Abstract
Clonal hematopoiesis of indeterminate potential (CHIP) is common in the elderly and has been reported to associate with accelerated epigenetic age (AgeAccel), especially intrinsic (ie, cell-type independent) AgeAccel and to a lesser degree extrinsic AgeAccel, which reflects the immune-cell composition of the peripheral blood. We investigated the association between CHIP occurrence and AgeAccel in 154 Danish twin pairs aged 73–90 years (mean 79), using both individual-level and intrapair analyses, the latter to control for shared genetic and environmental factors. Of 308 individuals, 116 carried a CHIP mutation. CHIP carriers had non-significantly increased AgeAccel compared with non-carriers; the strongest association was for the Intrinsic Epigenetic Age Acceleration (IEAA) estimator (CHIP carriers 1.4 years older, P = 0.052). In intrapair analyses, the extrinsic Hannum age estimator showed the strongest association (1.6 years older, P = 0.027). In mutation-specific analyses, TET2 mutations were associated with the extrinsic Hannum age estimator in both individual-level (3.0 years older, P = 0.003) and intrapair analyses (2.8 years older, P = 0.05). DNMT3A mutations were associated with IEAA in individual-level (1.9 years older, P = 0.034) but not intrapair analysis (0.9 years, P = 0.41). Analyses of logit-transformed variant allele frequency were generally consistent with these results. Together, these observations indicate that different factors may be driving the expansion of DNMT3A and TET2 clones, respectively. Finally, CHIP carriers accelerated in both the Hannum and the GrimAge age estimators did not have an increased mortality risk in our cohort followed for 22 years (HR = 1.02, P = 0.93), hence not replicating the stratification model proposed by Nachun et al.
INTRODUCTION
Clonal hematopoiesis of indeterminate potential (CHIP) is a commonly occurring phenomenon in the elderly characterized by propagation of blood cell clones carrying somatic mutations in genes related to myeloid cancers.1,2 CHIP may develop into clonal cytopenia of undetermined significance or overt myeloid malignancy, but CHIP has also been reported to associate with a range of age-related disorders and outcomes, most importantly all-cause mortality and cardiovascular disease.1,3,4
Two previous studies have demonstrated an association between CHIP detected by whole-genome sequencing and accelerated epigenetic age. First, Robertson et al5 found that CHIP was associated with several estimators of epigenetic age acceleration, especially the Intrinsic Epigenetic Age Acceleration (IEAA) estimator, which is designed to measure DNA methylation changes occurring independently of age-related changes to the immune-cell composition of the peripheral blood.6 Subsequently, Nachun et al7 replicated the association between CHIP and intrinsic epigenetic age and furthermore reported a less pronounced association with extrinsic epigenetic age estimators such as the Hannum and the Extrinsic Epigenetic Age Acceleration (EEAA) estimators, which also reflect DNA methylation changes that occur as a result of age-related shifts in immune-cell proportions (eg, decreased numbers of naive CD8+ T cells).6,8 Finally, Nachun et al7 also reported that a specific subset of CHIP carriers who had positive age acceleration in both the Hannum and GrimAge estimators (AgeAccelHG+) had an increased risk of all-cause mortality compared with CHIP carriers without AgeAccelHG+, as well as compared with individuals without CHIP regardless of AgeAccelHG status.
The aim of this study was to replicate and extend these findings in a cohort of elderly Danish twins.9 Compared with the previously published studies, the twin design enables us to control for confounding effects of familial factors, that is, shared genetic and environmental factors. Furthermore, our cohort is older or of similar age than the 2 previous studies, and CHIP was detected using error-corrected panel sequencing which allowed for more sensitive detection of low-frequency variants.
MATERIALS AND METHODS
Study population
The study population was comprised of 154 same-sex twin pairs (121 monozygotic and 33 dizygotic pairs) from the Longitudinal Study of Aging Danish Twins (LSADT).10 LSADT is a cohort sequential study of Danish twins aged 70 years or more, initiated in 1995.11 In 1997, whole blood samples were collected from 689 same-sex twins and the present study includes all the 308 twins for whom data regarding CHIP and DNAmAge were available.9,12 Mortality was assessed through January 1, 2020, for the present study population, except for 84 individuals as they had been selected for survival 10 years from baseline, leaving 224 individuals for survival analysis. At the end of follow-up, 220 (98%) were deceased. The median follow-up time was 8.39 years. Informed consent was obtained from all participants and the surveys and CHIP analyses were approved by The Regional Committees on Health Research Ethics for Southern Denmark (S-VF-20040241 and S-20170053). Consequently, the study was conducted in accordance with the Helsinki II declaration.
