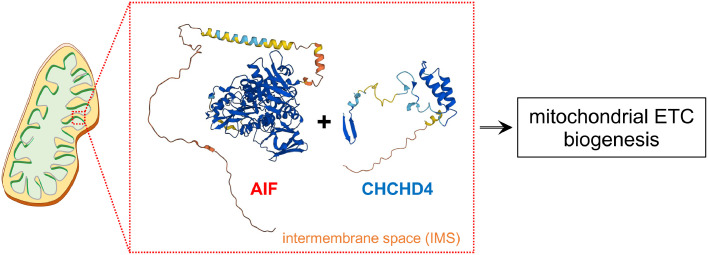
Summary
Apoptosis-inducing factor (AIF) is a mitochondrial intermembrane space flavoprotein with diverse functions in cellular physiology. In this regard, a large number of studies have elucidated AIF's participation to chromatin condensation during cell death in development, cancer, cardiovascular and brain disorders. However, the discovery of rare AIFM1 mutations in patients has shifted the interest of biomedical researchers towards AIF's contribution to pathogenic mechanisms underlying inherited AIFM1-linked metabolic diseases. The functional characterization of AIF binding partners has rapidly advanced our understanding of AIF biology within the mitochondria and beyond its widely reported role in cell death. At the present time, it is reasonable to assume that AIF contributes to cell survival by promoting biogenesis and maintenance of the mitochondrial oxidative phosphorylation (OXPHOS) system. With this review, we aim to outline the current knowledge around the vital role of AIF by primarily focusing on currently reported human diseases that have been linked to AIFM1 deficiency.
Keywords: Aapoptosis-inducing factor (AIF), CHCHD4, Mitochondria, Mitochondrial diseases, Oxidative phosphorylation (OXPHOS)
Introduction
Mitochondria are intracellular double-membrane organelles hosting a large number of biochemical reactions that critically influence cellular fate. The tightly regulated oxidation of carbon substrates converts nutrients into reduced cofactors (i.e., NADH and FADH2), which subsequently transfer electrons to the electron transport chain (ETC) to promote mitochondrial oxidative phosphorylation (OXPHOS) and ATP production. Equally important, mitochondria support the biosynthesis of molecules (e.g., nucleotides, lipids, amino acids and their derivatives) that are essential for cell survival and proliferation.1, 2, 3, 4 Given the critical role of mitochondria in both catabolic and anabolic processes, it should come as no surprise that lesions of mitochondrial components and/or proteins involved in mitochondrial bioenergetics can lead to human diseases.5,6 The impact of mitochondrial dysfunction is not restricted to inherited metabolic syndromes,7,8 but rather extends to age-related frailty, tumorigenesis, cardiovascular, metabolic and neurodegenerative diseases.9, 10, 11, 12, 13 For the large majority of human illnesses with clear signatures of mitochondrial dysfunction, patients may develop a variety of symptoms with variable age-onset, organ involvement and clinical outcomes that negatively impact their life quality and may shorten their lifespan expectancy. Despite the effort of the scientific community, therapeutic options remain limited and are not efficient disease modifiers.5,14, 15, 16, 17 To complicate even further the management and diagnosis of these heterogeneous pathologies, their molecular aetiology includes disease-causing lesions affecting either nuclear or mitochondrial genes.5,7 This loose genotype-phenotype relationship has important clinical implications, since mutations in different genes can impair mitochondrial function in a comparable manner, leading to apparently similar clinical features, as in the case of Leigh syndrome.18 The opposite scenario can also arise when several clinical manifestations are causally linked to one gene, which may encode pathogenic variants that may lead to early-onset multi-organ failure or to a tissue-specific degeneration eventually later in life.16,19 A good example of the latter case is the nuclear AIFM1 gene encoding the apoptosis-inducing factor (AIF), a mitochondrial oxidoreductase that has considerably attracted the attention of the scientific community because of its essential contribution to human pathophysiology.20, 21, 22, 23 As originally described,24 upon cell injury AIF is released from the mitochondria and translocates to the nucleus, where it binds endonucleases that promote chromatin condensation and DNA fragmentation. From this initial seminal work, the pro-death contribution of AIF has been implicated in a range of biological processes associated with human disorders,21,22 including stroke,25,26 metabolic syndromes dysfunction27 and neurodegenerative diseases, such as Parkinson's disease.28 However, over time it became evident that AIF has also a critical homeostatic role in mitochondrial bioenergetics, since it takes part in the ETC assembly,29, 30, 31 hence regulating OXPHOS and ATP production (Figure 1). In the present review, we will discuss AIF's participation in mitochondrial OXPHOS biogenesis and maintenance. We will not review the large literature describing AIF's contribution to cell death pathways, since this biological aspect has been extensively discussed elsewhere.20,32, 33, 34 To better understand the importance of AIF in human pathophysiology, we will draw attention to those inherited metabolic disorders that have been causally associated with AIF deficiency. We will outline the additional research efforts required to fully elucidate the mechanisms by which inherited AIFM1 mutations cause functional and/or structural AIF defects that lead to bioenergetically compromised mitochondria, which are unsuitable to support cellular homeostasis and, as a consequence, organismal health and survival.
Figure 1.
Within the IMS, membrane-embedded AIF interacts with CHCHD4 and promotes oxidative folding of newly imported polypeptides, including subunits of the mitochondrial respiratory chain complexes. AIF and CHCHD4 protein structure predictions were obtained using AlphaFold.130,131
From structural organization to functional insights
AIF (Gencode: ENST00000676229.1) is a nuclear-encoded polypeptide that is imported into the mitochondria, where it is proteolytically processed to an intermembrane space (IMS) protein tethered to the inner mitochondrial membrane (IMM)24,35 (Figure 1). AIF consists of at least six functionally defined regions: an N-terminal mitochondrial localization sequence (MLS) that is cleaved at Met54/Ala55 by mitochondrial processing peptidases; a hydrophobic transmembrane sequence (Asn66/Ser67-Ala84/Tyr85); three structural domains that coordinate the binding of one FAD and two NADH molecules; a C-terminal motif (Met481-Asp613) containing a Pro/Glu/Ser/Thr (PEST)-rich sequence and a proline-rich binding domain that may contribute to the physical association with molecular partners35, 36, 37 (Figures 1 and 2A). Phylogenetic analyses indicate that AIF belongs to an evolutionarily conserved sub-branch of oxidoreductases that includes also AMID/AIFM2/FSP1, AIFL/AIFM3 and the distal yeast homologues Nde1 and Nde2, with the latter two probably having evolved much before the divergence of eukaryotes.38 As an oxidoreductase, tissue-purified FAD-containing AIF exhibits NADH oxidase activities, which can compensate growth defects of bacteria lacking Complex I and NDH-2 by sustaining the electron flow of the residual respiratory chain to promote ATP biosynthesis.39 Soluble folded AIF molecules structurally resemble glutathione reductase (GR)-like proteins,36,37,40 although AIF possesses a β-hairpin (Thr190-Ser202) and a regulatory segment (or C-loop, Ala509-Asp559) that gives unique molecular properties. Upon mitochondrial import, the unfolded AIF precursor undergoes conformational changes that, along with the incorporation of one FAD molecule, establish its redox potential as clearly defined by several in vitro studies.40,41 The subsequent binding of one NADH molecule to the NADH-binding domain (Thr263-Gly399) stimulates rearrangement of several side chains, causing conformational changes in the regulatory peptide that enables the formation of a dimeric air-stable FADH2-NAD+ charge-transfer-complex relatively inefficient in electron transfer.40, 41, 42 The displacement of the regulatory segment exposes hydrophobic surfaces at the interface of monomeric AIF molecules, facilitating the remodeling of salt bridges (including those within the β-hairpin) and dimerization of reduced NADH-containing AIF molecules.40,43, 44, 45 The physical interaction with thioredoxins contributes to AIF's redox state 46,47 and may prevent oxidative modifications of cysteine residues, which is assumed to be a rate-determining step in the proteolytic processing and subsequent nuclear translocation of AIF in cells undergoing oxidative stress, apoptosis or calcium (Ca2+) overload.48,49 Following the reorganization of the regulatory peptide and the β-hairpin, three amino acid side chains (i.e., Trp196, Trp483 and Glu493) contribute to the stabilization of a second NADH that acts as an allosteric cofactor, further contributing to the redox properties of dimeric AIF.44,45 Thus, the incorporation of NADH into FAD-containing AIF molecules determines the ratio of monomeric and dimeric forms, rather than NAD(P)H oxidase activity as initially proposed.50 Based on this and other in vitro evidence, it seems that under normal conditions NADH binding may positively facilitate long-lived charge-transfer-complex formation and the subsequent enrichment of AIF dimers within the intermembrane space. Conversely, a decline of NADH availability may shift the monomer-dimer equilibrium to a state in which monomeric AIF represents the predominant form, potentially affecting AIF affinity toward the different substrates with which AIF physically interacts according to its subcellular localization.
Figure 2.
