, broad reaching, and thorough discussion of major factors to consider for the development of cardiovascular tissues from stem cell derived cardiomyocytes. In this study, our review will examine important considerations in undertaking a cardiovascular tissue engineering project and will present, interpret, and summarize some of the recent advancements in this field. Throughout, we review different forms of tissue engineered constructs, a discussion on cardiomyocyte sources, and an in-depth discussion of the fabrication and maturation procedures for tissue engineered heart constructs.
Impact statement
With advancements in cardiac differentiation protocols, the production of human induced pluripotent stem cell derived cardiomyocytes is becoming cost effective and routine in the laboratory setting. Monolayer based culture methods are rapidly being replaced by three-dimensional (3D) tissue engineered constructs, which are more representative of the heart geometry. In the review presented, we delve into important concepts and tissue engineering principles that should be considered when generating 3D cardiac constructs, interpreting data acquired from, and embarking on a 3D cardiac tissue-based research project.
Keywords: organoids, EHTs, iPSCs, cardiac tissue, tissue engineering
Introduction
Heart failure rates are increasing,1 and more than 650,000 deaths are associated with cardiovascular disease each year in the United States.2 Given that heart failure rates are rising, investigating heart failure is a major research focus.3 The multitude of factors influencing the heart presents a significant challenge in the development of treatments for cardiovascular disease and the study of cardiac pathogenesis. For example, emotional stress,4 kidney functionality,5 and gut microbiota6 have been identified as significant factors that can modulate heart function. With an improved scientific understanding of factors influencing the human heart, heart failure can be combated through treatment, prevention, and even tissue engineering based surgical therapy.
Developing a deeper scientific understanding of the human heart through direct experimentation is not feasible due to ethical concerns.7 In addition, human based research can be difficult to reproduce and can be costly.8 The difficulty in reproducibility originates from the high variability in patient lifestyle, age, comedication, comorbidity, and from costs associated with obtaining a high sample size.9 Furthermore, only half of clinical trials are tested for reproducibility, and half of those tested were shown to be reproducible.10
The most common method for investigating cardiac physiology is through the use of preclinical trials on biological models, with computational models arising recently as an inexpensive alternative.11 These biological models come in three general forms—animal models and cell- and tissue-scale engineered heart constructs. Such models are advantageous compared to human models in several ways, as they allow researchers to induce disease states synthetically or exert an increased degree of control over the physiology of the system (Fig. 1). Animal models have historically been used as biological models to parse out individual mechanisms in the heart. However, animal models imperfectly approximate human physiology due to the multitude of physiological differences between human and animal hearts.12,13 For example, the resting heart rate in a mouse is ∼500 to 700 beats per minute (bpm).14–18 The resting heart rate of an adult human is between 60 and 90 bpm.19–22 The considerable heart rate differences between mice and humans make the study of heart diseases such as arrhythmogenic cardiomyopathy difficult. The more expensive and logistically complicated canine and porcine models possess fewer physiological differences from human hearts, but these differences are non-negligible. Aside from having nearly double the resting heart rate of humans, both pigs and dogs are quadrupeds, giving them altered valvular anatomy and subtracting much of the gravitational components of venal drainage found in humans.12 Furthermore, the porcine cardiac electrical activity is conducted primarily by specialized cardiac muscle cells, whereas the human heart possesses fewer neural ganglion cell bodies and primarily propagates its action potentials myogenically.23 The differences between human and animal models become even starker at the transcriptomic level.24,25 Succinctly, even very accurate animal models are insufficient to negate the need for extensive human clinical trials.
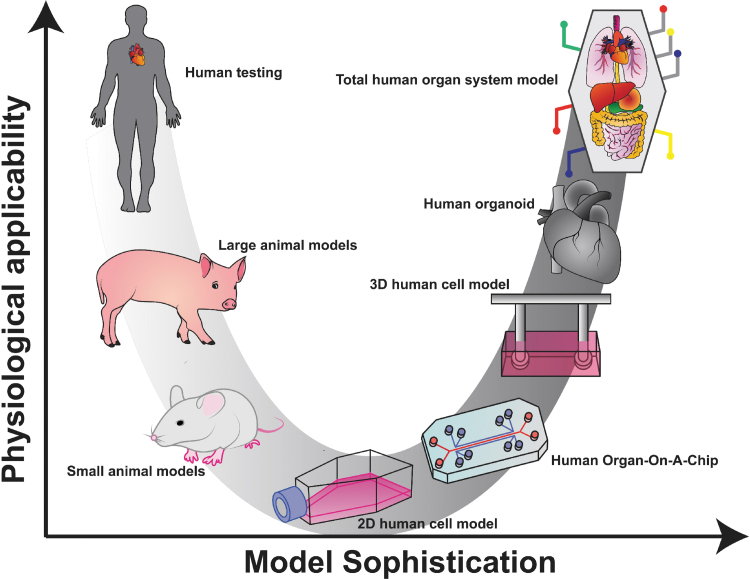
FIG. 1.
Biological models differ in applicability and sophistication. Biological models for the human body exist on a continuum of physiological applicability and model sophistication. Animal models are more sophisticated as they spare a human subject and are applicable in that they demonstrate the systemic effect of a treatment. Isolated cells, organ-on-a-chip, and 3D human cell models are more sophisticated approaches that allow for continuous observation and greater control over the stimuli provided to the heart cells but are incapable of recreating the influence of tissue-level and total systemic responses. Human heart organoids recreate tissue-level responses, and the idealized goal of a total organ system model would be capable of realizing total systemic responses. 2D, two-dimensional; 3D, three-dimensional. Color images are available online.
Because of the drawbacks of animal models for heart research, biologically engineered organ constructs are an attractive alternative. Cell-scale heart models have been under investigation since the 1990s and can be used to recreate the heart on a series of microfluidic chips.26 Microfluidic chips represent an “Organ-On-A-Chip,” which act as a platform to rapidly screen drugs for their impact on human heart cell contraction and metabolism.27 The concept of microfluidic chips is promising because alterations in human cardiomyocyte functionality can be detected in a high throughput manner with a relatively lower cost.26,28 However, microscale and monolayer-based systems do not allow for spatially separated three-dimensional (3D) coculturing or the direct observation of cardiac remodeling, both of which are significant contributors to cardiac tissue behavior.29 Tissue-scale heart constructs are a larger and more reagent intensive version of cell-scale heart constructs, but with the added benefit of allowing for 3D cell culture, observation of cardiac remodeling, and the coculture of multiple cell types in physiologically relevant spatial distribution.30–32 Furthermore, tissue-scale heart constructs can be used in surgical interventions for heart failure as heart tissue patches.33 Both cell- and tissue-scale heart models are under intensive investigation and have the potential to allow for relevant and high-throughput experimentation on human heart tissue to rapidly investigate disease pathogenesis and develop useful tissue engineered heart constructs.
Our review will outline the motivation for research using tissue engineered cardiac constructs, detail what considerations and techniques go into engineering cardiac constructs, and expound on the current discoveries and projects in cardiac tissue engineering.
Engineered Cardiac Constructs
Various cardiac constructs have been developed throughout the years (Fig. 2), and these constructs have been shown to produce more mature and physiologically accurate (addition of protein and collagen matrices)34 cardiac models compared to monolayer-based cultures. Specifically, 3D cardiac constructs have been reported to possess an enhanced cardiac ion channel density, faster upstroke velocity, increased catecholamine response,35 and matured membranous structure formation (T-tubule-like structures).36 These 3D cardiac constructs are assembled by various methods and may contain various cardiac cell types and matrices. A promising platform to assemble cardiac constructs is 3D bioprinting.37,38 Deposition of cardiac cell types and “bioinks” (permanent or dissolvable) spatially controls the construct architecture, cell type interactions formed, and the physiological relevance of the cardiac construct.39,40 This method has shown promise for use in anatomical studies, drug discovery/screening, and pathophysiological modeling.41–45
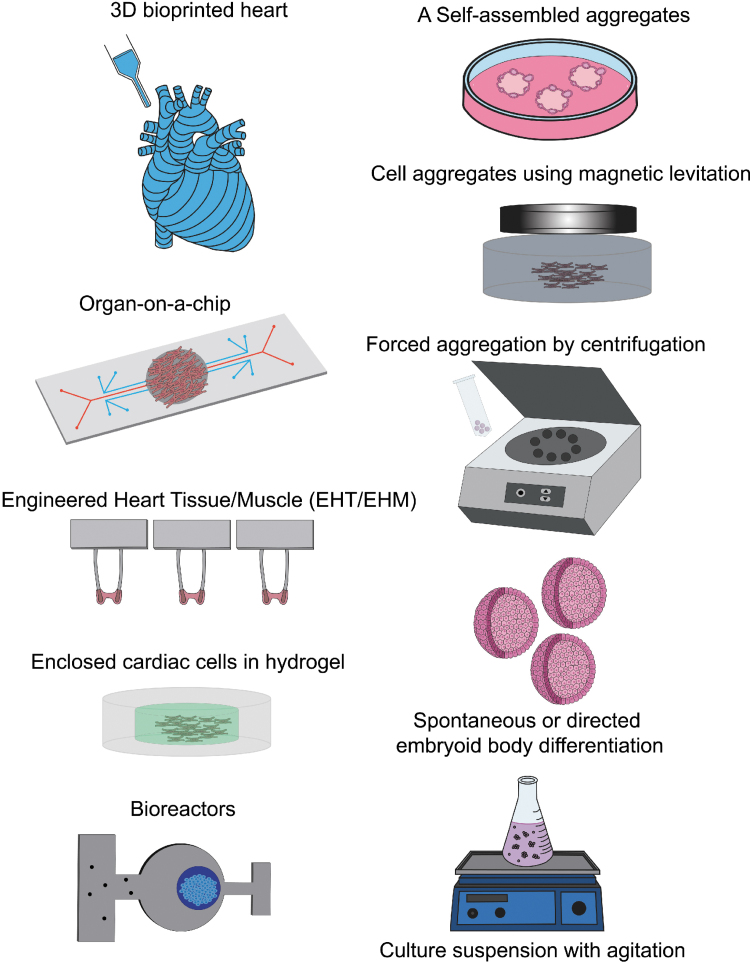
FIG. 2.
Three-dimensional induced pluripotent stem cell-derived cardiac constructs. A variety of 3D cardiac constructs have been developed to study cardiac physiology and to mimic the microenvironment of the heart. 3D bioprinted cardiac constructs promote higher spatial and anatomical accuracy with a mixture of various matrices and cardiac cells. Organ-on-a-chip and other microfluidic chips mimic complex human cardiac tissue at a miniaturized scale. EHTs/EHMs are optimized to measure the contractile force of cardiomyocytes mixed in a matrix and suspended between two posts. Hydrogels exhibit tissue-like properties to model the physiologically relevant cardiac microenvironment. Bioreactors mimic the fluid dynamics and nutrient need to assess cardiac constructs. Cardiac constructs can be generated using aggregation methods such as monolayer culture on a low adhesion surface matrix to spontaneously self-aggregate, magnetic levitation, forced aggregation using centrifugation, and spontaneous or directed cardiac differentiation in embryoid body form which can be produced at a larger scale using culture suspension with agitation (magnetic- or shaker based). Color images are available online.
Another method to generate 3D cardiac constructs is through self-aggregation. If cardiac constructs are generated by differentiating stem cells into cardiomyocytes, stem cells cultured in suspension will self-aggregate to form embryoid bodies (EBs). EBs can be expanding in suspension using a shaking platform or by magnetic agitation and directed to differentiation into beating cardiomyocyte clusters through growth factors and small molecule treatment further described below. Furthermore, cardiac cells cultured on a low adhering matrix will spontaneously self-aggregate.46–49 In addition, 3D cardiac constructs can also be generated using directed aggregation methods. Cells which have taken up magnetic nanoparticles can be forced to aggregate when a magnet is added below a culture well.50–52 Cells can also be forced to aggregate by being centrifuged into cone-shaped microwells.53
Various vessels support the culture environment of cardiac constructs. Aggregates enclosed within bioreactors which control the microenvironment of cells with a continuous flow of nutrient-rich media54 are shown to be more robust, have enhanced viability, and more uniform cell distribution and alignment.55 In addition, an overall improvement in cell distribution and morphology, cardiac protein expression, and tissue organization is seen with the use of perfusion in cardiac cell aggregates.56–58 Aggregates can also be molded into specific matrices or hydrogel to mimic the extracellular matrix environment of cardiac cells.59,60 Crosslinked 3D networks composed of hydrophilic polymers mimic the soft and flexible structures and water content of native tissues, which promote a more physiologically accurate cardiac model.61 Another vessel with a defined purpose involves the integration of microfluidic technology with living cells on slides to generate “organ-on-a-chip” systems. Organ-on-a-chip systems are focused on studying organ-specific interactions and replacing animal drug testing models.62
Finally, cardiac constructs such as engineered heart tissues or engineered heart muscles (EHTs/EHMs)63,64 have been developed with a focus on understanding cardiac contraction parameters and will serve to demonstrate a system that integrates many of the topics discussed in our article. The EHT system uses a convex mold to form molten agarose into 1 cm long rectangular cavities in each well of a 24-well plate. Two flexible posts are then suspended vertically in each well, and the cavity is filled with cardiomyocytes in a crosslinking matrix. After gelating, the resultant heart tissue is found to be strung between the two flexible posts and can be removed from the mold and placed in media, where it develops. Electrodes can be inserted on either side of the tissue to electrically pace the construct. Gravity tensions the tissue, promoting sarcomere alignment and causing contractions to pull the beams together. The resultant construct's contraction amplitude can be quantified into force units through the use of the two flexible beams attached to each end of the construct. Because the posts’ Young's modulus (E, units N/m2), length (L, units m), and cross-sectional moment of inertia (I, units m4) are known, video analysis that can measure the axial postdeflection (δ, units m) can be used to find the contraction force (F, units N or kg·m/s2) using the simple equation65 . The equation is one of the most widely used 3D tissue models that incorporates electrical and passive mechanical stimulation with contraction force transduction, with many applications having been explored since its inception in 2010.63 The system has also been modified extensively by investigators to include a variety of unique features, such as the addition of piezoelectric actuator for active mechanical stimulation, and a modular add-on that allows for the incremental increase of passive mechanical stimulation of the postbeams66,67 (Table 1).
