Abstract
Chemical modifications of RNA are associated with fundamental biological processes such as RNA splicing, export, translation, degradation, as well as human disease states such as cancer. However, the analysis of ribonucleoside modifications is hampered by the hydrophilicity of the ribonucleoside molecules. In this work, we used solid-phase permethylation to firstly efficiently derivatize the ribonucleosides and quantitatively analyze them by a liquid chromatography–tandem mass spectrometry (LC–MS/MS)-based method. We identified and quantified more than 60 RNA modifications simultaneously by ultrahigh-performance liquid chromatography coupled with triple quadrupole mass spectrometry (UHPLC-QqQ-MS) performed in the dynamic multiple reaction monitoring (dMRM) mode. The increased hydrophobicity of permethylated ribonucleosides significantly enhanced their retention, separation, and ionization efficiency, leading to improved detection and quantification. We further demonstrate That this novel approach is capable of quantifying cytosine methylation and hydroxymethylation in complex RNA samples obtained from mouse embryonic stem cells with genetic deficiencies in the ten-eleven translocation (TET) enzymes. The results match previously performed analyses and highlighted the improved sensitivity, efficacy, and robustness of the new method. Our protocol is quantitative and robust, and thus provides an augmented approach for comprehensive analysis of RNA modifications in biological samples.
Graphical Abstract
Introduction
Messenger RNAs (mRNAs) are transcribed from the genome and direct protein synthesis, which is highly regulated by a variety of noncoding RNAs, including transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs). Recently, chemical modifications deposited on RNA, during or after transcription, have been identified to contribute to regulation of gene expression, giving rise to the novel field of epitranscriptomics.1-3 Thus far, more than 150 RNA modifications, across multiple species, have been identified on the four canonical bases [adenosine (A), guanosine (G), cytidine (C), and uridine (U)].4 These RNA modifications are involved in fundamental processes, including cell differentiation and stress response.5 Additionally, RNA modifications are crucial for RNA folding, topology, stability, higher-order structure, alternative splicing, and protein translation.6, 7 Modification of rRNA and tRNA also contributes to the rate and fidelity of mRNA translation.8 Moreover, modifications facilitate recognition of RNA by RNA-binding proteins,9, 10 and dysregulation is often associated with human disease. For example, the altered modulation of N6-methyladenosine (m6A), one of the most prevalent RNA methylations, is linked to oncogenesis and progression of colorectal cancer.11
A continued limitation of understanding RNA modifications is the lack of analytical tools for comprehensive characterization and quantification. Sequencing-based techniques are powerful, athough they are unable to directly identify RNA modifications.12 Nanopore sequencing is able to directly analyze m6A, but lacks sensitivity and at the time being is not easily adaptable for other modifications.13 While indirect identification with modification-specific antibodies or chemical reagents has been robustly performed, these methods cannot comprehensively or quantitatively measure multiple types of RNA modifications and each mark requires a distinct workflow.14 Nuclear magnetic resonance (NMR) has also been applied to RNA modification analysis,15 however, its application is limited due to chemical shift overlaps and the resulting line broadening often lead to high background noise and signal loss.16
Recently, mass spectrometry (MS) has emerged as a powerful tool to analyze RNA modifications.17 Multiple modifications can be characterized in a single analysis using liquid chromatography (LC) separation and MS detection. Although promising, MS analysis of oligonucleotides is still far from established due to the limitations in RNA digestion, instrumentation, and software.18-20 One strategy is to digest RNA to mononucleosides, and then RNA modifications can be examined at the single ribonucleoside level.21-23 Reversed-phase chromatography, commonly used for LC-MS, however, is not optimal for native ribonucleosides due to the high polarity of these compounds. Other stationary phase materials such as porous graphitic carbon (PGC) and hydrophilic interaction liquid chromatography (HILIC) have been utilized; however, they are not ideal for profiling ribonucleoside modifications due to some issues such as compromised separations and retention.24 For example, N6-threonylcarbamoyladenosine (t6A) has shown difficult to be reliably analyzed using PGC.25 Additionally, chromatographic approaches using ion-pairing agents are not always compatible with mass spectrometry and also are potentially difficult to remove from the LC system.26
Instead of optimizing the stationary phase, several groups have worked on derivatization methods to alter the chemical and physical properties of ribonucleosides, increasing their compatibility and interactions with existing stationary phase materials. Patteson et al. developed a method using 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide to label pseudouridine (Ψ) residues.27 Huang et al. also demonstrated the derivatization of 5-methylcytosine (m5C) and its oxidation products using bromoacetonyl-containing reagents.28 These approaches can be used to monitor specific modifications based on the selectivity of derivatization reagents. Cai et al. converted ribonucleosides with cis–diol groups to acetonides using acetone, resulting in the identification of more than 50 modified ribonucleosides.29 Unfortunately, ribonucleosides containing 2'-O-methylation could not be identified due to the lack of cis–diol groups. Therefore, there currently is still an unmet need for broader methods to boost the detection of a wide class of RNA modifications.
