Abstract
To clarify the physiological and pathological roles of gut-liver-axis (GLA) in the human body, a GLA microphysiological system (GLA-MPS) holds great potential. However, in current GLA-MPSs, the importance of a physiologically relevant flow for gut and liver cells' cultivation is not fully addressed. In addition, the integration of individual organ perfusion, circulation flow, and organ tissue functions in a single device has not been achieved. Here, we introduce a GLA-MPS by integrating two cell-culture chambers with individually applied perfusion flows and a circulation channel with an on-chip pneumatic micropump under cell-culture chambers via a porous membrane for interconnecting them. We analyzed the fluid shear stress (FSS) with computational fluid dynamics simulations and confirmed that the physiologically relevant FSS could be applied to the gut (Caco-2) (8 × 10−3 dyn cm−2) and liver (HepG2) cells (1.2 × 10−7 dyn cm−2). Under the physiologically relevant flow, the Caco-2 and HepG2 cells in the GLA-MPS maintained a cell survival rate of 95% and 92%, respectively. Furthermore, the expression of functional proteins such as zonula occludens 1 (in Caco-2) and albumin (in HepG2) was enhanced. To demonstrate the GLA interaction, the inflammatory bowel disease was recapitulated by applying lipopolysaccharide for only Caco-2 cells. The inflammatory proteins, such as inducible nitric oxide synthase, were induced in Caco-2 and HepG2 cells. The presented GLA-MPS can be adapted as an advanced in vitro model in various applications for disease modeling associated with inter-tissue interactions, such as inflammatory disease.
I. INTRODUCTION
Gut-liver-axis (GLA) is a bidirectional interaction between the gut and liver mediated by dietary, genetic, and environmental factors1 that regulates the absorption/prevention of exogeneous agents at the gut barrier and metabolic processes in the liver.2,3 The disruption of GLA functions leads to liver disease progression and gastrointestinal disorders.4 Therefore, there are ongoing trends in the study of GLA for disease modeling and drug discovery. However, using experimental animal models does not yield clear information about GLA because they have a whole set of organs that show complicated interactions in addition to GLA. Therefore, it is necessary to establish an in vitro model to better understand GLA.
Microphysiological systems (MPSs) or organs-on-chips (OoCs) recapitulate pathophysiological conditions in vitro by utilizing micro/nanotechnology approaches for applications in drug discovery. The MPSs provide physiologically relevant cellular microenvironments (i.e., soluble factors, perfusion, extracellular matrices, tissue-like structures, and physical stimuli) for cells and organs,5 and interconnecting organs via flow channels and controls.6 These features are critical to investigating the absorption, distribution, metabolism, excretion, and toxicity (ADME-T) of the drug candidate and for also disease modeling.6 Therefore, MPSs are suitable for recapitulating GLA in vitro.
Thus far, several MPSs as an in vitro GLA model (GLA-MPS) have been reported, such as GLA for metabolism and inflammation axes.3,7–11 They showed better predictive capabilities of organ-to-organ interaction compared to normal static cell-culture models. However, there remain several concerns of flow perfusion control, organ functions, and inter-tissue interaction that need to be addressed for GLA-MPS. First, gut and liver cells have different physiologically relevant flow perfusion to be realized via a precise flow control in the chip. Second, gut and liver cells have cellular polarity, which should be functionalized in molecular transportation. Third, circulation flow needs to connect two organs for inter-tissue interactions. The current GLA-MPSs are based on two approaches: the cell-culture insert in the microtiter plates7,11 and microfluidic devices.12,13 For the cell-culture insert system, cellular uptake and transport across the cell barrier (e.g., endothelial cells and intestinal epithelial cells) can be studied with the porous membrane; under porous membranes, a circulation channel could be used to connect two organs and apply flow perfusion. However, it is difficult to apply an appropriate uniform perfusion flow for the surface of each organ within the insert precisely, because of its macroscopic scale in the millimeter range and lack of a perfusion controlling system. To express their organ cell functions, gut epithelial cells require physiological relevant fluid shear stress (FSS) from the perfusion flow.14,15 In contrast, using a microfluidic device allows the control of perfusion flow in the planar compartmentalized chambers and the interconnection of two or more tissue cells via a microfluidic channel with an on-chip micropump.12,13,16 These microfluidic devices have some issues to be addressed; for example, we can apply only one flow rate for the introduced tissues, although the gut and liver require different flow rates to execute their functions properly.17,18 Furthermore, the device cannot be applied to study organ molecular transport functions across the gut barrier because it does not have an apical to basal side transport structure such as a porous membrane substrate on the bottom channel.12 Thus, there is an urgent need to fulfill these requirements for establishing an advanced in vitro GLA model that (i) enables the application of an appropriate perfusion flow for the gut and liver, (ii) studies molecular transports across the gut barrier, and (iii) interconnects the gut and liver to study their interactions within a single microfluidic device.
Here, we present a GLA-MPS device that can achieve individual physiological flow perfusion for each organ in the cell-culture chamber, form barrier tissue structures on a porous membrane, and connect two organs with circulation flow. The device is fabricated with four-layered microfluidic structures such as a cell-culture layer, porous membrane, circulation layer, and control layer with an on-chip micropump structure. The FSS on cells is estimated by the computational fluid dynamic simulation. The Caco-2 intestinal epithelial and HepG2 hepatocyte-like cells are applied to their own physiologically relevant FSSs to enhance their functions. The circulation flow controlled by the micropump performed closed-loop circulation to interconnect the Caco-2 and HepG2 cell-culture chambers via a porous membrane. The lipopolysaccharide (LPS) and dexamethasone (Dex) were used to stimulate the GLA communications. This device allows modeling the GLA interaction as an in vitro model.
II. MATERIALS AND METHODS
A. Device concept and structure
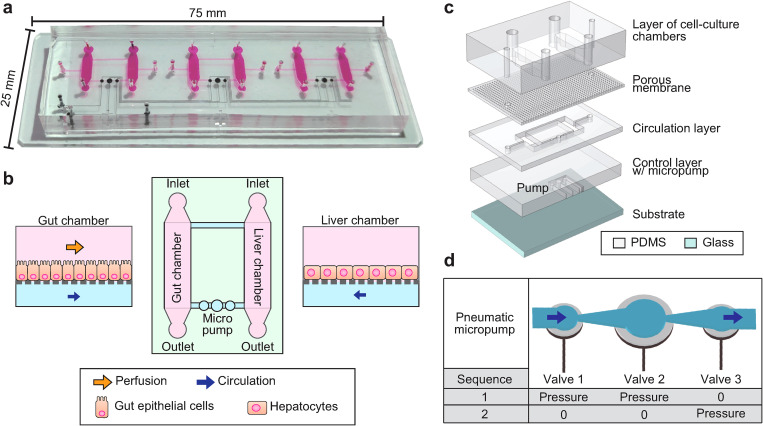
The GLA-MPS device was designed as shown in Figs. 1(a) and 1(b). The GLA-MPS device comprises four layers made of polydimethylsiloxane (PDMS) and a glass substrate [Fig. 1(c) and Fig. S1 in the supplementary material]. The four PDMS layers work as a cell-culture layer, porous membrane, circulation layer, and control layer integrated with a micropump. The PDMS meets the requirements as a structural material owing to its biocompatibility, gas permeability, and elasticity to realize micropumping.19 The cell-culture layer contains cell-culture chambers (length: 5.3 mm, width: 2 mm, height: 360 μm). A porous membrane has holes with a hole size of 5 μm, pitch of 20 μm, and thickness of 20 μm to prevent cell penetration through the holes and reduce the FSS for cells from the basal side. This membrane separates the cell-culture chamber and circulation channel to perform two different flows; soluble factors are transported via holes. An on-chip micropump was integrated into the control layer to perform closed-loop circulation. Furthermore, the external pump was connected to the inlet of the gut chamber to apply the FSS on the apical side of the gut epithelial cells.