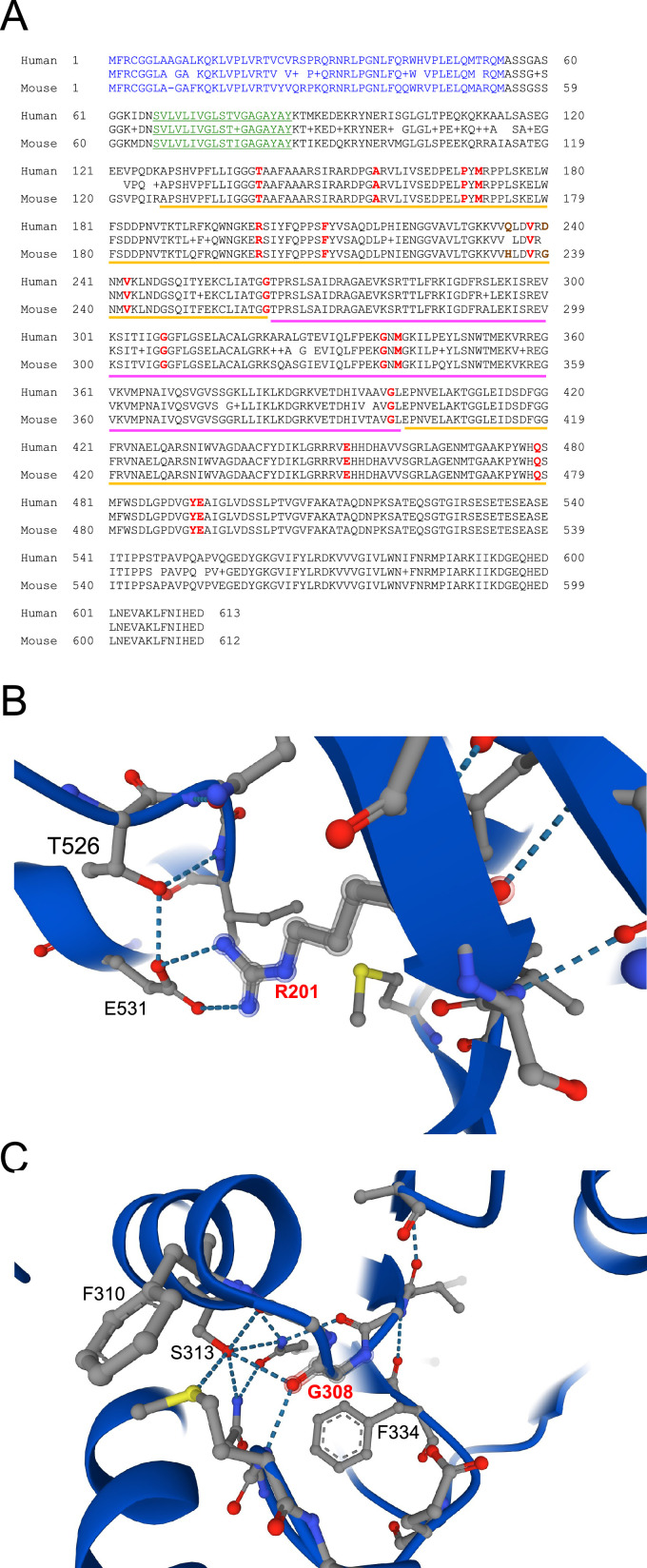
(A) Protein alignment of human and mouse AIF. The N-terminal MLS and the putative transmembrane sequence are highlighted in blue and green, respectively. The yellow and magenta lines highlight amino acids that are part of the FAD and NADH domains, respectively. In bold red, we report the evolutionarily conserved residues that are mutated in patients with mitochondrial diseases. Gln235 and Asp240 (in brown) have been linked to mitochondrial diseases, although they are not evolutionarily conserved across species. (B) Arg201 is predicted to form hydrogen bonds with Glu531 and its loss may compromise the folding of AIF regulatory peptide (Ala509-Asp559), with important consequence on AIF stability. (C) Gly308 is within the NADH-binding domain and is predicted to coordinate Ser313, contributing to the proper organization of the catalytic site and NADH stabilization by Phe310. Protein structure prediction was obtained using AlphaFold.130,131
AIF and mitochondrial bioenergetics: functional dependency on binding partners
The first evidence arguing that AIF may have a function beyond cell death came from the characterization of the Harlequin (Hq) mutant mice.51 As a spontaneous ecotropic proviral insertion within the intron 1 of the Aifm1 gene, the hypomorphic X-linked Hq mutation causes the reduction of AIF expression to approximately 20% of normal levels, leading to retinal and cerebellar degeneration associated with enhanced hydrogen peroxide sensitivity.51 While these initial observations could imply a putative role of AIF as a free radical scavenger, later seminal works demonstrated that AIF deficiency compromises the maintenance of the OXPHOS system.31,52 In this regard, the Hq mutation as well as Aifm1 knockout and/or downregulation significantly reduce the post-transcriptional expression of Complex I, and to a lesser extent, Complex III and IV subunits of the ETC, with a resulting impairment of the mitochondrial respiration in tumorigenic and ES cells.31 Consistently, AIF instability due to an amino acid deletion (i.e., Arg200: see below) leads to ETC defects, aberrant formation of mitochondrial respiratory supercomplexes and impaired mitochondrial bioenergetics in mice.53,54 The contribution of AIF to the mitochondrial OXPHOS system is evolutionarily conserved in invertebrates,48,55,56 further indicating that AIF's participation in mitochondrial bioenergetics has been acquired relatively early in the metazoan evolution. At least in mammals, mitochondrial defects can be almost completely rescued by the expression of a mitochondrially anchored AIF that is not proteolytically cleaved during cell death, suggesting that the proapoptotic role of AIF is dispensable for mitochondrial homeostasis.52 Mechanistically, AIF physically interacts and stabilizes the coiled-coil-helix-coiled-coil-helix domain containing 4 (CHCHD4),20,22,23,29,30 the mammalian homolog of yeast MIA40 regulating the formation of intramolecular disulfide bonds of newly imported polypeptides in the IMS (Figure 1).57, 58, 59, 60, 61 In line with prior experimental evidence,30,62 coevolution analyses and structural modelling indicate that the N-terminal region of CHCHD4 may form a β-hairpin that binds the C-terminal motif of AIF, with Tyr560 that is predicted to form hydrophobic interactions with two residues (Ile12 and Phe14) of CHCHD4.63 At the time of writing, it has been reported that AIF expression determines the balance between monomeric soluble CHCHD4 and its stable functional form associated with membrane-tethered AIF.62 Consequently, AIF knockout negatively influences CHCHD4 interaction with its substrates, including Complex I subunit NDUFS5. The subsequent cytosolic accumulation and proteasomal degradation of NDUFS5 causes the stalling of Complex I and, therefore, alters ETC biogenesis.62 Based on these compelling lines of evidence,20,22,23,62,64, 65, 66 AIF may now be considered a new component of the disulfide relay machinery that catalyzes the oxidative folding of nuclear encoded, cysteine-containing small precursor proteins imported in the IMS (Figure 1). However, AIF can physically interact also with subunits of the ETC, as shown for Complex IV using cross-linking mass-spectrometry profiling.67 Although it remains very unlikely that dimeric AIF can directly shuttle electrons to Complex IV, this new set of evidence may imply an additional stimulatory function of AIF in the molecular biogenesis and/or maturation of respiratory complexes. Apart from ETC subunits, AIF/CHCHD4 facilitates the IMS import of the C9orf72 protein, which enables C9orf72-dependent stabilization of the translocase of IMM domain containing 1 (TIMMDC1), a crucial chaperone protein taking part in Complex I biogenesis.68 In this regard, C9ORF72 haploinsufficiency favors the m-AAA metalloprotease AFG3L2-dependent degradation of TIMMDC1 in amyotrophic lateral sclerosis (ALS)/frontotemporal dementia (FTD) patient-derived tissues and induced pluripotent stem cells (iPSCs).68 These recent data suggest that AIF/CHCHD4-dependent mitochondrial biogenesis may contribute to sporadic ALS/FTD and possibly other human neurodegenerative diseases.
The influence of AIF on mitochondrial bioenergetics is however not limited to the IMS import and subsequent oxidative folding of ETC subunits that are required for the OXPHOS system biogenesis. Through the disulfide relay machinery, AIF may indirectly influence the import, folding and IMS localization of adenylate kinase 2 (AK2),69,70 an enzyme catalysing the reversible transfer of a terminal phosphate group between ATP and AMP to generate ADP as a necessary biochemical process to sustain Complex V/ATP synthase activity. Moreover, AIF expression may affect Ca2+ influx into the mitochondrial matrix because of an indirect modulation of Ca2+ flow through the mitochondrial calcium uniporter (MCU). In this regard, it was shown that CHCHD4 can bind the mitochondrial Ca2+ uptake protein 1 (MICU1) and stimulate the formation of an intermolecular disulfide bond between MICU1 and its paralog MICU2, resulting in aberrant regulation of mitochondrial Ca2+ uptake.69 In Drosophila and in mammalian cells, AIF deficiency may compromise IMS import and cristae junction localization of CHCHD3/MI19,56,71 thus disrupting mitochondrial contact site and crista organizing system (MICOS) complex to an extent that phenotypically resembles CHCHD4/MIA40 loss in HeLa cells and yeast.72, 73, 74 The impact of AIF deficiency on MICOS complex stability is not limited to one single subunit, but extends to others, such as coiled-coil-helix-coiled-coil-helix domain containing 10 (CHCHD10). In this regard, ALS-causing mutations alter the import and stability of CHCHD10 as a result of aberrant CHCHD4-dependent disulfide oxidation, ultimately leading to mitochondrial dysfunction due to cristae disruption.75 Although these are only a few examples of novel processes directly or indirectly linked to AIF, they provide a general snapshot of the downstream molecular consequences of AIF deficiency.
