Abstract
Under anaerobic conditions, structurally diverse aromatic compounds are catabolized by bacteria to form benzoyl-coenzyme A (benzoyl-CoA), the starting compound for a central reductive pathway for aromatic ring degradation. The structural genes required for the conversion of 4-hydroxybenzoate (4-HBA) to benzoyl-CoA by Rhodopseudomonas palustris have been identified. Here we describe a regulatory gene, hbaR, that is part of the 4-HBA degradation gene cluster. An hbaR mutant that was constructed was unable to grow anaerobically on 4-HBA. However, the mutant retained the ability to grow aerobically on 4-HBA by an oxygen-requiring pathway distinct from the anaerobic route of 4-HBA degradation. The effect of the HbaR protein on expression of hbaA encoding 4-HBA-CoA ligase, the first enzyme for 4-HBA degradation, was investigated by using hbaA::′lacZ transcriptional fusions. HbaR was required for a 20-fold induction of β-galactosidase activity that was observed with a chromosomal hbaA::′lacZ fusion when cells grown anaerobically on succinate were switched to anaerobic growth on succinate and 4-HBA. HbaR also activated expression from a plasmid-borne hbaA-′lacZ fusion when it was expressed in aerobically grown Pseudomonas aeruginosa cells, indicating that the activity of this regulator is not sensitive to oxygen. The deduced amino acid sequence of HbaR indicates that it is a member of the FNR-CRP superfamily of regulatory proteins. It is most closely related to transcriptional activators that are involved in regulating nitrate reduction. Previously, it has been shown that R. palustris has an FNR homologue, called AadR, that is also required for 4-HBA degradation. Our evidence indicates that AadR activates expression of hbaR in response to anaerobiosis and that HbaR, in turn, activates expression of 4-HBA degradation in response to 4-HBA as an effector molecule.
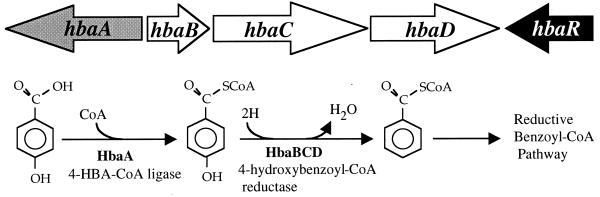
A general strategy used by microbes to degrade diverse aromatic compounds anaerobically is to first convert them to benzoyl-coenzyme A (benzoyl-CoA), a compound that is the starting point for a central pathway of aromatic ring reduction and cleavage (18). Peripheral reactions leading to benzoyl-CoA formation include modification and removal of benzene ring substituents, often via 4-hydroxybenzoate (4-HBA) or 4-hydroxybenzoyl-CoA as an intermediate. The conversion of 4-HBA to benzoyl-CoA is a relatively well-studied reaction sequence (Fig. 1). First, CoA is added to 4-HBA by 4-hydroxybenzoate-CoA ligase. Enzymes catalyzing this reaction have been purified from the purple nonsulfur bacterium Rhodopseudomonas palustris (16) and from the denitrifying species Thauera aromatica (4). The R. palustris gene encoding this enzyme, hbaA, has been cloned and sequenced (16). 4-Hydroxybenzoyl-CoA (4-HBA-CoA) is then dehydroxylated by 4-HBA-CoA reductase to yield benzoyl-CoA (17). The genes encoding the dehydroxylating reductase have been cloned from both R. palustris and T. aromatica, and sequenced, and this enzyme has been purified from T. aromatica (5).
FIG. 1.
The hba gene cluster for 4-HBA degradation. These genes are part of a 26-kb cluster of genes whose products are involved in anaerobic degradation of aromatic compounds by R. palustris (12). The roles of the hba gene products in the conversion of 4-HBA to benzoyl-CoA are also shown (16, 17).