FIG. 1.
Conceptual illustration of the gut-liver-axis (GLA)-MPS. (a) Photograph of the GLA-MPS with three sets of in vitro GLA and an integrated micropump. (b) Illustration of microfluidic flow channels for establishing GLA-MPS. Gut epithelial cells and liver hepatocytes are cultured in corresponding cell-culture chambers, applied optimal perfusion flow for each cell type, and interconnected by circulation flow. (c) Illustration of the GLA-MPS that comprises four PDMS layers and the bottom glass layer. (d) Schematic of the actuating micropump.
B. Micropump actuation for closed-loop circulation
An approach that uses a pneumatic micropump enabled integration with a microfluidic device. We fabricated a pneumatic micropump, which is a modified version of Quake's microvalve pump20 to perform closed-loop circulation [Fig. 1(d)]. The microvalve is integrated into the control layer, consisting of a flexible PDMS membrane and an underlying control channel. Membrane actuation is achieved by varying the positive hydraulic pressure from the control channel, bending toward the circulation channel. This micropump uses three adjacent microvalves of the membranes in combination with a larger size (diameter: 1 mm) and two smaller ones (diameter: 0.6 mm), connected to a diffuser and nozzle structure16 for increasing the one-directional flow by reducing backward flow. The purpose of this “two phase” actuating method is to simplify the actuating sequence, which reduces the efforts of frequency and pressure control from the pneumatic system. Here, circulatory perfusion flow is controlled by selectively releasing (“0”) and actuating (“pressure”) microvalves in the membrane “push-up” configuration. “sequence-1” represents actuating both valves 1 and 2 simultaneously, which keeps valve 3 free. Then, “sequence-2” represents the state where only valve 3 is actuating. The actuation time of the 1-mm microvalve is slower than that of the 0.6-mm microvalves because the larger volume of the control layer chamber leads to a high inertial mass.21 Thus, valve 2 is slower than that of valves 1 and 3, which leads to a peristaltic valve motion to generate output flow in one direction during actuation.
C. Chip fabrication
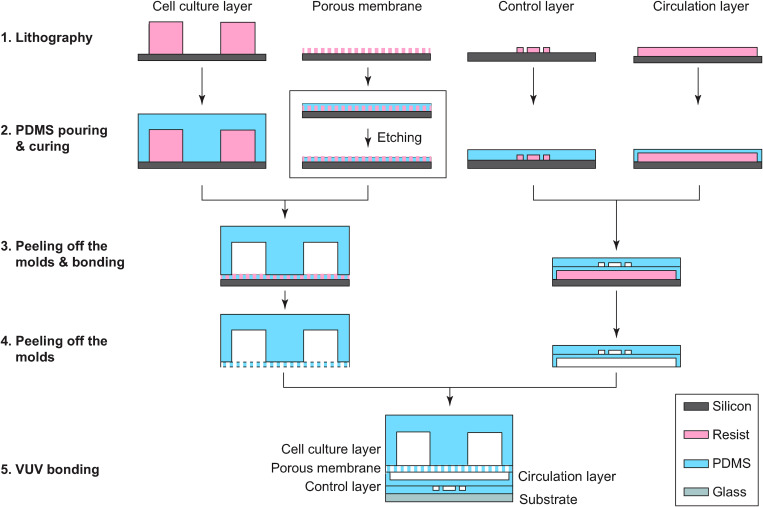
The GLA-MPS device was fabricated using multilayer soft-lithography (Fig. 2).22 For the resist mold of the cell-culture layer, a dry-film type negative photoresist (45 μm; TMMF S2045TM, Tokyo Ohka Kogyo) was laminated eight times to achieve a thickness of 360 μm; similarly, the TMMF was laminated two times to achieve a thickness of 90 μm for fabricating the resist mold of the circulation and control layers. The resist mold of the porous membrane layer was fabricated using a spin-coated 25-μm-thick negative photoresist (TMMR S2000TM, Tokyo Ohka Kogyo). UV exposure was performed using a mask aligner (MUM-1000 Series, Japan Science Engineering Co. Ltd.)23,24 with Cr-patterned glass masks for the corresponding molds of each layer (cell-culture layer: 1000 mJ cm−2, porous membrane: 1500 mJ cm−2, and circulation layer and control layers: 500 mJ cm−2). For the development, the resist molds were put in a propylene glycol monomethyl ether acetate (PGMEA) solution [PM Thinner, Tokyo Ohka Kogyo)] at 23 °C for 20 min.23,24 The PDMS base and curing agent (Sylgard 184, Dow Silicones) mixed in a ratio of 5:1 (w/w) for the porous PDMS membrane and 10:1 for other layers were poured onto resist molds and cured at 25 °C for 48 h. The designed membrane thicknesses were tuned by spin coating to 25 μm for the porous PDMS membrane and 60 μm for the pump membrane. The cured PDMS layer of the porous membrane was etched with a 5-μm thickness using the O2 plasma (power 100 W: pressure 50 Pa: flow rate 50 SCCM; FA-1, Samco Inc.) to open the through holes (Fig. S2 in the supplementary material). The cell-culture layer was bonded to the porous PDMS membrane, whereas the control layer was bonded to the circulation layer. The two assembled structures were peeled off from the substrate and bonded together using VUV surface activation (Min-Excimer SUS713, Ushio Inc.). Finally, the bonded PDMS layers were bonded on the glass substrate and cured in an oven at 45 °C for 24 h. Low-temperature bonding causes less deformations on the PDMS block, which prevents shrinkage and bending, and leads to flat surfaces for multilayer bonding.
FIG. 2.
Fabrication procedure of the GLA-MPS. (1) Each mold was fabricated with photolithography. (2) Pre-PDMS mixture was poured on the molds and cured. (3) Solidified PDMS structures of the cell-culture layer and control layer were peeled off from the molds and bonded to the porous membrane layer and circulation layer, respectively. (4) The bonded layers were peeled off from the mold, and then (5) the layers were assembled on the glass substrate using VUV bonding.
D. Evaluation of micropump flow rate
Microbead tracing (4.5 μm in diameter) was used for visualizing the culture medium flow to characterize the flow rate generated by the on-chip micropump; the flow distances of the beads were measured. Before the experiments, the microfluidic channel was coated with 1% (w/v) bovine serum albumin (BSA, Sigma-Aldrich) in PBS for 2 h at 25 °C to prevent the nonspecific adhesion of beads on flow channels and cell-culture chambers. The beads were suspended in a 1% (w/v) BSA solution at 1 × 106 beads ml−1; subsequently, a bead-suspended solution was introduced into the device. The movement of the microbeads was captured using a CMOS camera (EO-4010M, Edomond Optics) equipped with a microscope (GX53, Olympus); the bead velocities for flow velocity were analyzed using a custom-made MATLAB program (R2020b, MathWorks). The volumetric flow rates were calculated by multiplying the flow velocity by the cross-sectional area of the microfluidic channel.
E. Computational fluid dynamics for fluid shear stress
Computational fluid dynamics analysis was used to estimate the FSS of the cell surface using COMSOL Multiphysics (Ver. 5.6, COMSOL Inc.). The center cross section along the gut chamber perfusion direction was used in the calculation; the microhole structure of the porous membrane was incorporated. The cell-culture medium was considered an incompressible and homogeneous Newtonian fluid with conditions similar to water at 37 °C (density: 997 kg m−3, viscosity: 6.9 × 10−4 Pa s−1). A 10-μm-thick layer covered the upper side of the porous membrane with no direct flow across via the interior wall setting to model the cell layer. No-slip boundary conditions were applied to the channel walls. A laminar flow/stationary library was used to determine the shear rate ( ), and the FSS ( ) was calculated by multiplying the shear rate with the dynamic viscosity of water.
