Abstract
Medical institutions often revoke data access due to the privacy concern of patients. Federated Learning (FL) is a collaborative learning paradigm that can generate an unbiased global model based on collecting updates from local models trained by client’s data while keeping the local data private. This study aims to address the centralized data collection issue through the application of FL on brain tumor identification from MRI images. At first, several CNN models were trained using the MRI data and the best three performing CNN models were selected to form different variants of ensemble classifiers. Afterward, the FL model was constructed using the ensemble architecture. It was trained using model weights from the local model without sharing the client’s data (MRI images) using the FL approach. Experimental results show only a slight decline in the performance of the FL approach as it achieved 91.05% accuracy compared to the 96.68% accuracy of the base ensemble model. Additionally, same approach was taken for another slightly larger dataset to prove the scalability of the method. This study shows that the FL approach can achieve privacy-protected tumor classification from MRI images without compromising much accuracy compared to the traditional deep learning approach.
Keywords: MRI, Brain cancer, CNN ensemble, Deep learning, Voting ensemble, Federated learning
Introduction
Deep learning (DL) approaches to classify medical images usually require an extensive database, including the complete spectrum of feasible anatomies, pathologies, and image scans [1]. However, regulatory bodies such as GDPR (general data protection regulation) and HIPAA (Health insurance portability and accountability act) have legislated new laws to conserve user privacy while sharing data. Therefore, it has become challenging to develop extensive data collection since it usually comes alongside private metadata such as patient name, health information, or birth date [2]. We can use Federated Learning (FL) environment to address this privacy issue where each data controller is trained collaboratively on local data. At every communication round, clients such as mobile, edge devices, industries, and data centers assign updated model weights to a central server for aggregation while keeping local training data private.
A brain tumor is an unusual growth of tissues of the brain caused by genetic mutations [3]. Because of brain tumors, symptoms like headache, irregular vision, seizure, memory loss, personality changes, anxiety, depression, etc., can occur [4] depending on the location, type, and stage of the tumor. There are mainly two types of brain tumors [5]: One is the benign tumor, which is local and yet not cancerous. Another one is the malignant tumor, where cancerous cells are found and tend to metastasize, i.e., spread from one location to the other parts [6, 7]. Doctors or medical experts can manually examine tumors by observing the Magnetic Resonance Images (MRI), even though it is a time-consuming procedure. From the perspective of the patients, manual diagnosis is expensive, and expert doctors are often hard to come by, making it even difficult to make a timely diagnosis and early management. Meanwhile, many researchers have had been successful in making faster detection and recognition of brain tumors using different image processing and machine learning algorithms [8, 9]. Early detection of many lethal brain tumors like Meningioma, Glioblastoma, and Astrocytoma often increases the chance of patient survival as they can be treated efficiently at an early stage [10].
Various degrees of research and experiments have already been performed on brain tumor detection through the applications of machine learning algorithms [11–19]. Subashini et al. [14] proposed a model for detecting the grade of a tumor. In their method, the noise was removed from the image using a pulse-coupled neural network (PCNN) at the beginning. Then for segmentation, Fuzzy C-means was used to extract the features. Afterward, a naïve bayes classifier was used to classify the features and 91% accuracy was achieved in classification. Gaikwad et al. [15] classified tumors into normal, benign, and malignant classes. Principal component analysis (PCA) was initially applied to extract features, and classification was done by employing a probabilistic neural network. They used 70 samples to train their algorithm and tested it on 35 with an accuracy of 97.14%. Bahadure et al. [16] performed brain tumor detection using adaptive contrast enhancement based on the modified Sigmoid function to pre-process the magnetic resonance images (MRI) and skull stripping process to exclude non-brain tissues from the MRI image by combining thresholding with binary masking. Next, the pre-processed image was segmented by using a threshold again. Furthermore, the resulting binary image was processed using erosion so those white pixels could be removed that did not belong to the infected region. The result was considered to be a mask on the original image. Finally, they computed the gray level co-occurrence matrix and applied the support vector machine (SVM) to classify the extracted features with an accuracy of 96.51%.
All the studies mentioned so far were done using conventional ML, where there is a central server or a computing device that manages both data storage and training models. Due to this, all the data are needed to be collected in a central place before training. Aledhari et al. [11] discussed that the conventional machine learning techniques requires a model to learn from a vast amount of training samples, which is sometimes very difficult to collect due to privacy issues. But in FL environment, models get trained separately in local devices and the trained weights are transferred back to the main server for aggregation. Therefore, the central server does not receive any data other than the attributes of models such as parameters, gradients, weights, etc. Zhang et al. demonstrated that FL solves the issue of collecting medical datasets and highlighted the difficulties that are required to be attended [12]. In their research, they applied a slightly modified FL model to detect COVID-19 from medical diagnostic images with improved performance. The presented method was to determine the best models based on their performance, and the models aggregated automatically according to data training time.
In this proposed study, the FL technique was used for brain tumor identification while protecting data privacy. Firstly, slightly modified CNN architectures leveraging transfer learning were built to construct distinctive ensembles that can accurately detect a tumor. Afterward, the ensemble model was used to create an FL environment for tumor detection and classification. The contribution of the proposed model is to show that the usage of complex models in the federated environment does not sacrifice the results much. Therefore, in the medical domain, even if there are privacy issues in data collection, we can quickly develop robust classifiers using the FL approach. To best our knowledge, this is the only study that develops an FL environment based on a complex ensemble model incorporating transfer learning for identifying brain tumors using MRI images. The graphical abstract of our proposed study is depicted in Fig. 1.
Fig. 1.
The graphical abstraction of the proposed study to detect brain tumors from the MRI dataset. a The labeled dataset. b Six pre-trained CNN models named VGG16, Inception V3, VGG19, ResNet50, Xception & DenseNet121. c1 Made average CNN model with VGG16, Inception V3, DenseNet121 as they produced best results to detect brain tumor. c2 Calculated voting ensemble by using correct prediction from VGG16, Inception V3, DenseNet121. d Constructed a global model with average CNN model in FL as it performed best, deployed it to the local devices, sent back the updated weights, and finally, checked the performance for tumor detection with test data
The rest of the paper is organized as follows. Section 2 includes our proposed method. Section 3 is dedicated to results and analysis of the following approaches. Section 4 concludes the paper.
Proposed Method
The research method followed three main steps for the detection of brain tumor, (1) Data pre-processing for converting the images from NIfTI format to PNG format, labeling the tumor and separating them into the train, and validation sets, (2) Model training and analysis of results with CNN model architectures, making a model with the average of the best three CNN models and voting ensemble to find the best accuracy, (3) Application of federated learning by creating a central server and client site. The pre-processed data were used to train the model, and the test set was used to test the accuracy of CNN model architectures. Then, the result was compared with the average model’s performance and voting ensemble approach for selecting the global model in the federated method. The processes of the proposed model are shown in Fig. 2.
Fig. 2.
