Abstract
Purpose
Previous studies that identify putative genes associated with diabetic retinopathy are only focusing on specific clinical stages, thus resulting genes are not necessarily reflective of disease progression.
This study identified genes associated with the severity level of diabetic retinopathy using the likelihood-ratio test (LRT) and ordinal logistic regression (OLR) model, as well as to profile immune and retinal cell landscape in progressive diabetic retinopathy using a machine learning deconvolution approach.
Methods
This study used a published transcriptomic dataset (GSE160306) from macular regions of donors with different degrees of diabetic retinopathy (10 healthy controls, 10 cases of diabetes, 9 cases of nonproliferative diabetic retinopathy, and 10 cases of proliferative diabetic retinopathy or combined with diabetic macular edema). LRT and OLR models were applied to identify severity-associated genes. In addition, CIBERSORTx was used to estimate proportional changes of immune and retinal cells in progressive diabetic retinopathy.
Results
By controlling for gender and age using LRT and OLR, 50 genes were identified to be significantly increased in expression with the severity of diabetic retinopathy. Functional enrichment analyses suggested these severity-associated genes are related to inflammation and immune responses. CCND1 and FCGR2B are further identified as key regulators to interact with many other severity-associated genes and are crucial to inflammation. Deconvolution analyses demonstrated that the proportions of memory B cells, M2 macrophages, and Müller glia were significantly increased with the progression of diabetic retinopathy.
Conclusions
These findings demonstrate that deep analyses of transcriptomic data can advance our understanding of progressive ocular diseases, such as diabetic retinopathy, by applying LRT and OLR models as well as bulk gene expression deconvolution.
Keywords: transcriptome, diabetic retinopathy, scRNA-seq
Diabetic retinopathy (DR) is a progressive retinal complication of diabetes, causing significant visual impairment on a global scale.1 Microvascular lesions of the retina have been used to evaluate and classify the severity level of DR. There are two broad categories, including the early stage of nonproliferative diabetic retinopathy (NPDR) and the advanced stage of proliferative diabetic retinopathy (PDR).1 NPDR is manifested by increased vascular permeability and capillary occlusion, whereas PDR is characterized by retinal neovascularization in addition to NPDR features.2 An important additional category of DR is diabetic macular edema (DME), the most common cause of vision loss in patients with DR.2 Genetic studies, including candidate gene studies, linkage studies, powerful genomewide association studies (GWAS), and meta-analysis, have identified putative candidate gene or single nucleotide polymorphisms associated with DR.3,4 Although these studies provide critical information that helps to interpret the mechanisms of DR, they mostly study one specific type of DR, thus the resulting putative genes are not likely to be associated with DR progression.
In this study, we assess the macular transcriptomic data from donors with different severity levels of DR and define the genes changed with DR progression. The landscape of immune cells and retinal cells in relation to DR progression has also been established by using a machine learning deconvolution algorithm, CIBERSORTx.
Materials and Methods
Data Accession
RNA-Seq raw counts were obtained from Gene Expression Omnibus (GSE160306, accessed on March 28, 2022). Raw counts were converted to transcripts per million (TPM) in R (version 4.1.0) for downstream analyses. Samples from the human macular region were selected from the raw dataset using the subset function from the R package Seurat (version 4.0.5).5 Detailed clinical information, including patient gender, age, and disease stages, were accessed from GSE160310. Single-cell transcriptomes of the human neural retina were directly downloaded from Human Cell Atlas (HCA; https://www.humancellatlas.org/, E-MTAB-7316, accessed on January 15, 2022).6
Identification of Severity-Associated Genes in the Macular Region From Donors With DR at a Different Stage
To identify genes related to the progressive severity of DR, the selected RNA-Seq raw count was analyzed by DESeq2 (version 1.32.0),7 using the likelihood-ratio test (LRT), as we previously described.8 Briefly, gene expression was determined by three variables due to limited access to clinical labeling, consisting of gender, age, and disease status (severity). To simplify the design in the LRT, the progressive disease status was indexed using integer scaling: control-1, diabetic-2, NPDR-3, and NPDR/PDR+DME-4. Gender and age as covariants were regressed out in the LRT model, leaving disease status as the main factor to affect gene expression. The batch correction was performed using the removeBatchEffect function in limma (version 3.48.3).9 Significant severity-associated genes were determined by adjusted P < 0.05, controlled for gender and age. The resulting genes were then scaled to z-score and clustered using the degPatterns function from the R package DEGreport (version 1.28.0). Gene clusters with consistent and progressive changes were selected for further analyses.
To further screen significant severity-associated genes, the TPM values of the genes from the selected cluster were analyzed by R package MASS (version 7.3-54) and ordinal (version 2019.12-10) using the ordinal logistic regression (OLR) model.10 Using Cumulative Link Model function in OLR,10 we controlled for gender and age and identified input genes that were associated with disease severity. These severity-associated genes were further determined at P < 0.05. The resulting genes were represented in a forest plot and ranked according to their beta coefficient. A heatmap of severity-associated genes was generated by the heatmap function from the R package NMF (version 0.24.0).11
To assess the gene-gene correlation of severity-associated genes, the raw counts were processed by the Variance Stabilizing Transformation function in DESeq2.7 followed by statistical adjustment for gender and age using the Remove Batch Effect function in limma (version 3.48.3).9 The correlation heatmap was plotted using R package corrplot (version 0.92).12
Functional Enrichment Analysis and Gene Expression Visualization
Functional enrichment analyses (Gene Ontology Biological Processes) for significant severity-associated genes were performed with Metascape (https://metascape.org/gp/index.html#/main/step1),13 which provides more frequently updated bioinformatics analyses than DAVID.14
To visualize the expression of selected genes, the raw counts were processed by the Variance Stabilizing Transformation function in DESeq2.7 followed by statistical adjustment for gender and age using the Remove Batch Effect function in limma (version 3.48.3).9 Tukey boxplots were used to compare gene expression from patients with different DR statuses, with interquartile range boxes and 1.5 times interquartile range whiskers. The 2-tailed Mann-Whitney test was applied to assess pairwise comparisons in the plots, whereas the Kruskal-Wallis test was used to perform statistical comparison across all groups of different disease statuses.