In previous work on anaerobic 4-HBA degradation, we identified a transcriptional activator, AadR, that is required for anaerobic growth of R. palustris on 4-HBA and for expression of hbaA (9). AadR is a member of the FNR family of transcriptional regulators, and based on the presence of the conserved cysteine residues shown to be essential for sensing anaerobiosis by FNR (23), we have proposed that AadR functions in oxygen sensing (9). In addition to being involved in expression of hbaA, AadR is required for expression of the benzoyl-CoA reductase genes, badDEFG (11). Here we describe HbaR, a second transcriptional activator and member of the FNR-CRP superfamily that is involved in regulating the anaerobic degradation of 4-HBA in response to 4-HBA as an effector molecule.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used are described in Table 1. R. palustris cultures were grown anaerobically in defined mineral medium (21) prepared as described previously (19). Carbon sources were added at the time of inoculation to a final concentration of 3 mM except for succinate, which was added to 10 mM. Cultures were incubated at 30°C and illuminated with a 40-W incandescent light bulb for phototrophic growth or in the dark with shaking for aerobic growth. Escherichia coli cultures were routinely grown in Luria broth (LB) (7) at 37°C. For β-galactosidase activity assays, E. coli cultures were grown in LB containing 0.2% glucose, either aerobically with shaking or anaerobically in butyl-rubber-stoppered tubes incubated statically. Pseudomonas aeruginosa cultures were grown at 37°C in LB for routine cultivation and in defined minimal medium (25) with 10 mM succinate for cultures to be assayed for β-galactosidase activity. Antibiotics were used at the following concentrations (in micrograms per milliliter): for R. palustris, gentamicin (100) and kanamycin (100); for P. aeruginosa, kanamycin (100) and gentamicin (20); and for E. coli, ampicillin (100), gentamicin (10), kanamycin (100), and spectinomycin (100).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− λ−recA1 Δ(lacZYA-argF)U169 hsdR17 thi-1 gyrA96 supE44 endA1 relA1 φ80dlacZΔM15 | GIBCO-BRL |
| BL21(DE3) | Carries T7 RNA polymerase under the control of the lacUV5 promoter | 34 |
| S17-1 λpir | S17-1 lysogenized with λpir phage | 32 |
| RZ4500 | ΔlacZ | 6 |
| RZ8459 | ΔlacZ ΔfnrΩSpr | P. Kiley |
| P. aeruginosa PAO1 | Wild-type strain | A. Kropinsky |
| R. palustris strains | ||
| CGA009 | Wild-type strain; spontaneous Cmr derivative | 21 |
| CGA507 | hbaA::′lacZ | This study |
| CGA612 | hbaR | This study |
| CGA614 | hbaA::′lacZ hbaR | This study |
| Plasmids | ||
| pUC19 | High-copy-number cloning vector; Apr | 40 |
| pJQ200mp18 | Mobilizable suicide vector; sacB; Gmr | 28 |
| pCF116 | Mobilizable suicide vector; sacB; Kmr | 14 |
| pUCGM | Contains Gmr cassette | 31 |
| pBBR1MCS-2 | Broad-host-range vector; Kmr | 22 |
| pT7-7 | T7 promoter expression vector; Apr | 35 |
| pHRP309 | Reporter plasmid, contains ′lacZ | 26 |
| pHRP311 | pHRP309 with ΩSpr cassette cloned in front of ′lacZ | 26 |
| pHRP315 | Cohort vector for pHRP309 cloning system; Apr Spr | 26 |
| pHRP316 | Cohort vector for pHRP309 cloning system; Apr Spr | 26 |
| pPE604 | pUC19 with a 1.9-kb PCR product containing hbaR | This study |
| pPE604::Gmr | pPE604 with Gmr cassette from pUCGM at the AgeI site of hbaR | This study |
| pPE900 | pCF116 containing the hbaR::Gmr construct from pPE604::Gmr at the XbaI site | This study |
| pPE901 | pBBRMCS-2 with the hbaR-containing XbaI fragment from pPE604 | This study |
| pPE905 | pT7-7 with hbaR under the control of the T7 promoter | This study |
| pPE906 | pHRP309 with the ΩSpr and hbaR promoter region fused to ′lacZ; Spr Gmr | This study |
| pMD300 | Contains the 2.1-kb EcoRI fragment containing hbaA | 16 |
| pMD425 | pJQ200mp18 with a 2.1-kb hbaA-containing fragment | This study |
| pMD426 | pJQ200mp18 with a 2.1-kb hbaA-containing fragment interrupted with ′lacZ-Kmr at a unique SalI site | This study |
| pMD505 | pHRP309 with a ΩSpr and hbaA promoter region fused to ′lacZ; Spr Gmr | This study |
Apr, ampicillin resistance; Kmr, kanamycin resistance; Gmr, gentamicin resistance; Cmr, chloramphenicol resistance; Spr, spectinomycin resistance; MCS, multiple cloning site.
Cloning and DNA manipulation.
Standard protocols were used for cloning and transformations (30). Plasmid DNA was purified by using the QIAprep Spin Miniprep kit (Qiagen Inc., Chatsworth, Calif.). DNA fragments were purified from agarose gels by using the GeneClean spin kit from Bio 101 (La Jolla, Calif.). Chromosomal DNA was purified by using a variation on the method of Saito and Miura (29) as described previously (10).
Reporter plasmids pPE906 and pMD505 containing promoter-lacZ fusions were constructed by a two-step cloning procedure described previously (26). Briefly, promoter-containing DNA fragments were cloned directionally into ΩSpr-containing cohort vectors (pHRP315 or pHRP316). Fragments containing the ΩSpr cassette and promoter region were then cloned upstream of the promoterless lacZ gene (′lacZ) of pHRP309. The fusion of the promoter-containing fragments and ′lacZ were then confirmed by sequencing.
Strains with mutations in hbaR (CGA612 and CGA614) were generated by gene replacement with a cloned copy of hbaR that had been interrupted with a gentamicin resistance (Gmr) cassette. The mutagenesis construct was generated by cloning a Gmr cassette from pUCGM (31) into the unique AgeI site of hbaR in pPE604. The hbaR::Gmr construct was then cloned into pCF116 (14), which contains sacB for counterselection. The resulting plasmid, pPE900, was mated into R. palustris, and exconjugants were screened as described previously (10). The hbaA::′lacZ strain (CGA507) was generated by using a similar strategy. The hbaA-containing fragment from pMD300 (16) was cloned into pJQ200mp18 (28) to generate pMD425. The hbaA reading frame was then interrupted at a SalI site with a ′lacZ-Kmr cassette derived from pUTminiTn5-lacZ (8) to generate pMD426 and mated into R. palustris.