F. Cell culture
HepG2 human hepatocellular carcinoma and Caco-2 human colorectal adenocarcinoma cell lines were obtained from the American type culture collection (ATCC). Cells were maintained in the cell-culture medium containing Dulbecco's modified Eagle medium (DMEM) (Sigma-Aldrich) supplemented with 10% (v/v) fetal bovine serum (FBS, Cell Culture Bioscience), 1% (v/v) nonessential amino acids (NEAA, Thermo Fisher Scientific), and 1% (v/v) penicillin/streptomycin (P/S, Thermo Fisher Scientific) in a humidified incubator at 37 °C with 5% (v/v) CO2. HepG2 and Caco-2 cells were passaged with trypsin/EDTA (0.04%/0.03%[v/v]) solution every 3 and 7 days, respectively.
G. Cell culture on a chip
The chips were sterilized with 70% (v/v) ethanol and ultraviolet light in a biosafety cabinet at 25 °C for 40 min prior to the cell culture on the chips. Then, two chambers in the cell-culture-chamber layer were filled with a 1.3% (v/v) Matrigel hESC-qualified matrix (Corning) in DMEM/F12 (Sigma-Aldrich) at 25 °C for 2 h. The cell culture and circulation chambers were washed and filled with a cell-culture medium. The chips were incubated at 4 °C for 16 h to remove all air bubbles from the chips. The next day, after washing the chips with a cell-culture medium warmed at 37 °C, 3 μl of cell suspensions of Caco-2 and HepG2 cells (5 × 106 cells ml−1) were introduced into the corresponding cell-culture chamber chips; the cell density was 1800 cells mm−2. The chips were placed in a humidified incubator at 37 °C with 5% (v/v) CO2. After 24 h, the cell-culture medium was changed to remove the floating dead cells. For circulation and perfusion, the inlet of the gut culture chambers was connected to a syringe pump (KDS LEGATO 210, KD Scientific Inc.); the micropump was actuated for medium recirculation. For a static cell culture, the cell culture and circulation chamber layers were changed with the cell-culture medium every 6 h. The total cell culture on the chip was maintained for up to 10 days.
H. Live/dead cell assay
A cell viability assay was conducted using the NucleoCounter NC-200, Via1-CassetteTM (Chemometec). After the cell culture or treatment, cells in the chips were washed with PBS once by adding 50 μl PBS through the cell-culture chamber and the circulation channel. Then, 20 μl of the trypsin/EDTA solution was added through the cell-culture chambers and incubated at 37 °C for 10 min. After cell detachment, cells in both chambers were separately harvested into a 1-ml tube and diluted to 200 μl with the cell-culture medium. The homogeneous cell suspension was aspirated into a Via1-CassetteTM to count the number and ratio of dead/live cells.
I. Immunocytochemistry
After the cell culture and treatment, the cell-culture chambers and circulation layer were washed twice with PBS containing 50 μl of PBS. The cells were fixed with 4% paraformaldehyde (PFA, FUJIFILM Wako Pure Chemical) in PBS for 15 min at 25 °C and permeabilized with 0.1% (v/v) Triton X-100 in PBS for 30 min at 25 °C. The cells were then incubated in a blocking buffer (5% normal goat serum, Vector; 5% normal donkey serum, Wako; 3% BSA, Sigma-Aldrich; and 0.1% Tween-20, Nacalai Tesque, Inc., in PBS) at 4 °C for 16 h. After blocking, the cells were incubated with primary antibodies such as rabbit anti-human ZO-1 IgG (Thermo Fisher Scientific), mouse anti-human albumin IgG (Thermo Fisher Scientific), and mouse anti-human inducible nitric oxide synthase (iNOS) IgG (R&D Systems) in a blocking buffer at 4 °C for 16 h. After washing the primary antibodies with PBS-T twice, the cells were incubated with secondary antibodies such as Alexa Fluor 488-labeled donkey anti-rabbit IgG and Alexa Fluor 647-labeled donkey anti-mouse IgG H&L (Jackson ImmunoResearch) in a blocking buffer at 25 °C for 1 h. The cell nuclei were stained with 300 nM 4,6-diamidino-2-phenylindole (DAPI, Dojindo Laboratories). The DAPI solution (50 μl) was introduced into the chambers and incubated at 25 °C for 30 min; the chambers were washed with PBS.
J. Imaging acquisition
Confocal microscopic imaging was performed using an Andor Dragonfly 502 system (Andor, UK). This system is built on a Nikon ECLIPSE Ti2-E inverted fluorescence microscope (Nikon) and equipped with a Zyla 4.2 Plus scientific complementary metal oxide semiconductor (sCMOS) camera (Andor) driven by Fusion (Andor), a CFI Plan Apochromat Lambda 10× dry/0.45 NA (Nikon), motorized XY stage and stage piezo (Applied Scientific Instrumentation), and three lasers (405, 488, and 637 nm). For image acquisition, the lasers were set to 488 nm for ZO-1, 637 nm for albumin, and 405 nm for DAPI.
K. Imaging quantification
The imaging data quantification process was conducted using the CellProfilerTM (the Broad Institute of Harvard and MIT, version 4.07). To reduce the background staining effect, a mathematically subtracting of the background signal was applied in every image in every calculating cycle using the correctilluminationCalcuate and correctilluminationApply functions (Median filter, 50). Using IdentifyPrimaryObjects, DAPI images were selected and nuclei were identified. The parameters were set as follows: typical diameter of objects from 10 to 40; threshold smoothing scale of 0.5; threshold correction factor of 1; lower and upper bounds on the threshold of 0 and 1; method to distinguish clumped objects, intensity; method to draw dividing lines between clumped objects, intensity; default values were used for the other parameters. In IdentifySecondaryObjects, stained total cell images were selected, and nuclei, as the input objects, were selected. The parameters were set as follows: method to identify the secondary objects, propagation; threshold strategy, global; thresholding method, Otsu with two classes; threshold smoothing scale of 0; threshold correction factor of 1; lower and upper bounds on the threshold of 0 and 1; regularization of 0.05. Holes were filled in the identified objects. Objects touching the border of images were not discarded. In MeasureObjectIntensity, the intensity of nuclei and cells of identified objects was measured.
L. Permeability assay across cell barrier
FITC-dextran (4 kDa, Sigma-Aldrich) was used for the permeability assay.25 It was dissolved into a cell-culture medium at a concentration of 1 mg ml−1 as the working solution. The working solution was introduced into the gut cell-culture chamber and incubated for 4 h. The FITC-dextran in the medium of the cell-culture chamber and circulation channel was separately collected and measured using the Synergy HTX microplate reader (BioTek Instruments). The hydraulic conductivity Kdextran was calculated using
| (1) |
where A, C0, and dQ/dt represent the transporting surface area of the porous membrane (5.3 × 2 mm2), original concentration of the FITC-dextran in the gut apical chamber, and amount of FITC-dextran transported across the membrane over the respective time interval, respectively.
M. Cell treatment in GLA-MPS
LPS (Escherichia coli O111:B4, Sigma-Aldrich) and Dex (Wako) were used to treat Caco-2 cells. LPS and Dex were dissolved in a cell-culture medium at 1 mg ml−1 and 200 μM, respectively, as the treatment solutions. After 9 days of culturing in the GLA-MPS, only Caco-2 cells were treated with the treatment solutions for 4 days.