Here, we adapted permethylation derivatization as a novel method to advance detection of RNA modifications by MS. Permethylation, an efficient strategy for derivatizing a polar molecules, has been extensively utilized for glycan analysis.30 Permethylation of ribonucleosides was first attempted to generate volatile derivatives for gas chromatography–MS analysis,31, 32 however, the properties of these RNA derivatives have not been characterized by modern LC-MS. Without derivatization, unmodified and differentially methylated ribonucleosides (e.g., A, m6A, and Am) can often not be distinguished due to lack of chromatographic separation leading to mixed MS2 spectra. To combate these challanges, we established a solid-phase permethylation method using isotopically labeled iodomethane to derivatize ribonucleosides. The generated hydrophobic ribonucleosides were subsequently separated by C18-based reversed-phase liquid chromatography (RP-LC). Precursor and product ions were simultaneously detected and quantified using a triple quadrupole MS (QqQ MS) operated in the targeted dynamic multiple reaction monitoring (dMRM) mode. This method provides a few advantages: (i) the solid-phase permethylation allows for efficient labeling; (ii) the endogenously methylated ribonucleosides are distinguishable by precursor and/or product ions after this isotopic labeling; (iii) the permethylation product of endogenously methylated and unmodified ribonucleosides have the same retention time, allowing for improved quantification; (iv) the method allows for the differentiation of some ribonucleoside isomers (e.g., m3U and m5U) spontaneously, attributed to their unique precursor and product ions after permethylation; (v) the method increases sensitivity (sub-femtomole level) compared to underivatized ribonucleosides; and lastly, (vi) the predictability of the derivatization reaction facilites adaptability for the potential discovery of new RNA modifications in the future. To validate our method, we built a library of more than 60 transitions for distinct modifications using a combination of ribonucleoside standards and ribonucleosides digested from cell extracts. To further demonstrate the applicability of the method, we examined the cytosine modifications in wild type (WT) mouse embryonic stem cells (mESCs) and ten-eleven translocation (TET) methylcytosine dioxygenases-knockout (KO) mESCs. We observed lower abundance of 5-hydroxymethylcytosine (hm5C) in cells lacking one or more TET enzymes, which was consistent with prior experiments.33 In addition, our improved quantification revealed an expected, but previously unreported increase in m5C levels. This work demonstrates that permethylation as a derivatization strategy combined with mass spectrometry is a highly efficient approach for the accurate detection and quantification of RNA modifications in biological samples.
Experimental section
Samples and materials.
The ribonucleoside standards were purchased from Carbosynth (San Diego, CA). Iodomethane-d3, dichloromethane (DCM), zinc chloride (ZnCl2), sodium acetate (NaOAc), sodium hydroxide (NaOH) beads, nucleosides test mix, nuclease P1, phosphodiesterase I, and phosphodiesterase II were purchased from Sigma-Aldrich (St. Louis, MO). Anhydrous dimethyl sulfoxide (DMSO) was purchased from Biotium (Fremont, CA). Acetonitrile (ACN), formic acid, recombinant shrimp alkaline phosphatase, porous graphitic carbon (PGC) stage tips, and micro spin column were purchased from Thermo Scientific (Waltham, MA).
Culturing Mouse Embryonic Stem Cells (mESCs).
Tet2 KO and Tet1/2/3 triple KO mESC cell lines were cultured as previously described.33 All cells were cultured on gelatin-coated dishes in Knockout DMEM supplemented with 15% FBS, 0.1 mM MEM nonessential amino acids, 0.1 mM 2-mercaptoethanol, 1 mM L-glutamine, 0.5% penicillin streptomycin, 100 U/mL leukemia inhibitory factor (LIF), 3 μM CHIR99021, and 1 μM PD0325901. Cells were maintained in a humidified cell culture incubator with 5% CO2 at 37 °C.
Preparation of ribonucleosides from cell culture.
Total RNA was extracted with Direct-zol RNA Kit (Thermo Scientific, Waltham, MA) as described in the manufacture protocol. RNA samples (100ng) were digested into ribonucleosides with 5 mU/μL of nuclease P1, 5 mU/μL of recombinant shrimp alkaline phosphatase, 500 μU/μL of phosphodiesterase I, and 6.25 μU/μL of phosphodiesterase II in 20 μL of digestion buffer (1 mM ZnCl2, 30 mM NaOAc, pH 7.5) overnight at room temperature. The digested ribonucleosides were purified using PGC stage tips and dried in a Savant SpeedVac concentrator (Thermo Scientific, Waltham, MA).
Solid-phase permethylation of ribonucleosides.
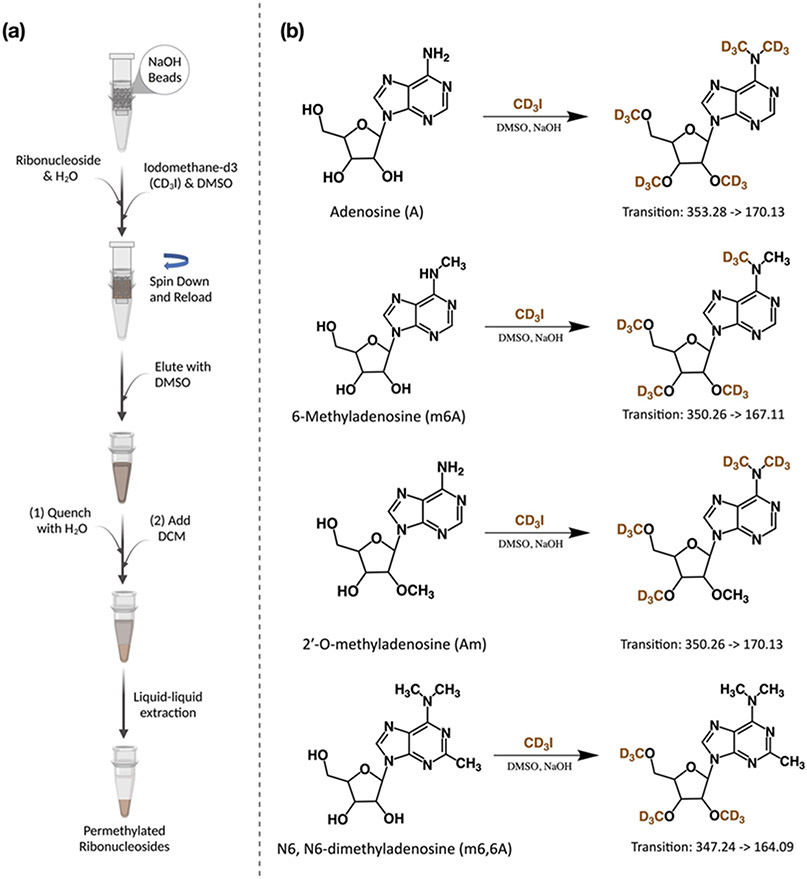
The digested ribonucleoside samples were permethylated using solid-phase permethylation, as previously described by Kang et al., with some optimization.34 Briefly, NaOH beads were packed into the empty spin columns (about 2 cm in height), and the beads were washed with 100 μL of DMSO twice. The purified and dried ribonucleoside samples were reconstituted in a mixture of 1 μL of water, 50 μL of DMSO, and 30 μL of iodomethane-d3. The samples were loaded into the spin column and spun down at 200 x g, followed by reloading the samples into the column four times. Next, 20 μL of iodomethane-d3 was added to the sample and incubated at room temperature for another 10 min. The column was washed with 50 μL of DMSO twice, 500 μL of ice-cold water was added into the sample, and the mixture was incubated at room temperature for at least 1 min to quench the permethylation reaction. Afterward, 300 μL of DCM was added, the liquid–liquid extraction was repeated at least five times for each sample, and the organic layer was dried using a Savant SpeedVac concentrator.
