Summary
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron BA.2 was a dominant circulating SARS-CoV-2 variant worldwide. Recent reports hint that BA.2 is similarly potent regarding antibody evasion but may be more transmissible than BA.1. The pathogenicity of BA.2 remains unclear and is of critical public health significance. Here we investigated the virological features and pathogenicity of BA.2 with in vitro and in vivo models. We show that BA.2 is less dependent on transmembrane protease serine 2 (TMPRSS2) for virus entry in comparison with BA.1 in vitro. In K18-hACE2 mice, BA.2 replicates more efficiently than BA.1 in the nasal turbinates and replicates marginally less efficiently in the lungs, leading to decreased body weight loss and improved survival. Our study indicates that BA.2 is similarly attenuated in lungs compared with BA.1 but is potentially more transmissible because of its better replication at the nasal turbinates.
Keywords: SARS-CoV-2, COVID-19, Omicron, BA.1, BA.2, replication, pathogenicity, TMPRSS2
Graphical abstract
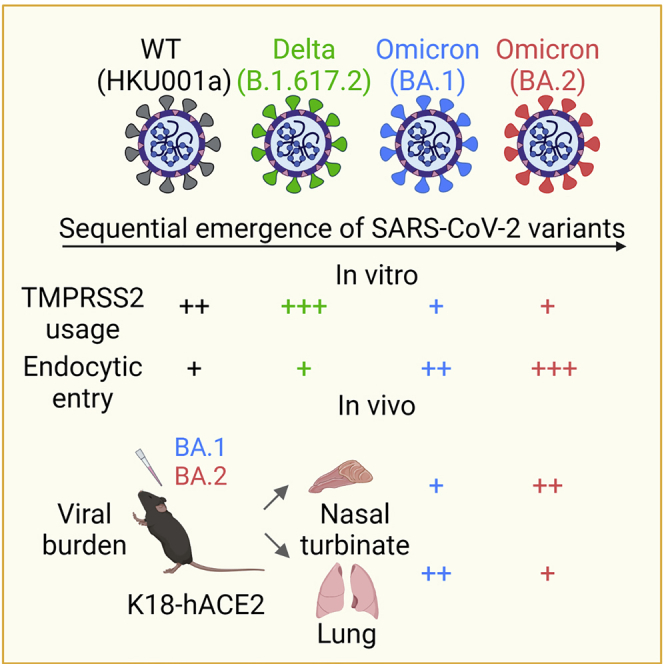
Highlights
-
•
BA.2 is less dependent on TMPRSS2 for virus entry compared with BA.1 in vitro
-
•
BA.2 depends more on endocytic entry than plasma membrane entry compared with BA.1
-
•
BA.2 replication is increased in NT and decreased in the lungs compared with BA.1
-
•
BA.2 is similarly attenuated in mouse lungs compared with BA.1
Chan et al. investigate the virological features and pathogenicity of BA.2. They find that BA.2 is less efficient regarding plasma membrane entry compared with BA.1. In K18-hACE2 mice, BA.2 replicates more efficiently in the nasal turbinates but less efficiently in the lungs compared with BA.1.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron BA.1 (PANGO lineage B.1.1.529.1) variant was first discovered in South Africa in late November 2021 and has caused an explosive upsurge of coronavirus disease 2019 (COVID-19) cases, quickly replacing the Delta (B.1.617.2) variant as the dominant circulating variant.1 BA.1 is characterized by a large number of changes in its spike protein, including 30 amino acid substitutions, three short deletions, and an insertion, compared with the ancestral SARS-CoV-2. As a result of these changes, BA.1 is highly immunoevasive to antibodies elicited by previous infection and vaccines, allowing the variant to cause reinfection and vaccine-breakthrough infection.2, 3, 4, 5 We6 and others7 have recently demonstrated that the pathogenicity of BA.1 is substantially attenuated in vivo compared to the wild-type (WT) SARS-CoV-2 and previous variants of concern (VOCs). In keeping with our animal model findings, BA.1 exhibited lower clinical severity in humans.8, 9, 10, 11, 12, 13 Mechanistically, we and others have shown that the attenuated pathogenicity of BA.1 is associated with reduced spike cleavage at the S1/S2 site, leading to inefficient usage of the host transmembrane protease, serine 2 (TMPRSS2) for entry.6,14,15
More recently, surveillance of SARS-CoV-2 evolution has revealed that Omicron BA.2 (B.1.1.529.2) has rapidly replaced BA.1 and BA.1.1 as the predominant circulating SARS-CoV-2 variant (Figure 1A), which may be due to its higher transmissibility compared with BA.1.16 Similar to BA.1, BA.2 demonstrates a robust capacity of antibody evasion.17,18 However, the virological features and intrinsic pathogenicity of BA.2 in comparison with that of BA.1 remain incompletely understood.
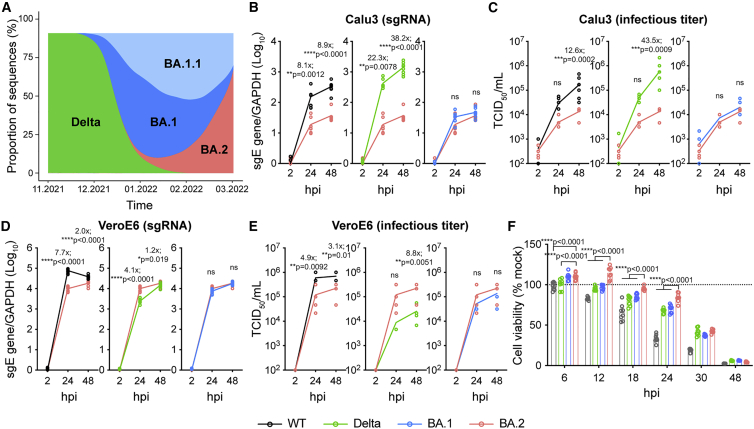
Figure 1.
Virus replication kinetics of BA.2
(A) The change in proportion of SARS-CoV-2 lineages deposited in GISAID from November 2021 to March 2022. The x axis indicates the collection date. The y axis indicates the proportion of the selected SARS-CoV-2 lineages.
