Abstract
The diagnosis of vitreoretinal lymphoma (VRL), a rare subtype of primary central nervous system lymphoma, is challenging. We aimed to investigate the mutational landscape of VRL by sequencing circulating tumor DNA (ctDNA) from aqueous humor (AH) and/or vitreous fluid (VF), as well as applying ctDNA sequencing to diagnosis and treatment monitoring. Baseline AH and/or VF specimens from 15 VRL patients underwent comprehensive genomic profiling using targeted next-generation sequencing. The molecular profiles of paired baseline AH and VF specimens were highly concordant, with comparable allele frequencies. However, the genetic alterations detected in cerebrospinal fluid ctDNA only partially overlapped with those from simultaneously collected AH/VF samples, with much lower allele frequencies. Serial post-treatment AH or VF samples were available for five patients and their changes in ctDNA allele frequency displayed a similar trend as the changes in interleukin-10 levels; an indicator of response to treatment. A cohort of 23 patients with primary central nervous system lymphoma was included as a comparison group for the genetic landscape and evaluations of the efficacy of ibrutinib. More MYD88 mutations, but fewer IRF4 mutations and CDKN2A/B copy number losses were observed in the baseline samples of primary central nervous system lymphoma than VRL patients. The objective response rate to ibrutinib treatment was much higher for patients with primary central nervous system lymphoma (64.7%, 11/17) than for those with VRL (14.3%, 1/7). In summary, we provide valuable clinical evidence that AH is a good source of tumor genomic information and can substitute VF. Moreover, molecular profiling of AH has clinical utility for the diagnosis of VRL and treatment monitoring.
Introduction
Vitreoretinal lymphoma (VRL) is a rare type of primary central nervous system lymphoma (PCNSL) which primarily involves the retina and vitreous.1-3 Based on the origins of the lymphoma, it can be classified as primary VRL, synchronous VRL with central nervous system or systemic involvement, or secondary VRL, which occurs as a site of relapsed lymphoma.4,5 Approximately 95% of primary VRL are histologically identified as diffuse large B-cell lymphomas, while at the molecular level, both PCNSL and primary VRL are classified into MCD/C5 subgroups of diffuse large B-cell lymphoma based on concurrent MYD88 (L265P) and CD79B mutations.6-8 Primary VRL is highly aggressive with an elevated mortality rate, and 65% to 90% of patients eventually develop brain parenchymal involvement. In contrast, 15% to 25% of patients with PCNSL have vitreoretinal involvement, either as synchronous VRL and PCNSL or secondary VRL.1 The diagnosis of VRL remains a challenge for clinicians as the symptoms of VRL can mimic those of posterior uveitis. Thus, patients are typically diagnosed with intermediate and/or posterior uveitis, with vitritis and/or subretinal infiltrates.1,9,10 The diagnosis of VRL is made on the basis of histological and molecular tests. First, cytology and flow cytometry of ocular samples are used to identify a monoclonal neoplastic B lymphocyte population. Second, elevated levels of interleukin (IL)-10 in aqueous humor (AH) and/or vitreoretinal fluid (VF), and the detection of MYD88 mutations in tumor cells can also be used as ancillary tests.11 However, cytomorphological evaluations often fail to detect lymphoma cells because of the limited sample volume, low cellularity of VF samples, and cell lysis during biopsies.12-14
Recently, several studies reported the genetic profiles of VRL using VF ctDNA sequencing.15-17 Vitreous biopsy is a common surgical procedure, but it is an invasive process and may cause severe complications. It has been reported that AH can serve as a source of liquid biopsy for molecular profiling in retinoblastomas, which is enriched in eye-specific tumor-derived cell-free DNA (cfDNA) and can be collected for repeated sampling.18 Thus, AH is an emerging tool for the diagnosis, prognosis, and treatment monitoring of tumors with ocular invasion.
The MYD88 L265P mutation is a primary oncogenic driver in PCNSL and was recently identified as a disease biomarker in VRL as well.17,19 MYD88 L265P continuously activates the nuclear factor kappa light-chain enhancer of activated B cells (NF-kB) by Bruton tyrosine kinase (BTK), promoting tumor proliferation. As ibrutinib, a BTK inhibitor, can cross the blood-brain barrier, ibrutinib-based combination therapy is widely used in PCNSL patients, with promising responses.20 However, the efficacy of ibrutinib in patients with VRL remains to be evaluated.
In this study, we sought to investigate the mutational landscape of VRL and the application of serial molecular profiling of AH/VF ctDNA for treatment monitoring. We also explored the differences in genomic profiles between VRL and PCNSL patients, and their responses to ibrutinib treatment.
Methods
Patients and the diagnosis of vitreoretinal lymphoma
A total of 15 patients who presented with severe vitreous opacity and/or diffuse yellow subretinal lesions were admitted to the Department of Medical Oncology at Sun Yatsen University Cancer Center (SYSUCC) between December 2018 and December 2020. VRL was diagnosed by two experienced specialists (DL and WS) at the Zhongshan Ophthalmic Center of the Sun Yat-Sen University (Guangzhou, China). The VRL diagnoses of three patients were confirmed by pathology studies using standard cytology and immunocytochemistry of B-cell markers, and clonality analyses for the presence of immunoglobulin heavy chain (IGH) rearrangements. The other 12 patients were diagnosed based on typical clinical manifestations, including elevation of the IL-10/IL-6 ratio and/or IGH rearrangements in AH/VF. All patients underwent further examinations, including positron emission tomography and computed tomography (PET/CT) or magnetic resonance imaging and seven also underwent lumbar puncture for cytology and cerebrospinal fluid (CSF) ctDNA sequencing.