N. Enzyme-linked immunosorbent assays
Enzyme-linked immunosorbent assay (ELISA) was performed to measure TNF-α secreted from the cells by following the manufacturer's instructions of RayBio Human TNF-α ELISA kit (RayBiotech). During 4-day stimulation, the cell-culture medium from cell-culture chambers and the circulation channel were collected, and the secreted TNF-α was measured at 450 nm absorbance by the Synergy HTX microplate reader (BioTek Instruments).
O. Statistical analysis
The Student's t-test, asymptotic Wilcoxon signed-rank test, and Tukey's multiple comparison test were conducted using R software (ver. 3.5.2; https://www.r-project.org/).
III. RESULTS AND DISCUSSIONS
A. Computational estimation of shear stress and flow rates in GLA-MPS
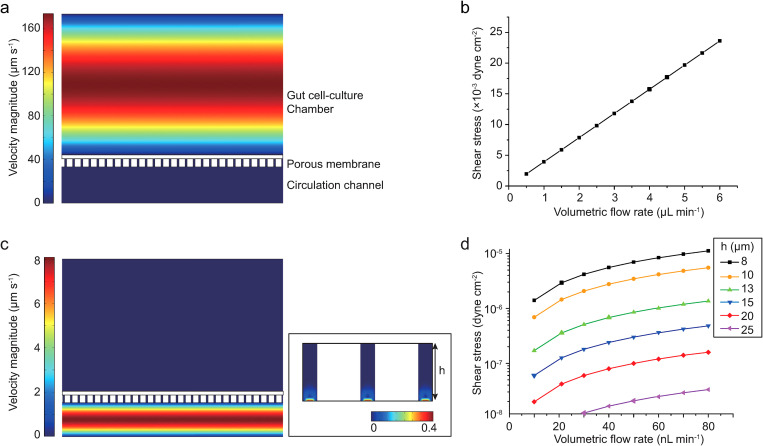
The FSS was one of the most important physical factors affecting the cell function and morphology,18,26 but the previous GLA-MPSs faced challenges in the control. The flow perfusion conditions should be investigated for each organ cell. In this device, we estimated the FSS and flow rates using COMSOL simulation to investigate whether the GLA-MPS provides proper FSS on cells (Fig. 3). When the medium perfusion in the gut chamber was performed from 2 to 5 μl min−1, the surface FSS on the apical side of cells had a linear relationship with the external pumping flow rate [Figs. 3(a) and 3(b)]. The mean FSS for the intestinal cells was in the range of 8 × 10−3–2 × 10−2 dyn cm−2, which was in the physiologically relevant range26,27 and could promote gut cell barrier formation and differentiation processes.28,29
FIG. 3.
Simulated perfusion flow in the GLA-MPS. (a) Simulated perfusion flow in the gut cell-culture chamber. (b) Simulated shear stress on the cell surface cultured on the porous membrane. (c) Simulated circulation flow in the circulation chamber. (d) Simulated shear stress by the circulation flow.
The FSS applied to cells from the basal side through the holes of the membrane was determined using the membrane thickness (h) and circulation flow rate [Figs. 3(c) and 3(d)]. The FSS was reduced dramatically with an increase in membrane thickness. In our setup parameters (20-μm-thick membrane), we found that the surface FSS (1 × 10−8–1.2 × 10−7 dyn cm−2) on the basal sides of cells by the circulation flow was considerably smaller than the FSS on the apical side of gut epithelial cells. Thus, in the effect of circulation flow, we not only considered the effects of FSSs of basal sides for the later cell experiments, but we also recognized that circulation flow would mediate communication between the gut and liver cells.
In recent results for the gut on a chip,17 the Caco-2 cells on the porous membrane experienced flow perfusion in the apical side or basal side, which showed an improvement in cell functions. In our device, by using the simulation model, the FSS on cells is well-designed and can be clearly used for investigating their effect on cells from both sides.
B. Evaluation of circulation flow by the micropump
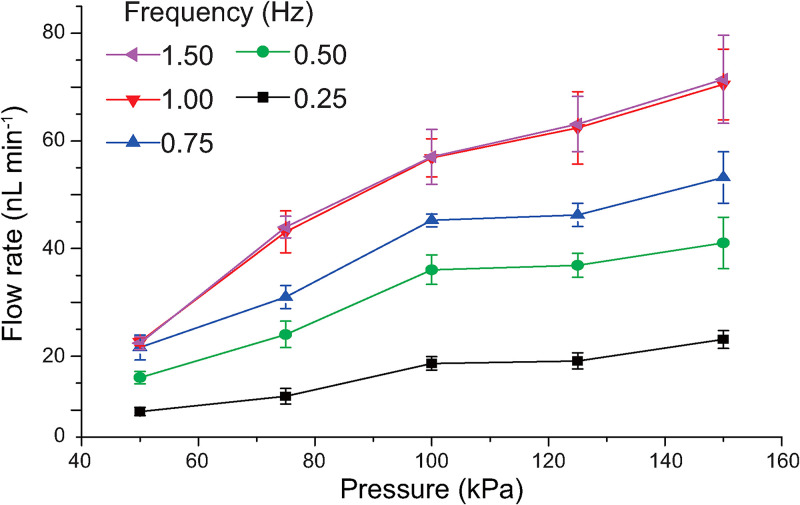
The micropump for circulation flow was controlled using the developed pneumatic actuating method to induce the flow circulation between two organ chambers. We investigated the circulation flow rates controlled by an on-chip micropump after the fabrication of the GLA-MPS (Fig. 4). The mean flow rate increased with driving pressure (0–150 kPa) and pumping frequency (0–1.5 Hz). Working frequencies over 1 Hz exhibited no significant differences in the generated flow rates; in contrast, the GLA-MPS had disassembled by applying over 150 kPa in the control layer for 14 days (data not shown). Therefore, the working pressure was set from 50 to 150 kPa to carry out cell culture and assay in the GLA-MPS; this gave flow rates from 22 to 61 nl min−1 for circulation at a 1-Hz working frequency.
FIG. 4.
Measured mean flow rate of the micropump for pressure-dependent actuation from 0.25 to 1.5 Hz frequency. Each dot represents the mean ± standard deviation (SD) (n = 10).
The flow rate requirement for inducing two organ interactions could be estimated from the exchange ratio of circulation flow rate (q) and flow volume (Q), i.e., q/Q. In this device, the microfluidic system has a circulation flow volume of 1.5 μl. The exchange ratio is 1.5%–9% in 1 min. In other GLA-MPSs,3,7 the exchange ratio is approximately 0.25%–1% in 1 min. Thus, our device realized a more intensified circulation flow between the gut and liver. For 9-day cell culture, this flow is sufficient to induce the interaction between two organs in the GLA-MPS device. Besides, the on-chip micropump could realize a fine range of flow rate adjustment, which is consistent with the aforementioned FSS simulation circulation flow rate range. Therefore, we could couple the circulation flow with gut perfusion flow to improve the cell function and induce the organ to organ interaction for cell-culture experiments.
C. GLA culture on a chip
Caco-2 gut intestinal30 and HepG2 hepatocyte-like cells31 were used and cultured in GLA-MPS to establish in vitro GLA. Prior to the cell culture, the surface of the porous membrane should be coated with Matrigel for cell attachment. By applying the pressure barriers of caplillary,32 Matrigel was only coated on the surface of the cell-culture chamber side (upper side), but not the surface of holes and circulation channel side (bottom side). This is to reduce cell attaching and migrating through the hole of the porous membrane (Fig. S3 in the supplementary material).