Like other mitochondrial proteins, AIF is a substrate of mitochondrial proteases, including mitochondrial ATP-dependent Lon protease 1 (LONP1) that binds and modulates AIF protein levels in a direct manner.76 Moreover, AIF may undergo posttranslational modifications with a modulatory effect on AIF stability and/or activity. In this regard, while calpain cleavage of AIF has been assumed to be a necessary step for AIF mitochondrial release and subsequent nuclear translocation,77 less is known about other posttranslational processes and their impact on mitochondrial bioenergetics. As an example of AIF posttranslational modifications, it was reported that the X-linked inhibitor of apoptosis (XIAP) ubiquitinates AIF at K255 and consequently regulates AIF pro-apoptotic features, rather than its proteasomal degradation or NADH oxidoreductase properties.78,79 On the other hand, OTU deubiquitinase 1 (OTUD1) directly deubiquitinates AIF at K244 and alters AIF binding to CHCHD4, resulting in impaired OXPHOS and swollen mitochondria lacking intact cristae.80 The competition between enzymes goes beyond AIF ubiquitination/deubiquitination rate, as exemplified by the antagonistic biological competition of XIAP with the mitochondrial phosphoglycerate mutase 5 (PGAM5),81 a Ser/Thr phosphatase that, through AIF interaction,69,82 is normally localized to the IMM. Since PGAM5 can contribute to mitochondrial dynamics and Wnt-dependent mitochondrial biogenesis,83,84 it is plausible to imagine a scenario in which AIF posttranslational modifications may alter the import and/or the release of AIF binding partners (e.g., PGAM5), with indirect consequences for mitochondrial bioenergetics. To date, this aspect remains poorly explored and future studies are warranted to experimentally address this hypothesis.
AIFM1 mutations and their impact on human pathophysiology
The first disease-causing mutation in the AIFM1 gene was reported in two male infants conceived by healthy related mothers (Figure 2A and Table 1). The X-linked genetic lesion was described as a trinucleotide deletion (c.601_603delAGA) in exon 5 causing the ablation of an Arg residue at position 201 (R201) of the full-length AIF polypeptide.85 Within a few months after birth, one of the patients developed involuntary and progressive neurological symptomatology, including axonal sensory and motor peripheral neuropathy that were confirmed at 15 months of age. Later, the young boy developed seizures and respiratory problems. His cousin showed a reduction of active movement before the first year of age, with progressive muscle wasting and weakness more evident in the following months.85 Structural in vitro analyses revealed that the soluble AIF (R201 del) variant shows aberrant charge-transfer-complex formation, weakened FAD binding and altered NADH oxidation compared to the wild type AIF protein.85,86 These changes in redox properties are linked to structural perturbations of the regulatory peptide folding, since Arg201 confers conformational stability through a salt bridge with Glu531, which influences the accessibility of the FAD-containing catalytic domain85,86 (Figure 2B). Although the FAD- and NADH-domains contain most of the mutational hotspots that lead to AIF dysfunction (Figure 2A and Table 1), the correlation between clinical outcomes and aberrant AIF structure is not as straightforward as expected. It is generally assumed that all the so far described disease-causing AIF variants are functionally unable to support OXPHOS biogenesis and/or other processes. However, current evidence suggests that some of the mutant AIF polypeptides are unstable or partially unfolded, whereas other variants have less obvious structural abnormalities. For example, the AIF (G308E) variant was found in three brothers (two infants and a fetus) who suffered from structural brain anomalies (i.e., choroids plexus cysts, bilateral brain ventriculomegaly and enlarged cisterna magna).87 After birth, the two patients displayed evident hypotonia, hypokinesia, seizures and neuromuscular symptoms, ultimately leading to premature death.87 The substitution of a Gly with a Glu at residue 308 hinders the coordination of the adenylate moiety within the NADH binding site (Figure 2C), with consequent inhibition of protein dimerization.42,86 Such a strong effect on AIF folding has not been reported for other disease-causing mutations that are predicted to be highly detrimental, as in the case of AIF (G262S), (G338E) and (V243L), which show only minor differences in terms of mRNA expression, NADH binding and/or protein stability.86 Despite these different profiles, all the three pathogenic AIF variants cause mitochondrial diseases. Specifically, the AIF (G262S) variant has been causally linked to Cowchock syndrome/X-linked recessive Charcot-Marie-Tooth disease-4 (CMTX4), with patients developing gait and limb ataxia, cognitive dysfunction, muscle wasting and weakness among other symptoms.88,89 As a consequence of impaired OXPHOS, it was reported that patients expressing the AIF (G338E) variant exhibit early onset mitochondrial encephalopathy,90 while children carrying AIF (V243L) develop progressive myopathy, ataxia and neuropathy within the first years of life.91 As for the other AIFM1 alleles reported to date, little is known about their in vitro biochemical properties, with their biological impact primarily determined by evidence obtained using patient-derived materials. Given these obvious limitations, one valuable option is to classify disease-causing AIFM1 mutations according to their clinical manifestations, with 6 AIF variants (P169L, R201del, V243L, G308E, G338E, Q479R) resulting in combined oxidative phosphorylation deficiency-6 (COXPD6) that may lead to encephalomyopathic disorder with onset before or immediately after birth (Table 1).85,87,90, 91, 92, 93 Using the same criteria based on reported clinical symptoms, it would be reasonable to include 7 AIF variants (T141I, A157V, F210L/S, G262S, M340T, E493V) in rare inherited X-linked Charcot-Marie-Tooth disease type 4, with pathological patterns that are highly variable in term of onset (childhood or early adolescence) and symptoms, the latter including slowly progressive axonal neuropathy associated with deafness and cognitive impairment, walking difficulties and gait ataxia (Table 1).88,94, 95, 96, 97, 98 As additional two groups, we can mention 4 AIF variants (Q235H, D237G/V, D240D synonymous substitution) that can give rise to a X-linked developmental disorder consisting of progressive skeletal and neurologic abnormalities,99, 100, 101, 102, 103 while the other 4 recently identified AIF mutants (G399S, E453Q, P488L, Y492H) have been linked to mitochondrial diseases with a variable spectrum of phenotypic outcomes (Table 1).104, 105, 106, 107 At the time of writing, a new case of fatal encephalomyopathy has been reported in an infant from a mother carrying a de novo AIFM1 intronic mutation that is supposed to alter the correct splicing of the precursor mRNA.108 Due to exon 11 skipping and the consequent amino acid frameshift, the predicted AIF variant is a truncated polypeptide of 361 amino acids that, if translated, is unstable and rapidly degraded.108 Finally, a large number of AIF variants has been associated with relatively mild syndromes primarily characterized by progressive auditory neuropathy and deafness109, 110, 111 (Table 1), which may indicate that these mutations have a weaker effect on the OXPHOS system compared to the first disease-causing AIFM1 allele identified more than a decade ago.85 Given the relevance of AIF in human pathophysiology, we envision that a better characterization of these clinically relevant AIF variants at biophysical, biochemical and cellular levels may help to further improve our understanding of the complex biology of AIF and its contribution to mitochondrial bioenergetics. Through this effort, in the near future it may be possible to develop novel concepts for urgently needed personalized therapeutic approaches that may help to ameliorate human illnesses associated with AIFM1 mutations.
Table 1.
List of AIFM1 mutations and associated clinical features of the reported cases.