The attempts followed in the proposed method for the detection of brain tumors
Data Description
The data was collected from the UK Data Service [20, 21] where 22 brain tumor patients were available. All the data were from the MRI images for surgical planning for brain tumor treatment and were in the shape of in-plane resolutions. There was information on the patients’ clinical data with unique MRI ID, treatment date, and necessary notes. The metadata contained pathological findings such as the location of the tumor, grey/white/CSF estimates, and tumor volumes. In the “Clinical Data,” there was a list of patient demographics, data on handedness, and various clinical test scores. Some sample of prepared data is presented in Fig. 3.
Fig. 3.
Sample images of brain tumor dataset (Axial T2 and Coronal slices)
Data Pre-processing
Data pre-processing is a machine learning technique that transforms raw data into an understandable or desired form [22]. Some factors were used for processing the MRI dataset. These factors are given below:
Image Extraction
Every file was in the 3D NIfTI (Neuroimaging Informatics Technology Initiative) format, which was used in neuroradiology research. In an RGB image, the pixel range was 0 to 255, but the collected 3D pictures had voxels meaning that they did not have any particular scale. So, 2D-shaped slices were extracted from the 3D file before doing further pre-processing. A library called ‘nibabel’ was used for the extraction that broke the 3D images into different slices and saved them into the desired path in PNG format [23, 24].
Data Labelling and Segmentation
After converting into 2D images, they were labeled as “Yes_Tumor” and “No_Tumor” according to the presence or absence of the tumor. Two sample pictures containing tumors and no tumors are displayed in Fig. 4.
Fig. 4.
Sample images containing no tumor & having a tumor
There was a total number of 2309 Axial T2 and Coronal images in the dataset. 70% of data was used for training and 30% of data was used for validation [25, 26] which means 1616 images were training data, 693 images were validation data. Putting a large portion of data into training helps the CNN models to overcome overfitting. A brief introduction is shown in Table 1.
Table 1.
A brief introduction to the MRI dataset
| Class | Training data | Validation data |
|---|---|---|
| No_Tumor | 1062 | 455 |
| Yes_Tumor | 554 | 238 |
Afterward, a normalization process was applied to rescale the images using the min-max scaling method, and the pixel values of the pictures ended up ranging between 0 and 1. The formula is:
| 2.1 |
Here, , is the ith normalized data, min(x) minimum value in the MRI dataset, and max(x) maximum value in MRI dataset.
CNN Architectures
In our proposed system, we imported VGG16, VGG19, Inception V3, ResNet50, DenseNet121, and Xception CNN models [27] using TensorFlow and Keras libraries [28, 29] . These models were already pre-trained on more than a million images from the ImageNet database [30], and we leveraged those previously learned weights to achieve faster training convergence. While the convolution layers were kept the same as the default architecture, the fully connected layers were dropped from the tail end of the model. These were designed to classify millions of classes that were not needed in our research. Next, the Max pooling layers were added to reduce the size of the feature maps, and all the pooled feature maps were flattened, which converted the 3D volume into a linear array. The output was passed to a dense layer consisting of 1024 neurons, and a 50% dropout layer was added after flatten layer, which prevented overfitting by randomly removing half of the neuron connection from our model during each training iteration [31]. For the dense layers, ReLU activation function was used and for the output layer, softmax activation function was used. We also used adam optimizer for the model and a learning rate of .001 was used. Through utilizing grid search, the CNN models were experimented with by examining every possible combination of the activation function, loss function, optimizer, and several neurons in a dense layer to achieve the best result. Figure 5 shows the general layer plot of the CNN models, and Figure 6 illustrates the graphical architecture of the proposed CNN model.
Fig. 5.

General layer plot of the CNN models
Fig. 6.
Graphical representation of CNN architecture to detect brain tumors
Model Training
The suggested models were trained by the labeled data, which were classified into two different classes. The “Yes_Tumor” class had 554 MRI slices with tumors, while the “No_Tumor” class had 1062 slices without any tumors. The model was built using Keras and Tensorflow 2.0 and used RTX 2080Ti as GPU for this experimental setup. The segmented images with a filter helped the models to learn key features of the tumor. The actual shape of the slices was , but it was resized into to train our model to leverage transfer learning using the default architecture. CNN model works better with a balanced dataset, but there were more images in the ‘No_Tumour’ class than in the ‘Yes_Tumor’ class in the dataset. Therefore, during training, more weights were given to each image in the ‘Yes_Tumor’ class. After the calculation, 1.46 weight was received for the ‘Yes_Tumor’ class and 0.76 for the ‘No_Tumor’ class. The equation of setting weights is shown in (2.2). Ten epochs were used for model training, the batch size was 16, and the step size was 101, which was obtained by dividing the total loaded data by the batch size. For further experiments, all the models were saved individually, right after the model training was done. Here, the and define the weight of the ‘Yes_Tumor’ class and ‘No_Tumour’ class accordingly. Basically, and here help us to understand the balance of the class distribution.
| 2.2 |
Data Augmentation
Data augmentation is a process that artificially generates more images while the primary input data remain the same [32]. It produces more variations of the training data that make the model training more robust and complete. The data augmentation parameters of our suggested method are available in Table 2 and some samples are displayed in Figure 7.
Table 2.
Data augmentation parameters of our research
| Augmentation Name | Value | Direction |
|---|---|---|
| Rotation range | 30 | Clockwise/Anti-clockwise |
| Width shift range | 0.1 | Left/Right |
| Height shift range | 0.1 | Top/Bottom |
| Shear range | 0.01 | Top/Bottom |
| X-axis zoom range | 0.9 | X-axis |
| Y-axis zoom range | 1.25 | Y-axis |
| Horizontal flip | True | Left/Right |
| Vertical flip | False | Top/Bottom |
Fig. 7.
Diagram of sample augmentation result of MRI brain tumor images
Model Validation
The 30% of data (693) were kept in the validation set for testing the models’ performance. While training the models, it was needed to estimate how well our model was learning per each iteration. So, from the unseen data, the accuracy was calculated by our experimented models to extract features for brain tumor detection using the previous learning experience. Based on the accuracy, the performance of the models was determined. If validation was not used, then there would be a high chance of the received result being biased after training.
Making Average CNN Model
The best three models were taken based on comparing the earlier specified parameters, and an average CNN model was designed. The same components (optimizer, loss function, number of epochs) were applied as previously mentioned for single CNN models to be built. The single model sometimes produces biased decisions and may cause an overfitting problem. The average CNN model uses the grouped weights to identify any object and this is how it overcomes these obstacles. The model architecture of the average CNN model is depicted in Fig. 8.
Fig. 8.

Descriptive architecture of the average ensemble
Note that the average CNN model was made with DenseNet121, VGG19, and Inception V3, the best three CNN models in our first experiment. The performance evaluation of the six CNN models is described in detail in the next chapter.