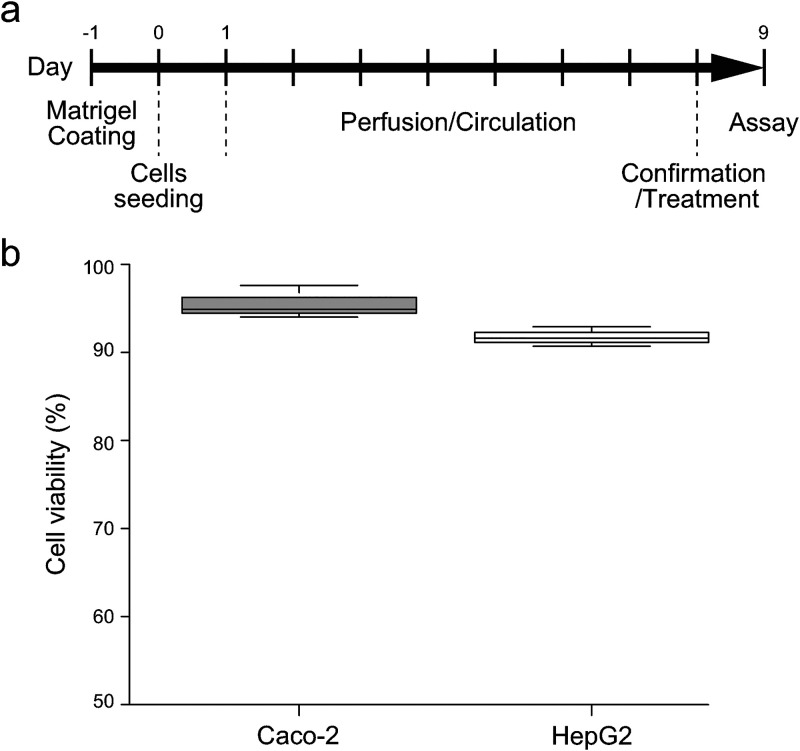
After culturing for 1 day under the static conditions for the cell attachment, only Caco-2 cells, but not HepG2 cells, were subjected to the perfusion flow, since the apical side of hepatocytes in the physiological situation is not subjected to the perfusion flow [Fig. 5(a)]. Both Caco-2 and HepG2 cells were maintained for eight days with the circulation flow in the circulation channel. After the culture, Caco-2 and HepG2 cells showed 95% and 92% cell viability, respectively [Fig. 5(b)]. It demonstrated that the cells could be maintained with relatively high cell viability in this device.
FIG. 5.
Cell culture and cell viability. (a) Experimental procedure of cell culture, perfusion, circulation, and treatments in the GLA-MPS. (b) Evaluation of cell viability of Caco-2 cells (2 μl min−1 of the perfusion flow; 61 nl min−1 of circulation flow) and HepG2 cells (without perfusion flow; 61 nl min−1 of circulation flow) in the GLA-MPS.
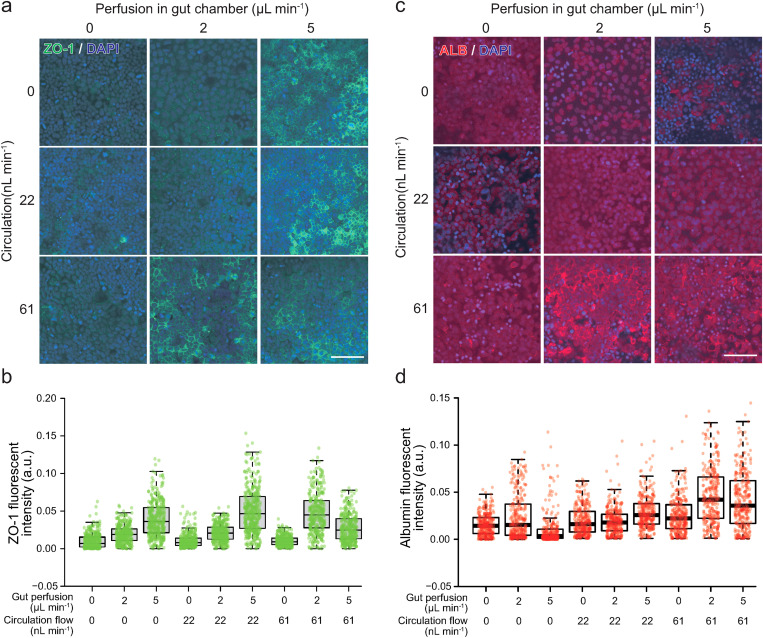
The expression of ZO-1 and albumin (ALB), as functional markers of the gut and liver, respectively, was observed by immunocytochemistry to confirm the functions of cells cultured in the GLA-MPS (Fig. 6). Caco-2 cells expressed ZO-1 tight-junction protein to form the gut barrier [Fig. 6(a)]; the increase in flow rates of perfusion flows in the gut cell-culture chamber induced the ZO-1 expression in the Caco-2 cells at 5 μl min−1 (FSS = 2 × 10−2 dyn cm−2), as shown in Fig. 6(b) and Table S1 in the supplementary material. Furthermore, the circulation flow did not change the ZO-1 expression in the Caco-2 cells without perfusion flow; instead, it induced the ZO-1 expression in addition to the perfusion flow [Fig. 6(b)]. In light of the simulation results, FSS by circulation flow was considerably smaller than that by perfusion flows; however, the cultured cells could receive the FSS of basal sides. The cell-culture medium in the circulation channel could accumulate molecules (i.e., growth factors, cytokines, metabolites, and lipids) and exosomes secreted from Caco-2 and HepG2 cells. Thus, the medium contained both autocrine and paracrine factors and facilitated the interactions of Caco-2 and HepG2 cells by the circulation flow. Thus, the induction of the ZO-1 expression could be the outcome of the combinations of FSS by perfusion/circulation flows and molecular transport.
FIG. 6.
Optimal perfusion and circulation flows enhanced the expression of ZO-1 in Caco-2 cells and ALB in HepG2 cells. (a) Immunofluorescent micrographs of the ZO-1 (green) expression in Caco-2 cells. (b) Quantitative single-cell profiling of the ZO-1 expression in Caco-2 cells. P values estimated by the Tukey's multiple comparison test are shown in Table S1 in the supplementary material. (c) Immunofluorescent micrographs of the ALB (red) expression in Caco-2 cells. (d) Quantitative single-cell profiling of the ALB expression in HepG2 cells. Centerlines of box plots indicate medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles. P values estimated by the Tukey's multiple comparison test are shown in Table S2 in the supplementary material. Nuclei were stained with DAPI. The scale bar represents 100 μm.
In the case of HepG2 cells, the HepG2 cells were not directly subjected to perfusion flow in the cell-culture chamber, since we focus to evaluate the effects of LPS from the circulation channel and there are numbers of reports describing the effects of perfusion flow on hepatocytes;33 also, increasing the gut cell-culture chamber perfusion flow rates did not improve the ALB expression in HepG2 cells [Fig. 6(c)], while the increase in circulation flow rates from 0 to 61 nl min−1 significantly induced the ALB expression in HepG2 cells [Fig. 6(d) and Table S2 in the supplementary material]. These results suggest that HepG2 cells facilitated hepatic functions such as ALB production by FSS and as a molecular transport in the circulation channel.