Ultrahigh-pressure liquid chromatography/triple quadrupole mass spectrometry (UHPLC/QqQ-MS) analysis.
Separation and characterization of the ribonucleosides were carried out on a Thermo Scientific Vanquish Flex binary UHPLC system coupled to a Thermo Scientific TSQ Altis QqQ mass spectrometer (Thermo Scientific, Waltham, MA). For the analysis, 2 μL of the sample was injected onto a Thermo Scientific Accucore Vanquish C18 column (150 × 2.1mm, 1.5 μm) and separated using a 20 min binary gradient with a constant flow rate of 0.2 mL/min at 60 °C. Mobile phase A was water with 0.1% formic acid, and mobile phase B was 80% ACN/water (v/v) with 0.1% formic acid. For analysis of permethylated ribonucleosides, the following binary gradient was used: 0–7 min, 20%–40% B; 7–10 min, 40–70% B; 10–11 min, 70%–99% B; 11–15 min, 99% B; 15–16 min, 99–20% B; and 16–20 min, 20% B. Samples were introduced into the mass spectrometer using an electrospray ionization (ESI) source operated in the positive ion mode at 3500 V. Nitrogen sheath gas, auxiliary gas, and sweep gas flow rates were set at 30, 5, and 2 psi, respectively. The ion transfer tube temperature and vaporizer temperature were set at 350 °C and 175 °C, respectively. The precursor ions were fragmented using collision-induced dissociation (CID) with optimized energy. Data acquired from the UHPLC/QqQ-MS was collected using Thermo Scientific Xcalibur software (v4.1), and data analysis was performed using Thermo Scientific FreeStyle software (v2.1).
nanoLC-MS/MS analysis.
The samples were characterized using an EASY-nLC™ 1200 system coupled to a Q Exactive mass spectrometer (ThermoFisher Scientific). 3 μL of the sample was injected, and the analytes were separated on a self-packed C18 column (3 μm, 0.150 mm × 250 mm) at a flow rate of 700 nL/min. Water containing 0.1% formic acid and 80% acetonitrile containing 0.1% formic acid were used as solvents A and B, respectively. MS spectra were collected with a mass scan range of m/z 200–600 in positive ionization mode. The filtered precursor ions in each MS spectrum were fragmented via high collisional dissociation (HCD) at 30% normalized collision energy (NCE) with nitrogen gas.
Results and discussions
Optimization of permethylation reaction.
During permethylation, all hydrogen atoms on the hydroxyl, amine, and carboxyl groups on ribonucleosides are replaced with methyl groups. Previously, NaOH dissolved in a DMSO solution was used for permethylation. The application of this method was limited due to low reaction efficiency. Kang et al. developed a solid-phase permethylation technique by packing sodium hydroxide beads in microspin columns, achieving high derivatization efficiency.34 This technique has extensively improved the characterization of glycans and glycoproteins. We employed this solid-phase-based technique to maximize the permethylation of the ribonucleoside samples (Figure 1a). The amount of water was found to be critical for the permethylation reaction. To determine the optimal conditions, the reaction was carried out using a mixture of the four canonical ribonucleoside standards (2.5 μg/mL each) dissolved in varying volumes of water. Reactions performed in the presence of 1 μL of water produced more permethylated ribonucleosides compared to reactions with 0.5 μL or 2 μL of water (reaction 1-3 in Figure S1a). We reasoned that the water improved the solubility of ribonucleosides in DMSO; however, excess water led to undesired side reactions. Mechref et al. demonstrated reaction efficiency could be improved by adding extra iodomethane in the middle of the reaction.35 As shown in Figure S1a, the yields of permethylated ribonucleosides increased significantly after the second aliquot of iodomethane-d3 was added.
Figure 1. Permethylation was used to derivatize the ribonucleosides.
(a) The workflow of the solid-phase permethylation method. (b) Example of permethylation of unmodified and methylated adenosine. Adenosine, 6-methyladenosine, and 2’-O-methyladenosine were labeled by the −CD3 group differently, and their unique precursor and product ions were monitored using UHPLC-QqQ-MS under the dMRM mode.
We also monitored the unreacted ribonucleosides extracted in the aqueous phase during the liquid-liquid extraction (Figure S1b). The results showed limited unreacted ribonucleosides in all four conditions, and the unreacted species were less abundant after adding the second aliquot of iodomethane-d3 (less than 0.005% for adenosine). Another concern was that the permethylation reaction may not fully replace all the active hydrogens, generating partially methylated products and hampering accurate quantification. For example, previous work had shown that adenosine had incomplete derivatization using the conventional in-solution method.36 To ensure that the permethylation goes to completion, we used LC-MS/MS to characterize the adenosine products after solid-phase derivatization. As shown in Figure S2, the signal for fully permethylated adenosine was exceedingly abundant, while the abundance of partially methylated adenosine was at the noise level, demonstrating high efficiency of the optimized solid-phase permethylation method (>99.9%). To this end, 1 μL of water and the addition of 20 μL of iodomethane-d3 was utilized for the complete derivatization of the ribonucleosides.