(B–E) Cells were challenged with SARS-CoV-2 WT, Delta, BA.1, or BA.2 at 0.5 MOI (Calu3) or 0.1 MOI (VeroE6). Cell lysates were harvested at the designated time points for quantification of the subgenomic RNA of the envelope (sgE) gene (n = 8) (B and D). Infectious viral particles were titrated with a 50% tissue culture infectious dose (TCID50) assay (n = 8) (C and E). The same BA.2 curves were used for statistical comparison between WT, Delta, and BA.1 (B–E).
(F) Cell viability of VeroE6-TMPRSS2 cells infected with the wild type (WT), Delta, BA.1, or BA.2 at 0.1 MOI was quantified at the designated time points (n = 8).
Data represents mean ± SD from the indicated number of biological repeats. Statistical significance was determined with two way-ANOVA (B–F). Data were obtained from three independent experiments. Each data point represents one biological repeat. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. ns, not statistically significant.
Results
Virological characteristics of BA.2 in vitro
To explore the virological characteristics of BA.2, we first compared the replication efficiency of BA.2 with that of SARS-CoV-2 WT, Delta, and Omicron BA.1 in Calu3 human lung epithelial cells, which predominantly support the TMPRSS2-mediated plasma membrane entry pathway for SARS-CoV-2 infection.19,20 By measuring the subgenomic envelope (sgE) gene, which represents replication intermediates (Figure 1B), and TCID50, which represents infectious virus titer (Figure 1C), our results suggested that replication of BA.2 was significantly attenuated in Calu3 cells in comparison with the WT and Delta but comparable with that of BA.1. In VeroE6 cells, which predominantly support the endocytic entry pathway for SARS-CoV-2 infection because of deficient TMPRSS2 expression,19,20 BA.2 replicated less efficiently than WT but more efficiently than Delta and was at a similar level compared with BA.1 (Figures 1D and 1E). BA.2 demonstrated a trend of poorer replication in Calu3 cells and better replication in VeroE6 cells in comparison with BA.1, although the differences were not statistically significant (Figures 1C and 1E). Next we evaluated the cytotoxicity of BA.2 in VeroE6-TMPRSS2 cells. Our results demonstrated that BA.2 infection resulted in significantly less cell damage compared with the WT, Delta, and BA.1 from 6–24 h post infection (hpi) (Figure 1F).
We and others recently demonstrated that BA.1 is attenuated in infecting and replicating in lung cells because of its reduced spike cleavage at the S1/S2, site leading to inefficient usage of TMPRSS2 for entry.6,14,15 To assess the efficiency of TMPRSS2 usage of BA.2, we overexpressed ACE2 with or without TMPRSS2 in 293T cells and challenged the cells with pseudoviruses bearing the spike proteins of SARS-CoV-2 WT, Delta, BA.1, or BA.2. Our results revealed that, although BA.1 is already defective in TMPRSS2 usage compared with the WT and Delta, BA.2 is less efficient in utilizing TMPRSS2 for entry than BA.1 (1.5-fold, p = 0.0186) (Figure 2A). In parallel, we evaluated pseudovirus entry of the WT, Delta, BA.1, and BA.2 in VeroE6 and VeroE6-TMPRSS2 cells. Our results similarly showed that BA.2 is less sensitive than BA.1 by additional expression of TMPRSS2 in VeroE6 cells (1.3-fold, p = 0.0334) (Figure 2B). We evaluated the effect of camostat, a pan-serine protease inhibitor, on pseudovirus entry of the WT, Delta, BA.1, and BA.2 in VeroE6-TMPRSS2 cells. Our data demonstrated that 1 μM of camostat significantly reduced entry of WT, Delta, and BA.1 pseudoviruses by 27.2% (p < 0.0001), 35.5% (p < 0.0001), and 21.4% (p < 0.0001), respectively, but did not significantly reduce pseudovirus entry of BA.2. At higher concentrations (25 and 50 μM), BA.2 was consistently less susceptible to camostat inhibition compared with WT, Delta, and BA.1 (Figure 2C).
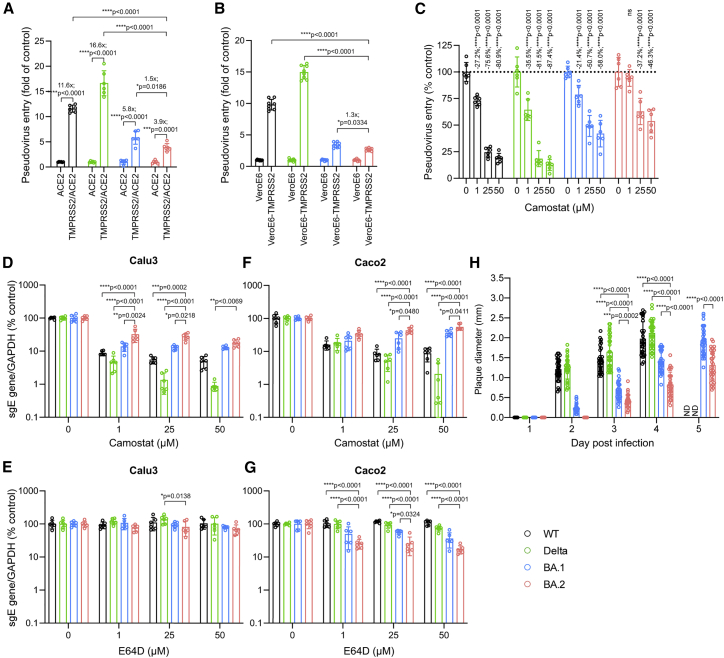
Figure 2.
Virological features of BA.2
(A) 293T cells were transfected with ACE2 or co-transfected with ACE2 and TMPRSS2, followed by transduction with pseudoviruses expressing the spike of SARS-CoV-2 WT, Delta, BA.1, or BA.2 at 24 h post transfection. Pseudovirus entry was quantified by measuring the luciferase signal (n = 6). Fold change in the luciferase signal was normalized to the mean luciferase readouts of cells with only ACE2 overexpression.
