Abstract
Expression of the breast cancer-associated gene 1 (BRCA1) in sporadic breast cancers is usually reduced, yet the underlying mechanisms remains elusive. To identify factors that are responsible for reduced BRCA1 expression, we screened 92 known transcription factors for their ability to regulate expression of BRCA1. Among several potential regulators, the Gli-Krueppel-related transcription factor Yin Yang 1 (YY1) showed the most dramatic transactivation of the BRCA1 promoter. YY1 binds to the promoter of BRCA1, and its overexpression resulted in increased expression of BRCA1 and a number of BRCA1 downstream genes. We further showed that overexpression of YY1 in cancer cells inhibited cell proliferation, foci formation and tumor growth in nude mice. To assess the clinical relevance between YY1 and BRCA1, we studied expression of YY1 and BRCA1 from human breast cancer samples and tissue arrays, and detected a significant positive correlation between the level of YY1 and BRCA1 expression in these cancers. Taken together, these findings suggest that YY1 is a key regulator of BRCA1 expression and may be causally linked to the molecular etiology of human breast cancer.
Keywords: BRCA1, breast cancer, promoter, nude mice, YY1
Introduction
Breast cancer is a major cause of cancer mortality in women and affects approximately one in eight women during their lifetime. Approximately 90% of breast cancers are sporadic while the remaining 10% are inheritable (Brody and Biesecker, 1998; Alberg et al., 1999; Eccles and Pichert, 2005; Zhang and Powell, 2005). Germline mutations in the breast cancer-associated gene 1 (BRCA1) have been detected in ~40% of familial breast cancer cases (reviewed in Brody and Biesecker (1998), Alberg et al. (1999) and Eccles and Pichert (2005)). Somatic mutations of BRCA1 are rarely detected in sporadic breast cancers; however, BRCA1 expression is low in most of these cancers (Turner et al., 2004b), suggesting that BRCA1 may be subjected to transcriptional regulation.
It is shown that promoters of many tumor suppressor genes are methylated in breast cancers, which accounts for the reduced expression of these genes (Oliveira et al., 2005; Giacinti et al., 2008; Rivenbark and Coleman, 2009). But, it turns out that the promoter of BRCA1 is methylated in only a small portion (~ 10%) of sporadic breast cancers (Birgisdottir et al., 2006). Thus, molecular mechanisms leading to reduced BRCA1 expression and/or decreased function in sporadic breast cancers remain unclear, suggesting that BRCA1 promoter silencing occurs by some other means in the majority of cases.
Several lines of evidence indicate that BRCA1 expression is subjected to both positive and negative regulation by transcription factors. Currently, several genes have been found to positively regulate expression of BRCA1 in cultured cells. This includes 53BP1 (Rauch et al., 2005), GA-binding protein ±/β (GABP-α/β; Atlas et al., 2000), cAMP-responsive element-binding protein (Atlas et al., 2001; Ghosh et al., 2008), E2F-1 (Wang et al., 2000), Caveolin-1 (Glait et al., 2006) and Hoxa9 (Gilbert et al., 2010). On the other hand, inhibitor of differentiation-4 (ID4; Beger et al., 2001), Ets-2 and SWI/SNF complex (Tan et al., 2003), GABP-α/β (MacDonald et al., 2007), high mobility group A1 (Baldassarre et al., 2003) and BP1, an isoform of DLX4 homeoprotein (Kluk et al., 2010) are identified as negative regulators of BRCA1 promoter activity. The BRCA1 promoter contains E2F DNA-binding sites that mediate transcriptional activation by E2F1 and repression by Rb (Wang et al. , 2000).
Yin Yang 1 (YY1) is a ubiquitously distributed transcription factor belonging to the Gli-Krueppel class of zinc-finger proteins (Park and Atchison, 1991; Shi et al., 1991). YY1 is a multifunctional protein and is involved in both repression and activation of many genes that have a role in various important biological processes (Shi et al., 1997; Gordon et al., 2006; Castellano et al., 2009). It was shown that YY1 positively regulates expression of several oncogenes, including E6 and E7, c-Myc, c-Fos and ErbB2 (Riggs et al., 1993; Lee et al., 1995a, b). YY1 also inactivates p53 activity through enhancing MDM2-mediated ubiquitination (Gronroos et al., 2004; Sui et al, 2004). It is, therefore, hypothesized that YY1 might act as an oncogene that promotes tumor growth and metastasis (Gordon et al., 2006).
On the other hand, YY1 serves as a positive regulator for a number of genes with tumor suppressor function, such as DnaJ-like heat shock protein 40, HLJ1 (Wang et al., 2005, 2007), p73 (Wu et al., 2007a) and p53 (Furlong et al., 1996) through various mechanisms. YY1 also acts as a negative regulator of cell growth through binding and inhibiting c-Myc function (Austen et al., 1998). Thus, it is conceivable that YY1 may regulate both oncogenes and tumor suppressor genes through different ways depending on the tissue context. Functions of YY1 have been studied in mice through gene targeting. However, the absence of YY1 results in peri-implantation lethality (Affar el et al., 2006), which makes it difficult to assess the role of YY1 in later stages of development and cancer formation.
To identify additional factors that may be responsible for reduced BRCA1 expression in sporadic breast cancers, we screened 92 transcription factors for their ability to regulate the BRCA1 promoter. Our data indicated that YY 1 is the most potent positive regulator of BRCA1 gene expression among several candidate genes examined and the expression of YY1 is causally linked to the molecular etiology of human breast cancer.
Results
Identification of YY1 as a positive regulator of BRCA1
The BRCA1 and the Near BRCA1 2 (NBR2) genes are separated by a short distance, with two promoters, α and β promoters, located in this region (Figure 1a). While both promoters are responsive to estrogen stimulation, promoter α is bi-directional and shared with the adjacent NBR2 gene (Xu et al., 1997a,b). To choose an appropriate construct for the screening of potential regulators of BRCA1 transcription, we generated two groups of BRCA1 promoter reporter constructs with varying lengths that cover both the α and β promoters (group 1, Figure 1a) or the α promoter only (group 2, Figure 1a). All constructs in group 2 had higher luciferase activities than their corresponding constructs in group 1 (Figure 1b), suggesting that there is a negative element in the β promoter. Constructs that contained additional sequences from −201 to −567 bp had markedly reduced activity, and the activity was reduced even further when the construct is longer (extended from −567 to −957 bp), suggesting that these regions also contain negative elements.
Figure 1.