Biological data
Data on CHIP mutations and DNA methylation levels
DNA sequencing was performed as described previously (see Hansen et al9 for details). In short, an Illumina TruSeq Custom Amplicon panel (Illumina, San Diego, CA), covering >95% of the mutations commonly associated with clonal hematopoiesis, was used for targeted sequencing. DNA was quantified with a Qubit fluorometer (Life Technologies, Carlsbad, CA) and 100–200 ng of DNA was applied for library preparation. Unique molecular identifiers, consisting of 6 random index nucleotides, were added to each sample before amplification to optimize variant calling and identification of low-level mutations. Libraries were pooled and sequenced using the Illumina NextSeq platform (Illumina) with a 300-cycle mid output kit, as specified by the manufacturer. Alignment was performed using the BWA-mem algorithm,13 and variant calling was conducted using Freebayes v.1.1.014 and VarDict v.1.5.1.15 In the present study, we explored CHIP mutations as defined by Steensma et al2 with a variant allele frequency (VAF) of 2% or above. A detailed description of the generation of the DNA sequencing data and variant calling can, furthermore, be found in the Supplementary Material of Hansen, et al.9
DNA methylation was measured using 2 Infinium HumanMethylation450K BeadChip datasets obtained as previously described.16 Methylation profiling was performed at 2 different occasions with 84 in the first batch and 224 in the second. Twin pairs were always run on the same array. Quality control (QC) and estimation of epigenetic age were performed individually for the 2 datasets (see Soerensen et al16 for details). Briefly, 500 ng DNA per sample was bisulfite converted using the EZ Methylation Gold kit (Zymo Research, Orange County, CA) before analysis with the Infinium HumanMethylation450K BeadChip (Illumina) using standard procedures. QC was performed with the MethylAid17 and Minfi18 R packages. Sample exclusion criteria: (1) <95% of probes with a detection P value <0.01, (2) samples failing the internal QC probes of the MethylAid, or (3) samples failing verification of sex by multidimensional scaling of the X chromosome probe values. No samples were removed during QC. Probe exclusion criteria were: (1) detection P value >0.01, (2) a raw intensity value of zero, (3) low bead count (<3 beads), (4) cross-reactive probes,19 or (5) measurement success rate <95%. Normalization was performed with Functional normalization20 using 4 principle components.
DNA methylation–derived estimates of biological age and cell counts
We calculated 6 different measures of epigenetic age: Horvath,21 Hannum,8 PhenoAge,22 GrimAge,23 IEAA, and EEAA.6 Of these 6 age estimators, Horvath and IEAA are considered intrinsic, that is, they reflect cell-type independent effects of aging on DNA methylation, while Hannum, PhenoAge, GrimAge, and EEAA are considered extrinsic measures, that is, they also reflect age-related changes to the immune-cell composition of the peripheral blood.6
The Horvath, Hannum, and PhenoAge age estimators were calculated as described in the original publications, while GrimAge and EEAA were calculated using scripts obtained from personal communication with Steve Horvath and Ake Lu (University of California). IEAA was calculated as described in the original publication using methylation-based cell counts obtained via the online DNA methylation age calculator (https://dnamage.genetics.ucla.edu/home, Horvath. 2013). Briefly, the IEAA is the residuals obtained by regressing chronological age and DNA methylation-derived estimates of cell counts, that is, plasma B cells (variable PlasmaBlast), CD8+CD28-CD45RA T cells (exhausted CD8+ T cells) (variable CD8pCD28nCD45Ran), naive CD8+ T cells (variable CD8_naive), CD4+ T cells (variable CD4T), natural killer cells (variable NK), monocytes (variable Mono), and granulocytes (variable Gran) onto the Horvath age estimator.6,21 The EEAA is obtained by regressing chronological age on to the BioAge4HAStatic variable, which is a modified version of the Hannum age estimator obtained as a weighted average of the Hannum age estimator and 3 estimated measures of age-associated blood cells, that is, naive (CD45RA+CCR7+) cytotoxic T cells, exhausted (CD28−CD45RA−) cytotoxic T cells and plasma B cells. For Horvath, Hannum, PhenoAge, and GrimAge, we defined epigenetic age acceleration (AgeAccel) as the residuals obtained from regressing DNAmAge on chronological age.
Statistical analyses
All analyses were performed using STATA16 (Stata Corporation, College Station, TX). All reported P values are 2-sided and not adjusted for multiple testing. We defined statistical significance as P < 0.05.
We first investigated the association between AgeAccel and CHIP mutations, and between AgeAccel and logit-transformed VAF considering each individual separately (ie, performing an individual-level analysis). The VAF values were logit-transformed to better reflect the growth rate of the CHIP clones and to obtain a distribution of the data more suitable for statistical analysis. The individual-level analyses were fitted as linear regressions with AgeAccel as the outcome variable and CHIP status, respectively, logit-transformed VAF as an exposure variable along with sex and DNA methylation batch as covariates. To account for dependency between twins in a pair, we used the Hubert-White-Sandwich (robust) estimator of variance, assuming independence between twin pairs (cluster function in STATA).