(B) VeroE6 and VeroE6-TMPRSS2 cells were transduced with pseudoviruses expressing the spike of WT, Delta, BA.1, or BA.2. Pseudovirus entry was quantified by measuring the luciferase signal (n = 8). Fold change in the luciferase signal was normalized to the mean luciferase readouts of VeroE6 cells.
(C) VeroE6-TMPRSS2 cells were pre-treated with 1, 25, or 50 μM camostat or DMSO for 2 h, followed by transduction with pseudoviruses expressing the spike of WT, Delta, BA.1, or BA.2 in the presence of camostat. Pseudovirus entry was quantified by measuring the luciferase signal of the cell lysates at 24 h post transduction (n = 6).
(D–G) Calu3 and Caco2 cells were pre-treated with 1, 25, or 50 μM camostat (D and F), E64D (E and G), or DMSO (D–G) for 2 h, followed by challenging the cells with authentic WT, Delta, BA.1, or BA.2. The amount of viral sgE RNA in the harvested cell lysates at 24 hpi was determined by qRT-PCR (n = 6).
(H) VeroE6-TMPRSS2 cells were infected with the WT, Delta, BA.1, or BA.2 and fixed with 4% paraformaldehyde at the designated time points, followed by crystal violet staining. Plaque diameters were measured by Adobe Photoshop. Plaque diameters for the WT and Delta at 5 dpi were too large to be measured.
See also Figures S1 and S2. Data represent mean ± SD from the indicated number of biological repeats. Statistical significance was determined with two way-ANOVA. Data were obtained from three independent experiments. Each data point represents one biological repeat. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. ND, not determined.
Next we evaluated the virus replication of authentic WT, Delta, BA.1, and BA.2 in Calu3 and Caco2 cells in the presence of camostat or E64D, an endosomal entry inhibitor of SARS-CoV-2. In Calu3 cells, BA.2 was significantly less susceptible to camostat inhibition than WT, Delta, and BA.1 at 1 μM and 25 μM (Figure 2D). E64D treatment in Calu3 cells is largely ineffective regarding entry of WT, Delta, BA.1, and BA.2 because SARS-CoV-2 entry is predominantly mediated by TMRPSS2 at the plasma membrane in this cell type (Figure 2E). In Caco2 cells, SARS-CoV-2 can enter via the plasma membrane and the endocytic pathway.21 In keeping with the findings from Calu3 cells, BA.2 was less sensitive to camostat inhibition in Caco2 cells compared with WT, Delta, and BA.1 (Figure 2F). In the presence of E64D, BA.2 was inhibited to a larger extent compared with WT, Delta, and BA.1 (Figure 2G). We next examined plaque formation and plaque size of WT-, Delta-, BA.1-, and BA.2-infected VeroE6-TMPRSS2 cells. Our data suggested that the plaques of BA.2 developed slower than those of the WT, Delta, and BA.1 and were not detected at 1–2 days post infection (dpi). At 3–5 dpi, the mean diameter of BA.2 plaques was significantly smaller than those of WT, Delta, and BA.1 (Figures 2H and S1). The smaller diameter of BA.2 plaques compared with those of BA.1 is also in keeping with the observation that cleavage of BA.2 spike protein is less efficient than that of BA.1 (Figure S2). Our in vitro results indicate that BA.2 is less dependent on TMPRSS2 for virus entry in comparison with BA.1.
Replication and pathogenicity of BA.2 in vivo
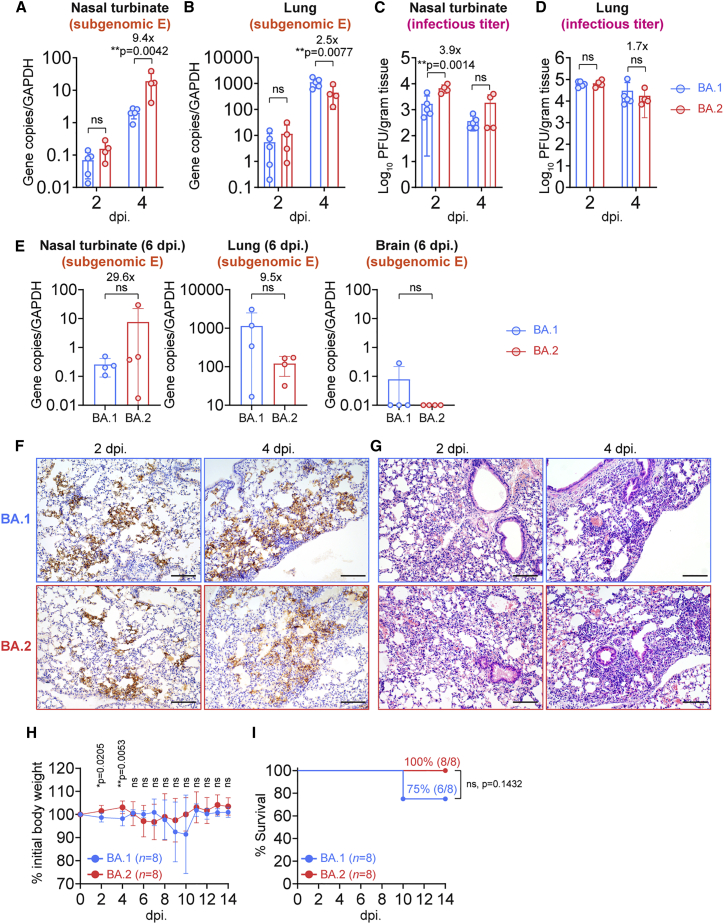
We and others have demonstrated recently that the pathogenicity of BA.1 is substantially attenuated in vivo compared with SARS-CoV-2 WT, Alpha, Beta, and Delta.6,7 To investigate the pathogenicity of BA.2 in vivo, we first compared virus propagation of BA.2 with that of BA.1 in the K18-hACE2 transgenic mouse model. To this end, 6- to 8-week-old K18-hACE2 transgenic mice were intranasally challenged with BA.2 or BA.1, and their tissues were collected for virological assessment. Our result demonstrated that BA.2 replicated more robustly than BA.1 in the nasal turbinate tissues of the infected mice. At 4 dpi, the sgE gene in nasal turbinate tissues of BA.2-infected mice was significantly (9.4-fold, p = 0.0042) higher than in BA.1-infected mice (Figure 3A). In contrast, our data revealed that BA.2 replicated less efficiently than BA.1 in the lung tissues of infected mice (Figure 3B), which is in keeping with the finding that BA.2 is less efficient in TMPRSS2 usage than BA.1 (Figures 2A–2H). At this time point, the sgE gene in lung tissues of BA.2-infected mice was significantly (2.5-fold, p = 0.0077) lower than in BA.1-infected mice (Figure 3B). Consistent with the sgE gene findings, the infectious virus titer in the nasal turbinate tissues was higher in BA.2-infected mice than in BA.1-infected mice (Figure 3C). In the lung samples, the infectious virus titer appeared to be lower in BA.2-infected mice compared with BA.1-infected mice, although the magnitude of the difference was small and did not reach statistical significance (Figure 3D). At 6 dpi, the trend of BA.2 replication compared with BA.1, including more efficient replication in the nasal turbinates and less efficient replication in the lungs, remained consistent (Figure 3E). Neither BA.2 nor BA.1 was readily detected in the mouse brains (Figure 3E). We next performed histopathological analysis of lung tissues from infected K18-hACE2 transgenic mice. We did not observe a significant difference in immunohistochemistry staining of viral antigen in the lungs; multi-focal expression of the viral nucleocapsid (N) protein was similarly observed in BA.2- and BA.1-infected mouse lungs on 2 dpi and 4 dpi (Figure 3F). Histological examination of BA.2-infected mouse lungs revealed localized inflammatory infiltrations and thickening of the alveolar septa (Figures 3G and S3), which was similar to BA.1-infected mouse lungs.