Voting Ensemble
At a different portion of the stated work, the previously picked best three CNN models were applied individually for the voting ensemble method. The iteration was done over each MRI scan of the validation dataset. The openCV library was used to make an image array to load them, and the image dimension was expanded. Rescaling image arrays divided by 255, which resulted in 0s and 1s, were used for training. Next, the deep CNN models were trained, and single prediction results were collected from each scaled image array. These were the binary value of “1” and “0,” where “1” indicated the tumor was present, and “0” implied no tumor was present. The theory was to count the majority votes from these three models. This meant that when we received two or more “1”s from the CNN models, it was considered as the tumor was present, and when two or more “0”s were present, it indicated no tumor. Note that the Keras library has a class attribute named ‘class_indices’ where the labeling of class indices (No_Tumor: 0, Yes_Tumor: 1) can be generated directly.
Federated Learning
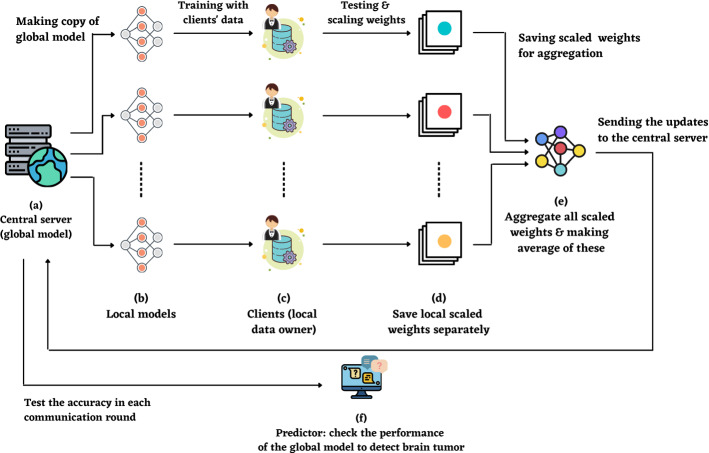
Federated Learning (FL) is a recently proposed machine learning technique, which tries to address the problem of strict regulations regarding data secrecy and limited supply of available data sets [11, 33, 34]. In federated learning, there are two ends; one is a central server, also known as a global server and another one is a client-end or local server. The central server has a global model, and the client-server contains local data (hold by clients on end devices). FL allows training a model without transferring the information, and the model gets trained on various decentralized end devices, which have data located in the local servers. It receives the updated weights from the local model trained by the client dataset.Therefore, it maintains the privacy that is very much required in medical data diagnosis [12, 35, 36]. The comparison was made between all individual CNN architectures, the average CNN model, voting ensemble, and the best of them was selected to build a global model in the central server. No previously trained CNN model was used here because the purpose of FL is that the global model can not be attached to any training data. Afterward, fifty copies of local models were created with the same weight as the global model. 50 clients were also created and located in the client-server, and all training data was distributed to them. Further, all the 50 local models were fitted with the 50 clients, and weights were scaled, averaged, and aggregated into an array. Finally, the aggregated weight (local models’ update) was sent back to the global model and tested to detect brain tumors using the validation data. Figure 9 exhibits the architecture of our FL model.
Fig. 9.
The model architecture of federated learning. a Global model (in central server) implemented with average CNN model. b 50 local models made as a copy of the global model (same weight). c 50 clients built with local data. d Training & testing with clients’ data, scaling the weights, and storing these into a variable. e Aggregating all different weights and making average. Last, sending the updated weight to the central server. The iteration went through 5 communication rounds. f For each communication round, the test accuracy of the global model is checked
Note that it was found that the average model achieved the best result compared to every other experimented method. Therefore, the global model in the FL algorithm was made with the average CNN model. The explanation of all the experiments is discussed in the next section.
Experimental Result Analysis
The following section includes our implementation’s final results and how good our models perform to detect brain tumors. There are presented precision, recall, F1 score, confusion matrix, accuracy, loss function values for each attempted model. Precision and recall were applied to validate the performance of classification, and the F1 score was used to examine as a single numerical analysis of a system’s completion [37]. The formulas are shown in Sects. 3.3, 3.4, and 3.5. The training accuracy is also called categorical accuracy. It means how well models are classifying the training dataset [38]. The model loss function is one of the most critical components of deep neural network [39]. It indicates how far the models are from the actual result. With the values of model loss, it can be determined that how bad CNN models are performing while predicting tumors from dataset [40]. The test accuracy is to evaluate the performance of models [41]. After training, the best performing CNN architectures were finalized based on the result values. Our primary aim was to get a higher test accuracy and a lower model loss function [42, 43]. All models were run for ten epochs with Adam as an optimizer with a learning rate of 0.00001. In the end, our work was summarised by providing a comparison with the other previously published states of art methods and clinical evaluations.
| 3.3 |
| 3.4 |
| 3.5 |
Performance Evaluation of 6 CNN Models
It has been discussed previously that six pre-trained CNN models: VGG16, VGG19, Inception V3, DenseNet121, ResNet50, and Xception were selected as our recommended model by removing the top and adding our unique layer combination. The classification report, confusion matrix, train, test, and model loss were presented to evaluate these models’ achievement. Based on these performances, the best three CNN models were picked for further experiments.
Classification Report of Six CNN Models
The precision, recall, and F1 score of our stated six CNN models are depicted in Table 3. The X-axis expresses the predicted labels (model output), while the Y-axis represents the actual labels (ground truth). All the models offered excellent results. After checking the precision value of No_Tumor class, the VGG19 model outperforms most of the other models as it received a 0.97 precision value, which means the model could predict the largest number of no tumor predictions that actually belong to the No_Tumor class. The VGG16 model predicts best for Yes_Tumor class as it obtained the highest precision value (0.95) for corrected yes tumor prediction associated with that class. The recall value of VGG16 is 0.98 in No_Tumor class means, from all No_Tumor examples in our MRI dataset, it made the most significant contribution in correctly predicting no tumor. To reduce the complexity of picking the best three CNN models, the F1 score was analyzed that considered precision and recall. After observing the tables, it becomes obvious that the best result came from VGG19 (0.95, 0.91), DenseNet121 (0.95, 0.91), and Inception V3 (0.95, 0.90) models as they scored the highest F1 score compare to other CNN models.
Table 3.
Classification report of six CNN models
| Class | VGG16 | VGG19 | Inception V3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | F1 score | Precision | Recall | F1 score | Precision | Recall | F1 score | |
| No_Tumor | 0.91 | 0.98 | 0.94 | 0.97 | 0.93 | 0.95 | 0.94 | 0.95 | 0.95 |
| Yes_Tumor | 0.95 | 0.82 | 0.88 | 0.87 | 0.95 | 0.91 | 0.91 | 0.89 | 0.90 |
| Class | DenseNet121 | ResNet50 | Xception | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Precision | Recall | F1 score | Precision | Recall | F1 score | Precision | Recall | F1 score | |
| No_Tumor | 0.94 | 0.97 | 0.95 | 0.93 | 0.94 | 0.94 | 0.90 | 0.97 | 0.94 |
| Yes_Tumor | 0.94 | 0.88 | 0.91 | 0.89 | 0.87 | 0.88 | 0.94 | 0.80 | 0.86 |
Confusion Matrix of Six CNN Models
The confusion matrix was plotted for the six CNN models to examine the models for each shuffle in Fig. 10. It shows all the models’ performances were very close to each other. In the No_Tumor class, the largest number of correct predictions received from the VGG16 model with 445 and VGG19 with 227 predicted Yes_Tumor sample, which was the best compared to other CNN models in that class. As it was needed to validate the performance of both classes in combination for selecting the best three models, the highest corrected prediction was achieved by the DenseNet121 model (650), VGG19 model (649), and Inception V3 model (645).