For intrapair analyses, we subsequently selected twin pairs discordant for CHIP mutation status or logit-transformed VAF. For mutation status, a linear regression with the intrapair difference in AgeAccel (ie, the AgeAccel value of the twin with a mutation minus the AgeAccel value of the co-twin without a mutation) was applied, simply investigating the intercept. For VAF, a linear regression model was applied with intrapair difference in logit-transformed VAF as the exposure and the intrapair difference in AgeAccel as the outcome. The intrapair differences were calculated by subtracting the value for the twin with the lowest VAF from the value for the co-twin with the highest VAF. As only same-sex twin pairs were included in the present study and as each twin pair was always analyzed on the same DNA methylation array, there is no intrapair variation in either sex or DNA methylation batch.
Finally, the associations between both CHIP and epigenetic age acceleration and increased risk of all-cause mortality are well established.5,7,24 Interestingly, it was previously reported that in the LSADT cohort, there was no significantly increased mortality risk among CHIP carriers, possibly due to the already highly advanced age and high CHIP prevalence in this cohort or a lack of statistical power.9 Since this result was published, Nachun et al7 showed that a combination of the Hannum and GrimAge age estimators together with CHIP status could identify individuals with highly increased mortality risk (specifically, individuals were defined as being double accelerated (termed AgeAccelHG+) if an individual was accelerated (ie, AgeAccel > 0) in both age estimators and individuals were defined as AgeAccelHG− if an individual was accelerated (ie, AgeAccel > 0) only in 1 of the 2 age estimators or was deaccelerated (ie, AgeAccel < 0) in both age estimators).7 We therefore aimed to investigate whether this set of predictors could also identify individuals with increased mortality in the present study population. Overall survival was analyzed with the Kaplan-Meier estimator using time since blood sample as the underlying time scale and with Cox proportional hazards regression using age as the timescale (delayed entry at blood collection) and adjusted for the sex by using sex-specific baseline hazards. To account for dependency between twins in each twin pair, we used the robust estimator of variance, assuming independence between pairs (cluster function). As the 84 individuals of the study population had been selected for survival 10 years from baseline, they were excluded from the survival analysis, leaving 224 individuals for analysis.
Finally, the previous study by Robertson et al5 included data on blood cell type proportions as covariates in their statistical models. In line with Nachun et al,7 we did not include cell counts in our statistical models due to the way the intrinsic and extrinsic epigenetic clocks are constructed, that is, with and without correction for cell counts. Nevertheless, expanding all our statistical models with adjustment for imputed cell counts led to very similar results and to the same conclusions as without cell counts (data not shown).
RESULTS
CHIP mutations and epigenetic age estimators in the twins
One hundred and sixteen individuals (37.7%) carried one or more CHIP mutations, while 47 (15.3%) and 52 (16.9%) carried a TET2 or DNMT3A mutation, respectively (Suppl. Table S1 and Suppl. Figures S1 and S2). All DNAmAges were significantly correlated with chronological age with correlation coefficients between 0.39 and 0.49 (all P < 1.30 × 10−7, Suppl. Figures S3 and S4). As expected, the Hannum and BioAge4HAStatic age estimators overestimated the chronological age (on average by 2.9–3.8 years), while the remaining age estimators underestimated the chronological age (on average by 0.7–7.6 years; Suppl. Table S1). As expected, AgeAccel was independent of chronological age (Suppl. Figures S5 and S6). Finally, males in general tended to be more accelerated than females; by 1.90 years for Hannum AgeAcceleration (P = 0.008), 2.82 years for EEAA (P = 0.003), 3.65 years for GrimAge AgeAcceleration (P = 4.16 × 10−7), and 0.75 years for Horvath AgeAcceleration (P = 0.364), while PhenoAge AgeAcceleration and IEAA reflected no difference (−0.05 years, P = 0.957, and −0.32, P = 0.679, respectively; Suppl. Table S2). All the positive effect sizes mentioned in the “Association analyses of CHIP and epigenetic age acceleration” section below regarding the association between CHIP mutation status and AgeAcceleration are larger than these sex differences in age acceleration.
Association analyses of CHIP and epigenetic age acceleration
Individual-level analyses of CHIP mutations and VAF
For all AgeAccel estimates, CHIP carriers had increased age acceleration compared with non-carriers in the individual-level analysis, although no estimates reached statistical significance (Table 1). The largest effect size was seen for IEAA (1.40 years mean difference between carriers and non-carriers, P = 0.052). We next investigated the same association for mutations in specific genes using 2 similar regression models restricted to individuals with either TET2 or DNMT3A mutations and individuals with no CHIP mutations. For TET2, we found that the extrinsic epigenetic age estimators were most strongly correlated with mutation status, most notably EEAA with TET2 mutation carriers being 3.61 years more accelerated than non-carriers (P = 0.004) and Hannum (3.04 years, P = 0.003). In contrast, DNMT3A mutations were most strongly associated with the intrinsic IEAA (1.93 years, P = 0.034), while the EEAA and Hannum estimators displayed negative, statistically insignificant associations of −1.13 years (P = 0.255) and −0.58 years (P = 0.456), respectively.