Figure 3.
Replication and pathogenicity of BA.2 in K18-hACE2 mice
6- to-8-week-old female and male K18-hACE2 transgenic mice were intranasally inoculated with 5 × 103 plaque-forming units (PFUs) BA.2 or BA.1. Nasal turbinates and lungs of infected mice were collected on 2, 4, or 6 dpi for viral burden determination (n = 4 for BA.2 on 2, 4, and 6 dpi; n = 5 for BA.1 on 2 and 4 dpi; n = 4 for BA.1 on 6 dpi). Brains of infected mice were collected on 6 dpi for viral burden determination (n = 4). Body weight and survival of the infected mice were monitored for 14 days (n = 8).
(A–E) Virological assessment of BA.2- and BA.1-infected K18-hACE2 mice. Subgenomic envelope (sgE) gene expression in nasal turbinates and lungs on 2 and 4 dpi was quantified with probe-specific qRT-PCR (A and B). Infectious viral titers in nasal turbinates and lungs on 2 and 4 dpi were quantified with plaque assays (C and D). SgE gene expression in nasal turbinates, lungs, and brains 6 dpi was quantified with probe-specific qRT-PCR (E).
(F) Representative images of immunohistochemistry staining for detection of N protein (brown) of SARS-CoV-2 in the lungs of infected mice (n = 5 for BA.1, n = 4 for BA.2).
(G) Representative images of hematoxylin and eosin (H&E) staining for detection of pathological changes in the lungs of infected mice (n = 5 for BA.1, n = 4 for BA.2). See also Figure S3.
(H and I) Body weight (H) and survival (I) of K18-hACE2 mice infected with BA.2 or BA.1.
Data represent mean ± SD from the indicated number of biological repeats. Statistical differences were determined with two-way ANOVA (A–D), two-tailed Student’s t test (E and H), or log rank (Mantel-Cox) test (I). Data were obtained from two independent experiments. Four to six sections were taken from each mouse for histology and immunochemistry analysis. Scale bars represent 100 μm. Each data point represents one biological repeat. ∗p < 0.05, ∗∗ p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
In keeping with the virological assessment findings of less efficient virus replication in the lungs, the body weight of BA.2-infected mice was significantly higher than that of BA.1-infected mice at early time points, including 2 dpi (p = 0.0205) and 4 dpi (p = 0.0053) (Figure 3H). Survival analysis demonstrated that, although 75% (6 of 8) of BA.1-infected K18-hACE2 mice survived the virus challenge, all (8 of 8) of the BA.2-infected mice survived the virus challenge with the same virus inoculum (Figure 3I).
Discussion
Omicron BA.2 has quickly replaced BA.1 as the predominant circulating SARS-CoV-2 variant worldwide. Recent reports have suggested that BA.2 is more transmissible than BA.116 and is similarly potent in antibody evasion.17,18 However, conclusive evidence is currently lacking regarding the virological features and intrinsic pathogenicity of BA.2 relative to that of BA.1. In this study, by combining a series of in vitro assays, we show that BA.2 is less dependent on TMPRSS2 for virus entry than BA.1. In K18-hACE2 mice, BA.2 replicates more efficiently than BA.1 in the nasal turbinates. In contrast, BA.2 replication in mouse lungs is marginally attenuated compared with BA.1, which results in less body weight loss and improved animal survival. Overall, the differences between BA.2 and BA.1 are small in magnitude compared with what we observed with BA.1 and previous variants,6 but the increased replication capacity of BA.2 in the nasal turbinates may explain the high transmissibility of BA.2. These findings are important for optimization of public health control measures of the ongoing COVID-19 pandemic.
Two recent studies have set out to explore the pathogenicity of BA.2.22,23 Yamasoba et al.23 compared the replication and pathogenicity of chimeric recombinant SARS-CoV-2 that encodes the S proteins of B.1.1, BA.1, and BA.2. Although they similarly observed reduced spike cleavage of BA.2 in comparison with BA.1 and B.1.1, they found that the pathogenicity of BA.2 is similar to that of the ancestral B.1.1 and higher than that of BA.1.23 The different findings between our studies are likely attributed to the non-spike substitutions between the genomes of BA.1 and BA.2, which warrant further investigations. Kawaoka et al.22 examined the pathogenicity of BA.2 in mice and hamsters.22 In BALB/c mice, they similarly showed that the virus titer in the lungs of BA.2-infected mice was lower than in BA.1-infected mice 5 dpi. In our study, we performed a side-by-side comparison of BA.2 and BA.1 by utilizing the K18-hACE2 mouse model, which has the advantage over the BALB/c mouse model of allowing survival analysis. By taking advantage of the higher sensitivity of the K18-hACE2 mouse model, we demonstrated a small but consistent difference between BA.2 and BA.1 in replication and pathogenicity.