Deconvolution Analyses of the Immune Cells and Retinal Cells From Donors With DR
The CIBERSORTx, a machine learning deconvolution algorithm that enables inference of cell-type-specific gene expression profiles,15 was applied to infer the estimated proportions of infiltrating immune cells or retinal cells of each bulk retinal transcriptome. We used the LM22 signature matrix to define 22 infiltrating immune cells.16 Normalized data (TPM value) of the transcriptome were uploaded to the CIBERSORTx web portal (https://cibersortx.stanford.edu/), with the algorithm run using LM22 signature matrix at 100 permutations with B-mode batch correction.
To infer the retinal cellular profile, we used a previously derived retinal signature matrix using the HCA scRNA-Seq dataset of the human retina.8 In brief, the retinal signature matrix contains rod, Müller glia, bipolar, cone, amacrine, microglia, ganglion, astrocytes, and horizontal cells. The retinal signature matrix was used to impute retinal cellular fraction from the bulk transcriptome at 100 permutations with S-mode batch correction. Statistical significance of cellular numbers associated with severity was assessed by an ordinal logistic regression model using the nonparametric Kruskal-Wallis test that allows for multifactorial designs. The estimated coefficients and P value were presented as a forest plot with 95% confidence intervals. The estimated proportions of cell types were adjusted for gender and age to assess the effects by disease severity by extracting the residuals from fitting a generalized linear model with variables.
Statistical Analyses
Detailed information of statistical analyses was described in various places, including results and figure legends, where the test methods, significance, P values, error bars, and coefficients were included.
Results
Identification of Severity-Associated Genes in the Macula of Patients With DR at Different Clinical Stages
We analyzed a publicly available transcriptional dataset of the macular region from the retina of patients at different stages of DR and healthy controls. A total of 39 macular RNA-Seq profiles from healthy controls (n = 10) and cases from early to late DR stage (n = 29) were analyzed (Fig. 1A). The patient groups were previously classified according to the severity of DR progression, including diabetic (n = 10), NPDR (n = 9), NPDR with DME (n = 7), and PDR with DME (n = 3).17 Of which, patients with NPDR with DME or PDR with DME were combined as one group of NPDR/PDR+DME (n = 10), as the previous study described.17
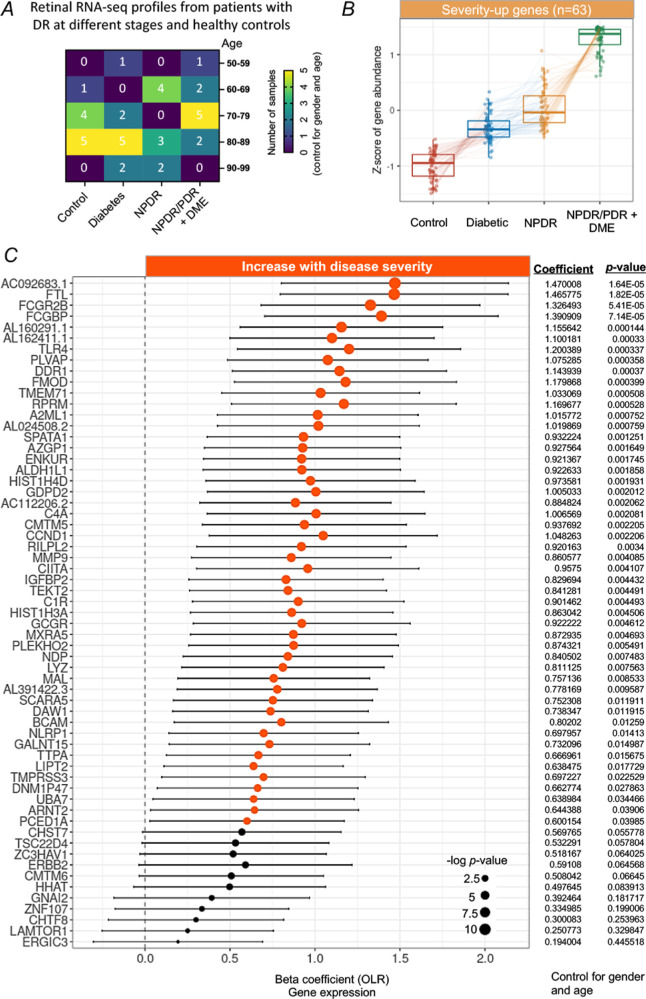
Figure 1.
Identification of severity-associated genes in the macula from patients with degrees of DR. (A) Demographics of the human macular RNA-Seq profiles, detailed by age and different degrees of DR. (B) Tukey boxplots (interquartile range [IQR] boxes with 1.5 × IQR whiskers) of severity-associated genes found in the macula of patients with DR. The expression level of severity-up genes increases with the severity of DR (left, n = 63). Severity-up genes were defined with DESeq2 two-sided likelihood-ratio test (adjusted P < 0.05) by controlling for gender and age. Adjusted gene expressions were shown as z-score. (C) Forest plot of severity-associated genes significantly correlated with DR severity (P < 0.05, n = 50). Positive coefficients indicate severity-associated genes increase in expression with DR severity. Statistical analyses of the correlation between the severity-associated genes and the DR progression were assessed by a nonparametric ordinal logistic regression model that controls for gender and age. Point sizes are scaled by statistical significance. Error bars represent 95% confidence intervals.