Table 1.
Individual-level Analysis of CHIP Mutation Status and Age Acceleration by Linear Regression Analysis
| AgeAccel | Coef. (95% CI) | P | |
|---|---|---|---|
| Any CHIP mutation (N = 308 individuals) | Horvath | 0.98 (−0.67 to 2.63) | 0.243 |
| IEAA | 1.40 (−0.01 to 2.81) | 0.052 | |
| Hannum | 1.04 (−0.25 to 2.33) | 0.112 | |
| EEAA | 1.06 (−0.56 to 2.69) | 0.197 | |
| PhenoAge | 0.51 (−1.32 to 2.35) | 0.582 | |
| GrimAge | 0.82 (−0.14 to 1.78) | 0.094 | |
| TET2 mutation (N = 239 individuals) | Horvath | 1.12 (−1.16 to 3.39) | 0.333 |
| IEAA | 1.09 (−0.84 to 3.02) | 0.266 | |
| Hannum | 3.04 (1.07 to 5.01) | 0.003 | |
| EEAA | 3.61 (1.17 to 6.06) | 0.004 | |
| PhenoAge | 1.61 (−0.77 to 3.98) | 0.183 | |
| GrimAge | 1.48 (0.15 to 2.81) | 0.029 | |
| DNMT3A mutation (N = 244 individuals) | Horvath | 1.03 (−0.86 to 2.93) | 0.283 |
| IEAA | 1.93 (0.15 to 3.72) | 0.034 | |
| Hannum | −0.58 (−2.11 to 0.95) | 0.456 | |
| EEAA | −1.13 (−3.09 to 0.82) | 0.255 | |
| PhenoAge | 0.01 (−2.35 to 2.37) | 0.992 | |
| GrimAge | 0.68 (−0.81 to 2.16) | 0.369 |
Statistically significant findings (P < 0.05) are indicated in bold.
AgeAccel = epigenetic age acceleration; CHIP = clonal hematopoiesis of indeterminate potential; Coef. = coefficient (beta value); EEAA = extrinsic epigenetic age acceleration; IEAA = intrinsic epigenetic age acceleration.
Based on the expectation that any predictor associated with CHIP mutations should also be associated with the size of the mutated clones, we next carried out regression analyses restricted to individuals with CHIP, or with TET2 or DNMT3A mutations using logit-transformed VAF (base 10) as the covariate of interest instead of mutation status. Although these analyses are conducted within the same cohort, these results are statistically independent from the analyses of mutation status, as all individuals of the study population are included in the analysis of mutation status (ie, carrying a mutation or not), while only mutation carriers are included in the analysis of VAF. All VAF increased with AgeAccel for all epigenetic clock, although it was not statistically significant for the PhenoAge and GrimAge clocks (Suppl. Figure S7). Investigating the association between AgeAccel and VAF, we found strong signals for all CHIP mutations, most notably for EEAA (7.97 years increase per 1-unit increase on logit-transformed VAF, P = 0.00012, Table 2). In gene-specific analyses, the results were consistent with our findings for the analyses of mutation status, with TET2 most strongly associated with EEAA (6.32 years, P = 0.039) and Hannum (4.85 years, P = 0.066). DNMT3A was most strongly associated with the intrinsic age estimators IEAA (9.57 years, P = 0.013), and Horvath (9.64 years, P = 0.015), but also displayed significant associations at smaller effect sizes for EEAA (6.45 years, P = 0.046) and Hannum (5.65 years, P = 0.032).
Table 2.
Individual-level Analysis of VAF and Age Acceleration by Linear Regression Analysis
| AgeAccel | Coef. (95% CI) | P | |
|---|---|---|---|
| Any CHIP mutation (N = 116 individuals) | Horvath | 5.87 (1.47 to 10.27) | 0.009 |
| IEAA | 4.72 (0.57 to 8.88) | 0.026 | |
| Hannum | 6.38 (3.11 to 9.66) | 0.00020 | |
| EEAA | 7.97 (4.03 to 11.90) | 0.00012 | |
| PhenoAge | 4.92 (−0.44 to 10.29) | 0.071 | |
| GrimAge | 1.84 (−0.78 to 4.47) | 0.166 | |
| TET2 mutation (N = 47 individuals) | Horvath | 4.11 (−1.33 to 9.54) | 0.135 |
| IEAA | 2.14 (−2.52 to 6.80) | 0.359 | |
| Hannum | 4.85 (−0.33 to 10.03) | 0.066 | |
| EEAA | 6.32 (0.32 to 12.33) | 0.039 | |
| PhenoAge | −0.16 (−6.22 to 5.90) | 0.958 | |
| GrimAge | −1.35 (−4.88 to 2.19) | 0.447 | |
| DNMT3A mutation (N = 52 individuals) | Horvath | 9.64 (1.97 to 17.31) | 0.015 |
| IEAA | 9.57 (2.09 to 17.04) | 0.013 | |
| Hannum | 5.65 (0.51 to 10.78) | 0.032 | |
| EEAA | 6.45 (0.11 to 12.78) | 0.046 | |
| PhenoAge | 8.24 (−0.84 to 17.32) | 0.074 | |
| GrimAge | 2.31 (−2.40 to 7.02) | 0.329 |
Statistically significant findings (P < 0.05) are indicated in bold.