Table 1.
Studies Utilizing the Engineered Heart Tissue Platform
| Verification of EHT platform | A review of the EHT platform and notable indications of accuracy in recreating realistic behavior of cardiomyocytes in monoculture. This review includes confirmation of accurate inotropic reactivity, orientation and structure, functionality, and cell morphology.239 |
| A verification that iPSC-derived cardiomyocytes, when adequately matured in the EHT system, have a physiologically accurate positive force frequency relationship.223 | |
| A confirmation that EHT matured cardiomyocytes in monoculture develop a resting membrane potential similar to the right or left ventricle in vivo. Outward potassium ion channel concentration in iPSC-CM-EHTs was also found to be comparable to human mature cardiomyocytes. iPSC-CM-EHTs did not have functioning acetylcholine activated potassium channels, indicating that they do not have a right ventricular phenotype. Action potential waveform indicates a left ventricular phenotype.240 | |
| Cell signal and drug screening performed | Evidence that miR-24 controls smooth muscle cell proliferation and vascularization, shown using the EHT platform.241 |
| Evidence that blocking miR-140-3p stops deterioration in cardiomyocytes under stress, shown using the EHT platform.242 | |
| Evidence that myosin binding protein C reduces the deterioration of cardiomyocytes under stress, shown using the EHT platform.243 | |
| Attempts at increasing physiological accuracy | A review of attempts to increase the physiological accuracy of tissue engineered heart constructs. Many of the constructs were not created using the EHT platform, but with functionally identical platforms. Briefly, mixing fibroblasts and endothelial cells increased contractility, and engineered heart constructs can be induced to pathological conditions when given the same stimuli as found in vivo.244 |
| A mixture of epicardial cells and cardiomyocytes improves contractility of the EHT system.245 | |
| Mechanical stimulation | A novel rack that has an inflexible steel rod that replaces one of the beams on each EHT beam pair. The steel rod is attached to a piezoelectric actuator which deflects the inflexible beam to stretch the tissue. This setup was also modified to fit a much larger EHT in a six-well plate. This setup improved contractility.67 |
| A magnetic-based system that uses one immovable post and one magnetic post. The magnetic post's stiffness is increased by bringing a magnet closer to it, which resists the cardiomyocyte contraction and increases the “afterload” perceived by the heart. This setup found that afterload tripled the force exerted by the heart.246 | |
| A modification of the EHT system that uses both a stiff beam and PDMS inserts that increase the force required to deform the beams. This allows for an analysis of how afterload impacts contractility.66 | |
| Similar systems | A system that uses independent beam sets instead of a rack of four.247 |
| A system that uses a solitary beam of cardiomyocytes in a custom well.248 | |
| A thin filament of cardiomyocytes suspended in a mold.217 | |
| A fibrous mesh of heart tissue. Coculture of cardiomyocytes and fibroblasts at a 7:3 ratio optimized contractility.249 |
CM, cardiomyocytes; EHT, engineered heart tissue; iPSC, induced pluripotent stem cell; PDMS, polydimethylsiloxane.
Fabrication Techniques
Various fabrication techniques exist to generate engineered cardiac constructs. One method, called molding, involves adding cells to a prepolymer solution that is then permitted to crosslink, forming a hydrogel that traps the cells within. Molding creates an immediately cellularized construct, although different protocols and scaffold choices may impact the distribution of cells. For instance, bovine collagen and rat tail collagen set at different rates due to differences in molecular weights, with bovine collagen setting so slowly that cells may settle out before gelation. The use of hydrogels with a suspension of cells requires a concave mold to shape the tissue, into which the uncrosslinked hydrogel mixture is poured to set. After the hydrogel gelates, the tissue is then removed from the concave mold, revealing a conjugate construct.
Another method for fabricating cellularized heart constructs called seeding involves fabricating an extracellular matrix in a particular shape, which is later populated with cells.68 The method of seeding may involve the use of decellularized cardiac tissue due to its physiological relevance or the generation of completely synthetic matrices using electrospraying,69 electrowriting,70 or electrospinning71 techniques due to their excellent material properties. The precise placement of aligned fibers that these techniques provide is particularly useful in cardiac tissue engineering, as aligned fibers have been shown to promote cardiomyocyte alignment, unidirectional action potentials along the fiber grain, and improved intracellular electrical coupling through the major gap junction protein, connexin 43.72 In addition, these techniques allow for a composite matrix, which is to say a scaffold with unique material properties in localized regions, akin to a skeleton. The scaffold allows for the implementation of desired material parameters on a tissue scale without compromising on material composition ideal on a cell scale.73 These unique attributes make electrospraying, electrospinning, and electrowriting excellent for specialized cardiac tissue engineering projects.
Another method of construct generation is the use of 3D printing. The three most well established forms of cellular 3D printing are printers that extrude “bio-ink” in stacked layers,74 “bio-ink-jet” printers that squirt tiny droplets of cells in stacked layers using heat or pressure,75,76 and the photolithography printers which use light to crosslink a solution containing cells and a photosensitive hydrogel precursor.77,78 These techniques allow for complex 3D shapes to be created, with the proper spatial distribution of matrix composition and cell lines. For example, myocardial patches with internal endothelial vascularization have been fabricated in a single process using 3D printing techniques.79
Engineered Cardiac Construct Uses
The predominant use of tissue engineered cardiac constructs is to model the composition and function of the human heart. Therefore, generating tissue engineered constructs as biological models involves the generation of cardiac cells in vitro to fabricate human cardiovascular tissues that precisely match the in vivo heart. Once constructed, stimuli can be introduced to help understand the mechanisms of disease and cardiac remodeling. The combination of various cell types, extracellular matrices, and soluble factors is necessary to replicate or approximate the heart's native tissue microenvironment.80 In tissue engineering, generating a construct that can support various heart proteins in combination with human pluripotent stem cells is advantageous since they can be differentiated into all human resident heart cells.
Another aim of cardiac tissue engineering is to fabricate surgical products for implantation to achieve an improved clinical outcome. The goal involves the study of how to maximize construct functionality and tissue integration. There are several loci of interest in the clinical field, including surgical patches, recellularized hearts, and total heart construction. Concisely, projects focused on the fabrication of surgical projects can be thought of as direct translational research.
While there are currently no Food and Drug Administration (FDA) approved tissue engineered surgical products for the heart, stem cell and cardiomyocyte injections have been in use for some time.81 However, some animal preclinical trials for tissue engineered heart constructs intended as surgical products are underway82–85 (Table 2). A detailed review has been written on the topic of heart patches.86 The distinction between “surgical product” and “modeling” oriented tissue engineering projects is important to keep in mind, as the end goal of each project informs the decisions made therein (Fig. 3). For instance, a project intended to produce a surgical product might make use of anatomically incorrect scaffold geometries such as honeycombs to improve contraction,87 with the intent of studying how to maximize force generation for the creation of more effective heart patches. Comparatively, a modeling project might make use of a coculture of cardiomyocytes and endothelial cells to more accurately recreate conditions in vivo.73
Table 2.
Projects Intended for Surgical Implantation
| Projects for implantation | The “BioVAD”, a pouch made of rat cardiomyocytes which fits over the heart. There was no substantial change in rat heart function after implantation.250 |
| A patch composed of rat neonatal heart cells, which was stitched onto the heart of a living rat with a myocardial infarction. This patch improved QRS amplitude stroke volume.251 | |
| A bioreactor that electrically paces myocardial patches derived from rat hearts.196 | |
| A honeycomb shaped heart patch to maximize contraction forces, made with rat neonatal cardiomyocytes.87 |
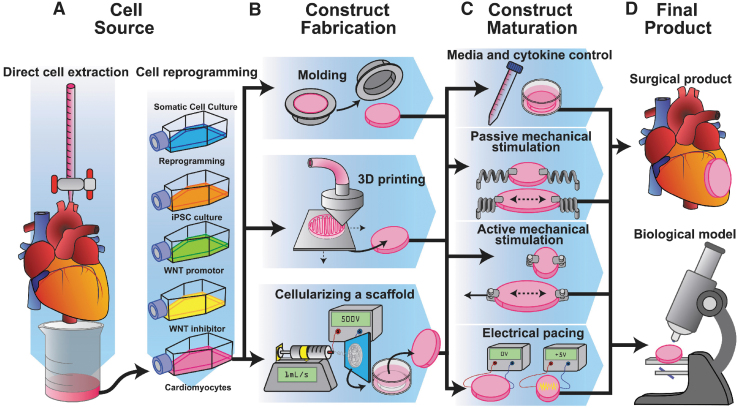
FIG. 3.
Techniques for cardiovascular tissue engineering. A summary of techniques for cell acquisition, construct fabrication, tissue maintenance, as well as the two major end products. (A) Heart cells are either retrieved from extracted heart tissue or generated through stem cell-based differentiations. (B) Isolated heart cells are then formed into a 3D construct through a variety of techniques. (C) 3D constructs are then matured through chemical, mechanical, and electrical stimulation. (D) The finalized product is then used to either model disease and study heart physiology or is used to form a useful heart construct for a patient. Color images are available online.
Engineered Cardiac Construct Components
Cell source
In a tissue engineered heart construct, cells are the most important component. There are two primary methods of gathering human heart cells—direct extraction from discarded tissue and the generation of cardiomyocytes through stem cell differentiation. Direct extraction yields more mature cardiomyocytes for use as a biological model and is typically performed on either discarded human tissue or from animal tissue.88–90 Cardiomyocytes that are extracted have been found to be large, rod shaped, and highly contractile.88,91 However, adult human cardiomyocytes are difficult to obtain.
An alternative is to generate human cardiomyocytes from stem cells.92–96 Human induced pluripotent stem cells (iPSCs) can be created by expressing four transcription factors into terminally differentiated cells to convert these cells into a pluripotent state.97 Once iPSCs have been generated, they can be expanded98 and differentiated into nearly any cell type (Fig. 3A). Pluripotent stem cells can be induced to differentiate into cardiomyocytes in a variety of ways, most notably through the use of growth factors.99–101 A growth factor based cardiac differentiation protocol using bone morphogenetic protein 4 (BMP4), basic fibroblast growth factor (FGF2), and activin is widely used to differentiate cardiomyocytes.102 However, a small molecule biphasic Wnt signaling modulation cardiac differentiation method103–105 is a fast, efficient, and cost-effective way to generate a high purity of iPSC derived cardiomyocytes. In addition, mechanical cues alone have been shown to differentiate cardiomyocytes through the integrin α and β signaling pathways.80,106,107 Several review articles have outlined the various methods used to differentiate and purify cardiomyocytes.108,109 One major advantage of iPSCs is the ability to generate stem cell derived cardiomyocytes from patients for use to model their patient-specific phenotype.110 Patient-specific phenotype models allow for the generation of virtually unlimited genetically identical cardiomyocytes from specific subjects for an intensive investigation into the exact genetic mechanisms of dysfunction for that phenotype.103,111
Cardiac tissues can also be differentiated in a more relevant manner through the use of a combination of mechanical and chemical cues in 3D in-situ differentiation, in which stem cells are added into a 3D matrix and provided with chemical signals to induce cardiac differentiation.112,113 3D in-situ cardiac differentiation may more accurately recapitulate the signal transduction pathway experienced by stem cells in utero and has been shown to produce differentiated tissues with spatially separated cardiac cell types.49
Matrix and composition
Due to the dynamic and kinetic nature of cardiovascular tissue, the cell density and protein matrix composition are important parameters to consider.114 While a hepatocyte might function on a rigid surface, a heart tissue's contraction would be less quantifiable (but not impossible to study115) if its force is exerted on a rigid surface. In addition, given the importance of the flexural, compressive, and tensile material properties of the heart, it is important to model the material composition found in the human heart (Fig. 4).
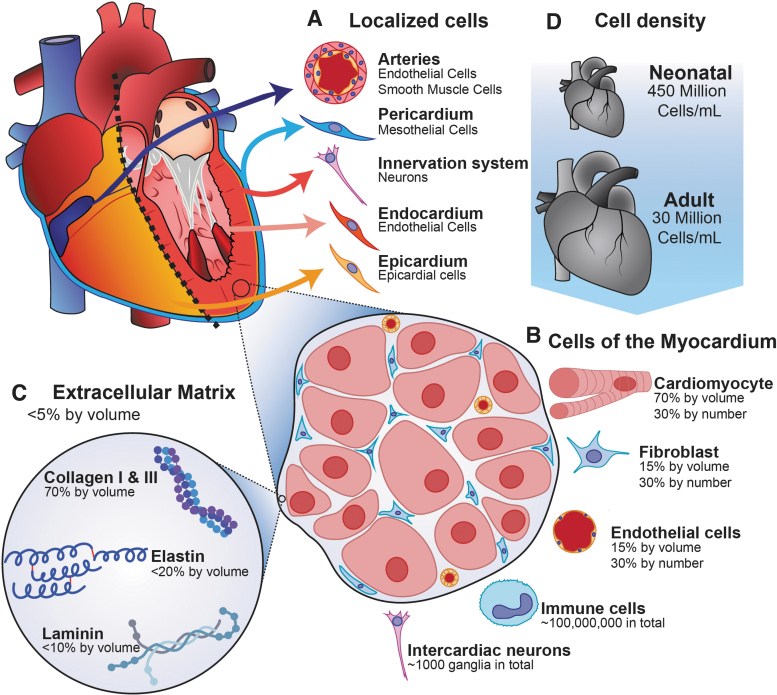
FIG. 4.
The tissue-level physiology of the human heart. The composition of the heart is varied based on position. (A) The heart has various subcomponents, including vasculature, the pericardium, neuronal innervation, and the endocardium. (C) Fibroblasts regulate the extracellular matrix of the heart, which is primarily composed of collagen I and III, elastin, and laminin. (B) The bulk of the heart is composed of the myocardium which by volume, is primarily composed of cardiomyocytes. The myocardium is heavily vascularized by capillaries, and other cell types are dispersed throughout. (D) The cell density of the human heart is highly variable, with neonatal human hearts having more than 10 times the cell density of mature hearts. Color images are available online.