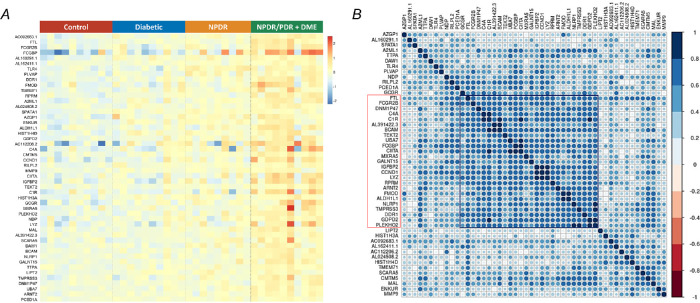
Gene expression levels in the retina were largely affected by age, gender, and disease stages.18 By controlling for age and gender, we identified a cluster of genes (n = 63) related to DR progression from the early to late stage (Fig. 1B) using an LRT.7 Of note, the expression level of these genes was increased with the severity of DR. We therefore defined them as severity-up genes. To further screen genes significantly changed with the severity of DR, we assessed the correlation between severity-up genes and the severity of DR by using the OLR model that controls for gender and age. Our results identified 50 genes progressively increasing their expression level with the severity of DR (Fig. 1C, hereafter referred to as severity-associated genes, P < 0.05). The remaining 13 severity-up genes were not significantly correlated to the severity of DR, thus excluding from the downstream analyses. A heatmap clearly demonstrated that the expression levels of 50 severity-associated genes were progressively increased with the severity of DR from the early (diabetic) to late stage (NPDR/PDR+DME), after adjustment for gender and sex (Fig. 2A). Correlation analysis among these genes in terms of expression level revealed a cluster of genes (n = 25) having the most interactions and strong correlations (|r| > 0.6) with other genes, suggesting their potentially important roles in the progression of DR (Fig. 2B).
Figure 2.
Expression profile and correlation analyses of severity-associated genes. (A) A heatmap of the expression level of 50 severity-associated genes in all samples across different groups, adjusted for gender and age. (B) Heatmap of the correlation matrix across 50 severity-associated genes. Pearson's correlation was calculated among the severity-associated genes to show the co-expression patterns of genes in the heatmap. Color key denotes the Pearson's correlation coefficient between genes. There is one clear cluster (highlighted in red and black) of severity-associated genes with a strong positive correlation, adjusted by gender and age.
Functional Enrichment Analyses of the Severity-Associated Genes and Identification of Key Genes Involved in the Progression of DR
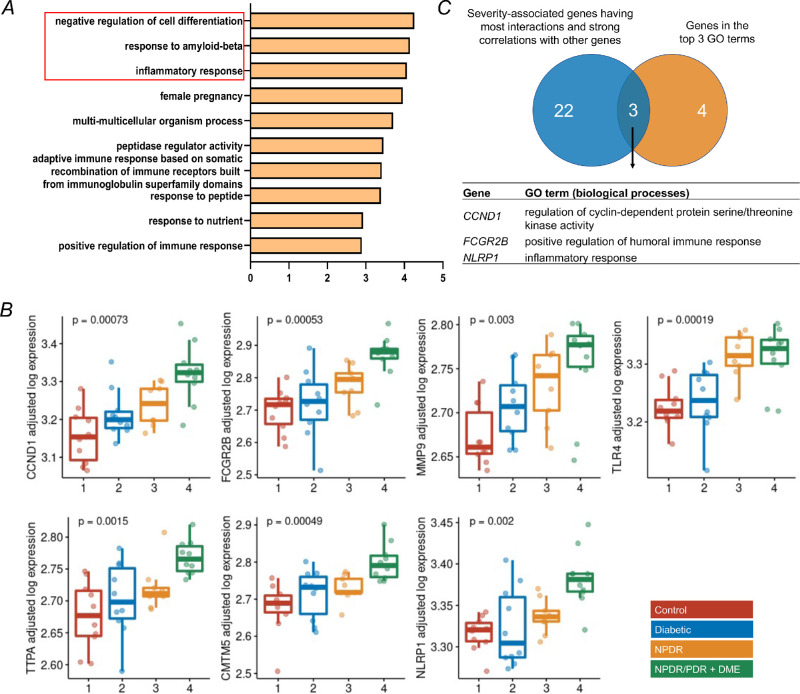
To determine the functional roles of the severity-associated genes in the pathogenesis of DR, functional enrichment analysis (Gene Ontology [GO] biological processes) was performed with Metascape. The results demonstrated significant enrichments mostly related to inflammation and immune response. Of which, negative regulation of cell differentiation, response to amyloid-beta, and inflammatory response rank top three among other GO terms (Fig. 3A). Genes involved in the top three GO terms were combined to identify key genes that contribute to the progression of DR development, resulting in seven genes with significant changes in expression with the severity of DR after controlling for gender and age (Fig. 3B), including cyclin D1 (CCND1), Fc gamma receptor IIb (FCGR2B), matrix metallopeptidase 9 (MMP9), toll like receptor 4 (TLR4), alpha tocopherol transfer protein (TTPA), CKLF like MARVEL transmembrane domain containing 5 (CMTM5), and NLR family pyrin domain containing 1 (NLRP1). To further identify key severity-associated genes involved in the progression of DR, we overlapped the cluster of severity-associated genes (n = 25) identified in Figure 2B with 7 genes identified in the functional enrichment analysis, leading to 3 genes shared by both clusters of genes (Fig. 3C), including CCND1, FCGR2B, and NLRP1. We focused on CCND1 and FCGR2B and investigated their role in immune cells and retinal cells during DR. Given that the expression pattern of NLRP1 was not consistently increased with the severity of DR, we therefore excluded it for further analyses.
Figure 3.
