Abstract
The tetranucleotide core recognition sequence (TTGC) of the sigma 54 promoter −12 recognition element was altered by random substitution. The resulting promoter mutants were characterized in vivo and in vitro. Deregulated promoters were identified, implying that this core element can mediate the response to enhancer-binding proteins. These promoters had in common a substitution at position −12 (consensus C), indicating its importance for keeping basal transcription in check. In another screen, nonfunctional promoters were identified. Their analysis indicated that positions −13 (consensus G) and −15 (consensus T) are important to maintain minimal promoter function. In vitro studies showed that the −13 and −15 positions contribute to closed-complex formation, whereas the −12 position has a stronger effect on recognition of the fork junction intermediate created during open-complex formation. Overall the data indicate that the −12 region core contains specific subsequences that direct the diverse RNA polymerase interactions required both to produce RNA and to restrict this RNA synthesis in the absence of activation.
Core sequences of promoters typically contain specific critical elements within 40 bp upstream from the transcription start site. There are often pairs of such sequences, which may have overlapping functions. Such pairs include the well known −10 and −35 sequences for the major bacterial promoters of Escherichia coli and related bacteria and TATA and initiator elements for major mammalian promoters (7, 21). Such elements may have multiple purposes. These include specifying the type of transcription components that will bind, contributing to the amount of mRNA that will be produced, and possibly contributing to the response to regulators. Although consensus sequences are simple to derive from analysis of databases, individual promoters rarely, if ever, match the consensus sequence. This suggests that core promoters sequences have evolved not just to give specific, abundant transcripts but to give physiologically appropriate amounts of mRNA. There have been relatively few studies on how these core sequences contribute to the regulation of RNA amounts.
Recently, we began to address this question in the case of promoters recognized by the sigma 54 form of bacterial RNA polymerase. Sigma 54 holoenzyme mediates enhancer-dependent transcription in bacteria (7, 13, 14, 19). The polymerase recognizes a pair of promoter elements termed the −12 and −24 elements (17, 18). The −24 element is always contacted when holoenzyme is bound, and it appears to be dominant in specifying promoter occupancy (11, 22, 30). However, the −12 element, with the central consensus sequence TTTGCA (29), also contributes to binding affinity (2, 26). The latter element may play a more complex role in RNA synthesis, beyond simply assisting in promoter recognition (2, 4, 15, 20, 24, 27, 29). Changes in the highly conserved GC doublet have long been known to have the potential to reduce binding affinity (1, 2) and transcription in vivo (4, 15, 16, 24, 29). However, we showed previously that the consensus −12 element did not specify the largest amount of RNA in vivo. Instead, changes in the upstream TTT consensus half of the element could increase transcription in conjunction with the wild-type downstream GCA half of the element (29).
Prior studies have suggested that the −12 region sequences can contribute to determining the level of basal transcription. One in vitro study showed that a nonconsensus sequence had a higher level of unregulated transcription than a consensus sequence (27). An in vivo study showed that a −12 region double mutation (D3) gave detectable RNA under conditions where the consensus promoter is fully repressed (29). Thus, the D3 mutation caused a defect in regulation, leading to leaky basal transcription. However, D3 was not a stronger promoter than the wild type under conditions of strong activation. These results imply that the −12 region can contribute not only to the specificity of transcription but also to its regulatory response.
As these prior studies used a limited series of site-directed mutants, it was not possible to explore the effects of the full range of DNA sequence changes on the function of the −12 region sequences. In the present study we used random mutagenesis within the core −12 sequences to make libraries of potential −12 region sequences. Two screens were used to attempt to identify mutants with different properties that might be specified by the −12 region. These screens were designed to find nucleotides that are most important for specifying minimal transcription and also those critical for proper regulation. The results identify nucleotides within this element that contribute to separate controls for regulatory response and production of RNA.
MATERIALS AND METHODS
Mutagenesis and screens.
The low-copy-number plasmid pRS415 (23), derived from pBR322, carries the lacZ gene. The new plasmid pRS415-M12 has lacZ expression driven by the consensus M12-glnHP2 promoter. This promoter contains the natural glnHP2 NtrC and integration host factor (IHF) binding sites and promoter elements except for the substitution of T to G (−14 in reference 6 [see reference 29]; renumbered −13 in this paper). Random mutagenesis of the −12 region and screening used pRS415-M12.
Two oligonucleotide mixtures were used for mutagenesis. The 33-mers contained glnH promoter sequences except for the TTGC from −15 to −12. Those four positions contained equal amounts of each of the four nucleotides. The two mixtures were used together to generate random substitutions within the TTGC sequence of the M12 promoter of pRS415-M12 by using a standard site-directed mutagenesis protocol (Stratagene). The resulting library was transformed into Epicurian Coli XL1-Blue supercompetent cells and was screened initially on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyryanoside)–Luria-Bertani medium (LB)–0.2% (NH4)2SO4–2% glucose plates. On these plates the parent consensus promoter M12 is white, whereas the bypass promoter D3 is blue, after overnight incubation at 37°C. Blue colonies were taken to be bypass mutants. Colonies that turned blue more slowly were not deemed to have passed the screen. White colonies were transferred to X-Gal–G-gln plates (500 ml of G-gln medium contains 5.25 g of K2HPO4, 2.25 g of KH2PO4, 7.5 g of agar, 10 ml of 20% glucose, 1.0 ml of thiamine [10.0 mg/ml], 0.215 ml of 1 M MgSO4, 1 g of l-glutamine, and 100 μg of ampicillin per ml). Colonies that remained white after overnight incubation at 37°C were taken to be nonfunctional mutants. Colonies that turned blue slowly were not deemed to have passed the screen. Promoter DNA from both types of colonies was sequenced by using standard dideoxy methods.