| AIF variants | Nucleotide changes | Reported clinical features | ||
| p.Thr141Ile | c.422C>T | Patients exhibited ataxia associated with variable degrees of hearing loss and neuropathy.94 | ||
| p.Ala157Val | c.470C>T | Progressive severe ataxia, polyneuropathy, atypical hemifacial spam and myoclonus.98 | ||
| p.Pro169Leu | c.506C>T | Severe multisystem pathology and metabolic acidosis resulting in an early demise (combined oxidative phosphorylation deficiency 6: COXPD6).92 | ||
| p.Met171Ile | c.513G>A | X-linked recessive Charcot-Marie-Tooth disease-4 (CMTX4).127 | ||
| p.Arg201del | c.del601-603 trinucleotide deletion | Severe mitochondrial encephalomyopathy; psychomotor regression; axonal sensory and motor peripheral neuropathy; muscle wasting; weakness (COXPD6).85 | ||
| p.Phe210Leu p.Phe210Ser |
c.630C>G c.629T>C |
Patients were diagnosed with a possible X-linked Charcot-Marie-Tooth disease-4 (CMTX4). Patients exhibited axonal polyneuropathy, muscle atrophy and weakness.97 Patients exhibited early-onset axonal polyneuropathy with progressive muscle weakness.128 |
||
| p.Gln235His | c.705G>C | X-linked spondylometaphyseal dysplasia with hypomyelinating leukodystrophy. Patients exhibited gait abnormalities; progressive skeletal and neurologic abnormalities; visual deficits; hypomyelination; skeletal dysplasia; mild cognitive dysfunction.99 | ||
| p.Asp237Val p.Asp237Gly |
c.710A>T c.710A>G |
X-linked hypomyelination with spondylometaphyseal dysplasia. Infants exhibited gait abnormalities and gradual deterioration of motor functions. Diffused hypomyelination included the cerebral white matter. Skeletal abnormalities were detected in all patients.99 X-linked spondylometaphyseal dysplasia with mental retardation. Patients exhibited progressive skeletal deformities, widespread hypomyelination, cerebral and cerebellar atrophy .99,100 |
||
| p.Asp240Asp | c.720C>T | X-linked hypomyelination with spondylometaphyseal dysplasia. Patients exhibited progressive skeletal and neurologic abnormalities, optic nerve atrophy and moderate hearing loss.99 | ||
| p.Val243Leu | c.727G>T | X-linked recessive severe encephalomyopathic disorder (COXPD6). The patient exhibited progressive myopathy, ataxia and neuropathy at childhood.91 | ||
| p.Gly262Ser | c.784G>A | In an adult patient, Gly(262) to Ser substitution was associated with slowly progressive mitochondrial disease. At childhood, the patient developed gait and limb ataxia, hearing loss and cognitive impairment. At puberty, the patient exhibited visual deficits, tremor, muscle wasting and weakness.95 | ||
| p.Gly308Glu | c.923G>A | Prenatal developmental brain abnormalities (ventriculomegaly: enlargement of lateral ventricles). Patients exhibited hypotonia, aberrant psychomotor development, seizures, muscle weakness and cardiac abnormalities.87 | ||
| p.Gly338Glu | c.1013G>A | X-linked early-onset mitochondrial encephalopathy. Patients exhibited hypotonia, muscle weakness, respiratory insufficiency, involuntary movements and motor axonal neuropathy (COXPD6).90 | ||
| p.Met340Thr | c.1019T>C | Patients exhibited ataxia associated with variable degrees of hearing loss and neuropathy .94,96 | ||
| p.Gly399Ser | c.1195G>A | Patients exhibited childhood-onset nonprogressive cerebellar ataxia, hearing loss, intellectual disability, peripheral neuropathy, mood and behaviour disorder.104 | ||
| p.Glu453Gln | c.1357G>C | Late-onset ataxic sensory neuropathy and hearing impairment in an adult male patient. Histopathological examination revealed neurogenic muscle atrophy. No ragged red fibres and cytochrome C oxidase-deficiency were detected in muscle biopsies.105 | ||
| p.Gln479Arg | c.1436A>G | In a male infant, Gln(479) to Arg substitution was associated with a severe, fatal, early-onset mitochondrial encephalomyopathy characterized by intractable seizures, polyneuropathy and myopathy (COXPD6).93 | ||
| p.Tyr492His | c.1474T>C | Mitochondrial disorders with spinal muscular atrophy-like pattern .92,107 | ||
| p.Glu493Val | c.1478A>T | Cowchock syndrome/X-linked recessive Charcot-Marie-Tooth disease-4 (CMTX4), with or without cerebellar ataxia; early-onset axonal sensorimotor neuropathy associated with hearing loss and cognitive impairment .88,89 | ||
| Mutations linked to auditory defects | ||||
| - p.Thr260Ala - - p.Ile304Met p.Leu333Phe p.Leu344Phe p.Ser349Gly p.Gly360Arg p.Arg422Trp p.Arg422Gln p.Arg430Cys p.Arg451Gln p.Ala465Val p.Ala472Val p.Pro475Leu p.Pro488Leu p.Val498Met p.Tyr560His p.Ile591Met |
c.547A>T c.778A>G c.881G>A c.890A>T c.912C>G c.997C>T c.1030C>T c.1045A>G c.1078G>C c.1264C>T c.1265G>A c.1288C>T c.1352G>A c.1394C>T c.1415C>T c.1424C>T c.1463C>T c.1492G>A c.1678T>C c.1773C>G |
X-linked deafness.109, 110, 111 Patients became symptomatic at different age. In a few cases, auditory neuropathy could be associated with other manifestations. Leu(344) to Phe substitution was the most common variant detected in patients. | ||
| Additional mutations identified in patients | ||||
| - | c.697-44T>G An intronic variant 44 bp from the 3’ splice site of exon 7 |
This mutation within intron 6 of the AIFM1 gene may alter mRNA splicing and potentially affect AIF expression. Clinical symptoms were reported in members of a family with diagnosed X-linked spondylometaphyseal dysplasia with hypomyelinating leukodystrophy. Gradual motor dysfunction, spasticity, ataxia, proximal weakness and join contractures were observed between 12 and 36 months of age .99,101 | ||
| - | c.1164+5G>A A variant in intron 11 resulting in aberrant splicing and frameshift |
This mutation is located at 5 bp of intron 11 and may cause aberrant splicing of AIFM1 precursor mRNA. Patient-derived cells express both wt mature mRNA and a transcript lacking exon 11. The predicted AIF variant is a truncated protein of 361 amino acids. Patients develop fatal encephalomyopathy.108 | ||
AIF deficient transgenic mice: preclinical models and their limitations
Based on the currently available clinical and biochemical evidence, it is reasonable to assume that disease-causing AIFM1 mutations are loss-of-function. If this scenario is accepted, transgenic AIF deficient mice become relevant bona fide models for preclinical studies of novel therapeutic interventions.
Since constitutive Aifm1 knockout results in embryonic lethality,112,113 the Hq mutant mouse model was initially a key tool to study AIF beyond its role in cell death51 (Table 2). With a reduction of Aifm1 expression (up to ∼80%) that may vary across different tissues, Hq mutant mice have been proposed as a model of Complex I deficiency.31,114 Apart from their smaller size compared to wild type littermates,51 Hq mutant mice develop skeletal muscle atrophy and signs of progressive retinal and cerebellar degeneration, the latter becoming phenotypically obvious between 3-4 months of age.51,115 Neuropathological changes include GFAP-positive astrogliosis in some tissues (e.g., thalamus, cerebellum), already at 2 months of age, while extensive microglia activation occurs at 3-4 months of age and is observed in the cerebellum concomitantly with the neurodegeneration of Purkinje cells and other neurons of the granular cell layer.115 In their original mixed genetic background, Hq mutant mice show variable phenotypic traits, including weight loss over time, baldness and locomotory defects linked to the severity of cerebellar ataxia.114 Hair loss is seen in most of the adult animals, while pelage abnormalities in the pups are associated with fragility of the hair cortex, possibly because of keratinization defects due to altered gene expression patterns associated with expression of retroviral elements from the Hq allele.116 Although these inter-individual phenotypic differences may not be suitable for conventional pharmacokinetic and toxicology studies, Hq mutant mice have been used in some settings to evaluate the disease-modifying effects of dietary interventions and FDA-approved drugs.54,117,118 Thus, Hq mutant mice may still represent a useful in vivo model of diseases linked to AIF deficiency, especially when the primary goal is to recapitulate the phenotypic heterogeneity associated with inherited mutations affecting mitochondrial function. However, while these aspects may come with advantages in certain experimental settings, they become limiting factors when consistent and reproducible outcomes are pre-requisites for molecular investigations in small-sized animal cohorts. To overcome this bottleneck, transgenic mice carrying floxed Aifm1 alleles have been generated and used to manipulate AIF expression in a tissue specific manner. Toward this end, Cre-mediated recombination of exon 7 was used to successfully disrupt AIF expression. Muscle-specific AIF KO mice exhibited severe progressive skeletal muscle atrophy, cardiomyopathy and body weight loss that are evident already at 2 months of age112 (Table 2). Similarly, Cre-mediated inactivation of the floxed exon 11 results in a premature stop codon that prevents the correct expression of a functional AIF protein.119 Consistent with a role in embryogenesis, loss of a single Aifm1 allele causes perinatal hydrocephaly in heterozygous AIF KO female mice119 (Table 2). Moreover, adult Aifm1+/- females exhibit an aberrant immune profile,119 consistent with previous evidence showing that cell-specific hematopoietic Aifm1 KO mice develop progressive hematopoietic stem cell exhaustion and consequently decreased number of all blood cell types.120 Thus, the use of mutant mice carrying a floxed Aifm1 allele has further expanded the investigation of AIF's contribution to biological processes in a tissue-specific manner and within certain experimental paradigms. In this regard, Aifm1 knockout in the skeletal muscle, liver or in hypothalamic proopiomelanocortin (POMC)-expressing neurons improves glucose homeostasis and prevents obesity of animals fed with high-fat diet,27,121 emphasizing how cell-specific mitochondrial dysfunction due to AIF deficiency can influence the global metabolism of a living multicellular organism.
Table 2.
list of Aifm1 deficient mouse strains and summary of their features.