Functional enrichment analyses of severity-associated genes and identification of key genes involved in the DR progression. (A) Functional enrichment analysis (gene ontology annotations of biological processes) of 50 severity-associated genes by Metascape. The top three biological processes highlighted in red were selected for down-stream analysis. Genes with significant changes in these selected annotations include CCND1, FCGR2B, MMP9, TLR4, TTPA, CMTN5, and NLRP1. (B) Tukey boxplots (interquartile range [IQR] boxes with 1.5 × IQR whiskers) showing the expression of CCND1, FCGR2B, MMP9, TLR4, TTPA, CMTN5, and NLRP1. Gene expression values are shown as log-transformed, controlled for gender and age. Statistical significance of difference was assessed by a two-sided Kruskal-Wallis test on the adjusted expression values. (C) Overlapping the clustered genes from Figure 2B (n = 25) and selected genes in Figure 3B (n = 7) resulted in 3 genes, including CCND1, FCGR2B, and NLRP1.
The Immune Cellular Landscape Changed in the Progression of DR
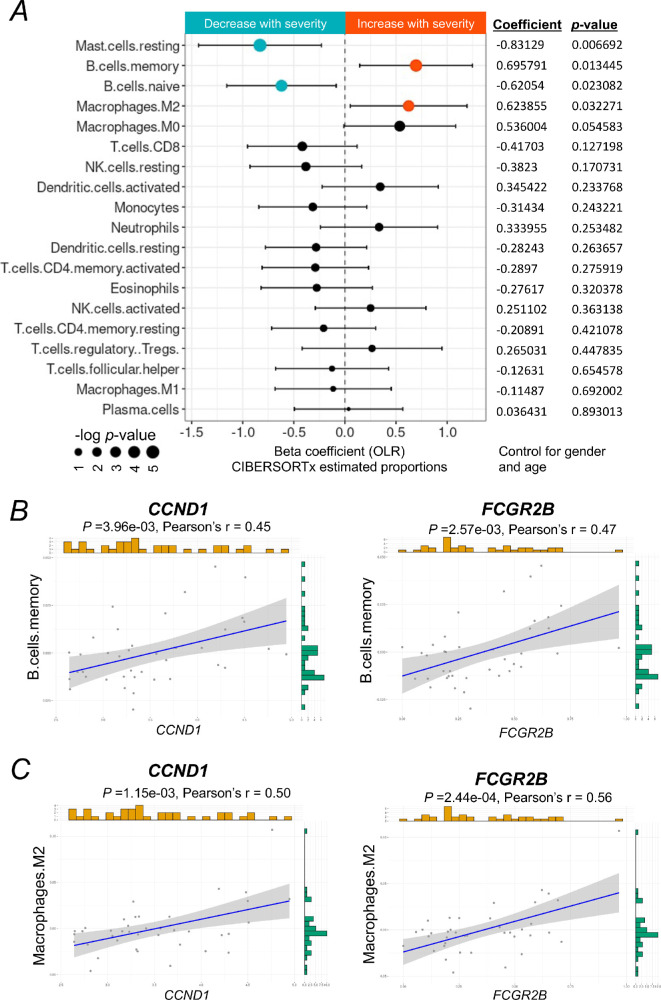
Given that severity-associated genes are strongly correlated to immune response, we next sought to understand the role of immune cells in the retina of patients with DR at different clinical stages. By using CIBERSORTx, we deconvoluted the macular transcriptomic data (Supplementary Fig. S1) with the signature matrix of immune cells, LM22.19 We further assessed what immune cell types change in proportions with the severity of DR using an OLR model. By controlling for gender and age, we demonstrated that estimated proportions of memory B cells and M2 macrophages were significantly increased with the progression of DR, whereas the estimated proportions of resting mast cells and naïve B cells were significantly decreased with the progression of DR (Fig. 4A, Supplementary Fig. S2). Other immune cell types were neither found in the macular tissue nor their estimated proportions were not significantly altered with the progression of DR.
Figure 4.
The immune cellular landscape of the macula of patients with DR and its relation to severity-associated genes. (A) Forest plot of estimated proportions of immune cells having a significant correlation with the severity of DR (P < 0.05). Positive or negative coefficients indicate the proportion of immune cells increases or decreases with the severity of DR. Statistical analyses of the correlation between the severity of DR and the proportions of immune cells were assessed by a nonparametric ordinal logistic regression model, adjusted for gender and age. Point sizes are scaled by statistical significance. Error bars represent 95% confidence intervals. (B, C) Analyses of the correlation between the expression level of CCND1 or FCGR2B and the estimated proportion of memory B cells or M2 macrophages, respectively, adjusted for gender and age. Pearson's correlation coefficient was used to test the strength of linear relationships between gene expression and the estimated proportion of specific cell types. Grey dots denote individual samples (n = 39). Blue lines denote regression lines.
We further studied the correlation between the expression level of two key genes identified above, CCND1 and FCGR2B, and the proportions of the altered immune cell types, including memory B cells and M2 macrophages, using a Pearson's regression analysis. Our results showed that expression levels of both CCND1 and FCGR2B have a moderate positive (0.4< r <0.6) correlation with the proportions of memory B cells (Fig. 4B) and M2 macrophages (Fig. 4C), respectively. These results suggest that CCND1 and FCGR2B may be involved in the regulation of immune responses during the progression of DR mainly through M2 macrophages.