Expression of HbaR in E. coli.
HbaR was expressed by using the T7 promoter system (3). DNA containing hbaR was PCR amplified and cloned into the NdeI and SmaI sites of pT7-7 (35) to generate pPE905. E. coli BL21(DE3) cells containing pPE905 were grown in LB medium to an A660 of approximately 0.25. Cells (1 ml) were harvested, washed, and resuspended in 1.0 ml of basal medium supplemented with 0.02% concentrations of each of 18 amino acids (no methionine or cysteine). After a 30-min incubation at 30°C, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 1 mM, and incubation was continued for 30 min. Rifampin was added to a final concentration of 0.5 mg/ml, and cells were incubated at 42°C for 10 min to allow rifampin to enter the cells. After an additional 30 min at 30°C, cells were incubated with 10 μCi of [35S]methionine for 2 min. Proteins were separated on sodium dodecyl sulfate–15% polyacrylamide gels.
An HbaR-histidine tag fusion protein (HbaR-His) was generated by cloning hbaR into the vector pET-16b (Novagen Inc., Madison, Wis.) and purified with the Novagen pET system. Extracts from E. coli BL21(DE3) cells expressing HbaR-His were loaded onto a 5-ml HiTrap chelating column (Pharmacia Biotech, Piscataway, N.J.) which had been charged with 400 mM NiSO4. HbaR-His was eluted from the column after a 4-min washing with 60% buffer containing 1 M imidazole–500 mM NaCl in triethanolamine buffer, pH 7.9.
Gel mobility shift assays.
Gel mobility shift assays were attempted by using several different protocols (15, 27). Binding buffers containing various salts at a range of concentrations were used. Target DNA fragments of different lengths were also tried.
Primer extension and reverse transcriptase PCR analysis.
Primer extension analysis was used to determine the transcriptional start sites of hbaR and hbaA. The avian myeloblastosis virus reverse transcriptase primer extension system was used according to the protocol supplied by the manufacturer (Promega Corp., Madison, Wis.). The primer used to map the hbaR start site was complementary to nucleotides 13 to 33 of hbaR. The primer used to map the hbaA start site was complementary to bases 147 to 164 of hbaA. Primer extension products were analyzed on a 6% polyacrylamide gel next to a sequence ladder generated by using the same primers. Sequencing reactions were performed with the fmol DNA sequencing system from Promega.
DNA sequencing and analysis.
DNA sequences were determined at the University of Iowa DNA Facility by using dye terminator cycle sequencing. The reactions were run on and analyzed with an Applied Biosystems model 373A stretch fluorescent automated sequencer. DNA sequences were analyzed with GENE Inspector, version 1.0.1 (Textco Inc., West Lebanon, N.H.). Similar sequences were identified from the SWISS-PROT 26 and GENPEP 78.0 databases by using the BLAST network service at the National Center for Biotechnology Information (Bethesda, Md.). The GAP program from the University of Wisconsin Genetics Computer Group software package, version 9.0, was used to make sequence comparisons and alignments. The AllAll program from the Computational Biochemistry Research Group (cbrg.inf.ethz.ch./section3_1.html) was used for phylogenetic analysis.
Enzyme assays.
4-HBA-CoA ligase activity was measured by using a spectrophotometric assay described previously (16). Briefly, reaction mixtures containing 50 mM Tris (pH 9.2), 5 mM MgCl2, 0.5 mM ATP, 0.8 mM reduced CoA, 0.25 mM 4-HBA, and cell extract were monitored for increase in absorbance at 330 nm due to formation of 4-HBA-CoA. Activity was calculated by using a millimolar extinction coefficient of 24.
β-Galactosidase activity in E. coli and P. aeruginosa cultures was measured by the method of Miller (24). For R. palustris cultures, β-galactosidase activity was measured by a variation of the method of Miller (24), as described previously (11). Briefly, logarithmically growing cells were harvested, washed in Z buffer, and sonicated. Cell extract and Z buffer were combined to a volume of 1 ml, and 0.2 ml of a 4-mg/ml solution of o-nitrophenylgalactopyranoside was added to start the reactions. The rate of increase in absorbance at 420 nm due to o-nitrophenol formation was measured spectrophotometrically. Activity was calculated by using a millimolar extinction coefficient of 4.5 for o-nitrophenol at 420 nm. Protein concentrations in cell extracts were determined by using the Bio-Rad (Richmond, Calif.) protein assay kit.
Nucleotide sequence accession number.
The DNA sequence of hbaR has been assigned GenBank accession no. AF172325.
RESULTS
Characteristics of hbaR.
