Abstract
RNA modifications, which are introduced post-transcriptionally, have recently been assigned pivotal roles in the regulation of spermatogenesis and embryonic development. However, the RNA modification landscape in human sperm is poorly characterized, hampering our understanding about the potential role played by RNA modification in sperm. Through our recently developed high-throughput RNA modification detection platform based on liquid chromatography with tandem mass spectroscopy, we are the first to have characterized the RNA modification signature in human sperm. The RNA modification signature was generated on the basis of 49 samples from participants, including 13 healthy controls, 21 patients with asthenozoospermia (AZS) and 15 patients with teratozoospermia (TZS). In total, we identified 13 types of RNA modification marks on the total RNA in sperm, and 16 types of RNA modification marks on sperm RNA fragments of different sizes. The levels of these RNA modifications on the RNA of patients with AZS or TZS were altered, compared to controls, especially on sperm RNA fragments >80 nt. A few types of RNA modifications, such as m1G, m5C, m2G and m1A, showed clear co-expression patterns as well as high linear correlations with clinical sperm motility. In conclusion, we characterized the RNA modification signature of human sperm and identified its correlation with sperm motility, providing promising candidates for use in clinical sperm quality assessment and new research insights for exploring the underlying pathological mechanisms in human male infertility syndromes.
Keywords: RNA modification, sperm RNA, epigenetic information, male infertility, asthenozoospermia, teratozoospermia, sperm motility
Introduction
Human reproductive health is essential to ensure the continuation of the human population. However, an estimated 15% of couples around the world suffer from infertility and experience difficulty with conception; in these couples, male factor infertility accounts for ∼40% of cases, a percentage which is still increasing (Krausz and Riera-Escamilla, 2018; Houston et al., 2021). Asthenozoospermia (AZS) and teratozoospermia (TZS) are common clinical symptoms of human male subfertility (Ventimiglia et al., 2016). AZS is characterized by reduced progressive motility of spermatozoa, while TZS is defined as abnormal sperm morphology. However, the molecular causes of AZS and TZS have not been defined. Only ∼4% of all men with infertility issues, and mainly with azoospermia, are diagnosed with genetic anomalies, and the pathological mechanisms involved in the majority (60–70%) of male infertility cases remain unexplained (Agarwal et al., 2021; Houston et al., 2021).
Spermatogenesis in the testicular seminiferous tubules is a complex process that is precisely controlled at the transcriptional, post-transcriptional and translational levels (Morgan et al., 2021). The end products of spermatogenesis, sperm, lose most of their cytoplasm and their histones are extensively replaced with protamines, leading to morphological structures that differ from those of somatic cells (Amaral et al., 2014; Teves and Roldan, 2022). Owing to the shut-down of transcription at the end of spermatogenesis and the reduction in cytoplasm, sperm was previously thought to lack RNA (Monesi, 1964; Monesi et al., 1978; Miller et al., 2005). However, with the development of next-generation sequencing, studies have shown that mature sperm contain a considerable amount of RNAs, including mRNAs, circular RNAs (cirRNAs), long noncoding RNAs (lncRNAs), fragmented rRNA and small noncoding RNAs (sncRNAs), which have been shown to play important regulatory roles in spermatogenesis, fertilization and early embryonic development (Chen et al., 2016a,b; Zhang et al., 2018). In recent years, sperm RNAs have also been associated with human semen characteristics and are dynamically altered in patients with AZS or TZS, suggesting potential applications in the diagnosis or treatment of male infertility (Jodar et al., 2012, 2013; Georgiadis et al., 2015; Jodar et al., 2015; Corral-Vazquez et al., 2019).
As a recently discovered post-transcriptional regulator (Frye et al., 2018), RNA modification has been extensively studied (Lewis et al., 2017; Barbieri and Kouzarides, 2020; Wiener and Schwartz, 2021). Over 170 types of RNA modification have been identified across the three kingdoms of life (Animalia, Plantae and Protista) (Frye et al., 2016). Some of these modifications enable cells to respond sensitively to various environmental stimuli and regulate cellular biological processes, such as the rate of mRNA translation and mRNA metabolism. RNA modifications also play active roles in regulating physiological and pathological processes, including tumorigenesis and intergenerational epigenetic inheritance of paternally acquired phenotypes (Chen et al., 2016b; Zhang et al., 2019). As the most common internal RNA modification in eukaryotic mRNA, N6-methyladenosine (m6A) has been extensively investigated and shown to play critical roles in spermatogenesis (Zheng et al., 2013; Lin et al., 2017; Zhong et al., 2021). The deletion of m6A writer proteins, such as methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14), in mouse germ cells, leads to the loss of m6A and the depletion of spermatogonial stem cells (SSCs) through the m6A-dependent translational dysregulation of transcripts required for SSC fate commitment (Lin et al., 2017; Xu et al., 2017). Moreover, ablation of the m6A reader proteins YTH domain containing 2 (YTHDC2) and YTH domain containing family protein 2 (YTHDF2) or the eraser protein AlkB Homolog 5 (ALKBH5) affects spermatogenesis and leads to male infertility in mice (Hsu et al., 2017; Qi et al., 2022), demonstrating the indispensable role of m6A in ensuring successful spermatogenesis. In addition to m6A, dysregulation of the 5-methylcytosine (m5C) and 2′-O-methylation modifications have also been reported to contribute to male infertility (Gui and Yuan, 2021). Lack of NOP2/Sun RNA methyltransferase 2 (NSUN2), an m5C methyltransferase, can block spermiogenesis at the pachynema stage (Hussain et al., 2013). Mutation of NOP2/Sun RNA methyltransferase family member (Nsun7), another member of the NSUN family, can also impair sperm motility, leading to pregnancy failure (Harris et al., 2007), and when La ribonucleoprotein 7 (Larp7), which is critical for establishing 2′-O-methylation of U6 on small nuclear RNA (snRNA) (Hasler et al., 2020), is deleted, the average testicular weight is reduced and abnormal sperm morphology is observed (Wang et al., 2020). However, RNA modification signatures of human sperm that are related to male infertility have not yet been identified, and whether changes in the levels of RNA modifications are associated with human AZS and TZS remains unclear.
In the present study, we focused on investigating the human sperm RNA modification signatures in patients with AZS and TZS using a liquid chromatography with tandem mass spectroscopy (LC-MS/MS)-based high-throughput RNA modification detection platform. The results revealed clear co-expression patterns and high linear correlations of RNA modification abundance with sperm motility. These findings offer new insights and a basis for exploring further the underlying pathogenic mechanisms in human male infertility syndromes.