This study was approved by the ethics committee of SYSUCC (approval n.: B2020-315-01) and was conducted in accordance with the Declaration of Helsinki. All participants provided written informed consent prior to sample collection.
Sample collection and DNA extraction
The median volume of AH/VF samples used for cfDNA extraction was 0.1 mL (range, 0.02–3.5 mL). Samples were transported to the laboratory at 4°C. cfDNA was extracted from the supernatants of AH/VF samples (1,800 g × 10 min) within 48 h of collection. DNA was extracted using a Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol and stored at -80°C until further analysis. Fragment distribution was determined on a Bioanalyzer 2100 using a High Sensitivity DNA Kit (Agilent Technologies, Santa Clara, CA, USA).
The tumor cell content of formalin-fixed, paraffin-embedded (FFPE) sections from 23 PCNSL patients was determined by a pathologist. Genomic DNA of the 23 PCNSL patients was purified from the FFPE slides using a QIAamp DNA FFPE Tissue Kit (Qiagen) and from oral swabs using a DNeasy Blood & Tissue Kit (Qiagen). This DNA was then quantified using a Nanodrop2000 (Thermo Fisher Scientific, Waltham, MA, USA). All DNA was quantified using the dsDNA HS Assay Kit on a Qubit 3.0 Fluorometer (Life Technologies, Carlsbad, CA, USA).
DNA sequencing and genomic mutational analyses
Targeted next-generation sequencing (NGS) was performed using a panel (HemasalusTM) of exons and splice sites of more than 400 genes that are recurrently mutated in B-cell lymphomas. The full list of genes included in the panel is provided in Online Supplementary Table S1. NGS was performed in a testing laboratory (Nanjing Geneseeq Technology, Inc., Nanjing, China) accredited by the Clinical Laboratory Improvement Amendments (CLIA) and the College of American Pathologists (CAP). The depth of coverage of the NGS panel was 500× for liquid biopsies. Sequencing libraries were prepared using the KAPA Hyper Prep Kit (KAPA Biosystems) and sequenced on a HiSeq 4000 NGS platform (Illumina).21 Sequencing data were processed as previously described.22 In brief, the data were first demultiplexed and the FASTQ file was subjected to quality control to remove low-quality data or N bases. Qualified reads were mapped to the reference human genome, hg19, using the Burrows-Wheeler Aligner. The Genome Analysis Toolkit (GATK 3.4.0) was used to perform local realignment around indels and base quality score recalibration. Picard was used to remove polymerase chain reaction duplicates. VarScan2 was used to detect single-nucleotide variants and insertion/deletion mutations. A mutant allele frequency cutoff of 1% was used for tissue samples and 0.3% for cfDNA samples. ADTEx was used to identify copy number variations with a normal human DNA sample, NA18535. A cutoff log2 ratio was set at ±0.6 for copy number changes (corresponding to a 1.5-fold copy number gain and 0.65-fold copy number loss).
Results
Overview of the patients with vitreoretinal lymphoma
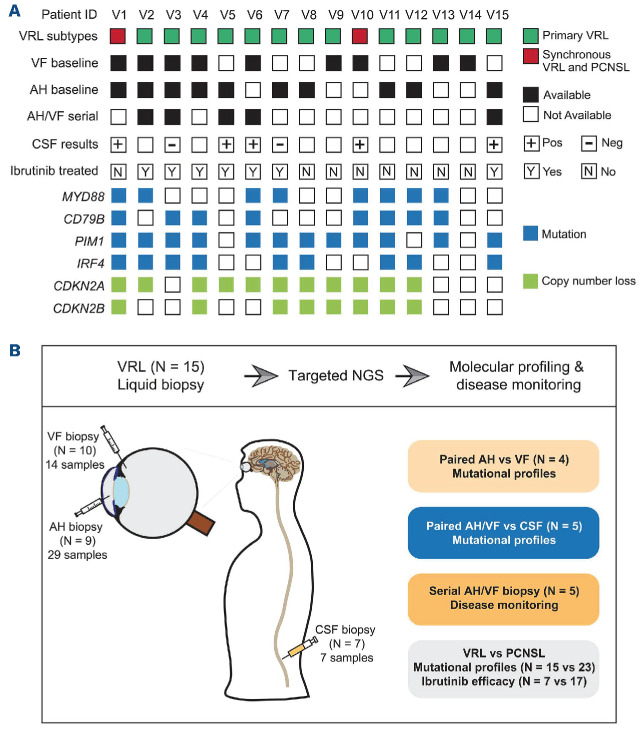
As shown in Table 1, a total of 15 VRL patients were enrolled in this study with a median age of 56 years (range, 38-68 years). Almost all patients (14/15) had binocular disease. Vitreous involvement was more frequent than subretinal infiltrates (11/15 vs. 8/15, respectively) and lymphoma was found at both locations in four patients. Thirteen individuals had primary VRL without brain involvement or histories of other systemic lymphomas at diagnosis, and the remaining two patients (V1 and V10) were diagnosed with synchronous VRL and PCNSL. Representative cytological and immunohistochemistry images of a VRL patient (V4) are shown in Online Supplementary Figure S1, with large, atypical cells positive for Ki67, CD20, and CD79A. Baseline AH and/or VF samples were collected from all patients, of whom five also underwent sequential sampling during their treatment for disease monitoring. The median concentration of cfDNA extracted from AH/VF samples was 0.5 ng/µL. A full list of liquid biopsy volumes and cfDNA concentrations is provided in Online Supplementary Table S2. Seven VRL patients received ibrutinib therapy, mainly for first- and second-line treatments (Figure 1A), while the remaining eight VRL patients were administered local ocular treatments (Table 1).