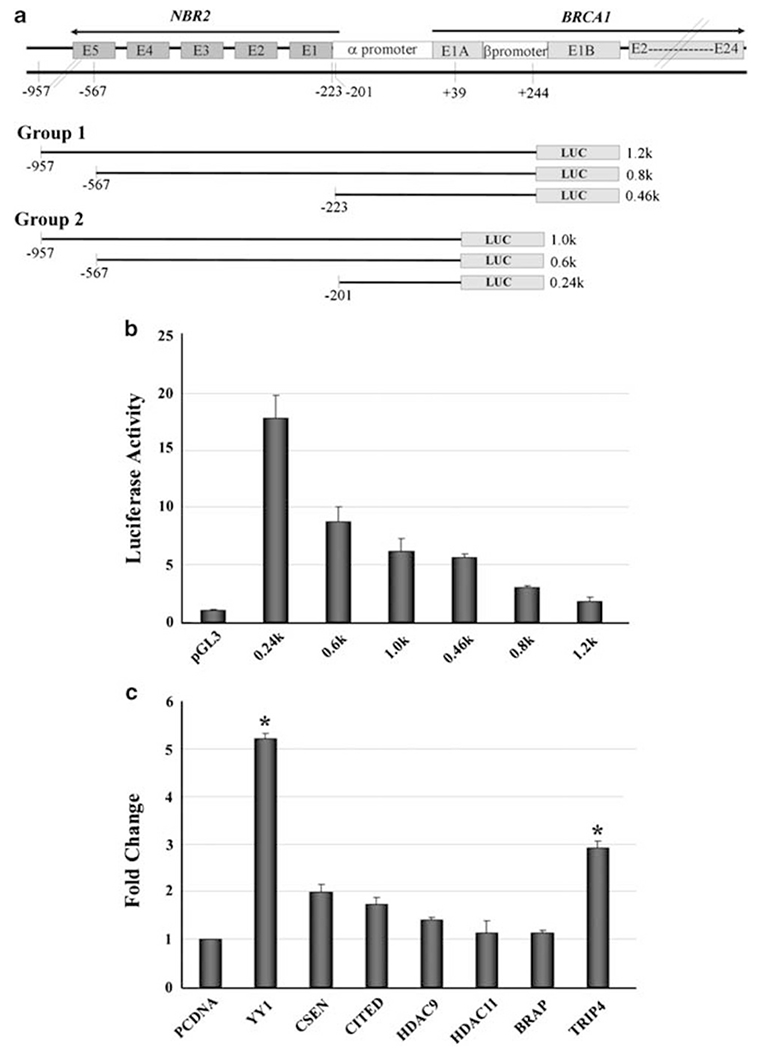
Identification of transcription factors that regulate BRCA1 expression. (a) Structures of BRCA1 and NRB promoter regions. Two groups of reporter constructs were used in this study. (b) Luciferase activities of BRCA1 reporter constructs in MCF-7 cells. (c) Fold change in luciferase activity of the 0.24-kb construct after transfection of YY1, CSEN, CITED, TRIP4 and HDAC9. Luciferase activity induced by YY1 and TRIP expression reached significant levels over pcDNA mock vector. *Student’s t test, P<0.05.
Using a BRCA1-0.24 kb construct that has the highest luciferase activity among all vectors, and a BRCA1-0.46 kb construct that has the highest luciferase activity among group 1 vectors, we screened 92 known transcription factors (Supplementary Table 1) for their ability to regulate BRCA1 using a reverse transfection in a 96-well format. Briefly, transcription factor cDNAs were pre-spotted on 96-well cell-culture plates as individual clones and then assayed by transfection with MCF-7 cells along with the BRCA1-0.24 kb, BRCA1-0.46 kb and PGL3 vector. There was significant increase of luciferase activity of both reporters (> 2.5-fold increase compared with a mock vector) after transfection of some of the transcription factors. This includes YY1, Cbp/p300-interacting transactivator, with Glu/Asp-rich CSEN (carboxy-terminal domain 1), calsenilin, presenilin-binding protein, EF-hand transcription factor (CITED), BRAP (BRCA1-associated protein), TRIP4 (thyroid hormone receptor interactor 4), histone deacetylase-9 (HDAC9) and HDAC11. To validate the data obtained from primary screening, we co-transfected these candidates individually into MCF-7 cells with the BRCA1-0.24kb reporter. We detected significantly increased luciferase activity after the transfection of YY1 and TRIP, reaching 5- and 3-fold, respectively, over the pcDNA mock vector (Figure 1c). Other candidates showed either no significant change (CSEN, CITED and HDAC9: 1.5- to 2-fold) or no change (HDAC11 and BRAP) (Figure 1c). Similar results were also obtained by transfecting these genes into HeLa cells (data not shown).
Our primary screening also showed that the expression of several transcription factors reduced luciferase activity of both reporters (> 2-fold decrease). This includes DNA binding 4 (ID4), GA-binding protein transcription factor, α subunit 60 kDa (GABP), PCAF (p300/CBP-associated factor), THRAP5 (thyroid hormone receptor-associated protein 5), DR1 (downreguator of transcription 1), and SMARCAD1 (SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1). Among them, previous investigations had identified ID4 (Beger et al., 2001) and GABP (MacDonald et al., 2007) as negative regulators of BRCA1 transcription. We found that the luciferase activity of the BRCA1-0.24 kb and BRCA1-0.46 kb constructs was reduced about 13- and 17-fold, respectively, upon transfection of ID4, and about 6- and 2-fold, respectively, upon transfection of GABP. These data confirmed the previous findings and provided a validation for the feasibility of our screening system for BRCA1 regulators. However, the study of these candidates that may serve as negative regulators of BRCA1 is beyond the scope of the current project and will be investigated separately in near future.
Because YY1 exhibited the highest transcriptional activity for BRCA1 reporters, we chose to further investigate the potential relationship between YY1 and BRCA1 in this study. To check if YY1 regulates endogenous BRCA1 expression, we expressed YY1 in three additional cell lines, HEK293, HeLa and U2OS. We showed that ectopic overexpression of YY1 increased the mRNA and protein levels of endogenous BRCA1 in both HEK293 and HeLa cells (Figures 2a and b and data not shown). U2OS cells contain a tetracycline (Tet)-off responsive YY1 construct (Sui et at., 2004), and Tet-mediated induction of YY1 also induced transcription of endogenous BRCA1 (Figure 2a). At the same time, BRCA1 protein level was elevated in both total lysate and nuclear extract in U2OS cells as detected by western blot analysis 48 h after withdrawing tetracycline (Figure 2c). Conversely, RNAi-mediated knockdown of YY1 using shRNA-based lentiviral infection decreased basal expression level of BRCA1 in MDA-MB-231 and T47D (Figure 2d). These data suggest that YY1 is capable of maintaining and upregulating the expression levels of BRCA1. Because levels of BRCA1 expression vary during cell-cycle progression (Jin et at., 1997; Ruffner and Verma, 1997; Okada and Ouchi, 2003), a caveat is whether altered expression of YY1 could cause profound perturb of distribution of the cell cycle, which secondarily affects BRCA1 expression. To investigate this, we analyzed cell-cycle distribution of two cell lines with ectopically overexpression of YY1 mediated by YY1 transfection (MDA-MB-231) or removal of Tet (U2SO). Our data indicated that YY1 overexpression in both cell lines induced G0-G1 population from 48.35 to 60.88% in MDA-MB-231 and 32.75–59.33% in U2SO cells, respectively, accompanied by a reduction of S and G2-M populations (Figure 2e). Because BRCA1 is expressed at higher levels in the S and G2-M phase (Wu et al., 2010) and our own observation (Supplementary Figure 1), these data rule out a possibility that the increased BRCA1 expression by YY1 overexpression is secondary to alterations in the cell cycle.
Figure 2.