Construction of the dMRM transitions.
Commercial ribonucleoside standards were employed to create the basic transitions, including A (adenosine), Am (2'-O-methyladenosine), m6A (N6-methyladenosine), t6A (N6-threonylcarbamoyladenosine), io6A [N6-(cis-hydroxyisopentenyl)adenosine], i6A (N6-isopentenyladenosine), I (inosine), C (cytidine), ac4C (N4-acetylcytidine), s2C (2-thiocytidine), m5C (5-methylcytidine), f5C (5-formylcytidine), G (guanosine), m7G (7-methylguanosine), U (uridine), m5U (5-methyluridine), s2U (2-thiouridine), D (dihydrouridine), m5D (5-methyldihydrouridine), and Ψ (pseudouridine). For analysis, permethylated ribonucleoside standards were prepared, and dMRM transitions were obtained by scanning their respective fragment ions using the product ion mode on the QqQ.
To distinguish between endogenous and derivatized chemical methylations, deuterium-labeled iodomethane was used. As shown in Figure 1b, five methyl-d3 (−CD3) groups replaced hydrogen atoms from A, including three from the hydroxyl group on the ribose ring and two from the amine group on the nucleobase, while m6A and Am were labeled with four molecules of −CD3. Notably and as expected, −CD3 groups labeled hydroxyl and amine groups on m6A and Am differently. For Am, two molecules of −CD3 on the ribose and two on the nucleobase replaced hydrogens, but three hydrogens on ribose and one hydrogen on the nucleobase were replaced by −CD3 for m6A. Only the hydrogens on ribose were replaced by −CD3 for N6,N6-dimethyladenosine (m6,6A). Similar to the underivatized form, the primary fragmentation of these permethylated ribonucleosides was produced after the neutral loss of the ribose ring. Hence, three different transitions were constructed: m/z 353.28 → 170.13 for A, m/z 350.26 → 167.11 for m6A, m/z 350.26 → 170.13 for Am, and m/z 347.24 → 164.09 for m6,6A. Notably, permethylation of m6A and m1A produced the same precursor ion (m/z 350.26) and product ion (m/z 167.11 in MS2). Fortunately, they could be differentiated by their (pseudo)-MS3 spectra, where the intensity ratio of the m/z 119.03 over m/z 120.04 was <1 and >1 for m6A and m1A, respectively (Figure S3). Importantly, unmodified and methylated ribonucleosides produced isotopically labeled products with the same retention time, allowing the transition list to be readily built. For example, the MRM transition for permethylated inosine (I) that was built upon its chemical standard can also be extended to create the transitions for methylated inosines, such as Im, m1I, and m1Im.
Permethylation also improved the ability to differentiate ribonucleoside isomers. For example, the isomer of canonical uridine, Ψ, is a crucial RNA modification and is involved in the regulation of gene expression.37 Various techniques have been developed for the analysis of Ψ, but the same precursor mass and poor separation of Ψ and U have hindered their applications.38 Notably, these two isomers generated different precursor and product ions after permethylation derivatization, suggesting that U and Ψ can be unambiguously characterized using our method. As mentioned previously, most fragments of ribonucleosides are generated after the loss of the ribose ring; however, Ψ is an exception due to the carbon-carbon linkage between the ribose and the nucleobase. To find the fragments of Ψ, we analyzed permethylated U and Ψ standards using a high-resolution Orbitrap MS. As shown in Figure S4a, U lost the methylated ribose ring (−180.14 Da) and yielded a unique reporter ion of m/z 130.07. Although m/z 149.11 is the most abundant fragment for U, it was not selected as it is generated from the cross-ring fragmentation of permethylated ribose which is produced by all other permethylated ribonucleosides. Additionally, the fragment ions at m/z 206.15 and 225.11 were uniquely produced by Ψ due to the cross-ring fragmentation (Figure S4b). Therefore, the transitions m/z 313.22 → 130.07 and m/z 330.25 → 206.15 were created for U and Ψ, respectively. Another issue with previous methods was distinguishing the two methylated uridine isomers (m5U and m3U). In our method, m5U had an additional −CD3 moiety labeled compared to m3U after permethylation, displaying differences in their precursor ion masses, product ion masses, and retention times.
We further monitored three adenosine-derived ribonucleoside standards, i6A, io6A, and t6A, which have more complicated chemical structures than methylated adenosine. Similar to the unmodified adenosine, the −CD3 groups labeled hydroxyl groups on the ribose and amines on the nucleobase on i6A (Figure S5a), while an extra −CD3 group reacted with io6A due to the presence of the hydroxyisopentenyl group (Figure S5b). Although permethylated io6A generated abundant i6A ion due to fragmentation of the −OCD3, both standards still generated high abundance fragments from ribose loss. Threonylcarbamoyl-modified adenosine, t6A, contains an acidic structure. The carboxyl group reacted with iodomethane-d3 and generated a product with m/z 532.38 as the precursor mass, which yielded an expected fragment at m/z 349.23 used for monitoring in positive ion mode (Figure S5c). One exception was positively charged m7G, where a hydroxyl group was added from the hydroxide, producing an additional site for methylation during permethylation (Figure S6a). As a result, m7G was monitored using m/z 435.37 → m/z 252.22 (Figure S6b). In general, −CD3 group can efficiently replace the hydrogens on hydroxyl, amine, and carboxyl groups on ribonucleosides. The resulting products after derivatization are straightforward to monitor for structurally complex RNA modifications. Overall, more than 60 transitions were monitored simultaneously as shown in Table S1, and the qualifying fragment ions of the permethylated ribonucleosides were also used to validate the method.