Only 2–4% of a healthy human heart's volume is composed of the extracellular matrix.116 The matrix found in the human ventricular myocardium is around 70% collagen types I, III, V, and VI,117 80% of which is type I and the remainder of which is primarily type III.118 The remaining 30% of the matrix is composed of elastin and dozens of other proteins and glycoproteins (like laminin and fibronectin) found in smaller amounts.119 Collagen is largely inert when interacting with cardiomyocytes in monoculture, although there is some amount of interaction between extracellular collagen and surface-bound proteins on the cardiomyocyte.120
The cellular composition of the heart is also critical to consider. Seventy percent of the heart's volume is composed of contracting cardiomyocytes, with the remaining 30% composed mostly of fibroblasts and endothelial cells, although all three cell types are found in roughly equal quantities due to fibroblasts and endothelial cells having a much smaller volume.121 Cardiac fibroblasts in coculture have been shown to improve cardiomyocyte force production,122 and endothelial cells in coculture with cardiomyocytes improve cardiac development, contractility, and rhythmicity by secreting factors such as nitric oxide and neuregulin.123 In addition, neurons and immune cells are mixed throughout in low density.124–126 In terms of cellular density, the adult heart has on average 28 ± 7.2 million cardiomyocytes/mL, with neonatal hearts possessing 430 ± 72 million cardiomyocytes/mL.127
Cardiomyocytes have some ability to remodel their matrix by secreting collagen, but the majority of matrix maintenance is controlled by both fibroblasts and endothelial cells.128 There are few (but not zero120) interactions between collagen and human cardiomyocytes, making cardiomyocytes in monoculture somewhat ambivalent if collagen was exchanged with another matrix material. Similarly inert but flexible polymers may be used as matrix components in surgical product-based projects. In contrast, there are some notable interactions between the noncardiomyocyte cells in the heart and the extracellular matrix within the cardiac tissue that cannot be overlooked when an accurate recreation of the heart is intended. For example, fibroblasts indirectly influence cardiac function by regulating the extracellular matrix of the heart, which is achieved by fabricating and breaking down the heart's extracellular matrix continuously.128 Chemical and physical signals modulate this dynamic balance, and maladaptive signals are the source of some forms of cardiomyopathy. For instance, excessive angiotensin II, aldosterone, and deoxycorticosterone are all signals which promote cardiac fibroblasts to fabricate an excessive amount of collagen, leading to ventricular fibrosis.118 Similarly, endothelial cells produce a significant amount of collagen in response to certain physical signals, particularly ventricular overload.129 Fibroblasts and endothelial cells also produce matrix metalloproteases (MMPs), which degrade collagen but not necessarily other polymers with different compositions. Therefore, if a consistent and robust model of cardiovascular dysfunction is intended, noncardiomyocyte–cardiomyocyte interactions must be included, necessitating the use of a scaffold composed primarily of collagen or perhaps using a scaffold intended to be wholly replaced with collagen by fibroblasts over time.
Another consideration is the importance of stiffness and viscoelasticity on tissue properties. Fibrosis is a pathogenic state that is the subject of a major field of cardiovascular research, during which collagen stiffens the heart. Stiffness impacts cardiomyocytes in several ways, including their action potential,130 metabolic activity,131 gene expression, and contractility.132 The parameter that signifies the stiffness of an elastic material is the Young's modulus, which describes the amount of force over an area (N/m2) required to achieve a certain amount of “stretch” (also known as engineering strain, a unitless ratio), resulting in a unit of N/m2, which is equivalent to the unit of Pascals (Pa). A Young's modulus of 9.5 ± 1.5 kPa has been observed in decellularized pig hearts along their cellular alignment and 3.2 ± 00.7 kPa perpendicular to cellular alignment.133 The strain at which the decellularized porcine extracellular matrix failed was 25%.134 Despite this, cellularized heart tissue was found to be much less stiff.133 Destructive material testing on the human heart is less common, as both human and porcine hearts have very similar matrix compositions.135 Nevertheless, noninvasive ultrasonic techniques show that healthy young human hearts have a Young's modulus of 2.5 kPa, increasing to 6 kPa by the age of 60.136 Both of these levels of stiffness can be achieved with a matrix by creating a dense enough scaffold by controlling the concentration (typically 1–10 mg/mL) of matrix polymer in the prematrix solution, although the addition of cells does alter the resulting stiffness by obstructing polymer–polymer interactions and introducing cell–polymer interactions.137 Another important consideration is the viscoelasticity of a scaffold, which is the sensitivity of a scaffold's stiffness to the rate at which it is physically manipulated. The viscoelasticity of a material typically is described using a generalized Maxwell model, a differential equation that can calculate the stress experienced by a material when it experiences a certain rate of stretch (strain rate). Several models exist that take viscoelasticity into account.138 The viscoelastic properties of collagen and other matrix substances have been quantified as well, which allow for the precise tuning of the mechanical properties of heart tissue constructs to match the kinetic material properties of the heart.137,139 Another important note is that many matrix compositions are characterized by their shear modulus (G) and its derivatives the elastic modulus (G′) and the viscous modulus (G″). These shear moduli are used instead of the Young's modulus due to how hard it is to grip and pull on low-density matrices. A commonly used collagen density for tissue engineering is 2 mg/mL, which has an elastic modulus (G′) of 40.12 ± 3.29 Pascals and a viscous modulus (G″) of 3.43 ± 0.33 Pascals at 49% strain.140 The material properties of the extracellular matrix of tissue engineered cardiac constructs have a significant impact on construct functionality and gene expression,141 most notably through the mitogen activated protein and extracellular signal-regulated kinase (MAPK and ERK) pathways.142,143 Specifically, these pathways are modulated by mechanosensitive surface proteins such as Cav1 and β1 integrin,144 and they are regulated by the mechanosensitive gene iex-1 (IER3).145 Through these mechanisms, cardiac tissue reacts to excessive stiffness, leading to functional modification146 or apoptosis.147 Furthermore, on the tissue scale, very dense matrices slow cardiomyocyte contraction and relaxation by physically dampening movement, which impacts performance.148
With these considerations in mind, there are many choices in matrix composition for use in cardiac tissue engineering, with the most common being collagen, fibrin clots, Matrigel and equivalent products, and various other molecularly tailored resorbable polymers (Tables 3 and 4). Technically all these materials are hydrogels, which is to say a cross-linked scaffold in low concentration that absorbs a large amount of fluid, to the extent that 90% or more of the resulting gel is water by volume.
Table 3.
Biologically Derived Scaffold Solutions for Three-Dimensional Cell Culture
| Polymer | Description | Example uses | Considerations |
|---|---|---|---|
| Fibrin | Thrombin and Fibrinogen can be mixed to rapidly form a crosslinked hydrogel. | EHM122 Heart patches252 EHTs63 |
Not immediately physiologically relevant to the heart's matrix. This material has an RGD charged amino acid sequence that readily allows cells to bind to its surface. |
| Collagens | Collagen stored under acidic conditions to prevent crosslinking is neutralized, mixed with cells, and heated to crosslink. | EHTs63 | Physiologically relevant |
| Matrigel | A combination of proteins, glycoproteins, and cytokines derived from lysed mouse sarcomas, producing a liquefied basement membrane. When heated above 14°C, it crosslinks and forms a hydrogel. Its major components are laminin and collagen IV. | EHTs63 Pacemaker cardiomyocytes with vascularization253 |
An imperfectly characterized substance that will vary from batch to batch. |
| Other biologically derived hydrogels | Hyaluronic acid, gelatin, chitosan, alginate, and dozens of other proteins, glycoproteins, and GAGs have been used to form hybrid biodegradable scaffolds. | Alginate gels254 Gelatin gels255 Chitosan gels256 |
These materials have tunable characteristics, such as pore size and stiffness, which make them useful for adding unusual properties, including timed biodegradation or extremely low cost. |
| Biologically modified hydrogels | The extracellular matrix is not perfectly uniform, and certain pathological and healthy conditions lead to postprocessing. Some of these modified proteins have superior properties for tissue engineering. | Glycated collagen with improved stiffness257 UV-denatured collagen with improved stiffness258 |
There are many permutations of hydrogel bases and modifications, some of which are highly specialized for specific projects. |
EHM, engineered heart muscle.
Table 4.
Engineered Solutions for Three-Dimensional Cell Culture
| Polymer name | Description | Example uses | Considerations |
|---|---|---|---|
| PLGA | A copolymer of lactic and glycolic acid, two monomers with different bond strengths. By changing the ratio of these two components, their hydrolysis rate can be tuned. PLGA has been used to form hard sponges at high densities and flexible hydrogels at low densities. | Electrospinning into filamentous scaffold, forming over dissolvable templates, crosslinking to form hydrogels259 | Intended as a robust initial matrix to be dissolved and wholly replaced over time. Used clinically in implants.260 |
| Poly (glycerol sebacate) | An engineered two-part polymer that has excellent biocompatibility and easily tunable degradation rates and stiffness. | Heart patches261 | Cells cannot be integrated into the scaffold during the formation process due to high temperatures and vacuum conditions. Cells must infiltrate into the scaffold postformation. |
| Photosetting hydrogels | PEGDA is an inert and biodegradable polymer that rapidly crosslinks when exposed to UV light and has been used to form 3D printed constructs through photolithographic printing.262,263 Other similar photosetting hydrogels exist.264,265 | Creation of vasculature in engineered cardiac tissue77 | The field of fabricating 3D printed biological constructs is rapidly evolving, so new and improved photosetting hydrogels are constantly being published. |
| PEG | A tunable biodegradable and largely inert polymer which is typically used for microscale tissue engineering, although it can be used as a surface treatment or used to fill up a more rigid scaffold. | High throughput microscale cardiac coculture with endothelial and fibroblasts for drug screening.266 PEG as a surface treatment.267 |
Typically used for 2D cell culture. |
| Modified biological hydrogels | Biological hydrogels improved using chemicals or postprocessing techniques. | Highly cross-linked collagen using a synthetic crosslinking compound.268 | Maintains physiological applicability of the base matrix, but with modified characteristics. |
2D, two-dimensional; PEG, polyethylene glycol; PEGDA, poly (ethylene glycol) diacrylate; PLGA, poly-lactic-glycolic-acid.
Engineered Cardiac Construct Maturation
As components of a highly metabolic, dynamic, and kinetic tissue, cardiomyocytes require a complex combination of culture conditions to mature in a physiologically relevant manner. The maturation has been investigated thoroughly, and the following section will highlight significant protocols that have been used to generate more mature cardiomyocytes. Nevertheless, immature cardiomyocytes have been used as models for embryonic hearts,149 and immature cardiomyocytes possess useful properties for surgical products.150,151 As such, cardiac tissue maturation protocols must be carefully chosen to suit the intended purpose of the construct.
Media
Media components influence human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) maturity, and various media compositions have been published to promote maturation.152–154 A common base media for cardiomyocyte culture and maturation is RPMI 1640 media containing the B27 supplement (a supplement containing various peptides, lipids, and cell viability components). However, various small molecules, amino acids, and hormones (dexamethasone, thyroid hormone [T3], and insulin-like growth factor 1 [IGF-1]152,155–161) have been identified to promote iPSC-CM maturation (Table 5). Given that cardiomyocytes in vivo primarily derive their energy from fatty acids,162 some studies emphasize the importance of fatty acid supplementation (most notably oleic, linolenic, and lipoic acid163) to improve the maturity of iPSC derived cardiomyocytes,162,163 and other groups have emphasized that culturing cardiomyocytes using a combination of fatty acids and high glucose concentration leads to a more mature gene expression profile and contraction activity.164–166 However, normal blood glucose levels are around 5 mM,167 and supraphysiologic glucose levels have been shown to induce adipogenesis and alter cardiomyocyte development.164–166 Furthermore, culturing and differentiating human pluripotent stem cells can be costly, and alternative, cost-effective media formulations exist to culture (B8) and differentiate iPSCs into cardiomyocytes (CDM3).103,168–170
Table 5.
Cardiac Cell Maturation Components
| Classification | Maturation component | Description |
|---|---|---|
| Culture supplement | B27 | B27 is a media supplement containing various hormones, lipids, and amino acids that promotes iPSC-CM maintenance and maturation.269 |
| Insulin | Insulin supports the growth and metabolism of myocardial cells.270 | |
| Hormone | Thyroid hormone (T3) | T3 increases contractile force, calcium release and reuptake, and cardiomyocyte size.155–157,271 |
| Dexamethasone | Dexamethasone, which is a glucocorticoid which enhances electromechanical maturation of iPSC-derived cardiomyocytes.152 | |
| Growth factor | IGF-1 | IGF-1 regulates contractility, metabolism, hypertrophy, and apoptosis in the heart.155 |
| Fatty acids | Oleic acid | Fatty acids improve contractile force, metabolism, and function of cardiac cells.163,271–273 |
| Palmitic acid/palmitate | ||
| Linoleic acid | ||
| Sodium L-lactate | ||
| Small molecules | Phosphodiesterase inhibitor (IBMX) | The addition of IBMX increases contractile activity and force.160,161 |
| HIF-1α inhibitor (FM19G1) | Increases fatty acid oxidation.272 | |
| PPARα agonist (WY-14643) | Facilitates mitochondrial metabolic maturation.272 | |
| mTOR inhibitor (Torin1) | Facilitates cardiomyocyte quiescence.274 | |
| Monosaccharide | Galactose | Addition of galactose to maturation media improves maturation speed and total oxidative capacity.271 |
| Amino acids | Taurine | Taurine is used for fat absorption, cardiomyocyte energetics, and as a pH buffer in the mitochondrial matrix for stabilization.152,275 |
| L-glutamine | L-glutamine promotes increased beating function and decreased apoptosis of cardiomyocytes.276 | |
| L-carnitine | Carnitine assists in transport of fatty acids through the mitochondrial membrane.163,277 | |
| Creatine | Creatine is an important temporal and spatial energy source.275,277 | |
| Genetic manipulation | Let-7 | Let-7 enhances cardiomyocyte size, sarcomere length, contraction force, and respiratory capacity.278 |
| microRNA-1 | miR-1 assists in facilitation of electrophysiological maturation.158,279 | |
| microRNA-499 | miR-499 promotes ventricular specification of human embryonic stem cells.158,279 | |
| Micropatterning | Sarcomeric alignment | Cardiomyocyte sarcomeric alignment and formation are improved after plated onto micropatterned matrices.22,280–283 |
IGF-1, insulin-like growth factor 1.