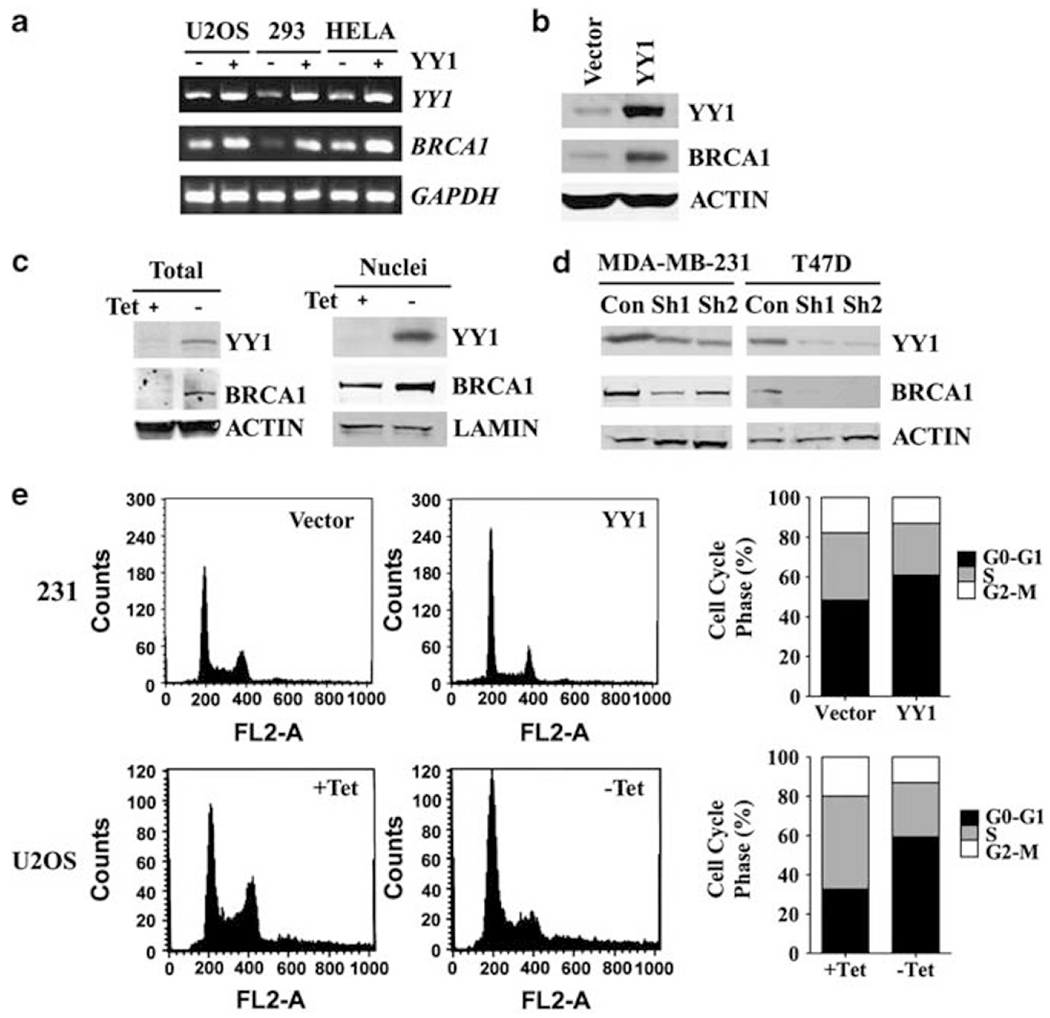
YY1 positively regulates BRCA1 expression. (a) YY1 positively regulated the endogenous BRCA1 expression as revealed by RT–PCR analysis in YY1-inducible U2OS cells (by tetracycline withdrawal, –Tet; Sui et al., 2004) and YY1 overexpression in HEK293 and HeLa cells. (b) Ectopic expression of YY1 in HeLa cells increased BRCA1 as revealed by western blot analysis. (c) Increased YY1 in U2OS cells induced BRCA1 protein level in the total protein lysates and nuclear extracts. (d) Reduced levels of YY1 and BRCA1 proteins in MDA-MB-231 and T47D breast cancer cell lines after YY1 knockdown by two lentiviral-based shRNA constructs that are specific to YY1 (Sh1 and Sh2) compared with shRNA control vector pLKO.1. (e) YY1 overexpression caused the increased G0-G1 population in the cell cycle in MDA-MB-231 and Tet-regulated U2OS cells.
YY1 upregulates BRCA1 expression by binding to its promoter
Our data so far suggest that YY1 may directly regulate BRCA1 expression. To define the essential regulatory element in the promoter of BRCA1 that interacts with YY1, we made serial deletion constructs from the BRCA1-0.24 kb construct (Figure 3a). Testing these serial deletion constructs in multiple cell lines, including MCF-7, HCC1937 and U2OS cells, we found that the element that responds to YY1 expression is located between 0.20 and 0.24 kb (−201 to −162) in MCF-7 cells. We also transfected these vectors into BRCA1-mutant HCC1937 cells and U2OS cells and obtained similar results (Figure 3a). We next used MatInspector (Genomatrix Software Inc., Ann Arbor, MI, USA) (version 5.0), which is a program that identifies DNA–protein interaction sites, to search these 40 nucleotides and identified a Gli-Krueppel-related transcription factor binding site, ccttttACGTcat (Figure 3b). Because YY1 is a member of the Gli-Krueppel-related transcription factors (Park and Atchison, 1991; Shi et al., 1991), we introduced two mutations in this site and found that both mutations significantly impaired induction of BRCA1 transcription by YY1 (Figure 3b).
Figure 3.
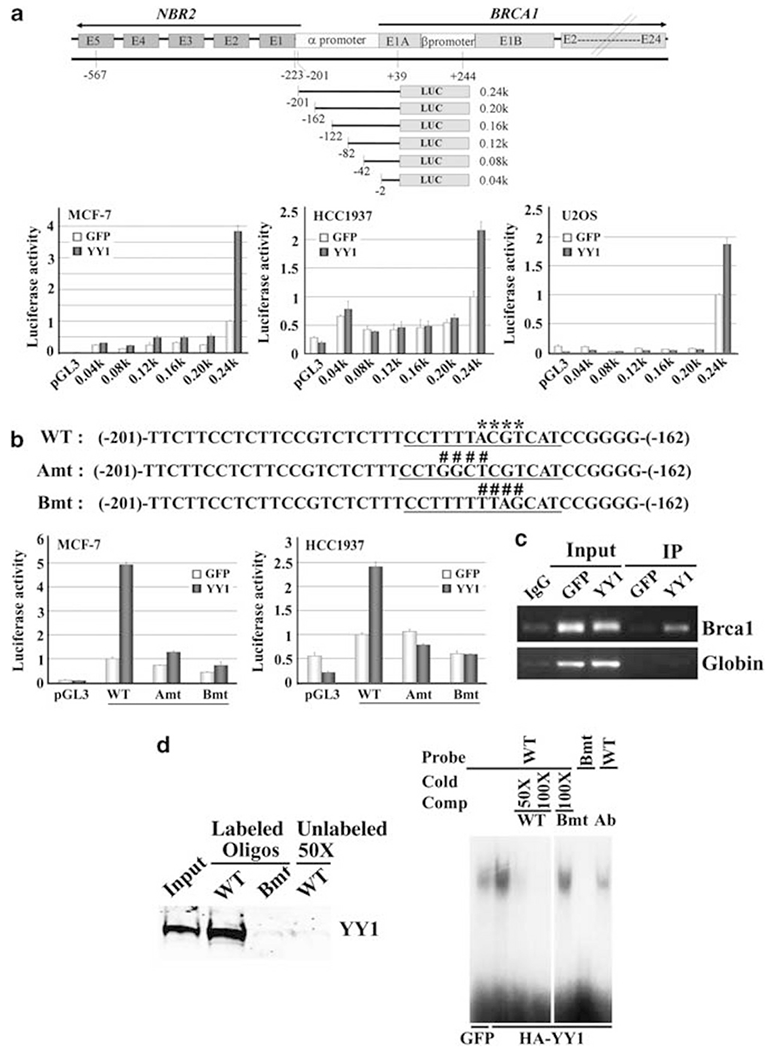
YY1 interacts with BRCA1 promoter and positively regulates its function. (a) Structures of BRCA1 and NRB promoter region and serial deletion from the 0.24-kb construct. Luciferase reporter assay of these constructs in MCF-7, HCC1937 and U2OS cells after co-transfection with constructs expressing either YY1 or GFP. (b) Wild-type (WT) and two mutant oligonucleotides (Amt and Bmt) within nucleotides −201 and −162. The putative Gli-Krueppel-related transcription factor YY1 binding site is underlined and the core sequence is marked with asterisks in the WT sequence. Both A and B mutations (Amt and Bmt, as marked with ####) diminished the induction of the BRCA1 promoter by YY1 in MCF-7 and HCC1937 cells. (c) ChIP assay to show YY1 binds to the BRCA1 promoter in MCF-7 cells transiently transfected with YY1. (d) Biotin-streptavidin pull-down assay (left side) and electrophoretic mobility shift assay (right side). Each reaction for biotin-streptavidin pull-down assay used 1 μg biotin-labeled oligonucleotide and 300 μg extract from HeLa cells transiently transfected with YY1. Electrophoretic mobility shift assay showing that the wild-type oligo, but not Bmt oligo, had reduced migration, which could be competed off in the presence of cold wild-type oligo, but not mutant oligo.