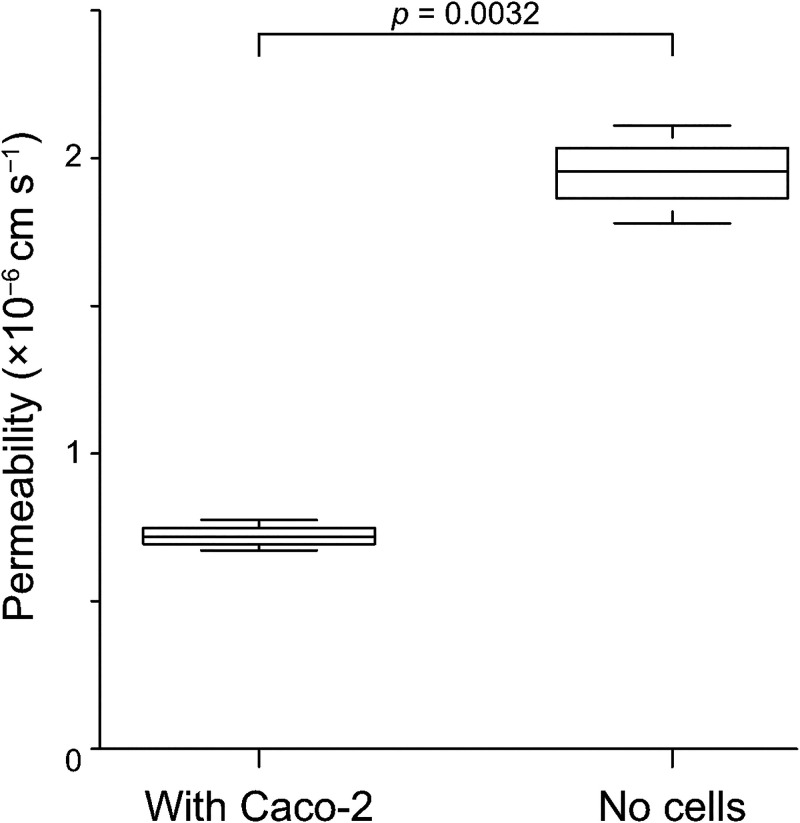
Thus, we selected the flow rates (gut perfusion flow: 2 μl min−1; circulation flow: 61 nl min−1) to establish in vitro GLA, which provided the highest expression of functional proteins in both Caco-2 and HepG2 cells among the tested conditions. Using this optimal condition, we tested the permeability of Caco-2 cells after the 9-day culture, and we confirmed the cell barrier function (Fig. 7). It indicated that Caco-2 cells formed an intact cell barrier to protect the transportation of 4-kDa FITC-dextran across the porous membrane.
FIG. 7.
Permeability assay to evaluate the Caco-2 barrier function by observing FITC-dextran transportation from the culture chambers to the circulation channel via the porous membrane (n = 3).
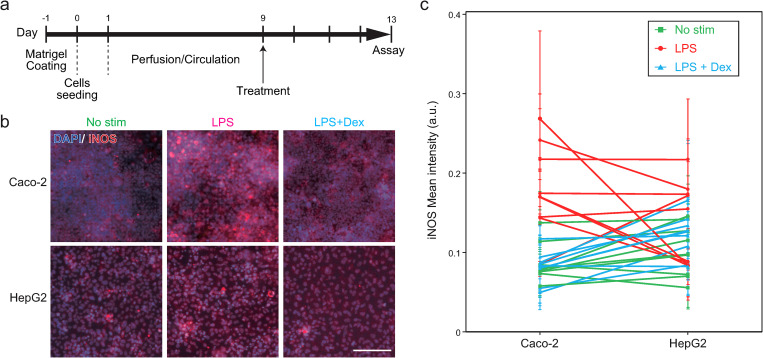
D. In vitro recapitulation of the inflammatory bowel disease in GLA-MPS
To demonstrate the applicability of the GLA-MPS for disease modeling, we selected to recapitulate inflammatory bowel disease (IBD)7 in vitro (Fig. 8). IBD is one of the chronic inflammatory intestinal disorders and is often caused by genetics, immune systems, microbiome, and environmental exposure, and their combinations. Importantly, IBD patients have a high risk of autoimmune liver disease via GLA,34 but the underlying mechanisms of IBD progression are not fully understood yet. The feature of the presented GLA-MPS has an individual addressability for each cell-culture chamber. This feature is advantageous for modeling and investigating multiple organ disease interactions such as IBD and liver disease.
FIG. 8.
In vitro recapitulation of IBD in the GLA-MPS. (a) Experimental procedure of cell culture and treatment. (b) Immunofluorescent micrographs of the iNOS (red) expression in Caco-2 and HepG2 cells with no stimulation (no stim), 1 mg ml−1 lPS treatment (LPS), and 1 mg ml−1 lPS with 200 μM Dex treatment (LPS + Dex). Nuclei were stained with DAPI. The scale bar represents 100 μm. (c) Quantification of the iNOS expression in individual Caco-2 and HepG2 cells based on the immunofluorescent micrographs shown in (b). Each bar and error bars represent the mean ± standard deviations. n = 3. P values of the Tukey's multiple comparison test are summarized in Table S3 in the supplementary material.
To recapitulate IBD using the GLA-MPS, we utilized the bacterial endotoxin, LPS,35 administrated in the Caco-2 culture chamber to induce gut inflammation. After the treatment with 1 mg ml−1 of LPS on Caco-2 cells for 4 days [Fig. 8(a)], most of the Caco-2 samples expressed iNOS, which is often observed in IBD;36 the comparison with non-treated Caco-2 cells is shown in Figs. 8(b) and 8(c), Tables S3 and S4 in the supplementary material. Following the inflammatory induction of Caco-2 cells, the iNOS expression was observed in four out of nine samples of HepG2 cells, while no change in the iNOS expression was observed in the other five samples. Despite the use of established cell lines, such as Caco-2 and HepG2 cell lines, the complexity of the experimental setup could have caused the observed individual differences in the LPS stimulation. In contrast, the treatment of 200 μM of dexamethasone (Dex), an anti-inflammation agent, on the Caco-2 cells during 1 mg ml−1 lPS treatment suppressed the iNOS expression in both Caco-2 and HepG2 cells. Thus, inflammatory responses of HepG2 cells were induced by the LPS-treated Caco-2 cells.
To identify the inflammatory mediators that induced inflammatory responses of HepG2 cells via the circulation channel, ELISA was performed for TNF-α, which was elevated in the gut of IBD models.36 However, TNF-α in the cultured medium was not observed because the amount of TNF-α within the circulation channel with a limited volume (1.5 μl) was lower than the detection limit of ELISA (Fig. S4 in the supplementary material). Therefore, highly sensitive methods are required to detect the mediators within such a small volume to identify the mediators that influence the interaction between Caco-2 and HepG2 cells via the circulation channel.
Gut microbiome37 plays critical roles in the pathophysiological scenarios in the gut; they can influence other organs, including liver,38,39 brain,40 and the immune system.41 Instead of liver cells, our MPS device allows the introduction of other tissue cells such as neurons, and we can investigate the interactions with the gut microbiome. Thus, our MPS is not limited to recapitulating only GLA, but it is also applicable to other organs and the microbiome.
Furthermore, the presented GLA-MPS allows a collection of conditioned cell-culture media from two cell-culture chambers and circulation channels separately for investigating the underlying molecular mechanisms of the GLA by biochemical assays such as ELISA and mass spectroscopy. Since these methods for the cell-culture medium are non-invasive for cultured cells, we will be able to determine the transitions of molecular contents in the medium during the experimental period to explore interesting molecular biomarkers. At the end of the experiments, GLA-MPS allows performing invasive cell-analytical methods such as immunocytochemistry, as shown in this paper, as well as multi-omics analysis (i.e., genomics, transcriptomics, proteomics, and metabolomics) to obtain deeper insights into normal and diseased GLA.
One advantage of using microfluidic technology for disease modeling is the minimization of the number of cells, such as the primary cells obtained from the patients. These cells are limited in their ability to recapitulate the patients' pathological conditions in vitro. The presented MPS requires only 1.5 × 104 cells for each cell-culture chamber, which is considerably smaller than those of the 24-well cell-culture inserts (3 × 105 cells). Therefore, we efficiently use precious patient samples, such as biopsy specimens. In the present study, we used widely used cell lines (Caco-2 and HepG2) that can provide the standard performance of the presented GLA-MPS. Since patient-derived primary cells could have large patient-to-patient variations, it would be beneficial to obtain such standard data sets with the use of commonly used cell lines.