In vitro transcription.
Standard one-round in vitro transcription was used. The activated transcription reaction mixture contained 75 nM NtrC, 100 nM sigma 54, 25 nM IHF (a gift of Steven Goodman), 5 nM supercoiled DNA template pBR-M12 or M12 derivatives, 10 mM carbamyl phosphate, 0.25 U of E. coli core RNA polymerase (Epicentre Technology), 0.5 mM GTP, 0.5 mM CTP, 4 μCi of [32P]UTP, 50 μM unlabeled UTP, and 3 mM ATP in transcription buffer (50 mM HEPES [pH 7.8], 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 50 ng of bovine serum albumin, and 3.5% polyethylene glycol), in a total reaction volume of 10 μl.
The reaction mixture was preincubated without GTP, UTP, and CTP for 20 min at 37°C, and then the missing nucleotides and heparin (final concentration, 100 μg/ml) were added. After 10 min the reactions were stopped by the addition of urea-saturated formamide dye and the mixtures were loaded on 6% denaturing polyacrylamide gels for electrophoresis. The data were analyzed with a phosphorimager.
In vitro bypass transcription was at 47°C with the same components, except without NtrC. All components were preincubated for 5 min to form closed complexes in the absence of nucleotides and heparin. The nucleoside triphosphates, including radioactive UTP, were then added (without heparin). After 20 min, the reaction mixtures were processed as described above.
Band shift analysis.
The band shift analysis was as described previously (10). Briefly, the DNA probes were prepared by annealing two complementary DNA strands. The bottom strand always contained the sequence from −29 to +1. The complementary top strands were different lengths, being truncated at either +1, −9, or −12. The annealing mixture contained 4 pmol of labeled bottom-strand DNA and 6 pmol of top strand in 10 mM HEPES (pH 7.9)–80 mM NaCl. The mixture was boiled for 2 min and gradually cooled to room temperature. Annealing was monitored by 10% polyacrylamide gel electrophoresis.
Band shift assay mixture contained 10 nM DNA and 15 nM RNA polymerase. The assays were initiated by mixing sigma 54 and core polymerase on ice in a molar ratio as 2.5:1 for 30 min. The 10-μl reaction mixtures also contained 6.0 ng of dI-dC per μl in 1× buffer (50 mM HEPES-HCl [pH 7.9], 100 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 0.05 μg of bovine serum albumin per ml, 2.8% polyethylene glycol 8000). The mixtures were incubated at 37°C for 10 min and subjected to 5% polyacrylamide gel electrophoresis, which was run at 350 V, while bathed in ice. After electrophoresis, the radioactive bands were visualized and analyzed with a phosphorimager.
RESULTS
We created libraries by targeting the −12 region for random substitutions (Fig. 1). Oligonucleotides were prepared with a mixture of all four nucleotides present in critical positions. The remaining positions correspond to the wild-type glnH promoter sequence. These oligonucleotides were used as mutagenic primers in standard site-directed mutagenesis protocols (see Materials and Methods). Thus, the collection of mutated glnH promoters would be expected to contain randomly selected nucleotides in the mutagenized positions.
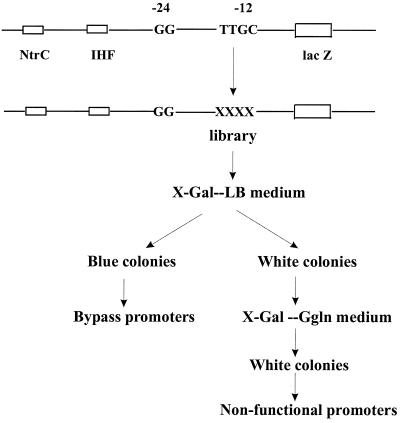
FIG. 1.
Scheme for isolation of −12 region mutations that allow deregulated expression (bypass) or lead to loss of expression (nonfunctional). The −12 region positions indicated by Xs were replaced randomly to create a library of promoters. Bypass mutants are blue on nonactivating LB. Nonfunctional mutants are white on activating G-gln medium. The plasmid contains the consensus M12 sequence in the context of the glnHP2 promoter with upstream IHF and NtrC sites.
Two libraries were created, one randomizing four nucleotides within the most highly conserved core of the −12 region (underlined in ATTTGCAT [29]) and one randomizing all eight nucleotides. The eight-nucleotide library gave an insufficient number of colonies, but the four-nucleotide library gave more than 4,000 colonies and therefore was subjected to screening. These four nucleotides are moderately well conserved; in a prior compilation the TTGC nucleotides are present in 11, 11, 15, and 16 of the 16 promoter sequences surveyed (29).