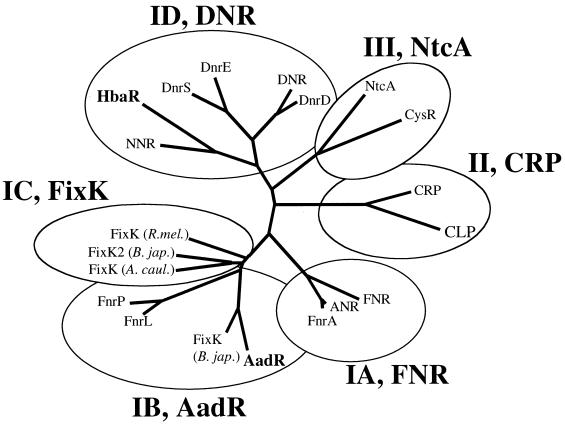
A 726-bp open reading frame designated hbaR was found next to the R. palustris genes encoding enzymes responsible for the conversion of 4-HBA to benzoyl-CoA (Fig. 1) (16, 17). HbaR was similar in its deduced amino acid sequence to members of the FNR-CRP superfamily of transcriptional regulators. It was most similar (52% similar, 42% identical) to NNR, a positive regulator of nitrite and nitric oxide reductase gene expression in Paracoccus denitrificans (37). The other proteins with the highest levels of amino acid sequence identity to HbaR were members of the DNR group of the FNR-CRP superfamily of transcriptional regulators, as described by Vollack et al. (39) (Fig. 2). HbaR had a low level of amino acid sequence similarity (36%) to the R. palustris protein AadR, an FNR-like regulator that contains the conserved cysteine residues shown to be involved in redox sensing by FNR (9, 23). HbaR does not contain these cysteine residues.
FIG. 2.
Phylogenetic tree of selected members of the FNR-CRP superfamily of transcriptional regulators constructed with the AllAll program from the Computational Biochemistry Research Group. Subdivisions shown are those proposed by Fischer (13) and recently modified by Vollack et al. (39). The proteins and their sources and accession numbers, respectively, are as follows: AadR, R. palustris, M92426; ANR, P. aeruginosa, P23926; CLP, Xanthomonas campestris, M58745; CRP, E. coli, U18997; CysR, Synechococcus sp., AAA73046; DNR, P. aeruginosa, D50019; DnrD, DnrE, and DnrS, P. stutzeri, AJ131715, AJ131716, and AJ131717, respectively; FixK (A. caul.), Azorhizobium caulinodans, P26488; FixK (B. jap.) and FixK2 (B. jap.), Bradyrhizobium japonicum, M86805 and CAA06287, respectively; FixK (R. mel.), Rhizobium meliloti, X15079; FNR, E. coli, P03019; FnrA, P. stutzeri, Z26044; FnrL, Rhodobacter capsulatus, U78309; FnrP, Paracoccus denitrificans, U34353; HbaR, R. palustris, AF172325; NNR, Paracoccus denitrificans, U17435; NtcA, Anabaena variabilis, Q05061.
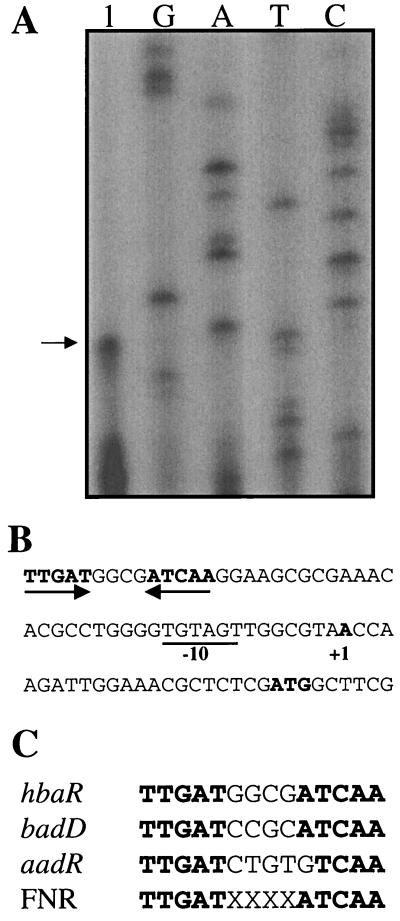
The transcriptional start site of hbaR was mapped by using primer extension to 23 bases upstream of the hbaR initiation codon (Fig. 3). The hbaR promoter region contained a sequence (5′-TGTAGT-3′) 6 bp upstream of the transcriptional start site that matched the E. coli ς70 consensus sequence (5′-TATAAT-3′) at four of six positions. The promoter region contained a 5-bp inverted repeat centered at −42.5 bases from the start site of transcription (Fig. 3B). The sequence of this repeat matched that of the consensus FNR binding site (33) as well as the inverted repeat previously proposed to be the binding site of AadR (9, 11).
FIG. 3.
(A) Mapping the hbaR start site by primer extension. Lane 1 contains the primer extension product from RNA isolated from cells grown anaerobically on succinate. A sequencing ladder generated with the same primer is shown. The arrow indicates the start site of transcription. (B) Nucleotide sequence of the hbaR promoter region showing the start site of transcription (+1), the putative −10 region (underlined), and the inverted repeat matching the consensus FNR binding site (inverted arrows). (C) Alignment of FNR boxes from R. palustris promoter regions (9, 11) and the consensus FNR binding sequence (33).