Materials and methods
Participant enrollment and human semen sample collection and analysis
A total of 49 men (22–49 years old) who underwent semen analysis at Xinqiao Hospital in Chongqing, China, were enrolled in this study. The semen samples were obtained through masturbation after 2–7 days of sexual abstinence. All samples were collected in sterile frustum cone-like cups, allowed to liquefy at 37°C for 30 min and subjected to diagnostic semen analysis. A whole sperm quality analysis was performed using an SCA-H-OIP system: an ‘SCA-H-OIP system’ is the SCA Scope (updated computer-assisted semen assessment (CASA)) from MICROPTIC (Microptic, Barcelona, Spain). Briefly, some areas under the microscope with a relatively uniform sperm distribution and no sperm overlap were randomly chosen for sperm motility and morphological assessment. When the sperm motility and morphological parameters reported by the SCA-H-OIP system were inconsistent with those observed under the microscope by laboratory staff, the sample was reanalyzed to confirm the accuracy of the analytical results. At least 200 spermatozoa were analyzed from each semen sample to generate the final semen analysis report. The semen analysis report was used to classify the semen sample into normozoospermia (NZS), AZS and TZS groups, according to the World Health Organization (2010) guidelines (Cooper et al., 2010) on sperm motility and morphological parameters, without additional cutoff value correction. Semen samples with a proportion of immotile sperm exceeding 60% and morphologically abnormal sperm over 96% were not included in this study. All experiments were performed in accordance with the principles set out in the World Medical Association Declaration of Helsinki (World Medical Association, 2013). This study was approved by the Institutional Ethics Committees of Xinqiao Hospital, and an informed consent form was signed by each participant.
Ejaculated human sperm purification
To reduce the viscosity of the collected samples, 5 ml PBS (with 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 1.8 mM KH2PO4, at pH 7.4) was added to the collected semen samples and the tubes were inverted to gently mix the contents before filtering with a 40 μM pore size cell strainer (Corning, Harrodsburg, KY, USA) on ice. After centrifugation at 800×g for 10 min at 4°C, the sperm were resuspended in 2 ml PBS, and 10 ml of somatic cell lysis buffer (0.1% sodium dodecyl sulphate and 0.5% Triton X-100 in diethylpyrocarbonate water) was added. The samples were incubated for 20 min on ice then centrifuged for 10 min at 4°C at 800×g, and the precipitated sperm were washed twice with 3 ml PBS to remove the somatic cell debris and then centrifuged again for 10 min at 4°C at 800×g. The purified sperm were lysed with 1 ml TRIzol (Invitrogen, Waltham, MA, USA) for RNA extraction.
Human sperm RNA extraction
The sperm RNA extraction procedure was based on the manufacturer’s instructions with slight modification. Briefly, the sperm were lysed in TRIzol for 2 h on ice. The lysate was pumped up and down several times (for homogenization) in a 1 ml sterile syringe with a 27 G needle and incubated on ice for another 2 h. This step was necessary to fully extract RNAs from the sperm, as previously reported (Chen et al., 2016a).
Determination of sperm purity by RT-PCR
To remove genomic DNA, sperm total RNA was incubated with RQ1 RNase-Free DNase I (Promega, Madison, AL, USA) for 30 min at 37°C in 10 μl of a buffer containing 40 mM Tris-HCl (pH 7.9), 10 mM NaCl, 6 mM MgCl2 and 10 mM CaCl2. The DNase was inactivated by heating for 10 min at 65°C in the presence of 2 mM EGTA (pH 8.0). Reverse transcription was conducted by using M-MuLV Reverse Transcriptase (NEB, Ipswich, MA, USA) according to the manufacturer’s instructions. Briefly, 2 μg of total RNA was converted into cDNA with Oligo-dT and the mRNA expression levels of the sperm-specific gene protamine-2 (PRM2) and the somatic cell-specific genes cadherin-1 (CDH1) and cadherin-2 (CDH2) (epithelial cell markers), CD4 (a leukocyte T-cell surface marker) and the germline-specific gene c-KIT were quantified by RT-PCR. The amplified PCR products were electrophoresed on a 1% agarose gel and stained with Gel Red (Promega, Madison, AL, USA). The data were analyzed with a Bio-Rad Gel imaging system. The primers used in the RT-PCR are listed in Supplementary Table SI.
Isolation of RNAs of different sizes from human sperm total RNA
Total sperm RNAs were separated by 15% urea-polyacrylamide gel electrophoresis (PAGE). RNAs of different sizes were excised from the gel and extracted as previously reported (Chen et al., 2016a). Briefly, 5 μg of total RNA from each sample was loaded on the gel and electrophoresed for 1 h in 1× Tris, borate and EDTA (TBE) buffer (Invitrogen, Waltham, MA, USA) at 200 V. The gel was then stained with 0.01% SYBR GOLD (Invitrogen, Waltham, MA, USA) and analyzed with a Bio-Rad imaging system. RNAs of different sizes (17–25, 25–50, 50–80, >80 nt) were excised and extracted from the gel, according to a low range single-strand RNA (ssRNA) ladder (NEB, Ipswich, MA, USA).
Preparation of mononucleotides for RNA modification detection
Sperm total RNA and different sized RNA fragments (17–25, 25–50, 50–80, >80 nt) were digested in 30 μl of enzymolysis system solution, which consisted of 3 μl of 10× reaction buffer (250 mM Tris-HCl, pH 8.0; 5 mM MgCl2 and 0.5 mg/ml BSA), 1 IU Benzonase (Sigma-Aldrich, St Louis, MO, USA), 0.2 IU alkaline phosphatase (Sigma-Aldrich, St Louis, MO, USA) and 0.05 IU phosphodiesterase I (Thermo Fisher Scientific, Grand Island, NY, USA). After 3 h of digestion at 37°C, the enzymolysis products were centrifuged at 13 000×g for 15 min on a Nanosep® 3K spin filter (Pall Corporation, Ann Arbor, MI, USA) to remove the enzymes and obtain the mononucleotides for LC-MS/MS analysis.
Ribonucleosides standards preparation and RNA modification quantification by LC-MS/MS
To quantify the RNA modifications, a multiple reaction monitoring (MRM) system was established based on LC-MS/MS (Acquity-UPLC I-class carrying a Xevo-TQ-S mass spectrometry system, Waters Corporation, Milford, MA, USA). Standardized ribonucleosides are used as LC-MS/MS standards in order to quantify the abundance of different RNA modification marks (Supplementary Fig. S1) in human sperm samples and these were prepared as previously described (Zhang et al., 2018). The original concentration of each ribonucleoside standard in the standard solution mixture is shown in Supplementary Table SII. In addition to the original full concentration, the mixture of standard solutions was diluted 2, 5, 25, 50, 200, 1000 and 2000 times, to obtain eight standard solutions of decreasing concentration (Supplementary Table SII). The standard curve for each RNA modification was obtained on the basis of concentration curve points that were corrected individually according to mass spectrometric signal.
LC-MS/MS data analysis
The raw data indicating the levels of RNA modification marks determined by LC-MS/MS were analyzed by the Mass Lynx (V1.4) software package (Waters Corporation, Milford, MA, USA). Each spectrum was examined and manually corrected when conspicuous outliers were identified. For example, when the autointegration-generated MRM peaks were not fully integrated, we performed manual integration to ensure the correct calculation of certain RNA modification marks. The percentage of each modified ribonucleoside was normalized to the total amount of quantified ribonucleosides with the same nucleobase in order to decrease/eliminate errors, which were caused by variation in amount of the sample loaded (Zhang et al., 2018). For example, the percentage of m1G = mole concentration (m1G)/mole concentration (m1G + m2G + G + Gm + m7G + G + 7G).