AgeAccel = epigenetic age acceleration; CHIP = clonal hematopoiesis of indeterminate potential; Coef. = coefficient (beta value); EEAA = extrinsic epigenetic age acceleration; IEAA = intrinsic epigenetic age acceleration; VAF = variant allele frequency.
In conclusion, our results are consistent with previous reports that CHIP is in general associated with increased epigenetic age acceleration. TET2 carriers had greater age acceleration for the extrinsic Hannum and EEAA estimators compared with the intrinsic estimators and conversely, the epigenetic age acceleration in DNMT3A carriers was mainly seen in the intrinsic Horvath and IEAA estimators compared with TET2 carriers, indicating that the 2 mutations more generally are associated with distinct types of epigenetic age acceleration.
Intrapair analyses of CHIP mutations and VAF
The observed associations between CHIP mutations and different epigenetic age estimators can either be results of a causal link between the 2 or they can arise from unmeasured, shared predisposing risk factors, either genetic or environmental. To attempt to eliminate the effects of unmeasured genetic and environmental confounders, we utilized the twin structure of our dataset to conduct intrapair analyses. We analyzed all pairs discordant for the outcome of interest, that is, pairs in which one twin carried a CHIP/TET2/DNMT3A mutation while the other did not, and pairs where both twins harbored a mutation but at different allele frequencies. In intrapair analyses of CHIP status, Hannum epigenetic age was positively associated with the presence of any CHIP mutation (1.62 years difference, P = 0.027, Table 3). For TET2 mutations, Hannum age showed a similar effect size, although not statistically significant (2.77 years, P = 0.052). Interestingly, the association between DNMT3A mutations and IEAA was absent in intrapair analysis (0.91 years, P = 0.410), indicating that the association observed in our individual-level analyses and by others7 may partly be the result of unmeasured confounding. Intrapair analyses of VAF showed similar results (ie, the co-twin with the highest VAF value tended to also have the highest AgeAccel), but were limited by very small sample sizes, especially for TET2 and DNMT3A mutations (Suppl. Table S3).
Table 3.
Intrapair Analysis of CHIP Mutation Status and Age Acceleration by Linear Regression Analysis in Twin Pairs Discordant for CHIP Mutation Status
| AgeAccel | Coef. (95% CI) | P | |
|---|---|---|---|
| Any CHIP mutation (N = 72 twin pairs) | Horvath | −0.11 (−1.83 to 1.61) | 0.898 |
| IEAA | 0.54 (−0.88 to 1.96) | 0.448 | |
| Hannum | 1.62 (0.19–3.06) | 0.027 | |
| EEAA | 1.64 (−0.19 to 3.47) | 0.078 | |
| PhenoAge | 0.87 (−1.49 to 3.22) | 0.465 | |
| GrimAge | 0.94 (−0.18 to 2.06) | 0.099 | |
| TET2 mutation (N = 30 twin pairs) | Horvath | −0.08 (−3.17 to 3.02) | 0.960 |
| IEAA | 0.90 (−1.46 to 3.25) | 0.443 | |
| Hannum | 2.77 (−0.03 to 5.57) | 0.052 | |
| EEAA | 2.71 (−0.86 to 6.29) | 0.131 | |
| PhenoAge | 1.51 (−3.34 to 6.35) | 0.529 | |
| GrimAge | 1.24 (−0.60 to 3.07) | 0.179 | |
| DNMT3A mutation (N = 33 twin pairs) | Horvath | −0.26 (−2.98 to 2.46) | 0.846 |
| IEAA | 0.91 (−1.31 to 3.13) | 0.410 | |
| Hannum | 0.77 (−1.60 to 3.13) | 0.514 | |
| EEAA | 0.62 (−2.50 to 3.74) | 0.689 | |
| PhenoAge | −0.59 (−4.72 to 3.54) | 0.774 | |
| GrimAge | 2.18 (0.05–4.30) | 0.045 |
Statistically significant findings (P < 0.05) are indicated in bold.
AgeAccel = epigenetic age acceleration; CHIP = clonal hematopoiesis of indeterminate potential; Coef. = coefficient (beta value); EEAA = extrinsic epigenetic age acceleration; IEAA = intrinsic epigenetic age acceleration.