Comparison with conventional ribonucleoside analysis.
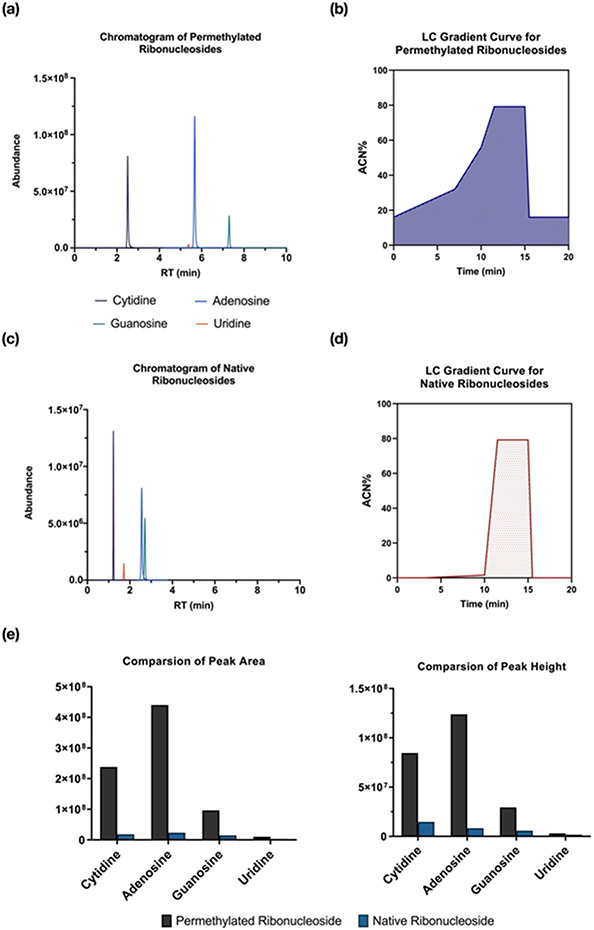
Ribonucleosides have similar structures, which contain a hydrophilic ribose and a nucleobase. Therefore, it is a challenge to profile and quantify these compounds by traditional RP-LC-MS methods. To evaluate the advantage of tderivatizing ribonucleosides compared to the underivatized forms, equal amounts of standards were injected into the QqQ instrument and analyzed. As shown in Figure 2a-b, all four unpermethylated ribonucleoside standards were eluted within 4 mins with less than 1% of ACN. Notably, these ribonucleosides had problems in peaks overlapping during the chromatographic analysis. For example, the more hydrophilic ribonucleosides, cytidine and uridine, had poor retention on C18 and eluted in 0% ACN, and often co-eluted with hydrophilic contaminants. Purines (adenosine and guanosine) were retained on the C18, but had poor chromatographic resolution. In contrast, all the permethylated ribonucleoside peaks were evenly distributed across the chromatogram and eluted with a higher percentage of ACN (~30%) (Figure 2c-d), enhancing chromatographic resolution of ribonucleosides using C18. The theoretical plate number was calculated based on retention time and full peak width (Table S2). The number of theoretical plates for conventional ribonucleoside analysis was determined to be around 4000. The plate number was greatly improved for permethylated ribonucleosides even when a much higher percentage of ACN was applied. The theoretical plate number was calculated to be >26000 for permethylated guanosine (G), and nearly 16000 for permethylated adenosine (A). This demonstrates that C18 can continue to be used as the stationary phase for nucleoside analysis without suffering from poor resolution and low retention.
Figure 2. Comparison of the analysis between permethylated and native ribonucleoside.
MRM chromatogram of (a) permethylated (d3) ribonucleoside standards and (c) native ribonucleoside standards. The LC gradient was applied for analyzing (b) permethylated (d3) canonical ribonucleosides and (d) the native forms. Higher ACN% (B%) was used for the analysis of permethylated (d3) ribonucleosides with improved separation. (e) The comparison of peak area and peak height between permethylated and native ribonucleosides.
Due to the hydrophilicity of the ribonucleosides, they normally have low ionization efficiencies leading to difficulties in quantification.39 It has been demonstrated that permethylation can enhance the signals of hydrophilic analytes.40 Indeed, the signal of permethylated ribonucleosides was greatly increased compared to the underivatized ribonucleosides (Figure 2 e and f). For example, the peak area of the adenosine increased by 20-fold. This is likely due to the high hydrophilicity of underivatized ribonucleosides, leading to lower proton affinity. At the same time, the incorporation of multiple methyl groups with increased hydrophobicity provided higher ionization efficiency in positive ionization mode. Uridine had the lowest ionization efficiency among four canonical ribonucleosides. However, the uridine signal after the permethylation increased 5-fold compared to the unpermethylated form. In general, the permethylation reaction improved the quantitative analysis of ribonucleosides with greater retention, separation, and sensitivity.
Method validation.
Quantitative analysis of canonical and modified ribonucleosides was performed to validate the derivatization method. The standard ribonucleoside mixtures were prepared by mixing the standards together and diluting solutions to a concentration range of 0.01–1000 ng/L. Working solutions with different dilution factors were derivatized according to the optimized solid-phase permethylation workflow. The calibration curve, linear regression coefficient (R2), linear range, limits of detection (LOD), limits of quantification (LOQ), and coefficients of variance (CV) for the permethylated ribonucleosides were calculated and are shown in Table S3. Adenosine had the highest response among all the standards due to its high ionization efficiency. All correlation coefficients for the analytes were between 0.992-0.998, indicating good linearity. Both LOD and LOQ values were at sub-femtomole levels, which are at an order of magnitude below the analysis of underivatized ribonucleosides.41 Notably, LOD and LOQ for dihydrouridine and 5-methyldihydrouridine were higher than other ribonucleosides, indicating lower detection sensitivity because dihydrouridine went through a pyrimidine ring-opening reaction under alkaline conditions and yielded by-products.42 The results suggest that this method is suitable for analyzing a broad spectrum of RNA modifications with low abundances in biological samples. Furthermore, the linear calibration ranges for the permethylated ribonucleosides demonstrated good linearity within three orders of magnitude, while the technical replicate CVs were about 4% for most ribonucleosides. Collectively, this method greatly improved the quantitative analysis of ribonucleosides after permethylation.21
Analysis of ribonucleosides from mESCs.