Statistical analysis
Statistical analyses were performed with one-way ANOVA and uncorrected Fisher’s least square difference for multiple comparisons of RNA modification levels by GraphPad Prism 8 (GraphPad Software Inc, San Diego, CA, USA). The correlation analyses were based on Pearson correlation coefficients computed with GraphPad Prism 8. Linear regression was performed to evaluate the correlation between sperm motility and RNA modification levels, and between sperm motility and different RNA modification marks, as determined using GraphPad Prism 8. A value of P < 0.05 was considered to be statistically significant.
Results
Participants and clinical semen characteristics
Human semen samples from 49 participants were collected by masturbation. No significant differences in semen volume or sperm concentration were observed among participants. The semen samples were categorized into three main groups on the basis of participant diagnosis. Specifically, 13 samples were from participants diagnosed with NZS; 21 samples were from participants diagnosed with AZS (proportion of immotile sperm exceeded 60%), and 15 samples were obtained from participants diagnosed with TZS (the proportion of sperm with abnormal morphology exceeded 96%). The AZS group was further classified into three clusters based on the percentage of sperm with progressive motility (PR) within a certain range: AZS I sperm showed a low level of motility defect (PR between ∼20% and 32%) (n = 8); AZS II sperm showed a moderate level of motility defect (PR between ∼10% and 20%) (n = 5) and AZS III sperm showed a high level of motility defect (PR <10%) (n = 2). All the clinical semen characteristics in this study are shown in Supplementary Table SIII.
RNA modification signature of human sperm total RNA
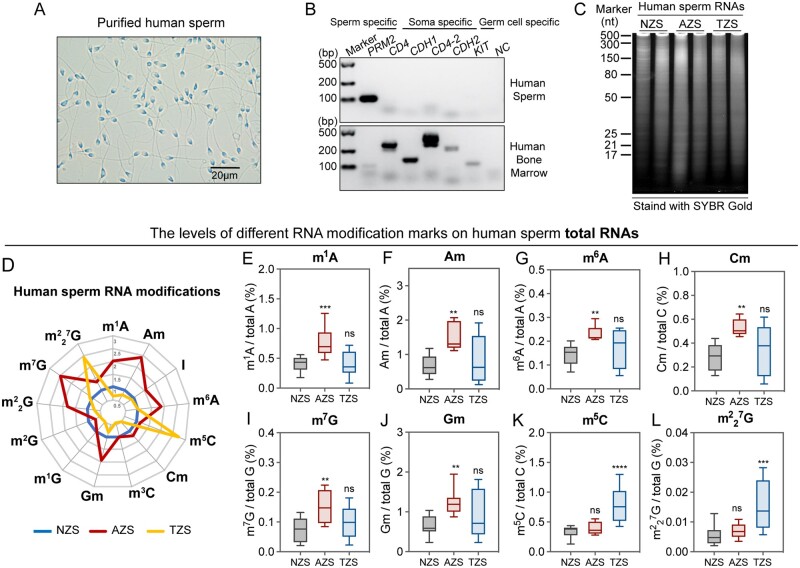
The purity of the human mature sperm isolated from the semen samples was evaluated at both the cellular and molecular levels. No visible somatic cell debris or nuclear contamination was observed in purified sperm samples (Fig. 1A). The mRNA expression level of CD4, CDH1 and CDH2 (markers of epithelial cells) was detected by RT-PCR to confirm that the samples were not contaminated with somatic cells, as well as the germline-specific cell marker c-KIT to eliminate the possibility of testicular germ cell contamination (Fig. 1B). A previous study reported that the majority of human sperm RNAs were actually fragmented (Krawetz, 2005), and our data confirmed this report by showing that the human sperm RNAs were evenly distributed on the 15% urea-PAGE gel, revealing a similar fragmented sperm RNA distribution pattern (Fig. 1C).
Figure 1.
Human sperm characters and RNA modifications in sperm total RNA. (A) Light microscope image of purified human sperm; (B) the purity of human sperm was evaluated by RT-PCR using specific molecular markers. PRM2, protamine 2; CD4, CD4 molecule; CDH1, cadherin 1; CD4-2, CD4 molecule; CDH2, cadherin 2; KIT, KIT proto-oncogene, receptor tyrosine kinase, also known as C-KIT; (C) the distribution of human sperm RNA after electrophoresis on a 15% urea-polyacrylamide gel, stained with SYBR GOLD. (D) Radar graph showing the comparison of RNA modification abundance in human sperm total RNA among NZS (n = 6), AZS (n = 6) and TZS (n = 5) samples; (E–L) the levels of RNA modification marks, such as (E) m1A, (F) Am, (G) m6A, (H) Cm, (I) m7G, (J) Gm, (K) m5C and (L) 7G in the total RNAs of human sperm from NZS, AZS and TZS groups. NZS, Normozoospermia; AZS, Asthenozoospermia; TZS, Teratozoospermia. Statistical analyses were performed with one-way ANOVA and uncorrected Fisher’s least significant difference (LSD) for multiple comparisons of RNA modification levels. **P < 0.01 (AZS versus NZS), ***P < 0.001 (AZS versus NZS; TZS versus NZS), ****P < 0.0001 (TZS versus NZS), ns, not significant (AZS versus NZS; TZS versus NZS). NZS, Normozoospermia; AZS, Asthenozoospermia; TZS, Teratozoospermia. The uncropped results for Fig. 1B and C are shown in Supplementary Fig. S9.
To characterize the RNA modification signature of total RNA in human sperm, 26 types of nucleobase standards (Supplementary Fig. S1) were used in our RNA modification detection platform based on LC-MS/MS. However, owing to the low abundance of several types of RNA modifications in this study, only 13 types of RNA modification marks were quantified in the total RNA of human sperm by LC-MS/MS. Mass spectrometry data analysis showed differences in the RNA modification signatures among the total RNAs of the NZS (n = 6), AZS (n = 6) and TZS (n = 5) groups (Fig. 1D), revealing a potential role of RNA modifications in regulating sperm quality. Among these RNA modification marks, eight showed a significantly different abundance in the AZS and TZS groups compared with the NZS group (Fig. 1E–L), but no significant difference was observed in other RNA modification marks among the different groups (Supplementary Fig. S2A–E). Specifically, the levels of m1A, Am, m6A, Cm, m7G and Gm in sperm total RNAs increased significantly in the AZS group (Fig. 1E–J), while that of m5C and 7G significantly increased in the TZS group (Fig. 1K and L), indicating that the RNA modification mark levels of human sperm were associated with male infertility syndrome.