Next, we investigated whether YY1 could bind to the BRCA1 promoter through this region. Chromatin immunoprecipitation (ChIP) assay showed that YY1 binds to the BRCA1 promoter in MCF-7 cells (Figure 3c). We also used the same condition to test another promoter (β-globin) and could not detect any obvious binding. Using a biotin-streptavidin pull-down assay, we observed that the wild-type oligo, but not the mutant oligo, could pull down YY1 and this interaction could be competed off by using an excess amount of unlabeled wild-type oligo (Figure 3d). Electrophoretic mobility shift assays showed that the wild-type oligo, but not the mutant oligo, showed reduced migration, which could be competed off in the presence of cold wild-type oligo, but not the mutant oligo (Figure 3d). The addition of an antibody to YY1 significantly diminished the interaction with the oligo (Figure 3d). These data suggest that YY1 binds to the BRCA1 promoter through this site.
Co-expression of YY1 and BRCA1 during mammary gland development
BRCA1 is expressed in mammary epithelial cells. If YY1 positively regulates BRCA1, they should co-express in same cells. To investigate this, we examined expression of YY1 and BRCA1 in the mouse mammary gland. Our western blot analysis revealed a significant change of both YY1 and BRCA1 along mammary cycle progression. Both YY1 and BRCA1 levels are low in virgin stage, significantly increased during the pregnancy and lactation stages and decreased in involution stage (Figure 4a). To view their expression at cellular level, we performed immunofluorescence staining and detected colocalization of them in mammary epithelial cells (Figure 4b). In pregnant and lactation stages, when levels of both proteins are high, these proteins are detected in the nucleus of all cells (Figure 4b, over 200 cells counted). In the virgin and involutions stages, when the levels of both proteins are low, our staining did not detect an obvious signaling (Figure 4b, over 200 cells counted). Finally, we performed siRNA-mediated knockdown experiment and found that siRNA specific to YY1, but not to control siRNA, in cultured cells significantly decreased expression of BRCA1 (Figure 4c). These data are consistent with our finding that YY1 positively regulates BRCA1.
Figure 4.
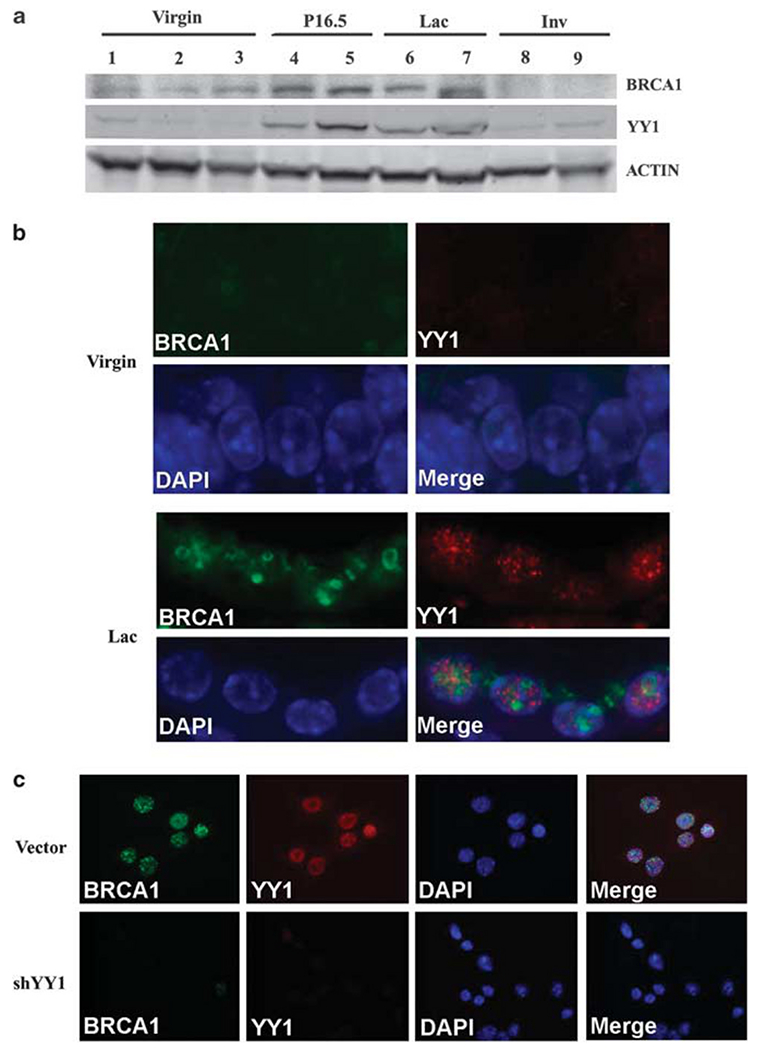
Colocalized expression of YY1 and BRCA1. (a) Expression of YY1 and BRCA1 during the mammary cycle of progression. Both proteins have higher levels in pregnant day 16.5 and lactation day 2. (b) Immunofluorescence staining revealed colocalization of BRCA1 and YY1 in the nucleus of the same mammary epithelial cells of lactation gland, while the expression levels of both proteins were beyond detection level of the same experimental condition. (c) Infection of lentivirus carrying YY1-specific shRNA, but not vector control lentivirus, in cultured mouse mammary tumor cell lines (MMTV-ras) reduces BRCA1 expression.
Ectopic overexpression of YY1 caused cell-cycle arrest and inhibited tumor growth
Positive regulation of BRCA1 implicates YY1 in tumorigenesis. To investigate this, we infected YY1 into several cancer cell lines and studied their growth both in vitro and in vivo. We found that overexpression of YY1 in MDA-MB-231, 92J (a cell line that was derived from MDA-MB-231 but formed more aggressive tumors in nude mice) and HeLa cells reduced cell proliferation as revealed by [3-(4,5-dimethylthiazo(-2-yl)-2.5-diphenyltetrazoliam bromide] (MTT) assay (Figure 5a). YY1-transfected cells also exhibited a significant reduction in the number and size of colonies compared with vector-transfected cells (Figures 5b and c). To investigate whether this effect is mediated by BRCA1, we infected YY1 lentivirus into MDA-MB-231 cells that carry a shRNA lentivirus specific for BRCA1 (about 90% knockdown) or a control vector (Figure 5d). Our data showed that the growth inhibition and colony formation induced by YY1 were blocked in the shBRCA1 cells (Figures 5a and b). A similar effect was also observed in MCF-7 cells (data not shown).
Figure 5.