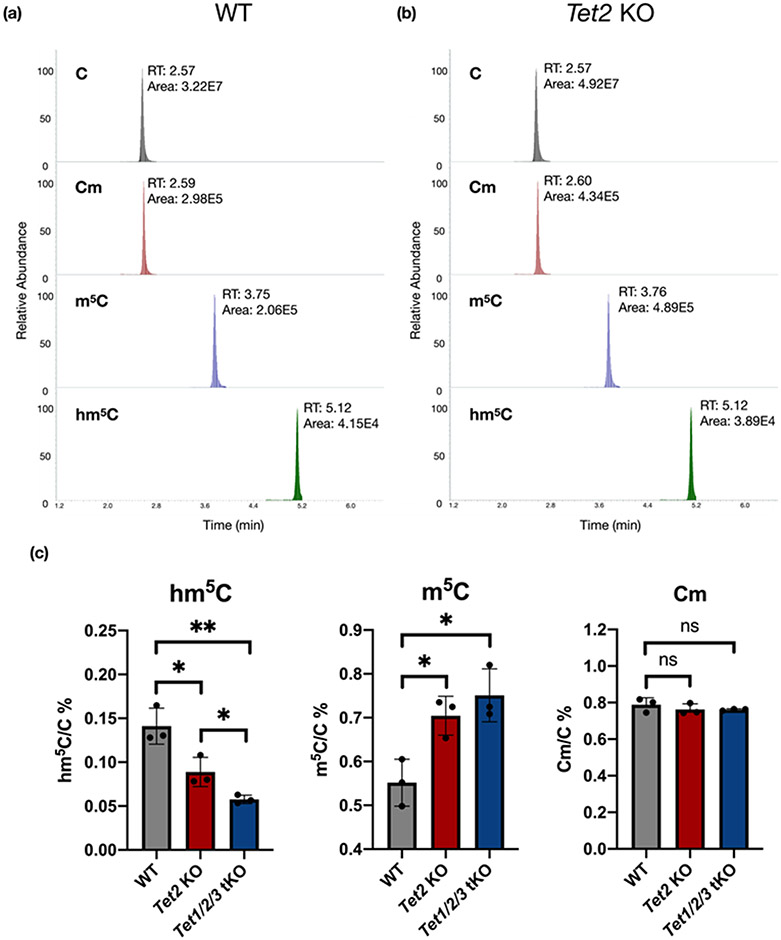
To demonstrate the applicability of the method, we quantified the levels of C, Cm, hm5C, and m5C in RNAs extracted from mESCs carrying knockout in genes encoding TET dioxygenases as well as wildtype (WT) controls. TET dioxygenases, including TET1, TET2, and TET3, catalyze the oxidation of 5-methylcytosine in DNA to form oxidized products, and thus play important roles in epigenetic regulation.43 Recently, He et al. demonstrated that TET2 is essential for converting m5C to hm5C on tRNAs, which regulates the formation of tRNA fragments.33 In this study, hm5C was substantially decreased in TET mutants measured by native ribonucleoside MS analysis. However, our previous analyses were not accurate or sensitive enough to detect the expected increase in the m5C level.
We repeated these measurements using the improved permethylation-based method (Figure 3a-b). Consistent with our published results, a significant decrease in hm5C level was detected in RNA samples obtained from TET2-deficient mESCs, which was even more pronounced in that from Tet1/2/3 triple KO (tKO) cells (Figure 3c).33 Notably, the levels of hm5C detected by our improved method were 10-fold higher than previous results from native ribonucleoside analyses. We were also able to detect the expected increase in m5C in both Tet2 KO and Tet1/2/3 tKO cells compared to WT cells (Figure 3d). As a control, the levels of Cm, not known to be targeted by TET enzymes, were not affected by these mutations (Figure 3e).
Figure 3. Application of the permethylation method for analyzing modified cytosine in WT and Tet-KO mESCs.
The example chromatography of C, Cm, m5C, and hm5C in purified total RNAs from (a) WT and (b) Tet2 KO cells. (c) The relative abundance of hm5C, m5C, and Cm. (p values were determined using two-tailed Student's t test for unpaired samples. Error bars represent mean ± s.d., n = 3, ** p < 0.01, **** p < 0.001, n.s. means no significance with p > 0.05.)
These observations not only in agreement with our previous findings, but also provided more accurate measurements, including the levels of m5C and Cm. The results exemplify the advantage of applying improved detection and quantification by permethylation to the analysis of ribonucleoside modifications.
Conclusions
The analysis of the RNA modifications has been greatly hindered by the high hydrophilicity of ribonucleotides, leading to low sensitivity and inaccurate quantification. In this study, we derivatized ribonucleosides to more hydrophobic molecules utilizing solid-phase permethylation. A dynamic MRM method was developed to provide a quantitative characterization of RNA modifications. The reproducible results suggest that this method will improve the ability to monitor ribonucleoside modifications on RNAs. Our high throughput method is capable of profiling more than 60 modified ribonucleosides in RNAs. The MRM transition list can be readily adapted for newly discovered RNA modifications as well. Similar to applications in glycan research, isotopically labeled iodomethane can be further applied as multiplexing reagents.44 Additionally, our protocol can be potentially adapted for detecting canonical and modified deoxyribonucleosides in DNA samples, facilitating both epigenetic and epitranscriptomic analysis. Finally, the quantitative results of ribonucleosides extracted from WT and Tet-KO mESCs demonstrate that this approach can detect subtle differences in low-abundant, but critical RNA modifications from biological samples. As ribonucleoside modifications are associated with various disease states, this new method could be used as a reliable platform for biomarker research and discovery in RNA-related fields.