RNA modification signatures in sperm RNA fragments of different sizes
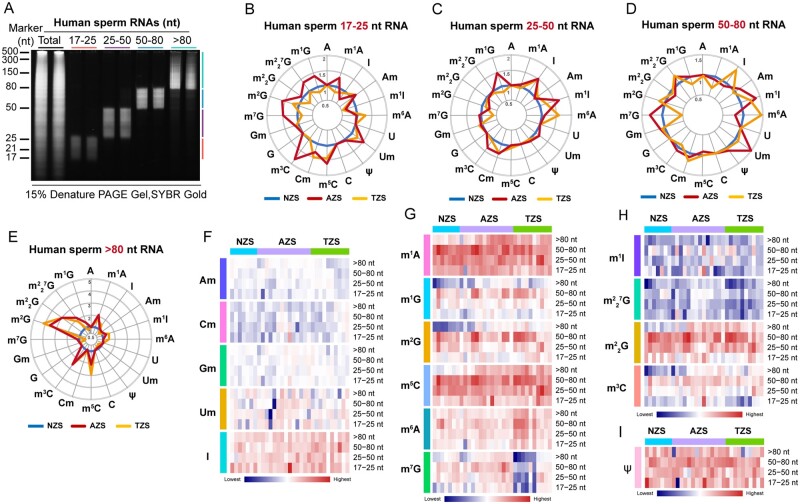
To investigate which part of the sperm RNA was critical for the observed alteration of RNA modification abundance in the AZS and TZS total RNAs, we separated human sperm total RNA into four types on the basis of RNA nucleotide length: 17–25 nt RNA (mostly microRNAs (miRNA)), 25–50 nt RNA (some tRNA-derived small RNAs (tsRNAs) and rRNA-derived small RNAs (rsRNAs)), 50–80 nt RNA (tRNA fragments and other ncRNAs) and >80 nt RNA (large RNA fragments) (Fig. 2A). As shown in Fig. 2B–E, 16 types of RNA modification marks were quantified in all sperm RNA fragments, and dynamic abundance variation of RNA modification marks in different RNA fragments were identified among the NZS (n = 7), AZS (n = 15) and TZS (n = 10) groups (Fig. 2F–I). The most dynamic alteration signature of these RNA modification marks in AZS, and TZS groups was found in >80 nt RNA fragments (Supplementary Fig. S3). Specifically, the levels of m5C, m3C, m7G, m2G, G, m1G and m1A marks were significantly higher in both the AZS and TZS groups than in the NZS group (Supplementary Fig. S3B–H), and the levels of m5C and m6A marks even higher in the TZS group (Supplementary Fig. S3B and I). Additionally, we found that the abundance of m7G and Am declined in the TZS group compared with the NZS group (Supplementary Fig. S3D and J). Taken together, RNA modification abundance variation in sperm RNA fragments >80 nt suggests that the sperm RNA modification marks are closely associated with AZS and TZS in human male infertility syndromes.
Figure 2.
The RNA modification signature of human sperm RNAs. (A) Sperm total RNA and RNA fragments of nucleotide length >80, 50–80, 25–50 and 17–25 nt after polyacrylamide gel electrophoresis (PAGE) on a 15% urea-polyacrylamide gel, stained with SYBR GOLD. (B–E) The radar graphs show the alteration of RNA modification abundance in human sperm RNA fragments of 17–25 nt (B), 25–50 nt (C), 50–80 nt (D) and >80 nt (E) among NZS (n = 7), AZS (n = 15) and TZS (n = 10) samples. The axes in the radar graphs represent the fold change in RNA modification level in AZS or TZS samples compared to NZS samples; (F–I) the heatmaps show the abundance of RNA modification marks, such as Am Cm Gm Um and I in (F), m1A, m1G, m2G, m5C, m6A and m7G in (G), m1I, 7G, G and m3C in (H) and Ψ in (I), in NZS (n = 7), AZS (n = 15) and TZS (n = 10) groups. NZS, Normozoospermia; AZS, Asthenozoospermia; TZS, Teratozoospermia. The uncropped gel for Fig. 2A is shown in Supplementary Fig. S9.
However, compared with the dynamic alteration of RNA modification abundance on sperm RNA fragments >80 nt (Supplementary Fig. S3), the RNA modification levels on the other sperm RNA fractions showed relatively more stable patterns (Supplementary Figs S4–S6), indicating the particular susceptibility to RNA modification of different sizes of sperm RNA fragments among the groups of men with fertility dysfunction. Our data reveal that the altered RNA modification signatures in sperm total RNA from patients with infertility were mainly related to changes in RNA modification abundance for RNA fragments >80 nt.
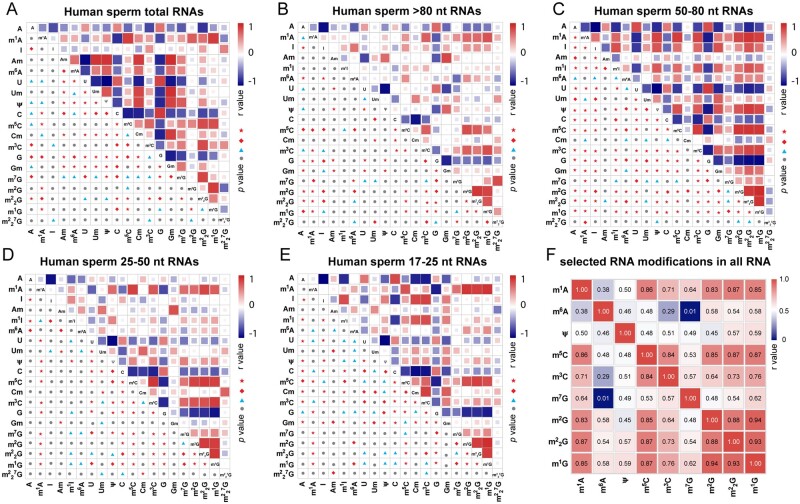
Linear correlation between different types of RNA modification in sperm RNA
Epigenetic marks, such as DNA methylation, histone marks and RNA modifications, exhibit definitive crosstalk patterns in the regulation of gene expression during embryonic development and human disease progression (Huang et al., 2019; Weinberg et al., 2019; Xu et al., 2021). However, whether the crosstalk among different RNA modification marks is extensive in human sperm RNAs, particularly in RNAs of different sizes, is unknown. Therefore, we analyzed the co-expression relations between different types of RNA modification marks on sperm total RNA and on sperm RNA fragments of different sizes in the NZS, AZS and TZS samples. Our data strongly suggest that many types of RNA modifications exhibit significantly linear co-dependent relations between total RNAs and RNA fragments of different sizes in human sperm (Fig. 3A–E). Among all RNA samples, we found that certain types of RNA modification followed a linear co-ordinated abundance pattern (Fig. 3F). Specifically, m1G showed a notable direct positive linear correlation with m1A, m5C, m3C, m2G and G, and all of these RNA modification marks showed strong relations with each other in all evaluated RNA samples, namely the total RNA and RNA fragments of different sizes in NZS, AZS and TZS groups (Supplementary Fig. S7). Notably, we found that m5C and m2G showed a much stronger positive linear correlation with each other in all groups (Fig. 3A–F and Supplementary Fig. S7I), consistent with our previous report (Zhang et al., 2018). Taken together, our present data support the idea that co-ordinated deposition of different RNA modifications marks on different types of RNA is prevalent, suggesting that crosstalk between RNA modification marks in human sperm RNA might be involved in regulating RNA metabolism associated with human infertility syndromes.