Fig. 10.
Confusion matrix of six CNN models
Training Accuracy, Model Loss and Test Accuracy of Six CNN Models
The comparison of training accuracy, model loss function, and test accuracy are sketched in Fig. 11 to explain the better performance of six CNN models. In the training accuracy graph, the comparison of training accuracy is shown between all of the 6 CNN architectures. Here, the lines of CNN models are going upward in every epoch, which indicated that the models were learning at a great rate on the training data. The blue line is marked as VGG19, which obtained the highest 97.38% training accuracy. After that, DenseNet121 and Inception V3 models gained 96.95% and 96.72% training accuracy, respectively. The comparison of CNN models’ loss functions is plotted in the middle one. DenseNet121 model achieved the lowest model loss value, which is 4.95%, VGG19 and VGG16 models came after this with a percentage of 5.01% and 9.27%. Thus, for learning the training dataset, the DenseNet121 model performed better than other CNN model architectures. However, every CNN model started from a high level but ended up at a lower value. The more time the models got trained, the lower loss functions was acquired [39]. The last figure exhibits the comparison between 6 CNN architectures for test accuracy. The graph is going up in each epoch for every CNN model indicating that our suggested model performed very well for detecting brain tumors. However, the Xception model, labeled as brown color, spiked up and down continuously determining that the model was not well fitted with the MRI dataset. The DenseNet121 model, which is marked as deep blue color, reached the highest 93.80% test accuracy. The other two best models are the VGG19 model (93.65%) and the Inception V3 model (93.07%). These figures demonstrate that our models were trained on the MRI dataset accurately. The models were not either under-fit or over-fit and detected tumors from the test dataset perfectly. These are the reasons why the models produced an excellent result in the classification report and confusion matrix.
Fig. 11.
Comparison of train, loss and test accuracies between CNN architectures
Performance Evaluation of Ensemble Methods
After evaluating the performance based on the classification report, confusion matrix, training accuracy, model loss, and test accuracy, DenseNet121, VGG19, and Inception V3 were selected as the best three models to detect brain tumors from MRI scans. An average CNN model and voting ensemble method were built with these models because sometimes a single or individual model may produce biased results. When the ensemble method was applied, several models’ combined weight worked together and generated a better result. In this sub-section, at first, the performance of the average CNN model is demonstrated based on three previously mentioned criteria. Then the experiment of the voting ensemble is explained.
Classification Report of Average CNN Model
The precision, recall, F1 score of the average CNN model is displayed in Table 4. Both No_Tumor and Yes_Tumor classes got 0.97 precision value. In recall, the model received 0.98 value for No_Tumor class and 0.94 value for Yes_Tumor class. These are outstanding results and were obtained from separate models previously, but in this average model, the highest value of precision and recall are attained together. The F1 score is 0.97 and 0.95 for the two classes, which is the best result in our experiment till now.
Table 4.
Classification report of average CNN model
| Class | Precision | Recall | F1 score |
|---|---|---|---|
| No_Tumor | 0.97 | 0.98 | 0.97 |
| Yes_Tumor | 0.97 | 0.94 | 0.95 |
Confusion Matrix of Average CNN Model
Figure 12 represents the confusion matrix of average CNN model. The total number of correct predictions is 670 (No_Tumor: 447, Yes_Tumor: 223), the biggest value for our experiment of brain tumor detection. This reflects a significant aspect of the applicability of the proposed model as it operates well on a diverse range of loads.
Fig. 12.

Confusion matrix of average CNN model
Training and Test Accuracy of Average CNN Model
The training and test accuracy of the average CNN model graph is plotted on Fig. 13. This graph explains the model has trained accurately on the MRI dataset. As well as this model achieves the best training and test accuracy, which are 98.2% and 96.68% sequentially. The model summary is exhibited in Table 5. Non-trainable parameters are the number of weights that remain constant while training the models. The ensemble’s improved results show that the combined architectures perform better for MRI classification tasks when a large volume of data is fed and transfer learning is utilized.
Fig. 13.

Training and test accuracy of average CNN model
Table 5.
Average CNN model’s summary
| Layer (type) | Output Shape | Parameter | Connected to |
|---|---|---|---|
| input_1 (Input Layer) | (None, 224, 224, 3) | 0 | |
| DenseNet121 (Sequential) | (None, 2) | 58416706 | input_1[0][0] |
| VGG19 (Sequential) | (None, 2) | 45717570 | input_1[0][0] |
| Inception_V3 (Sequential) | (None, 2) | 74234658 | input_1[0][0] |
| average (Average) | (None, 2) | 0 | DenseNet121[0][0], VGG19[0][0], Inception_V3[0][0] |
Experiment of Voting Ensemble
The voting ensemble is a unique work in our research. The result depends on the vote of the best three CNN model architectures. The best three individual CNN models: DenseNet121, VGG19, and Inception V3, were chosen from the previous results. After training these models with the same parameters discussed earlier, the normalized value of the test/validation dataset (0 & 1) was collected. Some build-in python libraries were applied for that, such as OpenCV, OS, and a class attribute called ‘class_indices,’ which helped us to generate the label of No_Tumor (0) and Yes_Tumor (1) automatically. After collecting the binary values of all test data, these were stored in an excel sheet, and the voting ensemble was calculated. The criteria were when at least two of the models were giving a “1” (yes) vote; the result was considered as “1” (yes); otherwise, “0” (no). It reduces model variance for detecting the brain tumor because our output was set based on the majority of votes of the best CNN models [40]. Some arbitrary results and the calculation of the voting ensemble method from our research are explained in Table 6. It is depicted that the accuracy of the individual CNN models’ results has changed a little bit from the previously achieved results (650, 649, 645). It is a widespread scenario that the accuracy does not remain the same in every training. However, the voting ensemble result is slightly lower than the average CNN model. The average CNN model uses the combined weight of the three trained models, but the calculation depends on the votes of the individual CNN models only.
Table 6.