Another consideration is that the importance of preserving the matrix that the cells inhabit is also through the use of media supplements. One common approach is supplementing media with the MMP inhibitor aprotinin171 to prevent noncardiomyocyte cells in coculture from substantially remodeling the matrix.172 However, matrix degradation and production in vivo are in a dynamic counterbalance, and interfering MMP may subsequently impact the physiological component of the construct.173
Electrical stimulation
To have a physiologically relevant cardiomyocyte tissue construct, it is important to encourage the cells to mature through electric and mechanical pacing. Pacing has been shown to improve the contractility,174 intracellular protein density, and transcriptome36,175,176 of cardiomyocytes. The biological basis for electrical pacing is to replicate a sudden charge differential between the inside and outside of a cardiomyocyte, which occurs at the beginning of an action potential. The interior of a mature human cardiomyocyte is typically polarized to −90 mV at rest, due to ion pumps keeping positive calcium ions in the sarcoplasmic reticulum or outside the cell. The initiation of contraction is accomplished through the depolarization of specialized cardiac pacemaker cells.
Cardiomyocytes in contact will spontaneously synchronize calcium channel depolarization and develop a sarcoplasmic reticulum, which may lead to rhythmic contraction within 30 days of culture.177–181 However, slight differences in stress and tissue geometry can alter the contraction frequency, which over time will alter tissue performance. For example, cardiac patches with a honeycombed structure permit improved cardiomyocyte alignment in comparison to disc-shaped patches, resulting in a more contractile tissue.182 To control the contraction differences, electrical pacing has been used, which recreates the conditions that lead to cardiomyocyte maturity in a noncontact manner. For projects intended to fabricate a surgical product, pacing is important for the integration of the resulting tissue into the native electrical pacing system in the recipient's heart.175,183,184 Similarly, projects intended to model the human heart must consider the development of pacemaker cells and the electric coupling of cardiomyocytes to the neuronal signal induction system found in vivo. Generally, electrical pacing for tissue engineering is performed by inserting an inert electrode composed of graphite or platinum into the media on either side of a tissue construct and by allowing a pulse of electrical current to pass through the tissue for a known duration (pulse width) and a known period between pulses (pulse frequency).185,186 The pulse generates an electric field that replicates the electrical field caused by ion movement during an action potential.187 Another method for electrical stimulation is the use of a salt bridge, which is the same concept but with a wet sponge separating the electrode from the target tissue.188 The salt bridge sponge allows current and certain ions to travel between the electrodes but prevents biomolecule aggregation on the electrodes. Both methods coordinate the tissue to repolarize and depolarize at a designated frequency, which “exercises” the construct.
As for the duration of the stimulation, a 1–2 ms pulse is sufficient for a 5 V/cm field strength over a 1 cm distance, with smaller electrical field strengths requiring larger pulse lengths.189 An important note is that a direct current passing between the two electrodes aggregates biomolecules on the surface of the electrodes, due to net or gross electrical charges on these molecules.190 A biphasic signal (a positive current that is then countered by an inverse current of equal duration, for a net current of zero) is sometimes used for stimulation to result in a net zero charge over the duration of the stimulation. However, the biphasic waveform is not physiologically accurate.191
Another consideration is the frequency of stimulation. Physiological heart rate ranges from 0.07 Hz (4 bpm) in diving whales to 20 Hz (1200 bpm) in hummingbirds, with humans ranging from 0.7 Hz (40 bpm) to 3.3 Hz (200 bpm). Higher stimulation frequencies can be used to investigate diseased states, representing the higher heart rate (tachycardia) required by weak tissues to pump the same amount of blood. Higher rates can also be used to “exercise” the heart to a hypertrophic state.
Stimulation intensity is often left undefined by researchers due to its complexity and the difficulty in directly measuring the intensity within the myocardium of a healthy human heart. Signal intensity for pacing is measured in units of Volts/cm (cm being the distance between electrodes) or milliamps, with relevant levels being between 0.1 and 10 V/cm192 and a reasonable level of continuous stimulation being 2–7 V/cm.175,193,194 Larger setups require larger voltages to achieve 5 V/cm, with greater distances between electrodes resulting in greater resistance and thus lower amperage and electric field strength. In terms of resistivity, the conductivity of saline solution is around 1.4 S/m or 0.7 Ω·m,195 giving saline a resistance equation of . As an example, a 1 cm spacing between two 1 cm2 rectangular electrodes in a 24 well plate would have an end-to-end resistance of around 70 Ω per well. If 24 wells were given similar electrodes, the effective area of the electrodes would increase 24 times, and the overall resistance would be 3 Ω. Each 70 Ω well draws 70 mA from the 5 V source, which is a reasonable stimulation intensity for a 1 ms pulse.194
One way to quantify the safety of electrical pacing is with the use of current density, which is the number of amperes passing through a unit area (mA/cm2), which can be reduced by increasing the surface area of the electrodes used. Current densities below 100 mA/cm2 are sufficient to pace cardiomyocytes.196 However, a major unmet need in tissue engineering investigation is a unified protocol for pacing cardiomyocyte tissue constructs, described using quantitative measurements of construct stimulation that are interchangeable between systems.
A final consideration is the impact of electrolysis, in which a strong electric current breaks down certain chemicals into smaller molecules. One major example of electrolysis is the rapid electrodegradation of phenol, which is the active component in the pH indicator used in most forms of media.197 Because electrolysis occurs, it is important to carefully decide how strong of a stimulus to use and what effect the stimulus will have on the media and its content.
Mechanical stimulation
The mechanical stimulation of heart tissue is also a significant contributor to engineered heart tissue development. The mechanical properties of the human heart change substantially as the heart matures (with stiffness increasing with age).198 The physical change in the stiffness of the heart is an important indicator of heart development, and the stiffness of tissue engineered cardiac constructs has an impact on construct functionality. Construct stiffness can be nondestructively quantified through physical tests of the material properties of the engineered tissue, including ultrasound and biopsies.199–201 Due to the conditions of cardiac tissue culturing, in vitro quantification of stiffness is typically accomplished by growing the tissue in specialized culture setups capable of measuring their material properties or through destructive testing.202 Cardiac tissue stiffens along its striations as it matures, and engineered tissues that undergo mechanical stimulation demonstrate substantial improvements in cardiac orientation.203,204 The mechanisms by which cardiac tissues detect mechanical stimulation involve the MAPK and ERK signaling pathways, which are modulated by stretch and cyclic stretch responsive proteins such as titin and titin-associated proteins.142,205–207
Mechanical stimulation has historically been performed on two-dimensional (2D) cell cultures of cardiomyocytes grown in monoculture on a flexible inert material, such as polydimethylsiloxane (PDMS). PDMS can be treated to increase the capacity of cells to adhere through a variety of techniques, including using the same plasma treatment used to create cell culture plates,208 functionalizing the surface with adhesive substances209 or washing the surface with polydopamine to increase the surface's wettability.210 Historically, mechanical stimulation has been achieved by modulating surface stiffness,211,212 stretching the material stepwise over time,213 or by dynamically actuating the material to simulate normal heart functionality.214
In recent years, 3D mechanical stimulation systems have become more common.215–217 These 3D systems allow for greater construct handleability, making dynamic mechanical stimulation a feasible endeavor. Dynamic mechanical stimulation achieves superior outcomes by many metrics.214 The quantification of dynamic mechanical stimulation has two forms, stress and strain. There are multiple forms of strain, with the most appropriate for cardiomyocytes being engineering strain which denotes the change in the length of a material in one direction. Engineering strain describes increased length (e.g., a 10% increase), whereas the strain rate of a system is the amount of strain given to a system over time (e.g., 10% over 1 s). Strain rate is important because the heart is a viscoelastic material, which makes the rate of applied strain significant in determining material behavior. Steel, for example, is not particularly viscoelastic and will snap at around the same tension whether it is stretched quickly or slowly. In contrast, biological tissue will snap easier if subjected to tension at a higher strain rate.218 Excessive strain rates may damage the tissue and may give incorrect measurements of cardiomyocyte contractility originating from the viscoelastic behavior of the extracellular matrix. The physiological origin of strain stimulation is the stretch experienced by cardiomyocytes during heart filling, and a strain stimulation of 10% has been used extensively for tissue engineered heart constructs.175
Stress, however, is the amount of tension or compression exerted on an area. A heart tissue can be stimulated with stress by giving it a set amount of tension to pull against or the construct can be stimulated by strain by stretching the tissue for a set amount. However, stress is an uncommon method of mechanical stimulation, as variability between constructs makes a unified stress stimulation protocol difficult, whereas strain is normalized to construct geometry. Both stress and strain can be used to tension a heart in one direction, which is important for proper cardiomyocyte sarcomere alignment.219 Some systems use a 2D stretch instead of a 1D tension, which creates an isotropic force that produces a radial cardiomyocyte orientation, as seen at the apex of the heart.220
Both mechanical and electrical stimulation can be used in conjunction to produce excellent systems capable of stimulating tissue engineered heart constructs in a physiologically applicable way.175
Engineered Cardiac Construct Function and Molecular Assessment
A final consideration is the analysis of tissue engineered heart constructs. The primary methods of analyzing a tissue engineered heart construct include electrophysiological, biochemical, and morphological analysis. The intended purpose of the construct greatly alters the importance of the various parameters outlined in the following section. For instance, modeling-based projects may attempt to maximize the transcriptomic and proteomic applicability of their construct, with little regard for the contraction strength of the construct. In contrast, surgical product-based projects may hope to maximize the contractility of their constructs and focus their transcriptome analysis on conduction and structural biomarkers.
Contractility analysis is a nondestructive test that is unique to each construct, with 2D constructs typically involving video analysis of the monolayer115 or specifically designed bioreactors capable of transducing contraction into a force value.221 3D constructs allow for direct measurement of contraction using force sensors or calibrated materials for force quantification.67,215,216 Contractility analysis can be used to demonstrate the maturation of the heart tissue by quantifying frequency, contraction duration and amplitude variance, and rate of contraction and relaxation. The action potential can also be analyzed nondestructively using patch clamp or calcium-sensitive dyes.122,222,223 Another method of analyzing the action potential of cardiac constructs is through voltage sensitive dyes and gene-encoded voltage indicators. With the use of membrane potential dyes, physiological changes such as muscle contraction and cell signaling can be more accurately measured and analyzed.224,225 Unfortunately, the unique physical shapes of different constructs make contractility data difficult to compare between systems, although 3D systems can divide construct force by construct cross-sectional area to quantify tissue level contraction strength. Destructive tests that isolate the contractile machinery allow for a standardized comparison between cardiomyocytes from different systems.226
Proteomic and transcriptomic analysis are also highly important in the quantification of heart construct functionality. Several biomarkers can be used to quantifiably compare constructs, establish a measurement of maturity (MYH6 levels compared to MYH7), cell population identity (CD31 for endothelial cells, PDGRFA for fibroblasts), and to assess for cardiomyocyte subtype heterogeneity (MYL2 for ventricular, SLN for atrial). Quantification of biomarkers allows for the systematic improvement of constructs into progressively more physiologically applicable models.
Furthermore, given that CRISPR/Cas9 technology has become more widespread, genetic manipulation of iPSCs is now becoming routine. It is now possible to directly compare cardiomyocytes from an iPSC line harboring cardiomyopathy causing mutations in direct comparison with isogenic corrected hiPSC-CMs. In addition, CRISPR/Cas9 can be used to fluorescently tag various cardiac proteins such as titin so contractile analysis can be performed easily.227 Furthermore, the use of CRISPRi and CRISPRa within iPSCs now presents the opportunity to turn on or off genes being studied either during differentiation or to assess end states of disease.228–230
Morphological analysis is primarily performed through the use of histology techniques such as immunohistochemistry to identify the location and concentration of substances of interest.231 Hematoxylin and eosin stains are used to visualize the extracellular matrix and cell density inexpensively, while fluorescent antibodies commonly used to inspect sarcomeric integrity and alignment include cardiac troponin T (TNNT2), actin (ACTB), myomesin (MYOM1), and actinin (ACTN1 and ACTN2).
Conclusion
The field of heart tissue engineered is exciting, relevant, and has the potential to revolutionize drug screening, cardiovascular surgical techniques, and eventually even human heart transplants. The general trends in cardiac tissue engineering are dependent on the project type. For modeling-based projects, the general trends are to develop techniques for differentiating a broader array of cell types, generate constructs with specific numbers of different cell types placed in a particular spatial distribution, and to improve the applicability of construct behavior using more relevant electrical and mechanical pacing techniques. For surgical product-based projects, the general trends are to develop more sophisticated and tuned matrix geometries and compositions, cell type mixes with greater contractility, and pacing techniques that achieve stronger contractions and capacity for electro-coupling with the existent heart. However, both types of projects suffer from a lack of standardization.
The future of cardiac tissue engineering must include a greater standardization of techniques to facilitate more rapid technology transfer between research groups. A standardized protocol for electrical and mechanical stimulation that takes electrode and construct geometry into account is needed, as is a standardized media composition schedule that optimizes cardiomyocyte development and maturation. Furthermore, a standardization of input factors and analytes, including cell count, maturity markers, cell origin, and matrix density, would help to isolate the influence of controlled variables. As for the future of tissue engineering projects, there is a substantial unmet need for a commercially available, electrically and mechanically paced, chambered, and macroscopically 3D construct generation system. Such a system would be able to include other important forms of stimulation for the heart, including surface shear and spatially separated cell layers.
While there have been significant advancements in assembling and functionally assessing cardiac constructs, it is still not possible to perfectly mimic the human cardiac cell populations (cardiac fibroblasts are derived from multiple developmental cellular lineages), develop a stable and functional tissue matrix,232 model chamber specific spatial pathophysiologies (fatty-fibro infiltration), establish robust and synchronous cardiac contraction,233 and engineer vascularized constructs at the thickness of the adult heart.234 The tissue engineering field is continuously evolving as the need to understand the complexities of cardiovascular diseases and disorders.235 Stepwise progress will eventually lead to a robust platform to understand and model various cardiomyopathies with the goal to create a functional engineered heart designed for transplantation.236,237 Although this is an active field of research, the future of engineered heart models is promising.238
Acknowledgment
The authors thank Sobhi Kazmouz for help in editing the article.