Figure 3.
Correlation analysis between the abundance of different RNA modification marks on human sperm total RNA and RNA fragments of different sizes. (A–E) Pearson correlation analysis for the abundance of different RNA modification marks on (A) sperm total RNAs (n = 17), (B) sperm RNA fragments >80 nt (n = 32), (C) sperm RNA fragments 50-80 nt (n = 32), (D) sperm RNA fragments 25–50 nt (n = 32) and (E) sperm RNA fragments 17–25 nt (n = 32). (F) Pearson correlation analysis for the abundance of selected RNA modification marks on total RNA and RNA fragments of different sizes (n = 145: total number of RNA modification results used in the correlation analysis). Pearson correlation coefficients were computed with GraphPad Prism 8, and Pearson r values are presented by heatmaps. The P values in (A–E) are shown on each cell as follows: ●, not significant; ▲, P < 0.05; ◆, P < 0.01; ★, P < 0.001. The detailed r values in (F) are presented in each cell.
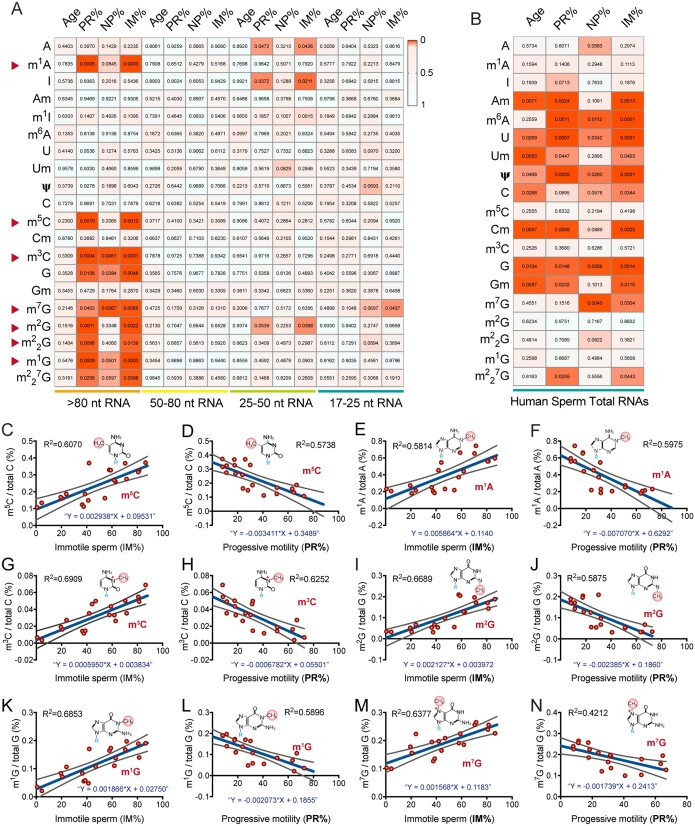
The sperm RNA modification signature is correlated with human sperm motility
To investigate the biological relevance of human sperm RNA modification levels in human male infertility syndromes, we performed a combination analysis of the RNA modification levels on sperm total RNA and RNA fragments with different sizes for the NZS and AZS samples with sperm motility scores. We found that the levels of m1A, m5C, m3C, m2G, G, m1G and m7G on the sperm RNA fragments >80 nt (n = 22) (Fig. 4A, Supplementary Table SIV), and Am, Cm, Um, Gm, pseudouridine (Ψ) and m6A on sperm total RNAs (n = 12) (Fig. 4B and Supplementary Table SIV) exhibited linear correlations with sperm motility parameters, which were positively associated with the percentage of immotile sperm and were negatively associated with the percentage of PR sperm (Fig. 4C–N). Since sperm motility and the cellular RNA modification signature may be affected by aging in humans (Johnson et al., 2015; Min et al., 2018), we analyzed the correlation of human age with sperm motility and RNA modification levels to exclude the effect of age on the correlation analyses between RNA modification abundance and sperm motility scores (Fig. 4A and B and Supplementary Fig. S8). Although we did not detect a significant correlation between human age (from 22 to 49 years) and sperm motility (Supplementary Fig. S8) or between human age and RNA modifications on sperm RNA fragments of different sizes (Fig. 4A), we indeed found that 2′-O-methylation (Am, Cm, Um and Gm) in human sperm total RNAs showed a significant linear correlation with human age (Fig. 4B), suggesting that the levels of 2′-O-methylation might be age-dependent in the total RNA of human sperm, which needs further investigation.
Figure 4.
Correlation analysis between human sperm RNA modification abundance and sperm motility. (A and B) The heatmaps show the Pearson correlation analyses between RNA modification abundance and human age, and between RNA modification abundance and sperm motility scores of NZS and AZS samples in (A) sperm RNA fragments of different sizes (n = 22) and in (B) sperm total RNAs (n = 12); P values are shown on each cell of the heat maps. Pearson correlation coefficients were computed with GraphPad Prism 8. (C–N) selected linear correlation analyses between RNA modification abundance and sperm motility scores for m5C, m1A, m3C, m2G, m1G and m7G, with human sperm motility of IM% (C, E, G, I, K, M) and PR% (D, F, H, J, L, N) in human sperm RNA fragments >80 nt (n = 20). Linear regressions were performed with GraphPad Prism 8. The linear equations R2 are shown in each panel. IM%, percentage of immotile sperm; PR%, percentage of progressive motility sperm; NP%, percentage of non-progressive sperm; NZS, Normozoospermia; AZS, Asthenozoospermia.
Discussion
Human male infertility is characterized by its complexity and heterogeneity, and sometimes is diagnosed on the basis of abnormal genetic components (such as chromosomal anomalies and gene copy number variations) or gene mutations (Krausz and Riera-Escamilla, 2018; Lotti and Maggi, 2018). However, the etiology and underlying pathological mechanisms of ∼50% of cases of clinical male factor infertility remain unexplained (Krausz and Riera-Escamilla, 2018), highlighting the urgent need to understand the pathological mechanisms involved and establish reliable diagnostic and treatment methods. Recently, researchers have been dedicated to investigating the genealogical tree of male infertility syndromes, including human AZS and TZS by whole-genome sequencing (Houston et al., 2021). Genetic screening has led to the identification of various gene mutations that can cause male infertility both in humans and mice; the mutated genes included FA complementation group M (FANCM) (Yin et al., 2019), dynein axonemal heavy chain 17 (DNAH17) (Zhang et al., 2020), Homo sapiens chromosome 14 open reading frame 39 (C14orf39/SIX6OS1) (Fan et al., 2021) and RAD51-associated protein 2 (RAD51AP2) (Ma et al., 2022). In the present work, we focused on illustrating the RNA modification landscape of human sperm to characterize the altered RNA modification signature in AZS and TZS patients. Notably, the hormone levels of the participants were not measured in this study, which might have affected the accuracy of the clinical diagnosis of male infertility syndrome. Moreover, since the participants in this study did not undergo genetic mutation screening, clarification of the pathologies of the different participants with similar semen analytic reports remains a challenge. Whether the altered RNA modification levels in human sperm RNA are associated with genetic mutations or abnormal hormone levels needs to be further investigated.