The amino acid substitutions that contribute to the observed differences between BA.2 and BA.1 are currently unknown. BA.2 and BA.1 shares 21 substitutions in spike compared with the ancestral WT SARS-CoV-2. However, BA.2 contains 8 unique changes that are not present in BA.1, including 3 substitutions (T19I, A27S, and V213G) and 1 deletion (24–26 deletion) in the N-terminal domain (NTD) and 4 substitutions (S371F, T376A, D405N, R408S) in the receptor-binding domain (RBD). BA.2 also possesses changes in other non-spike regions that may also contribute to the observed phenotypic differences with BA.1. The importance and function of these changes should be further investigated with reverse genetics.
Limitations of the study
In the current study, we used the K18-hACE2 mouse model to compare the pathogenicity of BA.1 and BA.2. Although this model has been widely used for SARS-CoV-2 pathogenicity studies, it is known that the introduced hACE2 may not be expressed at physiological levels across different tissues. In this regard, hamsters and WT mice can be used for in-parallel analyses for a more comprehensive conclusion regarding the in vivo pathogenicity of BA.1 and BA.2.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-SARS-CoV-2 N immune serum | In house | N/A |
| Anti-VSV-G (8G5F11) antibody | Kerafast | Cat#: EB0010; RRID:AB_2811223 |
| Rabbit anti-SARS-CoV-2 spike S2 antibody | Sino Biological | Cat#: 40590-T62 |
| Mouse β-actin antibody (clone AC-74) | Sigma | Cat#: A5316; RRID:AB_476743 |
| Goat anti-Rabbit IgG(H + L) Secondary Antibody,HRP | Thermo Fisher | Cat#: 31460; RRID:AB_228341 |
| Goat anti-Mouse IgG(H + L) Secondary Antibody,HRP | Thermo Fisher | Cat#: 31430; RRID:AB_228307 |
| Bacterial and virus strains | ||
| SARS-CoV-2 HKU001a | Chu et al., 2020 | GENBANK: MT230904 |
| SARS-CoV-2 B.1.617.2 (Delta) | Shuai et al., 2021 | GENBANK: OM212471 |
| SARS-CoV-2 BA.1.529.1 (Omicron BA.1) | Shuai et al., 2021 | GENBANK: OM212472 |
| SARS-CoV-2 BA.1.529.2 (Omicron BA.2) | Iketani et al., 2022 | GISAID: EPI_ISL_9845731 |
| Chemicals, peptides, and recombinant proteins | ||
| SuperSignal West Pico PLUS Chemiluminescent Substrate | Thermo Fisher | Cat#: 34580 |
| Camostat | MedChemExpress | Cat#: HY-13512 |
| E64D | MedChemExpress | Cat#: HY-100229 |
| VectaMount® Permanent Mounting Medium | Vector Laboratories | Cat#: H-5000-60 |
| Critical commercial assays | ||
| DAB (3,3′-diaminobenzidine) substrate kit | Vector Laboratories | Cat#: SK-4100 |
| CellTiter-Glo luminescent cell viability assay kit | Promega | Cat#: G7572 |
| Luciferase assay system | Promega | Cat#: E1501 |
| QIAsymphony RNA Kit | Qiagen | Cat#: 931636 |
| RNeasy Mini kit | Qiagen | Cat#: 74106 |
| QuantiNova Probe RT-PCR Kit | Qiagen | Cat#: 208354 |
| Experimental models: Cell lines | ||
| Human lung adenocarcinoma: Calu3 | ATCC | HTB-55 |
| Human colorectal adenocarcinoma: Caco2 | ATCC | HTB-37 |
| Human embryonic kidney: 293T | ATCC | CRL-3216 |
| African green monkey kidney (clone of Vero-76): VeroE6 | ATCC | CRL-1586 |
| African green monkey: VeroE6-TMPRSS2 | Japanese Collection of Research Bioresources (JCRB) Cell Bank | JCRB1819 |
| Experimental models: Organisms/strains | ||
| Mouse: K18-hACE2 C57BL/6J (2B6 Cg-Tg(K18-ACE2)2Prlmn/J) | The Jackson Laboratory | Strain#:034860 |
| Oligonucleotides | ||
| E_Sarbeco_F1, (SARS-CoV-2), forward primer: 5’- CGATCTCTTGTAGATCTGTTCTC -3’ |
Integrated DNA Technologies | Cat#: 10006889 |
| E_Sarbeco_R2 (SARS-CoV-2), reverse primer: 5’- ATATTGCAGCAGTACGCACACA -3’ |
Integrated DNA Technologies | Cat#: 10006891 |
| E_Sarbeco_P1 (FAM) Probe (SARS-CoV-2) : 5’- FAM- ACACTAGCCATCCTTACTGC GCTTCG -ZEN-IBHQ -3’ |
Integrated DNA Technologies | Cat#: 10006893 |
| Recombinant DNA | ||
| pCAGEN-huACE2 | This paper | NCBI Reference Sequence: NM_001371415.1 |
| pCMV3-huTMPRSS2 | Sino biological | Cat#: HG13070-CH |
| Software and algorithms | ||
| GraphPad Prism 8.0 | Dotmatics | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Other | ||
| EnSight Multimode Microplate Reader | Perkin Elmer | https://www.perkinelmer.com/product/ensight-instrument-hh34000000 |
| TissueLyser II | Qiagen | https://www.qiagen.com/us/products/human-id-and-forensics/automation/tissuelyser-ii/ |
| Olympus BX53 light microscope | Olympus | https://www.olympus-lifescience.com/en/microscopes/upright/bx53f2/ |
| Alliance Imager apparatus | Uvitec | https://www.uvitec.co.uk/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Hin Chu (hinchu@hku.hk).
Materials availability
Materials generated in this study will be available upon fulfilment of material transfer agreement (MTA).