In conclusion, the associations between TET2 mutations and the extrinsic measures of epigenetic age appear stable when using intrapair analyses to control for shared genetic and environmental factors between co-twins, but the associations between DNMT3A mutations and IEAA are weakened in intrapair analyses.
Replication of the AgeAccelHG estimator in relation to survival by CHIP status
Finally, we aimed to replicate the Hannum-GrimAge stratification model proposed by Nachun et al.7 We first analyzed the associations between CHIP status and overall survival: in CHIP carriers, the hazard ratio for death was 1.09 compared with non-carriers (95% CI = 0.84–1.43, P = 0.516). Subsequently, using the definition by Nachun et al,7 69 of the 224 individuals were found to be accelerated in both the Hannum and GrimAge age estimators (ie, to be AgeAccelHG+), yet the hazard ratio of death for these individuals was not different from the rest of the study population (HR = 1.10, 95% CI = 0.85–1.43, P = 0.460). Both these findings are contrary to Nachun et al,7 who reported borderline significance for mortality for CHIP carriers versus non-carriers, respectively, a significant association for mortality for AgeAccelHG+ versus others (see Suppl. Table S4, for comparison). Individually analyzing the Hannum and GrimAge age estimators, GrimAge age acceleration was associated with mortality risk (HR = 1.07, 95% CI = 1.03–1.10, P = 6.60 × 10−4), while Hannum age acceleration was not (HR = 1.001, 95% CI = 0.98–1.02, P = 0.862).
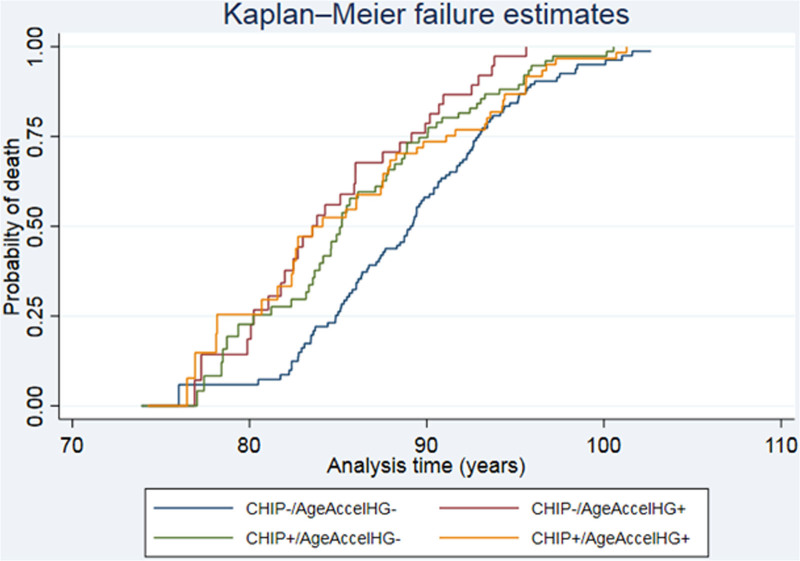
Furthermore, when subsequently stratifying by CHIP status as suggested by Nachun et al,7 39 of the 224 individuals carried a CHIP mutation and were AgeAccelHG+ (ie, they were CHIP+/AgeAccelHG+), but their hazard of death was not different from individuals without CHIP mutations and without double acceleration (ie, CHIP−/AgeAccelHG−, N = 101): HR = 1.03, 95% CI = 0.72–1.47, P = 0.877. When directly comparing this estimate to the estimate reported by Nachun et al7 (ie, HR = 2.90, 95% CI = 1.98–4.24), it was evident that the 2 CIs did not overlap, that is, the increased risk of mortality for CHIP carriers with double acceleration was not confirmed in the present study population. In contrast, the CI for our estimate for the CHIP−/AgeAccelHG+ group included the point estimate by Nachun et al7 (Suppl. Table S4). Finally, CHIP carriers of the present study population who were not double accelerated (ie, individuals being CHIP+/AgeAccelHG−, N = 54) or individuals without CHIP yet being double accelerated (ie, individuals being CHIP−/AgeAccelHG+, N = 30) revealed a significant or borderline significant association with increased mortality compared with individuals without CHIP mutations and without double acceleration (ie, individual being CHIP−/AgeAccelHG−, N = 101): HR = 1.34, 95% CI = 0.96–1.88, P = 0.086 for CHIP+/AgeAccelHG−, and HR = 1.58, 95% CI = 1.11–2.25, P = 0.012 for CHIP−/AgeAccelHG+. A Kaplan-Meier mortality curve for the AgeAccelHG estimator stratified by CHIP status is shown in Figure 1.
Figure 1.