Table 1.
Patients’ characteristics.
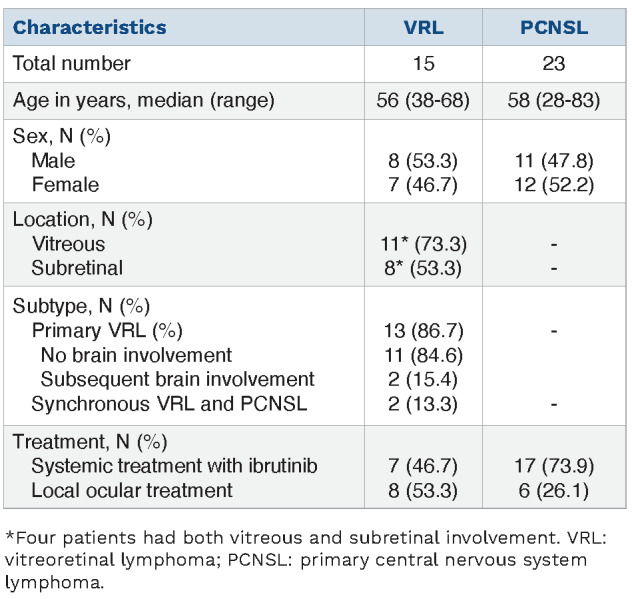
Figure 1.
Overview of the patients’ characteristics and study design. (A) The availability of samples from all enrolled patients, as well as ibrutinib treatment information are shown as indicated in the legend. The molecular features of the most prevalent gene alterations detected in baseline samples are also provided. (B) An illustration of sampling aqueous humor, vitreous fluid, and cerebrospinal fluid is shown on the left. The four aims of this study are listed on the right. ID: identity; VRL: vitreoretinal lymphoma; PCNSL: primary central nervous system lymphoma; AH: aqueous humor; VF: vitreous fluid; CSF: cerebrospinal fluid; NGS: next-generation sequencing.
High concordance of mutations detected in paired aqueous humor and vitreous fluid cell-free DNA samples
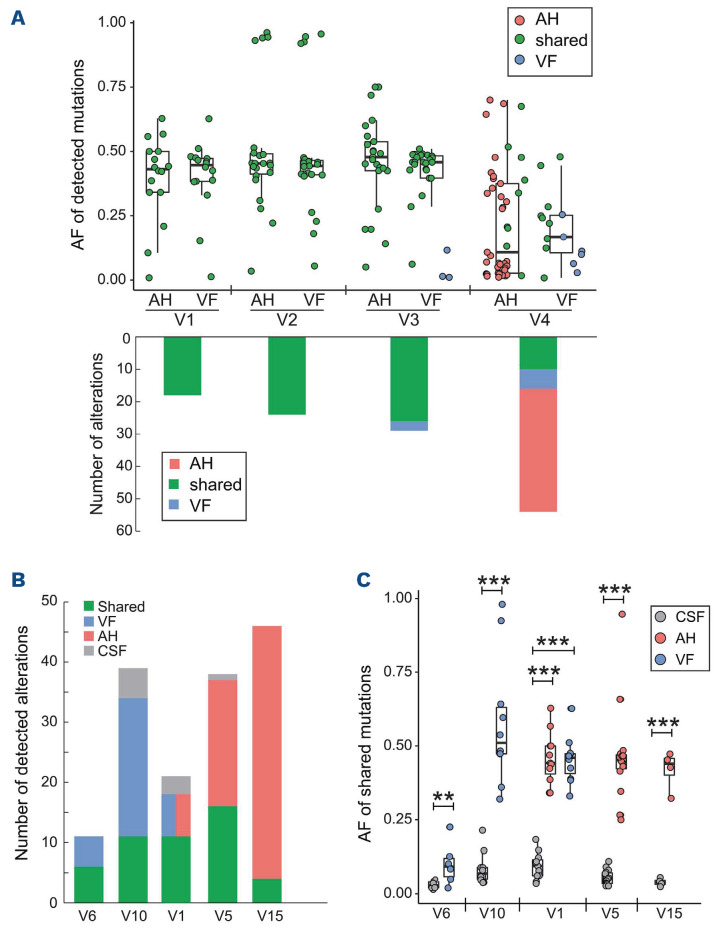
Baseline AH and/or VF samples were collected from all 15 VRL patients and underwent targeted NGS. Two-thirds of samples carried MYD88 L265P and/or CD79B mutations (Figure 1A). Other frequently mutated genes included PIM1 (80%), IRF4 (60%), and CDKN2A/B copy number losses (73.3% and 53.3%). A complete list of mutations in all samples is provided in Online Supplementary Table S3. Four patients (V1-V4) had matched baseline AH and VF samples that were subjected to sequencing. Two patients (V1 and V2) shared an identical mutational spectrum in the matched AH and VF samples; their allele frequencies were also comparable (Figure 2A). The concordance rates of mutations between AH and paired VF samples in the other two patients (V3 and V4) were 90% (26/29) and 60% (10/16), respectively. Thus, those results suggest that AH can serve as a surrogate for mutational analysis of VRL and is more easily accessible than VF biopsies.