Data and code availability
-
•
All data reported in this study will be shared upon request from the lead contact.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and subject details
Viruses
Wild type SARS-CoV-2 HKU-001a (GenBank: MT230904), B.1.617.2/Delta (GenBank: OM212471), BA.1 (GenBank: OM212472), and BA.2 (Global Initiative on Sharing Avian Influenza Data, GISAID: EPI_ISL_9845731) were isolated from laboratory-confirmed COVID-19 patients in Hong Kong.24,25 All variants of SARS-CoV-2 were cultured and titrated by plaque assays using VeroE6-TMPRSS2 cells. Sequences of all variants used in this study were confirmed with nanopore sequencing. In vivo and in vitro experiments with infectious SARS-CoV-2 were performed according to the approved standard operating procedures of the Biosafety Level 3 facility at Department of Microbiology, HKU.
Cells
Caco2, 293T, and VeroE6 were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Amarillo, Texas, USA) containing 10% fetal bovine serum, 100 units penicillin, and 100ug/mL streptomycin. Calu3 was maintained in DMEM/F12 (Gibco) containing 10% fetal bovine serum, 100 units penicillin, and 100ug/mL streptomycin. VeroE6-TMPRSS2 was cultured in DMEM supplemented with 10% fetal bovine serum, 100 units penicillin,100ug/mL streptomycin and 2% G418. All cells were cultured at 37°C in an incubator with 5% CO2. All cell lines used are routinely tested for mycoplasma and are maintained mycoplasma-free.
Mice
The use of animals was approved by the Committee on the Use of Live Animals in Teaching and Research of The University of Hong Kong. Heterogenous K18-hACE2 C57BL/6J mice (2B6.Cg-Tg(K18-ACE2)2Prlmn/J) were obtained from The Jackson Laboratory. 6- to 10-week-old male and female K18-hACE2 were used for all in vivo experiments. Animals were kept in cages with individual ventilation with 65% humidity and ambient temperature ranging between 21-23 °C with 12-hour-interval day/night cycle for housing and husbandry.
Method details
In vivo virus challenge in mice
For virus challenge in mice, K18-hACE2 transgenic mice were anaesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg), followed by intranasal inoculation with 5 × 103 PFU of Omicron BA.1 or BA.2 diluted in 20 μL DMEM for each mouse. Mice were sacrificed at 2, 4, and 6 dpi for harvesting nasal turbinate, lung, or brain tissues for virological assessment and histological examination as we previously described.6,26 Survival and body weight of the infected animals were monitored for 14 days or until death of the animal.
Histology and immunohistochemistry staining
Animal tissues were harvested and fixed with 10% neutral-buffered formalin. Nasal turbinates were decalcified with 15% formic acid for 7 days before being processed with the TP1020 Leica semi-enclosed benchtop tissue processor. IHC was performed with the DAB (3,3′-diaminobenzidine) substrate kit (Vector Laboratories) as we previously described.27 The in-house rabbit anti-SARS-CoV-2 N immune serum (1:5000) was used to detect viral antigen, followed by color development with the DAB substrate kit. The nuclei were detected with haematoxylin before the tissue sections was mounted with the VectaMount permanent mounting medium (Vector Laboratories). For H&E staining, tissue sections were stained with Gill’s haematoxylin and eosin-Y. Images were acquired with the Olympus BX53 light microscope. Four to five mice were sampled each group (as specified in the figure legends) and four to six sections from each animal were used for histology analysis.
Infectious virus titration by plaque assays and TCID50 assays
To quantify infectious viral titer with plaque assays, nasal turbinates and lung tissues harvested from infected mice were homogenized in 1 mL DMEM with Tissue Lyzer II (Qiagen, Germany) and clarified supernatants were 10-fold serially diluted and inoculated onto a monolayer of VeroE6-TMPSS2 cells for 2 h at 37°C. After inoculation, the cells were covered with 1% low-melting agarose in DMEM with 1% FBS. After four days, the cells were fixed by 4% paraformaldehyde, followed by staining with 0.5% crystal violet in 25% ethanol/distilled water for plaque visualization. For TCID50 assays, supernatants from cells infected with SARS-CoV-2 WT, Delta, BA.1, or BA.2 were harvested and 10-fold serially diluted before inoculated onto VeroE6-TMPRSS2 cells. Cytopathic effect (CPE) was observed at four days post infection for the quantification of the median tissue culture infectious dose.
Cell viability assays
VeroE6-TMPRSS2 cells were seeded in 96-well plate and infected with SARS-CoV-2 WT, Delta, Omicron BA.1, and Omicron BA.2 at 0.1 MOI. Cell viability was quantified by CellTiter-Glo luminescent cell viability assay kit (Promega, USA) at 6 h, 12 h, 18 h, 24 h, 30 h and 48 h post infection. Briefly, culture medium was discarded and replaced with 100 μL DMEM medium containing 1% FBS and 1% penicillin-streptomycin (5000 U/mL). Then 100 μL CellTiter-Glo reagent was added to each well and the plate was incubated for 10 min at room temperature. 100 μL reaction was taken to the opaque-walled 96-well plate for reading the luminescence, following manufacturer’s manual with the EnSight Multimode Microplate Reader (Perkin Elmer, USA) at the designated time points.
Pseudovirus entry assays
All variants of SARS-CoV-2-spike pseudoviruses were packaged as previously described.6,28 Briefly, 293T cells were transfected with different spikes with Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA). At 24 h post transfection, the cells were transduced with VSV-deltaG-firefly pseudotyped with VSV-G. At 2 h post transduction, the cells were washed three times by phosphate-buffered saline (PBS) and cultured in DMEM containing 1%FBS and anti-VSV-G (8G5F11) antibody (EB0010, kerafast, Boston, MA, USA) (1:1000). The pseudoviruses were then harvested at 16 h post transduction titrated with TCID50. For pseudovirus entry assays, target cells were grown in 96-well plates and inoculated with pseudoviruses carrying respective SARS-CoV-2 spike for 2 h and cultured in media containing 1% FBS for 24 h. The cells were washed and lysed in passive lysis buffer (E194A, Promega, USA), followed by addition of luciferase assay reagent for detection of luciferase (E1501, Promega, Madison, WI, USA) according to manufacturer’s instructions.