Screens (Fig. 1) were developed for loss of regulation and for loss of function. Two plate tests were used to assess whether sigma 54-dependent expression occurs under either activating or nonactivating conditions. The reference reporter in both screens is a plasmid containing the M12 promoter upstream from the beta-galactosidase gene; M12 is a glnHP2 derivative with a consensus −12 core sequence and is responsive to nitrogen availability through activator NtrC (6, 29).
The behavior of the M12 reporter plasmid in plate tests is consistent with properly regulated expression of the promoter. On G-gln plates, which are known to have low nitrogen availability, the transformed colonies were blue, indicative of promoter-directed beta-galactosidase expression. This is expected, as promoter expression is activated under these conditions. Below we use the loss of blue color on G-gln plates as an indicator of mutant promoters that have lost function. On nitrogen-rich LB plates, transformed colonies were white, as expected from the repression that accompanies the availability of excess nitrogen. Below we use the appearance of blue colonies on LB plates to indicate mutated promoters that have lost proper regulation.
Screen for promoters exhibiting unregulated basal transcription.
This screen for loss of proper regulation was tested for appropriateness by using the D3 mutant promoter (29). The D3 mutation is a double substitution within the tetranucleotide core sequence that will be subject to mutation in the screen. Although D3 has not previously been tested on plates, it was shown previously to yield detectable levels of mRNA in liquid LB (29). We found that D3 induced blue color under conditions in which the M12 wild-type parent remained white. We infer that the screen for blue colonies on LB–X-Gal plates can detect low levels of unregulated expression. We have previously termed mutants with this property bypass mutants (28, 29), and Fig. 1 outlines the protocol used to detect bypass promoter mutants.
A total of 184 of the 4,340 colonies obtained from mutagenesis showed obvious blue color after overnight incubation on X-Gal–LB plates. The 184 colonies were transferred to fresh LB–X-Gal plates and were found to maintain the blue color. As this number was sufficient for further analysis, colonies that turned blue more slowly were not analyzed further. Slightly more than half of the blue colonies (97 colonies) were sequenced, yielding 43 unique promoter sequences. None of these were wild type or contained just a single substitution; all 43 contained multiple substitutions restricted to the tetranucleotide that was targeted. Two promoters had double changes, 20 had triple changes, and 21 had quadruple changes. The D3 promoter sequence was one of the two double changes that passed the bypass screen. This D3 promoter is known to lose its bypass expression in a strain lacking sigma 54 (29). Thus, bypass expression should be sigma 54 dependent, a property consistent with the analysis of sequences and of in vitro transcription (see below).
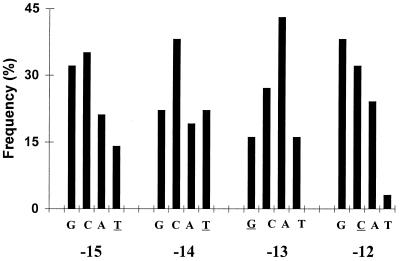
The frequency of mutation at each position was calculated and is presented in Fig. 2. A total of 148 nucleotide substitutions are represented in this collection. The numbering system used here (−15 to −12 for the sequence TTGC) differs from one used previously (29) to conform with systems used for other promoters (10). The most stunning result is that the C at position −12 (−12C) was not present in any of the 43 colonies. This 0% appearance of the consensus −12 nucleotide is far less than the 25% expected. The other consensus nucleotides were present at frequencies much closer to the 25% expected for random substitution, with frequencies of 16, 16, and 23% for −15T, −14T, and −13G, respectively. We infer that it is necessary to mutate the −12C in order to obtain bypass mutants.
FIG. 2.
Distribution of mutations obtained from the bypass library. The frequency of appearance of each nucleotide in each of the four promoter positions is shown. The consensus nucleotides are underlined.
The blue-white screen did not identify any promoters with single substitutions, even for −12C. This is consistent with prior study of site-directed −12C substitutions, which did not lead to detectable transcription in liquid LB (29). There were two double-substitution mutants that showed a bypass phenotype, and both contained a −12C substitution. One of these double substitutions had the same sequence as the D3 sequence from site-directed mutagenesis studies, which was shown previously to produce mRNA in a deregulated manner (29). The data indicate that in addition to the change of the −12C, at least one other substitution is required to obtain detectable unregulated transcription.
In order to see if changing −12C is strictly required, we made and screened another library. In this case only the trinucleotide TTG of the core tetranucleotide was subject to random substitution; the −12 position was held fixed as the wild-type C. If −12C substitution is required for unregulated transcription, no colonies in this library should pass the bypass screen. A total of 2,400 colonies were obtained, and 2 of these were blue. This 0.1% frequency is far less than the 4% obtained when the entire tetranucleotide core, including −12C, was targeted for mutagenesis. When the DNAs from these two colonies were sequenced, both turned out to have the −12C changed as well as to have other changes. This indicates that mutations within the TTG alone are unlikely to be sufficient for bypass transcription and supports the necessity for changes that must include substitution for the C at −12. The data in Fig. 2 show that there is a bias towards changing this C to T in the bypass library but that the substitution to thymine is not required. The key point is that the presence of the −12C is required to hold unregulated expression in check and that changing it is required to obtain −12 region-dependent bypass expression.
Screen for promoters of lowest function.