Disclosure Statement
No competing financial interests exist.
Funding Information
This review was funded by the Computational and Mathematical Modeling of Biomedical Systems training grant (Grant number 5T32GM084905-03), the National Institutes of Health under the National Heart, Lung, and Blood Institute (NHLBI) R00 HL128906 (J.M.C.), and the Steven M. Gootter Foundation.
References
- 1. Anderson, L.A., Goodman, R.A., Holtzman, D., et al. Aging in the United States: opportunities and challenges for public health. Am J Public Health 102, 393, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benjamin, E.J., Blaha, M.J., Chiuve, S.E., et al. Heart disease and stroke statistics-2017 update: a report from the American Heart Association. Circulation 135, e146, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davis, R.C., Hobbs, F.D., and Lip, G.Y.. ABC of heart failure. History and epidemiology. BMJ 320, 39, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Watkins, L., Blumenthal, J., Babyak, M., et al. Phobic anxiety and increased risk of mortality in coronary heart disease. Psychosom Med 72, 664, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Damman, K., and Testani, J.M.. The kidney in heart failure: an update. Eur Heart J 36, 1437, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tang, W.H., Kitai, T., and Hazen, S.L.. Gut microbiota in cardiovascular health and disease. Circ Res 120, 1183, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandrasekera, P.C., and Pippin, J.J.. The human subject: an integrative animal model for 21(st) century heart failure research. Am J Transl Res 7, 1636, 2015. [PMC free article] [PubMed] [Google Scholar]
- 8. Constantin, A. Human subject research: international and regional human rights standards. Health Hum Rights 20, 137 2018. [PMC free article] [PubMed] [Google Scholar]
- 9. Bøtker, H.E., Hausenloy, D., Andreadou, I., et al. Practical guidelines for rigor and reproducibility in preclinical and clinical studies on cardioprotection. Basic Res Cardiol 113, 39, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Niven, D.J., McCormick, T.J., Straus, S.E., et al. Reproducibility of clinical research in critical care: a scoping review. BMC Med 16, 26, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Garbern, J.C., Mummery, C.L., and Lee, R.T.. Model systems for cardiovascular regenerative biology. Cold Spring Harb Perspect Med 3, a014019, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Camacho, P., Fan, H., Liu, Z., et al. Large mammalian animal models of heart disease. J Cardiovasc Dev Dis 3, 30, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Camacho, P., Fan, H., Liu, Z., et al. Small mammalian animal models of heart disease. Am J Cardiovasc Dis 6, 70, 2016. [PMC free article] [PubMed] [Google Scholar]
- 14. McCauley, M.D., and Wehrens, X.H.. Animal models of arrhythmogenic cardiomyopathy. Dis Model Mech 2, 563, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Janssen, P.M., Biesiadecki, B.J., Ziolo, M.T., et al. The need for speed: mice, men, and myocardial kinetic reserve. Circ Res 119, 418, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Janssen, B., Debets, J., Leenders, P., et al. Chronic measurement of cardiac output in conscious mice. Am J Physiol Regul Integr Comp Physiol 282, R928, 2002. [DOI] [PubMed] [Google Scholar]
- 17. Georgakopoulos, D., and Kass, D.. Minimal force-frequency modulation of inotropy and relaxation of in situ murine heart. J Physiol 534, 535, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lujan, H.L., and DiCarlo, S.E.. Cardiac output, at rest and during exercise, before and during myocardial ischemia, reperfusion, and infarction in conscious mice. Am J Physiol Regul Integr Comp Physiol 304, R286, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mason, J.W., Ramseth, D.J., Chanter, D.O., et al. Electrocardiographic reference ranges derived from 79,743 ambulatory subjects. J Electrocardiol 40, 228, 2007. [DOI] [PubMed] [Google Scholar]
- 20. Gillum, R.F. Epidemiology of resting pulse rate of persons ages 25-74—data from NHANES 1971-74. Public Health Rep 107, 193, 1992. [PMC free article] [PubMed] [Google Scholar]
- 21. Kannel, W.B., Kannel, C., Paffenbarger, R.S.Jr., et al. Heart rate and cardiovascular mortality: the Framingham Study. Am Heart J 113, 1489, 1987. [DOI] [PubMed] [Google Scholar]
- 22. Avram, R., Tison, G.H., Aschbacher, K., et al. Real-world heart rate norms in the Health eHeart study. NPJ Digit Med 2, 58, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lelovas, P.P., Kostomitsopoulos, N.G., and Xanthos, T.T.. A comparative anatomic and physiologic overview of the porcine heart. J Am Assoc Lab Anim Sci 53, 432, 2014. [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao, Y., Sheng, Z., and Huang, J.. A systematic analysis of heart transcriptome highlights divergent cardiovascular disease pathways between animal models and humans. Mol Biosyst 8, 504, 2012. [DOI] [PubMed] [Google Scholar]
- 25. Anzai, T., Yamagata, T., and Uosaki, H.. Comparative transcriptome landscape of mouse and human hearts. Front Cell Dev Biol 8, 268, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu, Q., Liu, J., Wang, X., et al. Organ-on-a-chip: recent breakthroughs and future prospects. Biomed Eng Online 19, 9, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sosa-Hernández, J.E., Villalba-Rodríguez, A.M., Romero-Castillo, K.D., et al. organs-on-a-chip module: a review from the development and applications perspective. Micromachines (Basel) 9, 536, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ribas, J., Sadeghi, H., Manbachi, A., et al. Cardiovascular organ-on-a-chip platforms for drug discovery and development. Appl In Vitro Toxicol 2, 82, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mathur, A., Loskill, P., Shao, K., et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep 5, 8883, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Veldhuizen, J., Migrino, R.Q., and Nikkhah, M.. Three-dimensional microengineered models of human cardiac diseases. J Biol Eng 13, 29, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eyckmans, J., and Chen, C.S.. 3D culture models of tissues under tension. J Cell Sci 130, 63, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma, Z., Koo, S., Finnegan, M.A., et al. Three-dimensional filamentous human diseased cardiac tissue model. Biomaterials 35, 1367, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ye, L., Zimmermann, W.H., Garry, D.J., et al. Patching the heart: cardiac repair from within and outside. Circ Res 113, 922, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lemoine, M.D., Mannhardt, I., Breckwoldt, K., et al. Human iPSC-derived cardiomyocytes cultured in 3D engineered heart tissue show physiological upstroke velocity and sodium current density. Sci Rep 7, 5464, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Uzun, A.U., Mannhardt, I., Breckwoldt, K., et al. Ca(2+)-currents in human induced pluripotent stem cell-derived cardiomyocytes effects of two different culture conditions. Front Pharmacol 7, 300, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ronaldson-Bouchard, K., Ma, S.P., Yeager, K., et al. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emmert, M.Y., Hitchcock, R.W., and Hoerstrup, S.P.. Cell therapy, 3D culture systems and tissue engineering for cardiac regeneration. Adv Drug Deliv Rev 69–70, 254, 2014. [DOI] [PubMed] [Google Scholar]
- 38. Cui, H., Miao, S., Esworthy, T., et al. 3D bioprinting for cardiovascular regeneration and pharmacology. Adv Drug Deliv Rev 132, 252, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duan, B. State-of-the-art review of 3D bioprinting for cardiovascular tissue engineering. Ann Biomed Eng 45, 195, 2017. [DOI] [PubMed] [Google Scholar]
- 40. Murphy, S.V., and Atala, A.. 3D bioprinting of tissues and organs. Nat Biotechnol 32, 773, 2014. [DOI] [PubMed] [Google Scholar]
- 41. Rimann, M., and Graf-Hausner, U.. Synthetic 3D multicellular systems for drug development. Curr Opin Biotechnol 23, 803, 2012. [DOI] [PubMed] [Google Scholar]
- 42. Roth, A., and Singer, T.. The application of 3D cell models to support drug safety assessment: opportunities & challenges. Adv Drug Deliv Rev 69–70, 179, 2014. [DOI] [PubMed] [Google Scholar]
- 43. Liu, N., Ye, X., Yao, B., et al. Advances in 3D bioprinting technology for cardiac tissue engineering and regeneration. Bioact Mater 6, 1388, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mir, T.A., and Nakamura, M.. Three-dimensional bioprinting: toward the era of manufacturing human organs as spare parts for healthcare and medicine. Tissue Eng Part B Rev 23, 245, 2017. [DOI] [PubMed] [Google Scholar]
- 45. Mathur, A., Ma, Z., Loskill, P., et al. In vitro cardiac tissue models: current status and future prospects. Adv Drug Deliv Rev 96, 203, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lancaster, M.A., and Knoblich, J.A.. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125, 2014. [DOI] [PubMed] [Google Scholar]
- 47. Zuppinger, C. 3D cardiac cell culture: a critical review of current technologies and applications. Front Cardiovasc Med 6, 87, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xie, A.W., Binder, B.Y.K., Khalil, A.S., et al. Controlled self-assembly of stem cell aggregates instructs pluripotency and lineage bias. Sci Rep 7, 14070, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Drakhlis, L., Biswanath, S., Farr, C.-M., et al. Human heart-forming organoids recapitulate early heart and foregut development. Nat Biotechnol 39, 737, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haisler, W.L., Timm, D.M., Gage, J.A., et al. Three-dimensional cell culturing by magnetic levitation. Nat Protoc 8, 1940, 2013. [DOI] [PubMed] [Google Scholar]
- 51. Penland, N., Choi, E., Perla, M., et al. Facile fabrication of tissue-engineered constructs using nanopatterned cell sheets and magnetic levitation. Nanotechnology 28, 075103, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Souza, G.R., Molina, J.R., Raphael, R.M., et al. Three-dimensional tissue culture based on magnetic cell levitation. Nat Nanotechnol 5, 291, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ng, E.S., Davis, R.P., Azzola, L., et al. Forced aggregation of defined numbers of human embryonic stem cells into embryoid bodies fosters robust, reproducible hematopoietic differentiation. Blood 106, 1601, 2005. [DOI] [PubMed] [Google Scholar]
- 54. Radisic, M., Marsano, A., Maidhof, R., et al. Cardiac tissue engineering using perfusion bioreactor systems. Nat Protoc 3, 719, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kenar, H., Kose, G.T., Toner, M., et al. A 3D aligned microfibrous myocardial tissue construct cultured under transient perfusion. Biomaterials 32, 5320, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Maidhof, R., Tandon, N., Lee, E.J., et al. Biomimetic perfusion and electrical stimulation applied in concert improved the assembly of engineered cardiac tissue. J Tissue Eng Regen Med 6, e12, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Miklas, J.W., Nunes, S.S., Sofla, A., et al. Bioreactor for modulation of cardiac microtissue phenotype by combined static stretch and electrical stimulation. Biofabrication 6, 024113, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Frey, O., Misun, P.M., Fluri, D.A., et al. Reconfigurable microfluidic hanging drop network for multi-tissue interaction and analysis. Nat Commun 5, 4250, 2014. [DOI] [PubMed] [Google Scholar]
- 59. Zuppinger, C. 3D culture for cardiac cells. Biochim Biophys Acta 1863, 1873, 2016. [DOI] [PubMed] [Google Scholar]
- 60. Slaughter, B.V., Khurshid, S.S., Fisher, O.Z., et al. Hydrogels in regenerative medicine. Adv Mater 21, 3307, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Camci-Unal, G., Cuttica, D., Annabi, N., et al. Synthesis and characterization of hybrid hyaluronic acid-gelatin hydrogels. Biomacromolecules 14, 1085, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huh, D., Hamilton, G.A., and Ingber, D.E.. From 3D cell culture to organs-on-chips. Trends Cell Biol 21, 745, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hansen, A., Eder, A., Bönstrup, M., et al. Development of a drug screening platform based on engineered heart tissue. Circ Res 107, 35, 2010. [DOI] [PubMed] [Google Scholar]
- 64. Thavandiran, N., Hale, C., Blit, P., et al. Functional arrays of human pluripotent stem cell-derived cardiac microtissues. Sci Rep 10, 6919, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gere, J.M. Mechanics of Materials. 5th ed. Pacific Grove, CA: Brooks/Cole, 2001. [Google Scholar]
- 66. Leonard, A., Bertero, A., Powers, J.D., et al. Afterload promotes maturation of human induced pluripotent stem cell derived cardiomyocytes in engineered heart tissues. J Mol Cell Cardiol 118, 147, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mannhardt, I., Warncke, C., Trieu, H.K., et al. Piezo-bending actuators for isometric or auxotonic contraction analysis of engineered heart tissue. J Tissue Eng Regen Med 13, 3, 2019. [DOI] [PubMed] [Google Scholar]
- 68. Zhao, G., Zhang, X., Lu Tian, J., et al. Recent advances in electrospun nanofibrous scaffolds for cardiac tissue engineering. Adv Funct Mater 25, 5726, 2015. [Google Scholar]
- 69. Wang, J., Jansen, J.A., and Yang, F.. Electrospraying: possibilities and challenges of engineering carriers for biomedical applications-a mini review. Front Chem 7, 258, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dalton, P.D. Melt electrowriting with additive manufacturing principles. Curr Opin Biomed Eng 2, 49, 2017. [Google Scholar]
- 71. Xue, J., Wu, T., Dai, Y., et al. Electrospinning and electrospun nanofibers: methods, materials, and applications. Chem Rev 119, 5298, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kim, D.H., Lipke, E.A., Kim, P., et al. Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs. Proc Natl Acad Sci U S A 107, 565, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ramaciotti, C., McClellan, G., Sharkey, A., et al. Cardiac endothelial cells modulate contractility of rat heart in response to oxygen tension and coronary flow. Circ Res 72, 1044, 1993. [DOI] [PubMed] [Google Scholar]
- 74. Ahn, G., Min, K.H., Kim, C., et al. Precise stacking of decellularized extracellular matrix based 3D cell-laden constructs by a 3D cell printing system equipped with heating modules. Sci Rep 7, 8624, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Singh, A., Nikkhah, M., and Annabi, N. Organoids and Ex Vivo Tissue On-Chip Technologies. Volume 198. Amsterdam, The Netherlands: Elsevier, 2019. [Google Scholar]
- 76. Mirdamadi, E., Tashman, J.W., Shiwarski, D.J., et al. FRESH 3D bioprinting a full-size model of the human heart. ACS Biomater Sci Eng 6, 6453, 2020. [DOI] [PubMed] [Google Scholar]