Supplementary Material
Acknowledgements
The authors thank Juan J. Castillo and Siyu Chen (University of California, Davis) for their suggestions on the permethylation reaction. This work was supported by grants (No.: AI118891, CA196539 and AG031862) to B.A.G. from the NIH.
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Supporting Information
Detailed information on optimization of permethylation reaction (Figure S1), partially methylated adenosine (Figure S2), differentiation of m6A and m1A, fragmentation of U, Ψ, m7G, and modified adenosine (Figure S3-6), collision energy optimization (Figure S7), MRM transitions (Table S1), separation of underivatized and permethylated ribonucleosides (Table S2), and linearities of ribonucleoside standards (Table S3).
References
- 1.Li S; Mason CE, The Pivotal Regulatory Landscape of RNA Modifications. Annu. Rev. Genomics. Hum. Genet 2014, 15 (1), 127–150. [DOI] [PubMed] [Google Scholar]
- 2.Peer E; Rechavi G; Dominissini D, Epitranscriptomics: regulation of mRNA metabolism through modifications. Curr. Opin. Chem. Biol 2017, 41, 93–98. [DOI] [PubMed] [Google Scholar]
- 3.Xiong X; Yi C; Peng J, Epitranscriptomics: Toward A Better Understanding of RNA Modifications. Genom. Proteom. Bioinform 2017, 15 (3), 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boccaletto P; Machnicka MA; Purta E; Piątkowski P; Bagiński B; Wirecki TK; de Crécy-Lagard V; Ross R; Limbach PA; Kotter A; Helm M; Bujnicki JM, MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46 (D1), D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yi C; Pan T, Cellular Dynamics of RNA Modification. Acc. Chem. Res 2011, 44 (12), 1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuorto F; Liebers R; Musch T; Schaefer M; Hofmann S; Kellner S; Frye M; Helm M; Stoecklin G; Lyko F, RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol 2012, 19 (9), 900–905. [DOI] [PubMed] [Google Scholar]
- 7.Helm M, Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006, 34 (2), 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frye M; Harada Bryan T; Behm M; He C, RNA modifications modulate gene expression during development. Science 2018, 361 (6409), 1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nechay M; Kleiner RE, High-throughput approaches to profile RNA-protein interactions. Curr. Opin. Chem. Biol 2020, 54, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roundtree IA; Evans ME; Pan T; He C, Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169 (7), 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang Z; Hu Y; Hu J; Huang Y; Zheng S; Guo C, The crucial roles of N6-methyladenosine (m6A) modification in the carcinogenesis and progression of colorectal cancer. Cell Biosci. 2021, 11 (1), 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krogh N; Nielsen H, Sequencing-based methods for detection and quantitation of ribose methylations in RNA. Methods 2019, 156, 5–15. [DOI] [PubMed] [Google Scholar]
- 13.Garalde DR; Snell EA; Jachimowicz D; Sipos B; Lloyd JH; Bruce M; Pantic N; Admassu T; James P; Warland A; Jordan M; Ciccone J; Serra S; Keenan J; Martin S; McNeill L; Wallace EJ; Jayasinghe L; Wright C; Blasco J; Young S; Brocklebank D; Juul S; Clarke J; Heron AJ; Turner DJ, Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 2018, 15 (3), 201–206. [DOI] [PubMed] [Google Scholar]
- 14.Helm M; Motorin Y, Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet 2017, 18 (5), 275–291. [DOI] [PubMed] [Google Scholar]
- 15.Yoluç Y; Ammann G; Barraud P; Jora M; Limbach PA; Motorin Y; Marchand V; Tisné C; Borland K; Kellner S, Instrumental analysis of RNA modifications. Crit. Rev. Biochem. Mol. Biol 2021, 56 (2), 178–204. [DOI] [PubMed] [Google Scholar]
- 16.Becette Owen B; Zong G; Chen B; Taiwo Kehinde M; Case David A; Dayie TK, Solution NMR readily reveals distinct structural folds and interactions in doubly 13C- and 19F-labeled RNAs. Sci. Adv 6 (41), eabc6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wetzel C; Limbach PA, Mass spectrometry of modified RNAs: recent developments. Analyst 2016, 141 (1), 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutton JM; Guimaraes GJ; Annavarapu V; van Dongen WD; Bartlett MG, Current State of Oligonucleotide Characterization Using Liquid Chromatography–Mass Spectrometry: Insight into Critical Issues. J. Am. Soc. Mass Spectrom 2020, 31 (9), 1775–1782. [DOI] [PubMed] [Google Scholar]
- 19.Hagelskamp F; Borland K; Ramos J; Hendrick AG; Fu D; Kellner S, Broadly applicable oligonucleotide mass spectrometry for the analysis of RNA writers and erasers in vitro. Nucleic Acids Res. 2020, 48 (7), e41–e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wein S; Andrews B; Sachsenberg T; Santos-Rosa H; Kohlbacher O; Kouzarides T; Garcia BA; Weisser H, A computational platform for high-throughput analysis of RNA sequences and modifications by mass spectrometry. Nat. Commun 2020, 11 (1), 926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su D; Chan CTY; Gu C; Lim KS; Chionh YH; McBee ME; Russell BS; Babu IR; Begley TJ; Dedon PC, Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat. Protoc 2014, 9 (4), 828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregorova P; Sipari NH; Sarin LP, Broad-range RNA modification analysis of complex biological samples using rapid C18-UPLC-MS. RNA Biol. 2021, 18 (10), 1382–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssen KA; Xie Y; Kramer MC; Gregory BD; Garcia BA, Data-Independent Acquisition for the Detection of Mononucleoside RNA Modifications by Mass Spectrometry. J. Am. Soc. Mass Spectrom 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuytten R; Lemière F; Van Dongen W; Witters E; Esmans EL; Newton RP; Dudley E, Development of an On-Line SPE-LC–ESI-MS Method for Urinary Nucleosides: Hyphenation of Aprotic Boronic Acid Chromatography with Hydrophilic Interaction LC–ESI-MS. Anal. Chem 2008, 80 (4), 1263–1271. [DOI] [PubMed] [Google Scholar]