The original library, with the core TTGC tetranucleotide targeted, was also screened for promoters that directed the lowest expression. Colonies that were blue on LB–X-Gal plates were excluded from the screen. The remaining approximately 4,000 non-bypass colonies were transferred to G-gln–X-Gal plates, where activation causes the consensus M12 promoter to induce blue color formation. Seventy-two colonies were definitively white on these plates, and these were thus identified as being nonfunctional. Light blue colonies were not further analyzed.
Sequence analysis of plasmids from these 72 colonies revealed 37 unique promoter sequences. No single mutations were identified. This need for multiple mutations is consistent with prior experiments using single substitutions created by site-directed mutagenesis (29). In that case all of the single substitutions retained at least 30% of the M12 mRNA production level in liquid media under activating conditions. Four nonfunctional promoters contained double mutations, 22 contained triple mutations, and 11 contained quadruple mutations. Thus, a total of 118 nucleotide changes are represented in this collection. None of the 37 promoters had the same sequence as any of the 43 promoters that passed the screen for bypass expression.
The frequency with which each base appears in the nonfunctional library is displayed in Fig. 3. As there are 37 promoters, one would expect each substitution to be present in 9, based on random inclusion of the four nucleotides. The data indicate that there is a very strong bias against the −12C being mutated to a T; only 1 of the 37 promoters had this change. By contrast, all other possible substitutions in the four positions each occurred between 5 and 15 times, much closer to the expected 9 for random changes. Recall that the −12T, largely absent in this library, was strongly preferred among promoters that passed the bypass screen. Taken together, the data suggest that promoters with a −12C-to-T change have a high probability of being deregulated and thus giving some basal expression. This may be sufficient to yield an expression level that exceeds the level for indicating lack of function in this screen; that is, such colonies are likely to be blue.
FIG. 3.
Distribution of mutations obtained from the nonfunctional library. The frequency of appearance of each nucleotide in each of the four promoter positions is shown. The consensus nucleotides are underlined.
There are only minor differences in the frequencies with which each of the four positions are mutated in this nonfunctional library. The retention of the wild-type nucleotide was 14, 22, 16, and 32% for the −15T, −14T, −13G, and −12C, respectively. This indicates that the nonfunctional promoters have a stronger tendency to retain the −12C nucleotide and a greater tendency to substitute for the −15 and −13 nucleotides. The retention of the −12C has been discussed and probably relates to minimizing unregulated expression. The more frequent substitution at −15 and −13 may indicate that these positions are slightly more important in retention of minimal function.
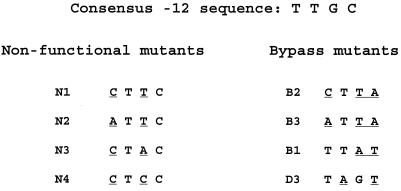
Further indications of which nucleotides are more important come from analysis of the individual nonfunctional promoters with the fewest sequence changes. Only four nonfunctional promoters had the minimum of two nucleotide changes (Fig. 4). All four of these promoters had substitutions in common positions, −15 and −13. This commonality supports the importance of these nucleotides, as weakly suggested by the analysis of the total spectrum of nonfunctional substitutions presented above.
FIG. 4.
−12 tetranucleotide sequences of the double-substitution mutants and two selected triple mutants. Substitutions within the consensus TTGC are underlined.
Overall, the analysis suggests that the −12C is required to prevent unregulated expression and that the −15T and the −13G are especially important in maintaining expression. Analysis of individual promoters lends further support to this view. Two of the nonfunctional promoters with double mutations (N1 and N2 in Fig. 4) are closely related to promoters from the bypass library (B2 and B3 in Fig. 4). Each nonfunctional mutant has a bypass partner with the same inactivating changes at both −15 and −13. However, each bypass partner has an additional change substituting for the −12C. This comparison suggests that the latter change could conceivably restore low-level unregulated expression even to a nonfunctional promoter.
In vitro transcription.
We tested several of the promoters from the library for transcription in vitro. The promoters were transferred into a vector with a downstream terminator to allow direct visualization of any RNA produced. They were transcribed in vitro by using activated NtrC and IHF in the same glnHP2 promoter context used for the in vivo selection. Six promoters were selected from the nonfunctional library. Four of these were the double mutants N1, N2, N3, and N4 (Fig. 4). These were chosen because they have the fewest substitutions and are thus more likely to show residual function in vitro; prior study of protein mutants showed that proteins testing as nonfunctional on plate tests could nonetheless show residual function in vitro (25). Two triple mutants (N5 and N6) were also chosen, as random examples of more highly mutated promoters.
Figure 5A shows the results of transcription of promoters from the nonfunctional library. The two triple mutants, N5 and N6, gave amounts of RNA that were at the limit of detection, less than 1% of the consensus M12 amount. Three of the four double mutants (N1, N3, and N4) gave RNA amounts that averaged 10% of the M12 level. One mutant (N2) gave substantial amounts of RNA, slightly more than half of the wild-type amount. Thus, five of the six nonfunctional mutants gave small amounts of mRNA in vitro, consistent with the lack of expression in vivo; double mutant N2 was the exception. Based on the very low level associated with the triple mutants, we expect that the large majority of promoters in this library, all triple or quadruple mutants, would show very low function in this test.
FIG. 5.