Figure 2.
Mutational comparisons between matched aqueous humor, vitreous fluid and cerebrospinal fluid samples. (A) Four pairs of baseline aqueous humor (AH) and vitreous fluid (VF) samples underwent targeted next-generation sequencing. The allele frequencies of shared and unique mutations detected in matched AH and VF samples are shown in the top panel. The number of alterations detected in each patient is shown at the bottom of the panel. Shared, AH-only, and VF-only alterations are colored green, red, and blue, respectively. (B) The number of alterations detected in cerebrospinal fluid (CSF) and AH/VF samples from five patients is as labeled in the legend. The brain involvement in patients V1 and V10 was confirmed by positron emission tomography/computed tomography examinations. (C) Allele frequencies of mutations present in both CSF and AH/VF samples. **P<0.05, ***P<0.01. AF: allele frequency.
In addition, to avoid unnecessary clinical injury to patients with binocular disease, AH/VF sampling is usually performed in a single eye. However, patient V5 underwent AH extraction from both eyes for sequencing. Overall, 32 mutations were identified in AH samples from either the left or right eye, over 80% (26/32) of which were detected in both samples, with similar allele frequencies (R2=0.899) (Online Supplementary Figure S2).
Figure 3.
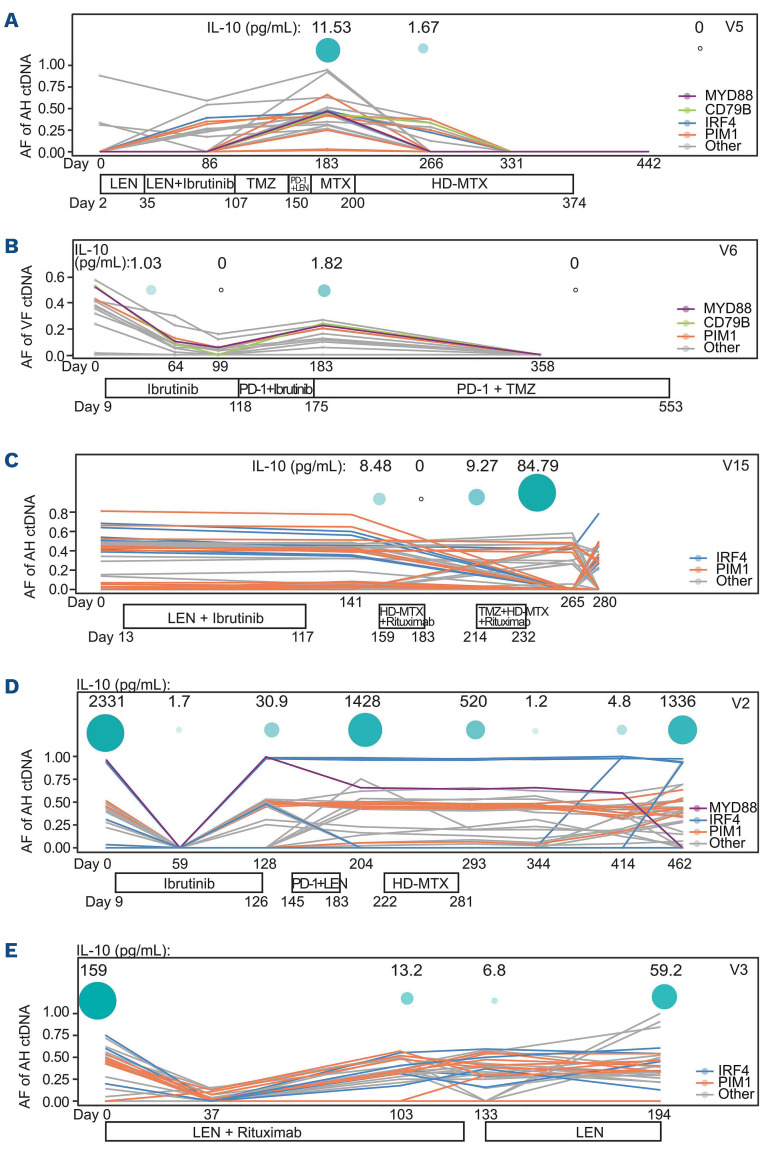
Dynamic profiling of circulating tumor DNA and interleukin-10 levels in aqueous humor or vitreous fluid samples. The treatment histories of five patients (identity numbers in the top right corner of each panel) with dynamic and sequential sampling of aqueous humor (AH) or vitreous fluid (VF) are shown at the bottom of each panel. The allele frequency changes to driver mutations and other alterations are shown in the line chart. Interleukin-10 levels (pg/mL) at each time point are indicated by the green circles at the top of each panel. AF: allele frequency; ctDNA: circulating tumor DNA; IL-10: interleukin.10; LEN: lenalidomide; TMZ: temozolomide; HD-MTX: high-dose methotrexate.