- 25.Sarin LP; Kienast SD; Leufken J; Ross RL; Dziergowska A; Debiec K; Sochacka E; Limbach PA; Fufezan C; Drexler HCA; Leidel SA, Nano LC-MS using capillary columns enables accurate quantification of modified ribonucleosides at low femtomol levels. RNA 2018, 24 (10), 1403–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J; Zhang Y; Wiegand R; Wang J; Bepler G; Li J, Quantitative analysis of intracellular nucleoside triphosphates and other polar metabolites using ion pair reversed-phase liquid chromatography coupled with tandem mass spectrometry. J. Chromatogr. B 2015, 1006, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patteson KG; Rodicio LP; Limbach PA, Identification of the mass-silent post-transcriptionally modified nucleoside pseudouridine in RNA by matrix-assisted laser desorption/ionization mass spectrometry. Nucleic Acids Res. 2001, 29 (10), e49–e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W; Lan M-D; Qi C-B; Zheng S-J; Wei S-Z; Yuan B-F; Feng Y-Q, Formation and determination of the oxidation products of 5-methylcytosine in RNA. Chem. Sci 2016, 7 (8), 5495–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S; Jin Y; Tang Z; Lin S; Liu H; Jiang Y; Cai Z, A novel method of liquid chromatography–tandem mass spectrometry combined with chemical derivatization for the determination of ribonucleosides in urine. Anal. Chim. Acta 2015, 864, 30–38. [DOI] [PubMed] [Google Scholar]
- 30.Ruhaak LR; Zauner G; Huhn C; Bruggink C; Deelder AM; Wuhrer M, Glycan labeling strategies and their use in identification and quantification. Anal. Bioanal. Chem 2010, 397 (8), 3457–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pantarotto C; Martini A; Belvedere G; Bossi A; Donnelli MC; Frigerio A, Application of gas chromatography—chemical ionization mass fragmentography in thed evaluation of bases and nucleoside analogues used in cancer chemotherapy. J. Chromatogr. A 1974, 99, 519–527. [DOI] [PubMed] [Google Scholar]
- 32.Von Minden DL; McCloskey JA, Mass spectrometry of nucleic acid components. N,O-Permethyl derivatives of nucleosides. J. Am. Chem. Soc 1973, 95 (22), 7480–7490. [DOI] [PubMed] [Google Scholar]
- 33.He C; Bozler J; Janssen KA; Wilusz JE; Garcia BA; Schorn AJ; Bonasio R, TET2 chemically modifies tRNAs and regulates tRNA fragment levels. Nat. Struct. Mol. Biol 2021, 28 (1), 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang P; Mechref Y; Klouckova I; Novotny MV, Solid-phase permethylation of glycans for mass spectrometric analysis. Rapid Commun. Mass Spectrom 2005, 19 (23), 3421–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou S; Wooding KM; Mechref Y, Analysis of Permethylated Glycan by Liquid Chromatography (LC) and Mass Spectrometry (MS). In High-Throughput Glycomics and Glycoproteomics: Methods and Protocols, Lauc G; Wuhrer M, Eds. Springer New York: New York, NY, 2017; pp 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolhun JJ; Wiebers JL, Mass spectra of nucleoside derivatives. Org. Mass Spectrom 1970, 3 (5), 669–681. [Google Scholar]
- 37.Zhao Y; Dunker W; Yu Y-T; Karijolich J, The Role of Noncoding RNA Pseudouridylation in Nuclear Gene Expression Events. Front. Bioeng. Biotechnol 2018, 6 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan M; Wang Y; Hu Y; Feng Y; Dai C; Wu J; Wu D; Zhang F; Zhai Q, A High-Throughput Quantitative Approach Reveals More Small RNA Modifications in Mouse Liver and Their Correlation with Diabetes. Anal. Chem 2013, 85 (24), 12173–12181. [DOI] [PubMed] [Google Scholar]
- 39.Lan M-D; Yuan B-F; Feng Y-Q, Deciphering nucleic acid modifications by chemical derivatization-mass spectrometry analysis. Chin. Chem. Lett 2019, 30 (1), 1–6. [Google Scholar]
- 40.Ruhaak LR; Xu G; Li Q; Goonatilleke E; Lebrilla CB, Mass Spectrometry Approaches to Glycomic and Glycoproteomic Analyses. Chem. Rev 2018, 118 (17), 7886–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang Z; Hu Y; Chen J; Xu K; Wang K; Zheng S; Guo C, Mass Spectrometry-Based Targeted Serum Monomethylated Ribonucleosides Profiling for Early Detection of Breast Cancer. Front. Mol. Biosci 2021, 8 (833). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Motorin Y; Muller S; Behm-Ansmant I; Branlant C, Identification of Modified Residues in RNAs by Reverse Transcription-Based Methods. In Methods Enzymol., Academic Press: 2007; Vol. 425, pp 21–53. [DOI] [PubMed] [Google Scholar]
- 43.Wu H; Zhang Y, Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011, 25 (23), 2436–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong X; Peng W; Yu C-Y; Zhou S; Donohoo KB; Tang H; Mechref Y, 8-plex LC–MS/MS Analysis of Permethylated N-Glycans Achieved by Using Stable Isotopic Iodomethane. Anal. Chem 2019, 91 (18), 11794–11802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.