Kaplan-Meier mortality estimates according to combined CHIP and AgeAccelHG status. CHIP± indicates the presence or absence of clonal hematopoiesis of indeterminate potential. AgeAccelHG± indicates whether individuals had positive age acceleration in both Hannum and GrimAge estimators (+) or in only one or neither estimators (−). The plot is based on a Cox proportional hazards model with delayed entry (origin date in the individuals’ birth date and entry date at date of blood sampling), hence the x-axis shown in the plot starts at blood sampling. One twin pair of the present study population died very soon after blood sampling; this twin pair was removed for the plot, as a gap in analysis time, would otherwise have distorted the dimensions of the plot. The estimates obtained for all individuals (N = 224) and leaving out the twin pair (N = 222) were identical (data not shown). AgeAccelHG = age acceleration defined by both the Hannum and GrimAge estimators; CHIP = clonal hematopoiesis of indeterminate potential.
DISCUSSION
The association between CHIP and epigenetic age acceleration is well-documented, but the results presented here allow us to draw several more nuanced conclusions.
First, previous reports have focused mainly on CHIP as a single entity and concluded that the intrinsic age estimators were more strongly associated with CHIP, while the extrinsic age estimators showed weaker associations.5,7 Although Nachun et al7 also found that DNMT3A mutations were more strongly associated with intrinsic than with extrinsic age acceleration, while the opposite was the case for TET2 mutations, this observation did not lead the authors to draw any conclusions about gene-specific divergent effects on intrinsic versus extrinsic age acceleration. In the present study, we confirm these divergent associations for TET2 and DNMT3A both in analyses of CHIP mutation status and of clone size, establishing that in the context of epigenetic aging CHIP should not be considered as a single, uniform entity.
As age-related phenomena, TET2 and DNMT3A mutations are different in one other important aspect, namely their age-specific incidences with TET2 mutations arising later in life than DNMT3A mutations.25 Many different causal explanations for the links between different mutations and types of epigenetic aging are possible, but especially the observation that TET2 mutations are accompanied by accelerated extrinsic aging indicates that an immunologically senescent hematopoietic microenvironment may be necessary for TET2 clones to expand. The observation that the associations between TET2 mutations and extrinsic epigenetic aging appeared stable when controlling for shared genetic and environmental factors in intrapair analyses is consistent with research showing that immunologic variation increases with age and is not driven by heritable influences.26
Conversely, one possible explanation for the link between DNMT3A mutations and intrinsic epigenetic aging (and especially IEAA) is the link between clonal expansion and stem cell proliferation rate, since IEAA is strongly associated with number of cell divisions in cultured fibroblasts.27 Heyde et al28 demonstrated that a likely driving factor behind clonal expansion is an increased hematopoietic stem cell proliferation rate which can result from, among other things, atherosclerosis and associated traits. Under this explanation, our finding that the association between DNMT3A and IEAA vanished in intrapair analyses is thus consistent with the large estimates of heritability for many of the traits associated with atherosclerosis and cardiovascular disease.29
The association between CHIP and adverse outcomes and especially attempts at identifying subsets of CHIP carriers at elevated risk of death or heart disease is particularly interesting at the moment due to the rising interest in both treatment strategies and monitoring regimens for individuals with CHIP.30–32 Nachun et al7 proposed that the combination of CHIP and AgeAccelHG+ could be used in future intervention trials to identify high-risk individuals.7 Contrary to Nachun et al,7 we found that the combination of CHIP and AgeAccelHG+ identified individuals who were not at elevated risk of death. It cannot be ruled out that his lack of replication is due to the smaller sample size of the present study population (N = 224) compared with Nachun et al7 (N = 3624). There are, however, 2 reasons why this is likely not the case. First, as mentioned above, the CIs of the estimated HRs do not overlap, that is, present study: 1.03 (95% CI = 0.72–1.47) versus Nachun et al7: 2.90 (95% CI = 1.98–4.24), indicating that lack of replication is not due to lack of statistical power. Second, the estimates are based on study populations with very different frequencies of CHIP+/AgeAccelHG+ individuals: 39 (17.4%) for the present study population versus 88 (2.4%) for the study population in Nachun et al.7 So even though the Nachun et al7 study population is 16 times larger than the present study population, it only holds about double the number of CHIP+/AgeAccelHG+ individuals. And, furthermore, the present study population are older than the population in the study by Nachun et al7; that is, the present study population has an age span of 73–90 years (mean age: 78.5, median age: 77.1), while the median ages of the sub-cohorts by Nachun et al7 spans 56–78 years of age, with the majority of the individuals being in their 60s at intake). This could indicate that being a CHIP+/AgeAccelHG+ individual likely reflects 2 different things in these study populations, and hence why the proposed stratification model might not be appropriate in the age range of the present cohort. Finally, the study by Nachun et al7 did, similarly to the study by Robertson et al,5 apply Noob normalization of the DNA methylation data, while functional normalization was used in the present study. Performing Noob normalization on the present data led to very similar results and to the same conclusions as presented above (Suppl. Tables S5–S9). Hence, the differences in findings do not appear to be due to differences in normalization methods.