In vitro transcription. (A) Transcription of bypass (B and D3) and nonfunctional (N) mutants in the presence of NtrC under standard conditions is shown at the top. Amounts of RNA compared to those for the M12 consensus promoter are shown at the bottom. (B) Transcription at 47°C in the absence of NtrC. Lanes M12, markers for fully activated transcription from the consensus promoter under standard conditions. The arrows denote the correct transcript.
Mutants screened from the bypass library were also tested by in vitro transcription. In this case, the reaction mixtures lacked activator NtrC to attempt to mimic bypass expression in vivo. Preliminary experiments under standard conditions did not show detectable transcription without the activator (not shown). It is known that bypass transcription in vitro can be weak and is enhanced by altering solution conditions, particularly by lowering the ionic strength and raising the temperature (27). We explored such alterations in conditions to see if the bypass mutants could be distinguished from the M12 parent and also from the nonfunctional mutants. One condition, the use of 47°C, allowed the bypass mutants to be distinguished from the M12 parent and from the nonfunctional mutants. The best signal under these suboptimal conditions (Fig. 5B, left panel) corresponds to 5% or less of the activated signal from the consensus promoter. However, the weak bypass signal was detectable at the four promoters selected from the bypass library and was greater than that of the M12 control and greater than the signal from mutants screened from the nonfunctional library (Fig. 5B, right panel). We infer that bypass promoters can be distinguished from the M12 and nonfunctional promoters in vitro but that their transcription is very weak.
The bypass signal for the D3 promoter is at least twofold weaker than that seen previously in vivo for the same promoter in liquid LB (29). Collectively, signals for these bypass promoters are much weaker than signals from in vitro transcription with bypass mutants of the sigma 54 protein (data not shown and reference 27). Bypass promoters are also not transcribed very well in the presence of activator in vitro (Fig. 5A). They have not been studied in vivo, except for the strong D3 promoter, which is not severely defective in activated transcription. Their in vitro defects are discussed below.
DNA binding.
Recently, the pathway for promoter recognition and melting has been analyzed by using band shift assays with a variety of promoter probes (9, 10, 12). It was proposed (10) that open-complex formation includes the following three sequential steps: (i) formation of a closed complex, (ii) formation of a junction complex that includes a structure with −12 intact and an adjacent single-stranded segment, and (iii) spreading of melting to include binding to downstream single-stranded DNA. Optimal regulated transcription is presumed to require the use of all three of these interactions sequentially. These studies suggested that low-level bypass transcription could be triggered by inactivation of the protein-DNA junction complex. That is, proper formation of a −12/−11 junction complex is presumed to be required to keep regulation intact. If it is not formed due to a defect in either the protein or the DNA partner, then melting might spread downstream inappropriately and give unregulated bypass transcription.
We tested these ideas with the promoter mutants isolated from the two screens. Three band shift probes are relevant (Fig. 6A). Probe T1 corresponds to double-stranded promoter DNA, and binding to it assesses closed-complex formation. Probe T12 corresponds to a tight binding fork junction, and binding to it helps assess the ability to recognize the appropriate −12/−11 fork junction. Probe T9 has its junction in a downstream location, and binding to it helps assess downstream single-strand binding (10). We used the promoter mutants identified here, in the forms of these three probes, to assess the consequences of mutation.
FIG. 6.
Band shift analysis of mutant promoters in the form of various probes. (A) The three types of probes: T1 is double-stranded DNA, T9 has a double-strand–single-strand junction at position −9/−8, and T12 has a double-strand–single-strand junction at position −12/−11. The asterisks indicate the positions of radioactive labeling. Each group of three mutant probes uses a unique labeled bottom strand. Thus, the extent of binding between mutants may not be compared directly and was calculated by comparison to free probe run in parallel (not shown). (B) Top, holoenzyme binding to the M12 consensus sequence and the indicated mutant promoters in the forms of the three probes. Bottom, Row T1 shows the fraction of each probe shifted, normalized to M12. Rows T12/T1 and T9/T1 show the relevant binding ratios for each promoter.
Figure 6B shows the use of each promoter, in the form of each of the three probes, in a band shift protocol with sigma 54 holoenzyme. We compare the results for the bypass mutants to those for the nonfunctional mutants N1, N3, and N4. Results for nonfunctional mutant N2 are also presented, but recall that this mutant is unique among the nonfunctional mutants in exhibiting substantial function in vitro.
The binding to each probe for each promoter was measured several times, and the average data are compiled at the bottom of Fig. 6B. Double-strand binding for each promoter is normalized to that for the M12 consensus parent (line T1). −12 junction binding was assessed by determining the ratio of binding of the T12 probe to that of the T1 probe; the normalization to T1 ensures that any loss in signal of T12 binding is not a consequence of interactions specific to closed-complex formation. The data for T9 binding are similarly normalized to assess single-strand binding independent of double-strand binding.
First, we consider the results for the bypass mutants. The data (Fig. 6B) show that they have the strongest defects in recognition of the −12 fork junction. That is, the ratio of binding to the T12 fork probe compared to binding to the T1 double-strand probe is approximately fivefold less than that for M12 parent for this group. The mutants have relatively minor defects in the other interactions; the extent of T1 binding and the T9/T1 ratio for this group are 40 to 100% of the value for the M12 consensus. In this regard they resemble the previously obtained bypass mutants of sigma 54 protein, which also are defective in recognition of this critical fork junction. The results are consistent with qualitative data obtained recently for the D3 promoter (10). We infer that this group of mutants is most defective in recognition of the −12/−11 fork junction.