Compared to that of genetic mutations, the involvement of regulatory epigenetic marks (including DNA methylation and histone modification, noncoding RNAs and RNA modifications) in regulating broad biological processes is reversible and more dynamic (Minton, 2017; Cavalli and Heard, 2019). Specifically, RNA modification-based epigenetic regulation has been shown to play important roles in post-transcriptional regulation in embryonic development, stem cell fate commitment and human tumorigenesis (Roundtree et al., 2017), and in the sensitivity of responses to environmental alterations and cellular internal stresses (Yi and Pan, 2011). To date, over 170 types of RNA modification have been identified in all three main kingdoms of life (Animalia, Plantae and Protista) (Boccaletto et al., 2018). In this study, we described the abundance landscape of 22 types of RNA modifications in different sperm RNA fractions and found multiple linear correlations between the abundance of different RNA modification marks, providing the opportunity to understand the regulatory network of RNA modification marks in human sperm. Recent studies have reported crosstalk and co-ordinated regulation patterns among different RNA modification marks, which have been implicated in regulating mRNA translational efficiency (Ontiveros et al., 2020), paternally acquired epigenetic inheritance (Zhang et al., 2018) and human tumorigenesis (Chen et al., 2021a), adding complexity and diversity to the transcriptional and translational regulation of multiple biological processes. Moreover, the numerous combinations of different RNA modification marks and the regulatory crosstalk between them might generate a highly complex network with multifaceted functions in the regulation of biological processes. However, the limited commercially available modified RNA nucleobase standards and the differing abundances of the RNA modification marks in sperm RNA have restricted our research aimed at generating a more comprehensive landscape of RNA modification in human sperm. In the present study, we focused on identifying the RNA modifications that correlated with abnormal sperm motility and sperm morphology, and the results showed that: the abundance of eight types of RNA modification marks (m1A, Am, m6A, Cm, m7G, Gm, m5C and 7G) was altered in the total RNA of AZS and TZS samples compared with healthy controls; and the levels of nine types of RNA modification marks (m5C, m3C, m7G, m2G, G, m1G, m1A, m6A and Am) were significantly altered in sperm RNA fragments >80 nt in AZS and TZS samples. Notably, RNA modifications, including m1A, m3C, m5C, m7G, m2G, G and m1G, exhibited a high linear correlation with sperm motility, which might suggest essential roles in regulating sperm motility. However, whether the relation between altered RNA modification abundance and impaired sperm quality is causal remain unclear. More screening through RNA modification-specific sequencing, such as m1A-seq and m6A-seq, is expected to provide information to determine any causality between observed RNA modification alterations and RNA expression levels. In addition, the exploration of various RNA modifications mapped to specific RNA sequences based on mass spectrometry or nanopore-based de novo RNA-sequencing are promising methods to interpret the potential biological mechanisms of RNA modification alterations in male infertility syndrome (Shi et al., 2022).
The distinct signature of sperm RNA modifications in human sperm might be the result of the diverse composition of RNAs in sperm of the AZS and TZS groups, such as fragmented tRNAs, rRNAs and mRNAs inherited from the previous stages of spermatogenesis. In fact, RNA modifications are closely associated with cellular RNA stability, especially for tRNAs and rRNAs (Shi et al., 2021). RNA modification marks, such as m5C on C38 and m1A on A58 of tRNA, protected tRNAs against cleavage into tsRNAs (Chen et al., 2021b), while YTHDF2 recognized m6A modified mRNAs to promote mRNA degradation (Hou et al., 2021). m5C has been reported to be involved in regulating spermiogenesis (Hussain et al., 2013), and dysregulation of the m6A led to impaired spermiogenesis and decreased sperm motility (Gui and Yuan, 2021). In order to determine whether and how the other types of aforementioned RNA modification marks are also involved in spermiogenesis or sperm quality control, more mechanistic investigations are needed. Nevertheless, this is the first study to demonstrate a linear correlation between RNA modification and human sperm motility, which might provide clues for identifying pathological mechanisms in AZS and TZS (beyond genetic causes) and lead to applications of sperm RNA modification analysis to clinical sperm quality assessment. However, owing to the highly technical nature of quantitative analysis of RNA modification, the clinical application of this technology might be limited in most infertility clinics. A simpler and more convenient method for multiple RNA modification quantitative analyses needs to be developed to promote clinical application of RNA modification in human sperm quality assessment.
Interestingly, we had previously found that RNA modification marks, such as m5C and m2G, were affected by a nutritionally unbalanced diet and were involved in transmitting paternally acquired metabolic disorders across generations in mice (Chen et al., 2016a; Zhang et al., 2018). In this study, we found significant alterations of m5C and m2G levels in the total RNA of human sperm and sperm RNA fragments in the AZS and TZS samples, which were closely associated with human sperm motility. More importantly, the levels of m5C and m2G marks also showed a high linear correlation in human sperm RNAs, indicating a conserved, coordinated regulatory mechanism between m5C and m2G in human and mice sperm RNAs. However, whether and how RNA modifications in human sperm are associated with paternally acquired epigenetic inheritance remains unclear and further investigation is needed.
Taken together, our data revealed the first RNA modification landscape identified in human sperm and revealed multiple linear correlations of RNA modification levels with sperm motility, providing a potential medical diagnostic methodology for human fertility assessment and new insights into the underlying epigenetic regulation mechanisms involved in AZS and TZS infertility.
Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Supplementary Material
Acknowledgements
The authors thank Dr Qi Chen at the University of California, Riverside, and Dr Tong Zhou at the University of Nevada, Reno School of Medicine, for their important suggestions on the correlation analysis between different types of RNA modifications. We appreciate the participants for their contribution in providing samples. The authors also thank the clinical staff of the Center for Reproductive & Genetic Medicine for their assistance in clinical data collection and semen sample preparation.
Authors’ roles
Y.Z. and X.Z. conceived the idea of this study and Y.Z. and X.S. designed this project. P.Z. and H.H. communicated with the participants and provided the clinical data on the sperm. X.S. and T.H. collected semen samples and isolated sperm. X.S. and H.G. contributed to sperm RNA purification and performed the RNA modification detection experiments. X.S. and H.G. also acquired RNA modification data and analyzed them under the supervision of Y.Z. and X.Z.; Y.Z. and X.S. wrote the manuscript with the help of H.G. X.Z. and D.T. critically revised the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the National Key R&D Program of China (2019YFA0802600), the National Natural Science Foundation of China (82022029 and 81971460) and the Natural Science Foundation of Chongqing (cstc2019jcyjjqX001) and Key Project in Clinical Medical Research of Army Medical University (2018XLC2018).