|
Strains (genotype) |
Reported phenotypes |
| Harlequin mutant mice (Aifm1Hq/Y in a mixed genetic background) 51 |
-Ectopic proviral insertion within the intron 1 of the Aifm1/Pdcd8 gene. -High variability of Aifm1 expression is observed in tissues across individual animals. -Within a few weeks after birth, Hq mutant animals exhibit weight loss, first signs of muscle wasting and a heterogeneous pelage paucity. -Between 3 and 6 months of age, hemizygous males (Hq/Y) and homozygous females (Hq/Hq) develop progressive cerebellar ataxia and blindness. -Neurodegeneration is evident in the cerebellum, although cortical, thalamic and striatal regions may be affected at a later time. -Astrogliosis and microglial hyperactivity are additional traits observed in Hq mutant brains. |
|
Aifm1flox/flox flox-exon 7-flox (C57BL/6J)112 |
-Aifm1 null embryos die during gestation between E8.5 and E10.5. -Muscle-specific Aifm1 KO leads to severe dilated cardiomyopathy and skeletal muscle atrophy. -Skeletal muscle-specific and liver-specific Aifm1 KO are resistant to high fat diet-induced obesity and display increased glucose tolerance. -Loss of AIF in POMC-expressing neurons alters neuronal activity and fatty acid metabolism, improving systemic glucose tolerance in obese mice. |
|
Aifm1flox/flox flox-exon 11-flox (C57BL/6J)119 |
-AIF ablation in hematopoietic stem cells alters the differentiation of blood cells, causing progressive pancytopenia in very young transgenic mice. -AIF deficiency impairs mitochondrial respiration and fatty acid metabolism, leading to aberrant thymocyte differentiation and fate. |
|
Aifm1(R200 del) knockin (C57BL/6J)54 |
-Aifm1(R200 del) KI mice develop early-onset myopathy without evident sign of neurodegeneration. -AIF (R200 del) variant is unstable and leads to OXPHOS defects associated with aberrant folate metabolism and hyperactive Akt/mTOR signaling. |
| Additional mouse models | Reported phenotypes |
|
Aifm1flox/flox flox-exon 3-flox (C57BL/6J)129 |
-Cre recombinase-mediated deletion of exon 3 induces the expression of an AIF variant (i.e., AIF3) lacking 81 amino acids (Gly35 to Ser115 of mouse AIF) that are part of the mitochondrial targeting sequences and downstream the hydrophobic transmembrane domain. -This mouse model is a conditional KO of wt Aifm1 and a gain-of-function for AIF3. Aifm1fl/Y; CamKIIαCre/tg males and Aifm1fl/fl; CamKIIαCre/tg females exhibit progressive neurodegeneration and die prematurely before adulthood. |
In an effort to develop a reliable model that could partially recapitulate the pathological profiles observed in patients expressing an AIF variant across tissues, our laboratory generated transgenic mice carrying the first identified disease-causing AIFM1 mutation in humans (i.e., human Arg 201 deletion85). This newly developed knockin mouse line expresses an AIF variant lacking R200 (R201 in human) and additional recognition elements that can enable tissue-specific Aifm1 knockout by using the phiC31 recombination system54 (Table 2). Aifm1(R200 del) KI mice are born at the expected Mendelian frequency and display early sign of skeletal muscle wasting associated with hind limb clasping and kyphosis. Consistent with previous in vitro evidence,85,86 posttranslational loss of AIF (R200 del) variant may lead to CHCHD4 deficiency in the skeletal muscle and other tissues.54 The loss of cytochrome c oxidase (COX)-positive muscle fibers correlates with impaired expression of ETC subunits. Contrary to Hq mutant mice, Aifm1(R200 del) KI mice do not exhibit cerebellar degeneration or other obvious lesions in the brain, despite comparable defects of the mitochondrial OXPHOS system.53,54 As a side note, Aifm1(R200 del) KI mice show metabolic signatures (e.g., aberrant folate metabolism) and increased Akt/mTOR signaling that are often observed in tissues from other mouse models of mitochondrial diseases.122, 123, 124, 125 Thus, the use of Aifm1(R200 del) KI mice may help the scientific community in the investigation of molecular processes associated with AIF deficiency, although additional in vitro and in vivo models may be required to fully recapitulate the pathological profiles observed in patients carrying AIF variants. Indeed, the use of transgenic mice may not be sufficient to explore how pathogenic AIF mutants can induce metabolic syndromes in humans. In support of this argument, AIF (Q235H) has been identified in infants with X-linked spondylometaphyseal dysplasia with hypomyelinating leukodystrophy.99 However, this substitution is a natural variant that occurs in rodents and does not cause obvious AIF instability (Figure 2A), further indicating possible limitations of eventual disease-causing Aifm1 mutant KI mice in fully recapitulating pathogenic processes observed in patients.
We look forward to future studies that may help to expand our understanding of disease-associated AIF variants. We hope that the currently available AIF mutant mice as well as future alternative in vitro and in vivo models may contribute to the development of novel therapeutic interventions that are urgently needed by patients and their families.
Conclusions and outstanding questions
The name apoptosis-inducing factor was initially used by Guido Kroemer and his team to describe a molecule that is released from the mitochondria upon dissipation of the mitochondrial membrane potential during caspase-dependent apoptosis.24,126 The expanding case reports on disease-causing AIFM1 mutations have further emphasized the importance of AIF function in human pathophysiology. On top of these important clinical aspects, an emerging array of interactors have placed AIF in a complex biological network with clear implications in mitochondrial bioenergetics, intracellular signaling and pathogenic processes. Despite three decades of studies, there are still many open questions that we consider relevant for both biology and medicine. Since AIF is an oxidoreductase, is there any substrate that undergoes AIF-dependent redox reactions? Alternatively, does AIF simply act as a sensor within the mitochondria, adjusting mitochondrial activity according to the needs of the cell? If so, which are the molecules that modify AIF activity? Is there any factor that, independently of CHCHD4, binds AIF and modulates mitochondrial bioenergetics? In the context of the different clinical settings, how can disease-causing AIF variants lead to different metabolic syndromes? Is AIF a good target for small bioactive molecules? Since the currently available treatments (e.g., riboflavin supplementation) do not lead to substantial improvements,85,92,94,105 a deeper understanding of molecular and mechanistic aspects associated with pathogenic AIF variants may help to dissect the complexity of human diseases linked to AIFM1 mutations. We look forward to new exciting studies that will add crucial pieces to the existing puzzle.
Search strategy and selection criteria
Original findings were searched in PubMed and Scopus. References from relevant articles were obtained using the following keywords, as single search term or in combination: “AIF”, “AIFM1”, “mitochondrial diseases”, “neurodegenerative diseases”, “metabolic diseases”, “oxidoreductase”, “therapeutic strategies”. Additional keywords may have been used in a few additional cases. Only articles published in English were considered. The final reference list was generated based on the relevance to the scope of this review.
Contributors
LW: literature search, text review and editing. ES: text review, editing and discussion. DE: text review, editing and discussion. DB: literature search, figures, text writing and editing. All authors read and approved the final version of the manuscript.
Declaration of interests
The authors declare no conflict of interest.
Acknowledgments
This research was supported by the DZNE institutional budget and the Helmholtz cross-program topic “Aging and Metabolic Programming (AMPro)”. DB is a member of the Excellence Cluster ImmunoSensation,2 which has received funds by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy–EXC2151–390873048. DB is a member of the Mitochondrial Dysfunction in Parkinson's Consortium (PD-MitoQUANT). PD-MitoQUANT has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No. 821522. This Joint Undertaking receives support from the European Union's Horizon 2020 research and innovation programme and EFPIA.
References
- 1.Khacho M, Harris R, Slack RS. Mitochondria as central regulators of neural stem cell fate and cognitive function. Nat Rev Neurosci. 2019;20(1):34–48. doi: 10.1038/s41583-018-0091-3. [DOI] [PubMed] [Google Scholar]
- 2.Spinelli JB, Haigis MC. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol. 2018;20(7):745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lisowski P, Kannan P, Mlody B, Prigione A. Mitochondria and the dynamic control of stem cell homeostasis. EMBO Rep. 2018;19(5) doi: 10.15252/embr.201745432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakrabarty RP, Chandel NS. Mitochondria as signaling organelles control mammalian stem cell fate. Cell Stem Cell. 2021;28(3):394–408. doi: 10.1016/j.stem.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell OM, Gorman GS, Lightowlers RN, Turnbull DM. Mitochondrial diseases: hope for the future. Cell. 2020;181(1):168–188. doi: 10.1016/j.cell.2020.02.051. [DOI] [PubMed] [Google Scholar]
- 6.Wallace DC. Mitochondrial genetic medicine. Nat Genet. 2018;50(12):1642–1649. doi: 10.1038/s41588-018-0264-z. [DOI] [PubMed] [Google Scholar]