- 77. Jang, J., Park, H.J., Kim, S.W., et al. 3D printed complex tissue construct using stem cell-laden decellularized extracellular matrix bioinks for cardiac repair. Biomaterials 112, 264, 2017. [DOI] [PubMed] [Google Scholar]
- 78. Hinton, T.J., Jallerat, Q., Palchesko, R.N., et al. Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci Adv 1, e1500758, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Noor, N., Shapira, A., Edri, R., et al. Tissue engineering: 3D printing of personalized thick and perfusable cardiac patches and hearts (Adv. Sci. 11/2019). Adv Sci (Weinh) 6, 1970066, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Oberwallner, B., Brodarac, A., Anić, P., et al. Human cardiac extracellular matrix supports myocardial lineage commitment of pluripotent stem cells. Eur J Cardiothorac Surg 47, 416; discussion 425, 2015. [DOI] [PubMed] [Google Scholar]
- 81. Nguyen, P.K., Rhee, J.W., and Wu, J.C.. Adult stem cell therapy and heart failure, 2000 to 2016: a systematic review. JAMA Cardiol 1, 831, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang, J., Zhu, W., Radisic, M., et al. Can we engineer a human cardiac patch for therapy? Circ Res 123, 244, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kadota, S., and Shiba, Y.. Pluripotent stem cell-derived cardiomyocyte transplantation for heart disease treatment. Curr Cardiol Rep 21, 73, 2019. [DOI] [PubMed] [Google Scholar]
- 84. Zimmermann, W.-H. Tissue engineered heart repair from preclinical models to first-in-patient studies. Curr Opin Physiol 14, 70, 2020. [Google Scholar]
- 85. Ogle, B.M., Bursac, N., Domian, I., et al. Distilling complexity to advance cardiac tissue engineering. Sci Transl Med 8, 342ps313, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Streeter, B.W., and Davis, M.E.. Therapeutic cardiac patches for repairing the myocardium. Adv Exp Med Biol 1144, 1, 2019. [DOI] [PubMed] [Google Scholar]
- 87. Engelmayr, G.C., Cheng, M., Bettinger, C.J., et al. Accordion-like honeycombs for tissue engineering of cardiac anisotropy. Nat Mater 7, 1003, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Coppini, R., Ferrantini, C., Aiazzi, A., et al. Isolation and functional characterization of human ventricular cardiomyocytes from fresh surgical samples. J Vis Exp 51116, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Guo, G.R., Chen, L., Rao, M., et al. A modified method for isolation of human cardiomyocytes to model cardiac diseases. J Transl Med 16, 288, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Louch, W.E., Sheehan, K.A., and Wolska, B.M.. Methods in cardiomyocyte isolation, culture, and gene transfer. J Mol Cell Cardiol 51, 288, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Roth, G.M., Bader, D.M., and Pfaltzgraff, E.R. Isolation and physiological analysis of mouse cardiomyocytes. J Vis Exp e51109, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Narsinh, K.H., Plews, J., and Wu, J.C.. Comparison of human induced pluripotent and embryonic stem cells: fraternal or identical twins? Mol Ther 19, 635, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Vazin, T., and Freed, W.J.. Human embryonic stem cells: derivation, culture, and differentiation: a review. Restor Neurol Neurosci 28, 589, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Bai, Q., Desprat, R., Klein, B., et al. Embryonic stem cells or induced pluripotent stem cells? A DNA integrity perspective. Curr Gene Ther 13, 93, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chaudhry, F., Isherwood, J., Bawa, T., et al. Single-cell RNA sequencing of the cardiovascular system: new looks for old diseases. Front Cardiovasc Med 6, 173, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Willems, E., Spiering, S., Davidovics, H., et al. Small-molecule inhibitors of the wnt pathway potently promote cardiomyocytes from human embryonic stem cell–derived mesoderm. Circ Res 109, 360, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Takahashi, K., and Yamanaka, S.. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663, 2006. [DOI] [PubMed] [Google Scholar]
- 98. Chatterjee, I., Li, F., Kohler, E.E., et al. Induced pluripotent stem (iPS) cell culture methods and induction of differentiation into endothelial cells. Methods Mol Biol 1357, 311, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Laco, F., Low, J.L., Seow, J., et al. Cardiomyocyte differentiation of pluripotent stem cells with SB203580 analogues correlates with Wnt pathway CK1 inhibition independent of p38 MAPK signaling. J Mol Cell Cardiol 80, 56, 2015. [DOI] [PubMed] [Google Scholar]
- 100. Monzen, K., Shiojima, I., Hiroi, Y., et al. Bone morphogenetic proteins induce cardiomyocyte differentiation through the mitogen-activated protein kinase kinase kinase TAK1 and cardiac transcription factors Csx/Nkx-2.5 and GATA-4. Mol Cell Biol 19, 7096, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Parikh, A., Wu, J., Blanton, R.M., et al. Signaling pathways and gene regulatory networks in cardiomyocyte differentiation. Tissue Eng Part B Rev 21, 377, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kattman, S.J., Witty, A.D., Gagliardi, M., et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8, 228, 2011. [DOI] [PubMed] [Google Scholar]
- 103. Burridge, P.W., Matsa, E., Shukla, P., et al. Chemically defined generation of human cardiomyocytes. Nat Methods 11, 855, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lian, X., Hsiao, C., Wilson, G., et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A 109, E1848, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Laflamme, M.A., Chen, K.Y., Naumova, A.V., et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol 25, 1015, 2007. [DOI] [PubMed] [Google Scholar]
- 106. Battista, S., Guarnieri, D., Borselli, C., et al. The effect of matrix composition of 3D constructs on embryonic stem cell differentiation. Biomaterials 26, 6194, 2005. [DOI] [PubMed] [Google Scholar]
- 107. Santoro, R., Perrucci, G.L., Gowran, A., et al. Unchain my heart: integrins at the basis of iPSC cardiomyocyte differentiation. Stem Cells Int 2019, 8203950, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ban, K., Bae, S., and Yoon, Y.S.. Current strategies and challenges for purification of cardiomyocytes derived from human pluripotent stem cells. Theranostics 7, 2067, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Fujita, J., Tohyama, S., Kishino, Y., et al. Concise review: genetic and epigenetic regulation of cardiac differentiation from human pluripotent stem cells. Stem Cells 37, 992, 2019. [DOI] [PubMed] [Google Scholar]
- 110. Musunuru, K., Sheikh, F., Gupta, R.M., et al. Induced pluripotent stem cells for cardiovascular disease modeling and precision medicine: a scientific statement from the American Heart Association. Circ Genom Precis Med 11, e000043, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kadari, A., Mekala, S., Wagner, N., et al. Robust generation of cardiomyocytes from human iPS cells requires precise modulation of BMP and WNT signaling. Stem Cell Rev Rep 11, 560, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kupfer, M.E., Lin, W.H., Ravikumar, V., et al. In situ expansion, differentiation, and electromechanical coupling of human cardiac muscle in a 3D bioprinted, chambered organoid. Circ Res 127, 207, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Branco, M.A., Cotovio, J.P., Rodrigues, C.A.V., et al. Transcriptomic analysis of 3D cardiac differentiation of human induced pluripotent stem cells reveals faster cardiomyocyte maturation compared to 2D culture. Sci Rep 9, 9229, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Boudou, T., Legant, W.R., Mu, A., et al. A microfabricated platform to measure and manipulate the mechanics of engineered cardiac microtissues. Tissue Eng Part A 18, 910, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Sala, L., van Meer, B.J., Tertoolen, L.G.J., et al. MUSCLEMOTION: a versatile open software tool to quantify cardiomyocyte and cardiac muscle contraction in vitro and in vivo. Circ Res 122, e5, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Soufen, H.N., Salemi, V.M., Aneas, I.M., et al. Collagen content, but not the ratios of collagen type III/I mRNAs, differs among hypertensive, alcoholic, and idiopathic dilated cardiomyopathy. Braz J Med Biol Res 41, 1098, 2008. [DOI] [PubMed] [Google Scholar]
- 117. Johnson, T.D., Hill, R.C., Dzieciatkowska, M., et al. Quantification of decellularized human myocardial matrix: a comparison of six patients. Proteomics Clin Appl 10, 75, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Weber, K.T., Sun, Y., Tyagi, S.C., et al. Collagen network of the myocardium: function, structural remodeling and regulatory mechanisms. J Mol Cell Cardiol 26, 279, 1994. [DOI] [PubMed] [Google Scholar]
- 119. Parker, K.K., and Ingber, D.E.. Extracellular matrix, mechanotransduction and structural hierarchies in heart tissue engineering. Philos Trans R Soc Lond B Biol Sci 362, 1267, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lu, Y.Y., Lin, Y.K., Kao, Y.H., et al. Collagen regulates transforming growth factor-β receptors of HL-1 cardiomyocytes through activation of stretch and integrin signaling. Mol Med Rep 14, 3429, 2016. [DOI] [PubMed] [Google Scholar]
- 121. Zhou, P., and Pu, W.T.. Recounting cardiac cellular composition. Circ Res 118, 368, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Tiburcy, M., Hudson, J.E., Balfanz, P., et al. Defined engineered human myocardium with advanced maturation for applications in heart failure modeling and repair. Circulation 135, 1832, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Brutsaert, D.L. Cardiac endothelial-myocardial signaling: its role in cardiac growth, contractile performance, and rhythmicity. Physiol Rev 83, 59, 2003. [DOI] [PubMed] [Google Scholar]
- 124. Hanna, P., Rajendran, P.S., Ajijola, O.A., et al. Cardiac neuroanatomy—imaging nerves to define functional control. Auton Neurosci 207, 48, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Bönner, F., Borg, N., Burghoff, S., et al. Resident cardiac immune cells and expression of the ectonucleotidase enzymes CD39 and CD73 after ischemic injury. PLoS One 7, e34730, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Pauza, D.H., Skripka, V., Pauziene, N., et al. Morphology, distribution, and variability of the epicardiac neural ganglionated subplexuses in the human heart. Anat Rec 259, 353, 2000. [DOI] [PubMed] [Google Scholar]
- 127. Bergmann, O., Zdunek, S., Felker, A., et al. Dynamics of cell generation and turnover in the human heart. Cell 161, 1566, 2015. [DOI] [PubMed] [Google Scholar]
- 128. Souders, C.A., Bowers, S.L., and Baudino, T.A.. Cardiac fibroblast: the renaissance cell. Circ Res 105, 1164, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Segers, V.F.M., Brutsaert, D.L., and De Keulenaer, G.W.. Cardiac remodeling: endothelial cells have more to say than just NO. Front Physiol 9, 382, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Boothe, S.D., Myers, J.D., Pok, S., et al. The effect of substrate stiffness on cardiomyocyte action potentials. Cell Biochem Biophys 74, 527, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Solomon, T., Choi Yu, S., Hool, L., et al. The role of extracellular matrix stiffness on cardiac metabolic activity. Heart Lung Circ 28, 2019; DOI: 10.1016/j.hlc.2019.06.140. [DOI] [Google Scholar]
- 132. Callaghan, N.I., and Lee, X.A.. Extracellular matrix stiffness affects contractility in adult rat cardiomyocytes: implications for dynamic nitric oxide signalling and calcium handling. J Physiol 595, 5759, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Wang, B., Tedder, M.E., Perez, C.E., et al. Structural and biomechanical characterizations of porcine myocardial extracellular matrix. J Mater Sci Mater Med 23, 1835, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Wang, B., Borazjani, A., Tahai, M., et al. Fabrication of cardiac patch with decellularized porcine myocardial scaffold and bone marrow mononuclear cells. J Biomed Mater Res A 94, 1100, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Blazeski, A., Kostecki, G.M., and Tung, L.. Engineered heart slices for electrophysiological and contractile studies. Biomaterials 55, 119, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Villemain, O., Correia, M., Mousseaux, E., et al. Myocardial stiffness evaluation using noninvasive shear wave imaging in healthy and hypertrophic cardiomyopathic adults. JACC Cardiovasc Imaging 12, 1135, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Raub, C.B., Putnam, A.J., Tromberg, B.J., et al. Predicting bulk mechanical properties of cellularized collagen gels using multiphoton microscopy. Acta Biomater 6, 4657, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Liu, W., and Wang, Z.. Current understanding of the biomechanics of ventricular tissues in heart failure. Bioengineering (Basel) 7, 2, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Xu, B., Li, H., and Zhang, Y.. Understanding the viscoelastic behavior of collagen matrices through relaxation time distribution spectrum. Biomatter 3, e24651, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Valero, C., Amaveda, H., Mora, M., et al. Combined experimental and computational characterization of crosslinked collagen-based hydrogels. PLoS One 13, e0195820, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Silva, A.C., Pereira, C., Fonseca, A.C.R.G., et al. Bearing my heart: the role of extracellular matrix on cardiac development, homeostasis, and injury response. Front Cell Dev Biol 8, 621644, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Gallo, S., Vitacolonna, A., Bonzano, A., et al. ERK: a key player in the pathophysiology of cardiac hypertrophy. Int J Mol Sci 20, 2164, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Turner, N.A., and Blythe, N.M.. Cardiac fibroblast p38 MAPK: a critical regulator of myocardial remodeling. J Cardiovasc Dev Dis 6, 27, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Yeh, Y.-C., Ling, J.-Y., Chen, W.-C., et al. Mechanotransduction of matrix stiffness in regulation of focal adhesion size and number: reciprocal regulation of caveolin-1 and β1 integrin. Sci Rep 7, 15008, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. De Keulenaer, G.W., Wang, Y., Feng, Y., et al. Identification of IEX-1 as a biomechanically controlled nuclear factor-kappaB target gene that inhibits cardiomyocyte hypertrophy. Circ Res 90, 690, 2002. [DOI] [PubMed] [Google Scholar]
- 146. Niu, L., Jia, Y., Wu, M., et al. Matrix stiffness controls cardiac fibroblast activation through regulating YAP via AT. J Cell Physiol 235, 8345, 2020. [DOI] [PubMed] [Google Scholar]