Explanation of some voting ensemble calculation
| DenseNet121 | VGG19 | Inception V3 | Voting ensemble |
|---|---|---|---|
| Counting majority of votes for “containing brain tumor” | |||
| 1 | 1 | 0 | 1 |
| 1 | 0 | 0 | 0 |
| 1 | 0 | 1 | 1 |
| 1 | 1 | 1 | 1 |
| 0 | 0 | 1 | 0 |
| 0 | 1 | 1 | 1 |
| ... | ... | ... | ... |
| ... | ... | ... | ... |
| Counting majority of votes for “containing no brain tumor” | |||
| 0 | 1 | 0 | 0 |
| 0 | 0 | 1 | 0 |
| 1 | 1 | 0 | 1 |
| 0 | 0 | 0 | 0 |
| 1 | 0 | 1 | 1 |
| 1 | 0 | 0 | 0 |
| ... | ... | ... | ... |
| ... | ... | ... | ... |
| Total corrected prediction | |||
| 652 | 647 | 642 | 665 |
| Total test data | |||
| 693 | 693 | 693 | 693 |
| Accuracy in percentage (%) | |||
| (652693)100 | (647693)100 | (642693)100 | (665693)100 |
| = 94.08% | = 93.36% | = 92.64% | = 95.96% |
Performance Evaluation of Federated Learning
After the previous comparison, it was observed that the average CNN model performs the best. Therefore, the average CNN model architecture was replicated to create the global model for the FL environment. Additionally, 50 local models were constructed using the global model’s weight. Fifty clients were built to gather all the training and validation/test data, all the local models were distributed and trained within them, and the local models’ accuracy was calculated. The data were not distributed to the clients equally; for example, there were 1616 training datasets, and as these were divided among 50 clients, some of the clients would get more than 32 (1616/50) images while some would get less. A scaling factor was created to reduce this bias, and the equation is shown in (3.6).
| 3.6 |
As the number of images per client can be estimated through this process, it reveals only a little bit of data information which is a drawback of the proposed model. As no data was transferred to the global model, the main principle of federated learning had been accomplished here. All of the individual local models’ weight was scaled by multiplying each of them with the scaling factor like (3.7) and all were stored into a variable.
| 3.7 |
Thus, the client who had more data received more weight. Note that, unlike the previously mentioned weight for "Yes_Tumor" & "No_Tumor", the scaled weights were applied in the local models here. Furthermore, all the scaled weights were aggregated by saving them in a list, and an average was measured. Lastly, this weight was transferred to the central server, and the previous weights of the global model got updated with these new weights. The iteration went through 5 communication rounds and tested the global model’s accuracy each time with the test dataset. The FL performance is expected to be high as the global model receives an update that is an aggregated average of 50 other clients. Every device can take benefits from other’s experiences, so the learning can be shared across all users. The three criteria of evaluating FL model for detecting brain tumor is demonstrated below:
Classification Report of Federated Learning
To analyze the performance of the suggested FL model, the calculation of precision, recall, and F1 score are shown in Table 7. The experimental results show that the FL performs 0.93 F1 scores for No_Tumor and 0.86 F1 scores for Yes_Tumor. After observing the table, this could be found that the other scores of precision and recall were lower than previously described models. One reason for this might be that FL used decentralized data and all of the data remained to the clients generating different predictions. Although the weights were aggregated, the wrong prediction from some local models probably pulled the accuracy down.
Table 7.
Classification report of federated learning model
| Class | Precision | Recall | F1 score |
|---|---|---|---|
| No_Tumor | 0.91 | 0.96 | 0.93 |
| Yes_Tumor | 0.91 | 0.82 | 0.86 |
Confusion Matrix of Federated Learning
The federated learning’s confusion matrix is illustrated in Figure 14 to estimate the model for each shuffle. It misclassified the highest number of Yes_Tumor sample (43) as negative which may cost the patients severely causing treatment delays. The total number of corrected predictions is 631 (No_Tumor: 447 & Yes_Tumor: 223), reflecting a worse performance in detecting brain tumors. It also indicates a possible performance drop while aggregating the weights. The drop in performance for FL is expected, as discussed in later sections.
Fig. 14.

Confusion matrix of federated learning
Training and Test Accuracy of Federated Learning
To validate the FL model’s model efficiency, the training and test accuracy was measured which is plotted in Fig. 15. The model is excellently trained with the MRI dataset, and it is not over-fit or under-fit. At the last epoch, 91.05% test accuracy was attained, which is relatively low because of the decentralized data. It could not extract deeper features from the MRI dataset. The algorithm maintains the clients’ data privacy as the global model does not receive any clients’ data besides the updated weight of local models. It is projected to be beneficial to use in the medical imaging diagnoses like MRI, X-rays, CT scans, etc.
Fig. 15.

Training and test accuracies of federated learning
Although the evaluation results are a bit low for the FL environment compared to the non-FL one, it makes sense since FL is a completely decentralized learning system. Decent result under the FL environment is naturally harder to achieve since it learns across a variety of clients and that introduces several hurdles. First of all, the distributed data across clients is non-IID, some client has more data while some have less. Therefore, not all clients have an equal contribution to the learned parameters and this introduces some severe bias in the system. Although clients with more data were given a higher weight, the bias problem could not be addressed entirely since it is a relatively weak weighting approach. Additionally, such weighted learning approach may introduce a different paradigm of security issues since the system administrators can easily figure out the differences of data volume between all the clients. Thus, if the data volume information is sensitive, clients may get exposed due to the weighted learning system. Furthermore, the class distribution across different clients also varies. Some clients might have a majority of non-tumor images and vice versa. This uneven class distribution also adds additional challenges to the learning procedure. Considering all these hurdles, a drop of near 4.5% accuracy is a relatively decent result. The drop in performance might be worth it since the data privacy issues are handled and FL can introduce a higher availability of data if the global model is open sourced for learning from a public distribution of clients. Of course, open sourcing the global model will require extensive security measures to handle data poisoning.
Analyzing the Best Model for Brain Tumor Detection
Federated learning (FL) preserves the privacy of the data by not sharing local data with the global model, and it does not compromise much performance regarding accuracy compared to average CNN model. The Receiver Operator Characteristic (ROC) curve of the FL method is exhibited in Fig. 16. This graph shows an exceptional result by gradually increasing the curve, which summarizes all of the information. The Y-axis is the true positive rate (TPR), indicating the proportion of Yes_Tumor samples that were correctly classified. The X-axis is the false-negative rate (FNR), reflecting the proportion of No_Tumor slices that were incorrectly categorized. There was also measured Area Under the Curve (AUC), which is approximately 0.908. The 0.9 to 1.0 range of AUC is considered to be an outstanding value for any model. The sensitivity and specificity values of the FL model are respectively 0.910 and 0.909. Sensitivity is the true positive rate that calculates the proportion of Yes_Tumor data that are correctly recognized, and specificity is the true negative rate that measures the ratio of No_Tumor images that are correctly identified. The FL model’s Dice similarity coefficient (DSC) value measures the automatic and manual segmentation [44] of a brain tumor which is 0.906. The formulas are given below:
| 3.8 |
| 3.9 |
| 3.10 |
Fig. 16.