- 7.Frazier AE, Thorburn DR, Compton AG. Mitochondrial energy generation disorders: genes, mechanisms, and clues to pathology. J Biol Chem. 2019;294(14):5386–5395. doi: 10.1074/jbc.R117.809194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeviani M, Viscomi C. Mitochondrial neurodegeneration. Cells. 2022;11(4) doi: 10.3390/cells11040637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zong WX, Rabinowitz JD, White E. Mitochondria and cancer. Mol Cell. 2016;61(5):667–676. doi: 10.1016/j.molcel.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang JY, Blum A, Liu J, Finkel T. The role of mitochondria in aging. J Clin Invest. 2018;128(9):3662–3670. doi: 10.1172/JCI120842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schon EA, Przedborski S. Mitochondria: the next (neurode)generation. Neuron. 2011;70(6):1033–1053. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camandola S, Mattson MP. Brain metabolism in health, aging, and neurodegeneration. EMBO J. 2017;36(11):1474–1492. doi: 10.15252/embj.201695810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzales MM, Garbarino VR, Pollet E, et al. Biological aging processes underlying cognitive decline and neurodegenerative disease. J Clin Invest. 2022;132(10) doi: 10.1172/JCI158453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viscomi C, Bottani E, Zeviani M. Emerging concepts in the therapy of mitochondrial disease. Biochim Biophys Acta. 2015;1847(6-7):544–557. doi: 10.1016/j.bbabio.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Avula S, Parikh S, Demarest S, Kurz J, Gropman A. Treatment of mitochondrial disorders. Curr Treatment Options Neurol. 2014;16(6):292. doi: 10.1007/s11940-014-0292-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorman GS, Chinnery PF, DiMauro S, et al. Mitochondrial diseases. Nat Rev Dis Primers. 2016;2:16080. doi: 10.1038/nrdp.2016.80. [DOI] [PubMed] [Google Scholar]
- 17.Pfeffer G, Horvath R, Klopstock T, et al. New treatments for mitochondrial disease-no time to drop our standards. Nat Rev Neurol. 2013;9(8):474–481. doi: 10.1038/nrneurol.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lake NJ, Compton AG, Rahman S, Thorburn DR. Leigh syndrome: one disorder, more than 75 monogenic causes. Ann Neurol. 2016;79(2):190–203. doi: 10.1002/ana.24551. [DOI] [PubMed] [Google Scholar]
- 19.Chinnery PF. Mitochondrial disease in adults: what's old and what's new? EMBO Mol Med. 2015;7(12):1503–1512. doi: 10.15252/emmm.201505079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrmann JM, Riemer J. Apoptosis inducing factor and mitochondrial NADH dehydrogenases: redox-controlled gear boxes to switch between mitochondrial biogenesis and cell death. Biol Chem. 2021;402(3):289–297. doi: 10.1515/hsz-2020-0254. [DOI] [PubMed] [Google Scholar]
- 21.Bano D, Prehn JHM. Apoptosis-inducing factor (AIF) in physiology and disease: the tale of a repented natural born killer. EBioMedicine. 2018;30:29–37. doi: 10.1016/j.ebiom.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinhardt C, Arena G, Nedara K, et al. AIF meets the CHCHD4/Mia40-dependent mitochondrial import pathway. Biochim Biophys Acta Mol Basis Dis. 2020;1866(6) doi: 10.1016/j.bbadis.2020.165746. [DOI] [PubMed] [Google Scholar]
- 23.Modjtahedi N, Tokatlidis K, Dessen P, Kroemer G. Mitochondrial proteins containing coiled-coil-helix-coiled-coil-helix (CHCH) domains in health and disease. Trends Biochem Sci. 2016;41(3):245–260. doi: 10.1016/j.tibs.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Susin SA, Lorenzo HK, Zamzami N, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397(6718):441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 25.Zhu C, Wang X, Huang Z, et al. Apoptosis-inducing factor is a major contributor to neuronal loss induced by neonatal cerebral hypoxia-ischemia. Cell Death Differ. 2007;14(4):775–784. doi: 10.1038/sj.cdd.4402053. [DOI] [PubMed] [Google Scholar]
- 26.Culmsee C, Zhu C, Landshamer S, et al. Apoptosis-inducing factor triggered by poly(ADP-ribose) polymerase and Bid mediates neuronal cell death after oxygen-glucose deprivation and focal cerebral ischemia. J Neurosci. 2005;25(44):10262–10272. doi: 10.1523/JNEUROSCI.2818-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pospisilik JA, Knauf C, Joza N, et al. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 2007;131(3):476–491. doi: 10.1016/j.cell.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, An R, Umanah GK, et al. A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science. 2016;354(6308) doi: 10.1126/science.aad6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer K, Buettner S, Ghezzi D, Zeviani M, Bano D, Nicotera P. Loss of apoptosis-inducing factor critically affects MIA40 function. Cell Death Dis. 2015;6:e1814. doi: 10.1038/cddis.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hangen E, Feraud O, Lachkar S, et al. Interaction between AIF and CHCHD4 regulates respiratory chain biogenesis. Mol Cell. 2015;58(6):1001–1014. doi: 10.1016/j.molcel.2015.04.020. [DOI] [PubMed] [Google Scholar]
- 31.Vahsen N, Cande C, Briere JJ, et al. AIF deficiency compromises oxidative phosphorylation. EMBO J. 2004;23(23):4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fatokun AA, Dawson VL, Dawson TM. Parthanatos: mitochondrial-linked mechanisms and therapeutic opportunities. Br J Pharmacol. 2014;171(8):2000–2016. doi: 10.1111/bph.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sevrioukova IF. Apoptosis-inducing factor: structure, function, and redox regulation. Antioxid Redox Signaling. 2011;14(12):2545–2579. doi: 10.1089/ars.2010.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hangen E, Blomgren K, Benit P, Kroemer G, Modjtahedi N. Life with or without AIF. Trends Biochem Sci. 2010;35(5):278–287. doi: 10.1016/j.tibs.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 35.Otera H, Ohsakaya S, Nagaura Z, Ishihara N, Mihara K. Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 2005;24(7):1375–1386. doi: 10.1038/sj.emboj.7600614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mate MJ, Ortiz-Lombardia M, Boitel B, et al. The crystal structure of the mouse apoptosis-inducing factor AIF. Nat Struct Biol. 2002;9(6):442–446. doi: 10.1038/nsb793. [DOI] [PubMed] [Google Scholar]
- 37.Ye H, Cande C, Stephanou NC, et al. DNA binding is required for the apoptogenic action of apoptosis inducing factor. Nat Struct Biol. 2002;9(9):680–684. doi: 10.1038/nsb836. [DOI] [PubMed] [Google Scholar]
- 38.Klim J, Gladki A, Kucharczyk R, Zielenkiewicz U, Kaczanowski S. Ancestral state reconstruction of the apoptosis machinery in the common ancestor of eukaryotes. G3 (Bethesda) 2018;8(6):2121–2134. doi: 10.1534/g3.118.200295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elguindy MM, Nakamaru-Ogiso E. Apoptosis-inducing factor (AIF) and its family member protein, AMID, are rotenone-sensitive NADH:ubiquinone oxidoreductases (NDH-2) J Biol Chem. 2015;290(34):20815–20826. doi: 10.1074/jbc.M115.641498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sevrioukova IF. Redox-linked conformational dynamics in apoptosis-inducing factor. J Mol Biol. 2009;390(5):924–938. doi: 10.1016/j.jmb.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Churbanova IY, Sevrioukova IF. Redox-dependent changes in molecular properties of mitochondrial apoptosis-inducing factor. J Biol Chem. 2008;283(9):5622–5631. doi: 10.1074/jbc.M709147200. [DOI] [PubMed] [Google Scholar]
- 42.Sorrentino L, Calogero AM, Pandini V, Vanoni MA, Sevrioukova IF, Aliverti A. Key role of the adenylate moiety and integrity of the adenylate-binding site for the NAD(+)/H binding to mitochondrial apoptosis-inducing factor. Biochemistry. 2015;54(47):6996–7009. doi: 10.1021/acs.biochem.5b00898. [DOI] [PubMed] [Google Scholar]
- 43.Brosey CA, Ho C, Long WZ, et al. Defining NADH-driven allostery regulating apoptosis-inducing factor. Structure. 2016;24(12):2067–2079. doi: 10.1016/j.str.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villanueva R, Ferreira P, Marcuello C, et al. Key residues regulating the reductase activity of the human mitochondrial apoptosis inducing factor. Biochemistry. 2015;54(33):5175–5184. doi: 10.1021/acs.biochem.5b00696. [DOI] [PubMed] [Google Scholar]
- 45.Ferreira P, Villanueva R, Martinez-Julvez M, et al. Structural insights into the coenzyme mediated monomer-dimer transition of the pro-apoptotic apoptosis inducing factor. Biochemistry. 2014;53(25):4204–4215. doi: 10.1021/bi500343r. [DOI] [PubMed] [Google Scholar]
- 46.Nakao LS, Everley RA, Marino SM, et al. Mechanism-based proteomic screening identifies targets of thioredoxin-like proteins. J Biol Chem. 2015;290(9):5685–5695. doi: 10.1074/jbc.M114.597245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lo SM, Martinez PA, Marques EF, et al. Oxidation of apoptosis-inducing factor (AIF) to disulfide-linked conjugates. Arch Biochem Biophys. 2020;692 doi: 10.1016/j.abb.2020.108515. [DOI] [PubMed] [Google Scholar]
- 48.Joza N, Galindo K, Pospisilik JA, et al. The molecular archaeology of a mitochondrial death effector: AIF in Drosophila. Cell Death Differ. 2008;15(6):1009–1018. doi: 10.1038/cdd.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chelko SP, Keceli G, Carpi A, et al. Exercise triggers CAPN1-mediated AIF truncation, inducing myocyte cell death in arrhythmogenic cardiomyopathy. Sci Transl Med. 2021;13(581) doi: 10.1126/scitranslmed.abf0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miramar MD, Costantini P, Ravagnan L, et al. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J Biol Chem. 2001;276(19):16391–16398. doi: 10.1074/jbc.M010498200. [DOI] [PubMed] [Google Scholar]