- 147. Piek, A., de Boer, R.A., and Silljé, H.H.. The fibrosis-cell death axis in heart failure. Heart Fail Rev 21, 199, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Engler, A.J., Carag-Krieger, C., Johnson, C.P., et al. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. J Cell Sci 121, 3794, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Evans, T. Embryonic stem cells as a model for cardiac development and disease. Drug Discov Today Dis Models 5, 147, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Boheler, K.R., Joodi, R.N., Qiao, H., et al. Embryonic stem cell-derived cardiomyocyte heterogeneity and the isolation of immature and committed cells for cardiac remodeling and regeneration. Stem Cells Int 2011, 214203, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Alperin, C., Zandstra, P.W., and Woodhouse, K.A.. Engineering cardiac healing using embryonic stem cell-derived cardiac cell seeded constructs. Front Biosci 12, 3694, 2007. [DOI] [PubMed] [Google Scholar]
- 152. Feyen, D.A.M., McKeithan, W.L., Bruyneel, A.A.N., et al. Metabolic maturation media improve physiological function of human iPSC-derived cardiomyocytes. Cell Rep 32, 107925, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Slaats, R.H., Schwach, V., and Passier, R.. Metabolic environment in vivo as a blueprint for differentiation and maturation of human stem cell-derived cardiomyocytes. Biochim Biophys Acta Mol Basis Dis 1866, 165881, 2020. [DOI] [PubMed] [Google Scholar]
- 154. Mills, R.J., Titmarsh, D.M., Koenig, X., et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc Natl Acad Sci U S A 114, E8372, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Birket, M.J., Ribeiro, M.C., Kosmidis, G., et al. Contractile defect caused by mutation in MYBPC3 revealed under conditions optimized for human PSC-cardiomyocyte function. Cell Rep 13, 733, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Yang, X., Rodriguez, M., Pabon, L., et al. Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells. J Mol Cell Cardiol 72, 296, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Chattergoon, N.N., Giraud, G.D., Louey, S., et al. Thyroid hormone drives fetal cardiomyocyte maturation. FASEB J 26, 397, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Fu, J.D., Rushing, S.N., Lieu, D.K., et al. Distinct roles of microRNA-1 and -499 in ventricular specification and functional maturation of human embryonic stem cell-derived cardiomyocytes. PLoS One 6, e27417, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Troncoso, R., Ibarra, C., Vicencio, J.M., et al. New insights into IGF-1 signaling in the heart. Trends Endocrinol Metab 25, 128, 2014. [DOI] [PubMed] [Google Scholar]
- 160. Jing, D., Parikh, A., and Tzanakakis, E.S.. Cardiac cell generation from encapsulated embryonic stem cells in static and scalable culture systems. Cell Transplant 19, 1397, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Saleem, U., Ismaili, D., Mannhardt, I., et al. Regulation of ICa,L and force by PDEs in human-induced pluripotent stem cell-derived cardiomyocytes. Br J Pharmacol 177, 3036, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Martínez, M., García, A., Luzardo, E., et al. Energetic metabolism in cardiomyocytes: molecular basis of heart ischemia and arrhythmogenesis. Vessel Plus 1, 130, 2017. [Google Scholar]
- 163. Yang, X., Rodriguez, M.L., Leonard, A., et al. Fatty acids enhance the maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cell Reports 13, 657, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Dongjian, H., Annet, L., Abir, Y., et al. Metabolic maturation of human pluripotent stem cell-derived cardiomyocytes by inhibition of HIF1α and LDHA. Circ Res 123, 1066, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Wasserman, D.H. Four grams of glucose. Am J Physiol Endocrinol Metab 296, E11, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Dyntar, D., Sergeev, P., Klisic, J., et al. High glucose alters cardiomyocyte contacts and inhibits myofibrillar formation. J Clin Endocrinol Metab 91, 1961, 2006. [DOI] [PubMed] [Google Scholar]
- 167. Sekine, M., Tomita, Y., Iguchi, A., et al. Investigation of fluctuations in blood glucose level due to dietary restrictions during impacted mandibular third molar extraction under intravenous sedation: effect of perioperative glucose administration. Oral Maxillofac Surg 24, 289, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Burridge, P.W., Holmstrom, A., and Wu, J.C.. Chemically defined culture and cardiomyocyte differentiation of human pluripotent stem cells. Curr Protoc Hum Genet 87, 21.3.1, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Tan, X., Dai, Q., Guo, T., et al. Efficient generation of transgene- and feeder-free induced pluripotent stem cells from human dental mesenchymal stem cells and their chemically defined differentiation into cardiomyocytes. Biochem Biophys Res Commun 495, 2490, 2018. [DOI] [PubMed] [Google Scholar]
- 170. Kuo, H.H., Gao, X., DeKeyser, J.M., et al. Negligible-cost and weekend-free chemically defined human iPSC culture. Stem Cell Reports 14, 256, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Coffin, S.T., and Gaudette, G.R.. Aprotinin extends mechanical integrity time of cell-seeded fibrin sutures. J Biomed Mater Res A 104, 2271, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Bax, N.A., van Marion, M.H., Shah, B., et al. Matrix production and remodeling capacity of cardiomyocyte progenitor cells during in vitro differentiation. J Mol Cell Cardiol 53, 497, 2012. [DOI] [PubMed] [Google Scholar]
- 173. Ugolini, G.S., Pavesi, A., Rasponi, M., et al. Human cardiac fibroblasts adaptive responses to controlled combined mechanical strain and oxygen changes in vitro. Elife 6, e22847, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Thavandiran, N., Dubois, N., Mikryukov, A., et al. Design and formulation of functional pluripotent stem cell-derived cardiac microtissues. Proc Natl Acad Sci U S A 110, E4698, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Stoppel, W.L., Kaplan, D.L., and Black, L.D.. Electrical and mechanical stimulation of cardiac cells and tissue constructs. Adv Drug Deliv Rev 96, 135, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Zhao, Y., Rafatian, N., Feric, N.T., et al. A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell 176, 913, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Kane, C., Couch, L., and Terracciano, C.M.. Excitation-contraction coupling of human induced pluripotent stem cell-derived cardiomyocytes. Front Cell Dev Biol 3, 59, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Robertson, C., Tran, D.D., and George, S.C.. Concise review: maturation phases of human pluripotent stem cell-derived cardiomyocytes. Stem Cells 31, 829, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Satin, J., Itzhaki, I., Rapoport, S., et al. Calcium handling in human embryonic stem cell-derived cardiomyocytes. Stem Cells 26, 1961, 2008. [DOI] [PubMed] [Google Scholar]
- 180. Louch, W.E., Koivumäki, J.T., and Tavi, P.. Calcium signalling in developing cardiomyocytes: implications for model systems and disease. J Physiol 593, 1047, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Hwang, H.S., Kryshtal, D.O., Feaster, T.K., et al. Comparable calcium handling of human iPSC-derived cardiomyocytes generated by multiple laboratories. J Mol Cell Cardiol 85, 79, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Nakane, T., Masumoto, H., Tinney, J.P., et al. Impact of cell composition and geometry on human induced pluripotent stem cells-derived engineered cardiac tissue. Sci Rep 7, 45641, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Coppen, S.R., Fukushima, S., Shintani, Y., et al. A factor underlying late-phase arrhythmogenicity after cell therapy to the heart: global downregulation of connexin43 in the host myocardium after skeletal myoblast transplantation. Circulation 118, S138, 2008. [DOI] [PubMed] [Google Scholar]
- 184. Shimizu, T., Yamato, M., Akutsu, T., et al. Electrically communicating three-dimensional cardiac tissue mimic fabricated by layered cultured cardiomyocyte sheets. J Biomed Mater Res 60, 110, 2002. [DOI] [PubMed] [Google Scholar]
- 185. Tandon, N., Cannizzaro, C., Figallo, E., et al. Characterization of electrical stimulation electrodes for cardiac tissue engineering. Conf Proc IEEE Eng Med Biol Soc 2006, 845, 2006. [DOI] [PubMed] [Google Scholar]
- 186. Milan, H., Bassani, R., and Bassani, J.. Testing electrode suitability for field stimulation of high-threshold biological preparations. Res Biomed Eng 31, 273, 2015. [Google Scholar]
- 187. Morrissette-McAlmon, J., Blazeski, A., Somers, S., et al. Adipose-derived perivascular mesenchymal stromal/stem cells promote functional vascular tissue engineering for cardiac regenerative purposes. J Tissue Eng Regen Med 12, e962, 2018. [DOI] [PubMed] [Google Scholar]
- 188. Tandon, N., Marsano, A., Cannizzaro, C., et al. Design of electrical stimulation bioreactors for cardiac tissue engineering. Conf Proc IEEE Eng Med Biol Soc 2008, 3594, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Cannizzaro, C., Tandon, N., Figallo, E., et al. Practical aspects of cardiac tissue engineering with electrical stimulation. Methods Mol Med 140, 291, 2007. [DOI] [PubMed] [Google Scholar]
- 190. Porous reduced graphene oxide modified electrodes for the analysis of protein aggregation. Part 1: Lysozyme aggregation at pH 2 and 7.4. Electrochim Acta 254, 375, 2017. [Google Scholar]
- 191. Pietronave, S., Zamperone, A., Oltolina, F., et al. Monophasic and biphasic electrical stimulation induces a precardiac differentiation in progenitor cells isolated from human heart. Stem Cells Dev 23, 888, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Song, B., Gu, Y., Pu, J., et al. Application of direct current electric fields to cells and tissues in vitro and modulation of wound electric field in vivo. Nat Protoc 2, 1479, 2007. [DOI] [PubMed] [Google Scholar]
- 193. Tandon, N., Cannizzaro, C., Chao, P.H., et al. Electrical stimulation systems for cardiac tissue engineering. Nat Protoc 4, 155, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Tandon, N., Marsano, A., Maidhof, R., et al. Optimization of electrical stimulation parameters for cardiac tissue engineering. J Tissue Eng Regen Med 5, e115, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Chen, M.T., Jiang, C., Vernier, P.T., et al. Two-dimensional nanosecond electric field mapping based on cell electropermeabilization. PMC Biophys 2, 9, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Visone, R., Talò, G., Lopa, S., et al. Enhancing all-in-one bioreactors by combining interstitial perfusion, electrical stimulation, on-line monitoring and testing within a single chamber for cardiac constructs. Sci Rep 8, 16944, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Piotrowska, G., and Pierozynski, B.. Electrodegradation of phenol through continuous electrolysis of synthetic wastewater on platinized titanium and stainless steel anodes. Int J Electrochem Sci 12, 4444, 2017. [Google Scholar]
- 198. Jacot, J.G., Martin, J.C., and Hunt, D.L.. Mechanobiology of cardiomyocyte development. J Biomech 43, 93, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Chin, I.L., Hool, L., and Choi, Y.S.. A review of in vitro platforms for understanding cardiomyocyte mechanobiology. Front Bioeng Biotechnol 7, 133, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Scuderi, G.J., and Butcher, J.. Naturally engineered maturation of cardiomyocytes. Front Cell Dev Biol 5, 50, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Kim, K., and Wagner, W.R.. Non-invasive and non-destructive characterization of tissue engineered constructs using ultrasound imaging technologies: a review. Ann Biomed Eng 44, 621, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Paez-Mayorga, J., Hernández-Vargas, G., Ruiz-Esparza Guillermo, U., et al. Bioreactors for cardiac tissue engineering. Adv Healthc Mater 8, 1701504, 2019. [DOI] [PubMed] [Google Scholar]
- 203. Majkut, S., Idema, T., Swift, J., et al. Heart-specific stiffening in early embryos parallels matrix and myosin expression to optimize beating. Curr Biol 23, 2434, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Dou, W., Wang, L., Malhi, M., et al. A microdevice platform for characterizing the effect of mechanical strain magnitudes on the maturation of iPSC-cardiomyocytes. Biosens Bioelectron 175, 112875, 2021. [DOI] [PubMed] [Google Scholar]
- 205. Asrih, M., Mach, F., Nencioni, A., et al. Role of mitogen-activated protein kinase pathways in multifactorial adverse cardiac remodeling associated with metabolic syndrome. Mediators Inflamm 2013, 367245, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Saucerman, J.J., Tan, P.M., Buchholz, K.S., et al. Mechanical regulation of gene expression in cardiac myocytes and fibroblasts. Nat Rev Cardiol 16, 361, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Linke, W.A. Sense and stretchability: the role of titin and titin-associated proteins in myocardial stress-sensing and mechanical dysfunction. Cardiovasc Res 77, 637, 2008. [DOI] [PubMed] [Google Scholar]
- 208. Razavi, M., and Thakor, A.S.. An oxygen plasma treated poly(dimethylsiloxane) bioscaffold coated with polydopamine for stem cell therapy. J Mater Sci Mater Med 29, 54, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Kuddannaya, S., Chuah, Y.J., Lee, M.H., et al. Surface chemical modification of poly(dimethylsiloxane) for the enhanced adhesion and proliferation of mesenchymal stem cells. ACS Appl Mater Interfaces 5, 9777, 2013. [DOI] [PubMed] [Google Scholar]
- 210. Chuah, Y.J., Koh, Y.T., Lim, K., et al. Simple surface engineering of polydimethylsiloxane with polydopamine for stabilized mesenchymal stem cell adhesion and multipotency. Sci Rep 5, 18162, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Wang, P.-Y., Yu, J., Lin, J.-H., and Tsai, W.-B.. Modulation of alignment, elongation and contraction of cardiomyocytes through a combination of nanotopography and rigidity of substrates. Acta Biomater 7, 3285, 2011. [DOI] [PubMed] [Google Scholar]
- 212. Forte, G., Pagliari, S., Ebara, M., et al. Substrate stiffness modulates gene expression and phenotype in neonatal cardiomyocytes in vitro. Tissue Eng Part A 18, 1837, 2012. [DOI] [PubMed] [Google Scholar]