Early detection of circulating tumor DNA in cerebrospinal fluid in patients with primary vitreoretinal lymphoma may predict parenchymal invasion of the brain
CSF samples were collected from seven patients for whom CSF cytology examinations were negative. Five of the patients were positive for CSF ctDNA and the median concentration of the extracted cfDNA from CSF samples was 1.1 ng/µL (Online Supplementary Table S2). The numbers of alterations in VF/AH and CSF samples are shown in Figure 2B, while the allele frequencies of the shared mutations are shown in Figure 2C. All five patients had significantly higher allele frequencies for the shared mutations in AH and/or VF samples than in CSF ctDNA. It is also of note that patients V1 and V10, who had synchronous PCNSL confirmed by PET/CT examinations, harbored a higher number of mutations detected only in CSF samples with significantly higher allele frequencies (P<0.01) than those of the other three patients (V5, V6, and V15). Notably, brain involvement developed in patients V5 and V6 after 9 and 19 months, respectively. Thus, positive CSF ctDNA detection might be an early indicator of brain involvement with a subset of alterations in patients with primary VRL.
Dynamic profiling of circulating tumor DNA in aqueous humor or vitreous fluid samples to monitor response to treatment
Of the seven VRL patients who received systemic treatment, five underwent a series of dynamic AH/VF sampling (4 underwent serial AH sampling and 1 underwent serial VF sampling) (Figure 3, Online Supplementary Table S4). AH or VF samples were collected for ctDNA detection throughout treatment to dynamically monitor the therapeutic responses. In addition, the IL-10 levels in AH/VF samples were also analyzed to assist with the response evaluation of VRL patients. For patients V5 and V6, the changes of IL-10 levels were highly concordant with the allele frequencies of AH/VF ctDNA (Figure 3A, B). For patient V15, all four IL-10 tests were performed within the interval of two rounds of AH ctDNA sequencing, which provided limited information about the correlation between the IL-10 levels and ctDNA allele frequencies (Figure 3C). The remaining two patients (V2 and V3) with extremely high levels of IL-10 at baseline showed dramatic decreases in post-treatment IL-10 levels and AH ctDNA allele frequencies (Figure 3D, E). These findings imply that AH/VF ctDNA profiling could assist in the evaluation of treatment efficacy, in addition to the use of IL-10 tests and eye examinations.
Comparisons of mutational landscape between patients with vitreoretinal lymphoma and primary central nervous system lymphoma
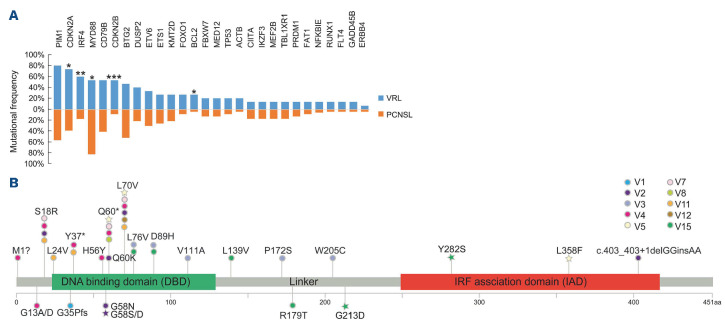
To further compare the mutational landscapes of VRL and PCNSL, we included another cohort of 23 PCNSL patients. When comparing the mutations detected in the baseline VRL AH/VF samples and PCNSL tumor biopsy specimens, we found that PIM1 mutations (80%) and CDKN2A copy number loss (73.3%) were the two most frequently observed alterations in VRL patients, but were underrepre-sented in the PCNSL cohort with frequencies of 56.5% and 39.1%, respectively (Online Supplementary Figure S3). Furthermore, the canonical oncogenic driver mutation, MYD88 L265P, was less frequently detected in VRL patients than in the the PCNSL cohort (53.3% vs. 82.6%, respectively, P=0.07) (Figure 4A). In contrast, the frequencies of IRF4 mutations and CDKN2B copy number loss were significantly higher in the VRL cohort (Figure 4A).
Figure 4.
Comparisons of mutation frequencies between patients with vitreoretinal lymphoma and primary central nervous system lymphoma. (A) The frequencies of the most prevalent mutations in patients with vitreoretinal lymphoma and primary central nervous system lymphoma are shown in the bar plot. *P<0.1; **P<0.05; ***P<0.01. (B) The locations of the mutations in the IRF4 gene are shown. Each patient is represented by a color, as indicated in the legend. The mutations detected in the baseline samples are represented by circles, while stars represent the mutations that were only detected in the post-treatment samples. VRL: vitreoretinal lymphoma; PCNSL: primary central nervous system lymphoma.