Characterization of an hbaR mutant.
An hbaR mutant (strain CGA612) was unable to grow on 4-HBA under anaerobic conditions, but it grew at wild-type rates on benzoate. The hbaR mutant also grew at wild-type rates on succinate and, like the wild-type parent strain, could grow aerobically on 4-HBA. The defect in anaerobic growth on 4-HBA was complemented in trans by a plasmid-borne copy of hbaR supplied on pPE901. The growth phenotype of this mutant suggested that hbaR is involved in regulating the enzymes that convert 4-HBA to benzoyl-CoA. The first step in this process is the addition of CoA to 4-HBA by 4-HBA-CoA ligase. The activity of 4-HBA-CoA ligase in extracts from wild-type cells grown anaerobically with 4-HBA (0.3 mM) plus succinate (10 mM) was 10.2 nmol/min/mg of protein, while the hbaR mutant had levels of 4-HBA-CoA ligase activity below the level of detection for the assay used (<0.25 nmol/min/mg of protein).
HbaR activates expression of 4-HBA-CoA ligase in response to 4-HBA.
To examine a possible regulatory role of HbaR, the hbaR mutation was introduced into an R. palustris strain containing a chromosomal hbaA::′lacZ fusion (CGA507). Levels of expression of hbaA, the gene encoding 4-HBA-CoA ligase, in wild-type and hbaR (CGA614) backgrounds were then compared. Cells with an intact hbaR gene had 20-fold-higher levels of hbaA expression when 4-HBA was present in anaerobic growth medium than when they were grown on succinate only. In contrast, expression of the hbaA::′lacZ fusion was not induced by 4-HBA in hbaR mutant cells (Table 2). This 20-fold activation by HbaR is consistent with the difference in 4-HBA-CoA ligase activity between the wild type and hbaR mutant.
TABLE 2.
Effects of hbaR on β-galactosidase activity expressed from a chromosomally encoded hbaA::′lacZ fusion in R. palustris
| Strain genotype | β-Galactosidase activitya
|
|||
|---|---|---|---|---|
| Anaerobic growth
|
Aerobic growth
|
|||
| 4-HBA + succinate | Succinate | 4-HBA | Succinate | |
| hbaA::′lacZ | 4,900 (240) | 240 (75) | 90 (5) | 140 (10) |
| hbaA::′lacZ hbaR | 120 (10) | 150 (10) | 70 (5) | 120 (10) |
Expressed as nanomoles of product formed per minute per milligram of protein. Values are the averages of three independent trials conducted in triplicate. Standard errors of the means are in parentheses.
In addition to being able to grow anaerobically on 4-HBA via the reductive benzoyl-CoA degradation pathway, R. palustris can grow aerobically on 4-HBA by using an oxygenase-mediated meta ring fission pathway (20). The aerobic pathway does not require 4-hydroxybenzoate-CoA ligase. Consistent with this, cells grown aerobically with 4-HBA exhibited very low levels of hbaA::′lacZ expression (Table 2). The hbaR mutation did not affect the levels of β-galactosidase activity in the aerobically grown cells.
HbaR is active under aerobic conditions.
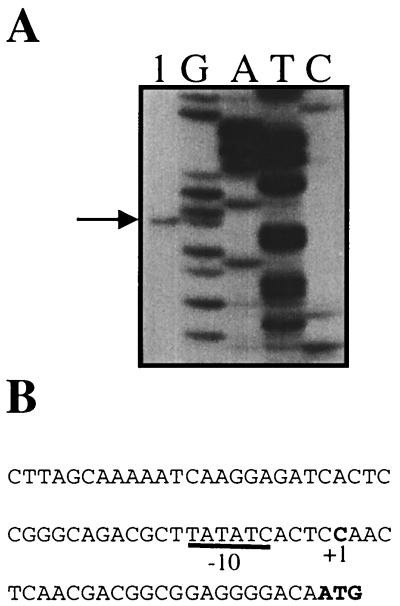
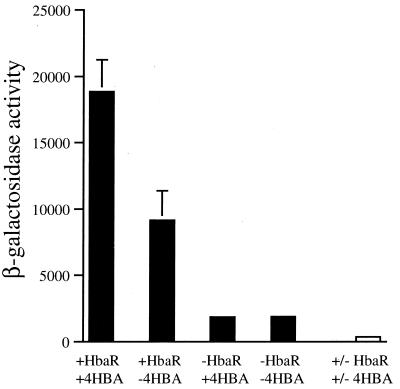
Primer extension analysis was used to identify the promoter region of hbaA (Fig. 4). The possibility that HbaR is active in the presence of oxygen was investigated by expressing hbaR in aerobically growing P. aeruginosa cells containing a reporter plasmid (pMD505) that had the hbaA promoter fused to a promoterless lacZ gene. Such cells exhibited a fivefold increase in β-galactosidase expression over the levels seen in the absence of hbaR (Fig. 5). When 4-HBA was present in the growth medium, levels of β-galactosidase activity increased twofold more over levels in cells grown on succinate only, for a net 10-fold induction. This suggests that HbaR acts directly at the hbaA promoter and activates expression in response to 4-HBA. In addition, this shows that, although hbaA is not expressed in aerobically growing R. palustris cells (Table 2), HbaR is able to activate transcription from the hbaA promoter in the presence of oxygen.