- 213. Rysä, J., Tokola, H., and Ruskoaho, H.. Mechanical stretch induced transcriptomic profiles in cardiac myocytes. Sci Rep 8, 4733, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Govoni, M., Muscari, C., Guarnieri, C., et al. Mechanostimulation protocols for cardiac tissue engineering. Biomed Res Int 2013, 918640, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Mannhardt, I., Breckwoldt, K., Letuffe-Brenière, D., et al. Human engineered heart tissue: analysis of contractile force. Stem Cell Reports 7, 29, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Tiburcy, M., Meyer, T., Liaw, N.Y., and Zimmermann, W.-H.. Generation of engineered human myocardium in a multi-well format. STAR Protoc 1, 100032, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Nunes, S.S., Miklas, J.W., Liu, J., et al. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods 10, 781, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Pislaru, C., Urban, M.W., Pislaru, S.V., et al. Viscoelastic properties of normal and infarcted myocardium measured by a multifrequency shear wave method: comparison with pressure-segment length method. Ultrasound Med Biol 40, 1785, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Vandenburgh, H.H., Solerssi, R., Shansky, J., et al. Mechanical stimulation of organogenic cardiomyocyte growth in vitro. Am J Physiol 270, C1284, 1996. [DOI] [PubMed] [Google Scholar]
- 220. Friedrich, O., Merten, A.L., Schneidereit, D., et al. 2D inplane cell stretch systems for studies of cardiac mechano-signaling. Front Bioeng Biotechnol 7, 55, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. van Meer, B.J., Tertoolen, L.G., and Mummery, C.L.. Concise review: measuring physiological responses of human pluripotent stem cell derived cardiomyocytes to drugs and disease. Stem Cells 34, 2008, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Verkerk, A.O., Veerman, C.C., Zegers, J.G., et al. Patch-clamp recording from human induced pluripotent stem cell-derived cardiomyocytes: improving action potential characteristics through dynamic clamp. Int J Mol Sci 18, 1873, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Saleem, U., Mannhardt, I., Braren, I., et al. Force and calcium transients analysis in human engineered heart tissues reveals positive force-frequency relation at physiological frequency. Stem Cell Reports 14, 312, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Bedut, S., Seminatore-Nole, C., Lamamy, V., et al. High-throughput drug profiling with voltage- and calcium-sensitive fluorescent probes in human iPSC-derived cardiomyocytes. Am J Physiol Heart Circ Physiol 311, H44, 2016. [DOI] [PubMed] [Google Scholar]
- 225. Liu, P., and Miller, E.W.. Electrophysiology, unplugged: imaging membrane potential with fluorescent indicators. Acc Chem Res 53, 11, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Woulfe, K.C., Ferrara, C., Pioner, J.M., et al. A novel method of isolating myofibrils from primary cardiomyocyte culture suitable for myofibril mechanical study. Front Cardiovasc Med 6, 12, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Sharma, A., Toepfer, C.N., Schmid, M., et al. Differentiation and contractile analysis of GFP-sarcomere reporter hiPSC-cardiomyocytes. Curr Protoc Hum Genet 96, 21.12.1, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Wienert, B., Wyman, S.K., Richardson, C.D., et al. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science 364, 286, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Liu, S.J., Horlbeck, M.A., Cho, S.W., et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, aah7111, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Mandegar, M.A., Huebsch, N., Frolov, E.B., et al. CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell 18, 541, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Simionescu, A., Tedder, M.E., Chuang, T.H., et al. Lectin and antibody-based histochemical techniques for cardiovascular tissue engineering. J Histotechnol 34, 20, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Vunjak-Novakovic, G., Tandon, N., Godier, A., et al. Challenges in cardiac tissue engineering. Tissue Eng Part B Rev 16, 169, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Zhang, Y.S., Aleman, J., Arneri, A., et al. From cardiac tissue engineering to heart-on-a-chip: beating challenges. Biomed Mater 10, 034006, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Chang, Y.C., Mirhaidari, G., Kelly, J., et al. Current challenges and solutions to tissue engineering of large-scale cardiac constructs. Curr Cardiol Rep 23, 47, 2021. [DOI] [PubMed] [Google Scholar]
- 235. Shafiee, A., and Atala, A.. Tissue engineering: toward a new era of medicine. Annu Rev Med 68, 29, 2017. [DOI] [PubMed] [Google Scholar]
- 236. Taylor, D.A. The future of tissue engineering in heart transplantation. Tex Heart Inst J 46, 73, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Fleischer, S., Feiner, R., and Dvir, T.. Cutting-edge platforms in cardiac tissue engineering. Curr Opin Biotechnol 47, 23, 2017. [DOI] [PubMed] [Google Scholar]
- 238. Nguyen, A.H., Marsh, P., Schmiess-Heine, L., et al. Cardiac tissue engineering: state-of-the-art methods and outlook. J Biol Eng 13, 57, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Eder, A., Vollert, I., Hansen, A., et al. Human engineered heart tissue as a model system for drug testing. Adv Drug Deliv Rev 96, 214, 2016. [DOI] [PubMed] [Google Scholar]
- 240. Horváth, A., Lemoine, M.D., Löser, A., et al. Low resting membrane potential and low inward rectifier potassium currents are not inherent features of hiPSC-derived cardiomyocytes. Stem Cell Reports 10, 822, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Fiedler, J., Stöhr, A., Gupta, S.K., et al. Functional microRNA library screening identifies the hypoxamir miR-24 as a potent regulator of smooth muscle cell proliferation and vascularization. Antioxid Redox Signal 21, 1167, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Werner, T.R., Kunze, A.C., Stenzig, J., et al. Blockade of miR-140-3p prevents functional deterioration in afterload-enhanced engineered heart tissue. Sci Rep 9, 11494, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Dutsch, A., Wijnker, P.J.M., Schlossarek, S., et al. Phosphomimetic cardiac myosin-binding protein C partially rescues a cardiomyopathy phenotype in murine engineered heart tissue. Sci Rep 9, 18152, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Mills, R.J., Voges, H.K., Porrello, E.R., and Hudson, J.E.. Disease modeling and functional screening using engineered heart tissue. Curr Opin Physiol 1, 80, 2018. [Google Scholar]
- 245. Bargehr, J., Ong, L.P., Colzani, M., et al. Epicardial cells derived from human embryonic stem cells augment cardiomyocyte-driven heart regeneration. Nat Biotechnol 37, 895, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Rodriguez, M.L., Werner, T.R., Becker, B., et al. A magnetics-based approach for fine-tuning afterload in engineered heart tissues. ACS Biomater Sci Eng 5, 3663, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Ronaldson-Bouchard, K., Yeager, K., Teles, D., et al. Engineering of human cardiac muscle electromechanically matured to an adult-like phenotype. Nat Protoc 14, 2781, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Turnbull, I.C., Karakikes, I., Serrao, G.W., et al. Advancing functional engineered cardiac tissues toward a preclinical model of human myocardium. FASEB J 28, 644, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Naito, H., Melnychenko, I., Didié, M., et al. Optimizing engineered heart tissue for therapeutic applications as surrogate heart muscle. Circulation 114, I72, 2006. [DOI] [PubMed] [Google Scholar]
- 250. Yildirim, Y., Naito, H., Didié, M., et al. Development of a biological ventricular assist device: preliminary data from a small animal model. Circulation 116, I16, 2007. [DOI] [PubMed] [Google Scholar]
- 251. Zimmermann, W.H., Melnychenko, I., Wasmeier, G., et al. Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts. Nat Med 12, 452, 2006. [DOI] [PubMed] [Google Scholar]
- 252. Roura, S., Gálvez-Montón, C., and Bayes-Genis, A.. Fibrin, the preferred scaffold for cell transplantation after myocardial infarction? An old molecule with a new life. J Tissue Eng Regen Med 11, 2304, 2017. [DOI] [PubMed] [Google Scholar]
- 253. Zhang, X., Guo, J.P., Chi, Y.L., et al. Endothelin-induced differentiation of Nkx2.5⁺ cardiac progenitor cells into pacemaking cells. Mol Cell Biochem 366, 309, 2012. [DOI] [PubMed] [Google Scholar]
- 254. Kuo, C.K., and Ma, P.X.. Ionically crosslinked alginate hydrogels as scaffolds for tissue engineering: part 1. Structure, gelation rate and mechanical properties. Biomaterials 22, 511, 2001. [DOI] [PubMed] [Google Scholar]
- 255. Kharaziha, M., Nikkhah, M., Shin, S.R., et al. PGS:Gelatin nanofibrous scaffolds with tunable mechanical and structural properties for engineering cardiac tissues. Biomaterials 34, 6355, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Jennings, J.A., Bumgardner, J.D., and Alexander, C.. Chitosan Based Biomaterials: Volume 2: Tissue Engineering and Therapeutics. Sawston, United Kingdom: Woodhead Publishing, 2017. [Google Scholar]
- 257. Gostynska, N., Shankar Krishnakumar, G., Campodoni, E., et al. 3D porous collagen scaffolds reinforced by glycation with ribose for tissue engineering application. Biomed Mater 12, 055002, 2017. [DOI] [PubMed] [Google Scholar]
- 258. Oliveira, S.M., Ringshia, R.A., Legeros, R.Z., et al. An improved collagen scaffold for skeletal regeneration. J Biomed Mater Res A 94, 371, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Kitsara, M., Agbulut, O., Kontziampasis, D., Chen, Y., and Menasché, P.. Fibers for hearts: a critical review on electrospinning for cardiac tissue engineering. Acta Biomater 48, 20, 2017. [DOI] [PubMed] [Google Scholar]
- 260. Gentile, P., Chiono, V., Carmagnola, I., et al. An overview of poly(lactic-co-glycolic) acid (PLGA)-based biomaterials for bone tissue engineering. Int J Mol Sci 15, 3640, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Rai, R., Tallawi, M., Barbani, N., et al. Biomimetic poly(glycerol sebacate) (PGS) membranes for cardiac patch application. Mater Sci Eng C Mater Biol Appl 33, 3677, 2013. [DOI] [PubMed] [Google Scholar]
- 262. Jang, J., Kim, T.G., Kim, B.S., et al. Tailoring mechanical properties of decellularized extracellular matrix bioink by vitamin B2-induced photo-crosslinking. Acta Biomater 33, 88, 2016. [DOI] [PubMed] [Google Scholar]
- 263. Jammalamadaka, U., and Tappa, K.. Recent advances in biomaterials for 3D printing and tissue engineering. J Funct Biomater 9, 22, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Kačarević, Ž., Rider, P.M., Alkildani, S., et al. An introduction to 3D bioprinting: possibilities, challenges and future aspects. Materials (Basel) 11, 2199, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Qasim, M., Haq, F., Kang, M.H., et al. 3D printing approaches for cardiac tissue engineering and role of immune modulation in tissue regeneration. Int J Nanomedicine 14, 1311, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Iyer, R.K., Chiu, L.L., and Radisic, M.. Microfabricated poly(ethylene glycol) templates enable rapid screening of triculture conditions for cardiac tissue engineering. J Biomed Mater Res A 89, 616, 2009. [DOI] [PubMed] [Google Scholar]
- 267. Moorthi, A., Tyan, Y.C., and Chung, T.W.. Surface-modified polymers for cardiac tissue engineering. Biomater Sci 5, 1976, 2017. [DOI] [PubMed] [Google Scholar]
- 268. Rowland, C.R., Lennon, D.P., Caplan, A.I., et al. The effects of crosslinking of scaffolds engineered from cartilage ECM on the chondrogenic differentiation of MSCs. Biomaterials 34, 5802, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Yang, L., Han, Y., Nilsson-Payant, B.E., et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell 27, 125, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Lian, X., Zhang, J., Zhu, K., et al. Insulin inhibits cardiac mesoderm, not mesendoderm, formation during cardiac differentiation of human pluripotent stem cells and modulation of canonical Wnt signaling can rescue this inhibition. Stem Cells 31, 447, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Correia, C., Koshkin, A., Duarte, P., et al. Distinct carbon sources affect structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Sci Rep 7, 8590, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Gentillon, C., Li, D., Duan, M., et al. Targeting HIF-1α in combination with PPARα activation and postnatal factors promotes the metabolic maturation of human induced pluripotent stem cell-derived cardiomyocytes. J Mol Cell Cardiol 132, 120, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Hu, D., Linders, A., Yamak, A., et al. Metabolic maturation of human pluripotent stem cell-derived cardiomyocytes by inhibition of HIF1alpha and LDHA. Circ Res 123, 1066, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Garbern, J.C., Helman, A., Sereda, R., et al. Inhibition of mTOR signaling enhances maturation of cardiomyocytes derived from human-induced pluripotent stem cells via p53-induced quiescence. Circulation 141, 285, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Horikoshi, Y., Yan, Y., Terashvili, M., et al. Fatty acid-treated induced pluripotent stem cell-derived human cardiomyocytes exhibit adult cardiomyocyte-like energy metabolism phenotypes. Cells 8, 1095, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Lin, Y.R., Li, C.J., Syu, S.H., et al. Early administration of glutamine protects cardiomyocytes from post-cardiac arrest acidosis. Biomed Res Int 2016, 2106342, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Wesselink, E., Koekkoek, W.A.C., Grefte, S., et al. Feeding mitochondria: potential role of nutritional components to improve critical illness convalescence. Clin Nutr 38, 982, 2019. [DOI] [PubMed] [Google Scholar]
- 278. Kuppusamy, K.T., Jones, D.C., Sperber, H., et al. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc Natl Acad Sci U S A 112, E2785, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Sluijter, J.P., van Mil, A., van Vliet, P., et al. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler Thromb Vasc Biol 30, 859, 2010. [DOI] [PubMed] [Google Scholar]
- 280. Ribeiro, A.J.S., Schwab, O., Mandegar, M.A., et al. Multi-imaging method to assay the contractile mechanical output of micropatterned human iPSC-derived cardiac myocytes. Circ Res 120, 1572, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Huethorst, E., Hortigon, M., Zamora-Rodriguez, V., et al. Enhanced human-induced pluripotent stem cell derived cardiomyocyte maturation using a dual microgradient substrate. ACS Biomater Sci Eng 2, 2231, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282. Salick, M.R., Napiwocki, B.N., Sha, J., et al. Micropattern width dependent sarcomere development in human ESC-derived cardiomyocytes. Biomaterials 35, 4454, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Agarwal, A., Farouz, Y., Nesmith, A.P., et al. Micropatterning alginate substrates for in vitro cardiovascular muscle on a chip. Adv Funct Mater 23, 3738, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]