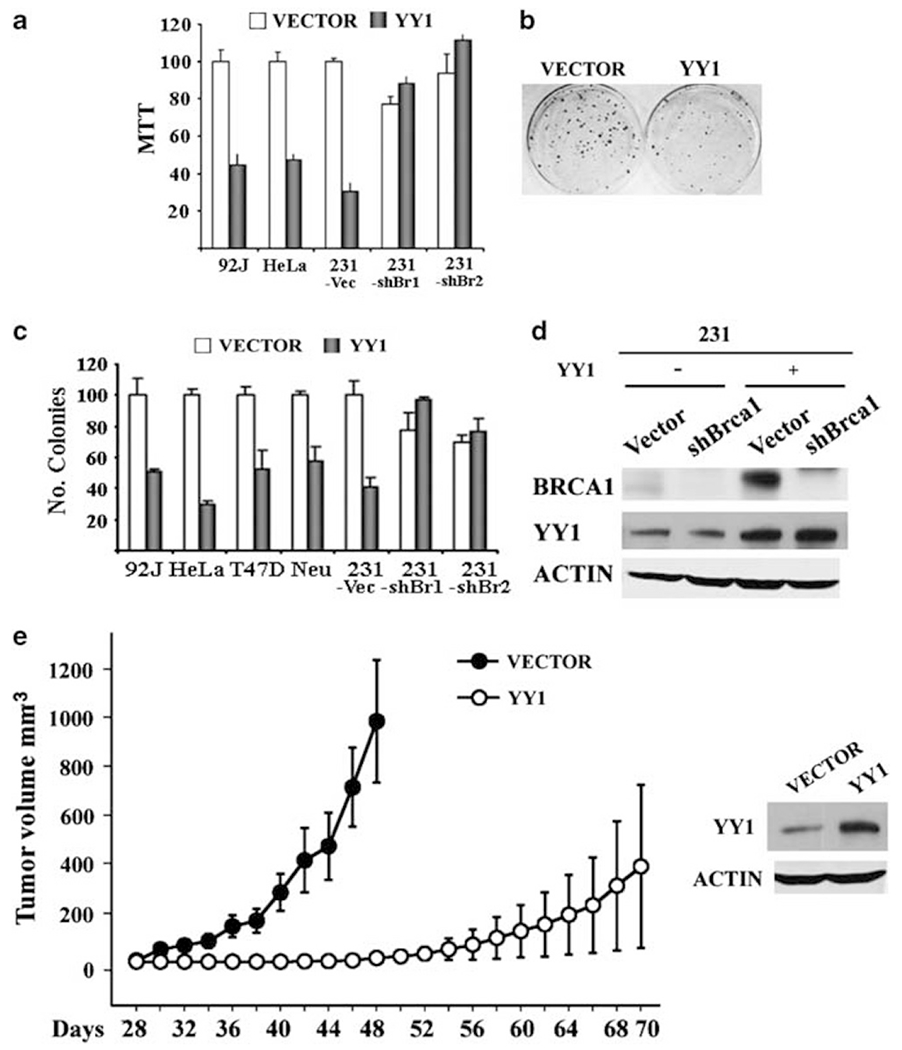
Ectopic overexpression of YY1 inhibits cancer cell growth both in vitro and in vivo. (a) Ectopic expression of YY1 reduces proliferation of BRCA1 wild-type cancer cells (92J, HeLa and MDA-MB-231-Vector), but not MDA-MB-231 cells with shRNA-mediated BRCA1 knockdown as revealed by MTT assay. (b, c) Expression of YY1 reduces foci formation in BRCA1 wild-type cancer cells (92J, HeLa, T47D, Neu and MDA-MB-231-Vector), but not shBRCA1-MDA-MB-231. An example of colonies formed in pCDH vector as compared with YY1 expression vector transformed MDA-MB-231 cells was shown in (b). (d) Western blot showing levels of YY1 in MDA-MB-231 cells before (vector control) and after ectopically overexpression of YY1. The MDA-MB-231 cells have been stably infected with lentiviruses carrying shRNA for BRCA1 or control virus. (e) Expression of YY1 in MDA-MB-231 cells inhibits tumor formation of these cells in nude mice. Three million cells were subcutaneously implanted for tumor formation. Levels of YY1 expression are shown in the right panel.
Next, we performed a xenograft tumor formation study in nude mice. Overexpression of YY1 strongly inhibited cancer formation of MDA-MB-231 xenografts (Figure 5e). In the vector-infected MDA-MB-231 cells, 11 out of 12 implantation sites developed tumors and tumors became visible 30 days after implantation and reached almost 1000 mm3 in tumor volume on the average 18 days later (Figure 5e). Whereas in the YY1-infected cells, 4 out of 12 implantation sites developed tumors and tumors became visible 52 days after implantation and reached ~400 mm3 in volume on the average 18 days later. A similar strong inhibitory effect of YY1 was also observed in 92J cells (data not shown). These data indicate that YY1 overexpression inhibits tumor cell proliferation in vitro; and reduced tumor onset and growth in vivo indicate that YY1 may serve as a tumor suppressor for breast cancer formation.
Expression of YY1 in human clinical breast cancers
Next, we measured the expression of YY1 in human breast cancers. We initially searched the Oncomine database and found that three studies showed reduced YY1 expression in breast cancers compared with normal tissue while another study showed the opposite data (Supplementary Figure 2). To obtain firsthand data regarding YY1 expression, we assessed YY1 protein levels as measured by tissue array between breast cancer (n = 42) and normal breast tissue (n = 19) samples. A majority, 95% (18/19) of normal tissues had higher YY1 levels than tumors, while 88% (37/42) of tumors contained undetectable or low levels of YY1 (Figures 6a and b). This observation is consistent with our data obtained earlier that overexpression of YY1 inhibits breast cancer cell proliferation.
Figure 6.
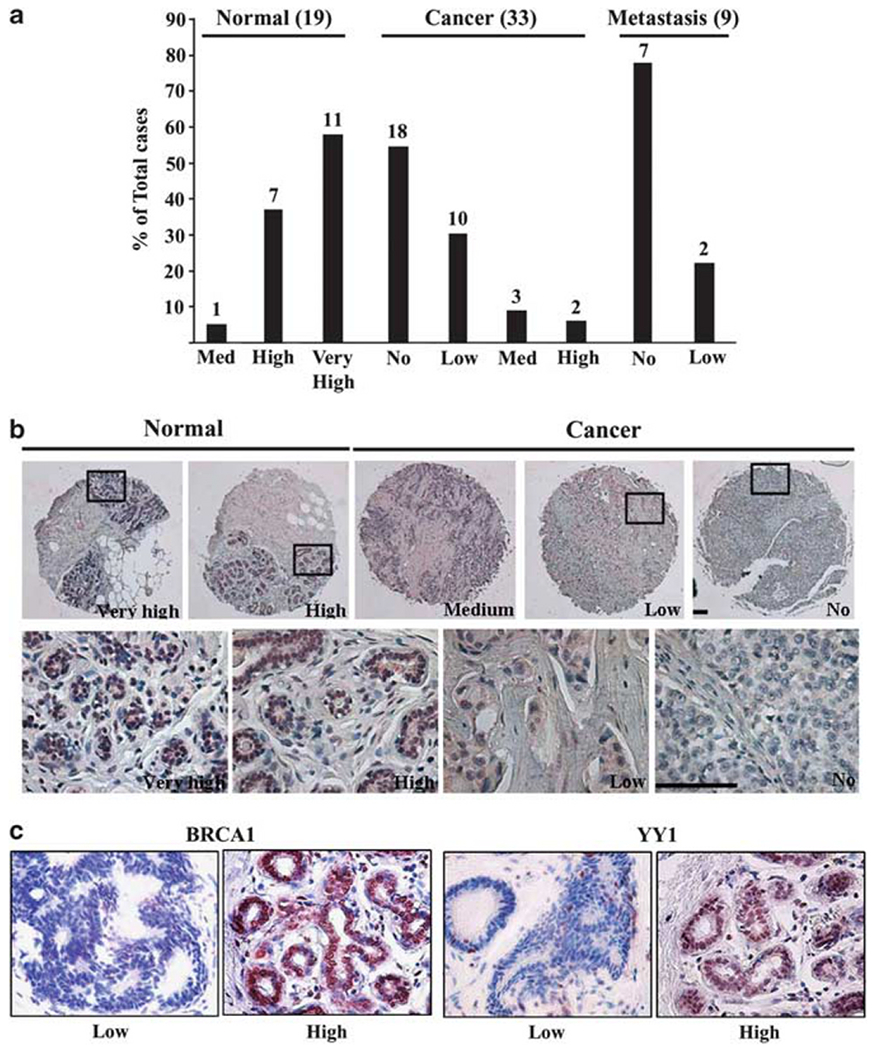
YY1 gene expression in human cancers. (a) Expression levels of YY1 protein in 42 (including 9 metastasis cases) breast cancer and 19 normal breast tissue samples measured by tissue array. YY1 and BRCA1 protein levels were detected by immunohistochemical staining using antibodies against YY1 and BRCA1, and intensity of staining was classified as no, low, medium, high or very high. (b) Immunohistochemical images. The boxed regions (enlarged below) show high levels of YY1 in normal epithelium and lower levels in cancers. Scale bars = 100 μm. (c) YY1 and BRCA1 protein levels in 46 human sporadic breast cancers. These cancer samples were reported previously (Man et al., 2005). Intensity of YY1 and BRCA1 staining of these cancers showed a positive correlation. Representative images with low and high expression of BRCA1 and YY1 were shown.