The Retinal Cellular Landscape Changed in the Progression of DR
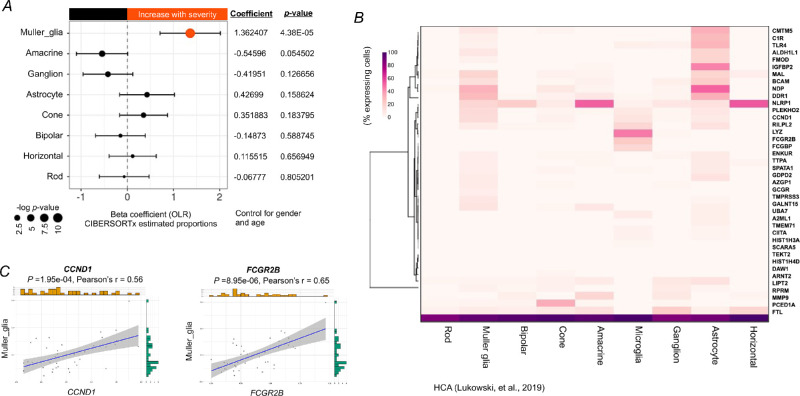
A previous study demonstrated that the retinal structure, particularly the retinal nerve fiber layer, is changed at the early onset of DR.20 However, quantitative changes in retinal cells during the progression of DR have not been reported. Here, we deconvoluted the macular transcriptomes with CIBERSORTx to profile the estimated proportions of each of retinal cell types, using the scRNA-seq data from HCA as a reference (Supplementary Fig. S3). We applied an OLR model to control for gender and age and evaluated if the severity of DR affects the proportions of retinal cell types. The results revealed that the proportions of Müller glia were significantly increased with the progression of DR, whereas the proportions of other retinal cell types were altered but without significant difference (Fig. 5A, Supplementary Fig. S4). To confirm this finding, we further analyzed the expression profile of the severity-associated genes (40 genes were found in the scRNA-Seq dataset) in the human neural retina by using our previous scRNA-Seq data. Of the major retinal cell types, our results showed that most of the severity-associated genes were expressed in a high percentage of Müller glial cells and astrocytes (Fig. 5B). Given the estimated proportion of astrocytes is not significantly changed, we suspect Müller glial cells play an important role in relation to the severity of DR.
Figure 5.
The cellular landscape of the macula of patients with DR and its relation to severity-associated genes. (A) Forest plot of estimated proportions of retinal cells having a significant correlation with the severity of DR (P < 0.05). Positive or negative coefficients indicate the proportion of retinal cells increases or decreases with the severity of DR, respectively. Statistical analyses of the correlation between the severity of DR and the proportions of retinal cells were assessed by a nonparametric ordinal logistic regression model, adjusted for gender and age. Point sizes are scaled by statistical significance. Error bars represent 95% confidence intervals. (B) Heatmap showing the percentage of retina cells expressing severity-associated genes (40 genes were found in the retinal scRNA-Seq dataset from the Human Cell Atlas), scaled by gene across the different cell types. (C) Analyses of the correlation between the expression level of CCND1 or FCGR2B and the estimated proportion of Müller glia, adjusted for gender and age. Pearson's correlation coefficient was used to test the strength of linear relationships between gene expression and the estimated proportion of Müller glia. Grey dots denote individual samples (n = 39). Blue lines denote regression lines.
Similarly, we investigated the relationship between the expression level of two key genes, CCND1 and FCGR2B, and the proportion of Müller glia in DR. Our results showed that the expression level of CCND1 and FCGR2B has a moderate positive (0.4< r <0.6) correlation with the proportion of Müller glia (Fig. 5C), indicating that these genes may be modulated to promote the increase of Müller glia with the severity of DR.
Discussion
Genetic studies, including linkage analyses, candidate gene association studies, and GWAS, have identified many risk loci or corresponding genes for DR.3,21,22 However, such studies tended to focus on one specific clinical stage of DR, particularly the advanced stage, PDR, and DME. Therefore, the resulting disease-causing genes from the genetic studies are not necessary to be associated with the progression of DR. Through the application of LRT on transcriptome from patients with different degrees of DR along with OLR modeling, we identified 50 genes that significantly increased in expression level from the early to late stage of DR (severity-associated genes) by controlling for gender and age. Among which, several severity-associated genes have been previously identified to be associated with DR in human or animal models, including TLR4,23 plasmalemma vesicle-associated protein (PLVAP),24 fibromodulin (Fmod),25 C4A,26 MMP9,27 insulin-like growth factor binding protein 2 (IGFBP2),28 and NLR Family Pyrin Domain Containing 1 (NLRP1),29 whereas none is reportedly increased with the progression of DR. Our results further supported those previous findings, indicating our transcriptome analyses are reliable and comparable to other genetic studies. Many severity-associated genes have not been reported in association with the progression of DR, interestingly, however, they are related to diabetes, such as alpha-2-Macroglobulin Like 1 (A2ML1),30 and C1r,31 suggesting their potential role in the pathogenesis of DR.
Inflammation is a nonspecific response of the immune system to harmful stimuli or stress that includes many functional and molecular mediators.32 Increasing evidence demonstrates that inflammation is crucial to the development of DR. Increased levels of a variety of inflammatory cytokines and chemokines were observed in serum33 and ocular samples (aqueous and vitreous) from patients with PDR.34 However, it remains unclear how inflammation takes part in the progression of DR due to the difficulty of sample collection. Our functional enrichment analyses revealed that the severity-associated genes are mainly involved in the negative regulation of cell differentiation, inflammation, and immune responses, suggesting these pathological processes are progressive during the development of DR.