In addition, we further investigated the mutational sites of IRF4 in VRL patients. Recurrent IRF4 mutations mainly occurred in the DNA binding domain with L70V, Q60* and S18R being the most frequent mutations (Figure 4B). Most IRF4 mutation sites were detected in primary and post-treatment AH/VF samples (e.g., V1-V3 and V15), while G58S/D, G213D, Y282S, and L358F were acquired after multiple lines of treatment (e.g., V2, V5, and V15).
In the VRL cohort, seven patients received ibrutinib, only one of whom (V2) achieved a partial response and had a significant improvement in visual acuity, leading to an overall response rate of 14% to ibrutinib (Online Supplementary Table S5). In contrast, 17 PCNSL patients received systemic treatment containing ibrutinib and the overall response rate was 65%, with nine complete responses and two partial responses. It should be noted that these patients received either ibrutinib monotherapy (PCNSL, 41%; VRL, 29%) or combined therapies (PCNSL, 59%; VRL, 71%); 29% (5/17) of the PCNSL patients and 57% (4/7) of the VRL patients received ibrutinib as first-line treatment. The diverse treatment regimens prevented a direct comparison of the efficacy of ibrutinib between VRL and PCNSL patients; however, there was a trend to the efficacy of ibrutinib being poorer in VRL patients.
Discussion
To our knowledge, this study was the first to investigate the molecular profiles of AH/VF ctDNA using a large NGS panel covering over 400 lymphoma-related genes in the Chinese population. The findings revealed the potential utility of analyzing AH ctDNA for the diagnosis of VRL, as well as for disease monitoring with serial sampling. The high concordance between AH and VF profiles indicated that AH could serve as a surrogate for VF liquid biopsies, as AH is more easily accessible. We also revealed differences in the molecular landscapes between VRL and PCNSL, and the suboptimal responses to ibrutinib in the VRL cohort.
Due to the low incidence of VRL, our study cohort was relatively small; however, this was a common limitation of other published studies as well.4 For instance, Lee et al. investigated the mutational signatures of primary VRL using whole-exon sequencing in an Asian cohort (n=9), finding that all patients carried MYD88 mutations. The frequencies of IRF4 and PIM1 mutations in that cohort were 44.4% and 88.9%, respectively, which were comparable to the frequencies observed in the current study (60% and 80%, respectively).16 However, in another study of 16 primary VRL patients analyzed using targeted NGS, only 18.8% harbored IRF4 mutations, and the frequencies of MYD88 and PIM1 mutations were both 68.8%.15 Notably, that study15 was conducted in a Western population, which might explain the different mutational frequencies. Bonzheim et al.15 also reported a similar frequency of CD79B mutations (43.8%) as that in our study (53.3%) and in a study by Yonese et al. (35%).23 Another small cohort study (n=8) detected MYD88 L265P in 75% of patients’ AH samples, with a consistency of 87.5% when compared to VF samples using polymerase chain reaction and pyrosequencing assays.19 Overall, the mutational frequencies reported in those studies were comparable to those observed in the current study, although factors including the small cohort sizes and different ethnic populations being studied should be taken into consideration.
IRF4 encodes a transcription factor that controls the differentiation of B, T, dendritic, and myeloid cells, and regulates various aspects of their respective immune responses.24 IRF4 is also an oncogene in some subtypes of diffuse large B-cell lymphoma and a tumor-suppressor in c-Myc-induced malignancies.25-29 IRF4 rearrangement and aberrant hypermutation were commonly characterized in lymphoproliferative malignancies, especially pediatric large B-cell lymphomas.30 In our study, the detected IRF4 mutation sites spanned the complete coding region, and especially affected the DNA-binding domain (DBD) and the IRF association domain (IAD).
CDKN2B (cyclin dependent kinase inhibitor 2B) is a tumor suppressor gene located adjacent to CDKN2A, both of which are frequently deleted in multiple tumors.31,32 In our VRL cohort, 73.3% and 53.3% of patients carried CDKN2A and CDKN2B deletions, respectively. In contrast, PCNSL patients showed much lower frequencies of deletions (CDKN2A: 39% and CDKN2B: 8.7%). These findings revealed the genetic differences between VRL and PCNSL, suggesting that more precise clinical management is needed for this rare and unique type of cancer.
There is a limited evidence-base to guide the treatment of VRL. There have been no randomized clinical trials directed at the treatment of VRL specifically, although some trials of treatments for PCNSL have included patients with VRL, facilitating limited subset analyses.33 Recently, a phase II clinical trial showed promising efficacy of ibrutinib monotherapy in relapsed or refractory PCNSL and VRL patients, independently of B-cell antigen receptor pathway alterations, including CD79A/B.34 Conversely, another study suggested that diffuse large B-cell lymphomas with mutated CD79B and MYD88 were more responsive to ibrutinib, while MYD88-only mutant tumors were likely to be ibrutinib-resistant.35 In our study, four PCNSL patients harboring concurrent CD79B and MYD88 mutations all achieved a complete response to ibrutinib treatment (Online Supplementary Table S5). The efficacy of ibrutinib treatment in our VRL cohort was suboptimal compared to that in PCNSL patients. There are a few possible explanations for this observation, including the fact that both ibrutinib monotherapy and combined therapies were included in this study; the line of treatment varied from first-line to third-line; and subset analyses were not performed because of the restricted cohort size. These limitations prevented a direct comparison of the efficacy of ibrutinib treatment between VRL and PCNSL patients; however, the findings should promote additional well-designed and comprehensive studies of the efficacy of ibrutinib in VRL patients in the future.