RNA extraction and real-time reverse-transcription polymerase chain reaction
The infected cells were lysed with 90 μL RLT buffer then viral RNA was extracted and eluted in 100 μL DNase/RNase-free water using QIAsymphony RNA Kit (931636, Qiagen, Germantown Road Germantown, MD, USA). Viral RNA from mice lung and nasal turbinate samples were extracted with the RNeasy Mini kit (74106, Qiagen). Viral subgenomic RNA was detected using primers targeting the subgenomic E gene with the QuantiNova Probe RT-PCR Kit (208354, Qiagen). The primer and probe sequences are available upon request.
Protease inhibitor treatment assay
The serine protease inhibitor, camostat, and the cysteine protease inhibitor, E64D, were purchased from MedChemExpress (Monmouth Junction, NJ, USA). Calu3, Caco2, or VeroE6-TMPRSS2 cells were seeded in 96-well plates and treated with DMSO, Camostat, or E64D at concentration of 1, 25, and 50μM for 2h. Calu3 and Caco2 cells were challenged with viruses at 0.1 MOI or 0.5 MOI. At 24 hpi, the cell lysates were lysed in 90 μL RLT buffer and RNA were extracted for qRT-PCR quantification of virus replication. For pseudovirus transduction, the cells were incubated with different spikes of pseudovirus, followed by luciferase signal measurement at 24 hpi.
Western blot analysis of spike cleavage
VeroE6 cells were seeded in 6-well plates and infected with BA.2 or BA.1 at 0.1 MOI. Cell lysates were harvested in 200 μL RIPA buffer (89901, Thermo Scientific ) at 24 h post infection for the analysis of spike processing. The samples were subjected to 8% of SDS-PAGE and transferred to the PDVF membranes, followed by blocked with 5% skim milk in PBS for 2h at room temperature and incubated with specific primary antibodies at 4°C overnight, followed by incubating with horseradish peroxidase (HRP) conjugated secondary antibodies (Thermo Fisher Scientific) for 1h at room temperature. The signal was developed using SuperSignal West Pico PLUS Chemiluminescent Substrate (34580, Thermo Scientific, USA) and detected using Alliance Imager apparatus (Uvitec, Cambridge, UK). Full-length spike and S2 was detected with a rabbit anti-SARS-CoV-2 spike S2 antibody (40590-T62, Sino Biological) (1:5000). Nucleocapid (N) was detected with an in-house rabbit anti-SARS-CoV-2 N immune serum (1:10000) and β-actin was detected with a β-actin antibody (clone AC-74, A5316, Sigma, USA) (1:5000).
Quantification and statistical analysis
Statistical comparison between two experimental groups were performed with unpaired two-tailed Student’s t-test. Comparison between three or more experimental groups was performed with one-way or two-way ANOVA. Survival of animals were compared with Log- rank (Mantel-Cox) test. Differences were considered statistically significant when p < 0.05. Data analysis was performed with Graphpad prism 8.0.
Acknowledgments
This work was partly supported by funding from the Health and Medical Research Fund (CID-HKU1-5, COVID1903010-Projects 7 and 14, 20190652, and 20190572), the Food and Health Bureau, the Government of the Hong Kong Special Administrative Region; the General Research Fund (17118621 and 17123920), Collaborative Research Fund (C7060-21G), and Theme-Based Research Scheme (T11-709/21-N), the Research Grants Council of the Hong Kong Special Administrative Region; Health@InnoHK, Innovation and Technology Commission, the Government of the Hong Kong Special Administrative Region; the National Natural Science Foundation of China Excellent Young Scientists Fund (Hong Kong and Macau) (32122001); the National Program on Key Research Project of China (2020YFA0707500 and 2020YFA0707504); the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance for the Department of Health of the Hong Kong Special Administrative Region Government, Sanming Project of Medicine in Shenzhen, China (SZSM201911014); the High Level-Hospital Program, Health Commission of Guangdong Province, China; the University of Hong Kong Li Ka Shing Faculty of Medicine Enhanced New Staff Start-up Fund; the University of Hong Kong Outstanding Young Researcher Award; the University of Hong Kong Research Output Prize (Li Ka Shing Faculty of Medicine); the Major Science and Technology Program of Hainan Province (ZDKJ202003); the research project of the Hainan Academician Innovation Platform (YSPTZX202004); the Hainan Talent Development Project (SRC200003); the Emergency Collaborative Project (EKPG22-01) of Guangzhou Laboratory; the Emergency COVID-19 Project (2021YFC0866100), Major Projects on Public Security, National Key Research and Development Program; and donations from the Shaw Foundation Hong Kong, Richard Yu and Carol Yu, May Tam Mak Mei Yin, Michael Seak-Kan Tong, the Providence Foundation Limited (in memory of the late Lui Hac Minh), Lee Wan Keung Charity Foundation Limited, Hui Ming, Hui Hoy and Chow Sin Lan Charity Fund Limited, the Chen Wai Vivien Foundation Limited, Hong Kong Sanatorium & Hospital, Chan Yin Chuen Memorial Charitable Foundation, Marina Man-Wai Lee, the Hong Kong Hainan Commercial Association South China Microbiology Research Fund, the Jessie & George Ho Charitable Foundation, Perfect Shape Medical Limited, Kai Chong Tong, Foo Oi Foundation Limited, Tse Kam Ming Laurence, Betty Hing-Chu Lee, Ping Cham So, and the Lo Ying Shek Chi Wai Foundation. The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report.
Author contributions
J.F.-W.C., B.H., Y.C., and H.S. contributed equally to this work. J.F.-W.C. and H.C. had roles in study design, data collection, data analysis, data interpretation, literature search, and writing of the manuscript. B.H., Y.C., H.S., H.L., J.S., Y.L., C.Y., J. Zhang, J.-C.H., Y.H., X.H., T.T.-T.Y., T.Z., W.L., J.-P.C., C.L., C.C.-Y.Y., A.J.Z., J. Zhou, S.Y., B.-.Z.Z., J.-D.H., and K.K.-W.T. performed the experiments and/or analyzed the data. J.F.-W.C., K.-Y.Y., and H.C. oversaw the conception and supervised the study. J.F.-W.C., K.-Y.Y., and H.C. provided funding support.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects. We worked to ensure diversity in experimental samples through the selection of the cell lines.