Conflict of interest
The authors declare no competing or financial interests.
Contributor Information
Huanping Guo, Medical Center of Hematology, The Second Affiliated Hospital of Army Medical University, Chongqing, China.
Xipeng Shen, Medical Center of Hematology, The Second Affiliated Hospital of Army Medical University, Chongqing, China.
Hua Hu, Center for Reproductive & Genetic Medical, The Second Affiliated Hospital of Army Medical University, Chongqing, China.
Peng Zhou, Center for Reproductive & Genetic Medical, The Second Affiliated Hospital of Army Medical University, Chongqing, China.
Tong He, Medical Center of Hematology, The Second Affiliated Hospital of Army Medical University, Chongqing, China; Laboratory Animal Center, Chongqing Medical University, Chongqing, China.
Lin Xia, Medical Center of Hematology, The Second Affiliated Hospital of Army Medical University, Chongqing, China.
Dongmei Tan, Laboratory Animal Center, Chongqing Medical University, Chongqing, China.
Xi Zhang, Medical Center of Hematology, The Second Affiliated Hospital of Army Medical University, Chongqing, China.
Yunfang Zhang, Medical Center of Hematology, The Second Affiliated Hospital of Army Medical University, Chongqing, China; Clinical and Translational Research Center of Shanghai First Maternity and Infant Hospital, Shanghai Key Laboratory of Signaling and Disease Research, Frontier Science Center for Stem Cell Research, School of Life Sciences and Technology, Tongji University, Shanghai, China.
Data Availability
The data related to this article are available in the article and in its supplementary information. The raw RNA modification mass spectrum data underlying this article will be shared on reasonable request to the corresponding author.
References
- Agarwal A, Baskaran S, Parekh N, Cho CL, Henkel R, Vij S, Arafa M, Panner Selvam MK, Shah R.. Male infertility. Lancet 2021;397:319–333. [DOI] [PubMed] [Google Scholar]
- Amaral A, Castillo J, Ramalho-Santos J, Oliva R.. The combined human sperm proteome: cellular pathways and implications for basic and clinical science. Hum Reprod Update 2014;20:40–62. [DOI] [PubMed] [Google Scholar]
- Barbieri I, Kouzarides T.. Role of RNA modifications in cancer. Nat Rev Cancer 2020;20:303–322. [DOI] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A et al MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 2018;46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G, Heard E.. Advances in epigenetics link genetics to the environment and disease. Nature 2019;571:489–499. [DOI] [PubMed] [Google Scholar]
- Chen H, Yao J, Bao R, Dong Y, Zhang T, Du Y, Wang G, Ni D, Xun Z, Niu X. et al. Cross-talk of four types of RNA modification writers defines tumor microenvironment and pharmacogenomic landscape in colorectal cancer. Mol Cancer 2021a;20:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016a;351:397–400. [DOI] [PubMed] [Google Scholar]
- Chen Q, Yan W, Duan E.. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet 2016b;17:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhang X, Shi J, Yan M, Zhou T.. Origins and evolving functionalities of tRNA-derived small RNAs. Trends Biochem Sci 2021b;46:790–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TG, , NoonanE, , Von EckardsteinS, , AugerJ, , BakerHG, , BehreHM, , HaugenTB, , KrugerT, , WangC, , Mbizvo MT. et al. World Health Organization reference values for human semen characteristics. Human Reproduction Update 2010;16:231–245. [DOI] [PubMed] [Google Scholar]
- Corral-Vazquez C, Salas-Huetos A, Blanco J, Vidal F, Sarrate Z, Anton E.. Sperm microRNA pairs: new perspectives in the search for male fertility biomarkers. Fertil Steril 2019;112:831–841. [DOI] [PubMed] [Google Scholar]
- Fan S, Jiao Y, Khan R, Jiang X, Javed AR, Ali A, Zhang H, Zhou J, Naeem M, Murtaza G. et al. Homozygous mutations in C14orf39/SIX6OS1 cause non-obstructive azoospermia and premature ovarian insufficiency in humans. Am J Hum Genet 2021;108:324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Harada BT, Behm M, He C.. RNA modifications modulate gene expression during development. Science 2018;361:1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M, Jaffrey SR, Pan T, Rechavi G, Suzuki T.. RNA modifications: what have we learned and where are we headed? Nat Rev Genet 2016;17:365–372. [DOI] [PubMed] [Google Scholar]
- Georgiadis AP, Kishore A, Zorrilla M, Jaffe TM, Sanfilippo JS, Volk E, Rajkovic A, Yatsenko AN.. High quality RNA in semen and sperm: isolation, analysis and potential application in clinical testing. J Urol 2015;193:352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui Y, Yuan S.. Epigenetic regulations in mammalian spermatogenesis: RNA-m6A modification and beyond. Cell Mol Life Sci 2021;78:4893–4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T, Marquez B, Suarez S, Schimenti J.. Sperm motility defects and infertility in male mice with a mutation in Nsun7, a member of the Sun domain-containing family of putative RNA methyltransferases. Biol Reprod 2007;77:376–382. [DOI] [PubMed] [Google Scholar]
- Hasler D, Meduri R, Bąk M, Lehmann G, Heizinger L, Wang X, Li Z-T, Sement FM, Bruckmann A, Dock-Bregeon A-C. et al. The Alazami syndrome-associated protein LARP7 guides U6 small nuclear RNA modification and contributes to splicing robustness. Mol Cell 2020;77:1014–1031.e13. [DOI] [PubMed] [Google Scholar]
- Hou G, Zhao X, Li L, Yang Q, Liu X, Huang C, Lu R, Chen R, Wang Y, Jiang B. et al. SUMOylation of YTHDF2 promotes mRNA degradation and cancer progression by increasing its binding affinity with m6A-modified mRNAs. Nucleic Acids Res 2021;49:2859–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston BJ, Riera-Escamilla A, Wyrwoll MJ, Salas-Huetos A, Xavier MJ, Nagirnaja L, Friedrich C, Conrad DF, Aston KI, Krausz C. et al. A systematic review of the validated monogenic causes of human male infertility: 2020 update and a discussion of emerging gene-disease relationships. Hum Reprod Update 2021;28:15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J. et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res 2017;27:1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, Chen Z, Deng X, Xiao G, Auer F. et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 2019;567:414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Tuorto F, Menon S, Blanco S, Cox C, Flores JV, Watt S, Kudo NR, Lyko F, Frye M.. The mouse cytosine-5 RNA methyltransferase NSun2 is a component of the chromatoid body and required for testis differentiation. Mol Cell Biol 2013;33:1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodar M, Kalko S, Castillo J, Ballesca JL, Oliva R.. Differential RNAs in the sperm cells of asthenozoospermic patients. Hum Reprod 2012;27:1431–1438. [DOI] [PubMed] [Google Scholar]