Next, we compared YY1 and BRCA1 protein levels in a group of 46 human sporadic breast cancers by immunohistochemical staining using antibodies against YY1 and BRCA1. Our data also revealed a positive correlation between YY1 and BRCA1 expression levels in these samples. An example of low and high expression of BRCA1 and YY1 was shown in Figure 6c. These data are consistent with the view that YY1 is a positive regulator of BRCA1 in cancers.
Discussion
In this study, we screened 92 transcription factors and found several of them, YY1, CSEN, CITED and a number of HDACs, act as potential transcription activators, while some others, including ID4, GABPA, PCAF, serve as potential repressors of BRCA1. Further studying these factors that either positively or negatively regulate BRCA1 expression should help to understand the mechanisms through which BRCA1 is downregulated in sporadic breast cancer.
It has been shown that somatic mutations of BRCA1 are rarely detected in sporadic breast cancers although many of these cancers exhibit low levels of BRCA1 expression (Turner et al., 2004b). Studies also revealed that BRCA1 null mutations resulted in growth disadvantage and cause apoptosis of the mutant cells (Deng, 2006; Drost and Jonkers, 2009). Therefore, it is conceivable that the malignant process evolves to reduce BRCA1 expression rather than mutate it to avoid cell lethality. This may be a primary reason that BRCA1 mutations are not found in sporadic cancers. However, the regulation of BRCA1 is quite complex. A previous study identified a 36-bp region (−198 to −162) in the α promoter that serves as a positive element for BRCA1 expression (Thakur and Croce, 1999). The YY1 response element identified here lies in this 36-bp positive element (Figure 3b). A transcriptional repressor is also found in an 83-bp region in the first intron of BRCA1 (Suen and Goss, 2001). Our study revealed one fragment (−201 to +39) in the BRCA1 promoter that contains a positive regulatory element and at least three regions that may contain repressor elements. Comparing the luciferase activity of three vectors in group 2, we found two regions (−957 to −567 and −567 to −201) that may contain repressor elements (Figures 1a and b), while a comparison between group 1 and group 2 vectors uncovered an additional negative element in a fragment (+39 to +244) that covers most of the β promoter. The β promoter has an important role in mediating a positive regulation of BRCA1 transcription by ERα (Xu et al., 1997b), the potential relationship between this repressor element on estrogen/ERα signaling in BRCA1 transcription is an interesting issue and should be further studied.
Notably, all genes, irrespective of their positive or negative action on BRCA1, revealed by this study seemed to execute their effect through the −201 to +39 region, suggesting that this region may contain both positive and negative elements. While this complex pattern of regulation of BRCA1 may also deserve special attention in a future study, we decided to focus on YY1, as it exhibits the strongest activity in regulating BRCA1. Our study revealed that YY1 positively regulates BRCA1 expression through binding to a Gli-Krueppel-related transcription factor binding site, ccttttACGTcat, that is located between −180 and −168. Furthermore, our data revealed that both genes are co-expressed in the same set of mammary epithelial cells and have a similar pattern of expression during mammary gland development. These data provide a basis for the regulation of BRCA1 by YY1 during mammary gland development.
A most notable finding in this study is that overexpression of YY1 inhibits tumor formation both in vitro and in vivo. YY1 is known to have an important role in many biological processes; however, its function in cancer remains controversial (Park and Atchison, 1991; Shi et al., 1991, 1997; Gordon et al., 2006; Castellano et al., 2009). Because YY1 positively regulates expression of several oncogenes, that is, E6 and E7, c-Myc, c-Fos and ErbB2 (Riggs et al., 1993; Lee et al., 1995a, b), and negatively regulates p53 stability (Gronroos et al., 2004; Sui et al., 2004), it is believed that YY1 acts as an oncogene (Gordon et al., 2006; Castellano et al., 2009). On the other hand, YY1 also inhibits cancer cell growth through binding and inhibiting c-Myc function (Austen et al., 1998), and positively regulates expression of a number of genes with tumor suppressor function such as HLJ1, p73 and p53 (Furlong et al., 1996; Wang et al., 2005, 2007; Wu et al., 2007a). These data suggest that YY1 can act as both a tumor suppressor and an oncogene, depending on tissue context and distribution of its downstream genes.
Several lines of evidence uncovered by this study indicate that YY1 may act as a tumor suppressor for breast cancer. First, YY1 binds to the promoter of BRCA1 and positively regulates the expression of this well-known tumor suppressor. Second, we have profiled the expression levels of YY1 and BRCA1 in 46 human breast cancers and a tissue array containing 42 breast cancers and 19 normal breast tissues. There was a significant positive correlation between the level of YY1 and BRCA1 expression in human breast tissues. Finally, we found that overexpression of YY1 inhibits tumor cell growth both in vitro and in vivo. These findings indicate that YY1 positively regulates BRCA1 gene expression and serves as a tumor suppressor in breast cancer formation.
It is unclear regarding the underlying mechanism of how YY1 may act as a tumor suppressor. Previous studies indicated that YY1 preferentially binds to a recombination-intermediate structure in vitro and forms a protein complex with INO80 to regulate genomic stability through homologous recombination-based repair (Wu et al., 2007b). YY1-deficient spermatocytes had increased double-strand break signals on chromosomes at the leptotene/zygotene stages of spermatogenesis that is accompanied by increased aneuploidy, and pachytene cell death, which are likely due to defects in DNA repair (Wu et al., 2009). These phenotypes are similar to those displayed by BRCA1-mutant mice that carry a targeted deletion of full-length BRCA1 (Xu et al., 2003; Turner et al., 2004a). As maintenance of genome integrity is critical for cancer suppression, we believe it may account for the tumor suppressor function of YY1.
In summary, we demonstrated that YY1 binds to the promoter of BRCA1, and serves as a positive regulator of BRCA1 expression. Our data also revealed that overexpression of YY1 in cancer cells inhibited foci formation and tumor growth in nude mice. Moreover, we detected a significant positive correlation between expression of YY1 and BRCA1 in human breast tissues, that is, cancers that have low levels of YY1 expression also have low levels of BRCA1. These data not only identify YY1 as a potential tumor suppressor, but also uncover an underlying mechanism of how BRCA1 expression is regulated in sporadic breast cancers.
Materials and methods
RT-PCR analysis
Total RNA from cells was extracted with STAT-60 following the manufacturer’s protocol (Tel-Test, Inc., Friendswood, TX, USA). Reverse transcription reactions were carried out using the first strand cDNA synthesis kit (Cat. 1483188; Roche, Indianapolis, IN, USA). One microgram of total RNA was used as template for each reaction. For PCR, the samples were heated to 94 °C for 2min, run through 22–31 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 10 min and then stopped at 4 °C. The primers for human BRCA1 are forward 5′-GAAGAAACCACCAAGGTCCA-3′ and reverse 5′-GGGATCTGGGGTATCAGGTA-3′. The primers for human YY1 are forward 5′-GGAGGAATACCTGGCATTGA-3′ and reverse 5′-GGTTGTTTTTGGCCTTAGCA-3′. The primers for internal control of human GAPDH are forward 5′-GGGAGCCAAAAGGGTCATCA-3′ and reverse 5′-TTTCTAGACGGCAGGTCAGGT-3′.