FIG. 4.
(A) Mapping the hbaA start site by primer extension. Lane 1 contains the primer extension product (arrow) from RNA isolated from cells grown anaerobically on 4-HBA. A sequencing ladder generated with the same primer is shown. (B) Nucleotide sequence of the hbaA promoter region showing the start site of transcription (+1) and a putative −10 region that matches the E. coli ς70 consensus (TATAAT) at four of six positions.
FIG. 5.
Expression of β-galactosidase activity from P. aeruginosa cells containing either the PhbaA-′lacZ reporter plasmid pMD505 (filled bars) or a promoterless negative control (pHRP311) (open bar) and either the HbaR-expressing plasmid pPE901 (+HbaR) or the vector pBBR1MCS-2 (−HbaR). Cells were grown aerobically on succinate in the presence or absence of 0.5 mM 4-HBA. β-Galactosidase activity is in Miller units. Error bars represent standard deviations.
Expression of HbaR in E. coli.
HbaR was expressed in E. coli cells from the plasmid pPE905 as described in Materials and Methods. The apparent molecular mass of the HbaR peptide (25 kDa) was close to that predicted based on the deduced amino acid sequence of HbaR (28.5 kDa). E. coli cell extracts containing overexpressed HbaR were tested to see if they would cause a shift in gel mobility of DNA fragments containing the hbaA promoter region. No change in the mobility of a fragment containing the hbaA promoter region was seen with these extracts or with extracts from cultures of R. palustris or P. aeruginosa expressing HbaR (data not shown). The addition of 4-HBA to the gel shift assay mixtures had no apparent effect on binding. An HbaR-His fusion peptide purified to near homogeneity was also tested in gel mobility shift assays. The purified peptide was mostly insoluble, precipitated out of solution easily, and bound nonspecifically to all DNA fragments tested including vector DNA (data not shown).
E. coli FNR can activate hbaR expression.
The hbaR promoter region has an inverted repeat centered at −42.5 bp from the start site of transcription that matches the FNR consensus binding site (Fig. 3B). To determine whether hbaR expression might be activated by an FNR-type regulator, a 226-bp fragment containing the hbaR promoter region was cloned in front of the promoterless lacZ gene in pHRP309 by using a two-step cloning system described previously (26). β-Galactosidase expression from the PhbaR-′lacZ plasmid (pPE906) was measured in wild-type and fnr mutant E. coli cells. In wild-type cells, the levels of PhbaR-′lacZ fusion expression were threefold higher when cells were grown anaerobically than when cells were grown in the presence of oxygen (Table 3). PhbaR-′lacZ activity was not induced in response to anaerobiosis in the fnr mutant strain of E. coli. This suggests that hbaR could also be regulated by an FNR-like protein in R. palustris. Similar experiments were done to examine the possible effect of FNR on hbaA expression. We found that β-galactosidase expression from a PhbaA-′lacZ plasmid present in E. coli was not influenced by the fnr mutation.
TABLE 3.
Effects of fnr on β-galactosidase activity expressed from a plasmid-borne PhbaR-′lacZ fusion in E. coli
| Strain | fnr status | β-Galactosidase activitya
|
|
|---|---|---|---|
| Anaerobic | Aerobic | ||
| RZ4500 | Wild type | 4,700 (190) | 1,700 (40) |
| RZ8495 | fnr mutant | 1,600 (90) | 1,200 (40) |
Expressed in Miller units. Values are the averages of at least two independent trials conducted in triplicate. Standard errors of the means are in parentheses.
DISCUSSION
Data presented here indicate that HbaR is a transcriptional activator that senses 4-HBA as an effector molecule and induces expression of hbaA, the gene encoding 4-HBA-CoA ligase and first enzyme of 4-HBA degradation. Although we were unable to demonstrate binding of HbaR to the hbaA promoter in vitro with gel mobility shift assays, experiments with HbaR expressed in P. aeruginosa indicate that HbaR activates expression of the hbaA promoter. These experiments also show that when expressed in P. aeruginosa, HbaR can activate expression from the hbaA promoter in the presence of oxygen (Fig. 5). In contrast, hbaA expression was not activated in aerobically grown R. palustris cells. This suggests that a second regulator is required for activation of hbaA expression in response to anaerobiosis. Our results showing that E. coli fnr influences hbaR expression and the fact that hbaR has an FNR binding site in its promoter region suggest that hbaR expression is activated in response to anaerobiosis in R. palustris by an FNR homologue. HbaR, in turn, activates hbaA expression in response to 4-HBA. The R. palustris FNR homologue in question is, presumably, AadR, since AadR is required for HbaA expression in this organism (9). This restriction of HbaR expression in R. palustris to anaerobically growing cells indicates that this organism must have a separate system for detecting 4-HBA under aerobic conditions and for activating the genes encoding enzymes of the aerobic 4-HBA degradation pathway.