- Jodar M, Selvaraju S, Sendler E, Diamond MP, Krawetz SA; Reproductive Medicine Network. The presence, role and clinical use of spermatozoal RNAs. Hum Reprod Update 2013;19:604–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodar M, Sendler E, Moskovtsev SI, Librach CL, Goodrich R, Swanson S, Hauser R, Diamond MP, Krawetz SA.. Absence of sperm RNA elements correlates with idiopathic male infertility. Sci Transl Med 2015;7:295re296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Dunleavy J, Gemmell NJ, Nakagawa S.. Consistent age-dependent declines in human semen quality: a systematic review and meta-analysis. Ageing Res Rev 2015;19:22–33. [DOI] [PubMed] [Google Scholar]
- Krausz C, Riera-Escamilla A.. Genetics of male infertility. Nat Rev Urol 2018;15:369–384. [DOI] [PubMed] [Google Scholar]
- Krawetz SA. Paternal contribution: new insights and future challenges. Nat Rev Genet 2005;6:633–642. [DOI] [PubMed] [Google Scholar]
- Lewis CJ, Pan T, Kalsotra A.. RNA modifications and structures cooperate to guide RNA-protein interactions. Nat Rev Mol Cell Biol 2017;18:202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou Q, Zhang KJ, Zhang X, Zhou Y, Zhang T. et al. Mettl3-/Mettl14-mediated mRNA N(6)-methyladenosine modulates murine spermatogenesis. Cell Res 2017;27:1216–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotti F, Maggi M.. Sexual dysfunction and male infertility. Nat Rev Urol 2018;15:287–307. [DOI] [PubMed] [Google Scholar]
- Ma H, Li T, Xie X, Jiang L, Ye J, Gong C, Jiang H, Fan S, Zhang H, Shi B. et al. RAD51AP2 is required for efficient meiotic recombination between X and Y chromosomes. Sci Adv 2022;8:eabk1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D, Ostermeier GC, Krawetz SA.. The controversy, potential and roles of spermatozoal RNA. Trends Mol Med 2005;11:156–163. [DOI] [PubMed] [Google Scholar]
- Min KW, Zealy RW, Davila S, Fomin M, Cummings JC, Makowsky D, McDowell CH, Thigpen H, Hafner M, Kwon SH. et al. Profiling of m6A RNA modifications identified an age-associated regulation of AGO2 mRNA stability. Aging Cell 2018;17:e12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton K. Stem cells: HSC function determined by epigenetic memory. Nat Rev Mol Cell Biol 2017;18:1. [DOI] [PubMed] [Google Scholar]
- Monesi V. Ribonucleic acid synthesis during mitosis and meiosis in the mouse testis. J Cell Biol 1964;22:521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monesi V, Geremia R, D'Agostino A, Boitani C.. Biochemistry of male germ cell differentiation in mammals: RNA synthesis in meiotic and postmeiotic cells. Curr Top Dev Biol 1978;12:11–36. [DOI] [PubMed] [Google Scholar]
- Morgan M, Kumar L, Li Y, Baptissart M.. Post-transcriptional regulation in spermatogenesis: all RNA pathways lead to healthy sperm. Cell Mol Life Sci 2021;78:8049–8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ontiveros RJ, Shen H, Stoute J, Yanas A, Cui Y, Zhang Y, Liu KF.. Coordination of mRNA and tRNA methylations by TRMT10A. Proc Natl Acad Sci USA 2020;117:7782–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M, Sun H, Guo Y, Zhou Y, Gu X, Jin J, Chen X, Wang F, Ma H, Guo X. et al. m6A reader protein YTHDF2 regulates spermatogenesis by timely clearance of phase‐specific transcripts. Cell Proliferation 2022;55:e13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C.. Dynamic RNA modifications in gene expression regulation. Cell 2017;169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhang Y, Tan D, Zhang X, Yan M, Zhang Y, Franklin R, Shahbazi M, Mackinlay K, Liu S. et al. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat Cell Biol 2021;23:424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhou T, Chen Q.. Exploring the expanding universe of small RNAs. Nat Cell Biol 2022;24:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves ME, Roldan ERS.. Sperm bauplan and function and underlying processes of sperm formation and selection. Physiol Rev 2022;102:7–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventimiglia E, Montorsi F, Salonia A.. Comorbidities and male infertility: a worrisome picture. Curr Opin Urol 2016;26:146–151. [DOI] [PubMed] [Google Scholar]
- Wang X, Li ZT, Yan Y, Lin P, Tang W, Hasler D, Meduri R, Li Y, Hua MM, Qi HT. et al. LARP7-mediated U6 snRNA modification ensures splicing fidelity and spermatogenesis in mice. Mol Cell 2020;77:999–1013.e6. [DOI] [PubMed] [Google Scholar]
- Weinberg DN, Papillon-Cavanagh S, Chen H, Yue Y, Chen X, Rajagopalan KN, Horth C, McGuire JT, Xu X, Nikbakht H. et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 2019;573:281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener D, Schwartz S.. The epitranscriptome beyond m(6)A. Nat Rev Genet 2021;22:119–131. [DOI] [PubMed] [Google Scholar]
- World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 2013;310:2191–2194. [DOI] [PubMed] [Google Scholar]
- Xu K, Yang Y, Feng G-H, Sun B-F, Chen J-Q, Li Y-F, Chen Y-S, Zhang X-X, Wang C-X, Jiang L-Y. et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res 2017;27:1100–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Li J, He C, Wen J, Ma H, Rong B, Diao J, Wang L, Wang J, Wu F. et al. METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature 2021;591:317–321. [DOI] [PubMed] [Google Scholar]
- Yi C, Pan T.. Cellular dynamics of RNA modification. Acc Chem Res 2011;44:1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Ma H, Hussain S, Zhang H, Xie X, Jiang L, Jiang X, Iqbal F, Bukhari I, Jiang H. et al. A homozygous FANCM frameshift pathogenic variant causes male infertility. Genet Med 2019;21:62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Ma H, Khan T, Ma A, Li T, Zhang H, Gao J, Zhou J, Li Y, Yu C. et al. A DNAH17 missense variant causes flagella destabilization and asthenozoospermia. J Exp Med 2020;217:e20182365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shi J, Rassoulzadegan M, Tuorto F, Chen Q.. Sperm RNA code programmes the metabolic health of offspring. Nat Rev Endocrinol 2019;15:489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Shi J, Tuorto F, Li X, Liu Y, Liebers R, Zhang L, Qu Y, Qian J. et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol 2018;20:535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013;49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong D, Chen M, Zhang L, Chen H, Shi D, Liu Q, Li H.. Aberrant regulation of RNA methylation during spermatogenesis. Reprod Dom Anim 2021;56:3–11. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data related to this article are available in the article and in its supplementary information. The raw RNA modification mass spectrum data underlying this article will be shared on reasonable request to the corresponding author.