This loss of junction recognition is also typical of protein bypass mutants (10). However, in one regard these bypass promoter mutants do not resemble the bypass protein mutants. The bypass protein mutant ΔN works in part by unmasking both double-strand and single-strand binding determinants (5, 10). However, these promoter bypass mutants do not show increased T1 or T9 binding compared to the M12 parent (Fig. 5B). Thus, the bypass promoters show only one of the two altered properties associated with bypass proteins, probably accounting for the weaker bypass transcription from the promoters compared to the proteins.
Next, we consider the results for the nonfunctional mutants. The three mutants, N1, N3, and N4, that transcribe poorly in vitro have common behavior in the band shift assays. By far the strongest deviation from the M12 parent is in T1 binding. This binding is three- to fivefold less than from that of M12. By contrast, the T12/T1 ratio for these three mutants is reduced by less than a factor of 2. Thus, this group of nonfunctional mutants has band shift properties distinguishable from those of the group of bypass mutants. Bypass mutants have their strongest defect in junction recognition, whereas nonfunctional mutants have their strongest defect in closed-complex formation. The only exception among the eight mutants studied is N2, which functioned well in vitro but not in vivo and has a defect in junction recognition.
All of the eight mutants retain a recognition component that includes the single-stranded DNA downstream from the −12 region consensus sequence. This is indicated by the stronger binding to the T9 probe than to the T1 probe for all eight mutants (Fig. 6B, bottom row). However, as discussed above, none of the promoter mutants show the even greater preference for the T9 probe that was observed with strong bypass mutants of the sigma 54 protein (10). Thus, the data indicate that single-strand binding is preserved in this collection of promoters that have various mutations in the −12 region consensus sequence. We infer that the −12 consensus sequence does not have a direct effect on the ability to recognize downstream single-stranded DNA, although indirect effects are discussed below.
DISCUSSION
These experiments have demonstrated multiple roles for the nucleotides within the core of the −12 element of sigma 54-dependent promoters. Two clear roles have been identified, and the contributions of individual nucleotides to these roles have been assessed. The most unexpected result is that the presence of a single consensus nucleotide is necessary for ensuring that unregulated transcription does not occur. Such an observation has not been made for the large sigma 70 family of promoters. In those cases, deregulation has been observed only when nonconsensus core elements are made consensus by mutation; this can make a promoter such as lacUV5 so strong as to eliminate the requirement for activator. Thus, the use of consensus sequences to restrict basal transcription appears to be unique for the enhancer-dependent sigma 54 promoters. The data also show the more expected role for the −12 element of determining induced RNA levels. In the discussion that follows, we elaborate on how the −12 region sequence appears to control both the level and the regulatory response of promoter-dependent RNA synthesis.
Regulatory response.
Each of the 43 bypass promoters identified in the genetic screen had a substitution for the −12 consensus C. Another experiment showed that if this C is held fixed, one cannot isolate bypass mutants by changing other core −12 region positions. Thus, the retention of C at −12 is critical for maintaining proper regulation. In support of this, we note that the −12C is the most conserved nucleotide in natural promoters (17, 29). Nonetheless, substitution at −12 alone is not sufficient to give detectable levels of unregulated transcription in vivo (29). At least one additional substitution is required. Although the deregulated transcription can be detected in vitro, it appears to be stronger in vivo.
A common property of the deregulated promoters is a defect in RNA polymerase recognition when binding is tested with a probe that mimics a physiologically relevant fork junction. Band shift experiments showed a fivefold-lowered binding to such probes (probes T12) when they contained substitutions that deregulated promoter expression in vivo. By contrast, the deregulated promoters were recognized fairly well when in the form of a double-stranded probe, which mimics closed complexes (probes T1). Thus, the common defect is specifically related to fork junction recognition. The deregulated promoters have as a common feature the loss of C at −12, and so this nucleotide would appear to be critical for fork junction binding. This is reasonable, as the −12C is within the physiologically relevant fork junction itself; it is the terminal base pair at the upstream fork of the open complexes formed at sigma 54 promoters.
Recent work has independently led to the speculation that recognition of this junction is critical for regulation (10). The idea is that sigma 54 needs to be directed to this junction in order to prevent inappropriate downstream DNA recognition and melting in the absence of activator. The present data support that speculation by demonstrating that promoters with deregulated expression have in common a mutation within the terminal base pair of the junction that likely inhibits its recognition. However, these promoter mutations do not enhance binding to the downstream single-stranded regions, as occurred with bypass mutations in the sigma 54 protein (5, 10). This correlates with the weaker transcription from the promoter mutants. Overall, the data suggest that proper regulation is enhanced by −12 region-directed recognition of the appropriate fork junction and by proper masking of the full single-strand binding region of the protein component.
Minimal promoter function and a perspective on the function of the −12 region core.