Published: August 29, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100743.
Supplemental information
References
- 1.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., Sominsky L.A., Clark J.J., Adelsberg D.C., Bielak D.A., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 4.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., San J.E., Cromer D., Scheepers C., Amoako D.G., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L., Iketani S., Guo Y., Chan J.F.W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 6.Shuai H., Chan J.F., Hu B., Chai Y., Yuen T.T., Yin F., Huang X., Yoon C., Hu J.C., Liu H., et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022 doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 7.Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., Darling T.L., Joshi A., Loeber S., Singh G., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussey H., Davies M.A., Heekes A., Williamson C., Valley-Omar Z., Hardie D., Korsman S., Doolabh D., Preiser W., Maponga T., et al. Assessing the clinical severity of the Omicron variant in the Western Cape Province, South Africa, using the diagnostic PCR proxy marker of RdRp target delay to distinguish between Omicron and Delta infections - a survival analysis. Int. J. Infect. Dis. 2022;118:150–154. doi: 10.1016/j.ijid.2022.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iuliano A.D., Brunkard J.M., Boehmer T.K., Peterson E., Adjei S., Binder A.M., Cobb S., Graff P., Hidalgo P., Panaggio M.J., et al. Trends in disease severity and health Care utilization during the early omicron variant period compared with previous SARS-CoV-2 high transmission periods - United States, december 2020-January 2022. MMWR Morb. Mortal. Wkly. Rep. 2022;71:146–152. doi: 10.15585/mmwr.mm7104e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolter N., Jassat W., Walaza S., Welch R., Moultrie H., Groome M., Amoako D.G., Everatt J., Bhiman J.N., Scheepers C., et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet. 2022;399:437–446. doi: 10.1016/S0140-6736(22)00017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L., Berger N.A., Kaelber D.C., Davis P.B., Volkow N.D., Xu R. COVID infection severity in children under 5 years old before and after Omicron emergence in the US. medRxiv. 2022 doi: 10.1101/2022.01.12.22269179. Preprint at. [DOI] [Google Scholar]
- 12.Nyberg T., Ferguson N.M., Nash S.G., Webster H.H., Flaxman S., Andrews N., Hinsley W., Bernal J.L., Kall M., Bhatt S., et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet. 2022 doi: 10.1016/S0140-6736(22)00462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewnard J.A., Hong V.X., Patel M.M., Kahn R., Lipsitch M., Tartof S.Y. Clinical outcomes associated with SARS-CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in southern California. Nat. Med. 2022 doi: 10.1038/s41591-022-01887-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng B., Abdullahi A., Ferreira I.A.T.M., Goonawardane N., Saito A., Kimura I., Yamasoba D., Gerber P.P., Fatihi S., Rathore S., et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts tropism and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki R., Yamasoba D., Kimura I., Wang L., Kishimoto M., Ito J., Morioka Y., Nao N., Nasser H., Uriu K., et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022;603:700–705. doi: 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyngse P.F. Transmission of SARS-CoV-2 omicron VOC subvariants BA.1 and BA.2: evidence from Danish households. medRxiv. 2022 doi: 10.1101/2022.01.28.22270044. Preprint at. [DOI] [Google Scholar]
- 17.Iketani S., Liu L., Guo Y., Liu L., Chan J.F.W., Huang Y., Wang M., Luo Y., Yu J., Chu H., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J., Collier A.R.Y., Rowe M., Mardas F., Ventura J.D., Wan H., Miller J., Powers O., Chung B., Siamatu M., et al. Neutralization of the SARS-CoV-2 omicron BA.1 and BA.2 variants. N. Engl. J. Med. 2022;386:1579–1580. doi: 10.1056/NEJMc2201849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koch J., Uckeley Z.M., Doldan P., Stanifer M., Boulant S., Lozach P.Y. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. EMBO J. 2021;40:e107821. doi: 10.15252/embj.2021107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawaoka Y., Uraki R., Kiso M., Iida S., Imai M., Takashita E., Kuroda M., Halfmann P., Loeber S., Maemura T., et al. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron/BA.2. Res Sq. 2022 doi: 10.21203/rs.3.rs-1375091/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamasoba D., Kimura I., Nasser H., Morioka Y., Nao N., Ito J., Uriu K., Tsuda M., Zahradnik J., Shirakawa K., et al. Virological characteristics of SARS-CoV-2 BA.2 variant. bioRxiv. 2022 doi: 10.1101/2022.02.14.480335. Preprint at. [DOI] [Google Scholar]
- 24.Chu H., Chan J.F.W., Yuen T.T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C.Y., Tsang J.O.L., Huang X., et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet. Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shuai H., Chan J.F.W., Yuen T.T.T., Yoon C., Hu J.C., Wen L., Hu B., Yang D., Wang Y., Hou Y., et al. Emerging SARS-CoV-2 variants expand species tropism to murines. EBioMedicine. 2021;73:103643. doi: 10.1016/j.ebiom.2021.103643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang B.Z., Shuai H., Gong H.R., Hu J.C., Yan B., Yuen T.T.T., Hu Y.F., Yoon C., Wang X.L., Hou Y., et al. Bacillus Calmette-Guerin-induced trained immunity protects against SARS-CoV-2 challenge in K18-hACE2 mice. JCI Insight. 2022;7:e157393. doi: 10.1172/jci.insight.157393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu H., Shuai H., Hou Y., Zhang X., Wen L., Huang X., Hu B., Yang D., Wang Y., Yoon C., et al. Targeting highly pathogenic coronavirus-induced apoptosis reduces viral pathogenesis and disease severity. Sci. Adv. 2021;7:eabf8577. doi: 10.1126/sciadv.abf8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu H., Hu B., Huang X., Chai Y., Zhou D., Wang Y., Shuai H., Yang D., Hou Y., Zhang X., et al. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat. Commun. 2021;12:134. doi: 10.1038/s41467-020-20457-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this study will be shared upon request from the lead contact.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.