HbaR is a member of the FNR-CRP superfamily of transcriptional regulators. This protein family has been divided into three classes based on sequence similarity to the reference proteins FNR, which activates gene expression in response to anaerobiosis, CRP, a regulator of catabolic functions, and NtcA, a regulator of genes involved in nitrogen and sulfur metabolism (13) (Fig. 2). The FNR class (which includes AadR of R. palustris) has been further subdivided into four groups based on the presence and spacing of the conserved cysteine residues required for assembly of the redox-sensitive iron-sulfur center. The sequence of HbaR places it in a group (DNR) of the FNR class that is comprised of proteins that lack the cysteine residues required for iron-sulfur center coordination and oxygen sensing (39). With the exception of HbaR, all the proteins within the DNR group are global regulators that control expression of genes involved in denitrification (1, 36–38).
A recently recognized theme among members of the FNR-CRP superfamily is the involvement of two members of the protein family in regulation of the same system. Transcriptional regulation of genes involved in denitrification in P. aeruginosa and Paracoccus denitrificans also involves control by two members of the FNR-CRP superfamily (2, 37). The involvement of multiple family members has also been demonstrated in Pseudomonas stutzeri, which has at least four FNR family members involved in regulating metabolic processes (39). In some cases, including the system we have described here, this multiplicity of regulators involves a regulatory cascade, with one protein acting as an oxygen sensor and, in turn, activating expression of a second regulator (2, 39). Hierarchical expression of two regulators that are members of the greater FNR family would be an effective strategy to prevent cross talk and provide regulation of relatively specialized target genes, as in the case of 4-HBA degradation, under conditions of oxygen deprivation. This study expands the range of functions regulated by the greater FNR-CRP superfamily to include aromatic carbon source utilization and expands the range of effectors sensed by members of this superfamily to include aromatic acids.
ACKNOWLEDGMENTS
This work was supported by the Division of Energy Biosciences, Department of Energy (grant DE-FG02-95ER20184), and by the U.S. Army Research Office (grant DAAG55-98-0188).
We thank Walter Zumft for helpful discussions and Marilyn Dispensa for help with some strain constructions.
REFERENCES
- 1.Arai H, Igarashi Y, Kodama T. Expression of the nir and nor genes for denitrification of Pseudomonas aeruginosa requires a novel CRP/FNR-related transcriptional regulator, DNR, in addition to ANR. FEBS Lett. 1995;371:73–76. doi: 10.1016/0014-5793(95)00885-d. [DOI] [PubMed] [Google Scholar]
- 2.Arai H, Kodama T, Igarashi Y. Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol Microbiol. 1997;25:1141–1148. doi: 10.1046/j.1365-2958.1997.5431906.x. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1990. [Google Scholar]
- 4.Biegert T, Altenschmidt U, Eckerskorn C, Fuchs G. Enzymes of anaerobic metabolism of phenolic compounds: 4-hydroxybenzoate-CoA ligase from a denitrifying Pseudomonas species. Eur J Biochem. 1993;213:555–561. doi: 10.1111/j.1432-1033.1993.tb17794.x. [DOI] [PubMed] [Google Scholar]
- 5.Breese K, Fuchs G. 4-Hydroxybenzoyl-CoA reductase (dehydroxylating) from the denitrifying bacterium Thauera aromatica. Prosthetic groups, electron donor, and genes of a member of the molybdenum-flavin-iron-sulfur proteins. Eur J Biochem. 1998;251:916–923. doi: 10.1046/j.1432-1327.1998.2510916.x. [DOI] [PubMed] [Google Scholar]
- 6.Choe M, Reznikoff W S. Anaerobically expressed Escherichia coli genes identified by operon fusion techniques. J Bacteriol. 1991;173:6139–6146. doi: 10.1128/jb.173.19.6139-6146.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 8.De Lorenzo V, Cases I, Herrero M, Timmis K N. Early and late responses of TOL promoters to pathway inducers: identification of postexponential promoters in Pseudomonas putida with lacZ-tet bicistronic reporters. J Bacteriol. 1993;175:6902–6907. doi: 10.1128/jb.175.21.6902-6907.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dispensa M, Thomas C T, Kim M-K, Perrotta J A, Gibson J, Harwood C S. Anaerobic growth of Rhodopseudomonas palustris on 4-hydroxybenzoate is dependent on AadR, a member of the cyclic AMP receptor protein family of transcriptional regulators. J Bacteriol. 1992;174:5803–5813. doi: 10.1128/jb.174.18.5803-5813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egland P G, Gibson J, Harwood C S. Benzoate-coenzyme A ligase, encoded by badA, is one of three ligases able to catalyze benzoyl-coenzyme A formation during anaerobic growth of Rhodopseudomonas palustris on benzoate. J Bacteriol. 1995;177:6545–6551. doi: 10.1128/jb.177.22.6545-6551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egland P G, Harwood C S. BadR, a new MarR family member, regulates anaerobic benzoate degradation by Rhodopseudomonas palustris in concert with AadR, an Fnr family member. J Bacteriol. 1999;181:2102–2109. doi: 10.1128/jb.181.7.2102-2109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egland P G, Pelletier D A, Dispensa M, Gibson J, Harwood C S. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc Natl Acad Sci USA. 1997;94:6484–6489. doi: 10.1073/pnas.94.12.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer H-M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuqua C W, Winans S C. A luxR-luxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garner M M, Revzin A. Gel retardation analysis of nucleic acids-protein interactions. In: Rickwood D, Hames B D, editors. Gel electrophoresis of nucleic acids. A practical approach. 2nd ed. New York, N.Y: IRL Press; 1990. pp. 201–223. [Google Scholar]