We further identified CCND1 and FCGR2B as two crucial genes involved in the inflammation in DR. CCND1 protein is the critical gatekeeping protein regulating the transition from G1 phase to S phase via the restriction point of the cell cycle.35 A higher mRNA expression level of CCND1 was observed in human diabetic islets by microarray and quantitative real-time PCR (qRT-PCR).36 Heightened hepatic expression of CCND1 was also found in animal models of obesity or diabetes due to hyperinsulinemia,37 suggesting a close relationship between CCND1 and diabetes. The original study with the macular transcriptomic data from patients with DR that we analyzed in the present study also found that CCND1 was one of the genes identified with significant changes in expression levels.17 Our correlation analysis among severity-associated genes confirmed that CCND1 has a strong interaction with most of other severity-associated genes, indicating its critical role in mediating or regulating different genes that together contributes to the progression of DR. FCGR2B protein is a low affinity receptor for the Fc region of immunoglobulin gamma complexes.38 FCGR2B involves many aspects of inflammatory and immune responses and the complex regulation of defense against infection.39 However, the regulation of FCGR2B expression is complicated depending on the cell types. For example, interferon-gamma, a signature proinflammatory cytokine, escalates FCGR2B mRNA and cell surface expression by lipopolysaccharide-stimulated B cells while reducing it on monocytes, a process keeping immune response away from antibody production to pathogen clearance.39 Increased levels of FCGR2B have been observed in the pancreata of autoantibody-positive at-risk individuals with type 1 diabetes versus controls,40 and in the retina of a rat model with type 1 diabetes induced by streptocotocin.31 Similar to CCND1, FCGR2B is also one of the severity-associated genes with substantial interaction with other genes. Altogether, our results are in line with previous findings and further confirmed both CCND1 and FCGR2B increase in mRNA expression with the severity of DR, suggesting their potential role as biomarkers to monitor the progression of DR.
In the early stages of DR, when blood-retinal barrier is intact, the immune system is mildly activated, and retinal microglia plays a major role in the para-inflammatory response, an important process to maintain retinal homeostasis.41 Consistent stimulation of damage-associated molecules induced by diabetes in the eyes cause maladaptation of the innate immune systems, ultimately promoting the development of DR.41 Circulating immune cells penetrate into the retina and result in chronic inflammation, leading to retina vascular and neuronal damage in the late stage.42 Despite the importance of the immune system in the pathogenesis of DR, few studies have investigated the changes in immune cell types over the progression of DR. Using the deconvolution method with CIBERSORTx, we systematically analyzed immune cellular profiles containing 22 immune cells and identified significant changes in proportions of resting mast cells, memory B cells, naïve B cells, and M2 macrophages in the macular region from donors with different degrees of DR. Mast cells are tissue-resident with heterogeneous phenotypes tuned by inflammatory stimuli.43 We suspect that inflammation in DR results in the phenotype change of mast cells from resting to activated, for which proportions of resting mast cells are significantly decreased over the severity of DR. Memory B cells are the populations of cells that provide long-term humoral immunity.44 A study revealed that chronic inflammation triggers an increase of activated memory B cells in patients with erythema nodosum leprosum, an inflammatory complication of leprosy.45 Given that DR is a progressive disease featured with chronic inflammation, it is reasonable to see increased and decreased proportions of memory B cells and naïve B cells, respectively, with the severity of DR. Macrophages are heterogeneous and feature various functions once activated.46 Our previous study demonstrated the significant increase of M2 macrophages in the retina of the rat model of retinal neovascularization and patients with PDR by deconvolution analysis.47 Our results are in line with previous findings that the proportions of M2 macrophages are increased with the severity of DR. Correlation analyses showed CCND1 and FCGR2B have a moderate positive correlation with memory B cells and M2 macrophages, suggesting both genes may be crucial in regulating activation of M2 macrophages.
Despite studies that have shown morphological or structural changes in retina cells from patients with DR, such as Müller glia and microglia cells,48,49 few assessed changes in proportions of each retinal cell type from patients with DR. Using CIBERSORTx, we, for the first time, quantified the proportions of all retinal cells in the macular region from patients with different degrees of DR. We revealed that the proportion of Müller glia is significantly increased with the severity of DR, whereas changes of the proportions of other retinal cells were observed but without statistical significance. Expression levels of CCND1 and FCGR2B are moderate positive and related to the proportions of Müller glia, indicating their importance in the activation of Müller glia. Müller glia is important to maintain retinal function and health because it spans the entire width of the retina and contacts every other cell type. It is believed that Müller glia becomes activated in DR, characterized by the increased expression of glial fibrillary acidic protein, a marker of reactive gliosis.49,50 However, some histological studies also reported Müller cells loss during the DR progress possibly due to the increased caspase-1 activity and IL-1β production following exposure to hyperglycemic conditions.50,51 More studies are required to determine the mechanisms of Müller glia activation and death to pinpoint whether all Müller glia are equally affected by hyperglycemia. Decreased number of astrocytes in the retina has been found in both patients with DME and rat models of patients with diabetes.52,53 Notably, our results showed an increased proportion of astrocytes, although it was not significantly correlated to the severity of DR. The DR progression is affected by multiple risk factors, such as age, gender, diabetes duration, fasting blood glucose, and glycosylated haemoglobin.54,55 In our deconvolution analyses, we only controlled for age and gender as covariants of gene expression due to limited access to clinical labeling, leaving other risk factors that potentially confound the regression analyses. Moreover, a relatively small sample size could be another factor that affects the deconvolution outcome. Therefore, future studies with a large sample size and enriched clinical labeling should be performed to validate our results.
There were several limitations of this study. First, the sample size in each group of the DR stage is relatively small, which likely affects the statistical power to identify a high number of severity-associated genes defined by LRT. In addition, the current study is based on in silico analysis of transcriptomic data. Future biological studies that investigate the functionality of CCND1 and FCGR2B in memory B cells and M2 macrophages, as well as Müller glia, would be important to understand their role in progressive DR.
Conclusion
The findings reveal a group of genes, particularly CCND1 and FCGR2B, significantly increase in expression with the severity of DR, by controlling for other factors affecting gene expression, such as gender and age. Moreover, proportions of memory B cells, M2 macrophages, and Müller glia are significantly increased with the severity of DR. Our analyses provide potential avenues for subsequent research to study the mechanisms of DR progression.