The ROC curve for brain tumor detection
In terms of model selection, we chose the best-performing model under the non-FL environment. Since the best performing model manages to classify tumors well, it is also expected to perform well under the FL environment. However, since data distribution wildly varies between a central versus distributed learning environment, choosing the best model might not be as straightforward. Ensemble neural networks are generally heavy models with lots of learnable parameters, and in our proposed method, a similar heavy model is distributed across clients with small chunks of data. Unless the FL environment is heavily regulated, there might be clients with as few as a couple of images. The large-scale ensemble model may introduce heavy overfitting across these smaller clients and, as a result, may affect the overall global performance. Weighing client contribution on the global learning based on data distribution obviously helps, but if there are many clients with tiny segments of data alongside a small number of clients with a larger volume of data, plenty of valuable data may get under-weighted. Therefore, there is room for further experimentation on the model selection. Based on the data distribution, a smaller Neural Network architecture for the FL environment might be more desirable in different use cases as it is less prone to overfitting and generally learns better from the small-scale data distribution. If there is any existing knowledge on the client distribution, it should be utilized to deploy the model architecture for the FL environment. In our proposed model, there were clients with a minimal amount of MRI images, but it did not hamper the learning of the ensemble model much. In general, as long as the number of very small clients is low, the smaller clients are weighted less, and there is a continuous stream of data from larger clients, the overfitted models from the smaller clients should get outweighed during the global averaging anyway. Therefore, the best-performing ensemble model was the obvious choice for us.
Ensemble Model and Federated Learning’s Results on an Extended Dataset
In order to verify the effectiveness of the procedure further, We have applied our ensemble, and federated learning on a different dataset [45]. This particular dataset is collected from Kaggle and is slightly larger than the previous dataset used, consisting of 3000 images, where 1500 are tumorous, and 1500 are non-tumorous. We divided it into a 70:30 ratio for training and validation sets. There was no third separate test set as we did not perform any hyperparameter optimization in this case, as we planned to keep the overall procedure exactly the same. The architecture and the setup of the ensemble model and the federated environment were kept the same as the initial setup applied for the previous dataset. On this extended dataset, the ensemble model achieved 95.89% validation accuracy, and federated learning(FL) reached 93.22% accuracy. The validation accuracy and result of the confusion matrix of the ensemble model are shown in Fig. 17 and FL in Fig. 18. We can also see that our models performed an excellent outcome in detecting brain tumors in this dataset set as well. This particular result shows that the proposed procedure scales well across other datasets too.
Fig. 17.
Extended dataset ensemble results
Fig. 18.
Extended dataset FL result
Comparison with Other States of the Art Methods
This sub-section includes a comparison of the previously published techniques for the MRI dataset with our suggested model. We showed the result of federated learning (FL) technique and the average CNN model in this comparison for classifying brain tumors. As depicted in Table 8, the different proposed methods are experimented with various parameters such as AUC, sensitivity, specificity, DSC, accuracy [46]. Note that only Afsara et al. [47] used the same MRI dataset as ours; other research worked on MRI datasets taken from different sources. Amin et al. [48] achieved a slightly higher result than our average CNN model, but the combination of various methods and calculated parameters we applied made our model unique. Approximately in all other cases, our model performed better. Afsara et al. showed in their methodology that 95.40% accuracy was achieved for binary brain tumor classification. They used three individual CNN architectures: VGG16, VGG19, and Inception V3. Our suggested approach shows that the average of CNN models and the federated learning method is more efficient and produces a better predictive result in brain tumor detection than using a single classifier or model.
Table 8.
Overall comparison among the proposed model and state-of-the-art methods on the MRI dataset
| Literature | AUC | Sensitivity | Specificity | DSC | Accuracy | Method used |
|---|---|---|---|---|---|---|
| Sudharani et al. [46] | N/A | 0.889 | 0.9 | N/A | 89.20% | Advanced morphological technique |
| Subashini et al. [14] | N/A | N/A | N/A | N/A | 91% | Naives Bayes |
| Abdel-Maksoud et al. [49] | N/A | N/A | N/A | N/A | 95.06% | K-means clustering, Fuzzy C-means |
| Nabizadeh and Kubat [50] | N/A | 0.77 | 0.81 | N/A | % | Statistical features |
| Amin et al. [48] | 0.98 | 0.92 | 0.98 | N/A | 97.1% | SVM classifier |
| Zhao and Jia [51] | N/A | N/A | N/A | N/A | 88% | CNN architecture |
| Zhao and Jia [38] | N/A | N/A | N/A | N/A | 90.0% | Multiscale CNNs |
| Li et al. [52] | N/A | 0.928 | 0.998 | 0.927 | N/A | Multi-CNNs |
| Yi et al. [53] | 0.997 | N/A | N/A | 0.785 | N/A | Inception V1, U-Net, and FL |
| Sheller et al. [54] | N/A | N/A | N/A | 0.852 | N/A | U-Net, and FL |
| Afsara et al. [47] | N/A | N/A | N/A | N/A | 95.40% | CNN architectures |
| Proposed (FL) | 0.908 | 0.910 | 0.909 | 0.906 | 91.05% | CNN, average CNN, voting ensemble and FL |
| Proposed (Average CNN) | 0.959 | 0.968 | 0.965 | 0.951 | 96.68% | CNN, average CNN, voting ensemble and FL |
Clinical Evaluation
The Federated learning (FL) model is prepared for use in the clinical area so that patients can be benefited by getting an early alert. Some MRI images were chosen randomly from the test dataset and given as input to the model to illustrate the performance of our attempted study. Figure 19 shows the correct results that the model produced. Again, our model did not obtain 100% accuracy. Among the given images for testing, there are a few images that the model predicted wrongly. One such example is depicted in Fig. 20. The picture clearly shows that there is a tumor, but our model predicted it as No_Tumor. However, the number of incorrect predictions by the model is comparatively low.
Fig. 19.
Some examples of correct brain tumor detection by the FL model from randomly chosen test images
Fig. 20.

An example of incorrect brain tumor detection by the FL model from a randomly chosen test image
In the end, it has been shown that our model performed greatly in identifying brain tumors from MRI images. This system detected brain tumors using four different techniques, and the combination of the techniques, that are discussed earlier, made this method unique. The dataset was split into 70:30, and models ran only for ten epochs. Even after that, the method achieved a better result than most of the previously published methods. Although the performance of the recommended approach is excellent, it is not ready to replace the traditional techniques of brain tumor detection yet. Sometimes the wrong predictions by the system may become very problematic for the patients. Nevertheless, it can help clinicians to make proper decisions regarding brain tumor detection.
Conclusion
It is hard to obtain medical diagnostic datasets for research purposes due to the privacy concern of the patients. For this reason, the data is usually insufficient to work with any ML models. This research has used CNN architecture, average CNN model, voting ensemble, and federated learning (FL) to solve these problems. The dataset contains Axial T2 and Coronal slices of MRI images. The method used six different types of CNN model architectures, which are VGG16, VGG19, Inception V3, ResNet50, DenseNet121, and Xception. These CNN architectures produced different results and accuracy while tested with the test dataset. A comparative analysis based on precision, recall, f1 score, train, model loss, test accuracy was determined, and the three best models: DenseNet121, VGG19, and Inception V3, were selected. An average model from these three models was designed, and the same parameters were measured. After that, the binary value of the test dataset was taken for the voting ensemble using previously trained CNN models, which represents “0” means “No tumor,” and “1” means the presence of a tumor. The test accuracy was calculated and compared with the average model for creating a global model for the next method. The FL technique was applied at the end, where a global model was made as a central server that receives updates from trained local models. The study showed average CNN model achieved better accuracy 96.68%, but FL preserves the privacy of data with 91.05% accuracy for detecting brain tumors. Furthermore, the whole procedure was applied on another dataset in order to prove the legitimacy of the method across different datasets.