- 16.Gibson J, Dispensa M, Fogg G C, Evans D T, Harwood C S. 4-Hydroxybenzoate-coenzyme A ligase from Rhodopseudomonas palustris: purification, gene sequence, and role in anaerobic degradation. J Bacteriol. 1994;176:634–641. doi: 10.1128/jb.176.3.634-641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson J, Dispensa M, Harwood C S. 4-Hydroxybenzoyl coenzyme A reductase (dehydroxylating) is required for anaerobic degradation of 4-hydroxybenzoate by Rhodopseudomonas palustris and shares features with molybdenum-containing hydroxylases. J Bacteriol. 1997;179:634–642. doi: 10.1128/jb.179.3.634-642.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harwood C S, Burchhardt G, Herrmann H, Fuchs G. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol Rev. 1999;22:439–458. [Google Scholar]
- 19.Harwood C S, Gibson J. Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl Environ Microbiol. 1988;54:712–717. doi: 10.1128/aem.54.3.712-717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegeman G D. The metabolism of p-hydroxybenzoate by Rhodopseudomonas palustris and its regulation. Arch Microbiol. 1967;59:143–148. doi: 10.1007/BF00406325. [DOI] [PubMed] [Google Scholar]
- 21.Kim M-K, Harwood C S. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol Lett. 1991;83:199–204. [Google Scholar]
- 22.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop II R M, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 23.Melville S B, Gunsalus R P. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J Biol Chem. 1990;265:18733–18736. [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 25.Ornston L N, Stanier R Y. The conversion of catechol and protocatechuate to β-ketoadipate by Pseudomonas putida. I. Biochemistry. J Biol Chem. 1966;241:3776–3786. [PubMed] [Google Scholar]
- 26.Parales R E, Harwood C S. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for Gram− bacteria. Gene. 1993;133:23–30. doi: 10.1016/0378-1119(93)90220-w. [DOI] [PubMed] [Google Scholar]
- 27.Parsek M R, Coco W M, Chakrabarty A M. Gel-shift assay and DNase I footprinting in analysis of transcriptional regulation of biodegradation genes. In: Adolph K W, editor. Molecular biological techniques, part A. Vol. 3. New York, N.Y: Academic Press; 1994. pp. 273–290. [Google Scholar]
- 28.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 29.Saito H, Miura K I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Schweizer H P. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–834. [PubMed] [Google Scholar]
- 32.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 33.Spiro S, Gaston K L, Bell A I, Roberts R E, Busby S J W, Guest J R. Interconversion of the DNA-binding specificities of two related transcriptional regulators, CRP and FNR. Mol Microbiol. 1990;4:1831–1838. doi: 10.1111/j.1365-2958.1990.tb02031.x. [DOI] [PubMed] [Google Scholar]
- 34.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 35.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Spanning R J M, De Boer A P N, Reijnders W N M, Spiro S, Westerhoff H V, Stouthamer A H, Van Der Oost J. Nitrite and nitric oxide reduction in Paracoccus denitrificans is under the control of NNR, a regulatory protein that belongs to the FNR family of transcriptional activators. FEBS Lett. 1995;360:151–154. doi: 10.1016/0014-5793(95)00091-m. [DOI] [PubMed] [Google Scholar]
- 37.Van Spanning R J M, De Boer A P N, Reijnders W N M, Westerhoff H V, Stouthamer A H, Van Der Oost J. FnrP and NNR of Paracoccus denitrificans are both members of the FNR family of transcriptional activators but have distinct roles in respiratory adaptation in response to oxygen limitation. Mol Microbiol. 1997;25:893–907. doi: 10.1046/j.1365-2958.1997.2801638.x. [DOI] [PubMed] [Google Scholar]
- 38.Van Spanning R J M, Houben E, Reijnders W N M, Spiro S, Westerhoff H V, Saunders N. Nitric oxide is a signal for NNR-mediated transcription activation in Paracoccus denitrificans. J Bacteriol. 1999;181:4129–4132. doi: 10.1128/jb.181.13.4129-4132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vollack K-U, Härtig E, Körner H, Zumft W G. Multiple transcription factors of the FNR family in denitrifying Pseudomonas stutzeri: characterization of four fnr-like genes, regulatory responses and cognate metabolic processes. Mol Microbiol. 1999;31:1681–1694. doi: 10.1046/j.1365-2958.1999.01302.x. [DOI] [PubMed] [Google Scholar]
- 40.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]