Code Availability
Analysis scripts will be made available upon request.
Supplementary Material
Acknowledgments
The authors thank Ryan D. Chow for sharing codes for data analyses at Github.
Supported by grants from the National Health and Medical Research Council of Australia (GNT1185600). The Centre for Eye Research Australia receives Operational Infrastructure Support from the Victorian Government.
Author contributions: J.-H.W. and G.-S.L. conceived and designed the study. J.-H.W. developed the analysis approach, performed all data analyses, and generated figures. J.-H.W., R.C.B.W., and G.-S.L. prepared the manuscript. G.-S.L. supervised the work.
Disclosure: J.-H. Wang, None; R.C.B. Wong, None; G.-S, Liu, None
References
- 1. Duh EJ, Sun JK, Stitt AW.. Diabetic retinopathy: current understanding, mechanisms, and treatment strategies. JCI Insight. 2017; 2: e93751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang W, Lo ACY.. Diabetic Retinopathy: Pathophysiology and Treatments. Int J Mol Sci. 2018; 19: 1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cho H, Sobrin L.. Genetics of diabetic retinopathy. Curr Diab Rep. 2014; 14: 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grassi MA, Tikhomirov A, Ramalingam S, Below JE, Cox NJ, Nicolae DL.. Genome-wide meta-analysis for severe diabetic retinopathy. Hum Mol Genet. 2011; 20: 2472–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stuart T, Butler A, Hoffman P, et al.. Comprehensive Integration of Single-Cell Data. Cell. 2019; 177: 1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lukowski SW, Lo CY, Sharov AA, et al.. A single-cell transcriptome atlas of the adult human retina. EMBO J. 2019; 38: e100811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang J-H, Wong RC, Liu G-S.. Retinal aging transcriptome and cellular landscape in association with the progression of age-related macular degeneration. medRxiv preprint, 10.1101/2022.04.03.22273375. [DOI] [PMC free article] [PubMed]
- 9. Ritchie ME, Phipson B, Wu D, et al.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Q, Shepherd BE, Li C, Harrell FE Jr. Modeling continuous response variables using ordinal regression. Stat Med. 2017; 36: 4316–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gaujoux R, Seoighe C.. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010; 11: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wei T, Simko V, Levy M, Xie Y, Jin Y, Zemla J. Corrplot: Visualization of a correlation matrix. R Package Version 073. 2013. Available at: https://rdrr.io/cran/corrplot/man/corrplot.html.
- 13. Zhou Y, Zhou B, Pache L, et al.. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019; 10: 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wadi L, Meyer M, Weiser J, Stein LD, Reimand J.. Impact of outdated gene annotations on pathway enrichment analysis. Nat Methods. 2016; 13: 705–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Newman AM, Steen CB, Liu CL, et al.. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019; 37: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Newman AM, Liu CL, Green MR, et al.. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015; 12: 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Becker K, Klein H, Simon E, et al.. In-depth transcriptomic analysis of human retina reveals molecular mechanisms underlying diabetic retinopathy. Sci Rep. 2021; 11: 10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ratnapriya R, Sosina OA, Starostik MR, et al.. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat Genet. 2019; 51: 606–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA.. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol. 2018; 1711: 243–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vujosevic S, Midena E.. Retinal layers changes in human preclinical and early clinical diabetic retinopathy support early retinal neuronal and Muller cells alterations. J Diabetes Res. 2013; 2013: 905058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Azzam SK, Osman WM, Lee S, et al.. Genetic Associations With Diabetic Retinopathy and Coronary Artery Disease in Emirati Patients With Type-2 Diabetes Mellitus. Front Endocrinol (Lausanne). 2019; 10: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Graham PS, Kaidonis G, Abhary S, et al.. Genome-wide association studies for diabetic macular edema and proliferative diabetic retinopathy. BMC Med Genet. 2018; 19: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang L, Wang J, Fang J, Zhou H, Liu X, Su SB.. High glucose induces and activates Toll-like receptor 4 in endothelial cells of diabetic retinopathy. Diabetol Metab Syndr. 2015; 7: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bosma EK, van Noorden CJF, Schlingemann RO, Klaassen I.. The role of plasmalemma vesicle-associated protein in pathological breakdown of blood-brain and blood-retinal barriers: potential novel therapeutic target for cerebral edema and diabetic macular edema. Fluids Barriers CNS. 2018; 15: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Freeman WM, Bixler GV, Brucklacher RM, et al.. Transcriptomic comparison of the retina in two mouse models of diabetes. J Ocul Biol Dis Infor. 2009; 2: 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shahulhameed S, Vishwakarma S, Chhablani J, et al.. A Systematic Investigation on Complement Pathway Activation in Diabetic Retinopathy. Front Immunol. 2020; 11: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kowluru RA, Zhong Q, Santos JM.. Matrix metalloproteinases in diabetic retinopathy: potential role of MMP-9. Expert Opin Investig Drugs. 2012; 21: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meyer-Schwickerath R, Pfeiffer A, Blum WF, et al.. Vitreous levels of the insulin-like growth factors I and II, and the insulin-like growth factor binding proteins 2 and 3, increase in neovascular eye disease. Studies in nondiabetic and diabetic subjects. J Clin Invest. 1993; 92: 2620–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Y, Liu C, Wan XS, Li SW.. NLRP1 deficiency attenuates diabetic retinopathy (DR) in mice through suppressing inflammation response. Biochem Biophys Res Commun. 2018; 501: 351–357. [DOI] [PubMed] [Google Scholar]