Western blot analysis, immunohistochemical and immunofluorescent staining
Western blot analysis was carried out according to standard procedures using ECL detection from Millipore (Billerica, MA, USA) or Licor (Lincoln, NE, USA) with antibodies against human YY1 and BRCA1 from Santa Cruz (Santa Cruz, CA, USA), β-Actin and Lamin A and α-Tubulin from Sigma (St Louis, MO, USA). For histology, mammary gland tissues were fixed in 10% formalin, blocked in paraffin wax, sectioned, stained with Hematoxylin and Eosin, and examined by light microscopy. Detection of primary antibodies was performed using the ZYMED Histomouse SP Kit (Zymed Laboratories Inc, South San Francisco, CA, USA) according to the manufacturer’s instructions. For human samples, an antibody against human BRCA1 was used from Calbiochem (La Jolla, CA, USA), and an antibody against mouse BRCA1 (Turner et al., 2004a) was used for mouse mammary gland tissue. Methanol-fixed cells or paraffin-fixed slides were used for immunofluorescent staining using methods described previously (Wang et al., 2004).
Transfection and luciferase assay
The transfections were carried out with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) and the luciferase activity was performed with the Dual-Luciferase Reporter Assay System and determined as following the manufacturer’s protocol (Promega, Madison, WI, USA). In the transfected cells, Renilla luciferase pRL-TK vector (Promega) was used as an internal control. All the data are from triplicates of three independent experiments.
ChIP assay
ChIP assays were performed as described previously (Wang et al., 2004). The antibody against YY1 was purchased from Santa Cruz. ChIP primers for the BRCA1 promoter are the following: forward 5′-TTTATGGCAAACTCAGGTAG-3′ and reverse 5′-CACGCCAGTACCCCAGAGCA-3′. Internal control primer for β-globin is the following: forward 5′-ATCTTCCTCCCACAGCTCCT-3′ and reverse 5′-TTTGCAGCCTCACCTTCTTT-3′.
Biotins-streptavidin pull-down assay
Wild-type and Bmt oligonucleotides containing biotin on the 5′ nucleotide of the sense strand were used in the pull-down assay. We incubated 1 μg of each double-stranded oligonucleotide with 300 μg of nuclear protein for 20 min at room temperature. We added 30 μl of poly(dI-dC) pre-absorbed streptavidin-agarose beads for 4h at 4 °C. The protein–DNA streptavidin-agarose complex was analyzed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Wang et al., 2004).
Electrophoretic mobility shift assay
The sequences of the wild-type and mutant oligonucleotides are the same as the sequences for the site-directed mutagenesis and biotin-streptavidin pull-down assay. These double-stranded DNA probes were end-labeled with (α-32P)dCTP using Klenow enzyme and performed as described previously (Lee et al., 2005).
MTT proliferation assays
In all, 10000 cells in the log phase of growth were plated in 24-well plates. The cells were treated with thiazolyl blue tetrazolium bromide (Sigma) working solution (0.5 mg/ml) for 1–3 h and with isopropanol for 30 min. The absorption was read with the Perkin-Elmer 1420 multi-label counter (Perkin-Elmer, Waltham, MA, USA).
Focus formation assay
Lentiviral YY1 virus supernatant was infected into several cell lines and 300–1000 cells were plated in 10 cm dishes. The colonies were grown for 1–3 weeks after which the colonies were fixed with methanol and stained with 1% Giemsa stain and the total number of foci was scored. Each experiment was performed in triplicate and each dish counted three times.
Nude mice injection
Lentiviral YY1 virus supernatant was infected into MDA-MB-231 or 92J cells and the cells (3 × 106 cells/injection) were inoculated subcutaneously bilaterally in the flanks and on the back of 6–8-week-old female nude mice (Jackson). Two sites per mouse and at least five mice were used for each group. Mice were observed every other day for tumor formation. The tumor size was measured with calipers when visible nodules were present. We calculated the tumor volume (V) by the formula V = 2/3 πrxryrz (r is radius and x, y, z refer to each axis and π = 3.14).
Clinical specimens
The tissue array of breast cancer samples was purchased from US Biomax (Rockville, MD, USA) (Cat. BR1002). YY1 and BRCA1 expression levels were detected with 46 human sporadic breast cancer tissues and the tissue array sample by immunohistochemical staining.
Statistical analyses
Student’s t-test (http://www.physics.csbsju.edu/stats/ttest.html) was used to compare differences between samples analyzed. Any P-value of <0.05 (P<0.05) is considered as statistically significant difference.
Supplementary Material
Acknowledgements
We thank members of Dr Deng laboratory for their critical discussion of this work and technical assistance. This research was supported by the Intramural Research Program of the National Institute of Diabetes, Digestive and Kidney Diseases, National Institutes of Health, USA.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S et al. (2006). Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol 26: 3565–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberg AJ, Lam AP, Helzlsouer KJ. (1999). Epidemiology, prevention, and early detection of breast cancer. Curr Opin Oncol 11: 435–441. [DOI] [PubMed] [Google Scholar]
- Atlas E, Stramwasser M, Mueller CR. (2001). A CREB site in the BRCA1 proximal promoter acts as a constitutive transcriptional element. Oncogene 20: 7110–7114. [DOI] [PubMed] [Google Scholar]
- Atlas E, Stramwasser M, Whiskin K, Mueller CR. (2000). GA-binding protein alpha/beta is a critical regulator of the BRCA1 promoter. Oncogene 19: 1933–1940. [DOI] [PubMed] [Google Scholar]
- Austen M, Cerni C, Luscher-Firzlaff JM, Luscher B. (1998). YY1 can inhibit c-Myc function through a mechanism requiring DNA binding of YY1 but neither its transactivation domain nor direct interaction with c-Myc. Oncogene 17: 511–520. [DOI] [PubMed] [Google Scholar]
- Baldassarre G, Battista S, Belletti B, Thakur S, Pentimalli F, Trapasso F et al. (2003). Negative regulation of BRCA1 gene expression by HMGA1 proteins accounts for the reduced BRCA1 protein levels in sporadic breast carcinoma. Mol Cell Biol 23: 2225–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beger C, Pierce LN, Kruger M, Marcusson EG, Robbins JM, Welcsh P et al. (2001). Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc Natl Acad Sci USA 98: 130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir V, Stefansson OA, Bodvarsdottir SK, Hilmarsdottir H, Jonasson JG, Eyfjord JE. (2006). Epigenetic silencing and deletion of the BRCA1 gene in sporadic breast cancer. Breast Cancer Res 8: R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody LC, Biesecker BB. (1998). Breast cancer susceptibility genes. BRCA1 and BRCA2. Medicine (Baltimore) 77: 208–226. [DOI] [PubMed] [Google Scholar]
- Castellano G, Torrisi E, Ligresti G, Malaponte G, Militello L, Russo AE et al. (2009). The involvement of the transcription factor Yin Yang 1 in cancer development and progression. Cell Cycle 8: 1367–1372. [DOI] [PubMed] [Google Scholar]
- Deng CX. (2006). BRCA1: cell cycle checkpoint, genetic instability, DNA damage response, and cancer evolution. Nucleic Acids Res 34: 1416–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost RM, Jonkers J. (2009). Preclinical mouse models for BRCA1-associated breast cancer. Br J Cancer 101: 1651–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles DM, Pichert G. (2005). Familial non-BRCA1/BRCA2-associated breast cancer. Lancet Oncol 6: 705–711. [DOI] [PubMed] [Google Scholar]