The device is fabricated using the microfabrication method with a four-layered planer microfluidic structure. Owing to this feature, another advantage is the capability to integrate electrical read-out sensors in cell-culture chambers and circulation channels, which allows for monitoring cell conditions and behaviors during cell culture and treatments in a real-time and non-invasive fashion.42 Such sensors will provide temporal information on tissue formation, disease progression, and drug treatments. In the case of the GLA, the sensor for measuring trans-epithelial electrical resistance (TEER) was beneficial for evaluating gut barrier formation and causing damage during disease progression. In this study, we used a fluorescent dye to investigate gut barrier function; however, the dye might also influence cellular functions. Until now, gut-on-a-chips integrated with TEER measurements have been reported.43,44 The designs and fabrication processes need to be further improved for better accuracy and reproducibility and for integration with multi-organ MPS. This integrative approach will lead to the advancement of MPS for a better understanding of disease mechanisms and applications in drug discovery.
IV. CONCLUSIONS
We developed a GLA-MPS platform that can (i) cultivate gut and liver cells separately with applied different perfusion flows/FSS to improve their cell viabilities and functionalities, (ii) form barrier tissue, and (iii) interconnect GLA via circulation channels. The device is a more distinctive platform than the current GLA-MPS, resembling the physiological flow perfusion and circulation of in vivo GLA. It is also suitable for demonstrating the individual accessibility to investigate the GLA interaction mediated by IBD. However, the GLA-MPS still has some limitations that need to be improved in the future. For example, the device can be integrated with the organoid structure, which could extend the gut and liver organ functions for tissue engineering and disease modeling. In addition, the device should have capabilities beyond FSS control and effectively control the molecular transport gradient across the barrier tissues in the microenvironment to understand the GLA interaction. Finally, our approach based on GLA-MPS has the potential for applications in understanding the GLA mechanisms associated with individual physiological flow perfusion and multi-organ interactions for disease modeling.
SUPPLEMENTARY MATERIAL
See the supplementary material (Fig. S1) for the mask pattern design of GLA-MPS. Figure S2 shows the microfabrication of the porous PDMS membrane. Figure S3 shows the single-side coating of Matrigel on the porous membrane in cell-culture chambers to prevent cell migration across the pores of the porous membrane by applying pressure barriers to the capillary. Figure S4 shows the ELISA testing results to measure the secreted TNF-α. Tables S1–S4 show the Tukey's multiple comparison test for ZO-1 of Caco-2, ALB of HepG2, iNOS of Caco-2, and iNOS of HepG2.
ACKNOWLEDGMENTS
This work was funded by the Japan Society for the Promotion of Science (Nos. 17H02083, 18KK0306, 19H02572, and 21H01728), the Ebara Hatakeyama Memorial Foundation, Japan Agency for Medical Research and Development (No. 17937667), and the LiaoNing Revitalization Talents Program (No. XLYC1902061). Part of this work was supported by the Nanotechnology Platform Project within MEXT, Japan, through the Kyoto University Nano Technology Hub. The authors thank the iCeMS Analysis Center for access to the analytical instruments. The authors would like to thank Editage (www.editage.jp) for the English language review. WPI-iCeMS is supported by the World Premier International Research Centre Initiative (WPI), MEXT, Japan.
AUTHOR DECLARATIONS
Conflict of Interest
The authors have no conflicts to disclose.
Author Contributions
Jiandong Yang: Conceptualization (equal); Data curation (lead); Formal analysis (lead); Investigation (lead); Methodology (lead). Satoshi Imamura: Conceptualization (equal); Data curation (lead); Formal analysis (lead); Investigation (equal); Methodology (lead); Writing – review & editing (equal). Yoshikazu Hirai: Conceptualization (lead); Data curation (lead); Formal analysis (lead); Funding acquisition (lead); Investigation (lead); Methodology (lead); Project administration (lead); Resources (lead); Software (equal); Supervision (lead); Validation (equal); Visualization (equal); Writing – original draft (equal); Writing – review & editing (equal). Toshiyuki Tsuchiya: Project administration (equal); Supervision (equal). Osamu Tabata: Funding acquisition (equal); Supervision (equal); Writing – review & editing (equal). Ken-ichiro Kamei: Conceptualization (equal); Data curation (equal); Formal analysis (equal); Funding acquisition (lead); Investigation (lead); Methodology (equal); Project administration (lead); Resources (equal); Software (equal); Supervision (lead); Validation (equal); Visualization (equal); Writing – original draft (lead); Writing – review & editing (lead).
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
REFERENCES
- 1.Albillos A., de Gottardi A., and Rescigno M., J. Hepatol. 72, 558 (2020). 10.1016/j.jhep.2019.10.003 [DOI] [PubMed] [Google Scholar]
- 2.Arakawa H., Sugiura S., Kawanishi T., Shin K., Toyoda H., Satoh T., Sakai Y., Kanamori T., and Kato Y., Lab Chip 20, 537 (2020). 10.1039/C9LC00884E [DOI] [PubMed] [Google Scholar]
- 3.Prot J. M., Maciel L., Bricks T., Merlier F., Cotton J., Paullier P., Bois F. Y., and Leclerc E., Biotechnol. Bioeng. 111, 2027 (2014). 10.1002/bit.25232 [DOI] [PubMed] [Google Scholar]
- 4.Tripathi A., Debelius J., Brenner D. A., Karin M., Loomba R., Schnabl B., and Knight R., Nat. Rev. Gastroenterol. Hepatol. 15, 397 (2018). 10.1038/s41575-018-0011-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang B., Korolj A., Lai B. F. L., and Radisic M., Nat. Rev. Mater. 3, 257 (2018). 10.1038/s41578-018-0034-7 [DOI] [Google Scholar]
- 6.Caplin J. D., Granados N. G., James M. R., Montazami R., and Hashemi N., Adv. Healthc. Mater. 4, 1426 (2015). 10.1002/adhm.201500040 [DOI] [PubMed] [Google Scholar]
- 7.Trapecar M., Communal C., Velazquez J., Maass C. A., Huang Y.-J., Schneider K., Wright C. W., Butty V., Eng G., Yilmaz O., Trumper D., and Griffith L. G., Cell Syst. 10, 223 (2020). 10.1016/j.cels.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen W. L. K., Edington C., Suter E., Yu J., Velazquez J. J., Velazquez J. G., Shockley M., Large E. M., Venkataramanan R., Hughes D. J., Stokes C. L., Trumper D. L., Carrier R. L., Cirit M., Griffith L. G., and Lauffenburger D. A., Biotechnol. Bioeng. 114, 2648 (2017). 10.1002/bit.26370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fowler S., Chen K., Duignan D., Gupta A., Hariparsad N., Kenny J. R., Lai G., Liras J., Phillips J. A., and Gan J., Lab Chip 20, 1179 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Sung J. H., Wang Y. I., Narasimhan Sriram N., Jackson M., Long C., Hickman J. J., and Shuler M. L., Anal. Chem. 91, 330 (2019). 10.1021/acs.analchem.8b05293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinha K., Nihei W., Nakamura H., Goto T., Kawanishi T., Ishida N., Yamazaki N., Imakura Y., Mima S., Inamura K., Arakawa H., Nishikawa M., Kato Y., Sakai Y., and Kimura H., Micromachines 12, 1007 (2021). 10.3390/mi12091007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J., Hirai Y., Iida K., Ito S., Trumm M., Terada S., Sakai R., Tsuchiya T., Tabata O., and Kamei K., BioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 13.Kamei K., Kato Y., Hirai Y., Ito S., Satoh J., Oka A., Tsuchiya T., Chen Y., and Tabata O., RSC Adv. 7, 36777 (2017). 10.1039/C7RA07716E [DOI] [Google Scholar]