- 30. Hatem G, Hjort L, Asplund O, et al.. Mapping the Cord Blood Transcriptome of Pregnancies Affected by Early Maternal Anemia to Identify Signatures of Fetal Programming. J Clin Endocrinol Metab. 2022; 107: 1303–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jargen P, Dietrich A, Herling AW, Hammes HP, Wohlfart P.. The role of insulin resistance in experimental diabetic retinopathy-Genetic and molecular aspects. PLoS One. 2017; 12: e0178658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell. 2010; 140: 771–776. [DOI] [PubMed] [Google Scholar]
- 33. Quevedo-Martinez JU, Garfias Y, Jimenez J, Garcia O, Venegas D, Bautista de Lucio VM.. Pro-inflammatory cytokine profile is present in the serum of Mexican patients with different stages of diabetic retinopathy secondary to type 2 diabetes. BMJ Open Ophthalmol. 2021; 6: e000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu F, Phone A, Lamy R, et al.. Correlation of Aqueous, Vitreous, and Plasma Cytokine Levels in Patients With Proliferative Diabetic Retinopathy. Invest Ophthalmol Vis Sci. 2020; 61: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen Y, Huang Y, Gao X, et al.. CCND1 Amplification Contributes to Immunosuppression and Is Associated With a Poor Prognosis to Immune Checkpoint Inhibitors in Solid Tumors. Front Immunol. 2020; 11: 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taneera J, Fadista J, Ahlqvist E, et al.. Expression profiling of cell cycle genes in human pancreatic islets with and without type 2 diabetes. Mol Cell Endocrinol. 2013; 375: 35–42. [DOI] [PubMed] [Google Scholar]
- 37. Luo C, Liang J, Sharabi K, et al.. Obesity/Type 2 Diabetes-Associated Liver Tumors Are Sensitive to Cyclin D1 Deficiency. Cancer Res. 2020; 80: 3215–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Willcocks LC, Carr EJ, Niederer HA, et al.. A defunctioning polymorphism in FCGR2B is associated with protection against malaria but susceptibility to systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2010; 107: 7881–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith KG, Clatworthy MR.. FcgammaRIIB in autoimmunity and infection: evolutionary and therapeutic implications. Nat Rev Immunol. 2010; 10: 328–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yip L, Fuhlbrigge R, Alkhataybeh R, Fathman CG.. Gene Expression Analysis of the Pre-Diabetic Pancreas to Identify Pathogenic Mechanisms and Biomarkers of Type 1 Diabetes. Front Endocrinol (Lausanne). 2020; 11: 609271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu H, Chen M.. Diabetic retinopathy and dysregulated innate immunity. Vision Res. 2017; 139: 39–46. [DOI] [PubMed] [Google Scholar]
- 42. Stem MS, Gardner TW.. Neurodegeneration in the pathogenesis of diabetic retinopathy: molecular mechanisms and therapeutic implications. Curr Med Chem. 2013; 20: 3241–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krystel-Whittemore M, Dileepan KN, Wood JG.. Mast Cell: A Multi-Functional Master Cell. Front Immunol. 2015; 6: 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Good-Jacobson KL, Tarlinton DM.. Multiple routes to B-cell memory. Int Immunol. 2012; 24: 403–408. [DOI] [PubMed] [Google Scholar]
- 45. Negera E, Walker SL, Bekele Y, Dockrell HM, Lockwood DN.. Increased activated memory B-cells in the peripheral blood of patients with erythema nodosum leprosum reactions. PLoS Negl Trop Dis. 2017; 11: e0006121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Corliss BA, Azimi MS, Munson JM, Peirce SM, Murfee WL.. Macrophages: An Inflammatory Link Between Angiogenesis and Lymphangiogenesis. Microcirculation. 2016; 23: 95–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang JH, Kumar S, Liu GS.. Bulk Gene Expression Deconvolution Reveals Infiltration of M2 Macrophages in Retinal Neovascularization. Invest Ophthalmol Vis Sci. 2021; 62: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Altmann C, Schmidt MHH.. The Role of Microglia in Diabetic Retinopathy: Inflammation, Microvasculature Defects and Neurodegeneration. Int J Mol Sci. 2018; 19: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mizutani M, Gerhardinger C, Lorenzi M.. Muller cell changes in human diabetic retinopathy. Diabetes. 1998; 47: 445–449. [DOI] [PubMed] [Google Scholar]
- 50. Lieth E, Barber AJ, Xu B, et al.. Glial reactivity and impaired glutamate metabolism in short-term experimental diabetic retinopathy. Penn State Retina Research Group. Diabetes. 1998; 47: 815–820. [DOI] [PubMed] [Google Scholar]
- 51. Kuser-Abali G, Ozcan F, Ugurlu A, Uysal A, Fuss SH, Bugra-Bilge K. SIK2 is involved in the negative modulation of insulin-dependent muller cell survival and implicated in hyperglycemia-induced cell death. Invest Ophthalmol Vis Sci. 2013; 54: 3526–3537. [DOI] [PubMed] [Google Scholar]
- 52. Haj Najeeb B, Simader C, Deak G, et al.. The Distribution of Leakage on Fluorescein Angiography in Diabetic Macular Edema: A New Approach to Its Etiology. Invest Ophthalmol Vis Sci. 2017; 58: 3986–3990. [DOI] [PubMed] [Google Scholar]
- 53. Rungger-Brandle E, Dosso AA, Leuenberger PM.. Glial reactivity, an early feature of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2000; 41: 1971–1980. [PubMed] [Google Scholar]
- 54. Tan CS, Gay EM, Ngo WK.. Is age a risk factor for diabetic retinopathy? Br J Ophthalmol. 2010; 94: 1268. [DOI] [PubMed] [Google Scholar]
- 55. Liu Y, Yang J, Tao L, et al.. Risk factors of diabetic retinopathy and sight-threatening diabetic retinopathy: a cross-sectional study of 13 473 patients with type 2 diabetes mellitus in mainland China. BMJ Open. 2017; 7: e016280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.