- Furlong EE, Rein T, Martin F. (1996). YY1 and NF1 both activate the human p53 promoter by alternatively binding to a composite element, and YY1 and E1A cooperate to amplify p53 promoter activity. Mol Cell Biol 16: 5933–5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Lu Y, Hu Y. (2008). A role of CREB in BRCA1 constitutive promoter activity and aromatase basal expression. Int J Biomed Sci 4: 260–265. [PMC free article] [PubMed] [Google Scholar]
- Giacinti L, Vici P, Lopez M. (2008). Epigenome: a new target in cancer therapy. Clin Ter 159: 347–360. [PubMed] [Google Scholar]
- Gilbert PM, Mouw JK, Unger MA, Lakins JN, Gbegnon MK, Clemmer VB et al. (2010). HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype. J Clin Invest 120: 1535–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glait C, Ravid D, Lee SW, Liscovitch M, Werner H. (2006). Caveolin-1 controls BRCA1 gene expression and cellular localization in human breast cancer cells. FEBS Lett 580: 5268–5274. [DOI] [PubMed] [Google Scholar]
- Gordon S, Akopyan G, Garban H, Bonavida B. (2006). Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25: 1125–1142. [DOI] [PubMed] [Google Scholar]
- Gronroos E, Terentiev AA, Punga T, Ericsson J. (2004). YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc Natl Acad Sci USA 101: 12165–12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Xu XL, Yang MC, Wei F, Ayi TC, Bowcock AM et al. (1997). Cell cycle-dependent colocalization of BARD1 and BRCA1 proteins in discrete nuclear domains. Proc Natl Acad Sci USA 94: 12075–12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluk BJ, Fu Y, Formolo TA, Zhang L, Hindle AK, Man YG et al. (2010). BP1, an isoform of DLX4 homeoprotein, negatively regulates BRCA1 in sporadic breast cancer. Int J Biol Sci 6: 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Galvin KM, See RH, Eckner R, Livingston D, Moran E et al. (1995a). Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev 9: 1188–1198. [DOI] [PubMed] [Google Scholar]
- Lee JS, See RH, Galvin KM, Wang J, Shi Y. (1995b). Functional interactions between YY1 and adenovirus E1A. Nucleic Acids Res 23: 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Kim YJ, Yoon WJ, Kim JI, Kim BG, Hwang YS et al. (2005). Dlx5 specifically regulates Runx2 type II expression by binding to homeodomain-response elements in the Runx2 distal promoter. J Biol Chem 280: 35579–35587. [DOI] [PubMed] [Google Scholar]
- MacDonald G, Stramwasser M, Mueller CR. (2007). Characterization of a negative transcriptional element in the BRCA1 promoter. Breast Cancer Res 9: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man YG, Fu SW, Schwartz A, Pinzone JJ, Simmens SJ, Berg PE. (2005). Expression of BP1, a novel homeobox gene, correlates with breast cancer progression and invasion. Breast Cancer Res Treat 90: 241–247. [DOI] [PubMed] [Google Scholar]
- Oliveira AM, Ross JS, Fletcher JA. (2005). Tumor suppressor genes in breast cancer: the gatekeepers and the caretakers. Am J Clin Pathol 124(Suppl): S16–S28. [DOI] [PubMed] [Google Scholar]
- Okada S, Ouchi T. (2003). Cell cycle differences in DNA damage-induced BRCA1 phosphorylation affect its subcellular localization. J Biol Chem 278: 2015–2020. [DOI] [PubMed] [Google Scholar]
- Park K, Atchison ML. (1991). Isolation of a candidate repressor/activator, NF-E1 (YY-1, delta), that binds to the immunoglobulin kappa 3’ enhancer and the immunoglobulin heavy-chain mu E1 site. Proc Natl Acad Sci USA 88: 9804–9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch T, Zhong X, Pfeifer GP, Xu X. (2005). 53BP1 is a positive regulator of the BRCA1 promoter. Cell Cycle 4: 1078–1083. [PubMed] [Google Scholar]
- Riggs KJ, Saleque S, Wong KK, Merrell KT, Lee JS, Shi Y et al. (1993). Yin-yang 1 activates the c-myc promoter. Mol Cell Biol 13: 7487–7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivenbark AG, Coleman WB. (2009). Epigenetic regulation of cystatins in cancer. Front Biosci 14: 453–462. [DOI] [PubMed] [Google Scholar]
- Ruffner H, Verma IM. (1997). BRCA1 is a cell cycle-regulated nuclear phosphoprotein. Proc Natl Acad Sci USA 94: 7138–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lee JS, Galvin KM. (1997). Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta 1332: F49–F66. [DOI] [PubMed] [Google Scholar]
- Shi Y, Seto E, Chang LS, Shenk T. (1991). Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67: 377–388. [DOI] [PubMed] [Google Scholar]
- Suen TC, Goss PE. (2001). Identification of a novel transcriptional repressor element located in the first intron of the human BRCA1 gene. Oncogene 20: 440–450. [DOI] [PubMed] [Google Scholar]
- Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P et al. (2004). Yin Yang 1 is a negative regulator of p53. Cell 117: 859–872. [DOI] [PubMed] [Google Scholar]
- Tan SH, Baker CC, Stunkel W, Bernard HU. (2003). A transcriptional initiator overlaps with a conserved YY1 binding site in the long control region of human papillomavirus type 16. Virology 305: 486–501. [DOI] [PubMed] [Google Scholar]
- Thakur S, Croce CM. (1999). Positive regulation of the BRCA1 promoter. J Biol Chem 274: 8837–8843. [DOI] [PubMed] [Google Scholar]
- Turner JM, Aprelikova O, Xu X, Wang R, Kim S, Chandramouli GV et al. (2004a). BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol 14: 2135–2142. [DOI] [PubMed] [Google Scholar]
- Turner N, Tutt A, Ashworth A. (2004b). Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer 4: 814–819. [DOI] [PubMed] [Google Scholar]
- Wang A, Schneider-Broussard R, Kumar AP, MacLeod MC, Johnson DG. (2000). Regulation of BRCA1 expression by the Rb-E2F pathway. J Biol Chem 275: 4532–4536. [DOI] [PubMed] [Google Scholar]
- Wang CC, Tsai MF, Dai TH, Hong TM, Chan WK, Chen JJ et al. (2007). Synergistic activation of the tumor suppressor, HLJ1, by the transcription factors YY1 and activator protein 1. Cancer Res 67: 4816–4826. [DOI] [PubMed] [Google Scholar]
- Wang CC, Tsai MF, Hong TM, Chang GC, Chen CY, Yang WM et al. (2005). The transcriptional factor YY1 upregulates the novel invasion suppressor HLJ1 expression and inhibits cancer cell invasion. Oncogene 24: 4081–4093. [DOI] [PubMed] [Google Scholar]
- Wang RH, Yu H, Deng CX. (2004). A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc Natl Acad Sci USA 101: 17108–17113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Hu YC, Liu H, Shi Y. (2009). Loss of YY1 impacts the heterochromatic state and meiotic double-strand breaks during mouse spermatogenesis. Mol Cell Biol 29: 6245–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Murai S, Kataoka K, Miyagishi M. (2007a). Cooperative regulation of p73 promoter by Yin Yang 1 and E2F1. Nucleic Acids Symp Ser (Oxf) 51: 347–348. [DOI] [PubMed] [Google Scholar]
- Wu S, Shi Y, Mulligan P, Gay F, Landry J, Liu H et al. (2007b). A YY1-INO80 complex regulates genomic stability through homologous recombination-based repair. Nat Struct Mol Biol 14: 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Sato K, Koike A, Nishikawa H, Koizumi H, Venkitaraman AR et al. (2010). HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res 70: 6384–6392. [DOI] [PubMed] [Google Scholar]
- Xu CF, Brown MA, Nicolai H, Chambers JA, Griffiths BL, Solomon E. (1997a). Isolation and characterisation of the NBR2 gene which lies head to head with the human BRCA1 gene. Hum Mol Genet 6: 1057–1062. [DOI] [PubMed] [Google Scholar]
- Xu CF, Chambers JA, Solomon E. (1997b). Complex regulation of the BRCA1 gene. J Biol Chem 272: 20994–20997. [DOI] [PubMed] [Google Scholar]
- Xu X, Aprelikova O, Moens P, Deng CX, Furth PA. (2003). Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development 130: 2001–2012. [DOI] [PubMed] [Google Scholar]
- Zhang J, Powell SN. (2005). The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Mol Cancer Res 3: 531–539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