The evaluation of disease progression in VRL patients, unlike that of patients with solid tumors, relies mainly on eye examinations and patients’ visual acuities, which are subjective rather than quantitative. Our results suggest that the changes in AH/VF ctDNA allele frequencies were relatively concordant with IL-10 levels in AH/VF. It is known that IL-10 is a clinical marker of VRL,36,37 so AH ctDNA monitoring might be an efficient and quantitative tool to evaluate treatment responses in patients with VRL. In addition, we found that the early detection of CSF ctDNA might be an indication of brain involvement, suggesting that patients with positive CSF ctDNA require close monitoring. Of note, although CSF has been accepted as a better surrogate than plasma for the molecular profiling of brain tumors.38,39 the allele frequencies of CSF ctDNA-derived mutations were significantly lower than those of AH/VF-derived aberrations. Furthermore, mutations detected in CSF samples represented only a subset of AH/VF-borne mutations. That observation suggested that lymphoma was likely to be of an ocular origin in those patients, but considering that CSF has a much higher dilution volume than AH/VF, caution in the interpretation of disease origin is warranted.
Several limitations of this study should be noted. First, the cohort size was restricted due to the rarity of VRL, and thus, further validation in larger cohorts is warranted. Second, the volume of AH/VF samples available for cfDNA extraction varied between patients, but sample volumes were usually small, leading to a limited amount of cfDNA for NGS. Therefore, ctDNA sequencing with AH/VF biopsies was technically demanding. Lastly, the efficacy profile of ibrutinib in VRL and PCNSL requires further investigation because of the heterogeneity and diverse treatment conditions between cohorts in this study. However, our collective findings revealed the molecular heterogeneity of VRL and PCNSL, and highlighted the clinical utility of serial AH/VF ctDNA profiling for the diagnosis and monitoring of VRL.
Supplementary Material
Acknowledgments
The authors would like to thank the patients and family members who gave their consent to presenting the data in this study, as well as the investigators and research staf involved in the study.
Funding Statement
Funding: This study was supported by the National Science & Technology Major Project (grant number: 2017ZX09304021).
References
- 1.Chan CC, Rubenstein JL, Coupland SE, et al. Primary vitreoretinal lymphoma: a report from an International Primary Central Nervous System Lymphoma Collaborative Group symposium. Oncologist. 2011;16(11):1589-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buggage RR, Chan CC, Nussenblatt RB. Ocular manifestations of central nervous system lymphoma. Curr Opin Oncol. 2001;13(3):137-142. [DOI] [PubMed] [Google Scholar]
- 3.Chan CC, Buggage RR, Nussenblatt RB. Intraocular lymphoma. Curr Opin Ophthalmol. 2002;13(6):411-418. [DOI] [PubMed] [Google Scholar]
- 4.Soussain C, Malaise D, Cassoux N. Primary vitreoretinal lymphoma: a diagnostic and management challenge. Blood. 2021;138(17):1519-1534. [DOI] [PubMed] [Google Scholar]
- 5.Salomao DR, Pulido JS, Johnston PB, Canal-Fontcuberta I, Feldman AL. Vitreoretinal presentation of secondary large B-cell lymphoma in patients with systemic lymphoma. JAMA Ophthalmol. 2013;131(9):1151-1158. [DOI] [PubMed] [Google Scholar]
- 6.Coupland SE, Hummel M, Stein H, Willerding G, Jahnke K, Stoltenburg-Didinger G. Demonstration of identical clonal derivation in a case of "oculocerebral" lymphoma. Br J Ophthalmol. 2005;89(2):238-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24(5):679-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan CC, Wallace DJ. Intraocular lymphoma: update on diagnosis and management. Cancer Control. 2004;11(5):285-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reichstein D. Primary vitreoretinal lymphoma: an update on pathogenesis, diagnosis and treatment. Curr Opin Ophthalmol. 2016;27(3):177-184. [DOI] [PubMed] [Google Scholar]
- 11.Dawson AC, Williams KA, Appukuttan B, Smith JR. Emerging diagnostic tests for vitreoretinal lymphoma: a review. Clin Exp Ophthalmol. 2018;46(8):945-954. [DOI] [PubMed] [Google Scholar]
- 12.Caraballo JN, Snyder MR, Johnston PB, et al. Vitreoretinal lymphoma versus uveitis: cytokine profile and correlations. Ocul Immunol Inflamm. 2014;22(1):34-41. [DOI] [PubMed] [Google Scholar]
- 13.Kimura K, Usui Y, Goto H, Japanese Intraocular Lymphoma Study Group. Clinical features and diagnostic significance of the intraocular fluid of 217 patients with intraocular lymphoma. Jpn J Ophthalmol. 2012;56(4):383-389. [DOI] [PubMed] [Google Scholar]
- 14.Davis JL, Miller DM, Ruiz P. Diagnostic testing of vitrectomy specimens. Am J Ophthalmol. 2005;140(5):822-829. [DOI] [PubMed] [Google Scholar]