- 51.Klein JA, Longo-Guess CM, Rossmann MP, et al. The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature. 2002;419(6905):367–374. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- 52.Cheung EC, Joza N, Steenaart NA, et al. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. EMBO J. 2006;25(17):4061–4073. doi: 10.1038/sj.emboj.7601276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bertan F, Wischhof L, Scifo E, et al. Comparative analysis of CI- and CIV-containing respiratory supercomplexes at single-cell resolution. Cell Reports Methods. 2021;1(1) doi: 10.1016/j.crmeth.2021.100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wischhof L, Gioran A, Sonntag-Bensch D, et al. A disease-associated Aifm1 variant induces severe myopathy in knockin mice. Mol Metab. 2018;13:10–23. doi: 10.1016/j.molmet.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Troulinaki K, Buttner S, Marsal Cots A, et al. WAH-1/AIF regulates mitochondrial oxidative phosphorylation in the nematode Caenorhabditis elegans. Cell Death Discov. 2018;4:2. doi: 10.1038/s41420-017-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murari A, Rhooms SK, Goparaju NS, Villanueva M, Owusu-Ansah E. An antibody toolbox to track complex I assembly defines AIF's mitochondrial function. J Cell Biol. 2020;219(10) doi: 10.1083/jcb.202001071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Banci L, Bertini I, Cefaro C, et al. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat Struct Mol Biol. 2009;16(2):198–206. doi: 10.1038/nsmb.1553. [DOI] [PubMed] [Google Scholar]
- 58.Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, Herrmann JM. The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol. 2007;179(3):389–395. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terziyska N, Lutz T, Kozany C, et al. Mia40, a novel factor for protein import into the intermembrane space of mitochondria is able to bind metal ions. FEBS Lett. 2005;579(1):179–184. doi: 10.1016/j.febslet.2004.11.072. [DOI] [PubMed] [Google Scholar]
- 60.Mesecke N, Terziyska N, Kozany C, et al. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121(7):1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 61.Chacinska A, Pfannschmidt S, Wiedemann N, et al. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23(19):3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salscheider SL, Gerlich S, Cabrera-Orefice A, et al. AIFM1 is a component of the mitochondrial disulfide relay that drives complex I assembly through efficient import of NDUFS5. EMBO J. 2022 doi: 10.15252/embj.2022110784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pei J, Zhang J, Cong Q. Human mitochondrial protein complexes revealed by large-scale coevolution analysis and deep learning-based structure modeling. Bioinformatics. 2022 doi: 10.1093/bioinformatics/btac527. [DOI] [PubMed] [Google Scholar]
- 64.Finger Y, Riemer J. Protein import by the mitochondrial disulfide relay in higher eukaryotes. Biol Chem. 2020;401(6-7):749–763. doi: 10.1515/hsz-2020-0108. [DOI] [PubMed] [Google Scholar]
- 65.Herrmann JM, Riemer J. Mitochondrial disulfide relay: redox-regulated protein import into the intermembrane space. J Biol Chem. 2012;287(7):4426–4433. doi: 10.1074/jbc.R111.270678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou ZD, Saw WT, Tan EK. Mitochondrial CHCHD-containing proteins: physiologic functions and link with neurodegenerative diseases. Mol Neurobiol. 2017;54(7):5534–5546. doi: 10.1007/s12035-016-0099-5. [DOI] [PubMed] [Google Scholar]
- 67.Hevler JF, Zenezeni Chiozzi R, Cabrera-Orefice A, Brandt U, Arnold S, Heck AJR. Molecular characterization of a complex of apoptosis-inducing factor 1 with cytochrome c oxidase of the mitochondrial respiratory chain. Proc Nat Acad Sci USA. 2021;118(39) doi: 10.1073/pnas.2106950118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang T, Liu H, Itoh K, et al. C9orf72 regulates energy homeostasis by stabilizing mitochondrial complex I assembly. Cell Metab. 2021;33(3):531–546.e9. doi: 10.1016/j.cmet.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petrungaro C, Zimmermann KM, Kuttner V, et al. The Ca(2+)- dependent release of the Mia40-induced MICU1-MICU2 dimer from MCU regulates mitochondrial Ca(2+) uptake. Cell Metab. 2015;22(4):721–733. doi: 10.1016/j.cmet.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 70.Finger Y, Habich M, Gerlich S, et al. Proteasomal degradation induced by DPP9-mediated processing competes with mitochondrial protein import. EMBO J. 2020;39(19) doi: 10.15252/embj.2019103889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bertan F, Wischhof L, Scifo E, et al. Comparative profiling of N-respirasomes predicts aberrant mitochondrial bioenergetics at single-cell resolution. Biorxiv. 2020 doi: 10.1101/2020.12.07.414730. [DOI] [Google Scholar]
- 72.Sakowska P, Jans DC, Mohanraj K, Riedel D, Jakobs S, Chacinska A. The oxidation status of Mic19 regulates MICOS assembly. Mol Cell Biol. 2015;35(24):4222–4237. doi: 10.1128/MCB.00578-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Darshi M, Trinh KN, Murphy AN, Taylor SS. Targeting and import mechanism of coiled-coil helix coiled-coil helix domain-containing protein 3 (ChChd3) into the mitochondrial intermembrane space. J Biol Chem. 2012;287(47):39480–39491. doi: 10.1074/jbc.M112.387696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Darshi M, Mendiola VL, Mackey MR, et al. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem. 2011;286(4):2918–2932. doi: 10.1074/jbc.M110.171975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lehmer C, Schludi MH, Ransom L, et al. A novel CHCHD10 mutation implicates a Mia40-dependent mitochondrial import deficit in ALS. EMBO Mol Med. 2018;10(6) doi: 10.15252/emmm.201708558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sheng X, Liu C, Yan G, et al. The mitochondrial protease LONP1 maintains oocyte development and survival by suppressing nuclear translocation of AIFM1 in mammals. EBioMedicine. 2022;75 doi: 10.1016/j.ebiom.2021.103790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Polster BM, Basanez G, Etxebarria A, Hardwick JM, Nicholls DG. Calpain I induces cleavage and release of apoptosis-inducing factor from isolated mitochondria. J Biol Chem. 2005;280(8):6447–6454. doi: 10.1074/jbc.M413269200. [DOI] [PubMed] [Google Scholar]
- 78.Lewis EM, Wilkinson AS, Davis NY, Horita DA, Wilkinson JC. Nondegradative ubiquitination of apoptosis inducing factor (AIF) by X-linked inhibitor of apoptosis at a residue critical for AIF-mediated chromatin degradation. Biochemistry. 2011;50(51):11084–11096. doi: 10.1021/bi201483g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wilkinson JC, Wilkinson AS, Galban S, Csomos RA, Duckett CS. Apoptosis-inducing factor is a target for ubiquitination through interaction with XIAP. Mol Cell Biol. 2008;28(1):237–247. doi: 10.1128/MCB.01065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luo Q, Wu X, Zhao P, et al. OTUD1 activates caspase-independent and caspase-dependent apoptosis by promoting AIF nuclear translocation and MCL1 degradation. Adv Sci (Weinh) 2021;8(8) doi: 10.1002/advs.202002874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lenhausen AM, Wilkinson AS, Lewis EM, et al. Apoptosis inducing factor binding protein PGAM5 triggers mitophagic cell death that is inhibited by the ubiquitin ligase activity of X-linked inhibitor of apoptosis. Biochemistry. 2016;55(23):3285–3302. doi: 10.1021/acs.biochem.6b00306. [DOI] [PubMed] [Google Scholar]
- 82.Holze C, Michaudel C, Mackowiak C, et al. Oxeiptosis, a ROS-induced caspase-independent apoptosis-like cell-death pathway. Nat Immunol. 2018;19(2):130–140. doi: 10.1038/s41590-017-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu B, Ma J, Li J, Wang D, Wang Z, Wang S. Mitochondrial phosphatase PGAM5 modulates cellular senescence by regulating mitochondrial dynamics. Nat Commun. 2020;11(1):2549. doi: 10.1038/s41467-020-16312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bernkopf DB, Jalal K, Bruckner M, et al. Pgam5 released from damaged mitochondria induces mitochondrial biogenesis via Wnt signaling. J Cell Biol. 2018;217(4):1383–1394. doi: 10.1083/jcb.201708191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghezzi D, Sevrioukova I, Invernizzi F, et al. Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor. Am J Hum Genet. 2010;86(4):639–649. doi: 10.1016/j.ajhg.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sevrioukova IF. Structure/function relations in AIFM1 variants associated with neurodegenerative disorders. J Mol Biol. 2016 doi: 10.1016/j.jmb.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 87.Berger I, Ben-Neriah Z, Dor-Wolman T, et al. Early prenatal ventriculomegaly due to an AIFM1 mutation identified by linkage analysis and whole exome sequencing. Mol Genet Metab. 2011;104(4):517–520. doi: 10.1016/j.ymgme.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 88.Rinaldi C, Grunseich C, Sevrioukova IF, et al. Cowchock syndrome is associated with a mutation in apoptosis-inducing factor. Am J Hum Genet. 2012;91(6):1095–1102. doi: 10.1016/j.ajhg.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cowchock FS, Duckett SW, Streletz LJ, Graziani LJ, Jackson LG. X-linked motor-sensory neuropathy type-II with deafness and mental retardation: a new disorder. Am J Med Genet. 1985;20(2):307–315. doi: 10.1002/ajmg.1320200214. [DOI] [PubMed] [Google Scholar]