Even though the current procedure and results look optimistic, there is still plenty of room for improvement. In the proposed paper, no measure was taken to address class distribution imbalance within the clients. The weighted average addresses the data distribution imbalance across the clients to some extent but can pose privacy issues in different use cases. Additionally, the datasets on which the experimental analysis was done could be larger. The ensemble model is a relatively large neural network architecture that may heavily overfit and degrade the global performance if many clients have tiny datasets. In the future, we aim to train the model with a more significant number of datasets. Image formatting remains a place for improvement to make the models more compatible to work with poor-quality images. We hope to use different feature extraction techniques, whereas to improve accuracy, more advanced algorithms of the FL aggregation method can be used. Furthermore, as genetic mutations cause brain cancers, we aim to work with DNA images in the future.
Footnotes
Md. Tanzim Reza, Mohammed Kaosar and Mohammad Zavid Parvez contributed equally to this work.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Moinul Islam, Email: moin.islamshawon@gmail.com.
Md. Tanzim Reza, Email: tanzim.reza@bracu.ac.bd.
Mohammed Kaosar, Email: mohammed.kaosar@murdoch.edu.au.
Mohammad Zavid Parvez, Email: zavid.parvez@acu.edu.au.
References
- 1.Rieke N, Hancox J, Li W, Milletari F, Roth HR, Albarqouni S, Bakas S, Galtier MN, Landman BA, Maier-Hein K, et al. The future of digital health with federated learning. NPJ Digit Med. 2020;3(1):1–7. doi: 10.1038/s41746-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grama M, Musat M, Muñoz-González L, Passerat-Palmbach J, Rueckert D, Alansary A (2020) Robust aggregation for adaptive privacy preserving federated learning in healthcare. arXiv:2009.08294
- 3.Bondy ML, Scheurer ME, Malmer B, Barnholtz-Sloan JS, Davis FG, Il’Yasova D, Kruchko C, McCarthy BJ, Rajaraman P, Schwartzbaum JA, et al. Brain tumor epidemiology: consensus from the brain tumor epidemiology consortium. Cancer. 2008;113(S7):1953–1968. doi: 10.1002/cncr.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong TS, Vera-Bolanos E, Acquaye AA, Gilbert MR, Ladha H, Mendoza T. The symptom burden of primary brain tumors: evidence for a core set of tumor-and treatment-related symptoms. Neuro Oncol. 2015;18(2):252–260. doi: 10.1093/neuonc/nov166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McFaline-Figueroa JR, Lee EQ. Brain tumors. Am J Med. 2018;131(8):874–882. doi: 10.1016/j.amjmed.2017.12.039. [DOI] [PubMed] [Google Scholar]
- 6.Kumar S, Dabas C, Godara S. Classification of brain mri tumor images: a hybrid approach. Procedia Comput Sci. 2017;122:510–517. doi: 10.1016/j.procs.2017.11.400. [DOI] [Google Scholar]
- 7.Bayen E, Laigle-Donadey F, Prouté M, Hoang-Xuan K, Joël M-E, Delattre J-Y. The multidimensional burden of informal caregivers in primary malignant brain tumor. Support Care Cancer. 2017;25(1):245–253. doi: 10.1007/s00520-016-3397-6. [DOI] [PubMed] [Google Scholar]
- 8.Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin P-M, Larochelle H. Brain tumor segmentation with deep neural networks. Med Image Anal. 2017;35:18–31. doi: 10.1016/j.media.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Devkota B, Alsadoon A, Prasad P, Singh A, Elchouemi A. Image segmentation for early stage brain tumor detection using mathematical morphological reconstruction. Procedia Comput Sci. 2018;125:115–123. doi: 10.1016/j.procs.2017.12.017. [DOI] [Google Scholar]
- 10.Seyfried TN, Flores R, Poff AM, D’Agostino DP, Mukherjee P. Metabolic therapy: a new paradigm for managing malignant brain cancer. Cancer Lett. 2015;356(2):289–300. doi: 10.1016/j.canlet.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Aledhari M, Razzak R, Parizi RM, Saeed F. Federated learning: a survey on enabling technologies, protocols, and applications. IEEE Access. 2020;8:140699–140725. doi: 10.1109/ACCESS.2020.3013541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, Zhou T, Lu Q, Wang X, Zhu C, Wang Z, Wang F (2020) Dynamic fusion based federated learning for covid-19 detection. arXiv:2009.10401 [DOI] [PMC free article] [PubMed]
- 13.Li Q, Wen Z, Wu Z, Hu S, Wang N, He B (2019) A survey on federated learning systems: vision, hype and reality for data privacy and protection. arXiv:1907.09693
- 14.Subashini MM, Sahoo SK, Sunil V, Easwaran S. A non-invasive methodology for the grade identification of astrocytoma using image processing and artificial intelligence techniques. Expert Syst Appl. 2016;43:186–196. doi: 10.1016/j.eswa.2015.08.036. [DOI] [Google Scholar]
- 15.Gaikwad SB, Joshi MS (2015) Brain tumor classification using principal component analysis and probabilistic neural network. Int J Comput Appl 120(3)
- 16.Bahadure NB, Ray AK, Thethi HP (2017) Image analysis for mri based brain tumor detection and feature extraction using biologically inspired bwt and svm. Int J Biomed Imaging [DOI] [PMC free article] [PubMed]
- 17.Zulpe N, Pawar V. Glcm textural features for brain tumor classification. Int J Comput Sci Issues (IJCSI) 2012;9(3):354. [Google Scholar]
- 18.Hanwat S, Chandra J. Convolutional neural network for brain tumor analysis using mri images. Int J Eng Technol (IJET) 2019;11:67–77. doi: 10.21817/ijet/2019/v11i1/191101022. [DOI] [Google Scholar]
- 19.Guo P, Wang P, Zhou J, Jiang S, Patel VM (2021) Multi-institutional collaborations for improving deep learning-based magnetic resonance image reconstruction using federated learning, pp 2423–2432 [DOI] [PMC free article] [PubMed]
- 20.Pernet C, Gorgolewski K, Ian W. A neuroimaging dataset of brain tumour patients. ReShare
- 21.Pernet CR, Gorgolewski KJ, Job D, Rodriguez D, Whittle I, Wardlaw J. A structural and functional magnetic resonance imaging dataset of brain tumour patients. Scientific Data. 2016;3(1):1–6. doi: 10.1038/sdata.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamiran F, Calders T. Data preprocessing techniques for classification without discrimination. Knowl Inf Syst. 2012;33(1):1–33. doi: 10.1007/s10115-011-0463-8. [DOI] [Google Scholar]
- 23.Singh G, Ansari M (2016) Efficient detection of brain tumor from mris using k-means segmentation and normalized histogram. IEEE, pp 1–6