- 15.Bonzheim I, Sander P, Salmeron-Villalobos J, et al. The molecular hallmarks of primary and secondary vitreoretinal lymphoma. Blood Adv. 2022. 6(3):1598-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J, Kim B, Lee H, et al. Whole exome sequencing identifies mutational signatures of vitreoretinal lymphoma. Haematologica. 2020;105(9):e458-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonzheim I, Giese S, Deuter C, et al. High frequency of MYD88 mutations in vitreoretinal B-cell lymphoma: a valuable tool to improve diagnostic yield of vitreous aspirates. Blood. 2015;126(1):76-79. [DOI] [PubMed] [Google Scholar]
- 18.Xu L, Kim ME, Polski A, et al. Establishing the clinical utility of ctDNA analysis for diagnosis, prognosis, and treatment monitoring of retinoblastoma: the aqueous humor liquid biopsy. Cancers (Basel). 2021;13(6):1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miserocchi E, Ferreri AJM, Giuffre C, et al. MYD88 L265P mutation detection in the aqueous humor of patients with vitreoretinal lymphoma. Retina. 2019;39(4):679-684. [DOI] [PubMed] [Google Scholar]
- 20.Lionakis MS, Dunleavy K, Roschewski M, et al. Inhibition of B cell receptor signaling by ibrutinib in primary CNS lymphoma. Cancer Cell. 2017;31(6):833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shu Y, Wu X, Tong X, et al. Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci Rep. 2017;7(1):583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z, Yang N, Ou Q, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor ssimertinib in non-small cell lung cancer patients. Clin Cancer Res. 2018;24(13):3097-3107. [DOI] [PubMed] [Google Scholar]
- 23.Yonese I, Takase H, Yoshimori M, et al. CD79B mutations in primary vitreoretinal lymphoma: diagnostic and prognostic potential. Eur J Haematol. 2019;102(2):191-196. [DOI] [PubMed] [Google Scholar]
- 24.Nam S, Lim JS. Essential role of interferon regulatory factor 4 (IRF4) in immune cell development. Arch Pharm Res. 2016;39(11):1548-1555. [DOI] [PubMed] [Google Scholar]
- 25.Klein U, Casola S, Cattoretti G, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7(7):773-782. [DOI] [PubMed] [Google Scholar]
- 26.De Silva NS, Simonetti G, Heise N, Klein U. The diverse roles of RF4 in late germinal center B-cell differentiation. Immunol Rev. 2012;247(1):73-92. [DOI] [PubMed] [Google Scholar]
- 27.Mittrucker HW, Matsuyama T, Grossman A, et al. Pillars article: requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997. 275: 540-543. J Immunol. 2017;199(11): 3717-3720. [PubMed] [Google Scholar]
- 28.Yang Y, Shaffer AL, 3rd, Emre NC, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell. 2012;21(6):723-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acquaviva J, Chen X, Ren R. IRF-4 functions as a tumor suppressor in early B-cell development. Blood. 2008;112(9):3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blunt MD, Koehrer S, Dobson RC, et al. The dual Syk/JAK inhibitor cerdulatinib antagonizes B-cell receptor and microenvironmental signaling in chronic lymphocytic leukemia. Clin Cancer Res. 2017;23(9):2313-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laharanne E, Chevret E, Idrissi Y, et al. CDKN2A-CDKN2B deletion defines an aggressive subset of cutaneous T-cell lymphoma. Mod Pathol. 2010;23(4):547-558. [DOI] [PubMed] [Google Scholar]
- 32.Tu Q, Hao J, Zhou X, et al. CDKN2B deletion is essential for pancreatic cancer development instead of unmeaningful codeletion due to juxtaposition to CDKN2A. Oncogene. 2018;37(1):128-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen DT, Houillier C, Choquet S, et al. Primary oculocerebral lymphoma: MTX polychemotherapy alone on intraocular disease control. Ophthalmology. 2016;123(9):2047-2050. [DOI] [PubMed] [Google Scholar]
- 34.Soussain C, Choquet S, Blonski M, et al. Ibrutinib monotherapy for relapse or refractory primary CNS lymphoma and primary vitreoretinal lymphoma: final analysis of the phase II 'proof-of-concept' iLOC study by the Lymphoma Study Association (LYSA) and the French Oculo-cerebral Lymphoma (LOC) network. Eur J Cancer. 2019;117:121-130. [DOI] [PubMed] [Google Scholar]
- 35.Wilson WH, Young RM, Schmitz R, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21(8):922-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cassoux N, Giron A, Bodaghi B, et al. IL-10 measurement in aqueous humor for screening patients with suspicion of primary intraocular lymphoma. Invest Ophthalmol Vis Sci. 2007;48(7):3253-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costopoulos M, Touitou V, Golmard JL, et al. ISOLD: a new highly sensitive interleukin score for intraocular lymphoma diagnosis. Ophthalmology. 2016;123(7):1626-1628. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, He JY, Cui JZ, et al. Detection of genes mutations in cerebrospinal fluid circulating tumor DNA from neoplastic meningitis patients using next generation sequencing. BMC Cancer. 2020;20(1):690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McEwen AE, Leary SES, Lockwood CM. Beyond the blood: CSF-derived cfDNA for diagnosis and characterization of CNS tumors. Front Cell Dev Biol. 2020;8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.