- 90.Diodato D, Tasca G, Verrigni D, et al. A novel AIFM1 mutation expands the phenotype to an infantile motor neuron disease. Eur J Hum Genet. 2016;24(3):463–466. doi: 10.1038/ejhg.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kettwig M, Schubach M, Zimmermann FA, et al. From ventriculomegaly to severe muscular atrophy: expansion of the clinical spectrum related to mutations in AIFM1. Mitochondrion. 2015;21C:12–18. doi: 10.1016/j.mito.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 92.Moss T, May M, Flanagan-Steet H, et al. Severe multisystem pathology, metabolic acidosis, mitochondrial dysfunction, and early death associated with an X-linked AIFM1 variant. Cold Spring Harb Mol Case Stud. 2021;7(3) doi: 10.1101/mcs.a006081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morton SU, Prabhu SP, Lidov HG, et al. AIFM1 mutation presenting with fatal encephalomyopathy and mitochondrial disease in an infant. Cold Spring Harb Mol Case Stud. 2017;3(2) doi: 10.1101/mcs.a001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heimer G, Eyal E, Zhu X, et al. Mutations in AIFM1 cause an X-linked childhood cerebellar ataxia partially responsive to riboflavin. Eur J Paediatr Neurol. 2018;22(1):93–101. doi: 10.1016/j.ejpn.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 95.Ardissone A, Piscosquito G, Legati A, et al. A slowly progressive mitochondrial encephalomyopathy widens the spectrum of AIFM1 disorders. Neurology. 2015;84(21):2193–2195. doi: 10.1212/WNL.0000000000001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bogdanova-Mihaylova P, Alexander MD, Murphy RP, et al. Clinical spectrum of AIFM1-associated disease in an Irish family, from mild neuropathy to severe cerebellar ataxia with colour blindness. J Peripher Nerv Syst. 2019;24(4):348–353. doi: 10.1111/jns.12348. [DOI] [PubMed] [Google Scholar]
- 97.Hu B, Wang M, Castoro R, et al. A novel missense mutation in AIFM1 results in axonal polyneuropathy and misassembly of OXPHOS complexes. Eur J Neurol. 2017;24(12):1499–1506. doi: 10.1111/ene.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Depierreux F, Alkan S. Atypical hemifacial spasm and myoclonus related to AIFM1 variant. Neuropediatrics. 2022;53(3):217. doi: 10.1055/s-0042-1744159. [DOI] [PubMed] [Google Scholar]
- 99.Miyake N, Wolf NI, Cayami FK, et al. X-linked hypomyelination with spondylometaphyseal dysplasia (H-SMD) associated with mutations in AIFM1. Neurogenetics. 2017;18(4):185–194. doi: 10.1007/s10048-017-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mierzewska H, Rydzanicz M, Bieganski T, et al. Spondyloepimetaphyseal dysplasia with neurodegeneration associated with AIFM1 mutation - a novel phenotype of the mitochondrial disease. Clin Genet. 2017;91(1):30–37. doi: 10.1111/cge.12792. [DOI] [PubMed] [Google Scholar]
- 101.Neubauer BA, Stefanova I, Hubner CA, et al. A new type of leukoencephalopathy with metaphyseal chondrodysplasia maps to Xq25-q27. Neurology. 2006;67(4):587–591. doi: 10.1212/01.wnl.0000230133.11951.75. [DOI] [PubMed] [Google Scholar]
- 102.Kimura-Ohba S, Kagitani-Shimono K, Hashimoto N, et al. A case of cerebral hypomyelination with spondylo-epi-metaphyseal dysplasia. Am J Med Genet A. 2013;161A(1):203–207. doi: 10.1002/ajmg.a.35686. [DOI] [PubMed] [Google Scholar]
- 103.Bieganski T, Dawydzik B, Kozlowski K. Spondylo-epimetaphyseal dysplasia: a new X-linked variant with mental retardation. Eur J Pediatr. 1999;158(10):809–814. doi: 10.1007/s004310051211. [DOI] [PubMed] [Google Scholar]
- 104.Pandolfo M, Rai M, Remiche G, Desmyter L, Vandernoot I. Cerebellar ataxia, neuropathy, hearing loss, and intellectual disability due to AIFM1 mutation. Neurol Genet. 2020;6(3):e420. doi: 10.1212/NXG.0000000000000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kawarai T, Yamazaki H, Yamakami K, et al. A novel AIFM1 missense mutation in a Japanese patient with ataxic sensory neuronopathy and hearing impairment. J Neurol Sci. 2020;409 doi: 10.1016/j.jns.2019.116584. [DOI] [PubMed] [Google Scholar]
- 106.Wang Q, Xingxing L, Ding Z, Qi Y, Liu Y. Whole exome sequencing identifies a novel variant in an apoptosis-inducing factor gene associated with X-linked recessive hearing loss in a Chinese family. Genet Mol Biol. 2019;42(3):543–548. doi: 10.1590/1678-4685-GMB-2018-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pronicka E, Piekutowska-Abramczuk D, Ciara E, et al. New perspective in diagnostics of mitochondrial disorders: two years' experience with whole-exome sequencing at a national paediatric centre. J Transl Med. 2016;14(1):174. doi: 10.1186/s12967-016-0930-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Peng Q, Ma K, Wang L, et al. Case report: a novel intronic mutation in AIFM1 associated with fatal encephalomyopathy and mitochondrial disease in infant. Front Pediatr. 2022;10 doi: 10.3389/fped.2022.889089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang H, Bing D, Li J, et al. High frequency of AIFM1 variants and phenotype progression of auditory neuropathy in a Chinese population. Neural Plast. 2020;2020 doi: 10.1155/2020/5625768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zong L, Guan J, Ealy M, et al. Mutations in apoptosis-inducing factor cause X-linked recessive auditory neuropathy spectrum disorder. J Med Genet. 2015;52(8):523–531. doi: 10.1136/jmedgenet-2014-102961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Elrharchi S, Riahi Z, Salime S, et al. Novel mutation in AIFM1 gene associated with X-linked deafness in a moroccan family. Hum Hered. 2020;85(1):35–39. doi: 10.1159/000512712. [DOI] [PubMed] [Google Scholar]
- 112.Joza N, Oudit GY, Brown D, et al. Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Mol Cell Biol. 2005;25(23):10261–10272. doi: 10.1128/MCB.25.23.10261-10272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brown D, Yu BD, Joza N, et al. Loss of Aif function causes cell death in the mouse embryo, but the temporal progression of patterning is normal. Proc Nat Acad Sci USA. 2006;103(26):9918–9923. doi: 10.1073/pnas.0603950103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Benit P, Goncalves S, Dassa EP, Briere JJ, Rustin P. The variability of the harlequin mouse phenotype resembles that of human mitochondrial-complex I-deficiency syndromes. PLoS One. 2008;3(9):e3208. doi: 10.1371/journal.pone.0003208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.El Ghouzzi V, Csaba Z, Olivier P, et al. Apoptosis-inducing factor deficiency induces early mitochondrial degeneration in brain followed by progressive multifocal neuropathology. J Neuropathol Exp Neurol. 2007;66(9):838–847. doi: 10.1097/NEN.0b013e318148b822. [DOI] [PubMed] [Google Scholar]
- 116.Hintze M, Griesing S, Michels M, et al. Alopecia in Harlequin mutant mice is associated with reduced AIF protein levels and expression of retroviral elements. Mamm Genome. 2021;32(1):12–29. doi: 10.1007/s00335-020-09854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Benit P, Pelhaitre A, Saunier E, et al. Paradoxical inhibition of glycolysis by pioglitazone opposes the mitochondriopathy caused by AIF deficiency. EBioMedicine. 2017;17:75–87. doi: 10.1016/j.ebiom.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schiff M, Benit P, El-Khoury R, Schlemmer D, Benoist JF, Rustin P. Mouse studies to shape clinical trials for mitochondrial diseases: high fat diet in Harlequin mice. PLoS One. 2011;6(12):e28823. doi: 10.1371/journal.pone.0028823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Delavallee L, Mathiah N, Cabon L, et al. Mitochondrial AIF loss causes metabolic reprogramming, caspase-independent cell death blockade, embryonic lethality, and perinatal hydrocephalus. Mol Metab. 2020;40 doi: 10.1016/j.molmet.2020.101027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cabon L, Bertaux A, Brunelle-Navas MN, et al. AIF loss deregulates hematopoiesis and reveals different adaptive metabolic responses in bone marrow cells and thymocytes. Cell Death Differ. 2018 doi: 10.1038/s41418-017-0035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Timper K, Paeger L, Sanchez-Lasheras C, et al. Mild impairment of mitochondrial OXPHOS promotes fatty acid utilization in POMC neurons and improves glucose homeostasis in obesity. Cell Rep. 2018;25(2):383–397.e10. doi: 10.1016/j.celrep.2018.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Khan NA, Nikkanen J, Yatsuga S, et al. mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab. 2017;26(2):419–428.e5. doi: 10.1016/j.cmet.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 123.Ising C, Koehler S, Brahler S, et al. Inhibition of insulin/IGF-1 receptor signaling protects from mitochondria-mediated kidney failure. EMBO Mol Med. 2015;7(3):275–287. doi: 10.15252/emmm.201404916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Johnson SC, Yanos ME, Kayser EB, et al. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342(6165):1524–1528. doi: 10.1126/science.1244360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van de Wal M, Adjobo-Hermans M, Keijer J, et al. Ndufs4 knockout mouse models of Leigh syndrome: pathophysiology and intervention. Brain. 2021 doi: 10.1093/brain/awab426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Susin SA, Zamzami N, Castedo M, et al. Bcl-2 inhibits the mitochondrial release of an apoptogenic protease. J Exp Med. 1996;184(4):1331–1341. doi: 10.1084/jem.184.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang B, Li X, Wang J, et al. A novel AIFM1 mutation in a Chinese family with X-linked charcot-marie-tooth disease type 4. Neuromuscul Disord. 2018;28(8):652–659. doi: 10.1016/j.nmd.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 128.Sancho P, Sanchez-Monteagudo A, Collado A, et al. A newly distal hereditary motor neuropathy caused by a rare AIFM1 mutation. Neurogenetics. 2017 doi: 10.1007/s10048-017-0524-6. [DOI] [PubMed] [Google Scholar]
- 129.Liu S, Zhou M, Ruan Z, et al. AIF3 splicing switch triggers neurodegeneration. Mol Neurodegener. 2021;16(1):25. doi: 10.1186/s13024-021-00442-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Varadi M, Anyango S, Deshpande M, et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50(D1):D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]