- 24.Lareyre F, Adam C, Carrier M, Dommerc C, Mialhe C, Raffort J. A fully automated pipeline for mining abdominal aortic aneurysm using image segmentation. Sci Rep. 2019;9(1):1–14. doi: 10.1038/s41598-019-50251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabria I, Gondra I. Mri segmentation fusion for brain tumor detection. Inform Fusion. 2017;36:1–9. doi: 10.1016/j.inffus.2016.10.003. [DOI] [Google Scholar]
- 26.Prajapati SJ, Jadhav KR. Brain tumor detection by various image segmentation techniques with introduction to non negative matrix factorization. Brain. 2015;4(3):600–603. [Google Scholar]
- 27.Rehman A, Naz S, Razzak MI, Akram F, Imran M. A deep learning-based framework for automatic brain tumors classification using transfer learning. Circ Syst Signal Process. 2020;39(2):757–775. doi: 10.1007/s00034-019-01246-3. [DOI] [Google Scholar]
- 28.Sawant A, Bhandari M, Yadav R, Yele R, Bendale MS (2018) Brain cancer detection from mri: a machine learning approach (tensorflow). Brain 5(04)
- 29.Ketkar N (2017) Introduction to keras, 97–111
- 30.Naseer A, Rani M, Naz S, Razzak MI, Imran M, Xu G. Refining Parkinson’s neurological disorder identification through deep transfer learning. Neural Comput Appl. 2020;32(3):839–854. doi: 10.1007/s00521-019-04069-0. [DOI] [Google Scholar]
- 31.Hussain S, Anwar SM, Majid M. Segmentation of glioma tumors in brain using deep convolutional neural network. Neurocomputing. 2018;282:248–261. doi: 10.1016/j.neucom.2017.12.032. [DOI] [Google Scholar]
- 32.Cheng J, Huang W, Cao S, Yang R, Yang W, Yun Z, Wang Z, Feng Q. Enhanced performance of brain tumor classification via tumor region augmentation and partition. PLoS ONE. 2015;10(10):0140381. doi: 10.1371/journal.pone.0140381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, Lu Q, Yu Q, Li Z, Liu Y, Lo SK, Chen S, Xu X, Zhu L. Blockchain-based federated learning for device failure detection in industrial iot. IEEE Internet Things J. 2020;8(7):5926–5937. doi: 10.1109/JIOT.2020.3032544. [DOI] [Google Scholar]
- 34.Sarma KV, Harmon S, Sanford T, Roth HR, Xu Z, Tetreault J, Xu D, Flores MG, Raman AG, Kulkarni R, et al. Federated learning improves site performance in multicenter deep learning without data sharing. J Am Med Inform Assoc. 2021;28(6):1259–1264. doi: 10.1093/jamia/ocaa341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aich S, Sinai NK, Kumar S, Ali M, Choi YR, Joo M-I, Kim H-C (2021) Protecting personal healthcare record using blockchain & federated learning technologies. IEEE, pp 109–112
- 36.Stripelis D, Ambite JL, Lam P, Thompson P (2021) Scaling neuroscience research using federated learning. IEEE, pp 1191–1195
- 37.Nicholson C (2019) Evaluation metrics for machine learning-accuracy, precision, recall, and f1 defined
- 38.Zhao L, Jia K (2016) Multiscale cnns for brain tumor segmentation and diagnosis. Comput Math Methods Med [DOI] [PMC free article] [PubMed]
- 39.Erden B, Gamboa N, Wood S (2017) 3d convolutional neural network for brain tumor segmentation. Stanford University, USA, Technical report, Computer Science
- 40.Sun L, Zhang S, Chen H, Luo L. Brain tumor segmentation and survival prediction using multimodal mri scans with deep learning. Front Neurosci. 2019;13:810. doi: 10.3389/fnins.2019.00810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rehman A, Khan MA, Saba T, Mehmood Z, Tariq U, Ayesha N (2020) Microscopic brain tumor detection and classification using 3d cnn and feature selection architecture. Microscopy Research and Technique [DOI] [PubMed]
- 42.Amin J, Sharif M, Yasmin M, Fernandes SL. Big data analysis for brain tumor detection: deep convolutional neural networks. Futur Gener Comput Syst. 2018;87:290–297. doi: 10.1016/j.future.2018.04.065. [DOI] [Google Scholar]
- 43.Dolz J, Desrosiers C, Wang L, Yuan J, Shen D, Ayed IB. Deep cnn ensembles and suggestive annotations for infant brain mri segmentation. Comput Med Imaging Graph. 2020;79:101660. doi: 10.1016/j.compmedimag.2019.101660. [DOI] [PubMed] [Google Scholar]
- 44.Amin J, Sharif M, Yasmin M, Fernandes SL. Big data analysis for brain tumor detection: deep convolutional neural networks. Futur Gener Comput Syst. 2018;87:290–297. doi: 10.1016/j.future.2018.04.065. [DOI] [Google Scholar]
- 45.Ahmed H (2020) Br35H Brain Tumor Detection. https://www.kaggle.com/datasets/ahmedhamada0/brain-tumor-detection/metadata?select=yes
- 46.Sudharani K, Sarma T, Prasad KS. Advanced morphological technique for automatic brain tumor detection and evaluation of statistical parameters. Procedia Technol. 2016;24:1374–1387. doi: 10.1016/j.protcy.2016.05.153. [DOI] [Google Scholar]
- 47.Afsara M, Reza RA, Fahmeda HF, Tanzim R, Md, R. Anisur, Mohammad ZP (2020) Detection of brain tumor and identification of tumor region using deep neural network on fmri images. In: The 19th international conference on machine learning and cybernetics (ICMLC) 2020
- 48.Amin J, Sharif M, Yasmin M, Fernandes SL (2017) A distinctive approach in brain tumor detection and classification using mri. Pattern Recognit Lett
- 49.Abdel-Maksoud E, Elmogy M, Al-Awadi R. Brain tumor segmentation based on a hybrid clustering technique. Egypt Inform J. 2015;16(1):71–81. doi: 10.1016/j.eij.2015.01.003. [DOI] [Google Scholar]
- 50.Nabizadeh N, Kubat M (2015) Brain tumors detection and segmentation in mr images: Gabor wavelet vs. statistical features. Comput Electr Eng 45:286–301
- 51.Zhao L, Jia K (2015) Deep feature learning with discrimination mechanism for brain tumor segmentation and diagnosis. In: 2015 International conference on intelligent information hiding and multimedia signal processing (IIH-MSP). IEEE, pp 306–309
- 52.Li M, Kuang L, Xu S, Sha Z. Brain tumor detection based on multimodal information fusion and convolutional neural network. IEEE Access. 2019;7:180134–180146. doi: 10.1109/ACCESS.2019.2958370. [DOI] [Google Scholar]
- 53.Yi L, Zhang J, Zhang R, Shi J, Wang G, Liu X. Su-net: an efficient encoder-decoder model of federated learning for brain tumor segmentation. New York: Springer; 2020. pp. 761–773. [Google Scholar]
- 54.Sheller MJ, Reina GA, Edwards B, Martin J, Bakas S. Multi-institutional deep learning modeling without sharing patient data: a feasibility study on brain tumor segmentation, 92–104. New York: Springer; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]