No single nucleotide was dominant in the results of the genetic screen for loss of promoter function. The data showed a slight bias towards mutating the −13 and −15 positions. These same two positions were jointly mutated in the four double-substitution mutants that passed the screen for nonfunctional mutants. The appearance of these double mutants may be understood in the context of a prior study of site-directed single-substitution mutants (29) and the deregulated mutants just discussed. In that study no single substitution was strongly defective, consistent with the requirement for double substitution indicated in the present study.
The four double mutants represent the minimal changes that lead to loss of expression as judged by the loss of blue color in lacZ promoter fusions. These four promoters had the sequence from −15 to −12 of NTNC, as only the T and C are in common. The retention of the consensus −14T and −12C in this set of loss-of-function mutants is explicable in terms of other data. First, as just discussed, substitution for the −12C can lead to unregulated expression, likely allowing some inappropriate use of DNA downstream from the −12 region. Had this substitution occurred, the ensuing unregulated expression may have been sufficient to lead to blue color. Therefore, one expects the −12C to be retained in the double mutants, as was observed. Second, the −14T that is retained was shown to be particularly repressive in the prior study of single-substitution mutants. Therefore, its presence would be expected to contribute to maintaining low levels of expression. Thus, the NTNC sequence would be expected to give low levels of regulated expression.
These interpretations may be viewed in context to give an overview of the role of the −12 region. Although the −24 region is dominant for recruitment of sigma 54 polymerase (11, 30), the −12 region contributes to enhance closed-complex formation (2). Lowering the level of closed complexes can lead to lowering of expression levels (see above) (8). Thus, the sequence of the −12 region likely contributes to expression, in part, by specifying the extent of promoter occupancy.
The two halves of the consensus central −12 element appear to have separate but overlapping roles. Each half responds differently when mutated; single substitutions at TT can raise transcription, and single substitutions at GC can lower it (29). With regard to the GC, as shown above, retention of the −12C is critical for proper regulation, in the sense of preventing leaky transcription. The data also are consistent with the −13G being important for allowing an efficient positive response to the regulator. That is, when transcription for the 11 promoters (Fig. 5B) is normalized to promoter binding (T1) (Fig. 6, bottom) to calculate the response per occupied promoter, the two highest ratios are for D3 and M12; these are the only 2 of the 11 promoters to retain the −13G. Thus, the downstream half element is required for providing regulation, with a −12C contributing to negative regulation and a −13G probably contributing to positive regulation. This view is consistent with the appearance of this GC doublet in 15 of the 16 promoters recently surveyed (29).
The role of the −15 and −14 consensus TT appears to be different. Single changes here can increase RNA levels (29) but when coupled with changes in the −13G can decrease expression. Neither thymine is particularly important for regulation, so a role in determining maximal transcription is suggested. Seven of the 16 promoters surveyed do not retain Ts at both positions (29), so obviously the TT is dispensable to obtain transcription. Taken together, the data suggest that the TT most likely plays a role in modulating expression levels, making them appropriate to the need for the specific operon that is controlled by the promoter.
Overall, the central TTGC of the −12 region of the collected promoters may be considered as being constructed of two interrelated halves. Both halves can contribute to the affinity of closed-complex formation (3). The C of the highly conserved GC contributes to directing the polymerase to the precise position at which the melted fork junction will be created during activation; in its absence the restriction is lost and deregulation can occur. The TT is highly variable among promoters, and this variation creates sequences with the potential to direct promoter-specific differences in RNA levels. Thus, the two halves together ensure that appropriate amounts of properly regulated RNA are produced. Consensus elements for promoters for other holoenzymes have not been analyzed at this level of detail. In general, roles in specifying amounts of RNA are generally discussed more prominently than roles in regulation. It would be interesting to learn if the features found in this study are particular to sigma 54 promoters or are generally applicable.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant GM35754.
We thank S. Goodman (USC) for IHF and F. Govantes and R. Gunsalus (UCLA) for plasmids.