- 14.Bein A., Shin W., Jalili-Firoozinezhad S., Park M. H., Sontheimer-Phelps A., Tovaglieri A., Chalkiadaki A., Kim H. J., and Ingber D. E., Cell. Mol. Gastroenterol. Hepatol. 5, 659 (2018). 10.1016/j.jcmgh.2017.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan H.-Y., Trier S., Rahbek U. L., Dufva M., Kutter J. P., and Andresen T. L., PLoS One 13, e0197101 (2018). 10.1371/journal.pone.0197101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mi S., Pu H., Xia S., and Sun W., Sens. Actuators A 301, 111704 (2020). 10.1016/j.sna.2019.111704 [DOI] [Google Scholar]
- 17.Shin W., Hinojosa C. D., Ingber D. E., and Kim H. J., IScience 15, 391 (2019). 10.1016/j.isci.2019.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du Y., Li N., Yang H., Luo C., Gong Y., Tong C., Gao Y., Lü S., and Long M., Lab Chip 17, 782 (2017). 10.1039/C6LC01374K [DOI] [PubMed] [Google Scholar]
- 19.Halldorsson S., Lucumi E., Gómez-Sjöberg R., and Fleming R. M. T., Biosens. Bioelectron. 63, 218 (2015). 10.1016/j.bios.2014.07.029 [DOI] [PubMed] [Google Scholar]
- 20.Unger M. A., Chou H. P., Thorsen T., Scherer A., and Quake S. R., Science 288, 113 (2000). 10.1126/science.288.5463.113 [DOI] [PubMed] [Google Scholar]
- 21.Lai H. and Folch A., Lab Chip 11, 336 (2011). 10.1039/C0LC00023J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai A., Altemose N., White J. A., and Streets A. M., J. Micromech. Microeng. 29, 107001 (2019). 10.1088/1361-6439/ab341e [DOI] [Google Scholar]
- 23.Hirai Y., Inamoto Y., Sugano K., Tsuchiya T., and Tabata O., J. Micromech. Microeng. 17, 199 (2006). 10.1088/0960-1317/17/2/003 [DOI] [Google Scholar]
- 24.Hirai Y., Takagi D., Anai S., Chihara Y., Tsuchiya T., Fujimoto K., Hirao Y., and Tabata O., Sens. Actuators B 213, 547 (2015). 10.1016/j.snb.2015.01.118 [DOI] [Google Scholar]
- 25.Frost T. S., Jiang L., Lynch R. M., and Zohar Y., Micromachines 10, 533 (2019). 10.3390/mi10080533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lentle R. G. and Janssen P. W. M., J. Comp. Physiol. B 178, 673 (2008). 10.1007/s00360-008-0264-x [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa T., Sato T., Mohit G., Imai Y., and Yamaguchi T., J. Theor. Biol. 279, 63 (2011). 10.1016/j.jtbi.2011.03.026 [DOI] [PubMed] [Google Scholar]
- 28.Kim H. J., Huh D., Hamilton G., and Ingber D. E., Lab Chip 12, 2165 (2012). 10.1039/c2lc40074j [DOI] [PubMed] [Google Scholar]
- 29.Delon L. C., Guo Z., Oszmiana A., Chien C.-C., Gibson R., Prestidge C., and Thierry B., Biomaterials 225, 119521 (2019). 10.1016/j.biomaterials.2019.119521 [DOI] [PubMed] [Google Scholar]
- 30.Sambuy Y., De Angelis I., Ranaldi G., Scarino M. L., Stammati A., and Zucco F., Cell Biol. Toxicol. 21, 1 (2005). 10.1007/s10565-005-0085-6 [DOI] [PubMed] [Google Scholar]
- 31.Lasli S., Kim H. J., Lee K. J., Suurmond C. A. E., Goudie M., Bandaru P., Sun W., Zhang S., Zhang N., Ahadian S., Dokmeci M. R., Lee J., and Khademhosseini A., Adv. Biosyst. 3, e1900104 (2019). 10.1002/adbi.201900104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wevers N. R., Van Vught R., Wilschut K. J., Nicolas A., Chiang C., Lanz H. L., Trietsch S. J., Joore J., and Vulto P., Sci. Rep. 6, 38856 (2016). 10.1038/srep38856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S. Y., Kim D., Lee S. H., and Sung J. H., APL Bioeng. 5, 041505 (2021). 10.1063/5.0061896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loftus E. V., Harewood G. C., Loftus C. G., Tremaine W. J., Harmsen W. S., Zinsmeister A. R., Jewell D. A., and Sandborn W. J., Gut 54, 91 (2005). 10.1136/gut.2004.046615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X.-X., Huang X.-L., Chen R.-R., Li T., Ye H.-J., Xie W., Huang Z.-M., and Cao G.-Z., Inflammation 42, 2215 (2019). 10.1007/s10753-019-01085-z [DOI] [PubMed] [Google Scholar]
- 36.Fort M. M., Mozaffarian A., Stöver A. G., Correia J. d. S., Johnson D. A., Crane R. T., Ulevitch R. J., Persing D. H., Bielefeldt-Ohmann H., Probst P., Jeffery E., Fling S. P., and Hershberg R. M., J. Immunol. 174, 6416 (2005). 10.4049/jimmunol.174.10.6416 [DOI] [PubMed] [Google Scholar]
- 37.Collins S. L. and Patterson A. D., Acta Pharm. Sin. B 10, 19 (2020). 10.1016/j.apsb.2019.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohtani N. and Kawada N., Hepatol. Commun. 3, 456 (2019). 10.1002/hep4.1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman S. L., Neuschwander-Tetri B. A., Rinella M., and Sanyal A. J., Nat. Med. 24, 908 (2018). 10.1038/s41591-018-0104-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller P. A., Schneeberger M., Matheis F., Wang P., Kerner Z., Ilanges A., Pellegrino K., del Mármol J., Castro T. B. R., Furuichi M., Perkins M., Han W., Rao A., Pickard A. J., Cross J. R., Honda K., de Araujo I., and Mucida D., Nature 583, 441 (2020). 10.1038/s41586-020-2474-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schluter J., Peled J. U., Taylor B. P., Markey K. A., Smith M., Taur Y., Niehus R., Staffas A., Dai A., Fontana E., Amoretti L. A., Wright R. J., Morjaria S., Fenelus M., Pessin M. S., Chao N. J., Lew M., Bohannon L., Bush A., Sung A. D., Hohl T. M., Perales M.-A., van den Brink M. R. M., and Xavier J. B., Nature 588, 303 (2020). 10.1038/s41586-020-2971-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soucy J. R., Bindas A. J., Koppes A. N., and Koppes R. A., IScience 21, 521 (2019). 10.1016/j.isci.2019.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyazaki T., Hirai Y., Kamei K., Tsuchiya T., and Tabata O., Electron. Commun. Jpn. 104, 285 (2021). 10.1002/ecj.12296 [DOI] [Google Scholar]
- 44.Henry O. Y. F., Villenave R., Cronce M. J., Leineweber W. D., Benz M. A., and Ingber D. E., Lab Chip 17, 2264 (2017). 10.1039/C7LC00155J [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See the supplementary material (Fig. S1) for the mask pattern design of GLA-MPS. Figure S2 shows the microfabrication of the porous PDMS membrane. Figure S3 shows the single-side coating of Matrigel on the porous membrane in cell-culture chambers to prevent cell migration across the pores of the porous membrane by applying pressure barriers to the capillary. Figure S4 shows the ELISA testing results to measure the secreted TNF-α. Tables S1–S4 show the Tukey's multiple comparison test for ZO-1 of Caco-2, ALB of HepG2, iNOS of Caco-2, and iNOS of HepG2.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.