AUTHOR CONTRIBUTIONS
MS and MT did conception and design of the study, performance of data and bioinformatics analyses, interpretation of results, project administration and writing of the original draft. JWH and JW did conception and design of the study, interpretation of results, review and editing of draft. KG and KC did conception and design of the study, interpretation of results, review and editing of draft and data acquisition. All authors have read and approved the final version of the manuscript.
DISCLOSURES
The authors have no conflicts of interest to disclose.
SOURCES OF FUNDING
MS is supported by the Fabrikant Vilhelm Pedersen og Hustrus Legat on recommendation by the Novo Nordisk Foundation. MT is supported by the Research Foundation at Rigshospitalet. The Danish Twin Registry is supported by the Odense University Hospital AgeCare program (Academy of Geriatric Cancer Research). MT, JH, JW, KG are supported by the Novo Nordisk Foundation (Novo Nordisk Foundation Center for Stem Cell Biology, DanStem; grant NNF17CC0027852) and by The Danish Cancer Society (Danish Research Center for Precision Medicine in Blood Cancer; grant 223-A13071-18-S68) and from Greater Copenhagen Health Science Partners (Clinical Academic Group in Blood Cancers). The Danish Twin Registry has been supported by The National Program for Research Infrastructure 2007 grant 09-063256 from the Danish Agency for Science Technology and Innovation, the Velux Foundation, and the National Institutes of Health, National Institute on Aging grant P01 AG08761.
Supplementary Material
Footnotes
MS and MT have contributed equally to this work.
Supplemental digital content is available for this article.
REFERENCES
- 1.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaiswal S, Natarajan P, Silver AJ, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zink F, Stacey SN, Norddahl GL, et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 2017;130:742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson NA, Hillary RF, McCartney DL, et al. Age-related clonal haemopoiesis is associated with increased epigenetic age. Curr Biol. 2019;29:R786–R787. [DOI] [PubMed] [Google Scholar]
- 6.Chen BH, Marioni RE, Colicino E, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY). 2016;8:1844–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nachun D, Lu AT, Bick AG, et al. Clonal hematopoiesis associated with epigenetic aging and clinical outcomes. Aging Cell. 2021;20:e13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen JW, Pedersen DA, Larsen LA, et al. Clonal hematopoiesis in elderly twins: concordance, discordance, and mortality. Blood. 2020;135:261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedersen DA, Larsen LA, Nygaard M, et al. The Danish Twin Registry: an updated overview. Twin Res Hum Genet. 2019;22:499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGue M, Christensen K. Social activity and healthy aging: a study of aging Danish twins. Twin Res Hum Genet. 2007;10:255–265. [DOI] [PubMed] [Google Scholar]
- 12.Debrabant B, Soerensen M, Christiansen L, et al. DNA methylation age and perceived age in elderly Danish twins. Mech Ageing Dev. 2018;169:40–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2012:1–9. [Published online]. http://arxiv.org/abs/1207.3907 and https://github.com/freebayes/freebayes. Accessed August 8, 2022.
- 15.Lai Z, Markovets A, Ahdesmaki M, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soerensen M, Li W, Debrabant B, et al. Epigenome-wide exploratory study of monozygotic twins suggests differentially methylated regions to associate with hand grip strength. Biogerontology. 2019;20:627–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Iterson M, Tobi EW, Slieker RC, et al. MethylAid: visual and interactive quality control of large Illumina 450k datasets. Bioinformatics. 2014;30:3435–3437. [DOI] [PubMed] [Google Scholar]
- 18.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YA, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the illumina infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fortin J, Labbe A, Lemire M, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10:573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY). 2019;11:303–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fransquet PD, Wrigglesworth J, Woods RL, et al. The epigenetic clock as a predictor of disease and mortality risk: a systematic review and meta-analysis. Clin Epigenetics. 2019;11:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brodin P, Jojic V, Gao T, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu AT, Xue L, Salfati EL, et al. GWAS of epigenetic aging rates in blood reveals a critical role for TERT. Nat Commun. 2018;9:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heyde A, Rohde D, McAlpine CS, et al. Increased stem cell proliferation in atherosclerosis accelerates clonal hematopoiesis. Cell. 2021;184:1348–1361.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lusis AJ, Fogelman AM, Fonarow GC. Genetic basis of atherosclerosis: part I: new genes and pathways. Circulation. 2004;110:1868–1873. [DOI] [PubMed] [Google Scholar]
- 30.Steensma DP, Bolton KL. What to tell your patient with clonal hematopoiesis and why: insights from two specialized clinics. Blood. 2020;136:1623–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bick AG, Pirruccello JP, Griffin GK, et al. Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation. 2020;141:124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svensson EC, Madar A, Campbell CD, et al. Abstract 15111: TET2-Driven Clonal hematopoiesis predicts enhanced response to canakinumab in the cantos trial: an exploratory analysis. Circulation. 2018;138(suppl_1):A15111. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.