REFERENCES
- 1.Buck M, Cannon W. Activator-independent formation of a closed complex between sigma 54-holoenzyme and nifH and nifU promoters of Klebsiella pneumoniae. Mol Microbiol. 1992;6:1625–1630. doi: 10.1111/j.1365-2958.1992.tb00887.x. [DOI] [PubMed] [Google Scholar]
- 2.Buck M, Cannon W. Mutations in the RNA polymerase recognition sequence of the Klebsiella pneumoniae nifH promoter permitting transcriptional activation in the absence of NifA binding to upstream activator sequences. Nucleic Acids Res. 1989;17:2597–2612. doi: 10.1093/nar/17.7.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck M, Cannon W. Specific binding of the transcription factor sigma-54 to promoter DNA. Nature. 1992;358:422–424. doi: 10.1038/358422a0. [DOI] [PubMed] [Google Scholar]
- 4.Buck M, Khan H, Dixon R. Site-directed mutagenesis of the Klebsiella pneumoniae nifL and nifH promoters and in vivo analysis of promoter activity. Nucleic Acids Res. 1985;13:7621–7638. doi: 10.1093/nar/13.21.7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon W, Gallegos M T, Casaz P, Buck M. Amino-terminal sequences of sigmaN (sigma54) inhibit RNA polymerase isomerization. Genes Dev. 1999;13:357–370. doi: 10.1101/gad.13.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claverie-Martin F, Magasanik B. Positive and negative effects of DNA bending on activation of transcription from a distant site. J Mol Biol. 1992;227:996–1008. doi: 10.1016/0022-2836(92)90516-m. [DOI] [PubMed] [Google Scholar]
- 7.Gralla J D, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1232–1245. [Google Scholar]
- 8.Guo Y, Gralla J D. DNA-binding determinants of sigma 54 as deduced from libraries of mutations. J Bacteriol. 1997;179:1239–1245. doi: 10.1128/jb.179.4.1239-1245.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo Y, Gralla J D. Promoter opening via a DNA fork junction binding activity. Proc Natl Acad Sci USA. 1998;95:11655–11660. doi: 10.1073/pnas.95.20.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo Y, Wang L, Gralla J D. A fork junction DNA-protein switch that controls promoter melting by the bacterial enhancer-dependent sigma factor. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh M, Gralla J D. Analysis of the N-terminal leucine heptad and hexad repeats of sigma 54. J Mol Biol. 1994;239:15–24. doi: 10.1006/jmbi.1994.1347. [DOI] [PubMed] [Google Scholar]
- 12.Kelly M T, Hoover T R. Mutant forms of Salmonella typhimurium sigma 54 defective in transcription initiation but not promoter binding activity. J Bacteriol. 1999;181:3351–3357. doi: 10.1128/jb.181.11.3351-3357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kustu S, North A K, Weiss D S. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem Sci. 1991;16:397–402. doi: 10.1016/0968-0004(91)90163-p. [DOI] [PubMed] [Google Scholar]
- 14.Magasanik B. The regulation of nitrogen utilization in enteric bacteria. J Cell Biochem. 1993;51:34–40. doi: 10.1002/jcb.240510108. [DOI] [PubMed] [Google Scholar]
- 15.Martin-Verstraete I, Debarbouille M, Klier A, Rapoport G. Mutagenesis of the Bacillus subtilis “−12, −24” promoter of the levanase operon and evidence for the existence of an upstream activating sequence. J Mol Biol. 1992;226:85–99. doi: 10.1016/0022-2836(92)90126-5. [DOI] [PubMed] [Google Scholar]
- 16.Merrick M, Chambers S. The helix-turn-helix motif of sigma 54 is involved in recognition of the −13 promoter region. J Bacteriol. 1992;174:7221–7226. doi: 10.1128/jb.174.22.7221-7226.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merrick M J. In a class of its own—the RNA polymerase sigma factor sigma 54 (sigma N) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 18.Morett E, Buck M. In vitro studies on the interactions of DNA polymerase-sigma 54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters. The role of NifA in the formation of an open promoter complex. J Mol Biol. 1989;210:65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- 19.Ninfa A J, Reitzer L J, Magasanik B. Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell. 1987;50:1039–1046. doi: 10.1016/0092-8674(87)90170-x. [DOI] [PubMed] [Google Scholar]
- 20.Ray L, Claverie-Martin F, Weglenski P, Magasanik B. Role of the promoter in activation of transcription by nitrogen regulator I phosphate in Escherichia coli. J Bacteriol. 1990;172:818–823. doi: 10.1128/jb.172.2.818-823.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 22.Sasse-Dwight S, Gralla J D. Role of eukaryotic-type functional domains found in the prokaryotic enhancer receptor factor sigma 54. Cell. 1990;62:945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- 23.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 24.Stigter J, Schneider M, de Bruijn F J. Azorhizobium caulinodans nitrogen fixation (nif/fix) gene regulation: mutagenesis of the nifA −24/−12 promoter element, characterization of a ntrA(rpoN) gene, and derivation of a model. Mol Plant Microbe Interact. 1993;6:238–252. doi: 10.1094/mpmi-6-238. [DOI] [PubMed] [Google Scholar]
- 25.Syed A, Gralla J D. Identification of an N-terminal region of sigma 54 required for enhancer responsiveness. J Bacteriol. 1998;180:5619–5625. doi: 10.1128/jb.180.21.5619-5625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tintut Y, Wang J T, Gralla J D. A novel bacterial transcription cycle involving sigma(54) Genes Dev. 1995;9:2305–2313. doi: 10.1101/gad.9.18.2305. [DOI] [PubMed] [Google Scholar]
- 27.Wang J T, Syed A, Gralla J D. Multiple pathways to bypass the enhancer requirement of sigma 54 RNA polymerase: roles for DNA and protein determinants. Proc Natl Acad Sci USA. 1997;94:9538–9543. doi: 10.1073/pnas.94.18.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J T, Syed A, Hsieh M, Gralla J D. Converting Escherichia coli RNA polymerase into an enhancer-responsive enzyme: role of an NH2-terminal leucine patch in sigma 54. Science. 1995;270:992–994. doi: 10.1126/science.270.5238.992. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Gralla J D. Multiple in vivo roles for the −12-region elements of sigma 54 promoters. J Bacteriol. 1998;180:5626–5631. doi: 10.1128/jb.180.21.5626-5631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong C, Tintut Y, Gralla J D. The domain structure of sigma 54 as determined by analysis of a set of deletion mutants. J Mol Biol. 1994;236:81–90. doi: 10.1006/jmbi.1994.1120. [DOI] [PubMed] [Google Scholar]